
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 07 September 2021
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.705249
 Ahmed H. El-Sappah1,2,3†
Ahmed H. El-Sappah1,2,3† Kuan Yan1,3†
Kuan Yan1,3† Qiulan Huang1,3,4†
Qiulan Huang1,3,4† Md. Monirul Islam5
Md. Monirul Islam5 Quanzi Li6,7
Quanzi Li6,7 Yu Wang1,3
Yu Wang1,3 Muhammad Sarwar Khan8
Muhammad Sarwar Khan8 Xianming Zhao1,3
Xianming Zhao1,3 Reyazul Rouf Mir9
Reyazul Rouf Mir9 Jia Li1,3*
Jia Li1,3* Khaled A. El-Tarabily10,11*
Khaled A. El-Tarabily10,11* Manzar Abbas1,3*
Manzar Abbas1,3*Gene silencing is a negative feedback mechanism that regulates gene expression to define cell fate and also regulates metabolism and gene expression throughout the life of an organism. In plants, gene silencing occurs via transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS). TGS obscures transcription via the methylation of 5′ untranslated region (5′UTR), whereas PTGS causes the methylation of a coding region to result in transcript degradation. In this review, we summarized the history and molecular mechanisms of gene silencing and underlined its specific role in plant growth and crop production.
Gene silencing is a molecular mechanism that knocks down the gene expression in plants both in nature and in response to external stimuli (Wassenegger, 2002). It plays a pivotal role in plant defense mechanisms by detecting an aberrant RNA via nonsense-mediated messenger RNA (mRNA) decay (NMD), which could be fatal in case of remains in the RNA pool of the cell. Gene silencing mechanisms in plants are transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS) (Kelly and Aramayo, 2007). TGS is a nuclear-localized mechanism, which quenches transcription by blocking a promoter region for the binding of transcriptional machinery (Vaucheret and Fagard, 2001). Different methods of TGS are RNA-directed DNA methylation (RdDM), genomic imprinting, paramutation, transposon silencing, transgene silencing, and position effect. Notably, TGS is predominantly responsible for transposon and transgene silencing, but PTGS plays a limited role in this silencing (Wakimoto, 1998).
Post-transcriptional gene silencing is a cytoplasm-localized phenomenon to precisely target and degrade mRNA transcripts of specific genes (Vaucheret et al., 2001). Different methods of PTGS are RNA interference (RNAi), clustered regularly interspaced short palindromic repeats (CRISPR/Cas9), and NMD (Zebec et al., 2016). Recently, several studies have been conducted on gene silencing by deploying RNAi, the virus-induced gene silencing (VIGS), and CRISPR/Cas9 to enhance the resistance of plants against pathogens, drought tolerance, and lingo-cellulose pathway engineering (Borrelli et al., 2018; Abbas et al., 2020). In plants, small RNAs (sRNAs), such as microRNA (miRNA) and small-interfering RNA (siRNA) play a key role in plant fight against pathogens (Axtell, 2013). Gene silencing causes the periodic knock down of gene expression at the mRNA or protein level. Gene silencing spatiotemporally controls the regulation of gene networks, which subsequently regulate developmental processes in plant metabolism, such as genome stability, the detoxification of plant waste, and allergens (Mirouze et al., 2009; Ito, 2013).
Position variegation or position effect is a kind of gene silencing that was first discovered in Drosophila melanogaster by Muller (1930), which opened up new avenues of studies, such as genetics and functional genomics, and subsequently paved the way for the exploration of other possible gene silencing mechanisms in different organisms. After 1 year, a new type of gene silencing phenomenon was discovered in petunia, namely “co-suppression” (Napoli et al., 1990). VIGS was unexpectedly discovered in a hit and trial experiment (Ruiz et al., 1998). An mRNA sequence which encodes color pigments was artificially designed and ligated in a vector of virus origin, which resulted in the synthesis of double-stranded RNA (dsRNA) with its complementary counterpart on introduction in to petunia plant, dsRNA molecules subsequently triggered RNA induced gene silencing complex (RISC) which cleaved all dsRNA molecules of that specific gene and resulted in albino phenotype.
A remarkable breakthrough in gene silencing research was the discovery of RNAi when Fire et al. (1998) introduced artificially designed single- and double-stranded unc-22-nt RNA molecules in Caenorhabditis elegans to observe phenotypic outcomes. As a surprise, albino phenotype was observed because both sense and antisense RNA strands were completely degraded (Fire et al., 1998). CRISPR is a robust gene silencing mechanism discovered in early 1993 by Francisco Mojica in prokaryotes (Nobel Prize) (Mojica et al., 2005), which was employed first time by Zhang et al. (2013) for genetic engineering in eukaryotes (Cong et al., 2013). A pictorial representation of the history of all silencing techniques has been illustrated in Figure 1. In this review, we comprehensively underlined the mechanism of gene silencing, molecular mechanisms behind gene silencing, and their pivotal roles in plant growth and crop production.
RNA-directed DNA methylation is a fundamental epigenetic gene silencing phenomenon almost found in all living organisms (Wassenegger et al., 1994). In RdDM, siRNAs transactivate the RISC of ∼21–24 nt, which regulates gene silencing via homologous DNA methylation. RISC is comprised of the following components: Argonaut (AGO) proteins, DNA methyltransferase (DNMT), chromatin remodeling complexes, and RNA polymerase IV-V (Mette et al., 2000; Sijen et al., 2001). The RdDM pathway is induced by the generation of dsRNA by transposable elements (TEs), transcribed inverted repeats (IR), viral replication intermediates, and RNA-directed RNA polymerase (RDR) (Mette et al., 2000; Sijen et al., 2001). DNA methylation predominantly occurs at the following nucleotide combinations of an RNA-DNA sequence homology: CG, CHG, and CHH (H is; A, C, or T) (Pélissier and Wassenegger, 2000). Both symmetric sequences, such as CG and CHG are methylated by methyltransferase 1 (MET1) and chromomethylase 3 (CMT3), whereas an asymmetric sequence of CHH is methylated by DNMTs, chromomethylase 2 (CMT2), and domains rearranged methyltransferase 2 (DRM2) (Huang and Zhu, 2014; Ito and Kakutani, 2014).
The RdDM pathway was originally discovered in Arabidopsis thaliana, which completes in two sequential steps: the biogenesis of ∼23–24 nt siRNAs and subsequent de novo methylation (Figure 2; Wierzbicki, 2012; Matzke et al., 2015). There are two types of pathways for the biogenesis of siRNAs: canonical and non-canonical, and the selection of either pathway is decided by polymerase IV, RDR2, and Dicer homolog 3 (DCL3). The initiation of the canonical pathway begins with the binding of DNA transcription factor 1/Sawadee homeodomain homolog 1 (DTF1/SHH1) to lysine 4 (K4) of non-methylated histone 3 (H3) and eventually causes the methylation of lysine 9 (K9), which stimulates the transcription of a specific DNA region due to an interaction between polymerase IV and RDR2 with the assistance of chromatin remodeling SNF2 domain-containing protein Classy 1 (CLSY1), and finally, the biogenesis of dsRNA is started (Enke et al., 2011; Haag et al., 2012; Du et al., 2014). By using the endonuclease activity of endo-ribonuclease DCL3, dsRNA molecules are cleaved to produce ∼23–24 nt siRNAs (Wu et al., 2012; Kanno et al., 2013). Approximately 23–24 nt mature siRNAs intercalate with AGO4 or AGO6 to induce methylation (Kanno et al., 2013).
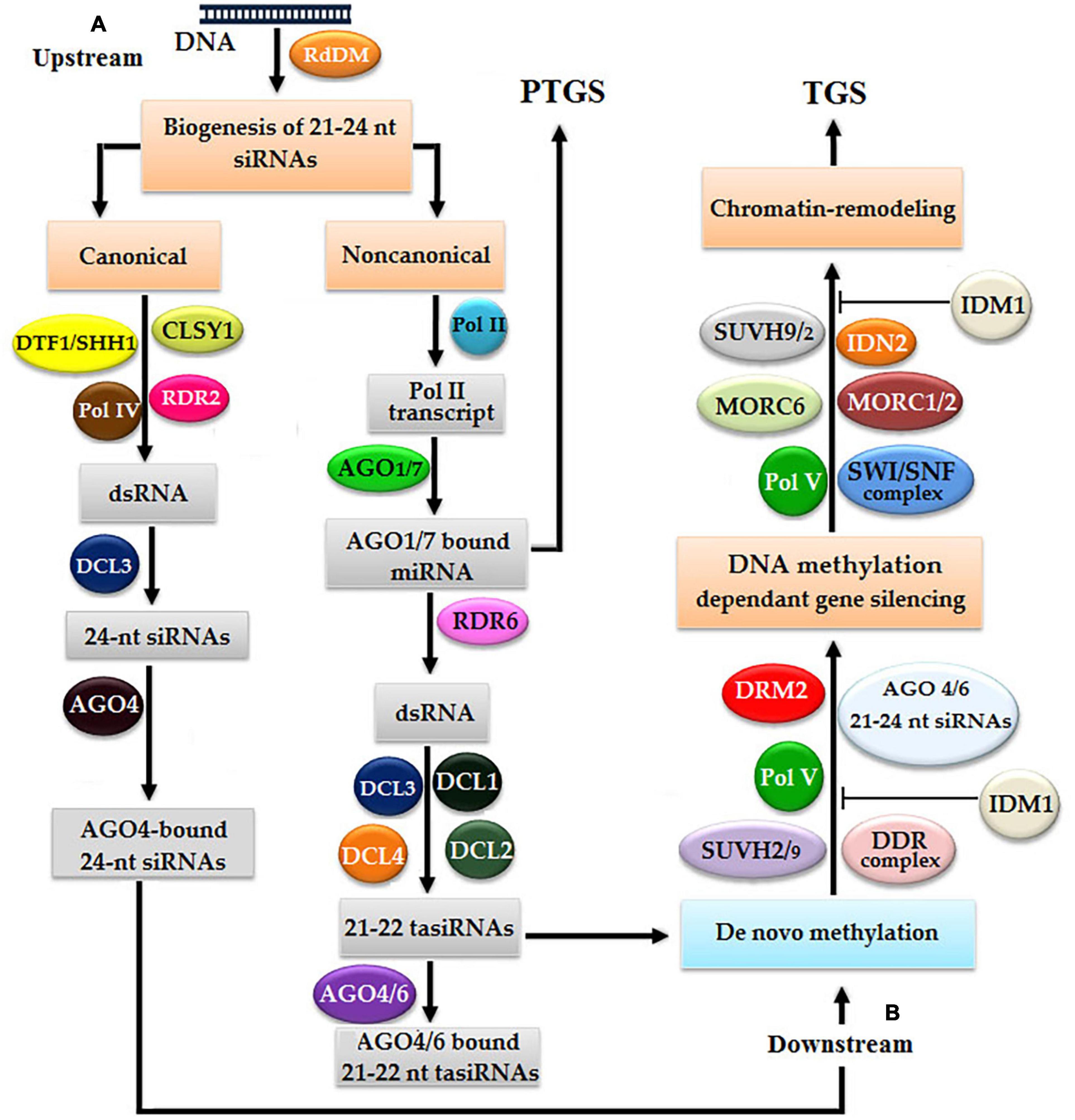
Figure 2. Arabidopsis models of RNA-directed DNA methylation (RdDM): double-stranded RNAs (dsRNAs) proceed into small-interfering RNAs (siRNAs) via canonical and non-canonical pathways. Both pathways undergo two steps: (A) in the first step, siRNAs are produced, which subsequently bind with argonaut (AGO) and (B) in the second step, DNA methylation leads to chromatin remodeling resulting in gene silencing via transcriptional gene silencing (TGS).
In the non-canonical pathway, non-coding RNAs of the telomere-associated satellite (TAS) gene are transcribed by polymerase II, which subsequently bind with AGO1 or AGO7. Some TAS transcripts are cleaved by miRNAs, and one cleaved RNA is transformed into dsRNA by RNA-dependent RNA polymerase 6 (RDR6), which acts as a substrate of Dicer-like 4 (DCL4) to finally produce ∼21–22 nt transacting siRNAs (ta-siRNAs) (Kanno et al., 2013). Finally, if ta-siRNAs make a complex with AGO4 or AGO6, then the TGS pathway is activated, contrastingly, if ta-siRNAs make a complex with AGO1 or AGO7, then the PTGS pathway is activated (Pontier et al., 2012; Marí-Ordóñez et al., 2013; Nuthikattu et al., 2013; Panda and Slotkin, 2013).
DNA methyltransferase is responsible for the methylation of DNA, which acts as a switch on/off of gene expression (Harris et al., 2018), and depends upon polymerase V, AGO4 bound ∼21–24-nt RNA complex, DNMT, DRM2, SRA-domain-containing proteins SUVH1, SUVH2, SU(VAR)3-9, and several other proteins (Fischer et al., 2006). SUVH proteins are predominantly responsible for the establishment of a heterochromatin chromatic domain using H3K9me. Arabidopsis harbor 10 members of the SUVH gene family, of which SUVH1, SUVH2, and SUVH4 have non-unanimous effects on heterochromatic methylation. SUV39H1 is involved in H3K9me of heterochromatin and along with Sirtuin 1 (SIRT1) regulates facultative heterochromatin (Vaquero et al., 2007). SUV39H1 binds with DNMTs 1 and 3a with the help of its PHD-like and HP1beta motif to regulate both global epigenetic modifications, such as DNA methylation and histone methylation (François et al., 2003). SUVH1 and SUVH3 are involved in the methylation of euchromatin by binding with DNAj1 and DNAj2, and act as transcriptional antisilencing factors (Harris et al., 2018). SUVH2 plays a key role in gene silencing in Arabidopsis by the heterochromatin formation, any mutation in SUVH2 resulted in the hypomethylation of DNA at chromocenter heterochromatin and its overexpression causes ectopic heterochromatization, which also resulted in severe loss in growth (Fischer et al., 2006).
The initiation of methylation starts with the initiation of transcription of the target DNA locus with the binding of polymerase V and methyl-DNA-binding proteins SUVH2/9, where the role of SUVH2 is dominant over SUVH9 (Johnson et al., 2014; Liu et al., 2014). SUVHs are comprised of a DDR complex [defective in meristem silencing 3 (DMS3), dopamine receptor D1 (DRD1), and DRM2], which mediates polymerase V functionality, enhances DNA methylation, and quenches the target gene expression (Law et al., 2011). Finally, chromatin remodeling takes place by SUVH2/9, while in this study, the role of SUVH9 is significant in the inhibition of IDM1. SUVH9 also mediates with SWI/SNF-dependent chromatin remodeling and development of a microconidia MORC6 complex (Zhu et al., 2013). Thus, the silencing of a target gene is accomplished with the end of chromatin remodeling (Figure 2; Matzke and Mosher, 2014).
Genomic imprinting is an epigenetic phenomenon in which alleles of the same gene express divergently depending upon the parent of origin like in alternative splicing (Feng et al., 2010). Genomic imprinting may affect the inactivation of entire chromosomes, such as paternal X-chromosome in marsupials (Cooper et al., 1971). Based on the dominance, imprinted genes are of two types, such as maternally expressed imprinted genes (MEGs) and paternally expressed imprinted genes (PEGs) (Garnier et al., 2008; Köhler et al., 2012). In plants, all dominant imprinted genes are expressed only in the endosperm of flowering plants (García-Aguilar and Gillmor, 2015). During endosperm development, multiple nuclear divisions are followed by the formation of distinct mitotic domains, which determine the peripheral endosperm (PEN), micropylar endosperm (MCE), or chalazal endosperm (CZE) (Boisnard-Lorig et al., 2001; Stangeland et al., 2003). Noticeably, the imprinted genes in plants and animals are only 2% and predominantly express in the CZE endosperm (Gehring, 2013). For example, paternally inherited Pr1 reciprocal kernel-color allele of maize displayed colorless or spotted kernels, whereas maternally inherited same allele displayed colored kernels.
Chromatin modifications have serious implications over the pattern of imprinted gene expression, such as methylated paternal allele remains transcriptionally silent in case of the demethylated maternal allele being transcriptionally active (Gehring et al., 2009). Genomic imprinting in A. thaliana got switched on due to a differential demethylation of DEMETER (DME) gene by DNA glycosylase, which dominantly expresses in female gametophyte (Choi et al., 2002; Schoft et al., 2011). The demethylation of DNA sequence repeats and TEs predominantly takes place by the removal of 5-methylcytosine. DMT is predominantly responsible for parental DNA methylation, and RdDM only occurs at MEG loci of parental allele (Figure 3; Köhler et al., 2012; Gehring, 2013; Zhang et al., 2013). H3K27me3 causes the suppression of the hypomethylated DNA of the maternal allele, but the polycomb repressive complex 2 (PRC2) interferes with the hypermethylated DNA of the parental allele at PGG to refrain it from the action of H3K27me3 resulted in the activation of the maternal allele (Figure 3; Weinhofer et al., 2010; Deleris et al., 2012; Makarevitch et al., 2013; Jermann et al., 2014).
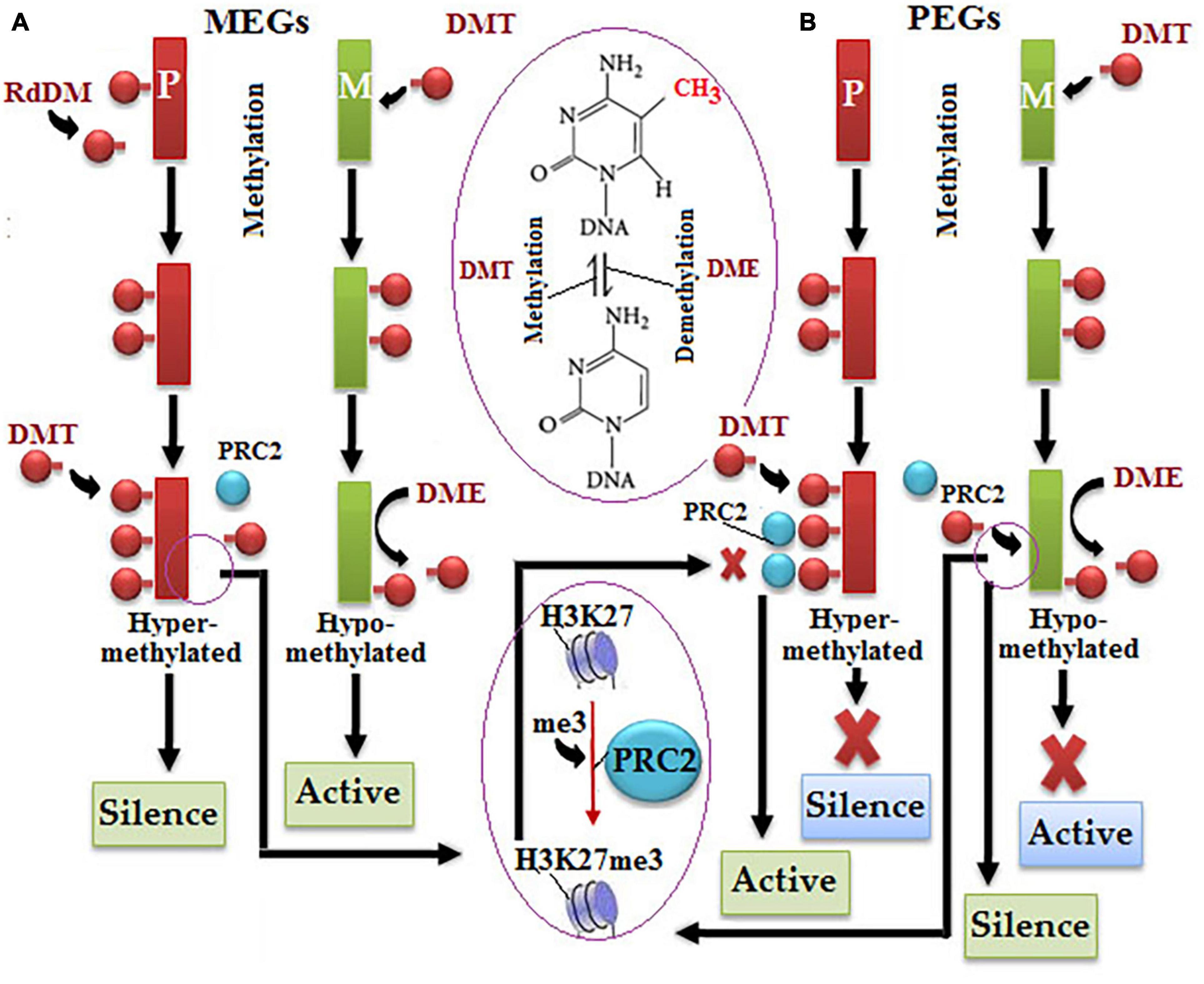
Figure 3. Molecular mechanism of alternative imprinting gene expression: (A) in the maternally expressed imprinted genes (MEGs) pathway, differential methylation of both maternal alleles occurs (green color), i.e., DEMETER (DME) downregulates DNA methylation by the hypomethylation process resulted in parental allele normal expression (red color), but increased methylation by DMT and polycomb repressive complex 2 (PRC2) causes histone methylation resulted in allele suppression, (B) in the paternally expressed imprinted genes (PEGs) pathway, the hypermethylated parental allele switches from the silent to active state under the action of PRC2 due to a halt in methylation by DMT, contrastingly hypomethylated maternal allele switches the active to silent state under the action of PRC2 via histone methylation.
Paramutation is an epigenetic phenomenon in which heritable changes in one allele are induced due to trans-interaction between two alleles at the same locus or different locus, which include DNA methylation and histone modifications (Chandler and Stam, 2004). The term “paramutation” was first coined by Alexander Brink to describe a puzzling phenomenon at the r1 locus in maize (Brink, 1956). A positive paramutagenic allele is capable of transforming the second allele from a paramutable state to a new paramutagenic allele state (Chandler and Alleman, 2008). Paramutation is a kind of trans-regulation mechanism, which falls under the category of TGS (Hollick et al., 2000; Wagner et al., 2008). Noticeably, the mechanism of paramutation is similar to genetic recombination, transposition effect, and other genetic mutations (Hale et al., 2007).
Three models are used to describe paramutation: RNA model, physical interaction, and RNA–physical interaction (Figure 4; Stam, 2009). In the RNA model, only paramutagenic repeats are first transcribed into mRNA and then catalyzed by RdRP to transform into dsRNA, and finally dsRNA is cleaved by Dicer-like protein into siRNA (Grewal and Jia, 2007; Slotkin and Martienssen, 2007; Zaratiegui et al., 2007). These methylated free-state siRNAs are responsible for RdDM, which resulted in the silencing of a paramutable allele (Bühler et al., 2006). In the physical model, a physical interaction is established between both paramutable and paramutagenic alleles with the help of a pairing protein complex and transform paramutagenic alleles into paramutable alleles (Gohl et al., 2008). In the last combo-model, RdDM is accompanied by the physical interaction between paramutable and paramutagenic alleles.
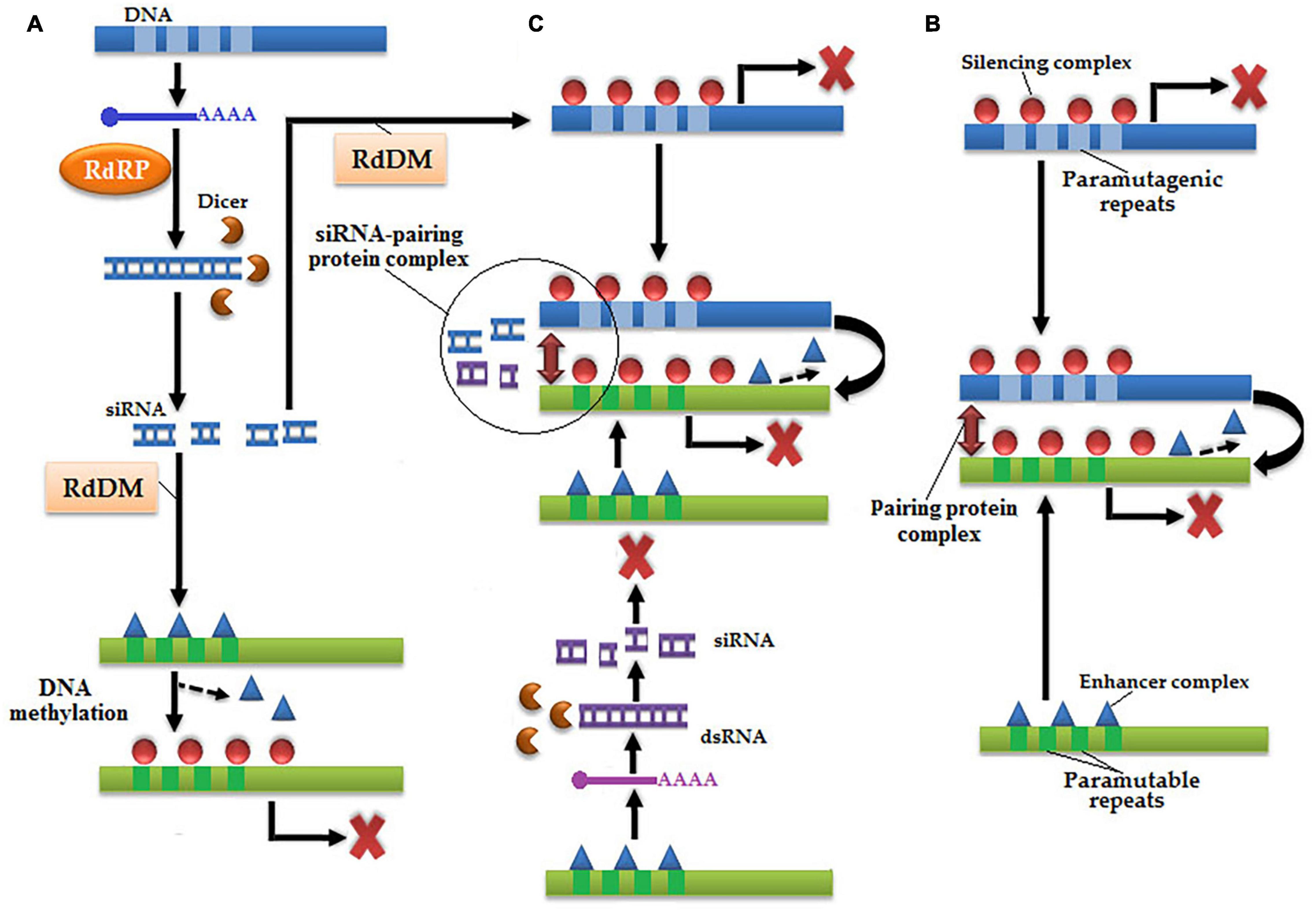
Figure 4. There are three models of paramutation: (A) RNA model, in which paramutagenic repeats of DNA are transcribed into dsRNA under the action of RdRP, which subsequently transformed into siRNA by a Dicer-like protein. Methylation of siRNA occurs via RdDM, which causes chromatin silencing of paramutable repeats by the addition of silencing complex inside enhancer complex resulted in the occurrence of paramutation. (B) Physical interaction, in which both paramutable and paramutagenic repeats physically interact by pairing protein complex and paramutation occurs epigenetically. (C) RNA-physical interaction, which is a combination of both abovementioned models, in which paramutable and paramutagenic repeats transcribed by RaRP into dsRNA, which are subsequently transformed into siRNA by a Dicer-like protein, and only cause chromatin silencing of paramutagenic sequence by RdDM. Subsequently, physical interaction is established via siRNA and pairing protein complex resulted in the modification of paramutable repeats.
Transposable elements are auto-replicative short DNA repeats that can translocate within the genome (Sun et al., 2015). The main classes of TEs are DNA transposons and retrotransposons (RTs), which are further divided into two subclasses, such as autonomous and sub-autonomous (Feschotte et al., 2002). Autonomous TEs can translocate by themselves while non-autonomous TEs are dependent on other TEs for their translocation. The family of RTs is comprised of long terminal repeats-RTs (LTR-RTs) (class I), non-LTR-RTs (class II), short interspersed nuclear elements (SINEs), and pseudogenes. LTR-RTs are 100 bp to >5 kb long major internal coding repeats found in the genome of fungi, plants, and protists. LTR-RTs harbor reverse transcriptase, integrase, protease, RNase H, and capsid/gag proteins, which are inevitable for their transposition. Based on their encoded gene products and degree of sequence similarity, LTRs are of the following types: Ty1-copia RTs, Ty3-gypsy RTs, BEL/pao family, terminal repeat RTs in miniature (TRIMs), and endogenous retroviruses (ERV) (Havecker et al., 2004).
The initiation of TE silencing occurs via the following two pathways, such as homology-dependent/identity-based and homology-independent/expression-based initiation of silencing (Figure 5; Fultz and Slotkin, 2017). In the upstream phase of a homology-dependent pathway, polymerase IV, RDR2, and DCL3 make a complex to produce 24-nt-long siRNAs from TEs associated with H3K9me (Nobuta et al., 2007; Huang et al., 2013; Law et al., 2013). In the downstream phase, a 24-nt-long siRNA molecule along with AGO4 or AGO6 protein interacts with polymerase V scaffold transcript resulting in transcriptional silencing of homologous TEs by the methylation of both DNA and H3K9me (Teixeira et al., 2009; Ito et al., 2011; Wierzbicki, 2012). Expression-dependent silencing of TEs is a kind of post-transcriptional silencing, which unleashes the synthesis of 21–22-nt-long siRNAs of target TEs with the help of miRNAs. Subsequently, the activation of the RNAi pathway helps in identifying and cleaving TE transcripts (Dunoyer et al., 2010; McCue et al., 2012).
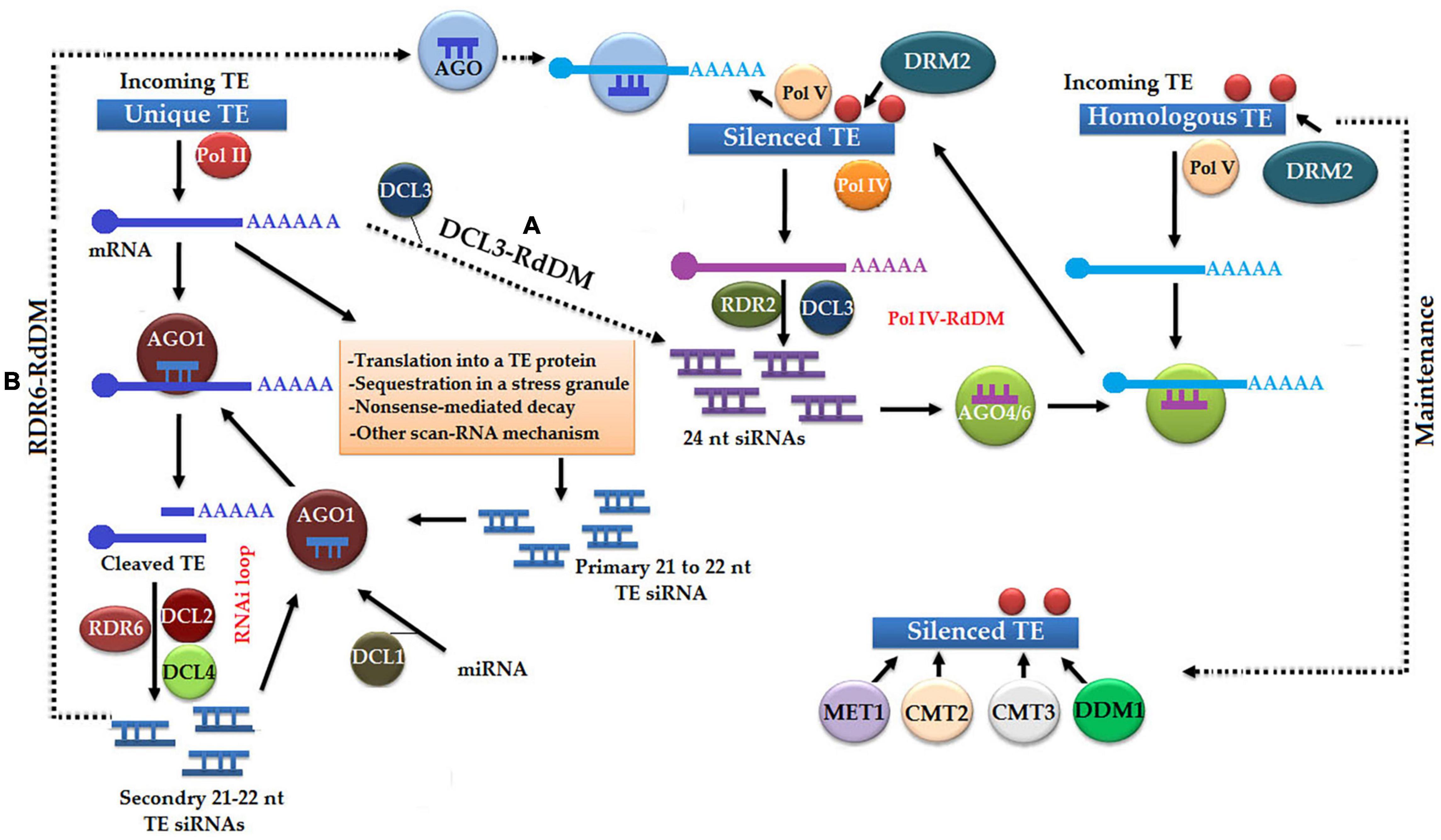
Figure 5. Homology-independent initiation of silencing is a primary pathway, which is unleashed with the expression of unique transposable elements (TEs) under the action of Pol II and messenger RNA (mRNA) undergoing post-transcriptional silencing via RNA interference (RNAi). In this pathway gene, silencing occurs by the following methods: (A) Dicer homolog 3- (DCL3-) RdDM, in which 24-nt siRNAs play a major role in a homology-dependent pathway, and (B) RNA dependent RNA polymerase 6- (RDR6-) RdDM, in which AGO protein-based silencing complex causes the silencing of Pol V transcripts. Homology-dependent pathway, in which TEs are transcribed by Pol IV and digested into 24-nt siRNAs by RDR2 and DCL3, which subsequently bind with AGO4 or AGO6 to become active and interact with polymerase V scaffolding transcript resulted in transcriptional TE silencing. Finally, silencing state is restored with the help of methyltransferase 1 (MET1), chromomethylase 2 (CMT2), chromomethylase 3 (CMT3), and DECREASE IN DNA METHYLATION 1 (DDM1).
Sometimes, the expression level of a successful transgene is not up to the mark due to: (a) the transgene insertion in heterochromatin or junk DNA (Tzfira et al., 2004) and (b) formation of complex T-DNA structures due to the binding of multiple integrated T-DNAs at a single locus (Gelvin, 2003). Transgene silencing occurs via: (a) PTGS triggered mechanisms by sense or antisense transgenes, IR, hairpin RNAs (hp-RNAs), and (b) VIGS (Figure 6; Wassenegger, 2002; Wang and Metzlaff, 2005). In transgene silencing, the first single-stranded sense RNA (ssRNA) becomes the dsRNA under the action of RDR6, RNA-binding suppressor of gene silencing 3 (SGS3), and RNA helicase (SDE3), and then cleaved by the RISC (Bond and Baulcombe, 2015). On the other hand, antisense ssRNA either directly hybridizes with endogenous ssRNA or indirectly hybridizes with the ssRNA of sense transgene to produce dsRNA to be finally cleaved by the RISC. Dicer-like enzymes (DCLs) determine the selection of pathways for the conversion of ssRNA to dsRNA, such as DCL3 being responsible for the selection of the TGS pathway and DCL4 for the PTGS pathway (Brodersen and Voinnet, 2006).
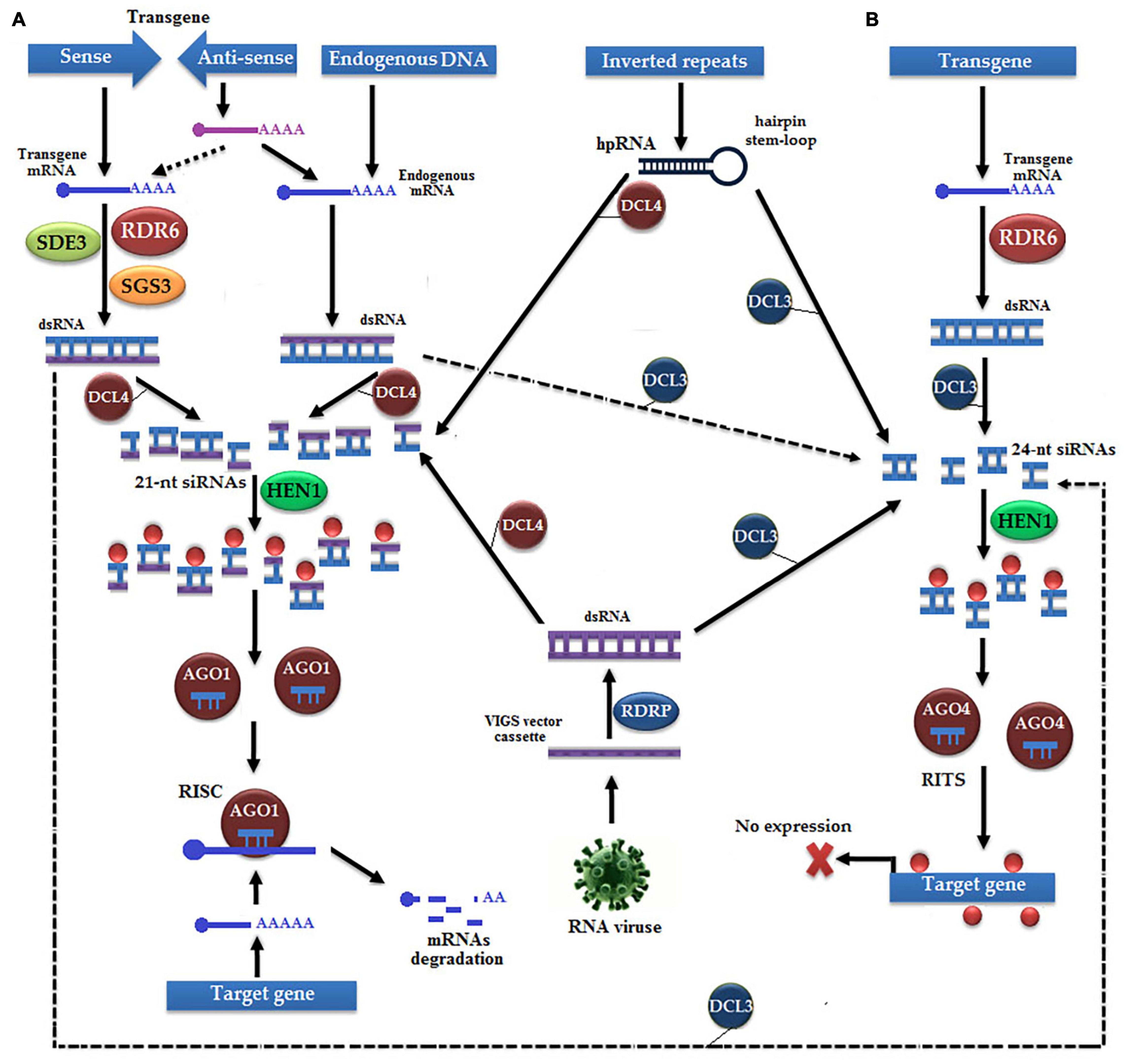
Figure 6. Transgene silencing occurs by the following two pathways: (A) post-transcriptional gene silencing (PTGS), which is initiated by sense or antisense transgenes, IR, hp-RNAs, and virus-induced gene silencing (VIGS). The sense strand is transcribed into mRNA, and transformed into dsRNA under the action of RDR6, suppressor of gene silencing 3 (SGS3), and RNA helicase (SDE3). Subsequently, dsRNA is either transformed into methylated 21-nt siRNAs by Dicer-like 4 (DCL4) and Hua Enhancer 1 (HEN1) or transformed into 24-nt siRNAs by AGO3 to cause gene silencing by the TGS pathway. The 21-nt siRNAs bind with AGO1 to make the RNA-induced gene silencing complex (RISC) that causes the degradation of mRNA of the target gene. Antisense strand is transcribed into mRNA, which either follows the same pathway with sense strand, or, like endogenous mRNA, transformed into dsRNA and bind with AGO to trigger the RISC. IR and VIGS follow either of the two available silencing pathways depending on the type of DCL. (B) TGS pathway, which is initiated with the transcription of transgene, dsRNA is formed under the action of RDR6, which is further cleaved into 24-nt siRNAs by DCL3, is methylated with HEN1, and make the RNA-induced transcriptional silencing (RITS) complex with the help of AGO4, which finally causes silencing.
In the PTGS pathway, DCL4 performs an exonuclease activity to cleave dsRNA into 21-nt siRNAs, which are subsequently methylated by sRNA-specific methyltransferase Hua Enhancer 1 (HEN1) (Li et al., 2005). Methylated siRNA binds with AGO1 to make the RISC, which cleaves the entire mRNA of transgene and results in no phenotype or transgene silencing (Vazquez et al., 2004). Surprisingly, in some cases of transgene silencing, both sense and antisense strands undergo the TGS pathway under the action of DCL3 proteins, such as multicopy transgene loci, hp-RNA, and VIGS (Mishiba et al., 2005). The first mRNA of the transgene is converted into dsRNA with the help of RDR2 protein, cleaved into 24-nt siRNAs by DCL3, methylated by HEN1 to become capable of binding with AGO4 to make the RNA-induced transcriptional silencing (RITS) complex that finally causes DNA methylation of the transgene (Wassenegger, 2002; Wang and Metzlaff, 2005).
Position effect is a variation in gene expression due to translocation or inversion as a result of crossing over, chromosomal aberration, and transgene insertion (Weiler and Wakimoto, 1995). Position effect variegation (PEV) and telomeric position effect (TPE) are the two reasons of gene silencing during position effect. The reasons for gene silencing in PEV are the translocation of a gene from euchromatin to heterochromatin and vice versa (Girton and Johansen, 2008), modification in nucleosome by histone methylation, deacetylation, or structural changes (Bannister et al., 2001), and a close distance between gene and heterochromatin (Ryan and Vandenbergh, 2002). Gene silencing in TPE occurs when the transgene is inserted within or nearby a telomeric region (Pedram et al., 2006). Mosaic repeats, such as TAS-like sequences play a key role in gene silencing (Doheny et al., 2008). Gene silencing in mosaic repeats entirely depends upon histone modifications, such as standard tri-methylation H3K4, H3K9, and H4K20 (Boivin et al., 2003; Andreyeva et al., 2005).
Epigenetic models that describe the molecular mechanism of PEV are cis-spreading and trans-interaction (Figure 7; Wakimoto, 1998). In the cis-spreading model, heterochromatin directly causes conformational changes in the packaging of euchromatin of transgene to refrain the binding of transcriptional machinery at a promoter resulted in transcriptional inhibition (Elgin, 1996). The cis-spreading model does not cover all aspects, such as some inserted genes induced variegation mode despite the long distance between the insertion site and breakpoint of heterochromatin (Henikoff and Dreesen, 1989; Weiler and Wakimoto, 1995). Trans-interactions or nuclear compartment is a comprehensive model to describe PEV, which means multiple interactions of different regions of heterochromatin and extensive chromosomal rearrangement resulted in gene silencing (Csink and Henikoff, 1996; Dernburg et al., 1996).
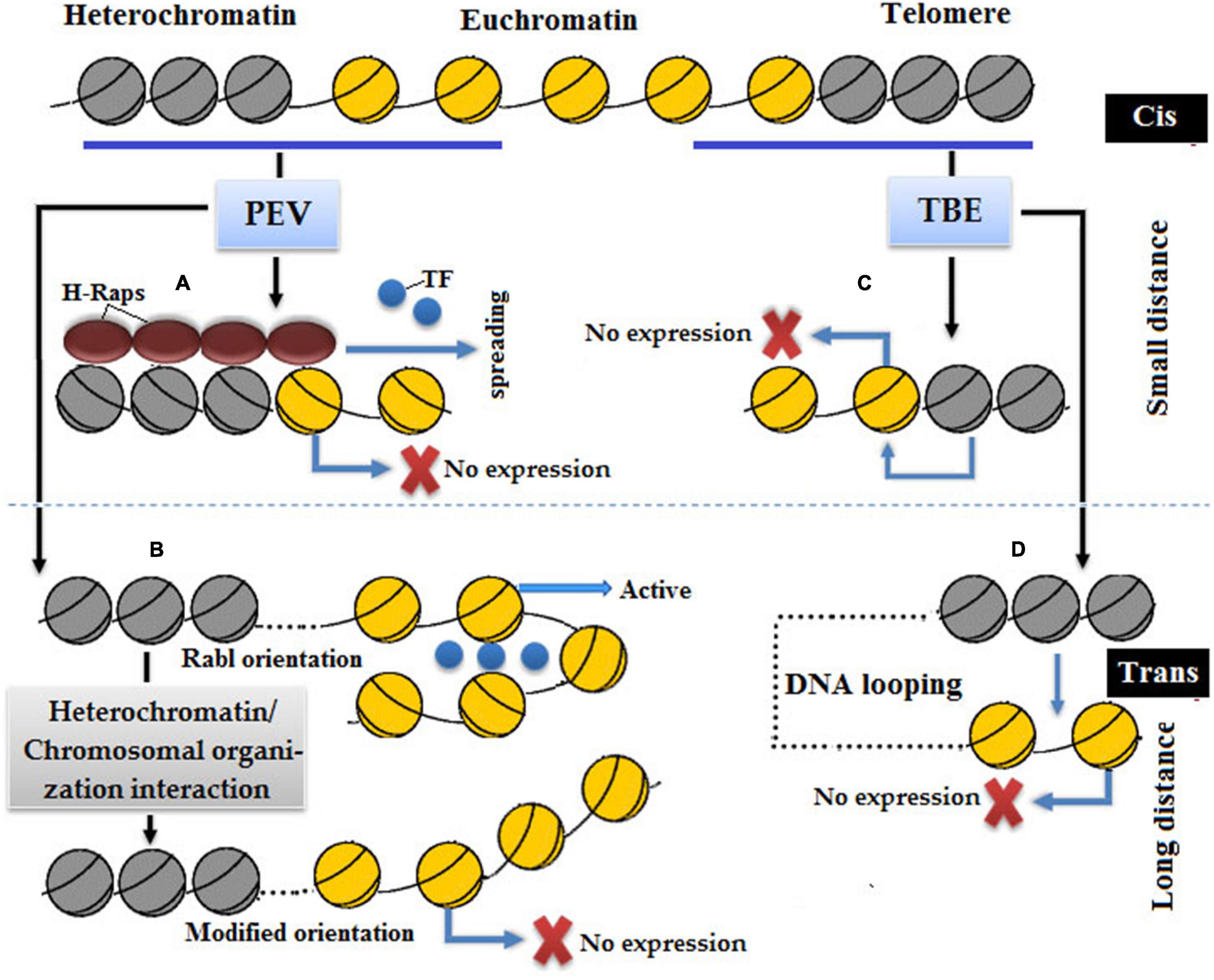
Figure 7. In position effect variegation (PEV) silencing: (A) cis-spreading model in which a repressor complex, namely, invading heterochromatin proteins H-Raps stop transcription by binding with a promoter of euchromatic gene. (B) Trans-interaction in which a new chromosomal orientation leads to silencing due to the loss of transcriptional machinery. In telomeric position effect (TPE) silencing, (C) cis-spreading model in which heterochromatin quenches adjacent genes resulted in gene silencing. (D) Trans-interaction in which distantly related euchromatin and heterochromatin come closer via DNA loop formation so that heterochromatin becomes capable of quenching the expression of euchromatin.
RNA interference is a homology-dependent gene silencing phenomenon that depends on dsRNA in gene silencing at the post-transcription level (Fire et al., 1998). The whole RNAi gene silencing factory consists of the following components: DCL, AGO, RDR, and dsRNA-binding domain (dsRBP) (Sabbione et al., 2019). sRNA biosynthesis is of primary importance in RNAi, which includes miRNAs and the following types of siRNAs: natural-antisense siRNA (nat-siRNA), ta-siRNA, heterochromatic siRNA (hc-siRNA), or repeated-associated siRNAs (ra-siRNAs) (Figure 8; Borges and Martienssen, 2015; Zheng et al., 2018). The cleavage of primary miRNAs (pri-miRNAs) into the precursor miRNAs (pre-miRNAs) is performed inside the nucleus under the action of DCL1, hyponastic leaves 1 (HYL1), and dsRBP. Then, pre-miRNA undergoes the second cleavage by DCL1 and HYL1 to finally produce miRNA duplex (Lu and Fedoroff, 2000; Vazquez et al., 2004). Subsequently, duplex miRNA is methylated by sRNA-specific methyltransferase HEN1 and exported to the cytoplasm with the help of exportin-5 ortholog HASTY (HST) (Park et al., 2002). Inside the cytoplasm, mature single-stranded miRNA binds with AGO1 to activate an RISC, which cleaves all homologous transcripts (Mallory and Vaucheret, 2006).
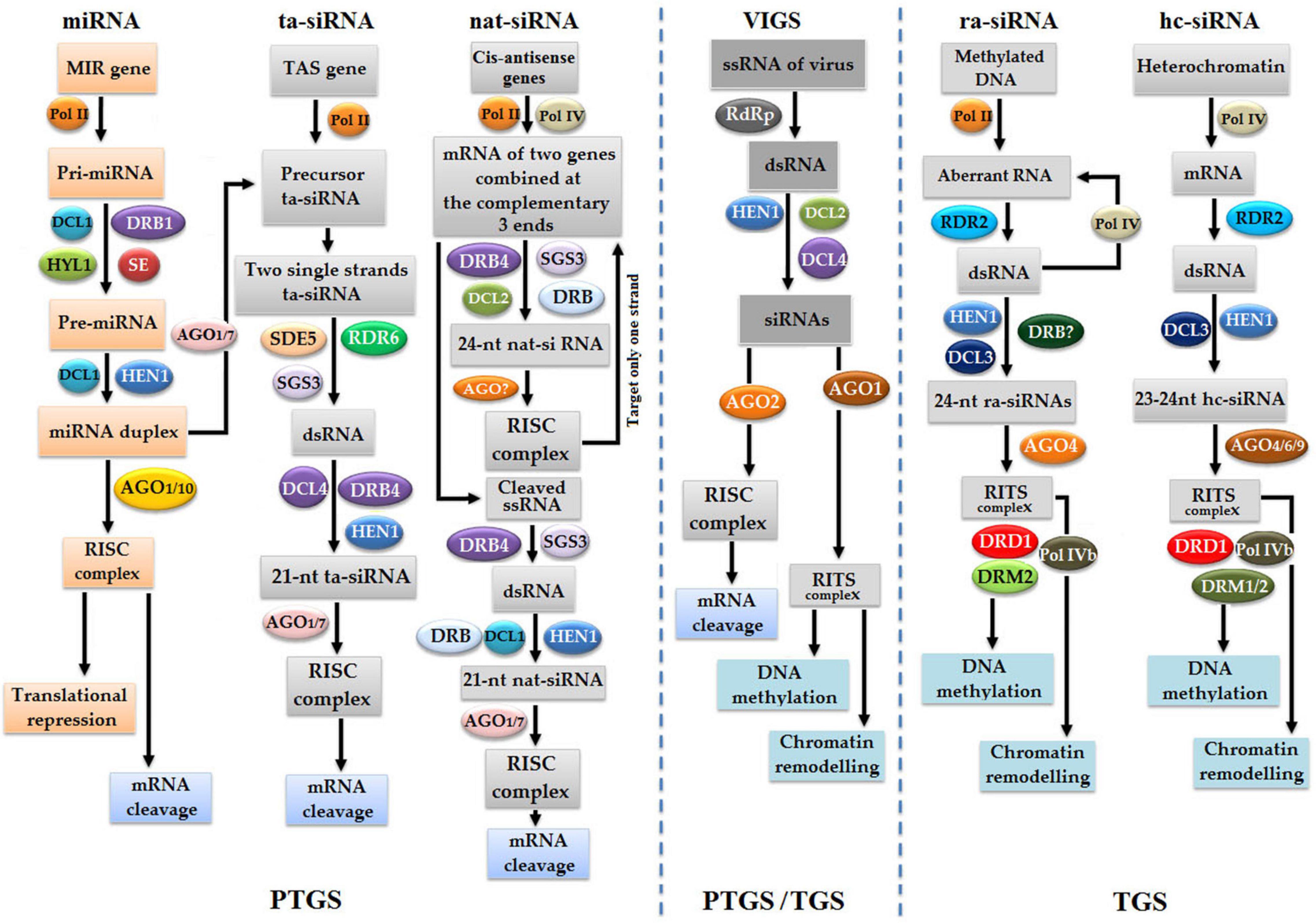
Figure 8. Naturally occurring gene silencing pathways in plants are PTGS gene silencing pathways, such as microRNA (miRNA), transacting siRNA (ta-siRNA), and natural-antisense siRNA (nat-siRNA), whereas TGS gene silencing pathways are repeated-associated siRNA (ra-siRNA) and heterochromatic siRNA (hc-siRNA). Meanwhile, VIGS is considered as artificial silencing methods that occurs via both TGS and PTGS pathways.
The second pathway of RNAi is dependent on the following long dsRNAs, nat-siRNA, ra-siRNA, ta-siRNA, and hc-siRNA, which could be exogenous due to viral infection or endogenous, such as transposons (Zheng et al., 2018). The nat-siRNA pathway is initiated with the transcription of natural-antisense gene pairs/overlapping genes distributed throughout the genome of plants. Overlapping genes are transcribed into two-three complementary mRNAs, which ligate each other to produce dsRNAs (Borsani et al., 2005). Subsequently, dsRNAs are cleaved by DCL2, SGS3, and RDR6 to generate 24-nt-long nat-siRNAs (Phillips et al., 2007), which bind AGO to activate the RISC to finally cleave out only cis-antisense mRNA strands. Trans-sense strand eventually becomes dsRNA under the action of RDR6 and SGS3, cleaved into 21-nt-long nat-siRNAs with the help of DCL1 and AGO1/7 to activate the RISC, and finally cleaves all homologous transcripts (Borsani et al., 2005).
Repeated-associated siRNAs and hc-siRNA are directly associated with methylation. In the ra-siRNA pathway, the promoter region of genes is methylated, so aberrant transcripts are produced with the help of RNA polymerase II, transformed into dsRNA under the action of RDR2, and follow further steps of gene silencing, or, dsRNA goes under the action of polymerase IVa to produce additionally aberrant RNA in a self-perpetuating loop (Xie et al., 2012). Subsequently, dsRNA is digested into 24-nt-long methylated ra-siRNAs under the action of DCL3 and HEN1, which binds with AGO4 to activate the (AGO4)-PolV complex or transcriptional silencing complex (RITS), causing the methylation of the complementary region of DNA with the help of SNF2-like chromatin remodeling proteins DRD1, DRM2, and alternative forms of polymerase Vb (Lippman et al., 2004; Xie et al., 2004). In the case of hc-siRNA, modified heterochromatin or DNA repeats are transcribed polymerase IV into mRNA, transformed into dsRNA under the action of RDR2, cleaved by DCL3 into 23–24-nt-long hc-siRNA (Chapman and Carrington, 2007; Matzke et al., 2009), binds with AGO6/9 to activate the RITC complex, and resulted in the TGS pathway (Chellappan et al., 2010; Fei et al., 2013). VIGS is another type of RNAi inbuilt defense system against viruses that can trigger both TGS and PTGS pathways of gene silencing in plants (Bartel, 2004; Burch-Smith et al., 2006; Lange, 2010).
CRISPR is a natural immune system of Streptococcus pyogenes against viruses based on camera unit (Jinek et al., Science). A copy of invading virus genome is saved in bacterial geneome namely CRISPR array to deter future viral attack, and Cas9 protein cleaves out viral genome to refrain it from hijacking bacterial genome and cause disease (Makarova et al., 2011), and Cas9 protein cleaves out viral genome. The CRISPR/Cas system is further divided into Class I-III, and CRISPR/Cas9 lies under the umbrella of bacteria-specific Class II (Figure 9; Heler et al., 2015; Wei et al., 2015). Class I of the type II Cas system is further divided into three major types: I, III, and IV, whereas class II is further divided into the following major types: II, V, and VI. Type I CRISPR/Cas system is widely distributed among bacteria and archaea, which is subdivided into six subtypes (A to F) (Sinkunas et al., 2011). Cas3 has been amplified in all subtypes except A, B, and D, with a few variations in the protein sequence (Sinkunas et al., 2011). The second type of Cas system is comprised of Cas1, Cas2, and Cas9, furthermore, Cas2 belongs to type II-A (Barrangou et al., 2007; Heler et al., 2015) and Cas4 belongs to type II-B (Li et al., 2014). Noticeably, type I and type II Cas systems depend on the following two factors for gene editing, (a) CRISPR RNA (crRNA) spacer and (b) protospacer adjacent motif (PAM) (Mojica et al., 2009).
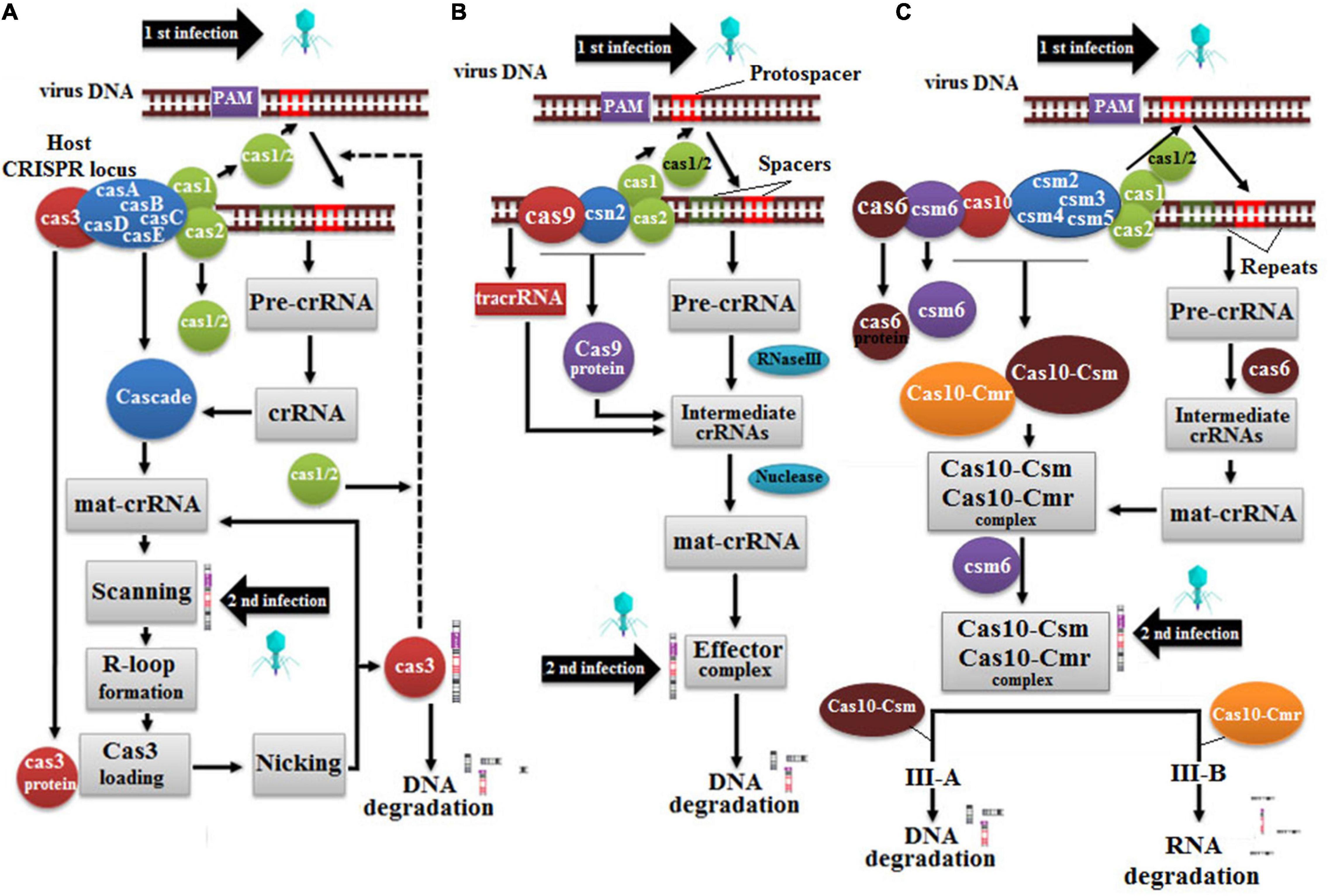
Figure 9. Cas protein mechanism is as follows: (A) Type I system depends on Cas3 protein, which recognizes new viral infection and counters it, (B) Type II system depends on Cas9 proteins and tracrRNA for the biogenesis of crRNA, and (C) Type III system depends on either Cas6 or Cas10 in which Csm targets DNA while Cmr targets viral RNA.
Generally, the CRISPR/Cas system is accomplished in three sequential steps: spacer acquisition, crRNA processing, and interference (Rath et al., 2015; Amitai and Sorek, 2016; Wang et al., 2016). On viral infection, Cas operon first expressed into a Cas1-Cas2 protein complex, which recognizes and makes a copy of protospacer of viral DNA, and integrates it between the repeated sequences of CRISPR array of the host genome. Subsequently, three CRISPR-related genes, trans-activating CRISPR RNA (tracr), and CRISPR array are transcribed into tracrRNA and pre-crRNA while the Cas9 gene encodes Cas9 protein. In the second step, tracrRNA anneals with the repeated sequence of pre-crRNA, and Cas9 protein binds with duplex. RNA transcript complex triggers RNase III enzyme to cleave pre-crRNA repeats (Jinek et al., 2012), nuclease finally produces the CRISPR/Cas9 complex by trimming extra nucleotides from 5́ end of pre-crRNA and leaving 20-nt-long spacer sequence (Liu et al., 2017). Finally, an interference step is kicked off, which activates the effector complex to recognize foreign DNA through its PAM site, such as 5́NGG3́. Subsequently, a spacer sequence of mature crRNA effector complex binds with the target sequence of viral DNA, which further activates RuvC and HNH domains of Cas9 protein (Gasiunas et al., 2012; Nishimasu et al., 2014). The RuvC domain cleaves non-target DNA strand, whereas the HNH domain cleaves the second strand to produce a blunt-end double-strand break in a 3-bp spacer region very next to the PAM site (Cong et al., 2013).
Nonsense-mediated decay is considered as one of the most important RNA surveillance pathways, which occur at the post-transcriptional level (Gloggnitzer et al., 2014). NMD performs the two primary functions: (a) the regulation of transcription and (b) protein expression (Maquat, 2005), and remodel a gene product from proteins by targeting premature termination codons (PTCs) (Bhuvanagiri et al., 2010; Schweingruber et al., 2013; Miller and Pearce, 2014). Occasionally, abnormal translation during the NMD pathway occurs due to two reasons: (a) the ribosome is unable to bind with an exon junction complex (EJC) and (b) the ribosome removes the EJC (Figure 10; Kurosaki et al., 2014). At the same time, the PABPC1 is too distant from the PTC, so UPF1 usually combines with the termination complex leading to an independent NMD pathway (Amrani et al., 2004; Ivanov et al., 2008). EJC model is common in mammal cells, in which ≥50–55 nt upstream of the exon-exon junction a PTC exists while EJC is downstream of termination codon (Nagy and Maquat, 1998; Thermann et al., 1998).
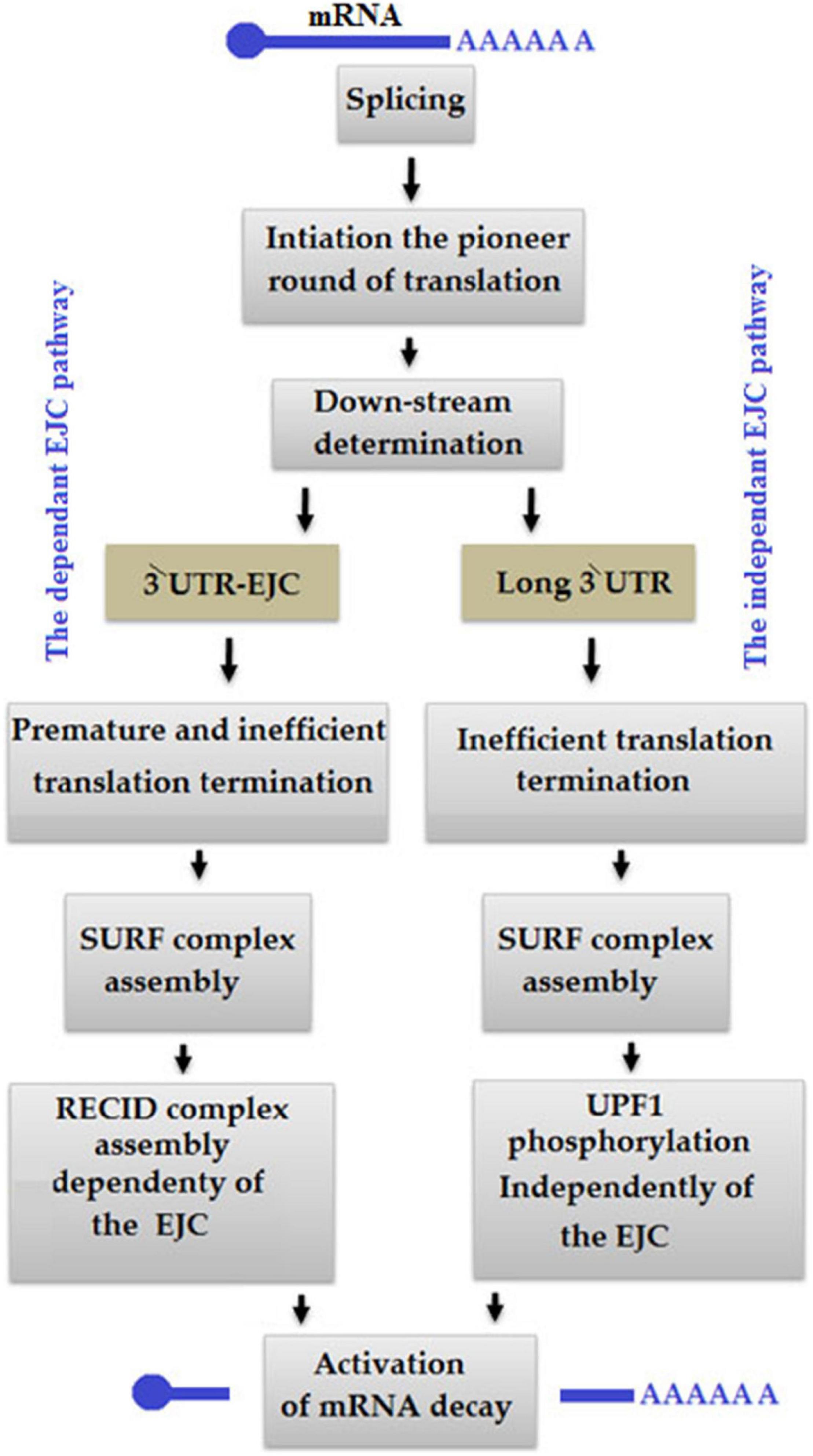
Figure 10. In an exon junction complex (EJC) pathway-dependent nonsense-mediated mRNA decay (NMD) model, the termination of translation occurs at the premature termination codon (PTC) level of ≥50–55 nt upstream of EJC, so that the ribosome is unable to dislocate it. In an EJC-independent pathway, PABPC1 distantly occurs from PTC to control eRF1-eRF3, which is related to the termination of translation.
In the NMD pathway, ribosome first binds with mRNA to initiate translation, removes existing EJC during the elongation of translation and stop at PTC, then eukaryotic release factors 1 and 3 of PABPC1 bind with the ribosome to make eRF1-eRF3 translation termination complex for translation termination (Kurosaki et al., 2014). Simultaneously, transient interaction between CBP80 and UPF1 enhances the attachment of serine/threonine kinase SMG1 complex to UPF1, which leads UPF1 to associate with eRF1 and eRF3 in the SURF complex (Kashima et al., 2006; Hwang et al., 2010). CBP80-UPF1 promotes SMG1-UPF1 binding to PTC-distal EJC in the DECID complex (Kashima et al., 2006). UPF1 is phosphorylated under the action of SMG1, which later causes mRNA decay and translational repression (Okada-Katsuhata et al., 2012; Durand et al., 2016).
Genetic engineering is a promising technology for interspecies and intraspecies gene transfer to get desired traits, such as biotic and abiotic stress resistance, biofortification, higher yield in plants, but a serious constraint is transgene silencing (Stam et al., 1997). Gene silencing is a natural defense mechanism of living organisms against invaders including transgene (Rajeev Kumar et al., 2015). It was first observed in transgenic petunia transformed by chalcone synthase-A (CHS-A) gene, in which PTGS-directed gene silencing resulted in unexpected loss of flower pigments (Napoli et al., 1990). In particular, transgene silencing occurs via co-suppression, endogenous homology gene-induced PTGS pathway, and endogenous heterologous gene-induced PTGS pathway (Hamilton and Baulcombe, 1999). The PTGS pathway is initially induced in specific tissues and then transmit signals to nearby tissues, it does not only induce gene silencing in homologous transgenes but also in viral genes (VIGS) (Hamilton and Baulcombe, 1999). Furthermore, transgene along with a large number of intrinsic direct repeats significantly increases the frequency of induction of inheritable PTGS (Ma and Mitra, 2002).
The group of genes, which play a role in the developmental processes of plants, such as primary growth, secondary growth, meristem growth, leaf morphogenesis, secondary root elongation, organogenesis, and flowering, are called developmental genes. From the whole transcriptome, few numbers of coding mRNAs are translated into proteins to perform a specific function in a cell (Berget et al., 1977; Chow et al., 1977; Djebali et al., 2012). Contrastingly, non-coding mRNAs are undesired because they are unable to be translated into protein but occasionally encode sRNAs (Rajagopalan et al., 2006). The sRNAs target transcriptional and post-transcriptional silencing of developmental genes to maintain transcriptional equilibrium to enhance the adaptation of plant (Dunoyer et al., 2010). Natural RNAi, such as siRNAs and miRNAs, play a significant role in plant tissue development (Voinnet, 2009) by controlling the expression of AGO1 and DCL1 genes (Xie et al., 2003). Moreover, miR168 and AGO1-derived siRNAs participate in feedback mechanisms to regulate the expression of the AGO1 gene (Mallory and Vaucheret, 2006; Rajagopalan et al., 2006). The regulation of gene expression in plants is performed by ta-siRNAs during the RNAi pathway (Peragine et al., 2004; Allen et al., 2005), by targeting mRNAs (Howell et al., 2007).
Genetic imprinting also affects the imprinting of developmental genes, such as angiosperms gene, which is involved in endosperm and seed size development (Bauer and Fischer, 2011; Haig, 2013). Genome-wide analysis of endosperm-related gene expression of rice revealed 162 MEGs and 95 PEGs relevant to imprinted differentially methylated loci, long non-coding RNAs (lncRNAs), miniature inverted-repeat TEs, and a few 21–22-nt-long siRNAs. One-half of PEGs and one-third of MEGs were related to nutrient metabolism and endosperm development, and thus represent grain yield quantitative trait loci (Yuan et al., 2017). Furthermore, few imprinted genes relevant to the transcription in endosperm cellularization and cell proliferation control seed size (Bauer and Fischer, 2011). Similarly, MEGs encoding transcription factor OsMADS13 regulate the seed size in rice (Li et al., 2011), and PRC2 and AGL62 regulate the seed size in Arabidopsis (Kang et al., 2008; Lu et al., 2012). Furthermore, few TFs responsible for seed development in maize and Arabidopsis are also regulated under MEGs (Asano et al., 2002; Luo et al., 2005; Fu and Xue, 2010; Xin et al., 2013).
Transposons or jumping genes are self-replicative short DNA sequences that can translocate within the genomes of the plant (Feschotte and Pritham, 2007; Sun et al., 2015). TEs are a severe threat to genome stability and are dealt with by TEs silencing (Kasschau et al., 2007; Slotkin and Martienssen, 2007), that is why a significant number of TEs remain silent in the plant genome (Okamoto and Hirochika, 2001). On the other hand, TEs can be beneficial in one population in the form of sRNAs (Ito et al., 2011; Parent et al., 2012), such as the activation of germinal cell RTs under stress stimulate the transcription of 24-nt-long siRNAs in Arabidopsis (Ito et al., 2011). TEs could be active or silent at a specific place and time within a genome, such as genes responsible for the silencing of pollen vegetative nuclei (VN) during seed development are downregulated, which were active during vegetative growth. Similarly, the final product of TEs from siRNAs enhances silencing mode in germ nuclei or sperm cell (SC) (Slotkin et al., 2009). Furthermore, TEs in double haploid rice plants cultured from anthers are reactivated after silencing as a result of somaclonal variations (Kikuchi et al., 2003). A relation exists between VN and SCs, which was observed in Arabidopsis and tobacco, causes the transcription of 21-nt-long siRNAs (McCormick, 2004).
Multiple factors influence genomic instability by DNA damaging or rearrangement, such as translocation and integration of TEs from one site to another within genome (Curcio and Derbyshire, 2003; Feschotte and Pritham, 2007). Transposons cause mutation within a genome, which are rectified under the PTGS pathway for genomic stability, and methylation also causes gene silencing resulted in a decrease in transposon activity. For example, the methylation of MET1 resulted in a decrease in PTGS activity and an increase in transposon activity (Ronemus et al., 1996; Morel et al., 2000; Kato et al., 2003), whereas a mutation in DECREASE IN DNA METHYLATION 1 (DDM1) causes a halt in a ratio of CG/non-CG methylation and increase the activity of transposons in Arabidopsis and C. elegans (Hirochika et al., 2000; Miura et al., 2001; Tsukahara et al., 2009). Similarly, three mutations rde2, rde3, and mut7 in the PTGS pathway resulted in a higher activity in transposons in C. elegans, and mut6 in Chlamydomonas also displayed the activation of transposons. In conclusion, TGS and PTGS pathways along with different siRNAs quench the activity of transposons to avoid genomic instability in plants (Mirouze et al., 2009; Ito, 2013).
Plants are a major source of nutrition for all living organisms, but a few plant species harbor health-hazardous toxins and allergens that should be detoxified (Johansson et al., 2004; Lee and Burks, 2006). Furthermore, 90% of all food allergens are found in the following eight types of foods: soybean, peanut, wheat, tree nuts, fish, egg, shellfish, and milk (Zuidmeer et al., 2008), and to date, there is no proper treatment of food allergy except precautionary measures (Sicherer and Sampson, 2006). The PTGS mechanism of gene silencing, such as RNAi is pivotal to detoxify toxins and allergens in rice and soybean (Gu et al., 2016). In rice, antisense mRNA strategy of gene expression leads to a loss in a 14–16-kDa allergenic protein motif (Sheehy et al., 1988; Tada et al., 1996), while soybean harbors Gly-m-Bd-60K, Gly-m-Bd-30K, and Gly-m-Bd-28K seed-specific allergens (Ogawa et al., 2000), and Gly-m-Bd-30K (P34) was completely detoxified using transgene-induced gene silencing (Herman et al., 2003). RNAi is a super-duper technique against toxins and allergens, for example, the knock down of 7-Nmethylxanthine methyltransferase gene (CaMXMT1) resulted in 70% reduced caffeine contents in transgenic plants (Ogita et al., 2003). Similarly, the knock down of cytochrome P450, CYP79D1, and CYP79D2 resulted in 90% loss in cyanogenic glucoside contents tubers of cassava (Manihot esculenta) (Siritunga and Sayre, 2003).
The downregulation of phytochelatin synthase gene (OsPCS1) in rice with the help of RNAi resulted in reduced accumulation of toxic metal Cd (Li et al., 2007), and nicotine demethylase gene knocks down in tobacco with the help of RNAi resulted in reduced nitrosamines [tobacco-specific N-nitrosamines (TSNAs)] carcinogen (Lewis et al., 2008). sRNAs successfully reduced terpenoid gossypol toxin contents in seeds and oil of cotton (Sunilkumar et al., 2006). Mal d1 allergen of apple (Malus domestica), Lyc e1 and Lyc e3 allergens of tomato, and Ara h2 allergen of peanut were significantly reduced with the help of RNAi (Gilissen et al., 2005; Le et al., 2006a, b; Dodo et al., 2008). The downregulation of the pathogenesis-related 10 (PR 10) gene with the help of RNAi resulted in decreased Dau c1.01 and Dau c1.02 allergens in patients (Peters et al., 2011).
Genes are always under a specific promoter that controls both the site and time of their expression, which is regulated by different kinds of gene silencing, i.e., transgene silencing. On exogenous or transgene expression, endogenous genes undergo non-symmetrical methylation by an RNA-chromatin mechanism resulted in the silencing of endogenous genes by the activation of the TGS pathway (Meyer et al., 1993; McGinnis et al., 2006). Furthermore, if a transgene induces the PTGS pathway, it can stop the expression of both exogenous and its homologous endogenous gene (Napoli et al., 1990; Van der Krol et al., 1990). The transgenic effect, in which both exogenous and endogenous genes are suppressed is known as co-suppression, which was observed in Petunia for the first time when CHS-A gene of pigmentation was overexpressed, which resulted in the suppression of both transgene and endogenous genes (Napoli et al., 1990).
Antisense and hp-RNA transgenes induced silencing in two endogenous genes in Arabidopsis resulted in more 4n than 2n plants due to a high level of methylation in tetraploid plants (Finn et al., 2011). The effect of gene silencing on the endogenous gene has also been observed in tobacco plants, in which the occurrence of co-suppression to nitrite reductase, nitrate reductase, and SAM synthase resulted in necrotic or chlorotic phenotypes (Palauqui et al., 1996). Comparatively, the transgene is more efficient against RNA silencing than endogenous genes in plants (Rajeev Kumar et al., 2015). RNAi-induced PTGS pathway of gene silencing also suppresses endogenous genes (Zhang et al., 2015). RNAi plays a significant role in silencing endogenous genes relevant to metabolic disease pathways, such as gluconeogenesis and phosphoenolpyruvate carboxykinase (PEPCK) enzyme (Zimmermann et al., 2006). PTGS enhances the gene silencing of endogenous genes, but bidirectional cytoplasmic RNA decay hinders its action on the endogenous gene (Zhang et al., 2015). NMD also plays a substantial role in the dynamic regulation of gene expression by controlling alternative splicing (Palusa and Reddy, 2010).
Biotic stress factors exert serious implications on plants including viruses, bacteria, fungi, insects, and nematodes (Romanel et al., 2012). Gene silencing enhances the immunity of plants, which plays a pivotal role to counter biotic stress. Plant viruses seriously affect plant growth, which resulted in a significant loss in yield. Although plant viruses do not directly affect animals and humans, losses in food quality and crop yield are noted. In this study, RNA silencing plays a key role to deter viral genome integration in the host genome by cleavage and protects the plant against several viruses (Pumplin and Voinnet, 2013). RNA silencing depends on sRNAs, which are further subdivided into two classes: siRNAs and miRNAs (Wang and Smith, 2016). siRNA performs a preferred antiviral activity, which induces gene silencing by transitive siRNA and defensive signal, but its limitation is that it remains inactive until the infection begins (Eamens et al., 2008).
Contrastingly, the expression of miRNAs is constitutive and directly targets the viral genome on entry inside the host cell to assure plant protection while siRNA indirectly activates the biogenesis of 22–24-nt-long siRNAs, which subsequently respond to viral infection (Simón-Mateo and García, 2011). For example, miR156 and miR164 express P1/HC-Pro turnip mosaic virus—(TuMV-) encoded RNA silencing suppressors in Arabidopsis (Kasschau et al., 2003). Noticeably, artificially designed miRNA (amiRNAs) also induced resistance against grapevine virus A in tobacco (Roumi et al., 2012). Recently, CRISPR/Cas9 and CRISPR/Cas13a are being employed in enhancing the resistance against both RNA and DNA viruses by mutating susceptible genes in the host (Chandrasekaran et al., 2016).
Bacterial pathogens cause severe diseases in the targeted plant organs, such as scabs, leaf spots, and cankers, which can be controlled by gene silencing. For example, crown gall disease caused by Agrobacterium is being controlled by targeting silencing viz and iaaM genes by RNAi (Dunoyer et al., 2006). miRNAs and siRNAs play a pivotal role in defense against bacterial infection, such as the regulation of an auxin signaling pathway by miR393, resulted in enhancing plant antibacterial PTI (Navarro et al., 2006), and miR167, miR393, and miR160 play a role against bacteria in tomato (Fahlgren et al., 2016). NMD is a kind of PTGS gene silencing pathway, which contains bacterial infection by enhancing innate immunity of plants against bacterial infection by controlling numerous TIR domain-containing, nucleotide-binding, leucine-rich repeat (TNL) immune receptor-encoding mRNAs (Gloggnitzer et al., 2014). CRISPR/Cas9 and its roles in enhancing plant resistance against bacteria (Zaynab et al., 2020), such as CRISPR/Cas9-induced OsSWEET13 rice mutants displayed enhanced immunity against bacterial blight (Zhou et al., 2015).
Fungi cause 70% of the total plant diseases including smut, rusts, and mildew (Dean et al., 2012). Host-pathogen interaction at the surface of the host cell is established via haustorium and resulted in an exchange of signal and nutrients (Panstruga, 2003). Gene silencing enhances plant resistance against a broad range of fungal pathogens by the transfer of siRNAs or silencing signals from the host to the pathogen (Duan et al., 2012). For example, Avra10 in wheat and barley is a host-induced gene that limits fungal pathogen Blumeria graminis due to silent point mutations (Nowara et al., 2010). Similarly, the overexpression of miR1138 in wheat countered infection caused by Puccinia graminis (Gupta et al., 2012), and the downregulation of miR1448 and miR482 in cotton resulted in severe Verticillium infection (Jagadeeswaran et al., 2009). Moreover, the confirmed roles of some miRNAs-like Md-miRln20 allow enhancing resistance in apple against Glomerella leaf spot (Zhang et al., 2019). CRISPR/Cas9 has a potential role in the activation of genes to confer the resistance against fungi, such as the transient expression of TcNPR3 via CRISPR/Cas9 resulted in conferring the resistance against Phytophthora tropicalis in Theobroma cacao (Fister et al., 2018).
RNA interference and CRISPR/Cas9 are being widely used in improving plant protection against insect pests, such as artificially designed dsRNAs that have been transformed to enhance plant resistance against Coleoptera and Lepidoptera (Price and Gatehouse, 2008). For example, the larval stage of cotton bollworm was reduced by transforming 22–24-nt-long artificially designed P450 monooxygenase genes, resulted in short feeding on plant tissues (Mao et al., 2007). Four wild populations of flour beetle (Tribolium castaneum) displayed that most of its variants harbor Cas9 target sites, and some of them are immune to drive and can be targeted (Drury et al., 2017). Nematodes also cause diseases and severe reduction of yield in many crops, which are being contained via gene silencing (El-Sappah et al., 2019). Approximately 30 miRNAs in Arabidopsis and 40 in soybean were differentially expressed during cyst nematodes infection (Hewezi et al., 2008), and miR159 plays a key role in gall and giant cell infection (Medina et al., 2017). RNAi is a robust technique for the development of plant resistance against nematode by reducing gall formation (Huang et al., 2006; Yadav et al., 2006).
An adverse effect of non-living surrounding factors on plants is known as abiotic stress, such as drought, heat, cold, light intensity, salinity, mineral deficiency, mineral toxicity, soil acidity, ozone, SO2, NO2, and higher CO2. Gene silencing via RNAi, CRISPR/Cas9, and miRNAs are promising to enhance plant resistance against the abovementioned abiotic stress conditions (Khraiwesh et al., 2012; Sun, 2012; Sunkar et al., 2012). Salinity is a critical abiotic factor significantly affecting crop productivity around the world (Bartels and Sunkar, 2005). Natural resistance, abiotic resistance genes, and miRNAs play a key role in plant tolerance against salinity (Ding and Zhu, 2009; Trindade et al., 2010). MiRNAs, miR159, miR160, miR167, miR169, miR393, and miR397 play a significant role in several plant species during salt stress (Sunkar and Zhu, 2004; Zhao et al., 2009; Gao et al., 2011; Kitazumi et al., 2015). During extended heat stress, miR398 plays a significant role in enhancing plant tolerance (Guan et al., 2013), whereas miR319 plays a significant role in extended cold tolerance (Thiebaut et al., 2019). In drought stress, miR160, miR167, miR169, and miR393 play a key role (Zhao et al., 2009; Sunkar, 2010), for example, miR171a, miR171b, and miR171c express in response to drought stress in potato (Hwang et al., 2011). MiRNAs regulate the cell wall to deter metal stress, for example, miR319, miR390, miR393, and miR398 express under Cu stress and miR160, miR164, and miR167 express under Cd stress (Yamasaki et al., 2007; Abdel-Ghany and Pilon, 2008; Huang et al., 2009) while miR390, miR168, miR156, miR162, miR166, and miR171 were downregulated and miR528 was upregulated (Ding et al., 2011).
The quality traits of any crop are shape, color, shelf life, nutritional value, etc., which can be improved by conventional breeding and modern molecular techniques (Saurabh et al., 2014). Noticeably, the biggest challenge during the quality trait improvement is the loss of other desired characteristics, which can only be overcome by a selective transformation of flexible regulatory genes, potentially providing more competence and accurate regulation in a targeted manner (Tang and Chu, 2017). Gene silencing by deploying genetic engineering techniques, such as RNAi and CRISPR/Cas9 play a fundamental role in the regulation of genes relevant to quality traits in different crops (Saurabh et al., 2014). A miRNA is also a promising tool for the improvement of quality traits in different crops at the post-transcriptional level, such as miR156 and miR397 control grain size, quality, and yield (Jiao et al., 2010; Si et al., 2016), miR159 regulates stem elongation and floral development (Tsuji et al., 2006), and miR160 plays a critical role in the growth, development, and immunity of rice (Li et al., 2014). miR164 plays a key role in lateral root development, and miR166 in leaf polarity in maize (Juarez et al., 2004; Li et al., 2012), whereas miR159 plays an important role in anthers development and heat response in wheat (Wang et al., 2012). miR172 plays an essential role in cleistogamous flowering and grain density in barley (Nair et al., 2010; Houston et al., 2013). miR156 regulates vegetative and reproductive growth (Silva et al., 2014), whereas miR159, miR167, and miR4376 regulate flowering in tomatoes (Buxdorf et al., 2010; Wang et al., 2011; Liu et al., 2014). Recently, CRISPR/Cas9 has emerged as a promising tool for the identification of new genes, and genetic modification to improve quality traits and yield (Wang et al., 2019). CRISPR/Cas9 was used to improve fruit size by regulating classical CLAVATA-WUSCHEL (CLV-WUS) stem cell circuit (Ma et al., 2015), and the identification of new genes relevant to malate contents and aluminum-activated malate transporter 9 (ALMT9) in tomatoes (Ye et al., 2017).
Tools in molecular biology have been developed for the identification, amplification, interspecies, and intraspecies transformation of desired genes, which predominantly rely on T-DNA activation and knockout libraries (Weigel et al., 2000). Comparatively, VIGS is efficient out of all available tools for the study of functional genomics (Cakir et al., 2010), due to (a) fast, (b) easy designing due to the independence of full-length cDNA, (c) transient gene silencing, (d) higher efficacy even in polyploid species, and (e) easy delivery (Cakir et al., 2010). RNAi is also being widely employed in studying functional genomics in many organisms without any limitation (Whitehurst et al., 2007; Smith et al., 2010). In plants, a lot of studies have been conducted with the help of RNAi to determine the function of different genes to improve plant resistance against biotic and abiotic stress, biofortification, lingo-cellulosic pathway engineering, and the improvement of quality traits. Furthermore, the discovery of CRISPR/Cas9 has revolutionized functional genomics exponentially (Liu et al., 2017).
Transposons translocate within the genome and pose severe threats to genomic stability, which are deterred by gene silencing (Slotkin and Martienssen, 2007). Gene silencing maintains the balance of transcripts to ensure the adaptation of a plant to environmental fluctuations (Dunoyer et al., 2010). Tissue development and finishing are entirely controlled by gene silencing (ti-siRNAs and miRNAs) via a negative feedback mechanism (Voinnet, 2009; Cheng et al., 2021). Gene silencing plays a key role in crop yield by controlling the expression of genes related to seed size, for example, the silencing of OsMADS13, and PRC2 and AGL62 regulate seed size in rice (Li et al., 2011) and Arabidopsis, respectively (Kang et al., 2008; Lu et al., 2012). Gene silencing plays a key role to deter pathogenicity caused by biotic stress factors, such as insects, nematodes, bacteria, viruses, and fungi, which cause severe loss in crop yield (Zaynab et al., 2020). Allergy is an incurable disease mainly caused by the ingestion of daily food items, and gene silencing is helpful in the detoxification of these allergens (Gu et al., 2016). Caffeine contents were decreased by 70% in tea by the silencing of the CaMXMT1 gene, cyanogenic contents were decreased by 90% by the silencing of cytochrome P450, CYP79D1, and CYP79D2 in cassava tubers (Ogita et al., 2003; Siritunga and Sayre, 2003), the silencing of OsPCS1 in rice resulted in a significant loss in toxic metal contents (Li et al., 2007), and nicotine demethylase gene knockdown in tobacco resulted in reduced carcinogen (Lewis et al., 2008). Gene silencing also downregulates toxins in the seeds and oil of cotton, in apples, tomatoes, and peanuts.
To achieve higher crop yield, the scientist had developed genetic transformation techniques to overcome biotic and abiotic stress factors. Gene silencing negatively affects genetic transformation by the silencing of transgenes (Stam et al., 1997), such as the CHS-A gene, which was transformed in petunia to get a dark brown color but albino phenotype was observed in transgenic plants (Napoli et al., 1990). RTs are responsible for the activation of germinal cells in Arabidopsis, which goes silent due to gene silencing and results in the loss of switch from vegetative to reproductive growth (Ito et al., 2011).
AE-S, MA, KE-T, JL, and XZ: conceptualization. AE-S: writing the original draft and drawing figures. MI, KY, YW, RM, MK, JL, MA, XZ, AE-S, KE-T, QH, and QL: editing and proofreading. AE-S and MA: writing the final manuscript. All the authors reviewed and approved the final submission.
This study was supported by the Sichuan Provincial Department of Science and Technology Project (Grant No. 18ZDYF0293), the Key Laboratory of Sichuan Province for Refining Sichuan Tea, and the Sichuan Province Tea Industry Group Co., Ltd.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are very grateful to the kind administration of Yibin University, Yibin, China for providing us such a prestigious and well-equipped platform for research and development. We are also grateful to the kind administration of the Ministry of Science and Technology of Sichuan province, China. Finally, KE-T would like to thank the library at Murdoch University-Australia for the valuable online resources and comprehensive databases.
Abbas, M., Peszlen, I., Shi, R., Kim, H., Katahira, R., Kafle, K., et al. (2020). Involvement of CesA4, CesA7-A/B and CesA8-A/B in secondary wall formation in Populus trichocarpa wood. Tree Physiol. 40, 73–89. doi: 10.1093/treephys/tpz020
Abdel-Ghany, S. E., and Pilon, M. (2008). MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283, 15932–15945. doi: 10.1074/jbc.M801406200
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Amitai, G., and Sorek, R. (2016). CRISPR–Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 14:67. doi: 10.1038/nrmicro.2015.14
Amrani, N., Ganesan, R., Kervestin, S., Mangus, D. A., Ghosh, S., and Jacobson, A. (2004). A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432, 112–118. doi: 10.1038/nature03060
Andreyeva, E. N., Belyaeva, E. S., Semeshin, V. F., Pokholkova, G. V., and Zhimulev, I. F. (2005). Three distinct chromatin domains in telomere ends of polytene chromosomes in Drosophila melanogaster Tel mutants. J. Cell Sci. 118, 5465–5477. doi: 10.1242/jcs.02654
Asano, T., Kunieda, N., Omura, Y., Ibe, H., Kawasaki, T., Takano, M., et al. (2002). Rice SPK, a calmodulin-like domain protein kinase, is required for storage product accumulation during seed development: phosphorylation of sucrose synthase is a possible factor. Plant Cell 14, 619–628. doi: 10.1105/tpc.010454
Axtell, M. J. (2013). Classification and comparison of small RNAs from plants. Ann. Rev. Plant Biol. 64, 137–159. doi: 10.1146/annurev-arplant-050312-120043
Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C., et al. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124. doi: 10.1038/35065138
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartels, D., and Sunkar, R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58. doi: 10.1080/07352680590910410
Bauer, M. J., and Fischer, R. L. (2011). Genome demethylation and imprinting in the endosperm. Curr. Opin. Plant Biol. 14, 162–167. doi: 10.1016/j.pbi.2011.02.006
Berget, S. M., Moore, C., and Sharp, P. A. (1977). Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. U.S.A. 74, 3171–3175. doi: 10.2307/66899
Bhuvanagiri, M., Schlitter, A. M., Hentze, M. W., and Kulozik, A. E. (2010). NMD: RNA biology meets human genetic medicine. Biochem. J. 430, 365–377. doi: 10.1042/BJ20100699
Boisnard-Lorig, C., Colon-Carmona, A., Bauch, M., Hodge, S., Doerner, P., Bancharel, E., et al. (2001). Dynamic analyses of the expression of the HISTONE::YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13, 495–509. doi: 10.2307/3871402
Boivin, A., Gally, C., Netter, S., Anxolabéhère, D., and Ronsseray, S. (2003). Telomeric associated sequences of Drosophila recruit polycomb-group proteins in vivo and can induce pairing-sensitive repression. Genetics 164, 195–208. doi: 10.1023/A:1022937713781
Bond, D. M., and Baulcombe, D. C. (2015). Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 112, 917–922. doi: 10.1073/pnas.1413053112
Borges, F., and Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Borrelli, V. M., Brambilla, V., Rogowsky, P., Marocco, A., and Lanubile, A. (2018). The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front. Plant Sci. 9:1245. doi: 10.3389/fpls.2018.01245
Borsani, O., Zhu, J., Verslues, P. E., Sunkar, R., and Zhu, J.-K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123, 1279–1291. doi: 10.1016/j.cell.2005.11.035
Brink, R. A. (1956). A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41:872. doi: 10.2307/2407003
Brodersen, P., and Voinnet, O. (2006). The diversity of RNA silencing pathways in plants. Trends Genet. 22, 268–280. doi: 10.1016/j.tig.2006.03.003
Bühler, M., Verdel, A., and Moazed, D. (2006). Tethering RITS to a nascent transcript initiates RNAi-and heterochromatin-dependent gene silencing. Cell 125, 873–886. doi: 10.1016/j.cell.2006.04.025
Burch-Smith, T. M., Schiff, M., Liu, Y., and Dinesh-Kumar, S. P. (2006). Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol. 142, 21–27. doi: 10.1104/pp.106.084624
Buxdorf, K., Hendelman, A., Stav, R., Lapidot, M., Ori, N., and Arazi, T. (2010). Identification and characterization of a novel miR159 target not related to MYB in tomato. Planta 232, 1009–1022. doi: 10.1007/s00425-010-1231-9
Cakir, C., Gillespie, M. E., and Scofield, S. R. (2010). Rapid determination of gene function by virus-induced gene silencing in wheat and barley. Crop Sci. 50, 77–84. doi: 10.2135/cropsci2009.10.0567
Chandler, V. L., and Stam, M. (2004). Chromatin conversations: mechanisms and implications of paramutation. Nat. Rev. Genet. 5, 532–544. doi: 10.1038/nrg1378
Chandler, V., and Alleman, M. (2008). Paramutation: epigenetic instructions passed across generations. Genetics 178:1839. doi: 10.1002/gepi.20302
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Chapman, E. J., and Carrington, J. C. (2007). Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 8, 884–896. doi: 10.1038/nrg2179
Chellappan, P., Xia, J., Zhou, X., Gao, S., Zhang, X., Coutino, G., et al. (2010). siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res. 38, 6883–6894. doi: 10.1093/nar/gkq590
Cheng, Y., Wang, L., Abbas, M., Huang, X., and Li, Q. (2021). MicroRNA319-mediated gene regulatory network impacts leaf development and morphogenesis in poplar. Forest. Res. 1:4. doi: 10.48130/FR-2021-0004
Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J. J., Goldberg, R. B., et al. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33–42. doi: 10.1016/S0092-8674(02)00807-3
Chow, L. T., Gelinas, R. E., Broker, T. R., and Roberts, R. J. (1977). An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12, 1–8. doi: 10.1016/0092-8674(77)90180-5
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Cooper, D., VandeBerg, J., Sharman, G., and Poole, W. (1971). Phosphoglycerate kinase polymorphism in kangaroos provides further evidence for paternal X inactivation. Nat. New Biol. 230, 155–157. doi: 10.1038/newbio230155a0
Csink, A. K., and Henikoff, S. (1996). Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature 381, 529–531. doi: 10.1038/381529a0
Curcio, M. J., and Derbyshire, K. M. (2003). The outs and ins of transposition: from mu to kangaroo. Nat. Rev. Mol. Cell Biol. 4, 865–877. doi: 10.1038/nrm1241
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2012.00822.x
Deleris, A., Stroud, H., Bernatavichute, Y., Johnson, E., Klein, G., Schubert, D., et al. (2012). Loss of the DNA methyltransferase MET1 induces H3K9 hypermethylation at PcGt target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet. 8:e1003062. doi: 10.1371/journal.pgen.1003062
Dernburg, A. F., Broman, K. W., Fung, J. C., Marshall, W. F., Philips, J., Agard, D. A., et al. (1996). Perturbation of nuclear architecture by long-distance chromosome interactions. Cell 85, 745–759. doi: 10.1016/S0092-8674(00)81240-4
Ding, Y. F., and Zhu, C. (2009). The role of microRNAs in copper and cadmium homeostasis. Biochem. Biophys. Res. Commun. 386, 6–10. doi: 10.1016/j.bbrc.2009.05.137
Ding, Y., Chen, Z., and Zhu, C. (2011). Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J. Exp. Bot. 62, 3563–3573. doi: 10.1093/jxb/err046
Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. doi: 10.1038/nature11233
Dodo, H. W., Konan, K. N., Chen, F. C., Egnin, M., and Viquez, O. M. (2008). Alleviating peanut allergy using genetic engineering: the silencing of the immunodominant allergen Ara h 2 leads to its significant reduction and a decrease in peanut allergenicity. Plant Biotechnol. J. 6, 135–145. doi: 10.1111/j.1467-7652.2007.00292.x
Doheny, J. G., Mottus, R., and Grigliatti, T. A. (2008). Telomeric position effect—a third silencing mechanism in eukaryotes. PLoS One 3:e3864. doi: 10.1371/journal.pone.0003864
Drury, D. W., Dapper, A. L., Siniard, D. J., Zentner, G. E., and Wade, M. J. (2017). CRISPR/Cas9 gene drives in genetically variable and nonrandomly mating wild populations. Sci. Adv. 3:e1601910. doi: 10.1126/sciadv.1601910
Du, J., Johnson, L. M., Groth, M., Feng, S., Hale, C. J., Li, S., et al. (2014). Mechanism of DNA methylation-directed histone methylation by KRYPTONITE. Mol. Cell 55, 495–504. doi: 10.1016/j.molcel.2014.06.009
Duan, C.-G., Wang, C.-H., and Guo, H.-S. (2012). Application of RNA silencing to plant disease resistance. Silence 3:5. doi: 10.1186/1758-907X-3-5
Dunoyer, P., Brosnan, C. A., Schott, G., Wang, Y., Jay, F., Alioua, A., et al. (2010). Retracted: an endogenous, systemic RNAi pathway in plants. EMBO J. 29, 1699–1712. doi: 10.1038/emboj.2010.65
Dunoyer, P., Himber, C., and Voinnet, O. (2006). Induction, suppression and requirement of RNA silencing pathways in virulent Agrobacterium tumefaciens infections. Nat. Genet. 38, 258–263. doi: 10.1038/ng1722
Durand, S., Franks, T. M., and Lykke-Andersen, J. (2016). Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nat. Commun. 7:12434. doi: 10.1038/ncomms12434
Eamens, A., Wang, M.-B., Smith, N. A., and Waterhouse, P. M. (2008). RNA silencing in plants: yesterday, today, and tomorrow. Plant Physiol. 147, 456–468. doi: 10.1104/pp.108.117275
Elgin, S. C. (1996). Heterochromatin and gene regulation in Drosophila. Curr. Opin. Genet. Dev. 6, 193–202. doi: 10.1016/S0959-437X(96)80050-5
El-Sappah, A. H., Mm, H. I., El-awady, H., Yan, S., Qi, S., Liu, J., et al. (2019). Tomato natural resistance genes in controlling the root-knot nematode. Genes 10:925. doi: 10.3390/genes10110925
Enke, R. A., Dong, Z., and Bender, J. (2011). Small RNAs prevent transcription-coupled loss of histone H3 lysine 9 methylation in Arabidopsis thaliana. PLoS Genet. 7:e1002350. doi: 10.1371/journal.pgen.1002350
Fahlgren, N., Hill, S. T., Carrington, J. C., and Carbonell, A. (2016). P-SAMS: a web site for plant artificial microRNA and synthetic trans-acting small interfering RNA design. Bioinformatics 32, 157–158. doi: 10.1093/bioinformatics/btv534
Fei, Q., Xia, R., and Meyers, B. C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25, 2400–2415. doi: 10.1105/tpc.113.114652
Feng, S., Jacobsen, S. E., and Reik, W. (2010). Epigenetic reprogramming in plant and animal development. Science 330, 622–627. doi: 10.1126/science.1190614
Feschotte, C., and Pritham, E. J. (2007). DNA transposons and the evolution of eukaryotic genomes. Ann. Rev. Genet. 41, 331–368. doi: 10.1146/annurev.genet.40.110405.090448
Feschotte, C., Jiang, N., and Wessler, S. R. (2002). Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3, 329–341. doi: 10.1038/nrg793
Finn, T. E., Wang, L., Smolilo, D., Smith, N. A., White, R., Chaudhury, A., et al. (2011). Transgene expression and transgene-induced silencing in diploid and autotetraploid Arabidopsis. Genetics 187, 409–423. doi: 10.1534/genetics.110.124370
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
Fischer, A., Hofmann, I., Naumann, K., and Reuter, G. (2006). Heterochromatin proteins and the control of heterochromatic gene silencing in Arabidopsis. J. Plant Physiol. 163, 358–368. doi: 10.1016/j.jplph.2005.10.015
Fister, A. S., Landherr, L., Maximova, S. N., and Guiltinan, M. J. (2018). Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao. Front. Plant Sci. 9:268. doi: 10.3389/fpls.2018.00268
François, F., Hurd, P. J., Rachel, D., and Tony, K. (2003). The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31, 2305–2312. doi: 10.1093/nar/gkg332
Fu, F. F., and Xue, H. W. (2010). Coexpression analysis identifies rice starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 154, 927–938. doi: 10.1101/gad.1413706
Fultz, D., and Slotkin, R. K. (2017). Exogenous transposable elements circumvent identity-based silencing, permitting the dissection of expression-dependent silencing. Plant Cell 29, 360–376. doi: 10.1105/tpc.16.00718
Gao, P., Bai, X., Yang, L., Lv, D., Pan, X., Li, Y., et al. (2011). Osa-MIR393: a salinity-and alkaline stress-related microRNA gene. Mol. Biol. Rep. 38, 237–242. doi: 10.1007/s11033-010-0100-8
García-Aguilar, M., and Gillmor, C. S. (2015). Zygotic genome activation and imprinting: parent-of-origin gene regulation in plant embryogenesis. Curr. Opin. Plant Biol. 27, 29–35. doi: 10.1016/j.pbi.2015.05.020
Garnier, O., Laoueillé-Duprat, S., and Spillane, C. (2008). Genomic imprinting in plants. Epigenetics 3, 14–20. doi: 10.1385/1-59259-211-2:327
Gasiunas, G., Barrangou, R., Horvath, P., and Siksnys, V. (2012). Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 109, E2579–E2586. doi: 10.1073/pnas.1208507109
Gehring, M. (2013). Genomic imprinting: insights from plants. Ann. Rev. Genet. 47, 187–208. doi: 10.1146/annurev-genet-110711-155527
Gehring, M., Bubb, K. L., and Henikoff, S. (2009). Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324, 1447–1451. doi: 10.1126/science.1171609
Gelvin, S. B. (2003). Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37. doi: 10.1128/MMBR.67.1.16-37.2003
Gilissen, L. J., Bolhaar, S. T., Matos, C. I., Rouwendal, G. J., Boone, M. J., Krens, F. A., et al. (2005). Silencing the major apple allergen Mal d 1 by using the RNA interference approach. J. Allergy Clin. Immunol. 115, 364–369. doi: 10.1016/j.jaci.2004.10.014
Girton, J. R., and Johansen, K. M. (2008). Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila. Adv. Genet. 61, 1–43. doi: 10.1016/S0065-2660(07)00001-6
Gloggnitzer, J., Akimcheva, S., Srinivasan, A., Kusenda, B., Riehs, N., Stampfl, H., et al. (2014). Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host 16, 376–390. doi: 10.1016/j.chom.2014.08.010
Gohl, D., Müller, M., Pirrotta, V., Affolter, M., and Schedl, P. (2008). Enhancer blocking and transvection at the Drosophila apterous locus. Genetics 178, 127–143. doi: 10.1534/genetics.107.077768
Grewal, S. I., and Jia, S. (2007). Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46. doi: 10.1038/nrg2008
Gu, M., Chen, A., Sun, S., and Xu, G. (2016). Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: what is missing? Mol. Plant 9, 396–416. doi: 10.1016/j.molp.2015.12.012
Guan, Q., Lu, X., Zeng, H., Zhang, Y., and Zhu, J. (2013). Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 74, 840–851. doi: 10.1111/tpj.12169
Gupta, O. P., Permar, V., Koundal, V., Singh, U. D., and Praveen, S. (2012). MicroRNA regulated defense responses in Triticum aestivum L. during Puccinia graminis f. sp. tritici infection. Mol. Biol. Rep. 39, 817–824. doi: 10.1007/s11033-011-0803-5
Haag, J. R., Ream, T. S., Marasco, M., Nicora, C. D., Norbeck, A. D., Pasa-Tolic, L., et al. (2012). In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol. Cell 48, 811–818. doi: 10.1016/j.molcel.2012.09.027
Haig, D. (2013). Kin conflict in seed development: an interdependent but fractious collective. Ann. Rev. Cell Dev. Biol. 29, 189–211. doi: 10.1146/annurev-cellbio-101512-122324
Hale, C. J., Stonaker, J. L., Gross, S. M., and Hollick, J. B. (2007). A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol. 5:e275. doi: 10.1371/journal.pbio.0050275
Hamilton, A. J., and Baulcombe, D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. doi: 10.1126/science.286.5441.950
Harris, C. J., Scheibe, M., Wongpalee, S. P., Liu, W., Cornett, E. M., Vaughan, R. M., et al. (2018). A DNA methylation reader complex that enhances gene transcription. Science 362, 1182–1186. doi: 10.1126/science.aar7854
Havecker, E. R., Gao, X., and Voytas, D. F. (2004). The diversity of LTR retrotransposons. Genome Biol. 5:225. doi: 10.1186/gb-2004-5-6-225
Heler, R., Samai, P., Modell, J. W., Weiner, C., Goldberg, G. W., Bikard, D., et al. (2015). Cas9 specifies functional viral targets during CRISPR–Cas adaptation. Nature 519, 199–202. doi: 10.1038/nature14245
Henikoff, S., and Dreesen, T. D. (1989). Trans-inactivation of the Drosophila brown gene: evidence for transcriptional repression and somatic pairing dependence. Proc. Natl. Acad. Sci. U.S.A. 86, 6704–6708. doi: 10.2307/34603
Herman, E. M., Helm, R. M., Jung, R., and Kinney, A. J. (2003). Genetic modification removes an immunodominant allergen from soybean. Plant Physiol. 132, 36–43. doi: 10.1104/pp.103.021865
Hewezi, T., Howe, P., Maier, T. R., and Baum, T. J. (2008). Arabidopsis small RNAs and their targets during cyst nematode parasitism. Mol. Plant Microbe Interact. 21, 1622–1634. doi: 10.1094/MPMI-21-12-1622
Hirochika, H., Okamoto, H., and Kakutani, T. (2000). Silencing of retrotransposons in Arabidopsis and reactivation by the ddm1 mutation. Plant Cell 12, 357–368. doi: 10.2307/3870941
Hollick, J. B., Patterson, G. I., Asmundsson, I. M., and Chandler, V. L. (2000). Paramutation alters regulatory control of the maize pl locus. Genetics 154, 1827–1838. doi: 10.1017/S0016672399004462
Houston, K., McKim, S. M., Comadran, J., Bonar, N., Druka, I., Uzrek, N., et al. (2013). Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proc. Natl. Acad. Sci. U.S.A. 110, 16675–16680. doi: 10.1073/pnas.1311681110
Howell, M. D., Fahlgren, N., Chapman, E. J., Cumbie, J. S., Sullivan, C. M., Givan, S. A., et al. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA-and tasiRNA-directed targeting. Plant Cell 19, 926–942. doi: 10.1105/tpc.107.050062
Huang, C. F., and Zhu, J. K. (2014). RNA splicing factors and RNA-directed DNA methylation. Biology 3, 243–254. doi: 10.3390/biology3020243
Huang, G., Allen, R., Davis, E. L., Baum, T. J., and Hussey, R. S. (2006). Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. U.S.A. 103, 14302–14306. doi: 10.1073/pnas.0604698103
Huang, S. Q., Peng, J., Qiu, C. X., and Yang, Z. M. (2009). Heavy metal-regulated new microRNAs from rice. J. Inorgan. Biochem. 103, 282–287. doi: 10.1016/j.jinorgbio.2008.10.019
Huang, Y., Kendall, T., and Mosher, R. A. (2013). Pol IV-dependent siRNA production is reduced in Brassica rapa. Biology 2, 1210–1223. doi: 10.3390/biology2041210
Hwang, E.-W., Shin, S.-J., Yu, B.-K., Byun, M.-O., and Kwon, H.-B. (2011). miR171 family members are involved in drought response in Solanum tuberosum. J. Plant Biol. 54, 43–48. doi: 10.1007/s12374-010-9141-8
Hwang, J., Sato, H., Tang, Y., Matsuda, D., and Maquat, L. E. (2010). UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol. Cell 39, 396–409. doi: 10.1016/j.molcel.2010.07.004
Ito, H. (2013). Small RNAs and regulation of transposons in plants. Genes Genet. Syst. 88, 3–7. doi: 10.1266/ggs.88.3
Ito, H., and Kakutani, T. (2014). Control of transposable elements in Arabidopsis thaliana. Chrom. Res. 22, 217–223. doi: 10.1007/s10577-014-9417-9
Ito, H., Gaubert, H., Bucher, E., Mirouze, M., Vaillant, I., and Paszkowski, J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119. doi: 10.1038/nature09861
Ivanov, P. V., Gehring, N. H., Kunz, J. B., Hentze, M. W., and Kulozik, A. E. (2008). Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 27, 736–747. doi: 10.1038/emboj.2008.17
Jagadeeswaran, G., Zheng, Y., Li, Y. F., Shukla, L. I., Matts, J., Hoyt, P., et al. (2009). Cloning and characterization of small RNAs from Medicago truncatula reveals four novel legume-specific microRNA families. New Phytol. 184, 85–98. doi: 10.1111/j.1469-8137.2009.02915.x
Jermann, P., Hoerner, L., Burger, L., and Schübeler, D. (2014). Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 111, E3415–E3421. doi: 10.1073/pnas.1400672111
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42:541. doi: 10.1038/ng.591
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial Immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Johansson, S., Bieber, T., Dahl, R., Friedmann, P. S., Lanier, B. Q., Lockey, R. F., et al. (2004). Revised nomenclature for allergy for global use: report of the nomenclature review committee of the world allergy organization, October 2003. J. Allergy Clin. Immunol. 113, 832–836. doi: 10.1016/j.jaci.2003.12.591
Johnson, L. M., Du, J., Hale, C. J., Bischof, S., Feng, S., Chodavarapu, R. K., et al. (2014). SRA-and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507, 124–128. doi: 10.1038/nature12931
Juarez, M. T., Kui, J. S., Thomas, J., Heller, B. A., and Timmermans, M. C. (2004). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88. doi: 10.1038/nature02363
Kang, I.-H., Steffen, J. G., Portereiko, M. F., Lloyd, A., and Drews, G. N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20, 635–647. doi: 10.1105/tpc.107.055137
Kanno, T., Yoshikawa, M., and Habu, Y. (2013). Locus-specific requirements of DDR complexes for gene-body methylation of TAS genes in Arabidopsis thaliana. Plant Mol. Biol. Rep. 31, 1048–1052. doi: 10.1007/s11105-012-0554-z
Kashima, I., Yamashita, A., Izumi, N., Kataoka, N., Morishita, R., Hoshino, S., et al. (2006). Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 20, 355–367. doi: 10.1101/gad.1389006
Kasschau, K. D., Fahlgren, N., Chapman, E. J., Sullivan, C. M., Cumbie, J. S., Givan, S. A., et al. (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5:e57. doi: 10.1371/journal.pbio.0050057
Kasschau, K. D., Xie, Z., Allen, E., Llave, C., Chapman, E. J., Krizan, K. A., et al. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217. doi: 10.1016/S1534-5807(03)00025-X
Kato, M., Miura, A., Bender, J., Jacobsen, S. E., and Kakutani, T. (2003). Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 13, 421–426. doi: 10.1016/S0960-9822(03)00106-4
Kelly, W. G., and Aramayo, R. (2007). Meiotic silencing and the epigenetics of sex. Chrom. Res. 15, 633–651. doi: 10.1007/s10577-007-1143-0
Khraiwesh, B., Zhu, J.-K., and Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819, 137–148. doi: 10.1016/j.bbagrm.2011.05.001
Kikuchi, K., Terauchi, K., Wada, M., and Hirano, H.-Y. (2003). The plant MITE mPing is mobilized in anther culture. Nature 421, 167–170. doi: 10.1038/nature01218
Kitazumi, A., Kawahara, Y., Onda, T. S., De Koeyer, D., and de los Reyes, B. G. (2015). Implications of miR166 and miR159 induction to the basal response mechanisms of an andigena potato (Solanum tuberosum subsp. andigena) to salinity stress, predicted from network models in Arabidopsis. Genome 58, 13–24. doi: 10.1139/gen-2015-0011
Köhler, C., Wolff, P., and Spillane, C. (2012). Epigenetic mechanisms underlying genomic imprinting in plants. Ann. Rev. Plant Biol. 63, 331–352. doi: 10.1146/annurev-arplant-042811-105514
Kurosaki, T., Li, W., Hoque, M., Popp, M. W.-L., Ermolenko, D. N., Tian, B., et al. (2014). A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev. 28, 1900–1916. doi: 10.1101/gad.245506.114
Lange, B. M. (2010). VIGS – genomics goes functional. Trends Plant Sci. 15, 1–4. doi: 10.1016/j.tplants.2009.09.002
Law, J. A., Du, J., Hale, C. J., Feng, S., Krajewski, K., Palanca, A. M. S., et al. (2013). Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498, 385–389. doi: 10.1038/nature12178
Law, J. A., Vashisht, A. A., Wohlschlegel, J. A., and Jacobsen, S. E. (2011). SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 7:e1002195. doi: 10.1371/journal.pgen.1002195
Le, L. Q., Lorenz, Y., Scheurer, S., Fötisch, K., Enrique, E., Bartra, J., et al. (2006a). Design of tomato fruits with reduced allergenicity by dsRNAi-mediated inhibition of ns-LTP (Lyc e 3) expression. Plant Biotechnol. J. 4, 231–242. doi: 10.1111/j.1467-7652.2005.00175.x
Le, L. Q., Mahler, V., Lorenz, Y., Scheurer, S., Biemelt, S., Vieths, S., et al. (2006b). Reduced allergenicity of tomato fruits harvested from Lyc e 1–silenced transgenic tomato plants. J. Allergy Clin. Immunol. 118, 1176–1183. doi: 10.1016/j.jaci.2006.06.031
Lee, L. A., and Burks, A. W. (2006). Food allergies: prevalence, molecular characterization, and treatment/prevention strategies. Ann. Rev. Nutr. 26, 539–565. doi: 10.1146/annurev.nutr.26.061505.111211
Lewis, R. S., Jack, A. M., Morris, J. W., Robert, V. J., Gavilano, L. B., Siminszky, B., et al. (2008). RNA interference (RNAi)-induced suppression of nicotine demethylase activity reduces levels of a key carcinogen in cured tobacco leaves. Plant Biotechnol. J. 6, 346–354. doi: 10.1111/j.1467-7652.2008.00324.x
Li, J. C., Guo, J. B., Xu, W. Z., and Ma, M. (2007). RNA interference-mediated silencing of phytochelatin synthase gene reduce cadmium accumulation in rice seeds. J. Integr. Plant Biol. 49, 1032–1037. doi: 10.1111/j.1672-9072.2007.00473.x
Li, H., Liang, W., Yin, C., Zhu, L., and Zhang, D. (2011). Genetic interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in specifying rice floral organ identities and meristem determinacy. Plant Physiol. 156, 263–274. doi: 10.1104/pp.111.172080
Li, J., Guo, G., Guo, W., Guo, G., Tong, D., Ni, Z., et al. (2012). miRNA164-directed cleavage of ZmNAC1 confers lateral root development in maize (Zea mays L.). BMC Plant Biol. 12:220. doi: 10.1186/1471-2229-12-220
Li, J., Yang, Z., Yu, B., Liu, J., and Chen, X. (2005). Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15, 1501–1507. doi: 10.1016/j.cub.2005.07.029
Li, M., Wang, R., Zhao, D., and Xiang, H. (2014). Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 42, 2483–2492. doi: 10.1093/nar/gkt1154
Lippman, Z., Gendrel, A.-V., Black, M., Vaughn, M. W., Dedhia, N., McCombie, W. R., et al. (2004). Role of transposable elements in heterochromatin and epigenetic control. Nature 430, 471–476. doi: 10.1038/nature02651
Liu, X., Wu, S., Xu, J., Sui, C., and Wei, J. (2017). Application of CRISPR/Cas9 in plant biology. Acta Pharm. Sin. B 7, 292–302. doi: 10.1016/j.apsb.2017.01.002
Liu, Z. W., Shao, C. R., Zhang, C. J., Zhou, J. X., Zhang, S. W., Li, L., et al. (2014). The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 10:e1003948. doi: 10.1371/journal.pgen.1003948
Lu, C., and Fedoroff, N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12, 2351–2365. doi: 10.2307/3871234
Lu, J., Zhang, C., Baulcombe, D. C., and Chen, Z. J. (2012). Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc. Natl. Acad. Sci. U.S.A. 109, 5529–5534. doi: 10.1073/pnas.1203094109
Luo, M., Dennis, E. S., Berger, F., Peacock, W. J., and Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 17531–17536. doi: 10.1073/pnas.0508418102
Ma, C., and Mitra, A. (2002). Intrinsic direct repeats generate consistent post-transcriptional gene silencing in tobacco. Plant J. 31, 37–49. doi: 10.1046/j.1365-313X.2002.01332.x
Ma, X., Nicole, M.-C., Meteignier, L.-V., Hong, N., Wang, G., and Moffett, P. (2015). Different roles for RNA silencing and RNA processing components in virus recovery and virus-induced gene silencing in plants. J. Exp. Bot. 66, 919–932. doi: 10.1093/jxb/eru447
Makarevitch, I., Eichten, S. R., Briskine, R., Waters, A. J., Danilevskaya, O. N., Meeley, R. B., et al. (2013). Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. Plant Cell 25, 780–793. doi: 10.1105/tpc.112.106427
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., Horvath, P., et al. (2011). Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 9, 467–477. doi: 10.1007/978-1-4939-2687-9_4
Mallory, A. C., and Vaucheret, H. (2006). Functions of microRNAs and related small RNAs in plants. Nat. Genet. 38, S31–S36. doi: 10.1038/ng1791
Mao, Y.-B., Cai, W.-J., Wang, J.-W., Hong, G.-J., Tao, X.-Y., Wang, L.-J., et al. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. doi: 10.1038/nbt1352
Maquat, L. E. (2005). Nonsense-mediated mRNA decay in mammals. J. Cell Sci. 118, 1773–1776. doi: 10.1242/jcs.01701
Marí-Ordóñez, A., Marchais, A., Etcheverry, M., Martin, A., Colot, V., and Voinnet, O. (2013). Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45, 1029–1039. doi: 10.1038/ng.2703
Matzke, M. A., and Mosher, R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408. doi: 10.1038/nrg3683
Matzke, M. A., Kanno, T., and Matzke, A. J. (2015). RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Ann. Rev. Plant Biol. 66, 243–267. doi: 10.1146/annurev-arplant-043014-114633
Matzke, M., Kanno, T., Daxinger, L., Huettel, B., and Matzke, A. J. (2009). RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21, 367–376. doi: 10.1016/j.ceb.2009.01.025
McCormick, S. (2004). Control of male gametophyte development. Plant Cell 16(Suppl. 1), S142–S153. doi: 10.1105/tpc.016659
McCue, A. D., Nuthikattu, S., Reeder, S. H., and Slotkin, R. K. (2012). Gene expression and stress response mediated by the epigenetic regulation of a transposable element small RNA. PLoS Genet. 8:e1002474. doi: 10.1371/journal.pgen.1002474
McGinnis, K. M., Springer, C., Lin, Y., Carey, C. C., and Chandler, V. (2006). Transcriptionally silenced transgenes in maize are activated by three mutations defective in paramutation. Genetics 173, 1637–1647. doi: 10.1534/genetics.106.058669
Medina, C., da Rocha, M., Magliano, M., Ratpopoulo, A., Revel, B., Marteu, N., et al. (2017). Characterization of microRNAs from Arabidopsis galls highlights a role for miR159 in the plant response to the root-knot nematode Meloidogyne incognita. New Phytol. 216, 882–896. doi: 10.1111/nph.14717
Mette, M., Aufsatz, W., Van der Winden, J., Matzke, M., and Matzke, A. (2000). Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19, 5194–5201. doi: 10.1093/emboj/19.19.5194
Meyer, P., Heidmann, I., and Niedenhof, I. (1993). Differences in DNA-methylation are associated with a paramutation phenomenon in transgenic petunia. Plant J. 4, 89–100. doi: 10.1046/j.1365-313X.1993.04010089.x
Miller, J. N., and Pearce, D. A. (2014). Nonsense-mediated decay in genetic disease: friend or foe? Mutat. Res. Rev. Mutat. Res. 762, 52–64. doi: 10.1016/j.mrrev.2014.05.001
Mirouze, M., Reinders, J., Bucher, E., Nishimura, T., Schneeberger, K., Ossowski, S., et al. (2009). Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461, 427–430. doi: 10.1038/nature08328
Mishiba, K., Nishihara, M., Nakatsuka, T., Abe, Y., Hirano, H., Yokoi, T., et al. (2005). Consistent transcriptional silencing of 35S-driven transgenes in gentian. Plant J. 44, 541–556. doi: 10.1111/j.1365-313X.2005.02556.x
Miura, A., Yonebayashi, S., Watanabe, K., Toyama, T., Shimada, H., and Kakutani, T. (2001). Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411, 212–214. doi: 10.1038/35075612
Mojica, F. J. M., Díez-Villaseñor, C., García-Martínez, J., and Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182. doi: 10.1007/s00239-004-0046-3
Mojica, F. J., Díez-Villaseñor, C., García-Martínez, J., and Almendros, C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. doi: 10.1099/mic.0.023960-0
Morel, J. B., Mourrain, P., Béclin, C., and Vaucheret, H. (2000). DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr. Biol. 10, 1591–1594. doi: 10.1016/S0960-9822(00)00862-9
Muller, H. J. (1930). Types of visible variations induced by X-rays in Drosophila. J. Genet. 22, 299–334. doi: 10.1007/BF02984195
Nagy, E., and Maquat, L. E. (1998). A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 23, 198–199. doi: 10.1016/S0968-0004(98)01208-0
Nair, S. K., Wang, N., Turuspekov, Y., Pourkheirandish, M., Sinsuwongwat, S., Chen, G., et al. (2010). Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc. Natl. Acad. Sci. U.S.A. 107, 490–495. doi: 10.1073/pnas.0909097107
Napoli, C., Lemieux, C., and Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2, 279–289. doi: 10.2307/3869076
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.1126088
Nishimasu, H., Ran, F. A., Hsu, P. D., Konermann, S., Shehata, S. I., Dohmae, N., et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949. doi: 10.1016/j.cell.2014.02.001
Nobuta, K., Venu, R., Lu, C., Beló, A., Vemaraju, K., Kulkarni, K., et al. (2007). An expression atlas of rice mRNAs and small RNAs. Nat. Biotechnol. 25, 473–477. doi: 10.1038/nbt1291
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Nuthikattu, S., McCue, A. D., Panda, K., Fultz, D., DeFraia, C., Thomas, E. N., et al. (2013). The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162, 116–131. doi: 10.1104/pp.113.216481
Ogawa, T., Samoto, M., and Takahashi, K. (2000). Soybean allergens and hypoallergenic soybean products. J. Nutr. Sci. Vitaminol. 46, 271–279. doi: 10.3177/jnsv.46.271
Ogita, S., Uefuji, H., Yamaguchi, Y., Koizumi, N., and Sano, H. (2003). Producing decaffeinated coffee plants. Nature 423:823. doi: 10.1038/423823a
Okada-Katsuhata, Y., Yamashita, A., Kutsuzawa, K., Izumi, N., Hirahara, F., and Ohno, S. (2012). N-and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5: SMG-7 during NMD. Nucleic Acids Res. 40, 1251–1266. doi: 10.1093/nar/gkr791
Okamoto, H., and Hirochika, H. (2001). Silencing of transposable elements in plants. Trends Plant Sci. 6, 527–534. doi: 10.1016/S1360-1385(01)02105-7
Palauqui, J. C., Elmayan, T., de Borne, F. D., Crété, P., Charles, C., and Vaucheret, H. (1996). Frequencies, timing, and spatial patterns of co-suppression of nitrate reductase and nitrite reductase in transgenic tobacco plants. Plant Physiol. 112, 1447–1456. doi: 10.1104/pp.112.4.1437
Palusa, S. G., and Reddy, A. S. (2010). Extensive coupling of alternative splicing of pre-mRNAs of serine/arginine (SR) genes with nonsense-mediated decay. New Phytol. 185, 83–89. doi: 10.1111/j.1469-8137.2009.03065.x
Panda, K., and Slotkin, R. K. (2013). Proposed mechanism for the initiation of transposable element silencing by the RDR6-directed DNA methylation pathway. Plant Signal. 8, 116–131. doi: 10.4161/psb.25206
Panstruga, R. (2003). Establishing compatibility between plants and obligate biotrophic pathogens. Curr. Opin. Plant Biol. 6, 320–326. doi: 10.1016/S1369-5266(03)00043-8
Parent, J. S., Martinez de Alba, A. E., and Vaucheret, H. (2012). The origin and effect of small RNA signaling in plants. Front. Plant Sci. 3:179. doi: 10.3389/fpls.2012.00179
Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495. doi: 10.1016/S0960-9822(02)01017-5
Pedram, M., Sprung, C. N., Gao, Q., Lo, A. W., Reynolds, G. E., and Murnane, J. P. (2006). Telomere position effect and silencing of transgenes near telomeres in the mouse. Mol. Cell. Biol. 26, 1865–1878. doi: 10.1128/MCB.26.5.1865-1878.2006
Pélissier, T., and Wassenegger, M. (2000). A DNA target of 30 bp is sufficient for RNA-directed DNA methylation. RNA 6, 55–65. doi: 10.1016/0093-691X(95)00276-E
Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L., and Poethig, R. S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes 18, 2368–2379. doi: 10.1103/PhysRevB.54.4762
Peters, S., Imani, J., Mahler, V., Foetisch, K., Kaul, S., Paulus, K. E., et al. (2011). Dau c 1.01 and Dau c 1.02-silenced transgenic carrot plants show reduced allergenicity to patients with carrot allergy. Transgenic Res. 20, 547–556. doi: 10.1007/s11248-010-9435-0
Phillips, J. R., Dalmay, T., and Bartels, D. (2007). The role of small RNAs in abiotic stress. FEBS Lett. 581, 3592–3597. doi: 10.1016/j.febslet.2007.04.007
Pontier, D., Picart, C., Roudier, F., Garcia, D., Lahmy, S., Azevedo, J., et al. (2012). NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in Arabidopsis. Mol. Cell 48, 121–132. doi: 10.1016/j.molcel.2012.07.027
Price, D. R., and Gatehouse, J. A. (2008). RNAi-mediated crop protection against insects. Trends Biotechnol. 26, 393–400. doi: 10.1016/j.tibtech.2008.04.004
Pumplin, N., and Voinnet, O. (2013). RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat. Rev. Microbiol. 11, 745–760. doi: 10.1038/nrmicro3120
Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20, 3407–3425. doi: 10.1101/gad.1476406
Rajeev Kumar, S., Anunanthini, P., and Ramalingam, S. (2015). Epigenetic silencing in transgenic plants. Front. Plant Sci. 6:693. doi: 10.3389/fpls.2015.00693
Rath, D., Amlinger, L., Rath, A., and Lundgren, M. (2015). The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie 117, 119–128. doi: 10.1016/j.biochi.2015.03.025
Romanel, E., Silva, T. F., Corrêa, R. L., Farinelli, L., Hawkins, J. S., Schrago, C. E., et al. (2012). Global alteration of microRNAs and transposon-derived small RNAs in cotton (Gossypium hirsutum) during Cotton leafroll dwarf polerovirus (CLRDV) infection. Plant Mol. Biol. 80, 443–460. doi: 10.1007/s11103-012-9959-1
Ronemus, M. J., Galbiati, M., Ticknor, C., Chen, J., and Dellaporta, S. L. (1996). Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273, 654–657. doi: 10.1126/science.273.5275.654
Roumi, V., Afsharifar, A., Saldarelli, P., Niazi, A., Martelli, G., and Izadpanah, K. (2012). Transient expression of artificial microRNAs confers resistance to Grapevine virusA in Nicotiana benthamiana. J. Plant Pathol. 94, 643–649. doi: 10.1186/1471-2229-12-198
Ruiz, M. T., Voinnet, O., and Baulcombe, D. C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10, 937–946. doi: 10.1105/tpc.10.6.937
Ryan, B. C., and Vandenbergh, J. G. (2002). Intrauterine position effects. Neurosci. Biobehav. Rev. 26, 665–678. doi: 10.1016/S0149-7634(02)00038-6
Sabbione, A., Daurelio, L., Vegetti, A., Talón, M., Tadeo, F., and Dotto, M. (2019). Genome-wide analysis of AGO, DCL and RDR gene families reveals RNA-directed DNA methylation is involved in fruit abscission in Citrus sinensis. BMC Plant Biol. 19:401. doi: 10.1186/s12870-019-1998-1
Saurabh, S., Vidyarthi, A. S., and Prasad, D. (2014). RNA interference: concept to reality in crop improvement. Planta 239, 543–564. doi: 10.1007/s00425-013-2019-5
Schoft, V. K., Chumak, N., Choi, Y., Hannon, M., Garcia-Aguilar, M., Machlicova, A., et al. (2011). Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc. Natl. Acad. Sci. U.S.A. 108, 8042–8047. doi: 10.1073/pnas.1105117108
Schweingruber, C., Rufener, S. C., Zünd, D., Yamashita, A., and Mühlemann, O. (2013). Nonsense-mediated mRNA decay—mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta 1829, 612–623. doi: 10.1016/j.bbagrm.2013.02.005
Sheehy, R. E., Kramer, M., and Hiatt, W. R. (1988). Reduction of polygalacturonase activity in tomato fruit by antisense RNA. Proc. Natl. Acad. Sci. U.S.A. 85, 8805–8809. doi: 10.1073/pnas.85.23.8805
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Sicherer, S. H., and Sampson, H. A. (2006). Food allergy. J. Allergy Clin. Immunol. 117, S470–S475. doi: 10.1016/j.jaci.2009.08.028
Sijen, T., Fleenor, J., Simmer, F., Thijssen, K. L., Parrish, S., Timmons, L., et al. (2001). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476. doi: 10.1016/S0092-8674(01)00576-1
Silva, G. F. F., Silva, E. M., da Silva Azevedo, M., Guivin, M. A. C., Ramiro, D. A., Figueiredo, C. R., et al. (2014). micro RNA 156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J. 78, 604–618. doi: 10.1111/tpj.12493
Simón-Mateo, C., and García, J. A. (2011). Antiviral strategies in plants based on RNA silencing. Biochim. Biophys. Acta 1809, 722–731. doi: 10.1016/j.bbagrm.2011.05.011
Sinkunas, T., Gasiunas, G., Fremaux, C., Barrangou, R., Horvath, P., and Siksnys, V. (2011). Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 30, 1335–1342. doi: 10.1038/emboj.2011.41
Siritunga, D., and Sayre, R. T. (2003). Generation of cyanogen-free transgenic cassava. Planta 217, 367–373. doi: 10.2307/23387818
Slotkin, R. K., and Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285. doi: 10.1038/nrg2072
Slotkin, R. K., Vaughn, M., Borges, F., Tanurdžić, M., Becker, J. D., Feijó, J. A., et al. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472. doi: 10.1016/j.cell.2008.12.038
Smith, J. A., White, E. A., Sowa, M. E., Powell, M. L., Ottinger, M., Harper, J. W., et al. (2010). Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc. Natl. Acad. Sci. U.S.A. 107, 3752–3757. doi: 10.1073/pnas.0914818107
Stam, M. (2009). Paramutation: a heritable change in gene expression by allelic interactions in trans. Mol. Plant 2, 578–588. doi: 10.1093/mp/ssp020
Stam, M., Mol, J. N., and Kooter, J. M. (1997). The silence of genes in transgenic plants. Ann. Bot. 79, 3–12. doi: 10.1006/anbo.1996.0295
Stangeland, B., Salehian, Z., Aalen, R., Mandal, A., and Olsen, O. A. (2003). Isolation of GUS marker lines for genes expressed in Arabidopsis endosperm, embryo and maternal tissues. J. Exp. Bot. 54, 279–290. doi: 10.1093/jxb/erg031
Sun, C., Feschotte, C., Wu, Z., and Mueller, R. L. (2015). DNA transposons have colonized the genome of the giant virus Pandoravirus salinus. BMC Biol. 13:38. doi: 10.1186/s12915-015-0145-1
Sun, G. (2012). MicroRNAs and their diverse functions in plants. Plant Mol. Biol. 80, 17–36. doi: 10.1007/s11103-011-9817-6
Sunilkumar, G., Campbell, L. M., Puckhaber, L., Stipanovic, R. D., and Rathore, K. S. (2006). “Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol,” in Proceedings of the National Academy of Sciences 103, 18054–18059. doi: 10.1073/pnas.0605389103
Sunkar, R. (2010). MicroRNAs with macro-effects on plant stress responses. Semin. Cell Dev. Biol. 21, 805–811. doi: 10.1016/j.semcdb.2010.04.001
Sunkar, R., and Zhu, J.-K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. doi: 10.1105/tpc.104.022830
Sunkar, R., Li, Y.-F., and Jagadeeswaran, G. (2012). Functions of microRNAs in plant stress responses. Trends Plant Sci. 17, 196–203. doi: 10.1016/j.tplants.2012.01.010
Tada, Y., Nakase, M., Adachi, T., Nakamura, R., Shimada, H., Takahashi, M., et al. (1996). Reduction of 14–16 kDa allergenic proteins in transgenic rice plants by antisense gene. FEBS Lett. 391, 341–345. doi: 10.1016/0014-5793(96)00773-9
Tang, J., and Chu, C. (2017). MicroRNAs in crop improvement: fine-tuners for complex traits. Nat. Plants 3:17077. doi: 10.1038/nplants.2017.77
Teixeira, F. K., Heredia, F., Sarazin, A., Roudier, F., Boccara, M., Ciaudo, C., et al. (2009). A role for RNAi in the selective correction of DNA methylation defects. Science 323, 1600–1604. doi: 10.1126/science.1165313
Thermann, R., Neu-Yilik, G., Deters, A., Frede, U., Wehr, K., Hagemeier, C., et al. (1998). Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 17, 3484–3494. doi: 10.1093/emboj/17.12.3484
Thiebaut, F., Hemerly, A. S., and Ferreira, P. C. G. (2019). A role for epigenetic regulation in the adaptation and stress responses of non-model plants. Front. Plant Sci. 10:246. doi: 10.3389/fpls.2019.00246
Trindade, I., Capitão, C., Dalmay, T., Fevereiro, M. P., and Dos Santos, D. M. (2010). miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 231, 705–716. doi: 10.1007/s00425-009-1078-0
Tsuji, H., Aya, K., Ueguchi-Tanaka, M., Shimada, Y., Nakazono, M., Watanabe, R., et al. (2006). GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 47, 427–444. doi: 10.1111/j.1365-313X.2006.02795.x
Tsukahara, S., Kobayashi, A., Kawabe, A., Mathieu, O., Miura, A., and Kakutani, T. (2009). Bursts of retrotransposition reproduced in Arabidopsis. Nature 461, 423–426. doi: 10.1038/nature08351
Tzfira, T., Li, J., Lacroix, B., and Citovsky, V. (2004). Agrobacterium T-DNA integration: molecules and models. Trends Genet. 20, 375–383. doi: 10.1016/j.tig.2004.06.004
Van der Krol, A. R., Mur, L. A., Beld, M., Mol, J., and Stuitje, A. R. (1990). Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2, 291–299. doi: 10.2307/3869077
Vaquero, A., Scher, M., Erdjument-Bromage, H., Tempst, P., Serrano, L., and Reinberg, D. (2007). SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 450, 440–444. doi: 10.1038/nature06268
Vaucheret, H., and Fagard, M. (2001). Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 17, 29–35. doi: 10.1016/S0168-9525(00)02166-1
Vaucheret, H., Béclin, C., and Fagard, M. (2001). Post-transcriptional gene silencing in plants. J. Cell Sci. 114, 3083–3091. doi: 10.1385/1-59259-775-0:117
Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A. C., et al. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16, 69–79. doi: 10.1016/j.molcel.2004.09.028
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. doi: 10.1016/j.cell.2009.01.046
Wagner, K. D., Wagner, N., Ghanbarian, H., Grandjean, V., Gounon, P., Cuzin, F., et al. (2008). RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev. Cell 14, 962–969. doi: 10.1016/j.devcel.2008.03.009
Wakimoto, B. T. (1998). Beyond the nucleosome: epigenetic aspects of position–effect variegation in Drosophila. Cell 93, 321–324. doi: 10.1016/S0092-8674(00)81159-9
Wang, H., La Russa, M., and Qi, L. S. (2016). CRISPR/Cas9 in genome editing and beyond. Ann. Rev. Biochem. 85, 227–264. doi: 10.1146/annurev-biochem-060815-014607
Wang, J., Mei, J., and Ren, G. (2019). Plant microRNAs: biogenesis, homeostasis, and degradation. Front. Plant Sci. 10:360. doi: 10.3389/fpls.2019.00360
Wang, M. B., and Smith, N. A. (2016). Satellite RNA pathogens of plants: impacts and origins—an RNA silencing perspective. Wiley Interdiscip. Rev. RNA 7, 5–16. doi: 10.1002/wrna.1311
Wang, M.-B., and Metzlaff, M. (2005). RNA silencing and antiviral defense in plants. Curr. Opin. Plant Biol. 8, 216–222. doi: 10.1016/j.pbi.2005.01.006
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44:950. doi: 10.1038/ng.2327
Wang, Y., Itaya, A., Zhong, X., Wu, Y., Zhang, J., van der Knaap, E., et al. (2011). Function and evolution of a MicroRNA that regulates a Ca2+-ATPase and triggers the formation of phased small interfering RNAs in tomato reproductive growth. Plant Cell 23, 3185–3203. doi: 10.1105/tpc.111.088013
Wassenegger, M. (2002). Gene silencing-based disease resistance. Transgenic Res. 11, 639–653. doi: 10.1023/A:1021130127700
Wassenegger, M., Heimes, S., Riedel, L., and Sänger, H. L. (1994). RNA-directed de novo methylation of genomic sequences in plants. Cell 76, 567–576. doi: 10.1016/0092-8674(94)90119-8
Wei, Y., Terns, R. M., and Terns, M. P. (2015). Cas9 function and host genome sampling in Type II-A CRISPR–Cas adaptation. Genes Dev. 29, 356–361. doi: 10.1101/gad.257550.114
Weigel, D., Ahn, J. H., Blázquez, M. A., Borevitz, J. O., Christensen, S. K., Fankhauser, C., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122, 1003–1014. doi: 10.1104/pp.122.4.1003
Weiler, K. S., and Wakimoto, B. T. (1995). Heterochromatin and gene expression in Drosophila. Ann. Rev. Genet. 29, 577–605. doi: 10.1109/TED.2006.874759
Weinhofer, I., Hehenberger, E., Roszak, P., Hennig, L., and Köhler, C. (2010). H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 6:e1001152. doi: 10.1371/journal.pgen.1001152
Whitehurst, A. W., Bodemann, B. O., Cardenas, J., Ferguson, D., Girard, L., Peyton, M., et al. (2007). Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 446, 815–819. doi: 10.1038/nature05697
Wierzbicki, A. T. (2012). The role of long non-coding RNA in transcriptional gene silencing. Curr. Opin. Plant Biol. 15, 517–522. doi: 10.1016/j.pbi.2012.08.008
Wu, L., Mao, L., and Qi, Y. (2012). Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol. 160, 990–999. doi: 10.1104/pp.112.200279
Xie, M., Ren, G., Zhang, C., and Yu, B. (2012). The DNA-and RNA-binding protein FACTOR of DNA METHYLATION 1 requires XH domain-mediated complex formation for its function in RNA-directed DNA methylation. Plant J. 72, 491–500. doi: 10.1111/j.1365-313X.2012.05092.x
Xie, Z., Johansen, L. K., Gustafson, A. M., Kasschau, K. D., Lellis, A. D., Zilberman, D., et al. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2:e104. doi: 10.1371/journal.pbio.0020104
Xie, Z., Kasschau, K. D., and Carrington, J. C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13, 784–789. doi: 10.1016/S0960-9822(03)00281-1
Xin, M., Yang, R., Li, G., Chen, H., Laurie, J., Ma, C., et al. (2013). Dynamic expression of imprinted genes associates with maternally controlled nutrient allocation during maize endosperm development. Plant Cell 25, 3212–3227. doi: 10.1105/tpc.113.115592
Yadav, B. C., Veluthambi, K., and Subramaniam, K. (2006). Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol. Biochem. Parasitol. 148, 219–222. doi: 10.1016/j.molbiopara.2006.03.013
Yamasaki, H., Abdel-Ghany, S. E., Cohu, C. M., Kobayashi, Y., Shikanai, T., and Pilon, M. (2007). Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 282, 16369–16378. doi: 10.1074/jbc.M700138200
Ye, J., Wang, X., Hu, T., Zhang, F., Wang, B., Li, C., et al. (2017). An InDel in the promoter of Al-ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell 29, 2249–2268. doi: 10.1105/tpc.17.00211
Yuan, J., Chen, S., Jiao, W., Wang, L., Wang, L., Ye, W., et al. (2017). Both maternally and paternally imprinted genes regulate seed development in rice. New Phytol. 216, 373–387. doi: 10.1111/nph.14510
Zaratiegui, M., Irvine, D. V., and Martienssen, R. A. (2007). Noncoding RNAs and gene silencing. Cell 128, 763–776. doi: 10.1016/j.cell.2007.02.016
Zaynab, M., Sharif, Y., Fatima, M., Afzal, M. Z., Aslam, M. M., Raza, M. F., et al. (2020). CRISPR/Cas9 to generate plant immunity against pathogen. Microb. Pathogen. 141:103996. doi: 10.1016/j.micpath.2020.103996
Zebec, Z., Zink, I. A., Kerou, M., and Schleper, C. (2016). Efficient CRISPR-mediated post-transcriptional gene silencing in a hyperthermophilic archaeon using multiplexed crRNA expression. G3 6, 3161–3168. doi: 10.1534/g3.116.032482
Zhang, X., Zhu, Y., Liu, X., Hong, X., Xu, Y., Zhu, P., et al. (2015). Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science 348, 120–123. doi: 10.1126/science.aaa2618
Zhang, Y., Zhang, Q., Hao, L., Wang, S., Wang, S., Zhang, W., et al. (2019). A novel miRNA negatively regulates resistance to Glomerella leaf spot by suppressing expression of an NBS gene in apple. Hortic. Res. 6:93. doi: 10.1038/s41438-019-0175-x
Zhang, Y.-C., Yu, Y., Wang, C.-Y., Li, Z.-Y., Liu, Q., Xu, J., et al. (2013). Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 31, 848–852. doi: 10.1038/nbt.2646
Zhao, B., Ge, L., Liang, R., Li, W., Ruan, K., Lin, H., et al. (2009). Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 10:29. doi: 10.1186/1471-2199-10-29
Zheng, X., Yang, L., Li, Q., Ji, L., Tang, A., Zang, L., et al. (2018). MIGS as a simple and efficient method for gene silencing in rice. Front. Plant Sci. 9:662. doi: 10.3389/fpls.2018.00662
Zhou, J., Peng, Z., Long, J., Sosso, D., Liu, B., Eom, J. S., et al. (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643. doi: 10.1111/tpj.12838
Zhu, Y., Rowley, M. J., Böhmdorfer, G., and Wierzbicki, A. T. (2013). A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol. Cell 49, 298–309. doi: 10.1016/j.molcel.2012.11.011
Zimmermann, T. S., Lee, A. C., Akinc, A., Bramlage, B., Bumcrot, D., Fedoruk, M. N., et al. (2006). RNAi-mediated gene silencing in non-human primates. Nature 441, 111–114. doi: 10.1038/nature04688
Keywords: transcriptional gene silencing, post-transcriptional gene silencing, genomics imprinting, paramutation, RNAi, CRISPR/Cas9
Citation: El-Sappah AH, Yan K, Huang Q, Islam MM, Li Q, Wang Y, Khan MS, Zhao X, Mir RR, Li J, El-Tarabily KA and Abbas M (2021) Comprehensive Mechanism of Gene Silencing and Its Role in Plant Growth and Development. Front. Plant Sci. 12:705249. doi: 10.3389/fpls.2021.705249
Received: 04 May 2021; Accepted: 10 August 2021;
Published: 07 September 2021.
Edited by:
Peng Zhang, Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Rajarshi Kumar Gaur, Deen Dayal Upadhyay Gorakhpur University, IndiaCopyright © 2021 El-Sappah, Yan, Huang, Islam, Li, Wang, Khan, Zhao, Mir, Li, El-Tarabily and Abbas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manzar Abbas, YWJiYXMyNDcyQGhvdG1haWwuY29t; Khaled A. El-Tarabily, a3RhcmFiaWx5QHVhZXUuYWMuYWU=; Jia Li, amlhLmxpMTZAYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.