
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol., 06 January 2025
Sec. Invertebrate Physiology
Volume 15 - 2024 | https://doi.org/10.3389/fphys.2024.1475489
This article is part of the Research TopicInsect Physiology Aspects of Environmentally Friendly Strategies for Crop Pests and Invertebrate Vectors Control, volume IIView all 11 articles
Background: Riptortus pedestris (Fabricius) (Hemiptera: Alydidae) is a major soybean pest throughout East Asia that relies on its advanced olfactory system for the perception of plant-derived volatile compounds and aggregation pheromones for conspecific and host plant localization. Odorant binding proteins (OBPs) facilitate the transport of odorant compounds across the sensillum lymph within the insect olfactory system, enabling their interaction with odorant receptors (ORs).
Methods: Real-time quantitative PCR (qRT-PCR) analyses, fluorescence-based competitive binding assays, and molecular docking analyses were applied to assess the expression and ligand-binding properties of OBP38 from R. peddestris.
Results: The qRT-PCR analyses revealed high levels of RpedOBP38 expression in the antennae without any apparent sex bias, and it was also highly expressed in the adult stage. Recombinant RpedOBP38 was prepared by expressing it in E. coli BL21 (DE3) followed by its purification with a Ni-chelating affinity column. RpedOBP38 was found to bind most strongly to trans-2-decenal (Ki = 7.440) and trans-2-nonenal (Ki = 10.973), followed by β-pinene, (+) -4-terpineol, carvacrol, methyl salicylate, and (-)-carvone. The 3D structure of RpedOBP38 contains six α-helices and three interlocked disulfide bridges comprising a stable hydrophobic binding pocket. In a final series of molecular docking analyses, several polar (e.g., His 94, Glu97) and nonpolar (e.g., Leu29, Ile59) residues were found to be involved in RpedOBP38-ligand binding.
Conclusion: These data support a role for RpedOBP38 in the perception of volatiles derived from host plants, providing important insight into the mechanisms that govern olfactory recognition in R. pedestris, thereby informing the development of ecologically friendly approaches to managing R. pedestris infestations.
The ability of insects to perceive pheromones, host-derived odorants, and the wide array of other peripheral chemical signals present in their surrounding environment is dependent on a complex olfactory system that ultimately shapes key physiological processes such as foraging, mating, and oviposition (Martin et al., 2011; Leal, 2013). The ability to accurately recognize and decipher these signals is thus vital for the ability of insects to survive and reproduce. Hydrophobic chemicals need to successfully penetrate the olfactory sensilla and the hydrophilic sensillum lymph in order to access the odorant receptors (ORs) present on sensory neuron surfaces, thereby triggering downstream signal transduction (Li et al., 2015; Zhou et al., 2022). To facilitate this process, specialized supporting cells produce odorant-binding proteins (OBPs), which are secreted into the olfactory sensillum lymph and play a vital role in the process of insect odorant reception (Leal, 2013; Paula et al., 2018). OBPs can selectively bind, solubilize, and transport odorant molecules as they diffuse into the sensillum lymph, thereby enabling the activation of ORs and associated downstream signaling pathways (Leal, 2013; Liu et al., 2023). Given the importance of OBPs during this initial stage of odorant reception, they hold great promise as molecular targets for pest control efforts and the development of superior integrated pest management (IPM) strategies (Zhou et al., 2010; Venthur and Zhou, 2018).
The OBPs produced by insects are low-molecular-weight (12–20 kDa) proteins approximately 100–200 amino acids in length that are water soluble and typically feature a ∼20 amino acid N-terminal signal peptide sequence (Ahmed et al., 2017; Li J. B. et al., 2022; Zeng et al., 2019). The 3D structures of classical OBPs are stabilized by three disulfide bridges formed by six conserved cysteine residues (Leal et al., 1999; Scaloni et al., 1999; Pelosi et al., 2014). The patterns of conserved cysteines have also been used to define four other classes of OBPs, including “Dimer” OBPs with two typical cysteines, “Minus-C” OBPs that lack 1-2 cysteines, “Plus-C″ OBPs with 2-3 extra cysteines, and “Atypical” OBPs with a long, atypical C-terminal domain (Zhou et al., 2010; Spinelli et al., 2012; Manoharan et al., 2013; Venthur et al., 2014; Zeng et al., 2019). Identified in 1981, the first characterized OBP in insects was found to be exclusively expressed in Antberaea polypbemus antennae, enabling male moths to detect a particular sex pheromone (trans-6, cis-11-hexadecadienyl acetate) such that they were able to locate conspecific females to engage in mating (Vogt and Riddiford, 1981). Advances in molecular biology and transcriptomic technologies have fueled the identification of a growing number of genes encoding OBPs in many orders of insects, including Coleoptera (e.g., 39 OBPs in Phyllotreta striolata, Xiao et al., 2023), Hemiptera (e.g., 49 OBPs in Riptortus pedestris, Li L. L. et al. (2022)), Diptera (e.g., 28 OBPs in Liriomyza trifolii, Zhang et al. (2022)), Lepidoptera (e.g., 31 OBPs in Chilo sacchariphagus, Liu et al. (2021a)), Hymenoptera (e.g., 21 OBPs in Apis mellifera, Forêt and Maleszka (2006)), Orthoptera (e.g., 22 OBPs in Locusta migratoria, Pelosi et al. (2018)). Experimental efforts have revealed that OBPs which are primarily expressed in the antennae of certain insects are capable of interacting with specific chemical ligands including host volatiles and pheromones (Zhang et al., 2020a; Rihani et al., 2021). AlepOBP6, for instance, is predominantly expressed in the antennae of male Athetis lepigone individuals and can recognize both maize-derived volatile compounds and sex hormones produced by conspecific females (Li J. B. et al., 2022). In Hippodamia variegate, both males and females exhibit high levels of HvarOBP5 expression in their antennae, thus enabling the perception of plant and prey-derived volatiles (Tang et al., 2023). The behavioral responses of Eupeodes corolla to the aphid alarm pheromone (E)-β-farnesene have been shown to be regulated by EcorOBP15 (Wang et al., 2022). Several OBPs have also been demonstrated to be expressed in other organs with or without primary chemosensory functions, including mouthpart palps (Pregitzer et al., 2018), labella (Sparks et al., 2014), legs (Hull et al., 2014), thorax (Zhang et al., 2018), and reproductive organs (Sun et al., 2012). These OBPs can facilitate a range of physiological functions including the recognition of taste compounds, the solubilization of nutrients, and the augmentation of resistance against insecticides (Pelosi et al., 2018).
Riptortus pedestris (Fabricius) (Hemiptera: Alydidae), known as the bean bug, is a serious agricultural pest species that is widely distributed throughout China, Japan, Korea, and other nations in East Asia (Jung and Lee, 2018; Jin et al., 2022). R. pedestris is a polyphagous pest species, feeding on over 30 different plants across 13 families (including Gramineae, Cruciferae, and other crop families), although they exhibit a particular preference for soybeans and other leguminous plants (Mainali et al., 2014; Ahn et al., 2020). Large numbers of these bean bugs typically infest soybean fields in the late flowering or early pod-growing stages and persistently feed on and damage these plants until harvest time (Endo et al., 2011). Soybean leaves, stems, pods, and flowers can be damaged by both R. pedestris adults and nymphs through their piercing and sucking behaviors, resulting in leaf rolling, stunted growth, and seed pods that are shriveled or empty, culminating in serious reductions in soybean quality and yield (Ahn et al., 2020). R. pedestris-associated soybean damage has recently emerged as a particularly serious problem in the Huang-Huai-Hai region of China (Li L. L. et al., 2022). Soybean plants in this region often suffer from the staygreen phenomenon that can be caused by R. pedestris feeding, which results in leaves that remain green, shriveled pods, and maturity stage seed abortion in soybean plants (Li et al., 2019; Dong et al., 2022). The control of R. pedestris has traditionally been achieved through the application of pyrethroids or other broad-spectrum insecticides (Gao et al., 2019; Guo et al., 2023). Such insecticide-based management practices, however, entail many potentially serious issues including environmental pollution, elevated levels of insecticide resistance, and inadequate efficacy owing to the highly mobile nature of these insects and their behavioral avoidance of insecticides (Bae et al., 2019; Zhu et al., 2022). There is thus a pressing need to develop new, ecologically friendly olfaction-based strategies for the control of R. pedestris infestations.
R. pedestris rely on their highly-developed antennae harboring abundant sensilla to detect both adult male-derived aggregation pheromone and host plant-derived volatiles, thus facilitating conspecific and host location efforts (Leal et al., 1995; Kim et al., 2016; Roh et al., 2021; Song et al., 2022). Li J. B. et al. (2022) previously analyzed the R. pedestris genome and identified 49 candidate RpedOBPs, including RpedOBP38, which exhibited high levels of expression in the antennae. The specific involvement of RpedOBP38 in the detection of host volatiles or other chemical signals, however, has yet to be documented. Accordingly, this study was devised to clarify the olfactory functions of RpedOBP38. To that end, the sequence of the RpedOBP38 gene was initially analyzed, after which RpedOBP38 expression was analyzed across a variety of tissues and developmental stages via real-time quantitative PCR (qRT-PCR). The binding affinity of RpedOBP38 for 36 volatiles (including 11 green leaf volatiles, 11 soybean volatiles, 10 volatiles associated with repellent activity, and 4 aggregation pheromone compounds) was characterized through a fluorescence binding assay. Lastly, homology modeling and molecular docking approaches were used to characterize the binding sites and key amino acids related to the ligand binding activity of RpedOBP38. Together, the results of these analyses provide a robust evidence base for the further molecular characterization of the mechanisms governing olfactory recognition in R. pedestris, thus supporting efforts to improve the integrated management of this economically significant pest species.
R. pedestris specimens were captured in July-August 2019 from soybean fields in Shijiazhuang, Hebei province, China. Adults and nymphs were reared as in prior reports (Guo et al., 2023). Briefly, these insects were housed at 26°C ± 1°C under 60% ± 5% relative humidity (RH) with a 16 h: 8 h (L:D) photoperiod in cages, and were fed dried seeds (variety Jidou 12) and soybean seedlings that were replaced every 5–7 days. Based on the study of Li L. L. et al. (2022), 3-day-old virgin male and female adults were processed to collect antennae (40 pairs), heads without antennae (from 10 individuals), thoraxes (from 4 individuals), abdomens (from 3 individuals), wings (from 40 individuals), and legs (from 20 individuals). In addition, antennae were collected from 2nd (200 pairs), 3rd (120 pairs), 4th (60 pairs), and 5th (60 pairs) instar nymphs, after which they were snap-frozen with liquid nitrogen and stored at −80°C.
TRIzol (TransGen, China) was used to extract RNA according to the manufacturer’s instructions, the quality of which was analyzed via 1.0% agarose gel electrophoresis and spectrophotometry with a NanoDrop™ 2000 instrument (Thermo Fisher Scientific, United States). Next, 1 μg of the extracted RNA was processed with All-in-One First-Strand cDNA Synthesis SuperMix (TransGen), and the resultant cDNA was stored at −20°C.
The SignalP 6.0 server (https://services.healthtech.dtu.dk/services/SignalP-6.0/) was used for signal peptide prediction, while ClustalX 2.0 was used for multiple alignment of the RpedOBP38 protein sequence and those of other Hemiptera OBPs, with GeneDoc (http://nrbsc.org/gfx/genedoc) being used for result visualization. The amino acid sequences of other hemipteran species were downloaded by accessing the NCBI website. MEGA7 was used to construct a phylogenetic tree with the neighbor-joining method and bootstrap testing (1,000 replicates). The Poisson correction method was employed when calculating evolutionary distance.
RpedOBP38 expression was validated via qRT-PCR with an ABI QuantStudio6 Q6 Real-Time PCR System (Applied Biosystems, CA, United States) using primers designed with Premier 6 and prepared by Sangon Biotech Co., Ltd (Beijing, China) (Supplementary Table S1). Individual 20 μL reactions comprised 1 μL of cDNA, 0.6 μL each of F/R primers (10 μM), 10 μL of 2 × FastFire qPCR PreMix (TianGen Biotech, Beijing, China), and 7.8 μL of ddH2O. Reaction settings were: 94°C for 30 s; 40 cycles of 94°C for 5 s, 55°C for 15 s, and 72°C for 10 s. Relative RpedOBP38 expression was assessed with the 2-△△Ct method, using EF1 and Actin as reference genes (Wang et al., 2023). Three independent biological replicates were analyzed per sample.
The RpedOBP38 open reading frame (ORF) lacking a signal peptide sequence was PCR amplified with TransStart® FastPfu PCR SuperMix (TransGen Biotech). Primers used to construct an RpedOBP38 expression vector were as follows: Forward: 5′-GATGAGGCGAAACAGATG-3′, Reverse: 5′-TCACTGTAGATCTTCAGTTCC-3’. Amplification settings were as follows: 95°C for 1 min; 35 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 1 min; 72°C for 5 min. The products of PCR amplification were ligated into the pEASY-Blunt E1 vector (TransGen Biotech) and transformed into E. coli Trans-T1. Sangon Biotech then sequenced and confirmed the amplified gene products, and positive recombinant pEASY-Blunt E1-RpedOBP38 plasmids were obtained for further use.
After transforming E. coli BL21 (DE3) with recombinant RpedOBP38 expression vectors, positive clones were isolated and used to initiate cultures in LB broth containing 50 μg/mL ampicillin that were incubated at 37°C and 220 rpm. When the OD600 reached 0.6, 1 mM of isopropyl β-D-thiogalactoside (IPTG) was added and bacteria were incubated under the same conditions for a further 6 h. Cells were then centrifuged (8,000 xg, 4°C) and resuspended in 20 mL of PBS (pH 7.0). Cells were then ultrasonically disrupted, and homogenates were centrifuged (14,000 rpm, 20 min, 4°C). The supernatants were then assessed via 12% SDS-PAGE separation. Target proteins from the supernatant fractions were applied to a Ni-chelating affinity column (GE, United States), which was subsequently equilibrated with 100 mM NaCl, 20 mM Tris-HCl, pH 7.9, and eluted using an ascending imidazole concentration series (50, 100, 150 and 200 mM). Dialysis was used to desalt the eluent, and target protein size and purity were assessed via SDS-PAGE. Recombinant protein concentrations were measured via Bradford assay.
Recombinant RpedOBP38 binding to putative chemical ligands was characterized with a microplate reader (BioTek Synergy H1, United States). Fluorescence intensity values at the excitation wavelength of 337 nm and a maximum fluorescence emission wavelength of 450 nm were plotted against the free concentration of ligand for the measurement of dissociation constants, selecting candidate ligands from among 36 volatile compounds that included 11 green leaf volatiles (Chen et al., 2018; Guo and Wang, 2019; Cheng et al., 2020; Hong et al., 2022; Tang et al., 2023; Zhu et al., 2023), 11 soybean volatiles (Wang et al., 2019a; Zhu et al., 2022), 10 repellent activity volatiles (Zhang et al., 2013; Zhang et al., 2014), and 4 aggregation pheromone compounds (Leal et al., 1995; Yasuda et al., 2007). HPLC-grade methanol was used for the dissolution of the probe N-phenyl-1-naphthylamine (1-NPN) and all ligands. The ability of RpedOBP38 to bind 1-NPN was assessed by using 10 mM PBS (pH 7.4) to prepare a 2 μM purified protein solution, titrating with 1 mM 1-NPN in methanol to prepare final concentrations from 2–20 μM. RpedOBP38 binding to each ligand was evaluated in a solution consisting of 2 μM purified protein and 1-NPN, followed by titration through the addition of ligands until no further decrease in the fluorescence intensity was observed. Ligands were independently replicated three times, and dissociation constants for each ligand were measured as follows: Ki = [IC50]/(1 + [1-NPN]/K1-NPN), where IC50 denotes the ligand concentration when the fluorescence intensity is half of the initial value [1-NPN] is the free 1-NPN concentration, and K1-NPN is the dissociation constant for the RpedOBP38/1-NPN complex (Campanacci et al., 2001). Based on the study of Cui et al. (2018), the strength of binding affinity could be indicated by Ki value, including very strong (Ki < 6 μM), strong (6 µM ≤ Ki < 22 µM), moderate (22 µM ≤ Ki < 40 µM) and weak (Ki > 40 µM).
RpedOBP38 tertiary structure modeling was performed with the I-TASSER server (Zheng et al., 2021), due to the <30% homology with the protein sequences in the Swiss-model server. The RpedOBP38 amino acid sequence was utilized as an input, utilizing the 10 template proteins exhibiting the highest sequence identity for the purposes of modeling (Supplementary Table S2). C-score values were used to choose the best model from among the top 5 (Supplementary Table S3), with C-scores generally falling in the [-5, 2] range, and higher scores being indicative of greater model confidence. For the chosen ligands, the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) was accessed to download 3D structures that were subsequently converted into the mol2 format with Open Babel GUI v.3.1.1 (O’Boyle et al., 2011). Molecular docking analyses of the interactions between RpedOBP38 and seven ligands were performed with AutoDock Vina (v.1.1.2) (Trott and Olson, 2010) using default parameters. PyMOL v.2.0 (Schrödinger, LLC) was used for the visualization of the molecular docking results, and interaction forces were examined with PLIP (https://plip-tool.biotec.tu-dresden.de/plipweb/plip/index).
RpedOBP38 expression was analyzed across various R. pedestris tissues and developmental stages using one-way ANOVA with Tukey’s multiple comparison test. p < 0.05 was selected as the cut-off for significance, and SPSS 20.0 (IBM 2011) was used for all statistical analyses, while GraphPad Prism 8.0 was used for figure generation.
RpedOBP38 cDNA sequences were downloaded from the R. pedestris genome (Li J. B. et al., 2022). The RpedOBP38 ORF was found to consist of 462 bp encoding a 153-amino-acid (aa) protein, with a 19-aa N-terminal signal peptide. This protein had a predicted molecular mass of 15.08 kDa and a predicted isoelectric point of 5.20. BLASTp similarity analyses revealed some level of sequence identity with OBPs from other Hemiptera species, including YsigOBP15 from Yemma signatus (43.42%), PstaOBP3 from Plautia stali (38.78%), HhalOBP15 from Halyomorpha halys (37.50%), TeleOBP5 from Tropidothorax elegans (36.23%), and NvirOBP20 from Nezara viridula (35.71%). Phylogenetic tree analyses revealed the clustering of RpedOBP38 and YsigOBP15 from Y. signatus (Figure 1A). Sequence alignment also revealed the presence of six conserved cysteine residues within RpedOBP38 (Figure 1B), consistent with its classification within the classical OBP family.
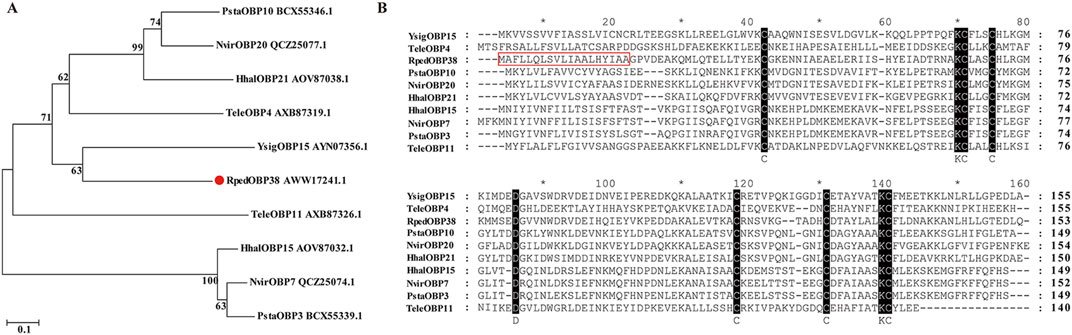
Figure 1. RpedOBP38 sequence characteristics. (A) Odorant-binding proteins (OBPs) from Riptortus pedestris and other hemipteran species were used to construct a phylogenetic tree. (B) RpedOBP38 alignment to OBP sequences from other hemipteran species. The signal peptide sequence is marked with a red box. Hhal: Halyomorpha halys, Nvir: Nezara viridula, Psta: Plautia stali, Tele: Tropidothorax elegans, Ysig: Yemma signatus.
RpedOBP38 expression across tissues and developmental stages was next characterized by qPCR, revealing significant differences in RpedOBP38 among tissues in both female (F5, 12 = 65.68, P < 0.001) and male (F5, 12 = 129.09, P < 0.001) adults, with the highest expression levels in the antennae of adult females and males, respectively (Figure 2A). Antennae RpedOBP38 expression levels rose with increasing developmental stages, with significant differences among stages (F5, 12 = 106.62, P < 0.001), and expression levels being highest in adult antennae. However, there was no significant sex difference for RpedOBP38 expression levels (Figure 2B).
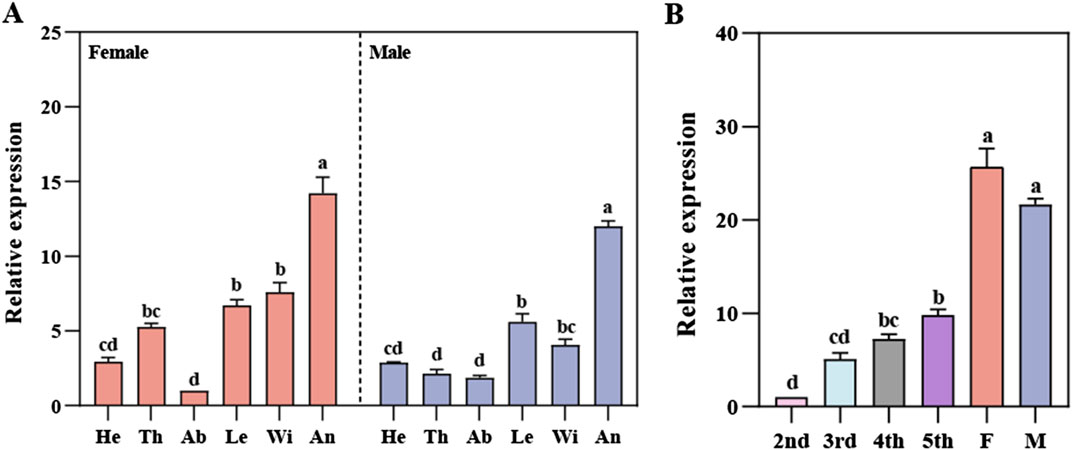
Figure 2. Relative RpedOBP38 expression analyses. (A) qRT-PCR analyses of RpedOBP38 mRNA levels in various tissues. He: heads, Th: thoraxes, Ab: abdomens, Le: legs, Wi: wings, An: antenna. (B) qRT-PCR analyses of RpedOBP38 mRNA levels in the antennae of Riptortus pedestris at different developmental stages. F: female, M: male. Significant differences are indicated by different lowercase letters (p < 0.05; Tukey’s HSD test).
After expressing recombinant RpedOBP38 in E. coli BL21 (DE3), it was purified, yielding a final recombinant RpedOBP38 concentration of 0.603 mg/mL. SDS-PAGE analyses confirmed a similar target protein size to the predicted size (Figure 3A). The ability of RpedOBP38 to bind 1-NPN was then assessed, revealing strong binding between RpedOBP38 and 1-NPN (dissociation constant [Kd]): 4.059 μmol/L). Binding curve analyses and Scatchard plots revealed the presence of a single binding site, indicating that 1-NPN was a highly suitable probe for subsequent binding analyses (Figure 3B).
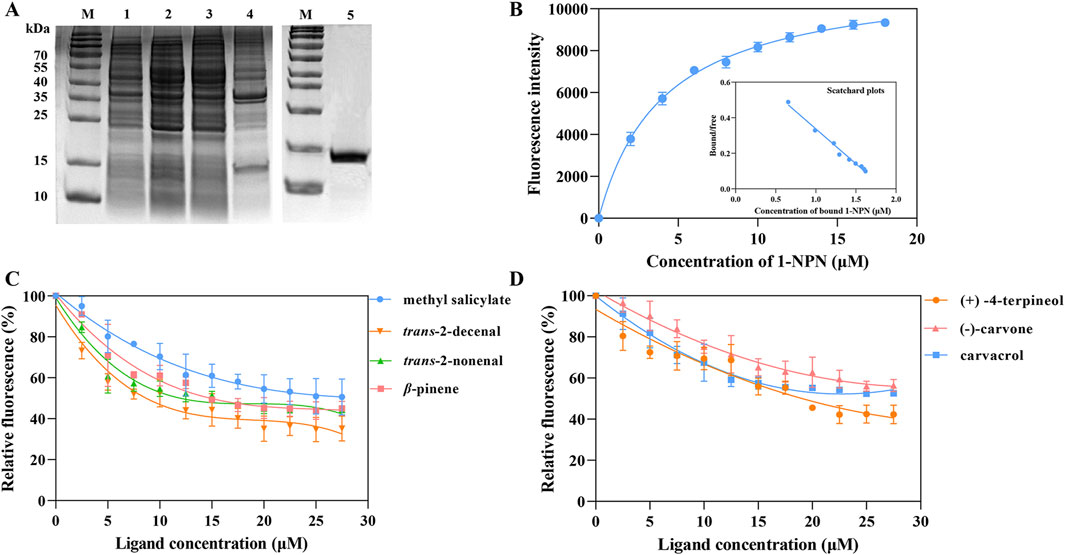
Figure 3. Characterization of RpedOBP38 ligand binding properties. (A) SDS-PAGE analyses pertaining to recombinant RpedOBP38 expression and purification. Lane 1: non-induced pEasy-Blunt E1-RpedOBP38; Lane 2: induced pEasy-Blunt E1-RpedOBP38; Lane 3: Supernatant of induced pEasy-Blunt E1-RpedOBP38; Lane 4: precipitation of induced pEasy-Blunt E1-RpedOBP38; Lane 5: purified RpedOBP38. (B) Binding curves and scatchard plots correspond to the interaction between the fluorescent probe 1-NPN and RpedOBP38. (C, D) Binding curves corresponding to interactions between RpedOBP38 and green leaf volatiles, soybean volatiles. (C) or repellent volatiles (D).
In total, 36 volatile compounds including 11 green leaf volatiles, 11 soybean volatiles, 10 volatiles associated with repellent activity, and 4 aggregation pheromone compounds were chosen for the evaluation of RpedOBP38 ligand binding. The resultant analyses demonstrated the ability of RpedOBP38 to strongly bind to the soybean volatiles trans-2-decenal (Ki = 7.440 μM), trans-2-nonenal (Ki = 10.973 μM) and methy salicylate (Ki = 21.065 μM) (Figure 3C; Table 1). It also exhibited strong or moderate binding to three volatiles associated with repellent activity ((+) -4-terpineol, Ki = 14.017 μM; carvacrol, Ki = 19.446 μM; (−)-carvone, Ki = 27.215 μM) (Figure 3D; Table 1). In contrast, it exhibited weak binding activity for the tested aggregation pheromone compounds (Ki > 40 μM) (Table 1).
When the I-TASSER server was used to construct 3D models of the structure of RpedOBP38, the first model among the top five generated models exhibited the highest C-score of −1.13 (Supplementary Table S3). This predicted RpedOBP38 model contained six α-helices designated α1 (Pro21-Glu42), α2 (Glu47-Ser55), α3 (Cys67-Gly75), α4 (Trp87-Glu97), α5 (Pro101-Ala113), and α6 (His124-Ala142) that were folded around a hydrophobic cavity. It also harbored three interlocking disulfide bridges formed by links between Cys39 in α1 and Cys71 in α3, Cys67 in α3 and Cys125 in α6, and Cys114 in α5 and Cys134 in α6, providing further stability to the hydrophobic structure of this protein (Figure 4A).
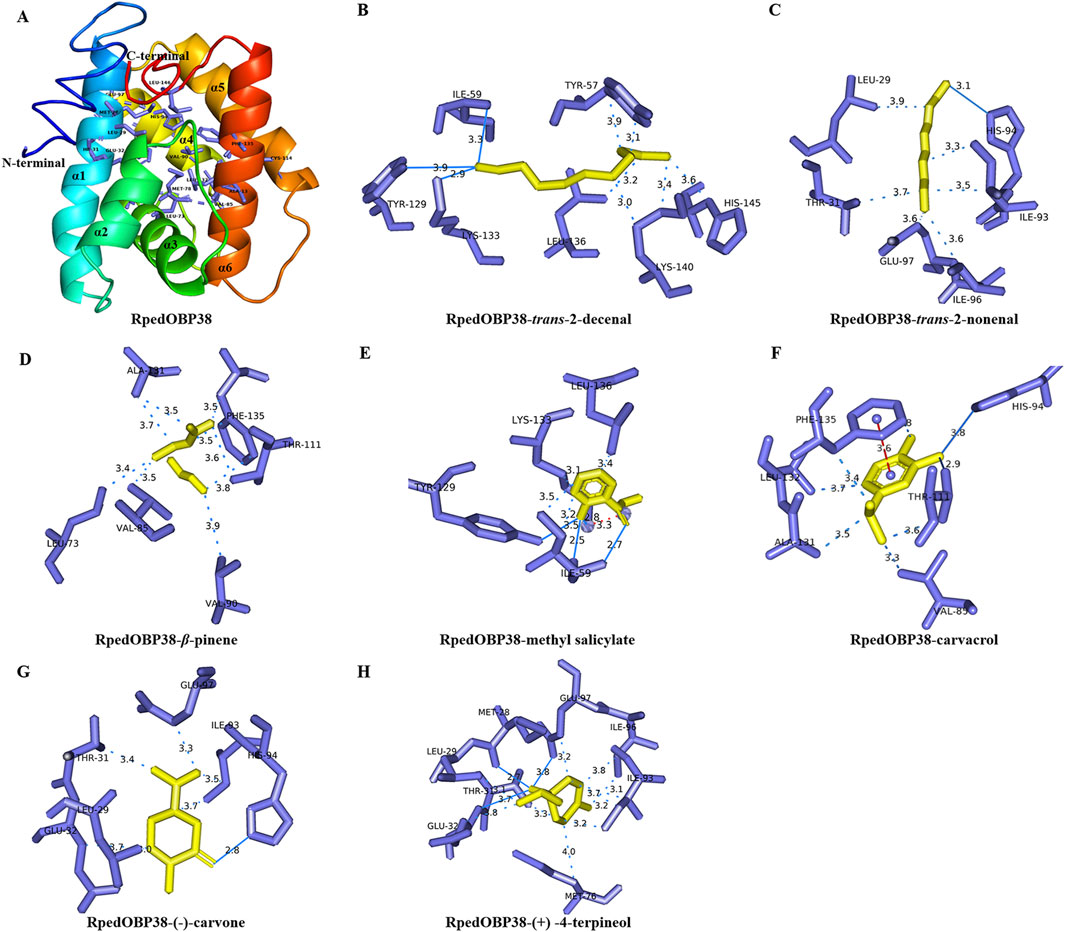
Figure 4. Molecular docking of ligands within the putative RpedOBP38 ligand binding pocket. (A) A structural model of RpedOBP38. The indicated amino acid residues correspond to key residues within the predicted RpedOBP38 pocket. (B–H) Molecular docking analyses for interactions between RpedOBP38 and trans-2-decenal (B), trans-2-nonenal (C), β-pinene (D), methyl salicylate (E), carvacrol (F) (−)-carvone (G), and (+) -4-terpineol (H). Hydrogen bonds are indicated with blue lines, while hydrophobic interactions are denoted using blue dashed lines, and π-stacking is represented using red dashed lines.
Based on the fluorescence competitive binding assays performed above, β-pinene, methyl salicylate, trans-2-nonenal, trans-2-decenal (+) -4-terpineol (−)-carvone, and carvacrol were chosen as target ligands for molecular docking analyses. All seven of these ligands exhibited negative binding energy values when interacting with RpedOBP3 ranging from −5.71 to −4.20 (Table 2). Hydrogen bonds, hydrophobic interactions, and π-stacking were all found to contribute to these RpedOBP38-ligand binding interactions (Figures 4B–H). Both polar (e.g., Lys133, Glu97, His 94, Thr31) and nonpolar (e.g., Ile59, Leu136, Val85, Phe135) residues within the hydrophobic RpedOBP38 cavity were found to contribute to intermolecular binding interactions. Some of these amino acid residues were found to bind to multiple ligands, including 10 (Glu32, Ile59, Val85, Ile96, Thr111, Tyr129, Ala131, Lys133, Phe135, and Leu136) that were able to bind to three ligands, and 5 (Leu29, Thr31, Ile93, His94, and Glu97) that were able to bind to three ligands (Table 2).
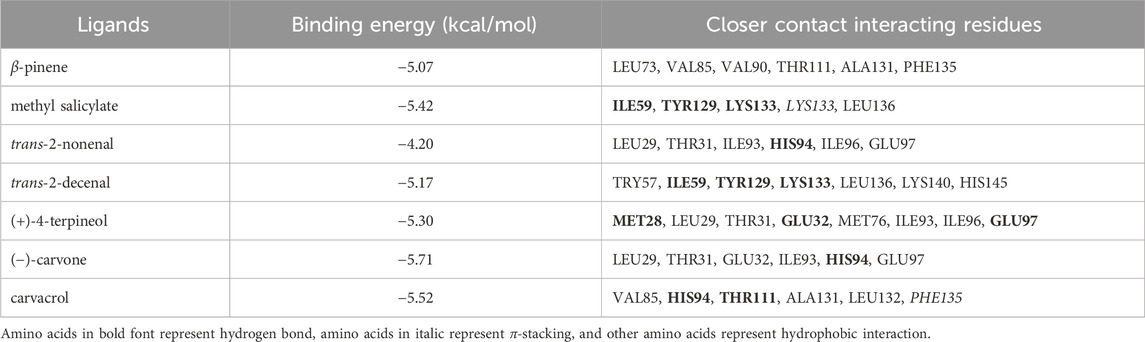
Table 2. Prediction of key amino acid residues involved in the docking of RpedOBP38 to different ligands.
OBPs have been identified across many insect species to date and have been confirmed to be integral to the recognition of exogenous chemical signals and the regulation of physiological activities (Venthur and Zhou, 2018). OBPs have been established as promising molecular targets when screening for odorous compounds with attractant or repellent properties, informing the development of push-pull pest control strategies (Zhang et al., 2021; Song et al., 2022; Zhu et al., 2022; Zhu et al., 2023). For instance, CquiOBP1 of Culex quinquefasciatus was used as a target to guide the successful synthesis of a blend of trimethylamine and nonanal through the combination of conventional and reverse chemical ecology methodological approaches (Leal et al., 2008). The ability of certain OBPs to bind to aphid alarm pheromone has also enabled the design and synthesis of novel (E)-β-farnesene analogs with repellent and insecticidal activity for Acythosiphon pisum (Sun et al., 2011). In light of the importance of OBPs and the rising demand for environmentally friendly approaches to managing pest species, OBPs have emerged as a research hotspot in the insect chemical ecology space.
Initial sequencing analyses performed in this study revealed that RpedOBP38 had 153 amino acids in length with a 19-aa N-terminal signal peptide and six conserved cysteine residues, consistent with its classification as a member of the classic OBP family (Pelosi et al., 2006; Brito et al., 2016). Phylogenetic analyses can be used to infer evolutionary relationships for particular genes across species, thereby informing functional analyses such that they have been widely used for characterizing insect OBPs (Chen et al., 2018; Zhang et al., 2022; Tang et al., 2023; Zhu et al., 2023). In this study, RpedOBP38 and YsigOBP15 from Y. signatus clustered together, suggesting their evolution from a shared ancestor and their potential for similar physiological functions. Analyzing the patterns of insect OBP expression across developmental stages and tissues is vital for the clarification of the physiological functions of these factors (Ju et al., 2014; Wang et al., 2019b; Li et al., 2020). In general, OBPs are likely to play a role in the recognition of chemical signals if they are expressed at high levels in antennae and other olfactory organs (Chen et al., 2018; Li et al., 2020; Hong et al., 2022; Zhang et al., 2022; Tang et al., 2023). Higher levels of RpedOBP38 expression were noted in the antennae relative to other tissues in this study, with no significant difference between females and males. This result was in line with a prior report by Li L. L. et al (2022), indicating that RpedOBP38 may play an important role in the recognition of host volatiles and/or aggregation pheromones by R. pedestris. Other groups have also reported similar outcomes. For instance, Huang et al. (2018) reported the specific expression of AipsOBP2 in Agrotis ipsilon antennae and found that it was capable of binding both host volatiles and sex pheromones. Cheng et al. (2020) additionally noted the strong binding of SmosOBP12, which was expressed at high levels in the antennae of female Sitodiplosis mosellana, to host volatiles derived from wheat including hexyl acetate and 3-hexanol. R. pedestris reportedly harbor many different olfactory sensors on their antennae (Kim et al., 2016), and are attracted to soybean-derived volatile compounds and aggregation pheromones released by conspecific males (Leal et al., 1995; Song et al., 2022). High levels of RpedOBP38 expression were also noted in the adult stage, suggesting its potential involvement as a mediator of soybean volatile and aggregation pheromone recognition in R. pedestris.
Given the role that OBPs play as carriers in the context of chemical communication in insects, there is a need to clarify the affinity of these compounds for exogenous organic factors including pheromones and host-derived odorants, thereby potentially offering insight into the structural features of cognate ligands to guide reverse chemical ecology studies (D'Onofrio et al., 2020). Fluorescence competitive binding have been established as a reliable means of assessing in vitro binding between OBPs and their ligands (He et al., 2019; D'Onofrio et al., 2020). This approach has been successfully implemented across various species of insects including Diaphorina citri (Liu et al., 2021b), Liromyza trifolii (Zhang et al., 2022), R. pedestris (Zhu et al., 2022), Bradysia odoriphaga (Zhu et al., 2023), and Hippodamia variegate (Tang et al., 2023). In this study, the ability of RpedOBP38 to bind to 11 green leaf volatiles, 11 soybean volatiles, 10 volatiles with repellent activity, and 4 aggregation pheromone compounds was assessed. In total, it was found to bind to three soybean volatiles (trans-2-decenal, trans-2-nonenal, methyl salicylate) and one green leaf volatile (β-pinene). Host plant volatiles have been shown to promote feeding, avoidance, oviposition, and a range of other behavioral responses (Anderson et al., 1993; Leal et al., 1994; Zhu et al., 2022). RpedOBP38 may thus play a role in the detection of soybean volatiles, although behavioral and RNA interference assays will be necessary to confirm this hypothesis. Zhu et al. (2022) previously demonstrated the ability of RpedOBP4 to bind other soybean volatiles including 1-hexanol and trans-2-hexenyl acetate, supporting the potential involvement of multiple OBPs in the process of host plant recognition in line with what has been reported by Li et al. (2017). RpedOBP38 was also able to bind less strongly to plant essential oil-derived volatiles with repellent activity ((+) -4-terpineol (−)-carvone, carvacrol) that exhibit high levels of repellency for various insect species (Quintana et al., 2009; Zhang et al., 2014). However, the binding affinity of RpedOBP38 for tested aggregation pheromones was low (Ki > 40 μM), suggesting that binding to these compounds may be primarily mediated by other chemosensory proteins including RpedCSP12 (Yin et al., 2023). Notably, RpedOBP38 exhibited distinct binding affinity levels for certain isomers as in the case of (+) -4-terpineol (Ki = 14.02 μM) and (−) -4-terpineol (Ki > 40 μM), or (−)-carvone (Ki = 27.22 μM) and (+)-carvone (Ki > 40 μM). Factors including carbon chain length, conformational changes, and structural features can thus likely shape RpedOBP38 binding affinity (Chen et al., 2018; Hong et al., 2022).
The physiological functions of a given protein are determined by its 3D structure, and insect OBPs generally harbor a hydrophobic cavity formed from multiple α-helices, with some of the amino acids therein facilitating interactions between these OBPs and their ligands (He et al., 2019; Zhang et al., 2020b; Yang et al., 2021; Zhu et al., 2023). Molecular modeling analyses performed herein revealed the presence of a hydrophobic binding pocket within RpedOBP38 that was stabilized by six α-helices and three interlocking disulfide bridges. This is consistent with similar reports for DcitOBP7 in Diaphorina citri (Liu et al., 2021a), and PyasOBP2 in Pachyrhinus yasumatsui (Hong et al., 2022), suggesting that they may engage in similar ligand-binding mechanisms. Molecular docking analyses revealed negative binding energy values for interactions between RpedOBP38 and seven analyzed ligands, implying strong protein-ligand interactions, consistent with the fluorescence competitive binding assay results. OBP-ligand binding is generally mediated by types of intermolecular forces including hydrogen bonds, van der Waals interactions, and hydrophobic interactions (Zhuang et al., 2014; Li et al., 2021; Hong et al., 2022). In this study, hydrogen bonds, hydrophobic interactions, and π-stacking were all found to shape RpedOBP38-ligand interactions, with molecular docking analyses also revealing the distribution of several polar (e.g., Lys133, Glu97, His 94, Thr31) and nonpolar (e.g., Ile59, Leu136, Val85, Phe135) residues within the RpedOBP38 hydrophobic pocking jointly contributing to such intermolecular binding. This aligns well with other reports for insect OBPs, including the Val114, Thr9, and Val111 residues in Grapholita Molesta OBP2 (Li et al., 2016), Tyr77, Ile41, Ala116, and Lys38 in Apbid Sitobion OBP9 (Ullah et al., 2020), Leu33, Phe8, Met76, IIe30, Tyr47, Asp29, and Lys120 in R. pedestris OBP4 (Zhu et al., 2022), and Lys43, His64, and Leu42 in H, variegate OBP5 (Tang et al., 2023). Some of these amino acids were found to be capable of binding to more than one ligand, including Leu29, Thr31, His94, Glu97, Ile59, and Lys133, in line with what has previously been described in both Athetis lepigone (Li L. L. et al., 2022) and R. pedestris (Zhu et al., 2022). These residues may thus be particularly important mediators of RpedOBP38-ligand binding, highlighting an opportunity for site-directed mutagenesis to validate this hypothesis in the future (Zhu et al., 2020).
In summary, these experiments revealed that RpedOBP38, which was highly expressed in the antennae of adult R. pedestris, is a classical OBP family member that clusters most closely with YsigOBP15 from Y. signatus. Fluorescence competitive binding analyses demonstrated the ability of RpedOBP38 to bind strongly to two soybean volatiles (trans-2-decenal, Ki = 7.440 μM; trans-2-nonenal, Ki = 10.973 μM; methyl salicylate, Ki = 21.065 μM) and to bind strongly or moderately to volatiles associated with repellent activity ((+) -4-terpineol, Ki = 14.017 μM; carvacrol, Ki = 19.456 μM; (−)-carvone, Ki = 27.215 μM). Through 3D modeling and molecular docking analyses, RpedOBP38 was found to harbor six α-helices that form a stable hydrophobic binding pocket, with the Leu29, Thr31, His94, Glu97, Ile59, and Lys133 amino acid residues all playing key roles in the ability of this OBP to bind its ligands. Together, these results offer further insight into the mechanisms that govern olfactory recognition in R. pedestris. In order to more deeply elucidate the function of RpedOBP38, future studies are planned to analyse the exact role of RpedOBP38 in the recognition of more green leaf volatiles and soybean volatiles using a combination of behavioural experiments, electrophysiological experiments, and RNA inference (Zhu et al., 2022). Furthermore, we attempt to use RpedOBP38 as a control target, devise ecologically friendly behavioural inhibitors to disrupt the feeding behavior of R. pedestris and thus improve the management of R. pedestris (Zhu et al., 2022).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The manuscript presents research on animals that do not require ethical approval for their study.
JG: Data curation, Formal Analysis, Funding acquisition, Visualization, Writing–original draft. PL: Methodology, Software, Validation, Visualization, Writing–original draft. XZ: Data curation, Formal Analysis, Software, Writing–original draft. JA: Data curation, Formal Analysis, Resources, Validation, Writing–review and editing. YL: Data curation, Validation, Writing–review and editing. TZ: Conceptualization, Investigation, Project administration, Writing–review and editing. ZG: Conceptualization, Funding acquisition, Investigation, Supervision, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (32302358), HAAFS Agriculture Science and Technology Innovation Project (2022KJCXZX-ZBS-4), Natural Science Foundation of Hebei Province (C2022301052), and Key research and Development Program of Hebei Province (22326513D).
We would like to thank all the reviewers who participated in the review during the preparation of this manuscript. We are also grateful to Hui-Fang Zhou from the Plant Protection Institute, Hebei Academy of Agricultural and Forestry Sciences, for her contribution in insect collection.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2024.1475489/full#supplementary-material
Ahmed T., Zhang T., Wang Z., He K., Bai S. (2017). Molecular cloning, expression profile, odorant affinity, and stability of two odorant-binding proteins in Macrocentrus cingulum Brischke (Hymenoptera: braconidae). Arch. Insect Biochem. 94 (2), e21374. doi:10.1002/arch.21374
Ahn J. J., Choi K. S., Koh S. (2020). Population parameters and growth of Riptortus pedestris (Fabricius) (Hemiptera: Alydidae) under elevated CO2 concentrations. Entomol. Res. 51 (1), 12–23. doi:10.1111/1748-5967.12479
Anderson P., Hilker M., Hansson B. S., Bombosch S., Klein B., Schildknecht H. (1993). Oviposition deterring components in larval frass of Spodoptera littoralis (Boisd.) (Lepidoptera: noctuidae): a behavioural and electrophysiological evaluation. J. Insect Physiol. 39 (2), 129–137. doi:10.1016/0022-1910(93)90104-Y
Bae S., Yi H., Yoon Y., Jang Y., Kim Y., Maharijan R. (2019). Attraction of stink bugs to rocket traps with different combinations of wing and landing board color. J. Asia-Pac. Entomol. 22 (1), 243–249. doi:10.1016/j.aspen.2019.01.007
Brito N. F., Moreira M., Melo A. C. A. (2016). A look inside odorant-binding proteins in insect chemoreception. J. Insect Physiol. 95 (6), 51–65. doi:10.1016/j.jinsphys.2016.09.008
Campanacci V., Krieger J., Bette S., Sturgis J. N., Lartigue A., Cambillau C., et al. (2001). Revisiting the specificity of Mamestra brassicae and Antheraea polyphemus pheromone-binding proteins with a fluorescence binding assay. J. Biol. Chem. 276 (23), 20078–20084. doi:10.1074/jbc.M100713200
Chen X. L., Li G. W., Xu X. L., Wu J. X. (2018). Molecular and functional characterization of odorant binding protein 7 from the oriental fruit moth Grapholita molesta (Busck) (Lepidoptera: tortricidae). Front. Physiol. 9, 1762. doi:10.3389/fphys.2018.01762
Cheng W. N., Zhang Y. D., Yu J. L., Liu W., Zhu-Salzman K. (2020). Functional analysis of odorant-binding proteins 12 and 17 from wheat blossom midge Sitodiplosis mosellana Géhin (Diptera: cecidomyiidae). Insects 11 (12), 891–906. doi:10.3390/insects11120891
Cui X. N., Liu D. G., Sun K. K., He Y., Shi X. Q. (2018). Expression profiles and functional characterization of two odorant-binding proteins from the apple buprestid beetle Agrilus mali (Coleoptera: buprestidae). J. Econ. Entomol. 11 (3), 1420–1432. doi:10.1093/jee/toy066
Dong Y. M., Huang X. G., Yang Y. X., Li J. F., Zhang M. Q., Shen H., et al. (2022). Characterization of salivary secreted proteins that induce cell death from Riptortus pedestris (Fabricius) and their roles in insect-plant interactions. Front. Plant Sci. 13, 912603. doi:10.3389/fpls.2022.912603
D'Onofrio C., Zaremska V., Zhu J., Knoll W., Pelosi P. (2020). Ligand-binding assays with OBPs and CSPs. Method. Enzymol. 642, 229–258. doi:10.1016/bs.mie.2020.05.006
Endo N., Wada T., Sasaki R. (2011). Seasonal synchrony between pheromone trap catches of the bean bug, Riptortus pedestris (Heteroptera: Alydidae) and the timing of invasion of soybean fields. Appl. Entomol. Zool. 46 (4), 477–482. doi:10.1007/s13355-011-0065-7
Forêt S., Maleszka R. (2006). Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 16 (11), 1404–1413. doi:10.1101/gr.5075706
Gao Y., Chen J. H., Shi S. S. (2019). Research progress on soybean stink bug (Riptortus pedestris). Chin. J. Oil Crop Sci. 41 (5), 804–815. doi:10.19802/j.issn.1007-9084.2019033
Guo H., Wang C. Z. (2019). The ethological significance and olfactory detection of herbivore-induced plant volatiles in interactions of plants, herbivorous insects, and parasitoids. Arthropod-Plant Inte 13 (2), 161–179. doi:10.1007/s11829-019-09672-5
Guo J. L., An J. J., Chang H., Li Y. F., Dang Z. H., Wu C., et al. (2023). The lethal and sublethal effects of lambda-cyhalothrin and emamectin benzoate on the soybean pest Riptortus pedestris (Fabricius). Toxics 11 (2), 971–989. doi:10.3390/toxics11120971
He P., Chen G. L., Li S., Wang J., Ma Y. F., Pan Y. F., et al. (2019). Evolution and functional analysis of odorant binding proteins in three rice planthoppers: Nilaparvata lugens, Sogatella furcifera, and Laodelphax striatellus. Pest Manag. Sci. 75 (6), 1606–1620. doi:10.1002/ps.5277
Hong B., Chang Q., Zhai Y. Y., Ren B. W., Zhang F. (2022). Functional characterization of odorant binding protein PyasOBP2 from the jujube bud weevil, Pachyrhinus yasumatsui (Coleoptera: Curculionidae). Front. Physiol. 13, 900752. doi:10.3389/fphys.2022.900752
Huang G. Z., Liu J. T., Zhou J. J., Wang Q., Dong J. Z., Zhang Y. J., et al. (2018). Expressional and functional comparisons of two general odorant binding proteins in Agrotis ipsilon. Insect biochem. molec. 98, 34–47. doi:10.1016/j.ibmb.2018.05.003
Hull J. J., Perera O. P., Snodgrass G. L. (2014). Cloning and expression profiling of odorant-binding proteins in the tarnished plant bug, Lygus lineolaris. Insect Mol. Biol. 23 (1), 78–97. doi:10.1111/imb.12064
Jin Y., Zhang W. D., Dong Y. M., Xia A. (2022). Feeding behavior of Riptortus Pedestris (Fabricius) on soybean: electrical penetration graph analysis and histological investigations. Insects 13 (6), 511–523. doi:10.3390/insects13060511
Ju Q., Li X., Jiang X. J., Qu M. J., Guo X. Q., Han Z. J., et al. (2014). Transcriptome and tissue-specific expression analysis of obp and csp genes in the Dark Black Chafer. Arch. Insect Biochem. 87 (4), 177–200. doi:10.1002/arch.21188
Jung M., Lee D. H. (2018). Characterization of overwintering behaviors and sites of bean bug, Riptortus pedestris (Hemiptera: Alydidae), under laboratory and field conditions. Environ. Entomol. 47 (5), 1280–1286. doi:10.1093/ee/nvy123
Kim J., Park K. C., Roh H. S., Kim J., Oh H. W., Kim J. A., et al. (2016). Morphology and distribution of antennal sensilla of the bean bug Riptortus pedestris (Hemiptera: Alydidae): antennal sensilla of R. pedestris. Microsc. Res. Tech. 79 (6), 501–511. doi:10.1002/jemt.22658
Leal W. S. (2013). Odorant Reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi:10.1146/annurev-ento-120811-153635
Leal W. S., Barbosa R. M. R., Xu W., Ishida Y., Syed Z., Latte N., et al. (2008). Reverse and conventional chemical ecology approaches for the development of oviposition attractants for Culex mosquitoes. PLoS ONE 3 (8), e3045. doi:10.1371/journal.pone.0003045
Leal W. S., Higuchi H., Mizutani N., Nakamori H., Kadosawa T., Ono M. (1995). Multifunctional communication in Riptortus clavatus (Heteroptera: Alydidae): conspecific nymphs and egg parasitoid Ooencyrtus nezarae use the same adult attractant pheromone as chemical cue. J. Chem. Ecol. 21 (7), 973–985. doi:10.1007/BF02033802
Leal W. S., Nikonova L., Peng G. (1999). Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 464 (1-2), 85–90. doi:10.1016/S0014-5793(99)01683-X
Leal W. S., Ono M., Hasegawa M., Sawada M. (1994). Kairomone from dandelion, Taraxacum officinale, attractant for scarab beetle Anomala octiescostata. J. Chem. Ecol. 20 (7), 1697–1704. doi:10.1007/BF02059891
Li D. X., Li C. B., Liu D. G. (2021). Analyses of structural dynamics revealed flexible binding mechanism for the Agrilus mali odorant binding protein 8 towards plant volatiles. Pest Manag. Sci. 77 (4), 1642–1653. doi:10.1002/ps.6184
Li G. W., Chen X. L., Li B. L., Zhang G. H., Li Y. P., Wu J. X. (2016). Binding properties of general odorant binding proteins from the oriental fruit moth, Grapholita molesta (Busck) (Lepidoptera: tortricidae). PLoS ONE 11 (5), e0155096. doi:10.1371/journal.pone.0155096
Li G. W., Du J., Li Y. P., Wu J. X. (2015). Identification of putative olfactory genes from the oriental fruit moth Grapholita molesta via an antennal transcriptome analysis. PLoS ONE 10 (11), e0142193. doi:10.1371/journal.pone.0142193
Li J. B., Yin M. Z., Yao W. C., Ma S., Dewer Y., Liu X. Z., et al. (2022a). Genome-wide analysis of odorant-binding proteins and chemosensory proteins in the bean bug Riptortus pedestris. Front. Physiol. 13, 949607. doi:10.3389/fphys.2022.949607
Li K., Zhang X. X., Guo J. Q., Penn H., Wu T. T., Li L., et al. (2019). Feeding of Riptortus pedestris on soybean plants, the primary cause of soybean staygreen syndrome in the Huang-Huai-Hai river basin. Crop J. 7 (3), 360–367. doi:10.1016/j.cj.2018.07.008
Li L. L., Huang J. R., Xu J. W., Yao W. C., Yang H. H., Shao L., et al. (2022b). Ligand-binding properties of odorant-binding protein 6 in Athetis lepigone to sex pheromones and maize volatiles. Pest Manag. Sci. 78 (1), 52–62. doi:10.1002/ps.6606
Li M. Y., Jiang X. Y., Qi Y. Z., Huang Y. J., Li S. G., Liu S. G. (2020). Identification and expression profiles of 14 odorant-binding protein genes from Pieris rapae (Lepidoptera: pieridae). J. Insect Sci. 20 (5), 2–10. doi:10.1093/jisesa/ieaa087
Li Z. Q., Zhang S., Cai X. M., Luo J. Y., Dong S. L., Cui J. J., et al. (2017). Three odorant binding proteins may regulate the behavioural response of Chrysopa pallens to plant volatiles and the aphid alarm pheromone (E)-β-farnesene. Insect Mol. Biol. 26 (3), 255–265. doi:10.1111/imb.12295
Liu J. B., Liu J., Yi J. Q., Mao Y. K., Li J. H., Sun D. L., et al. (2021). Transcriptome characterization and expression analysis of chemosensory genes in Chilo sacchariphagus (Lepidoptera Crambidae), a key pest of sugarcane. Front. Physiol. 12, 636353. doi:10.3389/fphys.2021.636353
Liu Q., Yin M. Z., Ma S., Gu N., Qian L. F., Zhang Y. N., et al. (2023). Ligand-binding properties of chemosensory protein 1 in Callosobruchus chinensis to mung bean volatiles. Pestic. Biochem. Phys. 192, 105394. doi:10.1016/j.pestbp.2023.105394
Liu X. Q., Jiang H. B., Fan J. Y., Liu T. X., Meng L. W., Liu Y., et al. (2021). An odorant-binding protein of Asian citrus psyllid, Diaphorina citri, participates in the response of host plant volatiles. Pest Manag. Sci. 77 (7), 3068–3079. doi:10.1002/ps.6352
Mainali B. P., Kim H. J., Yoon Y. N., Oh I. S., Bae S. D. (2014). Evaluation of different leguminous seeds as food sources for the bean bug Riptortus pedestris. J. Asia-Pacific Entomol. 17 (2), 115–117. doi:10.1016/j.aspen.2013.11.007
Manoharan M., Chong M. N. F., Vaïtinadapoulé A. G., Etienne F., Sowdhamini R., Bernard O. (2013). Comparative genomics of odorant binding proteins in Anopheles gambiae, Aedes aegypti, and Culex quinquefasciatus. Genome Biol. Evol. 5 (1), 163–180. doi:10.1093/gbe/evs131
Martin J. P., Beyerlein A., Dacks A. M., Reisenman C. E., Riffell J. A., Lei H., et al. (2011). The neurobiology of insect olfaction: sensory processing in a comparative context. Prog. Neurobiol. 95 (3), 427–447. doi:10.1016/j.pneurobio.2011.09.007
O’Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., Hutchison G. R. (2011). Open babel: an open chemical toolbox. J. cheminformatics 3 (1), 33. doi:10.1186/1758-2946-3-33
Paula D. P., Togawa R., Costa M., Grynberg P., Martins N. F., Andow D. (2018). Systemic and sex-biased regulation of OBP expression under semiochemical stimuli. Sci. Rep. 8 (1), 6035. doi:10.1038/s41598-018-24297-z
Pelosi P., Iovinella I., Zhu J., Wang G., Dani F. (2018). Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 93 (1), 184–200. doi:10.1111/brv.12339
Pelosi P., Mastrogiacomo R., Iovinella I., Tuccori E., Persaud K. C. (2014). Structure and biotechnological applications of odorant-binding proteins. Appl. Microbiol. Biot. 98 (1), 61–70. doi:10.1007/s00253-013-5383-y
Pelosi P., Zhou J., Ban L., Calvello M. (2006). Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 63 (14), 1658–1676. doi:10.1007/s00018-005-5607-0
Pregitzer P., Zielonka M., Eichhorn A. S., Jiang X., Krieger J., Breer H. (2018). Expression of odorant-binding proteins in mouthpart palps of the desert locust Schistocerca gregaria. Insect Mol. Biol. 28 (2), 264–276. doi:10.1111/imb.12548
Quintana L. S. N., Olivero-Verbel J., Stashenko E. (2009). Repellent activity of essential oils: a review. Bioresour. Technol. 101 (1), 372–378. doi:10.1016/j.biortech.2009.07.048
Rihani K., Ferveur J. F., Briand L. (2021). The 40-year mystery of insect odorant-binding proteins. Biomolecules 11 (4), 509–536. doi:10.3390/biom11040509
Roh G. H., Cha D. H., Park C. G. (2021). Olfactory attraction to aggregation pheromone is mediated by disti-flagellum of antennal segments in Riptortus pedestris. J. Asia-Pacific Entomol. 24 (1), 415–420. doi:10.1016/j.aspen.2021.01.005
Scaloni A., Monti M., Angeli S., Pelosi P. (1999). Structural analysis and disulfide-bridge pairing of two odorant-binding proteins from Bombyx mori. Biochem. Bioph. Res. Co. 266 (2), 386–391. doi:10.1006/bbrc.1999.1791
Song J. Y., Lee G., Jung J., Moon J. K., Kim S. G. (2022). Effect of soybean volatiles on the behavior of the bean bug, Riptortus pedestris. J. Chem. Ecol. 48 (2), 207–218. doi:10.1007/s10886-021-01343-1
Sparks J. T., Bohbot J. D., Dickens J. C. (2014). The genetics of chemoreception in the labella and tarsi of Aedes aegypti. Insect biochem. molec. 48 (1), 8–16. doi:10.1016/j.ibmb.2014.02.004
Spinelli S., Lagarde A., Iovinella I., Legrand P., Tegoni M., Pelosi P., et al. (2012). Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules. Insect biochem. molec. 42 (1), 41–50. doi:10.1016/j.ibmb.2011.10.005
Sun Y. F., Qiao H. L., Ling Y., Yang S. X., Rui C. H., Pelosi P., et al. (2011). New analogues of (E)-β-farnesene with insecticidal activity and binding affinity to aphid odorant-binding proteins. J. Agric. Food Chem. 59 (6), 2456–2461. doi:10.1021/jf104712c
Sun Y. L., Huang L. Q., Pelosi P., Wang C. Z. (2012). Expression in antennae and reproductive organs suggests a dual role of an odorant-binding protein in two sibling Helicoverpa species. PLoS ONE 7 (1), e30040. doi:10.1371/journal.pone.0030040
Tang H. Y., Xie J. X., Liu J. T., Khashaveh A., Liu X. X., Yi C. Q., et al. (2023). Odorant-binding protein HvarOBP5 in ladybird Hippodamia variegata regulates the perception of semiochemicals from preys and habitat plants. J. Agric. Food Chem. 71 (2), 1067–1076. doi:10.1021/acs.jafc.2c07355
Trott O., Olson A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31 (2), 455–461. doi:10.1002/jcc.21334
Ullah R. M. K., Quershi S. R., Adeel M. M., Abdelnabby H., Waris M. I., Duan S. G., et al. (2020). An odorant binding protein (SaveOBP9) involved in chemoreception of the wheat aphid Sitobion avenae. Int. J. Mol. Sci. 21 (21), 8331. doi:10.3390/ijms21218331
Venthur H., Mutis A., Zhou J., Quiroz A. (2014). Ligand binding and homology modelling of insect odorant-binding proteins. Physiol. Entomol. 39 (3), 183–198. doi:10.1111/phen.12066
Venthur H., Zhou J. (2018). Odorant receptors and odorant-binding proteins as insect pest control targets: a comparative analysis. Front. Physiol. 9, 1163. doi:10.3389/fphys.2018.01163
Vogt R. G., Riddiford L. M. (1981). Pheromone binding and inactivation by moth antennae. Nature 293 (5828), 161–163. doi:10.1038/293161a0
Wang B., Dong W. Y., Li H. M., D’Onofrio C., Bai P. H., Chen R. P., et al. (2022). Molecular basis of (E)-β-farnesene-mediated aphid location in the predator Eupeodes corolla. Curr. Biol. 32 (5), 951–962.e7. doi:10.1016/j.cub.2021.12.054
Wang J. Z., Peng G., Luo Y. Q., Tao J. (2019). Characterization and expression profiling of odorant-binding proteins in Anoplophora glabripennis Motsch. Gene 693 (1), 25–36. doi:10.1016/j.gene.2018.12.075
Wang L., Bi Y. D., Liu M., Li W., Liu M., Di S. F., et al. (2019). Identification and expression profiles analysis of odorant-binding proteins in soybean aphid, Aphis glycines (Hemiptera: aphididae). Insect Sci. 27 (5), 1019–1030. doi:10.1111/1744-7917.12709
Wang L. Y., Liu Q. Y., Guo P., Gao Z. L., Chen D., Zhang T., et al. (2023). Evaluation of reference genes for quantitative real-time PCR analysis in the bean bug, Riptortus pedestris (Hemiptera: Alydidae). Insects 14 (12), 960–973. doi:10.3390/insects14120960
Xiao Y., Sun L., Wu Y. H., Wang Q., Zhang Y. J., Jing X. F., et al. (2023). The larvae of Phyllotreta striolata share the same olfactory cues for locating Brassicaceae plant with conspecific adults. J. Pest Sci. 97 (2), 979–992. doi:10.1007/s10340-023-01690-w
Yang Y. T., Luo L., Tian L. X., Zhao C. W., Niu H. L., Hu Y. F., et al. (2021). Function and characterization analysis of BodoOBP8 from Bradysia odoriphaga (Diptera: sciaridae) in the recognition of plant volatiles and sex pheromones. Insects 12 (10), 879–891. doi:10.3390/insects12100879
Yasuda T., Mizutani N., Honda Y., Endo N., Yamaguchi T., Moriya S., et al. (2007). A supplemental component of aggregation attractant pheromone in the bean bug Riptortus clavatus (Thunberg) (Heteroptera: Alydidae), related to food exploitation. Appl. Entomol. Zool. 42 (1), 161–166. doi:10.1303/aez.2007.161
Yin M. Z., Li J. Q., Liu Q., Ma S., Hu Z. Z., Liu X. Z., et al. (2023). Binding properties of chemosensory protein 12 in Riptortus pedestris to aggregation pheromone (E)-2-hexenyl (Z)-3-hexenoate. Pestic. Biochem. Phys. 194, 105513. doi:10.1016/j.pestbp.2023.105513
Zeng Y., Yang Y. T., Wu Q. J., Wang S. L., Xie W., Zhang Y. J. (2019). Genome-wide analysis of odorant-binding proteins and chemosensory proteins in the sweet potato whitefly, Bemisia tabaci. Insect Sci. 26 (4), 620–634. doi:10.1111/1744-7917.12576
Zhang F. M., Merchant A., Zhao Z. B., Zhang Y. H., Zhang J., Zhang Q. W., et al. (2020). Characterization of MaltOBP1, a minus-c odorant-binding protein, from the Japanese pine sawyer beetle, Monochamus alternatus Hope (Coleoptera: cerambycidae). Front. Physiol. 11, 212. doi:10.3389/fphys.2020.00212
Zhang J., Luo D., Wu P., Li H. Z., Zhang H. Y., Zheng W. W. (2018). Identification and expression profiles of novel odorant binding proteins and functional analysis of OBP99a in Bactrocera dorsalis. Arch. Insect Biochem. 98 (1), e21452. doi:10.1002/arch.21452
Zhang Q. H., Schneidmiller R., Hoover D. (2013). Essential oils and their compositions as spatial repellents for pestiferous social wasps. Pest Manag. Sci. 69 (4), 542–552. doi:10.1002/ps.3411
Zhang Q. H., Schneidmiller R. G., Hoover D. R., Zhou G. J., Margaryan A., Bryant P. (2014). Essential oils as spatial repellents for the brown marmorated stink bug, Halyomorpha halys (Stål) (Hemiptera: pentatomidae). J. Appl. Entomol. 138 (7), 490–499. doi:10.1111/jen.12101
Zhang Q. K., Li Z. B., Chen D. K., Wu S. Y., Wang H. H., Li Y. L., et al. (2022). The molecular identification, odor binding characterization, and immunolocalization of odorant-binding proteins in Liriomyza trifolii. Pestic. Biochem. Phys. 181 (6), 105016. doi:10.1016/j.pestbp.2021.105016
Zhang X., Huang C., Wu Q., Yang N. W., Qian W. Q., Wan F. H. (2021). Advances in the study of general odorant binding proteins in insects. J. Biosaf. 30 (1), 11–19. doi:10.3969/j.issn.2095-1787.2021.01.003
Zhang X. Q., Yan Q., Li L. L., Xu J. W., Mang D. Z., Wang X. L., et al. (2020). Different binding properties of two general-odorant binding proteins in Athetis lepigone with sex pheromones, host plant volatiles and insecticides. Pestic. Biochem. Phys. 164, 173–182. doi:10.1016/j.pestbp.2020.01.012
Zheng W., Zhang C. X., Li Y., Pearce R., Bell E. W., Zhang Y. (2021). Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep. Methods 1, 100014. doi:10.1016/j.crmeth.2021.100014
Zhou J. J., Field L., He X. L. (2010). Insect odorant-binding proteins: do they offer an alternative pest control strategy? Outlooks Pest Manag. 21 (1), 31–34. doi:10.1564/21feb08
Zhou X., Wang Z., Cui G. C., Du Z. M., Qian Y. L., Yang S. M., et al. (2022). Binding properties of odorant-binding protein 4 of Tirathaba rufivena to areca catechu volatiles. Plants 11 (2), 167–178. doi:10.3390/plants11020167
Zhu J., Zaremska V., D'Onofrio C., Knoll W., Pelosi P. (2020). Site-directed mutagenesis of odorant-binding proteins. Method. Enzymol. 642, 301–324. doi:10.1016/bs.mie.2020.05.014
Zhu J. Q., Wang F., Zhang Y. J., Yang Y. T., Hua D. K. (2023). Odorant-binding protein 10 from Bradysia odoriphaga (Diptera: sciaridae) binds volatile host plant compounds. J. Insect Sci. 23 (1), 7–8. doi:10.1093/jisesa/iead004
Zhu X. Y., Li J. B., Liu L., Dewer Y., Zhang H., Zhang H. R., et al. (2022). Binding properties of odorant-binding protein 4 from bean bug Riptortus pedestris to soybean volatiles. Insect Mol. Biol. 31 (6), 760–771. doi:10.1111/imb.12802
Keywords: Riptortus pedestris, OBPs, soybean volatiles, fluorescence competitive binding, molecular docking
Citation: Guo J, Liu P, Zhang X, An J, Li Y, Zhang T and Gao Z (2025) Characterization of the ligand-binding properties of odorant-binding protein 38 from Riptortus pedestris when interacting with soybean volatiles. Front. Physiol. 15:1475489. doi: 10.3389/fphys.2024.1475489
Received: 04 August 2024; Accepted: 09 December 2024;
Published: 06 January 2025.
Edited by:
Ana Claudia A. Melo, Federal University of Rio de Janeiro, BrazilReviewed by:
Xue-Qing Yang, Shenyang Agricultural University, ChinaCopyright © 2025 Guo, Liu, Zhang, An, Li, Zhang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Zhang, Y2F1emh0QDE2My5jb20=; Zhanlin Gao, emJzMzA4QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.