
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 30 June 2020
Sec. Plant Cell Biology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00802
Chromosome painting is one of the key technologies in cytogenetic research, which can accurately identify chromosomes or chromosome regions. Oligonucleotide (oligo) probes designed based on genome sequences have both flexibility and specificity, which would be ideal probes for fluorescence in situ hybridization (FISH) analysis of genome structure. In this study, the bulked oligos of the two arms of chromosome seven of cotton were developed based on the genome sequence of Gossypium raimondii (DD, 2n = 2× = 26), and each arm contains 12,544 oligos. Chromosome seven was easily identified in both D genome and AD genome cotton species using the bulked chromosome-specific painting probes. Together with 45S ribosomal DNA (rDNA) probe, the chromosome-specific painting probe was also successfully used to correct the chromosomal localization of 45S rDNA in G. raimondii. The study reveals that bulked oligos specific to a chromosome is a useful tool for chromosome painting in cotton.
Identification of an individual chromosome is the foundation of cytogenetic research. In the early studies, chromosomes can be identified majorly by chromosome morphological characteristics (McClintock, 1929) and chromosome banding techniques (Comings, 1978). Since 1980s, the development of fluorescence in situ hybridization (FISH) has led to the development of different chromosome identification techniques owing to its higher sensitivity and resolution. Among them, chromosome painting is one of FISH techniques using chromosome-specific probes to detect specific chromosome regions or an entire chromosome (Pinkel et al., 1988). At early stage, chromosome painting using DNA probes prepared from flow-sorted or micro-dissected chromosomes was successfully used for chromosome identification, chromosome structure analysis, and comparative genomic analysis in human and other mammals with relatively small genomes (Breneman et al., 1993; Scherthan et al., 1994; Rens et al., 2006; Ferguson-Smith and Trifonov, 2007). However, the same type of probes could not work effectively in higher plants with large, complex genomes owing to the poor specificity of probes, which resulted from higher content of repetitive sequences (Fuchs et al., 1996; Lysak and Mandáková, 2013).
In the middle of 1990s, construction of bacterial artificial chromosome (BAC) system in plants enhanced the application of BAC-based FISH in plant cytogenetic research (Woo et al., 1994). Based on pooled BAC clones derived from a specific chromosome, the chromosome-specific painting technique was developed in certain plant species (Lysak et al., 2003, 2010; Ziolkowski et al., 2006; Mandáková and Lysak, 2008). This facilitated the studies on genome structure and evolution. However, this technique also shows some limitations in plant species with complex genome enriched in high repetitive sequences, which cannot guarantee the specificity of the selected BAC contigs.
Sequencing of a large amount of plant genomes and the development of massive parallel synthesis techniques make oligonucleotide (oligo)-based chromosome painting possible in plants. Han et al. (2015) developed a bioinformatic pipeline to select oligos specific to an individual chromosome of cucumber and first successfully applied this technique in plants. Following the application of oligo-based painting in cucumber, this technique has been quickly modified and applied in other plant species for chromosome identification and study on karyotyping, chromosome variations and evolution, such as in strawberry (Qu et al., 2017), Saccharum spontaneum (Meng et al., 2018), Solanum (He et al., 2018), rice (Hou et al., 2018; Liu et al., 2019), and maize (Albert et al., 2019). To further improve the efficiency, the technology was optimized, and a multiplex PCR-based oligo-painting strategy (MP-OP) was developed, which allows different double-strand-labeled probes to generate from the same oligo library with fluorophore-conjugated nested specific primers (Bi et al., 2019).
The cotton genus (Gossypium) is divided into eight monophyletic groups (A–G, and K) (2n = 2× = 26), and one allopolyploid lineage (AD) (2n = 4× = 52) (Wendel and Albert, 1992; Percival et al., 1999). Up to now, approximately 48 diploids and seven tetrapolyploid cotton species have been identified (Wang et al., 2018). It is believed that A and D diploids diverged 4–8 million years ago and re-hybridized about 1,2 million years ago to form allopolyploid AD lineage (Cronn et al., 2002). Two diploid species, Gossypium arboreum and Gossypium herbaceum, contain the A genome, whereas the genome D is present in 14 species (Wang et al., 2018). However, the donors of A and D genomes are still unclear. Based on the genetic and morphological evidence, some earlier studies suggested that the allopolyploid cotton is of monophyletic origin (Hutchinson et al., 1947; Wendel and Cronn, 2003), but some others took the point of polyphyletic origin (Johnson, 1975; Parks et al., 1975). Among D genome cotton species, Gossypium raimondii has been considered as the most possible paternal donor to the allopolyploids, although other species have been suggested (Wendel et al., 2010). The application of FISH technology using genome-DNA (gDNA) and specific repetitive sequence probes had provided strong evidences for the origin and evolution of the cotton genus (Liu et al., 2005; Xiao et al., 2017; Lu et al., 2018). As a widely used repetitive sequence, 45S ribosomal RNA gene (rDNA), is composed of 18S, 5.8S, and 28S rRNA encoding sequences connected by two internal transcribed spacers (ITS1 and ITS2) (Wendel et al., 1995). Highly repeated 45S rDNA units are arranged in tandem at one or several chromosomal loci (Chang et al., 2010). In G. raimondii, the 45S rDNA was located at the terminal region of chromosomes D502, D509, and D511 (corresponding to Chr.05, Chr.06, and Chr.07 of G. raimondii genome sequence map), which differ greatly with other D genome species (Gan et al., 2013). The sequence of ITS region (accession number U12718) amplified from G. raimondii (Wendel et al., 1995) had been used to blast against the G. raimondii genome sequence1. Result showed that the G. raimondii ITS was mapped on the 38,109,269–38,109,944 bp of chromosome seven with only one hit match, which is inconsistent with previous results in copy number and chromosome position (data unpublished). The distribution of 45S rDNA sites is one of the important references for the study of genome reorganization and phylogenetic analysis (Hanson et al., 1996; Wendel and Cronn, 2003; Chacón et al., 2012). Therefore, the accurate chromosome localization of 45S rDNA is crucial. In this study, we developed oligo pools of G. raimondii chromosome seven on the basis of the released whole genomic sequence for chromosome identification as well as 45S rDNA location verification, which would provide evidence for the study of the evolutionary relationship between cotton species.
Eleven cotton species were used in this study: four wild species of D genome cotton, Gossypium thurberi D1, Gossypium davidsonii D3–d, Gossypium klotzschianum D3–k, and G. raimondii D5 (DD, 2n = 2× = 26); two A genome cotton, Gossypium herbacium A1 and Gossypium arboretum A2 (AA, 2n = 2× = 26); and five AD genome cotton, Gossypium hirsutum AD1, Gossypium barbadense AD2, Gossypium tomentosum AD3, Gossypium mustelinum AD4, Gossypium darwinii, and AD5 (AADD, 4n = 4× = 52). All materials were grown in National Wild Cotton Nursery in Sanya, China. Root tips were harvested from the about 6-day seedlings planted in an incubator and pretreated by 25-ppm cycloheximide at 20°C for 80 min, then fixed in methanol–acetic acid (3:1), and stored at 4°C for 24 h. Metaphase chromosome preparations were prepared according to a previously reported method (Liu et al., 2016).
Chorus2 software2 was used to design the two oligo sets, and it was carried out following published procedure (Albert et al., 2019). Briefly, the G. raimondii genome sequence (Paterson et al., 2012)1 was divided into 45-nt oligos in step size of 5 nt, and short sequences were mapped to genome, and oligos mapped in two or more locations (with 75% of homology) were eliminated. Then, all respective sequences related oligos were filtered out from oligo set using ChorusNGSfilter.py and ChorusNGSselect.py in Chorus2 package.
Two oligo libraries were synthesized by Synbio Technologies (Suzhou, China). The oligo probes were prepared following the previous protocol (Liu et al., 2019). In brief, through four stages, that is, library amplification with high-fidelity enzyme (KAPA HiFi HotStart ReadyMix, KAPA Biosystems) and T7 in vitro transcription with MEGAshortscriptTM T7 kit (Invitrogen), reverse transcription using biotin- and digoxigenin-labeled RT primers (5′ dye/CGTGGTCGCGTCTCA), and enzymatic RNA removal. Then, the biotin-labeled or digoxigenin-labeled single-stranded oligo probes were directly used as FISH probes. The probe 45S rDNA was isolated using Plasmid Miniprep Kit (Biomiga) according to the handbook and were labeled with Biotin-Nick Translation Mix (Roche) according to the instructions of the manufacturer3.
FISH analysis of single-stranded oligo probes was performed according to a previous protocol (Han et al., 2015). Biotin- and digoxigenin-labeled probes were detected using streptavidin, Alexa FluorTM 488 conjugate (Invitrogen) and anti-digoxigenin Rhodamine Fab fragments (Roche), respectively. Chromosomes were counter-stained with 4′−6-diamidino-2-phenylindole (DAPI) in Vectashield antifading solution (Vector Laboratories) under a cover slip. Slides were examined under a Zeiss Imager M1 microscope. Images were captured and merged using MetaSystems Isis software with a CCD camera (MetaSystems CoolCube 1) attached to a Zeiss Imager M1 microscope. After the first-round FISH, slides were immersed in 1× phosphate-buffered saline (PBS) to remove the coverslips and were dehydrated in ethanol series (70, 90, and 100%, each for 5 min). Then the repeated FISH was performed according to a previous procedure (Cheng et al., 2001).
By using the developed bioinformatics pipeline (Albert et al., 2019), two sets of oligos were developed for the two arms of chromosome seven in G. raimondii on the basis of the genome assembly. The distribution of the two sets of oligos on chromosome 7 of G. raimondii genome sequence map is shown (Figure 1). Set 1 oligo pool (green) consists of 12,544 45-nt oligos spanning 6,710,744–12,163,021 bp of the G. raimondii chromosome 7, with the density of 2.3 oligos per kilobase. Set two oligo pool (red) consists of 12,544 45-nt oligos spanning 42,643,131–58,639,905 bp of the G. raimondii chromosome seven, with the density of 0.78 oligo per kilobase. To facilitate oligo sequence amplification, every 45-nt oligo contained consistent 5′-forward (T7 RNA polymerase promoter sequence) and 3′-reverse primers.
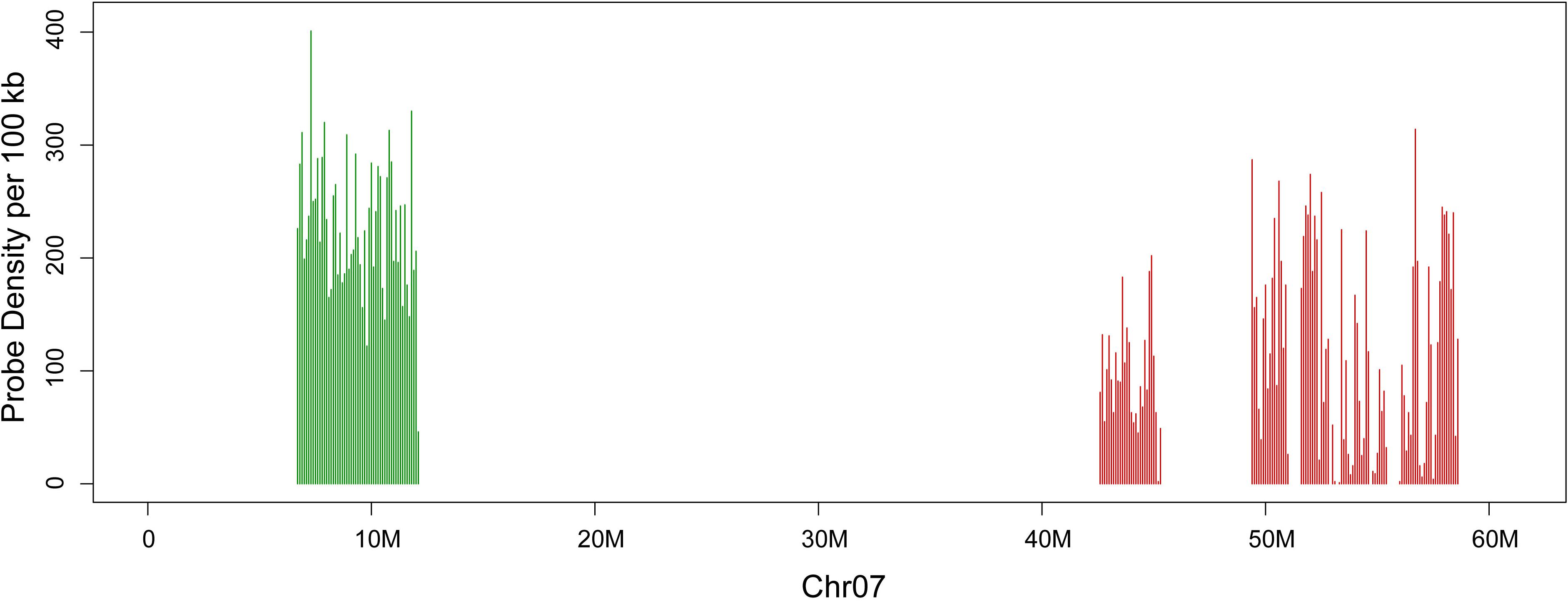
Figure 1. Distribution of two oligo sets on chromosome seven of Gossypium raimondii. The x-axis shows DNA sequence position on chromosome. The y-axis shows the number of oligos in 100-kb windows. Set 1 is in green, and set 2 is in red.
To test the sensitivity and reliability of the chromosome 7 oligo probes, set 1 (biotin-labeled) and set 2 (digoxigenin-labeled) oligo probes were hybridized to metaphase chromosomes of G. raimondii using double-color FISH. Both probes produced bright FISH signals on one arm of a pair of G. raimondii metaphase chromosomes, respectively. The FISH signal from set 2 oligo probe (red) had higher chromosome coverage than that of set 1 oligo probe (green). Additionally, owing to the lower resolution of metaphase FISH, no signal gap was viewed on the corresponding chromosomal region (Figure 2A).
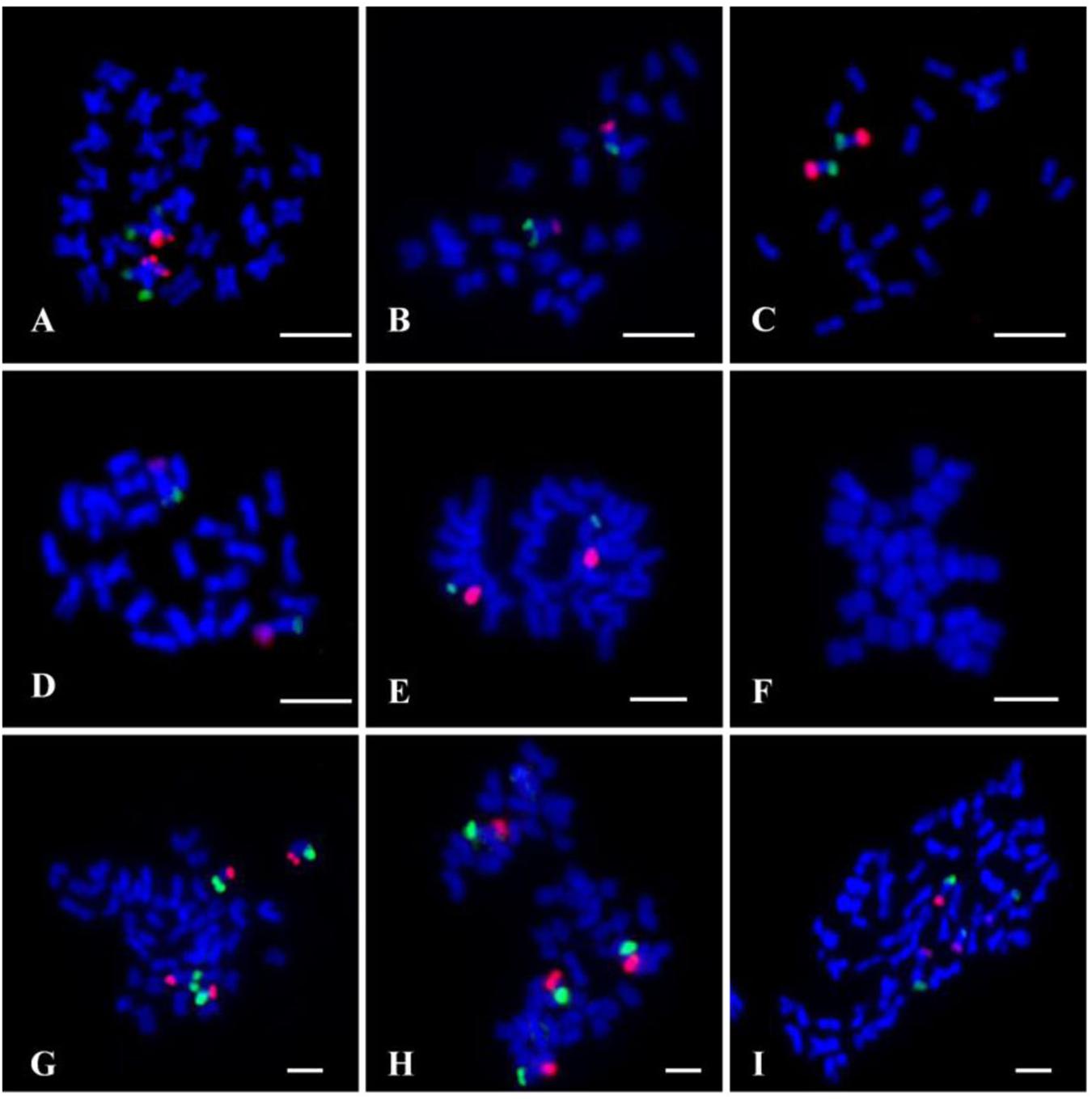
Figure 2. Fluorescence in situ hybridization (FISH) using the chromosome 7 oligo probes (set 1, green; set 2, red) on metaphase chromosomes of cotton. (A) Gossypium raimondii (D5). (B) Gossypium thurberi (D1). (C) Gossypium davidsonii (D3–d). (D) Gossypium klotzschianum (D3–k). (E) Gossypium herbacium (A1). (F) Gossypium arboretum (A2). (G) Gossypium hirsutum (AD1). (H) Gossypium barbadense (AD2). (I) Gossypium mustelinum (AD4). Chromosomes were stained with 4′,6-diamidino-2-phenylindole (DAPI). Bars, 5 μm.
To further verify if the chromosome 7-specific probes selected based on D genome of G. raimondii would work for other cotton species, both the two sets of oligo probes were also hybridized to metaphase chromosomes of D genome species (Gossypium thurberi D1, G. davidsonii D3–d, and G. klotzschianum D3–k), G. herbacium (A1) and Gossypium arboretum (A2) from A genome, and three AD genome cotton species (G. hirsutum AD1, G. barbadense AD2, and G. mustelinum AD4). The two oligo probes produced signals at the ends of two arms of a pair of D-genome chromosomes, with stronger signals in G. davidsonii than G. thurberi and G. klotzschianum (Figures 2B–D). Probes produced bright FISH signals on a pair of chromosomes of G. herbacium (Figure 2E). There were two pairs of chromosome 7 oligo signals in the three tetraploid cotton species (Figures 2G–I), indicating that the oligo probes had signals in both subgenome of tetraploid cotton. However, there was no signal on chromosome of G. arboretum (Figure 2F).
The chromosome 7 of G. raimondii genome sequence is homologous to the chromosome 11 (here named as D11) of tetraploid cotton; the former was based on a genetic map (Rong et al., 2004; Paterson et al., 2012), and the latter is based on the physical chromosomes (Wang et al., 2006). A 45S rDNA signal is typically located at the telomeric region on D11 in G. raimondii on the basis of the study of Gan et al. (2013). Theoretically, the chromosome 7-specific oligo developed from G. raimondii genome sequence should be collinear with one of 45S rDNA locus. To verify this, the chromosome 7-specific oligo probe and 45S rDNA probe were hybridized to the same chromosome preparation of G. raimondii (Figure 2A) by repeated FISH. The results indicated that three pairs of 45S rDNA signals (red) were detected on G. raimondii chromosomes, but none of them was collinear with the chromosome 7-specific oligo signals (green) (Figure 3A). Next, the chromosome 7-specific set 2 oligo probe and 45S rDNA probe were tested on other three D genome species. Four pairs of 45S rDNA signals (red) were detected on G. thurberi chromosomes, and a pair of 45S rDNA overlapped with chromosome 7 set 2 oligo (Figure 3B shown with arrows), which is consistent with a previous study in which there was a 45S rDNA locus on G. thurberi chromosome D11 (Gan et al., 2011). FISH results on G. davidsonii and G. klotzschianum showed no collinearity between chromosome 7-specific oligo and 45S rDNA (Figures 3C,D). This confirmed a previous study in which there were no 45S rDNA loci on chromosomes D11 of these two species (Gan et al., 2012).
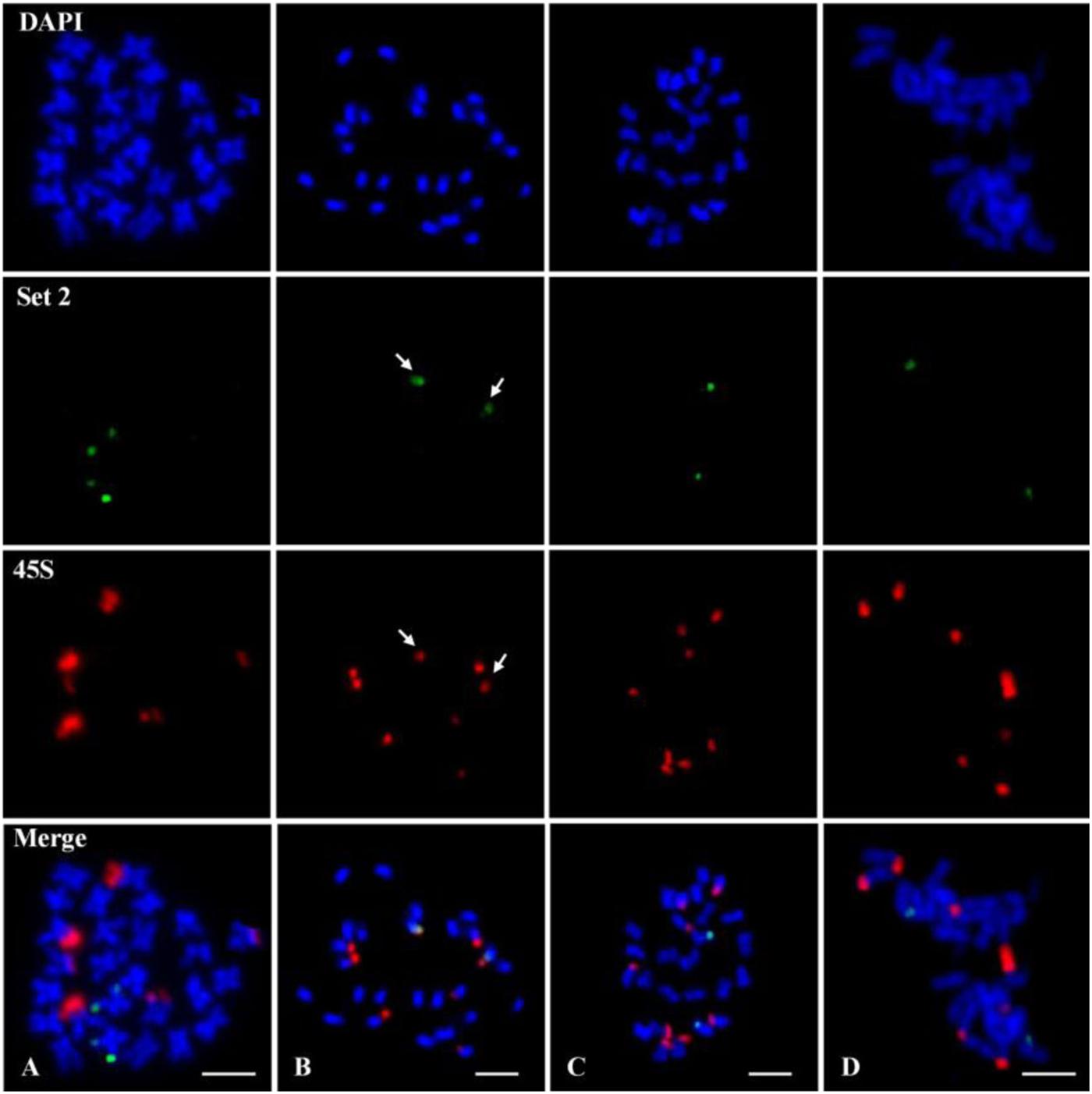
Figure 3. Fluorescence in situ hybridization (FISH) using the chromosome 7 set 2 oligo (green) probe and 45S ribosomal DNA (rDNA) (red) probe on metaphase chromosomes of four D genome cotton species. (A) Gossypium raimondii. (B) Gossypium thurberi. (C) Gossypium davidsonii. (D) Gossypium klotzschianum. The arrows in (B) show overlapped signals between 45S rDNA and set 2 oligo. Chromosomes were stained with 4′,6-diamidino-2-phenylindole (DAPI). Bars, 5 μm.
FISH using the chromosome 7 set 2 oligo (red) probe and 45S rDNA (green) probe was performed on metaphase chromosomes of five tetraploid cotton species. All five species showed clear two pairs of set 2 oligo signals on At11 and Dt11 homoeologous chromosomes (Figure 4, Set 2); each species had three pairs of 45S rDNA signals (Figure 4, 45S), but none of them located are on the same chromosome (Figure 4, Merge).
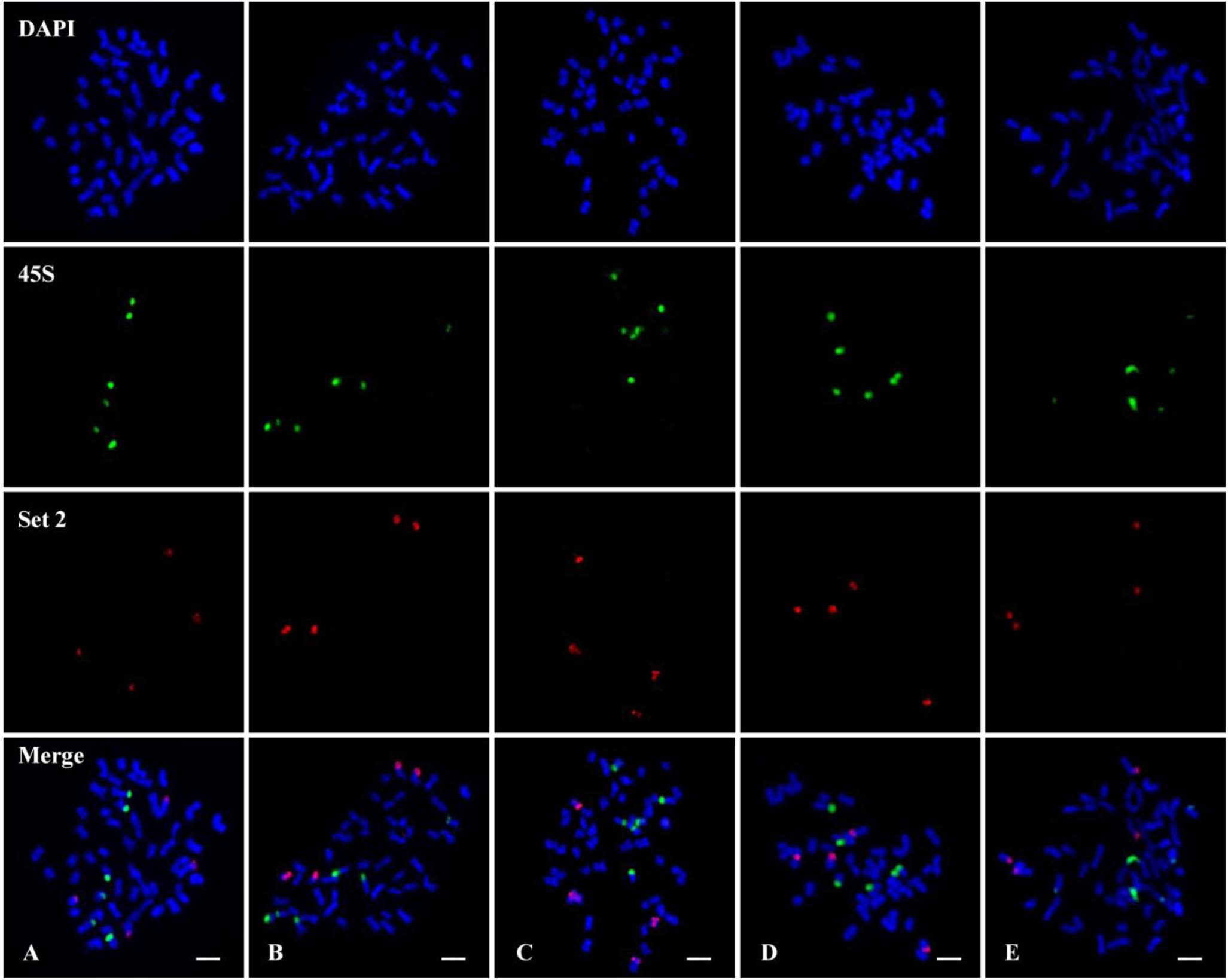
Figure 4. Fluorescence in situ hybridization (FISH) using the chromosome 7 set 2 oligo (red) probe and 45S ribosomal DNA (rDNA) (green) probe on metaphase chromosomes of five tetraploid cotton species. (A) Gossypium hirsutum (AD1). (B) Gossypium barbadense (AD2). (C) Gossypium tomentosum (AD3). (D) Gossypium mustelinum (AD4). (E) Gossypium darwinii (AD5). Chromosomes were stained with 4′,6-diamidino-2-phenylindole (DAPI). Bars, 5 μm.
FISH technique has become one of the most important techniques in plant molecular cytogenetics owing to its superiority of direct mapping of DNA sequences on cytological specimens (Jiang, 2019). During the application of FISH technology, significant efforts have been made to develop suitable probes to identify chromosomes of different cotton species, such as specific genome repetitive sequence probe for identification of specific chromosome region (Hanson et al., 1996; Liu et al., 2016), genomic DNA probe for genome-specific chromosome painting in polyploid species (Wu et al., 2013), and large-insert genomic DNA clone probe for individual chromosomes identification (Wang et al., 2006; Gan et al., 2011). However, the above-mentioned probes showed different limitations in the application process (Jiang and Gill, 2006). In particular, owing to the more than 60% repetitive sequences in cotton genome (Paterson et al., 2012; Hu et al., 2019), the FISH result of large-insert genomic DNA clone probe may be greatly affected by repetitive sequences, which may show low chromosome specificity. The bulked oligo probes designed based on specific chromosome sequence had demonstrated great superiority in resolution and versatility (Han et al., 2015; Qu et al., 2017; He et al., 2018; Meng et al., 2018). In this study, we developed two oligo probes of G. raimondii chromosome 7. Owing to the removal of repetitive sequences from the oligo sets, the two oligo probes produced clear signals without background noise on the chromosome 7 when hybridized to chromosomes of G. raimondii and its relative species (Figures 2A–D). At the same time, high proportions of repetitive sequences in the pericentromeric heterochromatic region of chromosomes (Liu et al., 2016; Lu et al., 2019) caused the disappearance of oligos in the middle region of the chromosome (Figure 1), which led to the signals of the oligo probes not diffusing the whole chromosome arms, and only appeared at the two end regions of chromosome (Figure 2). Moreover, the homoeologous chromosomes of G. raimondii chromosome 7 in allotetraploid cotton were unambiguously identified using the designed oligo probes (Figures 2G–I). Thus, bulked oligo probes are superior in flexibility, specificity, and repeatability than are the BAC-based probes, which will be useful for studying cotton karyotype evolution.
rRNA gene is highly a conserved repetitive sequence in the plant genome. Variation in the number and distribution of 45S rDNA signals can indicate genome reorganization and phylogenetic context (Hasterok et al., 2006, 2019; Wolny and Hasterok, 2009; Chacón et al., 2012). As an attractive model for studying the origin, evolution of polyploids, great efforts have been made using 45S rDNA as a probe to study the evolutional relationship of cotton (Hanson et al., 1996; Ji et al., 1999). In G. raimondii, the 45S rDNA was located at the terminal region of chromosomes D502, D509, and D511 (corresponding to Chr.05, Chr.06, and Chr.07 of G. raimondii genome sequence map) (Gan et al., 2013). That is, there is a 45S rDNA locus at the chromosome 7 of G. raimondii according to the chromosome number in the genome sequence. As a component of 45S rDNA, ITS sequence is highly repetitive in the nuclear genome and has become an important molecular marker in the study of phylogeny reconstruction in plant (Wendel et al., 1995). The blast result of ITS region of G. raimondii against the G. raimondii genome sequence is inconsistent with previous results in copy number and chromosome position (data unpublished). In this study, the result of co-location with the chromosome 7-specific oligo and 45S rDNA probes showed that no 45S rDNA was located at chromosome 7 (D511) of G. raimondii (Figure 3A). This is not consistent with previous studies too (Gan et al., 2013) but is in agreement with ITS blast result. Previous identification of 45S rDNA-bearing chromosomes was by means of BAC-FISH using the chromosome-specific BAC clone derived from G. hirsutum (AD1) (Gan et al., 2013). As allotetraploid, G. hirsutum contains high proportions of repetitive sequences, which would make it difficult for genome-derived BAC clones to produce chromosome-specific FISH signals by BAC-FISH (Jiang and Gill, 2006). Owing to the specificity and repeatability of the chromosome-specific oligo probe, it is most likely that no 45S rDNA locates on chromosome 7 (D511) of G. raimondii. This result will prompt us to reposition 45S rDNA loci of G. raimondii to investigate the evolutionary relationship of cotton.
The specificity and presence or absence of oligo signals between different species can reflect the differences in genetic relationships and genomic sequences (Liu et al., 2019). In present study, two pairs of chromosome 7 oligo signals were viewed in both subgenome of tetraploid cotton (Figures 2G–I). Among two A genome species, there was a pair of bright oligo signals on chromosomes of Gossypium herbacium (Figure 2E). However, there was no signal on chromosome of Gossypium arboretum (Figure 2F). That is, G. herbacium generated similar signals with A subgenome of tetraploid cotton, suggesting that G. herbaceum was more likely a closer relative to the A subgenome donor of tetraploid cotton than G. arboreum, which supported the views of the predecessors by means of visual evidences (Gerstel, 1953; Wendel et al., 2010).
Using a massive oligo synthesis strategy, we developed the bulked oligos of the two arms of chromosome 7 of G. raimondii. The results showed that the chromosome 7 was easily identified in both D genome and AD genome cotton species using the bulked chromosome-specific painting probes. The study reveals that bulked oligos specific to a chromosome is a useful tool for chromosome painting in cotton. Therefore, the oligo-FISH probes we established will be a powerful tool for studying chromosome variations and evolution in the genus Gossypium. Based on the established oligo-FISH system, more oligo probes can be designed to study karyotype structure and evolution in more sequenced cotton genomes.
All datasets generated for this study are included in the article/supplementary material.
RP, YL, and TZ conceived the study and drafted manuscript. YL, XW, and YW conducted the cytogenetic experiments. TZ designed the oligo probes. ZL, QL, and FL participated in the data analysis. All authors read and approved the final manuscript.
The research was supported by the National Key Research and Development Program of China (2018YFD0100300), Program for Innovative Research Team (in Science and Technology) in the University of Henan Province (20IRTSTHN021), Science and Technology Development Project of Henan Province (182102410041), and Science and Technology Development Project of Anyang City (2018-66-133).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to express our gratitude to Prof. Baohong Zhang (East Carolina University, United States) for editing the English text of a draft of this manuscript.
Albert, P. S., Zhang, T., Semrau, K., Rouillard, J. M., and Kao, Y. H. (2019). Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc. Natl. Acad. Sci. U.S.A. 116, 1679–1685. doi: 10.1073/pnas.1813957116
Bi, Y. F., Zhao, Q. Z., Yan, W. K., Li, M. X., Liu, Y. X., Cheng, C. Y., et al. (2019). Flexible chromosome painting based on multiplex PCR of oligonucleotides and its application for comparative chromosome analyses in Cucumis. Plant J. 102, 178–186. doi: 10.1111/tpj.14600
Breneman, J. W., Ramsey, M. J., Lee, D. A., Eveleth, G. G., Minkler, J. L., and Tucker, J. D. (1993). The development of chromosome-specific composite DNA probes for the mouse and their application to chromosome painting. Chromosoma 102, 591–598. doi: 10.1007/BF00352306
Chacón, J., Sousa, A., Baeza, C. M., and Renner, S. S. (2012). Ribosomal DNA distribution and a genus-wide phylogeny reveal patterns of chromosomal evolution in Alstroemeria (Alstroemeriaceae). Am. J. Bot. 99, 1501–1512. doi: 10.3732/ajb.1200104
Chang, K. D., Fang, S. A., Chang, F. C., and Chung, M. C. (2010). Chromosomal conservation and sequence diversity of ribosomal RNA genes of two distant Oryza species. Genomics 96, 181–190. doi: 10.1016/j.ygeno.2010.05.005
Cheng, Z. K., Presting, G. G., Buell, C. R., Wing, R. A., and Jiang, J. M. (2001). High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157, 1749–1757.
Comings, D. E. (1978). Mechanisms of chromosome banding and implications for chromosome structure. Annu. Rev. Genet. 12, 25–46. doi: 10.1146/annurev.ge.12.120178.000325
Cronn, R. C., Small, R. L., Haselkorn, T., and Wendel, J. F. (2002). Rapid diversification of the cotton genus (Gossypium: Malvaceae) revealed by analysis of sixteen nuclear and chloroplast genes. Am. J. Bot. 89, 707–725. doi: 10.3732/ajb.89.4.707
Ferguson-Smith, M. A., and Trifonov, V. (2007). Mammalian karyotype evolution. Nat. Rev. Genet. 8, 950–962. doi: 10.1038/nrg2199
Fuchs, J., Houben, A., Brandes, A., and Schubert, I. (1996). Chromosome painting in plants-a feasible technique? Chromosoma 104, 315–320. doi: 10.1007/bf00337219
Gan, Y. M., Chen, D., Liu, F., Wang, C. Y., Li, S. H., Zhang, X. D., et al. (2011). Individual chromosome assignment and chromosomal collinearity in Gossypium thurberi, G. trilobum and D subgenome of G. barbadense revealed by BAC-FISH. Genes. Genet. Syst. 86, 165–174. doi: 10.1266/ggs.86.165
Gan, Y. M., Liu, F., Chen, D., Wu, Q., Qin, Q., Wang, C. Y., et al. (2013). Chromosomal Locations of 5S and 45S rDNA in Gossypium genus and its phylogenetic implications revealed by FISH. PLoS One 8:e68207. doi: 10.1371/journal.pone.0068207
Gan, Y. M., Liu, F., Peng, R. H., Wang, C. Y., Li, S. H., Zhang, X. D., et al. (2012). Individual chromosome identification, chromosomal collinearity and genetic-physical integrated map in Gossypium darwinii and four D genome cotton species revealed by BAC-FISH. Genes. Genet. Syst. 87, 233–241. doi: 10.1266/ggs.87.233
Gerstel, D. U. (1953). Chromosomal translocations in interspecific hybrids of the genus Gossypium. Evolution 7, 234–244. doi: 10.2307/2405734
Han, Y. H., Zhang, T., Thammapichai, P., Weng, Y., and Jiang, J. M. (2015). Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 200, 771–779. doi: 10.1534/genetics.115.177642
Hanson, R. E., Islam-Faridi, M. N., Percival, E. A., Crane, C. F., Ji, Y. F., McKnight, T. D., et al. (1996). Distribution of 5S and 18S-28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and putative diploid ancestors. Chromosoma 105, 55–61. doi: 10.1007/BF02510039
Hasterok, R., Wang, K., and Jenkins, G. (2019). Progressive refinement of the karyotyping of Brachypodium genomes. New Phytol. doi: 10.1111/nph.16342 [Epub ahead of print].
Hasterok, R., Wolny, E., Hosiawa, M., Kowalczyk, M., Kulak-Ksiazczyk, S., Ksiazczyk, T., et al. (2006). Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Ann. Bot. 97, 205–216. doi: 10.1093/aob/mcj031
He, L., Braz, G. T., Torres, G. A., and Jiang, J. M. (2018). Chromosome painting in meiosis reveals pairing of specific chromosomes in polyploid Solanum species. Chromosoma 127, 505–513. doi: 10.1007/s00412-018-0682-9
Hou, L., Xu, M., Zhang, T., Xu, Z., Wang, W., Zhang, J., et al. (2018). Chromosome painting and its applications in cultivated and wild rice. BMC Plant Biol. 18:110. doi: 10.1186/s12870-018-1325-2
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748. doi: 10.1038/s41588-019-0371-5
Hutchinson, J. B., Silow, R. A., and Stephens, S. G. (1947). The Evolution Of Gossypium And The Differentiation Of The Cultivated Cottons. New York, NY: Oxford University Press.
Ji, Y., De Donato, M., Crane, C. F., Raska, W. A., and Islam-Faridi, M. N. (1999). New ribosomal RNA gene locations in Gossypium hirsutum mapped by meiotic FISH. Chromosoma 108, 200–207. doi: 10.1007/s004120050369
Jiang, J. M. (2019). Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosom. Res. 27, 153–165. doi: 10.1007/s10577-019-09607-z
Jiang, J. M., and Gill, B. S. (2006). Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49, 1057–1068. doi: 10.1139/g06-076
Johnson, B. L. (1975). Gossypium palmeri and a polyphyletic origin of the New World cottons. Bull. Torrey Bot. Club 102, 340–349. doi: 10.2307/2484760
Liu, S. H., Wang, K. B., Song, G. L., Wang, C. Y., Liu, F., Li, S. H., et al. (2005). Primary investigation on GISH-NOR in cotton. Chin. Sci. Bull. 50, 425–429. doi: 10.1360/982004-582
Liu, X., Sun, S., Wu, Y., Zhou, Y., Gu, S., Yu, H., et al. (2019). Dual-color oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J. 101, 112–121. doi: 10.1111/tpj.14522
Liu, Y. L., Peng, R. H., Liu, F., Wang, X. X., Cui, X. L., Zhou, Z. L., et al. (2016). A Gossypium BAC clone contains key repeat components distinguishing sub-genome of allotetraploidy cottons. Mol. Cytogenet. 9:27. doi: 10.1186/s13039-016-0235-y
Lu, H. J., Cui, X. L., Liu, Z., Liu, Y. L., Wang, X. X., Zhou, Z. L., et al. (2018). Discovery and annotation of a novel transposable element family in Gossypium. BMC Plant Biol. 18:307. doi: 10.1186/s12870-018-1519-7
Lu, H. J., Cui, X. L., Zhao, Y. Y., Magwang, R. O., Li, P. C., Cai, X. Y., et al. (2019). Identification of a genome-specific repetitive element in the Gossypium D genome. PeerJ 7:e27806v1. doi: 10.7287/peerj.preprints.27806v1
Lysak, M. A., and Mandáková, T. (2013). Analysis of plant meiotic chromosomes by chromosome painting. Methods Mol. Biol. 990, 13–24. doi: 10.1007/978-1-62703-333-6_2
Lysak, M. A., Mandáková, T., and Lacombe, E. (2010). Reciprocal and multi-species chromosome BAC painting in crucifers (Brassicaceae). Cytogenet. Genom. Res. 129, 184–189. doi: 10.1159/000312951
Lysak, M. A., Pecinka, A., and Schubert, I. (2003). Recent progress in chromosome painting of Arabidopsis and related species. Chromosom. Res 11, 195–204. doi: 10.1023/A:1022879608152
Mandáková, T., and Lysak, M. A. (2008). Chromosomal phylogeny and karyotype evolution in x = 7 Crucifer species (Brassicaceae). Plant Cell 20, 2559–2570. doi: 10.1105/tpc.108.062166
McClintock, B. (1929). Chromosome morphology in Zea mays. Science 69:629. doi: 10.1126/science.69.1798.629
Meng, Z., Zhang, Z., Yan, T., Lin, Q., Wang, Y., Huang, W., et al. (2018). Comprehensively characterizing the cytological features of Saccharum spontaneum by the development of a complete set of chromosome-specific oligo probes. Front. Plant Sci. 9:1624. doi: 10.3389/fpls.2018.01624
Parks, C. R., Ezell, W. L., Williams, D. E., and Dreyer, D. L. (1975). The application of flavonoid distribution to taxonomic problems in the genus Gossypium. Bull. Torrey Bot. Club 102, 350–361. doi: 10.2307/2484761
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Percival, A. E., Wendel, J. F., and Stewart, J. M. (1999). “Taxonomy and germplasm resources,” in Cotton; Origin, History Technology, And Production, eds W. C. Smith, and J. T. Cothren (New York, NY: Wiley), 33–63.
Pinkel, D., Landegent, J., Collins, C., Fuscoe, J., Segraves, R., Lucas, J., et al. (1988). Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc. Natl. Acad. Sci. U.S.A. 85, 9138–9142. doi: 10.1073/pnas.85.23.9138
Qu, M., Li, K., Han, Y., Chen, L., Li, Z., and Han, Y. (2017). Integrated karyotyping of woodland strawberry (Fragaria vesca) with oligopaint FISH probes. Cytogenet. Genome. Res. 153, 158–164. doi: 10.1159/000485283
Rens, W., Fu, B., O’brien, P. C., and Ferguson-Smith, M. (2006). Cross-species chromosome painting. Nat. Protoc. 1, 783–790. doi: 10.1038/nprot.2006.91
Rong, J., Abbey, C., Bowers, J. E., Brubaker, C. L., Chang, C., Chee, P. W., et al. (2004). A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166, 389–417. doi: 10.1534/genetics.166.1.389
Scherthan, H., Cremer, T., Arnason, U., Weier, H. U., and Lima-de-Faria, A. (1994). Comparative chromosome painting discloses homologous segments in distantly related mammals. Nat. Genet. 6, 342–347. doi: 10.1038/ng0494-342
Wang, K., Song, X., Han, Z., Guo, W., Yu, J. Z., Sun, J., et al. (2006). Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theor. Appl. Genet. 113, 73–80. doi: 10.1007/s00122-006-0273-7
Wang, K. B., Wendel, J. F., and Hua, J. P. (2018). Designations for individual genomes and chromosomes in Gossypium. Cotton Res. 1:3. doi: 10.1186/s42397-018-0002-1
Wendel, J. F., and Albert, V. A. (1992). Phylogenetics of the cotton genus (Gossypium L): character-state weighted parsimony analysis of chloroplast DNA restriction site data and its systematic and biogeographic implications. Syst. Bot. 17, 115–143. doi: 10.2307/2419069
Wendel, J. F., Brubaker, C. L., and Seelanan, T. (2010). “The origin and evolution of Gossypium,” in Physiology of Cotton, eds J. Stewart, D. M. Oosterhuis, J. J. Heitholt, and J. R. Mauney (Cham: Springer), 12–14.
Wendel, J. F., and Cronn, R. C. (2003). Polyploidy and evolutionary history of cotton. Adv. Agron. 78, 139–186. doi: 10.1016/s0065-2113(02)78004-8
Wendel, J. F., Schnabel, A., and Seelanan, T. (1995). Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc. Natl. Acad. Sci. U.S.A. 92, 280–284. doi: 10.1073/pnas.92.1.280
Wolny, E., and Hasterok, R. (2009). Comparative cytogenetic analysis of the genomes of the model grass Brachypodium distachyon and its close relatives. Ann. Bot. 104, 873–881. doi: 10.1093/aob/mcp179
Woo, S. S., Jiang, J. M., Gill, B. S., Paterson, A. H., and Wing, R. A. (1994). Construction and characterization of bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 22, 4922–4931. doi: 10.1093/nar/22.23.4922
Wu, Q., Liu, F., Li, S. H., Song, G. L., Wang, C. Y., Zhang, X. D., et al. (2013). Uniqueness of the Gossypium mustelinum genome revealed by GISH and 45S rDNA FISH. J. Integr. Plant Biol. 55, 654–662. doi: 10.1111/jipb.12084
Xiao, S. P., Chen, Y., Ke, X. S., Wang, Y. H., Wang, C. Y., Yang, L., et al. (2017). Genomic in situ hybridization analysis of the genetic relationship between wild-type diploid and cultivated allotetraploid Gossypium species. Cotton Sci. 29, 99–107. doi: 10.11963/issn.1002-7807.201701011
Keywords: Gossypium raimondii, oligonucleotide, fluorescence in situ hybridization, chromosome painting, 45S rDNA
Citation: Liu Y, Wang X, Wei Y, Liu Z, Lu Q, Liu F, Zhang T and Peng R (2020) Chromosome Painting Based on Bulked Oligonucleotides in Cotton. Front. Plant Sci. 11:802. doi: 10.3389/fpls.2020.00802
Received: 03 March 2020; Accepted: 19 May 2020;
Published: 30 June 2020.
Edited by:
Zhaojun Ding, Shandong University, ChinaReviewed by:
Shulan Fu, Sichuan Agricultural University, ChinaCopyright © 2020 Liu, Wang, Wei, Liu, Lu, Liu, Zhang and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Zhang, emhhbmd0YW9AeXp1LmVkdS5jbg==; Renhai Peng, YXlkeHByaEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.