- 1Graduate School of Life Science, Hokkaido University, Sapporo, Hokkaido, Japan
- 2Faculty of Science, Hokkaido University, Sapporo, Hokkaido, Japan
New transposon insertions are deleterious to genome stability. The RNA-directed DNA methylation (RdDM) pathway evolved to regulate transposon activity via DNA methylation. However, current studies have not yet clearly described the transposition regulation. ONSEN is a heat-activated retrotransposon that is activated at 37°C. The plant-specific SUPPRESSOR OF VARIEGATION 3–9 HOMOLOG (SUVH) family proteins function downstream of the RdDM pathway. The SUVH protein families are linked to TE silencing by two pathways, one through DNA methylation and the other through chromatin remodeling. In this study, we analyzed the regulation of ONSEN activity by SUVH2. We observed that ONSEN transcripts were increased; however, there was no transpositional activity in Arabidopsis suvh2 mutant. The suvh2 mutant produced siRNAs from the ONSEN locus under heat stress, suggesting that siRNAs are involved in suppressing transposition. These results provide new insights into the regulatory mechanisms of retrotransposons that involve siRNA in the RdDM pathway.
1 Introduction
Plants and animals must cope with various environmental stressors. Environmental stress can activate transposable elements (TEs), and new insertions are inherited by the next generation of germ cells (Grandbastien et al., 1997; Oliver and Greene, 2009) contributing to genome evolution in plants (Oliver and Greene, 2009; Bennetzen and Wang, 2014). TEs are categorized into two main classes based on their mode of transposition: class I and II transposons (Bourque et al., 2018). Class I transposons are known as retrotransposons, and their transposition activity requires an RNA intermediate. Long terminal repeat (LTR) and non-LTR retrotransposons (long interspersed nuclear elements [LINEs] and short interspersed nuclear elements [SINEs]) are the two main classes of retrotransposons (Wicker et al., 2007). The transposition activity of the LTR retrotransposon begins at the promoter within the 5’ LTR, where the TE is first transcribed into RNA by RNA polymerase II. In the cytoplasm, the RNA is reverse-transcribed into cDNA and reintroduced into the nucleus, and, finally, the cDNA is reincorporated into the host DNA (Havecker et al., 2004). In contrast, most class II elements accomplish transposition activity by excising themselves and thus jumping to a new gene location (Wells and Feschotte, 2020). TEs regulate gene activity by transpositional insertions into or close to genes, manifesting either through the direct disruption of gene transcription or by indirectly affecting gene expression through epigenetic modifications (Niu et al., 2022a; Berthelier et al., 2023).
TEs are regulated through epigenetic modifications, such as two representative epigenetic processes: DNA methylation and histone modification (Lippman et al., 2004; Slotkin and Martienssen, 2007; Mhiri et al., 2022). In plants, de novo DNA methylation is induced by RNA-directed DNA methylation (RdDM), mediated by small interfering RNAs (siRNAs) and scaffolding RNAs (Gallego-Bartolome, 2020). RdDM is classified into canonical and non-canonical pathways (Matzke and Mosher, 2014). In the canonical RdDM pathway, Pol IV (RNA polymerase IV) and Pol V (RNA polymerase Pol V) are essential for the RdDM process and serve the production of 24 nt siRNA and long non-coding RNA (lncRNAs) (Onodera et al., 2005; Wierzbicki et al., 2008). Previous reports have shown that there are ten different suppressors of variegation 3–9 homolog (SUVH) family proteins in Arabidopsis thaliana (Naumann et al., 2005), of which SUVH2 and SUVH9 are closely associated with DNA methylation activities in RdDM (Johnson et al., 2008). SUVH2 and SUVH9 bind to methylated DNA and promote the recruitment of Pol V to the target locus to produce long non-coding RNAs by interacting with the DDR (DRD1-DMS3-RDM1; defective in RNA-directed DNA methylation 1 [drd1], defective in meristem silencing 3 [DMS3], and RNA-directed DNA methylation 1 [RDM1]) and MORC (microrchidia) complex (Kanno et al., 2004; Johnson et al., 2008; Ream et al., 2009; Law et al., 2010; Zhong et al., 2012; Brabbs et al., 2013; Liu et al., 2014).
ONSEN is an LTR retrotransposon defined in A. thaliana and is usually activated by heat stress (Ito et al., 2011). Previous studies have shown that some RdDM pathway mutants subjected to heat stress show transcriptional activation of ONSEN (Hayashi et al., 2020; Niu et al., 2022b). siRNAs are involved in transcriptional repression processes, especially in TEs (Moses et al., 2010). In the nrpd1 mutant (siRNA biogenesis deficient mutant), ONSEN transcripts and extrachromosomal DNA levels increased, and transgenerational transposition was observed in the next generation (Ito et al., 2011). However, our understanding of ONSEN transposition repression has yet to be profound. This study focused on the contributions of SUVH2 and SUVH9 to the regulatory mechanism of ONSEN.
2 Materials and methods
2.1 Plant material
Arabidopsis thaliana accession Columbia (Col-0) was used as a wild-type control. The suvh2 (SALK_079574) and suvh9 (SALK_048033) mutants were obtained from the ABRC stock center, and the nrpd1 (C366150) mutant was obtained from Ohio State University. Suvh2 mutants were crossed with suvh9 or nrpd1 mutants to generate suvh2/9 and suvh2/nrpd1 double mutants, respectively. All mutants were of the Col-0 background.
2.2 Growth conditions
Seeds sterilized in a 1:1 solution of sodium hypochlorite and 0.04% Triton X-100 were planted in 0.5× Murashige and Skoog (MS) medium. Seeds were sowed at 4°C (dark conditions) for 2 days and then cultured in 0.5x MS medium or potting (soil: vermiculite = 1:7) at 21°C under continuous light.
2.3 Heat-stress treatment
Seven-day-old seedlings grown in 0.5× MS medium at 21°C under continuous light were subjected to heat stress at 37°C. For Southern blot experiments, we allowed them to recover to 21°C for two days before transferring them to the soil because the 7-day-old plants we analyzed would be weakened if transplanted directly into the soil immediately after heat treatment. Then, the seedlings were transferred to the soil at 21°C under continuous light. Progeny seeds were directly planted in the soil at 21°C under continuous light.
2.4 Southern blotting
DNA was extracted from 3-week-old plants using a Nucleon PhytoPure DNA Extraction Kit (Cytiva, Tokyo, Japan). The 2.4 µg of extracted DNA was processed with the restriction enzyme EcoRV overnight at 37°C, and the enzyme-treated DNA was purified by ethanol precipitation. DNA was electrophoresed on a 1% agarose gel for 24 h at 20 V. The DNA was transferred onto a Biodyne B Nylon Membrane (Thermo Fisher Scientific, Waltham, Massachusetts, USA) overnight. PCR product for the probe was generated by TaKaRa Ex Taq (TaKaRa, Kusatsu, Japan). The 577 bp of the predicted ORF of ONSEN was used as a probe. The PCR primers are shown in Supplementary Table 1. Hybridization signals were detected in a high SDS hybridization buffer using the Megaprime DNA Labeling System (Cytiva, Tokyo, Japan) with radioisotope-labeled probes.
2.5 Quantitative analysis
For qRT-PCR, total RNA was extracted using the TRI reagent (Sigma-Aldrich, St. Louis, Missouri, USA). After treatment with RQ1 Rnase-Free Dnase (Promega, Madison, Wisconsin, USA), RNA was reverse-transcribed using the ReverTra Ace qPCR RT Kit (TOYOBO, Osaka, Japan) to synthesize cDNA. The total DNA was extracted using the Nucleon PhytoPure DNA Extraction Kit (Cytiva, Tokyo, Japan) for qPCR. The Ct method was used to determine expression, and the expression of the 18S rRNA gene was used as an internal control (Schmittgen and Livak, 2008). qPCR amplification was performed with the appropriate primers (Supplementary Table 1).
2.6 DNA methylation analysis
The DNA used for bisulfite sequencing was obtained from 7-day-old seedlings. Total DNA was bisulfite-converted and desulfated using a MethylCode Bisulfite Conversion Kit (Invitrogen, Waltham, Massachusetts, USA). The target fragments were amplified by PCR (40 cycles, 98°C for 10 s, 55°C for 30 s, 72°C for 1.5 min), and the PCR products were cloned. PCR amplification was performed with the appropriate primers (Supplementary Table 1). Twenty-four clones were sequenced, and the results were analyzed using the MEGA-X software and the Cymate website to detect DNA methylation levels.
2.7 Formaldehyde-assisted isolation of regulatory elements
Seven-day-old seedlings (0.3 g) were cross-linked in a 1% formaldehyde solution (Thermo Fisher Scientific, Waltham, Massachusetts, USA) under vacuum for 10 min (2 min + 8 min). The cross-linking was quenched by adding glycine at a final concentration of 0.125 M for 5 min under vacuum. The samples were ground to powder with liquid nitrogen, and separation buffer 1 (0.4 M sucrose, 10 mM Tris-HCl, pH 8, 10 mM MgCl2, 5 mM β-mercaptoethanol, and protease inhibitor [Roche, Basel, Switzerland]) was added, followed by filtration using Miracloth (Millipore, Burlington, Massachusetts, USA) to remove cell debris. After centrifugation at 2000 g for 20 min at 4°C, precipitated nuclei were dispersed in isolation buffer 2 (0.25 M sucrose, 10 mM Tris-HCl, pH 8, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, and protease inhibitor [Roche, Basel, Switzerland]). After centrifugation at 13,000 rpm for 20 min at 4°C, the precipitated nuclei were re-dispersed in Isolation Buffer 2. Add Isolation Buffer 3 (0.25 M sucrose, 10 mM Tris-HCl, pH 8, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, and protease inhibitor [Roche, Basel, Switzerland]) to keep the solution stratified and centrifuge for 60 min at 4°C at 13000 rpm. The precipitates were resuspended using 0.3 mL of SDS nuclear lysis buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, 1% SDS, and protease inhibitor cocktail). The chromatin was sheared using Covaris M220 (Covaris, Woburn, Massachusetts, USA). Suspensions were centrifuged at 13,000 rpm for 10 min at 4°C to obtain a supernatant. DNA was purified using the phenol-chloroform method. The abundance of DNA fragments relative to the input DNA was determined using qPCR. qPCR amplification was performed with the appropriate primers (Supplementary Table 1). For input DNA treatment, 2 µL of supernatant was mixed with 198 µL of extraction buffer (10 mM Tris-HCl pH 8, 0.3 M NaCl, 5 mM EDTA, 0.5% SDS) and incubated overnight at 65°C to reverse formaldehyde cross-linking. The de-crosslinked DNA was purified using the phenol-chloroform method after treatment with Rnase and proteinase K. Input DNA was used as an internal control to identify the non-crosslinked portion of the condensed genomic region (NDR).
2.8 Cytology
The leaves of 7-day-old plants were fixed overnight with ethanol: acetic acid (3:1). Leaves were treated with the enzymes (boehmotoxin [Yakult Pharmaceutical Industries, Yakult Pharmaceutical Industries], pectinase [Kyowa Chemical Products, Tokyo, Japan], and cytochromes [Sigma-Aldrich, St. Louis, Missouri, USA] [1% (v/v) in citrate buffer]) at 37°C for 2 h. The leaves were placed on a slide on a heating plate at 45°C for 30 s, and the leaves were simultaneously torn with a needle to disperse the tissue in 45% acetic acid. After the acetic acid had evaporated, a fixative was added, and the slides were dried. Staining was performed by adding 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Newark, California, USA).
2.9 Northern blotting
Total RNA was extracted using the TRI reagent (Sigma-Aldrich, St. Louis, Missouri, USA). Low-molecular-weight RNA was purified by ethanol precipitation and dissolved in 100% formamide. RNA samples (1400ng) were denatured at 65°C for 5 min, and electrophoresis was performed on a 15% PAGE gel in 0.5× TBE buffer, run at 50 V for 20 min to make the dyes move at the same rate, and run at 200 V and 500 mA for 3 h. The RNA was transferred onto a Hybond-N+ hybridization membrane (Sigma-Aldrich, St. Louis, Missouri, USA) overnight and hybridized with a DIG-labeled RNA probe at 40°C overnight. We used synthetic oligonucleotides containing the T7 RNA polymerase promoter and the MRGA shortscript kit for generating probe (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The PCR primers for the probe are shown in Supplementary Table 1. DIG-labeled RNA probes were synthesized using the DIG RNA Labeling Mix (Roche, Basel, Switzerland). Hybridized signals were detected using anti-dig (Roche, Basel, Switzerland) and CDP Star (Roche, Basel, Switzerland) using a LAS3000.
3 Results
3.1 Deletion of SUVH2 results in massive activation of ONSEN transcription
We investigated transcript levels and copy numbers (extrachromosomal cDNA levels) of ONSEN in WT, suvh2, suvh9, and nrpd1. We observed that after 24 h of heat stress, the transcript level and copy number of ONSEN in suvh2 were significantly increased compared to those in the WT and exhibited approximately the same level as in nrpd1 (Figures 1A, B), suggesting that deletion of suvh2 released the transcriptional silencing of ONSEN. Consistent with previous findings, 48 h of heat treatment resulted in a further increase in ONSEN copy compared with 24 h of heat treatment (Figure 1C). In suvh9, ONSEN exhibited the same transcript levels and copy number as in the WT (Figures 1A, B). To determine the synergistic effect of SUVH2 and SUVH9 on the RdDM pathway, we investigated the transcript and copy number levels of ONSEN in the suvh2/9 double mutant. However, ONSEN did not exhibit higher transcript levels or copy numbers in suvh2/9 than in suvh2 (Figures 1D, E), suggesting that SUVH9 was not involved in the transcriptional repression of ONSEN by RdDM.
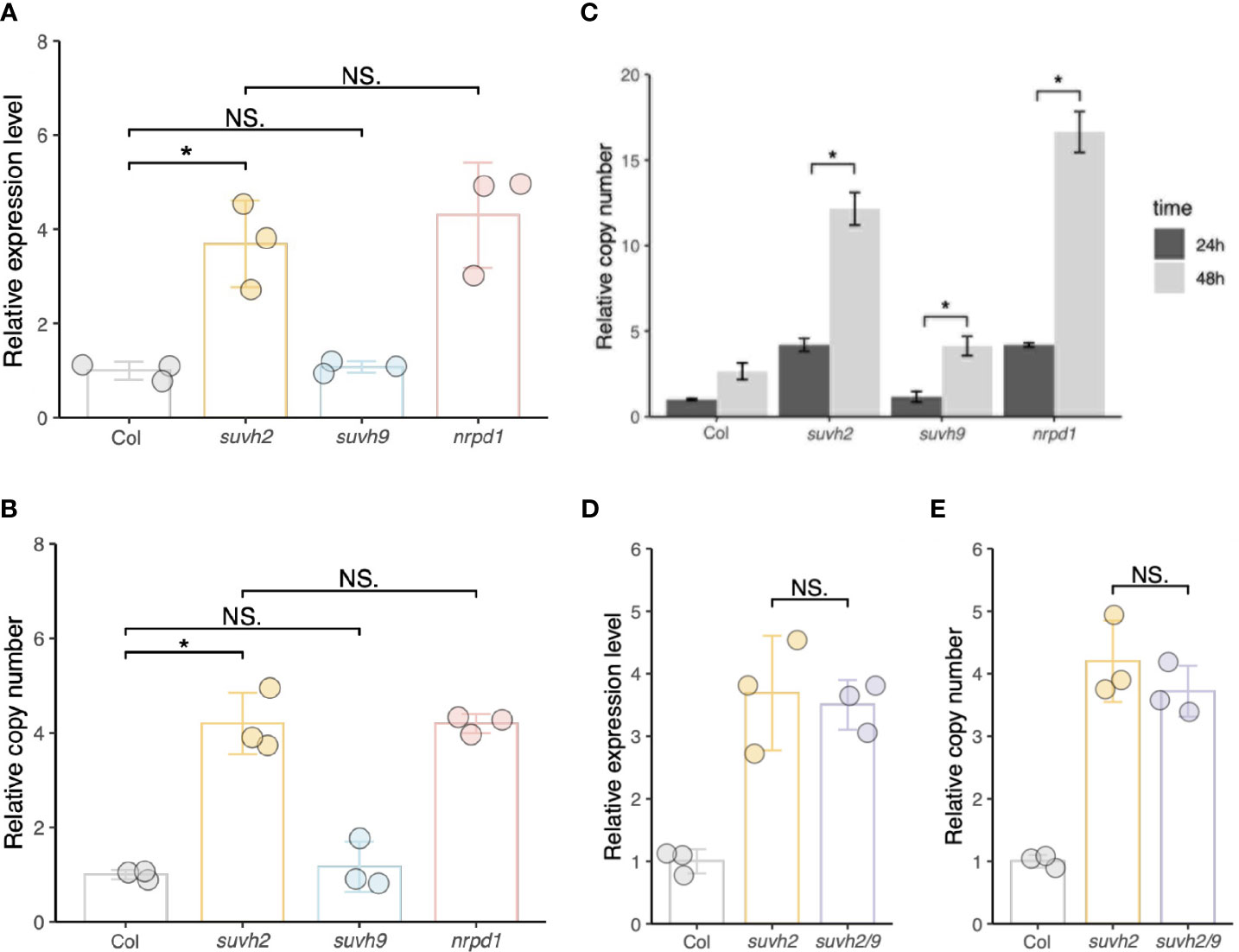
Figure 1 SUVH2 is essential for the transcriptional repression of ONSEN but not SUVH9. (A) and (B) Relative expression levels (A) and relative copy number (B) of ONSEN in wild-type, suvh2, suvh9, and nrpd1 mutants at 24 h heat stress. (C) Relative copy number of ONSEN in wild type, suvh2, suvh9, and nrpd1 at 24 h and 48 h heat stress. (D) and (E) Relative expression levels (D) and relative copy numbers (E) of ONSEN in wild-type, suvh2, and suvh2/9 mutants at 24 h heat stress. Asterisks indicate significant differences between the two groups (Student’s t-test, P ≤0.05). NS indicates no significant differences between the two groups (Student's t-test, P >0.05).
3.2 The transposition of ONSEN is infrequent in suvh2 mutants
To investigate whether the disruption of SUVH2 and SUVH9 affects ONSEN transposition, we investigated the transgenerational transposition of ONSEN in suvh2 and suvh9 mutants. Previous studies have shown that transposition frequency correlates with the duration of heat stress; suvh2 and suvh9 were subjected to heat stress for 24 and 48 h, respectively. Southern blot analysis showed that ONSEN transposition was observed in the suvh2 mutant after 48 h of heat stress. In contrast, no transgenerational transposition occurred after 48 h of heat stress in the suvh9 mutant (Figure 2A). This suggested that SUVH2 regulates ONSEN transposition. However, the frequency of ONSEN transposition in suvh2 was much lower than nrpd1 (Figure 2A). Because SUVH2 and SUVH9 are partially non-redundant in RdDM (Kuhlmann and Mette, 2012), we investigated ONSEN transposition in suvh2/9. The results showed that no new ONSEN insertion was observed in suvh2/9 after 24 h of heat treatment; however, a new insertion was observed after 48 h of heat stress (Figure 2B). The frequency of ONSEN transposition was low in suvh2/9 cells (Figure 2B), suggesting that SUVH2 and SUVH9 do not synergistically suppress ONSEN transposition. No ONSEN transposition was observed under heat stress in suvh9 (Figure 2A), suggesting that SUVH9 was not directly involved in suppressing ONSEN transposition. Although ONSEN cDNA accumulated in suvh2 after heat stress, it could not be inserted into new genomic loci. Next, we investigated the activity of ONSEN in suvh2/nrpd1 double mutants. In the suvh2/nrpd1 double mutant after 24 h of heat treatment, ONSEN exhibited a higher transposition frequency than suvh2 (Figure 2C). However, the transposition frequency is similar to that in nrpd1 (Figure 2C). Also, we analyzed the transposition and cDNA levels of ONSEN in suvh2/nrpd1 under 48h HS. The results showed that ONSEN in suvh2/nrpd1 after 48h heat treatment showed higher transposition frequency and cDNA levels. The copy number of ONSEN was significantly higher in suvh2/nrpd1 than in suvh2 or nrpd1 (Figure 2D). This suggests that SUVH2 and NRPD1 synergistically affect the regulation of ONSEN transcription, but not transposition.
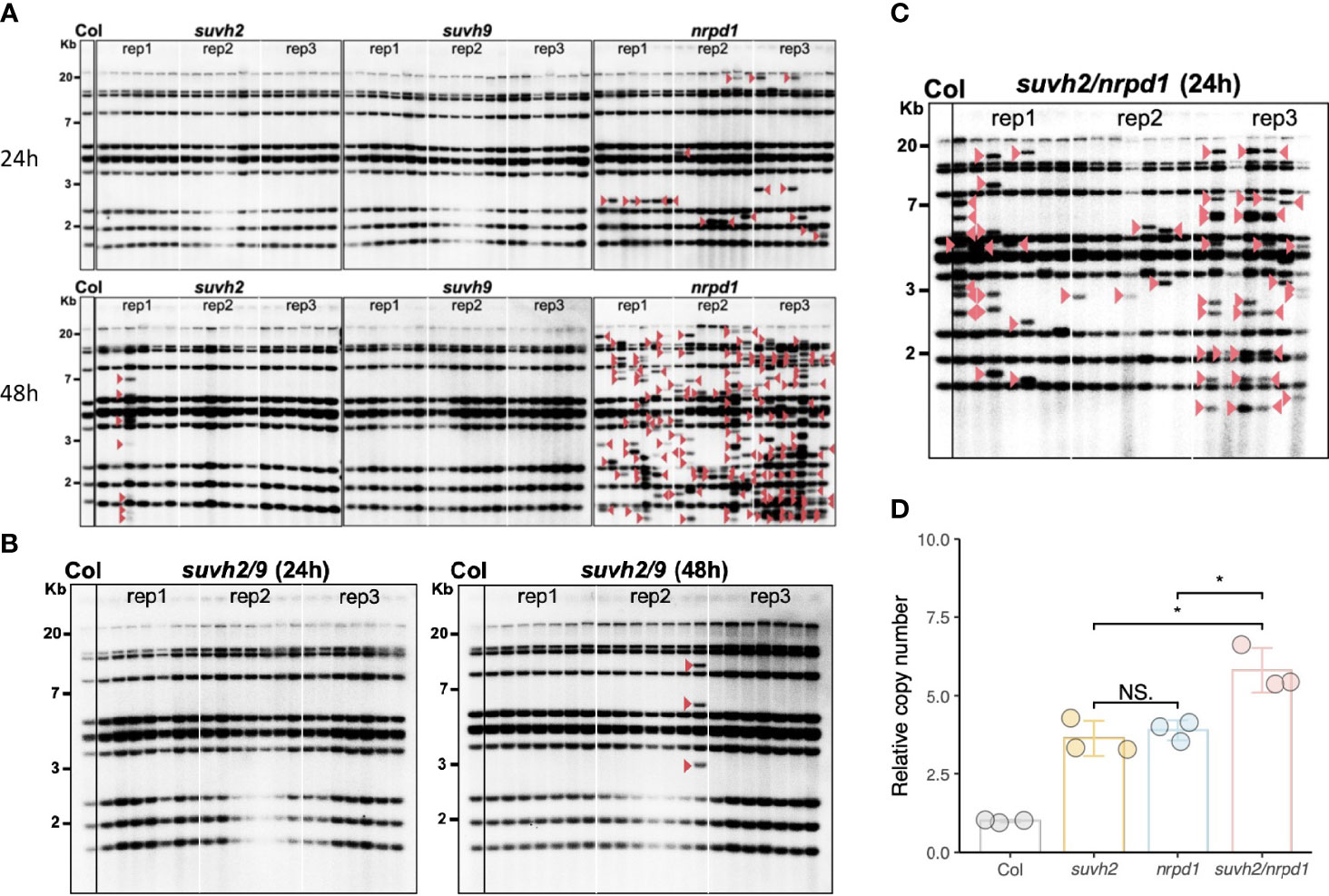
Figure 2 In suvh2 mutants, the frequency of ONSEN transposition is low. (A) Southern blot results show transgenerational transposition of ONSEN in suvh2, suvh9, and nrpd1 under heat stress of 24 h (top) or 48 h (bottom). (B) New insertions of ONSEN in suvh2/9 under heat stress conditions at 24 h (left) or 48 h (right). The leftmost lane of each graph is the WT under non-stress conditions. For each mutant, three plants were subjected to heat stress (24 h or 48 h), and seven progenies were considered for ONSEN transposition analysis. Red arrow head indicates ONSEN insertions. (C) The next generation of suvh2/nrpd1 mutants after 24 h heat stress was analyzed by Southern blot to examine the transgenerational transposition of ONSEN. The leftmost lane of each graph is the wild type under non-stress conditions. Three homozygous mutant plants were heat stressed, and seven progenies were considered for ONSEN transposition analysis for each individual. Red triangles indicate insertions of ONSEN. (D) Relative copy number of ONSEN in wild-type, suvh2, nrpd1, and suvh2/nrpd1 under 24 h of heat stress. Asterisks indicate significant differences between the two groups (Student’s t-test, P ≤0.05). NS indicates no significant differences between the two groups (Student’s t-test, P >0.05).
3.3 DNA hypomethylation was independent of the transposition of ONSEN in suvh2
SUVH2 and SUVH9 are essential for DNA methylation (Liu et al., 2014). To determine what suppresses ONSEN transposition in suvh2, we investigated DNA methylation levels of the ONSEN in the SUVH mutant. We observed that the level of CHH methylation in ONSEN was significantly reduced in suvh2 compared to WT (Figure 3; Table 1), suggesting that SUVH2 is essential for establishing CHH DNA methylation in ONSEN. This result was similar to that obtained for the single-copy SINE AtSN1 (Johnson et al., 2008). The loss of SUVH2 and SUVH9 resulted in lower CHH methylation levels (Figure 3; Table 1), suggesting that SUVH2 and SUVH9 have synergistic effects in regulating methylation. However, the level of CHH methylation in ONSEN was significantly higher in suvh9 mutants than in the WT (Figure 3; Table 1). The ONSEN promoter is present in the LTR region, and DNA methylation is present only in the CHH contexts. The CHH hypermethylation of ONSEN in suvh9 may be responsible for the inability of ONSEN to be transcribed at a high level. Heat stress did not change the DNA methylation levels of the ONSEN region in suvh2 (Supplementary Figure 1), which is consistent with our previous findings (Niu et al., 2022b). In addition, the DNA methylation pattern of ONSEN in suvh2 was nearly identical to that of nrpd1 (Figure 3), indicating that DNA hypomethylation released transcriptional silencing of ONSEN but did not affect transposition.
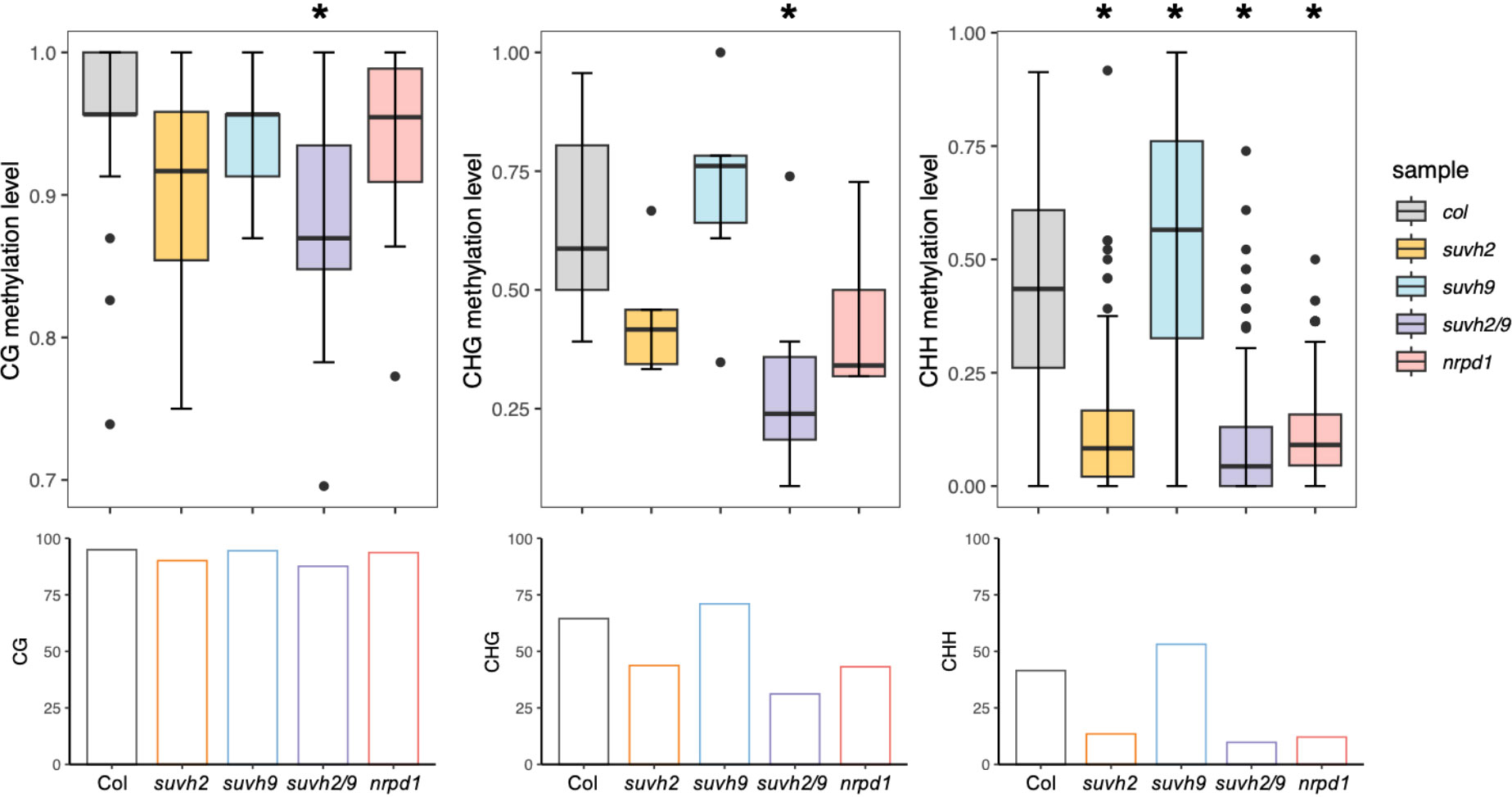
Figure 3 SUVH2 is essential for the establishment of DNA methylation on ONSEN. Box plots (top) showing CG (left), CHG (middle), and CHH (right, H = A, T, C) methylation levels of ONSEN (At1g11265) in wild-type, suvh2, suvh9, suvh2/9, and nrpd1 mutants under non-stress conditions. Bar graphs (bottom) show the average levels of CG (left), CHG (middle), and CHH (right, H = A, T, C) methylation of ONSEN in each mutant. Asterisks indicate significant differences compared to Col (Student’s t-test, P > 0.05).
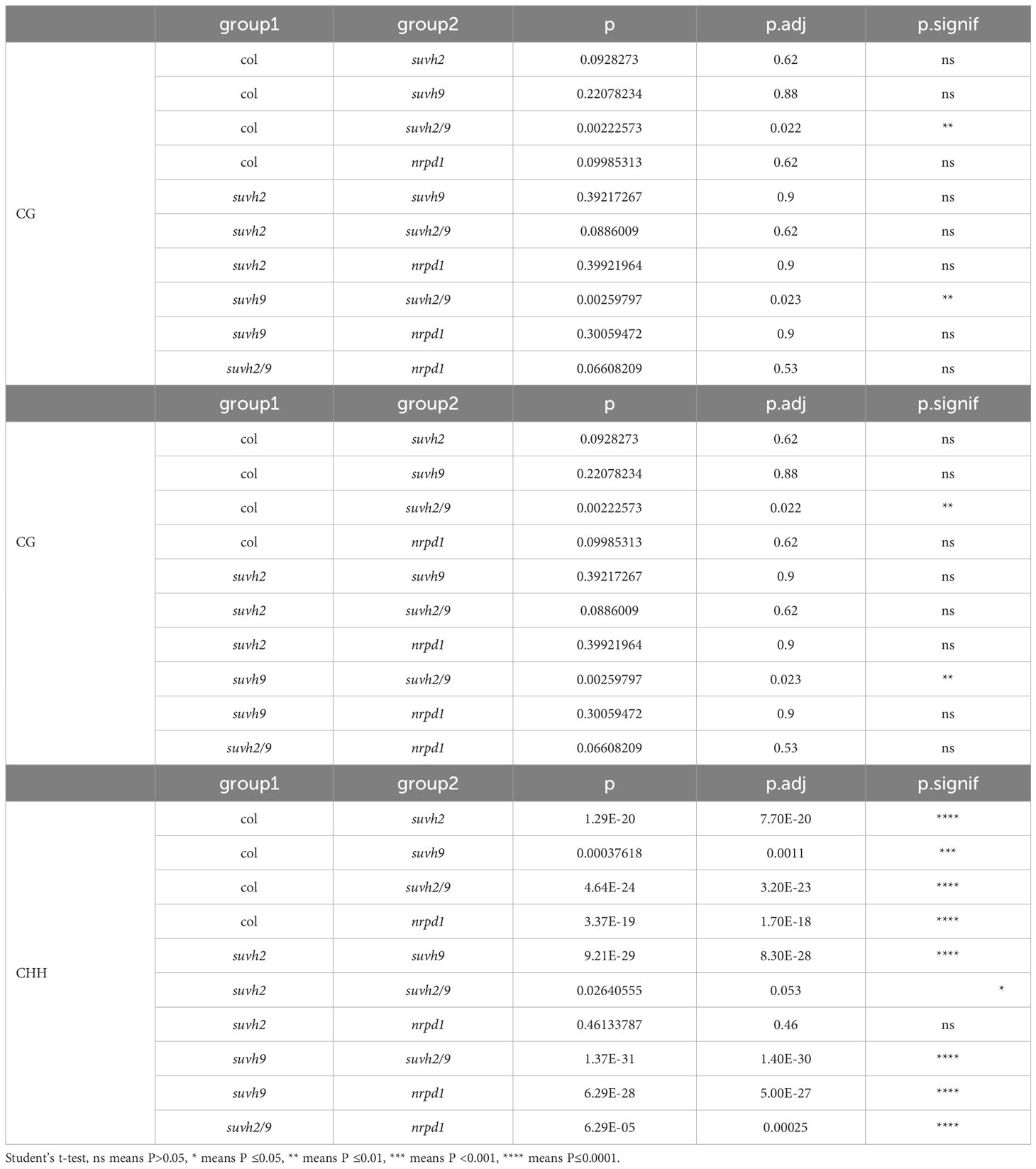
Table 1 Results of significant difference analysis of methylation data and asterisks indicate significant differences in Figure 3 (Student’s t-test, P < 0.05).
3.4 Chromatin condensation was independent of the activation of ONSEN
Chromatin repression causes transgene silencing in a non-DNA methylation-dependent manner. (Mittelsten Scheid et al., 2002). FAIRE-qPCR was used to examine the status of chromatin condensation in the LTR, or gene body region, of ONSEN. The levels of open chromatin in the LTR and gene body of ONSEN in suvh2 and suvh2/9 were much lower than those in nrpd1 (Figures 4A, B). Since the promoter of ONSEN exists within LTRs, we concluded that the open chromatin of ONSEN does not directly correlate with the transcriptional repression of ONSEN. The open chromatin of the ONSEN gene body region appeared to be lower in suvh2/nrpd1 double mutants than in nrpd1 (Figure 4B). As heat stress activates ONSEN, producing extrachromosomal cDNAs, FAIRE of the ONSEN region after heat treatment is challenging. DAPI staining was used to investigate the condensation of heterochromatin in the nuclei of the mutants. We categorized heterochromatin as condensed or dispersed (Figure 4C). Heat stress loosened the heterochromatin; dispersed nuclei showed similar proportions across mutants with or without heat stress treatment (Figures 4D, E). These results imply that the chromatin state may not affect ONSEN transcription or transposition.
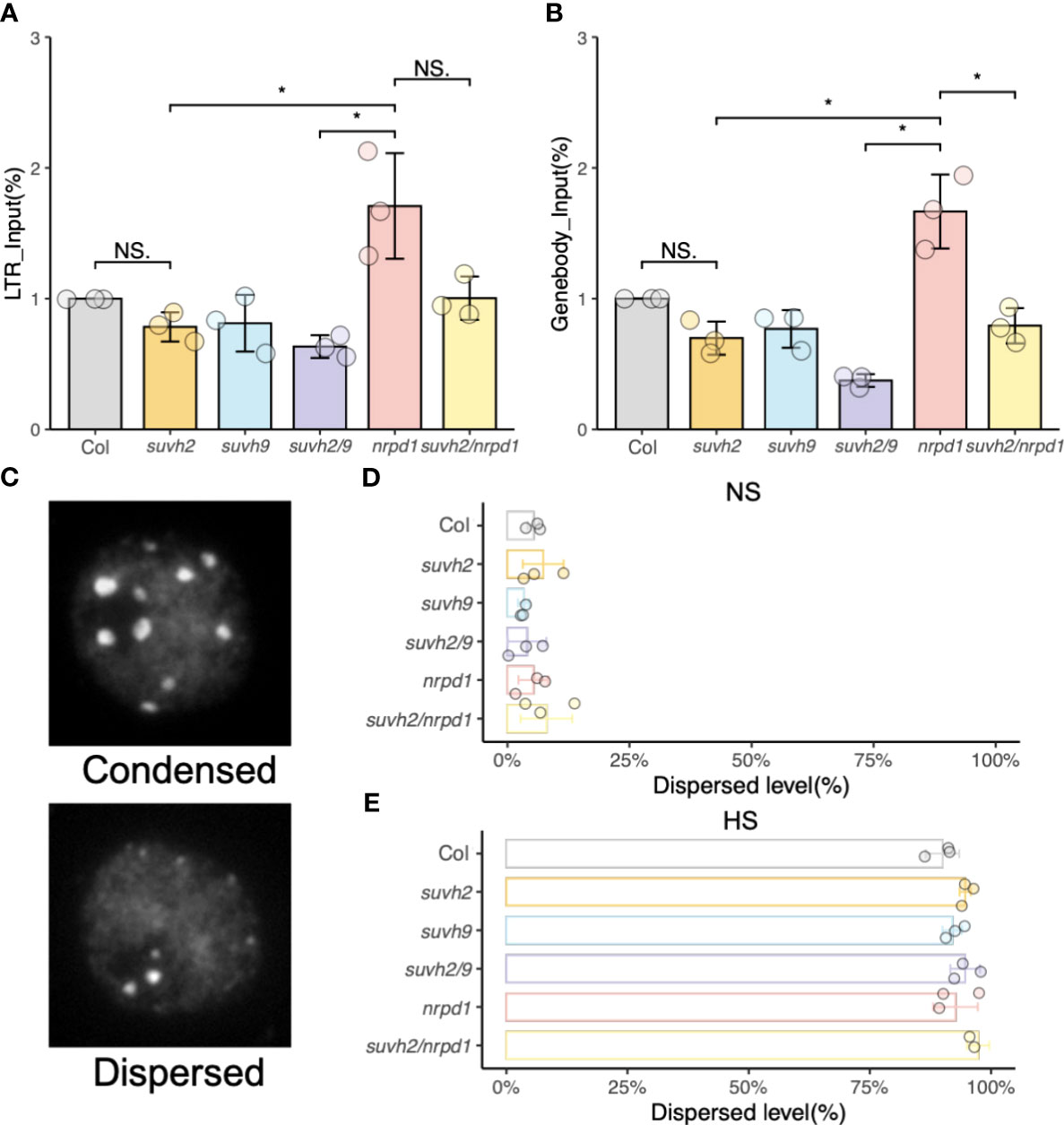
Figure 4 Analysis of open chromatin. (A) and (B) FAIRE-qPCR results show the level of open chromatin in a small region of LTR (A) and genebody (B) on ONSEN. (C) Representative images of nuclei under DAPI staining. (D) and (E) Percentage of chromatin in two (condensed or dispersed) states in wild-type, suvh2, suvh9, suvh2/9, nrpd1, and suvh2/nrpd1 under non-stress (NS) (C) and 48 h heat stress (HS) (D) (n = 150). Asterisks indicate a significant difference between the two groups (Student’s t-test, P <0.05). NS indicates no significant difference between the two groups (Student’s t-test, P ≥0.05).
3.5 The siRNA regulates ONSEN transcription and transposition in suvh2
Pol IV is a significant factor in siRNA synthesis, and siRNAs are involved in ONSEN’s transcriptional repression regulation. Therefore, we explored the siRNA accumulation of ONSEN in each mutant. First, we investigated the accumulation of siRNAs in the LTR of ONSEN in WT plants under non-stress and heat-stress conditions. The results showed that siRNA was not produced immediately after heat stress and gradually accumulated in recovery at 21 degrees (Figure 5A). siRNA was produced in the WT plants under non-stressed conditions, and the amount of siRNA gradually increased over time (Figure 5A).
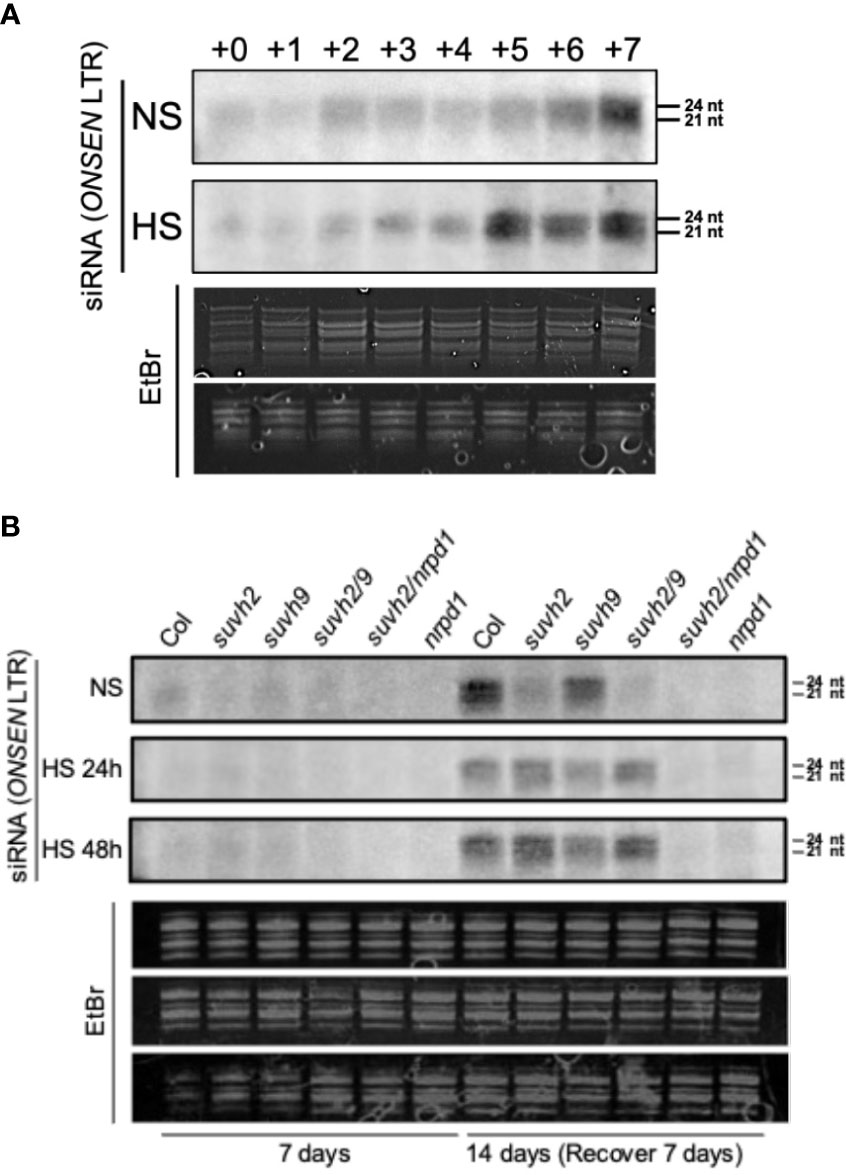
Figure 5 Northern blot analysis to investigate siRNA accumulation from the ONSEN LTR. (A) Levels of siRNA accumulation in WT under non-stress (NS) or heat stress (HS) conditions. The numbers above represent the accumulation levels of siRNAs in one-week-old seedlings placed under non-stress (NS) or heat stress (HS) conditions (37°C) for 24 h (+0) and recovery at normal conditions (21°C) for one day (+1), two days (+2)…seven days (+7) after heat stress treatment. (B) Accumulation levels of siRNAs in wild-type, suvh2, suvh9, suvh2/9, nrpd1, and suvh2/nrpd1 in 7-week-old seedlings treated with non-stress (NS) or heat stress (HS) (24 h or 48 h) and recovered at normal conditions (21°C) for seven days (+7) after treatment.
Because large amounts of siRNA were not produced immediately after heat stress, we investigated the accumulation levels of siRNA after heat stress and after 7 days of recovery. Accumulation of siRNA was observed in WT, suvh2, suvh9, and suvh2/9 plants recovered at 21 degrees for 7 days after both 24-h and 48-h heat treatments (Figure 5B). As expected, we did not detect siRNA production in plants deficient in NRPD1 (Figure 5B). This suggests that the deletion of SUVH2 and/or SUVH9 did not completely disrupt siRNA production, inhibiting ONSEN transposition. In contrast, accumulation of siRNAs was observed in suvh9 without heat stress, whereas siRNAs did not appear in suvh2 and suvh2/9 (Figure 5B). Because heat stress did not increase ONSEN transcription in suvh9 (Figure 1A), we suggest that siRNA produced under non-stress conditions may make it difficult for ONSEN to be transcribed when subjected to heat stress. Since the deletion of SUVH2 reduced the production of ONSEN siRNA under non-stress conditions (Figure 5B), thereby diminishing siRNA involvement in heat stress, which could lead to the disruption of the siRNA-dependent transcriptional repression of ONSEN, However, suvh2 mutant plants under heat stress showed similar amount of siRNAs to WT plants, and we propose that it is precisely the siRNAs produced after heat stress that result in the inhibition of ONSEN transposition in suvh2 mutants (Figure 6).
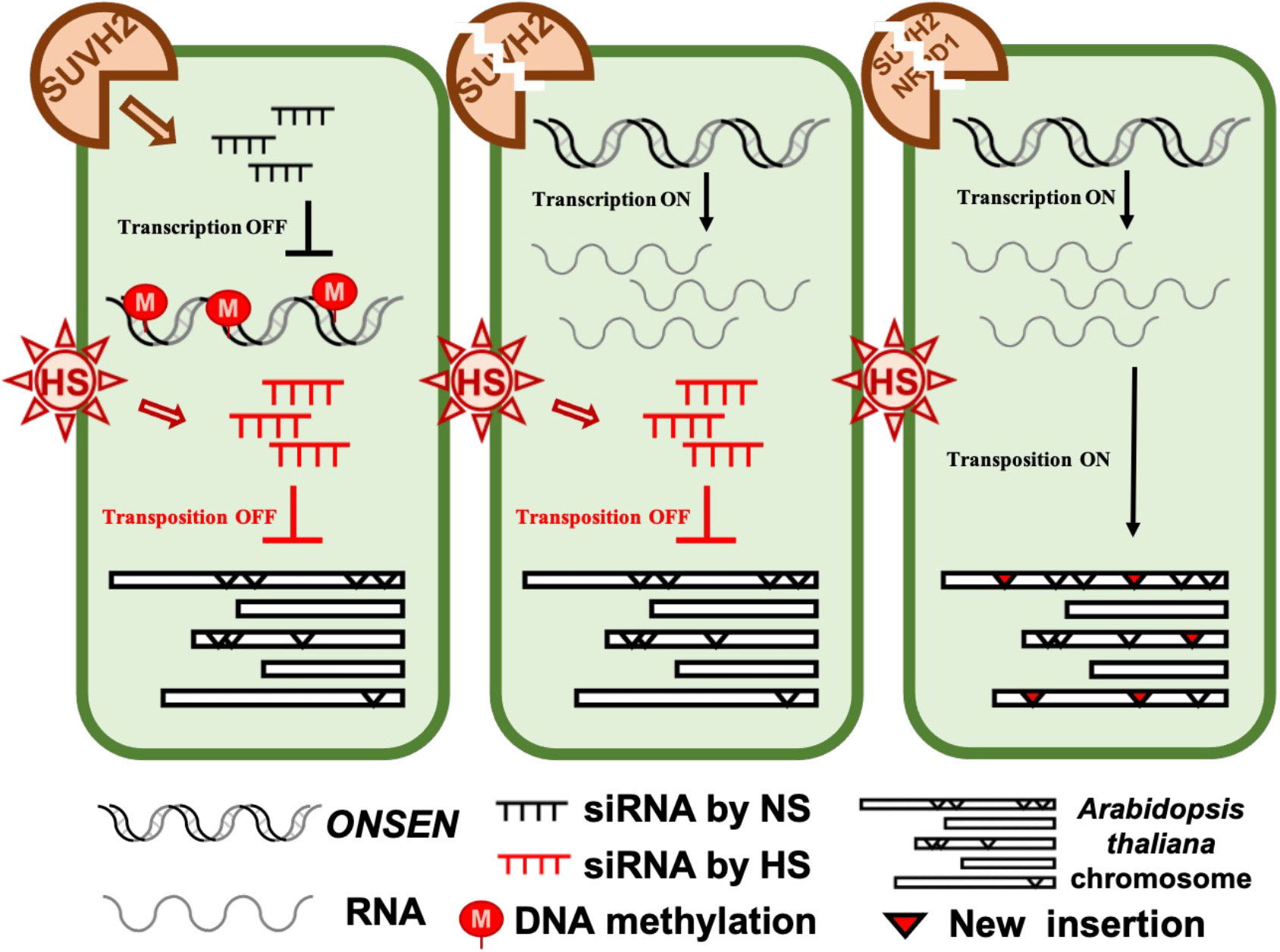
Figure 6 The model of SUVH2 involves ONSEN regulation. In the wild-type, intact RdDM can produce siRNAs to suppress ONSEN transcription. In the suvh2 mutant, the RdDM pathway is disrupted, releasing ONSEN transcription by heat stress. Under heat stress, plants bypass the SUVH2-dependent pathway to produce new siRNAs, and these siRNAs produced by heat stress prevent ONSEN from transposing into other sites on the chromosome. In the suvh2/nrpd1 double mutant, siRNA could not be synthesized under heat stress conditions releasing of transpositional silencing of ONSEN.
4 Discussion
RdDM is essential for plant resistance to biotic and abiotic stresses (Erdmann and Picard, 2020). Downstream of the RdDM pathway, SUVH2 and SUVH9 are significant for the association of Pol V with chromatin (Johnson et al., 2014). In this study, we investigated the inhibition of ONSEN activity by SUVH2 and SUVH9. Our results show that SUVH2 represses a large amount of ONSEN transcription. In contrast, SUVH9 did not transcriptionally repress ONSEN (Figure 2A). AtSN1, a SINE retrotransposon found in A. thaliana, is a model target for studying RdDM and its transcriptional silencing. Liu et al. proposed that SUVH2 and SUVH9 have redundant functions in transcriptional silencing and observed that AtSN1 expression was increased in the suvh2/9 double mutant and not in the suvh2 or suvh9 single mutant (Liu et al., 2014). This result differed from the expression of ONSEN, and we hypothesized that SUVH2 and SUVH9 are partially non-redundant when involved in the transcriptional repression of ONSEN. Another possibility is that SUVH9 is not involved in the transcriptional repression of ONSEN.
We observed the new ONSEN insertions in the progeny of the 48-h heat-stressed suvh2 (Figure 1A). In contrast, ONSEN transposition was suppressed in the plants after 24 h of heat stress (Figure 1A). The copy number of ONSEN was not reduced in suvh2 compared to that in nrpd1, in which a high frequency of ONSEN transposition occurred (Figures 1A, 2C). This may be due to the post-transcriptional regulation of ONSEN in suvh2.
SUVH2 and SUVH9 lack the SET post-structural domain, and although they do not have histone methyltransferase activity, they can participate in RdDM by binding methylated DNA through their SET and RING-associated (SAR) domains (Johnson et al., 2008). The level of DNA methylation in the LTR region of ONSEN was lost in SUVH2-deficient plants, especially CHH methylation (Figure 3). A related report on SDC showed that a lack of either suvh2 or suvh9 results in the loss of CHH methylation (Johnson et al., 2008). This contrasts with our finding that there was a slight increase instead of a decrease in DNA methylation in the ONSEN region of suvh9 (Figure 3). The high DNA methylation level of ONSEN in suvh9 is responsible for its low transcriptional activity. ONSEN showed a similar pattern of DNA methylation in suvh2 and nrpd1 (Figure 3). In conclusion, we suggest that the transcriptional activity of ONSEN depends on the level of DNA methylation.
Heterochromatin is enriched with transposable elements (Marsano and Dimitri, 2022). Although chromatin accessibility is associated with establishing DNA methylation (Zhong et al., 2021), it has been suggested that chromatin remodeling factors may occur independently of the DNA methylation process. For example, Morpheus Molecue 1 (MOM1) is required to silence repetitive heterochromatin sequences and is involved in epigenetic modification in a DNA methylation-independent manner (Amedeo et al., 2000). Similarly, AtMORC1 and AtMORC6 are involved in the RdDM pathway by controlling the decondensation of heterochromatin around filaments and not through DNA methylation (Moissiard et al., 2012). Jing et al. identified SUVH9 as a linker between MORC-mediated chromatin remodeling (Jing et al., 2016). We speculated that SUVH9 might be involved in silencing ONSEN via a non-DNA methylation pathway. Deletion of SUVH2 and/or SUVH9 did not result in open chromatin in the ONSEN LTR or gene body (Figures 4A, B). SUVH9 plays a minimal role in ONSEN silencing. In addition, the results of DAPI staining suggested that heat stress leads to the decondensation of heterochromatin. However, heat-induced decondensation of heterochromatin was not nprd1 mutant-specific (Figures 4D, E). These results suggest that heterochromatin decondensation does not directly affect the transgenerational transposition of ONSEN. Open chromatin may be essential for transposition activity. However, it has been challenging to determine open chromatin in the promoter region of ONSEN under HS because of the presence of cDNA made from ONSEN. Whether open chromatin is required for ONSEN insertion is also considered, and information on the insertion site is needed. Previous studies have shown that ONSEN in nrpd1 mutants exhibits random insertions (Ito et al., 2016). Investigating the open chromatin of heat-stressed random insertion sites is a challenge.
siRNAs are closely associated with gene silencing (Hamilton et al., 2002). Pol IV transcribes TE- and repeat-related genes as primary transcripts that are loaded by RDR2 (RNA-dependent RNA polymerase 2), DCL3 (Dicer-like 3), and AGO4 (ARGONAUTE 4) proteins to produce siRNAs (Ferrafiat et al., 2019). Transgenerational transposition of ONSEN often occurs in nrpd1 mutants (Ito et al., 2011; Ito et al., 2016). Northern blot analysis showed that, although ONSEN siRNAs were not observed in suvh2 and suvh2/9 under non-stress conditions, new siRNAs were synthesized after heat stress (Figure 5B). In contrast, deletion of SUVH9 did not disrupt siRNA synthesis under non-stress or heat-stress conditions. This indicated that the absence of SUVH2 affected the siRNA synthesis activity of ONSEN.
In conclusion, our results showed that SUVH2 is involved in the transcriptional silencing of ONSEN in a siRNA pathway-dependent manner. We provide evidence that SUVH2 and SUVH9 are functionally non-redundant when involved in regulating retrotransposon activity. In addition, plants can adapt their protective mechanisms to cope with the possibility of genetic disruption resulting from adversity.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
XN: Writing – original draft. ZG: Writing – original draft. HI: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare fnancial support was received for the research, authorship, and/or publication of this article. This work was supported by JSPS Grant-in-Aid for Scientific Research C (21K06008), JSPS Fund for the Promotion of Joint International Research A (Fostering Joint International Research A) (21KK0263), and Hokkaido University Ambitious Doctoral Fellowship.
Acknowledgments
We would like to thank Dr. Yoko Ikeda for her assistance in FAIRE analysis, Mr. Yusuke Nakada for his technical advice in Northern blotting, and Dr. Leandro Quadrana for his comments on transposition analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1355626/full#supplementary-material
References
Amedeo, P., Habu, Y., Afsar, K., Mittelsten Scheid, O., Paszkowski, J. (2000). Disruption of the plant gene MOM releases transcriptional silencing of methylated genes. Nature 405 (6783), 203–206. doi: 10.1038/35012108
Bennetzen, J. L., Wang, H. (2014). The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 65, 505–530. doi: 10.1146/annurev-arplant-050213-035811
Berthelier, J., Furci, L., Asai, S., Sadykova, M., Shimazaki, T., Shirasu, K., et al. (2023). Long-read direct RNA sequencing reveals epigenetic regulation of chimeric gene-transposon transcripts in Arabidopsis thaliana. Nat. Commun. 14 (1), 3248. doi: 10.1038/s41467-023-38954-z
Bourque, G., Burns, K. H., Gehring, M., Gorbunova, V., Seluanov, A., Hammell, M., et al. (2018). Ten things you should know about transposable elements. Genome Biol. 19 (1), 199. doi: 10.1186/s13059-018-1577-z
Brabbs, T. R., He, Z., Hogg, K., Kamenski, A., Li, Y., Paszkiewicz, K. H., et al. (2013). The stochastic silencing phenotype of Arabidopsis morc6 mutants reveals a role in efficient RNA-directed DNA methylation. Plant J. 75 (5), 836–846. doi: 10.1111/tpj.12246
Erdmann, R. M., Picard, C. L. (2020). RNA-directed DNA methylation. PloS Genet. 16 (10), e1009034. doi: 10.1371/journal.pgen.1009034
Ferrafiat, L., Pflieger, D., Singh, J., Thieme, M., Bohrer, M., Himber, C., et al. (2019). The NRPD1 N-terminus contains a Pol IV-specific motif that is critical for genome surveillance in Arabidopsis. Nucleic Acids Res. 47 (17), 9037–9052. doi: 10.1093/nar/gkz618
Gallego-Bartolome, J. (2020). DNA methylation in plants: mechanisms and tools for targeted manipulation. New Phytol. 227 (1), 38–44. doi: 10.1111/nph.16529
Grandbastien, M. A., Lucas, H., Morel, J. B., Mhiri, C., Vernhettes, S., Casacuberta, J. M. (1997). The expression of the tobacco Tnt1 retrotransposon is linked to plant defense responses. Genetica 100 (1-3), 241–252. doi: 10.1007/978-94-011-4898-6_24
Hamilton, A., Voinnet, O., Chappell, L., Baulcombe, D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21 (17), 4671–4679. doi: 10.1093/emboj/cdf464
Havecker, E. R., Gao, X., Voytas, D. F. (2004). The diversity of LTR retrotransposons. Genome Biol. 5 (6), 225. doi: 10.1186/gb-2004-5-6-225
Hayashi, Y., Takehira, K., Nozawa, K., Suzuki, T., Masuta, Y., Kato, A., et al. (2020). ONSEN shows different transposition activities in RdDM pathway mutants. Genes Genet. Syst. 95 (4), 183–190. doi: 10.1266/ggs.20-00019
Ito, H., Gaubert, H., Bucher, E., Mirouze, M., Vaillant, I., Paszkowski, J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472 (7341), 115–U151. doi: 10.1038/Nature09861
Ito, H., Kim, J. M., Matsunaga, W., Saze, H., Matsui, A., Endo, T. A., et al. (2016). A stress-activated transposon in arabidopsis induces transgenerational abscisic acid insensitivity. Sci. Rep. 6. doi: 10.1038/Srep23181
Jing, Y., Sun, H., Yuan, W., Wang, Y., Li, Q., Liu, Y., et al. (2016). SUVH2 and SUVH9 couple two essential steps for transcriptional gene silencing in arabidopsis. Mol. Plant 9 (8), 1156–1167. doi: 10.1016/j.molp.2016.05.006
Johnson, L. M., Du, J., Hale, C. J., Bischof, S., Feng, S., Chodavarapu, R. K., et al. (2014). SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507 (7490), 124–128. doi: 10.1038/nature12931
Johnson, L. M., Law, J. A., Khattar, A., Henderson, I. R., Jacobsen, S. E. (2008). SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PloS Genet. 4 (11), e1000280. doi: 10.1371/journal.pgen.1000280
Kanno, T., Mette, M. F., Kreil, D. P., Aufsatz, W., Matzke, M., Matzke, A. J. (2004). Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 14 (9), 801–805. doi: 10.1016/j.cub.2004.04.037
Kuhlmann, M., Mette, M. F. (2012). Developmentally non-redundant SET domain proteins SUVH2 and SUVH9 are required for transcriptional gene silencing in Arabidopsis thaliana. Plant Mol. Biol. 79 (6), 623–633. doi: 10.1007/s11103-012-9934-x
Law, J. A., Ausin, I., Johnson, L. M., Vashisht, A. A., Zhu, J. K., Wohlschlegel, J. A., et al. (2010). A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr. Biol. 20 (10), 951–956. doi: 10.1016/j.cub.2010.03.062
Lippman, Z., Gendrel, A. V., Black, M., Vaughn, M. W., Dedhia, N., McCombie, W. R., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430 (6998), 471–476. doi: 10.1038/nature02651
Liu, Z. W., Shao, C. R., Zhang, C. J., Zhou, J. X., Zhang, S. W., Li, L., et al. (2014). The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PloS Genet. 10 (1), e1003948. doi: 10.1371/journal.pgen.1003948
Marsano, R. M., Dimitri, P. (2022). Constitutive heterochromatin in eukaryotic genomes: A mine of transposable elements. Cells 11 (5), 761. doi: 10.3390/cells11050761
Matzke, M. A., Mosher, R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15 (6), 394–408. doi: 10.1038/nrg3683
Mhiri, C., Borges, F., Grandbastien, M. A. (2022). Specificities and dynamics of transposable elements in land plants. Biol. (Basel) 11 (4), 488. doi: 10.3390/biology11040488
Mittelsten Scheid, O., Probst, A. V., Afsar, K., Paszkowski, J. (2002). Two regulatory levels of transcriptional gene silencing in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99 (21), 13659–13662. doi: 10.1073/pnas.202380499
Moissiard, G., Cokus, S. J., Cary, J., Feng, S., Billi, A. C., Stroud, H., et al. (2012). MORC family ATPases required for heterochromatin condensation and gene silencing. Science 336 (6087), 1448–1451. doi: 10.1126/science.1221472
Moses, J., Goodchild, A., Rivory, L. P. (2010). Intended transcriptional silencing with siRNA results in gene repression through sequence-specific off-targeting. RNA 16 (2), 430–441. doi: 10.1261/rna.1808510
Naumann, K., Fischer, A., Hofmann, I., Krauss, V., Phalke, S., Irmler, K., et al. (2005). Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 24 (7), 1418–1429. doi: 10.1038/sj.emboj.7600604
Niu, C., Jiang, L., Cao, F., Liu, C., Guo, J., Zhang, Z., et al. (2022a). Methylation of a MITE insertion in the MdRFNR1-1 promoter is positively associated with its allelic expression in apple in response to drought stress. Plant Cell 34 (10), 3983–4006. doi: 10.1093/plcell/koac220
Niu, X., Chen, L., Kato, A., Ito, H. (2022b). Regulatory mechanism of a heat-activated retrotransposon by DDR complex in Arabidopsis thaliana. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1048957
Oliver, K. R., Greene, W. K. (2009). Transposable elements: powerful facilitators of evolution. Bioessays 31 (7), 703–714. doi: 10.1002/bies.200800219
Onodera, Y., Haag, J. R., Ream, T., Costa Nunes, P., Pontes, O., Pikaard, C. S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120 (5), 613–622. doi: 10.1016/j.cell.2005.02.007
Ream, T. S., Haag, J. R., Wierzbicki, A. T., Nicora, C. D., Norbeck, A. D., Zhu, J. K., et al. (2009). Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of. Mol. Cell 33 (2), 192–203. doi: 10.1016/j.molcel.2008.12.015
Schmittgen, T., Livak, K. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Slotkin, R. K., Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8 (4), 272–285. doi: 10.1038/nrg2072
Wells, J. N., Feschotte, C. (2020). A field guide to eukaryotic transposable elements. Annu. Rev. Genet. 23 (54), 539–561. doi: 10.1146/annurev-genet-040620-022145
Wicker, T., Sabot, F., Hua-Van, A., Bennetzen, J. L., Capy, P., Chalhoub, B., et al. (2007). A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8 (12), 973–982. doi: 10.1038/nrg2165
Wierzbicki, A. T., Haag, J. R., Pikaard, C. S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135 (4), 635–648. doi: 10.1016/j.cell.2008.09.035
Zhong, X., Hale, C. J., Law, J. A., Johnson, L. M., Feng, S., Tu, A., et al. (2012). DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat. Struct. Mol. Biol. 19 (9), 870–875. doi: 10.1038/nsmb.2354
Keywords: epigenetics, transposon, environmental stress, Arabidopsis thaliana, ONSEN
Citation: Niu X, Ge Z and Ito H (2024) Regulatory mechanism of heat-active retrotransposons by the SET domain protein SUVH2. Front. Plant Sci. 15:1355626. doi: 10.3389/fpls.2024.1355626
Received: 14 December 2023; Accepted: 22 January 2024;
Published: 08 February 2024.
Edited by:
Yezhang Ding, Berkeley Lab (DOE), United StatesReviewed by:
Ming-Bo Wang, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaYi Zhai, Berkeley Lab (DOE), United States
Copyright © 2024 Niu, Ge and Ito. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hidetaka Ito, hito@sci.hokudai.ac.jp
 Xiaoying Niu1
Xiaoying Niu1