- 1Suzhou Polytechnic Institute of Agriculture, Suzhou, China
- 2State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Institute of Vegetables, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 3Wulanchabu Academy of Agricultural and Husbandry Sciences, Wulanchabu, China
- 4Institute of Crops, Lishui Academy of Agricultural and Forestry Sciences, Lishui, China
- 5Jiangsu Coastal Area Institute of Agricultural Sciences, Yancheng, China
Pepper is widely grown all over the world, so it faces many abiotic stresses, such as drought, high temperature, low temperature, salt damage, and so on. Stresses causing the accumulation of reactive oxidative species (ROS) in plants are removed by antioxidant defense systems, and ascorbate peroxidase (APX) is an important antioxidant enzyme. Therefore, the present study performed genome-wide identification of the APX gene family in pepper. We identified nine members of the APX gene family in the pepper genome according to the APX proteins’ conserved domain in Arabidopsis thaliana. The physicochemical property analysis showed that CaAPX3 had the longest protein sequence and the largest molecular weight of all genes, while CaAPX9 had the shortest protein sequence and the smallest MW. The gene structure analysis showed that CaAPXs were composed of seven to 10 introns. The CaAPX genes were divided into four groups. The APX genes of groups I and IV were localized in the peroxisomes and chloroplasts, respectively; the group II genes were localized in the chloroplasts and mitochondria; and the group III genes were located in the cytoplasm and extracell. The conservative motif analysis showed that all APX genes in the pepper had motif 2, motif 3, and motif 5. The APX gene family members were distributed on five chromosomes (Chr. 2, 4, 6, 8, and 9). The cis-acting element analysis showed that most CaAPX genes contain a variety of cis-elements related to plant hormones and abiotic stress. RNA-seq expression analysis showed that the expression patterns of nine APXs were different in vegetative and reproductive organs at different growth and development stages. In addition, the qRT-PCR analysis of the CaAPX genes revealed significant differential expression in response to high temperature, low temperature, and salinity stresses in leaf tissue. In conclusion, our study identified the APX gene family members in the pepper and predicted the functions of this gene family, which would provide resources for further functional characterization of CaAPX genes.
Introduction
Plants will be subjected to various environmental stresses during their growth and development, comprising both abiotic stresses such as drought, freezing, high temperature, heavy metal, and salinity and biotic stresses such as viruses and fungi (Bhat et al., 2019; Raza et al., 2020). Environmental stress can directly or indirectly destroy the reactive oxygen species (ROS) balance in plants, causing a series of stress responses in crops. ROS has dual functions: on the one hand, an appropriate amount of ROS is necessary for organisms to participate in the regulation of signal networks as signaling molecules. On the other hand, excessive reactive oxygen species will cause certain damage to the cell membrane and organelles of crops, and when they exceed a certain limit, they will affect the growth and development of individual plants and even lead to their death, which must be removed as soon as possible (Davletova et al., 2005; Foyer and Shigeoka, 2011). Therefore, the balance between the production and removal of reactive oxygen species in cells is very important. Plants maintain the dynamic balance of reactive oxygen species through enzymatic or nonenzymatic mechanisms and form a complete antioxidant system in the process of evolution (Shigeoka et al., 2002; Maruta et al., 2016).
The clearance mechanism includes the enzymatic scavenging system, such as ascorbate peroxidase (APX), catalase (CAT), glutathione peroxidase (GPX), superoxide dismutase (SOD), and other antioxidant enzymes (Faize et al., 2011). APX is a very effective H2O2 scavenger and has the highest affinity for H2O2 among all H2O2 metabolic enzymes (Huang et al., 2017). Plant APX belongs to the type I heme peroxidase and copper oxidase families and can reduce H2O2 to O2 and H2O using ASA as an electron donor through the ascorbic acid (ASA)-glutathione (GSH) cycle (Asada, 1999).
Ascorbate peroxidase (APX) is a protease encoded by members of a multi-gene family. According to the subcellular localization of their proteins, they can be divided into four categories: cytoplasmic APX, peroxidase APX, chloroplast APX, and mitochondrial APX (Shigeru et al., 2002; Apel and Hirt, 2004). Eight members of the AtAPX gene family have been identified in Arabidopsis thaliana, including three in the cytoplasm (APX1, APX2, and APX6), three in the peroxidase (APX3, APX4, and APX5), and two in the chloroplast (sAPX both in the mitochondrial and chloroplast, tAPX in the thylakoid membrane) (Panchuk et al., 2002; Chew et al., 2003; Mittler et al., 2004; Narendra et al., 2006). The APX gene family of rice has eight genes, including two cytoplasmic isoforms (OsAPx1 and OsAPx2), two peroxidase isoforms (OsAPx3 and OsAPx4), two mitochondrial isoforms (OsAPx5 and OsAPx6), and two in the chloroplast (OsAPx7 and OsAPx8) (Teixeira et al., 2004; Teixeira et al., 2006; Hong et al., 2007; Ribeiro et al., 2017). Tomato has seven genes encoding APX isoforms: three cytoplasmic isoforms, two peroxidase isoforms, and two chloroplast isoforms (Najami et al., 2008). Recently, as more and more plant genomes have been sequenced, genome-wide identification and functional analysis of APX have been accomplished, such as upland cotton (Gossypium hirsutum Linn.) (Tao et al., 2018), sorghum (Sorghum bicolor L.) (Akbudak et al., 2018), maize (Qu et al., 2020), kiwifruit (Actinidia chinensis) (Liao et al., 2020), and so on.
The APX gene plays an important role in the growth and development of different plants and their response to environmental stress. The expression of AtAPX1 was significantly upregulated in various biological and abiotic stress responses (Zimmermann et al., 2004). APX2 plays an important role in regulating heat stress response at different stages of plant development (Schramm et al., 2006; Frank et al., 2009). Overexpression of the AtAPX3 gene in tobacco increased the protective effect of transgenic plants against oxidative stress (Wang et al., 1999). APX6 modulates the ROS signal cross-talk with hormone signals to properly execute the germination program in Arabidopsis, and the apx6 mutant exhibited a high level of ROS and reduced germination (Chen et al., 2014). Loss of function in OsAPX2 showed semi-dwarf seedlings, yellow–green leaves, leaf lesion mimics, and seed sterility (Zhang et al., 2013). Transgenic plants overexpressing the SbpAPX gene showed enhanced salt and drought stress tolerance compared to wild-type plants (Singh et al., 2014). Another study found that BcAPX genes were overexpressed in transgenic Arabidopsis, and the expression of APX and the APX activity in transgenic lines were higher than in nontransgenic (NT) plants under high temperatures (Chiang et al., 2015).
Pepper is an important solanaceous vegetable and is widely cultivated and eaten both as a vegetable and as a spice. However, pepper is particularly vulnerable to a number of biotic and abiotic stresses in production, such as pathogens, drought, and cold temperatures, which can easily cause a production drop. The APX gene, as an important class of antioxidant enzymes, has rarely been reported. In this study, APX gene family members were identified from the pepper genome database, and the physicochemical properties of APX gene members were analyzed by the bioinformatics method. The expression of APX gene members in different tissues was analyzed by downloading the RNA-seq data, and the gene expression under abiotic stress was analyzed by the qRT-PCR technique. This study provided the scientific basis for further exploring the molecular mechanism of APX gene family members in pepper under abiotic stress.
Materials and methods
Identification of the APX gene family in pepper
All of the pepper genome sequence data were downloaded from the Solanaceae Genomics Network (http://solgenomics.net/). The local database of the pepper genome sequences was constructed by the Bioedit7.0 software. In order to retrieve all the members of the APX family in pepper, two methods were employed, including the BLASTP algorithm and the Hidden Markov model (HMM). First, for BLASTP, we used 31 APX amino acid sequences from Arabidopsis thaliana, rice, tomato, and potato as an inquiry with the e-value set to 1e−10. The amino acid sequences of 23 APXs were retrieved from the PLAZA 4.0 (https://bioinformatics.psb.ugent.be/plaza/versions/) and Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html). We then used the HMM profile of the APX domain (Pfam: PF00141), which was from the Pfam database (http://pfam.xfam.org), as a query to detect this local pepper database. Finally, nine CaAPX genes were identified by merging the two methods in the pepper genome.
Characterization of APX gene family in pepper
Physicochemical properties of APX proteins, such as protein length, molecular weight, and isoelectric point were checked using the ExPASy server (www.expasy.org). The subcellular localization of the CaAPX proteins was predicted using the online Euk-mPLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/euk-multi-2/) (Chou and Shen, 2010). For the analysis of the structural feature, the Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn) online tool was adopted by comparing the CDS and genomic sequences of each APX gene (Hu et al., 2014). Conserved motif analyses of the CaAPX genes were then performed using the MEME tool (http://meme-suite.org/tools/meme), identifying a maximum of 10 motifs with a motif width range of 6 to 50 (Bailey et al., 2006). The chromosomal location of the CaAPX genes was determined from the SGN database. The map diagram showing the location of APX genes on chromosomes was drawn by implementing MapDraw V2.1 software.
Multiple sequence alignment and phylogenetic analysis
To determine the evolutionary relationship of the APX gene family, a phylogenetic tree was carried out among A. thaliana, Oryza sativa, Solanum lycopersicum, Solanum tuberosum, and Capsicum annuum protein sequences. The sequence alignment was constructed using Clustal X (version 1.8) and a neighbor-joining method with 1,000 bootstrap replicates in MEGA 7.0 software (Kumar et al., 2016). Any gaps or missing data in the sequence treatment were applied to partial deletion, and then branch lengths were assigned via the pairwise calculations of genetic distances.
Cis-element analysis in the CaAPX gene promoters
The 1-kb sequence upstream of the start code from each of the CaAPX genes was obtained from the SGN database. The cis-elements in the promoter regions of the individual gene were examined using the means of the PlantCARE server (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). The cis-element analysis figure was made by TBtools (Chen et al., 2020).
Expression analysis of CaAPX genes in diverse tissues
In this study, RNA-seq data from the NCBI database (http://pepperhub.hzau.edu.cn/) were used to investigate expression patterns of putative CaAPX genes in different tissues of cultivated pepper (Zunla-1). Different tissues in cultivated pepper included root, stem, leaf, bud, flower, and F-Dev1-9. The heatmap was formed using Multi Experiment Viewer (MeV) software (Howe et al., 2010).
Plant materials and stress treatments
This study’s samples used for stress treatment were collected from a typical cultivated variety, D50. The pepper plants were cultivated in a chamber under 14-h light (28°C to 30°C) and 10-h dark (18°C to 20°C) period conditions.
Seedlings at the six true leaf stages were used for all experiments. For cold treatment, seedlings were subjected to 4°C for 1, 3, 6, and 12 h. For heat shock treatment, seedlings were subjected to 42°C for 0.5, 1, 3, and 6 h. Plants were subjected to 26°C for control. For salt stress treatment, seedlings were subjected to 300 mM NaCl. Plants were subjected to sterile water for control. Three biological replicates for each treatment were carried out in the experiments. All of the samples were immediately frozen in liquid nitrogen and stored at −80°C for further experimentation.
RNA isolation and quantitative real-time PCR analysis
The transcription profiles of APX genes in pepper were performed through qRT-PCR analysis. All primers used in the experiment were designed by the Primer 5.0 software and listed in Supplementary Table S1.
The total RNA isolation and cDNA synthesis was performed using Trizol (Tiangen, Beijing, China) and DNase I (Transgen, Beijing, China) according to manufacturer instructions. In total, 1 μg of each of the treated RNA samples was reverse transcribed using PrimeScriptTM RT reagent Kit for qPCR (Transgen, Beijing, China). A 20-μl qRT-PCR reaction mixture, which contained 10 μl of SYBR Green I, 1 μl of forward primer, 1 μl of reverse primer, 1 μl of diluted cDNA, and 7 μl of sterile distilled H2O, was prepared. The mixture was subjected to the following program: 35 cycles of 3 min at 95°C, 30 s at 58°C, and 20 s at 72°C (ABI Real-Time PCR System, USA). GAPDH was used as an internal control (Cheng et al., 2017), and the fold change in gene expression was calculated using the 2−ΔΔCT method. Each qRT-PCR reaction was carried out using three biological and technical replicates.
Results
Identification of the APX gene family in the pepper genome
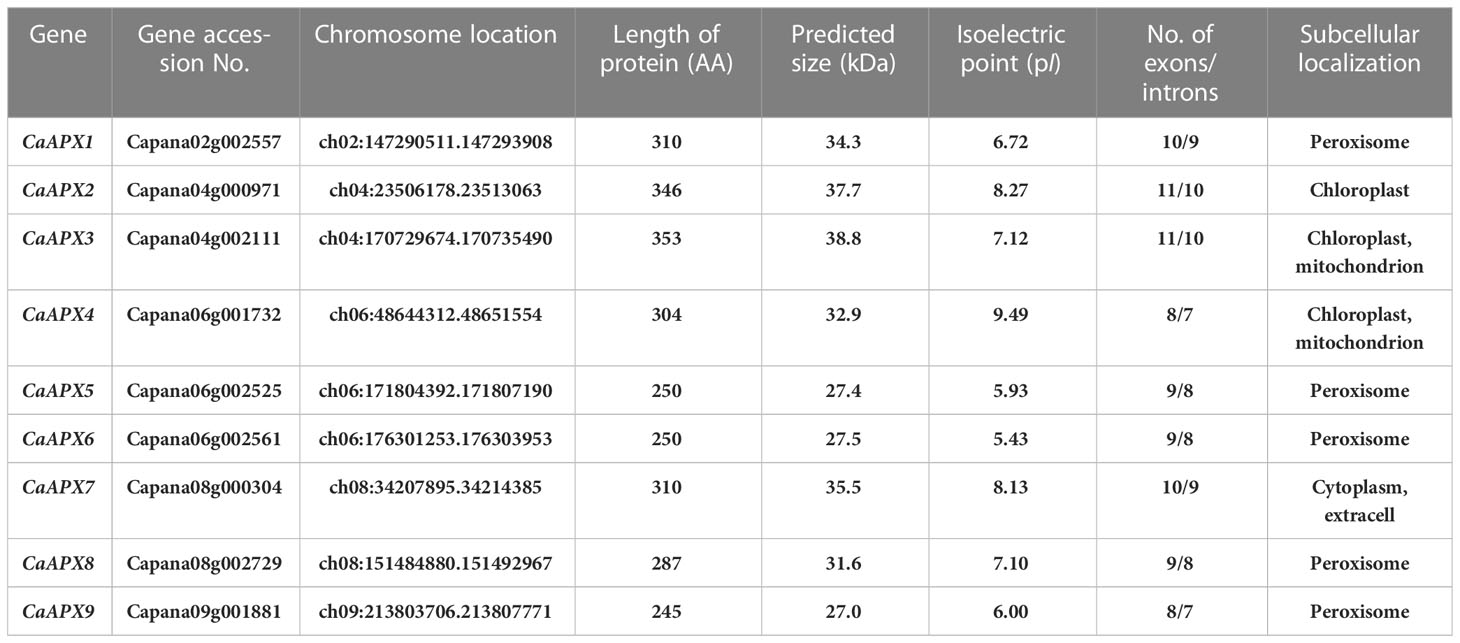
In the present study, a total of nine APX genes were recognized and designated as CaAPX1 to CaAPX9, respectively, based on BLASTP examinations against the pepper genome employing 31 APX proteins (eight AtAPX proteins, eight OsAPX proteins, seven SlAPX proteins, and eight StAPXs) as inquiries (Table 1). The comprehensive information of the nine CaAPX genes (e.g., molecular weights (MW), isoelectric points (pI), numbers of exons/introns, and subcellular localization) is shown in Table 1. CaAPX amino acid residue lengths ranged from 245 aa (CaAPX9) to 353 aa (CaAPX3), MW ranged from 27.0 (CaAPX9) to 38.8 kDa (CaAPX3), and pI ranged from 5.43 (CaAPX6) to 9.49 (CaAPX4). The number of exons ranged from eight to 11, with two genes comprising eight exons, three genes comprising nine exons, two genes comprising 10 exons, and two genes consisting of 11 exons (Table 1). The subcellular localization results predicted that five APX (CaAPX1, CaAPX5, CaAPX6, CaAPX8, and CaAPX9) proteins are located in the peroxisome, three proteins (CaAPX2, CaAPX3, and CaAPX4) are located in chloroplast, two proteins (CaAPX3 and CaAPX4) are also located in the mitochondrion, and only one protein (CaAPX7) is located in the cytoplasm and extracell (Table 1).
Phylogenetic analysis of APX genes
The 40 genes from five species (Arabidopsis thaliana, rice, tomato, potato, and pepper) were clustered into six groups (I–VI) (Figure 1). The nine CaAPX belonged to five groups in the phylogenetic tree, with the exception of group III. Group III only has one gene, StAPX3, including the lowest genes among the six groups. Group VI consisted of 13 genes, including the most genes among the six groups, which contained AtAPX1 and AtAPX2; OsAPX1 and OsAPX2; SlAPX1, SlAPX2, and SlAPX3; StAPX4, StAPX5, and StAPX7; and CaAPX5, CaAPX6, and CaAPX9. Groups IV, V, and VI exhibited five species of APX genes, while group I had CaAPX and AtAPX, group II had CaAPX, StAPX, and AtAPX, and group III only had StAPX.
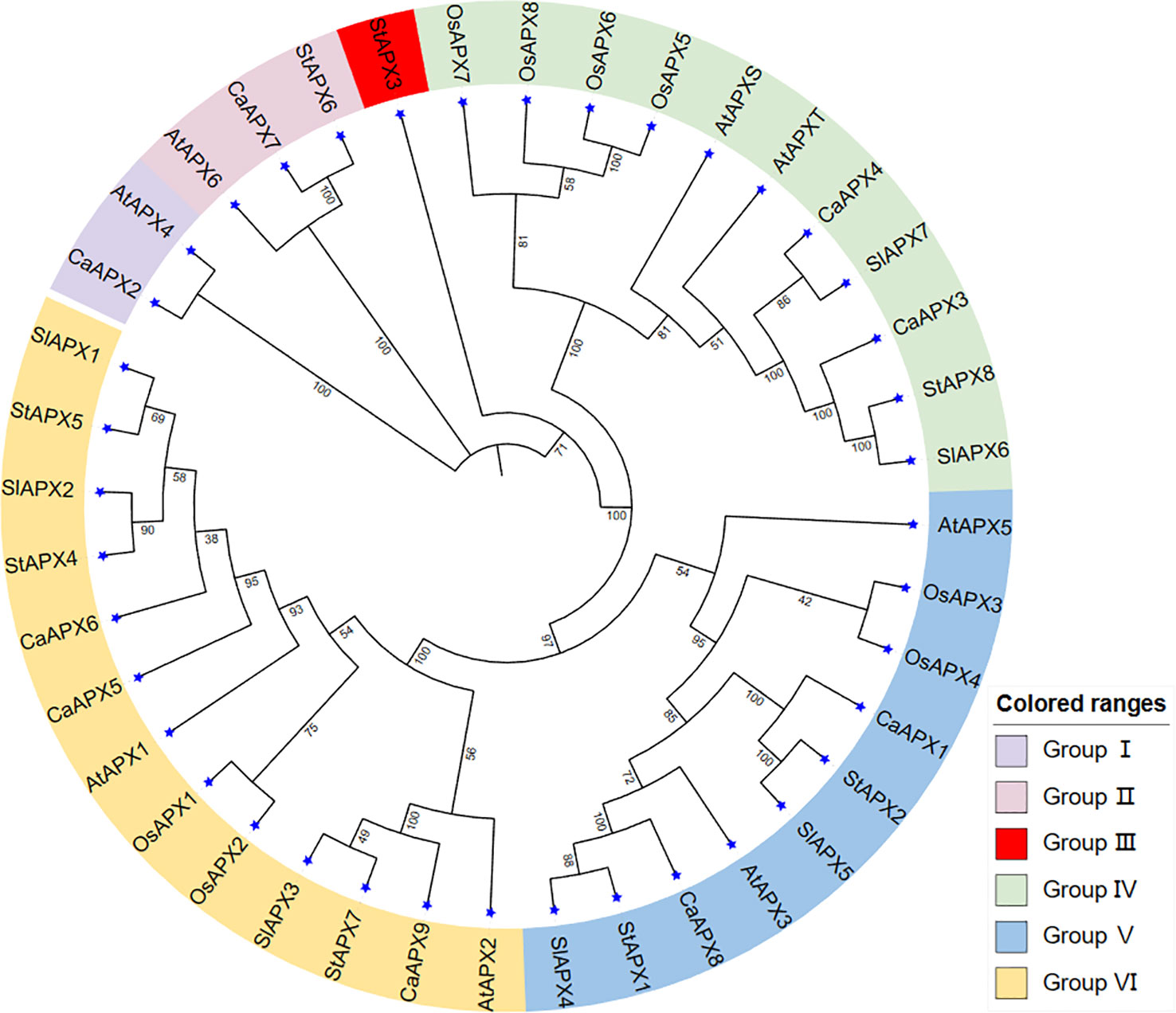
Figure 1 Phylogenetic relationships of APX gene family members in five species, including rice, tomato, potato, pepper, and Arabidopsis thaliana.
This study also found nine pairs of orthologous genes between the various species: CaAPX2 and AtAPX4; CaAPX7 and StAPX6; StAPX8 and SlAPX6; CaAPX4 and SlAPX7; StAPX1 and SlAPX4; StAPX2 and SlAPX4; StAPX7 and SlAPX3; StAPX4 and SlAPX2; and StAPX5 and SlAPX1, respectively. There are six pairs of the nine orthologous genes between the tomato and potato, and it may be noticed that StAPXs have a stronger phylogenetic link with the SlAPXs. There were also three pairs of paralogous genes discovered only in rice.
Gene structures and motif analysis of CaAPX genes
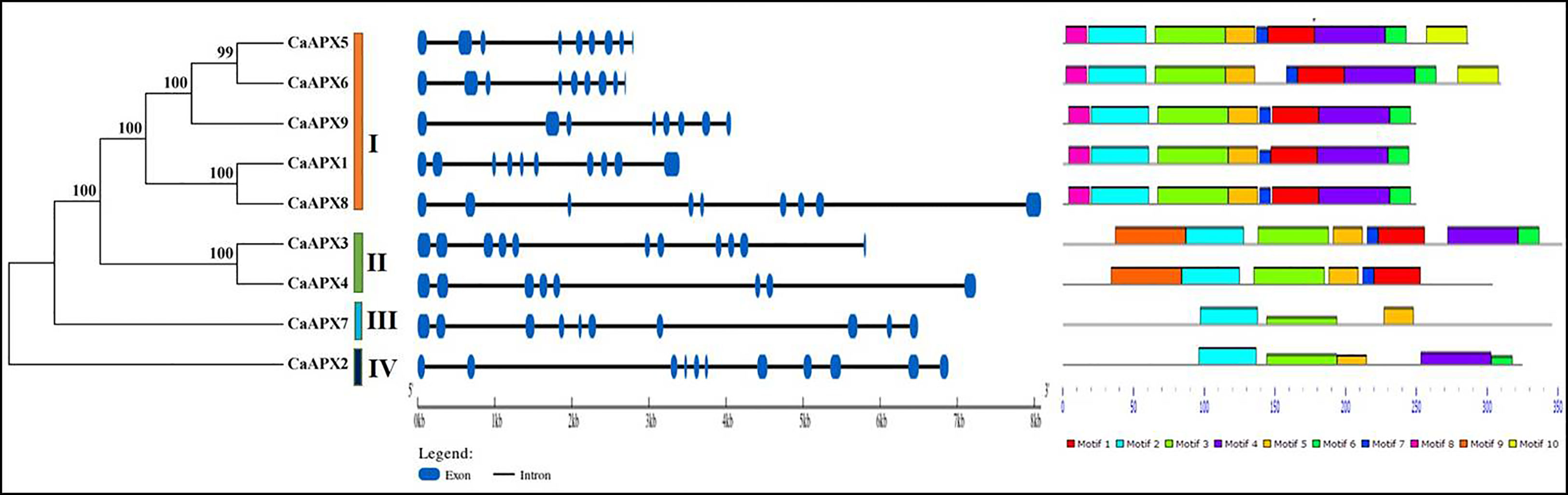
The gene structure of the CaAPX genes was guided to obtain insight into the evolution of the APX family genes. Moreover, the phylogenetic relationships of the APX proteins in pepper were identified using MEGA 7.0 software. It was to see whether the exon–intron distribution pattern and the phylogenetic tree were in compliance with each other. The results showed that the number of exons ranged from eight to 11, and the number of introns varied from seven to 10, as detailed in Figure 2. Group I includes seven to nine introns. Groups III and IV only have one gene, including nine and 10 introns, respectively. While CaAPX3 and CaAPX4 showed different exon–intron structures in group II.
The presence of conserved motifs was also identified in nine CaAPX genes using MEME. The conserved motif of the CaAPX genes explored 64 motifs that varied from three to nine. The motif distributed in the same group is highly similar. Group I includes eight to nine, and group II contains six to eight (Figure 2). Motifs 2, 3, and 5 were present in all CaAPX proteins, indicating that these motifs were conserved motifs and functional domains of the APX protein family in pepper. Motif 8 was only discovered in group I, and motif 9 was only present in group II. Overall, as shown in Figure 2, the closely related genes of CaAPX shared a similar pattern of motif distribution.
Chromosomal distributions of the CaAPX genes
The results showed that the CaAPX genes were distributed unevenly on five chromosomes, including chromosomes 2, 4, 6, 8, and 9. The other chromosomes did not carry the APX genes (Figure 3). A maximum of three genes (CaAPX4–CaAPX6) were localized on chromosome 6, which was followed by chromosome 4 (CaAPX2 and CaAPX3) and chromosome 8 (CaAPX7 and CaAPX8) with two genes. The remaining chromosomes (chromosomes 2 and 9) only consisted of one gene.
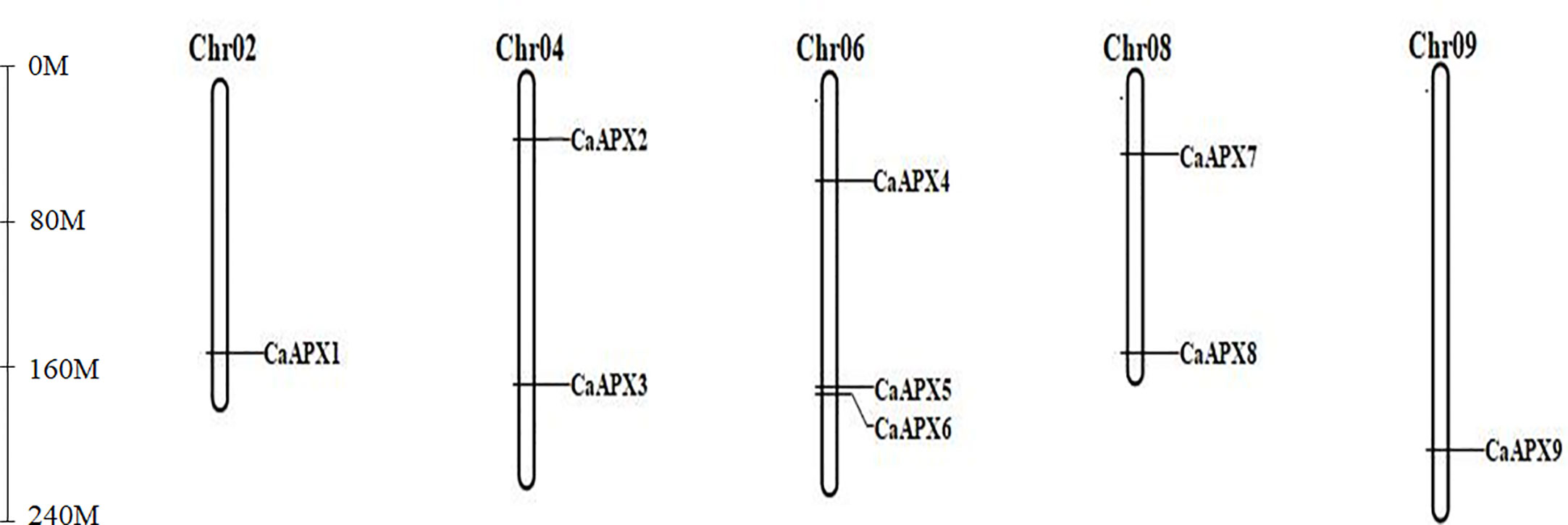
Figure 3 Chromosomal localization of CaAPX genes. Nine CaAPX genes were distributed unevenly on five chromosomes (Chr02, Chr04, Chr06, Chr08, and Chr09).
Cis-element in the promoters of CaAPX genes
To gain the gene functions and regulations of the CaAPX genes, cis-elements in CaAPX promoter regions were analyzed by searching a 1,000-bp upstream region from each individual CaAPX gene’s transcriptional activation site against the PlantCARE database. The results depicted that five major classes of cis-elements were present in the promoter region of all APX genes: light response; process-specific; environment-specific; plant tissue, and binding site, as shown in Figure 4. Among all of the genes except CaAPX3, 15 light response elements were found. The results that four phytohormone-correlated (ABA, MeJA, GA, auxin) responsive elements comprising CGTCA-motif, TGACG-motif, p-box, ABRE, and TGA-element were documented. The cis-acting regulatory elements involved in environment-specific such as drought (MBS), anoxia (ARE, GC-motif), and low-temperature responsive element (LTR) among all of the genes were found in the promoter region of the APX genes of pepper. In addition, four tissue-specific cis-elements and five binding site cis-elements were also identified.
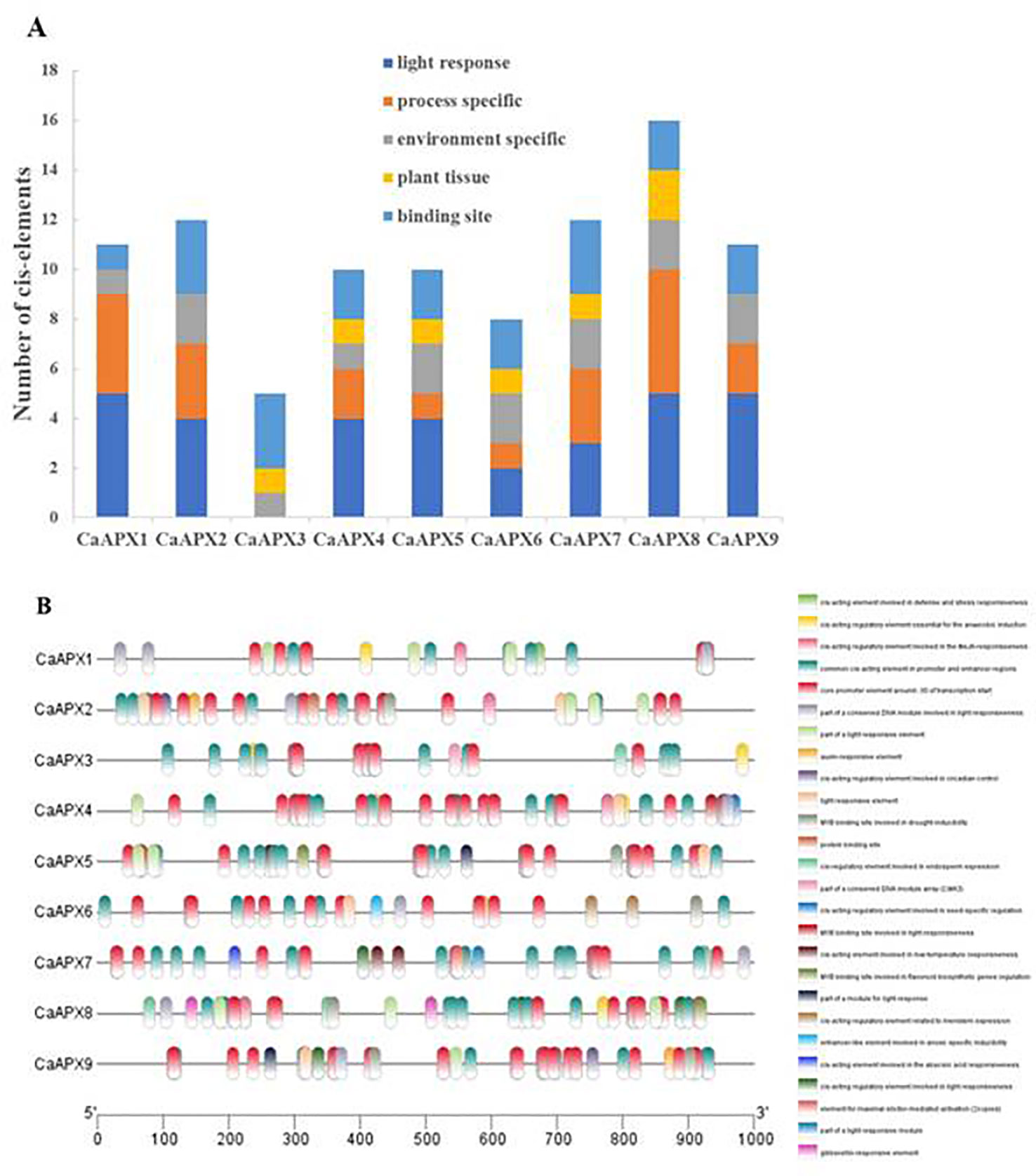
Figure 4 Predicted cis-elements in the promoter regions of CaAPX genes. (A) Number of different cis-elements. (B) The elements identified are in different colored boxes.
Expression profiles of CaAPX genes in different tissues
The tissue-specific expression profiles of CaAPX genes were investigated at various developmental stages of the pepper cultivar Zunla, as illustrated in Figure 5. Results of the study revealed that CaAPX8 exhibits higher expression levels among all tissues except F-Dev5, implicating the vital roles for pepper, whereas four genes (CaAPX1, CaAPX2, CaAPX3, and CaAPX4) showed lower expression in all of the tissues. CaAPX7 expressed higher values in ripe fruit (F-Dev7, F-Dev8, and F-Dev9), suggesting its participation in fruit development. Among those genes, it was observed that CaAPX5 had the highest expression levels during the bud stage. The rest of the genes (CaAPX6 and CaAPX9) with the higher expression level displayed tissue specificity in the root and F-Dev9.
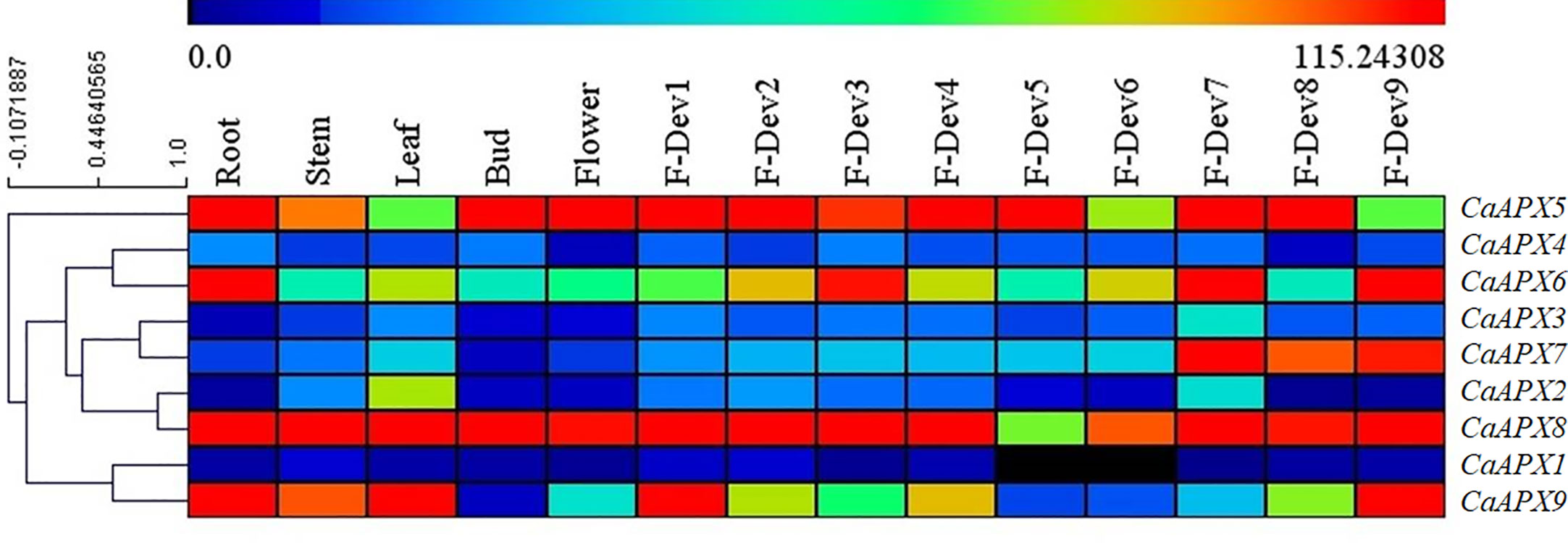
Figure 5 Expression profiles of CaAPX genes on RNA-seq in different pepper tissues. The one to nine stages of fruit development are presented in five early stages of color breaking (0–1-, 1–3-, 3–4-, and 4–5-cm-long fruit, and mature-green fruit), the color breaking stage (fruit began to turn red), and three late stages of color breaking (3, 5, and 7 days after color breaking), respectively.
Expression pattern of CaAPX genes in response to different abiotic stresses
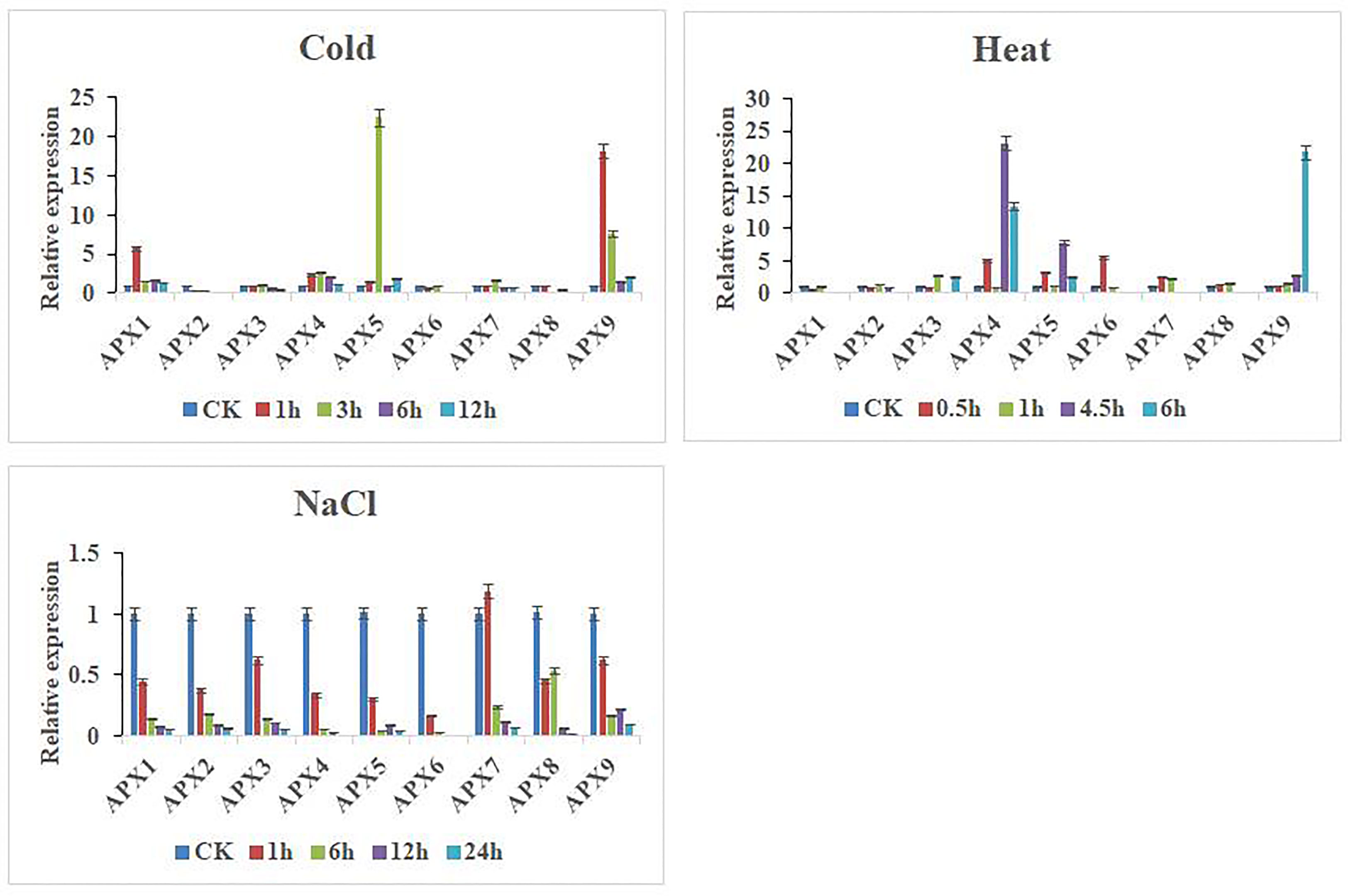
In this study, to further investigate the expression patterns of nine CaAPX genes differentially expressed under abiotic stresses, we performed qRT-PCR analysis on the cultivated variety, D50, under cold (4°C), heat (42°C), and NaCl (300 mM) treatment conditions (Figure 6). Under cold treatment, CaAPX1, CaAPX4, CaAPX5, and CaAPX9 were upregulated in response to cold stress at 1, 3, 6, and 12 h as compared to CK, while CaAPX2 showed continuous downregulation at all treatment points. CaAPX1 was upregulated (5.81-fold) at 1 h, CaAPX5 was upregulated (22.45-fold) at 3 h, and CaAPX9 was upregulated (18.13- and 7.62-fold) at 1 and 3 h, respectively, compared to CK. Under heat treatment, CaAPX9 showed a continuous upregulation response as compared to CK and showed maximum expression (21.80-fold) at 6 h, while CaAPX1 was downregulated at all treatment points. CaAPX4 was upregulated (5.04-, 23.13-, and 13.43-fold) at 0.5, 4.5, and 6 h, CaAPX5 was upregulated (3.24-, 7.81-, and 2.38-fold) at 0.5, 4.5, and 6 h, and CaAPX6 was upregulated (5.58-fold) at 0.5 h, respectively, compared to CK. Under NaCl treatment, most of the genes were downregulated, especially CaAPX1, CaAPX2, CaAPX3, CaAPX4, and CaAPX6, which were continuously downregulated at all treatment points, except CaAPX7, which showed a slightly higher expression at 1 h.
Discussion
APX is one of the key enzymes for active oxygen removal in plants, playing an important role in plant growth and development as well as stress response. With the development of sequenced genomes, APX gene families have been found in many plants, such as Arabidopsis (Panchuk et al., 2002), rice (Teixeira et al., 2004), upland cotton (Tao et al., 2018), tomato (Najami et al., 2008), and so on. According to the subcellular localization, APX can be divided into cytoplasmic APX, peroxisome APX, chloroplast APX, and mitochondrial APX, but the localization and quantity of APXs in cells are different in different species. Previous studies have shown that different types of APX genes have different functions. For example, chloroplast APX is mainly used to protect the photosynthetic system, while mitochondrial APX plays a positive role in removing hydrogen peroxide generated by fatty acid β oxidation (Renu et al., 2011; Andreia et al., 2014), the cytoplasmic OsAPX2 gene plays an important role in maintaining H2O2 homeostasis (Wu et al., 2018), silened peroxisome OsAPX4 can lead to premature aging of rice (Guan et al., 2010). In this work, nine APX genes were found in the pepper genome and divided into four subfamilies. The results of the phylogenetic tree showed that five genes of subfamily I were located in the peroxisome, two genes of subfamily II were located in chloroplasts/mitochondria, one gene of subfamily III was located in cytoplasm and extracell, and one member of subfamily IV was only located in chloroplasts, indicating that the close relatives may have had the same subcellular localization results (Figure 2). CaAPX3 and CaAPX4 of subfamily II were all located in two chloroplast/mitochondrial organelles, as well as in previous studies of PtrAPX1 and PtrAPX5 in Populus trichocarpa (Leng et al., 2021). CaAPX genes located in peroxisome all contain eight different conserved motifs and have eight to ten exon regions. Rice is a monocotyledonous plant, while Arabidopsis thaliana, tomato, potato, and pepper belong to the dicotyledonous family. Therefore, the phylogenetic tree of Arabidopsis thaliana, rice, tomato, potato, and pepper was constructed, and it was found that the APX gene of pepper is further related to the homologous evolution of rice.
The promoter cis-acting element plays an important role in regulating plant biological processes, such as participating in exogenous hormone induction and abiotic stress response (Yang et al., 2019). In this study, it was found that CaAPX genes contain hormone-induced related and stress regulatory elements, such as CGTCA-motif, TGACG-motif, TGA-element, ABRE, CGTCA-motif, TCA-element, ARE, MBS, and LTR, through the 1,000-bp upstream ATG sequence analysis of the CaAPX family members. Every CaAPX gene contains at least one process-specific and one environment-specific class, except CaAPX3, which has no hormone-induced cis-element, suggesting that CaAPXs play an important role in plant hormone and abiotic stress responses.
More and more evidence suggested that APX genes may exhibit different expression profiles in different organs/tissues and stress environments. Therefore, the tissue-specific expression of CaAPX genes was displayed in eight diverse developmental tissues using RNA-seq data (Figure 5), and these results are consistent with previous reports. For example, AtAPX1 is involved in many biological processes (Pnueli et al., 2003; Davletova et al., 2005) and is constitutionally higher expressed in plant roots, stems, leaves, and many other tissues (Zimmermann et al., 2004). The expression of AtAPX6 was very high at the seed postmaturity stage (Chen et al., 2014). Maize APXb1 is highly expressed in roots, shoots, and immature leaves, and APXb2 is abundantly expressed in leaves and reproductive organs (Qu et al., 2020). In our study, CaAPX8 and CaAPX5 presented higher expressions in most tissue, suggesting two genes involved in many biological processes. The expression levels of CaAPX1, CaAPX2, CaAPX3, and CaAPX4 exhibited very low levels during the most normal growth and development of pepper. These results indicate that the four genes may play a small role in the normal growth and development of plants. CaAPX7 expression in pepper increased significantly with fruit maturation. These results suggest that APX7 may play a key role in fruit development.
In this study, qRT-PCR was used to appraise the expression patterns of CaAPXs under low temperature, high temperature, and salt stress (Figure 6). Under the three abiotic stresses, the expression patterns of CaAPXs were different, CaAPX5 and CaAPX9 showed upregulated expression under low temperature, and CaAPX4 and CaAPX9 showed higher expression under high temperature, indicating that CaAPXs may play an important role in the abiotic stress response of pepper, which was consistent with previous research results (Sharma and Dubey, 2005; Koussevitzky et al., 2008). However, the expression levels of some genes showed a downregulated expression under stress. For example, under salt stress, the expression levels of all the CaAPXs except CaAPX7 at 1 h were downregulated at all times. It is speculated that the expression of these genes was inhibited after stress and that ROS is cleared by other genes or by other means. In addition, the expression patterns of individual genes were different under different stresses. The expression level of CaAPX1 was significantly upregulated in low-temperature stress but downregulated in high-temperature stress, suggesting that CaAPX genes played different functions under different stresses. The result agreed with earlier discoveries; for example, cAPX expression of Spinacia oleracea was upregulated under strong light and ultraviolet light, but the transcription level did not change under drought and salt stress. The transcription levels of mAPX, sAPX, and tAPX did not change after ABA, drought, strong light, and salt treatment (Yoshimura et al., 2000). The results of qRT-PCR showed that APX genes were induced to express under high temperatures, and CaAPX5 and CaAPX9 showed significantly upregulated expression every time. By constructing a phylogenetic tree, we found that CaAPX5, CaAPX9, and AtAPX2 are clustered into a cluster that is close in evolutionary relationship. Studies have shown that AtAPX2 plays a very important regulatory role in the process of heat stress response at different developmental stages of plants (Shi et al., 2001; Frank et al., 2009). These discoveries offered compelling evidence that APX genes play a well-maintained role in defending diverse plant species against abiotic stresses. In the future, we could validate the function of CaAPX genes using overexpression and the CRISPR/Cas system and provide a basis study for breeding pepper.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
Conceived and designed the experiments: XP and HW. Performed the experiments: JC, YX, JL, and YZ. Analyzed the data: XP, YX, LW, and JZ. Wrote the paper: XP, YX, and HW. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (32100302, 31960601, 32060446, 32260769), Young and middle-aged academic leaders of the “Qing Lan Project” in universities of Jiangsu Province (2022), Technology Cultivation Project of Suzhou Polytechnic Institute of Agriculture (PY2005), Jiangsu Provincial Natural Science Foundation of China (BK20211113), Natural Science Foundation of Inner Mongolia Autonomous Region of China (2021MS03106, 2021MS03071), Zhejiang Provincial Natural Science Foundation of China (LQ23C020004), and Basic Research Project of Ulanqab, Inner Mongolia Autonomous Region of China (2021JC202).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1189020/full#supplementary-material
References
Akbudak, M. A., Filiz, E., Vatansever, R., Kontbay, K. (2018). Genome-wide identification and expression profiling of ascorbate peroxidase (APX) and glutathione peroxidase (GPX) genes under drought stress in sorghum (Sorghum bicolor l.). J. Plant Growth Regul. 37 (3), 925–936. doi: 10.1007/s00344-018-9788-9
Andréia, C., Aurenivia, B., Fabricio, E. L. C., Claudia, M. B. A., Gisele, P., Mariana, S., et al. (2014). The knockdown of chloroplastic ascorbate peroxidases reveals its regulatory role in the photosynthesis and protection under photo-oxidative stress in rice. Plant Sci. 214, 74–87. doi: 10.1016/j.plantsci.2013.10.001
Apel, K., Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Asada, K. (1999). The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 (1), 601–639. doi: 10.1146/annurev.arplant.50.1.601
Bailey, T. L., Williams, N., Misleh, C., Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Bhat, J. A., Shivaraj, S., Singh, P., Navadagi, D. B., Tripathi, D. K., Dash, P. K., et al. (2019). Role of silicon in mitigation of heavy metal stresses in crop plants. Plants. 8, 71. doi: 10.3390/plants8030071
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Letnik, I., Hacham, Y., Dobrev, P., Ben-Daniel, B. H., Vanková, R., et al. (2014). ASCORBATE PEROXIDASE6 protects arabidopsis desiccating and germinating seeds from stress and mediates cross talk between reactive oxygen species, abscisic acid, and auxin. Plant Physiol. 166 (1), 370–383. doi: 10.1104/pp.114.245324
Cheng, Y., Pang, X., Wan, H. J., Ahammed, G. J., Yu, J., Yao, Z., et al. (2017). Identification of optimal reference genes for normalization of qPCR analysis during pepper fruit development. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01128
Chew, O., Whelan, J., Millar, A. H. (2003). Molecular definition of the ascorbate-glutathione cycle in arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 278 (47), 46869–46877. doi: 10.1074/jbc.M307525200
Chiang, C. M., Chien, H. L., Chen, L. O., Hsiung, T. C., Chiang, M. C., Chen, S. P. (2015). Overexpression of the genes coding ascorbate peroxidase from brassica campestris enhances heat tolerance in transgenic arabidopsis thaliana. Biol. Plant 59 (2), 305–315. doi: 10.1007/s10535-015-0489-y
Chou, K. C., Shen, H. B. (2010). A new method for predicting the subcellular localization of eukaryotic proteins with both single and multiple sites: euk-mPLoc 2.0. PloS One 5 (4), e9931. doi: 10.1371/journal.pone.0009931
Davletova, S., Rizhsky., L., Liang, H., Shengqiang, Z., Oliver, D. J., Coutu, J., et al. (2005). Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of arabidopsis. Plant Cell. 17 (1), 268–281. doi: 10.1105/tpc.104.026971
Faize, M., Burgos, L., Faize, L., Piqueras, A., Nicolas, E., Barba-Espin, G., et al. (2011). Involvement of cytosolic ascorbate peroxidase and Cu/Zn superoxide dismutase for improved tolerance against drought stress. J. Exp. Bot. 62 (8), 2599–2613. doi: 10.1093/jxb/erq432
Foyer, C. H., Shigeoka, S. (2011). Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 155 (1), 93–100. doi: 10.1104/pp.110.166181
Frank, G., Pressman, E., Ophir, R., Althan, L., Shaked, R., Freedman, M., et al. (2009). Transcriptional profiling of maturing tomato (Solanum lycopersicum l.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J. Exp. Bot. 60 (13), 3891–3908. doi: 10.1093/jxb/erp234
Guan, Q. J., Xia, D. X., Liu, S. K. (2010). OsAPX4 gene response to several environmental stresses in rice (Oryza sativa l.). Afr J. Biotechnol. 9 (36), 5908–5913. doi: 10.5897/AJB10.037
Hong, C. Y., Hsu, Y. T., Tsai, Y. C., Kao, C. H. (2007). Expression of ASCORBATE PEROXIDASE 8 in roots of rice (Oryza sativa l.) seedlings in response to NaCl. J. Exp. Bot. 58, 3273–3283. doi: 10.1093/jxb/erm174
Howe, E., Holton, K., Nair, S., Schlauch, D., Sinha, R., Quackenbush, J. (2010). Mev: multiexperiment viewer in biomedical informatics for cancer research. Ed. Ochs, M. F. (New York, NY: Springer US), 267–277.
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2014). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, L. P., Jia, J., Zhao, X. X., Zhang, M. Y., Huang, X. X., Ji, E., et al. (2017). The ascorbate peroxidase APX1 is a direct target of a zinc finger transcription factor ZFP36 and a late embryogenesis abundant protein OsLEA5 interacts with ZFP36 to co-regulate OsAPX1 in seed germination in rice. Biochem. Bioph Res. Co. 495 (1), 339–345. doi: 10.1016/j.bbrc.2017.10.128
Koussevitzky, S., Suzuki, N., Huntington, S., Armijo, L., Sha, W., Cortes, D., et al. (2008). Ascorbate peroxidase 1 plays a key role in the response of arabidopsis thaliana to stress combination. J. Biol. Chem. 283 (49), 34197–34203. doi: 10.1074/jbc.M806337200
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Leng, X., Wang, H. Z., Zhang, S., Qu, C. P., Yang, C. P., Xu, Z., et al. (2021). Identification and characterization of the APX gene family and its expression pattern under phytohormone treatment and abiotic stress in Populus trichocarpa. Genes. 12 (3), 334. doi: 10.3390/genes12030334
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Liao, G. L., Liu, Q., Li, Y. Q., Zhong, M., Huang, C. H., Jia, D. F., et al. (2020). Identification and expression profiling analysis of ascorbate peroxidase gene family in actinidia chinensis (Hongyang). J. Plant Res. 133, 715–726. doi: 10.1007/s10265-020-01206-y
Maruta, T., Sawa, Y., Shigeoka, S., Ishikawa, T. (2016). Diversity and evolution of ascorbate peroxidase functions in chloroplasts: more than just a classical antioxidant enzyme? Plant Cell Physiol. 57 (7), 1377–1386. doi: 10.1093/PCP/PCV203
Mittler, R., Vanderauwera, S., Gollery, M., Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9 (10), 490–498. doi: 10.1016/j.tplants.2004.08.009
Najami, N., Janda, T., Barriah, W., Kayam, G., Tal, M., Micha, G., et al. (2008). Ascorbate peroxidase gene family in tomato: its identification and characterization. Mol. Genet. Genomics 279, 171–182. doi: 10.1007/s00438-007-0305-2
Narendra, S., Venkataramani, S., Shen, G. X., Wang, J., Pasapula, V., Lin, Y., et al. (2006). The arabidopsis ascorbate peroxidase 3 is a peroxisomal membrane-bound antioxidant enzyme and is dispensable for arabidopsis growth and development. J. Exp. Bot. 57, 3033–3042. doi: 10.1093/jxb/erl060
Panchuk, I. I., Volkov, R. A., Schöffl, F. (2002). Heat stress- and heat shock transcription factor-dependent expression and activity of ascorbate peroxidase in arabidopsis. Plant Physiol. 129, 838–853. doi: 10.1104/pp.001362
Pnueli, L., Liang, H., Rozenberg, M., Mittler, R. (2003). Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient arabidopsis plants. Plant J. 34 (2), 187–203. doi: 10.1046/j.1365-313x.2003.01715.x
Qu, C. X., Wang, L., Zhao, Y. W., Liu, C. (2020). Molecular evolution of maize ascorbate peroxidase genes and their functional divergence. Genes. 11 (10), 1204. doi: 10.3390/genes11101204
Raza, A., Ashraf, F., Zou, X., Zhang, X., Tosif, H. (2020). “Plant adaptation and tolerance to environmental stresses: mechanisms and perspectives,” in Plant ecophysiology and adaptation under climate change: mechanisms and perspectives I (Singapore: Springer), 117–145.
Renu, K. C., Anjana, J., Vimal, K. S. (2011). Chloroplasts and mitochondria have multiple heat tolerant isozymes of SOD and APX in leaf and inflorescence in chenopodium album. Biochem. Biophys. Res. Commun. 412 (4), 522–525. doi: 10.1016/j.bbrc.2011.06.179
Ribeiro, C. W., Korbes, A. P., Garighan, J. A., Jardim-Messeder, D., Carvalho, F. E. L., Sousa, R. H. V. (2017). Rice peroxisomal ascorbate peroxidase knockdown affects ROS signaling and triggers early leaf senescence. Plant Sci. 263, 55–65. doi: 10.1016/j.plantsci.2017.07.009
Schramm, F., Ganguli, A., Kiehlmann, E., Englich, G., Walch, D., von Koskull-Döring, P. (2006). The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 60 (5), 759–772. doi: 10.1007/s11103-005-5750-x
Sharma, P., Dubey, R. S. (2005). Modulation of nitrate reductase activity in rice seedlings under aluminium toxicity and water stress: role of osmolytes as enzyme protectant. J. Plant Physiol. 162 (8), 854–864. doi: 10.1016/j.jplph.2004.09.011
Shi, W. M., Muramoto, Y., Ueda, A., Takabe, T. (2001). Cloning of peroxisomal ascorbate peroxidase gene from barley and enhanced thermotolerance by overexpressing in arabidopsis thaliana. Gene. 273 (1), 23–27. doi: 10.1016/S0378-1119(01)00566-2
Shigeoka, S., Ishikawa, T., Tamoi, M., Miyagawa, Y., Takeda, T., Yabuta, Y., et al. (2002). Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot 53(3772), 1305–1319. doi: 10.1093/jxb/53.372.1305
Shigeru, S., Takahiro, I., Masahiro, T., Yoshiko, M., Toru, T., Yukinori, Y., et al. (2002). Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53 (372), 1305–1319. doi: 10.1093/jxb/53.372.1305
Singh, N., Mishra, A., Jha, B. (2014). Over-expression of the peroxisomal ascorbate peroxidase (SbpAPX) gene cloned from halophyte salicornia brachiata confers salt and drought stress tolerance in transgenic tobacco. Mar. Biotechnol. 16 (3), 321–332. doi: 10.1007/s10126-013-9548-6
Tao, C. C., Jin, X., Zhu, L. P., Xie, Q. L., Wang, X. C., Li, H. B. (2018). Genome-wide investigation and expression profiling of APX gene family in gossypium hirsutum provide new insights in redox homeostasis maintenance during different fiber development stages. Mol. Genet. Genomics 293, 685–697. doi: 10.1007/s00438-017-1413-2
Teixeira, F. K., Menezes-Benavente, L., Galvão, V. C., Margis, R., Margis-Pinheiro, M. (2006). Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta. 224, 300–314. doi: 10.1007/s00425-005-0214-8
Teixeira, F. K., Menezes-Benavente, L., Margis, R., Margis-Pinheiro, M. (2004). Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: inferences from the rice genome. J. Mol. Evol. 59 (6), 761–770. doi: 10.1007/s00239-004-2666-z
Wang, J., Zhang, H., Allen, R. D. (1999). Overexpression of an arabidopsis peroxisomal ascorbate peroxidase gene in tobacco increases protection against oxidative stress. Plant Cell Physiol. 40 (7), 725–732. doi: 10.1093/oxfordjournals.pcp.a029599
Wu, B. M., Li, L., Qiu, T. H., Zhang, X., Cui, S. X. (2018). Cytosolic APX2 is a pleiotropic protein involved in H2O2 homeostasis, chloroplast protection, plant architecture and fertility maintenance. Plant Cell Rep. 37 (6), 833–848. doi: 10.1007/s00299-018-2272-y
Yang, Y. X., Ahammed, G. J., Wan, C. P., Liu, H. J., Chen, R. R., Zhou, Y. (2019). Comprehensive analysis of TIFY transcription factors and their expression profiles under jasmonic acid and abiotic stresses in watermelon. Int. J. Genomics 2314 (436), 1–13. doi: 10.1155/2019/6813086
Yoshimura, K., Yabuta, Y., Ishikawa, T., Shigeoka, S. (2000). Expression of spinach ascorbate peroxidase isoenzymes in response to oxidative stresses. Plant Physiol. 123 (1), 223–234. doi: 10.1104/pp.123.1.223
Zhang, Z. G., Zhang, Q., Wu, J. X., Zheng, X., Zheng, S., Sun, X. H., et al. (2013). Gene knockout study reveals that cytosolic ascorbate peroxidase 2(OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PloS One 8 (2), e57472. doi: 10.1371/journal.pone.0057472
Keywords: pepper, ascorbate peroxidase, bioinformatics analysis, gene expression, phylogenetic relationship
Citation: Pang X, Chen J, Xu Y, Liu J, Zhong Y, Wang L, Zheng J and Wan H (2023) Genome-wide characterization of ascorbate peroxidase gene family in pepper (Capsicum annuum L.) in response to multiple abiotic stresses. Front. Plant Sci. 14:1189020. doi: 10.3389/fpls.2023.1189020
Received: 18 March 2023; Accepted: 12 April 2023;
Published: 08 May 2023.
Edited by:
Xinyang Wu, China Jiliang University, ChinaReviewed by:
Guang-Long Wang, Huaiyin Institute of Technology, ChinaSi Ma, China Agricultural University, China
Copyright © 2023 Pang, Chen, Xu, Liu, Zhong, Wang, Zheng and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongjian Wan, d2FuaG9uZ2ppYW5Ac2luYS5jb20=; Jiaqiu Zheng, bmt5ODIzNkAxNjMuY29t
 Xin Pang1
Xin Pang1 Jun Chen
Jun Chen Jia Liu
Jia Liu Hongjian Wan
Hongjian Wan