
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Physiol. , 24 January 2014
Sec. Cardiac Electrophysiology
Volume 5 - 2014 | https://doi.org/10.3389/fphys.2014.00015
Atrial fibrillation (AF) is the most common sustained arrhythmia, especially in the elderly, and has a significant genetic component. Recently, several independent investigators have demonstrated a functional role for small non-coding RNAs (microRNAs) in the pathophysiology of this cardiac arrhythmia. This report represents a systematic and updated appraisal of the main studies that established a mechanistic association between specific microRNAs and AF, focusing both on the regulation of electrical and structural remodeling of cardiac tissue.
MicroRNAs (miRs) are an evolutionarily conserved class of small (~22 nucleotides) non-coding RNAs (Ambros, 2004; Gan et al., 2013), first discovered in Caenorhabditis elegans (Ruvkun and Giusto, 1989; Ruvkun et al., 1989). They represent a vital component of genetic regulation, existing in virtually all organisms, suggesting thereby a pivotal role in biological processes (Latronico and Condorelli, 2008; Thum et al., 2008). Indeed, miRs are important regulators of gene expression in numerous biological processes including cellular proliferation, differentiation, and tumorigenesis (Care et al., 2007; Dvinge et al., 2013; Shen et al., 2013; Song et al., 2013). Typically, miRs are regarded as negative regulators of gene expression that inhibit translation and/or promote mRNA degradation by base pairing to complementary sequences within the 3′-untranslated region (3′-UTR) of protein-coding mRNA transcripts (Van Rooij and Olson, 2012; Meijer et al., 2013). Generally, mRNA degradation accounts for the majority of miR activity (Guo et al., 2010). Hence, by altering levels of key regulators within complex genetic pathways, miRs provide a posttranscriptional level of control of homeostatic and developmental events (Callis et al., 2009; Yates et al., 2013).
It is estimated that miRs regulate over 60% of all protein-coding genes (Friedman et al., 2009; Akerman and Mukherjee, 2013; Leucci et al., 2013). Considering that a single miR can regulate multiple mRNAs and that each mRNA may be a target of multiple miRs, the possible pathways for miR-dependent regulation of protein abundance seem to be extremely complicated (Akerman and Mukherjee, 2013; Santulli and Totary-Jain, 2013). In this model, a biologic response would be expected only after co-expression of various miRs that cooperatively target different components of a functional network (Liu et al., 2012; Van Rooij and Olson, 2012) or are all required to sufficiently repress a single target (Lagos-Quintana et al., 2001; Kim, 2013).
Maturation of miRs involves a multi-stepped process (Bartel, 2004; Cullen, 2004) that starts from the transcription (mainly operated by RNA polymerase II) of single-stranded non-protein-coding RNAs, which are either transcribed as stand alone transcripts (intergenic miRs), often encoding various miRs, or generated by the processing of introns of protein-coding genes (intragenic or intronic miRs).
Transcription of intergenic miRs leads to the formation of primary miRs (pri-miR) with a characteristic hairpin or stem-loop structure (Denli et al., 2004), which are subsequently processed by the nuclear RNase III, Drosha (Zeng et al., 2005), and its partner proteins, among which there is the DiGeorge Syndrome Critical Region 8 (DGCR8, known as Pasha in invertebrates), named for its association with DiGeorge Syndrome (Shiohama et al., 2003; Roth et al., 2013), to become precursor miR s (pre-miR). On the other hand, intronic miRs are obtained by the regular transcription of their host genes and then spliced to form looped pre-miRs, bypassing thereby the Drosha pathway (Bartel, 2004; Ruby et al., 2007).
Pre-miRs are exported from the nucleus in the cytoplasm in a process involving the Ran-GTP-dependent shuttle Exportin-5 (Lund et al., 2004). Once in the cytosol, the pre-miR hairpin is cleaved by the RNase III enzyme Dicer (Saxena and Tabin, 2010; Marasovic et al., 2013), yielding a mature miR:miR* duplex about 22 nucleotides in length, which is subsequently incorporated into the protein complex called RNA-induced silencing complex (RISC) to form miRISC (Filipowicz et al., 2008; Wu et al., 2013). At this point, one of the double strands, the guide strand, is selected by the argonaute protein (Pfaff et al., 2013), the catalytically active RNase in the RISC complex, on the basis of the thermodynamic stability of the 5′ end. In particular, the strand with a less thermodynamically stable 5′ end is commonly chosen and loaded into the RISC complex (Siomi and Siomi, 2009), serving as a guide for mRISC to find its complementary motifs in the 3′-UTR of the target mRNA(s). Although either strand of the mature duplex may potentially act as a functional miR, only one strand is usually incorporated into the RISC where the miR and its mRNA target interact (Fabian and Sonenberg, 2012; Von Brandenstein et al., 2012). Such a binding inhibits the translation of the protein that the target mRNA encodes or promotes gene silencing via mRNA degradation (Latronico and Condorelli, 2008; Kallen et al., 2012; Papait et al., 2013).
In the human genome more than 1200 miR sequences have been identified, hitherto [miRTarBase, Release 4.5, version 20 (Hsu et al., 2011)], with over 50,000 miR-target interactions. Recently, several algorithms and bioinformatics websites, including TargetScan and miRWalk (Lewis et al., 2005; Dweep et al., 2011) have been developed to predict specific mRNA/miR interactions. However, miR-binding rules are quite complex and are not fully understood, resulting in a lack of consensus in the literature.
Given all these crucial features, miRs could represent an important way for the cell to establish intercellular (with other cells, via secreted miRs) and intracellular (among its own genes) communication.
Establishing direct cause-and-effect links between miRs and mRNA targets is essential to understanding the molecular mechanisms underlying disease and the subsequent development of targeted therapies (Ambros et al., 2003; Zacharewicz et al., 2013).
Atrial fibrillation (AF) is a highly prevalent disease with a significant genetic component (Den Hoed et al., 2013; Mahida, 2013; Santulli, 2013), considered the most common sustained arrhythmia, which can cause or exacerbate heart failure and represents an important risk factor for ischemic stroke (Fye, 2006; Conen et al., 2011; Santulli, 2012b; Santulli et al., 2013; Thomas and Sorrentino, 2014). AF represents the most commonly seen arrhythmia worldwide, especially in the geriatric population (Huikuri, 2008; Riley and Manning, 2011; Santulli et al., 2012c; Santulli and Iaccarino, 2013) and is associated with a substantially pronounced morbidity and mortality (Beyerbach and Zipes, 2004; Santulli, 2012a; Garg and Akoum, 2013; Menezes et al., 2013). From a pathophysiological point of view, AF is characterized by atrial electrical remodeling, mainly mediated by ion-channel alterations (Brundel et al., 2001; Santulli et al., 2012b; Xie et al., 2013) and structural remodeling (fibrosis and apoptosis), which favors arrhythmia recurrence and maintenance (Perino et al., 2011; Santulli and D'Ascia, 2012; Santulli et al., 2012b). A noticeable feature of the electrical remodeling associated with AF is the abbreviation of the effective refractory period favoring reentry (D'Ascia et al., 2010, 2011; Kapur and Macrae, 2013), primarily due to shortening of atrial action potential duration (APD).
Three potential models have been proposed to explain the pathophysiology of AF (Jalife, 2011), albeit the precise relationship of each of these conceptual frameworks to human AF remains under investigation (Vikman et al., 2005; Kapur and Macrae, 2013; Shah et al., 2013):
The focal mechanism theory suggests that AF is provoked by the rapid firing of single or multiple ectopic foci, and also proposes a functional role for continued ectopic firing in the maintenance of AF (Lee et al., 2013).
The single circuit re-entry theory of AF assumes the presence of a single dominant reentry circuit alongside with the fragmentation of emanating waves in the heterogeneous electrical substrate of normal atrial tissue (Zemlin and Pertsov, 2007; Kapur and Macrae, 2013).
The multiple wavelet theory of AF stands on the notion that multiple reentry circuits exist, with randomly propagating wavefronts that must find receptive tissue in order to persist (Haissaguerre et al., 2013).
Slowing of conduction velocities and shortening of the refractory period of atrial myocytes (both central features of the electrical remodeling seen in AF) might help to stabilize the arrhythmia by decreasing circuit size. Of course these mechanistic models are not mutually exclusive. They may coexist in a single subject at various stages in the pathogenesis of AF and each may be applicable to certain subgroups of AF patients (Lindgren et al., 2003; Brieger and Freedman, 2009; Ruwald et al., 2013). Theoretically, all the miRs that are directly or indirectly involved in one of these processes, which are eventually based on the regulation of structural or electrical remodeling (cardiac automaticity, ion channels, fibrosis, and apoptosis), could participate in AF induction or perpetuation.
The most common experimental approach to identify the specific miRs that play a role in a certain disease mainly consists of three phases: (1) Use a microarray matrix to recognize a list of miRs that are differentially expressed in subjects with the disease compared to control subjects (Frezzetti et al., 2011; Jayaswal et al., 2011); (2) assess the putative target site efficacy by using bioinformatics-based algorithms or other computational tools that score potential interactions between microRNAs and mRNAs (Witkos et al., 2011); (3) validate in vitro (or in vivo) the existence of an inverse correlation between the expression levels of the miR and protein levels of its target gene(s). Another biological validation could be also achieved using a reporter system or other assays to prove that the binding of the miR and the target mRNA occurs within a RISC complex (Ling et al., 2013).
Growing evidence demonstrates that miRs regulate several properties of cardiac physiology and excitability, including automaticity, Ca2+ handling, conduction, and repolarization (Grueter et al., 2012; Boon et al., 2013; Heymans et al., 2013; Latronico and Condorelli, 2013). In particular, recent reports have unveiled an essential role of miRs in regulating cardiac excitability and arrhythmogenesis (Callis et al., 2009; Shan et al., 2009). These studies have primarily focused on the two muscle-specific miRs, i.e., miR-1 and miR-133, which are among the most abundantly expressed miRs in the heart (Liang et al., 2007; Wang et al., 2011).
However, lately other ubiquitously distributed miRs, such as miR-328 have been shown to exhibit a strong arrhythmogenic potential (Lu et al., 2010). It is likely that multiple miRs contribute to controlling arrhythmogenicity of the heart and that different miRs are involved in different types of arrhythmias under different pathological conditions of the heart (Horie et al., 2012; Kochegarov et al., 2013; Qiao et al., 2013; Zhang et al., 2013a). The most important miRs so far implicated in the pathophysiology of AF, regulating both electrical and structural remodeling, are reported in Table 1, alongside with their target gene(s) and function(s).
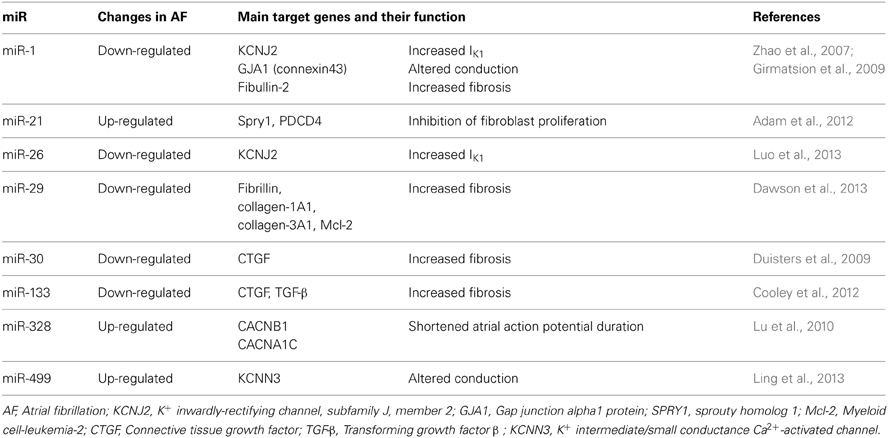
Table 1. miRs with an established role in the regulation of cardiac electrical and structural remodeling.
Although circulating miRs seem to be interesting candidates as biomarkers in AF patients, they still possess several important limitations (Quiat and Olson, 2013; Santulli and Totary-Jain, 2013). Indeed, at the moment there is no good natural stable housekeeping control for circulating miRs, which may result in strong variations, and they are generally present in low amounts in plasma and serum.
As mentioned above, arrhythmogenesis is essentially attributed to enhanced triggered activity, and several studies have attributed such activity to alterations in different ion channels, including a peculiar instability in Ca2+ handling (Ter Keurs and Boyden, 2007; Zhang et al., 2008; Latronico and Condorelli, 2009; Kushnir and Marks, 2010; Nivala et al., 2012) and modulation of the cardiac inwardly rectifying K+ current (IK1), which stabilizes the resting membrane potential, controls cardiac excitability, regulates late-phase repolarization, and is thereby responsible for shaping the initial depolarization and final repolarization of the action potential (Dhamoon and Jalife, 2005).
In a seminal study, the expression of miR-1 was demonstrated to be reduced by ~86% in tissue samples of AF patients (Girmatsion et al., 2009). This miR has been shown to be mechanistically critical in the pathophysiology of AF via targeting IK1 and connexin43 (Girmatsion et al., 2009), both considered master regulators of cardiac conduction (Delmar and Makita, 2012; Musa et al., 2013). Similarly, miR-26 has the potential to repress IK1 (Luo et al., 2013). Intriguingly, in the setting of ventricular arrhythmias miR-1 levels appear to be augmented (Anderson and Mohler, 2007; Yang et al., 2007). Thus, the different regulation of such a miR in ventricular and atrial tissue deserves further investigations (Anderson and Mohler, 2007; Yang et al., 2007; Santulli et al., 2012a). The recently proposed involvement of miR-1 in the modulation of Ca2+ handling proteins, including calmodulin, phospholamban, Na+/Ca2+ exchanger (NCX), sorcin, junctin, triadin, eventually resulting in shortened refractoriness of sarcoplasmic reticulum Ca2+ release further support the hypothesis of a pivotal functional role of such a miR in the pathogenesis of AF (Ali et al., 2012; Karakikes et al., 2013; Slagsvold et al., 2013; Tritsch et al., 2013; Zhang et al., 2013b).
Lu et al. (2010) demonstrated that miR-328 level was elevated in AF patients and identified as target genes CACNA1C and CACNB1, encoding L-type Ca2+ channel subunits. Thereby, increased levels of miR-328 reduce ICaL density and shorten APD, leading to an increased arrhythmogenic potential. Additionally, miR-223, miR-328, and miR-664 were found to be upregulated by >2 fold in AF samples (Lu et al., 2010), and further investigations are required to establish the molecular mechanism underlying such a change in the miR transcriptome.
Most recently, Ling and colleagues found a strong association between mir-499, which is significantly up-regulated in atrial tissue from AF patients, and KCNN3, the gene that encodes the small-conductance Ca2+-activated K+ channel 3 (SK3), possibly contributing to the electrical remodeling observed in AF (Ling et al., 2013)
Fibrosis is the hallmark of structural cardiac remodeling (Allessie et al., 2002; Nguyen et al., 2013). Structural changes in the atria of AF patients have been identified (Anyukhovsky et al., 2005) at the level of cardiomyocytes and extracellular matrix (ECM), which predominately includes collagen types I and III, fibronectin, laminin, fibromodulin, and entactin (Goudis et al., 2012). ECM remodeling represents a maladaptive response to changes in myocardial structure and function during the progression of cardiac disease (Siwik and Colucci, 2004; Dernellis and Panaretou, 2006). Degradation and deposition of ECM is a process dynamically regulated by a delicate balance (Goudis et al., 2012) between matrix metalloproteinases (MMPs) and tissue inhibitors of matrix metalloproteinases (TIMPs).
In addition to the regulation of key proteins involved in electrical remodeling, miR-1 also modulate cardiac fibrosis, through means of its target protein Fibullin-2, a secreted protein implicated in ECM remodeling (Karakikes et al., 2013). An intriguing role for miR-21 and its downstream target Sprouty (Spry1), a master regulator of fibroblast survival and growth factor secretion, controlling the extent of interstitial fibrosis (Thum et al., 2008), has been demonstrated both in AF patients, in which atria miR-21 is increased, and murine models (Adam et al., 2012). This miR might also be involved in the apoptosis process and in inflammation (Ando et al., 2013). Several groups have recently proposed miR-29 as a mechanistic contributor in AF, through means a regulation of several proteins involved in cardiac fibrosis and apoptosis (Straten and Andersen, 2010; Dawson et al., 2013; Hale and Levis, 2013).
Another potential therapeutic target to modulate fibrosis in AF is miR-30, which expression is down-regulated in AF. This miR directly interacts with the 3′ untranslated region of the connective tissue growth factor (CTGF), a key profibrotic protein (Duisters et al., 2009). The same group also proposed miR-133 as a modulator of CTGF protein levels (Duisters et al., 2009). Of interest, the same miR-133, which levels are reduced in AF patients (Cooley et al., 2012), is involved in the regulation of apoptosis and TGF-β signaling (Goette, 2009).
Mounting evidence demonstrates that miRs are becoming one of the most fascinating areas of biology, given their critical roles in fine-tuning of physiological processes and their deregulation in several disorders, including AF. The relative role of different miRs in AF may also depend on the underlying etiology of AF, as the rhythm is an end stage manifestation of multiple, distinct predisposing pathological changes. The functional role of miRs as direct or indirect post-transcriptional regulators of genes implied in electrical and/or structural remodeling strongly suggest that these miRs may serve as potential biomarkers or promising drug targets, in prevention, treatment, and management of AF.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Adam, O., Lohfelm, B., Thum, T., Gupta, S. K., Puhl, S. L., Schafers, H. J., et al. (2012). Role of miR-21 in the pathogenesis of atrial fibrosis. Basic Res. Cardiol. 107, 278. doi: 10.1007/s00395-012-0278-0
Akerman, A. W., and Mukherjee, R. (2013). MicroRNAs emerging as mediators of remodeling with atrial fibrillation. Heart Rhythm 10, 1010–1011. doi: 10.1016/j.hrthm.2013.03.021
Ali, R., Huang, Y., Maher, S. E., Kim, R. W., Giordano, F. J., Tellides, G., et al. (2012). miR-1 mediated suppression of Sorcin regulates myocardial contractility through modulation of Ca2+ signaling. J. Mol. Cell. Cardiol. 52, 1027–1037. doi: 10.1016/j.yjmcc.2012.01.020
Allessie, M., Ausma, J., and Schotten, U. (2002). Electrical, contractile and structural remodeling during atrial fibrillation. Cardiovasc. Res. 54, 230–246. doi: 10.1016/S0008-6363(02)00258-4
Ambros, V., Bartel, B., Bartel, D. P., Burge, C. B., Carrington, J. C., Chen, X., et al. (2003). A uniform system for microRNA annotation. RNA 9, 277–279. doi: 10.1261/rna.2183803
Anderson, M. E., and Mohler, P. J. (2007). MicroRNA may have macro effect on sudden death. Nat. Med. 13, 410–411. doi: 10.1038/nm0407-410
Ando, Y., Yang, G. X., Kenny, T. P., Kawata, K., Zhang, W., Huang, W., et al. (2013). Overexpression of microRNA-21 is associated with elevated pro-inflammatory cytokines in dominant-negative TGF-beta receptor type II mouse. J. Autoimmun. 41, 111–119. doi: 10.1016/j.jaut.2012.12.013
Anyukhovsky, E. P., Sosunov, E. A., Chandra, P., Rosen, T. S., Boyden, P. A., Danilo, P., et al. (2005). Age-associated changes in electrophysiologic remodeling: a potential contributor to initiation of atrial fibrillation. Cardiovasc. Res. 66, 353–363. doi: 10.1016/j.cardiores.2004.10.033
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Beyerbach, D. M., and Zipes, D. P. (2004). Mortality as an endpoint in atrial fibrillation. Heart Rhythm 1, B8–B18. discussion: B18–B19. doi: 10.1016/j.hrthm.2004.04.018
Boon, R. A., Iekushi, K., Lechner, S., Seeger, T., Fischer, A., Heydt, S., et al. (2013). MicroRNA-34a regulates cardiac ageing and function. Nature 495, 107–110. doi: 10.1038/nature11919
Brieger, D. B., and Freedman, S. B. (2009). Delirium cordis: can we predict the onset of atrial fibrillation? Lancet 373, 698–700. doi: 10.1016/S0140-6736(09)60415-3
Brundel, B. J., Van Gelder, I. C., Henning, R. H., Tuinenburg, A. E., Wietses, M., Grandjean, J. G., et al. (2001). Alterations in potassium channel gene expression in atria of patients with persistent and paroxysmal atrial fibrillation: differential regulation of protein and mRNA levels for K+ channels. J. Am. Coll. Cardiol. 37, 926–932. doi: 10.1016/S0735-1097(00)01195-5
Callis, T. E., Pandya, K., Seok, H. Y., Tang, R. H., Tatsuguchi, M., Huang, Z. P., et al. (2009). MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 119, 2772–2786. doi: 10.1172/JCI36154
Care, A., Catalucci, D., Felicetti, F., Bonci, D., Addario, A., Gallo, P., et al. (2007). MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13, 613–618. doi: 10.1038/nm1582
Conen, D., Chae, C. U., Glynn, R. J., Tedrow, U. B., Everett, B. M., Buring, J. E., et al. (2011). Risk of death and cardiovascular events in initially healthy women with new-onset atrial fibrillation. JAMA 305, 2080–2087. doi: 10.1001/jama.2011.659
Cooley, N., Cowley, M. J., Lin, R. C., Marasco, S., Wong, C., Kaye, D. M., et al. (2012). Influence of atrial fibrillation on microRNA expression profiles in left and right atria from patients with valvular heart disease. Physiol. Genomics 44, 211–219. doi: 10.1152/physiolgenomics.00111.2011
Cullen, B. R. (2004). Transcription and processing of human microRNA precursors. Mol. Cell 16, 861–865. doi: 10.1016/j.molcel.2004.12.002
D'Ascia, S. L., D'Ascia, C., Marino, V., Lombardi, A., Santulli, R., Chiariello, M., et al. (2011). Cardiac resynchronisation therapy response predicts occurrence of atrial fibrillation in non-ischaemic dilated cardiomyopathy. Int. J. Clin. Pract. 65, 1149–1155. doi: 10.1111/j.1742-1241.2011.02732.x
D'Ascia, S. L., Santulli, G., Liguori, V., Marino, V., Arturo, C., Chiariello, M., et al. (2010). Advanced algorithms can lead to electrocardiographic misinterpretations. Int. J. Cardiol. 141, e34–e36. doi: 10.1016/j.ijcard.2008.11.144
Dawson, K., Wakili, R., Ordog, B., Clauss, S., Chen, Y., Iwasaki, Y., et al. (2013). MicroRNA29: a mechanistic contributor and potential biomarker in atrial fibrillation. Circulation 127, 1466–1475, 1475e1–e28. doi: 10.1161/CIRCULATIONAHA.112.001207
Delmar, M., and Makita, N. (2012). Cardiac connexins, mutations and arrhythmias. Curr. Opin. Cardiol. 27, 236–241. doi: 10.1097/HCO.0b013e328352220e
Den Hoed, M., Eijgelsheim, M., Esko, T., Brundel, B. J., Peal, D. S., Evans, D. M., et al. (2013). Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat. Genet. 45, 621–631. doi: 10.1038/ng.2610
Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F., and Hannon, G. J. (2004). Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235. doi: 10.1038/nature03049
Dernellis, J., and Panaretou, M. (2006). Effects of C-reactive protein and the third and fourth components of complement (C3 and C4) on incidence of atrial fibrillation. Am. J. Cardiol. 97, 245–248. doi: 10.1016/j.amjcard.2005.08.027
Dhamoon, A. S., and Jalife, J. (2005). The inward rectifier current (IK1) controls cardiac excitability and is involved in arrhythmogenesis. Heart Rhythm 2, 316–324. doi: 10.1016/j.hrthm.2004.11.012
Duisters, R. F., Tijsen, A. J., Schroen, B., Leenders, J. J., Lentink, V., Van Der Made, I., et al. (2009). miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ. Res. 104, 170–178, 176p following 178. doi: 10.1161/CIRCRESAHA.108.182535
Dvinge, H., Git, A., Graf, S., Salmon-Divon, M., Curtis, C., Sottoriva, A., et al. (2013). The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 497, 378–382. doi: 10.1038/nature12108
Dweep, H., Sticht, C., Pandey, P., and Gretz, N. (2011). miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 44, 839–847. doi: 10.1016/j.jbi.2011.05.002
Fabian, M. R., and Sonenberg, N. (2012). The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 19, 586–593. doi: 10.1038/nsmb.2296
Filipowicz, W., Bhattacharyya, S. N., and Sonenberg, N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114. doi: 10.1038/nrg2290
Frezzetti, D., De Menna, M., Zoppoli, P., Guerra, C., Ferraro, A., Bello, A. M., et al. (2011). Upregulation of miR-21 by Ras in vivo and its role in tumor growth. Oncogene 30, 275–286. doi: 10.1038/onc.2010.416
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Fye, W. B. (2006). Tracing atrial fibrillation–100 years. N. Engl. J. Med. 355, 1412–1414. doi: 10.1056/NEJMp068059
Gan, Z., Rumsey, J., Hazen, B. C., Lai, L., Leone, T. C., Vega, R. B., et al. (2013). Nuclear receptor/microRNA circuitry links muscle fiber type to energy metabolism. J. Clin. Invest. 123, 2564–2575. doi: 10.1172/JCI67652
Garg, A., and Akoum, N. (2013). Atrial fibrillation and heart failure: beyond the heart rate. Curr. Opin. Cardiol. 28, 332–336. doi: 10.1097/HCO.0b013e32835fb710
Girmatsion, Z., Biliczki, P., Bonauer, A., Wimmer-Greinecker, G., Scherer, M., Moritz, A., et al. (2009). Changes in microRNA-1 expression and IK1 up-regulation in human atrial fibrillation. Heart Rhythm 6, 1802–1809. doi: 10.1016/j.hrthm.2009.08.035
Goette, A. (2009). Nicotine, atrial fibrosis, and atrial fibrillation: do microRNAs help to clear the smoke? Cardiovasc. Res. 83, 421–422. doi: 10.1093/cvr/cvp188
Goudis, C. A., Kallergis, E. M., and Vardas, P. E. (2012). Extracellular matrix alterations in the atria: insights into the mechanisms and perpetuation of atrial fibrillation. Europace 14, 623–630. doi: 10.1093/europace/eur398
Grueter, C. E., Van Rooij, E., Johnson, B. A., Deleon, S. M., Sutherland, L. B., Qi, X., et al. (2012). A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell 149, 671–683. doi: 10.1016/j.cell.2012.03.029
Guo, H., Ingolia, N. T., Weissman, J. S., and Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840. doi: 10.1038/nature09267
Haissaguerre, M., Hocini, M., Shah, A. J., Derval, N., Sacher, F., Jais, P., et al. (2013). Noninvasive panoramic mapping of human atrial fibrillation mechanisms: a feasibility report. J. Cardiovasc. Electrophysiol. 24, 711–717. doi: 10.1111/jce.12075
Hale, C. S., and Levis, W. R. (2013). MicroRNA-29 and an integrated understanding of atrial fibrillation. J. Drugs Dermatol. 12, 1083.
Heymans, S., Corsten, M. F., Verhesen, W., Carai, P., Van Leeuwen, R. E., Custers, K., et al. (2013). Macrophage MicroRNA-155 promotes cardiac hypertrophy and failure. Circulation 128, 1420–1432. doi: 10.1161/CIRCULATIONAHA.112.001357
Horie, T., Baba, O., Kuwabara, Y., Chujo, Y., Watanabe, S., Kinoshita, M., et al. (2012). MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE-/- mice. J. Am. Heart Assoc. 1:e003376. doi: 10.1161/JAHA.112.003376
Hsu, S. D., Lin, F. M., Wu, W. Y., Liang, C., Huang, W. C., Chan, W. L., et al. (2011). miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 39, D163–D169. doi: 10.1093/nar/gkq1107
Huikuri, H. V. (2008). Heart rate dynamics as a marker of vulnerability to atrial fibrillation. J. Cardiovasc. Electrophysiol. 19, 913–914. doi: 10.1111/j.1540-8167.2008.01197.x
Jalife, J. (2011). Deja vu in the theories of atrial fibrillation dynamics. Cardiovasc. Res. 89, 766–775. doi: 10.1093/cvr/cvq364
Jayaswal, V., Lutherborrow, M., Ma, D. D., and Yang, Y. H. (2011). Identification of microRNA-mRNA modules using microarray data. BMC Genomics 12:138. doi: 10.1186/1471-2164-12-138
Kallen, A. N., Ma, J., and Huang, Y. (2012). Does Lin28 antagonize miRNA-mediated repression by displacing miRISC from target mRNAs? Front. Genet. 3:240. doi: 10.3389/fgene.2012.00240
Kapur, S., and Macrae, C. A. (2013). The developmental basis of adult arrhythmia: atrial fibrillation as a paradigm. Front. Physiol. 4:221. doi: 10.3389/fphys.2013.00221
Karakikes, I., Chaanine, A. H., Kang, S., Mukete, B. N., Jeong, D., Zhang, S., et al. (2013). Therapeutic cardiac-targeted delivery of miR-1 reverses pressure overload-induced cardiac hypertrophy and attenuates pathological remodeling. J. Am. Heart Assoc. 2:e000078. doi: 10.1161/JAHA.113.000078
Kim, G. H. (2013). MicroRNA regulation of cardiac conduction and arrhythmias. Transl. Res. 161, 381–392. doi: 10.1016/j.trsl.2012.12.004
Kochegarov, A., Moses, A., Lian, W., Meyer, J., Hanna, M. C., and Lemanski, L. F. (2013). A new unique form of microRNA from human heart, microRNA-499c, promotes myofibril formation and rescues cardiac development in mutant axolotl embryos. J. Biomed. Sci. 20:20. doi: 10.1186/1423-0127-20-20
Kushnir, A., and Marks, A. R. (2010). The ryanodine receptor in cardiac physiology and disease. Adv. Pharmacol. 59, 1–30. doi: 10.1016/S1054-3589(10)59001-X
Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. doi: 10.1126/science.1064921
Latronico, M. V., and Condorelli, G. (2008). On the road to the definition of the cardiac miRNome in human disease states. J. Mol. Cell. Cardiol. 45, 162–164. doi: 10.1016/j.yjmcc.2008.05.018
Latronico, M. V., and Condorelli, G. (2009). RNA silencing: small RNA-mediated posttranscriptional regulation of mRNA and the implications for heart electropathophysiology. J. Cardiovasc. Electrophysiol. 20, 230–237. doi: 10.1111/j.1540-8167.2008.01357.x
Latronico, M. V., and Condorelli, G. (2013). MicroRNA-dependent control of the cardiac fibroblast secretome. Circ. Res. 113, 1099–1101. doi: 10.1161/CIRCRESAHA.113.302566
Lee, G., Kumar, S., Teh, A., Madry, A., Spence, S., Larobina, M., et al. (2013). Epicardial wave mapping in human long-lasting persistent atrial fibrillation: transient rotational circuits, complex wavefronts, and disorganized activity. Eur. Heart J. 35, 86–97. doi: 10.1093/eurheartj/eht267
Leucci, E., Patella, F., Waage, J., Holmstrom, K., Lindow, M., Porse, B., et al. (2013). microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 3:2535. doi: 10.1038/srep02535
Lewis, B. P., Burge, C. B., and Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. doi: 10.1016/j.cell.2004.12.035
Liang, Y., Ridzon, D., Wong, L., and Chen, C. (2007). Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8:166. doi: 10.1186/1471-2164-8-166
Lindgren, K. S., Pekka Raatikainen, M. J., Juhani Airaksinen, K. E., and Huikuri, H. V. (2003). Relationship between the frequency of paroxysmal episodes of atrial fibrillation and pulmonary venous flow pattern. Europace 5, 17–23. doi: 10.1053/eupc.2002.0285
Ling, T. Y., Wang, X. L., Chai, Q., Lau, T. W., Koestler, C. M., Park, S. J., et al. (2013). Regulation of the SK3 channel by microRNA-499–potential role in atrial fibrillation. Heart Rhythm 10, 1001–1009. doi: 10.1016/j.hrthm.2013.03.005
Liu, X., Jin, D. Y., McManus, M. T., and Mourelatos, Z. (2012). Precursor microRNA-programmed silencing complex assembly pathways in mammals. Mol. Cell 46, 507–517. doi: 10.1016/j.molcel.2012.03.010
Lu, Y., Zhang, Y., Wang, N., Pan, Z., Gao, X., Zhang, F., et al. (2010). MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation 122, 2378–2387. doi: 10.1161/CIRCULATIONAHA.110.958967
Lund, E., Guttinger, S., Calado, A., Dahlberg, J. E., and Kutay, U. (2004). Nuclear export of microRNA precursors. Science 303, 95–98. doi: 10.1126/science.1090599
Luo, X., Pan, Z., Shan, H., Xiao, J., Sun, X., Wang, N., et al. (2013). MicroRNA-26 governs profibrillatory inward-rectifier potassium current changes in atrial fibrillation. J. Clin. Invest. 123, 1939–1951. doi: 10.1172/JCI62185
Mahida, S. (2013). Transcription factors and atrial fibrillation. Cardiovasc. Res. doi: 10.1093/cvr/cvt261. [Epub ahead of print].
Marasovic, M., Zocco, M., and Halic, M. (2013). Argonaute and triman generate dicer-independent priRNAs and mature siRNAs to initiate heterochromatin formation. Mol. Cell. 52, 173–183. doi: 10.1016/j.molcel.2013.08.046
Meijer, H. A., Kong, Y. W., Lu, W. T., Wilczynska, A., Spriggs, R. V., Robinson, S. W., et al. (2013). Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 340, 82–85. doi: 10.1126/science.1231197
Menezes, A. R., Lavie, C. J., Dinicolantonio, J. J., O'Keefe, J., Morin, D. P., Khatib, S., et al. (2013). Atrial fibrillation in the 21st century: a current understanding of risk factors and primary prevention strategies. Mayo Clin. Proc. 88, 394–409. doi: 10.1016/j.mayocp.2013.01.022
Musa, H., Carlton, L., Klos, M., Vikstrom, K., Anumonwo, J., Jalife, J., et al. (2013). Arrhythmogenesis in a novel murine model with KCNJ2 mutation of familial atrial fibrillation. Heart Rhythm 10, 1749. doi: 10.1016/j.hrthm.2013.09.077
Nguyen, T. P., Qu, Z., and Weiss, J. N. (2013). Cardiac fibrosis and arrhythmogenesis: the road to repair is paved with perils. J. Mol. Cell. Cardiol. doi: 10.1016/j.yjmcc.2013.10.018. [Epub ahead of print].
Nivala, M., Ko, C. Y., Nivala, M., Weiss, J. N., and Qu, Z. (2012). Criticality in intracellular calcium signaling in cardiac myocytes. Biophys. J. 102, 2433–2442. doi: 10.1016/j.bpj.2012.05.001
Papait, R., Kunderfranco, P., Stirparo, G. G., Latronico, M. V., and Condorelli, G. (2013). Long noncoding RNA: a new player of heart failure? J. Cardiovasc. Transl. Res. 6, 876–883. doi: 10.1007/s12265-013-9488-6
Perino, A., Ghigo, A., Ferrero, E., Morello, F., Santulli, G., Baillie, G. S., et al. (2011). Integrating cardiac PIP3 and cAMP signaling through a PKA anchoring function of p110gamma. Mol. Cell 42, 84–95. doi: 10.1016/j.molcel.2011.01.030
Pfaff, J., Hennig, J., Herzog, F., Aebersold, R., Sattler, M., Niessing, D., et al. (2013). Structural features of Argonaute-GW182 protein interactions. Proc. Natl. Acad. Sci. U.S.A. 110, E3770–E3779. doi: 10.1073/pnas.1308510110
Qiao, A., Khechaduri, A., Kannan Mutharasan, R., Wu, R., Nagpal, V., and Ardehali, H. (2013). MicroRNA-210 decreases heme levels by targeting ferrochelatase in cardiomyocytes. J. Am. Heart Assoc. 2:e000121. doi: 10.1161/JAHA.113.000121
Quiat, D., and Olson, E. N. (2013). MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J. Clin. Invest. 123, 11–18. doi: 10.1172/JCI62876
Riley, A. B., and Manning, W. J. (2011). Atrial fibrillation: an epidemic in the elderly. Expert Rev. Cardiovasc. Ther. 9, 1081–1090. doi: 10.1586/erc.11.107
Roth, B. M., Ishimaru, D., and Hennig, M. (2013). The core microprocessor component DiGeorge syndrome critical region 8 (DGCR8) is a nonspecific RNA-binding protein. J. Biol. Chem. 288, 26785–26799. doi: 10.1074/jbc.M112.446880
Ruby, J. G., Jan, C. H., and Bartel, D. P. (2007). Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83–86. doi: 10.1038/nature05983
Ruvkun, G., Ambros, V., Coulson, A., Waterston, R., Sulston, J., and Horvitz, H. R. (1989). Molecular genetics of the Caenorhabditis elegans heterochronic gene lin-14. Genetics 121, 501–516.
Ruvkun, G., and Giusto, J. (1989). The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature 338, 313–319. doi: 10.1038/338313a0
Ruwald, A. C., Bloch Thomsen, P. E., Gang, U., Jorgensen, R. M., Huikuri, H. V., and Jons, C. (2013). New-onset atrial fibrillation predicts malignant arrhythmias in post-myocardial infarction patients-A Cardiac Arrhythmias and RIsk Stratification after acute Myocardial infarction (CARISMA) substudy. Am. Heart J. 166, 855–863.e853. doi: 10.1016/j.ahj.2013.08.017
Santulli, G. (2012a). Coronary heart disease risk factors and mortality. JAMA 307, 1137. doi: 10.1001/jama.2012.323
Santulli, G. (2012b). Thrombolysis outcomes in acute ischemic stroke patients with prior stroke and diabetes mellitus. Neurology 78, 840. doi: 10.1212/WNL.0b013e31824de51b
Santulli, G. (2013). Epidemiology of cardiovascular disease in the 21st century: updated numbers and updated facts. J. Cardiovasc. Dis. 1, 1–2.
Santulli, G., Ciccarelli, M., Trimarco, B., and Iaccarino, G. (2013). Physical activity ameliorates cardiovascular health in elderly subjects: the functional role of the beta adrenergic system. Front. Physiol. 4:209. doi: 10.3389/fphys.2013.00209
Santulli, G., and D'Ascia, C. (2012). Atrial remodelling in echocardiographic super-responders to cardiac resynchronization therapy. Heart 98, 517. doi: 10.1136/heartjnl-2012-301731
Santulli, G., D'Ascia, S., and D'Ascia, C. (2012a). Regarding the impact of left ventricular size on response to cardiac resynchronization therapy. Am. Heart J. 163, e11. doi: 10.1016/j.ahj.2012.01.001
Santulli, G., D'Ascia, S. L., and D'Ascia, C. (2012b). Development of atrial fibrillation in recipients of cardiac resynchronization therapy: the role of atrial reverse remodelling. Can. J. Cardiol. 28, 245.e217. doi: 10.1016/j.cjca.2011.11.001
Santulli, G., D'Ascia, S., Marino, V., and D'Ascia, C. (2012c). Atrial function in patients undergoing CRT. JACC Cardiovasc. Imaging 5, 124–125. doi: 10.1016/j.jcmg.2011.11.002
Santulli, G., and Iaccarino, G. (2013). Pinpointing beta adrenergic receptor in ageing pathophysiology: victim or executioner? Evidence from crime scenes. Immun. Ageing 10:10. doi: 10.1186/1742-4933-10-10
Santulli, G., and Totary-Jain, H. (2013). Tailoring mTOR-based therapy: molecular evidence and clinical challenges. Pharmacogenomics 14, 1517–1526. doi: 10.2217/pgs.13.143
Saxena, A., and Tabin, C. J. (2010). miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proc. Natl. Acad. Sci. U.S.A. 107, 87–91. doi: 10.1073/pnas.0912870107
Shah, A. J., Hocini, M., Xhaet, O., Pascale, P., Roten, L., Wilton, S. B., et al. (2013). Validation of novel 3-dimensional electrocardiographic mapping of atrial tachycardias by invasive mapping and ablation: a multicenter study. J. Am. Coll. Cardiol. 62, 889–897. doi: 10.1016/j.jacc.2013.03.082
Shan, H., Li, X., Pan, Z., Zhang, L., Cai, B., Zhang, Y., et al. (2009). Tanshinone IIA protects against sudden cardiac death induced by lethal arrhythmias via repression of microRNA-1. Br. J. Pharmacol. 158, 1227–1235. doi: 10.1111/j.1476-5381.2009.00377.x
Shen, J., Xia, W., Khotskaya, Y. B., Huo, L., Nakanishi, K., Lim, S. O., et al. (2013). EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 497, 383–387. doi: 10.1038/nature12080
Shiohama, A., Sasaki, T., Noda, S., Minoshima, S., and Shimizu, N. (2003). Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem. Biophys. Res. Commun. 304, 184–190. doi: 10.1016/S0006-291X(03)00554-0
Siomi, H., and Siomi, M. C. (2009). On the road to reading the RNA-interference code. Nature 457, 396–404. doi: 10.1038/nature07754
Siwik, D. A., and Colucci, W. S. (2004). Regulation of matrix metalloproteinases by cytokines and reactive oxygen/nitrogen species in the myocardium. Heart Fail. Rev. 9, 43–51. doi: 10.1023/B:HREV.0000011393.40674.13
Slagsvold, K. H., Rognmo, O., Hoydal, M. A., Wisloff, U., and Wahba, A. (2013). Remote ischemic preconditioning preserves mitochondrial function and influences myocardial microRNA expression in atrial myocardium during coronary bypass surgery. Circ. Res. doi: 10.1161/CIRCRESAHA.114.302751. [Epub ahead of print].
Song, S. J., Poliseno, L., Song, M. S., Ala, U., Webster, K., Ng, C., et al. (2013). MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell 154, 311–324. doi: 10.1016/j.cell.2013.06.026
Straten, P., and Andersen, M. H. (2010). The anti-apoptotic members of the Bcl-2 family are attractive tumor-associated antigens. Oncotarget 1, 239–245.
Ter Keurs, H. E., and Boyden, P. A. (2007). Calcium and arrhythmogenesis. Physiol. Rev. 87, 457–506. doi: 10.1152/physrev.00011.2006
Thomas, I. C., and Sorrentino, M. J. (2014). Bleeding risk prediction models in atrial fibrillation. Curr. Cardiol. Rep. 16, 432. doi: 10.1007/s11886-013-0432-9
Thum, T., Gross, C., Fiedler, J., Fischer, T., Kissler, S., Bussen, M., et al. (2008). MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456, 980–984. doi: 10.1038/nature07511
Tritsch, E., Mallat, Y., Lefebvre, F., Diguet, N., Escoubet, B., Blanc, J., et al. (2013). An SRF/miR-1 axis regulates NCX1 and Annexin A5 protein levels in the normal and failing heart. Cardiovasc. Res. 98, 372–380. doi: 10.1093/cvr/cvt042
Van Rooij, E., and Olson, E. N. (2012). MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat. Rev. Drug Discov. 11, 860–872. doi: 10.1038/nrd3864
Vikman, S., Lindgren, K., Makikallio, T. H., Yli-Mayry, S., Airaksinen, K. E., and Huikuri, H. V. (2005). Heart rate turbulence after atrial premature beats before spontaneous onset of atrial fibrillation. J. Am. Coll. Cardiol. 45, 278–284. doi: 10.1016/j.jacc.2004.10.033
Von Brandenstein, M., Richter, C., and Fries, J. W. (2012). MicroRNAs: small but amazing, and their association with endothelin. Life Sci. 91, 475–489. doi: 10.1016/j.lfs.2012.06.025
Wang, Z., Lu, Y., and Yang, B. (2011). MicroRNAs and atrial fibrillation: new fundamentals. Cardiovasc. Res. 89, 710–721. doi: 10.1093/cvr/cvq350
Witkos, T. M., Koscianska, E., and Krzyzosiak, W. J. (2011). Practical aspects of microRNA target prediction. Curr. Mol. Med. 11, 93–109. doi: 10.2174/156652411794859250
Wu, P. H., Isaji, M., and Carthew, R. W. (2013). Functionally diverse microRNA effector complexes are regulated by extracellular signaling. Mol. Cell 52, 113–123. doi: 10.1016/j.molcel.2013.08.023
Xie, W., Santulli, G., Guo, X., Gao, M., Chen, B. X., and Marks, A. R. (2013). Imaging atrial arrhythmic intracellular calcium in intact heart. J. Mol. Cell. Cardiol. 64, 120–123. doi: 10.1016/j.yjmcc.2013.09.003
Yang, B., Lin, H., Xiao, J., Lu, Y., Luo, X., Li, B., et al. (2007). The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 13, 486–491. doi: 10.1038/nm1569
Yates, L. A., Norbury, C. J., and Gilbert, R. J. (2013). The long and short of microRNA. Cell 153, 516–519. doi: 10.1016/j.cell.2013.04.003
Zacharewicz, E., Lamon, S., and Russell, A. P. (2013). MicroRNAs in skeletal muscle and their regulation with exercise, ageing, and disease. Front. Physiol. 4:266. doi: 10.3389/fphys.2013.00266
Zemlin, C. W., and Pertsov, A. M. (2007). Bradycardic onset of spiral wave re-entry: structural substrates. Europace 9(Suppl. 6), vi59–vi63. doi: 10.1093/europace/eum205
Zeng, Y., Yi, R., and Cullen, B. R. (2005). Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 24, 138–148. doi: 10.1038/sj.emboj.7600491
Zhang, R., Niu, H., Ban, T., Xu, L., Li, Y., Wang, N., et al. (2013a). Elevated plasma microRNA-1 predicts heart failure after acute myocardial infarction. Int. J. Cardiol. 166, 259–260. doi: 10.1016/j.ijcard.2012.09.108
Zhang, Y., Sun, L., Zhang, Y., Liang, H., Li, X., Cai, R., et al. (2013b). Overexpression of microRNA-1 causes atrioventricular block in rodents. Int. J. Biol. Sci. 9, 455–462. doi: 10.7150/ijbs.4630
Zhang, Y., Huang, Z. J., Dai, D. Z., Feng, Y., Na, T., Tang, X. Y., et al. (2008). Downregulated FKBP12.6 expression and upregulated endothelin signaling contribute to elevated diastolic calcium and arrhythmogenesis in rat cardiomyopathy produced by l-thyroxin. Int. J. Cardiol. 130, 463–471. doi: 10.1016/j.ijcard.2008.05.018
Keywords: atrial fibrillation, microRNA (miRNA), electrical remodeling, apoptosis, structural remodeling, electrophysiology, fibrosis
Citation: Santulli G, Iaccarino G, De Luca N, Trimarco B and Condorelli G (2014) Atrial fibrillation and microRNAs. Front. Physiol. 5:15. doi: 10.3389/fphys.2014.00015
Received: 03 December 2013; Accepted: 08 January 2014;
Published online: 24 January 2014.
Edited by:
Zhilin Qu, University of California, Los Angeles, USAReviewed by:
David R. Van Wagoner, Cleveland Clinic Lerner College of Medicine of Case Western Reserve University, USACopyright © 2014 Santulli, Iaccarino, De Luca, Trimarco and Condorelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gaetano Santulli, Columbia University Medical Center, College of Physicians & Surgeons, New York Presbyterian Hospital - Manhattan, 1150 Saint Nicholas Avenue, New York, 10032 NY, USA e-mail:Z3MyNjIwQGNvbHVtYmlhLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.