
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 18 August 2020
Sec. Plant Metabolism and Chemodiversity
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.01273
This article is part of the Research TopicNew Insights Into Seed Metabolites: From Research to ApplicationView all 8 articles
Predicted climate change is widely cited to significantly reduce yields of the major cereal crop species in a period where demand is rapidly rising due to a growing global population. This requires exhaustive research to develop genetic resources in order to address the expected production deficiencies which will largely be driven by abiotic stress. Modification of multiple genes is an approach that can address the predicted challenges; however, it is time-consuming and costly to modify multiple genes simultaneously. Transcription factors represent a group of proteins regulating multiple genes simultaneously and are therefore promising targets to concurrently improve multiple traits concurrently, such as abiotic stress tolerance and grain size (a contributor to yield). Many studies have identified the complex role that transcription factors of multiple families have contributed toward abiotic stress tolerance or grain size, although research addressing both simultaneously is in its infancy despite its potential significance for cereal crop improvement. Here we discuss the potential role that transcription factors may contribute toward improving cereal crop productivity under adverse environmental conditions and offer research objectives that need to be addressed before the modification of transcription factors becomes routinely used to positively manipulate multiple target traits.
Rice, maize, and wheat collectively represent the primary food source for over half of the world’s population and are thus crucially important for global food security. However, with increased demand from a growing global population and an increasing fluctuation in crop productivity driven by climatic variability, food security is becoming increasingly vulnerable. Modest increases of 1°C in average growing season temperature for instance has been predicted to reduce global wheat yields anywhere between 4 and 8%, less for rice but greater yield losses are expected for maize, barley, and sorghum (Challinor et al., 2014; Liu B. et al., 2016; Zhao C. et al., 2017; Abhinandan et al., 2018). Already average temperature increases between 1981 and 2002 of ~0.4°C have resulted in estimated combined annual yield losses of 42 Mt for wheat, barley, and maize (Lobell and Field, 2007). Alternatively, the frequency of drought is projected to intensify, and 20% of global agricultural land is affected by salinity stress that is expected to double by 2050 both of which are considered to be the two primary threats to future agriculture (Lesk et al., 2016; Majeed et al., 2019; Nutan et al., 2019b). Our ability to ensure food security in the face of these threats therefore lies in our ability to improve cereal yields which in turn are reflections of two major and genetically manipulable morphological components: (1) number of grains/m2 and, (2) individual grain weight (Xing and Zhang, 2010; Kennedy et al., 2016; Li et al., 2018; Ji et al., 2019). Individual grain weight is a reflection of grain size which is the culmination of complex biological processes and pathways controlled by polygenes acting pre- and post-anthesis to determine the maximal grain size that can be achieved (Ji et al., 2019). Addressing food security could therefore focus on increasing and/or maintaining grain size under adverse environmental conditions as a strategy to improve cereal yields.
In general, grain size is co-ordinately controlled by cell expansion and proliferation in the developing endosperm and floral tissues (lemma, palea) surrounding the developing grain, determining the ‘sink’ capacity of the grain (Li et al., 2018). In cereals, cell proliferation precedes cell expansion to some extent, beginning at fertilization and ceasing 15–25 days later (Evers and Millar, 2002; Farooq et al., 2014; Li et al., 2018; Bian, 2019). This period of grain development predetermines the maximum size of the grain along the longitudinal and transverse axes and is quite sensitive to abiotic stress; for example in wheat, drought decreases endosperm cell proliferation reducing this ‘sink’ capacity (Setter and Flannigan, 2001). Alternatively, cell expansion relates to the accumulation of dry matter (protein, carbohydrates, and lipids) in the developing grain which is related to photo-assimilate production and transport. This accumulation of dry matter is the major contributor to final grain size and weight and is reported to begin 5–7 days after fertilization and ceases at physiological maturity (Coventry et al., 2003; Sreenivasulu et al., 2004). Adverse conditions during cell expansion primarily damages and/or reduces the photosynthetic area reducing the production and translocation of photo-assimilates to developing grains. Optimization of source–sink pathways therefore represents a promising avenue towards contributing to grain size and weight improvement under both optimal and adverse environmental conditions.
In rice, maize, wheat, and barley thousands of quantitative trait loci (QTL) influencing grain size have collectively been detected, yet only a small fraction of underlying candidate genes have been functionally annotated using advanced molecular approaches such as gene cloning with the majority of studies in rice (Ayoub et al., 2002; Huang et al., 2013; Walker et al., 2013; Chen et al., 2016; Yu et al., 2017; Azizi et al., 2019; Li et al., 2019; Wang Q. et al., 2019; Watt et al., 2019). Broadly, the regulatory pathways involved in grain size regulation are represented by: hormone signaling, IKU pathway, G-protein signaling, ubiquitin–proteasome pathway, the mitogen-activated protein kinase pathway and transcription factors (Li and Li, 2016; Azizi et al., 2019). Genes underlying these pathways can be negative or positive regulators of grain size as a result of allele specific epistatic interactions which can substantially influence their effect; for example OsGLA1 confers a positive effect on grain length and weight driven by a single SNP (Wang T. et al., 2019). Alternatively, TaDA1 in wheat, a ubiquitin receptor negatively regulates grain size and weight by restricting cell proliferation in the maternal integuments via the ubiquitin-proteasome pathway (Liu et al., 2020).
Due to culinary preferences for different sized rice, the majority of grain size research has targeted this staple food crop, particularly the genetic engineering and functional analysis style research. Li et al. (2018) and Azizi et al. (2019) synthesize our current understanding of the pathways and genes involved in grain size regulation in rice which, due to the conservation of gene order and function between the major cereal crop species, is likely to reflect a similar genetic control of grain size in maize, wheat, and barley (Li et al., 2010; Richards et al., 2016). Despite our general understanding of the genetic control of grain size, it is the role of stress inducible transcription factors and their induction that offer promising alternative strategies to maintain and improve grain size and yield under adverse conditions primarily through their stimulation of numerous stress responsive genes (Nakashima et al., 2007; Nutan et al., 2019b).
The ability of a plant to perceive stressful conditions and subsequently respond by inducing stress responsive genes is triggered partly by transcription factors and their interaction with cis-acting promoter elements of genes in complex regulatory networks (Gujjar et al., 2014; Rahman et al., 2019). Transcriptional analyses have identified thousands of differentially expressed genes resulting from single and combined abiotic stresses indicating the complexity of stress response and gene expression regulation (Li et al., 2017; Xiong et al., 2017; Osthoff et al., 2019; Pradhan et al., 2019). Multiple transcription factor families have been implicated in abiotic stress response namely the: DREB (dehydration-responsive element binding), ABRE/ABF (ABA-responsive element), MYB (myeloblastosis), NAC, bZIP (basic leucine zipper), and WRKY gene families (Ambawat et al., 2013; Nuruzzaman et al., 2013; Gujjar et al., 2014; Rahman et al., 2019). Often a single transcription factor is able to induce gene expression in response to multiple abiotic stress conditions. The wheat TaNAC2-5A transcription factor, for example, is induced by drought, salt, cold, and abscisic acid (ABA) treatment. Overexpression of TaNAC2-5A in Arabidopsis simultaneously improved drought, salinity, and freezing tolerance (Mao et al., 2012). Interestingly, stress induced TaNAC2-5A activity enhanced the expression of DREB2A and ABI5 transcription factors. It has been shown that stress-induced and constitutive overexpression of DREB2A in wheat and barley improved tolerance to drought and cold stress due to increased expression of late embryogenesis abundant (LEA) genes encoding dehydrins and cold-responsive proteins that contribute to membrane stability as well as other DREB family genes, further indicating the complexity of stress response and regulatory control (Morran et al., 2011). Similarly, ZmSNAC1 was found to improve stress tolerance through reduced dehydration, possibly through the NAC-DREB-LEA regulatory module as evidenced in wheat, barley, Arabidopsis, and rice (Lu et al., 2012; Hong et al., 2016).
Aside from the well understood role transcription factors play in plant recognition and response to abiotic stress, numerous studies across the major cereal crop species, particularly rice, have dissected their involvement in grain size modulation (Table 1). They co-ordinate cell proliferation and expansion processes not only in the developing grain itself but also in the surrounding floral tissues, lemma, and palea which additionally limit grain size. The antagonistic behavior of two bHLH-type transcription factors (PGL1 and APG) for example, regulates cell elongation in the lemma/palea of rice by the heterodimerization of the two encoded proteins, regulating grain length (Heang and Sassa, 2012). A major gene designated WIDE AND THICK GRAIN 1 (WTG1) in rice encodes an otubain-like protease involved in the ubiquitin–proteasome pathway regulating grain size via cell expansion that is reportedly regulated by the transcription factors ABF1 and ABI5 (Huang et al., 2017; Li et al., 2018; Zhang et al., 2018). An ortholog in wheat (TaWTG1-7B) is highly correlated with the expression of ABF2 of the ABRE transcription factor family (Zhang et al., 2018).
The NAC transcription factor family is one of the largest, and numerous NAC genes have been implicated in the control of grain size via multiple pathways driven by the diversity of subdomains and their variable protein–protein interactions and DNA-binding activities (Olsen et al., 2005; Dwivedi et al., 2019). The OsMED15A-OsNAC024/025 regulatory module for example, positively regulates GW2, GW5 (negative regulators of grain width) and D11 (positive regulator of grain length) in rice through the interaction of the mediator tail subunit of OsMAD15A with the two NAC transcription factors that promote the recruitment of additional transcriptional machinery to the promoters of OsNAC024/025 targets (Figure 1) (Dwivedi et al., 2019). In barley, HvNAC005 transcriptional activity is promoted by the presence of a conserved C-terminal motif reportedly involved in protein–protein interaction suggesting the importance of these types of interactions for the assembly of basic transcriptional apparatus (Christiansen et al., 2016). A signal mediating protein phosphatase 2C (PP2C) is reported to interact with AtNAP, a homolog of HvNAC005; thus PP2C represents a possible mediator initiating transcription of HvNAC005 targets. Alternatively, the PP2C–SnRK2 ABA-responsive complex is reported to enhance bZIP transcription factor activity in Arabidopsis suggesting that this same signaling pathway is active in barley (Hirayama and Umezawa, 2010; Zhang and Gan, 2012).
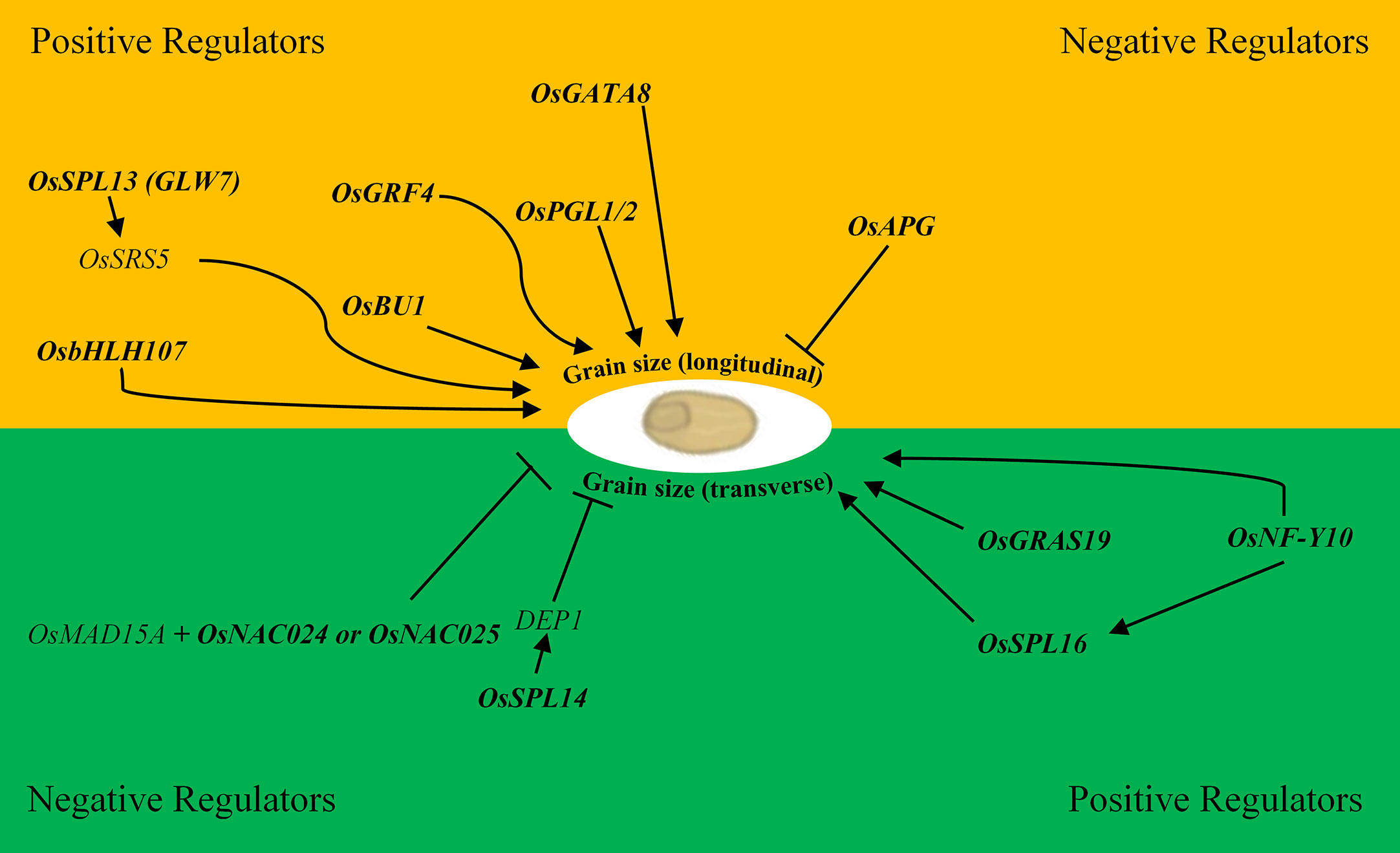
Figure 1 Positive and negative regulatory transcription factors of rice involved in grain size variation in the longitudinal (length; yellow) and transverse directions (width and thickness; green). Bold text represents transcription factors. Arrows represent a positive interaction, and blunt ends are negative interactions i.e. OsSPL16 positively regulates DEP1 which in-turn negatively regulates grain width.
Despite the diversity of transcription factors, their function, and their involvement in abiotic stress response and regulation of grain size, research marrying the two is surprisingly limited considering the contribution this knowledge could provide to improving cereal productivity. Overexpression of NACs for example, have significantly improved tolerance to drought, salinity, and cold stress in rice and maize (Nuruzzaman et al., 2013). OsNAC022 overexpression in rice was shown to significantly improve drought and salinity tolerance, although there was a significant negative effect on 1,000-grain weight, a reflection of grain size indicating the importance of identifying suitable transcription factors that can simultaneously improve both stress tolerance and grain size if transcription factors were going to be manipulated for trait improvement (Hong et al., 2016). This study however, drove overexpression via the constitutive maize ubiquitin promoter, an approach known to often confer undesirable phenotypes under optimal environmental conditions as was the case for constitutive overexpression of Ubi1:OsNAC6. It is possible that maintenance of 1,000-grain weight in this instance could have been achieved through the use of a stress inducible and/or tissue specific promoter coupled to OcNAC022 such as rice Wsi18 gene promoter which exhibits strong stress induced expression and elevated activity in developing grains specifically (Nakashima et al., 2007; Yi et al., 2011). A GATA-transcription factor, OsGATA8 has successfully been manipulated, and constitutive expression has been proven to improve drought and salinity tolerance while simultaneously increasing grain length and TGW with no undesirable phenotypes in rice and Arabidopsis transgenic lines (Nutan et al., 2019b). It was shown that OsGATA8 regulated genes involved in reactive-oxygen scavenging enzymes, chlorophyll-biosynthesis enzymes as well as other transcription factors such as OsDREB1A.
A WRKY transcription factor OsWRKY78 positively regulated grain width and was upregulated by ABA and salinity, but downregulated by cold (Zhang et al., 2011). OsSPL14 positively regulates DEP1 (Figure 1) in-turn influencing grain size, plant architecture, and yield (Jiao et al., 2010; Zhou and Luo, 2013; Stief et al., 2014; Zheng and Qu, 2015; Yue et al., 2017; Watt et al., 2019). Promisingly, OsSPL14 was recently shown to regulate OsTB1 which acted as a negative regulator of OsWRKY94 and suppressed the cold stress induced expression of OsMADS57 indicating the potential to manipulate OsSPL14 to simultaneously improve abiotic stress tolerance and grain size (Chen et al., 2018; Nutan et al., 2019a). The R2R3-subclass of the MYB transcription factor family is primarily involved in developmental processes and abiotic stress response compared to the other three subclasses suggesting there is potential to improve both characteristics by focusing on the manipulation of this subclass specifically (Ambawat et al., 2013; Hou et al., 2018). OsGAMYB for example, an R2R3-MYB transcription factor involved in gibberellic acid signalling, has been linked to both grain size variation and response to salinity stress possibly through miR159 induction in separate studies (Zhao Y. et al., 2017; Liu et al., 2019). This suggests that it may be possible to manipulate stress response and grain size co-ordinately by targeting transcription factors and/or components of the regulatory network such as microRNAs, the main targets of which are transcription factors (Zhou and Luo, 2013).
The relationship between grain size, yield and the negative effect abiotic stress has on these traits requires further research in order to address the expected threats associated with climate change. The complex regulatory pathways involved in grain size and abiotic stress response suggest that modification of a single transcription factor may offer potential strategies to improving grain size, yield, and abiotic stress tolerance simultaneously as observed by Nutan et al. (2019b). Research needs to address the combined effect of stress response and grain size to transcription factor manipulation to identify on a species-specific level, suitable candidates for trait improvement, something that can currently only be conferred based on a limited number of studies. As between Arabidopsis and tomato (Zhou and Luo, 2013), microRNAs which in-turn regulate transcription factors can confer either negative or positive effects on trait expression depending on the species background; in cereals this would necessitate further research, but the relative conservation of gene order and function between the major cereal crop species may enable rapid transferability of knowledge from one cereal species to another. However, care must be taken if a transcription factor is to be manipulated through microRNA modification due to the large gene networks these microRNAs target; miR156 for example, regulates a reported 11 OsSPL genes including OsSPL14 thus the manipulation of miR156 may confer a positive or negative phenotypic response depending on the SPL gene target and necessitates a greater level of understanding (Xie et al., 2006; Wang and Wang, 2015). In addition to alternative targets of transcription factor networks i.e. microRNAs, constitutive overexpression of transcription factors as a tool for trait improvement has been proven to induce negative pleiotropic effects on phenotypes in many instances, thus research should address trait response using stress induced and/or tissue specific promoters to improve commercial viability of certain transcription factor modifications.
CW came up with the initial concept and wrote the initial draft in conjunction with GZ and CL. All authors contributed to the article and approved the submitted version.
CW receives a postgraduate scholarship from the Grains Research and Development Corporation (GRDC), project code UMU1903-003RSX.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The corresponding author CW would like to thank all co-authors for their guidance. Additionally, all authors would like to thank the GRDC for funding.
Abhinandan, K., Skori, L., Stanic, M., Hickerson, N. M. N., Jamshed, M., Samuel, M. A. (2018). Abiotic stress signaling in wheat – An inclusive overview of hormonal interactions during abiotic stress responses in wheat. Front. Plant Sci. 9, 1–25. doi: 10.3389/fpls.2018.00734
Ambawat, S., Sharma, P., Yadav, N. R., Yadav, R. C. (2013). MYB transcription factor genes as regulators for plant responses: an overview. Physiol. Mol. Biol. Plants 19, 307–321. doi: 10.1007/s12298-013-0179-1
Ayoub, M., Symons, S. S., Edney, M. M., Mather, D. D. (2002). QTLs affecting kernel size and shape in a two-rowed by six-rowed barley cross. Theor. Appl. Genet. 105, 237–247. doi: 10.1007/s00122-002-0941-1
Azizi, P., Osman, M., Hanafi, M. M., Sahebi, M., Rafii, M. Y., Taheri, S., et al. (2019). Molecular insights into the regulation of rice kernel elongation. Crit. Rev. Biotechnol. 39, 904–923. doi: 10.1080/07388551.2019.1632257
Bian, J. (2019). Transcriptional dynamics of grain development in barley (Hordeum vulgare L.). Int. J. Mol. Sci. 20, 962–978. doi: 10.3390/ijms20040962
Challinor, A. J., Watson, J., Lobell, D. B., Howden, S. M., Smith, D. R., Chhetri, N. (2014). A meta-analysis of crop yield under climate change and adaptation. Nat. Clim. Change 4, 287–291. doi: 10.1038/nclimate2153
Che, R., Tong, H., Shi, B., Liu, Y., Fang, S., Liu, D., et al. (2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2, 1–7. doi: 10.1038/nplants.2015.1195
Chen, L., Xiong, G., Cui, X., Yan, M., Xu, T., Qian, Q., et al. (2013). OsGRAS19 may be a novel component involved in the brassinosteroid signaling pathway in rice. Mol. Plant 6, 988–991. doi: 10.1093/mp/sst027
Chen, J., Zhang, L., Liu, S., Li, Z., Huang, R., Li, Y., et al. (2016). The genetic basis of natural variation in kernel size and related traits using a four-way cross population in maize. PLoS One 11, 1–12. doi: 10.1371/journal.pone.0153428
Chen, L., Zhao, Y., Xu, S., Zhang, Z., Xu, Y., Zhang, J., et al. (2018). OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 218, 219–231. doi: 10.1111/nph.14977
Christiansen, M. W., Matthewman, C., Podzimska-Sroka, D., O’Shea, C., Lindemose, S., Møllegaard, N. E., et al. (2016). Barley plants over-expressing the NAC transcription factor gene HvNAC005 show stunting and delay in development combined with early senescence. J. Exp. Bot. 67, 5259–5273. doi: 10.1093/jxb/erw286
Coventry, S. J., Barr, A. R., Eglinton, J. K., McDonald, G. K. (2003). The determinants and genome locations influencing grain weight and size in barley. Aust. J. Agric. Res. 54, 1103–1115. doi: 10.1071/AR02194
Dwivedi, N., Maji, S., Waseem, M., Thakur, P., Kumar, V., Parida, S. K., et al. (2019). The Mediator subunit OsMED15a is a transcriptional co-regulator of seed size/weight–modulating genes in rice. Biochim. Biophys. Acta 1862, 1–17. doi: 10.1016/j.bbagrm.2019.194432
Evers, T., Millar, S. (2002). Cereal grain structure and development: Some implications for quality. J. Cereal Sci. 36, 261–284. doi: 10.1006/jcrs.2002.0435
Farooq, M., Wahid, A., Siddique, K. (2014). “Physiology of grain development in cereals,” in Handbook of Plant and Crop Physiology, 3rd ed. Ed. Pessarakli, M. (Boca Raton, FL: Taylor and Francis Group), 301–308.
Gujjar, R. S., Akhtar, M., Singh, M. (2014). Transcription factors in abiotic stress tolerance. Indian J. Plant Physiol. 19, 306–316. doi: 10.1007/s40502-014-0121-8
Heang, D., Sassa, H. (2012). An atypical bHLH protein encoded by POSITIVE REGULATOR OF GRAIN LENGTH 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breed. Sci. 62, 133–141. doi: 10.1270/jsbbs.62.133
Hirayama, T., Umezawa, T. (2010). The PP2C-SnRK2 complex: the central regulator of an abscisic acid signaling pathway. Plant Signal. Behav. 5, 160–163. doi: 10.4161/psb.5.2.10460
Hong, Y., Zhang, H., Huang, L., Li, D., Song, F. (2016). Overexpression of a stress-responsive nac transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 7, 1–19. doi: 10.3389/fpls.2016.00004
Hou, D., Cheng, Z., Xie, L., Li, X., Li, J., Mu, S., et al. (2018). The R2R3MYB Gene Family in Phyllostachys edulis: Genome-Wide Analysis and Identification of Stress or Development-Related R2R3MYBs. Front. Plant Sci. 9, 1–22. doi: 10.3389/fpls.2018.00738
Hu, J., Wang, Y., Fang, Y., Zeng, L., Xu, J., Yu, H., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8, 1455–1465. doi: 10.1016/j.molp.2015.07.002
Huang, R., Jiang, L., Zheng, J., Wang, T., Wang, H., Huang, Y., et al. (2013). Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci. 18, 218–226. doi: 10.1016/j.tplants.2012.11.001
Huang, K., Wang, D., Duan, P., Zhang, B., Xu, R., Li, N., et al. (2017). WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice. Plant J. 91, 849–860. doi: 10.1111/tpj.13613
Ji, X., Du, Y., Li, F., Sun, H., Zhang, J., Li, J., et al. (2019). The basic helix-loop-helix transcription factor, OsPIL15, regulates grain size via directly targeting a purine permease gene OsPUP7 in rice. Plant Biotechnol. J. 17, 1527–1537. doi: 10.1111/pbi.13075
Jia, S., Xiong, Y., Xiao, P., Wang, X., Yao, J. (2019). OsNF-YC10, a seed preferentially expressed gene regulates grain width by affecting cell proliferation in rice. Plant Sci. 280, 219–227. doi: 10.1016/j.plantsci.2018.09.021
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Kennedy, S. P., Bingham, I. J., Spink, J. H. (2016). Determinants of spring barley yield in a high-yield potential environment. J. Agric. Sci. 155, 60–80. doi: 10.1017/S0021859616000289
Lesk, C., Rowhani, P., Ramankutty, N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi: 10.1038/nature16467
Li, N., Li, Y. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. doi: 10.1016/j.pbi.2016.05.008
Li, Q., Li, L., Yang, X., Warburton, M. L., Bai, G., Dai, J., et al. (2010). Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol. 10, 143–158. doi: 10.1186/1471-2229-10-143
Li, P., Cao, W., Fang, H., Xu, S., Yin, S., Zhang, Y., et al. (2017). Transcriptomic profiling of the maize (Zea mays L.) leaf response to abiotic stresses at the seedling stage. Front. Plant Sci. 8, 1–13. doi: 10.3389/fpls.2017.00290
Li, N., Xu, R., Duan, P., Li, Y. (2018). Control of grain size in rice. Plant Reprod. 31, 1–15. doi: 10.1007/s00497-018-0333-6
Li, L., Li, X., Li, L., Schnable, J., Gu, R., Wang, J. (2019). QTL identification and epistatic effect analysis of seed size- and weight-related traits in Zea mays L. Mol. Breed. 39, 1–11. doi: 10.1007/s11032-019-0981-8
Lin, Z., Yan, J., Su, J., Liu, H., Hu, C., Li, G., et al. (2019). Novel OsGRAS19 mutant, D26, positively regulates grain shape in rice (Oryza sativa). Funct. Plant Biol. 46, 857–868. doi: 10.1071/FP18266
Liu, B., Asseng, S., Müller, C., Ewert, F., Elliott, J., Lobell, David, B., et al. (2016). Similar estimates of temperature impacts on global wheat yield by three independent methods. Nat. Clim. Change 6, 1130–1136. doi: 10.1038/nclimate3115
Liu, Q., Harberd, Nicholas, P., Fu, X. (2016). SQUAMOSA promoter binding protein-like transcription factors: Targets for improving cereal grain yield. Mol. Plant 9, 765–767. doi: 10.1016/j.molp.2016.04.008
Liu, C., Chen, K., Zhao, X., Wang, X., Shen, C., Zhu, Y., et al. (2019). Identification of genes for salt tolerance and yield-related traits in rice plants grown hydroponically and under saline field conditions by genome-wide association study. Rice 12, 88. doi: 10.1186/s12284-019-0349-z
Liu, H., Li, H., Hao, C., Wang, K., Wang, Y., Qin, L., et al. (2020). TaDA1, a conserved negative regulator of kernel size, has an additive effect with TaGW2 in common wheat (Triticum aestivum L.). Plant Biotechnol. J. 18, 1330–1342. doi: 10.1111/pbi.13298
Lobell, D. B., Field, C. B. (2007). Global scale climate–crop yield relationships and the impacts of recent warming. Environ. Res. Lett. 2, 1–17. doi: 10.1088/1748-9326/1082/1081/014002
Lu, M., Ying, S., Zhang, D.-F., Shi, Y.-S., Song, Y.-C., Wang, T.-Y., et al. (2012). A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep. 31, 1701–1711. doi: 10.1007/s00299-012-1284-2
Majeed, A., Muhammad, Z., Islam, S., Ahmad, H. (2019). Salinity imposed stress on principal cereal crops and employing seed priming as a sustainable management approach. Acta Ecol. Sin. 39, 280–283. doi: 10.1016/j.chnaes.2018.09.004
Mao, X., Zhang, H., Qian, X., Li, A., Zhao, G., Jing, R. (2012). TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 63, 2933–2946. doi: 10.1093/jxb/err462
Morran, S., Eini, O., Pyvovarenko, T., Parent, B., Singh, R., Ismagul, A., et al. (2011). Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 9, 230–249. doi: 10.1111/j.1467-7652.2010.00547.x
Nakashima, K., Tran, L.-S. P., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Nuruzzaman, M., Sharoni, A. M., Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4, 248–248. doi: 10.3389/fmicb.2013.00248
Nutan, K. K., Rathore, R. S., Tripathi, A. K., Mishra, M., Pareek, A., Singla-Pareek, S. L. (2019a). Integrating the dynamics of yield traits in rice in response to environmental changes. J. Exp. Bot. 71, 490–506. doi: 10.1093/jxb/erz364
Nutan, K. K., Singla-Pareek, S. L., Pareek, A. (2019b). The Saltol QTL-localized transcription factor OsGATA8 plays an important role in stress tolerance and seed development in Arabidopsis and rice. J. Exp. Bot. 71, 684–698. doi: 10.1093/jxb/erz368
Olsen, A. N., Ernst, H. A., Leggio, L. L., Skriver, K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Osthoff, A., Donà dalle Rose, P., Baldauf, J. A., Piepho, H.-P., Hochholdinger, F. (2019). Transcriptomic reprogramming of barley seminal roots by combined water deficit and salt stress. BMC Genomics 20, 325–339. doi: 10.1186/s12864-019-5634-0
Pradhan, S. K., Pandit, E., Nayak, D. K., Behera, L., Mohapatra, T. (2019). Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biol. 19, 352–369. doi: 10.1186/s12870-12019-11922-12878
Rahman, M.U., Munazza, I., Shanzay, Q., Shazia Anwer, B., Kausar, M. (2019). “Chapter 27 - Abiotic stress signaling in rice crop,” in Advances in Rice Research for Abiotic Stress Tolerance. Eds. Hasanuzzaman, M., Fujita, M., Nahar, K., Biswas, J. K. (Cambridge, UK: Woodhead Publishing), 551–569.
Richards, J., Chao, S., Friesen, T., Brueggeman, R. (2016). Fine mapping of the barley chromosome 6H net form net blotch susceptibility locus. G3: Genes Genomes Genet. 6, 1809–1818. doi: 10.1534/g3.116.028902
Sakuma, S., Lundqvist, U., Kakei, Y., Thirulogachandar, V., Suzuki, T., Hori, K., et al. (2017). Extreme suppression of lateral floret development by a single amino acid change in the VRS1 transcription factor. Plant Physiol. 175, 1720–1731. doi: 10.1104/pp.17.01149
Setter, T. L., Flannigan, B. A. (2001). Water deficit inhibits cell division and expression of transcripts involved in cell proliferation and endoreduplication in maize endosperm. J. Exp. Bot. 52, 1401–1408. doi: 10.1093/jexbot/52.360.1401
Sreenivasulu, N., Altschmied, L., Radchuk, V., Gubatz, S., Wobus, U., Weschke, W. (2004). Transcript profiles and deduced changes of metabolic pathways in maternal and filial tissues of developing barley grains. Plant J. 37, 539–553. doi: 10.1046/j.1365-313X.2003.01981.x
Stief, A., Altmann, S., Hoffmann, K., Pant, B. D., Scheible, W.-R., Bäurle, I. (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell. 26, 1792–1807. doi: 10.1105/tpc.114.123851
Uauy, C., Brevis, J. C., Dubcovsky, J. (2006). The high grain protein content gene Gpc-B1 accelerates senescence and has pleiotropic effects on protein content in wheat. J. Exp. Bot. 57, 2785–2794. doi: 10.1093/jxb/erl047
Walker, C. K., Ford, R., Munoz-Amatriam, M., Panozzo, J. F. (2013). The detection of QTLs in barley associated with endosperm hardness, grain density, grain size and malting quality using rapid phenotyping tools. Theor. Appl. Genet. 126, 2533–2551. doi: 10.1007/s00122-013-2153-2
Wang, H., Wang, H. (2015). The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant 8, 677–688. doi: 10.1016/j.molp.2015.01.008
Wang, S., Li, S., Liu, Q., Wu, K., Zhang, J., Wang, S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, Q., Sun, G., Ren, X., Du, B., Cheng, Y., Wang, Y., et al. (2019). Dissecting the genetic basis of grain size and weight in barley (Hordeum vulgare L.) by QTL and comparative genetic analyses. Front. Plant Sci. 10, 469. doi: 10.3389/fpls.2019.00469
Wang, T., Zou, T., He, Z., Yuan, G., Luo, T., Zhu, J., et al. (2019). GRAIN LENGTH AND AWN 1 negatively regulates grain size in rice. J. Integr. Plant Biol. 61, 1036–1042. doi: 10.1111/jipb.12736
Watt, C., Zhou, G., McFawn, L.-A., Chalmers, K. J., Li, C. (2019). Fine mapping of qGL5H, a major grain length locus in barley (Hordeum vulgare L.). Theor. Appl. Genet. 132, 883–893. doi: 10.1007/s00122-018-3243-y
Watt, C., Zhou, G., McFawn, L.-A., Li, C. (2020). Fine mapping qGL2H, a major locus controlling grain length in barley (Hordeum vulgare L.). Theor. Appl. Genet. 133, 2095–2103. doi: 10.1007/s00122-00020-03579-z
Wu, W., Liu, X., Wang, M., Meyer, R. S., Luo, X., Ndjiondjop, M.-N., et al. (2017). A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants 3, 1–7. doi: 10.1038/nplants.2017.1064
Xie, K., Wu, C., Xiong, L. (2006). Genomic Organization, Differential Expression, and Interaction of SQUAMOSA Promoter-Binding-Like Transcription Factors and microRNA156 in Rice. Plant Physiol. 142, 280–293. doi: 10.1104/pp.106.084475
Xing, Y., Zhang, Q. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. doi: 10.1146/annurev-arplant-042809-112209
Xiong, H., Guo, H., Xie, Y., Zhao, L., Gu, J., Zhao, S., et al. (2017). RNAseq analysis reveals pathways and candidate genes associated with salinity tolerance in a spaceflight-induced wheat mutant. Sci. Rep. 7, 1–13. doi: 10.1038/s41598-41017-03024-41590
Yang, X., Ren, Y., Cai, Y., Niu, M., Feng, Z., Jing, R., et al. (2018). Overexpression of OsbHLH107, a member of the basic helix-loop-helix transcription factor family, enhances grain size in rice (Oryza sativa L.). Rice 11, 1–12. doi: 10.1186/s12284-018-0237-y
Yang, C. C., Ma, J., Li, T., Luo, W., Mu, Y., Tang, H. P., et al. (2019). Structural organization and functional activity of the orthologous TaGLW7 genes in bread wheat (Triticum aestivum L.). Russ. J. Genet. 55, 571–579. doi: 10.1134/S1022795419050168
Yi, N., Oh, S.-J., Kim, Y. S., Jang, H.-J., Park, S.-H., Jeong, J. S., et al. (2011). Analysis of the Wsi18, a stress-inducible promoter that is active in the whole grain of transgenic rice. Transgenic Res. 20, 153–163. doi: 10.1007/s11248-010-9400-y
Yu, J., Xiong, H., Zhu, X., Zhang, H., Li, H., Miao, J., et al. (2017). OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol. 15, 28–46. doi: 10.1186/s12915-017-0365-7
Yue, E., Liu, Z., Li, C., Li, Y., Liu, Q., Xu, J.-H. (2017). Overexpression of miR529a confers enhanced resistance to oxidative stress in rice (Oryza sativa L.). Plant Cell Rep. 36, 1171–1182. doi: 10.1007/s00299-017-2146-8
Zhang, K., Gan, S.-S. (2012). An Abscisic Acid-AtNAP Transcription Factor-SAG113 Protein Phosphatase 2C Regulatory Chain for Controlling Dehydration in Senescing Arabidopsis Leaves. Plant Physiol. 158, 961–969. doi: 10.1104/pp.111.190876
Zhang, C.-Q., Xu, Y., Lu, Y., Yu, H.-X., Gu, M.-H., Liu, Q.-Q. (2011). The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 234, 541–554. doi: 10.1007/s00425-011-1423-y
Zhang, H., Ma, J., Liu, J., Mu, Y., Tang, H., Liu, Y., et al. (2018). Molecular characterization of the TaWTG1 in bread wheat (Triticum aestivum L.). Gene 678, 23–32. doi: 10.1016/j.gene.2018.08.010
Zhang, X., Guo, W., Du, D., Pu, L., Zhang, C. (2020). Overexpression of a maize BR transcription factor ZmBZR1 in Arabidopsis enlarges organ and seed size of the transgenic plants. Plant Sci. 292 1–10. doi: 10.1016/j.plantsci.2019.110378
Zhao, C., Liu, B., Piao, S., Wang, X., Lobell, D. B., Huang, Y., et al. (2017). Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. 114, 9326–9331. doi: 10.1073/pnas.1701762114
Zhao, Y., Wen, H., Teotia, S., Du, Y., Zhang, J., Li, J., et al. (2017). Suppression of microRNA159 impacts multiple agronomic traits in rice (Oryza sativa L.). BMC Plant Biol. 17, 215. doi: 10.1186/s12870-017-1171-7
Zheng, L.-L., Qu, L.-H. (2015). Application of microRNA gene resources in the improvement of agronomic traits in rice. Plant Biotechnol. J. 13, 329–336. doi: 10.1111/pbi.12321
Keywords: transcription factor, abiotic stress, grain size, rice, wheat
Citation: Watt C, Zhou G and Li C (2020) Harnessing Transcription Factors as Potential Tools to Enhance Grain Size Under Stressful Abiotic Conditions in Cereal Crops. Front. Plant Sci. 11:1273. doi: 10.3389/fpls.2020.01273
Received: 10 June 2020; Accepted: 05 August 2020;
Published: 18 August 2020.
Edited by:
Alma Balestrazzi, University of Pavia, ItalyReviewed by:
Abdelali Hannoufa, Agriculture and Agri-Food Canada (AAFC), CanadaCopyright © 2020 Watt, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Calum Watt, Y2FsdW13YXR0MTJAb3V0bG9vay5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.