
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 19 June 2018
Sec. Plant Biotechnology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00826
This article is part of the Research Topic Novel Insights into Algal Biology and Biotechnology View all 13 articles
To verify the function of malic enzyme (ME1), the ME1 gene was endogenously overexpressed in Phaeodactylum tricornutum. Overexpression of ME1 increased neutral and total lipid content and significantly increased saturated fatty acids (SFAs) and polyunsaturated fatty acids (PUFAs) in transformants, which varied between 23.19 and 25.32% in SFAs and between 49.02 and 54.04% in PUFAs, respectively. Additionally, increased ME1 activity was accompanied by elevated NADPH content in all three transformants, indicating that increased ME1 activity produced additional NADPH comparing with that of WT. These results indicated that ME1 activity is NADP-dependent and plays an important role in the NADPH levels required for lipid synthesis and fatty acid desaturation in P. tricornutum. Furthermore, our findings suggested that overexpression of endogenous ME1 represents a valid method for boosting neutral-lipid yield in diatom.
Malic enzyme (ME) is widespread in all kinds of organisms and catalyzes the reversible oxidative decarboxylation of malate to pyruvate, CO2, and NAD(P)H in the presence of a divalent metal ion (Drincovich et al., 2001). Malic enzymes are divided into three categories on the basis of the substrate specificity and coenzyme preference: NAD+(EC 1.1.1.38-39), NADP+-dependent (EC1.1.1.40), or dependent upon both cofactors. These enzymes localize to the cytoplasm, mitochondria or chloroplasts in eukaryotes (Tang et al., 2010); however, little is known about the molecular basis for cofactor selectivity of these enzymes (Chang and Tong, 2003).
In spite of decades of research, the physiological function of ME remains poorly understood, and its role might vary from organism to organism (Kendrick and Ratledge, 1992). ME plays a crucial part in the provision of NADPH to promote desaturation and elongation reactions leading to the formation of polyunsaturated fatty acids (PUFAs) in some oleaginous organisms (Kendrick and Ratledge, 1992; Ren et al., 2013; Ratledge, 2014; Lv et al., 2016). Previous studies also reported novel physiological functions associated with this enzyme, including renovating UV-induced damage in maize seedlings (Drincovich et al., 1998; Casati et al., 1998), affecting the development of chloroplasts by generating excessive reducing power in transgenic C3 plants (Takeuchi et al., 2000), lengthening the lifespan of Drosophila during the larval stage (Kim et al., 2015), keeping stable levels of TCA-cycle intermediates in the bacterium Sinorhizobium meliloti (Zhang et al., 2016), and acting as a potential target of cancer chemotherapy (Chang and Tong, 2003) and as an anaplerotic enzyme in Saccharomyces cerevisiae (Zelle et al., 2011) and Streptomyces coelicolor (Rodriguez et al., 2012).
Few studies have centered on the role of MEs in lipid accumulation, with those undertaken focusing mainly in plants and mammals; however, little is known about the role of these enzymes in microalgae. An NADP-dependent ME was proposed for diatoms, with possible function as a decarboxylase of releasing CO2 in chloroplasts of diatoms, similar to that in C4 plants (Granum et al., 2005). The gene encoding ME in Dunaliella parva was cloned and characterized, but further research on the function of this gene was not conducted (Shang et al., 2012). Kroth et al. (2008) reported that Phaeodactylum tricornutum appears to contain two mitochondrial ME that are either NAD- or NADP-dependent. One of the ME-encoding genes was overexpressed endogenously in P. tricornutum (PtME), revealing its predominant localization to the mitochondria, as well as its significant impact on promoting lipid accumulation; however no further studies were performed on the role of ME in increasing lipid content (Xue et al., 2015).
The marine diatom P. tricornutum represents a potential producer of biodiesel because of its rapid growth, lipid-accumulation capability, and the availability of genetic tools (Zaslavskaia et al., 2000). Therefore, it is possible to genetically manipulate the key genes involved in fatty acid synthesis in this alga to enhance characters to gain both high lipid and high biomass levels necessary for industrial production.
To verify ME function, the ME1 gene from P. tricornutum (different from that studied by Xue et al., 2015), which possesses a mitochondria presequence and might have a dinucleotide-binding site given for NADP (Kroth et al., 2008), was overexpressed in Escherichia coli (Lv et al., 2016) and in P. tricornutum (this study). The results of this study, as well as those previously published (Lv et al., 2016), indicated that ME1 from P. tricornutum is NADP-dependent and can supply enough NADPH for both fatty acid biosynthesis and desaturation in E. coli (Lv et al., 2016) and P. tricornutum (this study).
Wild-type (WT) P. tricornutum Bohlin (LAMB118), provided by Institute of Hydrobiology, Chinese Academy of Sciences, and zeocin-resistant colonies (named as PtME1-1, PtME1-2, and PtME1-3) were cultured in f/2 medium (Guillard, 1975) prepared with sterile seawater at 20 ± 1°C and under 37.50 μmol photons m-2 s-1, following a 12:12 photoperiod. Three cultures for every transformant and the WT strain (300-mL each) were cultivated axenically to monitor their growth. To set up growth curves, the optical density (OD) was measured at 750 nm every 2 days using a UV-3310 spectrophotometer (Hitachi, Tokyo, Japan) (Griffiths et al., 2011).
The ME1 gene was amplified by polymerase chain reaction (PCR) using cDNA from P. tricornutum (GenBank accession: XP_002177890.1) as the template and the primers PtME1-F (GGGGTACCATGATATCATCGGCGTGTCG) and PtME1-R (GCTCTAGACTAGTGGTGGTGGTGGTGGTGGATTGATATTTCTCGTTTTTCC). To generate the pPha-T1-ME1 recombinant plasmid, the amplified gene was inserted into the pPha-T1 (Zaslavskaia et al., 2000) plasmid using an fcpA promoter driving the ME1 gene, and the resistant strains were selected with ZeocinTM (Invitrogen, Carlsbad, CA, United States).
The recombinant plasmid (pPha-T1-ME1) was introduced into P. tricornutum by a Bio-Rad Biolistic PDS-1000/He particle-delivery system (Bio-Rad, Hercules, CA, United States), according to the methods described by Zaslavskaia et al. (2000) and Zhu et al. (2016). It should be pointed out that seawater f/2 medium was used for culturing in this study.
Total genomic DNA was isolated in the light of the method described by Watanabe et al. (1998). Transformants were screened by PCR using the gene-specific Sh-ble and PtME1-ble primers (Table 1), respectively. To prepare a digoxigenin-labeled probe for Southern blot analysis (Falciatore et al., 1999), the ble fragment was used as the template and genomic DNA was digested with EcoR I and KpnI, respectively. For western blot (Poulsen and Kröger, 2005), an anti-His-tag antibody (Bioss, Woburn, MA, United States) was used to detect the ME1 protein, and actin was served as the internal control. Cells were harvested by centrifugation at late exponential stage for assays.
RNA extraction and qPCR were performed as previously described (Zhu et al., 2016) using primers shown in Table 1. The histone H4 gene was served as the internal reference (Siaut et al., 2007). The 2-ΔΔCt method (Livak and Schmittgen, 2001) was used to analyze the ME1 expression and calculate relative ME1-transcript abundance.
Total lipids were extracted according to the method as described previously (Zhu et al., 2016). To detect cellular neutral-lipid content in P. tricornutum, BODIPY505/515 (Invitrogen) staining was carried out according to the protocol described by Cooper et al. (2010). Algal cells (106 cells/mL) were first treated with 2% dimethyl sulfoxide (DMSO) for 10 min at room temperature. A stock solution of 100 μg/mL BODIPY505/515 was prepared using anhydrous DMSO and added directly to algal solution to obtain a final BODIPY 505/515-labeling concentration of 0.87 μg/mL, followed by incubation in darkness for 10 min at room temperature. Stained cells were detected by their fluorescence intensity with flow cytometry (BD FACSVantage SE; BD Biosciences, Franklin Lakes, NJ, United States). Excitation and emission wavelengths were 485 nm and 535 nm, respectively. Quantitative comparison of neutral-lipid content between samples was obtained according to relative fluorescence-intensity values.
Fatty acid methyl esters were analyzed using gas chromatography (Agilent 6890 Series GC System; US10251016; Agilent, Santa Clara, CA, United States) as previously described (Zhu et al., 2014) after transmethylation according to a method described by Lepage and Roy (1984). Mixed external standards of fatty acids (Supelco 37, United States) were used to detect and determine the Fatty acid composition.
Malic enzyme activity in P. tricornutum was measured using an NADPH-ME kit (Solarbio, Beijing, China) according to manufacturer instructions. The optimum reaction system was prepared as followed: 50 mM, pH 7.5 Tris–HCl, 1 mM MgCl2, 0.5 mM NADP+, 10 mM L-malate. Soluble protein concentration was quantified using a Bradford assay kit (Genmed Scientifics, Shanghai, China). ME activity was determined by monitoring the change in absorbance at 1-min intervals continuously at 340 nm using a UV-3310 spectrophotometer (Hitachi). One unit of ME activity was defined as 1 μM NADPH generated by 1 mg protein per minute in the reaction system:
where A1 is the initial absorbance, A2 is the absorbance after the reaction, 6.22 represents the extinction coefficient per mM NADPH, t is the reaction time (1 min), l is the path length of the cuvette (1 cm), V1 is the total reaction volume (900 μL), V2 is the volume of ME solution (30 μL), and C is the concentration of protein (mg/mL).
NADPH content in P. tricornutum was analyzed with the Amplite Fluorimetric NADP/NADPH ratio assay kit (AAT Bioquest, Sunnyvale, CA, United States) according to manufacturer instructions. After lysis and ultrasonication extraction, microalgal samples were centrifuged at 8000 × g for 10 min at 4°C, and the supernatant was prepared for testing. Traditional NADPH assays are performed by monitoring the changes in NADPH absorption at 340 nm using a Synergy microplate reader (BioTek, Winooski, VT, United States). The excitation and emission wavelengths used were 540 nm and 590 nm, respectively. The concentration of NADPH was determined from the standard curve of NADPH.
One-way analysis of variance was used at a level of significance of P < 0.05 to calculate significant differences between treatments using SPSS 17.0 software (IBM, Armonk, NY, United States), and all data are reported as the mean ± standard deviation (three replicates were used, n = 3).
Similar growth curves were shown in Figure 1 for both the WT strain and transformants although all the resistant strains showed a slightly increased growth rate and did not differ significantly from that of the WT strain, which suggested that the transgene exerted a temperate influence on the growth of the transformants. In contrast to this study, Jiang et al. (2013) reported that depletion of ME1 or ME2 strongly impaired tumor-cell growth, and overexpression of these genes enhanced tumor-cell growth. By contrast, growth reductions in transgenic strains were reported by other studies (Li et al., 2010; Radakovits et al., 2011). Therefore, further research is needed to understand the underlying mechanism.
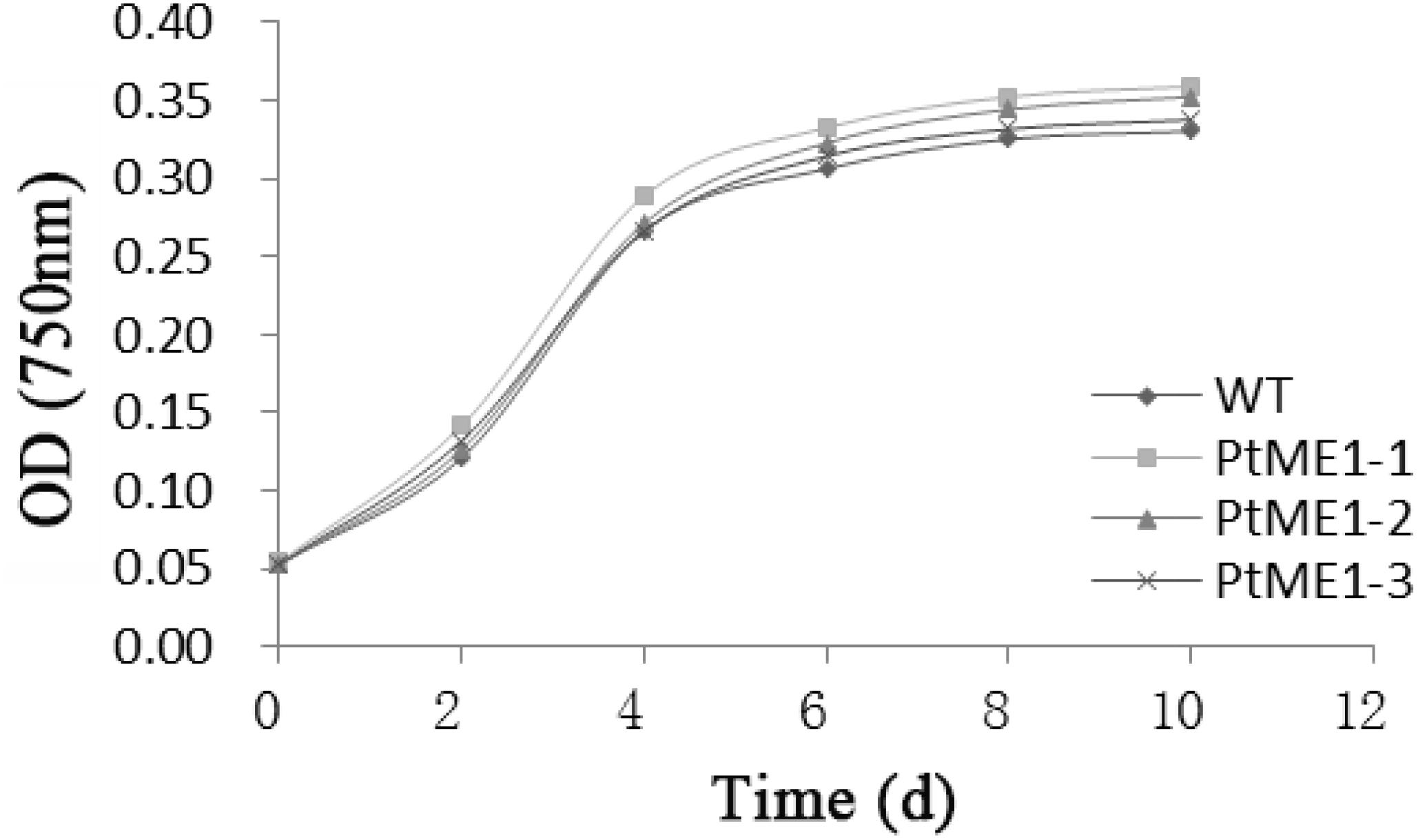
FIGURE 1. Growth curves of transgenic clones and the WT strain. Values represent averages of three replicates (n = 3).
Resistant clones were first selected by growth on f/2 solid medium (1% agar) with 100 μg/mL Zeocin. Further screening was conducted by PCR using the gene-specific Sh-ble and ME1-ble primers (Table 1). As shown in Figures 2A,B, all resistant clones presented the expected fragment sizes, but no DNA band was seen in the WT strain. To further verify gene integration, Southern blot analysis was performed in all three resistant strains and the WT strain. As shown in Figure 2C, all three resistant strains showed two or more hybridized bands detected with the ble probe digested using different restriction enzymes, indicating that exogenous sh-ble and endogenous ME1 had been integrated into the P. tricornutum genome, and that these clones were undoubted transformants. To detect target protein levels translated from the introduced ME1 gene in all the resistant strains, western blot was conducted with an anti-His-tag antibody. A cross-reacting band, the same size as expected, was shown in all the resistant strains, whereas this was not observed in the WT strain (Figure 2D).
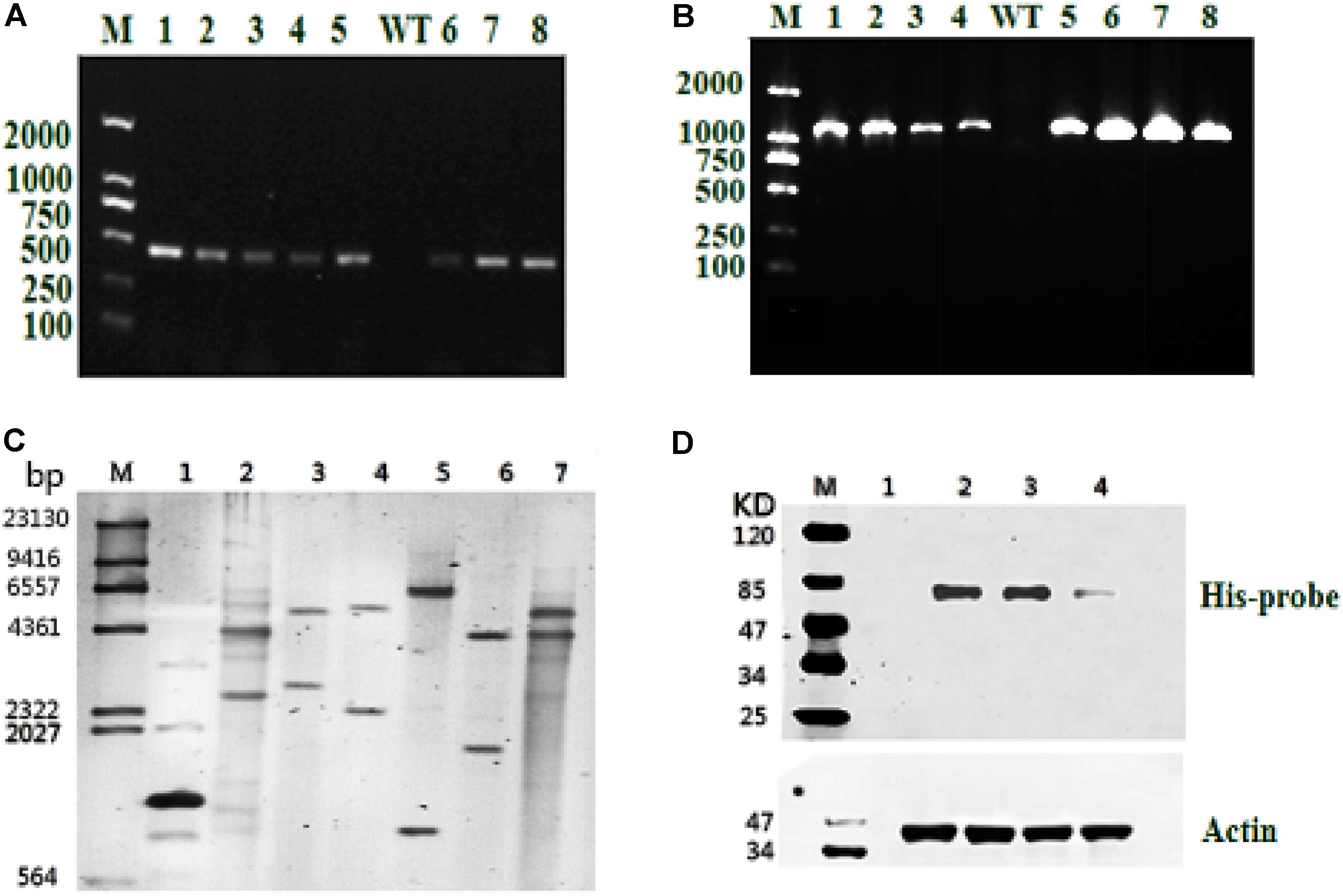
FIGURE 2. Molecular analysis of resistant clones. (A) PCR of the resistant gene Sh-ble in selected clones and the WT strain. M: DNA-size marker. (B) PCR performed using primers designed from tandem ME1 and ble sequences in the recombinant plasmid pPha-T1-ME1. The reference band is 1126 bp. M: DNA-size marker. (C) Southern blot analysis of EcoR I-digested products (lane 1: positive control represents the plasmid, pPha-T1; lanes 2–4: PtME1-1, PtME1-2, and PtME1-3) and Kpn I-digested products (lanes 5–7: PtME1-1, PtME1-2, and PtME1-3) from genomic DNA with a ble fragment used as a digoxigenin-labeled probe. (D) Western blot analysis using an anti-His-tag antibody. Actin was used as an internal control (lanes 2–4: PtME1-1, PtME1-2, and PtME1-3).
The impact of ME1 overexpression on ME1 mRNA levels was investigated by qPCR analysis in three resistant clones and the WT strain. As shown in Figure 3A, all transformants displayed elevated ME1-transcript abundance as compared with the WT strain. Additionally, the relative ME1-transcript abundance of the transformants was 6.09–11.87 time higher than that of the WT strain. Furthermore, the increase in ME1-transcript abundance was accompanied by enhanced ME1 activity (Figure 3B), which increased 1.52–1.81 fold relative to that observed in the WT strain. These results validated these resistant colonies as the expected transformants.
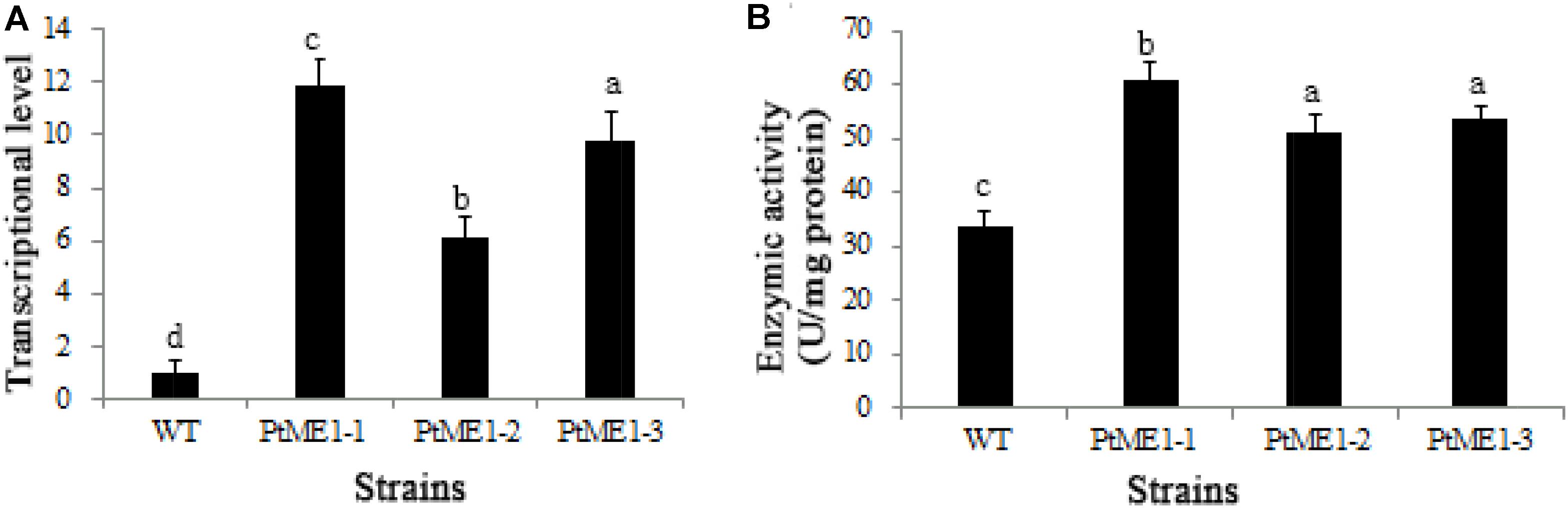
FIGURE 3. Molecular characterization of resistant clones. (A) Relative quantification of ME1 transcripts in the WT strain and the three Phaeodactylum tricornutum resistant clones. ME1-transcript abundance in the resistant clones was quantified with WT cells as the standard and normalized to endogenous histone H4 expression (n = 6). (B) NADP-ME activity (U/106 cells) in the WT strain and the three P. tricornutum transformants. Activity assays were performed on samples from the same experiments. Values represent averages of at least three replicates. Error bars indicate standard deviations. Values with different letters (a,b,c,d) indicate a significant difference between them (p < 0.05).
As shown in Figure 4A, the neutral-lipid content of all three transformants (PtME1-1, PtME1-2, and PtME1-3) increased significantly (P < 0.05; 33.33, 20.25, and 29.63%, respectively) as compared with that in the WT strain. Additionally, total lipid content of the three resistant clones was enhanced significantly, and that of the PtME1-1 transformant increased by 48.42% as compared with that of the WT strain (Figure 4B). These findings suggested that PtME1 overexpression induced the accumulation of neutral- and total lipids in P. tricornutum. Similarly, overexpression of two exogenous ME genes in M. circinelloides led to a 2.5-fold increase in lipid accumulation (Zhang et al., 2007). Moreover, PtME expressed in the green microalga Chlorella pyrenoidosa resulted in a 3.2-fold increase in neutral-lipid content relative to that observed in the WT strain, with total lipid content reaching 40.9% (dry cell weight) (Xue et al., 2016).
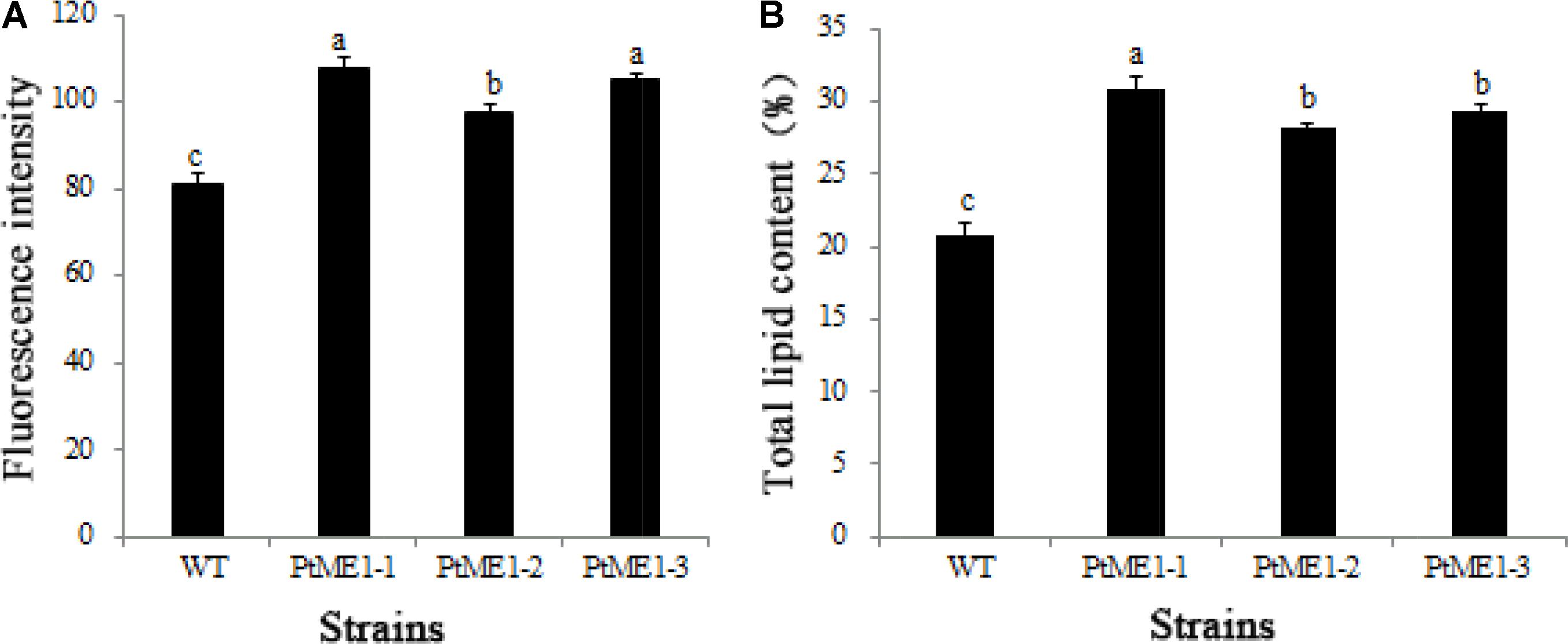
FIGURE 4. Neutral-lipid accumulation and total lipid contents. (A) Neutral-lipid and (B) total lipid contents in the WT strain and the three Phaeodactylum tricornutum transformants. Values represent averages of three replicates (n = 3). Values with different letters (a,b,c) indicate a significant difference between them (p < 0.05).
A previous study reported that overexpressing ME in E. coli led to a 4-fold increase in intracellular lipids by providing a high level of NADPH (Meng et al., 2011). In a previous study with different methods (the recombinant plasmid pHY-PtME instead of pPha-T1-ME1, transformation by electroporation rather than Biolistic PDS-1000/He particle-delivery system) to the present study, PtME overexpressed endogenously in P. tricornutum, resulting in a marked increase (2.5-fold) in total lipid content in transgenic strains, reaching a 57.8% dry cell weight and a similar growth rate to that observed in the WT strain (Xue et al., 2015). However, in that study, the authors did not confirm the NADP+-dependence of the ME-encoding gene, suggesting that it would not necessarily have produced NADPH. Therefore, the mechanisms associated with the increases in lipids remains unclear.
As shown in Table 2, Significant decreases in monounsaturated fatty acids (MUFAs) from 24.61 to 19.96% were detected in transgenic microalgae. With regard to saturated fatty acids (SFAs) and PUFAs, marked increases in the transgenic lines, from 23.19 to 25.32% and from 49.02 to 54.04%, respectively, were observed, revealing that overexpression of endogenous ME1 facilitated SFA and PUFA biosynthesis. In contrast to the present study, other PtMEs different from those in this study were overexpressed endogenously in P. tricornutum, resulting in an increase in MUFAs and a slight decrease in PUFAs in transgenic microalgae (Xue et al., 2015). Wynn and Ratledge (1997) reported that enhanced ME activity leads increases in the cytosolic NADPH pool, and that ME plays a vital role in the provision of NADPH for storage lipids synthesis by Aspergillus nidulans. These findings indicated that the function of ME in lipid biosynthesis involves supplying NADPH for fatty acid desaturation (Liang and Jiang, 2013).
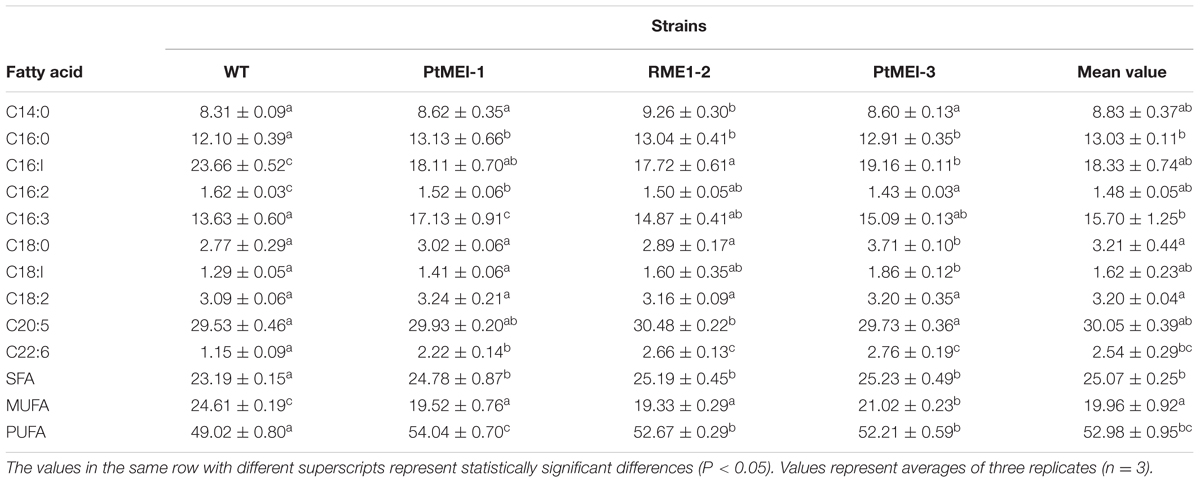
TABLE 2. Fatty acid composition of the WT strain and the three transformants (% of total fatty acids).
It is likely that the elevated ME activity generated NADPH for fatty acid desaturases and also increased fatty acid desaturase activity. (Zhang et al., 2007). The present study showed a 1.52- to 1.81-fold increase in ME1 activity (Figure 3B) as compared with that in the WT strain, which led to an increase in fatty acid desaturase activity and induced further accumulation of PUFAs in transgenic microalgae. A previous study showed that docosahexaenoic acid content increased significantly by adding ME based on the resulting elevation in ME activity and NADPH supply during a specific fermentation stage in Schizochytrium sp. HX-308 (Ren et al., 2009). Exogenously overexpressing ME in the green microalga C. pyrenoidosa results in a 34% increase in PUFAs in the transformed line, with the content of C16:3 and C18:3 lipids in the transgenic lines also elevated by 68.5 and 42.9%, respectively (Xue et al., 2016). Overexpressing ME1 in E. coli showed that C14:0, C16:0, C18:1, and total fatty acid contents were increased by 34.8, 69.9, 54.2, and 50.2%, respectively, and that the content of C16:1 lipids was elevated 5.6-fold as compared with that of controls (Lv et al., 2016). These findings suggested that PtME influenced fatty acid composition by regulating lipogenesis (Xue et al., 2016).
To verify whether ME1 overexpression can promote NADPH production, NADPH content was measured in the three transformants and the WT strain during cultivation on days 3, 6, and 9. As shown in Figure 5, the NADPH content of all algal strains increased markedly on day 6 and decreased significantly on day 9, indicating that NADPH could be used to produce other compounds, such as lipids, and expended during the late exponential-growth phase. However, the NADPH content of three transformants was clearly enhanced relative to that observed in the WT strain during the cultivation stage, increasing by 40.6, 17.8, and 30.0% on day 3, 49.3, 28.4, and 46.7% on day 6, and 80.3, 33.2, and 68.2% on day 9 for PtME1-1, PtME1-2, and PtME1-3, respectively, suggesting that ME1 overexpressing significantly improved NADPH biosynthesis in P. tricornutum. Furthermore, the increase in ME1 activity was accompanied by elevated NADPH content, indicating that increased ME1 activity produced additional NADPH.
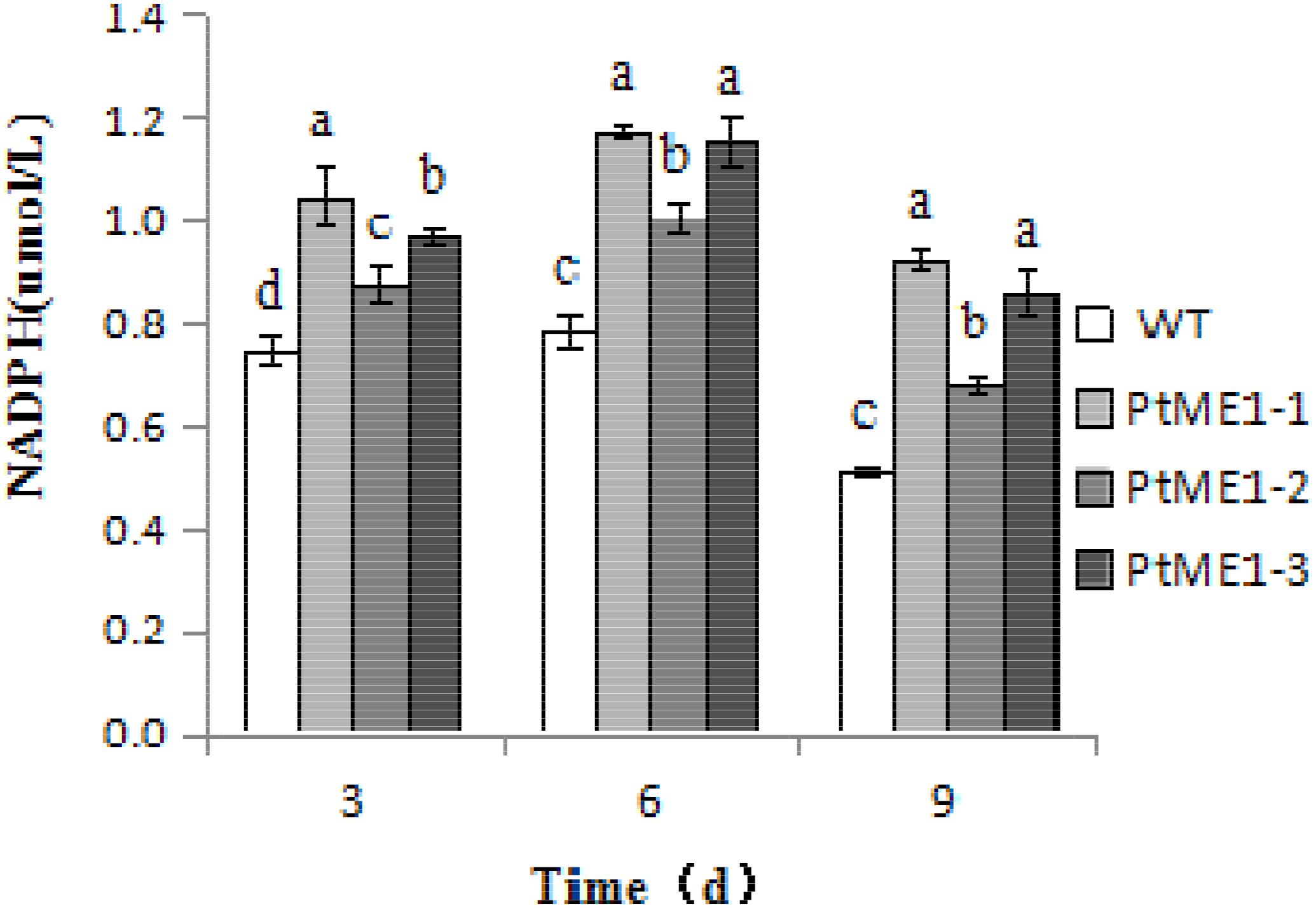
FIGURE 5. NADPH content in the WT strain and the three Phaeodactylum tricornutum transformants. Values represent averages of three replicates (n = 3). Values with different letters (a,b,c,d) indicate a significant difference between them (p < 0.05).
NADPH plays a crucial part in fatty acid accumulation in oleaginous organisms, with these organisms exhibiting a unique mechanism involving NADPH supply. Wynn et al. (1999) reported that ME was important for providing sufficient NADPH for lipid biosynthesis in the fungi, hypothesizing that lipid production was controlled by ME-mediated NADPH generation to fatty acid synthase activity, given that no other enzyme activity has displayed such a strong link with the content of lipid accumulation. A previous report indicated that ME represents the only NADPH source for fatty acid biosynthesis in the yeast Rhodosporidium toruloides (Evans and Ratledge, 1985). Furthermore, ME overexpression in E. coli resulted in increased lipid accumulation in engineered strains relative to controls due to two potentially linked results: increased NADPH and pyruvate levels (Meng et al., 2011).
Until now, no study reported whether ME was capable of providing NADPH in eukaryotic microalgae. Xue et al. (2015) reported that PtME overexpression significantly impacted the promotion of lipid accumulation, attaining a 57.8% increase in dry cell weight, along with a similar growth rate to that of the WT strain in P. tricornutum. That study also showed that PtME was predominantly localized to the mitochondria, although no further studies were performed on the role of ME in accumulating lipid content. The data obtained from this study along with our previously published work (Lv et al., 2016) suggested that ME1 plays an important role in NADPH supply for lipid synthesis and fatty acid desaturation in P. tricornutum (Wynn et al., 1999).
However, the role of ME in the provision of NADPH for lipid biosynthesis remains incompletely understood (Ratledge, 2014; Liang and Jiang, 2015). Ren et al. (2013) reported that the main source of NADPH might be from glucose-6-phosphate dehydrogenase at the early stage of fermentation, whereas ME was the main provider during the late stage in the oleaginous fungi Schizochytrium sp. HX-308.
Malic enzyme can promote NADPH production in oleaginous organisms; however, other some studies indicated that the enzyme cannot provide all of the NADPH required. In animal cells, although ME is important for generating NADPH for fatty acid biosynthesis (Ceddia et al., 2000), it is not the sole supplier, as 50% of the NADPH is from both glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase (Shimomura et al., 1998; Zhang et al., 2007). Pentose phosphate pathway related reactions seem to be the most likely way, in spite of a possibility that a cytosolic isocitrate dehydrogenase (ICDH) reaction with a mitochondrial ICDH function might also produce some NADPH in the reverse reaction (Ratledge, 2014).
NADPH production was not affected in the strain Saccharomyces cerevisiae overexpressing ME gene (lacking the mitochondria-target sequence) and exhibiting ME localization to the mitochondria due to transfer of NADPH across the mitochondrial membrane (Moreira dos Santos et al., 2004). The ME shunt proved to be diversified in this respect because of the possibility of outputting additional NADPH to the cytosol (Moreira dos Santos et al., 2004). Therefore, it is difficult to identify the correct ME gene involved in producing NADPH particularly for fatty acid biosynthesis (Zhang et al., 2007).
Two additional decarboxylating enzymes, which belong to the ME family, have been identified (27477 and 56501 represent NCBI predicted ME protein sequences), with both possessing mitochondria presequences in P. tricornutum (Kroth et al., 2008), suggesting that ME likely localizes to the mitochondria (Valenzuela et al., 2012). One of these enzymes (56501) has a dinucleotide-binding site given for NAD rather than NADP. Therefore, P. tricornutum appears to have two mitochondrial MEs that are either NAD- or NADP-dependent (Kroth et al., 2008). Further research is needed to determine ME1 localization.
These results along with those from other studies (Kroth et al., 2008; Xue et al., 2015) indicated that P. tricornutum possesses two MEs, one being an NAD cofactor (Xue et al., 2015) and the other NADP-dependent (this study).
In this study, endogenous ME1 was succeed in overexpressing in P. tricornutum, resulting in a significant increase in total lipid and PUFA content, and producing additional NADPH, thereby demonstrating that ME1 is NADP-dependent and plays a vital role in the supply of NADPH for lipid biosynthesis and desaturation of fatty acids in P. tricornutum. These findings suggested that overexpression of endogenous ME1 represents a valid method for boosting neutral-lipid yield in diatom.
B-HZ, R-HZ, and N-NL designed the experiments, analyzed and interpreted the data, and wrote the article. Y-SW participated in algal cultivation and analyzed the data. G-PY and K-HP supervised specific experiments and gave critical revisions of the article. All authors agreed on the manuscript.
This work was supported by The National Key Research and Development Program-China (2016YFB0601001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Prof. Matsuda (Kwansei-Gakuin University, Nishinomiya, Japan) for kindly offering the pPha-T1 plasmid and the three reviewers for their valuable suggestions.
Casati, P., Drincovich, M. F., Andreo, C. S., Donahue, R., and Edwards, G. E. (1998). UV-B, red and far-red light regulate induction of the C4 isoform of NADP-malic enzyme in etiolated maize seedlings. Aust. J. Plant Physiol. 25, 701–708. doi: 10.1071/PP98004
Ceddia, R. B., William, W. N., Lima, F. B., Flandin, P., Curi, R., and Giacobino, J. P. (2000). Leptin stimulates uncoupling protein-2 mRNA expression and Krebs cycle activity and inhibits lipid synthesis in isolated rat white adipocytes. Eur. J. Biochem. 267, 5952–5958. doi: 10.1046/j.1432-1327.2000.01664.x
Chang, G. G., and Tong, L. (2003). Structure and function of malic enzymes, a new class of oxidative decarboxylases. Biochemistry 42, 12721–12733. doi: 10.1021/bi035251+
Cooper, M. S., Hardin, W. R., Petersen, T. W., and Cattolico, R. A. (2010). Visualizing “green oil” in live algal cells. J. Biosci. Bioeng. 109, 198–201. doi: 10.1016/j.jbiosc.2009.08.004
Drincovich, M. F., Casati, P., and Andreo, C. S. (2001). NADP-malic enzyme from plants: a ubiquitous enzyme involved in different metabolic pathways. FEBS Lett. 490, 1–6. doi: 10.1016/S0014-5793(00)02331-0
Drincovich, M. F., Casati, P., Andreo, C. S., Donahue, R., and Edwards, G. E. (1998). UV-B induction of NADP-malic enzyme in etiolated and green maize seedlings. Plant Cell Environ. 21, 63–70. doi: 10.1046/j.1365-3040.1998.00240.x
Evans, C. T., and Ratledge, C. (1985). Possible regulatory roles of ATP: citrate lyase, malic enzyme, and AMPdeaminase in lipid accumulation by Rhodosporidium toruloides CBS 14. Can. J. Microbiol. 31, 1000–1005. doi: 10.1139/m85-189
Falciatore, A., Casotti, R., Leblanc, C., Abrescia, C., and Bowler, C. (1999). Transformation of nonselectable reporter genes in marine diatoms. Mar. Biotechnol. 1, 239–251. doi: 10.1007/PL00011773
Granum, E., Raven, J. A., and Leegood, R. C. (2005). How do marine diatoms fix 10 billion tonnes of anorganic carbon per year. Can. J. Bot. 83, 898–908. doi: 10.1139/b05-077
Griffiths, M. J., Garcin, C., VanHille, R. P., and Harrison, S. T. (2011). Interference by pigment in the estimation of microalgal biomass concentration by optical density. J. Microbiol. Met. 85, 119–123. doi: 10.1016/j.mimet.2011.02.005
Guillard, R. R. L. (1975). “Culture of phytoplankton for feeding marine invertebrates,” in Culture of Marine Invertebrate Animals, eds W. L. Smith and M. H. Chanley (New York, NY: Plenum Press), 29–60.
Jiang, P., Du, W. J., Mancuso, A., Wellen, K. E., and Yang, X. L. (2013). Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 493, 689–693. doi: 10.1038/nature11776
Kendrick, A., and Ratledge, C. (1992). Desaturation of polyunsaturated fatty acids in Mucor circinelloides and the involvement of a novel membrane-bound malic enzyme. Eur. J. Biochem. 209, 667–673. doi: 10.1111/j.1432-1033.1992.tb17334.x
Kim, G. H., Lee, Y. E., Lee, G. H., Cho, Y. H., Lee, Y. N., Jang, Y., et al. (2015). Overexpression of malic enzyme in the larval stage extends Drosophila lifespan. Biochem. Biophys. Res. Commun. 456, 676–682. doi: 10.1016/j.bbrc.2014.12.020
Kroth, P. G., Chiovitti, A., Gruber, A., Martin-Jezequel, V., Mock, T., Parker, M. S., et al. (2008). A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS One 3:1426. doi: 10.1371/journal.pone.0001426
Lepage, G., and Roy, C. C. (1984). Improved recovery of fatty acid through direct transesterification without prior extraction or purification. J. Lipid Res. 25, 1391–1396.
Li, Y., Han, D., Hu, G., Dauvillee, D., Sommerfeld, M., and Ball, S. (2010). Chlamydomonas starchless mutant defective in ADP-glucose pyrophosphorylase hyper-accumulates triacylglycerol. Metab. Eng. 12, 387–391. doi: 10.1016/j.ymben.2010.02.002
Liang, M. H., and Jiang, J. G. (2013). Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology. Prog. Lipid Res. 52, 395–408. doi: 10.1016/j.plipres.2013.05.002
Liang, Y. J., and Jiang, J. G. (2015). Characterization of malic enzyme and the regulation of its activity and metabolic engineering on lipid production. RSC Adv. 5, 45558–45570. doi: 10.1039/C5RA04635A
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lv, N. N., Zhu, B. H., Lu, L., Yang, G. P., and Pan, K. H. (2016). Overexpression of malic enzyme gene from Phaeodactylum tricornutum promotes fatty acids production in Escherichia coli. Period. Ocean Univ. China 46, 65–69.
Meng, X., Yang, J., Cao, Y., Li, L., Jiang, X., and Xu, X. (2011). Increasing fatty acid production in E. coli by simulating the lipid accumulation of oleaginous microorganisms. J. Ind. Microbiol. Biotechnol. 38, 919–925. doi: 10.1007/s10295-010-0861-z
Moreira dos Santos, M., Raghevendran, V., Kötter, P., Olssona, L., and Nielsen, J. (2004). Manipulation of malic enzyme in Saccharomyces cerevisiae for increasing NADPH production capacity aerobically in different cellular compartments. Metab. Eng. 6, 352–363. doi: 10.1016/j.ymben.2004.06.002
Poulsen, N., and Kröger, N. (2005). A new molecular tool for transgenic diatoms: control of mRNA and protein biosynthesis by an inducible promoter–terminator cassette. FEBS J. 272, 3413–3423. doi: 10.1111/j.1742-4658.2005.04760.x
Radakovits, R., Eduafo, P. M., and Posewitz, M. C. (2011). Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab. Eng. 13, 89–95. doi: 10.1016/j.ymben.2010.10.003
Ratledge, C. (2014). The role of malic enzyme as the provider of NADPH in oleaginous microorganisms: a reappraisal and unsolved problems. Biotechnol. Lett. 36, 1557–1568. doi: 10.1007/s10529-014-1532-3
Ren, L. J., Feng, Y., Li, J., Qu, L., and Huang, H. (2013). Impact of phosphate concentration on docosahexaenoic acid production and related enzyme activities in fermentation of Schizochytrium sp. Bioprocess Biosyst. Eng. 36, 1177–1183. doi: 10.1007/s00449-012-0844-8
Ren, L. J., Huang, H., Xiao, A. H., Lian, M., Jin, L. J., and Ji, X. J. (2009). Enhanced docosahexaenoic acid production by reinforcing acetyl-CoA and NADPH supply in Schizochytrium sp. HX-308. Bioprocess Biosyst. Eng. 32, 837–843. doi: 10.1007/s00449-009-0310-4
Rodriguez, E., Navone, L., Casati, P., and Gramajo, H. (2012). Impact of malic enzymes on antibiotic and triacylglycerol production in Streptomyces coelicolor. Appl. Environ. Microbiol. 78, 4571–4579. doi: 10.1128/AEM.00838-12
Shang, C. H., Zhu, S. N., Yuan, Z. H., and Wang, Z. M. (2012). Molecular cloning and characterization analysis of malic enzyme gene from Dunaliella parva. Adv. Mater. Res. 347, 2536–2540.
Shimomura, I., Shimano, H., Korn, B. S., Bashmakov, Y., and Horton, J. D. (1998). Nuclear sterol regulatory element-binding proteins activate genes responsible for the entire program of unsaturated fatty acid biosynthesis in transgenic mouse liver. J. Biol. Chem. 273, 35299–35306. doi: 10.1074/jbc.273.52.35299
Siaut, M., Heijde, M., Mangogna, M., Montsant, A., Coesel, S., and Allen, A. (2007). Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406, 23–35. doi: 10.1016/j.gene.2007.05.022
Takeuchi, Y., Akagi, H., Kamasawa, N., Osumi, M., and Honda, H. (2000). Aberrant chloroplasts in transgenic rice plants expressing a high level of maize NADP-dependent malic enzyme. Planta 211, 265–274. doi: 10.1007/s004250000282
Tang, W., Zhang, S. F., Tan, H. D., and Zhao, Z. K. (2010). Molecular cloning and characterization of a malic enzyme genefrom the oleaginous yeast Lipomyces starkeyi. Mol. Biotechnol. 45, 121–128. doi: 10.1007/s12033-010-9255-8
Valenzuela, J., Mazurie, A., Carlson, R. P., Gerlach, R., Cooksey, K. E., Peyton, B. M., et al. (2012). Potential role of multiple carbon fixation pathways during lipid accumulation in Phaeodactylum tricornutum. Biotechnol. Biofuels 5:40. doi: 10.1186/1754-6834-5-40
Watanabe, K. I., Ehara, M., Inagaki, Y., and Ohama, T. (1998). Distinctive origins of group I introns found in the COXI genes of three green algae. Gene 213, 1–7. doi: 10.1016/S0378-1119(98)00235-2
Wynn, J. P., Bin Abdul Hamid, A., and Ratledge, C. (1999). The role of malic enzyme in the regulation of lipid accumulation infilamentous fungi. Microbiology 145, 1911–1917. doi: 10.1099/13500872-145-8-1911
Wynn, J. P., and Ratledge, C. (1997). Malic enzyme is a major source of NADPH for lipid accumulation by Aspergillus nidulans. Microbiology 143, 253–257. doi: 10.1099/00221287-143-1-253
Xue, J., Niu, Y. F., Huang, T., Yang, W. D., Liu, J. S., and Li, H. Y. (2015). Genetic improvement of the microalga Phaeodactylum tricornutum for boosting neutral lipid accumulation. Metab. Eng. 27, 1–9. doi: 10.1016/j.ymben.2014.10.002
Xue, J., Wang, L., Zhang, L., Balamurugan, S., Li, D. W., Zeng, H., et al. (2016). The pivotal role of malicenzyme in enhancing oil accumulation in green microalga Chlorella pyrenoidosa. Microb. Cell Fact 15, 120–129. doi: 10.1186/s12934-016-0519-2
Zaslavskaia, L. A., Lippmeier, J. C., Kroth, P. G., Grossman, A. R., and Apt, K. E. (2000). Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J. Phycol. 36, 379–386. doi: 10.1046/j.1529-8817.2000.99164.x
Zelle, R. M., Harrison, J. C., Pronk, J. T., and van Maris, A. J. (2011). Anaplerotic role for cytosolic malic enzyme in engineered Saccharomyces cerevisiae strains. Appl. Environ. Microbiol. 7, 732–738. doi: 10.1128/AEM.02132-10
Zhang, Y., Adams, I. P., and Ratledge, C. (2007). Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 153, 2013–2025. doi: 10.1099/mic.0.2006/002683-0
Zhang, Y., Smallbone, L. A., diCenzo, G. C., Morton, R., and Finan, T. M. (2016). Loss of malic enzymes leads to metabolic imbalance and altered levels of trehalose and putrescine in the bacterium Sinorhizobium meliloti. BMC Microbiol. 16:163. doi: 10.1186/s12866-016-0780-x
Zhu, B. H., Shi, H. P., Yang, G. P., Lv, N. N., Yang, M., and Pan, K. H. (2016). Silencing UDP-glucose pyrophosphorylase gene in Phaeodactylum tricornutum affects carbon allocation. New Biotechnol. 33, 237–244. doi: 10.1016/j.nbt.2015.06.003
Keywords: malic enzyme, lipid, fatty acid, NADPH, Phaeodactylum tricornutum
Citation: Zhu B-H, Zhang R-H, Lv N-N, Yang G-P, Wang Y-S and Pan K-H (2018) The Role of Malic Enzyme on Promoting Total Lipid and Fatty Acid Production in Phaeodactylum tricornutum. Front. Plant Sci. 9:826. doi: 10.3389/fpls.2018.00826
Received: 10 February 2018; Accepted: 28 May 2018;
Published: 19 June 2018.
Edited by:
Jianhua Fan, East China University of Science and Technology, ChinaReviewed by:
Jianhui Zhang, North Carolina Central University, United StatesCopyright © 2018 Zhu, Zhang, Lv, Yang, Wang and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bao-Hua Zhu, emh1YmFvaHVhQG91Yy5lZHUuY24= Ke-Hou Pan, cWRraHBhbkAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.