- 1Industry and University Research Cooperation Demonstration Base in Fujian Province, College of Life Sciences, Ningde Normal University, Ningde, China
- 2State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, College of Agriculture, Guangxi University, Nanning, China
- 3College of Life Science, Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, Fujian Agriculture and Forestry University, Fuzhou, China
The WRKY transcription factors (TFs) are among the most diverse TF families of plants. They are implicated in various processes related to plant growth and stress response. Kenaf (Hibiscus cannabinus L.), an important fiber crop, has many applications, including the phytoremediation of saline-alkaline soil. However, the roles of WRKY TFs in kenaf are rarely studied. In the present study, 46 kenaf WRKY genes were genome-widely identified and characterized by gene structure, phylogeny and expression pattern analysis. Furthermore, the HcWRKY44 gene was functionally characterized in Arabidopsis under salinity and drought stresses. HcWRKY44 is a nuclear-localized protein that is positively induced by salinity and drought, with roots showing maximum accumulation of its transcripts. Under NaCl and abscisic acid (ABA) stress conditions, plants overexpressing HcWRKY44 had higher germination rates, better root growth and increased survival than control plants; however, it did not improve the ability to withstand drought stress. Moreover, ABA signaling genes (ABI1, ABI2, and ABI5), ABA-responsive genes (ABF4, RD29B, COR15A, COR47, and RD22), stress-related genes (STZ, P5CS, and KIN1), and ionic homeostasis-related genes (SOS1, AHA1, AHA2, and HKT1) were positively induced in HcWRKY44 transgenic plants under NaCl treatment. These results suggest that HcWRKY44 improved plant’s tolerance to salt stress but not osmotic stress through an ABA-mediated pathway. In summary, this study provides provided comprehensive information about HcWRKY genes and revealed that HcWRKY44 is involved in salinity tolerance and ABA signaling.
Introduction
Abiotic stresses, such as drought, salinity, and temperature extremes, are major environmental factors affecting plant growth and development (Zhu, 2016). To overcome these adverse conditions, plants have developed multiple defense strategies to adapt directly and/or indirectly. Conceptually, the adaptive responses could be classified into osmotic adjustment and basic defenses, including ion detoxification and growth regulation (Dang et al., 2013; Ding et al., 2014). During the stress responses, phytohormone abscisic acid (ABA) is essential in regulating osmotic responses and stress-responsive gene expression (Cutler et al., 2010; Ding et al., 2014; Zhu, 2016). From the perception of the ABA signal to the activation of the relevant protein kinases, phosphatases and transcription factors, the ABA signaling pathway has been thoroughly explored (Fujii and Zhu, 2009; Fujii et al., 2009; Cutler et al., 2010;Jiang et al., 2012; Luo et al., 2013; Ding et al., 2014). Many transcription factors have been reported to respond against osmotic stresses in both ABA-independent and ABA-dependent manner (Rushton et al., 2012; Ding et al., 2014; Zhu, 2016).
WRKY genes play a crucial role in responding to osmotic stress by indirectly and/or directly binding to the promoters of ABA-regulated genes such ABFs, ABI4, ABI5, and DREBs (Jiang et al., 2012; Bakshi and Oelmuller, 2014; Wani et al., 2021). In Arabidopsis, AtWRKY63 and AtWRKY57 are involved in regulating the ABA signaling pathway and enhancing plant tolerance to drought (Ren et al., 2010; Jiang et al., 2012). Similarly, OsWRKY24, OsWRKY45, OsWRKY72, and OsWRKY77 also improved salinity tolerance in rice through ABA signaling (Xie et al., 2005). In wheat, overexpression of TaWRKY2 and TaWRKY19 enhanced tolerance to salinity stress by directly binding to the regulatory elements of well-known ABA signaling genes DREB2A, RD29A, RD29B, and COR6.6 (Niu et al., 2012). Consistently, overexpression of GsWRKY20 and HcWRKY50 also improved drought tolerance by regulating ABA-mediated stomatal aperture (Luo et al., 2013; Niu et al., 2022).
Kenaf (Hibiscus cannabinus L.) is a diploid (2n = 36) herbaceous fiber crop belonging to the Malvaceae family. Kenaf was domesticated in Africa and is grown in the Asia-Pacific region as the third natural fiber species after cotton and jute (Chen P. et al., 2020; Zhang et al., 2020; Sim and Nyam, 2021). Kenaf can produce enormous amounts of fiber biomass with up to 100–150 t per hectare and grows quickly, reaching heights of 4–6 m over a 4-month growth period (Supplementary Figure 1) (Chen P. et al., 2020; Sim and Nyam, 2021). Kenaf has been widely applied in papermaking, building materials, bio-composites, animal feed, and recycled plastics due to its colossal fiber yield and biodegradable nature (Niu et al., 2015; Sim and Nyam, 2021). More importantly, kenaf performs well and has a high tolerance for drought, salinity, and barrenness (Danalatos and Archontoulis, 2010; Ramesh, 2016; Sim and Nyam, 2021). Therefore, kenaf could be used in phytoremediation of saline-alkali soil and/or as an osmotic-stress tolerant crop. However, its tolerance mechanism is still unclear, and how WRKY genes in kenaf regulate the tolerance remains obscure.
In this study, we performed a genome-wide identification of kenaf WRKY transcription factors and analyzed the gene structure, evolutionary relationship, and expression pattern of HcWRKYs. The results showed that the drought and salinity stress could positively induce HcWRKY44. Besides, HcWRKY44 overexpression plants improved the tolerance to salinity stress but not drought stress. We further revealed that HcWRKY44 increased plant tolerance to salinity stress through modulating the ABA signaling pathways.
Materials and methods
Identification and characterization of WRKY genes in Hibiscus cannabinus
The HcWRKY gene sequences were retrieved from the kenaf genome database1 (Zhang et al., 2020). The sequence data of a kenaf relative species, Gossypium hirsutum, and other species were acquired from the Phytozome v13 database. The HMM model of the WRKY domain (PF03106) was used as the query to search the kenaf genome database. The resulting candidate WRKY genes were further confirmed by the CDD program to verify the CXHX domain and WRKYGQK domain. The properties of HcWRKY proteins, such as amino acid length, molecular weight (MW), and isoelectric point (pI), were predicted using ExPASy-Compute pI/Mw tool as described previously (She et al., 2022).
Phylogenetic analysis, chromosome localization, and syntenic analysis
Multiple sequence alignment of Arabidopsis thaliana, G. hirsutum, and Hibiscus cannabinus WRKY proteins were conducted by MUSCLE, using the default setting parameters. The maximum number was 20, and minimum/maximum width was 6/50, and the results were visualized by Jalview software. The phylogenetic tree was generated by the MEGA 7.0 program using the ML method based on the JTT substitution model. The loci of HcWRKY genes were retrieved from the kenaf annotation GFF3 files and gene locations on the chromosomes were visualized by TBtools (Chen C. et al., 2020). MCScanX software was used for collinearity analysis and generating collinearity blocks with the threshold value of 1 × 10–5 (Wang et al., 2012). The collinearity block mapping within the kenaf, Arabidopsis, and cotton genome were visualized by CIRCOS software. Ka and Ks values of paralogous genes were estimated by the K-estmator program, and Ks value was used for estimating time of segmental duplication events according to the method descripted by Chen et al. (2014).
Plant materials and growth conditions
Kenaf (H. cannabinus L.) cultivar Fuhong 992 was used in this study. The seeds were washed three times in running water and then cultured under controlled conditions at 28°C for 16 h in light and 26°C for 8 h in the dark with a relative humidity of 65–75%. For stress treatments, 2 weeks old healthy seedlings were cultivated in a solution containing 200 mM NaCl for salt stress and 15%(w/v) PEG6000 for osmotic stress treatment, according to the previously described method (Niu et al., 2015). Afterward, healthy seedlings were selected, and leaves were harvested from each treatment for further analysis (Niu et al., 2016).
Arabidopsis thaliana ecotype (Col-0) was cultured in a walk-in growth chamber at 22°C under a 16 h light/8 h dark cycle. For the differential expression analysis of the ABA-related or stress-responsive marker genes, the WT and transgenic lines (2-week-old) were treated with 15% (w/v) PEG6000, 200 mM NaCl, and 100 mM ABA, respectively. After the treatment, samples were harvested and used for further analysis.
RNA extraction, qRT-PCR, and gene expression analysis
Total RNA was isolated from independently collected samples using the Ultrapure RNA kit (CW0597, Beijing, China). The cDNA was synthesized according to the instruction manual of the Reverse Transcription Kit (Pimerscript™ RT DRR037S TaKaRa, Japan), then used as PCR templates and/or qRT-PCR analysis. For sequencing, the amplified products were purified and ligated into the pMD18-T vector and then transformed into Escherichia coli DH5α cells. The qRT-PCR analysis was conducted using the qPCR SuperMix TransStart Top Green (TransGen, AQ132-11) on the Bio-Rad CFX-96 detection system with the following amplification programs of 94°C for 30 s, and 40 cycles of 94°C for 5 s, 60°C for 15 s, and a melting curve cycle from 65°C to 95°C. For normalization of HcWRKYs, HcTUBα (Niu et al., 2015) in kenaf and AtACT2 in Arabidopsis were used. The primers used in qRT-PCR are listed in Supplementary Table 1. Each reaction was performed in three independent biological and three technical replicates.
Vector construction, subcellular localization, and transgenic transformation
The full-length CDS of HcWRKY44 without terminator code was amplified from kenaf cDNA, and the PCR products were inserted into the pENTRTM/D-topo vector. After sequencing, the positive clones were selected and recombined into the destination vector pGWB605. Finally, the positive plasmids were transformed into Agrobacterium tumefaciens GV3101 strain, which was used for Arabidopsis transformation. The 4-week-old plants of A. thaliana ecotype (Col-0) were transformed by the vacuum infiltration method using the 35S:HcWRKY44-GFP and 35S:GFP constructs. The T1 transgenic seedlings were sprayed with 20 mg/L herbicide, and the positive T1 lines were selected. The T2 plants were further selected and separated, and the homozygous lines of T3 generations were used for the subsequent experimental analysis. For subcellular localization analysis, the 35S:HcWRKY44-GFP constructs and the control 35S:GFP vector were introduced into the epidermal leaves of Nicotiana benthamiana and incubated in the dark for 36–48 h. GFP fluorescence signals were checked using an Olympus confocal microscope (Olympus FV500, Olympus, Japan) under a 488 nm exciting wavelength.
Stress treatments and physiological indexes measurement
For the stress treatments, the seeds of the HcWRKY44-transgenic lines and wild-type plants were surfaced-sterilized and then germinated on 1/2 MS agar medium with 200 and 300 mM mannitol to mimic the drought stress and with 150 and 200 mM NaCl for the salinity stress. The physiological indexes, such as germination rate, cotyledon greening, root length and survival rate assays, were measured and the corresponding phenotypes were photographed according to the methods described by Niu et al. (2022). For the relative electrolyte leakage measurement, the methods were referred to the reference described by Zhang et al. (2019). All assays were performed in three independent replicates.
Results
Identification and sequence characteristic of HcWRKY genes
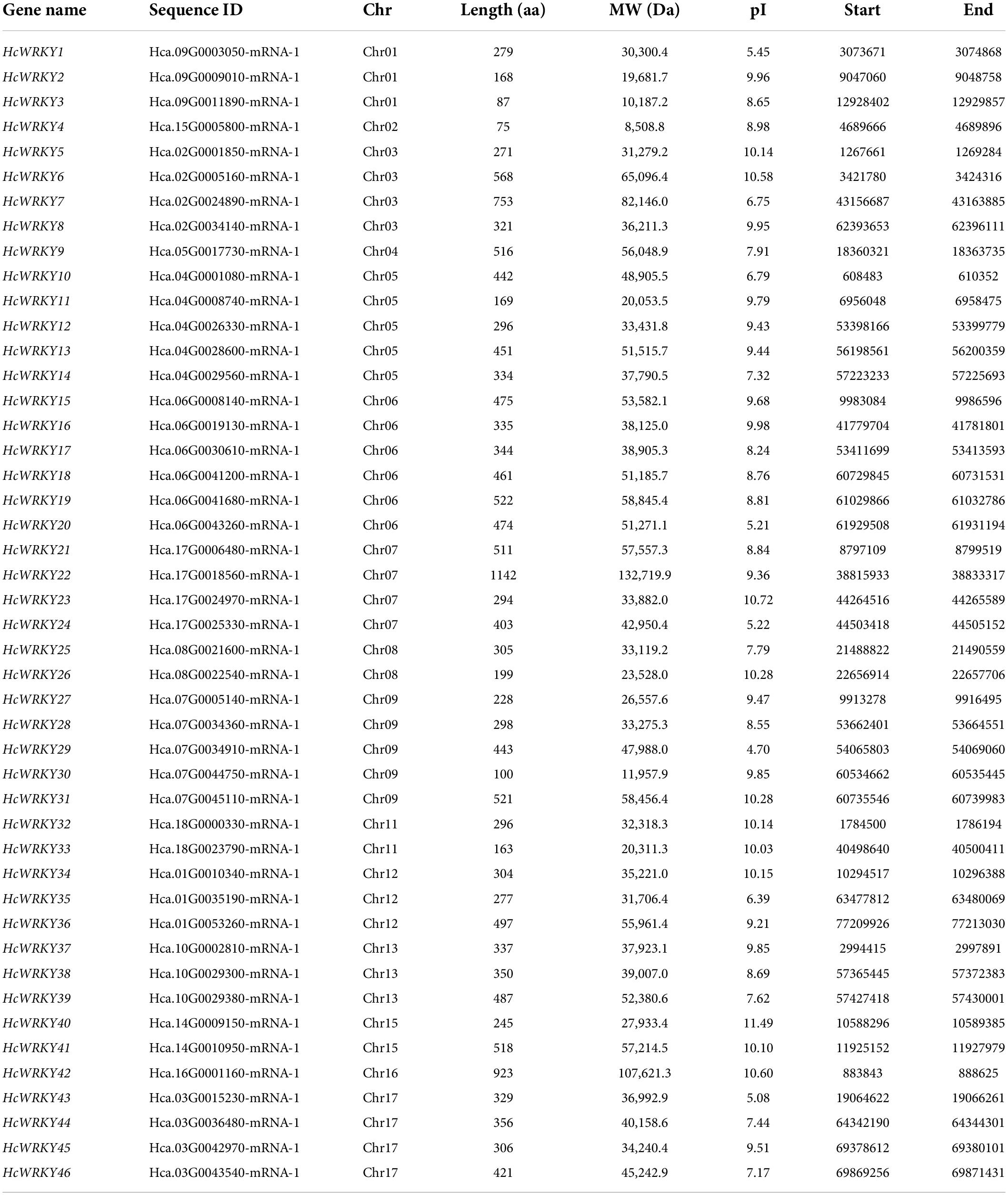
A HMMER-BLASTP-InterProScan module method was used to identify the sequences containing the PF03106 domain and acquire the WRKY gene sequences in the kenaf genome. A total of 46 WRKY genes were identified in the kenaf genome with the complete WRKY domain (Table 1). Based on the gene distribution information on the chromosomes, the identified genes were named from HcWRKY1 to HcWRKY46. The physicochemical analysis revealed that the amino acid length of HcWRKY ranged from 75 aa (HcWRKY4) to 1142 aa (HcWRKY22), and the corresponding protein MWs ranged from 8508.8 to 132719.9 Da, and the predicted theoretical pIs of HcWRKYs ranged from 4.70 (HcWRKY29) to 11.49 (HcWRKY40) (Table 1).
To further confirm these HcWRKY genes, the typical conserved WRKYGQK domain was searched and visualized. According to the conserved 60 amino acids of the WRKY domain, the HcWRKY proteins were classified into three groups, and each group was divided into different subgroups. For example, the group I could be divided into I-N and I-C subgroups, which possessed two WRKY domains and CX4C22–23HXH zinc finger structures. Group II was divided into five subgroups (II-a, II-b, II-c, II-d, and II-e), each containing 3, 6, 11, 5, and 8 members, with the structure of WRKYGQK and CX5C23HXH in the II-a, II-b, II-d, and II-e subgroups except for II-c subgroup. On the other hand, group III had 7 members with the zinc finger structure of WRKYGQK and CX7C23HXC at the C-terminal (Supplementary Figure 2). The WRKY proteins of kenaf generally had the same WRKYGQK domain as Arabidopsis, with the exception that the WRKYGQK domains of HcWRKY3 were changed into WRKYGKK and the QRKYGQK domains for HcWRKY35, as well as additional amino acid variations outside the WRKYGQK domains in HcWRKY9/19/28/27/30/37 (Supplementary Figure 2). These results indicated that those mutated HcWRKY genes might gain a novel function during evolution.
Phylogenetic analysis, gene structure, and synteny analysis
The gene structure analysis revealed that natural mutation occurred in HcWRKY genes of kenaf, i.e., some WRKYGQK domains were changed into WRKYGKK, WRKYGEK and WRKYGQE. To further investigate the divergence of HcWRKYs, a comparative phylogenetic tree was constructed using the maximum-likelihood (ML) method between 46 HcWRKYs and 71 AtWRKYs. Expectedly, the phylogenetic tree classified the WRKY genes into groups I, II, and III and group II was also divided into five subgroups (II-a, II-b, II-c, II-d, and II-e) (Figure 1), indicating that these HcWRKYs may share the similar functions with that in Arabidopsis (Figure 1).
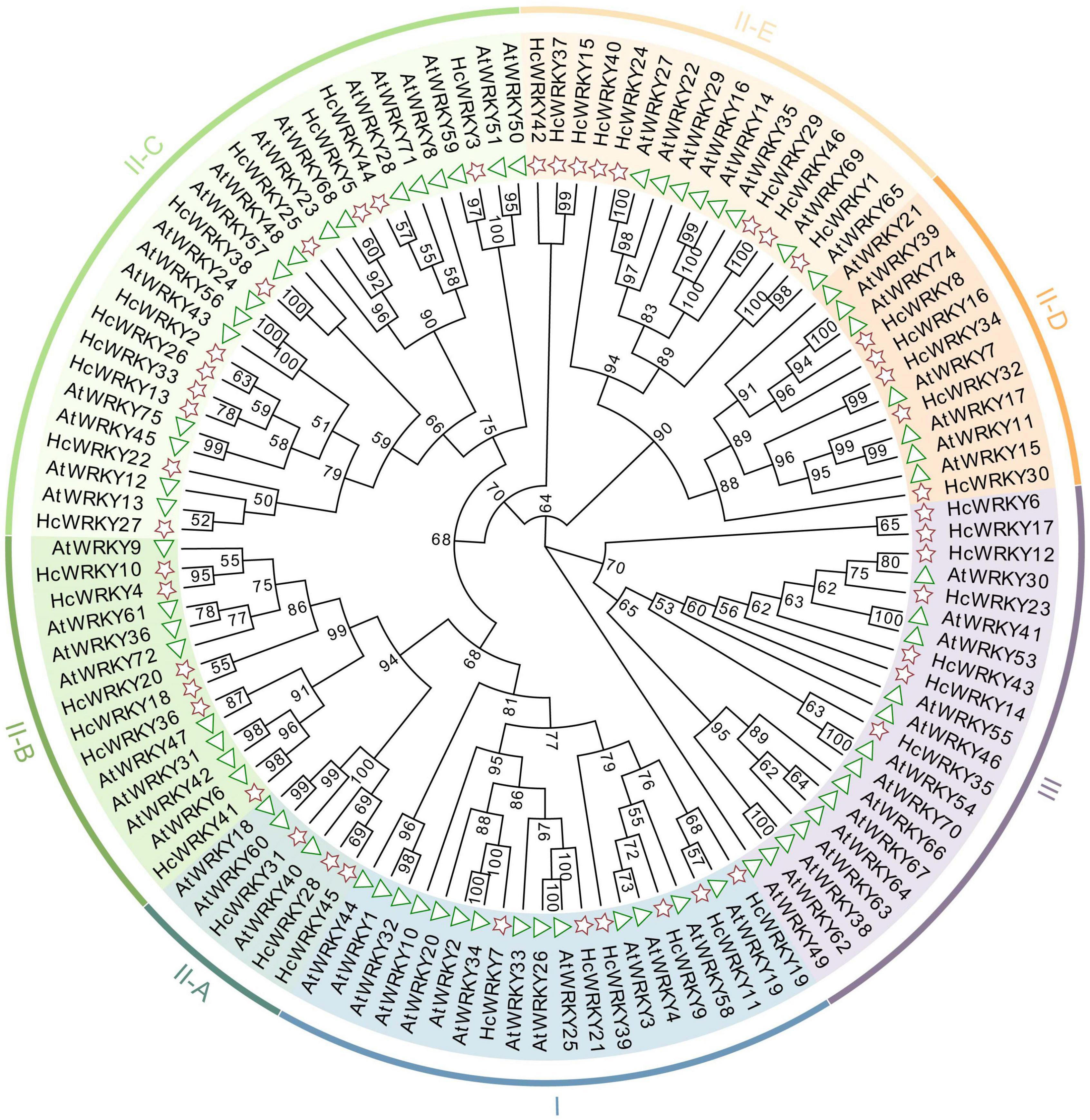
Figure 1. Phylogenetic tree analysis revealed the relationship of WRKY genes between kenaf and Arabidopsis. The unrooted phylogenetic tree was generated by the MEGA7.0 software using the neighbor-joining method, with bootstrap value was 1000 replicates. The different color arcs represent different groups and/or subgroups of WRKY genes. AtWRKY genes were represented by triangle, and HcWRKY genes were pentagon.
The HcWRKYs introns and exons investigation revealed a variable number of exons ranging from 1 to 18 (Figure 2). Group I HcWRKY genes had 1–5 introns, Group IIa had 4-6 introns, Group IIb, IIc, IId, IIe and III had 1-4 introns, with some exceptions for HcWRKY22 (24 introns), HcWRKY25 (5 introns), and HcWRKY10 (0 introns) (Figure 2). On the other hand, gene motifs analysis showed the similar motifs shared in the each subgroups. For example, motif 1, 2, 3, and 5 jointly possessed in Group I, motif 2, 4, and 5 possessed in Group IIb and IIe (Figure 2). These findings indicate that exon and intron numbers vary between groups but are nearly constant within the same group.
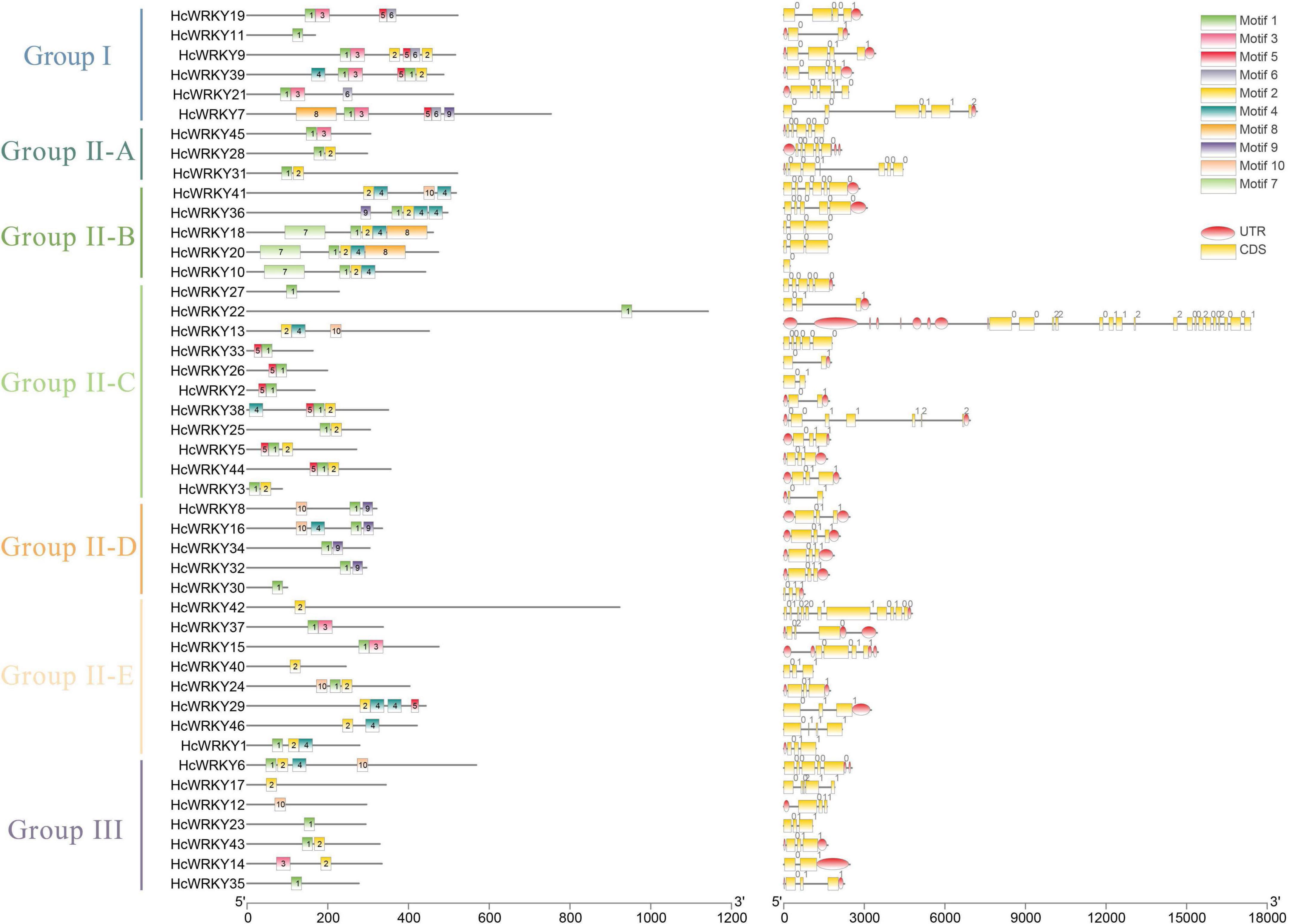
Figure 2. Conserved motifs and gene structures of the HcWRKY proteins. The left line shows the 10 conserved motifs of WRKY proteins were identified by the MEME program in kenaf, indicated by different colors and numbers. Motif 1 and motif 2 are essential components of the WRKY domain. The right line shows the exon-intron structure organization of the corresponding HcWRKY genes performed by the online tool GSDS. The red oval represents the UTR, the yellow rectangle represents the CDS, and the gray lines represent the introns.
The tandem and segment duplication events were also analyzed and identified using MCScanX software to understand the evolution of HcWRKY genes. The results showed 6 segmental duplication syntenic gene pairs (HcWRKY1/HcWRKY40, HcWRKY5/HcWRKY17, HcWRKY8/HcWRKY16, HcWRKY18HcWRKY20, HcWR KY28/HcWRKY45, HcWRKY29/HcWRKY46) (Figure 3A and Supplementary Table 2), and 11 tandem duplication gene pairs (HcWRKY1/2/3, HcWRKY5/6, HcWRKY13/14, HcWRKY18/19/20, HcWRKY23/24, HcWRKY25/26, HcWR KY28/29, HcWRKY30/31, HcWRKY38/39, HcWRKY40/41, HcWRKY45/46) in kenaf. In addition, the evolutionary dates of duplicated HcWRKY genes were also estimated using Ks as the proxy for time (Supplementary Table 2), the results showed that kenaf duplication events for kenaf 6 of 17 pairs occurred within the past 12.385–165.717 million years (Supplementary Table 2). These results suggested that during their evolution, both segmental and tandem duplications contributed to the gene expansion of HcWRKYs. In addition, the syntenic blocks were comparatively analyzed among H. cannabinus, A. thaliana, and G. hirsutum and 16 orthologous syntenic gene pairs were identified between H. cannabinus and A. thaliana (Figure 3B and Supplementary Table 3). Interestingly, one HcWRKY gene could match two or more HcWRKY genes, i.e., HcWRKY39 could align with AtWRKY3/4, and HcWRKY5 could match with AtWRKY23/68 (Figure 3B and Supplementary Table 3). For H. cannabinus and its most relative G. hirsutum, 64 syntenic orthologous gene pairs were found, and a similar phenomenon of one HcWRKY syntenic with two or three GhWRKYs was also observed (Figure 3B and Supplementary Table 4).
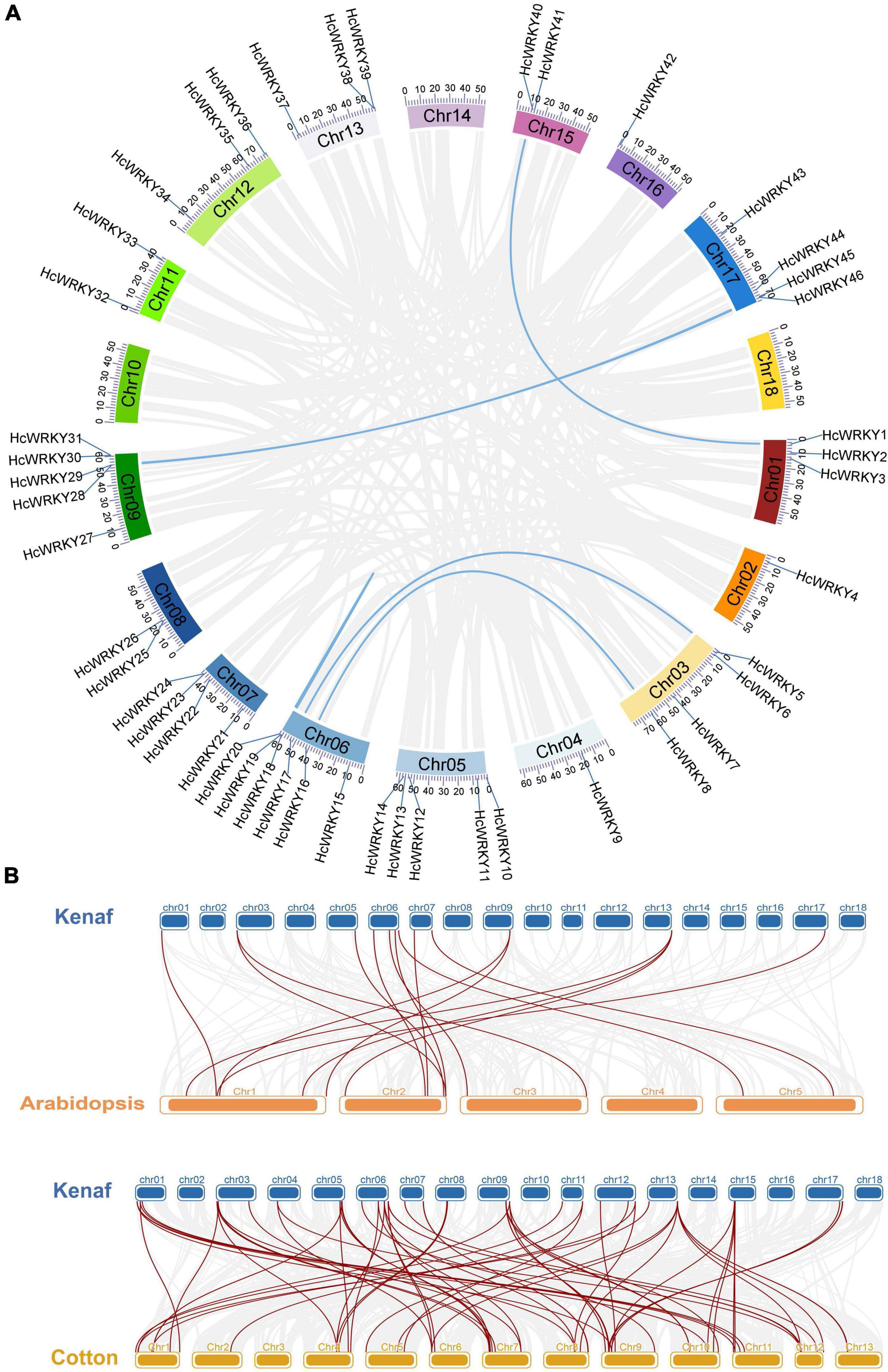
Figure 3. Collinearity analysis for all HcWRKYs. (A) HcWRKY genes mapped onto the corresponding chromosomes position of kenaf, as shown in different colors, were used for their collinearity analysis. (B) Synteny analysis among the Arabidopsis, kenaf and cotton.
Expression pattern analysis of HcWRKYs in different tissues and treatments
The expression pattern of genes is often associated with their specific gene function. Therefore, we examined the expression level of 20 HcWRKYs using qRT-PCR analysis to explore the HcWRKYs functions. The results for the various tissues revealed that the 20 HcWRKYs express differently in the roots, stems, leaves, and phloem. They were primarily expressed in the roots, except for HcWRKY2, HcWRKY14 and HcWRKY25, which were expressed strongly in the phloem (Supplementary Figure 3). These findings suggested that the majority of HcWRKYs are crucial for root growth.
To verify HcWRKYs role in environmental response, the expression levels of the 20 HcWRKYs were further examined under salinity and drought stresses. The results showed that the expression level of 20 HcWRKYs significantly changed against the salinity and drought stimuli (Figure 4 and Supplementary Figure 4). For the salinity stress, HcWRKYs positively responded after 12 h and HcWRKY7, HcWRKY11, HcWRKY15, HcWRKY16, HcWRKY22, HcWRKY26, HcWRKY27, and HcWRKY38 showed increased expression levels after salinity stress. While HcWRKY24 and HcWRKY25 showed decreased expression in response to salinity stress. Compared to control conditions, the expressions of HcWRKY13, HcWRKY15, HcWRKY25, HcWRKY33, and HcWRKY44 were lower at 6 h then it increased at 12 h and subsequently decreased at 24 h (Figure 4).
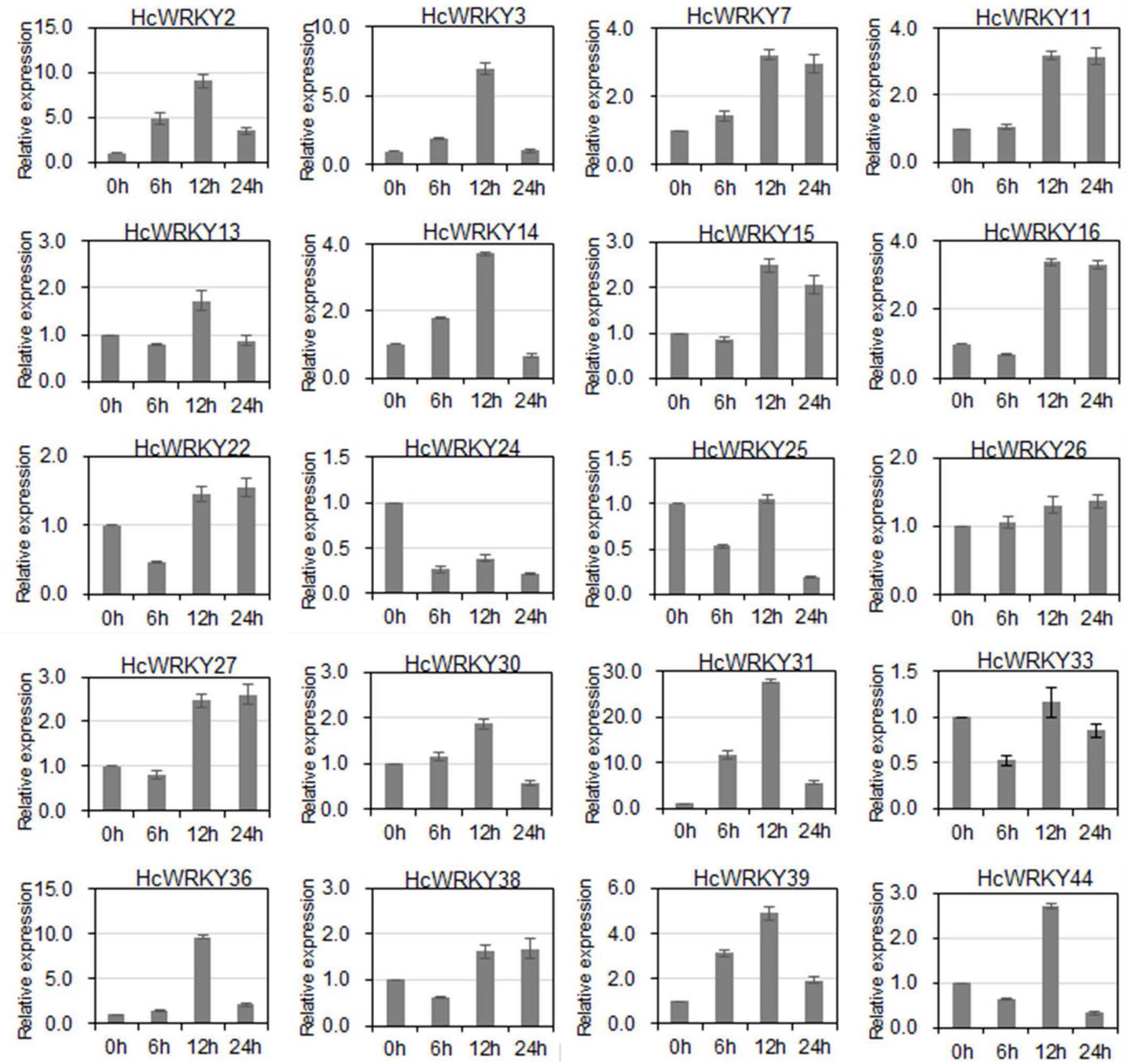
Figure 4. Expression profiles of 20 HcWRKY genes under NaCl stress. Twenty HcWRKY genes were cloned and selected for expression analysis under salinity stress, mimicked by 200 mM NaCl solution irrigation. After treatment, leaves were harvested and used for mRNA transcripts analysis by qRT-PCR. The 18S rRNA and TUBα gene was used as the standard control to normalize the qRT-PCR results. Each assay was replicated three times.
For the drought stress, most of HcWRKYs showed a negative expression pattern (Supplementary Figure 4). Despite having decreased expression compared to the control conditions (0 h), after drought stimuli, HcWRKY11, HcWRKY13, HcWRKY14, HcWRKY15, HcWRKY22, HcWRKY25, HcWRKY26, HcWRKY27, HcWRKY30, HcWRKY31, HcWRKY33, and HcWRKY39 transcripts demonstrated an increased expression pattern from 6 to 24 h. HcWRKY7, HcWRKY13, HcWRKY31 and HcWRKY39 expression decreased from 0 to 12 h. While HcWRKY16, HcWRKY24 and HcWRKY44 showed increased expression levels from 0 to 24 h under drought stress (Supplementary Figure 4). Altogether, these findings indicate that the HcWRKY genes respond to the salinity and drought treatments and display differential expression patterns in response to these stress (Figure 4 and Supplementary Figure 4).
HcWRKY proteins localize in cell nuclei
Three representative HcWRKY proteins (HcWRKY39, HcWRKY44, and HcWRKY43) out of each group were randomly chosen for subcellular localization studies to explore the functional properties of HcWRKY proteins. The HcWRKY-GFP vectors (Figure 5A) and control 35S-GFP were injected into the epidermal cells of N. benthamiana leaves. The results showed that the GFP signals of HcWRKY39/44/43-GFP were exclusively localized in the nuclei of epidermal cells of N. benthamiana (Figure 5B). In contrast, the control GFP protein was found in both the nucleus and cell membrane (Figure 5B). These results were coincided with previous studies that WRKY proteins functioned in the nuclear as transcription factors.
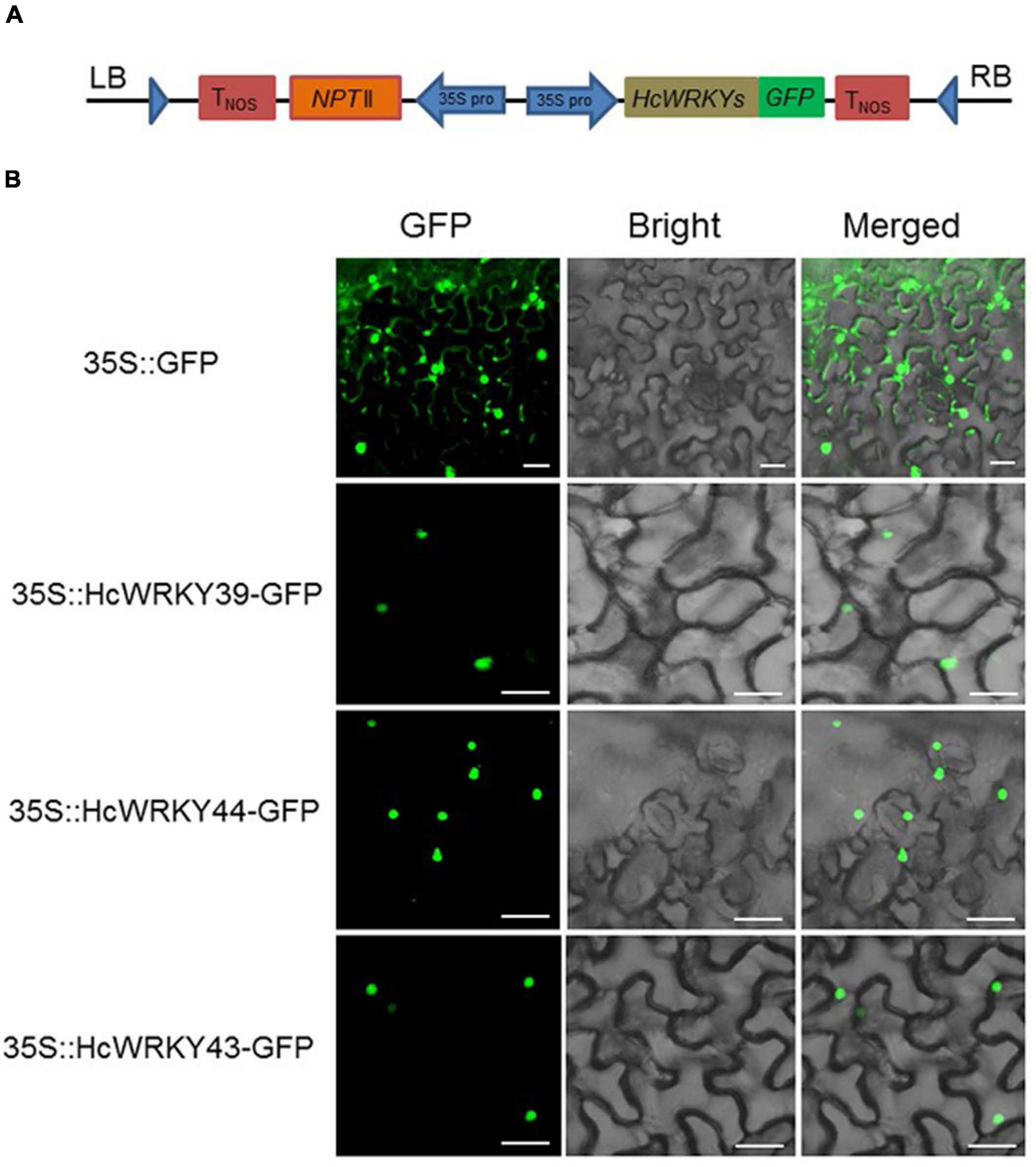
Figure 5. Subcellular localization of three HcWRKY proteins from each group. (A) A schematic representation of 35S:HcWRKYs-GFP fusion construct and 35S:GFP construct. The coding sequences of HcWRKY39/44/43 genes were fused with GFP expression vector. (B) Subcellular localization analysis was performed using the transient transformation in leaves of N. benthamiana, and GFP signals were checked by a confocal laser scanning microscope.
HcWRKY44 overexpression plants enhanced the salinity but not drought tolerance
The expression patterns of HcWRKYs revealed that the HcWRKY44 strongly induces by salinity and drought stresses (Figure 4 and Supplementary Figures 3, 4). This led us to study the roles of HcWRKY44 in response to osmotic stresses. Three independent transgenic lines of HcWRKY44, OE44-2#, OE44-3#, and OE44-4# (Supplementary Figure 5) were selected to investigate their performance against the salinity and drought stresses. The salinity and drought stresses were mimicked by using NaCl and mannitol irrigation, respectively. For the salinity stress, the results of germination rates showed no obvious difference between the HcWRKY44 overexpression lines and control plants on the 1/2 MS media. After salinity treatment, OE44-2#, OE44-3#, and OE44-4# lines significantly reduced the germination rate compared to the control, and transgenic lines showed no seed coat breakage (Figures 6A,B). However, transgenic lines showed a faster root growth rate than control plants, and they possessed longer root lengths and more lateral root numbers (Figures 6C,D). To further investigate the tolerance performance of HcWRKY44 in response to the salinity stress, the seedlings of 2-week-old transgenic lines were treated with different concentrations of NaCl solution. After treatment for 7 days, the leaves of transgenic and control plants turned yellow, wilted and showed no obvious difference. However, after being re-watered for 7 days, leaves of OE44-2#, OE44-3#, and OE44-4# transgenic lines returned green and thrived, while that of the control lines were yellow or even dry and wilted (Figure 6E). Eventually, the overexpression lines of HcWRKY44 showed a higher survival rate than those of the control plants (Figure 6F). Meanwhile, the relative electrolyte leakage was also checked to determine the plasma membrane permeability and lipid peroxidation under the salinity treatment. The results showed that the electrolyte leakage of all lines increased with the increased salinity concentrantions, and the OE44-2#, OE44-3#, and OE44-4# transgenic lines exhibited a significantly lower electrolyte leakage than the control lines under the salinity treatment (Figure 6G).
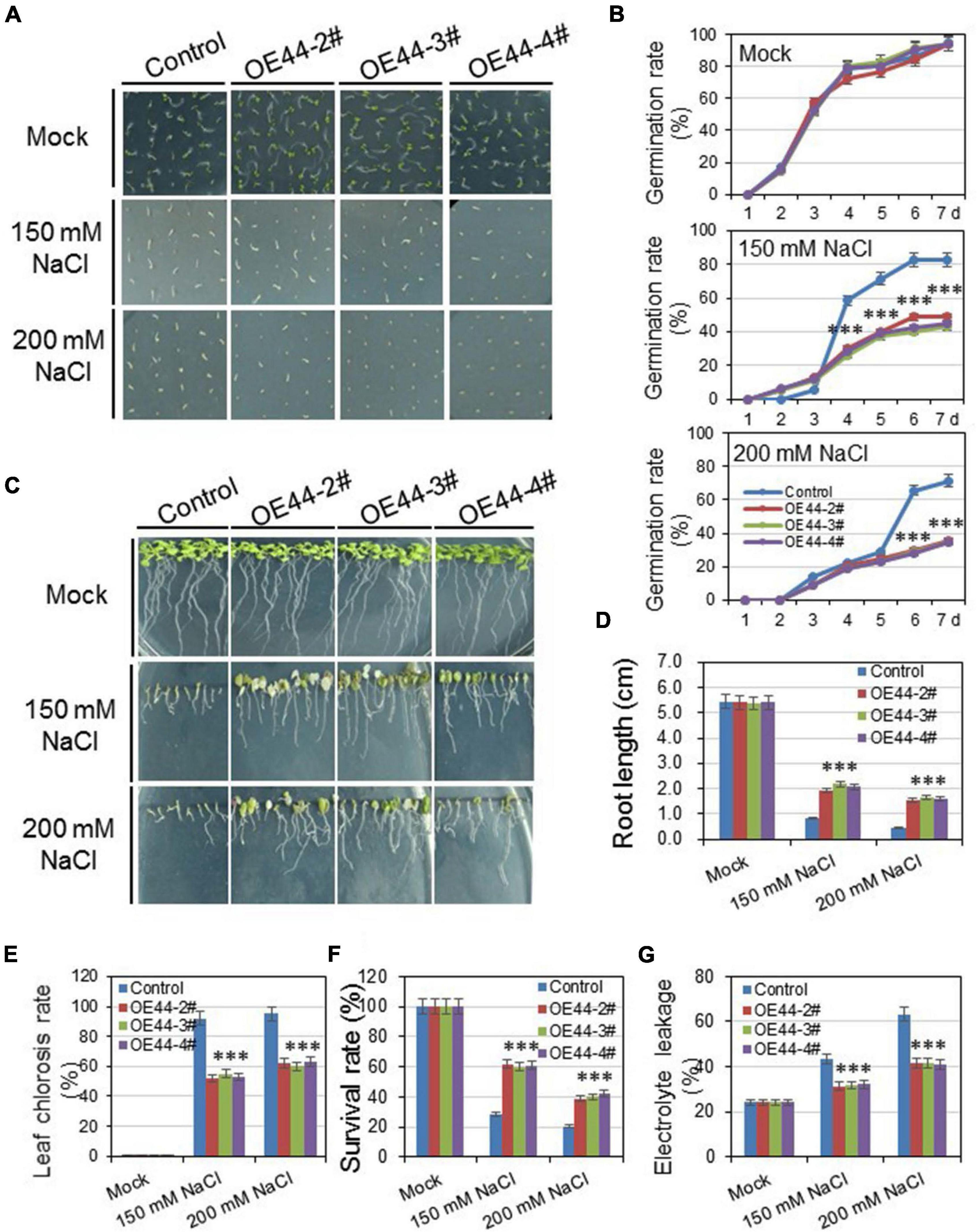
Figure 6. Overexpression of HcWRKY44 in Arabidopsis enhanced tolerance to salinity stress. (A,B) The seed germination rate was comparably analyzed between the HcWRKY44 transgenic lines and control lines on the 150 mM and 200 mM NaCl plates. The images were photographed at 7 d after germination. (C–F) The root length, leaf chlorosis rate, survival rate and electrolyte leakage were comparably analyzed between HcWRKY44 transgenic and control lines after treated with 150 and 200 mM NaCl. All the values represent the means of three independent biological replicates; error bars indicate the SD. *P < 0.05 and ***P < 0.01 represented the significant differences by Student’s t-test, respectively.
For the drought stress, similar indexes were investigated. The results showed no significant difference in germination rate between the transgenic lines and control plants under mannitol treatments. However, there was a slight difference under 300 mM mannitol treatment (Supplementary Figures 5B,C). The root length of HcWRKY44 transgenic and control lines was also investigated under different mannitol concentrations and no significant difference was observed between the overexpression lines and control plants (Supplementary Figures 5D,E). The performances of HcWRKY44 transgenic lines indicated that overexpression of HcWRKY44 enhance tolerance to the salinity stress but not to drought stress.
HcWRKY44 regulated plant’s salinity tolerance through the abscisic acid pathway
To investigate the roles of HcWRKY44 in ABA-mediated pathways, the germination rates and root lengths of HcWRKY44 transgenic lines and control plants were comparatively analyzed with and/or without ABA. On the control medium, both HcWRKY44 transgenic lines and control plants showed a similar growth tendency, with comparable germination rates, leaf greening and root lengths (Figure 7). On the contrary, when seedlings were grown on the ABA supplemented media, the HcWRKY44 transgenic lines (OE44-2#, OE44-3#, and OE44-4#) showed better performance than that of the control plants in germination rates and root lengths. The germination of HcWRKY44 transgenic lines (OE44-2#, OE44-3#, and OE44-4#) was much faster than the control lines in the presence of ABA (Figures 7A–C). The transgenic lines showed more open and green leaves than control lines when treated with different ABA concentrations (0.5 and 0.8 μM ABA) for 7 days (Figures 7B,C). Moreover, the root lengths of HcWRKY44 transgenic lines showed significantly improved growth than control plants (Figures 7D,E). On 10 and 20 μM ABA ABA concentrations, the root growth of control plants was significantly inhibited, but HcWRKY44 transgenic lines were slightly affected and had a significantly longer root length than control plants (Figures 7D,E). These results indicated that HcWRKY44 transgenic lines were resistant to ABA treatment.
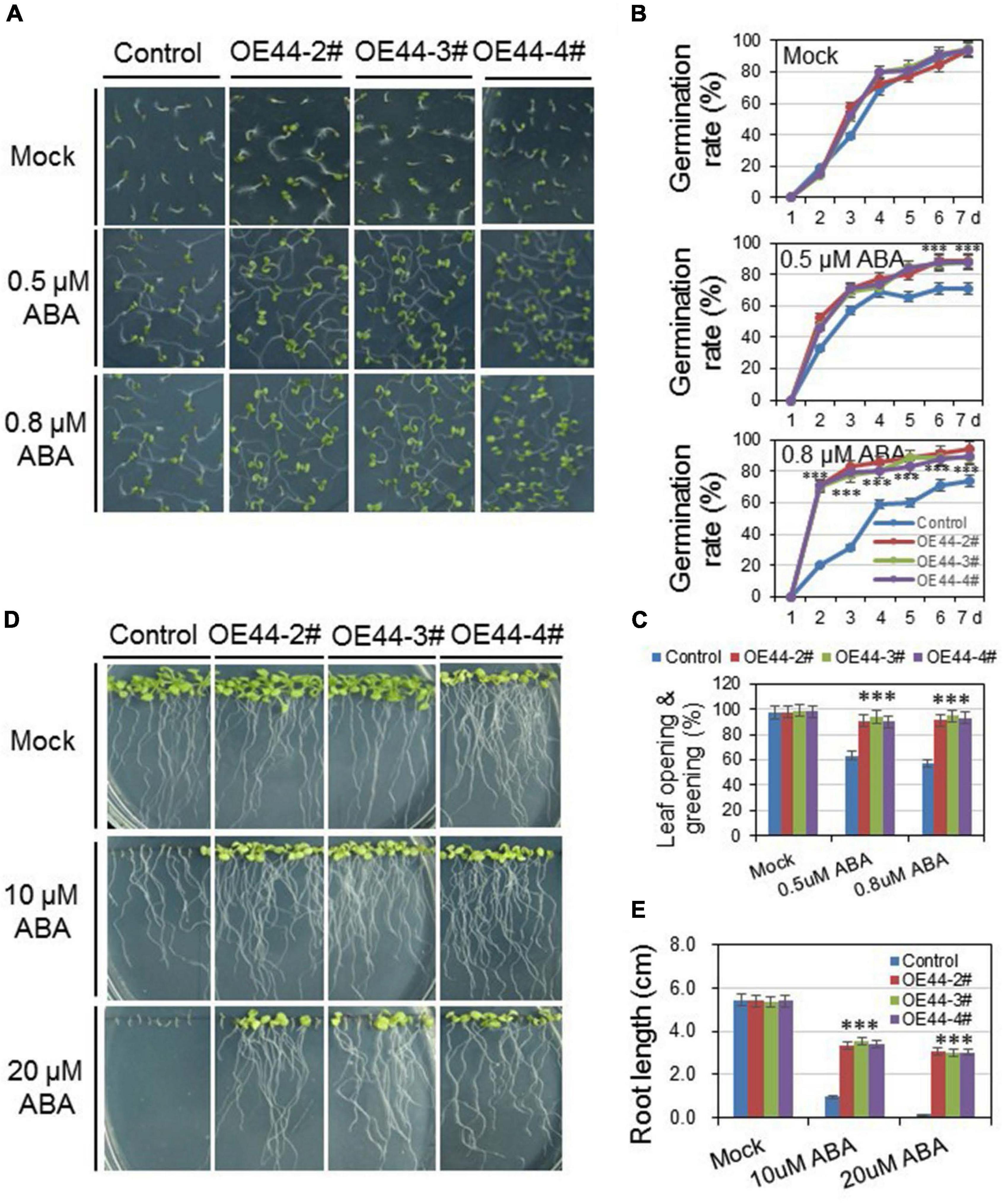
Figure 7. Overexpression of HcWRKY44 in Arabidopsis was insensitivity to ABA. (A–C) The seed germination rate and leaf opening and greening rate of HcWRKY44 transgenic lines and control lines were analyzed on 1/2 MS with or without ABA supplement. (D,E) Five-day-old seedlings grown on blank media were transferred onto new plates supplemented with 0, 10, and 20 μM ABA to investigate the ABA sensitivity between HcWRKY44 transgenic lines and control lines. All values are means (±SD) from three independent experiments. ***P < 0.01 represented the significant differences by Student’s t-test.
To further investigate the involvement of HcWRKY44 in ABA pathways, we comparably analyzed the expression profiles of ABA signaling related genes under salinity stress in the HcWRKY44 transgenic lines and control plants. After NaCl treatment for 7 days, the ABA-responsive genes, such as ABA insensitive 1 and 2 (ABI1, ABI2, and ABI5), ABA-responsive element binding factor 4 (ABF4), COR15A and COR47, were significantly up-regulated in these transgenic lines (OE44-2#, OE44-3#, and OE44-4#) (Figures 8A–F). Moreover, the stress-related marker genes, DREB2A, RD29B, STZ, P5CS, and KIN1, were also positively regulated in the transgenic lines (Figures 8G,H,J–L), while the RD22 gene was down-regulated (Figure 8I). These results further confirmed that HcWRKY44 improved the plant’s salinity tolerance through the ABA-mediated signaling pathways.
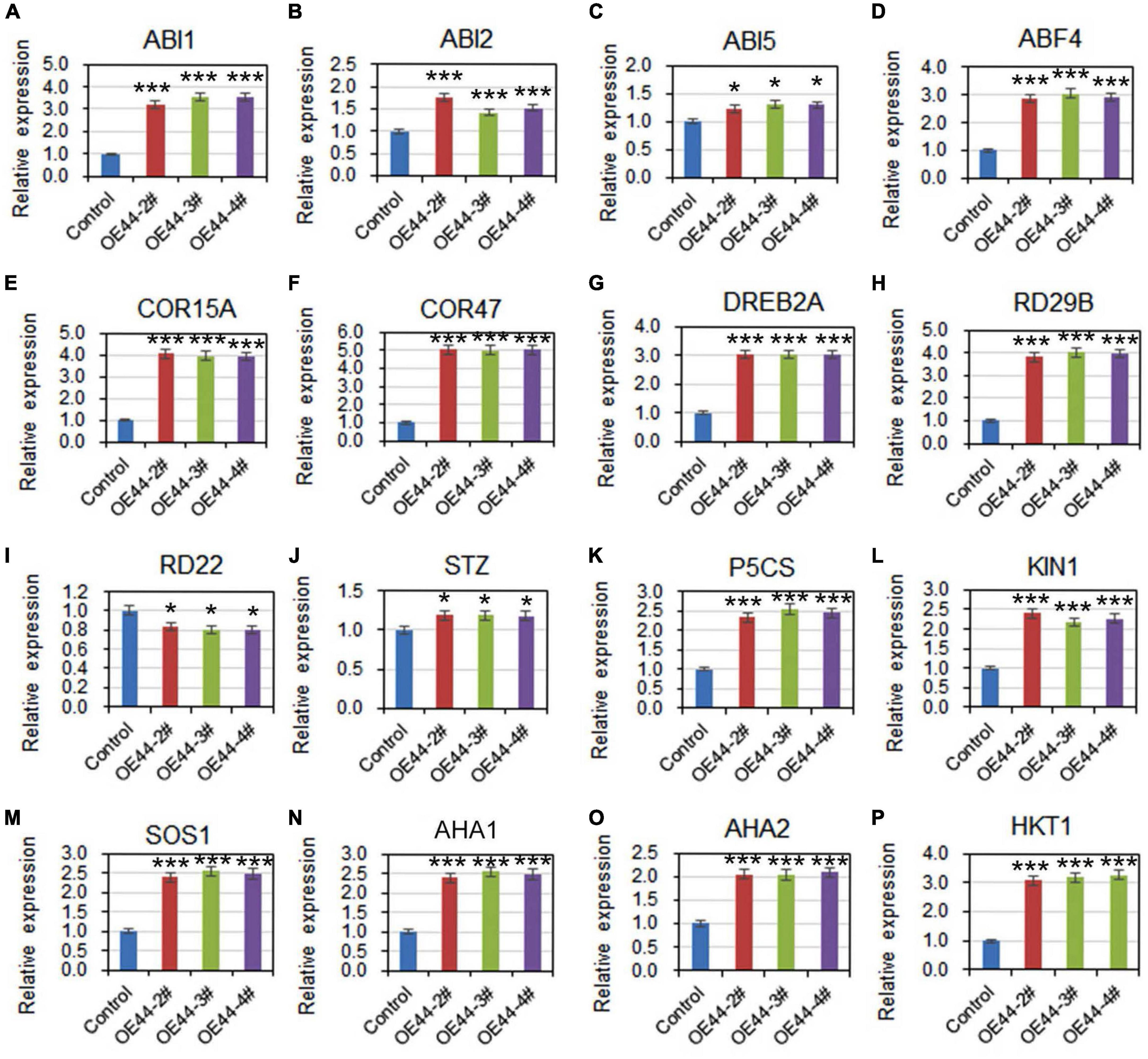
Figure 8. HcWRKY44 regulated the expression of ABA- and stress-related regulators. (A–F) ABA-mediated genes including ABI1, ABI2, ABI5, ABF4, COR15A, and COR47 were selected to analyze their expression in HcWRKY44 transgenic lines and control lines under salinity stress for 7 days. (G–L) Stress-responsive genes DREB2A, KIN1, RD29B, STZ, and P5CS were selected for evaluation of their expression level in the OE44-2#, OE44-3#, and OE44-4# lines and control lines. (M–P) Ionic homeotasis-related genes SOS1, AHA1, AHA2, and HKT1 were selected for evaluation of their expression level in the HcWRKY44 transgenic lines and control lines. Each value was the mean ± SD of three independent replicates. *P < 0.05 and ***P < 0.01 represent significant differences between different samples by Student’s t-test.
To determine HcWRKY44 improved the tolerance to salt stress but not osmotic stress, the transcript levels of Na+/H+ antiporter gene (SOS1), Na+/K+ homeostasis regulated genes (AHA1 and AHA2), and high affinity K+ transporter gene (HKT1) were further compared under salt treatment. The results showed that the mRNA transcripts of SOS1 (Figure 8M), AHA1 (Figure 8N), AHA2 (Figure 8O), and HKT1 (Figure 8P) were significantly enriched in the HcWRKY44 transgenic lines than that in the control lines (Figures 8M–P). These results confirmed that HcWRKY44 improved the plant’s tolerance to salt stress but not osmotic stress.
Discussion
The WRKY transcription factors are characterized by a highly conserved WRKYGQK heptapeptide domain and are implicated in various aspects of plant growth, development and stress responses (Rushton et al., 2010; Bakshi and Oelmuller, 2014; Wani et al., 2021). Here we identified 46 HcWRKY genes in the genome of kenaf. Compared to kenaf, its relative species cotton possesses 116 WRKY members. The difference in WRKY members might be due to kenaf’s poor genome assembly, which resulted in the lack of the entire WRKY domain in many HcWRKYs. The other possibility could be that kenaf did not experience whole genome duplication events unlike cotton. In general, the WRKY proteins of kenaf shared the typical WRKYGQK domain, and could be classified into three groups according to the types of WRKY domains and their zinc-finger domains. Gene structure analysis revealed natural mutations in HcWRKY genes, for instance, some WRKYGQK domains were altered to WRKYGKK, WRKYGEK and WRKYGQE (Supplementary Figure 2). This result coincided with the structural characteristics of WRKY genes of Arabidopsis, rice and wheat (Rushton et al., 2010; Jiang et al., 2012; Niu et al., 2012; Huang et al., 2021). The variations in WRKY gene architecture align with its functional diversity (Bakshi and Oelmuller, 2014; Zhu, 2016; Wani et al., 2021), and/or the acquisition of a novel function during the evolutionary progress. Moreover, gene duplication events, including tandem duplication, segmental duplication and whole-genome duplication, are significant factors influencing gene evolution (Qiao et al., 2018; She et al., 2022). This study found 17 (6 segmental and 11 tandem) duplication gene pairs in HcWRKY genes, and the evolutionary dates of these duplicated HcWRKY genes were also estimated using Ks as the proxy for time, the results showed that kenaf duplication events for kenaf 6 of 17 pairs occurred within the past 12.385-165.717 million years (Supplementary Table 2). This period is consistent with the speciation time of H. cannabinus that occurred 14–31 million years ago (MYA) (Zhang et al., 2020). Furthermore, several orthologous genes (64 gene pairs) were discovered between cotton and kenaf, indicating that the WRKY genes may have similar functions in different physiological processes.
In cotton, GhWRKY1 and GhWRKY6 mediated drought and salt tolerance by activating the ABA signaling pathway (Ullah et al., 2018; Hu et al., 2021). Phosphorylation of GhWRKY16 by MPK3-1 positively regulated fiber initiation and elongation (Wang et al., 2021). Interestingly, one HcWRKY gene could correspond with two or more GhWRKY genes, suggesting that these genes play a role in salinity and drought stress response as well as in fiber development. To investigate the functional roles of HcWRKY genes, the expression profiles of HcWRKY genes were performed across different tissues and stress conditions. Due to the lack of full-length sequence information and the poor genome assembly of kenaf, only 20 HcWRKY genes’ expression levels were availably obtained. The expression pattern suggested that most of the HcWRKY genes get enriched in root and are strongly induced by the salinity and drought stimuli (Figure 4 and Supplementary Figure 4). In contrast, the phloem of kenaf showed strong expression of HcWRKY2, HcWRKY14, and HcWRKY25 (Supplementary Figure 3). These findings suggested possible functions for HcWRKYs in response to salinity, drought conditions, and phloem fiber formation.
Expression pattern analysis revealed that HcWRKY genes were differencially regulated in response to salt and drought stress. Only HcWRKY7, HcWRKY16, and HcWRKY44 positively responded to salt and drought. Among these three genes, HcWRKY44 belongs to group II-c. Group II-c members are reported to be involved in abiotic stress response (Xie et al., 2005; Niu et al., 2012). Therefore, HcWRKY44 was selected as a candidate for further functional characterization. We transformed HcWRKY44 into Arabidopsis instead of kenaf because Arabidopsis plants have frequently been used in transgenic studies for gene function investigation for crops that are challenging for gene transformation (Niu et al., 2012, 2022; Luo et al., 2013; She et al., 2022). In transgenic lines, overexpression of HcWRKY44 increased salinity stress tolerance, as evidenced by longer roots, more lateral roots and a greater survival rate (Figure 6). However, HcWRKY44 overexpression lines did not respond to drought stress, and no noticeable difference was observed between the transgenic plants and control lines (Supplementary Figure 5).
Previous investigations have revealed that salinity stress is closely associated with ABA-mediated signaling pathways (Zou et al., 2004; Cutler et al., 2010; Bakshi and Oelmuller, 2014; Zhu, 2016). ABI1, ABI2, ABF4, COR15A, and COR47 were up-regulated in HcWRKY44 overexpression lines compared to control plants (Figures 8A–F). The upregulation of ABA-mediated genes is consistent with the ABA insensitive phenotype of HcWRKY44 transgenic lines in seed germination and seedling growth (Figure 7). It is worth nothing that the seed germination is regulated by a delicate balance between phytohormones GA and ABA (Cutler et al., 2010), the ABA signaling is regulated in the HcWRKY44 transgenic lines, which could be affected the seed germination. These results align with the previous studies that fine-regulation of ABA signal pathways could improve plant tolerance to salinity and drought stresses (Shen et al., 2006; Shang et al., 2010; Liu et al., 2012). Moreover, the stress-related marker genes were also positively regulated in these transgenic lines. For example, DREB2A, RD29B, STZ, P5CS, and KIN1, were positively regulated compared to control plants (Figures 8G,H,J–L). This result is consistent with previous studies that DREB2A overexpression could induce its target genes expression, including RD29A and RD29B, endowing plants with higher resistances to osmotic stress and finally improving plant’s tolerance to abiotic stresses (Sakuma et al., 2006; Jia et al., 2012; Gao et al., 2020). On the other hand, STZ, P5CS, and KIN1 were upregulated in HcWRKY44 transgenic plants under salinity stress. This is consistent with earlier findings that overexpression of the STZ gene can improve the resistance to abiotic stress (Mittler et al., 2006; Zhou et al., 2008). P5CS encodes delta-1-pyrroline-5-carboxylate synthetase, controlling proline biosynthesis and positively regulating the response of plants to salt stress (Szekely et al., 2008; Funck et al., 2020). ABA and osmotic stressors potentially activate KIN1 by binding to the dehydration-responsive element (DRE) motif in its promoter and increasing the plant’s tolerance to stress (Knight et al., 2004; Chu et al., 2018).
In addition, Na+ extrusion and K+ maintenance are critical for plants adapting to the salt stress (Zhu, 2003; Zhang et al., 2019). In our study, the ironic homeostasis-regulated genes SOS1, AHA1, AHA2, and HKT1 were significantly upregulated in the HcWRKY44 transgenic lines under salt treatment (Figures 8M–P), indicating that overexpression of HcWRKY44 gene could protect plant cells against Na+ excess through upregulation of ironic transporter genes. This result is in lines with the previous studies that salt tolerance in plants is closely related to the ability of Na+ extrusion and K+ maintenance (Shi et al., 2000; Rus et al., 2004; Sun et al., 2009; Zhang et al., 2019). These results confirmed that HcWRKY44 improved the plant’s tolerance to salt stress but not osmotic stress by regulating the ironic homeostasis-related genes.
Conclusion
The present study identified and characterized 46 WRKY transcription factor genes in the kenaf genome. Gene structure and phylogenetic analysis revealed that tandem and segment duplication might have facilitated the natural variation of HcWRKY genes. Expression pattern analysis revealed that HcWRKY2/14/25 play essential roles in the phloem of kenaf. Other HcWRKY members HcWRKY7/16/44 were enriched in roots and positively responded to the drought and salinity stresses. Furthermore, the HcWRKY44 gene was functionally characterized in Arabidopsis, and the results demonstrated overexpression of HcWRKY44 improves salinity tolerance via regulating ABA and stress-related genes. In summary, this present study provides comprehensive information about HcWRKY genes and reveals that HcWRKY44 is involved in salinity tolerance via the ABA signaling pathway.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MC and XN designed the research and wrote the manuscript. MC and ZS performed phylogenetic analysis and conducted the evolution analysis. MC, TL, ZW, and JQ performed the experiment and analyzed data. MA and XN revised the manuscript. All authors have read and approved the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (31800262 and 31501347), the Ningde Normal University Project (2020FZ04), China Postdoctoral Science Foundation (2018M632564), Natural Science Foundation of Guangxi (2022GXNSFAA035601), and Weng Hongwu Academic Innovation Research Fund of Peking University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.984233/full#supplementary-material
Footnotes
References
Bakshi, M., and Oelmuller, R. (2014). WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 9:e27700. doi: 10.4161/psb.27700
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, P., Chen, T., Li, Z., Jia, R., Luo, D., Tang, M., et al. (2020). Transcriptome analysis revealed key genes and pathways related to cadmium-stress tolerance in Kenaf (Hibiscus cannabinus L.). Ind. Crops Prod. 158:112970.
Chen, Y., Hao, X., and Cao, J. (2014). Small auxin up-regulated RNA (SAUR) gene family in maize: Identification, evolution, and its phylogenetic comparison with Arabidopsis, rice and sorghum. J. Integr. Plant Biol. 56, 133–150. doi: 10.1111/jipb.12127
Chu, M. X., Li, J. J., Zhang, J. Y., Shen, S. F., Li, C. N., Gao, Y. J., et al. (2018). AtCaM4 interacts with a Sec14-like protein, PATL1, to regulate freezing tolerance in Arabidopsis in a CBF-independent manner. J. Exp. Bot. 69, 5241–5253. doi: 10.1093/jxb/ery278
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679.
Danalatos, N. G., and Archontoulis, S. V. (2010). Growth and biomass productivity of kenaf (Hibiscus cannabinus, L.) under different agricultural inputs and management practices in central Greece. Ind. Crops Prod. 32, 231–240.
Dang, F. F., Wang, Y. N., Yu, L., Eulgem, T., Lai, Y., Liu, Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. doi: 10.1111/pce.12011
Ding, Z. J., Yan, J. Y., Xu, X. Y., Yu, D. Q., Li, G. X., Zhang, S. Q., et al. (2014). Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 79, 13–27. doi: 10.1111/tpj.12538
Fujii, H., Chinnusamy, V., Rodrigues, A., Rubio, S., Antoni, R., Park, S. Y., et al. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664.
Fujii, H., and Zhu, J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106, 8380–8385. doi: 10.1073/pnas.0903144106
Funck, D., Baumgarten, L., Stift, M., Von Wiren, N., and Schonemann, L. (2020). Differential Contribution of P5CS Isoforms to Stress Tolerance in Arabidopsis. Front. Plant Sci. 11:565134. doi: 10.3389/fpls.2020.565134
Gao, Y. F., Liu, J. K., Yang, F. M., Zhang, G. Y., Wang, D., Zhang, L., et al. (2020). The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol. Plant. 168, 98–117. doi: 10.1111/ppl.12978
Hu, Q., Ao, C. W., Wang, X. R., Wu, Y. F., and Du, X. Z. (2021). GhWRKY1-like, a WRKY transcription factor, mediates drought tolerance in Arabidopsis via modulating ABA biosynthesis. BMC Plant Biol. 21:458. doi: 10.1186/s12870-021-03238-5
Huang, S., Hu, L., Zhang, S., Zhang, M., Jiang, W., Wu, T., et al. (2021). Rice OsWRKY50 Mediates ABA-Dependent Seed Germination and Seedling Growth, and ABA-Independent Salt Stress Tolerance. Int. J. Mol. Sci. 22:8625. doi: 10.3390/ijms22168625
Jia, H. Y., Zhang, S. J., Ruan, M. Y., Wang, Y. L., and Wang, C. Y. (2012). Analysis and application of RD29 genes in abiotic stress response. Acta Physiol. Plant. 34, 1239–1250.
Jiang, Y., Liang, G., and Yu, D. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 5, 1375–1388.
Knight, H., Veale, E. L., Warren, G. J., and Knight, M. R. (2004). The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 16:2541. doi: 10.1105/tpc.11.5.875
Liu, Z. Q., Yan, L., Wu, Z., Mei, C., Lu, K., Yu, Y. T., et al. (2012). Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J. Exp. Bot. 63, 6371–6392. doi: 10.1093/jxb/ers293
Luo, X., Bai, X., Sun, X., Zhu, D., Liu, B., Ji, W., et al. (2013). Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 64, 2155–2169. doi: 10.1093/jxb/ert073
Mittler, R., Kim, Y., Song, L., Coutu, J., Coutu, A., Ciftci-Yilmaz, S., et al. (2006). Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580, 6537–6542. doi: 10.1016/j.febslet.2006.11.002
Niu, C. F., Wei, W., Zhou, Q. Y., Tian, A. G., Hao, Y. J., Zhang, W. K., et al. (2012). Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 35, 1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x
Niu, X., Chen, M., She, Z., Aslam, M., Qi, J., and Qin, Y. (2022). Ectopic Expression of Kenaf (Hibiscus cannabinus L.) HcWRKY50 Improves Plants’ Tolerance to Drought Stress and Regulates ABA Signaling in Arabidopsis. Agronomy 12:1176.
Niu, X., Chen, M., Tao, A., and Qi, J. (2016). Salinity and Drought Treatment Assays in Kenaf (Hibiscus cannabinus L.). Bio-Protocol 6:e1918.
Niu, X., Qi, J., Chen, M., Zhang, G., Tao, A., Fang, P., et al. (2015). Reference genes selection for transcript normalization in kenaf (Hibiscus cannabinus L.) under salinity and drought stress. PeerJ 3:e1347. doi: 10.7717/peerj.1347
Qiao, X., Yin, H., Li, L. T., Wang, R. Z., Wu, J. Y., Wu, J., et al. (2018). Different Modes of Gene Duplication Show Divergent Evolutionary Patterns and Contribute Differently to the Expansion of Gene Families Involved in Important Fruit Traits in Pear (Pyrus bretschneideri). Front. Plant Sci. 9:161. doi: 10.3389/fpls.2018.00161
Ramesh, M. (2016). Kenaf (Hibiscus cannabinus L.) fibre based bio-materials: A review on processing and properties. Prog. Mater. Sci. 78-79, 1–92.
Ren, X., Chen, Z., Liu, Y., Zhang, H., Zhang, M., Liu, Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. doi: 10.1111/j.1365-313X.2010.04248.x
Rus, A., Lee, B., Munoz-Mayor, A., Sharkhuu, A., Miura, K., Zhu, J. K., et al. (2004). AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 136, 2500–2511. doi: 10.1104/pp.104.042234
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11.
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258.
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. doi: 10.1105/tpc.105.035881
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
She, Z., Huang, X., Aslam, M., Wang, L., Yan, M., Qin, R., et al. (2022). Expression characterization and cross-species complementation uncover the functional conservation of YABBY genes for leaf abaxial polarity and carpel polarity establishment in Saccharum spontaneum. BMC Plant Biol. 22:124. doi: 10.1186/s12870-022-03501-3
Shen, Y. Y., Wang, X. F., Wu, F. Q., Du, S. Y., Cao, Z., Shang, Y., et al. (2006). The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443, 823–826.
Shi, H., Ishitani, M., Kim, C., and Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97, 6896–6901.
Sim, Y. Y., and Nyam, K. L. (2021). Hibiscus cannabinus L. (kenaf) studies: Nutritional composition, phytochemistry, pharmacology, and potential applications. Food Chem. 344:128582. doi: 10.1016/j.foodchem.2020.128582
Sun, J., Chen, S., Dai, S., Wang, R., Li, N., Shen, X., et al. (2009). NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiol. 149, 1141–1153. doi: 10.1104/pp.108.129494
Szekely, G., Abraham, E., Cselo, A., Rigo, G., Zsigmond, L., Csiszar, J., et al. (2008). Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53, 11–28. doi: 10.1111/j.1365-313X.2007.03318.x
Ullah, A., Sun, H., Hakim, Yang, X. Y., and Zhang, X. L. (2018). A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol. Plant. 162, 439–454. doi: 10.1111/ppl.12651
Wang, N. N., Li, Y., Chen, Y. H., Lu, R., Zhou, L., Wang, Y., et al. (2021). Phosphorylation of WRKY16 by MPK3-1 is essential for its transcriptional activity during fiber initiation and elongation in cotton (Gossypium hirsutum). Plant Cell 33, 2736–2752. doi: 10.1093/plcell/koab153
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucl. Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wani, S. H., Anand, S., Singh, B., Bohra, A., and Joshi, R. (2021). WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 40, 1071–1085. doi: 10.1007/s00299-021-02691-8
Xie, Z., Zhang, Z. L., Zou, X., Huang, J., Ruas, P., Thompson, D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. doi: 10.1104/pp.104.054312
Zhang, H., Deng, C., Yao, J., Zhang, Y., Zhang, Y., Deng, S., et al. (2019). Populus euphratica JRL mediates ABA response, ionic and ROS homeostasis in Arabidopsis under salt stress. Int. J. Mol. Sci. 20:815. doi: 10.3390/ijms20040815
Zhang, L., Xu, Y., Zhang, X., Ma, X., Zhang, L., Liao, Z., et al. (2020). The genome of kenaf (Hibiscus cannabinus L.) provides insights into bast fibre and leaf shape biogenesis. Plant Biotechnol. J. 18, 1796–1809. doi: 10.1111/pbi.13341
Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Zhu, J. K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6, 441–445.
Keywords: kenaf (Hibiscus cannabinus L.), WRKY, salinity, drought, ABA
Citation: Chen M, She Z, Aslam M, Liu T, Wang Z, Qi J and Niu X (2022) Genomic insights of the WRKY genes in kenaf (Hibiscus cannabinus L.) reveal that HcWRKY44 improves the plant’s tolerance to the salinity stress. Front. Plant Sci. 13:984233. doi: 10.3389/fpls.2022.984233
Received: 01 July 2022; Accepted: 01 August 2022;
Published: 18 August 2022.
Edited by:
Mehtab Muhammad Aslam, The Chinese University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Antt Htet Wai, Yangon University of Education (YUOE), MyanmarUmashankar Chandrasekaran, Shanghai Center for Plant Stress Biology, Shanghai Institute for Biological Sciences (CAS), China
Copyright © 2022 Chen, She, Aslam, Liu, Wang, Qi and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoping Niu, eHBuaXUwNjEzQDEyNi5jb20=
 Meixia Chen1
Meixia Chen1 Zeyuan She
Zeyuan She Mohammad Aslam
Mohammad Aslam Xiaoping Niu
Xiaoping Niu