- 1Biotechnology and Germplasm Resources Institute, Yunnan Academy of Agricultural Sciences, Yunnan Provincial Key Lab of Agricultural Biotechnology, Kunming, China
- 2College of Plant Protection, Yunnan Agricultural University, Kunming, China
Bacterial blight (BB) induced by Xanthomonas oryzae pv. oryzae (Xoo) is a devastating bacterial disease in rice. The use of disease resistance (R) genes is the most efficient method to control BB. Members of the nucleotide-binding domain and leucine-rich repeat containing protein (NLR) family have significant roles in plant defense. In this study, Xa47, a new bacterial blight R gene encoding a typical NLR, was isolated from G252 rice material, and XA47 was localized in the nucleus and cytoplasm. Among 180 rice materials tested, Xa47 was discovered in certain BB-resistant materials. Compared with the wild-type G252, the knockout mutants of Xa47 was more susceptible to Xoo. By contrast, overexpression of Xa47 in the susceptible rice material JG30 increased BB resistance. The findings indicate that Xa47 positively regulates the Xoo stress response. Consequently, Xa47 may have application potential in the genetic improvement of plant disease resistance. The molecular mechanism of Xa47 regulation merits additional examination.
Introduction
Crop production occurs in complex and changing ecological environments, and plants inevitably experience many biotic stresses, including attacks by fungi, bacteria, viruses, and insects (Savary et al., 2019; Zhang et al., 2020). To prevent onset of disease, plants evolved innate immune mechanisms to stop the invasion of pathogens primarily by recognizing pathogenic signals by various genes and associated gene products (Ahuja et al., 2012; Bednarek, 2012). When a pathogen invades, the first level of a plant defense system is activated, and pattern recognition receptors on plant cell membranes recognize pathogen/microbe-associated molecular patterns (PAMPs/MAMP), triggering PAMPs/MAMPs-induced immunity (PTI/MTI), which inhibits further pathogen spread (Tena et al., 2011). To interfere with or break plant PTI/MTI, pathogens secrete effectors or virulence factors that enter plant cells directly or indirectly to cause infection (Chen and Ronald, 2011). However, because of co-evolution with pathogens, the effectors can be recognized directly or indirectly by plant resistance proteins to trigger effector-triggered immunity (ETI), a second level of a plant immune system, which is also known as “gene-to-gene” disease resistance (Jones and Dangl, 2006; Dodds and Rathjen, 2010). Activation of ETI is usually accompanied by a burst of reactive oxygen species and a hypersensitive response (HR), among other actions, to prevent pathogen colonization (Spoel and Dong, 2012; Withers and Dong, 2017).
Rice is one of the most important agricultural crops and is consumed by more than half of the global population (Ainsworth, 2008). Rice production is often threatened by pathogens, and rice leaf blight (BB) caused by Xanthomonas oryzae pv. oryzae (Xoo) can cause 10% to 30% reductions in yield or even crop failure (Liu et al., 2014). Compared with chemical applications, cultivating resistant varieties is a more effective and environmentally friendly method to control BB in rice (Ke et al., 2017; Li et al., 2020; Zhang et al., 2020). The interaction between rice and Xoo has become an important model in analyzing plant disease resistance and bacterial pathogens (Niño-Liu et al., 2006). Clarifying the various molecular mechanisms of this interaction is of great importance to basic research and crop breeding (Jiang et al., 2020). To date, 12 genes against Xoo have been isolated: Xa1, Xa3/Xa26, Xa4, xa5, Xa7, Xa10, xa13, Xa21, Xa23, xa25, Xa27, and xa41 (Song et al., 1995; Yoshimura et al., 1998; Iyer and McCouch, 2004; Sun et al., 2004; Gu et al., 2005; Jiang et al., 2006; Xiang et al., 2006; Yang et al., 2006; Liu et al., 2011; Tian et al., 2014; Wang et al., 2015; Hutin et al., 2016; Hu et al., 2017; Chen et al., 2021). The genes encode multiple protein types and are an important in the rice–Xoo interaction. Notably, nine of the genes (Xa1, Xa7, Xa10, Xa23, Xa27, xa5, xa13, xa25, and xa41) are related to the transcription activator-like effectors (TALEs) secreted by the type III secretion system (T3SS) of pathogens in the resistance mechanism of host cells (Boch and Bonas, 2010; Jiang et al., 2020).
The TALEs are important pathogenic factors secreted by Xoo that enter the host nucleus with the assistance of T3SS and then bind to a specifically identified target gene promoters to regulate the transcription of downstream genes (Bogdanove et al., 2010). The promoter regions that bind to TALEs are effector binding elements (EBEs). The target genes to which TALEs bind can be either susceptibility genes that regulate host plant physiology to help a pathogen survive and increase colonization or disease resistance genes that mediate the production of plant ETI (Boch et al., 2014; Peng et al., 2019). For example, instance, the expression of the rice executor resistance (R) genes Xa7, Xa10, Xa23, and Xa27 leads to transcriptional activation by the TALEs AvrXa7/PthXo3, AvrXa10, AvrXa23, and AvrXa27, respectively, triggering a host-induced HR response to prevent Xoo colonization in rice and achieve strong disease resistance (Luo et al., 2021). Alternatively, Xoo TALEs (PthXo1, PthXo2, AvrXa7/Tal5/PthXo3/TalC) can directly target EBEs of the corresponding OsSWEET genes (Xa13/OsSWEET11/Os8N3, Xa25/OsSWEET13, and Xa41/OsSWEET14/Os11N3), thereby up-regulating the expression in rice, which supplies sufficient sugar for the growth of Xoo in host cells and increases susceptibility to BB (Yuan et al., 2009; Streubel et al., 2013; Zhou et al., 2015). Moreover, mutations in the EBEs of xa13, xa25, and xa41 interfere with Xoo TALE interactions, resulting in cryptic resistance to BB (Chu et al., 2006; Yang et al., 2006; Liu et al., 2011; Yuan et al., 2010; Hutin et al., 2015; Zhou et al., 2015). In addition, Xa1 and its alleles (Xa2, Xa14, Xa31, and Xa45) encode atypical nucleotide binding site-leucine-rich repeat (NBS-LRR) containing proteins that recognize multiple Xoo TALEs (PthXo1, Tal4, and Tal9d) and can confer resistance to BB in rice (Zhang et al., 2020). However, interfering TALEs (iTALEs), which are truncated to lack a transcriptional activation domain, can interfere with XA1, XA2, XA14, and XA45 to confer resistance to Xoo in rice (Zhang et al., 2020).
Previously, the gene Xa47 was localized in a 26.24-kb interval between molecular markers R13I14 and 13rbq-71 and was found to co-segregate with the InDel marker Hxjy-1 (Xing et al., 2021). Based on analyses of the Gramene database and the rice genome RGAP annotation database, LOC_Os11g46200 (in this study, xa47) was ultimately selected as the target gene of Xa47 (Xing et al., 2021). The gene Xa47 was originally discovered in the Yuanjiang common wild rice infiltration line material G252, but the gene has not been cloned. However, whether G252, a broad-spectrum, highly resistant rice germplasm material, is endowed with the resistance function of Xa47 is unknown. Therefore, in this study, the Xa47 gene was cloned, its function in improving resistance to BB was investigated, and its potential mechanism of resistance was examined.
Materials and methods
Plant materials and bacterial inoculation
The rice G252, a BC2F16 generation material of the Yuanjiang common wild rice infiltration line (IL), was the donor material for Xa47 cloning and gene editing. Indica varieties susceptible to Xoo strains PXO99 and PB included IR24,IRBB1, and IRBB14. Dr. Zaiquan Cheng (Biotechnology & Genetic Germplasm Institute, Yunnan Academy of Agricultural Sciences, Yunnan, China) kindly provided 80 rice landraces and 100 ILs. Transgenic plants were grown in an artificial climate chamber at 28°C for 12 h (light) and 20°C for 12 h (dark) with relative humidity of 65% to 80%. The other rice materials were grown in the field.
The Xoo strains included the Chinese strains YN18 (C1), YN1 (C2), GD414 (C3), HEN11 (C4), ScYc-b (C5), YN7 (C6), JS49-6 (C7), FuJ (C8), and YN24 (C9), the Filipino strain PXO99, and PB, a strain with Tal3a and Tal3b genes knocked out from PXO99 (Ji et al., 2016). All Xoo strains were grown at 28°C on nutrient agar medium (1 g/L yeast extract, 12 g/L sucrose, 18 g/L agar, 5 g/L peptone, 3 g/L beef extract). Cultured Xoo strains were eluted with sterile water, and concentrations of bacterial suspensions were configured to 3 × 108 colony forming units/mL (OD600 = 0.5). Rice plants at the gestation stage were inoculated with a Xoo strain using the sword leaf tip-clipping method. Disease lesion length (resistant: ≤6 cm; susceptible: >6 cm) was measured approximately 14 d after inoculation when disease development of the susceptible control material JG30 stabilized, as described previously (Chen et al., 2008). Each Xoo strain was tested in three replicates, each with three rice plants. A detailed description of the Xoo strains involved in this study can be found in Supplementary Table 1.
Cloning and sequence analysis of Xa47
The CTAB method was used to extract rice genomic DNA (Porebski et al., 1997). Based on the sequence of LOC_Os11g46200 (xa47), the primer pair Xa47-12F/R, with forward primer 5’-TGGTGCCTATACCTTCATTG-3’ and reverse primer 5’-AATTCGTCATGTTCTACTAGC-3’, was designed and used to clone the gene. PCR amplification of Xa47 was performed using primers Xa47-12F/R and G252 genomic DNA, and the PCR products were used for sequencing. Based on the sequencing results, the CDS sequence of Xa47 was obtained using SnapGene 6.0 software (GSL Biotech, USA). BLASTP1 (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for the Xa47 homologous gene search, and CD-search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to predict protein structural domains. Multiple sequence alignment and phylogenetic analysis was performed using DNAMAN8.0 (Lynnon Biosoft, USA) and (Kumar et al., 2016), and bootstrap testing was performed with 500 replicates using the neighbor-joining method.
Detection of Xa47 in different germplasm resources
The gene Xa47 was detected in 180 germplasm resources (Supplementary Tables 2, 3) using a molecular marker screening method combined with homologous cloning, in which the primers used were Hxjy-1F/R and Xa47-12F/R, in that order. In parallel, the rice materials were evaluated for resistance to strains C5 and C9 (described above).
Subcellular localization of XA47
An Eastep® Super Total RNA Isolation kit (Proegal, Shanghai, China) was used to extract total RNA from rice leaves. A HiScript® III RT SuperMix for qPCR kit (Vazyme Biotech Co., Ltd., Nanjing, China) was used to synthesize cDNA. Based on the Xa47 gene sequence and the pBE221-GFP expression vector, primers with double restriction enzyme sites XbaI and SalI were designed to amplify the Xa47 CDS sequence without a terminator. Polyethylene glycol mediated the transient expression of the XA47::GFP and pBE221-GFP constructs in rice protoplasts under the control of the 35S promoter. Sixteen hours after inoculation, resulting protoplasts were observed using a laser scanning confocal microscope (Nikon A1, Tokyo, Japan).
CRISPR-Cas9-based gene editing in rice
The target vector pH-ubi-cas9 and the entry vector pOs-sgRNA were generously provided by Dr. Liang Chen (College of Life Sciences, Xiamen University, China). The selection and design of sgRNA target sequences were based on the web tool CRISPR-P2.0 (http://crispr.hzau.edu.cn/CRISPR2/news.php) (He et al., 2021). The pOs-Xa47-sgRNA was obtained by ligation reaction of sgRNA with BsaI-digested pOs-sgRNA vector by T4 DNA Ligase (New England Biolabs Ltd., Beijing, China). Similarly, the Xa47-ubi-Cas9 recombinant vector was obtained by ligation reaction of pOs-Xa47-sgRNA vector and pH-ubi-cas9 vector by T4 Polynucleotide Kinase (New England Biolabs Ltd., Beijing, China). The Xa47-ubi-Cas9 vector was introduced into G252 by an Agrobacterium-mediated method (Zhang et al., 2016). All CRISPR plants were genotyped, target locus regions were amplified, and PCR products were Sanger sequenced.
35S::Xa47 expression vector construction and transformation
The Xa47-CDS containing the homologous sequence of pCAMBIA1305 was amplified using the appropriate double digestion sites NcoII and BstEI based on the CDS sequence of Xa47 and the pCAMBIA1305 expression vector. This approach used the primers Xa47-CDS-F/R and Xa47-35S-F/R (attached). The Xa47 was ligated to the pCAMBIA1305 vector in the presence of T4 DNA Ligase (TaKaRa, Dalian, China), resulting in the CaMV35S promoter-driven 35S::Xa47. An Agrobacterium-mediated approach was used to deliver the 35S::Xa47 expression vector into the indica rice variety JG30, which is sensitive to Xoo strains. All transgenic plants were screened for thaumatin resistance. The resistant transgenic plants were planted in greenhouse until maturity. The continuous PCR test for Xa47 was carried out. T2 homozygous transgenic plants were selected for further analysis.
Gene expression assays
From the inverted second leaves of wild-type and mutant lines (described above), RNA was isolated, and cDNA was synthesized. Reverse transcription quantitative PCR (RT-qPCR) using ChamQTM Universal SYBR® qPCR Master Mix (Vazyme Biotech Co., Ltd., Nanjing, China) was performed on a LightCycler® 96 platform (Roche, Shanghai, China). The rice Actin1 gene was used as the endogenous control. All reactions were conducted three times. The 2-△△Cq approach (Livak and Schmittgen, 2001) was used to examine relative expression. The primers used in this work are listed in Supplementary Table 4.
Statistical analyses
For statistical analysis with Student’s t-tests, SPSS 26.0 (IBM, USA) was used. To prepare figures, TBtools (Chen et al., 2020) and GraphPad Prism 8.0 software (GraphPad Software, China) were used.
Results
Cloning of Xa47
Previously, to isolate Xa47, an F2 population obtained by crossing the IL G252 with the susceptible cultivar 02428, which therefore included plants with or without Xa47, was inoculated with different strains of Xoo to trigger Xa47-mediated BB resistance in rice (Xing et al., 2021). After gene linkage and lesion length analyses, the Xa47 gene was localized to a 2.6-kb area on chromosome 11 between markers Hxjy-14 and 13rbq-71 (Figure 1A). In addition, Xa47 in G252 was identified as a dominant disease-resistance gene (Xing et al., 2021). According to the reference sequence of the rice cultivar Nipponbare (NPB), the NLR-encoding gene LOC_Os11g46200 was chosen as the target gene for Xa47 (Xing et al., 2021).
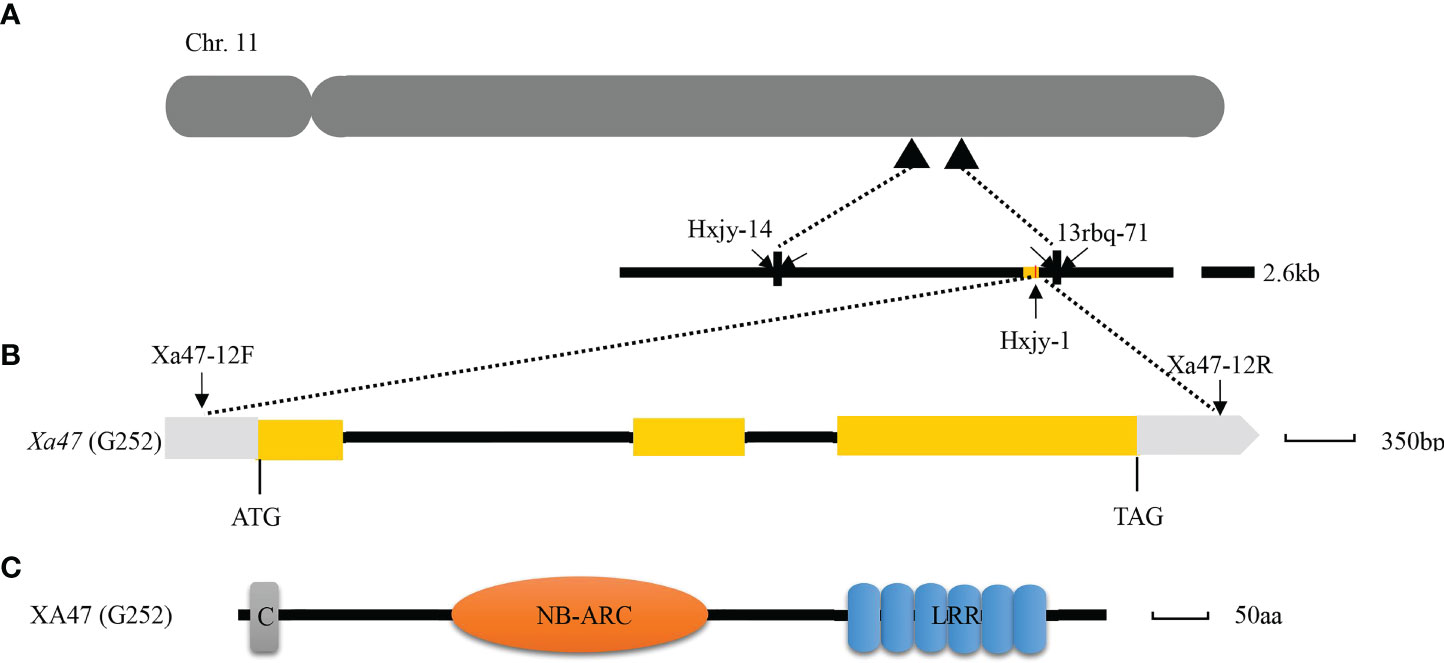
Figure 1 Map-based cloning of Xa47. (A) Physical mapping of Xa47 on chromosome 11 of rice based on the Nipponbare reference genome. (B) Structure of the Xa47 gene. Yellow rectangles indicate exons, and the black line indicates introns. (C) Prediction of the functional domain of XA47. C, Rx N-terminus; NB-ARC, NB-ARC domain; LRR, leucine-rich repeat protein.
Homologous cloning was used to clone Xa47 in sequential rice materials G252 and 02428 (Supplementary Figures 1A–C). Notably, the xa47 sequences in rice materials 02428 and NPB were consistent, whereas the Xa47 sequences in G252 differed from those sequences (Supplementary Figure 2). The gene Xa47 was 4,240 bp in length, had three exons and two introns (Figure 1B), and encoded 802 amino acids (aa) (Figure 1C). The protein XA47 (G252) contained a typical NB-ARC domain and a leucine-rich repeat (LRR) protein, consisting of 237 and 178 aa, respectively (Figure 1C). In addition, XA47 (G252) also contained an Rx N-terminal domain (Figure 1C). Further multisequence alignment and phylogenetic analysis showed that all homologous genes of Xa47 were distributed in graminaceous crops. Notably, XA47 (G252) was slightly more homologous to EAY84977.1 than to XA47 (NPB) (Supplementary Figure 3).
Xa47 is in some bacterial blight-resistant rice
The functional marker Hxjy-1 of Xa47 and homologous cloning were used to detect Xa47 in 180 rice materials. Twenty of 100 ILs contained Xa47 (Figures 2A, B and Supplementary Table 2), and eight of 80 rice landraces contained Xa47 (Figure 2C and Supplementary Table 3). To confirm whether the materials were resistant to BB, a disease assessment was performed. Almost all Xa47-containing materials showed resistance to C5 and C9 strains (Figure 2B).
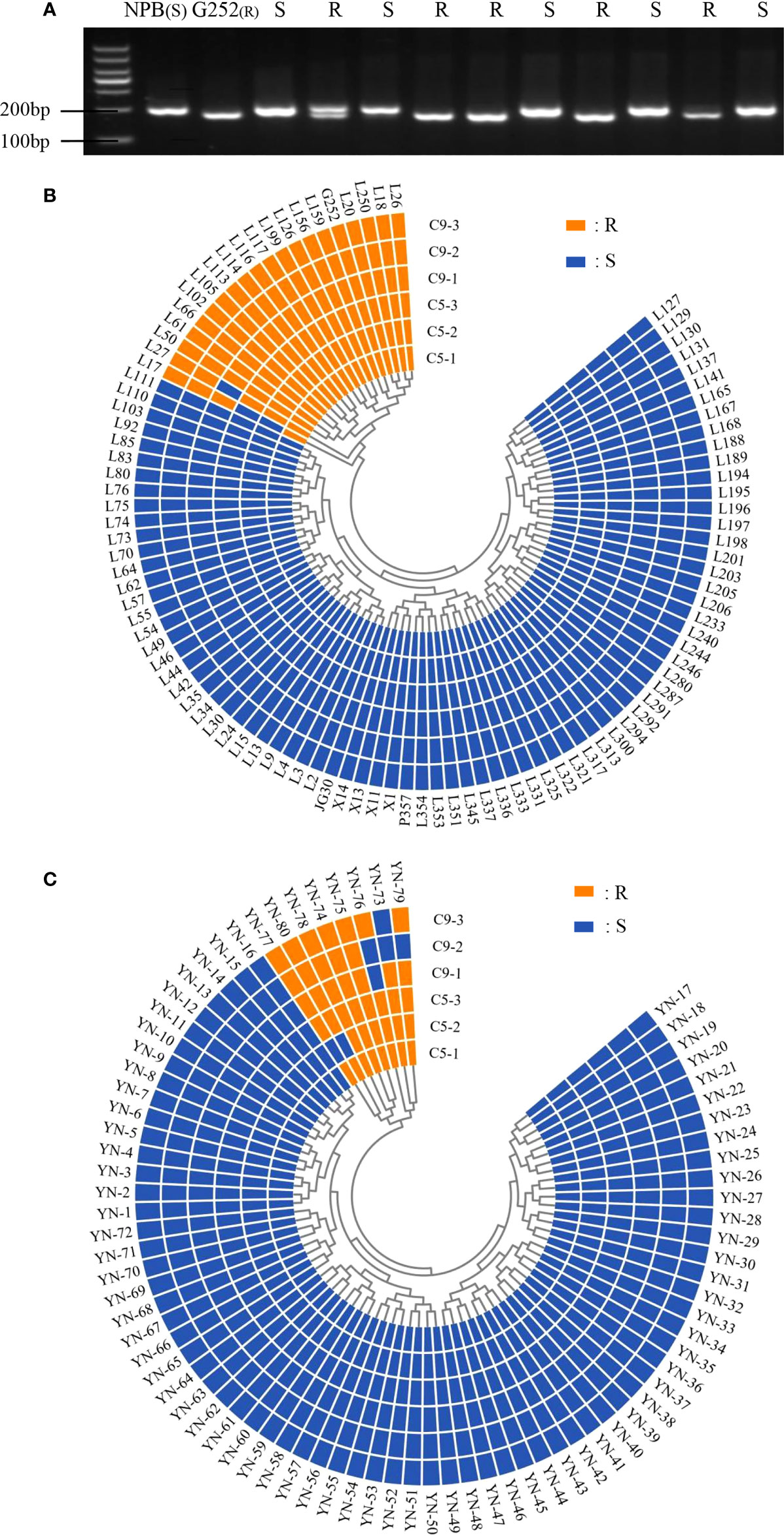
Figure 2 Identification of Xa47 in 180 rice varieties. (A) Detection of rice resistance gene Xa47 using the molecular marker Hxjy-1. R indicates Xa47 disease resistance allele fragment of 170 bp; S indicates Xa47 disease susceptibility allele fragment of 210 bp.NBP was the susceptible control material and G252 was the susceptible control material. (B) Reactions of 100 infiltration lines inoculated with C5 and C9 strains of Xanthomonas oryzae pv. oryza (Xoo). R: resistant (lesion length ≤ 6 cm); S: susceptible (lesion length > 6 cm). (C) Reactions of 80 Yunnan (YN) landraces inoculated with C5 and C9 strains of Xoo. R, resistant (lesion length ≤ 6 cm); S, susceptible (lesion length > 6 cm).
Xa47 is localized in the nucleus and cytoplasm
To determine the subcellular localization of XA47 in cells, a 35S::Xa47-GFP vector was constructed for transient expression in rice protoplasts. Laser scanning confocal microscopy showed that the fluorescence signal of cells transfected with 35S::GFP was uniformly distributed throughout cells, whereas the strong fluorescence signal of cells transfected with 35S::Xa47-GFP was distributed in the nucleus and cytoplasm. Thus, XA47 was localized in the nucleus and cytoplasm (Figure 3).
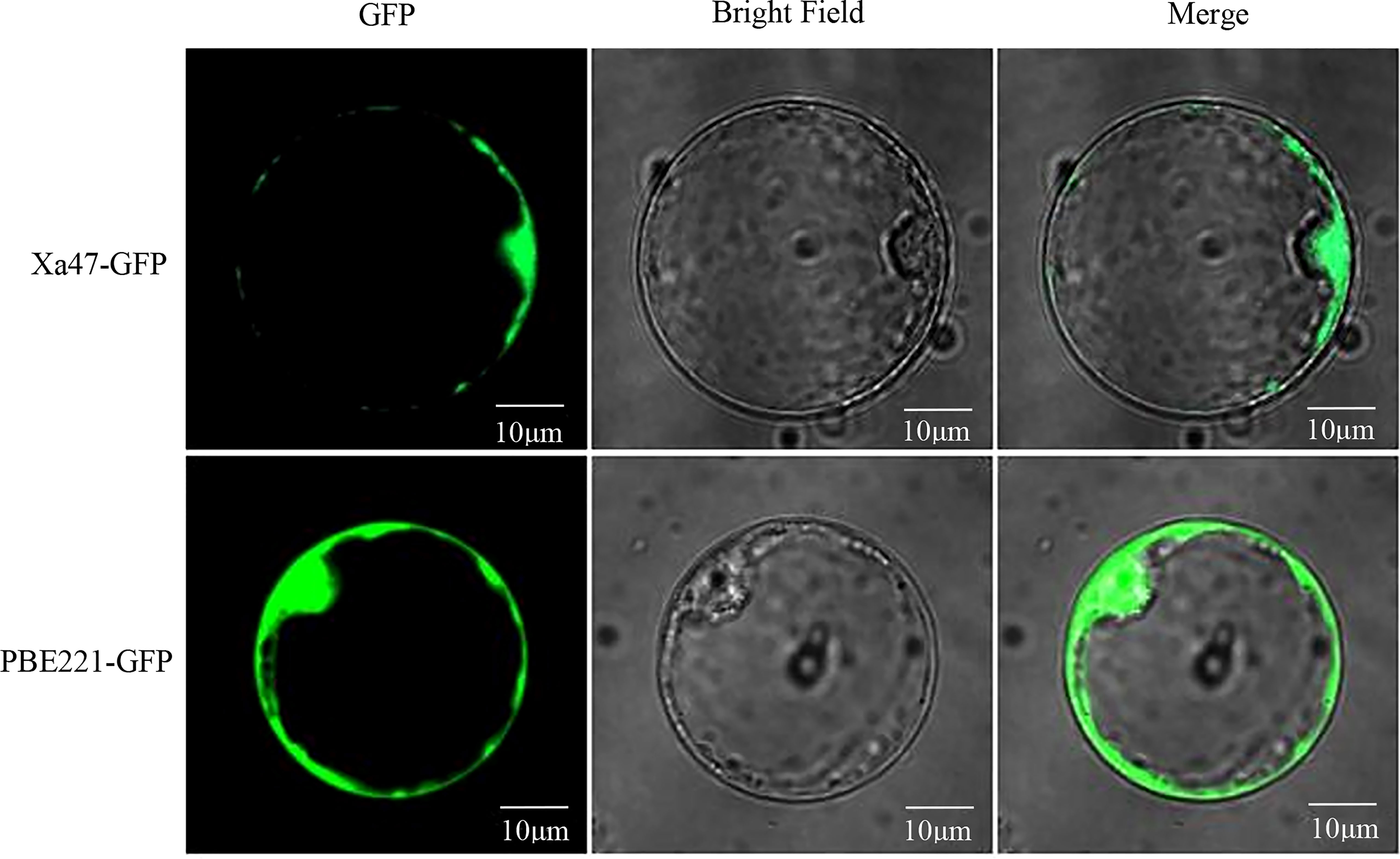
Figure 3 Subcellular localization of XA47 in rice protoplasts. Fluorescence detection of XA47-GFP fusion protein in rice protoplast cells with pBE221::GFP transformed rice protoplast cells as controls.
Loss-of-function mutations of Xa47 result in loss of resistance against Xanthomonas oryzae pv. oryzae
Previously, when locating the Xa47 gene, it was predicted to be a new R gene that conferred resistance to Xoo in rice (Xing et al., 2021). Therefore, a sgRNA was designed to specifically target the second exon of Xa47 (sequence ‘CAAGGTGCCGGAAAAAAGAA’), and it was cloned into a Ubi-Cas9 vector. When G252 was transformed with the constructed knockout vector, 30 T0 transgenic plants were obtained. The mutation sites were sequenced, and six mutations were identified in T0 transgenic plants. Two homozygous mutant lines were selected, one with two base pairs inserted at the target site (XCas9+2) and one with eight base pairs missing (XCas9-8), which were used in further molecular and phenotypic analyses (Figures 4A, B).
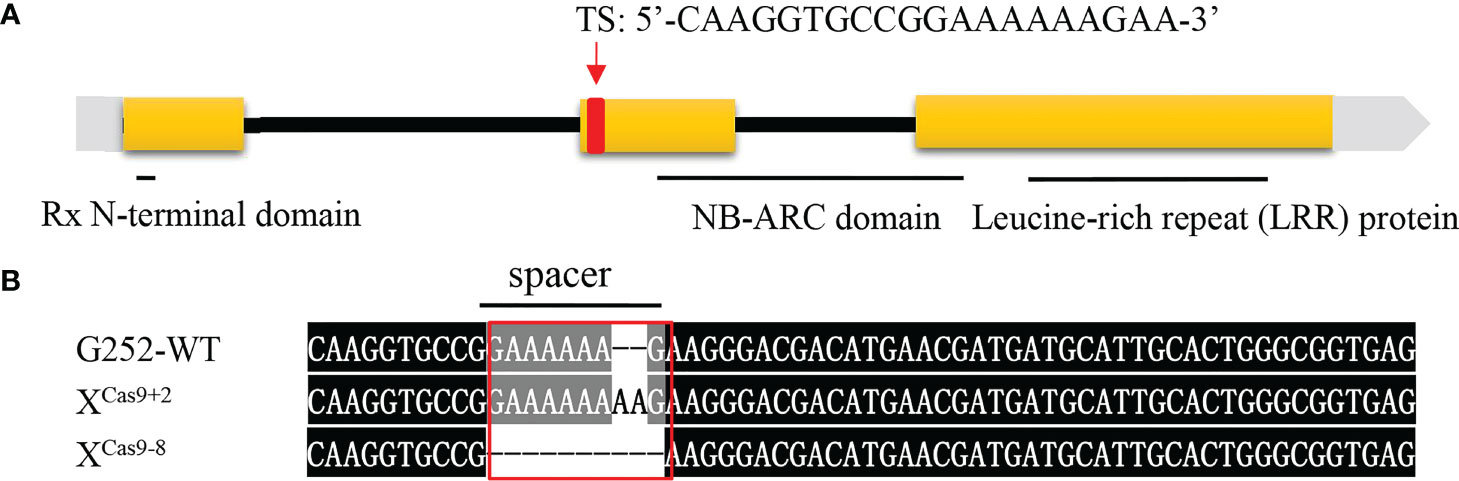
Figure 4 Xa47 knockout mutant lines. (A) Design of target site (TS) sequences. (B) Sequence mutations at the Xa47 target site in two T0-generation XCas9 rice plants. The TS sequence is from G252.
The XCas9 and G252 (WT) plants were grown to the late tillering stage, and leaf specimens were collected at 0, 24, 48, and 72 h after inoculation with Xoo and subjected to RT-qPCR. At each time point, the expression level of Xa47 in XCas9 lines was much lower than that in G252 (WT) lines, which further confirmed the successful knockdown of Xa47 in G252 (Figures 5A-D). Transcript levels of the defense-related marker genes OsNPR1, OsPR1a, and OsPR10 were also measured. At nearly all time points, expression levels of the three genes were considerably lower in XCas9 plants than in G252 plants, which suggested that the defense response was affected in mutant plants. Therefore, the two T1 Xcas9 mutants were inoculated with nine Xoo strains at the rice booting stage using the leaf tip-clipping method. At 14 d following inoculation, all XCas9 plants exhibited much longer lesions than those in G252 plants (Figure 6). Collectively, the findings revealed that XCas9 mutations caused Xa47 to lose its resistance to BB.
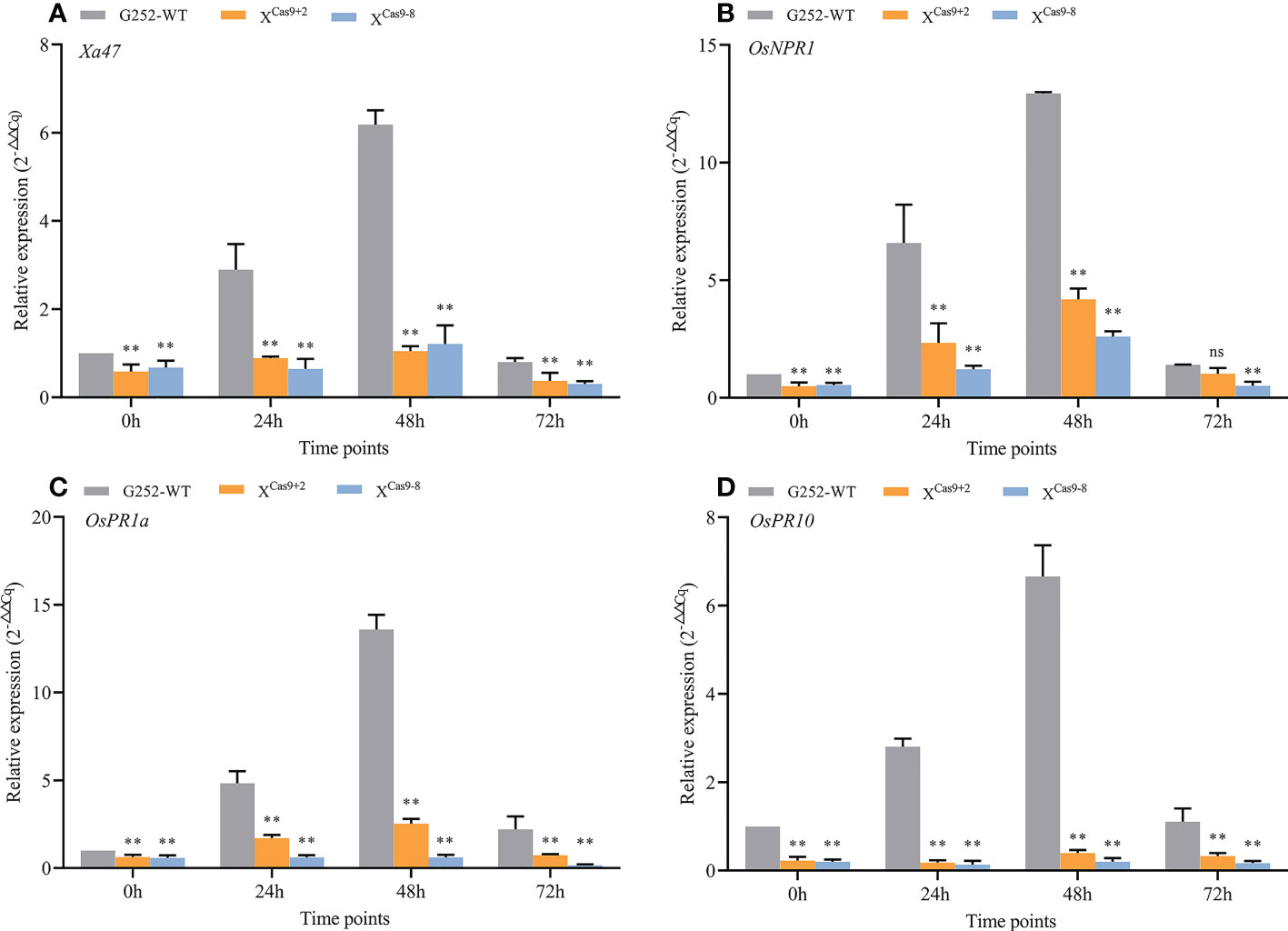
Figure 5 Expression of defense-related genes in XCas9 plants after inoculation with Xanthomonas oryzae pv. oryzae (Xoo). (A–D) Relative expression level of Xa47, OsNPR1, OsPR1a, and OsPR10 in the G252 and two XCas9 lines at 0, 24, 48, and 72 h post-inoculation with Xoo. Values are the mean ± SD (n = 3). Asterisks indicate significant differences compared with the wild type (WT), based on Student’s t-tests (**P < 0.01). Each experiment was performed with three biological replicates.
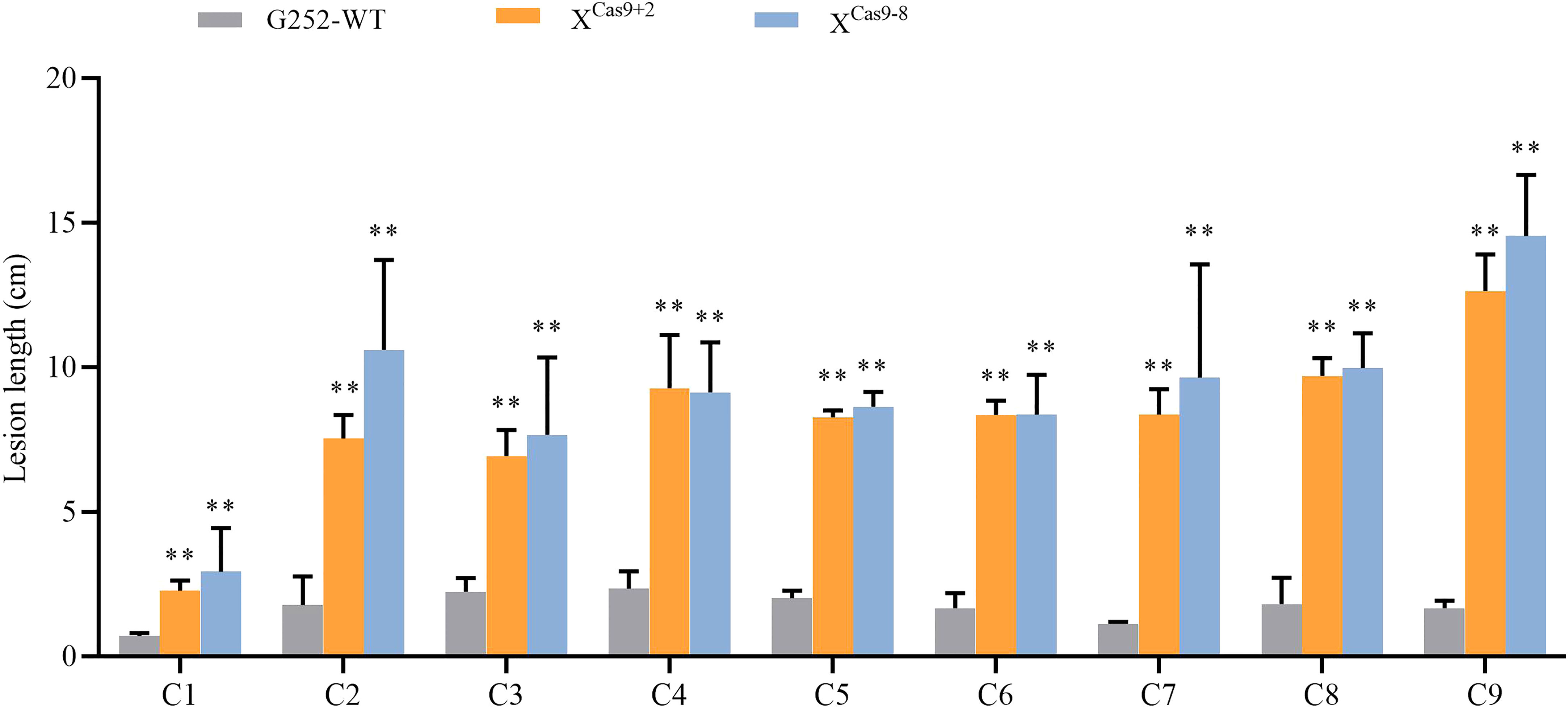
Figure 6 Lesion length of rice after infection with nine Xanthomonas oryzae pv. oryzae isolates (C1–C9). Values are the mean ± SD (n = 9). Asterisks indicate significant differences compared with the wild type (WT), based on Student’s t-tests (**P < 0.01). Each experiment was performed with three biological replicates.
Overexpression of Xa47 increases resistance of JG30 to bacterial blight
The indica rice cultivar JG30 is extremely vulnerable to Xoo because it lacks R genes for BB resistance. Notably, the Xa47 genotype in JG30 was consistent with that of NPB and 02428. To determine whether Xa47 could increase resistance in JG30 to BB, an expression vector 35S::Xa47 was constructed, and then, Xa47 was overexpressed in JG30. Overexpression (35S::Xa47-JG30-1 and 35S::Xa47-JG30-2) and JG30 (WT) plants were grown to the late tillering stage, and leaf specimens were collected at 0, 24, 48, and 72 h after inoculation with Xoo and subjected to RT-qPCR. At each time point, the expression level of Xa47 in 35S::Xa47-JG30 lines was much higher than that in JG30 lines, which confirmed the successful transformation of 35S::Xa47 in JG30 (Figures 7A-D). Transcript levels of the defense-related marker genes OsNPR1, OsPR1a, and OsPR10 were also measured. At nearly all time points, expression levels of the three genes were significantly up-regulated in 35S::Xa47-JG30 plants compared with JG30 plants. The data indicated that overexpression of Xa47 in rice could activate its defenses. Therefore, 35S::Xa47-JG30 plants were inoculated with nine Xoo strains at the rice booting stage using the leaf tip-clipping method. At 14 d following inoculation, all overexpression plants exhibited much shorter lesions than those of JG30 plants (Figure 8). Notably, the growth of 35S-Xa47-JG30 plants was similar to that of JG30-WT (Supplementary Figures 4A-H). Collectively, the findings revealed that overexpression of Xa47 increased the resistance of JG30 to BB.
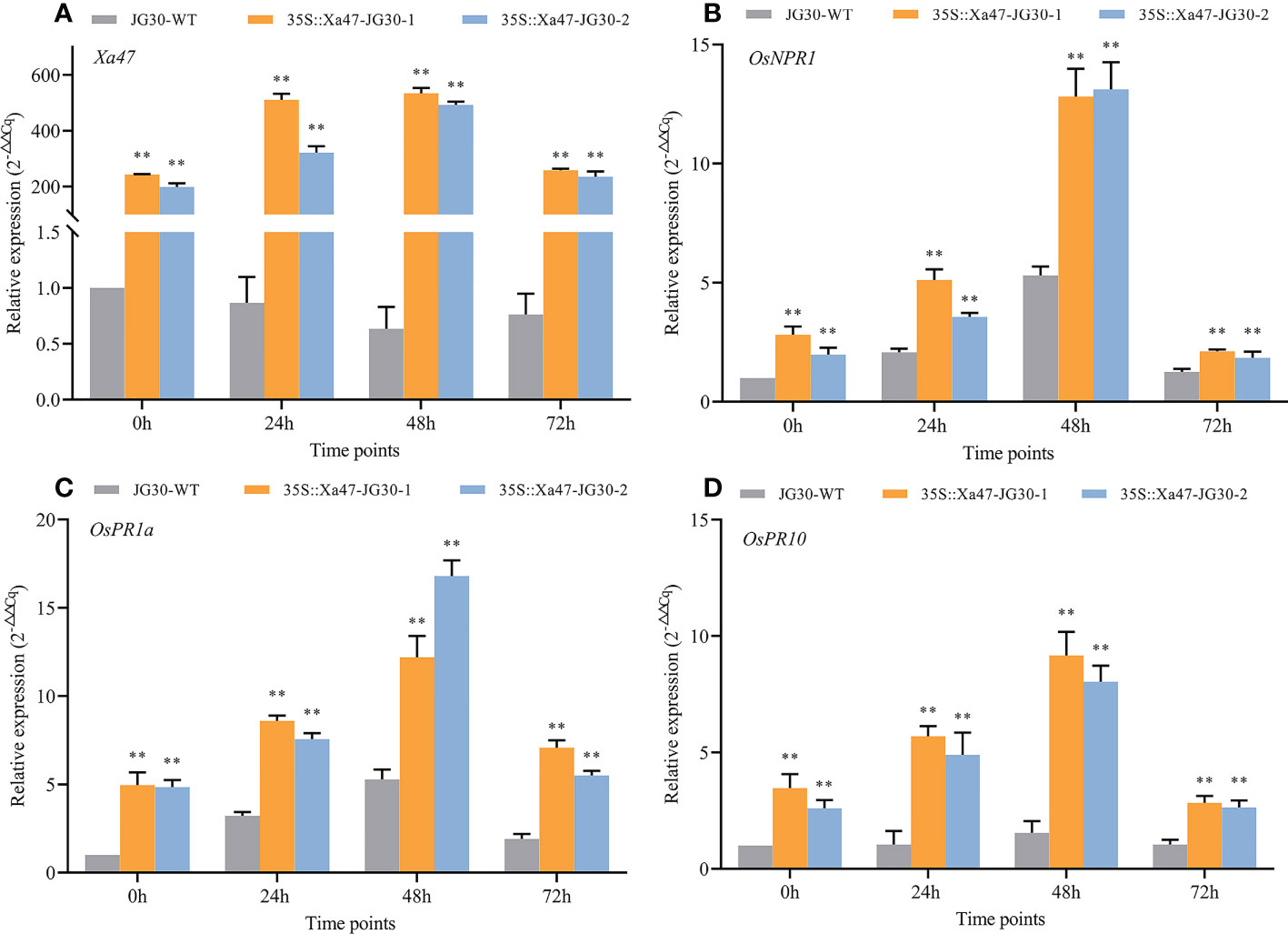
Figure 7 Expression of defense-related genes in 35S::Xa47-JG30 plants after inoculation with Xanthomonas oryzae pv. oryzae (Xoo). (A–D) Relative expression levels of Xa47, OsNPR1, OsPR1a, and OsPR10 in the G252 and two 35S::Xa47-JG30 lines at 0, 24, 48, and 72 h post-inoculation with Xoo. Values are the mean ± SD (n = 3). Asterisks indicate significant differences compared with the wild type (WT), based on Student’s t-tests (**P < 0.01). Each experiment was performed with three biological replicates.
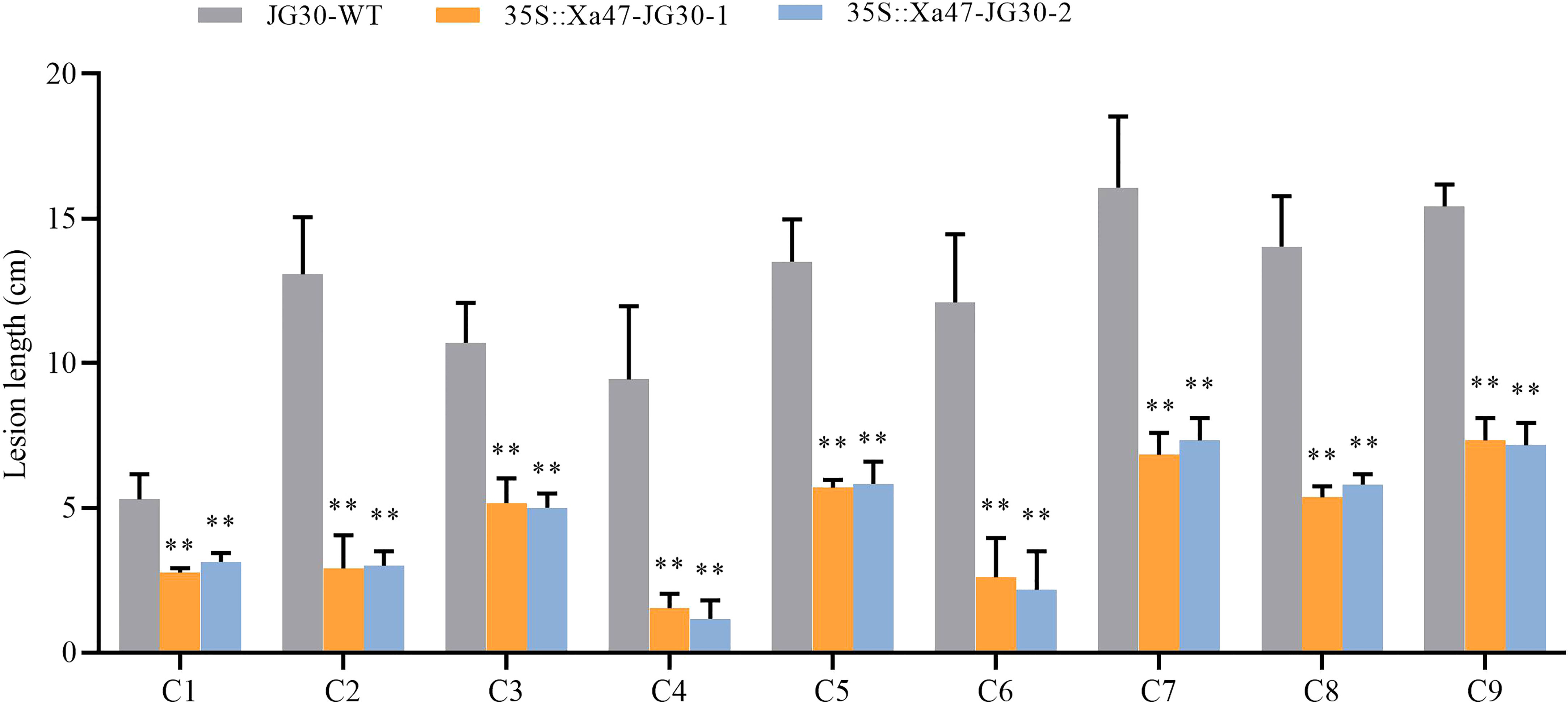
Figure 8 Lesion length of rice after infection with nine Xanthomonas oryzae pv. oryzae isolates (C1–C9). Values are the mean ± SD (n = 9). Asterisks indicate significant differences compared with the wild type (WT), based on Student’s t-tests (**P < 0.01). Each experiment was performed with three biological replicates.
Interfering transcription activator-like effectors reduce resistance mediated by Xa47
As NLR genes specific to rice resistance to BB, Xa1 and its alleles (Xa2, Xa14, Xa31, and Xa45) have identified various TALEs (PthXo1 and Tal4 and Tal9d) to induce resistance to Xoo. Nonetheless, certain iTALEs, such Tal3a and Tal3b in PXO99, reduce this resistance (Zhang et al., 2020). Notably, Xa47, a newly identified NLR gene in rice BB resistance after Xa1 and its alleles, also possessed a functional structural domain similar to that of Xa1. Nevertheless, XA47 had little sequence similarity to XA1 and XA2/XA31 and XA14 and XA45 proteins. To investigate whether Xa47 was influenced by iTALEs, PXO99 and PB (which does not secrete iTALEs) strains were used for identification. After inoculation with PXO99, JG30, 35S::Xa47-1 and 35S::Xa47-2, and XCas9+2 and XCas9-8 plants were as susceptible as IRBB1, IRBB2, and IR24 plants, but the opposite was observed for G252 (Figures 9A, B). By contrast, the resistance of 35S::Xa47-1 and 35S::Xa47-2 and G252 plants after PB inoculation was similar to that observed in IRBB1 and IRBB2 plants (Figures 9A, B). However, XCas9+2 and XCas9-8 and JG30 plants after PB inoculation were as susceptible as IR24 plants (Figures 9A, B). The findings suggested that iTALEs can also reduce Xa47-mediated resistance.
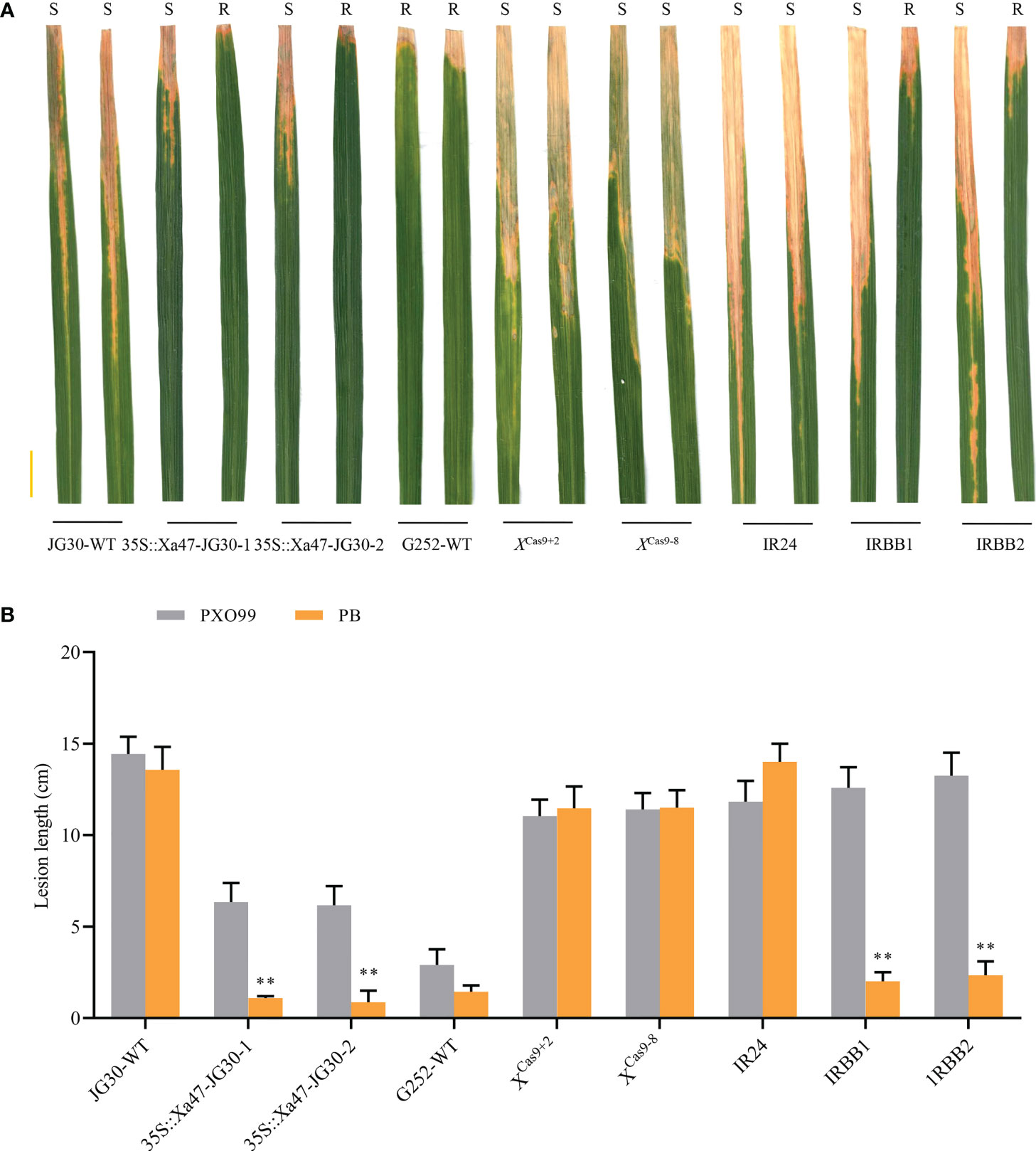
Figure 9 Xa47-mediated resistance is attenuated by interfering transcription activator-like effectors. (A) Photographs of infected leaves of different rice materials 2 weeks after inoculation with Xanthomonas oryzae pv. oryzae strains PXO99 and PB. Scale bar: 2 cm. Two leaves of each rice variety show disease spots after inoculation with PXO99 and PB. (B) Lesion lengths on infected leaves in (A). Values are the mean ± SD (n = 9). Asterisks indicate significant differences compared with the wild type (WT), based on Student’s t-tests (**P < 0.01). Each experiment was performed with three biological replicates.
Discussion
The R genes have essential roles in the competition between rice and Xoo, and the discovery of new R genes will be crucial to rice success. Nearly one-third of the Xa genes are localized on chr. 11, including Xa3/Xa26, Xa4, Xa10, Xa21, Xa22, Xa23, Xa30, Xa32, Xa35, Xa36, Xa39, Xa40, xa41, Xa43, xa44-1, xa44-2, Xa46, and Xa47 (Sun et al., 2004; Xiang et al., 2006; Tian et al., 2014; Wang et al., 2014; Kim et al., 2015; Zhang et al., 2015; Kim, 2018; Chen et al., 2020; Neelam et al., 2020; Xing et al., 2021). Even though many R genes have been identified, relatively few have been cloned or used in breeding. For example, Xa4, Xa21, and Xa23 have been used most frequently in breeding for disease resistance in hybrid rice (Balachiranjeevi et al., 2018). The primary cause of this problem is that most R genes have disadvantages, such as a limited resistance spectrum or insufficient resistance. To overcome such problems, the exploration for new Xa genes with broad-spectrum resistance must be continuous. In addition to identifying new Xa genes with broad-spectrum resistance, multiple Xa genes can be aggregated into the same variety to produce disease-resistant varieties (Dash et al., 2016; Wu et al., 2019). Additionally, rice variants with broad-spectrum resistance can be developed using gene editing technologies (Tao et al., 2021).
In this study, a new NLR gene, Xa47, was cloned from G252 (Figure 1C). The cloning of Xa47 increased the number of NLR genes for BB resistance to two (Xa1 and Xa47, not including alleles). Allelic variety in functional genes is prevalent in plants, and that variation is essential for plant evolution (Zhang et al., 2020). In rice, Xa1 and Xa2/Xa31, Xa14, and Xa45 are alleles of one another, and there are sequence differences among them. Notably, Xa47 and xa47 are alleles of one another, and there were sequence differences between them (Supplementary Figure 2). This result also indicated that Xa47 is variable at natural loci, which warrants additional study. Most plant R genes code for NLRs, which are composed of an N-terminal signaling domain, a central nucleotide-binding region (NBS), and a C-terminal leucine zipper repeat region (LRR) (Mermigka et al., 2020). The NLRs are split into two types based on the N-terminal structure. The TIR-NBS-LRR has a toll/interleukin receptor (TIR) structural domain, whereas the CC-NBS-LRR contains a coiled-coil (CC) motif (van Wersch et al., 2020). In addition, some NLRs contain other structural domains, such as kinase, WRKY, and BED finger domains (Le Roux et al., 2015; Kroj et al., 2016; Bailey et al., 2018). In this study, XA47 was a typical CC-NBS-LRR resistance protein (Figure 1C), whereas XA1 was not a typical CC-NBS-LRR resistance protein and contained other structural domains (Zhang et al., 2020). The results indicated that XA47 and XA1 have distinct functions in disease resistance.
The NLR genes are critical in plant immune systems, because they recognize specific pathogens and activate resistance responses (Lei and Li, 2020). The NLR gene-mediated disease resistance response has a dose effect (Howles et al., 2005). Simultaneously, NLR proteins can lead to activation of plant disease resistance responses (Tao et al., 2000). However, during normal plant development, NLR genes are strictly regulated to prevent induced self-activating reactions (Du et al., 2021), hence preventing abnormal plant growth and development. In this study, in the mutant lines overexpressing Xa47, cell death was not observed (Supplementary Figures 4A-H), and thus, the overexpression of Xa47 did not spontaneously activate the rice ETI response to the HR-like phenotype.
The gene OsNPR1 is essential for plant disease resistance, especially for resistance to rice bacterial blight (Chern et al., 2005). The OsNPR1 gene was expressed in response to Xoo at different times after inoculation in 35S::Xa47-JG30 plants, and OsNPR1 expression was significantly higher in 35S::Xa47-JG30 plants than in those of the wild type. Expression of the OsNPR1 gene in XCas9 plants was also induced by Xoo at different times after inoculation, but expression was significantly lower in XCas9 plants than in those of the wild type. Therefore, Xa47 can strongly activate the expression of OsNPR1 and thus stimulate OsNPR1-mediated disease resistance response in rice. Expression of pathogenesis related(PR) genes tends to promote plant cell death, hence inhibiting the spread of harmful microorganisms (Kim et al., 2011; Hu et al., 2017). Notably, both disease process-related genes OsPR1a and OsPR10 were significantly activated with significantly higher expression in the 35S::Xa47-JG30 strain than in the wild type when resisting infestation by Xoo (Figures 7A-D). Both OsPR1a and OsPR10 were also significantly activated with significantly higher expression in G252 than in XCas9 plants in response to Xoo infestation (Figures 5A-D). The results suggested that Xa47 regulates the resistance response in rice.
Identifying the level of resistance of an R gene to pathogens is also an assessment of the value and potential of the application of that gene (Feng et al., 2022). In this study, Xa47 was determined to be an R gene with broad-spectrum resistance by multiple methods. There were three essential observations: (1) Xa47 occurred in some BB-resistant rice; (2) loss-of-function mutations of Xa47 led to loss of resistance against Xoo; and (3) overexpression of Xa47 increased the resistance of JG30 to BB (Figures 2-7). China is a rice-producing nation, and most rice-growing regions are affected by Xoo (Lu et al., 2021). Representative Xoo strains, including C1, C2, C3, C4, C5, C6, C7, C8, and C9, have been discovered in the disease-endemic regions. Therefore, the use of isolates from diverse rice-growing regions in China to evaluate the resistance of rice lines to BB, particularly during gestation, was an important aspect of this work.
Previously, it was reported that iTALEs reduce Xa1-mediated resistance (Zhang et al., 2020). In this study, the resistance mediated by Xa47 was also inhibited by iTALEs. Notably, G252 showed resistance to PXO99 and PB strains. In 35S::Xa47-JG30 plants inoculated with PXO99 strains, lesion length was approximately 6.0 cm, whereas the lesion length in JG30 and IRBB1 plants was more than 10 cm (Figures 9A, B). The difference in resistance between G252 and 35S::Xa47-JG30 plants might be attributed to differences in genetic background. Moreover, the results indicated that Xa47-mediated resistance exceeds Xa1 resistance. The isolation and identification of Xa47 will not only reduce yield loss of rice due to Xoo infection but will also enable crop breeders to include Xa47 in breeding programs. Notably, 100 BC2F16 generations of Yuanjiang common wild rice IL materials, which possess a diverse antimicrobial spectrum, were available for selection of rice varieties. The varieties containing Xa47 (YN-80, YN-79, YN-78, YN-77, YN-76, YN-75, YN-74, and YN-73) that were isolated from 80 local rice varieties can be used as novel donors in rice breeding efforts. Therefore, the results of this study can help expedite the development of leaf blight-resistant cultivars in Yunnan Province and adjacent areas.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
ZC and LC designed the research. YL, XK, YZ, FY, DZ, CJ, LL, JL, TY and LW performed the experiments and analyzed the data. YL and LC wrote the manuscript. QZ, SX and BW revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Yunnan Yanlongan Academician Expert Workstation (no. 202005AF150032), the Yunnan Seed Seed Industry Joint Laboratory (no. 202205AR070001), the Yunnan Basic Research Special Project (no. 202001AT070015 and 202001AT070003), the Yunnan Science and Technology Talents and Platform Project (no. 2019HB034 and 202005AM070029), the Yunnan Young Top Talents Special Project (no. YNWR-QNBJ-2018-284), the National Natural Science Foundation Regional Project (no. 31960374), and the Central Guidance Local Science and Technology Development Fund Plan (no. 202207AB110012), respectively.
Acknowledgments
We are grateful for the Xoo strains that were provided by Dr. Gongyou Chen (College of Agriculture and Biology, Shanghai Jiaotong University, Shanghai, China).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1037901/full#supplementary-material
References
Ahuja, I., Kissen, R., Bones, A. M. (2012). Phytoalexins in defense against pathogens. Trends Plant Sci. 17, 73–90. doi: 10.1016/j.tplants.2011.11.002
Ainsworth, E. A. (2008). Rice production in a changing climate: A meta-analysis of responses to elevated carbon dioxide and elevated ozone concentration. Glob. Change Biol. 14, 1642–1650. doi: 10.1111/j.1365-2486.2008.01594.x
Bailey, P. C., Schudoma, C., Jackson, W., Baggs, E., Dagdas, G., Haerty, W., et al. (2018). Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 19, 23. doi: 10.1186/s13059-018-1392-6
Balachiranjeevi, C. H., Naik, S. B., Kumar, V. A., Harika, G., Mahadev, S. H. K., Hajira, S., et al. (2018). Marker-assisted pyramiding of two major, broad-spectrum bacterial blight resistance genes, Xa21 and Xa33 into an elite maintainer line of rice, DRR17B. PLoS One 13, e0201271. doi: 10.1371/journal.pone.0201271
Bednarek, P. (2012). Chemical warfare or modulators of defence responses-the function of secondary metabolites in plant immunity. Curr. Opin. Plant Biol. 15, 407–414. doi: 10.1016/j.pbi.2012.03.002
Boch, J., Bonas, U. (2010). Xanthomonas AvrBs3 family-type III effectors: Discovery and function. Annu. Rev. Phytopathol. 48, 419–436. doi: 10.1146/annurev-phyto-080508-081936
Boch, J., Bonas, U., Lahaye, T. (2014). TAL effectors-pathogen strategies and plant resistance engineering. New Phytol. 204, 823–832. doi: 10.1111/nph.13015
Bogdanove, A. J., Schornack, S., Lahaye, T. (2010). TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 13, 394–401. doi: 10.1016/j.pbi.2010.04.010
Chen, S., Huang, Z., Zeng, L., Yang, J., Liu, Q., and Zhu, X., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, S., Huang, Z., Zeng, L., Yang, J., Liu, Q., Zhu, X. (2008). High-resolutionmapping and gene prediction of Xanthomonas oryzae pv. oryzae resistance gene Xa7. Mol. Breeding 22, 433–441. doi: 10.1007/s11032-008-9187-1
Chen, X., Liu, P., Mei, L., X., Chen, L., Liu, H., et al. (2021). Xa7, a new executor R gene that confers durable and broad-spectrum resistance to bacterial blight disease in rice. Plant Commun. 2, 100143. doi: 10.1016/j.xplc.2021.100143
Chen, X., Ronald, P. C. (2011). Innate immunity in rice. Trends Plant Sci. 16, 451–459. doi: 10.1016/j.tplants.2011.04.003
Chen, S., Wang, C., Yang, J., Chen, B., Zhu, X. (2020). Identification of the novel bacterial blight resistance gene xa46(t) by mapping and expression analysis of the rice mutant h120. Sci. Rep. 10, 12642. doi: 10.1038/s41598-020-69639-y
Chern, M., Fitzgerald, H. A., Canlas, P. E., Navarre, D. A., Ronald, P. C. (2005). Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant Microbe Interact. 18, 511–520. doi: 10.1094/MPMI-18-0511
Chu, Z., Yuan, M., Yao, J., Ge, X., Yuan, B., Xu, C., et al. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20, 1250–1255. doi: 10.1101/gad.1416306
Dash, A. K., Rao, R. N., Rao, G. J., Verma, R. L., Katara, J. L., Mukherjee, A. K., et al. (2016). Phenotypic and marker-assisted genetic enhancement of parental lines of rajalaxmi, an elite rice hybrid. Front. Plant Sci. 13. doi: 10.3389/fpls.2016.01005
Dodds, P. N., Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Du, D., Zhang, C. W., Xing, Y. D., Lu, X., Cai, L. J., Yun, H., et al. (2021). The CC-NB-LRR OsRLR1 mediates rice disease resistance through interaction with OsWRKY19. Plant Biotechnol. J. 19, 1052–1064. doi: 10.1111/pbi.13530
Feng, Z., Li, M., Xu, Z., Gao, P., Wu, Y., Wu, K., et al. (2022). Development of rice variety with durable and broad-spectrum resistance to blast disease through marker-assisted introduction of pigm gene. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.937767
Gu, K., Yang, B., Tian, D., Wu, L., Wang, D., Sreekala, C., et al. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature . 435, 1122–1125. doi: 10.1038/nature03630
He, C., Liu, H., Chen, D., Xie, W. Z., Wang, M., Li, Y., et al. (2021). CRISPR-Cereal: a guide RNA design tool integrating regulome and genomic variation for wheat, maize and rice. Plant Biotechnol. J. 19, 2141–2143. doi: 10.1111/pbi.13675
Howles, P., Lawrence, G., Finnegan, J., McFadden, H., Ayliffe, M., Dodds, P., et al. (2005). Autoactive alleles of the flflax L6 rust resistance gene induce non-race-specifific rust resistance associated with the hypersensitive response. Mol. Plant Microbe Interact. 18, 570–582. doi: 10.1094/MPMI-18-0570
Hu, K., Cao, J., Zhang, J., Xia, F., Ke, Y., Zhang, H., et al. (2017). Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants 3, 17009. doi: 10.1038/nplants.2017.9
Hutin, M., Sabot, F., Ghesquiére, A., Koebnik, R., Szurek, B. (2015). A knowledge-based molecular screen uncovers a broad-spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 84, 694–703. doi: 10.1111/tpj.13042
Hutin, M., Sabot, F. O., Ghesquie`re, A., Koebnik, R., Szurek, B. (2016). A knowledge-based molecular screen uncovers a broadspectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 84, 694–703. doi: 10.1111/tpj.13042
Hu, L., Wu, Y., Wu, D., Rao, W. W., Guo, J. P., Ma, Y. H., et al. (2017). The coiled-coil and nucleotide binding domains of BROWN PLANTHOPPER RESISTANCE14 function in signaling and resistance against planthopper in rice. Plant Cell. 29, 3157–3185. doi: 10.1105/tpc.17.00263
Iyer, A. S., McCouch, S. R. (2004). The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol. Plant Microbe Interact. 17, 1348–1354. doi: 10.1094/MPMI.2004.17.12.1348
Jiang, G., Xia, Z., Zhou, Y., Wan, J., Li, D., Chen, R., et al. (2006). Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAg1. Mol. Genet. Genomics 275, 354–366. doi: 10.1007/s00438-005-0091-7
Jiang, N., Yan, J., Liang, Y., Shi, Y., He, Z., Wu, Y., et al. (2020). Resistance genes and their interactions with bacterial blight/leaf streak pathogens (Xanthomonas oryzae) in rice (Oryza sativa l.)- an updated review. Rice 13, 3. doi: 10.1186/s12284-019-0358-y
Ji, Z., Ji, C., Liu, B., Zou, L., Chen, G., Yang, B. (2016). Interfering TAL effectors of Xanthomonas oryzae neutralize r-gene-mediated plant disease resistance. Nat. Commun. 7, 13435. doi: 10.1038/ncomms13435
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Ke, Y., Deng, H., Wang, S. (2017). Advances in understanding broad-spectrum resistance to pathogens in rice. Plant J. 90, 738–748. doi: 10.1111/tpj.13438
Kim, S. M. (2018). Identification of novel recessive gene xa44(t) conferring resistance to bacterial blight races by QTL linkage analysis using an SNP chip. Theoret Appl. Genet. 131, 2733–2743. doi: 10.1007/s00122-018-3187-2
Kim, S. G., Kim, S. T., Wang, Y., Yu, S., Choi, I. S., Kim, Y. C., et al. (2011). The RNase activity of rice probenazole-induced protein1 (PBZ1) plays a key role in cell death in plants. Mol. Cells 31, 25–31. doi: 10.1007/s10059-011-0004-z
Kim, S. M., Suh, J. P., Qin, Y., Noh, T. H., Reinke, R. F., Jena, K. K. (2015). Identification and fine-mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races in rice (Oryza sativa l.). Theoret Appl. Genet. 128, 1933–1943. doi: 10.1007/s00122-015-2557-2
Kroj, T., Chanclud, E., Michel-Romiti, C., Grand, X., Morel, ,. J. B. (2016). Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 210, 618–626. doi: 10.1111/nph.13869
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Le, R. C., Huet, G., Jauneau, A., Camborde, L., Tremousaygue, D., Kraut, A., et al. (2015). A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell . 161, 1074–1088. doi: 10.1016/j.cell.2015.04.025
Lei, T., Li, X. (2020). Enzyme formation by immune receptors. Science 370, 1163–1164. doi: 10.1126/science.abf2833
Li, W., Deng, Y., Ning, Y., He, Z., Wang, G. L. (2020). Exploiting broad-spectrum disease resistance in crops: from molecular dissection to breeding. Annu. Rev. Plant Biol. 71, 575–603. doi: 10.1146/annurev-arplant-010720-022215
Liu, W., Liu, J., Triplett, L., Leach, J. E., Wang, G. L. (2014). Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 52, 213–241. doi: 10.1146/annurev-phyto-102313-045926
Liu, Q., Yuan, M., Zhou, Y., Li, X., Xiao, J., Wang, S. (2011). A paralog of the MtN3/saliva family recessively confers race-specific resistance to xanthomonas oryzae in rice. Plant Cell Environ. 34, 1958–1969. doi: 10.1111/j.1365-3040.2011.02391.x
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△Cq method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, J. L., Li, Q. L., Wang, C. C., Wang, M. M., Zeng, D., Zhang, F., et al. (2021). Identification of quantitative trait loci associated with resistance to xanthomonas oryzae pv. oryzae pathotypes prevalent in south China. Crop J. 10, 498–507. doi: 10.1016/j.cj.2021.05.009
Luo, D., Huguet-Tapia, J. C., Raborn, R. T., White, F. F., Brendel, V. P., Yang, B. (2021). The Xa7 resistance gene guards the susceptibility gene SWEET14 of rice against exploitation by bacterial blight pathogen. Plant Commun. 2, 100164. doi: 10.1016/j.xplc.2021.100164
Mermigka, G., Amprazi, M., Mentzelopoulou, A., Amartolou, A., Sarris, ,. P. F. (2020). Plant and animal innate immunity complexes: Fighting different enemies with similar weapons. Trends Plant Sci. 25, 80–91. doi: 10.1016/j.tplants.2019.09.008
Neelam, K., Mahajan, R., Gupta, V., Bhatia, D., Gill, B. K., Komal, R., et al. (2020). High-resolution genetic mapping of a novel bacterial glight resistance gene xa-45(t) identified from Oryza glaber-rima and transferred to oryza sativa. Theor. Appl. Genet. 133, 689–705. doi: 10.1007/s00122-019-03501-2
Niño-Liu, D., Ronald, P., Bogdanove, A. (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324. doi: 10.1111/j.1364-3703.2006.00344.x
Peng, Z., Hu, Y., Zhang, J., Huguet-Tapia, J. C., Block, A. K., Park, S., et al. (2019). Xanthomonas translucens commandeers the host rate-limiting step in ABA biosynthesis for disease susceptibility. Proc. Natl. Acad. Sci. 116, 20938–20946. doi: 10.1073/pnas.1911660116
Porebski, S., Bailey, L. G., Baum, B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant mol. Biol. Rep. 15, 8–15. doi: 10.1007/bf02772108
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., Nelson, A. (2019). The global burden of pathogens and pests onmajor food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Song, W., Wang, G., Chen, L., Kim, H. S., Pi, L., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. doi: 10.1126/science.270.5243.1804
Spoel, S. H., Dong, X. (2012). How do plants achieve immunity? defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. doi: 10.1038/nri3141
Streubel, J., Pesce, C., Hutin, M., Koebnik, R., Boch, J., Szurek, B. (2013). Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. Oryzae New Phytol. 200 (3), 808–819. doi: 10.1111/nph.12411
Sun, X., Cao, Y., Yang, Z., Xu, C., Li, X., Wang, S., et al. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37, 517–527. doi: 10.1046/j.1365-313x.2003.01976.x
Tao, H., Shi, X., He, F., Wang, D., Xiao, N., Fang, H., et al. (2021). Engineering broad-spectrum disease-resistant rice by editing multiple susceptibility genes. J. Integr. Plant Biol. 63, 1639–1648. doi: 10.1111/jipb.13145
Tao, Y., Yuan, F. H., Leister, R. T., Ausubel, F. M., Katagiri, F. (2000). Mutational analysis of the arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell. 12. doi: 10.1105/tpc.12.12.2541
Tena, G., Boudsocq, M., Sheen, J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14, 519–529. doi: 10.1016/j.pbi.2011.05.006
Tian, D., Wang, J., Zeng, X., Gu, K., Qiu, C., Yang, X., et al. (2014). The rice TAL effectordependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell. 26, 497–515. doi: 10.1105/tpc.113.119255
van Wersch, S., Tian, L., Hoy, R., Li, X. (2020). Plant NLRs: the whistleblowers of plant immunity. Plant Commun. 1, 100016. doi: 10.1016/j.xplc.2019.100016
Wang, C., Fan, Y., Zheng, C., Qin, T., Zhang, X., Zhao, K. (2014). High-resolution genetic mapping of rice bacterial blight resistance gene Xa23. Mol. Genet. Genom. 289, 745–753. doi: 10.1007/s00438-014-0848-y
Wang, C., Zhang, X., Fan, Y., Gao, Y., Zhu, Q., Zheng, C., et al. (2015). XA23 is an executor r protein and confers broad-spectrum disease resistance in rice. Mol. Plant 8, 290–302. doi: 10.1016/j.molp.2014.10.010
Withers, J., Dong, X. (2017). Post-translational regulation of plant immunity. Curr. Opin. Plant Biol. 38, 124–132. doi: 10.1016/j.pbi.2017.05.004
Wu, Y., Xiao, N., Chen, Y., Yu, L., Pan, C., Li, Y., et al. (2019). Comprehensive evaluation of resistance effects of pyramiding lines with different broad-spectrum resistance genes against magnaporthe oryzae in rice (Oryza sativa l.). Rice (N Y) 1, 12–11. doi: 10.1186/s12284-019-0264-3
Xiang, Y., Cao, Y., Xu, C., Li, X., Wang, S. (2006). Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinaselike protein, is the same as Xa26. Theor. Appl. Genet. 113, 1347–1355. doi: 10.1007/s00122-006-0388-x
Xing, J. X., Zhang, D. Y., Yin, F. Y., Zhong, Q. F., Wang, B., Xiao, S. Q., et al. (2021). Identification and fine-mapping of a new bacterial blight resistance gene, Xa47(t), in G252, an introgression line of yuanjiang common wild rice (Oryza rufipogon). Plant Dis. 105, 4106–4112. doi: 10.1094/PDIS-05-21-0939-RE
Yang, B., Sugio, A., White, ,. F. F. (2006). Os8N3 is a host diseasesusceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci.USA. 103, 10503–10508. doi: 10.1073/pnas.0604088103
Yoshimura, S., Yamanouchi, U., Katayose, Y., Toki, S., Wang, Z., Kono, I., et al. (1998). Expression of Xa1, a bacterial blight-resistancegene in rice, is induced by bacterial inoculation. Proc. Nat. Acad. Sci. U. S. A. 95, 1663–1668. doi: 10.1073/pnas.95.4.1663
Yuan, M., Chu, Z., Li, X., Xu, C., Wang, S. (2009). Pathogen-induced expressional loss of function is the key factor in race-specific bacterial resistance conferred by a recessive. R Gene Xa13 Rice Plant Cell Physiol. 50 (5), 947–955. doi: 10.1093/pcp/pcp046
Yuan, M., Chu, Z., Li, X., Xu, C., Wang, S. (2010). The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. Plant Cell. 22, 3164–3176. doi: 10.1105/tpc.110.078022
Zhang, Y., Li, J., Gao, C. X. (2016). Generation of stable transgenic rice (Oryza sativa l.) by Agrobacterium-mediated transformation. Curr. Protoc. Plant Biol. 1, 235–246. doi: 10.1002/cppb.20004
Zhang, B., Zhang, H., Li, F., Ouyang, Y., Yuan, M., Li, X., et al. (2020). Multiple alleles encoding atypical NLRs with unique central tandem repeats in rice confer resistance to Xanthomonas oryzae pv. oryzae. Plant Commun. b1, 100088. doi: 10.1016/j.xplc.2020.100088
Zhang, F., Zhuo, D. L., Huang, L. Y., Wang, W. S., Zhou, ,. Y. L. (2015). Xa39, a novel dominant gene conferring broad-spectrum resistance to Xanthomonas oryzae pv. oryzae in rice. Plant Pathol. 64, 568–575. doi: 10.1111/ppa.12283
Keywords: Xa47, NLR, Xanthomonas oryzae pv. oryzae, broad-spectrum resistance, bacterial blight
Citation: Lu Y, Zhong Q, Xiao S, Wang B, Ke X, Zhang Y, Yin F, Zhang D, Jiang C, Liu L, Li J, Yu T, Wang L, Cheng Z and Chen L (2022) A new NLR disease resistance gene Xa47 confers durable and broad-spectrum resistance to bacterial blight in rice. Front. Plant Sci. 13:1037901. doi: 10.3389/fpls.2022.1037901
Received: 06 September 2022; Accepted: 31 October 2022;
Published: 24 November 2022.
Edited by:
Andres Mäe, Estonian Crop Research Institute, EstoniaReviewed by:
Zhiqian Pang, Citrus Research and Education Center, Institute of Food and Agricultural Sciences, University of Florida, United StatesNannan Yang, NSW Government, Australia
Copyright © 2022 Lu, Zhong, Xiao, Wang, Ke, Zhang, Yin, Zhang, Jiang, Liu, Li, Yu, Wang, Cheng and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zaiquan Cheng, Y2hlbmd6cTk5QHNvdWh1LmNvbQ==; Ling Chen, Y2xAeWFhcy5vcmcuY24=
 Yuanda Lu
Yuanda Lu Qiaofang Zhong1
Qiaofang Zhong1 Li Liu
Li Liu