- 1Fujian Colleges and Universities Engineering Research Institute of Conservation and Utilization of Natural Bioresources, College of Forestry, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Key Laboratory of Orchid Conservation and Utilization of National Forestry and Grassland Administration at College of Landscape Architecture and Art, Fujian Agriculture and Forestry University, Fuzhou, China
TCP gene family are specific transcription factors for plant, and considered to play an important role in development and growth. However, few related studies investigated the TCP gene trait and how it plays a role in growth and development of Orchidaceae. In this study, we obtained 14 TCP genes (CgTCPs) from the Spring Orchid Cymbidium goeringii genome. The classification results showed that 14 CgTCPs were mainly divided into two clades as follows: four PCF genes (Class I), nine CIN genes and one CYC gene (Class II). The sequence analysis showed that the TCP proteins of C. goeringii contain four conserved regions (basic Helix-Loop-Helix) in the TCP domain. The exon−intron structure varied in the clade according to a comparative investigation of the gene structure, and some genes had no introns. There are fewer CgTCP homologous gene pairs compared with Dendrobium catenatum and Phalaenopsis equestris, suggesting that the TCP genes in C. goeringii suffered more loss events. The majority of the cis-elements revealed to be enriched in the function of light responsiveness, followed by MeJA and ABA responsiveness, demonstrating their functions in regulating by light and phytohormones. The collinearity study revealed that the TCPs in D. catenatum, P. equestris and C. goeringii almost 1:1. The transcriptomic data and real-time reverse transcription-quantitative PCR (RT−qPCR) expression profiles showed that the flower-specific expression of the TCP class II genes (CgCIN2, CgCIN5 and CgCIN6) may be related to the regulation of florescence. Altogether, this study provides a comprehensive analysis uncovering the underlying function of TCP genes in Orchidaceae.
Introduction
TCP transcription factors (TFs) compose a gene family particular to plant which relate to growth and development. The gene family was discovered by Cubas et al. (1999) and named according to the TEOSINTE BRANCHED 1 (TB1) gene (Zea mays), CYCLOIDEA (CYC) gene (Antirrhinum majus), and Proliferating Cell Factor gene (PCF) (Oryza sativus) (TCP). TCP genes are characterized by a basic helix-loop-helix motif of 59 amino acids (aa) named the TCP domain, which allows protein interaction and DNA binding (Kosugi & Ohashi, 1997; Cubas et al., 1999; Shang et al., 2022). TCP genes are mainly divided into Class I and Class II according to the loss of four amino acids in Class II basic domains (Martín-Trillo and Cubas, 2010). Class I, also called the TCP-P class, contains the PCF genes PCF1 and PCF2, which were discovered in rice and bind the promoter of the PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) gene, which is related to DNA replication, chromosome maintenance and cell cycle progression (Cubas et al., 1999). Class II was also known as the TCP-C class and contains the TB1, CYC, and CIN genes. The TB1 genes mainly regulate apical dominance and inflorescence development (Doebley et al., 1997; Dixon et al., 2018), the CYC genes mainly control floral bilateral symmetry development (Luo et al., 1999; Hileman, 2014), and the CIN genes are mostly involved in the development of lateral organ (Palatnik et al., 2003; Nath et al., 2003; Crawford et al., 2004; Walcher-Chevillet and Kramer, 2016). More non-model plant TCP genes were identified and analyzed in the genome era, promoting the comprehension of the structures and functions of this gene family.
TCP transcription factors are ancient proteins that exist in all plant phylostrata, except for unicellular algae (Navaud et al., 2007; Floyd & Bowman, 2007). This gene family contains only five to six members in moss, ferns, and lycophytes. With genome duplication, evolution and diversification, more than ten genes have been generated in angiosperms and gymnosperms, and more than 20 members have been generated in some model plant species (Riechmann et al., 2000; Cubas, 2002; Navaud et al., 2007; Mao et al., 2014). Two classes of genes exist in all plants that contain TCP genes, but the CYC/TB1 genes were not found in lycophytes or more ancient plants (Floyd & Bowman, 2007; Horn et al., 2015). Therefore, the CIN class is more ancient than CYC/TB1, and the CYC/TB1 clade may have originated in gymnosperms or angiosperms and is mainly involved in floral organ development (Palatnik et al., 2003; Koyama et al., 2007; Horn et al., 2015). The TB1 genes and homologs mainly control leaf axillary development in vegetable and reproductive organs of monocots (Hubbard et al., 2002; Lewis et al., 2008; Yuan et al., 2009; De Paolo et al., 2015). The CYC1/2/3 genes perform different functions in core eudicots (Howarth & Donoghue, 2006). The number and function of TCP genes varied and differentiated during plant evolution.
Orchidaceae is one of the most diverse families in vascular plants, containing about 28,000 species in 700 genera (Chase et al., 2015; Christenhusz & Byng, 2016; Zhang et al., 2017). Orchids grow in an extensive range of habitats worldwide, with high diversity in flower and vegetable morphology, life form, and pollination (Givnish et al., 2015; Chen et al., 2020a; Chen et al., 2021; Liu and Lan, 2022; Li et al., 2022). Although orchids have high biological and economic value, few studies investigating TCP TFs have been performed in orchids, and the floral morphology and reproductive development are still unknown. Lin et al. (2016) identified 23 TCP genes in Phalaenopsis equestris, focused on the development of ovules, and discovered that PePCF10 and PeCIN8 influence cell division to play crucial roles in ovule formation. Zhang et al. (2021a) identified 25 TCP genes in Dendrobium catenatum and concentrated on the TCP genes that control leaf development through the jasmonate-signaling pathway. DcaTCP4 and DcaTCP9 were found to possess varied expression patterns after 3 h of jasmonate treatment. Recently, some orchid genomes have been sequenced and analyzed (Sun et al., 2021; Yang et al., 2021; Ai et al., 2021; Li et al., 2022), and a TCP gene family analysis could be promoted.
To demonstrate the properties of TCP genes over the evolution of orchids, we undertook genome-wide identification, comparison, and expression investigations of TCP genes in Cymbidium goeringii (Spring Orchid or Chunlan) (Sun et al., 2021) in this work. The findings could offer novel understanding of the fundamental processes driving the growth and development of orchid organs and other flowering plants.
Materials and methods
Plant materials
Wild plants of C. goeringii were cultivated and collected from the Forest Orchid Garden greenhouse at Fujian Agriculture and Forestry University (Fuzhou, Fujian Province, China). The temperature of the greenhouse culture was approximately 25°C. The roots, stems, leaves and flowers (sepal, petal, lip and column) were used in this study. All samples were gathered, put in tubes, and frozen before being stored in an ultralow temperature refrigerator at −80°C.
Genome-wide identification and physicochemical properties of TCP genes
CgTCP (C. goeringii TCP gene) genes were identified in the C. goeringii genome (Sun et al., 2021), and five species (Arabidopsis thaliana, Oryza sativa, Ananas comosus, Phalaenopsis equestris, and Dendrobium catenatum) TCP proteins served as the queries. The genome and transcriptome data of C. goeringii were downloaded from the NCBI database1, and the Arabidopsis thaliana TCP proteins were downloaded from TAIR2, the rice TCP proteins download from Rice Genome Annotation Project (http://rice.uga.edu/), the Ananas comosus, Phalaenopsis equestris, and Dendrobium catenatum TCP proteins download from previous studies (Lin et al., 2016; Zhang et al., 2021a; Wang et al., 2022). BLAST and HMMER were used to identify the TCP genes. A BLAST table (E-value < 0.05) and sequence were acquired from TBtools (Chen et al., 2020b). The hidden Markov model (HMM) of the TCP domain (PF03634) was downloaded from the Pfam protein family database3, and the HMM profile was used to identify the TCP protein sequences through the Simple HMM Search in TBtools (Chen et al., 2020b). Combining the results of BLAST and Hmmsearch, all TCP genes were analyzed by NCBI Batch-CDD tools4, and the genes containing the entire TCP domain were retained. The ExPASy online tool5 (Artimo et al., 2012) used to predict physicochemical properties of the TCP genes, and the Plant-mPloc6 (Chou and Shen, 2010) used to predict subcellular localizations.
Motif and gene structure analysis of TCP genes
According to the gene location of the TCP genes in the C. goeringii genome, complete gene sequences were extracted. Complete gene sequences and coding sequences (CDSs) were prepared for the gene structure analysis in GSDS7 (Hu et al., 2015). The MEME8 online tool (Bailey et al., 2009) used the default parameters to examine the motifs of TCP genes, and the visualization of the results of the gene structure and motifs were combined using TBtools.
Phylogenetic analysis and classification of TCP genes
The TCP genes of C. goeringii were collected and identified, and the sequences were aligned by MUSCLE (Edgar, 2004) using MEGA 7 (Kumar et al., 2016). Alignment was also conducted in the TCP genes of C. goeringii, Arabidopsis thaliana, Phalaenopsis equestris (Lin et al., 2016) and Dendrobium catenatum (Zhang et al., 2021a). The phylogenetic analysis was performed by the maximum likelihood (ML) method using RA×ML (RAxML-HPC2 on XSEDE) in the CIPRES Science Gateway web server9 (Miller et al., 2010) with the Protein CAT model and GTR matrix with 1,000 bootstrap iterations. The Evolview (He et al., 2016) used to polish the output phylogenetic tree.
Collinearity analysis and mapping TCP genes on chromosomes
The protein sequences (PEPs), CDSs and annotation files (gff files) of the C. goeringii, D. catenatum and P. equestris genomes were prepared for a collinearity analysis and gene mapping. The TCP gene location information in the C. goeringii genome chromosomes was acquired in the gff file, and visualization was conducted by the R package ‘RIdeogram’ (Hao et al., 2020). JCVI v1.2.10 (https://pypi.org/project/jcvi/) was used in the collinearity analysis of C. goeringii, P. equestris and D. catenatum. First, the PEP, CDS and gff files were formatted. Second, the PEP files were aligned and analyzed to acquire the collinearity file. The anchor file was used in the visualization, and the TCP genes were highlighted in the collinearity map.
Prediction of Cis-acting elements
In total, 2000 bp upstream sequences of TCP genes were retrieved to investigate the regulatory functions in plant growth and development. The sequences were extracted by TBtools according to the gene locations in the annotation file. The cis-acting elements of the TCP genes were identified and annotated in the promoter regions by the online tool PlantCARE10 (Lescot et al., 2002). TBtools (Chen et al., 2020b) was used to display the findings of the cis-acting element counts and annotation.
Expression pattern analysis
RNA-Seq by Expectation Maximization (RSEM) (Li & Dewey, 2011) was employed for the transcript quantification and calculation of the fragments per kilobase per million mapped reads (FPKM) value of each gene in the transcriptomic analysis. The FPKM matrix and heatmaps were generated by TBtools (Chen et al., 2020b).
Roots, stems, leaves, and mature flowers (sepal, petal, lip, and column) from C. goeringii grown at the Forest Orchid Garden of Fujian Agriculture and Forestry University were taken for a quantitative real-time PCR (qRT-PCR) analysis to confirm the expression pattern of the TCP genes. Three replicates of each type of tissue were sampled in the analysis. The total RNA was extracted from the tissues by RNA Simple Plant Kit. The RNA concentration in each tissue was in the range of 34.3–398.9 ng/µl, with A260/280 values ranging from 2.01 to 2.14, indicating that the extracted RNA was of high quality. The primer tool in Geneious (Kearse et al., 2012) was employed to design specific PCR primers. In Supplementary Table 1, the gene-specific primers for the three candidate genes are given together with the associated internal reference genes. The cDNA synthesis and qPCR were carried out using the Vazyme/R223 and Yeasen/11202ES03 kits, respectively. RT−qPCR was carried out to confirm the precise expression of Class II genes in the roots, stems, leaves and flowers using CIN genes in C. goeringii (GL10643, GL16029 and GL28896). Three biological replicates and three technical replicates were used in each experiment. The relative gene expression was calculated using the 2−ΔΔCT method.
Gene ontology analysis
The protein file of the C. goeringii genome was used to search against the eggNOG 5.0 database using EggNOG-mapper v211 (Huerta-Cepas et al., 2019) for Gene Ontology (GO) functional annotation. Sequence similarity was used to predict function, sequence alignment was used to predict orthology, E-values and bit scores were used to filter out low-quality orthology alignments, and GO annotation terms associated with proteins involved in well-known biological processes were used to classify functions.
Results
Identification and protein traits of TCP genes
In total, 14 full-length TCP genes were identified from the C. goeringii genome. The Supplementary Data Sheet contained the complete protein sequences for TCP. InterproScan 5 (Jones et al., 2014) was used to confirm that all potential TCP genes encode the conserved TCP domain. The 14 TCP genes had an average length of 323 aa (range 169–572 aa), which was located on eight chromosomes. The average molecular weight (MW) was 35321.21 kDa (range 19216.05–63038.55 kDa). The grand average of hydrophilic (GRAVY) values were all negative in C. goeringii TCP proteins, suggesting genes with strong hydrophilicity. The average isoelectric point (pI) of the TCP proteins was 8.5 (range 6.60–10.55), indicating relatively strong acidity. The results of the subcellular location predictions revealed that all TCP proteins were found in the nucleus, except for one gene (GL08210), suggesting that they mostly function as TFs in the nucleus (Table 1).
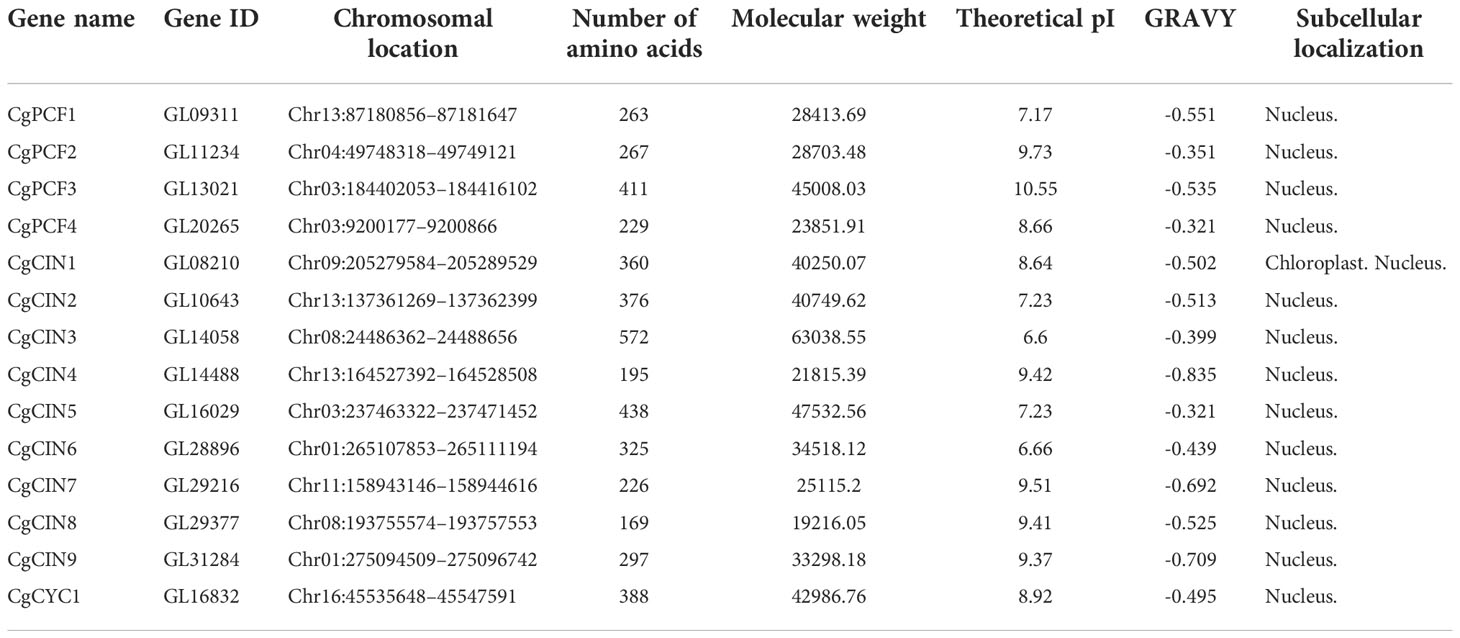
Table 1 Analysis of amino acid sequence characteristics of the CgTCP gene family in Cymbidium goeringii.
Phylogenetic analysis of the protein sequence
TCP genes of four species (Arabidopsis thaliana, Phalaenopsis equestris, Dendrobium catenatum and C. goeringii) were used to conduct the phylogenetic analysis. In total, 87 genes in four species were classified into two clades (Figure 1). In total, 14 genes of C. goeringii were divided into two clades, Class I (PCF genes) and Class II (CIN genes and CYC/TB1 genes). Similar to other orchids, three types of genes exist in C. goeringii as follows: four PCF genes, nine CIN genes and one CYC gene. We also performed the TCP proteins phylogenetic analysis of 18 different plant phylostrata groups species, results showed Class I and Class II (Class II-A, B, C) were divided (Supplementary Figure 1; Supplementary Table 2). The TCP protein of algae only found in Class I and orchids TCP proteins were scattered in different clades.
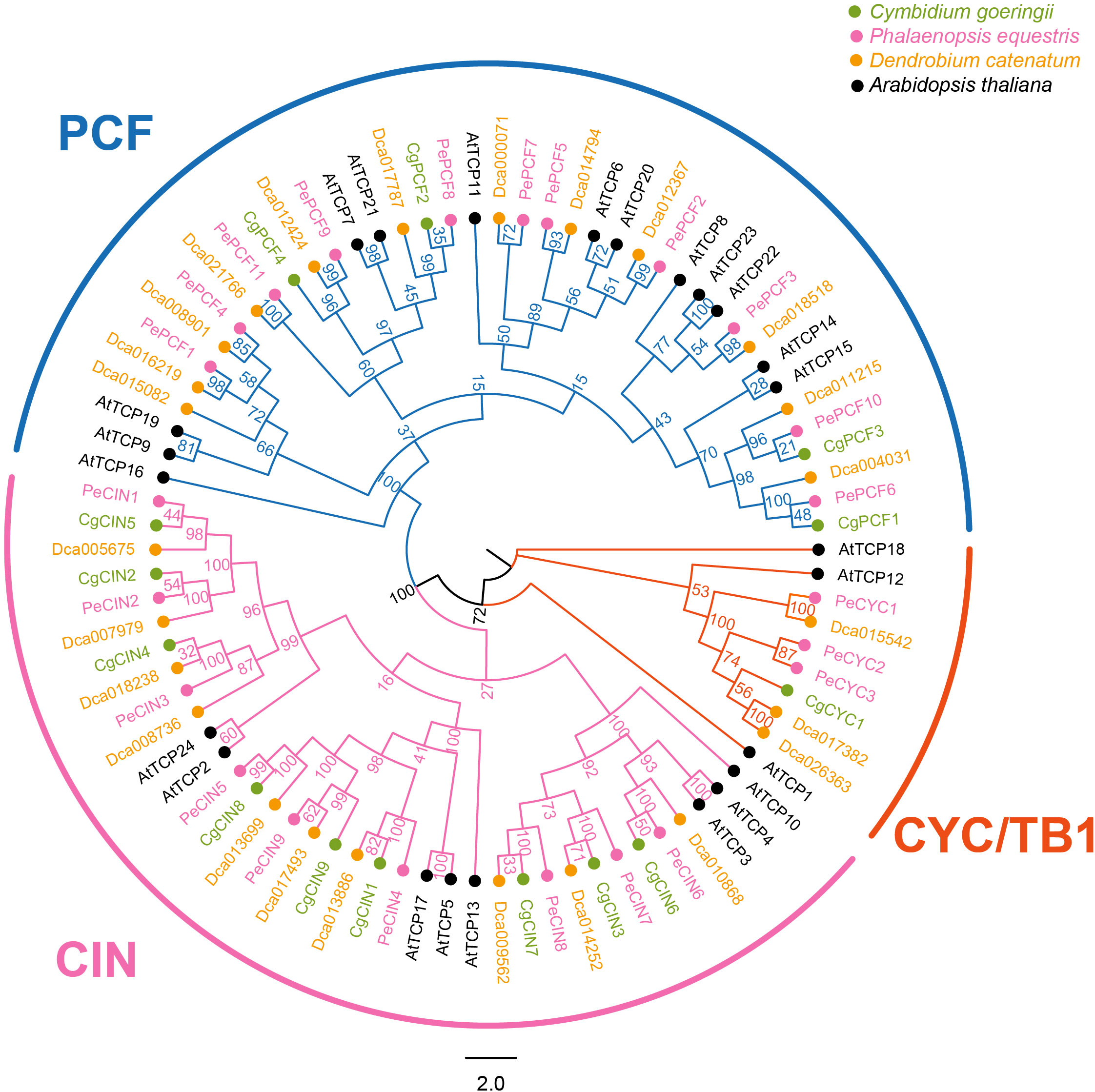
Figure 1 Phylogenetic tree based on the TCP protein sequences of Cymbidium goeringii, Phalaenopsis equestris, Dendrobium catenatum and Arabidopsis thaliana. Phylogenetic analysis indicated that the TCP gene family was classified into the following two clades: Class I (PCF) and Class II (CIN and CYC/TB1); there are two subclades in Class II. TCP protein sequences of C. goeringii are available in the Supplementary Data Sheet.
We also analyzed the sequence alignment of CgTCP genes. The TCP domain was present in all CgTCP genes, and the domain can be divided into four parts (Figure 2A, Supplementary Figure 2). The basic region was the most conserved, followed by two helix regions, and the loop region was more varied than the other regions. The TCP domain of the CgTCP proteins was similar to that in P. equestris and D. catenatum (Lin et al., 2016; Zhang et al., 2021a), indicating that the TCP domain is highly conserved in orchids. One CgTCP protein (CgCYC1) exited an R domain compared with CgCIN9 (Figure 2B), and the R domain was only present in the CYC gene, which is consistent with the phylogenetic results.
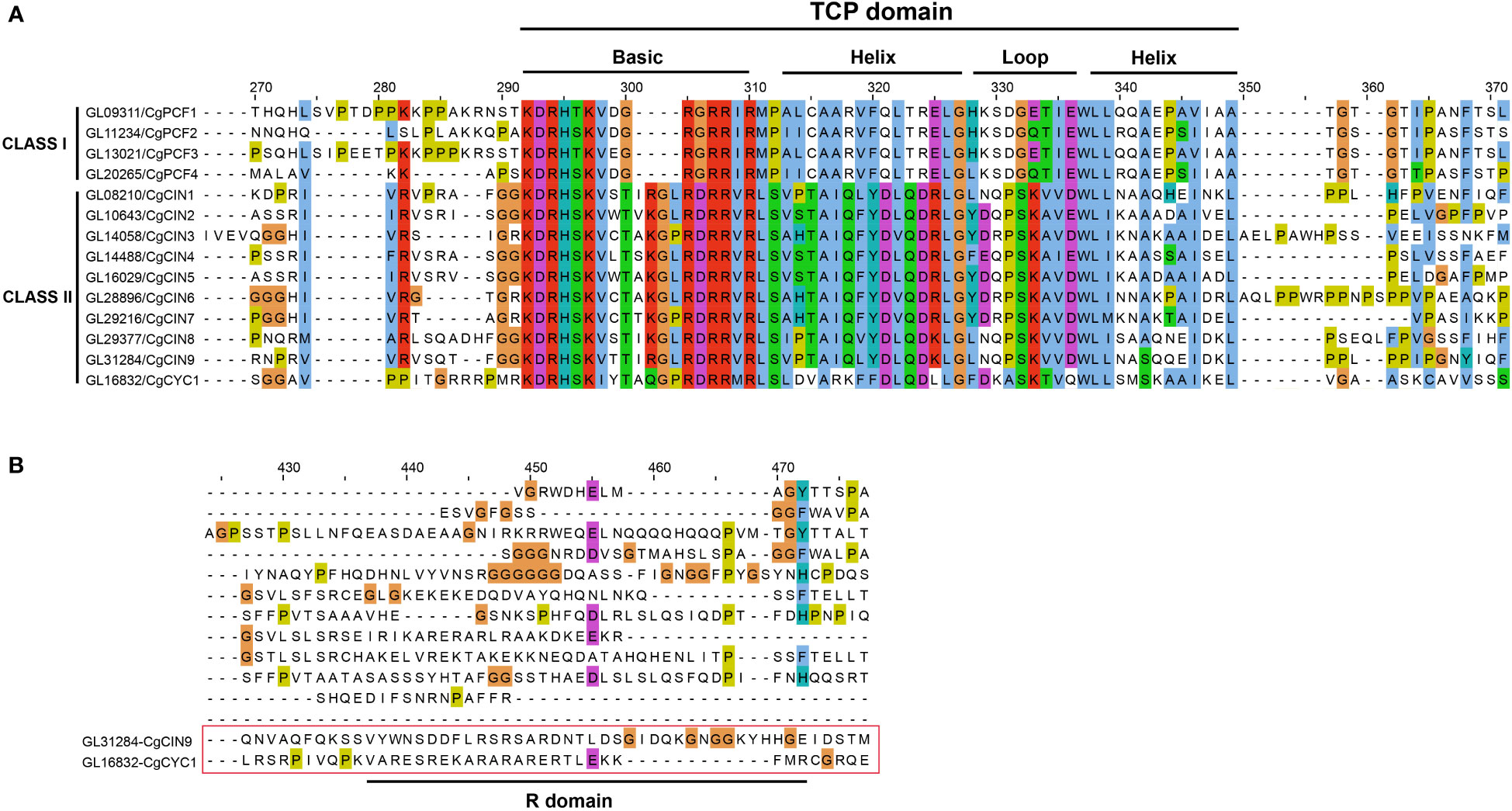
Figure 2 Multiple sequence alignment and protein sequence of the TCP domain. (A) Alignment of the TCP domain containing the Basic, Helix and Loop sequences for the predicted C. goeringii TCP proteins. (B) Alignment of the R-domain of the CIN and CYC genes. .
Collinearity analysis of TCP genes between orchids
The gene location on the chromosome of C. goeringii was examined, and 14 TCP genes were located on eight chromosomes (Figure 3A). In addition, a collinearity analysis was conducted among the TCP genes of C. goeringii, P. equestris, and D. catenatum. The collinear relationship among the TCP genes was examined to identify potential duplication events during TCP gene evolution in orchids. The results demonstrated a nearly 1:1 correspondence between all TCP genes in the three orchids, indicating minimal genomic rearrangements and TCP orthologs reshuffling after the lineages of Dendrobium and Cymbidium diverged (Figure 3B). The results showed that gene pseudogenization and loss might lead to a decrease in TCP in C. goeringii.
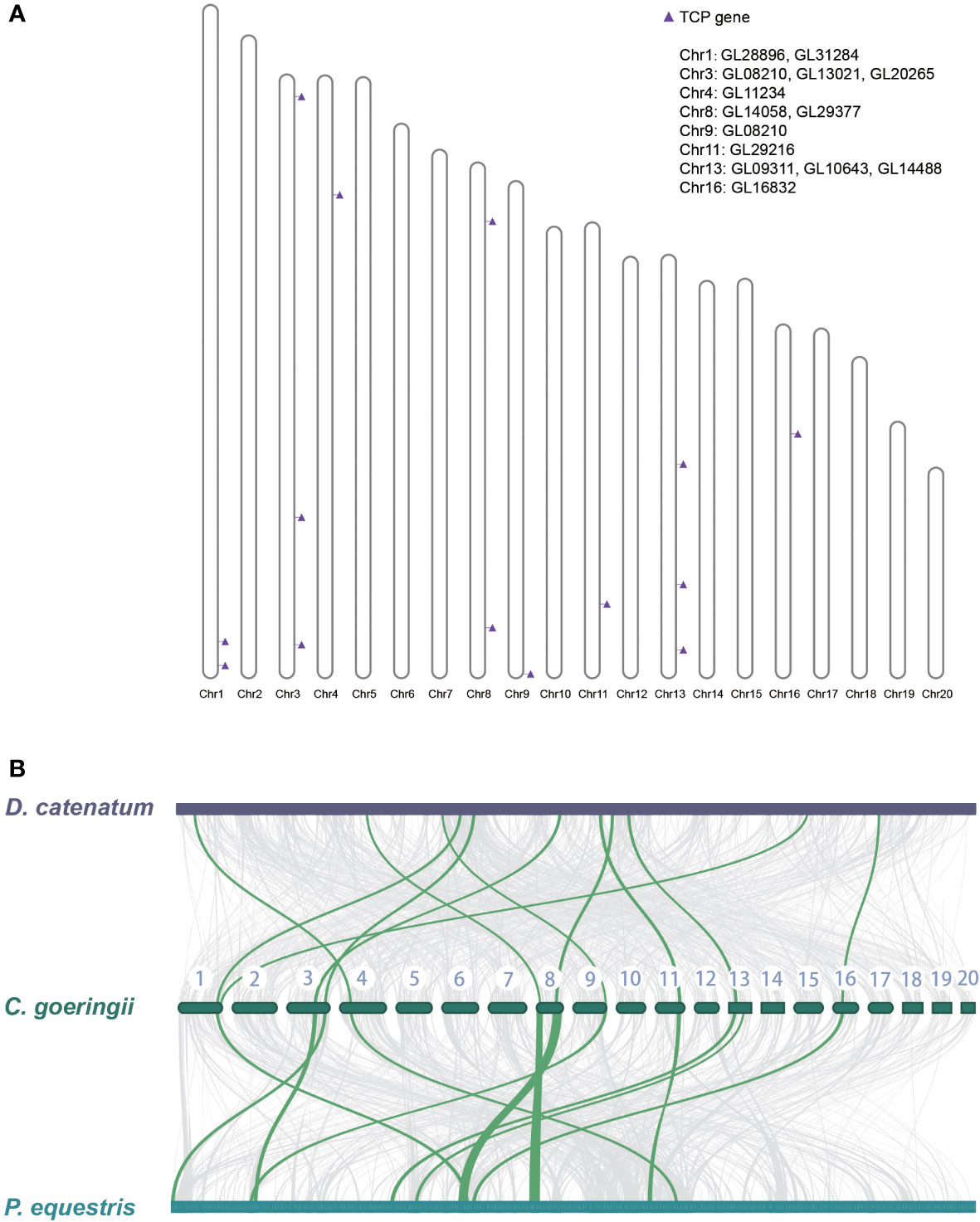
Figure 3 The TCP gene location on the chromosome of C. goeringii and collinearity between P. equestris and D. catenatum. (A) The chromosomal location of CgTCP genes. (B) The collinearity analysis suggested that the three orchids presented nearly one-to-one correspondence of TCP proteins.
Gene structure and motif analysis
To explore the gene structure of TCP genes in orchids, the intron–exon and up/downstream regulatory element distributions of C. goeringii, P. equestris and D. catenatum were analyzed (Figure 4A). The results revealed that the TCP family of C. goeringii had 1–5 exons and 0–4 introns, P. equestris had 1–3 exons and 0–2 introns, D. catenatum had 1–2 exons and 0–1 intron. Although similarity in the gene structure was found in each subclade, the TCP genes of C. goeringii have a high degree of variance in the intron length and exon numbers in comparison with A. thaliana, P. equestris and D. catenatum. In general, most Class I genes exhibited fewer exons than the Class II genes. Notably, in Class I, CgPCF1, CgPCF2 and CgPCF4 have no exons, whereas in Class II, CgCIN2 and CgCIN4 have no exons (Figure 4A).
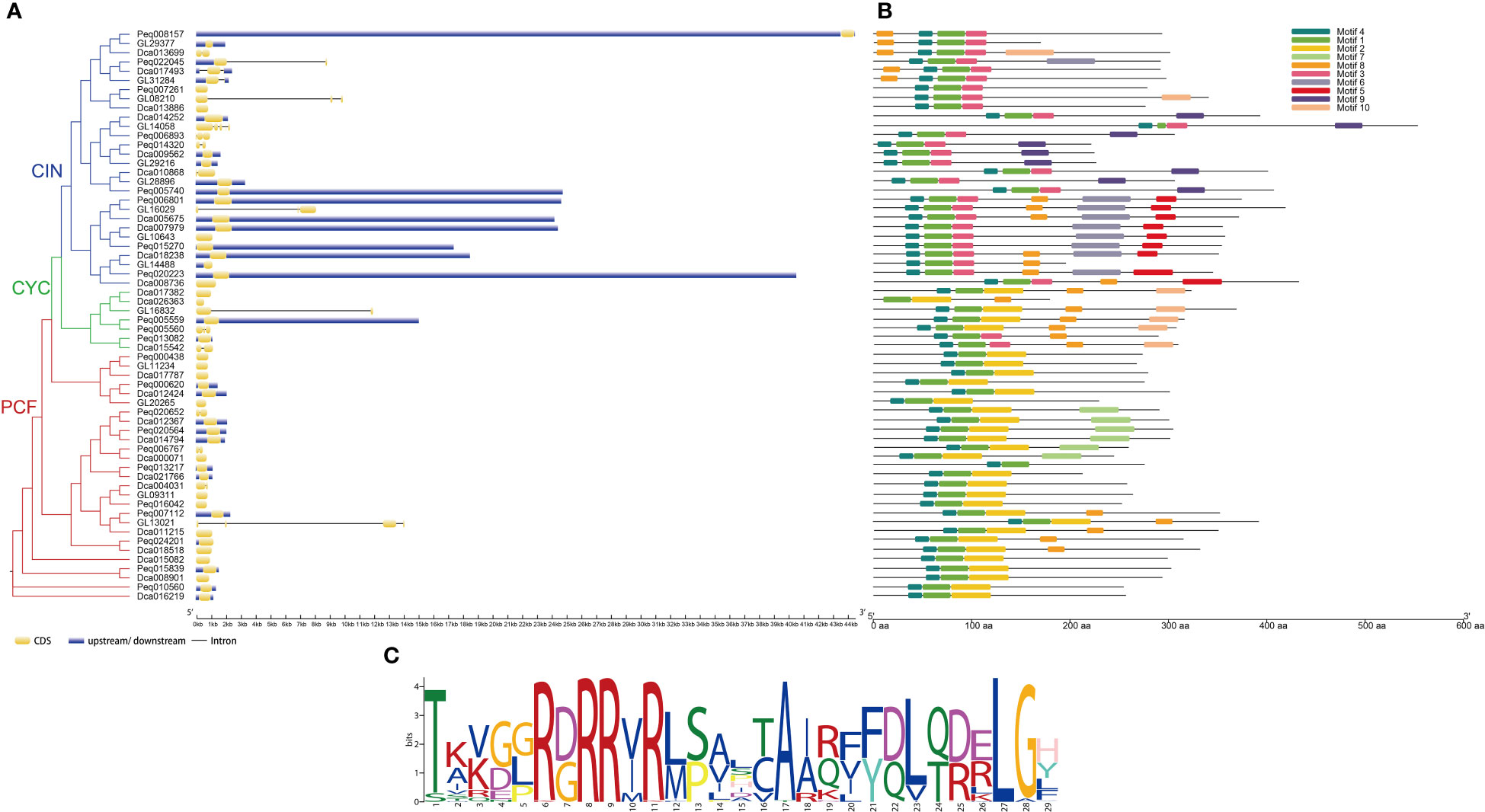
Figure 4 Gene structure, conserved motifs, and conserved domains of TCP genes. (A) Phylogenetic tree and gene structure of the TCP genes of C. goeringii, P. equestris and D. catenatum. (B) Predicted motifs of the TCP genes of C. goeringii, P. equestris and D. catenatum. (C) Sequence logo of motif 1, which encodes the TCP domain.
The motifs of the TCP proteins in C. goeringii, P. equestris and D. catenatum were analyzed by the MEME, and 10 motifs were set as upper bounds (Figure 4B). The number of TCP motifs both ranged from three to six in C. goeringii and D. catenatum, and ranged from two to six in P. equestris. Motif 1 encoded the TCP domains (Figure 4C). The PCF proteins all contain motif 1, motif 2 and motif 4, CIN protein all contain motif 1 and motif 3, the PCF proteins were more conserved compare with CIN and CYC proteins by motif analysis.
Cis-acting regulatory elements and GO enrichment analysis
The 2,000 bp upstream regions of C. goeringii, P. equestris and D. catenatum TCP genes were extracted for the identification of putative cis-elements and used to explore the promotor region functions. In total, 1644, 2250, 2440 cis-acting elements attributed to 26, 26, 29 types and 9, 9, 10 responsive functions were identified in C. goeringii, P. equestris and D. catenatum, respectively (Figure 5A and Supplementary Table 3). TATA-box made up the majority of these components (67.76%, 52.18%, 46.31%), followed by CAAT-box (11.25%, 12.80%, 15.49%) in C. goeringii, P. equestris and D. catenatum, respectively (Supplementary Table 4). The cis-element functions included phytohormone responsiveness to auxin, abscisic acid (ABA), gibberellin (GA), methyl jasmonate (MeJA) and salicylic acid; stress responsiveness, such as anoxic, anaerobic, drought and low-temperature stress; and growth and development elements, such as light response, cell cycle regulation, endosperm expression, meristem expression and circadian control (Figure 5A). Light responsiveness was the most prevalent element function in each TCP gene, suggesting the crucial functions that light plays in modulating TCP function throughout plant growth and development (Figure 5A). The second and third most prevalent types were MeJA-responsive and ABA-responsive elements, which were likewise broadly distributed in the majority of orchid TCPs (Supplementary Table 5). These results suggest that these elements may have roles in controlling these two phytohormones. We also discovered other components involved in meristem expression and meristem-specific activation, which is in line with the crucial functions played by TCP in the preservation of meristem homeostasis.
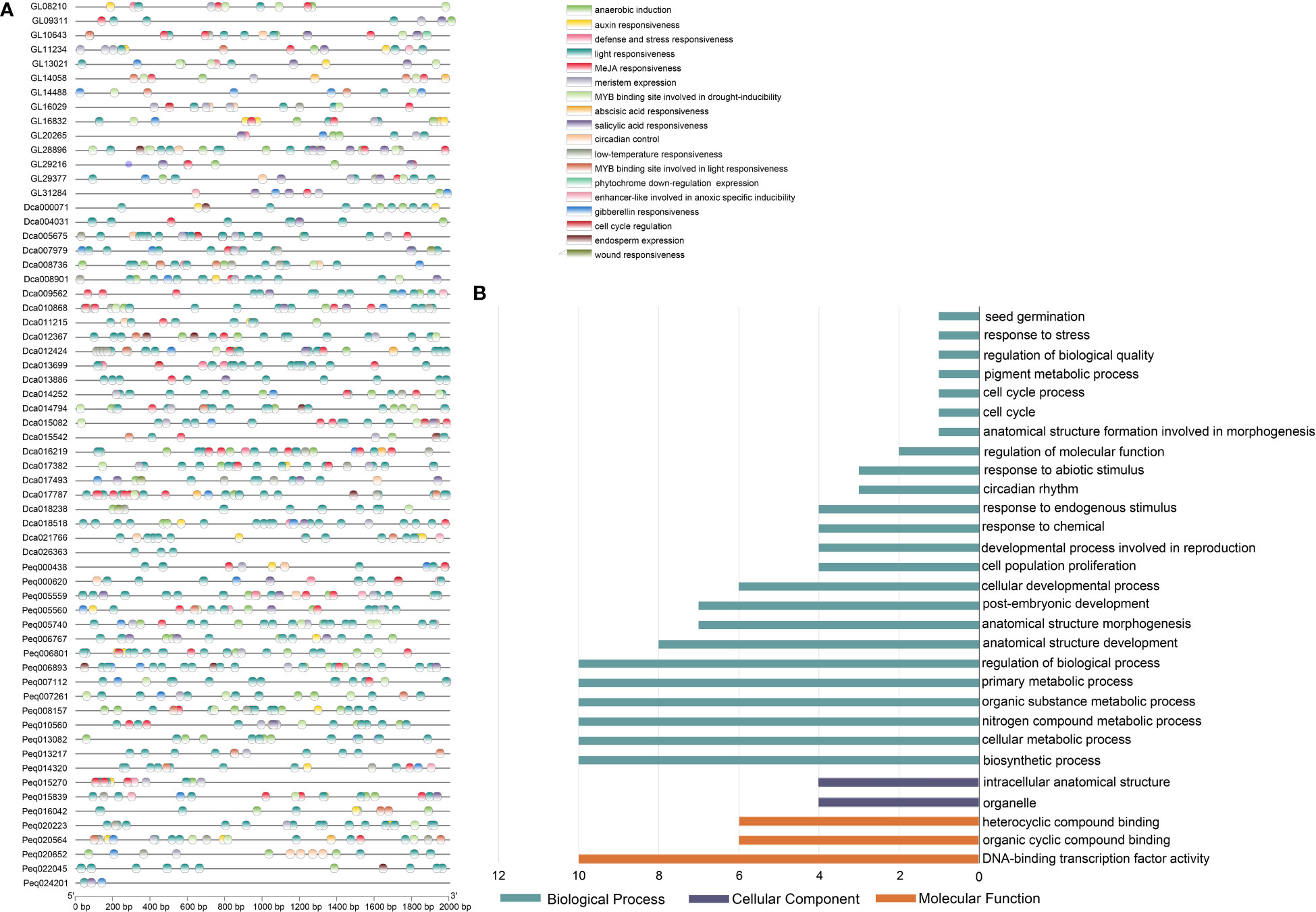
Figure 5 Cis-acting elements and GO enrichment analysis in the promoter regions of TCP genes. (A) Elements analysis of C. goeringii, P. equestris and D. catenatum, the same regulatory functions are presented in the same color. (B) Gene Ontology (GO) terms of C goeringii TCP genes; the detailed data are listed in Supplementary Table 5.
A GO analysis was performed to annotate the gene functional classifications of the TCP genes of C. goeringii and investigate the important biological processes (Figure 5B). As a result, the GO terms “cellular developmental process”, “intracellular anatomical structure” and “DNA-binding transcription factor activity” were the most related to plant growth and development in the GO ontologies “Biological Process,” “Cellular Component,” and “Molecular Function,”, respectively (Figure 5B and Supplementary Table 6). The findings are consistent with the fact that TCP is a premier regulator involved with a number of downstream transcriptional networks and functions mostly acts in the nucleus (Table 1). Several other terms, such as “circadian rhythm” and “seed germination”, are consistent with TCP’s function.
Expression pattern analysis of TCP genes
Based on transcriptome data from C. goeringii tissues, including flowers, leaves, stems, and roots, an expression analysis was carried out. The expression profile revealed that Class I TCP genes were expressed at lower levels in differentiating tissues and mature organs, while the CgPCF2 gene was expressed at moderate levels in various tissues (Figure 6A). Among the Class II genes, three CIN genes (CgCIN2, CgCIN5 and CgCIN6) exhibited high expression levels in flower organs (Figure 6A, Supplementary Table 7). In addition, the CgCYC1 gene was only expressed in the stem, indicating that the Class II TCP genes function in plant development.
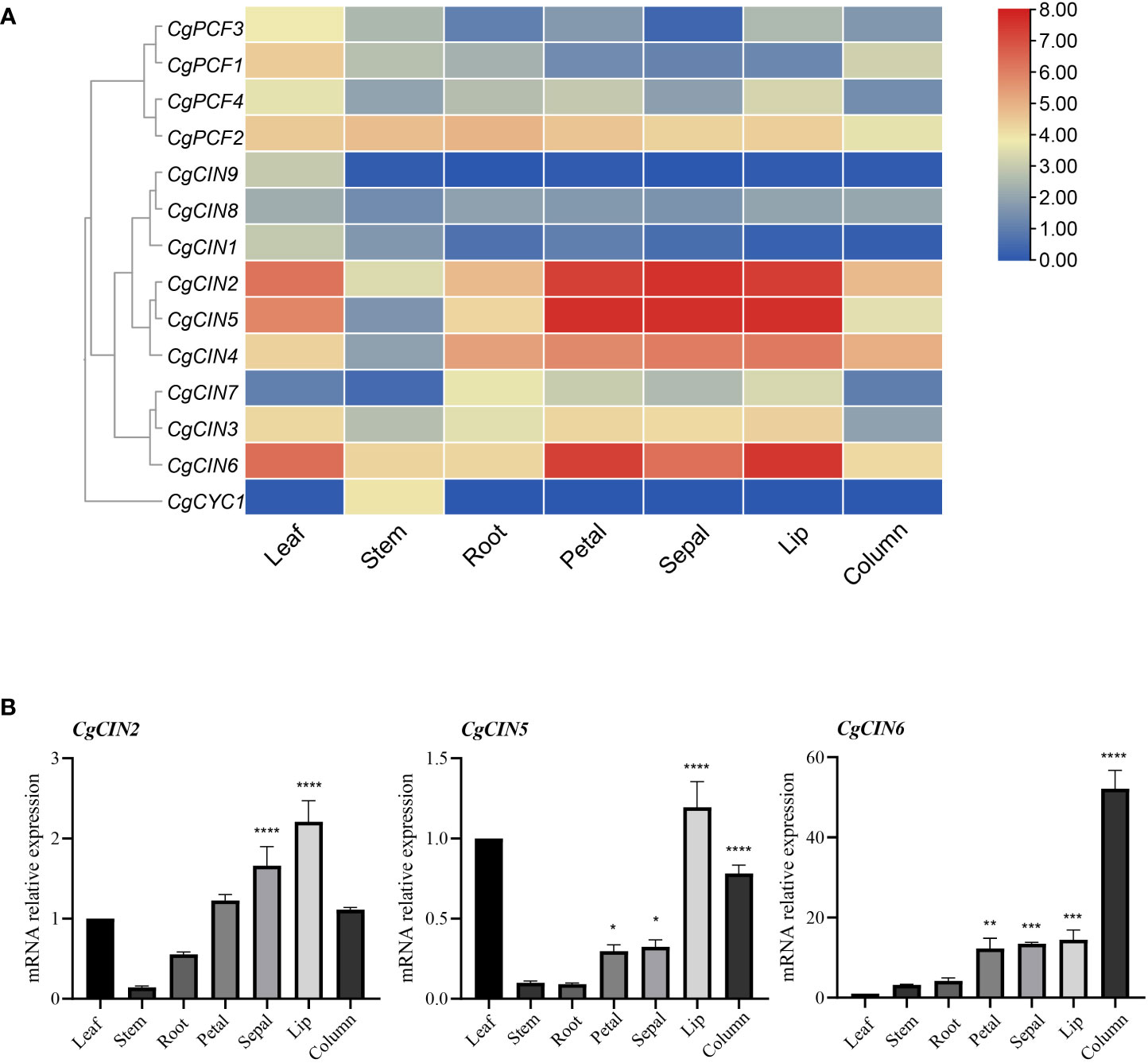
Figure 6 The expression pattern and RT−qPCR verification of TCP genes in seven tissues in C. goeringii. (A) Expression heatmap of TCP genes in seven tissues in C. goeringii. (B) Expression pattern of CgCIN2, CgCIN5 and CgCIN6 in seven tissues in C. goeringii by RT−qPCR. We performed the ANOVA multiple comparisons in the statistical test, star marks *, **, ***, and **** representing adjusted p < 0.05, p < 0.01, p < 0.001, and p < 0.0001, respectively (Supplementary Table 8).
Flowers showed strong expression of class II genes such CgCIN2, CgCIN5, and CgCIN6 (Figure 6A), indicating high expression and their potential significance in driving tissue differentiation. The gene expression of CIN homologs in C. goeringii was studied further in order to elucidate the precise functions of the Class II TCP genes by analyzing in the leaves, flowers, and stems using RT−qPCR (Figure 6B; Supplementary Table 8). The CgCIN2 showed high expression in the sepals and lips, the CgCIN5 showed high expression in the lips and columns, the CgCIN6 showed extremely high expression in columns, of three examined CIN genes were barely detected in the stems and roots (Figure 6B). To understand the underlying functions of the TCP gene in C. goeringii, a functional study of these two classes will be a crucial next step.
Discussion
As a traditional flowering plant, C. goeringii and their products have significantly contributed to the Chinese flower industry. However, the molecular biological mechanisms of orchid development have seldom been reported. TCP proteins are crucial for the development and growth of plants. Numerous plant species, including A. thaliana (Riechmann et al., 2000), Hypoxis decumbens (Madrigal et al., 2017), P. equestris (Lin et al., 2016), and D. catenatum (Zhang et al., 2021a), the TCP gene family have been discovered. However, the analysis of the TCP gene family in orchids was still insufficient. The TCP gene family of C. goeringii was identified using a variety of methods in this work, and the evolution of CgTCP proteins as well as their functional properties were examined.
Non-model plant gene family studies have greatly increased due to advances in whole-genome sequencing. This study identified 14 CgTCPs from C. goeringii and divided them into two main clades (Table 1 and Figure 1). Compared with other orchids, the number of TCP genes in C. goeringii was less than that in D. catenatum (25) and P. equestris (23) (Lin et al., 2016; Zhang et al., 2021a). C. goeringii may have suffered gene loss during evolution. The three orchids have similar numbers of CIN genes but different numbers of PCF and CYC genes. There are 11 PCF genes in both D. catenatum and P. equestris, four PCF genes in C. goeringii, three CYC genes in D. catenatum and P. equestris, and only one CYC gene in C. goeringii. The different TCP gene numbers may be related to the life form. The physical and chemical properties of CgTCP proteins were similar to those of D. catenatum and P. equestris. All CgTCP proteins were predicted to be located in the nucleus, except for CgCIN1, which is also located in the chloroplast and may contribute to new functions. Furthermore, we performed the evolution of TCP genes in orchids and other land plants (Supplementary Figure 1). Previous study suggested TCP gene family may origin from the common ancestor of Phragmoplastophyta, and expanded through the whole-genome duplication during the evolution (Liu et al., 2019; Wang et al., 2022). Our results supported that lower plants have a smaller number of TCP genes and TCP family has undergone expansion in the course of plant evolution. We also investigated the evolution of orchid TCP genes and found that almost all subfamilies of TCP are present in orchids, and D. catenatum possessed more members compared with other orchids. Comparing with other monocots, orchids Class II-A genes were expanded, PeCIN8 which located in Class II-A plays curial role in ovule development (Lin et al., 2016), the expanded genes may relate to orchid ovule unique development.
The conserved gene structure in the same clade of the TF gene family had been carried out in previous researches (Wang et al., 2021; Ke et al., 2021; Zhang et al., 2022; Chen et al., 2022). This study revealed that C. goeringii has an exon–intron structure similar to that of Camellia sinensis and Prunus mume (Zhou et al., 2016; Shang et al., 2022) (Figure 4). Almost all sequenced orchids have been reported to have long introns (Cai et al., 2015; Zhang et al., 2016; Zhang et al., 2017; Yuan et al., 2018; Ai et al., 2021; Sun et al., 2021; Zhang et al., 2021b; Li et al., 2022), but in CgTCPs, only four genes have long introns, and eight CgTCPs have no introns. The motif analysis of C. goeringii, P. equestris and D. catenatum indicated that class I, which contains motifs 1, 2, and 4, was conserved and that class II, which contains different motifs in different clades, was more variable (Figure 4). The conservatism of Class I genes may result in the crucial role in plants, which mostly exist in all phylostrata plants (Supplementary Figure 1). The CgTCP alignment was also analyzed in this study, and the results showed that the TCP domain and R domain were conserved in orchids (Figure 2).
Gene location in scaffolds and chromosomes and genomic comparison are closely related to the gene structure and function (Ren et al., 2000). The gene location analysis results showed that CgTCPs were scattered on eight chromosomes, and no gene tandem repeats were found in C. goeringii (Figure 3A). The genomic comparisons among D. catenatum, P. equestris and C. goeringii in chromosomes or scaffolds showed that their TCP genes were in an almost 1:1 correspondence, supporting that no duplication events occurred in CgTCPs (Figure 3B). In addition, P. equestris exhibited a more syntenic relationship between its scaffold or chromosome than D. catenatum, indicating that there were few chromosomal structure variations after the two species diverged.
Studies of promoter regions that regulate gene expression at the transcriptional level by cis-acting elements promote our basic understanding of gene regulation and expand the toolbox of available promoters (Hernandez-Garcia and Finer, 2014). A series of functional types of regulatory elements in the promoter regions have been identified in TCP genes, including the following categories: growth and development elements, stress-responsive elements and phytohormone-responsive elements (Figure 5). We identified a large proportion of light responsiveness elements in the promotor region, indicating that orchids TCP genes are regulated by the light signal to work in concert with plant growth and development (Supplementary Table 4). Furthermore, CgTCPs perform important functions regulated by phytohormones, which contain many elements responsive to MeJA, ABA, GA, and auxin. Based on previous studies, the TCP3, TCP14 and TCP15 genes regulate cell expansion and differentiation by auxin (Koyama et al., 2010; Ferrero et al., 2021), and CgTCPs may interact with auxin by a similar mechanism. In the inflorescence shoot apex and embryo root apical meristem, GA-induced DELLA protein degradation releases TCP genes to stimulate shoot elongation and seed germination (Nicolas and Cubas, 2016). In leaves, TCP-CIN genes interact with the CK receptor HISTIDINE KINASE 4 (AHK4)/CRE1 (CYTOKININ RESPONSE 1) and can promote the expression of the CK response genes ARR7 and ARR15 (Das Gupta et al., 2014). In seeds, the TCP14 gene could promote germination by regulating ABA signaling (Tatematsu et al., 2008). The cis-element analysis and GO analysis indicate that the TCP genes are mainly regulated by phytochromes and light and play an important role in different plant tissues of growth and development.
TCP transcription factors control key vegetation and reproductive developmental processes and participate in the growth patterns of meristems and organs (Nicolas & Cubas, 2016). Class II TCP genes contain genes mainly involved in lateral organ development (Crawford et al., 2004; Ori et al., 2007). In this study, we found and verified that CgCIN2, CgCIN5 and CgCIN6 were highly expressed in various flower components (sepal, petal, lip and column) and relatively highly expressed in leaves (Figure 6; Supplementary Table 8). Liu et al. (2017) found that the downregulation of five class II TCP genes may be related to the delayed flowering phenotype of Arabidopsis mutants. We speculate that CgCIN2, CgCIN5 and CgCIN6 may produce a marked effect on the regulation of florescence.
Conclusion
TCP transcription factor proteins are known to play a crucial role in many aspects of plant growth and development. Here, we identified 14 TCP genes of C. goeringii and classified them into two clades by a phylogenetic analysis, with more members found in Class II. The sequence alignment, motif, gene structure and collinearity analyses indicated that CgTCPs were conserved and that no gene duplication events occurred in C. goeringii. The cis-element and GO analyses indicated that CgTCPs are regulated by light and various phytochromes. Furthermore, the expression pattern and RT−qPCR analyses suggested that CgCIN2, CgCIN5 and CgCIN6 may produce a marked effect on the regulation of florescence. Altogether, this study presents a comprehensive analysis of the structure and expression pattern of TCP genes in C. goeringii. These results provide a reference for further understanding how TCP genes participate in plant vegetative and reproductive growth and development and explain the plasticity of plant morphogenesis.
Data availability statement
The datasets presented in this study can be found in Figshare, https://doi.org/10.6084/m9.figshare.21548541.
Author contributions
The research was created and planned by SL and Z-JL. The data analysis was completed by D-KL and M-HL, who also prepared the paper. The analysis was carried out experimentally by CZ, XZ, QZ, and DZ. The plant materials were gathered by SK and YL. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Forestry Peak Discipline Construction Project of Fujian Agriculture and Forestry University (72202200205) and National Key Research and Development Program of China (2019YFD1000400).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1068969/full#supplementary-material
Supplementary Data Sheet | Sequences of Cymbidium goeringii TCP proteins.
Supplementary Figure 1 | The phylogenetic tree of TCP genes based on 18 plant species.
Supplementary Figure 2 | The entire map of the sequence alignment presentation of Cymbidium goeringii TCP proteins.
Supplementary Table 1 | Prime used in RT−qPCR.
Supplementary Table 2 | The TCP protein sequences of 18 plant species.
Supplementary Table 3 | Detailed information of the cis-acting element prediction.
Supplementary Table 4 | Statistics of the quantity and proportion of different types of cis-acting elements.
Supplementary Table 5 | Statistics of cis-acting elements involved responsive functions.
Supplementary Table 6 | GO annotation of the TCP genes of C. goeringii.
Supplementary Table 7 | Expression pattern data in seven tissues of C. goeringii.
Supplementary Table 8 | Statistical analysis using ANNOVA multiple comparisons for qPCR expression levels between reproductive organs and vegetable organs.
References
Ai, Y., Li, Z., Sun, W., Chen, J., Zhang, D., Ma, L., et al. (2021). The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res 8, 255. doi: 10.1038/s41438-021-00683-z
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res 40, 597–603. doi: 10.1093/nar/gks400
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi: 10.1093/nar/gkp335
Cai, J., Liu, X., Vanneste, K., Proost, S., Tsai, W. C., Liu, K. W., et al. (2015). The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 47, 65–72. doi: 10.1038/ng.3149
Chase, M. W., Cameron, K. M., Freudenstein, J. V., Pridgeon, A. M., Salazar, G., van den Berg, C., et al. (2015). An updated classification of orchidaceae. Bot. J. Linn. Soc 177, 151–174. doi: 10.1111/boj.12234
Chen, J., Bi, Y., Wang, Q., Liu, D. K., Zhang, D., Ding, X., et al. (2022). Genome-wide identification and analysis of anthocyanin synthesis-related R2R3-MYB genes in Cymbidium goeringii. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1002043
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020b). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y. Y., Hsiao, Y. Y., Chang, S., Zhang, D., Lan, S. R., Liu, Z. J., et al. (2020a). Genome-wide identification of YABBY genes in orchidaceae and their expression patterns in Phalaenopsis orchid. Genes 11, 1–17. doi: 10.3390/genes11090955
Chen, Y. Y., Hsiao, Y. Y., Li, C. I., Yeh, C. M., Mitsuda, N., Yang, H., et al. (2021). The ancestral duplicated DL/CRC orthologs, PeDL1 and PeDL2 , function in orchid reproductive organ innovation. J. Exp. Bot. 72, 5442–5461. doi: 10.1093/jxb/erab195
Chou, K. C., Shen, H.B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PloS One 5, e11335. doi: 10.1371/journal.pone.0011335
Christenhusz, Byng, J. W. (2016). The number of known plants species in the word and its annual increase. Phytotaxa 261, 201–217. doi: 10.11646/phytotaxa.261.3.1
Crawford, B. C. W., Nath, U., Carpenter, R., Coen, E. S. (2004). Cincinnata controls both cell differentiation and growth in petal lobes and leaves of antirrhinum. Plant Physiol. 135, 244–253. doi: 10.1104/pp.103.036368
Cubas, P., Lauter, N., Doebley, J., Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Das Gupta, M., Aggarwal, P., Nath, U. (2014). CINCINNATA in Antirrhinum majus directly modulates genes involved in cytokinin and auxin signaling. New Phytol. 204, 901–912. doi: 10.1111/nph.12963
De Paolo, S., Gaudio, L., Aceto, S. (2015). Analysis of the TCP genes expressed in the inflorescence of the orchid Orchis italica. Sci. Rep. 5, 1–11. doi: 10.1038/srep16265
Dixon, L. E., Greenwood, J. R., Bencivenga, S., Zhang, P., Cockram, J., Mellers, G., et al. (2018). TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum). Plant Cell 30, 563–581. doi: 10.1105/tpc.17.00961
Doebley, J., Stec, A., Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. doi: 10.1038/386485a0
Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ferrero, L. V., Gastaldi, V., Ariel, F. D., Viola, I. L., Gonzalez, D. H. (2021). Class I TCP proteins TCP14 and TCP15 are required for elongation and gene expression responses to auxin. Plant Mol. Biol. 105, 147–159. doi: 10.1007/s11103-020-01075-y
Floyd, S. K., Bowman, J. L. (2007). The ancestral developmental tool kit of land plants. Int. J. Plant Sci. 168, 1–35. doi: 10.1086/509079
Givnish, T. J., Spalink, D., Ames, M., Lyon, S. P., Hunter, S. J., Zuluaga, A., et al. (2015). Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. Biol. Sci. 282, 20151553. doi: 10.1098/rspb.2015.1553
Hao, Z., Lv, D., Ge, Y., Shi, J., Weijers, D., Yu, G., et al. (2020). RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput. Sci. 6, 1–11. doi: 10.7717/peerj-cs.251
Hernandez-Garcia, C. M., Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 217–218, 109–119. doi: 10.1016/j.plantsci.2013.12.007
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241. doi: 10.1093/NAR/GKW370
Hileman, L. C. (2014). Bilateral flower symmetry - how, when and why? Curr. Opin. Plant Biol. 17, 146–152. doi: 10.1016/j.pbi.2013.12.002
Horn, S., Pabõn-Mora, N., Theuß, V. S., Busch, A., Zachgo, S. (2015). Analysis of the CYC/TB1 class of TCP transcription factors in basal angiosperms and magnoliids. Plant J. 81, 559–571. doi: 10.1111/tpj.12750
Howarth, D. G., Donoghue, M. J. (2006). Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. U. S. A. 103, 9101–9106. doi: 10.1073/pnas.0602827103
Hubbard, L., McSteen, P., Doebley, J., Hake, S. (2002). Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162, 1927–1935. doi: 10.1093/genetics/162.4.1927
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Ke, Y. J., Zheng, Q. D., Yao, Y. H., Ou, Y., Chen, J. Y., Wang, M. J., et al. (2021). Genome-wide identification of the myb gene family in Cymbidium ensifolium and its expression analysis in different flower colors. Int. J. Mol. Sci. 22, 13245. doi: 10.3390/ijms222413245
Kosugi, S., Ohashi, Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9, 1607–1619. doi: 10.1105/tpc.9.9.1607
Koyama, T., Furutani, M., Tasaka, M., Ohme-Takagi, M. (2007). TCP Transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in arabidopsis. Plant Cell 19, 473–484. doi: 10.1105/tpc.106.044792
Koyama, T., Mitsuda, N., Seki, M., Shinozaki, K., Ohme-Takagi, M. (2010). TCP Transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in arabidopsis. Plant Cell 22, 3574–3588. doi: 10.1105/tpc.110.075598
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7 : molecular evolutionary genetics analysis version 7 . 0 for bigger datasets. Mol. Evol. Genet. Anal. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Lewis, J. M., Mackintosh, C. A., Shin, S., Gilding, E., Kravchenko, S., Baldridge, G., et al. (2008). Overexpression of the maize Teosinte Branched1 gene in wheat suppresses tiller development. Plant Cell Rep. 27, 1217–1225. doi: 10.1007/s00299-008-0543-8
Li, B., Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Li, M., Liu, K., Li, Z., Lu, H., Ye, Q., Zhang, D., et al. (2022). Genomes of leafy and leafless Platanthera orchids illuminate the evolution of mycoheterotrophy. Nat. Plants 8, 373–388. doi: 10.1038/s41477-022-01127-9
Lin, Y., Chen, Y., Hsiao, Y., Shen, C., Hsu, J., Yeh, C., et al. (2016). Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. J. Exp. Bot. 67, 5051–5066. doi: 10.1093/jxb/erw273
Liu, J., Cheng, X., Liu, P., Li, D., Chen, T., Gu, X., et al. (2017). MicroRNA319-regulated TCPs interact with FBHs and PFT1 to activate CO transcription and control flowering time in Arabidopsis. PloS Genet. 13, 1–22. doi: 10.1371/journal.pgen.1006833
Liu, Z. J., Lan, S. (2022). The evolutionary mechanisms of mycoheterotrophic orchids. Nat. Plants 8, 328–329. doi: 10.1080/0965431042000183978
Liu, M. M., Wang, M. M., Yang, J., Wen, J., Guo, P. C., Wu, Y. W., et al. (2019). Evolutionary and comparative expression analyses of TCP transcription factor gene family in land plants. Int. J. Mol. Sci. 20, 1–24. doi: 10.3390/ijms20143591
Luo, D., Carpenter, R., Copsey, L., Vincent, C., Clark, J., Coen, E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99, 367–376. doi: 10.1016/s0092-8674(00)81523-8
Madrigal, Y., Alzate, J. F., Pabón-Mora, N. (2017). Evolution and expression patterns of TCP genes in asparagales. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00009
Mao, Y., Wu, F., Yu, X., Bai, J., Zhong, W., He, Y. (2014). microRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 164, 710–720. doi: 10.1104/pp.113.228007
Martín-Trillo, M., Cubas, P. (2010). TCP Genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Miller, M. A., Pfeiffer, W., Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gatew. Comput. Environ. Work. GCE. doi: 10.1109/GCE.2010.5676129
Nath, U., Crawford, B. C. W., Carpenter, R., Coen, E. (2003). Genetic control of surface curvature. Science 299, 1404–1407. doi: 10.1126/science.1079354
Navaud, O., Dabos, P., Carnus, E., Tremousaygue, D., Hervé, C. (2007). TCP Transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33. doi: 10.1007/s00239-006-0174-z
Nicolas, M., Cubas, P. (2016). TCP Factors: New kids on the signaling block. Curr. Opin. Plant Biol. 33, 33–41. doi: 10.1016/j.pbi.2016.05.006
Ori, N., Cohen, A. R., Etzioni, A., Brand, A., Yanai, O., Shleizer, S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39, 787–791. doi: 10.1038/ng2036
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., et al. (2000). Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309. doi: 10.1126/science.290.5500.2306
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C. Z., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Shang, X., Han, Z., Zhang, D., Wang, Y., Qin, H., Zou, Z. (2022). Genome-wide analysis of the TCP gene family and their expression pattern analysis in tea plant (Camellia sinensis). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.840350
Sun, Y., Chen, G., Huang, J., Liu, D., Xue, F., Chen, X., et al. (2021). The Cymbidium goeringii genome provides insight into organ development and adaptive evolution in orchids. Ornam. Plant Res. 1, 10. doi: 10.48130/OPR-2021-0010
Tatematsu, K., Nakabayashi, K., Kamiya, Y., Nambara, E. (2008). Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 53, 42–52. doi: 10.1111/j.1365-313X.2007.03308.x
Walcher-Chevillet, C. L., Kramer, E. M. (2016). Breaking the mold: understanding the evolution and development of lateral organs in diverse plant models. Curr. Opin. Genet. Dev. 39, 79–84. doi: 10.1016/j.gde.2016.06.005
Wang, J. L., Wang, H. W., Cao, Y. N., Kan, S. L., Liu, Y. Y. (2022). Comprehensive evolutionary analysis of the TCP gene family: further insights for its origin, expansion, and diversification. front. Plant Sci. 13. doi: 10.3389/fpls.2022.994567
Wang, Q. Q., Zhu, M. J., Yu, X., Bi, Y. Y., Zhou, Z., Chen, M. K., et al. (2021). Genome-wide identification and expression analysis of terpene synthase genes in Cymbidium faberi. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.751853
Yang, F., Gao, J., Wei, Y., Ren, R., Zhang, G., Lu, C., et al. (2021). The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 19, 2501–2516. doi: 10.1111/pbi.13676
Yuan, Z., Gao, S., Xue, D. W., Luo, D., Li, L. T., Ding, S. Y., et al. (2009). RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 149, 235–244. doi: 10.1104/pp.108.128231
Yuan, Y., Jin, X., Liu, J., Zhao, X., Zhou, J., Wang, X., et al. (2018). The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 9, 1615. doi: 10.1038/s41467-018-03423-5
Zhang, D., Lan, S., Yin, W.-L., Liu, Z.-J. (2022). Genome-wide identification and expression pattern analysis of LRR-RLK gene family in apple. Front. Plant Sci. 13, 901089. doi: 10.3864/j.issn.0578-1752.2021.14.015
Zhang, G. Q., Liu, K. W., Li, Z., Lohaus, R., Hsiao, Y. Y., Niu, S. C., et al. (2017). The Apostasia genome and the evolution of orchids. Nature 549, 379–383. doi: 10.1038/nature23897
Zhang, L., Li, C., Yang, D., Wang, Y., Yang, Y., Sun, X. (2021a). Genome-wide analysis of the tcp transcription factor genes in Dendrobium catenatum lindl. Int. J. Mol. Sci. 22, 10269. doi: 10.3390/ijms221910269
Zhang, G. Q., Xu, Q., Bian, C., Tsai, W. C., Yeh, C. M., Liu, K. W., et al. (2016). The Dendrobium catenatum lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 6, 19029. doi: 10.1038/srep19029
Zhang, Y., Zhang, G., Zhang, D., Liu, X., Xu, X., Sun, W., et al. (2021b). Chromosome-scale assembly of the Dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic. Res. 8, 183. doi: 10.1038/s41438-021-00621-z
Keywords: genome-wide analysis, TCP gene family, evolution, expression pattern, Cymbidium goeringii
Citation: Liu D-K, Zhang C, Zhao X, Ke S, Li Y, Zhang D, Zheng Q, Li M-H, Lan S and Liu Z-J (2022) Genome-wide analysis of the TCP gene family and their expression pattern in Cymbidium goeringii. Front. Plant Sci. 13:1068969. doi: 10.3389/fpls.2022.1068969
Received: 13 October 2022; Accepted: 21 November 2022;
Published: 08 December 2022.
Edited by:
Raju Datla, Global Institute for Food Security (GIFS), CanadaReviewed by:
Chen Zhu, South China Botanical Garden (CAS), ChinaJing Cai, Northwestern Polytechnical University, China
Copyright © 2022 Liu, Zhang, Zhao, Ke, Li, Zhang, Zheng, Li, Lan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siren Lan, bGt6eEBmYWZ1LmVkdS5jbg==; Zhong-Jian Liu, empsaXVAZmFmdS5lZHUuY24=
 Ding-Kun Liu
Ding-Kun Liu Cuili Zhang
Cuili Zhang Xuewei Zhao
Xuewei Zhao Shijie Ke1
Shijie Ke1 Yuanyuan Li
Yuanyuan Li Diyang Zhang
Diyang Zhang Qinyao Zheng
Qinyao Zheng Siren Lan
Siren Lan Zhong-Jian Liu
Zhong-Jian Liu