- 1College of Agriculture, Institute of Entomology, Yangtze University, Jingzhou, China
- 2College of Life Sciences, Yangtze University, Jingzhou, China
- 3Guangxi Key Laboratory of Biology for Crop Diseases and Insect Pests, Guangxi Academy of Agricultural Sciences, Nanning, China
Callose synthase plays an essential role in plant growth and development and in response to all sorts of stresses through regulating callose formation. However, few research about the function and mechanism of the insect resistance of callose synthase genes have been reported in cotton. In this study, a cotton callose synthase gene GhCalS5 was cloned, and its function and mechanism of resistance to cotton aphids were analyzed. The expression of GhCalS5 was significantly upregulated in both, leaves and stems of cotton plants at 48 h after cotton aphid infestation and in the leaves of cotton plants at 24 h after salicylic acid treatment. The overexpression of GhCalS5 enhanced cotton resistance to cotton aphids. Expectedly silencing of GhCalS5 reduced cotton resistance to cotton aphids. Overexpression of GhCalS5 enhanced callose formation in cotton leaves. Our results suggest that GhCalS5 is involved in cotton resistance against cotton aphids by influencing callose formation.
Introduction
Callose is a β-1, 3-glucan polymer synthesized by callose synthases in higher plants (Wang et al., 2021). It exists in multiple places in plant cells. For example, callose is transiently deposited in the cell plate at the cytokinesis stage and is a vital component of the transient cell wall surrounding pollen mother cells, the four microspores after meiosis, and the pre-cell wall of the growing pollen tube tip (Mccormick, 1993; Hong et al., 2001). Callose is also present in sieve plates under normal plant growth and developmental conditions but can accumulate quickly and plugs the sieve pores under stress (Xie et al., 2011). More importantly, callose is deposited between the plasma membrane and the pre-existing cell wall at sites of pathogen attack (Voigt, 2014). In addition, callose is also synthesized in the neck region of plasmodesmata during abiotic and biotic stresses (Zavaliev et al., 2011). Callose synthase (CalS) is also referred to as glucan synthase-like (GLS; Richmond and Somerville, 2000; Verma and Hong, 2001). Twelve AtCalSs in Arabidopsis thaliana have been described (Hong et al., 2001), as well as 12 CsCalSs in Citrus sinensis (Granato et al., 2019), 32 BnCalSs in Brassica napus (Liu et al., 2018), 8 VvCalSs in Vitis vinifera (Yu et al., 2016), 7 HvCalSs in Hordeum vulgare (Schober et al., 2009), 15 BraCalSs in Chinese cabbage (Pu et al., 2019), and 27, 28, 16, and 15 GhCalSs, respectively, in Gossypium hirsutum, G. barbadense, G. raimondii, and G. arboreum (Feng et al., 2021) have been reported.
Callose synthases play crucial roles in plants by regulating callose deposition. Firstly, it is involved in normal plant growth and development of plants. For example, callose synthase GSL7 is necessary for inflorescence growth in Arabidopsis (Paul Barratt et al., 2011). A callose synthase Tie-dyed2 mutant of Zea mays displays variegated green and yellow leaves (Slewinski et al., 2012). Secondly, callose synthases are also influenced by multiple abiotic stress. For instance, when maize leaf tip was stimulated by low temperature, action potential was generated in the phloem, intercellular movement from mesophyll to sheath cells decreased significantly, callose content in leaves increased, and the transport of photosynthates in phloem decreased (Fromm et al., 2013). A. thaliana AtCalS7 (AtGSL7), AtCalS8 (AtGSL4), and AtCalS12 (AtGSL5/PMR4) are associated with callose synthesis upon mechanical wounding (Cui and Lee, 2016). Additionally, callose synthases are also affected by various types of biotic stress. Recently, callose synthase family genes have been found to play an essential role in citrus defense response to Candidatus Liberibacter asiaticus, and a novel demonstration of RNAi in citrus reveals the importance of citrus callose synthase in defense against Xanthomonas citri subsp. citri (Enrique et al., 2011; Granato et al., 2019). Transient expression of the A. thaliana callose synthase PMR4 increases penetration resistance to powdery mildew in barley (Blümke et al., 2013), and this pathogen-induced callose deposition functions as a chemical and physical defense mechanism by reinforcing plant cell wall (Ellinger and Voigt, 2014). In addition to induction by plant disease invasion, rapid deposition of callose at the point of attempted penetration was observed after insect feeding (Hao et al., 2008; Kuśnierczyk et al., 2008) or nematode infestation (Hofmann et al., 2010). When attacked by piercing insects, plants activate callose deposition, which hinders phloem’s flow, discourages phloem feeding, and this is the key resistance mechanism in rice varieties resistant to brown planthopper (Nilaparvata lugens; Hao et al., 2008). An AtCalS7 gene has been associated with the synthesis of callose in the sieve plates of phloem in response to stresses resulting in the phloem blockage (Paul Barratt et al., 2011; Xie et al., 2011). Aphids and silver whitefly nymphs have been reported to induce callose in plants upon infestation (Botha and Matsiliza, 2004; Kempema et al., 2007). Meanwhile, callose synthase is mediated by all signaling pathways, and different biological regulatory processes also involve hormones. For example, abscisic acid (ABA) increased rice callose synthase activity, promoted callose deposition, and improved its resistance to N. lugens (Liu et al., 2017). Arabidopsis callose synthase 1 (CalS1) and CalS8 have been identified as the key genes involved in altering plasmodesmal permeability upon mechanical wounding stress (Dong et al., 2008). While previous studies have focused on pathogen-induced callose, little research has been reported on herbivore-induced callose.
Herbivore feeding accounts for > 20% of net plant productivity in vegetation systems, including the loss of foliage, sap, and root feeding (Agrawal, 2011). These losses occur despite increased pesticide use, highlighting the need to develop sustainable approaches for pest control with less reliance on chemical inputs. Cotton aphid (Aphis gossypii) is the most problematic pest to growers in cotton production. They insert their stylets into plant tissues and probe until they contact a phloem vessel to feed on phloem sap, resulting in the plant’s retarded growth, giving rise to deformed leaves, and the infestation may result in the death of young plants if it occurs early in the season. Survival of plants upon aphid attack depends on a dynamic defense system involving structural barriers (including plant callose deposition) and attraction of natural enemies of target pests in nature (Sánchez-Sánchez and Morquecho, 2017). Therefore, the cotton gene GhCalS5 was cloned in this study, and the gene function and mechanism in cotton response to cotton aphids were determined through overexpressing and silencing methods, respectively. Our results will help reveal that the GhCalS5 gene has a role in cotton plant response to aphid infestation.
Materials and methods
Plant materials, growth conditions, and aphid rearing
Red-leaf cotton (G. hirsutum), a cotton aphid-resistant variety, was provided by the Cotton Research Institute of Chinese Academy of Agricultural Sciences. The seeds of red-leaf cotton were planted directly in the nutritional soil (peat soil: perlite: vermiculite = 4:1:1) in 30-cm-diameter plastic pots. The plants were kept and grown in a growth room with 75% relative humidity and photoperiod 16 h light/8 h darkness at 25°C. Cotton aphids were collected from the laboratory cotton field at Yangtze University, cultured on Coker 312, a variety with weak resistance to aphids, and maintained in an environment chamber at 25°C with a long day photoperiod (16 h light and 8 h darkness). Active adult aphids of the same age and size were used in all the experiments.
Sequence analysis of GhCalS5
The coding sequence (CDS) of GhCalS5 from the leaf transcriptome data of red-leaf cotton induced by cotton aphids was amplified as a cotton cDNA template, using gene-specific primer pair for GhCalS5 (Supplementary Table S1). The PCR products were linked to the pMD 18 T cloning vector (Takara) and transformed into Escherichia coli strain DH5α. Positive clones were inoculated for sequencing of GhCalS5. The obtained sequence was used as a query in a BLASTp program1 in the National Center for Biotechnology Information (NCBI), and the protein sequences with the highest homology to GhCalS5 from other plant species were downloaded. The theoretical isoelectric point and molecular weight of GhCalS5 protein were obtained using Lasergene 7.1 software. The alignment of callose synthase proteins from different species was carried out using the DNAMAN, and the phylogenetic tree was constructed by the software MEGA11 with the maximum likehood method. Numbers on the branches represent bootstrap values (1,000 replicates).
Vector construction and transformation of cotton
The PCR products were first cloned into the pMD-18T cloning vector, and the intermediate vector pMD18-T-GhCalS5 was then digested by Xba1 and BamH1. The resultant digestion fragment was further inserted into the corresponding sites of the overexpression vector pBI121 containing Cauliflower mosaic virus (CaMV) 35S promoter to generate pBI121-GhCalS5. The tobacco rattle virus (TRV)-based VIGS system was used to silence GhCalS5 expression in cotton. The pMD-18T-GhCalS5-digested fragment was inserted in the corresponding sites in the plasmid pTRV2 to generate the virus gene-silencing vector TRV: GhCalS5. The plasmids of TRV1, TRV:00 (the negative control with empty vector), TRV: GhCalS5, pBI121 (the negative control with empty vector), and pBI121-GhCalS5 were transformed into Agrobacterium strain GV3101 via the freeze–thaw method (Degefu and Hanif, 2003), respectively. Recombinant bacteria were separately grown overnight from pre-cultures at 28°C and 200 rpm in an orbital shaker using LB medium (supplemented with appropriate antibiotics), pelleted by centrifugation, and resuspended in MMA (10 mM MES, 10 mM MgCl2, and 100 mM acetosyringone; Norkunas et al., 2018). The GV3101 strain harboring TRV1 was mixed with those harboring TRV: GhCalS5 at a 1:1 ratio. The mixture and three A. tumefaciens harboring TRV:00, pBI121-GhCalS5, or pBI121 were infiltrated into the cotyledons of 8-day-old cotton using a needless syringe, respectively.
RNA extraction and quantitative RT-PCR
Total RNA was extracted from cotton roots, leaves, and stems using a Fast pure plant total RNA isolation kit (Vazyme, Nanjing, China) according to the manufacturer’s instructions. The quality of RNA was verified after separation on agarose gel by ethidium bromide staining and quantified using a NanoPhotometer N50 (Implen). 2 μg total RNA of each sample was reverse transcribed into cDNA using the PrimeScript™RT Reagent Kit (TaKaRa, Shiga, Japan) according to the manufacturer’s instructions. The cotton ubiquitin gene GhUBI1 (accession number: EU604080) was used as the reference gene. The fold change of the relative genes was quantified using the 2–∆∆CT method (Livak and Schmittgen, 2001). Each sample had three biological repeats, which were repeated three times.
Expression patterns of GhCalS5 under different treatments in cotton
In order to detect expression patterns of GhCalS5 in cotton tissues upon aphid infestation, the tissue samples from cotton roots, stems, and leaves of cotton cotyledons of 14 days were harvested at 24, 48, and 72 h after aphid infestation or no-cotton aphid infestation. Cotton cotyledons of 14 days seedlings were sprayed with SA (5 mmol/l) solution as a foliar spray until runoff and collected at 24, 48, and 72 h post spray. Three independent tests with three biological replicates were performed.
Host choice assay
The host choice test of cotton aphids was performed in 9-cm-diameter Petri dishes. The cotyledons were detached from GhCalS5-overexpressing, pBI121, and wild-type cotton plants, and their petioles were wrapped with a wet cotton ball. Each group of cotyledons was symmetrically placed on the inner edge of the Petri dish. A total of 20 apterous adults of cotton aphids were put at the center of the Petri dish and counted after 24, 48, and 72 h. The host no-choice assay was performed in vivo; 20 apterous adults of cotton aphids were inoculated on each of the 9-day-old GhCalS5-overexpressing, pBI121 (control), and wild-type cotton plants or 15-day-old GhCalS5-silenced plants-silenced, TRV:00, and wild-type cotton plants; and the number of aphids was counted at 24, 48, and 72 h or 7–22 days after feeding, respectively. Each treatment was replicated three times.
Determination of aphid feeding activity
The feeding activity of cotton aphids can be determined by measuring the amount of honeydew produced. Split Whatman filter papers were placed under each plant on GhCalS5-overexpressing, pBI121, and wild-type cotton plants or GhCalS5-silenced plants, TRV: 00, and wild-type cotton plants; 20 apterous aphid adults were put on each plant. A plastic membrane was placed under the filter paper to avoid the absorbance of water from the soil. Whatman filter papers were used to collect honeydew after 24, 48, and 72 h for GhCalS5-overexpressing, pBI121, and wild-type cotton plants and 8 days, 15 days, and 22 days for GhCalS5-silenced plants, and TRV: 00, and wild-type cotton plants, respectively. Whatman filter papers were soaked in 0.1% (w/v) ninhydrin solution in acetone and dried in a 65°C oven for 30 min, and purple spots were shown when honeydew was stained by ninhydrin (Kim and Jander, 2007). To quantify the honeydew stains, stained filter papers were cut into pieces and extracted with 1 ml of 90% (v/v) methanol for 1 h at 4°C with continuous agitation. The supernatant absorbance was measured at 500 nm after centrifugation at 6,000× g for 1 min. 90% methanol was used as a blank. Each treatment was replicated four times.
Cotton leaf callose analysis
Aniline blue was used to stain callose deposition, according to (Zhang et al., 2014). In short, the cotyledons leaves from GhCalS5-overexpressing, pBI121, and wild-type cotton plants and GhCalS5-silenced plants, TRV: 00, and wild-type cotton plants were put in 5 ml lactophenol solution: phenol, glycerol (100%), lactic acid, distilled water, and absolute ethanol (1:1:1:1:8 v/v), and then immersed by the vacuum pump. Leaves were incubated in a water bath at 65°C for 1 h. Cotton leaves were soaked in 0.01% aniline blue staining solution (0.15M K2HPO4, pH 9.5). Callose deposition was visualized under Nikon fluorescent microscopy equipped with an ultraviolet lamp (330–380 nm). Callose deposits were quantified by a protocol previously described by Jin and Mackey (2017). Each treatment was replicated four times.
Statistical analysis
The data were obtained from three independent technical or biological replicates per treatment and presented as the mean ± standard error. All data were subjected to analysis of variance (ANOVA). Mean differences were estimated by Tukey’s honestly significant difference (HSD) using statistical software SPSS 26.0 (SPSS Inc., Chicago, IL, United States of America) and GraphPad Prism 8.0 software. Statistical significance was considered when the p-value was less than 0.05.
Results
GhCalS5 sequence analysis
The sequence analysis showed that GhCalS5 CDS was made up of 1,851 bp. Its encoded protein comprises 616 amino acids, its relative molecular mass is 42.58 kDa, and its theoretical isoelectric point is 9.689 (Supplementary File S1). The maximum likehood method was used to analyze the phylogenetic relationships between GhCalS5 and callose synthase from other plant species. From the phylogenetic tree, the amino acid sequence of GhCalS5 was located in a clade where GhCalS5.1 and GaCalS5-like.2, and GhCalS5-like, were tightly clustered. Other homologous proteins were clustered into other large clusters (Figure 1). The close relationship between GhCalS5, GhCalS5.1, GaCalS5-like.2, and GhCalS5-like suggests having a similar function (Supplementary Figure S1).
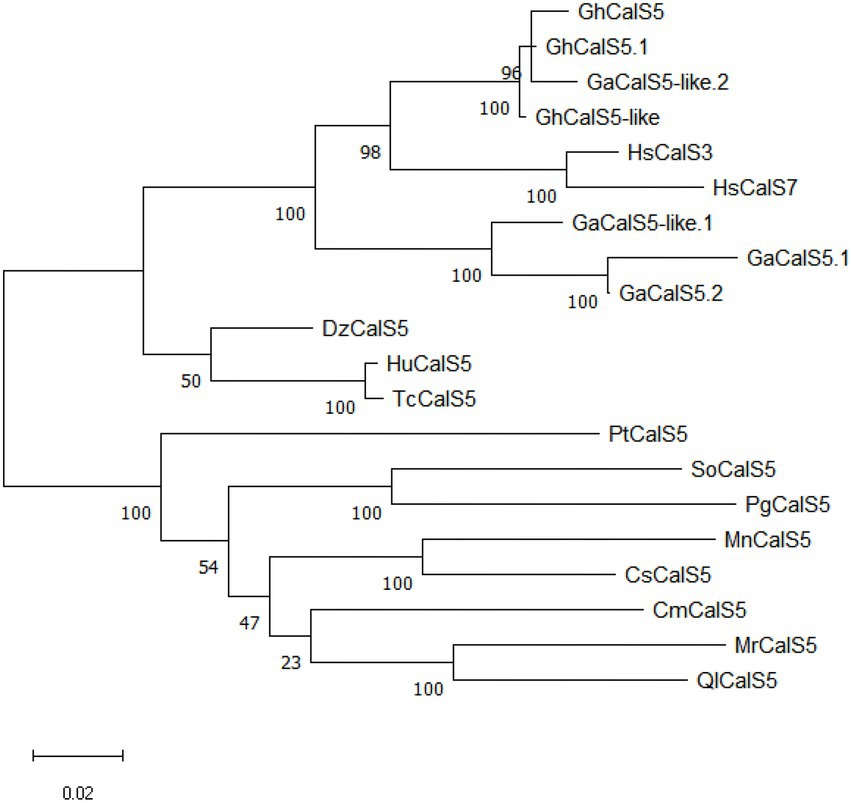
Figure 1. Phylogenetic analysis of GhCalS5 protein and CalS proteins of other plant species. The phylogenetic tree was constructed by the maximum likehood method using MEGA11 software. The accession numbers of sequence data were as follows: GhCalS5.1, Gossypium hirsutum CalS5.1 [XP_016676296.2]; GaCalS5.1, Gossypium arboreum CalS5.1[KHG21671.1]; GaCalS5-like.1, Gossypium austral CalS5-like.1 [KAA3479749.1]; GaCalS5.2, G. arboreum CalS5.2 [KHG21670.1]; HsCalS3, Hibiscus syriacus CalS3[KAE8662811.1]; HsCalS7, H. syriacus CalS7 [KAE8680723.1]; DzCalS5, Durio zibethinus CalS5 [XP_022750939.1]; HuCalS5, Herrania umbratica CalS5 [XP_021300964.1]; TcCalS5, Theobroma cacao CalS5 [EOX92541.1]; MnCalS5, Morus notabilis CalS5 [EXB36810.1]; SoCalS5, Syzygium oleosum CalS5[XP_030440758.1]; CsCalS5, Cannabis sativa CalS5[XP_030499529.1]; MrCalS5, Morella rubra CalS5[KAB1210572.1]; PgCalS5, Punica granatum CalS5[XP_031399619.1]; PtCalS5, Populus trichocarpa CalS5[XP_024456702.1]; CmCalS5, Cucurbita maxima CalS5[XP_023007270.1]; QlCalS5, Quercus lobata CalS5[XP_030973950.1]; GaCalS5-like.2, G. arboreum CalS5-like.2 [XP_017647032.1]; GhCalS5-like,G. hirsutum CalS5-like[XP_016698828.1].
GhCalS5 expression is induced by aphid and SA
The qPCR results showed that GhCalS5 was expressed in root, stem, and leaves of G. hirsutum after cotton aphid feeding, salicylic acid (SA) treatment, or no-treatment (control; Figure 2). Compared with control, no significance was observed in the expression level of GhCalS5 in root at 24, 48, and 72 h after aphid feeding (Figure 2A). The GhCalS5 transcript was significantly upregulated at 48 h after aphid feeding in stems. However, no significant difference was detected in the expression level at 24 and 72 h between cotton aphid and no-aphid feeding (Figure 2B). The GhCalS5 expression level was significantly upregulated at 48 h after aphid feeding in leaves, but no significant difference was detected at 24 and 72 h between cotton aphid feeding treatment and control (Figure 2C). In addition, the expression level of GhCalS5 was very significantly upregulated in the leaves at 24 h after SA treatment, but no significant differences were detected at 48 and 72 h between SA treatment and control (Figure 2D). These results suggested that cotton aphid feeding or SA treatment could induce a transient upregulation of GhCalS5 expression in cotton tissues.
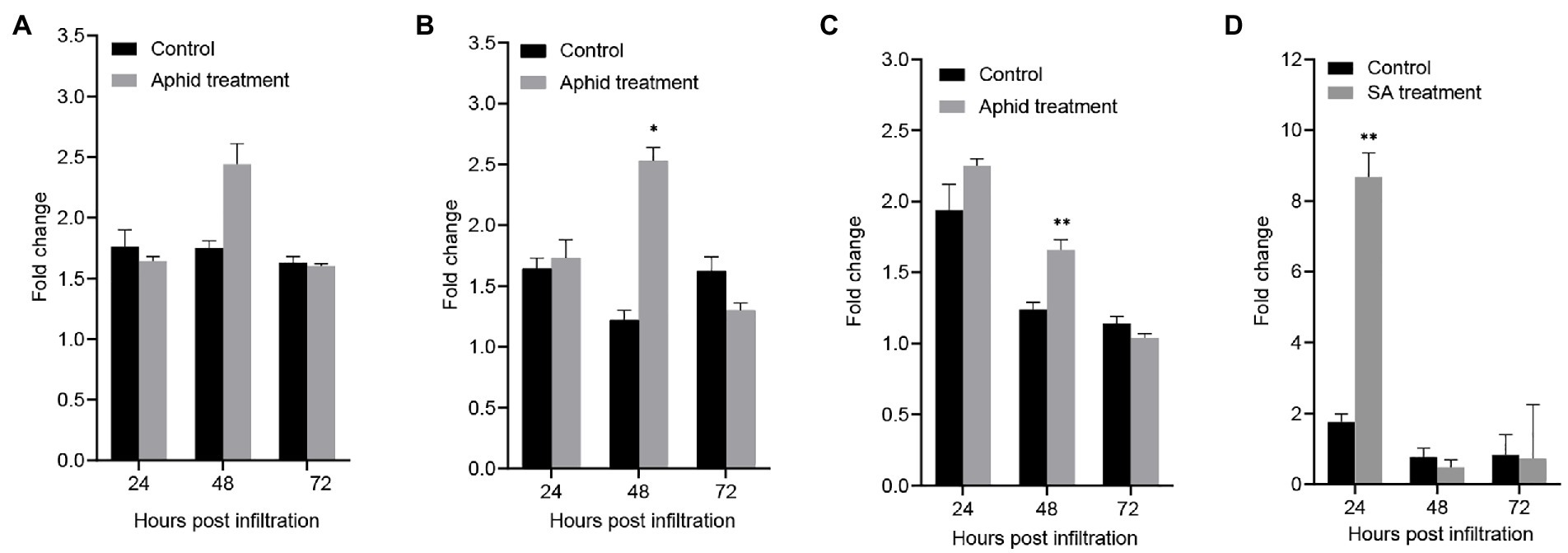
Figure 2. Expression of GhCalS5 in cotton plants after cotton aphid treatment and after SA treatment. (A) Root after aphid treatment, (B) stem after aphid treatment, (C) leaf after aphid treatment (D) leaf after SA treatment. Error bars represent the standard deviation of measurements from three biological replicates. Number of plants used = 3 per treatment. Independent Student’s t-tests analyzed data. Error bars represent the SD of three biological replicates. Statistical significance for treatment effects is marked (*p < 0.05) or (**p < 0.01).
Overexpression of GhCalS5 enhances cotton resistance to cotton aphids
To understand the involvement of GhCalS5 in cotton resistance to aphids, GhCalS5 was overexpressed using the transient expression method, in which Agrobacterium strain harboring pBI121 and pBI121-GhCalS5 were infiltrated into cotton cotyledons. After 24, 48, and 72 h of infiltration, the transcript level of GhCalS5 in cotton plants was significantly higher in GhCalS5-overexpressing plants than in control plants (pBI121) and wild-type, respectively (Figure 3A). To validate the role of GhCalS5 in cotton response to aphid damage, aphid performance on wild-type, pBI121, and pBI121—GhCalS5 plants was analyzed via host choice assay. In the no-choice assay, the population of aphids was significantly low in GhCalS5-overexpressing cotton plants at 48 and 72 h after infiltration compared to the wild-type and control plants, suggesting that the overexpression of GhCalS5 in cotton plants reduced aphid offspring (Figure 3B). Additionally, the choice assay showed that aphids preferred feeding on the wild-type leaves and control leaves than on leaves from GhCalS5-overexpressing plants significantly at 48 and 72 h after post infiltration (Figure 3C). These results indicate that GhCalS5 overexpression in cotton leaves improves cotton resistance against aphids. Then, we investigated the effects of GhCalS5 gene overexpression on aphid honeydew excretion. Honeydew produced by aphids at 24, 48, and 72 h after feeding was quantified. However, no significant difference was detected in the honeydew absorbance in all plants after aphid feeding (Figures 3D,E).
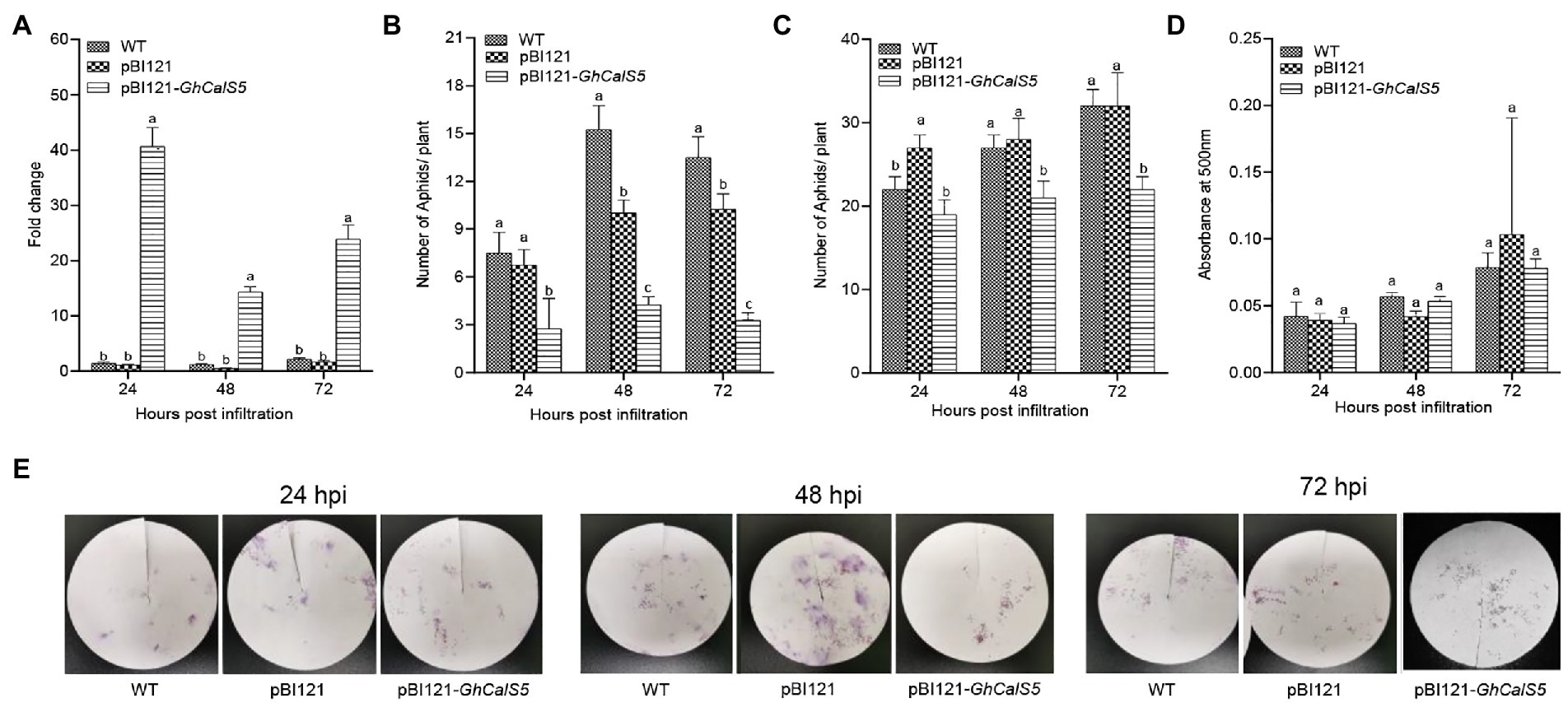
Figure 3. Overexpression GhCalS5 inhibited the population of cotton aphids in cotton plants. (A) transient expression of GhCalS5 in cotton leaves, (B) choice assay, (C) no-choice assay, (D) absorbance levels of aphid honeydew at 500 nm, (E) aphid honeydew deposition staining by ninhydrin. HPI, hours post infiltration. Number of plants used = 3 per treatment. One-way ANOVA analyzed data. Error bars represents the SD of three biological replicates. Different letters indicate significant differences (p < 0.05) based on Tukey’s HSD test.
Silencing of GhCalS5 increases cotton susceptibility to cotton aphids
To further explore the role of GhCalS5 in cotton response to aphids, the GhCalS5 gene expression was silenced using the VIGS method. The transcriptional levels of GhCalS5 in GhCalS5-silenced plants were significantly low compared to the wild-type and TRV: 00 plants at 8 and 15 days after injecting (Figure 4A). To further investigate the involvement of GhCalS5 in plant response to cotton aphids, a no-choice assay test was performed on GhCalS5-silenced plants, control (TRV: 00), and wild-type plants. The population of aphids was higher in GhCalS5-silenced plants than in the TRV: 00 and wild-type plants. At 15 days after A. tumefaciens infiltration, the number of cotton aphids elevated rapidly on GhCalS5-silenced plants and reached an average of 473 aphids per plant at 22 d, while the number of cotton aphids on wild plants reached an average of 256 aphids per plant (Figure 4B). Additionally, we also determined the aphid feeding activity after silencing the GhCalS5 gene in cotton plants. The honeydew absorbance was the same 8 days after A. tumefaciens infiltration and gradually increased. It was significantly higher in GhCalS5-silenced plants than wild-type cotton at 15 days and remained elevated up to 22 days after A. tumefaciens infiltration (Figures 4C,D). These results suggest that silencing GhCalS5 enhances cotton susceptibility to aphids.
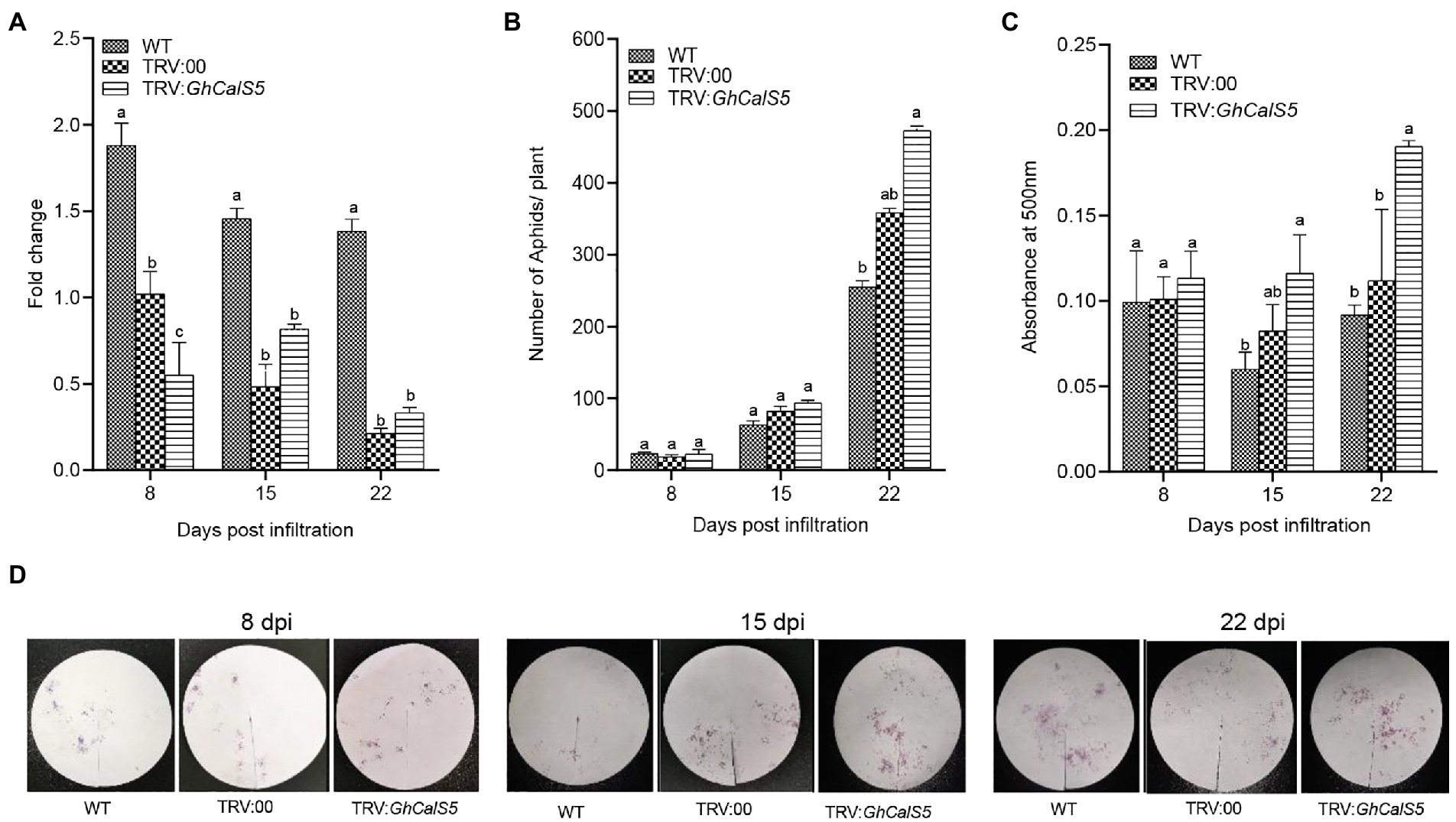
Figure 4. Silencing expression of GhCalS5 increased aphid population and feeding in cotton plants. (A) fold change in transcription of GhCalS5 in cotton leaves, (B) aphid no-choice assay, (C) absorbance levels of aphid honeydew at 500 nm (D) aphid honeydew deposition staining by ninhydrin. DPI, days post infiltration. Number of plants used = 3 per treatment. One-way ANOVA analyzed data. Error bars represents the SD of three biological replicates. Different letters indicate significant differences (p < 0.05) based on Tukey’s HSD test.
GhCalS5 is involved in callose formation in cotton response to cotton aphid damage
Callose is considered an essential index of resistance to aphids, and its formation is closely related to callose synthase. To clarify callose deposition in GhCalS5-overexpressing plants, we visualized callose deposits in leaves through fluorescence microscopy. Callose deposits were higher on leaves from GhCalS5-overexpressing plants than in wild-type (WT) and control plants (pBI121; Figures 5A,B). These results correlated with GhCalS5 transcript level after GV3101 infiltration and suggest that the overexpression of GhCalS5 increases callose deposition in the leaves of cotton plants. Furthermore, we observed callose deposition in cotton leaves after silencing GhCalS5 in cotton. Few callose deposits accumulated on leaves from GhCalS5-silenced plants, no significant difference in leaf disk callose deposits from wild-type plants. The results indicate that the silencing of GhCalS5 in cotton reduces the number of callose deposits on cotton leaves (Figures 5A,C). In short, these results demonstrated that GhCalS5 is involved in cotton in response to cotton aphid damage through callose formation.
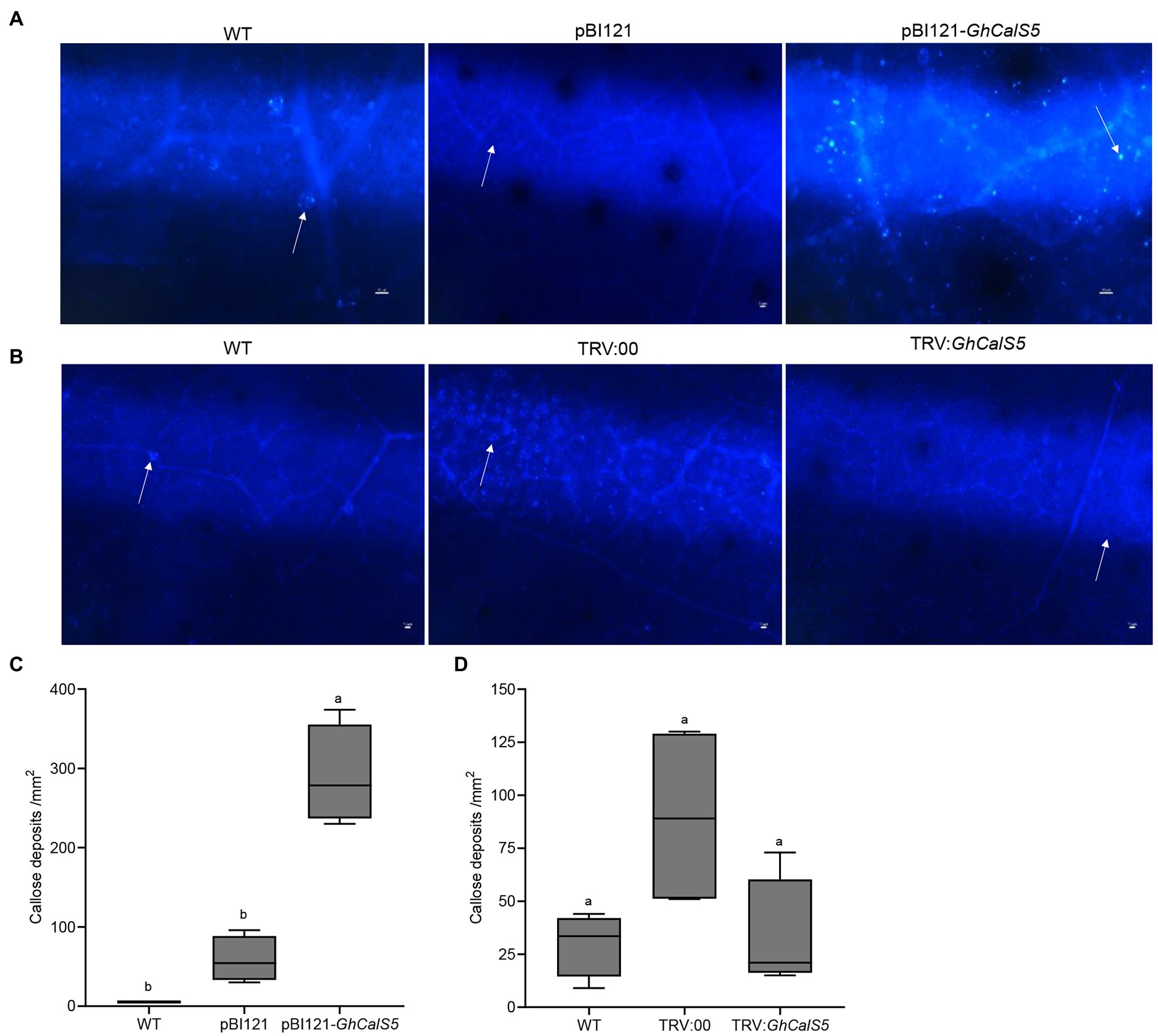
Figure 5. Callose deposition and quantification in GhCalS5-overexpressed or silenced cotton leaves. (A) Callose deposition in cotton leaves in WT, pBI121 (control), GhCalS5-overexpressed. (B) Callose deposition in cotton leaves in TRV: 00 and GhCalS5-silenced plants. (B) Callose deposits quantification in WT, pBI121 (control), and GhCalS5-overexpressed using ImageJ software. Callose deposits quantification in (C) WT, pBI121 (control), pBI121-GhCalS5 plants (D) WT, TRV: 00 (control), TRV:GhCalS5 plants using ImageJ software. One-way ANOVA analyzed data. Error bars represents the SD of three biological replicates. Different letters indicate significant differences (p < 0.05) based on Tukey’s HSD test. Values are shown per mm2 from the leaf of four independent plants. Scar bar: 50 μm.
Discussion
Biotic and abiotic stresses in nature often challenge plants. Plants have gradually developed various defined and complex mechanisms to respond to these damages during a long period of coevolution (Erb and Reymond, 2019). Callose deposition is induced in plant tissue as a defense mechanism response to both biotic and abiotic stresses. Several studies have reported that callose deposition is induced by biotic and abiotic stress (Jacobs et al., 2003; Hao et al., 2008; Li et al., 2018; O’Lexy et al., 2018; Hunter et al., 2019). However, the function and mechanism of callose formation in the plant defense response to insect pests have still not been studied in depth (Liu et al., 2017). Therefore, we performed a study on the role of callose synthase gene GhCalS5 in cotton response to aphid infestation.
Callose has been reported to play a role in plant defense response against insect pests. For instance, the Russian wheat aphid’s (Diuraphis noxia) sustained phloem feeding could reduce transport in Triticum aestivum by influencing callose formation. However, the mechanism of callose formation is not described (Botha and Matsiliza, 2004). The Arabidopsis transcriptome analysis in response to the nymphs of Bemisia tabaci type B showed that callose synthase gene RNAs accumulated and callose deposition was observed in SLWF-infested tissue, suggesting that callose synthase is involved in callose formation in A. thaliana (Kempema et al., 2007). In addition, exogenous ABA suppressed β-1,3-glucanase and induced synthase activity, and promoted callose deposition (Liu et al., 2017). Upon aphid attack in the pepper plant, callose synthase genes were induced in the leaves (Sun et al., 2018). However, there are few reports on the function of callose synthase genes in cotton insect resistance that have been made. In this study, we cloned a cotton callose synthase gene GhCalS5 and demonstrated that the GhCalS5 gene was upregulated in cotton plants at 48 h after cotton aphid infestation (Figures 2B,C). The results confirmed that GhCalS5 is involved in cotton response to cotton aphid attack. Moreover, the GhCalS5 transcript was upregulated significantly at 24 h after SA treatment (Figure 2D), suggesting that the GhCalS5 transcript is related to SA, which was consistent with the results that direct exogenous application of SA improve plasmodesmata closure and callose deposition (Wang et al., 2013).
Upon aphid infestation, plant defense response is often reflected by reduced non-preference (antixenosis) in a choice test and by reduced offspring production (antibiosis) in a no-choice test (Lei et al., 2014). In this study, when aphids were given a choice of host plants (Figure 3B), approximately twice as many cotton aphids preferred to feed on wild-type than GhCalS5 overexpressed plants. Secondly, aphids are likely to have low fecundity and die early due to difficulties taking up nutrients when feeding on plants containing phloem-based resistance (Sun et al., 2018). This is in line with our observations that the overexpression of GhCalS5 resulted in an impaired aphid population after 72 h post infiltration in the no-choice tests assay (Figure 3C). To further validate the role of GhCalS5 in cotton response to aphid attack, GhCalS5-silenced plants were exposed to aphid feeding. The population of aphid feeding on GhCalS5-silenced plants was higher than the aphids feeding on wild-type plants, and aphid feeding on GhCalS5-silenced plants excreted more honeydew than aphid feeding on wild-type, which indicates that GhCalS5 silence contributed to feeding of cotton aphids (Figure 4).
Callose plays an important role in aphid resistance in plants. Previous studies have shown that brown planthopper’s (BPH) feeding on rice resistant cultivars was interrupted due to callose deposition in the sieve plates, BPH preferred to feed on the susceptible variety of plants (Hao et al., 2008). We conclude that the preference of aphids to feed on wild-type plants and the reduced population were due to callose deposition in the leaves in GhCalS5-overexpressing plants (Figure 3). To confirm this hypothesis, callose deposits were stained in cotton leaves. Callose deposition in GhCalS5 overexpressed plants was significantly more than that in WT plants (Figures 5A,B), suggesting that the overexpression of GhCalS5 could promote callose deposition. Depleting DIMBOA-Glc reserves and inducing HDMBA-Glc production by caterpillar feeding in maize induces aphid susceptibility by suppressing callose (Meihls et al., 2013). Aphids grew better on maize cultivars with low DIMBOA-Glc and high HDMBOA-Glc levels, which coincides with low callose inducibility in the genotypes (Maag et al., 2016). In this study, we found a slight difference in callose deposits between the GhCalS5-silenced and WT plants (Figures 5A,C). The low callose deposition level in these plants may have resulted in an increased population of aphid feeding on GhCalS5-silenced plants. Of course, further research is needed.
In summary, GhCalS5 could be involved in controlling aphids in cotton plants. However, there is still a research gap on the detailed mechanisms through which callose acts during defenses. Therefore, there is a need to unceasingly study the callose synthase genes to understand their function in plant-herbivore interaction better.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
JZ and YY designed the research. NITM, ZH, and HZ performed the experiments with the assistance of YZ, XL, YuW, YiW, TL, and JL. JZ, NITM, and XW analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (grant no. 31471783) and the Foundation of Guangxi Key Laboratory of Biology for Crop Diseases and Insect Pests (grant no. 20–065-30-KF-01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materials
The Supplementary materials for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.892630/full#supplementary-material
Footnotes
References
Agrawal, A. A. (2011). Current trends in the evolutionary ecology of plant defence. Funct. Ecol. 25, 420–432. doi: 10.1111/j.1365-2435.2010.01796.x
Blümke, A., Somerville, S. C., Voigt, C. A., Blümke, A., Somerville, S. C., and Voigt, C. A. (2013). Transient expression of the Arabidopsis thaliana callose synthase PMR4 increases penetration resistance to powdery mildew in barley. Adv. Biosci. Biotechnol. 4, 810–813. doi: 10.4236/abb.2013.48106
Botha, C. E. J., and Matsiliza, B. (2004). Reduction in transport in wheat (Triticum aestivum) is caused by sustained phloem feeding by the Russian wheat aphid (Diuraphis noxia). South African J. Bot. 70, 249–254. doi: 10.1016/S0254-6299(15)30242-8
Cui, W., and Lee, J. Y. (2016). Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat. Plants 2, 1–9. doi: 10.1038/nplants.2016.34
Degefu, Y., and Hanif, M. (2003). Agrobacterium-tumefaciens-mediated transformation of Helminthosporium turcicum, the maise leaf-blight fungus. Arch. Microbiol. 180, 279–284. doi: 10.1007/s00203-003-0589-5
Dong, X., Hong, Z., Chatterjee, J., Kim, S., and Verma, D. P. S. (2008). Expression of callose synthase genes and its connection with Npr1 signaling pathway during pathogen infection. Planta 229, 87–98. doi: 10.1007/s00425-008-0812-3
Ellinger, D., and Voigt, C. A. (2014). Callose biosynthesis in arabidopsis with a focus on pathogen response: what we have learned within the last decade. Ann. Bot. 114, 1349–1358. doi: 10.1093/aob/mcu120
Enrique, R., Siciliano, F., Favaro, M. A., Gerhardt, N., Roeschlin, R., Rigano, L., et al. (2011). Novel demonstration of RNAi in citrus reveals importance of citrus callose synthase in defence against Xanthomonas citri subsp. citri. Plant Biotechnol. J. 9, 394–407. doi: 10.1111/j.1467-7652.2010.00555.x
Erb, M., and Reymond, P. (2019). Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 70, 527–557. doi: 10.1146/annurev-arplant-050718-095910
Feng, J., Chen, Y., Xiao, X., Qu, Y., Li, P., Lu, Q., et al. (2021). Genome-wide analysis of the CalS gene family in cotton reveals their potential roles in fiber development and responses to stress. Peer J. 9:e12557. doi: 10.7717/peerj.12557
Fromm, J., Hajirezaei, M. R., Becker, V. K., and Lautner, S. (2013). Electrical signaling along the phloem and its physiological responses in the maise leaf. Front. Plant Sci. 4:239. doi: 10.3389/fpls.2013.00239
Granato, L. M., Galdeano, D. M., D’Alessandre, N. D. R., Breton, M. C., and Machado, M. A. (2019). Callose synthase family genes plays an important role in the Citrus defense response to Candidatus Liberibacter asiaticus. Eur. J. Plant Pathol. 155, 25–38. doi: 10.1007/s10658-019-01747-6
Hao, P., Liu, C., Wang, Y., Chen, R., Tang, M., Du, B., et al. (2008). Herbivore-induced callose deposition on the sieve plates of rice: An important mechanism for host resistance. Plant Physiol. 146, 1810–1820. doi: 10.1104/pp.107.111484
Hofmann, J., Youssef-Banora, M., De Almeida-Engler, J., and Grundler, F. M. W. (2010). The role of callose deposition along plasmodesmata in nematode feeding sites. Mol. Plant Microbe Interact. 23, 549–557. doi: 10.1094/MPMI-23-5-0549
Hong, Z., Delauney, A. J., and Verma, D. P. S. (2001). A cell plate-specific callose synthase and its interaction with phragmoplastin. Plant Cell 13, 755–768. doi: 10.1105/tpc.13.4.755
Hunter, K., Kimura, S., Rokka, A., Tran, H. C., Toyota, M., Kukkonen, J. P., et al. (2019). CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDα1. Plant Physiol. 180, 2004–2021. doi: 10.1104/pp.19.00560
Jacobs, A. K., Lipka, V., Burton, R. A., Panstruga, R., Strizhov, N., Schulze-Lefert, P., et al. (2003). An Arabidopsis Callose synthase, GSL5, is required for wound and papillary Callose formation. Plant Cell 15, 2503–2513. doi: 10.1105/tpc.016097
Jin, L., and Mackey, D. M. (2017). Measuring callose deposition, an indicator of cell wall reinforcement, during bacterial infection in Arabidopsis. Methods Mol. Biol. 1578, 195–205. doi: 10.1007/978-1-4939-6859-6_16
Kempema, L. A., Cui, X., Holzer, F. M., and Walling, L. L. (2007). Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol. 143, 849–865. doi: 10.1104/pp.106.090662
Kim, J. H., and Jander, G. (2007). Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J. 49, 1008–1019. doi: 10.1111/j.1365-313X.2006.03019.x
Kuśnierczyk, A., Winge, P., Jørstad, T. S., Troczyńska, J., Rossiter, J. T., and Bones, A. M. (2008). Towards global understanding of plant defence against aphids - timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell Environ. 31, 1097–1115. doi: 10.1111/j.1365-3040.2008.01823.x
Lei, J., Finlayson, S. A., Salzman, R. A., Shan, L., and Zhu-Salzman, K. (2014). BOTRYTIS-INDUCED KINASE1 modulates Arabidopsis resistance to green peach aphids via PHYTOALEXIN DEFICIENT4. Plant Physiol. 165, 1657–1670. doi: 10.1104/pp.114.242206
Li, B., Förster, C., Robert, C. A. M., Züst, T., Hu, L., Machado, R. A. R., et al. (2018). Convergent evolution of a metabolic switch between aphid and caterpillar resistance in cereals. Sci. Adv. 4. doi: 10.1126/sciadv.aat6797
Liu, J., Du, H., Ding, X., Zhou, Y., Xie, P., and Wu, J. (2017). Mechanisms of callose deposition in rice regulated by exogenous abscisic acid and its involvement in rice resistance to Nilaparvata lugens Stål (Hemiptera: Delphacidae). Pest Manag. Sci. 73, 2559–2568. doi: 10.1002/ps.4655
Liu, F., Zou, Z., and Dilantha Fernando, W. G. (2018). Characterisation of callose deposition and analysis of the callose synthase gene family of Brassica napus in response to Leptosphaeria maculans. Int. J. Mol. Sci. 19, 3796. doi: 10.3390/ijms19123769
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Maag, D., Köhler, A., Robert, C. A. M., Frey, M., Wolfender, J. L., Turlings, T. C. J., et al. (2016). Highly localised and persistent induction of Bx1-dependent herbivore resistance factors in maise. Plant J. 88, 976–991. doi: 10.1111/tpj.13308
Meihls, L. N., Handrick, V., Glauser, G., Barbier, H., Kaur, H., Haribal, M. M., et al. (2013). Natural variation in maise aphid resistance is associated with 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell 25, 2341–2355. doi: 10.1105/tpc.113.112409
Norkunas, K., Harding, R., Dale, J., and Dugdale, B. (2018). Improving agroinfiltration-based transient gene expression in Nicotiana benthamiana. Plant Methods 141, 1–14. doi: 10.1186/s13007-018-0343-2
O’Lexy, R., Kasai, K., Clark, N., Fujiwara, T., Sozzani, R., and Gallagher, K. L. (2018). Exposure to heavy metal stress triggers changes in plasmodesmatal permeability via deposition and breakdown of callose. J. Exp. Bot. 69, 3715–3728. doi: 10.1093/jxb/ery171
Paul Barratt, D. H., Kölling, K., Graf, A., Pike, M., Calder, G., Findlay, K., et al. (2011). Callose synthase GSL7 is necessary for normal phloem transport and inflorescence growth in Arabidopsis. Plant Physiol. 155, 328–341. doi: 10.1104/pp.110.166330
Pu, Y., Hou, L., Guo, Y., Ullah, I., Yang, Y., and Yue, Y. (2019). Genome-wide analysis of the callose enzyme families of fertile and sterile flower buds of the Chinese cabbage (Brassica rapa L. ssp. pekinensis). FEBS Open Bio. 9, 1432–1449. doi: 10.1002/2211-5463.12685
Richmond, T. A., and Somerville, C. R. (2000). The cellulose synthase superfamily [2]. Plant Physiol. 124, 495–498. doi: 10.1104/pp.124.2.495
Sánchez-Sánchez, H., and Morquecho-Contreras, A. (2017). Chemical Plant Defense Against Herbivores. Herbivores 1–28. doi: 10.5772/67346
Schober, M. S., Burton, R. A., Shirley, N. J., Jacobs, A. K., and Fincher, G. B. (2009). Analysis of the (1,3)-beta-D-glucan synthase gene family of barley. Phytochemistry 70, 713–720. doi: 10.1016/j.phytochem.2009.04.002
Slewinski, T. L., Frank Baker, R., Stubert, A., and Braun, D. M. (2012). Tie-dyed2 encodes a callose synthase that functions in vein development and affects symplastic trafficking within the phloem of maise leaves. Plant Physiol. 160, 1540–1550. doi: 10.1104/pp.112.202473
Sun, M., Voorrips, R. E., Steenhuis-Broers, G., van’t Westende, W., and Vosman, B. (2018). Reduced phloem uptake of Myzus persicae on an aphid resistant pepper accession. BMC Plant Biol. 18, 1–14. doi: 10.1186/s12870-018-1340-3
Verma, D. P. S., and Hong, Z. (2001). Plant callose synthase complexes. Plant Mol. Biol. 476, 693–701. doi: 10.1023/a:1013679111111
Voigt, C. A. (2014). Callose-mediated resistance to pathogenic intruders in plant defense-related papillae. Front. Plant Sci. 5:168. doi: 10.3389/fpls.2014.00168
Wang, Y., Li, X., Fan, B., Zhu, C., and Chen, Z. (2021). Regulation and function of defense-related callose deposition in plants. Int. J. Mol. Sci. 22, 1–15. doi: 10.3390/ijms22052393
Wang, X., Sager, R., Cui, W., Zhang, C., Lu, H., and Lee, J. Y. (2013). Salicylic acid regulates Plasmodesmata closure during innate immune responses in Arabidopsis. Plant Cell 25, 2315–2329. doi: 10.1105/tpc.113.110676
Xie, B., Wang, X., Zhu, M., Zhang, Z., and Hong, Z. (2011). CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J. 65, 1–14. doi: 10.1111/j.1365-313X.2010.04399.x
Yu, Y., Jiao, L., Fu, S., Yin, L., Zhang, Y., and Lu, J. (2016). Callose synthase family genes involved in the grapevine defense response to downy mildew disease. Phytopathology 106, 56–64. doi: 10.1094/PHYTO-07-15-0166-R
Zavaliev, R., Ueki, S., Epel, B. L., and Citovsky, V. (2011). Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma 248, 117–130. doi: 10.1007/s00709-010-0247-0
Keywords: GhCalS5, cotton, callose, plant response, aphid resistance
Citation: Mbiza NIT, Hu Z, Zhang H, Zhang Y, Luo X, Wang Y, Wang Y, Liu T, Li J, Wang X, Zhang J and Yu Y (2022) GhCalS5 is involved in cotton response to aphid attack through mediating callose formation. Front. Plant Sci. 13:892630. doi: 10.3389/fpls.2022.892630
Edited by:
Carlos Augusto Avila, Texas A&M Research, United StatesReviewed by:
Vamsi J. Nalam, Colorado State University, United StatesFreddy Ibanez-Carrasco, Texas A&M AgriLife Research and Extension Center at Weslaco, United States
Copyright © 2022 Tanatsiwa Mbiza, Hu, Zhang, Zhang, Luo, Wang, Wang, Liu, Li, Wang, Zhang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianmin Zhang, MzcxOTk1OTY2QHFxLmNvbQ==; Yonghao Yu, eXhwMTEyN0AxNjMuY29t
 Natasha Isabel Tanatsiwa Mbiza
Natasha Isabel Tanatsiwa Mbiza Zongwei Hu1
Zongwei Hu1 Jianmin Zhang
Jianmin Zhang