
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 April 2022
Sec. Plant Nutrition
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.859395
This article is part of the Research Topic Advanced Insights into Plant Rhizosphere Functionality from the Perspective of Declining Soil Fertility Status in the Era of Climate Change View all 6 articles
Malus sieversii grows on the slopes of the Tianshan Mountains in Xinjiang where the difference in daily temperature is significant. In recent years, the rhizosphere soil health of Malus sieversii has been severely impacted by anthropogenic disturbance and pathogenic infestation. The soil nutrient content and soil microorganism diversity are the main components of soil health. Low temperature has negative effects on soil bacterial community structure by inhibiting the accumulation of carbon and nitrogen. However, the effects of temperature and nitrogen application on soil carbon and nitrogen accumulation and the bacterial community composition in the rhizosphere soil of Malus sieversii are unclear. We set two temperature levels, i.e., low temperature (L) and room temperature (R), combined with no nitrogen (N0) and nitrogen application (N1) to explore the response of plant carbon and nitrogen uptake, rhizosphere soil carbon and nitrogen accumulation and bacterial community composition to temperature and nitrogen fertilization. At the same temperature level, plant 13C abundance (P-Atom13C), plant 15N absolute abundance (P-Con15N), soil 15N abundance (S-Atom15N) and soil urease, protease and glutaminase activities were significantly higher under nitrogen application compared with the no-nitrogen application treatment. The bacterial community diversity and richness indices of the apple rhizosphere soil in the N1 treatment were higher than those in the N0 treatment. The relative abundances of Actinobacteria, Rhodopseudomonas, and Bradyrhizobium were higher in the LN1 treatment than in the LN0 treatment. Redundancy analysis (RDA) showed that plant 13C absolute abundance (P-Con13C) and plant 15N absolute abundance (P-Con15N) were the main factors affecting the soil bacterial community composition. In summary, Nitrogen application can alleviate the effects of low temperature stress on the soil bacterial community and is of benefit for the uptakes of carbon and nitrogen in Malus sieversii plants.
Carbon and nitrogen are important nutrients necessary for plant growth, development, and metabolism and are also important factors limiting soil productivity (Chen et al., 2016; Gu et al., 2019). Carbon and nitrogen nutrition have a direct impact on the formation of photosynthetic products, mineral element uptake, and fruit development (Rodriguez-Lovelle and Gaudillère, 2002; Huang et al., 2015; Sivanandhan et al., 2015). The plant carbon pool will be shifted after nitrogen input (Zhou et al., 2021). Thus, it is necessary to further understand the processes of carbon and nitrogen fixation, allocation and transfer in the plant–soil system (Wang et al., 2019; Wang F. et al., 2020). The capacity of plant photosynthetic carbon fixation to nitrogen input varies across ecosystems, and plant carbon and nitrogen contents and photosynthetic carbon fixation capacity change accordingly (Sheel et al., 2012; Yang et al., 2018). A previous study showed that the δ13C values of the upper leaves of Lolium perenne L. and Trifolium repens L. increased rapidly after 2 days of urea addition, while after 12 days, the δ13C values decreased (Ambus et al., 2007). Experiments conducted in Californian chaparral showed a significant increase in aboveground carbon and nitrogen storage rates after 4–5 years of nitrogen application (Vourlitis and Hentz, 2016). Furthermore, a study in a pure Larix principis-rupprechtii plantation in northern China showed that nitrogen addition can alter soil enzyme activities and further affect soil carbon turnover through microbial regulation (Wu et al., 2019). Temperature is the limiting factor affecting the growth and respiration of soil microorganisms and enzyme dynamics (Steinweg et al., 2012; Zhong et al., 2021). Different ambient temperatures have different effects on the soil carbon and nitrogen cycles as well as plant development and growth (Dierig et al., 2006; Hatfield and Prueger, 2015; Tulina, 2019; Cruz-Paredes et al., 2021; Hasi et al., 2021). Seasonal low temperature or diurnal variation can significantly affect soil carbon and nitrogen nutrient turnover efficiency (Kurihara et al., 2018). Zhang incubated plants for 16 weeks at four temperatures (10, 15, 20, and 25°C) and discovered that temperature can alter plant metabolism and photosynthesis, as well as the compositions and concentrations of carbon and nitrogen sources, thereby influencing plant δ13C and δ15N signatures (Zhang P. et al., 2021).
The plant root zone is a soil microzone where plant and microbial communication is highly active (Rzehak et al., 2022). The root system of plants can secrete various microbially beneficial vitamins, enzymes, plant growth regulators, and amino acids (Mommer et al., 2016; Chamkhi et al., 2021), which in turn have an impact on the species, number and distribution of rhizosphere microorganisms (Vives-Peris et al., 2020). The interrelationship of plants, soil, and microorganisms maintains the function of the soil ecosystem (Nihorimbere et al., 2011; Lozano et al., 2014; Purahong et al., 2018; Sun et al., 2018). Soil microorganisms play an important role in the agricultural response to changing ecological environment due to their various nutrient cycles and soil carbon sequestration (Basu et al., 2020; Chen et al., 2021b). The study of the composition of soil microbial communities not only contributes to a more in-depth understanding of the ecological process, but it also has important implications for the conservation of wild resources (Samuel, 2014; Rigg et al., 2016; Shao et al., 2020). Changes in the abundance and diversity of bacteria and in the structural and compositional characteristics of the community can affect soil fertility and the sustainable productivity of fruit forests (Brussaard et al., 2007; Bhat, 2013). Bacteria are an important community of soil microorganism, involved in processes such as nutrient cycling, litter degradation, and soil fertility changes (Cao H. et al., 2021). Nitrogen input has been shown to alter soil nitrogen cycling processes, affecting soil nitrification and denitrification (Li et al., 2010; Yang et al., 2020) and leading to changes in the soil bacterial community structure (Fierer et al., 2021; Xiao et al., 2021). Studies have shown that nitrogen application can significantly increase bacterial abundance (Liu et al., 2020) and alter fungal-to-bacterial ratios (Chinta et al., 2021; Li et al., 2021c), thereby changing the soil microbial community structure and affecting ecosystem biogeochemical cycles (Yu et al., 2021). Nonetheless, numerous studies have found that nitrogen application reduces soil bacterial abundance and diversity (Wang C. et al., 2018; Castellano-Hinojosa et al., 2020; Wang W. et al., 2020). Currently, the effect of nitrogen input on soil bacterial diversity and community composition is still controversial.
Malus sieversii (Ledeb.) M. Roem., also known as Tienshan or Xinjiang wide apple, is an important wild fruit tree resource in China (Sitpayeva et al., 2020). It is the original ancestor of the world’s cultivated apples (Harris et al., 2002; Chen et al., 2007) and is listed as a second-class priority plant in China and a national biodiversity priority species (Fu and Chin, 1992). The genetic resources of M. sieversii are rich and diverse and are of great value in the conservation and utilization of germplasm resources (Wiedow et al., 2004; Wang N. et al., 2018). There are numerous links between plant and soil microbial diversity, and plant species and microbial diversity both play important roles in maintaining ecosystem stability and health (Zak et al., 2003; Gabriele et al., 2017; Rawat et al., 2020). Numerous scholars have conducted systematic studies on the response of diversity to environmental changes and investigated the feedback mechanisms of plant species and microbial diversity (Bouasria et al., 2012; Oliveira et al., 2012; Jia et al., 2021; Li et al., 2021a). Currently, it is extremely difficult to replace new populations of M. sieversii with live seedlings under natural conditions (Liu and Dong, 2018). Therefore, it is particularly important to study the response of soil carbon and nitrogen allocation to temperature and nitrogen fertilization and their microbial mechanisms. In this study, 13C and 15N isotope dual-labeling technology and Illumina NovaSeq high-throughput sequencing technology were used to explore differences in the rhizosphere soil carbon and nitrogen distribution and bacterial community diversity. This study provides fundamental information for the dynamic balance of rhizosphere soil ecology in M. sieversii, thereby providing new insights into plant–soil–microbe interactions that can be harnessed for M. sieversii seedlings breeding and germplasm conservation.
Our experiment was conducted in the Key Laboratory of Special Fruits and Vegetables Cultivation Physiology and Germplasm Resources Utilization of Xinjiang Production and Construction Corps of Shihezi University, Xinjiang Uygur Autonomous Region, China. The M. sieversii seeds were treated with low-temperature lamination at 4°C for 90 days under dark conditions. On November 11, 2019, 200 seeds with consistent germination growth were selected and sown in 50-cell seedling trays containing a mixed substrate with peat–vermiculite–apple orchard soil (volume ratio 3:1:0.2, and the peat–vermiculite mixture was autoclaved). One plant was grown per cell, and each cell of the tray measured 4 cm in length, 4 cm in width and 10 cm in height. M. sieversii seeds were incubated in an artificial climate chamber (RXZ-300B, Ningbo Jiangnan Instrument Co., Ningbo, China). The culture conditions were as follows: temperature 25°C, relative humidity 70–80%, darkness during the germination period, light intensity 134 μmol m–2 s–1 during the seedling emergence period, and a 12-h:12-h light-dark cycle. Individual, healthy and uniform seedlings (when they had 7–8 true leaves) were selected for isotope labeling and low temperature treatment. The seedlings were watered once every 3 days during planting period as needed (Nagakura et al., 2004; Fernandez-Going et al., 2013).
A solution containing 320 mg CO(15NH4)2 (abundance of 10.16%) was dissolved in water and was applied to burrowing trays on January 3, 2020. Nitrogen labeling was performed 7 days after 13C pulse labeling, and labeling was performed in a transparent agricultural film labeling chamber (Figure 1). The seal of the marker chamber was checked before marking. A syringe was used to inject 1 mL of HCl solution at a concentration of 1 mol/L into a test tube containing 0.6 g of Ba13CO3 (abundance of 98%). Two nitrogen fertilizer treatments were set up, i.e., the N1 treatment (urea applied at a fertilizer to substrate ratio of 0.43 g kg−1; N1) and N0 treatment (0 g kg−1; N0). The temperature was set at two levels, i.e., the L treatment (5°) and R treatment (25°). The experiment was designed based on a completely randomized design with four treatment groups (LN0, LN1, RN0, and RN1) and three replications per treatment.
Samples were collected on the 7th day of the low temperature and nitrogen application treatments. Three M. sieversii seedlings were randomly selected from each of the four treatments, the aboveground parts were cut off, and the plants were destructively removed. Large clods of soil around the root system were removed, and the soil was gently shaken off the root surface of the plants. A portion of the collected fresh samples was directly packed into sterile plastic bags and stored at −80°C in the refrigerator for soil DNA extraction. The other part was mixed well and dried naturally, ground through a 0.25 mm sieve and placed in plastic bags for the determination of soil enzyme activity and soil 13C and 15N abundance. Three additional M. sieversii seedlings with essentially uniform growth were randomly selected for destructive sampling in each treatment. The samples were rinsed in the order of water, detergent, water, 1% hydrochloric acid and then three times with deionized water, after which they were dried at 105°C for 30 min, followed by drying at 80°C to a constant weight (Zhang R. et al., 2021). The dried samples were ground through a 0.25 mm sieve and stored in plastic bags for determination of the 13C and 15N abundance of the plants (Yan et al., 2020).
The 13C and 15N abundances were measured using a DELTA V advantage isotope ratio mass spectrometer and were analyzed by the China Academy of Forestry Sciences Stable Isotope Laboratory. The formulas were as follows: P-Atom13C (or S-Atom13C) = (δ13C + 1,000) × RPDB/[(δ13C + 1,000) × RPDB + 1,000] × 100, P-Con13C (or S-Con13C) = P-Atom13C (or S-Atom13C) × 0.01 × C% × 0.01 × 1,000, P-Atom15N (or S-Atom15N) = (δ15N + 1,000) × RPDB/[(δ15N + 1,000) × RPDB + 1,000] × 100, P-Con15N (or S-Con15N) = P-Atom15N (or S-Atom15N) × 0.01 × N% × 0.01 × 1,000, and δ13C (or δ15N) = (Rs/RPDB − 1) × 1,000, where δ13C is the amount of 13C assimilate that was fixed (‰); δ15N is the amount of 15N assimilate that was fixed (‰); RS is the ratio of 13C to 12C (or the ratio of 15N to 14N); RPDB is the standard ratio of carbon isotopes, i.e., 0.0112372 (or the standard ratio of nitrogen isotopes, i.e., 0.0036765); P-Atom13C and P-Atom15N are plant 13C abundance and plant 15N abundance (%), respectively, which refer to 13C and 15N as percentages of total carbon and nitrogen of the plant samples, respectively; S-Atom13C and S-Atom15N are soil 13C abundance and soil 15N abundance (%), respectively, which refer to 13C and 15N as percentages of total carbon and nitrogen of the soil samples; P-Con13C and P-Con15N are plant 13C absolute abundance and plant 15N absolute abundance (mg g−1), respectively, which refer to the amount (mg) of 13C and 15N contained in one gram of the plant sample; S-Con13C and S-Con15N are soil 13C absolute abundance and soil 15N absolute abundance (mg g−1), respectively, which refer to the amount (mg) of 13C and 15N contained in one gram of the soil sample; and C% and N% are the amount (g) of total carbon and nitrogen contained in 100 g of sample.
Determination of rhizosphere soil urease (EC3.5.1.5) and catalase (EC1.11.1.6) activities according to the method described by Guan (1986). Urease activity (EC3.5.1.5) was measured by the colorimetric analysis of sodium phenate-sodium hypochlorite, and the activity was expressed as micrograms of NH3-N in 1 g of soil after 24 h (μg g–1 d–1). Catalase activity (EC1.11.1.6) was evaluated using the potassium permanganate titration volume method, and the activity was expressed as milliliters of 0.1 mol L−1 potassium permanganate in 1 g of soil after 20 min (ml g–1 20 min–1). Protease activity (EC 3.4.2.21-24) was determined according to the method developed by Ladd and Butler (1972), and the activity was expressed as micrograms of glycine in 1 g of soil after 24 h (μg g–1 d–1). Glutaminase activity (EC3.5.1.2) was assayed using a glutaminase kit (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) with the specification of 50 tubes/24 samples. The method was visible spectrophotometry, and 1 g of soil-catalyzed glutamine production of 1 μmol L−1 ammonia per day at 37°C was defined as one enzyme activity unit (U g−1) (Sakai et al., 2022).
The genomic DNA of the samples was extracted using the SDS method (Nanasato et al., 2018). The purity and concentration of DNA were subsequently examined using agarose gel electrophoresis. An appropriate amount of sample DNA was placed in a centrifuge tube, and the sample was diluted to 1 ng μL−1 using sterile water. To ensure amplification efficiency and accuracy, PCR amplification of the V4 region gene fragment was performed using primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) and high-fidelity polymerase (Carini et al., 2016). The PCR mixture (30 μl) contained 15 μL of Phusion Master Mix, 3 μL of each primer and 10 μL of DNA template (5–10 ng). The amplification program consisted of predenaturation at 98°C for 1 min, 30 cycles (denaturation at 98°C for 10 s, annealing at 50°C for 30 s, extension at 72°C for 30 s), and a final extension step at 72°C for 5 min. The PCR products were extracted from 2% agarose gel, and the target bands were recovered using a gel recovery kit (QIAGEN China Co., Ltd. Guangzhou, China) (Abdel-Ghany et al., 2016). The products were then assayed for quantification and mixing, and library construction was performed after mixing and purification. The qualified libraries were sequenced using an Illumina NovaSeq6000 (Illumina, San Diego, CA, United States) (Modi et al., 2021).
The data of each sample were split from the downstream data based on barcode sequences and PCR amplification primer sequences. The sequences of barcodes and primers were intercepted and then spliced and filtered using FLASH (Magoc and Salzberg, 2011) and QIIME (Caporaso et al., 2010). The chimeric sequences were removed from these sequences to obtain the final valid data. OTUs were obtained by clustering the sequences with 97% similarity among the valid sequences of all samples using Uparse software (Haas et al., 2011). The SSUrRNA database (Wang et al., 2007) of SILVA132 (Edgar, 2013) was subsequently consulted for species annotation of OTU sequences, and diversity index, species classification and abundance analysis were carried out. In addition, redundancy analysis (RDA) was used to identify key environmental factors that significantly influenced changes in bacterial communities between treatment groups.
Alpha diversity analysis (including Shannon, Simpson, Chao1, and Ace indices) was performed using QIIME (Version 1.9.1). Significant differences between treatments were evaluated by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test using SPASS 20.0 (SPSS Inc., Chicago, IL, United States). Origin 2021 (Origin Software, Inc. Guangzhou, China) was used for plotting. Redundancy analysis (RDA) was used to examine the relationship between the 13C and 15N abundance of plants and rhizosphere soil and the rhizosphere soil bacterial community compositions with the CANOCO 5.0 software (Microcomputer Power, Ithaca, NY, United States).
Temperature and nitrogen application treatments significantly affected P-Atom13C, P-Con13C, P-Atom15N, P-Con15N, S-Atom13C, S-Con13C, S-Atom15N, and S-Con15N (Supplementary Table 1). P-Atom13C and P-Atom15N were significantly and positively correlated with S-Atom13C, S-Con13C, S-Atom15N and S-Con15N (P < 0.05) (Supplementary Table 5). P-Atom13C, P-Con13C, and S-Con13C of the RN1 treatment were significantly higher than those of the RN0 treatment (P < 0.05) (Figures 2A,B,F). There was no significant difference between P-Con13C, S-Atom13C, and S-Con13C of the LN0 treatment and LN1 treatment (P > 0.05) (Figures 2B,E,F). P-Atom15N and P-Con15N of the RN1 treatment were significantly higher than those of the LN1 treatment by 69.05% and 105.06%, respectively (Figures 2C,D). There was no significant difference between P-Atom15N, P-Con15N, S-Atom15N, and S-Con15N in the LN0 treatment and RN0 treatment (P > 0.05) (Figures 2C,D,G,H). S-Atom15N and S-Con15N of the RN1 treatment were significantly lower than those of LN1 by 38.25 and 49.63%, respectively (P < 0.05) (Figures 2G,H). Both S-Atom15N and S-Con15N had the following treatment rankings: LN1 > RN1 > LN0, RN0.
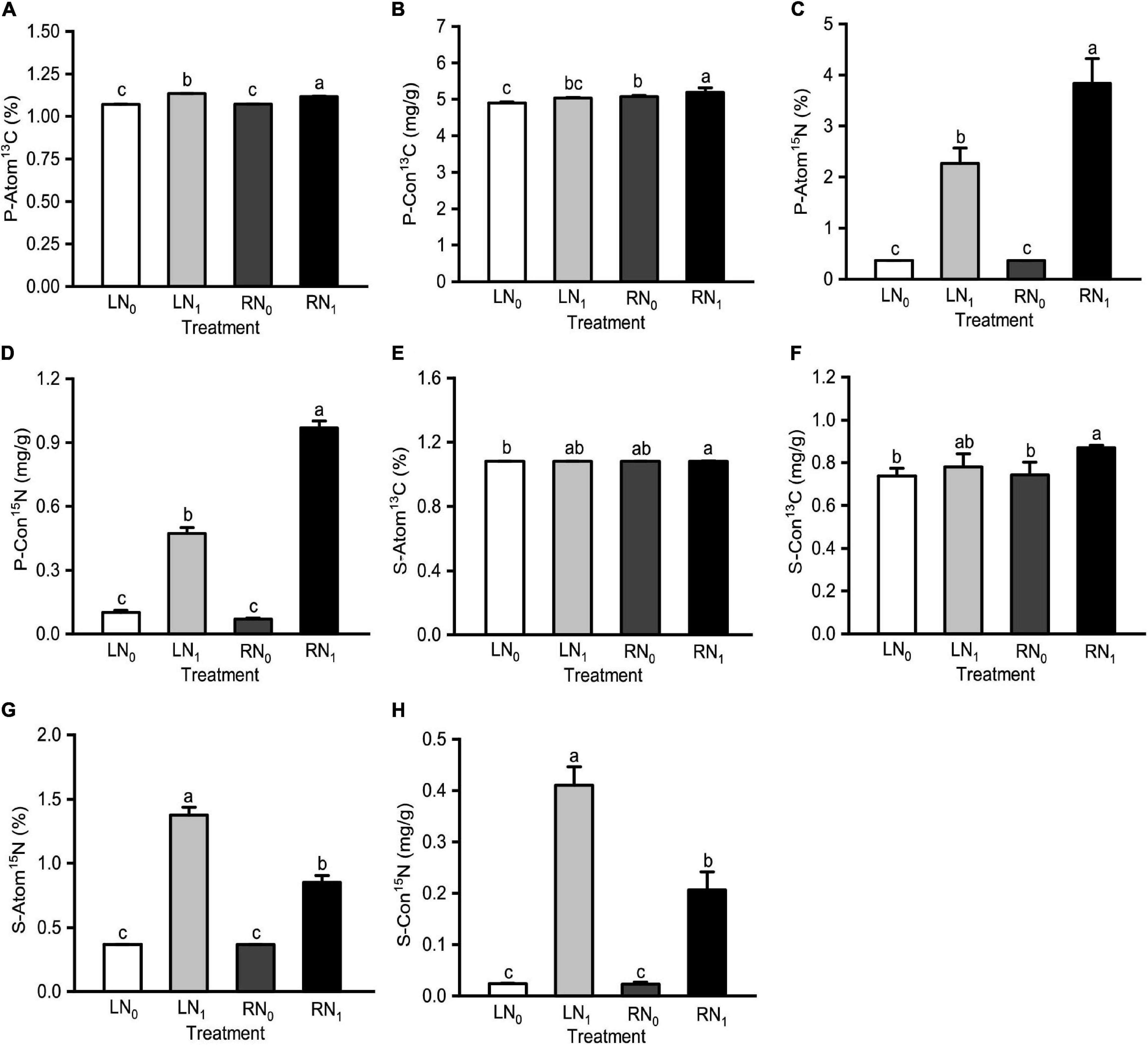
Figure 2. Comparisons of (A) plant 13C abundance (P-Atom 13C), (B) plant 13C absolute abundance (P-Con13C), (C) plant 15N abundance (P-Atom15N), (D) plant 15N absolute abundance (P-Con15N), (E) soil 13C abundance (S-Atom 13C), (F) soil 13C abundance (S-Con13C), (G) soil 15N absolute abundance (S-Atom15N), and (H) soil 15N absolute abundance (S-Con15N) among different treatments. L and R were low and room temperature treatments, N0 and N1 were non-nitrogen and nitrogen treatments. Values were shown as means ± standard deviations (SD, n = 3). Different lowercase letters were indicated statistically significant differences between the four treatments at 0.05 level.
There were significant differences in the rhizosphere soil urease, protease, and glutaminase catalase activities between the different temperature and nitrogen application treatments (Supplementary Table 1). At the same temperature level, the urease activity was significantly higher in the nitrogen application treatment group (LN1, RN1) than in the no-nitrogen treatment group (LN0, RN0) (P < 0.05); at the same nitrogen level, the urease activity was significantly higher in the room temperature treatment group (RN0, RN1) than in the low temperature treatment group (LN0, LN1) (P < 0.05) (Figure 3A). The urease activity of the RN0 and RN1 treatments was significantly higher than that of the LN0 and LN1 treatments by 3.49% and 21.95%, respectively (P < 0.05). The trends of the protease and glutaminase activities in each treatment were consistent with that of the urease activity (Figures 3B,C). Protease and glutaminase activities were significantly increased by 28.84% and 34.18% in the LN1 treatment compared to the LN0 treatment (P < 0.05). The catalase activity of the LN1 and RN1 treatments was significantly higher than that of the LN0 and RN0 treatments by 46.05 and 42.55%, respectively (P < 0.05) (Figure 3D).
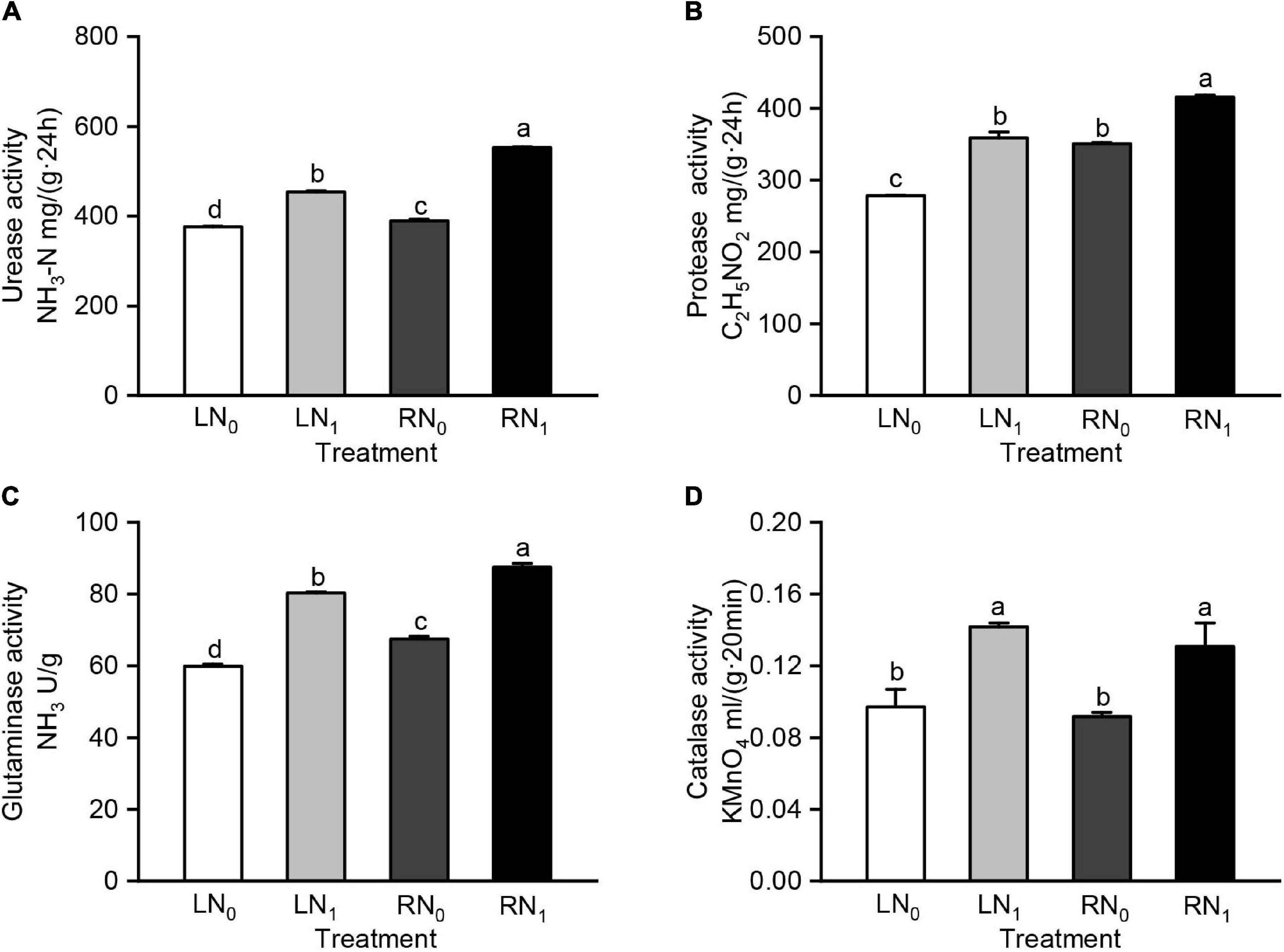
Figure 3. Comparisons of (A) soil urease activity, (B) soil protease activity, (C) soil glutaminase activity, and (D) soil catalase activity among different treatments. L and R were low and room temperature treatments, N0 and N1 were non-nitrogen and nitrogen treatments. Values were shown as means ± standard deviations (SD, n = 3). Different lowercase letters were indicated statistically significant differences between the four treatments at 0.05 level.
After sequencing quality filtering of the base-called sequences, a total of 738,893 high-quality sequences were obtained for the bacteria. A total of 3,148 operational taxonomic units (OTUs) at 97% similarity were obtained from the rhizosphere soil (Table 1). Temperature and nitrogen application treatments had significant effects on the Shannon, Simpson, Chao1, and Ace indices of the rhizosphere soil bacterial community of M. sieversii (Supplementary Table 1). Bacterial community diversity indices (Shannon and Simpson indices) showed that the Shannon index of the N1 treatment was significantly higher than that of N0 treatment (P < 0.05), and the Simpson index of the LN0 treatment was significantly higher than that of the other three treatments (P < 0.05). The Shannon index of the RN1 treatment was significantly higher than that of the other treatments (P < 0.05), and the Simpson index of the RN1 treatment was the lowest (0.976), indicating the highest soil bacterial community diversity in the RN1 treatment. Bacterial community richness indices (Chao1 and Ace indices) showed that the Chao1 index of the LN1 and RN1 treatments was significantly higher than that of LN0 and RN0 treatments (P < 0.05), and the Ace index of the RN1 treatment was significantly higher than that of the LN0, LN1, and RN0 treatments by 21.85, 10.64, and 17.70%, respectively (P < 0.05).
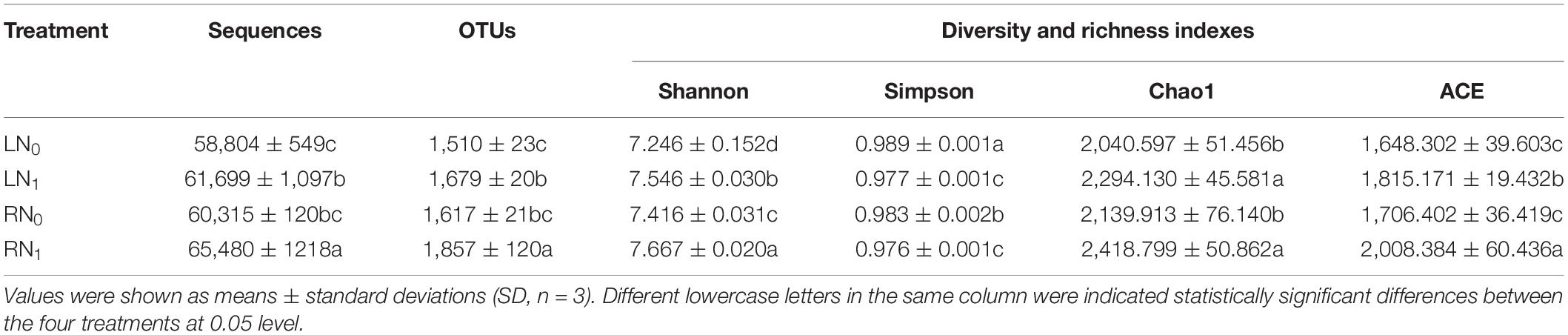
Table 1. Effect of temperature and nitrogen application on alpha diversity index in rhizosphere soil bacterial communities of M. sieversii.
At the phylum level, a total of 33 bacterial phyla were obtained, and 9 dominant phyla were obtained (Figure 4A). Among them, the dominant phyla (relative abundance > 5%) were Proteobacteria, Bacteroidetes, Acidobacteria, and Verrucomicrobia. The relative abundances of Proteobacteria, Bacteroidetes, Acidobacteria, and Verrucomicrobia were 62.05–72.59%, 10.89–14.23%, 4.13–6.11%, and 2.92–5.95%, respectively, accounting for 86.37–90.53% of all phyla. The average relative abundance of the other five phyla only accounted for 9.47–13.63% of the total bacterial community. Further analysis of bacterial phyla with relative abundances greater than 1% showed that temperature and nitrogen application treatments at the bacterial phylum level had significant effects on Proteobacteria, Acidobacteria, Verrucomicrobia, and Actinobacteria in the rhizosphere soil (Supplementary Tables 1, 2). The relative abundance of Acidobacteria was significantly higher in the RN0 treatment than in the LN0 treatment (P < 0.05). The relative abundance of Verrucomicrobia in the RN1 treatment was 103.45, 84.38, and 47.50% higher than that in the LN0, LN1, and RN0 treatments, respectively. The relative abundance of Actinobacteria in the LN1 treatment was significantly higher than that in the RN0 and RN1 treatments by 58.82% and 170.00%, respectively (P < 0.05). At the genus level, a total of 404 bacterial genera were obtained. Fifteen dominant genera with relative abundances greater than 0.5% were obtained in each sample (Figure 4B). The two most abundant bacterial genera in the LN0 treatment were Asticcacaulis (11.14%) and Devosia (5.83%). The relative abundances of Asticcacaulis (7.07%) and Rhizobacter (4.68%) were highest in the LN1 treatment. Moreover, the relative abundance of Asticcacaulis was highest in both the RN0 and RN1 treatments, i.e., 7.28 and 9.55%, respectively. At the species level, a total of 237 bacterial species were obtained. The dominant species (relative abundance > 5%) were Mesorhizobium_ciceri, Acidobacteria_bacterium_SCN_69-37, and bacterium_TG149. Other bacterial species had the largest proportion, with an average relative abundance of 96.76% (Figure 4C).
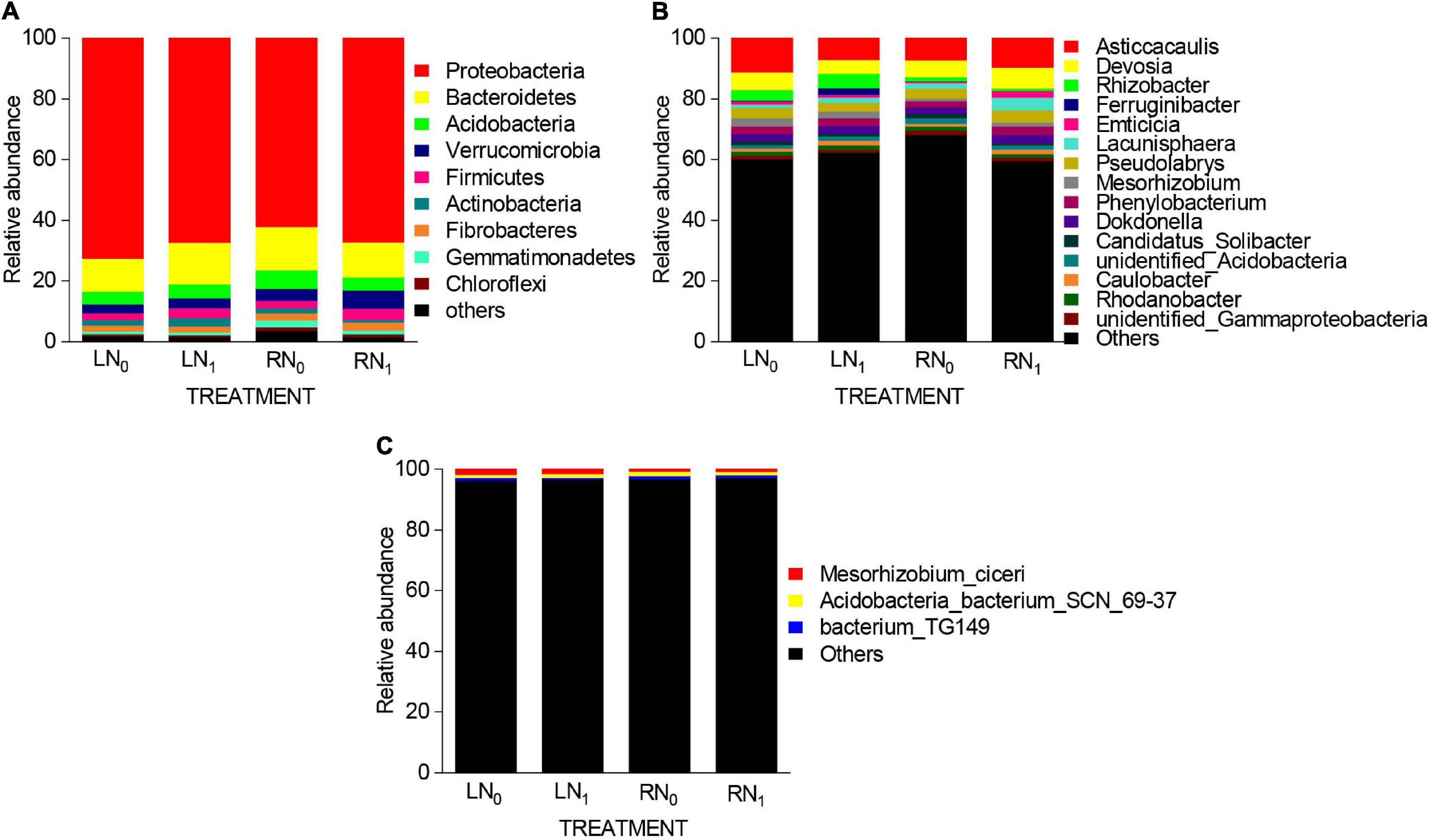
Figure 4. Relative abundance of primary (A) bacterial phyla (relative abundance ≥ 0.5%), (B) bacterial genera (relative abundance ≥ 0.5%), and (C) bacterial species (relative abundance ≥ 0.5%) present in the rhizosphere soil bacterial communities of the different treatments.
The relative abundances of Rhodopseudomonas, Methylibium, Pseudomonas, and Bradyrhizobium differed significantly between the different temperature and nitrogen fertilization treatments (Supplementary Table 1). The relative abundances of Rhodopseudomonas and Methylibium were significantly higher in the nitrogen application treatment groups (LN1, RN1) than in the non-nitrogen application treatment groups (LN0, RN0) at the same temperature level (Table 2). The relative abundances of Rhodopseudomonas and Methylibium in the LN1 treatment were significantly higher than those in the LN0 treatment by 75.00 and 129.03%, respectively (P < 0.05). The relative abundances of Rhodopseudomonas and Methylibium in the RN1 treatment were significantly higher than those in the RN0 treatment by 120.00% and 178.72%, respectively (P < 0.05). The relative abundance of Pseudomonas and Bradyrhizobium was higher in the nitrogen treatment group (LN1, RN1) than in the non-nitrogen treatment group (LN0, RN0) at the same temperature level. The relative abundance of Bradyrhizobium was significantly higher in the LN1 and RN1 treatments than in the LN0 and RN0 treatments by 63.27% and 81.36%, respectively (P < 0.05).
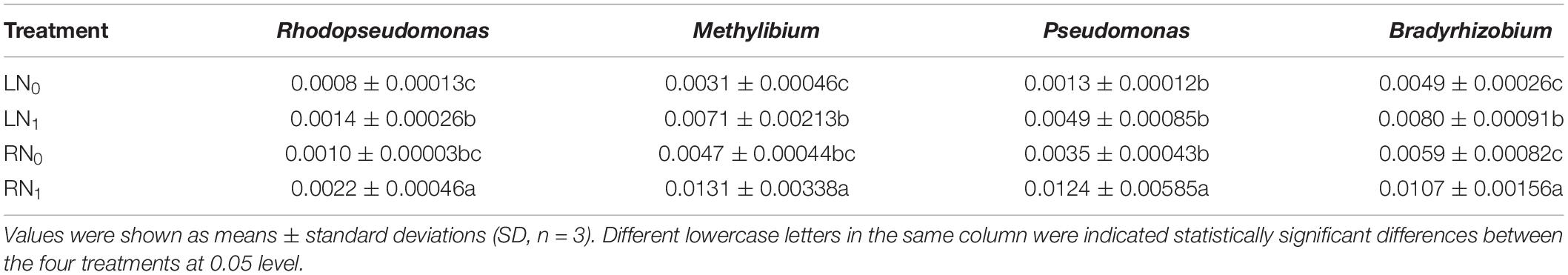
Table 2. Effect of temperature and nitrogen application on significantly different carbon- and nitrogen-fixing bacterial genera in the rhizosphere soil.
The relationship between plant 13C and 15N abundance, soil 13C and 15N abundance and bacterial phyla (relative abundance > 0.5%) in rhizosphere soil was analyzed by RDA. Considering the 13C and 15N abundance of M. sieversii plants and rhizosphere soil as environmental variables, axes 1 and 2 explained 46.86% and 17.72%, respectively, of the total variation (Figure 5A). P-Atom15N, P-Con15N, and S-Atom13C were negatively correlated with Proteobacteria, Bacteroidetes, Actinobacteria, Gemmatimonadetes, and Chloroflexi (P ≥ 0.05) (Figure 5A and Supplementary Table 3). P-Atom15N and P-Con15N were significantly and positively correlated with Verrucomicrobia and Firmicutes (P < 0.05). The RDA showed that the bacterial communities were differentially influenced by 13C and 15N abundance. The contribution of P-Con15N was 28.00%, which was the environmental factor with the largest contribution.
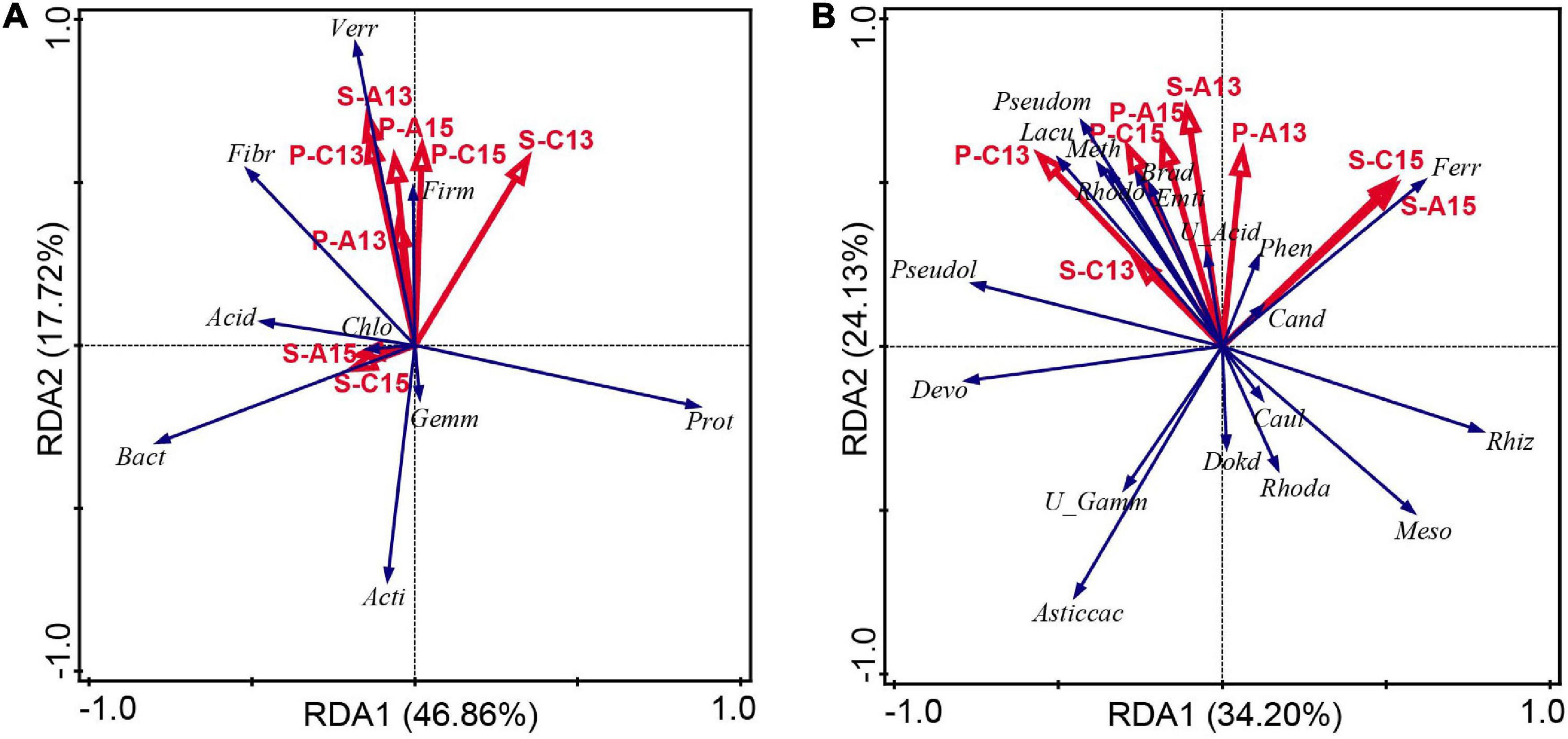
Figure 5. Redundancy analysis of (A) dominant bacterial phyla (relative abundance ≥ 0.5%) and (B) dominant bacterial genera (relative abundance ≥ 0.5%) and significantly different bacterial genera related to metabolism of carbon and nitrogen across all of the soil samples. Phyla and genera are indicated by blue vectors and environmental variables are represented by red vectors. The positions and lengths of the arrows indicate the directions and strengths, respectively, of the effects of variables on bacterial communities. Abbreviations in panel (A), Acid, Acidobacteria; Acti, Actinobacteria; Bact, Bacteroidetes; Chlo, Chloroflexi; Fib, Fibrobacteres; Firm, Firmicutes; Gemm, Gemmatimonadetes; P-A13, P-Atom13C; P-A15, P-Atom15N; P-C13, P-Con13C; P-C15, P-Con15N; Prot, Proteobacteria; S-A13, S-Atom13C; S-A15, S-Atom15N; S-C13, S-Con13C; S-C15, S-Con15N; Verr, Verrucomicrobia. Abbreviations in panel (B), Asti, Asticcacaulis; Brad, Bradyrhizobium; Cand, Candidatus_Solibacter; Caul, Caulobacter; Devo, Devosia; Dokd, Dokdonella; Emti, Emticicia; Ferr, Ferruginibacter; Lacu, Lacunisphaera; Meso, Mesorhizobium; Meth, Methylibium; P-A13, P-Atom13C; P-A15, P-Atom15N; P-C13, P-Con13C; P-C15, P-Con15N; Pseudol, Pseudolabrys; Pesudom, Pseudomonas; Phen, Phenylobacterium; Rhiz, Rhizobacter; Rhoda, Rhodanobacter; Rhodo, Rhodopseudomonas; S-A13, S-Atom13C; S-A15, S-Atom15N; S-C13, S-Con13C; S-C15, S-Con15N; U_Acid, unidentified_Acidobacteria; U_Gamm, unidentified_Gammaproteobacteria.
The relationship between plant 13C and 15N abundance, soil 13C and 15N abundance and bacterial genera in rhizosphere soil was analyzed by RDA. Considering the 13C and 15N abundance of M. sieversii plants and rhizosphere soil as environmental variables, axes 1 and 2 explained 34.20% and 24.13%, respectively, of the total variation (Figure 5B). P-Atom13C, P-Con13C, P-Atom15N, and P-Con15N showed highly significant positive correlations with the carbon and nitrogen metabolism-related bacterial genera Rhodopseudomonas, Methylibium, and Bradyrhizobium (P < 0.01) (Figure 5B and Supplementary Table 4). S-Atom13C and S-Con13C were significantly positively correlated with Rhodopseudomonas, Methylibium, Pseudomonas, and Bradyrhizobium (P < 0.05). S-Atom13C, S-Con13C, S-Atom15N, and S-Con15N were significantly positively correlated with Caulobacter (P < 0.05). RDA also showed that P-Con13C (24.60%) and S-Atom15N (24.40%) were the two factors with the highest contribution.
Carbon and nitrogen metabolism are the two most important metabolic processes in plants, and they are very closely related (Nunes-Nesi et al., 2010; Zhang et al., 2018; Zhong et al., 2021). Carbon metabolism provides carbon and energy for nitrogen metabolism, which in turn provides enzymes and photosynthetic pigments for carbon metabolism, both of which together regulate the material and energy metabolic processes in plants (Zhang et al., 2014; Ding et al., 2017). The photosynthetic carbon sequestration capacity of plants in different ecosystems responds in different ways to nitrogen inputs (Zhu et al., 2021). In this study, 13C and 15N dual-labeled isotope tracing techniques revealed that the 13C abundance and 13C absolute abundance of M. sieversii plants with high 15N abundance and 15N absolute abundance were also at higher levels in the treatments, indicating that appropriate nitrogen levels can promote the allocation and functioning of carbon assimilates. Zhao et al. (2021) showed that increased nitrogen fertilizer application increased photosynthetic carbon accumulation in wheat by 11–20% during 62 consecutive days of 13CO2 labeling. In this study, the 13C abundance and 13C absolute abundance of plants and soil showed roughly the same distribution pattern at different temperatures and nitrogen levels. The 13C abundance and 13C absolute abundance of the nitrogen treatment groups (LN1 and RN1) were significantly higher than those of the non-nitrogen treatment groups (LN0 and RN0) (P < 0.05) (Figure 1). P-Atom13C and P-Atom15N were significantly and positively correlated with S-Atom13C, S-Con13C, S-Atom15N, and S-Con15N (P < 0.05) (Supplementary Table 5). The S-Atom15N and S-Con15N values of the soil in the LN1 treatment were the highest and were significantly higher than those in the RN1 treatment. These results indicate that at the same temperature level, M. sieversii plants had a strong ability to exchange with soil under room temperature and nitrogen application, which facilitated the uptake of carbon and nitrogen by seedlings. More photosynthetic products made by the leaves were transported downward to the soil, which provided the material basis for root growth and development, thus alleviating the effects of low temperature stress (Jiang et al., 2015; Cornic, 2022).
Soil enzymes are an important indicator of soil biological activity, and all biochemical activities in soil are performed under the action of soil enzymes (Utobo and Tewari, 2015; Nannipieri et al., 2018). Soil enzyme activity is influenced by soil temperature, soil nutrients, microbial community, fertilization, and other factors (Cheng et al., 2013; Díaz et al., 2021; Levakov et al., 2021; Tan et al., 2021). The activities of nitrogen cycle enzymes such as urease, protease, glutaminase, and catalase varied significantly under different nitrogen fertilizer treatments (Cao et al., 2014; He et al., 2021; Li et al., 2021b). Seasonal low temperatures or diurnal variations in temperature can have a significant impact on soil enzymes (Viswanathan and Krishnan, 1962; Cao R. et al., 2021). In a field experiment with a winter temperature range of 0.5–2.0°C, the activities of soil catalase, urease, and phosphatase were reduced by 0.08–1.20 mL g–1, 0.004–0.019 mg g–1, and 0.10–0.25 mg kg–1, respectively (Xiao et al., 2012). In this study, the soil urease, protease, glutaminase, and catalase activities were higher in the R treatment than in the L treatment, and the soil urease, glutaminase, and catalase activities was significantly higher in the N1 treatment than in the N0 treatment (P < 0.05) (Figure 2). These results indicate low temperature significantly reduced the soil urease, protease, and glutaminase activities, while nitrogen application mitigated the effect of low temperature on the activities of nitrogen metabolism related enzymes.
Soil microorganisms are sensitive to environmental changes, and their composition and activity are influenced by a variety of factors including fertilizations, climate, and plant type (Dong et al., 2014; Soman et al., 2017; Grosso et al., 2018; Hu et al., 2019). Nitrogen fertilizer is an important factor that affects soil microbial communities in many agricultural systems (Wang L. et al., 2021; Zhang X. et al., 2021; Hu et al., 2022). In this study, the bacterial community diversity and richness of the apple rhizosphere soil in the nitrogen application treatment were higher than those without nitrogen treatment. The dominant phyla (relative abundance > 0.5%) of soil bacteria in the different treatments were Proteobacteria, Bacteroidetes, Acidobacteria, and Verrucomicrobia, followed by Firmicutes and Gemmatimonadetes (Figure 3). This result is similar to the dominant bacterial taxa obtained by Joa et al. (2014) and Wu et al. (2020). The higher abundance of the phyla Acidobacteria and Verrucomicrobia in the soil of the room temperature treatment group indicated that the application of nitrogen at room temperature could provide a good survival environment for Acidobacteria and Verrucomicrobia. The major reason is that nitrogen fertilization provides mineral elements for plant growth, promotes the growth and substance secretion of plant root organs, and accordingly increases the physiological activity of the root system (Hamm et al., 2016; Chen et al., 2020). Therefore, nitrogen application increased the relative abundance of Acidobacteria and Verrucomicrobia, which are closely related to the rhizosphere effect.
Microbial photosynthesis plays an important role in agricultural soils, and increased fertilizer application can significantly affect soil carbon decomposition and CO2 emissions (Xun et al., 2016; Carrara et al., 2018; Tian et al., 2019). The microorganisms involved in CO2 fixation are gram-negative bacteria, with the main dominant group being Proteobacteria (Li et al., 2020; Wang X. et al., 2021). The Alphaproteobacteria phylum mainly includes some typical carbon-fixing genera, such as Rhodopseudomonas and Methylibium (Liao et al., 2020; Chen et al., 2021a). Bradyrhizobium is a parthenogenic nitrogen-fixing bacterium that supports nutrient growth by depleting soil resources through fertilizer application (Li et al., 2019). In addition, this genus is found in Alphaproteobacteria, and it is usually classified as a eutrophic organism (Jabir et al., 2021). Short-term applications of nitrogen fertilizer can increase the abundance of biological nitrogen-fixing bacteria, and these microbial communities may use the resources in the fertilizer to support their own nutritional growth (Karlidag et al., 2007; Vitousek et al., 2013; Liu et al., 2022). Our results were consistent with the findings of the previous studies mentioned above. In this study, the relative abundances of Rhodopseudomonas and Methylibium were higher in the R treatment than in the L treatment at the same level of nitrogen application, and the relative abundance of Bradyrhizobium was significantly higher in the N1 treatment than in the N0 treatment at the same temperature level (P < 0.05) (Table 2). These results indicate that applying nitrogen fertilizer at the appropriate temperature can increase the number of soil carbon- and nitrogen-fixing bacterial genera. This study did not investigate the role of carbon- and nitrogen-fixing bacteria in the rhizosphere soil material cycle. It is necessary to further quantify the carbon fixation and nitrification characteristics of Rhodopseudomonas, Methylibium, and Bradyrhizobium, which are more responsive to low temperature and nitrogen application than other bacterial genera.
Soil environmental factors have an effect on the soil microbial community (Liu et al., 2021; Wang M. et al., 2021; Yin et al., 2021). Several studies have shown that soil microbial community composition is influenced by NO3–, soil organic carbon, and soil nitrogen content (Chen et al., 2021c; Liu et al., 2021; Ren et al., 2021). Shen et al. (2015) discovered that the structure of the bacterial community was significantly correlated with soil total carbon, total nitrogen, C:N ratio, and dissolved organic carbon. In this study, P-Atom15N and P-Con15N showed a significant positive correlation with Verrucomicrobia and Firmicutes (P < 0.05) (Figure 5A and Supplementary Table 3), and P-Atom13C, P-Con13C, P-Atom15N, and P-Con15N showed a highly significant positive correlation with the carbon and nitrogen metabolism-related genera Rhodopseudomonas, Methylibium, and Bradyrhizobium (P < 0.01) (Figure 5B and Supplementary Table 4). Therefore, plant carbon and nitrogen accumulation are key factors affecting the diversity and structure of the rhizosphere soil bacterial community in M. sieversii. The improvement of nutrient uptake by plants may be related to the mechanisms produced by rhizosphere soil microorganisms (Jacoby et al., 2017; Jing et al., 2021).
In conclusion, nitrogen application altered rhizosphere soil bacterial communities by influencing soil carbon and nitrogen accumulation as well as enzyme activities related to nitrogen metabolism. Furthermore, nitrogen application aided in the diversification and richness of the bacterial community, as well as the aggregation of carbon- and nitrogen-fixing bacterial genera (Rhodopseudomonas, Methylibium, and Bradyrhizobium) in the rhizosphere soil. RDA suggested that P-Con13C and P-Con15N were the key variables regulating the composition of the rhizosphere soil bacterial communities in M. sieversii. This study creates a suitable soil environment for M. sieversii roots from the perspectives of soil carbon and nitrogen cycling and microbial ecology, which has important practical significance for the breeding of M. sieversii seedlings and the conservation of M. sieversii germplasm resources. In the future, more emphasis could be placed on the role and function of carbon- and nitrogen-fixing bacteria in the rhizosphere soil material cycle of M. sieversii.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
HZ, SY, and KY planned and designed the study. HZ performed experiments and wrote original draft. FP and FZ commented on data interpretation and the whole manuscript. All authors contributed to the study and approved the final manuscript.
This study was supported by the National Key Research and Development Program (2018YFD1000200), the Natural Science Foundation of China (31760550), the Crops Major Scientific and Technological Project Plan (2016AA002), the Agricultural Science and Technology Tackling Project in Shihezi City of Bashi (2017HZ05), and the Transformation Project of Scientific and Technological Achievements of the Xinjiang Production and Construction Corps (2020BA006).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to express our sincere thank to the reviewers and issue editor of the journal for their valuable comments, suggestions, and revisions of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.859395/full#supplementary-material
Abdel-Ghany, S. E., Hamilton, M., Jacobi, J. L., Ngam, P., Devitt, N., Schilkey, F., et al. (2016). A survey of the sorghum transcriptome using single-molecule long reads. Nat. Commun. 7:11706. doi: 10.1038/ncomms11706
Ambus, P., Petersen, S. O., and Soussana, J. F. (2007). Short-term carbon and nitrogen cycling in urine patches assessed by combined carbon-13 and nitrogen-15 labelling. Agr. Ecosyst. Environ. 121, 84–92. doi: 10.1016/j.agee.2006.12.007
Basu, S., Kumar, G., Chhabra, S., and Prasad, R. (2020). “Role of soil microbes in biogeochemical cycle for enhancing soil fertility,” in New and Future Developments in Microbial Biotechnology and Bioengineering, ed. A. Rodrigues (Amsterdam: Elseiver), 149–157. doi: 10.1016/B978-0-444-64325-4.00013-4
Bhat, A. K. (2013). Preserving microbial diversity of soil ecosystem: a key to sustainable productivity. Int. J. Curr. Microbiol. App. Sci. 2, 85–101. doi: 10.1038/s41598-021-97373-6
Bouasria, A., Mustafa, T. De Bello, F., Zinger, L., Lemperiere, G., Geremia, R. A., et al. (2012). Changes in root-associated microbial communities are determined by species-specific plant growth responses to stress and disturbance. Eur. J. Soil Biol. 52, 59–66. doi: 10.1016/j.ejsobi.2012.06.003
Brussaard, L., de Ruiter, P. C., and Brown, G. G. (2007). Soil biodiversity for agricultural sustainability. Agr. Ecosyst. Environ. 121, 233–244. doi: 10.1016/j.agee.2006.12.013
Cao, H., Du, Y., Gao, G., Rao, L., Ding, G., and Zhang, Y. (2021). Afforestation with Pinus sylvestris var. mongolica remodelled soil bacterial community and potential metabolic function in the Horqin Desert. Glob. Ecol. Conserv. 29:e01716. doi: 10.1016/j.gecco.2021.e01716
Cao, R., Yang, W., Chang, C., Wang, Z., Wang, Q., Li, H., et al. (2021). Differential seasonal changes in soil enzyme activity along an altitudinal gradient in an alpine-gorge region. Appl. Soil Ecol. 166:104078. doi: 10.1016/j.apsoil.2021.104078
Cao, X., Liu, H. F., Hao, J. C., and She, C. M. (2014). The influence of Hg on soil enzyme activity under different nitrogen levels. Adv. Mat. Res. 955–959, 3701–3704. doi: 10.4028/www.scientific.net/AMR.955-959.3701
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carini, P., Marsden, P. J., Leff, J. W., Morgan, E. E., Strickland, M. S., and Fierer, N. (2016). Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2:16242. doi: 10.1038/nmicrobiol.2016.242
Carrara, J. E., Walter, C. A., Hawkins, J. S., Peterjohn, W. T., Averill, C., and Brzostek, E. R. (2018). Interactions among plants, bacteria, and fungi reduce extracellular enzyme activities under long-term N fertilization. Glob. Chang. Biol. 24, 2721–2734. doi: 10.1111/gcb.14081
Castellano-Hinojosa, A., Correa-Galeote, D., González-López, J., and Bedmar, E. J. (2020). Effect of nitrogen fertilisers on nitrous oxide emission, nitrifier and denitrifier abundance and bacterial diversity in closed ecological systems. Appl. Soil Ecol. 145:103380. doi: 10.1016/j.apsoil.2019.103380
Chamkhi, I., El Omari, N., Balahbib, A., El Menyiy, N., Benali, T., and Ghoulam, C. (2021). Is the rhizosphere a source of applicable multi-beneficial microorganisms for plant enhancement? Saudi J. Biol. Sci. 29, 1246–1259. doi: 10.1016/j.sjbs.2021.09.032
Chen, B., Yang, H., Song, W., Liu, C., Xu, J., Zhao, W., et al. (2016). Effect of N fertilization rate on soil alkali-hydrolyzable N, subtending leaf N concentration, fiber yield, and quality of cotton. Crop J. 4, 323–330. doi: 10.1016/j.cj.2016.03.006
Chen, L. L., Zhu, Z. B., Guo, Q. S., Tang, S. Q., Huang, Z. G., Zhong, W. H., et al. (2020). Growth response of Monochasma savatieri to nitrogen, phosphorus, and potassium in the absence of a host plant: essential demand for nutrients of root hemiparasitic plants. J. Plant Nutr. 43, 444–455. doi: 10.1080/01904167.2019.1683580
Chen, Q., Hu, Y., Hu, A., Niu, B., Yang, X., Jiao, H., et al. (2021b). Shifts in the dynamic mechanisms of soil organic matter transformation with nitrogen addition: from a soil carbon/nitrogen-driven mechanism to a microbe-driven mechanism. Soil Biol. Biochem. 160:108355. doi: 10.1016/j.soilbio.2021.108355
Chen, K., He, R., Wang, L., Liu, L., Huang, X., Ping, J., et al. (2021a). The dominant microbial metabolic pathway of the petroleum hydrocarbons in the soil of shale gas field: carbon fixation instead of CO2 emissions. Sci. Total Environ. 807:151074. doi: 10.1016/j.scitotenv.2021.151074
Chen, Q., Liu, Z., Zhou, J., Xu, X., and Zhu, Y. (2021c). Long-term straw mulching with nitrogen fertilization increases nutrient and microbial determinants of soil quality in a maize–wheat rotation on China’s Loess Plateau. Sci. Total Environ. 775:145930. doi: 10.1016/j.scitotenv.2021.145930
Chen, X., Feng, T., Zhang, Y., He, T., Feng, J., and Zhang, C. (2007). Genetic diversity of volatile components in Xinjiang wild apple (Malus sieversii). J. Genet. Genomics 34, 171–179. doi: 10.1016/s1673-8527(07)60018-6
Cheng, D. D., Zhao, G. Z., Liu, Y. Q., and Hao, S. Q. (2013). Influences of soil temperature and moisture on nutrients release of polymeric slow release fertilizer and soil enzyme activity. J. Soil Water Conserv. 27, 216–225. doi: 10.13870/j.cnki.stbcxb.2013.06.025
Chinta, Y. D., Uchida, Y., and Araki, H. (2021). Roles of soil bacteria and fungi in controlling the availability of nitrogen from cover crop residues during the microbial hot moments. Appl. Soil Ecol. 168:104135. doi: 10.1016/j.apsoil.2021.104135
Cornic, G. (2022). Effects of Temperature on Photosynthesis, Encyclopedia of the Environment, [Online ISSN 2555-0950]. Available Online at: https://www.encyclopedie-environnement.org/en/life/effects-temperature-on-photosynthesis/ (accessed February 15, 2022).
Cruz-Paredes, C., Tájmel, D., and Rousk, J. (2021). Can moisture affect temperature dependences of microbial growth and respiration? Soil Biol. Biochem. 156:108223. doi: 10.1016/j.soilbio.2021.108223
Díaz, F. J., Sanchez-Hernandez, J. C., and Notario, J. S. (2021). Effects of irrigation management on arid soils enzyme activities. J. Arid. Environ. 185:104330. doi: 10.1016/j.jaridenv.2020.104330
Dierig, D. A., Adam, N. R., Mackey, B. E., Dahlquist, G. H., and Coffelt, T. A. (2006). Temperature and elevation effects on plant growth, development, and seed production of two Lesquerella species. Ind. Crops Prod. 24, 17–25. doi: 10.1016/j.indcrop.2005.10.004
Ding, N., Chen, Q., Zhu, Z., Peng, L., Ge, S., and Jiang, Y. (2017). Effects of crop load on distribution and utilization of 13C and 15N and fruit quality for dwarf apple trees. Sci. Rep. 7:14172. doi: 10.1038/s41598-017-14509-3
Dong, W., Zhang, X., Dai, X., Fu, X., Yang, F., Liu, X., et al. (2014). Changes in soil microbial community composition in response to fertilization of paddy soils in subtropical China. Appl. Soil Ecol. 84, 140–147. doi: 10.1016/j.apsoil.2014.06.007
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–8. doi: 10.1038/nmeth.2604
Fernandez-Going, B., Even, T., and Simpson, J. (2013). The effect of different nutrient concentrations on the growth rate and nitrogen storage of watercress (Nasturtium officinale R. Br.). Hydrobiologia 705, 63–74. doi: 10.1007/s10750-012-1380-x
Fierer, N., Wood, S. A., and Bueno de Mesquita, C. P. (2021). How microbes can, and cannot, be used to assess soil health. Soil Biol. Biochem. 153:108111. doi: 10.1016/j.soilbio.2020.108111
Fu, L. G., and Chin, C. M. (1992). China Plant Red Data Book: Rare and Endangered Plants [in Chinese]. Beijing: Science Press.
Gabriele, B., Martina, K., Daria, R., Henry, M., Rita, G., and Kornelia, S. (2017). Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol. Ecol. 93:fix050. doi: 10.1093/femsec/fix050
Grosso, F., Iovieno, P., Alfani, A., and De Nicola, F. (2018). Structure and activity of soil microbial communities in three Mediterranean forests. Appl. Soil Ecol. 130, 280–287. doi: 10.1016/j.apsoil.2018.07.007
Gu, X. B., Cai, H. J., Du, Y. D., and Li, Y. N. (2019). Effects of film mulching and nitrogen fertilization on rhizosphere soil environment, root growth and nutrient uptake of winter oilseed rape in northwest China. Soil Till. Res. 187, 194–203. doi: 10.1016/j.still.2018.12.009
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hamm, A. C., Tenuta, M., Krause, D. O., Ominski, K. H., Tkachuk, V. L., and Flaten, D. N. (2016). Bacterial communities of an agricultural soil amended with solid pig and dairy manures, and urea fertilizer. Appl. Soil Ecol. 103, 61–71. doi: 10.1016/j.apsoil.2016.02.015
Harris, S. A., Robinson, J. P., and Juniper, B. E. (2002). Genetic clues to the origin of the apple. Trends Genet. 18, 426–430. doi: 10.1016/s0168-9525(02)02689-6
Hasi, M., Zhang, X., Niu, G., Wang, Y., Geng, Q., Quan, Q., et al. (2021). Soil moisture, temperature and nitrogen availability interactively regulate carbon exchange in a meadow steppe ecosystem. Agric. For. Meteorol. 304–305:108389. doi: 10.1016/j.agrformet.2021.108389
Hatfield, J. L., and Prueger, J. H. (2015). Temperature extremes: effect on plant growth and development. Weather Clim. Extreme 10, 4–10. doi: 10.1016/j.wace.2015.08.0011
He, W., Zhang, M., Jin, G., Sui, X., Zhang, T., and Song, F. (2021). Effects of nitrogen deposition on nitrogen-mineralizing enzyme activity and soil microbial community structure in a Korean pine plantation. Microb. Ecol. 81, 410–424. doi: 10.1007/s00248-020-01595-6
Hu, A., Nie, Y. X., Yu, G. R., Han, C. H., He, J. H., He, N. P., et al. (2019). Diurnal temperature variation and plants drive latitudinal patterns in seasonal dynamics of soil microbial community. Front. Microbiol. 10:674. doi: 10.3389/fmicb.2019.00674
Hu, Y., Chen, M., Yang, Z., Cong, M., Zhu, X., and Jia, H. (2022). Soil microbial community response to nitrogen application on a swamp meadow in the arid region of central Asia. Front. Microbiol. 12:797306. doi: 10.3389/fmicb.2021.797306
Huang, W. J., Zhou, G. Y., Liu, J. X., Zhang, D. Q., Liu, S. Z., Chu, G. W., et al. (2015). Mineral elements of subtropical tree seedlings in response to elevated carbon dioxide and nitrogen addition. PLoS One 10:e0120190. doi: 10.1371/journal.pone.0120190
Jabir, T., Vipindas, P. V., Jesmi, Y., Divya, P. S., Adarsh, B. M., Nafeesathul Miziriya, H. S., et al. (2021). Influence of environmental factors on benthic nitrogen fixation and role of sulfur reducing diazotrophs in a eutrophic tropical estuary. Mar. Pollut. Bull. 165, 112126–112126. doi: 10.1016/j.marpolbul.2021.112126
Jacoby, R., Peukert, M., Succurro, A., Koprivova, A., and Kopriva, S. (2017). The role of soil microorganisms in plant mineral nutrition-current knowledge and future directions. Front. Plant Sci. 8:1617. doi: 10.3389/fpls.2017.01617
Jia, Y., Zhai, G., Zhu, S., Liu, X., Schmid, B., Wang, Z., et al. (2021). Plant and microbial pathways driving plant diversity effects on soil carbon accumulation in subtropical forest. Soil Biol. Biochem. 161:108375. doi: 10.1016/j.soilbio.2021.108375
Jiang, Z. Z., Zhu, H. G., Zhang, Q., Song, B. G., Meng, L. J., and Yang, D. G. (2015). Progress of influence of low temperature on plant photosynthesis. Crops 03, 23–28. doi: 10.16035/j.issn.1001-7283.2015.03.005
Jing, H., Li, J., Yan, B., Wei, F., Wang, G., and Liu, G. (2021). The effects of nitrogen addition on soil organic carbon decomposition and microbial C-degradation functional genes abundance in a Pinus tabulaeformis forest. Forest Ecol. Manag. 489:119098. doi: 10.1016/j.foreco.2021.119098
Joa, J. H., Weon, H. Y., Hyun, H. N., Jeun, Y. C., and Koh, S. W. (2014). Effect of long-term different fertilization on bacterial community structures and diversity in citrus orchard soil of volcanic ash. J. Microbiol. 52, 995–1001. doi: 10.1007/s12275-014-4129-6
Karlidag, H., Esitken, A., Turan, M., and Sahin, F. (2007). Effects of root inoculation of plant growth promoting rhizobacteria (PGPR) on yield, growth and nutrient element contents of leaves of apple. Sci. Hortic. 114, 16–20. doi: 10.1016/j.scienta.2007.04.013
Kurihara, H., Ikeda, N., and Umezawa, Y. (2018). Diurnal and seasonal variation of particle and dissolved organic matter release by the coral Acropora tenuis. PeerJ 6:e5728. doi: 10.7717/peerj.5728
Ladd, J. N., and Butler, J. H. A. (1972). Short-term assays of soil proteolytic enzyme activities using proteins and dipeptide derivatives as substrates. Soil Biol. Biochem. 4, 19–30. doi: 10.1016/0038-0717(72)90038-7
Levakov, I., Ronen, Z., Siebner, H., and Dahan, O. (2021). Continuous in-situ measurement of free extracellular enzyme activity as direct indicator for soil biological activity. Soil Biol. Biochem. 163:108448. doi: 10.1016/j.soilbio.2021.108448
Li, L. J., Zeng, D. H., Yu, Z. A., Fan, Z. P., and Mao, R. (2010). Soil microbial properties under N and P additions in a semi-arid, sandy grassland. Biol. Fertil. Soils 46, 653–658. doi: 10.1007/s00374-010-0463-y
Li, M., Xu, J., Jiang, Z., and Li, Q. (2020). Molecular understanding of autotrophic CO2-fixing bacterial communities in composting based on RuBisCO genes analysis. J. Biotechnol. 320, 36–43. doi: 10.1016/j.jbiotec.2020.06.010
Li, Y., Wu, Z., Dong, X., Jia, Z., and Sun, Q. (2019). Variance in bacterial communities, potential bacterial carbon sequestration and nitrogen fixation between light and dark conditions under elevated CO2 in mine tailings. Sci. Total Environ. 652, 234–242. doi: 10.1016/j.scitotenv.2018.10.253
Li, Y., Yuan, L., Xue, S., Liu, B., and Jin, G. (2021c). Artificial root exudates excite bacterial nitrogen fixation in the subsurface of mine soils. Appl. Soil Ecol. 157:103774. doi: 10.1016/j.apsoil.2020.103774
Li, J., Chen, Q., Li, Q., Zhao, C., and Feng, Y. (2021a). Influence of plants and environmental variables on the diversity of soil microbial communities in the Yellow River Delta Wetland, China. Chemosphere 274:129967. doi: 10.1016/j.chemosphere.2021.129967
Li, Y., Wang, C., Gao, S., Wang, P., Qiu, J., and Shang, S. (2021b). Impacts of simulated nitrogen deposition on soil enzyme activity in a northern temperate forest ecosystem depend on the form and level of added nitrogen. Eur. J. Soil Biol. 103:103287. doi: 10.1016/j.ejsobi.2021.103287
Liao, H., Qin, F., Wang, K., Zhang, Y., Hao, X., Chen, W., et al. (2020). Long-term chemical fertilization-driving changes in soil autotrophic microbial community depresses soil CO2 fixation in a Mollisol. Sci. Total Environ. 748:141317. doi: 10.1016/j.scitotenv.2020.141317
Liu, M., Zhang, W., Wang, X., Wang, F., Dong, W., Hu, C., et al. (2020). Nitrogen leaching greatly impacts bacterial community and denitrifiers abundance in subsoil under long-term fertilization. Agr. Ecosyst. Environ. 294:106885. doi: 10.1016/j.agee.2020.106885
Liu, S., Wang, Z., Niu, J., Dang, K., Zhang, S., Wang, S., et al. (2021). Changes in physicochemical properties, enzymatic activities, and the microbial community of soil significantly influence the continuous cropping of Panax quinquefolius L. (American ginseng). Plant Soil 463, 427–446. doi: 10.1007/s11104-021-04911-2
Liu, X., Lu, X., Zhao, W., Yang, S., Wang, J., Xia, H., et al. (2022). The rhizosphere effect of native legume Albizzia julibrissin on coastal saline soil nutrient availability, microbial modulation, and aggregate formation. Sci. Total Environ. 806:150705. doi: 10.1016/j.scitotenv.2021.150705
Liu, Z. Q., and Dong, H. G. (2018). Spatial distribution and survival status of apple seedlings in wild apple forest in Xinjiang – A case study of Xinyuan county. Xingjiang Agr. Sci. Tech. 5, 37–41. doi: 10.3969/j.issn.1007-3574.2018.05.018
Lozano, Y. M., Hortal, S., Armas, C., and Pugnaire, F. I. (2014). Interactions among soil, plants, and microorganisms drive secondary succession in a dry environment. Soil Biol. Biochem. 78, 298–306. doi: 10.1016/j.soilbio.2014.08.007
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Modi, A., Vai, S., Caramelli, D., and Lari, M. (2021). “The illumina sequencing protocol and the NovaSeq 6000 System,” in Bacterial Pangenomics, Vol. 2242, eds A. Mengoni, G. Bacci, and M. Fondi (New York: Humana), 15–42. doi: 10.1007/978-1-0716-1099-2_2
Mommer, L., Kirkegaard, J., and van Ruijven, J. (2016). Root-Root interactions: towards a rhizosphere framework. Trends Plant Sci. 21, 209–217. doi: 10.1016/j.tplants.2016.01.009
Nagakura, J., Shigenaga, H., Akama, A., and Takahashi, M. (2004). Growth and transpiration of Japanese cedar (Cryptomeria japonica) and Hinoki cypress (Chamaecyparis obtusa) seedlings in response to soil water content. Tree Physiol. 24, 1203–1208. doi: 10.1093/treephys/24.11.1203
Nanasato, Y., Uenoyama, R., Tomooka, K., Kato, A., and Akashi, K. (2018). Nuclear isolation and purification using SDS/urea (NIPSU) method for efficient and rapid extraction of high-purity genomic DNAs from Jatropha curcas L: a comparative analysis of DNA isolation protocols. Afr. J. Biotechnol. 17, 981–988. doi: 10.5897/AJB2017.16325
Nannipieri, P., Trasar-Cepeda, C., and Dick, R. P. (2018). Soil enzyme activity: a brief history and biochemistry as a basis for appropriate interpretations and meta-analysis. Biol. Fertil. Soils 54, 11–19. doi: 10.1007/s00374-017-1245-6
Nihorimbere, V., Ongena, M., Smargiassi, M., and Thonart, P. (2011). Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnol. Agron. Soc. 15, 327–337. doi: 10.1071/EN10115
Nunes-Nesi, A., Fernie, A. R., and Stitt, M. (2010). Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 3, 973–996. doi: 10.1093/mp/ssq049
Oliveira, V., Gomes, N., Correia, A., Henriques, I., Marques, B., and Lillebø, A. (2012). Impact of sampling depth and plant species on local environmental conditions, microbiological parameters and bacterial composition in a mercury contaminated salt marsh. Mar. Pollut. Bull. 64, 263–271. doi: 10.1016/j.marpolbul.2011.11.020
Purahong, W., Orrù, L., Donati, I., Perpetuini, G., Cellini, A., Lamontanara, A., et al. (2018). Plant microbiome and its link to plant health: host species, organs and Pseudomonas syringae pv. actinidiae infection shaping bacterial phyllosphere communities of kiwifruit plants. Front. Plant Sci. 9:1563. doi: 10.3389/fpls.2018.01563
Rawat, J., Yadav, N., and Pande, V. (2020). “Role of rhizospheric microbial diversity in plant growth promotion in maintaining the sustainable agrosystem at high altitude regions,” in Recent Advancements in Microbial Diversity, eds S. de Mandal and P. Bhatt (Cambridge: Academic Press), 147–196. doi: 10.1016/B978-0-12-821265-3.00007-4
Ren, J., Liu, X., Yang, W., Yang, X., Li, W., Xia, Q., et al. (2021). Rhizosphere soil properties, microbial community, and enzyme activities: short-term responses to partial substitution of chemical fertilizer with organic manure. J. Environ. Manage. 299:113650. doi: 10.1016/j.jenvman.2021.113650
Rigg, J. L., Offord, C. A., Singh, B. K., Anderson, I. C., Clarke, S., and Powell, J. R. (2016). Variation in soil microbial communities associated with critically endangered Wollemi pine affects fungal, but not bacterial, assembly within seedling roots. Pedobiologia 9, 61–71. doi: 10.1016/j.pedobi.2016.02.002
Rodriguez-Lovelle, B., and Gaudillère, J. P. (2002). Carbon and nitrogen partitioning in either fruiting or non-fruiting grapevines: effects of nitrogen limitation before and after veraison. Aust. J. Grape Wine Res. 8, 86–94. doi: 10.1111/j.1755-0238.2002.tb00216.x
Rzehak, T., Praeg, N., and Illmer, P. (2022). A standardized and miniaturized method to investigate rhizosphere microorganisms, with a focus on methanogenic archaea and methanotrophic bacteria. Pedobiologia 90:150775. doi: 10.1016/j.pedobi.2021.150775
Sakai, K., Sato, Y., Okada, M., and Yamaguchi, S. (2022). Enhanced activity and stability of protein-glutaminase by Hofmeister effects. Mol. Catal. 517:112054. doi: 10.1016/j.mcat.2021.112054
Samuel, M. (2014). Is the Thermodynamic Efficiency of Soil Microbial Communities Related to Ecosystem Maturity and Stress?. Bedford: Cranfield University.
Shao, T., Zhao, J., Liu, A., Long, X., and Rengel, Z. (2020). Effects of soil physicochemical properties on microbial communities in different ecological niches in coastal area. Appl. Soil Ecol. 150:103486. doi: 10.1016/j.apsoil.2019.103486
Sheel, B., Marie-Charlotte, N., and Wardle, D. A. (2012). Response of photosynthetic carbon gain to ecosystem retrogression of vascular plants and mosses in the boreal forest. Oecologia 169, 661–672. doi: 10.1007/s00442-011-2246-z
Shen, C., Ni, Y., Liang, W., Wang, J., and Chu, H. (2015). Distinct soil bacterial communities along a small-scale elevational gradient in alpine tundra. Front. Microbiol. 6:582. doi: 10.3389/fmicb.2015.00582
Sitpayeva, G. T., Kudabayevà, G., Dimeyeva, L. A., Gemejiyeva, N. G., and Vesselova, P. V. (2020). Crop wild relatives of Kazakhstani Tien Shan: flora, vegetation, resources. Plant Divers. 42, 19–32. doi: 10.1016/j.pld.2019.10.003
Sivanandhan, G., Theboral, J., Dev, G. K., Selvaraj, N., Manickavasagam, M., and Ganapathi, A. (2015). Effect of carbon and nitrogen sources on in vitro flower and fruit formation and withanolides production in Withania somnifera (L.) Dunal. Indian J. Exp. Biol. 53, 177–183. doi: 10.1016/j.joca.2012.02.579
Soman, C., Li, D. F., Wander, M. M., and Kent, A. D. (2017). Long-term fertilizer and crop-rotation treatments differentially affect soil bacterial community structure. Plant Soil 413, 145–159. doi: 10.1007/s11104-016-3083-y
Steinweg, J. M., Dukes, J. S., and Wallenstein, M. D. (2012). Modeling the effects of temperature and moisture on soil enzyme activity: linking laboratory assays to continuous field data. Soil Biol. Biochem. 55, 85–92. doi: 10.1016/j.soilbio.2012.06.015
Sun, C., Liu, G., and Xue, S. (2018). Interaction between plant competition and rhizospheric bacterial community influence secondary succession of abandoned farmland on the loess plateau of China. Front. Plant Sci. 9:898. doi: 10.3389/fpls.2018.00898
Tan, X., Nie, Y., Ma, X., Guo, Z., Liu, Y., Tian, H., et al. (2021). Soil chemical properties rather than the abundance of active and potentially active microorganisms control soil enzyme kinetics. Sci. Total Environ. 770:144500. doi: 10.1016/j.scitotenv.2020.144500
Tian, J., Dungait, J. A. J., Lu, X., Yang, Y., Hartley, I. P., Zhang, W., et al. (2019). Long-term nitrogen addition modifies microbial composition and functions for slow carbon cycling and increased sequestration in tropical forest soil. Glob. Chang. Biol. 25, 3267–3281. doi: 10.1111/gcb.14750
Tulina, A. S. (2019). Effects îf temperature, moisture ànd straw incorporation în organic matter mineralization dynamics ànd soil carbon ànd nitrogen pools. Agrokhimiya 3, 3–18. doi: 10.1134/s0002188119030141
Utobo, E. B., and Tewari, L. (2015). Soil enzymes as bioindicators of soil ecosystem status. Appl. Ecol. Env. Res. 13, 147–169. doi: 10.15666/aeer/1301_147169
Viswanathan, P. N., and Krishnan, L. (1962). Diurnal variations in some enzymes of carbohydrate metabolism in tapioca leaves. Plant Physiol. 37, 283–287. doi: 10.2307/4259913
Vitousek, P. M., Menge, D., Reed, S. C., and Cleveland, C. C. (2013). Biological nitrogen fixation: rates, patterns and ecological controls in terrestrial ecosystems. Philos. Trans. R. Soc. B Biol. Sci. 368:20130119. doi: 10.1098/rstb.2013.0119
Vives-Peris, V., de Ollas, C., Gomez-Cadenas, A., and Perez-Clemente, R. M. (2020). Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17. doi: 10.1007/s00299-019-02447-5
Vourlitis, G. L., and Hentz, C. S. (2016). Impacts of chronic N input on the carbon and nitrogen storage of a postfire Mediterranean-type shrubland. J. Geophys. Res. Biogeosci. 121, 385–398. doi: 10.1002/2015JG003220
Wang, B., Gong, J., Zhang, Z., Yang, B., Liu, M., Zhu, C., et al. (2019). Nitrogen addition alters photosynthetic carbon fixation, allocation of photoassimilates, and carbon partitioning of Leymus chinensis in a temperate grassland of Inner Mongolia. Agric. For. Meteorol. 279:107743. doi: 10.1016/j.agrformet.2019.107743
Wang, C., Liu, D., and Bai, E. (2018). Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition. Soil Biol. Biochem. 120, 126–133. doi: 10.1016/j.soilbio.2018.02.003
Wang, F., Sha, J., Chen, Q., Xu, X., Zhu, Z., Ge, S., et al. (2020). Exogenous abscisic acid regulates distribution of 13C and 15N and Anthocyanin Synthesis in ‘Red Fuji’ apple fruit under high nitrogen supply. Front. Plant Sci. 10:1738. doi: 10.3389/fpls.2019.01738
Wang, L., Xin, J., Nai, H., and Zheng, X. (2021). Effects of different fertilizer applications on nitrogen leaching losses and the response in soil microbial community structure. Environ. Technol. Innov. 23:101608. doi: 10.1016/j.eti.2021.101608
Wang, M., Sun, H., Xu, L., and Xu, Z. (2021). Bacterial diversity in tea plant (Camellia sinensis) rhizosphere soil from Qinling Mountains and its relationship with environmental elements. Plant Soil 460, 403–415. doi: 10.1007/s11104-020-04822-8
Wang, N., Jiang, S., Zhang, Z., Fang, H., Xu, H., Wang, Y., et al. (2018). Malus sieversii: the origin, flavonoid synthesis mechanism, and breeding of red-skinned and red-fleshed apples. Hortic. Res. 5:70. doi: 10.1038/s41438-018-0084-4
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, W., Wang, N., Dang, K., Dai, W., Guan, L., Wang, B., et al. (2020). Long-term nitrogen application decreases the abundance and copy number of predatory myxobacteria and alters the myxobacterial community structure in the soil. Sci. Total Environ. 708:135114. doi: 10.1016/j.scitotenv.2019.135114
Wang, X., Li, W., Xiao, Y., Cheng, A., Shen, T., Zhu, M., et al. (2021). Abundance and diversity of carbon-fixing bacterial communities in karst wetland soil ecosystems. Catena 204:105418. doi: 10.1016/j.catena.2021.105418
Wiedow, C., Dehmer, K. J., and Geibel, M. (2004). Molecular diversity in populations of Malus sieversii (Leded.) Roem. Acta Hortic. 663, 539–544. doi: 10.17660/ActaHortic.2004.663.94
Wu, L., Jiang, Y., Zhao, F., He, X., Liu, H., and Yu, K. (2020). Increased organic fertilizer application and reduced chemical fertilizer application affect the soil properties and bacterial communities of grape rhizosphere soil. Sci. Rep. 10:9568. doi: 10.1038/s41598-020-66648-9
Wu, R., Cheng, X. Q., Zhou, W. S., and Han, H. R. (2019). Microbial regulation of soil carbon properties under nitrogen addition and plant inputs removal. PeerJ 7:e7343. doi: 10.7717/peerj.7343
Xiao, G., Zhang, Q., Bi, J., Zhang, F., and Luo, C. (2012). The relationship between winter temperature rise and soil fertility properties. Air Soil Water Res. 5:15. doi: 10.4137/ASWR.S8599
Xiao, R., Ran, W., Hu, S., and Guo, H. (2021). The response of ammonia oxidizing archaea and bacteria in relation to heterotrophs under different carbon and nitrogen amendments in two agricultural soils. Appl. Soil Ecol. 158:103812. doi: 10.1016/j.apsoil.2020.103812
Xun, W. B., Zhao, J., Xue, C., Zhang, G. S., Ran, W., Wang, B. R., et al. (2016). Significant alteration of soil bacterial communities and organic carbon decomposition by different long-term fertilization management conditions of extremely low-productivity arable soil in South China. Environ. Microbiol. 18, 1907–1917. doi: 10.1111/1462-2920.13098
Yan, G. Y., Han, S. J., Zhou, M. X., Sun, W. J., Huang, B. B., Wang, H. L., et al. (2020). Variations in the natural 13C and 15N abundance of plants and soils under long-term N addition and precipitation reduction: interpretation of C and N dynamics. For. Ecosyst. 7:49. doi: 10.1186/s40663-020-00257-w
Yang, W. T., Miao, J. Q., Wang, X. W., Xu, J. C., Lu, M. J., and Li, Z. X. (2018). Corn-soybean intercropping and nitrogen rates affected crop nitrogen and carbon uptake and C:N ratio in upland red soil. J. Plant Nutr. 41, 1890–1902. doi: 10.1080/01904167.2018.1476540
Yang, X., Ni, K., Shi, Y., Yi, X., Ji, L., Ma, L., et al. (2020). Heavy nitrogen application increases soil nitrification through ammonia-oxidizing bacteria rather than archaea in acidic tea (Camellia sinensis L.) plantation soil. Sci. Total Environ. 717:137248. doi: 10.1016/j.scitotenv.2020.137248
Yin, X., Peñuelas, J., Xu, X., Sardans, J., Fang, Y., Wiesmeier, M., et al. (2021). Effects of addition of nitrogen-enriched biochar on bacteria and fungi community structure and C, N, P, and Fe stoichiometry in subtropical paddy soils. Eur. J. Soil. Biol. 106:103351. doi: 10.1016/j.ejsobi.2021.103351
Yu, X. Y., Zhu, Y. J., Wang, B., Liu, D., Bai, H., Jin, L., et al. (2021). Effects of nitrogen addition on rhizospheric soil microbial communities of poplar plantations at different ages. For. Ecol. Manag. 494:119328. doi: 10.1016/j.foreco.2021.119328
Zak, D. R., Holmes, W. E., White, D. C., Peacock, A. D., and Tilman, D. (2003). Plant diversity, soil microbial communities, and ecosystem function: are there any links? Ecology 84, 2042–2050. doi: 10.1890/02-0433
Zhang, C. C., Zhou, C. Z., Burnap, R. L., and Peng, L. (2018). Carbon/Nitrogen metabolic balance: lessons from Cyanobacteria. Trends Plant Sci. 23, 1116–1130. doi: 10.1016/j.tplants.2018.09.008
Zhang, D., Yan, F., Sun, Z., Zhang, Q., Xue, S., and Cong, W. (2014). On-line modeling intracellular carbon and energy metabolism of Nannochloropsis sp. in nitrogen-repletion and nitrogen-limitation cultures. Bioresour. Technol. 164, 86–92. doi: 10.1016/j.biortech.2014.04.083
Zhang, P., Kong, X., Bakker, E. S., Xu, J., and Zhang, M. (2021). Temperature affects carbon and nitrogen stable isotopic signatures of aquatic plants. Aquat. Sci. 83:39. doi: 10.1007/s00027-021-00794-8
Zhang, R., Xu, C., Bao, Z. L., Xiao, R., Chen, X. D., Xiao, W., et al. (2021). Auxin alters sodium ion accumulation and nutrient accumulation by playing protective role in salinity challenged strawberry. Plant Physiol. Biochem. 164, 1–9. doi: 10.1016/j.plaphy.2021.04.008
Zhang, X., Teng, Z., Zhang, H., Cai, D., Zhang, J., Meng, F., et al. (2021). Nitrogen application and intercropping change microbial community diversity and physicochemical characteristics in mulberry and alfalfa rhizosphere soil. J. For. Res. 32, 2121–2133. doi: 10.1007/s11676-020-01271-y
Zhao, Y., Liang, C., Shao, S., Li, J., Xie, H., Zhang, W., et al. (2021). Interactive effects of elevated CO2 and nitrogen fertilization levels on photosynthesized carbon allocation in a temperate spring wheat and soil system. Pedosphere 31, 191–203. doi: 10.1016/S1002-0160(20)60056-X
Zhong, C., Jian, S. F., Chen, D. L., Huang, X. J., and Miao, J. H. (2021). Organic nitrogen sources promote andrographolide biosynthesis by reducing nitrogen metabolism and increasing carbon accumulation in Andrographis paniculata. Plant Physiol. Biochem. 164, 82–91. doi: 10.1016/j.plaphy.2021.04.016
Zhou, G., Terrer, C., Hungate, B., Van Gestel, N., Zhou, X., and van Groenigen, K. J. (2021). Nitrogen availability controls plant carbon storage with warming. Res. Sq. [Preprint]. doi: 10.21203/rs.3.rs-432920/v2
Keywords: temperature stress, rhizosphere soil, carbon and nitrogen isotope, bacterial community, Malus sieversii
Citation: Zhang H, Phillip FO, Wu L, Zhao F, Yu S and Yu K (2022) Effects of Temperature and Nitrogen Application on Carbon and Nitrogen Accumulation and Bacterial Community Composition in Apple Rhizosphere Soil. Front. Plant Sci. 13:859395. doi: 10.3389/fpls.2022.859395
Received: 21 January 2022; Accepted: 28 February 2022;
Published: 04 April 2022.
Edited by:
Anoop Kumar Srivastava, Indian Council of Agricultural Research (ICAR), IndiaReviewed by:
Ram Awadh Ram, Central Institute for Subtropical Horticulture, IndiaCopyright © 2022 Zhang, Phillip, Wu, Zhao, Yu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kun Yu, eXVrdW40MTBAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.