
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 September 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.970716
This article is part of the Research Topic Molecular crosstalk between oilseed crops and pathogens View all 10 articles
Sclerotinia sclerotiorum, a worldwide distributed fungal pathogen, causes serious adverse effects on the yield and seed quality of rapeseed. Polygalacturonase-inhibiting proteins (PGIPs) can protect the cell wall from degradation by pathogen-secreted polygalacturonases (PGs). The present study found several PGIPs from Oryza sativa, especially OsPGIP6 and 3 have much higher inhibitory activities to SsPGs than BnPGIP2 from Brassica napus. Among them, OsPGIP1, 4, 6 can significantly elevate the resistance of transgenic Arabidopsis to S. sclerotiorum. Subsequently, OsPGIP1, 3, 4, 6 were subjected to SSR resistance assay in transgenic rapeseed plants. Among which, OsPGIP6 showed the highest resistance to S. sclerotiorum. At 48 h after detached leaves inoculation, the lesion area of OE-OsPGIP6 rapeseed plants is only 17.93% of the non-transgenic line, and 22.17, 21.32, 52.78, 56.47%, compared to OE-BnPGIP2, OE-OsPGIP1, OE-OsPGIP2, OE-OsPGIP4, respectively. Furthermore, the lesion area of OE-OsPGIP6 reached 10.11% compared to WT at 72 hpi. Also, the lesion length on the stem of OE-OsPGIP6 plants was reduced by 36.83% compared to WT. These results reveal that OsPGIP family, especially OsPGIP6, has a great potential in rapeseed S. sclerotiorum-resistance breeding.
Sclerotinia sclerotiorum (Lib.) de Bary, one of the worldwide distributed necrotrophic fungal pathogens, can infect more than 400 plant species, including some important economic crops, such as rapeseed, soybean, sunflower, and so on, among which rapeseed (Brassica napus L.), as one of the most important oil crops worldwide, deserves great attention (Bolton et al., 2006). Studies show that Sclerotinia stem rot (SSR) caused by S. sclerotiorum can lead to severe yield loss,10%−70% yield loss per year, and results in low oil quality due to a decrease in oil content, erucic acid content, glucosinolate, and protein content. The S. sclerotiorum attack host during all growth stages especially in the flowering period, causing white watery lesions at the stem, siliques, and leaf, which consequently affect the host physiological activities. For a long time, the obtained breeding materials were still compromised in further agricultural applications (Zhao and Wang, 2004; Liu et al., 2005; Mei et al., 2015). In addition, the pathogenesis mechanisms of SSR still remain elusive. Current research on the pathogenesis of SSR and the crop genetic improvement for SSR resistance mainly focuses on mining major genes associated with innate defense signaling pathways. Rice, as a typical model plant, have gained great attention, since it lives in high temperature and humidity and is susceptible to fungal infection. Therefore, the rice-derived resistance genes may have an important potential in fighting against fungal disease for other plants.
Till now, some breakthroughs have been made in the study of the pathogenicity and effector of S. sclerotiorum. Researchers proposed that the acidic environment rather than oxalic acid is the key factor in determining the virulence of S. sclerotiorum, namely PH-dependent theory (Liang et al., 2015; Xu et al., 2015, 2018). S. sclerotiorum can detoxify reactive oxygen species (ROS) by regulating the “copper ion import/transport” pathway. And the hosts competitively absorb copper ions in the lesion to restrict S. sclerotiorum (Dingid et al., 2020). In the face of the Brassicaceae plant's glucosinolate-myrosinase system, S. sclerotiorum metabolize isothiocyanates effectively via hydrolysis to amines that are not toxic to the fungus (Chen et al., 2020). In addition to the above studies, many other effectors involved in the pathogenicity of S. sclerotiorum have also been reported. For example, the SsCP1, a cerato-platanin protein, targeting host PR1 in apoplast facilitates the infection of S. sclerotiorum (Yang et al., 2018). The SsITL targets a chloroplast-localized calcium-sensing receptor (CAS) inhibiting SA accumulation during the early stage of infection and contributes to the virulence of S. sclerotiorum (Tang et al., 2020). The interaction between S. sclerotiorum and hosts remains to be further clarified.
It is well-known that during the early infection stage, S. sclerotiorum secretes a series of cell wall degrading enzymes (CWDEs) such as Polygalacturonases (PGs) to facilitate its colonization in the hosts. PGs can degrade the structural polysaccharides between the mesophytic layer and the primary cell wall of plants (Bolton et al., 2006). To fight against the PGs, hosts secrete polygalacturonase-inhibiting proteins (PGIPs) as the defender, which can specifically bind to the active site of PGs to inhibit their activities (Vorwerk et al., 2004; Shanmugam, 2005). Besides, PGIPs can attenuate the degradation of oligosaccharides which are able to elicit defense responses, and thus prolong the protection of the plant against pathogens (Mishra et al., 2012).
PGIPs have been widely studied with a transgenic approach in many species including Arabidopsis (Ferrari et al., 2012), rice (Lu et al., 2012; Wang et al., 2015; Feng et al., 2016), wheat (Janni et al., 2013), tobacco (Joubert et al., 2006), rapeseed (HuangFu et al., 2014; Wang et al., 2018), apple (Oelofse et al., 2006), cabbage (Hwang et al., 2010), tomato (Schacht et al., 2011), kiwi (Szankowski et al., 2003), grapevine (Szankowski et al., 2003; Richter et al., 2006; Nguema-Ona et al., 2013; Zhang et al., 2021), mulberry (Hu et al., 2012), and mungbean (Chotechung et al., 2016). The results suggest that PGIPs play crucial roles in plant fungi resistance. Therefore, PGIPs are considered effective candidate genes against S. sclerotiorum. Rice is an important crop as well as a model plant around the world. It is worth noting that rice is often cultivated in high-temperature and high-humidity environments, which makes it easier to confront with fungi. However, few studies have been carried out to discover and identify gene resources in rice that fight against fungal disease for other crops, such as oilseed rape. As for OsPGIPs, OsPGIP1, OsPGIP2, and OsPGIP4 have been identified to be involved in the defense against sheath blight and bacterial leaf streak, respectively, in transgenic plants (Wang et al., 2015; Feng et al., 2016). And another research showed that OsPGIP2 confers S. sclerotiorum resistance in B. napus through increased activation of defense mechanisms (Wang et al., 2018). Therefore, researchers have proposed that increasing the expression of OsPGIPs by transgenic technology might correspondingly increase the resistance to pathogens in transgenic lines.
In the present study, five OsPGIPs (OsPGIP1, OsPGIP2, OsPGIP3, OsPGIP4, OsPGIP6) and BnPGIP2 were functionally identified for their resistance against S. sclerotiorum both in vitro and in vivo. OsPGIPs showed obvious inhibitory activities toward PGs from S. sclerotiorum, and remarkably OsPGIP6 and OsPGIP3 exhibited high-level inhibitory activities. The results of pathogenicity assays showed that overexpressing OsPGIP2, OsPGIP3, OsPGIP4, and OsPGIP6 in rapeseed plants conferred significantly elevated resistance to S. sclerotiorum, and the S. sclerotiorum resistances were consistent with the SsPGs inhibitory activities. Among these genes, OsPGIP6 showed the highest resistance to S. sclerotiorum in both pathogenicity assays and PGIP activity assays, suggesting their potential values in crop breeding for S. sclerotiorum resistance, and a batch of high-quality transgenic resistance materials represented by OE-OsPGIP6-3 have been screened in this study.
The total genomic DNA of Nipponbare and B. napus was used as a template to amplify OsPGIP1, OsPGIP2, OsPGIP3, OsPGIP4, OsPGIP6, and BnPGIP2 (accession numbers: AM180652, AM180653, AM180654, AM180655, NM-001068720, and EU142024, respectively) with specific primers. The PCR-generated fragments were inserted into T-vector and confirmed by sequencing with primers SP6 and T7. Sequence-confirmed clones containing the target genes were digested by corresponding restriction enzymes and, respectively, cloned into prokaryotic expression vector pMAL-c2X.
Positive recombinant plasmids were, respectively, transformed into E. coli transetta (DE3), and then grown at 37°C. When the cultures reached an OD600 of 0.5, protein expression was induced by the addition of IPTG to 0.1 mM at 16°C for 20 h. Cells collected by centrifugation at 12,000 rpm for 1 min at 4°C were re-suspended in pre-cooled Hepes buffer (pH 7.0) and then broken by ultrasonication and centrifuged at 12,000 rpm for 10 min at 4°C. The supernatant and sediment were analyzed by SDS-PAGE.
For Western blot analysis, soluble proteins with different OsPGIP-MBP or MBP control were, respectively, transferred to PVDF membranes after SDS-PAGE. Western blot analysis was performed with Anti-MBP Monoclonal Antibody (New England Biolabs, E8032, 1:10,000 working dilution) as the primary antibody and HRP-conjugated murine antibody (1:10,000 working dilution) as the second antibody according to standard protocols.
PGs from S. sclerotiorum were isolated according to a previous study. PGIP activities were determined by reducing end-group analysis with the 3, 5-Dinitrosalicylic Acid (DNS) method (HuangFu et al., 2014). Ten units of PG from S. sclerotiorum (strain Ep-1PNA367) were mixed with different prokaryotically expressed proteins and were then incubated for 30 min at 25°C. Then the mixtures were, respectively, added with polygalacturonic acid (2.5 mg/ml) and incubated for 1 h at 30°C. The experiment was performed in triplicate.
Each PGIP was amplified with specific primers. Sequence-confirmed clones containing the target genes were digested by corresponding restriction enzymes and, respectively, cloned into pCHF3. The positive clones were transformed into the Agrobacterium tumefaciens strain GV3101 by electroporation. Subsequently, all the constructs were introduced into Arabidopsis (ecotype Columbia-0) and rapeseed (Westar) by Agrobacterium-mediated transformation (Chotechung et al., 2016). The transformation procedures were carried out as previously described (Janni et al., 2013). Subsequently, PCR analysis was performed to determine the presence of the target genes in transgenic plants with the specific primers 35S-F and 35S-R.
The total RNAs of the transgenic plants were extracted and reverse-transcribed as described previously (Lu et al., 2012). Reverse transcription PCR (RT-PCR), as well as quantitative real-time PCR (qRT-PCR), were performed according to the same reference. The relative expression levels were determined using method (Schmittgen and Livak, 2001).
The PCR-positive transformed plants and untransformed controls were grown in the field to the 9–10 leaf stage. And pathogenicity assay was performed with detached leaves as previously described (HuangFu et al., 2014). The sixth-eighth leaf of each transgenic plant was removed for detached leaf inoculation with S. sclerotiorum in a greenhouse. Maintaining a relatively high humidity is a key factor in this experiment. The leaf lesions were taken pictures and measured at 48 h and 72 h post-inoculation (hpi). Each treatment was performed with three independent biological replicates. And lesion sizes were measured using Image J. Normally growing rapeseed were screened for stem inoculation of S. scleroterum. Agar discs (8 mm in diameter) were excised from the edges of growing fungal colonies, which were then affixed onto rapeseed stems 50 cm from the ground. Disease severity was assessed by measuring lesion length per pathogen infection spot at 120 hpi. Inoculation, observation, and disease resistance were only determined for plants located in the center of blocks to avoid marginal effects.
In order to characterize inhibitory activities of OsPGIPs including OsPGIP1, OsPGIP2, OsPGIP3, OsPGIP4, and OsPGIP6 to PGs from S. sclerotiorum, the OsPGIPs:MBP in-frame gene fusions were, respectively, expressed in E.coli Rosetta (DE3) cell (Figure 1). The SDS-PAGE results suggested that highly active fusion protein could be obtained in the supernatant (Figure 1A). Western blot assay further proved the target proteins were successfully obtained (Figure 1B).
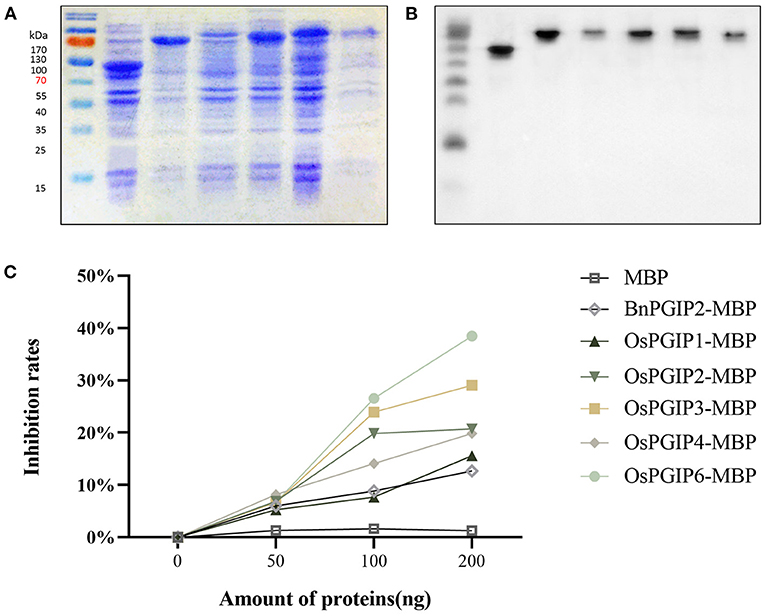
Figure 1. OsPGIPs show PG inhibitory activity of S. sclerotiorum in vitro. (A) Supernatant protein of prokaryotic expression. Expression of OsPGIPx in E. coli. M, marker; Lane 1, 20 h after IPTG inducement with MBP; Lane 2–6, 20 h after IPTG inducement with OsPGIP-MBP (Lane 2: OsPGIP1-MBP; Lane 3: OsPGIP2-MBP; Lane 4: OsPGIP3-MBP; Lane 5: OsPGIP4-MBP; Lane 6: OsPGIP6-MBP). (B) The corresponding immune signals of target proteins in (A) were generated by Western blot. Western blot was performed with Anti-MBP Monoclonal Antibody (1:10,000 working dilution) as primary antibody and HRP-conjugated murine antibody (1:10,000 working dilution) as second antibody. (C) OsPGIP activity assays. Variation of S. sclerotiorum PG activity with increasing amounts (0–200 ng) of different prokaryotically expressed proteins. Ten units of PG and 2.5 mg/ml polygalacturonic acid were used in the assays. In vitro inhibition of enzyme activity by purified PGIP protein equal the reduction percentage of D-GA in the reaction.
The PGs isolated from S. sclerotiorum were used for OsPGIPs activity assay. In vitro activity assays showed that SsPGs inhibitory activities of different PGIPs are diverse (Figure 1C). PG inhibitory rate of OsPGIP6 is high up to 26.56%, followed by OsPGIP3, OsPGIP2, OsPGIP4, OsPGIP1, and BnPGIP2 in 100 ng purified protein treatment. The PGs inhibitory rate of each PGIP has increased as the amount of purified protein did. When elevated to 200 ng, the inhibition rates of different PGIPs were increased to 38.48, 29.06, 20.75, 19.86, 15.59, and 12.67%, respectively. The results indicate that the five OsPGIPs can significantly reduce the activity of PGs hydrolyzing polygalacturonic acid in vitro. Notably, OsPGIP6 performed outstandingly.
Subsequently, the five OsPGIPs were subjected to in vivo resistance assay. Their expression levels in various tissues and organs throughout the entire life cycle of rice were shown in Figure 2A. The construct containing OsPGIPs under the control of 35S promoter were, respectively, transformed into Arabidopsis thaliana (ecotype Columbia-0) (Figure 2B). Three overexpression transgenic lines of each OsPGIP were obtained after semi-quantitative PCR (Figure 2C) and quantitative real-time PCR (qRT-PCR) (Figure 2D) identification. The target gene was expressed at high levels in all transgenic lines, and we performed inoculation assay in these OE-OsPGIPs lines.
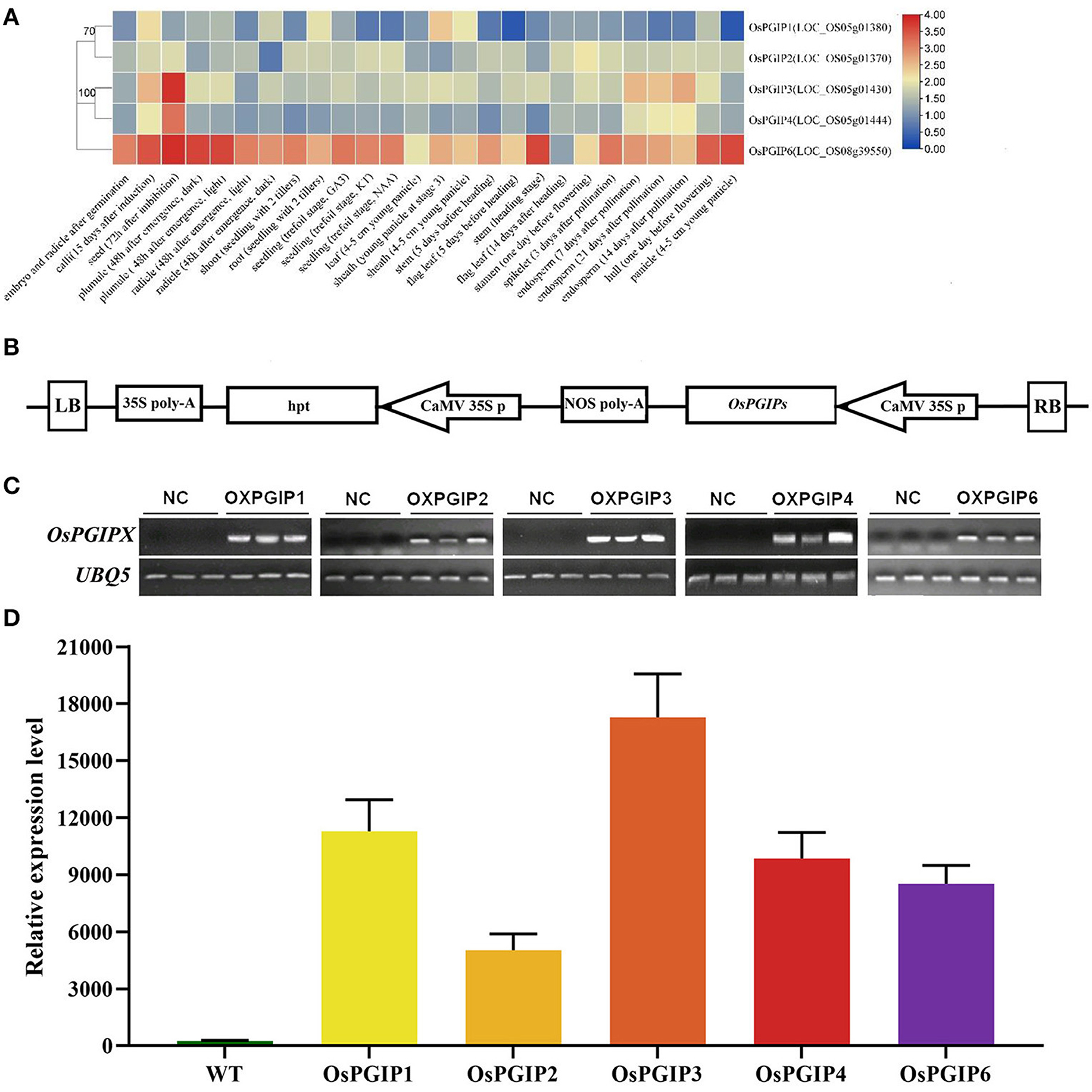
Figure 2. The expression analysis of OsPGIPs in rice and transgenic Arabidopsis plants. (A) The expression of OsPGIPs in various tissues and organs throughout the entire life cycle of rice. The color scale (representing |lg signal values|) is shown on the right. (B) Schemes of recombinant plasmids for overexpression. The expression level of each target gene was analyzed by RT-PCR (C) and qRT-PCR (D) with three independent transgenic lines. NC, negative control, Error bars indicate SE based on three independent biological replicates.
To determine the in vivo functions of the five OsPGIPs in S. sclerotiorum-plant interaction, the leaves detached from OsPGIP transgenic plants (OE-OsPGIPs) were, respectively, subjected to pathogenicity assay (Figure 3). 36 h after the inoculation of S. sclerotiorum, the infection phenotypes of the leaves were obviously diverse among different transgenic plants. The leaves of OE-OsPGIP1 and OE-OsPGIP4 showed the highest resistance to this pathogen; OE-OsPGIP2 and OE-OsPGIP6 also exhibited resistance, but it was lower than that of OE-OsPGIP1 and OE-OsPGIP4; while OE-OsPGIP3 showed no obviously elevated resistance. Furthermore, the sizes of the spreading lesions on the leaves from OsPGIPx transgenic plants (Figure 3B) indicated that their resistance to S. sclerotiorum followed the order of OE-OsPGIP4, OE-OsPGIP1, OE-OsPGIP2, OE-OsPGIP6, OE-OsPGIP3. Meanwhile, OE-OsPGIP4, OE-OsPGIP1, OE-OsPGIP2, and OE-OsPGIP6 showed significantly elevated resistance compared to the non-transgenic plants. These results also suggested that the S. sclerotiorum resistance of the transgenic plants overexpressing different OsPGIPs was in accordance with their inhibitory activities to PGs from this pathogen.
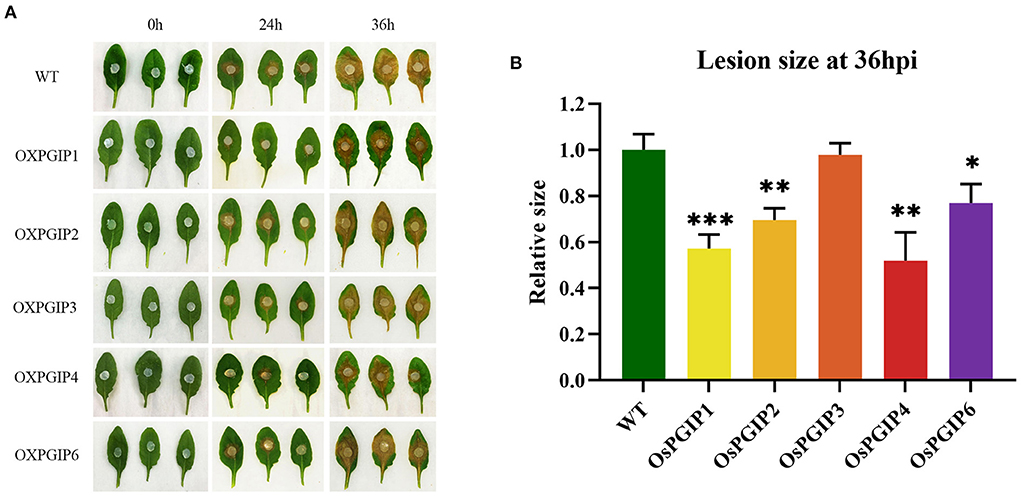
Figure 3. S. sclerotiorum resistance assay in OsPGIPx transgenic Arabidopsis. (A) Phenotypes of pathogenicity assays on detached leaves from different transgenic plants at 0 h, 24 h and 36 h after inoculation with S. sclerotiorum. (B) Statistical analysis of relative lesion sizes on the inoculated leaves at 36 hpi. Significant differences were showed by: ***P < 0.001, **P < 0.01, *P < 0.05. Error bars indicate SE based on three independent biological replicates.
Quantitative real-time PCR (qRT-PCR) was used to detect the expression levels of target genes in transgenic rapeseed plants. Ten independent biological replicates in T0 generation of each PGIP transgenic rapeseeds were subjected to expression analysis (Figure 4). Based on the results, five individual transgenic families of each PGIP with representative expression levels were selected for S. sclerotiorum resistance assay, their seed yields per plant were shown in Supplementary Figure 1.
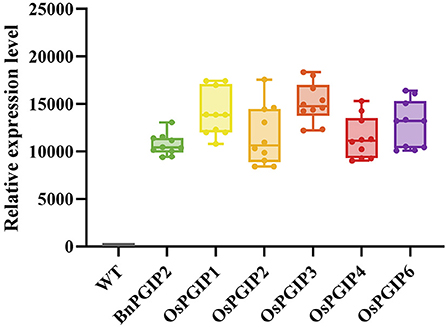
Figure 4. Relative expression of OsPGIPx in transgenic rapeseed plants. Ten individual plants in T0 generation of each PGIP transgenic rapeseeds were subjected to expression analysis.
The resistance of S. sclerotiorum was identified by leaf inoculation at the 9–10 leaf stage and detached stem inoculation at the flowering stage. The results told that 48 and 72 h after inoculation with S. sclerotiorum, the transgenic plant lesion areas were smaller than that of the wild-type material significantly. In the detached leaf inoculation, the transgenic lines, OE-OsPGIP6 and OE-OsPGIP3, showed a greatly improved resistance to SSR, especially OE-OsPGIP6. The lesion area of transgenic lines OE-OsPGIP6 and OE-OsPGIP3 are 43.63 and 57.64 mm2, respectively, while the non-transgenic line presented a lesion area of 243.36 mm2 (Figure 5) at 48 hpi. At 72 hpi, the lesion area of OE-OsPGIP6 was even reduced to 10.11% compared to the non-transgenic lines. In addition, some other transgenic strains also showed enhanced S. sclerotiorum resistance by delaying pathogen infection. When living leaves were inoculated with S. sclerotiorum, the transgenic lines, OE-OsPGIP6 and OE-OsPGIP1, showed the best resistance to SSR, with a lesion area reduction of 88.52 and 84.38% at 72 hpi (Figure 6). In the green-pod stage, the resistance of S. sclerotiorum was further identified by detached stem inoculation. The experimental results showed that the resistances of the detached stems from OE-OsPGIP2, OE-OsPGIP3, OE-OsPGIP4, and OE-OsPGIP6 to S. sclerotiorum were significantly higher than that of the wild-type after 120 h inoculation with S. sclerotiorum, and the lesion length caused by S. sclerotiorum infection was significantly lower, with a reduction of 24.27, 25.33, 28.47, and 36.83%, respectively (Figure 7).
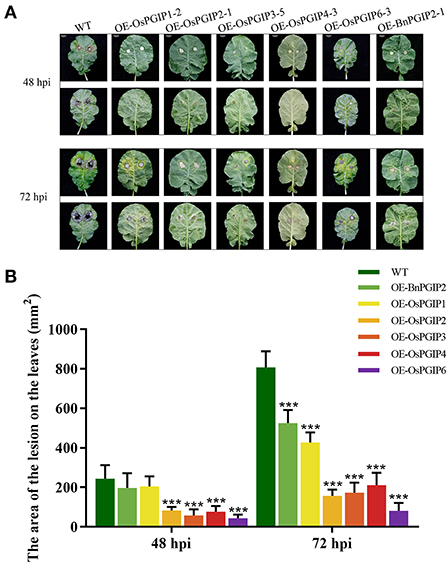
Figure 5. OsPGIPx overexpression conferred resistance to S. sclerotiorum in detached leaves inoculation. (A) Detached leaves of overexpression lines and non-transgenic line inoculated with 10-mm agar plugs of S. sclerotiorum hyphae at 22°C. Images were taken at 48 and 72 h post-inoculation (hpi). Scale bars are 20 mm. (B) Lesion on detached leaves at 48 and 72 hpi. Significant differences were showed by: ***P < 0.001. Error bars indicate SE based on five independent biological replicates.
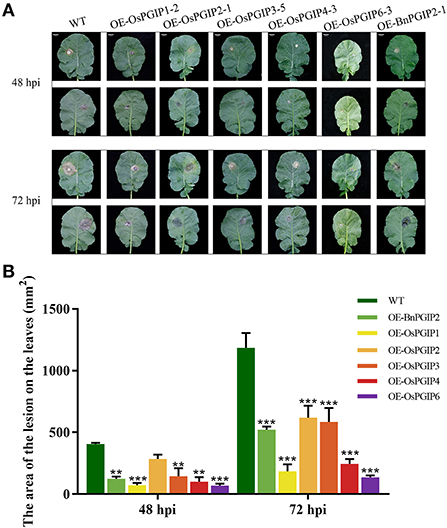
Figure 6. OsPGIPx overexpression conferred resistance to S. sclerotiorum in Living leaves inoculation. (A) Living leaves of overexpression lines and non-transgenic line inoculated with 10-mm agar plugs of S. sclerotiorum hyphae at 22°C. Lesion on living leaves at 48 and 72 hpi. Scale bars are 20 mm. (B) Lesion on living leaves at 48 and 72 hpi. Significant differences were showed by: ***P < 0.001, **P < 0.01. Error bars indicate SE based on five independent biological replicates.
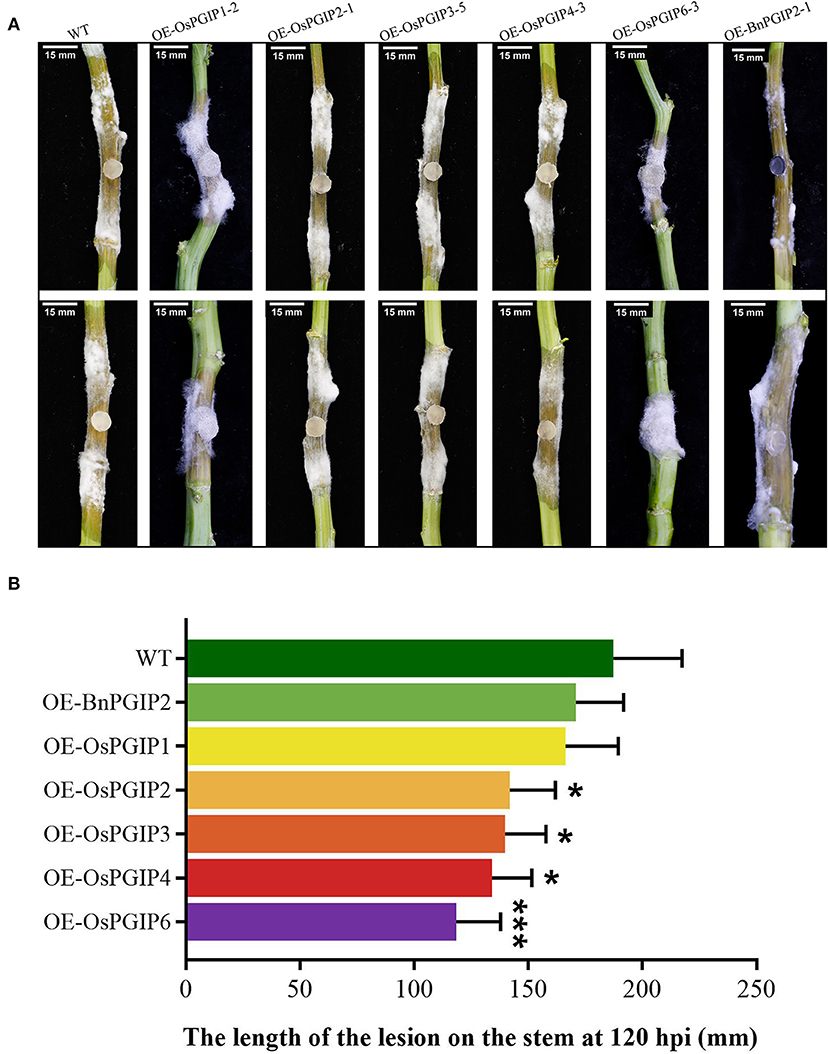
Figure 7. OsPGIPx overexpression conferred resistance to S. sclerotiorum in detached stem inoculation. (A) Stems of transgenic lines and non-transgenic line 120 h after inoculation with S. sclerotiorum. Scale bars are 15 mm. (B) Lesion on stem at 120 hpi. Significant differences were showed by: ***P < 0.001, *P < 0.05. Error bars indicate SE based on five independent biological replicates.
In nature, rice live in high temperature and high humidity environments which are more susceptible to fungi. Considering that a dozen PGIPs have been found in B. napus while few have resistance to S. sclerotiorum, we speculated that the PGIPs family from rice may have more ideal resistance potential to fighting against SSR (Bashi et al., 2013; HuangFu et al., 2014). BnPGIP2 was selected for its superior performance in BnPGIPs against S. sclerotiorum (Wang et al., 2021). Our study confirms this hypothesis, which shows that OsPGIPs have better S. sclerotiorum resistance than BnPGIP2 when introduced in B. napus, providing a new implication for rapeseed S. sclerotiorum-resistance breeding.
Although OsPGIP has been reported for its resistance to S. sclerotiorum, the candidate gene was only confined to OsPGIP2 (Wang et al., 2018). In this study, the performance of the living leaves of OsPGIP2-overexpressing rapeseed plants was not ideal compared to other OsPGIPs, which may be related to the functional specificity of different PGIPs. Therefore, the resistance gene screened by a detached test could not meet the needs of the application. Here, we found that rapeseeds overexpressing OsPGIP6 performed much better than OsPGIP2 in S. sclerotiorum invasion. Moreover, OsPGIP6-transgenic plants showed stable high resistances to S. sclerotiorum in different inoculation experiments. Hence, OsPGIP6 is a valuable S. sclerotiorum-resistance gene both in theory and applications.
The present study demonstrates that the PGIPs family from rice can effectively elevate the resistance of transgenic plants to SSR. By in vitro assays, the PG inhibitory activities of OsPGIPs were all evaluated. Subsequently, these OsPGIPs were overexpressed in Arabidopsis for pathogenicity assays. Pathogenicity assays of transgenic plants showed that OsPGIP1, OsPGIP2, OsPGIP4, and OsPGIP6 could significantly improve the resistance of Arabidopsis to S. sclerotiorum.
The PGIP activity assays showed that the inhibitory activities of different OsPGIPs to S. sclerotiorum PGs were widely divergent. OsPGIP6 and OsPGIP3 showed strong inhibitory activities, while OsPGIP1 showed relatively low inhibitory activity. The results are also consistent with the finding of previous reports, PGIPs are diverse in their inhibition specificities and potentials. Hence, it can be inferred that despite the conservation of PGIPs family across different species during evolution, each PGIP within the family is specific, especially in the domain involved in the interaction with PGs. In addition, five OsPGIPs in Arabidopsis and rapeseed showed inconsistent resistances to S. sclerotiorum, which may be caused by the complex polyploidy genome of B. napus. As an allotetraploid plant, a more complicated defense system could further improve or alter the effects of resistance genes.
The PGIP-PG interaction is a classic immune evolutionary system model for analyzing the resistant-protein and pathogenic factors. Pathogens produced varieties of PGs to enhance aggressivity, while the hosts have evolved corresponding resistant proteins to minimize harmlessness. Therefore, PGIPs are diverse in their inhibition specificities and potentials. Combined with the results, it can be inferred that OsPGIP6 has a high potential in preventing S. sclerotiorum from degrading the plant cell wall. The specific molecular mechanism needs to be elucidated in further study.
The results of living leaves pathogenicity assays showed that the S. sclerotiorum resistance of OsPGIP2 transgenic plants was obviously lower than that of OsPGIP4 transgenic plants, while the PG-inhibitory activity of OsPGIP2 was almost the same as that of OsPGIP4. This phenotype might arise from the less elevation of expression levels of the target gene in the OsPGIP2-overexpressing line compared with the OsPGIP4-overexpressing line. Our previous report also showed that the resistance of transgenic plants is related to the expression levels of PGIP (Wang et al., 2015). Since the potential PG-inhibitory ability of OsPGIP2 is like that of OsPGIP4, it can be inferred that if the expression level of OsPGIP2 in transgenic plants is further elevated, the resistance of these plants will be further improved.
In this study, S. sclerotiorum-resistant rapeseed plants were obtained by introducing the rice-derived resistance genes, OsPGIPs, into B. napus, proved by in vitro and in vivo pathogenicity assays. Different OsPGIPs have diverse potential in S. sclerotiorum resistance. OsPGIP6-overexperssing lines showed vastly lesion reduction compared to the non-transgenic line, also other OsPGIPs- and BnPGIP- transgenic lines. Overall, our results show that OsPGIPs could provide substantial S. sclerotiorum resistance in rapeseeds and facilitate SSR-resistance crop breeding.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
YZ, YL, RW, MY, SL, JJ, WW, and MW contributed to design of the study. MY and SL performed the data analysis and completed the original draft. JJ provided partial resources. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (2016YFD0100202), the National Key Research and Development Plan (2018YFD0100501), Sichuan Province (Grants 2021YJ0296, 2021YFYZ0018, and 2022ZDZX0015), and People's Republic of China. This research was also supported by the Fundamental Research Funds for the Central Universities (SCU2019D013).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.970716/full#supplementary-material
Bashi, Z. D., Rimmer, S. R., Khachatourians, G. G., and Hegedus, D. D. (2013). Brassica napus polygalacturonase inhibitor proteins inhibit Sclerotinia sclerotiorum polygalacturonase enzymatic and necrotizing activities and delay symptoms in transgenic plants. Can. J. Microbiol. 59,79–86. doi: 10.1139/cjm-2012-0352
Bolton, M. D., Thomma, B. P. H. J., and Nelson, B. D. (2006). Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. doi: 10.1111/j.1364-3703.2005.00316.x
Chen, J., Ullah, C., Reichelt, M., Beran, F., Yang, Z. L., Hammerbacher, A., et al. (2020). The phytopathogenic fungus Sclerotinia sclerotiorum detoxifies plant glucosinolate hydrolysis products via an isothiocyanate hydrolase. Nat. Commun. 11, 3090. doi: 10.1038/s41467-020-16921-2
Chotechung, S., Somta, P., Chen, J., Yimram, T., Chen, X., and Srinives, P. (2016). A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: bruchidae) resistance in mungbean (Vigna radiata). Theor. Appl. Genet. 129, 1673–1683. doi: 10.1007/s00122-016-2731-1
Dingid, Y., Chai, Y., Mei, J., Yang, W., Mao, Y., Yan, B., et al. (2020). Sclerotinia sclerotiorum utilizes host-derived copper for ROS detoxification and infection. PLoS Pathog. 16, e1008919. doi: 10.1371/journal.ppat.1008919
Feng, C., Zhang, X., Wu, T., Yuan, B., Ding, X., Yao, F., et al. (2016). The polygalacturonase-inhibiting protein 4 (OsPGIP4), a potential component of the qBlsr5a locus, confers resistance to bacterial leaf streak in rice. Planta 243, 1297–1308. doi: 10.1007/s00425-016-2480-z
Ferrari, S., Sella, L., Janni, M., de Lorenzo, G., Favaron, F., and D'Ovidio, R. (2012). Transgenic expression of polygalacturonase-inhibiting proteins in Arabidopsis and wheat increases resistance to the flower pathogen Fusarium graminearum. Plant Biol. 14, 31–38. doi: 10.1111/j.1438-8677.2011.00449.x
Hu, D., Dai, R., Wang, Y., Zhang, Y., Liu, Z., Fang, R., et al. (2012). Molecular cloning, sequence analysis, and expression of the polygalacturonase-inhibiting protein (PGIP) gene in mulberry. Plant Mol. Biol. Rep. 30, 176–186. doi: 10.1007/s11105-011-0324-3
HuangFu, H., Guan, C., Jin, F., and Yin, C. (2014). Prokaryotic expression and protein function of Brassica napus PGIP2 and its genetic transformation. Plant Biotechnol. Rep. 8, 171–181. doi: 10.1007/s11816-013-0307-y
Hwang, B. H., Bae, H., Lim, H. S., Kim, K. B., Kim, S. J., Im, M. H., et al. (2010). Overexpression of polygalacturonase-inhibiting protein 2 (PGIP2) of Chinese cabbage (Brassica rapa ssp. pekinensis) increased resistance to the bacterial pathogen Pectobacterium carotovorum ssp. carotovorum. Plant Cell Tissue Org. 103, 293–305. doi: 10.1007/s11240-010-9779-4
Janni, M., Bozzini, T., Moscetti, I., Volpi, C., and D'Ovidio, R. (2013). Functional characterisation of wheat Pgip genes reveals their involvement in the local response to wounding. Plant Biol. 15, 1019–1024. doi: 10.1111/plb.12002
Joubert, D. A., Slaughter, A. R., Kemp, G., Becker, J. V. W., Krooshof, G. H., Bergmann, C., et al. (2006). The grapevine polygalacturonase-inhibiting protein (VvPGIP1) reduces Botrytis cinerea susceptibility in transgenic tobacco and differentially inhibits fungal polygalacturonases. Transgenic Res. 15, 687–702. doi: 10.1007/s11248-006-9019-1
Liang, X., Liberti, D., Li, M., Kim, Y., Hutchens, A., Wilson, R., et al. (2015). Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 16, 559–571. doi: 10.1007/s41348-021-00509-z
Liu, S., Wang, H., Zhang, J., Fitt, B. D. L., Xu, Z., Evans, N., et al. (2005). In vitro mutation and selection of doubled-haploid Brassica napus lines with improved resistance to Sclerotinia sclerotiorum. Plant Cell Rep. 24, 133–144. doi: 10.1007/s00299-005-0925-0
Lu, L., Zhou, F., Zhou, Y., Fan, X., Ye, S., Wang, L., et al. (2012). Expression profile analysis of the polygalacturonase-inhibiting protein genes in rice and their responses to phytohormones and fungal infection. Plant Cell Rep. 31, 1173–1187. doi: 10.1007/s00299-012-1239-7
Mei, J., Liu, Y., Wei, D., Wittkop, B., Ding, Y., Li, Q., et al. (2015). Transfer of sclerotinia resistance from wild relative of Brassica oleracea into Brassica napus using a hexaploidy step. Theor. Appl. Genet. 128, 639–644. doi: 10.1007/s00122-015-2459-3
Mishra, A. K., Sharma, K., and Misra, R. S. (2012). Elicitor recognition, signal transduction and induced resistance in plants. J. Plant Interact. 7, 95–120. doi: 10.1080/17429145.2011.597517
Nguema-Ona, E., Moore, J. P., Fagerström, A. D., Fangel, J. U., Willats, W. G. T., Hugo, A., et al. (2013). Overexpression of the grapevine PGIP1 in tobacco results in compositional changes in the leaf arabinoxyloglucan network in the absence of fungal infection. BMC Plant Biol. 13, 46. doi: 10.1186/1471-2229-13-46
Oelofse, D., Dubery, I. A., Meyer, R., Arendse, M. S., Gazendam, I., Berger, D. K., et al. (2006). Apple polygalacturonase inhibiting protein1 expressed in transgenic tobacco inhibits polygalacturonases from fungal pathogens of apple and the anthracnose pathogen of lupins. Phytochemistry 67, 255–263. doi: 10.1016/j.phytochem.2005.10.029
Richter, A., Jacobsen, H. J., de Kathen, A., de Lorenzo, G., Briviba, K., Hain, R., et al. (2006). Transgenic peas (Pisum sativum) expressing polygalacturonase inhibiting protein from raspberry (Rubus idaeus) and stilbene synthase from grape (Vitis vinifera). Plant Cell Rep. 25, 1166–1173. doi: 10.1007/s00299-006-0172-z
Schacht, T., Unger, C., Pich, A., and Wydra, K. (2011). Endo- and exopolygalacturonases of Ralstonia solanacearum are inhibited by polygalacturonase-inhibiting protein (PGIP) activity in tomato stem extracts. Plant Physiol. Biochem. 49, 377–387. doi: 10.1016/j.plaphy.2011.02.001
Schmittgen, T. D., and Livak, K. J. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Shanmugam, V. (2005). Role of extracytoplasmic leucine rich repeat proteins in plant defence mechanisms. Microbiol. Res. 160, 83–94. doi: 10.1016/j.micres.2004.09.014
Szankowski, I., Briviba, K., Fleschhut, J., Schönherr, J., Jacobsen, H. J., and Kiesecker, H. (2003). Transformation of apple (Malus domestica Borkh.) with the stilbene synthase gene from grapevine (Vitis vinifera L.) and a PGIP gene from kiwi (Actinidia deliciosa). Plant Cell Rep. 22, 141–149. doi: 10.1007/s00299-003-0668-8
Tang, L. G., Yang, G. G., Ma, M., Liu, X. F., Li, B., Xie, J. T., et al. (2020). An effector of a necrotrophic fungal pathogen targets the calcium-sensing receptor in chloroplasts to inhibit host resistance. Mol. Plant Pathol. 21, 686–701. doi: 10.1111/mpp.12922
Vorwerk, S., Somerville, S., and Somerville, C. (2004). The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci. 9, 203–209. doi: 10.1016/j.tplants.2004.02.005
Wang, R., Lu, L., Pan, X., Hu, Z., Ling, F., Yan, Y., et al. (2015). Functional analysis of OsPGIP1 in rice sheath blight resistance. Plant Mol. Biol. 87, 181–191. doi: 10.1007/s11103-014-0269-7
Wang, Z., Wan, L., Xin, Q., Chen, Y., Zhang, X., Dong, F., et al. (2018). Overexpression of OsPGIP2 confers Sclerotinia sclerotiorum resistance in Brassica napus through increased activation of defense mechanisms. J. Exp. Bot. 69, 3141–3155. doi: 10.1093/jxb/ery138
Wang, Z. R., Wan, L., Zhang, X. H., Xin, Q., Song, Y. X., Hong, D. F., et al. (2021). Interaction between Brassica napus polygalacturonase inhibition proteins and Sclerotinia sclerotiorum polygalacturonase: implications for rapeseed resistance to fungal infection. Planta 253, 34. doi: 10.1007/s00425-020-03556-2
Xu, L., Li, G., Jiang, D., and Chen, W. (2018). Sclerotinia sclerotiorum: an evaluation of virulence theories. Annu. Rev. of Phytopathol. 56, 311–338. doi: 10.1146/annurev-phyto-080417-050052
Xu, L., Xiang, M., White, D., and Chen, W. (2015). pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate-minus mutants of Sclerotinia sclerotiorum. Environ. Microbiol. 17, 2896–2909. doi: 10.1111/1462-2920.12818
Yang, G., Tang, L., Gong, Y., Xie, J., Fu, Y., Jiang, D., et al. (2018). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. N. Phytol. 217, 739–755. doi: 10.1111/nph.14842
Zhang, Q., Yan, Q., Yuan, X., Lin, Y., Chen, J., Wu, R., et al. (2021). Two polygalacturonase-inhibiting proteins (VrPGIP) of Vigna radiata confer resistance to bruchids (Callosobruchus spp.). J. Plant Physiol. 153376, 258–259. doi: 10.1016/j.jplph.2021.153376
Keywords: Sclerotinia sclerotiorum, Brassica napus, PGIP, PG inhibitory activity, Arabidopsis thaliana
Citation: Yin M, Wang R, Li S, Luo M, Wei W, Wang M, Jiang J, Lin Y and Zhao Y (2022) High Sclerotinia sclerotiorum resistance in rapeseed plant has been achieved by OsPGIP6. Front. Plant Sci. 13:970716. doi: 10.3389/fpls.2022.970716
Received: 16 June 2022; Accepted: 06 July 2022;
Published: 15 September 2022.
Edited by:
Cunmin Qu, Southwest University, ChinaReviewed by:
Yujie Fang, Yangzhou University, ChinaCopyright © 2022 Yin, Wang, Li, Luo, Wei, Wang, Jiang, Lin and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Zhao, emhhb3l1bkBzY3UuZWR1LmNu; Yongjun Lin, eW9uZ2p1bmxpbkBtYWlsLmh6YXUuZWR1LmNu; Jun Jiang, amlhbmdqdW4yMjdAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.