- 1Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
- 2National Key Facility of Crop Gene Resources and Genetic Improvement, Beijing, China
Soil salinity is a major environmental stress that restricts the growth and yield of crops. Mining the key genes involved in the balance of rice salt tolerance and yield will be extremely important for us to cultivate salt-tolerance rice varieties. In this study, we report a WUSCHEL-related homeobox (WOX) gene, quiescent-center-specific homeobox (OsQHB), positively regulates yield-related traits and negatively regulates salt tolerance in rice. Mutation in OsQHB led to a decrease in plant height, tiller number, panicle length, grain length and grain width, and an increase in salt tolerance. Transcriptome and qPCR analysis showed that reactive oxygen species (ROS) scavenging-related genes were regulated by OsQHB. Moreover, the osqhb mutants have higher ROS-scavenging enzymes activities and lower accumulation of ROS and malondialdehyde (MDA) under salt stress. Thus, our findings provide new insights into the role of rice WOX gene family in rice development and salt tolerance, and suggest that OsQHB is a valuable target for improving rice production in environments characterized by salt stress.
Introduction
Soil salinity is one of the most widespread and significant soil problems worldwide, as salt hampers plant growth and development and hinders crop yield. As the primary source of food for more than half of the world population, rice is affected by salinity stress in varying degrees throughout all the developmental stages (Nam et al., 2015; Saini et al., 2018; He et al., 2019). Especially, rice is more sensitive to salinity during early seedling growth and flowering than other growth stages (Nam et al., 2015; Qin et al., 2019). Therefore, the growth ability of rice seedlings under salt stress conditions is considered an indicator of salt tolerance, and improving the salt tolerance of rice seedlings could increase the utilization of saline-alkali land and alleviate the world food crisis. Salt tolerance in rice is a polygenic trait controlled by quantitative trait loci (Ismail and Horie, 2017; Qin et al., 2020). Dissecting the key genes involved in rice salt tolerance and yield is an important objective to accelerating rice breeding.
Reactive oxygen species (ROS) is a versatile signal molecule that could be rapidly induced by a variety of environmental stresses (Miller et al., 2010; Lv et al., 2018). Generally, low concentrations of ROS function as signal molecules to regulate many biological processes, whereas high concentrations of ROS damage proteins, lipids, DNA, and carbohydrates (Miller et al., 2010; Xu et al., 2018; Yang and Guo, 2018). Therefore, maintaining an appropriate level of ROS is essential for plant growth and development. To avoid excessive accumulation of ROS and cause oxidative damage to cells, plants have evolved defense systems that include ROS-scavenging enzymes, such as superoxide dismutase (SOD), ascorbate peroxidase (APX), glutathione peroxidase (GPX), and catalase (CAT) (Foyer and Noctor, 2005). Various studies have shown that salt stress leads to excessive accumulation of ROS, and enhancing the activity of ROS-scavenging enzymes can improve the salt tolerance of plants (Lu Z. et al., 2007; Saini et al., 2018). For example, overexpression of OsAPXa or OsAPXb in Arabidopsis enhances tolerance to salinity stress (Lu Z. et al., 2007). Knockdown GPX1 leads to enhanced photosynthesis impairment in response to salinity in rice (Lima-Melo et al., 2016). Transgenic rice plants overexpression APX exhibit reduced ROS accumulation and enhanced salt tolerance (Teixeira et al., 2006; Hong et al., 2007; Zhang et al., 2013).
The WUSCHEL related homeobox (WOX) gene family is one of plant homeobox (HB) transcription factor families (Yang et al., 2017). WOX genes have been shown to function in coordinate gene transcriptional related to shoot and root meristem establishment and organogenesis (Zhao et al., 2009; Hu and Xu, 2016; Cheng et al., 2018). In Arabidopsis, AtWOX5 is specifically expressed in the quiescent center (QC) and acts as a key regulator of the root stem cell population (Sarkar et al., 2007; Forzani et al., 2014; Savina et al., 2020). AtWOX9 is essential to maintaining cell division and preventing premature differentiation in vegetative shoot apical meristem and early embryogenesis (Wu et al., 2005, 2007; Skylar et al., 2010). In rice, OsWOX3A modulates lateral root development and root hair formation through regulating auxin-transport gene expression (Yoo et al., 2013; Cho et al., 2016). OsWOX4 is involved in meristem maintenance and acts as a key regulator in early leaf development and primary root elongation (Ohmori et al., 2013; Yasui et al., 2018; Chen et al., 2020). OsWOX11 promotes crown root and shoot development by modulating cell proliferation in crown root meristem and shoot apical meristem (Zhao et al., 2009; Zhou et al., 2017; Cheng et al., 2018). Quiescent-center-specific homeobox (OsQHB), a homolog of AtWOX5, is involved in specification and maintenance of QC cell in root apical meristem and controlling lateral root primordium size (Kamiya et al., 2003; Kawai et al., 2022). All these studies suggest that WOX genes play an important role in regulating plant development.
As sessile organisms, plants must cope with various stresses in their environment to ensure the optimal combination of proliferation and survival. Accumulating studies show that WOX genes are associated with plant abiotic stress responses (Cheng et al., 2016; Liu H. et al., 2021; Wang L. Q. et al., 2021). In poplar, PagWOX11/12a positively regulates drought and salt tolerance by enhancing ROS scavenging capacity (Liu R. et al., 2021; Wang L. Q. et al., 2021). Knock-down of GhWOX4 in cotton decreases the drought tolerance (Sajjad et al., 2021). In rice, overexpression of OsWOX11 enhances drought tolerance by promoting root hair growth and development (Cheng et al., 2016). In addition, several rice WOX genes are responsive to salt stress (Cheng et al., 2014), implying that WOX genes might involve in improving the salt tolerance of rice. In the present study, we demonstrate that OsQHB coordinately regulates salt tolerance and yield of rice. Transcriptome analysis and physiological and biochemical indices show that OsQHB negatively regulates salt tolerance by improving ROS scavenging capacity. Thus, our researches enrich the functions of WOX genes in rice, and precisely manipulation of OsQHB could be useful to improve rice yield under salt stress.
Results
OsQHB Is a Nuclear Localization Protein and Mainly Expressed in Root and Shoot Apical Meristem
WOX transcription factors play important roles in key developmental processes and in response to different abiotic stresses (Cheng et al., 2016; Jha et al., 2020; Wang L. Q. et al., 2021). Rice genome contains at least 13 WOX genes (Zhang et al., 2010). Among them, OsQHB encodes 200 amino acids including a homeodomain (HD) with 66 amino acids at the N-terminal (Supplementary Figure 1A). Phylogenetic analysis of rice and Arabidopsis WOX families indicated that OsQHB showed the highest similarity to AtWOX5 (Supplementary Figure 1B and Supplementary Table 1), which is specifically expression in the QC cells and is essential for stem cell maintenance in different meristems (Sarkar et al., 2007; Pi et al., 2015). β-glucuronidase (GUS) staining analysis using transgenic rice plants harboring OsQHB promoter-GUS construct showed that the transcription of OsQHB was detected in QC and stele of the root apexes, stem base, and crown root primordium (Figures 1A–D). Subsequently, the subcellular localization of OsQHB was investigated by fusing the OsQHB coding sequence with GFP and transiently expressed in rice protoplasts. The fluorescent signals of OsQHB-GFP fusion protein were found in the nucleus, as revealed by co-localization with the nuclear marker DAPI (4′,6-diamidino-2-phenylidone) (Yu et al., 2013; Figure 1E). These results suggest that OsQHB might be involved in the maintenance of the QC cells through a mechanism similar to that of AtWOX5.
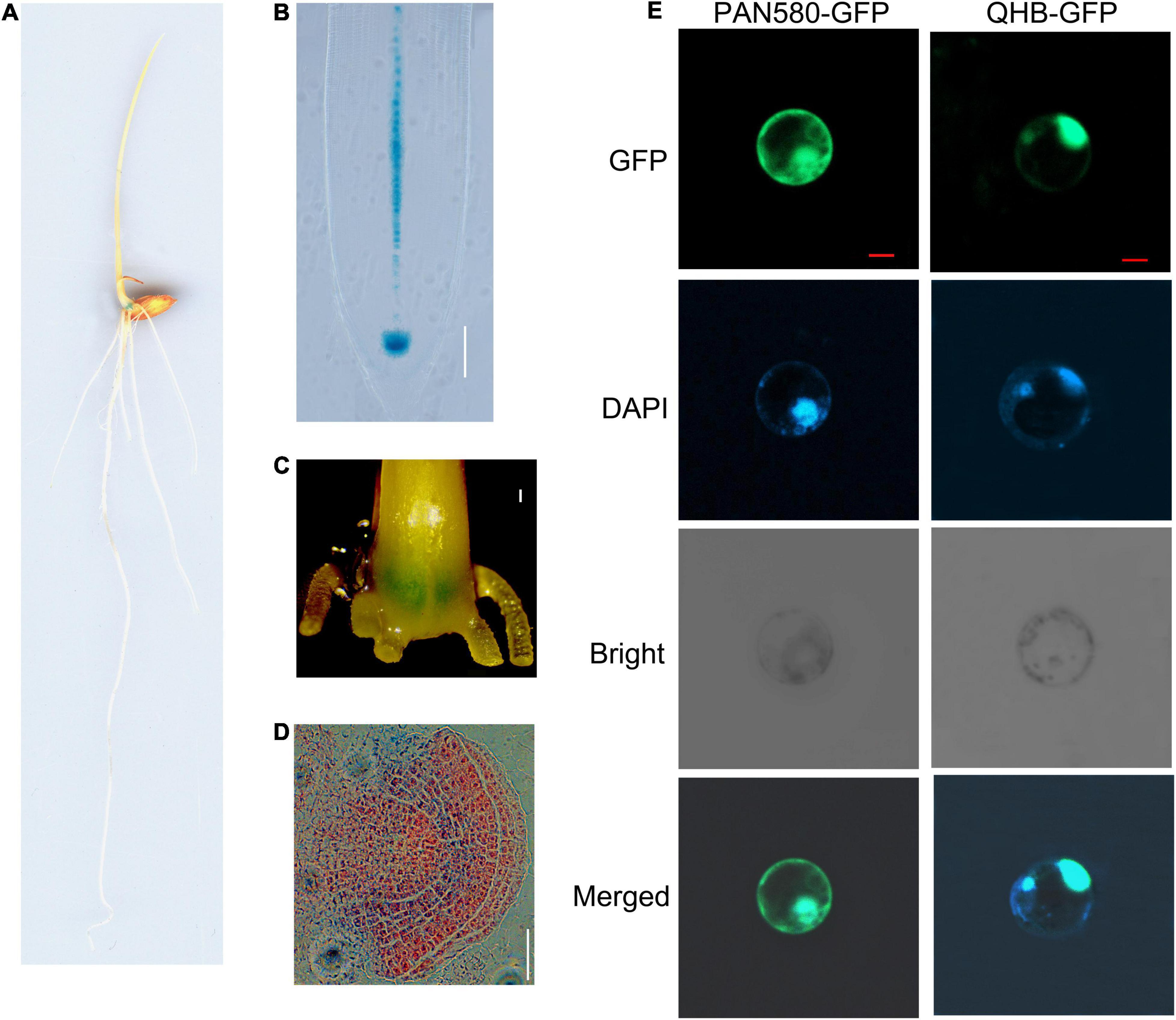
Figure 1. OsQHB expression and protein subcellular localization. (A–D) The tissue-specific expression of OsQHB revealed by promoter-GUS analysis. 3-d-old OsQHBp-GUS transgenic seedlings was used for GUS staining. At least 10 samples were observed for each organ and representative ones are presented. (A) 3-d-old seedling, (B) Primary root, bar = 100 μm, (C) Stem base, Bar = 200 μm. (D) crown root primordium. Bar = 200 μm. (E) Subcellular localization of OsQHB in rice protoplasts. Protoplasts were derived from etiolated shoots. Fluorescence from GFP and DAPI was detected by confocal microscopy. Merged indicates co-localization of OsQHB with DAPI. Bar = 5 μm. DAPI, 4′,6-diamidino-2-phenylindole.
OsQHB Positively Regulates Yield-Related Traits
To study the function of OsQHB, we generated knockout mutants in the Nipponbare (NIP) background via CRISPR-Cas9 mediated genome editing and isolated three alleles for further investigation. The osqhb-1 and osqhb-2 mutant plants contained a 5-bp and 8-bp deletion, whereas the osqhb-3 mutant plants contained a 1-bp insertion, in the coding region of the target gene, correspondingly, leading to a frame shift in the open reading frame and the generation of a premature stop codon (Supplementary Figures 2A,B). We also generated overexpression (OE) lines containing the coding region of OsQHB under the control of the CaMV35S promoter, the increased expression of the target gene was confirmed by qPCR (Supplementary Figure 2C). Although the expression of OsQHB was significantly increased in OsQHB-OE plants, whereas the OsQHB protein was increased slightly in OsQHB-OE plants (Supplementary Figure 2D), suggesting that OsQHB may be rapidly degraded by the ubiquitin system or the proteasome. To verify this hypothesis, we treated OsQHB-OE plants with MG132 (an inhibitor of the ubiquitin proteasome system) and then detected the OsQHB protein levels. The results showed that MG132 treatment significantly increased the accumulation of OsQHB protein (Supplementary Figure 2D), indicating that OsQHB is rapidly degraded by the ubiquitin proteasome system, and implying that OsQHB is essential for plant development and plants must maintain low levels of OsQHB protein to ensure normal growth.
Next, we investigated the agronomic traits of OsQHB overexpressed plants and osqhb mutants. At maturation stage, the osqhb mutants were shorter and the OsQHB-OE plants were slightly taller than wild-type plants (Figures 2A,B). The number of effective tiller and panicle length in wild-type and OsQHB-OE plants were identical, whereas osqhb mutants produced less effective tiller and smaller panicle than the wild type (Figures 2C–E). The total number of grains per panicle was significantly reduced in osqhb mutants, whereas slightly increased in OsQHB-OE plants, compared to that of the wild type (Figure 2F). All these analyses indicate that modulation of OsQHB expression affects yield-related traits in rice.
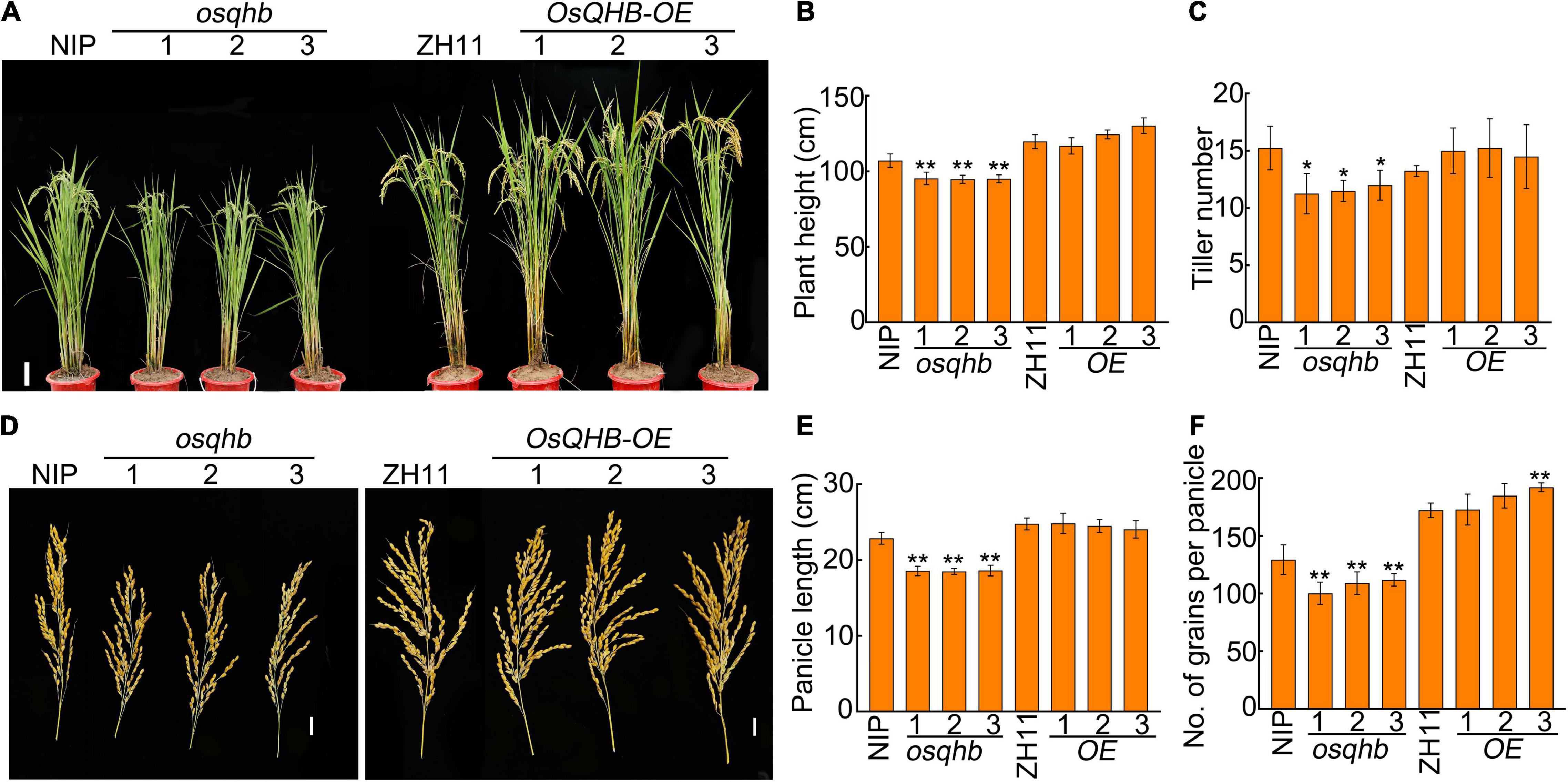
Figure 2. Mutation in OsQHB reduced plant height, tiller number, panicle length, and number of grains per panicle. (A) Phenotypic comparison of field-grown wild-type and OsQHB transgenic plants. Bar = 10 cm. (B,C) Plant height (B) and tiller number (C) of field-grown wild-type and OsQHB transgenic plants. Values are means ± SD (n ≥ 20). * and ** indicates significant difference compared to NIP at P < 0.05 and P < 0.01 by Student’s t-test. (D) Panicle phenotypes of wild-type and OsQHB transgenic plants. Bar = 2 cm. (E,F) Panicle length (E) and number of grains per panicle (F) of wild-type and OsQHB transgenic plants. Each value is average of 20 plants. Bars indicate SD. ** indicates significant difference compared to NIP or ZH11 at P < 0.01 by Student’s t-test.
The grain size was also examined in osqhb mutants and OsQHB-OE plants using well-filled grains. The grain length and grain width was significantly reduced in osqhb mutants, whereas increased in OsQHB-OE plants, compared with those in the wild type (Figures 3A–C). The thousand-grain weight of various plants was further measured. The results showed that osqhb mutants had significant reductions, whereas the OsQHB-OE plants had significant increases in this parameter (Figure 3D). These results indicate that OsQHB positively regulates the grain size and thousand-grain weight in rice, implying that manipulation of OsQHB could be useful to improve rice yield.
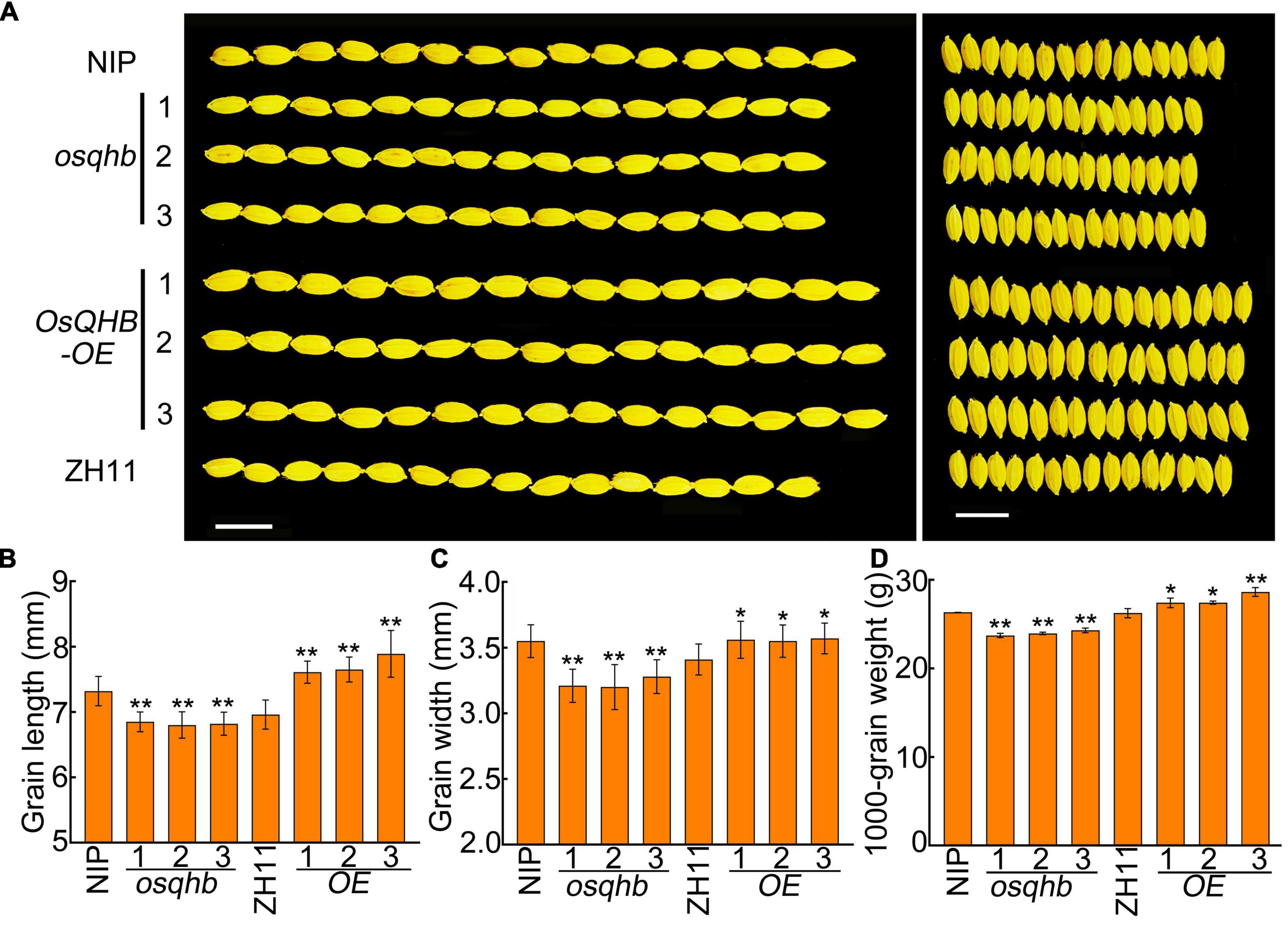
Figure 3. OsQHB positively regulates grain size. (A) Phenotypic comparison of well-filled grains of wild-type and OsQHB transgenic plants. Bar = 1 cm. (B–D) Grain length (B), grain width (C) and 1,000-grain weight (D) from well-filled grains. Values are means ± SD of three replicates. * and ** indicates significant difference compared to NIP or ZH11 at P < 0.05 and P < 0.01 by Student’s t-test.
OsQHB Is Involved in Abiotic Stress and Hormone Response
The promoter sequence of OsQHB contains many putative hormone- and stress-response cis-elements, such as ABRE element (4 hits), AuxRR core (2 hits), P-box (1 hit), G-box (6 hits), and MYB recognition site (1 hit) (Figure 4A). ABRE element is associated with ABA response (Nakashima and Yamaguchi-Shinozaki, 2013), G-box and MYB recognition site with drought response (Ezer et al., 2017), AuxRR core with auxin response, and P-box with gibberellin response (Sakai et al., 1996; Hwang et al., 2010; Wang Y. et al., 2011; Ambawat et al., 2013; Wei et al., 2013; Zhang et al., 2020; Dutta et al., 2021). These elements are contained in the promoter of OsQHB suggests that OsQHB might be involved in modulating hormone response and stress tolerance.
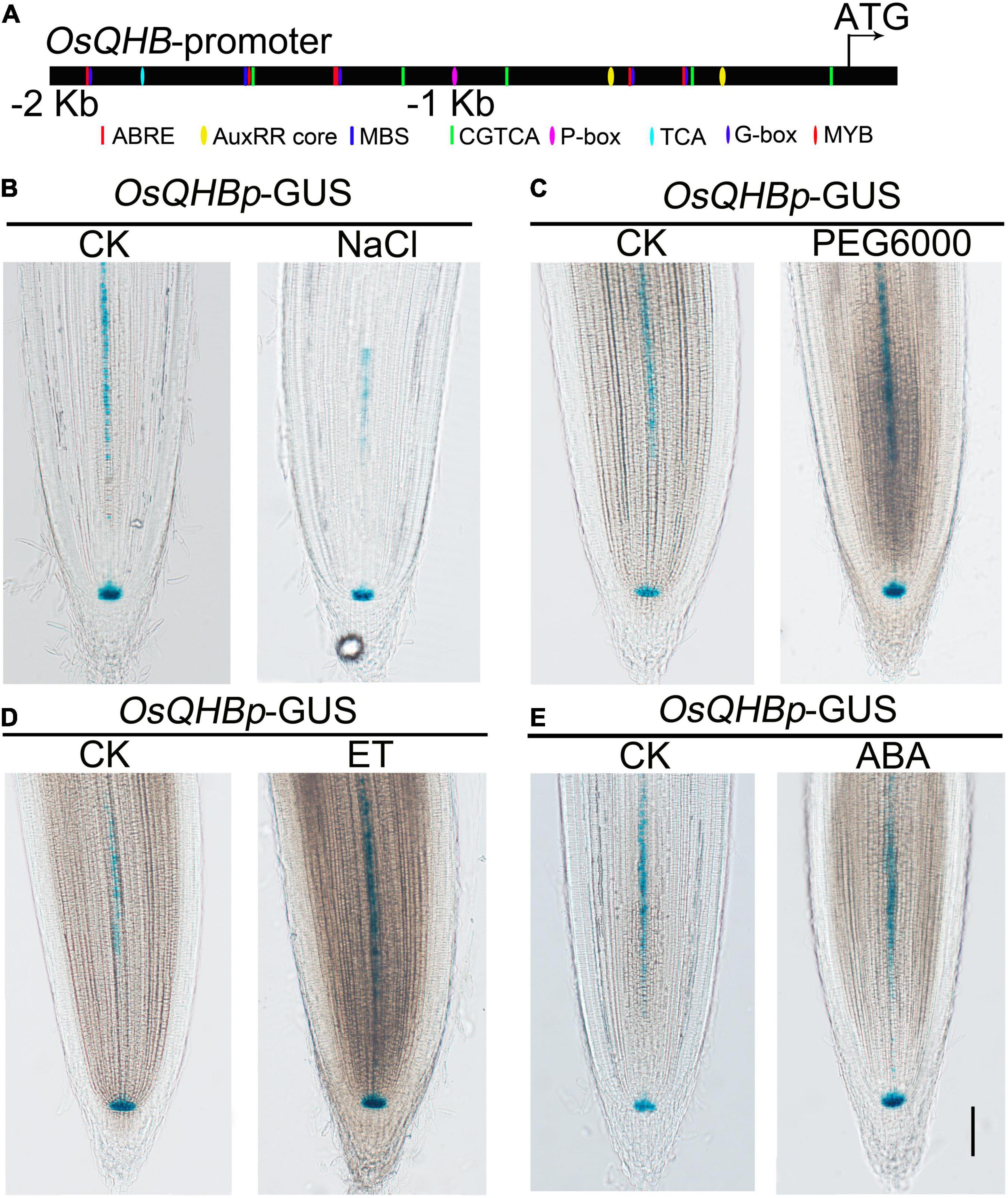
Figure 4. Expression of OsQHB under stress and hormone treatments. (A) Distribution of major stress-related cis-elements in the promoter region of OsQHB. (B–E) GUS staining of 3-d-old transgenic lines contained OsQHBp-GUS treated with 100 mM NaCl (B), 10% PEG6000 (C), 10 μL/L Ethylene (D), or 1 μm ABA (E) treatment. Bar = 100 μm.
Furthermore, we examined the responses of OsQHB to various abiotic stresses and hormone treatments by β-glucuronidase (GUS) staining. Considering that OsQHB is mainly expressed in root tips, we selected root tips for GUS staining. Our results showed that the GUS staining was significantly repressed by NaCl and weakly induced by osmotic stress (PEG6000) and ethylene, but apparently not affected by abscisic acid (ABA) treatment, respectively (Figures 4B–E). Numerous studies have shown that ABA and ethylene play important roles in regulating salt and drought tolerance (Pan et al., 2012; Liang et al., 2019; Liu C. Y. et al., 2019; Luo et al., 2020). Collectively, these results indicate that OsQHB might be associated with ethylene- and ABA-related environmental stimuli in rice.
OsQHB Negatively Regulates Salt Tolerance Through Modulating ROS Scavenging
Salt stress is a major environmental problem globally by affecting plant growth and causing crop production. Increasing numbers of studies have shown that the WOX gene family plays a role in salt tolerance (Wang L. Q. et al., 2021). As salt represses OsQHB transcription, we hypothesized that OsQHB is involved in regulating salt tolerance in rice. To verify this, we treated wild type, osqhb, and OsQHB-OE seedlings with NaCl. After 10 days of treatment with 120 mM NaCl, osqhb seedlings were obviously less affected than NIP seedlings by salt stress, whereas OsQHB-OE seedlings were all wilted to death (Figure 5A). After recovery for 10 days in non-salt conditions, the survival rate of osqhb seedlings was significantly higher, and that of OsQHB-OE seedlings was significantly lower, than that of the wild-type seedlings (Figures 5B,C). These results indicate that OsQHB negatively regulates the response of rice to salt stress at seedling stage.
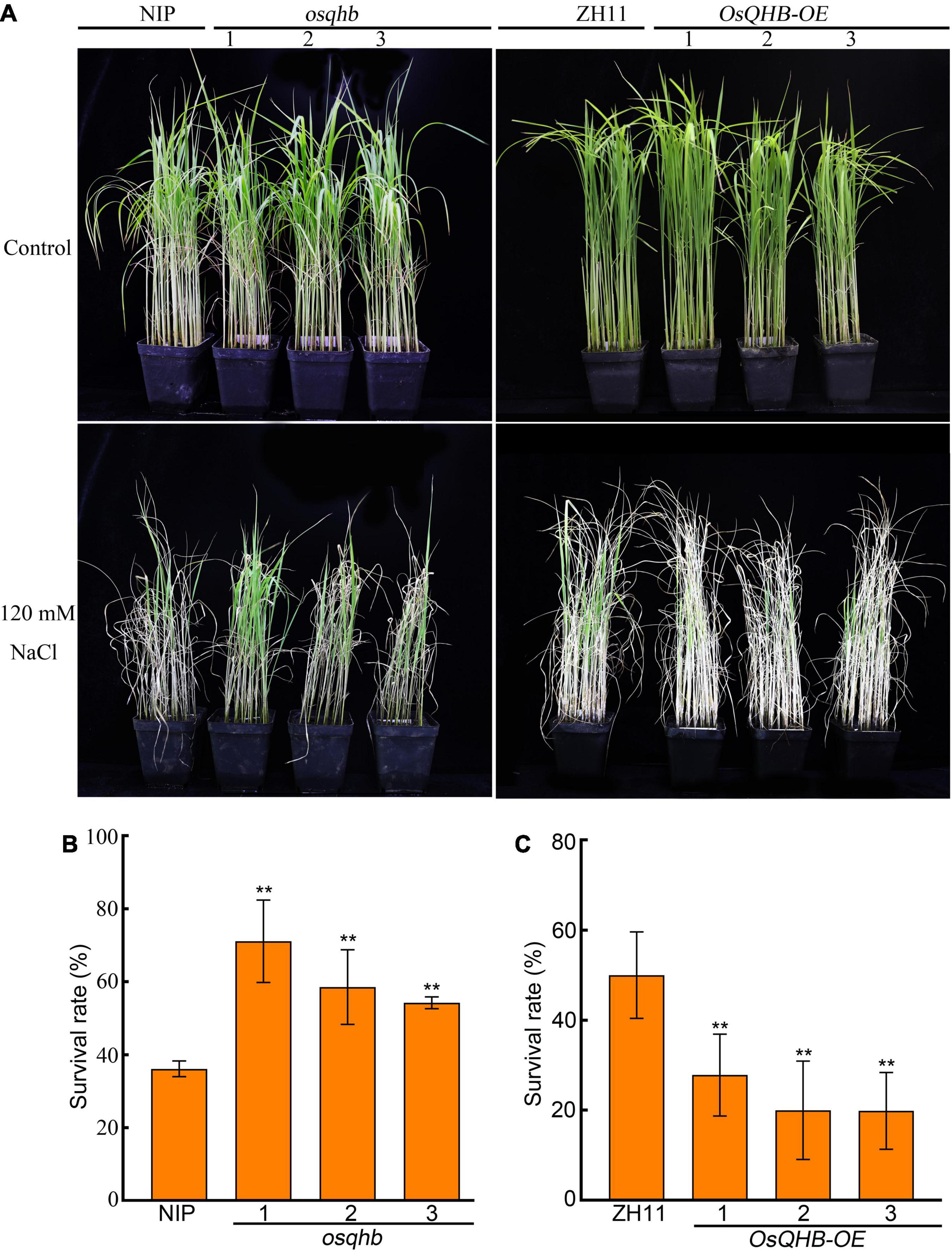
Figure 5. OsQHB negatively regulates salt tolerance of rice. (A) Phenotypes of wild-type and OsQHB transgenic plants under salt stress. 3-week-old wild-type and OsQHB transgenic plants were treated with or without 120 mM NaCl for 10 days. (B,C) Survival rates of the plants shown in panel (A) after recovery for 10 days. Approximately 50–60 seedlings were used per experiment. Data are the mean ± SD of 3 biological replicates. ** indicates significant difference compared to NIP or ZH11 at P < 0.01 by Student’s t-test.
To elucidate the molecular mechanisms regulated by OsQHB, we compared the transcriptomes of wild type, osqhb, and OsQHB-OE seedlings using transcriptome deep sequencing (RNA-seq). Totally 1079 differential expressed genes (DEGs) were identified in osqhb mutants, including 525 up-regulated DEGs and 554 down-regulated DEGs (Figure 6A and Supplementary Table 2). Gene Ontology (GO) enrichment analysis showed that these OsQHB-regulated DEGs were involved in stress response, regulation of metabolic and cellular biosynthetic process, ion transport, and kinase activity (Figure 6B), indicating that OsQHB is involved in a variety of biological processes and molecular functions, including those associated with abiotic stress responses.
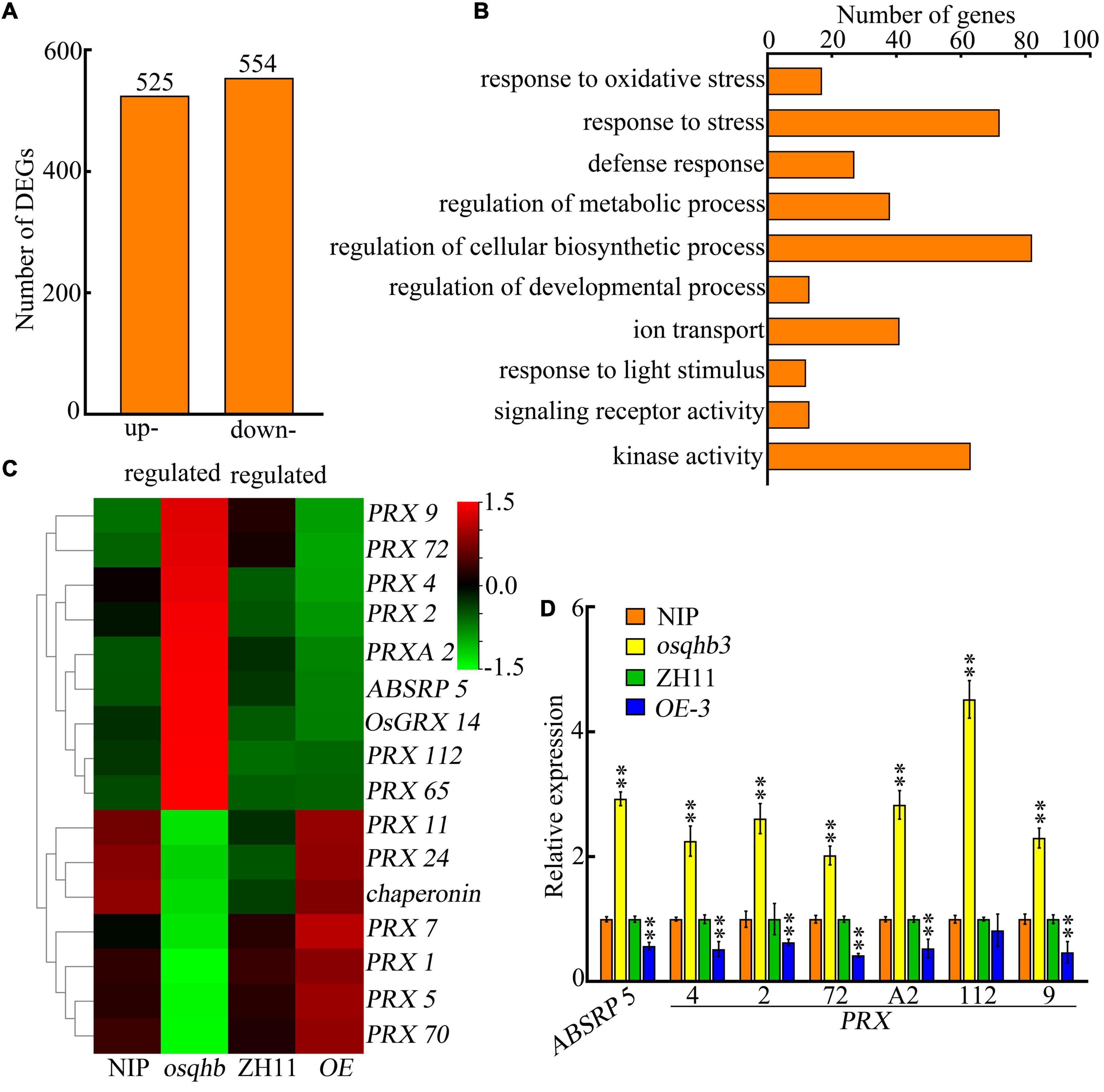
Figure 6. Transcriptome analysis reveals differentially expressed genes (DEGs) in OsQHB transgenic plants. (A) Number of DEGs up- and down-regulated in osqhb mutants. (B) Gene ontology (GO) analysis of OsQHB-regulated genes. (C) Heat map of the microarray expression profiles of ROS scavenging genes in OsQHB transgenic plants. (D) Quantitative real-time PCR (qPCR) analysis of ROS scavenging genes in 3-d-old wild-type and OsQHB transgenic seedlings. OsActin1 was used as an internal control. Three biological replicates were performed with similar results, and a representative experiment is shown. Samples were collected from three independent experiments. Bars indicate ± SD. The asterisks indicate significant differences compared to NIP or ZH11 (**P < 0.01, Student’s t-test).
Previous studies have shown that salt stress leads to over-production of ROS, and enhanced ROS scavenging capacity can improve salt tolerance in plants (Steffens, 2014; Xu et al., 2018; Yang and Guo, 2018). Thus we analyzed the expression of ROS scavenging-related genes in transcriptome data and found that multiple peroxidase (PRX) genes were regulated by OsQHB (Figure 6C and Supplementary Table 2), this regulation was further confirmed by qPCR (Figure 6D). PRX genes play diverse roles in plant physiology by scavenging ROS (Liu R. et al., 2021). These results indicate that OsQHB regulates the expression of ROS scavenging-related genes, which could further affect ROS accumulation.
To investigate whether ROS levels were altered in wild type, osqhb, and OsQHB-OE seedlings under salt treatment, we measured ROS levels in wild type, osqhb, and OsQHB-OE seedling leaf. Nitro blue tetrazolium (NBT) staining showed that the osqhb mutants accumulated less O2– in the leaves compared with wild-type plants under salt stress (Figure 7A). On the contrary, the OsQHB-OE plants accumulated much more O2– in the leaves compared with wild-type plants under salt stress (Figure 7A). Further determination of H2O2 content showed that the H2O2 content of osqhb mutants was lower, while that of OsQHB-OE plants was higher, than that of wild type after salt treatment (Figure 7B). Malondialdehyde (MDA) is an important product of lipid peroxidation, which represents the degree of oxidative damage to the plant cell (Qin et al., 2016). Salt stress significantly increased the MDA content in wild-type (Figure 7C). This tendency was weakened in osqhb mutants, but enhanced in OsQHB-OE plants (Figure 7C). These results indicate that knocking out OsQHB leads to the reduction of ROS accumulation under salt stress, thereby improving the salt tolerance of rice seedlings.
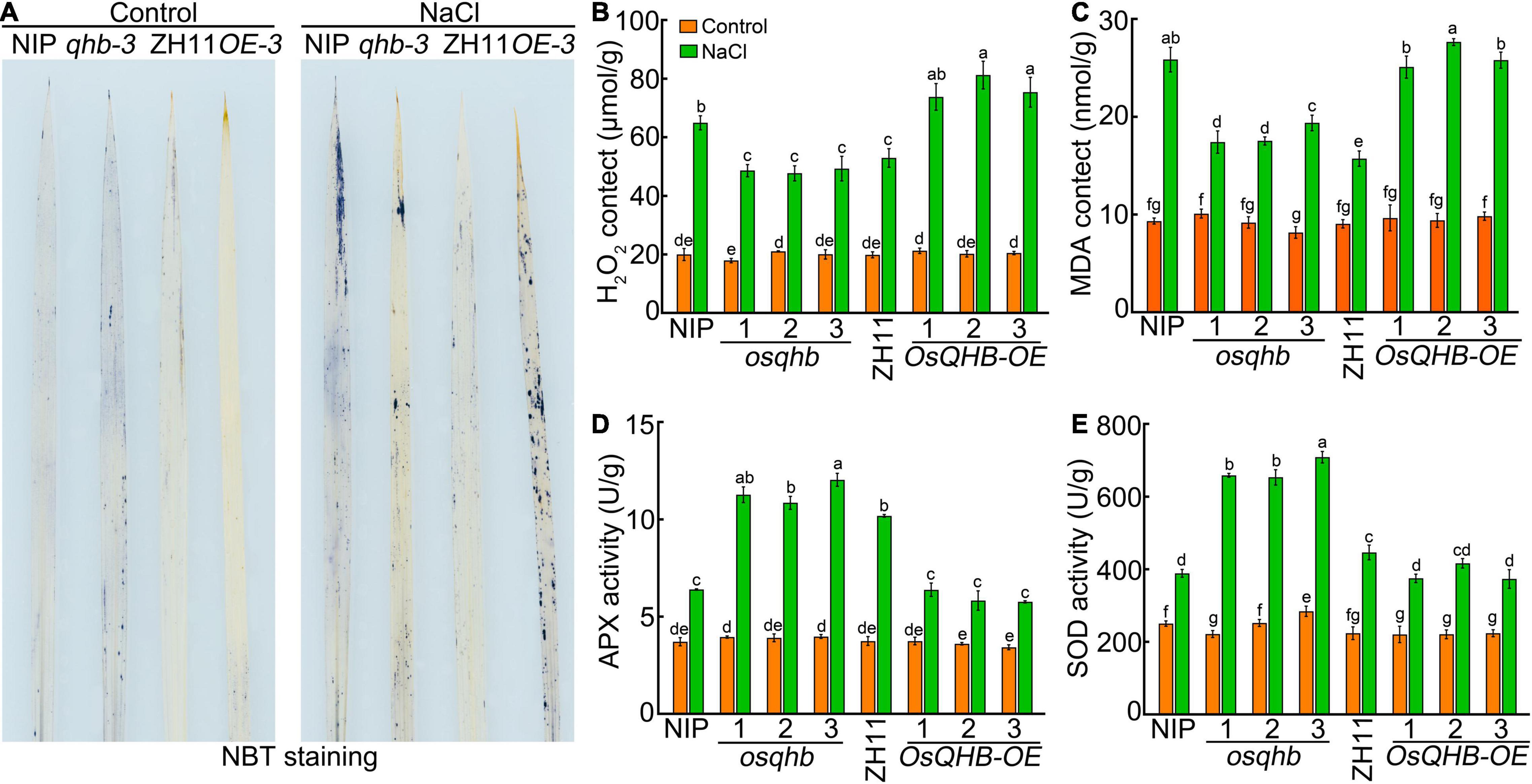
Figure 7. Loss of OsQHB function enhanced the ROS scavenging capability under salt stress. (A) NBT staining of 3-week-old wild-type and OsQHB transgenic plant leaves treated with or without 120 mM NaCl treatment for 3 days. (B,C) Hydrogen peroxide (H2O2) (B) and malondialdehyde (MDA) (C) contents in leaves of 3-week-old wild-type and OsQHB transgenic plants treated with or without 120 mM NaCl treatment for 3 days. (D,E) Ascorbate peroxidase (APX) (D) and Superoxide dismutase (SOD) (E) activities in leaves of 3-week-old wild-type and OsQHB transgenic plants treated with or without 120 mM NaCl treatment for 3 days. For (B) to panel (E), the data are shown as mean ± SD. n = 3 biological replicates. Statistically significant differences are indicated by different letters in panels (B–E) (P < 0.05, one-way ANOVA with Tukey’s test).
Salt tolerance is tightly related to the activities of ROS-scavenging enzymes. Therefore, we measured the activities of ROS-scavenging enzymes after exposed to NaCl. As shown in Figures 7D,E, after exposed to salinity stress, the activities of antioxidant enzymes in all the three genotypes were significantly increased, whereas the osqhb mutants showed a higher antioxidant enzymes activities and the OsQHB-OE lines showed a lower antioxidant enzymes activities, as compared with wild-type plants (Figures 7D,E), indicating that the OsQHB mutation leads to enhancement of salt tolerance in rice seedlings by enhancing the activities of antioxidant enzymes.
Discussion
Maintaining stable, high yields under fluctuating environmental conditions is a long-standing goal of crop improvement. However, due to internal trade-off mechanisms in plants, high yields and high biotic/abiotic resistance are usually incompatible (Deng et al., 2017). Therefore, dissecting genes that simultaneously regulate grain size and salt resistance is a promising strategy for improving both grain yield and salt tolerance in crop. In the present study, we demonstrated that OsQHB positively regulates yield-related traits but negatively regulates salt tolerance in rice. Further investigations showed that OsQHB regulates salt tolerance by enhancing ROS scavenging capacity. Our findings deepen our understanding of the functions of rice WOX gene family in response to salt stress and provide useful genes for improving yield and salt tolerance in rice.
WOX is a transcription factor family that plays a critical role in plant growth and development (Zhao et al., 2009; Hu and Xu, 2016; Chen et al., 2020). Accumulating studies show that WOX genes are associated with plant abiotic stress responses (Cheng et al., 2016; Liu H. et al., 2021; Wang L. Q. et al., 2021). In poplar, PagWOX11/12a positively regulates drought resistance through modulating ROS scavenging (Liu R. et al., 2021). Here, we showed that mutation in OsQHB weakened the accumulation of ROS under salt stress. Correspondingly, the osqhb mutants exhibited higher ROS-scavenging enzyme activity after salt treatment. Transcriptome and qPCR analysis showed that ROS scavenging-related genes were regulated by OsQHB, indicating that OsQHB improves rice salt tolerance by enhancing ROS scavenging capability through possibly regulating ROS scavenging-related genes expression. These findings deepen our understanding of the role of WOX genes in salt resistance. Further investigations will be required to determine how ROS scavenging-related genes are regulated by OsQHB to enhance salt tolerance.
The root system architecture of crops can affect their production, particularly in abiotic stress conditions (Uga et al., 2013; Kitomi et al., 2020). Numerous studies have shown that WOX gene family improves crop resistance to abiotic stress by modulating root system architecture (Cheng et al., 2016; Wang L. Q. et al., 2020). In Arabidopsis, AtWOX5 is specifically expressed in QC and acts as a root stem cell organizer (Sarkar et al., 2007). Loss of AtWOX5 function in the root stem cell niche causes terminal differentiation of columella stem cells (Sarkar et al., 2007). OsQHB is a homolog of AtWOX5, suggesting that OsQHB and AtWOX5 may have similar functions. Previous studies have shown that OsQHB is involved in the specification and maintenance of QC cells in the root apical meristem, and overexpression of OsQHB leads to abnormal root formation (Kamiya et al., 2003). In this study, our results show that OsQHB is localized in the nuclear and specifically expressed in QC cells of the root and stem base, which is consistent with previous reports (Kamiya et al., 2003). Stem base is where the crown root initiates (Zhao et al., 2009), and our results show that OsQHB is also expressed at the crown root primordial, indicating the important role of OsQHB in rice root development. Moreover, OsQHB negatively regulates salt tolerance in rice seedlings by enhancing ROS scavenging capability, suggesting that OsQHB could be useful for the controlled breeding of root system architectures that are adapted to the salinity conditions. Subsequent studies should focus on dissecting the function and regulatory network of OsQHB in root development.
Rice is considered to be a salt-sensitive species and salt-activated inhibition of plant growth may represent a positive mechanism to help plants adapt to salinity stress (Chinnusamy et al., 2005; Achard et al., 2006; Wang J. et al., 2020), this means that the development of superior crop cultivars with both high yields and high abiotic stress resistance is a tremendous challenge for crop breeding. Identifying key genes involved in the balance of rice salt tolerance and yield and discovering favorable alleles for these genes could be used to enhance both yield and abiotic stress resistance in rice. In this study, our results showed that knocking out OsQHB enhances the salt tolerance of rice seedling by improving ROS scavenging capability. Moreover, mutation in OsQHB significantly reduced yield-related traits, whereas overexpression of OsQHB had no obvious effect, likely due to rapidly degradation of OsQHB protein by the ubiquitin proteasome system, as revealed by MG132 treatment. Therefore, precisely manipulation of OsQHB represents a promising strategy for improving both grain yield and stress tolerance in rice. Further study should focus on mining favorable alleles of OsQHB and elucidating the molecular mechanism by which OsQHB is degraded.
Taken together, our research presents new insights into the roles of OsQHB in improving yield and salt tolerance in rice, and dissecting the underlying mechanism and mining favorable alleles of OsQHB will facilitate the practical use of OsQHB in breeding superior crop cultivars with both high yields and high salt resistance.
Materials and Methods
Plant Material and Growth Conditions
OsQHBp-GUS transgenic line was described previously (Ni et al., 2014). osqhb allelic mutants in the Nipponbare (NIP) background were generated by using CRISPR/Cas9 (Xu et al., 2017). Briefly, the target region 5′-GGAGCAGGTGAAGGTCCTGA-3′ was introduced into the pHUN4c12 vector backbone, and the recombinant vector was transformed into Agrobacterium strain EHA105-pSOUP for rice transformation. To generate the overexpression transgenic plants, the coding sequence of OsQHB was amplified by PCR and cloned in-frame with a Flag-tag into pCAMBIA1307 under the control of the CaMV 35S promoter (Wang J. et al., 2013). The recombinant plasmids were introduced separately into NIP or Zhonghua 11 (ZH11) via Agrobacterium-mediated transformation.
Rice seeds were imbibed in Petri dishes with sterile distill water at 37°C for 48 h. The germinated seeds were sown in a bottomless 96-well plate in a container of Yoshida’s culture solution (Cui et al., 2015) or in growing trays filled with soil. The plants were grown in a growth chamber under a 14 h light (30°C)/10 h dark (25°C) photoperiod, with a light intensity of ∼150 μmol m–2s–1 (white light) and 60% relative humidity. To assess salt-tolerance of rice seedlings, 3-week-old seedlings were treated with 120 mM NaCl solution for 10 days, and then transferred to water without NaCl for an additional 10 days. Subsequently, the survival rate of rice seedlings was counted (surviving seedlings indicated plants with green leaves).
For propagation and investigation of agronomic traits, the plants were cultivated at the Experimental Station of the Chinese Academy of Agricultural Sciences in Beijing during the natural growing seasons.
Phylogenetic and Promotor Analysis
Arabidopsis WOX proteins sequences were available at the Arabidopsis genome sequence database1 and rice WOX proteins sequences were downloaded from the National Center for Biotechnology Information (NCBI) database2. Full-length protein sequences of all WOX proteins were aligned by multiple sequence alignment program of the Clustal X, and then phylogenetic tree was constructed by MEGA 6 software according to the following parameters: neighbor-joining method, 500 bootstraps (Tamura et al., 2007).
For promoter analysis, the promotor sequence (2,000 bp upstream of ATG) of OsQHB was uploaded to the PlantCARE database3 to identify stress related cis-elements as previously reported (Yang and Guo, 2018).
Quantitative Real-Time PCR and β-Glucuronidase Staining
Total RNA was extracted from leaf tissues using an Ultrapure RNA Kit (CWBIO, China) according to the manufacturer’s instructions. ∼2 μg total RNA was reverse transcribed using HiScript II Q RT SuperMix (Vazyme, China) according to the manufacturer’s instructions. qPCR was performed on an optical 96-well plate with Bio-Rad iQ5 system using SYBR Green Real-time PCR Master Mix reagent (Vazyme, China) as previously described (Zhang et al., 2012). The rice Actin1 gene was used as an internal standard to normalize gene expression. The primers used for qPCR are listed in Supplementary Table 3.
For GUS staining, 3-d-old OsQHBp-GUS seedlings were immersed in GUS staining buffer (50 mM sodium phosphate, pH 7.0; 10 mM EDTA; 0.5 mM K3[Fe (CN)6]; 0.5 mM K4[Fe (CN)6]; 0.1% [v/v] Triton X-100; and 1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid). The samples were vacuumed in a vacuum pump for 30 min, and then stained in 37°C for 2 d. After dehydrated in 75% (v/v) ethanol to remove the chlorophyll, the root tip was sectioned and photographed under a stereoscopic microscope.
Subcellular Location
To determine the subcellular localization of OsQHB, the full coding sequence of OsQHB was cloned into the pAN580-GFP vector under the control of the CaMV 35S promoter. Rice protoplasts prepared from etiolated shoots were transfected with OsQHB-GFP (Jiang et al., 2018). After incubated at 28°C for 16–20 h, equal volumes of DAPI (4′,6-diamidino-2-phenylidone) staining solution and protoplast suspension were mixed, and the fluorescence signal was observed with a confocal laser scanning microscope.
Nitro Blue Tetrazolium Assay for the Superoxide Anion
The leaves of 3-week-old seedlings were immersed in 20 mM phosphate buffer (pH 6.1) containing 2 mM NBT. After vacuumed in pump for 1 h, the samples were stained in 37°C for 12 h and then transferred to 75% (v/v) ethanol to stop the reaction. The material was photographed under a light stereomicroscope.
Superoxide Dismutase and Ascorbate Peroxidase Activity Measurement
Ascorbate peroxidase and SOD activities were determined according to the instruction of the superoxide dismutase assays kits (Solarbio, China) and the ascorbate peroxidase assays kits (Solarbio, China). In brief, 0.1 g of normal or salt-stressed rice leaves from 3-week-old seedlings were harvested, ground in liquid nitrogen, and extracted by extraction buffer. After centrifuged at 8,000 g (SOD) for 10 min or 13,000 g (APX) for 20 min, the supernatant was used for SOD and APX activity measurement. The absorbance values were measured at 560 and 290 nm, respectively. SOD and APX activities were calculated from the formula provided with the SOD and APX assay kit (Solarbio, China).
Measurement of Hydrogen Peroxide and Malondialdehyde Contents
H2O2 content was determined using the method described previously (Wang Y. et al., 2019). Briefly, 0.1 g rice leaves from 3-week-old seedlings with or without salt treatment were harvested and homogenized in 1 ml cold acetone. Then, H2O2 content was determined using a hydrogen peroxide assay kit (Solarbio, China) according to the manufacturer’s instructions.
To measure MDA, 0.1 g of normal or salt-stressed rice leaves were homogenized in 1 ml 0.1% (w/v) trichloroacetic acid (TCA) followed by centrifugation at 8,000 g for 10 min at 4°C. Four volumes of 0.5% (w/v) thiobarbituric acid (TBA) in 20% (w/v) TCA were added to one volume of supernatant; the mixture was incubated at 100°C for 1 h. The reaction was terminated by incubating the mixture on ice for 15 min, and the absorbance was measured by spectrophotometry at 450 nm, 532 and 600 nm. The content of MDA was calculated according to the formula provided in the MDA assay kit (Solarbio, China).
Transcriptome Analysis
The primary roots of 3-d-old OsQHB overexpression line, osqhb mutant, and wild-type seedlings were collected and extracted using the TRIzol method (TIANGEN BIOTECH, China) and treated with RNase-free DNase I (TaKaRa, Japan). mRNA was purified from total RNA using poly-T oligo-attached magnetic beads and then subjected to RNA-seq library construction for the transcriptome experiments using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, United States). Multiplex paired-end adapters were used for multiplex libraries. The library quality was assessed on the Agilent Bioanalyzer 2100 system. The library preparations were sequenced on an Illumina Hiseq 4000 platform by the Beijing Allwegene Technology Company Limited (Beijing, China) and paired-end 150 bp reads were generated. After removing adaptor and low-quality reads, clean reads were mapped to rice genome MSU7.0 using TopHat, and analyzed using Cufflinks according to Trapnell et al. (2012). Poisson-dispersion model of fragment was used to conduct statistical analysis (FDR < 0.05) and responsive genes were identified by fragments per kilobase per million reads (FPKM) requiring more than twofold change between two samples. Three biological replicates were used, and their repeatability and correlation were evaluated by the Pearson’s Correlation Coefficient (Schulze et al., 2012). The transcriptome data from this study can be found in the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under the accession number PRJNA801123.
Primer Sequences
The primers used in this study are shown in Supplementary Table 3.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under the accession number PRJNA801123.
Author Contributions
JZ: performed the research, data curation and writing-original draft. JQ: performed the research. RQ: analyzed and discussed the data. JW: analyzed and discussed the data. RH: project administration, writing-review and editing. HQ: conceptualization, supervision, designed, and writing-review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by National Natural Science Foundation of China grants 32030079, 31801445, and 31871551 to RH and HQ; National Key R&D Program of China grant 2020YFE0202300 to RQ; and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences to JW and RH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Ke-Ke Yi from the Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences for kindly sharing the OsQHBp-GUS seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.848891/full#supplementary-material
Footnotes
- ^ http://www.Arabidopsis.org/
- ^ http://www.ncbi.nlm.nih.gov/protein
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science. 311, 91–94. doi: 10.1126/science.1118642
Ambawat, S., Sharma, P., Yadav, N. R., and Yadav, R. C. (2013). MYB transcription factor genes as regulators for plant responses: an overview. Physiol. Mol. Biol. Plants 19, 307–321. doi: 10.1007/s12298-013-0179-1
Chen, R., Xu, N., Yu, B., Wu, Q., Li, X., Wang, G., et al. (2020). The WUSCHEL-related homeobox transcription factor OsWOX4 controls the primary root elongation by activating OsAUX1 in rice. Plant Sci. 298:110575. doi: 10.1016/j.plantsci.2020.110575
Cheng, S., Huang, Y., Zhu, N., and Zhao, Y. (2014). The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene 549, 266–274. doi: 10.1016/j.gene.2014.08.003
Cheng, S., Tan, F., Lu, Y., Liu, X., Li, T., Yuan, W., et al. (2018). WOX11 recruits a histone H3K27me3 demethylase to promote gene expression during shoot development in rice. Nucleic Acids Res. 46, 2356–2369. doi: 10.1093/nar/gky017
Cheng, S., Zhou, D. X., and Zhao, Y. (2016). WUSCHEL-related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development. Plant Signal. Behav. 11:e1130198. doi: 10.1080/15592324.2015.1130198
Chinnusamy, V., Jagendorf, A., and Zhu, J. K. (2005). Understanding and improving salt tolerance in plants. Crop Sci. 45, 437–448. doi: 10.2135/cropsci2005.0437
Cho, S. H., Kang, K., Lee, S. H., Lee, I. J., and Paek, N. C. (2016). OsWOX3A is involved in negative feedback regulation of the gibberellic acid biosynthetic pathway in rice (Oryza sativa L.). J. Exp. Bot. 67, 1677–1687. doi: 10.1093/jxb/erv559
Cui, L. G., Shan, J. X., Shi, M., Gao, J. P., and Lin, H. X. (2015). DCA1 acts as a transcriptional co-activator of DST and contributes to drought and salt tolerance in rice. PLoS Genet. 11:e1005617. doi: 10.1371/journal.pgen.1005617
Deng, Y., Zhai, K., Xie, Z., Yang, D., Zhu, X., Liu, J., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965. doi: 10.1126/science.aai8898
Dutta, M., Saha, A., Moin, M., and Kirti, P. B. (2021). Genome-wide identification, transcript profiling and bioinformatic analyses of GRAS transcription factor genes in rice. Front. Plant Sci. 12:777285. doi: 10.3389/fpls.2021.777285
Ezer, D., Shepherd, S. J. K., Brestovitsky, A., Dickinson, P., Cortijo, S., Charoensawan, V., et al. (2017). The G-box transcriptional regulatory code in Arabidopsis. Plant Physiol. 175, 628–640. doi: 10.1104/pp.17.01086
Forzani, C., Aichinger, E., Sornay, E., Willemsen, V., Laux, T., Dewitte, W., et al. (2014). WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche. Curr. Biol. 24, 1939–1944. doi: 10.1016/j.cub.2014.07.019
Foyer, C. H., and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17, 1866–1875. doi: 10.1105/tpc.105.033589
He, Y., Yang, B., He, Y., Zhan, C., Cheng, Y., Zhang, J., et al. (2019). A quantitative trait locus, qSE3, promotes seed germination and seedling establishment under salinity stress in rice. Plant J. 97, 1089–1104. doi: 10.1111/tpj.14181
Hong, C. Y., Hsu, Y. T., Tsai, Y. C., and Kao, C. H. (2007). Expression of ASCORBATE PEROXIDASE 8 in roots of rice (Oryza sativa L.) seedlings in response to NaCl. J. Exp. Bot. 58, 3273–3283. doi: 10.1093/jxb/erm174
Hu, X., and Xu, L. (2016). Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 172, 2363–2373. doi: 10.1104/pp.16.01067
Hwang, J. E., Hong, J. K., Lim, C. J., Chen, H., Je, J., Yang, K. A., et al. (2010). Distinct expression patterns of two Arabidopsis phytocystatin genes, AtCYS1 and AtCYS2, during development and abiotic stresses. Plant Cell Rep. 29, 905–915. doi: 10.1007/s00299-010-0876-y
Ismail, A. M., and Horie, T. (2017). Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 68, 405–434. doi: 10.1146/annurev-arplant-042916-040936
Jha, P., Ochatt, S. J., and Kumar, V. (2020). WUSCHEL: a master regulator in plant growth signaling. Plant Cell Rep. 39, 431–444. doi: 10.1007/s00299-020-02511-5
Jiang, Y., Cheng, K., Zhao, X., and Quyang, Y. (2018). Isolation and transformation of rice protoplasts. Plant Cell Rep. 13, 394–396. in Chinese. doi: 10.21769/BioProtoc.1010125
Kamiya, N., Nagasaki, H., Morikami, A., Sato, Y., and Matsuoka, M. (2003). Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J. 35, 429–441. doi: 10.1046/j.1365-313x.2003.01816.x
Kawai, T., Shibata, K., Akahoshi, R., Nishiuchi, S., Takahashi, H., Nakazono, M., et al. (2022). WUSCHEL-related homeobox family genes in rice control lateral root primordium size. Proc. Natl. Acad. Sci. U. S. A. 119:e2101846119. doi: 10.1073/pnas.2101846119
Kitomi, Y., Hanzawa, E., Kuya, N., Inoue, H., Hara, N., Kawai, S., et al. (2020). Root angle modifications by the DRO1 homolog improve rice yields in saline paddy fields. Proc. Natl. Acad. Sci. U. S. A. 117, 21242–21250. doi: 10.1073/pnas.2005911117
Liang, S. S., Xiong, W., Yin, C. C., Xie, X. D., Jin, Y. J., Zhang, S. J., et al. (2019). Overexpression of OsARD1 improves submergence, drought, and salt tolerances of seedling through the enhancement of ethylene synthesis in rice. Front. Plant Sci. 10:1088. doi: 10.3389/fpls.2019.01088
Lima-Melo, Y., Carvalho, F. E., Martins, M. O., Passaia, G., Sousa, R. H., Neto, M. C., et al. (2016). Mitochondrial GPX1 silencing triggers differential photosynthesis impairment in response to salinity in rice plants. J. Integr. Plant Biol. 58, 737–748. doi: 10.1111/jipb.12464
Liu, C. Y., Li, J., Zhu, P. P., Yu, J., Hou, J. M., Wang, C. H., et al. (2019). Mulberry EIL3 confers salt and drought tolerances and modulates ethylene biosynthetic gene expression. Peerj 7:e6391. doi: 10.7717/peerj.6391
Liu, H., Dong, S., Li, M., Gu, F., Yang, G., Guo, T., et al. (2021). The Class III peroxidase gene OsPrx30, transcriptionally modulated by the AT-hook protein OsATH1, mediates rice bacterial blight-induced ROS accumulation. J. Integr. Plant Biol. 63, 393–408. doi: 10.1111/jipb.13040
Liu, R., Wang, R., Lu, M. Z., and Wang, L. Q. (2021). WUSCHEL-related homeobox gene PagWOX11/12a is involved in drought tolerance through modulating reactive oxygen species scavenging in poplar. Plant Signal. Behav. 16:1866312. doi: 10.1080/15592324.2020.1866312
Lu, Z., Liu, D., and Liu, S. (2007). Two rice cytosolic ascorbate peroxidases differentially improve salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 26, 1909–1917. doi: 10.1007/s00299-007-0395-7
Luo, X. Y., Li, C., He, X., Zhang, X. L., and Zhu, L. F. (2020). ABA signaling is negatively regulated by GbWRKY1 through JAZ1 and ABI1 to affect salt and drought tolerance. Plant Cell Rep. 39, 181–194. doi: 10.1007/s00299-019-02480-4
Lv, B., Tian, H., Zhang, F., Liu, J., Lu, S., Bai, M., et al. (2018). Brassinosteroids regulate root growth by controlling reactive oxygen species homeostasis and dual effect on ethylene synthesis in Arabidopsis. PLoS Genet. 14:e1007144. doi: 10.1371/journal.pgen.1007144
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., and Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Nakashima, K., and Yamaguchi-Shinozaki, K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Nam, M. H., Bang, E., Kwon, T. Y., Kim, Y., Kim, E. H., Cho, K., et al. (2015). Metabolite profiling of diverse rice germplasm and identification of conserved metabolic markers of rice roots in response to long-term mild salinity stress. Int. J. Mol. Sci. 16, 21959–21974. doi: 10.3390/ijms160921959
Ni, J., Shen, Y., Zhang, Y., and Wu, P. (2014). Definition and stabilisation of the quiescent centre in rice roots. Plant Biol. 16, 1014–1019. doi: 10.1111/plb.12138
Ohmori, Y., Tanaka, W., Kojima, M., Sakakibara, H., and Hirano, H. Y. (2013). WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice. Plant Cell 25, 229–241. doi: 10.1105/tpc.112.103432
Pan, Y., Seymour, G. B., Lu, C. G., Hu, Z. L., Chen, X. Q., and Chen, G. P. (2012). An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 31, 349–360. doi: 10.1007/s00299-011-1170-3
Pi, L., Aichinger, E., van der Graaff, E., Llavata-Peris, C. I., Weijers, D., Hennig, L., et al. (2015). Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell 33, 576–588. doi: 10.1016/j.devcel.2015.04.024
Qin, H., Li, Y., and Huang, R. (2020). Advances and challenges in the breeding of salt-tolerant rice. Int. J. Mol. Sci. 21:8385. doi: 10.3390/ijms21218385
Qin, H., Wang, J., Chen, X., Wang, F., Peng, P., Zhou, Y., et al. (2019). Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 223, 798–813. doi: 10.1111/nph.15824
Qin, H., Wang, Y., Wang, J., Liu, H., Zhao, H., Deng, Z., et al. (2016). Knocking down the expression of GMPase gene OsVTC1-1 decreases salt tolerance of rice at seedling and reproductive stages. PLoS One 11:e0168650. doi: 10.1371/journal.pone.0168650
Saini, S., Kaur, N., and Pati, P. K. (2018). Reactive oxygen species dynamics in roots of salt sensitive and salt tolerant cultivars of rice. Anal. Biochem. 550, 99–108. doi: 10.1016/j.ab.2018.04.019
Sajjad, M., Wei, X., Liu, L., Li, F., and Ge, X. (2021). Transcriptome analysis revealed GhWOX4 intercedes myriad regulatory pathways to modulate drought tolerance and vascular growth in cotton. Int. J. Mol. Sci. 22:898. doi: 10.3390/ijms22020898
Sakai, T., Takahashi, Y., and Nagata, T. (1996). Analysis of the promoter of the auxin-inducible gene, parC, of tobacco. Plant Cell Physiol. 37, 906–913. doi: 10.1093/oxfordjournals.pcp.a029038
Sarkar, A. K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., et al. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. doi: 10.1038/nature05703
Savina, M. S., Pasternak, T., Omelyanchuk, N. A., Novikova, D. D., Palme, K., Mironova, V. V., et al. (2020). Cell dynamics in WOX5-overexpressing root tips: the impact of local auxin biosynthesis. Front. Plant Sci. 11:560169. doi: 10.3389/fpls.2020.560169
Schulze, S. K., Kanwar, R., Golzenleuchter, M., Therneau, T. M., and Beutler, A. S. (2012). SERE: single-parameter quality control and sample comparison for RNA-Seq. BMC Genomics 13:524. doi: 10.1186/1471-2164-13-524
Skylar, A., Hong, F., Chory, J., Weigel, D., and Wu, X. (2010). STIMPY mediates cytokinin signaling during shoot meristem establishment in Arabidopsis seedlings. Development 137, 541–549. doi: 10.1242/dev.041426
Steffens, B. (2014). The role of ethylene and ROS in salinity, heavy metal, and flooding responses in rice. Front. Plant Sci. 5:685. doi: 10.3389/fpls.2014.00685
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Teixeira, F. K., Menezes-Benavente, L., Galvao, V. C., Margis, R., and Margis-Pinheiro, M. (2006). Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta 224, 300–314. doi: 10.1007/s00425-005-0214-8
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. doi: 10.1038/ng.2725
Wang, J., Qin, H., Zhou, S., Wei, P., Zhang, H., Zhou, Y., et al. (2020). The Ubiquitin-binding protein OsDSK2a mediates seedling growth and salt responses by regulating gibberellin metabolism in rice. Plant Cell 32, 414–428. doi: 10.1105/tpc.19.00593
Wang, L. Q., Li, Z., Wen, S. S., Wang, J. N., Zhao, S. T., and Lu, M. Z. (2020). WUSCHEL-related homeobox gene PagWOX11/12a responds to drought stress by enhancing root elongation and biomass growth in poplar. J. Exp. Bot. 71, 1503–1513. doi: 10.1093/jxb/erz490
Wang, J., Yu, Y., Zhang, Z., Quan, R., Zhang, H., Ma, L., et al. (2013). Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 25, 625–636. doi: 10.1105/tpc.112.106880
Wang, L. Q., Wen, S. S., Wang, R., Wang, C., Gao, B., and Lu, M. Z. (2021). PagWOX11/12a activates PagCYP736A12 gene that facilitates salt tolerance in poplar. Plant. Biotechnol. J. 19, 2249–2260. doi: 10.1111/pbi.13653
Wang, Y., Liang, C., Meng, Z., Li, Y., Abid, M. A., Askari, M., et al. (2019). Leveraging atriplex hortensis choline monooxygenase to improve chilling tolerance in cotton. Environ. Exp. Bot. 162, 364–373. doi: 10.1016/j.envexpbot.2019.03.012
Wang, Y., Liu, G. J., Yan, X. F., Wei, Z. G., and Xu, Z. R. (2011). MeJA-inducible expression of the heterologous JAZ2 promoter from Arabidopsis in Populus trichocarpa protoplasts. J. Plant Dis. Protect. 118, 69–74. doi: 10.1007/Bf03356384
Wei, Q., Cao, H. M., Li, Z. R., Kuai, B. K., and Ding, Y. L. (2013). Identification of an AtCRN1-like chloroplast protein BeCRN1 and its distinctive role in chlorophyll breakdown during leaf senescence in bamboo (Bambusa emeiensis ‘Viridiflavus’). Plant Cell Tissue Org Cult. 114, 1–10. doi: 10.1007/s11240-013-0298-y
Wu, X., Chory, J., and Weigel, D. (2007). Combinations of WOX activities regulate tissue proliferation during Arabidopsis embryonic development. Dev. Biol. 309, 306–316. doi: 10.1016/j.ydbio.2007.07.019
Wu, X., Dabi, T., and Weigel, D. (2005). Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr. Biol. 15, 436–440. doi: 10.1016/j.cub.2004.12.079
Xu, N., Chu, Y., Chen, H., Li, X., Wu, Q., Jin, L., et al. (2018). Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging. PLoS Genet. 14:e1007662. doi: 10.1371/journal.pgen.1007662
Xu, R., Wei, P., and Yang, J. (2017). Use of CRISPR/Cas genome editing technology for targeted mutagenesis in rice. Methods Mol. Biol. 1498, 33–40. doi: 10.1007/978-1-4939-6472-7_3
Yang, Y., and Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, Z., Gong, Q., Qin, W., Yang, Z., Cheng, Y., Lu, L., et al. (2017). Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 17:113. doi: 10.1186/s12870-017-1065-8
Yasui, Y., Ohmori, Y., Takebayashi, Y., Sakakibara, H., and Hirano, H. Y. (2018). WUSCHEL-RELATED HOMEOBOX4 acts as a key regulator in early leaf development in rice. PLoS Genet. 14:e1007365. doi: 10.1371/journal.pgen.1007365
Yoo, S. C., Cho, S. H., and Paek, N. C. (2013). Rice WUSCHEL-related homeobox 3A (OsWOX3A) modulates auxin-transport gene expression in lateral root and root hair development. Plant Signal. Behav. 8:e25929. doi: 10.4161/psb.25929
Yu, Y., Wang, J., Zhang, Z., Quan, R., Zhang, H., Deng, X. W., et al. (2013). Ethylene promotes hypocotyl growth and HY5 degradation by enhancing the movement of COP1 to the nucleus in the light. PLoS Genet. 9:e1004025.
Zhang, H. L., Zhao, X. J., Zhang, J. P., Yang, B., Yu, Y. H., Liu, T. F., et al. (2020). Functional analysis of an anthocyanin synthase gene StANS in potato. Sci. Hortic. 272:10956910.
Zhang, X., Zong, J., Liu, J., Yin, J., and Zhang, D. (2010). Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 52, 1016–1026. doi: 10.1111/j.1744-7909.2010.00982.x
Zhang, Z., Wang, J., Zhang, R., and Huang, R. (2012). The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 71, 273–287.
Zhang, Z., Zhang, Q., Wu, J., Zheng, X., Zheng, S., Sun, X., et al. (2013). Gene knockout study reveals that cytosolic ascorbate peroxidase 2(OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS One 8:e57472. doi: 10.1371/journal.pone.0057472
Zhao, Y., Hu, Y., Dai, M., Huang, L., and Zhou, D. X. (2009). The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21, 736–748. doi: 10.1105/tpc.108.061655
Keywords: OsQHB, salt stress, ROS scavenging, yield, rice
Citation: Zhou J, Qiao J, Wang J, Quan R, Huang R and Qin H (2022) OsQHB Improves Salt Tolerance by Scavenging Reactive Oxygen Species in Rice. Front. Plant Sci. 13:848891. doi: 10.3389/fpls.2022.848891
Received: 05 January 2022; Accepted: 30 March 2022;
Published: 04 May 2022.
Edited by:
Pasqualina Woodrow, University of Campania Luigi Vanvitelli, ItalyReviewed by:
Sung-Hwan Cho, University of Missouri, United StatesChao Wang, Northeast Forestry University, China
Copyright © 2022 Zhou, Qiao, Wang, Quan, Huang and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Qin, cWluaHVhQGNhYXMuY24=
 Jiahao Zhou
Jiahao Zhou Jinzhu Qiao
Jinzhu Qiao Juan Wang
Juan Wang Ruidang Quan1,2
Ruidang Quan1,2 Rongfeng Huang
Rongfeng Huang Hua Qin
Hua Qin