
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 13 May 2021
Sec. Plant Biotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fpls.2021.682953
This article is part of the Research Topic Engineering the Plant Biofactory for the Production of Biologics and Small-Molecule Medicines, Volume 2 View all 13 articles
 Konlavat Siriwattananon1,2
Konlavat Siriwattananon1,2 Suwimon Manopwisedjaroen3
Suwimon Manopwisedjaroen3 Balamurugan Shanmugaraj4
Balamurugan Shanmugaraj4 Kaewta Rattanapisit4
Kaewta Rattanapisit4 Supaporn Phumiamorn5
Supaporn Phumiamorn5 Sompong Sapsutthipas5
Sompong Sapsutthipas5 Sakalin Trisiriwanich5
Sakalin Trisiriwanich5 Eakachai Prompetchara6,7
Eakachai Prompetchara6,7 Chutitorn Ketloy6,7
Chutitorn Ketloy6,7 Supranee Buranapraditkun6,8
Supranee Buranapraditkun6,8 Wassana Wijagkanalan9
Wassana Wijagkanalan9 Kittipan Tharakhet6
Kittipan Tharakhet6 Papatsara Kaewpang6
Papatsara Kaewpang6 Kantinan Leetanasaksakul10
Kantinan Leetanasaksakul10 Taratorn Kemthong11
Taratorn Kemthong11 Nutchanat Suttisan11
Nutchanat Suttisan11 Suchinda Malaivijitnond11
Suchinda Malaivijitnond11 Kiat Ruxrungtham6,8
Kiat Ruxrungtham6,8 Arunee Thitithanyanont3
Arunee Thitithanyanont3 Waranyoo Phoolcharoen1,2*
Waranyoo Phoolcharoen1,2*The emergence of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected global public health and economy. Despite the substantial efforts, only few vaccines are currently approved and some are in the different stages of clinical trials. As the disease rapidly spreads, an affordable and effective vaccine is urgently needed. In this study, we investigated the immunogenicity of plant-produced receptor-binding domain (RBD) of SARS-CoV-2 in order to use as a subunit vaccine. In this regard, RBD of SARS-CoV-2 was fused with Fc fragment of human IgG1 and transiently expressed in Nicotiana benthamiana by agroinfiltration. The plant-produced RBD-Fc fusion protein was purified from the crude extract by using protein A affinity column chromatography. Two intramuscular administration of plant-produced RBD-Fc protein formulated with alum as an adjuvant have elicited high neutralization titers in immunized mice and cynomolgus monkeys. Further it has induced a mixed Th1/Th2 immune responses and vaccine-specific T-lymphocyte responses which was confirmed by interferon-gamma (IFN-γ) enzyme-linked immunospot assay. Altogether, our results demonstrated that the plant-produced SARS-CoV-2 RBD has the potential to be used as an effective vaccine candidate against SARS-CoV-2. To our knowledge, this is the first report demonstrating the immunogenicity of plant-produced SARS-CoV-2 RBD protein in mice and non-human primates.
In December 2019, the unknown pneumonia cases have been first reported in Wuhan, Hubei Province, China, which were initially reported to be caused by novel coronavirus (nCoV-2019) and later named as SARS-CoV-2. The disease condition associated with it is referred as Coronavirus Disease (COVID-19). The virus was closely related to the severe acute respiratory syndrome coronavirus (SARS-CoV) that cause massive outbreak in 2002–2003 (Amanat and Krammer, 2020; Li Q. et al., 2020; Malik et al., 2020; Quinlan et al., 2020; She et al., 2020; Singhal, 2020). In short time, the virus spreads rapidly to more than 200 countries (Malik et al., 2020; She et al., 2020). As of April 2021, more than 130 million confirmed cases with a toll of more than 2.9 million deaths were globally reported and the number of infected patients has still been exponentially increasing daily (World Health Organization, 2021). Furthermore, only few vaccines are currently approved. Thus, the development of affordable effective vaccine or therapeutics is highly essential to control and prevent the infection.
SARS-CoV-2 belongs to the family Coronaviridae in the genera of Betacoronavirus, which are known to infect mammals. Coronaviruses (CoVs) are enveloped and single-stranded positive sense RNA viruses (Banerjee et al., 2019; Amanat and Krammer, 2020; Li H. et al., 2020; Rabi et al., 2020; Yuki et al., 2020). The CoV genome consists of 6–11 open reading frames (ORFs) encoding for non-structural polyproteins and structural proteins. The SARS-CoVs have four major structural proteins such as spike (S) surface glycoprotein, membrane (M) protein, envelope (E) protein, and nucleocapsid (N) protein, which are essential for viral assembly and infection (Bosch et al., 2003; Masters, 2006; Banerjee et al., 2019; Li H. et al., 2020; Malla et al., 2020; Rabi et al., 2020; Yuki et al., 2020). S glycoprotein plays a major role in viral attachment to host cells during the viral infection and cleaved by the host proteases into S1 and S2 subunits. SARS-CoV-2 infection starts with pre-fusion of receptor binding domain (RBD) located on the S1 subunit to host receptor, angiotensin converting enzyme 2 (ACE2 receptor) and followed by S2 subunit post-fusion, leading to viral RNA penetration into host cells (Li et al., 2003; Li, 2016; Yuan et al., 2017; Walls et al., 2019; Rabi et al., 2020; Shanmugaraj et al., 2020c; Yuki et al., 2020; Zhou et al., 2020). In addition, RBD was known to elicit potent immune response and considered as prime target for eliciting of host neutralizing antibodies (Wang et al., 2018; Li H. et al., 2020; Smith et al., 2020). Furthermore, previous studies demonstrated that the sera isolated from animals immunized with inactivated SARS-CoV virus significantly neutralize the virus by inhibiting the binding of RBD with ACE2 receptor which proved that the antibodies targeting the RBD domain could prevent the virus infection (He et al., 2004, 2005a,2005b; Zhu et al., 2013; Li H. et al., 2020).
Currently, recombinant proteins are produced mainly by bacterial fermentation or mammalian cell cultures, which still have many limitations including high production costs, immunogenicity profile, and pathogen contamination (Kelley, 2009; Phoolcharoen et al., 2011; Gomes et al., 2016; Kodati et al., 2016; Fuenmayor et al., 2017; Kaur et al., 2018; Rattanapisit et al., 2019a). Plant expression system is considered as the cost-effective platform for the production of vaccine antigens, diagnostic reagents, valuable biopharmaceuticals such as therapeutic immunoglobulins, human enzymes, and human growth factors (Gleba et al., 2005; Miao et al., 2008; Phoolcharoen et al., 2011; Ahmad et al., 2019; Rattanapisit et al., 2019a,b, 2021). Plant expression system offers several advantages in terms of rapidity, flexibility, post-translational modification of recombinant proteins, safety due to lack of animal pathogen, toxin contamination and scalability compared to other available conventional systems (Vitale and Denecke, 1999; Ma et al., 2003; Phoolcharoen et al., 2011; Shahid and Daniell, 2016; Bellucci et al., 2017). Hence, plant platform can be considered as an alternative platform for economical production of commercially viable biopharmaceuticals and vaccines especially for developing countries during pandemic situation (Phoolcharoen et al., 2011; Shanmugaraj et al., 2020a).
In this study, we produced SARS-CoV-2 RBD-Fc fusion protein by fusing SARS-CoV-2 RBD to the Fc domain of human IgG1 at the C-terminus and cloned into geminiviral vector for expression in Nicotiana benthamiana plants. The plant-produced RBD-Fc showed specific binding with both human embryonic kidney 293 (HEK293) and Chinese hamster ovary (CHO) cells produced ACE2 protein. Further the plant-produced RBD-Fc was shown to be immunogenic and significantly boosted a humoral and cell-mediated immune response in both mice and cynomolgus macaques (Macaca fascicularis). Our results demonstrated that this plant-produced protein has the potential for use as a vaccine candidate against SARS-CoV-2.
The RBD of SARS-CoV-2 (SARS-CoV-2 RBD) (Genbank accession number: YP_009724390.1; F318-C617) was designed to anneal with Fc region of human immunoglobulin G1 (IgG1) (GenBank accession number: 4CDH_A) by peptide linker 3XGGGGs at the C-terminus (Figure 1). The nucleotide sequence of SARS-CoV-2 RBD was codon optimized and commercially synthesized (Genewiz, Suzhou, China) with XbaI and BamHI restriction sites for cloning with the Fc region, that contains BamHI and SacI restriction sites at the 5′ and 3′ ends, respectively. The SARS-CoV-2 RBD and human Fc region were ligated into a geminiviral vector (pBYR2eK2Md; pBYR2e) (Chen et al., 2011; Diamos and Mason, 2018) using XbaI and SacI restriction sites to construct the plant expression vector pBYR2e-SARS-CoV-2 RBD-Fc. The murine leader sequence (Shanmugaraj et al., 2020b) and ER retention signal peptide (SEKDEL) was included at N-terminus and C-terminus of the gene construct, respectively (Figure 1).
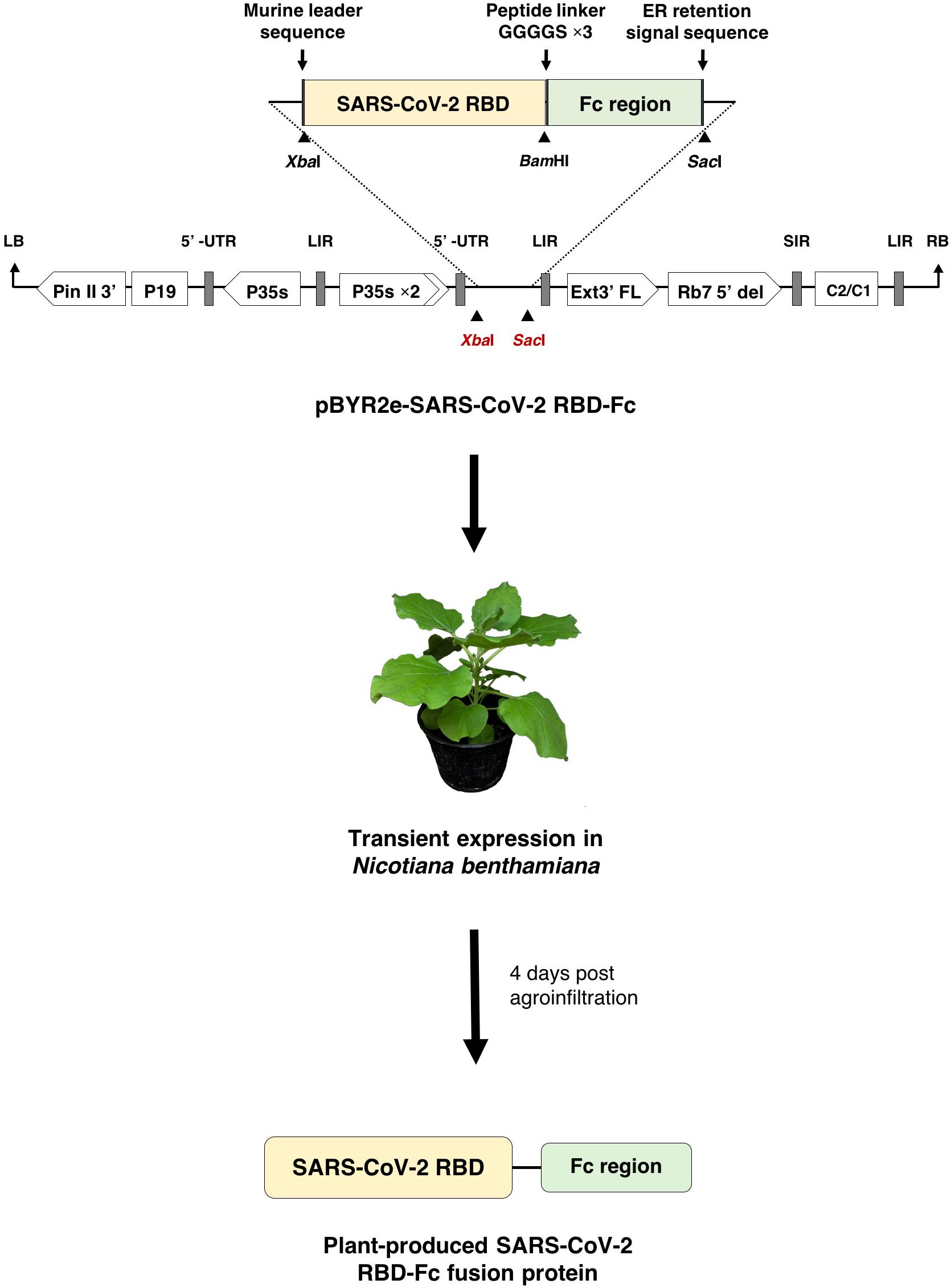
Figure 1. Diagrammatic representation showing the T-DNA of plant expression vector pBYR2e-SARS-CoV-2 RBD-Fc and the overview of transient expression in N. benthamiana plants. RB and LB, left and right borders of the T-DNA used in Agrobacterium DNA delivery into plant cells; Pin II 3’, the terminator from potato proteinase inhibitor II gene; P19, the RNA silencing suppressor from Tomato Bushy Stunt Virus (TBSV); P35s, 35s promoter from Cauliflower Mosaic Virus (CaMV); P35s × 2, 35s promoter from CaMV with duplicated enhancer; Ext3’ FL, 3’ region of tobacco extension gene; Rb7 5’ del, tobacco RB7 promoter; SIR, short intergenic region of Bean Yellow Dwarf Virus (BeYDV); LIR, long intergenic region of BeYDV; C2/C1, Rep/RepA gene from BeYDV encoding for replication initiation protein (Rep) and RepA.
The plant expression vector pBYR2e-SARS-CoV-2 RBD-Fc was transformed into Agrobacterium tumefaciens strain GV3101 cells by electroporation (MicroPulser, Bio-Rad, United States). The recombinant Agrobacterium clones were confirmed by polymerase chain reaction (PCR) using the RBD gene-specific primers. Agrobacterium containing pBYR2e-SARS-CoV-2 RBD-Fc was resuspended with 1xinfiltration buffer (10 mM 2-(N-morpholino) etanesulfonic acid (MES), 10 mM MgSO4, at pH 5.5) to get final OD600 of 0.2 prior to agroinfiltration. The Agrobacterium suspension was injected into the adaxial side of 6-week-old N. benthamiana leaves. The infiltrated plants were maintained in an optimal 16-h light/8-h dark condition at 28°C and harvested after 4 days post infiltration (dpi).
The infiltrated leaves were harvested and extracted with 1xPBS (phosphate-buffered saline: 137 mM NaCl, 2.68 mM KCl, 10.1 mM Na2HPO4, 1.76 mM KH2PO4 pH 7.4) and clarified by centrifugation at 26,000 g for 45 min at 4°C. The supernatant was filtered by using 0.45 μm S-Pak membrane (Merck, Massachusetts, United States). The clarified supernatant was purified by affinity column chromatography with protein-A beads (Expedeon, Cambridge, United Kingdom). The purified column was equilibrated and washed by 1xPBS pH 7.4 followed by elution with 0.1 M glycine buffer pH 3. Subsequently, the pH of the eluted proteins was neutralized by using Tris-HCl pH 8.8. The purified SARS-CoV-2 RBD-Fc was concentrated by using Amicon® ultracentrifugal filter (Merck, Massachusetts, United States) and filtered through 0.22 μm syringe filter (Merck, Massachusetts, United States). The purified plant-produced SARS-CoV-2 RBD-Fc fusion protein was analyzed by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting under reducing and non-reducing conditions. The SARS-CoV-2 RBD-Fc samples were subjected to 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis and stained by Coomassie staining solution for protein visualization. For western blot analysis, the separated proteins were transferred to nitrocellulose membrane (Biorad, United States) and detected with a 1:5,000 sheep anti-human gamma chain-HRP conjugate antibody diluted in 1xPBS (The Binding Site, United Kingdom). The yield of purified plant-produced RBD-Fc fusion protein was estimated by direct ELISA assay using human IgG (Abcam, United Kingdom) as protein standard. The samples were probed by using anti-human gamma chain-HRP fusion (The Binding Site, United Kingdom) at the dilution of 1:1,000 in 1xPBS. A 3,3,5,5′-Tetramethylbenzidine (TMB) solution (Promega, United States) was added into the plate as a colorimetric developer followed by the addition of 1M H2SO4. The absorbance at 450 nm was read using a 96-well plate reader (Molecular Devices, United States).
The purified proteins were separated on SDS-PAGE. The targeted protein band was excised and sent to National Center for Genetic Engineering and Biotechnology, Pathum Thani, Thailand for LC-MS analysis. The protein was enzymatically digested with trypsin and injected into Hybrid quadrupole Q-Tof impact IITM (Bruker Daltonics Ltd., Hamburg, Germany) equipped with a Nano-captive spray ion source was coupled to a nanoLC system: Ultimate 3000 LC System (Thermo Fisher Scientific, United States). Equipment operation was controlled by Compass 1.9 software (Bruker Daltonics Ltd., Hamburg, Germany). The resulting MS/MS spectra were searched using the Mascot Sever (Matrix Science) against SwissProt database. For Mascot searches, the peptide mass tolerance was set at 0.6 Da and the fragment mass tolerance was set at 1.2 Da.
The binding activity of plant-produced SARS-CoV-2 RBD-Fc fusion protein to ACE2 protein was demonstrated by ELISA. Briefly, 96-well ELISA plate was coated by 100 ng of two different ACE2 protein derived either from HEK293-cells (Abcam, United Kingdom) or CHO-cells (InvivoGen, California, United States). For blocking, 5% skim milk in 1xPBS was added into the wells and incubated for 2 h at 37°C. After blocking, the plate was washed three times with 1xPBST (1xPBS plus 0.05% Tween-20) and incubated with various concentrations of plant-produced SARS-CoV-2 RBD-Fc fusion protein in 1xPBS. The SARS-CoV-2 RBD proteins in the wells were detected by addition of 1: 100 dilution of plant-produced anti-SARS-CoV-2 (H4) mAb (Shanmugaraj et al., 2020b) and followed by a 1:1,000 dilution of anti-human Kappa chain-HRP fusion (SouthernBiotech, United States) in 1xPBS for 1 h at 37°C. For colorimetric development, a TMB solution (Promega, United States) was added into the wells followed by addition of 1M H2SO4 for terminating the enzymatic reaction. The absorbance at 450 nm was measured using a 96-well microplate reader (Molecular Devices, United States).
Mice immunization protocols were approved by the Institutional Animal Care and Use Committee, Faculty of Medicine, Chulalongkorn University (Protocol review No. 012/2563). Seven-week-old female ICR mice (n = 5 per group) were intramuscularly (IM) immunized via., anterior tibialis with 10 μg of plant-produced SARS-CoV-2 RBD-Fc fusion protein without adjuvant or formulated with 0.1 mg alum (InvivoGen, California, United States) on days 0 and 21. Mice sera were collected prior to the first immunization (pre-bleed, day 0) and 14 days post-vaccination to assess the SARS-CoV-2-specific antibody response. The mice were sacrificed on day 35 (14 days after second booster) to collect the spleen for quantitative measurement of SARS-CoV-2 RBD-specific T-cell responses.
For non-human primate immunogenicity studies, all procedures were reviewed and approved by the National Primate Research Center of Thailand-Chulalongkorn University (NPRCT-CU) Animal Care and Use Committee (Protocol review No. 207512) and the facility has been AAALAC International Accredited (1752). Thirteen SPF juvenile cynomolgus macaques (Macaca fascicularis), aged between 2.5 and 3.5 years old, and body weight between 2.18 and 3.17 kg, were assigned into three groups in which the control group (n = 3) was immunized by PBS adjuvanted with 0.5 mg alum, and other two groups (n = 5 per group) were administered with 25 and 50 μg of plant-produced SARS-CoV-2 RBD-Fc fusion protein along with 0.5 mg alum adjuvant, respectively. Monkeys were received two intramuscular injections on day 0 and 21. The blood samples were collected on day 0 (pre-immunization) and 14 days after each vaccination (days 14 and 35) to assess SARS-CoV-2 RBD specific IgG, neutralizing antibody and cell-mediated immune responses.
96-well plate was coated with 100 ng of SARS-CoV-2 spike protein RBD derived from Sf9 insect cells (GenScript, United States) and incubated overnight at 4°C. Then, the wells were blocked with 5% skim milk powder in 1xPBS for 2 h at 37°C. Subsequently, the animal sera were twofold serially diluted with 1xPBS starting at 1:100 was loaded on the wells and incubated for 2 h at 37°C. Goat anti-mouse IgG HRP conjugate antibody (Jackson ImmunoResearch, Pennsylvania, United States) and goat anti-monkey IgG HRP conjugation (Abcam, United Kingdom) diluted 1:2,000 in 1xPBS were added into the wells for detecting mouse and monkey antibodies, respectively, and the plate was incubated for 2 h at 37°C. TMB substrate (Promega, United States) was added into the plates for colorimetric development. The enzymatic reactions were terminated by adding 1M H2SO4. The absorbance was measured at 450 nm using a microplate reader (BMG Labtech, Germany). Between each step, the plates were washed by 1xPBST for three times. For mouse IgG1 and IgG2a analysis, the mice sera were twofold serially diluted with 1xPBS starting 1:100 in the same fashion and detected by 1:2,000 goat anti-mouse IgG1 (HRP) and goat anti-mouse IgG2a heavy chain (HRP) antibody (Abcam, United Kingdom), respectively, diluted in 1xPBS. The endpoint titer of IgG1 and IgG2a were also computed for monitoring Th2 and Th1 lymphocyte responses, respectively. The endpoint titers were determined as the highest dilution of immunized sera, which had A450 more than cut off calculated from A450 of pre-immunized sera in the dilution of 1:100 in 1xPBS (Frey et al., 1998). All experiments were performed in duplicates and 1xPBS was used as a control. The statistical analyses of immunological data were performed using GraphPad Prism software version 8.0. Statistical significance was calculated by two-way analysis of variance (ANOVA). All data in each group were compared by using Tukey’s multiple comparisons test and the values of p < 0.05 were considered as statistically significant.
Microneutralization assay was performed using Vero E6 cell line and live SARS-CoV-2 virus isolated from a COVID-19 patient and conducted in a certified biosafety level 3 facility of Microbiology department, Faculty of Science, Mahidol University, Bangkok, Thailand. The cells were plated in 96-well plate at 1 × 104 cells/well in DMEM (Dulbecco’s Modified Eagle’s medium supplemented with 10% heat-inactivated FBS, 100 U/mL of penicillin and 0.1 mg/mL of streptomycin) and incubated for overnight. The immunized sera and positive convalescent serum from COVID-19 patient were heat-inactivated at 56°C for 30 min. The immunized sera were twofold serially diluted in duplicates and incubated with 100 of 50% tissue culture infective dose (TCID50) of the SARS-CoV-2 virus in DMEM at 37°C for 1 h. Virus control at 100TCID50 and uninfected cell control wells were included in all plates. Subsequently, the mixture of diluted serum and virus was transferred to the cell monolayer and incubated at 37°C with 5% CO2 for 2 days. After incubation, the cells were washed once with 1xPBS and then fixed and permeabilized with ice-cold 1:1 methanol/acetone fixative solution at 4°C for 20 min. The cells were washed 3 times with 1xPBST and blocked with 2% BSA at room temperature (RT) for 1 h. After washing, the viral nucleocapsid was detected using 1:5,000 of SARS-CoV/SARS-CoV-2 nucleocapsid (N) monoclonal antibody (SinoBiological, United States) and incubated at 37°C for 1 h followed by adding 1:2,000 of HRP-conjugated goat anti-rabbit polyclonal antibody (Dako, Denmark) diluted with 1xPBS and incubated at 37°C for 1h. The KPL SureblueTM TMB substrate (SeraCare, United States) was added for colorimetric development. Then the reaction was stopped by the addition of 1N HCl. The absorbance at 450 and 620 nm was read by a SunriseTM microplate reader (Tecan, Switzerland). The absorbance differences between 450 and 620 nm (A450–A620) of diluted samples were compared with the 50% specific signal of the cut point, which was calculated by the following equation to determine the potent neutralization titers of the immunized sera.
Where AVirus control and Acell control are the average of A450-A620 of virus control wells and cell control wells, respectively. The neutralizing titers were defined as the reciprocal highest dilution providing the average of A450–A620 of the diluted serum well more than the cut point. The neutralizing antibody titers of each experimental group were compared by GraphPad Prism 8.0 using Mann-Whitney test. The significant differences were considered when p < 0.05.
For mouse splenocyte preparation, the spleen cells were aseptically plated in the petri-dish and dissociated into single-cell suspension using needle#21 for 2–3 times. The separated splenocytes were cultured in R5 medium (RPMI1640 with 100 U/mL penicillin, 100 U/mL streptomycin, 5% heat-inactivated fetal bovine serum (FBS, Gibco, United States) and 2-mercaptoethanol) and centrifuged at 1,200 g 4°C for 5 min. Subsequently, splenocytes were lysed with 1xACK lysis buffer, added R5 medium and collected the cell pellet by centrifugation at 1,200 g 4°C for 5 min. Finally, splenocytes were counted and adjusted for using in ELISpot assay. The IFN-γ secreting cells were quantified by using mouse IFN-γ ELISpot assay kit (Mabtech, Stockholm, Sweden). Briefly, splenocytes were resuspended at 5 × 106 cells/mL in R5 medium. The 96-well plates (Millipore, Bedford, MA, United States) were coated with 500 ng of anti-mouse IFN-γ (AN18) monoclonal antibody (mAb) (Mabtech, Stockholm, Sweden) in 1xPBS at 37°C with 5% CO2 for 3 h. Then, the plates were washed six times with 1xPBS and blocked with R10 medium at RT for 1 h. A quantity of 5 × 105 splenocytes per well were activated by lyophilized SARS-CoV-2 peptide pools (BioNet-Asia, Thailand, and Mimotopes, Australia) at a final concentration of 2 μg/mL at 37°C with 5% CO2 for 40 h. R5 medium and concanavalin A (ConA) were used as negative and positive controls, respectively. After incubation, the plates were washed six times with 1xPBST followed by three times with 1xPBS. Then, the plates were incubated with anti-mouse IFN-γ-biotinylated mAb (Mabtech, Stockholm, Sweden) diluted in 1xPBS at RT for 3 h. After washing, streptavidin-alkaline phosphatase (ALP: Mabtech, Stockholm, Sweden) was added and incubated at RT for 1 h. The substrate solution (5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium; BCIP/NBT) were added into the wells and incubated until distinct spots emerge. Reactions were stopped by washing extensively in tap water and rinsing the underside of membrane. Inspect and count spots were performed with an ELISpot reader (ImmunoSpot® Analyzer, United States). Results were expressed as spot-forming cells (SFCs)/106 splenocytes. The positive responses were defined as > 50 SFCs/106 splenocytes over the background signal. The result analyses were conducted using Kruskal-Wallis test in GraphPad Prism 6.0. All p-values < 0.05 were defined as significant.
For peripheral blood mononuclear cells (PBMCs) preparation, the cells were isolated by density gradient separation using Isoprep (Robbins Scientific Corporation, CA). Briefly, after removal of plasma, EDTA-treated whole blood was diluted (1:1) with RPMI1640 medium containing 2 mM L-Glutamine (Gibco, United States) and layered over Isoprep. Samples were then centrifuged at 1,200 g for 30 min. The PBMC layer was harvested and washed twice with RPMI1640. Then, the cells were resuspended in R10 medium (RPMI1640 supplemented with 100 U/mL of penicillin, 100 U/mL of streptomycin and 10% heat-inactivated fetal bovine serum (FBS, Gibco, United States) for applying in ELISpot assay. The antigen-specific cells secreting monkey IFN-γ were enumerated by using monkey IFN-γ ELISpot assay kit (Mabtech, Stockholm, Sweden). Briefly, PBMCs were stimulated with SARS-CoV-2 spike peptides pools (BioNet-Asia, Thailand, and Mimotopes, Australia) at 37°C with 5% CO2 for 40 h. R5 medium and phytohemagglutinin (PHA) were served as negative and positive control, respectively. The secreted monkey IFN-γ were detected by anti-monkey IFN-γ-biotinylated mAb (Mabtech, Stockholm, Sweden) and followed by addition of ALP solution (Mabtech, Stockholm, Sweden). For spot development, BCIP/NBT-plus substrate solution was added into the wells and incubated until distinct spots emerge. The spots were inspected and counted by ELISpot reader (ImmunoSpot® Analyzer, Germany). Results were expressed as spot-forming cells (SFCs)/106 PBMCs following the subtraction of the negative control. The positive responses were defined > 100 SFCs/106 PBMCs over the background. Statistical analyses were presented using GraphPad Prism 8.0. Comparison of frequencies of populations in each group was made using Friedman and Mann-Whitney tests. All p-values < 0.05 were defined as significant.
The nucleotide sequence of RBD of SARS-CoV-2 was codon-optimized and fused with Fc region of human IgG1 at the C terminus of the RBD gene construct. The codon-optimized SARS-CoV-2 RBD-Fc fusion gene was cloned into the geminiviral plant expression vector pBYR2e. For expression of RBD-Fc fusion protein in plants, N. benthamiana plants were infiltrated with Agrobacterium containing pBYR2e-SARS-CoV-2 RBD-Fc (Figure 1). The leaves infiltrated with Agrobacterium containing pBYR2e-SARS-CoV-2 RBD-Fc showed significant phenotypic necrosis compared to the leaves infiltrated by Agrobacterium without the plant expression vector (Figure 2A).
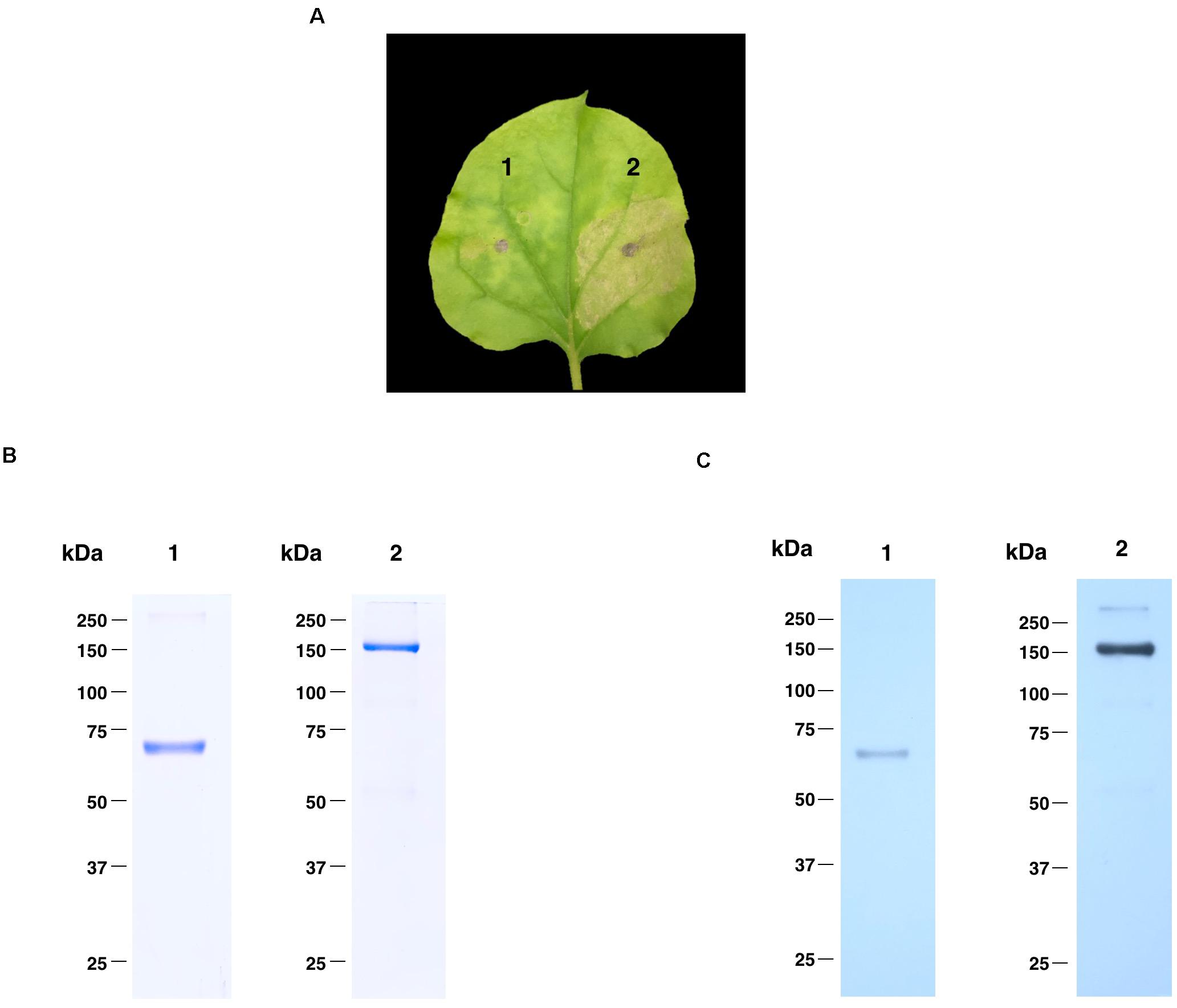
Figure 2. Expression profiles of plant-produced SARS-CoV-2 RBD-Fc fusion protein. Phenotype of leaf infiltrated with Agrobacterium control (1) and Agrobacterium containing pBYR2e-SARS-CoV-2 RBD-Fc (2) after 4 dpi (A). SDS-PAGE analysis of plant-produced SARS-CoV-2 RBD-Fc fusion protein stained with Coomassie staining (B) and western blot of plant-produced SARS-CoV-2 RBD-Fc fusion protein probed with anti-human gamma-HRP conjugate antibody (C). Lane 1 and 2, purified plant-produced SARS-CoV-2 RBD-Fc fusion protein under reducing and non-reducing condition, respectively.
Plant-produced SARS-CoV-2 RBD-Fc fusion protein was extracted and purified from crude extract by single-step protein A affinity chromatography. The purified SARS-CoV-2 RBD-Fc fusion protein was concentrated and filtered by using 0.22 μm syringe filter. The purity of the purified plant-produced SARS-CoV-2 RBD-Fc was analyzed by SDS-PAGE gel stained with Coomassie blue staining under reducing and non-reducing condition. The purity of SARS-CoV-2 RBD-Fc fusion protein was found to be higher than 90% based on the visual inspection of a Coomassie-stained SDS gel (Figure 2B). The SARS-CoV-2 RBD-Fc fusion protein was further analyzed by western blot probed with anti-human gamma chain-HRP conjugate antibody. The protein band corresponding to the molecular weight of 65 kDa was observed in reducing condition (Figure 2C; lane 1). In addition, the plant-produced SARS-CoV-2 RBD-Fc fusion protein under non-reducing condition was observed at approximately 150 kDa (Figure 2C; lane 2), which implies that the SARS-CoV-2 RBD-Fc fusion protein could be linked by disulfide bond into dimeric form. The expression level of plant-produced SARS-CoV-2 RBD-Fc was quantified by ELISA and found to be 25 μg/g fresh weight. The authenticity of purified plant-produced SARS-CoV-2 RBD-Fc fusion protein was further confirmed by using a high-resolution LC-TOF MS/MS analysis as shown in Supplementary Document.
The binding of plant-produced SARS-CoV-2 RBD-Fc fusion protein was confirmed by ELISA by using commercial HEK293 and CHO-produced ACE2 protein as the capture reagent. The various dilutions of purified plant-produced SARS-CoV-2 RBD-Fc was incubated with commercial ACE2 proteins. For detection of SARS-CoV-2 RBD, anti-SARS-CoV-2 (H4) mAb (Shanmugaraj et al., 2020b) and anti-human kappa chain-HRP conjugate antibody were added into the wells. The results showed that plant-produced SARS-CoV-2 RBD-Fc fusion protein produced saturable binding to both commercial ACE2 proteins with substantially high affinity in comparison to PBS control (Figure 3), which confirms the authenticity of plant-produced SARS-CoV-2 RBD-Fc.
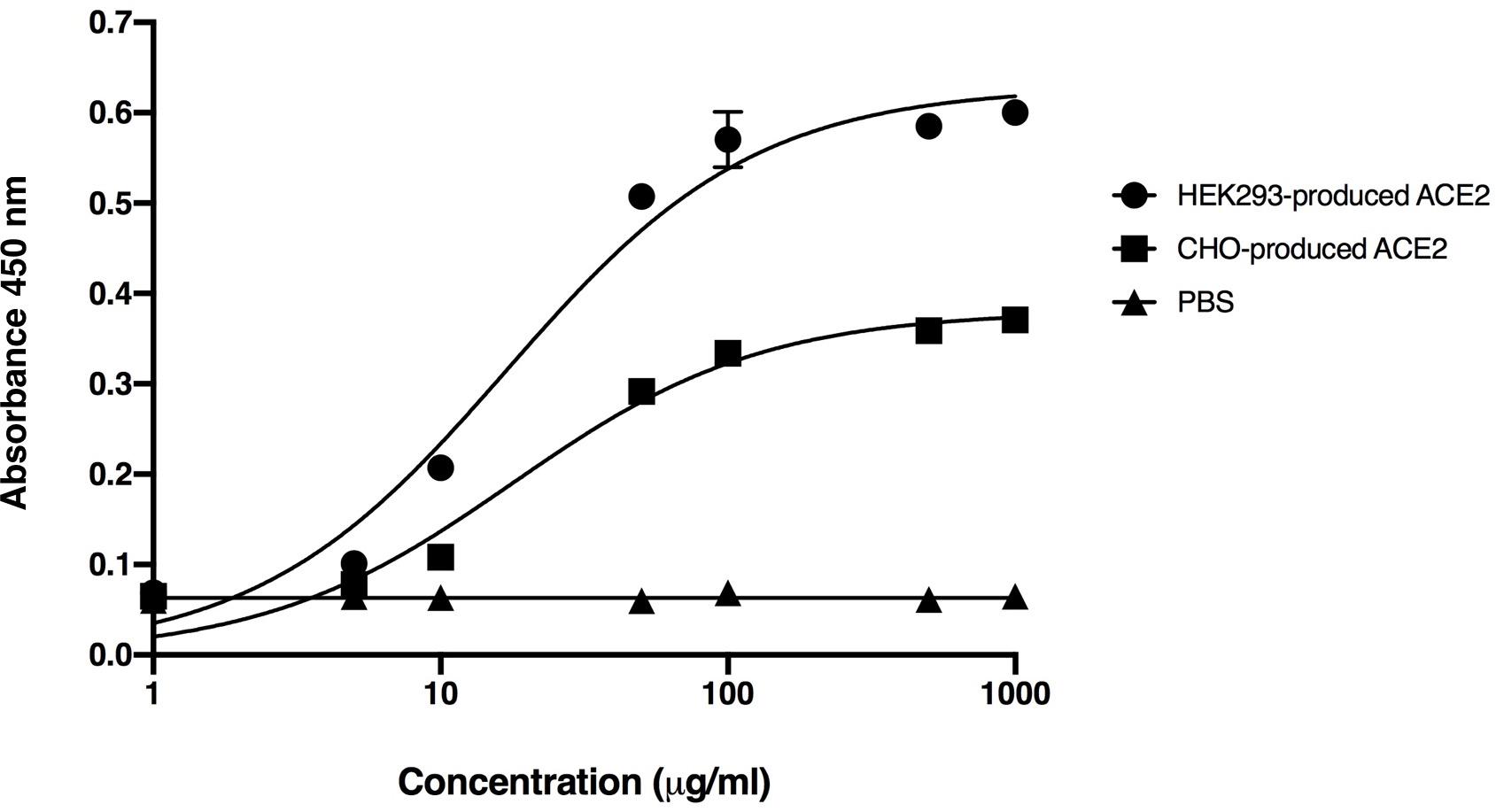
Figure 3. Binding activity of the plant-produced SARS-CoV-2 RBD-Fc with the commercial angiotensin-converting enzyme 2 (ACE2 proteins) derived from HEK293 and CHO cells analyzed by ELISA. PBS was used as negative control. Data are presented as mean ± standard deviation (SD) of triplicates in each sample dilution.
Mice immunogenicity was assessed in 7-week-old female Mlac:ICR mice by immunizing intramuscularly on days 0 and 21 with 10 μg of plant-produced SARS-CoV-2 RBD-Fc fusion protein with or without alum adjuvant. Mice sera were collected on days 0, 14, and 35 (Figure 4A). SARS-CoV-2 RBD-specific antibodies were evaluated by ELISA using commercial SARS-CoV-2 RBD-His protein produced from Sf9 cells as a capture antigen. The SARS-CoV-2 RBD-specific immune responses were observed after first immunization of plant-produced SARS-CoV-2 RBD-Fc alone, whilst a slightly increased specific-mouse total IgG response was observed in SARS-CoV-2 RBD-Fc immunized with alum. All mice immunized with plant-produced RBD-Fc elicited significantly higher antibody titer after second booster compared with the control group (Figure 4B). Plant-produced SARS-CoV-2 RBD-Fc was found to be immunogenic, while the addition of alum adjuvant could significantly improve its immunogenicity. In addition, we appraised the levels of SARS-CoV-2 RBD-specific IgG1 and IgG2a subclasses, which are indicators of Th2 and Th1 lymphocyte responses in mice, respectively. The results demonstrated that plant-produced SARS-CoV-2 RBD-Fc induced both SARS-CoV-2 RBD-specific IgG1 (Figure 4C) and IgG2a (Figure 4D) with the IgG1 bias. The in vitro neutralizing ability of the immunized sera was evaluated. The SARS-CoV-2 RBD-Fc without adjuvant induced neutralizing antibody against SARS-CoV-2 after the second dose at a dilution of 1:1,280 (Figure 4E). Interestingly, mice sera immunized by SARS-CoV-2 RBD-Fc adjuvanted with alum displayed maximum SARS-CoV-2 neutralization with a dilution more than 1:10,240 (Figure 4E). IFN-γ levels of splenocytes isolated from mice was tested by IFN-γ ELISpot assay. The results showed that plant-produced RBD-Fc elicited IFN-γ secretion which was statistically significant compared with mock control group (Figure 4F).
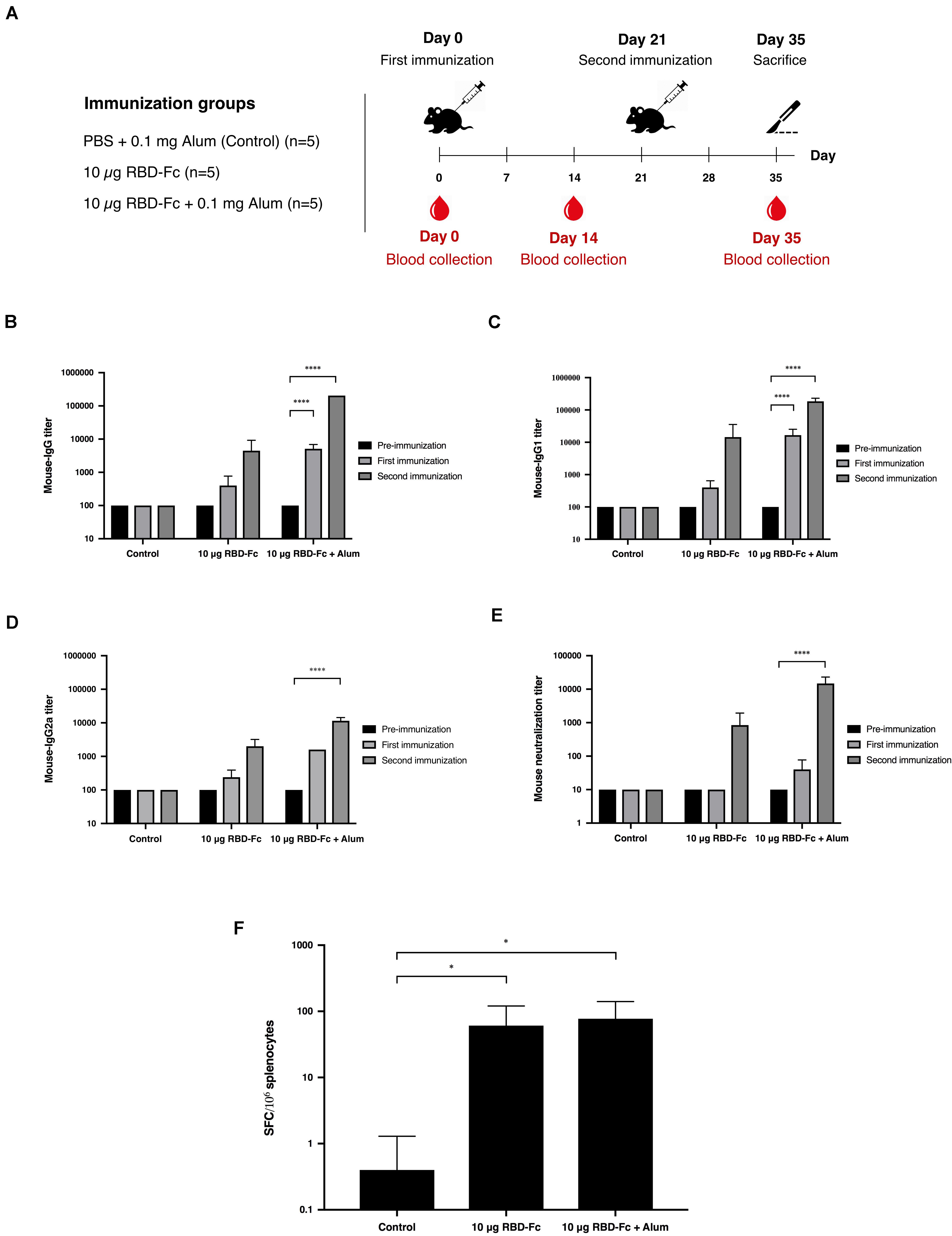
Figure 4. Immunogenic studies in mice. Schematic representation of immunization protocol and sample collection. Groups of mice (five mice per each group) were intramuscularly immunized with 10 μg of SARS-CoV-2 RBD-Fc fusion protein alone or with alum adjuvant, followed by booster dose at 21 days after first immunization. Mice sera were collected on day 0 (pre-bleed) and day 14 post-immunization (A). Titers of SARS-CoV-2 RBD-specific total IgG (B), IgG1 (C), and IgG2a (D) in the immunized sera collected on day 0, 14, and 35 were analyzed by indirect ELISA using Sf9-produced SARS-CoV-2 RBD-His as the capture antigen. Potent neutralizing antibody titers in mice sera were tested by in vitro microneutralization assay using Vero E6 cell line and live SARS-CoV-2 (E). The functional profiles of SARS-CoV-2 RBD-specific T-cell responses expressing in mouse splenocytes immunized with plant-produced SARS-CoV-2 RBD-Fc adjuvanted with alum were determined by mouse ELISpot assay (F). Data presented as mean ± SD of the endpoint titers in each mice vaccination group (n = 5). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Cynomolgus macaques (Macaca fascicularis) were intramuscularly immunized with 25 and 50 μg of plant-produced SARS-CoV-2 RBD-Fc with the presence of alum on day 0 and 21. Monkey sera were collected on day 0, 14, and 35 (Figure 5A). Plant-produced SARS-CoV-2 RBD-Fc protein adjuvanted with alum was capable of inducing dose-independent SARS-CoV-2 RBD-specific IgG antibodies in monkeys after first and second immunization with the dilution 1:800, and 1:51,200, respectively (Figure 5B). Specifically, the microneutralization assay was performed by using Vero E6 cell line to evaluate the level of neutralizing antibodies against live SARS-CoV-2. Sera collected 14 days after the first immunization (day 14) showed neutralizing activity against the SARS-CoV-2 and increased at day 35 (14 days after the second immunization) with the neutralization titer approximately 5,120 (Figure 5C). In addition, cell-mediated immune responses in cynomolgus monkeys were monitored by IFN-γ ELISpot assay by using peripheral blood mononuclear cells isolated from immunized sera on day 35. The specific IFN-γ expression was detected in monkeys immunized with two doses of plant-produced SARS-CoV-2 RBD-Fc, and a significant difference from the control group was detected compared to immunized monkeys (Figure 5D).
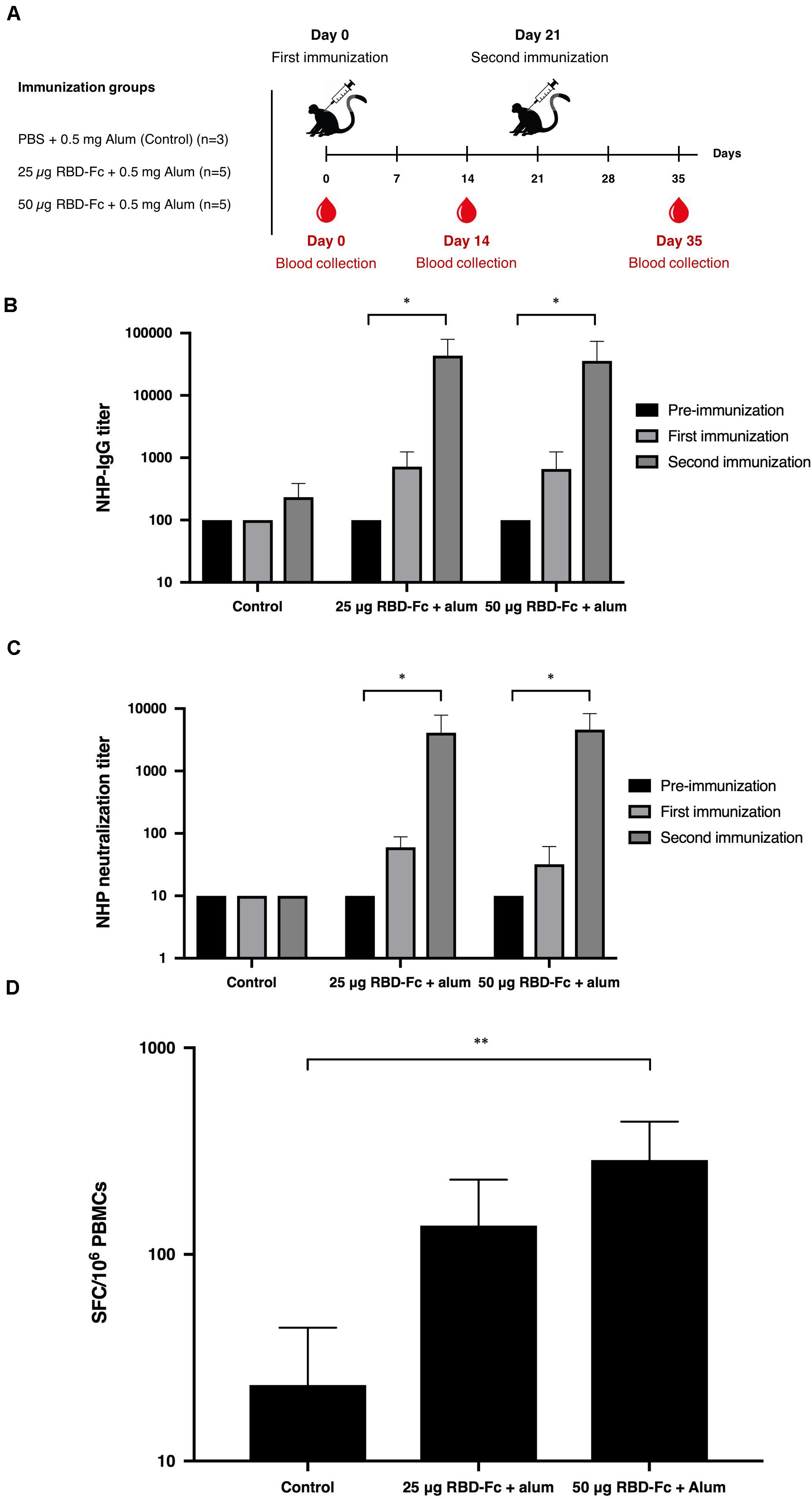
Figure 5. Immunogenic studies in non-human primates (Macaca fascicularis). Experimental design of immunogenicity studies in non-human primates. Thirteen juvenile-adult non-human primates were separated into 3 groups; Control group was immunized with PBS adjuvanted by alum (n = 3) and two experimental groups were immunized with 25 and 50 μg SARS-CoV-2 RBD-Fc along with alum adjuvant (n = 5 per group). All non-human primates were intramuscularly injected either with PBS or plant-produced RBD-Fc for 3 weeks interval (on day 0 and 21). The sera were collected on day 0 and 14 after each boost (A). Serum specific IgG response in non-human primates were determined by ELISA (B). Virus neutralizing titer of RBD immunized non-human primate sera against live SARS-CoV-2 were evaluated (C). The functional profiles of SARS-CoV-2 RBD-specific T-cell responses expressing in non-human primate peripheral blood mononuclear cells immunized with plant-produced RBD-Fc adjuvanted with alum on day 14 after second immunization (D). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
The recent emergence of coronavirus diseases (COVID-19) in China is responsible for the current global pandemic and public health crisis (Li Q. et al., 2020; Malik et al., 2020; Singhal, 2020; Yuki et al., 2020). Few vaccines are currently approved for human use. Hence, it is highly essential to develop safe, effective vaccines and therapeutics against this infection to prevent its spread. Recently, several groups have predicted and assessed the immunogenicity potential of SARS-CoV-2 related proteins and showed that SARS-CoV-2 S protein is the suitable candidate for recombinant vaccine development as it can elicit potent immune response and is the major target of neutralizing antibodies. The receptor-binding domain (RBD) of SARS-CoV-2 plays a key role in viral attachment and entry into the host cells by interaction with the ACE2 receptor in the host cells (Li et al., 2003; Lei et al., 2020; Quinlan et al., 2020; Rabi et al., 2020; Shanmugaraj et al., 2020c; Sun et al., 2020; Xie et al., 2020; Yuki et al., 2020). Particularly, RBD domain has multiple conformational epitopes, which can induce host immune responses and highly potent neutralizing antibodies (He et al., 2005a,b; Zhu et al., 2013; Wang et al., 2018; Smith et al., 2020). Hence, RBD domain in the S protein is considered as a potential target and could serve as a potent immunogen for developing the possible SARS-CoV-2 vaccines.
Since 1980s, plants have been utilized for the development of highly valuable biopharmaceuticals either for human or veterinary applications. Plant-based expression system offers several key advantages over conventional systems in terms of production speed, cost, and safety (Basaran and Rodríguez-Cerezo, 2008; Paul and Ma, 2011; Krenek et al., 2015; Yao et al., 2015; Burnett and Burnett, 2019; Shanmugaraj et al., 2020a; Daniell et al., 2021). Remarkably, plant-based expression system can produce large amounts of recombinant antigens in a relatively short time period within few weeks after making the gene construct (D’Aoust et al., 2008, 2010; Rybicki, 2009; Vézina et al., 2009; Pillet et al., 2016; Rattanapisit et al., 2020; Ward et al., 2020). The concept of plant-produced biopharmaceuticals and vaccines has been assessed and well explored by number of research groups worldwide. Several plant-produced therapeutics (Kizhner et al., 2015; Ma et al., 2015; Mor, 2015; Prevail Ii Writing Group et al., 2016) and vaccines (Yuki et al., 2013; Hendin et al., 2017; Donini and Marusic, 2019) are currently in pipeline for clinical trials and approval, Notably plant-produced Glucocerebrosidase enzyme (ElelysoTM) for the treatment of type I Gaucher’s disease has been approved by US Food and Drug Administration (Fox, 2012) and tobacco-derived seasonal flu VLP vaccine from Medicago Inc., is currently in final stages of clinical trial (Ward et al., 2021). Hence, plant-based expression could be an alternative option for rapid production of emergency vaccines or therapeutic antibodies (Iyappan et al., 2018; Diego-Martin et al., 2020; Rattanapisit et al., 2020; Shanmugaraj and Phoolcharoen, 2021; Siriwattananon et al., 2021).
The geographical distribution of virus is increasing rapidly and global concern on this pandemic demands an affordable and scalable protein production platform that can produce recombinant proteins relatively in short time with much reduced cost. Hence in this study, we have demonstrated the rapid production of SARS-CoV-2 RBD-Fc fusion protein in N. benthamiana plants that could be used as a potential vaccine candidate for prevention of SARS-CoV-2 infection.
Significant efforts have been made by the scientific community across the world to develop the effective vaccine against SARS-CoV-2. Plant-derived vaccine candidates for other coronaviruses such as SARS and porcine epidemic diarrhea virus are shown to elicit potent immunogenic response in animal studies (Kang et al., 2005; Pogrebnyak et al., 2005). Earlier studies showed that full length S protein-based vaccine could cause liver damage or enhance virus infection. RBD-based vaccines formulated with alum was shown to elicit high level of protective immunity in animal experiments (He et al., 2006; Du et al., 2007; Jiang et al., 2012; Chen et al., 2017, 2020a,2020b). Hence based on the available data on the immunogenicity of SARS-CoV-2 proteins, we have chosen RBD for plant expression.
The presence of Fc domain in fusion protein offers favorable characteristics such as improving the expression and secretion of the recombinant proteins, improving protein solubility and stability (Huang, 2009; Czajkowsky et al., 2012; Yang et al., 2018). Moreover, Fc domain increases the serum half-life and prolongs therapeutic protein activities due to pH-dependent binding to the neonatal Fc receptor (FcRn) leading to prevention of protein degradation in endosomes as well as reduces renal clearance rate due to larger molecular weight of protein (Jazayeri and Carroll, 2008; Suzuki et al., 2010; Carter, 2011; Rath et al., 2015; Rosales-Mendoza et al., 2017; Yang et al., 2018). Fc region have been used as a fusion protein partner for several recombinant proteins such as receptors, ligands, enzymes, and soluble cytokines for therapeutic applications (Strohl and Knight, 2009; Li and Ravetch, 2011; Czajkowsky et al., 2012; Rattanapisit et al., 2019c; Liu et al., 2020). In addition to the mentioned advantages, Fc region is used as a tagged protein for facilitating the effective purification of recombinant protein by protein A chromatography that can provide high purity of SARS-CoV-2 RBD-Fc which can be visualized on the Coomassie-stained SDS-PAGE compared to crude extract sample as shown in previous studies (Rattanapisit et al., 2019c; Siriwattananon et al., 2021). Hence, we engineered SARS-CoV-2 RBD by fusing with Fc region of human IgG1 in order to use it as a subunit vaccine against SARS-CoV-2.
The SARS-CoV-2 RBD-Fc fusion protein was transiently expressed as a soluble protein in plants. The results showed that the expression of SARS-CoV-2 RBD-Fc was achieved rapidly within 4 days post infiltration with necrosis signal was observed on the infiltrated leaves (Figure 2A). The recombinant protein was purified from the plant crude extracts by affinity column chromatography and used for further studies.
The plant-produced SARS-CoV-2 RBD-Fc apparently showed effective binding activity with commercial ACE2 proteins produced from HEK293 and CHO cells (Figure 3). This data indicated that the SARS-CoV-2 RBD protein folded correctly in plants and produced authentic antigen. The immunogenicity of plant-produced SARS-CoV-2 RBD-Fc was tested in mice and cynomolgus monkeys using alum as an adjuvant. Alum stimulates the innate immunity, particularly presenting the antigen to major histocompatibility complex (MHC) class II, CD40 and CD86 or inducing the Th2 responses to mediate B-cell differentiation and elicit the antigen specific-IgG1 isotype (Marrack et al., 2009; Exley et al., 2010; Zhang et al., 2016). Furthermore, alum is having good safety profile and has been used as an adjuvant in several currently available licensed vaccines to enhance the immune response of the antigen (HogenEsch et al., 2018).
Mice administered with two doses of plant-derived SARS-CoV-2 RBD-Fc protein formulated with alum as adjuvant developed the neutralizing immune response (Figure 4E). The results confirmed the immunogenicity of plant-produced recombinant SARS-CoV-2 RBD protein. Mice immunized with RBD with alum showed the higher titer of neutralizing antibodies compared with the mice immunized with SARS-CoV-2 RBD alone. The analysis of the mouse specific-IgG subtypes suggested that plant-produced SARS-CoV-2 RBD induced a mixed Th1/Th2-specific immune responses. Further the efficacy of plant-produced SARS-CoV-2 RBD-Fc fusion protein was investigated in cynomolgus monkeys by administering the SARS-CoV-2 RBD-Fc with alum as adjuvant. The results confirmed that the plant-produced SARS-CoV-2 RBD-Fc could induce neutralizing antibodies in monkeys (Figure 5C). To accomplish the capability of plant-produced SARS-CoV-2 RBD-Fc in induction of cell-mediated immune responses, mouse splenocytes and monkey peripheral blood mononuclear cells were collected 14 days after second immunization (Figures 4F, 5D). The IFN-γ-expressing T cells were analyzed by ELISPOT assay. Plant-produced SARS-CoV-2 RBD-Fc without alum induced SARS-CoV-2-specific T-cell responses, as evidenced by significant IFN-γ expression compared with the control (Figure 4F). Addition of alum adjuvant did not significantly increase the number of IFN-γ in the animals. These results suggested that plant-produced SARS-CoV-2 RBD-Fc itself could induce T-cell responses.
These results clearly showed that the plant-expressed SARS-CoV-2 RBD-Fc fusion protein maintains their authentic structure and retains its antigenicity. Our results are consistent with those of previous studies which showed that the vaccine antigens expressed in N. benthamiana elicited potent immune responses in animal experiments (Zheng et al., 2009; He et al., 2014). In consistent with earlier reports on the immunogenicity of RBD of SARS-CoV, our study showed that neutralizing antibodies induced by plant produced RBD of SARS-CoV-2 suppress SARS-CoV-2 infection in vitro (Bisht et al., 2005; Kapadia et al., 2005; Du et al., 2010). Our data showed the potential of plant-produced subunit vaccine candidate as the effective SARS-CoV-2 vaccine.
In conclusion, our study demonstrated that it was feasible to produce SARS-CoV-2 RBD protein in N. benthamiana plants by transient expression system. Further plant-produced recombinant protein was shown to be immunogenic in mice and non-human primates. The vaccine elicited both humoral and cell mediated immune responses suggesting the potential of plant- produced RBD as the effective vaccine against SARS-CoV-2. To our knowledge, this is the first report demonstrating the immunogenicity of plant-produced SARS-CoV-2 RBD protein in mice and non-human primates. Collectively this proof of concept study demonstrated that the plant-produced SARS-CoV-2 proteins could possibly be further developed as candidate vaccines for early stage clinical development.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee, Faculty of Medicine and the National Primate Research Center of Thailand-Chulalongkorn University (NPRCT-CU) Animal Care and Use Committee, Chulalongkorn University, Bangkok, Thailand.
KiR, AT, and WP designed all experiments. KS, BS, and KaR performed protein expression, protein purification, ELISA, and protein preparation for animal experiments. KS, SMan, SP, SS, ST, EP, CK, SB, WW, KT, and PK performed vaccination and immunogenic studies in mice. KS, SMan, SP, SS, ST, SB, WW, TK, NS, and SMal performed vaccination and immunogenic studies in non-human primates. KL performed LC-MS analysis. All authors analyzed the data and contributed to manuscript preparation.
This research was funded by Baiya Phytopharm Co., Ltd. and TSRI Fund to SM and WP (CU_FRB640001_01_33_2). The authors declare that Baiya Phytopharm Co., Ltd. was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
WP from Chulalongkorn University is a founder/shareholder of Baiya Phytopharm Co., Ltd. BS and KR are employed by Baiya Phytopharm Co., Ltd., Thailand. WW is employed by BioNet-Asia Co., Ltd., Thailand.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank the Department of Disease Control, Ministry of Public Health, Thailand for providing clinical specimens for the viral isolate and sera from a COVID-19 survivor. We would like to thank the 100th anniversary Chulalongkorn University (KS) for doctoral scholarship.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.682953/full#supplementary-material
Ahmad, A. R., Kaewpungsup, P., Khorattanakulchai, N., Rattanapisit, K., Pavasant, P., and Phoolcharoen, W. (2019). Recombinant human dentin matrix protein 1 (hDMP1) expressed in Nicotiana benthamiana potentially induces osteogenic differentiation. Plants 8:566. doi: 10.3390/plants8120566
Amanat, F., and Krammer, F. (2020). SARS-CoV-2 vaccines: status report. Immunity 52, 583–589. doi: 10.1016/j.immuni.2020.03.007
Banerjee, A., Kulcsar, K., Misra, V., Frieman, M., and Mossman, K. (2019). Bats and Coronaviruses. Viruses 11:41. doi: 10.3390/v11010041
Basaran, P., and Rodríguez-Cerezo, E. (2008). Plant molecular farming: opportunities and challenges. Crit. Rev. Biotechnol. 28, 153–172. doi: 10.1080/07388550802046624
Bellucci, M., Marchis, F. D., and Pompa, A. (2017). The endoplasmic reticulum is a hub to sort proteins toward unconventional traffic pathways and endosymbiotic organelles. J. Exp. Bot. 69, 7–20. doi: 10.1093/jxb/erx286
Bisht, H., Roberts, A., Vogel, L., Subbarao, K., and Moss, B. (2005). Neutralizing antibody and protective immunity to SARS coronavirus infection of mice induced by a soluble recombinant polypeptide containing an N-terminal segment of the spike glycoprotein. Virology 334, 160–165. doi: 10.1016/j.virol.2005.01.042
Bosch, B. J., Zee, R. V. D., Haan, C. A. M. D., and Rottier, P. J. M. (2003). The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77, 8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003
Burnett, M. J. B., and Burnett, A. C. (2019). Therapeutic recombinant protein production in plants: Challenges and opportunities. Plants People Planet 2, 121–132. doi: 10.1002/ppp3.10073
Carter, P. J. (2011). Introduction to current and future protein therapeutics: a protein engineering perspective. Exp. Cell Res. 317, 1261–1269. doi: 10.1016/j.yexcr.2011.02.013
Chen, Q., He, J., Phoolcharoen, W., and Mason, H. S. (2011). Geminiviral vectors based on bean yellow dwarf virus for production of vaccine antigens and monoclonal antibodies in plants. Hum. Vaccin. 7, 331–338. doi: 10.4161/hv.7.3.14262
Chen, W.-H., Chag, S. M., Poongavanam, M. V., Biter, A. B., Ewere, E. A., Rezende, W., et al. (2017). Optimization of the production process and characterization of the yeast-expressed SARS-CoV recombinant receptor-binding domain (RBD219-N1), a SARS vaccine candidate. J. Pharm. Sci. 106, 1961–1970. doi: 10.1016/j.xphs.2017.04.037
Chen, W.-H., Strych, U., Hotez, P. J., and Bottazzi, M. E. (2020a). The SARS-CoV-2 vaccine pipeline: an overview. Curr. Trop. Med. Rep. 3, 1–4. doi: 10.1007/s40475-020-00201-6
Chen, W.-H., Tao, X., Agrawal, A., Algaissi, A., Peng, B.-H., Pollet, J., et al. (2020b). Yeast-expressed SARS-CoV recombinant receptor-binding domain (RBD219-N1) formulated with alum induces protective immunity and reduces immune enhancement. bioRxiv [preprint] doi: 10.1101/2020.05.15.098079
Czajkowsky, D. M., Hu, J., Shao, Z., and Pleass, R. J. (2012). Fc-fusion proteins: new developments and future perspectives. EMBO Mol. Med. 4, 1015–1028. doi: 10.1002/emmm.201201379
D’Aoust, M.-A., Couture, M. M.-J., Charland, N., Trépanier, S., Landry, N., Ors, F., et al. (2010). The production of hemagglutinin-based virus-like particles in plants: a rapid, efficient and safe response to pandemic influenza. Plant Biotechnol. J. 8, 607–619. doi: 10.1111/j.1467-7652.2009.00496.x
D’Aoust, M.-A., Lavoie, P.-O., Couture, M. M.-J., Trépanier, S., Guay, J.-M., Dargis, M., et al. (2008). Influenza virus-like particles produced by transient expression in Nicotiana benthamiana induce a protective immune response against a lethal viral challenge in mice. Plant Biotechnol. J. 6, 930–940. doi: 10.1111/j.1467-7652.2008.00384.x
Daniell, H., Jin, S., Zhu, X. G., Matthew, A., Gitzendanner, Soltis, D. E., et al. (2021). Green giant—a tiny chloroplast genome with mighty power to produce high−value proteins: history and phylogeny. Plant Biotechnol. J. 19, 430–447. doi: 10.1111/pbi.13556
Diamos, A. G., and Mason, H. S. (2018). High-level expression and enrichment of norovirus virus-like particles in T plants using modified geminiviral vectors. Protein Expr. Purif. 151, 86–92. doi: 10.1016/j.pep.2018.06.011
Diego-Martin, B., González, B., Vazquez-Vilar, M., Selma, S., Mateos-Fernández, R., Gianoglio, S., et al. (2020). Pilot production of SARS-CoV-2 related proteins in plants: a proof of concept for rapid repurposing of indoor farms into biomanufacturing facilities. Front. Plant Sci. 11:612781. doi: 10.3389/fpls.2020.612781
Donini, M., and Marusic, C. (2019). Current state-of-the-art in plant-based antibody production systems. Biotechnol. Lett. 41, 335–346. doi: 10.1007/s10529-019-02651-z
Du, L., Zhao, G., Chan, C. C., Li, L., He, Y., Zhou, Y., et al. (2010). A 219-mer CHO-expressing receptor-binding domain of SARS-CoV S protein induces potent immune responses and protective immunity. Viral. Immunol. 23, 211–219. doi: 10.1089/vim.2009.0090
Du, L., Zhao, G., He, Y., Guo, Y., Zheng, B.-J., Jiang, S., et al. (2007). Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine 25, 2832–2838. doi: 10.1016/j.vaccine.2006.10.031
Exley, C., Siesjö, P., and Eriksson, H. (2010). The immunobiology of aluminium adjuvants: how do they really work? Trends Immunol. 31, 103–109. doi: 10.1016/j.it.2009.12.009
Fox, J. L. (2012). First plant-made biologic approved. Nat. Biotechnol. 30:472. doi: 10.1038/nbt0612-472
Frey, A., Canzio, J. D., and Zurakowski, D. (1998). A statistically defined endpoint titer determination method for immunoassays. J. Immunol. Methods 221, 35–41. doi: 10.1016/s0022-1759(98)00170-7
Fuenmayor, J., Gòdia, F., and Cervera, L. (2017). Production of virus-like particles for vaccines. N. Biotechnol. 39, 174–180. doi: 10.1016/j.nbt.2017.07.010
Gleba, Y., Klimyuk, V., and Marillonnet, S. (2005). Magnifection–a new platform for expressing recombinant vaccines in plants. Vaccine 23, 2042–2048. doi: 10.1016/j.vaccine.2005.01.006
Gomes, A. R., Munivenkatappa, S., Belamaranahally, B., Veeregowda, M., and Balamurugan, V. (2016). An overview of heterologous expression host systems for the production of recombinant proteins. Adv. Anim. Vet. Sci. 4, 346–356. doi: 10.14737/journal.aavs/2016/4.7.346.356
Prevail Ii Writing Group, Multi-National Prevail Ii Study Team, Davey, R. T. Jr., Dodd, L., Proschan, M. A., Neaton, J., et al. (2016). A randomized, controlled trial of ZMapp for ebola virus infection. N. EngI. J. Med. 375, 1448–1456. doi: 10.1056/nejmoa1604330
He, J., Peng, L., Lai, H., Hurtado, J., Stahnke, J., and Chen, Q. (2014). A plant-produced antigen elicits potent immune responses against West Nile virus in mice. Biomed. Res. Int. 2014:952865. doi: 10.1155/2014/952865
He, Y., Li, J., Li, W., Lustigman, S., Farzan, M., and Jiang, S. (2006). Cross-neutralization of human and palm civet severe acute respiratory syndrome coronaviruses by antibodies targeting the receptor-binding domain of spike protein. J. Immunol. 176, 6085–6092. doi: 10.4049/jimmunol.176.10.6085
He, Y., Lu, H., Siddiqui, P., Zhou, Y., and Jiang, S. (2005a). Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J. Immunol. 174, 4908–4915. doi: 10.4049/jimmunol.174.8.4908
He, Y., Zhou, Y., Siddiqui, P., and Jianga, S. (2004). Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem. Biophys. Res. Commun. 325, 445–454. doi: 10.1016/j.bbrc.2004.10.052
He, Y., Zhu, Q., Liu, S., Zhou, Y., Yang, B., Li, J., et al. (2005b). Identification of a critical neutralization determinant of severe acute respiratory syndrome (SARS)-associated coronavirus: importance for designing SARS vaccines. Virology 334, 74–82. doi: 10.1016/j.virol.2005.01.034
Hendin, H. E., Pillet, S., Lara, A. N., Wu, C.-Y., Charland, N., Landry, N., et al. (2017). Plant-made virus-like particle vaccines bearing the hemagglutinin of either seasonal (H1) or avian (H5) influenza have distinct patterns of interaction with human immune cells in vitro. Vaccine 35, 2592–2599. doi: 10.1016/j.vaccine.2017.03.058
HogenEsch, H., O’Hagan, D. T., and Fox, C. B. (2018). Optimizing the utilization of aluminum adjuvants in vaccines: you might just get what you want. NPJ Vaccines 3:51. doi: 10.1038/s41541-018-0089-x
Huang, C. (2009). Receptor-Fc fusion therapeutics, traps, and MIMETIBODY technology. Curr. Opin. Biotechnol. 20, 692–699. doi: 10.1016/j.copbio.2009.10.010
Iyappan, G., Shanmugaraj, B. M., Inchakalody, V., Ma, J. K. C., and Ramalingam, S. (2018). Potential of plant biologics to tackle the epidemic like situations - case studies involving viral and bacterial candidates. Int. J. Infect. Dis. 73:363. doi: 10.1016/j.ijid.2018.04.4236
Jazayeri, J. A., and Carroll, G. J. (2008). Fc-based cytokines: prospects for engineering superior therapeutics. Biodrugs 22, 11–26. doi: 10.2165/00063030-200822010-00002
Jiang, S., Bottazzi, M. E., Du, L., Lustigman, S., Tseng, C.-T. K., Curti, E., et al. (2012). Roadmap to developing a recombinant coronavirus S protein receptor-binding domain vaccine for severe acute respiratory syndrome. Exp. Rev. Vacc. 11, 1405–1413. doi: 10.1586/erv.12.126
Kang, T.-J., Kim, Y.-S., Jang, Y.-S., and Yang, M.-S. (2005). Expression of the synthetic neutralizing epitope gene of porcine epidemic diarrhea virus in tobacco plants without nicotine. Vaccine 23, 2294–2297. doi: 10.1016/j.vaccine.2005.01.027
Kapadia, S. U., Rose, J. K., Lamirande, E., Vogel, L., Subbarao, K., and Robertsc, A. (2005). Long-term protection from SARS coronavirus infection conferred by a single immunization with an attenuated VSV-based vaccine. Virology 340, 174–182. doi: 10.1016/j.virol.2005.06.016
Kaur, J., Kumar, A., and Kaur, J. (2018). Strategies for optimization of heterologous protein expression in E. coli: roadblocks and reinforcements. Int. J. Biol. Macromol. 106, 803–822. doi: 10.1016/j.ijbiomac.2017.08.080
Kelley, B. (2009). Industrialization of mAb production technology: the bioprocessing industry at a crossroads. MABS 1, 443–452. doi: 10.4161/mabs.1.5.9448
Kizhner, T., Azulay, Y., Hainrichson, M., Tekoah, Y., Arvatz, G., Shulman, A., et al. (2015). Characterization of a chemically modified plant cell culture expressed human a-Galactosidase-A enzyme for treatment of Fabry disease. Mol. Genet Metab. 114, 259–267. doi: 10.1016/j.ymgme.2014.08.002
Kodati, B., Dorbha, S., and Kunaparaju, R. K. (2016). Heterologus protein expression in different host systems. J. Chem. Pharm. Res. 8, 1068–1074.
Krenek, P., Samajova, O., Luptovciak, I., Doskocilova, A., Komis, G., and Samaj, J. (2015). Transient plant transformation mediated by Agrobacterium tumefaciens: principles, methods and applications. Biotechnol. Adv. 33, 1024–1042. doi: 10.1016/j.biotechadv.2015.03.012
Lei, C., Qian, K., Li, T., Zhang, S., Fu, W., Ding, M., et al. (2020). Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 11:2070. doi: 10.1038/s41467-020-16048-4
Li, F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 3, 237–261. doi: 10.1146/annurev-virology-110615-042301
Li, F., and Ravetch, J. V. (2011). Inhibitory Fcγ receptor engagement drives adjuvant and anti-tumor activities of agonistic CD40 antibodies. Science 333, 1030–1034. doi: 10.1126/science.1206954
Li, H., Liu, S.-M., Yu, X.-H., Tang, S.-L., and Tang, C.-K. (2020). Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int. J. Antimicrob. Agents 55:105951. doi: 10.1016/j.ijantimicag.2020.105951
Li, Q., Guan, X., Wu, P., Wang, X., Zhou, L., Tong, Y., et al. (2020). Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 382, 1199–1207. doi: 10.1056/NEJMoa2001316
Li, W., Moore, M. J., Vasilieva, N., Sui, J., Wong, S. K., Berne, M. A., et al. (2003). Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454. doi: 10.1038/nature02145
Liu, Z., Xu, W., Xia, S., Gu, C., Wang, X., Wang, Q., et al. (2020). RBD-Fc-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal. Transduct. Target. Ther. 5:282. doi: 10.4172/2167-1079.1000255
Ma, J. K.-C., Drake, P. M. W., and Christou, P. (2003). The production of recombinant pharmaceutical proteins in plants. Nat. Rev. Genet. 4, 794–805. doi: 10.1038/nrg1177
Ma, J. K.-C., Drossard, J., Lewis, D., Altmann, F., Boyle, J., Christou, P., et al. (2015). Regulatory approval and a first-in-human phase I clinical trial of a monoclonal antibody produced in transgenic tobacco plants. Plant Biotechnol. J. 13, 1106–1120. doi: 10.1111/pbi.12416
Malik, Y. S., Sircar, S., Bhat, S., Sharun, K., Dhama, K., Dadar, M., et al. (2020). Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet. Q. 40, 68–76. doi: 10.1080/01652176.2020.1727993
Malla, A., Shanmugaraj, B., and Ramalingam, S. (2020). Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Emerging Zoonotic Respiratory Pathogen in Humans. J. Pure. Appl. Microbiol. 14, 931–936. doi: 10.22207/JPAM.14.SPL1.30
Marrack, P., McKee, A. S., and Munks, M. W. (2009). Towards an understanding of the adjuvant action of aluminium. Nat. Rev. Immunol. 9, 287–293. doi: 10.1038/nri2510
Miao, Y., Ding, Y., Sun, Q.-Y., Xu, Z.-F., and Jiang, L. (2008). Plant bioreactors for pharmaceuticals. Biotechnol. Genet. Eng. Rev. 25, 363–380. doi: 10.5661/bger-25-363
Mor, T. S. (2015). Molecular pharming’s foot in the FDA’s door: protalix’s trailblazing story. Biotechnol. Lett. 37, 2147–2150. doi: 10.1007/s10529-015-1908-z
Paul, M., and Ma, J. K. C. (2011). Plant−made pharmaceuticals: leading products and production platforms. Biotechnol. Appl. Biochem. 58, 58–67. doi: 10.1002/bab.6
Phoolcharoen, W., Bhoo, S. H., Lai, H., Ma, J., Arntzen, C. J., Chen, Q., et al. (2011). Expression of an immunogenic Ebola immune complex in Nicotiana benthamiana. Plant Biotechnol. J. 9, 807–816. doi: 10.1111/j.1467-7652.2011.00593.x
Pillet, S., Aubin, É, Trépanier, S., Bussière, D., Dargis, M., Poulin, J.-F., et al. (2016). A plant- derived quadrivalent virus like particle influenza vaccine induces cross-reactive antibody and T cell response in healthy adults. Clin. Immunol. 168, 72–87. doi: 10.1016/j.clim.2016.03.008
Pogrebnyak, N., Golovkin, M., Andrianov, V., Spitsin, S., Smirnov, Y., Egolf, R., et al. (2005). Severe acute respiratory syndrome (SARS) S protein production in plants: development of recombinant vaccine. Proc. Natl. Acad. Sci. U. S. A. 102, 9062–9067. doi: 10.1073/pnas.0503760102
Quinlan, B. D., Mou, H., Zhang, L., Guo, Y., He, W., Ojha, A., et al. (2020). The SARS-CoV-2 receptor-binding domain elicits a potent neutralizing response without antibody-dependent enhancement. bioRxiv [preprint] doi: 10.1101/2020.04.10.036418
Rabi, F. A., Zoubi, M. S. A., Kasasbeh, G. A., Salameh, D. M., and Al-Nasser, A. D. (2020). SARS-CoV-2 and coronavirus disease 2019: what we know so far. Pathogens 9:231. doi: 10.3390/pathogens9030231
Rath, T., Baker, K., Dumont, J. A., Peters, R. T., Jiang, H., Qiao, S.-W., et al. (2015). Fc-fusion proteins and FcRn: structural insights for longer-lasting and more effective therapeutics. Crit. Rev. Biotechnol. 35, 235–254. doi: 10.3109/07388551.2013.834293
Rattanapisit, K., Chao, Z., Siriwattananon, K., Huang, Z., and Phoolcharoen, W. (2019a). Plant-produced anti-enterovirus 71 (EV71) monoclonal antibody efficiently protects mice against EV71 infection. Plants 8:12. doi: 10.3390/plants8120560
Rattanapisit, K., Phakham, T., Buranapraditkun, S., Siriwattananon, K., Boonkrai, C., Pisitkun, T., et al. (2019b). Structural and in vitro functional analyses of novel plant-produced anti-Human PD1 antibody. Sci. Rep. 9:15205. doi: 10.1038/s41598-019-51656-1
Rattanapisit, K., Shanmugaraj, B., Manopwisedjaroen, S., Purwono, P. B., Siriwattananon, K., Khorattanakulchai, N., et al. (2020). Rapid and scalable production of functional anti-coronavirus monoclonal antibody CR3022 in plants. Sci. Rep. 10:17698. doi: 10.21203/rs.3.rs-27160/v1
Rattanapisit, K., Srifa, S., Kaewpungsup, P., Pavasant, P., and Phoolcharoen, W. (2019c). Plant-produced recombinant Osteopontin-Fc fusion protein enhanced osteogenesis. Biotechnol. Rep. 21:e00312. doi: 10.1016/j.btre.2019.e00312
Rattanapisit, K., Yusakul, G., Shanmugaraj, B., Kittirotruji, K., Suwatsrisakul, P., Prompetchara, E., et al. (2021). Plant-produced recombinant SARS-CoV-2 receptor-binding domain; an economical, scalable biomaterial source for COVID-19 diagnosis. Biomater. Transl. 2, 43–49. doi: 10.3877/cma.j.issn.2096-112X.2021.01.006
Rosales-Mendoza, S., Nieto-Gómez, R., and Angulo, C. (2017). A perspective on the development of plant-made vaccines in the fight against Ebola virus. Front. Immunol. 8:252. doi: 10.3389/fimmu.2017.00252
Rybicki, E. P. (2009). Third international conference on plant-based vaccines and antibodies. Expert. Rev. Vacc. 8, 1151–1155. doi: 10.1586/erv.09.85
Masters, S. P. (2006). The molecular biology of coronaviruses. Adv. Virus Res. 66, 193–292. doi: 10.1016/s0065-3527(06)66005-3
Shahid, N., and Daniell, H. (2016). Plant-based oral vaccines against zoonotic and non-zoonotic diseases. Plant Biotechnol. J. 14, 2079–2099. doi: 10.1111/pbi.12604
Shanmugaraj, B., Bulaon, C. J. I., and Phoolcharoen, W. (2020a). Plant molecular farming: A viable platform for recombinant biopharmaceutical production. Plants 9:842. doi: 10.3390/plants9070842
Shanmugaraj, B., and Phoolcharoen, W. (2021). Addressing demand for recombinant biopharmaceuticals in the COVID-19 era. Asian Pac. J. Trop. Med. 14, 49–51.
Shanmugaraj, B., Rattanapisit, K., Manopwisedjaroen, S., Thitithanyanont, A., and Phoolcharoen, W. (2020b). Monoclonal antibodies B38 and H4 produced in Nicotiana benthamiana neutralize SARS-CoV-2 in vitro. Front. Plant Sci. 11:589995. doi: 10.3389/fpls.2020.589995
Shanmugaraj, B., Siriwattananon, K., Wangkanont, K., and Phoolcharoen, W. (2020c). Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19). Asian Pac. J. Allergy Immunol. 38, 10–18. doi: 10.12932/ap-200220-0773
She, J., Jiang, J., Ye, L., Hu, L., Bai, C., and Song, Y. (2020). 2019 novel coronavirus of pneumonia in Wuhan, China: emerging attack and management strategies. Clin. Transl. Med. 9:19. doi: 10.1186/s40169-020-00271-z
Singhal, T. (2020). A review of coronavirus disease-2019 (COVID-19). Indian J. Pediatr. 87, 281–286. doi: 10.1007/s12098-020-03263-6
Siriwattananon, K., Manopwisedjaroen, S., Kanjanasirirat, P., Purwono, P. B., Rattanapisit, K., Shanmugaraj, B., et al. (2021). Development of plant-produced recombinant ACE2-Fc fusion protein as a potential therapeutic agent against SARS-CoV-2. Front. Plant Sci. 11:604663. doi: 10.3389/fpls.2020.604663
Smith, T. R. F., Patel, A., Ramos, S., Elwood, D., Zhu, X., Yan, J., et al. (2020). Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 11:2601. doi: 10.1038/s41467-020-16505-0
Strohl, W. R., and Knight, D. M. (2009). Discovery and development of biopharmaceuticals: current issues. Curr. Opin. Biotechnol. 20, 668–672. doi: 10.1016/j.copbio.2009.10.012
Sun, X., Wang, T., Cai, D., Hu, Z., Chen, J. A., Liao, H., et al. (2020). Cytokine storm intervention in the early stages of COVID-19 pneumonia. Cytokine Growth Factor Rev. 53, 38–42. doi: 10.1016/j.cytogfr.2020.04.002
Suzuki, T., Ishii-Watabe, A., Tada, M., Kobayashi, T., Kanayasu-Toyoda, T., Kawanishi, T., et al. (2010). Importance of neonatal FcR in regulating the serum half-life of therapeutic proteins containing the Fc domain of human IgG1: a comparative study of the affinity of monoclonal antibodies and Fc-fusion proteins to human neonatal FcR. J. Immunol. 184, 1968–1976. doi: 10.4049/jimmunol.0903296
Vézina, L.-P., Faye, L., Lerouge, P., D’Aoust, M.-A., Marquet-Blouin, E., Burel, C., et al. (2009). Transient co-expression for fast and high-yield production of antibodies with human-like N-glycans in plants. Plant Biotechnol. J. 7, 442–455. doi: 10.1111/j.1467-7652.2009.00414.x
Vitale, A., and Denecke, J. R. (1999). The endoplasmic reticulum-gateway of the secretory pathway. Plant Cell 11, 615–628. doi: 10.1105/tpc.11.4.615
Walls, A. C., Xiong, X., Park, Y.-J., Tortorici, M. A., Snijder, J., Quispe, J., et al. (2019). Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176, 1026–1039. doi: 10.1016/j.cell.2018.12.028
Wang, L., Shi, W., Chappell, J. D., Joyce, M. G., Zhang, Y., Kanekiyo, M., et al. (2018). Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on the middle east respiratory syndrome coronavirus spike glycoprotein to avoid neutralization escape. J. Virol. 92:10. doi: 10.1128/jvi.02002-17
Ward, B. J., Gobeil, P., Séguin, A., Atkins, J., Boulay, I., Charbonneau, P.-Y., et al. (2020). Phase 1 trial of a candidate recombinant virus-like particle vaccine for COVID-19 disease produced in plants. medRxiv [preprint] doi: 10.1101/2020.11.04.20226282
Ward, B. J., Séguin, A., Couillard, J., Trépanier, S., and Landry, N. (2021). Phase III: Randomized observer-blind trial to evaluate lot-to-lot consistency of a new plant-derived quadrivalent virus like particle influenza vaccine in adults 18-49 years of age. Vaccine 39, 1528–1533. doi: 10.1016/j.vaccine.2021.01.004
World Health Organization (2021). Weekly epidemiological update - 6 March 2021 [Online]. Availableonline at: : https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19—6-april-2021 [accessed on Apr 11, 2021]
Xie, P., Ma, W., Tang, H., and Liu, D. (2020). Severe COVID-19: a review of recent progress with a look toward the future. Front. Publ. Health 8:189. doi: 10.3389/fpubh.2020.00189
Yang, C., Gao, X., and Gong, R. (2018). Engineering of Fc fragments with optimized physicochemical properties implying improvement of clinical potentials for Fc-based therapeutics. Front. Immunol. 8:1860. doi: 10.3389/fimmu.2017.01860
Yao, J., Weng, Y., Dickey, A., and Wang, K. Y. (2015). Plants as factories for human pharmaceuticals: applications and challenges. Int. J. Mol. Sci. 16, 28549–28565. doi: 10.3390/ijms161226122
Yuan, Y., Cao, D., Zhang, Y., Ma, J., Qi, J., Wang, Q., et al. (2017). Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 8:15092. doi: 10.1038/ncomms15092
Yuki, K., Fujiogi, M., and Koutsogiannaki, S. (2020). COVID-19 pathophysiology: a review. Clin. Immunol. 215:108427. doi: 10.1016/j.clim.2020.108427
Yuki, Y., Mejima, M., Kurokawa, S., Hiroiwa, T., Takahashi, Y., Tokuhara, D., et al. (2013). Induction of toxin-specific neutralizing immunity by molecularly uniform rice-based oral cholera toxin B subunit vaccine without plant-associated sugar modification. Plant Biotechnol. J. 11, 799–808.
Zhang, N., Channappanavar, R., Ma, C., Wang, L., Tang, J., Garron, T., et al. (2016). Identification of an ideal adjuvant for receptor-binding domain-based subunit vaccines against Middle East respiratory syndrome coronavirus. Cell. Mol. Immunol. 13, 180–190. doi: 10.1038/cmi.2015.03
Zheng, N., Xia, R., Yang, C., Yin, B., Li, Y., Duan, C., et al. (2009). Boosted expression of the SARS-CoV nucleocapsid protein in tobacco and its immunogenicity in mice. Vaccine 27, 5001–5007. doi: 10.1016/j.vaccine.2009.05.073
Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., et al. (2020). Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. Nature 579, 270–273. doi: 10.1038/s41586-020-2012-7
Keywords: COVID-19, SARS-CoV-2, receptor-binding domain, Nicotiana benthamiana, plant-produced recombinant protein, Fc fusion protein, subunit vaccine
Citation: Siriwattananon K, Manopwisedjaroen S, Shanmugaraj B, Rattanapisit K, Phumiamorn S, Sapsutthipas S, Trisiriwanich S, Prompetchara E, Ketloy C, Buranapraditkun S, Wijagkanalan W, Tharakhet K, Kaewpang P, Leetanasaksakul K, Kemthong T, Suttisan N, Malaivijitnond S, Ruxrungtham K, Thitithanyanont A and Phoolcharoen W (2021) Plant-Produced Receptor-Binding Domain of SARS-CoV-2 Elicits Potent Neutralizing Responses in Mice and Non-human Primates. Front. Plant Sci. 12:682953. doi: 10.3389/fpls.2021.682953
Received: 19 March 2021; Accepted: 15 April 2021;
Published: 13 May 2021.
Edited by:
Ryo Matsuda, The University of Tokyo, JapanReviewed by:
Chiara Lico, Italian National Agency for New Technologies, Energy and Sustainable Economic Development (ENEA), ItalyCopyright © 2021 Siriwattananon, Manopwisedjaroen, Shanmugaraj, Rattanapisit, Phumiamorn, Sapsutthipas, Trisiriwanich, Prompetchara, Ketloy, Buranapraditkun, Wijagkanalan, Tharakhet, Kaewpang, Leetanasaksakul, Kemthong, Suttisan, Malaivijitnond, Ruxrungtham, Thitithanyanont and Phoolcharoen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Waranyoo Phoolcharoen, V2FyYW55b28uUEBjaHVsYS5hYy50aA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.