- Key Laboratory of Sustainable Forest Ecosystem Management-Ministry of Education, College of Forestry, Northeast Forestry University, Harbin, China
Neuropeptides function as central neuromodulators and circulating hormones that modulate insect behavior and physiology. Leucokinin (LK) is an intercellular signaling molecule that mediates many physiological and behavioral processes. However, the functions of LK associated with environmental stress and feeding behavior in the fall webworm, Hyphantria cunea, is little known. Our primary objective is to understand the function of LK and LK receptor (LKR) neuroendocrine system in H. cunea. In the present study, the results showed that LK/LKR are expressed at different developmental stages and in various tissues of H. cunea. A candidate receptor–ligand pairing for LK was identified in the larval transcriptome of H. cunea. In a heterologous expression system, the calcium assay was used to demonstrate that LKR is activated by HcLKs in a dose-dependent manner, with 50% effective concentration (EC50) values of 8.44–90.44nM. Knockdown of HcLK and HcLKR by microinjecting target-specific dsRNA leads to several effects in H. cunea, including feeding promotion, increase in resistance to desiccation and starvation stress, and regulation of water homeostasis. The transcript levels of HILP2 (except in the LK knockdown group), HILP5, and HILP8 increased, whereas those of HILP3, HILP4, and HILP6 decreased; HILP1, HILP2 (in the LK knockdown group), and HILP7 gene expression was not influenced after LK and LKR knockdown. Variations in mRNA expression levels in insulin-like peptide genes in the knockdown larvae suggest an essential role of these genes in survival in H. cunea. To our knowledge, the present study is the first comprehensive study of LK and LKR – from gene to behavior – in H. cunea.
Introduction
As the central neuromodulators and circulating hormones, neuropeptides orchestrate insect behavior and physiology. The complex hormonal and neuronal regulatory mechanisms maintain the metabolic homeostasis, which balance the food intake, energy expenditure, and nutrient storage in insects (Murphy and Bloom, 2006; Baker and Thummel, 2007; Leopold and Perrimon, 2007; Woods and D’Alessio, 2008; Teleman, 2010; Dalamaga et al., 2013; de Araujo et al., 2013; Vogt and Bruning, 2013). Mechanisms of feeding and metabolism have been explored in depth in Drosophila melanogaster (Baker and Thummel, 2007; Itskov and Ribeiro, 2013; Owusu-Ansah and Perrimon, 2014; Padmanabha and Baker, 2014), and it is known that food ingestion and metabolic homeostasis are mediated by several peptide hormones (Wu et al., 2003, 2005; Melcher and Pankratz, 2005; Géminard et al., 2006; Bharucha et al., 2008; Al-Anzi et al., 2010; Cognigni et al., 2011; Hergarden et al., 2012; Söderberg et al., 2012; Itskov and Ribeiro, 2013). Insect food ingestion is associated with a balance of water and ions (Coast et al., 2002; Dow and Davies, 2006; Dow, 2009). Thus, it is likely that insect diuretic hormones collaborate with the hormones released after food intake to regulate satiety, metabolism, and energy reallocation.
Kinins (leucokinins) in insects have a highly conserved C-terminal pentapeptide sequence – Phe-Xaa-Xbb-Trp-Gly-NH2, where Xaa represents Tyr, His, Ser, or Asn; Xbb may be Ala but is generally Ser or Pro (Holman et al., 1990, 1999). Insect leucokinins (LKs) are multifunctional peptides acting as neurohormones and neurotransmitters, which regulate diuresis, sleep, metabolism, response to ionic stress, food intake, and taste responsiveness (Terhzaz et al., 1999; Radford et al., 2002; Al-Anzi et al., 2010; Cognigni et al., 2011; López-Arias et al., 2011; Kwon et al., 2016; Zandawala et al., 2018a,b; Yurgel et al., 2019). In D. melanogaster, LK acts in vitro on stellate cells of the renal tubules to trigger fluid secretion, which is produced by a small set of neurons and neurosecretory cells in the central nervous system (CNS; de Haro et al., 2010). Leucokinins aid fluid excretion by increasing the secretion of primary urine by the Malpighian tubules and contracting the hindgut. Together with insulin signaling, the LK neuropeptide regulates stress tolerance and metabolism in D. melanogaster (Zandawala et al., 2018a).
The fall webworm Hyphantria cunea Drury (Lepidoptera: Noctuidae), a worldwide forest pest that originated in North America, was first reported in China in 1979 (Rong et al., 2003; Zhang et al., 2008). To alleviate the damage caused by H. cunea, various control strategies have been developed, such as natural predation, microbial intervention, and insecticide usage (Beckage, 2008). Because neuropeptides are regulators of critical life processes in insects and are highly specific, they are the potential targets in the development of green insecticides. The present study aims to understand the neuroendocrine pathways regulating the key physiological processes in pest insects for screening the potential analogs. The leucokinin signaling system has been studied in several other insect species; however, localization and functional roles of leucokinin in H. cunea remain unknown.
In this study, we first investigated the function of the LK ligand and receptor signaling system in H. cunea. Subsequently, we determined the transcript levels of the LK and LK receptor (LKR) genes under starvation to examine whether this signaling system was affected by the feeding behavior of H. cunea. LK gene knockdown via RNAi was used to further examine the potential relationship between LK signaling and the feeding behavior of H. cunea. We demonstrate that LK signaling regulates starvation stress and feeding.
Materials and Methods
Insects
Hyphantria cunea eggs and artificial diets were obtained from the Research Institute of Forest Ecology, Environment and Protection, Chinese Academy of Forestry (Beijing, China). Eggs were incubated at 25°C until hatching, and larvae were fed on artificial diets in 250ml transparent plastic bottles, which were maintained at 25±1°C with a 16:8h light:dark photoperiod.
Molecular Cloning and Plasmid Construction
Reverse transcription PCR was initially used to validate the sequences of H. cunea LK and LKR transcripts from the H. cunea genome database. The LK and LKR genes were cloned using the following thermal conditions: 94°C for 3min; followed by 35cycles of 94°C for 30s, 60°C for 30s, and 72°C for 1min; then a final extension at 72°C for 10min. The PCR product was sub-cloned into pMD18-T vector (TaKaRa, Japan) and then verified sequences. The primers used for the PCR cloning of HcLK and Hyphantria cunea leucokinin receptor (HcLKR) are presented in Table 1. The PCR products were directly cloned into the pcDNA-3.1-myc-His vector. The recombinant vectors were verified by sequencing.
Analysis of LK and LKR
The deduced amino acid sequences of LK and LKR orthologs were obtained from GenBank using BLAST searches (blastx and tblastx). Multiple alignment of the amino acid sequences was performed using the ClustalX2 program and BioEdit. A phylogenetic tree was constructed using the neighbor-joining (NJ) method in MEGA 5.0 with 1,000 bootstrap replicates (Tamura et al., 2011). Signal peptides were predicted using Signal P 4.1 Server (Mccarthy et al., 2004), and transmembrane domains were predicted using TMHMM server v2.02 (Sonnhammer et al., 1998). The presence of N-glycosylation sites in predicted protein sequences was assessed using NetNGlyc 1.01, and the generation of sequence logos for the C-terminal motifs of LK proteins was created by Weblogo (Crooks et al., 2004).
Cell Culture and Transfection
The human embryonic kidney 293 (HEK293) cell line was cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum (FBS) and 4mM L-glutamine (Invitrogen) at 37°C in a humidified incubator containing 5% CO2. HEK293 cells were transfected with LKR cDNA plasmid constructs using Effectene transfection reagent (Qiagen) according to the manufacturer’s instructions. Two days after transfection, stably expressing cells were selected by the addition of 800mg/L G418.
Intracellular Calcium Assay
To investigate the interaction between the LKR and LKs in H. cunea, the response of the LKR to chemically synthesized LKs was examined using the Ca2+ imaging assay. A fluorescent Ca2+-sensitive probe, Fura-4/AM (Beyotime, Shanghai, China), was used to detect the intracellular cytosolic calcium signals according to the manufacturers’ instructions. In brief, HEK293 cells stably expressing LKR were washed twice with phosphate-buffered saline and were suspended at 5×106 cells/ml in Hanks’ balanced salt solution. The cells were then loaded with 2μl Fura-4/AM for 20min and washed twice with HBSS buffered medium. Then, cells were stimulated with 0.1 and 1μM HcLKs (HcLK-1, HcLK-2, and HcLK-3) chemically synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Each 96-well plate was transferred into a Multi-Mode Microplate Reader (Varioskan Flash Beckman XL-70 F; Thermo Fisher Scientific Inc. Waltham, MA) to monitor the Fluo-4 fluorescence. The excitation wavelength was 485nm, and fluorescence emission was detected at 520nm. Various concentrations of receptor ligands were added when Fluo-4 fluorescence had reached a stable value in each well. The changes in Fluo-4 fluorescence were recorded automatically. Dose–response curves for putative agonists were established in at least three independent experiments.
RNA Interference
A 463-bp dsRNA representing the H. cunea LK-encoding gene sequence and a 505-bp dsRNA representing the H. cunea LKR-encoding gene sequence were synthesized using the MEGAscript T7 high-yield transcription kit (Ambion) according to the manufacturers’ protocol. The dsRNA was purified with phenol/chloroform followed by ethanol precipitation. The dsRNA of the enhanced green fluorescent protein gene (pEGFP-N1 plasmid as template, WP_031943942.1, 507-bp dsRNA) was employed as a control. A 2μg/μl dsRNA solution (1μl) was microinjected into the penultimate posterior abdominal section of individual seventh instar H. cunea larvae using an injection needle (MICROLITERTM #65 with 33-gauge needle, Hamilton Co., Reno, NV, United States) under ice anesthesia (Sun et al., 2016). Control H. cunea larvae were microinjected with the EGFP dsRNA. Microinjected H. cunea larvae were allowed to recover for 2h at room temperature and then reared on an artificial diet under a 16:8h light: dark photoperiod at 25±1°C. After 72 and 96h, LK and LKR mRNA levels in the dsRNA-treated seventh instar H. cunea larvae were measured by qRT-PCR technology.
Bioassays
To measure water content, the larvae treated with dsEGFP, dsLK, and dsLKR for 48h were dehydrated at 80°C until a constant weight. Ten H. cunea larvae were weighed before and after dehydration using a Mettler MT5 analytical microbalance (Columbus, OH, United States). Water content was calculated as the difference between the fresh and dry weight. Each replicate contained 10 H. cunea larvae, and the experiment was performed in triplicate.
To study survival under desiccation and starvation, the H. cunea larvae treated with dsRNA were kept in empty vials or vials containing cotton ball with sterile water, respectively. Ten H. cunea larvae were used per replicate, and the experiment was performed in triplicate. The survival was recorded every 24h until all the H. cunea larvae were dead. The vials were placed in an incubator at 25±1°C under normal photoperiod conditions (16:8h light: dark).
Food Intake Assay
On day of the seventh instar stage, H. cunea larvae were microinjected with dsRNA (LK and LKR dsRNA or EGFP dsRNA) and then returned to transparent plastic vials and starved for 24h. After a subsequent 4-day feeding period, the appetite of the larvae was checked by measuring the amount of artificial diet eaten by individual larvae during 24h. The weight of the artificial diet was measured before and after H. cunea larva feeding. Three biological replicates were included for each experiment, and for each biological replicate, 10 H. cunea larvae were kept in transparent plastic vials. The vials were placed in an incubator at 25°C under normal photoperiod conditions (16:8h light: dark).
Quantitative Real-Time Reverse Transcription PCR
The RNA was extracted from H. cunea eggs, first to seventh instar larvae, pupae, adults, and tissue samples using the RNeasy Mini Kit (Qiagen, Valencia, CA, United States). The tissues – head, silk glands, midgut, epidermis, testis, ovary, Malpighian tubules, and fat body – were collected from larvae on day 1 of the seventh instar stage. cDNA was synthesized using the total RNA (0.5μg) and the PrimeScript®RT Reagent Kit with gDNA Eraser (Perfect Real Time, TaKaRa, Japan), according to the manufacturer’s protocol. The mRNA levels of LK, LKR and insulin-like peptide (ILP) genes were assessed using RT-qPCR with a SYBR Green kit (Toyobo, Osaka, Japan) and MJ Opticon™2 machine (Bio-Rad, Hercules, CA, United States). The reaction mixture (20μl) was composed of SYBR Green Real-time PCR Master Mix (10μl; Toyobo), nuclease-free water (7μl), gene-specific primers (1μl, 0.5μM; Table 1), and cDNA template (2μl; equivalent to 50ng of total RNA). RPL13 and EF-1α were used as internal reference genes (Sun et al., 2019). The conditions for RT-qPCR reactions were as follows: 1cycle at 95°C for 30s, followed by 45cycles at 95°C for 12s, 60°C for 30s, 72°C for 40s, and 82°C for 1s for plate reading. The purity of the amplified products was analyzed by melting curve analysis. qRT-PCR was performed in using independent biological repeats in triplicate to ensure the reproducibility of the results. The expression levels of the clones were calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Statistical Analysis
Statistical analysis was performed using SPSS (v17.0, SPSS Inc., Chicago, Illinois). One-way ANOVA was performed using Prism 8.0 (GraphPad Software, La Jolla, CA, United States). Value of p<0.05 was considered to indicate statistical significance for all experiments performed in the present study.
Results
HcLK and HcLKR Analyses
The sequences of LK and LKR genes were identified using transcriptome and genome analysis (Sun et al., 2019; Wu et al., 2019). The LK gene contains a 1,014bp open reading frame (ORF), which encodes a signal peptide (23 residues). The three mature peptide sequences comprise six (YFSPWGamide, HcLK-1), seven (VRFSPWGamide, HcLK-2), and eight (KVKFSAWGamide, HcLK-3) amino acid residues, respectively. The mature peptide cleavage site is a combination of lysine (K) and arginine (R) and has an amidation site “G” (Figure 1A). LK proteins from H. cunea and other insects showed very high sequence similarity (Figure 1B). Phylogenetic analysis revealed that HcLK and LKs from other insect species were clustered in a single group and that HcLK is most closely related to the Danaus plexippus plexippus homologs (Figure 1C).
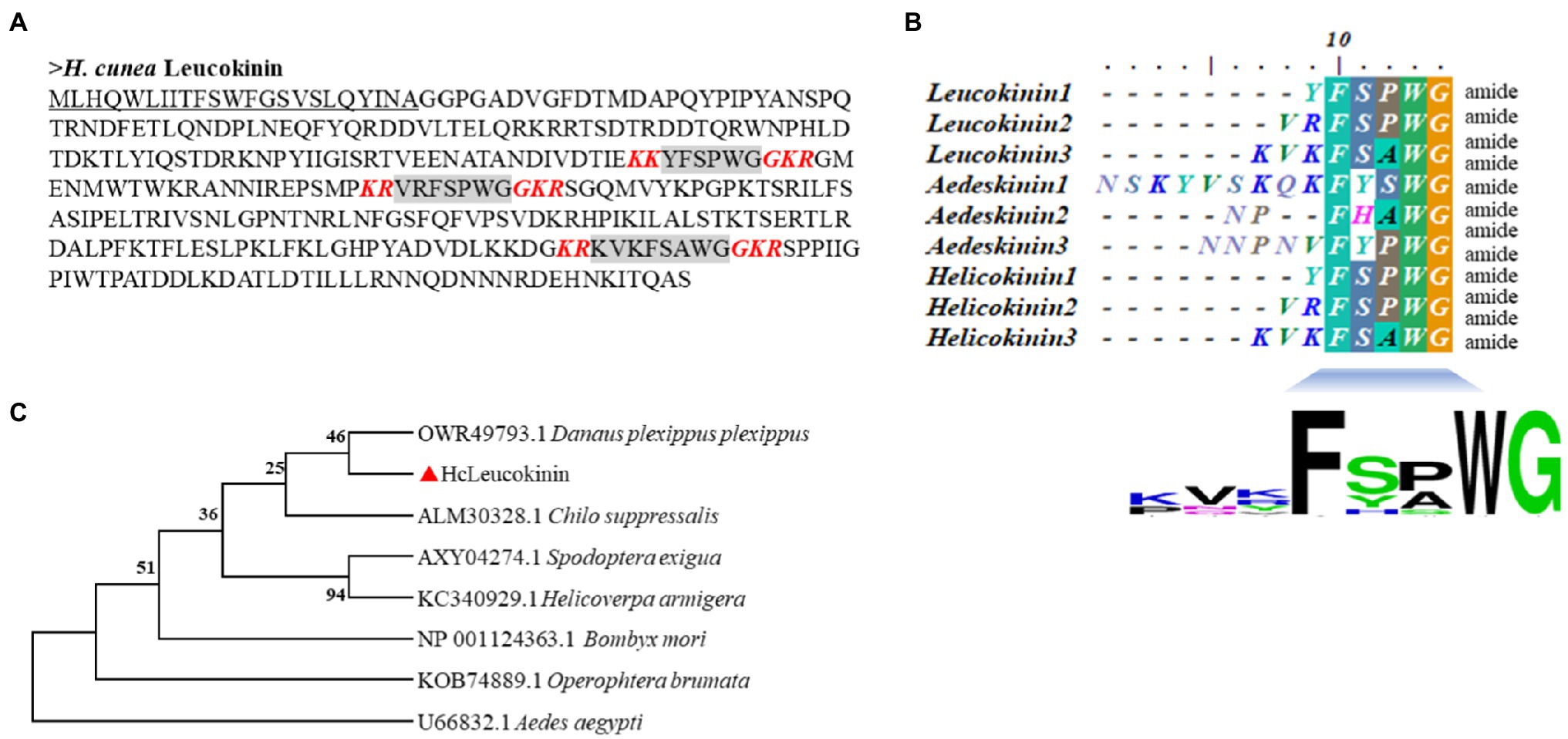
Figure 1. Comparison of the amino acid sequences of the Hyphantria cunea leucokinin (HcLK) precursor with those of other insect species. (A) shows the deduced amino acid sequence of HcLK. The underlined part of the sequence indicates the N-terminal putative signal peptide, and the gray part indicates the predicted HcLK peptide. The red bold and italic letters indicate the predicted amidation signals with dibasic cleavage sites. (B) shows an alignment of the consensus sequences of H. cunea, Aedes aegypti, and Helicoverpa zea putative, mature LK peptides. The calculated consensus logo is shown at the bottom. (C) shows cluster analysis of HcLK in various arthropods.
The full-length HcLKR cDNA consists of 2,186 nucleotides; the predicted ORF encodes 485 amino acids (Figure 2A). The ORF contains an ATG initiation codon, an upstream 608bp 5′ untranslated region (UTR), and a termination (TAA) codon followed by a 120bp 3′ UTR (Figure 2A). The HcLKR protein contains the characteristic seven transmembrane domains (TM, Figure 1B, TMHMM 2.0 server), with a typical signature of rhodopsin-like G protein-coupled receptor (GPCR; Figure 2A). The amino acid residues at positions 49–73, 82–104, 120–142, 163–179, 215–238, 266–291, and 306–331 represented TMI, TMII, TMIII, TMIV, TMV, TMVI, and TMVII, respectively. The predicted three-dimensional model of HcLKR showed a characteristic structure, with seven TM segments with α-helices (TM-I to TM-VII) linked by three intracellular and three extracellular loops, an extracellular amino terminus, and an intracellular carboxyl terminus (Figure 2B). Likewise, the Pfam analysis predicted seven transmembrane passes, and six conserved cysteine residues in the N-terminal extracellular domain. Six potential N-glycosylation sites were predicted for the N-terminal extracellular domain (NetNGlyc 1.0 server). Multiple amino acid sequence alignment between HcLKR and other LKRs showed high overall amino acid homology in the seven transmembrane domains (Figure 2B).
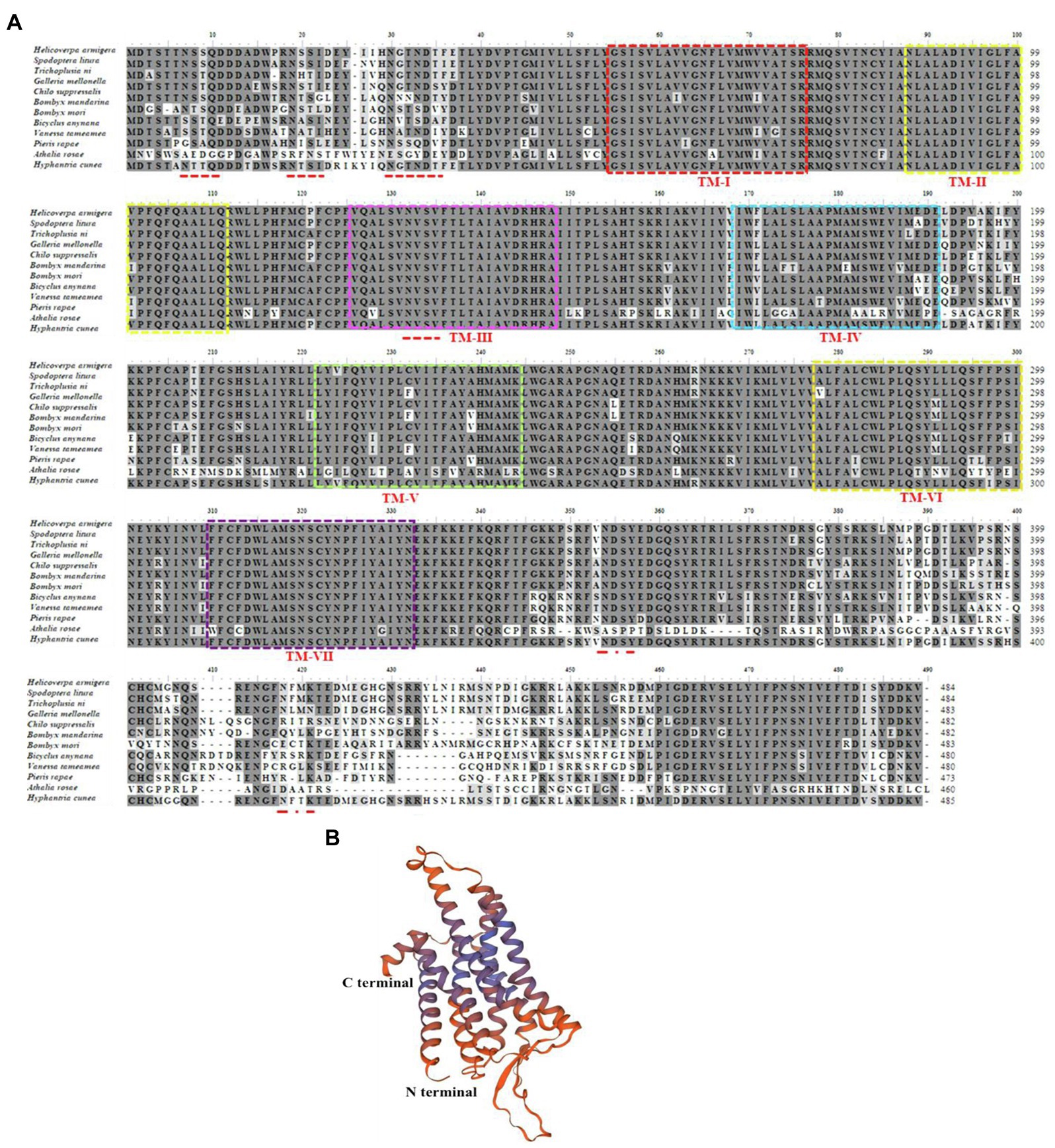
Figure 2. Amino acid sequence alignment of leucokinin receptor (LKR) in Hyphantria cunea with functionally deorphanized homologs in other insects (A) and predicted three-dimensional structure of mature HcLKR (B). LKR sequences from the following species were used: Helicoverpa armigera XP_021201003.1, Spodoptera litura XP_022816525.1, Trichoplusia ni XP_026729237.1, Galleria mellonella XP_026751254.1, Chilo suppressalis ALM88319.1, Bombyx mandarina XP_028041362.1, Bombyx mori NP_001127721.1, Bicyclus anynana XP_023939936.1, Vanessa tameamea XP_026486140.1, Pieris rapae XP_022117160.1, and Athalia rosae XP_012267620.1. For ease of interpretation, identical residues are shaded black and conserved substitutions are shaded gray. The seven predicted transmembrane regions for all LKRs are marked with boxes of different colors. Putative N-glycosylation sites on the extracellular N–terminal domain of H. cunea LKR are indicated by red lines.
Developmental and Tissue-Specific Expressions of HcLK and HcLKR
The tissue-specific and developmental mRNA profiles of HcLK and HcLKR in H. cunea were quantified using RT-qPCR (Figure 3). Compared with that at the egg stage, the transcript level of HcLK in the first instar larvae was the highest (1.81-fold that in eggs) and that in the seventh instar larvae was the lowest (0.47-fold that in eggs). The expression level of HcLK in the hindgut was 24.45-fold of that in the head (Figures 3A,B). The expression of HcLK in silk gland, foregut, Malpighian tubules, testis, and ovary was 0.24-fold, 0.74-fold, 0.52-fold, 1.87-fold, and 0.38-fold of that in head tissue, respectively, and did not differ significantly. The HcLKR expression in the first and fifth larval stages was similar but significantly higher than that at other instar stages (p<0.05, Figure 3C). Compared with that in the head, the transcript level of HcLKR in the hindgut was the highest (32.96-fold that in the head and that in the fat body was the lowest 0.0004-fold that in the head). The HcLKR expression in the epidermis, silk gland, foregut, Malpighian tubules, ovary, and testis was 0.0004–1.16-fold that in the head (Figure 3D).
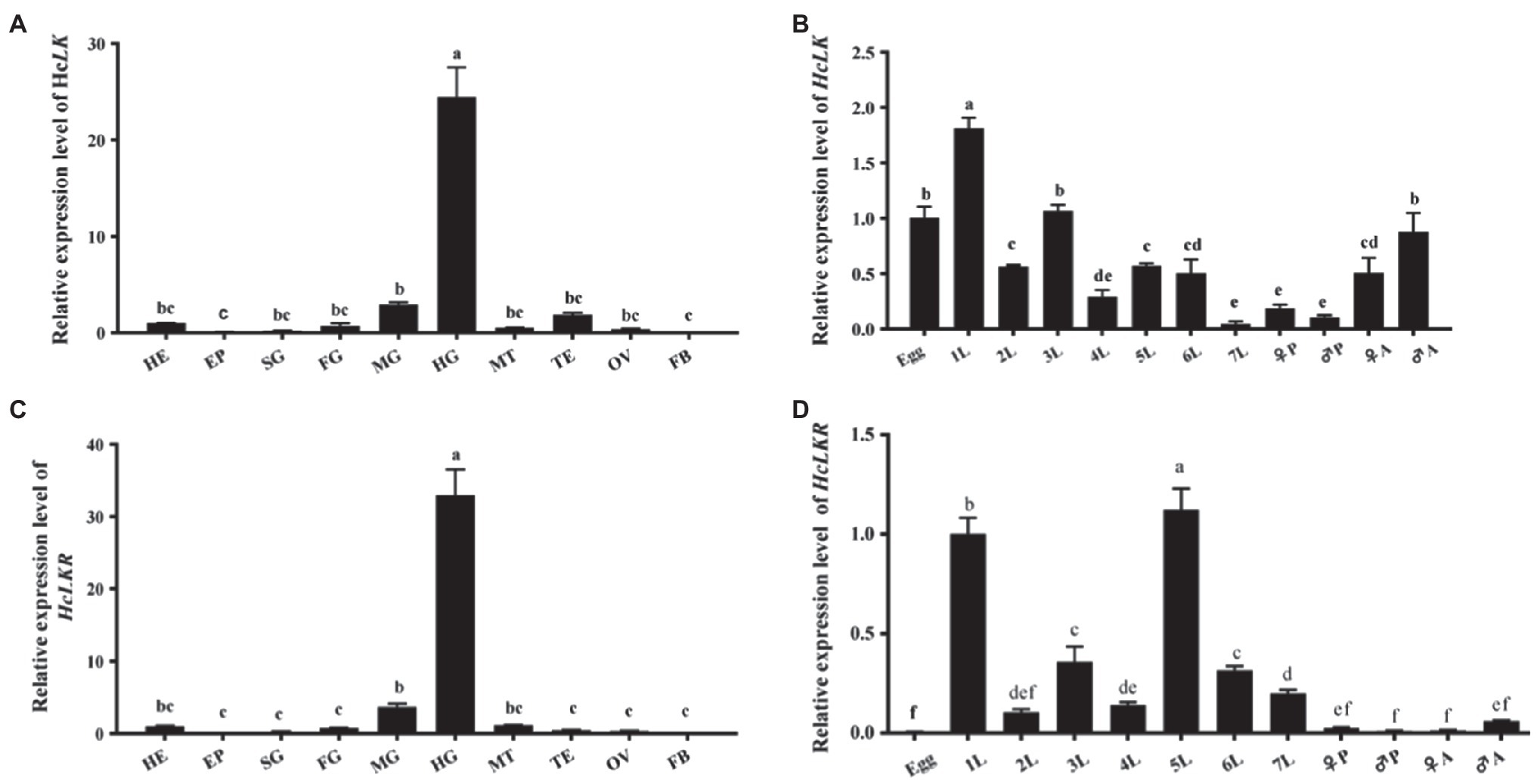
Figure 3. mRNA expression levels of Hyphantria cunea leucokinin and leucokinin receptor (HcLK and HcLKR) genes in Hyphantria cunea. (A) HcLK and (C) HcLKR mRNA expression levels in different tissues. Head (HE), epidermis (EP), silk gland (SG), foregut (FG), midgut (MG), hindgut (HG), Malpighian tubules (MT), testis (TE), ovary (OV), and fat body (FB) were dissected from H. cunea larvae on day 1 of the seventh instar stage; (B) HcLK and (D) HcLKR mRNA expression levels at various developmental stages. Each value is presented as mean±SD (n=3). Different lowercase letters (a–f) represent significant difference of HcLK and HcLKR levels among various tissues and developmental stages determined by one-way ANOVA followed by Tukey’s multiple comparison test (p<0.05).
Functional Activation of HcLKR
The ORF of the HcLKR was inserted into the expression vector pcDNA3.1-Myc-His to construct a recombinant plasmid for stable expression. The HcLK gene encodes a 338-amino acid polypeptide (Figure 1A), which is a precursor of three LKs – LK-1–3 (Figure 4A). Notably, HEK293 cells expressing HcLKR responded to all HcLKs at a concentration of 1μM. The dose response of LKR to LKs was further investigated (Figure 4B). Of the three tested LKs, LK-2, and LK-3 stimulated LKR at lower concentrations, with EC50 values of 28.0 and 8.44nM, respectively, whereas LK-1 showed a lower activity (EC50 values: 90.44nM).
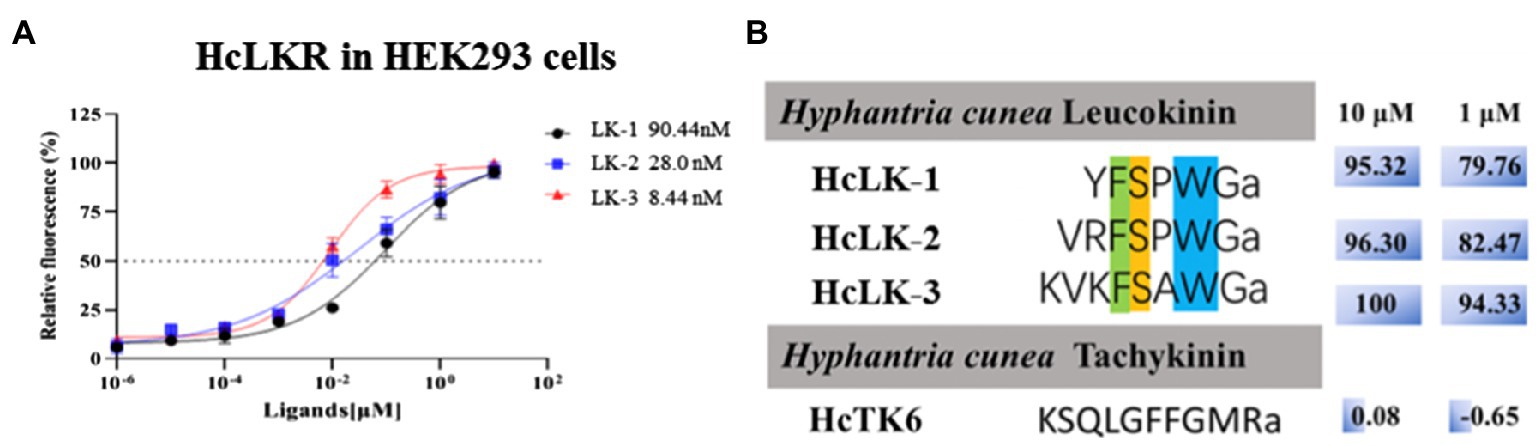
Figure 4. Ligand–receptor specificity of the Hyphantria cunea leucokinin receptor (HcLKR). (A) shows concentration-dependent response curves for HcLKR expressed in HEK293 cells induced by HcLKs. (B) shows ligand activity calculated on the basis of relative activity compared with the highest response of the receptor for HcLK-3.
Functions of HcLK and HcLKR by RNAi
Considering the induction of HcLK and HcLKR mRNA expression by starvation stress, we investigated whether HcLK and HcLKR gene expression in the systemic silence plays a functional role in organismal stress tolerance employing knockdown of HcLK and HcLKR via dsRNA microinjection. The HcLK and HcLKR knockdown larvae showed ~80% lower LK and LKR mRNA levels than the control dsEGFP larvae after 96h (Figures 5A,B). Next, the survival of H. cunea RNAi larvae was investigated following desiccation and starvation stress.
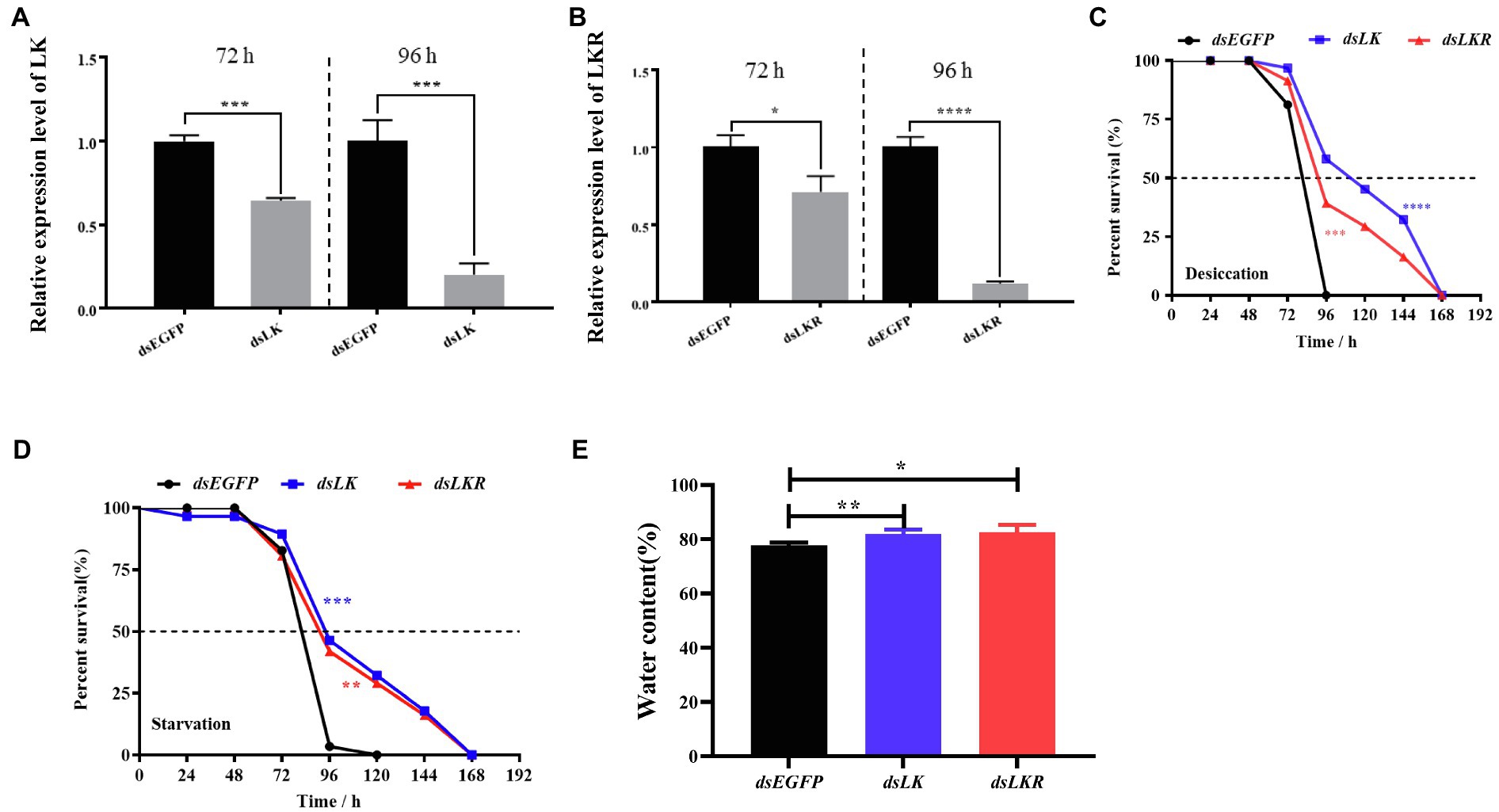
Figure 5. Effects of LK and LKR knockdown on seventh instar Hyphantria cunea larvae. (A) and (B) show effects of RNAi on gene expression of LK and LKR at 72 and 96h, respectively. Microinjection with double-stranded (ds) RNA targeting enhanced green fluorescent protein (EGFP) was used as a negative control. Values (mean±SD) are based on three biological replicates; each replicate contained pooled samples from four larvae. p values were calculated by unpaired t-test (*p<0.05, ***p<0.001, and ****p<0.0001). (C) and (D) show the survival rate of H. cunea larvae with LK or LKR knockdown under desiccation and starvation, respectively. Data are presented as survival curves, and the error bars represent SE [**p<0.01, ***p<0.001, and ****p<0.0001, as assessed by Log-rank (Mantel-Cox) test]. (E) shows water content of H. cunea larvae with LK and LKR knockdown. *p<0.05 and **p<0.01 as assessed by one-way ANOVA followed by Tukey’s multiple comparison test.
Under desiccation and starvation stress, HcLK and HcLKR RNAi larvae survived longer than control larvae (Figures 5C,D). To determine whether the difference in survival rates of larvae stems from changes in water content, the water content in H. cunea larvae microinjected with dsEGFP, dsLK, and dsLKR were assayed after 48h of desiccation treatment. As expected, H. cunea larvae with dsLK and dsLKR silencing contained more water than those in control dsEGFP group (Figure 5E).
The expression of ILP genes in H. cunea was altered in dsLK and dsLKR larvae after 48h of starvation. Significant effects on HILP transcription were observed only for HILP2 (except dsLK treatment), HILP3, HILP4, HILP5, HILP6, and HILP8. The transcript levels of HILP2, HILP5, and HILP8 in the dsLKR larvae were significantly higher (1.31–4.42-fold) than those in the dsEGFP group. However, the transcript levels of HILP3, HILP4, and HILP6 in the dsLK larvae were significantly lower (0.33–0.52-fold) than those in the dsEGFP group (Figure 6). Complex results were also observed when LKR and LK were knocked down in H. cunea larvae, the LK signal negatively regulated HILP3, HILP4, and HILP6 expression but positively regulated HILP5 and HILP8 expression and played no significant regulatory role in HILP1 and HILP7 expression (Figure 6).
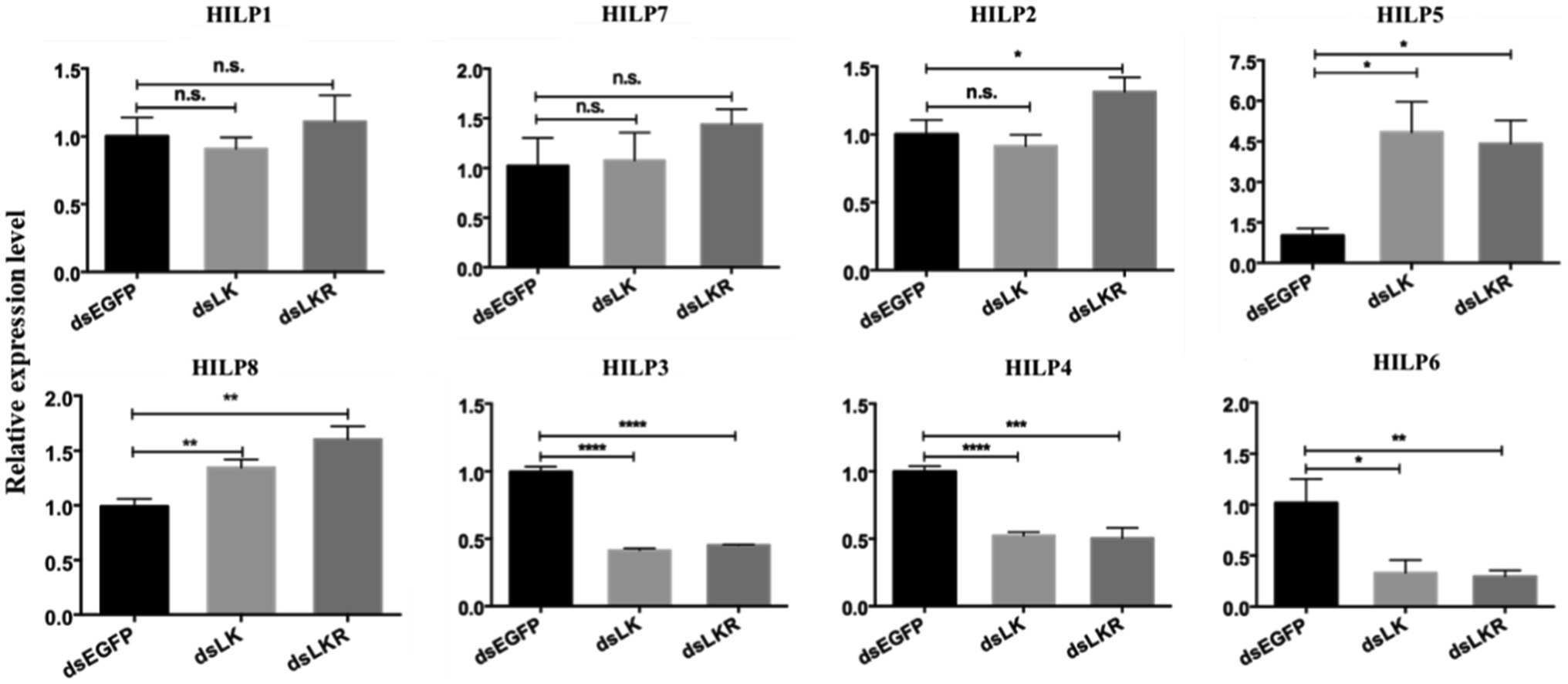
Figure 6. Effects of HcLK and HcLKR knockdown on insulin expression and starvation resistance. Values (mean±SD) are based on three biological replicates; each replicate contained pooled samples from four larvae. *p<0.05, **p<0.01, ***p<0.001, and ****p<0.001 as assessed by unpaired t-test. Values marked with “ns” are not significantly different (p>0.05; t test; n=3).
HcLK and HcLKR Knockdown Promoting Feeding Behavior
Our results suggested that LK signaling is associated with starvation stress. Thus, the HcLK and HcLKR knockdown mutants were found to affect food intake over different periods. The food intake of larvae microinjected with dsHcLK and dsHcLKR after starvation for 1day was significantly different from that of larvae microinjected with dsEGFP (Figure 7). During the feeding time tested, the food intake of dsHcLK and dsHcLKR larvae was significantly higher than that of the control dsEGFP larvae. The food intake of dsHcLK and dsHcLKR larvae on the day 1 was 1.61- and 1.62-fold higher than that of the control dsEGFP larvae, respectively (Figure 7). On day 4 of feeding, the food intake of dsHcLK and dsHcLKR larvae was 1.26- and 1.66-fold higher than that of the control dsEGFP larvae, respectively.
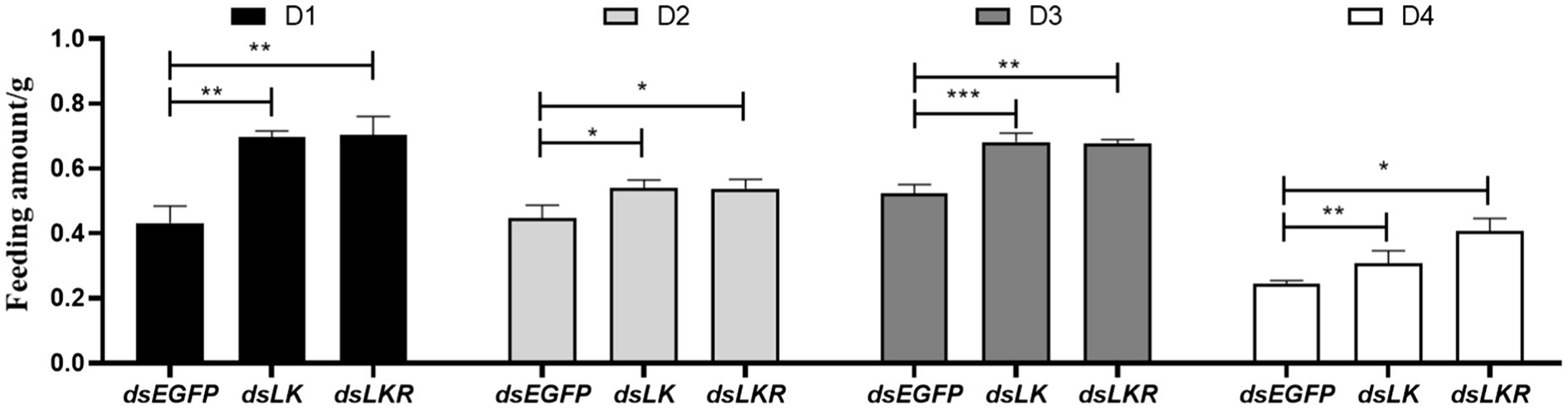
Figure 7. Effects of LK and LKR gene silencing on feeding behavior in H. cunea. Microinjection with ds RNA targeting EGFP was used as a negative control. Values (mean±SD) are based on three biological replicates. Each replicate contained pooled samples from 10 larvae. *p<0.05, **p<0.01, and ***p<0.001 as assessed by unpaired t-test (n=3).
Discussion
Leucokinin, a multifunctional peptide acting as a neurohormone and neurotransmitter, is primarily synthesized in the CNS. Only a single LK gene was identified in D. melanogaster (Terhzaz et al., 1999). However, a single LK gene was first identified in H. cunea; which shares a similar typical structure of the LK family. Specifically, three putative LK proteins (LK-1–3) in H. cunea (HcLK-1–3) possess the general C-terminal motif sequence FxyWGamide (Veenstra et al., 1997). HcLK-1–3 showed high similarity with helicokinins 1–3 of Helicoverpa zea. The LKs are highly conserved between H. cunea and H. zea (Figure 1B). The HcLK genes were expressed in various tissues of H. cunea, especially highly expressed in the midgut and hindgut, as has been demonstrated in several insect species (Terhzaz et al., 1999; Kwon et al., 2016). In Grapholita molesta, LK was predominately expressed in the gut and FB (Cheng et al., 2021), whereas in Chilo suppressalis, LK were predominately expressed in the CNS and gut (Xu et al., 2016). Seven transmembrane domains involved in GPCR ligand binding and receptor activation are functionally conserved in HcLKR, which contains amino acid motifs typical of the GPCR family (Marco et al., 2013). Moreover, the isolated HcLKR was highly analogous to other LK receptors in various insect species. HcLKR was mostly expressed in the midgut and hindgut, as previously reported in Aedes aegypti and D. melanogaster (Kwon et al., 2016; Zandawala et al., 2018a,b). This phenomenon corresponds with the main function of LK in diuresis and ion transport (Gonzalez et al., 2012). The insect hindgut is the main organ of the excretory system. The highest expression levels of HcLK and HcLKR genes in the hindgut suggest a conserved function of the LK signaling system in the regulation of diuresis and ion transport (Coast et al., 2002; Dow and Davies, 2006; Nässel and Winther, 2010).
The intracellular Ca2+ levels were performed to determine the binding between HcLK peptides and HcLKR because Ca2+ acts as a second messenger for LKR signal transduction. Pharmacological data demonstrate that HcLKR was strongly activated by HcLK peptides in a concentration-dependent manner. Our results are consistent with the previously reported pharmacological characterization of LKR in D. melanogaster (Terhzaz et al., 1999; Radford et al., 2002).
The LK signaling system has been demonstrated to be involved in food intake, metabolism, and stress in insects (Al-Anzi et al., 2010; Liu et al., 2015; Zandawala et al., 2018a,b). Feeding or starvation affects the expression of LK and LKR in D. melanogaster (Zandawala et al., 2018a,b). Cannell et al. (2016) showed that, in D. melanogaster, starvation increases the epithelial LKR gene expression, and Malpighian tubule stellate cell-specific knockdown of LKR significantly reduces starvation tolerance. Zandawala et al. (2018b) showed that targeted knockdown of LKR in abdominal ganglion LK neurons using the CRISPR/Cas9 technology significantly increased starvation tolerance in in D. melanogaster. LKR mutation and targeted knockdown of LKR in insulin-producing cells of Drosophila altered the expression of ILPs and increased starvation resistance (Zandawala et al., 2018b). Yurgel et al. (2019) reported that the LK neuropeptide plays an essential role in the metabolic regulation of sleep. Moreover, the activity of LK neurons is modulated by feeding; decreased activity is observed in response to glucose, whereas increased activity is observed under starvation conditions. In the present study, our results showed that LK or LKR knockdown increased the water content in H. cunea and extended survival during desiccation and starvation. Under desiccation conditions, the survival rate of H. cunea larvae was improved by deletion of LK/LKR signaling, which promotes water retention. The findings confirm that the LK signaling system plays a vital role in the regulation of water homeostasis and the resistance to desiccation and starvation. The LK likely plays a regulatory role during starvation; however, its detailed functions remain to be identified. Moreover, HcLK and HcLKR knockdown increased the transcript levels of HILP2 (except in the dsLK larvae), HILP5, and HILP8 and decreased the transcript levels of HILP3, HILP4, and HILP6. However, HcLK and HcLKR knockdown had little effect on the transcript levels of HILP1, HILP7, and HILP2 (except in the dsLKR treatment group). The LK/LKR system in H. cunea could be used to control H. cunea by synthesizing leucokinin analogs. However, the potential regulatory role of LK and LKR in the transcription of ILPs in H. cunea needs to be further studied.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
LS and CC designed the research and wrote the manuscript. HM, YG, and ZW performed the experiments and analyzed the data. CC revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by grants from the National Natural Science Foundation of China (32171791 and 31700570), the Fundamental Research Funds for the Central Universities (2572019CG04), the Natural Science Foundation of Heilongjiang (YQ2021C007), Heilongjiang Postdoctoral funds (LBH-Q20064), and Heilongjiang Touyan Innovation Team Program (Tree Genetics and Breeding Innovation Team).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Al-Anzi, B., Armand, E., Nagamei, P., Olszewski, M., Sapin, V., Waters, C., et al. (2010). The leucokinin pathway and its neurons regulate meal size in Drosophila. Curr. Biol. 20, 969–978. doi: 10.1016/j.cub.2010.04.039
Baker, K. D., and Thummel, C. S. (2007). Diabetic larvae and obese flies—emerging studies of metabolism in Drosophila. Cell Metab. 6, 257–266. doi: 10.1016/j.cmet.2007.09.002
Bharucha, K. N., Tarr, P., and Zipursky, S. L. (2008). A glucagon-like endocrine pathway in Drosophila modulates both lipid and carbohydrate homeostasis. J. Exp. Biol. 211, 3103–3110. doi: 10.1242/jeb.016451
Cannell, E., Dornan, A. J., Halberg, K. A., Terhzaz, S., Dow, J. A. T., and Davies, S. A. (2016). The corticotropin-releasing factor-like diuretic hormone 44 (DH44) and kinin neuropeptides modulate desiccation and starvation tolerance in Drosophila melanogaster. Peptides 80, 96–107. doi: 10.1016/j.peptides.2016.02.004
Cheng, J., Yang, X., Tian, Z., Shen, Z., Wang, X., Zhu, L., et al. (2021). Coordinated transcriptomics and peptidomics of central nervous system identify neuropeptides and their G protein-coupled receptors in the oriental fruit moth Grapholita molesta. Comp. Biochem. Physiol. Part D Genomics Proteomics 40:100882. doi: 10.1016/j.cbd.2021.100882
Coast, G. M., Orchard, I., Phillips, J. E., and Schooley, D. A. (2002). “Insect diuretic and antidiuretic hormones,” in Advances in Insect Physiology. ed. P. D. Evans (London: Academic Press), 279–409.
Cognigni, P., Bailey, A. P., and Miguel-Aliaga, I. (2011). Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell Metab. 13, 92–104. doi: 10.1016/j.cmet.2010.12.010
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Dalamaga, M., Chou, S. H., Shields, K., Papageorgiou, P., Polyzos, S. A., and Mantzoros, C. S. (2013). Leptin at the intersection of neuroendocrinology and metabolism: current evidence and therapeutic perspectives. Cell Metab. 18, 29–42. doi: 10.1016/j.cmet.2013.05.010
de Araujo, I. E., Lin, T., Veldhuizen, M. G., and Small, D. M. (2013). Metabolic regulation of brain response to food cues. Curr. Biol. 23, 878–883. doi: 10.1016/j.cub.2013.04.001
de Haro, M., AI-Ramahi, I., Benito-Sipos, J., López-Arias, B., Veenstra, J. A., and Herrero, P. (2010). Detailed analysis of leucokinin-expressing neurons and their candidate functions in the Drosophila nervous system. Cell Tissue Res. 339, 321–326. doi: 10.1007/s00441-009-0890-y
Dow, J. A. (2009). Insights into the Malpighian tubule from functional genomics. J. Exp. Biol. 212, 435–445. doi: 10.1242/jeb.024224
Dow, J. A., and Davies, S. A. (2006). The Malpighian tubule: rapid insights from post-genomic biology. J. Insect Physiol. 52, 365–378. doi: 10.1016/j.jinsphys.2005.10.007
Géminard, C., Arquier, N., Layalle, S., Bourouis, M., Slaidina, M., Delanoue, R., et al. (2006). Control of metabolism and growth through insulin-like peptides in Drosophila. Diabetes 55, S5–S8. doi: 10.2337/db06-S001
Gonzalez, A., Cordomí, A., Caltabiano, G., and Pardo, L. (2012). Impact of helix irregularities on sequence alignment and homology modeling of G protein-coupled receptors. Chembiochem 13, 1393–1399. doi: 10.1002/cbic.201200189
Hergarden, A. C., Tayler, T. D., and Anderson, D. J. (2012). Allatostatin-A neurons inhibit feeding behavior in adult Drosophila. Proc. Natl. Acad. Sci. U. S. A. 109, 3967–3972. doi: 10.1073/pnas.1200778109
Holman, G. M., Nachman, R. J., and Coast, G. M. (1999). Isolation, characterization and biological activity of a diuretic myokinin neuropeptide from the housefly, Musca domestica. Peptides 20, 1–10. doi: 10.1016/S0196-9781(98)00150-8
Holman, G. M., Nachman, R. J., and Wright, M. S. (1990). Comparative aspects of insect myotropic peptides. In Progress in Comparative Endocrinology. (eds.) A. Epple, Scanes, C. G., and M. H. Stetson New York: Wiley-Liss. 35–39.
Itskov, P. M., and Ribeiro, C. (2013). The dilemmas of the gourmet fly: the molecular and neuronal mechanisms of feeding and nutrient decision making in Drosophila. Front. Neurosci. 7:12. doi: 10.3389/fnins.2013.00012
Kwon, H., Ali Agha, M., Smith, R. C., Nachman, R. J., Marion-Poll, F., and Pietrantonio, P. V. (2016). Leucokinin mimetic elicits aversive behavior in mosquito Aedes aegypti (L.) and inhibits the sugar taste neuron. Proc. Natl. Acad. Sci. U. S. A. 113, 6880–6885. doi: 10.1073/pnas.1520404113
Leopold, P., and Perrimon, N. (2007). Drosophila and the genetics of the internal milieu. Nature 450, 186–188. doi: 10.1038/nature06286
Liu, Y. T., Luo, J. N., Carlsson, M. A., and Nässel, D. R. (2015). Serotonin and insulin-like peptides modulate leucokinin-producing neurons that affect feeding and water homeostasis in Drosophila. J. Comp. Neurol. 523, 1840–1863. doi: 10.1002/cne.23768
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López-Arias, B., Dorado, B., and Herrero, P. (2011). Blockade of the release of the neuropeptide leucokinin to determine its possible functions in fly behavior: chemoreception assays. Peptides 32, 545–552. doi: 10.1016/j.peptides.2010.07.002
Marco, H. G., Simek, P., Clark, K. D., and Gäde, G. (2013). Novel adipokinetic hormones in the kissing bugs Rhodnius prolixus, Triatoma infestans, Dipetalogaster maxima and Panstrongylus megistus. Peptides 41, 21–30. doi: 10.1016/j.peptides.2012.09.032
Mccarthy, M., Duff, P., Muller, H. L., and Randell, C. (2004). Prediction of proprotein convertase cleavage sites. Protein Eng. Des. Sel. 17, 107–112. doi: 10.1093/protein/gzh013
Melcher, C., and Pankratz, M. J. (2005). Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 3:e305. doi: 10.1371/journal.pbio.0030305
Murphy, K. G., and Bloom, S. R. (2006). Gut hormones and the regulation of energy homeostasis. Nature 444, 854–859. doi: 10.1038/nature05484
Nässel, D. R., and Winther, A. M. E. (2010). Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 92, 42–104. doi: 10.1016/j.pneurobio.2010.04.010
Owusu-Ansah, E., and Perrimon, N. (2014). Modeling metabolic homeo-stasis and nutrient sensing in Drosophila: implications foraging and metabolic diseases. Dis. Model. Mech. 7, 343–350. doi: 10.1242/dmm.012989
Padmanabha, D., and Baker, K. D. (2014). Drosophila gains traction as a repurposed tool to investigate metabolism. Trends Endocrinol. Metab. 25, 518–527. doi: 10.1016/j.tem.2014.03.011
Radford, J. C., Davies, S. A., and Dow, J. A. (2002). Systematic G-protein-coupled receptor analysis in Drosophila melanogaster identifies a leucokinin receptor with novel roles. J. Biol. Chem. 277, 38810–38817. doi: 10.1074/jbc.M203694200
Rong, J., Xie, B. Y., Li, X. H., Gao, Z. X., and Li, D. M. (2003). Research progress on the invasive species, Hyphantria cunea. Entomol. Knowl. 40, 13–18.
Söderberg, J. A., Carlsson, M. A., and Nässel, D. R. (2012). Insulin-producing cells in the Drosophila brain also express satiety-inducing cholecystokinin-like peptide, drosulfakinin. Front. Endocrinol. 3:109. doi: 10.3389/fendo.2012.00109
Sonnhammer, E. I. L., von Heijne, G., and Krogh, A. (1998). “A hidden Markov model for predicting transmembrane helices in protein sequences”, In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. (eds.) J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen June 28–July 01; (Menlo Park, CA: AAAI Press), 175–182.
Sun, L., Liu, P., Sun, S., Yan, S., and Cao, C. (2019). Transcriptomic analysis of interactions between Hyphantria cunea larvae and nucleopolyhedro virus. Pest Manag. Sci. 75, 1024–1033. doi: 10.1002/ps.5212
Sun, L., Wang, Z. Y., Wu, H. Q., Liu, P., Zou, C. S., Xue, X. T., et al. (2016). Role of ocular albinism type 1 (OA1) GPCR in Asian gypsy moth development and transcriptional expression of heat-shock protein genes. Pestic. Biochem. Physiol. 126, 35–41. doi: 10.1016/j.pestbp.2015.07.004
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Teleman, A. A. (2010). Molecular mechanisms of metabolic regulation by insulin in Drosophila. Biochem. J. 425, 13–26. doi: 10.1042/BJ20091181
Terhzaz, S., O'Connell, F. C., Pollock, V. P., Kean, L., Davies, S. A., Veenstra, J. A., et al. (1999). Isolation and characterization of a leucokinin-like peptide of Drosophila melanogaster. J. Exp. Biol. 202, 3667–3676. doi: 10.1242/jeb.202.24.3667
Veenstra, J. A., Pattillo, J. M., and Petzel, D. H. (1997). A single cDNA encodes all three Aedes leucokinins, which stimulate both fluid secretion by the Malpighian tubules and hindgut contractions. J. Biol. Chem. 272, 10402–10407. doi: 10.1074/jbc.272.16.10402
Vogt, M. C., and Bruning, J. C. (2013). CNS insulin signaling in the control of energy homeostasis and glucose metabolism—from embryo to old age. Trends Endocrinol. Metab. 24, 76–84. doi: 10.1016/j.tem.2012.11.004
Woods, S. C., and D’Alessio, D. A. (2008). Central control of body weight and appetite. J. Clin. Endocrinol. Metab. 93, S37–S50. doi: 10.1210/jc.2008-1630
Wu, Q., Wen, T., Lee, G., Park, J. H., Cai, H. N., and Shen, P. (2003). Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron 39, 147–161. doi: 10.1016/S0896-6273(03)00396-9
Wu, N. N., Zhang, S. F., Li, X. W., Cao, Y. H., Liu, X. J., Wang, Q. H., et al. (2019). Fall webworm genomes yield insights into rapid adaptation of invasive species. Nat. Ecol. Evol. 3, 105–115. doi: 10.1038/s41559-018-0746-5
Wu, Q., Zhang, Y., Xu, J., and Shen, P. (2005). Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 102, 13289–13294. doi: 10.1073/pnas.0501914102
Xu, G., Gu, G. X., Teng, Z. W., Wu, S. F., Huang, J., Song, Q. S., et al. (2016). Identification and expression profiles of neuropeptides and their G protein-coupled receptors in the rice stem borer Chilo suppressalis. Sci. Rep. 6:28976. doi: 10.1038/srep28976
Yurgel, M. E., Kakad, P., Zandawala, M., Nässel, D. R., Godenschwege, T. A., and Keene, A. C. (2019). A single pair of leucokinin neurons are modulated by feeding state and regulate sleep-metabolism interactions. PLoS Biol. 17:e2006409. doi: 10.1371/journal.pbio.2006409
Zandawala, M., Marley, R., Davies, S. A., and Nässel, D. R. (2018a). Characterization of a set of abdominal neuroendocrine cells that regulate stress physiology using colocalized diuretic peptides in Drosophila. Cell Mol. Life Sci. 75, 1099–1115. doi: 10.1007/s00018-017-2682-y
Zandawala, M., Yurgel, M. E., Texada, M. J., Liao, S. F., Rewitz, K. F., Keene, A. C., et al. (2018b). Modulation of Drosophila post-feeding physiology and behavior by the neuropeptide leucokinin. PLoS Genet. 14:e1007767. doi: 10.1371/journal.pgen.1007767
Keywords: G-protein-coupled receptor, Hyphantria cunea, leucokinin, RNA interference, gene function
Citation: Sun L, Ma H, Gao Y, Wang Z and Cao C (2021) Functional Identification and Characterization of Leucokinin and Its Receptor in the Fall Webworm, Hyphantria cunea. Front. Physiol. 12:741362. doi: 10.3389/fphys.2021.741362
Edited by:
Natraj Krishnan, Mississippi State University, United StatesReviewed by:
Tanja Angela Godenschwege, Florida Atlantic University, United StatesGang Xu, Yangzhou University, China
Copyright © 2021 Sun, Ma, Gao, Wang and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanwang Cao, Y2h1YW53YW5nY2FvQG5lZnUuZWR1LmNu
 Lili Sun
Lili Sun Heting Ma
Heting Ma Chuanwang Cao
Chuanwang Cao