- 1The State Key Laboratory of Subtropical Silviculture, Zhejiang A & F University, Hangzhou, China
- 2School of Geography and Remote Sensing, Nanjing University of Information Science and Technology, Nanjing, China
- 3Institute of Biotechnology, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
- 4Zhejiang Provincial Collaborative Innovation Center for Bamboo Resources and High-efficiency Utilization, Zhejiang A & F University, Hangzhou, China
14-3-3 Proteins are a family of highly conserved regulatory molecules expressed in all eukaryotic cells and regulate a diverse set of biological responses in plants. However, their functions in flowering of Phyllostachys violascens are poorly understood. In this study, four non-𝜀 Pv14-3-3 genes from P. violascens were identified and named PvGF14b, PvGF14c, PvGF14e, and PvGF14f. qRT-PCR analyses revealed that PvGF14b and PvGF14e exhibited widely expressed in all tested bamboo tissues. PvGF14b was highest expression in root and lowest in immature leaf. Whereas PvGF14c and PvGF14f showed tissue-specific expression. PvGF14c was mainly expressed in immature and mature leaves. PvGF14f was highest expression in mature leaves. These four genes were not significantly differentially expressed in mature leaf before bamboo flowering and during flower development. PvGF14b and PvGF14c were not induced by circadian rhythm. PvGF14c displayed subcellular localization in the cytoplasm and PvFT in nucleus and cytoplasm. Yeast two-hybrid screening and bimolecular fluorescence complementation confirmed the interaction between PvGF14c and PvFT. The overexpression of PvGF14b, PvGF14c, and PvGF14e significantly delayed flowering time in transgenic Arabidopsis under long-day condition. These findings suggested that at least three PvGF14 genes are involved in flowering and may act as a negative regulator of flowering by interacting with PvFT in bamboo.
Introduction
In higher plants, flowering transition time is a major factor affecting the adaptability of plants to their environment. Flowering time in higher plants are controlled by complex signaling pathways, such as photoperiod, autonomous flowering, vernalization, and gibberellin pathways, which are independent yet interconnected with one another, eventually forming a complex network (Johanson et al., 2000; Kim et al., 2009). SUPPRESSOR OF OVER-EXPRESSION OF CONSTANS1 (SOC1), FLOWERING LOCUS T (FT), and LEAFY (LFY) are considered integrating factors in these pathways (Moon et al., 2005) and are located downstream of FLOWERING LOCUS C (FLC) and CONSTANS (CO) genes, which regulate plant flowering time by integrating different flowering signals. The FT protein, which is synthesized in leaf blade and migrated along phloem tissue, plays a critical role in the shoot apical meristem (SAM). FT binds to 14-3-3 protein, and then interacts with transcription factor, FLOWERING LOCUS D (FD), thereby promoting the flowering of plants (Taoka et al., 2011). Whereas, overexpression of OsGF14c, an isoform of the rice 14-3-3 gene family, delayed strongly flowering time (Purwestri et al., 2009).
In eukaryotes, the 14-3-3 protein family comprises highly conserved proteins that exist as multiple isoforms in many species. Seven 14-3-3 genes are present in mammals (Arner and Holmgren, 2006), eight in rice (Chen et al., 2006), and 13 in Arabidopsis (Yao et al., 2007). According to homologous sequence evolution analysis, 14-3-3 proteins in plants can be divided into 𝜀 and non-𝜀 groups: the latter is plant specific (Chung et al., 1999; DeLille et al., 2001; Ferl et al., 2002). The members of the 14-3-3 family exhibit different expression patterns and appear to have varied functions. For example, BdGF14d, a non-𝜀 14-3-3 gene in Brachypodium distachyon, can enhance salt stress tolerance (He et al., 2017). OsGF14b, a non-𝜀 14-3-3 protein in rice, positively regulates panicle blast resistance while negatively regulating leaf blast resistance (Liu Q. et al., 2016). Whereas, overexpression of OsGF14c negatively regulate flowering time in rice (Purwestri et al., 2009).
Previous studies showed that 14-3-3 proteins are involved in the regulation of flowering in many plants (Mayfield et al., 2007; Paul et al., 2008; Purwestri et al., 2009). They interact with FT in Arabidopsis and SP (SELF-PRUNING, ortholog of FT) in tomato (Pnueli et al., 2001; Zhang, 2012). In rice, 14-3-3 proteins act as intracellular receptors for florigen Hd3a (Heading date 3a), an ortholog of Arabidopsis FT (Taoka et al., 2011). Hd3a is localized in the cytoplasm and nucleus, whereas OsGF14c is mainly found in the cytoplasm. Hd3a–GF14c interaction is mainly detected in the cytoplasm, indicating that Hd3a cytoplasmic retention is increased by interacting with GF14c (Taoka et al., 2011). Thus, overexpression of OsGF14c plants display a delayed flowering phenotype, suggesting that OsGF14c acts as a negative regulator of flowering by interacting with Hd3a (Purwestri et al., 2009).
Similar to rice, bamboo plants belong to Gramineae but exhibit different flowering characteristics. The flowering cycle of bamboo generally lasts 3–120, and most kinds of bamboo undergoing flowering die (de Carvalho et al., 2013). However, the cause of bamboo flowering remains unclear. Phyllostachys violascens sprouts in early stages, produces high yields, provides good taste, offers remarkable economic benefits. As such, this species is well known for shoot production in China. P. violascens belong to sporadically flowering bamboos, which exhibit either a flowering with dying pattern or a flowering without dying pattern. The flowering of bamboo leading to death reduces the production of bamboo shoots and causes economic loss. Thus, many research groups tried to uncover the flowering mechanism of bamboo plants by genomes and transcriptomes (Zhang et al., 2012; Peng et al., 2013; Gao et al., 2014; Shih et al., 2014; Zhao et al., 2014). In addition, numerous genes and miRNAs related to bamboo flowering were reported (Lin et al., 2010; Gao et al., 2015; Zhao et al., 2015; Guo et al., 2016a; Liu S. et al., 2016; Zheng et al., 2017). Our group reported previously that PvFT played an important role in the induction of bamboo flowering (Guo et al., 2016b). However, no proteins interacting with PvFT have been identified in bamboo. In the present study, four non-𝜀 14-3-3 genes and their expression profiles in P. violascens were identified. The interaction of PvGF14c and PvFT in P. violascens were determined. Meanwhile, overexpression of PvGF14b, PvGF14c, and PvGF14e was shown to delay the flowering time of transgenic Arabidopsis under long-day (LD) condition.
Materials and Methods
Plant Materials
Phyllostachys violascens samples were harvested from the Bamboo Garden of the Zhejiang A & F University. To detect the expression of Pv14-3-3 during the flowering period, we collected leaf samples every 10 days from April 2 to May 31. According to our observation, the plants maintained flowering from mid-March to mid-May. The leaves from the flowering P. violascens were collected every 3 h for nine times to analyze the diurnal expression of the target genes. To determine the tissue-specific expression, we sampled mature leaves, immature leaves, stems, roots, and spikelets. All samples were stored at -80°C until further experiment. Wild-type (ecotype Columbia) and transgenic plants of Arabidopsis were cultivated in a controlled temperature room under 22°C with 16-h light/8-h dark cycle.
Isolation of Four Pv14-3-3 Genes
Total RNA was extracted from the leaves by using the Trizol method. First-strand cDNA was synthesized using a Super ScriptTM kit (Invitrogen, United States) according to the manufacturer’s instructions. Genomic DNA was isolated by the modified CTAB method from leaves (Guo et al., 2016b). According to 14-3-3 homologous genes of rice, BLAST was performed against Phyllostachys heterocycla cDNA database (Peng et al., 2013), and eight Ph14-3-3 genes were identified. Of these genes, four candidate genes that shared more than 85% amino acid sequence identity with OsGF14c. Four primers used to amplify Pv14-3-3s were designed on the basis of four candidate genes of P. heterocycla (Table 1). The full-length cDNA and DNA sequences of four non-𝜀 Pv14-3-3s of P. violascens were amplified through PCR with the designed primers (Table 1). The thermocycling conditions were as follows: initialization at 94°C for 3 min; 32 cycles of denaturation at 94°C for 30 s, annealing at 50–60°C, and extension at 72°C for 50 s; and final extension at 72°C for 10 min. The amplified fragments were separated through electrophoresis on 1.5% (m/v) agarose gel and stained with ethidium bromide.
Bioinformatics Analysis
The open reading frames of four Pv14-3-3s were confirmed using the ORF Finder1. The exon–intron organization of four Pv14-3-3s was revealed using a gene structure display server2. The protein domains of four Pv14-3-3s genes were predicted using InterPro3. The Molecular weights and isoelectric points of 14-3-3 proteins were predicted using ExPASy program4. The phylogenetic tree of 14-3-3 proteins from P. violascens, P. heterocycla, Arabidopsis, and Oryza sativa was constructed using the neighbor-joining method in MEGA 5.05 with bootstrap values obtained from 1000 replications.
Expression Analysis of Four Pv14-3-3s Genes
Total RNAs were extracted from the collected samples by using the Trizol method. First-strand cDNA was synthesized using a Super ScriptTM kit (Invitrogen, United States). The qRT-PCR primers were designed according to the non-conserved region of full-length cDNA sequences of four Pv14-3-3s. The NTB gene was used as the internal control gene (Table 1; Fan et al., 2013). The qRT-PCR analysis was performed using CFX96TM Real-Time PCR Detection System (Bio-Rad). Reactions were performed in 20-μl mixtures consisting of 10 μl 2 × SYBR Premix Ex Taq II Mix (Takara), 0.5 μl each of forward or reverse primer, 0.5 μl cDNA template (100 ng/μl), and 8 μl double distilled H2O. The program was 95°C for 30 s, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s. The data were analyzed by the 2-ΔΔCt method. Three independent replicates were performed for each experiment.
Subcellular Localization
The full-length sequence of PvGF14c or PvFT were inserted at the C- and N-terminus of CaMV35S-GFP vector, forming PvGF14c-GFP or PvFT-GFP fusion protein, transient expression. The transient expression of the PvGF14-GFP or PvFT-GFP fusions in onion epidermal cells was made by the particle bombardment method (Wang et al., 1988). A confocal laser scanning microscope was used to observe the onion epidermal cells.
Plasmid Construction and Transformation
The full-length ORF of Pv14-3-3s was attached to CaMV35S-GFP to form the plasmid 35S::Pv14-3-3s. Afterward, the floral dipping method mediated by Agrobacterium tumefaciens strain GV3101 was performed to convert the plasmid into wild-type Arabidopsis plants. The transgenic Arabidopsis seeds were chosen using 50 mg/ml hygromycin in 0.5× solid medium. Positive transgenic plants were further identified through PCR. Flowering time was measured using lines with typical phenotypes. The expression level of four Pv14-3-3s genes in transgenic and WT Arabidopsis were analyzed by qRT-PCR using gene-specific primers (Table 1).
Yeast Two-Hybrid (Y2H) Assay
The yeast two-hybrid (Y2H) assay was conducted following a previously reported method (Sun et al., 2015). The ORF of PvFT and PvGF14c were respectively cloned into the pGBKT7 BD vector and pGADT7 AD eukaryotic expression vectors for the swapping experiment. The Y2H was performed according to the BD Matchmaker Library Construction & Screening Kits instructions (Clontech, Palo Alto, CA, United States).
Bimolecular Fluorescence Complementation
The ORF sequence of PvGF14c was inserted into pSAT4-cEYFP-C1-B (nYFP) vector. The ORF sequence of PvFT or AtFT was inserted into pSAT1 nEYFP - C1 (cYFP) vector, respectively. Subsequently, these vectors were introduced into DH5α. And then, the purified plasmids were introduced into the onion epidermal cells following the protocol in Section “Subcellular Localization.”
Statistical Analysis
Statistical analysis was performed using SPSS 19.0 (SPSS Inc., Chicago, IL, United States). Differences were analyzed with one-way ANOVA followed by Tukey’s range test. Significance was accepted at the level of P < 0.01.
Results
Cloning of Four Pv14-3-3 Genes in P. violascens
We performed a BLAST 14-3-3 homologous genes of rice against P. heterocycla cDNA database and identified eight Ph14-3-3 genes named PhGF14a–h according to the nomenclature of rice. PhGF14a–h showed 77, 88, 96, 83, 91, 86, 58, and 44% homolog to OsGF14c, respectively (Table 2). Phylogenetic analysis showed that P. heterocycla contained two 𝜀 and six non-𝜀 14-3-3 isoforms (Figure 1A). Four candidate genes shared more than 85% of amino acid sequence identity with OsGF14c. Four primers for cloning Pv14-3-3s were designed according to four candidate genes of P. heterocycla, and four Pv14-3-3s genes from P. violascens were cloned and named PvGF14b, PvGF14c, PvGF14e, and PvGF14f, respectively. All of information about four non-𝜀 Pv14-3-3 genes is shown in Table 3. The four Pv14-3-3 proteins contain 256–262 amino acid residues, and their molecular weight range was from 28.9 to 29.8 kDa. Their isoelectric points were from 4.70 to 4.86. The exon–intron structural analysis reveals that four non-𝜀 Pv14-3-3 genes contain five exons and four introns (Figure 1B). To understand the genetic relationships of Pv14-3-3s in plants, we performed a phylogenetic analysis of Arabidopsis, O. sativa, P. violascens, and P. heterocycla (Table 4). The results indicated that Pv14-3-3s were closely related to Ph14-3-3s, as revealed by their occurrence in the same branch (Figure 1A), and four Pv14-3-3s genes belonged to the non-𝜀 group. Furthermore, we performed a multiple sequence alignment of the amino acid sequence of four Pv14-3-3s genes and demonstrated that four 14-3-3 proteins of P. violascens were highly conserved. Some sequences were completely conserved among four proteins. Eight amino acid residues in the N-terminal and 18 amino acid residues in the C-terminal exhibited large differences, suggesting that the various termini could provide different functions in each protein by interacting with a series of target proteins. Four non-𝜀 Pv14-3-3 proteins in P. violascens consisted of the following nine antiparallel α-helices: αC and αD have the longest sequence of amino acids with 30–34 amino acid residues; αA–D form dimers at the interface; αC, αE, αG, and αL constitute conservative peptides with a structure similar to 14-3-3s in other plants (Figure 1C).
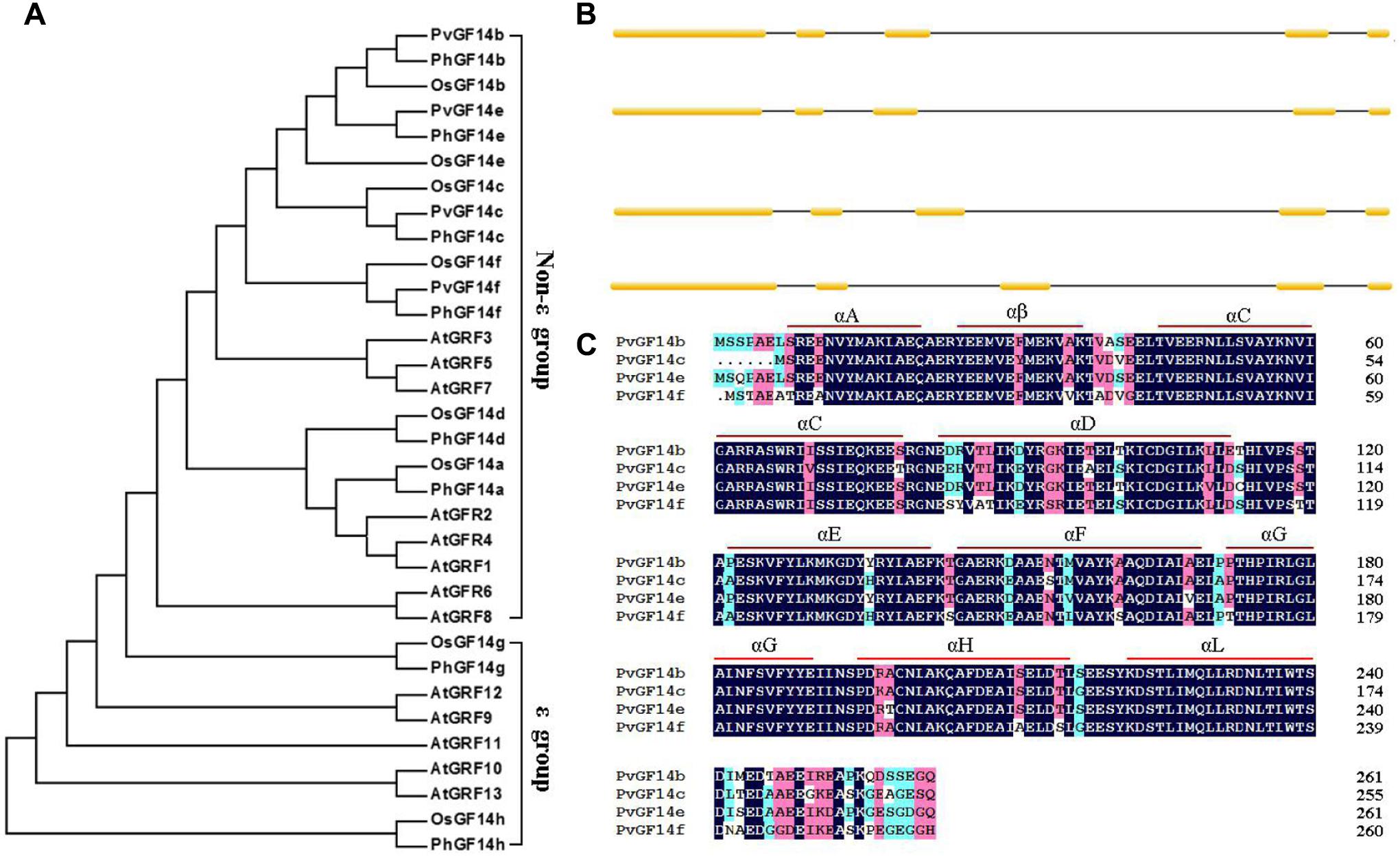
FIGURE 1. Phylogenetic and gene structure analyses of Pv14-3-3s. (A) Phylogenetic tree of 14-3-3s from P. violascens, P. heterocycla, O. sativa, and Arabidopsis plants constructed using MAGA5.0. The registration number 14-3-3 family genes were shown in Table 4. (B) Gene structures of Pv14-3-3s. [scale=0.30]img001 represent the exons; [scale=0.80]img002 represent the introns. (C) Amino acid sequence alignments of four Pv14-3-3s.
Analysis on the Expression Feature of Four non-𝜀 Pv14-3-3s of P. violascens
To examine the biological functions of four non-𝜀 Pv14-3-3s genes, qRT-PCR was performed to analyze the temporal expression and tissue-specific expression of 14-3-3 genes of P. violascens. The results showed that PvGF14b and PvGF14e were constitutive expression, whereas PvGF14c and PvGF14f exhibited tissue specificity (Figures 2A–D). The highest expression level of PvGF14f was observed in the stem. Its expression was undetectable in the immature leaves and in the roots (Figure 2D). The highest expression level of PvGF14c was found in the leaves. Conversely, a low expression level was recorded in the root, stem, and spikelet (Figure 2B). The samples of mature leaves collected were used to detect the expression levels of four Pv14-3-3s genes before bamboo flowering and during flowering development. The expression levels of each Pv14-3-3s genes did not significantly different between these two development stages (Figures 2E–H). Before bamboo flowering, the relative expression level of three Pv14-3-3s genes, PvGF14b, PvGF14c, and PvGF14f were similar and higher than it of PvGF14e. Furthermore, the data showed that PvGF14b and PvGF14c were expressed in 24 h with highest at 11 am, but without obvious difference between day and night, suggesting that PvGF14b and PvGF14c were not shown to have diurnal rhythm expression (Figures 2I,J).
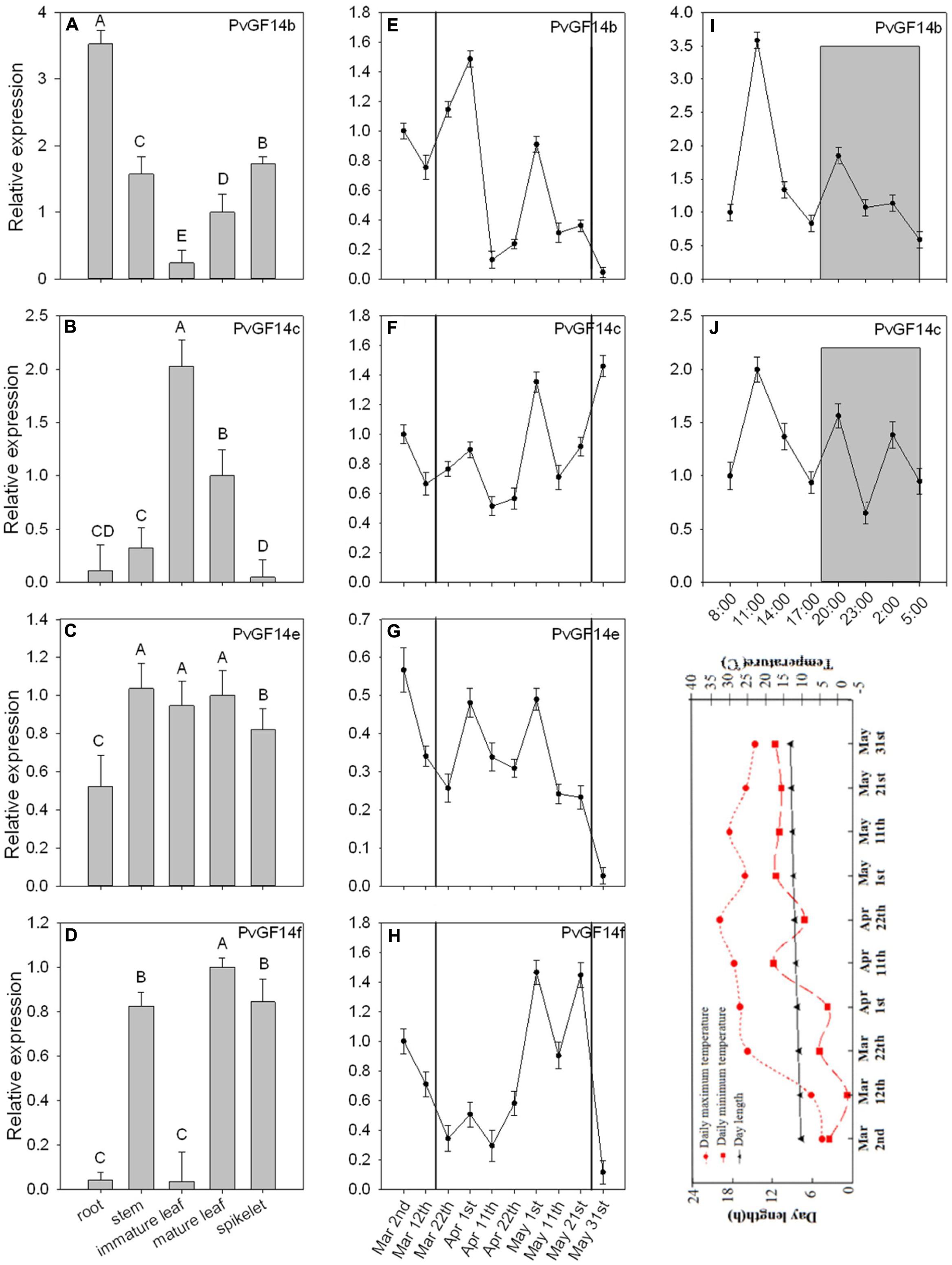
FIGURE 2. Expression analysis of four Pv14-3-3 genes in P. violascens. (A–D) Relative expression of four Pv14-3-3 genes in different tissues. (E–H) Temporal expression of four Pv14-3-3 genes in leaves during different development stages. The period between black solid lines represents flowering time. (I,J) Daily expression of PvGF14b and PvGF14c. Dusk and dawn are at 17:00 and 5:30. Gray areas indicate darkness. Gray shading denotes the dark/night portion of 24 h cycle. Expression of the PvNTB gene was used as an internal control. Error bar indicates standard deviation. A, B indicate highly significant differences (P < 0.01). The image appeared below the part “J” is rends in daylength and temperature during the period of collecting samples.
The Influence of Ectopic Expression of Four Pv14-3-3 Genes on Flowering Time in Transgenic Arabidopsis
To demonstrate whether four 14-3-3 genes can influence flowering in plants, we constructed the overexpression vectors 35S::PvGF14b, 35S::PvGF14c, 35S::PvGF14e, and 35S::PvGF14f which were transformed to Arabidopsis, respectively, and got 31 T1 GF14b-ox plants, 42 T1 GF14c-ox plants, 17 T1 GF14e-ox plants, and 15 T1 GF14f-ox plants. At last, four independent lines of GF14b-ox, GF14e-ox, and GF14f-ox in the T2 generation, as well as four independent lines of GF14c-ox in the T3 generation were grown under greenhouse condition for further analysis. 35S::PvGF14b, 35S::PvGF14c and 35S::PvGF14e delayed flowering in transgenic Arabidopsis (Figures 3–5). Whereas, PvGF14f-overexpressing plants showed the similar flowering time as that of the wild-type (Figure 6). The number of rosette leaves of PvGF14b, PvGF14c, and PvGF14e transgenic plants was 2–5, 5–7, and 3–6 more than that of the wild-type, respectively (Figures 3–5). The bolting time in 35S::PvGF14b, 35S::PvGF14c and 35S::PvGF14e transgenic Arabidopsis plants was 3–4, 6–8, and 4–6 days later than that in the wild-type (Figures 3–5).
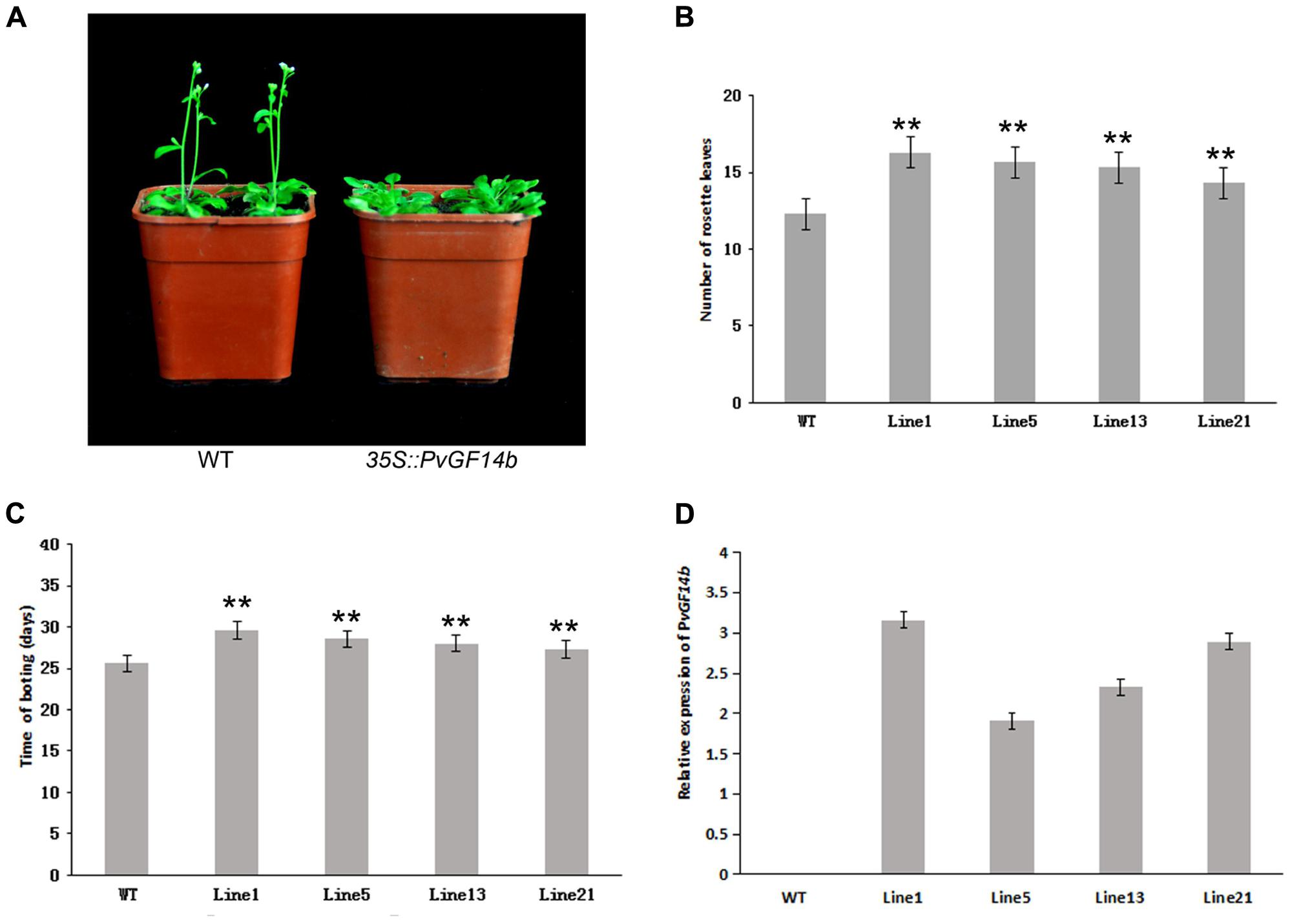
FIGURE 3. Phenotype analysis of 35S::PvGF14b Arabidopsis plants under LD conditions. (A) Phenotype differences in flowering between 35S::PvGF14b transgenic plants and wild-type plants. Photographs were taken at 26-day-old wild-type plants. (B) The number of rosette leaves in T2 35S::PvGF14b in transgenic plants (n = 20) and wild-type plants. (C) Bolting time in T2 35S::PvGF14b transgenic plants (n = 20) and wild-type plants. (D) qRT-PCR expression analysis of PvGF14b. Asterisks “∗∗” indicate significant differences in comparison with the WT at P < 0.01.
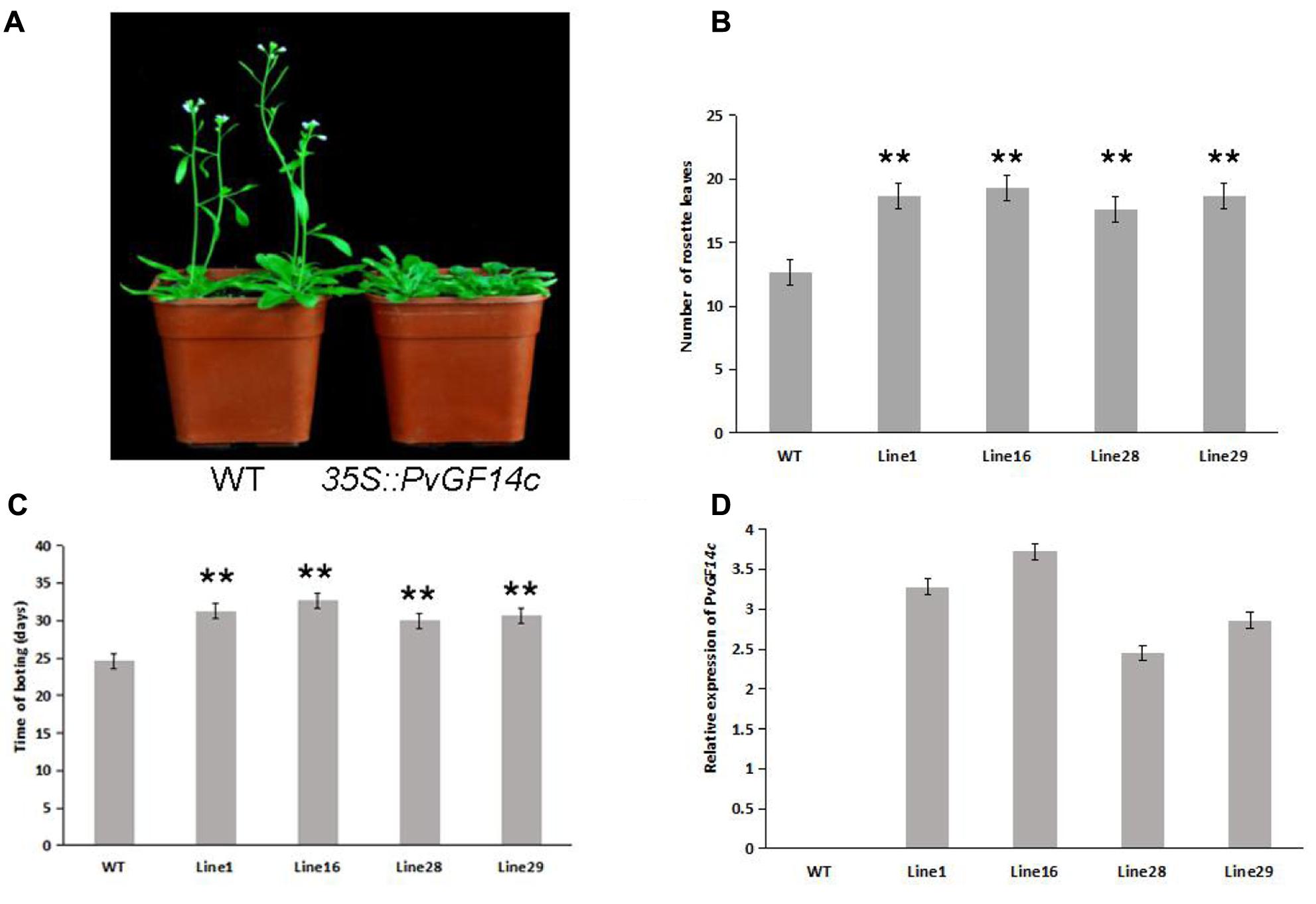
FIGURE 4. Phenotype analysis of 35S::PvGF14c Arabidopsis plants under LD conditions. (A) Phenotype differences in flowering between 35S::PvGF14c transgenic plants and wild-type plants. Photographs were taken at 28-day-old wild-type plants. (B) The number of rosette leaves in T3 35S::PvGF14c in transgenic plants (n = 24) and wild-type plants. (C) Bolting time in T3 35S::PvGF14c transgenic plants (n = 24) and wild-type plants. (D) qRT-PCR expression analysis of PvGF14c. Asterisks “∗∗” indicate significant differences in comparison with the WT at P < 0.01.
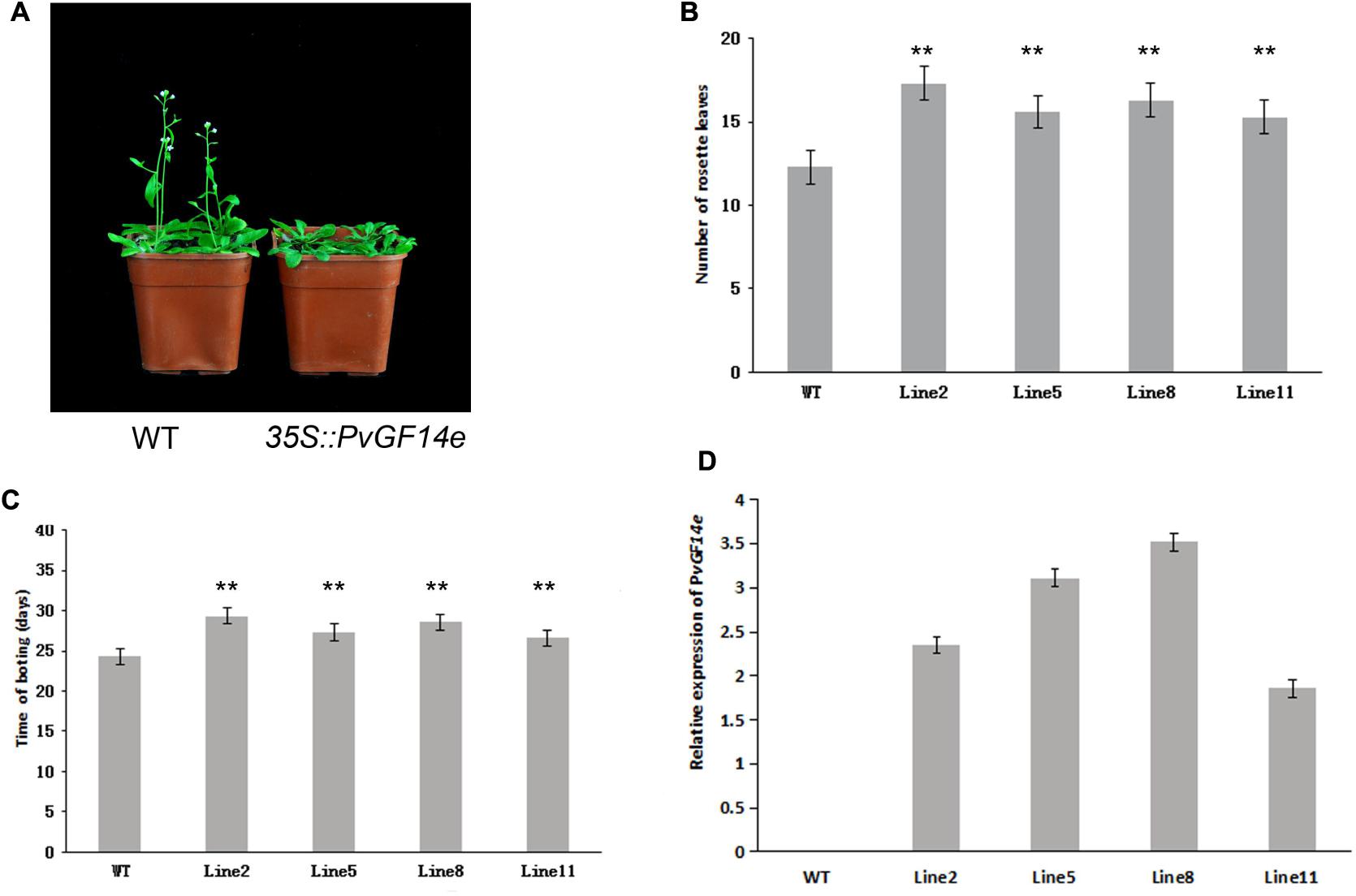
FIGURE 5. Phenotype analysis of 35S::PvGF14e Arabidopsis plants under LD conditions. (A) Phenotype differences in flowering between 35S::PvGF14e transgenic plants and wild-type plants. Photographs were taken at 25-day-old wild-type plants. (B) The number of rosette leaves in T2 35S::PvGF14e in transgenic plants (n = 20) and wild-type plants. (C) Bolting time in T2 35S::PvGF14e transgenic plants (n = 20) and wild-type plants. (D) qRT-PCR expression analysis of PvGF14e. Asterisks “∗∗” indicate significant differences in comparison with the WT at P < 0.01.
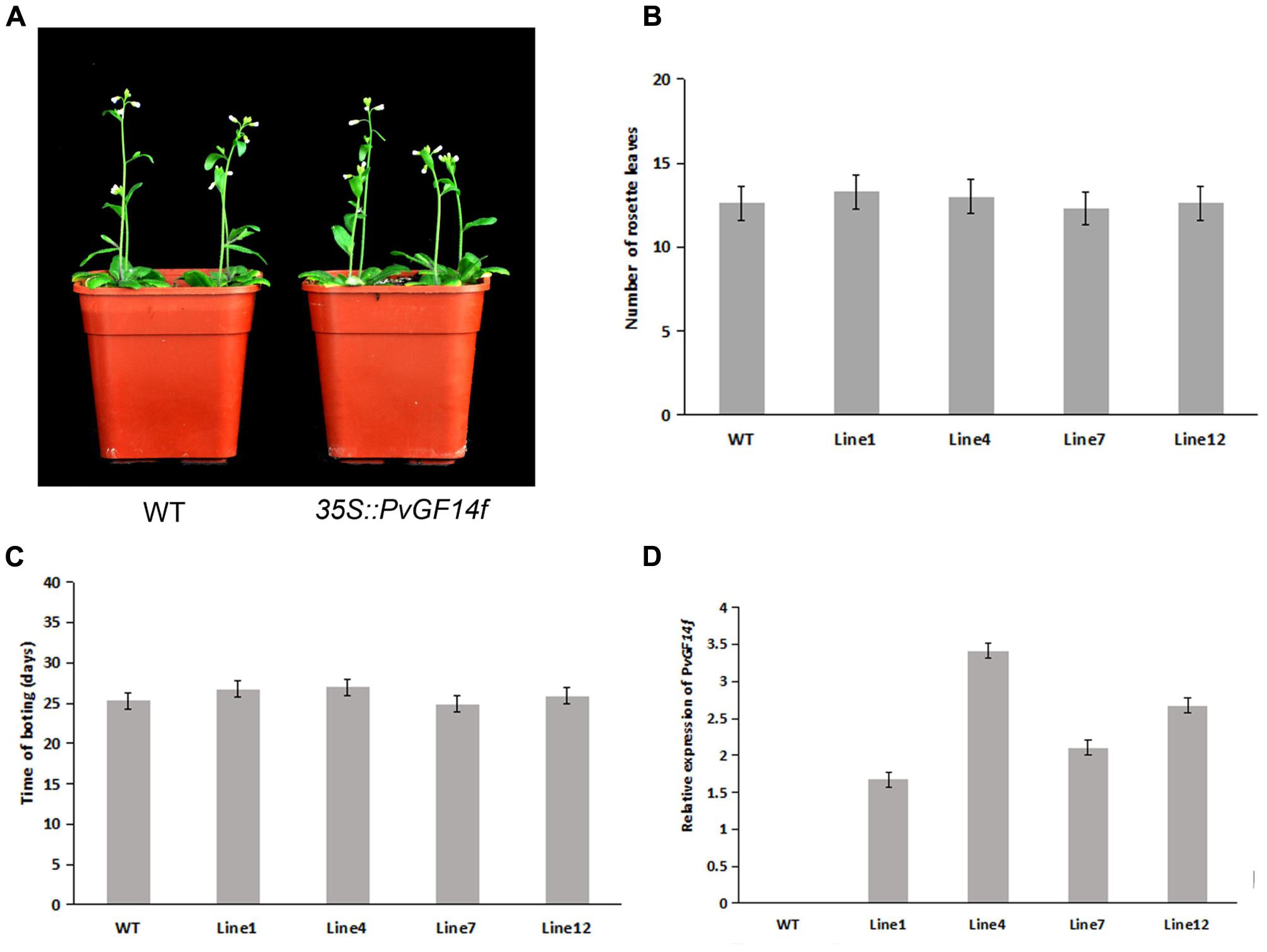
FIGURE 6. Phenotype analysis of 35S::PvGF14f Arabidopsis plants under LD conditions. (A) Phenotype differences in flowering between 35S::PvGF14f transgenic plants and wild-type plants. Photographs were taken at 27-day-old wild-type plants. (B) The number of rosette leaves in T2 35S::PvGF14f in transgenic plants (n = 20) and wild-type plants. (C) Bolting time in T2 35S::PvGF14f transgenic plants (n = 20) and wild-type plants. (D) qRT-PCR expression analysis of PvGF14f.
To determine whether the flowering time in transgenic Arabidopsis had relation with the expression of examined genes, we detected the transcript levels of PvGF14b, PvGF14e, and PvGF14f in T2 lines of 35S::PvGF14b, 35S::PvGF14e, and 35S::PvGF14f Arabidopsis, as well as PvGF14c expression level in T3 lines of 35S::PvGF14c Arabidopsis, respectively. The data revealed that expression levels of examined genes in different transgenic lines were significantly increased (Figures 3–6). The flowering time was positively associated with the expression of PvGF14c in transgenic Arabidopsis plants (Figure 3D). However, there was no correlation between other three PvGF14s mRNA abundance and the flowering time (Figures 4D, 5D, 6D).
Subcellular Localization of PvGF14c
Previous studies showed that OsGF14c predominantly localized in the cytoplasm and OsGF14c-ox plants were delayed by 5–20 days in flowering relative to wild-type plants (Purwestri et al., 2009). In addition, based on the data above, we found that the effect of PvGF14c on flowering time of transgenic plants was stronger than other three PvGF14s genes (Figures 3–6) and phylogenetic tree showed that PvGF14c was closest related to OsGF14c (Figure 1). Therefore, we built PvGF14c-GFP fusion constructs driven by CaMV35S promoter to analyze the intracellular localization of PvGF14c and to investigate the molecular mechanism of PvGF14c. These constructs were introduced into onion epidermal cells for transient expression. As expected, the empty GFP signals were ubiquitously distributed throughout the cells (Figure 7A). PvGF14c-GFP fusion proteins were localized in the cytoplasm (Figure 7B).
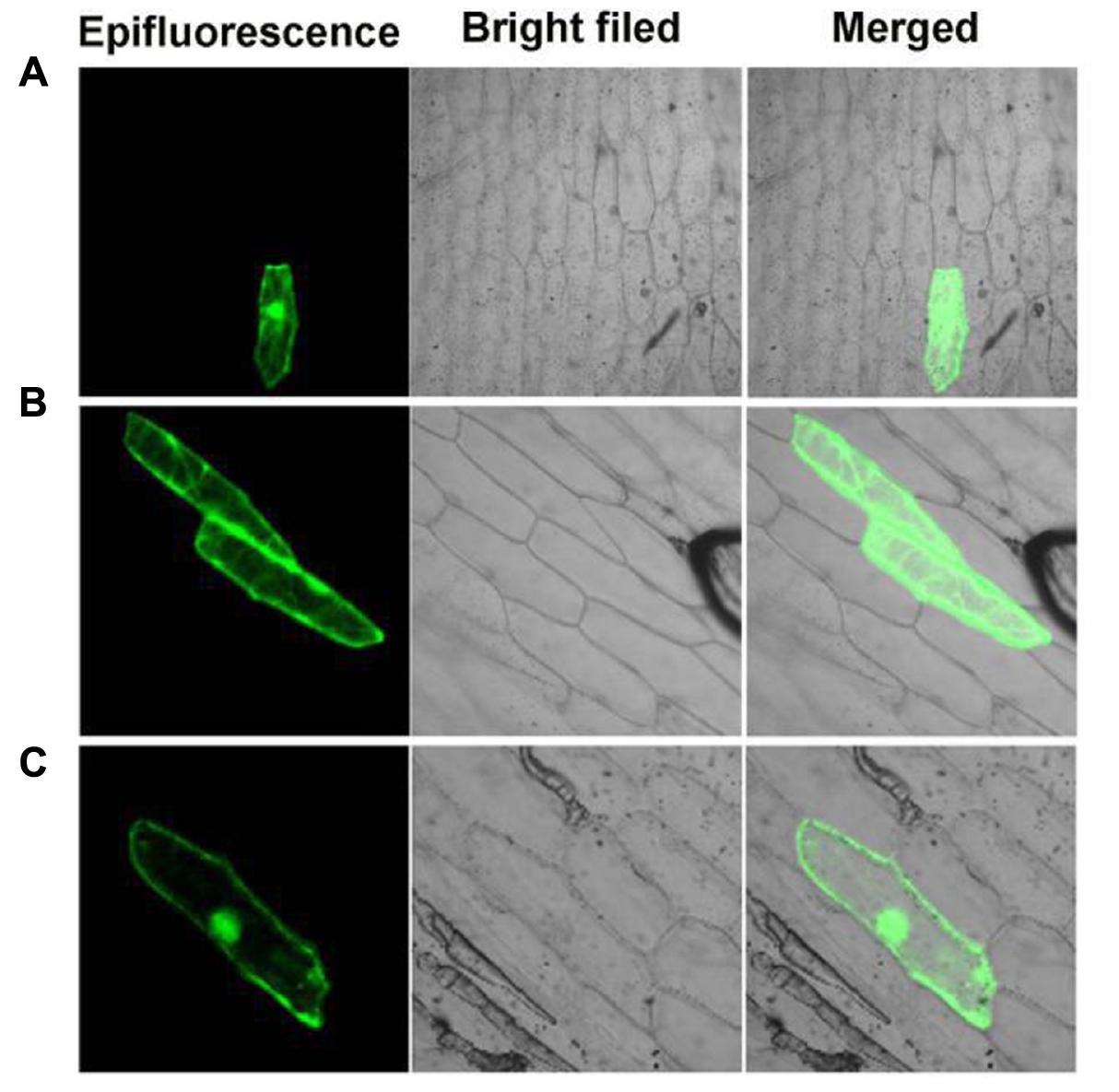
FIGURE 7. Subcellular localization of PvGF14c and PvFT protein. (A) Cells bombarded with 35S::GFP. (B) Cells bombarded with PvGF14c-GFP. (C) Cells bombarded with PvFT-GFP.
Interaction between PvGF14c and PvFT Proteins
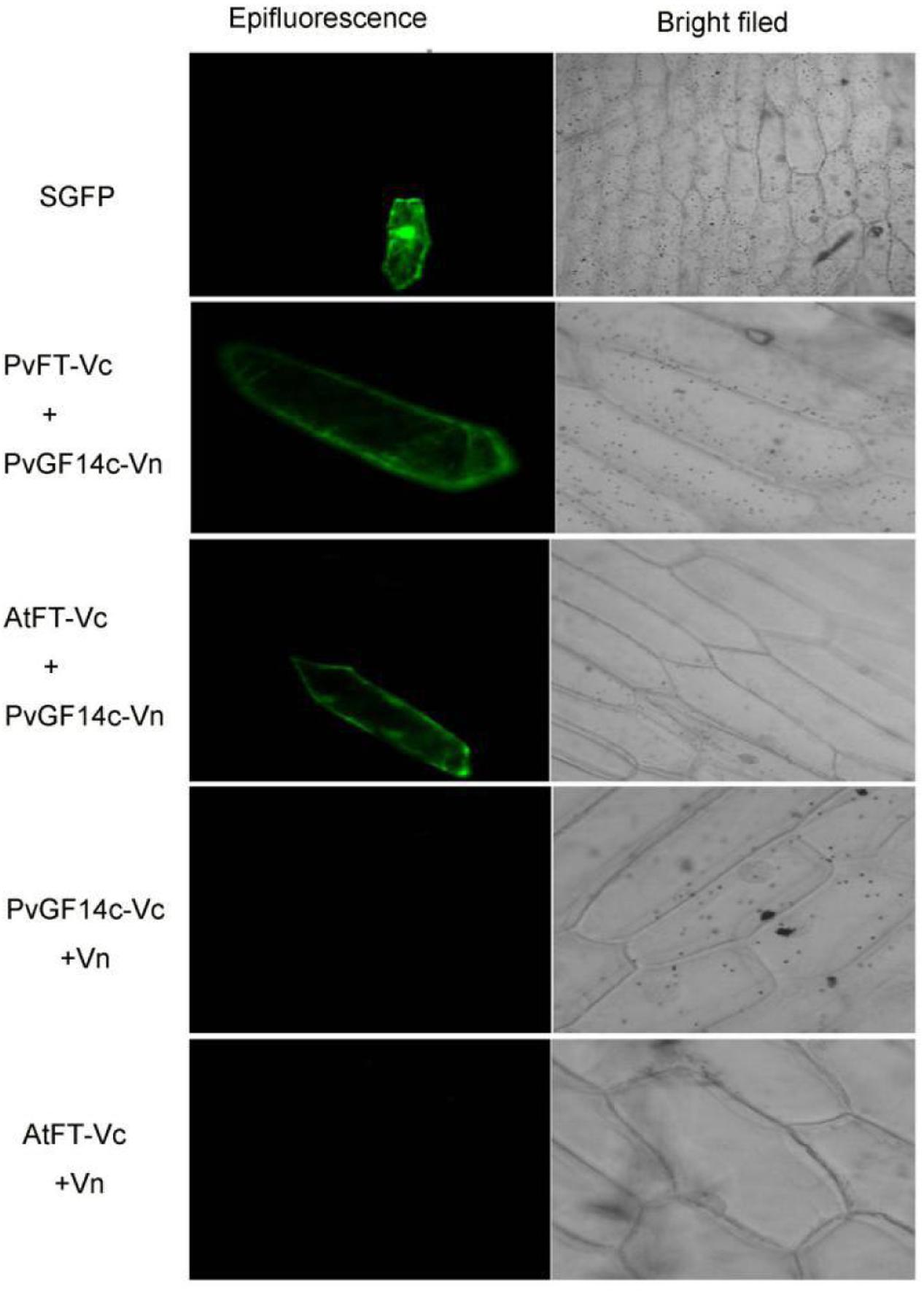
Previous presentation showed that the binding of 14-3-3 to Hd3a is important for flowering and OsGF14c interacted with Hd3a by Y2H screen and bimolecular fluorescence complementation (BiFC; Purwestri et al., 2009; Taoka et al., 2011). Thus, Y2H screening was performed to identify the relationship between PvGF14c and PvFT. The results demonstrated that PvGF14c could interact with PvFT protein as either a bait or a prey in P. violascens (Figure 8). Meanwhile, we used BiFC to determine the interaction of PvFT and PvGF14c in onion cell. As shown in Figure 9, the PvFT and PvGF14c interaction was detected in the cytoplasm. However, PvFT alone was detected in cytoplasm and nucleus (Figure 7C).
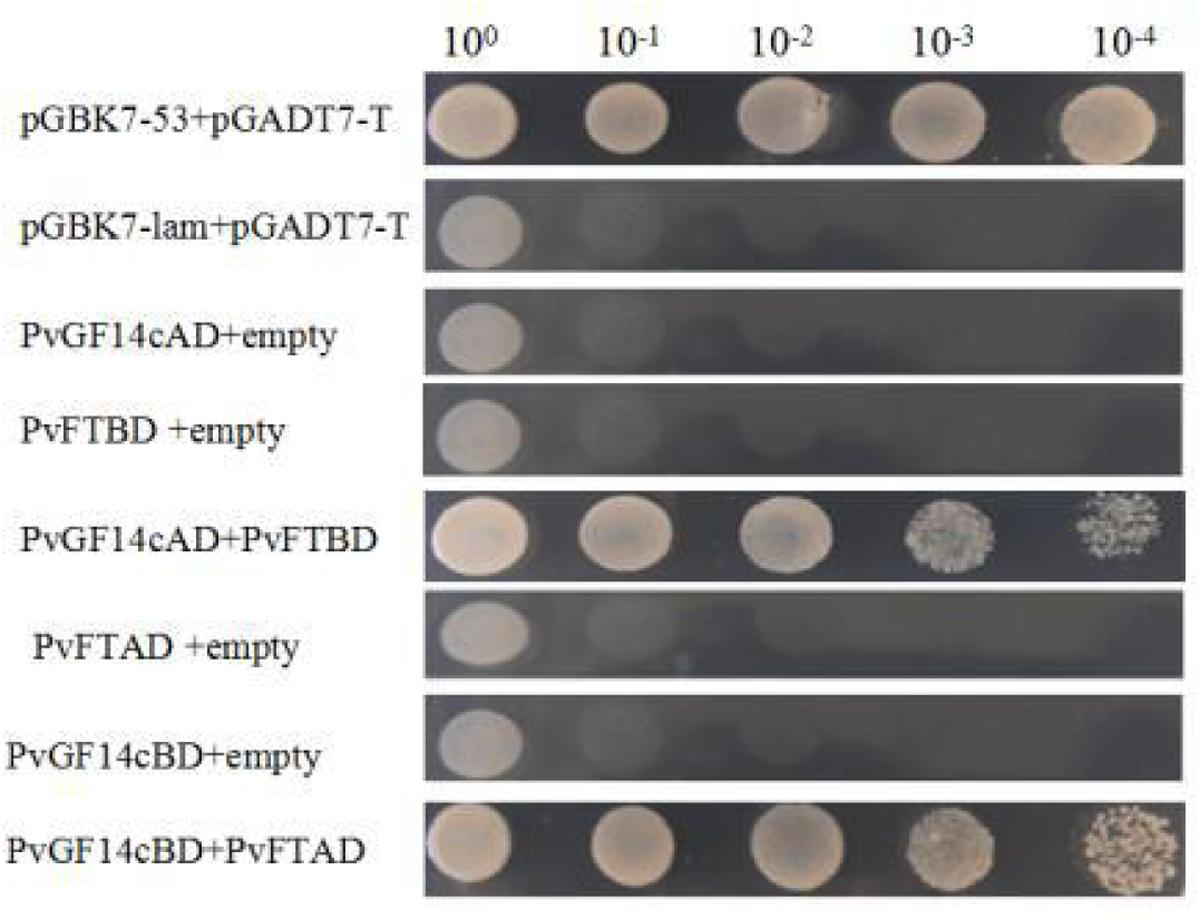
FIGURE 8. Identification of the interaction between PvGF14c protein and PvFT protein in yeast. The interaction between pGBKT7-53 and pGADT7-T was the positive protein–protein control, whereas the interaction between pGBKT7-lam and pGADT7-T was the negative protein–protein control.
The PvGF14c and AtFT Complex Is Localized in the Cytoplasm
Purwestri et al. (2009) presented that the interaction of Hd3a and OsGF14c resulted in Hd3a retaining in cytoplasmic and delayed flowering time in OsGF14c-ox. Our data showed that PvGF14c-ox plants also displayed the delayed flowering phenotype. Therefore, we determined the distribution of PvGF14c and AtFT in vivo by using BiFC. As shown in Figure 9, the PvGF14c and AtFT complex was located in cytoplasm.
Discussion
Plant 14-3-3 proteins act in a phosphorylation network and play an important role in plant growth and development. 14-3-3 Proteins interact with FT in Arabidopsis, with SP in tomato, and Hd3a in rice (Pnueli et al., 2001; Purwestri et al., 2009; Zhang, 2012). Studies on overexpression transgenic plants and mutant plants have revealed that OsGF14c functions as a negative regulator of flowering (Purwestri et al., 2009). However, the exact role of 14-3-3 proteins in the regulation of flowering in P. violascens has not yet to be reported.
In this study, four 14-3-3 genes, which share more than 85% amino acid sequence identity with OsGF14c, were identified in P. heterocycla and synthesized from P. violascens. The molecular masses and isoelectric points of four Pv14-3-3 proteins were consistent with the values of 14-3-3 families in reported plants (Xu and Shi, 2006; Li and Dhaubhadel, 2011; Qin et al., 2016). In Arabidopsis, the non-𝜀 group comprises four exons and three introns, introns insert position quite conservative (Wu et al., 1997). By comparison, the 𝜀 group generally includes six to seven exons, and the genetic structures of the 𝜀 group are different from those of the non-𝜀 group (DeLille et al., 2001). But the exon–intron structure analysis revealed that the four Pv14-3-3s consisted of five exons and four introns (Figure 1B), which are similar to non-𝜀 14-3-3s in rice (Chen et al., 2006).
Phylogenetic analysis indicated that four Pv14-3-3s proteins all belonged to the non-𝜀 group. Pv14-3-3s were closely related to Ph14-3-3s, and they gathered in the same branch. Meanwhile, Pv14-3-3s were closely related to non-𝜀 14-3-3s of O. sativa. The conservation of non-𝜀 14-3-3s between P. violascens and O. sativa indicated that the functions of four non-𝜀 Pv14-3-3s may be similar to those of non-𝜀 14-3-3s in O. sativa. Especially, PvGF14c is the highest identity with the OsGF14c which act as intercellular receptor for florigen, and OsGF14c-overexpressing plants display a delay flowering phenotype (Purwestri et al., 2009; Taoka et al., 2011). Thus, the regulation of flowering by Pv14-3-3s in P. violascens should be further investigated.
To examine the biological functions of four Pv14-3-3s, we conducted temporal-spatial expression analysis and observed that the expression pattern of these four genes were diverse, and there were various degree of expression in leaf. The mRNA of PvGF14c accumulated strongest in leaf relative to other tissues (Figure 2B). In addition, their expression levels did not differ before bamboo flowering and during flowering development (Figures 2E–H). The expression of PvGF14c and PvGF14b showed no diurnal changes, suggesting that the expression of these two genes were independent of photoperiod during plant development (Figures 2I,J).
In rice, GF14c-overexpressing plants displayed late flowering time and the knockout mutants exhibited early flowering under short-day conditions (Purwestri et al., 2009). In our study, PvGF14c-overexpressing Arabidopsis plants underwent late flowering under LD conditions, which is similar to OsGF14c delaying strongly flowering time (Purwestri et al., 2009). Considering that the 14-3-3 protein family were highly conserved, we also determine whether three other 14-3-3 proteins could regulate flowering. The results showed that PvGF14b and PvGF14e overexpression delayed slightly flowering time in transgenic Arabidopsis. Whereas, PvGF14f-ox plants showed the similar flowering time as that of the wild-type (Figures 3–6).
14-3-3 Isoforms bind to many kinds of proteins including regulators of flowering time, such as CO (Mayfield et al., 2007), and florigen FT (Pnueli et al., 2001). In rice, 14-3-3 proteins act as intercellular receptors for Hd3a which is localized in the cytoplasm and nucleus, whereas GF14c are mainly found in the cytoplasm. Hd3a–GF14b and Hd3a–GF14c interactions were detected mainly in the cytoplasm (Taoka et al., 2011). In wild-type of rice, Hd3a protein transported from leaves to the SAM is first received by OsGF14c protein in the cytoplasm, and then Hd3a–14-3-3 complex is translocated to the nucleus, where it forms a larger protein complex, named florigen activation complex, with OsFD1, then induces transcription of OsMADS15, leading to flowering (Taoka et al., 2011, 2013). 14-3-3 Proteins regulate their interaction proteins through a variety of mechanisms such as altering their subcellular localization (Roberts, 2003). The delayed flowering phenotype in GF14c-ox plants can be explained by the increased cytoplasmic retention of Hd3a by OsGF14c which are highly expressed in plants (Purwestri et al., 2009; Taoka et al., 2011). In our study, PvGF14s-ox plants exhibit delayed flowering. The interaction of PvGF14c and PvFT in P. violascens as well as PvGF14c and AtFT complex locating in cytoplasm were determined. These experiments hinted that high expression of PvGF14c gene regulate the import of FT into the nucleus in transgenic Arabidopsis. The floral induction could be attenuated by the cytoplasmic retention of FT by PvGF14s.
In flowering regulation, whether Pv14-3-3 proteins bind to PvFT and PvFD, forming a complex, and participate in the molecular machinery of flowering regulation is unknown, and the molecular mechanism of PvGF14-3-3s in regulating flowering should be further investigated.
Author Contributions
XG, BL, and GX conceived and designed the experiments. BL, GX, ZW, BM, and XL conducted and analyzed the experiments. KL helped to analyze the data. BL and XG wrote the paper. All authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the following grants: National Natural Science Foundation of China (grant no. 30901155), Natural Science Foundation of Zhejiang Province (grant no. Y307499).
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/gorf
- ^ http://gsds.cbi.pku.edu.cn
- ^ http://www.ebi.ac.uk/
- ^ http://web.expasy.org/compute_pi
- ^ www.megasoftware.net
References
Arner, E. S., and Holmgren, A. (2006). The thioredoxin system in cancer-introduction to a thematic volume of seminars in cancer biology. Semin. Cancer Biol. 16:419. doi: 10.1016/j.semcancer.2006.10.001
Chen, F., Li, Q., Sun, L., and He, Z. (2006). The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res. 13, 53–63.
Chung, H. J., Sehnke, P. C., and Ferl, R. J. (1999). The 14-3-3 proteins:cellular regulators of plant metabolism. Trends Plant Sci. 4, 367–371. doi: 10.1016/S1360-1385(99)01462-4
de Carvalho, A. L., Nelson, B. W., Bianchini, M. C., Plagnol, D., Kuplich, T. M., and Daly, D. C. (2013). Bamboo-dominated forests of the southwest amazon: detection, spatial extent, life cycle length and flowering waves. PLOS ONE 8:e54852. doi: 10.1371/journal.pone.0054852
DeLille, J. M., Sehnke, P. C., and Ferl, R. J. (2001). The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 126, 35–38. doi: 10.1104/pp.126.1.35
Fan, C., Ma, J., Guo, Q., Li, X., Hui, W., and Lu, M. (2013). Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLOS ONE 8:e56573. doi: 10.1371/journal.pone.0056573
Ferl, R. J., Manak, M. S., and Reyes, M. F. (2002). The 14-3-3s. Genome Biol. 3, reviews3010.1–reviews3010.7. doi: 10.1186/gb-2002-3-7-reviews3010
Gao, J., Ge, W., Zhang, Y., Cheng, Z., Li, L., Hou, D., et al. (2015). Identification and characterization of microRNAs at different flowering developmental stages in moso bamboo (Phyllostachys edulis) by high-throughput sequencing. Mol. Genet. Genomics 290, 2335–2353. doi: 10.1007/s00438-015-1069-8
Gao, J., van Kleeff, P. J., Oecking, C., Li, K. W., Erban, A., Kopka, J., et al. (2014). Light modulated activity of root alkaline/neutral invertase involves the interaction with 14-3-3 proteins. Plant J. 80, 785–796. doi: 10.1111/tpj.12677
Guo, X. Q., Guan, Y., Xiao, G. H., Xu, Z. E., Yang, H. Y., and Fang, W. (2016a). Isolation, characterization, and expression of an Indeterminate1 gene, BmID1, from bamboo (Bambusa multiplex). J. Plant Biochem. Biotechnol. 25, 30–39.
Guo, X. Q., Wang, Y., Wang, Q., and Xu, Z. E. (2016b). Molecular characterization of FLOWERING LOCUS T (FT) genes from bamboo (Phyllostachys violascens). J. Plant Biochem. Biotechnol. 25, 168–178. doi: 10.1007/s13562-015-0322-x
He, Y., Zhang, Y., Chen, L., Wu, C., Luo, Q., Zhang, F., et al. (2017). A member of the 14-3-3 gene family in Brachypodium distachyon, BdGF14d, confers salt tolerance in transgenic tobacco plants. Front. Plant Sci. 8:340. doi: 10.3389/fpls.2017.00340
Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347. doi: 10.1126/science.290.5490.344
Kim, D. H., Doyle, M. R., Sung, S., and Amasino, R. M. (2009). Vernalization: winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25, 277–299. doi: 10.1146/annurev.cellbio.042308.113411
Li, X., and Dhaubhadel, S. (2011). Soybean 14-3-3 gene family: identification and molecular characterization. Planta 233, 569–582. doi: 10.1007/s00425-010-1315-6
Lin, X., Chow, T., Chen, H., Liu, C., Chou, S., Huang, B., et al. (2010). Understanding bamboo flowering based on large-scale analysis of expressed sequence tags. Genet. Mol. Res. 9, 1085–1093. doi: 10.4238/vol9-2gmr804
Liu, Q., Yang, J., Zhang, S., Zhao, J., Feng, A., Yang, T., et al. (2016). OsGF14b positively regulates panicle blast resistance but negatively regulates leaf blast resistance in rice. Mol. Plant Microbe Interact. 29, 46–56. doi: 10.1094/MPMI-03-15-0047-R
Liu, S., Ma, T., Ma, L., and Lin, X. (2016). Ectopic expression of PvSOC1, a homolog of SOC1 from Phyllostachys violascens, promotes flowering in Arabidopsis and rice. Acta Physiol. Plant. 38, 1–9. doi: 10.1007/s11738-016-2186-7
Mayfield, J. D., Folta, K. M., Paul, A. L., and Ferl, R. J. (2007). The 14-3-3 proteins mu and upsilon influence transition to flowering and early phytochrome response. Plant Physiol. 145, 1692–1702.
Moon, J., Lee, H., Kim, M., and Lee, I. (2005). Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 46, 292–299. doi: 10.1093/pcp/pci024
Paul, A. L., Folta, K. M., and Ferl, R. J. (2008). 14-3-3 proteins, red light and photoperiodic flowering: a point of connection? Plant Signal. Behav. 3, 511–555.
Peng, Z. H., Lu, Y., Li, L., Zhao, Q., Feng, Q., Gao, Z. M., et al. (2013). The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 45, 456–461. doi: 10.1038/ng.2569
Pnueli, L., Gutfinger, T., Hareven, D., Ben-Naim, O., Ron, N., Adir, N., et al. (2001). Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. Plant Cell 13, 2687–2702.
Purwestri, Y. A., Ogaki, Y., Tamaki, S., Tsuji, H., and Shimamoto, K. (2009). The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant Cell Physiol. 50, 429–438. doi: 10.1093/pcp/pcp012
Qin, C., Cheng, L., Shen, J., Zhang, Y., Cao, H., Lu, D., et al. (2016). Genome-wide identification and expression analysis of the 14-3-3 family genes in Medicago truncatula. Front. Plant Sci. 7:320. doi: 10.3389/fpls.2016.00320
Roberts, M. R. (2003). 14-3-3 proteins find new partners in plant cell signalling. Trends Plant Sci. 8, 218–223. doi: 10.1016/S1360-1385(03)00056-6
Shih, M. C., Chou, M. L., Yue, J. J., Hsu, C. T., Chang, W. J., Ko, S. S., et al. (2014). BeMADS1 is a key to delivery MADSs into nucleus in reproductive tissues-De novo characterization of Bambusa edulis transcriptome and study of MADS genes in bamboo floral development. BMC Plant Biol. 14:179. doi: 10.1186/1471-2229-14-179
Sun, T., Wang, Y., Wang, M., Li, T., Zhou, Y., Wang, X., et al. (2015). Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum). BMC Plant Biol. 15:269. doi: 10.1186/s12870-015-0657-4
Taoka, K., Ohki, I., Tsuji, H., Furuita, K., Hayashi, K., Yanase, T., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476, 332–335. doi: 10.1038/nature10272
Taoka, K., Ohki, I., Tsuji, H., Kojima, C., and Shimamoto, K. (2013). Structure and function of florigen and the receptor complex. Trends Plant Sci. 18, 286–294. doi: 10.1016/j.tplants.2013.02.002
Wang, Y. C., Klein, T. M., Fromm, M., Cao, J., Sanford, J. C., and Wu, R. (1988). Transient expression of foreign genes in rice, wheat and soybean cells following particle bombardment. Plant Mol. Biol. 11, 433–439. doi: 10.1007/BF00039024
Wu, K., Rooney, M. F., and Ferl, R. J. (1997). The Arabidopsis 14-3-3 multigene family. Plant Physiol. 114, 1421–1431.
Xu, W. F., and Shi, W. M. (2006). Expression profiling of the 14-3-3 gene family in response to salt stress and potassium and iron deficiencies in young tomato (Solanum lycopersicum) roots: analysis by real-time RT-PCR. Ann. Bot. 98, 965–974. doi: 10.1093/aob/mcl189
Yao, Y., Du, Y., Jiang, L., and Liu, J. Y. (2007). Molecular analysis and expression patterns of the 14-3-3 gene family from Oryza sativa. J. Biochem. Mol. Biol. 40, 349–357. doi: 10.5483/BMBRep.2007.40.3.349
Zhang, L. (2012). Genetic map of Triticum turgidum Based on a Hexaploid Wheat Population Without Genetic Recombination for D Genome and SDG8, FT Function Synergistically to Regulate Plant Architecture and Flowering Time. Ya’an: Sichuan Agricultural University.
Zhang, X.-M., Zhao, L., Larson-Rabin, Z., Li, D.-Z., and Guo, Z.-H. (2012). De novo sequencing and characterization of the floral transcriptome of Dendrocalamus latiflorus (Poaceae: Bambusoideae). PLOS ONE 7:e42082. doi: 10.1371/journal.pone.0042082
Zhao, H., Peng, Z. H., Fei, B., Li, L., Hu, T., Gao, Z. M., et al. (2014). Bamboo GDB: a bamboo genome database with functional annotation and an analysis platform. Database 2014:bau006. doi: 10.1093/database/bau006
Zhao, X. Y., Wang, X. Y., Zhao, L., Zhang, X. M., Chen, S. Y., Ma, P. F., et al. (2015). Investigating the MicroRNAomes of two developmental phases of Dendrocalamus latiflorus (Poaceae: Bambusoideae) inflorescences. Plant Mol. Biol. Rep. 33, 1141–1155. doi: 10.1007/s11105-014-0808-z
Keywords: Phyllostachys violascens, Pv14-3-3s, PvGF14c, flowering, ectopic expression
Citation: Li B, Xiao G, Luo K, Wang Z, Mao B, Lin X and Guo X (2018) Overexpression of PvGF14c from Phyllostachys violascens Delays Flowering Time in Transgenic Arabidopsis. Front. Plant Sci. 9:105. doi: 10.3389/fpls.2018.00105
Received: 18 October 2017; Accepted: 19 January 2018;
Published: 14 February 2018.
Edited by:
Sagadevan G. Mundree, Queensland University of Technology, AustraliaReviewed by:
Hak Soo Seo, Seoul National University, South KoreaKeqiang Wu, National Taiwan University, Taiwan
Copyright © 2018 Li, Xiao, Luo, Wang, Mao, Lin and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqin Guo, eHFndW9AemFmdS5lZHUuY24=
† These authors have contributed equally to this work.
 Bingjuan Li
Bingjuan Li Guohui Xiao1†
Guohui Xiao1† Xiaoqin Guo
Xiaoqin Guo