- 1College of Plant Protection, Hunan Agricultural University, Changsha, China
- 2State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Heat shock proteins are molecular chaperones that are involved in numerous normal cellular processes and stress responses, and heat shock factors are transcriptional activators of heat shock proteins. Heat shock factors and heat shock proteins are coordinated in various biological processes. The regulatory function of heat shock factors in the expression of genes encoding heat shock proteins (Hsps) has been documented in some model insects, however, the role of transcription factors in modulating Hsps in other insects is still limited. In this study, one heat shock factor gene (AhHsf) was isolated and its two potential target genes (AhHsp70 and AhsHsp21) were confirmed from Agasicles hygrophila. AhHsf sequence analysis indicated that it belongs to the Hsfs gene family. RT-qPCR showed that expression levels of heat shock factors and of two heat shock proteins significantly increased under heat stress. Injection with double-stranded Hsf RNA in freshly emerged adult beetles significantly inhibited expression of AhHsp70 and AhsHsp21, shortened the adult survival, drastically reduced egg production, and ultimately led to a decrease in fecundity. RNA interference (RNAi)-mediated suppression of AhHsp70 or AhsHsp21 expression also significantly affected expression of AhHsf. Our findings revealed a potential transcriptional function of AhHsf to regulate expression of AhHsp70 and AhsHsp21, which may play a key role in A. hygrophila thermotolerance. Our results improve our understanding of the molecular mechanisms of the AhHsf - AhHsps signaling pathway in A. hygrophila.
Introduction
Organisms respond to elevated temperatures and to several chemical and physiological stressors by increasing the synthesis of heat shock proteins (Hsp) (Wu, 1995; Tatar et al., 1997; Feder and Hofmann, 1999; Kristensen et al., 2003; Sørensen et al., 2003). Inducible expression of heat shock genes is a response to a plethora of stress signals (Lis and Wu, 1993; Morimoto, 1993; Wu, 1995) that are triggered by (1) abiotic stressors such as irradiation, temperature, salinity, and drought and (2) biotic stressors such as natural enemies and pathogen invasion (Lindquist and Craig, 1988). The heat shock response was first described by Ritossa (1962) who observed an induction of specific chromosome puffs on the polytene chromosomes in Drosophila melanogaster following heat or chemical treatment; this constitutes an efficient defense system against detrimental effects of protein denaturation that are predominantly associated with heat stress. Subsequent studies elucidated the nature of induced RNAs and proteins at a molecular level and led to the isolation and characterization of Hsp genes (Lindquist, 1986; Lindquist and Craig, 1988; Nover, 1991; Lü et al., 2014; Deng et al., 2018; Jin et al., 2020a). Central to the heat shock response is the induction of Hsps that effectively counteract the effects of stress by stabilizing, re-folding, or degrading denatured proteins (Parsell and Lindquist, 1993).
Stress-induced transcription requires activation of a heat shock factor protein (Hsf) (Lis and Wu, 1993; Morimoto, 1993; Wu, 1995; Voellmy, 2004) that binds to the heat shock promoter element (HSE) (Pelham, 1982). The heat shock response element is composed of three contiguous inverted repeats of a 5-base-pair (bp) sequence whose consensus was defined as nGAAn (Amin et al., 1988; Xiao and Lis, 1988) and was recently revised to AGAAn (Cunniff and Morgan, 1993; Fernandes et al., 1994; Kroeger and Morimoto, 1994). Hsf is present in a latent state under normal conditions and typically occurs as an inactive protein (Zhong et al., 1996), however, Hsf is activated in response to heat stress (Nair et al., 1996; Voellmy, 2004; Marchler and Wu, 2014), and activation of Hsf generally occurs in three stages: (a) thermally induced formation of homologous trimers and hyperphosphorylation, (b) transfer to the nucleus and recognition, and (c) binding to HSE domain sequences of specific protein genes (Sarge et al., 1993; Morimoto, 1993; Jolly et al., 1999; Cohen, 2000; Scharf et al., 2012). Sequencing of Hsf genes of numerous species has provided crucial information for understanding the heat shock signaling pathway (Schuetz et al., 1991; Wisniewski et al., 1996; Gonsalves et al., 2011). A considerable amount of studies has been conducted on how Hsfs are changed in response to heat stress, however, the mechanism by which these particular stress signals are transduced to Hsf genes remain unclear. Previous studies observed up-regulation of the gene Hsp70 in response to long-term cold exposure, indicating activation of Hsfs (Burton et al., 1988; Goto and Kimura, 1998; Sejerkilde et al., 2003), and Burton et al. (1988) observed increased survival of D. melanogaster larvae receiving a mild heat treatment prior to cold stress, suggesting cross-resistance to different stressors.
Stress response mechanisms following high temperature knock down (Huey et al., 1992) are likely to be closely associated with heat stress responses, thus it is reasonable to speculate that Hsps may exert a protective effect. Previous studies showed that expression of heat shock proteins increases survival in D. melanogaster (McColl et al., 1996).
Alligator weed [Alternanthera philoxeroides (Mart.) Griesb. (Amaranthaceae)] originates from South America and has become highly invasive in numerous countries in North and Central America, Australia, and Asia (Julien et al., 1995; Buckingham, 1996; Xu et al., 2003; Zhang et al., 2016). In China, this species was first introduced as a forage crop in the late 1930s, and it subsequently spread to the country’s southern regions (Wang et al., 2005; Ye et al., 2010). A. philoxeroides is a well-adapted species, and it can reproduce both sexually and asexually, although the seeds are often not viable (Julien et al., 1995). It causes economic and ecological problems in almost all its newly invaded habitats (Maddox and Rhyne, 1975). The flea beetle Agasicles hygrophila Selman & Vogt (Coleoptera: Chrysomelidae) has shown to be an efficient biological control agent of A. philoxeroides after investigations by the United States Department of Agriculture in South America from 1960 to 1974 (Zeiger, 1967; Spencer and Coulson, 1976; Coulson, 1977; Buckingham, 1996; Zhao et al., 2016). A. hygrophila was first introduced in China in 1986, and it effectively helped control A. philoxeroides spreading in the southern regions of China (Wang, 1989; Pan et al., 2006). This was also the most successful case of biological control of exotic weeds in China (Ma et al., 2003). However, field surveys showed that the population density of A. hygrophila typically decreases markedly from July to September during midsummer in Hunan province, China, leading reduced inhibition of A. philoxeroides growth (Li et al., 2011). In addition, our analysis of temperature data from 2003 to 2013 in Changsha City showed that in May and June, the frequency of high ambient temperatures over 36°C was only 0.29 and 3.3%, respectively. In July and August, the frequencies of temperatures over 36°C were 42.5 and 32.1%, respectively; the daily maximum temperatures often exceeded 39°C. In September, the frequency of high temperatures over 36°C was still high [Jin et al., 2020b (Accepted); Supplementary Table S1].
Previous studies reveal that the three Hsp genes play important roles for thermotolerance in A. hygrophila (Jin et al., 2020a), however, the respective molecular mechanisms affecting thermotolerance are poorly understood. In the present study, we identified a heat shock factor (AhHsf) and two of its potential downstream target genes, AhHsp70 and AhsHsp21 via gene functional verification. Reverse-transcription quantitative polymerase chain reactions (RT-qPCR) and RNA interference (RNAi) techniques were used to assess the effect of Hsf on expression of two Hsp genes and fecundity of A. hygrophila. Our study helps improve the understanding of the mechanisms of thermotolerance in A. hygrophila at a molecular level, understand the adaptability of A. hygrophila to temperature changes, and predict the efficacy of bio-control using A. hygrophila in the face of climate change.
Materials and Methods
Host Plants and Experimental Insects
Rhizomes and roots of A. philoxeroides were collected from a pond at the Institute of Plant Protection, Hunan Academy of Agricultural Sciences, China, and were planted in plastic vessels (30 × 30 × 30 cm) containing sterilized soil. A. philoxeroides plants were placed in a greenhouse of the Langfang Experimental Station, Chinese Academy of Agricultural Sciences, Hebei province, China, and were watered daily.
Adult specimens of A. hygrophila were collected in June 2017 from a field in Changsha (28°11′49″N, 112°58′42″E), Hunan province, China. Numerous specimens were collected using the sweeping method and were then applied to the experimental plants in the laboratory at the Chinese Academy of Agricultural Sciences, Beijing, China. Plants were grown at a temperature of 26 ± 1°C, 80 ± 5% relative humidity, and under a photoperiod of 12:12 h light:darkness (Guo et al., 2011). The gender of the experimental insects was determined by the presence of a groove at the end of the abdomen (< 12 h after eclosing); this groove was present in males and absent in females (Supplementary Figure S1). Groups of five females and five males were each placed in a cylindrical box of 8 cm diameter and 12 cm height which contained fresh A. philoxeroides stems.
Sample Collection
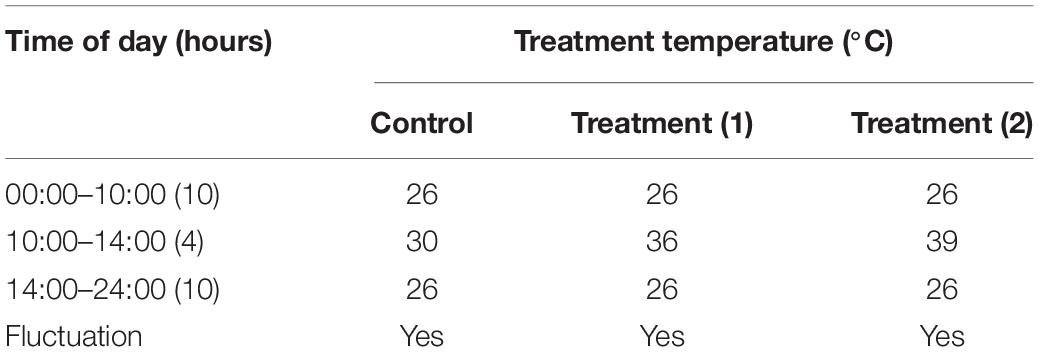
Newly hatched (<12 h) adult A. hygrophila specimens were used in the experiments. Each group of five pairs was placed in one cylindrical box as detailed above which was then covered with gauze. The effects at each temperature was studied using 10 boxes of adult A. hygrophila beetles. The temperature tolerance test was set as described in Table 1. Insects were exposed to 30, 36, or 39°C for 4 h (10:00 a.m. to 2:00 p.m.) each day using a constant temperature incubator (PRX-450D-30, Saifu, China). A control group was exposed to 30°C. To analyze the expression of AhHsp70 and AhsHsp21, five females and five males were collected from each temperature group every day for a week. A total of 50 pairs of adults for each temperature treatment were used and three replicates were performed for each treatment. The sampled individuals were immediately frozen in liquid nitrogen and subsequently stored in a −80°C-freezer (DW-86L628, Haier special electric appliance co. Ltd., Tsingtao) until further analyses.
RNA Isolation, cDNA Synthesis, and Gene Cloning
TRIzol (Trizol reagent Invitrogen, United States) reagent was used to extract total RNA from sampled insects according to the manufacturer’s instructions. Isolated RNA was stored at −80°C until first-strand cDNA synthesis was performed using a commercial reverse transcription kit (AT341-02, TransGen Biotech, China). Full-length cDNA of AhHsp70 and AhsHsp21 of A. hygrophila was produced by RT-PCR and rapid amplification of cDNA-ends (RACE)-PCR, and the resulting sequences were submitted to NCBI (GenBank accession numbers: MN138034 and MN163038, respectively, Supplementary Figures S2, S3; Jin et al., 2020a). We used A. hygrophila transcriptome data to obtain expressed sequence tags (ESTs) that showed similarities to other insect Hsfs. Polymerase chain reaction (PCR) primers were designed using primer 5.0 (Supplementary Table S2). The PCR reaction was performed following the procedure of Jin et al. (2020a); PCR products were cloned into a pEASY-T3 vector (TransGen, Beijing, China) and were then sequenced.
Sequence Analysis Identification of AhHsf cDNA
The cDNAs of AhHsf were used as query sequences to search for other Hsfs in GenBank using the BLAST software available at the NCBI website1. Sequence identification analyses were carried out using MEGA6 or vector NTI software.
Relative Quantitative Real Time PCR
Heat shock-regulated gene expression was analyzed using RT-qPCR and TransStart Green qPCR SuperMix Kit with SYBR (Transgen, Beijing, China) with an ABI Prism 7500 Real Time PCR System (Applied Biosystems, NYC, United States). Gene-specific primers for Hsf gene amplification by primer 5.0 are shown in Supplementary Table S1. PCR reactions were performed using a total reaction volume of 20 μL comprising 10 μL 2 × TransStart® Tip Green qPCR SuperMix (TransGen Biotech, China), 0.4 μL Passive Reference Dye II, 0.4 μL of each of gene-specific primer pair, 1 μL cDNA template, and 7.8 μL ddH2O. Each experiment comprised three biological and three technical replicates. Relative expression levels of the target molecule was determined using the Cp method according to the mathematical model of Pfaffl (Fleige et al., 2006), using β-actin as an internal reference. Aimplified to 2–△△ Ct as follows:
△△ Ct = (Cp target-Cp reference) treatment–(Cp target-Cp reference) control
Double-Stranded AhHsf, AhHsp70, and AhsHsp21 RNA Synthesis and RNAi
dsRNA was synthesized from the AhHsf, AhHsp70 and AhsHsp21 cDNA from A. hygrophila using gene-specific primers, and EGFP (GenBank Accession No. AIR08541.1) dsRNA was synthesized as a negative control. A T7 promoter as described by Ghanim et al. (2007) was added to the 5′-end of each primer. The promoter sequence was 5′-TAATACGACTCACTATAGGG-3′. PCR products were purified using an AxyPrepTM DNA Gel Extraction Kit (Axygen, Silicon Valley, United States) according to the manufacturer’s instructions. PCR products were stored at −80°C prior to synthesis of dsRNA.
dsRNA was synthesized using the HiScribeTM T7 Quick High Yield RNA Synthesis Kit (New England BioLabs, Ipswich, MA, United States; #E2050S) following the manufacturer’s protocol. The concentration of dsRNA was measured using a NanoVue spectrophotometer (GE Healthcare, Germany), and purity was tested by electrophoresis using a 1.0% agarose gel. To ensure that the injection volume between the control group and treatment group was consistent, the concentration of all synthesized dsRNA was adjusted to 8,000 ng/μL; dsRNA solution (125 nL) was injected in the abdomen of the insects (with a final dsRNA amount of 1.0 μg). Where subsequent experiments were not conducted immediately, dsRNA was stored at −80°C.
Expression Analysis of AhHsf After Injection With dsAhHsf
Newly emerged adult A. hygrophila females (< 12 h after eclosing) were collected for dsRNA injection using a PLI-100 Pico-Injector (Harvard Apparatus, Holliston, MA, United States) with an MP-255 Micromanipulator (Sutter, Novato, CA, United States) using an Olympus stereomicroscope. dsAhHsf solution (125nL; with a final dsRNA amount of 1.0 μg) was injected in the insects’ abdomen. After injection, the insects were kept under standard conditions, as described above. Six adults were collected from each temperature treatment and used for expression analysis of AhHsf genes every day for 1 week. The control group individuals were injected with dsRNAs of EGFP.
Knockdown of AhHsf Affecting Expression of Two Potential Target AhHsps
To confirm the specificity of RNAi, dsRNA solution (the final amount of dsRNA reached 1.0 μg) for AhHsf was injected into insects in the penultimate abdomen of newly emerged adult female beetles. Each experiment was performed three times and a total of 50 of adults were used for each temperature treatment. After injection, these beetles were subjected to heat-shock at each of the three temperatures (30, 36, or 39°C) for 4 h from 10:00 to 14:00 in a constant-temperature incubator (RPX-450, NUNON, Beijing) daily. Five females were collected and used for expression analysis of the two AhHsp genes by using gene-specific PCR primers at each temperature every day for a week. The blank control was injected with dsRNAs of EGFP. β-actin gene primers were used as an internal control to monitor equal loading of cDNA for analyses of transcription levels.
Effects of dsAhHsp70 or dsAhsHsp21 Injection on AhHsf Expression
dsRNA solution for AhHsp70, AhsHsp21 or AhHsp70 and AhsHsp21 was injected into insects in the penultimate abdomen of newly emerged adult female beetles. Each experiment was repeated three times and a total of 50 of adults were used for each treatment. After injection, the expression of Ahhsf was analyzed by qRT-PCR. The blank control was injected with dsEGFP.
Effects of RNAi on Fecundity of A. hygrophila
After injection with dsAhHsf, one newly emerged (< 12 h following eclosion) adult female and one male of A. hygrophila were placed together in a Petri dish (with a diameter of 9 cm) containing fresh alligator weed leaves, and they were kept under standard conditions as described above; a total of 15 pairs of adults were used for each temperature treatment and three replicates were performed for each treatment. To ensure the survival of 15 females for later experimental observation after injection, we injected about 50 females for our experiment. The blank control was injected with dsEGFP.
Effects of Injection of dsRNA on A. hygrophila Longevity
Newly hatched adult A. hygrophila females and males (< 12 h) were collected for dsAhHsf injection (30 females and 30 males per treatment). After injection, one female and one male were kept under standard conditions, as described above, for visual monitoring of phenotype changes and further analyses. Individual survival was recorded daily until the last specimen died. The control was injected with dsEGFP and replicated three times for each treatment.
Statistical Analyses
Statistical analyses were performed using SAS software (v8) for Microsoft Windows; data are shown as means ± standard deviation. Experimental data were checked for normality and homoscedasticity, and where needed, were arcsine square-root- or log-transformed before analysis. Target gene expression levels were analyzed using a one-way analysis of variance (ANOVA; SAS Institute Inc., 1996, United States), and a least significant difference test was used to test differences across data. Statistical significance is reported at p < 0.05. Genes expression levels and fecundity of A. hygrophila after injection with dsRNA were analyzed by Student’s t-test. Data on adult survival rates were analyzed using a log-rank (Mantel-Cox) test (Mantel, 1985), and a survival curve was constructed using GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA, United States). When the p-values were 0.05 or lower, they were considered significant.
Results
Sequence Analysis of AhHsf
We produced a 792-bp partial cDNA sequence of AhHsf by RT-PCR from A. hygrophila, which was deposited at the NCBI database (GenBank accession numbers: MT133904) (Figure 1). AhHsf sequence analysis indicated that it belongs to the Hsfs gene family (Supplementary Figure S4) by BLAST software available at the NCBI website1.
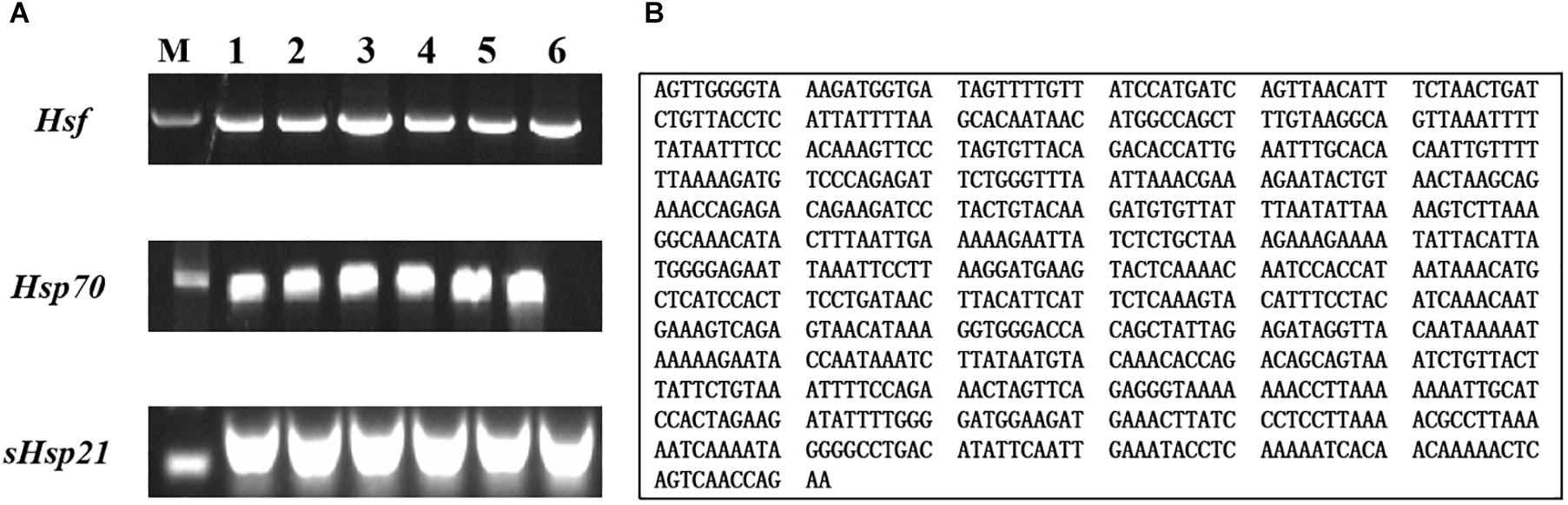
Figure 1. Gel electrophoretogram and nucleotide sequences of AhHsf from A. hygrophila. (A) Gel electrophoretogram; (B) Nucleotide sequences of Hsf. M: M stands for Trans2KTM DNA marker; 1, 2, 3, 4, 5, and 6 gel lanes represent gel electrophoretogram at different annealing temperatures with the same gene primer. Lane 1, 52°C; Lane 2, 54°C; Lane 3, 56°C; Lane 4, 58°C; Lane 5, 60°C; Lane 6, 62°C; Hsf, Hsp70, and sHsp21 respectively represent the gel electrophoretogram of PCR amplification sequences of AhHsf, AhHsp70, and AhsHsp21.
Expression of the AhHsf Gene in A. hygrophila Under Heat Stress
The standard curves of the AhHsf gene and the housekeeping gene produced a correlation coefficient of 0.998 and 1.000, respectively and the amplification efficiency was 104.852 and 96.265, respectively (Supplementary Figure S5).
The expression levels of AhHsf mRNAs in A. hygrophila under 30, 36, and 39°C temperature treatments were determined using relative quantitative real time PCR. qPCR analysis showed that the expression of the AhHsf gene was induced by 4-h heat treatments. Significant up-regulation of AhHsf expression was observed when the temperature was further increased from 30 to 39°C. AhHsf expression was significantly higher in the 36 and 39°C temperature treatment groups than in the control group kept at 30°C during the 6 days of observation [1d: F(2, 8) = 80.60, p = 0.0038; 2d: F(2, 8) = 188.89, p = 0.0013; 3d: F(2, 8) = 775.06, p = 0.0005; 4d: F(2, 8) = 767.06, p = 0.0001; 5d: F(2, 8) = 565.15, p = 0.0009; and 6d: F(2, 8) = 491.12, p = 0.0001] (Figure 2). The expression of β-actin was used as an internal control.
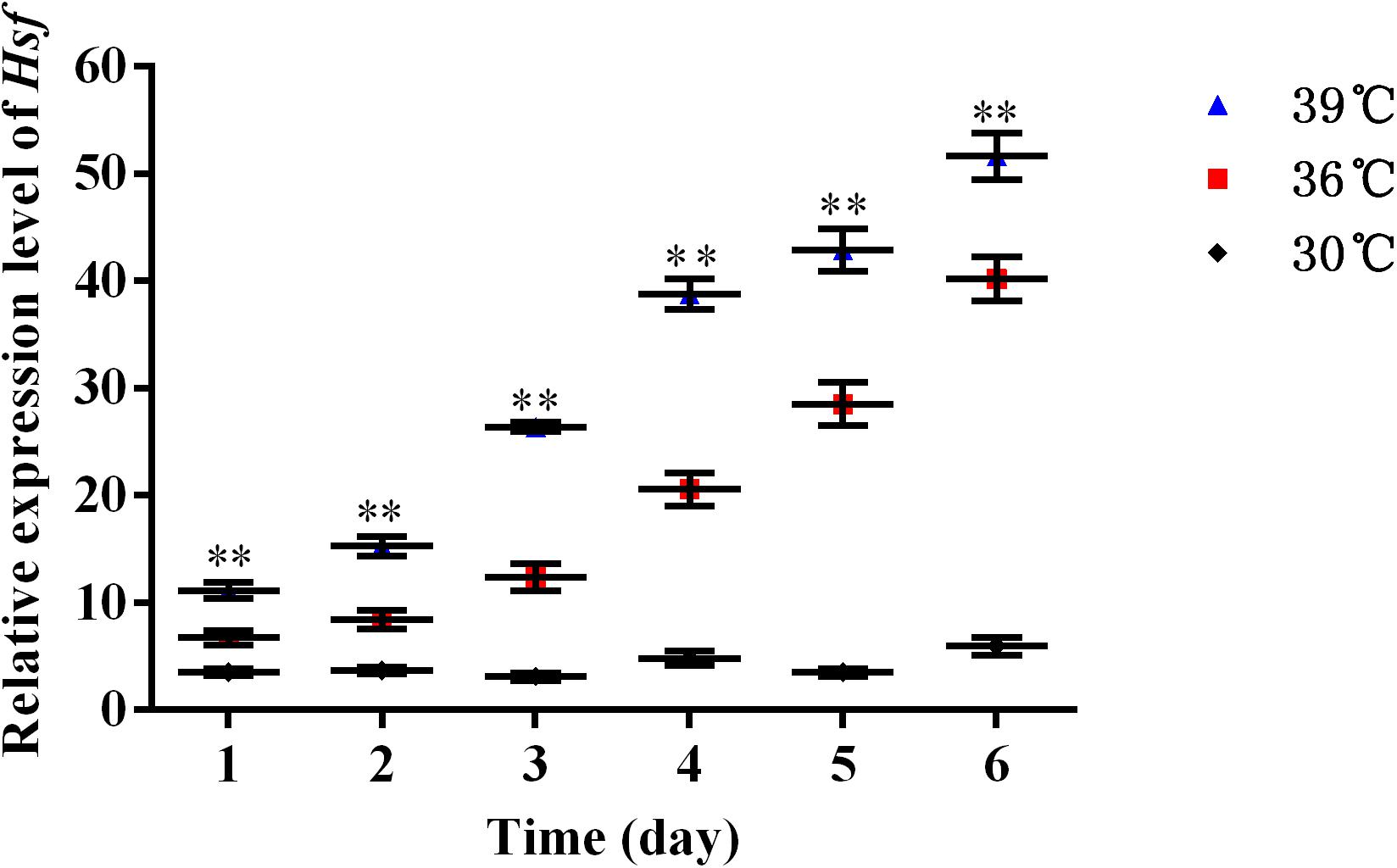
Figure 2. A high temperature of 30, 36, and 39°C induced expression profiles of AhHsf in A. hygrophila. Relative mRNA levels were analyzed using the 2–△△Ct method. The figure shows data from three replicates that were analyzed by one-way ANOVA followed by the least significant difference (LSD) test. All values are shown as the mean ± SD. ∗P < 0.05, significant; ∗∗P < 0.01, extremely significant. ns, not significant.
AhHsf and Two AhHsp Genes Expression Levels After Injection With dsAhHsf
qPCR assays showed that injection with dsAhHsf significantly inhibited endogenous expression of AhHsf mRNA at each time point of sampling (1, 2, 3, 4, 5, and 6 days). From 1 to 6 days after injection, the expression level of AhHsf mRNA decreased significantly when injected with dsAhHsf compared to that when injected with dsEGFP at 30°C and 36°C (A: 1d: t = 43.29, p < 0.05; 2d: t = 16.79, p < 0.05; 3d: t = 35.47, p < 0.05; 4d: t = 28.68, p < 0.05; 5d: t = 12.80, p < 0.05; and 6d: t = 26.54, p < 0.05. B: 1d: t = 30.61, p < 0.05; 2d: t = 15.68, p < 0.05; 3d: t = 18.04, p < 0.05; 4d: t = 14.41, p < 0.05; 5d: t = 24.32, p < 0.05; and 6d: t = 28.10, p < 0.05) (Figures 3A,B). At 39°C, from day 2–6, after the injection with dsAhHsf, AhHsf mRNA level was significantly decreased compared to that in the control group (2d: t = 20.10, p < 0.05; 3d: t = 12.51, p < 0.05; 4d: t = 15.58, p < 0.05; 5d: t = 57.62, p < 0.05; and 6d: t = 19.80, p < 0.05) (Figure 3C), However, on the first day, there was no significant difference in the AhHsf mRNA expression level between the samples injected with dsAhHsf and dsEGFP (1d: t = −0.72, p = 0.5135) (Figure 3C).
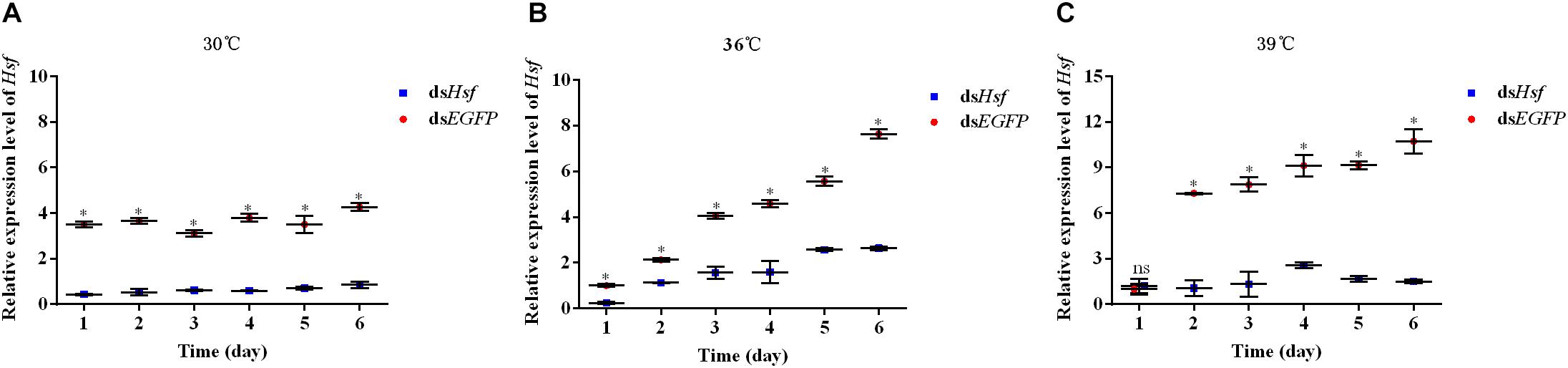
Figure 3. (A) Relative expression levels of AhHsf after injection of dsHsf under the 30°C condition; (B) Relative expression levels of AhHsf after injection of dsHsf under the 36°C condition; (C) Relative expression levels of AhHsf after injection of dsHsf under the 39°C condition. Relative expression levels of AhHsf after injection of dsRNA into freshly emerged A. hygrophila adults (n = 3). Relative mRNA levels were normalized to those of the CoxI gene and analyzed using the 2–△△Ct method. All values are shown as the mean ± SD. The data were analyzed by Student’s t-test. ∗P < 0.05. ns, not significant. dsEGFP RNA was used as the control.
RT-qPCR analyses of RNA isolated one to 6 days after injection showed that dsRNA suppressed the transcription levels of the target genes AhHsp70 and AhsHsp21 (A: Hsp70: t = −9.06, p < 0.05; sHsp21: t = −6.35, p < 0.05. B: Hsp70: t = −10.41, p < 0.05; sHsp21: t = −14.33, p < 0.05. C: Hsp70: t = −17.68, p < 0.05; sHsp21: t = −5.30, p < 0.05) (Figure 4). The results revealed that these two AhHsps could be regulated by AhHsf at high temperatures.
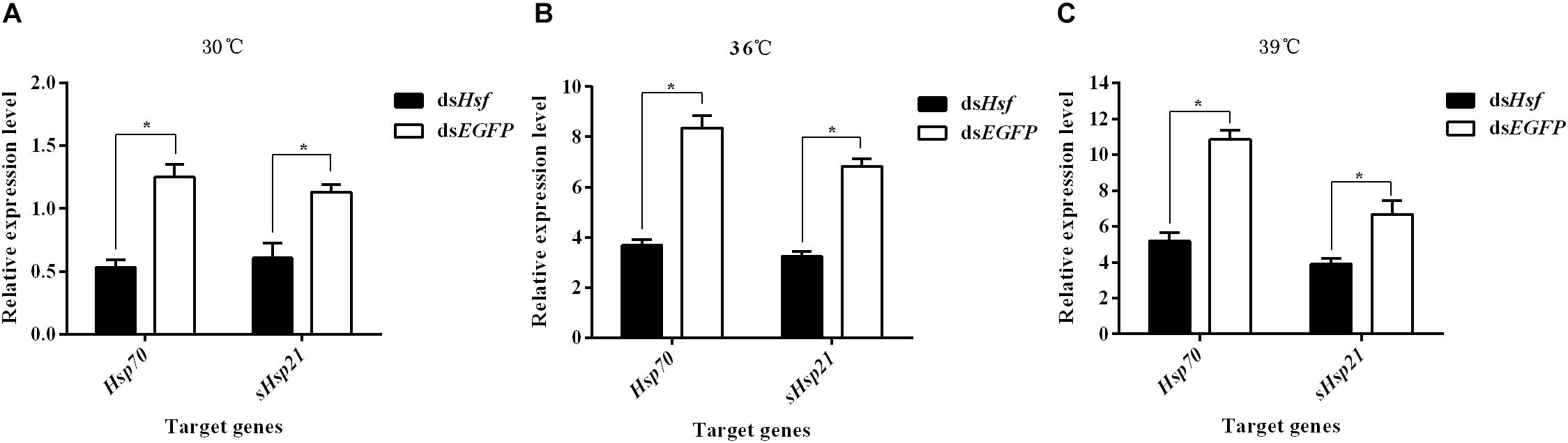
Figure 4. (A) Relative expression levels of AhHsp70, and AhsHsp21 after injection of dsAhHsf under the 30°C condition; (B) Relative expression levels of AhHsp70, and AhsHsp21 after injection of dsAhHsf under the 36°C condition; (C) Relative expression levels of AhHsp70, and AhsHsp21 after injection of dsAhHsf under the 39°C condition. Relative expression levels of AhHsp70, and AhsHsp21 after injection of dsAhHsf into freshly emerged female A. hygrophila adults. All values are shown as the mean ± SD. The values show data from three replicates that were analyzed using Student’s t-test. Different amounts of dsEGFP were injected as a control. * P < 0.05. ns, not significant.
Effects of dsAhHsp70, AhsHsp21, or Their Combination Injection on Expression of AhHsf Genes
qPCR assays showed that AhHsf expression in dsAhHsp70, dsAhsHsp21, or dsAhHsp70 & AhsHsp21- injected samples decreased significantly at sampled compared to that in dsEGFP - injected samples (A: t = −13.94, p < 0.05. B: t = −9.14, p < 0.05. C: t = −16.77, p < 0.05) (Figure 5). These results probably suggest that the downstream genes AhHsp70 and AhsHsp21can also influence the expression of upstream gene AhHsf through feedback regulation.
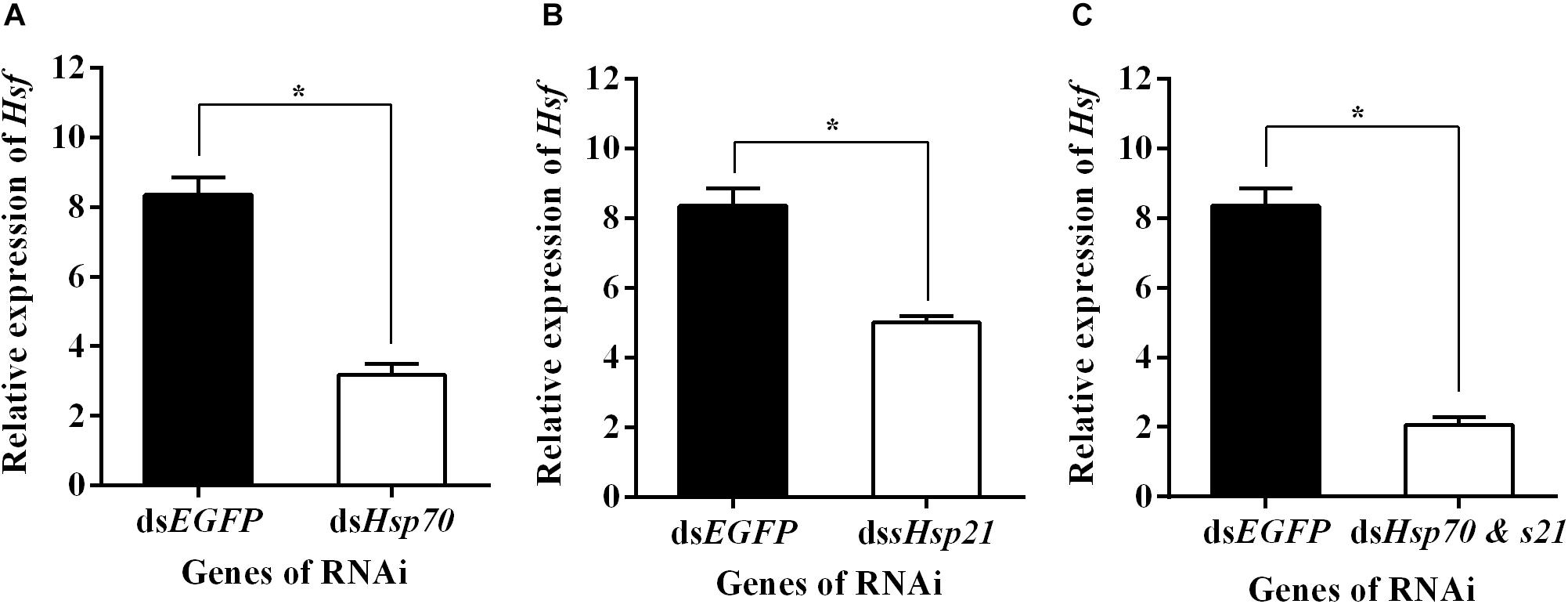
Figure 5. (A) Relative expression levels of AhHsf after injection of dsAhHs70; (B) Relative expression levels of AhHsf after injection of dsAhHs21; (C) Relative expression levels of AhHsf after injection of dsAhHs70 and sHsp21. Relative expression levels of AhHsf after injection of dsAhHs70, dsAhsHsp21, or dsAhHs70 and AhsHsp21 into freshly emerged female A. hygrophila adults. All values are shown as the mean ± SD. The values show data from three replicates that were analyzed using Student’s t-test. Different amounts of dsEGFP were injected as a control. * P < 0.05. ns, not significant.
Knockdown of AhHsf Affects Fecundity and Survival of A. hygrophila
The number of eggs produced was significantly lower in individuals injected with dsAhHsf than those injected with dsEGFP (30°C: t = −20.64, p < 0.05. 36°C: t = −14.63, p < 0.05. 39°C: t = −3.66, p < 0.05) (Figure 6). This decrease was more pronounced in the 39°C treatment group, where female fecundity was close to zero (t = −3.66, p < 0.05) (Figure 6). Results on fecundity confirmed that RNA interference of AhHsf expression significantly inhibits reproduction in A. hygrophila.
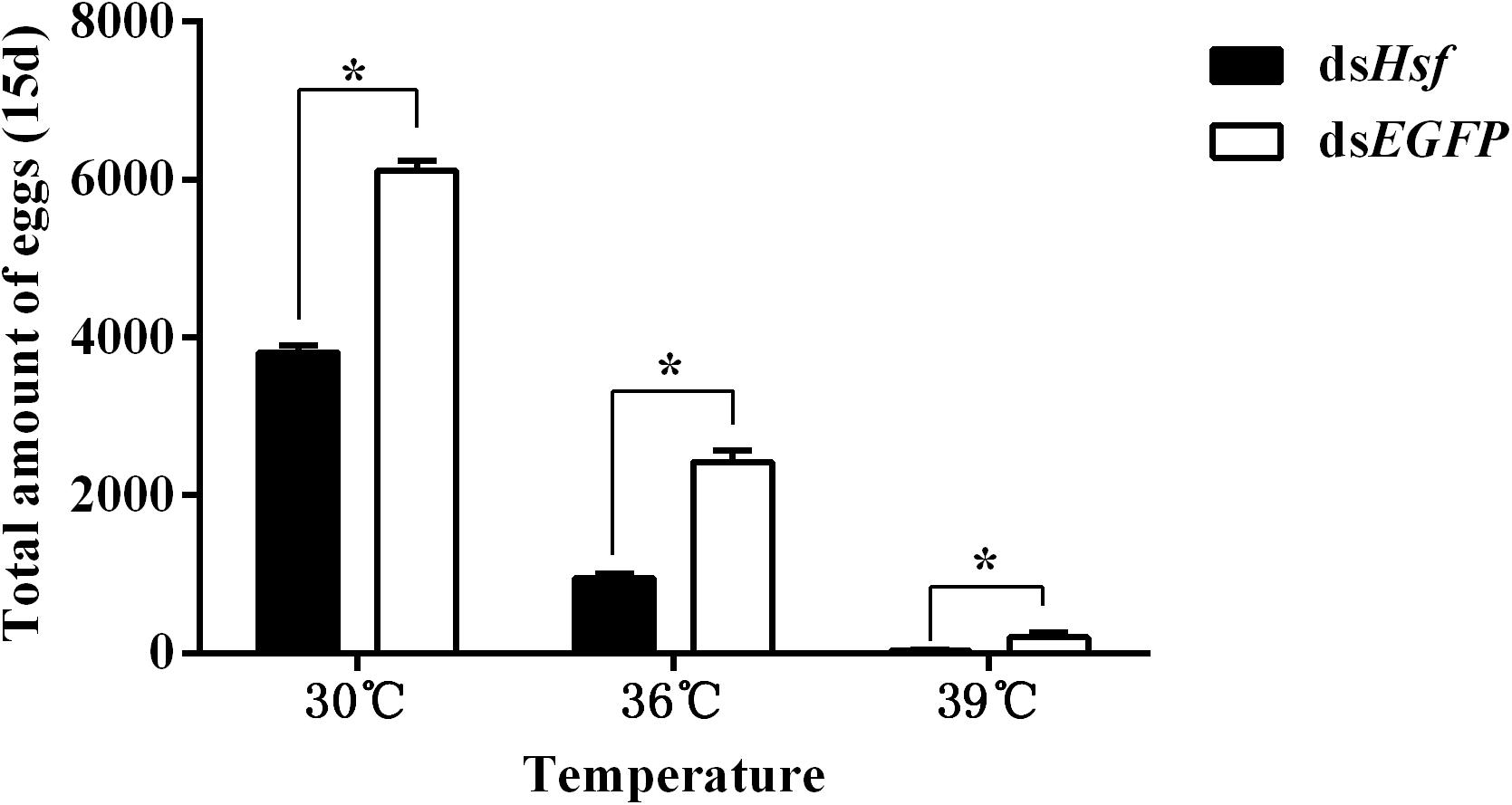
Figure 6. Fecundity of A. hygrophila after injection of dsAhHsf. Different amounts of dsEGFP were injected as a control. A total of 15 pairs of adults were used for each temperature treatment and three replicates were performed for each treatment. All values are shown as the mean ± SD. Data were analyzed using Student’s t-test. *P < 0.05. ns, not significant.
Silencing of the AhHsf gene significantly reduced the survival of A. hygrophila in the group in which the temperature was increased, compared to that in the control group where when temperature was 30°C (females −30°C: χ2 = 2.027, p = 0.1545; 36°C: χ2 = 9.093, p = 0.0026; 39°C: χ2 = 10.65, p = 0.0011; males −30°C: χ2 = 2.642, p = 0.1041; 36°C: χ2 = 4.551, p = 0.0329; 39°C: χ2 = 5.954, p = 0.0147) (Figure 7).
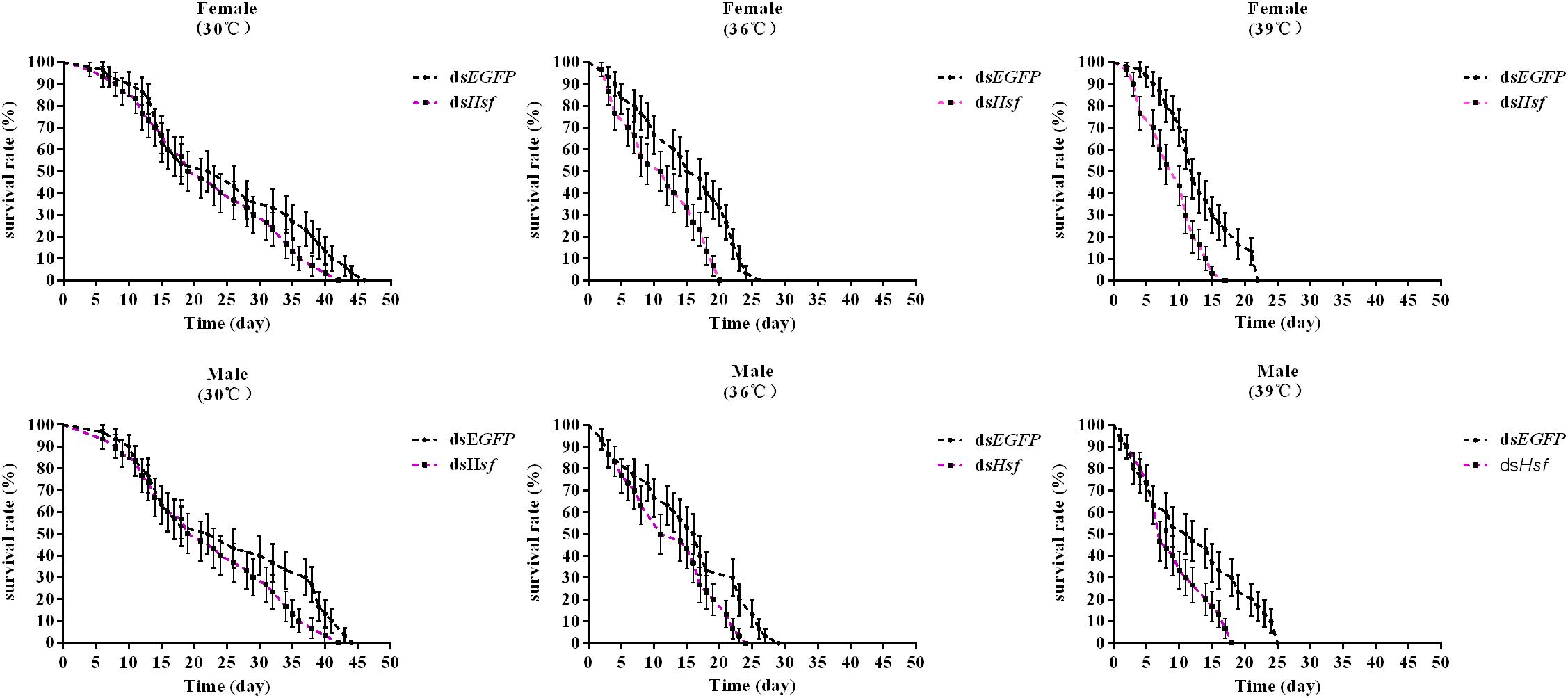
Figure 7. The effect of adult survival of A. hygrophila after injection of dsAhHsf (n = 3). Vertical bars represent standard errors of the mean. Data on the adult survival rate were analyzed using the log-rank (Mantel-Cox) test, and a survival curve was constructed using GraphPad Prism 6.
Discussion
Previous studies showed that Hsfs play a central role in remodeling the chromatin structure of Hsps promoter via constitutive interactions with its high-affinity binding site, the HSE (Westwood et al., 1991; Peteranderl and Nelson, 1992; Rabindran et al., 1993; Sarge et al., 1993; Erkine et al., 1999). The interaction between Hsfs and HSE is also critical for stimulating both basal (non-induced) and induced transcription (Erkine et al., 1999). Considering the complexity of the Hsf gene family, knockout of Hsf genes is necessary to determine their respective functional role and biological importance (Kumar et al., 2009). In the present study, injection with dsAhHsf significantly decreased the expression of AhHsp70 and AhsHsp21 in A. hygrophila, compared with the control group that was injected with dsEGFP. In addition, the expression of AhHsf also decreased significantly after injection with dsAhHsp70, dsAhsHsp21 or dsAhHsp70 & AhsHsp21- injected compared to the control group that was injected with dsEGFP. It has been reported in other studies (Sato et al., 1998; Ayté et al., 2001) that downstream genes could also influence the expression of upstream genes through feedback regulation. Ayté et al. (2001) reported that the cell-cycle-regulated transcriptional expression of the cyclin cig2 gene is dependent on the regulation of the transcription factor Mlu1 cell – cycle box binding factor (MBF) in yeast. However, the deletion of Mlu1 cell – cycle box (MCB) elements in the cig2 promoter perturbed the expression of not only cig2 but also of other MBF – dependent genes, indicating that Cig2p feedback could regulate the activity of the transcription factor, MBF; therefore, AhHsf is involved in the regulation of the heat shock response. This outcome is similar to that reported for D. melanogaster, where expression of Hsf was affected by heat shock treatment, which produced marked effects on the heat shock response (Sørensen et al., 2007).
In freshly emerged A. hygrophila females, injection with dsAhHsf significantly reduced expression of the genes AhHsp70 and AhsHsp21, and the treatment also reduced fecundity of A. hygrophila female and longevity of adult specimens in general. Our results, which were in corroboration with those of D. melanogaster, demonstrated a potential connection that AhHsf expression was involved in the up-regulation of AhHsp70 and AhsHsp21 in response to high temperatures. Previous studies showed that Nielsen et al. (2005) performed a study on the role of Hsf activation for resistance to heat, cold, and high-temperature knock-down and reported that the induction of stress genes was regulated by Hsf, such as Hsps. In addition, our finding, which showed a correlation between the AhHsp and AhHsf activities, was consistent with the observations of Edwards et al. (1992), in which a qualitative correlation was seen in humans between levels of Hsp70 induced in response to heat shock and potentiation by heat shock of progesterone receptor-mediated CAT gene expression; thus, the levels of Hsp genes are largely determined by the activity of Hsf. However, the precise mechanism by which increased temperature mediates the AhHsf-AhHsp signaling pathway is not entirely clear and requires further studies.
Previous studies showed that increased temperature elicits activation of a conserved pathway involving heat-shock transcription factor HSF, which enhances the heat shock response (Schuetz et al., 1991; Abravaya et al., 1992; Baler, 1992; Schlesinger and Ryan, 1993; Mosser et al., 1993; Erkine et al., 1999; Singh and Aballay, 2006; Kumar et al., 2009; Gonsalves et al., 2011). Moreover, Li et al. (2019) identified a novel heat shock transcription factor, REVEILLE 4/8, in Arabidopsis which regulates heat shock-induced gene expression; our study revealed that the heat shock factor in A. hygrophila showed a similar expression pattern. The expression levels of AhHsf and the two potential target AhHsps increased with the increasing heat shock temperature. Therefore, we inferred that the AhHsf gene plays an important role in the heat shock response of A. hygrophila.
Conclusion
In this study, we isolated and identified a heat shock factor gene (AhHsf) and its two potential downstream genes AhHsp70 and AhsHsp21 from A. hygrophila. We used RT-qPCR and RNAi technology to detect the effects of AhHsf on the expression levels of two potential downstream AhHsp genes, fecundity of A. hygrophila females, and adult longevity. Our results showed that injection with dsAhHsf significantly inhibited the expression of AhHsp70 and AhsHsp21 mRNAs and decreased fecundity of A hygrophila female and adult longevity. Our qPCR assays showed that AhHsf expression in dsAhHsp70, dsAhsHsp21, or dsAhHsp70 & AhsHsp21- injected samples decreased significantly compared to that in dsEGFP-injected samples. Therefore, our findings provide evidence that AhHsf is involved in regulating the transcriptional expression of AhHsp70 and AhsHsp21 and plays a key role in thermotolerance in A. hygrophila. These results may improve our understanding of the molecular mechanisms of the AhHsf- AhHsp signaling pathways in A. hygrophila.
Data Availability Statement
The datasets generated for this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Author Contributions
JJ, JG, ZZ, and FW conceived and designed the experiments. JJ conducted the experiments, bioinformatic analyses, RT-PCR, qPCR, and RNAi. JJ, ZZ, and YL contributed to data analyses. JJ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the programs from the National Natural Science Foundation of China (Grant No. 31572068) and the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (Grant No. 2015BAD08B03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.562204/full#supplementary-material
Footnotes
References
Abravaya, K., Myers, M. P., Murphy, S. P., and Morimoto, R. I. (1992). The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Gene Dev. 6, 1153–1164. doi: 10.1101/gad.6.7.1153
Amin, J., Ananthan, J., and Voellmy, R. (1988). Key features of heat shock regulatory elements. Mol. Cell Biol. 8, 3761–3769. doi: 10.1128/MCB.8.9.3761
Ayté, J., Schweitzer, C., Zarzov, P., Nurse, P., and DeCaprio, J. A. (2001). Feedback regulation of the MBF transcription factor by cyclin Cig2. Nat. Cell Biol. 3, 1043–1050. doi: 10.1038/ncb1201-1043
Baler, R. (1992). Heat shock gene regulation by nascent polypeptides and denatured proteins: hsp70 as a potential autoregulatory factor. J. Cell Biol. 117, 1151–1159. doi: 10.1083/jcb.117.6.1151
Buckingham, G. R. (1996). Biological control of alligator weed, Alternanthera philoxeroides, the world’s first aquatic weed success story. Castanea 61, 232–243.
Burton, V., Mitchell, H. K., Young, P., and Petersen, N. S. (1988). Heat shock protection against cold stress of Drosophila melanogaster. Mol. Cell Biol. 8, 3550–3552. doi: 10.1128/mcb.8.8.3550
Cohen, P. (2000). The regulation of protein function by multisite phosphorylation–a 25 year update. Trends Biochem. Sci. 25, 596–601. doi: 10.1016/s0968-0004(00)01712-6
Coulson, J. R. (1977). Biological Control of Alligator Weed, 1959–1972. A Review and Evaluation. Technical Bulletin, Agricultural Research Service, United States Department of Agriculture.
Cunniff, N. F., and Morgan, W. D. (1993). Analysis of heat shock element recognition by saturation mutagenesis of the human HSP70.1 gene promoter. J. Biol. Chem. 268, 8317–8324. doi: 10.1111/j.1432-1033.1993.tb17832.x
Deng, Y., Hu, Z., Chai, Z., and Tang, Y. Z. (2018). Molecular cloning of heat shock protein 60 (Hsp60) and 10 (Hsp10) genes from the cosmopolitan and harmful dinoflagellate Scrippsiella trochoidea and their differential transcriptions responding to temperature stress and alteration of life cycle. Mar. Biol. 166:7. doi: 10.1007/s00227-018-3455-3
Edwards, D. P., Estes, P. A., Fadok, V. A., Bona, B. J., Oñate, S., Nordeen, S. K., et al. (1992). Heat shock alters the composition of heteromeric steroid receptor complexes and enhances receptor activity in vivo. Biochemistry 31, 2482–2491. doi: 10.1021/bi00124a007
Erkine, A. M., Magrogan, S. F., Sekinger, E. A., and Gross, D. S. (1999). Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro. Mol. Cell Biol. 19, 1627–1639. doi: 10.1128/MCB.19.5.3929
Feder, M. E., and Hofmann, G. E. (1999). Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282. doi: 10.1146/annurev.physiol.61.1.243
Fernandes, M., Xiao, H., and Lis, J. T. (1994). Fine structure analyses of the Drosophila and Saccharomyces heat shock factor-heat shock element interactions. Nucleic Acids Res. 22, 167–173. doi: 10.1093/nar/22.2.167
Fleige, S., Walf, V., Huch, S., Prgomet, C., Sehm, J., and Pfaffl, M. W. (2006). Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnol. Lett. 28, 1601–1613. doi: 10.1007/s10529-006-9127-2
Ghanim, M., Kontsedalov, S., and Czosnek, H. (2007). Tissue-specific gene silencing by RNA interference in the whitefly Bemisia tabaci (Gennadius). Insect. Biochem. Mol. 37, 732–738. doi: 10.1016/j.ibmb.2007.04.006
Gonsalves, S. E., Moses, A. M., Razak, Z., Robert, F., and Westwood, J. T. (2011). Whole-genome analysis reveals that active heat shock factor binding sites are mostly associated with non-heat shock genes in Drosophila melanogaster. PLoS One 6:e15934. doi: 10.1371/journal.pone.0015934
Goto, S. G., and Kimura, M. T. (1998). Heat and cold shock responses and temperature adaptations in subtropical and temperate species of Drosophila. J. Insect Physiol. 44, 1233–1239. doi: 10.1016/S0022-1910(98)00101-2
Guo, J. Y., Fu, J. W., Xian, X. Q., Ma, M. Y., and Wan, F. H. (2011). Performance of Agasicles hygrophila (Coleoptera: Chrysomelidae), a biological control agent of invasive alligator weed, at low non-freezing temperatures. Biol. Invasions 14, 1597–1608. doi: 10.1007/s10530-010-9932-3
Huey, R. B., Crill, W. D., Kingsolver, J. G., and Weber, K. E. (1992). A method for rapid measurement of heat or cold resistance of small insects. Funct. Ecol. 6, 489–494. doi: 10.2307/2389288
Jin, J. S., Zhao, M. T., Wang, Y., Zhou, Z. S., Wan, F. H., and Guo, J. Y. (2020a). Induced thermotolerance and expression of three key heat shock protein genes (Hsp70, Hsp21, and sHsp21) and their roles in the high temperature tolerance of alligator weed flea beetle Agasicles hygrophila. Front. Physiol. 10:1593. doi: 10.3389/fphys.2019.01593
Jin, J. S., Zhao, M. T., Zhang, H., Liu, Y. R., Wan, F. H., Zhou, Z. S., et al. (2020b). Experimental evidence that extreme temperatures may contribute to a sharp decrease in the Agasicles hygrophila population in Hunan during the summer. Entomol. Gen.
Jolly, C., Usson, Y., and Morimoto, R. I. (1999). Rapid and reversible relocalization of heat shock factor 1 within seconds to nuclear stress granules. Proc. Natl. Acad. Sci. U.S.A. 96, 6769–6774. doi: 10.1073/pnas.96.12.6769
Julien, M. H., Skarratt, B., and Maywald, G. F. (1995). Potential geographical distribution of alligator weed and its biological control by Agasicles hygrophila. J. Aquat Plant Manage. 33, 55–60. doi: 10.1086/297279
Kristensen, T. N., Sørensen, J. G., and Loeschcke, V. (2003). Mild heat stress at a young age in Drosophila melanogaster leads to increased Hsp70 synthesis after stress exposure later in life. J. Genet. 82, 89–94. doi: 10.1007/bf02715811
Kroeger, P. E., and Morimoto, R. I. (1994). Selection of new HSF1 and HSF2 DNA-binding sites reveals difference in trimer cooperativity. Mol. Cell Biol. 14, 7592–7603. doi: 10.1128/MCB.14.11.7592
Kumar, M., Busch, W., Birke, H., Kemmerling, B., Nurnberger, T., and Schoffl, F. (2009). Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Mol. Plant. 2, 152–165. doi: 10.1093/mp/ssn095
Li, B., Gao, Z., Liu, X., Sun, D., and Tang, W. (2019). Transcriptional profiling reveals a time-of-day specific role of REVEILLE 4/8 in regulating the first wave of heat shock-induced gene expression in Arabidopsis. Plant Cell 31, 2353–2369. doi: 10.1105/tpc.19.00519
Li, Y. N., Fu, J. W., and Guo, J. Y. (2011). Effects of release density on the population dynamics of the biocontrol agent, Agasicles hygrophila (Coleoptera: Chrysomelidae). J. Biosafety 20, 275–280.
Lindquist, A. S., and Craig, E. A. (1988). The heat-shock proteins. Annu. Rev. Genet. 22, 631–677. doi: 10.1146/annurev.ge.22.120188.003215
Lindquist, S. L. (1986). The heat-shock response. Annu. Rev. Biochem. 55, 1151–1191. doi: 10.1146/annurev.bi.55.070186.005443
Lis, J., and Wu, C. (1993). Protein traffic on the heat shock promoter: parking, stalling, and trucking alo. ngCell 74, 1–4. doi: 10.1016/0092-8674(93)90286-Y
Lü, Z. C., Wang, L. H., Zhang, G. F., Wan, F. H., Guo, J. Y., Yu, H., et al. (2014). Three heat shock protein genes from Bactrocera (Tetradacus) minax Enderlein: gene cloning, characterization, and association with diapause. Neotrop. Entomol. 43, 362–372. doi: 10.1007/s13744-014-0216-y
Ma, R. Y., Wang, R., and Ding, J. Q. (2003). Classical biological control of exotic weeds. Acta Ecol. Sin. 12, 2677–2688.
Maddox, D. M., and Rhyne, M. (1975). Effects of induced host-plant mineral deficiencies on attraction, feeding, and fecundity of the alligator weed flea beetle. Environ. Entomol. 4, 682–686. doi: 10.1093/ee/4.5.682
Mantel, N. (1985). Propriety of the Mantel–Haenszel variance for the log rank test. Biometrika 72, 471–472. doi: 10.1093/biomet/72.2.471
Marchler, G., and Wu, C. (2014). Modulation of Drosophila heat shock transcription factor activity by the molecular chaperone DROJ1. EMBO J. 20, 499–509. doi: 10.1093/emboj/20.3.499
McColl, G., Hoffmann, A. A., and Mckechnie, S. W. (1996). Response of two heat genes to selection for knockdown heat resistance in Drosophila melanogaster. Genetics 143, 1615–1627. doi: 10.1111/j.1365-2443.1996.tb00017.x
Morimoto, R. I. (1993). Cells in stress: transcriptional activation of heat shock genes. Science 259, 1409–1410. doi: 10.1126/science.8451637
Mosser, D. D., Duchaine, J., and Massie, B. (1993). The DNA-binding activity of the human heat shock transcription factor is regulated in vivo by hsp70. Mol. Cell Biol. 13, 5427–5438. doi: 10.1128/MCB.13.9.5427
Nair, S. C., Toran, E. J., Rimerman, R. A., Hjermstad, S., Smithgall, T. E., and Smith, D. F. (1996). A pathway of multi-chaperone interactions common to diverse regulatory proteins: estrogen receptor, Fes tyrosine kinase, heat shock transcription factor Hsf1, and the aryl hydrocarbon receptor. Cell Stress Chaperon. 1, 237–250. doi: 10.2307/1601686
Nielsen, M. M., Overgaard, J. S., Rensen, J. G., et al. (2005). Role of HSF activation for resistance to heat, cold and high-temperature knock-down. J. Insect Physiol. 51, 1320–1329. doi: 10.1016/j.jinsphys.2005.08.002
Nover, L. (1991). HSFs and HSPs–a stressful program on transcription factors and chaperones. Stress proteins and the heat shock response, sponsored by cold spring harbor laboratory, cold spring harbor, NY USA, April 29-May 2. 1991. New Biol. 3, 855–859.
Pan, X. Y., Geng, Y., Zhang, W. J., Li, B., and Chen, J. K. (2006). The influence of abiotic stress and phenotypic plasticity on the distribution of invasive Alternanthera philoxeroides along a riparian zone. Acta Oecol. 30, 333–341. doi: 10.1016/j.actao.2006.03.003
Parsell, D. A., and Lindquist, S. (1993). The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu. Rev. Genet. 27, 437–496. doi: 10.1146/annurev.ge.27.120193.002253
Pelham, H. R. (1982). A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell 30, 517–528. doi: 10.1016/0092-8674(82)90249-5
Peteranderl, R., and Nelson, H. C. (1992). Trimerization of the heat shock transcription factor by a triple-stranded alpha-helical coiled-coil. Biochemistry 31, 12272–12276. doi: 10.1021/bi00163a042
Rabindran, S. K., Haroun, R. I., Clos, J., Wisniewski, J., and Wu, C. (1993). Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science 259, 230–234. doi: 10.1126/science.8421783
Ritossa, F. M. (1962). A new puffing pattern induced by temperature shock and DNP in Drosophila. Cell. Mol. Life Sci. 18, 571–573. doi: 10.1007/BF02172188
Sarge, K. D., Murphy, S. P., and Morimoto, R. I. (1993). Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell Biol. 13, 1392–1407. doi: 10.1128/MCB.13.6.3838
Sato, M., Hata, N., Asagiri, M., Nakaya, T., Taniguchi, T., and Tanaka, N. (1998). Positive feedback regulation of type IIFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441, 106–110. doi: 10.1016/s0014-5793(98)01514-2
Scharf, K. D., Berberich, T., Ebersberger, I., and Nover, L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta 1819, 104–119. doi: 10.1016/j.bbagrm.2011.10.002
Schlesinger, M. J., and Ryan, C. (1993). An ATP and hsc70 dependent oligomerization of nascent heat-shock factor (HSF) polypeptide suggests that HSF itself could be a “sensor” for the cellular stress response. Protein Sci. 2, 1356–1360. doi: 10.1002/pro.5560020819
Schuetz, T. J., Gallo, G. J., Sheldon, L., Tempst, P., and Kingston, R. E. (1991). Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc. Natl. Acad. Sci. U.S.A. 88, 6911–6915. doi: 10.1073/pnas.88.16.6911
Sejerkilde, M., Sørensen, J. G., and Loeschcke, V. (2003). Effects of cold and heat hardening on thermal resistance in Drosophila melanogaster. J. Insect Physiol. 49, 719–726. doi: 10.1016/s0022-1910(03)00095-7
Singh, V., and Aballay, A. (2006). Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc. Natl. Acad. Sci. U.S.A. 103, 13092–13097. doi: 10.1073/pnas.0604050103
Sørensen, J. G., Kristensen, T. N., Kristensen, K. V., and Loeschcke, V. (2007). Gender specific effects of heat induced hormesis in Hsf-deficient. Exp. Gerontol. 42, 1123–1129. doi: 10.1016/j.exger.2007.09.001
Sørensen, J. G., Loeschcke, V., and Kristensen, T. N. (2003). The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 6, 1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x
Spencer, N. R., and Coulson, J. R. (1976). The biological control of alligatorweed, Alternanthera philoxeroides, in the United States of America. Aquat. Bot. 2, 177–190. doi: 10.1016/0304-3770(76)90019-x
Tatar, M., Khazaeli, A. A., and Curtsinger, J. W. (1997). Chaperoning extended life. Nature 390:30. doi: 10.1038/36237
Voellmy, R. (2004). Transcriptional regulation of the metazoan stress protein response. Prog. Nucleic Acid Res. 78, 143–185. doi: 10.1016/S0079-6603(04)78004-6
Wang, B. R., Li, W. G., and Wang, J. B. (2005). Genetic diversity of Alternanthera philoxeroides in China. Aquat. Bot. 81, 277–283. doi: 10.1016/j.aquabot.2005.01.004
Wang, R. (1989). “Biological control of weeds in China: a status report,” in Proceedings of the Seventh International Symposium on Biological Control of Weeds, Rome, 689–693.
Westwood, J. T., Clos, J., and Wu, C. (1991). Stress-induced oligomerization and chromosomal relocalization of heat shock factor. Nature 353, 822–827. doi: 10.1038/353822a0
Wisniewski, J., Orosz, A., Allada, R., and Wu, C. (1996). The C-terminal region of Drosophila heat shock factor (HSF) contains a constitutively functional transactivation domain. Nucleic Acids Res. 24, 367–374. doi: 10.1093/nar/24.2.367
Wu, C. (1995). Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 11, 441–469. doi: 10.1146/annurev.cb.11.110195.002301
Xiao, H., and Lis, J. T. (1988). Germline transformation used to define key features of heat shock response elements. Science 239, 1139–1142. doi: 10.1126/science.3125608
Xu, C. Y., Zhang, W. J., Fu, C. Z., and Lu, B. R. (2003). Genetic diversity of alligator weed in China by RAPD analysis. Biodivers. Conserv. 12, 637–645. doi: 10.1023/a:1022453129662
Ye, W. H., Li, J., Cao, H. L., and Ge, X. J. (2010). Genetic uniformity of Alternanthera philoxeroides in South China. Weed Res. 43, 297–302. doi: 10.1046/j.1365-3180.2003.00346.x
Zeiger, C. F. (1967). Biological control of alligatorweed with Agasicles n. sp. in Florida. Hyacinth. Control. J. 6, 31–34.
Zhang, W., Ma, L., Xiao, H., Xie, B., Smagghe, G., Guo, Y., et al. (2016). Molecular characterization and function analysis of the vitellogenin receptor from the cotton bollworm. PLoS One 11:e0155785. doi: 10.1371/journal.pone.0155785
Zhao, M. T., Wang, Y., Zhou, Z. S., Wang, R., Guo, J. Y., and Wan, F. H. (2016). Effects of periodically repeated heat events on reproduction and ovary development of Agasicles hygrophila (Coleoptera: Chrysomelidae). J. Econ. Entomol. 109, 1586–1594. doi: 10.1093/jee/tow093
Keywords: Agasicles hygrophila, heat shock protein, heat shock factor, RNAi, thermotolerance
Citation: Jin J, Li Y, Zhou Z, Zhang H, Guo J and Wan F (2020) Heat Shock Factor Is Involved in Regulating the Transcriptional Expression of Two Potential Hsps (AhHsp70 and AhsHsp21) and Its Role in Heat Shock Response of Agasicles hygrophila. Front. Physiol. 11:562204. doi: 10.3389/fphys.2020.562204
Received: 14 May 2020; Accepted: 25 August 2020;
Published: 15 September 2020.
Edited by:
Guy Smagghe, Ghent University, BelgiumCopyright © 2020 Jin, Li, Zhou, Zhang, Guo and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianying Guo, Z3VvamlhbnlpbmdAY2Fhcy5jbg==; Fanghao Wan, d2FuZmFuZ2hhb0BjYWFzLmNu
 Jisu Jin
Jisu Jin Youzhi Li
Youzhi Li Zhongshi Zhou
Zhongshi Zhou Hong Zhang
Hong Zhang Jianying Guo2*
Jianying Guo2* Fanghao Wan
Fanghao Wan