
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 13 October 2023
Sec. Plant Breeding
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1274392
This article is part of the Research TopicThe Role of Water Stress and Soil Texture on Plant Roots Anatomy, Architecture, and SenescenceView all 5 articles
Identifying loci for root system architecture (RSA) traits and developing available markers are crucial for wheat breeding. In this study, RSA-related traits, including total root length (TRL), total root area (TRA), and number of root tips (NRT), were evaluated in the Doumai/Shi4185 recombinant inbred line (RIL) population under hydroponics. In addition, both the RILs and parents were genotyped using the wheat 90K single-nucleotide polymorphism (SNP) array. In total, two quantitative trait loci (QTLs) each for TRL (QTRL.caas-4A.1 and QTRL.caas-4A.2), TRA (QTRA.caas-4A and QTRA.caas-4D), and NRT (QNRT.caas-5B and QNRT.caas-5D) were identified and each explaining 5.94%–9.47%, 6.85%–7.10%, and 5.91%–10.16% phenotypic variances, respectively. Among these, QTRL.caas-4A.1 and QTRA.caas-4A overlapped with previous reports, while QTRL.caas-4A.2, QTRA.caas-4D, QNRT.caas-5B, and QNRT.caas-5D were novel. The favorable alleles of QTRL.caas-4A.1, QTRA.caas-4A, and QTRA.caas-5B were contributed by Doumai, whereas the favorable alleles of QTRL.caas-4A.2, QTRA.caas-4D, and QTRA.caas-5D originated from Shi 4185. Additionally, two competitive allele-specific PCR (KASP) markers, Kasp_4A_RL (QTRA.caas-4A) and Kasp_5D_RT (QNRT.caas-5D), were developed and validated in 165 wheat accessions. This study provides new loci and available KASP markers, accelerating wheat breeding for higher yields.
Wheat production is influenced seriously by abiotic stresses. Breeding higher-yielding and more stable accessions under abiotic stress is a crucial objective in modern wheat breeding (Bai et al., 2013; Alahmad et al., 2019; Adeleke et al., 2020). Root system architecture (RSA) traits, which contribute to the shape of the root system, are pivotal for wheat agronomic performance and play vital roles in plant development (Kabir et al., 2015; Li et al., 2015; Kulkarni et al., 2017; Alemu et al., 2021). Optimizing RSA traits is important not only for abiotic stress tolerance but also for efficient nutrient and water acquisition (Gupta et al., 2017; Griffiths et al., 2022). RSA traits primarily include root length, surface area, and the number of root tips, which influence the major components and spatial arrangement of root systems, significantly affecting water and nutrient uptake (Atkinson et al., 2015; Maccaferri et al., 2016; Liu et al., 2019; Li et al., 2020; Ma et al., 2022; Maqbool et al., 2022).
Previous breeding programs predominantly focused on aboveground traits such as disease resistance, grain quality, and harvest index, while the application of RSA traits has been limited due to the complexity of phenotypic evaluation (Rogers and Benfey, 2015; Roselló et al., 2019; Mehrabi et al., 2020; Rufo et al., 2020; Salarpour et al., 2020; Ober et al., 2021; Pariyar et al., 2021; Saini et al., 2021). Previous studies have indicated that the RSA traits are influenced by environmental factors and controlled by minor genes (Xie et al., 2017; Soriano and Alvaro, 2019; Maqbool et al., 2022). To accelerate the progress of breeding for RSA-related traits, it is imperative to identify the significant associated genomic regions (Soriano and Alvaro, 2019; Yang et al., 2021a). Nowadays, with the advancement of high-throughput genotyping, such as re-sequence and SNP assay (Wang et al., 2014), genome-wide linkage mapping has been widely employed to elucidate the genetic basis of complex traits. Over the past two decades, numerous QTLs have been identified for RSA-related traits (Maccaferri et al., 2016; Xie et al., 2017; International Wheat Genome Sequencing Consortium (IWGSC) et al., 2018; Ye et al., 2018; Soriano and Alvaro, 2019; Yang et al., 2021a; Yang et al., 2021b; Xu et al., 2023).
In this study, we conducted linkage mapping for RSA traits using the wheat 90K assays in a biparental recombinant inbred line (RIL) population derived from the Doumai/Shi 4185 cross. The primary objective of this study is to uncover the genetic basis of these traits and develop available KASP markers for improving wheat RSA.
Doumai is a derivative line of Hesheng 2, which originate from Yuanfeng 6 by Cobalt-60 radiation, whereas Shi 4185 is a widely grown winter wheat cultivar. The 262 F2:6 RILs derived from the Doumai/Shi 4185 were used for evaluating RSA-related traits. A hydroponic experiment was conducted with three replicates in a greenhouse. There were 20 seeds from each line that were surface sterilized with10% H2O2 for 20 min. Subsequently, the cleaned seeds were placed in Petri dishes with moist filter paper. When the coleoptiles reached about 2 cm in length, wheat seedlings were transferred to plastic trays (53 × 27 cm) containing Hoagland’s nutrient solution. Plastic trays were kept in 25°C with a 16-h light and 8-h darkness cycle. After 3 weeks, roots were evaluated for RSA-related traits (Table S1). Additionally, a diverse panel of 165 wheat cultivars primarily originating from the Yellow-Huai Wheat Region were also assessed for TRL, TRA, and NRTRAA traits to validate the effectiveness of competitive allele-specific PCR (KASP) markers.
Three RSA-related traits, namely, total root length (TRL), total root area (TRA), and number of root tips (NRT), were assessed using the WinRHIZO root analysis system (LA6400XL). The roots were arranged systematically in a dish and scanned using the Expression 11000XL. Subsequently, the images were imported into WinRHIZO software and analyzed using a fixed threshold parameter of 40. Each accession was scored on five plants for RSA traits, and means of three replicates were obtained. Basic statistical analyses and frequency distributions were conducted using SAS v9.3 (http://www.sas.com).
Both RILs and parents were genotyped using the wheat 90K SNP arrays (80,547 SNPs) by CapitalBio Corporation. SNPs with missing data >20% or minor allele frequency (MAF) <0.5 were filtered for further analysis. The filtered SNPs were then analyzed using the BIN function of IciMapping v4.2 (Meng et al., 2015) and grouped into bin markers, which were used to construct a linkage map employing the regression mapping algorithm by JoinMap v4.0. The linkage maps have been reported by Wen et al. (2017) and Li et al. (2018).
The inclusive composite interval mapping (ICIM) method using IciMapping v4.1 (Meng et al., 2015) was applied in this study. The logarithm of odds (LOD) threshold for declaring significant QTL was set as 2.62 based on 1000 permutation. The physical positions of SNPs were based on the IWGSC v1.0.
SNPs flanking QTL with higher PVE (QTRL.caas-4A.1, QTRA.caas-4A, and QNRT.caas-5D) were converted to KASPs (Rasheed et al., 2016) and designed using PolyMarker (http://www.polymarker.info/) (Table S2). The 384-well plates were read on PHERA starplus SNP (BMG Labtech GmbH, Ortenberg, Germany) and the genotype analysis was carried out using KlusterCaller (LGC, Hoddesdon, UK). All the KASPs were validated by 165 cultivars from the Yellow and Huai Wheat Region (Liu et al., 2019).
To identify candidate genes involved in the QTL for RSA-related traits detected in the Doumai/Shi 4185 RIL population, the genes located in the LD block region around the peak SNP ( ± 3.0 Mb kb based on previous LD decay analysis) of each QTL were extracted from the wheat IWGSC v1.1 annotation (https://wheat.pw.usda.gov/GG3/). Genes, excluding hypothetical proteins, transposon proteins, and retrotransposon proteins with SNPs in the coding region, were considered as candidate genes. Quantitative real-time PCR (qRT-PCR) was conducted to test expression differences of the candidate genes between Doumai and Shi 4185. The roots were sampled for RNA extraction after phenotyping. RNA was extracted using the TRIzol method, and the cDNA was synthesized with the HiScript II 1st Strand cDNA Synthesis Kit. Primers were designed using the Primer Premier V5.0. PCR was conducted with a mixture of 20 μl, including 2 μl cDNA, 10 μl ChamQ Universal SYBR qPCR Master Mix, and 0.4 μl of each primer. qRT-PCR was conducted in the ABI StepOnePlus Real-Time PCR System with Tower, and the gene expression level was analyzed by the 2–ΔΔCT method. All assays were performed in two biological replicates and three technical replicates. TaActin1 was used as the internal control to normalize the expression levels of different samples.
All three RSA-related traits exhibited continuous and significantly wide variation across the 262 RILs (Figure S1). The means of TRL, TRA, and NRT were 74.8 cm (range: 40.2 cm–111.9 cm), 10.4 cm2 (range: 6.9 cm2–15.2 cm2), and 328.2 root tips (range: 94.7–772.0). The standard deviation and coefficient of variation for TRL, TRA, and NRT were 18.1 cm (24.2%), 1.7 cm2 (16.4%), and 126.4 (38.5%), respectively. A significant correlation was observed between TRL, TRA, and NRT, with the correlation coefficient of 0.603 (P < 0.05) between TRL and TRA, 0.371 (P < 0.05) between TRL and NRT, and 0.312 (P < 0.05) between TRA and NRT.
In total, 11,012 SNP markers were employed for the construction of genetic linkage maps spanning 2030.0 cM (Wen et al., 2017; Li et al., 2018). Two QTLs for TRL were detected on chromosomes 4A, referred to as QTRL.caas-4A.1 (BobWhite_c20306_147-Tdurum_contig54973_1510) and QTRL.caas-4A.2 (BS00059454_51-Kukri_c19883_816), respectively. These QTLs explained 5.94% (additive effect: 5.17) and 9.45% (additive effect: −7.12) of the total phenotypic variances (Table 1; Figure 1). The favorable allele of QTRL.caas-4A.1 was contributed by Doumai, while the favorable allele of QTRL.caas-4A.2 was contributed by Shi 4185. Two QTL for TRA were detected on chromosomes 4A and 4D, named QTRA.caas-4A (BS00041735_51-BS00021715_51) and QTRL.caas-4D (Ex_c6665_1067-wsnp_J-D_rep_c51623_35119179), respectively, explaining 7.10% (additive effect: 0.40) and 6.85% (additive effect: −0.39) of the total phenotypic variance. The favorable allele of QTRA.caas-4A TRA was contributed by Doumai, while the favorable allele of QTRA.caas-4D was originated from Shi4185, respectively. Two QTLs for NRT were detected on chromosomes 5B and 5D and named as QNRT.caas-5B (wsnp_Ex_rep_c67320_65870601-Tdurum_contig10191_996) and QNRT.caas-5D (IAAV6218-Kukri_c46526_103), respectively, explaining 5.91% (additive effect: 26.7) and 10.16% (additive effect: 39.1) of the total phenotypic variances. The favorable alleles of QNRT.caas-5B and QNRT.caas-5D were contributed by Doumai and Shi 4185, respectively (Table 1).
Three QTL (QTRL.caas-4A.1, QTRA.caas-4A, and QNRT.caas-5D) with higher phenotypic effects were used to develop KASP markers. Although attempts were made to develop a KASP marker for QTRA.caas-4A, it was unable to effectively distinguish between the two parental genotypes in the RIL population. Therefore, the marker did not yield conclusive results. Consequently, two KASP markers, Kasp_4A_RL (QTRL.caas-4A, Bobwhite_c20306_147, 702.3 Mb) and Kasp_5D_RT (QNRT.caas-5D, Kukri_c46526_103, 454.1 Mb), were successfully developed based on the tightly linked SNP markers (Table 2; Figure 2). A total of 165 diverse cultivars were used to verify the effectiveness of the two KASP markers. For Kasp_4A_RL, the favorable allele (TT, account for 75.7%, mean TRL: 276.6 mm) exhibited higher TRL compared to the unfavorable allele (CC, 6.7%, mean TRL: 230.6 mm) at the P = 0.05 level (Table S3). For Kasp_5D_RT, the favorable allele (GG 88.5%, mean NRT: 226.7) showed higher NRT than unfavorable allele (AA, 5.5%, mean NRT: 172.5) at the P = 0.05 level (Table S4).
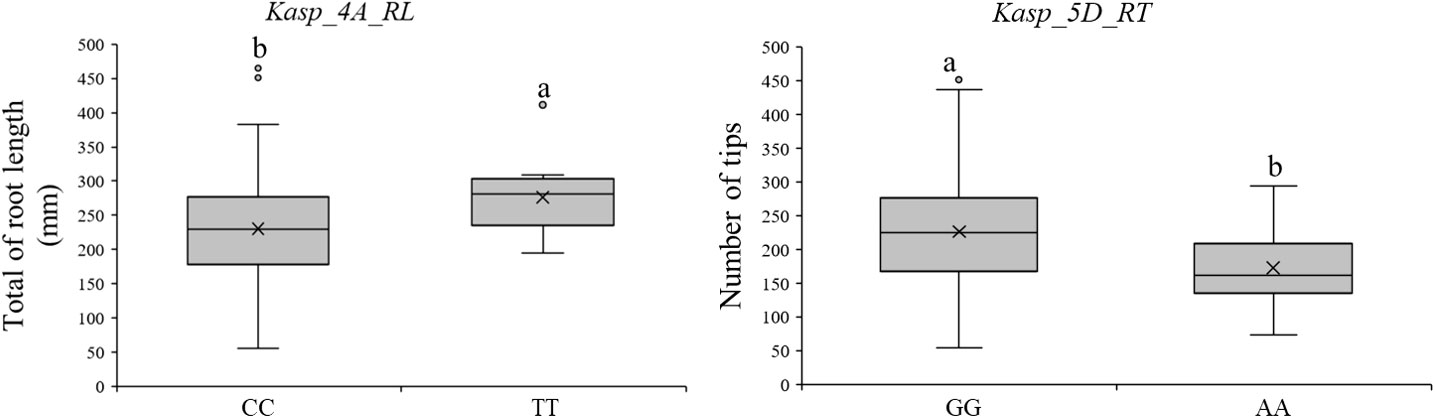
Figure 2 Validating the efficiency of KASP markers for discriminating RSA-related traits. Different lowercase letters means significant different at p<0.05 level.
In total, seven candidate genes were selected, primarily involved in the biological metabolism of plant hormones, cellulose, and the ubiquitin pathway (Table 3). Among these candidates, TraesCS4A01G296500 (QTRA.caas-4A) encoded an ethylene-regulated nuclear protein (ERT2)-like protein; TraesCS4A01G436700 (QTRL.caas-4A.1) encoded a calcium-binding protein kinase (CDPK)-related kinase; TraesCS4A01G456300 (QTRL.caas-4A.1) encoded a cellulose synthase-like protein; TraesCS4A01G469400 (QTRL.caas-4A.2) encoded an F-box protein; and TraesCS4D01G092000 (QTRA.caas-4D) encoded an ABC transporter, whereas both the TraesCS5D01G378400 and TraesCS5D01G380200 for QNRT.caas-5D encoded E3 ubiquitin-protein ligase. The expressions of the seven candidate genes in Doumai and Shi 4185 were detected using qRT-PCR. Of these, TraesCS4A01G469400, TraesCS4D01G092000, and TraesCS5D01G380200 showed no significant differences between the parents, whereas TraesCS4A01G296500, TraesCS4A01G436700, TraesCS4A01G456300, and TraesCS5D01G378400 showed more than 1.4–3.9-fold higher expression in Doumai compared to Shi4185 (Figure S2).
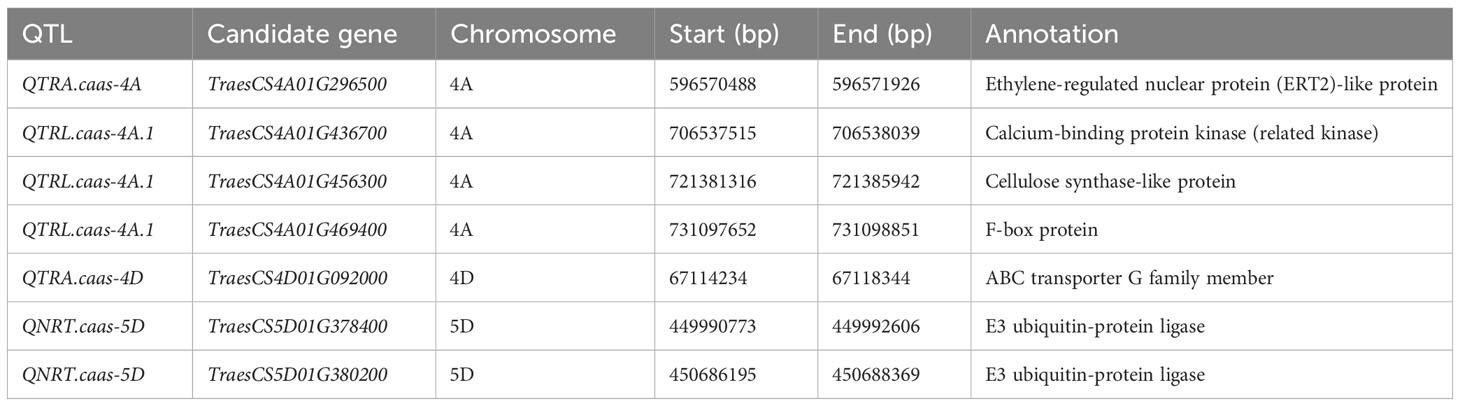
Table 3 The candidate genes for RSA-related traits identified in the Doumai/Shi 4185 RIL population.
Optimization of crop root systems has long been proposed. However, genetic improvement of crop roots has been rarely attempted (Maccaferri et al., 2016; Yang et al., 2021a). A comprehensive understanding of the genetic basis of RSA traits would facilitate the optimization of root systems under nutrient deficiency (Gu et al., 2016; Xie et al., 2017; Yang et al., 2021a; Yang et al., 2021b). In this study, we identified two QTLs each for TRL (QTRL.caas-4A.1 and QTRL.caas-4A.2), TRA (QTRA.caas-4A and QTRA.caas-4D), and NRT (QTRA.caas-5B and QTRA.caas-5D) TRA. Each of these QTLs accounted for 5.94%–9.47%, 6.85%–7.10%, and 5.91%–10.16% of the phenotypic variance, respectively.
Yang et al. (2021b) have identified five QTLs for root length (chromosomes 1A, 2B, 3B, and 7D), three QTLs for root tips (chromosomes 4A, 5A, and 7D), and nine QTLs for root surface (chromosomes 1A, 3A, 4B, and 4D) by linkage mapping in 198 doubled haploid lines of the Yangmai 16/Zhongmai 895 cross. Of the QTLs identified by Yang et al. (2021b), 4A (17.03 Mb–17.05 Mb) and 4D (16.64 Mb–30.66 Mb and 100.69 Mb–108.96 Mb) are different with the loci identified in our study (4A: 594.2 Mb–602.9 Mb, 702.3 Mb–721.3 Mb, and 731.4 Mb–732.5 Mb; 4D: 65.1 Mb–69.9 Mb). Liu et al. (2020) have evaluated 10 RSA-related traits at the seedling stage in 111 F9 RIL lines and have identified 19 QTLs mainly distributed on chromosomes 1A, 2B, 2D, 3A, 3B, 3D, 5A, and 5D. Of these, the 5D loci (415.8 Mb–457.6 Mb) overlapped with theQNRT.caas-5D (449.4 Mb–454.1 Mb) identified in this study. Alemu et al. (2021) have identified 38 QTLs from 167 historical landraces and 25 modern cultivars by GWAS. Of these, the loci on 4A (IWB21309, 17.01 Mb) and 5B (IWB8808, 701.5 Mb) for RSA traits were also different with the loci identified in this study (4A: 4A: 594.2 Mb–602.9 Mb, 702.3 Mb–721.3 Mb, and 731.4 Mb–732.5 Mb; 5B: 74.3–78.9). Furthermore, Saini et al. (2021) have identified six meta-QTLs for root-related traits on chromosome 4A by meta-analysis; MQTL4A.1 (651.78 Mb–705.73 Mb) and MQTL4A.6 (600.04 Mb–691.14 Mb) significantly associated with root length, root surface, and root tips overlapped with QTRL.caas-4A.1 (702.3 Mb–721.3 Mb) and QTRA.caas-4A (594.2 Mb–602.9 Mb). Xu et al. (2023) also identified 25 QTLs for root and shoot-related traits in 142 RILs derived from Xiaoyan 54 and Jing 411 cross, mainly located on chromosomes 1A, 3A, 4A, and 5B and different with the loci identified in this study. Previous studies have indicated that genes associated with plant height and vernalization may also have certain effects on root system establishment. Furthermore, multiple genes related to plant height and vernalization are found on chromosomes 4D (Rht2/Rht10 19.18 Mb, SVP3-4D/BM1-4D 469.46 Mb, Vrn2-4D/ZCCT1-4D 509.43 Mb), 5B (TaDEP1-5B 381.54 Mb, Vrn1-5B, 577.00 Mb, Q-5B 658.75 Mb), and 5D (TaDEP1-5D 329.11 Mb, Rht23 524.96 Mb, and Vrn1-5D 470.00 Mb) (Xu et al., 2023). Based on physical position, the RSA-related genes identified in this study (QTRA.caas-4D 65.1 Mb–69.9 Mb, QNRT.caas-5B 74.3 Mb–78.9 Mb, QNRT.caas-5D 449.6 Mb–454.1 Mb) are different from the reported wheat genes associated with plant height and vernalization. Therefore, they represent novel genes associated with RSA-related traits. Above all, compared with the previous results and meta-analysis, QTRL.caas-4A.2, QTRL.caas-4D, QNRT.caas-5B, and QNRT.caas-5D were novel.
In our study, we conducted linkage mapping for agronomic traits in the Doumai/Shi 4185 RIL population (Li et al., 2018) and identified several regions associated with both RSA-related traits and agronomic traits. Specifically, we found that QTRL.caas-4A.1 (702.3 Mb–721.3 Mb) co-located influencing QTL clusters related to thousand kernel weight (QTKW.caas-4AL.1, 708.6 Mb, IWB42202) (Li et al., 2018). Additionally, the loci for RSA traits on chromosomes 5D (IAAV6218, 449.6 Mb–454.1 Mb) co-located with regions affecting QTL clusters related to yield, including SN, KNS, TKW, FLW, and KL (IWB61072-IWB49479, 382.9 Mb–465.6 Mb) (Li et al., 2018). These results indicated that RSA traits loci could also be targeted to enhance yield potential and stability.
Four genes involved in the biological metabolism of plant hormones, cellulose, and the ubiquitin pathway were identified as high confidence candidate genes. Among these, TraesCS4A01G436700 of QTRL.caas-4A.1 encodes the CDPK-related kinase. In plants, CDPKs play a critical role in various signaling pathways and are pivotal in root growth and development (Yip Delormel and Boudsocq, 2019; Dekomah et al., 2022). Silencing CDPK in Medicago truncatula led to a significant reduction in root hair growth and cell length (Ivashuta et al., 2005). TRA Another candidate gene for QTRL.caas-4A.1 is TraesCS4A01G456300, encoding a cellulose synthase-like protein, which holds importance in plant growth-related and stress-responsive activities (Karas et al., 2021; Lou et al., 2022). TraesCS4A01G296500, associated with QTRA.caas-4AS, encodes an ethylene-regulated nuclear protein (ERT2)-like protein. Ethylene, a simple small molecule, plays a vital role in signal transduction, cell differentiation, division, and elongation (Qin et al., 2019). Ethylene has also been reported to influence traits related to RSA, leading to root hair and cluster root formation (Xu et al., 2020). Additionally, TraesCS5D01G380200, linked to QNRT.caas-5D, encodes an E3 ubiquitin-protein ligase, which plays essential roles in plant development (Chen et al., 2018) and is crucial for primary root growth and shoot development (Ramaiah et al., 2022).
Although conventional breeding has contributed to the enhancement of root system, the selection process is time-consuming and less efficient due to the challenges in field measurements of RSA traits (Rasheed et al., 2017). KASP offers a cost-effective, flexible, and highly accurate approach for MAS breeding. In this study, Kasp_4A_RL and Kasp_5D_RT were successfully developed based on tightly linked SNP markers, proving to be valuable tools for MAS in breeding programs. Additionally, accessions with more favorable alleles, superior RSA traits and appropriate agronomic traits, such as Yumai 18, Jinhe 9123, Yumai 34, Bainong 3217, Lumai 6, Jinmai 61, Lumai 5, Lumai 23, and Yumai 57 are recommended as parental lines for improvement of RSA traits.
In this study, we identified six QTLs associated with wheat RSA traits and successfully developed two KASP markers that can be utilized in wheat breeding programs aimed at achieving higher and more stable yields. This research serves as a foundational step towards gene cloning and enhancement of wheat root systems.
All datasets generated for this study are included in the article or Supplementary Material; further inquiries can be directed to the first author.
YJ: Data curation, Funding acquisition, Writing – original draft. YW: Data curation, Software, Validation, Writing – original draft, Writing – review & editing. JL: Software, Validation, Writing – review & editing. FW: Formal Analysis, Resources, Software, Investigation, Writing – review & editing. XQ: Investigation, Project administration, Writing – review & editing. PL: Funding acquisition, Supervision, Validation, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the Key Project of Shandong Natural Science Foundation (ZR2020KC042); Shandong Agricultural Seeds Engineering Project (2021LZGC013); and Agricultural Science and Technology Innovation Project of Shandong (2020CXGC01080517).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1274392/full#supplementary-material
Supplementary Figure 1 | Frequency distributions for RSA related traits in the Doumai/Shi 4185 RIL population. TRL: total root length; TRA: total root surface area; NRT: number of root tips.
Supplementary Figure 2 | qRT-PCR of the seven candidate genes identified in Doumai/Shi 4185 RIL population.
Adeleke, E., Millas, R., McNeal, W., Faris, J., Taheri, A. (2020). Variation analysis of root system development in wheat seedlings using root phenotyping system. Agronomy 1, 206. doi: 10.3390/agronomy10020206
Alahmad, S., El Hassouni, K., Bassi, F. M., Dinglasan, E., Youssef, C., Quarry, G., et al. (2019). A major root architecture QTL responding to water limitation in durum wheat. Front. Plant Sci. 10, 436. doi: 10.3389/fpls.2019.00436
Alemu, A., Feyissa, T., Maccaferri, M., Sciara, G., Tuberosa, R., Ammar, K., et al. (2021). Genome-wide association analysis unveils novel QTLs for seminal root system architecture traits in Ethiopian durum wheat. BMC Genomics 22, 1–16. doi: 10.1186/s12864-020-07320-4
Atkinson, J. A., Wingen, L. U., Griffiths, M., Pound, M. P., Gaju, O., Foulkes, M. J., et al. (2015). Phenotyping pipeline reveals major seedling root growth QTL in hexaploid wheat. J. Exp. Bot. 66, 2283–2292. doi: 10.1093/jxb/erv006
Bai, C., Liang, Y., Hawkesford, M. J. (2013). Identification of QTL associated with seedling root traits and their correlation with plant height in wheat. J. Exp. Bot. 64, 1745–1753. doi: 10.1093/jxb/ert041
Chen, H., Ma, B., Zhou, Y., He, S. J., Tang, S. Y., Lu, X., et al. (2018). E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of Aux/IAA protein. P Natl. Acad. Sci. U.S.A. 115, 4513–4518. doi: 10.1073/pnas.1719387115
Dekomah, S. D., Bi, Z., Dormatey, R., Wang, Y., Haider, F. U., Sun, C., et al. (2022). The role of CDPKs in plant development, nutrient and stress signaling. Front. Genet. 13, 996203. doi: 10.3389/fgene.2022.996203
Griffiths, M., Atkinson, J. A., Gardiner, L. J., Swarup, R., Pound, M. P., Wilson, M. H., et al. (2022). Identification of QTL and underlying genes for root system architecture associated with nitrate nutrition in hexaploid wheat. J. Integr. Agr. 21, 917–932. doi: 10.1016/S2095-3119(21)63700-0
Gu, R., Chen, F., Long, L., Cai, H., Liu, Z., Yang, J., et al. (2016). Enhancing phosphorus uptake efficiency through QTL-based selection for root system architecture in maize. J. Genet. Genomics 43, 663–672. doi: 10.1016/j.jgg.2016.11.002
Gupta, P. K., Balyan, H. S., Gahlaut, V. (2017). QTL analysis for drought tolerance in wheat: present status and future possibilities. Agronomy 7, 5. doi: 10.3390/agronomy7010005
International Wheat Genome Sequencing Consortium (IWGSC), Appels, R., Eversole, K., Stein, N., Feuillet, C., Keller, B., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, 7191. doi: 10.1126/science.aar7191
Ivashuta, S., Liu, J., Liu, J., Lohar, D. P., Haridas, S., Bucciarelli, B., et al (2005). RNA interference identifies acalcium-dependent protein kinase involved in Medicago truncatula root development. Plant Cell, 17, 2911–2921. doi: 10.1105/tpc.105.03539
Kabir, M. R., Liu, G., Guan, P., Wang, F., Khan, A. A., Ni, Z., et al. (2015). Mapping QTL associated with root traits using two different populations in wheat (Triticum aestivum L.). Euphytica 206, 175–190. doi: 10.1007/s10681-015-1495-z
Karas, B. J., Ross, L., Novero, M., Amyot, L., Shrestha, A., Inada, S., et al. (2021). Intragenic complementation at the Lotus japonicus CELLULOSE SYNTHASE-LIKE D1 locus rescues root hair defects. Plant Physiol. 186, 2037–2050. doi: 10.1093/plphys/kiab204
Kulkarni, M., Soolanayakanahally, R., Ogawa, S., Uga, Y., Selvaraj, M. G., Kagale, S. (2017). Drought response in wheat: key genes and regulatory mechanisms controlling root system architecture and transpiration efficiency. Front. Chem. 5, 106. doi: 10.3389/fchem.2017.00106
Li, P., Chen, F., Cai, H., Liu, J., Pan, Q., Liu, Z., et al. (2015). A genetic relationship between nitrogen use efficiency and seedling root traits in maize as revealed by QTL analysis. J. Exp. Bot. 66, 3175–3188. doi: 10.1093/jxb/erv127
Li, T., Ma, J., Zou, Y., Chen, G., Ding, P., Zhang, H., et al. (2020). Quantitative trait loci for seeding root traits and the relationships between root and agronomic traits in common wheat. Genome 63, 27–36. doi: 10.1139/gen-2019-0116
Li, F., Wen, W., He, Z., Liu, J., Jin, H., Cao, S., et al. (2018). Genome-wide linkage mapping of yield-related traits in three Chinese bread wheat populations using high-density SNP markers. Theor. Appl. Genet. 131, 1903–1924. doi: 10.1007/s00122-018-3122-6
Liu, P., Jin, Y., Liu, J., Liu, C., Yao, H., Luo, F., et al. (2019). Genome-wide association mapping of root system architecture traits in common wheat (Triticum aestivum L.). Euphytica 215, 1–12. doi: 10.1007/s10681-019-2452-z
Liu, R., Wu, F., Xin, Y., Yu, L., Wang, Z., Liu, S., et al. (2020). Quantitative trait loci analysis for root traits in synthetic hexaploid wheat under drought stress conditions. J. Integr. Agr. 19, 1947–1960. doi: 10.1016/S2095-3119(19)62825-X
Lou, H., Tucker, M. R., Shirley, N. J., Lahnstein, J., Yang, X., Ma, C., et al. (2022). The cellulose synthase-like F3 (CslF3) gene mediates cell wall polysaccharide synthesis and affects root growth and differentiation in barley. Plant J. 110, 1681–1699. doi: 10.1111/tpj.15764
Ma, J., Zhao, D., Tang, X., Yuan, M., Zhang, D., Xu, M., et al. (2022). Genome-wide association study on root system architecture and identification of candidate genes in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 23, 1843. doi: 10.3390/ijms23031843
Maccaferri, M., El-Feki, W., Nazemi, G., Salvi, S., Canè, M. A., Colalongo, M. C., et al. (2016). Prioritizing quantitative trait loci for root system architecture in tetraploid wheat. J. Exp. Bot. 67, 1161–1178. doi: 10.1093/jxb/erw039
Maqbool, S., Ahmad, S., Kainat, Z., Khan, M. I., Maqbool, A., Hassan, M. A., et al. (2022). Root system architecture of historical spring wheat cultivars is associated with alleles and transcripts of major functional genes. BMC Plant Biol. 22, 590. doi: 10.1186/s12870-022-03937-7
Mehrabi, A. A., Pour-Aboughadareh, A., Mansouri, S., Hosseini, A. (2020). Genome-wide association analysis of root system architecture features and agronomic traits in durum wheat. Mol. Breed. 40, 1–17. doi: 10.1007/s11032-020-01136-6
Meng, L., Li, H., Zhang, L., Wang, J. (2015). QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Ober, E. S., Alahmad, S., Cockram, J., Forestan, C., Hickey, L. T., Kant, J., et al. (2021). Wheat root systems as a breeding target for climate resilience. Theor. Appl. Genet. 134, 1645–1662. doi: 10.1007/s00122-021-03819-w
Pariyar, S. R., Nagel, K. A., Lentz, J., Galinski, A., Wilhelm, J., Putz, A., et al. (2021). Variation in root system architecture among the founder parents of two 8-way MAGIC wheat populations for selection in breeding. Agronomy 11, 2452. doi: 10.3390/agronomy11122452
Qin, H., He, L., Huang, R. (2019). The coordination of ethylene and other hormones in primary root development. Front. Plant Sci. 10, 874. doi: 10.3389/fpls.2019.00874
Ramaiah, M., Jain, A., Yugandhar, P., Raghothama, K. G. (2022). ATL8, a RING E3 ligase, modulates root growth and phosphate homeostasis in Arabidopsis. Plant Physiol. Bioch. 179, 90–99. doi: 10.1016/j.plaphy.2022.03.019
Rasheed, A., Hao, Y., Xia, X., Khan, A., Xu, Y., Varshney, R. K., He, Z. (2017). Crop breedingchips and genotyping platforms: progress, challenges, and perspectives. Mol. Plant. 10, 1047–1064. doi: 10.1016/j.molp.2017.06.008
Rasheed, A., Wen, W., Gao, F., Zhai, S., Jin, H., Liu, J., et al. (2016). Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 129, 1843–1860. doi: 10.1007/s00122-016-2743-x
Rogers, E. D., Benfey, P. N. (2015). Regulation of plant root system architecture: implications for crop advancement. Curr. Opin. Biotech. 32, 93–98. doi: 10.1016/j.copbio.2014.11.015
Roselló, M., Royo, C., Sanchez-Garcia, M., Soriano, J. M. (2019). Genetic dissection of the seminal root system architecture in mediterranean durum wheat landraces by genome-wide association study. Agronomy 9, 364. doi: 10.3390/agronomy9070364
Rufo, R., Salvi, S., Royo, C., Soriano, J. M. (2020). Exploring the genetic architecture of root-related traits in mediterranean bread wheat landraces by genome-wide association analysis. Agronomy 10, 613. doi: 10.3390/agronomy10050613
Saini, D. K., Chopra, Y., Pal, N., Chahal, A., Srivastava, P., Gupta, P. K. (2021). Meta-QTLs, ortho-MQTLs and candidate genes for nitrogen use efficiency and root system architecture in bread wheat (Triticum aestivum L.). Physiol. Mol. Biol. Pla. 27, 2245–2267. doi: 10.1007/s12298-021-01085-0
Salarpour, M., Pakniyat, H., Abdolshahi, R., Heidari, B., Razi, H., Afzali, R. (2020). Mapping QTL for agronomic and root traits in the Kukri/RAC875 wheat (Triticum aestivum L.) population under drought stress conditions. Euphytica 216, 1–19. doi: 10.1007/s10681-020-02627-5
Soriano, J. M., Alvaro, F. (2019). Discovering consensus genomic regions in wheat for root-related traits by QTL meta-analysis. Sci-Rep. 9, 1–14. doi: 10.1038/s41598-019-47038-2
Wang, S., Wong, D., Forrest, K., Allen, A., Chao, S., Huang, B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. doi: 10.1111/pbi.12183
Wen, W., He, Z., Gao, F., Liu, J., Jin, H., Zhai, S., et al. (2017). A high-density consensus map of common wheat integrating four mapping populations scanned by the 90K SNP array. Front. Plant Sci. 8, 1389. doi: 10.3389/fpls.2017.01389
Xie, Q., Fernando, K. M., Mayes, S., Sparkes, D. L. (2017). Identifying seedling root architectural traits associated with yield and yield components in wheat. Ann. Bot-London 119, 1115–1129. doi: 10.1093/aob/mcx001
Xu, D., Jia, C., Lv, X., Yang, T., Qin, H., Wang, Y., et al. (2023). In silico curation of QTL-rich clusters and candidate gene identification for plant height of bread wheat. Crop J. doi: 10.1016/j.cj.2023.05.007
Xu, Y., Yang, Y., Wu, S., Liu, D., Ren, Y. (2023). QTL mapping for root traits and their effects on nutrient uptake and yield performance in common wheat (Triticum aestivum L.). Agriculture 13, 210. doi: 10.3390/agriculture13010210
Xu, P., Zhao, P. X., Cai, X. T., Mao, J. L., Miao, Z. Q., Xiang, C. B. (2020). Integration of jasmonic acid and ethylene into auxin signaling in root development. Front. Plant Sci. 11, 271. doi: 10.3389/fpls.2020.00271
Yang, M., Wang, C., Hassan, M. A., Li, F., Xia, X., Shi, S., et al. (2021b). QTL mapping of root traits in wheat under different phosphorus levels using hydroponic culture. BMC Genomics 22, 1–12. doi: 10.1186/s12864-021-07425-4
Yang, M., Wang, C., Hassan, M., Wu, Y., Xia, X., Shi, S., et al. (2021a). QTL mapping of seedling biomass and root traits under different nitrogen conditions in bread wheat (Triticum aestivum L.). J. Integr. Agr. 20, 1180–1192. doi: 10.1016/S2095-3119(20)63192-6
Ye, H., Roorkiwal, M., Valliyodan, B., Zhou, L., Chen, P., Varshney, R. K., et al. (2018). Genetic diversity of root system architecture in response to drought stress in grain legumes. J. Exp. Bot. 69, 3267–3277. doi: 10.1093/jxb/ery082
Keywords: bread wheat, marker-assisted selection, quantitative trait locus (QTL), root system architecture (RSA), single-nucleotide polymorphism (SNP)
Citation: Jin Y, Wang Y, Liu J, Wang F, Qiu X and Liu P (2023) Genome-wide linkage mapping of root system architecture-related traits in common wheat (Triticum aestivum L.). Front. Plant Sci. 14:1274392. doi: 10.3389/fpls.2023.1274392
Received: 08 August 2023; Accepted: 22 September 2023;
Published: 13 October 2023.
Edited by:
Dongcheng Liu, Hebei Agricultural University, ChinaReviewed by:
Ana M. Casas, Spanish National Research Council (CSIC), SpainCopyright © 2023 Jin, Wang, Liu, Wang, Qiu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jindong Liu, bGl1amluZG9uZ0BjYWFzLmNu; Peng Liu, bGl1cDlAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.