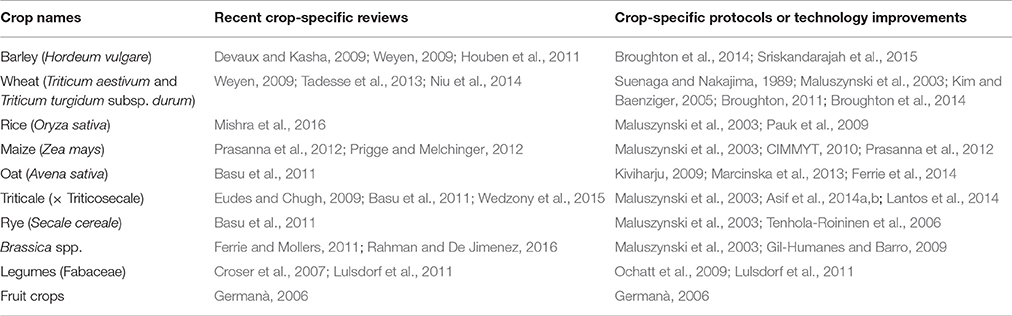
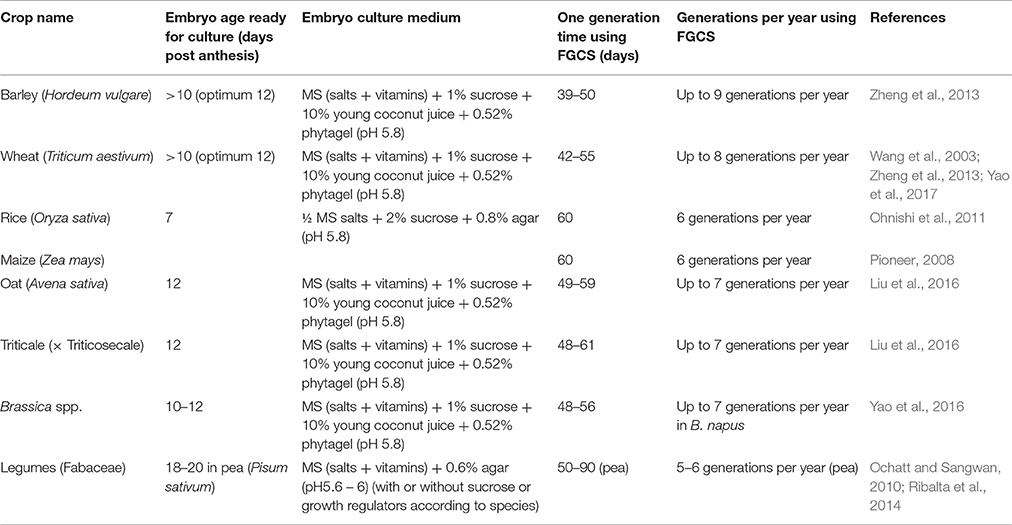
- 1Faculty of Science, UWA School of Agriculture and Environment, University of Western Australia, Perth, WA, Australia
- 2The UWA Institute of Agriculture, University of Western Australia, Crawley, WA, Australia
- 3Hebei Centre of Plant Genetic Engineering, Institute of Genetics and Physiology, Hebei Academy of Agricultural and Forestry Sciences, Shijiazhuang, China
- 4Inner Mongolia Academy of Agriculture and Animal Husbandry Sciences, Huhhot, China
- 5Hebei Province Wheat Engineering Technical Research Center, Shijiazhuang Academy of Agricultural Sciences, Shijiazhuang, China
- 6InterGrain Pty. Ltd., Bibra Lake, WA, Australia
- 7SuperSeeds Technologies Pty. Ltd., Perth, WA, Australia
- 8Commonwealth Scientific and Industrial Research Organisation Agriculture and Food, St. Lucia, QLD, Australia
Production of pure lines is an important step in biological studies and breeding of many crop plants. The major types of pure lines for biological studies and breeding include doubled haploid (DH) lines, recombinant inbred lines (RILs), and near isogenic lines (NILs). DH lines can be produced through microspore and megaspore culture followed by chromosome doubling while RILs and NILs can be produced through introgressions or repeated selfing of hybrids. DH approach was developed as a quicker method than conventional method to produce pure lines. However, its drawbacks of genotype-dependency and only a single chance of recombination limited its wider application. A recently developed fast generation cycling system (FGCS) achieved similar times to those of DH for the production of selfed pure lines but is more versatile as it is much less genotype-dependent than DH technology and does not restrict recombination to a single event. The advantages and disadvantages of the technologies and their produced pure line populations for different purposes of biological research and breeding are discussed. The development of a concept of complete in vitro meiosis and mitosis system is also proposed. This could integrate with the recently developed technologies of single cell genomic sequencing and genome wide selection, leading to a complete laboratory based pre-breeding scheme.
Introduction
High yield and quality with tolerance to biotic and abiotic stresses are the major breeding objectives of crop breeding and also of interest in genetic studies. Crop breeding programmes are frequently based on pure lines. These require 5–6 generations of inbreeding and selection after crossing to generate inbred homozygous lines suitable for extensive evaluation. This process is the reason for the time taken, often 11–13 years between making crosses to releasing cultivars. Similarly, the production of pure lines from segregating populations is also necessary to identify genes for a trait of interest.
The developed pure lines provide permanent populations allowing phenotyping of traits of the same genotypes at different times and under different environmental conditions. Both geneticists and breeders have been seeking ways to accelerate the process of pure line production. Doubled haploids (DH) are a successful technique for this purpose as it dramatically reduces the time for the production of homozygous pure lines. Since the first report of haploid production in 1920s, the DH, both the range of species and efficiency of production, has been greatly improved and is routinely used in many crop breeding programs (Asif, 2013; Li et al., 2013). Another system for pure line production is a fast generation cycling system (FGCS). This has been recently developed and applied in crop breeding (Wang et al., 1999, 2003; Ochatt et al., 2002; Zheng et al., 2013; Forster et al., 2014; Liu et al., 2016; Yao et al., 2016, 2017). FGCS shortens each generation cycle by culturing young embryos and managing plants to greatly reduce the time from seed to the seed of the next generation. The key difference between DH and FGCS technology lies in the greater opportunity for genetic recombination to occur in FGCS as it goes through more generations.
This review summarizes: (1) the general process of DH and FGCS technologies; (2) the applications of the produced pure line populations in genetic studies and crop breeding; (3) the constraints and opportunities of DH and FGCS technologies; and (4) the possible futures for technology improvement in fast development of pure lines.
DH Technology
DH generates homozygous lines by doubling chromosomes of haploid plants generated from either egg or sperm cells (Figure 1). There have been many reviews of DH technology in plants, covering a range of species, protocol improvements, recent concepts, and applications in genetic study and breeding (Maluszynski et al., 2003; Forster et al., 2007; Touraev et al., 2009; Dunwell, 2010; Forster and Thomas, 2010; Murovec and Bohanec, 2012; Asif, 2013; Dwivedi et al., 2015).
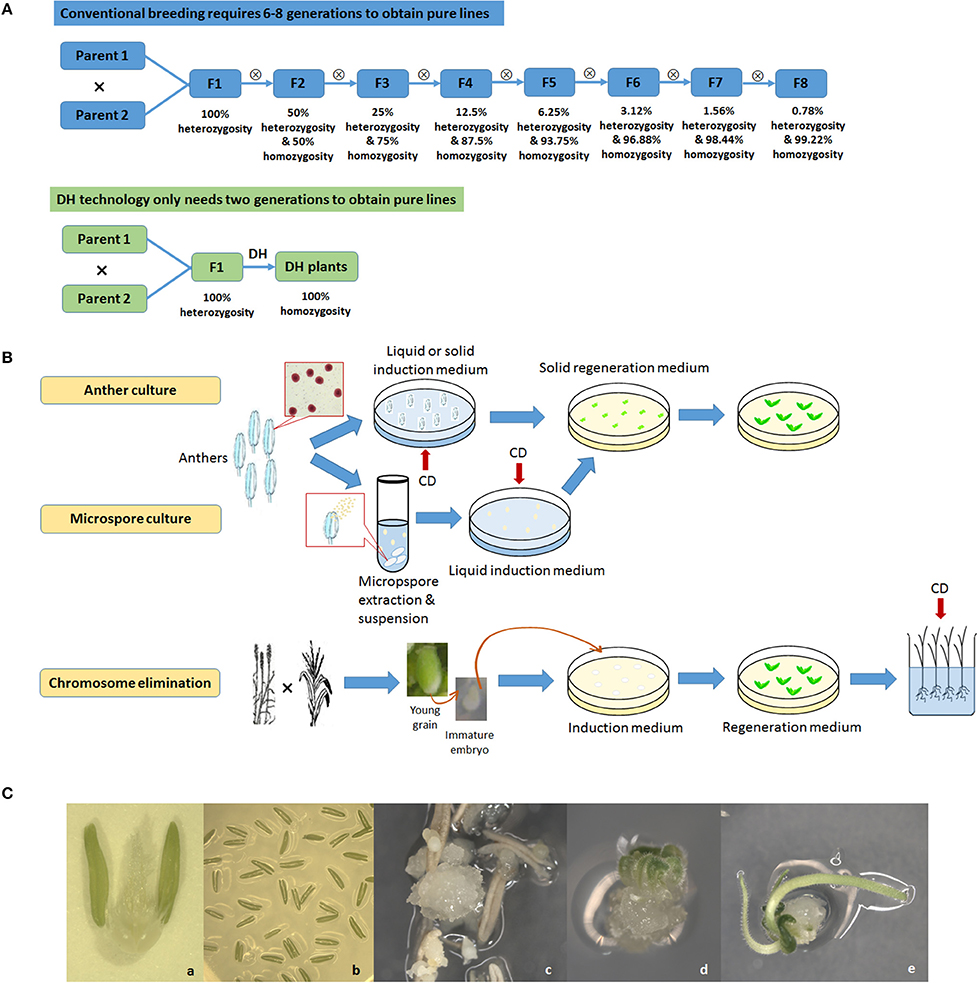
Figure 1. Doubled haploid (DH) technology. (A), Comparison between conventional breeding and DH technology; (B), Diagram of three major DH technologies adopted in crop breeding: anther culture, microspore culture and chromosome elimination. CD, Chromosome doubling with chemical treatment; (C), Anther culture in wheat. a, Anthers on a flower; b, Anthers cultured on medium; c, Callus formation; d, Initiation of differentiation; e, Differentiation of roots and shoots.
There are three widely used methods to produce DHs. androgenesis uses sperm cell microspore culture or another culture. Microspore culture uses microspores, preferably at the uni-nucleated stage, isolated from young anthers and then cultured in vitro where they develop into either embryoids or callus tissue that is regenerated into plantlets. Another culture uses developing anthers that are directly excised from flower buds at a critical stage and cultured in vitro where the microspores within the anthers develop into callus or embryoids that can be regenerated into plantlets. Chemical (usually colchicine) treatments for chromosome doubling may occur at either in vitro culture stage by using media containing the chemical to directly generate DH embryoids or callus or at a later stage on the regenerated haploid plantlets. The former is now the most often used methodology. The second method is gynogenesis, using ovary or ovule culture. The ovary or ovule is carefully dissected from the flower, a labor-intensive step that limits its wide adoption in crop breeding. The third method is chromosome elimination, where the target species is crossed to a distant related relative and the embryos produced are cultured or rescued in vitro. The chromosomes of the distant relative are eliminated, giving rise to haploid plantlets having only chromosomes from the target species (Jensen, 1983; Houben et al., 2011). The chromosomes of the regenerated haploid plantlets are then chemically doubled. Chromosome elimination has been successfully used in barley (Hordeum vulgare) crossed with Hordeum bulbosum (Kasha and Kao, 1970) and wheat (Triticum aestivum) crossed with maize (Zea mays; Laurie and Bennett, 1988; Inagaki and Tahir, 1990). In maize, a specific Stock-6 based intraspecific system (Coe, 1959) has been extensively exploited and has generated a majority of commercial DH maize cultivars. The genetic basis underlying the haploid induction in maize has been elucidated in recent studies (Hu et al., 2016; Gilles et al., 2017; Kelliher et al., 2017).
The advantages and disadvantages of these three methods and their optimizations have been extensively discussed (Reed, 2004; Bohanec, 2009; Germanà, 2011; Asif, 2013). Another culture is relatively easy to operate, and often quicker to produce DH lines compared to other methodologies as anther walls have positive effects on promoting culture growth. The disadvantage is that plantlets may originate from haploid microspore as well as from diploid anther wall tissue. Microspore culture eliminates this problem and is ideal for genetic manipulations as it starts from single cells that can be better regulated. Technological advances have now enabled microspore culture to achieve reasonably high yields in some crop genotypes, such as, canola, barley, and wheat (Gil-Humanes and Barro, 2009; Broughton et al., 2014). The efficiency of this technique, however, is highly genotype-dependent and its success rate may be low in many plant species (Cegielska-Taras et al., 2015). Chromosome elimination has an advantage of relatively easy laboratory protocols, but also has the issue of low efficiency. In addition, it requires an extra crossing procedure, and its chromosome doubling is usually applied to the regenerated haploid plants which involves direct contact with a large amount of hazardous chemicals.
New ideas for DH production have included centromere-mediated genome elimination to create a haploid inducer line (GFPtailswap cenH3−/−) in Arabidopsis (Ravi and Chan, 2010) and maize (Kelliher et al., 2016) for haploid production followed by chromosome doubling. Another method involving the use of unreduced gametes referred to “synthesizing DH (SynDH) technology” that employs interspecific hybridization between tetraploid and diploid lines followed by spontaneous polyploidization to obtain hexaploid plants has been described in wheat (Zhang et al., 2011).
The advantage of DH over conventional breeding methods is that DH achieves complete homozygosity in one generation. This enables significant shortening of time to the production of pure lines. Complete homozygosity allows more precise phenotyping and allows accurate gene-trait association in genetic mapping and gene function studies. The single cell cultures can also be used as targets for cell biology and genetic engineering studies. DH technology has been successfully developed and improved many crops (Table 1), in which barley and rapeseed are among the most responsive. However, there are constraints to the use of DH routinely for breeding and genetic study. Cottons and many legume species are recalcitrant to DH technology and despite major effort there have been few successes, while some others species suffer from high costs and low efficiency of DH technology. The major factors influencing the efficiency of DH production include species and genotype dependency (Ei-Hennawy et al., 2011; Murovec and Bohanec, 2012), a high proportion of albinism (Kumari et al., 2009; Makowska and Oleszczuk, 2014; Sriskandarajah et al., 2015), high frequencies of clones via androgenesis (Oleszczuk et al., 2014), and genome instability such as, aneuploidy due to somaclonal variation (Oleszczuk et al., 2011; Wedzony et al., 2015). These problems may jeopardize both breeding and genetic studies. In addition, DH requires skilled personnel and tissue culture facilities, which may not be available and DH only allows one or two opportunities of recombination, as DH lines are usually generated from F1 or sometimes F2 plants, limiting the diversity of the DH lines.
FGCS Technology
FGCS combines embryo culture with plant management to greatly reduce generation time (Figure 2). It involves two steps in each generation. Firstly, plants are grown in a controlled environment where irrigation and nutrient managements accelerate the vegetative growth and flower differentiation. Secondly, young embryos are cultured reducing the time required for seed maturation (Wang et al., 1999, 2003). At this step, the removing of endosperm promotes germination because of embryos' easy absorption of the readily available sucrose in the medium (Tezuka et al., 2012) and the detachment of possible inhibitors in the endosperm (Chawla, 2002). A fully in vitro protocol for FGCS has also been reported for some plants including protein legumes, Arabidopsis and wheat etc. (Ochatt et al., 2002; Ochatt and Sangwan, 2008, 2010; Yao et al., 2017).
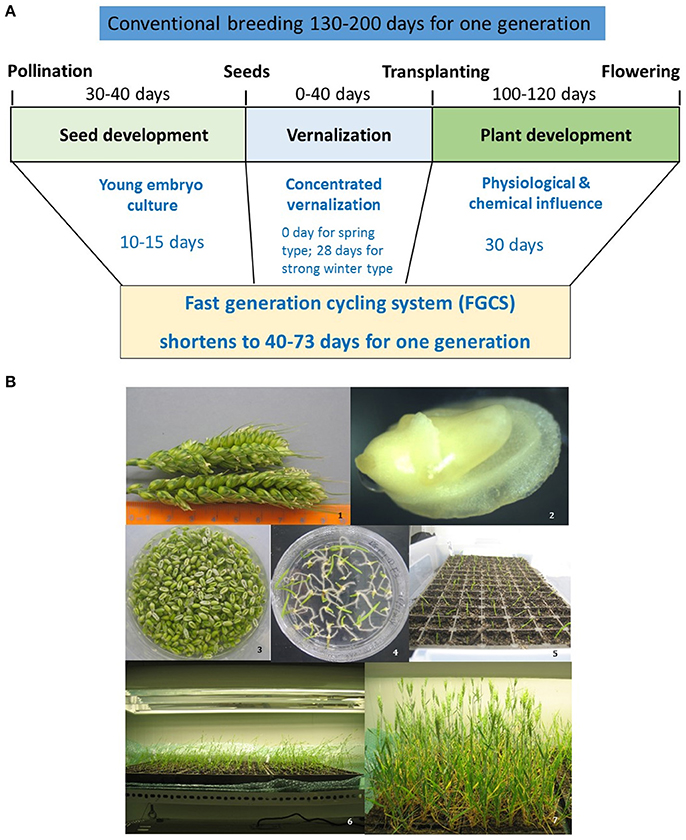
Figure 2. Fast generation cycling system (FGCS). (A), Comparison of one generation time between conventional breeding and FGCS in crops. (B), FGCS in wheat from young grains to flowering. 1, Yong wheat spikes collected for embryo culture; 2, An embryo dissected from a young wheat grain; 3, Young grains ready for embryo culture; 4, Germinated embryos in a plate; 5, Plantlets transferred to soil; 6, Plants grown into three-leaf stage; 7 Plants at flowering stage.
Several advantages for FGCS have been reported: (1) it reduces the time from crossing to variety release as generation time for RILs and introgression are dramatically reduced; (2) it does not reduce the number of meiotic events where recombination occurs, providing the same recombination as traditional breeding procedures, which is beneficial for both crop breeding and high resolution genetic mapping or fine mapping studies; (3) selection can be incorporated in any generation and NILs can be developed using the heterogeneous inbred family (HIF) selection (Tuinstra et al., 1997); and (4) it is less genotype-dependent.
Factors influencing the efficiency of FGCS could exist in both embryo culture (Lentini et al., 1988; Raghavan, 2003) and plant management (Ausín et al., 2005; Riboni et al., 2014). Variation of embryo culture between crops occurs with embryo age and culture medium used (Table 2). The optimum embryo age for culture varies from as early as 7 days post anthesis (dpa) in rice to as late as 18–20 dpa in peas. The embryo culture medium generally contains basal salts (macro- and micro-nutrients), vitamins, sucrose, and medium solidifier, or supplemented with young coconut juice depending on crop species. As an energy source, sucrose plays an important role in physiology of seed germination. Young liquid endosperm of green coconuts is recognized as an important organic supplement for embryo culture in many plants (Molnár et al., 2011). It contains a number of amino acids, organic acids, nucleic acids, vitamins, plant hormones, sugar alcohols, and some unidentified substances that may be responsible for promoting embryo growth. Environmental factors affecting vegetative growth and flower differentiation include light, temperature, irrigation, and nutrient solutions. Optimizing these are crucial in determining the efficiency of FGCS (Whitman et al., 1998; Cerdán and Chory, 2003; Cummings et al., 2007). Environmental stress may also accelerate flowering (Kumar et al., 2012). Warm temperatures and/or moderate drought can act as environmental cue to flowering in some plants (Riboni et al., 2014). Increased light intensity or photoperiod duration (such as, constant light) accelerate flowering in Arabidopsis (Kumar et al., 2012). Extreme examples of environmental stress leading to fast flowering occurs in some desert annual plants with no juvenile stage. Atmospheric CO2 concentration and tiller removal also affect FGCS in rice (Ohnishi et al., 2011). For soil nutrients, nitrogen usually delays reproduction and phosphorous and potassium usually promote reproduction (Wang et al., 2003). It has also been reported that small pot size with limited nutrient supply and limited irrigation prevent tiller production in cereal plants, shortening the vegetative period thus promoting flowering (Forster et al., 2014). Shortening the juvenile phase may result in early maturity (soybean and rice). The duration of juvenility can influence flowering regardless of other physiological controls of flowering (Atwell et al., 1999). To speed up the generation cycle, a balance between short and sufficient vegetative growth is needed to achieve development but must be balanced with enough biomass accumulation for flower initiation and seed set.
Other factors affecting FGCS include seed dormancy and vernalization. These affect seed/embryo germination and plant flowering, respectively. Embryo age has an important role in the efficiency of embryo culture where uniform and rapid germination is ideal but may be staggered by the effects of seed dormancy (Lefebvre et al., 2006; Hilhorst, 2007; Lee et al., 2010; Kang et al., 2015).
Early embryo culture can be carried out without waiting for full seed development and embryos can be harvested before dormancy sets in. When young embryos are dissected at a stage suitable for culturing on artificial medium, this could effectively circumvent seed dormancy. Although exactly when dormancy develops during seed development is unclear, it is considered to be initiated at the stage when food reserves start to accumulate, which is after the embryo develops into the heart stage (Bentsink and Koornneef, 2008). In cereal crops, seed dormancy could develop before the hard dough stage, as reported in wheat (Lan et al., 2005). Among the different types of seed dormancy, non-deep physiological dormancy is common in many crop species. It is determined by physiological factors in the embryo and surrounding layers including the endosperm and the testa. Embryos excised from seeds with such dormancy will usually germinate normally and treatments of gibberellins (GA), scarification, stratification, or a period of dry storage can break the dormancy (Atwell et al., 1999; Linkies et al., 2010; Willis et al., 2014).
Long season winter crops are important but their generation time is long due to their requirement for vernalization. These crops require 25 to at least 40 days of vernalization. This limits the efficiency of marker development and trait recombination in these crops. FGCS can be applied to winter crops by vernalizing germinated embryos before transplanting them into soil. With this procedure up to five generations of winter wheat per year have been achieved (Wang et al., 1999, 2003). In our study applying FGCS in winter wheat it was possible to reduce generation time by bypassing the vernalization requirement. Vernalization promotes flowering by causing an epigenetic shut off of FLOWERING LOCUS C (FLC) gene coding for a repressor of flowering. The status of the epigenetic modification can be changed. This is known as “memory” and “resetting,” with the former referring to the mitotic memory system of the histone modifications triggered by vernalization. This allows flowering to occur and reprogrammes the epigenetic states to their default position in the next generation, ensuring flowering occurs at an optimal time in every generation (Sheldon et al., 2008; Choi et al., 2009; Dennis and Peacock, 2009; Oliver et al., 2009; Crevillén et al., 2014; Woods, 2014). When FGCS was applied to winter wheat varieties, it was found that older embryos (more than 20 dpa) required vernalization to initiate flowering whereas plants generated from younger embryos (15 dpa) flowered without vernalization (Qin and Wang, 2002). The memory of the vernalized state may be maintained in the young embryos before the resetting of the system as the embryos aged. Screening a wide range of winter wheat varieties is needed in future studies so that the optimum age of embryo development can be determined to ensure that the generated plants flower without vernalization.
FGCS plants can be grown entirely in soil or in vitro as found in FGCS of protein legume crops (Ochatt et al., 2002). The earliest use of FGCS entirely in soil was in Brassica where rapid flowering species were used and grown at high densities in a controlled environment (constant temperature and light conditions) allowing up to 10 generations per year in B. rapa (Williams and Hill, 1986). Entirely in vitro FGCS is to complete a generation cycle from dissecting and germinating young embryo to inducing flowering and seed set totally under in vitro conditions, which is repeated until the desired generation (Yao et al., 2016). Plant growth regulators are sometimes needed for this system, as reported in Pisum and Vigna (Ochatt and Sangwan, 2008; Ribalta et al., 2014).
FGCS is cost effective as it saves time and space and is comparable to DH in regards of achieving pure lines with desired homozygosity. Single seed descent (SSD) is usually adopted in FGCS for developing RIL populations in crops, through continuous selfing from the F2 generation until the desired level of homozygosity is reached. For high resolution genetic analysis, a large F2 population is needed to ensure that later generation population size is representative and suitable for any statistically sound genetic analysis. Therefore, F1 plants must be grown under optimum conditions ensuring the maximum number of F2 seeds or embryos before FGCS can be applied. DH also require optimum conditions for the F1 plants to produce large numbers of healthy anthers/microspores/ovaries needed for DH production. The time taken for growing F1 plants is therefore similar for both DH and FGCS. From F1 flowering stage, production of DH lines can be achieved within a year or one and half years for many crops, whereas the time taken for FGCS to produce RILs (F6 or F8 depending on required use) from F1 embryos can also be achieved within a similar timeframe in the same crops (Liu et al., 2016). In FGCS, plants are grown in small pots or tray cells which require significantly less space and labor cost for large population development than that of the field conditions in conventional breeding. Being less genotype-dependent as shown in previous studies (Liu et al., 2016; Yao et al., 2016), FGCS can also provide savings compared to DH in crops where recalcitrant genotypes occur, for example, some oat genotypes having low DH efficiency with low haploid embryo production rates and haploid plant regeneration rates needed to be compensated by plating more anthers/microspores or embryos to raise the yield of DH production, which increased the labor and cost (Kiviharju et al., 2005; Marcinska et al., 2013).
FGCS is especially attractive in species or genotypes where DH lines are difficult to produce. Successful applications of FGCS were reported in several major crops, which significantly shortened the generation time and enabled 6–9 generations per year in the crops that would otherwise only allowed up to 1–3 generations per year by conventional methods (Table 2).
Application of Pure Line Populations in Genetic Studies and Breeding
Unlike early generation segregation populations such as, F2 and backcross lines, DH lines, RILs, and NILs are pure line populations because they are homozygous that can be multiplied and reproduced without genetic changes occurring (Semagn et al., 2006). They can be repeatedly used in trials, which is important for estimates of trait heritability and for genetic analyses such as, mapping and gene function studies. Each of the approaches has advantages and disadvantages when used in genetic studies and breeding.
DH Lines
DH lines are complete homozygotes and contain two identical sets of chromosomes/genes. They are ideal for estimating quantitative trait locus (QTL) × environment (E) interactions as complete homozygosity allows better estimates of trait means and allows more precise selection over locations and years. The expected genotypic segregation ratio is 1:1, irrespective of whether the marker is dominant or codominant. DH plants are fully fertile and if suitable can be used as parents or released as a cultivar by breeding programs. DHs have been widely used for cultivar development, genetic mapping, mutagenesis, and gene function studies (Ferrie and Mollers, 2011; Hussain et al., 2012).
However, distorted segregation ratios can be observed, reducing the accuracy of genetic maps. This may be due to several causes: (1) genetic factors due to gametic or zygotic selection for pollen tube competition, preferential fertilization, chromosome translocation, etc. (Liu et al., 2010); (2) the genotype-dependency of DH, i.e., the different responses of the cross parents to DH method (Tanhuanpää et al., 2008); (3) somaclonal variation arising during DH production resulting in aneuploid production (Oleszczuk et al., 2011); and (4) high frequencies of clones via androgenesis (Oleszczuk et al., 2014).
Haploids have long been proved to be invaluable materials for basic genetic studies and they can be used for quick generation of quadruple, quintuple, sextuple, or higher order multiple mutant combinations, production of homozygous maternal gametophyte lethal mutants, and detection of gene conversion events during meiosis. (Wijnker et al., 2013; Ravi et al., 2014; Fulchar and Riha, 2016). A new breeding concept “reverse breeding” using DH has recently been proposed and successfully demonstrated in Arabidopsis, in which the approach also enabled the quick generation of a series of chromosome substitution lines (Wijnker et al., 2012).
DH lines produced from chromosome doubling of pollen/egg derived haploid plants of F1 plants only have one recombination opportunity in the first generation. To increase recombination, sometime F2 pollen/egg is used instead of F1's for haploid production. The costs involved in establishing a reliable protocol for a wide range of genotypes is also a limiting factor restricting the general application of DH to crop breeding. The centromere mediated genome elimination method reported in Arabidopsis and maize (Ravi and Chan, 2010; Kelliher et al., 2016) is particularly attractive because it is easy to adopt (only one crossing of the target genotype with the haploid inducer line is needed) and it produces haploid seeds without the necessity of embryo rescue (Ravi et al., 2014). Although it has not yet been successfully employed in other species probably due to the lack of cenH3 knockouts, the recent development of CRISPR-based genome edition targeting specific genes might be the solution to finally translate this technology into other crops (Britt and Kuppu, 2016).
RILs
RILs are the products of successive inbreeding over several generations to develop true breeding lines. In self-pollinated crops, it is usually achieved by a SSD approach which involves continuous selfing of individuals from an initial F2 population until the desired level of homozygosity is achieved. F8 RILs are often used for genetic studies, where 99% homozygosity is expected (Seymour et al., 2012). As a pure line population, RILs can also be replicated over locations and years and shared among different research groups. As with DHs, the expected genetic segregation ratio of RILs and the overall frequency of alleles for both dominant and codominant marker are 1:1. RILs are often used to map traits that differ between the parental lines. As a product of many meiotic cycles, RILs are expected to have higher recombination rate than DH, which is not only important for QTL mapping but also very useful to identify tightly linked markers. RILs are difficult to develop in crops that exhibit high levels of inbreeding depression (Madhusudhana, 2015). For these cross-pollinated crops, RIL development can be achieved by sib-mating but takes longer due to the many crosses involved. Being nearly homozygous, RILs are similar to DH lines and can be used to estimate QTL × E interaction and additive and epistatic effects of multiple loci contributions to trait expressions, but not dominant effects that require analysis of segregating populations such as, F2 or early backcross populations.
Given the fact that RILs are not complete homozygotes and each RIL may harbor confounding genetic variation with the threat of residual recombination in further generations, they may be less statistically powerful than DH for analyzing effects of particular loci. Distorted segregation can also be observed in RILs due to genetic factors resulting in gametic and zygotic selection (Liu et al., 2010).
Several studies compared DH and RIL populations for linkage map construction and QTL analysis. Somers and Humphreys (2009) found that wheat RILs had nearly twice the number of breakpoints per chromosome compared to DHs. This is consistent with theoretical expectation as RILs pass through more meiotic events in development and should have greater recombination creating more breakpoints along the chromosomes. Sorrells et al. (2011) also reported that the wheat reference population SynOpRILs showed a higher frequency of segregation than the SynOpDH lines. Zhao et al. (2010, 2013) observed significantly larger transgressive segregations for most agronomic traits, and higher QTL × E interactions in RIL compared to DH populations. The common thread in these studies is that the QTLs identified for the same trait are mostly different by using DH and RIL populations, even when the DHs and RILs were derived from the same cross. This was attributed to the quantitative traits being controlled by large numbers of QTLs that were sensitive to environmental effects (Zhao et al., 2013).
In Australia, cereals, oilseeds, and grain legumes are produced on a large scale for human consumption and livestock feed. Most of these cultivated crops are spring-type and self-pollinated that are especially suitable for developing RILs using FGCS.
NILs
NILs are pairs of lines having identical genetic background but contrasting at the targeted genomic region. NIL iso-lines segregate primarily on the targeted locus/gene while all other genes affecting the trait of interest are the same. Thus, any phenotypic difference between the iso-lines can be assigned to the targeted locus/gene. This allows the conversion of a quantitative trait into a Mendelian factor, making detailed mapping and gene discovery possible. NILs can be developed by repeated selfing using the HIF method (Tuinstra et al., 1997) or by repeated backcrossing (Figure 3). An HIF population is developed from a cross between two inbred lines. A progeny with target heterozygous locus is chosen by marker selection in F2 population. This is selfed and then from F3 on, the progenies that are heterozygous at the target locus are selected in each generation. By these processes, the genetic background becomes homozygous whereas the desired locus remains heterozygous. In the F8 generation, the heterozygotes are selfed to produce two NILs that are homozygous and contrasting (either positive or negative) at the targeted locus (Ma et al., 2012; Habib et al., 2016).
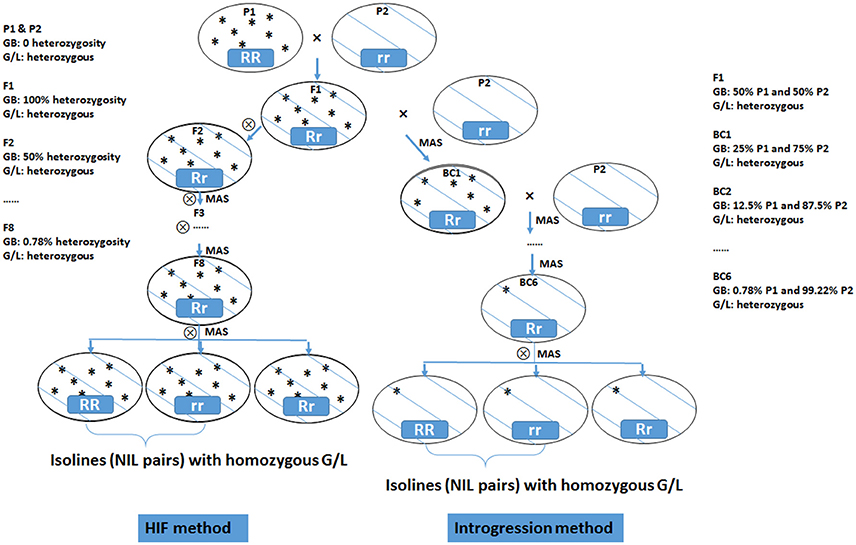
Figure 3. Diagram of heterogeneous inbred family (HIF) method and introgression method for developing near isogenic lines (NILs) from two inbred parental lines. GB, genetic background; G/L, target gene/locus, with RR representing dominant allele and rr representing recessive allele; MAS, marker assisted selection.
Using a backcross approach with marker selection for the trait of interest introgressed from the donor parent, it is possible to isolate the introgressed character from the donor genome in the recurrent parent. The introgression line is then selfed to generate the NIL pairs with or without the targeted locus. When screening with molecular markers, NILs are only polymorphic to the locus of interest.
NILs allow allelic variation at a targeted locus to be assessed enabling a detailed study of specific gene action at the physiological and developmental level (Moeller et al., 2014; Farré et al., 2016). Tagging traits are essential for functional genomics studies. NILs can also be used for the fine mapping of a gene, paving the way for gene isolation using positional cloning and allowing the development of tightly-linked genetic markers. The comparison of mRNA and protein expression in the NIL pairs can lead to the identification of candidate genes. If available, differentially expressed genes can be sequenced and mapped to a reference genome, to determine whether they are within a particular QTL chromosomal region (Ma et al., 2014; Barrero et al., 2015; Brechenmacher et al., 2015).
Preliminary QTL mapping by using DH or RIL populations has only limited resolution. Markers obtained from such studies may not be reliably used in tagging the targeted loci. Variations in genetic backgrounds of the mapping populations may interfere with the accurate phenotyping of a targeted trait. Thus, lines containing the same targeted allele may exhibit different phenotypes (Zheng et al., 2015). NIL derived fine mapping populations with fixed genetic backgrounds, except for the targeted genomic region, are suitable for gene discovery and for developing tightly linked markers. Fine mapping populations can be derived from NILs using heterozygous lines in advanced generations (for example, F8 or BC7) during development of the NILs. Usually a single heterozygous plant is used to be further advanced two or three more generations, to generate a large population allowing more recombination events within the targeted QTL region. The fine mapping population can then be phenotyped to establish the presence or absence of the QTL in that particular region, and ultimately reduce the QTL to a single or a few genes which can then be tested using functional genomics (Kooke et al., 2012; Zheng et al., 2015).
Like RILs, NILs require many generations to develop. Using FGCS can reduce the time needed for this. In FGCS, a moderate environmental stress level is used to promote reproduction whilst at the same time allowing enough vegetative growth for seed production. The volume of potting mix, temperature, and light conditions all affect the stress level and therefore affect the durations of the plant generation cycle as well as the number of seeds produced per plant. For NIL development, a larger number of progenies in each generation is required than is the case for RIL where only one seed is needed, hence a slightly larger potting soil volume is necessary to produce more seeds for each line, allowing selection for heterozygous or homozygous lines from them. Although this reduces stress level which delays flowering, a reasonably short generation cycle can still be achieved using the FGCS (Zheng et al., 2013).
NILs can only be used to study one locus at a time. The candidate locus typically comes from preliminary QTL mapping studies by using RILs or DH lines (Table 3). For crops with a large genome size, such as, wheat, many generations are needed to advance the lines to near-homozygosity allowing precise genetic mapping of the gene. Linkage drag, the simultaneous transfer of closely linked undesired genetic factor together with the targeted locus, can be a potential problem in the development of NILs (Madhusudhana, 2015).
Potential New Approaches
Fast development of pure lines using DH or FGCS can be improved. In DH, genotype-dependency for haploid production is a major obstacle for its wide application. Research aimed at achieving DH production independent of the genotypes will improve the efficiency of the technology. The synthesizing DH methodology is only suitable for (allo)polyploids that can produce viable unreduced restitution gametes (Zhang et al., 2011). The CenH3 based haploid production system reported in Arabidopsis and maize (Ravi and Chan, 2010; Kelliher et al., 2016) is promising, but has not yet been successfully used in other crops.
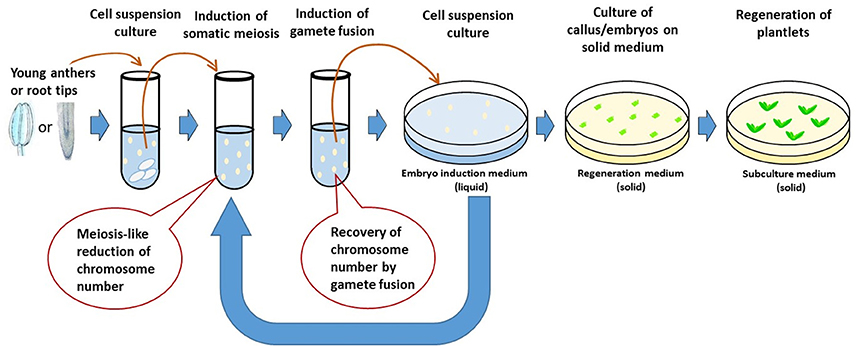
De La Fuente et al. (2013) proposed a concept of rapid generation cycling of meiosis and mitosis. The procedure uses plant cell culture system. Single somatic cells are induced to undergo meiosis to generate gametes which can be artificially genome-doubled to produce DH lines; or the gametes can be fused to form diploid cells which, equivalent to a zygote, can be regenerated to produce plants or be induced again to undergo new meiosis to start a new generation cycle. The procedure can be repeated for many generations in an in vitro system. Marker assisted genomic selection will be needed for evaluation and selection of the gametes or zygotes in each generation. The system, if successful, allows increased efficiency of pure line production and genetic introgression. It not only shortens each generation cycle, but also eliminates the process of multi-generation crosses. Major technical challenges involved in developing such a system include somatic meiosis induction, cell fusion, and regeneration plants from single cells. Although somatic meiosis (Yoshida and Yamaguchi, 1973; Chen et al., 2000, 2001), gamete fusion (Faure et al., 1994; Ponya et al., 2004; Uchiumi et al., 2007; Kranz and Scholten, 2008) and single cell regeneration (Vasil, 1983; Yang et al., 1991; Birnbaum and Sánchez Alvarado, 2008; Ikeuchi et al., 2013) have been reported in plants, there are few reports of a single crop species undergoing all these processes successfully. In addition, the efficiency (how quickly and how many normal gametes or zygotes can be produced) of each step remains unclear. Therefore, the concept of in vitro meiosis and mitosis has not yet realized in practice.
Interestingly, each of the necessary steps involved in the in vitro meiosis and mitosis system (IVMMS) has been reported as successful in wheat, for example, cell culture in suspension (Yang et al., 1991; Biesaga-Kos'Cielniak et al., 2008; Dong et al., 2010), somatic meiosis (Knott, 1956), gamete fusion (Kranz and Lorz, 1993; Zhou et al., 2002; Ge et al., 2006) and regeneration from somatic zygotes (Li et al., 1992; Kovács et al., 1995; Xia and Chen, 1996), which may be integrated to realize the concept of IVMMS in the crop (Figure 4). As somatic meiosis was reported mostly in either root tip cells (Mehar and Dhiman, 1986) or pre-meiotic cells (Knott, 1956), root tip cells and pollen mother cells can be used and treated with different combinations of somatic meiosis inducers (e.g., purine or pyrimidine derivatives or chloramphenicol) to induce somatic meiosis. The resulting cell culture can then be treated [e.g., with polyethylene glycol (PEG) or electric shock] to induce somatic fertilization. The fused cells may either be regenerated to plants or be induced again and again to complete more generations of meiosis and mitosis before regeneration to pure line plants. Cytogenetic studies at each of the step allows chromosome number of the cells to be determined, thus identifying the efficiency of somatic meiosis and somatic fertilization.
The IVMMS technology, if successful, can be integrated into breeding programs and make a new step-change for accelerating the breeding process. The plants resulting from IVMMS are expected to go through the similar recombination events, thus will have similar diversity, as those developed from conventional breeding. Along with the advent of new technologies, such as, single cell genomic sequencing (Macaulay and Voet, 2014) and genome wide selection (Goddard and Hayes, 2007; Morrell et al., 2012), IVMMS may allow a complete laboratory pre-breeding scheme to be established in major crops. Genome-wide selection is advantageous for predicting breeding value with high accuracy, as variation in most important traits are determined by many genes. Genomic sequencing of single cell progenies produced in IVMMS will allow the selection to be done completely in vitro, without the need to grow the plant lines. The IVMMS technology could also shed light into plant evolution. Genome-wide sequencing in the resulting progenies from IVMMS could reveal details about the evolutionary mechanisms in the plants that drive genetic divergence in the progenies that share a common original parent.
High density marker maps have been developed in some crops (Li et al., 2016; Punnuri et al., 2016), which makes genomic selection possible. Together with fast development of pure lines, the efficiency of crop genetic studies and breeding are expected to be significantly improved in the future.
Author Contributions
GY and HL conceived the ideas and developed the draft. HW, ZL, YW, DM, JH, and CL commented and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the Yitpi Foundation and COGGO for their partial financial support toward the publication of this manuscript.
References
Asif, M. (2013). Progress and Opportunities of Doubled Haploid Production. Cham: Springer International Publishing.
Asif, M., Eudes, F., Randhawa, H., Amundsen, E., and Spaner, D. (2014a). Induction medium osmolality improves microspore embryo genesis in wheat and triticale. In vitro Cell. Dev. Biol. Plant 50, 121–126. doi: 10.1007/s11627-013-9545-5
Asif, M., Eudes, F., Randhawa, H., Amundsen, E., and Spaner, D. (2014b). Phytosulfokine alpha enhances microspore embryo genesis in both triticale and wheat. Plant Cell Tissue Organ Cult. 116, 125–130. doi: 10.1007/s11240-013-0379-y
Atwell, B. J., Kriedemann, P. E., and Tumbull, C. G. N. (1999). Plants in Action: Adaptation in Nature, Performance in Cultivation. Melbourne, VIC: Macmillan Education Australia Pty Ltd.
Ausin, I., Alonso-Balanco, C., and Martínez-Zapater, J. (2005). Environmental regulation of flowering. Int. J. Dev. Biol. 49, 689–705. doi: 10.1387/ijdb.052022ia
Barrero, J. M., Cavanagh, C., Verbyla, K. L., Tibbits, J. F., Verbyla, A. P., Huang, B. E., et al. (2015). Transcriptomic analysis of wheat near-isogenic lines identifies Pm19-A1 and A2 as candidates for a major dormancy Qtl. Genome Biol 16:93. doi: 10.1186/s13059-015-0665-6
Basu, S. K., Datta, M., Sharma, M., and Kumar, A. (2011). Haploid production technology in wheat and some selected higher plants. Aust. J. Crop Sci. 5, 1087–1093.
Bentsink, L., and Koornneef, M. (2008). Seed dormancy and germination. Arabidopsis Book 6:E0119. doi: 10.1199/tab.0119
Biesaga-Kos'Cielniak, J., Kos'Cielniak, J., Filek, M., and Janeczco, A. (2008). Rapid production of wheat cell suspension cultures directly from immature embryos. Plant Cell Tissue Organ Cult. 94, 139–147. doi: 10.1007/s11240-008-9397-6
Birnbaum, K. D., and Sánchez Alvarado, A. (2008). Slicing across kingdoms: regeneration in plants and animals. Cell 132, 697–710. doi: 10.1016/j.cell.2008.01.040
Bohanec, B. (2009). “Doubled haploids via gynogenesis,” in Advances in Haploid Production in Higher Plants, eds A. Touraev, B. P. Forster and S. M. Jain (Dordrecht: Springer Science + Business Media B. V.), 35–44
Brechenmacher, L., Nguyen, T. H., Zhang, N., Jun, T., Xu, D., Mian, M. A. R., et al. (2015). Identification of soybean proteins and genes differentially regulated in near isogenic lines differing in resistance to aphid infestation. J. Proteome Res. 14, 4137–4146. doi: 10.1021/acs.jproteome.5b00146
Britt, A. B., and Kuppu, S. (2016). Cenh3: an emerging player in haploid inudction technology. Front. Plant Sci. 7:357. doi: 10.3389/fpls.2016.00357
Broughton, S. (2011). The application of n-butanol improves embryo and green plant production in anther culture of australian wheat (Triticum aestivum L.) genotypes. Crop Pasture Sci. 62, 813–822. doi: 10.1071/CP11204
Broughton, S., Sidhu, P. K., and Davies, P. A. (2014). In vitro culture for doubled haploids: tools for molecular breeding. Methods Mol. Biol. 1145, 167–189. doi: 10.1007/978-1-4939-0446-4_14
Cegielska-Taras, T., Szala, L., Matuszczak, M., Babula-Skowronska, D., Mikolajczyk, K., Poplawska, W., et al. (2015). Doubled haploids as a material for biotechnological manipulation and as a modern tool for breeding oilseed rape (Brassica napus L.). Biotechnologia 96, 7–18. doi: 10.5114/bta.2015.54169
Cerdán, P. D., and Chory, J. (2003). Regulation of flowering time by light quality. Nature 423, 881–885. doi: 10.1038/nature01636
Chawla, H. S. (2002). Introduction to Plant Biotechnology, 2nd Edn. Enfield, NH: Science Publishers Inc.
Chen, Y., Zhang, L., Geng, Y., and Cheng, Z. (2001). Meiosis-like reduction during somatic embryogenesis of Arabidopsis thaliana. In vitro Cell. Dev. Biol. Plant 37, 654–657. doi: 10.1007/s11627-001-0114-y
Chen, Y., Zhang, L., Zhou, Y., Geng, Y., and Chen, Z. (2000). Inducing somatic meiosis-like reduction at high frequency by caffeine in root-tip cells of vicia faba. Mutation Res. 452, 67–72. doi: 10.1016/S0027-5107(00)00045-2
Choi, J., Hyun, Y., Kang, M. J., In Yun, H., Yun, J. Y., Lister, C., et al. (2009). Resetting and regulation of flowering locus c expression during Arabidopsis reproductive development. Plant J. 57, 918–931. doi: 10.1111/j.1365-313X.2008.03776.x
CIMMYT (2010). Doubled Haploids: A Simple Method To Improve Efficiency Of Maize Breeding. 2016. Available online at: www.Youtube.Com/Watch?V=V2joeuzjjrg (Accessed September 18, 2017).
Coe, E. H. (1959). A line of maize with high haploid frequency. Am. Nat. 93, 381–382 doi: 10.1086/282098
Crevillén, P., Yang, H., Cui, X., Greeff, C., Trick, M., Qiu, Q., et al. (2014). Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515, 587–590. doi: 10.1038/nature13722
Croser, J. S., Lulsdorf, M. M., Davies, P. A., Clarke, H. J., Bayliss, K. L., Mallikarjuna, N., et al. (2007). Toward doubled haploid production in the fabaceae: progress, constraints, and opportunities. Crit. Rev. Plant Sci. 25, 139–157. doi: 10.1080/07352680600563850
Cummings, I. G., Reid, J. B., and Koutoulis, A. (2007). Red to far-red ratio correction in plant growth chambers–growth responses and influence of thermal load on garden pea. Physiol. Plant. 131, 171–179. doi: 10.1111/j.1399-3054.2007.00958.x
De La Fuente, G. N., Frei, U. K., and Lübberstedt, T. (2013). Accelerating plant breeding. Trends Plant Sci. 18, 667–672. doi: 10.1016/j.tplants.2013.09.001
Dennis, E. S., and Peacock, W. J. (2009). Vernalization in cereals. J. Biol. 8:57. doi: 10.1186/jbiol156
Devaux, P., and Kasha, K. J. (2009). “Overview of barley doubled haploid production,” in Advances in Haploid Production in Higher Plants, eds A. Touraev, B. P. Forster, and S. M. Jain (Dordrecht: Springer Science + Business Media B.V.), 47–64.
Dong, J., Bowra, S., and Vincze, E. (2010). The development and evaluation of single cell suspension from wheat and barley as a model system; a first step towards functional genomics application. BMC Plant Biol. 10:239. doi: 10.1186/1471-2229-10-239
Dunwell, J. M. (2010). Haploids in flowering plants: origins and exploitation. Plant Biotechnol. J. 8, 377–424. doi: 10.1111/j.1467-7652.2009.00498.x
Dwivedi, S. L., Britt, A. B., Tripathi, L., Sharma, S., Upadhyaya, H. D., and Ortiz, R. (2015). Haploids: constraints and opportunities in plant breeding. Biotechnol. Adv. 33, 812–829. doi: 10.1016/j.biotechadv.2015.07.001
Ei-Hennawy, M. A., Abdalla, A. F., Shafey, S. A., and Al-Ashkar, I. M. (2011). Production of doubled haploid wheat lines (Triticum aestivum L.) using anther culture technique. Ann. Agric. Sci. 56, 63–72. doi: 10.1016/j.aoas.2011.05.008
Eudes, F., and Chugh, A. (2009). An “Overview of triticale doubled haploids,” in Advances in Haploid Production in Higher Plants, eds A. Touraev, B. P. Forster, and S. M. Jain (Dordrecht: Springer Science + Business Media B.V), 87–96.
Farré, A., Sayers, L., Leverington-Waite, M., Goram, R., Orford, S., Wingen, L., et al. (2016). Application of a library of near isogenic lines to understand context dependent expression of qtl for grain yield and adaptive traits in bread wheat. BMC Plant Biol. 16:161. doi: 10.1186/s12870-016-0849-6
Faure, J. E., Digonnet, C., and Dumas, C. (1994). An in vitro system for adhesion and fusion of maize gametes. Science 23, 1598–1600. doi: 10.1126/science.263.5153.1598
Ferrie, A. M. R., and Mollers, C. (2011). Haploids and doubled haploids in Brassica Spp. for genetic and genomic research. Plant Cell Tissue Organ Cult. 104, 375–386. doi: 10.1007/s11240-010-9831-4
Ferrie, A. M. R., Irmen, K. I., Beattie, A. D., and Rossnagel, B. G. (2014). Isolated microspore culture of oat (Avena sativa L.) for the production of doubled haploids: effect of pre-culture and post-culture conditions. Plant Cell Tissue Organ Cult. 116, 89–96. doi: 10.1007/s11240-013-0385-0
Forster, B. P., and Thomas, W. T. B. (2010). “Doubled haploids in genetics and plant breeding,” in Plant Breeding Reviews, Vol. 25, ed J. Janick (Oxford, UK: John Wiley & Sons), 57–88. doi: 10.1002/9780470650301.ch3
Forster, B. P., Haberle-Bors, E., Kasha, K. J., and Touraev, A. (2007). The resurgence of haploids in higher plants. Trends Plant Sci. 12, 368–375. doi: 10.1016/j.tplants.2007.06.007
Forster, B. P., Till, B. J., Ghanim, A. M. A., Huynh, H. O. A., Burstmayr, H., and Caligari, P. D. S. (2014). Accelerated plant breeding. Cab Reviews 9, 1–16. doi: 10.1079/PAVSNNR20149043
Fulchar, N., and Riha, K. (2016). Using centromere mediated genome elimination to elucidate the functional redundancy of candidate telomere binding proteins in Arabidopsis thaliana. Front. Genet. 6:349. doi: 10.3389/fgene.2015.00349
Ge, T. M., Lin, X. H., Qin, F. L., Yu, S. W., and Yu, Y. J. (2006). Protoplast electrofusion between common wheat (Triticum aestivum L.) and Italian ryegrass (Lolium Multiflorum Lam.) and regeneration of mature cybrids. In vitro Cell. Dev. Biol. Plant 42, 179–187. doi: 10.1079/IVP2005742
Germanà, M. A. (2006). Doubled haploid production in fruit crops. Plant Cell Tissue Organ Cult. 86, 131–146. doi: 10.1007/s11240-006-9088-0
Germanà, M. A. (2011). Anther culture for haploid and doubled haploid production. Plant Cell Tissue Organ Cult. 104, 283–300. doi: 10.1007/s11240-010-9852-z
Gil-Humanes, J., and Barro, F. (2009). “Production of doubled haploids in Brassica,” in Advances In Haploid Production In Higher Plants, eds A. Touraev, B. P. Forster, and S. M. Jain (Dordrecht: Springer Science + Business Media B.V.), 65–73.
Gilles, L. M., Khaled, A., Laffaire, J. B., Chaignon, S., Gendrot, G., Laplaige, J., et al. (2017). Loss of pollen-specific phospholipase not like dad triggers gynogenesis in maize. EMBO J. 36, 707–717. doi: 10.15252/embj.201796603
Goddard, M. E., and Hayes, B. J. (2007). Genomic selection. J. Anim. Breed. Genet. 124, 323–330. doi: 10.1111/j.1439-0388.2007.00702.x
Habib, A., Shabala, S., Shabala, L., Zhou, M. X., and Liu, C. J. (2016). Near-isogenic lines developed for a major Qtl on chromosome arm 4hl conferring fusarium crown rot resistance in barley. Euphytica 209, 555–563. doi: 10.1007/s10681-015-1623-9
Hilhorst, H. W. M. (2007). “Definitions and hypotheses of seed dormancy,” in Annual Plant Reviews Seed Development, Dormancy and Germination, Vol. 27, eds K. J. Bradford and H. Nonogaki (Oxford: Blackwell), 50–71.
Houben, A., Sanei, M., and Pickering, R. (2011). Barley doubled-haploid production by uniparental chromosome elimination. Plant Cell Tissue Organ Cult. 104, 321–327. doi: 10.1007/s11240-010-9856-8
Hu, H., Schrag, T. A., Peis, R., Unterseer, S., Schipprack, W., and Chen, S. (2016). The genetic basis of haploid induction in maize identified with a novel genome-wide association method. Genetics 202, 1267–1276. doi: 10.1534/genetics.115.184234
Hussain, B., Kha, M., Ali, Q., and Shaukat, S. (2012). Double haploid production is the best method for genetic improvement and genetic studies of wheat. Int. J. Agro Vet. Med. Sci. 6, 216–228. doi: 10.5455/ijavms.169
Ikeuchi, M., Sugimoto, K., and Iwase, A. (2013). Plant callus: mechanisms of induction and repression. Plant Cell 25, 3159–3173. doi: 10.1105/tpc.113.116053
Inagaki, M. N., and Tahir, M. (1990). Comparison of haploid production frequencies in wheat varieties crossed with Hordeum bulbosum L. and Maize. Jpn. J. Breed. 40 209–216. doi: 10.1270/jsbbs1951.40.209
Jensen, J. (1983). “Producing haploid plants by chromosome elimination,” in Cell and Tissue Culture Techniques For Cereal Crop Improvement, (Beijing: Science Press), 55–80.
Kang, J., Yim, S., Choi, H., Kim, A., Lee, K. P., Lopez-Moloina, L., et al. (2015). Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 6:8113. doi: 10.1038/ncomms9113
Kasha, K. J., and Kao, K. N. (1970). High frequency haploid production in barley (Hordeum vulgate L.). Nature 225, 874–876. doi: 10.1038/225874a0
Kelliher, T., Starr, D., Richbourg, L., Chintamanani, S., Delzer, B., Nuccio, M. L., et al. (2017). Matrilineal, a sperm-specific phospholipase, triggers maize haploid induction. Nature 542, 105–109. doi: 10.1038/nature20827
Kelliher, T., Starr, D., Wang, W., McCuiston, J., Zhong, H., Nuccio, M. L., et al. (2016). Maternal haploids are preferentially induced by Cenh3-tailswap transgenic complementation in maize. Front. Plant Sci. 7:414. doi: 10.3389/fpls.2016.00414
Kim, K., and Baenziger, P. S. (2005). A simple wheat haploid and doubled haploid production system using anther culture. In vitro Cell. Devel. Biol. Plant 4, 22–27. doi: 10.1079/IVP2004594
Kiviharju, E. M. (2009). “Anther culture derived doubled haploids in oat,” in Advances in Haploid Production in Higher Plants, eds A. Touraev, B. P. Forster, and S. M. Jain (Dordrecht: Springer Science + Business Media B. V.), 171–178.
Kiviharju, E., Moisander, S., and Laurilaand, J. (2005). Improved green plant regeneration rates from oat anther culture and the agronomic performance of some Dh lines. Plant Cell Tissue Organ Cult. 81, 1–9. doi: 10.1007/s11240-004-1560-0
Knott, D. R. (1956). A case of somatic reduction in a premeiotic cell in wheat. Can. J. Bot. 34, 831–832. doi: 10.1139/b56-065
Kooke, R., Wijinker, E., and Keurentjes, J. J. (2012). Backcross populations and near isogenic lines. Methods Mol. Biol. 871, 3–16. doi: 10.1007/978-1-61779-785-9_1
Kovács, M., Barnabás, B., and Kranz, E. (1995). Electro-fused isolated wheat (Triticum aestivum L.) gametes develop into multicellular structures. Plant Cell Rep. 15, 170–180. doi: 10.1007/BF00193715
Kranz, E., and Lorz, H. (1993). In vitro fertilization with isolated, single gametes results in zygotic embryogenesis and fertile maize plants. Plant Cell 5, 739–746. doi: 10.1105/tpc.5.7.739
Kranz, E., and Scholten, S. (2008). In vitro fertilization: analysis of early post-fertilization development using cytological and molecular techniques. Sex. Plant Reprod. 21, 67–77. doi: 10.1007/s00497-007-0060-x
Kumar, A. A., Mishra, P., Kumari, K., and Panigrahi, K. C. (2012). Environmental stress influencing plant development and flowering. Front. Biosci. S4, 1315–1324. doi: 10.2741/s333
Kumari, M., Clarke, H. J., Small, I., and Siddique, K. H. M. (2009). Albinism in plants: a major bottleneck in wide hybridization, androgenesis and doubled haploid culture. Crit. Rev. Plant Sci. 28, 393–409. doi: 10.1080/07352680903133252
Lan, X., Wei, Y., Liu, D., Yan, Z., and Zheng, Y. (2005). Inheritance of seed dormancy in tibetan semi-wild wheat accession Q1028. J. Appl. Genet. 46, 133–138. Available online at: http://jag.igr.poznan.pl/2005-Volume-46/2/pdf/2005_Volume_46_2-133-138.pdf
Lantos, C., Bona, L., Boda, K., and Pauk, J. (2014). Comparative analysis of in vitro anther- and isolated microspore culture in hexaploid triticale (× Triticosecale Wittmack) for androgenic parameters. Euphytica 197, 27–37. doi: 10.1007/s10681-013-1031-y
Laurie, D. A., and Bennett, M. D. (1988). The production of haploid wheat plants from wheat x maize crosses. Theor. Appl. Genet. 76, 393–397. doi: 10.1007/BF00265339
Lee, K. P., Piskurewicz, U., Turecková, V., Strnad, M., and Lopez-Molina, L. (2010). A seed coat bedding assay shows that Rgl2- dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc. Natl. Acad. Sci. U.S.A. 107, 19108–19113. doi: 10.1073/pnas.1012896107
Lefebvre, V., North, H., Frey, A., Sotta, B., Seo, M., Okamoto, M., et al. (2006). Functional analysis of Arabidopsis Nced6 and Nced9 genes indicates that aba synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45, 309–319. doi: 10.1111/j.1365-313X.2005.02622.x
Lentini, Z., Mussell, H., Mutschler, M. A., and Earle, E. D. (1988). Ethylene generation and reversal of ethylene effects during development in vitro of rapid-cycling Brassica campestris L. Plant Sci. 54, 75–81. doi: 10.1016/0168-9452(88)90057-X
Li, H., Singh, R. P., Braun, H., Pfeiffer, W. H., and Wang, J. (2013). Doubled haploids versus conventional breeding in CIMMYT wheat breeding programs. Crop Sci. 53, 74–83. doi: 10.2135/cropsci2012.02.0116
Li, H., Vikram, P., Singh, R. P., Kilian, A., Carling, J., Song, J., et al. (2016). A high density Gbs map of bread wheat and its application for dissecting complex disease resistance traits. BMC Genomics 16:216. doi: 10.1186/s12864-015-1424-5
Li, Z., Xia, G., and Chen, H. (1992). Somatic embryo genesis and plant regeneration from protoplast isolated from embryo genic cell suspension of wheat (Triticum aestivum). Plant Cell Tissue Organ Cult. 28, 79–85. doi: 10.1007/BF00039918
Linkies, A., Graeber, K., Knight, C., and Leubner-Metzger, G. (2010). The evolution of seeds. New Phytol. 186, 817–831. doi: 10.1111/j.1469-8137.2010.03249.x
Liu, H., Zwer, P., Wang, H., Liu, C., Lu, Z., Wang, Y., et al. (2016). A fast generation cycling system for oat and triticale breeding. Plant Breed. 135, 574–579. doi: 10.1111/pbr.12408
Liu, X., Guo, L., You, J., Liu, X., He, Y., Yuan, J., et al. (2010). Progress of segregation distortion in genetic mapping of plants. Res. J. Agron. 4, 78–83. doi: 10.3923/rjagr.2010.78.83
Lulsdorf, M. M., Croser, J. S., and Ochatt, S. (2011). “Androgenesis and doubled-haploid production in food legumes,” in Biology and Breeding of Food Legumes, eds A. Pratap and J. Kumar (Oxfordshire: CAB International), 159–177.
Ma, J., Stiller, J., Zhao, Q., Feng, Q., Cavanagh, C., Wang, P., et al. (2014). Transcriptome and allele specificity associated with A 3bl locus for fusarium crown rot resistance in bread wheat. PLoS ONE 9:E113309. doi: 10.1371/journal.pone.0113309
Ma, J., Yan, G. J., and Liu, C. J. (2012). Development of near-isogenic lines for a major Qtl on 3bl conferring fusarium crown rot resistance in hexaploid wheat. Euphytica 183, 147–152. doi: 10.1007/s10681-011-0414-1
Macaulay, I. C., and Voet, T. (2014). Single cell genomics: advances and future perspectives. PLoS Genet. 10:E1004126. doi: 10.1371/journal.pgen.1004126
Madhusudhana, R. (2015). “Linkage mapping,” in Sorghum Molecular Breeding, eds R. Madhusudhana, P. Rajendrakumar, and J. V. Patil (New Delhi: Springer), 47–70.
Makowska, K., and Oleszczuk, S. (2014). Albinism in barley androgenesis. Plant Cell Rep. 33, 385–392. doi: 10.1007/s00299-013-1543-x
Maluszynski, M., Kasha, K. J., Forster, B. P., and Szarejko, I. (2003). Doubled Haploid Production in Crop Plants: A Manual. Dordrecht: Kluwer Academic Publishers.
Marcinska, I., Nowakowska, A., Skrzypek, E., and Czyczylo-Mysza, I. (2013). Production of double haploids in oat (Avena sativa L.) by pollination with maize (Zea mays L.). Central Eur. J. Biol. 8, 306–313. doi: 10.2478/s11535-013-0132-2
Mehar, P. N., and Dhiman, N. (1986). Induced meiotic reductions in root-tips. Cytogolia 51, 439–472.
Mishra, R., Jwala, G., and Rao, N. (2016). In-vitro androgenesis in rice: advantages, constraints and future prospects. Rice Sci. 23, 57–68. doi: 10.1016/j.rsci.2016.02.001
Moeller, C., Evers, J. B., and Rebetzke, G. (2014). Canopy architectural and physiological characterization of near-isogenic wheat lines differing in the tiller inhibition gene tin. Front. Plant Sci. 5:617. doi: 10.3389/fpls.2014.00617
Molnár, Z., Virág, E., and Ördög, V. (2011). Natural substances in tissue culture media of higher plants. Acta Biologica Szegediensis 55, 123–127. Available online at: https://www2.sci.u-szeged.hu/ABS/2011/Acta%20HP/55123.pdf
Morrell, P. L., Buckler, E. S., and Ross-Ibarra, J. (2012). Crop genomics: advances and applications. Nat. Rev. Genet. 13, 85–96. doi: 10.1038/nrg3097
Murovec, J., and Bohanec, B. (2012). “Haploids and doubled haploids in plant breeding,” in Plant Breeding, ed I. Abdurakhmonov (Rijeka: Intech), 87–106.
Niu, Z., Jiang, A., Hammad, W. A., Oladzadabbasabadi, A., Xu, S., Mergoum, M., et al. (2014). Review of doubled haploid production in durum and common wheat through wheat × maize hybridization. Plant Breed. 133, 313–320. doi: 10.1111/pbr.12162
Ochatt, S. J., and Sangwan, R. S. (2008). In vitro shortening of generation time in Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 93, 133–137. doi: 10.1007/s11240-008-9351-7
Ochatt, S. J., and Sangwan, R. S. (2010). In vitro flowering and seed set: acceleration of generation cycles,” in Plant Cell Culture: Essential Methods, eds M. R. Davey and P. Anthony (Chichester: John Wiley & Sons), 97–110.
Ochatt, S., Pech, C., Grewal, R., Conreux, C., Lulsdorf, M., and Jacas, L. (2009). Abiotic stress enhances androgenesis from isolated microspores of some legume species (Fabaceae). J. Plant Physiol. 166, 1314–1328. doi: 10.1016/j.jplph.2009.01.011
Ochatt, S. J., Sangwan, R. S., Marget, P., Ndong, Y. A., Rancillac, M., and Perney, P. (2002). New Approaches towards the shortening of generation cycles for faster breeding of protein legumes. Plant Breed. 121, 436–440. doi: 10.1046/j.1439-0523.2002.746803.x
Ohnishi, T., Yoshino, M., Yamakawa, H., and Kinoshita, T. (2011). The biotron breeding system: a rapid and reliable procedure for genetic studies and breeding in rice. Plant Cell Physiol. 52, 1249–1257. doi: 10.1093/pcp/pcr066
Oleszczuk, S., Tyrka, M., and Zimny, J. (2014). The origin of clones among androgenic regenerants of hexaploid triticale. Euphytica 198, 325–336. doi: 10.1007/s10681-014-1109-1
Oleszczuk, S., Zimny, J., Rabiz-Swider, J., and Lukaszewski, A. J. (2011). Aneuploidy among androgenic progeny of hexaploid triticale (× Triticosecale Wittmack). Plant Cell Rep. 30, 575–586. doi: 10.1007/s00299-010-0971-0
Oliver, S. N., Finnegan, E. J., Dennis, E. S., Peacock, W. J., and Trevaskis, B. (2009). Verbalization-induced flowering in cereals is associated with changes in histone methylation at the Vernalaization1 gene. Proc. Natl. Acad. Sci. U.S.A. 106, 8386–8391. doi: 10.1073/pnas.0903566106
Pauk, J., Jancso, M., and Simon-Kiss, I. (2009). “Rice doubled haploids and breeding,” in Advances in Haploid Production in Higher Plants, eds A. Touraev, B. P. Forster and S. M. Jain (Dordrecht: Springer Science + Business Media B. V), 189–197.
Pioneer (2008). Fast Corn Technology. Available online at: https://www.pioneer.com/home/site/about/news-media/media-kits/fast-corn-technology (Accessed October 10, 2017).
Ponya, Z., Kristo, ‘F. Z., Ciampolini, F., Faleri, C., and Cresti, M. (2004). Structural change in the endoplasmic reticulum during the in situ development and in vitro fertilisation of wheat egg cells. Sex. Plant Reprod. 17, 177–188. doi: 10.1007/s00497-004-0226-8
Prasanna, B. M., Chaikam, V., and Mahuku, G. (2012). Doubled Haploid Technology In Maize Breeding: Theory and Practice. Mexico: DF: CIMMYT.
Prigge, V., and Melchinger, A. E. (2012). Production of haploids and doubled haploids in maize. Methods Mol. Biol. 877, 161–172. doi: 10.1007/978-1-61779-818-4_13
Punnuri, S. M., Wallace, J. G., Knoll, J. E., Hyma, K. E., Mitchell, S. E., Buckler, E. S., et al. (2016). Development of a high-density linkage map and tagging leaf spot resistance in pearl millet using genotyping-by-sequencing markers. Plant Genome 9, 1–13. doi: 10.3835/plantgenome2015.10.0106
Qin, J., and Wang, H. (2002). The effectiveness of vernalization of immature embryos of winter wheat. Acta Agric. Bor. Sin. 17:143.
Raghavan, V. (2003). One hundred years of zygotic embryo culture investigations. In vitro Cell. Dev. Biol. Plant 39, 437–442. doi: 10.1079/IVP2003436
Rahman, M., and De Jimenez, M. M. (2016). Behind the scenes of microspore-based double haploid development in Brassica napus: a review. J. Plant Sci. Mol. Breed. 5, 1–9. doi: 10.7243/2050-2389-5-1
Ravi, M., and Chan, S. W. (2010). Haploid plants produced by centromere-mediated genome elimination. Nature 464, 615–618. doi: 10.1038/nature08842
Ravi, M., Marimuthu, M. P., Tan, E. H., Maheshwari, S., Henry, I. M., Marin-Rodriguez, B., et al. (2014). A haploid genetics toolbox for Arabidopsis thaliana. Nat. Commun. 5:5334. doi: 10.1038/ncomms6334
Reed, S. M. (2004). “Haploid cultures,” in Plant Development and Biotechnology, eds R. N. Trigiano and D. J. Gray (Boca Raton, FL: Crc Press, Taylor & Francis Group), 225–234.
Ribalta, F. M., Croser, J. S., Erskine, W., Finnegan, P. M., Lulsdorf, M. M., and Ochatt, S. J. (2014). Antigibberellin-induced reduction of internode length favors in vitro flowering and seed-set in different pea genotypes. Biol. Plant. 58, 39–46. doi: 10.1007/s10535-013-0379-0
Riboni, M., Test, A. R., Massimo, G., Tonelli, C., and Conti, L. (2014). Environmental stress and flowering time. Plant Signal. Behav. 9:E29036. doi: 10.4161/psb.29036
Semagn, K., Bjørnstad, A., and Ndjiondjop, M. N. (2006). Principles, requirements and prospects of genetic mapping in plants. Afr. J. Biotechnol. 5, 2569–2587. Available online at: http://www.academicjournals.org/AJB; http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.1029.6534&rep=rep1&type=pdf
Seymour, D. K., Filiault, D. L., Henry, I. M., Monson-Miller, J., Ravi, M., Pang, A., et al. (2012). Rapid creation of arabidopsis doubled haploid lines for quantitative trait locus mapping. Proc. Natl. Acad. Sci. U.S.A. 109, 4227–4232. doi: 10.1073/pnas.1117277109
Sheldon, C. C., Hills, M. J., Lister, C., Dean, C., Dennis, E. S., and Peacock, W. J. (2008). Resetting of flowering locus c expression after epigenetic repression by vernalziation. Proc. Natl. Acad. Sci. U.S.A. 105, 2214–2219. doi: 10.1073/pnas.0711453105
Somers, D. J., and Humphreys, G. (2009). “State of Qtl detection and marker-assisted selection in wheat improvement,” in Wheat Science and Trade, ed B. G. Carver (Ames, IA: Wiley-Blackwell), 312.
Sorrells, M. E., Gustafson, J. P., Somers, D., Chao, S., Benscher, D., Guedira-Brown, G., et al. (2011). Reconstruction of the synthetic w7984 × opata m85 wheat reference population. Genome Biol. 54, 875–882. doi: 10.1139/g11-054
Sriskandarajah, S., Sameri, M., Lerceteau-Kohler, E., and Westerbergh, A. (2015). Increased recovery of green doubled haploid plants from barley anther culture. Crop Sci. 55, 2806–2812. doi: 10.2135/cropsci2015.04.0245
Suenaga, K., and Nakajima, K. (1989). Efficient production of haploid wheat (Triticum aestivum) though crosses between japanese wheat and maize (Zea Mays). Plan Cell Rep. 8, 263–266. doi: 10.1007/BF00274125
Tadesse, W., Tawkaz, S., Inagaki, M. N., Picard, E., and Baum, M. (2013). Methods and Applications Of Doubled Haploid Technology In Wheat Breeding. Syria: Icarda, Aleppo.
Tanhuanpää, P., Kalendar, R., Schulman, A. H., and Kiviharju, E. (2008). The first doubled haploid linkage map for cultivated oat. Genome 51, 560–569. doi: 10.1139/G08-040
Tenhola-Roininen, T., Immonen, S., and Tanhuanpaa, P. (2006). Rye doubled haploids as a research and breeding tool - a practical point of view. Plant Breed. 125, 584–590. doi: 10.1111/j.1439-0523.2006.01296.x
Tezuka, T., Yokoyama, H., Tanaka, H., Shiozaki, S., and Oda, M. (2012). Seed and embryo germination in Ardisia crenata. J. Bot. 2012:679765. doi: 10.1155/2012/679765
Touraev, A., Forster, B. P., and Jain, S. M. (2009). Advances In Haploid Production In Higher Plants, Vol. 346. Dordrecht: Springer Science + Business Media B. V.
Tuinstra, M. R., Ejeta, G., and Goldsbrough, P. B. (1997). Heterogeneous Inbred Family (Hif) analysis: a method for developing near-isogenic lines that differ At quantitative trait loci. Theor. Appl. Genet. 95, 1005–1011. doi: 10.1007/s001220050654
Uchiumi, T., Uemura, I., and Okamoto, T. (2007). Establishment of an in vitro fertilization system in rice (Oryza sativa L.). Planta 226, 581–589. doi: 10.1007/s00425-007-0506-2
Vasil, I. K. (1983). “Regeneration of plants from single cells of cereals and grasses,” in Journal of Plant Physiology, eds P. F. Lurquin and A. Kleinhofs (New York, NY: Plenum Press), 233–252.
Wang, H., Wang, Y., and Zhao, H. (2003). How to accelerate the process of plant genetic modification. J. Hebei Agric. Sci. 7, 50–56.
Wang, H., Xie, X., Sun, G., Zhao, Y., Zhao, H., Chai, J., et al. (1999). Fast Breeding Technique to Achieve Many Generations a Year in Plants. Vol. Patent Cn 1262031ahebei, China.
Wedzony, M., Zur, I., Krzewska, M., Dubas, E., Szechynska-Hebda, M., and Wasek, I. (2015). Doubled haploids in triticale,” in Triticale, ed F. Eudes (Cham: Springer Science + Business Media), 111–128.
Weyen, J. (2009). “Barley and wheat doubled haploids in breeding,” in Advances in Haploid Production in Higher Plants, eds A. Touraev, B. P. Forster, and S. M. Jain (Dordrecht: Springer Science + Business Media B.V.), 179–187.
Whitman, C. M., Heins, R. D., Cameron, A. C., and Carlson, W. H. (1998). Lamp type and irradiance level for daylight extensions influence flowering of Campanula carpatica ‘Blue Clips’, Coreopsis grandiflora ‘Early Sunrise’, and Coreopsis verticillata ‘Moonbeam’. J. Am. Soc. Hortic. Sci. 123, 802–807.
Wijnker, E., Velikkakam Jame, G., Ding, J., Becker, F., Klasen, J. R., Rawat, V., et al. (2013). The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. Elife 2:E01426 doi: 10.7554/eLife.01426
Wijnker, E., Van Dun, K., De Snoo, C. B., Lelivelt, C. L., Keurentjes, J. J., Naharudin, N. S., et al. (2012). Reverse breeding in Arabidopsis thaliana generates homozygous parental lines from a heterozygous plant. Nat. Genet. 44, 467–470. doi: 10.1038/ng.2203
Williams, P. H., and Hill, C. B. (1986). Rapid-cycling populations of Brassica. Science 232, 1385–1389. doi: 10.1126/science.232.4756.1385
Willis, C. G., Baskin, C. C., Baskin, J. M., Auld, J. R., Venable, D. L., Cavender-Bares, J., et al. (2014). The evolution of seed dormancy: environmental cues, evolutionary hubs, and diversification of the seed plants. New Phytol. 203, 300–309. doi: 10.1111/nph.12782
Woods, D. P. (2014). Memory of the vernalized state in plants including the model grass Brachypodium distachyon. Front. Plant Sci. 5:99. doi: 10.3389/fpls.2014.00099
Xia, G., and Chen, H. (1996). Plant regeneration from intergeneric somatic hybridization between Triticum aestivum L. and Leymus chinensis (Trin.) Tzvel. Plant Sci. 120, 197–203. doi: 10.1016/S0168-9452(96)04492-5
Yang, Y. M., He, D. G., and Scott, K. J. (1991). Establishment of embryo genic suspension cultures of wheat by continuous callus selection. Aust. J. Plant Physiol. 18, 445–452. doi: 10.1071/PP9910445
Yao, Y., Zhang, P., Liu, H., Lu, Z., and Yan, G. (2017). A fully in vitro protocol towards large scale production of recombinant inbred lines in wheat (Triticum aestivum L.). Plant Cell Tissue Organ Cult. 128, 655–661. doi: 10.1007/s11240-016-1145-8
Yao, Y., Zhang, P., Wang, H., Lu, Z., Liu, C., Liu, H., et al. (2016). How to advance up to seven generations of canola (Brassica napus L.) per annum for the production of pure line populations. Euphytica 209, 113–119. doi: 10.1007/s10681-016-1643-0
Yoshida, H., and Yamaguchi, H. (1973). Arrangement and association of somatic chromosomes induced by chloramphenicol in barley. Chromosoma 43, 399–407. doi: 10.1007/BF00406746
Zhang, L., Zhang, L., Luo, J., Chen, W., Hao, M., Liu, B., et al. (2011). Synthesizing double haploid hexaploid wheat populations based on a spontaneous alloploidization process. J. Genet. Genomics 38, 89–94. doi: 10.1016/j.jcg.2011.01.004
Zhao, X., Qin, Y., Jia, B., Kim, S., Lee, H., Eun, M., et al. (2010). Comparison and analysis of main effects, epistatic effects, and Qtl × environment interactions of Qtls for agronomic traits using Dh and Rils populations in rice. J. Crop Sci. Biotechnol. 13, 235–241. doi: 10.1007/s12892-010-0076-x
Zhao, X., Qin, Y., Jia, B., Kim, S., Lee, H., Eun, M., et al. (2013). Comparison and analysis of qtls, epistatic effects and Qtl × environment interactions for yield traits using Dh and rils populations in rice. J. Integr. Agric. 12, 198–208. doi: 10.1016/S2095-3119(13)60219-1
Zheng, Z., Ma, J., Stiller, J., Zhao, Q., Feng, Q., Choulet, F., et al. (2015). Fine mapping a large-effect Qtl conferring fusarium crown rot resistance on the long arm of chromosome 3b in hexaploid wheat. BMC Genomics 16:850. doi: 10.1186/s12864-015-2105-0
Keywords: selfed pure lines, recombinant inbred lines (RILs), near isogenic lines (NILs), doubled haploid (DH), fast generation cycling system (FGCS), crop breeding
Citation: Yan G, Liu H, Wang H, Lu Z, Wang Y, Mullan D, Hamblin J and Liu C (2017) Accelerated Generation of Selfed Pure Line Plants for Gene Identification and Crop Breeding. Front. Plant Sci. 8:1786. doi: 10.3389/fpls.2017.01786
Received: 10 August 2017; Accepted: 02 October 2017;
Published: 24 October 2017.
Edited by:
Chengdao Li, Murdoch University, AustraliaReviewed by:
Ravi Maruthachalam, Indian Institute of Science Education and Research, Thiruvananthapuram, IndiaZhanguo Xin, Plant Stress and Germplasm Development Unit, USDA-ARS, United States
Copyright © 2017 Yan, Liu, Wang, Lu, Wang, Mullan, Hamblin and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guijun Yan, guijun.yan@uwa.edu.au
†These authors have contributed equally to this work.
 Guijun Yan
Guijun Yan