
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 December 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1308354
The GDP-D-mannose pyrophosphorylase (GMP) and microtubule severing enzyme KATANIN (KTN) are crucial for wood formation. Although functional identification has been performed in Arabidopsis, few comprehensive studies have been conducted in forest trees. In this study, we discovered 8 CcGMP and 4 CcKTN genes by analyzing the whole genome sequence of Corymbia citriodora. The chromosomal location, genome synteny, phylogenetic relationship, protein domain, motif identification, gene structure, cis-acting regulatory elements, and protein-interaction of CcGMP and CcKTN were all investigated. KTN has just one pair of segmentally duplicated genes, while GMP has no duplication events. According to gene structure, two 5’ UTRs were identified in CcGMP4. Furthermore, there is no protein-interaction between KTN and GMP. Based on real-time PCR, the expression of most genes showed a positive connection with DBH diameters. In addition, the expression of CcGMP4 and CcKTN4 genes were greater in different size tree, indicating that these genes are important in secondary xylem production. Overall, this findings will enhance our comprehension of the intricacy of CcGMP&CcKTN across diverse DBHs and furnish valuable insights for future functional characterization of specific genes in C. citriodora.
Wood (secondary xylem) originates from vascular cambium (Figures 1A, B), and its formation goes through a series of developmental processes, including secondary xylem mother cell differentiation, cell expansion, mass deposition of secondary walls and pit formation, programmed cell death, and heartwood formation (Plomion et al., 2001; Wang et al., 2021). The cambium consists of two meristem protocells, namely, fusiform initials and ray initials (Figure 1C). In angiosperms, fusiform initials mainly form parenchyma, ducts, and fibers, while ray initials produce rays, which mainly transport nutrients (Larson, 2012). At present, the genes related to vascular cambium in forest wood have been widely studied, including vascular cambium cell division and differentiation, cell wall synthesis (Ye and Zhong, 2015; Wang et al., 2021; Guo et al., 2022; Kim et al., 2022), etc. Therefore, given the role of vascular cambium in the secondary growth of trees, uncovering more genes related to vascular cambium is key to gaining insight into wood formation.
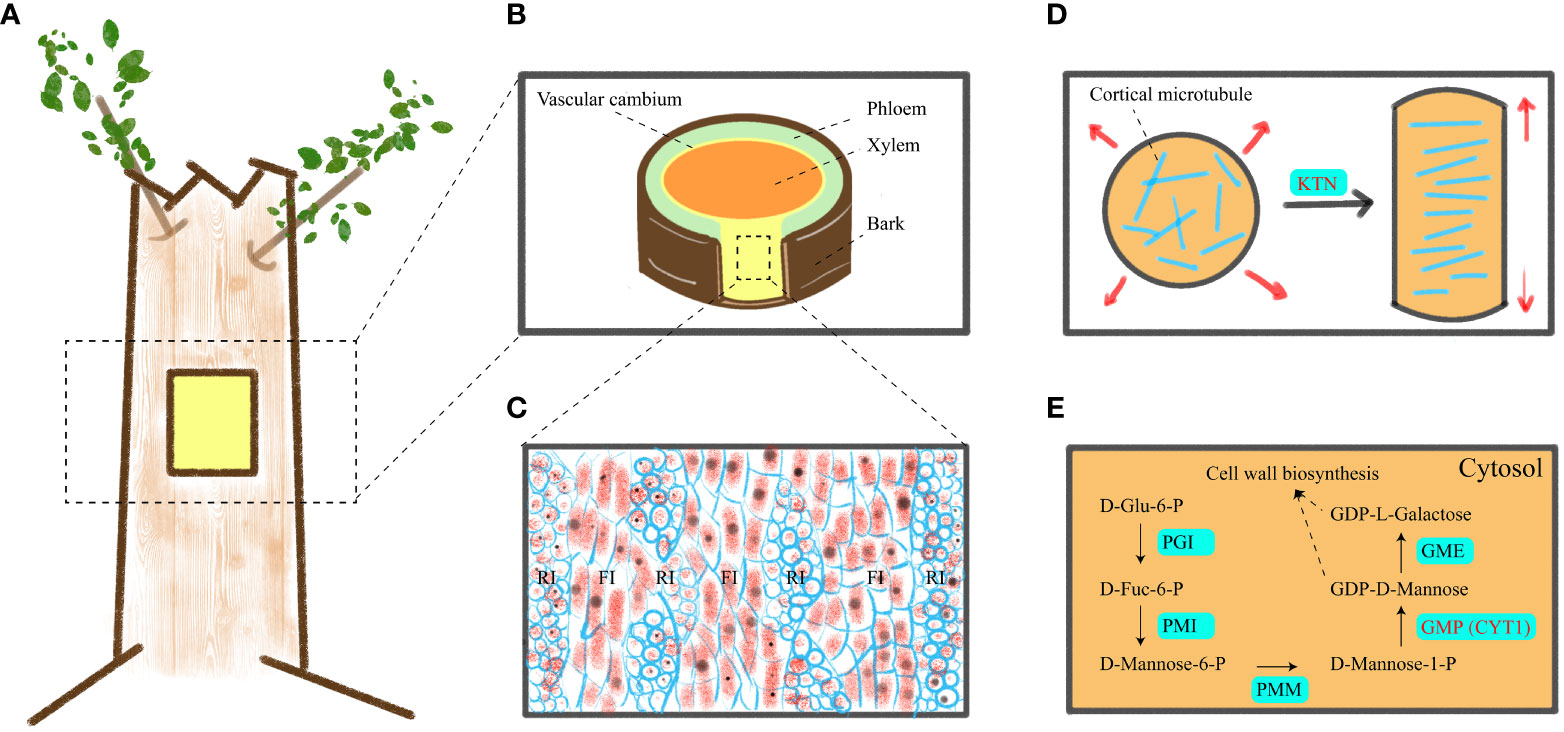
Figure 1 A schematic of vascular cambium, cell elongation mechanism and GDP-D-mannose synthesis. (A) Location of materials. (B) Transverse cutting of the stem. (C) Tangential longitudinal section of the cambium zone in a stem, revealing its anatomical structure. (D) Transverse cMT arrays support anisotropic elongation of plant cells. (E) Mechanism of GDP-D-mannose synthesis in cytoplasm. RI, FI, PGI, PMI, PMM, GMP, and GME indicated ray initials, fusiform initials, phosphoglucose isomerase, phosphomannose isomerase, phosphomannomutase, GDP-D-mannose pyrophosphorylase and GDP-D-mannose 3′, 5′ epimerase, respectively.
Wood consists of secondary cell walls and is composed mainly of three polymers: cellulose; Hemicellulose; and lignin. Cellulose consists of β-1, 4-linked chains of glucose units. It is synthesized by cellulose synthase complex (CSC) outside the plasma membrane. CSC moves within the plasma membrane and connects glucose subunits to each other to form cellulose microfibril. The movement path of CSC in the plasma membrane is mainly guided by cortical microtubules (cMT) (Paredez et al., 2006). Hemicellulose is composed of various chains, including glucomannan that links glucose and mannose residues via β-1,4 bonds (Lerouxel et al., 2006). Therefore, it can be seen that cMT and mannose play pivotal roles in wood formation (Mellerowicz and Sundberg, 2008).
Cytoskeleton and cell wall polysaccharides play an important physiological role in wood formation. Cortical microtubules control anisotropic cell expansion by directing the direction of cellulose microfibril (Baskin, 2001; Baskin, 2005). Transverse cMT arrangement causes cell growth (Figure 1D), while longitudinal cMT arrangement causes cell thickening (Kato et al., 2022). Furthermore, cMT are closely associated with the cell wall, and members of the cortical microtubule-associated protein family play a crucial role in regulating secondary cell wall synthesis during both secondary wall deposition and pit formation (Pesquet et al., 2010). The absence of genes encoding microtubule-associated proteins, such as CORD1 (Cortical Microtubule Disordering 1), resulted in smaller pits within secondary cell walls (Sasaki et al., 2017). In Arabidopsis, MOR1 (Microtubule organization 1), ABS6 (Abnormal shoot 6), SAV4 (Shade avoidance 4), KTN1 (Katanin 1), PP2A (Protein phosphatase 2A) genes are involved in the physiological activities of cMT (Li et al., 2021; Ren et al., 2022; Liu and Yu, 2023). However, these genes have been less studied in forest trees. Mannose is a constituent of the plant cell wall and is widely distributed in various plants. In the Smirnoff-Wheeler pathway (Figure 1E), GDP-D-mannose and L-galactose are utilized for cell wall metabolism, with GDP-D-mannose being synthesized by GDP-D-mannose pyrophosphorylase (GMP) (Lukowitz et al., 2001; Smirnoff et al., 2001). Additionally, GDP-D-mannose participates in ascorbic acid (Vitamin C) biosynthesis (Liu et al., 2022) and plays a crucial role in plant stress resistance. In previous studies, GMP activity is also affected by KJC1 (KONJAC 1), KJC2, CYT1 (Cytokinesis defective 1), AMR1L1 (Ascorbate Mannose pathway Regulator 1 Like 1), CSN5B (COP9 signalosome subunit 5B), in which CYT1 is closely related to cellulose synthesis (Lukowitz et al., 2001; Wang et al., 2013; Sawake et al., 2015; Ma et al., 2022). However, the mechanism of KTN1 and CYT1 genes in wood formation is still unclear, so we selected these two genes for in-depth study to identify whether they are related to wood formation.
KTN1 is a heterodimer microtubule (MT) severing protein that uses energy from ATP hydrolysis to generate internal breaks along MT. Katanin p60 is one of the two subunits with ATPase and MT binding and severing activity, which is critical for cMT ordered and anisotropic cell expansion (Li et al., 2021), while the p80 subunit is responsible for targeting Katanin to certain subcellular sites (Burk et al., 2007). Microtubules are closely related to cellulose complex enzymes (Crowell et al., 2009; Lei et al., 2014), in which cellulose complex enzymes mainly control cell wall synthesis, while cMT are believed to alter lipid composition in the plasma membrane, thus affecting cellulose synthesis (Fujita et al., 2012). In addition, cMT also regulate the speed of cellulose synthase (Bischoff et al., 2011) and the movement track of cellulose synthase (Paredez et al., 2006). Microtubule severing protein Katanin can change the direction of cMT by cutting microtubules, thus indirectly affecting cell wall synthesis and changing cell morphology (Lindeboom et al., 2013). Among all individuals related to mutations in microtubule severing enzymes in A. thaliana, the majority exhibit cMT arrangement disorders, cell elongation defects, hypocotyl shortening, growth retardation and plant dwarfism (Burk et al., 2001; Webb et al., 2002; Bouquin et al., 2003). Notably, ktn mutants in A. thaliana have a significant impact on cellulose microfibrils (Burk et al., 2001), while overexpression of KTN leads to abnormal secondary walls (Zhou et al., 2007). The main economic component of forest is wood, and MT is closely related to the formation of wood (Pesquet and Lloyd, 2011). It can be seen that KTN1 gene has certain physiological significance for wood formation.
GMP1 (also known as VTC1、CYT1) is associated with mannose synthesis, encoding GDP-D-mannose pyrophosphorylase (Conklin et al., 1999), which is a key gene in the complex process of ascorbic acid (AsA) synthesis (Terzaghi and De Tullio, 2022). In A. thaliana, cyt mutants cause incomplete cell walls and excessive callose accumulation (Nickle and Meinke, 1998), especially the depletion of significant amounts of mannose from the cell wall (Lukowitz et al., 2001). Deletion of genes encoding GMP has also been found in rice, resulting in decreased mannose and galactose, as well as decreased cellulose synthase (Lamanchai et al., 2022). Overexpression of GMP gene has also been reported in various species, most of which are related to abiotic stress studies. It has been discovered that overexpression of GMP in A. thaliana compensates for the sensitivity of myb30 and myc2-2 mutants to submergence, and improves antioxidant biosynthesis (Yuan et al., 2017; Xie et al., 2023). Furthermore, overexpression of GMP increases AsA accumulation in rice, strawberry and soybean (Qin et al., 2016; Xue et al., 2018; Lin et al., 2021). Although GMP is highly expressed in the vascular tissues of trees (xylem is more highly expressed than phloem) (Nairn et al., 2008), the relationship between GMP gene and wood formation needs further study.
Corymbia citriodora exhibits rapid growth (de Araujo et al., 2021) and high wood density (Massuque et al., 2022), making it a widely cultivated species (Hung et al., 2016; de Souza et al., 2020). Its timber is also commercially valuable. In comparison to other Eucalyptus trees, C. citriodora can be utilized as premium-grade charcoal for steel production. (Couto et al., 2015; Massuque et al., 2022). Taken together, a larger diameter at breast height (DBH) of C. citriodora can bring more economic benefits. However, we found that trees of the same species and age showed different DBH, among which the number of C. citriodora with larger DBH was relatively small. Therefore, it is crucial to investigate whether the expression levels of KTN and GMP genes are associated with wood formation at different stages of DBH.
Nevertheless, no comprehensive investigation into the GMP&KTN genes of C. citriodora has been documented to date. However, with the unveiling of the C. citriodora genome (Healey et al., 2021), it is now possible to systematically scrutinize the potential functions of these genes in this species. In this study, 8 CcGMP and 4 CcKTN genes were identified from this species, two of which (CcGMP4 and CcKTN4) are essential for wood formation. The results of our study will provide a basis for further functional identification of CcGMP&CcKTN genes in C. citriodora.
C. citriodora (Phytozome genome ID 507) and A. thaliana (447) gene sequence files, gene annotation files, protein sequences, and coding DNA sequence files were downloaded from the Phytozome (https://phytozome-next.jgi.doe.gov/) website. The protein sequences of A. thaliana CYT1 (At2g39770) and KTN1 (At1g80350) were compared with the C. citriodora genome to screen out candidate genes on the NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) website. Among them, all the selected GMP genes were subsequently identified, while the KTN genes were further artificially screened by using TBtools (Chen et al., 2020) to construct evolutionary trees. Putative GMP&KTN sequences were uploaded to Pfam (http://pfam.xfam.org/) and SMART (http://smart.embl-heidelberg.de/) databases to identify sequences protein domains (AtCYT1: PF00483, CL11394; AtKTN1: PF0004, CL38936). Due to the involvement of splice variants in the candidate genes, specific primers were designed by cutting a portion of the gene sequences based on the splicing pattern to distinguish different splice isoforms and analyze their expression levels. The protein sequences of CcGMP&CcKTN in the attached table (Table S1) can be referred to. Subsequently, the molecular weights (MWs), isoelectric points (pI), instability index (II), protein length and grand average of hydropathicity (GRAVY) of CcGMP&CcKTN proteins were calculated in the ExPasy website (https://web.expasy.org/protparam/). The subcellular location of CcGMP&CcKTN proteins was predicted by WoLF PSORT (https://www.genscript.com/wolf-psort.html?src=leftbar).
Chromosomal locations of all CcGMP&CcKTN family genes were confirmed using the C. citriodora genome annotation file downloaded from Phytozome and mapped using the TBtools. C. citriodora gene sequence files were used to extract GC ratio and N ratio by Tbtools (parameters: window size is 10000; window overlap is 1000). The MCScanX software (Wang et al., 2012) (default parameters) was used to perform inter-species synteny analysis between C. citriodora and two representative plants, A. thaliana and E. grandis, as well as intra-species synteny analysis of C. citriodora. Homology relationships were also visualized by Tbtools software. Finally, the collinearity corresponding to CcKTN&CcGMP was highlighted.
GMP&KTNs from A. thaliana, O. sativa and E. grandis were used for the classification of GMP&KTNs family in C. citriodora. These putative GMP&KTNs of other pattern plants were obtained by AtCYT1&AtKTN1 blast to their whole protein sequences. Subsequently, these putative protein sequences were submitted to the CDD (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) for identification of the conserved domain (search for Pfam). ClustalW program (Thompson et al., 2002) (version 2.1; http://www.clustal.org/) was used for multiple sequence alignment, and TBtools was used to trim the data after sequence alignment. An unrooted maximum likelihood (ML) phylogenetic tree based on GMP&KTNs protein sequence was constructed by using the MEGA11.0 program (Tamura et al., 2021), with 1,000 bootstrap replicates under J+G+I+F model. The resulted ML tree was visualized in Chiplot (https://www.chiplot.online/tvbot.html). The subgroups were named following available information in Arabidopsis and rice (https://www.arabidopsis.org/; https://bis.zju.edu.cn/ricenetdb/).
Conserved protein domains of the CcGMP&CcKTNs were blasted in the NCBI-CDD website (search for CDD). The CcGMP&CcKTN family gene structures were displayed by comparing the coding and genomic sequences with the TBtools. The obtained conserved motifs of the CcGMP&CcKTN family proteins were analyzed through the online website MEME (https://meme-suite.org/). Motif sequences were identified by InterPro online tool (https://www.ebi.ac.uk/interpro/).
The promoter sequences of 2,000 bp upstream of each CcGMP&CcKTN gene-coding region were extracted from the genome data. The PlantCARE online program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to search for assumed cis-acting elements. The cis-elements were annotated and visualized in a figure through the construction of a physical gene map using TBtools. The number of cis-elements associated with plant growth and development, phytohormone responsive, stress responsive and light responsive were also counted.
To reveal the structure and function of the CcGMP&CcKTNs protein interaction network, we performed protein interaction analysis based on known protein sequences from E. grandis. The protein sequences of CcGMP&CcKTNs were submitted to the STRING website (https://cn.string-db.org/) for analysis of protein interactions, and the predicted GO annotation results were visualized using R language.
Cambium of 6.5-year-old C. citriodora with large (20.7–23.6 cm), medium (14.8–15.8 cm), and small (9.6–10.8 cm) diameters was collected at breast height (DBH) between 10 a.m. and 1 p.m. The DBH of different sizes were treated as three groups with six samples per treatment. All samples were collected and frozen in liquid nitrogen, then stored at −80 °C until used.
Total RNA was isolated using the Trelief RNAprep Pure Plant Plus Kit (Tsingke Biotech, Beijing China) according to the manufacturer’s instructions. Then, the RNA was used for first-strand cDNA synthesis with reverse transcriptase and the reverse transcription was performed using 1 mg of total RNA and PrimeScrip RT reagent Kit with gDNA Eraser (TaKaRa, Tokyo). Subsequently, cDNA was used as a template for qRT-PCR analysis using Premier 5.0 primers based on C. citriodora internal reference gene sequences (Table S2). The specificity of primers to their target genes was evaluated on the website EnsemblPlants (http://plants.ensembl.org/hordeum_vulgare/info/index). All of the primers were synthesized by TSINGKE Biotechnology Co., Ltd. (Beijing, China). A CFX96 real-time PCR system (Bio-RAD, Laboratories, Hercules, CA, USA) was used for qRT-PCR analysis. Each reaction mixture contained TransStart Top Green qPCR SuperMix 10 μL, cDNA template 1.5 μL, upstream primer 0.4 μL, downstream primer 0.4 μL, and sterile distilled water 7.7 μL. The reaction conditions were as follows: ① 94 °C, 30 s; ② 94 °C, 5 s; ③ 60 °C, 15 s; ④ 72 °C, 10 s. The analysis was completed after 40 cycles of reactions ②–④.
The relative expression levels of each treatment were calculated by the 2−ΔΔCT method, and the mean ± standard deviation (SD) values were calculated from four independent biological replicates. The inner reference gene actin was utilized for data normalization, with CcGMP7&CcKTN2 and the small DBH group serving as external controls. Differences in gene expression levels were analyzed via analysis of variance (ANOVA) followed by Student’s t-test in R software. After expression analysis, function verification of differentially expressed genes was performed based on GO annotation results.
This study used the protein sequences of CYT1 and KTN1 genes in A. thaliana to identify homologous genes in C. citriodora using the BLAST method. After analyzing the conserved domains and removing redundant sequences, a total of 8 putative CcGMP genes and 4 CcKTN genes were identified, and the original splicing was manually trimmed. For convenience, we named the 8 CcGMP and 4 CcKTN genes based on their chromosomal location. The detailed information of all the studied genes was listed (Figure 2).
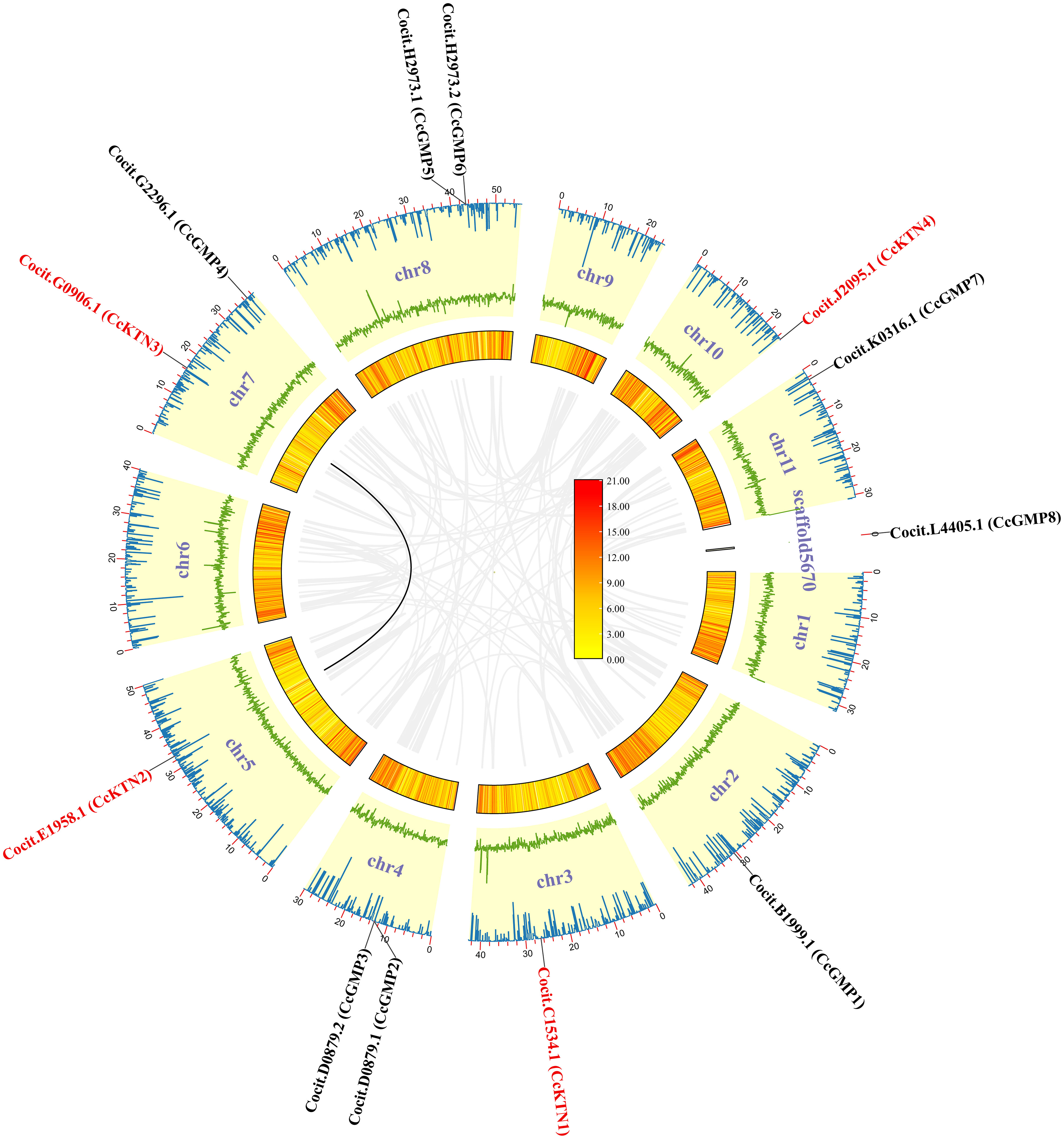
Figure 2 CcGMP&CcKTNs chromosomal localization and duplication events in the C. citriodora genome. From the inner to outer: gray line indicates all synteny blocks; black line highlights the collinear of CcGMP&CcKTNs pairs; heatmap for gene density profile; green line plot for GC ratio; blue line plot for N ratio.
According to our analysis (Table 1), the predicted protein length, MW, pI, and IIV of the CcGMPs ranged from 257 to 726 AA, 28.31 to 81.62 kDa, 4.30 to 8.56, and 30.29 to 44.27, respectively. Additionally, CcGMP4 exhibited the highest degree of homology to AtCYT1 protein in the protein alignment. The predicted subcellular localization of CcGMPs (except for splice form CcGMP3, 6) were in the cytoplasm. Furthermore, CcGMP3, 4, and 7 are hydrophobic proteins, while the others are hydrophilic.
Moreover, we also predicted that the length of CcKTN proteins ranged from 432 to 658 AA, with a MW of 47.93 to 72.94 kDa and a pI of 6.32 to 8.82. The IIV range from 30.29 to 44.27, with CcKTN4 being the most stable protein. Furthermore, CcKTN4 showed the highest homology with AtKTN1. All 4 CcKTNs are hydrophilic. The subcellular localization prediction of CcKTN was not entirely reliable. CcKTN1 may be localized in either chloroplasts or the nucleus, while CcKTN3 is likely to be found in the nucleus and CcKTN4 could potentially reside in either mitochondria or the cytoplasm. In general, microtubule severing regulation tends to occur within the cytoplasm, such as AtKTN1.
To produce visual representations of the chromosomal positions of CcGMP&CcKTN genes, an analysis of the genomic distribution in chromosomes was conducted to determine their physical locations. This information was then used to generate the graphics (Figure 2). The result revealed that all CcGMP&CcKTNs, except for CcGMP8, were unevenly distributed on chromosomes in C. citriodora. Due to the important effect on functional differentiation and gene expansion, gene duplication events among CcGMP&CcKTNs were also investigated. In this study, one pair of segmental duplicated genes (CcKTN2/3) was revealed and distributed (Figure 2). To determine the selection constraints on the duplicated CcKTN2/3, we estimated the Ka/Ks ratio using the Tbtools and found that the value of the Ka/Ks ratio of CcKTN2/3 was less than 1 (Table S3). These results indicated that these duplicated genes underwent strong purification/negative selection pressure, and almost no mutation occurred after duplication. To better understand the composition and structure of C. citriodora genome sequence, N ratio, GC ratio, and gene density were used for analysis. N ratio value indicated that the quality of C. citriodora genome sequence is poor and there may be missing, mismatched, or other abnormal sequences. GC content analysis results showed that the average GC content of every ten thousand bases is 0.4. Some segments of chromosomes 3, 8, and 6 have a higher GC content. Gene density distribution showed that most of the genes are concentrated at one or both ends of the chromosome.
Based on a comparative micro-syntenic map of C. citriodora versus two representative plant species (including A. thaliana and E. grandis), we found 11 pairs of GMP&KTN orthologous genes between C. citriodora and E. grandis, and 5 pairs between C. citriodora and A. thaliana (Figure 3). According to information collected on the TAIR website (https://www.arabidopsis.org/), CcGMP4, 7 and CcKTN4 may be involved in cellulose synthesis. Detailed genetic pairs are shown in Table S4.
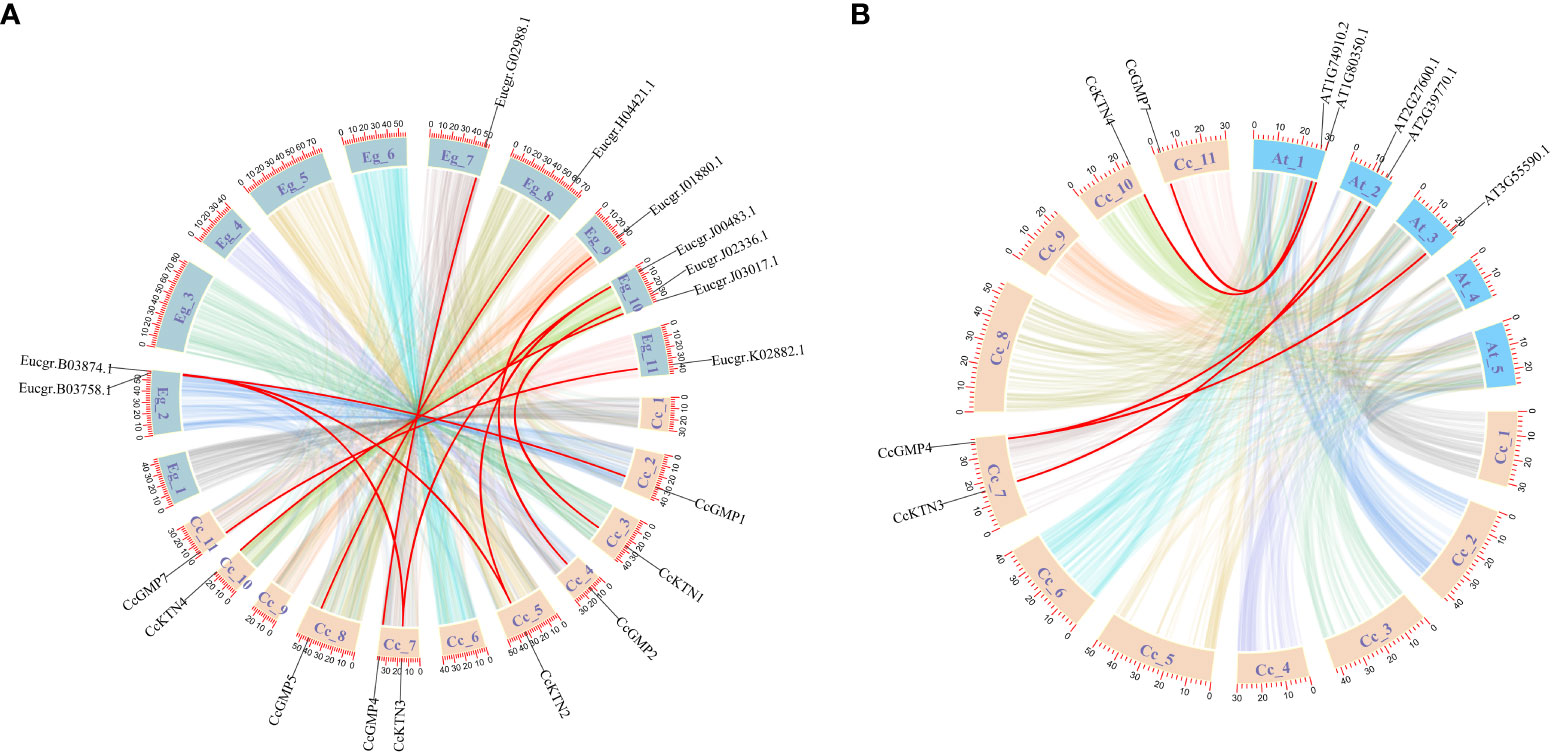
Figure 3 Synteny analysis of CcGMP&CcKTN genes between C. citriodora and two representative plant species (A. thaliana and E. grandis). (A) Syntenic relationship between C. citriodora and Eucalyptus grandis. (B) Syntenic relationship between C. citriodora and Arabidopsis thaliana. The colorful lines at the bottom indicate the collinear blocks within the C. citriodora and other plant genomes. The red lines indicate the pairs of CcGMP&CcKTN genes.
The protein sequences of GMP&KTNs from 4 species O. sativa, A. thaliana, E. grandis, and C. citriodora were used to investigate the phylogenetic relationship. An unrooted cladogram suggested that these GMP or KTNs were clustered into 4 categories and these categories contained members from both monocot and dicot plants except GMP-Group I (Figures 4, 5). According to previous studies on O. sativa and A. thaliana, we can speculate the function of GMP&KTNs in C. citriodora.
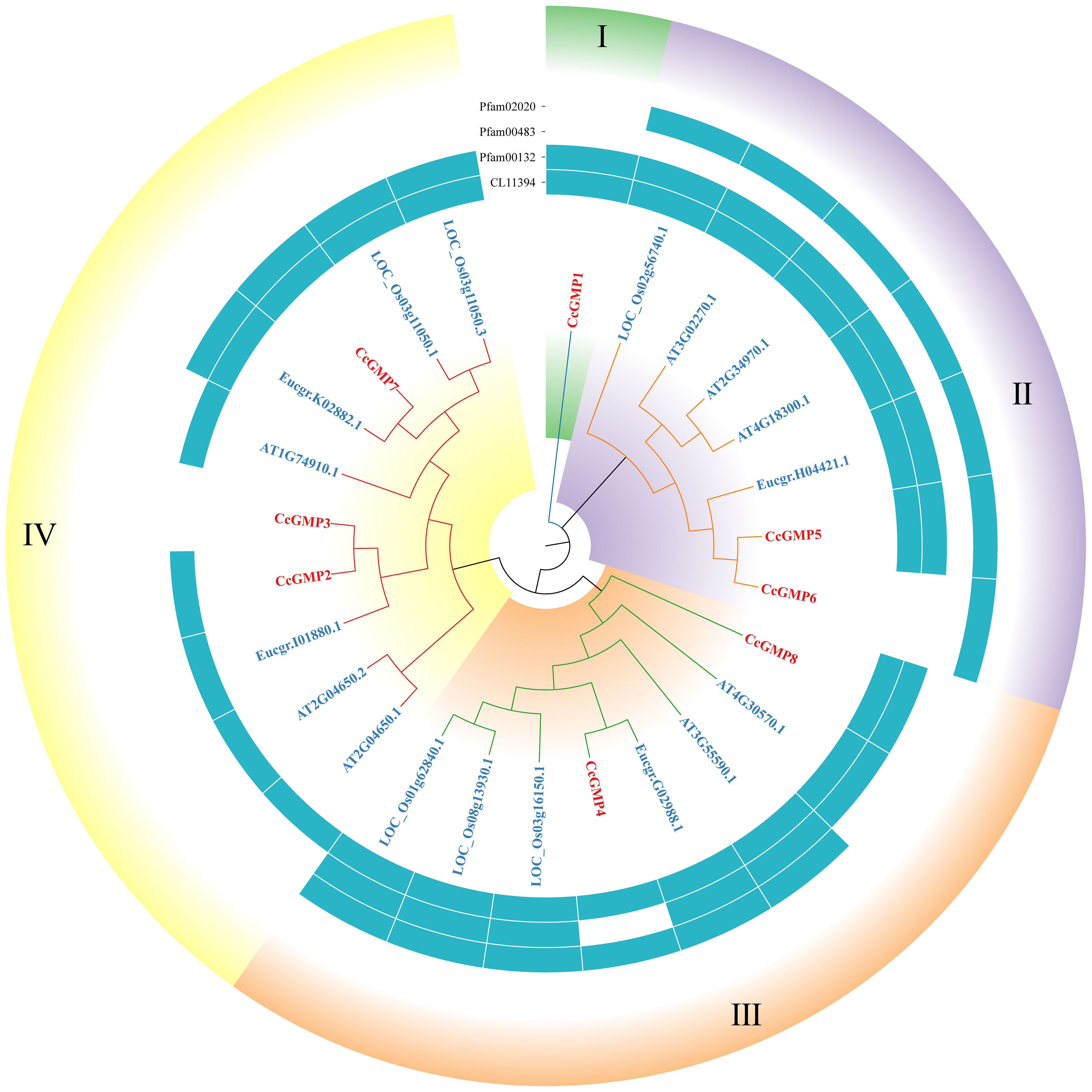
Figure 4 Phylogenetic tree of GMPs. Groups I-IV represent the classification of genes in the evolutionary tree. Conservative domains in the corresponding genes are represented by rectangular blocks in the color of teal.
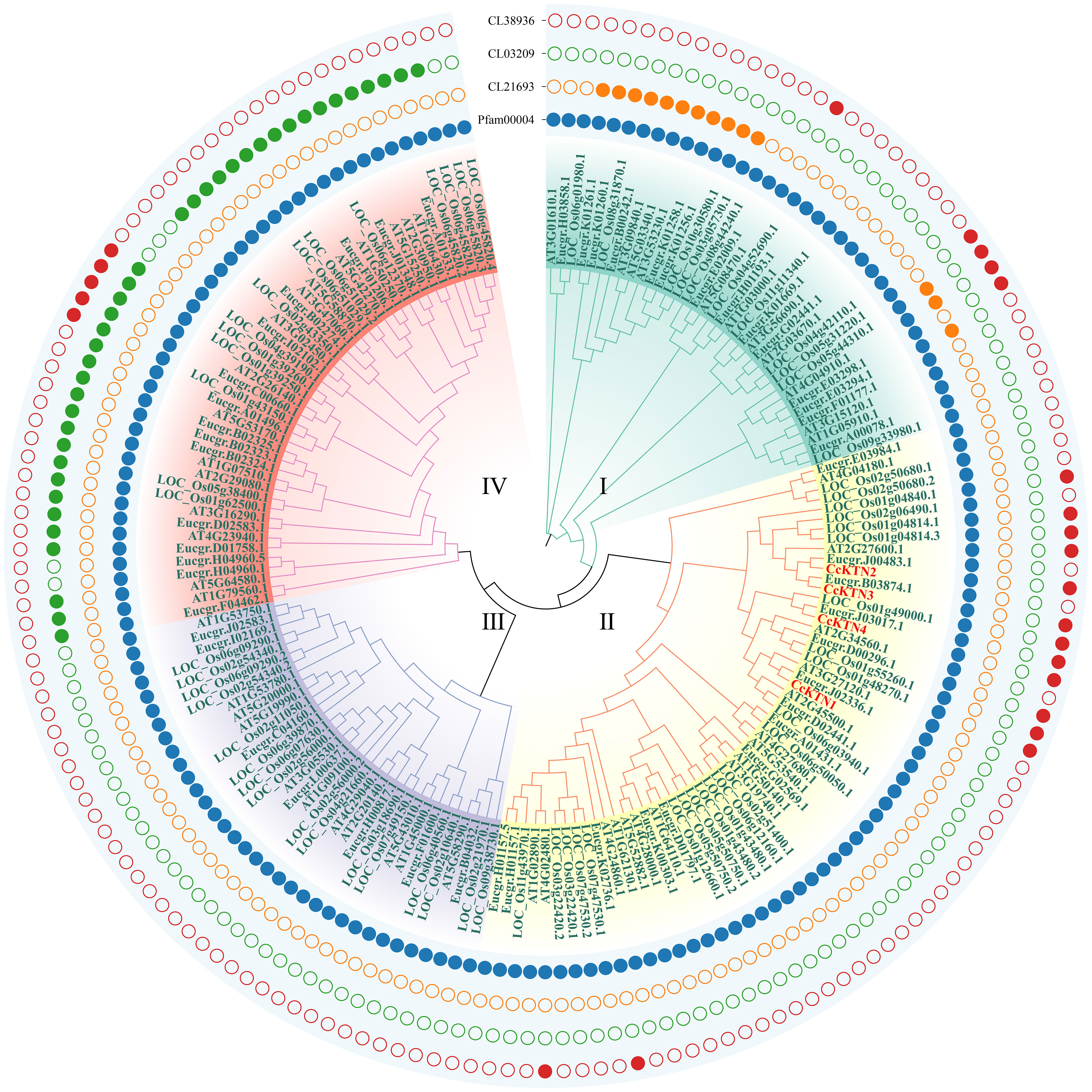
Figure 5 Phylogenetic tree of KTNs. Groups I-IV represent the classification of genes in the evolutionary tree. Solid and hollow circles correspond to the presence or absence of protein domains.
Four conserved domains had been identified in GMPs (Figure 4), namely pfam02020 (W2 domain), pfam00483 (NTP transferase), pfam00132 (Hexapep transferase), and CL11394. Except for the splicing forms CcGMP3 and 6, all other genes shared the CL11394 domain that involved in Glycosyltransferases (GTs). The group II of GMPs contained pfam02020, which mainly mediates RNA transcription and processing. Arabidopsis genes within this group may play a role in root development and floral organ differentiation. In contrast, Arabidopsis genes belonging to groups III and IV lack the W2 domain but are involved in regulating AsA and mannose formation. The group III of genes mostly contains the conserved domain pfam00483, which mainly transfers nucleotides onto phosphor sugars, and rice genes involved in this group regulate the synthesis of mannose-1-phosphate guanylyltransferase (MPG). Therefore, based on the similarity of sequence and domain, we classified CcGMPs into four groups and postulated that CcGMP2, 4, 7 and 8 are implicated in cell wall synthesis.
In addition, we also classified the genes related to KTN (Figure 5). All KTNs contained pfam00004 (ATPase family associated with various cellular activities, AAA), which related to performing chaperone-like functions. Only some genes in group I had cl21693 (Cell division cycle protein, CDC). Except group III some genes of the other groups contained cl38936 domain (P-loop_NTPase Superfamily), which is involved in diverse cellular functions. Most of the genes in group IV contain the cl03209 domain (Peptidase_M41 Superfamily), which may influence metallopeptidase activity. According to the clustering results, CcKTNs were all grouped into one category. In order to more accurately infer which gene was involved in cell wall synthesis, we verified by sequence similarity with Arabidopsis. According to the TAIR website, AtKTN1 and At2G34560 are involved in cell wall biosynthesis, so CcKTN4 may participate in the same process.
The evolutionary relationship of CcKTN&CcGMPs was investigated by constructing a phylogenetic tree based on their amino acid sequences. CcKTN&CcGMP proteins were respectively classified into 2 and 4 subfamilies in this phylogenetic tree, which is basically consistent with the results shown in Figure 6A based on phylogenetic analysis of the 4 plant species.
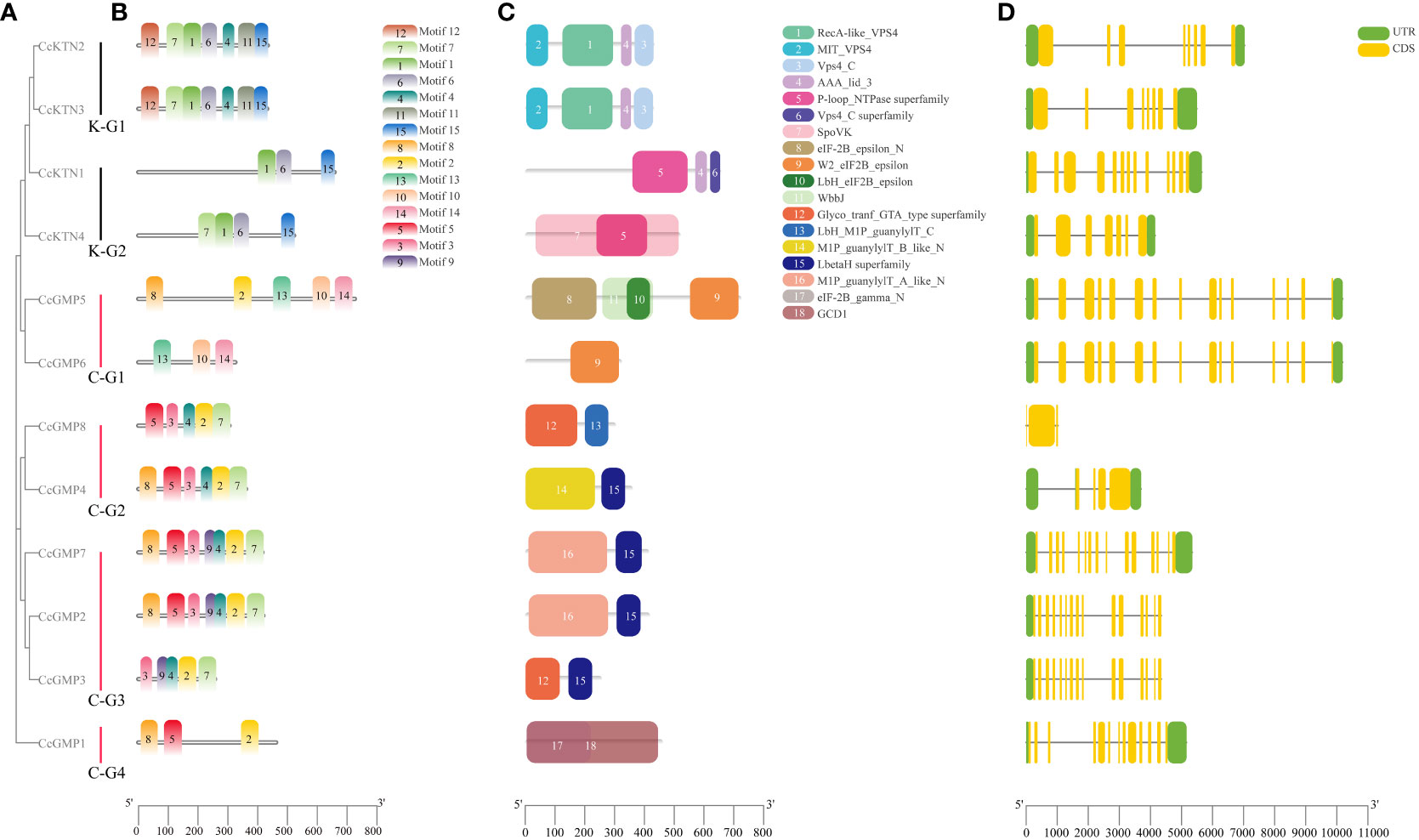
Figure 6 Motif, protein domain and gene structure of CcGMP&CcKTNs. (A) Phylogenetic relationship of the CcGMP&CcKTN gene family. CcKTN gene was divided into two groups and the CcGMP gene into four groups. (B) Protein motif composition of CcGMP&CcKTNs. (C) Conserved protein domains of CcGMP&CcKTNs. The domains and motifs were boxed in different colors. (D) Exon-intron structure of CcGMP&CcKTNs. CDS, UTR and intron are represented by yellow, green boxes and black lines, respectively.
To further investigate the diversity of the CcKTN&CcGMP genes, we analyzed their protein motifs using the MEME online server. Fifteen conserved motifs were identified, i.e., motifs 1 to 15 (Figure 6B). A detailed information of these protein motifs was presented in Table S5. Among the 12 gene products, CcKTN1 and CcGMP1, 5, and 6 lacked motif7. CcKTN genes shared the same motif1, 6, and 15. Except for CcGMP6, all genes of CcGMP shared the same motif2. In the InterPro website results, Motif3 and motif5 are potentially associated with nucleotide-diphospho-sugar transferases that participate in cell wall synthesis and these motifs are commonly found in CcGMPs, except for CcGMP5 and CcGMP6.
The conserved domains of CcKTN&CcGMPs were investigated (Figure 6C). K-G1 shares the same domain, where domains1, 2, and 3 belong to the vacuolar protein sorting-associated protein 4 (VPS4) family. The VPS4 protein family is responsible for the decomposition and recycling of plasma membrane proteins in cells and plays an important role in cell metabolism and homeostasis maintenance. Domain4 plays an important role in the function of ATPase and can influence ATP hydrolysis and energy release by regulating the conformation of ATPase. K-G2 also has the same domain4. The presence of domain5 makes proteins usually have one or more nucleotide binding sites, which can bind to ATP, GTP, CTP, UTP and other nucleotides and release energy through hydrolysis of ATP or GTP. Notably, the SpoVK (The stage V sporulation protein K) domain in CcKTN4 may be involved in cell wall synthesis.
There are three eukaryotic translation initiation factor (eIF) 2B epsilon subunit related domains in C-G1. W2_eIF2B_epsilon domain mainly interacts with eIF2B alpha, beta and gamma subunits to form a complete eIF2B complex. The eIF-2B_epsilon_N interacts with other proteins to regulate the function of eIF2B epsilon subunits. Lbh_eIF2B_epsilon is mainly involved in the formation and translation of eIF2B complex. Therefore, CcGMP5, 6 may be involved in translation regulation. Both C-G2 and C-G3 (except CcGMP8) genes shared the domain of LbetaH Superfamily, which plays important roles in a variety of biological processes, such as lipid metabolism, cell signal transduction, gene transcription and cell proliferation. Both C-G2 and C-G3 (excluding CcGMP3) are present in the (mannose-1-phosphate) M1P family, which is involved in the synthesis of GMP that plays a crucial role in maintaining cell wall integrity, morphogenesis and viability. Moreover, the GCD1 domain of C-G4 facilitates the synthesis of NDP-sugar pyrophosphorylase which contributes to cell wall synthesis.
To provide more valuable information on CcKTN&CcGMP genes, the gene structure is shown in Figure 6D. Exon numbers vary from 3 to 15, with CcGMP2, 3, 5, 6, and 7 genes having the highest number of exons. Apart from the spliced copies CcGMP3 and CcGMP6, different CcGMP genes ranged in length from 1,025 to 10,192 bp, while different CcKTN genes ranged in length from 3,759 to 4,351 bp. We found that the absence of 5’ and 3’ untranslated regions (UTR) in the CcGMP8 gene may be caused by incomplete annotation information of the gene, or the mRNA produced by the gene may start directly from the start codon. Also, two 5’ UTRs were identified in CcGMP4. While some genes exhibit multiple 5’ UTRs, their translational efficiency, stability and other characteristics are likely to be impacted. Further investigation is required to determine whether these abnormalities affect gene expression.
To understand the genetic functions and regulatory mechanisms of CcGMP&CcKTNs, the majority of cis-elements in their promoter regions were analyzed. The 2,000 bp sequence upstream of CcGMP&CcKTNs was obtained as the putative promoter. The PlantCARE tool was utilized to conduct a scan of the promoter region of CcGMP&CcKTNs in search of potential shared cis-elements. Then, to more clearly express the specificity of some genes, we categorized cis-elements based on their functions related to plant growth and development, phytohormone response, stress response, and light response.
The number of cis-elements of different genes is detailed in Table S6. A total of 259 cis-elements were identified, of which 99 were photo-responsive, followed by 78 were related to plant hormone regulation. According to the different functions of each cis-element, we screened them and made drawings with TBtools. The results showed that the cis-elements regulating the light response were distributed in all genes (Figure 7), with the largest number of 14 in CcKTN3. It can be observed that the distribution of functional cis-elements in CcGMP8 is limited, which may be attributed to incomplete gene annotation.
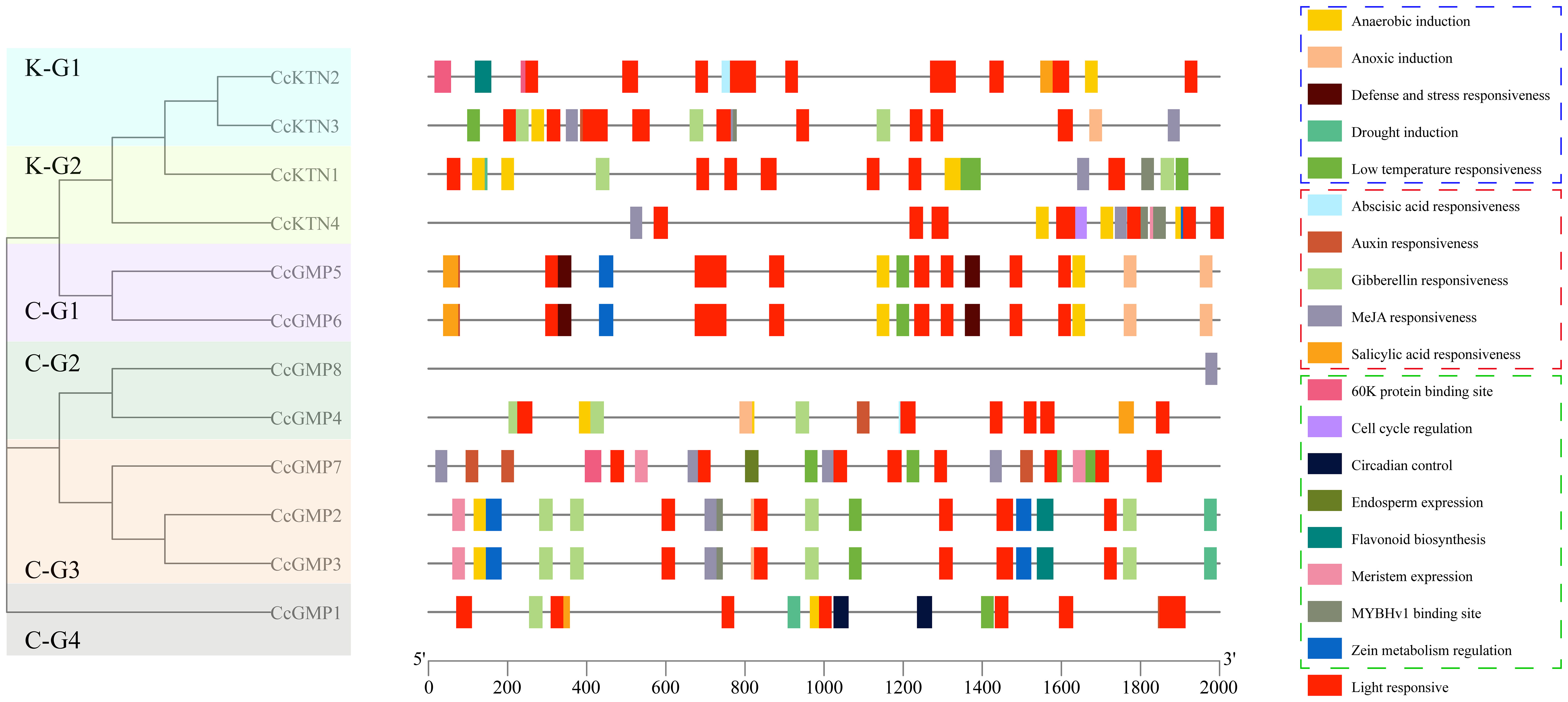
Figure 7 Cis-regulatory elements within the 2,000 bp upstream regions of putative CcGMP&CcKTN genes, arranged by the phylogenetic tree. Cis-elements involved in stress response, phytohormone response, and plant growth and development are represented by dotted boxes of blue, red, and green, respectively.
We delved deeper into the distinctiveness of each gene. Apart from light response and stress elements, only CcGMP1 gene was implicated in physiological control. Only CcGMP7 was involved in endosperm specific expression and possessed a 60K protein binding site. This implied that CcGMP7 plays a role in the plant’s immune response, facilitating its defense against viral infections.
Among CcKTN genes, only CcKTN4 was involved in meristem expression, cell cycle regulation and zein metabolism regulation. CcKTN2 was solely involved in flavonoid biosynthesis and harbored the 60K protein binding site. Therefore, except for CcGMP8, most genes may exert an influence on growth.
With the exception of CcGMP2 and CcGMP8, all other proteins within the CcGMPs were found to interact with one another, as depicted in the Figure S1. Notably, CcGMP1 displayed robust reliability in interacting with CcGMP5 and CcGMP6, while CcGMP4 had strong interaction reliability with CcGMP3 and CcGMP7. In addition, two E. grandis homologous genes were introduced to link the relationships of CcGMP1, 5, 6 and CcGMP3, 4, 7. These two Eucalyptus genes had strong reliability in interacting with CcGMP1, 5, 6 and both Eucalyptus genes were involved in eIF-related functions. In the CcKTN proteins, CcKTN1, 2 and 3 exhibited mutual interaction, whereas only a low level of reliability is observed in the interaction between CcKTN4 and CcKTN1. In addition, we found no interaction between GMP and KTN proteins.
We studied different sizes of DBH separately to find differentially expressed genes (Figure 8). In small DBH, the expression levels of CcGMP4 and CcGMP6 genes were significantly higher than those of reference genes. The expression of CcKTN4 was higher than that of the reference gene, but the difference was not significant. In medium DBH, the expression levels of CcGMP4 and CcGMP6 genes were higher than those of reference genes, but the difference was not significant, and the expression levels of CcKTN genes were significantly lower than those of reference genes. In large DBH, CcGMP4 was significantly larger than the reference gene, and CcKTN4 was larger than the reference gene, but the difference was not significant. There was a large difference in the mean value of data between CcGMP6 and CcGMP7, but no significant difference in the T-test results between the two groups, which may be due to too little biological duplication, resulting in a large variance. Therefore, CcGMP4, CcGMP6 and CcKTN4 can be regarded as differential genes for further analysis.
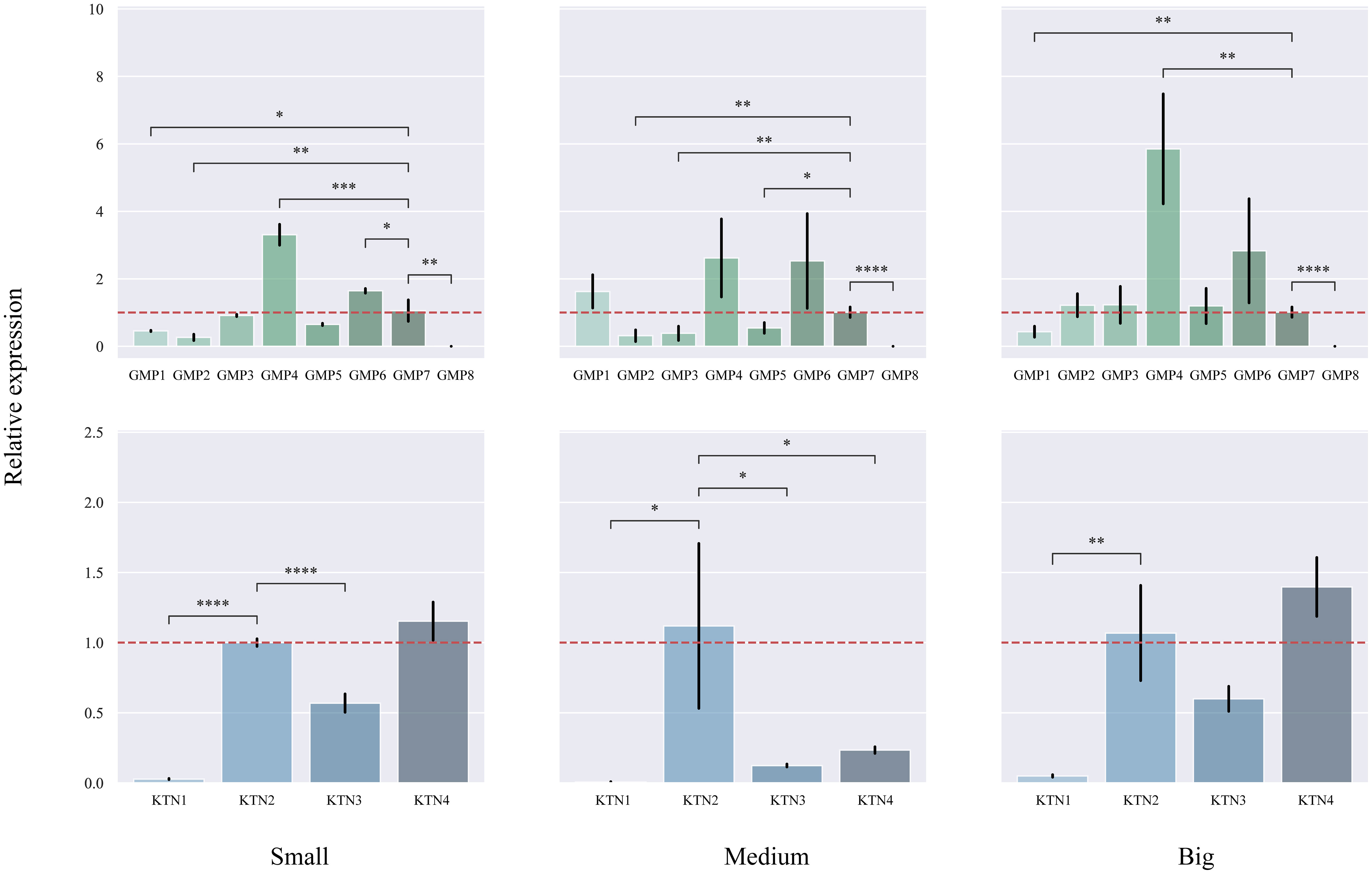
Figure 8 Quantitative real-time PCR (qRT-PCR) analyses in different genes. The CcGMP&CcKTN gene family was expressed in small, medium, and large DBH, respectively. Each treatment was controlled by CcGMP7&CcKTN2 (relative expression level was close to 1). The two red dotted lines are located at 1 and −1 on the Y-axis. Error bars indicate the standard deviation of four biological replicates. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
In order to investigate the correlation between DBH and gene expression, we analyzed the changes in different genes based on varying DBH values (Figure 9). Among the CcGMP genes, most exhibited an upward trend with increasing DBH, particularly CcGMP2. Additionally, while the expression of CcGMP1 was highest in trees with medium DBH, it decreased in those with larger diameters. With regards to CcKTN genes, all except CcKTN2 showed a similar increase. Notably, the expression level of CcKTN2 was extremely unstable at medium DBH, while the expression of other CcKTN genes was relatively stable. Therefore, further discussion is required to determine whether this variability is attributed to the characteristics of the CcKTN2 gene itself or the biological processes involved.
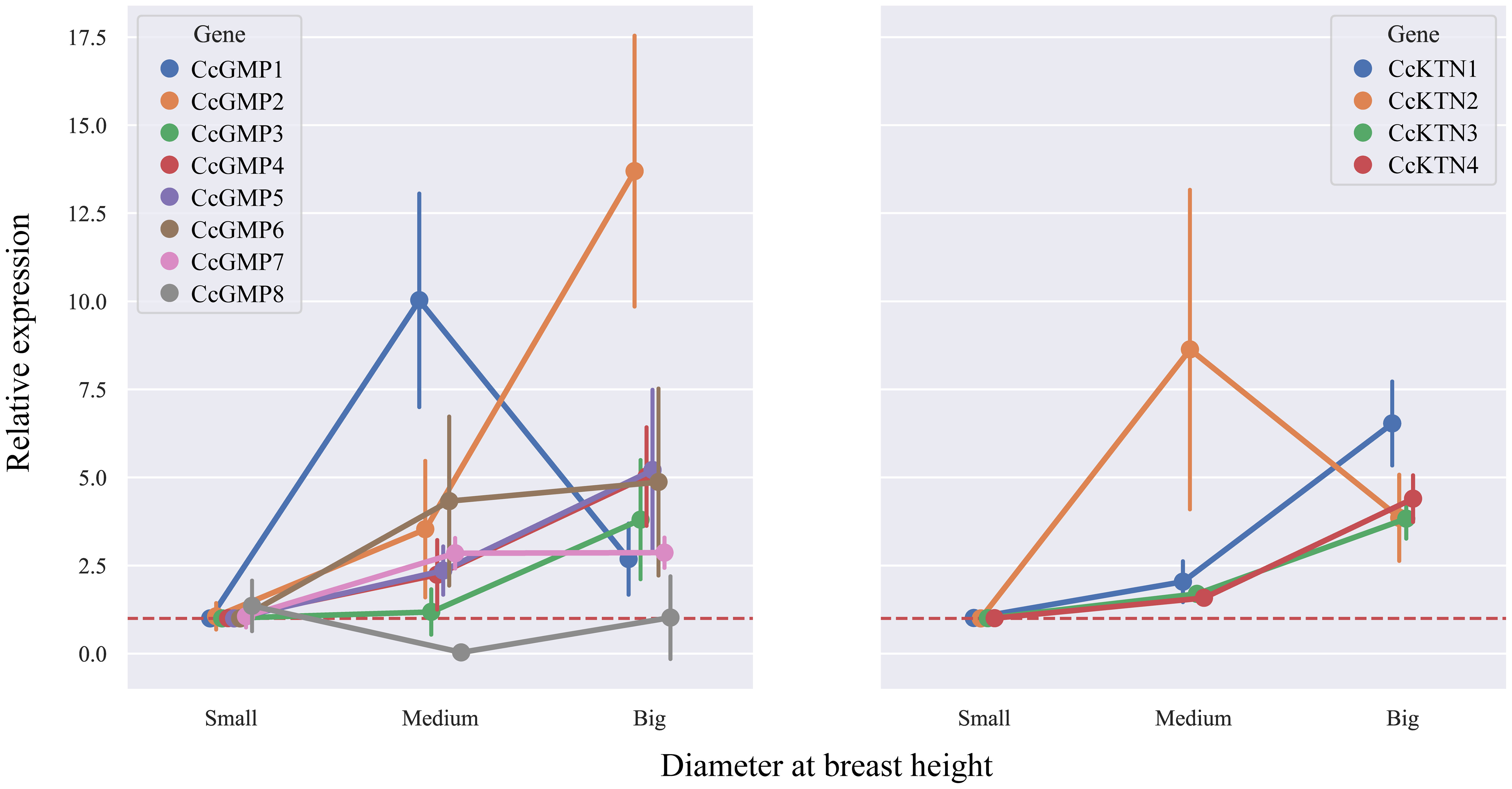
Figure 9 The relative expression in different DBH of CcGMPs and CcKTNs. Each gene was controlled by a small DBH (expression level close to 1). Error bars indicate standard deviation of four biological replicates.
From the expression results, differentially expressed genes (CcGMP4, CcGMP6, CcKTN4), abnormal genes (CcGMP1, CcKTN2) and reference genes (CcGMP7, CcKTN2) were selected for function prediction. Cellular component of these genes was located in cytoplasm (Table S7). Among the CcGMP1, 4, 6, and 7 genes, all the genes exhibited the molecular function of nucleotide transferase activity and participated in the biological process of nitrogen compound synthesis and organic compound synthesis in cells (Figure S2A). Except CcGMP7, all of them were involved in the molecular functions of organic and heterocyclic compounds. The genes CcGMP4 and CcGMP7 were involved in the biological process of mannose synthesis; CcGMP1 and CcGMP6 were active as guanylate exchange factors; CcGMP1 and CcGMP6 were involved in cytoplasmic transcription initiation factors; and CcGMP1 were involved in cell development and immune cell conduction. It is evident that the expression level of CcGMP1 is excessively high in medium DBH and low in large DBH (Figure 9B), which may be associated with cellular development and autoimmunity. In addition, mannose synthesis is present together at different DBH. Unexpectedly, a discrepancy was observed in the expression of CcGMP6, which is solely an artificially truncated transcript and contains only the W2 domain that is associated with transcription initiation factors. Although CcGMP5 also possessed this domain along with other domains, its expression level is not as high as that of CcGMP6. It can be inferred that the presence of other domains pfam00132 or Wbbj (involved in acetyltransferase) affects the expression level of CcGMP5.
In CcKTN2 and CcKTN4, all genes were involved in the molecular functions of ATPase activity and organic and heterocyclic compounds (Figure S2B). Among them, CcKTN4 was implicated in the molecular function of microtubule severing, as well as the biological processes of cell development and cytoplasmic microtubule organization. CcKTN2 was involved in the biological processes of endosomal and vacuolar tissues.
Previous studies were rarely related to biological information of GMP and KTN. In this paper, a total of 8 CcGMP genes and 4 CcKTN genes were identified. In expression analysis, CcGMP8 appeared to have no effect on DBH growth. It was reported that the FaGMP4 gene expression of strawberries of different varieties was different, and the FaGMP4 of one variety was low in the fruit development stage, resulting in its low AsA content (Lin et al., 2021). Therefore, we speculated that trees with small DBH had relatively low AsA content.
In addition, it was obvious that the CcGMP4 gene exhibited a higher level of expression compared to other genes, particularly in different DBH. Following closely is CcGMP6, which is an artificially truncated transcript based on CcGMP5 and contains pfam domain pfam02020. This domain is associated with guanine nucleotide exchange factor (Koonin, 1995), and eIF-2B is conserved across eukaryotes, primarily serving as an exchange factor between GTP and GDP while also acting as a key regulatory factor in mRNA translation (Wortham and Proud, 2015). However, it needs to be further verified whether the plant itself can translate proteins containing this domain. In the interaction with E. grandis protein, it was observed that the corresponding homologous protein could be identified. The GO annotation results revealed that this homologous protein possesses guanylate exchange factor activity and transcription factor initiation binding, implying the potential interaction of CcGMP6 with other proteins to form complex protein-DNA or protein-protein feedback loops regulating gene expression levels.
The paralogous genes of CcGMP4 were At3G55590.1 and At2G39770.1. These two genes belong to the key genes of synthetic GMP, but At3G55590.1 cannot compensate for the functional defect of At2G39770.1 (Qin et al., 2008). Thus, The function of the At3G55599.1 gene remains elusive, and further research is needed to fully understand its role. GMP catalyzed the conversion of D-mannose-1-P to GDP-D-mannose, which is a precursor of AsA synthesis. Studies have shown that there is a close relationship between AsA and non-cellulosic cell wall polysaccharide biosynthesis in plants (Gilbert et al., 2009). However, plants lacking At2G39770.1 tend to exhibit incomplete cell walls and insufficient cellulose content (Nickle and Meinke, 1998; Lukowitz et al., 2001). It means that CcGMP4 may increase the content of mannose in stem cell wall and maintain the intact plant cell wall.
In the multi-species evolutionary tree, CcGMP4 contains the conserved domains CL11394 and Pfam00483 of PFAM, while the genes homologous to AtCYT in all species share CL11394. This family containing CL11394 domain has the function of glycosyltransferases (GTs). Most of the GTs that catalyze the formation of carbohydrate in plant cell walls have not been biochemically characterized (Brown et al., 2012), mainly because the low abundance of these enzymes in plants makes them difficult to purify in sufficient quantities for enzymatic characterization (Amos and Mohnen, 2019). This also means that GTs that are homologous to AtCYT need to be further studied. In addition, The presence of the pfam00483 domain causes CcGMP4 to have Nucleotidyltransferase activity (Jensen and Reeves, 1998). Furthermore, The presence of M1P_guanylylT_B_like_N on CcGMP4 protein also verified its GMP function. This enzyme is extremely important for the integrity of cell walls (Jiang et al., 2008), which also indicates that CcGMP4 cannot be ignored in the process of cambium cell differentiation.
In addition to domain information, the prediction results of the CcGMP4 promoter suggest that it may have function related to cell wall synthesis. In the cis-element analysis results, CcGMP4 has elements related to gibberellin, which can promote cell division in cambium to a certain extent (Jeon et al., 2016).
As the reference gene, the annotation result of CcGMP7 also has the effect of GMP. Its paralogous gene, At1G74910.2 (KJC1), is homologous to human GMPPA (GMP alpha subunits) and likely interacts with AtCYT1 protein to positively regulate GMP activity, thereby promoting the synthesis of GDP-D-mannose (Zheng et al., 2021). Among them, the mannose content of vct1-1 was significantly lower than that of kjc mutant (Sawake et al., 2015; Nishigaki et al., 2021), indicating that CcGMP7 and CcGMP4 had a mutually reinforcing effect, and CcGMP4 regulated mannose-synthesis in a large proportion. The GO annotation results for CcGMP7 and CcGMP4 differed in that the former lacked binding activity with organic and heterocyclic compounds, which may account for its lower expression level compared to the latter. In the comparison of GMP protein sequences (Figure S3), it can be inferred that CcGMP7 and CcGMP4 are members of GMPPA and GMPPB (GMP beta subunits), respectively, based on the conserved motif (Sawake et al., 2015). It has been reported that genes belonging to GMPPB possess the ability to synthesize GDP-D-mannose (Yi et al., 2021). In the medium DBH, the expression of CcGMP1 was higher than that of CcGMP7. It is possible that CcGMP1 gene is related to cell development, or that it contains an important GCD1 (Glucosylation Defective 1) domain (Hill and Struhl, 1988), or that there may be some unknown factors leading to the overexpression of CcGMP1.
In addition to the promoting effect of GDP-D-mannose on cell wall synthesis, microtubule also has a certain promoting effect on the accumulation of cell wall polysaccharides (Paredez et al., 2006), which provides an idea for selective breeding. The expression level of CcKTN2 was higher in medium DBH, but lower in other DBH than that of CcKTN4. CcKTN2 and CcKTN3 were a pair of segmental duplicated genes, and the paralogous gene of CcKTN3 was At2G27600.1 (AtSKD1). This gene encodes a SKD1 (Suppressor of K+ Transport Growth Defect1) homolog and involved in multivesicular endosome function (Herberth et al., 2012). In the multi-species evolutionary tree, At2G27600.1 is relatively close to CcKTN2 and CcKTN3, with a conservative domain VPS4 (Vacuolar protein sorting 4) at the C-terminal, which does not cut off microtubules but rather disassembles protein complexes involved in membrane transport (Walker et al., 1982). TESCRT (Endosomal Sorting Complex Required for Transport) controls cell expansion and development in A. thaliana, and associated genes have N-terminal microtubule interaction domains (MIT) (Reyes et al., 2014). Notably, the MIT domain is also present in CcKTN2 and CcKTN3 (Figure 6B). In GO annotation, both CcKTN2 and CcKTN3 had ATPase activity, endosome organization and vacuole organization function. They have strong interaction in protein interaction prediction. In the vascular cambium, both the fusiform and ray initials cells are highly vacuolized and both have genes associated with cell wall synthesis (Goue et al., 2008). We hypothesize that CcKTN2 and CcKTN3 may regulate the fusion of multivesicular bodies into vacuoles in medium DBH, thereby facilitating material transport and metabolism while maintaining vacuolar integrity (Shahriari et al., 2010). In addition, among the cis-elements of CcKTN2 and CcKTN3 promoters, only CcKTN2 has a 60K protein, which is reported to interact with ESCRT and regulate its localization and function in the cell, thus affecting plant cell division and development (Katsiarimpa et al., 2011). It is concluded that CcKTN2 plays an important role in plant cell development, but its overexpression level at medium DBH needs to be verified.
CcKTN4 is a gene of notable significance, exhibiting higher expression levels than CcKTN2 in both small and large DBH trees. The paralogous gene of CcKTN4 was At1G803501, which primarily encodes the p60 subunit and participates in Katanin, a microtubule-severing enzyme (Burk et al., 2001). CcKTN4 holds the domain SpoVK (stage V sporulation protein K), which plays an important role in spore formation and division of bacteria (Fan et al., 1992; Hilbert and Piggot, 2004). However, the domains of bacteria and eukaryotes are different. There are some similarities between SpoVK and the ATPase in the AAA+ protein family. SpoVK forms a complex with other proteins, and this complex may be similar to the ATPase complex in the AAA+ protein family, which also plays a role in spore formation in bacteria (Eichenberger et al., 2001; Elsholz et al., 2017). The AAA+ ATPases are enzymes containing a P-loop NTPase domain, and function as molecular chaperones (Iyer et al., 2004). P-loop NTPase domains typically exhibit two conserved sequence characteristics, namely motif GxxxxGK[ST] (Walker A) and hhhh[DE] (Walker B) (Walker et al., 1982), both of which are conserved in the KTN genes (Figure S4). This domain is ubiquitous in bacteria and eukaryotes (Koonin et al., 2000), and its role in plants is very extensive, involving many biological processes in plants, including cell division (Yu et al., 2016), apoptosis (Lelpe et al., 2004), cellular immunity (Bonardi et al., 2011), etc. Similarly, Katanin also plays a role in cell division in plants (Panteris et al., 2011) and can even promote the elongation of plant cells (Chen et al., 2022). It can be seen that the increase of CcKTN4 expression with DBH may be precisely because of the large number of cells and continuous cell division.
In the field of wood property research, genes associated with cellulose synthesis are a crucial research topic. Previous studies have demonstrated that cyt and ktn mutants exhibit specific effects on cellulose production (Li et al., 2014). This study highlights the significance of CcGMP4&CcKTN4 as a key gene influencing eucalyptus wood size. CcGMP4 is capable of regulating mannose synthesis, thereby indirectly promoting cell wall synthesis, while CcKTN4 can influence the arrangement of cortical microtubules and thus indirectly regulate cell wall synthesis. These findings suggest that CcGMP4&CcKTN4 plays a crucial role in governing growth and wood quality. Therefore, CcGMP4&CcKTN4 can be used for gene knockout or overexpression to further explore the mechanism of wood formation, or through gene editing technology to achieve regulation and optimization of wood formation process.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
CW: Conceptualization, Data curation, Investigation, Project administration, Writing – original draft. JL: Methodology, Project administration, Writing – review & editing. WH: Software, Visualization, Writing – review & editing. AH: Investigation, Writing – review & editing. WL: Data curation, Writing – review & editing. YL: Investigation, Resources, Writing – review & editing. YO: Conceptualization, Methodology, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by program for scientific research start-upfunds of Guangdong Ocean University; and National Key R&D Program of China for the 14th Five-year Plan (No.: 2022YFD2200203).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1308354/full#supplementary-material
Amos, R. A., Mohnen, D. (2019). Critical review of plant cell wall matrix polysaccharide glycosyltransferase activities verified by heterologous protein expression. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00915
Baskin, T. I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: a review and a model. Protoplasma 215 (1-4), 150–171. doi: 10.1007/bf01280311
Baskin, T. I. (2005). Anisotropic expansion of the plant cell wall. Annu. Rev. Cell Dev. Biol. 21, 203–222. doi: 10.1146/annurev.cellbio.20.082503.103053
Bischoff, V., Desprez, T., Mouille, G., Vernhettes, S., Gonneau, M., Hofte, H. (2011). Phytochrome regulation of cellulose synthesis in Arabidopsis. Curr. Biol. 21 (21), 1822–1827. doi: 10.1016/j.cub.2011.09.026
Bonardi, V., Tang, S. J., Stallmann, A., Roberts, M., Cherkis, K., Dangl, J. L. (2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. U. S. A. 108 (39), 16463–16468. doi: 10.1073/pnas.1113726108
Bouquin, T., Mattsson, O., Naested, H., Foster, R., Mundy, J. (2003). The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J. Cell Sci. 116 (5), 791–801. doi: 10.1242/jcs.00274
Brown, C., Leijon, F., Bulone, V. (2012). Radiometric and spectrophotometric in vitro assays of glycosyltransferases involved in plant cell wall carbohydrate biosynthesis. Nat. Protoc. 7 (9), 1634–1650. doi: 10.1038/nprot.2012.089
Burk, D. H., Liu, B., Zhong, R. Q., Morrison, W. H., Ye, Z. H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13 (4), 807–827. doi: 10.1105/tpc.13.4.807
Burk, D. H., Zhong, R. Q., Ye, Z. H. (2007). The katanin microtubule severing protein in plants. J. Integr. Plant Biol. 49 (8), 1174–1182. doi: 10.1111/j.1672-9072.2007.00544.x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y., Liu, X., Zhang, W., Li, J., Liu, H., Yang, L., et al. (2022). MOR1/MAP215 acts synergistically with katanin to control cell division and anisotropic cell elongation in Arabidopsis. Plant Cell 34 (8), 3006–3027. doi: 10.1093/plcell/koac147
Conklin, P. L., Norris, S. R., Wheeler, G. L., Williams, E. H., Smirnoff, N., Last, R. L. (1999). Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 96 (7), 4198–4203. doi: 10.1073/pnas.96.7.4198
Couto, A. M., Trugilho, P. F., Napoli, A., Lima, J. T., da Silva, J. R. M., Protasio, T. D. (2015). Quality of charcoal from Corymbia and Eucalyptus produced at different final carbonization temperatures. Scientia Forestalis 43 (108), 817–831. doi: 10.18671/scifor.v43n108.7
Crowell, E. F., Bischoff, V., Desprez, T., Rolland, A., Stierhof, Y. D., Schumacher, K., et al. (2009). Pausing of golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21 (4), 1141–1154. doi: 10.1105/tpc.108.065334
de Araujo, M. J., Lee, D. J., Tambarussi, E. V., de Paula, R. C., da Silva, P. H. M. (2021). Initial productivity and genetic parameters of three Corymbia species in Brazil: designing a breeding strategy. Can. J. For. Res. 51 (1), 25–30. doi: 10.1139/cjfr-2019-0438
de Souza, B. M., Freitas, M. L. M., Sebbenn, A. M., Gezan, S. A., Zanatto, B., Zulian, D. F., et al. (2020). Genotype-by-environment interaction in Corymbia citriodora (Hook.) KD Hill, & LAS Johnson progeny test in Luiz Antonio, Brazil. For. Ecol. Manage. 460, 117855. doi: 10.1016/j.foreco.2019.117855
Eichenberger, P., Fawcett, P., Losick, R. (2001). A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis. Mol. Microbiol. 42 (5), 1147–1162. doi: 10.1046/j.1365-2958.2001.02660.x
Elsholz, A. K. W., Birk, M. S., Charpentier, E., Turgay, K. (2017). Functional diversity of AAA plus protease complexes in Bacillus subtilis. Front. Mol. Biosci. 4. doi: 10.3389/fmolb.2017.00044
Fan, N., Cutting, S., Losick, R. (1992). Characterization of the Bacillus-subtilis sporulation gene SPOVK. J. Bacteriol. 174 (3), 1053–1054. doi: 10.1128/jb.174.3.1053-1054.1992
Fujita, M., Lechner, B., Barton, D. A., Overall, R. L., Wasteneys, G. O. (2012). The missing link: Do cortical microtubules define plasma membrane nanodomains that modulate cellulose biosynthesis? Protoplasma 249, S59–S67. doi: 10.1007/s00709-011-0332-z
Gilbert, L., Alhagdow, M., Nunes-Nesi, A., Quemener, B., Guillon, F., Bouchet, B., et al. (2009). GDP-d-mannose 3,5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J. 60 (3), 499–508. doi: 10.1111/j.1365-313X.2009.03972.x
Goue, N., Lesage-Descauses, M. C., Mellerowicz, E. J., Magel, E., Label, P., Sundberg, B. (2008). Microgenomic analysis reveals cell type-specific gene expression patterns between ray and fusiform initials within the cambial meristem of Populus. New Phytol. 180 (1), 45–56. doi: 10.1111/j.1469-8137.2008.02556.x
Guo, Y. Y., Xu, H. M., Wu, H. Y., Shen, W. W., Lin, J. X., Zhao, Y. Y. (2022). Seasonal changes in cambium activity from active to dormant stage affect the formation of secondary xylem in Pinus tabulaeformis Carr. Tree Physiol. 42 (3), 585–599. doi: 10.1093/treephys/tpab115
Healey, A. L., Shepherd, M., King, G. J., Butler, J. B., Freeman, J. S., Lee, D. J., et al. (2021). Pests, diseases, and aridity have shaped the genome of Corymbia citriodora. Commun. Biol. 4 (1), 13. doi: 10.1038/s42003-021-02009-0
Herberth, S., Shahriari, M., Bruderek, M., Hessner, F., Muller, B., Hulskamp, M., et al. (2012). Artificial ubiquitylation is sufficient for sorting of a plasma membrane ATPase to the vacuolar lumen of Arabidopsis cells. Planta 236 (1), 63–77. doi: 10.1007/s00425-012-1587-0
Hilbert, D. W., Piggot, P. J. (2004). Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68 (2), 234–23+. doi: 10.1128/mmbr.68.2.234-262.2004
Hill, D. E., Struhl, K. (1988). Molecular characterization of GCD1, a yeast gene required for general control of amino acid biosynthesis and cell-cycle initiation. Nucleic Acids Res. 16 (19), 9253–9265. doi: 10.1093/nar/16.19.9253
Hung, T. D., Brawner, J. T., Lee, D. J., Meder, R., Dieters, M. J. (2016). Genetic variation in growth and wood-quality traits of Corymbia citriodora subsp variegata across three sites in south-east Queensland, Australia. South. Forests: J. For. Sci. 78 (3), 225–239. doi: 10.2989/20702620.2016.1183095
Iyer, L. M., Leipe, D. D., Koonin, E. V., Aravind, L. (2004). Evolutionary history and higher order classification of AAA plus ATPases. J. Struct. Biol. 146 (1-2), 11–31. doi: 10.1016/j.jsb.2003.10.010
Jensen, S. O., Reeves, P. R. (1998). Domain organisation in phosphomannose isomerases (types I and II). Biochim. Et Biophys. Acta-Protein Structure Mol. Enzymol. 1382 (1), 5–7. doi: 10.1016/s0167-4838(97)00122-2
Jeon, H. W., Cho, J. S., Park, E. J., Han, K. H., Choi, Y. I., Ko, J. H. (2016). Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol. J. 14 (4), 1161–1170. doi: 10.1111/pbi.12484
Jiang, H. C., Ouyang, H. M., Zhou, H., Jin, C. (2008). GDP-mannose pyrophosphorylase is essential for cell wall integrity, morphogenesis and viability of Aspergillus fumigatus. Microbiology-Sgm 154, 2730–2739. doi: 10.1099/mic.0.2008/019240-0
Kato, S., Murakami, M., Saika, R., Soga, K., Wakabayashi, K., Hashimoto, H., et al. (2022). Suppression of cortical microtubule reorientation and stimulation of cell elongation in Arabidopsis hypocotyls under microgravity conditions in space. Plants-Basel 11 (3), 12. doi: 10.3390/plants11030465
Katsiarimpa, A., Anzenberger, F., Schlager, N., Neubert, S., Hauser, M. T., Schwechheimer, C., et al. (2011). The Arabidopsis deubiquitinating enzyme AMSH3 interacts with ESCRT-III subunits and regulates their localization. Plant Cell 23 (8), 3026–3040. doi: 10.1105/tpc.111.087254
Kim, M. H., Bae, E. K., Lee, H., Ko, J. H. (2022). Current understanding of the genetics and molecular mechanisms regulating wood formation in plants. Genes 13 (7), 13. doi: 10.3390/genes13071181
Koonin, E. V. (1995). Multidomain organization of eukaryotic guanine-nucleotide exchange translation initiation-factor eIF-2b subunits revealed by analysis of conserved sequence motifs. Protein Sci. 4 (8), 1608–1617. doi: 10.1002/pro.5560040819
Koonin, E. V., Wolf, Y. I., Aravind, L. (2000). Protein fold recognition using sequence profiles and its application in structural genomics. Adv. Protein Chem. Vol. 54, 245–275. doi: 10.1016/s0065-3233(00)54008-x
Lamanchai, K., Salmon, D. L., Smirnoff, N., Sutthinon, P., Roytrakul, S., Leetanasaksakul, K., et al. (2022). OsVTC1-1 RNAi mutant with reduction of ascorbic acid synthesis alters cell wall sugar composition and cell wall-associated proteins. Agronomy-Basel 12 (6), 20. doi: 10.3390/agronomy12061272
Larson, P. R. (2012). The vascular cambium: development and structure. (Berlin:Springer). doi: 10.1007/978-3-642-78466-8
Lei, L., Li, S. D., Bashline, L., Gu, Y. (2014). Dissecting the molecular mechanism underlying the intimate relationship between cellulose microfibrils and cortical microtubules. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00090
Lelpe, D. D., Koonin, E. V., Aravind, L. (2004). STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: Multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 343 (1), 1–28. doi: 10.1016/j.jmb.2004.08.023
Lerouxel, O., Cavalier, D. M., Liepman, A. H., Keegstra, K. (2006). Biosynthesis of plant cell wall polysaccharides - a complex process. Curr. Opin. Plant Biol. 9 (6), 621–630. doi: 10.1016/j.pbi.2006.09.009
Li, S., Bashline, L., Lei, L., Gu, Y. (2014). Cellulose synthesis and its regulation. Arabidopsis Book 12, e0169. doi: 10.1199/tab.0169
Li, Y. F., Deng, M., Liu, H. F., Li, Y., Chen, Y., Jia, M., et al. (2021). ABNORMAL SHOOT 6 interacts with KATANIN 1 and SHADE AVOIDANCE 4 to promote cortical microtubule severing and ordering in Arabidopsis. J. Integr. Plant Biol. 63 (4), 646–661. doi: 10.1111/jipb.13003
Lin, Y. X., Zhang, J. H., Wu, L. T., Zhang, Y. T., Chen, Q., Li, M. Y., et al. (2021). Genome-wide identification of GMP genes in Rosaceae and functional characterization of FaGMP4 in strawberry (Fragaria x ananassa). Genes Genomics 43 (6), 587–599. doi: 10.1007/s13258-021-01062-7
Lindeboom, J. J., Nakamura, M., Hibbel, A., Shundyak, K., Gutierrez, R., Ketelaar, T., et al. (2013). A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science 342 (6163), 1202–120+. doi: 10.1126/science.1245533
Liu, Y. H., Wang, H., Liu, J. X., Shu, S., Tan, G. F., Li, M. Y., et al. (2022). AgGMP encoding GDP-D-mannose pyrophosphorylase from celery enhanced the accumulation of ascorbic acid and resistance to drought stress in Arabidopsis. Peerj 10, 15. doi: 10.7717/peerj.12976
Liu, X., Yu, F. (2023). New insights into the functions and regulations of MAP215/MOR1 and katanin, two conserved microtubule-associated proteins in Arabidopsis. Plant Signaling Behav. 18 (1), 2171360. doi: 10.1080/15592324.2023.2171360
Lukowitz, W., Nickle, T. C., Meinke, D. W., Last, R. L., Conklin, P. L., Somerville, C. R. (2001). Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 98 (5), 2262–2267. doi: 10.1073/pnas.051625798
Ma, S. Y., Li, H. X., Wang, L., Li, B. Y., Wang, Z. Y., Ma, B. Q., et al. (2022). F-box protein MdAMR1L1 regulates ascorbate biosynthesis in apple by modulating GDP-mannose pyrophosphorylase. Plant Physiol. 188 (1), 653–669. doi: 10.1093/plphys/kiab427
Massuque, J., Sanchez, J., Loureiro, B. A., Setter, C., Lima, M. D. R., da Silva, P. H. M., et al. (2022). Evaluating the potential of non-commercial Eucalyptus spp. and Corymbia spp. for bioenergy in Brazil. Bioenergy Res. 16(3), 1592–1603. doi: 10.1007/s12155-022-10502-5
Mellerowicz, E. J., Sundberg, B. (2008). Wood cell walls: Biosynthesis, developmental dynamics and their implications for wood properties. Curr. Opin. Plant Biol. 11 (3), 293–300. doi: 10.1016/j.pbi.2008.03.003
Nairn, C. J., Lennon, D. M., Wood-Jones, A., Nairn, A. V., Dean, J. F. D. (2008). Carbohydrate-related genes and cell wall biosynthesis in vascular tissues of loblolly pine (Pinus taeda). Tree Physiol. 28 (7), 1099–1110. doi: 10.1093/treephys/28.7.1099
Nickle, T. C., Meinke, D. W. (1998). A cytokinesis-defective mutant of Arabidopsis (cyt1) characterized by embryonic lethality, incomplete cell walls, and excessive callose accumulation. Plant J. 15 (3), 321–332. doi: 10.1046/j.1365-313X.1998.00212.x
Nishigaki, N., Yoshimi, Y., Kuki, H., Kunieda, T., Hara-Nishimura, I., Tsumuraya, Y., et al. (2021). Galactoglucomannan structure of Arabidopsis seed-coat mucilage in GDP-mannose synthesis impaired mutants. Physiol. Plant 173 (3), 1244–1252. doi: 10.1111/ppl.13519
Panteris, E., Adamakis, I. D. S., Voulgari, G., Papadopoulou, G. (2011). A role for katanin in plant cell division: microtubule organization in dividing root cells of fra2 and lue1 Arabidopsis thaliana mutants. Cytoskeleton 68 (7), 401–413. doi: 10.1002/cm.20522
Paredez, A. R., Somerville, C. R., Ehrhardt, D. W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312 (5779), 1491–1495. doi: 10.1126/science.1126551
Pesquet, E., Korolev, A. V., Calder, G., Lloyd, C. W. (2010). The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells. Curr. Biol. 20 (8), 744–749. doi: 10.1016/j.cub.2010.02.057
Plomion, C., Leprovost, G., Stokes, A. (2001). Wood formation in trees. Plant Physiol. 127 (4), 1513–1523. doi: 10.1104/pp.127.4.1513
Qin, H., Deng, Z. A., Zhang, C. Y., Wang, Y. Y., Wang, J., Liu, H., et al. (2016). Rice GDP-mannose pyrophosphorylase OsVTC1-1 and OsVTC1-3 play different roles in ascorbic acid synthesis. Plant Mol. Biol. 90 (3), 317–327. doi: 10.1007/s11103-015-0420-0
Qin, C., Qian, W. Q., Wang, W. F., Wu, Y., Yu, C. M., Jiang, X. H., et al. (2008). GDP-mannose pyrophosphorylase is a genetic determinant of ammonium sensitivity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 105 (47), 18308–18313. doi: 10.1073/pnas.0806168105
Ren, H. B., Rao, J. Q., Tang, M., Li, Y. X., Dang, X., Lin, D. S. (2022). PP2A interacts with KATANIN to promote microtubule organization and conical cell morphogenesis. J. Integr. Plant Biol. 64 (8), 1514–1530. doi: 10.1111/jipb.13281
Reyes, F. C., Buono, R. A., Roschzttardtz, H., Di Rubbo, S., Yeun, L. H., Russinova, E., et al. (2014). A novel endosomal sorting complex required for transport ( ESCRT) component in Arabidopsis thaliana controls cell expansion and development. J. Biol. Chem. 289 (8), 4980–4988. doi: 10.1074/jbc.M113.529685
Sasaki, T., Fukuda, H., Oda, Y. (2017). CORTICAL MICROTUBULE DISORDERING1 is required for secondary cell wall patterning in xylem vessels. Plant Cell 29 (12), 3123–3139. doi: 10.1105/tpc.17.00663
Sawake, S., Tajima, N., Mortimer, J. C., Lao, J., Ishikawa, T., Yu, X. L., et al. (2015). KONJAC1 and 2 are key factors for GDP-mannose generation and affect l-ascorbic acid and glucomannan biosynthesis in Arabidopsis. Plant Cell 27 (12), 3397–3409. doi: 10.1105/tpc.15.00379
Shahriari, M., Keshavaiah, C., Scheuring, D., Sabovljevic, A., Pimpl, P., Hausler, R. E., et al. (2010). The AAA-type ATPase AtSKD1 contributes to vacuolar maintenance of Arabidopsis thaliana. Plant J. 64 (1), 71–85. doi: 10.1111/j.1365-313X.2010.04310.x
Smirnoff, N., Conklin, P. L., Loewus, F. A. (2001). Biosynthesis of ascorbic acid in plants: A renaissance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 437–43+. doi: 10.1146/annurev.arplant.52.1.437
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Terzaghi, M., De Tullio, M. C. (2022). The perils of planning strategies to increase vitamin C content in plants: Beyond the hype. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1096549
Thompson, J. D., Gibson, T. J., Higgins, D. G. (2002). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinf. 2 (3), 1–22. doi: 10.1002/0471250953.bi0203s00
Walker, J. E., Saraste, M., Runswick, M. J., Gay, N. J. (1982). Distantly related sequences in the alpha-subunits and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1 (8), 945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x
Wang, D., Chen, Y., Li, W., Li, Q. Z., Lu, M. Z., Zhou, G. K., et al. (2021). Vascular Cambium: The source of wood formation. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.700928
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi: 10.1093/nar/gkr1293
Wang, J., Yu, Y. W., Zhang, Z. J., Quan, R. D., Zhang, H. W., Ma, L. G., et al. (2013). Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 25 (2), 625–636. doi: 10.1105/tpc.112.106880
Webb, M., Jouannic, S., Foreman, J., Linstead, P., Dolan, L. (2002). Cell specification in the Arabidopsis root epidermis requires the activity of ECTOPIC ROOT HAIR 3 - a katanin-p60 protein. Development 129 (1), 123–131. doi: 10.1242/dev.129.1.123
Wortham, N. C., Proud, C. G. (2015). eIF2B: Recent structural and functional insights into a key regulator of translation. Biochem. Soc Trans. 43, 1234–1240. doi: 10.1042/bst20150164
Xie, L. J., Wang, J. H., Liu, H. S., Yuan, L. B., Tan, Y. F., Tan, W. J., et al. (2023). MYB30 integrates light signals with antioxidant biosynthesis to regulate plant responses during postsubmergence recovery. New Phytol. 237 (6), 2238–2254. doi: 10.1111/nph.18674
Xue, C. C., Xu, J. Y., Wang, C., Guo, N., Hou, J. F., Xue, D., et al. (2018). Molecular cloning and functional characterization of a soybean GmGMP1 gene reveals its involvement in ascorbic acid biosynthesis and multiple abiotic stress tolerance in transgenic plants. J. Integr. Agric. 17 (3), 539–553. doi: 10.1016/s2095-3119(17)61727-1
Ye, Z. H., Zhong, R. Q. (2015). Molecular control of wood formation in trees. J. Exp. Bot. 66 (14), 4119–4131. doi: 10.1093/jxb/erv081
Yi, Y. Q., Liu, L. L., Zhou, W. Y., Peng, D. Y., Han, R. C., Yu, N. J. (2021). Characterization of GMPP from Dendrobium huoshanense yielding GDP-D-mannose. Open Life Sci. 16 (1), 102–107. doi: 10.1515/biol-2021-0015
Yu, Q. Q., Tian, H. Y., Yue, K., Liu, J. J., Zhang, B., Li, X. G., et al. (2016). A P-Loop NTPase regulates quiescent center cell division and distal stem cell identity through the regulation of ROS homeostasis in Arabidopsis root. PloS Genet. 12 (9), 21. doi: 10.1371/journal.pgen.1006175
Yuan, L. B., Dai, Y. S., Xie, L. J., Yu, L. J., Zhou, Y., Lai, Y. X., et al. (2017). Jasmonate regulates plant responses to postsubmergence reoxygenation through transcriptional activation of antioxidant synthesis. Plant Physiol. 173 (3), 1864–1880. doi: 10.1104/pp.16.01803
Zheng, L. Q., Liu, Z., Wang, Y., Yang, F., Wang, J. R., Huang, W. J., et al. (2021). Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis. Nat. Struct. Mol. Biol. 28 (5), 443–44+. doi: 10.1038/s41594-021-00591-9
Keywords: KATANIN, GDP-D-mannose pyrophosphorylase, Corymbia citriodora, expression analysis, wood
Citation: Wang C, Luo J, He W, Huang A, Lu W, Lin Y and Ou Y (2023) Genome-wide identification and expression analysis of GDP-D-mannose pyrophosphorylase and KATANIN in Corymbia citriodora. Front. Plant Sci. 14:1308354. doi: 10.3389/fpls.2023.1308354
Received: 06 October 2023; Accepted: 05 December 2023;
Published: 18 December 2023.
Edited by:
Shiming Liu, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Peng Wang, Chinese Academy of Tropical Agricultural Sciences, ChinaCopyright © 2023 Wang, Luo, He, Huang, Lu, Lin and Ou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuduan Ou, b3V5dWR1YW5AMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.