- 1Anhui Engineering Research Center for Eco-agriculture of Traditional Chinese Medicine, Anhui Dabieshan Academy of Traditional Chinese Medicine, College of Biological and Pharmaceutical Engineering, West Anhui University, Lu’an, China
- 2Department of Plant Science, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
- 3College of pharmacy, Anhui University of Chinese Medicine, Hefei, China
Introduction: KNOX plays a pivotal role in governing plant growth, development, and responses to diverse abiotic and biotic stresses. However, information on the relationship between the KNOX gene family and expression levels under different treatments in Dendrobium is still limited.
Methods: To address this problem, we first used bioinformatics methods and revealed the presence of 19 KNOX genes distributed among 13 chromosomes in the Dendrobium huoshanense genome. Through an analysis of phylogenetic relationships, these genes were classified into three distinct clades: class I, class II, and class M. Our investigation included promoter analysis, revealing various cis-acting elements associated with hormones, growth and development, and abiotic stress responses. Additionally, qRT-PCR experiments were conducted to assess the expression patterns of DhKNOX genes under different treatments, including ABA, MeJA, SA, and drought.
Results: The results demonstrated differential expression of DhKNOX genes in response to these treatments, thereby highlighting their potential roles in stress adaptation.
Discussion: Overall, our results contribute important insights for further investigations into the functional characterization of the Dendrobium KNOX gene family, shedding light on their roles in plant development and stress responses.
Introduction
The KNOTTED1-like homeobox (KNOX) is a big transcription factor family that plays a key role in the regulation of plant growth and development (Hamant and Pautot, 2010). KNOX proteins generally have four conserved domains, which are as follows: KNOX1, KNOX2, ELK, and HOX (Gao et al., 2015). The KNOX gene family is ubiquitous, from lower plant algae and bryophytes to higher seed plants (Gao et al., 2015). Based on the structural characteristics, expression mode, and phylogenetic relationship of the KNOX gene, researchers usually divide it into two subfamilies: class I and class II (Furumizu et al., 2015). Different types of KNOX genes exhibit distinct expression patterns and biological functions. Class I KNOX proteins can promote cell proliferation, and their members have a significant role in the formation and maintenance of the plant stem tip meristem (SAM) in angiosperms. In addition, the class I KNOX gene is expressed in the apical meristem, while its expression level decreases in lateral buds (axillary buds), and mutants cannot maintain apical meristem (Gao et al., 2015). Moreover, the class I KNOX gene is also involved in other development processes, such as compound leaf formation, internode elongation, inflorescence structure, and the establishment of vascular tissue connections between parasitic plants and their hosts (Alakonya et al., 2012; Cheng et al., 2019). Therefore, the regulatory function of the class I KNOX gene has been involved in various developmental processes during the development of the stem system.
In higher plants, class II KNOX genes have relatively broad tissue specificity, while the expression of class I KNOX genes is more limited. Class II KNOX gene is expressed in differentiated organs, including leaves, stems, flowers, and roots; its involvement in some developmental processes not only in vascular tissue but also in seed coat development and inflorescence stem in plants (Gao et al., 2015). In Arabidopsis, class II genes, such as KNAT3, KNAT4, and KNAT5 play a role in lateral organ differentiation due to functional redundancy. The class II KNOX family has relatively few studies due to the lack of phenotypes of known mutants. Among class II KNOX genes, KNAT7 has received the most attention and is known to play a role in the transcriptional network regulating secondary cell wall biosynthesis (Furumizu et al., 2015; Gao et al., 2015). However, there are relatively few reports on the function of the class II KNOX gene in plants, which is mainly related to the formation of secondary cell walls. For example, the synergistic effect of KNAT7 and KNAT3 affects the deposition of secondary cell walls in plants, thereby altering the mechanical support strength of Arabidopsis stems (Wang S. M. et al., 2020); class KNATM is a special KNOX gene subfamily of dicotyledons. Research on the KNATM gene of Arabidopsis showed that this gene is involved in regulating leaf polarity and leaf shape (Magnani and Hake, 2008).
Other studies have shown that KNOX is involved in a variety of developmental processes, mainly by influencing hormone metabolism and signal transduction pathways (Bolduc and Hake, 2009). KNOX activates the biosynthesis of plant cytokinin (Yanai et al., 2005). For instance, MtKNOX3 activates the biological isopentenyl transferase (IPT) gene of cytokinin in apples, regulates the development of nodules, and activates the biosynthesis of cytokinin during nodulation (Azarakhsh et al., 2015). KNOX protein can inhibit the production of gibberellin (GA), and KNOX negatively regulates the accumulation of GA by controlling the abundance of GA2 oxidase (Bolduc and Hake, 2009). In addition, the KNOX protein also participates in other hormone signal transduction pathways. KNOX changes the abundance of proteins related to auxin transporter signal transduction components, thus regulating tomato abscission (Ma et al., 2015). During the germination and early seedling development of Arabidopsis, KNAT3 interacts with BELL-like homeodomain (BLH) proteins and cooperatively regulates the ABA response (Dachan et al., 2013).
Dendrobium huoshanense is a perennial epiphytic herb belonging to the Orchidaceae family, and it holds the status of a nationally protected variety of traditional Chinese medicine. This plant species possesses significant medicinal value (Liu et al., 2018; Wang et al., 2022), which is considered the best all Dendrobium species due to its significant immune regulation, anti-inflammatory, and anticancer activities (Lin et al., 2014; Zha et al., 2017). In addition, clinical studies have found that polysaccharides from Dendrobium huoshanense have a significant inhibitory effect on the apoptosis of human lens epithelial cells (Zha et al., 2017). The continuous improvement of the genome information of D. huoshanense (Han et al., 2020) will provide an opportunity to analyze the KNOX gene, which plays significant roles in plant growth and development, as well as the response of Dendrobium to different environmental stress conditions. Although KNOX proteins have been reported in Arabidopsis and maize, there is limited research on the identification and expression analysis of KNOX gene family members in Dendrobium. In this study, a bioinformatics approach was employed to identify the KNOX gene in Dendrobium. The analysis combined the characteristics of the KNOX gene family as well as the basic physical and chemical properties, chromosome distribution, evolutionary relationship, gene structure, cis-acting elements, and expression patterns under different stress conditions. These findings lay the foundation for further investigation into the function and regulatory mechanisms of the KNOX gene in Dendrobium plants.
Materials and methods
Plant materials and growth conditions
Tissue-cultured seedlings of D. huoshanense were subjected to a sterilization process and then planted on Murashige and Skoog (MS) media. This cultivation occurred in the tissue culture room with long-day conditions (16 h light, 20°C/8 h dark) of West Anhui University’s Anhui Engineering Technology Research Center of Plant Cell Engineering, located in Lu’an City, Anhui Province. The methodology employed in this study was based on previously published articles (Cao et al., 2019). Subsequently, 100 μM MeJA (Aladdin, Shanghai of China), 100 μM SA (Aladdin, Shanghai of China), and 100 μM ABA (Aladdin, Shanghai of China) were added after 0.22 μM microporous filtration. Referring to the method of Cao et al. (2015), 20% PEG6000 was added to the culture medium to simulate drought stress. The leaves were sampled at 1 h, 4 h, and 16 h after treatment. There were three biological replicates per set of samples, and each sample was collected individually for each induction treatment and immediately stored at −80°C to facilitate RNA isolation. In addition, untreated D. huoshanense leaves were used as the control group for comparative purposes.
Identification of D. huoshanense KNOX gene family
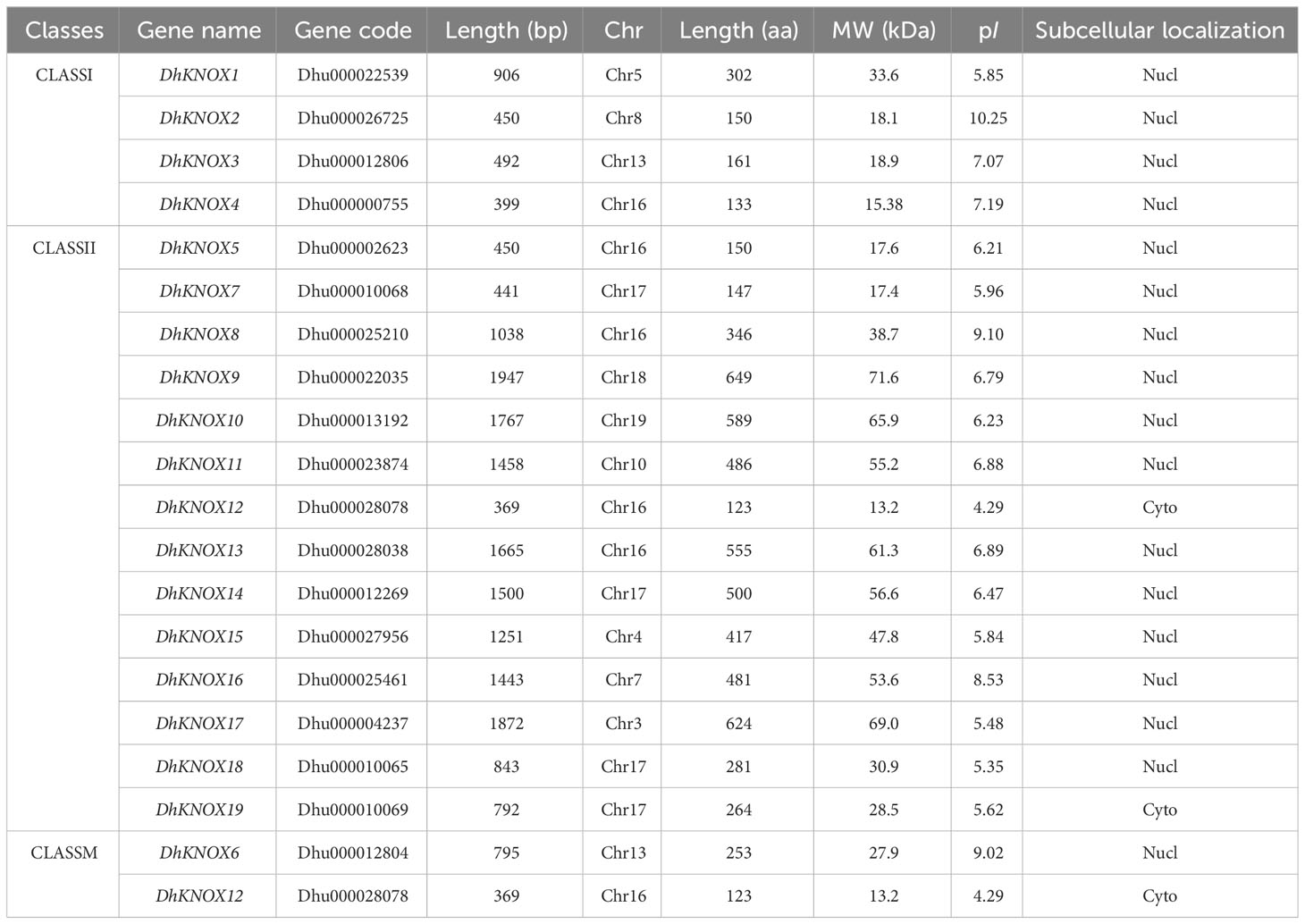
Using the BLAST tool available on the Dendrobium genome (https://ftp.cngb.org/pub/CNSA/data3) information website, a comparative analysis was conducted based on the KNOX protein sequence in Arabidopsis as the reference sequence (Furumizu et al., 2015), with a BLASTp (E-value = 0.001) and other default parameters. The objective was to identify candidate KNOX genes within the Dendrobium genome. Furthermore, the conserved KNOX1 (PF03790) or KNOX2 (PF03791) domains should be selected for further analysis using the structural properties that were identified using the Pfam website (Cheng et al., 2019). Subsequently, the isoelectric point and molecular weight of the KNOX gene family members of D. huoshanense were analyzed by ProtParam (https://web.expasy.org/protoparam/) (Finn et al., 2013). We predict of subcellular localization of the KNOX protein on the website WoLF PSORT (https://wolfpsort.hgc.jp/).
Construction of the phylogenetic tree and analysis of gene structures
To elucidate the evolutionary relationship of the DhKNOX gene, we employed MEGA 5.2 software to construct a KNOX gene phylogenetic tree with 1,000 bootstrap values (Tamura et al., 2011). The GenBank accession codes were used to construct the phylogenetic tree (Supplementary Table S1). According to the original annotation file in GFF3 format, the CDS of DhKNOXs was used for gene structure analysis and visualized with TBtools (Chen et al., 2018).
Chromosome mapping of DhKNOX gene family members
The chromosome location information of the KNOX gene family members was obtained from the Dendrobium genome annotation file. The KNOX gene information from the Dendrobium genome database was utilized to perform chromosome localization using the MapDraw V2.1 software (Lee et al., 2013).
Analysis of cis-acting elements of DhKNOX gene
TBtools software was used to obtain the 2,000-bp promoter region upstream of the CDS sequence of each KNOX gene Dendrobium from the genome database and to analyze cis-acting elements in the KNOX gene promoter. The obtained promoter region sequence was submitted to the PlantCARE database to predict cis-acting elements (Cheng et al., 2019). We implement the prediction of protein interactions among members of the DhKNOX gene family through the STRING website (https://cn.string-db.org) (Yang et al., 2022).
Identification of conserved motifs
The sequence alignment of KNOX proteins was performed using ClustalX software (Cao et al., 2016). Additionally, the conserved motifs of KNOX proteins were identified by the MEME online tools (Bailey et al., 2015).
RNA isolation and qRT-PCR analysis
The total RNA from D. huoshanense leaves was extracted using a plant RNA extraction kit (Tiangen). qRT-PCR was used to detect the expression level of the KNOX gene at different times after treatment. The internal control gene was used as a reference (Fan et al., 2016). The specific primers for each gene are shown in Supplementary Table S2. The qRT-PCR reaction system was 20 µL, and the reaction procedure was 95°C for 3 min, 95°C for 10 s, 60°C for 30 s, and 45 cycles. The melting (65°C~95°C) program is finally run after signal acquisition. Three biological repeats were set for each sample, and each biological duplicate sample was tested three times. Finally, the relative gene expression of KNOX genes was calculated using 2−ΔΔCt methods (Livak and Schmittgen, 2001).
Results
Identification and characterization of Dendrobium KNOX family members
The query sequence used for this analysis consisted of the KNOX family members from Arabidopsis thaliana, totaling nine sequences. Based on this query, 19 nonredundant KNOX family members were identified from the genome of Dendrobium. These newly identified members were named DhKNOX1 to DhKNOX19. Table 1 presents various characteristics of the DhKNOX genes in Dendrobium. The coding region lengths of the DhKNOX genes range from 369 to 1,872 bp, while the corresponding coding amino acid lengths range from 123 to 624. The molecular weight of the DhKNOX proteins spans from 18.81 to 69.0 kD, and their isoelectric points range from 4.29 to 10.25. Through the examination of predicted subcellular localization, it was ascertained that the nucleus emerges as the predominant site for the subcellular localization of the majority of DhKNOX genes. This observation is in consonance with their established functions as transcription factors.
Multiple sequence alignment of DhKNOX proteins
Multiple sequence alignments revealed that the KNOX protein sequence of Dendrobium has four relatively conserved domains (Figure 1). The KNOX1 and KNOX2 domains are located at the N-terminus of the protein, while ELK and HOX are located at the C-terminus of the protein. The HOX domain demonstrates the highest level of conservation and exhibits the typical structural characteristics found in the TALE homeobox protein superfamily. In addition, the conserved domains of KNOX1, KNOX2, and ELK exhibit notable conservation. For instance, the conserved domain KNOX2 displays a highly conserved E-L-D amino acid sequence, while the conserved domain ELK features a highly conserved E-L-K amino acid sequence.
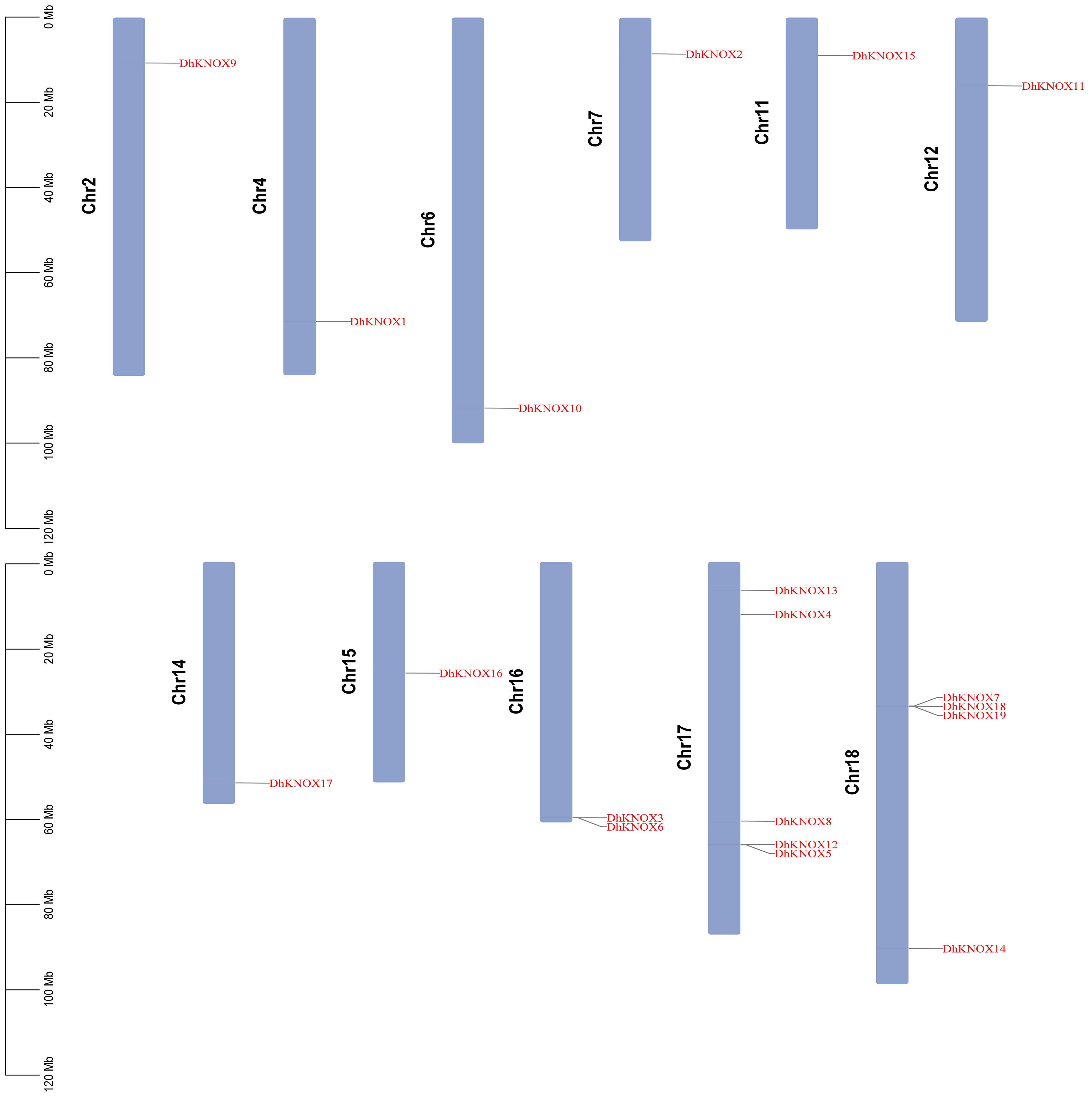
Chromosome mapping of the KNOX gene in Dendrobium
To elucidate the distribution of individual DhKNOX members within the Dendrobium genome, a distribution map was generated. This map visually depicts the arrangement of the 19 DhKNOX genes across each specific chromosome of Dendrobium, as shown in Figure 2. The 19 DhKNOX genes have been clearly located on chromosomes and are distributed on 11 different chromosomes. The distribution of DhKNOX genes across the 11 chromosomes of Dendrobium is widely scattered. Specifically, chromosomes 16, 17, and 18 harbor two, five, and four DhKNOX genes, respectively, while the remaining eight chromosomes each contain one DhKNOX gene. Furthermore, a notable observation is that the majority of DhKNOX genes are located toward the ends of the chromosomes.
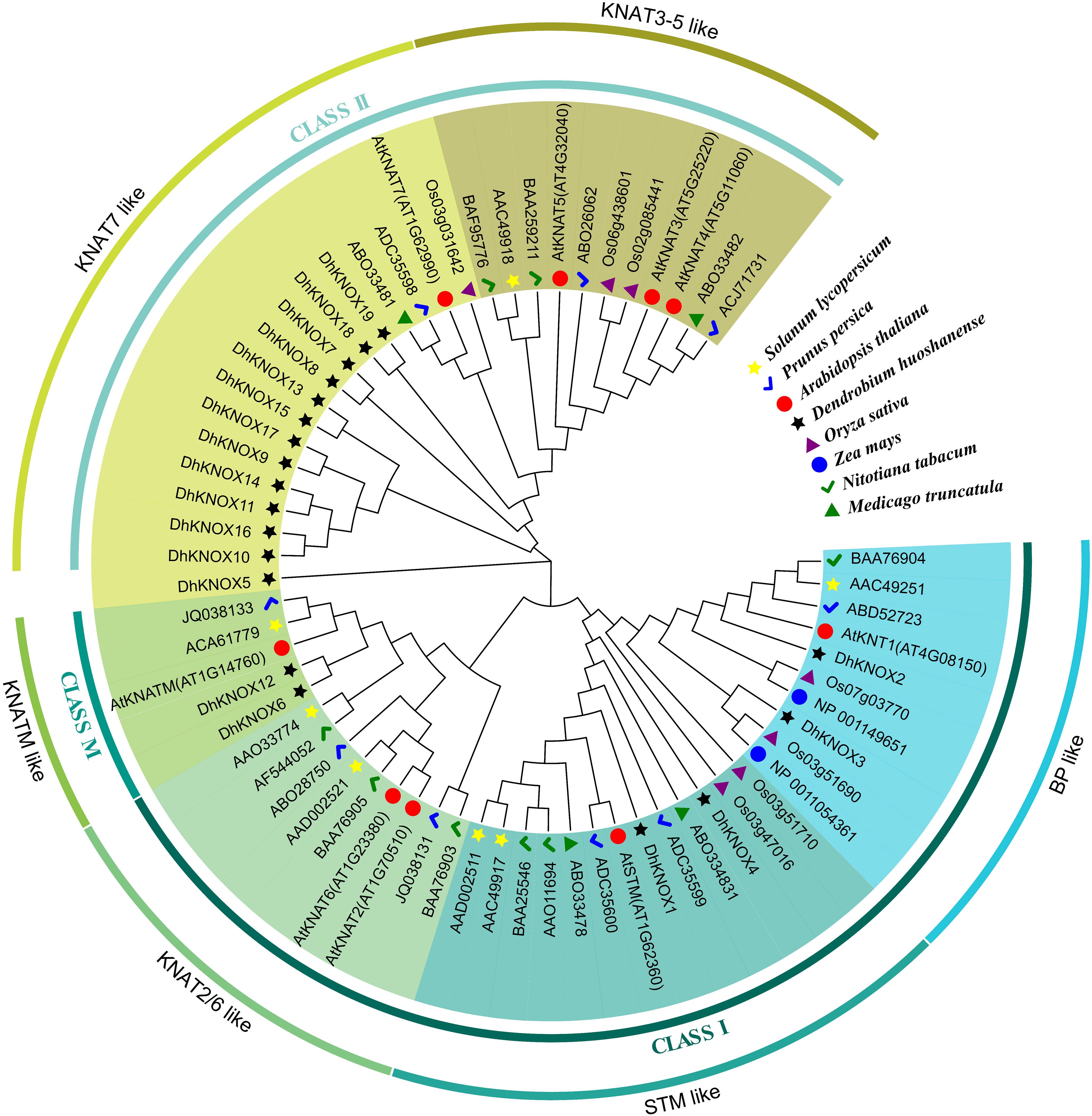
Phylogenetic analysis of the DhKNOX gene
The KNOX protein sequences of well-known model plants such as Arabidopsis and rice, along with the KNOX protein sequence of D. huoshanense were constructed using MEGA 5.2 software (Figure 3). The results showed that these KNOX genes were divided into three categories: class I, class II, and class KNATM. We further analyzed and found that these proteins include six subfamilies: class I includes three subfamilies, class II consists of two subfamilies, and class M contains only the KNATM-like subfamily. In these three large families, most DhKNOX genes are distributed in class II. Among the DhKNOX proteins, both the BP-like subfamily and STM-like subfamily contains two DhKNOX members. While there is no member of the KNAT2/6-like subfamily, the other 13 DhKNOX proteins are classified as belonging to the KNAT7-like subfamily. Previous research reported that class I KNOX genes are related to lignin metabolism (Cheng et al., 2019). In our study, DhKNOX2 and DhKNOX3 belong to the BP-like subfamily and speculated that they may have similar biological functions. Interestingly, Scofield et al. (2014) demonstrated in Arabidopsis that STM-like proteins can regulate the expression level of the AtKNAT1/BP gene. DhKNOX1 and DhKNOX4 belong to the same subfamily, so we speculate that they may have similar biological functions.
Analysis of DhKNOX gene structure and conserved domain
To gain deeper insights into the Dendrobium KNOX gene family, we utilized the intraspecific phylogenetic tree (Figure 4A) and protein-conserved domain distribution (Figure 4B), as well as the gene structure of DhKNOX genes (Figure 4C). Subsequently, employing the MEME website (Figure 4B), we identified 10 conserved motifs across the 19 DhKNOX proteins. Significantly, almost all DhKNOX proteins encompass at least one conserved domain, such as motif1, motif3, motif4, and motif10. This observation reinforces the reliability of the DhKNOX gene screening results. However, it is worth mentioning that DhKNOX6 and DhKNOX12 in class M are exceptions, lacking conserved domains. Furthermore, from an evolutionary perspective, the DhKNOX genes within the same subfamily and exhibiting close relationships also demonstrate consistency in gene structure composition and protein domain allocation, which supports their close evolutionary relationships.
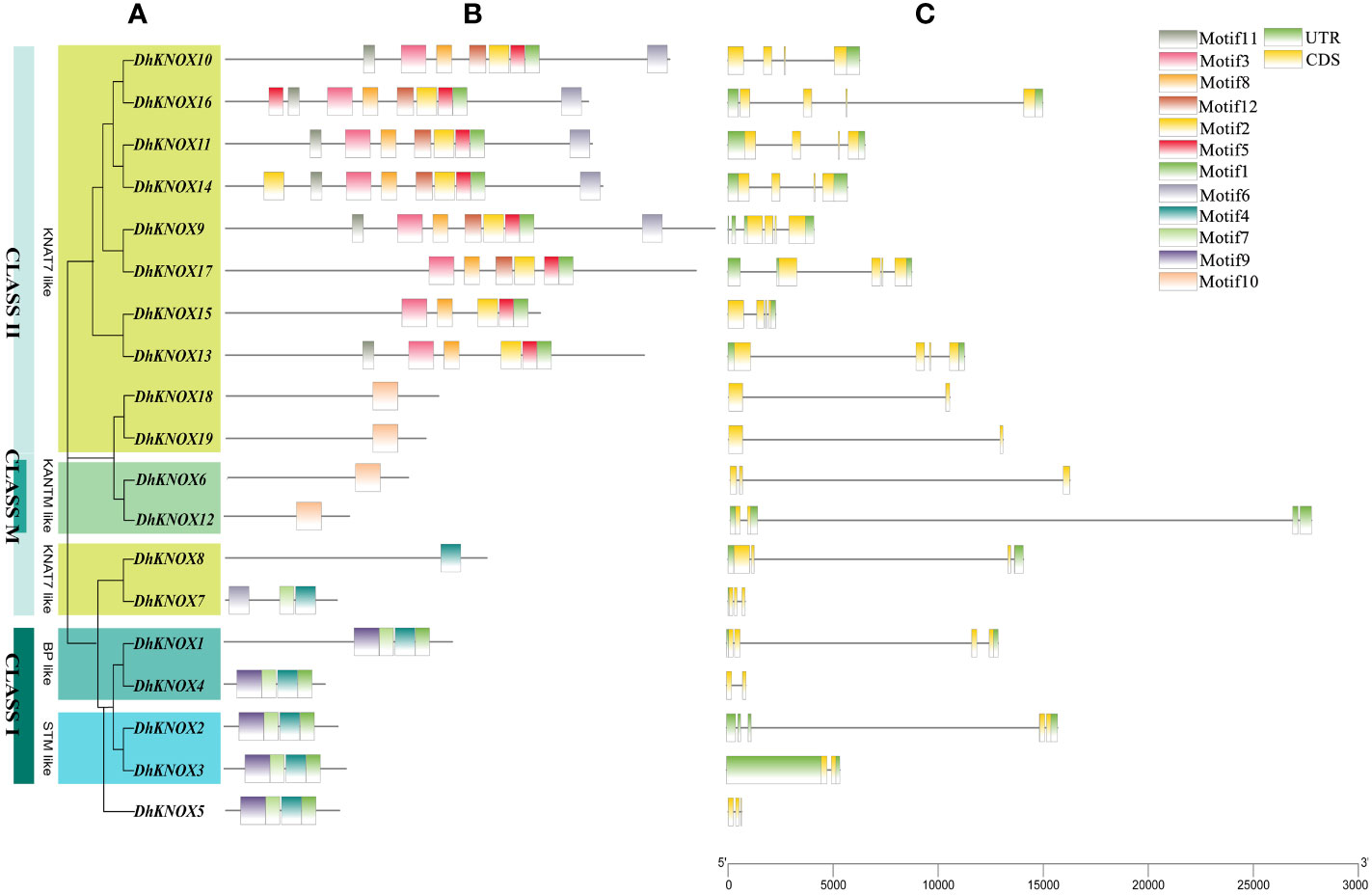
Figure 4 The gene structure and protein-conserved domain of the DhKNOX gene family. (A) Phylogenetic tree of DhKNOX genes. (B) Exon-intron structure of DhKNOX members. (C) Conserved motifs of DhKNOX proteins.
The findings indicated that all DhKNOX genes comprised both exons and introns, and the DhKNOX gene family predominantly exhibited a pattern of four to six exons and three to five introns. The number of gene exons in class I was between three and six, while almost all genes in class II contained five or six exons (Figure 4C). Among the DhKNOX family members, DhKNOX3, DhKNOX4, DhKNOX18, and DhKNOX19 contain only one intron. Most DhKNOX members have three introns. Generally, closely related members have similar genetic structures, such as DhKNOX11/DhKNOX14 and DhKNOX15/DhKNOX13. It is noteworthy that except for DhKNOX5, DhKNOX9, and DhKNOX15, class I genes contain a long intron (Figure 4C), which is consistent with the structural characteristics of the class I KNOX gene family (Morimoto et al., 2005).
Analysis of cis-acting elements in DhKNOX gene gamily promoters
To analyze the potential expression regulation mechanism of DhKNOX gene family members, we identified the cis-acting elements of each member (Figure 5). For the identified cis-acting elements, the elements related to plant hormones, plant growth, participation in stress responses, and light responses were selected for analysis. First, based on the 2,000-bp promoter sequence, stress-related elements, including LTR, MBS, ARE, and other elements, were identified, and transcriptional regulatory-related elements, including MYB, W-box, and MYB-also, were found. In addition, we discovered and validated a couple of elements related to plant hormones, such as P-box, TGA-element, ABRE, and other elements. It is worth noting that the most common motif is the cis-acting element of the motif related to ABA reactivity, accounting for 36% of the hormone response motifs scanned. Cis-acting elements of the TGACG motif related to MeJA reactivity accounted for 13%. In addition, we found that the TCA element in response to SA, among the 19 DhKNOX gene promoters, appeared 29 times and accounted for 10%. The MBS element was related to drought stress, which accounted for 12%. These findings suggest that the transcription of DhKNOX genes may be affected by these hormones and drought stress.
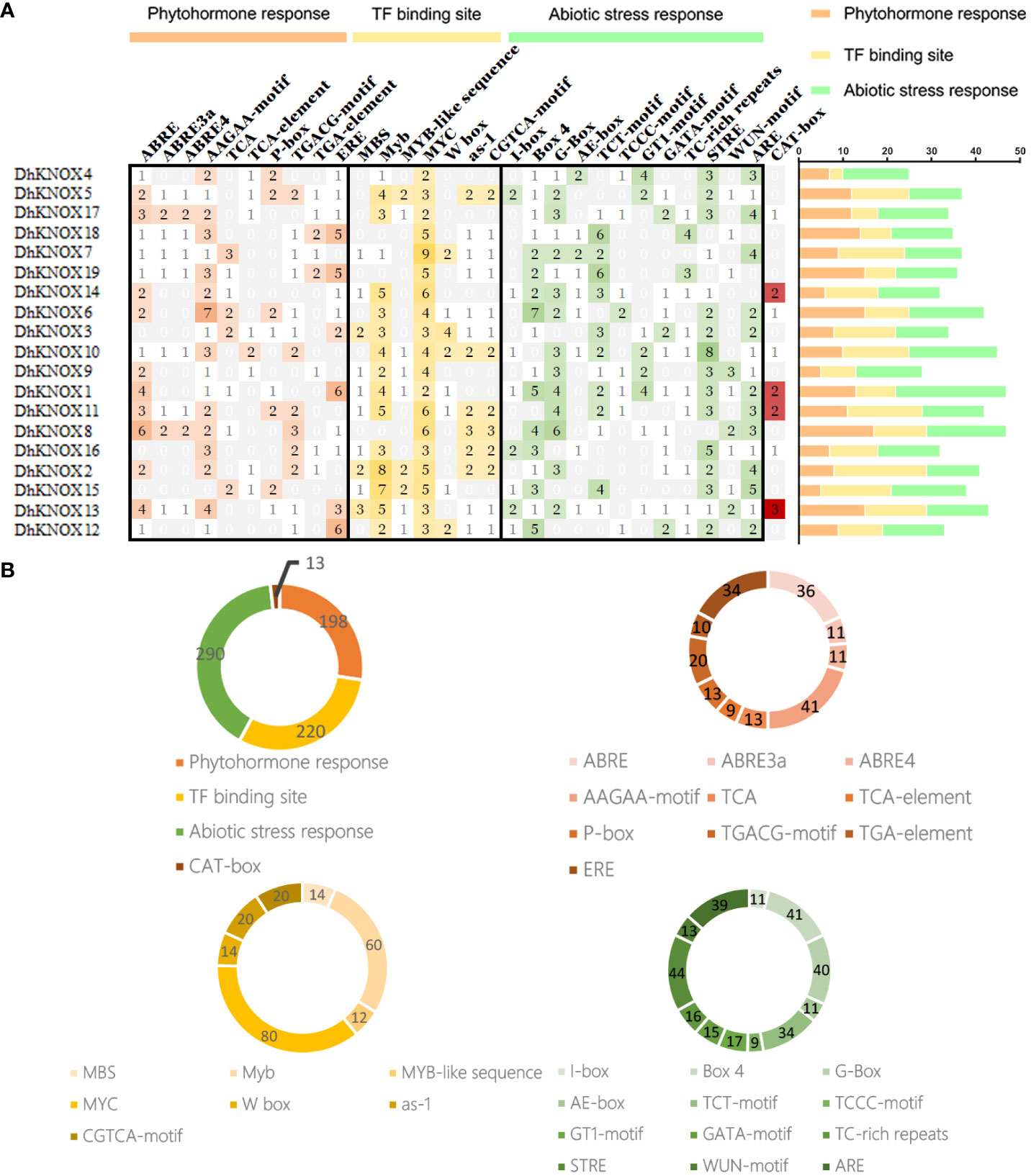
Figure 5 Analysis of cis-acting elements of the KNOX genes in D. huoshanense. (A) The different colors and numbers of the grid indicated the numbers of different promoter elements in these KNOX genes. The different colored histogram represented the sum of the cis-acting elements in each category. (B) Pie charts of different sizes indicated the ratio of each promoter element in each category, respectively.
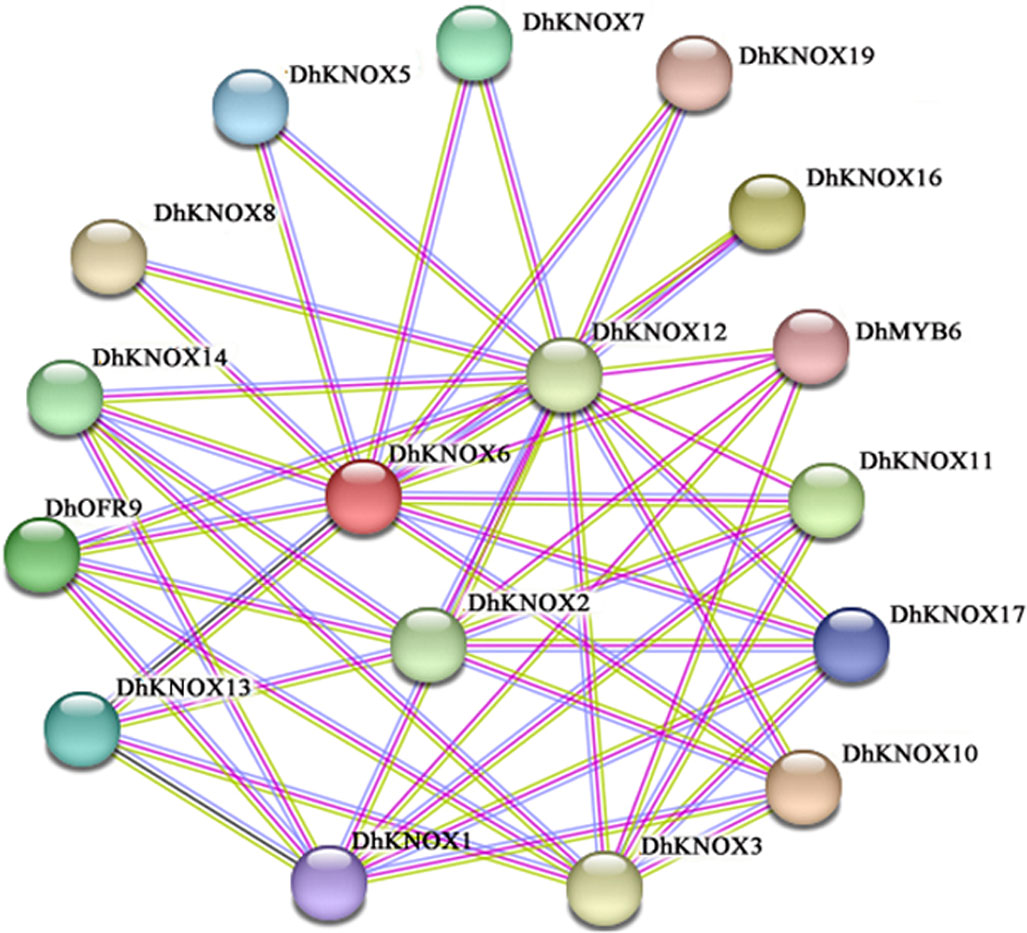
Prediction of protein interactions of DhKNOX gene family members
We conducted protein interaction predictions among the members of the DhKNOX gene family by using the STRING website (Figure 6). The results revealed a significant number of interactions among the family members. We found protein-to-protein interactions among 15 members of the DhKNOX family, including DhKNOX1, DhKNOX2, DhKNOX3, DhKNOX5, and DhKNOX19. There are 17 nodes and 42 sets of interaction relationships in the interaction network. DhKNOX2, DhKNOX6, and DhKNOX12 are located at the center of the entire protein interaction network and can interact with the proteins DhOFP9 and DhMYB6. In summary, the members of the DhKNOX gene family have the ability to form dimers or polymers through protein interactions and to carry out transcriptional regulatory functions.
Expression analysis of the DhKNOX genes under hormonal treatments
Various abiotic stresses may affect the growth and development of plants and ultimately affect the regulation of a series of stress-related genes (Grallath et al., 2005). Consequently, it becomes imperative to clarify the regulatory pathway and identify pivotal regulatory factors implicated in hormone responses in D. huoshanense. To better understand the changes in the expression level of the DhKNOX gene under various hormone treatments, we performed qRT-PCR experiments to analyze their expression patterns under MeJA, ABA, and SA treatments.
After treatment with MeJA, we found that the expression of 10 DhKNOX genes was downregulated to different degrees (Figure 7). The results showed that DhKNOX1, DhKNOX2, DhKNOX3, DhKNOX4, DhKNOX5, DhKNOX6, and DhKNOX12 were obviously significantly and rapidly upregulated at all time points. Among these DhKNOX genes, we found that the highest expression levels of DhKNOX1, DhKNOX2, DhKNOX3, DhKNOX4, and DhKNOX5 occurred 16 h after treatment. The expression of one DhKNOX6 and DhKNOX12 gene peaked at 4 h. Notably, it was found that four genes, DhKNOX1, DhKNOX2, DhKNOX3, and DhKNOX4, were all clustered in class I, and DhKNOX6 and DhKNOX12 were clustered in class II. Moreover, eight DhKNOX genes presented reduced expression levels to different degrees. Interestingly, we found that the expression of two DhKNOX genes (DhKNX13 and DhKNOX15) was not obviously changed at any time point, and the two DhKNOX genes lacked the TGACG motif.
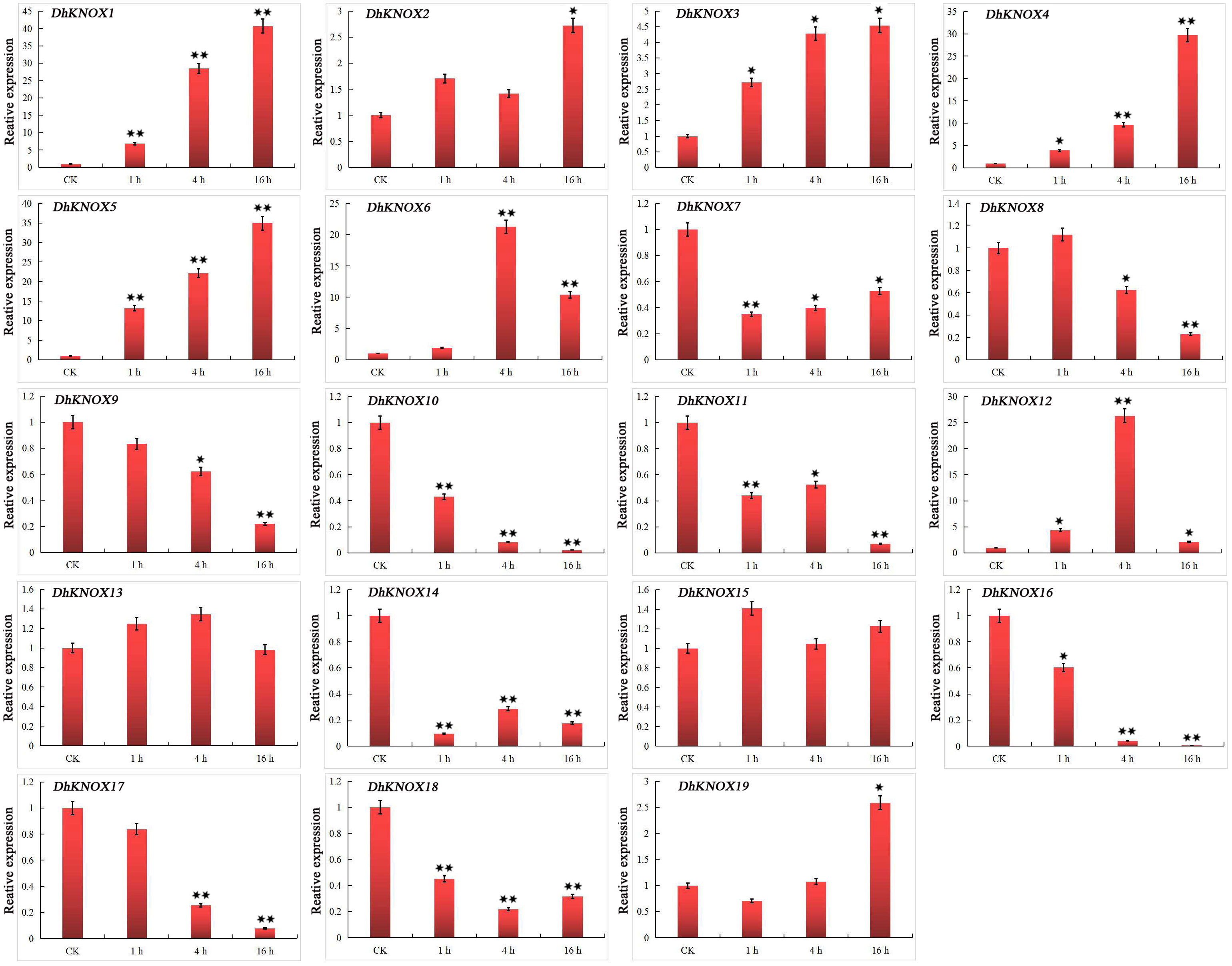
Figure 7 Expression patterns of DhKNOX genes in response to MeJA hormone treatments. The x-axis indicated the time course of each stress treatment, and the y-axis represented the relative expression level. The expression data of each gene at 0 h were used as a control sample to show the relative expression level. The data are presented as the mean values ± standard deviation of three biological replicates. Significant differences were determined by ANOVA combined with a post-hoc test (Student’s t-test). An asterisk indicates significant differences (*p < 0.05; **p < 0.01).
ABA induced the transcription of 19 DhKNOX genes. The expression levels of 17 DhKNOX genes increased to varying degrees (Figure 8). Among them, 11 DhKNOX genes were significantly upregulated at the last time point (16 h), such as DhKNOX2 and DhKNOX3, which were upregulated by more than 100- and 500-fold, respectively. Interestingly, we found that the expression of two DhKNOX genes was clustered in class I. However, we found that DhKNOX2 had only two ABREs, and DhKNOX3 lacked the ABREs. In addition, two DhKNOX genes (DhKNOX6 and DhKNOX12) clustered in class II, and their expression levels decreased significantly.
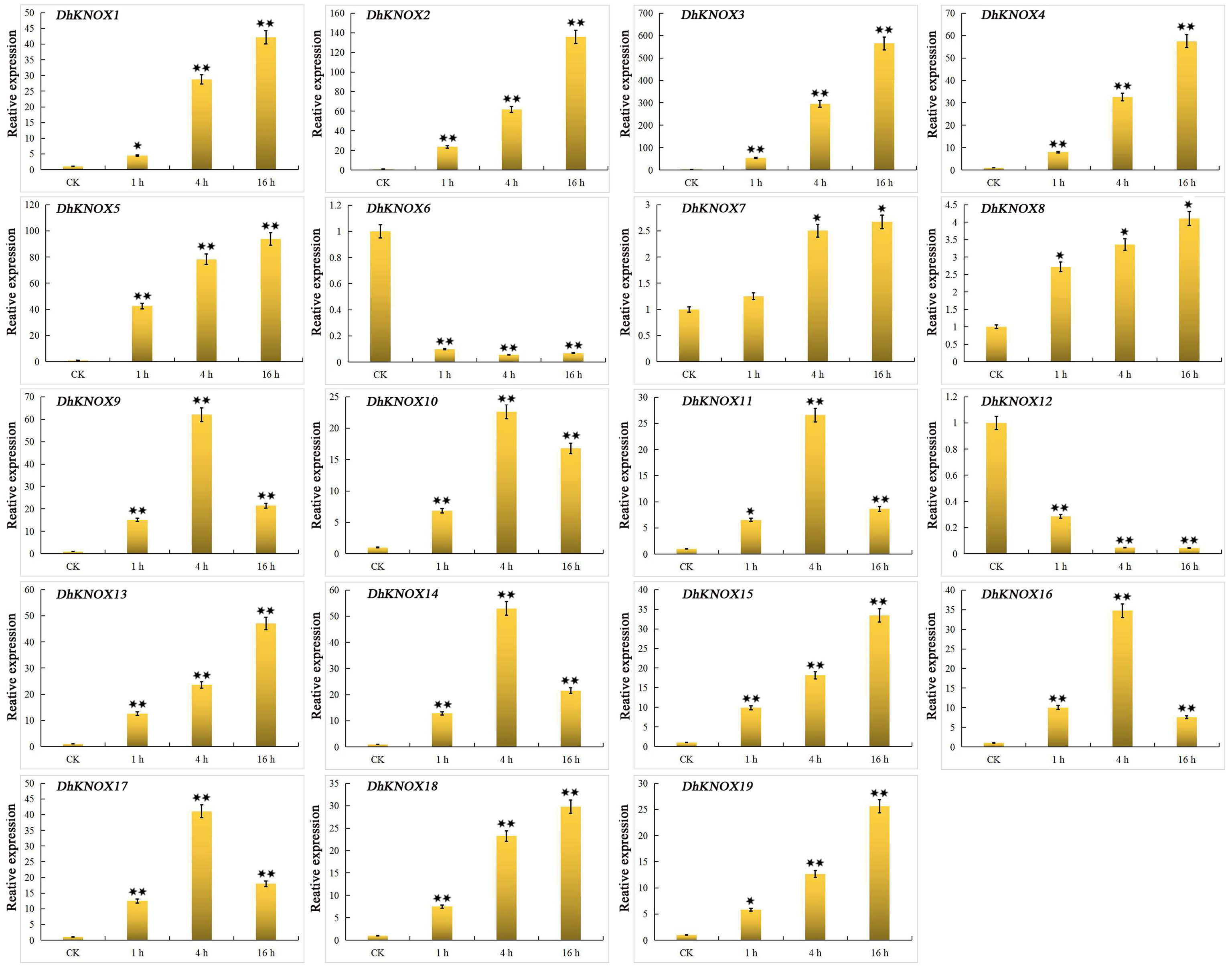
Figure 8 Expression patterns of DhKNOX genes in response to ABA hormone treatments. The x-axis indicated the time course of each stress treatment, and the y-axis represented the relative expression level. The expression data of each gene at 0 h were used as a control sample to show the relative expression level. The data are presented as the mean values ± standard deviation of three biological replicates. Significant differences were determined by ANOVA combined with a post-hoc test (Student’s t-test). An asterisk indicates significant differences (*p < 0.05; **p < 0.01).
Under SA treatment, in leaves, the expression level of 17 DhKNOX genes increased significantly at three treatment time points (1 h, 4 h, and 16 h) (Figure 9). We further found that the highest expression levels of 12 DhKNOX genes occurred after 16 h of treatment. The expression pattern of most other DhKNOX genes showed a trend of increasing first and then decreasing, and DhKNOX15 and DhKNOX18 had the highest expression levels after 1 h. Four of 17 genes reached their highest expression level after 4 h of SA treatment and 4 h but decreased after 16 h, such as DhKNOX2, DhKNOX13, DhKNOX16, and DhKNOX19. Of these four DhKNOX genes, only DhKNOX2 was clustered in class I and had one TCA element. Notably, DhKNOX13, DhKNOX16, and DhKNOX19 were clustered in class I, and all lacked TCA elements. Only two DhKNOX genes were obviously rapidly and significantly downregulated at all time points.
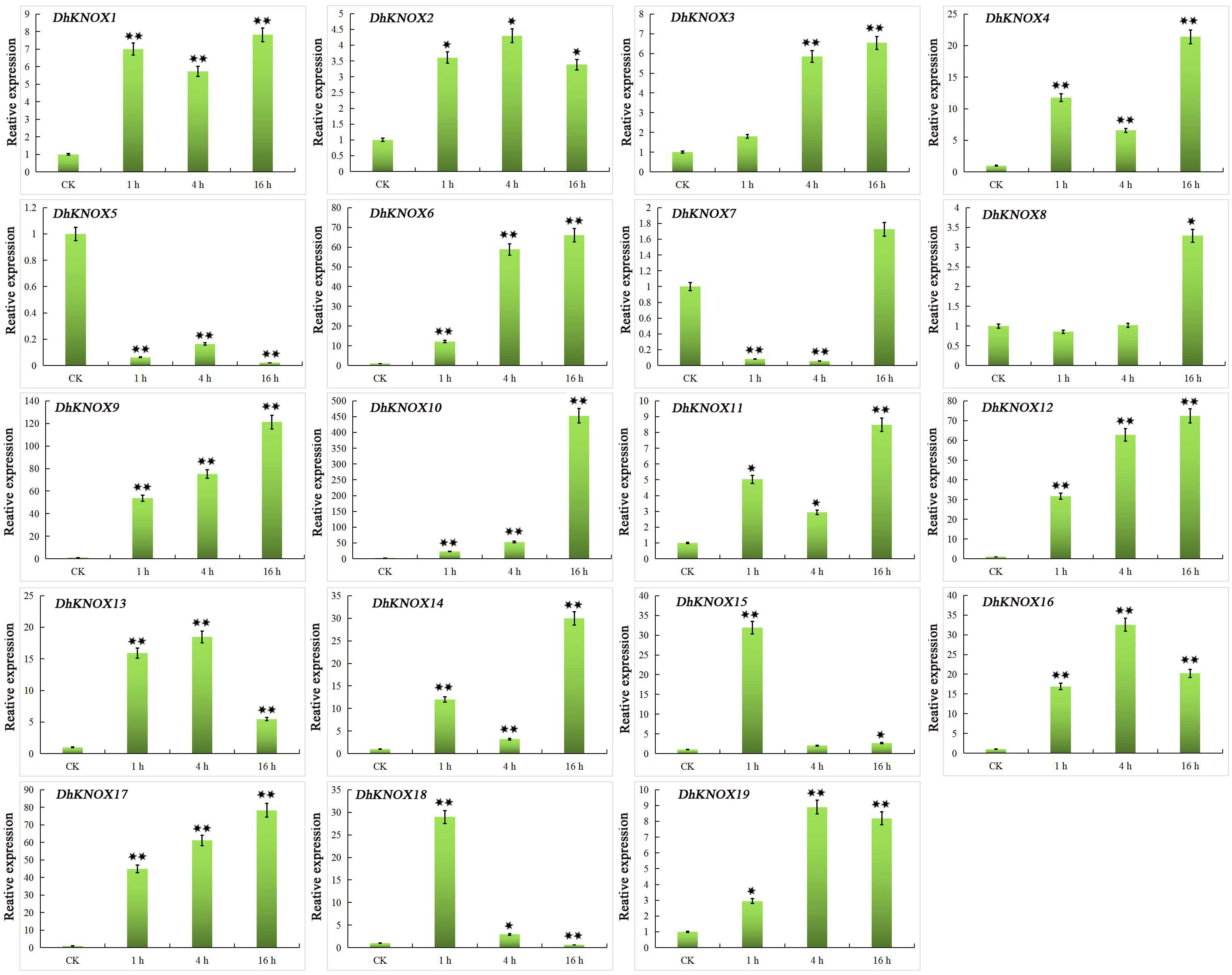
Figure 9 Expression patterns of DhKNOX genes in response to SA hormone treatments. The x-axis indicated the time course of each stress treatment, and the y-axis represented the relative expression level. The expression data of each gene at 0 h were used as a control sample to show the relative expression level. The data are presented as the mean values ± standard deviation of three biological replicates. Significant differences were determined by ANOVA combined with a post-hoc test (Student’s t-test). An asterisk indicates significant differences (*p < 0.05; **p < 0.01).
Expression analysis of the DhKNOX gene under PEG-mediated drought conditions
The expression level of most DhKNOX genes showed a significant increase or decrease under 20% PEG treatment, as shown in Figure 10. We further found that the expression levels of the five genes were clustered in class I, class II, and class III and gradually increased with the prolongation of treatment time, and the expression level of DhKNOX13 increased by 120 times compared to the control group, which contains three MBS elements. In addition, the expression levels of four genes, DhKNOX4, DhKNOX5, DhKNOX6, and DhKNOX18, did not show significant changes, and this gene promoter does not contain MBS elements. Additionally, the expression of DhKNOX7, DhKNOX8, and DhKNOX9 was consistently suppressed under drought treatment. Moreover, three DhKNOX genes (DhKNOX14, DhKNOX15, and DhKNOX16) were clustered in class I, and the expression level of these genes first increased and then decreased, reaching a maximum at 12 h. Furthermore, only two DhKNOX genes were clustered in class I, and the expression of these two genes varied irregularly.
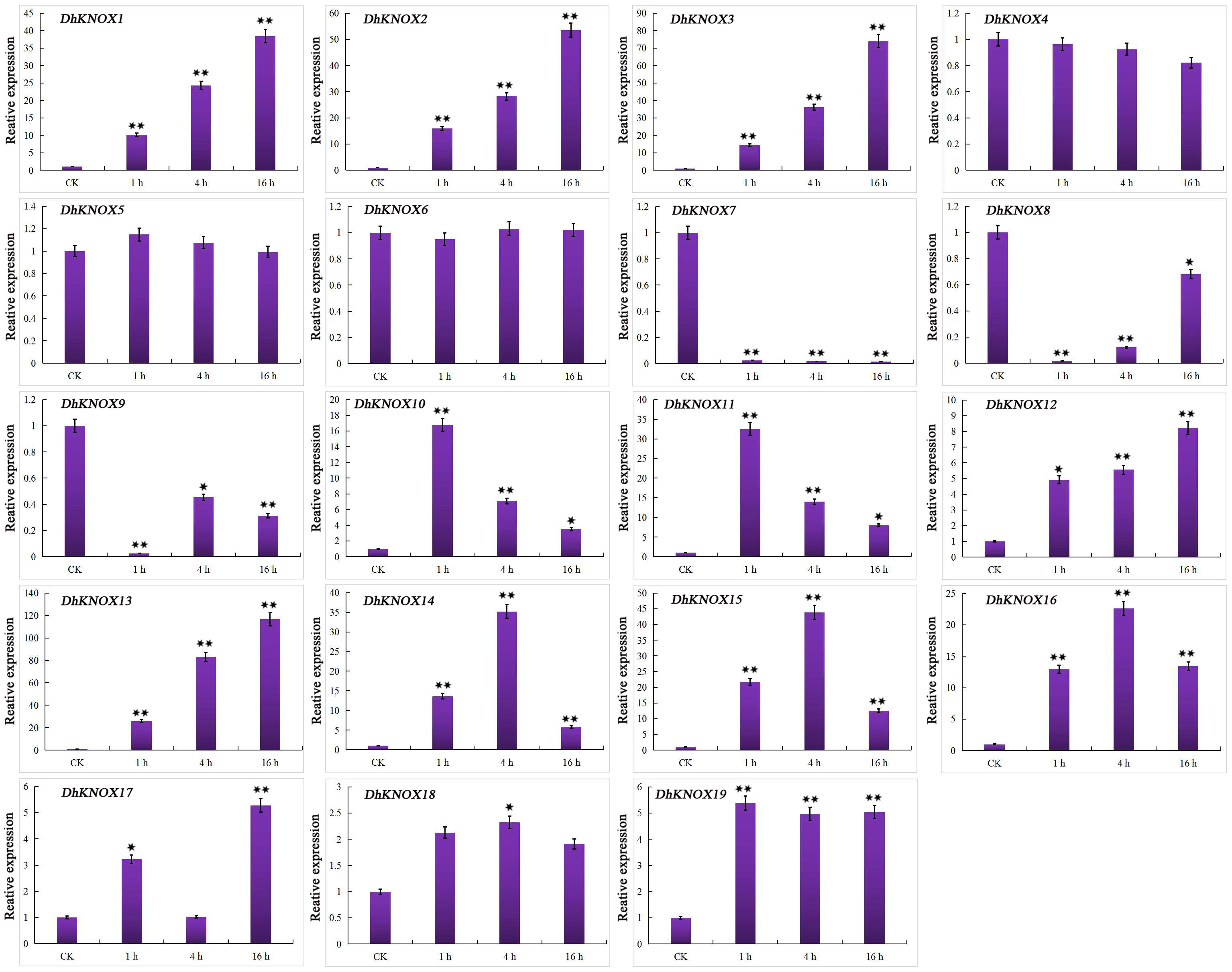
Figure 10 Expression patterns of DhKNOX genes in response to PEG treatments. The x-axis indicated the time course of each stress treatment, and the y-axis represented the relative expression level. The expression data of each gene at 0 h were used as a control sample to show the relative expression level. The data are presented as the mean values ± standard deviation of three biological replicates. Significant differences were determined by ANOVA combined with a post-hoc test (Student’s t-test). An asterisk indicates significant differences (*p < 0.05; **p < 0.01).
Discussion
Transcription factors are unique to plants and serve a critical function in regulating plant growth and development (Huang et al., 2012; Cao et al., 2017). However, limited research has been conducted on the KNOX transcription factor family of D. huoshanense. The ongoing enhancement of Dendrobium genome data offers an avenue to investigate whether the KNOX gene assumes pivotal roles in plant growth, development, and Dendrobium’s reaction to diverse environmental stress conditions.
A composite evolutionary tree was constructed by combining 19 DhKNOX proteins in Dendrobium with 68 KNOX protein sequences from different species. The class I subfamily can be divided into BP-like, STM-like, and KNAT2/6-like subfamilies; class II can be divided into KNAT3-5-like and KNAT7-like subfamilies. Functional studies showed that BP and STM play regulatory roles in the biosynthesis of lignin and the development of the secondary cell wall (Cheng et al., 2019). In this study, DhKNOX5, DhKNOX7, and DhKNOX19 in KNAT7-like in Dendrobium may also be involved in regulating the synthesis of secondary walls. Furthermore, DhKNOX2 and DhKNOX3 of the 19 DhKNOX members in Dendrobium and BP in Arabidopsis converge into the same branch of BP-like evolution, and DhKNOX3 and DhKNOX4 converge into the same group with STM. We hypothesized that two genes belonging to these two subfamilies are involved in the regulation of lignin synthesis.
Considering the functional significance of conserved domains within the KNOX protein, we focused on investigating the KNOX protein domains in Dendrobium. We discovered that most members of the same subgroup have at least one KNOX protein with the same motifs, such as DhKNOX6/DhKNOX12 and DhKNOX18/DhKNOX19. In addition, 19 DhKNOX genes contain different numbers of exons or introns, which indicates that the KNOX gene family of Dendrobium has great diversity. Furthermore, research has found that the gene intron can usually be well preserved during evolution (Wang et al., 2015). This may explain the functional differences and diversity of closely related DhKNOX genes, such as DhKNOX2 and DhKNOX3, as well as DhKNOX7 and DhKNOX 8 (Figure 4C).
It has been observed that the regulation of gene expression levels involves the coordination of various cis-acting elements (Cao et al., 2018; Soliman and Meyer, 2019). In our research, the DhKNOX promoter also contains hormone response elements, including the CGTCA motif and ABRE motif. Therefore, the DhKNOX family may be regulated by these plant hormones or even by hormone-mediated growth and development or stress regulation (Figure 5). Furthermore, the study of the gene function of this family in different plants also corroborated the results of the existence of many hormone-related elements in the cis-element of the promoter. For example, the KNOX gene in P. paten can promote cytokinin biosynthesis through the isopentenyl transferase gene PpIPT3 (Coudert et al., 2019) and LcKNAT1 in Litchi chinensis can regulate exfoliation by regulating ethylene biosynthesis (Zhao et al., 2020) and participate in the regulation of gibberellin function (Sakamoto et al., 2001; Hay et al., 2002).
In plants, genes that are commonly linked to hormone stress are responsible for generating stress responses. These responses are regulated and/or mediated by diverse hormone signaling pathways (Walther et al., 2007; Cao et al., 2019). For example, the promoter regions of the KNOX gene in Cymbidium and Punica granatum contain abundant MeJA and ABA-responsive cis-elements, as well as auxin and gibberellin components, respectively (Wang Y. Y. et al., 2020; Zhang Q. P. et al., 2021), and the expression level of the KNOX gene can be influenced by changes in exogenous 6-BA, IAA, and KT signals in Gossypium hirsutum (Zhang X. H. et al., 2021). In our study, many common cis-acting elements, such as MBS, LTR, HSF, ERE, and ABRE, were identified in the promoter region of DhKNOX. These KNOX genes contain at least hormone-acting elements, which indicates that the expression level of these genes will change under the action of hormones.
To deeply understand the hormone response mechanism of the D. huoshanense KNOX gene, qRT-PCR experiments under different hormone treatments, such as MeJA, ABA, and SA, in leaves (Figures 6–8). The DhKNOX gene then showed significant differential expression patterns under treatment with these three hormones. These treatments strongly upregulated some DhKNOX genes, indicating that these genes may play a key role in the hormone response of D. huoshanense. For example, overexpression of MdKNOX19 in apples increases the sensitivity of apple callus to ABA (Jia et al., 2021). While eight DhKNOX genes presented reduced expression levels to different degrees under MeJA treatment (Figure 6). Similar findings have been observed in other plants. For instance, DhKNOX2 and DhKNOX3 exhibited notably elevated expression levels under ABA (more than 100 and 500 times, respectively, the CK level) (Figure 7). Similarly, DhKNOX9 and DhKNOX10 demonstrated substantial upregulation under SA, exceeding 100 and 400 times, respectively, compared to the control (Figure 8). These three genes may be more sensitive to ABA and SA hormones of D. huoshanense plant growth and development. Among these DhKNOX genes, a majority possess conserved domains, and it is worth noting that all DhKNOX genes demonstrated hormone responsiveness regardless of the presence of conserved domains. These results suggested that gene-coding proteins with KNOXI, KNOXII, ELK, or HOM domains may play a role in the stress response. In addition, we found that the DhKNOX gene is sensitive to different hormones to varying degrees. These results provide evidence that KNOX members can participate in the response to abiotic stress. We conjecture that these three hormones may directly or indirectly regulate the transcription level of DhKNOX. In the future, we want to explore the influence of exogenous hormones on the regulation of DhKNOX expression level in order to determine whether the growth and development of Dendrobium can be altered by regulating exogenous hormones.
In summary, there is a certain correlation between hormone response and plant resistance to abiotic stress. For example, gene expression patterns related to ethylene suggest that ethylene may indirectly participate in the induction of dormant genes, thereby enhancing the cold resistance of P. mume (Li et al., 2021). This phenomenon is also found in the TALE gene expression mode, where expression is not only regulated by some hormones but is also sometimes affected by some abiotic stress (Tsuda and Hake, 2015; Niu and Fu, 2022). The expression of the pear KNOX gene family can regulate drought stress, especially the transcription level of PbKNOX7/13, which is significantly increased under drought stress, while the transcription level of PbKNOX5/16 is significantly reduced under drought stress (Liu et al., 2022). Notably, the expression of DhKNOX7, DhKNOX8, and DhKNOX9 remained consistently suppressed during drought treatment. This observed reduction in transcription levels could potentially contribute to enhancing the resistance of D. huoshanense to drought stress. However, a comprehensive and systematic investigation is warranted to fully comprehend the implications of these downregulated gene expressions in bolstering D. huoshanense’s resilience.
Conclusions
In our study, we identified and characterized 19 DhKNOX genes from the Dendrobium genome. These genes were classified into three major families based on their sequence similarities and phylogenetic relationships. Through promoter analysis, we discovered that several cis-acting elements were associated with phytohormone signaling, growth, and development, as well as stress responses in the DhKNOX gene family. Moreover, we conducted a comprehensive analysis of the expression patterns of these 19 DhKNOX genes under various hormonal treatments, including SA, ABA, MeJA, and drought. Our findings shed light on the important role of DhKNOX genes in regulating morphogenesis, growth, and development, particularly in response to abiotic stress conditions. This study serves as a foundation for further investigations into the precise mechanisms and functions of KNOX genes in the growth, development, and morphogenesis of Dendrobium.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Files, further inquiries can be directed to the corresponding author/s.
Author contributions
GL: Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. MM: Formal Analysis, Software, Writing – review & editing. GW: Formal Analysis, Writing – original draft. CC: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – original draft. CS: Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Software, Supervision, Writing – original draft, Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (32202442), Anhui Provincial University Research Projects (2023AH052637), Startup fund for high-level talents of West Anhui University (WGKQ2021079), the High-level Talents Research Initiation Fund of West Anhui University (WGKQ2022025), Anhui Dabieshan Academy of Traditional Chinese Medicine (TCMADM-2023-03), and the Demonstration Experiment Training Center of Anhui Provincial Department of Education (2022sysx033).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1258533/full#supplementary-material
Abbreviations
KNOX, KNOTTED1-like homeobox; ABA, abscisic acid; MeJA, methyl jasmonate; SA, salicylic acid; PEG, PED6000; SAM, stem tip meristem; GA, gibberellin; MS, Murashige and Skoo; qRT-PCR, real-time PCR; h, hour.
References
Alakonya, A., Kumar, R., Koenig, D., Kimura, S., Townsley, B., Runo, S., et al. (2012). Interspecific RNA interference of SHOOT MERISTEMLESS-like disrupts cuscuta pentagona Plant parasitism. Plant Cell. 24 (7), 3153–3166. doi: 10.1105/tpc.112.099994
Azarakhsh, M., Kirienko, A. N., Zhukov, V. A., Lebedeva, M. A., Dolgikh, E. A., Lutova, L. A. (2015). KNOTTED1-LIKE HOMEOBOX 3: a new regulator of symbiotic nodule development. J. Exp. Bot. 66, 7181–7195. doi: 10.1093/jxb/erv414
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W46. doi: 10.1093/nar/gkv416
Bolduc, N., Hake, S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21, 1647–1658. doi: 10.1105/tpc.109.068221
Cao, Y. P., Han, Y. H., Li, D. H., Lin, Y., Cai, Y. P. (2016). MYB Transcription Factors in Chinese Pear (Pyrus bretschneideri Rehd.): Genome-Wide Identification, Classification, and Expression Profiling during Fruit Development. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00577
Cao, Y. P., Han, Y. H., Meng, D. D., Abdullah, M., Li, D. H., Jin, Q., et al. (2018). Systematic analysis and comparison of the PHD-Finger gene family in Chinese pear (Pyrus bretschneideri) and its role in fruit development. Funct. Integrat. Genom. 18, 519–531. doi: 10.1007/s10142-018-0609-9
Cao, Y. P., Han, Y., Meng, D. D., Li, D. H., Jiao, C. Y., Jin, Q., et al. (2017). B-BOX genes: genome-wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri Rehd.). BMC Plant Biol. 17 (1), 156. doi: 10.1186/s12870-017-1105-4
Cao, Y. P., Meng, D. D., Han, Y. H., Chen, T. Z., Jiao, C. Y., Chen, Y., et al. (2019). Comparative analysis of B-BOX genes and their expression pattern analysis under various treatments in Dendrobium offificinale. BMC Plant Biol. 19, 245. doi: 10.1186/s12870-019-1851-6
Cao, Z. M., Zhang, Z. X., Hou, D. S. (2015). Identification and Evaluation of Drought Resistance of Mung Bean during Germination under PEG-6000 Solution Stress. Hebei Agricul. Sci. 19 (3), 27–31. doi: 10.3969/j.issn.1000-4440.2022.05.004
Chen, C., Xia, R., Chen, H. (2018). TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. Bio Rxiv 289660. doi: 10.1101/bio.Rxiv289660
Cheng, X., Li, M. L., Abdullah, M., Li, G. H., Zhang, J. Y., Muhammad, A. M., et al. (2019). In Silico Genome-Wide Analysis of the Pear (Pyrus bretschneideri) KNOX family and the functional characterization of PbKNOX1, an Arabidopsis BREVIPEDICELLUS orthologue gene, involved in cell wall and lignin biosynthesis. Front. Genet. 10, 632. doi: 10.3389/fgene.2019.00632
Coudert, Y., Novak, O., Harrison, C. J. (2019). A KNOX-ey-tokinin regulatory module predates the origin of indetermi-nate vascular plants. Curr. Biol. 9 (16), 2743–2750. doi: 10.1016/j.cub.2019.06.083
Dachan, K., Young-Hyun, C., Hojin, R., Kim, Y. H., Kim, T. H., Hwang, D. (2013). BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J. 75, 755–766. doi: 10.1111/tpj.12236
Fan, H., Wu, Q., Wang, X., Wu, L., Cai, Y., Lin, Y. (2016). Molecular cloning and expression of 1-deoxy-d-xylulose-5-phosphate synthase and 1-deoxy d-xylulose-5-phosphate reductoisomerase in Dendrobium offiffifficinale. Plant Cell Tiss. Organ Cult. 125, 381–385. doi: 10.1007/s11240-016-0945-1
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2013). Pfam: the protein families database. Nucleic Acids Res. 42 (D1), D222–D230. doi: 10.1093/nar/gkm960
Furumizu, C., Alvarez, J. P., Sakakibara, K. (2015). Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PloS Genet. 11 (2), e1004980. doi: 10.1371/journal.pgen.1004980
Gao, J., Yang, X., Zhao, W., Lang, T., Samuelsson, T. (2015). Evolution, diversification, and expression of KNOX proteins in plants. Front. Plant Sci. 6, 882. doi: 10.3389/fpls.2015.00882
Grallath, S., Weimar, T., Meyer, A., Gumy, C., Suter-Grotemeyer, M., Neuhaus, J. M., et al. (2005). The AtProT family. Compatible solute transporters with similar substrate specificity but differential expression patterns. Plant Physiol. 137 (1), 117–126. doi: 10.1104/pp.104.055079
Hamant, O., Pautot, V. (2010). Plant development: a TALE story. C. R. Biol. 333 (4), 371–381. doi: 10.1016/j.crvi.2010.01.015
Han, B. X., Jing, Y., Dai, J., Zheng, T., Gu, F. L., Zhao, Q., et al. (2020). A chromosome-level genome assembly of Dendrobium huoshanense using long reads and Hi-C data. Genome Biol. Evol. 12 (12), 2486–2490. doi: 10.1093/gbe/evaa215
Hay, A., Kaur, H., Philips, A. (2002). The gibberellin pathway mediates KNOTTEDl-type homeobox function in plants with different body plans. Curr. Biol. 12 (18), 1557–15651. doi: 10.1016/S0960-9822(02)01125-9
Huang, J., Zhao, X., Weng, X., Wang, L., Xie, W. (2012). The rice B-box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis. PloS One 7 (10), e48242. doi: 10.1371/journal.pone.0048242
Jia, P., Xing, L. B., Zhang, C. G., Zhang, D., Ma, J. J., Zhao, C. P., et al (2021). MdKNOX19, a class II knotted-like transcription factor of apple, plays roles in ABA signalling /sensitivity by targeting ABI5 during organ development. Plant Sci. 302. doi: 10.1016/j.plantsci.2020.110701
Lee, T. H., Tang, H., Wang, X., Paterson, A. H. (2013). PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41 (D1), 1152–1158. doi: 10.1093/nar/gks1104
Li, P., Zheng, T. C., Zhuo, X. K., Zhang, M., Yong, X., Li, L. L. (2021). Photoperiod and temperature-mediated control of the ethylene response and winter dormancy induction in Prunus mume. Hortic. Plant J. 7, 232–242. doi: 10.1016/j.hpj.2021.03.005
Lin, J., Chang, Y. J., Yang, W. B., Yu, A. L., Wang, C. H. (2014). The multifaceted effects of polysaccharides isolated from Dendrobium huoshanense on immune functions with the induction of interleukin-1 receptor antagonist (IL-1ra) in monocytes. PloS One 9, e94040. doi: 10.1371/journal.pone.0094040
Liu, L., Han, R., Yu, N., Zhang, W., Xing, L., Xie, D., et al. (2018). method for extracting high-quality total RNA from plant rich in polysaccharides and polyphenols using Dendrobium huoshanense. PloS One 13, e0196592. doi: 10.1371/journal.pone.0196592
Liu, J., Zhang, C., Han, J., Fang, X., Xu, H., Liang, C. (2022). Genome-wide analysis of KNOX transcription factors and expression pattern of dwarf-related KNOX genes in pear. Front. Plant Sci. 13, 806765. doi: 10.3389/fpls.2022.806765
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Ma, C., Meir, S., Xiao, L., Tong, J. H., Liu, Q., Reid, M. S., et al. (2015). A KNOTTED1-LIKE HOMEOBOX protein regulates abscission in tomato by modulating the auxin pathway. Plant Physiol. 167, 844–853. doi: 10.1104/pp.114.253815
Magnani, E., Hake, S. (2008). KNOX lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20 (4), 875–887. doi: 10.1105/tpc.108.058495
Morimoto, R., Kosugi, T., Nakamura, C., Takumi, S. (2005). Intragenic diversity and functional conservation of the three homoeologousloci of the KN1-type homeobox gene knox1 in common wheat. Plant Mol. Biol. 57 (6), 907–924. doi: 10.1007/s11103-005-3247-2
Niu, X. L., Fu, D. Q. (2022). The roles of BLH transcription factors in plant development and environmental response. Int. J. Mol. Sci. 23, 3731. doi: 10.3390/ijms23073731
Sakamoto, T., Kamiya, N., Ueguchi-Tanaka, M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15 (5), 581–590. doi: 10.1101/gad.867901
Scofield, S., Dewitte, W., Murray, J. A. H. (2014). STM sustains stem cell function in the Arabidopsis shoot apical meristem and controls KNOX gene expression independently of the transcriptional repressor AS1. Plant Signal Behav. 9, e28934. doi: 10.4161/psb.28934
Soliman, E. R. S., Meyer, P. (2019). Responsiveness and adaptation to salt stress of the redox-responsive transcription factor 1 (RRTF1) gene are controlled by its promoter. Mol. Biotechnol. 61, 254–260. doi: 10.1007/s12033-019-00155-9
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 (10), 2731–2739. doi: 10.1093/molbev/msr121
Tsuda, K., Hake, S. (2015). Diverse functions of KNOX transcription factors in the diploid body plan of plants. Curr. Opin. Plant Biol. 27, 91–96. doi: 10.1016/j.pbi.2015.06.015
Walther, D., Brunnemann, R., Selbig, J. (2007). The regulatory code for transcriptional response diversity and its relation to genome structural properties in A. thaliana. PloS Genet. 3 (2), e11. doi: 10.1371/journal.pgen.0030011
Wang, Y. J., Dai, J., Chen, R., Song, C., Wei, P. P., Cai, Y. P., et al. (2022). miRNA-based drought regulation in the important medicinal plant Dendrobium huoshanense. J. Plant Growth Regul. 41, 1099–1108. doi: 10.1007/s00344-021-10366-7
Wang, Q., Liu, J., Wang, Y., Zhao, Y., Jiang, H., Cheng, B. (2015). Systematic analysis of the maize PHD-finger gene family reveals a subfamily involved in abiotic stress response. Int. J. Mol. Sci. 16, 23517–23544. doi: 10.3390/ijms161023517
Wang, S. M., Yamaguchi, M., Grienenberger, E., Martone, P. T., Samuels, A. L., Mansfifield, S. D. (2020). The Class II KNOX genes KNAT3 and KNAT7 work cooperatively to inflfluence deposition of secondary cell walls that provide mechanical support to Arabidopsis stems. Plant J. 101 (2), 293–309. doi: 10.1111/tpj.14541
Wang, Y. Y., Zhao, Y. J., Yan, M., Zhao, H. L., Zhang, X. H., Yuan, Z. H. (2020). Genome-wide identification and expression analysis of TALE gene family in pomegranate (Punica granatum L.). Agronomy 10, 829. doi: 10.3390/agronomy10060829
Yanai, O., Shani, E., Dolezal, K., Tarkowski, P., Sablowski, R., Sandberg, G., et al. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15 (17), 1566–1571. doi: 10.1016/j.cub.2005.07.060
Yang, Q. Q., Yuan, C. Q., Cong, T. C., Wang, J., Zhang, Q. X. (2022). Genome-wide identification of three-amino-acid-loop extension gene family and their expression profile under hormone and abiotic stress treatments during stem development of Prunus mume. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1006360
Zha, X. Q., Deng, Y. Y., Li, X. L., Wang, J. F., Pan, L. H., Luo, J. P. (2017). The core structure of a Dendrobium huoshanense polysaccharide required for the inhibition of human lens epithelial cell apoptosis. Carbohydr. Polym. 155, 252–260. doi: 10.1016/j.carbpol.2016.08.087
Zhang, Q. P., Zhang, D. Y., Yu, K., Ji, J. J., Liu, N., Zhang, Y. P. (2021). Frequent germplasm exchanges drive the high genetic diversity of Chinese cultivated common apricot germplasm. Hortic. Res. 8, 215. doi: 10.1038/s41438-021-00650-8
Zhang, X. H., Zhao, J. J., Wu, X. Y., Hu, G. H., Fan, S. L., Ma, Q. F. (2021). Evolutionary relationships and divergence of KNOTTED1-like family genes involved in salt tolerance and development in cotton (Gossypium hirsutum L.). Front. Plant Sci. 12, 774161. doi: 10.3389/fpls.2021.774161
Keywords: KNOX, abiotic stress, gene expression, D. huoshanense, genome
Citation: Li G, Manzoor MA, Wang G, Chen C and Song C (2023) Comparative analysis of KNOX genes and their expression patterns under various treatments in Dendrobium huoshanense. Front. Plant Sci. 14:1258533. doi: 10.3389/fpls.2023.1258533
Received: 14 July 2023; Accepted: 11 September 2023;
Published: 04 October 2023.
Edited by:
Elsayed Nishawy, Desert Research Center, EgyptReviewed by:
Xiujun Zhang, Chinese Academy of Sciences (CAS), ChinaYuan Wei, Jiangsu University, China
Copyright © 2023 Li, Manzoor, Wang, Chen and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cunwu Chen, Y3Vud3VjaGVuQDEyNi5jb20=; Cheng Song, bGFubmlhbzgxMjMyOTIxOEAxNjMuY29t
†These authors have contributed equally to this work
 Guohui Li
Guohui Li Muhammad Aamir Manzoor
Muhammad Aamir Manzoor Guoyu Wang3
Guoyu Wang3 Cheng Song
Cheng Song