
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 16 June 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.901128
This article is part of the Research Topic Physiological, Molecular and Genetic Mechanisms of Abiotic Stress Tolerance in Tropical Crops View all 8 articles
 Feifei An1,2†
Feifei An1,2† Xinhui Xiao1†
Xinhui Xiao1† Ting Chen3
Ting Chen3 Jingjing Xue1
Jingjing Xue1 Xiuqin Luo1
Xiuqin Luo1 Wenjun Ou1
Wenjun Ou1 Kaimian Li1
Kaimian Li1 Jie Cai1*
Jie Cai1* Songbi Chen1*
Songbi Chen1*The basic helix-loop-helix (bHLH) proteins are a large superfamily of transcription factors, and play a central role in a wide range of metabolic, physiological, and developmental processes in higher organisms. However, systematic investigation of bHLH gene family in cassava (Manihot esculenta Crantz) has not been reported. In the present study, we performed a genome-wide survey and identified 148 MebHLHs genes were unevenly harbored in 18 chromosomes. Through phylogenetic analyses along with Arabidopsis counterparts, these MebHLHs genes were divided into 19 groups, and each gene contains a similar structure and conserved motifs. Moreover, many cis-acting regulatory elements related to various defense and stress responses showed in MebHLH genes. Interestingly, transcriptome data analyses unveiled 117 MebHLH genes during postharvest physiological deterioration (PPD) process of cassava tuberous roots, while 65 MebHLH genes showed significantly change. Meanwhile, the relative quantitative analysis of 15 MebHLH genes demonstrated that they were sensitive to PPD, suggesting they may involve in PPD process regulation. Cyanogenic glucosides (CGs) biosynthesis during PPD process was increased, silencing of MebHLH72 and MebHLH114 showed that linamarin content was significantly decreased in the leaves. To summarize, the genome-wide identification and expression profiling of MebHLH candidates pave a new avenue for uderstanding their function in PPD and CGs biosynthesis, which will accelerate the improvement of PPD tolerance and decrease CGs content in cassava tuberous roots.
The basic helix-loop-helix (bHLH) proteins are a large superfamily of transcription factors, and widely distributed in plants, fungi and animals (Steidl et al., 2000; Carretero-Paulet et al., 2010). The bHLH domain contains a basic region and a HLH (helix-loop-helix) region, and the N-terminal basic region directly followed by the HLH domain (Sharker et al., 2020). More than 50% of bHLHs in plant possess a conserved HER motif in order to binding and regulating their target genes (Toledo-Ortiz et al., 2003). The basic region, is a DNA-binding region that allows HLH proteins to bind to E-box (CANNTG) (Massari and Murre, 2000). Basic helix-loop-helix transcription factors play crucial roles in plant growth and development including morphogenesis (Gangappa and Kumar, 2017; Ballester et al., 2021), iron homeostasis (Li et al., 2019), flower and fruit development (Sun K. et al., 2019), stomatal initiation (Raissig et al., 2016), root vascular cell proliferation (Ohashi-Ito and Fukuda, 2016), grain yield (Luo et al., 2013), secondary metabolites biosynthesis, such as anthocyanin (Zhao R. et al., 2020; Li et al., 2021), also in the stress tolerance, for instance, drought (Verma et al., 2020; Zhao Q. et al., 2020), salt (Liu et al., 2021; Yu C. et al., 2021), cold (Luo et al., 2020; Shen et al., 2021), heavy metal toxicity (Sun W. et al., 2019), and osmotic stress (Yang et al., 2016). Despite some bHLHs functions have been elaborated, most plant bHLHs functions are still unclear, especially in tuber crops.
Cassava (Manihot esculenta Crantz) is a staple food in the tropics and sub-tropics regions, which has high starch content and strong adaptability (An et al., 2019a). However, cassava roots storage is limited by postharvest physiological deterioration (PPD), it begins in 24 hours and then the roots became unpalatable and unmarketable rapidly (Zainuddin et al., 2017). PPD is a wound response of cassava roots during the harvest time, which is not due to microbial infection (Udogu et al., 2021). Accompanied with PPD occurs, an oxidative burst is initiated, and then following with the accumulation of secondary metabolites (Reilly et al., 2004). The process of PPD involved in reactive oxygen species (ROS), calcium signal transduction, cyanogenic glucosides biosynthesis, starch degradation, phenylpropanoid biosynthesis, N-glycosylation modification and programmed cell death (PCD) (Owiti et al., 2011; Djabou et al., 2017; Qin et al., 2017; An et al., 2019b). Many bHLHs are involved in abiotic stress processes in plants, however, its role in PPD process is still unknown. In the present study, the bHLH genes of cassava (MebHLH) was systematically identified and characterized, also compared with the bHLHs homology of Arabidopsis thaliana. Sequence comparison revealed that the presence and distribution of duplicated genes occurred among MebHLH genes. To identify MebHLH candidate genes associated with PPD resistance, the expression patterns of MebHLH genes were analyzed using transcriptome data of different PPD materials, and then the gene expression profiling of selected MebHLH candidate genes were verified by qRT-PCR. In addition, the cyanogenic glycosides content was detected during PPD process, and the function of MebHLHs in cyanogenic glycosides biosynthesis was confirmed. This study provides comprehensive information about cassava bHLH genes family, as well as a basis for further analysis of novel MebHLH candidate genes, which may be useful for improving PPD resistance and cyanogenic glycosides biosynthesis in cassava.
The whole genome sequences (v. 8.0) of cassava were downloaded from the phytozome database1. And the Hidden Markov Model (HMM) profile of bHLH (PF0001) was retrieved from Pfam (Finn et al., 2016)2. The HMMER program (Johnson et al., 2010) was used to search for bHLH protein in cassava genome, then all the putative proteins were further confirmed by the Pfam and SMART database (Letunic et al., 2021)3. The MW (molecular weight) and pI (theoretical isoelectric point) of these identified bHLH protein were predicted by ExPASy (Artimo et al., 2012)4. Finally, according to their locations on the chromosomes, we named the sequences with complete bHLH domains in order.
The gene structures were performed by the CDS program and DNA sequences of MebHLH genes were visualized using the GSDS2.0 (Hu et al., 2015)5. The locations of these MebHLH genes were determined by querying the cassava genome. Additionally, chromosomal mapping was constructed by MapChart program (v. 2.32) (Voorrips, 2002). In addition, the online software MEME (Bailey et al., 2015)6 was used to identify the conserved motifs among all MebHLH genes, all the parameters were default setting except the number of motifs was set to 10.
The 1,500 bp upstream sequences of transcription start site ATG were extracted as the promoter sequence and screened them using PlantCare (Lescot et al., 2002)7 and PLACE (Higo et al., 1999)8 databases to identify cis-acting regulatory elements.
MEGA X (Kumar et al., 2018)9 was used to multiple sequence alignment analysis of bHLH domain sequences. A phylogenetic tree was constructed with the neighbor-joining method and the parameters were poisson correction, pairwise deletion, and 1,000 bootstrap replicates. Gene duplication analysis mainly adopted two criteria, that is, the aligned sequence length was more than 75% of the long gene, and the aligned region similarity was greater than 75% (Vatansever et al., 2016). Clustal X (V.2.0) program (Thompson et al., 1997) was used to compare the coding sequences of repeated genes, and KaKs was used to calculate the non-synonymous replacement rate (Ka) and synonymous replacement rate (Ks). Calculator package (Zhang et al., 2006) via model averaging. The approximate date of duplication events (million years ago, Mya) was estimated by the formula T D Ks = 2λ × 10–6, on the basis of molecular clock rate of 2.6 × 10–9 substitutions/synonymous site for cassava. Circos program (Krzywinski et al., 2009) was used to illustrate the relationships of duplicated genes. The synteny of bHLH genes between cassava and A. thaliana was detected by MCScanX (Wang et al., 2012).
The tuberous roots of South China 9 (SC9) were harvested after planting for 10 months, and then cultivated in a chamber at 26 °C and a 16/8 h photoperiod (day/night). After 3 days, the tuberous roots with different PPD degree were selected as materials for transcriptome analysis. The degree of PPD was classified into 4 levels, in which No PPD was marked as I and PPD score was 0, extremely slight PPD was marked as II and PPD score was 3.2%, slight PPD was marked as III and PPD score was 10.6%, severe PPD was marked as IV and PPD score was 32.5% (Figure 6A), PPD score was according to Pahmawati et al., 2022. All samples were collected and frozen in liquid nitrogen immediately and stored at −80°C for further analysis. The platform for transcriptome sequencing was Illumina (HiSeq X-Ten). Three biological repeats were implemented. The heat maps were drawn using the heatmap software Mev.
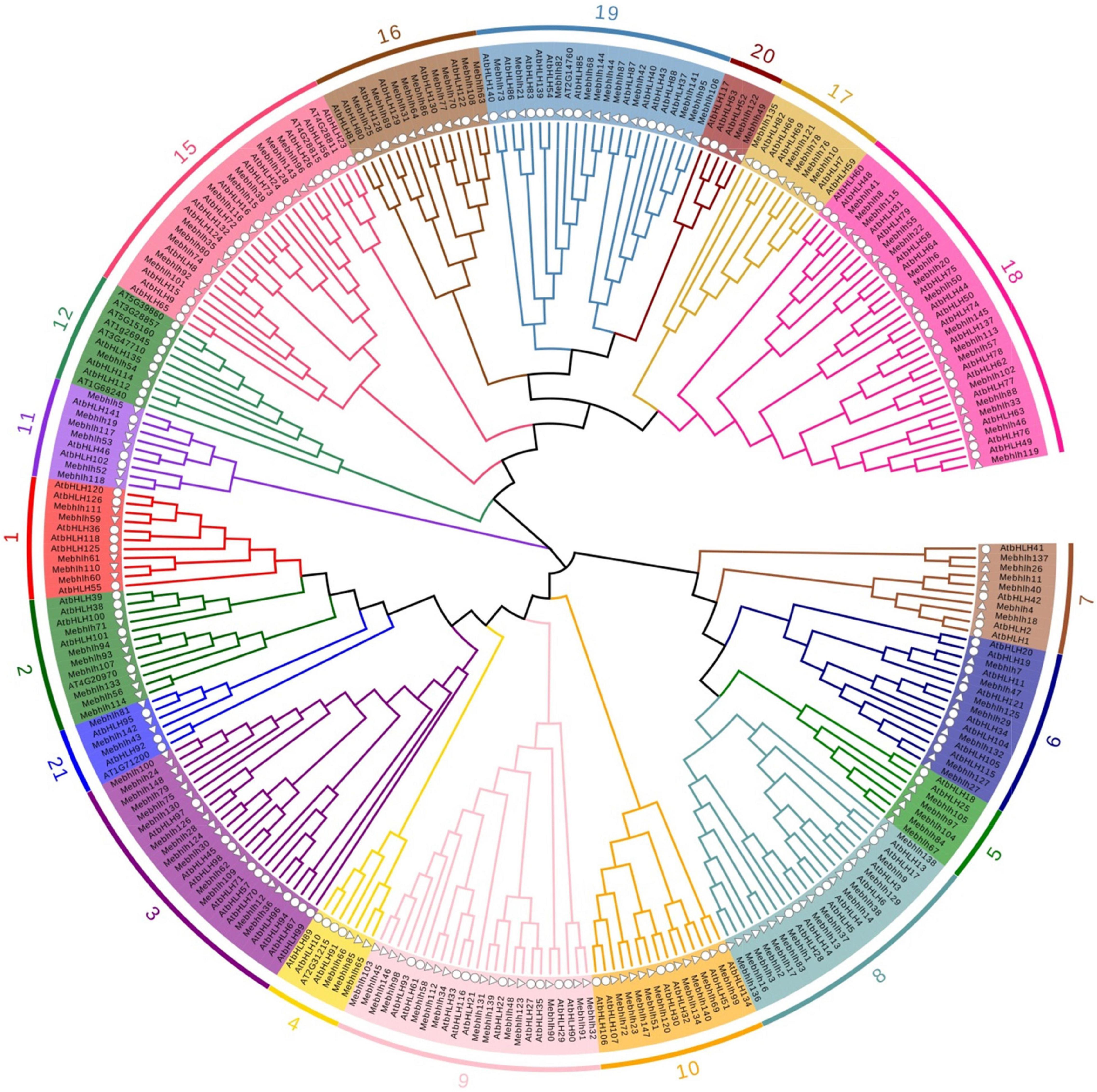
Figure 1. Phylogenetic tree of the 148 MebHLH proteins. Multiple sequence alignment of bHLH domain sequences of M. esculenta and A. thaliana was performed using Clustal W. MEGA X was used to construct the neighbor-joining (NJ) tree with 1,000 bootstrap replicates. Δ indicate bHLHs in M. esculenta, indicate bHLHs in A. thaliana.
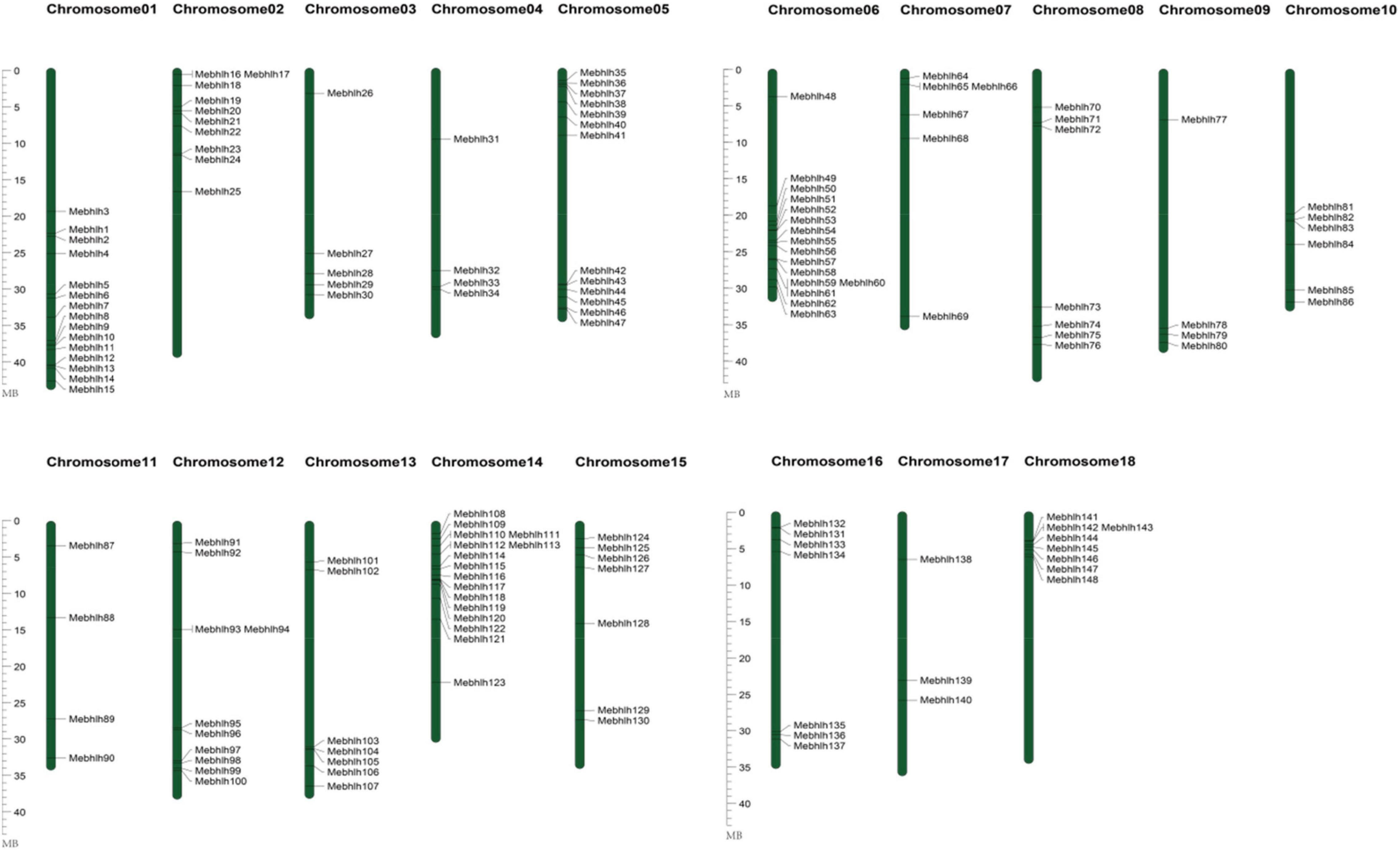
Figure 2. Chromosomal locations of the MebHLH genes. The color gradient provided a scale from 0 to 100% for assessing the magnitude of the parameters. Blue bars denote the Manihot esculenta chromosome. Scale bar on the left indicates the chromosome lengths (Mb).
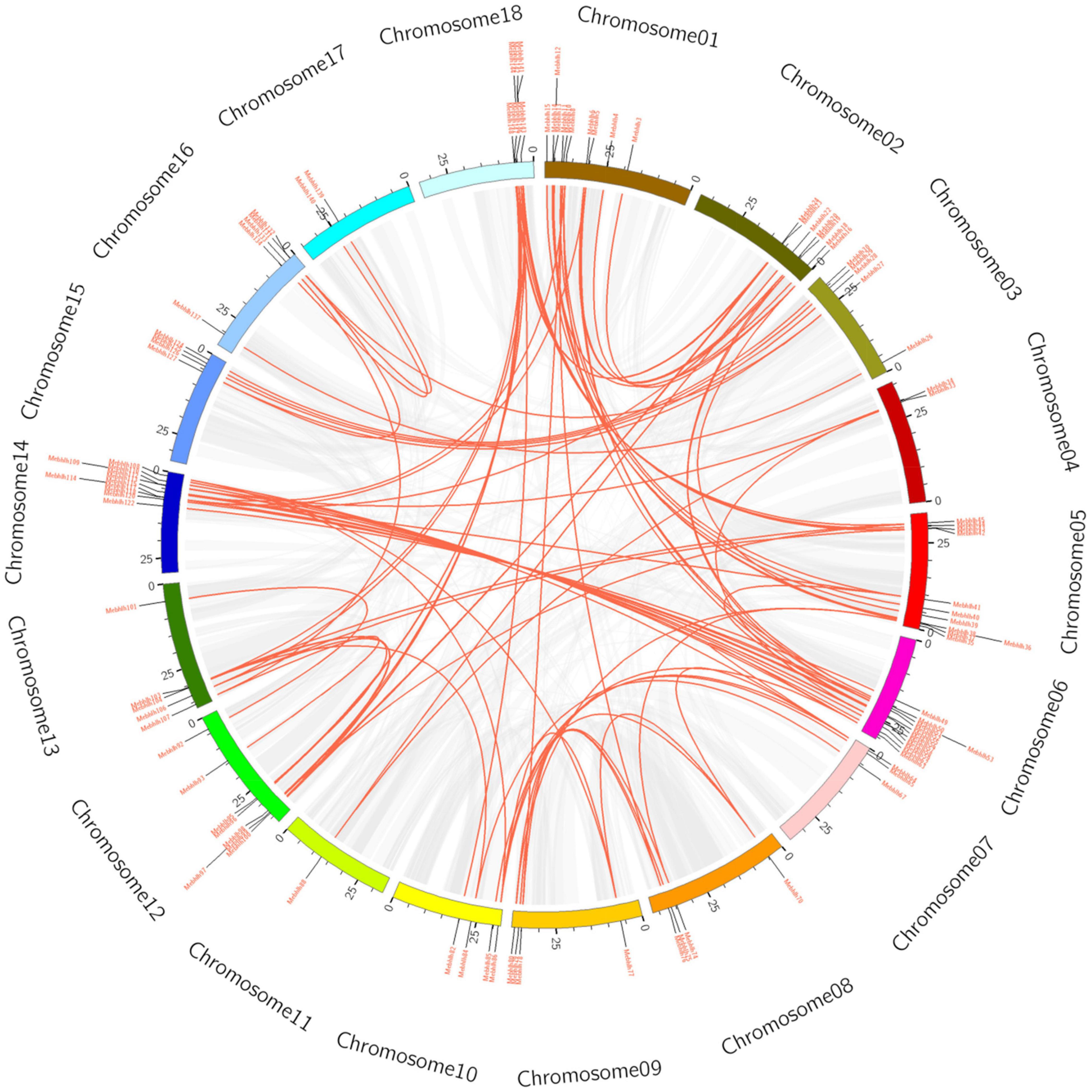
Figure 3. Circos diagram of the MebHLH duplication pairs in M. esculenta. MebHLH duplication pairs are linked with red lines. Scale bar marked on the chromosome indicating chromosome lengths (Mb).
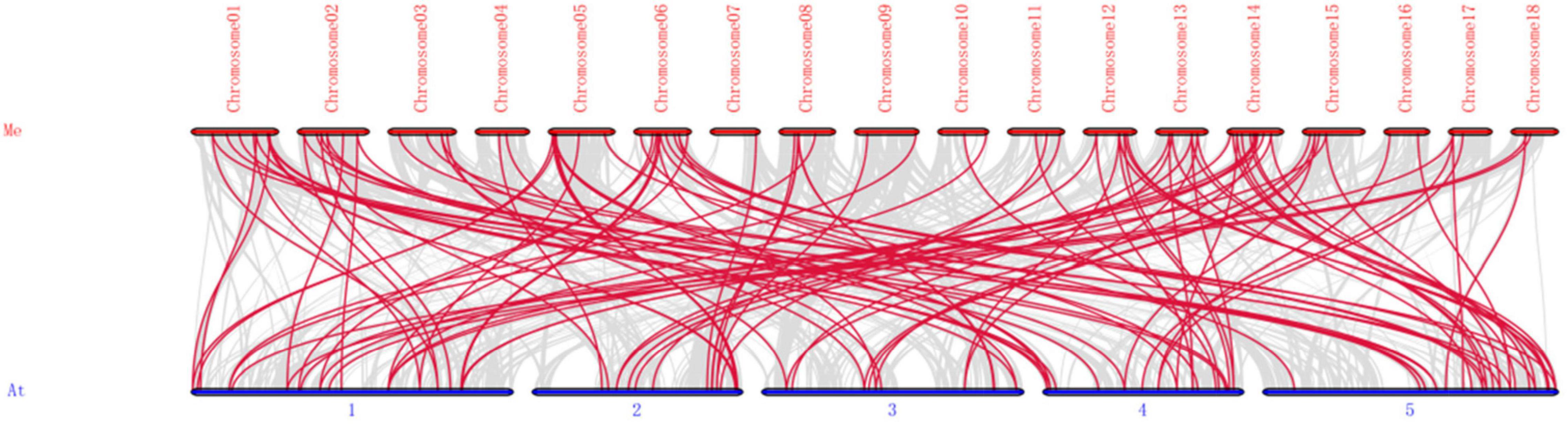
Figure 4. Synteny analysis of bHLH genes between M. esculenta and A. thaliana. Me indicates M. esculenta, At indicates A. thaliana. Gray lines in the background represent the collinear blocks within the genomes of M. esculenta and A. thaliana, while the red lines show the collinear bHLH gene pairs.
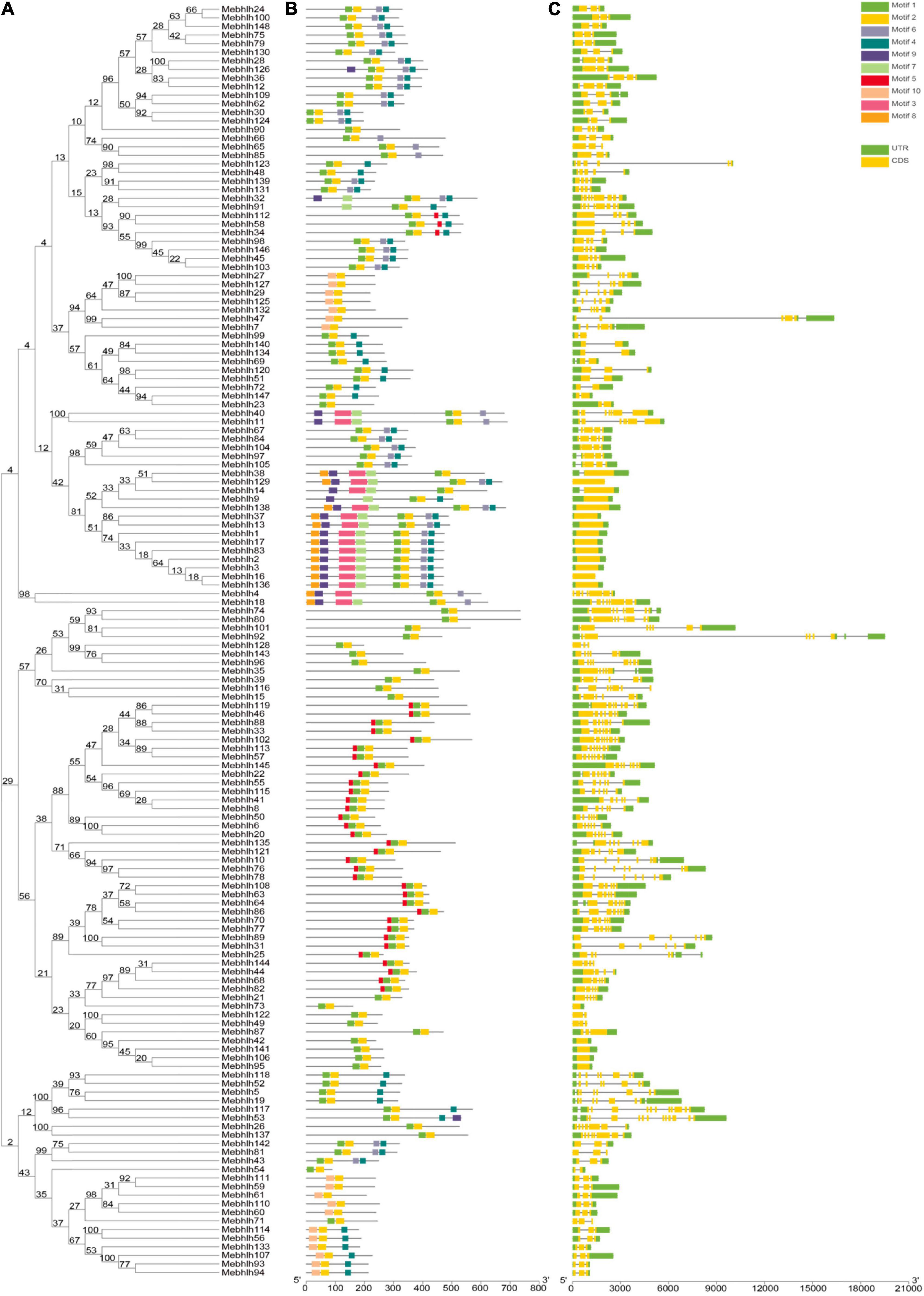
Figure 5. Putative conserved motifs and gene structures of the MebHLH genes. (A) Multiple sequence alignment of bHLH domain sequences of M. esculenta was performed using ClustalW. The neighbor-joining (NJ) tree was constructed using MEGA X with 1,000 bootstrap replicates. (B) Conserved motif. MEME analysis revealed the conserved motifs of the MebHLH proteins. The colored boxes on the right denote 10 motifs. (C) Gene structure. The yellow boxes, black lines, and green boxes represent exon, intron, and UTR (untranslated region), respectively.
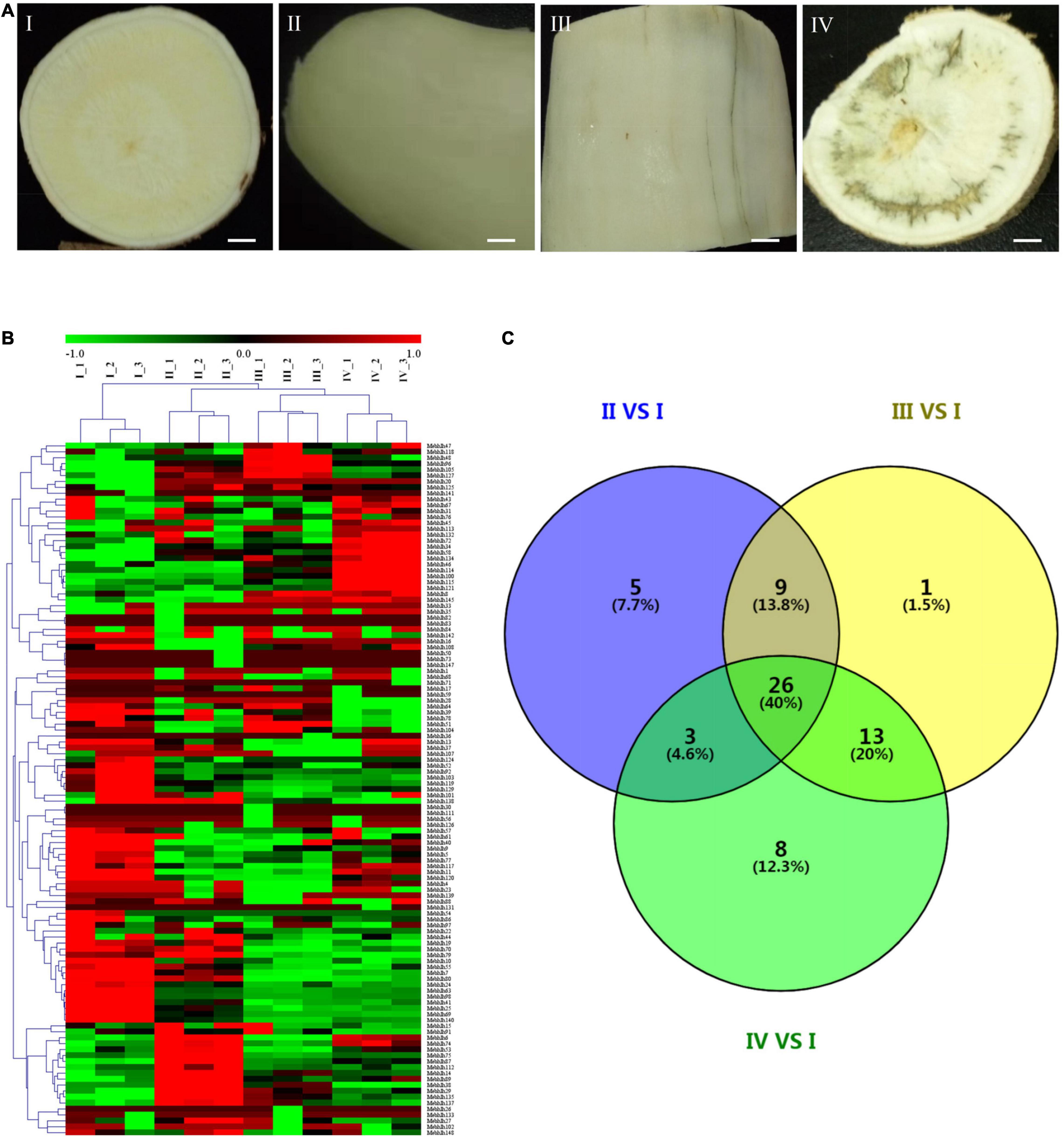
Figure 6. Expression profiles of MebHLH genes in tuberous roots during PPD process. (A) Different PPD degree samples collected from SC9 tuberous roots was classified into 4 levels. I, PPD score was 0, II, PPD score was 3.2%, III, PPD score was 10.6%, IV, PPD score was 32.5%. Bars = 5 mm. (B) The heat map was generated by Mev software. The expression data were gene-wise normalized and hierarchically clustered with average linkage. The bar on the right of the heat map indicates relative expression values. Values 1, 0, -1 represent high, intermediated, and low expression, respectively. (C) Venn diagram of differential expression MebHLHs in three comparison pairs. The sample with 0 PPD score was used as control.
Cyanogenic Glucosides (CGs) including linamarin and lotaustralin in cassava tuberous roots were measured as described in Wang et al., 2022, while CGs in cassava leaves were detected with minor modification. The HPLC-ELSD conditions are as follows: Waters Atlantic C18 column (4.6 mm x 150 mm, 5 μm), The column temperature is 25°C, the binary gradient elution is adopted, and the mobile phase A is 80% acetonitrile/water (V/V) containing 0.1% formic acid (V/V), B is 0.1% (V/V) aqueous formic acid solution. At the beginning, A is 10%, B is 90%, and at 15 min, both A and B are 50%; at 18 min, A is 100%, and maintained for 2 min; at 22 min, A changes 10%, and B% is 90%; balance the column for 5 min, and inject the next sample. The flow rate is 0.6 mL/min; drift tube temperature is 95°C, atomization temperature is 80°C, air flow rate is 2.0 L/min; gain is 3; injection volume is 10 μL. The contents of two CGs in the sample were calculated by external standard method. Linamarin and lotaustralin content (mg/kg) = C * V/1000M. C-the concentration of Linamarin or lotaustralin found in the standard curve, (mg/L); V-the total volume of the extract of the sample, (L); M-the mass of the sample, (kg).
Total RNA was extracted from 200 mg of tuberous roots using a RNAprep Pure Plant plus Kit (Tiangen, China). One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, China) was used for first-strand cDNA synthesis. The qPCR reactions were performed in 10 μL volume in thermocycler (Thermo Fisher Scientific Inc., Göteborg, Sweden). The qPCR primers were shown in Supplementary Table 1. Reference gene of MeActin was used as an internal reference. All experiments of each gene were performed in triplicate per sample. The formula 2–△△Ct method was used to calculate the relative gene expressions.
Full-length MebHLH72 and MebHLH114 were cloned into the transient expression vector pNC-Green-SubN and pNC-Green-SubC to generate 35S::GFP-MebHLH72 and 35S::GFP-MebHLH114 recombinant vector, 35S:: MebHLH72- GFP and 35S:: MebHLH114 - GFP recombinant vector, respectively (Yan et al., 2020). Then, Agrobacterium tumefaciens strain GV3101-psoup was cotransformed with the recombinant plasmid, and transiently expressed in Nicotiana benthamiana leaves. pNC-Green-SubN and pNC-Green-SubC were used as the positive control individually. The results were observed by laser scanning confocal microscope (TCS SP8, Leica) after 3 days.
For vector construction, 300 bp MebHLH72, MebHLH114 and 300 bp MeChlD DNA fragments were cloned into pCsCMV-NC as described by Tuo et al., 2021. The primers are listed in Supplementary Table 2. pCsCMV-ChlD300 as a positive control, which was a yellowing VIGS phenotype in the leaves. The recombinant plasmid co-transformed in A. tumefaciens strain GV3101-psoup, and cultured at 28 °C in chamber before resuspending in 10 mM MES, 10 mM MgCl2 and 20 mM acetosyringone. The preparations were injected into the back of cassava leaves using syringe, and grown in greenhouse (Yan et al., 2021). The expression level of the homolog genes that involved in CGs biosynthesis in MebHLH72 and MebHLH114 silenced lines were also detected, and the qPCR primers were shown in Supplementary Table 1.
Statistical Product and Service Solution program (version 20)(SPSS Inc., Chicago, IL, United States) was used for all statistical analysis. One-way ANOVA (Tukey) was conducted for the expression level and CGs in cassava tuberous roots and leaves comparisons, respectively.
A number of 148 bHLH proteins were characterized from M. esculenta and named them from MebHLH1 to MebHLH148. 148 MebHLHs all contained the bHLH domain (PF0001) based on SMART tests and Pfam analysis. MebHLH proteins lengths are ranged from 89 (MebHLH54) to 735 (MebHLH74) amino acids, MW from 10.06 to 78.27 kDa, and pI from 4.36 to 10.32. Subcellular localization of MebHLHs was predicted by WoLF PSORT10. Among the 148 MebHLH proteins, four were predicted to be cytoplasmic proteins (MebHLH99, 133, 56 and 114), three were located in the chloroplast (MebHLH54, 30 and 44), one was mitochondrial protein (MebHLH124); one was plasma membrane protein (MebHLH66); one was extracellular protein (MebHLH87), and the rest of MebHLHs were localized in the nucleus. More detailed information including bHLH name, gene accession, chromosome locus, protein length, MW, pI of all identified MebHLH proteins were shown in Supplementary Table 3.
The bHLH domain sequences of MebHLH proteins and AtbHLH proteins were used to construct a phylogenetic tree by using neighbor-joining method. The results indicated that the 148 MebHLHs were classified into 19 groups according to the groups in Arabidopsis (Figure 1). Group 18 contained the highest bHLH members with 16 MebHLHs, followed by group 8 and group 9 with 14 MebHLHs. Group 12 had the least bHLH members with only one MebHLH. Additionally, a total of 40 MebHLHs members were clustered in group 2, group 6, group 9, group 12 and group 15, respectively, which play important roles in Arabidopsis. These results suggest that bHLH members in these groups of cassava could have the similar functions with those members in Arabidopsis.
To investigate the chromosomal distribution of the MebHLH genes, the DNA sequence of each MebHLH was obtained using blastn in cassava genome database. 148 MebHLH were mapped on 18 chromosomes (Figure 2). Distribution of MebHLH genes on 18 chromosomes was uneven. For instance, chromosome 06 and 14 contained the highest number (16) of bHLH genes, however, chromosome 11 contained the least number (3) of bHLH genes. Interestingly, many MebHLH genes were clustered in a short distance, such as the top of chromosome 14 and the bottom of chromosome 01 and chromosome 06.
To reveal the expansion mechanism of the MebHLH gene family, blastn and the CDS of all MebHLH genes was performed. Totally, 91 pairs (105 MebHLH genes) of segmental duplications were identified (Supplementary Table 4), seven pairs of tandem duplications (MebHLH16/MebHLH17, MebHLH59/MebHLH60, MebHLH60/MebHLH61, MebHLH65/MebHLH66, MebHLH93/MebHLH94, MebHLH104/MebHLH105 and MebHLH110/MebHLH111) (Figure 3). Base on the Ka and Ks of each duplicated MebHLH gene pair, the Ka/Ks value of each gene pair was also calculated, gene pairs MebHLH60/MebHLH61 (Ka/Ks > 1) may evolve under positive selection after duplication, other six gene pairs were less than 1, which indicated these genes had evolved under purifying selection.
To detect the synteny of bHLH genes, a collinearity analysis between M. esculenta and A. thaliana using MCScanX were performed. As a result, 116 paired collinearity relationships between 83 MebHLH and 78 AtbHLH genes were founded (Figure 4 and Supplementary Table 5).
The exon/intron organization and conserved motifs were analyzed of all MebHLH genes (Figure 5 and Supplementary Figure 1). The exons number was ranged from 1 to 12, although MebHLH1, MebHLH2, MebHLH3, MebHLH9, MebHLH13, MebHLH14, MebHLH16, MebHLH17, MebHLH37, MebHLH38, MebHLH42, MebHLH73, MebHLH83, MebHLH95, MebHLH106, MebHLH129, MebHLH136, MebHLH138, MebHLH141 contained only one exon, MebHLH117 contained 11 exons, and MebHLH53 contained 12 exons, which is the highest number of all genes. Additionally, MebHLH genes in same group had similar gene structure.
Ten conserved motifs were identified among the 148 MebHLH proteins. Notably, the members with high similarity in the same group shared a common motif composition. For instance, MebHLH24 and MebHLH100 were found to contain four motifs. It indicates that these two genes may have a similar function. MebHLH proteins (128/148, 86.49%) contain 2-4 conserved motifs. However, 11 MebHLH proteins (MebHLH129, MebHLH138, MebHLH37, MebHLH13, MebHLH1, MebHLH17, MebHLH83, MebHLH2, MebHLH3, MebHLH16, MebHLH136) contain eight motif and 5 MebHLH proteins (MebHLH32, MebHLH40, MebHLH11, MebHLH38 and MebHLH4) contain six motifs, 3 MebHLH proteins (MebHLH126, MebHLH14 and MebHLH9) contain five motifs, only MebHLH18 contain one seven motifs.
PlantCARE and PLACE database were used to characterize the cis-acting regulatory elements of each MebHLH promoter within 1,500 bp from transcription start site. Many kinds of cis-acting regulatory elements were existed in MebHLH (Supplementary Figure 2 and Supplementary Table 6). For instance, the common regulatory elements such as CAAT-box and TATA-box were presented in all 148 MebHLH genes. Furthermore, cis-acting regulatory elements related to hormone responses were also identified. Such as AuxRR-core and TGA-element, MYB which involved in abscisic acid (ABA) responsiveness; P-box and TATC-box, gibberellin responsiveness; CGTCA-motif and ERE, MeJA responsive elements. Interestingly, the promoter of some MebHLH genes also contained elements related to biotic and abiotic responses, including wounding (WUN-motif), cold (DRE and LTR), drought (MBS), pathogen defense (TC-rich repeat). In summary, MebHLH genes could be regulated by diverse hormone and environment.
The expression levels of MebHLHs in tuberous roots during PPD were analyzed using RNA-seq data. The heatmap showed that compared with no PPD control, 117 (79.05%) MebHLHs were exhibited differences in their expression levels in response to PPD (Figure 6B). 43 MebHLHs showed significant differences in comparison with group II/I, 49 MebHLHs showed significant differences in comparison with group III/I, and 50 MebHLHs in comparison with group IV/I (Supplementary Table 7). Among them, 26 MebHLHs exhibited in all three comparison groups, 5 MebHLHs exhibited only in comparison with group II/I, 8 MebHLHs exhibited in comparison with group IV/I, and only MebHL68 exhibited in comparison with group III/I (Figure 6C).
CGs biosynthesis was associated with PPD, therefore the CGs content including linamarin and lotaustralin in tuberous roots during PPD were also detected. It showed that linamarin content in sample C was significantly increased more than other samples, while lotaustralin was not detected in all samples (Table 1).
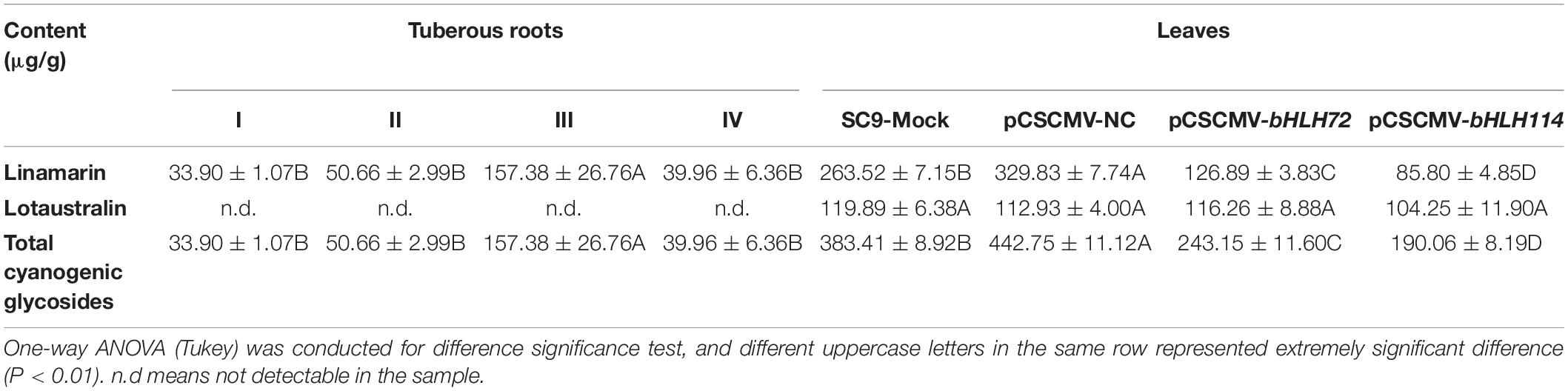
Table 1. Cyanogenic glucosides content of tuberous roots in four PPD samples and cassava leaves infected with pCsCMV-bHLH72 and pCsCMV-bHLH114.
Based on the RNA-seq data, we identified 15 up-regulated genes during PPD, and quantitative real-time PCR (qRT-PCR) for the 15 candidate MebHLHs in four stages were performed. It showed that the expression of 12 MebHLHs were rise remarkably accompanied with PPD process, and they gradually decreased in the flowing stage (Figure 7), these results showed no significant difference with RNA-seq, where they were up-regulated compared with no PPD control. However, the expression levels of MebHLH86, MebHLH89 in subfamily 16 and MebHLH115 in subfamily 18 were less than no PPD control, it was not consistent with the RNA-seq results, that may be caused by different methods.
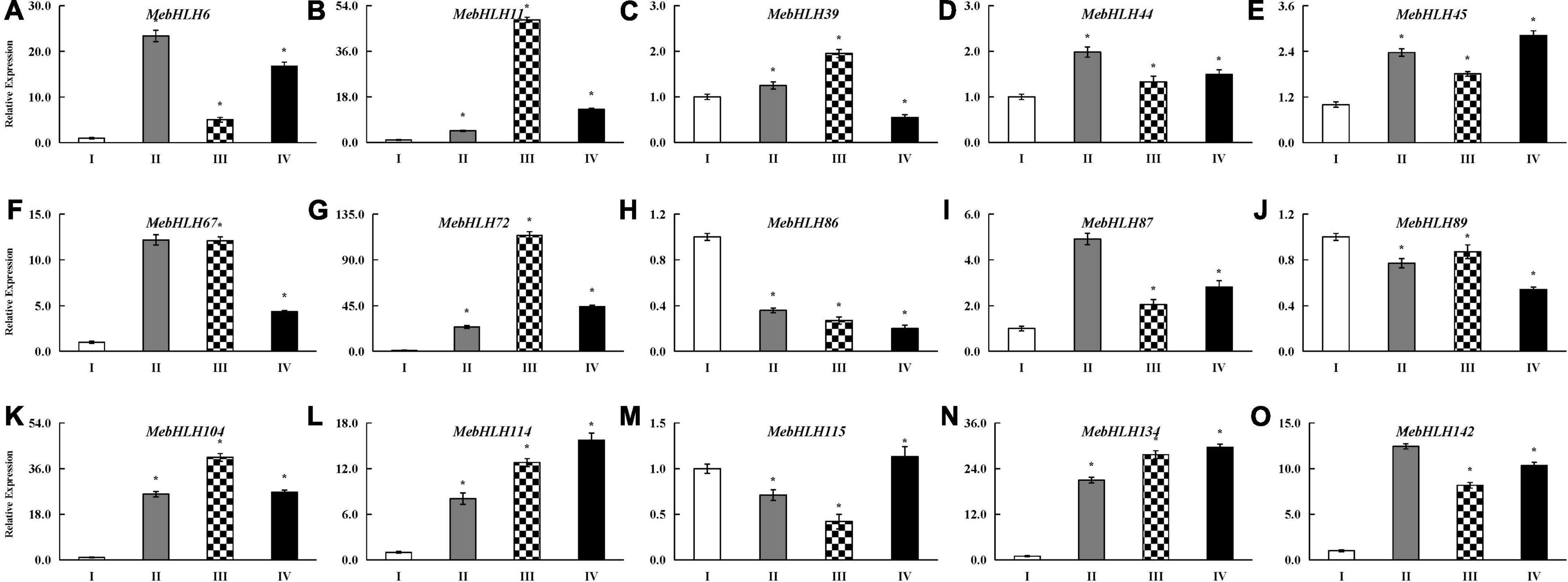
Figure 7. qRT-PCR results for MebHLHs under PPD process. qRT-PCR was used to analyze the expression profiles of 15 up-regulated MebHLH genes according to RNA-seq. PPD degrees I-IV were described in Figure 6A, (A–O) indicates the 15 MebHLH genes tested, respectively. The relative expression levels are normalized to MeActin. The data represent the mean of three biological replicates. The x-axis represents four materials from different PPD stage. The y-axis represents the relative expression levels of MebHLHs. All data are the Means ± SE of three independent experiments. Error bars indicate the standard deviation.
We investigated the subcellular localization of MebHLH72 and MebHLH114 proteins in planta. Consistent with a previous study, GFP-fused MebHLH72 protein was localized in nucleus, and GFP-fused MebHLH114 protein was localized in cytoplasm even in tobacco leaves (Figure 8A), indicating their similar localization with prediction in Supplementary Table 3. To study the in vivo roles of MebHLHs in cassava CGs biosynthesis, two gene-silenced cassava lines were constructed (pCsCMV-bHLH72 where MebHLH72 was silenced, and pCsCMV-bHLH114 where MebHLH114 was silenced), the phenotypes between MebHLHs-silenced lines and control plants were shown in Figure 8B. When the transcript levels of MebHLH72 and MebHLH114 were evaluated in MebHLH-silenced lines, the transcripts level of targeted MebHLHs were drastically reduced compared with pCsCMV-NC and Mock (Figure 8C). We compared the CGs content including linamarin and lotaustralin of MebHLHs-silenced lines in leaves with that of control plants, the linamarin content was significantly decreased in the leaves of both MebHLH72- and MebHLH114-silenced. However, the lotaustralin content was no significantly differences in leaves of MebHLH-silenced lines compare with that of control plants (Table 1). In addition, the expression level of the homolog genes that involved in CGs biosynthesis, MeCYP79D1, MeCYP79D2 and MeCYP71E were also detected in MebHLH72 and MebHLH114 silenced lines (Figure 8D). And the relative expression of all three genes was significantly reduced in two silenced lines. Therefore, MebHLH72 and MebHLH114 may be candidate genes involved in CGs biosynthesis.
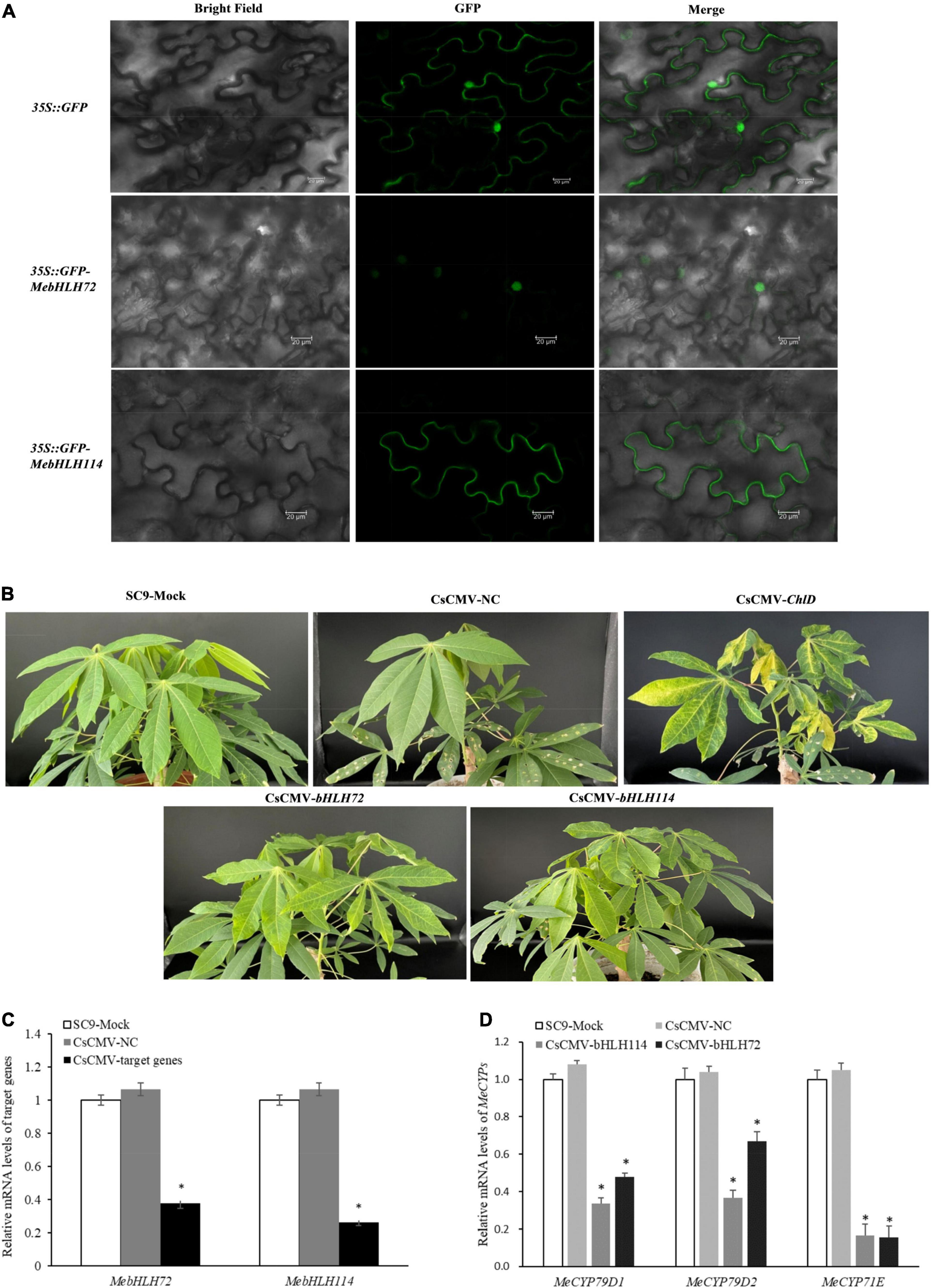
Figure 8. Subcellular localization and Silencing of MebHLH72, MebHLH114 using the pCsCMV-bHLH72 and pCsCMV-bHLH114. (A) Subcellular localization of MebHLH72 and MebHLH114 in tobacco leaves. Bars = 20 μm. (B) Silencing phenotypes in M. esculenta using the pCsCMV-bHLH72 and pCsCMV-bHLH114 at 40 dpi. SC9-Mock means no infected plant, pCsCMV-NC means infected with non-target control CsCMV-NC. (C) qRT-PCR analyses of MebHLH72 and MebHLH114 mRNA expression in M. esculenta infected with pCsCMV-bHLH72 and pCsCMV-bHLH114. The no infected plant was used as control. Three independent experiments were performed and each included six plants per treatment group. Error bars indicate the standard deviation. (D) qRT-PCR analyses of MeCYP79D1, MeCYP79D2 and MeCYP71E mRNA expression in M. esculenta infected with pCsCMV-bHLH72 and pCsCMV-bHLH114. The no infected plant was used as the control. Three independent experiments were performed and each included six plants per treatment group. Error bars indicate the standard deviation.
bHLHs, one of the largest transcription factor families, have been investigated in various plants species. In this study, 148 bHLH gene family members were identified in cassava genome, 136 bHLH proteins in Arabidopsis were used to construct phylogenetic tree, As an illustration of the size of the bHLHs family in plants, 164 bHLHs have been found in Arabidopsis (Carretero-Paulet et al., 2010), while there are 180 bHLHs in Rice (Xiong et al., 2005), 225 bHLHs in Wheat (Guo and Wang, 2017), 190 bHLHs in Tobacco (Rushton et al., 2008), 191 bHLHs in Grapes (Jaillon et al., 2007), 102 bHLHs in Walnut (Zhao et al., 2021), 159 bHLHs in Tomato (Sun et al., 2015). Figure 2 showed that 148 MebHLH genes were unevenly distributed on 18 chromosomes, and some MebHLH genes were clustered on top or bottom of chromosomes. Based on phylogenetic analysis, 148 bHLHs were classified into 19 subfamilies according to the subfamilies in Arabidopsis (Figure 1; Toledo-Ortiz et al., 2003), which was probably due to the methods and sequences were adopted (Carretero-Paulet et al., 2010; Song et al., 2013).
Gene duplication plays an essential role in genome amplification, species evolution and gene family evolution (Lynch and Conery, 2000). Gene duplication is including three main types: whole genome duplication, tandem duplication, and segmental duplication (Yu et al., 2005). In this study, 91 pairs of segmental duplications were identified (Supplementary Table 4), 7 pairs of tandem duplications in cassava (Figure 3). Tandem duplicated genes were less common relatively, this was consistent with results in barley and tobacco (Ke et al., 2020; Bano et al., 2021). These results implied that segmental duplication is the main style in cassava genome. Duplicated genes may exhibit functional diversity, and the mechanisms could be identified by sequencing. Only one pair of tandem duplicated genes (MebHLH60/MebHLH61) was uncovered in cassava with Ka/Ks > 1, which suggest that positive selection was occurred on MebHLH60/MebHLH61.
Cis-acting elements were analyzed in this study and a large number of hormone response elements were founded (AuxRR-core and TGA-element, ABRE, CGTCA-motif, TGA-element), stress response elements (WUN-motif, LTR, ARE, TC-rich repeat and MBS) (Supplementary Figure 2 and Supplementary Table 6). It implied that MebHLH genes may be involved in various stress responses and could be induced by many hormones. For example, exogenous MeJA could enhance the activity of bHLH promoters (Chen et al., 2019; Yu Z. et al., 2021), and then participating in the regulation of plant physiological processes.
As one of the most numerous TFs in plant, the bHLH family has many members and various functions. bHLHs can regulate plant resistances to diverse stresses including cold, drought, salt and wound (Guo et al., 2021; Qian et al., 2021). Postharvest physiological deterioration is one of the major constraints to commercial production and utilization of cassava (Saravanan et al., 2016), it was induced by wounding, which initiates from wound sites and triggers an oxidative burst of the superoxide radical (O2–) within 15 min, and further production of ROS (Zidenga et al., 2012). This was supported by earlier transcriptome data, 20% of the genes were involved in ROS turnover during PPD process (Reilly et al., 2007), and overexpression of superoxide dismutase (SOD) and catalase (CAT) would delayed PPD occurrence of cassava tuberous roots (Xu et al., 2013). As well, many previous reports showed that flavonoids synthesized in plant could effectively eliminate ROS to enhance the tolerance in adverse environment (Wang et al., 2016; Qin et al., 2020). Interestingly, some bHLHs were confirmed to be associated with flavonoids synthesis (Zhao et al., 2019; Aslam et al., 2020) and belongs to subfamilies 2 (Guan et al., 2013; Geng and Liu, 2018; Geng et al., 2019; Waseem et al., 2019; Zhao R. et al., 2020). Especially, experimental data showed that bHLHs bound to the promoter of antioxidant genes such as POD, SOD through E-box or G-box (Liu et al., 2013; Geng and Liu, 2018). Most bHLHs in subfamilies 1, 2, 5 and 15 could bind the E-box and the G-box (Yao et al., 2018; Xi et al., 2021). In our study, the majority of PPD responsive MebHLHs are from subfamilies 2, 5, 10, 15, 16 and 18. And the expression profile of some genes related to flavonoids synthesis and ROS scavenging were significantly difference in the RNA-seq data, such as MeANR, MeF3H, MeANS associated with flavonoids synthesis, and MePOD, MeCAT, MeAPX involved in ROS scavenging (Supplementary Figure 3A). Furthermore, all these genes have G-box in their promoters (Supplementary Figure 3B). qRT-PCR results of 15 up-regulated MebHLHs during PPD showed that MebHLH114 in subfamilies 2, MebHLH67 and MebHLH104 in subfamilies 5, MebHLH72 and MebHLH134 in subfamilies 10 were up-regulated compared with no PPD control, it was consistent with the RNA-seq results. Given all of that, it is reasonable to hypothesize that bHLHs in subfamilies 2, 5 and 15 could bind to E-box or G-box of target genes related with ROS scavenging to further regulate the PPD process. However, these hypotheses still need to be verified.
CGs play an important role in plant defense response, and its correlation with PPD has been confirmed (Reilly et al., 2007). The linamarin content increased in cassava tuberous roots of sample III (Table 1), and linamarase protein were accumulated at four days after PPD initiation (Owiti et al., 2011). The increase of linamarin content could be due to the up-regulation of the genes CYP79D1, CYP79D2, and CYP71E involved in CGs biosynthesis (Jørgensen et al., 2011). LjbHLH7 can directly activate the expression of CYP79D3 gene by binding to the G-box sequence of its promoter (Chen et al., 2022). In our study, the linamarin content significantly decreased in the leaves when MebHLH72 and MebHLH114 were silenced individually. The relative expression of CYP79D1, CYP79D2, and CYP71E were decreased in MebHLH72 and MebHLH114 silenced lines (Figure 8D), and all three genes have two G-box in their promoters (Supplementary Figure 3C). It means that MebHLH72 and MebHLH114 were involved in CGs biosynthesis, probably through combing with G-box of gene promoter which participates in CGs biosynthesis. As transcript factors, most of them were localized in nucleus, however, MebHLH114 (fusion with n/c-GFP) was localized in cytoplasm in this study (Figure 8A and Supplementary Figure 4). Previous report indicated that subcellular localization of AtbHLH039 was also localized at the cell periphery and only a rather weak presence in the nucleus (Trofimov et al., 2019). Recent research suggested that the transport factors will be altered the subcellular localization when environment changed or post translational modification occurred (Wang et al., 2021). Therefore, we inferred that the subcellular localization of MebHLH114 may be changed during the PPD process.
In this study, a genome-wide analysis of MebHLH gene family was characterized with particular focus on their response to cassava PPD and CGs biosynthesis. A total of 148 MebHLH genes were identified and characterized. The chromosomal distribution, gene structure, gene duplication, cis-elements, covered motif and expression profiles of MebHLHs were analyzed. Moreover, transcriptome data analyses unveiled that 65 MebHLH genes may play a crucial roles in PPD of tuberous root. During PPD, CGs biosynthesis was increased, linamarin content was significantly decreased in the leaves of cassava plants with silenced MebHLH72 and MebHLH114. Overall, the results will provide a new insight for cassava engineering programs, including improved PPD tolerance and decreased CGs content.
The datasets presented in this study can be found in online repositories. The names of the repository and accession numbers can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA841274.
FA: conceptualization and original draft preparation. XX: software, review and editing, and visualization. TC: methodology and project administration. JX: validation form analysis. XL: data curation. WO: investigation. KL: resources. SC and JC: modification and supervision. WO, KL, and SC: funding acquisition. All authors have agreed to the published version of the manuscript.
This work was financially supported by the National Key Research and Development Program of China (No. 2019YFD1000500), Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No. 1630032022007), Key Research and Development Program of Hainan (No. ZDYF2020101), and earmarked fund for China Agriculture Research System (CARS-11-HNLKM).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.901128/full#supplementary-material
An, F., Chen, T., Li, Q. X., Qiao, J., Zhang, Z., Carvalho, L. J. C. B., et al. (2019a). Protein cross-interactions for efficient photosynthesis in the cassava cultivar SC205 relative to its wild species. J. Agric. Food Chem. 32, 8746–8755. doi: 10.1021/acs.jafc.9b00046
An, F., Baker, M. R., Qin, Y., Chen, S., and Li, Q. X. (2019b). Relevance of Class I α−Mannosidases to cassava postharvest physiological deterioration. ACS Omega 4, 8739–8746. doi: 10.1021/acsomega.8b03558
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Aslam, M. Z., Lin, X., Li, X., Yang, N., and Chen, L. (2020). Molecular cloning and functional characterization of CpMYC2 and CpBHLH13 transcription factors from Wintersweet (Chimonanthus praecox L.). Plants 9:785. doi: 10.3390/plants9060785
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Ballester, P., Martinez-Godoy, M. A., Ezquerro, M., Navarrete-Gomez, M., Trigueros, M., Rodriguez-Concepcion, M., et al. (2021). A transcriptional complex of NGATHA and bHLH transcription factors directs stigma development in Arabidopsis. Plant Cell 33, 3645–3657. doi: 10.1093/plcell/koab236
Bano, N., Patel, P., Chakrabarty, D., and Bag, S. K. (2021). Genome-wide identification, phylogeny, and expression analysis of the bHLH gene family in tobacco (Nicotiana tabacum). Physiol. Mol. Biol. Plants 27, 1747–1764. doi: 10.1007/s12298-021-01042-x
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martinez-Garcia, J. F., Bilbao-Castro, J. R., and Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Chen, C., Liu, F., Zhang, K., Niu, X., Zhao, H., Liu, Q., et al. (2022). MeJA-responsive bHLH transcription factor LjbHLH7 regulates cyanogenic glucoside biosynthesis in Lotus japonicus. J. Exp. Bot. 73, 2650–2665. doi: 10.1093/jxb/erac026
Chen, Y., Wang, Y., Lyu, P., Chen, L., Shen, C., and Sun, C. (2019). Comparative transcriptomic analysis reveal the regulation mechanism underlying MeJA-induced accumulation of alkaloids in Dendrobium officinale. J. Plant Res. 132, 419–429. doi: 10.1007/s10265-019-01099-6
Djabou, A. S. M., Carvalho, L. J. C. B., Li, Q. X., Niemenak, N., and Chen, S. (2017). Cassava postharvest physiological deterioration: a complex phenomenon involving calcium signaling, reactive oxygen species and programmed cell death. Acta Physiol. Plant 39:91. doi: 10.1007/s11738-017-2382-0
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acid Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Gangappa, S. N., and Kumar, S. V. (2017). DET1 and HY5 control PIF4-mediated thermosensory elongation growth through distinct mechanisms. Cell Rep. 18, 344–351. doi: 10.1016/j.celrep.2016.12.046
Geng, J., and Liu, J. H. (2018). The transcription factor CsbHLH18 of sweet orange functions in modulation of cold tolerance and homeostasis of reactive oxygen species by regulating the antioxidant gene. J Exp. Bot. 69, 2677–2692. doi: 10.1093/jxb/ery065
Geng, J., Wei, T., Wang, Y., Huang, X., and Liu, J. H. (2019). Overexpression of PtrbHLH, a basic helix-loop-helix transcription factor from Poncirus trifoliata, confers enhanced cold tolerance in pummelo (Citrus grandis) by modulation of H2O2 level via regulating a CAT gene. Tree Physiol. 39, 2045–2054. doi: 10.1093/treephys/tpz081
Guan, Q., Wu, J., Yue, X., Zhang, Y., and Zhu, J. (2013). A Nuclear calcium-sensing pathway is critical for gene regulation and salt stress tolerance in Arabidopsis. PLoS Genet. 9:e1003755. doi: 10.1371/journal.pgen.1003755
Guo, J., Sun, B., He, H., Zhang, Y., Tian, H., and Wang, B. (2021). Current understanding of bHLH transcription factors in plant abiotic stress tolerance. Int. J. Mol. Sci. 22:4921. doi: 10.3390/ijms22094921
Guo, X., and Wang, J. (2017). Global identification, structural analysis and expression characterization of bHLH transcription factors in wheat. BMC Plant Bio. 17:90. doi: 10.1186/s12870-017-1038-y
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hu, B., Jin, J., Guo, A., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jaillon, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., and Casagrande, A. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Johnson, L. S., Eddy, S. R., and Portugaly, E. (2010). Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatic 11:431. doi: 10.1186/1471-2105-11-431
Jørgensen, K., Morant, A. V., Morant, M., Jensen, N. B., Olsen, C. E., Kannangara, R., et al. (2011). Biosynthesis of the cyanogenic glucosides linamarin and lotaustralin in cassava: isolation, biochemical characterization, and expression pattern of CYP71E7, the oxime-metabolizing cytochrome P450 enzyme. Plant Physiol. 155, 282–292. doi: 10.1104/pp.110.164053
Ke, Q., Tao, W., Li, T., Pan, W., Chen, X., Wu, X., et al. (2020). Genome-wide Identification, evolution and expression analysis of Basic Helix-loop-helix (bHLH) gene family in Barley (Hordeum vulgare L.). Curr. Genomics 21, 624–644. doi: 10.2174/1389202921999201102165537
Krzywinski, M., Schein, J. E., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, M., Sun, L., Gu, H., Cheng, D., Guo, X., Chen, R., et al. (2021). Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in spine grapes (Vitis davidii). Sci. Rep. 11:6863. doi: 10.1038/s41598-021-85754-w
Li, Y., Sui, X., Yang, J., Xiang, X., and Liu, D. (2019). A novel bHLH transcription factor, NtbHLH1, modulates iron homeostasis in tobacco (Nicotiana tabacum L.). Biochem. Bioph. Res. Commun. 522, 233–239. doi: 10.1016/j.bbrc.2019.11.063
Liu, R., Cui, B., Jia, T., and Song, J. (2021). Role of Suaeda salsa SsNRT2.1 in nitrate uptake under low nitrate and high saline conditions. Plant Physiol. Biochem. 159, 171–178. doi: 10.1016/j.plaphy.2020.12.024
Liu, W. W., Tai, H. H., Li, S. S., Gao, W., Zhao, M., and Xie, C. X. (2013). bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 201, 1192–1204. doi: 10.1111/nph.12607
Luo, J., Liu, H., Zhou, T., Gu, B., Huang, X., Shangguan, Y., et al. (2013). An-1 encodes a basic Helix-loop-Helix protein that regulates awn development, grain size, and grain number in Rice. Plant Cell 25, 3360–3376. doi: 10.1105/tpc.113.113589
Luo, P., Li, Z., Chen, W., Xing, W., Yang, J., and Cui, Y. (2020). Overexpression of RmICE1, a bHLH transcription factor from Rosa multiflora, enhances cold tolerance via modulating ROS levels and activating the expression of stress-responsive genes. Environ. Exp. Bot. 178:104160. doi: 10.1016/j.envexpbot.2020.104160
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Massari, M. E., and Murre, C. (2000). Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell Biol. 20, 429–440. doi: 10.1128/MCB.20.2.429-440.2000
Ohashi-Ito, K., and Fukuda, H. (2016). Functional mechanism of bHLH complexes during early vascular development. Curr. Opin. Plant Biol. 33, 42–47. doi: 10.1016/j.pbi.2016.06.003
Owiti, J., Grossmann, J., Gehrig, P., Dessimoz, C., Laloi, C., Hansen, M. B., et al. (2011). iTRAQ-based analysis of changes in the cassava root proteome reveals pathways associated with post-harvest physiological deterioration. Plant J. Cell Mol. Biol. 67, 145–156. doi: 10.1111/j.1365-313X.2011.04582.x
Pahmawati, R. S., Khumaida, N., Ardie, S. W., Sukma, D., and Arsono, S. (2022). Effects of harvest period, storage, and genotype on postharvest physiological deterioration responses in cassava. Biodiversitas 23, 100–109. doi: 10.13057/biodiv/d230113
Qian, Y., Zhang, T., Yu, Y., Gou, L., Yang, J., Xu, J., et al. (2021). Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses. Front. Plant Sci. 12:677611. doi: 10.3389/fpls.2021.677611
Qin, L., Sun, L., Wei, L., Yuan, J., Kong, F., and Zhang, Y. (2020). Maize SRO1e represses anthocyanin synthesis via regulating MBW complex in response to abiotic stress. Plant J. 105, 1010–1025. doi: 10.1111/tpj.15083
Qin, Y., Djabou, A. S. M., An, F., Li, K., Li, Z., Yang, L., et al. (2017). Proteomic analysis of injured storage roots in cassava (Manihot esculenta Crantz) under postharvest physiological deterioration. PLoS One 12:e0174238. doi: 10.1371/journal.pone.0174238
Raissig, M. T., Abrash, E., Bettadapur, A., Vogel, J. P., and Bergmann, D. C. (2016). Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc. Natl. Acad. Sci. U.S.A. 113, 8326–8331. doi: 10.1073/pnas.1606728113
Reilly, K., Bernal, D., Cortés, D. F., Gómez-Vásquez, R., Tohme, J., and Beeching, J. R. (2007). Towards identifying the full set of genes expressed during cassava post-harvest physiological deterioration. Plant Mol. Biol. 64, 187–203. doi: 10.1007/s11103-007-9144-0
Reilly, K., Gomez-Vasquez, R., Buschmann, H., Tohme, J., and Beeching, J. R. (2004). Oxidative stress responses during cassava post-harvest physiological deterioration. Plant Mol. Biol. 56, 625–641. doi: 10.1007/s11103-005-2271-6
Rushton, P. J., Bokowiec, M. T., Han, S. C., Zhang, H. B., Brannock, J. F., and Chen, X. F. (2008). Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 147, 280–295. doi: 10.1104/pp.107.114041
Saravanan, R., Ravi, V., Stephen, R., Thajudhin, S., and George, J. (2016). Post-harvest physiological deterioration of cassava (Manihot esculenta) - A review. Indian J. Agric. Sci. 86, 1383–1390.
Sharker, M. R., Sukhan, Z. P., Kim, S. C., Lee, W. K., and Kho, K. H. (2020). Identification, characterization, and expression analysis of a serotonin receptor involved in the reproductive process of the Pacific abalone. Haliotis discus hannai. Mol. Biol. Rep. 47, 555–567. doi: 10.1007/s11033-019-05162-2
Shen, T., Wen, X., Wen, Z., Qiu, Z., Hou, Q., Li, Z., et al. (2021). Genome-wide identification and expression analysis of bHLH transcription factor family in response to cold stress in sweet cherry (Prunus avium L.). Sci. Hortic. 279:109905. doi: 10.1016/j.scienta.2021.109905
Song, X., Huang, Z., Duan, W., Ren, J., Liu, T., Li, Y., et al. (2013). Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genomics 289, 77–91. doi: 10.1007/s00438-013-0791-3
Steidl, C., Leimeister, C., Klamt, B., Maier, M., Nanda, I., Dixon, M., et al. (2000). Characterization of the human and mouse HEY1, HEY2, and HEYL genes: cloning, mapping, and mutation screening of a new bHLH gene family. Genomics 66, 195–203. doi: 10.1006/geno.2000.6200
Sun, H., Fan, H., and Ling, H. (2015). Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Plant Biol. 16:9. doi: 10.1186/s12864-014-1209-2
Sun, K., Wang, H., and Xia, Z. (2019). The maize bHLH transcription factor bHLH105 confers manganese tolerance in transgenic tobacco. Plant Sci. 280, 97–109. doi: 10.1016/j.plantsci.2018.11.006
Sun, W., Jin, X., Chen, H., and Liu, M. (2019). Basic helix-loop-helix (bHLH) gene family in Tartary buckwheat (Fagopyrum tataricum): genome-wide identification, phylogeny, evolutionary expansion and expression analyses. Int. J. Biol. Macromol. 155, 1478–1490. doi: 10.1016/j.ijbiomac.2019.11.126
Thompson, J., Gibson, T., Plewniak, F., Jeanmougin, F., and Higgins, D. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Toledo-Ortiz, G., Huq, E., and Quail, P. H. (2003). The Arabidopsis basic/helixloop-helix transcription factor family. Plant Cell 15, 1749–1770. doi: 10.1105/tpc.013839
Trofimov, K., Lvanov, R., Eutebach, M., Acaroglu, B., Mohr, I., Bauer, P., et al. (2019). Mobility and localization of the iron deficiency-induced transcription factor bHLH039 change in the presence of FIT. Plant Direct. 3, 1–11. doi: 10.1002/pld3.190
Tuo, D., Zhou, P., Yan, P., Cui, H., Liu, Y., Wang, H., et al. (2021). A cassava common mosaic virus vector for virus-induced gene silencing in cassava. Plant Methods 17:74. doi: 10.1186/s13007-021-00775-w
Udogu, O. F., Omosun, G., and Njoku, D. N. (2021). Comparative evaluation of physiological post-harvest root deterioration, total carotenoids, starch content and dry matter of selected cassava cultivars. Nig. Agric. J. 52, 219–226.
Vatansever, R., Koc, I., Ozyigit, I. I., Sen, U., Uras, M. E., Anjum, N. A., et al. (2016). Genome-wide identification and expression analysis of sulfate transporter (SULTR) genes in potato (Solanum tuberosum L.). Planta 244, 1167–1183. doi: 10.1007/s00425-016-2575-6
Verma, D., Jalmi, S. K., Bhagat, P. K., Verma, N., and Sinha, A. K. (2020). A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J. 287, 2560–2576. doi: 10.1111/febs.15157
Voorrips, R. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, F., Zhu, H., Kong, W., Peng, R., Liu, Q., and Yao, Q. (2016). The Antirrhinum AmDEL gene enhances flavonoids accumulation and salt and drought tolerance in transgenic Arabidopsis. Planta 244, 59–73. doi: 10.1007/s00425-016-2489-3
Wang, Q., Lin, L., Zhang, Z., Yu, H., Xu, H., and Yang, X. (2022). Establishment and application of a method for detecting the cyanogenic glycoside in sweet cassava root and its products. Sci. Techno. Food Ind. 43, 271–278. doi: 10.13386/j.issn1002-0306.2021040260
Wang, R., Wang, R., Liu, M., Yuan, W., Zhao, Z., Liu, X., et al. (2021). Nucleocytoplasmic trafficking and turnover mechanisms of BRASSINAZOLE RESISTANT1 in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 118:e2101838118. doi: 10.1073/pnas.2101838118
Wang, Y., Tang, H., DeBarry, J., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Waseem, M., Rong, X., and Li, Z. (2019). Dissecting the role of a basic helix-loop-helix transcription factor, SlbHLH22, under salt and drought stresses in transgenic Solanum lycopersicum L. Front. Plant Sci. 10:734. doi: 10.3389/fpls.2019.00734
Xi, H. C., He, Y. J., and Chen, H. Y. (2021). Functional characterization of SmbHLH13 in anthocyanin biosynthesis and flowering in eggplant. Hortic. Plant J. 7, 73–80. doi: 10.1016/j.hpj.2020.08.006
Xiong, Y., Liu, T., Tian, C., Sun, S., Li, J., and Chen, M. (2005). Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol. Biol. 59, 191–203. doi: 10.1007/s11103-005-6503-6
Xu, J., Duan, X., Yang, J., Beeching, J. R., and Zhang, P. (2013). Enhanced reactive oxygen species scavenging by overproduction of superoxide dismutase and catalase delays postharvest physiological deterioration of cassava storage roots. Plant Physiol. 161, 1517–1528. doi: 10.1104/pp.112.212803
Yan, P., Zeng, Y., Shen, W., Tuo, D., Li, X., and Zhou, P. (2020). Nimble cloning: a simple, versatile, and efficient system for standardized molecular cloning. Front. Bioeng. Biotech. 7:460. doi: 10.3389/fbioe.2019.00460
Yan, Y., Wang, P., Lu, Y., Bai, Y., Wei, Y., Liu, G., et al. (2021). MeRAV5 promotes drought stress resistance in cassava by modulating hydrogen peroxide and lignin accumulation. Plant J. 107, 847–860. doi: 10.1111/tpj.15350
Yang, T. R., Yao, S. F., Hao, L., Zhao, Y. Y., Lu, W. J., and Xiao, K. (2016). Wheat bHLH-type transcription factor gene TabHLH1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-associated pathway. Plant Cell Rep. 35, 2309–2323. doi: 10.1007/s00299-016-2036-5
Yao, P., Sun, Z., Li, C., Zhao, X., Li, M., Deng, R., et al. (2018). Overexpression of Fagopyrum tataricum FtbHLH2 enhances tolerance to cold stress in transgenic Arabidopsis. Plant Physiol. Biochem. 125, 85–94. doi: 10.1016/j.plaphy.2018.01.028
Yu, C., Yan, M., Dong, H., Luo, J., Ke, Y., Guo, A., et al. (2021). Maize bHLH55 functions positively in salt tolerance through modulation of AsA biosynthesis by directly regulating GDP-mannose pathway genes. Plant Sci. 302:110676. doi: 10.1016/j.plantsci.2020.110676
Yu, J., Wang, J., Lin, W., Li, S., Li, H., Zhou, J., et al. (2005). The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3:e38. doi: 10.1371/journal.pbio.0030038
Yu, Z., Zhang, G., Teixeira da Silva, J. A., Zhao, C., and Duan, J. (2021). The methyl jasmonate-responsive transcription factor DobHLH4 promotes DoTPS10, which is involved in linalool biosynthesis in Dendrobium officinale during floral development. Plant Sci. 309:110952. doi: 10.1016/j.plantsci.2021.110952
Zainuddin, I. M., Fathoni, A., Sudarmonowati, E., Beeching, J. R., Gruissem, W., and Vanderschuren, H. (2017). Cassava post-harvest physiological deterioration: from triggers to symptoms. Postharvest Biol. Technol. 142, 115–123. doi: 10.1016/j.postharvbio.2017.09.004
Zhang, Z., Li, J., Zhao, X., Wang, J., Wong, G., and Yu, J. (2006). KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinf. 4, 259–263. doi: 10.1016/S1672-0229(07)60007-2
Zhao, Q., Fan, Z., Qiu, L., Che, Q., and Wang, Y. (2020). MdbHLH130, an apple bHLH transcription factor, confers water stress resistance by regulating dtomatal closure and ROS homeostasis in transgenic tobacco. Front. Plant Sci. 11:543696. doi: 10.3389/fpls.2020.543696
Zhao, R., Song, X., Yang, N., Chen, L., Xiang, L., Liu, X. Q., et al. (2020). Expression of the subgroup IIIf bHLH transcription factor CpbHLH1 from Chimonanthus praecox (L.) in transgenic model plants inhibits anthocyanin accumulation. Plant Cell Rep. 39, 891–907. doi: 10.1007/s00299-020-02537-9
Zhao, W., Liu, Y., Li, L., Meng, H., Yang, Y., and Dong, Z. (2021). Genome-wide identification and characterization of bHLH transcription factors related to anthocyanin biosynthesis in red walnut (Juglans regia L.). Front. Genet. 12:632509. doi: 10.3389/fgene.2021.632509
Zhao, Y., Zhang, Y., Liu, H., Zhang, X., Ni, R., and Wang, P. (2019). Functional characterization of a liverworts bHLH transcription factor involved in the regulation of bisbibenzyls and flavonoids biosynthesis. BMC Plant Biol. 19:497. doi: 10.1186/s12870-019-2109-z
Keywords: cassava, bHLH gene family, transcription factors, postharvest physiological deterioration, cyanogenic glucosides
Citation: An F, Xiao X, Chen T, Xue J, Luo X, Ou W, Li K, Cai J and Chen S (2022) Systematic Analysis of bHLH Transcription Factors in Cassava Uncovers Their Roles in Postharvest Physiological Deterioration and Cyanogenic Glycosides Biosynthesis. Front. Plant Sci. 13:901128. doi: 10.3389/fpls.2022.901128
Received: 21 March 2022; Accepted: 09 May 2022;
Published: 16 June 2022.
Edited by:
Jacobo Arango, International Center for Tropical Agriculture (CIAT), ColombiaReviewed by:
Xinxin Li, Fujian Agriculture and Forestry University, ChinaCopyright © 2022 An, Xiao, Chen, Xue, Luo, Ou, Li, Cai and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Songbi Chen, c29uZ2JpY2hlbkBob3RtYWlsLmNvbQ==; Jie Cai, Y2FpamllQGNhdGFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.