- 1School of Biotechnology, Sher-e-Kashmir University of Agricultural Sciences and Technology of Jammu, Jammu, India
- 2Department of Botany, School of Life Sciences, Central University of Jammu, Samba, Jammu & Kashmir, India
- 3Infectious Diseases Division, Council of Scientific and Industrial Research (CSIR)-Indian Institute of Integrative Medicine (CSIR-IIIM), Jammu, India
- 4Advance Center for Horticulture Research, Udheywala, Sher-e-Kashmir University of Agricultural Sciences and Technology of Jammu, Jammu & Kashmir, India
- 5Division of Biochemistry, Indian Council of Agricultural Research (ICAR), Indian Agricultural Research Institute, New Delhi, India
- 6Division of Integrated Farming System, Central Arid Zone Research Institute, Indian Council of Agricultural Research (ICAR), Jodhpur, India
- 7Department of Nutrition, Centre for Applied Soil Science and Biology of the Segura (CEBAS), of the Spanish National Research Council (CSIC), Murcia, Spain
- 8Department of Agriculture and Forest Sciences, University of Tuscia, Viterbo, Italy
Salinity or salt stress has deleterious effects on plant growth and development. It imposes osmotic, ionic, and secondary stresses, including oxidative stress on the plants and is responsible for the reduction of overall crop productivity and therefore challenges global food security. Plants respond to salinity, by triggering homoeostatic mechanisms that counter salt-triggered disturbances in the physiology and biochemistry of plants. This involves the activation of many signaling components such as SOS pathway, ABA pathway, and ROS and osmotic stress signaling. These biochemical responses are accompanied by transcriptional modulation of stress-responsive genes, which is mostly mediated by salt-induced transcription factor (TF) activity. Among the TFs, the multifaceted significance of WRKY proteins has been realized in many diverse avenues of plants’ life including regulation of plant stress response. Therefore, in this review, we aimed to highlight the significance of salinity in a global perspective, the mechanism of salt sensing in plants, and the contribution of WRKYs in the modulation of plants’ response to salinity stress. This review will be a substantial tool to investigate this problem in different perspectives, targeting WRKY and offering directions to better manage salinity stress in the field to ensure food security.
1 Introduction
Salinity stress is a foremost abiotic constraint that affects agricultural yields worldwide (Sanwal et al., 2022a). Nearly 20% (~310 million hectares) of the total irrigated land (1,500 million hectares) and 2% under dry land agriculture (~30 million hectares), across the world, have degraded due to high salts (Sanwal et al., 2022b). In India, it is estimated that ~10% of additional area is getting spoiled by salts every year and 2.1% (6.74 million ha) of total geographical area in India has already become salt affected (Kumar and Sharma, 2020). Plants’ reaction to environmental cues involve coordinated morphological, biochemical, and physiological responses, regulated by stress-responsive genes. Particularly with respect to high-saline conditions, genes related to synthesis and regulation of secondary metabolites, ion homeostasis, reactive oxygen species, salt overly sensitive (SOS) pathway, abscisic acid signaling, transcription factors (TFs), and mitogen-activated protein kinases (MAPK) are essential (Tuteja, 2007; Sytar et al., 2018). In fact, these mechanisms are also fundamental during chemical priming-based salt stress alleviation (Srivastava et al., 2021; Srivastava et al., 2022a; Mishra et al., 2023). Regulation of gene expression of associated pathways by TFs in response to various environmental triggers constitutes a basic regulatory mechanism of plants (Buscaill and Rivas, 2014). TFs comprise a significant portion of plant genome and are represented by many gene families such as NAC, AP2, MYB, and WRKY, which are reported to offer multifaceted impact on plant development and growth and regulate plants’ fitness against environmental constraints (Srivastava et al., 2022b; Chowdhary et al., 2023). WRKY proteins are among the important TFs involved in plants defense against several abiotic and biotic stimuli (Chen F. et al., 2017). These proteins are also known to be associated with different developmental and physiological processes in plants like seed and embryo development, trichome development, senescence, dormancy, and many metabolic pathways, and their role in mitigation of stress is widely studied (Eulgem et al., 2000; Pandey and Somssich, 2009; Chen et al., 2012; Yu et al., 2016a; Kang et al., 2021; Wani et al., 2021).
In plants, WRKY proteins constitute one of the biggest families of TFs, characterized by WRKYGQK DNA binding motif, which binds to W box (TTGACC/T) of the promoters (Eulgem et al., 2000; Rushton et al., 2010). Since their discovery in 1994, from sweet potato (Ishiguro and Nakamura, 1994, named as SPF1), WRKYs were thought to be exclusive to the plant kingdom. Later, Zhang and Wang in 2005 reported the presence of one copy of WRKY gene in Giardia lamblia (primitive protozoan), Dictyosteliium discoideum (slime mold), and Chlamydomonas reinhaidtii (green alga). With their origin in early eukaryotes, these genes have duplicated many times to evolve as an expanded super family of transcriptional regulators in land plants, viz., Oryza sativa L. ssp. indica (Ross et al., 2007), Saccharum spontaneum (Li et al., 2020b), Medicago sativa (Ma et al., 2021), and Glycine max (Yin et al., 2013), where their numbers reach hundreds. With this expansion in number, the WRKY superfamily has also been specified into three major sub-groups, namely, WRKY I, II, and III, based on the number of WRKY domains and Zn finger structure. The expansion of WRKY family in higher plants is due to segmental duplication events and subsequent divergent selection among the subgroups (Yin et al., 2013), which also diversify the functional prospects of WRKY protein family. WRKY genes are completely absent in kingdom Monera, Fungi, and Animalia (Zhang and Wang, 2005).
WRKY gene expression has been found to be induced in pathogenic conditions and other chemical and physical stresses (cold, heat, salinity, wounding, oxidative stress, and nutrition deficiency; Eulgem et al., 2000). Though the exact mechanisms of WRKY proteins are not well understood, it is reported that these factors repress or activate expression of other stress-responsive genes that ultimately confer protective effects. WRKY proteins are also known to regulate abscisic acid, ethylene, salicylic acid, and jasmonic acid signaling pathways, which mediate plant response to several stress conditions (Bakshi and Oelmüller, 2014) and are thus responsible for effective signal cross-talk and multifold regulations. Many investigations related to functional characterization of WRKYs have also suggested their contribution towards attainment of tolerance against abiotic stress like drought, heat, salt, and cold, and also offer resistance to pathogenic infections (Kumar et al., 2016; Gao et al., 2018; Shi et al., 2018; Wang et al., 2018; Gao et al., 2020; Yang et al., 2020; Kang et al., 2021). Moreover, WRKYs are also reported to regulate plant specialized metabolism (Mishra et al., 2013; Schluttenhofer and Yuan, 2015; Singh et al., 2017; Srivastava et al., 2017; Zhang et al., 2021).
Considering the significance of WRKY in plants’ life, many excellent reviews on general account of WRKY have been published (Eulgem et al., 2000; Rushton et al., 2010; Chen F. et al., 2017; Jiang et al., 2017; Wani et al., 2021), yet a judicial compilation of its role in individual stress is not much attempted. Nonetheless, several studies have been conducted in recent years to investigate its regulatory role in plant growth and development, and stress management, including salinity. The current review gives a comprehensive view on the WRKY-mediated plant response to salinity stress management and the associated mechanisms. The text discusses the impact of salinity stress and salt stress-related signaling mechanisms in plants, followed by a brief understanding of the WRKY gene family, their structure, and major classes in plant genome. Furthermore, it also highlights the various WRKY candidates involved in various stresses with a focus on salt stress tolerance and associated mechanism in plants.
2 Salinity stress and its impact on crop plants
The abiotic stresses decrease the yield, survival, and biomass of food crops by 70%, posing a serious risk to world food security (Ahmad et al., 2012; Parihar et al., 2015; Li et al., 2020a; Yoon et al., 2020; Ma et al., 2021). Salinity is one of the most serious constraints to crop development and productivity (Park et al., 2016). Among abiotic stress, the fraction of irrigated land affected by salt in different regions ranges from 9% to 34% with an average of 20% in the world (Table 1, cf. FAO-ITPS-GSP 2015). Salinity stress is the detrimental effect of excess elements like Na+ and Cl− on plants (Parihar et al., 2015; Isayenkov and Maathuis, 2019). In addition, salinity is naturally complemented by secondary stresses like oxidative stress due to generation of ROS (Isayenkov, 2012; Mishra et al., 2017; Yang and Guo, 2018; Isayenkov and Maathuis, 2019). Based on its cause, salinity is categorized as primary or secondary (Kumar and Sharma, 2020). Primary (natural) salinity is developed due to the accumulation of salts during long-term natural processes (weathering of parent materials and inland oceanic salt deposition by wind/rain) in soil or groundwater. Contrary to this, secondary salinity involves various human interventions resulting in the alteration of soil–water equilibrium (Manchanda and Garg, 2008). Common examples of such human activities are deforestation, replacement of perennial crops with annual ones, irrigation with highly saline water, or inadequate drainage.
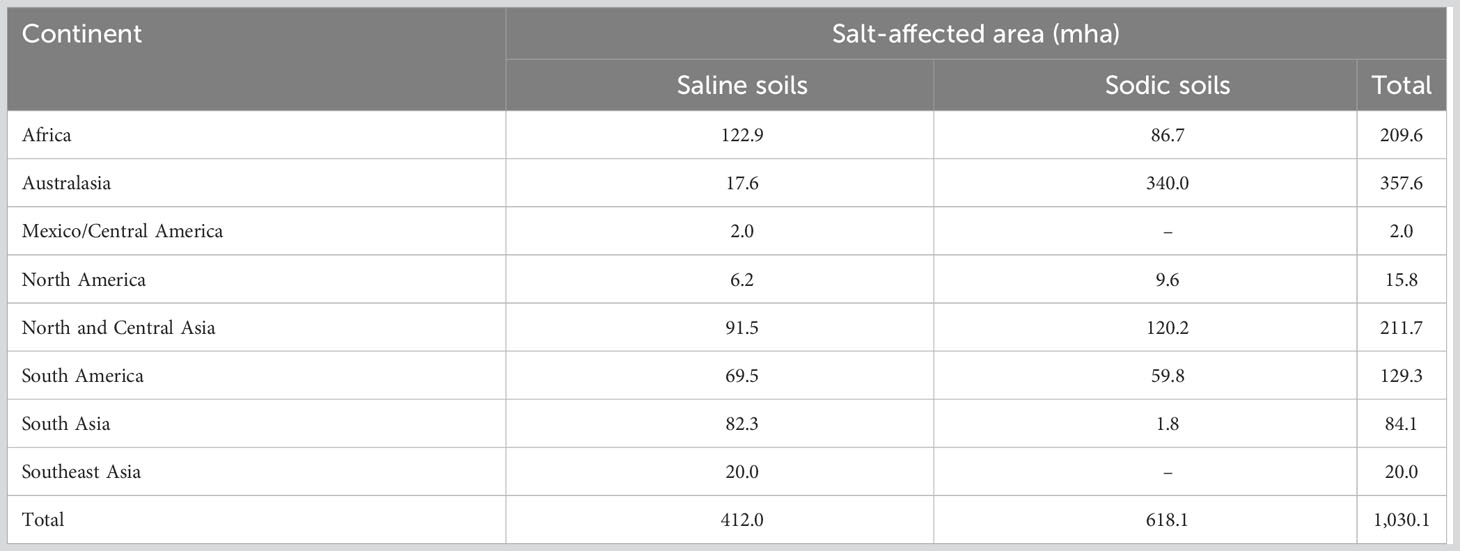
Table 1 Salt-affected soils in various regions of the world (cf. FAO-ITPS-GSP 2015).
Soil salinity is not a recent phenomenon; however, the issue has been accentuated as a result of agricultural activities such as intensive irrigation, poor water management, deforestation, and excessive use of pesticide and chemical fertilizers (Zhu, 2001; Tuteja, 2007; Gupta et al., 2022). It affects almost all the stages of growth and development in plants, from seed germination to blooming and seed maturation, thereby causing a significant loss in the crop yield (Singh et al., 2015; Srivastava et al., 2022a). Excess salt concentrations in the soil primarily affect ion balance in plants and create hyper osmotic stress and secondarily affect the accumulation of harmful ions, which results in poor or delayed germination and post-germination growth abnormalities (Majeed et al., 2019). It has been reported that a high Na+ concentration outside the plant cell has a negative impact on intracellular K+ influx, which is required for plant growth (Kumar and Sharma, 2020). Similarly, calcium and magnesium uptake by plants is also negatively impacted by high sodium content in saline soil. A disturbance in calcium uptake can lead to weakened cell walls, reduced enzyme activities, and altered signaling processes. Magnesium is critical for chlorophyll synthesis as well as production and transport of photoassimilates. During germination stage, salinity impairs the physiological function of seeds, which has a detrimental effect on seed germination and results in a general decrease in plant leaf area, biomass, yield, and root and shoot length (Zörb et al., 2019). Furthermore, it is known to cause various metabolic and physiological changes, depending on rigorousness and stress duration, and eventually reduces crop production (Figure 1A). The inhibitory effect of salinity on plant development involves reduction of water potential, disturbance of ion homeostasis, and associated cellular toxicity (Greenway and Munns, 1980; Isayenkov and Maathuis, 2019). In addition, it is also associated with numerous alterations in their physiology, such as hindering plant roots’ capacity to absorb water and essential minerals, reduction in the stomatal conductance, photosynthesis, and the inability for ROS detoxification, thereby inhibiting growth and development in plants (Abdallah et al., 2016; Ren et al., 2022; James et al., 2011; Gupta and Huang, 2014; Gulzar et al., 2019). Furthermore, the salinity-mediated oxidative stress causes accumulation of ROS such as superoxide anion, hydrogen peroxide, and the hydroxyl radicals, particularly in chloroplasts and mitochondria that damage cell membranes, proteins, lipids, and nucleic acids and may even lead to programmed cell death (Isayenkov, 2012; Mishra et al., 2017; Yang and Guo, 2018; Isayenkov and Maathuis, 2019).
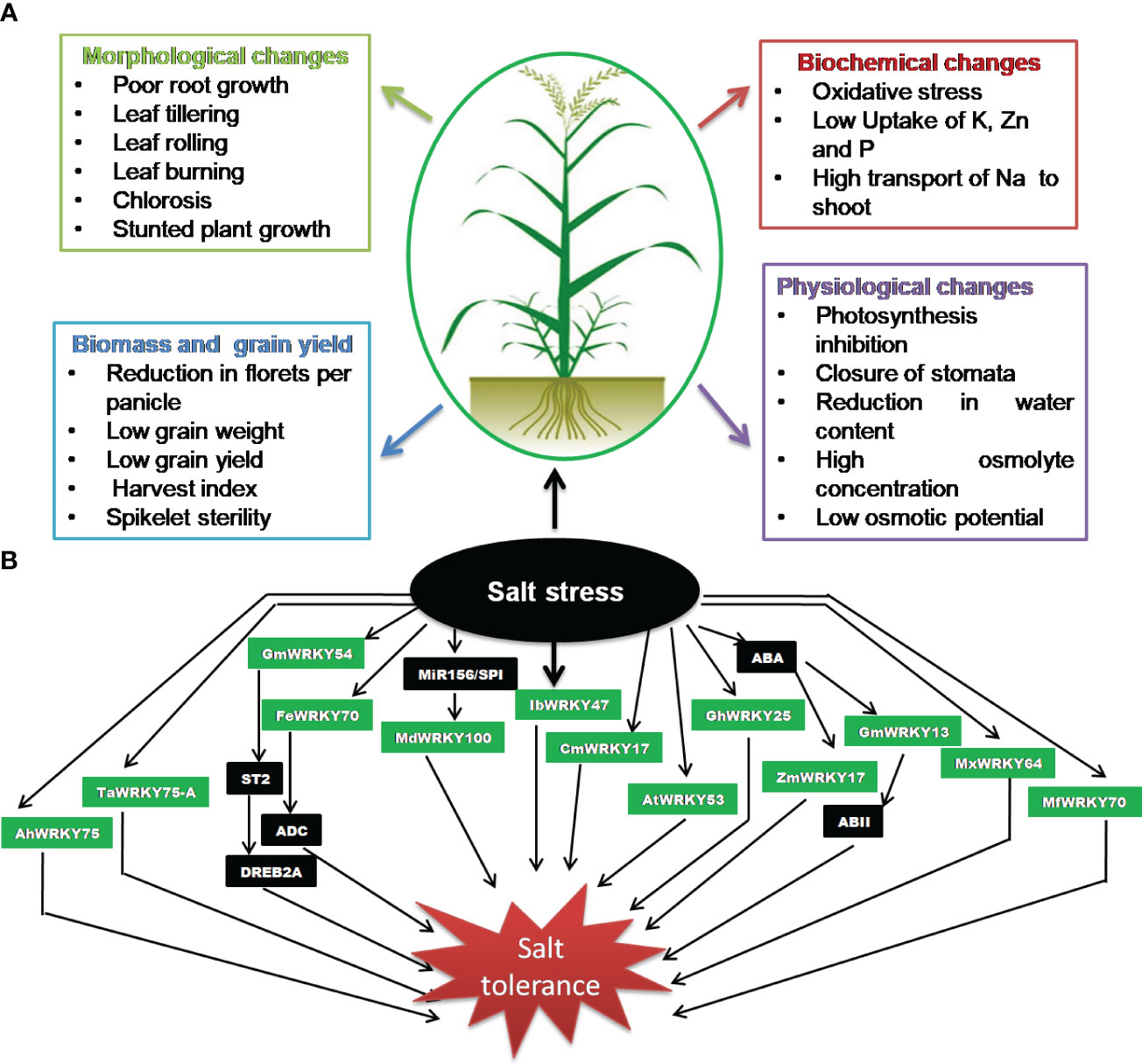
Figure 1 Salinity stress. (A) Impact on crop plants. (B) Significance of WRKY transcription factors (TFs).
3 Salt stress signaling pathway in plants
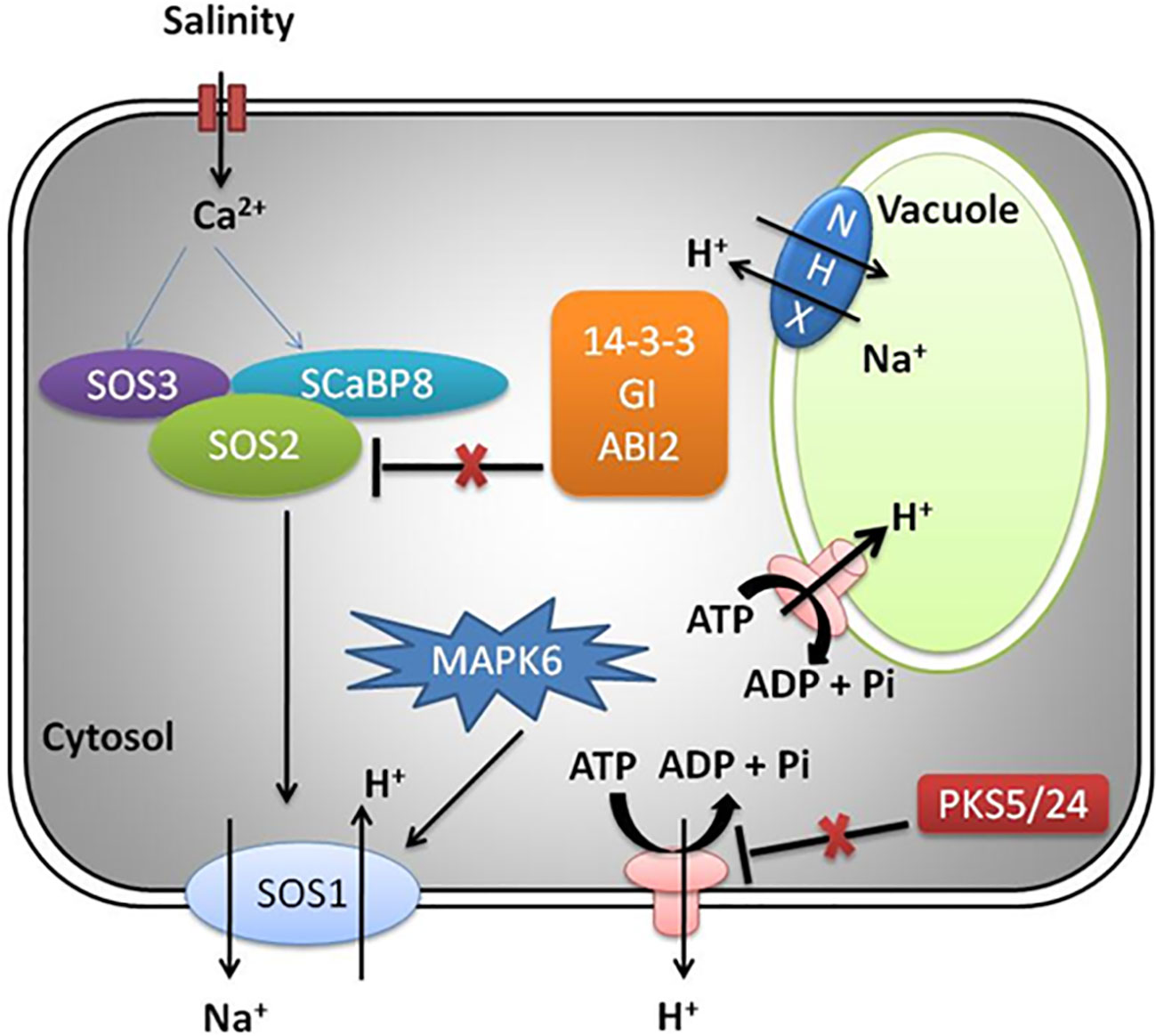
Plants differ widely in Na+ tolerance, and based on their capacity to tolerate salt stress, they are physiologically classified as glycophytes (low salinity tolerance) and halophytes (high salinity tolerance). The former (citrus, tomato, etc.) usually require fresh water and exhibit growth inhibition even under mild salinity. Citrus crops, therefore, showed signs of destruction and could not produce fruit and seeds even below 100 mM NaCl, whereas the halophytes can sustain and grow under elevated or high NaCl conditions (200 mM) (Flowers and Colmer, 2008; Flowers et al., 2010). Some plants such as Atriplex, Rhizophora, and Suaeda can even grow up to 1,000 mM NaCl (Ushakova et al., 2005; Park et al., 2016). Though the knowledge about sensor or receptor of Na+ is not known (Yang and Guo, 2018), it has been noticed that the ionic or osmotic stress may lead to increased cytosolic Ca2+ concentration (Kiegle et al., 2000; Choi et al., 2014). Furthermore, salinity treatment activates salt overly sensitive (SOS) pathway, abscisic acid (ABA) pathway, ROS signaling, and osmotic stress signaling (Yang and Guo, 2018).
One of the adaptive responses for cellular sustenance during salt stress is to retain ion homeostasis. This can be achieved by maintaining cytoplasmic K+/Na+ ratio by lowering Na+ and increasing K+ in the cytoplasm (Niu et al., 1995; Serrano et al., 1999), which involves Na+ uptake restriction, Na+ efflux enhancement, and Na+ compartmentalization in vacuole. Some of the specific transport system for Na+ and K+ uptake includes the low-affinity K+ channel (AKT1, Arabidopsis K+ Transporter1), the high-affinity K+ channel (HKT1, high-affinity K+ transporter 1), and the voltage-independent channel (Blumwald et al., 2000; Tuteja, 2007; Yang and Guo, 2018). Among these, HKT1 serves as a critical player in the improvement of tolerance to salinity by reducing Na+ accumulation in shoots, thereby avoiding Na+ toxicity in the leaves (Horie et al., 2005; Ren et al., 2005; Platten et al., 2006; Horie et al., 2009; Moller et al., 2009). Moreover, the contribution of Na+/Ca2+ exchanger-like proteins is also known to be prominent in ionic homeostasis (Mishra et al., 2021).
The Na+ efflux mechanism is well characterized in Arabidopsis by genetic screening of SOS mutants exposed to salinity stress and reviewed in detail as presented in Figure 2 (Yang and Guo, 2018). The SOS pathway exports Na+ ion from cells and involves activation of SOS2 (serine/threonine protein kinase) and SOS1 (Na+ antiporter) (Lin et al., 2009). The other players include helix E-loop-helix-F (EF-hand) calcium binding proteins (SOS3) and SCaBP8/CBL10, which recognizes high salt concentration and induction of cytosolic calcium signals (Liu and Zhu, 1998; Ishitani et al., 2000; Zhu, 2016). Under the influence of salt-induced cytoplasmic calcium induction, SOS3/SCaBP8 interact and induce SOS2 (Ishitani et al., 2000; Quan et al., 2007; Lin et al., 2009). The 14-3-3, GIGANTEA (GI), and ABA-INSENSITIVE 2 (ABI2) protein (phosphatase 2C) under non-saline (normal) conditions inhibit SOS pathway by interaction with SOS2, thereby repressing its kinase activity (Kim et al., 2013; Zhou et al., 2014; Yang and Guo, 2018). During salt stress, the 26S proteasome pathway degrades 14-3-3 and GI proteins. Additionally, PKS5 activity is also repressed, leading to normal functioning of PM H+-ATPase activity (Yang et al., 2010; Kim et al., 2013; Tan et al., 2016).
Na+ partitioning is also one of the adaptive responses that reduce cytoplasmic ionic toxicity, a mechanism conserved in glycophytes and halophytes (Blumwald et al., 2000; Hasegawa et al., 2000). Additionally, the abiotic stress including salinity leads to the generation of osmolytes, which can lower the water loss under short-term osmotic stress and enhances cell turgor during long-term osmotic stress (Apse and Blumwald, 2002). Furthermore, the osmotic stress also influences the regulation of enzymatic activities related to salt response.
The significance of ABA has also been observed in salinity stress. ABA induction under salt stress activates sucrose non-fermenting 1-related protein kinase 2 (SnRK2) kinase activities (Krzywinska et al., 2016); however, some evidence also indicated the ABA signaling-independent SnRK2 activation (Boudsocq et al., 2007; Zhang et al., 2011a; Zhang et al., 2016a). Additionally, it has been noticed that stimulation of salt stress led to the regulation of many stress-responsive genes, demonstrating correlation with osmotic stress. A study conducted by Sewelam et al. (2014) demonstrated induction of 932 genes under salt stress, out of which 435 overlap with transcripts induced by osmotic stress. Furthermore, 367 genes were found downregulated, in which 154 repressed genes were noted to overlap with osmotic stress (Sewelam et al., 2014).
The osmolytes can be grouped under several categories, viz., charged metabolites like proline, choline-O-sulfate, betaine, and glycine betaine; polyols like mannitol, glycerol, and myo-inositol; sugars such as fructose; complex sugars like fructans, raffinose, and trehalose; and ions such as K+ (Yang and Guo, 2018). Though these metabolites are accumulated in various plant species, few are specific to certain taxonomic categories. In addition, salt also induces the secondary stress response due to ROS generation (Ahmad and Prasad, 2011). ROS at low concentration functions as a signal; however, at high concentration, it has damaging effects over biomolecules (Miller et al., 2010; Gupta and Huang, 2014; Mishra et al., 2017). Therefore, tight regulation of ROS metabolism is a very important aspect for sustenance of normal plant growth under stress conditions. Furthermore, some small molecules act as signals, triggering downstream salt stress response (Yang and Guo, 2018), thereby improving salt tolerance, viz., proline (Khedr et al., 2003), carbon monoxide (Xie et al., 2008), phosphatidic acid (Yu et al., 2010), hydrogen sulfide (Christou et al., 2013; Srivastava et al., 2022a), γ-aminobutyric acid (Srivastava et al., 2021a), and melatonin (Liang et al., 2015; Wei et al., 2015; Mishra et al., 2023).
4 WRKY transcription factor family
TFs regulate expression of genes involved in diverse biological processes. More than 1,000 TF genes have been identified in angiosperms, which can be divided into 58 families depending on their DNA binding domains (Zhang et al., 2011b). WRKY is one of the most numerous TF families in plants involved in many signaling webs of several biological processes including specialized metabolism and stress tolerance (Rushton et al., 2010; Mishra et al., 2013; Kumar et al., 2016; Jiang et al., 2017). Being a TF, its predominant function is transcriptional modulation of genes by its repressor and activator (derepressor) activity. Since its initial reports (Ishiguro and Nakamura, 1994; Rushton et al., 1996), this protein family had been explored in several different plants that includes lower groups, eudicots, and monocots, and many excellent reviews are available mentioning its wide functional diversity (Eulgem et al., 2000; Rushton et al., 2010; Jiang et al., 2017). The investigations include model plants as well as several crops of high commercial significance (Chen F. et al., 2017). The development of sequencing technology has also triggered genome-wide investigation of imperative plant genes and many plant genomes have also been explored for the WRKY TFs (Table 2), which are mostly accompanied with expression study under diverse developmental, stress, and phyto-hormone treatment conditions (Kumar et al., 2016).
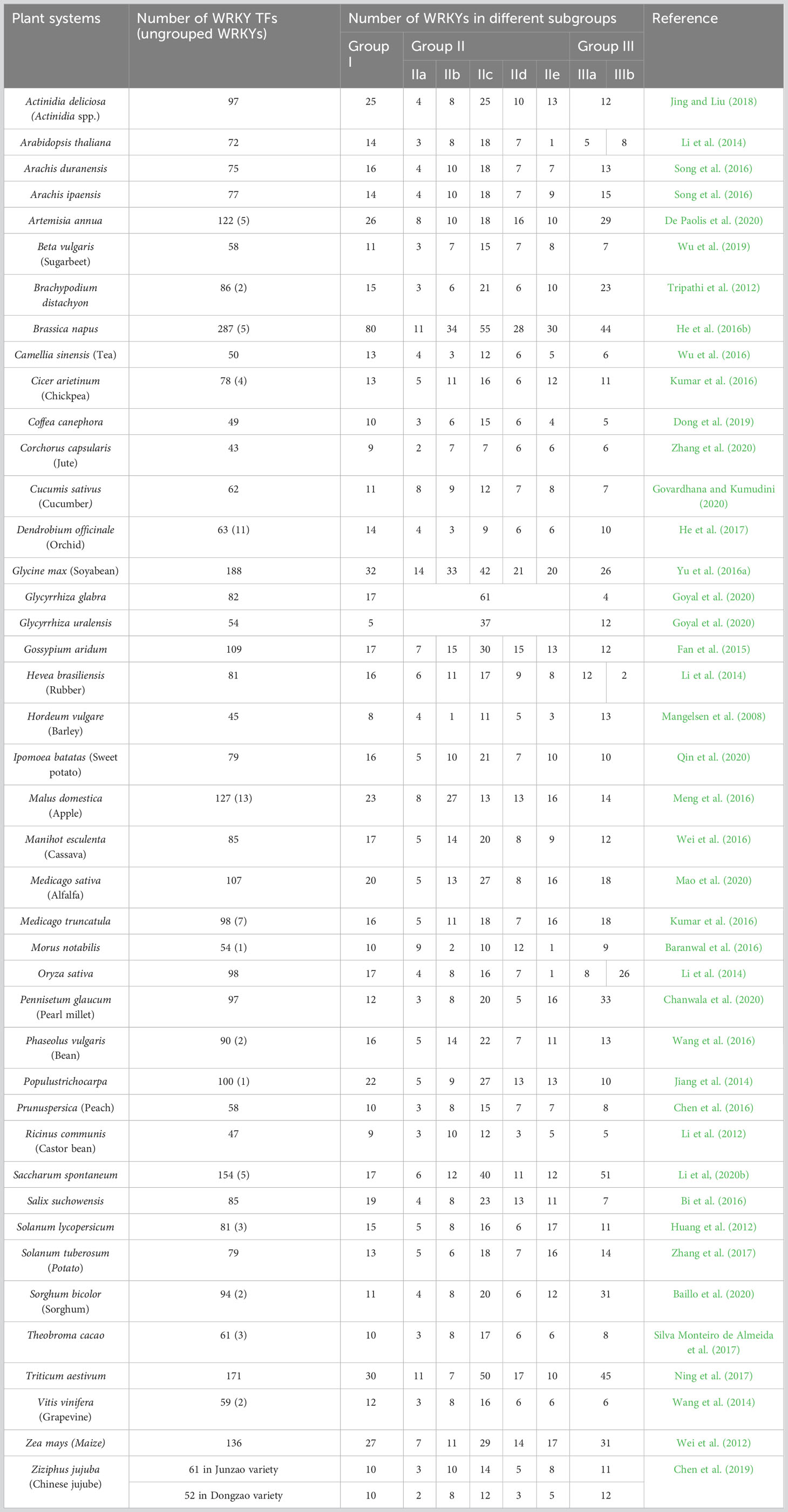
Table 2 Plant system explored for WRKY gene family survey and their representation in different sub-groups.
Currently, the scope of the WRKY family has achieved a broader perspective. In reference to functional diversity, the WRKYs are associated with numerous functions in plants including germination, growth and development, flowering, senescence, carbohydrate synthesis, and secondary metabolite synthesis (Yu et al., 2016b; Jiang et al., 2017; Yu et al., 2018). In numerous studies, it has been reported that WRKY TFs enhance tolerance to salinity stress (Lv et al., 2020; Zhu et al., 2020), drought stress (He et al., 2016a; Wang et al., 2018), heat stress (He et al., 2016a; Wang et al., 2018), chilling stress (Zhang et al., 2016b), heavy metal stress (Sheng et al., 2019), and biotic stress (Cheng and Wang, 2014; Bai et al., 2018) in plants.
Structurally, WRKY proteins consist of 60-amino-acid-long highly conserved WRKY domains. These WRKY domains are made up of four β-strand structures and a C-terminal zinc binding Cystine/Histidine finger motif (Eulgem, 2006; Rushton et al., 2010). The β-strand at the N-terminal contains a conserved stretch of seven amino acids also referred to as “WRKY Signature”, usually composed of “WRKYGQK”, while some WRKY variants, viz., WRKYGEK, WRKYGKK, WRICGQK, WRMCGQK, WKKYGQK, WIKYGQK, WKRYGQK, WSKYEQK, WRKYSEK, WRRYGQK, WSKYGOK, WVKYGQK, WRICGQK, and WRMCGQK, have also been reported in this family (Jiang et al., 2017). The hepta-peptide stretch is considered essential for WRKY binding to the gene promoters [at specific location referred as W-Box–(T)TGAC(C/T)], and hence, alterations in this pattern could lead to changes in their DNA binding ability (Chen F. et al., 2017). W-box components are typical in plant genomes and are made up of a conserved GAC core, a downstream pyrimidine (C/T) residue, and an upstream thymine residue. Although the core aids in WRKY binding, the neighboring residues provide specificity for recognition of a given W-box by a specific factor. For effective binding of WRKYs, more than one W-box can occur in proximity. Certain WRKY are also known to regulate gene expression by binding to elements other than W-box, which includes WT-box (GGACTTTC), WK-box (TTTTCCAC), PRE4-element (TGCGCTT), and SURE-element (TAAAGATTACTAATAGGAA) (Phukan et al., 2016; Chen F. et al., 2017). Other domains also exist among some members of WRKY, including nuclear localization signals (NLS), calmodulin binding sites (CBS), proline-rich region, nucleotide-binding site, leucine-rich repeat, toll interleukin-1 receptor (TIR), NAC (NAM, ATAF1/2 and CUC2) domain, SQUAMOSA promoter binding protein (SBP) domain, ubiquitin-like protease domain, paired amphipathic helix (PAH) domain, ATP-grasp, and other structures. These systems may provide additional functional benefits to WRKY TFs (Eulgem, 2006; Chen F. et al., 2017; Jiang et al., 2017).
The WRKY TFs have been classified into three groups (Table 2) depending on number of WRKY domains (WDs) and pattern of Zn finger motifs. Group I is composed of two WDs with C2H2-type zinc-finger motif, whereas group II has one WD with C2H2-type zinc-finger motif. Group III also possesses single WD like group II, but possesses C2HC-type zinc-finger motif (Kumar et al., 2016). Furthermore, phylogenetic analysis, conserved domain structures, and intron position of the WDs demonstrated further sub-grouping of WRKY TFs (Eulgem et al., 2000; Zhang and Wang, 2005; Kumar et al., 2016). Group II WRKYs are subdivided into five subgroups, namely, IIa, IIb, IIc, IId, and IIe. Group III WRKYs are also composed of two subgroups, namely, IIIa and IIIb (Wu et al., 2005; Zhu et al., 2013). The WRKY domain at the C-terminus of group I proteins is thought to be necessary for DNA binding activity and exhibits similarity to the WRKY domains of group II and group III proteins. WRKY TFs are further classified into two types: R-type and V-type WRKYs, based on the position of intron. The R-type WRKY has a splicing site between the first and second Gs of the AGG codon (arginine), while the V-type WRKY has a splicing site after the valine codon, which is located after the sixth amino acid from the second cysteine residue of the zinc-finger motif (Jiang et al., 2017).
5 WRKYs mediated transcriptional modulation, its interacting partners, and significance under a stressed environment
WRKYs function as either activators or repressors in a variety of molecular processes. They act in an auto-regulated or cross-regulated manner by interacting with other WRKY members or different proteins such as MAP kinases, calmodulin, histone deacetylases, 14-3-3 proteins, and VQ proteins (Rushton et al., 2010; Chi et al., 2013; Phukan et al., 2016). Sometimes, a single WRKY may exhibit several responses, while several WRKYs may also work together to mediate a particular response (Phukan et al., 2016).
Various transcriptional, post-transcriptional, post-translational, and proteasome-mediated mechanisms are known for regulating expression and downstream activation of WRKY in normal and stressed conditions. A zinc-finger protein, Zat12, induced by various abiotic stimuli (salinity, drought, and wounding) was reported to regulate the expression of AtWRKY25 (Mittler et al., 2006). Certain MYB TFs also regulate the expression of WRKYs (Ishida et al., 2007). Transcription of many WRKYs is also regulated by signal molecules. PTI [pathogen-associated molecular patterns (PAMPs)-triggered immunity]- and ETI (effector-triggered immunity)-mediated activation of WRKYs has been observed under several biotic stresses. NaCl treatment induced the expression of WRKY25 and WRKY33 in A. thaliana, and their overexpression increased tolerance to salinity stress (Jiang and Deyholos, 2009). Similarly, overexpression of GmWRKY54 in A. thaliana increased the plant’s tolerance to salt stress. Salt stress also led to accumulation of OsWRKY54 in rice, which, in turn, regulated the expression of OsHKT1;5 by binding to the W-box motif in its promoter. Extensive similarities and cross-talk exist between salinity and drought stress responses in plants (Golldack et al., 2014). WRKY46, WRKY54, and WRKY70 together interact with BES1 to regulate brassinosteroid-mediated drought response (Chen J. et al., 2017). DREB TFs are considered as master regulators in drought response. Regulation of DREBs by TaWRKY19 (Niu et al., 2012) and GhWRKY59 (Jin et al., 2017) is another example of crosstalk between TFs mediating salt response. On the other hand, there are also examples where WRKYs function differently in salt and drought stress. For instance, overexpression of GhWRKY25 in Nicotiana benthamiana increases salinity tolerance but negatively impacted drought tolerance and sensitivity to fungal pathogen. Expression of WRKY was also reported to be controlled by miRNAs at the post-transcriptional level (Phukan et al., 2016). Interactions of histone deactelyases (HDAC), histone demethylase, and histone methyl transferases with WRKY revealed the non-genetic regulation of WRKYs in plants (Chi et al., 2013; Phukan et al., 2016). Histone deactylase-19 removes acetyl groups from histone tails and downregulates the expression of AtWRKY38 and AtWRKY62 (Kim et al., 2008). The linker histone H1 MaHIS1 interacts with MaWRKY1 and functionally coordinates to influence stress responses and ripening in banana fruit (Wang et al., 2012b). Flowering Locus D (FLD) brings about histone modifications of WRKY 29 and WRKY6 gene promoters and, thus, epigenetically regulates their SAR (systemic-acquired resistance)-induced expression (Singh et al., 2014). Chloroplast- and mitochondria-mediated retrograde inter-organelle signaling to the nucleus regulates several WRKY factors (Hammargren et al., 2008; Shang et al., 2010). Furthermore, phosphorylation by kinases is also known to modulate the expression and functioning of WRKY TFs. MAPK regulates the expression of OsWRKY45 and provides resistance to various pathogenic infections in rice. Responses to bacterial and fungal infections are also modulated by AtWRKY22 and AtWRKY29 through the MAPK pathway (Göhre et al., 2012). WRKYs in tobacco interact with MAPK cascade pathways in plant defense against whiteflies (Yao et al., 2021). Proteasome-mediated degradation also maintains the level of WRKYs under various stressed and non-stressed conditions. UPS (ubiquitin proteasome system) is known to degrade OsWRKY45 at normal un-diseased state in plants, whereas the pathogenic invasions inhibit proteasomes and accumulate OsWRKY45 (Matsushita et al., 2013; Phukan et al., 2016).
6 WRKYs and crop improvement for salt tolerance involve multiple responses
WRKYs play promising roles during plant signaling and are extensively reported for their contributions in abiotic and biotic stress (Li et al., 2020a; Wani et al., 2021). Nevertheless, current advances do divulge the vast significance of WRKY proteins for regulation of plant abiotic stress tolerance (Huang and Amee, 2021; Xiang et al., 2021). Researchers have employed specific WRKY TFs to create transgenics with improved stress tolerance traits (Table 3), because of their regulatory effects on stress-responsive genes clusters (Banerjee and Roychoudhury, 2015). Understanding of the signaling cascades that lead to the activation and interaction of the WRKY proteins with other signaling proteins, and the regulation of downstream target genes are crucial in the choice of WRKY genes for engineering stress tolerance in plants.
Salinity stress is a key abiotic stress that affects agricultural productivity, mostly in semi-arid and arid areas. WRKYs are known to play a critical role in the regulation of plant salt stress responses (Figure 1B; Table 3). WRKY has been observed as both a positive (Han et al., 2021; Xiang et al., 2021; Ye et al., 2021; Zhu et al., 2021) and a negative regulator (Huang and Amee, 2021) for salinity stress. In a study, 47 WRKY genes were reported to respond to salinity stress in wheat (Hassan et al., 2019), which demonstrated the significance of WRKY during salinity stress. The STZ (zinc finger protein STZ/ZAT10) protein associated to ZPT2 (zinc finger protein) is known for downregulating the deactivation of other TFs and, therefore, functions as an inhibitor of transcription. Zhou et al. (2008) reported that the STZ expression is inhibited by GmWRKY54 in G. max, thus inducing response to salt stress via the positive regulation of DREB2A-mediated pathway (Zhou et al., 2008). In another study, Gong et al. (2015) demonstrated that FcWRKY70 is involved in upregulating expression of arginine decarboxylase (ADC), resulting in plant salinity tolerance. The miR156/SPL is involved in modulation of tolerance to salinity stress by upregulating MdWRKY100 in Malus domestica (Ma et al., 2021). The SbWRKY50 directly binds to SOS1 and HKT1 promoter and participated in plant salt response by regulating ion homeostasis in Sorghum bicolor (Song et al., 2020). The salt tolerance in transgenic Arabidopsis-overexpressing peanut AhWRKY75 (Zhu et al., 2021) involved the upregulation of genes associated with ROS scavenging activity and improved antioxidant system (SOD, POD, and catalase). Furthermore, the significantly lower accumulation of malondialdehyde and superoxide anion content was also noticed in transgenic plants (Zhu et al., 2021). Similar observation was also noticed in transgenic Arabidopsis overexpressing Myrothamnus flabellifolia MfWRKY70-mediated salt tolerance (Xiang et al., 2021). The transgenic plants demonstrated the positive regulation of stress-associated genes such as P5CS, NCED3, and RD29A.
The salinity (and drought) tolerance in the ectopically expressed TaWRKY75-A in Arabidopsis integrated jasmonic acid biosynthetic pathways (Ye et al., 2021). In contrast, an increased expression level of GhWRKY25 increases the salinity stress tolerance in upland cotton, whereas transgenic tobacco plant showed comparatively lower drought stress tolerance, signifying that the WRKY exhibited different regulatory effects in response to diverse stress conditions (Liu et al., 2016). Shen et al. (2015) revealed that the antioxidant enzyme activity is enhanced during salt-induced H2O2 and cytosolic Ca2+ stimulation in Populus euphratica, thus improving salt stress tolerance. Salinity stress response has been shown to be largely related to ABA-induced WRKY gene expression (Li et al., 2020a). Various reports have demonstrated that ABA and NaCl when applied exogenously can also induce WRKY expression like AtWRKY25 and AtWRKY33 in Arabidopsis (Jiang and Deyholos, 2009), OsWRKY72 in rice (Song et al., 2010), GbWRKY1 in Gossypium barbadense (Luo et al., 2020), and VpWRKY3 (Zhu et al., 2012) and VpWRKY1/2 (Li et al., 2010) in grape. Functional studies of WRKYs towards salt stress tolerance have been compiled in Table 3, which also explains the pathways regulated during WRKY-mediated tolerance to the salinity stress.
Additionally, WRKYs are also known as negative regulators of salt stress tolerance trait in plants (Zhou et al., 2008; Huang and Amee, 2021). The inhibition of salt stress tolerance via regulation of the DNA binding and transcriptional activity of WRKY53 was reported by Arabidopsis RPD3-like histone deacetylase HDA9 (Zheng et al., 2020). Li et al. (2015) reported overexpression of Chrysanthemum CmWRKY17 in Arabidopsis, which resulted in higher sensitivity towards salt stress. The study reported that stress resistance-related genes in wild-type plants showed higher expression against stress compared to transgenic Arabidopsis, demonstrating that CmWRKY17 may be implicated in negative regulation of salinity stress in Chrysanthemum (Li et al., 2015). Similarly, salinity sensitivity was also observed in CdWRKY50 overexpressing Arabidopsis. The CdWRKY50 can also bind to the AtDREB2A promoter, thereby regulating its expression (Huang and Amee, 2021). In G. max, ABI1 could be the downstream target gene of GmWRKY13. Transgenic studies in Arabidopsis exhibited that the overexpression of GmWRKY13 enhanced ABI1 expression; however, plants were found to be less tolerant to salt stress (Zhou et al., 2008). Overexpression of ZmWRKY17 in Arabidopsis demonstrated an inhibitory result on exogenous ABA treatment, ensuing comparatively lower tolerance to high salinity (Cai et al., 2017).
Although the literature strongly supported this function of WRKY in salinity stress, there are certain missing links that need reasonable research, viz., How does salt stress cause WRKY induction? Is this generalized or specific to plant/members of WRKY gene family? Does post-translational modification of WRKY impact its functionality during salt stress? Does the homo- and heterodimerization of WRKY influence its behavior during salinity? How does the WRKY-mediated metabolite regulation influence its role in salt stress mitigation?
7 Conclusion and future directions
The ultimate solution to ensure crop production potential is to incorporate tolerance traits into the plants. The significant impact of salinity stress over crop production is an urgent challenge to ensure sustainable crop production to feed the global population. Salinity stress significantly deteriorates the crop production potential throughout the globe, due to its larger effect on plant physiology and biochemistry, thus ultimately leading to significant agricultural loss. Plants differ significantly in terms of their tolerance to salinity and have the capacity to sense this stress through the SOS pathway, which involves many candidate proteins. Among several tolerance mechanisms to address salinity stress-mediated crop loss, the utility of TF-mediated tolerance is well documented. Being one of the major TF families, WRKY plays a significant role in plants at several avenues including stress tolerance (plant fitness to environmental constraints). Over the years, scientists have revealed that WRKY TFs not only contribute to growth and development in plants, but also exhibit complex regulatory networks and mechanism implicated in various stresses. Since crops generally face different stresses and WRKYs play crucial roles during stress response, further detailed studies on WRKY genes are needed to specify their unique functions. So far, characterization of WRKY is considered, and many plants have been established as a model to support the significance of WRKY in salt tolerance. Furthermore, the underlying mechanism is also explored at few instances (Table 3) but broader validation is needed. In addition, genomics has facilitated exploration of this protein family in many crops and newer studies are continuously enriching this data. Furthermore, such investigation offers a broader perspective as the researcher can individually target most promising WRKYs out of close putative candidates. Moreover, earlier research work over WRKY gene functions was mostly focused on transcriptomics and functional predictions, while further applications of genetic confirmation integrated with novel tools help to speed up the research regarding studies related to WRKY neo-functionalization. Further characterization of the downstream genes that are regulated through WRKY is still a challenge. Such research explorations will help to elucidate the regulatory networks involved in stress response in plants. Furthermore, non-coding RNAs (ncRNA) and epigenetic modifications entailed in the WRKY TFs regulation must be investigated in future research. By integrating multiomics methods such as genomics, transcriptomics, proteomics, and metabolomics, TFs have been investigated and further modified utilizing genome editing tools such as CRISPR/Cas systems to improve plant tolerance to various abiotic stresses such as salt stress. In-depth studies of TFs will possibly enhance our ability to improve the stress tolerance in crop plants to achieve food security at the global level. Finally, using WRKY TFs to monitor stress-tolerant plant cultivars and enhance stress resistance in plants will considerably help to improve quality and yield in the perspective of climate change and food security.
Author contributions
GR, VS, MC, and SG: Framing the concept and writing; SM, RC, MC, and MM: Manuscript writing; AC, RK, PK, and PR: prepared the figures and tables; FP-A, RK, MC, and PK: Manuscript correction; SG, MC, GC, FP-A, PK, and VS: modified and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research work was partially funded by MUR in the frame of the project ‘Digitali, Intelligenti, Verdi e Sostenibili (D.I.Ver.So)’ managed by DAFNE of Tuscia University.
Acknowledgments
SG thankfully acknowledges the Council of Scientific and Industrial Research (CSIR) Grant Number MLP110006.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ABA, abscisic acid; AKT, Arabidopsis potassium transporter; Ca2+, calcium; Cl−, chloride; K+, potassium; MAPK, mitogen-activated protein kinase; Mg+, magnesium; Na+, sodium; ROS, reactive oxygen species; SOS, salt overly sensitive; TF, transcription factor.
References
Abdallah, B. S., Aung, B., Amyot, L., Lalin, I., Lachâal, M., Karray-Bouraoui, N., et al. (2016). Salt stress (NaCl) affects plant growth and branch pathways of carotenoid and flavonoid biosyntheses in Solanum nigrum. Acta Physiol. Plant 38, 1–13.
Agarwal, P., Dabi, M., Sapara, K. K., Joshi, P. S., Agarwal, P. K. (2016). Ectopic expression of JcWRKY transcription factor confers salinity tolerance via salicylic acid signaling. Front. Plant Sci. 7, 1541. doi: 10.3389/fpls.2016.01541
Ahmad, P., Kumar, A., Ashraf, M., Akram., N. A. (2012). Salt-induced changes in photosynthetic activity and oxidative defense system of three cultivars of mustard (Brassica juncea L.). Afr. Journ.of Biotech. 11 (11), 2694–2703. doi: 10.5897/AJB11.3203
Ahmad, P., Prasad, M. N. V. (2011). “Abiotic stress responses in plants: metabolism, productivity and sustainability,” (Springer Science & Business Media). doi: 10.1007/978-1-4614-0634-1
Apse, M. P., Blumwald, E. (2002). Engineering salt tolerance in plants. Curr. Opin. Biotech. 13 (2), 146–150. doi: 10.1016/S0958-1669(02)00298-7
Bai, Y., Sunarti, S., Kissoudis, C., Visser, R. G., van der Linden, C. (2018). The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front. Plant Sci. 9, 801. doi: 10.3389/fpls.2018.00801
Baillo, E. H., Hanif, M. S., Guo, Y., Zhang, Z., Xu, P., Algam, S. A. (2020). Genome-wide Identification of WRKY transcription factor family members in sorghum (Sorghum bicolor (L.) moench). PLoSone 15 (8), e0236651. doi: 10.1371/journal.pone.0236651
Bakshi, M., Oelmüller, R. (2014). WRKY transcription factors: Jack of many trades in plants. Plant Signaling behavi. 9 (2), e27700. doi: 10.4161/psb.27700
Banerjee, A., Roychoudhury, A. (2015). WRKY proteins: signaling and regulation of expression during abiotic stress responses. Sci. World Journ. 2015, 807560. doi: 10.1155/2015/807560
Baranwal, V. K., Negi, N., Khurana, P. (2016). Genome-wide identification and structural, functional and evolutionary analysis of WRKY components of mulberry. Sci. Rep. 6, 30794. doi: 10.1038/srep30794
Bi, C., Xu, Y., Ye, Q., Yin, T., Ye, N. (2016). Genome-wide identification and characterization of WRKY gene family in Salix suchowensis. PeerJ. 4, e2437. doi: 10.7717/peerj.2437
Blumwald, E., Aharon, G. S., Apse, M. P. (2000). Sodium transport in plant cells. Biochim. Biophys. Acta (BBA)-Biomembranes 1465 (1-2), 140–151. doi: 10.1016/S0005-2736(00)00135-8
Boudsocq, M., Droillard, M.-J., Barbier-Brygoo, H., Laurière, C. (2007). Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol.Biol. 63 (4), 491–503. doi: 10.1007/s11103-006-9103-1
Buscaill, P., Rivas, S. (2014). Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 20, 35–46. doi: 10.1016/j.pbi.2014.04.004
Cai, R., Dai, W., Zhang, C., Wang, Y., Wu, M., Zhao, Y., et al. (2017). The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta. 246 (6), 1215–1231. doi: 10.1007/s00425-017-2766-9
Chanwala, J., Satpati, S., Dixit, A., Parida, A., Giri, M. K., Dey, N. (2020). Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC Genomics 21 (1), 1–16. doi: 10.1186/s12864-020-6622-0
Chen, X., Chen, R., Wang, Y., Wu, C., Huang, J. (2019). Genome-wide identification of WRKY transcription factors in Chinese jujube (Ziziphus jujuba Mill.) and their involvement in fruit developing, ripening, and abiotic stress. Genes. 10 (5), 360. doi: 10.3390/genes10050360
Chen, F., Hu, Y., Vannozzi, A., Wu, K., Cai, H., Qin, Y., et al. (2017). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36 (5-6), 11–335. doi: 10.1080/07352689.2018.1441103
Chen, J., Nolan, T. M., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in Brassinosteroid-regulated plant growth and drought responses. Plant Cell 29 (6), 1425–1439. doi: 10.1105/tpc.17.00364
Chen, L., Song, Y., Li, S., Zhang, L., Zou, C., Yu, D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 1819 (2), 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Chen, M., Tan, Q., Sun, M., Li, D., Fu, X., Chen, X., et al. (2016). Genome-wide identification of WRKY family genes in peach and analysis of WRKY expression during bud dormancy. Mol. Gen. Genom. 291 (3), 1319–1332. doi: 10.1007/s00438-016-1171-6
Cheng, H., Wang, S. (2014). WRKY-type transcription factors: a significant factor in rice-pathogen interactions. Scientia Sin. Vitae. 44 (8), 784–793. doi: 10.1360/052014-97
Chi, Y., Yang, Y., Zhou, Y., Zhou, J., Fan, B., Yu, J. Q., et al. (2013). Protein–protein interactions in the regulation of WRKY transcription factors. Mol. Plant 6 (2), 287–300. doi: 10.1093/mp/sst026
Choi, W. G., Toyota, M., Kim, S. H., Hilleary, R., Gilroy, S. (2014). Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. 111 (17), 6497–6502. doi: 10.1073/pnas.1319955111
Chowdhary, A. A., Mishra, S., Mehrotra, S., Upadhyay, S. K., Bagal, D., Srivastava, V. (2023). Plant transcription factors: an overview of their role in plant life. Plant Transcription Factors, 3–20. doi: 10.1016/B978-0-323-90613-5.00003-0
Christou, A., Manganaris, G. A., Papadopoulos, I., Fotopoulos, V. (2013). Hydrogen sulfide induces systemic tolerance to salinity and non-ionic osmotic stress in strawberry plants through modification of reactive species biosynthesis and transcriptional regulation of multiple defence pathways. J.Exp.Bot. 64 (7), 1953–1966. doi: 10.1093/jxb/ert055
De Paolis, A., Caretto, S., Quarta, A., Di Sansebastiano, G.-P., Sbrocca, I., Mita, G., et al. (2020). Genome-wide identification of WRKY genes in artemisia annua: Characterization of a putative ortholog of atWRKY40. Plants. 9 (12), 1669. doi: 10.3390/plants9121669
Dong, X., Yang, Y., Zhang, Z., Xiao, Z., Bai, X., Gao, J., et al. (2019). Genome-wide identification of WRKY genes and their response to cold stress in Coffea canephora. Forests 10 (4), 335. doi: 10.3390/f10040335
Dong, Q., Zheng, W., Duan, D., Huang, D., Wang, Q., Liu, C., et al. (2020). MdWRKY30, a group IIa WRKY gene from apple, confers tolerance to salinity and osmotic stresses in transgenic apple callus and Arabidopsis seedlings. Plant Sci. 299, 110611. doi: 10.1016/j.plantsci.2020.110611
Eulgem, T. (2006). Dissecting the WRKY web of plant defense regulators. PLoS Pathog. 2 (11), e126. doi: 10.1371/journal.ppat.0020126
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 (5), 199–206. doi: 10.1016/S1360-1385(00)01600-9
Fan, X., Guo, Q., Xu, P., Gong, Y., Shu, H., Yang, Y., et al. (2015). Transcriptome-wide identification of salt-responsive members of the WRKY gene family in Gossypium aridum. PLoS One 10 (5), e0126148. doi: 10.1371/journal.pone.0126148
Flowers, T. J., Colmer, T. D. (2008). Salinity tolerance in halophytes. New Phytologist. 179, 945–963. doi: 10.1111/j.1469-8137.2008.02531.x
Flowers, T. J., Galal, H. K., Bromham, L. (2010). Evolution of halophytes: multiple origins of salt tolerance in land plants. Funct. Plant Biol. 37 (7), 604–612. doi: 10.1071/FP09269
Gao, Y. F., Liu, J. K., Yang, F. M., Zhang, G. Y., Wang, D., Zhang, L., et al. (2020). The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Phys. Plant 168 (1), 98–117. doi: 10.1111/ppl.12978
Gao, H., Wang, Y., Xu, P., Zhang, Z. (2018). Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 9, 99. doi: 10.3389/fpls.2018.00997
Göhre, V., Jones, A. M., Sklenář, J., Robatzek, S., Weber, A. P. (2012). Molecular crosstalk between PAMP-triggered immunity and photosynthesis. Mol. Plant Microbe Interact. 25, 1083–1092. doi: 10.1094/MPMI-11-11-0301
Golldack, D., Li, C., Mohan, H., Probst, N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00151
Gong, X., Zhang, J., Hu, J., Wang, W., Wu, H., Zhang, Q., et al. (2015). FcWRKY 70, a WRKY protein of F ortunella crassifolia, functions in drought tolerance and modulates putrescine synthesis by regulating arginine decarboxylase gene. Plant Cell environment. 38 (11), 2248–2262. doi: 10.1111/pce.12539
Govardhana, M., Kumudini, B. S. (2020). In-silico analysis of cucumber (Cucumis sativus L.) Genome for WRKY transcription factors and cis-acting elements. Comput. Biol. Chem. 85, 107212. doi: 10.1016/j.compbiolchem.2020.107212
Goyal, P., Manzoor, M. M., Vishwakarma, R. A., Sharma, D., Dhar, M. K., Gupta, S. (2020). A comprehensive transcriptome-wide identification and screening of WRKY gene family engaged in abiotic stress in Glycyrrhiza glabra. Scient.Rep. 10 (1), 1–18. doi: 10.1038/s41598-019-57232-x
Greenway, H., Munns, R. (1980). Mechanisms of salt tolerance in nonhalophytes. Annu. Rev. Plant Physiol. 31 (1), 149–190. doi: 10.1146/annurev.pp.31.060180.001053
Gulzar, N., Mir, M. Y., Hamid, S. (2019). Effect of nitrogen and phosphorus on the growth and redox homeostasis of salt-stressed mustard plants. Asian J.Plant Sci. 18, 52–59. doi: 10.3923/ajps.2019.52.59
Gupta, A., Bano, A., Rai, S., Mishra, R., Singh, M., Sharma, S., et al. (2022). Mechanistic insights of plant-microbe interaction towards drought and salinity stress in plants for enhancing the agriculture productivity. Plant Stress 4, 100073.
Gupta, B., Huang, B. (2014). Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genomics 2014, 1–18. doi: 10.1155/2014/701596
Hammargren, J., Rosenquist, S., Jansson, C., Knorpp, C. (2008). A novel connection between nucleotide and carbohydrate metabolism in mitochondria: sugar regulation of the Arabidopsis nucleoside diphosphate kinase 3a gene. Plant Cell Rep. 27, 529–534. doi: 10.1007/s00299-007-0486-5
Han, D., Han, J., Xu, T., Li, T., Yao, C., Wang, Y., et al. (2021). Isolation and preliminaryfunctional characterization of MxWRKY64, a new WRKY transcription factor gene from Malus xiaojinensis. In Vitro Cell. Dev. Biology-Plant. 57 (2), 202–213. doi: 10.1007/s11627-021-10171-7
Han, D., Hou, Y., Wang, Y., Ni, B., Li, Z., Yang, G. (2018). Overexpression of a Malus baccata WRKY transcription factor gene (MbWRKY5) increases drought and salt tolerance in transgenic tobacco. Can. J. Plant Sci. 99 (2), 173–183. doi: 10.1139/cjps-2018-0053
Han, D., Zhou, Z., Du, M., Li, T., Wu, X., Yu, J., et al. (2020). Overexpression of a Malus xiaojinensis WRKY transcription factor gene (MxWRKY55) increased iron and high salinity stress tolerance in Arabidopsis thaliana. In Vitro Cell. Dev. Biology-Plant. 56 (5), 600–609. doi: 10.1007/s11627-020-10129-1
Hasegawa, P. M., Bressan, R. A., Zhu, J. K., Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Biol. 51 (1), 463–499. doi: 10.1146/annurev.arplant.51.1.463
Hassan, S., Lethin, J., Blomberg, R., Mousavi, H., Aronsson, H. (2019). In silico based screening of WRKY genes for identifying functional genes regulated by WRKY under salt stress. Comput. Biol. Chem. 83, 107131. doi: 10.1016/j.compbiolchem.2019.107131
He, C., da Silva, J., Tan, J., Zhang, J., Pan, X., Li, M., et al. (2017). A genome-wide identification of the WRKY family genes and a survey of potential WRKY target genes in Dendrobium officinale. Sci. Rep. 7 (1), 1–14. doi: 10.1038/s41598-017-07872-8
He, Y., Mao, S., Gao, Y., Zhu, L., Wu, D., Cui, Y., et al. (2016b). Genome-wide identification and expression analysis of WRKY transcription factors under multiple stresses in. Brassica napus.PLoS One 11 (6), e0157558. doi: 10.1371/journal.pone.0157558
He, G. H., Xu, J. Y., Wang, Y. X., Liu, J. M., Li, P. S., Chen, M., et al. (2016a). Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Bio. 16 (1), 1–16. doi: 10.1186/s12870-016-0806-4
Horie, T., Hauser, F., Schroeder, J. I. (2009). HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 14 (12), 660–668. doi: 10.1016/j.tplants.2009.08.009
Horie, T., Motoda, J., Kubo, M., Yang, H., Yoda, K., Horie, R., et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 44 (6), 928–938. doi: 10.1111/j.1365-313X.2005.02595.x
Huang, X., Amee, M. (2021). Bermudagrass CdWRKY50 gene negatively regulates plants’ response to salt stress. Environ. Exp. Bot. 188, 104513. doi: 10.1016/j.envexpbot.2021.104513
Huang, S., Gao, Y., Liu, J., Peng, X., Niu, X., Fei, Z., et al. (2012). Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom 287 (6), 495–513. doi: 10.1007/s00438-012-0696-6
Isayenkov, S. (2012). Physiological and molecular aspects of salt stress in plants. Cytology Genet. 46 (5), 302–318. doi: 10.3103/S0095452712050040
Isayenkov, S. V., Maathuis, F. J. (2019). Plant salinity stress: many unanswered questions remain. Front. Plant Sci. 10, 80. doi: 10.3389/fpls.2019.00080
Ishida, T., Hattori, S., Sano, R., Inoue, K., Shirano, Y., Hayashi, H., et al. (2007). Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell. 19 (8), 2531–2543. doi: 10.1105/tpc.107.052274
Ishiguro, S., Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 244 (6), 563–571. doi: 10.1007/BF00282746
Ishitani, M., Liu, J., Halfter, U., Kim, C.-S., Shi, W., Zhu, J. K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell. 12 (9), 1667–1677. doi: 10.1105/tpc.12.9.1667
James, R. A., Blake, C., Byrt, C. S., Munns, R. (2011). Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1; 4 and HKT1; 5), decrease Na+ accumulation in bread wheat leaves under saline and waterlogged conditions. J. Exp. Bot. 62 (8), 2939–2947. doi: 10.1093/jxb/err003
Jia, H., Wang, C., Wang, F., Liu, S., Li, G., Guo, X. (2015). GhWRKY68 reduces resistance to salt and drought in transgenic Nicotiana benthamiana. PLoS One 10 (3), e0120646. doi: 10.1371/journal.pone.0120646
Jiang, Y., Deyholos, M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69 (1-2), 91–105. doi: 10.1007/s11103-008-9408-3
Jiang, Y., Duan, Y., Yin, J., Ye, S., Zhu, J., Zhang, F., et al. (2014). Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp.Bot. 65 (22), 6629–6644. doi: 10.1093/jxb/eru381
Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J., Zhang, J. (2017). WRKY transcription factors in plant responses to stresses. JournalInteg.plant Biol. 59 (2), 86–101. doi: 10.1111/jipb.12513
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45 (D1), D1040–D1045. doi: 10.1093/nar/gkw982
Jing, Z., Liu, Z. (2018). Genome-wide identification of WRKY transcription factors in Kiwifruit (Actinidia spp.) and analysis of WRKY expression in responses to biotic and abiotic stresses. Genes &Genomics. 40 (4), 429–446. doi: 10.1007/s13258-017-0645-1
Kang, D. R., Zhu, Y., Li, S. L., Ai, P. H., Khan, M. A., Ding, H. X., et al. (2021). Transcriptome analysis of differentially expressed genes in chrysanthemum MET1 RNA interference lines. Physiol. Mol. Biol. Plants 27, 1455–1468. doi: 10.1007/s12298-021-01022-1
Khedr, A. H. A., Abbas, M. A., Wahid, A. A. A., Quick, W. P., Abogadallah, G. M. (2003). Proline induces the expression of salt-stress-responsive proteins and may improve the adaptation of Pancratium maritimum L. @ to salt-stress. J. Exp. Bot. 54 (392), 2553–2562. doi: 10.1093/jxb/erg277
Kiegle, E., Moore, C. A., Haseloff, J., Tester, M. A., Knight, M. R. (2000). Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 23 (2), 267–278. doi: 10.1046/j.1365-313x.2000.00786.x
Kim, W. Y., Ali, Z., Park, H. J., Park, S. J., Cha, J. Y., Perez-Hormaeche, J., et al. (2013). Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Comm.s 4 (1), 1–13. doi: 10.1038/ncomms2357
Kim, K. C., Lai, Z., Fan, B., Chen, Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 20 (9), 2357–2371. doi: 10.1105/tpc.107.055566
Krzywinska, E., Kulik, A., Bucholc, M., Fernandez, M. A., Rodriguez, P. L., Dobrowolska, G. (2016). Protein phosphatase type 2C PP2CA together with ABI1 inhibits SnRK2. 4 activity and regulates plant responses to salinity. Plant Signaling &Behavior. 11 (12), e1253647. doi: 10.1080/15592324.2016.1253647
Kumar, P., Sharma, P. (2020). Soil salinity and food security in India. Front. Sustain Food Syst. 4, 533781. doi: 10.3389/fsufs.2020.533781
Kumar, K., Srivastava, V., Purayannur, S., Kaladhar, V. C., Cheruvu, P. J., Verma, P. K. (2016). WRKY domain-encoding genes of a crop legume chickpea (Cicer arietinum): comparative analysis with Medicago truncatula WRKY family and characterization of group-III gene(s). DNA Res. 23 (3), 225–239. doi: 10.1093/dnares/dsw010
Li, H. L., Guo, D., Yang, Z. P., Tang, X., Peng, S. Q. (2014). Genome-wide identification and characterization of WRKY gene family in Hevea brasiliensis. Genomics. 104 (1), 14–23. doi: 10.1016/j.ygeno.2014.04.004
Li, Z., Hua, X., Zhong, W., Yuan, Y., Wang, Y., Wang, Z., et al. (2020b). Genome-wide identification and expression profile analysis of WRKY family genes in the autopolyploid Saccharum spontaneum. Plant Cell Physiol. 61 (3), 616–630. doi: 10.1093/pcp/pcz227
Li, W., Pang, S., Lu, Z., Jin, B. (2020a). Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants. 9 (11), 1515. doi: 10.3390/plants9111515
Li, P., Song, A., Gao, C., Wang, L., Wang, Y., Sun, J., et al. (2015). Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic Chrysanthemum and Arabidopsis plants. Plant Cell Rep. 34 (8), 1365–1378. doi: 10.1007/s00299-015-1793-x
Li, H., Xu, Y., Xiao, Y., Zhu, Z., Xie, X., Zhao, H., et al. (2010). Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata. Planta 232 (6), 1325–1337. doi: 10.1007/s00425-010-1258-y
Li, H. L., Zhang, L. B., Guo, D., Li, C. Z., Peng, S. Q. (2012). Identification and expression profiles of the WRKY transcription factor family in Ricinus communis. Gene. 503 (2), 248–253. doi: 10.1016/j.gene.2012.04.069
Liang, Q. Y., Wu, Y. H., Wang, K., Bai, Z. Y., Liu, Q. L., Pan, Y. Z., et al. (2017). Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic Chrysanthemum. Sci. Rep. 7 (1), 1–10. doi: 10.1038/s41598-017-05170-x
Liang, C., Zheng, G., Li, W., Wang, Y., Hu, B., Wang, H., et al. (2015). Melatonin delays leaf senescence and enhances salt stress tolerance in rice. J. Pineal Res. 59 (1), 91–101. doi: 10.1111/jpi.12243
Lin, H., Yang, Y., Quan, R., Mendoza, I., Wu, Y., Du, W., et al. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell. 21 (5), 1607–1619. doi: 10.1105/tpc.109.066217
Liu, X., Song, Y., Xing, F., Wang, N., Wen, F., Zhu, C. (2016). GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma. 253 (5), 1265–1281. doi: 10.1007/s00709-015-0885-3
Liu, J., Zhu, J.-K. (1998). A calcium sensor homolog required for plant salt tolerance. Science. 280 (5371), 1943–1945. doi: 10.1126/science.280.5371.1943
Luo, X., Li, C., He, X., Zhang, X., Zhu, L. (2020). ABA signaling is negatively regulated by GbWRKY1 through JAZ1 and ABI1 to affect salt and drought tolerance. Plant Cell Rep. 39 (2), 181–194. doi: 10.1007/s00299-019-02480-4
Lv, B., Wu, Q., Wang, A., Li, Q., Dong, Q., Yang, J., et al. (2020). A WRKY transcription factor, FtWRKY46, from Tartary buckwheat improves salt tolerance in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 147, 43–53. doi: 10.1016/j.plaphy.2019.12.004
Ma, Y., Xue, H., Zhang, F., Jiang, Q., Yang, S., Yue, P., et al. (2021). The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. Plant Biotechnol. J. 19 (2), 311–323. doi: 10.1111/pbi.13464
Majeed, A., Muhammad, Z. (2019). Salinity: a major agricultural problem—causes, impacts on crop productivity and management strategies. Plant abiotic stress tolerance: Agronomic, molecular and biotechnological approaches, 83–99.
Manchanda, G., Garg, N. (2008). Salinity and its effects on the functional biology of legumes. Acta Physiologiae Plant 30 (5), 595–618. doi: 10.1007/s11738-008-0173-3
Mangelsen, E., Kilian, J., Berendzen, K. W., Kolukisaoglu, U. H., Harter, K., Jansson, C., et al. (2008). Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genom. 9 (1), 194. doi: 10.1186/1471-2164-9-194
Mao, P., Jin, X., Bao, Q., Mei, C., Zhou, Q., Min, X., et al. (2020). WRKY transcription factors in medicago sativa L.: Genome-wide identification and expression analysis under abiotic stress. DNA Cell Biol. 39 (12), 2212–2225. doi: 10.1089/dna.2020.5726
Matsushita, A., Inoue, H., Goto, S., Nakayama, A., Sugano, S., Hayashi, N., et al. (2013). Nuclear ubiquitin proteasome degradation affects WRKY 45 function in the rice defense program. Plant J. 73 (2), 302–313. doi: 10.1111/tpj.12035
Meng, D., Li, Y., Bai, Y., Li, M., Cheng, L. (2016). Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiol. Biochem. 103, 71–83. doi: 10.1016/j.plaphy.2016.02.006
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salInity stresses. Plant Cell &Environ. 33 (4), 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Mishra, S., Bagal, D., Chowdhary, A. A., Mehrotra, S., Rai, G. K., Gandhi, S. G., et al. (2023). Signal crosstalk of phytomelatonin during salinity stress tolerance in plants. Plant Growth Regul. 101, 1–17. doi: 10.1007/s10725-023-01011-2
Mishra, S., Lhamo, S., Chowdhary, A. A., Mehrotra, S., Srivastava, V. (2021). The Na+/Ca2+ exchanger-like proteins from plants: an overview. Calcium Transport Elements in Plants, 143–155. doi: 10.1016/B978-0-12-821792-4.00008-4
Mishra, S., Srivastava, V., Mehrotra, S., Quadri, S. N. (2017). “The regulation of plant development: cross-talk of reactive oxygen species and plant hormones,” in Revisiting the role of reactive oxygen species (ros) in plants: ros boon or bane for plants (John Wiley & Sons Ltd).
Mishra, S., Triptahi, V., Singh, S., Phukan, U. J., Gupta, M. M., Shanker, K., et al. (2013). Wound induced tanscriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum. PLoS One 8 (1), e52784. doi: 10.1371/journal.pone.0052784
Mittler, R., Kim, Y., Song, L., Coutu, J., Coutu, A., Ciftci-Yilmaz, S., et al. (2006). Gain-and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580 (28-29), 6537–6542. doi: 10.1016/j.febslet.2006.11.002
Moller, I. S., Gilliham, M., Jha, D., Mayo, G. M., Roy, S. J., Coates, J. C., et al. (2009). Shoot Na+ exclusion and increased salinity tolerance engineered by cell type–specific alteration of Na+ transport in Arabidopsis. Plant Cell. 21 (7), 2163–2178. doi: 10.1105/tpc.108.064568
Ning, P., Liu, C., Kang, J., Lv, J. (2017). Genome-wide analysis of WRKY transcription factors in wheat (Triticum aestivum L.) and differential expression under water deficit condition. PeerJ 5, e3232. doi: 10.7717/peerj.3232
Niu, X., Bressan, R. A., Hasegawa, P. M., Pardo, J. M. (1995). Ion homeostasis in NaCl stress environments. Plant Physiol. 109 (3), 735. doi: 10.1104/pp.109.3.735
Niu, C. F., Wei, W., Zhou, Q. Y., Tian, A. G., Hao, Y. J., Zhang, W. K., et al. (2012). Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell &Environ. 35 (6), 1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x
Pandey, S. P., Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150 (4), 1648–1655. doi: 10.1104/pp.109.138990
Parihar, P., Singh, S., Singh, R., Singh, V. P., Prasad, S. M. (2015). Effect of salinity stress on plants and its tolerance strategies: a review. Environ. Sci. pollut. Res. 22 (6), 4056–4075. doi: 10.1007/s11356-014-3739-1
Park, H. J., Kim, W. Y., Yun, D. J. (2016). A new insight of salt stress signaling in plant. Molecules Cells 39 (6), 447. doi: 10.14348/molcells.2016.0083
Phukan, U. J., Jeena, G. S., Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7, 760. doi: 10.3389/fpls.2016.00760
Platten, J. D., Cotsaftis, O., Berthomieu, P., Bohnert, H., Davenport, R. J., Fairbairn, D. J., et al. (2006). Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 11 (8), 372–374. doi: 10.1016/j.tplants.2006.06.001
Qin, Z., Hou, F., Li, A., Dong, S., Wang, Q., Zhang, L. (2020). Transcriptome-wide identification of WRKY transcription factor and their expression profiles under salt stress in sweetpotato (Ipomoea batatas L.). Plant Biotechnol. Rep. 14 (5), 599–611. doi: 10.1007/s11816-020-00635-4
Qin, Y., Tian, Y., Liu, X. (2015). A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res.Comm. 464 (2), 428–433. doi: 10.1016/j.bbrc.2015.06.128
Quan, R., Lin, H., Mendoza, I., Zhang, Y., Cao, W., Yang, Y., et al. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell. 19 (4), 1415–1431. doi: 10.1105/tpc.106.042291
Ren, G., Yang, P., Cui, J., Gao, Y., Yin, C., Bai, Y., et al. (2022). Multiomics analyses of two sorghum cultivars reveal the molecular mechanism of salt tolerance. Front. Plant Sci. 13, 1640. doi: 10.3389/fpls.2022.886805
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., Chao, D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37 (10), 1141–1146. doi: 10.1038/ng1643
Ross, C. A., Liu, Y., Shen, Q. J. (2007). The WRKY gene family in rice (Oryza sativa). J. Int. Plant Biol. 49 (6), 827–842. doi: 10.1111/j.1744-7909.2007.00504.x
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15 (5), 247–258. doi: 10.1016/j.tplants.2010.02.006
Rushton, P. J., Torres, J. T., Parniske, M., Wernert, P., Hahlbrock, K., Somssich, I. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15 (20), 5690–5700. doi: 10.1002/j.1460-2075.1996.tb00953.x
Sanwal, S. K., Kesh, H., Kumar, A., Dubey, B. K., Khar, A., Rouphael, Y., et al. (2022b). Salt tolerance potential in onion: Confirmation through physiological and biochemical traits. Plants 11, 3325. doi: 10.3390/plants11233325
Sanwal, S. K., Kumar, P., Kesh, H., Gupta, V. K., Kumar, A., Kumar, A., et al. (2022a). Salinity stress tolerance in potato cultivars: Evidence from physiological and biochemical traits. Plants 11, 1842. doi: 10.3390/plants11141842
Schluttenhofer, C., Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167 (2), 295–306. doi: 10.1104/pp.114.251769
Serrano, R., Mulet, J. M., Rios, G., Marquez, J. A., De Larrinoa, I. F., Leube, M. P., et al. (1999). A glimpse of the mechanisms of ion homeostasis during salt stress. J. Exp. Bot. 50, 1023–1036. doi: 10.1093/jxb/50.Special_Issue.1023
Sewelam, N., Oshima, Y., Mitsuda, N., Ohme-Takagi, M. (2014). A step towards understanding plant responses to multiple environmental stresses: a genome-wide study. Plant Cell Environ. 37 (9), 2024–2035. doi: 10.1111/pce.12274
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 22, 1909–1935. doi: 10.1105/tpc.110.073874
Shen, Z., Yao, J., Sun, J., Chang, L., Wang, S., Ding, M., et al. (2015). Populus euphratica HSF binds the promoter of WRKY1 to enhance salt tolerance. Plant Science. 235, 89–100. doi: 10.1016/j.plantsci.2015.03.006
Sheng, Y., Yan, X., Huang, Y., Han, Y., Zhang, C., Ren, Y., et al. (2019). The WRKY transcription factor, WRKY 13, activates PDR8 expression to positively regulate cadmiun tolerance in Arabisopsis. Plant Cell Environment. 42 (3), 891–903. doi: 10.1111/pce.13457
Shi, W. Y., Du, Y. T., Ma, J., Min, D. H., Jin, L. G., Chen, J., et al. (2018). The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. Int. J.Mol.Sci. 19 (12), 4087. doi: 10.3390/ijms19124087
Silva Monteiro de Almeida, D., Oliveira Jordão do Amaral, D., Del-Bem, L.-E., Bronze dos Santos, E., Santana Silva, R. J., Peres Gramacho, K., et al. (2017). Genome-wide identification and characterization of cacao WRKY transcription factors and analysis of their expression in response to witches' broom disease. PLoS One 12 (10), e0187346. doi: 10.1371/journal.pone.0187346
Singh, A. K., Kumar, S. R., Dwivedi, V., Rai, A., Pal, S., Shasany, A. K., et al. (2017). A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 215 (3), 1115–1131. doi: 10.1111/nph.14663
Singh, V., Roy, S., Singh, D., Nandi, A. K. (2014). Arabidopsis flowering locus D influences systemic-acquired-resistance-induced expression and histone modifications of WRKY genes. J. Bioscienc. 39 (1), 119–126. doi: 10.1007/s12038-013-9407-7
Singh, R. P., Jha, P., Jha, P. N. (2015). The plant-growth-promoting bacterium Klebsiella sp. SBP-8 confers induced systemic tolerance in wheat (Triticum aestivum) under salt stress. J. Plant Physiol. 184, 57–67. doi: 10.1016/j.jplph.2015.07.002
Song, Y., Chen, L., Zhang, L., Yu, D. (2010). Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J. Biosci. 35 (3), 459–471. doi: 10.1007/s12038-010-0051-1
Song, Y., Li, J., Sui, Y., Han, G., Zhang, Y., Guo, S., et al. (2020). The sweet sorghum SbWRKY50 is negatively involved in salt response by regulating ion homeostasis. Plant Mol. Biol. 102 (6), 603–614. doi: 10.1007/s11103-020-00966-4
Song, H., Wang, P., Lin, J. Y., Zhao, C., Bi, Y., Wang, X. (2016). Genome-wide identification and characterization of WRKY gene family in peanut. Front. Plant Sci. 7, 534. doi: 10.3389/fpls.2016.00534
Srivastava, V., Mehrotra, S., Verma, P. K. (2017). Biotechnological interventions for production of therapeutic secondary metabolites using hairy root cultures of medicinal plants. Eds. Dubey, S. K., Pandey, A., Sangwan, R. S.. Current Developments in Biotechnology and Bioengineering, Elsevier (ISBN 9780444636614), pP259–282. doi: 10.1016/B978-0-444-63661-4.00012-8
Srivastava, V., Chowdhary, A. A., Verma, P. K., Mehrotra, S., Mishra, S. (2022a). Hydrogen sulfide-mediated mitigation and its integrated signaling crosstalk during salinity stress. Physiologia Plantarum 174 (1), e13633. doi: 10.1111/ppl.13633
Srivastava, V., Mishra, S., Chowdhary, A. A., Lhamo, S., Mehrotra, S. (2021). The γ-aminobutyric acid (GABA) towards abiotic stress tolerance. Compatible solutes Eng. Crop Plants facing Climate Change, 171–187. doi: 10.1007/978-3-030-80674-3_7
Srivastava, V., Mishra, S., Mehrotra, S., Upadhyay, S. K. (Eds.) (2022b). Plant transcription factors: Contribution in development, metabolism, and environmental stress. (Elsevier). doi: 10.1016/C2020-0-04071-5
Sytar, O., Mbarki, S., Zivcak, M., Brestic, M. (2018). “The involvement of different secondary metabolites in salinity tolerance of crops,” in Salinity responses and tolerance in plants, vol. 2. (Cham: Springer), 21–48. doi: 10.1007/978-3-319-90318-7_2
Tan, T., Cai, J., Zhan, E., Yang, Y., Zhao, J., Guo, Y., et al. (2016). Stability and localization of 14-3-3 proteins are involved in salt tolerance in Arabidopsis. Plant Mol.Biol. 92 (3), 391–400. doi: 10.1007/s11103-016-0520-5
Tripathi, P., Rabara, R. C., Langum, T. J., Boken, A. K., Rushton, D. L., Boomsma, D. D., et al. (2012). The WRKY transcription factor family in Brachypodium distachyon. BMC Genom. 13 (1), 270. doi: 10.1186/1471-2164-13-270
Tuteja, N. (2007). “Mechanisms of high salinity tolerance in plants,” Methods in enzymology (Elsevier) 428, 419–438. doi: 10.1016/S0076-6879(07)28024-3
Ushakova, S., Kovaleva, N., Gribovskaya, I., Dolgushev, V., Tikhomirova, N. (2005). Effect of NaCl concentration on productivity and mineral composition of Salicornia europaea as a potential crop for utilization NaCl in LSS. Adv. Space Res. 36 (7), 1349–1353. doi: 10.1016/j.asr.2004.09.017
Wang, J. N., Kuang, J. F., Shan, W., Chen, J., Xie, H., Lu, W. J., et al. (2012). Expression profiles of a banana fruit linker histone H1 gene MaHIS1 and its interaction with a WRKY transcription factor. Plant Cell Rep. 31 (8), 1485–1494. doi: 10.1007/s00299-012-1263-7
Wang, C. T., Ru, J. N., Liu, Y. W., Li, M., Zhao, D., Yang, J. F., et al. (2018). Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. M. Sci. 19 (10), 3046. doi: 10.3390/ijms19103046
Wang, N., Xia, E. H., Gao, L. Z. (2016). Genome-wide analysis of WRKY family of transcription factors in common bean, Phaseolus vulgaris: chromosomal localization, structure, evolution and expression divergence. Plant Gene. 5, 22–30. doi: 10.1016/j.plgene.2015.11.003
Wang, L., Zhu, W., Fang, L., Sun, X., Su, L., Liang, Z., et al. (2014). Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 4 (1), 1–14. doi: 10.1186/1471-2229-14-103
Wani, S.H., Anand, S., Singh, B., Bohra, A., Joshi, R. (2021). WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 40, 1071–1085.
Wei, K. F., Chen, J., Chen, Y. F., Wu, L. J., Xie, D. X. (2012). Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 19 (2), 153–164. doi: 10.1093/dnares/dsr048
Wei, W., Li, Q. T., Chu, Y. N., Reiter, R. J., Yu, X. M., Zhu, D. H., et al. (2015). Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J.Exp. Bot. 66 (3), 695–707. doi: 10.1093/jxb/eru392
Wei, Y., Shi, H., Xia, Z., Tie, W., Ding, Z., Yan, Y., et al. (2016). Genome-wide identification and expression analysis of the WRKY gene family in cassava. Front. Plant Sci. 7, 25. doi: 10.3389/fpls.2016.00025
Wu, K. L., Guo, Z. J., Wang, H. H., Li, J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26. doi: 10.1093/dnares/12.1.9
Wu, G. Q., Li, Z. Q., Cao, H., Wang, J. L. (2019). Genome-wide identification and expression analysis of the WRKY genes in sugar beet (Beta vulgaris L.) under alkaline stress. PeerJ 7, e27810v1. doi: 10.7287/peerj.preprints.27810v1
Wu, Z. J., Li, X. H., Liu, Z. W., Li, H., Wang, Y. X., Zhuang, J. (2016). Transcriptome-wide identification of Camellia sinensis WRKY transcription factors in response to temperature stress. Mol. Genet. Genom. 291 (1), 255–269. doi: 10.1007/s00438-015-1107-6
Wu, M., Liu, H., Han, G., Cai, R., Pan, F., Xiang, Y. (2017). A moso bamboo WRKY gene PeWRKY83 confers salinity tolerance in transgenic Arabidopsis plants. Sci. Repo. 7 (1), 1–16. doi: 10.1038/s41598-017-10795-z
Xiang, X. Y., Chen, J., Xu, W. X., Qiu, J. R., Song, L., Wang, J. T., et al. (2021). Dehydration-induced WRKY transcriptional factor mfWRKY70 of myrothamnus flabellifolia enhanced drought and salinity tolerance in arabidopsis. Biomolecules. 11 (2), 327. doi: 10.3390/biom11020327
Xie, Y., Ling, T., Han, Y., Liu, K., Zheng, Q., Huang, L., et al. (2008). Carbon monoxide enhances salt tolerance by nitric oxide-mediated maintenance of ion homeostasis and up-regulation of antioxidant defence in wheat seedling roots. Plant Cell &Environ. 31 (12), 1864–1881. doi: 10.1111/j.1365-3040.2008.01888.x
Xu, Z., Raza, Q., Xu, L., He, X., Huang, Y., Yi, J., et al. (2018). Gm WRKY49 a salt responsive nuclear protein, improved root length and governed better salinity tolerance in transgenic Arabidopsis. Front. Plant Sci. 9, 809. doi: 10.3389/fpls.2018.00809
Yan, H., Jia, H., Chen, X., Hao, L., An, H., Guo, X. (2014). The cotton WRKY transcription factorGhWRKY 17 functions in drought and stress in transgenic Nicotiana benthamiana through ABA signalling and modulation of reactive oxygen species production. Plant Cell Physiol. 55 (12), 2060–2076. doi: 10.1093/pcp/pcu133
Yang, Z., Chi, X., Guo, F., Jin, X., Luo, H., Hawar, A., et al. (2020). SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J. Plant Physiol. 246, 153142. doi: 10.1016/j.jplph.2020.153142
Yang, Y., Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217 (2), 523–539. doi: 10.1111/nph.14920
Yang, Y., Qin, Y., Xie, C., Zhao, F., Zhao, J., Liu, D., et al. (2010). The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell. 22 (4), 1313–1332. doi: 10.1105/tpc.109.069609
Yao, D. M., Zou, C., Shu, Y. N., Liu, S. S. (2021). WRKY Transcription Factors in Nicotiana tabacum Modulate Plant Immunity against Whitefly via Interacting with MAPK Cascade Pathways. Insects. 12 (1), 16. doi: 10.3390/insects12010016
Ye, H., Qiao, L., Guo, H., Guo, L., Ren, F., Bai, J., et al. (2021). Genome-wide identification of wheat WRKY gene family reveals that TaWRKY75-A is referred to drought and salt resistances. Front. Plant Sci. 12, 812. doi: 10.3389/fpls.2021.663118
Yin, G., Xu, H., Xiao, S., Qin, Y., Li, Y., Yan, Y., et al. (2013). The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMCPlant Biol. 13 (1), 1–19. doi: 10.1186/1471-2229-13-148
Yoon, Y., Seo, D. H., Shin, H., Kim, H. J., Kim, C. M., Jang, G. (2020). The Role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy 10 (6), 788. doi: 10.3390/agronomy10060788
Yu, Y., Liu, Z., Wang, L., Kim, S. G., Seo, P. J., Qiao, M., et al. (2016b). WRKY 71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 85 (1), 96–106. doi: 10.1111/tpj.13092
Yu, L., Nie, J., Cao, C., Jin, Y., Yan, M., Wang, F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188 (3), 762–773. doi: 10.1111/j.1469-8137.2010.03422.x
Yu, Y., Wang, L., Chen, J., Liu, Z., Park, C. M., Xiang, F. (2018). WRKY71 acts antagonistically against salt-delayed flowering in Arabidopsis thaliana. Plant Cell Physiol. 59 (2), 414–422. doi: 10.1093/pcp/pcx201
Yu, Y., Wang, N., Hu, R., Xiang, F. (2016a). Genome-wide identification of soybean WRKY transcription factors in response to salt stress. Springerplus 5 (1), 920. doi: 10.1186/s40064-016-2647-x
Zhang, L., Chen, C., Xie, F., Hua, Q., Zhang, Z., Zhang, R., et al. (2021). A Novel WRKY Transcription Factor HmoWRKY40 Associated with Betalain Biosynthesis in Pitaya (Hylocereus monacanthus) through Regulating HmoCYP76AD1. Intern.J.Mol. Sci. 22 (4), 2171. doi: 10.3390/ijms22042171
Zhang, H., Jin, J., Tang, L., Zhao, Y., Gu, X., Gao, G., et al. (2011b). PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 39 (suppl_1), D1114–D1117. doi: 10.1093/nar/gkq1141
Zhang, H., Li, W., Mao, X., Jing, R., Jia, H. (2016a). Differential activation of the wheat SnRK2 family by abiotic stresses. Front. Plant Sci. 7, 420. doi: 10.3389/fpls.2016.00420
Zhang, H., Mao, X., Jing, R., Chang, X., Xie, H. (2011a). Characterization of a common wheat (Triticum aestivum L.) TaSnRK2. 7 gene involved in abiotic stress responses. J. Exp. Bot. 62 (3), 975–988. doi: 10.1093/jxb/erq328
Zhang, L., Wan, X., Xu, Y., Niyitanga, S., Qi, J., Zhang, L. (2020). De novo assembly of transcriptome and genome-wide identification reveal GA 3 stress-responsive WRKY transcription factors involved in fiber formation in jute (Corchorus capsularis). BMC Plant Bio. 20 (1), 1–15. doi: 10.1186/s12870-020-02617-8
Zhang, Y., Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC evolutionary Biol. 5 (1), 1–12. doi: 10.1186/1471-2148-5-1
Zhang, C., Wang, D., Yang, C., Kong, N., Shi, Z., Zhao, P., et al. (2017). Genome-wide identification of the potato WRKY transcription factor family. PLoS One 12 (7), e0181573. doi: 10.1371/journal.pone.0181573
Zhang, Y., Yu, H., Yang, X., Li, Q., Ling, J., Wang, H., et al. (2016b). CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physio. Biochem. 108, 478–487. doi: 10.1016/j.plaphy.2016.08.013
Zhao, K., Zhang, D., Lv, K., Zhang, X., Cheng, Z., Li, R., et al. (2019). Functional characterization of poplar WRKY75 in salt and osmotic tolerance. Plant Sci. 289, 110259. doi: 10.1016/j.plantsci.2019.110259
Zheng, Y., Ge, J., Bao, C., Chang, W., Liu, J., Shao, J., et al. (2020). Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response. Mol. Plan.t 13 (4), 598–611. doi: 10.1016/j.molp.2019.12.011
Zhou, H., Lin, H., Chen, S., Becker, K., Yang, Y., Zhao, J., et al. (2014). Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell. 26 (3), 1166–1182. doi: 10.1105/tpc.113.117069
Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biot.J. 6 (5), 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Zhu, J. K. (2001). Plant salt tolerance. Trends Plant Sci. 6 (2), 66–71. doi: 10.1016/S1360-1385(00)01838-0
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell. 167 (2), 313–324. doi: 10.1016/j.cell.2016.08.029
Zhu, D., Hou, L., Xiao, P., Guo, Y., Deyholos, M. K., Liu, X. (2019). VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant science. 280, 132–142. doi: 10.1016/j.plantsci.2018.03.018
Zhu, H., Jiang, Y., Guo, Y., Huang, J., Zhou, M., Tang, Y., et al. (2021). A novel salt inducible WRKY transcription factor gene, AhWRKY75, confers salt tolerance in transgenic peanut. PlantPhysiology Biochem. 160, 175–183. doi: 10.1016/j.plaphy.2021.01.014
Zhu, X., Liu, S., Meng, C., Qin, L., Kong, L., Xia, G. (2013). WRKY transcription factors in wheat and their induction by biotic and abiotic stress. Plant Mol. Bio. Rep. 31 (5), 1053–1067. doi: 10.1007/s11105-013-0565-4
Zhu, Z., Shi, J., Cao, J., He, M., Wang, Y. (2012). VpWRKY3, a biotic and abiotic stress-related transcription factor from the Chinese wild Vitis pseudoreticulata. Plant Cell Rep. 31 (11), 2109–2120. doi: 10.1007/s00299-012-1321-1
Zhu, H., Zhou, Y., Zhai, H., He, S., Zhao, N., Liu, Q. (2020). A novel sweetpotato WRKY transcription factor, IbWRKY2, positively regulates drought and salt tolerance in transgenic Arabidopsis. Biomolecules. 10 (4), 506. doi: 10.3390/biom10040506
Keywords: abiotic stress, ABA signaling, transcription factors, food security, ROS, SOS pathway
Citation: Rai GK, Mishra S, Chouhan R, Mushtaq M, Chowdhary AA, Rai PK, Kumar RR, Kumar P, Perez-Alfocea F, Colla G, Cardarelli M, Srivastava V and Gandhi SG (2023) Plant salinity stress, sensing, and its mitigation through WRKY. Front. Plant Sci. 14:1238507. doi: 10.3389/fpls.2023.1238507
Received: 11 June 2023; Accepted: 31 August 2023;
Published: 04 October 2023.
Edited by:
Sushil Satish Chhapekar, University of Missouri, United StatesReviewed by:
Sheikh Mansoor, Jeju National University, Republic of KoreaFoad Fatehi, Payame Noor University, Iran
Viswanathan Satheesh, Iowa State University, United States
Copyright © 2023 Rai, Mishra, Chouhan, Mushtaq, Chowdhary, Rai, Kumar, Kumar, Perez-Alfocea, Colla, Cardarelli, Srivastava and Gandhi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gyanendra Kumar Rai, Z2tyYWk3NUBnbWFpbC5jb20=; Vikas Srivastava, dmlrYXNzcml2YXN0YXZhMjVAZ21haWwuY29t, dmlrYXMuYm90QGN1amFtbXUuYWMuaW4=; Mariateresa Cardarelli, dGNhcmRhcmVAdW5pdHVzLml0; Sumit G. Gandhi, c3VtaXRAaWlpbS5yZXMuaW4=
†These authors have contributed equally to this work and share first authorship
 Gyanendra Kumar Rai
Gyanendra Kumar Rai Sonal Mishra
Sonal Mishra Rekha Chouhan
Rekha Chouhan Muntazir Mushtaq
Muntazir Mushtaq Aksar Ali Chowdhary
Aksar Ali Chowdhary Pradeep K. Rai
Pradeep K. Rai Ranjeet Ranjan Kumar
Ranjeet Ranjan Kumar Pradeep Kumar
Pradeep Kumar Francisco Perez-Alfocea
Francisco Perez-Alfocea Giuseppe Colla
Giuseppe Colla Mariateresa Cardarelli
Mariateresa Cardarelli Vikas Srivastava
Vikas Srivastava Sumit G. Gandhi
Sumit G. Gandhi