
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 06 March 2024
Sec. Plant Abiotic Stress
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1349064
N-Acetylglucosamine (GlcNAc), a fundamental amino sugar moiety, is essential for protein glycosylation, glycolipid, GPI-anchor protein, and cell wall components. Uridine diphosphate-GlcNAc (UDP-GlcNAc), an active form of GlcNAc, is synthesized through the hexosamine biosynthesis pathway (HBP). Although HBP is highly conserved across organisms, the enzymes involved perform subtly distinct functions among microbes, mammals, and plants. A complete block of HBP normally causes lethality in any life form, reflecting the pivotal role of HBP in the normal growth and development of organisms. Although HBP is mainly composed of four biochemical reactions, HBP is exquisitely regulated to maintain the homeostasis of UDP-GlcNAc content. As HBP utilizes substrates including fructose-6-P, glutamine, acetyl-CoA, and UTP, endogenous nutrient/energy metabolites may be integrated to better suit internal growth and development, and external environmental stimuli. Although the genes encoding HBP enzymes are well characterized in microbes and mammals, they were less understood in higher plants in the past. As the HBP-related genes/enzymes have largely been characterized in higher plants in recent years, in this review we update the latest advances in the functions of the HBP-related genes in higher plants. In addition, HBP’s salvage pathway and GlcNAc-mediated two major co- or post-translational modifications, N-glycosylation and O-GlcNAcylation, are also included in this review. Further knowledge on the function of HBP and its product conjugates, and the mechanisms underlying their response to deleterious environments might provide an alternative strategy for agricultural biofortification in the future.
Plants, as sessile organisms, frequently suffer from deleterious environmental stimuli. Many cellular metabolic processes, such as carbohydrates, amino acids, lipids, and energy metabolism, are influenced by different developmental stages and abiotic stresses (Van Zelm et al., 2020; Mansour and Hassan, 2022). In response to developmental changes and external challenges, plants have evolved sophisticated mechanisms to better suit plant growth and environmental changes by integrating their internal metabolic status and optimizing metabolic reprogramming. One of these metabolic processes is the so-called hexosamine biosynthesis pathway (HBP), which utilizes fructose-6-phosphate (Fru-6-P), glutamine, acetyl-coenzyme A (acetyl-CoA), and uridine triphosphate (UTP) as substrates to synthesize uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc) (Figure 1). As the metabolic flux through HBP integrates glycolysis, amino acid, lipid, and nucleic acid pathways to maintain their balance and keep UDP-GlcNAc homeostasis, HBP may function as a metabolic integrator or hub for sensing nutrients (Buse, 2006; Chiaradonna et al., 2018) to link cellular nutrients/or energy signals and external cues. The HBP flux that generates UDP-GlcNAc is primarily regulated by the rate-limiting enzyme glutamine:fructose-6-phosphate amidotransferase (GFAT) activity and the obligatory substrate of O-linked GlcNAc transferase (OGT). The increased flux through HBP might be linked to insulin resistance, the vascular complications of diabetes, and cancer formation in mammals (Buse, 2006; Chiaradonna et al., 2018).
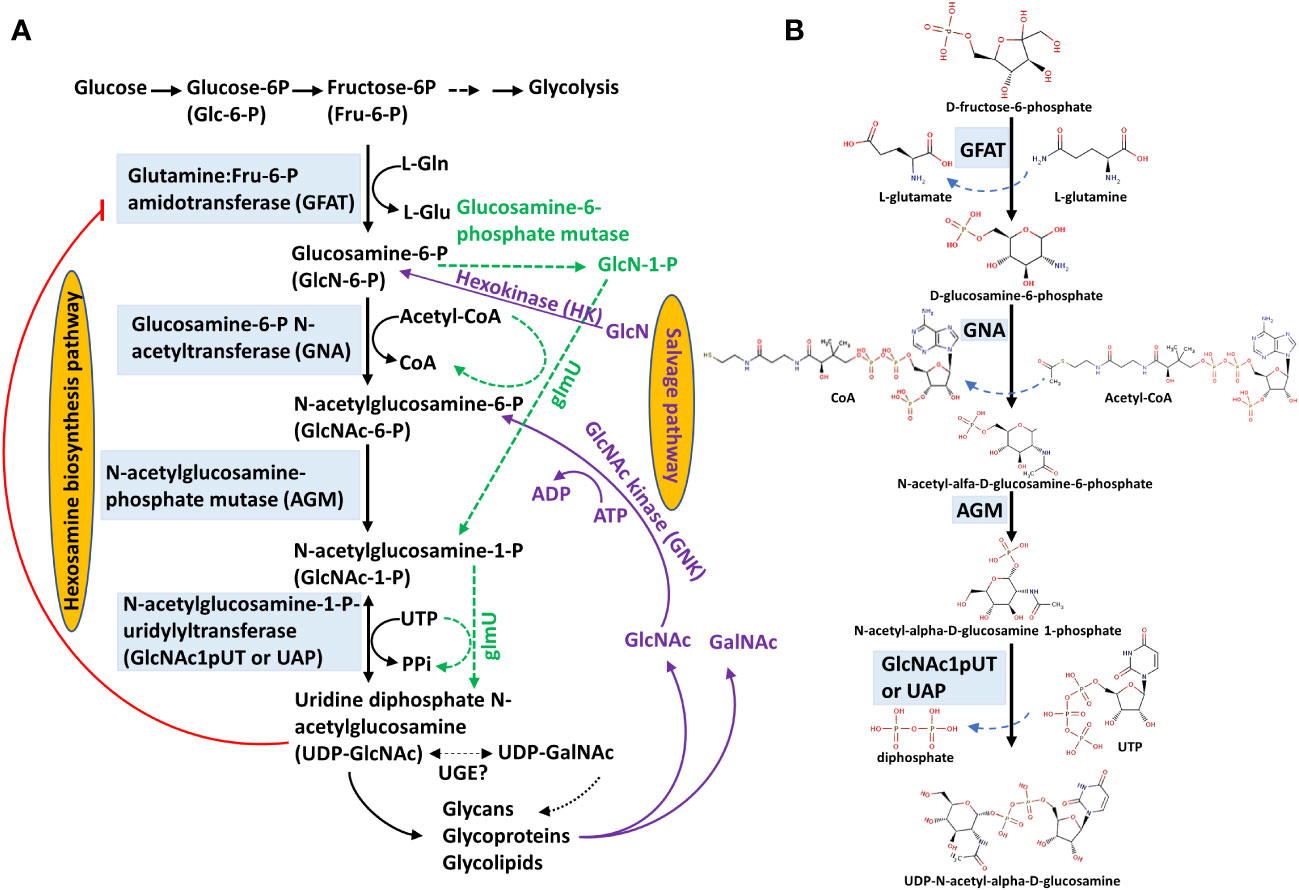
Figure 1 UDP-GlcNAc biosynthesis through hexosamine biosynthesis and salvage pathways. (A) Hexosamine biosynthesis and salvage pathways. The hexosamine biosynthesis pathway (HBP) is composed of four reactions catalyzed sequentially by glutamine:Fru-6-P amidotransferase (GFAT), glucosamine-6-P N-acetyltransferase (GNA), N-acetylglucosamine-phosphate mutase (AGM) and N-acetylglucosamine-1-P uridylyltransferase (GlcNAc1pUT or UAP) to synthesize uridine diphosphate N-acetylglucosamine (UDP-GlcNAc). UDP-GlcNAc is presumably interconverted to UDP-N-acetylgalactosamine (UDP-GalNAc) by an uncharacterized UDP-Glc-4-epimerase (UGE) in plants. In the salvage pathway (purple lines), GlcN is used and converted to GlcN-6-P catalyzed by a hexokinase (HK), followed by entering HBP to form UDP-GlcNAc. In addition, GlcNAc can be converted to GlcNAc-6-P catalyzed by an N-acetylglucosamine kinase (GNK); GlcNAc-6-P further enters the HBP to form UDP-GlcNAc. The green dashed lines represent HBP in prokaryotes. This HBP diagram is modified from Furo et al. (2015) and Chen et al. (2022). (B) Biochemical structures of HBP. These chemical structures are derived from the BRENDA database (https://www.brenda-enzymes.org/fulltext.php?overall=1).
UDP-GlcNAc, the active form of GlcNAc, is a fundamental amino sugar moiety essential for the glycosylation of proteins and lipids (Ebert et al., 2018), glycosylphosphatidylinositol (GPI)-anchor proteins (Lalanne et al., 2004), a cell wall component of chitin in yeast, and an exoskeleton of arthropods (Maia, 1994; Kato et al., 2002; Arakane et al., 2011). UDP-GlcNAc is synthesized through an HBP that involves four consecutive reactions orderly catalyzed by a GFAT (Hassid et al., 1959; Durand et al., 2008), a glucosamine-6-P N-acetyltransferase (GNA) (Vetting et al., 2005; Wang et al., 2008a), an N-acetylglucosamine-phosphate mutase (AGM)/or phosphoacetylglucosamine mutase (Mio et al., 2000), and an N-acetylglucosmine-1-P-uridylyltransferase (GlcNAc1pUT, Yang et al., 2010) or a UDP-N-acetylglucosamine pyrophosphorylase (UAP; Wang et al., 2015, Wang et al., 2021). These processes are essential for cell growth and stress response and are conserved across organisms (Milewski et al., 2006). Dysfunction of HBP enzymes frequently causes severe phenotypes (Schimmelpfeng et al., 2006); moreover, a complete block of HBP normally results in lethality in yeast, mammals, and plants (Milewski et al., 2006; Chen et al., 2014; Pantaleon, 2015; Vu et al., 2019; Jia et al., 2023).
In the past, the function and regulation of HBP have been more intensively studied in microbes and mammals than in plants. The application of HBP products or intermediates through dietary treatment has been proposed as a therapy for human genetic disorders (Paneque et al., 2023). Elegant reviews have been recently reported on the function and regulation of HBP, which are primarily stressed in microbes (Wyllie et al., 2022) and eukaryotes of mammals (Paneque et al., 2023); however, an overview of HBP functions in plants is lacking. As research progress has greatly advanced in plant HBP study in recent years, in this review, we update our current knowledge of HBP function in plants and its stress responses. In addition, we provide an overview of the salvage pathway of HBP and the targets of UDP-GlcNAc in two major co- or post-translational modifications, N-linked glycosylation (N-glycosylation) and O-linked β-N-acetylglucosamine (O-GlcNAcylation). Further knowledge of HBP function and its response to abiotic stress may provide an alternative strategy to manipulate plant growth and tolerance to abiotic stress.
The first committed step of HBP is the transamination of D-fructose-6-phosphate from L-glutamine to form D-glucoamine-6-phosphate and L-glutamate, catalyzed by an L-glutamine:D-fructose-6-phosphate amidotransferase (GFAT; EC2.6.1.16) (Figure 1), also known as glucosamine-6-phosphate synthase (GlcN6P synthase). GFAT acts as the rate-limiting enzyme in the de novo HBP in fungi and animals (Olchowy et al., 2007; Walter et al., 2020; Paneque et al., 2023). Based on its origins in prokaryotes and lower or higher eukaryotes, the GFAT-encoded gene has been termed Glms, GFA, or GFAT (Durand et al., 2008). The function of GFAT is conserved among organisms, including microorganisms, mammals, and plants (Milewski et al., 2006). Yeast GFA1 activity is inhibited by UDP-GlcNAc and this inhibition is noncompetitive. In the pathogenic yeast Candida albicans, GFA activity increases during the yeast-to-mycelium morphological transformation, ensuring that UDP-GlcNAc production is increased when more amino sugars are needed in mycelium cells (Milewski et al., 1999, Milewski et al., 2006). GFA1 is the primary target molecule of methylmercury in Saccharomyces cerevisiae and yeast cells overexpressing GFA1 confer resistance to methylmercury, an important environmental pollutant that causes neurological toxicity in mammals (Miura et al., 1999; Naganuma et al., 2000).
In plants, GFAT activity was first described by Hassid et al. (1959), and GFAT activity from mung bean Phaseolus aureus was partially purified and characterized (Vessal and Hassid, 1972). The Arabidopsis genome only contains a single copy of the GFAT gene (At3g24090), termed GFAT1, and its expression is primarily restricted to mature pollen grains (Wang et al., 2008b; Vu et al., 2019). Nevertheless, Arabidopsis GFAT1 transcripts are also detectable in roots, flowers, and siliques by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) (Jia et al., 2023). The loss-of-function AtGFAT1 displays defects in a polar deposition of pectin and callose in the pollen wall, leading to inactivation of pollen activity; thus, the knockout mutant Atgfat1-2 is lethal. In contrast, the knockdown mutant Atgfat1 or GFAT1 RNAi lines show reductions in glucosamine (GlcN) and UDP-GlcNAc levels in association with the reduced protein N-glycosylation but increased sensitivity of tunicamycin, an ER stress inducer agent. The RNAi lines also impair vegetative and reproductive development and display partial sterility. The abnormal phenotypes observed in Atgfat1 can be largely rescued by the exogenous application of GlcN (Vu et al., 2019). It was reported that GlcN inhibits Arabidopsis hypocotyl elongation due to the induction of reactive oxygen species (ROS). Arabidopsis transgenic plants overexpress E. coli glucosamine-6-phosphate deaminase (NagB) to scavenge endogenous GlcN and confer tolerance to oxidative, drought, and cold stresses. Moreover, overexpression of E. coli GlmS in Arabidopsis promotes cell death at an early stage (Chu et al., 2010).
The second enzyme in the HBP pathway is D-Glucosamine-6-phosphate N-acetyltransferase (GNA; EC 2.3.1.4), which converts GlcN-6-phosphate and acetyl-CoA to N-acetylglucosamine-6-phosphate (GlcNAc-6P) and CoA (Figure 1). GNA is a single-copy gene in the genome of most characterized organisms characterized. For example, the yeast S. cerevisiae gene (YFL017C) was demonstrated to exhibit GNA activity and is thus designated as ScGNA1 (Mio et al., 1999). Additionally, the Arabidopsis genome also contains one GNA (AtGNA, At5g15770), the expression of which is ubiquitous in all organs (Riegler et al., 2012) and shows a slightly diurnal expression pattern (Usadel et al., 2008). In contrast to Arabidopsis, rice possesses two GNAs, including OsGNA1 (LOC_Os09g31310) and OsGNA (LOC_Os02g48650). OsGNA1 is highly expressed in root tissues (Jiang et al., 2005) but OsGNA is less characterized (Riegler et al., 2012) and has low expression levels in all tissues as revealed by the rice eFP browser (Jain et al., 2007). Based on the transient expression of the AtGNA-GFP fused protein in Arabidopsis protoplasts, its subcellular localization is primarily in the endoplasmic reticulum (ER) (Riegler et al., 2012), This result supports the role of UDP-GlcNAc, the end product of the HBP pathway, in protein glycosylation and synthesis of the GPI anchor in the ER. It was observed that deletion of yeast ScGNA1 or AfGNA1 and the loss-of-function of AtGNA by a T-DNA insertion (AtGNA1-2 and AtGNA1-3), resulting in a complete block of GlcNAc production, is lethal (Mio et al., 1999; Riegler et al., 2012; Lockhart et al., 2020); this phenotype is similar to that obtained for the knockout mutants of Arabidopsis in GFAT, phospho-N-acetylglucosamine mutase or the double mutant glcnac.ut1/glcnac.ut2 (Chen et al., 2014; Vu et al., 2019; Jia et al., 2023). This result also reflects the vital role of UDP-GlcNAc in plant growth. Although AtGNA has a low protein sequence identity to Homo sapiens HsGNA (~39.1%) and S. cerevisiae ScGNA (~35.0%), this protein crystal structure at 1.5 Å resolution exhibited very high structural similarity to these two orthologs (Riegler et al., 2012).
An EMS-mutagenized missense mutation in Arabidopsis GNA, known as lignescens (lig), causes plant growth defects and ectopic lignin accumulation under high temperature (28°C) conditions. Compared to the wild type, the lig mutant plants exhibit lower levels of UDP-GlcNAc than the wild type, accompanied by defects in N-linked protein glycosylation, ER stress, and unfolded protein response (UPR). Supporting evidence reveals the upregulation of BiP3 expression, an ER stress marker, under high-temperature conditions and treatments with the ER stress-inducing agents, tunicamycin, and DTT, resulting in plants with phenotypes that mimic the lig mutant. Moreover, exogenous application of UDP-GlcNAc, GlcNAc, or GalNAc rescues the high-temperature sensitivity and ectopic accumulation of lignin observed in the gna/lig mutants. Thus, dysfunction of GNA causes a high-temperature-dependent defect in UDP-GlcNAc biosynthesis, which further affects N-linked protein glycosylation and lignin accumulation, mostly through the UPR (Nozaki et al., 2012).
The function of rice OsGNA1 was also reported by Jiang et al. (2005). Osgna1 is a T-DNA insertion mutant that shows lower levels of UDP-GlcNAc and defects in N-linked protein glycosylation, as well as a reduction in O-linked glycosylation activity. The short-root phenotype of Osgna1 is temperature-sensitive, particularly at 25°C, which can be largely rescued by a high temperature of 32°C. This low temperature-sensitive response in rice may be opposite to that of the Arabidopsis Atgna mutant, which shows greater sensitivity to high temperature. This discrepancy remains to be investigated in the future. These short roots observed in Osgna1 are linked to defects in mitochondrial dehydrogenase activity, root viability, cell shape, and microtubule stability. The latter may result from a defect in O-linked glycosylation of microtubule-associated proteins (Jiang et al., 2005).
N-acetylglucosamine-phosphate mutase (AGM; EC 5.4.2.3) or phosphoacetylglucosamine mutase catalyzes the isomerization of N-acetylglucosamine-6-P (GlcNAc-6-P) into N-acetylglucosamine-1-P (GlcNAc-1-P) (Figure 1). The growth of the yeast ScAGM deletion mutant (Scagm) cannot progress through five cell cycles. Overexpression of ScAGM may complement the growth defect of a phosphoglucomutase (PGM) double deletion mutant (pgm1/pgm2); however, overexpression of ScPGM2, a major PGM, cannot restore the growth of Scagm1 deletion mutant cells. These data suggested that the different hexosephosphate mutases of S. cerevisiae share partially overlapping substrate specificities but they have distinct physiological functions (Hofmann et al., 1994). In mice, severely reduced AGM1/PGM3 activity causes lethality during embryonic development, whereas mutated mice with partial AGM1/PGM3 activity do not perish but display severe syndromes, such as sterility (Greig et al., 2007). Human patients with mutations in PGM3/AGM1 will die in early infancy or have congenital immune system defects, developmental delays, and neurocognitive disorders (Ben-Khemis et al., 2017).
The Arabidopsis AGM gene (At5g18070) was first identified by selecting for complementation of Escherichia coli UV-sensitive mutants, and the identified gene was termed DNA-DAMAGE-REPAIR/TOLERANCE 101 (DRT101). The N-terminus of AGM/DRT101 contains an amino acid region similar to the chloroplast transit peptide, suggesting its possible subcellular localization in chloroplasts (Pang et al., 1993). Arabidopsis AGM shares 38 to 44% amino acid identity with Homo sapiens, S. cerevisiae, and Aspergillus fumigatus, and their protein structures are highly conserved. Although two members of the Arabidopsis α-D-phosphoglucosamine mutase family, At5g17530 and At1g70820, are phylogenetically similar to AtAGM, only AtAGM functions in the isomerization of GlcNAc-1-P and GlcNAc-6-P. AtAGM has promiscuous substrates and catalyzes the interconversion of GlcNAc-1-P and GlcNAc-6-P and Glc-1-P and Glc-6-P; the catalytic reaction by AtAGM requires divalent cations, such as Mg2+ or Mn2+ (Jia et al., 2023).
Based on the RT-qPCR analyses, AtAGM is highly expressesed in the roots, flowers, and siliques, similar to the AtGFAT expression pattern. Moreover, unlike other HBP enzymes present in the cytosol or ER membrane surface, overexpression of the 35S::AGM-GFP transgene in the Atagm background, i.e., Atagm-OE, reveals AtAGM localization in the cytosol, cytomembrane, chloroplasts, and mitochondria (Jia et al., 2023).
Similar to other HBP mutants, the homozygous knockout mutants, such as Atagm2 (SAIL_187_F01) are lethal; however, the knockdown mutants, Atagm1 (SALK_039132C) and Atagm2 (+/-) can survive. The expression of the AtAGM gene in both Atagm1 and Atagm2 (+/-) is greatly reduced, and these mutants show a ~40% reduction in UDP-GlcNAc content compared to wild-type plants. Interestingly, overexpression of AtAGM in the Atagm background, i.e., Atagm-OE, does not increase UDP-GlcNAc contents; this likely results from feedback inhibition of UDP-GlcNAc, which affects the glutaminase function of GFAT (Olchowy et al., 2007; Walter et al., 2018; Vu et al., 2019). Thus, exogenous tunicamycin impairs UDP-GlcNAc inhibition and enhances AtGFAT activity, leading to increasingly higher levels of UDP-GlcNAc in Atagm-OE plants than in the wild-type plants (Jia et al., 2023). Thus, HBP is exquisitely regulated to maintain UDP-GlcNAc homeostasis, which plays a critical role in normal plant growth and development. Although these knockdown mutants display no conceivable phenotype, they show more vigorous growth than the wild type and Atagm-OE at maturity under normal growth conditions. This vigorous growth observed in the mutants presumably results from high chlorophyll contents that enhance photosynthetic capability. Moreover, these mutants show temperature-dependent (28°C) growth defects, including short roots and germination delay. Temperature-sensitive phenotypes can be abolished by exogenous UDP-GlcNAc (Jia et al., 2023). These data suggest that a small amount of UDP-GlcNAc is sufficient for normal plant growth, which is also observed in mouse embryonic fibroblasts (Boehmelt et al., 2000). However, plants need more UDP-GlcNAc when adapting to abiotic stress, and the mutant plants, such as Atagm1 and Atagm2 (+/-), cannot produce adequate UDP-GlcNAc under deleterious environments, leading to stress-induced growth defects.
Total protein blots stained with concanavalin A (ConA) lectin revealed that glycoproteins show no significant difference, whereas the N-glycan composition varies among wild type, Atagm2 (+/-), and Atagm-OE. Moreover, an obvious impairment of O-GlcNAcylation is observed in the Atagm mutants. The temperature-sensitive growth defects are primarily linked to the impairment of protein O-GlcNAcylation but not N-glycosylation because the O-GlcNAcylation deficient mutants Atsecs, in which O-GlcNAc transferase (OGT, Figure 2) is defective, also display temperature-sensitive phenotypes; however, no significant phenotype was observed in the N-glycosylation deficient mutant Atstt3a, in which oligosaccharyltransferase (OST, Figure 2) is defective (Jia et al., 2023).
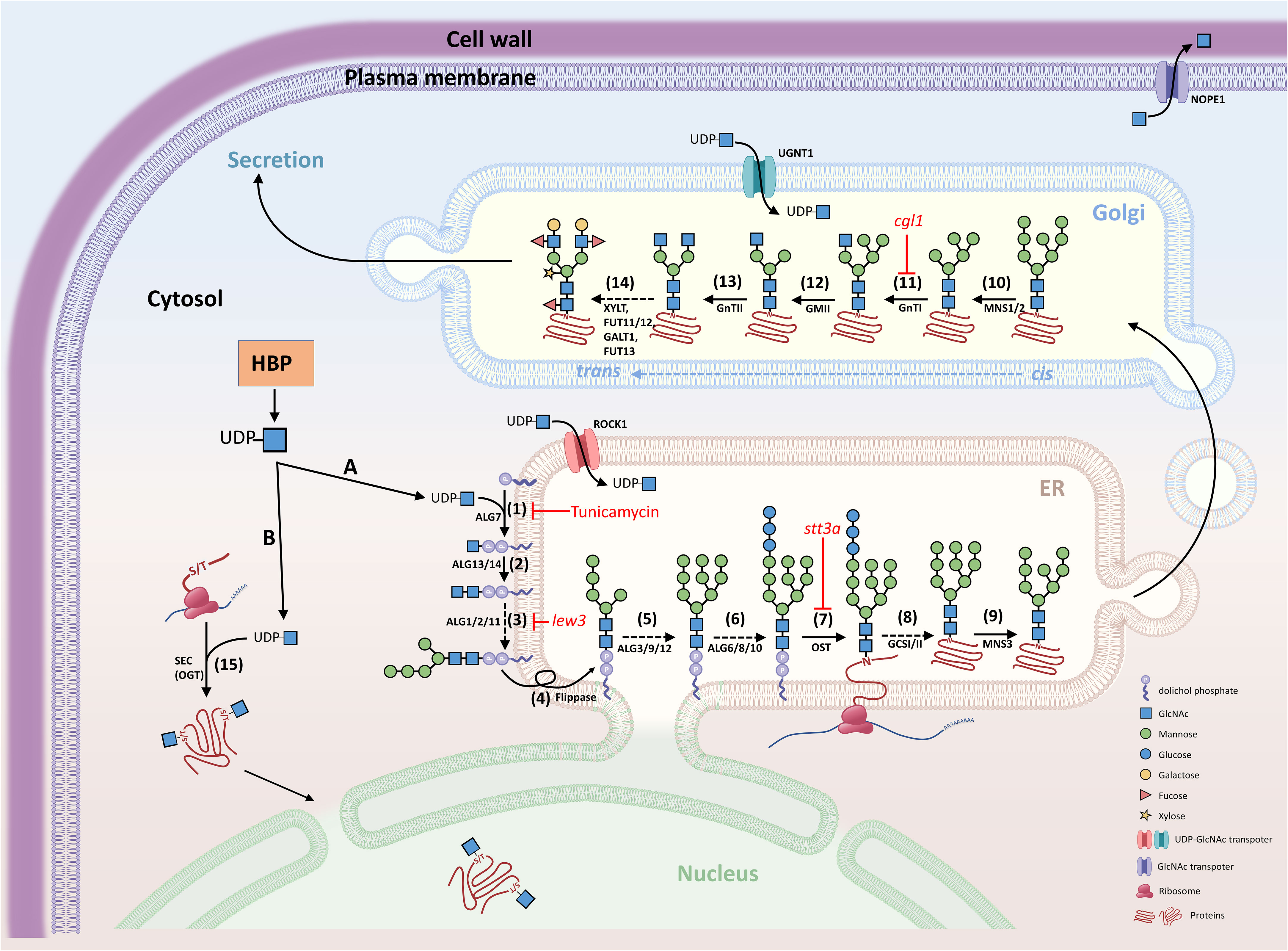
Figure 2 Schematic diagram of N-glycosylation and O-GlcNAcylation. GlcNAc is the fundamental amino sugar moiety essential for N-glycosylation and GlcNAcylation. (A) N-glycosylation. UDP-GlcNAc is generated by the hexosamine biosynthesis pathway (HBP) and provides GlcNAc for the initial biosynthesis of oligosaccharide precursors at the cytosolic side of the ER. The oligosaccharide precursor (Man5GlcNAc2-PP-Dol) enters the ER lumen for N-glycan modification and N-glycosylation of proteins. Complex and hybrid N-glycan processing occurs in the Golgi apparatus. Proteins with mature N-glycans will be secreted to their destinations. (B) O-GlcNAcylation. UDP-GlcNAc also provides the GlcNAc molecular unit directly to the Ser/Thr amino acids of proteins localized in the cytosol and nucleus. (1) Asparagine-linked glycosylation (ALG) enzyme ALG7, a UDP-N-acetylglucosamine:dolichol phosphate N-acetylglucosamine-1-P transferase; (2) ALG13 and ALG14, UDP-N-acetylglucosamine transferase subunits; (3) ALG1/2/11, mannosyltransferases; (4) Flippase-like protein; (5) ALG3/9/12, mannosyltransferases; (6) ALG6/8/10, glucosyltransferases; (7) OST, oligosaccharyltransferase complex; (8) GCSI/II, glucosidases; (9) MNS3, ER-α-mannosidase I; (10) MNS1/2, Golgi-α-mannosidase I; (11) GnTI, β-(1->2)-N-acetylglucosaminyltransferase I or COMPLEX GLYCAN LESS 1 (CGL1); (12) GMII, Golgi α-mannosidase II; (13) GnTII, β-(1->2)-N-acetylglucosaminyltransferase II; (14) XYLT, β-(1->2)-xylosyltransferase; FUT11/12, core α-(1->3)-fucosyltransferases; GALT1, β-(1->3)-galactosyltransferase 1; FUT13, α-(1->4)-fucosyltransferase; (15) SEC, SECRET AGENT (O-GlcNAc transferase, OGT); ROCK1, REPRESSOR OF CYTOKININ DEFICIENCY 1; UGNT1, UDP-GlcNAc transporter; NOPE1, NO PERCEPTION 1. The nomenclature of enzymes is generally based on the report by Strasser et al. (2021).
The last reaction of HBP is the uridylation of GlcNAc-1-P into UDP-GlcNAc by N-acetylglucosamine-1-P uridylyltransferase (GlcNAc1pUT), named after a forward catalytic reaction (Yang et al., 2010) or UDP-N-acetylglucosamine-1-P pyrophosphorylase (UAP; EC 2.7.7.23), named after a reverse reaction (Figure 1). Although the biosynthesis of UDP-GlcNAc in prokaryotes and eukaryotes is similar, their catalytic specificity is diverse (Mengin-Lecreulx and Van Heijenoort, 1993). In bacteria, GlmU is a bifunctional enzyme that exhibits both the phosphoglucosamine acetyltransferase and UDP-N-acetylglucosamine pyrophosphorylase activities; however, these two enzymatic activities are encoded by distinct essential genes in eukaryotes. Thus, GlmU catalyzes the acetylation of GlcN-1-P into GlcNAc-1-P followed by the uridylation of GlcNAc-1-P into UDP-GlcNAc (Figure 1, green dashed line). Inactivation of the GlmU gene reduces glycoprotein synthesis, leading to changes in cell shape and lysis changes (Mengin-Lecreulx and Van Heijenoort, 1993). In yeast, a null mutation of yeast UAP1/QRI1 is lethal, which mainly shows swollen and lysed cell shapes (Mio et al., 1998). In Drosophila melanogaster, the cabrio/mummy mutant derived from EMS mutagenesis loses DmUAP function and exhibits defects in dorsal closure, central nervous system, and embryo development (Schimmelpfeng et al., 2006). Two human UAPs (AGX1 and AGX2) were identified with only a 17-amino acid difference and these UAPs were derived from alternative splicing and led to preferential substrate specificity in GalNAc-1-P and GlcNAc-1-P, respectively (Wang-Gillam et al., 1998; Peneff et al., 2001).
In Arabidopsis, two UAPs termed GlcNAc1pUT1 and GlcNAc1pUT2 are encoded by GlcNA.UT1 and GlcNA.UT2, respectively. They were first cloned and their biochemical specificity was characterized by Yang et al. (2010). In general, GlcNAc1pUT1 uses GlcNAc-1-P or GalNAc-1-P as substrates together with UTP to form UDP-GlcNAc or UDP-GalNAc and PPi. This uridylation activity is similar to that of human AGX1. GlcNAc1pUT2 has broader substrate specificities and may utilize Glc-1-P as a substrate in addition to GlcNAc-1-P and GalNAc-1-P. Thus, the enzymatic activity of AtGlcNAc1pUT2 is closer to that of yeast UAP1/QRI1 (Mio et al., 1998) and rice OsUAP1/SPL29 (Wang et al., 2015). The substrate specificity between GlcNAc1pUT1 and GlcNAc1pUT2 is likely related to their protein structures, which share a similar fold but vary in some loop regions. The biochemical assay also indicated that Arabidopsis GlcNAc1pUTs require divalent ions (such as Mg2+ or Mn2+) for their enzymatic activity. Gel-filtration analysis revealed the monomer structure of the native GlcNAc1pUT1 protein (Yang et al., 2010), which is different from the dimer structure of human AGX1 (Wang-Gillam et al., 1998) and yeast UAP1/QRI1 (Milewski et al., 2006). Although GlcNAc1pUT1 and human AGX1 share 32% protein sequence identity, their three-dimensional protein structure models display a conserved catalytic fold and key conserved motifs (Yang et al., 2010).
AtGlcNAc1pUT1 may utilize both UDP-GlcNAc and UDP-GalNAc as substrates (Yang et al., 2010). UDP-GalNAc has been found in several plant species, including squash (Tolstikov and Fiehn, 2002) and dahlia tubers (Gonzalez and Pontis, 1963). In barley, UDP-Glc 4-epimerase or UDP-Gal 4-epimerase (UGE; EC 5.1.3.2) catalyzes the interconversion of UDP-Glc and UDP-Gal; the enzyme can also reversibly catalyze UDP-GlcNAc and UDP-GalNAc (Zhang et al., 2006). Although several UGE genes have been cloned in plants, such as peas, Arabidopsis, and the endospermous legume guar (Dörmann and Benning, 1996; Lake and Slocum, 1998; Joersbo et al., 1999), the GalNAc targets of glycans and glycoproteins and their physiological significance remain to be further examined. As UDP-GlcNAc and UDP-GalNAc contain the same molecular mass, they cannot be distinguished by mass spectrometry analysis. Specific HPLC analysis can separate these two hexosamines obtained from Arabidopsis tissues (Nozaki et al., 2012). In general, UDP-GlcNAc is more abundant than UDP-GalNAc in plant tissues under normal or temperature-stress conditions (Nozaki et al., 2012).
Mutation of Arabidopsis GlcNAc.UT1 or GlcNAc.UT2 shows no conceivable phenotype, whereas the double mutant is lethal (Chen et al., 2014), reflecting functional redundancy and the pivotal role of these genes in normal plant growth and development. The heterozygous double mutant GlcNA.UT1/glcna.ut1 glcna.ut2/glcna.ut2 obtained from the F2 segregating population following reciprocal crosses of glcna.ut1 and glcna.ut2, displays sterility. Furthermore, this heterozygous double mutant reveals the aberrant transmission of (glcna.ut1, glcna.ut2) gametes, which is consistent with the defects in male gametophytes during late vacuolation (or pollen mitosis I stage) and in female gametophytes during the uninucleate embryo sac stage. Interestingly, one normal allele of GlcNA.UT2 in the glcna.ut1/glcna.ut1 GlcNA.UT2/glcna.ut2 mutant has normal gamete transmission of (glcna.ut1, glcna.ut2) and gametophytic development, except that the development of numerous embryos is arrested during the early globular stage (Chen et al., 2014). Thus, GlcNA1pUT1 and GlcNA1pUT2 differentially regulate gametophytic and embryonic development, which may be associated with their spatiotemporal expression, subtle difference in GlcNAc1pUTase activity, and metabolic complementation (Bonhomme et al., 1998). To further study Arabidopsis GlcNA.UT function, the RNAi transgenic plants, termed iU1s, were generated by RNA interference of GlcNA.UT1 expression in the glcna.ut2 null mutant background. The iU1 transgenic plants resemble the heterogeneous double mutant GlcNA.UT1/glcna.ut1 glcna.ut2/glcna.ut2 showing sterility under normal growth conditions. The iU1s possess normal levels of hexosamine (UDP-GlcNAc and UDP-GalNAc) compared to the wild type under normal growth conditions, whereas they show reduced hexosamine biosynthesis, altered protein N-glycosylation, and an unfolded protein response under salt-stressed conditions. Moreover, the iU1s confer slat hypersensitivity, including delay of seed germination and early seedling establishment, in association with the induction of ABA biosynthesis and its signal networks under salt stress. Furthermore, microarray analysis data support the upregulation of genes involved in ABA (such as NCED3, ABI5, and ABCG25) and salt stress responses (such as RD29A, RD29B, and DREB2A) (Chen et al., 2022).
Likewise, rice GlcNA.UTs termed UAP1 or SPOTTED LEAF 29 (SPL29) and UAP2 (Wang et al., 2015, Wang et al., 2021) can catalyze GlcNAc-1-P and GalNAc-1-P as substrates to form UDP-GlcNAc (Xiao et al., 2018; Wang et al., 2021). OsUAP1/SPL29 irreversibly catalyzes the decomposition of uridine 5’-diphosphoglucose (UDPG) to form UTP and Glc-1-P. The loss-of-function Osuap1/spl29 mutant accumulates UDPG, which may be involved in ROS accumulation, early leaf senescence, plant cell death (PCD), and leaf lesion mimics (or defense response) (Wang et al., 2015; Xiao et al., 2018). It remains unknown whether Arabidopsis GlcNAc1pUTs may use UDPG as a substrate similar to rice. As spotted leaves are a lesion-mimic phenotype of the hypersensitive response, the Osuap1/spl29 mutant causes induction of the defense response by upregulation of defense-responsive genes and bacterial blight resistance. In addition, early leaf senescence and defense response enhancement are linked to the accumulation of jasmonic acid, abscisic acid, and reactive oxygen species (ROS) in Osuap1/spl29 mutant plants (Wang et al., 2015). OsUAP2 overexpression may rescue Osuap1/spl29 mutant phenotypes, reflecting that they share functional redundancy. OsUAP2 is primarily expressed in the early leaf development and OsUAP1/SPL29 at the whole leaf developmental stages, and both genes synergistically regulate rice leaf development and protect them from early senescence (Wang et al., 2021). Thus, unlike Arabidopsis single mutant glcna.ut1 or glcna.ut2, which show no conceivable phenotype, the single Osuap1/spl29 mutant displays early senescence and lesion-mimic spotted leaves, presumably indicating that rice plants are more sensitive to the defense response. The functions of HBP-related genes/proteins are summarized in Table 1.
It was reported that NO PERCEPTION 1 (NOPE1) acts as the GlcNAc transporter localized in the plasma membrane of the root tissues of rice and maize. NOPE1 transports GlcNAc into the rhizosphere, where it serves as a molecular signal to enhance branching hyphae of arbuscular mycorrhiza (AM) and benefit the symbiosis between AMs and host plants (Nadal et al., 2017). Arabidopsis genome contains UDP-GlcNAc transporters, one was termed UDP-GlcNAc transporter (UGNT1; At4g32272), in the Golgi membrane, which transports UDP-GlcNAc from the cytosol to the Golgi to initiate complex glycan processing. The Atugnt1 null mutant plants lack complex and hybrid N-glycans, and the N-glycopeptides primarily contain high-mannose structures. Moreover, AtUGNT1 is also needed for the biosynthesis of GlcNAc-containing glycosyl inositol phosphorylceramides (GIPCs) (Ebert et al., 2018). Another transporter for UDP-GlcNAc and UDP-GalNAc is the REPRESSOR OF CYTOKININ DEFICIENCY 1 (ROCK1, At5g65000), which is localized in the ER membrane and involved in ER quality control. The Atrock1 mutant reduces the activity of cytokinin oxidases/dehydrogenases (CKXs, cytokinin-degrading enzymes) and impairs the cytokinin-deficient mutant phenotype. Although the N-glycosylation of CKX1 is not affected in Atrock1, the stability of CKX1 is enhanced in the mutant (Niemann et al., 2015) (Figure 2). The function of ROCK1 in providing UDP-GlcNAc for the ER lumen remains unknown because GlcNAc conjugates in the ER have not been uncovered to date. One possibility is that UDP-GlcNAc in the ER may be transported to the Golgi apparatus for complex glycan modification (Ebert et al., 2018).
In addition to de novo biosynthesis through HBP, UDP-GlcNAc can be generated by the salvage pathway. In mammals, GlcN and GlcNAc can be retrieved from environmental resources and the degradation of glycans or glycoconjugates. These salvage nutrients can be used as dietary supplements to benefit from the treatment of disorders linked to glycosylation. GlcN can be phosphorylated to form GlcN-6-P by hexokinase and then enters the HBP to produce UDP-GlcNAc (Figure 1, purple line) (Kornfeld, 1967; Krug et al., 1984). In Arabidopsis, the exogenous application of GlcN to Atgfat1 mutant plants may rescue mutant phenotypes, indicating that the GlcN can be converted into GlcN-6-P, which further enters HBP to form UDP-GlcNAc (Vu et al., 2019).
GlcNAc can be phosphorylated to form GlcNAc-6-P by GlcNAc kinase (GNK) or N-acetylglucosamine kinase (NAGK) (Gindzieński et al., 1974; Allen and Walker, 1980; Hinderlich et al., 2000; Berger et al., 2002; Ryczko et al., 2016). This intermediate GlcNAc-6-P further enters the HBP to form UDP-GlcNAc (Figure 1, purple line). In mammals, NAGK is needed for embryonic mouse development (Dickinson et al., 2016). Deletion of NAGK increases de novo hexosamine biosynthesis; conversely, glutamine deprivation inhibits de novo HBP but triggers the NAGK-dependent salvage pathway in pancreatic ductal adenocarcinoma (PDAC) (Campbell et al., 2021), suggesting that cross-talk occurs between the salvage and de novo HBP. In higher plants, the GNK was first identified and characterized in Arabidopsis by Furo et al. (2015). Arabidopsis GNK (At1g30540) and human NAGK proteins share high structural conservation, particularly in GlcNAc and ATP binding domains. The kinase activity of AtGNK was confirmed by an enzymatic activity assay in vitro through recombinant AtGNK protein. Substrate analysis further supports that AtGNK exhibits high specificity for GlcNAc and less specificity for GalNAc. Furthermore, although the null mutant Atgnk shows no conceivable phenotype, the mutant plants reveal lower levels of UDP-GlcNAc than the wild type and are insensitive to the exogenous application of GlcNAc (Furo et al., 2015). The GlcNAc salvage pathway is also observed in the Atgna/lig mutant, which is defective in the conversion of GlcN-6-P to GlcNAc-6-P and leads to a reduction in UDP-GlcNAc levels, high-temperature sensitivity, and ectopic accumulation of lignin. Exogenous application of GlcNAc rescues the Atgna/lig mutant phenotypes and increases the UDP-GlcNAc content (Nozaki et al., 2012). Therefore, the Atgna/lig mutant fails to convert GlcN6-P to GlcNAc-6P; however, exogenous GlcNAc can be catalyzed by AtGNK to form GlcNAc-6-P, which further enters the HBP to produce UDP-GlcNAc. Similarly, wild-type plants (Col-o) produce more UDP-HexNAc (UDP-GlcNAc and UDP-GalNAc) by exogenous GlcNAc under normal and salt-stressed conditions (Chen et al., 2022). The coexistence of HBP and salvage pathways may finetune the homeostasis of UDP-GlcNAc contents in plants in response to nutrient fluctuations and environmental stimuli.
Asparagine (Asn- or N-) glycosylation is among the most common co- or post-translational modifications, which is essential for plant growth and stress responses and is conserved across eukaryotes (Banerjee et al., 2007; Bao and Howell, 2017; Nagashima et al., 2018). N-glycosylation regulates protein folding, transport, sorting, degradation, and intracellular signaling (Helenius and Aebi, 2001; Molinari, 2007; Aebi, 2013; Lannoo and Van Damme, 2015; Shin et al., 2018). Most secreted and membrane-associated proteins are N-glycosylated proteins (N-glycoproteins), and they are involved in a wide range of cellular processes, including cell wall biosynthesis (Jose-Estanyol and Puigdomenech, 2000), pollination (Hancock et al., 2005), pathogen defense (Pearce et al., 2007), and cell-to-cell communication (Taoka et al., 2007). Biosynthesis of N-glycan occurs in multiple subcellular compartments, including the cytosol, endoplasmic reticulum (ER) lumen, and Golgi apparatus (Pattison and Amtmann, 2009). Initially, N-glycan is formed as an oligosaccharide precursor on a lipid-linked carrier, dolichol pyrophosphate (PP-Dol), on the cytosolic side of the ER membrane (Figure 2). Two GlcNAc molecules are first transferred to PP-Dol by GlcNAc-1-phosphotransferase (GPT) or asparagine-linked glycosylation (ALG) enzyme ALG7 and the ALG13/14. Subsequently, five mannose (Man) residues are added by mannosyltransferases, ALG1/2/11, to form Man5GlcNAc2-PP-Dol (Burda and Aebi, 1999; Strasser et al., 2021). This oligosaccharide precursor is then flipped to face the ER lumen for further modification (Pattison and Amtmann, 2009; Strasser et al., 2021).
In the ER lumen, four more Man and three Glc residues are sequentially added to form the core oligosaccharide Glc3Man9GlcNAc2-PP-Dol, which is assembled by a series of membrane-bound mannosyltransferases (ALG3/9/12) and glycosyltransferases (ALG6/8/10) (Snider et al., 1980; Helenius and Aebi, 2001; Nagashima et al., 2018; Strasser et al., 2021). N-glycosylation occurs in the ER lumen by transferring the core oligosaccharide to Asn in the Asn-X-Ser/Thr motif (X, any amino acid except Pro) of a nascent peptide, which is mediated by an oligosaccharyltransferase (OST) complex (Burda and Aebi, 1999; Pattison and Amtmann, 2009; Strasser, 2016). The N-linked Glc3Man9GlcNAc2 glycan is further processed by the sequential removal of three Glc residues by glucosidase I and II (GCSI and GCSII) (Trombetta and Parodi, 2003; Nagashima et al., 2018; Strasser et al., 2021), and a Man residue is removed by the ER-α-mannosidase I (MNS3; Liebminger et al., 2009). The correctly folded glycoproteins leave the ER and move into the Golgi apparatus for further complex and hybrid N-glycan modification (Strasser, 2016).
In the Golgi, the first N-glycan processing is carried out by α-1,2-mannosidase I (MNS1/2), which removes three Man residues from Man8GlcNAc2 to form Man5GlcNAc2, the product for the subsequent complex and hybrid N-glycan processing. The formation of complex and hybrid N-glycan is initiated by the N-ACETYLGLUCOSAMINYL TRANSFERASE I (GnTI)-mediated addition of the GlcNAc residue to the α-1,3-linked Man of the Man5GlcNAc2 to form GlcNAcMan5GlcNAc2 (von Schaewen et al., 1993; Strasser et al., 1999). Subsequently, alternative processing pathways can occur in plants (Bencúr et al., 2005). In the canonical pathway, two Man residues are cleaved from GlcNAcMan5GlcNAc by Golgi-α-mannosidase II (GMII), followed by GnTII-mediated addition of another GlcNAc residue to the α1,6-linked Man to form GlcNAc2Man3GlcNAc2. Afterward, Xyl, Fuc, and two Gal are added to the acceptor substrate GlcNAc2Man3GlcNAc2, which are catalyzed by XylT (xylosyltransferase), FUT11/12 (fucosyltransferases), and GALT1 (galactosyltransferase), respectively. Finally, FUT13 (α-(1->,4)-fucosyltransferase) transfers a Fuc residue to the α-(1->4)-linked GlcNAc to complete the Lewis A-type structure, which is a trisaccharide structure (Figure 2) (Strasser, 2016; Strasser et al., 2021). The resulting products could be secreted to their destinations. Golgi-resident GnTI is the key enzyme in complex and hybrid N-glycan processing. The Arabidopsis complex glycan less 1 (cgl1) mutant, which is defective in GnTI activity, lacks complex and hybrid N-glycans and exhibits reduced N-glycosylation efficiency (von Schaewen et al., 1993; Strasser et al., 2005; Frank et al., 2008; Farid et al., 2013). However, the cgl1 mutant displays no apparent phenotype under normal growth conditions but confers salt hypersensitivity (Kang et al., 2008). In contrast to the Arabidopsis cgl1 mutant, the rice gnt1 mutant displays severe phenotypes showing arrest in postseedling development, defects in cell wall biosynthesis, and reduced cytokinin signaling (Fanata et al., 2013). The mechanisms that cause these markedly different phenotypes between Arabidopsis cgl1 and rice gnt1 remain to be illustrated in the future.
Interruption with N-glycan biosynthesis at any step by mutation of genes or treatments of pharmaceutical drugs, such as tunicamycin and DTT, will lead to incomplete N-glycans and affect N-glycosylated proteins (Pattison and Amtmann, 2009). Unfolded or misfolded proteins will accumulate in the ER and result in ER stress; eventually, the unfolded protein response (UPR) is activated to enhance the capacity for protein folding, increase the ER quality control, impair general protein translation, and maintain ER homeostasis (Bao and Howell, 2017; Yu et al., 2022). Defects in N-glycan processing may impair plant growth and stress responses or cause lethality (Lane et al., 2001; Koiwa et al., 2003; Lerouxel et al., 2005; Zhang et al., 2009; Fanata et al., 2013; Bao and Howell, 2017; Nagashima et al., 2018). Despite the significance of N-glycosylation, most studies in the past have focused on the core N-glycan formation in the ER lumen and the modification of complex N-glycans on glycoproteins in the Golgi apparatus. The effect of cytosolic oligosaccharide precursor production on plant growth and stress response is less addressed. As mentioned above, HBP generates UDP-GlcNAc to provide GlcNAc donors and initiate oligosaccharide precursor production on the cytosolic side of the ER. Defects in HBP enzymes may reduce UDP-GlcNAc levels, impair N-linked glycosylation, and alter plant growth under normal (Wang et al., 2015; Xiao et al., 2018) or abiotic stress conditions (Zhang et al., 2009; Nozaki et al., 2012; Chen et al., 2022). Moreover, a complete block of HBP normally leads to lethality (Chen et al., 2014; Vu et al., 2019; Jia et al., 2023).
It has been reported that N-glycan processing mutants alter the abiotic stress responses, such as salt stress. The staurosporine and temperature sensitive 3a (stt3a) mutant, in which a catalytic subunit of the OST complex in the ER is defective, and leaf wilting 3 (lew3), a mutant that lacks α-1,2-mannosyltransferase, induce UPR-mediated BiP gene expression and enhance salt stress sensitivity (Koiwa et al., 2003; Zhang et al., 2009; Jiao et al., 2020). However, Arabidopsis complex glycan 1 (cgl1), a mutant that lacks GnTI activity, shows a deprived complex and hybrid N-glycans and confers salt hypersensitivity (Frank et al., 2008; Kang et al., 2008). Unlike stt3a, which shows a UPR response, the cgl1 mutant fails to induce a UPR response. Thus, the UPR is likely not the major player that enhances salt hypersensitivity in the mutants with defective N-glycan processing. Furthermore, the mutation of UDP-GlcNAc transporter 1 (UGNT1) leads to deprived complex and hybrid N-glycan in the Golgi apparatus and does not increase salt hypersensitivity (Ebert et al., 2018). These data suggest that mature complex N-glycans are not the major factor leading to salt hypersensitivity. It was generally proposed that mutants defective in N-glycan processing in the ER lumen or Golgi apparatus might alter a different set of glycoprotein and/or glycolipid functions, which further integrate to alter plant growth and abiotic stress response. Compared to the Arabidopsis stt3a mutant showing short root elongation under salt stress, glcna.ut mutants, such as the RNAi knockdown mutants iU1s, display normal root elongation under salt stress (Chen et al., 2022). Although the UPR response is induced and N-linked glycosylation is impaired in iU1 mutants, these mutants exhibit salt hypersensitivity in terms of delayed seed germination and early seedling establishment, the phenotypes of which are different from those of stt3a mutant plants. The stt3a mutant, such as stt3a-2, also displays a higher stomatal density and transpiration rate in association with low endogenous ABA and auxin (IAA) levels. Thus, stt3a mutant plants are more sensitive to salt and drought stresses. These mutant phenotypes are correlated with the underglycosylation of β-glucosidase (AtBG1), catalyzing the conversion of conjugated ABA or IAA to its active form (Jiao et al., 2020). Consistently, exogenous application of ABA or IAA to stt3a-2 may partially rescue the mutant phenotypes. In contrast, the GlcNA.UT knockdown lines iU1s, reveal higher levels of ABA under salt stress conditions (Chen et al., 2022). Thus, although stt3a and iU1 affect the N-glycosylation of proteins, they could use different mechanisms in response to salt stress. It is conceivable that GlcNAc1pUTs produce UDP-GlcNAc not only for N-glycan synthesis in the ER lumen and maturation in the Golgi apparatus but also for the O-GlcNAcylation of primarily cytosolic and nuclear proteins (Figure 2). Thus, in addition to N-glycan processing, HBP has a wider range of effects on plant growth and abiotic stress response through diverse GlcNAc targets or conjugates.
O-GlcNAcylation is the addition of O-linked N-acetylglucosamine (O-GlcNAc) to the serine (Ser) and threonine (Thr) residues of nucleocytoplasmic and mitochondrial proteins (Hu et al., 2009; Ma et al., 2022), which was first reported by Torres and Hart (1984). In contrast to N-linked glycosylation, which involves the attachment of complex glycans to proteins for the secretary pathway, O-GlcNAcylation involves the direct addition of a single GlcNAc residue to the Ser/Thr residues of proteins, which primarily occurs in the cytosol or nucleus (Figure 2). O-GlcNAcylation is also among the most common co- or post-translational modifications and is conserved across organisms (Joshi et al., 2018; Ma et al., 2022). O-GlcNAcylated proteins are involved in most aspects of cellular functions including metabolism, transcriptional regulation, signaling, cell cycle regulation, protein trafficking, protein-protein interaction, and cell structure (Wells et al., 2001; Love and Hanover, 2005; Hart et al., 2011; Liu et al., 2022). In mammals, dysregulation of O-GlcNAcylation may be linked to chronic disorders, including the occurrence and progression of cancer (Slawson and Hart, 2011; Singh et al., 2015), diabetic complications (Peterson and Hart, 2016), neurodegeneration (Hart et al., 2011; Gong et al., 2012), and cardiovascular diseases (Wang et al., 2023), and the immune system (Golks and Guerini, 2008). Thus, manipulating O-GlcNAcylation may be a potential strategy for cancer therapy (Lu et al., 2022).
O-GlcNAcylated proteins are usually phosphorylated. As O-GlcNAcylation and phosphorylation are dynamic reactions that cycle rapidly, both post-translational modifications compete with the same Ser/Thr sites or modify nearby/or distant sites to show complex interplay and coordinate protein stability and function in response to external stimuli (Slawson and Hart, 2003; Wang et al., 2008c; Butkinaree et al., 2010; Zeidan and Hart, 2010; Hart et al., 2011; Martínez-Turiño et al., 2018; Xu et al., 2019). For instance, O-GlcNAcylation and phosphorylation coexist in the capsid protein (CP) of the plum pox virus (PPV). Although O-GlcNAcylation of PPV CP is not needed for virus viability, it increases viral infection (Pérez Jde et al., 2013; Martínez-Turiño et al., 2018). Moreover, vernalization increases the O-GlcNAc modification of nuclear TaGRP2 (a repressor in vernalization) and the phosphorylation of VER2 (an activator in vernalization); both modified proteins antagonistically regulate the expression of TaVRN1 to mediate flowering in winter wheat (Xiao et al., 2014; Xu et al., 2019).
The first public bioinformatics resource of O-GlcNAcylated proteins was established by Wang et al. (2011), in which approximately 1240 proteins are potentially O-GlcNAcylated. Later, over 1000 O-GlcNAcylated proteins were uncovered in different studies of mammalian cells (Trinidad et al., 2012; Hahne et al., 2013). Recently, with more improved techniques, over 5000 O-GlcNAcylated proteins were identified using human models (Wulff-Fuentes et al., 2021). In Arabidopsis, Xu et al. (2017) identified 262 proteins with O-GlcNAcylation. Among them, the O-GlcNAcylated and O-fucosylated protein AtACINUS is involved in ABA sensitivity through alternative splicing of HIGH LEVEL OF BETA-AMYLASE ACTIVITY 1 (HBA1) and ABA HYPERSENSITIVE 1 (ABH1), negative regulators of ABA signaling, and in flowering through transcriptional regulation of the floral repressor FLOWERING LOCUS C (FLC) (Bi et al., 2021). In addition, a total of 168 O-GlcNAcylated proteins were found in winter wheat (Xu et al., 2019); these proteins perform functions in metabolism, response to stimuli, cellular processing, signal transduction, and transcriptional regulation. Thus, the total number of identified proteins of O-GlcNAcylation is far lower in plants than in mammalian cells.
O-GlcNAcylation is catalyzed by O-GlcNAc transferase (OGT) (Figure 2). Phylogenetic analysis revealed that metazoans contain a single OGT, whereas vascular plants and moss have two homologs of OGTs (Olszewski et al., 2010). Considering that GlcNAc is needed for O-GlcNAcylation and UDP-GlcNAc, the donor of GlcNAc, is synthesized through HBP, HBP might perform crosstalk with O-GlcNAcylation to optimize nutrient status and O-GlcNAcylation cycling. In Drosophila, protein O-GlcNAcylation displays a circadian rhythm mediated by the HBP enzyme GFAT and the O-GlcNAcylation enzymes, OGT and O-GlcNAcase (OGA), an enzyme removing GlcNAc from O-GlcNAcylated proteins (Liu et al., 2021). The Arabidopsis knockdown mutant Atagm reduces UDP-GlcNAc production and shows a temperature-dependent growth defect that is associated with the impairment of protein O-GlcNAcylation (Jia et al., 2023). In Arabidopsis, two OGT homologs, SECRET AGENT (SEC) and SPINDLY (SPY), catalyze O-GlcNAcylation and O-linked fucosylation, respectively (Hartweck et al., 2002; Zentella et al., 2016, Zentella et al., 2017). The Arabidopsis null sec mutant only displays a subtle phenotype (Hartweck et al., 2002), but the spy mutant shows an apparent GA response, indicating that SPY acts as a negative regulator of GA signaling (Wilson and Somerville, 1995; Jacobsen et al., 1996). Moreover, the sec/spy double mutant is lethal, with defects in gamete and seed development that are similar to the knockout OGT mutants in mice and Drosophila, in which embryonic lethality occurs (Shafi et al., 2000; Gambetta et al., 2009). These data indicate that although SEC and SPY have overlapping functions involved in GA signaling, they also have distinct roles and may play a synergistic function in plant growth and development (Hartweck et al., 2002, Hartweck et al., 2006; Zentella et al., 2016, Zentella et al., 2017). Later, it was reported that the Arabidopsis sec mutant displays an early-flowering phenotype, which is associated with the inhibition of O-GlcNAcylation of ARABIDOPSIS HOMOLOG OF TRITHORAX1 (ATX1), a histone lysine methyltransferase (HKMT). The impaired activity of ATX1 reduces histone H3 lysine 4 trimethylation (H3K4me3) of the FLC gene, a negative regulator of flowering (Xing et al., 2018). The Arabidopsis DELLA protein RGA (REPRESSOR OF ga1-3), a master negative regulator of the GA response, is O-GlcNAcylated by SEC; this suppresses the interactions of RGA with other key transcription factors, such as PIFs, BZR1, and JAZ1, which are involved in light, brassinosteroid, and jasmonate signalings, respectively (Zentella et al., 2016). In addition to DELLA proteins, several important transcription factors involved in plant hormone signaling are O-GlcNAcylated, such as ARFs, TCPs, EIN2, and ABF3, which are involved in the signaling of auxin, cytokinin, ethylene, and ABA, respectively (Xu et al., 2017). Compared to mammals, numerous proteins of O-GlcNAcylation in plants remain to be uncovered, and further characterization of these modified proteins will shed light on the significance of O-GlcNAcylation biology.
UDP-GlcNAc biosynthesis through HBP is essential for the glycosylation of proteins and lipids (Ebert et al., 2018). Thus, the endogenous levels of UDP-GlcNAc levels intimately affect the glycosylation of proteins and lipids. For example, partial loss-of-function mutations in HBP-related genes normally reduce UDP-GlcNAc levels and impair N-glycosylation and/or O-GlcNAcylation of proteins (Jiang et al., 2005; Nozaki et al., 2012; Vu et al., 2019; Chen et al., 2022; Jia et al., 2023). Interestingly, these knockdown mutants largely display no apparent phenotype under normal growth conditions; however, these mutants exhibit stress-induced growth defects. This indicates that a small amount of UDP-GlcNAc is sufficient to maintain normal plant growth but more UDP-GlcNAc levels and protein glycosylation are needed for plants to adapt to deleterious environments. Most N-glycoproteins are membrane-associated and secreted proteins. Thus, changes in N-glycosylation through adverse environments or mutations of genes involved in HBP and N-glycan processing may alter glycoprotein functions in cell wall biosynthesis and integrity and membrane-associated proteins, resulting in altered sensitivities to biotic, such as bacterial blight tolerance (Wang et al., 2015), abiotic stresses, such as drought, salt, cold, and high temperature (Jiang et al., 2005; Xiao et al., 2018; Nozaki et al., 2012; Vu et al., 2019; Chen et al., 2022; Jia et al., 2023), or phytohormones, such as ABA, auxin, and JA (Zhang et al., 2009; Fanata et al., 2013; Wang et al., 2015; Jiao et al., 2020; Chen et al., 2022). Defects in N-glycoproteins might also cause the accumulation of unfolded or misfolded proteins in ER, leading to ER stress and further induction of UPR to enhance protein folding capacity and diminish ER stress (Bao and Howell, 2017; Yu et al., 2022). Therefore, ER stress or UPR can be observed in the mutations of HBP-related genes, such as GFAT, GNA, and GlcNA.UTs/or UAP/SPL29 (Xiao et al., 2018; Nozaki et al., 2012; Vu et al., 2019; Chen et al., 2022) and N-glycan processing mutants, stt3a and lew3 (Koiwa et al., 2003; Zhang et al., 2009). As UDP-GlcNAc is also essential for O-GlcNAcylation, defects in UDP-GlcNAc biosynthesis through HBP, such as AGM, or mutation of O-GlcNAcylation-related genes, such as SEC, might affect the functions of O-GlcNAcylated proteins, such as ATX1 and DELLA proteins, which further change temperature-dependent growth defects and cellular signalings, such as phytohormones ABA, GA, auxin, CK, and JA (Zentella et al., 2016; Jia et al., 2023), vernalization (Xiao et al., 2014; Xu et al., 2019), and viral infection (Pérez Jde et al., 2013; Martínez-Turiño et al., 2018). Hexosamine biosynthesis and related pathways in response to stresses are summarized in Figure 3.
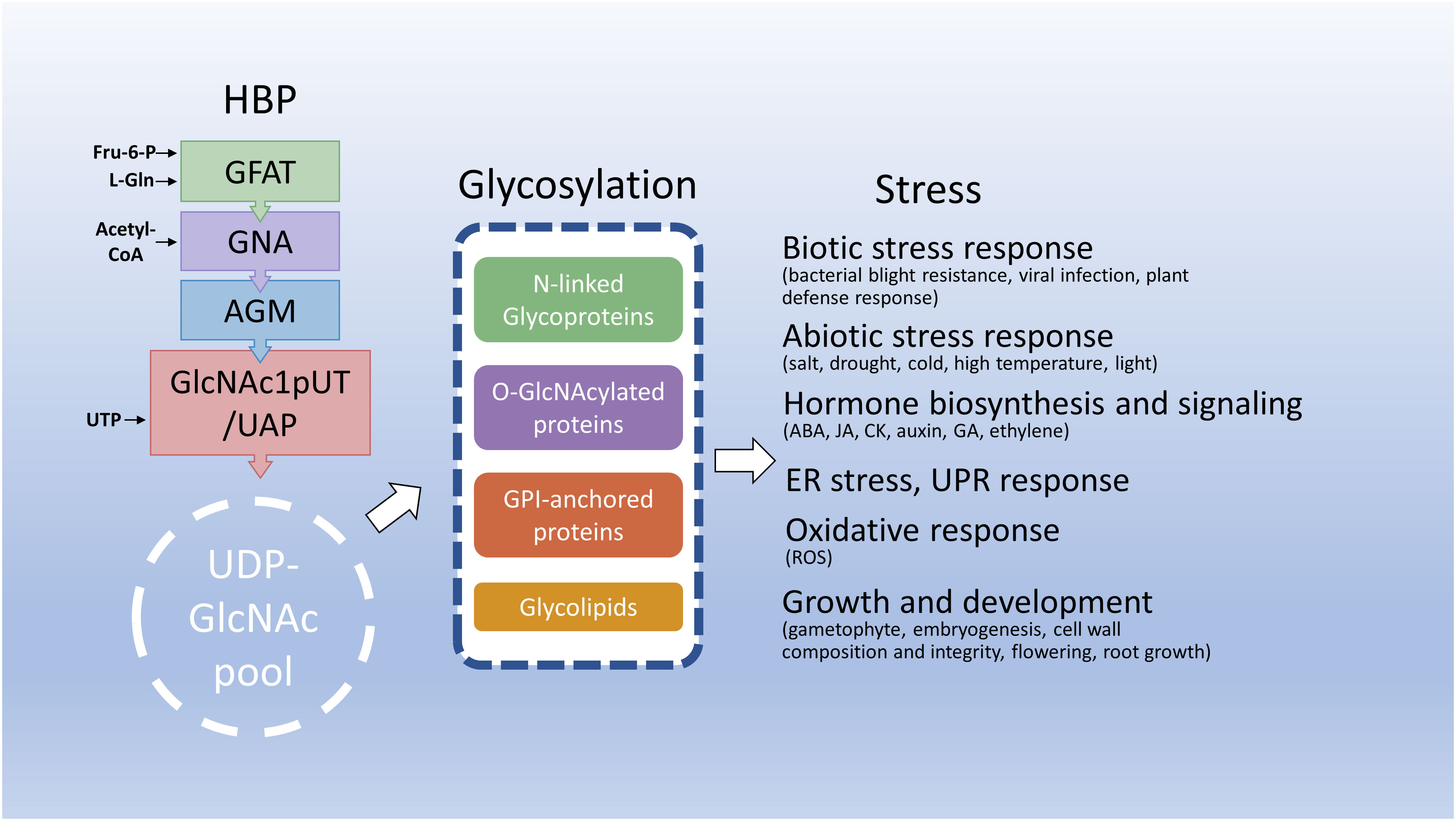
Figure 3 Hexosamine biosynthesis and related pathways in response to stresses. This diagram depicts that the HBP integrates several key metabolites to synthesize UDP-GlcNAc, an essential amino sugar moiety of glycosylation of proteins and lipids. Under stress conditions, HBP integrates endogenous metabolites and energy status to maintain UDP-GlcNAc homeostasis and reprogram metabolic pathways including glycosylation to benefit plant adaptation to deleterious environments. Fru-6-P, fructose-6-phosphate; L-Gln, L-glutamine; CoA, coenzyme A; UTP, uridine triphosphate; ABA, abscisic acid; JA, jasmonic acid; CK, cytokinin; GA, gibberellic acid.
Although the HBP is considered a minor side pathway of glycolysis, it integrates the endogenous nutrient status of plants and rewires the metabolic programs to improve plant development and adaption to environmental challenges. Thus, HBP serves as a metabolic integrator or sensor to fine-tune the nutrient balance and maintain UDP-GlcNAc homeostasis. Dysfunction of HBP often causes severe phenotypes or even lethality. Research progress on HBP in plants has been much slower than that in microbes and mammals. To date, despite HBP’s studies having made a great step in plants, several aspects remain to be further explored in the future.
1. HBP-related enzymatic proteins, such as GlcNAc1pUT2, UAP1/SPL29, and AtAGM, often have multiple substrates and products (or intermediates). The functions of these products’ targets or conjugates remain to be illustrated.
2. In addition to the cytosol and ER surface of the cytosolic side, enzymes, such as AGM and GlcNAc1pUT1, have several subcellular localizations, such as nuclei and organelles. It remains to be determined whether these proteins perform additional functions in addition to their involvement in the HBP.
3. How nutrient availability and environmental conditions control the HBP flux needs to be further examined in plants.
4. The total proteins of N-glycosylation and O-GlcNAcylation were underestimated in plants compared to mammals. Thus, high-throughput analysis of more GlcNAc-conjugated proteins needs to be performed, and the functions of these modified proteins remain to be characterized.
5. In addition to phosphorylation, O-GlcNAcylation sites of proteins can also compete with other post-translational modifications. The biological functions of these modified proteins also need to be unraveled in the future.
A better understanding of the functions of HBP, GlcNAc conjugates, and the mechanisms by which HBP responds to abiotic stress will reveal possible strategies to modify HBP in the biofortification of agriculture in the future.
Y-HC: Data curation, Writing – original draft. W-HC: Conceptualization, Funding acquisition, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Ministry of Science and Technology (MOST), Taipei, Taiwan (MOST105-2311-B-001-073; MOST110-2311-B-001-030-MY3).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aebi, M. (2013). N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 1833, 2430–2437. doi: 10.1016/j.bbamcr.2013.04.001.
Allen, M. B., Walker, D. G. (1980). Kinetic characterization of N-acetyl-D-glucosamine kinase from rat liver and kidney. Biochem. J. 185, 577–582. doi: 10.1042/bj1850577.
Arakane, Y., Baguinon, M. C., Jasrapuria, S., Chaudhari, S., Doyungan, A., Kramer, K. J., et al. (2011). Both UDP N-acetylglucosamine pyrophosphorylases of Tribolium castaneum are critical for molting, survival and fecundity. Insect Biochem. Mol. Biol. 41, 42–50. doi: 10.1016/j.ibmb.2010.09.011.
Banerjee, S., Vishwanath, P., Cui, J., Kelleher, D. J., Gilmore, R., Robbins, P. W., et al. (2007). The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc. Natl. Acad. Sci. U.S.A. 104, 11676–11681. doi: 10.1073/pnas.0704862104.
Bao, Y., Howell, S. H. (2017). The unfolded protein response supports plant development and defense as well as responses to abiotic stress. Front. Plant Sci. 8, 344. doi: 10.3389/fpls.2017.00344.
Bencúr, P., Steinkellner, H., Svoboda, B., Mucha, J., Strasser, R., Kolarich, D., et al. (2005). Arabidopsis thaliana beta1,2-xylosyltransferase: An unusual glycosyltransferase with the potential to act at multiple stages of the plant N-glycosylation pathway. Biochem. J. 388, 515–525. doi: 10.1042/BJ20042091
Ben-Khemis, L., Mekki, N., Ben-Mustapha, I., Rouault, K., Mellouli, F., Khemiri, M., et al. (2017). A founder mutation underlies a severe form of phosphoglutamase 3 (PGM3) deficiency in Tunisian patients. Mol. Immunol. 90, 57–63. doi: 10.1016/j.molimm.2017.06.248.
Berger, M., Chen, H., Reutter, W., Hinderlich, S. (2002). Structure and function of N-acetylglucosamine kinase. Identification of two active site cysteines. Eur. J. Biochem. 269, 4212–4218. doi: 10.1046/j.1432-1033.2002.03117.x.
Bi, Y., Deng, Z., Ni, W., Shrestha, R., Savage, D., Hartwig, T., et al. (2021). Arabidopsis ACINUS is O-glycosylated and regulates transcription and alternative splicing of regulators of reproductive transitions. Nat. Commun. 12, 945. doi: 10.1038/s41467-021-20929-7.
Boehmelt, G., Wakeham, A., Elia, A., Sasaki, T., Plyte, S., Potter, J., et al. (2000). Decreased UDP-GlcNAc levels abrogate proliferation control in EMeg32-deficient cells. EMBO J. 19, 5092–5104. doi: 10.1093/emboj/19.19.5092.
Bonhomme, S., Horlow, C., Vezon, D., De Laissardière, S., Guyon, A., Férault, M., et al. (1998). T-DNA mediated disruption of essential gametophytic genes in Arabidopsis is unexpectedly rare and cannot be inferred from segregation distortion alone. Mol. Gen. Genet. 260, 444–452. doi: 10.1007/s004380050915.
Burda, P., Aebi, M. (1999). The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426, 239–257. doi: 10.1016/S0304-4165(98)00127-5.
Buse, M. G. (2006). Hexosamines, insulin resistance, and the complications of diabetes: current status. Am. J. Physiol. Endocrinol. Metab. 290, E1–e8. doi: 10.1152/ajpendo.00329.2005.
Butkinaree, C., Park, K., Hart, G. W. (2010). O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim. Biophys. Acta 1800, 96–106. doi: 10.1016/j.bbagen.2009.07.018.
Campbell, S., Mesaros, C., Izzo, L., Affronti, H., Noji, M., Schaffer, B. E., et al. (2021). Glutamine deprivation triggers NAGK-dependent hexosamine salvage. Elife 10, e62644. doi: 10.7554/eLife.62644.sa2.
Chen, Y. H., Shen, H. L., Chou, S. J., Sato, Y., Cheng, W. H. (2022). Interference of Arabidopsis N-acetylglucosamine-1-P uridylyltransferase expression impairs protein N-glycosylation and induces ABA-mediated salt sensitivity during seed germination and early seedling development. Front. Plant Sci. 13, 903272. doi: 10.3389/fpls.2022.903272.
Chen, Y. H., Shen, H. L., Hsu, P. J., Hwang, S. G., Cheng, W. H. (2014). N-acetylglucosamine-1-P uridylyltransferase 1 and 2 are required for gametogenesis and embryo development in Arabidopsis thaliana. Plant Cell Physiol. 55, 1977–1993. doi: 10.1093/pcp/pcu127.
Chiaradonna, F., Ricciardiello, F., Palorini, R. (2018). The nutrient-sensing hexosamine biosynthetic pathway as the hub of cancer metabolic rewiring. Cells 7, 53. doi: 10.3390/cells7060053.
Chu, S. H., Noh, H., Kim, K. H., Hong, S., Lee, H. (2010). Enhanced drought tolerance in Arabidopsis via genetic manipulation aimed at the reduction of glucosamine-induced ROS generation. Plant Mol. Biol. 74, 493–502. doi: 10.1007/s11103-010-9691-7.
Dickinson, M. E., Flenniken, A. M., Ji, X., Teboul, L., Wong, M. D., White, J. K., et al. (2016). High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514. doi: 10.1038/nature19356.
Dörmann, P., Benning, C. (1996). Functional expression of uridine 5'-diphospho-glucose 4-epimerase (EC 5.1.3.2) from Arabidopsis thaliana in Saccharomyces cerevisiae and Escherichia coli. Arch. Biochem. Biophys. 327, 27–34. doi: 10.1006/abbi.1996.0088
Durand, P., Golinelli-Pimpaneau, B., Mouilleron, S., Badet, B., Badet-Denisot, M. A. (2008). Highlights of glucosamine-6P synthase catalysis. Arch. Biochem. Biophys. 474, 302–317. doi: 10.1016/j.abb.2008.01.026.
Ebert, B., Rautengarten, C., Mcfarlane, H. E., Rupasinghe, T., Zeng, W., Ford, K., et al. (2018). A Golgi UDP-GlcNAc transporter delivers substrates for N-linked glycans and sphingolipids. Nat. Plants 4, 792–801. doi: 10.1038/s41477-018-0235-5.
Fanata, W. I., Lee, K. H., Son, B. H., Yoo, J. Y., Harmoko, R., Ko, K. S., et al. (2013). N-glycan maturation is crucial for cytokinin-mediated development and cellulose synthesis in Oryza sativa. Plant J. 73, 966–979. doi: 10.1111/tpj.12087.
Farid, A., Malinovsky, F. G., Veit, C., Schoberer, J., Zipfel, C., Strasser, R. (2013). Specialized roles of the conserved subunit OST3/6 of the oligosaccharyltransferase complex in innate immunity and tolerance to abiotic stresses. Plant Physiol. 162, 24–38. doi: 10.1104/pp.113.215509.
Frank, J., Kaulfürst-Soboll, H., Rips, S., Koiwa, H., Von Schaewen, A. (2008). Comparative analyses of Arabidopsis complex glycan1 mutants and genetic interaction with staurosporin and temperature sensitive3a. Plant Physiol. 148, 1354–1367. doi: 10.1104/pp.108.127027.
Furo, K., Nozaki, M., Murashige, H., Sato, Y. (2015). Identification of an N-acetylglucosamine kinase essential for UDP-N-acetylglucosamine salvage synthesis in Arabidopsis. FEBS Lett. 589, 3258–3262. doi: 10.1016/j.febslet.2015.09.011.
Gambetta, M. C., Oktaba, K., Müller, J. (2009). Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325, 93–96. doi: 10.1126/science.1169727.
Gindzieński, A., Glowacka, D., Zwierz, K. (1974). Purification and properties of N-acetylglucosamine kinase from human gastric mucosa. Eur. J. Biochem. 43, 155–160. doi: 10.1111/j.1432-1033.1974.tb03395.x
Golks, A., Guerini, D. (2008). The O-linked N-acetylglucosamine modification in cellular signalling and the immune system. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 9, 748–753. doi: 10.1038/embor.2008.129.
Gong, C. X., Liu, F., Iqbal, K. (2012). O-GlcNAc cycling modulates neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 109, 17319–17320. doi: 10.1073/pnas.1215395109.
Gonzalez, N. S., Pontis, H. G. (1963). Uridine diphosphate fructose and uridine diphosphate acetylgalactosamine from dahlia tubers. Biochim. Biophys. Acta 69, 179–181. doi: 10.1016/0006-3002(63)91243-5.
Greig, K. T., Antonchuk, J., Metcalf, D., Morgan, P. O., Krebs, D. L., Zhang, J. G., et al. (2007). Agm1/Pgm3-mediated sugar nucleotide synthesis is essential for hematopoiesis and development. Mol. Cell Biol. 27, 5849–5859. doi: 10.1128/MCB.00802-07.
Hahne, H., Sobotzki, N., Nyberg, T., Helm, D., Borodkin, V. S., Van Aalten, D. M., et al. (2013). Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry. J. Proteome Res. 12, 927–936. doi: 10.1021/pr300967y.
Hancock, C. N., Kent, L., Mcclure, B. A. (2005). The stylar 120 kDa glycoprotein is required for S-specific pollen rejection in Nicotiana. Plant J. 43, 716–723. doi: 10.1111/j.1365-313X.2005.02490.x
Hart, G. W., Slawson, C., Ramirez-Correa, G., Lagerlof, O. (2011). Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858. doi: 10.1146/annurev-biochem-060608-102511.
Hartweck, L. M., Genger, R. K., Grey, W. M., Olszewski, N. E. (2006). SECRET AGENT and SPINDLY have overlapping roles in the development of Arabidopsis thaliana L. Heyn. J. Exp. Bot. 57, 865–875. doi: 10.1093/jxb/erj071.
Hartweck, L. M., Scott, C. L., Olszewski, N. E. (2002). Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161, 1279–1291. doi: 10.1093/genetics/161.3.1279.
Hassid, W. Z., Neufeld, E. F., Feingold, D. S. (1959). Sugar nucleotides in the interconversion of carbohydrates in higher plants. Proc. Natl. Acad. Sci. U.S.A. 45, 905–915. doi: 10.1073/pnas.45.7.905.
Helenius, A., Aebi, M. (2001). Intracellular functions of N-linked glycans. Science 291, 2364–2369. doi: 10.1126/science.291.5512.2364.
Hinderlich, S., Berger, M., Schwarzkopf, M., Effertz, K., Reutter, W. (2000). Molecular cloning and characterization of murine and human N-acetylglucosamine kinase. Eur. J. Biochem. 267, 3301–3308. doi: 10.1046/j.1432-1327.2000.01360.x.
Hofmann, M., Boles, E., Zimmermann, F. K. (1994). Characterization of the essential yeast gene encoding N-acetylglucosamine-phosphate mutase. Eur. J. Biochem. 221, 741–747. doi: 10.1111/j.1432-1033.1994.tb18787.x.
Hu, Y., Suarez, J., Fricovsky, E., Wang, H., Scott, B. T., Trauger, S. A., et al. (2009). Increased enzymatic O-GlcNAcylation of mitochondrial proteins impairs mitochondrial function in cardiac myocytes exposed to high glucose. J. Biol. Chem. 284, 547–555. doi: 10.1074/jbc.M808518200.
Jacobsen, S. E., Binkowski, K. A., Olszewski, N. E. (1996). SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 93, 9292–9296. doi: 10.1073/pnas.93.17.9292.
Jain, M., Nijhawan, A., Arora, R., Agarwal, P., Ray, S., Sharma, P., et al. (2007). F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143, 1467–1483. doi: 10.1104/pp.106.091900.
Jia, X., Zhang, H., Qin, H., Li, K., Liu, X., Wang, W., et al. (2023). Protein O-GlcNAcylation impairment caused by N-acetylglucosamine phosphate mutase deficiency leads to growth variations in Arabidopsis thaliana. Plant J. 114, 613–635. doi: 10.1111/tpj.16156.
Jiang, H., Wang, S., Dang, L., Chen, H., Wu, Y., Jiang, X., et al. (2005). A novel short-root gene encodes a glucosamine-6-phosphate acetyltransferase required for maintaining normal root cell shape in rice. Plant Physiol. 138, 232–242. doi: 10.1104/pp.104.058248.
Jiao, Q., Chen, T., Niu, G., Zhang, H., Zhou, C., Hong, Z. (2020). N-glycosylation is involved in stomatal development by modulating the release of active abscisic acid and auxin in Arabidopsis. J. Exp. Bot. 71, 5865–5879. doi: 10.1093/jxb/eraa321.
Joersbo, M., Pedersen, S. G., Nielsen, J. E., Marcussen, J., Brunstedt, J. (1999). Isolation and expression of two cDNA clones encoding UDP-galactose epimerase expressed in developing seeds of the endospermous legume guar. Plant Sci. 142, 147–154. doi: 10.1016/S0168-9452(99)00012-6.
Jose-Estanyol, M., Puigdomenech, P. (2000). Plant cell wall glycoproteins and their genes. Plant Physiol. Biochem. 38, 97–108. doi: 10.1016/S0981-9428(00)00165-0.
Joshi, H. J., Narimatsu, Y., Schjoldager, K. T., Tytgat, H. L. P., Aebi, M., Clausen, H., et al. (2018). Snapshot: O-glycosylation pathways across kingdoms. Cell 172, 632–632.e632. doi: 10.1016/j.cell.2018.01.016.
Kang, J., Frank, J., Kang, C., Kajiura, H., Vikram, M., Ueda, A., et al. (2008). Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. U.S.A. 105, 5933–5938. doi: 10.1073/pnas.0800237105.
Kato, N., Dasgupta, R., Smartt, C. T., Christensen, B. M. (2002). Glucosamine:fructose-6-phosphate aminotransferase: gene characterization, chitin biosynthesis and peritrophic matrix formation in Aedes aEgypti. Insect Mol. Biol. 11, 207–216. doi: 10.1046/j.1365-2583.2002.00326.x.
Koiwa, H., Li, F., Mccully, M. G., Mendoza, I., Koizumi, N., Manabe, Y., et al. (2003). The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15, 2273–2284. doi: 10.1105/tpc.013862.
Kornfeld, R. (1967). Studies on L-glutamine D-fructose 6-phosphate amidotransferase. I. Feedback inhibition by uridine diphosphate-N-acetylglucosamine. J. Biol. Chem. 242, 3135–3141. doi: 10.1016/S0021-9258(18)95943-0.
Krug, E., Zweibaum, A., Schulz-Holstege, C., Keppler, D. (1984). D-glucosamine-induced changes in nucleotide metabolism and growth of colon-carcinoma cells in culture. Biochem. J. 217, 701–708. doi: 10.1042/bj2170701.
Lake, M. R., Slocum, R. D. (1998). Molecular cloning and characterization of a UDP-glucose 4-epimerase gene (gaZE) and its expression in pea tissues. Plant Physiol. Biochem. 36, 555–562. doi: 10.1016/S0981-9428(98)80002-8.
Lalanne, E., Honys, D., Johnson, A., Borner, G. H., Lilley, K. S., Dupree, P., et al. (2004). SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16, 229–240. doi: 10.1105/tpc.014407.
Lane, D. R., Wiedemeier, A., Peng, L., Höfte, H., Vernhettes, S., Desprez, T., et al. (2001). Temperature-sensitive alleles of RSW2 link the KORRIGAN endo-1,4-beta-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 126, 278–288. doi: 10.1104/pp.126.1.278.
Lannoo, N., Van Damme, E. J. (2015). Review/N-glycans: The making of a varied toolbox. Plant Sci. 239, 67–83. doi: 10.1016/j.plantsci.2015.06.023.
Lerouxel, O., Mouille, G., Andème-Onzighi, C., Bruyant, M. P., Séveno, M., Loutelier-Bourhis, C., et al. (2005). Mutants in DEFECTIVE GLYCOSYLATION, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. Plant J. 42, 455–468. doi: 10.1111/j.1365-313X.2005.02392.x.
Liebminger, E., Hüttner, S., Vavra, U., Fischl, R., Schoberer, J., Grass, J., et al. (2009). Class I α-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21, 3850–3867. doi: 10.1105/tpc.109.072363.
Liu, X., Blaženović, I., Contreras, A. J., Pham, T. M., Tabuloc, C. A., Li, Y. H., et al. (2021). Hexosamine biosynthetic pathway and O-GlcNAc-processing enzymes regulate daily rhythms in protein O-GlcNAcylation. Nat. Commun. 12, 4173. doi: 10.1038/s41467-021-24301-7.
Liu, Y., Hu, Y. J., Fan, W. X., Quan, X., Xu, B., Li, S. Z. (2022). O-GlcNAcylation: The underestimated emerging regulators of skeletal muscle physiology. Cells 11, 1789. doi: 10.3390/cells11111789.
Lockhart, D. E. A., Stanley, M., Raimi, O. G., Robinson, D. A., Boldovjakova, D., Squair, D. R., et al. (2020). Targeting a critical step in fungal hexosamine biosynthesis. J. Biol. Chem. 295, 8678–8691. doi: 10.1074/jbc.RA120.012985.
Love, D. C., Hanover, J. A. (2005). The hexosamine signaling pathway: deciphering the "O-GlcNAc code". Sci. STKE 2005, re13. doi: 10.1126/stke.3122005re13
Lu, Q., Zhang, X., Liang, T., Bai, X. (2022). O-GlcNAcylation: an important post-translational modification and a potential therapeutic target for cancer therapy. Mol. Med. 28, 115. doi: 10.1186/s10020-022-00544-y.
Ma, J., Hou, C., Wu, C. (2022). Demystifying the O-GlcNAc code: A systems view. Chem. Rev. 122, 15822–15864. doi: 10.1021/acs.chemrev.1c01006.
Maia, J. C. (1994). Hexosamine and cell wall biogenesis in the aquatic fungus Blastocladiella emersonii. FASEB J. 8, 848–853. doi: 10.1096/fasebj.8.11.8070634.
Mansour, M. M. F., Hassan, F. A. S. (2022). How salt stress-responsive proteins regulate plant adaptation to saline conditions. Plant Mol. Biol. 108, 175–224. doi: 10.1007/s11103-021-01232-x.
Martínez-Turiño, S., Pérez, J. J., Hervás, M., Navajas, R., Ciordia, S., Udeshi, N. D., et al. (2018). Phosphorylation coexists with O-GlcNAcylation in a plant virus protein and influences viral infection. Mol. Plant Pathol. 19, 1427–1443. doi: 10.1111/mpp.12626
Mengin-Lecreulx, D., Van Heijenoort, J. (1993). Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J. Bacteriol 175, 6150–6157. doi: 10.1128/jb.175.19.6150-6157.1993.
Milewski, S., Gabriel, I., Olchowy, J. (2006). Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast 23, 1–14. doi: 10.1002/yea.1337.
Milewski, S., Kuszczak, D., Jedrzejczak, R., Smith, R. J., Brown, A. J., Gooday, G. W. (1999). Oligomeric structure and regulation of Candida albicans glucosamine-6-phosphate synthase. J. Biol. Chem. 274, 4000–4008. doi: 10.1074/jbc.274.7.4000.
Mio, T., Yabe, T., Arisawa, M., Yamada-Okabe, H. (1998). The eukaryotic UDP-N-acetylglucosamine pyrophosphorylases. Gene cloning, protein expression, and catalytic mechanism. J. Biol. Chem. 273, 14392–14397. doi: 10.1074/jbc.273.23.14392.
Mio, T., Yamada-Okabe, T., Arisawa, M., Yamada-Okabe, H. (1999). Saccharomyces cerevisiae GNA1, an essential gene encoding a novel acetyltransferase involved in UDP-N-acetylglucosamine synthesis. J. Biol. Chem. 274, 424–429. doi: 10.1074/jbc.274.1.424.
Mio, T., Yamada-Okabe, T., Arisawa, M., Yamada-Okabe, H. (2000). Functional cloning and mutational analysis of the human cDNA for phosphoacetylglucosamine mutase: Identification of the amino acid residues essential for the catalysis. Biochim. Biophys. Acta 1492, 369–376. doi: 10.1016/S0167-4781(00)00120-2.
Miura, N., Kaneko, S., Hosoya, S., Furuchi, T., Miura, K., Kuge, S., et al. (1999). Overexpression of L-glutamine:D-fructose-6-phosphate amidotransferase provides resistance to methylmercury in Saccharomyces cerevisiae. FEBS Lett. 458, 215–218. doi: 10.1016/S0014-5793(99)01158-8.
Molinari, M. (2007). N-glycan structure dictates extension of protein folding or onset of disposal. Nat. Chem. Biol. 3, 313–320. doi: 10.1038/nchembio880.
Nadal, M., Sawers, R., Naseem, S., Bassin, B., Kulicke, C., Sharman, A., et al. (2017). An N-acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize. Nat. Plants 3, 17073. doi: 10.1038/nplants.2017.73.
Naganuma, A., Miura, N., Kaneko, S., Mishina, T., Hosoya, S., Miyairi, S., et al. (2000). GFAT as a target molecule of methylmercury toxicity in Saccharomyces cerevisiae. FASEB J. 14, 968–972. doi: 10.1096/fasebj.14.7.968.
Nagashima, Y., Von Schaewen, A., Koiwa, H. (2018). Function of N-glycosylation in plants. Plant Sci. 274, 70–79. doi: 10.1016/j.plantsci.2018.05.007.
Niemann, M. C. E., Bartrina, I., Ashikov, A., Weber, H., Novák, O., SpÍchal, L., et al. (2015). Arabidopsis ROCK1 transports UDPGlcNAc/UDPGalNAc and regulates ER protein quality control and cytokinin activity. Proc. Natl. Acad. Sci. U.S.A. 112, 291–296. doi: 10.1073/pnas.1419050112.
Nozaki, M., Sugiyama, M., Duan, J., Uematsu, H., Genda, T., Sato, Y. (2012). A missense mutation in the glucosamine-6-phosphate N-acetyltransferase-encoding gene causes temperature-dependent growth defects and ectopic lignin deposition in Arabidopsis. Plant Cell 24, 3366–3379. doi: 10.1105/tpc.112.102806.
Olchowy, J., Gabriel, I., Milewski, S. (2007). Functional domains and interdomain communication in Candida albicans glucosamine-6-phosphate synthase. Biochem. J. 404, 121–130. doi: 10.1042/BJ20061502.
Olszewski, N. E., West, C. M., Sassi, S. O., Hartweck, L. M. (2010). O-GlcNAc protein modification in plants: Evolution and function. Biochim. Biophys. Acta 1800, 49–56. doi: 10.1016/j.bbagen.2009.11.016.
Paneque, A., Fortus, H., Zheng, J., Werlen, G., Jacinto, E. (2023). The hexosamine biosynthesis pathway: Regulation and function. Genes (Basel) 14, 933. doi: 10.3390/genes14040933.
Pang, Q., Hays, J. B., Rajagopal, I., Schaefer, T. S. (1993). Selection of Arabidopsis cDNAs that partially correct phenotypes of Escherichia coli DNA-damage-sensitive mutants and analysis of two plant cDNAs that appear to express UV-specific dark repair activities. Plant Mol. Biol. 22, 411–426. doi: 10.1007/BF00015972.
Pantaleon, M. (2015). The role of hexosamine biosynthesis and signaling in early development. Adv. Exp. Med. Biol. 843, 53–76. doi: 10.1007/978-1-4939-2480-6_3
Pattison, R. J., Amtmann, A. (2009). N-glycan production in the endoplasmic reticulum of plants. Trends Plant Sci. 14, 92–99. doi: 10.1016/j.tplants.2008.11.008.
Pearce, G., Siems, W. F., Bhattacharya, R., Ryan, C. A. (2007). Three hydroxyproline-rich glycopeptides derived from a single petunia polyprotein precursor activate defensin I, a pathogen defense response gene. J. Biol. Chem. 282, 17777–17784. doi: 10.1074/jbc.M701543200.
Peneff, C., Ferrari, P., Charrier, V., Taburet, Y., Monnier, C., Zamboni, V., et al. (2001). Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc(Gal)NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. EMBO J. 20, 6191–6202. doi: 10.1093/emboj/20.22.6191.
Pérez Jde, J., Udeshi, N. D., Shabanowitz, J., Ciordia, S., Juárez, S., Scott, C. L., et al. (2013). O-GlcNAc modification of the coat protein of the potyvirus plum pox virus enhances viral infection. Virology 442, 122–131. doi: 10.1016/j.virol.2013.03.029.
Peterson, S. B., Hart, G. W. (2016). New insights: A role for O-GlcNAcylation in diabetic complications. Crit. Rev. Biochem. Mol. Biol. 51, 150–161. doi: 10.3109/10409238.2015.1135102.
Riegler, H., Herter, T., Grishkovskaya, I., Lude, A., Ryngajllo, M., Bolger, M. E., et al. (2012). Crystal structure and functional characterization of a glucosamine-6-phosphate N-acetyltransferase from Arabidopsis thaliana. Biochem. J. 443, 427–437. doi: 10.1042/BJ20112071.
Ryczko, M. C., Pawling, J., Chen, R., Abdel Rahman, A. M., Yau, K., Copeland, J. K., et al. (2016). Metabolic reprogramming by hexosamine biosynthetic and Golgi N-glycan branching pathways. Sci. Rep. 6, 23043. doi: 10.1038/srep23043.
Schimmelpfeng, K., Strunk, M., Stork, T., Klämbt, C. (2006). Mummy encodes an UDP-N-acetylglucosamine-dipohosphorylase and is required during Drosophila dorsal closure and nervous system development. Mech. Dev. 123, 487–499. doi: 10.1016/j.mod.2006.03.004.
Shafi, R., Iyer, S. P., Ellies, L. G., O'donnell, N., Marek, K. W., Chui, D., et al. (2000). The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739. doi: 10.1073/pnas.100471497.
Shin, Y. J., Vavra, U., Veit, C., Strasser, R. (2018). The glycan-dependent ERAD machinery degrades topologically diverse misfolded proteins. Plant J. 94, 246–259. doi: 10.1111/tpj.13851.
Singh, J. P., Zhang, K., Wu, J., Yang, X. (2015). O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 356, 244–250. doi: 10.1016/j.canlet.2014.04.014.
Slawson, C., Hart, G. W. (2003). Dynamic interplay between O-GlcNAc and O-phosphate: the sweet side of protein regulation. Curr. Opin. Struct. Biol. 13, 631–636. doi: 10.1016/j.sbi.2003.08.003.
Slawson, C., Hart, G. W. (2011). O-GlcNAc signalling: implications for cancer cell biology. Nat. Rev. Cancer 11, 678–684. doi: 10.1038/nrc3114.
Snider, M. D., Sultzman, L. A., Robbins, P. W. (1980). Transmembrane location of oligosaccharide-lipid synthesis in microsomal vesicles. Cell 21, 385–392. doi: 10.1016/0092-8674(80)90475-4.
Strasser, R. (2016). Plant protein glycosylation. Glycobiology 26, 926–939. doi: 10.1093/glycob/cww023.
Strasser, R., Mucha, J., Schwihla, H., Altmann, F., Glössl, J., Steinkellner, H. (1999). Molecular cloning and characterization of cDNA coding for beta1,2N-acetylglucosaminyltransferase I (GlcNAc-TI) from Nicotiana tabacum. Glycobiology 9, 779–785. doi: 10.1093/glycob/9.8.779.
Strasser, R., Seifert, G., Doblin, M. S., Johnson, K. L., Ruprecht, C., Pfrengle, F., et al. (2021). Cracking the “Sugar Code”: A snapshot of N- and O-glycosylation pathways and functions in plants cells. Front. Plant Sci. 12, 640919. doi: 10.3389/fpls.2021.640919.
Strasser, R., Stadlmann, J., Svoboda, B., Altmann, F., Glossl, J., Mach, L. (2005). Molecular basis of N-acetylglucosaminyltransferase I deficiency in Arabidopsis thaliana plants lacking complex N-glycans. Biochem. J. 387, 385–391. doi: 10.1042/BJ20041686.
Taoka, K., Ham, B. K., Xoconostle-Cázares, B., Rojas, M. R., Lucas, W. J. (2007). Reciprocal phosphorylation and glycosylation recognition motifs control NCAPP1 interaction with pumpkin phloem proteins and their cell-to-cell movement. Plant Cell 19, 1866–1884. doi: 10.1105/tpc.107.052522.
Tolstikov, V. V., Fiehn, O. (2002). Analysis of highly polar compounds of plant origin: combination of hydrophilic interaction chromatography and electrospray ion trap mass spectrometry. Anal. Biochem. 301, 298–307. doi: 10.1006/abio.2001.5513.
Torres, C. R., Hart, G. W. (1984). Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317. doi: 10.1016/S0021-9258(17)43295-9.
Trinidad, J. C., Barkan, D. T., Gulledge, B. F., Thalhammer, A., Sali, A., Schoepfer, R., et al. (2012). Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell Proteomics 11, 215–229. doi: 10.1074/mcp.O112.018366.
Trombetta, E. S., Parodi, A. J. (2003). Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 19, 649–676. doi: 10.1146/annurev.cellbio.19.110701.153949.
Usadel, B., Bläsing, O. E., Gibon, Y., Retzlaff, K., Höhne, M., Günther, M., et al. (2008). Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 146, 1834–1861. doi: 10.1104/pp.107.115592.
Van Zelm, E., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005.
Vessal, M., Hassid, W. Z. (1972). Partial purification and properties of l-glutamine d-fructose 6-phosphate amidotransferase from Phaseolus aureus. Plant Physiol. 49, 977–981. doi: 10.1104/pp.49.6.977.
Vetting, M. W., de Calvarho, L. P. S., Yu, M., Hegde, S. S., Magnet, S., Roderick, S. L., et al. (2005). Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochim. Biophys. 433, 212–226. doi: 10.1016/j.abb.2004.09.003.
von Schaewen, A., Sturm, A., O’Neill, J., Chrispeels, M. (1993). Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 102, 1109–1118. doi: 10.1104/pp.102.4.1109.
Vu, K. V., Jeong, C. Y., Nguyen, T. T., Dinh, T. T. H., Lee, H., Hong, S. W. (2019). Deficiency of AtGFAT1 activity impairs growth, pollen germination and tolerance to tunicamycin in Arabidopsis. J. Exp. Bot. 70, 1775–1787. doi: 10.1093/jxb/erz055.
Walter, L. A., Batt, A. R., Darabedian, N., Zaro, B. W., Pratt, M. R. (2018). Azide- and alkyne-bearing metabolic chemical reporters of glycosylation show structure-dependent feedback inhibition of the hexosamine biosynthetic pathway. Chembiochem 19, 1918–1921. doi: 10.1002/cbic.201800280.
Walter, L. A., Lin, Y. H., Halbrook, C. J., Chuh, K. N., He, L., Pedowitz, N. J., et al. (2020). Inhibiting the hexosamine biosynthetic pathway lowers O-GlcNAcylation levels and sensitizes cancer to environmental stress. Biochemistry 59, 3169–3179. doi: 10.1021/acs.biochem.9b00560.
Wang, Z., Gucek, M., Hart, G. W. (2008c). Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl. Acad. Sci. U.S.A. 105, 13793–13798. doi: 10.1073/pnas.0806216105.
Wang, J., Liu, X., Liang, Y. H., Li, L. F., Su, X. D. (2008a). Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1. FEBS Lett. 582, 2973–2978. doi: 10.1016/j.febslet.2008.07.040.
Wang, J., Torii, M., Liu, H., Hart, G. W., Hu, Z. Z. (2011). dbOGAP - an integrated bioinformatics resource for protein O-GlcNAcylation. BMC Bioinf. 12, 91. doi: 10.1186/1471-2105-12-91.
Wang, Z., Wang, Y., Hong, X., Hu, D., Liu, C., Yang, J., et al. (2015). Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) induces early leaf senescence and defence responses in rice. J. Exp. Bot. 66, 973–987. doi: 10.1093/jxb/eru456.
Wang, Z., Wang, Q., Wei, L., Shi, Y., Li, T., Hu, K., et al. (2021). UDP-N-acetylglucosamine pyrophosphorylase 2 (UAP2) and 1 (UAP1) perform synergetic functions for leaf survival in rice. Front. Plant Sci. 12, 685102. doi: 10.3389/fpls.2021.685102.
Wang, H. F., Wang, Y. X., Zhou, Y. P., Wei, Y. P., Yan, Y., Zhang, Z. J., et al. (2023). Protein O-GlcNAcylation in cardiovascular diseases. Acta Pharmacol. Sin. 44, 8–18. doi: 10.1038/s41401-022-00934-2.
Wang, Y., Zhang, W. Z., Song, L. F., Zou, J. J., Su, Z., Wu, W. H. (2008b). Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148, 1201–1211. doi: 10.1104/pp.108.126375.
Wang-Gillam, A., Pastuszak, I., Elbein, A. D. (1998). A 17-amino acid insert changes UDP-N-acetylhexosamine pyrophosphorylase specificity from UDP-GalNAc to UDP-GlcNAc. J. Biol. Chem. 273, 27055–27057. doi: 10.1074/jbc.273.42.27055.
Wells, L., Vosseller, K., Hart, G. W. (2001). Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science 291, 2376–2378. doi: 10.1126/science.1058714.
Wilson, R. N., Somerville, C. R. (1995). Phenotypic suppression of the gibberellin-insensitive mutant (gai) of arabidopsis. Plant Physiol. 108, 495–502. doi: 10.1104/pp.108.2.495.
Wulff-Fuentes, E., Berendt, R. R., Massman, L., Danner, L., Malard, F., Vora, J., et al. (2021). The human O-GlcNAcome database and meta-analysis. Sci. Data 8, 25. doi: 10.1038/s41597-021-00810-4
Wyllie, J. A., Mckay, M. V., Barrow, A. S., Soares Da Costa, T. P. (2022). Biosynthesis of uridine diphosphate N-Acetylglucosamine: An underexploited pathway in the search for novel antibiotics? IUBMB Life 74, 1232–1252. doi: 10.1002/iub.2664.
Xiao, J., Xu, S., Li, C., Xu, Y., Xing, L., Niu, Y., et al. (2014). O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat. Commun. 5, 4572. doi: 10.1038/ncomms5572.
Xiao, G., Zhou, J., Lu, X., Huang, R., Zhang, H. (2018). Excessive UDPG resulting from the mutation of UAP1 causes programmed cell death by triggering reactive oxygen species accumulation and caspase-like activity in rice. New Phytol. 217, 332–343. doi: 10.1111/nph.14818.
Xing, L., Liu, Y., Xu, S., Xiao, J., Wang, B., Deng, H., et al. (2018). Arabidopsis O-GlcNAc transferase SEC activates histone methyltransferase ATX1 to regulate flowering. EMBO J. 37, e98115. doi: 10.15252/embj.201798115.
Xu, S. L., Chalkley, R. J., Maynard, J. C., Wang, W., Ni, W., Jiang, X., et al. (2017). Proteomic analysis reveals O-GlcNAc modification on proteins with key regulatory functions in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, E1536–e1543. doi: 10.1073/pnas.1610452114.
Xu, S., Xiao, J., Yin, F., Guo, X., Xing, L., Xu, Y., et al. (2019). The protein modifications of O-GlcNAcylation and phosphorylation mediate vernalization response for flowering in winter wheat. Plant Physiol. 180, 1436–1449. doi: 10.1104/pp.19.00081.
Yang, T., Echols, M., Martin, A., Bar-Peled, M. (2010). Identification and characterization of a strict and a promiscuous N-acetylglucosamine-1-P uridylyltransferase in Arabidopsis. Biochem. J. 430, 275–284. doi: 10.1042/BJ20100315.
Yu, C. Y., Cho, Y., Sharma, O., Kanehara, K. (2022). What's unique? The unfolded protein response in plants. J. Exp. Bot. 73, 1268–1276. doi: 10.1093/jxb/erab513.
Zeidan, Q., Hart, G. W. (2010). The intersections between O-GlcNAcylation and phosphorylation: implications for multiple signaling pathways. J. Cell Sci. 123, 13–22. doi: 10.1242/jcs.053678.
Zentella, R., Hu, J., Hsieh, W. P., Matsumoto, P. A., Dawdy, A., Barnhill, B., et al. (2016). O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev. 30, 164–176. doi: 10.1101/gad.270587.115.
Zentella, R., Sui, N., Barnhill, B., Hsieh, W. P., Hu, J., Shabanowitz, J., et al. (2017). The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nat. Chem. Biol. 13, 479–485. doi: 10.1038/nchembio.2320.
Zhang, M., Henquet, M., Chen, Z., Zhang, H., Zhang, Y., Ren, X., et al. (2009). LEW3, encoding a putative alpha-1,2-mannosyltransferase (ALG11) in N-linked glycoprotein, plays vital roles in cell-wall biosynthesis and the abiotic stress response in Arabidopsis thaliana. Plant J. 60, 983–999. doi: 10.1111/j.1365-313X.2009.04013.x.
Keywords: N-acetylglucosamine, hexosamine biosynthesis pathway, salvage pathway, N-glycosylation, O-GlcNAcylation, abiotic stress
Citation: Chen Y-H and Cheng W-H (2024) Hexosamine biosynthesis and related pathways, protein N-glycosylation and O-GlcNAcylation: their interconnection and role in plants. Front. Plant Sci. 15:1349064. doi: 10.3389/fpls.2024.1349064
Received: 04 December 2023; Accepted: 05 February 2024;
Published: 06 March 2024.
Edited by:
Muthusamy Ramakrishnan, Nanjing Forestry University, ChinaReviewed by:
Maksym Danchenko, Slovak Academy of Sciences, SlovakiaCopyright © 2024 Chen and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wan-Hsing Cheng, d2hjaGVuZ0BnYXRlLnNpbmljYS5lZHUudHc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.