- 1Faculty of Information Engineering and Automation, Kunming University of Science and Technology, Kunming, China
- 2Yunnan Key Laboratory of Artificial Intelligence, Kunming, China
Incorporating medical text annotations compensates for the quality deficiencies of image data, effectively overcoming the limitations of medical image segmentation. Many existing approaches achieve high-quality segmentation results by integrating text into the image modality. However, these approaches require matched image-text pairs during inference to maintain their performance, and the absence of corresponding text annotations results in degraded model performance. Additionally, these methods often assume that the input text annotations are ideal, overlooking the impact of poor-quality text on model performance in practical scenarios. To address these issues, we propose a novel generative medical image segmentation model, Cap2Seg (Leveraging Caption Generation for Enhanced Segmentation of COVID-19 Medical Images). Cap2Seg not only segments lesion areas but also generates related medical text descriptions, guiding the segmentation process. This design enables the model to perform optimal segmentation without requiring text input during inference. To mitigate the impact of inaccurate text on model performance, we consider the consistency between generated textual features and visual features and introduce the Scale-aware Textual Attention Module (SATaM), which reduces the model’s dependency on irrelevant or misleading text information. Subsequently, we design a word-pixel fusion decoding mechanism that effectively integrates textual features into visual features, ensuring that the text information effectively supplements and enhances the image segmentation task. Extensive experiments on two public datasets, MosMedData+ and QaTa-COV19, demonstrate that our method outperforms the current state-of-the-art models under the same conditions. Additionally, ablation studies have been conducted to demonstrate the effectiveness of each proposed module. The code is available at https://github.com/AllenZzzzzzzz/Cap2Seg.
1 Introduction
COVID-19 has rapidly become a global epidemic since the early 2020s Benvenuto et al. [1]. Within 6 months of the outbreak, over 1.5 million cases of COVID-19 had been reported worldwide, with more than 92,000 deaths Organization et al. [2]. Clinically, reverse transcription polymerase chain reaction (RT-PCR) is the standard method for diagnosing COVID-19. Still, it has drawbacks, such as a high false-negative rate Chan et al. [3] and an inability to provide information about the patient’s condition. Computed tomography (CT), due to its convenience and ability to display the three-dimensional structure of the lungs, has been considered an essential complement to RT-PCR testing for the early diagnosis of COVID-19, especially in the follow-up assessment and evaluation of disease progression Raoof and Volpi [4]. Consequently, the automatic segmentation of lung infections in CT scans using computer vision techniques has garnered widespread attention from clinical researchers Shi et al. [5].
With the advent of deep learning, medical image segmentation has become a hot topic in computer vision researchZhu et al. [6]. This task focuses on identifying pixel features of anatomical or pathological regions from the background of medical images and applying these features to the image segmentation process Liu et al. [7]; Zhu et al. [8]. Consequently, many deep learning systems have been proposed for COVID-19 infection detection Ronneberger et al. [9]; Zhou et al. [10], achieving state-of-the-art performance Wang et al. [11]; Fan et al. [12]. Figure 1A illustrates that the encoder-decoder architecture is a more commonly used approach. In this architecture, the encoder is responsible for extracting image features, while the decoder restores these features to the original image size and produces the final segmentation results.
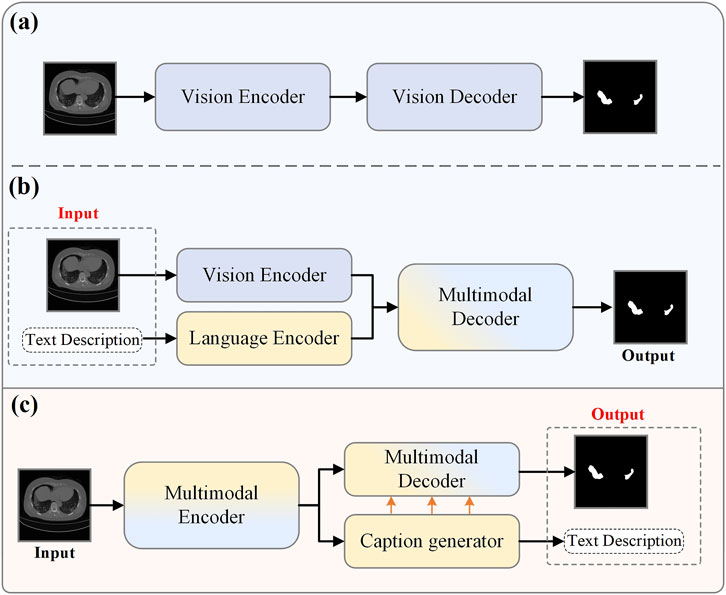
Figure 1. Current medical image segmentation models. (A) Traditional medical image segmentation. (B) Vision-Language multimodal medical image segmentation. (C) Our proposed model in this paper.
However, the aforementioned traditional pixel-wise supervised automatic segmentation methods based on deep learning neglect the semantic information in medical reports. Medical reports often contain information about the lesion areas, such as size and quantity, which can complement image data and provide additional supervisory signals for diagnosis Monajatipoor et al. [13]. Vision-language models have been extensively researched recently and achieved remarkable results in cross-modal tasks. Consequently, many studies have begun exploring combining textual information from medical reports with the segmentation process to improve segmentation accuracy Li et al. [14]; Chen et al. [15]; Huemann et al. [16]; Tomar et al. [17]. As shown in Figure 1B, a typical multimodal medical image segmentation research workflow first relies on two specially designed encoders to extract visual and language features separately. These extracted features are then integrated using a specific fusion strategy and processed through a network decoder intended explicitly for multimodality to obtain the segmentation results.
Although vision-language models have shown promising performance in the segmentation field, they face two significant challenges in practical applications within the medical domain. Firstly, these methods Li et al. [14]; Huemann et al. [16]; Wen et al. [18], trained using image-text pairs, often experience performance degradation during inference if the text is unavailable. This creates a dependency on image-text pairs. In real-world scenarios, this form of inference frequently contradicts the process of the model independently assisting clinical diagnosis: it is usually challenging to obtain textual information from medical reports before the doctor completes the diagnosis Li et al. [19]; Yu et al. [20]. This means that if the model relies on these finalized reports to enhance its performance, it is essentially duplicating the diagnosis already made by the doctor rather than providing an independent auxiliary diagnosis. This dependency significantly diminishes the model’s auxiliary value and deviates from its original purpose of independently aiding medical diagnosis. Secondly, existing vision-language models Wen et al. [21] often focus solely on effectively combining text and visual modalities, neglecting text accuracy’s impact on model performance. Inaccurate text can mislead the model and negatively affect its performance. In practical applications, medical reports may contain errors due to various factors. Effectively handling this imperfect textual information and preventing it from impairing model performance is also a significant challenge.
In summary, there are two main challenges: 1. How to address the model’s dependency on image-text pairs during the inference stage; 2. How to mitigate the impact of text accuracy on model performance. To solve the first challenge, we propose the Cap2Seg model, as shown in Figure 1C. This model combines the image captioning task and requires only a lesion image as input to simultaneously output segmentation results and corresponding text descriptions, successfully eliminating the model’s dependency on image-text pair data. Considering that some generated texts may sometimes deviate from actual medical reports and potentially affect segmentation performance, we designed a Scale-aware Textual Attention Module (SATaM) and a semantic consistency loss (SCloss) function to address the second challenge. These two mechanisms work together to ensure that the attention of the generated language features is focused on the lesion areas, effectively avoiding misleading the model with biased generated texts. Additionally, we introduced a Language-Aware Visual Decoder (LAVD), which effectively integrates multi-scale language features with visual features and decodes them, significantly improving the overall quality of the segmentation results. Our contributions are summarized as follows.
(1) The proposed Cap2Seg combines caption generation with lesion area segmentation, generating related medical text descriptions simultaneously. Leveraging the generated textual information to supplement the segmentation task effectively improves the accuracy of medical image segmentation. This eliminates the model’s dependency on image-text pairs and provides additional references for clinical diagnosis.
(2) The SATaM optimizes the quality of language features and enhances the model’s ability to handle textual biases, thereby improving overall robustness. Concurrently, the Language-Aware Visual Decoder (LAVD) effectively integrates visual and linguistic features, significantly improving segmentation quality.
(3) Experiments conducted on two publicly available COVID-19 datasets demonstrate that our proposed method outperforms most state-of-the-art models in segmentation performance.
The remainder of this paper is organized as follows: Section 2 provides a comprehensive review and summary of previous research related to our work. Section 3 describes the architecture of the proposed network. In Section 4, we present and analyze the experimental results. Finally, in Section 5, we conclude our work.
2 Related works
This section reviews and summarizes previous relevant studies related to our work, focusing on Visual-Language image segmentation, image captioning, and multi-task learning.
2.1 Visual-language image segmentation
In recent years, multimodal segmentation techniques that combine visual and language modalities have garnered extensive attention. Hu et al. Hu et al. [22] pioneered using textual descriptions to assist image segmentation, sparking further research into effectively integrating visual and textual information to enhance segmentation results. Broadly, this task can be categorized into two types: referring image segmentation in natural scenes and image segmentation in medical contexts.
2.1.1 Referring image segmentation
In applications within natural settings, early studies Liu et al. [23]; Li et al. [24]; Shi et al. [25]; Ye et al. [26] focused on developing more effective techniques for extracting and merging visual and linguistic features. Liu et al. Liu et al. [23] introduced a multimodal Long Short-Term Memory network specifically designed to process and fuse multimodal features of each word. Shi et al. Shi et al. [27] proposed a keyword-aware network that, while extracting text features, assigns higher weights to keywords, thereby improving the model’s ability to recognize text-indicated objects. The introduction of attention mechanisms paved new ways for effective cross-modal feature fusion. Ye et al. Ye et al. [26] employed non-local blocks Wang et al. [28] to design a cross-modal self-attention module for integrating features across modalities. Similarly, other studies Chen et al. [29]; Hu et al. [30]; Shi et al. [27]; Chen et al. [31] utilized various attention mechanisms to process and integrate cross-modal features. Unlike these later fusion approaches, LAVT Yang et al. [32] achieved an early fusion of linguistic and visual features at the intermediate layers of a Transformer network, enhancing cross-modal alignment and the model’s integration of visual and linguistic information. With the significant rise of CLIP Radford et al. [33] in the multimodal field, some research began to explore using contrastive learning to represent cross-modal data, such as LSeg Li et al. [34] and GroupViT Xu et al. [35]. These studies leveraged the advanced representational capabilities of CLIP in multimodal scenarios, effectively enhancing image segmentation efficiency and accuracy and demonstrating exceptional capabilities in zero-shot inference scenarios. Further research has focused on the role of text structure in enhancing multimodal information processing. Yu et al. Yu et al. [36] and Huang et al. Huang et al. [37] utilized sentence structure knowledge to capture concepts within multimodal features, such as categories, attributes, and relationships. Hui et al. Hui et al. [38] used syntactic structures between words to guide multimodal context aggregation. Ding et al. Ding et al. [39] introduced a dynamic query generation module capable of dynamically producing multiple queries based on the input text to accommodate diverse linguistic scenarios, making multimodal information fusion more targeted and specific.
2.1.2 Medical image segmentation
In the medical field, Li et al. Li et al. [14] proposed the LViT model, a hybrid of CNNs and Transformers, which incrementally integrates medical text annotations into the image segmentation process to compensate for the quality deficiencies of image data. Unlike LViT, Bi-VLGM Chen et al. [15] emphasizes maintaining consistency within modal features and uses a visual-language graph matching module to handle the category-severity relationships between visual and text features, enabling the segmentation model to learn valuable representations selectively. Other studies Huang et al. [40]; Zhang et al. [41]; Huemann et al. [16]; Dai et al. [42] have used more flexible medical reports for segmentation. ConTEXTualNet Huemann et al. [16] employs attention mechanisms to decode image features based on text in medical reports, guiding the model to focus on text-related image pixels. Some methods Tomar et al. [17], even without available medical reports or texts, utilize auxiliary classification tasks to embed textual attributes (size and number) during encoding. This approach enables the network to adapt to various sizes and numbers of polyp cases, thereby enhancing segmentation performance. However, existing state-of-the-art methods Li et al. [14]; Chen et al. [15] still rely on matched medical text and image data during the inference stage to achieve optimal performance. Their performance may suffer when only image input is available without corresponding text. In contrast, the Cap2Seg model proposed in this study requires only one image to achieve optimal performance during inference.
2.2 Image captioning
Image captioning, which aims to produce natural language descriptions based on static visual content Vinyals et al. [43]; Ghandi et al. [44], represents a challenging cross-modal translation task Zhang et al. [45]; Yu et al. [46]. This task demonstrates particular application value in the medical field Li et al. [47]; Hou et al. [48]; Wang et al. [49]. For instance, Li et al. Li et al. [47] developed a Knowledge-driven Encoding, Retrieval, and Paraphrasing (KERP) model to improve medical image descriptions. Our research focuses not on designing a new captioning model per se but on employing image caption generation as an auxiliary module. To the best of our knowledge, this study is the first attempt to explore caption generation in medical image segmentation.
2.3 Multi-task learning
Multi-task learning aims to enhance the performance of individual or multiple tasks by jointly training related tasks, utilizing the correlations and shared information between them for mutual benefit. For example, Wu et al. Wu et al. [50] introduced the CGG framework, which combines image caption generation and referring image segmentation tasks. This framework employs caption generation loss to supervise the model, improving image segmentation quality. Similarly, Sun et al.‘s PFOS model Sun et al. [51], which integrates the tasks of Referring Expression Comprehension and Generation, leverages cross-attention and multimodal fusion mechanisms to boost overall model performance significantly. Moreover, Zhang et al. Zhang et al. [52] demonstrated significant performance improvements in medical image analysis by combining gastric cancer segmentation with lymph node classification tasks, effectively managing the inter-task relationships and heterogeneity through multi-scale features and refined attention mechanisms. Following this concept, Cap2Seg merges the functions of image caption generation and medical image segmentation. The goal is to utilize the generated textual annotations as supplementary information to the image modality, thereby enhancing the performance of the segmentation task.
3 Proposed method
This section elaborates on the proposed method, encompassing four components: the Multimodal Synergistic Dual-Flow Encoder (MSDFE) module, the Multimodal Semantic Enhancement and Captioning Module (MSECM), the Scale-aware Textual Attention Module (SATaM), and the Language-Aware Visual Decoder (LAVD).
3.1 Overview
The caption-driven multimodal COVID-19 segmentation framework proposed in this paper is illustrated in Figure 2A. This framework addresses two primary tasks: medical image captioning and medical image lesion segmentation. The MSDFE module initially processes the input image, mapping it into a multimodal space that combines visual and textual data, thereby providing a comprehensive set of features for both tasks. The MSECM then refines these features to enhance their relevance to each task. Concurrently, the SATaM and Semantic Consistency Loss (SCloss) are employed to apply focused attention to the textual features, thereby minimizing the model’s reliance on non-relevant or potentially misleading information and concentrating efforts on lesion areas. Finally, the LAVD integrates and upsamples the textual and visual features to produce the final segmentation results. In summary, our proposed framework leverages the synergistic effects of multitask learning to exploit the rich complementary information contained in generated text annotations, thereby enhancing the segmentation quality of COVID-19 and providing additional textual diagnostic support.
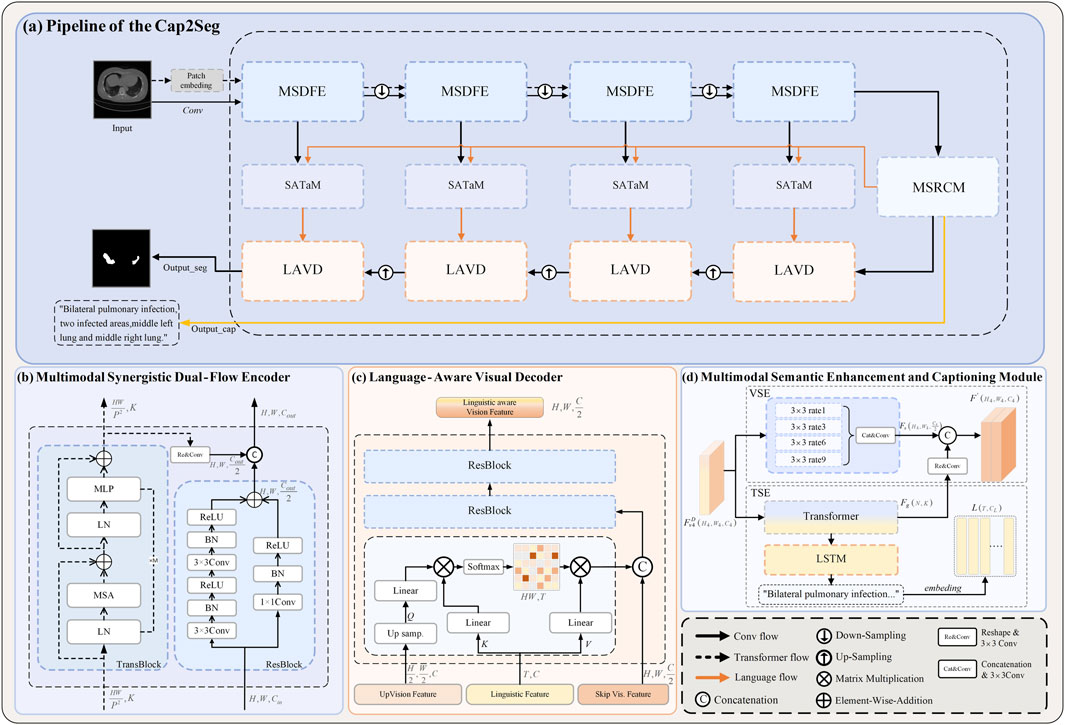
Figure 2. Overall framework of the proposed method. (A) The leading network comprises the following components: (B) The multimodal synergistic Dual-Flow Encoder (MSDFE) module, (C) the Multimodal Semantic Enhancement and Captioning Module (MSECM), the Scale-aware Textual Attention Module (SATaM), and (D) the Language-Aware Visual Decoder (LAVD).
3.2 Multimodal synergistic dual-flow encoder
Given the high variability in the shape, size, and location of COVID-19 infection-related issues, and the requirement for the Cap2Seg model to perform both image segmentation and image captioning tasks, extracting richer features from the input images is crucial. Convolutional Neural Networks (CNN) can accumulate spatial information of images, focusing on capturing local information such as the texture and contours of lesion areas. At the same time, the self-attention mechanism can explore long-range dependencies in images, focusing on capturing global information. To fully extract diverse features, this paper proposes a Multimodal Synergistic Dual-Flow Encoder (MSDFE), which combines the strengths of CNN and Transformer. As shown in Figure 2B, MSDFE consists of two parallel feature extraction branches: the first branch is the “trans flow” processed by
Specifically, The
In this context,
As for
Subsequently, the embedded feature
Herein,
Throughout the encoding process, the MSDFE module is configured with four instances. Initially, in the first two MSDFE modules, each branch functions independently, extracting features without interacting with each other. In the latter two modules, the outputs of these branches are amalgamated and then transferred to the next “conv flow” stage, facilitating collaborative learning. Specifically, in these later stages, the output from the “trans flow”
In this equation,
3.3 Multimodal semantic enhancement and captioning module
To leverage the multimodal features
We utilize atrous Chen et al. [57] convolution in the VSE to refine the multimodal features. Atrous convolution extends the receptive field by adjusting the dilation rate, allowing it to capture broader contextual information. Specifically, we use different dilation rates (1, 3, 6, 9) to ensure effective information acquisition across various scales. This multi-scale information capture enhances the specificity of visual features for segmentation tasks, providing a solid foundation for achieving accurate segmentation results. Furthermore, due to the Transformer’s strong ability to model the two modalities, we integrate the output of TSE into VSE, forming a comprehensive feature set
In these equations,
In the TSE, as illustrated in Equation 9, the Transformer module is utilized to optimize the multimodal features, with its self-attention mechanism enabling extensive context capture from within the image. This enhances feature coherence and provides a solid foundation for generating text closely related to the image. We employ a lightweight Long Short-Term Memory (LSTM) network Hochreiter and Schmidhuber [58] as the caption generator for the subsequent generation of medical image captions. This network comprises several interconnected LSTM units, enabling it to effectively process sequential data, which is crucial for generating coherent and informative medical texts. To quantitatively assess the accuracy of the generated texts, we use the cross-entropy loss function
In this formula,
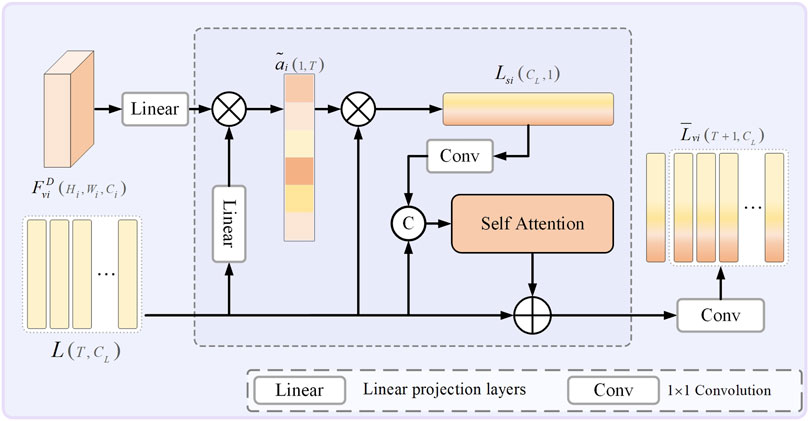
3.4 Scale-aware textual attention module
To mitigate the impact of variances between model-generated texts and labeled descriptions in a minority of samples, which may compromise the model’s segmentation performance, this research has integrated a Scale-aware Textual Attention Module (SATaM). This module exploits multimodal features
Herein,
Here,
Furthermore, before each skip connection within the model, the SATaM produces four adaptively scale-aware sentence features,
The introduction of SCloss ensures that each SATaM can effectively share insights and impose constraints on one another. This mechanism enables the linguistic features
3.5 Language-aware visual decoder
To optimize the decoding phase of segmentation, we have implemented a Language-Aware Visual Decoder (LAVD). This module is specifically designed to enable more effective integration of features, thereby facilitating the subsequent up-sampling steps. As shown in Figure 2C, the designated input features consist of the decoding features
Within this framework,
In our approach, the LAVD is set to 4, and after four iterations of up-sampling,
3.6 Overall loss functions
The overall training loss is divided into two main components: segmentation loss
The overall loss function is formulated in Equation 29:
Within this construct,
4 Experimental
This section comprehensively evaluates our Cap2Seg network using the QaTa-COV19 and MosMedData + datasets. Each experiment is meticulously described, and the results are rigorously analyzed.
4.1 Implementation details
This study’s methodology was executed on an NVIDIA RTX 4080 using PyTorch. The optimization of model parameters was carried out with an AdamW optimizer that includes a weight decay of 0.0001. Following Li et al. [14], the initial learning rates were configured at 3e-4 for the QaTa-COV19 dataset and 1e-3 for the MosMedData + dataset; due to the differing data sizes of the datasets, batch sizes were specifically configured at 4 for the QaTa-COV19 dataset and 8 for the MosMedData + dataset. The hyperparameters
Here, A and B denote the labels and segmentation predictions, respectively.
4.2 Datasets
The study utilized two primary public datasets: QaTa-COV19 Degerli et al. [61] and MosMedData + Morozov et al. [62]. The QaTa-COV19 dataset comprises 9,258 chest X-ray images of COVID-19, each with a
4.3 Results and analysis
We first validated the effectiveness of our method on the MosMedData + dataset and compared it with existing methods under three different conditions. The first condition is that no text modality is used as auxiliary input during inference, corresponding to the “w/o Text” column in the table. The second condition involves using generated medical text annotations to assist segmentation during inference, as shown in the “Generated Text” column in the table. These annotations are generated by Cap2Seg at its optimal performance and are used as inputs for other models. A detailed qualitative evaluation of these generated texts is provided in Section 4.4. The third condition is that real labeled medical text annotations assist segmentation during inference, corresponding to the “Ground Truth Text” column in the table. Since our model does not use any text input during inference and the model generates the auxiliary text, our method falls under the first two conditions. Therefore, we perform inference only under these two conditions and compare it with existing methods. We compared our method with mainstream text-guided image segmentation methods Yang et al. [32]; Li et al. [14]; Huemann et al. [16]; Tomar et al. [17] and some state-of-the-art segmentation methods Ronneberger et al. [9]; Zhou et al. [10]; Oktay et al. [63]; Katore and Thanekar [64]; Chen et al. [65]; Cao et al. [66]; Zhao et al. [67]. The corresponding comparison results are listed in Table 1, with the best results highlighted in bold.
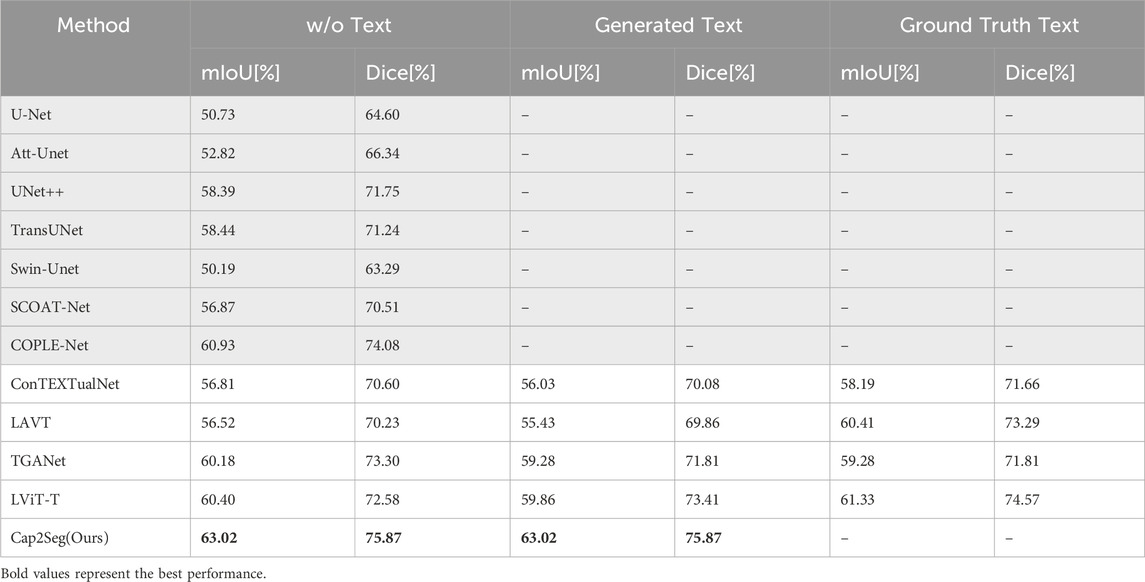
Table 1. Compares the state-of-the-art segmentation methods on the MOSMEDDATA + dataset. GRAY-SHADED methods exclude text input, while others include text input.
Our findings reveal that Cap2Seg substantially exceeded the performance of existing approaches in the three conditions outlined above. It is important to note that when using generated annotations with discrepancies from real text annotations for segmentation assistance, Li et al. [14]; Huemann et al. [16]; Tomar et al. [17] that did not account for this issue generally saw reduced performance. Nevertheless, Cap2Seg effectively addressed and mitigated this issue. Specifically, using generated medical text annotations, Cap2Seg increased the mIoU score by
In further evaluations conducted on the QaTa-COV19 dataset, the quantitative comparisons of our Cap2Seg are detailed in Table 2. Specifically, Cap2Seg achieved a mean Intersection over Union (mIoU) of
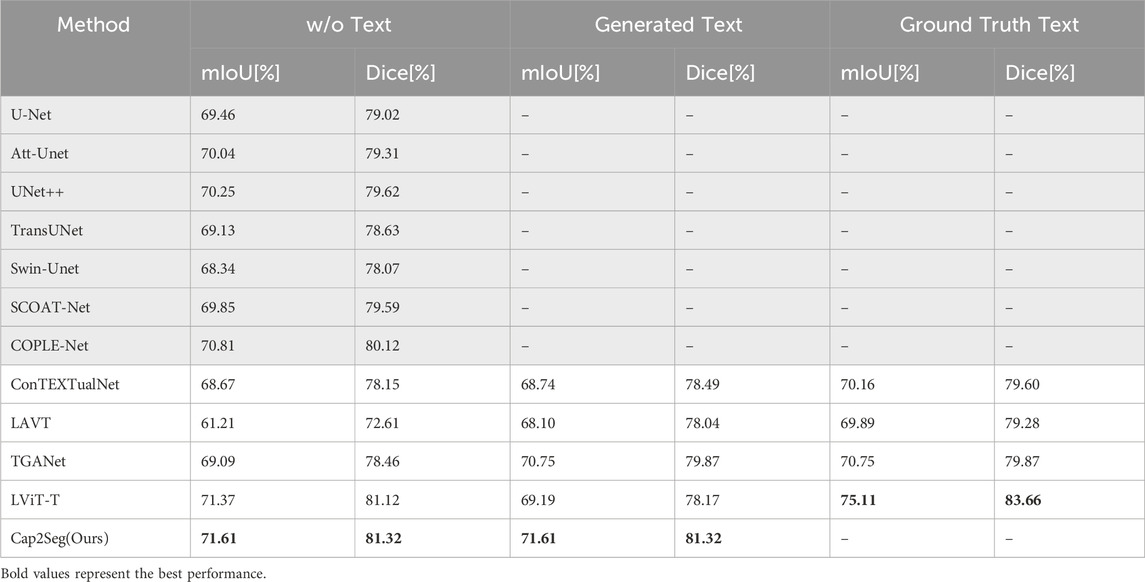
Table 2. Compares the state-of-the-art segmentation methods on the QaTa-COV19 dataset. GRAY-SHADED methods exclude text input, while others include text input.
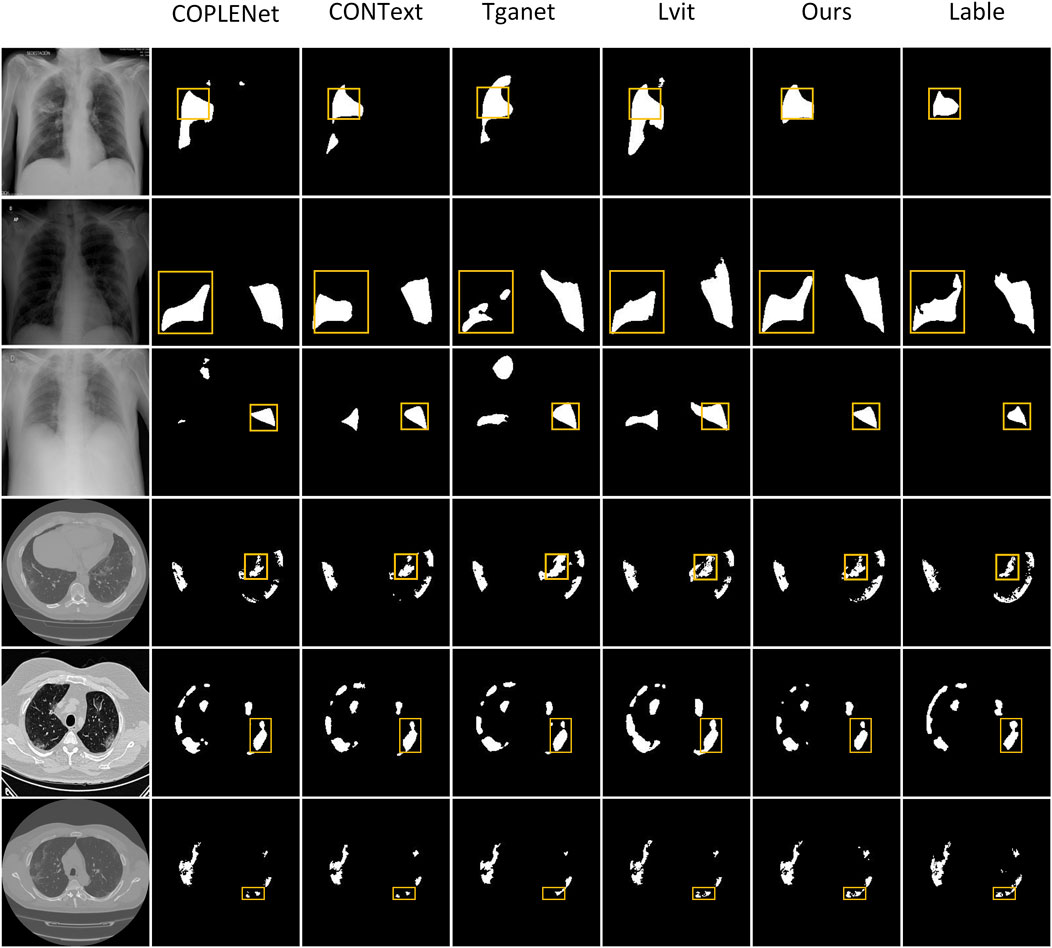
4.4 Visual comparison of segmentation results
We have conducted visual qualitative assessments of our Cap2Seg method on the MosMedData+ and QaTa-COV19 datasets, benchmarking it against current methodologies. As illustrated in Figure 4, segmentation inaccuracies are noticeable in the outputs from CopleNet Katore and Thanekar [64], ConTEXTualNet Huemann et al. [16], TGANet Tomar et al. [17], and LViT Li et al. [14] across the first, third and fourth rows, where these methods exhibit erroneous segmentation zones. In contrast, our approach effectively delineates the primary regions of lesions. Moreover, the sixth row demonstrates that while existing methods struggle with identifying lesion peripheries and finer details, Cap2Seg excels in recognizing these critical features, showcasing our network’s enhanced capability to capture lesion-specific areas accurately. The visual evidence indicates that our method achieves comparable or superior segmentation results relative to other models.
4.5 Ablation study
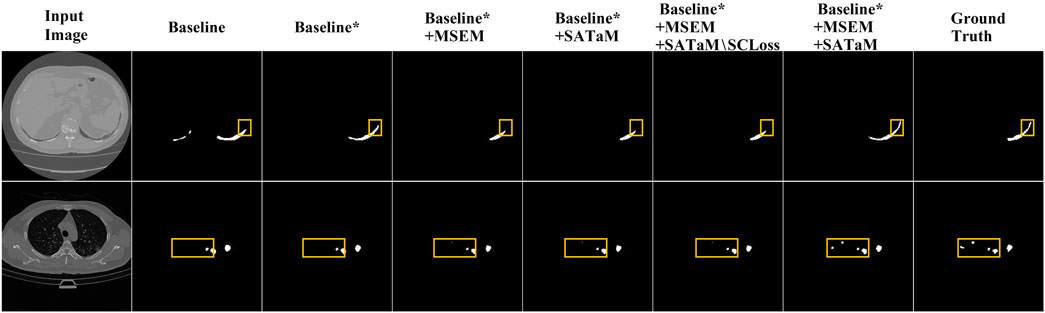
The proposed method is structured around three principal components: MSDFE, MSECM, and SATaM, with SATaM integrating SCloss, a feature proven effective in our analysis. The following ablation experiments were conducted to evaluate the efficacy of each component individually. MSDFE, MSECM, and SATaM were initially removed from our model to create a baseline. These components were then incrementally reintroduced to assess their contributions. This methodology was validated using the results from the MosMedData + dataset, summarized in Table 3, which indicated that the gradual reintroduction of these modules allowed our complete model to achieve an optimal mIoU score of
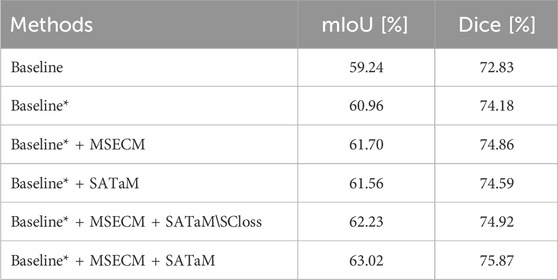
Table 3. Ablation study on the MOSMEDDATA + dataset. “Baseline” represents the utilization OF CNN as the encoder. “Baseline*” indicates the employment of the MSDFE introduced in this paper as the encoder. “SATaM
4.5.1 Effectiveness of MSDFE
In this study, the MSDFE module extracts a comprehensive set of multimodal features, effectively tackling complex cross-modal challenges. To ascertain the efficacy of this approach, we analyzed the segmentation performance differences between the “Baseline” and “Baseline*“. According to the results in Table 3, “Baseline*” reached mIoU and Dice scores of
Furthermore, the study delved into the effects of interactions between two designated sub-modules,
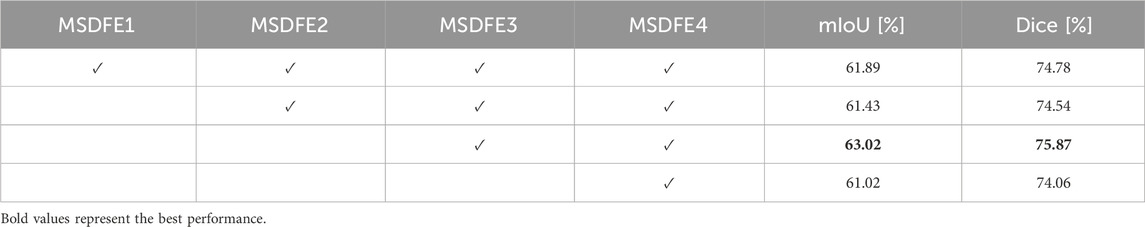
Table 4. Impact of interaction between two submodules in different MSDFE modules during the encoding stage. ‘✓’ indicates interaction within the current MSDFE.
4.5.2 Effectiveness of MSECM
The proposed MSECM module finely tunes the encoded multimodal features to enhance their task specificity. This adjustment results in two more features aligned with the intended tasks. To evaluate the effectiveness of MSECM, we analyzed the data presented in Table 3. The introduction of MSECM improved the mIoU and Dice scores of “Baseline* + MSECM” by
4.5.3 Effectiveness of SATaM
SATaM assigns attention weights related to visual information to linguistic features, thus enhancing their focus on crucial lesion areas and ensuring a tight linkage between linguistic characteristics and these areas. Consequently, this reduces the model’s attention to irrelevant or misleading textual features, enhancing its segmentation capabilities. The significant improvements in segmentation performance with the inclusion of SATaM are evident in Figure 5, validating its utility. Table 3 shows that “Baseline* + MSECM + SATaM” achieved increases of
5 Conclusion
This paper proposes Cap2Seg, a network that combines image segmentation and caption generation tasks. The introduction of the MSECM effectively coordinates both tasks, enhancing multi-task learning efficiency. The SATaM reduces the model’s reliance on irrelevant or misleading textual information, while the LAVD effectively fuses textual features with visual features. By generating text to guide the segmentation task, Cap2Seg fully leverages the potential of textual annotations, thereby improving the quality and accuracy of COVID-19 image segmentation. It eliminates the dependency on image-text pairs and provides additional textual references for clinical diagnosis. Extensive experimental results confirm the proposed method’s effectiveness and superiority over existing approaches. Ablation experiments also validate the efficacy of each core component of the proposed model. However, it is essential to acknowledge that although our method has achieved satisfactory results in image segmentation, we still face challenges in accurately generating specific keywords in a few samples, affecting the segmentation performance when dealing with complex lesion images. Additionally, due to the scarcity of paired medical image and text datasets, our method has only been validated on two COVID-19 datasets. Currently, we have not fully resolved the challenge. In future research, we will expand our study to more types of disease datasets. Meanwhile, we plan to optimize the caption generation module to improve the model’s ability to capture key lesion areas and explore more effective feature interaction and fusion strategies. These improvements and extensions will help further enhance the practicality and accuracy of our method in segmentation tasks.
Data availability statement
Publicly available datasets were analyzed in this study. The QaTa-COV19 dataset can be found at https://www.kaggle.com/datasets/aysendegerli/qatacov19-dataset, and the MosMedData+ dataset can be found at https://www.kaggle.com/datasets/maedemaftouni/covid19-ct-scan-lesion-segmentation-dataset.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
WZ: Methodology, Software, Writing–original draft, Writing–review and editing. FL: Funding acquisition, Resources, Supervision, Writing–review and editing. YD: Conceptualization, Formal Analysis, Writing–review and editing. PF: Data curation, Visualization, Writing–review and editing. ZC: Validation, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (Nos. 62362045), the Basic Research Project of Yunnan Province (Nos. 202401AT070412).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphy.2024.1439122/full#supplementary-material
References
1. Benvenuto D, Giovanetti M, Salemi M, Prosperi MCF, Flora CD, Alcantara LCJ, et al. The global spread of 2019-ncov: a molecular evolutionary analysis. Pathog Glob Health (2020) 114:64–7. doi:10.1080/20477724.2020.1725339
2. World Health Organization and others Coronavirus disease 2019 (COVID-19): situation report, 73, (2020). World Health Organization.
3. Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet (London, England) (2020) 395:514–23. doi:10.1016/s0140-6736(20)30154-9
4. Raoof S, Volpi A. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Radiology (2020). 296(1), 172–180. Radiological Society of North America.
5. Shi W, Peng X, Liu T, Cheng Z, Lu H, Yang S, et al. A deep learning-based quantitative computed tomography model for predicting the severity of covid-19: a retrospective study of 196 patients. Ann Translational Med (2020) 9:216. doi:10.21037/atm-20-2464
6. Zhu Z, He X, Qi G, Li Y, Cong B, Liu Y. Brain tumor segmentation based on the fusion of deep semantics and edge information in multimodal mri. Inf Fusion (2022) 91:376–87. doi:10.1016/j.inffus.2022.10.022
7. Liu Y, Shi Y, Mu F, Cheng J, Chen X. Glioma segmentation-oriented multi-modal mr image fusion with adversarial learning. IEEE CAA J Autom Sinica (2022) 9:1528–31. doi:10.1109/jas.2022.105770
8. Zhu Z, Wang Z, Qi G, Mazur N, Yang P, Liu Y. Brain tumor segmentation in mri with multi-modality spatial information enhancement and boundary shape correction. Pattern Recognition (2024) 153:110553. doi:10.1016/j.patcog.2024.110553
9. Ronneberger O., Fischer P., Brox T. (2015). U-net: convolutional networks for biomedical image segmentation, 234, 41. doi:10.1007/978-3-319-24574-4_28
10. Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. Unet++: a nested u-net architecture for medical image segmentation. Deep Learning in medical image Analysis and multimodal Learning for clinical decision support: 4th international workshop, dlmia 2018, and 8th international workshop, ML-CDS 2018, held in conjunction with miccai 2018, Proceedings 4 (2018) 3–11. Springer.
11. Wang G, Liu X, Li C, Xu Z, Ruan J, Zhu H, et al. A noise-robust framework for automatic segmentation of covid-19 pneumonia lesions from ct images. IEEE Trans Med Imaging (2020) 39:2653–63. doi:10.1109/tmi.2020.3000314
12. Fan D-P, Zhou T, Ji G-P, Zhou Y, Chen G, Fu H, et al. Inf-net: automatic covid-19 lung infection segmentation from ct images. IEEE Trans Med Imaging (2020) 39:2626–37. doi:10.1109/tmi.2020.2996645
13. Monajatipoor M, Rouhsedaghat M, Li LH, Chien A, Kuo C-CJ, Scalzo F, et al. Berthop: an effective vision-and-language model for chest x-ray disease diagnosis. In: 2021 IEEE/CVF International Conference on Computer Vision Workshops (ICCVW); Oct. 17 2021; China (2021). p. 3327–36.
14. Li Z, Li Y, Li Q, Zhang Y, Wang P, Guo D, et al. Lvit: language meets vision transformer in medical image segmentation. IEEE Trans Med Imaging (2022) 43:96–107. doi:10.1109/tmi.2023.3291719
15. Chen W, Liu J, Yuan Y. Bi-vlgm: Bi-level class-severity-aware vision-language graph matching for text guided medical image segmentation. (2023). ArXiv abs/2305.12231
16. Huemann Z, T Xin, Hu J, Bradshaw TJ. Contextual net: a multimodal vision-language model for segmentation of pneumothorax. Journal of Imaging Informatics in Medicine. (2024). Springer. 1–12.
17. Tomar NK, Jha D, Bagci U, Ali S. Tganet: text-guided attention for improved polyp segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention. (2022) 151–160. Springer.
18. Wen Y, Chen L, Qiao L, Deng Y, Chen H, Zhang T, et al. Let’s find fluorescein: cross-modal dual attention learning for fluorescein leakage segmentation in fundus fluorescein angiography. In: 2021 IEEE International Conference on Multimedia and Expo (ICME); July 5-9, 2021; China, 67 (2021). p. 1–6. doi:10.1109/icme51207.2021.9428108
19. Li Y, Luo L, Lin H, Chen H, Heng P-A. Dual-consistency semi-supervised learning with uncertainty quantification for covid-19 lesion segmentation from ct images. In: International Conference on Medical Image Computing and Computer-Assisted Intervention (2021). p. 199–209. doi:10.1007/978-3-030-87196-3_19
20. Yu X, Wang J, Hong Q-Q, Teku R, Wang S, Zhang Y. Transfer learning for medical images analyses: a survey. Neurocomputing (2022) 489:230–54. doi:10.1016/j.neucom.2021.08.159
21. Wen Y, Chen L, Qiao L, Deng Y, Chen H, Zhang T, et al. Fleak-seg: automated fundus fluorescein leakage segmentation via cross-modal attention learning. IEEE MultiMedia (2022) 29:114–23. doi:10.1109/mmul.2022.3142986
22. Hu R, Rohrbach M, Darrell T (2016). Segmentation from natural language expressions. ArXiv abs/1603.06180
23. Liu C, Lin ZL, Shen X, Yang J, Lu X, Yuille AL. Recurrent multimodal interaction for referring image segmentation. In: 2017 IEEE International Conference on Computer Vision (ICCV); 22-29 Oct. 2017; China (2017). p. 1280–9.
24. Li R, Li K, Kuo Y-C, Shu M, Qi X, Shen X, et al. Referring image segmentation via recurrent refinement networks. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; 18-23 June 2018; USA (2018). p. 5745–53.
25. Shi X, Chen Z, Wang H, Yeung DY, Wong W-K, chun Woo W. Convolutional lstm network: a machine learning approach for precipitation nowcasting. Neural Inf Process Syst (2015, 28).
26. Ye L, Rochan M, Liu Z, Wang Y. Cross-modal self-attention network for referring image segmentation. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 15-20 June 2019; China (2019). p. 10494–503.
27. Shi H, Li H, Meng F, Wu Q. Key-word-aware network for referring expression image segmentation. In: European Conference on Computer Vision (2018). p. 38–54. doi:10.1007/978-3-030-01231-1_3
28. Wang X, Girshick RB, Gupta AK, He K. Non-local neural networks. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; 18-23 June 2018; China (2017). p. 7794–803.
29. Chen D-J, Jia S, Lo Y-C, Chen H-T, Liu T-L. See-through-text grouping for referring image segmentation. In: 2019 IEEE/CVF international conference on computer vision (ICCV), 18-23 June 2018; China, (2019). p. 7453–62.
30. Hu Z, Feng G, Sun J, Zhang L, Lu H. Bi-directional relationship inferring network for referring image segmentation. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 13-19 June 2020; China (2020). p. 4423–32.
31. Chen D-J, Hsieh H-Y, Liu T-L. Referring image segmentation via language-driven attention. In: 2021 IEEE International Conference on Robotics and Automation (ICRA); 2021 May 30; China (2021). p. 13997–4003.
32. Yang Z, Wang J, Tang Y, Chen K, Zhao H, Torr PHS. Lavt: language-aware vision transformer for referring image segmentation. In: 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 18-24 June 2022; USA (2021). p. 18134–44.
33. Radford A, Kim JW, Hallacy C, Ramesh A, Goh G, Agarwal S, et al. Learning transferable visual models from natural language supervision. In: International Conference on Machine Learning (2021).
34. Li B, Weinberger KQ, Belongie SJ, Koltun V, Ranftl R (2022). Language-driven semantic segmentation. ArXiv abs/2201.03546
35. Xu J, Mello SD, Liu S, Byeon W, Breuel T, Kautz J, et al. Groupvit: semantic segmentation emerges from text supervision. In: 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 18-24 June 2022; USA (2022). p. 18113–23.
36. Yu L, Lin ZL, Shen X, Yang J, Lu X, Bansal M, et al. Mattnet: modular attention network for referring expression comprehension. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; 18-23 June 2018; USA (2018). p. 1307–15.
37. Huang S, Hui T, Liu S, Li G, Wei Y, Han J, et al. Referring image segmentation via cross-modal progressive comprehension. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 13-19 June 2020; Germany (2020). p. 10485–94.
38. Hui T, Liu S, Huang S, Li G, Yu S, Zhang F, et al. Linguistic structure guided context modeling for referring image segmentation. In: European Conference on Computer Vision (2020). p. 59–75. doi:10.1007/978-3-030-58607-2_4
39. Ding H, Liu C, Wang S, Jiang X. Vlt: vision-language transformer and query generation for referring segmentation. IEEE Trans Pattern Anal Machine Intelligence (2022) 45:7900–16. doi:10.1109/tpami.2022.3217852
40. Huang S-C, Shen L, Lungren MP, Yeung S. Gloria: a multimodal global-local representation learning framework for label-efficient medical image recognition. In: 2021 IEEE/CVF International Conference on Computer Vision (ICCV); 10-17 Oct. 2021; China (2021). p. 3922–31.
41. Zhang Y, Jiang H, Miura Y, Manning CD, Langlotz C. Contrastive learning of medical visual representations from paired images and text. Machine Learning for Healthcare Conference (2020), 2–25. PMLR.
42. Dai L, Fang R, Li H, Hou X, Sheng B, Wu Q, et al. Clinical report guided retinal microaneurysm detection with multi-sieving deep learning. IEEE Trans Med Imaging (2018) 37:1149–61. doi:10.1109/tmi.2018.2794988
43. Vinyals O, Toshev A, Bengio S, Erhan D. Show and tell: a neural image caption generator. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 7-12 June 2015; China (2014). p. 3156–64.
44. Ghandi T, Pourreza HR, Mahyar H. Deep learning approaches on image captioning: a review. ACM Comput Surv (2022) 56:1–39. doi:10.1145/3617592
45. Zhang W, Ying Y, Lu P, Zha H. Learning long- and short-term user literal-preference with multimodal hierarchical transformer network for personalized image caption. In: AAAI Conference on Artificial Intelligence, 34 (2020). p. 9571–8. doi:10.1609/aaai.v34i05.6503
46. Yu J, Li J, Yu Z, Huang Q. Multimodal transformer with multi-view visual representation for image captioning. IEEE Trans Circuits Syst Video Technology (2019) 30:4467–80. doi:10.1109/tcsvt.2019.2947482
47. Li Y, Liang X, Hu Z, Xing EP. Knowledge-driven encode, retrieve, paraphrase for medical image report generation. (2019) 33, 6666, 73. doi:10.1609/aaai.v33i01.33016666
48. Hou D, Zhao Z, Liu Y, Chang F, Hu S. Automatic report generation for chest x-ray images via adversarial reinforcement learning. IEEE Access (2021) 9:21236–50. doi:10.1109/access.2021.3056175
49. Wang F, Liang X, Xu L, Lin L. Unifying relational sentence generation and retrieval for medical image report composition. IEEE Trans Cybernetics (2020) 52:5015–25. doi:10.1109/tcyb.2020.3026098
50. Wu J, Li X, Ding H, Li X, Cheng G, Tong Y, et al. Betrayed by captions: joint caption grounding and generation for open vocabulary instance segmentation. In: 2023 IEEE/CVF International Conference on Computer Vision (ICCV); October 2 - 6, 2023; China (2023). p. 21881–91.
51. Sun M, Suo W, Wang P, Zhang Y, Wu Q. A proposal-free one-stage framework for referring expression comprehension and generation via dense cross-attention. IEEE Trans Multimedia (2023) 25:2446–58. doi:10.1109/tmm.2022.3147385
52. Zhang Y, Li H, Du J, Qin J, Wang T, Chen Y, et al. 3d multi-attention guided multi-task learning network for automatic gastric tumor segmentation and lymph node classification. IEEE Trans Med Imaging (2021) 40:1618–31. doi:10.1109/tmi.2021.3062902
53. Ioffe S, Szegedy L. Batch normalization: accelerating deep network training by reducing internal covariate shift. (2015). ArXiv abs/1502.03167
54. Nair V, Hinton GE. Rectified linear units improve restricted Boltzmann machines. In: International Conference on Machine Learning (2010).
55. Dosovitskiy A. An image is worth 16x16 words: transformers for image recognition at scale. (2020), ArXiv abs/2010.11929.
56. Vaswani A, Shazeer NM, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Neural Inf Process Syst (2017).
57. Chen L-C, Papandreou G, Schroff F, Adam H. Rethinking atrous convolution for semantic image segmentation. (2017), ArXiv abs/1706.05587.
58. Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput (1997) 9:1735–80. doi:10.1162/neco.1997.9.8.1735
59. Thomas E, Jogi PS, Kumar S, Horo A, Niyas S, Vinayagamani S, et al. Multi-res-attention unet: a cnn model for the segmentation of focal cortical dysplasia lesions from magnetic resonance images. IEEE J Biomed Health Inform (2020) 25(5), :1724–34.
60. Ouyang T, Yang S, Gou F, Dai Z, Wu J Rethinking u-net from an attention perspective with transformers for osteosarcoma mri image segmentation. Comput Intelligence Neurosci (2022) 2022:1–17. doi:10.1155/2022/7973404
61. Degerli A, Kiranyaz S, Chowdhury MEH, Gabbouj M Osegnet: operational segmentation network for covid-19 detection using chest x-ray images. In: 2022 IEEE International Conference on Image Processing (ICIP); 16-19 Oct. 2022; USA (2022). p. 2306–10.
62. Morozov S, Andreychenko AE, Pavlov NA, Vladzymyrskyy A, Ledikhova NV, Gombolevskiy VA, et al. Mosmeddata: chest ct scans with covid-19 related findings. (2020), ArXiv abs/2005.06465.
63. Oktay O, Schlemper J, Folgoc LL, Lee MJ, Heinrich MP, Misawa K, et al. Attention u-net: learning where to look for the pancreas. (2018), ArXiv abs/1804.03999.
64. Katore MK, Thanekar PS. A noise-resilient framework for automatic covid-19 pneumonia lesions segmentation from ct images. Int J Adv Res Sci Commun Technology (2022) 324–30. doi:10.48175/ijarsct-3746
65. Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, et al. Transunet: transformers make strong encoders for medical image segmentation. (2021), ArXiv abs/2102.04306.
66. Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, et al. Swin-unet: unet-like pure transformer for medical image segmentation. In: ECCV workshops (2021)
67. Zhao S, Li Z, Chen Y, Zhao W, Xie X, Liu J, et al. Scoat-net: a novel network for segmenting covid-19 lung opacification from ct images. Pattern Recognition (2020) 119:108109. doi:10.1016/j.patcog.2021.108109
Keywords: COVID-19, vision-language, multi-task learning, medical image segmentation, medical image captioning
Citation: Zhao W, Li F, Diao Y, Fan P and Chen Z (2024) Cap2Seg: leveraging caption generation for enhanced segmentation of COVID-19 medical images. Front. Phys. 12:1439122. doi: 10.3389/fphy.2024.1439122
Received: 27 May 2024; Accepted: 07 October 2024;
Published: 21 October 2024.
Edited by:
Haoyu Chen, University of Oulu, FinlandReviewed by:
Yang Yang, Yunnan Normal University, ChinaTanmoy Chakraborty, Sharda University, India
Zhiqin Zhu, Chongqing University of Posts and Telecommunications, China
Shireen Y. Elhabian, The University of Utah, United States
Copyright © 2024 Zhao, Li, Diao, Fan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fan Li, NDc4MjYzODIzQHFxLmNvbQ==
 Wanlong Zhao1,2
Wanlong Zhao1,2 Fan Li
Fan Li