
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 09 May 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.888425
This article is part of the Research Topic Insights in Plant Biotechnology: 2021 View all 22 articles
 Tawni Bull1,2
Tawni Bull1,2 Richard Michelmore1,3*
Richard Michelmore1,3*
In vitro plant regeneration involves dedifferentiation and molecular reprogramming of cells in order to regenerate whole organs. Plant regeneration can occur via two pathways, de novo organogenesis and somatic embryogenesis. Both pathways involve intricate molecular mechanisms and crosstalk between auxin and cytokinin signaling. Molecular determinants of both pathways have been studied in detail in model species, but little is known about the molecular mechanisms controlling de novo shoot organogenesis in lettuce. This review provides a synopsis of our current knowledge on molecular determinants of de novo organogenesis and somatic embryogenesis with an emphasis on the former as well as provides insights into applying this information for enhanced in vitro regeneration in non-model species such as lettuce (Lactuca sativa L.).
Plants have evolved a remarkable ability to regenerate tissues from differentiated organs, which involves the conversion of one cell type to others. Such plasticity provides the ability to regenerate whole organs and plants via dedifferentiation of cells and reprogramming of cell fates. There are three main types of regeneration: (1) Tissue regeneration, (2) de novo organogenesis, and (3) somatic embryogenesis (Xu and Huang, 2014; Sugimoto et al., 2019). Bryophytes have high capacity for tissue regeneration; for example, Marchantia spp. are capable of regenerating new meristems within their thallus (Yasui et al., 2019). However, vascular plants follow different regeneration pathways, which include de novo organogenesis or somatic embryogenesis (Figure 1). De novo organogenesis involves the regeneration of whole organs that did not previously exist. There are two types of de novo organogenesis: direct and indirect regeneration. Direct regeneration involves the development of organs directly from explants, whereas indirect regeneration involves an intermediate undifferentiated callus phase. For example, some plants, such as Jatropha curcas and succulents of the Cactaceae and Crassulaceae families (Preece, 2003; Severino et al., 2011), are capable of direct regeneration of new roots and shoots from stem cuttings. In contrast, many plants, such as lettuce, exhibit indirect organogenesis and regenerate shoots from calli (Michelmore and Eash, 1985). Somatic embryogenesis involves the regeneration of embryo or embryo-like structures from somatic cells, which can develop into a whole plant. In all forms of regeneration, cells must undergo dedifferentiation or transdifferentiation (reprogramming) into a more totipotent cell, ultimately changing the fate of the progenitor cell.
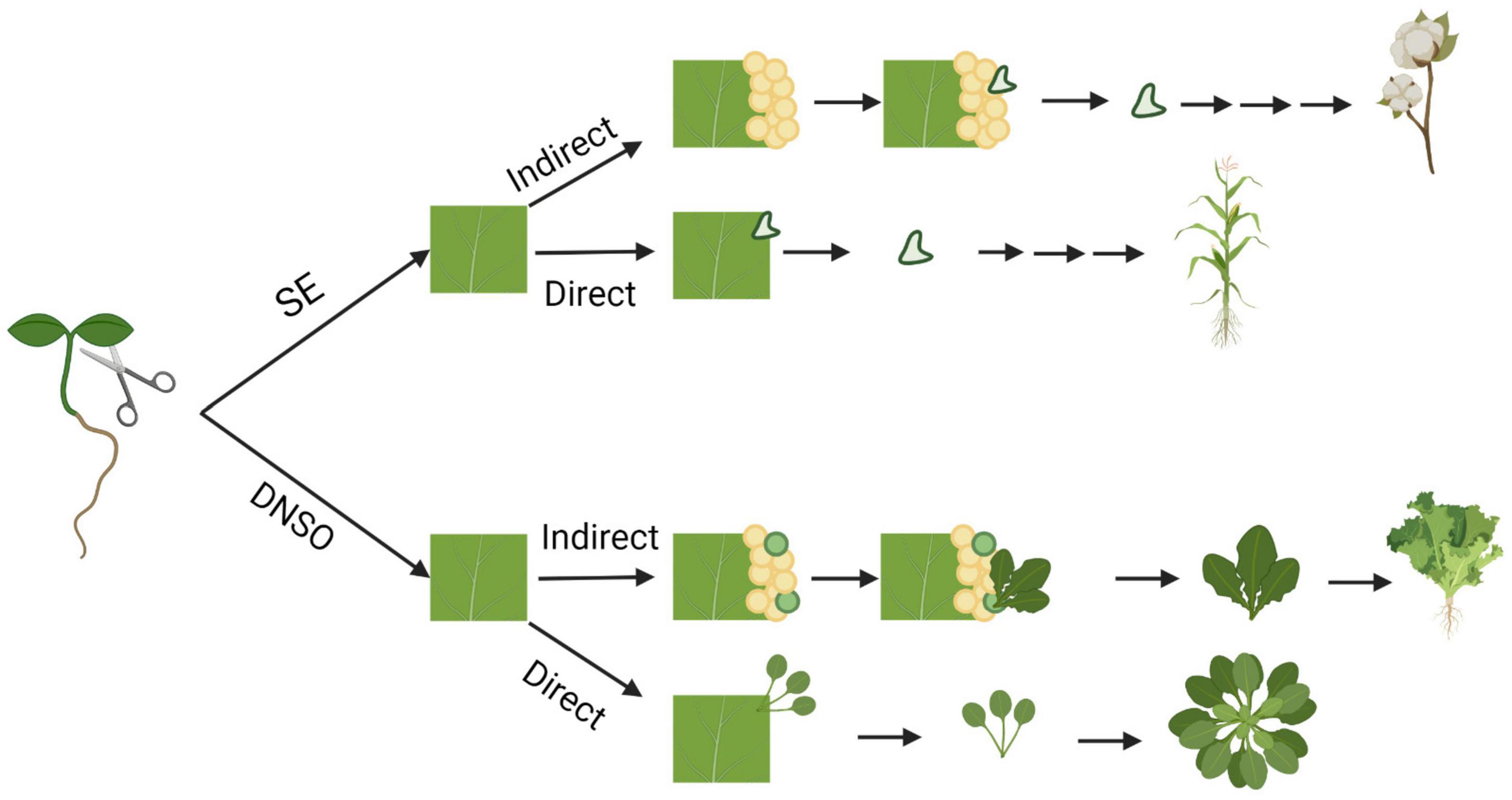
Figure 1. Pathways of in vitro regeneration of vascular plants. Somatic embryogenesis (SE) and de novo shoot organogenesis (DNSO) can occur directly on the explant or indirectly with the formation of pluripotent callus as an intermediate step. Species that are capable of regeneration for each pathway are represented from top to bottom: cotton, maize, Arabidopsis, and lettuce. Figure created using BioRender (https://biorender.com/).
Plant tissue culture and totipotency were first proposed by Haberlandt in 1902 (Krikorian and Berquam, 1969; Thorpe, 2007), who attempted to culture isolated photosynthetic leaf cells. Although this proved unsuccessful, it was the start of many decades of work on developing and improving plant tissue culture methods for multiple plant species. These failed experiments led to the development of root cultures using root tip cells in tomato and bud cultures. In 1904, embryo culture was first successful when embryos of crucifers (Brassicaceae) were isolated aseptically and grown in culture (Norstog, 1979). The first “true” plant tissue cultures were obtained on Knop’s medium from cambial tissues of sycamore maple (Acer psuedoplatanus) by Gautheret in 1934. This approach was optimized by additions of auxin, indole acetic acid (IAA), and B vitamins. This resulted in tissues that could be grown indefinitely in culture and the regeneration of both roots and shoots (Gautheret, 1934, 1935, 1939). The previous studies, however, used explant tissues that already contained meristematic cells. It was not until 1948 that methods were developed to induce roots and shoots from non-meristematic explants (Skoog and Tsui, 1948). This drastically increased the number of species that could be studied using in vitro culture systems (Miller et al., 1955; Skoog and Miller, 1957), and led to the recognition of the importance of exogenous ratios of cytokinin and auxin in culture medium. The differing ratios were shown to affect cell fate transition to either rooting or shooting from callus cells (Skoog and Miller, 1957), where high ratios of auxin to cytokinin promoted root regeneration, low ratios of auxin to cytokinin promoted shoot regeneration, and intermediate levels promoted proliferation of callus tissues. From the early to mid-1900s, research helped develop common plant tissue culture methods and media still used today (van Overbeek et al., 1941; Gautheret, 1942, 1955; Nobe’court, 1955). The earliest plant tissue culture media were based on nutrient necessities of whole plants, with the most common being Knop’s solution (Loomis and Schull, 1937). Numerous studies were conducted to optimize culture medium and in 1962, Murashige and Skoog reported a medium (MS salts) containing salt concentrations 25 times higher than those in Knop’s solution; in particular this resulted in much higher concentrations of NO3− and NH4+. The development of MS salts is still considered to be a major breakthrough in tissue culture because MS salts are still commonly used in plant tissue culture. The combination of exogenous plant hormones and appropriate salts allowed the study of basic plant biology questions about cell behavior, genetic improvement, disease biology, germplasm conservation, and clonal propagation.
Plant tissue culture to achieve in vitro regeneration was originally used to answer fundamental questions in plant biology but has since evolved to be foundational for genetic improvement, micropropagation, genetic engineering, and biotechnology (Michelmore et al., 1987; Zhang et al., 2006; Loberant and Altman, 2010; Xu and Huang, 2014; Chokheli et al., 2020). However, in vitro regeneration is not possible for all plant species and regeneration is very genotype dependent. Therefore, studying the molecular determinants of plant regeneration and exploiting these signaling pathways for improved in vitro regeneration of those recalcitrant genotypes and species is important. This review provides a synopsis of our current understanding of the pathways involved in de novo organogenesis and somatic embryogenesis. We focus on what is known of the molecular determinants of indirect de novo shoot organogenesis, which is the mode of regeneration in lettuce (Lactuca sativa L.). Finally, we describe future directions for improvement of in vitro regeneration of lettuce and other Compositae species.
Recently, many advances have been made toward understanding the cellular and molecular mechanisms that underlie plant regeneration (Xu and Huang, 2014; Ikeuchi et al., 2016; Sugimoto et al., 2019). Each of the regeneration processes described above have been studied in detail in model plants such as Arabidopsis thaliana. Each process entails a complex of molecular players involved in signaling and developmental pathways that regulate the dedifferentiation (somatic embryogenesis) or reprogramming (de novo organogenesis) of cells.
Callus formation is the first step in indirect organogenesis. Based on morphology, calli are thought to be the result of the dedifferentiation of cells to form totipotent cells. Callus can originate from the initiation of multiple pathways that contain some overlap in gene expression (Fehér, 2019) and can be auxin or wound induced (Fehér, 2019). In Arabidopsis, auxin induced calli resemble pluripotent cells similar to root tip cells at the molecular level and originate from pluripotent pericycle cells located adjacent to xylem poles (Atta et al., 2009; Sugimoto et al., 2010; Fehér, 2019). Root cell-like, auxin-induced callus follows a similar pathway as lateral root formation. In contrast, wound-induced callus does not involve players of lateral root formation, but rather occurs via upregulation of cytokinin signaling (Iwase et al., 2011a,b; Ikeuchi et al., 2017). Due to the similarity of gene expression patterns during callus formation with other developmental pathways some consider it a form of transdifferentiation rather than dedifferentiation (Fehér, 2019).
Many genes and transcription factors that are involved in lateral root development are also critical players in auxin-induced callus formation (Figure 2). For example, the LATERAL ORGAN BOUNDARIES (LBD) family of genes, such as LBD16, 17, 18, and 29, are critical to both lateral root formation and callus production (Fan et al., 2012; Feng et al., 2012; Xu et al., 2012; Lee H.W. et al., 2019). Ectopic expression of LBD genes led to the spontaneous formation of callus without exogenous applications of auxin and cytokinin, and repression of LBD16 showed inhibited callus formation (Fan et al., 2012). In lateral root formation, LBD16 and LBD29 are positively regulated by AUXIN RESPONSE FACTOR7 (ARF7) and ARF19, which provides evidence that ARFs are also involved in callus formation (Okushima et al., 2007). Furthermore, JUMONJI C DOMAIN CONTAINING PROTEIN 30 (JMJ30) interacts with ARF7 and ARF19 and directly binds to cis elements of LBD16 and LBD29, promoting their expression (Lee et al., 2018). Other key players in both lateral root and callus formation are ABERRANT LATERAL ROOT FORMATION 4 (ALF4) and SOLITARY ROOT/IAA14 (SLR/IAA14). ALF4 is involved in the earliest divisions of pericycle cells during lateral root formation. In alf4 mutants, callus-forming capability was lost in multiple tissues (DiDonato et al., 2004; Sugimoto et al., 2010). It was later shown that ALF4 is targeted for downregulation by CALLUS FORMATION RELATED-1 (CRF-1), which encodes an enzyme involved in very long chain fatty acid (VLCFA) biosynthesis (Shang et al., 2016). Another molecule involved in VLCFA biosynthesis is the AP2 transcription factor, PUCHI, which is also a key regulator controlling cell proliferation in lateral root primordia; puchi-1 mutants resulted in both defective and disorganized lateral root and callus formation further indicating a link between these pathways (Trinh et al., 2019). SLR is a member of the auxin signaling protein family Aux/IAA, and slr-1 mutants in A. thaliana were defective in both lateral root and callus formation (Fukaki et al., 2002; Shang et al., 2016). The functions of these genes and transcription factors provides evidence that callus formation and lateral root development have similar underlying mechanisms. In addition, callus formation can be initiated via a wound-induced signaling pathway and activation of a cytokinin response. Transcription factors involved during wound-induced callus formation include APETALA2/Ethylene Responsive Factor (AP2/ERF)-type transcription factors, WOUND-INDUCED DEDIFFERENTIATION1 (WIND1), and homologs (Iwase et al., 2011a,b, 2013). In Arabidopsis, expression of WIND1 and homologs are upregulated upon wounding and promote pluripotent callus formation at cut sites (Iwase et al., 2011a,b). Expression of Arabidopsis WIND1 was also shown to induce callus formation in other species such as rapeseed, tomato, and tobacco (Iwase et al., 2013). A transcriptome analysis showed WIND1 activates over 2,000 genes involved in multiple pathways including wound-induced cellular reprogramming and vascular formation (Iwase et al., 2021).
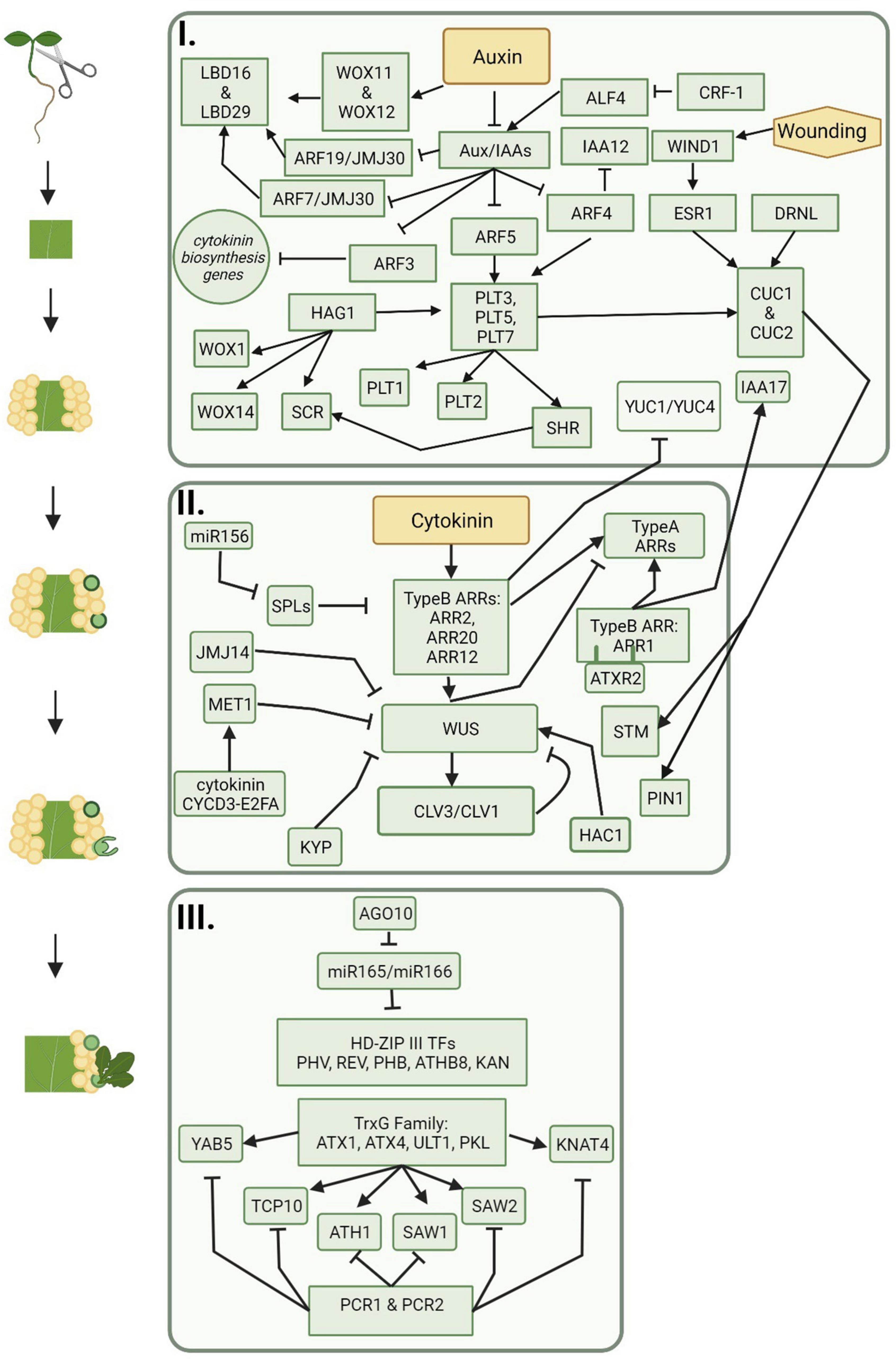
Figure 2. The progression of molecular players during indirect de novo shoot organogenesis. Callus is formed on auxin rich medium and includes signaling pathways represented in box one. Shoot promersitems and meristematic centers are formed on cytokinin rich medium and include signaling pathways represented in box two. Shoot regeneration follows meristem formation and is represented by the signaling pathways included in box three. Figure created using BioRender (https://biorender.com/).
Among the genes upregulated by WIND1 are those encoding for other AP2/ERF-type transcription factors including PLETHORA (PLT) genes (Kareem et al., 2015; Iwase et al., 2021). PLT genes work through the auxin signaling pathway, are often transcribed in response to auxin accumulation, and are activated downstream of ARF7 and ARF19 (Aida et al., 2004; Hofhuis et al., 2013). PLT3, PLT5, and PLT7 upregulate PLT1 and PLT2, which are important players in the regulation of lateral root formation, root apical meristem maintenance (RAM), and callus pluripotency (Xu et al., 2006; Durgaprasad et al., 2019). In Arabidopsis, PLT1 is also upregulated by JANUS through the recruitment of RNA Polymerase II to the root meristem (Xiong et al., 2020). In addition to root meristem maintenance, PLT proteins play important roles in conjunction with BABYBOOM/PLT4 (BBM/PLT4) in early embryogenesis (described further in section “Somatic Embryogenesis”), and activate regeneration responses in shoot organs by regulating the shoot promoting factors CUPPED-SHAPED COTYLEDON1 (CUC1) and CUC2 (Radhakrishnan et al., 2020). PLT-CUC2 together work through the auxin biosynthesis pathway and are essential for proper distribution and repolarization of auxin through PIN-FORMED (PIN) proteins (described further in section “De novo Root Organogenesis”) (Kareem et al., 2015; Shimotohno et al., 2018; Radhakrishnan et al., 2020).
Callus formation also involves epigenetic regulators. One regulator, HISTONE ACETYLTRANSFERASE OF THE GNAT/MYST SUPERFAMILY 1 (HAG1), also known as A. thaliana GENERAL CONTROL NON-REPRESSED 5 (AtGCN5), acts upstream of PLT1 and PLT2 (Kornet and Scheres, 2009; Kim et al., 2018). HAG1 also epigenetically upregulates root meristem genes WUSHCEL RELATED HOMEOBOX 5 (WOX5), WOX14, and SCARECROW (SCR) by acetylating the N terminus of histone 3. HAG1 is further involved in determining the root–shoot axis in embryo development and is a regulator of floral meristem activity (Kim et al., 2018). The RAM gene, ROOT CLAVATA-HOMOLOG 1 (RCH1), is also highly expressed in callus (Sugimoto et al., 2010), providing further evidence of homologies between lateral root development and callus formation. Although initiation of callus can follow multiple pathways, this provides further evidence that each pathway contains overlapping players.
De novo root organogenesis is the process by which adventitious roots are formed from detached plant tissues such as leaves and stems. Multiple studies have investigated the regeneration of the RAM in A. thaliana (Tian et al., 2002; Casamitjana-Martínez et al., 2003; Galinha et al., 2007; de Smet et al., 2008; Müller and Sheen, 2008; Perilli et al., 2012). The quiescent center (QC) is the site of stem cell maintenance of the RAM that is regenerated after QC ablation or entire removal of the root tip; polar transportation of auxin driven by PIN-FORMED (PIN) proteins results in auxin accumulation in cells adjacent to the damaged QC cells, which drives the reprogramming to new QC cells (van den Berg et al., 1997; Wildwater et al., 2005).
One of the key molecular players in root organogenesis is auxin. In Arabidopsis, auxin accumulates at cut sites, which induces expression of the homeobox transcription factors WOX11 and WOX12 (Liu et al., 2014). WOX11 and WOX12 directly upregulate WOX5, LBD16, and LBD29, marking the first step in cell differentiation and the formation of root meristems (Goh et al., 2012; Liu et al., 2014; Hu and Xu, 2016). Auxin accumulation at wound sites in Arabidopsis drives the expression of PLT genes (as seen in callus formation), which will in turn upregulate SHORT ROOT (SHR) (Kareem et al., 2015). The SHR proteins will localize to the nucleus, inducing the expression of SCR; SHR and SCR are both involved in QC identity and radial patterning (van den Berg et al., 1997; Wildwater et al., 2005). SCR and PLT work together with plant-specific teosinte-branched cycloidea PNCP (TCP) in PLT-TCP-SCR complexes to promote the organization of PIN proteins and expression of WOX5 in new meristem QC cells (Xu et al., 2006; Shimotohno et al., 2018). Root primordia formation is inhibited in shr, plt1, and plt2 mutants, indicating that these genes play an important role during root formation from root founder cells (Bustillo-Avendaño et al., 2018).
Shoot organogenesis may occur with direct regeneration of a shoot from an explant or indirect regeneration from a callus (Figure 1). Because a callus seems to resemble root tip cells rather than shoot cells at the molecular level, callus cells must undergo changes in gene expression that push the cells toward shoot development rather than root development. Shoot regeneration has been studied extensively in plants; however, while many genes and hormones have been identified as important players in the process (Figure 2), the detailed molecular interactions and pathways are unclear (reviewed in Xu et al., 2006; Su and Zhang, 2014; Xu and Huang, 2014; Ikeuchi et al., 2016; Lardon and Geelen, 2020).
Regeneration of shoots from callus requires the formation of a primary meristem or a shoot apical meristem (SAM) (Figure 3). Similar to the RAM, the SAM contains a population of pluripotent stem cells that give rise to all aboveground organs of a plant. The undifferentiated state of the organizing center (OC), which is similar to the RAM QC, and surrounding stem cells is maintained by a feedback mechanism between WUSHEL (WUS) and the signaling peptide CLAVATA3 (CLV3) (Sarkar et al., 2007). Leaves and other lateral organs arise from the peripheral regions of the SAM and the stem arises from the basal cells, called the rib zone. The SAM also contains the central zone, which consists of a stem cell pool that will replenish cells in the peripheral and rib zones that have further differentiated (Bowman and Eshed, 2000; Kwiatkowska, 2004). Unlike auxin accumulation in the RAM, the SAM contains high levels of cytokinins. Organization of auxin and cytokinin in cells help promote differentiation of pluripotent cells to either shoot or root cells.
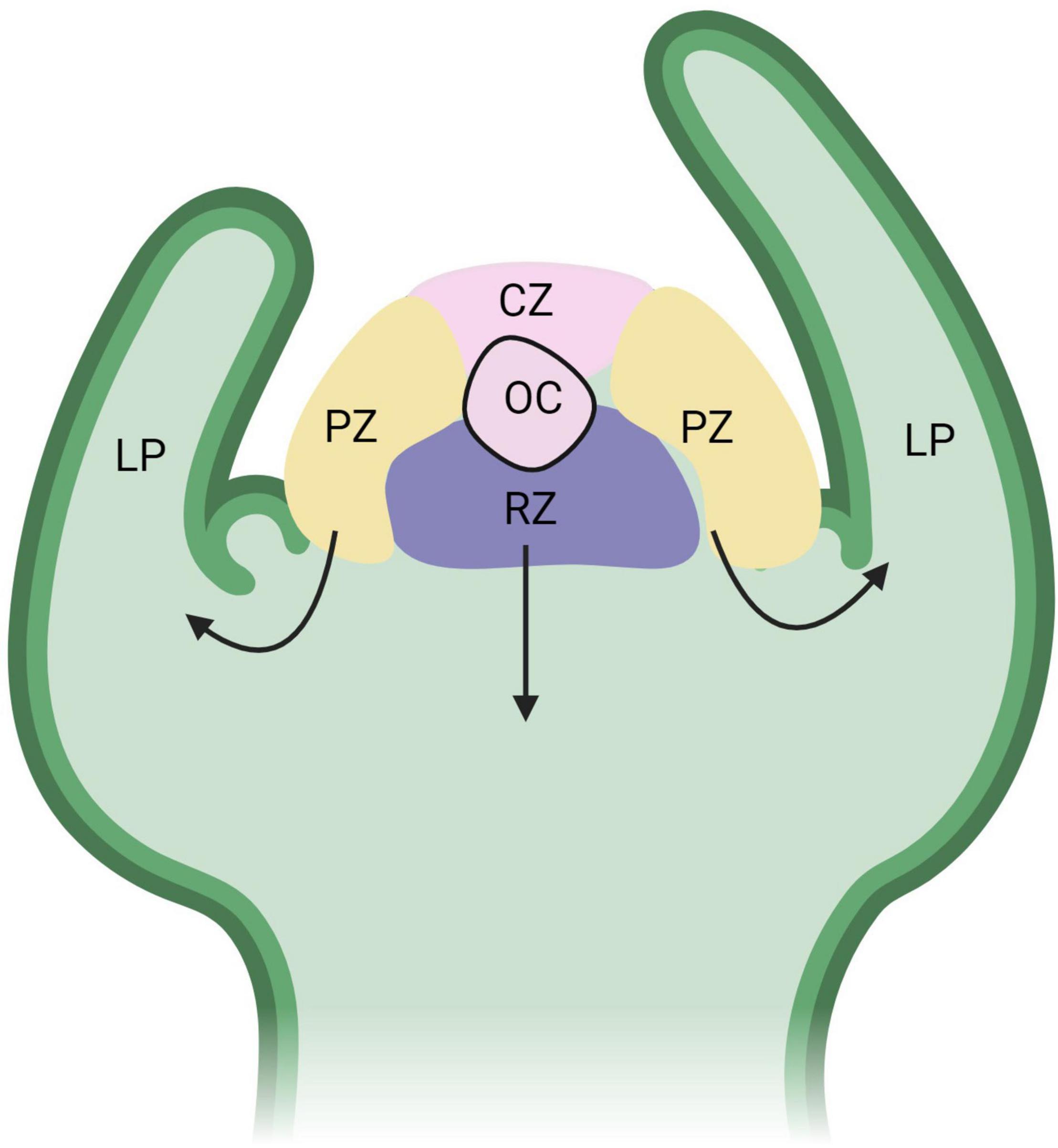
Figure 3. Functional domains of the shoot apical meristem (SAM). The organizing center (OC) is part of the central zone (CZ), which consists of a stem cell pool that replenishes cells to the peripheral zone (PZ) and rib zone (RB). The black arrows represent the direction of differentiating cells from the PZ to form leaf primordia (LP) and the RZ to form the stem. WUS expression is high in the OC and is regulated by CLV3/CLV1 from the CZ in a negative feedback loop. Figure created using BioRender (https://biorender.com/).
Shoot regeneration from callus occurs in four stages: (1) Formation of a pluripotent callus, (2) shoot promeristem formation, (3) shoot progenitor development, and (4) shoot regeneration (Shin et al., 2020). The development of a pluripotent callus cell mass (section “Synopsis of Studies on the Regeneration of Lettuce”) that highly expresses the No Apical Meristem/A. thaliana activating factor (NAC) transcription factor genes, CUC1 and CUC2 (Gordon et al., 2007), transitions into promeristems composed of a primary meristem of actively dividing cells. Within the callus CUC2 expression marks pre-meristematic regions by promoting cell proliferation and leading to the localized upregulation of a key shoot meristem regulator, SHOOT MERSITEMLESS (STM), and PIN1. As seen in de novo root organogenesis, PIN1 proteins polarly localize, denoting areas of cellular reprogramming toward promeristematic cells (Gordon et al., 2007). Both STM and PIN1 aid in the development of radial patterning as STM marks the promeristem and PIN1 marks primordia (Gordon et al., 2007). Because PIN1 proteins are important players in both promeristem formation and root de novo organogenesis, this suggests that auxin transport is important for both shoot and root meristem patterning.
Proper regulation and distribution of CUC1, CUC2, and WUS are critical for shoot progenitor cells. These NAC transcription factors are subject to upstream regulation during shoot promeristem formation. AP2/ERF-type transcription factors, ENHANCER OF SHOOT REGENERATION 1 (ESR1)/DORNROSCHEN (DRN) and ESR2/DRN-LIKE (DRNL) participate in upstream regulation of CUC genes by actively binding to the promoter and inducing expression (Banno et al., 2001; Kirch et al., 2003; Ikeda et al., 2006). Mutants of esr1, esr2, and esr1 esr2 show a reduction in shoot regeneration. This is likely due to improper regulation of CUC1 and CUC2 (Matsuo et al., 2011). WIND1 also upregulates ESR1 by directly binding to the vascular-responsive motifs in the ESR1 promoter (Iwase et al., 2017), suggesting that WIND1 is important in multiple plant regeneration processes. PLT5 and PLT7, which are induced during callus production, also influence the expression of CUC1 and CUC2 (Kareem et al., 2015). This further suggests that the molecular players and pathways involved in shoot regeneration are intertwined.
WUSCHEL (WUS) is a key regulator of the SAM and is upregulated during shoot regeneration. Expression of WUS is an important part of the conversion of a promeristem to a shoot progenitor as it represses cell division, cell elongation, and auxin-induced expression. This directs cell fate toward shoot development rather than root development. Ectopic expression of AtWUS results in de novo meristem formation and organogenesis in multiple plant species including Arabidopsis (Gallois et al., 2002; Negin et al., 2017), rice (Victorathisayam and Sridevi, 2020), and cotton (Bouchabké-Coussa et al., 2013). WUS expression is restricted to high cytokinin domains, while CUC2 expression tends to be restricted to low cytokinin and high auxin domains. This is consistent with high expression of CUC2 during induction of callus on media using higher concentrations of auxin (Daimon et al., 2003; Kareem et al., 2015). Regulation of WUS is subject to epigenetic regulation. METHYLTRANSFERASE1 (MET1), KRYPTONITE (KYP), histone acetyl transferase1 (HAC1), and JMJ14 are all required for proper expression of WUS, SAM organization, and shoot development (Li et al., 2011; Ishihara et al., 2019). MET1 is induced by the cytokinin-CYCD3-E2FA module, which represses WUS expression, allowing cells to retain callus identity rather than transitioning to shoot cells. However, in later stages of de novo shoot organogenesis, MET1 is spatially regulated, allowing for an increase in WUS expression in the inner cell layers of the callus (Liu et al., 2018). Previously, it was thought that WUS expression in the inner callus cell layers is directly activated by the cytokinin-responsive Type B ARABIDOPSIS RESPONSE REGULATORS (ARRS), ARR1, ARR2, ARR10, and ARR12 (Dai et al., 2017). However, a recent study showed that ARR1 is a strong inhibitor of callus formation and shoot regeneration. This occurs through indirect repression of CLV3 by competitive binding with ARR12 (Liu et al., 2020). ARR1 also indirectly represses WUS by inducing expression of the auxin response repressor gene INDOLE-3-ACETIC ACID INDUCIBLE17 (IAA17) (Liu et al., 2020). In addition, Type-B ARRs negatively regulate the expression of the auxin biosynthetic genes YUCCA1 (YUC1) and YUC4 (Meng et al., 2017). This results in indirect upregulation of WUS expression. Although it has been known for decades that auxin and cytokinin signaling is important for plant regeneration, these findings further untangle the underlying mechanisms of the signaling pathways.
Eukaryotic stem cells tend to have open chromatin states, while differentiated cells tend to have closed chromatin states (Shchuka et al., 2015). Epigenetic controls include Trithorax group (trxG) and Polycomb Group (PcG) proteins. The A. thaliana trxG, ATXR2, interacts with ARR1 and methylates the Type A ARRs, ARR5 and ARR7, marking them for increased transcription. This leads to a repression of cytokinin signaling and a reduction in de novo shoot organogenesis (Lee et al., 2021). PcG protein complexes, specifically POLYCOMB REPRESSIVE COMPLEX1 (PRC1) and PRC2, are chromatin modifiers and bind to Polycomb Response Elements (PRE) to keep genes transcriptionally repressed in order to fine-tune the balance between cell proliferation and cell differentiation (Köhler and Hennig, 2010). PRC2 suppresses leaf identity via H3K27me3 of leaf identity genes. PRC2 is also involved in callus formation as PRC2 mutants curly leaf swinger (clf swn) and embryonic flower2 (emf2) are incapable of developing callus from leaf and cotyledon explants but retained the ability to form callus in root explants (He et al., 2012). This suggests PCR2 represses leaf identify genes, allowing for the transition to root-like callus cells. Other instances of epigenetic regulation during the early stages of regeneration include gene priming by LYSINE-SPECIFIC DEMETHYLASE 1-LIKE 3 (LDL3), which involves the elimination of methylation of lysine 4 on histone 3 (H3K4me2) during callus formation. This indirectly promotes the expression of genes that are involved in shoot progenitor development (Ishihara et al., 2019).
Regulatory microRNA, miR156, plays a role in activating cytokinin signaling by targeting SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL). SPL genes control transitions in shoot development—juvenile-to-adult and vegetative-to-reproductive—by binding to and regulating Type-B ARRs, decreasing shoot regenerative capacity with age (Xu et al., 2015, 2016). miRNA156 expression is higher in younger tissues, which partially explains why younger explant tissue (i.e., cotyledons) is more amenable to in vitro regeneration, when compared to more mature tissue types. Type B ARRs and WUS also regulate the Type A ARRs, ARR7 and ARR15, which negatively regulate cytokinin signaling (Buechel et al., 2010).
After proper development of shoot progenitor cells, activation of leaf identity genes will lead to the development of leaf tissues and leaf emergence. Two important players involved in shoot regeneration are miR165 and miR166, both of which target HD-ZIP III transcription factor genes PHABULOSA (PHB), PHAVOLUTA (PHV), REVOLUTA (REV), KANADI (KAN), and ARABIDOPSIS THALIANA HOMEOBOX GENE 8 (ATHB8) (Shin et al., 2020). PHB, PHV, REV, and KAN function in radial leaf patterning (abaxial vs. adaxial), and phb, phv, rev, and kan mutants show a transition of abaxial leaf fates into adaxial leaf fates as well as altered auxin gradients (McConnell et al., 2001; Emery et al., 2003; Zhou et al., 2019). ATHB8 and SHR expression activate simultaneously and lead to leaf vein precursor cells (Gardiner et al., 2010). An RNA-induced silencing complex, ARGONAUTE10 (AGO10), helps sequester and repress the activity of miR165 and miR166. This indirectly promotes the activity of these leaf identity genes. Interestingly, accumulation of miR165/166 in overexpressing Arabidopsis mutants resulting in less HD-ZIP III transcription factor activity in shoot progenitor cells, increased the overall shoot regeneration (Xue et al., 2017). This suggests that leaf identity genes work to suppress in vitro transition from meristematic cells into shoot cells. In addition, AGO10 is repressed by LBD12, resulting in reduced apical meristem size (Ma et al., 2017). Leaf identity genes are also subject to epigenetic regulation. TrxG proteins, ATX1, ATX4, ULTRAPETALA1 (ULT1), and PICKLE (PKL), act as antagonists of PCR1 and PCR2 to activate transcription of leaf identity genes, which will aid in the development of leaves from shoot progenitor cells (Köhler and Hennig, 2010). In A. thaliana, ATX4 protein tri-methylates histone 3 (H3K4me3) to increase the expression of the shoot identity genes ARABIDOPSIS THALIANA HOMEOBOX GENE 1 (ATH1), KNOTTED1-LIKE HOMEOBOX (KNOX) GENE 4 (KNAT4), SAWTOOTH 1 (SAW1), SAW2, TCP FAMILY TRANSCRIPTION FACTOR 10 (TCP10), and YABBY 5 (YAB5) (Lee K. et al., 2019).
As elaborated above, de novo shoot regeneration is controlled by a complex network of genetic and epigenetic factors. Although we are gaining a more detailed understanding of the molecular players involved in this network via forward and reverse genetic approaches, there is clearly more information to discover involving interactions between these genetic, epigenetic, and hormone signaling pathways.
Formation of embryogenic callus results from acquisition of a new cell fate through expression of embryonic regulators. Similar to organogenic calli, embryogenic calli have been observed to originate from cells surrounding vascular tissue (pre-procambial cells) (de Almeida et al., 2012). Endogenous application of plant growth regulators such as auxin and cytokinin have been shown to induce proliferation of embryonic tissues in some species, such as soybean and cotton (Raza et al., 2020; Elhiti and Stasolla, 2022). This is similar to auxin-induced callus formation suggesting upregulation of ARFs such as ARF7 and ARF19 are also requirements for the formation of embryonic callus. Furthermore, LEAFY COTYLEDON1 (LEC1) and LEC2 genes are major embryonic regulators that control embryo maturation and development (Gaj et al., 2005). LEC1 overexpression induced embryogenic callus formation in Arabidopsis; however, lec1 and lec2 mutants resulted in the development of fewer somatic embryos via only indirect somatic embryogenesis (Gaj et al., 2005). This suggests that LEC1 is sufficient, but not necessary to the formation of embryogenic callus. Overexpression of the MADS-box transcription factor, AGAMOUS-LIKE 15 (AGL15), induced embryogenic callus-like structures on SAMs and extended embryonic development from callus in Arabidopsis (Harding et al., 2003). Expression of specific genes and presence of proteins have been observed in embryogenic callus, but not observed in non-embryogenic callus. The MADS-box gene, CUS1, whose amino acid sequence is highly similar to Arabidopsis AGL1 and AGL5 amino acid sequences, was detected in embryogenic callus during somatic embryogenesis in cucumber (Filipecki et al., 1997). Additionally, in sugar cane, unique proteins were identified during embryonic callus formation including proteins related to metabolic activity and stress (Schuabb Heringer et al., 2015). Induction of somatic embryogenesis and formation of proembyrogenic masses on calli (section “Somatic Embryogenesis”) involves different molecular players than formation of promeristems during organogenesis, but differences between embryogenic calli and organogenic calli formation, if any, are still not well characterized.
A second type of in vitro regeneration is somatic embryogenesis. Somatic embryogenesis results when a differentiated somatic cell undergoes molecular changes and genetic/epigenetic reprogramming resulting in the formation of a bipolar somatic embryo. In tissue culture, somatic embryogenesis can be induced in response to the addition of exogenous plant growth regulators or the introduction of stressful conditions. Similar to de novo organogenesis, somatic embryogenesis may originate directly at wound sites of explants or indirectly from embryogenic callus (Quiroz-Figueroa et al., 2006). Species tend to regenerate either through de novo organogenesis (e.g., tomato, lettuce, pepper) or somatic embryogenesis (e.g., cotton, wheat, rice) but rarely both (e.g., chickpea, purple coneflower) (Ozias-akins and Vasil, 1982; Michelmore et al., 1987; Rueb et al., 1994; Murthy et al., 1996; Choffe et al., 2000; Leelavathi et al., 2004; Heidmann et al., 2011; Sun et al., 2015).
Regulators and genetic determinants of embryo initiation are not well understood, although auxin signaling and accumulation are thought to play a major role. In tissue culture, addition of auxin is used to induce somatic embryogenesis by exposure of explants to high levels of auxin immediately followed by a transfer to auxin-free medium (Méndez-Hernández et al., 2019). This allows for the formation of auxin gradients within the developing embryos—areas of high auxin promote WUS expression, which denote areas of future SAM development as mentioned previously (Ikeuchi et al., 2016). In Arabidopsis, several ARFs are both up and downregulated during the first steps of somatic embryogenesis, and multiple arf mutants showed inhibited somatic embryogenesis (Wójcikowska and Gaj, 2017). SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES (SERKs), specifically SERK1 in Arabidopsis, are upregulated in embryonic callus and are continually expressed from megasporogenesis until the heart stage of the embryonic development (Hecht et al., 2001). Other genes, such as auxin-responsive gene EgIAA9 from Elaeis guineensis, have been shown to be upregulated during somatic embryogenesis initiation (Ooi et al., 2012).
The transcription factor BABY BOOM (BBM) and the LEC1-AB13-FUS3-LEC2 (LAFL) complex are master regulators of somatic embryogenesis (Horstman et al., 2017; Jones et al., 2019). BBM encodes an AINTEGUMENTA-LIKE (AIL) AP2/ERF and directly regulates all LAFL genes. LAFL genes are also regulated by a BBM-like protein, PLT2 (Horstman et al., 2017). The LAFL gene group consists of the LEC transcription factor genes, including LEC1, LEC2, and FUSCA3 (FUS3), and the ABA signaling transcription factor, ABSCISIC ACID INSENSITIVE 3 (ABI3). Somatic embryogenesis events are shown to significantly decrease in lec mutants (Gaj et al., 2005), and the overexpression of LEC2 led to an increase in the expression of auxin biosynthesis genes IAA30, YUC2, YUC4, and YUC10 (Stone et al., 2008; Junker et al., 2012), suggesting that LEC genes and the LAFL complex help promote auxin activity. LEC2 also induces the expression of LEC1, LEAFY COTYLEDON 1-LIKE (L1L), ABI3, and FUS3. Another transcription factor, AGL15, has been shown to directly regulate LAFL (Zheng et al., 2009) and promote the expression of the AP2/ERF gene At5g61590 (Zheng et al., 2013). At5g61590 is a relative of the Medicago truncatula SOMATIC EMBRYO-RELATED FACTOR 1 (MtSERF1), which is essential for somatic embryogenesis (Mantiri et al., 2008). Recently, another MADS-box transcription factor, AGL18, was identified as an active regulator in somatic embryogenesis in Arabidopsis (Paul et al., 2022). Overexpression of AGL18 resulted in an increase in somatic embryogenesis, while a decrease was observed in agl18 mutants; agl15 agl18 double mutants resulted in even less frequent development of somatic embryos. While the functions of AGL15 and AGL18 transcription factors were relatively redundant, different gene targets for each transcription factor were present and an AGL15/AGL18 regulatory loop was identified. This provides evidence that AGL18 may act in conjunction with AGL15 during somatic embryogenesis. Along with BBM, LAFL, and AGL15, the ectopic expression of WUS, PLT4/BBM, PLT5/EMBRYMAKER, MYB118, and RWP-RK DOMAIN-CONTAINING4 (RKD4)/GROUNDED (GRD) leads to the induction of somatic embryogenesis in Arabidopsis (Lotan et al., 1998; Boutilier et al., 2002; Harding et al., 2003; Gallois et al., 2004; Waki et al., 2011).
The master regulators work with other transcription factors to balance auxin, gibberellin (GA), and abscisic acid (ABA) signaling. In particular, the balance of GA and ABA has a major role in controlling cell identity in the developing embryo. Embryonic cells have been shown to have a higher ratio of GA to ABA than somatic cells (Yamaguchi et al., 2001; Mitchum et al., 2006; Hu et al., 2008). The LAFL transcription factors, LEC1, LEC2, FUS3, and AGL15, downregulate GA biosynthesis genes (Curaba et al., 2004; Zheng et al., 2009), while FUS3 positively regulates the ABA pathway (Gazzarrini et al., 2004). LEC1 and LEC2 promote the expression of auxin biosynthesis genes (Braybrook et al., 2006; Junker et al., 2012), and AGL15 negatively regulates the auxin response genes, ARF6, ARF8, and TRANSPORT INHIBITOR RESPONSE1 (TIR1) (Zheng et al., 2016). LEC1 and AGL15 positively regulate ABI3. Generally, these transcription factors work to negatively regulate GA biosynthesis and positively regulate ABA and auxin biosynthesis, transitioning cells from embryonic cells (high GA/ABA ratios) into differentiated somatic cells (low GA/ABA ratios). MYB-family transcription factors, MYB118 and MYB115, also play important roles in somatic embryogenesis. These transcription factors promote the expression of LEC1; overexpression of both resulted in the formation of somatic embryos on root explants (Wang et al., 2008). The micro RNA miR396 is associated with somatic embryogenesis induction and helps control PLT1 and PLT2 (Szczygieł-Sommer and Gaj, 2019). Other evidence suggests that AGL15 forms protein complexes with SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES (SERKs), which are induced in response to auxin (Zheng et al., 2009). Ethylene has also been shown to impact somatic embryogenesis because interactions between ETHYLENE RESPONSE FACTOR 002 (ERF022) and LEC2, and the involvement of other AP2/ERF transcription factors have been observed (Zheng et al., 2013; Xu and Huang, 2014; Horstman et al., 2017). Reprogramming of somatic cells to form embryos and then back to differentiated somatic cells requires multiple hormone signaling pathways to work together.
Genomic DNA methylation patterns change during development. In mature tissues, LEC1, LEC2, and AGL15 are hypermethylated in somatic cells, while hypomethylation has been seen of similar genes (e.g., SERKs, LEC2, WUS) in embryonic calli (Fraga et al., 2012). This suggests that somatic embryogenesis and genes involved in embryonic cell to somatic cell transition is subject to epigenetic regulation as the repression of embryonic genes leads to the development of mature and differentiated tissues. There is conflicting evidence for the role of DNA methylation in somatic embryogenesis. In some studies, the demethylation agent 5-azacitidine strongly inhibited embryogenesis in Medicago truncatula and Arabidopsis (Santos and Fevereiro, 2002; Grzybkowska et al., 2018), while in other plants, such as in Coco nucifera and Acca sellowiana, its application increased embryogenesis (Fraga et al., 2012; Osorio-Montalvo et al., 2020). This suggests that differential DNA methylation is required for successful somatic embryogenesis but its effects are highly genotype and species dependent.
Two critical regulatory epigenetic pathways, PcG and PKL, are involved in the epigenetic regulation of genes during somatic embryogenesis. As in shoot organogenesis, the PRC2-mediated H3K27 methylation, part of the PcG pathway, is involved in the repression of LEC1, LEC2, and FUS3, aiding in the transition from embryonic to somatic cells (Makarevich et al., 2006). The Repressive LEC2 Element (RLE) in the LEC2 promoter recruits PCR2 for methylation and repression of LEC2 in somatic cells (Berger et al., 2011). Evidence supporting this includes an increase in somatic embryogenesis of Arabidopsis in vegetative tissue depleted of PRC2 (Mozgová et al., 2017). PRC2 has also been shown to negatively regulate other important regulators of plant regeneration including WOX5, WOX11, WUS, and STM. PKL encodes for a chromatin remodeling factor, which serves to rearrange nucleosome positions in order to regulate gene expression. Multiple studies have demonstrated that pkl mutants show an increase in the ectopic expression of LEC1, LEC2, and FUS3, resulting in embryonic traits in somatic tissues (Ogas et al., 1997; Henderson et al., 2004). This suggests that PKL plays a role in negatively regulating embryonic genes in somatic tissues. However, the specific molecular mechanism by which PKL works is still unclear.
Signaling peptides are important players in plant development. One family of signaling peptides, CLAVATA/ENDOSPERM SURROUNDING REGION (CLE), has central roles in modulating stem cell differentiation during plant growth and development (Katsir et al., 2011). These peptides are post-translationally processed and contain a signal peptide targeting the protein for secretion, where it is used for cell-to-cell communication (Yamaguchi et al., 2016). In A. thaliana, there are 32 CLE peptides including CLV1, CLV2, and CLV3. CLV3 is secreted from cells and interacts with CLV1, a leucine-rich repeat receptor kinase, to maintain stem cell populations in the apical meristem (Clark et al., 1995; Hirakawa et al., 2008). In clv1 and clv3 mutants, plants develop enlarged shoot and floral apical meristems, suggesting improper signaling disruption to maintenance of stem cell populations (Clark et al., 1995). WUS promotes cell proliferation and division and upregulates CLV1-CLV3. This results in the downregulation of WUS by CLV1-CLV3 in a negative feedback loop. This feedback mechanism produces and maintains a constant stem cell pool (Mayer et al., 1998; Brand et al., 2000). Manipulating either WUS, CLV1, and/or CLV3 could lead to larger stem cell pools and greater potential for cell division. This in conjunction with downstream molecular players, such as CUC genes, PLT genes, or SPL, and could potentially lead to more and faster plant regeneration. However, this would require careful orchestration of the key regulators.
Growth Regulating Factors (GRF) are a transcription factor family that regulates many aspects of plant growth and development including leaf, stem, root, seed development, flowering, regulation of stress, and plant longevity. The first GRF, Os-GRF1, was identified two decades ago during a differential expression study of responses of deep-water rice to GA (van der Knaap et al., 2000). GRFs have now been identified in many plant species, where typically 8–20 different GRF genes are present in each genome (Omidbakhshfard et al., 2015). GRFs form complexes with their cofactor, GRF-interacting Factors (GIF), and will bind to cis-regulatory elements of different developmental genes in plants (Kim, 2019). For example, AtGRF7 binds to the promoter of the AP2/ERF gene Dehydration responsive element binding protein2A (DREB2A) and represses gene expression in leaf veins (Kim et al., 2012). In Arabidopsis, GRFs have been shown to be expressed in leaf and root tissue where prolific cell growth is occurring and tend to decrease with plant age (Kim et al., 2003; Lee et al., 2009; Hewezi et al., 2012; Szczygieł-Sommer and Gaj, 2019).
GRF proteins are post-transcriptionally regulated by miR396 throughout the course of plant development; miR396 recognizes and binds to GRF, resulting in degradation or translational arrest. Expression of miR396 occurs at low levels in leaf primordia that gradually increase throughout organ development and maturity (Rodriguez et al., 2010). Expression of AtGRF2 is restricted to specific portions of the leaf during development through antagonistic expression of miR396 (Rodriguez et al., 2010). In rice, miR396 mutants resulted in an upregulation of multiple GRF genes, in particular GRF3. These mutants also produced plants with longer leaves and shorter internodes (Miao et al., 2020). Because of their involvement in organ development, GRF and miR396 are potential targets for increasing in vitro regeneration.
GRFs regulate players important for in vitro regeneration. GRF proteins from rice, OsGRF3 and OSGRF10 repress promoter activity of a KNOX gene, Oskn2 (Kuijt et al., 2014). In the same study, barley GRF, BGRF1, repressed Hooded/Bkn3, a barley KNOX gene, and overexpression of OsGRF10, AtGRF4, AtGRF5, and AtGRF6 repressed activity of KNAT2 in Arabidopsis. In addition, overexpression of OsGRF3 and OsGRF10 induced root and shoot formation on primary tillers of rice (Kuijt et al., 2014). Because regulation of KNOX genes is necessary for cell identity transitions from meristem cells to mature organ cells (Hake et al., 1995, 2004; Tsuda et al., 2011), the reported functions of these GRFs demonstrate the potential importance of GRFs in both organogenesis and somatic embryogenesis. An RNA-seq study in rice showed upregulation of OsGRF6 resulted in an increase in expression of the auxin biosynthesis gene, OsYUCCA-like, and signaling genes, OsARF2, OsARF7, OsARF11 (Gao et al., 2015). In addition, altered expression of GRF and GIF affect root growth through regulation of PLT1, PLT2, and SCR (Ercoli et al., 2018). In Arabidopsis, the double mutant gif1/an3 gif2 and the triple mutant gif1/an3 gif2 gif3 both showed the formation of a disorganized QC and larger RAM, while overexpression of GRF3 with a mutated miRNA binding site (rGRF3) resulted in smaller meristems (Ercoli et al., 2018). These studies provide evidence that GRFs and GIFs are upstream regulators of molecular determinants involved in callus formation and shoot meristem identity, giving altered expression of GRFs and GIFs the potential to increase regeneration in plants.
GRFs and GIFs have now been shown to enhance regeneration capacity and rates in plants. Ectopic expression of AtGRF5 and orthologs increased callus production in canola and shoot organogenesis in sugar beet, soybean, and sunflower; also, ectopic expression of the maize GRF5 ortholog increased formation of embryogenic calli indicating that GRFs regulate multiple in vitro regeneration pathways (Kong et al., 2020). In addition, transformation with a chimeric GRF-GIF gene fusion can increase the rate and number of regenerates in wheat, rice, and citrus (Debernardi et al., 2020). Independent transformations and co-transformations of multiple wheat GRFs fused with GIFs were studied including GRF4, GRF5, GIF1, GIF2, and GIF3; the chimeric transgene composed of a fusion between GRF4 and GIF1 (GRF4-GIF1) resulted in the highest frequency of regeneration in wheat among all combinations of GRFs and GIFs tested. In addition to increased regeneration, shoot regeneration and transgenesis in wheat was successful without the use of cytokinins in the culture medium. Furthermore, regeneration could be induced from leaf explants rather than immature embryos. The efficacy of chimeric transgene was also tested in the dicotyledonous species, Citrus, using the Citrus and Vitis GRF4 and GIF1 homologs (Debernardi et al., 2020). Furthermore, the use of the microRNA insensitive rGRF4-GIF resulted in greater stimulation of regeneration in wheat, rice, and Citrus. This is a major breakthrough and will be exploited for the regeneration of recalcitrant species and cultivars, leading to a likelihood of higher transformation rates.
Lettuce, Lactuca sativa L. (Compositae), is a dicotyledonous plant that can be regenerated by indirect de novo shoot organogenesis (Figure 4) and was a model for early studies of regeneration (reviewed in Michelmore and Eash, 1985). Some genotypes regenerate readily on a variety of media formulations and growth regulators; however, some lettuce genotypes are recalcitrant to regeneration. Lettuce is also amenable to Agrobacterium-mediated transformation (Michelmore et al., 1987). Protocols for high efficiency, genotype-independent regeneration of lettuce are required in order to fully benefit from biotechnological approaches, including genome editing, for crop improvement. Given differences in regeneration rates of different genotypes and the wealth of knowledge from model species described above, top-down and bottom-up approaches to the molecular basis of regeneration in lettuce could lead to protocols for enhanced regeneration of multiple genotypes.
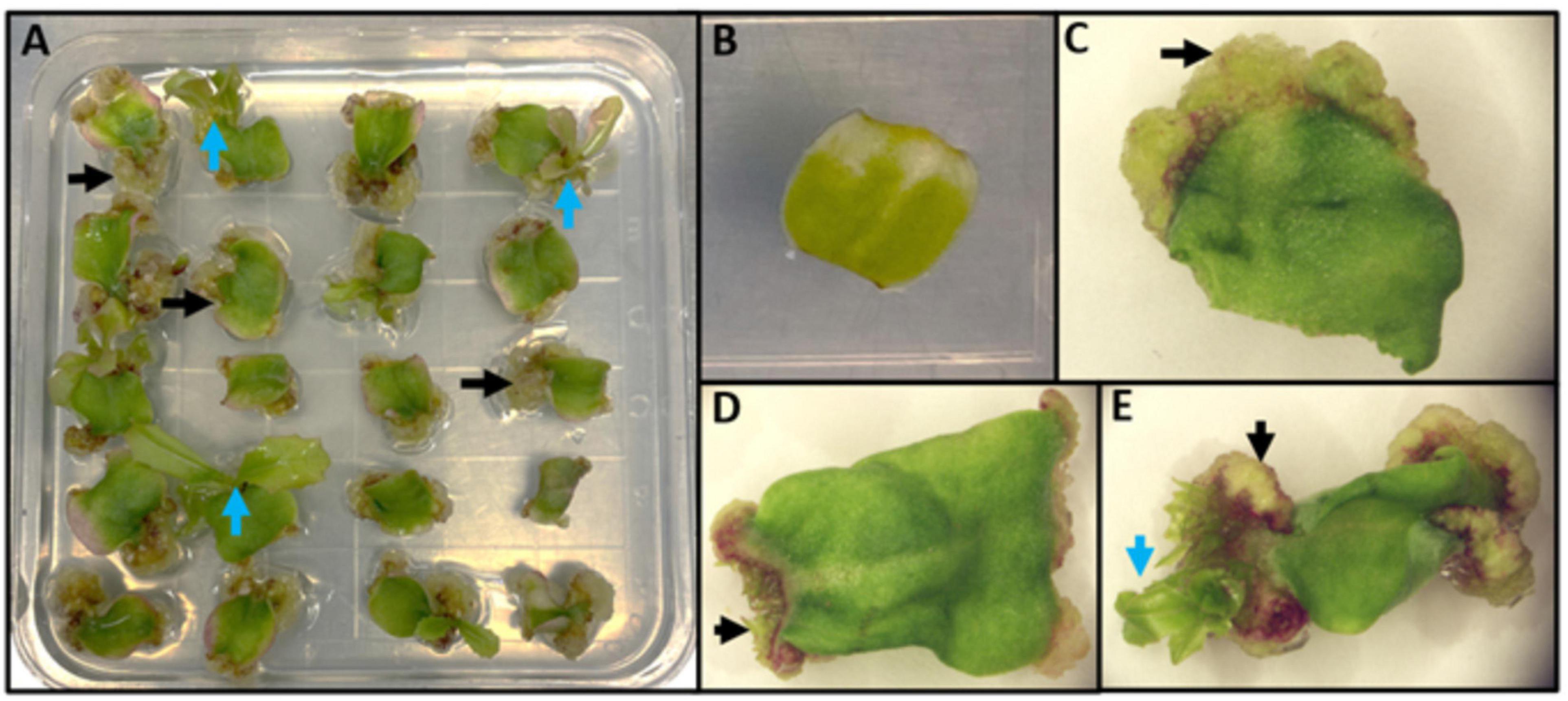
Figure 4. Representation of indirect de novo shoot organogenesis in lettuce. (A) A plate of 20 explants undergoing indirect de novo shoot regeneration. Black arrows represent friable callus formation at the wounded end of explants; blue arrows represent shoot regeneration from calli. (B) An explant before callus formation. (C) An explant during callus formation (black arrow). (D) First organized growth from callus (black arrow). (E) Indirect shoot regeneration (blue arrow) from callus (black arrow).
Lettuce regeneration has been studied for many decades. Lettuce was among the first plants to be tested for regeneration. The first studies on in vitro regeneration of lettuce failed to produce shoots from leaves of L. sativa and L. canadensis (LaRue, 1933, 1936). Later, regeneration of lettuce shoots was successful with the addition of adenine and kinetin to the growth medium (Doerschug and Miller, 1967). In this study, the regenerative capability of hypocotyl, cotyledon, and mature leaf explants was tested on the same base medium with different combinations of IAA, kinetin, and adenine, and cotyledons were shown to be the most effective explant source for shoot regeneration. In the same study, kinetin was effective at promoting the transition from callus formation to shoot regeneration (Doerschug and Miller, 1967). This suggested that in lettuce, as shown in other plant species, high levels of cytokinin promotes the formation of shoot meristems that results from the transition of cell fate from root-like callus cells to shoot cells. Later studies focused on the optimization of factors influencing lettuce regeneration, including media formulations, plant growth regulator use, light requirements, temperature, explant type, and genotype (Doerschug and Miller, 1967; Kadkade and Seibert, 1977; Koevary, 1978; Sasaki, 1979, 1982; Alconero, 1983; Webb et al., 1984; Michelmore and Eash, 1985). Light intensity and photoperiod were shown to be also important for lettuce regeneration; cotyledon explants developed well-formed shoots with a 16-h photoperiod but significantly fewer shoots formed in the dark; additionally, the presence of red light doubled the number of buds and shoots (Kadkade and Seibert, 1977). In aggregate, callus formation occurred on all lettuce cultivars studied when using both auxins and cytokinins in the culture medium, although there were differences between genotypes. Shoot regeneration was elicited when the medium contained cytokinins with little or no auxins. Although mature leaves and hypocotyls showed regenerative capabilities, cotyledons were the most amenable explant source for regeneration.
Indirect de novo shoot organogenesis in lettuce involves cell divisions of spongy, palisade, and epidermal cells. A cytohistological study of adventitious bud formation from cotyledon explants revealed initial divisions of spongy and palisade cells followed by divisions of epidermal cells to form tetrads (Nuti Ronchi and Gregorini, 1970). Callus was formed from the division of mesophyll cells and inward proliferation of epidermal cells. Subsequently, adventitious buds arose from one or two epidermal cells, which led to the formation and organization of shoot apical meristems. This study provided the timeline and steps that occur during organogenesis; however, the tools were not available to study the underlying genetic and molecular constituents responsible for the changes in cell anatomy and transition of cell fate, particularly epidermal cells to meristematic centers.
Like most plant species, regenerative capacity is highly dependent on genotype and there is considerable variation in regenerative capacity among lettuce cultivars (Michelmore et al., 1987; Curtis et al., 1994; Ampomah-Dwamena et al., 1997; Mohebodini et al., 2011). There is no significant correlation to regeneration efficiency and morphological group (i.e., crisphead, butterhead, cos, and leaf). In a side-by-side study, highly regenerating genotypes included Bambino (crisphead), Iceberg (crisphead), Cobham Green (butterhead), Sweet Butter (butterhead), Simpson Elite (leaf), Rosalita (cos), and Paris White (cos); recalcitrant genotypes included Oak Leaf (leaf), Royal Oak Leaf (leaf), Sangria (crisphead), and Mainspring (butterhead) (Ampomah-Dwamena et al., 1997). Generation of stable transgenics of lettuce relies on Agrobacterium-mediated transformation and in vitro regeneration. Therefore, it is important to understand and identify the genetic and molecular players to increase regeneration in order to manipulate recalcitrant lettuce varieties.
There have been few studies on the molecular determinants of regeneration in lettuce. A dominant mutation of the ethylene receptor ETR1-1 was shown to inhibit shoot regeneration in lettuce (Kim and Botella, 2004). Lettuce cultivars LEI26 and Seagreen were transformed using Agrobacterium-mediated transformation for the introduction of GUS under the control of the CaMV 35S constitutive promoter and the mutated ethylene receptor etr1-1 under the control of a leaf senescence-specific promoter, sag12. Transformations with 35S:GUS showed high regenerative potential with 85% of explants developing shoots, while the introduction of sag12:etr1-1 significantly reduced regenerative potential with only 2.86% of explants producing shoots. Explants transformed with sag:etr1-1 also stimulated root formation directly from cotyledon explants without the formation of callus (Kim and Botella, 2004). This suggests that ethylene responses are important in in vitro lettuce regeneration in which inhibiting ethylene receptors promotes root formation and inhibits callus and shoot formation. This is consistent with observations of other ethylene response factors during in vitro regeneration, such as the early expression of AP2/ERF transcription factors during callus formation and the involvement of ERF022 activity during somatic embryogenesis (Iwase et al., 2011a,b; Zheng et al., 2013; Xu and Huang, 2014; Horstman et al., 2017).
Data is limited for lettuce on the effects of the pathways and molecular determinants described in other species. A recent study examined the chronological expression of homeobox genes during in vitro regeneration of lettuce (Farina et al., 2021). Gene expression profiles of lettuce homologs to the homeobox WOX family transcription factor genes WUS (LsWUS1L and LsWUS2L) and the KNOTTED1-LIKE homeobox family transcription factor gene ST-M (LsSTM), were examined in cotyledon explants over 12 days on inductive medium. A time course analysis showed a steady increase of expression of LsWUS1; in early days of culture, increased expression of LsWUS2L correlated with the formation of poorly vacuolated cells with large nuclei in the explants. Expression of LsSTM1L also drastically increased in early days of culture, followed by a later decrease, suggesting that it helps recruit proteins and regulates expression of genes needed for the initiation of regeneration in lettuce (Farina et al., 2021). This parallels patterns of WUS and STM expression observed early in plant regeneration, specifically during the formation of shoot promeristems and meristematic centers from callus in Arabidopsis (Daimon et al., 2003; Zhang et al., 2017). This is also consistent with the essential role WUS plays in maintaining the stem cell pool that is critical for proper SAM function (Sarkar et al., 2007). The CCAAT-binding transcription factors, LEC1 and LEC2, play a major role in development and maturation of embryos (see sections “Embryogenic Callus Formation and Somatic Embryogenesis”). Nothing has been reported for homologs of LEC1 and LEC2 in lettuce. It would be interesting to overexpress homologs of these transcription factors in lettuce to determine if this results in enhanced regeneration as in Arabidopsis, tobacco, and cassava (Gaj et al., 2005; Guo et al., 2013; Brand et al., 2019). Similarly, over-expression of CUC1 and CUC2 as well as PLT genes (see sections “Organogenic Callus Formation, De novo Root Organogenesis, and De novo Shoot Organogenesis”) may also result in enhanced regeneration of lettuce as in Arabidopsis (Ikeda et al., 2006; Matsuo et al., 2009; Kareem et al., 2015).
MADS-box transcription factors, particularly AGL15 and AGL18, are major molecular players involved in in vitro regeneration (see sections “Embryogenic Callus Formation and Somatic Embryogenesis”). There are at least 82 MADS-box encoding genes in lettuce (Ning et al., 2019), most of which have been studied in relation to flowering time and floral development (reviewed in Han et al., 2021). Of these 82 genes, 23 encoded for M-type genes of the type 1 lineage and 59 floral genes of the type II lineage containing a MIKC domain. Within the type II MADs-box genes, 10 belonged to the AGL15 subfamily which contained homologs of Arabidopsis and tomato AGL15 genes. Currently, no work has been reported on the role of lettuce MADs-box genes during in vitro regeneration. The 10 genes identified in the AGL15 subfamily should be characterized for their roles in regeneration in lettuce; it should be tested whether over expression of ALG15 results in increased somatic embryogenesis as in Arabidopsis (Paul et al., 2022).
There are 15 GRF genes in lettuce and their chromosomal locations, gene structure, conserved motifs, and expression patterns have been characterized (Zhang et al., 2021). One GRF gene was studied in detail. LsaGRF5 showed low expression in leaves and roots with high expression in reproductive buds, suggesting an important function in flower development. The GRF regulator, miR396a, had high expression in mature flowers and stems and low expression in reproductive buds. These data suggest that high levels of LsaGRF5 expression in young tissues is coincident with actively dividing cells; as the cells and tissues mature, LsaGRF5 becomes downregulated by miR396a; this is similar to what is observed in other species (see section “Growth Regulating Factors as Agents for Increased Regeneration”). Furthermore, overexpression of LsaGRF5 resulted in larger leaf size, while overexpression of miR396a resulted in smaller leaf size (Zhang et al., 2021). However, none of the 15 GRF genes have been characterized for their effects on regeneration in lettuce. Given the success of GRF or GRF-GIF fusions with other species (see section “Growth Regulating Factors as Agents for Increased Regeneration”), it is likely that similar enhanced rates of regeneration and transformation will be reported soon.
The underlying processes of plant regeneration all involve cell fate transition by reprogramming gene expression. The several pathways involved in plant development and regeneration are complex. Although each pathway has unique molecular players, many of the key regulators overlap and have important functions in each. Auxin and cytokinin signaling pathways play a major role in regulating multiple regenerative pathways and accompany the genome-wide switch in gene expression profile during the early stages of regeneration. Other phytohormones, such as GA, ABA, and ethylene, also contribute to plant regeneration and cell fate transition.
Many of the players and regulators important for in vitro regeneration have been studied in model species, such as Arabidopsis, but have not been functionally characterized in non-model species such as lettuce. The complete genome sequence of L. sativa (Reyes-Chin-Wo et al., 2017) has provided useful genic targets for modification by genome editing. Currently, genome editing of lettuce requires Agrobacterium-mediated transformation, which requires in vitro regeneration; therefore, studying molecular determinants and understanding pathways controlling regeneration in lettuce has great value. Identifying orthologs of genes discussed in this review and then characterizing them in other systems, such as lettuce, will help form a more generalized understanding of in vitro regeneration in plants. Further studies on identification of recalcitrant varieties, quantitative trait locus analyses on varieties with varying regenerative capabilities, and expression profiles during in vitro regeneration could provide insight into other genes regulated during in vitro regeneration of lettuce. Understanding these pathways in lettuce will allow for a better understanding of the pathways in other important crops, particularly within the Compositae family such as sunflower, artichoke, safflower, and many ornamentals.
RM and TB conceived the idea for the manuscript. TB conducted the literature review and drafted the manuscript and figures. Both authors reviewed the final manuscript and approved submission.
This work was supported the USDA NIFA Specialty Crop Research Initiative (SCRI) (Grant Nos. 2015-51181-24283 and 2021-51181-35903) to RM.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Allen Van Deynze for providing input and Elizabeth Georgian for assistance in editing this manuscript.
Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., et al. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120. doi: 10.1016/j.cell.2004.09.018
Alconero, R. (1983). Regeneration of plants from cell suspensions of Lactuca saligna, Lactuca sativa, and Lactuca serriola. HortScience 18, 305–307.
Ampomah-Dwamena, C., Conner, A. J., and Fautrier, A. G. (1997). Genotypic response of lettuce cotyledons to regeneration in vitro. Sci. Horticult. 71, 137–145. doi: 10.1016/s0304-4238(97)00098-8
Atta, R., Laurens, L., Boucheron-Dubuisson, E., Guivarc’h, A., Carnero, E., Giraudat-Pautot, V., et al. (2009). Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 57, 626–644. doi: 10.1111/j.1365-313X.2008.03715.x
Banno, H., Ikeda, Y., Niu, Q.-W., and Chua, N.-H. (2001). Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13:2609. doi: 10.1105/tpc.010234
Berger, N., Dubreucq, B., Roudier, F., Dubos, C., and Lepiniec, L. (2011). Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 Involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell 23:4065. doi: 10.1105/tpc.111.087866
Bouchabké-Coussa, O., Obellianne, M., Linderme, D., Montes, E., Maia-Grondard, A., Vilaine, F., et al. (2013). Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 32, 675–686. doi: 10.1007/s00299-013-1402-9
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L., et al. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749. doi: 10.1105/tpc.001941
Bowman, J. L., and Eshed, Y. (2000). Formation and maintenance of the shoot apical meristem. Trends Plant Sci. 5, 110–115. doi: 10.1016/s1360-1385(00)01569-7
Brand, A., Quimbaya, M., Tohme, J., and Chavarriaga-Aguirre, P. (2019). Arabidopsis LEC1 and LEC2 orthologous genes are key regulators of somatic embryogenesis in cassava. Front. Plant Sci. 10:673. doi: 10.3389/fpls.2019.00673
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. doi: 10.1126/science.289.5479.617
Braybrook, S. A., Stone, S. L., Park, S., Bui, A. Q., Le, B. H., Fischer, R. L., et al. (2006). Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 3468–3473. doi: 10.1073/pnas.0511331103
Buechel, S., Leibfried, A., To, J. P. C., Zhao, Z., Andersen, S. U., Kieber, J. J., et al. (2010). Role of A-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur. J. Cell Biol. 89, 279–284. doi: 10.1016/j.ejcb.2009.11.016
Bustillo-Avendaño, E., Ibáñez, S., Sanz, O., Sousa Barros, J. A., Gude, I., Perianez-Rodriguez, J., et al. (2018). Regulation of hormonal control, cell reprogramming, and patterning during de novo root organogenesis. Plant Physiol. 176, 1709–1727. doi: 10.1104/pp.17.00980
Casamitjana-Martínez, E., Hofhuis, H. F., Xu, J., Liu, C. M., Heidstra, R., and Scheres, B. (2003). Root-specific CLE19 overexpression and the so1l/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13, 1435–1441. doi: 10.1016/s0960-9822(03)00533-5
Choffe, K. L., Victor, J. M. R., Murch, S. J., and Saxena, P. K. (2000). In vitro regeneration of Echinacea purpurea L.: direct somatic embryogenesis and indirect shoot organogenesis in petiole culture. In Vitro Cell. Dev. Biol. Plant 36, 30–36. doi: 10.1007/s11627-000-0008-4
Chokheli, V. A., Dmitriev, P. A., Rajput, V. D., Bakulin, S. D., Azarov, A. S., Varduni, T. V., et al. (2020). Recent development in micropropagation techniques for rare plant species. Plants 9:1733. doi: 10.3390/plants9121733
Clark, S. E., Running, M. P., and Meyerowitz, E. M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067. doi: 10.1242/dev.121.7.2057
Curaba, J., Moritz, T., Blervaque, R., Parcy, F., Raz, V., Herzog, M., et al. (2004). AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 136, 3660. doi: 10.1104/pp.104.047266
Curtis, I. S., Power, J. B., Blackhall, N. W., de Laat, A. M. M., and Davey, M. R. (1994). Genotype-independent transformation of lettuce using Agrobacterium tumefaciens. J. Exp. Bot. 45, 1441–1449. doi: 10.1093/jxb/45.10.1441
Dai, X., Liu, Z., Qiao, M., Li, J., Li, S., and Xiang, F. (2017). ARR12 promotes de novo shoot regeneration in Arabidopsis thaliana via activation of WUSCHEL expression. J. Integr. Plant Biol. 59, 747–758. doi: 10.1111/jipb.12567
Daimon, Y., Takabe, K., and Tasaka, M. (2003). The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol. 44, 113–121. doi: 10.1093/pcp/pcg038
de Almeida, M., Vieira De Almeida, C., Mendes, E., Gilvano, G., Brondani, E., and Fiori De Abreu-Tarazi, M. (2012). Pre-procambial cells are niches for pluripotent and totipotent stem-like cells for organogenesis and somatic embryogenesis in the peach palm: a histological study. Plant Cell Rep. 31, 1495–1515. doi: 10.1007/s00299-012-1264-6
de Smet, I., Vassileva, V., de Rybel, B., Levesque, M. P., Grunewald, W., van Damme, D., et al. (2008). Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322, 594–597. doi: 10.1126/science.1160158
Debernardi, J. M., Tricoli, D. M., Ercoli, M. F., Hayta, S., Ronald, P., Palatnik, J. F., et al. (2020). A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38, 1274–1279. doi: 10.1038/s41587-020-0703-0
DiDonato, R. J., Arbuckle, E., Buker, S., Sheets, J., Tobar, J., Totong, R., et al. (2004). Arabidopsis ALF4 encodes a nuclear-localized protein required for lateral root formation. Plant J. 37, 340–353. doi: 10.1046/j.1365-313x.2003.01964.x
Doerschug, M. R., and Miller, C. O. (1967). Chemical control of adventitious organ formation in Lactuca sativa explants. Am. J. Bot. 54, 410–413. doi: 10.1002/j.1537-2197.1967.tb10658.x
Durgaprasad, K., Roy, M. V., Venugopal M, A., Kareem, A., Raj, K., Willemsen, V., et al. (2019). Gradient expression of transcription factor imposes a boundary on organ regeneration potential in plants. Cell Rep. 29, 453–463.e3. doi: 10.1016/j.celrep.2019.08.099
Elhiti, M., and Stasolla, C. (2022). Transduction of signals during somatic embryogenesis. Plants 11:178. doi: 10.3390/plants11020178
Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A., et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774. doi: 10.1016/j.cub.2003.09.035
Ercoli, M. F., Ferela, A., Debernardi, J. M., Perrone, A. P., Rodriguez, R. E., and Palatnik, J. F. (2018). GIF transcriptional coregulators control root meristem homeostasis. Plant Cell 30, 347–359. doi: 10.1105/tpc.17.00856
Fan, M., Xu, C., Xu, K., and Hu, Y. (2012). LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 22, 1169–1180. doi: 10.1038/cr.2012.63
Farina, P., Fambrini, M., Pugliesi, C., and Viviani, A. (2021). Expression of homeobox genes during in vitro culture of Lactuca sativa. Plant Biosyst. 155, 609–621. doi: 10.1080/11263504.2020.1762793
Fehér, A. (2019). Callus, dedifferentiation, totipotency, somatic embryogenesis: what these terms mean in the era of molecular plant biology? Front. Plant Sci. 10:536. doi: 10.3389/fpls.2019.00536
Feng, Z., Zhu, J., Du, X., and Cui, X. (2012). Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta 236, 1227–1237. doi: 10.1007/s00425-012-1673-3
Filipecki, M. K., Sommer, H., and Malepszy, S. (1997). The MADS-box gene CUS1 is expressed during cucumber somatic embryogenesis. Plant Sci. 125, 63–74. doi: 10.1016/s0168-9452(97)00056-3
Fraga, H. P. F., Vieira, L. N., Caprestano, C. A., Steinmacher, D. A., Micke, G. A., Spudeit, D. A., et al. (2012). 5-Azacytidine combined with 2,4-D improves somatic embryogenesis of Acca sellowiana (O. Berg) Burret by means of changes in global DNA methylation levels. Plant Cell Rep. 31, 2165–2176. doi: 10.1007/s00299-012-1327-8
Fukaki, H., Tameda, S., Masuda, H., and Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29, 153–168. doi: 10.1046/j.0960-7412.2001.01201.x
Gaj, M. D., Zhang, S., Harada, J. J., and Lemaux, P. G. (2005). Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222, 977–988. doi: 10.1007/s00425-005-0041-y
Galinha, C., Hofhuis, H., Luijten, M., Willemsen, V., Blilou, I., Heidstra, R., et al. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057. doi: 10.1038/nature06206
Gallois, J.-L., Nora, F. R., Mizukami, Y., and Sablowski, R. (2004). WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 18, 375–380. doi: 10.1101/gad.291204
Gallois, J.-L., Woodward, C., Reddy, G. V., and Sablowski, R. (2002). Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development 129, 3207–3217. doi: 10.1242/dev.129.13.3207
Gao, F., Wang, K., Liu, Y., Chen, Y., Chen, P., Shi, Z., et al. (2015). Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat. Plants 2:15196. doi: 10.1038/nplants.2015.196
Gardiner, J., Donner, T. J., and Scarpella, E. (2010). Simultaneous activation of SHR and ATHB8 expression defines switch to preprocambial cell state in Arabidopsis leaf development. Dev. Dyn. 240, 261–270. doi: 10.1002/dvdy.22516
Gautheret, R. J. (1934). Culture du tissus cambial. Comptes Rendus Hebdomadaires Se’ances de l’Acad. Sci. 15, 2195–2196.
Gautheret, R. J. (1939). Sur la possibilite’ de re’aliser la culture inde’finie des tissus de tubercules de carotte. Comptes Rendus Hebdomadaires Se’ances de l’Acad. Sci. 208, 118–120.
Gautheret, R. J. (1942). He’te’ro-auxines et cultures de tissus ve’ge’taux. Bull. Soc. Chimie Biol. 24, 13–21.
Gautheret, R. J. (1955). Sur la variabilite’ des proprie’te’s physiologiques des cultures de tissues ve’ge’taux. Rev. Gen. Bot. 65, 5–112.
Gazzarrini, S., Tsuchiya, Y., Lumba, S., Okamoto, M., and McCourt, P. (2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7, 373–385. doi: 10.1016/j.devcel.2004.06.017
Goh, T., Joi, S., Mimura, T., and Fukaki, H. (2012). The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139, 883–893. doi: 10.1242/dev.071928
Gordon, S. P., Heisler, M. G., Reddy, G. V., Ohno, C., Das, P., and Meyerowitz, E. M. (2007). Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134, 3539–3548. doi: 10.1242/dev.010298
Grzybkowska, D., Morończyk, J., Wójcikowska, B., and Gaj, M. D. (2018). Azacitidine (5-AzaC)-treatment and mutations in DNA methylase genes affect embryogenic response and expression of the genes that are involved in somatic embryogenesis in Arabidopsis. Plant Growth Regul. 85, 243–256. doi: 10.1007/s10725-018-0389-1
Guo, F., Liu, C., Xia, H., Bi, Y., and Zhao, C. (2013). Induced expression of AtLEC1 and AtLEC2 differentially promotes somatic embryogenesis in transgenic tobacco plants. PLoS One 8:e71714. doi: 10.1371/journal.pone.0071714
Hake, S., Char, B. R., Chuck, G., Foster, T., Long, J., and Jackson, D. (1995). Homeobox genes in the functioning of plant meristems. Philos. Trans. R. Soc. B 350, 45–51. doi: 10.1098/rstb.1995.0136
Hake, S., Smith, H. M. S., Holtan, H., Magnani, E., Mele, G., and Ramirez, J. (2004). The role of KNOX genes in plant development. Annu. Rev. Cell Dev. Biol. 20, 125–151. doi: 10.1146/annurev.cellbio.20.031803.093824
Han, R., Truco, M. J., Lavelle, D. O., and Michelmore, R. W. (2021). A composite analysis of flowering time regulation in lettuce. Front. Plant Sci. 12:632708. doi: 10.3389/fpls.2021.632708
Harding, E. W., Tang, W., Nichols, K. W., Fernandez, D. E., and Perry, S. E. (2003). Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-like 15. Plant Physiol. 133:653. doi: 10.1104/pp.103.023499
He, C., Chen, X., Huang, H., and Xu, L. (2012). Reprogramming of H3K27me3 Is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 8:e1002911. doi: 10.1371/journal.pgen.1002911
Hecht, V., Vielle-Calzada, J.-P., Hartog, M. V., Schmidt, E. D. L., Boutilier, K., Grossniklaus, U., et al. (2001). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture 1. Plant Physiol. 127, 803–816. doi: 10.1104/pp.010324
Heidmann, I., de Lange, B., Lambalk, J., Angenent, G. C., and Boutilier, K. (2011). Efficient sweet pepper transformation mediated by the BABY BOOM transcription factor. Plant Cell Reports 30, 1107–1115. doi: 10.1007/s00299-011-1018-x
Henderson, J. T., Li, H. C., Rider, S. D., Mordhorst, A. P., Romero-Severson, J., Cheng, J. C., et al. (2004). PICKLE Acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol. 134, 995–1005. doi: 10.1104/pp.103.030148
Hewezi, T., Maier, T. R., Nettleton, D., and Baum, T. J. (2012). The Arabidopsis MicroRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol. 159, 321–335. doi: 10.1104/pp.112.193649
Hirakawa, Y., Shinohara, H., Kondo, Y., Inoue, A., Nakanomyo, I., Ogawa, M., et al. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. U.S.A. 105, 15208–15213. doi: 10.1073/pnas.0808444105
Hofhuis, H., Laskowski, M., Du, Y., Prasad, K., Grigg, S., Pinon, V., et al. (2013). Phyllotaxis and rhizotaxis in arabidopsis are modified by three PLETHORA transcription factors. Curr. Biol. 23, 956–962. doi: 10.1016/j.cub.2013.04.048
Horstman, A., Li, M., Heidmann, I., Weemen, M., Chen, B., Muino, J. M., et al. (2017). The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 175, 848–857. doi: 10.1104/pp.17.00232
Hu, J., Mitchum, M. G., Barnaby, N., Ayele, B. T., Ogawa, M., Nam, E., et al. (2008). Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell 20, 320–336. doi: 10.1105/tpc.107.057752
Hu, X., and Xu, L. (2016). Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis 1. Plant Physiol. 172, 2363–2373. doi: 10.1104/pp.16.01067
Ikeda, Y., Banno, H., Niu, Q.-W., Howell, S. H., and Chua, N.-H. (2006). The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 47, 1443–1456. doi: 10.1093/pcp/pcl023
Ikeuchi, M., Iwase, A., Rymen, B., Lambolez, A., Kojima, M., Takebayashi, Y., et al. (2017). Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol. 175, 1158–1174. doi: 10.1104/pp.17.01035
Ikeuchi, M., Ogawa, Y., Iwase, A., and Sugimoto, K. (2016). Plant regeneration: cellular origins and molecular mechanisms. Development 143, 1442–1451. doi: 10.1242/dev.134668
Ishihara, H., Sugimoto, K., Tarr, P. T., Temman, H., Kadokura, S., Inui, Y., et al. (2019). Primed histone demethylation regulates shoot regenerative competency. Nat. Commun. 10, 1–15. doi: 10.1038/s41467-019-09386-5
Iwase, A., Harashima, H., Ikeuchi, M., Rymen, B., Ohnuma, M., Komaki, S., et al. (2017). WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis. Plant Cell 29, 54–69. doi: 10.1105/tpc.16.00623
Iwase, A., Kondo, Y., Laohavisit, A., Takebayashi, A., Ikeuchi, M., Matsuoka, K., et al. (2021). WIND transcription factors orchestrate wound-induced callus formation, vascular reconnection and defense response in Arabidopsis. New Phytol. 232, 734–752. doi: 10.1111/nph.17594
Iwase, A., Mitsuda, N., Ikeuchi, M., Ohnuma, M., Koizuka, C., Kawamoto, K., et al. (2013). Arabidopsis WIND1 induces callus formation in rapeseed, tomato, and tobacco. Plant Signal. Behav. 8:e27432. doi: 10.4161/psb.27432
Iwase, A., Mitsuda, N., Koyama, T., Hiratsu, K., Kojima, M., Arai, T., et al. (2011a). The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 21, 508–514. doi: 10.1016/j.cub.2011.02.020
Iwase, A., Ohme-Takagi, M., and Sugimoto, K. (2011b). WIND1: A key molecular switch for plant cell dedifferentiation. Plant Signal. Behav. 6, 1943–1945. doi: 10.4161/psb.6.12.18266
Jones, T., Lowe, K., Hoerster, G., Anand, A., Wu, E., Wang, N., et al. (2019). Maize transformation using the morphogenic genes. Methods Mol. Biol. 1864, 81–93. doi: 10.1007/978-1-4939-8778-8_6
Junker, A., Mönke, G., Rutten, T., Keilwagen, J., Seifert, M., Thi, T. M. N., et al. (2012). Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 71, 427–442. doi: 10.1111/j.1365-313X.2012.04999.x
Kadkade, P., and Seibert, M. (1977). Phytochrome-regulated organogenesis in lettuce tissue culture. Nature 270, 49–50. doi: 10.1038/270049a0
Kareem, A., Durgaprasad, K., Sugimoto, K., Du, Y., Pulianmackal, A. J., Trivedi, Z. B., et al. (2015). PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 25, 1017–1030. doi: 10.1016/j.cub.2015.02.022
Katsir, L., Davies, K. A., Bergmann, D. C., and Laux, T. (2011). Peptide signaling in plant development. Curr. Biol. 21, R356–R364.
Kim, J., Yang, W., Forner, J., Lohmann, J. U., Noh, B., and Noh, Y. (2018). Epigenetic reprogramming by histone acetyltransferase HAG1/AtGCN5 is required for pluripotency acquisition in Arabidopsis. EMBO J. 37:e98726. doi: 10.15252/embj.201798726
Kim, J. H. (2019). Biological roles and an evolutionary sketch of the GRF-GIF transcriptional complex in plants. BMB Rep. 52:227. doi: 10.5483/BMBRep.2019.52.4.051
Kim, J. H., and Botella, J. R. (2004). Etr1-1 gene expression alters regeneration patterns in transgenic lettuce stimulating root formation. Plant Cell 78, 69–73. doi: 10.1023/b:ticu.0000020396.64257.c1
Kim, J. H., Choi, D., and Kende, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365-313x.2003.01862.x
Kim, J.-S., Mizoi, J., Kidokoro, S., Maruyama, K., Nakajima, J., Nakashima, K., et al. (2012). Arabidopsis GROWTH-REGULATING FACTOR7 functions as a transcriptional repressor of abscisic acid-and osmotic stress-responsive genes. including DREB2A W. Plant Cell 24, 3393–3405. doi: 10.1105/tpc.112.100933
Kirch, T., Simon, R., Grünewald, M., and Werr, W. (2003). The DORNRÖSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem cell fate and lateral organ development. Plant Cell 15:694. doi: 10.1105/tpc.009480
Köhler, C., and Hennig, L. (2010). Regulation of cell identity by plant polycomb and trithorax group proteins. Curr. Opin. Genet. Dev. 20, 541–547. doi: 10.1016/j.gde.2010.04.015
Kong, J., Martin-Ortigosa, S., Finer, J., Orchard, N., Gunadi, A., Batts, L. A., et al. (2020). Overexpression of the transcription factor GROWTH-REGULATING FACTOR5 improves transformation of dicot and monocot species. Front. Plant Sci. 11:572319. doi: 10.3389/fpls.2020.572319
Kornet, N., and Scheres, B. (2009). Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21, 1070–1079. doi: 10.1105/tpc.108.065300
Krikorian, A. D., and Berquam, D. L. (1969). Plant cell and tissue cultures: the role of HABERLANDT 1. Bot. Rev. 35, 59–67.
Kuijt, S. J. H., Greco, R., Agalou, A., Shao, J., ‘t Hoen, C. C. J., Övernäs, E., et al. (2014). Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 164, 1952–1966. doi: 10.1104/pp.113.222836
Kwiatkowska, D. (2004). Structural integration at the shoot apical meristem: models, measurements, and experiments. Am. J. Bot. 91, 1277–1293. doi: 10.3732/ajb.91.9.1277
Lardon, R., and Geelen, D. (2020). Natural variation in plant pluripotency and regeneration. Plants 9, 1–28. doi: 10.3390/plants9101261
LaRue, C. D. (1933). Regeneration in mutilated seedlings. Proc. Natl. Acad. Sci. U.S.A. 19, 53–63. doi: 10.1073/pnas.19.1.53
LaRue, C. D. (1936). Tissue culture of spermatophytes. Proc. Natl. Acad. Sci. U.S.A. 22, 201–209. doi: 10.1073/pnas.22.4.201
Lee, B. H., Ko, J. H., Lee, S., Lee, Y., Pak, J. H., and Kim, J. H. (2009). The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151, 655–668. doi: 10.1104/pp.109.141838
Lee, H. W., Cho, C., Pandey, S. K., Park, Y., Kim, M. J., and Kim, J. (2019). LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 19:46. doi: 10.1186/s12870-019-1659-4
Lee, K., Park, O. S., Choi, C. Y., and Seo, P. J. (2019). ARABIDOPSIS TRITHORAX 4 facilitates shoot identity establishment during the plant regeneration process. Plant Cell Physiol. 60, 826–834. doi: 10.1093/pcp/pcy248
Lee, K., Park, O. S., Go, J. Y., Yu, J., Han, J. H., Kim, J., et al. (2021). Arabidopsis ATXR2 represses de novo shoot organogenesis in the transition from callus to shoot formation. Cell Rep. 37:109980. doi: 10.1016/j.celrep.2021.109980
Lee, K., Park, O. S., and Seo, P. J. (2018). JMJ30-mediated demethylation of H3K9me3 drives tissue identity changes to promote callus formation in Arabidopsis. Plant J. 95, 961–975. doi: 10.1111/tpj.14002
Leelavathi, S., Sunnichan, V. G., Kumira, R., Vijaykanth, G. P., Bhatnagar, R. K., and Reddy, V. S. (2004). A simple and rapid Agrobacterium-mediated transformation protocol for cotton (Gossypium hirsutum L.): Embryogenic calli as a source to generate large numbers of transgenic plants. Plant Cell Rep. 22, 465–447. doi: 10.1007/s00299-003-0710-x
Li, W., Liu, H., Cheng, Z. J., Su, Y. H., Han, H. N., Zhang, Y., et al. (2011). DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet. 7:e1002243. doi: 10.1371/journal.pgen.1002243
Liu, H., Zhang, H., Dong, Y. X., Hao, Y. J., and Zhang, X. S. (2018). DNA METHYLTRANSFERASE1-mediated shoot regeneration is regulated by cytokinin-induced cell cycle in Arabidopsis. New Phytol. 217, 219–232. doi: 10.1111/nph.14814
Liu, J., Sheng, L., Xu, Y., Li, J., Yang, Z., Huang, H., et al. (2014). WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26, 1081–1093. doi: 10.1105/tpc.114.122887
Liu, Z., Dai, X., Li, J., Liu, N., Liu, X., Li, S., et al. (2020). The type-B cytokinin response regulator ARR1 inhibits shoot regeneration in an ARR12-dependent manner in Arabidopsis. Plant Cell 32, 2271–2291. doi: 10.1105/tpc.19.00022
Loberant, B., and Altman, A. (2010). “Micropropagation of plants,” in Encyclopedia of Industrial Biotechnology: Bioprocess, Bioseparation, and Cell Technology, ed. M. C. Flickinger (Hoboken, NJ: John Wiley & Sons, Inc.), 1–17. doi: 10.1007/s11120-017-0473-9
Loomis, W. G., and Schull, C. A. (1937). Methods in Plant Physiology. New York, NY: McGraw-Hull, 58.
Lotan, T., Ohto, M. A., Matsudaira Yee, K., West, M. A. L., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 Is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. doi: 10.1016/s0092-8674(00)81463-4
Ma, W., Wu, F., Sheng, P., Wang, X., Zhang, Z., Zhou, K., et al. (2017). The LBD12-1 transcription factor suppresses apical meristem size by repressing argonaute 10 expression. Plant Physiol. 173:801. doi: 10.1104/pp.16.01699
Makarevich, G., Leroy, O., Akinci, U., Schubert, D., Clarenz, O., Goodrich, J., et al. (2006). Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7, 947–952. doi: 10.1038/sj.embor.7400760
Mantiri, F. R., Kurdyukov, S., Lohar, D. P., Sharopova, N., Saeed, N. A., Wang, X. D., et al. (2008). The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in Medicago truncatula. Plant Physiol. 146, 1622–1636. doi: 10.1104/pp.107.110379
Matsuo, N., Makino, M., and Banno, H. (2011). Arabidopsis ENHANCER OF SHOOT REGENERATION (ESR)1 and ESR2 regulate in vitro shoot regeneration and their expressions are differentially regulated. Plant Sci. 181, 39–46. doi: 10.1016/j.plantsci.2011.03.007
Matsuo, N., Mase, H., Makino, M., Takahashi, H., and Banno, H. (2009). Identification of ENHANCER OF SHOOT REGENERATION 1-upregulated genes during in vitro shoot regeneration. Plant Biotechnol. 26, 385–393. doi: 10.5511/plantbiotechnology.26.385
Mayer, K. F. X., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the arabidopsis shoot meristem. Cell 95, 805–815. doi: 10.1016/s0092-8674(00)81703-1
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. doi: 10.1038/35079635
Méndez-Hernández, H. A., Ledezma-Rodríguez, M., Avilez-Montalvo, R. N., Juárez-Gómez, Y. L., Skeete, A., Avilez-Montalvo, J., et al. (2019). Signaling overview of plant somatic embryogenesis. Front. Plant Sci. 10:77. doi: 10.3389/fpls.2019.00077
Meng, W. J., Cheng, Z. J., Sang, Y. L., Zhang, M. M., Rong, X. F., Wang, Z. W., et al. (2017). Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29, 1357–1372. doi: 10.1105/tpc.16.00640
Miao, C., Wang, D., He, R., Liu, S., and Zhu, J. K. (2020). Mutations in MIR396e and MIR396f increase grain size and modulate shoot architecture in rice. Plant Biotechnol. J. 18, 491–501. doi: 10.1111/pbi.13214
Michelmore, R., Marsh, E., Seely, S., and Landry, B. (1987). Transformation of lettuce (Lactuca sativa) mediated by Agrobacterium tumefaciens. Plant Cell Rep. 6, 439–442.
Michelmore, R. W., and Eash, J. A. (1985). “Lettuce,” in The Handbook of Plant Cell Culture, Vol. 4, eds D. A. Evans, W. R. Sharp, and P. V. Ammirato (New York, NY: Macmillan) 512–551.
Miller, C., Skoog, F., von Saltza, M. H., and Strong, F. M. (1955). Kinetin, a cell division factor from desoxyribonucleic acid. J. Am. Chem. Soc. 77:1392. doi: 10.1021/ja01610a105
Mitchum, M. G., Yamaguchi, S., Hanada, A., Kuwahara, A., Yoshioka, Y., Kato, T., et al. (2006). Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 45, 804–818. doi: 10.1111/j.1365-313X.2005.02642.x
Mohebodini, M., Javaran, M. J., and Alizadeh, H. (2011). Effects of genotype, explant age and growth regulators on callus induction and direct shoot regeneration of Lettuce (Lactuca sativa L.). Aust. J. Crop Sci. 5, 92–95. doi: 10.33899/rjs.2012.44416
Mozgová, I., Muñoz-Viana, R., and Hennig, L. (2017). PRC2 represses hormone-induced somatic embryogenesis in vegetative tissue of Arabidopsis thaliana. PLoS Genet. 13:e1006562. doi: 10.1371/journal.pgen.1006562
Müller, B., and Sheen, J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094–1097. doi: 10.1038/nature06943
Murthy, B. N. S., Victor, J., Singh, R. P., Fletcher, R. A., and Praveen, K. S. (1996). In vitro regeneration of chickpea (Cicer arietinum L.): stimulation of direct organogenesis and somatic embryogenesis by thidiazuron. Plant Growth Regul. 19, 233–240. doi: 10.1007/bf00037796
Negin, B., Shemer, O., Sorek, Y., and Williams, L. E. (2017). Shoot stem cell specification in roots by the WUSCHEL transcription factor. PLoS One 12:e0176093. doi: 10.1371/journal.pone.0176093
Ning, K., Han, Y., Chen, Z., Luo, C., Wang, S., Zhang, W., et al. (2019). Genome-wide analysis of MADS-box family genes during flower development in lettuce. Plant Cell Environ. 42, 1868–1881. doi: 10.1111/pce.13523
Nobe’court, P. (1955). Variations de la morphologie et de la structure de cultures de tissues ve’ge’taux. Berichte Schweizerische Bot. Gesellschaft 65, 475–480.
Norstog, K. (1979). “Embryo culture as a tool in the study of comparative and developmental morphology,” in Plant Cell and Tissue Culture, eds W. R. Sharp, P. O. Larsen, E. F. Paddock, and V. Raghavan (Columbus, OH: Ohio State Univ. Press), 179–202. doi: 10.1016/0145-305x(87)90019-x
Nuti Ronchi, V., and Gregorini, G. (1970). Histological study of adventitious bud formation on Lactuca sativa cotyledons cultured in vitro. Giornale Bot. Italiano 104, 443–455. doi: 10.1080/11263507009426517
Ogas, J., Cheng, J. C., Sung, Z. R., and Somerville, C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277, 91–94. doi: 10.1126/science.277.5322.91
Okushima, Y., Fukaki, H., Onoda, M., Theologis, A., and Tasaka, M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19, 118–130. doi: 10.1105/tpc.106.047761
Omidbakhshfard, M. A., Proost, S., Fujikura, U., and Mueller-Roeber, B. (2015). Growth-Regulating Factors (GRFs): a small transcription factor family with important functions in plant biology. Mol. Plant 8, 998–1010. doi: 10.1016/j.molp.2015.01.013
Ooi, S. E., Choo, C. N., Ishak, Z., and Ong-Abdullah, M. (2012). A candidate auxin-responsive expression marker gene, EgIAA9, for somatic embryogenesis in oil palm (Elaeis guineensis Jacq.). Plant Cell Tissue Organ Cult. 110, 201–212. doi: 10.1007/s11240-012-0143-8
Osorio-Montalvo, P., De-la-Peña, C., Oropeza, C., Nic-Can, G., Córdova-Lara, I., Castillo-Castro, E., et al. (2020). A peak in global DNA methylation is a key step to initiate the somatic embryogenesis of coconut palm (Cocos nucifera L). Plant Cell Rep. 39, 1345–1357. doi: 10.1007/s00299-020-02568-2
Ozias-akins, P., and Vasil, I. K. (1982). Plant Regeneration from Cultured Immature embryos and inflorescences Triticum aestivum L. (Wheat): evidence for somatic embryogenesis. Protoplasma 110, 95–105. doi: 10.1007/bf01281535
Paul, P., Joshi, S., Tian, R., Diogo Junior, R., Chakrabarti, M., and Perry, S. E. (2022). The MADS-domain factor AGAMOUS-Like18 promotes somatic embryogenesis. Plant Physiol. 188, 1617–1631. doi: 10.1093/plphys/kiab553
Perilli, S., di Mambro, R., and Sabatini, S. (2012). Growth and development of the root apical meristem. Curr. Opin. Plant Biol. 15, 17–23. doi: 10.1016/j.pbi.2011.10.006
Preece, J. E. (2003). A century of progress with vegetative plant propagation. Hortscience 38, 1015–1025. doi: 10.1094/PDIS-07-14-0679-FE
Quiroz-Figueroa, F. R., Rojas-Herrera, R., Galaz-Avalos, R. M., and Loyola-Vargas, V. M. (2006). Embryo production through somatic embryogenesis can be used to study cell differentiation in plants. Plant Cell Tissue Organ Cult. 86, 285–301. doi: 10.1007/s11240-006-9139-6
Radhakrishnan, D., Shanmukhan, A. P., Kareem, A., Aiyaz, M., Varapparambathu, V., Toms, A., et al. (2020). A coherent feed-forward loop drives vascular regeneration in damaged aerial organs of plants growing in a normal developmental context. Development (Cambridge) 147:dev185710. doi: 10.1242/dev.185710
Raza, G., Singh, M. B., and Bhalla, P. L. (2020). Somatic embryogenesis and plant regeneration from commercial Soybean cultivars. Plants 9:38. doi: 10.3390/plants9010038
Reyes-Chin-Wo, S., Wang, Z., Yang, X., Kozik, A., Arikit, S., Song, C., et al. (2017). Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 8, 1–11. doi: 10.1038/ncomms14953
Rodriguez, R. E., Mecchia, M. A., Debernardi, J. M., Schommer, C., Weigel, D., and Palatnik, J. F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137, 103–112. doi: 10.1242/dev.043067
Rueb, S., Leneman, M., Schilperoort, R. A., and Hensgens, L. A. M. (1994). Efficient plant regeneration through somatic embryogenesis from callus induced on mature rice embryos (Oryza sativa L.). Plant Cell Tissue Organ Cult. 36, 259–264. doi: 10.1007/bf00037729
Santos, D., and Fevereiro, P. (2002). Loss of DNA methylation affects somatic embryogenesis in Medicago truncatula. Plant Cell Tissue Organ Cult. 70, 155–161.
Sarkar, A. K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., et al. (2007). Conserved factors regulate signaling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. doi: 10.1038/nature05703
Sasaki, H. (1979). Physiological and morphological studies on development of vegetable crops VI. Effect of several auxins, cytokinins and cytokinin-ribosides on the adventitious bud formation of lettuce hypocotyl tissue cultured in vitro. Physiol. Morphol. Stud. Dev. Veg. Crops 48, 67–72.
Sasaki, H. (1982). Effect of temperature and light on adventitious bud formation of lettuce hypocotyl tissue cultured in vitro. J. Jpn Soc. Horticult. 51, 187–194. doi: 10.2503/jjshs.51.187
Schuabb Heringer, A., Barroso, T., Ferreira Macedo, A., Santa-Catarina, C., Martins, G. H., Souza, F., et al. (2015). Label-free quantitative proteomics of embryogenic and non-embryogenic callus during sugarcane somatic embryogenesis. PLoS One 10:e0127803. doi: 10.1371/journal.pone.0127803
Severino, L. S., Lima, R. L. S., Lucena, A. M. A., Freire, M. A. O., Sampaio, L. R., Veras, R. P., et al. (2011). Propagation by stem cuttings and root system structure of Jatropha curcas. Biomass Bioenergy 35, 3160–3166. doi: 10.1016/j.biombioe.2011.04.031
Shang, B., Xu, C., Zhang, X., Cao, H., Xin, W., and Hu, Y. (2016). Very-long-chain fatty acids restrict regeneration capacity by confining pericycle competence for callus formation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 5101–5106. doi: 10.1073/pnas.1522466113
Shchuka, V. M., Malek-Gilani, N., Singh, G., Langroudi, L., Dhaliwal, N. K., Moorthy, S. D., et al. (2015). Chromatin dynamics in lineage commitment and cellular reprogramming. Genes 6, 641–661. doi: 10.3390/genes6030641
Shimotohno, A., Heidstra, R., Blilou, I., and Scheres, B. (2018). Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules. Genes Dev. 32, 1085–1100. doi: 10.1101/gad.314096.118
Shin, J., Bae, S., and Seo, P. J. (2020). De novo shoot organogenesis during plant regeneration. J. Exp. Bot. 71, 63–72. doi: 10.1093/jxb/erz395
Skoog, F., and Miller, C. O. (1957). Chemical regulation of growth and organ formation in plant tissue cultures in vitro. Symp. Soc. Exp. Biol. 11, 118–131.
Skoog, F., and Tsui, C. (1948). Chemical control of growth and bud formation in tobacco stem segments and callus cultured in vitro. Am. J. Bot. 35, 782–787. doi: 10.1002/j.1537-2197.1948.tb08148.x
Stone, S. L., Braybrook, S. A., Paula, S. L., Kwong, L. W., Meuser, J., Pelletier, J., et al. (2008). Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 3151–3156. doi: 10.1073/pnas.0712364105
Su, Y. H., and Zhang, X. S. (2014). The hormonal control of regeneration in plants. Curr. Top. Dev. Biol. 108, 35–69. doi: 10.1016/B978-0-12-391498-9.00010-3
Sugimoto, K., Jiao, Y., and Meyerowitz, E. M. (2010). Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18, 463–471. doi: 10.1016/j.devcel.2010.02.004
Sugimoto, K., Temman, H., Kadokura, S., and Matsunaga, S. (2019). To regenerate or not to regenerate: factors that drive plant regeneration. Curr. Opin. Plant Biol. 47, 138–150. doi: 10.1016/j.pbi.2018.12.002
Sun, S., Kang, X. P., Xing, X. J., Xu, X. Y., Cheng, J., Zheng, S. W., et al. (2015). Agrobacterium-mediated transformation of tomato (Lycopersicon esculentum L. cv. Hezuo 908) with improved efficiency. Biotechnol. Biotechnol. Equip. 29, 861–868. doi: 10.1080/13102818.2015.1056753
Szczygieł-Sommer, A., and Gaj, M. D. (2019). The miR396–GRF regulatory module controls the embryogenic response in Arabidopsis via an auxin-related pathway. Int. J. Mol. Sci. 20:5221. doi: 10.3390/ijms20205221
Tian, Q., Uhlir, N. J., and Reed, J. W. (2002). Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14, 301–319. doi: 10.1105/tpc.010283
Trinh, D. C., Lavenus, J., Goh, T., Boutté, Y., Drogue, Q., Vaissayre, V., et al. (2019). PUCHI regulates very long chain fatty acid biosynthesis during lateral root and callus formation. Proc. Natl. Acad. Sci. U.S.A. 116, 14325–14330. doi: 10.1073/pnas.1906300116
Tsuda, K., Ito, Y., Sato, Y., and Kurata, N. (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23, 4368–4381. doi: 10.1105/tpc.111.090050
van den Berg, C., Willemsen, V., Hendriks, G., Weisbeek, P., and Scheres, B. (1997). Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390, 287–289. doi: 10.1038/36856
van der Knaap, E., Kim, J. H., and Kende, H. (2000). A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122:695. doi: 10.1104/pp.122.3.695
van Overbeek, J., Conklin, M. E., and Blakeslee, A. F. (1941). Factors in coconut milk essential for growth and development of very young Datura embryos. Science 94, 350–351. doi: 10.1126/science.94.2441.350
Victorathisayam, T., and Sridevi, G. (2020). Ectopic expression of WUSCHEL (AtWUS) gene alters plant growth and development in. Rice Plant 8, 43–53. doi: 10.11648/j.plant.20200803.11
Waki, T., Hiki, T., Watanabe, R., Hashimoto, T., and Nakajima, K. (2011). The Arabidopsis RWP-RK Protein RKD4 triggers gene expression and pattern formation in early embryogenesis. Curr. Biol. 21, 1277–1281. doi: 10.1016/j.cub.2011.07.001
Wang, X., Niu, Q. W., Teng, C., Li, C., Mu, J., Chua, N. H., et al. (2008). Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 19, 224–235. doi: 10.1038/cr.2008.276
Webb, D. T., Torres, L. D., and Fobert, P. (1984). Interactions of growth regulators, explant age, and culture environment controlling organogenesis from lettuce cotyledons in vitro. Can. J. Bot. 62, 586–590. doi: 10.1139/b84-088
Wildwater, M., Campilho, A., Perez-Perez, J. M., Heidstra, R., Blilou, I., Korthout, H., et al. (2005). The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123, 1337–1349. doi: 10.1016/j.cell.2005.09.042
Wójcikowska, B., and Gaj, M. D. (2017). Expression profiling of AUXIN RESPONSE FACTOR genes during somatic embryogenesis induction in Arabidopsis. Plant Cell Rep. 36, 843–858. doi: 10.1007/s00299-017-2114-3
Xiong, F., Zhang, B.-K., Liu, H.-H., Wei, G., Wu, J.-H., Wu, Y.-N., et al. (2020). Transcriptional regulation of PLETHORA1 in the root meristem through an importin and its two antagonistic cargos. Plant Cell 32, 3812–3824. doi: 10.1105/tpc.20.00108
Xu, J., Hofhuis, H., Heidstra, R., Sauer, M., Friml, J., and Scheres, B. (2006). A molecular framework for plant regeneration. Science 311, 385–388. doi: 10.1126/science.1121790
Xu, K., Liu, J., Fan, M., Xin, W., Hu, Y., and Xu, C. (2012). A genome-wide transcriptome profiling reveals the early molecular events during callus initiation in Arabidopsis multiple organs | Elsevier Enhanced Reader. Genomics 100, 116–124. doi: 10.1016/j.ygeno.2012.05.013
Xu, L., and Huang, H. (2014). Genetic and epigenetic controls of plant regeneration. Curr. Top. Dev. Biol. 108, 1–33. doi: 10.1016/B978-0-12-391498-9.00009-7
Xu, M., Hu, T., Zhao, J., Park, M.-Y., Earley, K. W., Wu, G., et al. (2016). Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 12:e1006263. doi: 10.1371/journal.pgen.1006263
Xu, Z., Zhang, C., Ge, X., Wang, N., Zhou, K., Yang, X., et al. (2015). Construction of a high-density linkage map and mapping quantitative trait loci for somatic embryogenesis using leaf petioles as explants in upland cotton (Gossypium hirsutum L.). Plant Cell Rep. 34, 1177–1187. doi: 10.1007/s00299-015-1776-y
Xue, T., Dai, X., Wang, R., Wang, J., Liu, Z., and Xiang, F. (2017). ARGONAUTE10 inhibits in vitro shoot regeneration via repression of miR165/166 in Arabidopsis thaliana. Plant Cell Physiol. 58, 1789–1800. doi: 10.1093/pcp/pcx117
Yamaguchi, S., Kamiya, Y., and Sun, T. P. (2001). Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 28, 443–453. doi: 10.1046/j.1365-313x.2001.01168.x
Yamaguchi, Y. L., Ishida, T., and Sawa, S. (2016). CLE peptides and their signaling pathways in plant development. J. Exp. Bot. 67, 4813–4826.
Yasui, Y., Tsukamoto, S., Sugaya, T., Nishihama, R., Wang, Q., Kato, H., et al. (2019). GEMMA CUP_ASSOCIATED MYB1, an ortholog of axillary meristem regulators is essential in vegetative reproduction in Marchantia polyphorma. Curr. Biol. 29, 3986–3995. doi: 10.1016/j.cub.2019.10.004
Zhang, B., Tong, Y., Luo, K., Zhai, Z., Liu, X., Shi, Z., et al. (2021). Identification of GROWTH-REGULATING FACTOR transcription factors in lettuce (Lactuca sativa) genome and functional analysis of LsaGRF5 in leaf size regulation. BMC Plant Biol. 21:485. doi: 10.1186/s12870-021-03261-6
Zhang, X., Henriques, R., Lin, S.-S., Niu, Q.-W., and Chua, N.-H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhang, Y., Jiao, Y., Jiao, H., Zhao, H., and Zhu, Y. (2017). Two-step functional innovation of the stem-cell factors WUS/WOX5 during plant evolution. Mol. Biol. Evol. 34, 640–653. doi: 10.1093/molbev/msw263
Zheng, Q., Zheng, Y., Ji, H., Burnie, W., and Perry, S. E. (2016). Gene Regulation by the AGL15 transcription factor reveals hormone interactions in somatic embryogenesis. Plant Physiol. 172, 2374–2387. doi: 10.1104/pp.16.00564
Zheng, Q., Zheng, Y., and Perry, S. E. (2013). AGAMOUS-Like15 promotes somatic embryogenesis in Arabidopsis and soybean in part by the control of ethylene biosynthesis and response. Plant Physiol. 161, 2113–2127. doi: 10.1104/pp.113.216275
Zheng, Y., Ren, N., Wang, H., Stromberg, A. J., and Perry, S. E. (2009). Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21, 2563–2577. doi: 10.1105/tpc.109.068890
Keywords: regeneration, organogenesis, lettuce, somatic embryogenesis, WUSCHEL, Lactuca sativa (L.)
Citation: Bull T and Michelmore R (2022) Molecular Determinants of in vitro Plant Regeneration: Prospects for Enhanced Manipulation of Lettuce (Lactuca sativa L.). Front. Plant Sci. 13:888425. doi: 10.3389/fpls.2022.888425
Received: 02 March 2022; Accepted: 31 March 2022;
Published: 09 May 2022.
Edited by:
Goetz Hensel, Heinrich Heine University Düsseldorf, GermanyReviewed by:
Tatjana Cosic, University of Belgrade, SerbiaCopyright © 2022 Bull and Michelmore. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard Michelmore, cndtaWNoZWxtb3JlQHVjZGF2aXMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.