
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 01 February 2024
Sec. Plant Proteomics and Protein Structural Biology
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1344142
Chitinases degrade chitin, a linear homopolymer of β-1,4-linked N-acetyl-D-glucosamine (GlcNAc) residues found in the cell walls of fungi and the exoskeletons of arthropods. They are secreted by the roots into the rhizosphere, a complex and dynamic environment where intense nutrient exchange occurs between plants and microbes. Here we modeled, expressed, purified, and characterized Zea mays and Oryza sativa root chitinases, and the chitinase of a symbiotic bacterium, Chitinophaga oryzae 1303 for their activities with chitin, di-, tri-, and tetra-saccharides and Aspergillus niger, with the goal of determining their role(s) in the rhizosphere and better understanding the molecular mechanisms underlying plant-microbe interactions. We show that Zea mays basic endochitinase (ZmChi19A) and Oryza sativa chitinase (OsChi19A) are from the GH19 chitinase family. The Chitinophaga oryzae 1303 chitinase (CspCh18A) belongs to the GH18 family. The three enzymes have similar apparent KM values of (20-40 µM) for the substrate 4-MU-GlcNAc3. They vary in their pH and temperature optima with OsChi19A activity optimal between pH 5–7 and 30–40°C while ZmChi19A and CspCh18A activities were optimal at pH 7-9 and 50–60°C. Modeling and site-directed mutation of ZmChi19A identified the catalytic cleft and the active residues E147 and E169 strategically positioned at ~8.6Å from each other in the folded protein. Cleavage of 4-MU-GlcNAc3 was unaffected by the absence of the CBD but diminished in the absence of the flexible C-terminal domain. However, unlike for the soluble substrate, the CBD and the newly identified flexible C-terminal domain were vital for inhibiting Aspergillus niger growth. The results are consistent with the involvement of the plant chitinases in defense against pathogens like fungi that have chitin exoskeletons. In summary, we have characterized the functional features and structural domains necessary for the activity of two plant root chitinases that are believed to be involved in plant defense and a bacterial chitinase that, along with the plant chitinases, may participate in nutrient recycling in the rhizosphere.
Agricultural crops have great economic importance and with the worldwide population increase, the current crop production rate is not sufficient to feed the future human population (Pingali, 2012; Kc et al., 2018). Crops suffer attack by pathogens including fungi, bacteria, and viruses with fungi alone causing 26 – 30% of the yield losses for crops like wheat, sugar beet and cotton (Roy et al., 2021) and 35% to 40% of the damage in maize, potato and rice (Kc et al., 2018). Maize (Zea mays) and rice (Oryza sativa) are among the most important food crops globally, and their growth and productivity are greatly influenced by the rhizosphere (Roy et al., 2021; Yim et al., 2022; USDA, 2023). Thus, a large increase in crop yield with the consequent alleviation of food insecurity for millions of people can be achieved by successfully addressing the challenges of plant stress such as induced by fungal infection (Pingali, 2012; Haldar and Sengupta, 2015; Rizzo et al., 2021).
Chitinases are hydrolytic enzymes that degrade chitin, a straight-chain homopolymer of β-1,4-linked N-acetyl-D-glucosamine (GlcNAc) units found in arthropod exoskeletons and some fungi cell walls (Martínez-Caballero et al., 2014; Horiuchi et al., 2016; Oyeleye and Normi, 2018; Wang et al., 2019). By breaking down chitin, chitinases inhibit fungal growth and release essential nutrients that plants can use for growth and development. Chitinases are expressed by a variety of organisms, including fungi, bacteria, archaea, viruses, animals, and plants. They are classified into the GH18, GH19 and GH20 families based on the CAZy database (Oyeleye and Normi, 2018). GH18 chitinases are widely distributed in eukaryotes and prokaryotes (Ju et al., 2016; Wang et al., 2019; Renaud et al., 2023) while GH19 chitinases are mostly found in plants.
Plant roots secrete chitinases into the rhizosphere (Haldar and Sengupta, 2015; Wang et al., 2019; Yim et al., 2022), which is a complex and dynamic environment where intense nutrient exchange occurs between plants and microbes with important consequences for plant growth, health, and productivity (Haldar and Sengupta, 2015). As part of the plant’s defense response, chitinases can lyse pathogens directly or indirectly by weakening their cell walls. The expression of root chitinases is also influenced by rhizosphere microbes such as fungi and bacteria, which can activate or suppress the synthesis of these enzymes. This connection is bidirectional, with root chitinases influencing the quantity and diversity of rhizosphere microorganisms and rhizosphere bacteria influencing the expression and activity of root chitinases (Haldar and Sengupta, 2015; Roy et al., 2021; Yim et al., 2022).
To better understand the biochemical and molecular features of chitinases that function in the soil, we have identified chitinases that are expressed by Zea mays and Oryza sativa roots (Vega-Arreguín et al., 2009; The, 2018) and found in their root exudates (Alexandrov et al., 2009; Ma et al., 2010) These plant chitinases and a chitinase from the genus Chitinophaga, which includes a number of soil-dwelling bacterial species, were expressed, and characterized. Phylogenetic analysis and molecular modeling identified the class and structure of each. Although the GH19 plant chitinases possess a chitin binding domain, which is absent from the bacterial chitinase, the three chitinases have similar activity on colloidal chitin and similar kinetic parameters assessed by a trimeric saccharide substrate. pH and temperature optima and stabilities are like those reported for other chitinases. Studies of truncated versions of the Zea mays chitinase identified a C-terminal domain which, like the chitin binding domain, is not required for cleaving short oligosaccharides, but is required for cleaving colloidal chitin and for attacking the fungal cell wall. These chitinases have potential for industrial application and will provide meaningful biomarkers for tracking plant root activity in situ in response to stresses such as fungal invasion.
The fluorogenic soluble substrates used for enzymatic assays: 4-Methylumbelliferyl β-D-N,N′-diacetylchitobioside (Cat#53643-12-2), 4-Methylumbelliferyl β-D-N,N′,N′′-triacetylchitotrioside (Cat#M5639-5MG) and 4-Methylumbelliferyl β-D-N,N’,N”,N’’’-Tetraacetylchitotetraoside (Cat#53643-14-4) were purchased from Cayman Chemicals (Ann Arbor, MA, USA), Sigma Aldrich (St. Louis, MO, USA) and Toronto Research Chemicals (Toronto, ON, Canada) respectively in powder form and dissolved in appropriate solvents to make stock solutions according to the manufacturer’s instructions. Colloidal chitin for chitinase activity comparisons was prepared from chitin (from shells of lobster, crab, or shrimp) (Cat#1398-61-4, J61206) by dissolving 5 g of powdered chitin in 250 mL of cold concentrated HCl or 85% phosphoric acid and allowed to rest at 4 °C for 24 h. The resulting suspension was passed through layers of cheese cloth to remove chunks, then placed on layers of filter papers and washed with cold tap water until pH of the rinse was ~7.0 (tested with pH paper). The paste was then stored at 4 °C to be weighed and resuspended in desired buffers when needed. DNS (3,5 Dinitrosalicylic acid, 98%, Cat#609-99-4), used for reaction termination to quantify the reducing sugar released from chitinase activity reactions, was purchased from Fisher Scientific (Waltham, MA, USA). TALON Metal Affinity Resin (Cat#635502) purchased from TakaraBio (San Jose, CA, USA) and Ni-NTA affinity Resin (Cat#R90115) purchased from Thermofisher Scientific (Waltham, MA, USA) were used for protein purifications. Other chemical reagents were of analytical grade or higher purity and were obtained from Sigma-Aldrich (St. Louis, MO, USA). Single and multiple site-directed mutagenesis of the chitinases were conducted with the GeneArt® Site-Directed Mutagenesis PLUS Kit (Cat #A14604, Thermofisher Scientific, Waltham, MA) using the AccuPrime™ Pfx DNA Polymerase (Cat#12344-024, Thermofisher Scientific, Waltham, MA). The protein standards used to determine the protein molecular weights were the broad range color prestained protein standard (10-250 kDa) (Cat#P7719S) from New England Biolabs (NEB) (Ipswich, MA) and the broad range spectra multicolor (Product# 26634) purchased from Thermofisher Scientific (Waltham, MA). Oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, IA), The sequences of all oligonucleotides used in this study are listed in Supplementary Table S1.
Candidate maize (Zea mays L.) chitinase (Basic Endochitinase A, ZmChi19A) was selected from stress response proteins identified to be secreted into the root mucilage (Ma et al., 2010). The nucleotide sequences of maize ZmChi19A, rice (Oryza sativa) root chitinase, OsChi19A, and symbiotic bacteria Chitinophaga oryzae 1303 chitinase (CspCh18A) were retrieved from the nucleotide database of the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) (Alexandrov et al., 2009).
Identified gene sequences were translated using the Expasy translate tool (https://web.expasy.org/translate/) while the protein parameters were obtained using the Expasy ProtParam tool (https://web.expasy.org/protparam/) (Gasteiger et al., 2007). Phylogenic analysis was carried out to determine the evolutionary relationship between the two plant chitinases as well as the bacterial chitinase (http://www.phylogeny.fr/) (Dereeper et al., 2008). Multiple sequence alignment was performed using the ClustalW algorithm (https://www.ebi.ac.uk/Tools/msa/clustalo/) (Sievers and Higgins, 2014), while conserved motifs of the protein sequences were analyzed using (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi ) (Lu et al., 2020). The prediction of the signal peptide sequence was performed using the signal-5.0 application server at https://services.healthtech.dtu.dk/services/SignalP-5.0/. To predict N- and O-glycosylation sites, the servers NetNGlyc 1.0 (https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0) (Gupta and Brunak, 2001) and NetOGlyc 4.0 (https://services.healthtech.dtu.dk/service.php?NetOGlyc-4.0) (Steentoft et al., 2013) were used respectively.
Prediction of protein structures and mobility were done using AlphaFold2 (Jumper et al., 2021). The surface charge distribution and structural alignment of the modeled protein structures was determined using the Poisson Boltzmann tool at https://server.poissonboltzmann.org/ (Jurrus et al., 2018) and https://zhanglab.ccmb.med.umich.edu/TM-align/ (Zhang and Skolnick, 2005) respectively. Structure visualization, analysis, and representations were done using PyMOL and ChimeraX softwares (Pettersen et al., 2004).
To predict the catalytic residues, docking of the structures with a chitin substrate was done with HADDOCK, SWISSDOCK and CBDOCK-2 (Grosdidier et al., 2011; Honorato et al., 2021; Liu et al., 2022). The chitin substrate was obtained from the PDB structure 6BN0 (Hurlburt et al., 2018).
The ZmChi19A coding sequence was amplified from the maize root cDNA and cloned into the pET28b vector for E. coli expression. The pET28b expression vector had been modified to incorporate an N-terminal 10x-His and SUMO solubility tag. Overhangs of the forward and reverse primers were designed to contain the BamHI and HindIII restriction sites respectively. Gene block fragments of OsChi19A and CspCh18A were cloned into the pET28a vector for E. coli expression (Horiuchi et al., 2016). Overhangs of the forward primers were designed to contain the XbaI restriction site, and the reverse primers were designed to contain the XhoI restriction site. Gene sequence of OsChi19A was also codon modified for optimal expression in E. coli. All forward primers were designed to produce the Tobacco Etch Virus (TEV) protease site (ENLYFQG) at the N-terminus to provide options of cutting off the tags after expression and purification. Primers were also designed to truncate the signal peptides located on the N-terminal of the gene sequences. The ZmChi19A(E147A, E169A) mutant was prepared using a designed Gene Block fragment from Integrated DNA Technologies (IDT, Coralville, IA), while truncations of the CBD (ZmChi19AΔCBD) and flexible C-terminal (ZmChi19A_A328*) were achieved by PCR amplification of the cDNA using primers designed to make the truncations, PCR amplification of the truncated cDNA, followed by subsequent and insertion between the Xho1 and Xba1 sites of the expression vector. CspCh18A(D161A, E163A) mutant was prepared by site-directed mutagenesis (Wang et al., 2019) using the GeneArt® Site-Directed Mutagenesis PLUS Kit (Cat #A14604, Thermofisher Scientific, Waltham, MA) and AccuPrime™ Pfx DNA Polymerase (Cat#12344-024, Thermofisher Scientific, Waltham, MA). The numbers assigned to the mutant amino acid residues correspond to the positions of the amino acids in the complete translated protein, starting with methionine and including the signal sequence. Restriction enzymes used were purchased from New England Biolabs (NEB, Ipswich, MA). T4 DNA Ligase was obtained from Promega (Cat#C126A, Madison, WI). Chemically competent cells used for cloning (E. coli 10G & DH5α) were purchased from Lucigen (Cat. # 60107-1, Middleton, WI) and Thermofisher Scientific (Cat#12297-016) respectively. For protein expression, plasmids were retransformed into OverExpress chemically competent cells C43(DE3) (Cat. # 60446-1, Lucigen, Middleton, WI) and Rosetta-gamiTM2(DE3)pLysS Chemically Competent Cells were from Novagen (Cat#71432-3, Sigma-Aldrich, St. Louis, MO). All the recombinant plasmids were verified by Sanger sequencing at the Iowa State University (ISU) DNA facility. Gene sequences and primers used for cloning can be found in the Supplementary Data.
Competent cells harboring the expression plasmids were grown at 37°C with shaking at 250 rpm in 1000 ml of Luria-Bertani broth. When the cell culture reached an OD600 of 0.4, the temperature was lowered to 18°C, incubated for 15 mins and protein expression was induced by adding IPTG to a final concentration of 1.0 mM. Cells expressing all but ZmChi19AΔCBD were incubated for 18 h at 18°C and ZmChi19AΔCBD-transformed cells were incubated at 10 °C for 72 h. Induced cells were harvested by centrifugation, resuspended in lysis buffer (12.5 mL of 25 mM Tris-HCl pH 7.4, 300 mM NaCl, 0.1 mM EDTA) (Deshmukh et al., 2023) and rapidly frozen in liquid nitrogen. Cells were lysed by thawing and incubating for 30 min with 1 mg/mL of lysozyme and then sonicated for 15 s a total of five times. Solubilized proteins were collected by centrifugation at 15,000 x g for 30 min to obtain the crude lysates (Horiuchi et al., 2016; Singappuli-Arachchige et al., 2022). OsChi19A was further purified from inclusion bodies in the pellet and refolded using a refolding buffer (50 mM HEPES, 0.4 M L-Arginine, 6.3 mM GSH, 3.7 mM GSSG, 2 mM EDTA, pH 7.0). Crude lysate was loaded onto a TALON or Ni-NTA column with a lysate:resin ratio of 10:1 (v/v). The affinity resin was incubated on a shaker for 18 h at 4°C. Unbound proteins were removed as a flow-through fraction and the resin was washed five times with washing buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 20 mM imidazole). The proteins of interest were eluted in fractions with volumes of 1 mL using elution buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, and 250 mM imidazole) (Wang et al., 2019) Dialysis into storage buffer (50mM Tris-HCl pH 7.4, 150 mM NaCl) was performed using a dialysis bag to eliminate imidazole. The proteins were concentrated from the elution buffer using Amicon Ultra centrifuge filter units (30,000 Da cutoff, Millipore, Burlington, MA). Glycerol was added to the purified protein to a final concentration of 25% and proteins were stored at -80°C. The concentrations of proteins were estimated from absorption at 280 nm determined by a NanoDrop spectrophotometer (#ND-1000, Thermo Scientific/Gibco, Waltham, MA) and using an extinction coefficient calculate for each protein based on its amino acid content and the presence of free SH groups (Gasteiger et al., 2007) and by the Bradford assay (Quick Start Bradford Dye reagent 1X, Cat#500-0205, Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. Reducing and non-reducing SDS polyacrylamide gel electrophoresis (SDS-PAGE) for proteins were performed as previously described using 10 or 12% acrylamide gels (Nilsen-Hamilton et al., 1980). The gels were run for 2 h at constant current of 25 mA (Nilsen-Hamilton and Hamilton, 1987). All purified recombinant proteins were verified by Liquid Chromatography Mass Spectrometry (LC-MS) at the Iowa State University (ISU) Protein facility. Approximations for the purities of proteins used in this study are shown in Supplementary Table S2.
Size exclusion chromatography was performed 4 °C in an AKTA FPLC system with a pre- packed Superose 12 10/300GL (separation range: 1 kDa to 300 kDa; GE Healthcare, Cat#17517301, Waukesha, WI) with a flow rate of 0.2 mL/min. The inner dimensions of the column were 10x300-310 mm with a bed volume of 24 mL. Prior to being loaded on the column, samples were dialyzed against the column buffer (20 mM Tris, 100 mM KCl, pH 7.9) then centrifuged at 15,900 RCF at 4 °C for 30 min.
The overall secondary structures of purified proteins were investigated at 25 °C using a J-810 circular dichroism (CD) spectropolarimeter (Jasco, Hachioji, Tokyo, Japan). CD spectra were collected from 170 to 270 nm at a scanning rate of 200 nm/min with a path length of 0.1 cm (Figueroa et al., 2016; Giudice et al., 2017).
Enzyme kinetics assays were carried out with the fluorogenic substrate 4-Methylumbelliferyl β-DN,N′,N′′-triacetylchitotrioside [4-MU-(GlcNAc)3] substrate at concentrations from 5 to 100 µM. All enzymes used for determining the kinetic parameters were FPLC-purified. The optimum temperature for the chitinases was measured at temperatures ranging from 0 to 100 °C at pH 8.0 for ZmChi19A and CspCh18A and pH 5.0 for OsChi19A. To test for thermostability, protein samples were preincubated at temperatures ranging from 0 to 100 °C for 30 minutes, and the residual activity measured at 50 °C. The optimum pH was determined at 50 °C in buffers ranging from pH 2 to 13 in buffers A (25 mM sodium-citrate, pH 2 - 6), buffers B (25 mM Tris buffer, pH 7 and 8) and buffers C (25 mM Sodium-Carbonate, pH 9 – 13). To determine pH stability, the enzymes were preincubated at 0 °C for 30 minutes in buffers A-C with a pH range of 2.0–13.0 and the residual activity was measured at 50 °C in pH 8.0 (buffer B) for ZmChi19A and CspCh18A and pH 5.0 (buffer A) for OsChi19A (Horiuchi et al., 2016). For assays involving variations of pH and temperature, the 4-MU-(GlcNAc)3 was prepared in the identified buffer with the appropriate pH for the assay. The reactions were stopped by the addition of 100 µL of 1 M glycine/NaOH stop buffer to a final concentration of 1 mM glycine and fluorescence was measured immediately (Steentoft et al., 2013). The enzyme activity was calculated using an experimentally determined calibration curve to convert the change in fluorescence to the concentration of free 4-methylumbelliferone (4-MU) released. Fluorescence readings for all assays were measured at excitation 350 nm and emission 460 nm.
Colloidal chitin chitinolytic activities were assessed by incubating with 10 mg/mL colloidal chitin in 25 mM Tris-HCl buffer, pH 8.0 at 50 °C for 5 mins, supernatants were collected and mixed with 4 times volume of DNS, followed by heating at 95 °C. The resulting mixtures were further diluted 1:1 with distilled water, 100 uL of the final solution was added to a 96-well plate and the absorbance measured at 540 nm. All measurements were performed at room temperature in Falcon™ 96-well plates (Catalog# 351172 or 353948, Thermofisher Scientific, Waltham, MA) and read with a Synergy II plate reader (The Lab World Group, Hudson, MA) or Varian-Cary Eclipse Fluorescence Spectrophotometer (American Laboratory Trading, San Diego, CA) to obtain fluorescence spectra. All determinations were performed in triplicate and at least twice independently. For each incubation time, mixtures containing the same components (except for the protein) as the control condition with the SUMO protein, which provided the blank values (averages of triplicates) that were subtracted from the average value obtained in the presence of chitinases. The activity is expressed as µmol 4MU/min/mmol protein (Horiuchi et al., 2016).
A modified disk growth test was conducted to test the antifungal activity of wild-type and mutant chitinases. Fungal spores were resuspended in 25 mM Sodium Acetate, pH 5, imaged, and counted using ImageJ or Fiji software (National Institutes of Health, Bethesda, MD), and diluted to ~2.5 to ~5 spores/µL in 25 mM Sodium Acetate, pH 5 with the stated concentration of enzyme. The mixtures were incubated at 37°C for 30 min. Sample and spore mixtures were then applied to 5 mm sterile paper disks placed on potato dextrose agar in a 9 cm diameter Petri dish supplemented with 25 mg/mL chloramphenicol to inhibit bacterial growth. The plates were incubated at room temperature in a dedicated cell culture hood and photographed at 12 h intervals (Alastruey-Izquierdo et al., 2015; (Puškárová et al., 2017).
All experiments were performed in triplicates and error bars for standard deviation are shown in the figures. where Se = standard deviation of the sample and Sb = standard deviation of the subtracted blank. The blank values were derived from buffer alone or SUMO in buffer measured in the same experiment and subtracted from all values before fitting. All binding isotherms were fit to the Langmuir equation where A = the change in absorption at 450 nm per min with Amax being the maximum values and S = substrate concentration. Fittings and estimated of statistical significance were performed by Sigmaplot. All reported values for KM passed the Normality (Shapiro-Wilk) and the Constant Variance (Spearman Rank Correlation) tests. One unit (U) of chitinase activity represents 1μmol of 4-MU released by enzyme from 4MU-GlcNAc3 per min under reaction conditions.
The maize, rice chitinases and bacterial chitinases and some mutant versions were expressed in E. coli, purified, and analyzed by SDS-PAGE (Figure 1A). For most experiments a fusion protein of ZmChi19A with an N-terminal SUMO solubility tag was used due to the limited solubility of untagged ZmChi19A. Apparent molecular masses calculated from the Rfs after electrophoresis by SDS-PAGE were within 12% of those predicted from their amino acid sequences for all proteins (Supplementary Table S3).
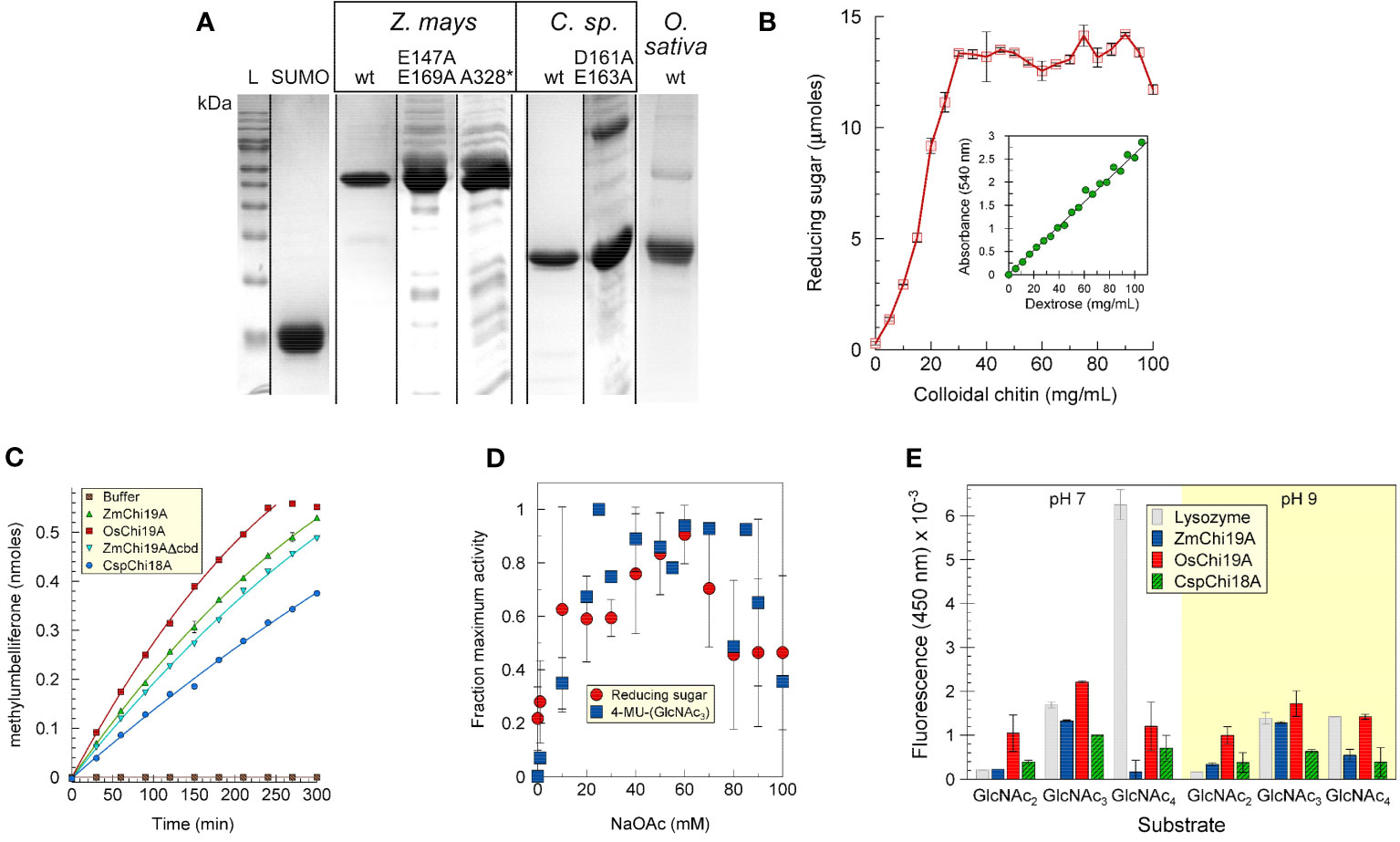
Figure 1 Purification and initial characterization of native chitinases and mutant forms. (A) SDS-PAGE analysis of purified SUMO, ZmChi19A and mutants, CspChi18A and mutants, and OsChi19A. Lane L, protein mass markers. (B) ZmChi19A chitin cleavage activity as a function of substrate concentration (inset: standard curve). (C) Enzyme activity as a function of time. (D) Effect of salt concentration on enzyme activity. (E) Substrate preferences of lysozyme (gray), ZmChi19A (blue), OsChi19A (red) and CspChi18A (hatched green) using 4-MU-GlcNac2, 4-MU-GlcNac3, and 4-MU-GlcNAc4 as substrates. The graphs show the averaged data from three (B, E) and two (D) experiment performed in duplicate or triplicate (means ± SD). (C) shows the results from one experiment performed in triplicate for which there is one other experiment showing similar results.
The ability to cleave colloidal chitin was demonstrated for SUMO-ZmChi19A (Figures 1B, Supplementary Figure S1) and CspCh18A (Supplementary Figure S1). A trimeric N-acetyl-glutamine substrate linked to methylumbelliferone (4-MU-GlcNAc3) was also a substrate for these chitinases (Figure 1C). The salt dependency of ZmChi19A activity was similar when tested with chitin or 4-MU-GlcNAc3 as substrates (Figure 1D). To determine the optimal length of substrate, we compared the activities of ZmChi19A and CspCh18A with 4-MU-GlcNAc3 substrates consisting of different multiples of N-acetyl-D-glucosamine. The GH19 chitinases, ZmChi19A and OsChi19A, prefer the 4-MU-GlcNAc3 at pH 7 and 9 (Figure 1E). Consequently, 4-MU-GlcNAc3 was used as the substrate for further enzymatic analysis.
A phylogenetic tree created from the multiple alignment of the maize, rice and bacterial chitinases (ZmChi19A, OsChi19A and CspCh18A respectively) predict that the maize and rice chitinases followed a similar evolutionary path but both lack orthology with the bacteria chitinase (Figure 2A). Tertiary structure predictions of the GH19 family chitinases, ZmChi19A and OsChi19A, identified the chitin-binding domain linked to an α-helix rich lysozyme-like catalytic domain with a deep cleft and a flexible C-terminal domain (Figures 2C–G). CspCh18A belongs to the GH18 family that is characterized by a catalytic region consisting of a triosephosphate isomerase (TIM) barrel (β/α) domain (Figures 2H, I). Despite both being GH19 chitinases, ZmChi19A and OsChi19A show contrasting surface charge distributions with the OsChi19A surface dominated by negatively charged amino acid residues (Figure 2G) and ZmChi19A having mostly positively charged surface residues on both the catalytic and chitin-binding domain (Figure 2E). All three chitinases have a predominantly negatively charged catalytic cleft (Figures 2E, G, I). The similar surface charge distributions of CspCh18A and ZmChi19A differ greatly from that of OsChi19A, which is highly negatively charged (Figure 3G).
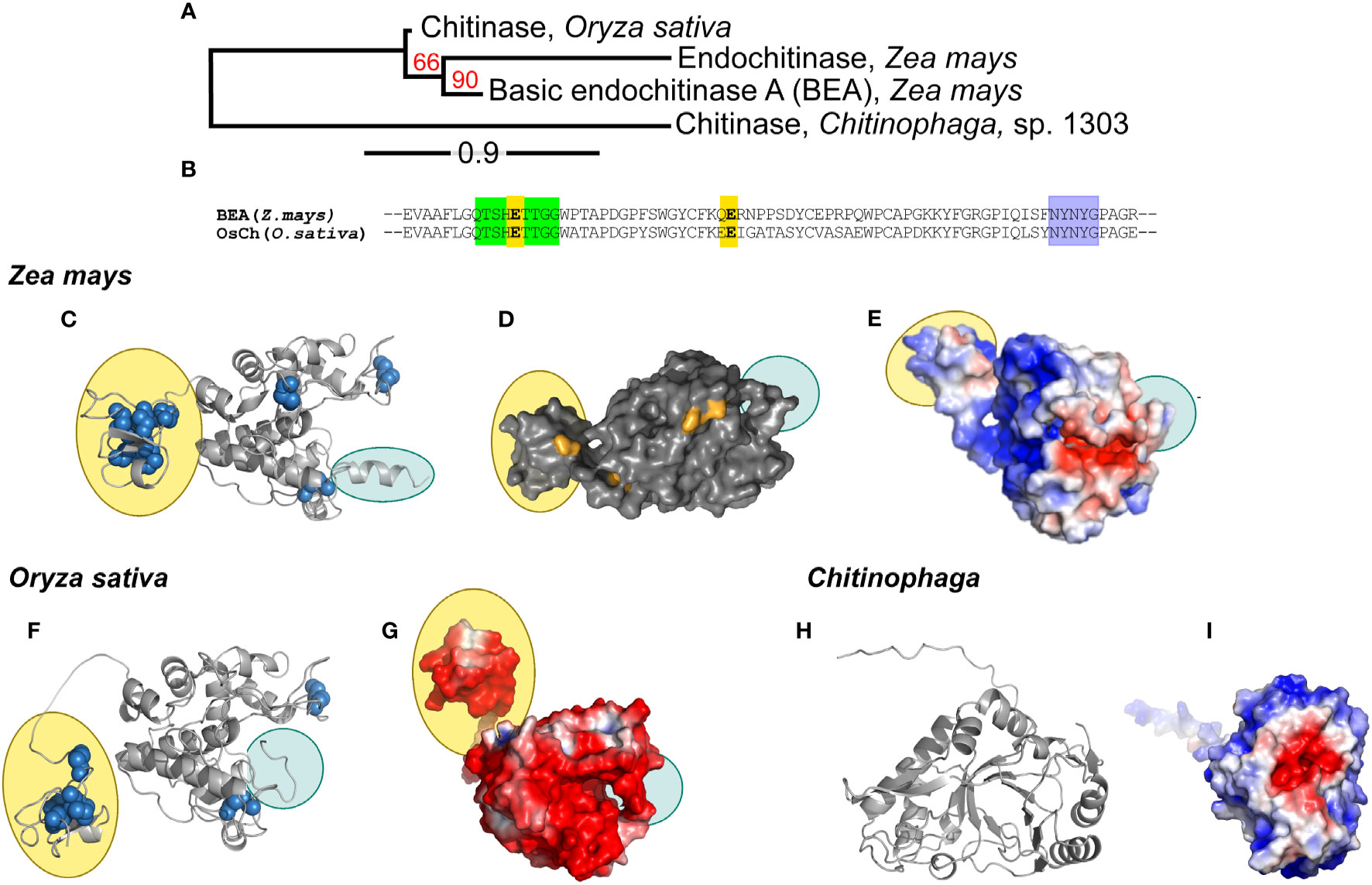
Figure 2 Phylogenetic analysis, conserved domains, structures, and surface charge distribution of rhizosphere associated chitinases. (A) Phylogenetic analysis of ZmChi19A, OsChi19A and CspChi18A. The red numbers on the branches are the probabilities of sharing a common ancestor determined by the bootstrap branch support. The line segment with the number ‘0.9’ shows the length of branch that represents the amount of evolutionary genetic change over time. (B) Sequence alignment of ZmChi19A and OsChi19A showing the conserved domains (in green and purple) and potential catalytic residues (in yellow). AlphaFold derived structures of (C) ZmChi19A, secondary structure, (D) ZmChi19A, surface structure showing potential glycosylation sites (in orange), (E) ZmChi19A, electrostatic surface view, (F) OsChi19A showing disulfide bridges (in blue spheres), (G) OsChi19A, electrostatic surface view, (H) CspChi18A, secondary structure, (I) CspChi18A, electrostatic surface view. All electrostatic surface views show positively charged regions and negatively charged regions in blue and red respectively. Yellow-filled ovals identify the chitin binding domains (CBD) and blue-filled ovals identify the C-terminal flexible domains (CTD). Supplementary Figure S2 compares the predicted glycosylation sites on the two plant chitinases.
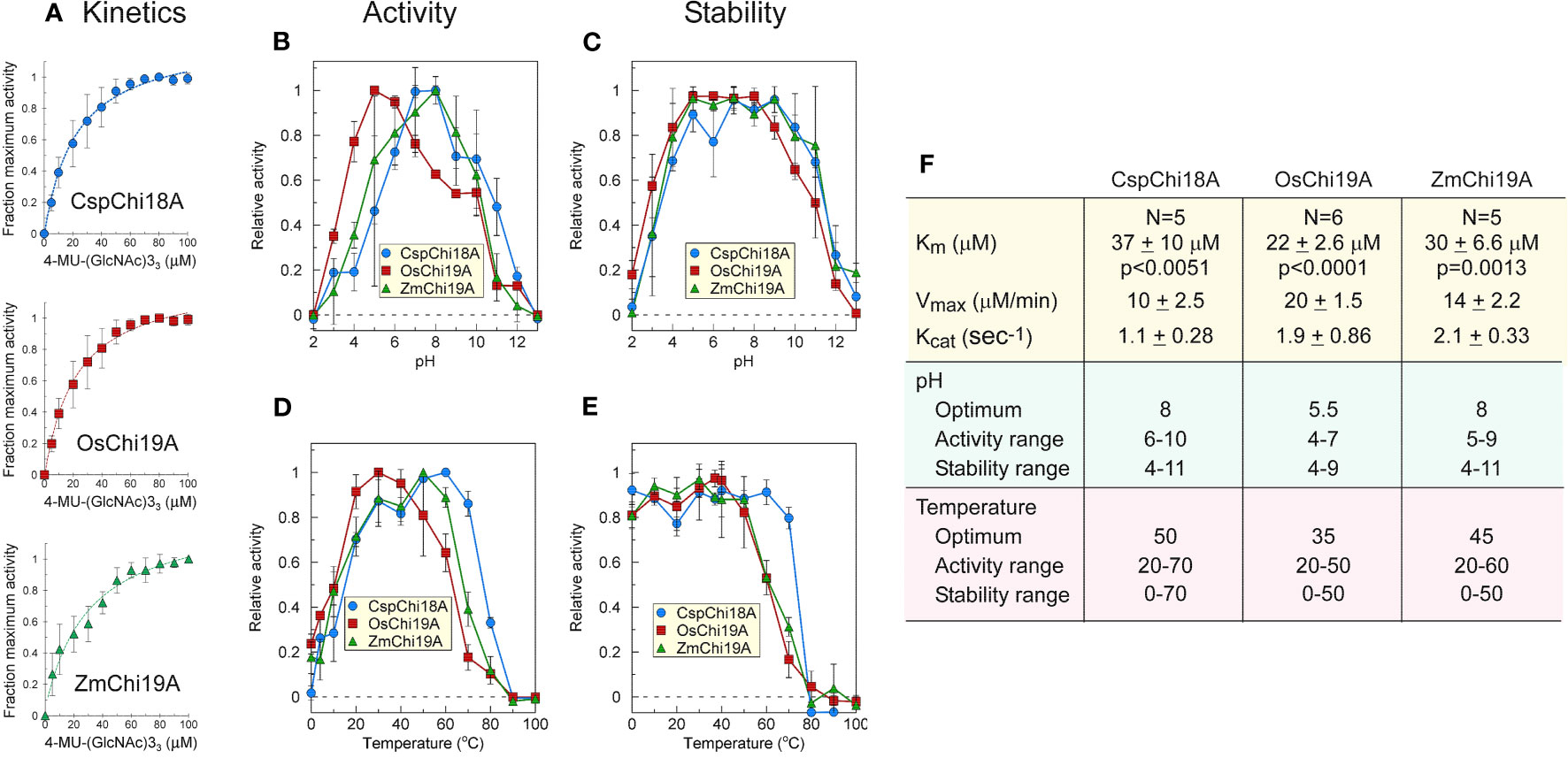
Figure 3 Catalytic activities of ZmChi19A, OsChi19A and CspChi18A using the soluble substrate (NAG)3-MUF. (A) Enzyme activity as a function of substrate concentration. Each curve shows the average of 5-6 independent experiments that were performed with 2 independently expressed and purified preparations of each enzyme (B) Effect of pH on chitinase activity. (C) Effect of pH on chitinase stability. (D) Effect of temperature on chitinase activity. (E) Effect of temperature on chitinase stability. (F) Table of derived kinetic constants. Data for B-E were from 2 independent experiments with enzymes from different expressed and purified preparations, each performed in triplicate. All values shown are the means ± SD.
Kinetic parameters (KM, Vmax, and Kcat) were similar for all three chitinases (Figures 3A, F), although they were determined under different conditions (50°C, pH 8 for SUMO-ZmChi19A and CspCh18A, and 40 °C, pH 5 for OsChi19A). ZmChi19A and CspCh18A showed a high pH optimum and retained at least 70% activity over a wide pH range (Figures 3B, F), whereas the pH optimum for OsChi19A was lower with a smaller pH range over which it retained at least 70% of the maximum activity (Figures 3B, F). ZmChi19A and CspCh18A were also stable (with at least 70% activity retained) to incubation for one hour over a larger pH range than OsChi19A (Figures 3C, F). In addition to being more stable at higher temperatures (Figures 3E, F), ZmChi19A and CspCh18A had higher temperature optima for catalysis than OsChi19A (Figures 3D, F). All three enzymes were stable to cold temperatures (Figure 3E).
To investigate the residues essential for catalysis of the GH19 and GH18 chitinases, we modeled the structure of ZmChi19A and CspCh18A in complex with the substrate (chitin, (GlcNAc)6) by molecular docking. In the modeled structures, (GlcNAc)6 is bound in the negatively charged substrate cleft with the average distance of substrate to the predicted catalytic residues being 3.1 Å (Figures 4A, D). In ZmChi19A, the scissile glycosidic bond is sandwiched between the side chain of the conserved and predicted catalytic residues Glu147 and Glu169, which are 8.6 Å apart (Figures 2B, 4B). In CspCh18A, Asp161 and Glu163 were identified as potential catalytic residues (Figures 4D, E). To test the predicted catalytic roles, ZmChi19A and CspCh18A were produced with alanine substitutions for these residues and the activities of these mutant proteins were compared with the wild-type enzymes using both 4-MU-GlcNAc3 and colloidal chitin (Figures 4C, F). The mutants of ZmChi19A and CspCh18A were also folded with AlphaFold and aligned with the native enzyme to evaluate the effect of the mutations on enzyme structures. These comparisons showed that the folded structures of all mutants were within 1.5 RMSD of the relevant native structure (Supplementary Figure S3). ZmChi19A(E147A,E169A) showed almost complete loss of activity, which supports a key role of these residues in catalysis. Loss of the chitin binding domain or the C-terminal domain had little effect on the ability of ZmChi19A to cleave the soluble substrate but effectively eliminated the ability to cleave colloidal chitin (Figure 4C). By contrast, CspCh18A(D161A,E163A) retained ~60% of the activity of the native enzyme in enzymatic assays using either 4-MU-GlcNAc3 and colloidal chitin, which suggests a potential ancillary role in catalysis (Figure 4F).
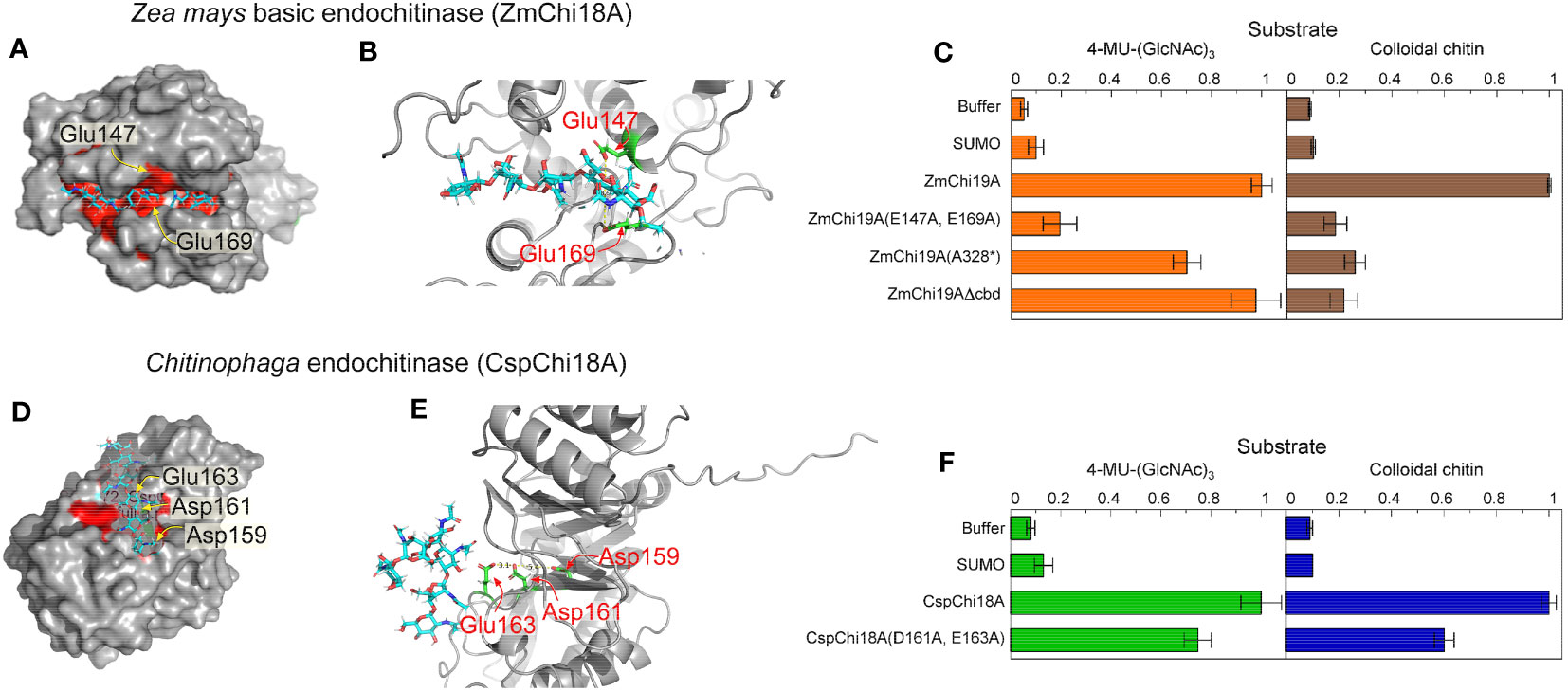
Figure 4 Amino acid residues in GH19 and GH18 chitinases for substrate binding and catalysis. The substrate (GlcNAc)6 is shown as cyan sticks in all models. Model structures were created with alphFold (A) Surface structure of ZmChi19A with the catalytic site in red and predicted catalytic residues labeled, (B) Structure of the ZmChi19A catalytic site with predicted catalytic residues in green. (C) Activities of the wild-type ZmChi19A and its mutants against substrates 4-MU-(GlcNAc)3 and colloidal chitin. (D) Surface structure of the CspChi18A catalytic site in red with predicted catalytic residues labeled. (E) Structure of the CspChi18A catalytic site with predicted catalytic residues in green. (F) Activities of the wild-type CspCh and its mutant against substrates 4-MU-(GlcNAc)3 and colloidal chitin. The graphs show the average results from 6 (4-MU-(GlcNAc)3) and 2 (chitin) experiments, each performed independently in triplicate. The background of fluorescence with SUMO was not subtracted from all values to show the value of the SUMO control compared with buffer and other activities. Whereas the activity of ZmChi19A and CspChi18A are shown relative to the native activities, the rate of chitin cleavage by CspChi18A is 27% of the rate of cleavage by BEA when both are normalized to the baseline rate of SUMO.
The three chitinases and their mutated and truncated versions were tested for their activity against the fungus Aspergillus niger (Figure 5). Only the native (full-length) ZmChi19A inhibited the fungal growth. This result and their requirement for colloidal chitin cleavage shows that, although the CBD and flexible C-terminal domain are not necessary for catalysis, they are essential for anti-fungal activity, which requires chitin cleavage.
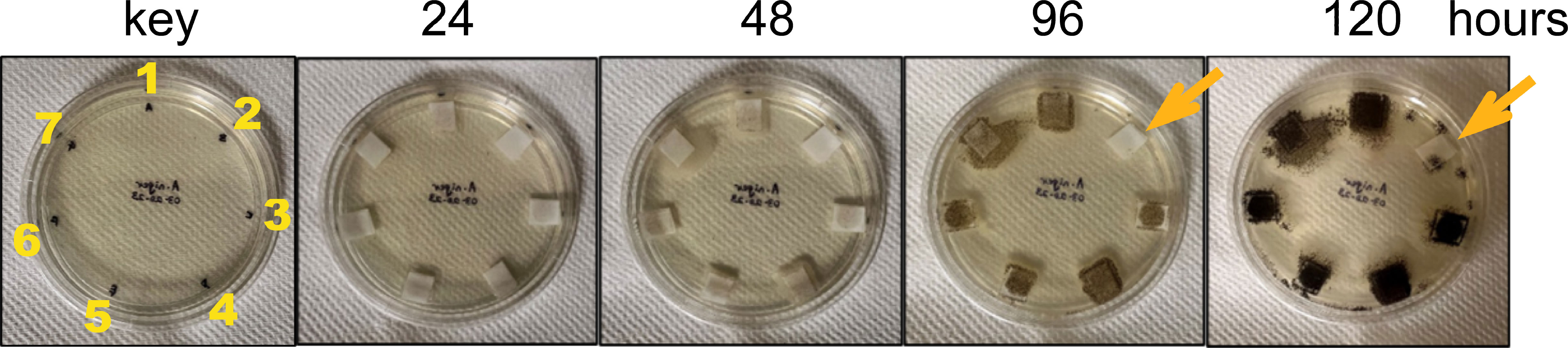
Figure 5 Antifungal activity assay of purified recombinant native and mutant chitinases towards Aspergillus niger. Starting as spores (~5/filter), Aspergillus niger was incubated with 50 ug (0.5 mg/mL) of enzyme at room temperature (23-24 °C) over a period of 5 days. 1) SUMO, 2) SUMO-ZmChi19A, 3) SUMO-ZmChi19A(E147A,E169A), 4) SUMO-ZmChi19A(A328*), 5) ZmChi19AΔCBD, 6) CspChi18A, and 7) CspChi18A(D161A,E163A). This data is representative of the results obtained in six independently performed experiments.
Based on sequence similarity, plant chitinases are classified into seven classes (I–VII). Classes represented in the GH19 family are I, II, IV, VI and VII, while classes III and V are members of the GH18 family. ZmChi19A and OsChi19A are maize and rice root endochitinases respectively, and members of the GH19 family to which most plant chitinases belong (Oyeleye and Normi, 2018; Zhou et al., 2023), while CspCh18A is a symbiotic bacterial endochitinase in the GH18 family (Oyeleye and Normi, 2018; Haxim et al., 2022). GH18 and GH19 chitinases commonly use different catalytic mechanisms, producing different configuration of products (Funkhouser and Aronson, 2007; Oliveira et al., 2020). All GH18 chitinases are characterized by a catalytic region that consists of a triosephosphate isomerase (TIM) barrel (β/α)8 domain, while the catalytic domain of family GH19 is an α-helix rich lysozyme-like domain characterized by a deep cleft (Wang et al., 2019). Class I endochitinases are characterized by the presence of an N-terminal hevein-like chitin-binding domain (CBD) (Horiuchi et al., 2016) and a C-terminal catalytic domain (CatD), which are connected by a short linker that varies in length and amino acid composition. The ZmChi19A and OsChi19A chitinases have a CBD and thus are classified as type I.
Despite having similar structures and conserved domains ZmChi19A and OsChi19A have contrasting surface charge distributions and different pH optima, which might be adaptations to different environments. Maize plants are C4 plants (Bellasio et al., 2023) more suited to hot climates while rice plants are C3 plants which grow in cool environments (Bellasio et al., 2019; Osinde et al., 2023). Oryza sativa normally grows emerged in water (Yan et al., 2023), whereas Zea mays grows under more dry conditions on land. Differences in the charge distributions of ZmChi19A and OsChi19A may be adaptations to specific ecological niches, lifestyles, or host-pathogen interactions (Vanhoye et al., 2004; Garcia-Moreno, 2009; Gerland et al., 2020; Kim et al., 2020; Panja et al., 2020; Di Savino et al., 2021; Doan et al., 2022; Vallina Estrada et al., 2023).
ZmChi19A and CspCh18A degraded colloidal chitin (OsChi19A was not tested) and all three chitinases degraded soluble chitin substrates, which suggests that they may play an important role in chitin recycling and defense against harmful fungi. The recombinant enzymes were tested for substrate preference and their kinetic parameters were determined using the preferred substrate, 4-MU-(GlcNAc)3. The obtained data were in the range of those reported for several chitinases as summarized in Supplementary Table S4. For example, the KM values for ZmChi19A, OsChi19A and CspCh18A were 30, 22 and 37 uM respectively, which are similar to 33 uM for a barley chitinase (Kuo et al., 2008), 42 uM for a S. marcescens chitinase (Honda et al., 2003), and 49 uM for an Aspergillus niger chitinase (van Munster et al., 2015). Higher KM values were reported for chitinases from rubber (Hevea brasiliensis) and toxic plant (weed) Ipomoea carnea (Patel et al., 2009; Sukprasirt and Wititsuwannakul, 2014, Supplementary Table S4).
The recombinant enzymes were also characterized for pH and temperature optima and stabilities. The optimum pH and temperature ranges over which these chitinases were stable were in the range of those reported for other reported plant, fungi and bacterial chitinases (Patel et al., 2009; Sukprasirt and Wititsuwannakul, 2014; Horiuchi et al., 2016; Thimoteo et al., 2017; Wang et al., 2019; Rajninec et al., 2020, Supplementary Table S4). The pH optimum and pH range for OsChi19A activity are consistent with those reported for other plant chitinases (Patel et al., 2009; Sukprasirt and Wititsuwannakul, 2014; Horiuchi et al., 2016; Sierra-Gómez et al., 2019; Wang et al., 2019). However, unlike other reported plants and bacterial chitinases, all of which have reported pH optima between 4.5 and 6, ZmChi19A and CspCh18A were most active at pH 8.
The conserved catalytic residues of GH19 chitinases Glu147 and Glu169 were confirmed to play a key role in catalysis by ZmChi19A due to their mutagenesis resulting in the complete loss of enzymatic activity against colloidal chitin, the soluble trimeric saccharide and Aspergillus niger. The identification of catalytic residues in CspCh18A based on the conservation of sequence with other GH18 chitinases combined with molecular docking of the substrate to the alphaFold modeled CspCh18A suggested that Asp161 and Glu163 were potential catalytic residues (Wang et al., 2019). However, with both residues mutated, the enzyme retained ~60% of its activity. Thus, these residues are not critical for catalysis.
Modeling by alphaFold identified the flexible C-terminal domain (CTD), which proved to be essential for ZmChi19A to cleave colloidal chitin and inhibit fungal growth. This is the first report showing the importance of the CTD in the role of plant chitinases. Only the CBD has been previously shown to be required for activity (Onaga and Taira, 2008; Huang et al., 2009; Yokoyama et al., 2009). We propose that the CTD might provide a second site of interaction with chitin in addition to the CBD or hold chitin in the catalytic cleft for enzymatic degradation, thereby enhancing the efficiency of chitin breakdown in the soil. In another example, a mobile cap domain identified in a family IV esterase from sorghum rhizosphere microbiome was proposed to regulate substrate access (Distaso et al., 2023).
The ability of Zea mays ZmChi19A to inhibit the growth of the pathogenic fungus, Aspergillus niger, is consistent with the hypothesis that GH19 chitinases like ZmChi19A become part of the plants’ defense arsenal against harmful soil fungi when released by plant roots (Huang et al., 2009; Yokoyama et al., 2009; Haldar and Sengupta, 2015). By contrast, the lack of ability of the plant root associated GH18 bacteria chitinase to inhibit Aspergillus niger growth is consistent with the hypothesis that it is responsible for downstream chitin degradation rather than being part of an initial defense again fungal pathogens in the rhizosphere (Huang et al., 2009; Roy et al., 2021; Chandra et al., 2022; Kotb et al., 2023). However, as we have only investigated the effects of these chitinases on one fungus in this study, it may also be that the bacterial chitinase is effective against some but not all fungi. Further investigation is required to fully understand the potential roles of these chitinases in plant defense.
In addition to their involvement in plant defense, plant root chitinases may play a role in plant growth and development by digesting chitin in the rhizosphere and releasing nutrients (Chandra et al., 2022). Chitinase activity in the rhizosphere can release nitrogen from chitin-containing residues, such as dead insects or fungal hyphae, providing more nitrogen to plants and potentially reduce the need for synthetic fertilizers.
Further understanding the mechanisms underlying plant-microbe interactions and roles of root chitinases in rhizosphere ecology can have many positive impacts on agriculture including to 1) support the development of more ecologically friendly and sustainable farming techniques, 2) decrease reliance on chemical pesticides and fertilizers, 3) improve nutrient cycling, and 4) support the development of transgenic crops with enhanced chitinase activity as an alternative to chemical fungicides (Oyeleye and Normi, 2018; Wang et al., 2019). This knowledge will also contribute to biotechnology applications, including the design of more effective chitin-degrading enzymes for the industrial processing of chitin (Krolicka et al., 2018; Karnaouri et al., 2019). They can also be developed as in situ biomarkers (Meirinho et al., 2016) in studies to understand root behavior during plant stress and disease so as to improve overall plant health and crop productivity (Yim et al., 2022).
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
SS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. OZ: Conceptualization, Investigation, Supervision, Writing – review & editing. MN-H: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the U.S. Department of Energy, Office of Science, Biological and Environmental Research (BER) through the Ames Laboratory. The Ames Laboratory is operated for the U.S. Department of Energy by Iowa State University under Contract No. DE-AC02-07CH11358.
We thank Dr. Maxwell McReynolds and Prof. Larry Halverson of the department of Plant Pathology and Microbiology, Iowa State University for providing the maize root cDNA used for ZmChi19A gene amplification and the fungal pathogens used in the antifungal assays respectively. We also thank Lee Bendickson for helping in the operation of the FPLC equipment in purification of protein samples, Samuel Ocasio-Rivera for his contribution to the site-directed mutagenesis of chitinase genes, and Dr. Pierre Palo for providing the plasmid used in cloning.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1344142/full#supplementary-material
Alastruey-Izquierdo, A., Melhem, M. S., Bonfietti, L. X., Rodriguez-Tudela, J. L.. (2015). Susceptibility Test for Fungi: Clinical and Laboratorial Correlations In Medical Mycology. Rev Inst Med Trop Sao Paulo 57 (Suppl 19), 57–64. doi: 10.1590/s0036-46652015000700011
Alexandrov, N. N., Brover, V. V., Freidin, S., Troukhan, M. E., Tatarinova, T. V., Zhang, H., et al. (2009). Insights into corn genes derived from large-scale cDNA sequencing. Plant Mol. Biol. 69 (1-2), 179–194. doi: 10.1007/s11103-008-9415-4
Bellasio, C., Farquhar, G. D., Bellasio, C. (2019). A leaf-level biochemical model simulating the introduction of C 2 and C 4 photosynthesis in C 3 rice : gains , losses and metabolite fluxes. New Phytol. 223 (1), 150–156. doi: 10.1111/nph.15787
Bellasio, C., Stuart-Williams, H., Farquhar, G. D., Flexas, J. (2023). C 4 maize and sorghum are more sensitive to rapid dehydration than C 3 wheat and sunflower. New Phytol. 240(6), 2239–2252. doi: 10.1111/nph.19299
Chandra, K., Roy Chowdhury, A., Chatterjee, R., Chakravortty, D. (2022). GH18 family glycoside hydrolase Chitinase A of Salmonella enhances virulence by facilitating invasion and modulating host immune responses. PloS Pathog. 18 (4), e1010407. doi: 10.1371/journal.ppat.1010407
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36 (Web Server issue), W465–W469. doi: 10.1093/nar/gkn180
Deshmukh, F. K., Ben-Nissan, G., Olshina, M. A., Füzesi-Levi, M. G., Polkinghorn, C., Arkind, G., et al. (2023). Allosteric regulation of the 20S proteasome by the Catalytic Core Regulators (CCRs) family. Nat. Commun. 14 (1), 3126. doi: 10.1038/s41467-023-38404-w
Di Savino, A., Foerster, J. M., Ullmann, G. M., Ubbink, M. (2021). The charge distribution on a protein surface determines whether productive or futile encounter complexes are formed. Biochemistry 60 (10), 747–755. doi: 10.1021/acs.biochem.1c00021
Distaso, M., Cea-Rama, I., Coscolín, C., Chernikova, T. N., Tran, H., Ferrer, M., et al. (2023). The mobility of the cap domain is essential for the substrate promiscuity of a family IV esterase from sorghum rhizosphere microbiome. Appl. Environ. Microbiol. 89 (1), e0180722. doi: 10.1128/aem.01807-22
Doan, L. C., Dahanayake, J. N., Mitchell-Koch, K. R., Singh, A. K., Vinh, N. Q. (2022). Probing adaptation of hydration and protein dynamics to temperature. ACS Omega 7 (25), 22020–22031. doi: 10.1021/acsomega.2c02843
Figueroa, M., Vandenameele, J., Goormaghtigh, E., Valerio-Lepiniec, M., Minard, P., Matagne, A., et al. (2016). Biophysical characterization data of the artificial protein Octarellin V.1 and binding test with its X-ray helpers. Data Brief 8, 1221–1226. doi: 10.1016/j.dib.2016.07.036
Funkhouser, J. D., Aronson, N. N., Jr. (2007). Chitinase family GH18: evolutionary insights from the genomic history of a diverse protein family. BMC Evol. Biol. 7, 96. doi: 10.1186/1471-2148-7-96
Garcia-Moreno, B. (2009). Adaptations of proteins to cellular and subcellular pH. J. Biol. 8 (11), 98. doi: 10.1186/jbiol199
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2007). “Protein identification and analysis tools on the expasy server,” in The proteomics protocols handbook. Ed. Walker, J. M. (Humana Press). Available at: www.springer.com.
Gerland, L., Friedrich, D., Hopf, L., Donovan, E. J., Wallmann, A., Erdmann, N., et al. (2020). pH-dependent protonation of surface carboxylate groups in PsbO enables local buffering and triggers structural changes. Chembiochem 21 (11), 1597–1604. doi: 10.1002/cbic.201900739
Giudice, R. D., Nilsson, O., Domingo-Espín, J., Lagerstedt, J. O. (2017). Synchrotron radiation circular dichroism spectroscopy reveals structural divergences in HDL-bound apoA-I variants. Sci. Rep. 7 (1), 13540. doi: 10.1038/s41598-017-13878-z
Grosdidier, A., Zoete, V., Michielin, O. (2011). SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39 (Web Server issue), W270–W277. doi: 10.1093/nar/gkr366
Gupta, R., Brunak, S. (2001). Prediction of glycosylation across the human proteome and the correlation to protein function. Biocomputing 322, 310–322. doi: 10.1142/9789812799623_0029
Haldar, S., Sengupta, S. (2015). Plant-microbe cross-talk in the rhizosphere: insight and biotechnological potential. Open Microbiol. J. 9, 1–7. doi: 10.2174/1874285801509010001
Haxim, Y., Kahar, G., Zhang, X., Si, Y., Waheed, A., Liu, X., et al. (2022). Genome-wide characterization of the chitinase gene family in wild apple ( Malus sieversii ) and domesticated apple ( Malus domestica ) reveals its role in resistance to Valsa Mali. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1007936
Honda, Y., Kitaoka, M., Tokuyasu, K., Sasaki, C., Fukamizo, T., Hayashi, K. (2003). Kinetic studies on the hydrolysis of N-acetylated and N-deacetylated derivatives of 4-methylumbelliferyl chitobioside by the family 18 chitinases ChiA and ChiB from Serratia marcescens. J. Biochem. 133 (2), 253–258. doi: 10.1093/jb/mvg031
Honorato, R. V., Koukos, P. I., Jiménez-García, B., Tsaregorodtsev, A., Verlato, M., Giachetti, A., et al. (2021). Structural biology in the clouds: the WeNMR-EOSC ecosystem. Front. Mol. Biosci. 8. doi: 10.3389/fmolb.2021.729513
Horiuchi, A., Aslam, M., Kanai, T., Atomi, H. (2016). A structurally novel chitinase from the chitin-degrading hyperthermophilic archaeon thermococcus chitonophagus. Appl. Environ. Microbiol. 82 (12), 3554–3562. doi: 10.1128/aem.00319-16
Huang, C. J., Guo, S.-H., Chung, S.-C., Lin, Y.-J., Chen, C.-Y. (2009). Analysis of the involvement of chitin-binding domain of ChiCW in antifungal activity , and engineering a novel chimeric chitinase with high enzyme and antifungal activities analysis of the involvement of chitin-binding domain of ChiCW in antifungal activity , and engineering a novel chimeric chitinase with high enzyme and antifungal activities. J. Microbiol. Biotechnol. 19 (10), 1169–1175.
Hurlburt, N. K., Chen, L. H., Stergiopoulos, I., Fisher, A. J. (2018). Structure of the Cladosporium fulvum Avr4 effector in complex with (GlcNAc)6 reveals the ligand-binding mechanism and uncouples its intrinsic function from recognition by the Cf-4 resistance protein. PloS Pathog. 14 (8), e1007263. doi: 10.1371/journal.ppat.1007263
Ju, Y., Wang, X., Guan, T., Peng, D., Li, H. (2016). Versatile glycoside hydrolase family 18 chitinases for fungi ingestion and reproduction in the pinewood nematode Bursaphelenchus xylophilus. Int. J. Parasitol. 46 (12), 819–828. doi: 10.1016/j.ijpara.2016.08.001
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi: 10.1038/s41586-021-03819-2
Jurrus, E., Engel, D., Star, K., Monson, K., Brandi, J., Felberg, L. E., et al. (2018). Improvements to the APBS biomolecular solvation software suite. Protein Sci. 27 (1), 112–128. doi: 10.1002/pro.3280
Karnaouri, A., Antonopoulou, I., Zerva, A., Dimarogona, M., Topakas, E., Rova, U., et al. (2019). Thermophilic enzyme systems for efficient conversion of lignocellulose to valuable products: Structural insights and future perspectives for esterases and oxidative catalysts. Bioresour Technol. 279, 362–372. doi: 10.1016/j.biortech.2019.01.062
Kc, K. B., Dias, G. M., Veeramani, A., Swanton, C. J., Fraser, D., Steinke, D., et al. (2018). When too much isn't enough: Does current food production meet global nutritional needs? PloS One 13 (10), e0205683. doi: 10.1371/journal.pone.0205683
Kim, S., Sureka, H. V., Kayitmazer, A. B., Wang, G., Swan, J. W., Olsen, B. D. (2020). Effect of protein surface charge distribution on protein-polyelectrolyte complexation. Biomacromolecules 21 (8), 3026–3037. doi: 10.1021/acs.biomac.0c00346
Kotb, E., Alabdalall, A. H., Alghamdi, A. I., Ababutain, I. M., Aldakeel, S. A., Zuwaid, S. K. A., et al. (2023). Screening for chitin degrading bacteria in the environment of Saudi Arabia and characterization of the most potent chitinase from Streptomyces variabilis Am1. Sci. Rep. 13 (1), 11723. doi: 10.1038/s41598-023-38876-2
Krolicka, M., Hinz, S. W. A., Koetsier, M. J., Joosten, R., Eggink, G., Van Den Broek, L. A. M., et al. (2018). Chitinase chi1 from myceliophthora thermophila C1, a thermostable enzyme for chitin and chitosan depolymerization. J. Agric. Food Chem. 66 (7), 1658–1669. doi: 10.1021/acs.jafc.7b04032
Kuo, C. J., Liao, Y. C., Yang, J. H., Huang, L. C., Chang, C. T., Sung, H. Y. (2008). Cloning and characterization of an antifungal class III chitinase from suspension-cultured bamboo ( Bambusa oldhamii ) cells. J. Agric. Food Chem. 56 (23), 11507–11514. doi: 10.1021/jf8017589
Liu, Y., Yang, X., Gan, J., Chen, S., Xiao, Z. X., Cao, Y. (2022). CB-Dock2: improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 50 (W1), W159–w164. doi: 10.1093/nar/gkac394
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48 (D1), D265–d268. doi: 10.1093/nar/gkz991
Ma, W., Muthreich, N., Liao, C., Franz-Wachtel, M., Schütz, W., Zhang, F., et al. (2010). The mucilage proteome of maize (Zea mays L.) primary roots. J. Proteome Res. 9 (6), 2968–2976. doi: 10.1021/pr901168v
Martínez-Caballero, S., Cano-Sánchez, P., Mares-Mejía, I., Díaz-Sánchez, A. G., Macías-Rubalcava, M. L., Hermoso, J. A., et al. (2014). Comparative study of two GH19 chitinase-like proteins from Hevea brasiliensis, one exhibiting a novel carbohydrate-binding domain. FEBS J. 281 (19), 4535–4554. doi: 10.1111/febs.12962
Meirinho, S. G., Dias, L. G., Peres, A. M., Rodrigues, L. R. (2016). Voltammetric aptasensors for protein disease biomarkers detection: A review. Biotechnol. Adv. 34 (5), 941–953. doi: 10.1016/j.bioteChadv.2016.05.006
Nilsen-Hamilton, M., Hamilton, R. (1987). Detection of proteins induced by growth regulators. Methods Enzymology 147, 427–444. doi: 10.1016/0076-6879(87)47132-2
Nilsen-Hamilton, M., Shapiro, J. M., Massoglia, S. L., Hamilton, R. T. (1980). Selective stimulation by mitogens of incorporation of 35S-methionine into a family of proteins released into the medium by 3T3 cells. Cell 20 (1), 19–28. doi: 10.1016/0092-8674(80)90230-5
Oliveira, S. T., Azevedo, M. I. G., Cunha, R. M. S., Silva, C. F. B., Muniz, C. R., Monteiro-Júnior, J. E., et al. (2020). Structural and functional features of a class VI chitinase from cashew (Anacardium occidentale L.) with antifungal properties. Phytochemistry 180, 112527. doi: 10.1016/j.phytochem.2020.112527
Onaga, S., Taira, T. (2008). A new type of plant chitinase containing LysM domains from a fern ( Pteris ryukyuensis ): Roles of LysM domains in chitin binding and antifungal activity. Glycobiology 18 (5), 414–423. doi: 10.1093/glycob/cwn018
Osinde, C., Sobhy, I. S., Wari, D., Dinh, S. T., Hojo, Y., Osibe, D. A., et al. (2023). Comparative analysis of sorghum ( C4 ) and rice ( C3 ) plant headspace volatiles induced by artificial herbivory. Plant Signaling Behav. 18(1), 2243064. doi: 10.1080/15592324.2023.2243064
Oyeleye, A., Normi, Y. M. (2018). Chitinase: diversity, limitations, and trends in engineering for suitable applications. Biosci. Rep. 38 (4), BSR2018032300. doi: 10.1042/bsr20180323
Panja, A. S., Maiti, S., Bandyopadhyay, B. (2020). Protein stability governed by its structural plasticity is inferred by physicochemical factors and salt bridges. Sci. Rep. 10 (1), 1822. doi: 10.1038/s41598-020-58825-7
Patel, A. K., Singh, V. K., Yadav, R. P., Moir, A. J., Jagannadham, M. V. (2009). ICChI, a glycosylated chitinase from the latex of Ipomoea carnea. Phytochemistry 70 (10), 1210–1216. doi: 10.1016/j.phytochem.2009.07.005
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25 (13), 1605–1612. doi: 10.1002/jcc.20084
Pingali, P. L. (2012). Green revolution: impacts, limits, and the path ahead. Proc. Natl. Acad. Sci. U.S.A. 109 (31), 12302–12308. doi: 10.1073/pnas.0912953109
Puškárová, A., Bučková, M., Kraková, L., Pangallo, D., Kozics, K. (2017). The antibacterial and antifungal activity of six essential oils and their cyto/genotoxicity to human HEL 12469 cells. Sci. Rep. 7 (1), 8211. doi: 10.1038/s41598-017-08673-9
Rajninec, M., Jopcik, M., Danchenko, M., Libantova, J. (2020). Biochemical and antifungal characteristics of recombinant class I chitinase from Drosera rotundifolia. Int. J. Biol. Macromol 161, 854–863. doi: 10.1016/j.ijbiomac.2020.06.123
Renaud, S., Dussutour, A., Daboussi, F., Pompon, D. (2023). Characterization of chitinases from the GH18 gene family in the myxomycete Physarum polycephalum. Biochim. Biophys. Acta Gen. Subj 1867 (6), 130343. doi: 10.1016/j.bbagen.2023.130343
Rizzo, D. M., Lichtveld, M., Mazet, J. A. K., Togami, E., Miller, S. A. (2021). Plant health and its effects on food safety and security in a One Health framework: four case studies. One Health Outlook 3, 6. doi: 10.1186/s42522-021-00038-7
Roy, S., Chakraborty, A., Chakraborty, R. (2021). Understanding the potential of root microbiome influencing salt-tolerance in plants and mechanisms involved at the transcriptional and translational level. Physiol. Plant 173 (4), 1657–1681. doi: 10.1111/ppl.13570
Sierra-Gómez, Y., Rodríguez-Hernández, A., Cano-Sánchez, P., Gómez-Velasco, H., Hernández-Santoyo, A., Siliqi, D., et al. (2019). A biophysical and structural study of two chitinases from Agave tequilana and their potential role as defense proteins. FEBS J. 286 (23), 4778–4796. doi: 10.1111/febs.14993
Sievers, F., Higgins, D. G. (2014). Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 1079, 105–116. doi: 10.1007/978-1-62703-646-7_6
Singappuli-Arachchige, D., Feng, S., Wang, L., Palo, P. E., Shobade, S. O., Thomas, M., et al. (2022). The magnetosome protein, mms6 from magnetospirillum magneticum strain AMB-1, is a lipid-activated ferric reductase. Int. J. Mol. Sci. 23 (18), 10305. doi: 10.3390/ijms231810305
Steentoft, C., Vakhrushev, S. Y., Joshi, H. J., Kong, Y., Vester-Christensen, M. B., Schjoldager, K. T., et al. (2013). Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32 (10), 1478–1488. doi: 10.1038/emboj.2013.79
Sukprasirt, P., Wititsuwannakul, R. (2014). A chitinolytic endochitinase and β-N-acetylglucosaminidase-based system from Hevea latex in generating N-acetylglucosamine from chitin. Phytochemistry 104, 5–11. doi: 10.1016/j.phytochem.2014.04.001
The, C. C. (2018). Ten years of CAZypedia: a living encyclopedia of carbohydrate-active enzymes. Glycobiology 28 (1), 3–8. doi: 10.1093/glycob/cwx089
Thimoteo, S. S., Glogauer, A., Faoro, H., De Souza, E. M., Huergo, L. F., Moerschbacher, B. M., et al. (2017). A broad pH range and processive chitinase from a metagenome library. Braz. J. Med. Biol. Res. 50 (1), e5658. doi: 10.1590/1414-431x20165658
USDA (2023) Rice Sector at a Glance. Available at: https://www.ers.usda.gov/topics/crops/rice/rice-sector-at-a-glance/ (Accessed November 2023).
Vallina Estrada, E., Zhang, N., Wennerström, H., Danielsson, J., Oliveberg, M. (2023). Diffusive intracellular interactions: On the role of protein net charge and functional adaptation. Curr. Opin. Struct. Biol. 81, 102625. doi: 10.1016/j.sbi.2023.102625
Vanhoye, D., Bruston, F., El Amri, S., Ladram, A., Amiche, M., Nicolas, P. (2004). Membrane association, electrostatic sequestration, and cytotoxicity of Gly-Leu-rich peptide orthologs with differing functions. Biochemistry 43 (26), 8391–8409. doi: 10.1021/bi0493158
van Munster, J. M., Sanders, P., Ten Kate, G. A., Dijkhuizen, L., Van Der Maarel, M. J. (2015). Kinetic characterization of Aspergillus Niger chitinase CfcI using a HPAEC-PAD method for native chitin oligosaccharides. Carbohydr Res. 407, 73–78. doi: 10.1016/j.carres.2015.01.014
Vega-Arreguín, J. C., Ibarra-Laclette, E., Jiménez-Moraila, B., Martínez, O., Vielle-Calzada, J. P., Herrera-Estrella, L., et al. (2009). Deep sampling of the Palomero maize transcriptome by a high throughput strategy of pyrosequencing. BMC Genomics 10, 299. doi: 10.1186/1471-2164-10-299
Wang, Y. J., Jiang, W. X., Zhang, Y. S., Cao, H. Y., Zhang, Y., Chen, X. L., et al. (2019). Structural insight into chitin degradation and thermostability of a novel endochitinase from the glycoside hydrolase family 18. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02457
Yan, Y., Ryu, Y., Dechant, B., Li, B., Kim, J. (2023). Dark respiration explains nocturnal stomatal conductance in rice regardless of drought and nutrient stress. Plant Cell Environ. 46(12), 3748–3749. doi: 10.1111/pce.14710
Yim, B., Ibrahim, Z., Rüger, L., Ganther, M., Maccario, L., Sørensen, S. J., et al. (2022). Soil texture is a stronger driver of the maize rhizosphere microbiome and extracellular enzyme activities than soil depth or the presence of root hairs. Plant Soil 478 (1), 229–251. doi: 10.1007/s11104-022-05618-8
Yokoyama, S., Iida, Y., Kawasaki, Y., Minami, Y., Watanabe, K., Yagi, F. (2009). The chitin-binding capability of Cy-AMP1 from cycad is essential to antifungal activity. J. Pept. Sci. 15 (7), 492–497. doi: 10.1002/psc.1147
Zhang, Y., Skolnick, J. (2005). TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33 (7), 2302–2309. doi: 10.1093/nar/gki524
Keywords: chitinase, hydrolases, chitin-binding domain, C-terminal domain, anti-fungal activity, rhizosphere
Citation: Shobade SO, Zabotina OA and Nilsen-Hamilton M (2024) Plant root associated chitinases: structures and functions. Front. Plant Sci. 15:1344142. doi: 10.3389/fpls.2024.1344142
Received: 25 November 2023; Accepted: 05 January 2024;
Published: 01 February 2024.
Edited by:
Benedetta Mattei, University of L’Aquila, ItalyReviewed by:
Fredy Albuquerque Silva, Universidade Federal de Viçosa, BrazilCopyright © 2024 Shobade, Zabotina and Nilsen-Hamilton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marit Nilsen-Hamilton, bWFyaXRAaWFzdGF0ZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.