
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 06 July 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.918437
This article is part of the Research Topic Genomics and disease resistance in wheat and maize View all 16 articles
Wheat stripe rust is an important foliar disease that affects the wheat yield globally. Breeding for resistant wheat varieties is one of the most economically and environmentally effective ways to control this disease. The common wheat (Triticum aestivum L.) cultivar “Pascal” exhibited susceptibility to stripe rust at the seedling stage but it showed high resistance to stripe rust at the adult plant stage over 20 years in Gansu, a hotspot of the disease in northwestern China. To understand the genetic mechanism of stripe rust resistance in this cultivar, a 55K SNP array was used to analyze the two parents and the 220 recombinant inbred lines (RILs) derived from the cross of “Huixianhong” × “Pascal.” We detected three new stripe rust adult plant resistance (APR) quantitative trait locus (QTL) contributed by Pascal, viz. QYr.gaas-1AL, QYr.gaas-3DL, and QYr.gaas-5AS, using the inclusive composite interval mapping method. They were flanked by SNP markers AX-111218361—AX-110577861, AX-111460455—AX-108798599, and AX-111523523—AX-110028503, respectively, and explained the phenotypic variation ranging from 11.0 to 23.1%. Bulked segregant exome capture sequencing (BSE-Seq) was used for fine mapping of QYr.gaas-1AL and selection of candidate genes, TraesCS1A02G313700, TraesCS1A02G313800, and TraesCS1A02G314900 for QYr.gaas-1AL. KASP markers BSE-1A-12 and HXPA-3D for QYr.gaas-1AL and QYr.gaas-3DL were developed for breeders to develop durable stripe rust-resistant wheat varieties.
Wheat stripe rust, also known as yellow rust, caused by the air-borne fungus Puccinia striiformis f. sp. tritici (Pst), is considered the primary biotic threat to wheat production globally (Todorovska et al., 2009). In 2017, 88% of the world’s wheat production was susceptible to stripe rust, causing yield losses estimated to be 1 billion USD (Brandt et al., 2021), and a countrywide epidemic affected about 5.5 million ha of wheat in China (Zeng et al., 2022). It shows regional characteristics and occurs frequently in the southwestern and northwestern of China, especially in the southeastern Gansu, a stripe rust hotspot in northwestern of China (Bai et al., 2012). The management of stripe rust includes the deployment of resistant wheat and the use of fungicide, and breeding resistant wheat cultivars is the most effective and economic strategy for controlling stripe rust (Wen et al., 2008).
Genetic resistance to stripe rust in wheat can be broadly divided into two phenotypically, mechanistically, and genetically distinct categories: (a) seedling resistance (all-stage resistance, ASR), which is detected at the seedling stage but is also expressed at all developmental stages. It usually shows a major effect and is race specific (Roberts and Caldwell, 1970; Lin and Chen, 2007); (b) adult plant resistance (APR), which is expressed at the adult plant stages and appears to be durable. Some APR genes are less characterized and are difficult to select phenotypically (Ren R. et al., 2012).
To date, most identified stripe rust resistance genes conferring “seedling resistance” encode classic nucleotide-binding site leucine-rich repeat (NBS-LRR) R proteins that recognize effectors and trigger a defense response to resist disease. While it is easy to overcome the recognition of a single classical R gene via a single genetic variation in an avirulence gene (Steuernagel et al., 2016; Schwessinger, 2017). The major epidemics causing severe economic losses have occurred in various regions of the world including United States, China, and Australia (Nsabiyera et al., 2018) due to virulence on seedling resistance genes including Yr2, Yr6, Yr9, Yr17, and Yr27 (Wellings and McIntosh, 1990; Nsabiyera et al., 2018). Only a few seedling stripe rust resistance genes (i.e., Yr5 and Yr15) are still effective to some Pst races in China, which drives the demand for persistent disease-resistant cultivars.
Adult plant resistance is generally known as more durable than ASR, since a single genetic variation appears insufficient to overcome this type of resistance in the asexual stage of Pst (Schwessinger, 2017). In general, APR delays infection and production of spore, leading to slow rusting phenotypes instead of complete immunity. This type of resistance gene usually encodes allele-specific protein variants that are molecularly unrelated to NBS-LRR proteins. In the case of these genes, Yr18 and Yr46 encode two types of transporters (Krattinger et al., 2009; Moore et al., 2015). In addition, Yr36, a chloroplast-localized kinase WKS1 regulating, reactive oxygen species production (Ellis et al., 2014; Gou et al., 2017). Since the durability of R genes is usually defined by the global genetic diversity of the pathogen, the combination of APR genes with ASR genes is the best way for a wheat breeder to develop the durable stripe rust resistant wheat variety at all plant stages. To date, more than 83 Yr genes or alleles (Yr1–Yr83) have been formally named in wheat (Li et al., 2020; Draz et al., 2021). Most of them are race-specific ASR genes, and only a few are APR genes. Lr34/Yr18/Pm38/Sr57 (Dyck, 1991), Lr46/Yr29/Pm39/Sr58 (Singh et al., 1998), and Lr67/Yr46/Pm46/Sr55 (Herrera-Foessel et al., 2011) confer pleiotropic APR to stripe rust, leaf rust, powdery mildew, and stem rust. QTL analysis has been widely used to dissect complex traits through identifying the genomic location and effects of genes contributing to quantitative variation (Young, 1996). Over the past 20 years, more than 320 stripe rust resistance QTLs have been reported using different molecular markers, including diversity array technology (DArT), single sequence repeats (SSRs), and single-nucleotide polymorphisms (SNPs) (Lan et al., 2017); the genetic locations of these QTLs are continually being refined through fine mapping studies (Lan et al., 2017; Draz et al., 2021).
Various approaches can be used to identify genetic loci and genes in wheat. High-density SNP markers using gene-chip technology provide a superior approach for QTL mapping due to their high-throughput, efficiency, allele specificity, and high-resolution capacity (Yu et al., 2011; Draz et al., 2021). The Axiom wheat 55K SNP array with 53,063 SNP probes was selected from the wheat 660K SNP array, which is considered to be more appropriate for wheat genetic research (Ren et al., 2018; Huang et al., 2019). Based on the advantages of lower costs, higher accuracy, and its medium density, this SNP array has been widely used in QTL identification in wheat (Huang et al., 2019; Zhang et al., 2019). Also, whole-genome exome sequencing (WES) has been successfully applied for the identification of genetic loci and isolating genes in wheat (Henry et al., 2014; Mo et al., 2018). For instance, candidate natural variants were identified using a 110 Mb exome capture assay at the Yr6 locus responsible for stripe rust resistance (Gardiner et al., 2016). A BSA-based exome capture sequencing pipeline for rapid gene cloning has been developed by Chengdu Tcuni Technology, which is an alternative method to effectively sequence coding regions with low cost in wheat (Dong et al., 2020). Therefore, SNP chip and WES approaches can be the effective strategies to find new genetic loci and candidate genes associated with important traits in wheat.
The Italian common winter wheat cv. Pascal (Lan Yin 1), introduced to Gansu Province of China in 1995, showed a high level of APR to stripe rust in the field but was susceptible to CYR32 and CYR33 at the seedling stage. However, the seedling gene locations and inheritance of APR to stripe rust in Pascal were not clear. Thus, the objectives of this study were to (1) map seedling stripe rust resistance gene in the Huixianhong/Pascal F5 RIL population; (2) identify the APR to stripe rust in the same population; (3) determine the interaction effect between the identified resistance loci on stripe rust in the adult plant stage; and (4) develop molecular markers of new and stable resistance QTL for the wheat breeder to breed durable stripe rust-resistant wheat variety.
A population of 220 F5 RILs was developed from a cross between “Huixianhong” and “Pascal.” “Pascal” is a stripe rust-resistant parent, whereas the cultivar “Huixianhong” is highly susceptible to stripe rust in the field. Huixianhong was widely used as a susceptible spreader and negative control in the genetic analysis of wheat stripe rust resistance. A single seed descend method was used to develop the F5 RIL population.
The parents were evaluated for their reaction to stripe rust at the seedling stage using the Pst races CYR32 and CYR33 under greenhouse conditions at Huazhong Agricultural University, and the reaction to CYR34 was tested in the Gansu Academy of Agricultural Sciences. The races of the pathogen were inoculated by spraying urediniospores suspended in mineral oil using an atomizer when wheat seedlings reached the two-leaf stage. The inoculated plants were left in an open area for 20 min to facilitate the evaporation of the oil and then placed in the dew chamber of 7°C for 24 h and back to the greenhouse. The infection types (ITs) were recorded approximately 2 weeks post-inoculation, based on a 0–9 scale modified from McNeal et al. (1971), and ITs with the scale of “0, 1, 2, 3, 4,” “5, 6,” and “7, 8, 9” are categorized as resistant, intermediate, and susceptible groups, respectively.
The parents and RIL population were evaluated for APR to stripe rust in Qingshui Experimental Station of Gansu Academy of Agricultural Sciences during the 2017–2018 and the 2018–2019 growing seasons (GS2018 and GS2019), and in 2018–2019 growing season in Pidu Experimental Station of Sichuan Academy of Agricultural Sciences (SC2019).
Field plots consisted of 1.5-m rows planted with approximately fifty seeds of each line, and we used randomized complete block design with two replicates of the RIL population in each location. “Jinmai 47” was planted at every tenth plot of experimental materials and around the experiment as a susceptible check and spreader to ensure the maximum inoculum for disease development. Disease severity (DS) for the RILs and their parents in each environment was recorded one time or two times following the modified Cobb’s scale (Peterson et al., 1948).
In 2018, we took the first rating when the susceptible check Huixianhong showed approximately 80% DS around 12 June, and the second rating about a week later when the DS of the susceptible check reached 90–100% in Gansu. In the other two environments, we investigated the phenotype once when the DS of susceptible check reached 90–100%. Combined with the natural infection, a mixture of Pst races CYR32, CYR33, and CYR34 was used to inoculate the spreaders at the jointing growth stage (around 2 months after planting). The lme4 package in Rstudio1 was used to calculate the best linear unbiased prediction (BLUP).
SAS 9.2 software (SAS Institute, Cary, NC) was used to calculate the correlations of final disease severity (FDS) for stripe rust in each season. The analysis of variance (ANOVA) was used to assess the significant effect of a single stripe rust resistance QTL and the interaction effect for multiple stripe rust resistance QTLs with the FDS values in each environment.
Deoxyribonucleic acid was extracted from about 20 plants of each line and parents using the cetyltrimethylammonium bromide (CTAB) method (Chatterjee et al., 2002). The 55K SNP array genotyping platform was used to genotype the parents and RIL population. A total of 55,000 SNP markers were obtained, of which 9,475 SNP markers were selected to make linkage groups by removing markers of distorted segregation (p < 0.001), monomorphic markers, and markers with more than 30% missing data. Genetic linkage groups were established using Joinmap 4.1 (Van, 2011) with a logarithm of odds (LODs) threshold of 10.0. MapChart (Voorrips, 2002) was used to draw genetic linkage maps. Software IciMapping 4.1 (Meng et al., 2015) was used to detect FDS-related QTL in each environment, BLUP and mean of final disease severity (MFDS) across each season to obtain the significant QTL position, LOD scores, phenotypic variance explained (PVE), and the additive effect of each locus. About one-thousand permutations were used to calculate LOD scores for each trait. The physical positions of identified QTL were compared with those previously reported on the same chromosome arm under different wheat lines, based on the wheat genome of (International Wheat Genome Sequencing Consortium [IWGSC], 2018).
The bulked segregant exome capture sequencing (BSE-Seq) was used for identifying causal mutations or candidate genes in this study. First, the genomic DNA of 33 extremely resistance (low FDS) lines and 43 extremely susceptible (high FDS) lines from the Huixianhong/Pascal F5 population was extracted by CTAB method (Chatterjee et al., 2002) and then bulked in an equal amount of each line to generate the resistant-type bulked DNA pool and susceptible-type bulked DNA pool separately. The genomic DNA of Huixianhong and Pascal was also extracted using the same method as above. Therefore, the resistant-type bulked DNA pool, susceptible-type bulked DNA pool, resistant-type parent DNA, and susceptible-type parent DNA were used to Exome Capture Sequencing, which was developed by Chengdu Tcuni Technology.
The Euclidean Distance (ED) algorithm is a method using sequencing data to find significantly different markers between bulks and evaluates the region associated with the target trait (Hill et al., 2013).
In the formula, A, T, C, and G mut are the frequencies of A, T, C, and G bases in the mutant bulk, respectively; A, T, C, and G wt are the frequencies of A, T, C, and G bases in the wild-type bulk, respectively. Theoretically, the ED values of other loci should tend to 0 except for the target trait-related sites between the two mixed pools.
Both Huixianhong and Pascal were susceptible after being inoculated by CYR32, CYR33, and CYR34 at the seedling stage (IT varied from 78 to 8) (Figures 1A,B). However, the MFDS for Huixianhong and Pascal were 100 and 3%, respectively, in the adult plant stage (Figures 1C,D) over three environments. The frequency distributions of stripe rust severity among F5 RILs showed a normal distribution (Figure 2), indicating a typical quantitative character for controlling stripe rust resistance in the Huixianhong/Pascal population. Pearson correlation coefficients (r) of stripe rust severity ranged from 0.66 to 0.85 over the environments (Table 1). In addition, it was estimated that there were around 3–4 stripe rust resistance genes with additive effects in the Huixianhong/Pascal RIL population based on the qualitative assessment of the Mendelian genetic segregation ratios (Table 2).

Figure 1. The two parents Huixianhong (left) and Pascal (right) showed susceptible against stripe rust race CYR32 (A) and CYR33 (B) at the seedling stage. The response of susceptible parent Huixianhong (C) and the resistant parent Pascal (D) in the adult plant stage under Gansu province field test.
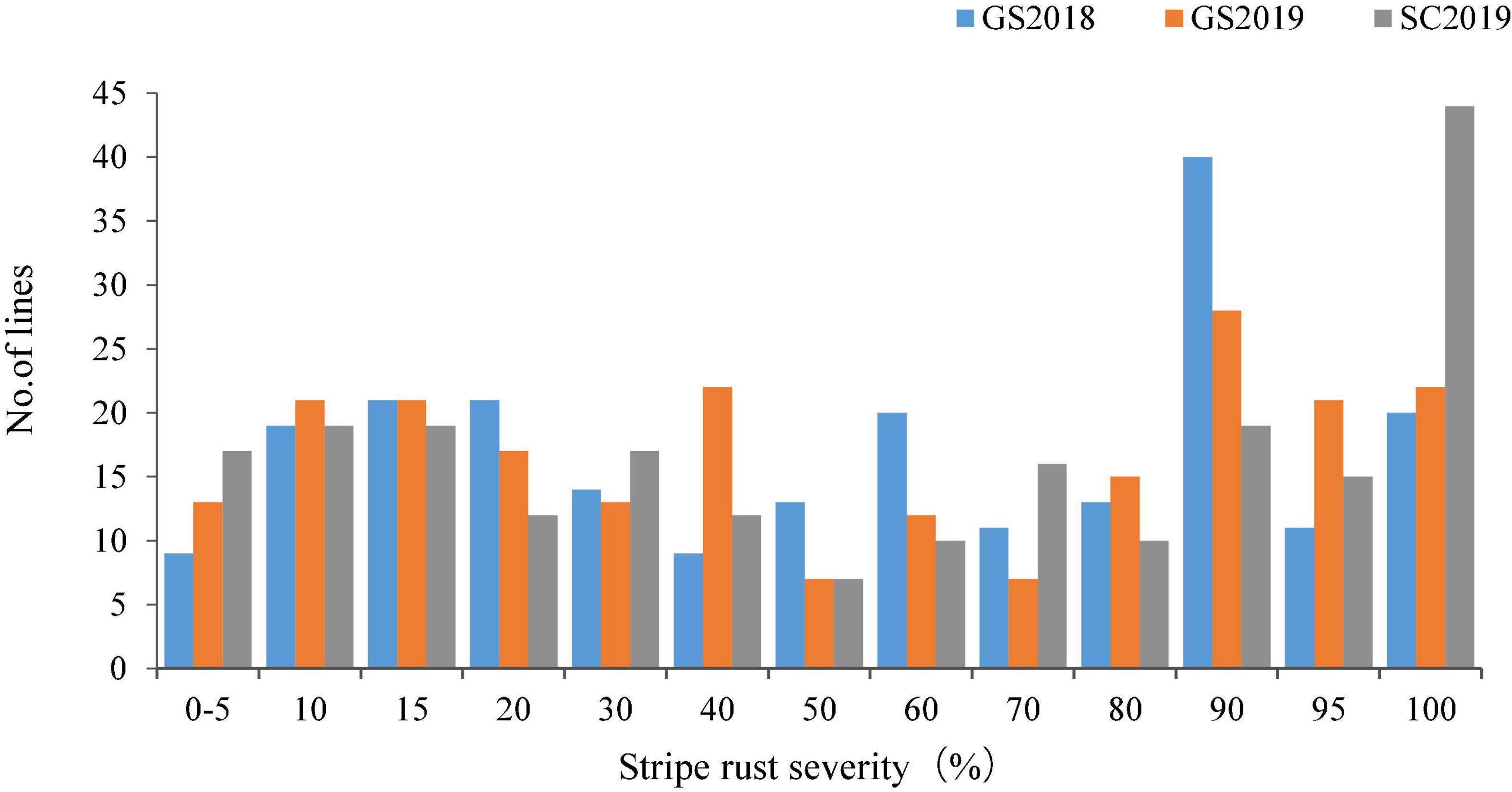
Figure 2. Frequency distributions of 220 Huixianhong/Pascal F5 recombinant inbred lines (RILs) for final disease severity of stripe rust in field traits at Gansu Province during 2017–2018 (GS2018), 2018–2019 (GS2019), and at Sichuan Province during 2018–2019 (SC2019).
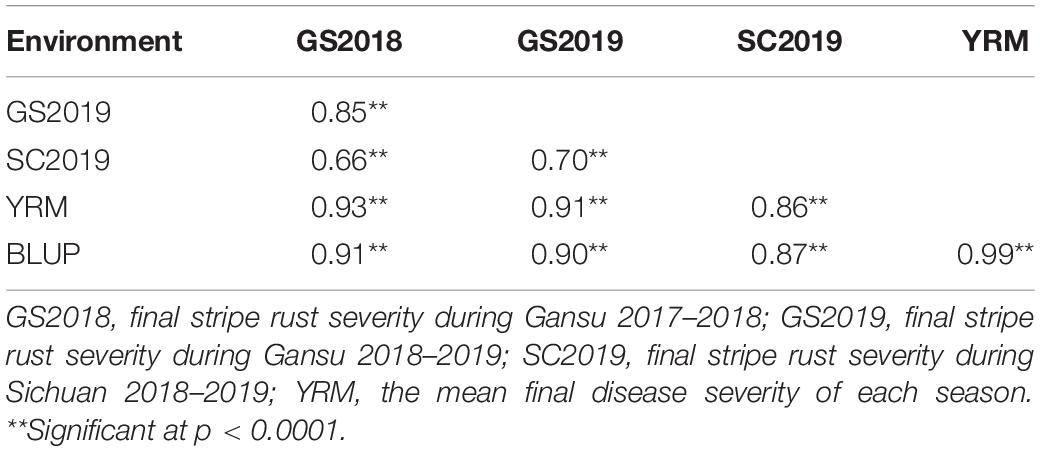
Table 1. Phenotypic Pearson’s correlations for stripe rust (GS2018, GS2019, SC2019) in the Huixianhong/Pascal F5 RIL population using the final disease severity for each season.
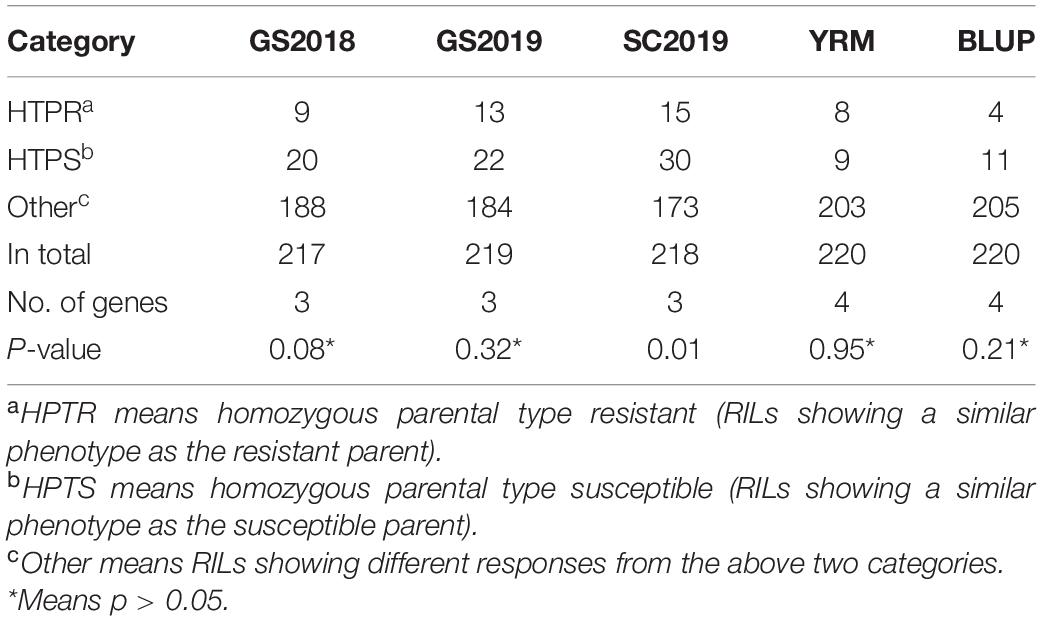
Table 2. The number of resistance genes that confer adult plant resistance to stripe rust calculated by Mendelian segregation ratios in Huixianhong/Pascal recombinant inbred lines population.
A genetic linkage map was constructed with a total of 9,475 markers and developed 28 linkage groups on the 21 wheat chromosomes. A total of three QTLs were identified for stripe rust resistance from “Pascal” and they were mapped on wheat chromosomes 1AL (QYr.gaas-1AL), 3DL (QYr.gaas-3DL), and 5AS (QYr.gaas-5AS) (Table 3). QYr.gaas-1AL was flanked by SNP markers AX-111218361 and AX-110577861 and located at 505.3–507.9 Mb based on IWGSC RefSeq v1.0 genome sequence information. It was stably detected in all tested environments with PVE values ranging from 11.3 to 23.1% (Table 3). QYr.gaas-3DL was closely linked to markers AX-108798599, AX-109580758, and AX-111460455. It was located around 354.6 Mb based on IWGSC RefSeq v1.0 genome sequence information and was also consistently detected in all three stripe rust field experiments explaining 16.1–20.6% total stripe rust variation (Table 3). The third QTL, QYr.gaas-5AS, was flanked by SNP markers AX-111523523 and AX-110028503 and mapped at the interval of 59.1–62.2 Mb based on IWGSC 1.0 with PVE values ranging from 11.0 to 17.3% (Figure 3C and Table 3).
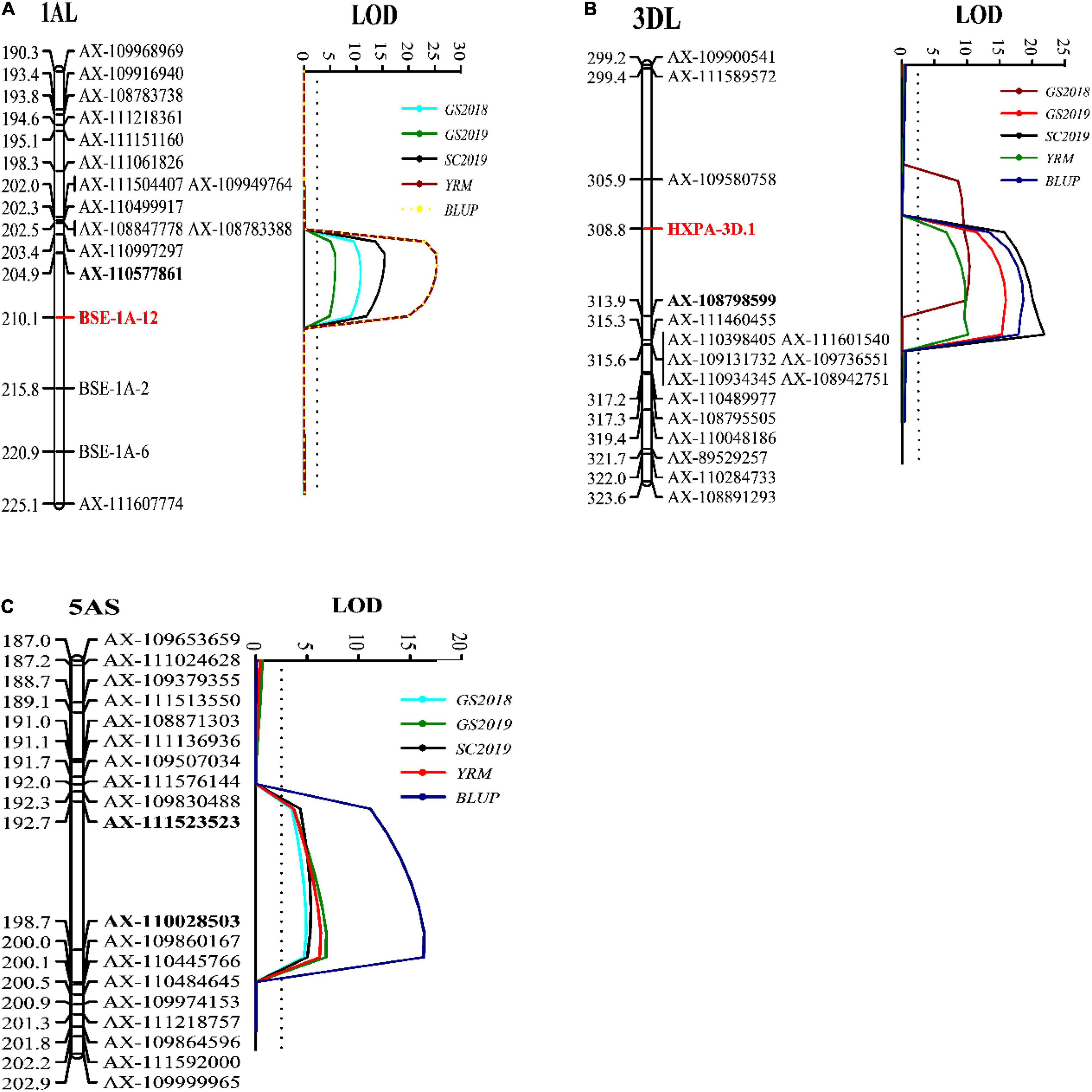
Figure 3. Likelihood plots of quantitative trait loci (QTL) for adult plant resistance to stripe rust on chromosomes 1AL (A) after adding three KASP markers BSE-1A-2, BSE-1A-6, and BSE-1A-12, 3DL (B) after adding KASP marker HXPA-3D and 5AS (C) identified by IciMapping 4.1 in the Huixianhong/Pascal RIL population. The significant LOD threshold was based on 1,000 permutations. Positions (in cM) of the molecular markers along chromosomes are shown on the vertical axes using cumulated genetic distances. KASP markers BSE-1A-2, BSE-1A-6, and BSE-1A-12 near QYr.gaas-1AL were designed based on the SNP of BSE-Seq in the interval of original marker AX-111218361—AX-110577861. New marker BSE-1A-12 and SNP marker AX-110577861 were closely linked with QYr.gaas-1AL identified in the present study. we converted SNP marker AX-109580758 as a KASP marker named HXPA-3D to detect 220 RIL families. KASP marker HXPA-3D was closely linked with QYr.gaas-3DL identified in the present study. GS2018, final stripe rust severity during Gansu 2017–2018; GS2019, final stripe rust severity, during Gansu 2018–2019; SC2019, final stripe rust severity during Sichuan 2018–2019; YRM, the mean final disease severity of each season; BLUP, best linear unbiased prediction.
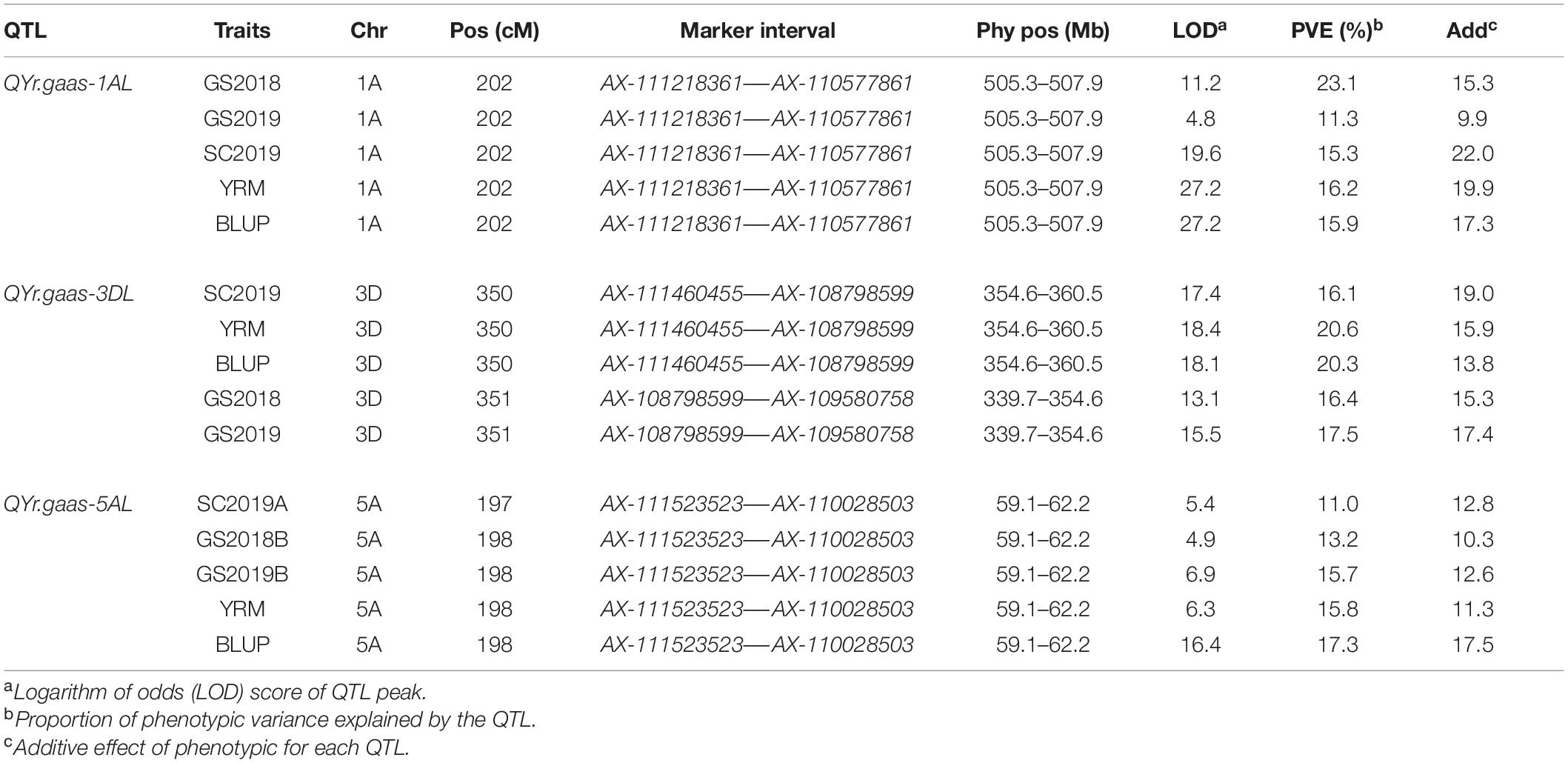
Table 3. Position and effect of quantitative trait loci (QTL) that were detected for adult plant resistance (APR) to stripe rust (YR) in the test years, using each final disease severity, the mean of final disease severity (MFDS), best linear unbiased prediction (BLUP) for the Huixianhong/Pascal RIL population.
Based on the flanking molecular markers of detected three stripe rust resistance QTLs, we divided the F5 RILs into 8 groups. The presence of parental alleles for each QTL was determined by markers inferred factorial combinations of the three stripe rust resistance QTLs.
There was a significantly different disease response between RILs carrying either QYr.gaas-1AL or QYr.gaas-3DL and RILs without any resistance loci (p < 0.0001), which explained 24.2 and 19.3% of the stripe rust variation, respectively (Table 4). The lines carrying QYr.gaas-5AS also showed a significantly different disease response from those without any resistance locus, but this locus only explained 8.5% of the stripe rust variation. This indicates that this third locus imparts a minor effect on stripe rust resistance than the other two loci (Table 4). A significant interaction (p < 0.0001) between QYr.gaas-1AL and QYr.gaas-3DL was observed across three environments as well as MFDS, which explained 3.3% of stripe rust variation based on MFDS. There were no significant differences among the average FDS of RILs carrying combinations of QYr.gaas-1AL*Qyr.gaas-5AS, Qyr.gaas-3DL*Qyr.gaas-5AS, and QYr.gaas-1AL*QYr.gaas-3DL*QYr.gaas-5AS (p > 0.05); however, the average FDS of RILs carrying combinations of three resistance QTLs was significantly lower than the average FDS of RILs carrying other combinations of two resistance QTLs (p < 0.0001) (Table 4).
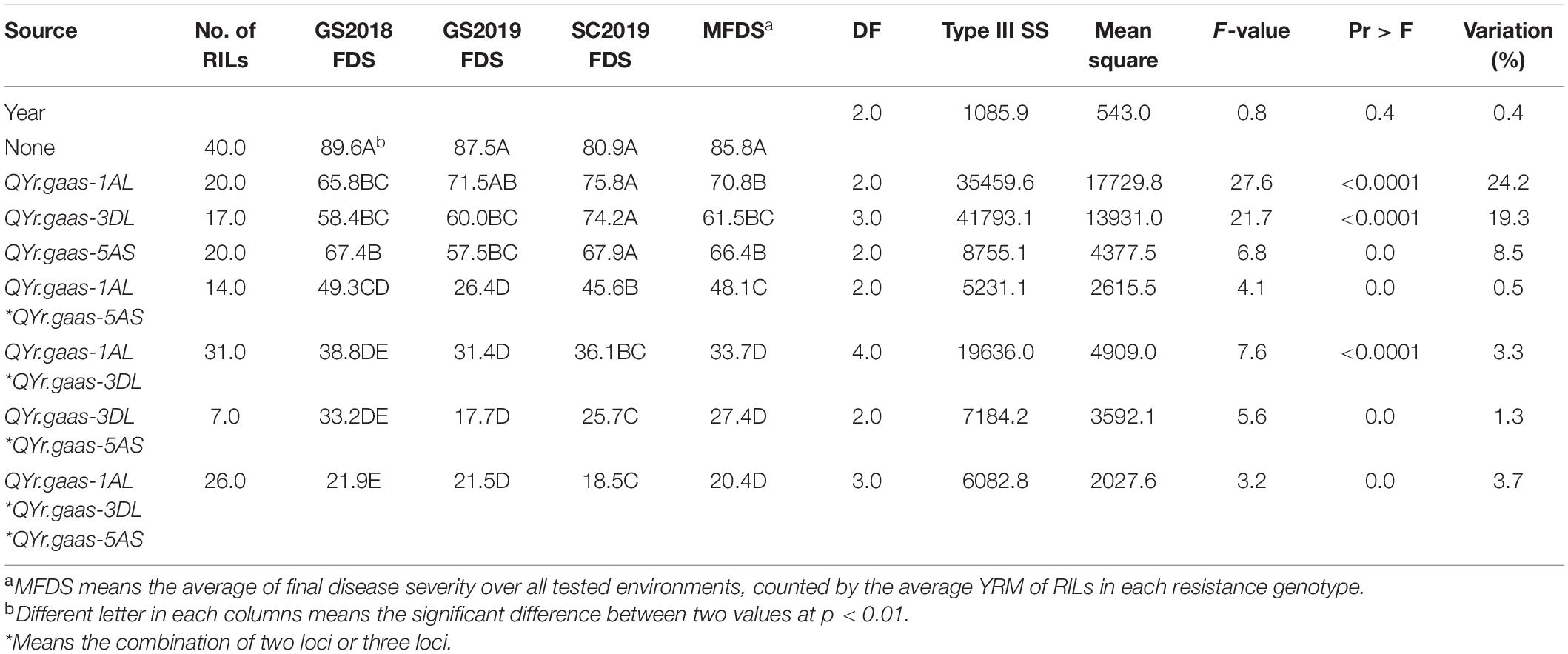
Table 4. Three-way factorial analysis of variance (ANOVA) of final disease severity (FDS) for stripe rust (YR) resistance loci, using years as blocks.
We filtered the data obtained by BSE-Seq with ref_Freq > 0.3, minDepth > 5 and ED = top 0.05 through the ED algorithm, and the results showed that there was a clear peak on chromosome 1AL, which is roughly around 506 Mb (Figures 4A,B). The SNPs between R bulk (same as resistance cultivar Pascal) and S bulk (same as susceptible cultivar Huixianhong) were filtered by AF < 0.2 or AF > 0.8, and the significant SNPs were found in a region of ∼506 Mb on chromosome 1A (Figures 4C,D). This result is also consistent with our mapping result, which indicates significant differences between two resistant and susceptible bulks and showed a high correlation between this region and stripe rust. Unfortunately, we failed to find high confidence SNPs on chromosome 3D through BSE-Seq, which may be related to the collection stage of materials or different process of related gene expression.
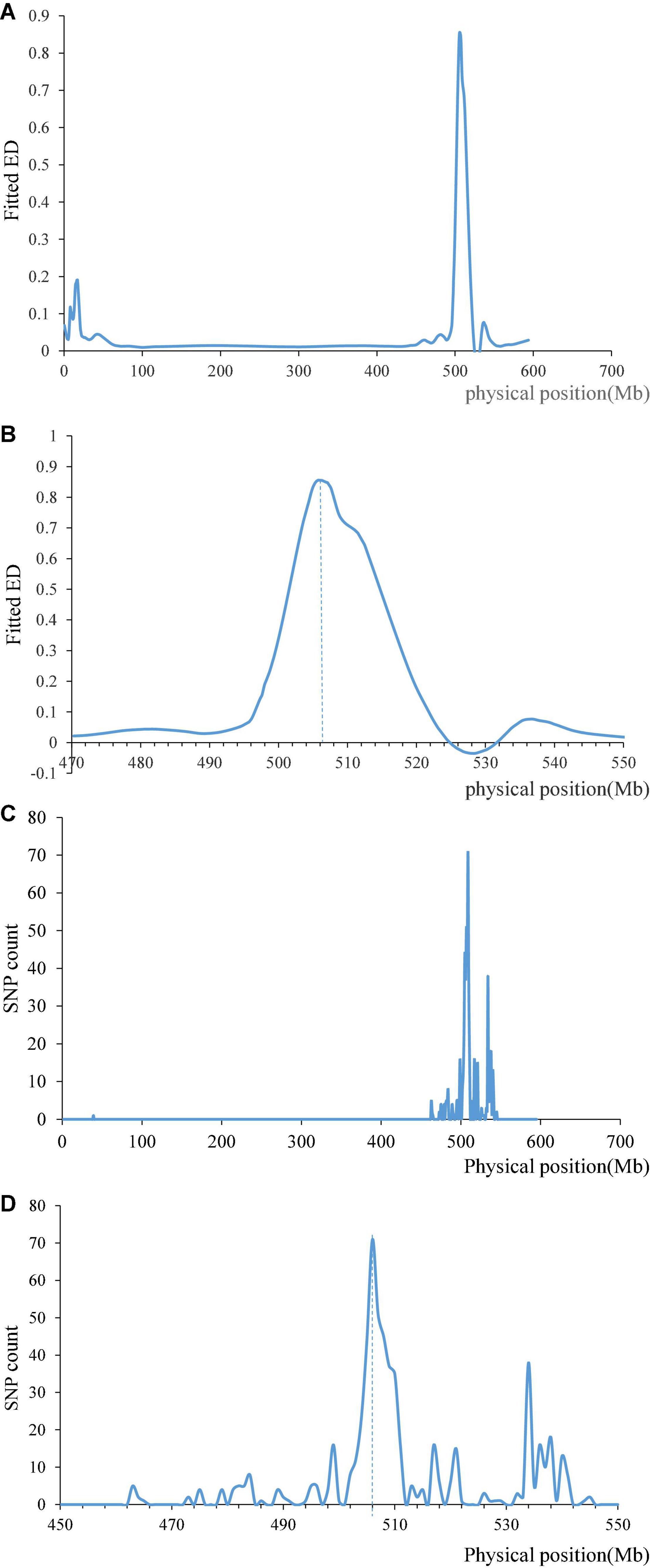
Figure 4. Results of the BSE-Seq analysis method. Fitted ED on chromosome 1A (A) and ED after zooming in the area near the maximum (B). SNP count between R bulk same as Pascal and S bulk same as Huixianhong on chromosome 1A (C), and SNP count after zooming in the area near the maximum (D).
For fine mapping QYr.gaas-1AL, we developed three Kompetitive allele-specific PCR (KASP) markers near QYr.gaas-1AL based on the SNP of BSE-Seq. QYr.gaas-1AL was 0.5–3.5 cM away from KASP marker BSE-1A-12 (Supplementary Table 1 and Figure 3A), which covered a physical interval of 505.3– 506.8 Mb on chromosome 1A. There were 18 high confidence genes in this interval, whereas 13 genes have SNPs on their exon regions. TraesCS1A02G313700 has 14 SNPs on its exon regions which encodes a dentin sialophosphoprotein-like protein. TraesCS1A02G313800 and TraesCS1A02G314900 encode early light-induced protein and metacaspase, respectively, which have 13 and 10 SNPs on their exon regions. The three candidate genes might have strong relationship with QYr.gaas-1AL.
Since QYr.gaas-3DL contributed stripe rust resistance in Pascal significantly, we converted SNP marker AX-109580758 to a KASP marker and combined it with the phenotypic data for QTL mapping in 220 RIL families. The KASP marker HXPA-3D was 1.5–3.5 cm away from QYr.gaas-3DL (Figure 3B). This fact suggests that KASP marker HXPA-3D can be used as an effective marker to detect QYr.gaas-3DL for developing durable stripe rust resistant wheat varieties.
A total of two KASP markers BSE-1A-12 and HXPA-3D for two stable loci were used to genotype a collection of 153 Chinese and global germplasm resources. In the global collection, QYr.gaas-1AL appears more frequently (69%), as compared to the Chinese wheat germplasm in which QYr.gaas-1AL appeared in 62%. Similarly, QYr.gaas-3DL is very common in foreign germplasm (75%), and it is significantly lower in Chinese wheat varieties and breeding lines (60%) (Supplementary Table 2). Our results suggest that Pascal could be a useful source of effective resistance against stripe rust in China.
“Pascal” was conferred by two recessive genes effective against two Pst races CYR 31 and CYR 32, respectively, at the seedling stage (Cao et al., 2006). In this study, we found both parents, Huixianhong and Pascal, susceptible to three Pst races of CYR32, CYR33, and CYR34 at the seedling stage (IT varied from 7 to 8). Most of the stripe rust resistance genes conferring seedling resistance can easily lose their effect since selection pressure favors the virulence in the pathogen population, which could explain that Pascal does not provide seedling resistance to stripe rust in this study. The constant evolution of pathogens and thus the breakdown of seedling resistance make our effort to continuously identify the sources of adult plant resistance increasingly important. We mapped three adult plant resistance QTLs for stripe rust in the Huixianhong/Pascal RIL population, and Pascal showed stable resistant over the last 50 years. There were five stripe rust resistance genes on chromosome 3D, viz. Yr45 (Li et al., 2011), Yr49 (Ellis et al., 2014), Yr66 (Bariana et al., 2022), Yr71 (Bariana et al., 2016), and Yr73 (Dracatos et al., 2016). Yr45, Yr71, and Yr73 were mapped on the 3DL, and the other two genes were mapped on 3DS. Yr45 was discovered from a common spring wheat “PI181314,” flanked by RGAP markers wgp118, wgp115, and two SSR markers Xwmc656 and Xbarc6 which were mapped 11.7 and 12.6 cM proximal to Yr45. We tried to test the two parents using markers wgp115, Xwmc656, and Xbarc6 (Li et al., 2011). However, none of them are polymorphic, and the physical interval of Yr45 was from 476.9 to 514.9 Mb on chromosome 3D, whereas QYr.gaas-3DL was located around 354.6 Mb on the same chromosome. Besides, Yr45 is an all-stage resistant gene, whereas QYr.gaas-3DL was an APR locus in wheat, and therefore, Yr45 could be different from QYr.gaas-3DL. Yr71 is an adult plant resistance gene mapped on 3DL in the Australian cultivar Sunco, and the physical position of its flanking marker gwm114b was at around 603.4 Mb (Bariana et al., 2016). KASP marker Yr71_16434, 1.8 cM distal to Yr71, was at 608 Mb on chromosome 3D. There was no polymorphic for Xgwm114b and Yr71_16434A in the “Huixianhong/Pascal” RILs population when we tried to detect the population, whereas QYr.gaas-3DL was located near the centromeric region of chromosome 3D. Another gene Yr73, a complementary gene on 3DL in the Australian cv. Avocet R, conferred seedling resistance to stripe rust. These results indicate that QYr.gaas-3DL is likely different from genes Yr71 and Yr73.
The QTL QYr/Lr.cim-3DC for stripe rust near the centromeric region of chromosome 3D was identified in common wheat line UC1110 (Lan et al., 2017). It provided both of the leaf rust and stripe rust resistance in the population. QYr.gaas-3DL was also located near the centromeric region of chromosome 3D, but we did not detect the effect on leaf rust. Qyrsicau-3DL, a high-confidence APR locus was detected in a genome-wide association study (GWAS) in Sichuan wheat (Ye et al., 2019). The SNP marker AX-109329567 closely linked with Qyrsicau-3DL was located at 595 Mb on chromosome 3D, which was about 240 Mb away from QYr.gaas-3DL. Huang et al. (2021) identified an APR QTL (QYrsn.nwafu-3DL) on 3DL explaining 5.8–12.2% of the phenotypic variation from Shannong 33 (SN33). The physical position of its flanking marker AX-109582945 was around 407.4 Mb on chromosome 3D, which is around 52 Mb away from QYr.gaas-3DL. Furthermore, QYrsn.nwafu-3DL was present in 7.4% of a panel of 420 current Chinese wheat cultivars, which was much lower than the presence of QYr.gaas-3DL in a collection of 153 Chinese germplasm resources in our studies. Therefore, QYrsn.nwafu-3DL could be different from the APR resistance QYr.gaas-3DL.
The stripe rust resistance genes and QTL located on chromosome 5AS in the previous studies are extensive. A minor effect QTL (QPst.jic-5A) for field stripe rust resistance on chromosome 5AS was identified in a stripe rust susceptible winter wheat cv. Brigadier (Jagger et al., 2011). QPst.jic-5A was located between marker Xwmc752 and Xgwm786, placing it near the centromere on the short arm of chromosome 5A. Since QYr.gaas-5AS was mapped at the interval of 59.1–62.2 Mb and far away from the centromere of chromosome 5A, QPst.jic-5A could be different from QYr.gaas-5AS. A major QTL, QYr.cau-5AS, exhibited 24.1–35.3% of PVE in winter wheat AQ24788-83 at the tillering growth stage (Quan et al., 2013). This QTL was flanked by markers Xcfa2250 and Xwmc705, which were located around 245 Mb, suggesting this QTL was different from QYr.gaas-5AS.
QYr.cim-5AS was a minor APR QTL derived from Avocet and mapped near the SSR marker Xcfa2104 and Xbarc186 (Ren et al., 2017). The physical position of those SSR markers was around 22–46 Mb, placing near QYr.gaas-5AS in the present studies. However, it only explained 4.0–5.1% of YR variation, but QYr.gaas-5AS had PVE values ranging from 11.0 to 17.3%. It is possible that QYr.gaas-5AS is different from this QTL. However, this needs to be confirmed by allelism test in the future. Qyrsicau-5AS, a minor APR QTL with a PVE of 7.1–8.1%, were detected by the GWAS in Sichuan wheat (Ye et al., 2019). The SNP marker AX-111623511 linked with Qyrsicau-5AS was located at 101 Mb on chromosome 5A, which was about 40 Mb from QYr.gaas-5AS, indicating QYrsicau-5AS could be different from the APR resistance QYr.gaas-5AS. Therefore, QYr.gaas-5AS is possibly a new QTL for APR to stripe rust.
Previous studies have detected several QTLs for stripe rust on chromosome 1AL in wheat. A minor APR QTL QYr.sgi-1A was located on chromosome 1A in the population of Kariega/Avocet S, contributing 6–12% to the phenotypic variance, and flanked by markers s15m19D and s23m18E, but was inconsistently detected across environments (Ramburan et al., 2004). Unfortunately, we failed to obtain the physical location of this QTL. Whereas QYr.gaas-1AL had PVE values ranging from 11.3 to 23.1%, it was stable across all environments in our studies, indicating that QTL QYr.sgi-1A was unlikely to be the same locus as QYr.gaas-1AL. Bariana et al. (2010) identified QYr.sun-1A controlling APR to stripe rust in Kukri/Janz-derived doubled haploid (DH) population, which explained 6–7% phenotypic variation for stripe rust. This QTL was flanked by marker Xgwm164 and its physical position was around 280.6 Mb. But QYr.gaas-1AL was located in the interval of 505.3–507.9 Mb, which indicated that QYr.gaas-1AL was different from QYr.sun-1A.
Ren Y. et al. (2012) identified a QTL (QYr.caas-1AL) contributed APR to stripe rust in German spring wheat cultivar Naxos. It was located in marker interval XwPt-2406–Xwmc59 at 575.4 Mb, which was roughly 70 Mb away from QYr.caas-1AL. Besides, QYr.caas-1AL only explained 8.2% of the phenotypic variance in one environment, which showed lower effect and stability than QYr.gaas-1AL. QYrid.ui-1A, a minor high-temperature adult plant (HTAP) resistance QTL from the hard red winter wheat germplasm IDO444, were significant only for IT in single environment (Chen et al., 2012). QYr.gaas-1AL had high effect of stripe rust resistance and were stable across all environments. Gebrewahid et al. (2020) identified an APR QTL on chromosome 1A in spring wheat Fuyu 3 designated QYr.hebau-1AL, which was located on the interval of AX-109403007 to AX-110502416 around 286.6–289.3 Mb. This QTL should be different from QYr.gaas-1AL based on a physical distance of about 220 Mb between them. Grover et al. (2022) identified an ASR locus Yraci in the European winter wheat cultivar “Acienda” on the distal end of wheat chromosome 1A. Besides, two SNPs AX-95162217 and AX-94540853 linked with Yraci were mapped in the bin at 54.04 cM on chromosome 1A, whereas QYr.gaas-1AL covered a physical interval of 505.3–506.8 Mb on chromosome 1A was far away from Yraci. Therefore, Yraci and QYr.gaas-1AL were not the same QTL for strip rust. Further research is needed to determine the novelty of QYr.gaas-1AL from Pascal.
Combining BSA and the exome sequence strategy could accelerate gene mapping, especially in wheat with a large and complex genome. BSE-Seq is helpful for the construction of a linkage map across the whole genome, and it could be easily used to identify the linked interval regardless of the multiple gene copies and obtain most of the variations existing in the coding regions of genes (Dong et al., 2020). Using bulked segregant analysis and the exome sequence strategy, Mo et al. (2018) identified a clear peak region on chromosome 4BS associated with increased plant height. Harrington et al. (2019) identified a locus controlling an environmentally dependent chlorosis phenotype in the Durum wheat cv. Kronos. Martinez et al. (2020) finely mapped a novel TaMKK3 allele conferring the wheat ERA8 ABA-hypersensitive germination phenotype in a wheat backcross population. In our studies, BSE-Seq helped us to confirm the reality of QTL and select candidate genes in QTL QYr.gaas-1AL region. TraesCS1A02G313700, TraesCS1A02G313800, and TraesCS1A02G314900 had more than 10 SNPs on their exon regions. Of these genes, TraesCS1A02G314900 encodes metacaspase, which has been reported to modulate autophagy to confine cell death to the target cells during Arabidopsis vascular xylem differentiation (Escamez et al., 2016), which suggests that this gene may be a strong candidate related to disease resistance. Further research is needed to determine the relationship between these genes and stripe rust resistance.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
BB initiated the project, designed the experiment, and contributed to phenotype data. CL designed the experiment and finalized the manuscript. ZL assisted in the data analysis and contributed to drafting the manuscript. HW and XD performed the sample preparation and DNA extraction. LW and JD contributed to the part of phenotype data in the field. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (grant no. 31860379), Key R&D Program of Gansu Province (grant no. 21YF5NA146), the National Natural Science Foundation of China (grant no. 31860378), International Cooperation and Exchange of the National Natural Science Foundation of China (grant no. 31861143010), and the Hubei Hongshan Laboratory (grant no. 2022hspy001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.918437/full#supplementary-material
Bai, B., Ren, Y., Xia, X., Du, J., and Zhou, G. (2012). Mapping of quantitative trait loci for adult plant resistance to stripe rust in German wheat cultivar Ibis. J. Integr. Agric. 11, 528–536. doi: 10.1016/S2095-3119(12)60039-2
Bariana, H., Bansal, U., Schmidt, A., Lehmensiek, J., Kaur, H., Miah, N., et al. (2010). Molecular mapping of adult plant stripe rust resistance in wheat and identification of pyramided QTL genotypes. Euphytica 176, 251–260. doi: 10.1007/s10681-010-0240-x
Bariana, H., Forrest, K., Qureshi, N., Miah, H., Hayden, M., and Bansal, U. (2016). Adult plant stripe rust resistance gene Yr71 maps close to Lr24 in chromosome 3D of common wheat. Mol. Breed. 36:98. doi: 10.1007/s11032-016-0528-1
Bariana, H., Kant, L., Qureshi, N., Forrest, K., Miah, H., and Bansal, U. (2022). Identification and characterisation of stripe rust resistance genes Yr66 and Yr67 in wheat cultivar VL Gehun 892. Agronomy 12:318. doi: 10.3390/agronomy12020318
Brandt, K., Chen, X., Tabima, J., See, D., Vining, K., and Zemetra, R. (2021). QTL analysis of adult plant resistance to stripe rust in a winter wheat recombinant inbred population. Plants 10:572. doi: 10.3390/plants10030572
Cao, S., Xie, Z., Jin, S., Jin, S., Jia, Q., Luo, H., et al. (2006). Inheritance analysis of resistance wheat cultivar Pascal to stripe rust. J. Gansu Agric. Univ. 2, 39–41. doi: 10.1016/S1872-2040(06)60047-9
Chatterjee, A., Moulik, S., Majhi, P., and Sanyal, S. (2002). Studies on surfactant-biopolymer interaction. I. Microcalorimetric investigation on the interaction of cetyltrimethylammonium bromide (CTAB) and sodium dodecylsulfate (SDS) with gelatin (Gn), lysozyme (Lz) and deoxyribonucleic acid (DNA). Biophys. Chem. 98, 313–327. doi: 10.1016/s0301-4622(02)00107-2
Chen, J., Chu, C., Souza, E., Guttieri, M., Chen, X., Xu, S., et al. (2012). Genome-wide identification of QTL conferring high-temperature adult-plant (HTAP) resistance to stripe rust (Puccinia striiformis f. sp. Tritici) in wheat. Mol. Breed. 29, 791–800. doi: 10.1007/s11032-011-9590-x
Dong, C., Zhang, L., Chen, Z., Xia, C., Gu, Y., Wang, J., et al. (2020). Combining a new exome capture panel with an effective varBScore algorithm accelerates BSA-Based gene cloning in wheat. Front. Plant Sci. 11:1249. doi: 10.3389/fpls.2020.01249
Dracatos, P., Zhang, P., Park, R., McIntosh, R., and Wellings, C. (2016). Complementary resistance genes in wheat selection ‘Avocet R’ confer resistance to stripe rust. Theor. Appl. Genet. 129, 65–76. doi: 10.1007/s00122-015-2609-7
Draz, I., Serfling, A., Muqaddasi, Q., and Röder, M. (2021). Quantitative trait loci for yellow rust resistance in spring wheat doubled haploid populations developed from the German Federal ex situ genebank genetic resources. Plant Genome 14:e20142. doi: 10.1002/tpg2.20142
Dyck, P. (1991). Genetics of adult-plant leaf rust resistance in ‘chinese spring’ and ‘sturdy’ wheats. Crop Sci. 31, 309–311.
Ellis, J., Lagudah, E., Spielmeyer, W., and Dodds, P. (2014). The past, present and future of breeding rust resistant wheat. Front. Plant Sci. 5:641. doi: 10.3389/fpls.2014.00641
Escamez, S., André, D., Zhang, B., Bollhöner, B., Pesquet, E., and Tuominen, H. (2016). METACASPASE9 modulates autophagy to confine cell death to the target cells during Arabidopsis vascular xylem differentiation. Biol. Open 5, 122–129. doi: 10.1242/bio.015529
Gardiner, L., Bansept-Basler, P., Olohan, L., Joynson, R., Brenchley, R., Hall, N., et al. (2016). Mapping-by-sequencing in complex polyploid genomes using genic sequence capture: a case study to map yellow rust resistance in hexaploid wheat. Plant J. 87, 403–419. doi: 10.1111/tpj.13204
Gebrewahid, T., Zhang, P., Zhou, Y., Yan, X., Xia, X., He, Z., et al. (2020). QTL mapping of adult plant resistance to stripe rust and leaf rust in a Fuyu 3/Zhengzhou 5389 wheat population. Crop J. 8, 655–665. doi: 10.1016/j.cj.2019.09.013
Gou, J., Li, K., Wu, K., Wang, X., Lin, H., Cantu, D., et al. (2017). Wheat stripe rust resistance protein WKS1 reduces the ability of the thylakoid-associated ascorbate peroxidase to detoxify reactive oxygen species. Plant Cell. 27, 1755–1770. doi: 10.1105/tpc.114.134296
Grover, G., Sharma, A., Mackay, I., Srivastava, P., Kaur, S., Kaur, J., et al. (2022). Identification of a novel stripe rust resistance gene from the European winter wheat cultivar ‘Acienda’: a step towards rust proofing wheat cultivation. PLoS One 17:e0264027. doi: 10.1371/journal.pone.0264027
Harrington, S., Cobo, N., Karafiátová, M., Doležel, J., Borrill, P., and Uauy, C. (2019). Identification of a dominant chlorosis phenotype through a forward screen of the Triticum turgidum cv. Kronos TILLING population. Front. Plant Sci. 10:963. doi: 10.3389/fpls.2019.00963
Henry, I., Nagalakshmi, U., Lieberman, M., Ngo, K., Krasileva, K., Vasquez-Gross, H., et al. (2014). Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and next-generation sequencing. Plant Cell 26, 1382–1397. doi: 10.1105/tpc.113.121590
Herrera-Foessel, S., Lagudah, E., Huerta-Espino, J., Hayden, M., Bariana, H., Singh, D., et al. (2011). New slow-rusting leaf rust and stripe rust resistance genes Lr67 and Yr46 in wheat are pleiotropic or closely linked. Theor. Appl. Genet. 122, 239–249. doi: 10.1007/s00122-010-1439-x
Hill, J., Demarest, B., Bisgrove, B., Gorsi, B., Su, Y., and Yost, H. (2013). MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 23, 687–697. doi: 10.1101/gr.146936.112
Huang, S., Liu, S., Zhang, Y., Xie, Y., Wang, X., Jiao, H., et al. (2021). Genome-wide wheat 55K SNP-based mapping of stripe rust resistance loci in wheat cultivar Shaannong 33 and their alleles frequencies in current Chinese wheat cultivars and breeding lines. Plant Dis. 105, 1048–1056. doi: 10.1094/PDIS-07-20-1516-RE
Huang, S., Wu, J., Wang, X., Mu, J., Xu, Z., Zeng, Q., et al. (2019). Utilization of the genomewide wheat 55K SNP array for genetic analysis of stripe rust resistance in common wheat line P9936. Phytopathology 109, 819–827. doi: 10.1094/PHYTO-10-18-0388-R
International Wheat Genome Sequencing Consortium [IWGSC] (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Jagger, L., Newell, C., Berry, S., MacCormack, R., and Boyd, L. (2011). The genetic characterisation of stripe rust resistance in the German wheat cultivar Alcedo. Theor. Appl. Genet. 122, 723–733. doi: 10.1007/s00122-010-1481-8
Krattinger, S., Lagudah, E., Spielmeyer, W., Singh, R., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Lan, C., Hale, I., Herrera-Foessel, S., Basnet, B., Randhawa, M., Huerta-Espino, J., et al. (2017). Characterization and mapping of leaf rust and stripe rust resistance loci in hexaploid wheat lines UC1110 and PI610750 under Mexican environments. Front. Plant Sci. 8:1450. doi: 10.3389/fpls.2017.01450
Li, J., Dundas, I., Dong, C., Li, G., Trethowan, R., Yang, Z., et al. (2020). Identification and characterization of a new stripe rust resistance gene Yr83 on rye chromosome 6R in wheat. Theor. Appl. Genet. 133, 1095–1107. doi: 10.1007/s00122-020-03534-y
Li, Q., Chen, X., Wang, M., and Jing, J. (2011). Yr45, a new wheat gene for stripe rust resistance on the long arm of chromosome 3D. Theor. Appl. Genet. 122, 189–197. doi: 10.1007/s00122-010-1435-1
Lin, F., and Chen, X. (2007). Genetics and molecular mapping of genes for race specific and all-stage resistance and non-specific high temperature adult-plant resistance to stripe rust in spring wheat cultivar Alpowa. Theor. Appl. Genet. 114, 1277–1287. doi: 10.1007/s00122-007-0518-0
Martinez, S., Shorinola, O., Conselman, S., See, D., Skinner, D., Uauy, C., et al. (2020). Exome sequencing of bulked segregants identified a novel TaMKK3-A allele linked to the wheat ERA8 ABA-hypersensitive germination phenotype. Theor. Appl. Genet. 133, 719–736. doi: 10.1007/s00122-019-03503-0
McNeal, F., Konzak, C., Smith, E., Tate, W., and Russell, T. (1971). A Uniform System for Recording and Processing. Washington, DC: Agricultural Research Service, 34–121.
Meng, L., Li, H., Zhang, L., and Wang, J. (2015). QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Mo, Y., Howell, T., Vasquez-Gross, H., Haro, L., Dubcovsky, J., and Pearce, S. (2018). Mapping causal mutations by exome sequencing in a wheat TILLING population: a tall mutant case study. Mol. Genet. Genomics 293, 463–477. doi: 10.1007/s00438-017-1401-6
Moore, J., Herrera-Foessel, S., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Nsabiyera, V., Bariana, H., Qureshi, N., Wong, D., Hayden, M., and Bansal, U. (2018). Characterisation and mapping of adult plant stripe rust resistance in wheat accession Aus27284. Theor. Appl. Genet. 131, 1459–1467. doi: 10.1007/s00122-018-3090-x
Peterson, R., Campbell, A., and Hannah, A. (1948). A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 26c, 496–500. doi: 10.1139/cjr48c-033
Quan, W., Hou, G., Chen, J., Du, Z., Lin, F., Guo, Y., et al. (2013). Mapping of QTL lengthening the latent period of Puccinia striiformis in winter wheat at the tillering growth stage. Eur. J. Plant Pathol. 136, 715–727. doi: 10.1007/s10658-013-0201-z
Ramburan, V., Pretorius, Z., Louw, J., Boyd, L., Smith, P., Boshoff, W., et al. (2004). A genetic analysis of adult plant resistance to stripe rust in the wheat cultivar Kariega. Theor. Appl. Genet. 108, 1426–1433. doi: 10.1007/s00122-003-1567-7
Ren, R., Wang, M., Chen, X., and Zhang, Z. (2012). Characterization and molecular mapping of Yr52 for high-temperature adult-plant resistance to stripe rust in spring wheat germplasm PI 183527. Theor. Appl. Genet. 125, 847–857. doi: 10.1007/s00122-012-1877-8
Ren, T., Hu, Y., Tang, Y., Li, C., Yan, B., Ren, Z., et al. (2018). Utilization of a wheat 55K SNP array for mapping of major QTL for temporal expression of the tiller number. Front. Plant Sci. 9:333. doi: 10.3389/fpls.2018.00333
Ren, Y., He, Z., Li, J., Lillemo, M., Wu, L., Bai, B., et al. (2012). QTL mapping of adult-plant resistance to stripe rust in a population derived from common wheat cultivars Naxos and Shanghai 3/Catbird. Theor. Appl. Genet. 125, 1211–1221. doi: 10.1007/s00122-012-1907-6
Ren, Y., Singh, R., Basnet, B., Lan, C., Huerta-Espino, J., Lagudah, E., et al. (2017). Identification and mapping of adult plant resistance loci to leaf rust and stripe rust in common wheat cultivar kundan. Plant Dis. 101, 456–463. doi: 10.1094/PDIS-06-16-0890-RE
Roberts, J., and Caldwell, R. (1970). General resistance slow mildewing to Erysiphe graminis f. sp. tritici in knox wheat. Phytopathology 60:1310. doi: 10.1094/Phyto-60-1831
Schwessinger, B. (2017). Fundamental wheat stripe rust research in the 21st century. New Phytol. 213, 1625–1631. doi: 10.1111/nph.14159
Singh, R., Mujeeb-Kazi, A., and Huerta-Espino, J. (1998). Lr46: a gene conferring slow-rusting resistance to leaf rust in wheat. Phytopathology 88, 890–894. doi: 10.1094/PHYTO.1998.88.9.890
Steuernagel, B., Periyannan, S., Hernández-Pinzón, I., Witek, K., Rouse, M., Yu, G., et al. (2016). Rapid cloning of diseaseresistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34, 652–655.
Todorovska, E., Christov, N., Slavov, S., Christova, P., and Vassilev, D. (2009). Biotic stress resistance in wheat—breeding and genomic selection implications. Biotechnol. Equip. 23, 1417–1426. doi: 10.2478/V10133-009-0006-6
Van, O. (2011). Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res. 93, 343–349. doi: 10.1017/S0016672311000279
Voorrips, R. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wellings, C., and McIntosh, R. (1990). Puccinia striiformis f. sp. Tritici in Australasia: pathogenic changes during the first 10 years. Plant Pathol. 39, 316–325. doi: 10.1111/j.1365-3059.1990.tb02509.x
Wen, W., Li, G., He, Z., Yang, W., Xu, M., and Xia, X. (2008). Development of an STS marker tightly linked to Yr26 against wheat stripe rust using the resistance gene-analog polymorphism (RGAP) technique. Mol. Breed. 22, 507–515. doi: 10.1007/s11032-008-9194-2
Ye, X., Li, J., Cheng, Y., Yao, F., Long, L., Yu, C., et al. (2019). Genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. Tritici) in Sichuan wheat. BMC Plant Biol. 19:147. doi: 10.1186/s12870-019-1764-4
Young, N. (1996). QTL mapping and quantitative disease resistance in plants. Annu. Rev. Phytopathol. 34, 479–501. doi: 10.1146/annurev.phyto.34.1.479
Yu, H., Xie, W., Wang, J., Xing, Y., Xu, C., Li, X., et al. (2011). Gains in QTL detection using an ultra-high density SNP map based on population sequencing relative to traditional RFLP/SSR markers. PLoS One 6:e17595. doi: 10.1371/journal.pone.0017595
Zeng, Q., Zhao, J., Wu, J., Zhan, G., Han, D., and Kang, Z. (2022). Wheat stripe rust and integration of sustainable control strategies in China. Front. Agr. Sci. Eng. 9, 37–51. doi: 10.15302/J-FASE-2021405
Keywords: genetic mapping, quantitative traits, adult plant resistance, stripe rust, wheat
Citation: Bai B, Li Z, Wang H, Du X, Wu L, Du J and Lan C (2022) Genetic Analysis of Adult Plant Resistance to Stripe Rust in Common Wheat Cultivar “Pascal”. Front. Plant Sci. 13:918437. doi: 10.3389/fpls.2022.918437
Received: 12 April 2022; Accepted: 09 June 2022;
Published: 06 July 2022.
Edited by:
Cheng Liu, Shandong Academy of Agricultural Sciences, ChinaReviewed by:
Jindong Liu, Institute of Crop Sciences (CAAS), ChinaCopyright © 2022 Bai, Li, Wang, Du, Wu, Du and Lan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Bai, YmFpYmluZ2Fhc0Bnc2Fnci5jbg==; Caixia Lan, Y3hsYW5AbWFpbC5oemF1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.