
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 06 January 2023
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.1053699
Salt stress is one of the significant environmental stressors that severely affects plant growth and development. Plant responses to salt stress involve a series of biological mechanisms, including osmoregulation, redox and ionic homeostasis regulation, as well as hormone or light signaling-mediated growth adjustment, which are regulated by different functional components. Unraveling these adaptive mechanisms and identifying the critical genes involved in salt response and adaption are crucial for developing salt-tolerant cultivars. This review summarizes the current research progress in the regulatory networks for plant salt tolerance, highlighting the mechanisms of salt stress perception, signaling, and tolerance response. Finally, we also discuss the possible contribution of microbiota and nanobiotechnology to plant salt tolerance.
Soil salinization is one of the most adverse environmental stressors, severely limiting plant growth and development and threatening agricultural production worldwide. In addition to naturally occurring soil salinization, the situation is even exacerbated by excessive chemical fertilizers and soil amendments, improper irrigation practices, and the effect of seawater exposure (Munns and Tester, 2008; Sahab et al., 2021). It is estimated that crop production on at least 20% of global irrigated land is impaired. About 2 million ha (~1%) of world agricultural land is under accelerating salinization each year (Tuteja, 2007). Salt stress causes plant growth inhibition, abnormal development, and metabolic disturbance (van Zelm et al., 2020). The detrimental effects of elevated salinity on plants include (i) osmotic stress as sodium accumulates in the soil and (ii) ionic stress (Figure 1) (Yang and Guo, 2018a; Yang and Guo, 2018b; Ludwiczak et al., 2021). Osmotic stress caused by hyperosmotic soil solution disrupts plant cell turgor. In contrast, ionic stress is characterized by disordered sodium (Na+)/potassium (K+) balance inside the cell, disrupting various metabolic and physiological processes (Zhang et al., 2018). These two processes were reported to be temporally and spatially separated in plants during the salt stress response (van Zelm et al., 2020). However, there exists a significant overlap between osmotic and ionic stress in both early and downstream signaling (Geng et al., 2013; van Zelm et al., 2020). In addition, the elevated content of reactive oxygen species (ROS) in plants is also observed upon salt exposure (Figure 1) (Yang and Guo, 2018a). Electrons leaked from the electron transport chain (ETC) can react with O2 during aerobic metabolism to produce ROS (Møller, 2001). The toxic level of ROS seriously disrupts normal cellular metabolism through the oxidation of macromolecules like lipids, proteins, and nucleic acids, resulting in severe oxidative damage (Apel and Hirt, 2004; Miller et al., 2010).
In general, there are two plant types regarding their distinct salt tolerance. Some plants, termed halophytes, naturally grow in or even adapt to the saline environment with more than 200 mM NaCl (Duarte et al., 2014). By contrast, most other plant species are salt-sensitive glycophytes, and their growth and development are adversely inhibited by soil salinization (Assaha et al., 2017). Due to their sessile lifestyle, plants have evolved with sophisticated but effective strategies with considerable plasticity in morphology, physiology and metabolism to deal with multiple environmental stimuli (Genoud and Métraux, 1999; Fraire-Velázquez et al., 2011). Therefore, a series of signaling pathways have been established in plants in response to salt stress, including the sensory mechanisms, the networks that mediate osmotic adjustment, redox balance and ionic homeostasis, and other stress response mechanisms (Figure 1) (Zhao et al., 2021). In this review, we briefly summarize the new findings of salt stress responses in plants, focusing on recent advances in salt-induced signal perception and transduction. Phytohormone and light signal are essential for plant development and stress response, and some of their regulatory elements in have been also found to play essential roles in coordinating salt stress response in plants (van Gelderen et al., 2018; Kovacs et al., 2019; Waadt et al., 2022). Here we also summarize the mechanisms of phytohormone and light signal-mediated salt tolerance of plants. Finally, we discuss the possible roles of plant microbiota in plant salt tolerance. Understanding the mechanisms of plant salt tolerance provides new opportunities for engineering salt-tolerant plants even grown in saline regions, making it a promising way to keep agricultural productivity and ecological security worldwide.
Plants might sense the alteration of salt concentration in the extracellular space and the change of mechanical effects on cellular structures caused by salt stress, triggering a series of signal transduction processes in plants (Figure 2). The initial salt stress signals mainly include excess apoplastic Na+, cytosolic Ca2+ ([Ca2+]cyt) level increase, ROS accumulation, and 3,5-cyclic guanosine monophosphate (cGMP) production (Shabala et al., 2015; Park et al., 2016a). Under saline condition, excess Na+ enters plant roots through nonselective cation channels (NSCCs), which mediate toxic sodium (Na+) influx into the cell across the plasma membrane (PM) (Demidchik and Tester, 2002; Demidchik and Maathuis, 2007). Cyclic nucleotide-gated channels (CNGCs) are the main NSCCs, which are implicated in ionic homeostasis during salt response. The initial salt-induced signals might contribute to regulating NSCCs in plants during salt response. NSCCs can be blocked by Ca2+ (Leng et al., 2002; Demidchik and Maathuis, 2007; Demidchik et al., 2018). The apoplastic Ca2+ concentration in root cells is probably in the region of 0.2-0.4 mM (Legué et al., 1997), which is enough to reduce NSCCs-mediated flux by 30-50% (Essah et al., 2003). The remaining flux can be further diminished by channel blockers like Gd3+ and La3+ (Demidchik and Maathuis, 2007). Other channels and transporters may also contribute to salt stress perception, but their regulatory role in sodium sensing and import in planta is debated (van Zelm et al., 2020; Wang et al., 2022).
Calcium signal functions as an essential secondary messenger, and salt stress leads to rapid and transient [Ca2+]cyt elevations to trigger salt tolerance responses (Knight et al., 1991; Knight et al., 1997). During initial salt response, calcium signal plays a vital role in osmosensing within seconds of exposure to osmotic stress caused by the saline conditions. REDUCED HYPEROSMOLALITY- INDUCED [Ca2+]cyt INCREASE1 (OSCA1) was initially identified as a hyperosmolality-activated Ca2+-permeable cation channel responsible for [Ca2+]cyt increase, might be a potential osmosensor in plants (Yuan et al., 2014). A Ca2+-responsive phospholipid-binding BONZAI (BON) protein was recently reported to mediate hyperosmotic stress tolerance by positively regulating osmotic stress-induced [Ca2+]cyt increase, ABA accumulation, and gene expression, indicating a possible role of the membrane-associated Ca2+-responsive BON proteins in osmotic sensing and signaling (Chen et al., 2020). Considering that salt stress triggers osmotic stress, it is possible that the aforementioned Ca2+ transporters and Ca2+-responsive BON protein might participate in initial salt sensing and signaling by mediating Ca2+ signal in plants. Recently, the Arabidopsis thaliana (Arabidopsis) PM glycosyl inositol phosphorylceramide (GIPC) sphingolipids was found to function as a sensor to sense Na+ level and regulate salt stress response by gating ionic stress-induced Ca2+ signaling (Jiang et al., 2019). GIPC sphingolipid biosynthesis is catalyzed by the protein of monocation-induced Ca2+ increases1 (MOCA1), and extracellular Na+ ions can bind GIPC sphingolipids to gate PM Ca2+ influx channels. Consistently, the moca1 mutant is hypersensitive to salt stress and lacks cation-evoked Ca2+ spikes and waves (Jiang et al., 2019).
High salinity stress rapidly triggers H2O2 bursts in plant cells, which might function as essential salt stress signal (Wang et al., 2020). HYDROGEN-PEROXIDE-INDUCED CA2+ INCREASES1 (HPCA1) functions as a H2O2 sensor that perceives the stress-induced extracellular H2O2 burst and generates increased [Ca2+]cyt under stress stimuli (Wu et al., 2020). Mathematical modeling showed that under salt stress, crosstalk between ROS and Ca2+ signaling is necessary to spread the Ca2+ signal between cells (Evans et al., 2016). Accordingly, the atrbohd atrbohf double mutant, which is hypersensitive to salt stress, exhibited a reduction in cytosolic free Ca2+ and PM Ca2+ influx (Ma et al., 2012). Importantly, Jiang et al., showed that lack of AtrbohF causes hypersensitivity of shoots to soil salinity with high Na+ accumulation in root vasculature cells and xylem sap (Jiang et al., 2012). AtrbohF-mediated ROS production in root vasculature contributes to Na+ concentration limitation, thus protecting shoot cells from transpiration-dependent excess Na+ delivery (Jiang et al., 2012). Moreover, the Ca2+ signaling complex CBL1/9-CIPK26 interacts with and phosphorylates AtRbohF (Drerup et al., 2013). Thus, in the early stages of salt stress, ROS and Ca2+ signals work together to affect ionic homeostasis in plants.
The rise of cellular cGMP can be detected within seconds after applying salinity and osmotic stress (Donaldson et al., 2004). Furthermore, cGMP inhibits Na+ influx in several plant species (Maathuis and Sanders, 2001; Essah et al., 2003; Rubio et al., 2003), while it can regulate transcription of various genes related to salinity stress and promote K+ uptake (Maathuis, 2006; 2014; Isner and Maathuis, 2016). Importantly, cGMP has a negative effect on the inward Na+ flux acrried by NSCCs (Maathuis and Sanders, 2001).
Peptide ligands have also emerged as essential mediators of cell-to-cell communication during plant growth and stress responses, possibly through the action of their PM-localized receptors, RECEPTOR-LIKE KINASEs (RLKs) (Gancheva et al., 2019; Xie et al., 2022). For instance, the defense-related peptide PLANT ELICITOR PEPTIDE AtPep3 and its receptor PEP1 RECEPTOR 1 (PEPR1) are implicated in plant salt response, and PEPR1 loss of function nearly abolishes AtPep3-induced salt resistance in Arabidopsis (Nakaminami et al., 2018). Recently, Zhou et al., reported that the RECEPTOR-LIKE KINASE 7 (RLK7), another PM receptor kinase in Arabidopsis, can be recognized by a secreted peptide, PAMP-INDUCED SECRETED PEPTIDE 3 (PIP3), and form an active ligand-receptor signaling cascade that modulates plant salt tolerance in Arabidopsis by activating MPK3/MPK6 cascade (Zhou et al., 2022). Furthermore, LEUCINE-RICH REPEAT EXTENSINs, RAPID ALKALINIZATION FACTOR peptides and FERONIA (FER), a member from the Catharanthus roseus RLK1-like (CrRLK1L) family in Arabidopsis, may form a functional module that connects salt stress-induced cell-wall alterations to salt stress responses (Zhao et al., 2018). In addition, FER-dependent signaling may elicit a cell-specific Ca2+ signal to maintain cell wall integrity during salt stress and root growth recovery after salt exposure (Feng et al., 2018). Although some peptides are implicated in plant salt response, upregulation of their coding genes and the maturation of these peptides during salt stress response might depend on the initial signals such as Ca2+.
The immediate problem plants face is dehydration due to osmotic stress caused by high salinity, and plants can initiate osmotic adjustment to maintain cell volume and turgor under salt stress (Figures 1, 2) (van Zelm et al., 2020). The osmotic adjustment is a process by which plants enhance their water availability by synthesizing compatible solutes, known as osmolytes, in their cells. The osmolytes mainly include organic substances and inorganic ions, and proline, glycine betaine, and soluble carbohydrates, have been proven effectively regulate osmotic pressure by lowering the osmotic potential in the cytosolic compartment, thus preventing cellular dehydration during salt stress (Yang and Guo, 2018a; Jogawat, 2019). Due to its strong hydration ability, proline accumulation is an effective strategy to prevent protein dehydration and denaturation under osmotic stress in plants (Liang et al., 2013). Data showed that treatment with proline to two contrasting cultivars of Brassica juncea could alleviate the adverse effects of salinity on photosynthesis and seed yield (Wani et al., 2019). Furthermore, proline can function as an O2 quencher, thus removing excessive ROS produced under stress (Rejeb et al., 2014; Rehman et al., 2021). Glycine betaine, a water-soluble amphoteric quaternary ammonium compound, is also an essential osmotic regulator in higher plants, playing a vital role in stress alleviation (Ashraf and Foolad, 2007). Exogenous application of glycine betaine can mitigate salt-induced damage in maize seedlings (Hossain et al., 2021; Bai et al., 2022). Non-structural carbohydrates, such as glucose, sucrose, fructan, and starch, are also found to accumulate in plants under salt stress and function in osmotic adjustment, therefore enhancing the salt tolerance of plants (Munns and Tester, 2008; Li and Sheen, 2016; Wang et al., 2021). Significantly, carbohydrates also act as sugar signaling in plant response to an environmental stimulus (Bhattacharya and Kundu, 2020), making it another layer of regulation in plant salt tolerance. Apart from organic solutes, inorganic ions have been demonstrated as significant contributors to osmotic adjustment during saline conditions. Sodium and chloride are toxic to plants, but salt-tolerant plants might take them as essential osmolytes to maintain the external osmolarity to avoid osmotic imbalance and sustain growth (Munns and Tester, 2008; Tanveer and Shah, 2017; Hussain et al., 2021). Plants enhance their environmental adaptability by accumulating inorganic ions like K+ under salt stress. It has been revealed that K+ is an influential contributor to osmotic regulation (Wang et al., 2013; Kumari et al., 2021), making it a crucial element for plant growth and stress response.
ROS in plant cells mainly includes oxygen radicals, like superoxide (*O2-), hydroxyl radical (*OH), and some non-radicals, such as hydrogen peroxide (H2O2), singlet oxygen (1O2), and ozone (O3) (Gill and Tuteja, 2010). The generation sites of ROS in salt-stressed plants mainly include chloroplasts, mitochondria, apoplast, and peroxisomes (Sharma et al., 2012). ROS might damage cells at excess levels while act as crucial signaling molecules essential for stress signaling at lower concentrations (Figure 2) (D'Autréaux and Toledano, 2007; Sewelam et al., 2016). The interaction between ROS and ethylene (ET) has been identified as the primary signal that mediates salinity stress in rice (Steffens, 2014). Excessive accumulation of ROS induced by salt stress is toxic to plants and causes oxidative damage to cellular constituents, leading to cell death (Ahanger et al., 2017; Ye et al., 2021). A high level of ROS (especially H2O2) leads to DNA damage and distorts genomic stability (Figure 1) (Lin et al., 2020). To detoxify ROS generated by salt stress, plants have evolved a set of antioxidant strategies, mainly including enzymatic (Racchi, 2013) and non-enzymatic (Khazaei and Aghaz, 2017) systems to protect cells from oxidative damage (Figure 1). Enzymatic systems include a set of superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and ascorbate peroxidase (APX). Numerous studies have shown that salt-tolerant species improve their salt tolerance by enhancing the antioxidant defense system under salt stress (Souana et al., 2020; Challabathula et al., 2022). Non-enzymatic antioxidants, such as glutathione (GSH), ascorbate (AsA), monodehydroascorbate reductase (MDHAR), and dehydroascorbate reductase (DHAR), can effectively scavenge some highly toxic ROS in plants (Maurya, 2020). Data showed that GSH and AsA levels increase with an elevated level of *O2- and H2O2 in Camellia sinensis (L.) exposed to 300 mM NaCl stress (Li et al., 2019). Enhanced activities of MDHAR and DHAR enzymes were reported to be induced by salt stress, thus decreasing cell membrane damage in sugar beet M14 (Li et al., 2020a). Consequently, plant antioxidant defense is proficient in maintaining ROS balance for salt tolerance. Importantly, as the interaction between O2 and reduced ETC components leads to ROS production, one effective strategy to reduce toxic ROS level is to prevent ETC over-reduction (Maxwell et al., 1999). The mitochondrial alternative oxidases (AOXs) participate in electron overflow when the cytochrome ETC chain is saturated with electrons due to impaired electron transport under challenging conditions, preventing further reduction of ubiquinone and stabilizing the whole ETC (Millenaar et al., 1998; Vanlerberghe, 2013). Our recent work demonstrates that SIZ1-mediated SUMOylation of R2R3-MYB transcription factor MYB30 modulates plant salt tolerance through the action of AOX1a. MYB30 binds the promoter of AOX1a and upregulates its expression in response to salt stress to maintain the cellular redox homeostasis through enhanced alternative respiration pathway (Gong et al., 2020).
Under salinity stress, excess toxic sodium ions (Na+) enter and accumulate in plant cells, disrupting ion homeostasis, especially Na+/K+ balance (Figures 1, 2) (Munns and Tester, 2008; Zhao et al., 2021). Keeping ionic homeostasis is a prerequisite for plant growth during salt stress, since disordered ionic homeostasis leads to the disruption of cellular metabolism (Zhu, 2003; Amin et al., 2021). Plants have developed sophisticated and effective mechanisms to keep optimal levels of Na+ by removing or vacuolar compartmentalizing Na+ from the cytoplasm, and a variety of carrier and channel proteins, symporters, and antiporters participate in this process (Figures 1, 2) (Tester and Davenport, 2003; Munns and Tester, 2008; Wu, 2018). On the other hand, K+ retention in the cytosol is esstisal for Na+/K+ balance during salt response (Yang et al., 2014; Yang and Guo, 2018a). The Na+/H+ antiporters that transport Na+ in exchange for H+ achieve the regulation of Na+ levels in the cytoplasm in plant cells. The PM-localized Na+/H+ antiporters can transport Na+ to the apoplast, and the vacuole-localized Na+/H+ antiporters are responsible for vacuolar sequestration of Na+ (Qiu et al., 2004; Keisham et al., 2018; Akhter et al., 2022). The increased accumulation of Na+ in vacuoles might also act as an osmoticum, enhancing salt tolerance ability (Solis et al., 2021). NHXs are putative Na+/H+ exchangers that transport Na+ from the cytoplasm to the vacuole, holding plant resistance to salt stress (Yokoi et al., 2002; Su et al., 2020). Several reports showed that overexpression of NHX confers salinity tolerance in many plant species. For example, constitutive overexpression of AtNHX1 significantly increases salt tolerance in rice (Fukuda et al., 2004), wheat (Xue et al., 2004), tomato (Zhang and Blumwald, 2001), and cotton (He et al., 2005). Taken together, Na+ exclusion, vacuolar Na+ sequestration, and K+ retention in the cytosol are essential for plant salt tolerance (Figure 2).
The Salt Overly Sensitive (SOS) regulatory pathway plays a pivotal role in regulating ionic homeostasis through modulating the activity of Na+/H+ antiporters under salt stress (Figure 2) (Yang et al., 2009; Ji et al., 2013). The SOS pathway effectively maintains the Na+ homeostasis by transporting excess Na+ from the cytosol to the apoplast, thus preventing the accumulation of Na+ to toxic levels (Halfter et al., 2000; Yang et al., 2009; Quintero et al., 2011). The SOS signaling pathway includes Salt Overly Sensitive-1 (SOS1), a PM Na+/H+ antiporter, the serine/threonine protein kinase SOS2, and two calcium sensors, SOS3 and SCaBP8/CBL10 (SOS3-like calcium-binding protein 8) (Yang and Guo, 2018a). When grown in normal condition, SOS pathway is “off” via the action of 14-3-3 proteins and GIGANTEA (GI) which interact with SOS2 and repress its kinase activity (Kim et al., 2013; Zhou et al., 2014). High salinity initiates a calcium signal that activates the SOS pathway (Figure 2). Under salt stress, 14-3-3 proteins are released from SOS2 and degraded through proteasomal pathways (Tan et al., 2016); SOS3/SCaBP8 protein perceives the increased [Ca2+]cyt, recruits SOS2 to the PM, and activates its activity (Halfter et al., 2000; Quan et al., 2007; Lin et al., 2009). Subsequently, the activated SOS2 phosphorylates SOS1, thus enhancing the transport activity of SOS1 and transporting Na+ from cytosol to apoplast (Quintero et al., 2011). In addition, SOS3/SCaBP8-SOS2 module might also positively regulate the activities of other transporters involved in ionic homeostasis like vacuolar Na+/H+ exchanger (NHX) (Zhao et al., 2021). Remarkably, SOS1-mediated Na+ exclusion in plants during salt response is regulated by MITOGEN-ACTIVATED PROTEIN KINASE (MAPK) signaling pathways (Figure 2). For instance, MITOGEN-ACTIVATED PROTEIN KINASE3 (MPK3) and MPK6 physically interact with and phosphorylates SOS1 and salt stress-induced PA (Yu et al., 2010), and thus mediate salt response and suppresse Na+ accumulation via in shoots (Ji et al., 2013). Phosphatase MAP KINASE PHOSPHATASE1 (MKP1) can be phosphorylated by MPK6 (Park et al., 2011), and MKP1 exhibits a negative effect on MPK3/6 activity (Bartels et al., 2010; Besteiro et al., 2011). A recent study revealed that the mkp1 mutation improves salt tolerance by restraining Na+ accumulation in shoots (Uchida et al., 2022). The salt tolerance in mkp1 might be attributed to the activation of SOS1 via the elevation of MPK6.
Besides Arabidopsis thaliana, SOS genes have been identified in many other plants, such as Triticum aestivum L. (Jiang et al., 2021) and Oryza sativa (Kumar et al., 2012). Overexpression of SOS genes could improve salt tolerance by regulating ionic homeostasis (Baghour et al., 2019; Gupta et al., 2021). Importantly, it is worth noting that the entry of Na+ across the tonoplast membrane or PM is driven by the proton motive force established by proton pumps in the tonoplast or PM. Activation of H+-ATPase (Conde et al., 2011) and H+ pyrophosphatases (Maeshima, 2000) generates such proton motive force across the PM, thus activating most of the ion and metabolite transport. Vacuolar H+-ATPase (V-ATPase) is the most prevailing H+ pump in the plant cells (Dietz et al., 2001). Studies have revealed that enhancing the expression level of V-ATPase could improve salt tolerance (Zhang et al., 2012). Interestingly, both V-ATPases and PPase are also thought to be regulated by the SOS components (Batelli et al., 2007; Silva and Gerós, 2009).
Under salinity stress, excessive Na+ leads to K+ loss in plant cells (Park et al., 2016a; Zhao et al., 2021). The transporters HAK/KT/KUP play an essential role in maintaining Na+/K+ homeostasis during salt stress, which is involved in enhancing K+ absorption and reducing Na+ accumulation inside the cells (Almeida et al., 2017). In rice, OsHAK1 dominates the Na+-sensitive high affinity K+ uptake system (Chen et al., 2017). Constitutive expression of OsHAK5 in BY2 cells enhances K+ accumulation under saline condition and confers salt tolerance in these cells (Horie et al., 2011; Yang et al., 2014). OsHAK5 might mainly function in shoot tissues and its overexpression leads 43-115% increase in K+/Na+ ratio compared to WT plants in shoot but not root (Yang et al., 2014). OsHAK21 is also reported to mediate K+ absorption across the PM and play an essential role in maintaining the Na+/K+ homeostasis in rice under salt stress (Shen et al., 2015). The mutant of oshak21 accumulates less K+ and considerably more Na+ in both shoots and roots compared with the wild type. Research also suggests that Arabidopsis NADPH oxidases ARABIDOPSIS THALIANA RESPIRATORY BURST OXIDASE HOMOLOG D (AtrbohD) and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis under salinity stress possibly regulating inward K+ currents under both normal and salt stress conditions (Ma et al., 2012).
Phytohormones are small chemicals that play an essential role in plant growth and development. Evidence indicates that phytohormones mediate various stress resistance, such as salt, osmotic, drought, cold, and pathogen stress (Carvalho et al., 2013; Verma et al., 2016; Yu et al., 2020; Waadt et al., 2022). Numerous studies have shown that plant hormone signaling plays integrated and sophisticated roles at different vegetative stages, in different tissues, or under various environmental stimuli (Ku et al., 2018; Waadt et al., 2022). How plant hormones, including abscisic acid (ABA), brassinosteroid (BR), ethylene (ET), gibberellin (GA), salicylic acid (SA), and jasmonic acid (JA), mediate salinity signals to regulate plant salt stress tolerance is briefly summarized here (Figure 3).
ABA functions as an essential central integrator to activate adaptive signaling cascades during the salt stress response in plants. Under abiotic stresses, including salinity and water deficit, endogenous ABA levels increase rapidly, and enhanced ABA signaling activates sucrose nonfermenting 1-related protein kinases (SnRK2s) (Zhu, 2016). SnRK2s are the central components in ABA signaling networks and play critical roles in ion transport, osmoregulation, ROS production, gene transcription, and the closing of stomata (Yang G et al., 2019). Stomata are the primary place for plant transpiration, and ABA-regulated stomatal opening and closing are critical for plants to respond to salt stress. OST1/SnRK2.6 interacts with and phosphorylates specific ion channels, such as the potassium channel KAT1 and the slow anion channel SLAC1, to mediate K+ efflux and anion currents in guard cells, thus enhancing stomatal closure during salt and osmotic stress (Brandt et al., 2015). SnRK2.2/2.3/2.6 phosphorylate and positively control various ABA-responsive element (ABRE)-binding protein/ABRE-binding factor (AREB/ABF) transcription factors, further regulating osmotic stress response in plants (Cai et al., 2017). ABA-activated SnRK2s also regulate osmotic stress tolerance by controlling the BAM1- and AMY3-dependent degradation of starch into sugar and sugar-derived osmolytes (Thalmann et al., 2016). It is deserved to determine whether similar mechanisms are involved in the osmotic regulation during salt stress in plants.
Salt stress leads to the increase of [Ca2+]cyt (Yang Y et al., 2019). Ca2+ signaling plays a real-time and influential role in response to salinity stress. ABA effectively helps plants survive salt stress by integrating with the versatile second messenger Ca2+ via provoking PM-bound channels or releasing Ca2+ from intracellular Ca2+ pools (Edel and Kudla, 2016). The damage to the cell wall caused by Na+ can be perceived by the kinase FER, which mediates salt stress signaling by increasing [Ca2+]cyt. In contrast, ABA flexibly controls FER activity through the dephosphorylation of ABA INSENSITIVE 2 (ABI2) (Chen et al., 2016). In addition, ABA-activated SnRKs can phosphorylate the membrane-bound NADPH oxidase AtrbohF, modulating ROS homeostasis in plant response to high salinity (Szymańska et al., 2019). A recent study has demonstrated that salt-induced ABA and Ca2+ signaling can fine-tune AtrbohF activity by activating SnRK2.6 and CIPK11/26 signaling modules (Han et al., 2019). Together, ABA, Ca2+ and ROS exhibit complicated signaling crosstalk to control plant resistance to salt stress.
BRs are a class of steroid phytohormones in plants and play pivotal roles in plant growth, development, and response to adverse stresses (Ahanger et al., 2018; Nolan et al., 2020). BRs have been widely reported to improve salt stress tolerance in a range of plants, including Arabidopsis, rice (Oryza sativa), tomato (Lycopersicon esculentum), and mustard (Brassica napus) (Özdemir et al., 2004; Wani et al., 2019; Jia et al., 2021). BR can significantly inhibit ROS generation by enhancing antioxidant capacity under saline conditions (Fariduddin et al., 2013; Li S. et al., 2020). Exogenous treatment with 24-epibrassinolide (EBL), an active by-product from brassinolide biosynthesis, effectively improves salt tolerance in soybean through regulating enzymatic antioxidants and osmolyte accumulation (Soliman et al., 2020). The exogenous application of BR has been reported to improve photosynthetic efficiency in different plant species. In a recent study, EBL application could alleviate the detrimental effects of salt stress on chloroplasts and photosynthesis in Robinia pseudoacacia L. seedlings (Yue et al., 2019). Moreover, exogenous BR application could also relieve salt toxicity by regulating the activity of Na+/H+ antiporters and NHX (Su et al., 2020). These results highlight the potential role of BR in plant salt resistance.
BR-induced enhanced tolerance to salinity is closely associated with BR signaling. When extracellular BR hormones directly bind to one of its membrane-localized receptors, BRASSINOSTEROID-INSENSITIVE 1 (BRI1), BRI1-LIKE 1 (BRL1) or BRL3, and the coreceptor BAK1(SERK3) in Arabidopsis, an efficient phosphorylation cascade, to relay BR signals to BRI1-EMS-SUPPRESSOR1 (BES1) and BRASSINAZOLE-RESISTANT1 (BZR1) family TFs, therefore controlling BR-regulated gene expression (He et al., 2000; Nolan et al., 2020). Salt stress leads to root growth inhibition in plants due to a reduced level of BZR1 in the nucleus and the repression of BR signaling (Srivastava et al., 2020). However, exogenous BR application can even partially enhance salt-induced growth inhibition (Liu et al., 2014; Guedes et al., 2021). Studies indicate that overexpression of vascular BR receptor BRL3 promotes the accumulation of osmoprotectant metabolites, including proline and sugars, which play essential roles in osmoregulation under salt stress (Fàbregas et al., 2018). Overexpression of SERK2, an interacting partner of BR receptor in rice, significantly enhances grain size and salt stress resistance (Dong et al., 2020). The accumulation of SERK2 induced by salt stress confers early BR signaling on the PM to enhance the salt stress response. SlBZR1, a BZR/BES TF in tomatoes, positively regulates BR signaling and salt stress tolerance in tomatoes and Arabidopsis (Jia et al., 2021). BIN2 is a critical negative component of BR signaling, which also acts as an essential molecular switch to balance plant growth recovery and salt stress response in Arabidopsis; however, BR signaling might not be implicated in BIN2-SOS2 module during salt response and growth recovery regulation (Li et al., 2020b). Importantly, multilayer crosstalks between BR and ABA have been observed. BIN2 activates ABA signaling through the phosphorylation of SnRK2.2 and SnRK2.3 (Cai et al., 2014). In contrast, phosphatases ABI1 and ABI2, two major negative players in ABA signaling, can mediate the dephosphorylation of BIN2, abolishing the activity of BIN2 and enhancing the transduction of the BR signaling pathway (Wang et al., 2018). It is possible that BR and ABA together control plant growth and salt stress response
ET and GA have been also reported to be involved in plant salt stress response. Plants rapidly generate gaseous ET under salt stress (Zhang et al., 2016). Increased endogenous ET or treatment with ACC, an ET precursor, both can enhance plant salt tolerance (Tao et al., 2015; Gharbi et al., 2017). ET promotes plant salt tolerance by maintaining the homeostasis of Na+/K+ and reducing ROS by inducing antioxidant defense (Yang et al., 2013; Wang et al., 2020). ET signaling is essential in plant salt tolerance. Loss of function of ET receptor ETHYLENE RESPONSE 1(ETR1) and ETHYLENE INSENSITIVE 4 (EIN4) or CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1), a negative regulator of ET signaling, confers enhancement of salt tolerance (Wilson et al., 2014; Dubois et al., 2018), while loss-of-function mutants of ETHYLENE INSENSITIVE 3 (EIN3) and EIN3-LIKE 1 (EIL1), two ET-activated TFs, exhibited higher salt sensitivity in contrast to wild-type plants (Peng et al., 2014). As a well-known regulator of seed germination, GA positively regulates plant growth (Sun and Gubler, 2004). By contrast, reduced bioactive GA levels or signaling after germination is required for plant tolerance to salt stress (Magome et al., 2004; Magome et al., 2008). Consistently, the GA-deficient mutant ga1-3 exhibits remarkable tolerance to salt stress (Magome et al., 2004); the growth of seedlings lacking GA signaling repressors GAI, RGA, RGL1, and RGL2 is less inhibited by salt stress compared with the corresponding wild-type plants (Achard et al., 2006; Achard et al., 2008). Moreover, overexpression of the GA catabolic gene CYP71D8L improves rice tolerance to salinity stress by affecting GA homeostasis (Zhou et al., 2020). The phytohormone SA is also repeatedly reported to take part in salt tolerance. For example, the exogenous application of SA together with nitric oxide (NO) significantly alleviates the NaCl-mediated oxidative damage in Vigna angularis by enhancing the synthesis of osmotic substances and improving photosynthesis (Ahanger et al., 2019). A recent study showed that priming the seed germination of Leymus chinensis in SA solution relieves salt-induced osmotic damage by accumulating K+ (Hongna et al., 2021). It might be due to the improved ATP content and H+-ATPase activity in the membrane of root cells (Ghassemi-Golezani and Farhangi-Abriz, 2018). In addition, SA might also crosstalk with other hormones, such as ABA, ET, and GA, which are closely correlated with the activation of osmotic adjustment and maintenance of ionic homeostasis (Khan et al., 2014; Jayakannan et al., 2015).
As a stress-related hormone, JA has been also found to be involved in salt-induced growth inhibition (Valenzuela et al., 2016). Salt stress induces the expression of JA biosynthesis-related genes in leaves and roots, leading to increased JA production (Du et al., 2013; Delgado et al., 2021). Exogenous application of JA significantly alleviates salt-induced damage by increasing the antioxidative enzyme activities and maintaining Na+/K+ balance (Qiu et al., 2014; Gao et al., 2021). JA signaling plays an essential role in plant salt tolerance. The crucial component activating JA signaling, MYC2, is implicated in salt-mediated JA-dependent inhibition of cell elongation in the elongation zone of Arabidopsis primary roots (Valenzuela et al., 2016; Verma et al., 2020). Additionally, MYC2 contributes to salt tolerance by regulating the proline biosynthesis gene in Arabidopsis (Verma et al., 2020). Jasmonate ZIM-domain (JAZ) proteins are the core components of the JA signaling pathway, and their roles in plant salt stress response have been characterized in many species. A recent study has shown that GaJAZ1 interacts with GaMYC2 to inhibit the expression of downstream genes, increasing salt tolerance in Gossypium hirsutum (Zhao et al., 2020). On the other hand, overexpression of CYP94C2b, a cytochrome P450 family protein involving JA catabolism, enhanced viability under salt conditions and delayed the salt stress-induced leaf senescence in rice (Kurotani et al., 2015).
The light signaling networks in plants begin with the perception of light signals, ultimately leading to changes in plant development and stress response (van Gelderen et al., 2018). Emerging evidence shows that light signaling is vital in shaping plant salt stress response (Figure 3) (Carvalho et al., 2011; Kovacs et al., 2019). For example, light signaling can affect salt stress-induced transcriptional memory response of P5CS1-mediated proline accumulation in Arabidopsis (Feng et al., 2016). In addition, PHYTOCHROME-INTERACTING FACTOR 4 (PIF4), a negative regulator of the phytochrome signaling pathway, negatively regulates plant salt tolerance by downregulating the expression of stress tolerance genes (Leivar and Quail, 2011; Sakuraba et al., 2017). CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), a master of the light signaling pathway, also regulates salt stress tolerance. Salt treatment can promote the translocation of COP1 to the cytosol; the cop1 mutants exhibited a significantly impaired resistance to salt stress than the wild-type plants at the germination and seedling stages (Yu et al., 2016). A recent study further confirmed that COP1 controls plant salt stress tolerance by modulating sucrose content (Kim et al., 2022). Moreover, constitutive nuclear-localized ELONGATED HYPOCOTYL 5 (HY5), a bZIP family TF acting as a critical regulator in light signaling and seedling development (Cluis et al., 2004; Gangappa and Botto, 2016), can promote proline biosynthesis by connecting light and salt stress signals (Kovacs et al., 2019). Recently, a study on tobacco suggests that NtHY5 enhances salt stress tolerance by positively regulating light-mediated flavonoid biosynthesis (Singh D et al., 2022).
Circadian clock regulates many physiological and developmental processes in plants, and its phase and period are adjusted by light, temperature, and nutrient input (Greenham and McClung, 2015; Greenwood and Locke, 2020). It has been found that salt tolerance is also regulated by the circadian clock via modulating the expression of salt-responsive genes like RD29A and SOS1 (Park et al., 2016b). The protein abundance of PM Na+/H+ antiporter SOS1 appears to occur in a diurnal cycle (Park et al., 2016b). Interestingly, a recent work revealed that SOS1 specifically functions as a salt-specific circadian clock regulator via GI in Arabidopsis. SOS1 directly interacts with GI in a salt-dependent manner and stabilizes this protein to sustain a proper clock period under saline conditions for the homeostasis of the salt response under high or daily fluctuating salt levels (Figure 3) (Cha et al., 2022). The regulatory role of light signals in plant salt tolerance needs to be clarified next.
Plants host a diverse community of microorganisms on and inside organs such as roots and leaves, collectively termed the plant microbiota (Vandenkoornhuyse et al., 2015; Dastogeer et al., 2020). Accumulating evidence indicates that plant microbiota plays a vital role in plant adaptation and resistance to saline soil (Ha-Tran et al., 2021; Chialva et al., 2022). Notably, multiple groups of root-associated microbes, including plant-growth-promoting rhizobacteria (PGPR) and endophytic bacteria, are essential to improve plant tolerance to high salinity (Figure 3) (Qin et al., 2016; Vives-Peris et al., 2018). PGPR alleviates the toxicity of salt stress on plants mainly by regulating ionic homeostasis, accumulating osmolytes, activating antioxidant capacity, and enhancing essential nutrient uptake (Santos et al., 2018; Ha-Tran et al., 2021; Shabaan et al., 2022). For instance, a recent study suggested that the bacterial strain E. cloacae PM23 mediated salt tolerance in maize by modulating plant physiology, antioxidant defense, and compatible solute accumulation (Ali et al., 2022).
Furthermore, PGPR can produce ACC deaminase, which reduces the excessive ET production in plants caused by salt stress. Plants with reduced ET level would finally cope with salt-induced growth inhibition by associating with ACC deaminase-producing microbes (Glick et al., 2007; Barnawal et al., 2014; Misra and Chauhan, 2020). Studies also indicate that PGPR can improve plant salt tolerance by producing a wide range of phytohormones as signal molecules in the rhizospheric region (Khan et al., 2020; Jalmi and Sinha, 2022). For instance, it has been revealed that an Algerian Sahara PGPR named strain Pp20 confers maize root tolerance to salt stress via producing and secreting plant growth-promoting hormone indole-3-acetic acid (IAA) and ACC deaminase (Zerrouk et al., 2019). In addition, the endophytic bacteria penetrating into the plant root cells possess similar functions in improving salt tolerance compared with PGPR (Sgroy et al., 2009; Yaish et al., 2015). For example, ACC deaminase-containing endophytic bacteria can ameliorate salt stress in Pisum sativum through reduced oxidative damage and induction of antioxidative defense systems (Sofy et al., 2021). Remarkably, some root-associated fungal endophytes are also shown to improve plant salt tolerance in terms of growth, ion homeostasis, and osmoregulation (Rodriguez et al., 2009; Bouzouina et al., 2021; Moghaddam et al., 2021). Taken together, both rhizospheric and endophytic bacteria can be employed as effective and eco-friendly adjuncts to promote plant tolerance to salinity (Figure 3).
In recent years, the plant nanobiotechnology approach has shown great potential to modulate plant stress response (Hofmann et al., 2020; Li et al., 2022). Nanotechnology is the application of small-sized materials with a basic structure of 1–100 nm (Farokhzad and Langer, 2009). A variety of nanomaterials (NMs) have been reported to enhance plant salt tolerance for growth under saline condition (Almutairi, 2016; Zulfiqar and Ashraf, 2021). For instance, some metal-based nanoparticles, cerium oxide nanoparticles, silica nanoparticles, titanium dioxide nanoparticles, and zinc oxide nanoparticles, can improve salt resistance in multiple plant species (Newkirk et al., 2018; Gaafar et al., 2020; Liu et al., 2021). NMs enhance plant salt tolerance mainly by improving plant photosynthesis performance, promoting ROS detoxification, and maintaining ionic homeostasis and resoring osmotic balance (Newkirk et al., 2018; Liu et al., 2021). Compared with the non-nanoparticle control, the application of cerium oxide nanoparticles significantly improved cotton salt tolerance by b maintaining cytosolic Na+/K+ ratio (Liu et al., 2021). In addition, zinc oxide nanoparticles have been shown to enhanc salt tolerance in seedlings by improving photosynthetic pigments and antioxidative systems (Singh A et al., 2022). Remarkably, exogenous application of biocompatible poly (acrylic acid)-coated cerium oxide nanoparticles can improve the production of gasious signaling molecules (i.e., NO), therefore maintaining the redox and ionic homeostasis in rice under salt stress (Zhou et al., 2021). Although the underlying mechanisms need to be furtherly elucidated, nanobiotechnology could be a promising approach to increase crop yield in saline soils by enhancing plant salt tolerance.
Over the past decades, much progress has been made in understanding how plants respond and adapt to salt stress. Plants have evolved various regulatory mechanisms to cope with the damages caused by excessive saline ions in the soil. Osmotic adjustment, redox, and ionic homeostasis regulation, and metabolic adjustment are the significant factors associated with plant salt tolerance (Figure 1). To cope with salt stress, plants have to rapidly and effectively perceive changes in Na+ levels and osmotic pressure caused by salt stress. Different sensors mediate stress-signaling sensing, which relays stress signals to secondary messengers that activate signaling cascades and downstream regulatory networks via multiple hormone-mediated signaling pathways. The mechanisms of plant salt response involve a variety of signaling components, transcription factors, and functional genes that directly mediate ionic homeostasis, osmoregulation, and antioxidation (Figures 2, 3). The phytohormone and light signals also mediate salt stress response in plants. Plant microbiota might also contribute to plant resistance. Exploring the molecular mechanisms of plant salt tolerance remains a great challenge. Many salt-responsive new genes still need to be annotated via advanced biotechnologies. The current knowledge of the salt-responsive molecular mechanisms in plants, from salt sensing and signaling to the development of adaptive tolerance mechanisms, still requires further studies. To date, the integration of multi-omics techniques and physiological phenotyping has proven to be a fast and effective method for probing the regulatory mechanism of plant salt tolerance (Song et al., 2020; Pazhamala et al., 2021). In particular, identification of upstream components regulating salt stress sensing is of paramount importance. Furthermore, the crosstalk between salt stress signaling networks and phytohormones still requires further investigation. Together, these findings provide valuable knowledge for breeding salt-tolerant crops through biotechnological approaches in the future.
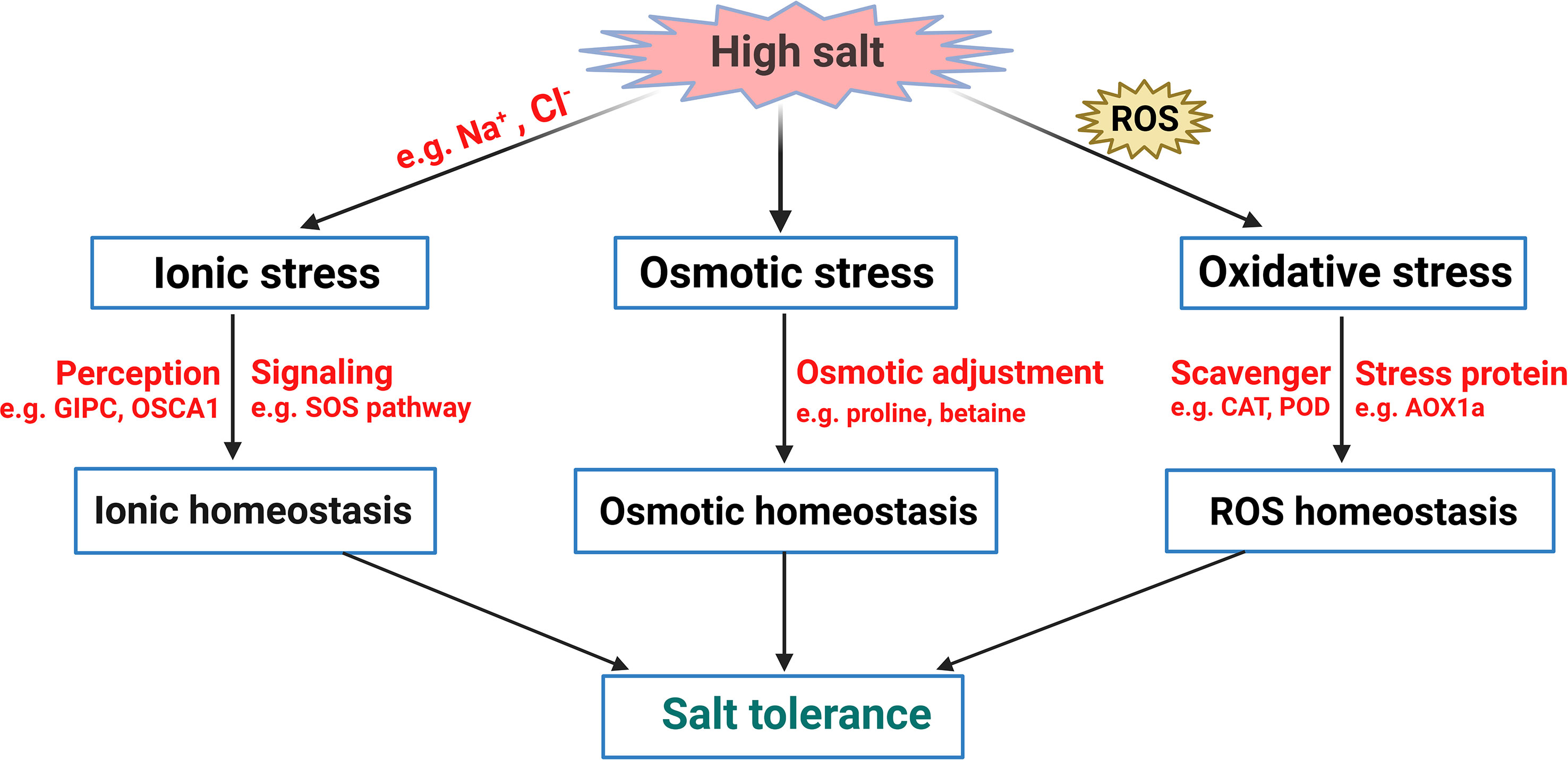
Figure 1 A simplified model of plant salt stress response. Salt stress primarily causes osmotic stress, oxidative stress, and ionic stress. By sensing such stresses, plants activate effective stress signaling networks to accumulate substances for osmotic adjustment, and to maintain ionic and redox homeostasis, leading to salt tolerance in plants.
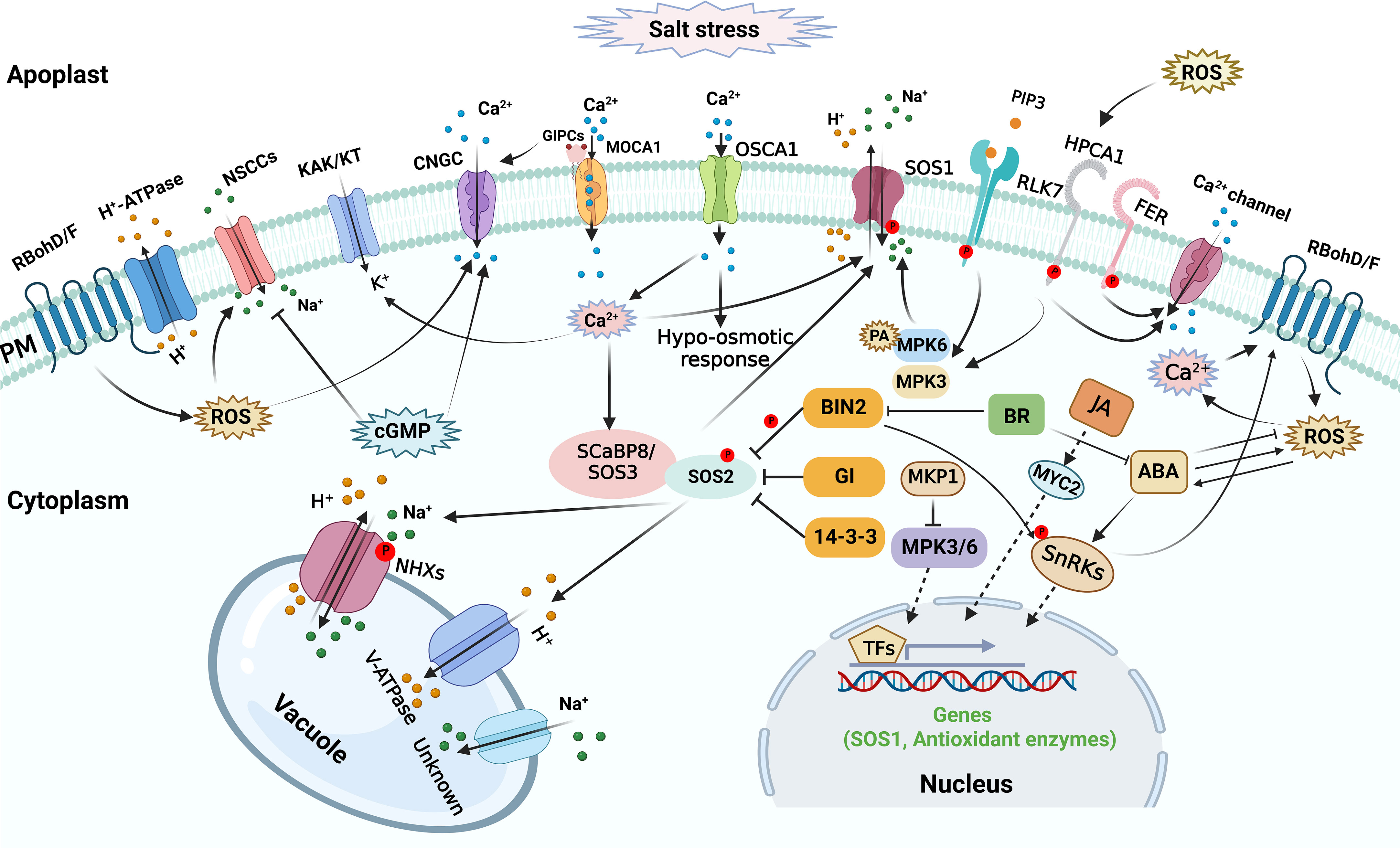
Figure 2 Salt stress sensing and signaling in plant cells. Osmotic alternation and Na+ import trigger a rise of cytosolic secondary messengers, which are sensed by specific sensors or receptors, therefore multiple signaling pathways involved in a variety of components are activated to maintain ionic balance and osmotic homeostasis or to regulate osmotic stress response. The arrows and bars indicate positive and negative regulation, whereas solid lines and dashed lines indicate direct regulation and indirect regulation, respectively.
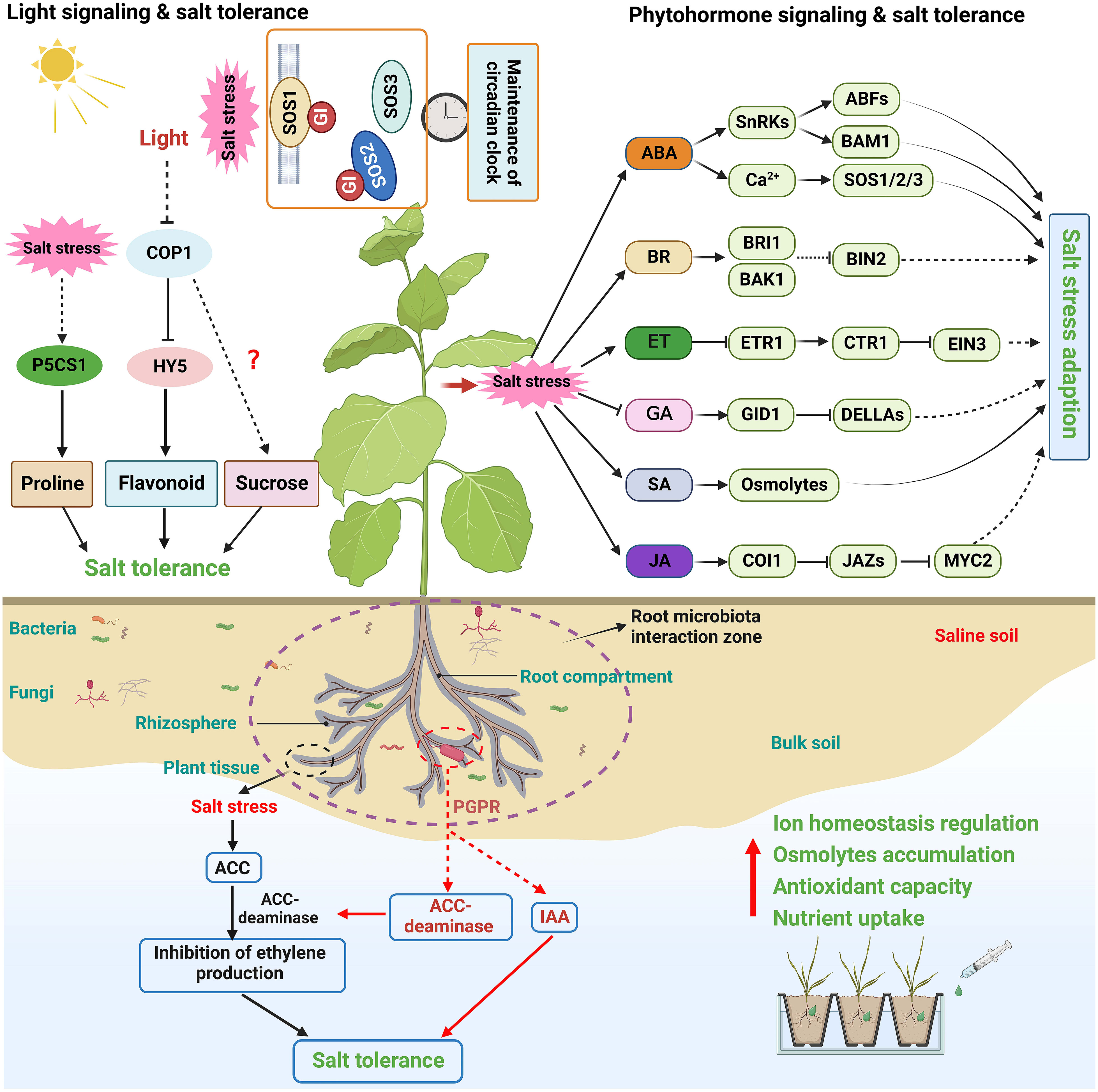
Figure 3 Schematic diagram of phytohormone, light signal, and microbiota-mediated plant salt tolerance. Phytohormone and light signal are essential for plant development and stress response, and some of their regulatory elements in have been also found to play essential roles in coordinating salt stress response in plants. Plant hormone signaling plays integrated and sophisticated roles at different vegetative stages, in different tissues, or under various environmental stimuli. Light signaling networks in plants begin with the perception of light signals, and are vital in shaping plant salt stress response. Plant microbiota plays a vital role in plant adaptation and resistance to saline soil. PGPR is essential to improve plant tolerance to high salinity, possibly by regulating ionic homeostasis, accumulating osmolytes, activating antioxidant capacity, and enhancing essential nutrient uptake.
FX and HZ wrote this manuscript and prepared the illustrations. All the authors contributed to the discussion and agreed to the published version of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (Grants 32170295 and 31870241 to HZ; Grant 32260074 to FX), the Youth Science Foundation of Xinjiang Uygur Autonomous Region (Grant 2022D01C94 to FX).
We sincerely apologize to those authors for not being able to cite their works in this review due to space limitations.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. doi: 10.1126/science.1118642
Achard, P., Renou, J.-P., Berthomé, R., Harberd, N. P., Genschik, P. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18 (9), 656–660. doi: 10.1016/j.cub.2008.04.034
Ahanger, M. A., Ashraf, M., Bajguz, A., Ahmad, P. (2018). Brassinosteroids regulate growth in plants under stressful environments and crosstalk with other potential phytohormones. J. Plant Growth Regul. 37 (4), 1007–1024. doi: 10.1007/s00344-018-9855-2
Ahanger, M. A., Aziz, U., Alsahli, A. A., Alyemeni, M. N., Ahmad, P. (2019). Influence of exogenous salicylic acid and nitric oxide on growth, photosynthesis, and ascorbate-glutathione cycle in salt stressed vigna angularis. Biomolecules 10 (1), 42. doi: 10.3390/biom10010042
Ahanger, M. A., Tomar, N. S., Tittal, M., Argal, S., Agarwal, R. (2017). Plant growth under water/salt stress: ROS production; antioxidants and significance of added potassium under such conditions. Physiol. Mol. Biol. Plants 23 (4), 731–744. doi: 10.1007/s12298-017-0462-7
Akhter, A., Bibi, G., Rasti, N., Rasheed, H., Noor, Z., Hussain, J. (2022). “Na+ sensing, transport, and plant salt tolerance,” in Sustainable plant nutrition under contaminated environments. sustainable plant nutrition in a changing world Cham: Springer, 257–285. doi: 10.1007/978-3-030-91499-8
Ali, B., Wang, X., Saleem, M. H., Hafeez, A., Afridi, M. S., Khan, S., et al. (2022). PGPR-mediated salt tolerance in maize by modulating plant physiology, antioxidant defense, compatible solutes accumulation and bio-surfactant producing genes. Plants 11 (3), 345. doi: 10.3390/plants11030345
Almeida, D. M., Oliveira, M. M., Saibo, N. J. M. (2017). Regulation of na+ and k+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 40, 326–345. doi: 10.1590/1678-4685-gmb-2016-0106
Almutairi, Z. M. (2016). Effect of nano-silicon application on the expression of salt tolerance genes in germinating tomato ('Solanum lycopersicum'L.) seedlings under salt stress. Plant Omics 9 (1), 106–114.
Amin, I., Rasool, S., Mir, M. A., Wani, W., Masoodi, K. Z., Ahmad, P. (2021). Ion homeostasis for salinity tolerance in plants: A molecular approach. Physiologia Plantarum 171 (4), 578–594. doi: 10.1111/ppl.13185
Apel, K., Hirt, H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signaling transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Ashraf, M., Foolad, M. R. (2007). Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 59 (2), 206–216. doi: 10.1016/j.envexpbot.2005.12.006
Assaha, D. V., Ueda, A., Saneoka, H., Al-Yahyai, R., Yaish, M. W. (2017). The role of na+ and k+ transporters in salt stress adaptation in glycophytes. Front. Physiol. 8, 509. doi: 10.3389/fphys.2017.00509
Baghour, M., Gálvez, F. J., Sánchez, M. E., Aranda, M. N., Venema, K., Rodríguez-Rosales, M. P. (2019). Overexpression of LeNHX2 and SlSOS2 increases salt tolerance and fruit production in double transgenic tomato plants. Plant Physiol. Biochem. 135, 77–86. doi: 10.1016/j.plaphy.2018.11.028
Bai, M., Zeng, W., Chen, F., Ji, X., Zhuang, Z., Jin, B., et al. (2022). Transcriptome expression profiles reveal response mechanisms to drought and drought-stress mitigation mechanisms by exogenous glycine betaine in maize. Biotechnol. Lett. 44 (3), 367–386. doi: 10.1007/s10529-022-03221-6
Barnawal, D., Bharti, N., Maji, D., Chanotiya, C. S., Kalra, A. (2014). ACC deaminase-containing arthrobacter protophormiae induces NaCl stress tolerance through reduced ACC oxidase activity and ethylene production resulting in improved nodulation and mycorrhization in pisum sativum. J. Plant Physiol. 171 (11), 884–894. doi: 10.1016/j.jplph.2014.03.007
Bartels, S., Besteiro, M. A. G., Lang, D., Ulm, R. (2010). Emerging functions for plant MAP kinase phosphatases. Trends Plant Sci. 15 (6), 322–329. doi: 10.1016/j.tplants.2010.04.003
Batelli, G., Verslues, P. E., Agius, F., Qiu, Q., Fujii, H., Pan, S., et al. (2007). SOS2 promotes salt tolerance in part by interacting with the vacuolar h+-ATPase and upregulating its transport activity. Mol. Cell. Biol. 27 (22), 7781–7790. doi: 10.1128/MCB.00430-07
Besteiro, G. M. A., Bartels, S., Albert, A., Ulm, R. (2011). Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-b stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J. 68 (4), 727–737. doi: 10.1111/j.1365-313X.2011.04725.x
Bhattacharya, S., Kundu, A. (2020). “Sugars and sugar polyols in overcoming environmental stresses,” in Protective chemical agents in the amelioration of plant abiotic stress: Biochemical and molecular perspectives, 71–101. doi: 10.1002/9781119552154.ch4
Bouzouina, M., Kouadria, R., Lotmani, B. (2021). Fungal endophytes alleviate salt stress in wheat in terms of growth, ion homeostasis and osmoregulation. J. Appl. Microbiol. 130 (3), 913–925. doi: 10.1111/jam.14804
Brandt, B., Munemasa, S., Wang, C., Nguyen, D., Yong, T., Yang, P. G., et al. (2015). Calcium specificity signaling mechanisms in abscisic acid signal transduction in arabidopsis guard cells. Elife 20 (4), e10328. doi: 10.7554/eLife.10328
Cai, S., Chen, G., Wang, Y., Huang, Y., Marchant, D. B., Wang, Y., et al. (2017). Evolutionary conservation of ABA signaling for stomatal closure. Plant Physiol. 174 (2), 732–747. doi: 10.1104/pp.16.01848
Cai, Z., Liu, J., Wang, H., Yang, C., Chen, Y., Li, Y., et al. (2014). GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in arabidopsis. Proc. Natl. Acad. Sci. 111 (26), 9651–9656. doi: 10.1073/pnas.1316717111
Carvalho, R. F., Campos, M. L., Azevedo, R. A. (2013). The role of phytochromes in stress tolerance. J. Integr. Plant Biol. 53 (12), 920–929. doi: 10.1007/978-1-4614-6108-1_12
Cha, J. Y., Kim, J., Jeong, S. Y., Shin, G. I., Ji, M. G., Hwang, J. W., et al. (2022). The Na+/H+ antiporter SALT OVERLY SENSITIVE 1 regulates salt compensation of circadian rhythms by stabilizing GIGANTEA in arabidopsis. Proc. Natl. Acad. Sci. 119 (33), e2207275119. doi: 10.1073/pnas.2207275119
Challabathula, D., Analin, B., Mohanan, A., Bakka, K. (2022). Differential modulation of photosynthesis, ROS and antioxidant enzyme activities in stress-sensitive and-tolerant rice cultivars during salinity and drought upon restriction of COX and AOX pathways of mitochondrial oxidative electron transport. J. Plant Physiol. 268, 153583. doi: 10.1016/j.jplph.2021.153583
Chen, K., Gao, J., Sun, S., Zhang, Z., Yu, B., Li, J., et al. (2020). BONZAI proteins control global osmotic stress responses in plants. Curr. Biol. 30 (24), 4815–4825.e4. doi: 10.1016/j.cub.2020.09.016
Chen, G., Liu, C., Gao, Z., Zhang, Y., Jiang, H., Zhu, L., et al. (2017). OsHAK1, a high-affinity potassium transporter, positively regulates responses to drought stress in rice. Front. Plant Sci. 1 (8), 1885. doi: 10.3389/fpls.2017.01885
Chen, J., Yu, F., Liu, Y., Du, C., Li, X., Zhu, S., et al. (2016). FERONIA interacts with ABI2-type phosphatases to facilitate signaling crosstalk between abscisic acid and RALF peptide in arabidopsis. Proc. Natl. Acad. Sci. 113 (37), E5519–E5527. doi: 10.1073/pnas.1608449113
Chialva, M., Lanfranco, L., Bonfante, P. (2022). The plant microbiota: composition, functions, and engineering. Curr. Opin. Biotechnol. 73, 135–142. doi: 10.1016/j.copbio.2021.07.003
Cluis, C. P., Mouchel, C. F., Hardtke, C. S. (2004). The arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 38 (2), 332–347. doi: 10.1111/j.1365-313X.2004.02052.x
Conde, A., Chaves, M. M., Gerós, H. (2011). Membrane transport, sensing and signaling in plant adaptation to environmental stress. Plant Cell Physiol. 52 (9), 1583–1602. doi: 10.1093/pcp/pcr107
D'Autréaux, B., Toledano, M. B. (2007). ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8 (10), 813–824. doi: 10.1038/nrm2256
Dastogeer, K. M., Tumpa, F. H., Sultana, A., Akter, M. A., Chakraborty, A. (2020). Plant microbiome–an account of the factors that shape community composition and diversity. Curr. Plant Biol. 23, 100161. doi: 10.1016/j.cpb.2020.100161
Delgado, C., Mora-Poblete, F., Ahmar, S., Chen, J. T., Figueroa, C. R. (2021). Jasmonates and plant salt stress: Molecular players, physiological effects, and improving tolerance by using genome-associated tools. Int. J. Mol. Sci. 22 (6), 3082. doi: 10.3390/ijms22063082
Demidchik, V., Maathuis, F. J. (2007). Physiological roles of nonselective cation channels in plants: From salt stress to signalling and development. New Phytol. 175 (3), 387–404. doi: 10.1111/j.1469-8137.2007.02128.x
Demidchik, V., Shabala, S., Isayenkov, S., Cuin, T. A., Pottosin, I. (2018). Calcium transport across plant membranes: Mechanisms and functions. New Phytol. 220 (1), 49–69. doi: 10.1111/nph.15266
Demidchik, V., Tester, M. (2002). Sodium fluxes through nonselective cation channels in the plasma membrane of protoplasts from arabidopsis roots. Plant Physiol. 128 (2), 379–387. doi: 10.1104/pp.010524
Dietz, K.-J., Tavakoli, N., Kluge, C., Mimura, T., Sharma, S., Harris, G., et al. (2001). Significance of the V-type ATPase for the adaptation to stressful growth conditions and its regulation on the molecular and biochemical level. J. Exp. Bot. 52 (363), 1969–1980. doi: 10.1093/jexbot/52.363.1969
Donaldson, L, Ludidi, N, Knight, MR, Gehring, C, Denby, K. (2004). Salt and osmotic stress cause rapid increases in Arabidopsis thaliana cGMP levels. FEBS letters 569 (1–3), 317–20. doi: 10.1016/j.febslet.2004.06.016
Dong, N., Yin, W., Liu, D., Zhang, X., Yu, Z., Huang, W., et al. (2020). Regulation of brassinosteroid signaling and salt resistance by SERK2 and potential utilization for crop improvement in rice. Front. Plant Sci. 11, 621859. doi: 10.3389/fpls.2020.621859
Drerup, M. M., Schlücking, K., Hashimoto, K., Manishankar, P., Steinhorst, L., Kuchitsu, K., et al. (2013). The calcineurin b-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the arabidopsis NADPH oxidase RBOHF. Mol. Plant 6 (2), 559–569. doi: 10.1093/mp/sst009
Duarte, B., Sleimi, N., Caçador, I. (2014). Biophysical and biochemical constraints imposed by salt stress: learning from halophytes. Front. Plant Sci. 5, 476. doi: 10.3389/fpls.2014.00746
Dubois, M., Van den Broeck, L., Inze, D. (2018). The pivotal role of ethylene in plant growth. Trends Plant Sci. 23 (4), 311–323. doi: 10.1016/j.tplants.2018.01.003
Du, H., Liu, H., Xiong, L. (2013). Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 4, 397. doi: 10.3389/fpls.2013.00397
Edel, K. H., Kudla, J. (2016). Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 33, 83–91. doi: 10.1016/j.pbi.2016.06.010
Essah, P. A., Davenport, R., Tester, M. (2003). Sodium influx and accumulation in arabidopsis. Plant Physiol. 133 (1), 307–318. doi: 10.1104/pp.103.022178
Evans, M. J., Choi, W.-G., Gilroy, S., Morris, R. J. (2016). A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress. Plant Physiol. 171 (3), 1771–1784. doi: 10.1104/pp.16.00215
Fàbregas, N., Lozano-Elena, F., Blasco-Escámez, D., Tohge, T., Martínez-Andújar, C., Albacete, A., et al. (2018). Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 9 (1), 1–13. doi: 10.1038/s41467-018-06861-3
Fariduddin, Q., Khalil, R. R., Mir, B. A., Yusuf, M., Ahmad, A. (2013). 24-epibrassinolide regulates photosynthesis, antioxidant enzyme activities and proline content of Cucumis sativus under salt and/or copper stress. Environ. Monit. Assess. 185 (9), 7845–7856. doi: 10.1007/s10661-013-3139-x
Farokhzad, O. C., Langer, R. (2009). Impact of nanotechnology on drug delivery. ACS Nano 3 (1), 16–20. doi: 10.1021/nn900002m
Feng, W., Kita, D., Peaucelle, A., Cartwright, H. N., Doan, V., Duan, Q., et al. (2018). The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 28 (5), 666–675.e5. doi: 10.1016/j.cub.2018.01.023
Feng, X. J., Li, J. R., Qi, S. L., Lin, Q. F., Jin, J. B., Hua, X. J. (2016). Light affects salt stress-induced transcriptional memory of P5CS1 in arabidopsis. Proc. Natl. Acad. Sci. 113 (51), E8335–E8343. doi: 10.1073/pnas.1610670114
Fraire-Velázquez, S., Rodríguez-Guerra, R., Sánchez-Calderón, L. (2011). “Abiotic and biotic stress response crosstalk in plants,” in Abiotic stress response in plants–physiological, biochemical and genetic perspectives IntechOpen, 3–26. doi: 10.5772/1762
Fukuda, A., Nakamura, A., Tagiri, A., Tanaka, H., Miyao, A., Hirochika, H., et al. (2004). Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol. 45 (2), 146–159. doi: 10.1093/pcp/pch014
Gaafar, R., Diab, R., Halawa, M., Elshanshory, A., El-Shaer, A., Hamouda, M. (2020). Role of zinc oxide nanoparticles in ameliorating salt tolerance in soybean. Egyptian J. Bot. 60 (3), 737–747. doi: 10.21608/ejbo.2020.26415.1475
Gancheva, M. S., Malovichko, Y. V., Poliushkevich, L. O., Dodueva, I. E., Lutova, L. A. (2019). Plant peptide hormones. Russian J. Plant Physiol. 66 (2), 171–189. doi: 10.1134/S1021443719010072
Gangappa, S. N., Botto, J. F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9 (10), 1353–1365. doi: 10.1016/j.molp.2016.07.002
Gao, Z., Gao, S., Li, P., Zhang, Y., Ma, B., Wang, Y. (2021). Exogenous methyl jasmonate promotes salt stress-induced growth inhibition and prioritizes defense response of nitraria tangutorum bobr. Physiologia Plantarum 172 (1), 162–175. doi: 10.1111/ppl.13314
Geng, Y., Wu, R., Wee, C. W., Xie, F., Wei, X., Chan, P. M., et al. (2013). A spatio-temporal understanding of growth regulation during the salt stress response in arabidopsis. Plant Cell 25 (6), 2132–2154. doi: 10.1105/tpc.113.112896
Genoud, T., Métraux, J.-P. (1999). Crosstalk in plant cell signaling: structure and function of the genetic network. Trends Plant Sci. 4 (12), 503–507. doi: 10.1016/S1360-1385(99)01498-3
Gharbi, E., Martínez, J.-P., Benahmed, H., Lepoint, G., Vanpee, B., Quinet, M., et al. (2017). Inhibition of ethylene synthesis reduces salt-tolerance in tomato wild relative species solanum chilense. J. Plant Physiol. 210, 24–37. doi: 10.1016/j.jplph.2016.12.001
Ghassemi-Golezani, K., Farhangi-Abriz, S. (2018). Foliar sprays of salicylic acid and jasmonic acid stimulate h+-ATPase activity of tonoplast, nutrient uptake and salt tolerance of soybean. Ecotoxicology Environ. Saf. 166, 18–25. doi: 10.1016/j.ecoenv.2018.09.059
Gill, S. S., Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48 (12), 909–930. doi: 10.1016/j.plaphy.2010.08.016
Glick, B. R., Cheng, Z., Czarny, J., Duan, J. (2007). “Promotion of plant growth by ACC deaminase-producing soil bacteria,” in New perspectives and approaches in plant growth-promoting rhizobacteria research Springer, 329–339. doi: 10.1007/s10658-007-9162-4
Gong, Q., Li, S., Zheng, Y., Duan, H., Xiao, F., Zhuang, Y., et al. (2020). SUMOylation of MYB30 enhances salt tolerance by elevating alternative respiration via transcriptionally upregulating AOX1a in arabidopsis. Plant J. 102 (6), 1157–1171. doi: 10.1111/tpj.14689
Greenham, K., McClung, C. R. (2015). Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16 (10), 598–610. doi: 10.1038/nrg3976
Greenwood, M., Locke, J. C. (2020). The circadian clock coordinates plant development through specificity at the tissue and cellular level. Curr. Opin. Plant Biol. 53, 65–72. doi: 10.1016/j.pbi.2019.09.004
Guedes, F. R. C. M., Maia, C. F., da Silva, B. R. S., Batista, B. L., Alyemeni, M. N., Ahmad, P., et al. (2021). Exogenous 24-epibrassinolide stimulates root protection, and leaf antioxidant enzymes in lead stressed rice plants: Central roles to minimize Pb content and oxidative stress. Environ. pollut. 280, 116992. doi: 10.1016/j.envpol.2021.116992
Gupta, B. K., Sahoo, K. K., Anwar, K., Nongpiur, R. C., Deshmukh, R., Pareek, A., et al. (2021). Silicon nutrition stimulates salt-overly sensitive (SOS) pathway to enhance salinity stress tolerance and yield in rice. Plant Physiol. Biochem. 166, 593–604. doi: 10.1016/j.plaphy.2021.06.010
Halfter, U., Ishitani, M., Zhu, J.-K. (2000). The arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. 97 (7), 3735–3740. doi: 10.1073/pnas.97.7.3735
Han, J. P., Köster, P., Drerup, M. M., Scholz, M., Li, S., Edel, K. H., et al. (2019). Fine-tuning of RBOHF activity is achieved by differential phosphorylation and Ca2+ binding. New Phytol. 221 (4), 1935–1949. doi: 10.1111/nph.15543
Ha-Tran, D. M., Nguyen, T. T. M., Hung, S.-H., Huang, E., Huang, C.-C. (2021). Roles of plant growth-promoting rhizobacteria (PGPR) in stimulating salinity stress defense in plants: A review. Int. J. Mol. Sci. 22 (6), 3154. doi: 10.3390/ijms22063154
He, Z., Wang, Z.-Y., Li, J., Zhu, Q., Lamb, C., Ronald, P., et al. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288 (5475), 2360–2363. doi: 10.1126/science.288.5475.2360
He, C., Yan, J., Shen, G., Fu, L., Holaday, A. S., Auld, D., et al. (2005). Expression of an arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field. Plant Cell Physiol. 46 (11), 1848–1854. doi: 10.1093/pcp/pci201
Hofmann, T., Lowry, G. V., Ghoshal, S., Tufenkji, N., Brambilla, D., Dutcher, J. R., et al. (2020). Technology readiness and overcoming barriers to sustainably implement nanotechnology-enabled plant agriculture. Nat. Food 1 (7), 416–425. doi: 10.1038/s43016-020-0110-1
Hongna, C., Leyuan, T., Junmei, S., Xiaori, H., Xianguo, C. (2021). Exogenous salicylic acid signal reveals an osmotic regulatory role in priming the seed germination of leymus chinensis under salt-alkali stress. Environ. Exp. Bot. 188, 104498. doi: 10.1016/j.envexpbot.2021.104498
Horie, T., Sugawara, M., Okada, T., Taira, K., Kaothien-Nakayama, P., Katsuhara, M., et al. (2011). Rice sodium-insensitive potassium transporter, OsHAK5, confers increased salt tolerance in tobacco BY2 cells. J. Bioscience Bioengineering 111 (3), 346–356. doi: 10.1016/j.jbiosc.2010.10.014
Hossain, A., Pramanick, B., Bhutia, K. L., Ahmad, Z., Moulick, D., Maitra, S., et al. (2021). “Emerging roles of osmoprotectant glycine betaine against salt-induced oxidative stress in plants: A major outlook of maize (Zea mays l.),” in Frontiers in plant-soil interaction (Elsevier), 567–587. doi: 10.1016/B978-0-323-90943-3.00015-8
Hussain, S., Hussain, S., Ali, B., Ren, X., Chen, X., Li, Q., et al. (2021). Recent progress in understanding salinity tolerance in plants: Story of Na+/K+ balance and beyond. Plant Physiol. Biochem. 160, 239–256. doi: 10.1016/j.plaphy.2021.01.029
Isner, J-C, Maathuis, FJ (2016). cGMP signalling in plants: from enigma to main stream. Functional Plant Biology 45 (2), 93–101. doi: 10.1071/FP16337
Jalmi, S. K., Sinha, A. K. (2022). Ambiguities of PGPR-induced plant signaling and stress management. Front. Microbiol. 13, 899563. doi: 10.3389/fmicb.2022.899563
Jayakannan, M., Bose, J., Babourina, O., Rengel, Z., Shabala, S. (2015). Salicylic acid in plant salinity stress signalling and tolerance. Plant Growth Regul. 76 (1), 25–40. doi: 10.1007/s10725-015-0028-z
Jiang, C, Belfield, EJ, Mithani, A, Visscher, A, Ragoussis, J, Mott, R, et al (2012). ROS‐mediated vascular homeostatic control of root‐to‐shoot soil Na delivery in Arabidopsis. The EMBO Journal 31 (22), 4359–4370. doi: 10.1038/emboj.2012.273
Jiang, W., Pan, R., Buitrago, S., Wu, C., Abou-Elwafa, S. F., Xu, Y., et al. (2021). Conservation and divergence of the TaSOS1 gene family in salt stress response in wheat (Triticum aestivum l.). Physiol. Mol. Biol. Plants 27 (6), 1245–1260. doi: 10.1007/s12298-021-01009-y
Jiang, Z., Zhou, X., Tao, M., Yuan, F., Liu, L., Wu, F., et al. (2019). Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572 (7769), 341–346. doi: 10.1038/s41586-019-1449-z
Jia, C., Zhao, S., Bao, T., Zhao, P., Peng, K., Guo, Q., et al. (2021). Tomato BZR/BES transcription factor SlBZR1 positively regulates B.R. signaling and salt stress tolerance in tomato and arabidopsis. Plant Sci. 302, 110719. doi: 10.1016/j.plantsci.2020.110719
Ji, H., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A., Li, X. (2013). The salt overly sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 6 (2), 275–286. doi: 10.1093/mp/sst017
Jogawat, A. (2019). “Osmolytes and their role in abiotic stress tolerance in plants,” in Molecular plant abiotic stress: biology and biotechnology Wiley, 91–104. doi: 10.1002/9781119463665.ch5
Keisham, M., Mukherjee, S., Bhatla, S. C. (2018). Mechanisms of sodium transport in plants-progresses and challenges. Int. J. Mol. Sci. 19 (3), 647. doi: 10.3390/ijms19030647
Khan, M. I. R., Asgher, M., Khan, N. A. (2014). Alleviation of salt-induced photosynthesis and growth inhibition by salicylic acid involves glycinebetaine and ethylene in mungbean (Vigna radiata l.). Plant Physiol. Biochem. 80, 67–74. doi: 10.1016/j.plaphy.2014.03.026
Khan, N., Bano, A., Ali, S., Babar, M. (2020). Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 90 (2), 189–203. doi: 10.1007/s10725-020-00571-x
Khazaei, M., Aghaz, F. (2017). Reactive oxygen species generation and use of antioxidants during in vitro maturation of oocytes. Int. J. Fertility Sterility 11 (2), 63. doi: 10.22074/ijfs.2017.4995
Kim, J. Y., Lee, S. J., Min, W. K., Cha, S., Song, J. T., Seo, H. S. (2022). COP1 controls salt stress tolerance by modulating sucrose content. Plant Signaling Behav. 17 (1), 2096784. doi: 10.1080/15592324.2022.2096784
Kim, W-Y, Ali, Z, Park, HJ, Park, SJ, Cha, J-Y, Perez-Hormaeche, J, et al (2013). Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nature communications 4 (1), 1–13. doi: 10.1038/ncomms2357
Knight, M. R., Campbell, A. K., Smith, S. M., Trewavas, A. J. (1991). Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352 (6335), 524–526. doi: 10.1038/352524a0
Knight, H., Trewavas, A. J., Knight, M. R. (1997). Calcium signalling in arabidopsis thaliana responding to drought and salinity. Plant J. 12 (5), 1067–1078. doi: 10.1046/j.1365-313X.1997.12051067.x
Kovacs, H., Aleksza, D., Baba, A. I., Hajdu, A., Kiraly, A. M., Zsigmond, L., et al. (2019). Light control of salt-induced proline accumulation is mediated by ELONGATED HYPOCOTYL 5 in arabidopsis. Front. Plant Sci. 10, 1584. doi: 10.3389/fpls.2019.01584
Kumari, S., Chhillar, H., Chopra, P., Khanna, R. R., Khan, M. I. R. (2021). Potassium: A track to develop salinity tolerant plants. Plant Physiol. Biochem. 167, 1011–1023. doi: 10.1016/j.plaphy.2021.09.031
Kumar, G., Kushwaha, H. R., Purty, R. S., Kumari, S., Singla-Pareek, S. L., Pareek, A. (2012). Cloning, structural and expression analysis of OsSOS2 in contrasting cultivars of rice under salinity stress. Genes Genomes Genomics 6 (1), 34–41.
Kurotani, K.-i., Hayashi, K., Hatanaka, S., Toda, Y., Ogawa, D., Ichikawa, H., et al. (2015). Elevated levels of CYP94 family gene expression alleviate the jasmonate response and enhance salt tolerance in rice. Plant Cell Physiol. 56 (4), 779–789. doi: 10.1093/pcp/pcv006
Ku, Y. S., Sintaha, M., Cheung, M. Y., Lam, H. M. (2018). Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 19 (10), 3206. doi: 10.3390/ijms19103206
Legué, V., Blancaflor, E., Wymer, C., Perbal, G., Fantin, D., Gilroy, S. (1997). Cytoplasmic free Ca2+ in arabidopsis roots changes in response to touch but not gravity. Plant Physiol. 114 (3), 789–800. doi: 10.1104/pp.114.3.789
Leivar, P., Quail, P. H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16 (1), 19–28. doi: 10.1016/j.tplants.2010.08.003
Leng, Q., Mercier, R. W., Hua, B.-G., Fromm, H., Berkowitz, G. A. (2002). Electrophysiological analysis of cloned cyclic nucleotide-gated ion channels. Plant Physiol. 128 (2), 400–410. doi: 10.1104/pp.010832
Liang, X., Zhang, L., Natarajan, S. K., Becker, D. F. (2013). Proline mechanisms of stress survival. Antioxid Redox Signal 19 (9), 998–1011. doi: 10.1089/ars.2012.5074
Li, Y., Hu, J., Qi, J., Zhao, F., Liu, J., Chen, L., et al. (2022). Improvement of leaf k+ retention is a shared mechanism behind CeO2 and Mn3O4 nanoparticles improved rapeseed salt tolerance. Stress Biol. 2 (1), 1–15. doi: 10.1007/s44154-022-00065-y
Li, J., Li, H., Yang, N., Jiang, S., Ma, C., Li, H. (2020a). Overexpression of a monodehydroascorbate reductase gene from sugar beet M14 increased salt stress tolerance. Sugar Technol. 23 (1), 45–56. doi: 10.1007/s12355-020-00877-0
Lin, H., Yang, Y., Quan, R., Mendoza, I., Wu, Y., Du, W., et al. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in arabidopsis. Plant Cell 21 (5), 1607–1619. doi: 10.1105/tpc.109.066217
Lin, Y.-J., Yu, X.-Z., Li, Y.-H., Yang, L. (2020). Inhibition of the mitochondrial respiratory components (Complex I and complex III) as stimuli to induce oxidative damage in oryza sativa l. under thiocyanate exposure. Chemosphere 243, 125472. doi: 10.1016/j.chemosphere.2019.125472
Li, L., Sheen, J. (2016). Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 33, 116–125. doi: 10.1016/j.pbi.2016.06.018
Liu, J., Gao, H., Wang, X., Zheng, Q., Wang, C., Wang, X., et al. (2014). Effects of 24-epibrassinolide on plant growth, osmotic regulation and ion homeostasis of salt-stressed canola. Plant Biol. 16 (2), 440–450. doi: 10.1111/plb.12052
Liu, J., Li, G., Chen, L., Gu, J., Wu, H., Li, Z. (2021). Cerium oxide nanoparticles improve cotton salt tolerance by enabling better ability to maintain cytosolic K+/Na+ ratio. J. Nanobiotechnology 19 (1), 1–16. doi: 10.1186/s12951-021-00892-7
Li, J., Yang, Y., Sun, K., Chen, Y., Chen, X., Li, X. (2019). Exogenous melatonin enhances cold, salt and drought stress tolerance by improving antioxidant defense in tea plant (Camellia sinensis (L.) o. kuntze). Molecules 24 (9), 1826. doi: 10.3390/molecules24091826
Li, S., Zheng, H., Lin, L., Wang, F., Sui, N. (2020). Roles of brassinosteroids in plant growth and abiotic stress response. Plant Growth Regul. 93 (1), 29–38. doi: 10.1007/s10725-020-00672-7
Li, J., Zhou, H., Zhang, Y., Li, Z., Yang, Y., Guo, Y. (2020b). The GSK3-like kinase BIN2 is a molecular switch between the salt stress response and growth recovery in arabidopsis thaliana. Dev. Cell 55 (3), 367–380. e366. doi: 10.1016/j.devcel.2020.08.005
Ludwiczak, A., Osiak, M., Cárdenas-Pérez, S., Lubińska-Mielińska, S., Piernik, A. (2021). Osmotic stress or ionic composition: Which affects the early growth of crop species more? Agronomy 11 (3), 435. doi: 10.3390/agronomy11030435
Møller, I. M. (2001). Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Biol. 52 (1), 561–591. doi: 10.1146/annurev.arplant.52.1.561
Maathuis, F. J., Sanders, D. (2001). Sodium uptake in arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol. 127 (4), 1617–1625. doi: 10.1104/pp.010502
Maathuis, F. J. (2006). cGMP modulates gene transcription and cation transport in Arabidopsis roots. Plant J. 45 (5), 700–711. doi: 10.1111/j.1365-313X.2005.02616.x
Maathuis, F. J., Ahmad, I, Patishtan, J (2014). Regulation of Na+ fluxes in plants. Front Plant Sci 5, 467. doi: 10.3389/fpls.2014.00467
Maeshima, M. (2000). Vacuolar h+-pyrophosphatase. Biochim. Biophys. Acta (BBA)-Biomembranes 1465 (1-2), 37–51. doi: 10.1016/S0005-2736(00)00130-9
Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., Oda, K. (2004). Dwarf and delayed-flowering 1, a novel arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 37 (5), 720–729. doi: 10.1111/j.1365-313X.2003.01998.x
Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., Oda, K. (2008). The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in arabidopsis. Plant J. 56 (4), 613–626. doi: 10.1111/j.1365-313X.2008.03627.x
Maurya, A. K. (2020). “Oxidative stress in crop plants,” in Agronomic crops (Springer), 349–380. doi: 10.1007/978-981-15-0025-1_18
Maxwell, D. P., Wang, Y., McIntosh, L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. 96 (14), 8271–8276. doi: 10.1073/pnas.96.14.8271
Ma, L., Zhang, H., Sun, L., Jiao, Y., Zhang, G., Miao, C., et al. (2012). NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in arabidopsis under salt stress. J. Exp. Bot. 63 (1), 305–317. doi: 10.1093/jxb/err280
Millenaar, F. F., Benschop, J. J., Wagner, A. M., Lambers, H. (1998). The role of the alternative oxidase in stabilizing the in vivo reduction state of the ubiquinone pool and the activation state of the alternative oxidase. Plant Physiol. 118 (2), 599–607. doi: 10.1104/pp.118.2.599
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33 (4), 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Misra, S., Chauhan, P. S. (2020). ACC deaminase-producing rhizosphere competent bacillus spp. mitigate salt stress and promote zea mays growth by modulating ethylene metabolism. 3 Biotech. 10 (3), 119. doi: 10.1007/s13205-020-2104-y
Moghaddam, M. S. H., Safaie, N., Soltani, J., Hagh-Doust, N. (2021). Desert-adapted fungal endophytes induce salinity and drought stress resistance in model crops. Plant Physiol. Biochem. 160, 225–238. doi: 10.1016/j.plaphy.2021.01.022
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nakaminami, K., Okamoto, M., Higuchi-Takeuchi, M., Yoshizumi, T., Yamaguchi, Y., Fukao, Y., et al. (2018). AtPep3 is a hormone-like peptide that plays a role in the salinity stress tolerance of plants. Proc. Natl. Acad. Sci. 115 (22), 5810–5815. doi: 10.1073/pnas.1719491115
Newkirk, G. M., Wu, H., Santana, I., Giraldo, J. P. (2018). Catalytic scavenging of plant reactive oxygen species in vivo by anionic cerium oxide nanoparticles. J. Visualized Experiments 26 (138), e58373. doi: 10.3791/58373
Nolan, T. M., Vukasinovic, N., Liu, D., Russinova, E., Yin, Y. (2020). Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32 (2), 295–318. doi: 10.1105/tpc.19.00335
Özdemir, F., Bor, M., Demiral, T., Türkan, İ (2004). Effects of 24-epibrassinolide on seed germination, seedling growth, lipid peroxidation, proline content and antioxidative system of rice (Oryza sativa l.) under salinity stress. Plant Growth Regul. 42 (3), 203–211. doi: 10.1023/B:GROW.0000026509.25995.13
Park, H. J., Kim, W. Y., Yun, D. J. (2016a). A new insight of salt stress signaling in plant. Molecules Cells 39 (6), 447–459. doi: 10.14348/molcells.2016.0083
Park, H. J., Qiang, Z., Kim, W.-Y., Yun, D.-J. (2016b). Diurnal and circadian regulation of salt tolerance in arabidopsis. J. Plant Biol. 59 (6), 569–578. doi: 10.1007/s12374-016-0317-8
Park, H. C., Song, E. H., Nguyen, X. C., Lee, K., Kim, K. E., Kim, H. S., et al. (2011). Arabidopsis MAP kinase phosphatase 1 is phosphorylated and activated by its substrate AtMPK6. Plant Cell Rep. 30 (8), 1523–1531. doi: 10.1007/s00299-011-1064-4
Pazhamala, L. T., Kudapa, H., Weckwerth, W., Millar, A. H., Varshney, R. K. (2021). Systems biology for crop improvement. Plant Genome 14 (2), e20098. doi: 10.1002/tpg2.20098
Peng, J., Li, Z., Wen, X., Li, W., Shi, H., Yang, L., et al. (2014). Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in arabidopsis. PloS Genet. 10 (10), e1004664. doi: 10.1371/journal.pgen.1004664
Qin, Y., Druzhinina, I. S., Pan, X., Yuan, Z. (2016). Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Adv. 34 (7), 1245–1259. doi: 10.1016/j.biotechadv.2016.08.005
Qiu, Q.-S., Guo, Y., Quintero, F. J., Pardo, J. M., Schumaker, K. S., Zhu, J.-K. (2004). Regulation of vacuolar Na+/H+ exchange in arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 279 (1), 207–215. doi: 10.1074/jbc.M307982200
Qiu, Z., Guo, J., Zhu, A., Zhang, L., Zhang, M. (2014). Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicology Environ. Saf. 104, 202–208. doi: 10.1016/j.ecoenv.2014.03.014
Quan, R., Lin, H., Mendoza, I., Zhang, Y., Cao, W., Yang, Y., et al. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect arabidopsis shoots from salt stress. Plant Cell 19 (4), 1415–1431. doi: 10.1105/tpc.106.042291
Quintero, F. J., Martinez-Atienza, J., Villalta, I., Jiang, X., Kim, W.-Y., Ali, Z., et al. (2011). Activation of the plasma membrane Na/H antiporter salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory c-terminal domain. Proc. Natl. Acad. Sci. 108 (6), 2611–2616. doi: 10.1073/pnas.1018921108
Racchi, M. L. (2013). Antioxidant defenses in plants with attention to prunus and citrus spp. Antioxidants 2 (4), 340–369. doi: 10.3390/antiox2040340
Rehman, A. U., Bashir, F., Ayaydin, F., Kóta, Z., Páli, T., Vass, I. (2021). Proline is a quencher of singlet oxygen and superoxide both in in vitro systems and isolated thylakoids. Physiologia Plantarum 172 (1), 7–18. doi: 10.1111/ppl.13265
Rejeb, K. B., Abdelly, C., Savouré, A. (2014). How reactive oxygen species and proline face stress together. Plant Physiol. Biochem. 80, 278–284. doi: 10.1016/j.plaphy.2014.04.007
Rodriguez, R., White, J., Arnold, A. E., Redman, R. S. (2009). Fungal endophytes: Diversity and functional roles. New Phytol. 182 (2), 314–330. doi: 10.1111/j.1469-8137.2009.02773.x
Rubio, F, Flores, P, Navarro, JM, Martınez, V (2003). Effects of Ca2+, K+ and cGMP on Na+ uptake in pepper plants. Plant Sci. 165 (5), 1043–1049. doi: 10.1016/S0168-9452(03)00297-8
Sahab, S., Suhani, I., Srivastava, V., Chauhan, P. S., Singh, R. P., Prasad, V. (2021). Potential risk assessment of soil salinity to agroecosystem sustainability: Current status and management strategies. Sci. Total Environ. 764, 144164. doi: 10.1016/j.scitotenv.2020.144164
Sakuraba, Y., Bulbul, S., Piao, W., Choi, G., Paek, N. C. (2017). Arabidopsis EARLY FLOWERING3 increases salt tolerance by suppressing salt stress response pathways. Plant J. 92 (6), 1106–1120. doi: 10.1111/tpj.13747
Santos, A., Silveira, J., Bonifacio, A., Rodrigues, A. C., Figueiredo, M. (2018). Antioxidant response of cowpea co-inoculated with plant growth-promoting bacteria under salt stress. Braz. J. Microbiol. 49, 513–521. doi: 10.1016/j.bjm.2017.12.003
Sewelam, N., Kazan, K., Schenk, P. M. (2016). Global plant stress signaling: reactive oxygen species at the cross-road. Front. Plant Sci. 7, 187. doi: 10.3389/fpls.2016.00187
Sgroy, V., Cassán, F., Masciarelli, O., Del Papa, M. F., Lagares, A., Luna, V. (2009). Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte prosopis strombulifera. Appl. Microbiol. Biotechnol. 85 (2), 371–381. doi: 10.1007/s00253-009-2116-3
Shabaan, M., Asghar, H. N., Zahir, Z. A., Zhang, X., Sardar, M. F., Li, H. (2022). Salt-tolerant PGPR confer salt tolerance to maize through enhanced soil biological health, enzymatic activities, nutrient uptake and antioxidant defense. Front. Microbiol. 13, 901865. doi: 10.3389/fmicb.2022.901865
Shabala, S., Wu, H., Bose, J. (2015). Salt stress sensing and early signalling events in plant roots: Current knowledge and hypothesis. Plant Sci. 241, 109–119. doi: 10.1016/j.plantsci.2015.10.003
Sharma, P., Jha, A. B., Dubey, R. S., Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 217037. doi: 10.1155/2012/217037
Shen, Y., Shen, L., Shen, Z., Jing, W., Ge, H., Zhao, J., et al. (2015). The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ. 38 (12), 2766–2779. doi: 10.1111/pce.12586
Silva, P., Gerós, H. (2009). Regulation by salt of vacuolar h+-ATPase and h+-pyrophosphatase activities and Na+/H+ exchange. Plant Signaling Behav. 4 (8), 718–726. doi: 10.4161/psb.4.8.9236
Singh, A., Sengar, R. S., Rajput, V. D., Minkina, T., Singh, R. K. (2022). Zinc oxide nanoparticles improve salt tolerance in rice seedlings by improving physiological and biochemical indices. Agriculture 12 (7), 1014. doi: 10.3390/agriculture12071014
Singh, D., Sinha, H., Singh, N., Dwivedi, S., Trivedi, P. K. (2022). Tobacco HY5, NtHY5, positively regulates flavonoid biosynthesis and enhances salt stress tolerance. bioRxiv. doi: 10.1101/2022.01.05.475064
Sofy, M. R., Aboseidah, A. A., Heneidak, S. A., Ahmed, H. R. (2021). ACC deaminase containing endophytic bacteria ameliorate salt stress in pisum sativum through reduced oxidative damage and induction of antioxidative defense systems. Environ. Sci. pollut. Res. 28 (30), 40971–40991. doi: 10.1007/s11356-021-13585-3
Soliman, M., Elkelish, A., Souad, T., Alhaithloul, H., Farooq, M. (2020). Brassinosteroid seed priming with nitrogen supplementation improves salt tolerance in soybean. Physiol. Mol. Biol. Plants 26 (3), 501–511. doi: 10.1007/s12298-020-00765-7
Solis, C. A., Yong, M. T., Venkataraman, G., Milham, P., Zhou, M., Shabala, L., et al. (2021). Sodium sequestration confers salinity tolerance in an ancestral wild rice. Physiologia Plantarum 172 (3), 1594–1608. doi: 10.1111/ppl.13352
Song, Q., Joshi, M., Joshi, V. (2020). Transcriptomic analysis of short-term salt stress response in watermelon seedlings. Int. J. Mol. Sci. 21 (17), 6036. doi: 10.3390/ijms21176036
Souana, K., Taïbi, K., Abderrahim, L. A., Amirat, M., Achir, M., Boussaid, M., et al. (2020). Salt-tolerance in Vicia faba l. is mitigated by the capacity of salicylic acid to improve photosynthesis and antioxidant response. Scientia Hortic. 273, 109641. doi: 10.1016/j.scienta.2020.109641
Srivastava, M., Srivastava, A. K., Orosa-Puente, B., Campanaro, A., Zhang, C., Sadanandom, A. (2020). SUMO conjugation to BZR1 enables brassinosteroid signaling to integrate environmental cues to shape plant growth. Curr. Biol. 30 (8), 1410–1423. e1413. doi: 10.1016/j.cub.2020.01.089
Steffens, B. (2014). The role of ethylene and ROS in salinity, heavy metal, and flooding responses in rice. Front. Plant Sci. 5, 685. doi: 10.3389/fpls.2014.00685
Sun, T.-p., Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55 (1), 197–223. doi: 10.1146/annurev.arplant.55.031903.141753
Su, Q., Zheng, X., Tian, Y., Wang, C. (2020). Exogenous brassinolide alleviates salt stress in malus hupehensis rehd. by regulating the transcription of NHX-type na+ (K+)/H+ antiporters. Front. Plant Sci. 11, 38. doi: 10.3389/fpls.2020.00038
Szymańska, K. P., Polkowska-Kowalczyk, L., Lichocka, M., Maszkowska, J., Dobrowolska, G. (2019). SNF1-related protein kinases SnRK2.4 and SnRK2.10 modulate ROS homeostasis in plant response to salt stress. Int. J. Mol. Sci. 20 (1), 143. doi: 10.3390/ijms20010143
Tan, T, Cai, J, Zhan, E, Yang, Y, Zhao, J, Guo, Y, et al (2016). Stability and localization of 14-3-3 proteins are involved in salt tolerance in Arabidopsis. Plant Mol Biol 92 (3), 391–400. doi: 10.1007/s11103-016-0520-5
Tanveer, M., Shah, A. N. (2017). An insight into salt stress tolerance mechanisms of chenopodium album. Environ. Sci. pollut. Res. 24 (19), 16531–16535. doi: 10.1007/s11356-017-9337-2
Tao, J., Chen, H., Ma, B., Zhang, W., Chen, S., Zhang, J. (2015). The role of ethylene in plants under salinity stress. Front. Plant Sci. 6, 1059. doi: 10.3389/fpls.2015.01059
Tester, M., Davenport, R. (2003). Na+ tolerance and na+ transport in higher plants. Ann. Bot. 91 (5), 503–527. doi: 10.1093/aob/mcg058
Thalmann, M., Pazmino, D., Seung, D., Horrer, D., Nigro, A., Meier, T., et al. (2016). Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 28 (8), 1860–1878. doi: 10.1105/tpc.16.00143
Tuteja, N. (2007). “Mechanisms of high salinity tolerance in plants,” in Osmosensing and osmosignaling. methods in enzymology Elsevier, 419–438. doi: 10.1016/S0076-6879(07)28024-3
Uchida, K., Yamaguchi, M., Kanamori, K., Ariga, H., Isono, K., Kajino, T., et al. (2022). MAP KINASE PHOSPHATASE1 promotes osmotolerance by suppressing PHYTOALEXIN DEFICIENT4-independent immunity. Plant Physiol. 189 (2), 1128–1138. doi: 10.1093/plphys/kiac131
Valenzuela, C. E., Acevedo-Acevedo, O., Miranda, G. S., Vergara-Barros, P., Holuigue, L., Figueroa, C. R., et al. (2016). Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in arabidopsis primary root. J. Exp. Bot. 67 (14), 4209–4220. doi: 10.1093/jxb/erw202
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206 (4), 1196–1206. doi: 10.1111/nph.13312
van Gelderen, K., Kang, C., Pierik, R. (2018). Light signaling, root development, and plasticity. Plant Physiol. 176 (2), 1049–1060. doi: 10.1104/pp.17.01079
Vanlerberghe, G. C. (2013). Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 14 (4), 6805–6847. doi: 10.3390/ijms14046805
van Zelm, E., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Verma, D., Jalmi, S. K., Bhagat, P. K., Verma, N., Sinha, A. K. (2020). A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in arabidopsis. FEBS J. 287 (12), 2560–2576. doi: 10.1111/febs.15157
Verma, V., Ravindran, P., Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16, 86. doi: 10.1186/s12870-016-0771-y
Vives-Peris, V., Gomez-Cadenas, A., Perez-Clemente, R. M. (2018). Salt stress alleviation in citrus plants by plant growth-promoting rhizobacteria pseudomonas putida and novosphingobium sp. Plant Cell Rep. 37 (11), 1557–1569. doi: 10.1007/s00299-018-2328-z
Waadt, R., Seller, C. A., Hsu, P. K., Takahashi, Y., Munemasa, S., Schroeder, J. I. (2022). Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23 (10), 680–694. doi: 10.1038/s41580-022-00479-6
Wang, Y., Diao, P., Kong, L., Yu, R., Zhang, M., Zuo, T., et al. (2020). Ethylene enhances seed germination and seedling growth under salinity by reducing oxidative stress and promoting chlorophyll content via ETR2 pathway. Front. Plant Sci. 11, 1066. doi: 10.3389/fpls.2020.01066
Wang, C. F., Han, G. L., Yang, Z. R., Li, Y. X., Wang, B. S. (2022). Plant salinity sensors: current understanding and future directions. Front. Plant Sci. 13, 859224. doi: 10.3389/fpls.2022.859224
Wang, H., Tang, J., Liu, J., Hu, J., Liu, J., Chen, Y., et al. (2018). Abscisic acid signaling inhibits brassinosteroid signaling through dampening the dephosphorylation of BIN2 by ABI1 and ABI2. Mol. Plant 11 (2), 315–325. doi: 10.1016/j.molp.2017.12.013
Wang, B., Zhang, J., Pei, D., Yu, L. (2021). Combined effects of water stress and salinity on growth, physiological, and biochemical traits in two walnut genotypes. Physiologia Plantarum 172 (1), 176–187. doi: 10.1111/ppl.13316
Wang, M., Zheng, Q., Shen, Q., Guo, S. (2013). The critical role of potassium in plant stress response. Int. J. Mol. Sci. 14 (4), 7370–7390. doi: 10.3390/ijms14047370
Wani, A. S., Ahmad, A., Hayat, S., Tahir, I. (2019). Epibrassinolide and proline alleviate the photosynthetic and yield inhibition under salt stress by acting on antioxidant system in mustard. Plant Physiol. Biochem. 135, 385–394. doi: 10.1016/j.plaphy.2019.01.002
Wilson, R. L., Kim, H., Bakshi, A., Binder, B. M. (2014). The ethylene receptors ETHYLENE RESPONSE1 and ETHYLENE RESPONSE2 have contrasting roles in seed germination of arabidopsis during salt stress. Plant Physiol. 165 (3), 1353–1366. doi: 10.1104/pp.114.241695
Wu, F, Chi, Y, Jiang, Z, Xu, Y, Xie, L, Huang, F, et al (2020). Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis. Nature 578 (7796), 577–581. doi: 10.1038/s41586-020-2032-3
Wu, H. (2018). Plant salt tolerance and na+ sensing and transport. Crop J. 6 (3), 215–225. doi: 10.1016/j.cj.2018.01.003
Xie, H., Zhao, W., Li, W., Zhang, Y., Hajný, J., Han, H. (2022). Small signaling peptides mediate plant adaptions to abiotic environmental stress. Planta 255 (4), 1–12. doi: 10.1007/s00425-022-03859-6
Xue, Z.-Y., Zhi, D.-Y., Xue, G.-P., Zhang, H., Zhao, Y.-X., Xia, G.-M. (2004). Enhanced salt tolerance of transgenic wheat (Tritivum aestivum l.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf na+. Plant Sci. 167 (4), 849–859. doi: 10.1016/j.plantsci.2004.05.034
Yaish, M. W., Antony, I., Glick, B. R. (2015). Isolation and characterization of endophytic plant growth-promoting bacteria from date palm tree (Phoenix dactylifera l.) and their potential role in salinity tolerance. Antonie Van Leeuwenhoek 107 (6), 1519–1532. doi: 10.1007/s10482-015-0445-z
Yang, Q., Chen, Z.-Z., Zhou, X.-F., Yin, H.-B., Li, X., Xin, X.-F., et al. (2009). Overexpression of SOS (Salt overly sensitive) genes increases salt tolerance in transgenic arabidopsis. Mol. Plant 2 (1), 22–31. doi: 10.1093/mp/ssn058
Yang, Y., Guo, Y. (2018a). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217 (2), 523–539. doi: 10.1111/nph.14920
Yang, Y., Guo, Y. (2018b). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60 (9), 796–804. doi: 10.1111/jipb.12689
Yang, Y., Wu, Y., Ma, L., Yang, Z., Dong, Q., Li, Q., et al. (2019). The Ca2+ sensor SCaBP3/CBL7 modulates plasma membrane h+-ATPase activity and promotes alkali tolerance in arabidopsis. Plant Cell 31 (6), 1367–1384. doi: 10.1105/tpc.18.00568
Yang, G., Yu, Z., Gao, L., Zheng, C. (2019). SnRK2s at the crossroads of growth and stress responses. Trends Plant Sci. 24 (8), 672–676. doi: 10.1016/j.tplants.2019.05.010
Yang, T., Zhang, S., Hu, Y., Wu, F., Hu, Q., Chen, G., et al. (2014). The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 166 (2), 945–959. doi: 10.1104/pp.114.246520
Yang, L., Zu, Y.-G., Tang, Z.-H. (2013). Ethylene improves arabidopsis salt tolerance mainly via retaining k+ in shoots and roots rather than decreasing tissue na+ content. Environ. Exp. Bot. 86, 60–69. doi: 10.1016/j.envexpbot.2010.08.006
Ye, C., Zheng, S., Jiang, D., Lu, J., Huang, Z., Liu, Z., et al. (2021). Initiation and execution of programmed cell death and regulation of reactive oxygen species in plants. Int. J. Mol. Sci. 22 (23), 12942. doi: 10.3390/ijms222312942
Yokoi, S., Quintero, F. J., Cubero, B., Ruiz, M. T., Bressan, R. A., Hasegawa, P. M., et al. (2002). Differential expression and function of arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 30 (5), 529–539. doi: 10.1046/j.1365-313X.2002.01309.x
Yuan, F., Yang, H., Xue, Y., Kong, D., Ye, R., Li, C., et al. (2014). OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in arabidopsis. Nature 514 (7522), 367–371. doi: 10.1038/nature13593
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25 (11), 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Yue, J., You, Y., Zhang, L., Fu, Z., Wang, J., Zhang, J., et al. (2019). Exogenous 24-epibrassinolide alleviates effects of salt stress on chloroplasts and photosynthesis in robinia pseudoacacia l. seedlings. J. Plant Growth Regul. 38 (2), 669–682. doi: 10.1007/s00344-018-9881-0
Yu, L., Nie, J., Cao, C., Jin, Y., Yan, M., Wang, F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in arabidopsis thaliana. New Phytol. 188 (3), 762–773. doi: 10.1111/j.1469-8137.2010.03422.x
Yu, Y., Wang, J., Shi, H., Gu, J., Dong, J., Deng, X. W., et al. (2016). Salt stress and ethylene antagonistically regulate nucleocytoplasmic partitioning of COP1 to control seed germination. Plant Physiol. 170 (4), 2340–2350. doi: 10.1104/pp.15.01724
Zerrouk, I., Rahmoune, B., Khelifi, L., Mounir, K., Baluska, F., Ludwig-Müller, J. (2019). Algerian Sahara PGPR confers maize root tolerance to salt and aluminum toxicity via ACC deaminase and IAA. Acta Physiologiae Plantarum 41 (6), 1–10. doi: 10.1007/s11738-019-2881-2
Zhang, H.-X., Blumwald, E. (2001). Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 19 (8), 765–768. doi: 10.1038/90824
Zhang, M., Fang, Y., Liang, Z., Huang, L. (2012). Enhanced expression of vacuolar h+-ATPase subunit e in the roots is associated with the adaptation of broussonetia papyrifera to salt stress. PloS One 7 (10), e48183. doi: 10.1371/journal.pone.0048183
Zhang, Y., Lv, Y., Jahan, N., Chen, G., Ren, D., Guo, L. (2018). Sensing of abiotic stress and ionic stress responses in plants. Int. J. Mol. Sci. 19 (11), 3298. doi: 10.3390/ijms19113298
Zhang, M., Smith, J. A. C., Harberd, N. P., Jiang, C. (2016). The regulatory roles of ethylene and reactive oxygen species (ROS) in plant salt stress responses. Plant Mol. Biol. 91 (6), 651–659. doi: 10.1007/s11103-016-0488-1
Zhao, G., Song, Y., Wang, Q., Yao, D., Li, D., Qin, W., et al. (2020). Gossypium hirsutum salt tolerance is enhanced by overexpression of g. arboreum JAZ1. Front. Bioengineering Biotechnol. 8, 157. doi: 10.3389/fbioe.2020.00157
Zhao, C., Zayed, O., Yu, Z., Jiang, W., Zhu, P., Hsu, C.-C., et al. (2018). Leucine-rich repeat extensin proteins regulate plant salt tolerance in arabidopsis. Proc. Natl. Acad. Sci. 115 (51), 13123–13128. doi: 10.1073/pnas.1816991115
Zhao, S., Zhang, Q., Liu, M., Zhou, H., Ma, C., Wang, P. (2021). Regulation of plant responses to salt stress. Int. J. Mol. Sci. 22 (9), 4609. doi: 10.3390/ijms22094609
Zhou, J., Li, Z., Xiao, G., Zhai, M., Pan, X., Huang, R., et al. (2020). CYP71D8L is a key regulator involved in growth and stress responses by mediating gibberellin homeostasis in rice. J. Exp. Bot. 71 (3), 1160–1170. doi: 10.1093/jxb/erz491
Zhou, H, Lin, H, Chen, S, Becker, K, Yang, Y, Zhao, J, et al. (2014). Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26 (3), 1166–1182. doi: 10.1105/tpc.113.117069
Zhou, H., Wu, H., Zhang, F., Su, Y., Guan, W., Xie, Y., et al. (2021). Molecular basis of cerium oxide nanoparticle enhancement of rice salt tolerance and yield. Environ. Science: Nano 8 (11), 3294–3311. doi: 10.1039/D1EN00390A
Zhou, H., Xiao, F., Zheng, Y., Liu, G., Zhuang, Y., Wang, Z., et al. (2022). PAMP-INDUCED SECRETED PEPTIDE 3 modulates salt tolerance through RECEPTOR-LIKE KINASE 7 in plants. Plant Cell 34 (2), 927–944. doi: 10.1093/plcell/koab292
Zhu, J.-K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6 (5), 441–445. doi: 10.1016/S1369-5266(03)00085-2
Zhu, J.-K. (2016). Abiotic stress signaling and responses in plants. Cell 167 (2), 313–324. doi: 10.1016/j.cell.2016.08.029
Keywords: salt stress, salt perception, salt response and tolerance, osmoregulation, ionic homeostasis, hormone mediation, light signaling, plant microbiota
Citation: Xiao F and Zhou H (2023) Plant salt response: Perception, signaling, and tolerance. Front. Plant Sci. 13:1053699. doi: 10.3389/fpls.2022.1053699
Received: 26 September 2022; Accepted: 05 December 2022;
Published: 06 January 2023.
Edited by:
Yasunari Fujita, Japan International Research Center for Agricultural Sciences (JIRCAS), JapanReviewed by:
Honghong Wu, Huazhong Agricultural University, ChinaCopyright © 2023 Xiao and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huapeng Zhou, emhvdWh1YXBlbmdAc2N1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.