- 1Department of Botany, Hansraj College, University of Delhi, Delhi, India
- 2Excellent Team for Mitigation (ETM), Faculty of Forestry and Wood Sciences, Czech University of Life Sciences Prague, Prague, Czechia
- 3Department of Zoology, Deshbandhu College, University of Delhi, New Delhi, India
- 4Jagdish Chandra Bose Center for Plant Genomics, Hansraj College, University of Delhi, Delhi, India
- 5Delhi School of Climate Change and Sustainability, Institution of Eminence, Maharishi Karnad Bhawan, University of Delhi, Delhi, India
Biotic stresses threaten to destabilize global food security and cause major losses to crop yield worldwide. In response to pest and pathogen attacks, plants trigger many adaptive cellular, morphological, physiological, and metabolic changes. One of the crucial stress-induced adaptive responses is the synthesis and accumulation of plant secondary metabolites (PSMs). PSMs mitigate the adverse effects of stress by maintaining the normal physiological and metabolic functioning of the plants, thereby providing stress tolerance. This differential production of PSMs is tightly orchestrated by master regulatory elements, Transcription factors (TFs) express differentially or undergo transcriptional and translational modifications during stress conditions and influence the production of PSMs. Amongst others, microRNAs, a class of small, non-coding RNA molecules that regulate gene expression post-transcriptionally, also play a vital role in controlling the expression of many such TFs. The present review summarizes the role of stress-inducible TFs in synthesizing and accumulating secondary metabolites and also highlights how miRNAs fine-tune the differential expression of various stress-responsive transcription factors during biotic stress.
1 Introduction
Being sedentary, plants are continually defied by various environmental stresses, orchestrated into declination of their growth and yield. Biotic stresses include infection by various organisms such as bacteria, fungi, nematodes, viruses, and herbivory by insect pests. Plants continue to live with these stresses, decreasing productivity and yield loss (approximately 35% loss due to biotic stress) (Foyer et al., 2016). During 2020, due to decreased plant productivity, the United Nations (UN) has decided to acknowledge the year as an International year of plant health to motivate research in the area of plant science (Roux and Ham, 2020). It has also been estimated that food production will have to be increased by approximately 60% to feed the growing population of estimated 10 billion people. The primary reason for this loss in yield is increased pathogens and pests encountering crop plants due to abrupt climate change Jha et al., 2022. Plants have progressed numerous responses to shield themselves and counter-attacks of far-ranging pests and pathogens (Brading et al., 2000). Thus, plants tend to foray into a balance in their response and existential methods against biotic stress, which show deleterious effects (Iqbal et al., 2021; Kumar et al., 2020). The plant defensive response affecting the mechanisms at the molecular level has been explained profoundly (Cheng et al., 2012; Wang and Wang, 2019). However, the reasons for this diversity in metabolite production during biotic stress are not abundant.
An array of diverse metabolites is produced by plants, which can be either vital (primary metabolites) or non-vital (secondary or specialized metabolites), affecting the fundamental processes of growth and development (Obata, 2019). Plant Secondary metabolites (PSMs) are multifunctional metabolites, characteristically intricate in plant defenses, and environmental communication, especially during stress response. Plants’ diversified machinery for plant defense not only allow them to survive against stressors but also influence PSM accumulation (Pagare et al., 2015). The Biosynthesis of PSMs in response to stressful environments is controlled at the transcriptome level by numerous genes and transcription factors (TFs). TFs are sequence-specific DNA binding proteins that identify and bind specifically to the cis-regulatory sequences of the promoter regions of the targeted genes which may activate or repress their expression levels in response to developmental and other environmental cues (Patra et al., 2013). TFs in plants encode up to 10% of the total genes at diverse stages, thereby regulating signal-mediated gene expression. TF families, namely WRKY, MYB, NAC, and AP2/ERF, are crucial regulators of various genes, which contribute to the model choice for genetic engineering to boost the immunity of plants against diverse stress stipulations (Baillo et al., 2019).
During pest and pathogenic attacks, the concentration of PSMs alters due to the differential activity of TFs. miRNAs are one of the prime regulators which control the activity of TFs at the post-transcriptional level. Small, non-coding riboregulators known as miRNAs (20–24 nucleotides) also regulate eukaryotic gene expression through base pairing the complementary mRNA. The miRNA genes are transcribed by RNA polymerase II to produce an imperfect hairpin structure called pri-miRNAs. These pri-miRNAs are further processed by DICER-LIKE 1 (DCL1) to generate 70-100 nucleotides long hairpin structures called pre-miRNAs. The same DCL1 converts pre-miRNAs to mature miRNAs following their loading into the RISC complex, these mature miRNAs bind to mRNAs to cleave and silence the genes (Jones-Rhoades et al., 2006). miRNAs are recognized not only to regulate various plant processes related to their growth and development but also to control biosynthesis and accumulation of secondary metabolites during biotic stress (Bulgakov and Avramenko, 2015).
The investigation on the role of miRNA’s in regulation of transcription factors during biotic and abiotic stress has been extensively studied (Javed et al., 2020; Šečić et al., 2021). There are reported studies of various transcription factors involved in the regulation of synthesis and accumulation of PSMs (Pagano et al., 2021; Jan et al., 2021b). However, in this article, we have updated knowledge of the present understanding of TFs responsible for regulating plant secondary metabolites on biotic stress. We have also emphasized how these TFs are regulated by regulators such as miRNAs, which are directing gene expression at the post-transcriptional and translational levels. The production of secondary metabolites by maneuvering the role of miRNA in various economically important crops may improve the profitability of agriculture and food industries.
2 Plant secondary metabolites
Plants produce thousands of specialized organic compounds, divided conventionally into two broad forms, i.e., primary and secondary metabolites (Obata, 2019). Primary metabolites portray a significant role in growth and development. In contrast, secondary metabolites are structurally diverse compounds that have been reported to be involved in plant defense (Pagare et al., 2015). They are known to contribute to the survival and health of plants and play a pivotal role in protecting them against abiotic (UV light, temperature, heavy metals, drought, salinity, etc.) as well as biotic stresses (herbivores, phytopathogens, fungi, bacteria, etc.) (Tiwari and Rana, 2015). It has been well-established that they show pharmaceutical properties (Verpoorte, 1998). Besides, it has also been reported that PSMs have been used commercially as agrochemicals in flavoring, fragrance, biopesticides, dyes, and food additives.
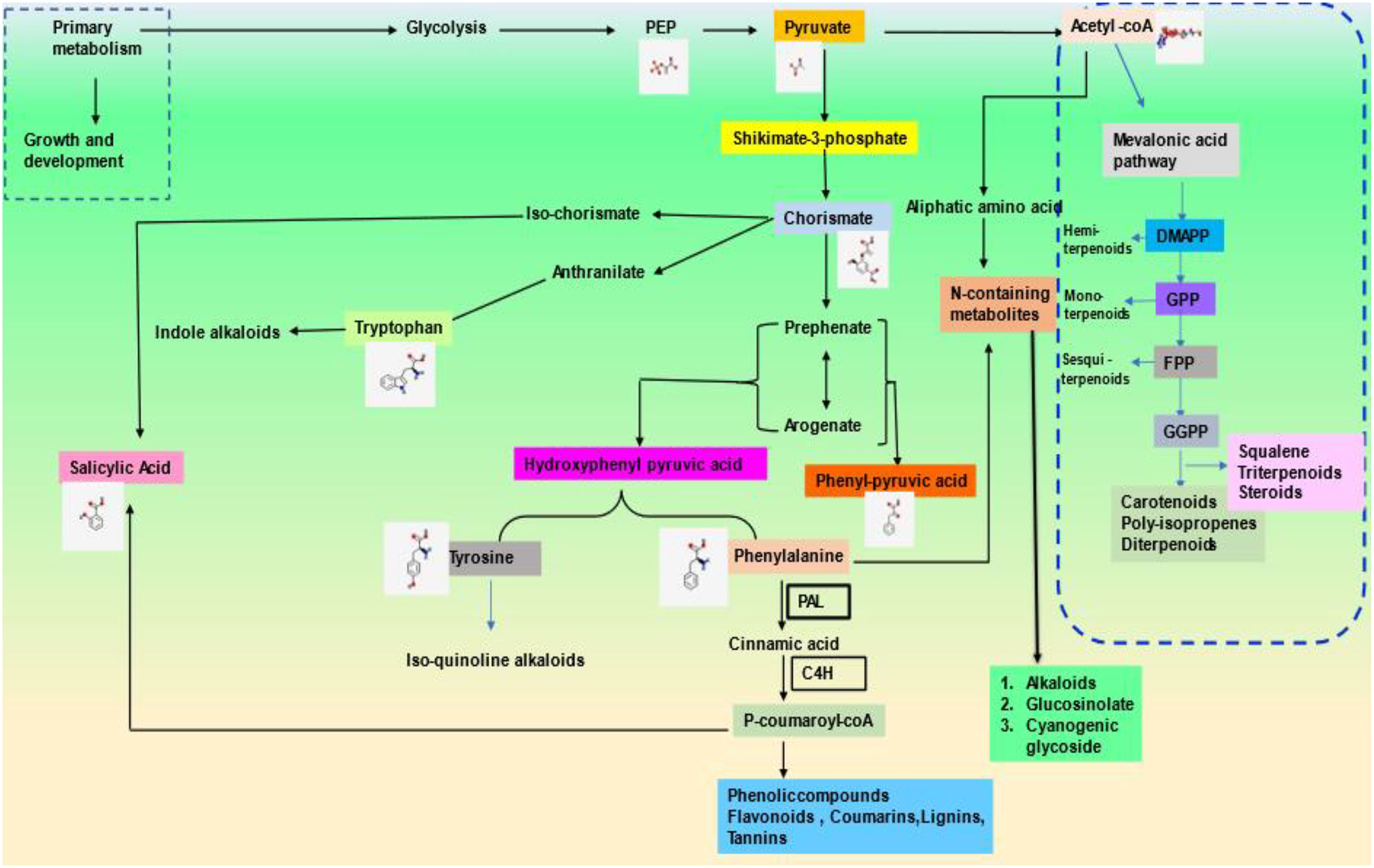
According to the studies on biochemical structure composition, PSMs have been broadly classified into two sets: nitrogen-containing (majorly alkaloids) and non-nitrogen-containing molecules (terpenes and phenols) (Patra et al., 2013). Plant-specialized metabolites (PSM) are chemically diverse in nature. The precursors for their biosynthesis are generated by either glycolysis or shikimic acid pathways, eventually leading to diversification based on cell types and environmental factors (Costa et al., 2012). PSMs have a much-limited presence in the plant kingdom. It has been noticed that they are recurrently found either in only one plant species or a taxonomically related genus (Jamwal et al., 2018). More than 100,000 SMs are produced in plants through different pathways, and their quality and quantity are mostly influenced by temperature and other biotic and abiotic factors (Meena et al., 2017).
Terpenes are the most diverse known PSMs synthesized from acetyl-CoA and glycolytic intermediates. They are formed by the fusion of 5 carbon units named isoprene and have a branched backbone. Terpenes consist of monoterpenes, diterpenes, sesquiterpene, triterpenes, tetraterpenes, and polyterpenes, which have significant importance in defense against pathogens and herbivory. Its biosynthesis includes two major pathways: the mevalonic acid pathway & methylerythritol phosphate pathway (MEP Pathway), which occur in plastids and are known to produce both isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), respectively, which are the basic unit of terpene synthesis (Figure 1) (Khare et al., 2020). Some terpenes are identified as hormones (e.g., gibberellins, a diterpene; brassinosteroids, triterpenes) and have crucial roles in growth and development. Terpenes, such as limonene and menthol, defend against herbivores (Ahmed et al., 2017; Lin et al., 2017). Another example is abietic acid, a class of diterpenes obtained from leguminous plants and pine trees that exhibit an antipathogenic effect (Lewis et al., 1997).
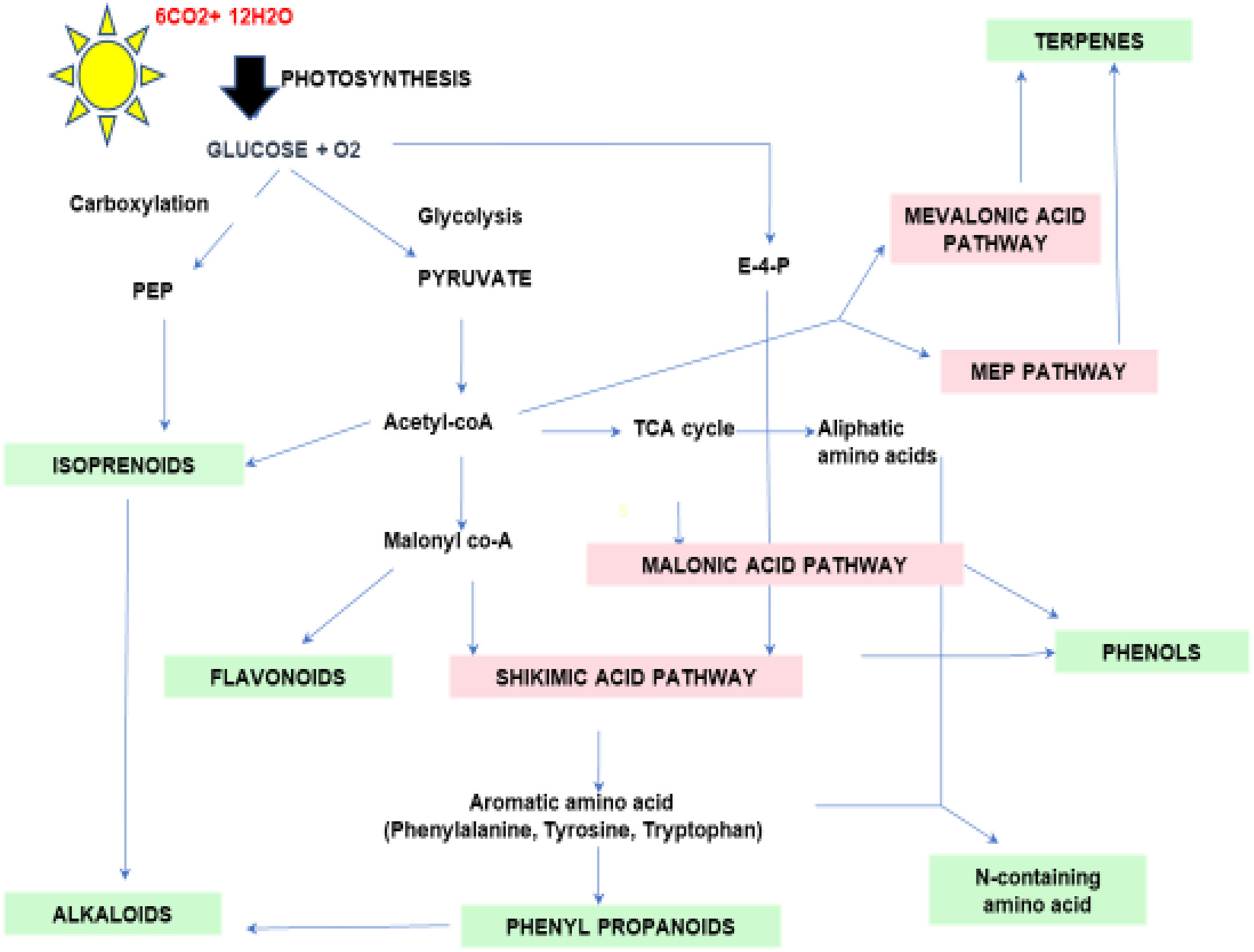
Figure 1 Schematic diagram of secondary metabolic biosynthesis pathway. Phosphenol pyruvate (PEP), Erythrose-4-phosphate (E-4-P), and Tricarboxylic acid (TCA) cycle.
Phenolics are ubiquitous compounds with a hydroxyl group attached to an aromatic ring. It can be simple or complex; simple phenolics such as, (gallic acid, caffeic acid, etc.) and polyphenols (stilbenes, flavonoids, etc.) have several curative properties (Jan et al., 2021b). They have significantly evolved the modern medicine system, be it cancer or major diseases by showing properties like anti-inflammation, anti-cancerous, and many more. Besides, it also shows antioxidative properties against oxidative damage in plants due to harmful UV rays. In addition, the plant’s pigmentation of flowers and leaves is due to a complex phenol molecule named anthocyanin. Flavonoids, however, play an essential role in pollination to attract pollinators in exchange for rewards such as nectar and seed dispersal (Winkel, 2004; Crozier et al., 2006; Butelli et al., 2008). There are two major biosynthetic pathways: shikimic acid and malonic acid pathway, where phenylalanine ammonia-lyase (PAL) and chalcone synthase (CHS) are the key enzymes involved in the phenolic compound synthesis, which is produced under various stress conditions (Khare et al., 2020) (Figure 2). Flavonoids are the most common class of secondary metabolites that can be found in edible plant parts like grains, fruits, and vegetables. The phenylpropanoid pathway is the synthesis pathway and are derivatives of 2-phenylbenzyl-pyrone (Roy et al., 2022; Tajammal et al., 2022). Among flavonoids, luteolin, apigenin, quercetin, kaempferol, and many more have been isolated and reported to exhibit defense against pathogens, fungi, and other biotic stress (Kumar and Pandey, 2013; Li et al., 2019; Hassani et al., 2020).
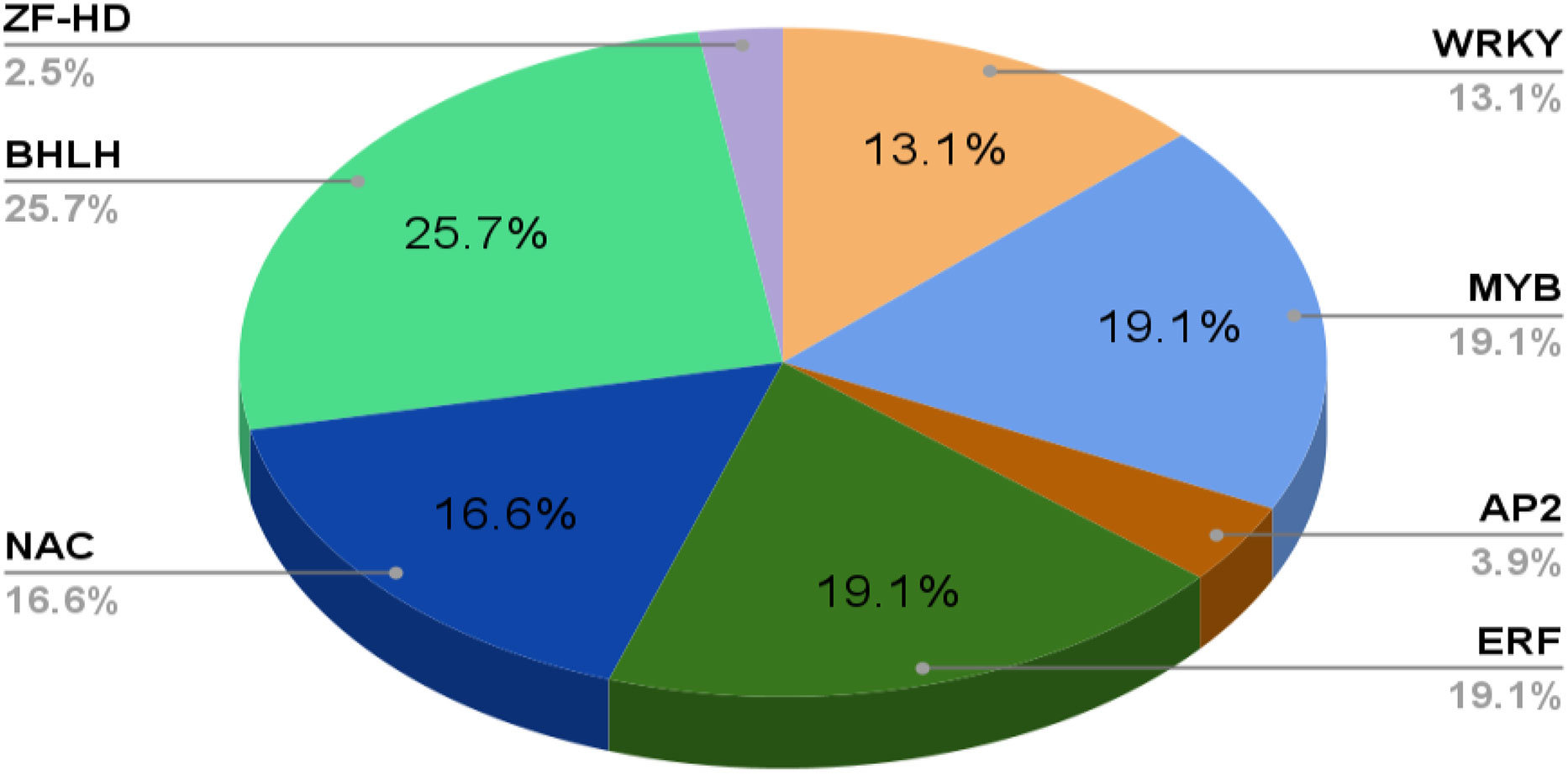
Figure 2 Schematic representation of the general pathways leading to various pathways of secondary metabolites biosynthesis such as phenols, terpenoids and N- containing secondary meabolites. PEP: Phosphoenol pyruvate, DMAPP : Dimethylallyl pyrophosphate, GPP: Geranyl pyrophosphate, FPP: Farnesyl pyrophosphate, GGPP: Geranylgeranyl diphosphate PAL : Phenylalanine ammonia-lyase, C4H:cinnamate-4-hydroxylase.
Nitrogen-containing PSMs contain nitrogen molecules in the structure, and precursors include amino acids such as lysine, tyrosine, tryptophan, etc. It mainly includes alkaloids, cyanogenic glucosides & non-protein amino acids. Alkaloids such as morphine, berberine, vinblastine, and scopolamine have pharmaceutical properties, whereas cocaine, caffeine, and nicotine have sedative and stimulant properties. Most alkaloids are toxic, such as pyrrolizidine alkaloids (PAs) help in defense against microbial infection and herbivory. For example, monocrotaline obtained from Crotalaria, pyrrolizidine from Panax quinquefolius, and senecionine from Senecio jacobaea is used as antiherbivore (Irmer et al., 2015; Graser and Hartmann 2000; Stella et al., 2018). Non-protein amino acids, however, do not incorporate into proteins; besides, they move freely, and acts as a protective defense in plants such as mimosine, citrulline, pipecolic acid, and canavanine obtained from Mimosa, Cucumis, Calliandra, and legumes respectively against various kind of stresses such as herbivory and pathogen (Yokota et al., 2002; Moulin et al., 2006; Mazis et al., 2011; Emendack et al., 2018). Cyanogenic glucosides are part of glycosides that break down to produce volatile poisonous substances like HCN that deter feeding by insects and other herbivores. Various plant families possess cyanogenic glucosides viz., Gramineae, Rosaceae, and Leguminosae. For example, amygdalin, dhurrin, linamarin, and lotaustralin extracted from Prunus sp, Sorghum, and Lotus japonicum has been premeditated for their role against drought and as antiherbivore (Mazid et al., 2011; Lai et al., 2015; Emendack et al., 2018; Yadav et al., 2022).
Sulfur-containing secondary metabolites are trivial groups of PSMs that include around 200 compounds. Sulfur-containing PSMs include glucosinolates (GSL), glutathione, glycosphingolipid, phytoalexins, allinin, thionins, and defensins (Venditti and Bianco, 2020). They are directly linked with the defense against microbial pathogens. They are known for their diverse biochemical structures and means of action, which have been reported to give plants a versatile array of chemical defenses to counter various potential enemies (Khare et al., 2020). Glucosinolates are N and S-containing glucosides that are active against unfavorable predators, competitors, and parasites (Bloem et al., 2007). Glucoraphanin, a glucosinolate from Brassica oleracea, is used majorly against UV stress (Coleto et al., 2017). Phytoalexins have antimicrobial activities that activate the defense system against these pathogens (Ziegler and Facchini, 2008). For example, brassinin, wasalexins, and camalexin obtained from higher plants show defense against pests and pathogens (Thomma et al., 2002; Abdalla and Mühling, 2019; Rodriguez-Salazar et al., 2017).
PSMs production is, however, not only limited to biotic stress rather an extensive production of PSMs have also been observed during abiotic stress. Drought, flood, and extreme temperature along with heavy metals, and chemicals have a much serious impact on plants (Jha et al., 2019). Solanum lycopersicum has been reported to show high activity of phytol (a diterpenoid) upon heat stress (Spicher et al., 2017). Further, other unfavorable condition like cold, is known to slow down the major chemical reactions in plants and under severe circumstances often lead to the death of plants. Pinoresinol, a lignin helps in alleviation during cold stress in almost all land plants (Griffith and Yaish, 2004). Similarly, drought or extreme salinity has reported drastic effects on both physiological and chemical processes (Aftab, 2019, Jan et al., 2021b). Thus, a huge literature supports the effect of abiotic stress on secondary metabolite production.
Since ancient times, the collection of secondary metabolites synthesized from diverse plants has been employed as medicinal ingredients to treat human illnesses. Certain plant species have been discovered to contain a range of active metabolites that participate in the treatment of multiple chronic human diseases, notably cancer, cardiovascular issues, and others. Because they possess antibacterial, antifeedant, and parasiticidal capabilities, as well as being less harmful and inexpensive. Hypericum perforatum is employed for its anti-depressant, anti-cancer, anti-inflammatory, anti-viral, and anti-bacterial qualities. This plant exhibits antidepressant drugs including fluoxetine and sertraline as well as other metabolites such as flavonoids, hyperforin, hypericin, and xanthones that increase its medical potential (Shakya, 2016). The Ipomoea batata L. is a popular food all over the world. Because it contains so many vitamins and phytochemicals, it has several positive impacts on human health. These phytochemicals have anti-cancer, anti-diabetic, anti-inflammatory, and antioxidant properties. Additionally, beta-carotene, a precursor of vitamin A that aids in the treatment of night blindness and vitamin A insufficiency, is present in sweet potatoes (Ghasemzadeh et al., 2016). There are several secondary metabolites in Caparis spinosa that contribute to enhancing biomarkers for diabetes and cardiovascular disease (Zhang and Ma, 2018). The use of secondary metabolites in contemporary medicine may open up new avenues for research into the identification and isolation of the desired pharmacologically active lead chemical in the drug discovery process.
3 Transcription factors regulating the biosynthesis of PSMs
Transcription factors (TFs) are class of proteins majorly responsible for regulating gene expression by binding to specific cis-acting elements in the promoter region of target genes. Generally, it has two domains, a DNA binding domain (DBD) and transcriptional activation or repression domain, known to regulate varied cellular processes. mRNA, which comprises 6% of the total genome, is considered the central point of control. In Arabidopsis thaliana, over 1600 TFs have been isolated and studied in detail (Gong et al., 2004). Based on the DBD, transcription factors have been named WRKY, bHLH, MYC, MYB, NAC, AP2/ERF, bZIP, and others.
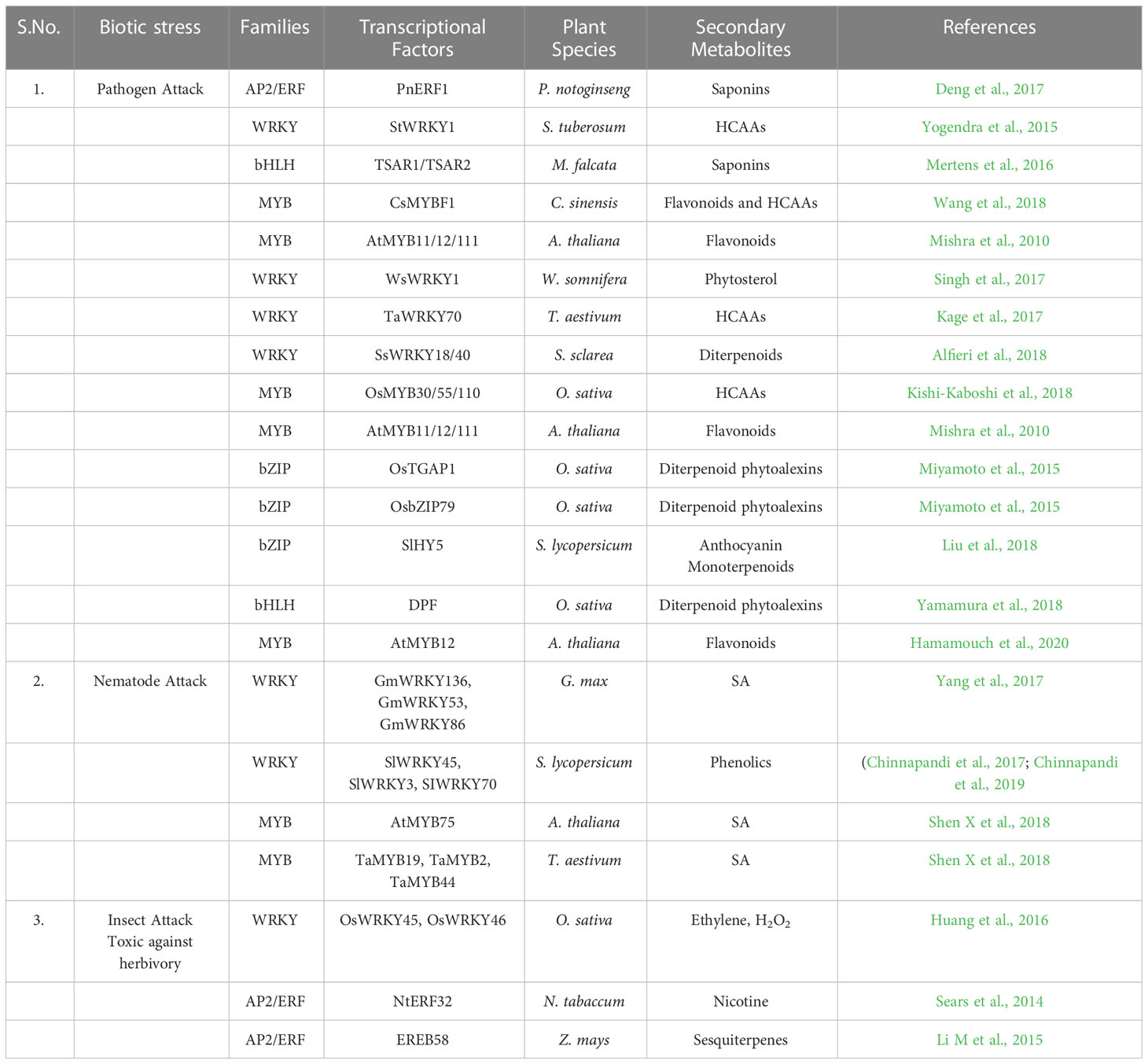
TFs have been categorized into various families and superfamilies, which seem to have a distinct role in regulating the stress response, i.e., both abiotic and biotic, a few of which are unique in plants. According to the information on specialized databases such as the Transcription factor database (TFDB), the plants contain more than 2000 genes encoding TFs in the genome. It has been reported in the literature that there are more than 63 families of TFs and about 22 regulators present (Pérez-Rodríguez et al., 2010). Out of these, there are majorly nine Transcription factor families reported recently which have a significant regulatory role in stress tolerance in plants (Chacón-Cerdas et al., 2020) which are identified as ARF, AP2/ERF-ERF, AP2/ERF-DREB, bZIP, BELL, TCP, NAC, WRYK, and ZFP (Table 1; Figure 3).
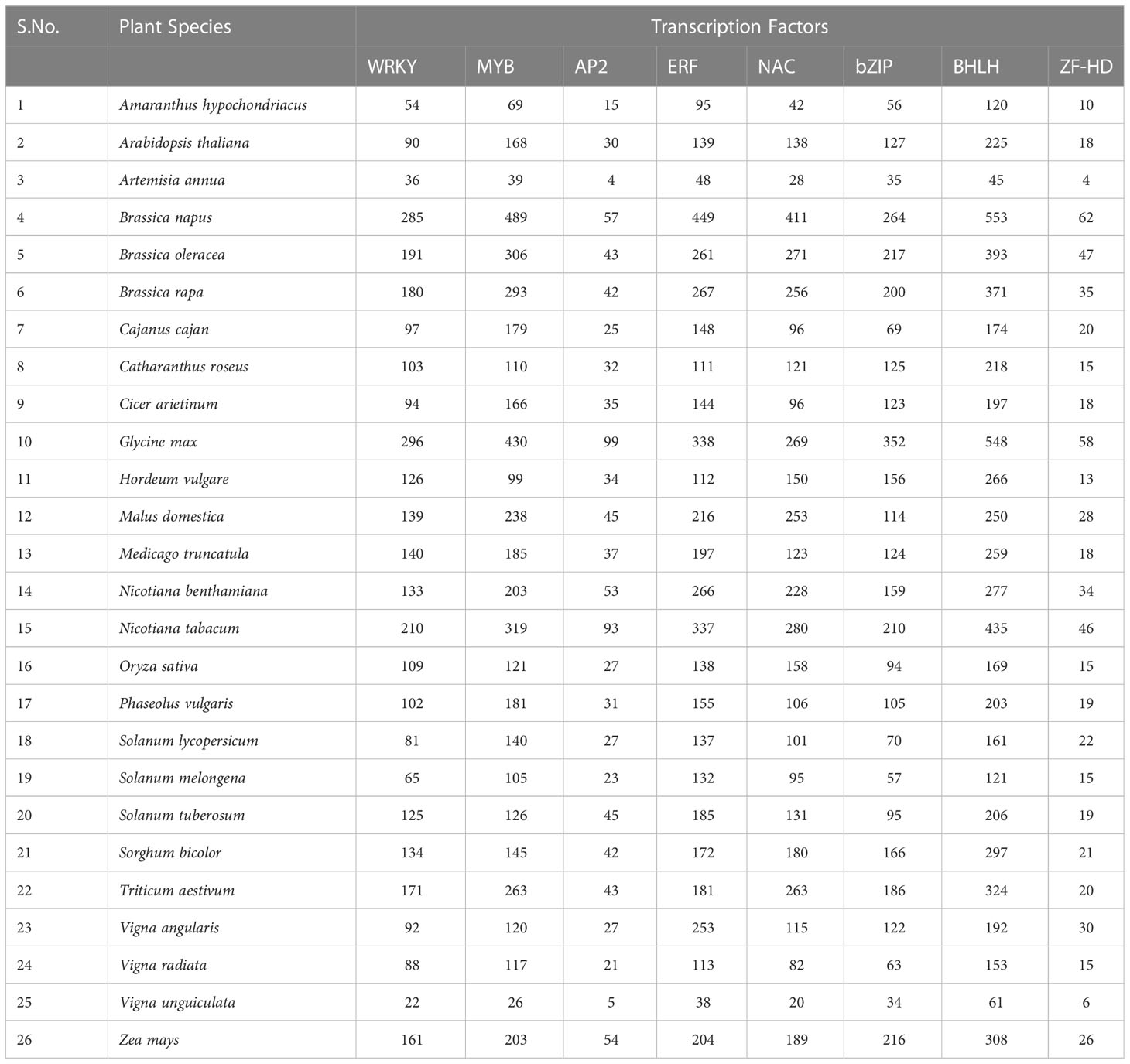
Table 1 Number of Genes encoding for Transcription factors involved in plant defense (Summarized from Transcription Factor Database, 2022).
This review focuses on the role of six TFs, namely, WRKY, NAC, AP2/ERF, MYB, bZIP, and bHLH, that are majorly intricated in regulating and synthesizing secondary metabolites under biotic stress in economically important crops. A few reviews have suggested the diverse role of TFs in regulating plant defense, but there has been a meager understanding of the role of TFs in biosynthesis and regulation of PSMs under biotic stress and how miRNAs regulate them. Therefore, this review has been planned to focus on transcription factors involved in PSMs biosynthesis during plant defense and their regulation by miRNAs in both model plants and economically important crops.
3.1 WRKY TF
WRKY is the foremost characterized class of plant TF, which regulates various plant processes related to development, physiology, metabolism and plant defense (Chen et al., 2017). From the discovery of WRKY in Ipomea batata in the 1990s to the diversified presence in many of the eukaryotic organisms, be it fungi, amoebae, or higher eukaryotic plants (Ülker and Somssich, 2004; Yamasaki et al., 2005; Zhang and Wang, 2005; Rushton et al., 2010). The genome-wide characterization and analysis in model plants stipulate a widely authenticated and accepted classification system of the WRKY TF family specific to plants. It consists of 60 amino acids in their highly conserved DBD (called WRKY domain) (Eulgem et al., 2000). The WRKY domain comprises the WRKYGQK motif at N- terminus, a highly conserved motif, and a zinc-finger region at C -terminus binding to W-box (TTG ACT/C), which shows higher affinity towards DNA binding, although other binding sites also have been reported (Ciolkowski et al., 2008; van Verk et al., 2008; Li et al., 2010; Schmutz et al., 2010; Ishiguro et al., 2011; Brand et al., 2013; Rinerson et al., 2015; Cia et al., 2018). The number of genes encoding for the WRKY family varies amongst plants being 171 WRKY genes in wheat,161 in maize, 94 in chickpea, 171 in rice, etc (Table 1).
WRKY TFs have a diverse role in various plant processes, such as development, germination, seed dormancy and were first reviewed in 2010 (Rushton et al., 2010). Since then, its role has been thoroughly studied and experimented with. However, its significance in regulating stress response is of prime importance. It has been known to regulate and interconnect multiple environmental stimulations (Rushton et al., 2010; Rushton et al., 2012; Schluttenhofer and Yuan, 2015; Banerjee and Roy choudhury, 2015; Phukan et al., 2016). There has been a detailed review of WRKY TFs in various abiotic stresses such as cold (Zou et al., 2010; Guo et al., 2015; Dai et al., 2016; Yu et al., 2016), drought (Tripathi et al., 2014; He et al., 2016; Liu J et al., 2016; Chu et al., 2015; Li R et al., 2015; Raineri et al., 2015; Yan et al., 2015; Wang Y et al., 2016; Wani et al., 2018; Jha et al., 2019), salt (Wang L et al., 2014; Ding et al., 2015; Ma et al., 2015; Bai et al., 2018), osmotic stress (Mao et al., 2011; Li H et al., 2013; Marchive et al., 2013; Phukan et al., 2016; Jiang et al., 2017; Li et al., 2020), heat (Grover et al., 2013; Cai et al., 2015; He et al., 2016; Wang Y et al., 2016; Ohama et al., 2017; Sita et al., 2017; Cheng Z et al., 2021) and many more in economically important crops.
Many previous studies have described that plants harbour evolutionarily conserved defense mechanisms against pests (Peng et al., 2011; Sun et al., 2015; Kloth et al., 2016; Yang et al., 2017; Jha et al., 2020) and pathogens such as fungus (Turck et al., 2004; Guo et al., 2011; Inoue et al., 2013; Dang et al., 2014; Zhang, 2014), and bacteria (Deslandes et al., 2002; Kim et al., 2008) (Table 2). Most WRKY TF have been known to show negative regulation towards stress, while only a few exhibit positive responses during stress (Kim et al., 2008; Xing et al., 2008).
WRKY TFs have been reported to regulate the biosynthesis of several secondary metabolites like phenols, lignin, flavonoids, tannins etc., and their inducible expression analysis revealed a role in regulating defense-related PSM biogenesis (Guillaumie et al., 2010; Wang et al., 2010; Grunewald et al., 2012; Phukan et al., 2016). For instance, WRKY3 and WRKY6 have been reported for their role in the biosynthesis of volatile terpenes in tobacco (Skibbe et al., 2008). Artemisinin, an isolated sesquiterpenoid lactone majorly used as an antimalarial drug, is extracted from Artemisia annua (Meshnick et al., 1996); it has shown upregulation in the presence of WRKY1 (Asadollahi et al., 2008). Similarly, AaWRKY17 is reported to positively regulate artemisinin synthesis in providing resistance against Pseudomonas syringae (Chen et al., 2021).
Another PSM, Hydroxycinnamic acid amide (HCAA), is a phenol derived from the phenylpropanoid pathway, which is majorly involved in lignin biosynthesis and is derived from phenylalanine (Humphreys et al., 1999; Vogt, 2010; Rio et al., 2021). WRKY1 in late blight-infected potato plants binds to its promoter sites (Yogendra et al., 2015). Similarly, associated genes like ACT, DGK, and GL1 are activated in wheat by TaWRKY70, upon encountering fungi (Kage et al., 2017). Red rot caused by Fusarium in barley, HvWRKY23 promotes the expression of genes like CHS, HCT and PAL, which further induces HCAA biosynthesis (Karre et al., 2019). Likewise, StWRKY8 enhances the expression level of COR2, NCS, and TyDC genes majorly involved in the production of benzyl iso-quinoline alkaloids in potatoes; against bacterial attack (Yogendra et al., 2017). In addition, SsWRKY18, SsWRKY40, and SsMYC2 play a regulatory role in the production of abietane-type diterpenes in Salvia sclarea, which exhibits antimicrobial (Alfieri et al., 2018).
Phytoalexins, another secondary metabolite, belonging to the stilbene family, are considered significant regulators of plant defense (Jiang et al., 2010; Ahuja et al., 2011). Resveratrol in grapes has been the first reported class of phytoalexins (Lanz et al., 1991). VvWRKY8 has been found to modulate the biosynthesis of resveratrol in grapes which ultimately can be engineered to enhance resistivity (Jeandet et al., 2019). It was shown to negatively regulate the stilbene synthase gene (Jiang et al., 2019). ZmWRKY79 in maize caused increased phytoalexin accumulation under biotic stress conditions (Fu et al., 2017). Similarly, GaWRKY is responsible for the increased production of gossypol, which shows anti-feeding properties in cotton (Xu et al., 2004). WsWRKY1, in Withania, has been reported to significantly reduce phytosterol accumulation, resulting in decreased resistance to bacteria, fungi, and insects (Singh et al., 2017). In addition, SsWRKY18, SsWRKY40, and SsMYC2 show regulation in accumulating abietane (diterpene) in Salvia sclarea, which exhibits antimicrobial properties (Alfieri et al., 2018). Another example CjWRKY1 from Coptis japonica controls berberine biosynthesis (Kato-Noguchi and Takeshita, 2013).The regulatory roles of WRKY TFs production have been explored extensively. This information can be employed to engineer biotic stress resistance in transgenics.
3.2 MYB
MYB (Myeloblastosis) TFs are essential and functionally diverse proteins in eukaryotes (Jia et al., 2004). It is widely studied in plants, highlighting its importance in the system. Scientific investigations on Zea mays led to the isolation of the first MYB-TF in 1980s, which was homologous to the MYB proto-oncogene present in animals (Paz-Ares et al., 1987). MYB proteins typically have two distinct regions: N-terminus with a conserved DNA binding MYB domain, and a variable C-terminus making these TFs functionally diverse (Frampton et al., 1991). MYB domains generally contain 1–4 imperfect repeats of 50–53 amino acid residues each. Each repeat contains three α-helices, of which the second and third helices are responsible for specific gene recognition (Ogata et al., 1996). The third helix is the central part that makes contact with the major groove of DNA (Jia et al., 2004). Based on the number of repeats and their position, MYB TFs are categorized into four classes:1R (R1/2, R3-MYB), 2R (R2R3-MYB), 3R (R1R2R3-MYB), and 4R (R1/R2-like repeats) (Dubos et al., 2010).
Various growth and developmental processes in plants involve MYB TFs, such as flower color development in Glycine max (Takahashi et al., 2013) and signal transduction in A. thaliana, Zea mays, Oryza sativa, and cassava (Bakhshi and Arakawa, 2006; Raffaele et al., 2006; Liao et al., 2016). MYB TFs also control cell cycle and morphogenesis in various plants (Joaquin and Watson, 2003; Cominelli and Tonelli, 2009; Baumann et al., 2007). In addition to these diverse roles, MYB has also been well documented to regulate secondary metabolite synthesis in many plants, for instance, Camellia sinensis, A. thaliana, and Medicago truncatula (Verdier et al., 2012; Ambawat et al., 2013; Patra et al., 2013; Liu et al., 2015; Nguyen et al., 2016; Zhao et al., 2022). This production and regulation of secondary metabolites help combat enemies during biotic stress. Whenever any pathogen or insect attacks a plant, it tends to fortify its cell walls and membranes to halt the invasion (Table 2) (Kim et al., 2003; Kim et al., 2013). Several MYB factors are shown that control lignin biosynthesis (Zhu et al., 2020; Xiao et al., 2021). CmMYB15 in Chrysanthemum morifolium binds to AC elements of lipid synthesis gene promoter to regulate lignin biosynthesis during aphid invasion. Overexpressing CmMYB15 led to a reduction in aphid proliferation (An et al., 2019). MdMYB30, a His MYB-TF was found to improve disease resistance by regulating wax biosynthesis in apples (Zhang et al., 2019). In some plants, secondary metabolites like phytoalexins are produced to inhibit the growth of pathogens. Sorghum MYB TF y1 (yellow seed 1) protects maize from Colletotrichum sublineolum infection by synthesizing 3-deoxyanthocyanidin phytoalexins (Ibraheem et al., 2015; Chen et al., 2018). VdMYB1, a member of the R2R3-MYB TF, induce a defense response in the grapevine against Erysiphe necator fungal infection by activating stilbene synthase 2 gene (VdSTS2) (Yu et al., 2019).
MYB TFs are also reported to modulate the production of essential PSMs such as flavonoids, glycolipids, hydroxycinnamic acid amides (HCAAs), and proanthocyanidins during biotic stress. AtMYB29 and AtMYB76 are correlated to the synthesis of aliphatic glucosinolate in Arabidopsis. On the other hand, AtMYB34, AtMYB51, and AtMYB122 are responsible for indole glucosinolate (IG) accumulation which is related to tryptophan biosynthesis genes (CYP79B2, CYP79B3, and CYP83B1) expression (Dubos et al., 2010). Arabidopsis triple mutant MYB-34/MYB-51/MYB122 has shown increased Plectosphaerella cucumerina susceptibility due to reduced IG levels (Frerigmann et al., 2016). MYBs are well known to control anthocyanin production. In Arabidopsis thaliana, AtMYB113, AtMYB114, AtMYB75, and AtMYB90 modulate phenylpropanoid pathway to alter anthocyanin levels (Gonzalez et al., 2008). Similarly, in Asiatic hybrid lily plants, anthocyanin can be regulated by MYB6 and MYB12 (Yamagishi et al., 2010). In citrus, CsMYBF1 upregulates chalcone synthase (CHS) gene during certain flavonoid production (Liu J et al., 2016). In Camellia sinensis, CsMYB2/CsMYB26 have been found to promote flavonoid accumulation by binding to CsF30H and CsLAR gene promoters, thus improving resistivity against Exobasidium vexans, the causative agent of blister blight (Nisha et al., 2018). It was reported that Arabidopsis AtMYB11, AtMYB12, and AtMYB111 play an essential role in flavonoid production and that heterologous expression of AtMYB TFs can be heterologously expressed to increase flavonoid content in several other plants (Wang Y et al., 2016).
Besides significant positive regulatory roles, AtMYB75 has a negative impact on flavonoid synthesis, which results in reduction of kaempferol-3,7-dirhamnoside. This reduction makes the plants susceptible to insect attack (Onkokesung et al., 2014). In rice, phenylpropanoid pathway genes are upregulated by MYB30, MYB55 and MYB110, which accounts for increased HCCAs content (Kishi-Kaboshi et al., 2018). In comparison, the proanthocyanidin biosynthesis genes are downregulated by VvMYBC2-L1 in grapes, negatively regulating proanthocyanidin biosynthesis (Huang et al., 2014). Moreover, enhanced proanthocyanidins can elevate resistance against wounding in Rosa rugosa by the action of RrMYB5 and RrMYB10 (Shen Y et al., 2019). In Poplar trees, PtMYB115 helps in enhancing resistance during Dothiorella gregaria infection by binding to ANR1 & LAR3 genes to elevate proanthocyanidin levels (Wang et al., 2017). Given the ample information in this field, it is worth discussing how MYB TFs regulate these PSMs’ production, which would help to generate generate future stress-tolerant crops of economic value (Ambawat et al., 2015; Dubos et al., 2010).
3.3 AP2/ERF
AP2/ERFs (APETALA2/Ethylene Response Factor) consist of an AP2 DNA binding domain consisting of 40–70 conserved amino acids, which was first discovered in floral homeotic gene APETALA2 (AP2) of A. thaliana (Feng et al., 2005; Nakano et al., 2014). It is categorized into four subfamilies based on the variation of conserved domains: AP2, DREB, RAV, and ERF. Reportedly AP2 comprises two AP2 domains, DREB contains one AP2 and an A-subfamily, RAV consists of one AP2 and a B3 domain, while ERF has one AP2 and a B-subfamily (Zhou et al., 2016). In addition to imposing transcriptional and post-translational control upon growth, developmental, hormonal, and stress-responsive genes, AP2/ERFs transcription factors also govern biosynthesis and regulation of secondary metabolites (Mizoi et al., 2012; Licausi et al., 2013; Gibbs et al., 2015; Chandler, 2018, Xie et al., 2019).
Scientists have also reported the role of ERF TFs in plant defense against different pathogens, which can be employed to enhance pathogen resistance in crops by engineering pathways regulating specialized secondary metabolites. In Catharanthus roseus, ORCA1 and ORCA2 are two AP2/ERF proteins that bind to the promoters of biosynthetic genes of terpenoid indole alkaloid (TIA) (Menke et al., 1999). JA-induced ORCA3-TF binding to the JERE element of the promoter then induces two TIA biosynthetic genes encoding tryptophan decarboxylase and strictosidine synthase (vander Fits and Memelink, 2000). ORCA3, 4, and 5 in C. roseus interact with each other and act on MAPK to regulate TIA biogenesis (Paul et al., 2017; Paul et al., 2020). These TIAs, like catharantins, are involved in fungal and insect resistance by protecting the leaf surface of C. roseus from pathogenic infections and insect infestation (Roepke et al., 2010).
Steroidal glycoalkaloids (SGAs) are cytotoxic, which benefits plants to resist pathogen and insect invasion (Friedman, 2002; Friedman, 2006; Itkin et al., 2013; Nakayasu et al., 2017). In tobacco and C. roseus, GLYCOALKALOID METABOLISM 9 (GAME9) is found to regulate the biosynthesis of SGAs. Modifications in gene expression for SGA production and mevalonate pathway due to overexpression and knockdown, respectively. (Cárdenas et al., 2016). GAME9/JRE4 bind to the promoter region of SGA biosynthetic gene, which results in the upregulation of GAME, CAS, HMGR, SGT, and C5-SD genes (Thagun et al., 2016). GAME9 is reported to be a regulator of SGA biosynthesis to provide Spodoptera litura resistance in tomatoes (Nakayasu et al., 2018). Similar SGAs accumulation and regulation were discussed in tomatoes and potatoes (Cárdenas et al., 2016; Thagun et al., 2016; Nakayasu et al., 2018).
Nicotine, a tobacco alkaloid, is a repellent that facilitates plant protection against herbivores (Steppuhn et al., 2004). The AP2/ERF gene has been reported to be responsible for nicotine biosynthesis in tabacco. Accumulating nicotine biosynthesis results in positive action of NtERF189 and ORC1 on the associated genes, and NtMYC2, a JA inducer, also has the same effect (Shoji and Hashimoto, 2011; De Boer et al., 2011). Expression of putrescine N-methyltransferase coding gene, NtPMT1a, involved in nicotine biosynthesis, was found to be downregulated by NtERF32 (Sears et al., 2014). ERF located at NIC-2 locus regulates biosynthesis in tobacco of nicotine against herbivory (Shoji et al., 2010 and De Boer et al., 2011), NtORC1/ERF221 and NtJAP1/ERF10 in tobacco have been positively regulated by PMT gene which synthesizes nicotine and pyridine alkaloids (De Sutter et al., 2005; Thagun et al., 2016).
Taxol, an anticancer and plant defense compound, provides resistance against Phytophthora capsici infection (Young et al., 1992). TcERF12 and TcERF15, are regulators of taxol biosynthesis, the former having adverse effects while the latter having positive action (Zhang et al., 2015). Another class of antimicrobial metabolites is the hydroxycinnamic acid amides (HCAAs), which confer resistance to many fungi like Alternaria brassicicola and Botrytis cinerea on plants (Meraj et al., 2020). ORA59, another AP2/ERF TF, governs HCAA production by controlling AtACT (agmatine coumaryl transferase) (Li et al., 2018).
Saponins have been shown to have a variety of functions, including serving as phytoprotectants (Papadopoulou et al., 1999; Avato et al., 2006). In Panax notoginseng, PnERF1 was discovered to bind to the saponin biosynthesis gene (HMGR, FPS, DS, SS) promoters that increase total saponin content in overexpressing lines (Deng et al., 2017). Furthermore, lignin is an essential defensive substance as a fundamental component of the plant cell wall. GbERF1 prevents Verticillium dahliae infection in Gossypium barbadense by increasing lignin production (Guo et al., 2016). Genome-wide & transcriptome analysis confirmed that gypenosides (a triterpenoid saponin) in Gynostemma are regulated under biotic stress (Xu et al., 2020). Similarly, in Solanum melongena, Smechr0902114, and Smechr1102075, which belong to ERF TF, were reported to regulate anthocyanin biosynthesis (Li et al., 2021).
Several ERFs have been known to activate the transcription of various disease-responsive genes such as PR (pathogenesis-related), glucanase, chitinase, etc. They have also been reported in the regulation of abiotic stress (Yi et al., 2004; Seo et al., 2010; Fukao and Mito et al., 2011; Fahad et al., 2017; Li Z et al., 2015; Verma et al., 2016; Chen et al., 2017; Nolan et al., 2017; Van den Broeck et al., 2017; Ye et al., 2017; Bechtold and Field, 2018; Chen et al., 2021) and biotic stress (Lorenzo et al., 2003; Zarei et al., 2011; Moffat et al., 2012, Zhang et al., 2009; Liu et al., 2011; Liu et al., 2012; Müller and Munné-Bosch, 2015; Abiri et al., 2017). Many elicitors (Dzhavakhiya and Shcherbakova, 2007) originating from yeast, fungi, and bacteria have been lately studied to induce the synthesis of PSMs (Dzhavakhiya et al., 2007; Hu et al., 2003; Sang et al., 2010; Almagro et al., 2012; Awad et al., 2014; Naik and Al-Khayri, 2016; Jan et al., 2021a).
3.4 NAC
NAC [NAM (no apical meristem)], ATAF1/2, and CUC2 (cup-shaped cotyledon) are TF families that are quite connected to plant stress response (Ahuja et al., 2010). NAC proteins consist of a highly diverse C-terminal transcription regulatory (TR) domain, a conserved N-terminal binding domain, and a transmembrane domain (Puranik et al., 2012; Tweneboah and Oh, 2017). Functional genomics studies have revealed that NAC genes are well distributed in different plant species. According to the latest Transcription Factor Database (TFDB) records, 138 non-redundant NAC genes exist in the model plant, i.e., A. thaliana, around 263 in Triticum aestivum, 158 in Oryza sativa and 189 in Zea mays and many others (Table 1).
The NAC gene families show extensive regulation under environmental stresses (Nakashima et al., 2012; Nuruzzaman et al., 2013). Many investigations on these lines revealed the role of NAC TFs during abiotic stress; however, in recent years, biotic stress has also gained attention among plant scientists. It has also been reported that NAC regulates defense responses against microbe and insect attacks (Puranik et al., 2012). During pathogen attack, NAC induces hypersensitive response in essential crops such as Glycine max (Faria et al., 2011), sorghum (Zhang et al., 2013), wheat (Saad et al., 2013), barley (Chen et al., 2013), rice (Liu et al., 2018) and other plants (Zhou et al., 2018). PSMs production during stress is one of the prime defense mechanisms controlled by NAC TFs. Among them, phytoalexins are low molecular mass PSMs produced during pathogen attacks (Ahuja et al., 2012). In A. thaliana, ANAC042 confers plant resistivity to Alternaria brassicicola attack by binding to the promoter of camalexin biosynthetic genes (Saga et al., 2012; Duan et al., 2017). Camalexin (3’-thiazol-2’-yl-indole) is a well-known primary phytoalexin released after microbial infection in many plants (Nguyen et al., 2022). Glyceollin, another isoflavonoid-derived phytoalexin from soybean is positively regulated by GmNAC42-1 which imparts resistivity to plants (Jahan et al., 2019).
NAC TFs also regulate ROS homeostasis during stress conditions by regulating certain PSMs. The Arginine decarboxylase gene (ADC) controls putrescine biosynthesis, which can regulate ROS homeostasis. In Poncirus trifoliata, PtrNAC72 was found to regulate the expression of ADC (Wu et al., 2016). Furthermore, glutathione biosynthesis is regulated by MfNAC in Medicago falcate, which controls the expression of glyoxalase-1 (GLO1) gene (Duan et al., 2017).
In Hevea brasiliensis, latex biosynthesis is governed by HbNAC1, which shows binding affinity towards the CACG motif present in the promoter region of small rubber protein. This may have a potential role in healing wounds caused by herbivores (Cao et al., 2017). Many NAC TFs have also been found to regulate specific genes involved in defense systems negatively. From Norway spruce (Picea abies), PaNAC03 has been reported to regulate flavonoid biosynthesis genes like CHS, LAR3 negatively, and F3’H, consequently providing resistance to Heterobasidion annosum (Danielsson et al., 2011). Similarly, ANACO32 negatively regulates anthocyanin biosynthesis genes like DFR, LODX, and ANS (Mahmood et al., 2016). Flavonoids and anthocyanin have been well-documented as important secondary metabolites produced during the plant-pathogen arms race (Shah and Smith, 2020, Sivankalyani et al., 2016).
3.5 bZIP
The basic-leucine-zipper (bZIP) proteins, a class of TFs, are crucial regulators of plant growth and development. It consists of a conserved bZIP domain composed of contiguous α-helix. It consists of a primary region (18 amino acids), NLS, and sequence-specific DNA binding region at the N-terminus, which is highly conserved, followed by leucine zipper (heptad repeats of hydrophobic amino acids) at the C-terminus, which is less conserved (Jakoby et al., 2002; Nijhawan et al., 2008; Landschulz et al., 1988; Ellenberger et al., 1992; Furihata et al., 2006; Liao et al., 2008). The bZIP members are identified in many species, such as rice (94), wheat (186), maize (216), Arabidopsis (127), and so on (Table 1; Figure 3).
bZIP TFs are widely associated with plant stresses (Alves et al., 2013; Li et al., 2017). However, its function has been extensively studied in regulating abiotic stress (Banerjee and Roy choudhury, 2017; Baillo et al., 2019) such as drought, chilling, heat, osmosis, high salinity, etc., (Golldack et al., 2014). They are more diverse in plants as compared to microorganisms and animals (Maillet et al., 2021). The role of bZIP under biotic stress such as herbivory, wounding, pathogen attack, etc., and the plant’s protection via phytohormone signaling pathways or hypersensitive responsive defense mechanisms have been remotely studied (Zhang H et al., 2017; Joo et al., 2019). The molecular regulation of bZIP during a pathogen attack has been very well reviewed by Alves et al., 2013. Similarly, the regulatory roles of bZIP against resistance to pathogens have already been reported (Noman et al., 2017). For example, four bZIP genes have been reported in Glycine max (GmbZIPE1, GmbZIPE2, GmbZIP105, and GmbZIP62) that defend plants against Asian soybean rust (ASR), which in turn produces PSMs (Alves et al., 2015). In cassava, it has been revealed that Xanthomonas axonopodis pv. manihotis induces MebZIP3 and MebZIP5 upon infection (Li et al., 2017).
Okada et al., 2009 have shown that OsTGAP1 in rice binds to the promoter of OsKSL4 and OsCPS4 and enhances terpenoid phytoalexins accumulation, improving resistivity against blast disease. Some specific bZIP proteins are also involved in pharmaceutically important SMs. SmbZIP20 and SmbZIP7 are known to regulate tanshinone in Salvia miltiorrhiza (Cao et al., 2018). Similarly, AabZIP1 regulates artemisinin synthesis in Artemisia annua (Shen Q et al., 2019). Li et al., 2017 have engineered cassava bacterial blight-resistant plants by overexpressing MebZIP3 and MebZIP5. There are many reports explaining the role of bZIP in plant defense against pathogens, but their importance during plant defense against insects and nematode attacks is still less explored; these areas have great potential for further exploration.
3.6 bHLH TFs
bHLH (basic-helix-loop-helix) TFs are one of the TF families found in eukaryotes, mainly plants (Carretero-Paulet et al., 2010). It consists of two connected regions the N-terminal region (basic in nature) followed by the HLH domain (40-50 amino acid residues) and a highly conserved HER motif (Atchley et al., 1999; Massari and Murre, 2000; Toledo-Ortiz et al., 2003) that binds to DNA and regulates transcription both positively and negatively (Yang et al., 2020). It was first reported in Zea mays for regulating abiotic stress response (Ludwig et al., 1989).
Many studies have deciphered the role of bHLH concerning environmental stresses (Babitha et al., 2015; Jin et al., 2016; Qian et al., 2021). However, reports of the role of bHLH in biotic stress are scarce (Meraj et al., 2020). bHLH has been known to regulate hormonal signaling pathways, especially Jasmonic acid that triggers plant immunity (Tian et al., 2015). They have also been found to regulate stress responses by controlling the production of secondary metabolites like flavonoids, anthocyanins, glucosinolates (GLs), phytoalexins, etc. (Sun et al., 2018).
Generally, bHLH interacts with MYB TFs to control specific gene expression. It has been explicitly reported in Arabidopsis, bHLH-04 to 06 interaction with MYB51 regulates the biosynthesis of GLs and the biosynthesis of anthocyanin and flavonoids through the phenylpropanoid pathway (Frerigmann et al., 2014). Similarly, GLABRA3 (GL3), Enhancer of GLABRA3 (EGL3), and Transparent Testa 8 (TT8) proteins of the bHLH family were found to activate anthocyanin biosynthetic genes after binding to MYB transcriptional complex (Dubos et al., 2010). MYC2, another bHLH member, has regulatory roles in jasmonic acid signaling by binding to Jasmonic acid responsive element (JARE) called ORCA-3, which acts as a promoter and enhances alkaloid biosynthesis (Zhang et al., 2011). MdMYC2 in Arabidopsis upregulates certain anthocyanin-related genes, promoting sesquiterpene synthesis (Nemesio-Gorriz et al., 2017). Similarly, in rice, diterpenoid phytoalexin factor (DPF) has been seen to regulate the production of diterpenoid phytoalexin, which could have a potential role during pathogen attack (Yamamura et al., 2015).
In Nicotiana benthamiana, NbbHLH1, NbbHLH2, and NbbHLH3 have been implicated in nicotine biosynthesis through virus-induced gene silencing (VIGS) by binding to the promoter of putrescine N-methyltransferase (Todd et al., 2010). HMGR activation in Medicago truncatula is accountable for saponin synthesis, which TSAR1 and TSAR2 regulate. (Mertens et al., 2016). Avenacin, an important saponin, has been found to check the fungal growth in plants. As mutants of Avena strigosa deficient in avenacin become susceptible to Gaeumannomyces graminis var. tritici infection (Papadopoulou et al., 1999). An IAA-LEUCINE RESISTANT3 (ILR3) has been reported to have a significant role against nematode infestation by regulating aliphatic GLs production (Samira et al., 2018). Although many stress-related TFs have been reported, the exact molecular mechanism of TFs regulating these PSMs is still a research hotspot.
4 MiRNAs - master regulators of plant defense response
As discussed earlier, microRNAs (miRNAs) are a class of small (20-24 nucleotide long), non-coding RNAs that play a presiding part in regulating gene expression post-transcriptionally through degrading their target mRNA or halting translational machinery (D. Baulcombe, 2004). During these years, the miRNA has gained massive attention by scientists due to its widespread role in plant development and stress tolerance (Yang T et al., 2007). Emerging evidences suggest its potential role in regulating several defense-related genes during biotic stress (Ruiz-Ferrer and Voinnet, 2009; Kumar, 2014; Šečić et al., 2021). Typically, miRNAs target specific genes that are directly or indirectly involved in any defense pathway when a plant faces pest and pathogen attack. During stress, many miRNAs have been found to target genes involved in the production of defense-related compounds like secondary metabolites (Owusu Adjei et al., 2021). Meanwhile, miRNAs have also been reported to control many transcription factors that control the production of defense products (Javed et al., 2020). Thus, miRNAs are crucial factor to be emphasized during stress as they also regulate the regulators i.e, TFs.
4.1 MiRNAs involved in PSM production
Secondary metabolites are one of the prime plant products used by plants to combat pest and pathogen attacks. Thus, differential regulation of these SMs is quite obvious during stress. miRNAs have been found to chiefly regulate these PSMs (Owusu Adjei et al., 2021). Kettles et al. (2013) have shown that miRNA mediates the regulation of secondary metabolites like camalexin, which provides resistivity to Arabidopsis against green peach aphids. miR393 is well documented to be involved in plant defense by shifting resources to the glucosinolate pathway from the camalexin pathway resulting in the production of the most effective PSMs (Robert-Seilaniantz et al., 2011). It has also been found that the flg22 application leads to the surge of miR393, suggesting involvement in defense.
miR169 is known for its participation in anthocyanin biosynthesis in Piper nigrum by targeting UGT79B1, an anthocyanidin 3-O-glucoside 2’’-Oxylosyltransferase enzyme; along with 73 other miRNAs indicating a significant role in regulating metabolic pathway during biotic stress (Ding et al., 2021). Additionally, miR828 and miR858 target repressors of MYB, which leads to the activation of anthocyanin biosynthesis pathway and accumulation of anthocyanin and flavonol.VvMYB114 is targeted by miR828 and miR858 that represses various enzymes such as DFR (Dihydroflavonol-4-reductase) and UFGT (UDP-glucose: flavonoid 3-O-glucosyltransferase) in grapes (Tirumalai et al., 2019). Sometimes miRNAs can also target certain genes to regulate PSMs production during biotic stresses. The squamosal promoter-binding protein-like (SPL) 9 gene can be targeted by miRNA156, which contributes to Arabidopsis’ stress resistance by promoting the massive accumulation of anthocyanins (Cui et al., 2014).
4.2 MiRNAs regulating TFs
TFs play a prime role during any defense response, thus being the preferred target of miRNA to bring changes at the molecular level (Li and Zhang, 2016). Many reports suggest the role of miRNA in controlling various stress-responsive TFs (Lima et al., 2012; Koroban et al., 2016; Samad et al., 2017). Additionally, a sufficient number of miRNAs have been documented to regulate TFs, such as MYB, NAC, WRKY, ERF, ARF, and bHLH, which have been previously discussed to control the production of secondary metabolites. In wheat, miR164 has been found to negatively interact with NAC21/22, which ultimately enhances the susceptibility to stripe rust brought on by the pathogen, Puccinia striiformis (Zheng et al., 2016).
Similarly, to target leaf spot fungus, Alternaria alternate, Md-miRNA395 and Md-miRNA156ab were reported to target MdWRKY26 and MdWRKYN1, respectively (Zhang Q et al., 2017). Sly-miR1127 is known to regulate slyWRKY75 during Botrytis cinerea infection in tomato plants (López-Galiano et al., 2017). Magnaporthe oryzae stimulated the expression of miR5819 and miR5075 in rice, and it was discovered that these miRNAs specifically aimed at OsbZIP38 and OsbZIP27, respectively. PvAP2-ERF genes in Phaseolus vulgaris L. have also been revealed to be the target of miRNAs from other plant species, with miR156, miR172, and miR838 possibly being involved in the regulation of ERF/AP2 (Kavas et al., 2015). During rhizobial infection, the concentration of miR160 and miR164 that influence ARF (ARF17, ARF10, and ARF16) and NAC respectively changed (Basel Khraiwesh et al., 2012; H. Li et al., 2010).
4.3 MiRNAs orchestrating PSMs productions through TFs
Since our focus is to deliberate on regulating PSMs biosynthesis by TFs, it is worthwhile to include how miRNAs modulate PSMs biosynthesis. During any biotic stress, many of the secondary metabolites are differentially regulated. Several miRNAs have been reported to target transcription factors to control secondary metabolite biosynthetic pathways (Figure 4) (Jiang et al., 2021). Xia et al., 2012 reported that miR828 targets MYBs to control anthocyanin production. Similarly, miR858-targeted MYBs were associated with lignification, anthocyanin production, and stress responses (Xia et al., 2012). These lignification and anthocyanin production are a well-known mechanisms for wall fortification during pathogen attacks (Shah and Smith, 2020). AtMYB111, AtMYB11, and AtMYB12, transcriptional activators of genes associated with flavonoid biosynthesis, are negatively regulated by miR858. AtMYB111, AtMYB11, and AtMYB12 positively modulated the defense response against Plectosphaerella cucumerina in Arabidopsis (Camargo Rairez et al., 2017).
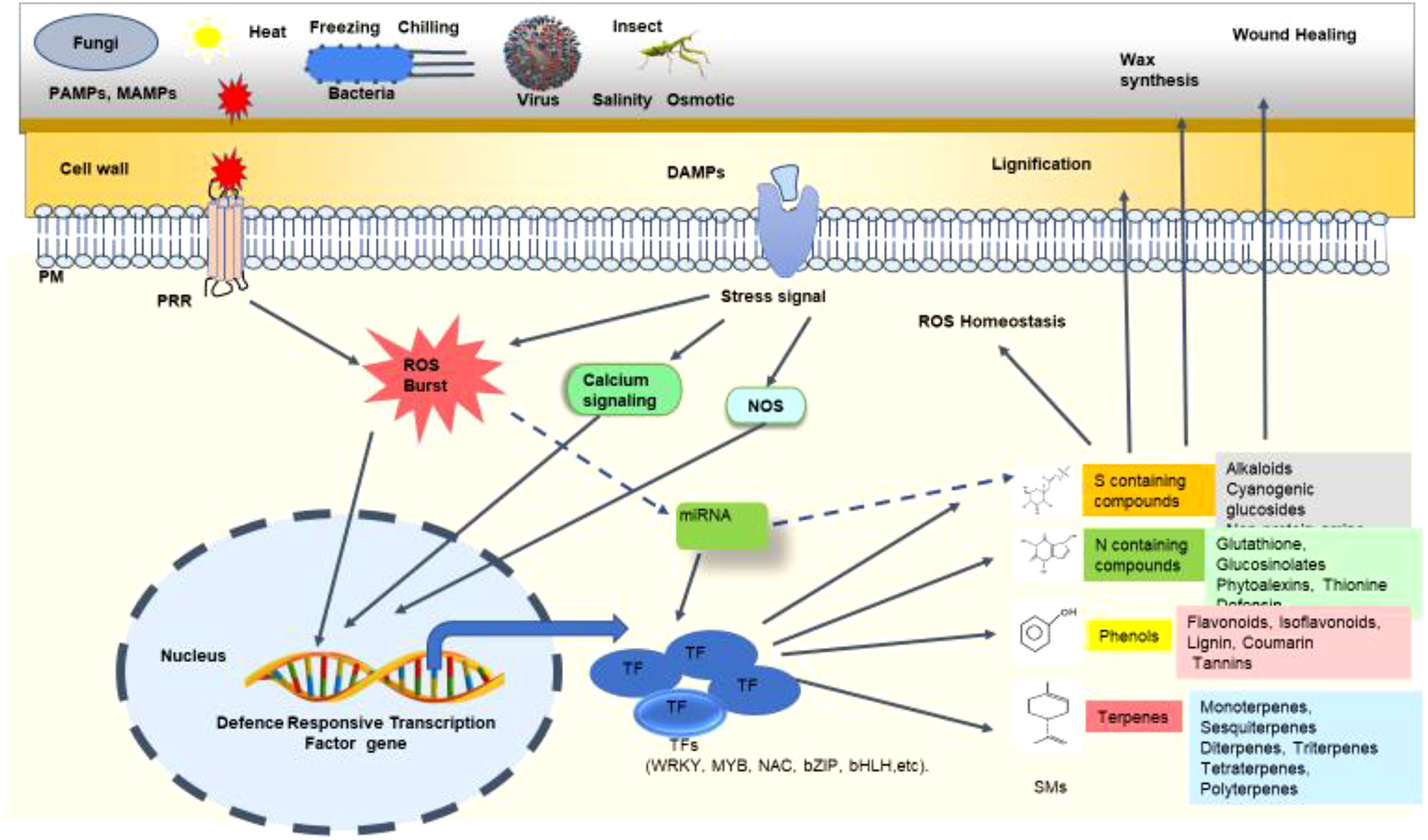
Figure 4 Schematic representation of the mechanism of transcription factors (TFs) regulating secondary metabolites (SMs) biosynthesis under biotic stress. At the time of biotic stress such as fungal, bacterial, viral, or insect attack, various pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) are recognized by the cell wall and membrane receptors like pattern recognition receptors (PRRs). Upon recognition, various stress signals are induced in the form of reactive oxygen species (ROS) burst, nitric oxide (NO) signalling, calcium signalling, etc. These stress signals ultimately trigger the expression of defense-responsive TF genes. These TFs like WRKY, MYB, NAC, bZIP, bHLH, etc. regulate the production of different secondary metabolites in the plant. These SMs protect the plant through lignification, wax synthesis, wound healing, ROS homeostasis, and many other mechanisms. Certain microRNAs (miRNAs) are also involved in controlling TF expression which in turn regulates SM production. The dashed line indicates the exact mechanism of SM production governed by miRNA, which is yet to be deciphered.
Khan et al., 2020 have also demonstrated the interaction of miR858 with MYB1 and concluded that miRNA mimic technique could enhance resistivity against fungal attacks in Artemisia annua. The geometrid Ectropis oblique poses a significant threat to the tea plant Camellia sinensis L. (Wang et al., 2014). During E. oblique attack, can-miR171 was reported to interact with bHLH, ultimately contributing to the production of secondary metabolites related to defense (Jeyaraj et al., 2017).
Although an elaborated study has been done in understanding the role of miRNA that regulates transcription factors, there are a few literature that supports the role of miRNA in regulating PSMs. But a correlation of miRNAs regulating TFs that regulate SM production during biotic stress is focused in the present review, which will aid in administering the production of SMs by controlling either the pathway directly or via the TFs. Thus, combining the molecular research to industrial engineering opens a whole bunch of opportunity to future researchers and industrialists that use secondary metabolites in production of various products such as medicines and cosmetics.
5 Conclusion and future perspective
Since plants are sessile, they have evolved complex regulatory mechanisms to respond to environmental stresses. In topical years, there has been a substantial upsurge in the importance of metabolic adaptation in plants under hostile environmental conditions. PSMs are natural products produced in plants when exposed to potential enemies. There are many transcription factors, WRKY, bHLH, bZIP, NAC, MYB, and AP2/ERF, that mediate the production and regulation of secondary metabolites related to plant defense against pests and pathogen attacks. Therefore, it is crucial to focus on the regulators such as miRNAs that control TFs and understanding their defensive role in plants under biotic constraints. TFs play a role in regulating PSMs fine-tune the expression of these TFs, and miRNas can, synchronize the synthesis and functional regulation of SMs during any pathogenic attack. This information can be utilized in future to modulate the production of SMs in any desired crop creating an enhanced variety in terms of resistivity, medicinal value or yield. In future, specific technologies, like miRNA mimicking, can be widely used to develop advanced SMs-producing plants to aid crops to combat stress and also to the pharmaceutical industries to produce desired amounts of medicinally important SMs. Since ancient times, plant extracts from a diverse range of medicinal plants have been used in the treatment of different diseases, owing to the presence of specific PSMs. With the recent increase in inclination towards traditional medicine (Chinese medicine and Ayurveda) and plant products for treating various diseases like diabetes, respiratory problems, arthritis, malaria and even certain cancers, the research on PSMs has gained momentum (Seca and Pinto, 2019). A thorough understanding of molecular pathways governing SM production can thus aid in enhanced SM producing engineered plants.
Author contributions
AS and IS conceptualized and supervised the study. AS, MK, IS, and AR contributed to the investigation. MK wrote the original draft. AS, IS, and AR contributed to reviewing, editing, and visualization. AS, IS, and AR contributed to the formal analysis. AS, IS, and AR contributed to funding acquisition. AS and IS contributed to resources. All authors contributed to the article and approved the submitted version.
Funding
AS was supported by SERB (Grant no. ECR/2017/002478). AR was supported by grant No. CZ.02.1.01/0.0/0.0/15_003/0000433, “EXTEMIT – K project,” and by grant No. CZ.02.1.01/0.0/0.0/16_019/0000803 financed by the Operational Program Research, Development and Education (OP RDE).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdalla, M. A., Mühling, K. H. (2019). Plant-derived sulfur-containing natural products produced as a response to biotic and abiotic stresses: A review of their structural diversity and medicinal importance. J. Appl. Bot. Food Qual 92, 204–215. doi:?10.5073/JABFQ.2019.092.029
Abiri, R., Shaharuddin, N. A., Maziah, M., Yusof, Z. N. B., Atabaki, N., Sahebi, M., et al. (2017). Role of ethylene and the APETALA 2/ethylene response factor superfamily in rice under various abiotic and biotic stress conditions. Environ. Exp. Bot. 134, 33–44. doi: 10.1016/j.envexpbot.2016.10.015
Aftab, T. (2019). A review of medicinal and aromatic plants and their secondary metabolites status under abiotic stress. J. Medicinal Plants 7 (3), 99–106.
Ahmed, E. F., Elkhateeb, W. A., Taie, H. A., Rateb, M. E., Fayad, W. (2017). Biological capacity and chemical composition of secondary metabolites from representatives Japanese lichens. J. Appl. Pharm. Sci. 7 (1), 098–103. doi: 10.7324/JAPS.2017.70113
Ahuja, I., de Vos, R. C., A.M. and Hall, R. D. (2010). Plant molecular stress responses face climate change. Trends Plant Sci. 15 (12), 664–674. doi: 10.1016/j.tplants.2010.08.002
Ahuja, I., Kissen, R., Bones, A. M. (2012). Phytoalexins in defense against pathogens. Trends Plant Sci. 17 (2), 73–90. doi: 10.1016/j.tplants.2011.11.002
Ahuja, I., Rohloff, J., Bones, A. M. (2011). “Defence mechanisms of brassicaceae: implications for plant-insect interactions and potential for integrated pest management,” in Sustainable agriculture, vol. 2. (Dordrecht: Springer), 623–670.
Alfieri, M., Vaccaro, M. C., Cappetta, E., Ambrosone, A., De Tommasi, N., Leone, A. (2018). Coactivation of MEP-biosynthetic genes and accumulation of abietane diterpenes in salvia sclarea by heterologous expression of WRKY and MYC2 transcription factors. Sci. Rep. 8 (1), 1–13. doi: 10.1038/s41598-018-29389-4
Almagro, L., Gutierrez, J., Pedreño Sottomayor, M.A. M. (2014). Synergistic and additive influence of cyclodextrins and methyl jasmonate on the expression of the terpenoid indole alkaloid pathway genes and metabolites in C atharanthus roseus cell cultures. Plant Cell, Tissue and Organ Culture (PCTOC), 119, pp.543–551.
Alves, M. S., Dadalto, S. P., Gonçalves, A. B., De Souza, G. B., Barros, V. A., Fietto, L. G. (2013). Plant bZIP transcription factors responsive to pathogens: a review. Int. J. Mol. Sci. 14 (4), 7815–7828. doi: 10.3390/ijms14047815
Alves, M. S., Soares, Z. G., Vidigal, P. M., Barros, E. G., Poddanosqui, A. M., Aoyagi, L. N., et al. (2015). Differential expression of four soybean bZIP genes during phakopsora pachyrhizi infection. Funct. Integr. Genomics 15 (6), 685–696. doi: 10.1007/s10142-015-0445-0
Ambawat, S., Sharma, P., Yadav, N. R., Yadav, R. C. (2013). MYB transcription factor genes as regulators for plant responses: an overview. Physiol. Mol. Biol. Plants 19 (3), 307–321. doi: 10.1007/s12298-013-0179-1
An, J. P., Zhang, X. W., You, C. X., Bi, S. Q., Wang, X. F., Hao, Y. J. (2019). Md WRKY 40 promotes wounding-induced anthocyanin biosynthesis in association with md MYB 1 and undergoes md BT 2-mediated degradation. New Phytol. 224 (1), 380–395. doi: 10.1111/nph.16008
Asadollahi, M. A., Maury, J., Møller, K., Nielsen, K. F., Schalk, M., Clark, A., et al. (2008). Production of plant sesquiterpenes in saccharomyces cerevisiae: effect of ERG9 repression on sesquiterpene biosynthesis. Biotechnol. bioengineering 99 (3), 666–677. doi: 10.1002/bit.21581
Atchley, W. R., Terhalle, W., Dress, A. (1999). Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J. Mol. Evol. 48 (5), 501–516. doi: 10.1007/PL00006494
Avato, P., Bucci, R., Tava, A., Vitali, C., Rosato, A., Bialy, Z., et al. (2006). Antimicrobial activity of saponins from medicago sp.: structure-activity relationship. Phytotherapy Research: Int. J. Devoted to Pharmacol. Toxicological Eval. Natural Product Derivatives 20 (6), 454–457. doi: 10.1002/ptr.1876
Awad, V., Kuvalekar, A., Harsulkar, A. (2014). Microbial elicitation in root cultures of taverniera cuneifolia (Roth) arn. for elevated glycyrrhizic acid production. Ind. Crops Products 54, 13–16. doi: 10.1016/j.indcrop.2013.12.036
Babitha, K. C., Vemanna, R. S., Nataraja, K. N., Udayakumar, M. (2015). Overexpression of EcbHLH57 transcription factor from eleusine coracana l. @ in tobacco confers tolerance to salt, oxidative and drought stress. PloS One 10 (9), e0137098.
Bai, Y., Sunarti, S., Kissoudis, C., Visser, R. G., van der Linden, C. (2018). The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front. Plant Sci. 9, 801. doi: 10.3389/fpls.2018.00801
Baillo, E. H., Kimotho, R. N., Zhang, Z., Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 10 (10), p.771. doi: 10.3390/genes10100771
Bakhshi, D., Arakawa, O. (2006). Induction of phenolic compounds biosynthesis with light irradiation in the flesh of red and yellow apples. J. Appl. Horticulture 8 (2), 101–104. doi: 10.37855/jah.2006.v08i02.23
Banerjee, A., Roy choudhury, A. (2015). WRKY proteins: signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015.
Banerjee, A., Roy choudhury, A. (2017). Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254 (1), 3–16. doi: 10.1007/s00709-015-0920-4
Baumann, K., Perez-Rodriguez, M., Bradley, D., Venail, J., Bailey, P., Jin, H., et al. (2007). Control of cell and petal morphogenesis by R2R3 MYB transcription factors. doi: 10.1242/dev.02836
Bechtold, U., Field, B. (2018). Molecular mechanisms controlling plant growth during abiotic stress. J. Exp. Bot. 69 (11), 2753–2758. doi: 10.1093/jxb/ery157
Bloem, E., Haneklaus, S., Schnug, E. (2007). Changes in the sulphur and glucosinolate content of developing pods and seeds of oilseed rape (Brassica napus l.) in relation to different cultivars. Landbauforschung Volkenrode 57 (4), 297.
Brading, P. A., Hammond-Kosack, K. E., Parr, A., Jones, J. D. (2000). Salicylic acid is not required for cf-2-and cf-9-dependent resistance of tomato to cladosporium fulvum. Plant J. 23 (3), 305–318. doi: 10.1046/j.1365-313x.2000.00778.x
Brand, L. H., Fischer, N. M., Harter, K., Kohlbacher, O., Wanke, D. (2013). Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 41 (21), 9764–9778. doi: 10.1093/nar/gkt732
Bulgakov, V. P., Avramenko, T. V. (2015). New opportunities for the regulation of secondary metabolism in plants: focus on microRNAs. Biotechnol. Lett. 37 (9), 1719–1727. doi: 10.1007/s10529-015-1863-8
Butelli, E., Titta, L., Giorgio, M., Mock, H. P., Matros, A., Peterek, S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26 (11), 1301–1308. doi: 10.1038/nbt.1506
Cai, H., Yang, S., Yan, Y., Xiao, Z., Cheng, J., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66 (11), 3163–3174. doi: 10.1093/jxb/erv125
Camargo Ramírez, R., San Segundo, B., Tolrá Pérez, R. (2017). Function of microRNAs in plant innate immunity. Available at: https://ddd.uab.cat/record/186986.
Cao, W., Wang, Y., Shi, M., Hao, X., Zhao, W., Wang, Y., et al. (2018). Transcription factor SmWRKY1 positively promotes the biosynthesis of tanshinones in salvia miltiorrhiza. Front. Plant Sci. 9, 554. doi: 10.3389/fpls.2018.00554
Cao, Y., Zhai, J., Wang, Q., Yuan, H., Huang, X. (2017). Function of hevea brasiliensis NAC1 in dehydration-induced laticifer differentiation and latex biosynthesis. Planta 245 (1), 31–44. doi: 10.1007/s00425-016-2589-0
Cárdenas, P. D., Sonawane, P. D., Pollier, J., Vanden Bossche, R., Dewangan, V., Weithorn, E., et al. (2016). GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat. Commun. 7 (1), 1–16.
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martínez-García, J. F., Bilbao-Castro, J. R., Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153 (3), 1398–1412. doi: 10.1104/pp.110.153593
Chacón-Cerdas, R., Barboza-Barquero, L., Albertazzi, F. J., Rivera-Méndez, W. (2020). Transcription factors controlling biotic stress response in potato plants. Physiol. Mol. Plant Pathol. 112, 101527. doi: 10.1016/j.pmpp.2020.101527
Chandler, J. W. (2018). Class VIIIb APETALA2 ethylene response factors in plant development. Trends Plant Sci. 23 (2), 151–162. doi: 10.1016/j.tplants.2017.09.016
Chen, Y. H., Cao, Y. Y., Wang, L. J., Li, L. M., Yang, J., Zou, M. X. (2018). Identification of MYB transcription factor genes and their expression during abiotic stresses in maize. Biol. plantarum 62 (2), 222–230. doi: 10.1007/s10535-017-0756-1
Chen, H. C., Chien, T. C., Chen, T. Y., Chiang, M. H., Lai, M. H., Chang, M. C. (2021). Overexpression of a novel ERF-x-type transcription factor, OsERF106MZ, reduces shoot growth and tolerance to salinity stress in rice. Rice 14 (1), 1–18. doi: 10.1186/s12284-021-00525-5
Chen, F., Hu, Y., Vannozzi, A., Wu, K., Cai, H., Qin, Y., et al. (2017). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36 (5-6), 311–335. doi: 10.1080/07352689.2018.1441103
Chen, Y. J., Perera, V., Christiansen, M. W., Holme, I. B., Gregersen, P. L., Grant, M. R., et al. (2013). The barley HvNAC6 transcription factor affects ABA accumulation and promotes basal resistance against powdery mildew. Plant Mol. Biol. 83 (6), 577–590. doi: 10.1007/s11103-013-0109-1
Cheng, F., Li, W., Zhou, Y., Shen, J., Wu, Z., Liu, G., et al. (2012). admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. doi: 10.1021/ci300367a
Cheng, Z., Luan, Y., Meng, J., Sun, J., Tao, J., Zhao, D. (2021). WRKY transcription factor response to high-temperature stress. Plants 10 (10), .2211. doi: 10.3390/plants10102211
Chinnapandi, B., Bucki, P., Braun Miyara, S. (2017). SlWRKY45, nematode-responsive tomato WRKY gene, enhances susceptibility to the root knot nematode; M. javanica infection. Plant Signaling Behav. 12 (12), e1356530. doi: 10.1080/15592324.2017.1356530
Chinnapandi, B., Bucki, P., Fitoussi, N., Kolomiets, M., Borrego, E., Braun Miyara, S. (2019). Tomato SlWRKY3 acts as a positive regulator for resistance against the root-knot nematode meloidogyne javanica by activating lipids and hormone-mediated defense-signaling pathways. Plant Signaling Behav. 14 (6), 1601951. doi: 10.1080/15592324.2019.1601951
Chu, X., Wang, C., Chen, X., Lu, W., Li, H., Wang, X., et al. (2015). The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana. PloS One 10 (11), e0143022. doi: 10.1371/journal.pone.0143022
Cia, M. C., de Carvalho, G., Azevedo, R. A., Monteiro-Vitorello, C. B., Souza, G. M., Nishiyama-Junior, M. Y., et al. (2018). Novel insights into the early stages of ratoon stunting disease of sugarcane inferred from transcript and protein analysis. Phytopathology 108 (12), 1455–1466. doi: 10.1094/PHYTO-04-18-0120-R
Ciolkowski, I., Wanke, D., Birkenbihl, R. P., Somssich, I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68 (1), 81–92. doi: 10.1007/s11103-008-9353-1
Coleto, I., de la Peña, M., Rodríguez-Escalante, J., Bejarano, I., Glauser, G., Aparicio-Tejo, P. M., et al. (2017). Leaves play a central role in the adaptation of nitrogen and sulfur metabolism to ammonium nutrition in oilseed rape (Brassica napus). BMC Plant Biol. 17 (1), 1–13. doi: 10.1186/s12870-017-1100-9
Cominelli, E., Tonelli, C. (2009). A new role for plant R2R3-MYB transcription factors in cell cycle regulation. Cell Res. 19 (11), 1231–1232. doi: 10.1038/cr.2009.123
Costa, P., Gonçalves, S., Valentão, P., Andrade, P. B., Coelho, N., Romano, A. (2012). Thymus lotocephalus wild plants and in vitro cultures produce different profiles of phenolic compounds with antioxidant activity. Food Chem. 135 (3), 1253–1260. doi: 10.1016/j.foodchem.2012.05.072
Crozier, A., Jaganath, I. B., Clifford, M. N. (2006). Phenols, polyphenols and tannins: an overview. Plant secondary metabolites: Occurrence structure role Hum. diet 1, 1–25.
Cui, L. G., Shan, J. X., Shi, M., Gao, J. P., Lin, H. X. (2014). The miR156-SPL 9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80 (6), 1108–1117. doi: 10.1111/tpj.12712
Dai, X., Wang, Y., Zhang, W. H. (2016). OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 67 (3), 947–960. doi: 10.1093/jxb/erv515
Dang, F., Wang, Y., She, J., Lei, Y., Liu, Z., Eulgem, T., et al. (2014). Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of capsicum annuum, positively regulates tobacco resistance to ralstonia solanacearum infection. Physiologia plantarum 150 (3), 397–411. doi: 10.1111/ppl.12093
Danielsson, M., Lundén, K., Elfstrand, M., Hu, J., Zhao, T., Arnerup, J., et al. (2011). Chemical and transcriptional responses of Norway spruce genotypes with different susceptibility to heterobasidion s infection. BMC Plant Biol. 11 (1), 1–15. doi: 10.1186/1471-2229-11-154
De Boer, K., Tilleman, S., Pauwels, L., Vanden Bossche, R., De Sutter, V., Vanderhaeghen, R., et al. (2011). APETALA2/ETHYLENE RESPONSE FACTOR and basic helix–loop–helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J. 66 (6), 1053–1065. doi: 10.1111/j.1365-313X.2011.04566.x
Deng, L. L., Yuan, D., Zhou, Z. Y., Wan, J. Z., Zhang, C. C., Liu, C. Q., et al. (2017). Saponins from Panax japonicus attenuate age-related neuroinflammation via regulation of the mitogen-activated protein kinase and nuclear factor kappa b signaling pathways. Neural Regeneration Res. 12 (11), 1877.
Deslandes, L., Olivier, J., Theulières, F., Hirsch, J., Feng, D. X., Bittner-Eddy, P., et al. (2002). Resistance to ralstonia solanacearum in arabidopsis thaliana is conferred by the recessive RRS1-r gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. 99 (4), 2404–2409. doi: 10.1073/pnas.032485099
De Sutter, V., Vanderhaeghen, R., Tilleman, S., Lammertyn, F., Vanhoutte, I., Karimi, M., et al. (2005). Exploration of jasmonate signalling via automated and standardized transient expression assays in tobacco cells. Plant J. 44 (6), 1065–1076. doi: 10.1111/j.1365-313X.2005.02586.x
Ding, M., Chen, J., Jiang, Y., Lin, L., Cao, Y., Wang, M., et al. (2015). Genome-wide investigation and transcriptome analysis of the WRKY gene family in gossypium. Mol. Genet. Genomics 290 (1), 151–171. doi: 10.1007/s00438-014-0904-7
Ding, Y., Mao, Y., Cen, Y., Hu, L., Su, Y., Ma, X., et al. (2021). Small RNA sequencing reveals various microRNAs involved in piperine biosynthesis in black pepper (Piper nigrum l.). BMC Genomics 22 (1), 1–12. doi: 10.1186/s12864-021-08154-4
Duan, M., Zhang, R., Zhu, F., Zhang, Z., Gou, L., Wen, J., et al. (2017). A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress. Plant Cell 29 (7), 1748–1772. doi: 10.1105/tpc.17.00044
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in arabidopsis. Trends Plant Sci. 15 (10), 573–581. doi: 10.1016/j.tplants.2010.06.005
Dzhavakhiya, V. G., Ozeretskovskaya, O. L., Zinovyeva, S. V. (2007). “Immune response,” in Comprehensive and molecular phytopathology (Elsevier), 265–314.
Dzhavakhiya, V. G., Shcherbakova, L. A. (2007). “Creation of disease-resistant plants by gene engineering,” in Comprehensive and molecular phytopathology (Elsevier), 439–466.
Ellenberger, T. E., Brandl, C. J., Struhl, K., Harrison, S. C. (1992). The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein-DNA complex. Cell 71 (7), 1223–1237. doi: 10.1016/S0092-8674(05)80070-4
Emendack, Y., Burke, J., Laza, H., Sanchez, J., Hayes, C. (2018). Abiotic stress effects on sorghum leaf dhurrin and soluble sugar contents throughout plant development. Crop Sci. 58 (4), 1706–1716. doi: 10.2135/cropsci2018.01.0059
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 (5), 199–206. doi: 10.1016/S1360-1385(00)01600-9
Fahad, S., Bajwa, A. A., Nazir, U., Anjum, S. A., Farooq, A., Zohaib, A., et al. (2017). Crop production under drought and heat stress: plant responses and management options. Frontiers in Plant Science (CRC Press), 1147.
Faria, J. A., Reis, P. A., Reis, M. T., Rosado, G. L., Pinheiro, G. L., Mendes, G. C., et al. (2011). The NAC domain-containing protein, GmNAC6, is a downstream component of the ER stress-and osmotic stress-induced NRP-mediated cell-death signaling pathway. BMC Plant Biol. 11 (1), 1–14. doi: 10.1186/1471-2229-11-129
Feng, J. X., Liu, D. I., Pan, Y. I., Gong, W., Ma, L. G., Luo, J. C., et al. (2005). An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol. 59 (6), 853–868. doi: 10.1007/s11103-005-1511-0
Foyer, C. H., Lam, H. M., Nguyen, H. T., Siddique, K. H., Varshney, R. K., Colmer, T. D., et al. (2016). Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2 (8), 1–10. doi: 10.1038/nplants.2016.112
Frampton, J., Gibson, T. J., Ness, S. A., Döderlein, G., Graf, T. (1991). Proposed structure for the DNA-binding domain of the myb oncoprotein based on model building and mutational analysis. Protein Engineering Design Selection 4 (8), 891–901. doi: 10.1093/protein/4.8.891
Frerigmann, H., Berger, B., Gigolashvili, T. (2014). bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in. doi: 10.1104/pp.114.240887
Frerigmann, H., Piślewska-Bednarek, M., Sánchez-Vallet, A., Molina, A., Glawischnig, E., Gigolashvili, T., et al. (2016). Regulation of pathogen-triggered tryptophan metabolism in arabidopsis thaliana by MYB transcription factors and indole glucosinolate conversion products. Mol. Plant 9 (5), 682–695. doi: 10.1016/j.molp.2016.01.006
Friedman, M. (2002). Tomato glycoalkaloids: role in the plant and in the diet. J. Agric. Food Chem. 50 (21), 5751–5780. doi: 10.1021/jf020560c
Friedman, M. (2006). Potato glycoalkaloids and metabolites: roles in the plant and in the diet. J. Agric. Food Chem. 54 (23), 8655–8681. doi: 10.1021/jf061471t
Fu, R., Zhang, M., Zhao, Y., He, X., Ding, C., Wang, S., et al. (2017). Identification of salt tolerance-related microRNAs and their targets in maize (Zea mays l.) using high-throughput sequencing and degradome analysis. Front. Plant Sci. 8, 864.
Furihata, T., Maruyama, K., Fujita, Y., Umezawa, T., Yoshida, R., Shinozaki, K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. 103 (6), 1988–1993. doi: 10.1073/pnas.0505667103
Ghasemzadeh, A., Talei, D., Jaafar, H. Z., Juraimi, A. S., Mohamed, M. T. M., Puteh, A., et al (2016). Plant-growth regulators alter phytochemical constituents and pharmaceutical quality in Sweet potato (Ipomoea batatas L.). BMC complementary and alternative medicine 16, 1–13.
Gibbs, D. J., Conde, J. V., Berckhan, S., Prasad, G., Mendiondo, G. M., Holdsworth, M. J. (2015). Group VII ethylene response factors coordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol. 169 (1), 23–31. doi: 10.1104/pp.15.00338
Golldack, D., Li, C., Mohan, H., Probst, N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5, 151. doi: 10.3389/fpls.2014.00151
Gong, Y., Hounsa, A., Egal, S., Turner, P. C., Sutcliffe, A. E., Hall, A. J., et al. (2004). Postweaning exposure to aflatoxin results in impaired child growth: a longitudinal study in Benin, West Africa. Environ. Health Perspect. 112 (13), 1334–1338. doi: 10.1289/ehp.6954
Gonzalez, A., Zhao, M., Leavitt, J. M., Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in arabidopsis seedlings. Plant J. 53 (5), 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Griffith, M., Yaish, M. W. (2004). Antifreeze proteins in overwintering plants: a tale of two activities. Trends Plant Sci. 9 (8), 399–405. doi: 10.1016/j.tplants.2004.06.007
Grover, A., Mittal, D., Negi, M., Lavania, D. (2013). Generating high temperature tolerant transgenic plants: achievements and challenges. Plant Sci. 205, 38–47. doi: 10.1016/j.plantsci.2013.01.005
Grunewald, W., De Smet, I., Lewis, D. R., Löfke, C., Jansen, L., Goeminne, G., et al. (2012). Transcription factor WRKY23 assists auxin distribution patterns during arabidopsis root development through local control on flavonol biosynthesis. Proc. Natl. Acad. Sci. 109 (5), 1554–1559. doi: 10.1073/pnas.1121134109
Guillaumie, S., Mzid, R., Méchin, V., Léon, C., Hichri, I., Destrac-Irvine, A., et al. (2010). The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol. Biol. 72 (1), 215–234. doi: 10.1007/s11103-009-9563-1
Guo, W., Jin, L., Miao, Y., He, X., Hu, Q., Guo, K., et al. (2016). An ethylene response-related factor, GbERF1-like, from gossypium barbadense improves resistance to verticillium dahliae via activating lignin synthesis. Plant Mol. Biol. 91 (3), 305–318. doi: 10.1007/s11103-016-0467-6
Guo, R., Yu, F., Gao, Z., An, H., Cao, X., Guo, X. (2011). GhWRKY3, a novel cotton (Gossypium hirsutum l.) WRKY gene, is involved in diverse stress responses. Mol. Biol. Rep. 38 (1), 49–58. doi: 10.1007/s11033-010-0076-4
Guo, D., Zhang, J., Wang, X., Han, X., Wei, B., Wang, J., et al. (2015). The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in arabidopsis. Plant Cell 27 (11), 3112–3127. doi: 10.1105/tpc.15.00829
Hamamouch, N., Winkel, B. S., Li, C., Davis, E. L. (2020). Modulation of Arabidopsis flavonol biosynthesis genes by cyst and root-knot nematodes. Plants 9 (2), 253. doi: 10.3390/plants9020253
Hassani, D., Fu, X., Shen, Q., Khalid, M., Rose, J. K., Tang, K. (2020). Parallel transcriptional regulation of artemisinin and flavonoid biosynthesis. Trends Plant Sci. 25 (5), 466–476. doi: 10.1016/j.tplants.2020.01.001
He, Y., Mao, S., Gao, Y., Zhu, L., Wu, D., Cui, Y., et al. (2016). Genome-wide identification and expression analysis of WRKY transcription factors under multiple stresses in Brassica napus. PloS One 11 (6), e0157558. doi: 10.1371/journal.pone.0157558
Hu, Y., Chen, L., Ha, S., Gross, B., Falcone, B., Walker, D., et al. (2003). Crystal structure of the MurG: UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc. Natl. Acad. Sci. 100 (3), 845–849. doi: 10.1073/pnas.0235749100
Huang, Y., Li, M. Y., Wu, P., Xu, Z. S., Que, F., Wang, F., et al. (2016). Members of WRKY group III transcription factors are important in TYLCV defense signaling pathway in tomato (Solanum lycopersicum). BMC Genomics 17 (1), 1–18. doi: 10.1186/s12864-016-3123-2
Huang, Y. F., Vialet, S., Guiraud, J. L., Torregrosa, L., Bertrand, Y., Cheynier, V., et al. (2014). A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol. 201 (3), 795–809. doi: 10.1111/nph.12557
Humphreys, J. M., Hemm, M. R., Chapple, C. (1999). New routes for lignin biosynthesis defined by biochemical characterization of recombinant ferulate 5-hydroxylase, a multifunctional cytochrome P450-dependent monooxygenase. Proc. Natl. Acad. Sci. 96 (18), 10045–10050. doi: 10.1073/pnas.96.18.10045
Ibraheem, F., Gaffoor, I., Tan, Q., Shyu, C. R., Chopra, S. (2015). A sorghum MYB transcription factor induces 3-deoxyanthocyanidins and enhances resistance against leaf blights in maize. Molecules 20 (2), 2388–2404. doi: 10.3390/molecules20022388
Inoue, H., Hayashi, N., Matsushita, A., Xinqiong, L., Nakayama, A., Sugano, S., et al. (2013). Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein–protein interaction. Proc. Natl. Acad. Sci. 110 (23), 9577–9582. doi: 10.1073/pnas.1222155110
Iqbal, Z., Iqbal, M. S., Hashem, A., E.F. and Ansari, M. I. (2021). Plant defense responses to biotic stress and its interplay with fluctuating dark/light conditions. Front. Plant Sci. 12, 631810. doi: 10.3389/fpls.2021.631810
Irmer, S., Podzun, N., Langel, D., Heidemann, F., Kaltenegger, E., Schemmerling, B., et al. (2015). New aspect of plant–rhizobia interaction: alkaloid biosynthesis in crotalaria depends on nodulation. Proc. Natl. Acad. Sci. 112 (13), 4164–4169. doi: 10.1073/pnas.1423457112
Ishiguro, K. I., Kim, J., Fujiyama-Nakamura, S., Kato, S., Watanabe, Y. (2011). A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep. 12 (3), 267–275. doi: 10.1038/embor.2011.2
Itkin, M., Heinig, U., Tzfadia, O., Bhide, A. J., Shinde, B., Cardenas, P. D., et al. (2013). Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341 (6142), 175–179.
Jahan, M. A., Harris, B., Lowery, M., Coburn, K., Infante, A. M., Percifield, R. J., et al. (2019). The NAC family transcription factor GmNAC42–1 regulates biosynthesis of the anticancer and neuroprotective glyceollins in soybean. BMC Genomics 20 (1), 1–21. doi: 10.1186/s12864-019-5524-5
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in arabidopsis. Trends Plant Sci. 7 (3), 106–111. doi: 10.1016/S1360-1385(01)02223-3
Jamwal, K., Bhattacharya, S., Puri, S. (2018). Plant growth regulator mediated consequences of secondary metabolites in medicinal plants. J. Appl. Res. medicinal aromatic Plants 9, 26–38. doi: 10.1016/j.jarmap.2017.12.003
Jan, R., Aaqil Khan, M., Asaf, S., Park, J. R., Lee, I. J., Kim, K. M. (2021a). Flavonone 3-hydroxylase relieves bacterial leaf blight stress in rice via overaccumulation of antioxidant flavonoids and induction of defense genes and hormones. Int. J. Mol. Sci. 22 (11), 6152. doi: 10.3390/ijms22116152
Jan, R., Asaf, S., Numan, M., Kim, K. M. (2021b). Plant secondary metabolite biosynthesis and transcriptional regulation in response to biotic and abiotic stress conditions. Agronomy 11 (5), 968. doi: 10.3390/agronomy11050968
Javed, T., Shabbir, R., Ali, A., Afzal, I., Zaheer, U., Gao, S. J. (2020). Transcription factors in plant stress responses: Challenges and potential for sugarcane improvement. Plants 9 (4), 491. doi: 10.3390/plants9040491
Jeandet, P., Clément, C., Cordelier, S. (2019). Regulation of resveratrol biosynthesis in grapevine: new approaches for disease resistance? J. Exp. Bot. 70 (2), 375–378. doi: 10.1093/jxb/ery446
Jeyaraj, A., Liu, S., Zhang, X., Zhang, R., Shangguan, M., Wei, C. (2017). Genome-wide identification of microRNAs responsive to ectropis oblique feeding in tea plant (Camellia sinensis l.). Sci. Rep. 7 (1), 1–16. doi: 10.1038/s41598-017-13692-7
Jha, U. C., Bohra, A., Jha, R., Parida, S. K. (2019). Salinity stress response and ‘omics’ approaches for improving salinity stress tolerance in major grain legumes. Plant cell reports 38, 255–277.
Jha, U. C., Bohra, A., Nayyar, H. (2020). Advances in “omics” approaches to tackle drought stress in grain legumes. Plant Breed. 139 (1), 1–27.
Jha, U. C., Nayyar, H., Parida, S. K., Siddique, K. H. (2022). “Horse gram, an underutilized climate-resilient legume: Breeding and genomic approach for improving future genetic gain,” in Developing climate resilient grain and forage legumes (Singapore: Springer), 167–178.
Jia, L., Clegg, M. T., Jiang, T. (2004). Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 134 (2), 575–585. doi: 10.1104/pp.103.027201
Jiang, X., Cao, Y., Wang, Y., Liu, L., Shen, F., Wang, R. (2010). A unique approach to the concise synthesis of highly optically active spirooxazolines and the discovery of a more potent oxindole-type phytoalexin analogue. J. Am. Chem. Soc. 132 (43), 15328–15333. doi: 10.1021/ja106349m
Jiang, S., Cui, J. L., Li, X. K. (2021). MicroRNA-mediated gene regulation of secondary metabolism in plants. Crit. Rev. Plant Sci. 40 (6), 459–478. doi: 10.1080/07352689.2022.2031674
Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J., Zhang, J. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59 (2), 86–101. doi: 10.1111/jipb.12513
Jiang, J., Xi, H., Dai, Z., Lecourieux, F., Yuan, L., Liu, X., et al. (2019). VvWRKY8 represses stilbene synthase genes through direct interaction with VvMYB14 to control resveratrol biosynthesis in grapevine. J. Exp. Bot. 70 (2), 715–729. doi: 10.1093/jxb/ery401
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., et al. (2016). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, gkw982. doi: 10.1093/nar/gkw982
Joaquin, Á., Watson, R. J. (2003). Cell cycle regulation by the b-myb transcription factor. Cell. Mol. Life Sci. CMLS 60 (11), 2389–2401. doi: 10.1007/s00018-003-3037-4
Jones-Rhoades, M. W., Bartel, D. P., Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. doi: 10.1146/annurev.arplant.57.032905.105218
Joo, J., Lee, Y. H., Song, S. I. (2019). OsbZIP42 is a positive regulator of ABA signaling and confers drought tolerance to rice. Planta 249 (5), 1521–1533. doi: 10.1007/s00425-019-03104-7
Kage, U., Yogendra, K. N., Kushalappa, A. C. (2017). TaWRKY70 transcription factor in wheat QTL-2DL regulates downstream metabolite biosynthetic genes to resist Fusarium graminearum infection spread within spike. Sci. Rep. 7 (1), 1–14. doi: 10.1038/srep42596
Karre, S., Kumar, A., Yogendra, K., Kage, U., Kushalappa, A., Charron, J. B. (2019). HvWRKY23 regulates flavonoid glycoside and hydroxycinnamic acid amide biosynthetic genes in barley to combat fusarium head blight. Plant Mol. Biol. 100 (6), 591–605. doi: 10.1007/s11103-019-00882-2
Kato-Noguchi, H., Takeshita, S. (2013). Contribution of a phytotoxic compound to the allelopathy of ginkgo biloba. Plant Signaling Behav. 8 (11), e26999. doi: 10.4161/psb.26999
Kavas, M., Kizildogan, A., Gökdemir, G., Baloglu, M. C. (2015). Genome-wide investigation and expression analysis of AP2-ERF gene family in salt tolerant common bean. EXCLI J. 14, 1187. doi: 10.17179/excli2015-600
Kettles, G. J., Drurey, C., Schoonbeek, H. J., Maule, A. J., Hogenhout, S. A. (2013). Resistance of a rabidopsis thaliana to the green peach aphid, m yzus persicae, involves camalexin and is regulated by micro RNA s. New Phytol. 198 (4), 1178–1190. doi: 10.1111/nph.12218
Khan, S., Ali, A., Saifi, M., Saxena, P., Ahlawat, S., Abdin, M. Z. (2020). Identification and the potential involvement of miRNAs in the regulation of artemisinin biosynthesis in A. annua. Sci. Rep. 10 (1), 1–11. doi: 10.1038/s41598-020-69707-3
Khare, S., Singh, N. B., Singh, A., Hussain, I., Niharika, K., Yadav, V., et al. (2020). Plant secondary metabolites synthesis and their regulations under biotic and abiotic constraints. J. Plant Biol. 63 (3), 203–216. doi: 10.1007/s12374-020-09245-7
Khraiwesh, B., Zhu, J. K., Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 1819 (2), 137–148. doi: 10.1016/j.bbagrm.2011.05.001
Kim, D. O., Jeong, S. W., Lee, C. Y. (2003). Antioxidant capacity of phenolic phytochemicals from various cultivars of plums. Food Chem. 81 (3), 321–326. doi: 10.1016/S0308-8146(02)00423-5
Kim, S. S., Ko, Y. J., Jang, J. Y., Lee, T., Lim, M. H., Park, S. Y., et al. (2008). Isolation and expression analysis of Brassica rapa WRKY 7. Plant Pathol. J. 24 (4), 478–481. doi: 10.5423/PPJ.2008.24.4.478
Kim, W. C., Ko, J. H., Kim, J. Y., Kim, J., Bae, H. J., Han, K. H. (2013). MYB 46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J. 73 (1), 26–36. doi: 10.1111/j.1365-313x.2012.05124.x
Kishi-Kaboshi, M., Seo, S., Takahashi, A., Hirochika, H. (2018). The MAMP-responsive MYB transcription factors MYB30, MYB55 and MYB110 activate the HCAA synthesis pathway and enhance immunity in rice. Plant Cell Physiol. 59 (5), 903–915. doi: 10.1093/pcp/pcy062
Kloth, K. J., Wiegers, G. L., Busscher-Lange, J., van Haarst, J. C., Kruijer, W., Bouwmeester, H. J., et al. (2016). AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. J. Exp. Bot. 67 (11), 3383–3396. doi: 10.1093/jxb/erw159
Koroban, N. V., Kudryavtseva, A. V., Krasnov, G. S., Sadritdinova, A. F., Fedorova, M. S., Snezhkina, A. V., et al. (2016). The role of microRNA in abiotic stress response in plants. Mol. Biol. 50 (3), 337–343. doi: 10.1134/S0026893316020102
Kumar, R. (2014). Role of microRNAs in biotic and abiotic stress responses in crop plants. Appl. Biochem. Biotechnol. 174 (1), 93–115. doi: 10.1007/s12010-014-0914-2
Kumar, S., Pandey, A. K. (2013). Chemistry and biological activities of flavonoids: an overview. Sci. World J, 2013.
Kumar, N., Srivastava, P., Vishwakarma, K., Kumar, R., Kuppala, H., Maheshwari, S. K., et al. (2020). The rhizobium–plant symbiosis: state of the art. Plant Microbe symbiosis, 1–20. doi: 10.1007/978-3-030-36248-5_1
Lai, D., Pičmanová, M., Abou Hachem, M., Motawia, M. S., Olsen, C. E., Møller, B. L., et al. (2015). Lotus japonicus flowers are defended by a cyanogenic β-glucosidase with highly restricted expression to essential reproductive organs. Plant Mol. Biol. 89 (1), 21–34. doi: 10.1007/s11103-015-0348-4
Landschulz, W. H., Johnson, P. F., McKnight, S. L. (1988). The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240 (4860), 1759–1764.
Lanz, T., Tropf, S., Marner, F. J., Schröder, J., Schröder, G. (1991). The role of cysteines in polyketide synthases. site-directed mutagenesis of resveratrol and chalcone synthases, two key enzymes in different plant-specific pathways. J. Biol. Chem. 266 (15), 9971–9976.
Lewis, W. J., Van Lenteren, J. C., Phatak, S. C., Tumlinson Iii, J. H. (1997). A total system approach to sustainable pest management. Proc. Natl. Acad. Sci. 94 (23), 12243–12248. doi: 10.1073/pnas.94.23.12243
Li, H., Deng, Y., Wu, T., Subramanian, S., Yu, O. (2010). Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 153 (4), 1759–1770. doi: 10.1104/pp.110.156950
Li, X., Fan, S., Hu, W., Liu, G., Wei, Y., He, C., et al. (2017). Two cassava basic leucine zipper (bZIP) transcription factors (MebZIP3 and MebZIP5) confer disease resistance against cassava bacterial blight. Front. Plant Sci. 8, 2110. doi: 10.3389/fpls.2017.02110
Li, H., Gao, Y., Xu, H., Dai, Y., Deng, D., Chen, J. (2013). ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis. Plant Growth Regul. 70 (3), 207–216. doi: 10.1007/s10725-013-9792-9
Li, D., He, Y., Li, S., Shi, S., Li, L., Liu, Y., et al. (2021). Genome-wide characterization and expression analysis of AP2/ERF genes in eggplant (Solanum melongena l.). Plant Physiol. Biochem. 167, 492–503. doi: 10.1016/j.plaphy.2021.08.006
Li, W., Pang, S., Lu, Z., Jin, B. (2020). Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 9 (11), p.1515. doi: 10.3390/plants9111515
Li, T., Wang, Y. H., Liu, J. X., Feng, K., Xu, Z. S., Xiong, A. S. (2019). Advances in genomic, transcriptomic, proteomic, and metabolomic approaches to study biotic stress in fruit crops. Crit. Rev. Biotechnol. 39 (5), 680–692. doi: 10.1080/07388551.2019.1608153
Li, Z. G., Xie, L. R., Li, X. J. (2015). Hydrogen sulfide acts as a downstream signal molecule in salicylic acid-induced heat tolerance in maize (Zea mays l.) seedlings. J. Plant Physiol. 177, 121–127. doi: 10.1016/j.jplph.2014.12.018
Li, M. Y., Xu, Z. S., Huang, Y., Tian, C., Wang, F., Xiong, A. S. (2015). Genome-wide analysis of AP2/ERF transcription factors in carrot (Daucus carota l.) reveals evolution and expression profiles under abiotic stress. Mol. Genet. Genomics 290 (6), 2049–2061.
Li, C., Zhang, B. (2016). MicroRNAs in control of plant development. J. Cell. Physiol. 231 (2), 303–313. doi: 10.1002/jcp.25125
Li, R., Zhang, J., Li, J., Zhou, G., Wang, Q., Bian, W., et al. (2015). Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. Elife 4, e04805. doi: 10.7554/eLife.04805.021
Li, J., Zhang, K., Meng, Y., Hu, J., Ding, M., Bian, J., et al. (2018). Jasmonic acid/ethylene signaling coordinates hydroxycinnamic acid amides biosynthesis through ORA 59 transcription factor. Plant J. 95 (3), 444–457. doi: 10.1111/tpj.13960
Liao, W., Yang, Y., Li, Y., Wang, G., Peng, M. (2016). Genome-wide identification of cassava R2R3 MYB family genes related to abscission zone separation after environmental-stress-induced abscission. Sci. Rep. 6 (1), 1–12. doi: 10.1038/srep32006
Liao, Y., Zou, H. F., Wei, W., Hao, Y. J., Tian, A. G., Huang, J., et al. (2008). Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic arabidopsis. Planta 228 (2), 225–240. doi: 10.1007/s00425-008-0731-3
Licausi, F., Ohme-Takagi, M., Perata, P. (2013). APETALA 2/Ethylene responsive factor (AP 2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 199 (3), 639–649. doi: 10.1111/nph.12291
Lima, J. C. D., Loss-Morais, G., Margis, R. (2012). MicroRNAs play critical roles during plant development and in response to abiotic stresses. Genet. Mol. Biol. 35, 1069–1077. doi: 10.1590/S1415-47572012000600023
Lin, Y., Qasim, M., Hussain, M., Akutse, K. S., Avery, P. B., C.K. and Wang, L. (2017). The herbivore-induced plant volatiles methyl salicylate and menthol positively affect growth and pathogenicity of entomopathogenic fungi. Sci. Rep. 7 (1), 1–11. doi: 10.1038/srep40494
Liu, J., Li, J., Wang, H., Fu, Z., Liu, J., Yu, Y., et al (2011). Identification and expression analysis of ERF transcription factor genes in petunia during flower senescence and in response to hormone treatments. Experimental Botany 62 (2), 825–840.
Liu, J., Chen, X., Liang, X., Zhou, X., Yang, F., Liu, J., et al. (2016). Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol. 171 (2), 1427–1442. doi: 10.1104/pp.15.01921
Liu, D., Chen, X., Liu, J., Ye, J., Guo, Z. (2012). The rice ERF transcription factor OsERF922 negatively regulates resistance to magnaporthe oryzae and salt tolerance. J. Exp. Bot. 63 (10), 3899–3911. doi: 10.1093/jxb/ers079
Liu, C., Long, J., Zhu, K., Liu, L., Yang, W., Zhang, H., et al. (2016). Characterization of a citrus R2R3-MYB transcription factor that regulates the flavonol and hydroxycinnamic acid biosynthesis. Sci. Rep. 6 (1), 1–16. doi: 10.1038/srep25352
Liu, J., Osbourn, A., Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8 (5), 689–708. doi: 10.1016/j.molp.2015.03.012
Liu, Q., Yan, S., Huang, W., Yang, J., Dong, J., Zhang, S., et al. (2018). NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol. Biol. 98 (4), 289–302. doi: 10.1007/s11103-018-0768-z
López-Galiano, M. J., González-Hernández, A. I., Crespo-Salvador, O., Rausell, C., Real, M. D., Escamilla, M., et al. (2018). Epigenetic regulation of the expression of WRKY75 transcription factor in response to biotic and abiotic stresses in solanaceae plants. Plant Cell Rep. 37 (1), 167–176. doi: 10.1007/s00299-017-2219-8
Lorenzo, O., Piqueras, R., Sánchez-Serrano, J. J., Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 (1), 165–178. doi: 10.1105/tpc.007468
Ludwig, S. R., Habera, L. F., Dellaporta, S. L., Wessler, S. (1989). Lc, a member of the maize r gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. 86 (18), 7092–7096. doi: 10.1073/pnas.86.18.7092
Ma, J., Lu, J., Xu, J., Duan, B., He, X., Liu, J. (2015). Genome-wide identification of WRKY genes in the desert poplar Populus euphratica and adaptive evolution of the genes in response to salt stress. Evolutionary Bioinf. 11, EBO–S22067. doi: 10.4137/EBO.S22067
Mahmood, K., Xu, Z., El-Kereamy, A., Casaretto, J. A., Rothstein, S. J. (2016). The arabidopsis transcription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses. Front. Plant Sci. 7, 1548. doi: 10.3389/fpls.2016.01548
Maillet, A., Bouju-Albert, A., Roblin, S., Vaissié, P., Leuillet, S., Dousset, X., et al. (2021). Impact of DNA extraction and sampling methods on bacterial communities monitored by 16S rDNA metabarcoding in cold-smoked salmon and processing plant surfaces. Food Microbiol. 95, 103705. doi: 10.1016/j.fm.2020.103705
Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., Zhang, S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in. Arabidopsis. Plant Cell 23 (4), 1639–1653. doi: 10.1105/tpc.111.084996
Marchive, C., Léon, C., Kappel, C., Coutos-Thévenot, P., Corio-Costet, M. F., Delrot, S., et al. (2013). Over-expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PloS One 8 (1), e54185. doi: 10.1371/journal.pone.0054185
Massari, M. E., Murre, C. (2000). Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20 (2), 429–440. doi: 10.1128/MCB.20.2.429-440.2000
Mazid, M., Khan, T. A., Mohammad, F. (2011). Role of secondary metabolites in defense mechanisms of plants. Biol. Med. 3 (2), 232–249.
Mazis, G. A., Sakellariou, V. I., Kontos, F., Zerva, L., Spyridonos, S. G. (2011). Recurrent fluctuant mass of the wrist and forearm associated with chronic tenosynovitis by Mycobacterium kansasii. Orthopedics 34 (5), e117–e120. doi: 10.3928/01477447-20110317-23
Meena, K. K., Sorty, A. M., Bitla, U. M., Choudhary, K., Gupta, P., Pareek, A., et al. (2017). Abiotic stress responses and microbe-mediated mitigation in plants: the omics strategies. Front. Plant Sci. 8, 172. doi: 10.3389/fpls.2017.00172
Menke, F. L., Champion, A., Kijne, J. W., Memelink, J. (1999). A novel jasmonate-and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene str interacts with a jasmonate-and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 18 (16), 4455–4463. doi: 10.1093/emboj/18.16.4455
Meraj, T. A., Fu, J., Raza, M. A., Zhu, C., Shen, Q., Xu, D., et al. (2020). Transcriptional factors regulate plant stress responses through mediating secondary metabolism. Genes 11 (4), p.346. doi: 10.3390/genes11040346
Mertens, J., Pollier, J., Vanden Bossche, R., Lopez-Vidriero, I., J.M. and Goossens, A. (2016). The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol. 170 (1), 194–210. doi: 10.1104/pp.15.01645
Meshnick, S. R., Taylor, T. E., Kamchonwongpaisan, S. (1996). Artemisinin and the antimalarial endoperoxides: from herbal remedy to targeted chemotherapy. Microbiological Rev. 60 (2), 301–315. doi: 10.1128/mr.60.2.301-315.1996
Mishra, B. B., Moura-Alves, P., Sonawane, A., Hacohen, N., Griffiths, G., Moita, L. F., et al. (2010). Mycobacterium tuberculosis protein ESAT-6 is a potent activator of the NLRP3/ASC inflammasome. Cell. Microbiol. 12 (8), 1046–1063. doi: 10.1111/j.1462-5822.2010.01450.x
Miyamoto, K., Nishizawa, Y., Minami, E., Nojiri, H., Yamane, H., Okada, K. (2015). Overexpression of the bZIP transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells. J. Plant Physiol. 173, 19–27. doi: 10.1016/j.jplph.2014.09.001
Mizoi, J., Shinozaki, K., Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 1819 (2), 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Moffat, C. S., Ingle, R. A., Wathugala, D. L., Saunders, N. J., Knight, H., Knight, M. R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against botrytis cinerea in arabidopsis. PloS One 7 (4), e35995. doi: 10.1371/journal.pone.0035995
Moulin, M., Deleu, C., Larher, F., Bouchereau, A. (2006). The lysine-ketoglutarate reductase–saccharopine dehydrogenase is involved in the osmo-induced synthesis of pipecolic acid in rapeseed leaf tissues. Plant Physiol. Biochem. 44 (7-9), 474–482. doi: 10.1016/j.plaphy.2006.08.005
Müller, M., Munné-Bosch, S. (2015). Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 169 (1), 32–41. doi: 10.1104/pp.15.00677
Naik, P. M., Al-Khayri, J. M. (2016). “Abiotic and biotic elicitors-role in secondary metabolites production through in vitro culture of medicinal plants,” in Abiotic and biotic stress in plants–recent advances and future perspectives (Rijeka: InTech), 247–277.
Nakano, T., Fujisawa, M., Shima, Y., Ito, Y. (2014). The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. J. Exp. Bot. 65 (12), 3111–3119. doi: 10.1093/jxb/eru154
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 1819 (2), 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nakayasu, M., Shioya, N., Shikata, M., Thagun, C., Abdelkareem, A., Okabe, Y., et al. (2018). JRE 4 is a master transcriptional regulator of defense-related steroidal glycoalkaloids in tomato. Plant J. 94 (6), 975–990. doi: 10.1111/tpj.13911
Nakayasu, M., Umemoto, N., Ohyama, K., Fujimoto, Y., Lee, H. J., Watanabe, B., et al. (2017). A dioxygenase catalyzes steroid 16α-hydroxylation in steroidal glycoalkaloid biosynthesis. Plant Physiol. 175 (1), 120–133. doi: 10.1104/pp.17.00501
Nemesio-Gorriz, M., Blair, P. B., Dalman, K., Hammerbacher, A., Arnerup, J., Stenlid, J., et al. (2017). Identification of Norway spruce MYB-bHLH-WDR transcription factor complex members linked to regulation of the flavonoid pathway. Front. Plant Sci. 8, 305. doi: 10.3389/fpls.2017.00305
Nguyen, D., Rieu, I., Mariani, C., van Dam, N. M. (2016). How plants handle multiple stresses: hormonal interactions underlying responses to abiotic stress and insect herbivory. Plant Mol. Biol. 91 (6), 727–740. doi: 10.1007/s11103-016-0481-8
Nguyen, N. H., Trotel-Aziz, P., Clément, C., Jeandet, P., Baillieul, F., Aziz, A. (2022). Camalexin accumulation as a component of plant immunity during interactions with pathogens and beneficial microbes. Planta 255 (6), 1–17. doi: 10.1007/s00425-022-03907-1
Nijhawan, A., Jain, M., Tyagi, A. K., Khurana, J. P. (2008). Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 146 (2), p.333. doi: 10.1104/pp.107.112821
Nisha, S. N., Prabu, G., Mandal, A. K. A. (2018). Biochemical and molecular studies on the resistance mechanisms in tea [Camellia sinensis (L.) o. kuntze] against blister blight disease. Physiol. Mol. Biol. Plants 24 (5), 867–880. doi: 10.1007/s12298-018-0565-9
Nolan, T., Chen, J., Yin, Y. (2017). Cross-talk of brassinosteroid signaling in controlling growth and stress responses. Biochem. J. 474 (16), 2641–2661. doi: 10.1042/BCJ20160633
Noman, A., Liu, Z., Aqeel, M., Zainab, M., Khan, M. I., Hussain, A., et al. (2017). Basic leucine zipper domain transcription factors: the vanguards in plant immunity. Biotechnol. Lett. 39 (12), 1779–1791. doi: 10.1007/s10529-017-2431-1
Nuruzzaman, M., Sharoni, A. M., Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4, 248. doi: 10.3389/fmicb.2013.00248
Obata, T. (2019). Metabolons in plant primary and secondary metabolism. Phytochem. Rev. 18 (6), 1483–1507. doi: 10.1007/s11101-019-09619-x
Ogata, K., Kanei-Ishii, C., Sasaki, M., Hatanaka, H., Nagadoi, A., Enari, M., et al. (1996). The cavity in the hydrophobic core of myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol. 3 (2), 178–187. doi: 10.1038/nsb0296-178
Ohama, N., Sato, H., Shinozaki, K., Yamaguchi-Shinozaki, K. (2017). Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22 (1), 53–65. doi: 10.1016/j.tplants.2016.08.015
Okada, A., Okada, K., Miyamoto, K., Koga, J., Shibuya, N., Nojiri, H., et al. (2009). OsTGAP1, a bZIP transcription factor, coordinately regulates the inductive production of diterpenoid phytoalexins in rice. J. Biol. Chem. 284 (39), 26510–26518. doi: 10.1074/jbc.M109.036871
Onkokesung, N., Reichelt, M., van Doorn, A., Schuurink, R. C., van Loon, J. J., Dicke, M. (2014). Modulation of flavonoid metabolites in arabidopsis thaliana through overexpression of the MYB75 transcription factor: role of kaempferol-3, 7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae. J. Exp. Bot. 65 (8), 2203–2217. doi: 10.1093/jxb/eru096
Owusu Adjei, M., Zhou, X., Mao, M., Rafique, F., Ma, J. (2021). MicroRNAs roles in plants secondary metabolism. Plant Signaling Behav. 16 (7), 1915590.
Pagano, L., Rossi, R., Paesano, L., Marmiroli, N., Marmiroli, M. (2015). miRNA regulation and stress adaptation in plants. Environmental and Experimental Botany 184, 104369.
Pagare, S., Bhatia, M., Tripathi, N., S. and Bansal, Y. K. (2015). Secondary metabolites of plants and their role: Overview. Curr. Trends Biotechnol. Pharm. 9 (3), 293–304.
Papadopoulou, K., Melton, R. E., Leggett, M., Daniels, M. J., Osbourn, A. E. (1999). Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. 96 (22), 12923–12928. doi: 10.1073/pnas.96.22.12923
Patra, B., Schluttenhofer, C., Wu, Y., Pattanaik, S., Yuan, L. (2013). Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 1829 (11), 1236–1247. doi: 10.1016/j.bbagrm.2013.09.006
Paul, P., Singh, S. K., Patra, B., Liu, X., Pattanaik, S., Yuan, L. (2020). Mutually regulated AP2/ERF gene clusters modulate biosynthesis of specialized metabolites in plants. Plant Physiol. 182 (2), 840–856. doi: 10.1104/pp.19.00772
Paul, P., Singh, S. K., Patra, B., Sui, X., Pattanaik, S., Yuan, L. (2017). A differentially regulated AP 2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in catharanthus roseus. New Phytol. 213 (3), 1107–1123. doi: 10.1111/nph.14252
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P. A., Saedler, H. (1987). The regulatory c1 locus of zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6 (12), 3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x
Peng, X. X., TANG, X. K., ZHOU, P. L., HU, Y. J., DENG, X. B., Yan, H. E., et al. (2011). Isolation and expression patterns of rice WRKY82 transcription factor gene responsive to both biotic and abiotic stresses. Agric. Sci. China 10 (6), 893–901. doi: 10.1016/S1671-2927(11)60074-6
Pérez-Rodríguez, P., Riano-Pachon, D. M., Corrêa, L. G. G., Rensing, S. A., Kersten, B., Mueller-Roeber, B. (2010). PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 38 (suppl_1), D822–D827. doi: 10.1093/nar/gkp805
Phukan, U. J., Jeena, G. S., Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7, 760. doi: 10.3389/fpls.2016.00760
Puranik, S., Sahu, P. P., Srivastava, P. S., Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17 (6), 369–381. doi: 10.1016/j.tplants.2012.02.004
Qian, Y., Zhang, T., Yu, Y., Gou, L., Yang, J., Xu, J., et al. (2021). Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses. Front. Plant Sci. 12, 1143. doi: 10.3389/fpls.2021.677611
Raffaele, S., Rivas, S., Roby, D. (2006). An essential role for salicylic acid in AtMYB30-mediated control of the hypersensitive cell death program in Arabidopsis. FEBS Lett. 580 (14), 3498–3504. doi: 10.1016/j.febslet.2006.05.027
Raineri, J., Wang, S., Peleg, Z., Blumwald, E., Chan, R. L. (2015). The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress. Plant Mol. Biol. 88 (4), 401–413. doi: 10.1007/s11103-015-0329-7
Rinerson, C. I., Rabara, R. C., Tripathi, P., Shen, Q. J., Rushton, P. J. (2015). The evolution of WRKY transcription factors. BMC Plant Biol. 15 (1), 1–18. doi: 10.1186/s12870-015-0456-y
Robert-Seilaniantz, A., MacLean, D., Jikumaru, Y., Hill, L., Yamaguchi, S., Kamiya, Y., et al. (2011). The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J. 67 (2), 218–231. doi: 10.1111/j.1365-313X.2011.04591.x
Rodriguez-Salazar, J., Moreno, S., Espín, G. (2017). LEA proteins are involved in cyst desiccation resistance and other abiotic stresses in Azotobacter vinelandii. Cell Stress Chaperones 22 (3), 397–408. doi: 10.1007/s12192-017-0781-1
Roepke, J., Salim, V., Wu, M., Thamm, A. M., Murata, J., Ploss, K., et al. (2010). Vinca drug components accumulate exclusively in leaf exudates of Madagascar periwinkle. Proc. Natl. Acad. Sci. 107 (34), 15287–15292. doi: 10.1073/pnas.0911451107
Roux, J., Ham, H. (2020). International year of plant health: a focus on tree health. South. Forests: J. For. Sci. 82 (3), iii–iiv. doi: 10.2989/20702620.2020.1852790
Roy, A., Khan, A., Ahmad, I., Alghamdi, S., Rajab, B. S., Babalghith, A. O., et al. (2022). Flavonoids a bioactive compound from medicinal plants and its therapeutic applications. BioMed. Res. Int. 2022.
Ruiz-Ferrer, V., Voinnet, O. (2009). Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60 (1), 485–510. doi: 10.1146/annurev.arplant.043008.092111
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15 (5), 247–258. doi: 10.1016/j.tplants.2010.02.006
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10 (1), 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Saad, A. S. I., Li, X., Li, H. P., Huang, T., Gao, C. S., Guo, M. W., et al. (2013). A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci. 203, 33–40. doi: 10.1016/j.plantsci.2012.12.016
Saga, H., Ogawa, T., Kai, K., Suzuki, H., Ogata, Y., Sakurai, N., et al. (2012). Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. Mol. Plant-Microbe Interact. 25 (5), 684–696. doi: 10.1094/MPMI-09-11-0244
Samad, A. F., Sajad, M., Nazaruddin, N., Fauzi, I. A., Murad, A. M., Zainal, Z., et al. (2017). MicroRNA and transcription factor: key players in plant regulatory network. Front. Plant Sci. 8, 565. doi: 10.3389/fpls.2017.00565
Samira, R., Li, B., Kliebenstein, D., Li, C., Davis, E., Gillikin, J. W., et al. (2018). The bHLH transcription factor ILR3 modulates multiple stress responses in arabidopsis. Plant Mol. Biol. 97 (4), 297–309. doi: 10.1007/s11103-018-0735-8
Sang, M. K., Kim, J. G., Kim, K. D. (2010). Biocontrol activity and induction of systemic resistance in pepper by compost water extracts against. Phytophthora capsici. Phytopathol. 100 (8), 774–783. doi: 10.1094/PHYTO-100-8-0774
Schluttenhofer, C., Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167 (2), 295–306. doi: 10.1104/pp.114.251769
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. nature 463 (7278), 178–183.
Sears, M. T., Zhang, H., Rushton, P. J., Wu, M., Han, S., Spano, A. J., et al. (2014). NtERF32: a non-NIC2 locus AP2/ERF transcription factor required in jasmonate-inducible nicotine biosynthesis in tobacco. Plant Mol. Biol. 84 (1), 49–66. doi: 10.1007/s11103-013-0116-2
Seca, A. M., Pinto, D. C. (2019). Biological potential and medical use of secondary metabolites. Medicines 6 (2), 66. doi: 10.3390/medicines6020066
Šečić, E., Kogel, K. H., Ladera-Carmona, M. J. (2021). Biotic stress-associated microRNA families in plants. J. Plant Physiol. 263, 153451. doi: 10.1016/j.jplph.2021.153451
Seo, Y. J., Park, J. B., Cho, Y. J., Jung, C., Seo, H. S., Park, S. K., et al. (2010). Overexpression of the ethylene-responsive factor gene BrERF4 from brassica rapa increases tolerance to salt and drought in Arabidopsis plants. Molecules Cells 30 (3), 271–277. doi: 10.1007/s10059-010-0114-z
Shah, A., Smith, D. L. (2020). Flavonoids in agriculture: Chemistry and roles in, biotic and abiotic stress responses, and microbial associations. Agronomy 10 (8), p.1209. doi: 10.3390/agronomy10081209
Shakya, A. K. (2016). Medicinal plants: Future source of new drugs. Int. J. Herbal Med. 4 (4), 59–64.
Shen, Q., Huang, H., Zhao, Y., Xie, L., He, Q., Zhong, Y., et al. (2019). The transcription factor Aabzip9 positively regulates the biosynthesis of artemisinin in artemisia annua. Front. Plant Sci. 10, 1294. doi: 10.3389/fpls.2019.01294
Shen, Y., Sun, T., Pan, Q., Anupol, N., Chen, H., Shi, J., et al. (2019). Rr MYB 5-and rr MYB 10-regulated flavonoid biosynthesis plays a pivotal role in feedback loop responding to wounding and oxidation in Rosa rugosa. Plant Biotechnol. J. 17 (11), 2078–2095. doi: 10.1111/pbi.13123
Shen, X. J., Wang, Y. Y., Zhang, Y. X., Guo, W., Jiao, Y. Q., Zhou, X. A. (2018). Overexpression of the wild soybean R2R3-MYB transcription factor GsMYB15 enhances resistance to salt stress and Helicoverpa armigera in transgenic Arabidopsis. Int. J. Mol. Sci. 19 (12), p.3958. doi: 10.3390/ijms19123958
Shoji, T., Hashimoto, T. (2011). Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol. 52 (6), 1117–1130. doi: 10.1093/pcp/pcr063
Shoji, T., Kajikawa, M., Hashimoto, T. (2010). Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22 (10), 3390–3409. doi: 10.1105/tpc.110.078543
Singh, A. K., Kumar, S. R., Dwivedi, V., Rai, A., Pal, S., Shasany, A. K., et al. (2017). A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 215 (3), 1115–1131. doi: 10.1111/nph.14663
Sita, K., Sehgal, A., Hanumantha Rao, B., Nair, R. M., Vara Prasad, P. V., Kumar, S., et al. (2017). Food legumes and rising temperatures: effects, adaptive functional mechanisms specific to reproductive growth stage and strategies to improve heat tolerance. Front. Plant Sci. 8, 1658. doi: 10.3389/fpls.2017.01658
Sivankalyani, V., Feygenberg, O., Diskin, S., Wright, B., Alkan, N. (2016). Increased anthocyanin and flavonoids in mango fruit peel are associated with cold and pathogen resistance. Postharvest Biol. Technol. 111, 132–139. doi: 10.1016/j.postharvbio.2015.08.001
Skibbe, M., Qu, N., Galis, I., Baldwin, I. T. (2008). Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 20 (7), 1984–2000. doi: 10.1105/tpc.108.058594
Spicher, L., Almeida, J., Gutbrod, K., Pipitone, R., Dörmann, P., Glauser, G., et al. (2017). Essential role for phytol kinase and tocopherol in tolerance to combined light and temperature stress in tomato. J. Exp. Bot. 68 (21-22), 5845–5856. doi: 10.1093/jxb/erx356
Stella, N. A., Brothers, K. M., Callaghan, J. D., Passerini, A. M., Sigindere, C., Hill, P. J., et al. (2018). An IgaA/UmoB family protein from serratia marcescens regulates motility, capsular polysaccharide biosynthesis, and secondary metabolite production. Appl. Environ. Microbiol. 84 (6), e02575–e02517. doi: 10.1128/AEM.02575-17
Steppuhn, A., Gase, K., Krock, B., Halitschke, R., Baldwin, I. T., Levine, M. (2004). Nicotine's defensive function in nature. PloS Biol. 2 (8), e217. doi: 10.1371/journal.pbio.0020217
Sun, X. C., Gao, Y. F., Li, H. R., Yang, S. Z., Liu, Y. S. (2015). Over-expression of SlWRKY39 leads to enhanced resistance to multiple stress factors in tomato. J. Plant Biol. 58 (1), 52–60. doi: 10.1007/s12374-014-0407-4
Sun, X., Wang, Y., Sui, N. (2018). Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 503 (2), 397–401. doi: 10.1016/j.bbrc.2018.07.123
Tajammal, A., Siddiqa, A., Irfan, A., Azam, M., Hafeez, H., Munawar, M. A., et al. (2022). Antioxidant, molecular docking and computational investigation of new flavonoids. J. Mol. Structure 1254, 132189. doi: 10.1016/j.molstruc.2021.132189
Takahashi, R., Yamagishi, N., Yoshikawa, N. (2013). A MYB transcription factor controls flower color in soybean. J. Heredity 104 (1), 149–153. doi: 10.1093/jhered/ess081
Thagun, C., Imanishi, S., Kudo, T., Nakabayashi, R., Ohyama, K., Mori, T., et al. (2016). Jasmonate-responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant Cell Physiol. 57 (5), 961–975. doi: 10.1093/pcp/pcw067
Thomma, B. P., Cammue, B. P., Thevissen, K. (2002). Plant defensins. Planta 216 (2), 193–202. doi: 10.1007/s00425-002-0902-6
Tian, Z., He, Q., Wang, H., Liu, Y., Zhang, Y., Shao, F., et al. (2015). The potato ERF transcription factor StERF3 negatively regulates resistance to Phytophthora infestans and salt tolerance in potato. Plant Cell Physiol. 56 (5), 992–1005. doi: 10.1093/pcp/pcv025
Tirumalai, V., Swetha, C., Nair, A., Pandit, A., Shivaprasad, P. V. (2019). miR828 and miR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes. J. Exp. Bot. 70 (18), 4775–4792. doi: 10.1093/jxb/erz264
Tiwari, R., Rana, C. S. (2015). Phytomedicine for the diabetes: A traditional approach. Ann. Phytomed 4 (1), 108–110.
Todd, A. T., Liu, E., Polvi, S. L., Pammett, R. T., Page, J. E. (2010). A functional genomics screen identifies diverse transcription factors that regulate alkaloid biosynthesis in Nicotiana benthamiana. Plant J. 62 (4), 589–600. doi: 10.1111/j.1365-313X.2010.04186.x
Toledo-Ortiz, G., Huq, E., Quail, P. H. (2003). The arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 (8), 1749–1770. doi: 10.1105/tpc.013839
Tripathi, P., Rabara, R. C., Rushton, P. J. (2014). A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239 (2), 255–266. doi: 10.1007/s00425-013-1985-y
Turck, F., Zhou, A., Somssich, I. E. (2004). Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related gene PcPR1-1 in parsley. Plant Cell 16 (10), 2573–2585. doi: 10.1105/tpc.104.024810
Tweneboah, S., Oh, S. K. (2017). Biological roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in solanaceous crops. J. Plant Biotechnol. 44 (1), 1–11. doi: 10.5010/JPB.2017.44.1.001
Ülker, B., Somssich, I. E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7 (5), 491–498. doi: 10.1016/j.pbi.2004.07.012
Van den Broeck, L., Dubois, M., Vermeersch, M., Storme, V., Matsui, M., Inzé, D. (2017). From network to phenotype: the dynamic wiring of an Arabidopsis transcriptional network induced by osmotic stress. Mol. Syst. Biol. 13 (12), p.961. doi: 10.15252/msb.20177840
van der Fits, L., Memelink, J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289 (5477), 295–297. doi: 10.1126/science.289.5477.295
van Verk, M. C., Pappaioannou, D., Neeleman, L., Bol, J. F., Linthorst, H. J. (2008). A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 146 (4), 1983–1995. doi: 10.1104/pp.107.112789
Venditti, A., Bianco, A. (2020). Sulfur-containing secondary metabolites as neuroprotective agents. Curr. medicinal Chem. 27 (26), 4421–4436. doi: 10.2174/0929867325666180912105036
Verdier, J., Zhao, J., Torres-Jerez, I., Ge, S., Liu, C., He, X., et al. (2012). MtPAR MYB transcription factor acts as an on-switch for proanthocyanidin biosynthesis in Medicago truncatula. Proc. Natl. Acad. Sci. 109 (5), 1766–1771. doi: 10.1073/pnas.1120916109
Verma, V., Ravindran, P., Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16 (1), 1–10. doi: 10.1186/s12870-016-0771-y
Verpoorte, R. (1998). Exploration of nature's chemodiversity: the role of secondary metabolites as leads in drug development. Drug Discovery Today 3 (5), 232–238. doi: 10.1016/S1359-6446(97)01167-7
Wang, H., Avci, U., Nakashima, J., Hahn, M. G., Chen, F., Dixon, R. A. (2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. 107 (51), 22338–22343. doi: 10.1073/pnas.1016436107
Wang, L., Ran, L., Hou, Y., Tian, Q., Li, C., Liu, R., et al. (2017). The transcription factor MYB115 contributes to the regulation of proanthocyanidin biosynthesis and enhances fungal resistance in poplar. New Phytol. 215 (1), 351–367. doi: 10.1111/nph.14569
Wang, Y., Shu, Z., Wang, W., Jiang, X., Li, D., Pan, J., et al. (2016). CsWRKY2, a novel WRKY gene from camellia sinensis, is involved in cold and drought stress responses. Biol. plantarum 60 (3), 443–451. doi: 10.1007/s10535-016-0618-2
Wang, Z., Yu, Q., Shen, W., El Mohtar, C. A., Zhao, X., Gmitter, F. G. (2018). Functional study of CHS gene family members in citrus revealed a novel CHS gene affecting the production of flavonoids. BMC Plant Biol. 18 (1), 1–13.
Wang, J., Wang, X. (2019). Structural equation modeling: Applications using mplus (John Wiley & Sons).
Wang, L., Zhu, W., Fang, L., Sun, X., Su, L., Liang, Z., et al. (2014). Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 14 (1), 1–14. doi: 10.1186/1471-2229-14-103
Wani, S. H., Tripathi, P., Zaid, A., Challa, G. S., Kumar, A., Kumar, V., et al. (2018). Transcriptional regulation of osmotic stress tolerance in wheat (Triticum aestivum l.). Plant Mol. Biol. 97 (6), 469–487. doi: 10.1007/s11103-018-0761-6
Winkel, B. S. (2004). Metabolic channeling in plants. Annu. Rev. Plant Biol. 55, 85. doi: 10.1146/annurev.arplant.55.031903.141714
Xia, R., Zhu, H., An, Y. Q., Beers, E. P., Liu, Z. (2012). Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 13 (6), 1–18. doi: 10.1186/gb-2012-13-6-r47
Xiao, R., Zhang, C., Guo, X., Li, H., Lu, H. (2021). MYB transcription factors and its regulation in secondary cell wall formation and lignin biosynthesis during xylem development. Int. J. Mol. Sci. 22 (7), p.3560. doi: 10.3390/ijms22073560
Xie, Z., Nolan, T., Jiang, H., Tang, B., Zhang, M., Li, Z., et al. (2019). The AP2/ERF transcription factor TINY modulates brassinosteroid-regulated plant growth and drought responses in arabidopsis. Plant Cell 31 (8), 1788–1806. doi: 10.1105/tpc.18.00918
Xing, D. H., Lai, Z. B., Zheng, Z. Y., Vinod, K. M., Fan, B. F., Chen, Z. X. (2008). Stress-and pathogen-induced arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol. Plant 1 (3), 459–470. doi: 10.1093/mp/ssn020
Xu, Y. H., Wang, J. W., Wang, S., Wang, J. Y., Chen, X. Y. (2004). Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-a. Plant Physiol. 135 (1), 507–515. doi: 10.1104/pp.104.038612
Xu, S., Yao, S., Huang, R., Tan, Y., Huang, D. (2020). Transcriptome-wide analysis of the AP2/ERF transcription factor gene family involved in the regulation of gypenoside biosynthesis in gynostemma pentaphyllum. Plant Physiol. Biochem. 154, 238–247. doi: 10.1016/j.plaphy.2020.05.040
Yadav, M., Singh, IK, Singh, A. (2022). Dhurrin: A naturally occurring phytochemical as a weapon against insect herbivores. Phytochemistry (2022). 205, 113483. doi: 10.1016/j.phytochem.2022.113483
Yamagishi, M., Shimoyamada, Y., Nakatsuka, T., Masuda, K. (2010). Two R2R3-MYB genes, homologs of petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol. 51 (3), 463–474. doi: 10.1093/pcp/pcq011
Yamamura, C., Mizutani, E., Okada, K., Nakagawa, H., Fukushima, S., Tanaka, A., et al. (2015). Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. Plant J. 84 (6), 1100–1113. doi: 10.1111/tpj.13065
Yamamura, Y., Taguchi, Y., Ichitani, K., Umebara, I., Ohshita, A., Kurosaki, F., et al. (2018). Characterization of ent-kaurene synthase and kaurene oxidase involved in gibberellin biosynthesis from Scoparia dulcis. J. Natural Medicines 72 (2), 456–463. doi: 10.1007/s11418-017-1168-4
Yamasaki, K., Kigawa, T., Inoue, M., Tateno, M., Yamasaki, T., Yabuki, T., et al. (2005). Solution structure of an arabidopsis WRKY DNA binding domain. Plant Cell 17 (3), 944–956. doi: 10.1105/tpc.104.026435
Yan, Y., Jia, H., Wang, F., Wang, C., Liu, S., Guo, X. (2015). Overexpression of GhWRKY27a reduces tolerance to drought stress and resistance to Rhizoctonia solani infection in transgenic nicotiana benthamiana. Front. Physiol. 6, 265. doi: 10.3389/fphys.2015.00265
Yang, T., Xue, L., An, L. (2007). Functional diversity of miRNA in plants. Plant Sci. 172 (3), 423–432. doi: 10.1016/j.plantsci.2006.10.009
Yang, Y., Yu, T. F., Ma, J., Chen, J., Zhou, Y. B., Chen, M., et al. (2020). The soybean bZIP transcription factor gene GmbZIP2 confers drought and salt resistances in transgenic plants. Int. J. Mol. Sci. 21 (2), 670. doi: 10.3390/ijms21020670
Yang, Y., Zhou, Y., Chi, Y., Fan, B., Chen, Z. (2017). Characterization of soybean WRKY gene family and identification of soybean WRKY genes that promote resistance to soybean cyst nematode. Sci. Rep. 7 (1), 1–13. doi: 10.1038/s41598-017-18235-8
Ye, Y., Ding, Y., Jiang, Q., Wang, F., Sun, J., Zhu, C. (2017). The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 36 (2), 235–242. doi: 10.1007/s00299-016-2084-x
Yi, S. Y., Kim, J. H., Joung, Y. H., Lee, S., Kim, W. T., Yu, S. H., et al. (2004). The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in arabidopsis. Plant Physiol. 136 (1), 2862–2874. doi: 10.1104/pp.104.042903
Yogendra, K. N., Kumar, A., Sarkar, K., Li, Y., Pushpa, D., Mosa, K. A., et al. (2015). Transcription factor StWRKY1 regulates phenylpropanoid metabolites conferring late blight resistance in potato. J. Exp. Bot. 66 (22), 7377–7389. doi: 10.1093/jxb/erv434
Yogendra, K. N., Sarkar, K., Kage, U., Kushalappa, A. C. (2017). Potato NAC43 and MYB8 mediated transcriptional regulation of secondary cell wall biosynthesis to contain phytophthora infestans infection. Plant Mol. Biol. Rep. 35 (5), 519–533. doi: 10.1007/s11105-017-1043-1
Yokota, A., Kawasaki, S., Iwano, M., Nakamura, C., Miyake, C., Akashi, K. (2002). Citrulline and DRIP-1 protein (ArgE homologue) in drought tolerance of wild watermelon. Ann. Bot. 89 (7), 825–832. doi: 10.1093/aob/mcf074
Young, D. H., Michelotti, E. L., Swindell, C. S., Krauss, N. E. (1992). Antifungal properties of taxol and various analogues. Experientia 48 (9), 882–885. doi: 10.1007/BF02118425
Yu, Y., Guo, D., Li, G., Yang, Y., Zhang, G., Li, S., et al. (2019). The grapevine R2R3-type MYB transcription factor VdMYB1 positively regulates defense responses by activating the stilbene synthase gene 2 (VdSTS2). BMC Plant Biol. 19 (1), 1–15. doi: 10.1186/s12870-019-1993-6
Yu, Y., Wang, N., Hu, R., Xiang, F. (2016). Genome-wide identification of soybean WRKY transcription factors in response to salt stress. Springerplus 5 (1), 1–15. doi: 10.1186/s40064-016-2647-x
Zarei, A., Körbes, A. P., Younessi, P., Montiel, G., Champion, A., Memelink, J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1. 2 promoters in. Arabidopsis. Plant Mol. Biol. 75 (4), 321–331. doi: 10.1007/s11103-010-9728-y
Zhang, Y. (2014). Identification and characterization of the grape WRKY family. BioMed. Res. Int. 2014.
Zhang, G., Chen, M., Li, L., Xu, Z., Chen, X., Guo, J., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60 (13), 3781–3796.
Zhang, H., Hedhili, S., Montiel, G., Zhang, Y., Chatel, G., Pré, M., et al. (2011). The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 67 (1), 61–71. doi: 10.1111/j.1365-313X.2011.04575.x
Zhang, H., Huang, Y., Zhang, H., Huang, Y. (2013). Genome-wide survey and characterization of greenbug induced nac transcription factors in sorghum [Sorghum bicolor (L.) moench]. Plant Anim. Genome. doi: 10.1007/s11103-015-0382-2
Zhang, M., Li, S., Nie, L., Chen, Q., Xu, X., Yu, L., et al. (2015). Two jasmonate-responsive factors, TcERF12 and TcERF15, respectively act as repressor and activator of tasy gene of taxol biosynthesis in Taxus chinensis. Plant Mol. Biol. 89 (4), 463–473. doi: 10.1007/s11103-015-0382-2
Zhang, Q., Li, Y., Zhang, Y., Wu, C., Wang, S., Hao, L., et al. (2017). Md-miR156ab and md-miR395 target WRKY transcription factors to influence apple resistance to leaf spot disease. Front. Plant Sci. 8, 526. doi: 10.3389/fpls.2017.00526
Zhang, H., Ma, Z. F. (2018). Phytochemical and pharmacological properties of capparis spinosa as a medicinal plant. Nutrients 10 (2), 116. doi: 10.3390/nu10020116
Zhang, W., Shu, C., Chen, Q., Cao, J., Jiang, W. (2019). The multi-layer film system improved the release and retention properties of cinnamon essential oil and its application as coating in inhibition to penicillium expansion of apple fruit. Food Chem. 299, 125109. doi: 10.1016/j.foodchem.2019.125109
Zhang, Y., Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC evolutionary Biol. 5 (1), 1–12. doi: 10.1186/1471-2148-5-1
Zhang, H., Wei, C., Yang, X., Chen, H., Yang, Y., Mo, Y., et al. (2017). Genome-wide identification and expression analysis of calcium-dependent protein kinase and its related kinase gene families in melon (Cucumis melo l.). PloS One 12 (4), e0176352. doi: 10.1371/journal.pone.0176352
Zhao, Y., Zhang, G., Tang, Q., Song, W., Gao, Q., Xiang, G., et al. (2022). EbMYBP1, a R2R3-MYB transcription factor, promotes flavonoid biosynthesis in erigeron breviscapus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.946827
Zheng, Q., Bao, C., Guo, W., Li, S., Chen, J., Chen, B., et al. (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7 (1), 1–13. doi: 10.1038/ncomms11215
Zhou, W., Qian, C., Li, R., Zhou, S., Zhang, R., Xiao, J., et al. (2018). TaNAC6s are involved in the basal and broad-spectrum resistance to powdery mildew in wheat. Plant Sci. 277, 218–228. doi: 10.1016/j.plantsci.2018.09.014
Zhou, Y., Sun, W., Chen, J., Tan, H., Xiao, Y., Li, Q., et al. (2016). SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci. Rep. 6 (1), 1–12. doi: 10.1038/srep22852
Zhu, L., Guan, Y., Zhang, Z., Song, A., Chen, S., Jiang, J., et al. (2020). CmMYB8 encodes an R2R3 MYB transcription factor which represses lignin and flavonoid synthesis in chrysanthemum. Plant Physiol. Biochem. 149, 217–224. doi: 10.1016/j.plaphy.2020.02.010
Ziegler, J., Facchini, P. J. (2008). Alkaloid biosynthesis: metabolism and trafficking. Annu. Rev. Plant Biol. 59, 735. doi: 10.1146/annurev.arplant.59.032607.092730
Keywords: pest attack, pathogen infection, transcription factors, miRNAs, plant specialized/secondary metabolites
Citation: Kajla M, Roy A, Singh IK and Singh A (2023) Regulation of the regulators: Transcription factors controlling biosynthesis of plant secondary metabolites during biotic stresses and their regulation by miRNAs. Front. Plant Sci. 14:1126567. doi: 10.3389/fpls.2023.1126567
Received: 18 December 2022; Accepted: 06 February 2023;
Published: 02 March 2023.
Edited by:
Vikas Srivastava, Central University of Jammu, IndiaReviewed by:
Danial Hassani, East China Normal University, ChinaPravin Prakash, National Botanical Research Institute (CSIR), India
Copyright © 2023 Kajla, Roy, Singh and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Archana Singh, YXJjaGFuYXNpbmdoQGhyYy5kdS5hYy5pbg==; Indrakant K. Singh, aWtzaW5naEBkYi5kdS5hYy5pbg==; Amit Roy, Um95QGZsZC5jenUuY3o=
 Mohini Kajla
Mohini Kajla Amit Roy
Amit Roy Indrakant K. Singh
Indrakant K. Singh Archana Singh
Archana Singh