
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 June 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.907635
This article is part of the Research Topic The Use of Setaria Species as a Molecular Genetic Model to Characterize C4 Plant Biology View all 5 articles
Potassium (K+) is one of the indispensable elements in plant growth and development. The Shaker K+ channel protein family is involved in plant K+ uptake and distribution. Foxtail millet (Setaria italica), as an important crop, has strong tolerance and adaptability to abiotic stresses. However, no systematic study focused on the Shaker K+ channel family in foxtail millet. Here, ten Shaker K+ channel genes in foxtail millet were identified and divided into five groups through phylogenetic analysis. Gene structures, chromosome locations, cis-acting regulatory elements in promoter, and post-translation modification sites of Shaker K+ channels were analyzed. In silico analysis of transcript level demonstrated that the expression of Shaker K+ channel genes was tissue or developmental stage specific. The transcription levels of Shaker K+ channel genes in foxtail millet under different abiotic stresses (cold, heat, NaCl, and PEG) and phytohormones (6-BA, BR, MJ, IAA, NAA, GA3, SA, and ABA) treatments at 0, 12, and 24 h were detected by qRT-PCR. The results showed that SiAKT1, SiKAT3, SiGORK, and SiSKOR were worth further research due to their significant responses after most treatments. The yeast complementation assay verified the inward K+ transport activities of detectable Shaker K+ channels. Finally, we found interactions between SiKAT2 and SiSNARE proteins. Compared to research in Arabidopsis, our results showed a difference in SYP121 related Shaker K+ channel regulation mechanism in foxtail millet. Our results indicate that Shaker K+ channels play important roles in foxtail millet and provide theoretical support for further exploring the K+ absorption mechanism of foxtail millet under abiotic stress.
As the main abiotic stresses, drought, salt, and temperature stresses severely affect plant growth and development. Potassium (K+) is one of the essential nutrients for plants and is involved in many plant physiological processes, including plant cell turgor maintenance, stomata movement, photosynthesis, carbon assimilation, and osmotic balance (Wang et al., 2013). K+ is also recognized as a key limiting factor in agricultural production. Moreover, many reports have suggested the protective role of K+ under abiotic stress in different species (Gong et al., 2020; Ma et al., 2020).
In plants, there are two main mechanisms for K+ absorption, the high-affinity K+ absorption mechanism mediated by transporters and the low-affinity K+ absorption mechanism mediated by K+ channels (Raddatz et al., 2020). The K+ channels are divided into three families, including Shaker, TPK, and Kir-like K+ channel families (Lebaudy et al., 2007). Among them, the Shaker K+ channel is the most extensively studied gene family and has been identified in many plant species, including Arabidopsis (Dreyer and Uozumi, 2011), rice (Amrutha et al., 2007; Hwang et al., 2013), maize (Bauer et al., 2000), poplar (Zhang et al., 2010), sweet potato (Jin et al., 2021), and soybean (Feng et al., 2021). Shaker K+ channels are involved in K+ uptake from soil and transport among different tissues. Their subunits share similar conserved structural features. All of them contain six transmembrane α-helices called S1–S6. They form a functional channel as homo- or hetero-tetrameric assemblies, which mediates the transport of K+ across the membrane (Dreyer and Blatt, 2009). In Arabidopsis, the nine Shaker K+ channel proteins can be divided into five groups (Pilot et al., 2003b). Group I (KATs) and Group II (AKT1, AKT5, and AKT6) are inward rectification channels whose opening are regulated by the negative membrane voltage. Group V (SKOR and GORK) is the outward rectification channel whose opening requires positive membrane voltages. Group III (AKT2) belongs to the weak inward rectification channel, and Group IV contains a silent subunit KC1 (Lebaudy et al., 2007).
The role of the Shaker K+ channel in plant abiotic stress responses has been well studied in many plant species. In Arabidopsis, most Shaker K+ channels are involved in the opening and closing of stomata under drought stress (Hosy et al., 2003; Lebaudy et al., 2008). The cold and salt stress-induced expression of GORK leads to disruption of cytosolic Na+/K+ ratio and suppression of plant K+-dependent metabolism (Becker et al., 2003; Chen et al., 2007). Over-expressing OsAKT1 in rice elevates the tissue K+ content and improves plant drought tolerance (Ahmad et al., 2016). A similar phenomenon has been reported for HvAKT1 over-expression in barley (Feng et al., 2020). ZMK1, a homolog of AKT1 in Zea May, mediates K+ uptake in coleoptile cells and its expression is induced by auxin (Philippar et al., 1999, 2006). Over-expressing GmAKT1 of soybean in Arabidopsis enhances root length and K+ concentration under drought and salt stress, suggesting that GmAKT1 plays a role in soybean response to such stresses (Feng et al., 2021). The GORK channel associated K+ efflux and the channel’s sub-cellular location in the guard cell is regulated by ABA, a drought-related phytohormone, through the OPEN STOMATA 1 (OST1) kinase involved signaling system (Chen et al., 2021). Ooi et al. have found that direct GORK-ABA interaction increases the K+ efflux current (Ooi et al., 2017). The inward rectification channel KAT1 is also regulated by ABA (Sutter et al., 2007). Recent reports suggest that KAT1 interacts with the membrane traffic related SNARE family protein SYP121, which regulates the ABA related channel trafficking and directly induces the channel activity through protein-protein interaction (Sutter et al., 2006; Honsbein et al., 2009). Further research reports that another SNARE protein, VAMP721, the partner of SYP121 during membrane fusion, also binds with K+ channels but regulates the channel activity in an opposite way (Zhang et al., 2015). The expression of SNARE protein is up-regulated by abiotic stresses (Kwon et al., 2020; Wang et al., 2021). Thus, the SNARE related regulation of K+ channels might be the key to understanding the mechanism of K+-dependent plant abiotic tolerance.
Setaria italica is a Poaceae crop with strong environmental adaptability. The high abiotic stress tolerance of foxtail millet leads to its broad cultivation area in Asia and Africa (He et al., 2015; Peng and Zhang, 2020). In 2012, the foxtail millet genome was sequenced, making it possible to identify genes and study their functions (Bennetzen et al., 2012; Zhang et al., 2012; Tan et al., 2022). Furthermore, its relatively small genome makes foxtail millet a promising C4 model plant (He et al., 2015). Previous research from our group has analyzed the role of SNARE proteins under drought stress in foxtail millet (Wang et al., 2021). The high-affinity K+ transporters in foxtail millet have also been analyzed (Zhang et al., 2018). However, little research has focused on the Shaker K+ channels in foxtail millet and their role in stress response. Therefore, we identified the Shaker K+ channel genes from the foxtail millet database and obtained ten members. These genes were further analyzed via phylogeny, conserved motifs and domain, promoter, and tissue expression patterns. Then, quantitative real-time PCR (qRT-PCR) was used to analyze the transcription levels of Shaker K+ channel genes in foxtail millet under different abiotic stresses and phytohormone treatments. The K+ transport activities of these genes were verified by yeast complementation assay. In addition, we also investigated the interaction between Shaker K+ channel proteins and SNARE proteins in foxtail millet. These analyses explored the mechanism of Shaker K+ channel-related K+ absorption in foxtail millet and provided directions for future research.
The Shaker K+ channel gene sequences of Arabidopsis and rice were extracted from the Arabidopsis Information Resource1 and the Rice Genome Annotation Project.2 These sequences were blasted in the Phytozome database (Phytozome 13, Setaria italica v2.2) (Goodstein et al., 2012) to obtain the candidate shaker K+ channel coding sequences. Then the Arabidopsis channel sequence was submitted to the Pfam database3 to obtain the hidden Markov model (HMM) profile of the Shaker K+ channel conserved domain, which were queried against the candidate sequences of Shaker K+ channels from foxtail millet. The protein sequences of these candidate genes were further submitted to the SMART program.4 Finally, the sequences with a complete channel conserved domain were selected for subsequent analysis. Based on the protein sequence, the theoretical isoelectric point (PI), molecular weight (MW), and amino acid composition of the Shaker K+ channels from foxtail millet were analyzed by ExPASY5 (Gasteiger et al., 2003). Their subcellular localization was predicted by WoLF PSORT6 (Horton et al., 2007). The protein transmembrane region of the Shaker K+ channel was predicted via the TMHMM 2.07 (Krogh et al., 2001).
The amino acid sequences of Shaker K+ channels from foxtail millet, Arabidopsis (Cao et al., 1995), and rice (Hwang et al., 2013) were homologously aligned by the ClustalX software (Thompson et al., 2003) with default parameters. Based on this comparison, MEGA7.0 (Kumar et al., 2016) software was used to construct the phylogenetic tree of the Shaker K+ channels with the following parameters: Poisson model and pairwise deletion, Bootstrap 1,000 repetitions. The gene annotations of candidate Shaker K+ channel genes were extracted from the Phytozome database (Phytozome 13, Setaria italica v2.2) (Goodstein et al., 2012), from which the position of the genes on the chromosome was obtained, and the physical map was drawn using the MapChart software (Voorrips, 2002).
The conserved protein motifs of Shaker K+ channels from foxtail millet were predicted by the online Multiple Em for Motif Elicitation (MEME) program8 (Bailey et al., 2009), with the following parameters: the number of repetition = any, the maximum number of motifs = 10. These genes’ CDS and genome sequences were downloaded from the Phytozome database and submitted to Gene Structure Display Server online program (GSDS9) (Hu et al., 2015) for gene structure analysis.
The 2,000 bp upstream sequences of Shaker K+ channel gene in foxtail millet were extracted from the Phytozome database and submitted to the PlantCARE to obtain the cis-elements in the promoter regions10 (Lescot et al., 2002).
To study the potential expression patterns of Shaker K+ channels from foxtail millet at different tissues and developmental stages, the fragments per kilobase of the exon model per million mapped (FPKM) values of these genes were obtained from the GeneAtlas v1 Tissue Sample (Phytozome 13), including the data of etiolated seeding (5 days), germ shoot (6 days), shoot (1 week), leaf [different leaf (1, 2, 3, 4, 5, 6) at 2 weeks], panicle stage 1 and 2, and root (10 days). These data were submitted to TBtools (Chen et al., 2020) for expression profile mapping.
The foxtail millet cultivars “Jingu21,” “Longgu16,” and “Jigu39” were used in the present study. Seeds are obtained from Prof. Lizhen Zhang’s lab (School of Life science, Shanxi University, China). The plant seeds were grown in a tray containing vermiculite and nutrient soil at a ratio of 1:1 and cultivated under greenhouse conditions (16 h light/8 h dark at 23–26°C, 50,000 Lux light, and 30–50% relative humidity). When the seeds were germinated and grown to the two-leaf stage (4 days), select seedlings with consistent growth stages were transferred to plastic pots with three plants per pot. For temperature stress, after another 14 days, “Jingu21” seedlings were separately subjected to heat (40°C day/32°C night) and cold (4°C) stress in a constant temperature incubator for 0, 12, and 24 h. For other abiotic stresses, 10 days old “Jingu21” seedlings were transferred to 1/2 MS liquid medium and cultured for another 4 days, then subjected to salt (150 mmol/l, 200 mmol/l NaCl), PEG6000 (10%, 15%), ABA (100 μmol/l), 6-BA (75 μmol/l), IAA (10 μmol/l), NAA (10 nmol/l), BR (100 μmol/l), GA3 (1 mmol/l), MJ (100 μmol/l), and SA (10 mmol/l) treatments for 0, 12, and 24 h. There were three replicates for each stress treatment. The leaves were harvested. In addition, the leaves and roots of “Jingu21,” “Longgu16,” and “Jigu39” seedlings grown for 14 days under normal conditions were also collected. All samples were immediately frozen in liquid nitrogen and stored at −80°C for further RNA extraction.
Total RNA was extracted from samples using TransZol™ UP Plus RNA Kit (TransGen Biotech, Beijing, China), according to the manufacturer’s instructions. The first-strand cDNA templates were synthesized using EasyScript ® One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. The total volume of the qRT-PCR reaction system was 10 μl, which included 5 μl of TransStart ® Tip Green qPCR SuperMix (TransGen Biotech, Beijing, China), 1 μl of diluted cDNA template, 0.8 μl of upstream and downstream primers (10 μmol/l each), and 3.2 μl of RNase free ddH2O. The qRT-PCR thermal cycler program included 94°C for 30 s, followed by 45 cycles at 94°C for 15 s and 58°C for 30 s. All primers were synthesized by Shanghai Sangon Biotech (As shown in Supplementary Table 1), wherein SiAct2 (Seita.8G043100) was used as the internal reference. Each experiment included three technical replicates and three biological replicates.
The Shaker K+ channel protein sequences were submitted to NetOGlyc 4.011 for N-glycosylation site prediction. The NetPhos 3.112 was used to predict protein phosphorylation sites (including Serine, Threonine, and Tyrosine). The positive result was confirmed when the score was higher than 0.5.
The presence of abscisic acid (ABA) interaction domain (D543–G575) (Ooi et al., 2017) and guanine nucleotide-binding proteins (G-protein) binding motif (E165–Y169) (Deupi et al., 2012; Adem et al., 2020) in the protein sequence of Arabidopsis AtGORK (At5G37500) have been reported. Due to the high sequence similarity between GORK and SKOR, it is worth predicting whether similar binding sites exist in other plants, especially foxtail millet. The GORK and SKOR amino-acid sequences of Arabidopsis (GORK, At5G37500; SKOR, At3G02850), tobacco (GORK, XP_016457756; SKOR, XP_016460239), foxtail millet (GORK, Seita.7G111600; SKOR, Seita.4G110300), rice (GORK, Os04g36740; SKOR, Os06g14030), sorghum (GORK, Sobic.006G093400; SKOR, Sobic.010G102800), and wheat (GORK, Traes_2BL_C71ACBCED; SKOR, Traes_7DS_965990A2F) were extracted from the Phytozome and NCBI database, and multiple sequence alignments were performed using the ClustalX software.
Open reading frames for SiVAMP721 (Seita.6G010600), SiSNAP33 (Seita.9G493300), SiSYP121 (Seita.9G063400), and Shaker K+ channel genes were amplified from “Jingu21” with gene-specific primers including Gateway attachment sites (attB1/attB2; as shown in Supplementary Table 1). These amplified products were inserted into the pDONR207 vector to obtain entry clones by BP reactions using BP Clonase II (Invitrogen) and verified via sequencing (Sangon Biotech, Shanghai, China).
For the mating-based Split-Ubiquitin System (mbSUS) assay, destination clones for Shaker K+ channel genes were used as bait and recombined in the pMetYC-Dest vector. SiSNAREs as prey were recombined in the pNX35-Dest vector (Grefen et al., 2009). These clones were generated using LR Clonase II (Invitrogen) by LR reaction according to the manufacturer’s instructions.
For yeast complementation assay, destination clones for all Shaker K+ channels were recombined in padh1-Dest vector by LR reaction. The padh1-Dest vector was created by replacing TRP1 in pZMT-Dest (Grefen, 2014) with LEU1. The sequence containing LEU1 with a NdeI site at the 3′ and a NruI site at the 5′ termini was produced by DNA synthesis (Sangon Biotech, Shanghai, China) and digested by NdeI and NruI (NEB). The backbone from pZMT-Dest was amplified using primers BZ-S-NruI-ZMT and BZ-A-NdeI-ZMT. T4 DNA ligase (NEB) was used to carry out ligation. The subsequent transformation and selection were in ccdB-survival cells (TransGen Biotech, Beijing, China). The vector sequence was verified by restriction endonuclease digestion and sequencing. As shown in Figure 6A, the core expression area of padh1-Dest contains a Gateway cassette (attR1/attR2) with a C-terminal Myc tag under the control of the adh1 promoter.
The full-length coding sequences of Shaker K+ channels in foxtail millet were inserted into yeast expression vector padh1-Dest and transformed into the yeast mutant strain R5421 (MATα ura3-52 leu2 trk1Δ his3Δ200 his4-15 trk2Δ1::pCK64; Weidi Biotech, Shanghai, China), in which two K+ transporters, TRK1 and TRK2, were deleted (Li et al., 2014). Wild-type yeast strain R757 was used as the positive control. The yeast complementation experiment was performed as described previously (Li et al., 2014; Han et al., 2016). A commercial Myc antibody (Abcam) was used in immunoblot analysis to verify protein expression.
The mbSUS assays were performed as described before (Grefen et al., 2009; Horaruang and Zhang, 2017). Bait destination clones were transformed into yeast strain THY.AP4 and prey clones were transformed into THY.AP5. Pools of 10–15 yeast colonies were selected and inoculated in selective media (CSM–LM for bait in THY.AP4 and CSM–MTU for prey in THY.AP5) and grown at 28°C, 180 rpm overnight. The liquid cultures were harvested and resuspended in the yeast peptone dextrose (YPD) medium for yeast mating. Equal aliquots of yeast containing bait and prey were mixed in sterile PCR tubes, dropped on the YPD plate, and cultured at 28°C overnight. The colonies were transferred from the YPD plate onto the CSM–LMTU plate and incubated at 28°C for 2–3 days. Diploid colonies were selected and inoculated in liquid CSM–LMTU medium and grown at 180 rpm and 28°C overnight. After that, yeast was harvested by centrifugation and resuspended in sterile water. Serial dilutions at OD600 = 1 and 0.1 in water were dropped (5 μl per spot) on CSM–AHLMTU plates with added Met. Plates were incubated at 28°C, and images were taken after 3 days. Yeast was also dropped on CSM–LMTU control plates to confirm mating efficiency and cell density, and growth was imaged after 24 h incubation at 28°C. To verify the expression of bait and prey, yeast was harvested and extracted for protein immunoblot analysis using commercial HA antibody (Abcam) for prey and commercial VP16 antibody (Abcam) for bait.
In the present study, the statistical analysis was reported as means ± SE with significance determined by Student’s t-test or ANOVA in the SigmaPlot v.11.2 (Systat Software).
The coding sequences of Shaker K+ channels from Arabidopsis and rice were used to blast in the Phytozome foxtail millet database, and 12 candidate genes were obtained. Then, the Arabidopsis Shaker K+ channel protein sequences were submitted to the Pfam database (see footnote 3) to recognize the channel domains (PF00520.31, PF00027.29, and PF11834.8). The protein sequences of these candidate genes were submitted to the SMART program, and 10 genes were identified as Shaker K+ channels in foxtail millet (Table 1). The foxtail millet Shaker K+ channel proteins were 505–903 amino acids in length, and molecular weight was distributed from 58.16 to 98.76 kDa. The isoelectric point of Shaker K+ channel proteins ranged from 6.23 to 9.08. Subcellular localization predictions indicated that all Shaker K+ channel proteins were localized on the plasma membrane with 4–6 trans-membrane segments (Table 1).
To further explore the evolutionary relationship of Shaker K+ channels among different species, the protein sequences from foxtail millet, Arabidopsis, and rice were used to construct a phylogenetic tree. As shown in Figure 1, the Shaker K+ channel genes from foxtail millet can be classified into five groups based on their phylogenetic relationships and functional divergences (Pilot et al., 2003b; Hwang et al., 2013). Group I and II were inward rectifying channels. There were SiAKT1, SiAKT2 in Group I and SiKAT1, SiKAT2, and SiKAT3 in Group II. Only SiAKT2/3 belongs to Group III. Group IV, the regulatory or silent channel, included SiKC1a and SiKC1b. Group V was the outward rectifying channel, including SiGORK and SiSKOR. Similar groups of Shaker K+ channels from different species suggested that this protein family was evolutionary conserved. Compared to Arabidopsis, proteins from foxtail millet were more similar to their orthologous in rice.
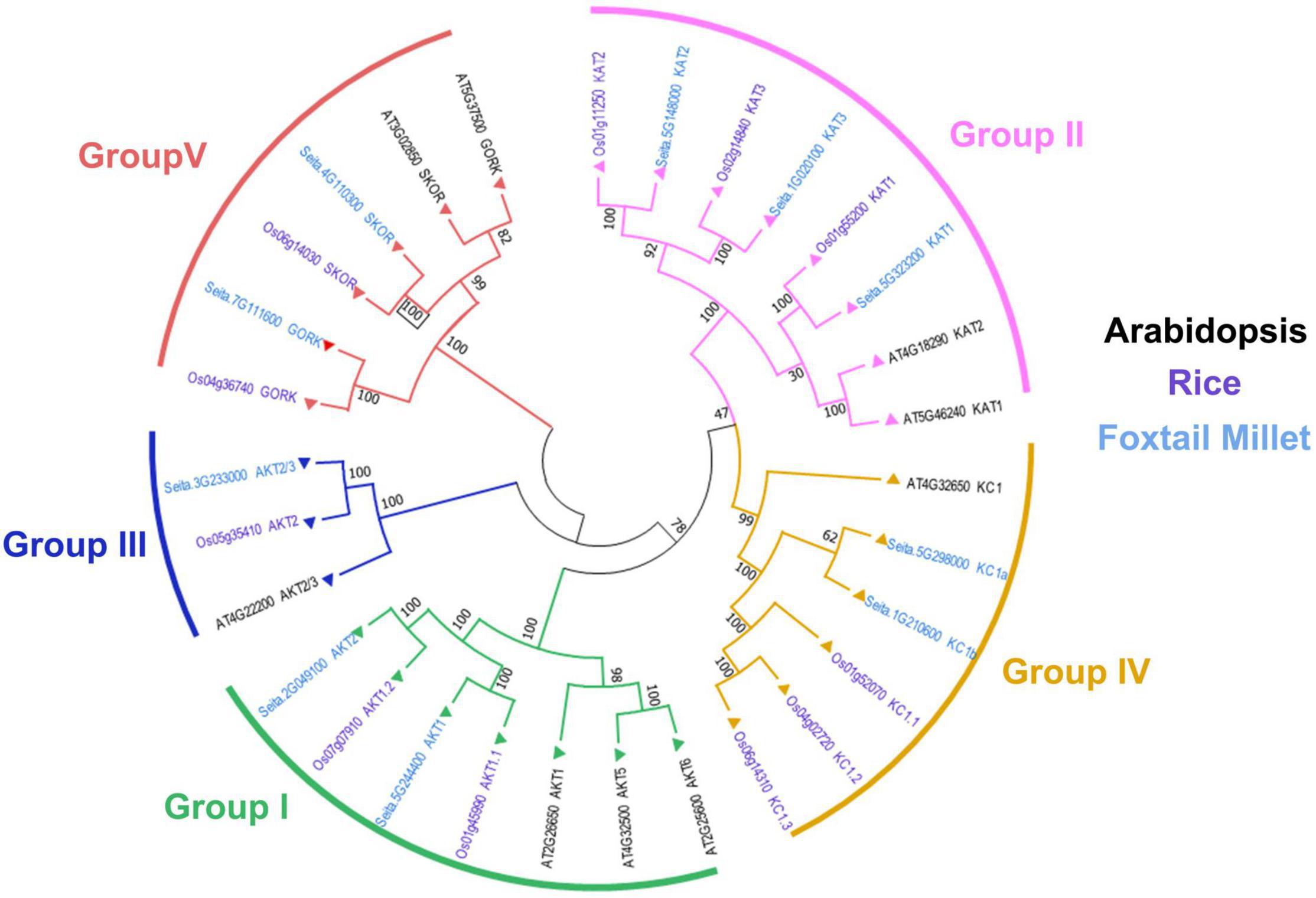
Figure 1. Phylogenetic analysis and classification of Shaker K+ channel proteins from foxtail millet (light blue), Arabidopsis (black), and rice (purple). Shaker K+ channels are divided into five distinct groups denoted by different colored lines.
Shaker K+ channel genes were distributed on 6 out of 9 chromosomes of foxtail millet (Supplementary Figure 1). Chromosome 1 contained 2 channel genes (SiKAT3 and SiKC1b). Only one gene on chromosome 2, 3, 4, and 7. Chromosome 5 contained the greatest number of Shaker K+ channel genes (4 genes).
Next, the gene structural diversity and protein motif distribution of Shaker K+ channels in foxtail millet were explored. As shown in Figure 2 at left, ten conserved motifs were detected in Shaker K+ channel proteins by MEME. All channel proteins contained conserved motifs 1–8. Proteins in Group I, III, and V contained two copies of motifs 10. Motif 9 were missing in SiSKOR and SiKC1b. Channels in the same group containing similar conserved motifs also supported our classification based on the phylogenetic tree.
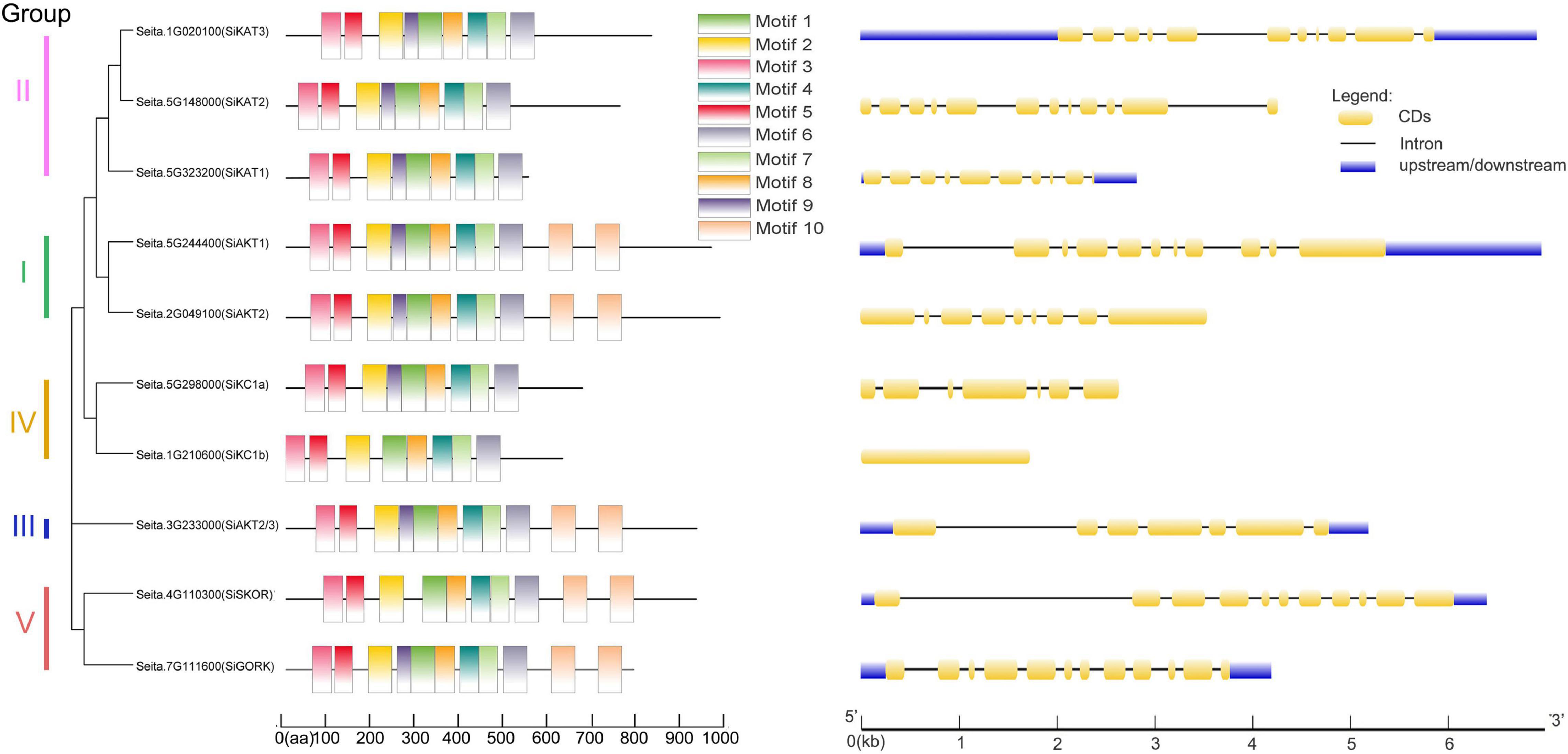
Figure 2. Protein motifs and gene structures of Shaker K+ channels from foxtail millet. (Left) In the Shaker K+ channel proteins, 10 motifs were identified by the MEME tool, represented by different colors (1–10) and depicted by TBtools. (Right) The exon-intron structure of these Shaker K+ channel genes was predicted by GSDS 2.0. The yellow boxes represent the coding sequence (CDs), the black lines represent introns, and the blue boxes represent up/downstream untranslated regions (UTR).
As shown on the right side of Figure 2, the same group of genes contained similar numbers of introns and exons. For example, Group I, II, and V contained 8–11 introns, and group III and IV contained 6 introns except for SiKC1b, which contained no intron. The differences in gene structures indicated the evolutionary and functional differences in the Shaker K+ channel family and provided additional evidence to support phylogenetic grouping.
To further understand the regulatory mechanism of the Shaker K+ channel genes, the 2,000 bp promoter regions upstream of them were used for cis-elements analysis. As shown in Supplementary Table 2, 68 elements were identified. Based on the functional differences, important cis-acting elements were divided into three major groups: plant growth and development related, phytohormone responsiveness related, and abiotic/biotic stresses related cis-acting elements. As shown in Supplementary Figure 2, circadian element (Anderson et al., 1994) was only found in SiKC1a, indicating that this gene may be related to photoperiod regulation. The RY element is a seed-specific promoter which mediates initial transcriptional activation during embryo mid maturation (Lelievre et al., 1992; Guerriero et al., 2009). This cis-acting element was only predicted in SiKAT3. In addition, promoter elements related to plant growth, such as A-box, CAT-box, and O2-site, were found in 3, 6, and 6 Shaker K+ channel genes, respectively. Abscisic acid (ABA) response elements (ABREs) (Hobo et al., 1999) were identified in 6 Shaker K+ channel genes. The SiKAT1 had 9 ABRE elements, while SiAKT2/3 had 8. The Methyl jasmonate (MJ)-responsive elements CGTCA-motif and TGACG-motif (Wang et al., 2019) were found in 9 genes. The SiAKT1 had the largest number of these two elements (6 for each). Other phytohormone-related cis-acting elements were identified in different genes, including AuxRR-core, TGA-element (auxin-responsive element) (Ulmasov et al., 1997), TCA-element (salicylic acid (SA) responsiveness; Zhang R. X. et al., 2017), GARE-motif, P-Box (gibberellin (GA3) responsive element; Washida et al., 1999), and ERE (ethylene-responsive element; Fujimoto et al., 2000), suggesting Shaker K+ channel genes were under the regulation of different phytohormones. Many cis-acting elements involved in plant stress response were also found in the promotor region of Shaker K+ channel genes. MYB and MYC, related to drought stress and ABA regulation (Abe et al., 1997), were predicted in all genes. Moreover, the MYC element was found in the largest number in Group II. The G-Box element was involved in chlorophyll synthesis (Abe et al., 1997) and was the most abundant in SiKAT3. The WUN-motif, a wound-responsive element (Hayashi et al., 2003), was only found in SiAKT1 and SiSKOR. In conclusion, cis-acting element analysis showed that most Shaker K+ channel genes responded to different phytohormones and environmental stresses.
Protein phosphorylation and asparagine (N)-linked glycosylation are two important post-translational modifications that affect the protein stability, protein subcellular location, and protein-protein interaction in plants (Xu et al., 2019). They are crucial for plant response to environmental stresses (Jiao et al., 2020). As shown in Supplementary Figure 3, these two kinds of post-translational modifications in Shaker K+ channel proteins from foxtail millet were predicted. SiKAT3 and SiAKT1 contained the highest number of glycosylation sites (five sites). SiKC1a, SiKC1b, and SiSKOR have three sites. SiAKT2 and SiAKT2/3 were predicted to have two sites, and the remaining proteins contained only 1 glycosylation site. Regarding the predicted phosphorylation sites, the Shaker K+ channel proteins in foxtail millet ranged from 40 to 79. There were 79 sites in AKTs (SiAKT1 and SiAKT2), while 40 and 45 were predicted in SiKC1a and SiKC1b, respectively.
The plant hormone ABA plays an important role in plant response to abiotic stress, especially drought stress (Danquah et al., 2014; Fahad et al., 2015). Previous reports have shown that the GORK of Arabidopsis harbors potential ABA interaction sites (GORKN558, K559, Y562, R565). The mutation of K559 and Y562 to Ala significantly reduces the outward K+ current of GORK, while the other two mutations of N558A and R565A lead to the complete loss of GORK’s channel function (Ooi et al., 2017). The relationship between outward rectifying K+ channel SKOR and GORK is close in evolution. Therefore, we compared the GORK and SKOR protein sequences of foxtail millet, Arabidopsis, tobacco, and other plants (rice, sorghum, and wheat). As shown in Supplementary Figure 3C, these four amino acid sites existed on the GORK protein of most species. Compared to the sequence of GORK in Arabidopsis, only N558 changed to Ser in SiGORK, while the N558 and K559 changed to Lys-Asn in SiSKOR. Considering that Asn and Ser are polar amino acids with uncharged R groups, it is still possible that SiGORK contains a potential ABA binding site.
To analyze the expression pattern of Shaker K+ channel genes in different tissues, publicly available foxtail millet RNA-seq data (GeneAtlas v1 Tissue Sample from Phytozome 13) was used. This dataset included information from various tissues, including etiolated seeding at 5 days, germ shoot at 6 days, shoot at 1 week, different leaf (1–6) at 2 weeks, panicle stage 1 and 2, and root at 10 days. As shown in Figure 3, the SiAKT1 was expressed highly in various tissues, especially in leaf and root, suggesting its role in K+ uptake and transport between tissues. The SiKAT3 could be found in all tissues above ground. The SiAKT2/3 was expressed highly in panicle. The two genes in group V (SiGORK and SiSKOR) were highly expressed in roots. The SiKAT1, SiAKT2, SiKC1a, and SiKC1b were almost not detected in all tissues.
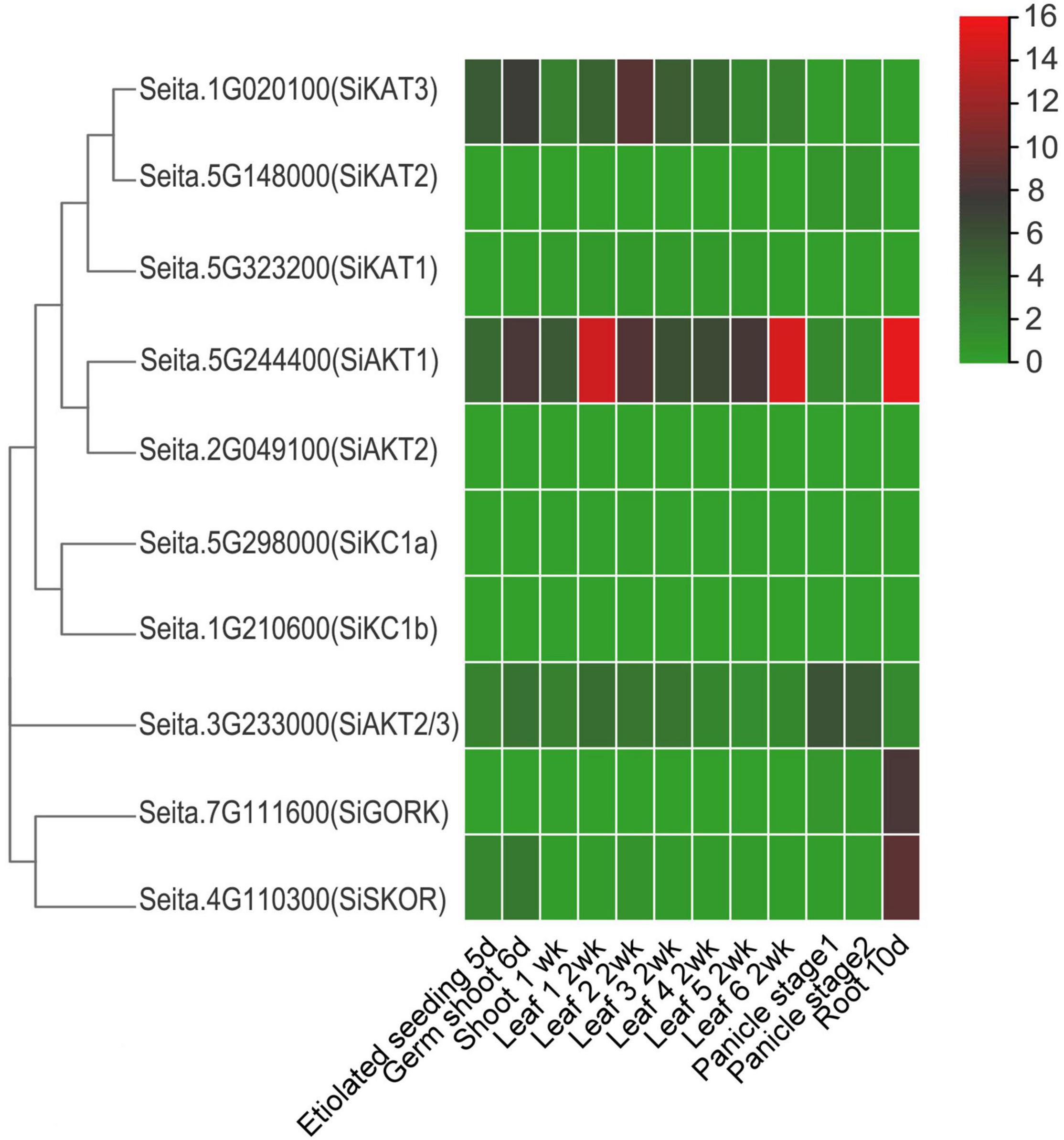
Figure 3. Expression profile of Shaker K+ channel genes in different tissues and developmental stages of foxtail millet. Gene expression data is downloaded from the GeneAtlas v1 Tissue Sample (Phytozome 13), including etiolated seeding (5 days), germ shoot (6 days), shoot (1 week), leaf (different leaf at 2 weeks), panicle stage 1 and 2, and root (10 days). The color scale represents expression levels (FPKM value) from high (red) to low (green color).
To further study the role of Shaker K+ channels in foxtail millet stress resistance, 14-days-old seedlings of “Jingu21” were subjected to different abiotic treatments (cold, heat, NaCl, and PEG). The seedlings under temperature stresses did not show any significant difference after 24 h (Supplementary Figure 4). However, after 24 h of other abiotic stresses, the leaves of seedlings turned yellow and wrinkled, especially under high levels of salt and PEG treatments. Then the leaf samples were harvested. The expression patterns of channel genes were detected by qRT-PCR. The transcription levels of four genes (SiKAT1, SiAKT2, SiKC1a, and SiKC1b) were not detectable, which was consistent with the pattern shown in Figure 3. Considering that many genes were expressed with circadian rhythms, we also tested the transcription of these channels without abiotic stresses. The results were shown in Supplementary Figure 5. There were no significant changes between 0 and 12 or 24 h in either soil culture (A) or liquid medium culture (B).
As shown in Figure 4, under cold stress (4°C), the transcription levels of SiAKT1, SiAKT2/3, SiKAT2, SiSKOR, and SiGORK increased significantly after 24 h treatment. The SiSKOR’s transcription level was up-regulated more after 12 h cold treatment than after 24 h. Under hot stress (40°C day/32°C night), SiAKT1 was up-regulated after 24 h treatment. The transcription levels of SiSKOR and SiGORK increased strongly after 12 h treatment. Under salt stress (150 mmol/l, 200 mmol/l NaCl) or PEG (10 and 15%) treatments, transcription of Shaker K+ channel genes in foxtail millet varied. The transcription level of SiAKT1 decreased under both treatments. SiAKT2/3 was up-regulated after 24 h treatment of PEG. For SiKAT2, 200 mmol/l salt stress induced its transcription at 12 h; 10% PEG treatment decreased its expression at 12 h, but significantly up-regulated it after 24 h. SiKAT3 was strongly induced by 10% PEG treatment at 12 h. At 24 h, this gene was induced by both 10 and 15% PEG treatments. SiGORK decreased under PEG treatment in both time points, while SiSKOR was up-regulated by salt stress and 10% PEG treatments after 24 h.
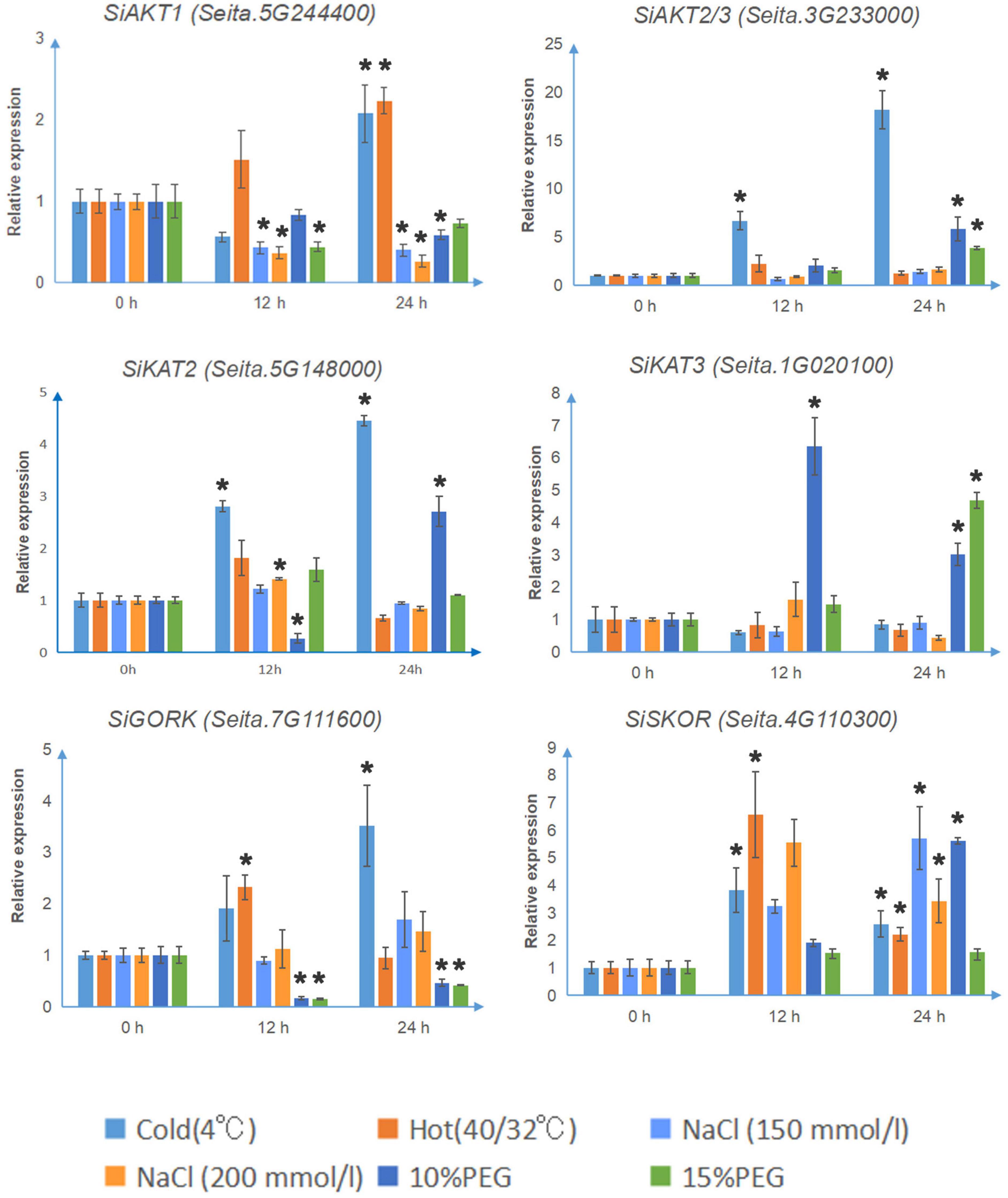
Figure 4. The relative transcript levels of Shaker K+ channel genes in foxtail millet under different abiotic stresses. The qRT-PCR analyses were used to access transcript levels of Shaker K+ channel genes from foxtail millet with or without 12 and 24 h cold (4°C), hot (40°C day/32°C night), salt (150 mmol/l, 200 mmol/l NaCl), and osmotic stress (10 and 15% PEG) treatments. Each bar represents the mean ± SE normalized to SiAct2 (Seita.8G043100). All samples were run in three biological and three technical replicates. Asterisk indicates that the gene expression under stress has a significant difference compared with the control at 0 h (*p < 0.05).
Considering that the promoters of Shaker K+ channel genes in foxtail millet contain many phytohormone-related elements (As shown in Supplementary Figure 2), the transcription patterns of channel genes under different phytohormone treatments were analyzed. After 24 h of phytohormone treatments, the “Jingu21” seedlings did not show any significant difference (Supplementary Figure 4). Again, four genes (SiKAT1, SiAKT2, SiKC1a, and SiKC1b) were not detected under treatments.
Most Shaker K+ channel genes contain the MeJA-responsive elements CGTCA-motif and TGACG-motif (Supplementary Figure 2). As shown in Figure 5, under MJ treatment, the transcription levels of all 4 detectable inward K+ channel genes were up-regulated. MJ suppressed both SiGORK and SiSKOR. Auxin treatment induced expression of SiAKT1, SiKAT3, and SiSKOR, but decreased the level of SiKAT2 and SiGORK. Gibberellin treatment also increased the transcription of SiAKT1, SiKAT3, and SiSKOR, but only inhibited the expression of SiAKT2/3. In the case of salicylic acid treatment, the transcription of SiAKT2/3 and SiKAT3 were down-regulated. SiSKOR was significantly inhibited at 12 h but induced at 24 h under SA treatment. Six channel genes contained the ABRE element. Among them, SiAKT2/3 and SiKAT3 were induced by ABA treatment, while the level of SiAKT1 was suppressed at 12 h. We also tested the response of channel genes to 6-BA and BR. The former induced transcription of all six detectable channels except SiKAT2. The latter up-regulated SiAKT2/3 and SiGORK but suppressed SiSKOR and SiAKT1 at 12 h.
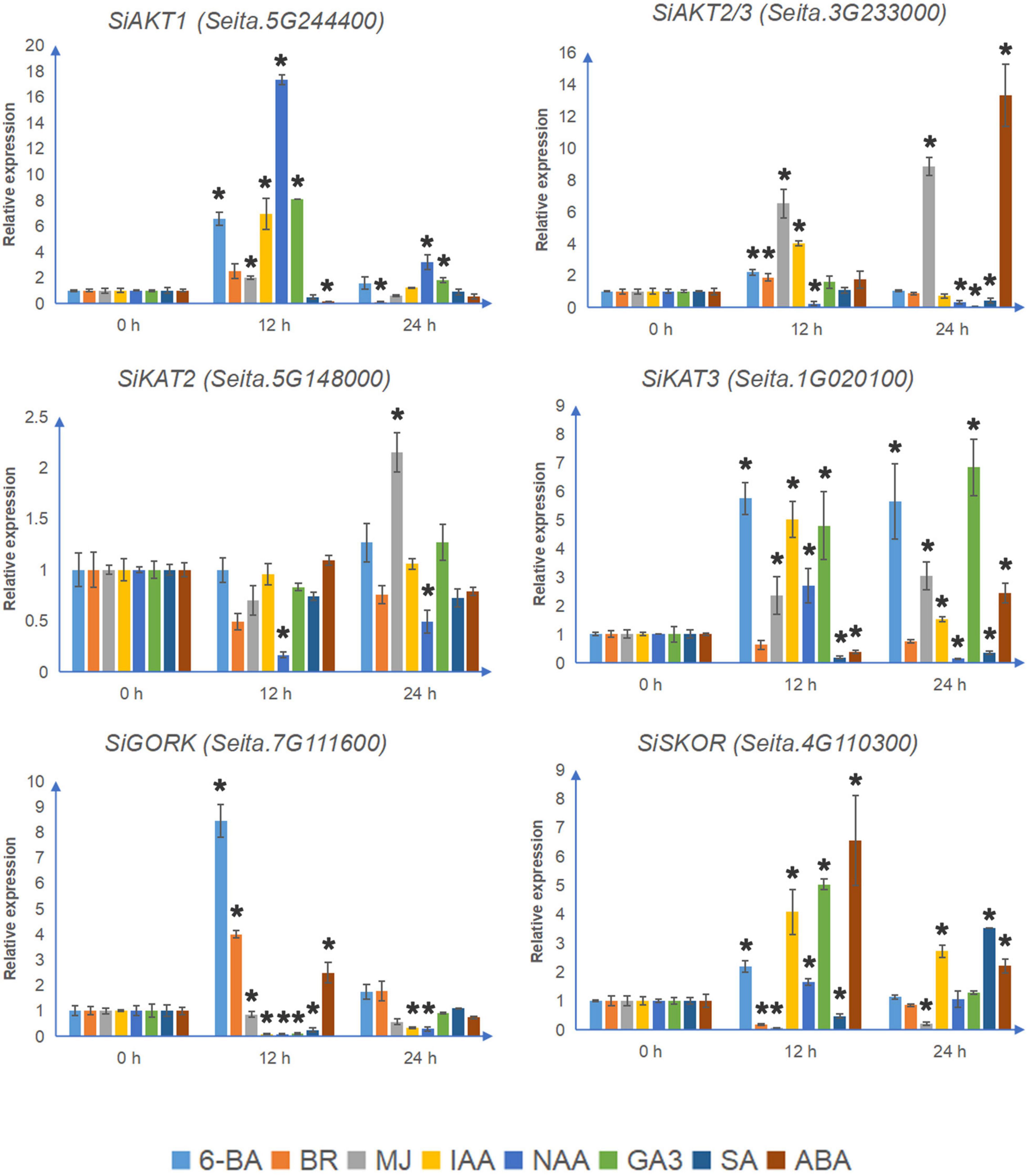
Figure 5. The relative transcript levels of Shaker K+ channel genes in foxtail millet under different phytohormone treatments. The qRT-PCR analyses were used to access transcript levels of Shaker K+ channel genes from foxtail millet with or without 12 and 24 h ABA (100 μmol/l), 6-BA (75 μmol/l), IAA (10 μmol/l), NAA (10 nmol/l), BR (100 μmol/l), GA3 (1 mmol/l), MJ (100 μmol/l), and SA (10 mmol/l) treatments. Each bar represents the mean ± SE normalized to SiAct2 (Seita.8G043100). All samples were run in three biological and three technical replicates. Asterisk indicates that the gene expression under stress has a significant difference compared with the control at 0 h (*p < 0.05).
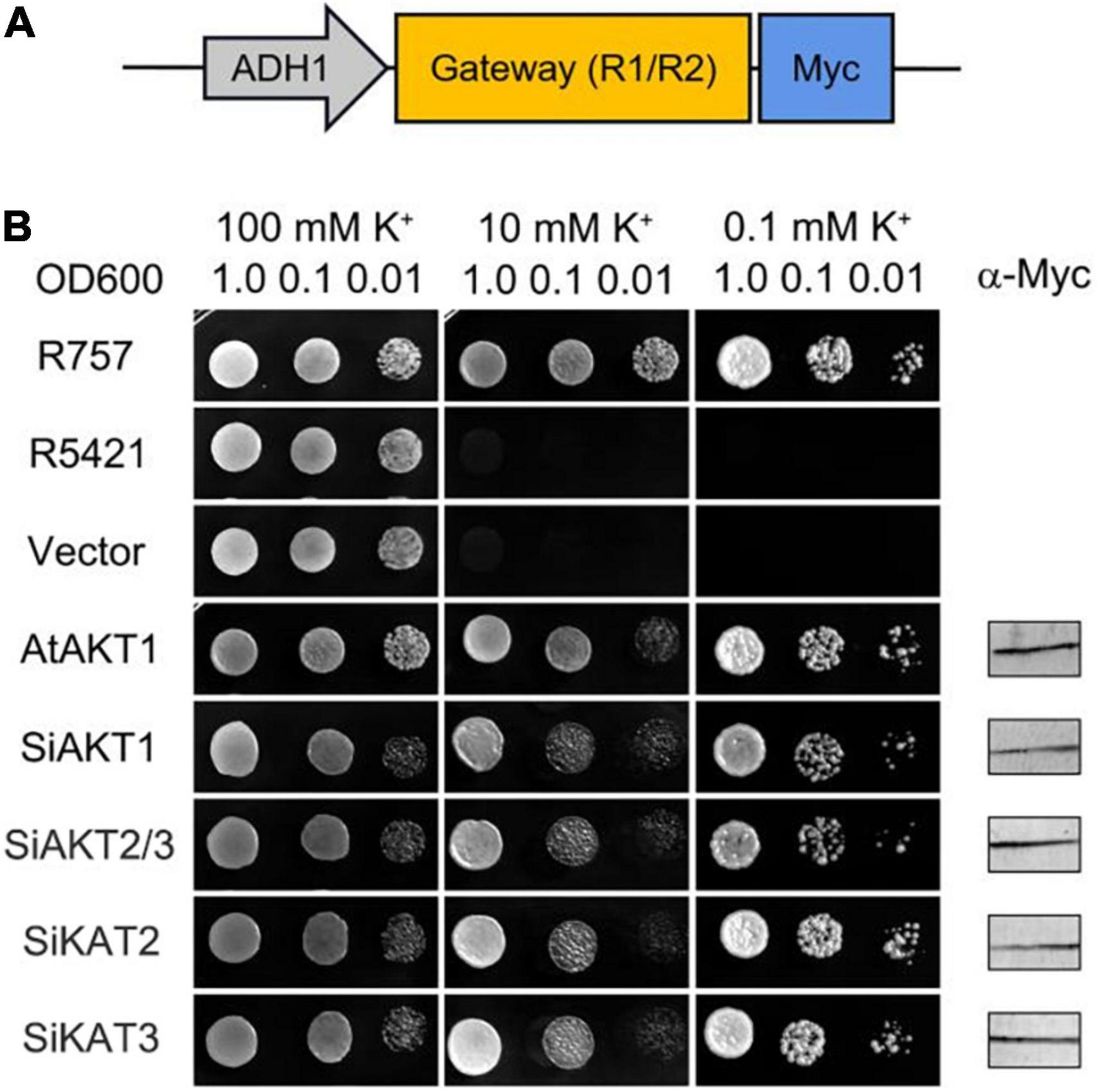
Figure 6. Shaker K+ channels from foxtail millet mediate K+ Transport in Yeast. (A) Schematic of the padh1-Dest vector. (B) AtAKT1 and Shaker K+ channels from foxtail millet complement the K+ uptake-deficient yeast mutant R5421. The yeast strain R757 was used as a control. Immunoblot analysis (5 μg total protein/lane) of yeast was performed using a commercial α-Myc antibody.
Under different abiotic stresses or phytohormone treatments, the transcription of SiKC1s was still missing. Whether this is a phenomenon that only existed in “Jingu21?” We performed the RT-PCR analysis in the other two foxtail millet cultivars “Longgu16” and “Jigu39,” to answer this question. One of the results was shown in Supplementary Figure 6. In either roots or leaves of different cultivars, we could not detect the expression of SiKC1s.
Taken together, SiAKT1, SiKAT3, SiGORK, and SiSKOR were more worth further research due to their significant changes under most stresses and phytohormone treatments.
The inward K+ transport activities of four detectable Shaker K+ channels in foxtail millet were tested in the K+-deficient yeast mutant strain R5421 (trk1Δ, trk2Δ), in which two K+ transporter genes (TRK1 and TRK2) were deleted (Gaber et al., 1988; Li et al., 2014). Shaker K+ channel genes were inserted into the padh1-Dest vector and transformed into R5421. AtAKT1 (At2g26650) from Arabidopsis was used as the positive control. The wild-type yeast strain R757 was used as the growth control. The yeast growth assays were performed on a CSM medium with different concentrations of K+. Figure 6 showed one of three independent experiments which yielded similar results. Under high K+ concentration (100 mmol/l), all yeast grew well. Under low K+ concentration (0.1 mmol/l), there was no growth recovery of R5421 and R5421 containing the empty vector. The K+ channels from foxtail millet or Arabidopsis rescued R5421 yeast growth, suggesting that the Shaker K+ channels we tested had similar K+ uptake function as the AKT1 channel in Arabidopsis.
Previous reports have shown that ion channels are regulated by SNARE proteins (Karnik et al., 2017; Zhang et al., 2020). In Arabidopsis, SYP121 and VAMP721 form the SNARE complex during membrane fusion. They interact with KC1 and KAT1 K+ channels and regulate the channel activity in an opposite way (Honsbein et al., 2009; Zhang et al., 2015). SNAP33 also interacts with K+ channels and regulates channel activity with SYP121 and VAMP721 (Waghmare et al., 2019). We analyzed foxtail millet SNARE-Shaker K+ channel interaction by mbSUS assay. SiKAT1 was almost undetected in all tissues (Figure 3). Thus, it could not be cloned. SiKAT3 failed to transform into yeast. In this assay, Shaker K+ channel SiKAT2 was fused with C-terminal halves of the ubiquitin (Cub) and used as bait, while SiSYP121, SiVAMP721, and SiSNAP33 were fused with N-terminal halves of the ubiquitin (Nub) and used as prey. The protein-protein interaction will reassemble ubiquitin, release trans-activator, and activate reporter genes, allowing yeast growth on selective media. This method has been previously used in membrane protein interaction analysis (Honsbein et al., 2009; Grefen et al., 2010; Zhang et al., 2015, 2019; Zhang B. et al., 2017). Figure 7 showed that diploid yeast expressing SiKAT2 with SiVAMP721 or SiSNAP33 grew well on selective media, even on the media containing 50 μmol/l Met (which suppresses bait protein expression). However, diploid yeast growth was not recovered with SiSYP121 as prey. This result demonstrated the selective interaction between the Shaker K+ channel and SNARE proteins in foxtail millet.
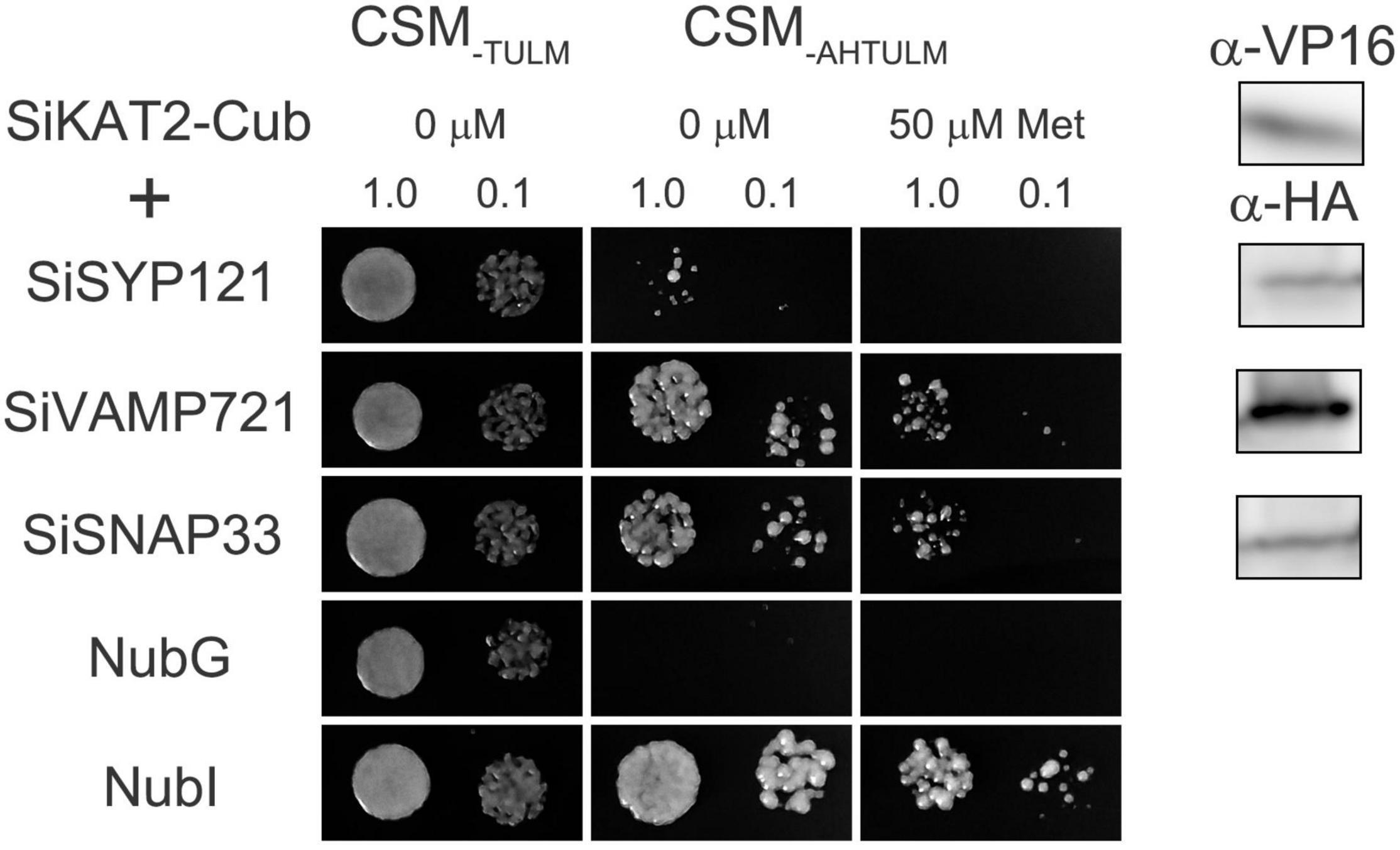
Figure 7. SiKAT2 selective interact with SiSNARE proteins. Yeast mating-based split-ubiquitin assay for the interaction of SiSYP121, SiVAMP721, and SiSNAP33 with SiKAT2-Cub as baits. After several attempts, SiKAT3 failed to transform into yeast. Yeast diploids were created with NubG fusion constructs of each of the SiSNARE proteins together with controls (NubG, negative; NubI, positive) spotted (left to right) on CSM medium without Leu, Trp, Ura, and Met (CSM–TULM) to verify mating, CSM medium without Leu, Trp, Ura, Met, Ade, and His (CSM–AHTULM) to verify adenine- and His-independent growth, and on CSM–LTUMAH with the addition of Met to verify interaction at lower K+ channel-Cub expression levels. Diploid yeast was dropped at 1.0 and 0.1 OD600 in each case. Immunoblot analysis (5 μg total protein/lane) of the haploid yeast used for mating (right) using commercial α-HA antibody for the SiSNARE fusions and α-VP16 antibody for the K+ channel fusions.
Potassium (K+) is involved in stress resistance in higher plants (Wang et al., 2013). Shaker K+ channels are highly conserved voltage-dependent ion channels in plants and play an important role in K+ absorption and transport. Investigations focused on this family of genes have been performed in many species (Bauer et al., 2000; Amrutha et al., 2007; Dreyer and Uozumi, 2011; Jin et al., 2021). The current study identified 10 Shaker K+ channel family members in foxtail millet. In 14-days-old seedlings of “Jingu21,” six of 10 Shaker K+ channels were detectable. Their inward K+ transport activities were verified in yeast. The qRT-PCR analysis showed that the transcriptions of channels responded to different abiotic stresses and phytohormone treatments. Compared to their homologous in Arabidopsis, there were different selective interactions between SNARE proteins and Shaker K+ channel SiKAT2 in foxtail millet.
Previous research suggested that Shaker K+ channels are highly conserved in plants (Dreyer and Blatt, 2009). In the present study, Shaker K+ channel genes from foxtail millet were classified into five groups as reported in other species (Table 1 and Figure 1). Further analysis showed that channel genes of each group shared similar motifs and gene structures (Figure 2). Based on the phylogenetic analysis, the channel genes from foxtail millet were more similar to their orthologous genes in rice than that in Arabidopsis, suggesting the diversification of Shaker K+ channels happened before the separation of monocots and dicots in evolution and was consistent with the previous report (Pilot et al., 2003b). The five-group classification of channels is based on their functional diversity (Pilot et al., 2003b; Dreyer and Blatt, 2009). We predicted each group’s K+ transport activity in foxtail millet based on their classification.
To identify the roles of Shaker K+ channels in abiotic stress response, we first analyzed the cis-acting elements in their promoter region. As shown in Supplementary Figure 2, most of the genes contained MYB and MYC elements, suggesting that the Shaker K+ channels in foxtail millet were regulated by drought stress. We also found various hormone response elements, including ABA, auxin, GA3, MeJA, and ethylene. Various cis-acting elements existed in Shaker K+ channel genes, even in the same group, suggesting the diversity of these genes’ expression in response to environmental stresses.
Expression profiling of Shaker K+ channels was also performed using previously released RNA-seq data (Figure 3). Shaker K+ channel genes showed various expression levels in different tissues. SiAKT1 was expressed in all tissues, especially in roots, supporting its role in K+ absorbing from the soil as its orthologous genes in other species (Véry and Sentenac, 2003; Li et al., 2014). In Arabidopsis, KC1 in group IV is silent when expressed alone but function as a general regulator of channel activity when it forms hetero-tetramer with other channel proteins (Duby et al., 2008; Jeanguenin et al., 2011). Interestingly, according to the RNA-seq data, the SiKC1s could not be detected in all tissues. One explanation of this result is that the expression of some K+ channels is only induced under stress (Zhang et al., 2018; Jin et al., 2021). We will go back to this point later.
Glycosylation and phosphorylation modifications play vital roles in protein functions. The potential post-translational modifications of Shaker K+ channel proteins in foxtail millet were also predicted. Our results showed that SiAKT1 and SiAKT2 had the largest number of modification sites (Supplementary Figure 3). Previous reports have shown that AtAKT1 was activated by phosphorylation, which depends on the calcineurin B-like 1 (CBL1) with the CBL-interacting protein kinase (CIPK 23) (Xu et al., 2006; Honsbein et al., 2009). Such phosphorylation activating might also happen in SiAKTs and other Shaker K+ channels in foxtail millet.
As mentioned above, to investigate whether Shaker K+ channels in foxtail millet, especially SiKC1s, were regulated by abiotic stresses or phytohormone treatments, we performed the qRT-PCR analysis in 14-days-old seedlings of “Jingu21” under different treatments (cold, heat, NaCl, PEG, ABA, 6-BA, IAA, NAA, BR, GA3, MJ, and SA). “Jingu21” was one of the cultivars produced by radiation-induced mutagenesis in breeding programs (He et al., 2015). This cultivar shows good quality, high yield, and drought stress-tolerant than the other cultivars (Wang et al., 2021). Our results showed that four Shaker K+ channel genes (SiKAT1, SiAKT2, SiKC1a, and SiKC1b) were not detected in “Jingu21” under different treatments (Figures 4, 5). This finding supports the RNA-seq data shown in Figure 3. In Arabidopsis, KC1 forms the functional channel with inward Shaker K+ channel subunit AKT1. It performs as an important regulator of inward K+ current by negatively shifting the channel activation threshold, which helps to limit the K+ leakage under low K+ condition (Honsbein et al., 2009; Li et al., 2014). To investigate whether the missing SiKC1s’ transcription was related to the cultivar, we performed the RT-PCR analysis in the other two foxtail millet cultivars, “Longgu16” and “Jigu39.” Our previous research has shown that “Longgu16” was more sensitive to drought stress than “Jingu21” (Wang et al., 2021). The results here showed that the transcription of SiKC1s could not be detected in different cultivars of foxtail millet (Supplementary Figure 6). The undetectable SiKC1s left a question mark in the present study. KC1 was up-regulated by salt stress and K+ deficiency in Arabidopsis (Pilot et al., 2003a). The transcription level of SiKC1s under low K+ stress was worth testing in future research. SiAKT1 might form a functional channel with other Shaker K+ channel proteins in foxtail millet was another explanation for the missing SiKC1s here. SiAKT2/3 is found in most tissues (Figure 3). It is worth co-expressing SiAKT2/3 with SiAKT1 and detecting the channel gating by the electrophysiological experiment. In rice, OsAKT1 is not regulated by the KC1-like gene (Fuchs et al., 2005; Li et al., 2014). Thus, Li et al. found that OsAKT1 alone could restrain the K+ leakage under low K+ (Li et al., 2014). OsAKT1 is affected by some other mechanisms (like phosphorylation and vitamin B6), which might help to modulate K+ uptake together (Lee et al., 2007; Xia et al., 2014). It is worth further investigating whether similar mechanisms happened in foxtail millet.
As shown in Figure 4, the other remaining genes displayed their regulation pattern under different treatments, consistent with findings from previous reports on Arabidopsis (Pilot et al., 2003a) and sweet potato (Jin et al., 2021). In Arabidopsis, AKT1 is involved in a major pathway for K+ uptake at the root epidermis (Geiger et al., 2009). SiAKT1 induced by temperature stress and decreased under salt and osmotic stress revealed the role of K+ absorption in plant adaptation to environmental conditions. GORK plays a central role in K+ loss from the cytosol, and the cytosol K+ change is an important second messenger in plant stress response (Adem et al., 2020). In foxtail millet, we found SiGORK was induced by cold stress but inhibited by osmotic stress, suggesting GORK is the key to understanding the foxtail millet response mechanism to these stresses.
As shown in Figure 5, Shaker K+ channel genes in foxtail millet responded variant to different phytohormone treatments, supported by previous reports in other species (Philippar et al., 1999; Pilot et al., 2003a; Jin et al., 2021). ZMK1 in maize is the homology to AKT1 in Arabidopsis. This channel gene was induced by auxin (NAA) treatments and involved in coleoptile elongation (Philippar et al., 1999, 2006). In the present study, we found that NAA significantly increased the transcription of SiAKT1. Considering the wide expression of SiAKT1 in various tissues (Figure 3), this gene might play an important role in auxin-related plant growth. ABA serves as an important endogenous messenger in abiotic stress responses in plant. SKOR is regulated by different abiotic stress and decreased under ABA treatment in Arabidopsis and sweet potato (Pilot et al., 2003a; Jin et al., 2021). This channel is involved in the K+ secretion into the xylem sap (Johansson et al., 2006; Dreyer and Blatt, 2009). Our analysis revealed that SiSKOR was up-regulated by all stresses except ABA treatment. The divergent pattern of SiSKOR between foxtail millet and other species suggests the role of K+ redistribution among tissues in foxtail millet stress-tolerant. Previous reports have also shown that ABA does not affect the transcription level of GORK but induces plasma membrane depolarization and activates GORK in guard cells, leading to stomatal closure (Thiel et al., 1992; Hosy et al., 2003; Pilot et al., 2003a). In our result, SiGORK was only induced by ABA at 12 h. In 2017, Ooi et al. has reported that direct GORK-ABA interaction enhances K+-efflux current through GORK (Ooi et al., 2017). So, we performed a protein sequence alignment between GORK and SKOR from different species. We found that the important sites for ABA docking existed in most species, especially foxtail millet (Supplementary Figure 3), suggesting these outward channels in foxtail millet might also be under the regulation of ABA through directly binding.
As shown in Figure 6, the yeast complementation helped to confirm that the detectable inward K+ channels had K+ transport activity. This result supported the five-group classification of Shaker K+ channels in foxtail millet.
Previous reports have shown that KAT1 interacts with SNARE proteins, including SYP121, VAMP721, and SNAP33 in Arabidopsis, and these SNARE proteins co-modulate the channel activities (Honsbein et al., 2009; Karnik et al., 2017; Waghmare et al., 2019). We found that SiKAT2 interacted with SiVAMP721 and SiSNAP33, suggesting similar SNARE-K+ channel interactions in different species. However, we did not find the interaction of SiKAT2 with SiSYP121 (Figure 7). In Arabidopsis, when VAMP721 negatively regulates the channel activity, the binding of SYP121 with K+ channel promotes channel activity, such SYP121-VAMP721 balance is essential for K+ uptake and stomata movement (Honsbein et al., 2009, 2011; Zhang et al., 2015). SiSYP121 is close in evolution to SYP121 of Arabidopsis, and their sequences are highly similar (Wang et al., 2021). Surprisingly, there was no interaction between SiSYP121 and SiKAT2. There might be other SiSNARE proteins that take the role of positive K+ channel regulation. Identifying the difference between foxtail millet and Arabidopsis for SNARE-related K+ channel regulation will help to reveal the mechanism of K+ uptake and stress tolerance of foxtail millet.
SiKAT3 was expressed in R5421 yeast used in the complementation assay, but it failed to be expressed in THY.AP4 yeast, which was used in the mbSUS assay. We speculate that the codon bias might affect the expression of SiKAT3 in THY.AP4. There were differences between R5421 and THY.AP4 in genotype (Grefen et al., 2009; Li et al., 2014). Codon bias affects proteins’ expression level, localization, time rhythm, and function (Zhou et al., 2013; Engel et al., 2021). Grefen et al. have optimized the codons of AKT1 of Arabidopsis and transferred it into THY.AP4 yeast to get better protein expression (Grefen and Blatt, 2012).
In conclusion, we identified ten Shaker K+ channel genes in foxtail millet and analyzed their roles in plant stress responses. This work will facilitate further research focused on the biological roles of Shaker K+ genes in foxtail millet, which is beneficial to realize the combination of theoretical research in model plants and application research in crops.
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
BZ, LZ, PY, and YG designed the experiments. YG, HW, XW, and ML performed the experiments. BZ, HW, YG, and LZ analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by the open funding of State Key Laboratory of Sustainable Dryland Agriculture, Shanxi Agricultural University (No. YJHZKF2108), National Natural Science Foundation of China (No. 32000210), Research Project Supported by Shanxi Scholarship Council of China (No. 2021-015), and National Key Research and Development Program of China (No. 2020YFD1001401) to BZ. The Innovative Projects for graduate education in Shanxi province (No. 2021Y131) to HW. The National Key Research and Development Program of China (No. 2020YFD1001405) to LZ. The Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (No. 2019L0104), National Natural Science Foundation of China (No. 32101384), and Science Foundation for Youths of Shanxi Province (No. 20210302124150) to PY. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Prof. Yi Wang (China Agricultural University, China) for providing us the yeast strain R757. We also thank Prof. Mike Blatt (University of Glasgow, United Kingdom) for the gift of pZMT-Dest vector.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.907635/full#supplementary-material
Supplementary Figure 1 | Chromosomal localization of Shaker K+ channel genes from foxtail millet. The location information of 10 Shaker K+ channel genes on the chromosome was obtained from the Phytozome database (Setaria italica v2.2). Different colors on the nine chromosomes (chr1–chr9) indicate different channel groups.
Supplementary Figure 2 | Cis-acting element analysis of the promoter regions of Shaker K+ channel genes from foxtail millet. The number of each cis-acting element in the promoter regions (2,000 bp upstream of the translation start site) of Shaker K+ channel genes was shown. Based on the functional annotations, the cis-acting elements were classified into three major classes: plant growth and development, phytohormone responsiveness, and abiotic/biotic stresses related cis-acting elements.
Supplementary Figure 3 | Prediction of post-translational modification sites and ABA binding sites in the amino acid sequences of Shaker K+ channel proteins from foxtail millet. (A) N-glycosylation site; (B) phosphorylation site; (C) ABA binding sites in GORK and SKOR.
Supplementary Figure 4 | The phenotype of “Jingu21” seedlings after 24 h treatment of different abiotic stresses (A) or phytohormones (B).
Supplementary Figure 5 | The transcript levels of six Shaker K+ channels did not change without abiotic stresses. (A) Seedlings of “Jingu 21” grow in soil (control for cold or hot treatments). (B) Seedlings of “Jingu 21” grow in liquid medium (control for salt, PEG, and phytohormone treatments). The Student’s t-test was used to analyze statistical significance. Compared to that at 0 h, the transcript levels of Shaker K+ channels at 12 and 24 h showed no difference (P > 0.05).
Supplementary Figure 6 | The transcripts of SiKC1a and SiKC1b could not be detected by RT-PCR in different cultivates. The transcripts of SiKC1a (Seita.5G298000) and SiKC1b (Seita.1G210600) were tested by RT-PCR in 14 days old seedlings of “Jingu21,” “Longgu16,” and “Jigu39.” R means root, and L means leaf. SiAKT1 was used as positive control. SiAct2 was used as the internal reference. In all three cultivates, there are similar strong bands for SiAKT1, supporting the RNA-seq analysis in Figure 3.
Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. doi: 10.1105/tpc.9.10.1859
Adem, G. D., Chen, G., Shabala, L., Chen, Z. H., and Shabala, S. (2020). GORK channel: a master switch of plant metabolism? Trends Plant Sci. 25, 434–445. doi: 10.1016/j.tplants.2019.12.012
Ahmad, I., Mian, A., and Maathuis, F. J. M. (2016). Overexpression of the rice AKT1 potassium channel affects potassium nutrition and rice drought tolerance. J. Exp. Bot. 67, 2689–2698. doi: 10.1093/jxb/erw103
Amrutha, R. N., Sekhar, P. N., Varshney, R. K., and Kishor, P. B. K. (2007). Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci. 172, 708–721.
Anderson, S. L., Teakle, G. R., Martino-Catt, S. J., and Kay, S. A. (1994). Circadian clock- and phytochrome-regulated transcription is conferred by a 78 bp cis-acting domain of the Arabidopsis CAB2 promoter. Plant J. 6, 457–470. doi: 10.1046/j.1365-313x.1994.6040457.x
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Bauer, C. S., Hoth, S., Haga, K., Philippar, K., Aoki, N., and Hedrich, R. (2000). Differential expression and regulation of K+ channels in the maize coleoptile: molecular and biophysical analysis of cells isolated from cortex and vasculature. Plant J. 24, 139–145. doi: 10.1046/j.1365-313x.2000.00844.x
Becker, D., Hoth, S., Ache, P., Wenkel, S., Roelfsema, M. R. G., Meyerhoff, O., et al. (2003). Regulation of the ABA-sensitive Arabidopsis potassium channel gene GORK in response to water stress. FEBS Lett. 554, 119–126. doi: 10.1016/s0014-5793(03)01118-9
Bennetzen, J. L., Schmutz, J., Wang, H., Percifield, R., Hawkins, J., Pontaroli, A. C., et al. (2012). Reference genome sequence of the model plant setaria. Nat. Biotechnol. 30, 555–561. doi: 10.1038/nbt.2196
Cao, Y., Ward, J. M., Kelly, W. B., Ichida, A. M., Gaber, R. F., Anderson, J. A., et al. (1995). Multiple genes, tissue specificity, and expression-dependent modulation contribute to the functional diversity of potassium channels in Arabidopsis thaliana. Plant Physiol. 109, 1093–1106. doi: 10.1104/pp.109.3.1093
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Ding, Y., Yang, Y., Song, C., Wang, B., Yang, S., et al. (2021). Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 63, 53–78. doi: 10.1111/jipb.13061
Chen, Z., Zhou, M., Newman, I. A., Mendham, N. J., Zhang, G., and Shabala, S. (2007). Potassium and sodium relations in salinised barley tissues as a basis of differential salt tolerance. Funct. Plant Biol. 34, 150–162. doi: 10.1071/FP06237
Danquah, A., de Zelicourt, A., Colcombet, J., and Hirt, H. (2014). The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32, 40–52. doi: 10.1016/j.biotechadv.2013.09.006
Deupi, X., Edwards, P., Singhal, A., Nickle, B., Oprian, D., Schertler, G., et al. (2012). Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc. Natl. Acad. Sci. U. S. A. 109, 119–124. doi: 10.1073/pnas.1114089108
Dreyer, I., and Blatt, M. R. (2009). What makes a gate? The ins and outs of Kv-like K+ channels in plants. Trends Plant Sci. 14, 383–390. doi: 10.1016/j.tplants.2009.04.001
Duby, G., Hosy, E., Fizames, C., Alcon, C., Costa, A., Sentenac, H., et al. (2008). AtKC1, a conditionally targeted shaker-type subunit, regulates the activity of plant K+ channels. Plant J. 53, 115–123. doi: 10.1111/j.1365-313X.2007.03324.x
Engel, A. J., Kithil, M., Langhans, M., Rauh, O., Cartolano, M., Van Etten, J. L., et al. (2021). Codon bias can determine sorting of a potassium channel protein. Cells 10:1128. doi: 10.3390/cells10051128
Fahad, S., Hussain, S., Matloob, A., Ahmed Khan, F., Khaliq, A., Saud, S., et al. (2015). Phytohormones and plant responses to salinity stress: a review. Plant Growth Regul. 75, 391–404.
Feng, C., He, C., Wang, Y., Xu, H., Xu, K., Zhao, Y., et al. (2021). Genome-wide identification of soybean shaker K+ channel gene family and functional characterization of GmAKT1 in transgenic Arabidopsis thaliana under salt and drought stress. J. Plant Physiol. 266:153529. doi: 10.1016/j.jplph.2021.153529
Feng, X., Liu, W., Cao, F., Wang, Y., Zhang, G., Chen, Z.-H., et al. (2020). Overexpression of HvAKT1 improves drought tolerance in barley by regulating root ion homeostasis and ROS and NO signaling. J. Exp. Bot. 71, 6587–6600. doi: 10.1093/jxb/eraa354
Fuchs, I., Stölzle, S., Ivashikina, N., and Hedrich, R. (2005). Rice K+ uptake channel OsAKT1 is sensitive to salt stress. Planta 221, 212–221. doi: 10.1007/s00425-004-1437-9
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Gaber, R. F., Styles, C. A., and Fink, G. R. (1988). TRK1 encodes a plasma membrane protein required for high-affinity potassium transport in Saccharomyces cerevisiae. Mol. Cell. Biol. 8, 2848–2859. doi: 10.1128/mcb.8.7.2848-2859.1988
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Geiger, D., Becker, D., Vosloh, D., Gambale, F., Palme, K., Rehers, M., et al. (2009). Heteromeric AtKC1AKT1 channels in Arabidopsis roots facilitate growth under K+-limiting conditions. J. Biol. Chem. 284, 21288–21295. doi: 10.1074/jbc.M109.017574
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-estrella, L. R., Xu, G., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. Life Sci. 336, 1–40. doi: 10.1007/s11427-020-1683-x
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, 1178–1186. doi: 10.1093/nar/gkr944
Grefen, C. (2014). The split-ubiquitin system for the analysis of three- component interactions. Arabidopsis Protocol. 2014, 659–678. doi: 10.1007/978-1-62703-580-4_34
Grefen, C., and Blatt, M. R. (2012). Do calcineurin B-like proteins interact independently of the serine threonine kinase CIPK23 with the K+ channel AKT1? Lessons learned from a menage a trois. Plant Physiol. 159, 915–919. doi: 10.1104/pp.112.198051
Grefen, C., Chen, Z., Honsbein, A., Donald, N., Hills, A., and Blatt, M. R. (2010). A novel motif essential for SNARE interaction with the K+ channel KC1 and channel gating in Arabidopsis. Plant Cell 22, 3076–3092. doi: 10.1105/tpc.110.077768
Grefen, C., Obrdlik, P., and Harter, K. (2009). The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS). Methods Mol. Biol. 2009, 217–233. doi: 10.1007/978-1-59745-289-2_14
Guerriero, G., Martin, N., Golovko, A., Sundström, J. F., Rask, L., and Ezcurra, I. (2009). The RY/Sph element mediates transcriptional repression of maturation genes from late maturation to early seedling growth. New Phytol. 184, 552–565. doi: 10.1111/j.1469-8137.2009.02977.x
Han, M., Wu, W., Wu, W., and Wang, Y. (2016). Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+ -limited conditions. Mol. Plant 9, 437–446. doi: 10.1016/j.molp.2016.01.012
Hayashi, T., Kobayashi, D., Kariu, T., Tahara, M., Hada, K., Kouzuma, Y., et al. (2003). Genomic cloning of ribonucleases in nicotiana glutinosa leaves, as induced in response to wounding or to TMV-infection, and characterization of their promoters. Biosci. Biotechnol. Biochem. 67, 2574–2583. doi: 10.1271/bbb.67.2574
He, L., Zhang, B., Wang, X., Li, H., and Han, Y. (2015). Foxtail millet: nutritional and eating quality, and prospects for genetic improvement. Front. Agric. Sci. Eng. 2:124–133.
Hobo, T., Asada, M., Kowyama, Y., and Hattori, T. (1999). ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 19, 679–689. doi: 10.1046/j.1365-313x.1999.00565.x
Honsbein, A., Blatt, M. R., and Grefen, C. (2011). A molecular framework for coupling cellular volume and osmotic solute transport control. J. Exp. Bot. 62, 2363–2370. doi: 10.1093/jxb/erq386
Honsbein, A., Sokolovski, S., Grefen, C., Campanoni, P., Pratelli, R., Paneque, M., et al. (2009). A tripartite SNARE-K+ channel complex mediates in channel-dependent K+ nutrition in Arabidopsis. Plant Cell 21, 2859–2877. doi: 10.1105/tpc.109.066118
Horaruang, W., and Zhang, B. (2017). Mating based split-ubiquitin assay for detection of protein interactions. Bio. Protocol. 7, 1–14. doi: 10.21769/BioProtoc.2258
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, 585–587. doi: 10.1093/nar/gkm259
Hosy, E., Vavasseur, A., Mouline, K., Dreyer, I., Gaymard, F., Porée, F., et al. (2003). The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc. Natl. Acad. Sci. U.S.A. 100, 5549–5554. doi: 10.1073/pnas.0733970100
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hwang, H., Yoon, J., Kim, H. Y., Min, M. K., Kim, J. A., Choi, E. H., et al. (2013). Unique features of two potassium channels, OsKAT2 and OsKAT3, expressed in rice guard cells. PLoS One 8:1–14. doi: 10.1371/journal.pone.0072541
Jeanguenin, L., Alcon, C., Duby, G., Boeglin, M., Chérel, I., Gaillard, I., et al. (2011). AtKC1 is a general modulator of Arabidopsis inward shaker channel activity. Plant J. 67, 570–582. doi: 10.1111/j.1365-313X.2011.04617.x
Jiao, Q. S., Niu, G. T., Wang, F. F., Dong, J. Y., Chen, T. S., Zhou, C. F., et al. (2020). N-glycosylation regulates photosynthetic efficiency of arabidopsis thaliana. Photosynthetica 58, 72–79.
Jin, R., Zhang, A., Sun, J., Chen, X., Liu, M., Zhao, P., et al. (2021). Identification of shaker K+ channel family members in sweetpotato and functional exploration of IbAKT1. Gene 768:145311. doi: 10.1016/j.gene.2020.145311
Johansson, I., Wulfetange, K., Porée, F., Michard, E., Gajdanowicz, P., Lacombe, B., et al. (2006). External K+ modulates the activity of the Arabidopsis potassium channel SKOR via an unusual mechanism. Plant J. 46, 269–281. doi: 10.1111/j.1365-313X.2006.02690.x
Karnik, R., Waghmare, S., Zhang, B., Larson, E., Lefoulon, C., Gonzalez, W., et al. (2017). Commandeering channel voltage sensors for secretion, cell turgor, and volume control. Trends Plant Sci. 22, 81–95. doi: 10.1016/j.tplants.2016.10.006
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kwon, C., Lee, J., and Yun, H. S. (2020). SNAREs in plant biotic and abiotic stress responses. Mol. Cells 25, 501–508. doi: 10.14348/molcells.2020.0007
Lebaudy, A., Vavasseur, A., Hosy, E., Dreyer, I., Leonhardt, N., Thibaud, J. B., et al. (2008). Plant adaptation to fluctuating environment and biomass production are strongly dependent on guard cell potassium channels. Proc. Natl. Acad. Sci. U.S.A. 105, 5271–5276. doi: 10.1073/pnas.0709732105
Lebaudy, A., Véry, A. A., and Sentenac, H. (2007). K+ channel activity in plants: genes, regulations and functions. FEBS Lett. 581, 2357–2366. doi: 10.1016/j.febslet.2007.03.058
Lee, S. C., Lan, W.-Z., Kim, B.-G., Li, L., Cheong, Y. H., Pandey, G. K., et al. (2007). A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc. Natl. Acad. Sci. U.S.A. 104, 15959–15964. doi: 10.1073/pnas.0707912104
Lelievre, J. M., Oliveira, L. O., and Nielsen, N. C. (1992). 5’-CATGCAT-3’ elements modulate the expression of glycinin genes. Plant Physiol. 98, 387–391. doi: 10.1104/pp.98.1.387
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, J., Yu, L., Qi, G. N., Li, J., Xu, Z. J., Wu, W. H., et al. (2014). The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 26, 3387–3402. doi: 10.1105/tpc.114.123455
Ma, W., Yang, G., Xiao, Y., Zhao, X., and Wang, J. (2020). ABA-dependent K+ flux is one of the important features of the drought response that distinguishes catalpa from two different habitats. Plant Signal. Behav. 15:1735755. doi: 10.1080/15592324.2020.1735755
Ooi, A., Lemtiri-Chlieh, F., Wong, A., and Gehring, C. (2017). Direct modulation of the guard cell outward-rectifying potassium channel (GORK) by abscisic acid. Mol. Plant 10, 1469–1472. doi: 10.1016/j.molp.2017.08.010
Peng, R., and Zhang, B. (2020). Foxtail millet: a new model for C4 plants. Trends Plant Sci. 26, 199–201. doi: 10.1016/j.tplants.2020.12.003
Philippar, K., Büchsenschütz, K., Edwards, D., Löffler, J., Lüthen, H., Kranz, E., et al. (2006). The auxin-induced K+ channel gene Zmk1 in maize functions in coleoptile growth and is required for embryo development. Plant Mol. Biol. 61, 757–768. doi: 10.1007/s11103-006-0047-2
Philippar, K., Fuchs, I., Lüthen, H., Hoth, S., Bauer, C. S., Haga, K., et al. (1999). Auxin-induced K+ channel expression represents an essential step in coleoptile growth and gravitropism. Proc. Natl. Acad. Sci. U.S.A. 96, 12186–12191. doi: 10.1073/pnas.96.21.12186
Pilot, G., Pratelli, R., Gaymard, F., Meyer, Y., and Sentenac, H. (2003b). Five-group distribution of the shaker-like K+ channel family in higher plants. J. Mol. Evol. 56, 418–434. doi: 10.1007/s00239-002-2413-2
Pilot, G., Gaymard, F., Mouline, K., Chérel, I., and Sentenac, H. (2003a). Regulated expression of Arabidopsis shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol. Biol. 51, 773–787. doi: 10.1023/a:1022597102282
Raddatz, N., Morales de los Ríos, L., Lindahl, M., Quintero, F. J., and Pardo, J. M. (2020). Coordinated transport of nitrate, potassium, and sodium. Front. Plant Sci. 11:1–18. doi: 10.3389/fpls.2020.00247
Sutter, J.-U., Campanoni, P., Tyrrell, M., and Blatt, M. R. (2006). Selective mobility and sensitivity to SNAREs is exhibited by the Arabidopsis KAT1 K+ channel at the plasma membrane. Plant Cell 18, 935–954. doi: 10.1105/tpc.105.038950
Sutter, J. U., Sieben, C., Hartel, A., Eisenach, C., Thiel, G., and Blatt, M. R. (2007). Abscisic acid triggers the endocytosis of the arabidopsis KAT1 K+ channel and its recycling to the plasma membrane. Curr. Biol. 17, 1396–1402. doi: 10.1016/j.cub.2007.07.020
Tan, P., Du, X., Shang, Y., Zhu, K., Joshi, S., Kaur, K., et al. (2022). Ion transporters and their exploration for conferring abiotic stress tolerance in plants. Plant Growth Regul. 96, 1–23.
Thiel, G., MacRobbie, E. A. C., and Blatt, M. R. (1992). Membrane transport in stomatal guard cells: the importance of voltage control. J. Membr. Biol. 126, 1–18. doi: 10.1007/BF00233456
Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2003). Multiple sequence alignment using clustalw and clustalx. Curr. Protoc. Bioinforma. 2003, 1–22. doi: 10.1002/0471250953.bi0203s00
Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T. J. (1997). Creation of a highly active synthetic auxRE. Society 9, 1963–1971.
Véry, A.-A., and Sentenac, H. (2003). Molecular mechanisms and regulation of K+ transport in higher plants. Ann. Rev. Plant Biol. 54, 575–603. doi: 10.1146/annurev.arplant.54.031902.134831
Voorrips, R. E. (2002). Mapchart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Waghmare, S., Lefoulon, C., Zhang, B., Liliekyte, E., Donald, N., and Blatt, M. R. (2019). K+ channel-SEC11 binding exchange regulates SNARE assembly for secretory traffic. Plant Physiol. 181, 1096–1113. doi: 10.1104/pp.19.00919
Wang, H., Hao, D., Wang, X., Zhang, H., Yang, P., Zhang, L., et al. (2021). Genome-wide identification and expression analysis of the SNARE genes in foxtail millet (setaria italica) reveals its roles in drought stress. Plant Growth Regul. 95, 355–369.
Wang, M., Zheng, Q., Shen, Q., and Guo, S. (2013). The critical role of potassium in plant stress response. Int. J. Mol. Sci. 14, 7370–7390. doi: 10.3390/ijms14047370
Wang, Y., Salasini, B. C., Khan, M., Devi, B., Bush, M., Subramaniam, R., et al. (2019). Clade i tgacg-motif binding basic leucine zipper transcription factors mediate blade-on-petiole-dependent regulation of development. Plant Physiol. 180, 937–951. doi: 10.1104/pp.18.00805
Washida, H., Wu, C. Y., Suzuki, A., Yamanouchi, U., Akihama, T., Harada, K., et al. (1999). Identification of cis-regulatory elements required for endosperm expression of the rice storage protein glutelin gene GluB-1. Plant Mol. Biol. 40, 1–12. doi: 10.1023/a:1026459229671
Xia, J., Kong, D., Xue, S., Tian, W., Li, N., Bao, F., et al. (2014). Nitric oxide negatively regulates AKT1-mediated potassium uptake through modulating vitamin B6 homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 16196–16201. doi: 10.1073/pnas.1417473111
Xu, J., Li, H. D., Chen, L. Q., Wang, Y., Liu, L. L., He, L., et al. (2006). A Protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360. doi: 10.1016/j.cell.2006.06.011
Xu, S., Xiao, J., Yin, F., Guo, X., Xing, L., Xu, Y., et al. (2019). The protein modifications of O-GlcNAcylation and phosphorylation mediate vernalization response for flowering in winter wheat. Plant Physiol. 180, 1436–1449. doi: 10.1104/pp.19.00081
Zhang, B., Karnik, R., Alvim, J., Donald, N., and Blatt, M. R. (2019). Dual sites for sec11 on the snare syp121 implicate a binding exchange during secretory traffic. Plant Physiol. 180, 228–239. doi: 10.1104/pp.18.01315
Zhang, B., Karnik, R., Waghmare, S., Donald, N., and Blatt, M. R. (2017). VAMP721 conformations unmask an extended motif for K+ channel binding and gating control. Plant Physiol. 173, 536–551. doi: 10.1104/pp.16.01549
Zhang, B., Karnik, R., Wang, Y., Wallmeroth, N., Blatt, M. R., and Grefen, C. (2015). The Arabidopsis R-SNARE VAMP721 interacts with KAT1 and KC1 K+ channels to moderate K+ current at the plasma membrane. Plant Cell 27, 1697–1717. doi: 10.1105/tpc.15.00305
Zhang, B., Wang, H., and Zhang, Y. (2020). SNARE proteins and their role in plant ion channel regulation. Plant Growth Regul. 92, 443–453.
Zhang, G., Liu, X., Quan, Z., Cheng, S., Xu, X., Pan, S., et al. (2012). Genome sequence of foxtail millet (setaria italica) provides insights into grass evolution and biofuel potential. Nat. Biotechnol. 30, 549–554. doi: 10.1038/nbt.2195
Zhang, H., Xiao, W., Yu, W., Yao, L., Li, L., Wei, J., et al. (2018). Foxtail millet SiHAK1 excites extreme high-affinity K+ uptake to maintain K+ homeostasis under low K+ or salt stress. Plant Cell Rep. 37, 1533–1546. doi: 10.1007/s00299-018-2325-2
Zhang, H., Yin, W., and Xia, X. (2010). Shaker-like potassium channels in populus, regulated by the CBL-CIPK signal transduction pathway, increase tolerance to low-K+ stress. Plant Cell Rep. 29, 1007–1012. doi: 10.1007/s00299-010-0886-9
Zhang, R. X., Qin, L. J., and Zhao, D. G. (2017). Overexpression of the OsIMP gene increases the accumulation of inositol and confers enhanced cold tolerance in tobacco through modulation of the antioxidant enzymes’activities. Genes (Basel). 8:179. doi: 10.3390/genes8070179
Keywords: shaker K+ channel, foxtail millet (Setaria italica), gene regulation, abiotic stress, phytohormone
Citation: Zhang B, Guo Y, Wang H, Wang X, Lv M, Yang P and Zhang L (2022) Identification and Characterization of Shaker K+ Channel Gene Family in Foxtail Millet (Setaria italica) and Their Role in Stress Response. Front. Plant Sci. 13:907635. doi: 10.3389/fpls.2022.907635
Received: 30 March 2022; Accepted: 11 May 2022;
Published: 09 June 2022.
Edited by:
Christopher Peter Grof, The University of Newcastle, AustraliaReviewed by:
Akila Chandra Sekhar, Yogi Vemana University, IndiaCopyright © 2022 Zhang, Guo, Wang, Wang, Lv, Yang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ben Zhang, benzhang@sxu.edu.cn; Lizhen Zhang, lizhen@sxu.edu.cn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.