- 1Department of Abdominal Oncology, Sichuan Clinical Research Center for Cancer, Sichuan Cancer Hospital and Institute, Sichuan Cancer Center, University of Electronic Science and Technology of China, Chengdu, China
- 2Information Technology Center, West China Hospital of Sichuan University, Chengdu, China
- 3Information Technology Center, West China Sanya Hospital of Sichuan University, Sanya, China
- 4Xiangya School of Pharmaceutical Sciences, Central South University, Changsha, China
- 5College of pharmacy, Chengdu Medical College, Chengdu, China
- 6Department of Oncology, Sichuan Clinical Research Center for Cancer, Sichuan Cancer Hospital and Institute, Sichuan Cancer Center, University of Electronic Science and Technology of China, Chengdu, China
- 7Department of Radiation Oncology, Radiation Oncology Key Laboratory of Sichuan Province, Sichuan Clinical Research Center for Cancer, Sichuan Cancer Hospital and Institute, Sichuan Cancer Center, University of Electronic Science and Technology of China, Chengdu, China
Non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancer cases and remains one of the leading causes of cancer-related mortality worldwide. The high mortality rate is primarily driven by delayed diagnosis, rapid metastasis, and frequent recurrence. Tumor-derived exosomes (TEXs) have emerged as critical mediators in NSCLC progression, offering valuable insights into the tumor microenvironment. Exosomes are small membrane vesicles that facilitate intercellular communication and transport bioactive molecules, including proteins, RNAs, and DNAs, thereby reflecting the genetic complexity of tumors. These exosomes play a key role in promoting tumor metastasis, epithelial-mesenchymal transition (EMT), neovascularization, drug resistance, and immune evasion, all of which are pivotal in the development of NSCLC. This review explores the diverse roles of TEXs in NSCLC progression, focusing on their involvement in pre-metastatic niche formation, tissue metastasis, and immune modulation. Specifically, we discuss the roles of exosome-associated RNAs and proteins in NSCLC, and their contribute to tumor growth and metastasis. Furthermore, we explore the potential of TEXs as biomarkers for NSCLC, emphasizing their application in diagnosis, prognosis, and prediction of resistance to targeted therapies and immunotherapies.
1 Introduction
NSCLC is the most common type of lung malignancy, accounting for approximately 80%–85% of all diagnosed lung cancer cases (Reck et al., 2022). Over the past decade, the incidence of NSCLC has declined from 46.4 to 40.9 per 100,000 cases, while the incidence in patients under 65 years old has shown an upward trend. Moreover, over 60% of patients are diagnosed at an advanced stage, with a 5-year survival rate falling below 15% (Siegel et al., 2022). Epidemiological studies have shown that approximately 40% of NSCLC patients experience recurrence after surgery (Han B. et al., 2022), and the 5-year survival rate for metastatic NSCLC patients is 7% (Rizwan et al., 2022). These statistics emphasize the urgent need for improved early diagnostic accuracy and enhanced treatment monitoring to improve prognosis and survival outcomes. Hence, identifying critical molecular players in NSCLC progression and metastasis, as well as discovering highly sensitive and specific biomarkers, is crucial for early diagnosis, prognostic assessment, and predicting treatment responses.
Approximately 20% of cancer patients are unable to undergo tissue biopsy due to patient conditions, technical limitations, or the inherent heterogeneity of tumor tissues, which hampers diagnostic accuracy (Rolfo et al., 2021; Shields et al., 2022). Non-invasive diagnostic methods, especially liquid biopsy, may become powerful tools for tumor diagnosis and identifying tumor biomarkers (Bertoli et al., 2023; Ma et al., 2024). Liquid biopsy enables the isolation and analysis of circulating cell-free DNA, RNA, and proteins from blood or other body fluids in cancer patients, offering a simplified, more convenient, and better-tolerated diagnostic method (Levy et al., 2016; Molina-Vila et al., 2016).
Exosomes, classified as members of the extracellular vesicle family, were firstly recognized in the early 1980s as vesicles (Nazarenko, 2020; Zhang Y. et al., 2019). These vesicles are released by a wide variety of normal and malignant cells (Whiteside TheresaL., 2016; Zhang et al., 2015). Exosomes have been successfully isolated and purified from various bodily fluids, including blood, encompassing urine, saliva, pleural effusion, ascites, breast milk, and bronchoalveolar lavage fluid, which have potential application prospects in liquid biopsy (Conde-Vancells et al., 2008). Recent research has primarily focused on the roles of exosomes in tumor diagnosis, disease monitoring, and evaluating treatment efficacy (Padinharayil and George, 2024; Ren et al., 2024). Exosomes also play pivotal roles in carcinogenesis and tumor progression, including intercellular signaling, metastasis, drug resistance, and immune suppression (Ren et al., 2024; Hamid et al., 2025; Li Dongqi et al., 2024). Our review highlights the critical role of exosomes in NSCLC progression and metastasis, while exploring their potential as non-invasive biomarkers for early detection and disease monitoring.
2 The functions of TEXs
Exosomes are active nanovesicles, composed of lipid bilayers, with diameters ranging from 40 to 150 nm. They originate from multivesicular bodies and are released into the extracellular space through fusion with the plasma membrane of various normal and tumor cells (Gurung et al., 2021; Liu et al., 2020; Daßler-Plenker et al., 2020). Initially, exosomes were believed to function solely as cellular waste disposal units, responsible for eliminating unwanted molecules within cells (Pan et al., 1985). However, recent studies have demonstrated that these vesicles play more complex roles, including transmitting biological information to neighboring cells and significantly contributing to carcinogenesis and tumor progression (Li Junshu et al., 2024; Agrawal et al., 2024; Oh et al., 2024). Exosomes facilitate the exchange of genetic material via autocrine, paracrine, and endocrine pathways within the cellular environment (Krylova and Feng, 2023; Arya et al., 2024). They deliver their contents through three primary mechanisms: fusion with the plasma membrane, resulting in the release of their internal contents; endocytosis; and interaction with cell surface receptors (Krylova and Feng, 2023). Thus, exosomes are considered a novel mode of cellular signaling. Exosomes can be identified using techniques such as nanoparticle tracking analysis (NTA), resistance pulse sensing (RPS), transmission electron microscopy (TEM), and flow cytometry (FCM) (Yuana et al., 2013; Xie et al., 2019). Methods for isolating and purifying exosomes include ultrafiltration, ultracentrifugation, immunoprecipitation, precipitation, and density gradient centrifugation (Xie et al., 2019; Mortezaee et al., 2022). The contents of exosomes vary depending on the type of secreting cell and include DNA, RNA, proteins, metabolic products, lipids, and cell membrane proteins (Kalluri and LeBleu, 2020; van der Pol et al., 2012; Najafi, 2022).
In normal human blood, approximately 200 trillion exosomes can be identified, whereas the blood of cancer patients contains approximately 400 trillion exosomes (Kalluri, 2016). TEXs have been successfully isolated from various bodily fluids, including urine, saliva, pleural effusion, ascites, breast milk, and bronchoalveolar lavage fluid (Conde-Vancells et al., 2008), highlighting the propensity of cancer cells to produce exosomes in higher concentrations. This suggests their potential as innovative tumor biomarkers. Furthermore, TEXs play a crucial role in the progression of malignant tumors and the development of distant metastases. TEXs regulate the tumor microenvironment, promote angiogenesis and EMT, enhance intercellular signaling, increase tumor cell invasiveness, and foster the establishment of a pre-metastatic niche that promotes distant metastasis (Ren et al., 2024; Hamid et al., 2025). Notably, TEXs influence immune regulation by affecting intercellular communication, immune activation, immune surveillance, antigen expression, and immune suppression (Greening et al., 2015; Dong et al., 2025). TEXs can also carry tumor-associated antigens, potentially impairing the efficacy of immunotherapy (Whiteside TheresaL., 2016). In addition, exosomes serve as key mediators in the resistance signaling pathways of malignant tumor cells, facilitating the transmission of resistance signals in response to targeted therapies (Zhang et al., 2015). Thus, TEXs may hold significant clinical potential for guiding diagnosis, predicting metastasis, evaluating treatment response, and understanding resistance mechanisms in malignant tumors.
3 Related exosome factors involved in NSCLC progression and metastasis
In malignant tumors, exosomes play a pivotal role in tumor progression and metastasis by modulating immune responses, promoting angiogenesis and influencing EMT (Ridder et al., 2015; Can et al., 2025). TEXs facilitate the evasion of immune surveillance by transferring specific proteins to recipient cells, thereby altering the functional behavior of immune cells and promoting tumor progression (Whiteside TheresaL., 2016). For instance, heat shock protein 72 (HSP72) carried by TEXs enhances the immunosuppressive capability of myeloid-derived suppressor cells (MDSCs) via a STAT3-dependent pathway, contributing to immune tolerance of tumor cells (Chalmin et al., 2010). TEXs also inhibit T cell proliferation and induce apoptosis by activating the FAS/FASL signaling pathway, exerting an immunosuppressive effect (Kim et al., 2005). Additionally, TEXs suppress natural killer (NK) cell activity and interfere with monocyte differentiation (Whiteside T. L., 2016). Tumor exosomes can also deliver epidermal growth factor receptor (EGFR) to host macrophages, inhibiting the production of type I interferons and thereby reducing the overall immune response in cancer patients (Gao et al., 2018).
Angiogenesis is critical for providing the blood supply necessary for tumor growth and metastasis (Goudar and Vlahovic, 2008). In the peripheral tissues surrounding malignant tumors, an equilibrium exists between pro-angiogenic and anti-angiogenic factors, which regulate the angiogenesis process. However, malignant tumors are characterized by a predominance of pro-angiogenic proteins, leading to the promotion of neovascularization (Olejarz et al., 2020; Liu et al., 2023). Tumor-derived angiogenic factors, along with other components in the tumor microenvironment, stimulate the proliferation and migration of endothelial cells, thereby facilitating new blood vessel formation to meet the tumor’s nutritional demands (Gasparics and Sebe, 2022).
EMT involves biochemical changes in epithelial cells, leading to the acquisition of mesenchymal phenotypes characterized by increased migratory and invasive capacities, as well as elevated production of extracellular matrix (ECM) components (Gasparics and Sebe, 2022; Greco et al., 2022; Zhang et al., 2022). EMT facilitates tumor metastasis by reducing intercellular adhesion among differentiated epithelial cells, allowing tumor cells to move either individually or collectively (Zhang et al., 2022; Han J. et al., 2022; Kim, 2022). Exosomes have been shown to contribute to both neovascularization and EMT in tumor cells (Lin et al., 2022; Song et al., 2022; Amicone et al., 2022). Here, we summarize the exosomal signaling factors involved in regulating the progression and metastasis of NSCLC.
3.1 Role of exosomal RNA in promoting cancer progression in NSCLC
Exosomal RNA is produced via the endocytosis process within the cell and primarily consists of three distinct classes of non-coding RNA: microRNAs (miRNAs) (Kogure et al., 2011), long non-coding RNAs (lncRNAs) (Lee et al., 2017; Min et al., 2018), and circular RNAs (circRNAs) (Andrey et al., 2011). Notably, studies have shown significantly higher expression levels of exosomal RNA in cancer patients compared to healthy individuals (Tang et al., 2024; Yi et al., 2024; Yue et al., 2024). These exosomal RNAs play pivotal roles in regulating key processes involved in tumor progression, including immune modulation, angiogenesis, metastasis, and drug resistance, contributing to the overall dynamics of the tumor microenvironment.
MiRNAs are a class of non-coding RNA (ncRNA) molecules, approximately 22 nucleotides in length, that regulate gene expression by binding to the 3′untranslated region or open reading frame of target messenger RNA (mRNA) (Saliminejad et al., 2019). In NSCLC, specific miRNA profiles are closely associated with tumor behavior and treatment response, and these miRNAs can be clinically detected by extracting exosomes from body fluids (Shanehbandi et al., 2023; Janpipatkul et al., 2021). Tumor cells release distinct miRNAs into the extracellular space, which are transported via exosomes circulating in the bloodstream (Du et al., 2018). Moreover, tumor cells produce exosomes in particularly high quantities under hypoxic conditions, where they play a crucial role in angiogenesis (Sandúa et al., 2021; Meng et al., 2019). Under hypoxic conditions, exosomes released by lung cancer cells show a significant upregulation of miR-23a, leading to the accumulation of hypoxia-inducible factor-1α (HIF-1α) in endothelial cells. This, in turn, increases tumor angiogenesis and vascular permeability, thereby promoting metastasis (Hsu et al., 2017). Another study also found that miR-619-5p is transferred to exosomes from NSCLC cells under hypoxic conditions, promoting tumor angiogenesis by inhibiting RCAN1.4 (Kim et al., 2020). Additionally, miR-3157-3p is transported from NSCLC cells to vascular endothelial cells through exosomes, targeting the vascular endothelial growth factor (VEGF)/matrix metalloproteinase 2 (MMP2)/MMP9 pathway to enhance the formation of new blood vessels (Ma et al., 2021). Additionally, miR-3157-3p is transported from NSCLC cells to vascular endothelial cells through exosomes, targeting the VEGF/MMP2/MMP9 pathway to enhance the formation of new blood vessels (Gu et al., 2021). Exosomal miR-34c-3p upregulates integrin α2β1, enhancing the invasive and migratory capacities of NSCLC cells (Huang et al., 2020).
LncRNAs are a class of RNA molecules that exceed 200 nucleotides in length (Bridges et al., 2021). Exosome-associated lncRNAs play key roles in tumor progression by regulating metastasis, stem cell maintenance, drug resistance, and the tumor microenvironment (Lin et al., 2023). One of the most studied lncRNAs in NSCLC is the metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), which is highly expressed in the serum of NSCLC patients and promotes tumor migration by inhibiting apoptosis and shortening the cell cycle (Zhang et al., 2017). Elevated levels of lnc-UFC1 have been detected in the exosomes of NSCLC patient serum, and the increase in UFC1 levels is associated with NSCLC invasion (Zang et al., 2020). Another study found that exosomes in the plasma of metastatic NSCLC patients show elevated levels of the lncRNAstem cell inhibitory RNA transcript (SCIRT), which is linked to survival in metastatic NSCLC (Wang et al., 2021). Interestingly, lncRNA SCIRT does not directly promote cancer progression but selectively sorts miR-665 into TEXs in a hnRNAPA1-dependent manner. Subsequently, exosomes enriched with miR-665 directly impact the enhancement of NSCLC invasion and migration capabilities by targeting the Notch downstream transcription factor HEYL. Furthermore, exosomal lncRNAs such as HOTAIR (Chen L. et al., 2021) and NEAT1 (Hussain et al., 2024), frequently upregulated in NSCLC, regulate cellular responses to external stimuli like hypoxia and oxidative stress, common in the tumor microenvironment. By acting as scaffolds for chromatin-modifying complexes, these lncRNAs promote tumor progression and chemoresistance. Targeting these lncRNAs in exosomes could provide a novel strategy to prevent metastasis and improve treatment outcomes in NSCLC.
CircRNAs are formed by back-splicing and have a unique covalent closed-loop structure, providing stability within cells and enabling them to regulate gene expression and affect biological functions (Zhou et al., 2020). circSATB2 promotes the progression of NSCLC and is upregulated in serum exosomes derived from cancer patients. Serum exosomal circSATB2 in patients with metastatic cancer suggests its potential role as a tumor biomarker for NSCLC (Zhang N. et al., 2020). Exosomes secreted by NSCLC repress the function of CD8+ T cells and contribute to resistance to anti-programmed cell death protein-1 (anti-PD1)immunotherapy (Chen et al., 2021b). In addition to circSATB2, other circRNAs, such as circ_0001946 (Zhang et al., 2021), circPVT1 (Huang et al., 2021), and circHIPK3 (Siedlecki et al., 2024), have also been associated with NSCLC. These circRNAs contribute to the initiation and progression of lung cancer by modulating various signaling pathways and gene expressions. They show promise as potential biomarkers for early diagnosis, prognosis assessment, and targeted therapies.
3.2 The functional proteins exosomes in NSCLC
In addition to ncRNAs, exosome proteins have also been considered as key molecules mediating the metastatic phenotype of NSCLC cells. Exosome proteins are mainly membrane transport and fusion proteins, such as annexins, RAB5/RAB7, and TSG101 (Théry et al., 2002). Among the most widely recognized exosome membrane proteins are the tetraspanins, including CD9, CD63, and CD81 (Rana and Zöller, 2011), which are overexpressed on the surface of exosomes and regulates cell-cell interactions, thereby influencing tumor behavior and progression (Zhang W. et al., 2019). These proteins facilitate the exchange of cellular information between the tumor and the surrounding microenvironment, aiding in metastasis and immune evasion.
A key protein involved in NSCLC metastasis is hepatocyte growth factor (HGF), which is enriched in exosomes derived from highly metastatic tumor cells. Exosomal HGF plays a pivotal role in promoting EMT and facilitating cancer cell migration and invasion. It achieves this by activating the c-Met receptor on non-metastatic cells, thereby triggering downstream signaling pathways that drive metastasis (Qiao et al., 2019). Similarly, exosomal proteins derived from the serum of patients with advanced malignancies have been shown to increase the expression of vimentin (VIM) and enhance the metastatic phenotype in recipient cells. This suggests that exosome-mediated protein transfer plays a significant role in promoting the EMT process, which is essential for metastasis (Rahman et al., 2016).
Exosomal proteins also contribute to tumor progression by modulating the immune response. For instance, exosomes secreted by NSCLC cells can interfere with CD8+ T cell function, promoting immune evasion (Chen et al., 2021c). These exosomes carry proteins that inhibit T cell activation and cytotoxicity, allowing the tumor to escape immune surveillance. This mechanism contributes to the resistance of NSCLC to immunotherapy, such as anti-PD1 treatments, by dampening the immune response against tumor cells (Rahman et al., 2016). In addition, exosomes facilitate tumor angiogenesis by delivering pro-angiogenic factors such as VEGF and MMPs to endothelial cells. The transfer of EGFR via exosomes to endothelial cells activates the mitogen-activated protein kinases (MAPK) and protein kinase B (AKT) signaling pathways, which, in turn, upregulate VEGF expression and enhance blood vessel formation (Al-Nedawi et al., 2009).
In conclusion, exosomal proteins in NSCLC play a central role in facilitating tumor progression, metastasis, and immune evasion. By influencing various signaling pathways, these proteins promote the transition from localized tumor growth to widespread metastatic disease. As such, exosomal proteins have significant potential as biomarkers for NSCLC diagnosis and prognosis, as well as therapeutic targets for inhibiting metastasis and enhancing the effectiveness of immunotherapies.
4 Exosome-mediated pre-metastatic niche formation and tissue metastasis
The target organs for malignant tumor metastasis are not selected randomly. In 1889, the “seed and soil” hypothesis was proposed, suggesting that certain tumor cells, referred to as “seeds,” have a propensity to metastasize to specific organs, termed as “soil.” Tumor cells can only successfully metastasize when the environment, or “soil,” is conducive to their growth (Paget, 1889). Despite extensive research, the specificity of organ targeting in tumor metastasis remains one of the most profound mysteries in the field. Recent studies have shown that exosomes play a crucial role in this process, facilitating the establishment of pre-metastatic niches before direct contact between the primary tumor and the distant organ (Milane et al., 2015; Su et al., 2021).
Exosomes facilitate tumor metastasis through various mechanisms. They contribute to the establishment of pre-metastatic niches by transferring molecular signals that prime distant organs for tumor cell colonization. Additionally, exosomes promote EMT, enhancing tumor cell motility and invasiveness. They also play a crucial role in angiogenesis and increase vascular permeability, facilitating tumor cell dissemination via the bloodstream. Moreover, exosomes contribute to immune modulation by suppressing antitumor immune responses, thus enabling immune evasion (Luo et al., 2023; Zhao et al., 2021). These coordinated actions enhance the metastatic potential of tumor cells, supporting their colonization in secondary sites (Figure 1).
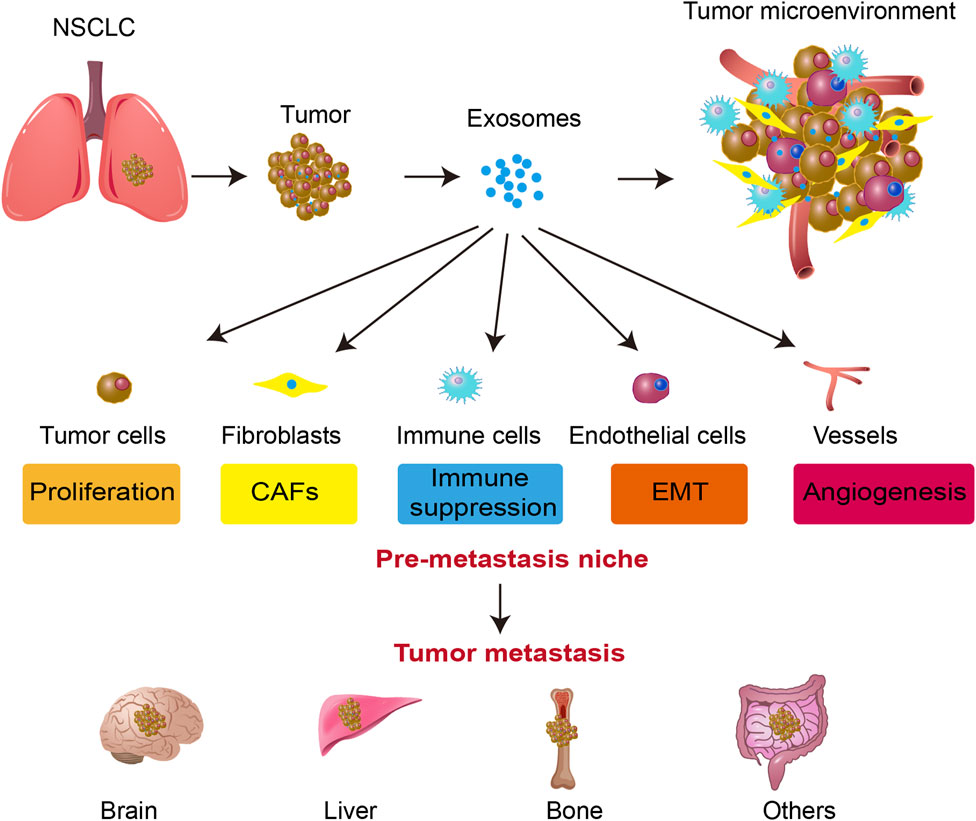
Figure 1. Exosome-mediated tumor progression and metastasis in NSCLC. Exosomes secreted by tumor cells influence various components of the tumor microenvironment, including fibroblasts (CAFs), immune cells, endothelial cells, and blood vessels. Exosomes facilitate tumor cell proliferation, immune suppression, EMT, and angiogenesis. These processes contribute to the formation of a pre-metastatic niche, ultimately leading to tumor metastasis to distant organs such as the brain, liver, and bone. The figure highlights the critical role of exosomes in promoting NSCLC progression and metastatic spread.
The organotropism of different types of metastatic tumor cells showed significant differences (Obenauf and Massagué, 2015), which are related to the integration of TEXs (Hoshino et al., 2015a). Proteomic analysis reveals that exosomes isolated from tumor cells originating from distinct organs exhibit distinct integrin expression patterns. Specifically, integrin α6β4 and α6β1 are associated with lung metastasis, whereas integrin αvβ5 is correlated with liver metastasis. The disruption of integrin α6β4 and αvβ5 expression has been shown to diminish the uptake of exosomes by recipient organ cells, thereby reducing lung and liver metastasis, respectively (Hoshino et al., 2015b).
The main sites of NSCLC metastasis are the bone, brain, and liver (Wood et al., 2014). Recent research has conclusively demonstrated the pivotal role that exosomes play in establishing a pre-metastatic immune microenvironment conducive to brain metastasis. The biological distribution of exosomes secreted by tumors was analyzed, found a tissue-specific fusion of integrins with T cells, which in turn facilitates organ-specific colonization and the formulation of a pre-metastatic niche tailored specifically for brain metastasis (Xu et al., 2020). In addition, CEMIP + exosomes secreted by tumors are absorbed by brain cells and microglia, leading to the enhanced expression of pro-inflammatory cytokines, which are encoded by genes such as PTGS2, TNF, and CCL/CXCL, thereby promoting cerebrovascular remodeling and metastasis (Rodrigues et al., 2019).
Additionally, exosomes derived from NSCLC cells treated with transforming growth factor (TGF-β) contain high levels of lnc-MMP2-2. This lncRNA stimulates MMP2 expression, positively correlating with tumor cell invasiveness and vascular permeability, further promoting metastasis (Wu et al., 2018; Valadi et al., 2007; Liao et al., 2015; Chen et al., 2013; Tang et al., 2016). A recent investigation has revealed that lnc-MMP1-2 disrupts the tight junctions present between human brain microvascular endothelial cells. Additionally, it has been observed to induce EMT and enhance the permeability of the blood-brain barrier, allowing tumor cells to penetrate the brain in the circulatory system (Wu et al., 2021).
Except for brain metastasis, bone metastasis is also a prevalent form of distant metastasis observed in NSCLC. Exosomes extracted from peripheral blood of NSCLC patients with bone metastasis exhibit a significant upregulation of SOX2 overlapping transcript (SOX2-OT), which is closely associated with lower overall survival rates. SOX2-OT increases RAC1 expression by targeting miR-194-5p to promote bone metastasis of NSCLC (Ni et al., 2021).
5 Exosomes as biomarkers of NSCLC
Conventional biomarkers, such as carcinoembryonic antigen (CEA), epithelial cell adhesion molecules (EPCAM), and EGFR, can be found in lung tissue, tumor-draining pulmonary blood, and bone marrow samples used for diagnosing NSCLC. However, these techniques are challenging to samples and cause much discomfort to patients (Rehulkova et al., 2023; Mederer et al., 2022). “Liquid biopsy” represents a non-invasive or minimally invasive approach to disease detection, utilizing molecular diagnostic techniques as its foundation (Ma et al., 2024). Recently this technology, which has become a research hotspot, utilizes bodily fluids such as blood, bronchial alveolar fluid, urine, pleural effusion, ascites, breast milk, and saliva from cancer patients to detect circulating biomarkers of tumors and obtain relevant genetic information about the disease (Buszka et al., 2022). It possesses the capability to identify tumors at an earlier stage than imaging techniques, rendering it a suitable tool for the early diagnosis of tumors.
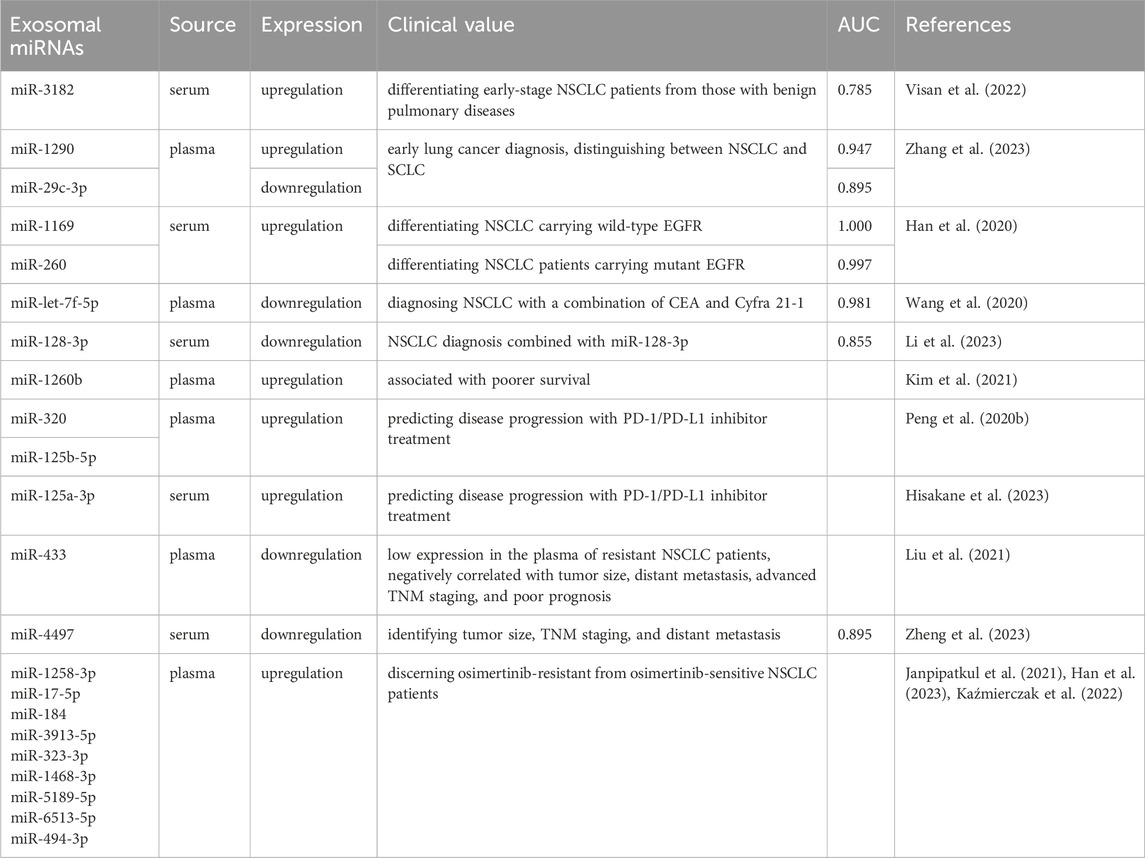
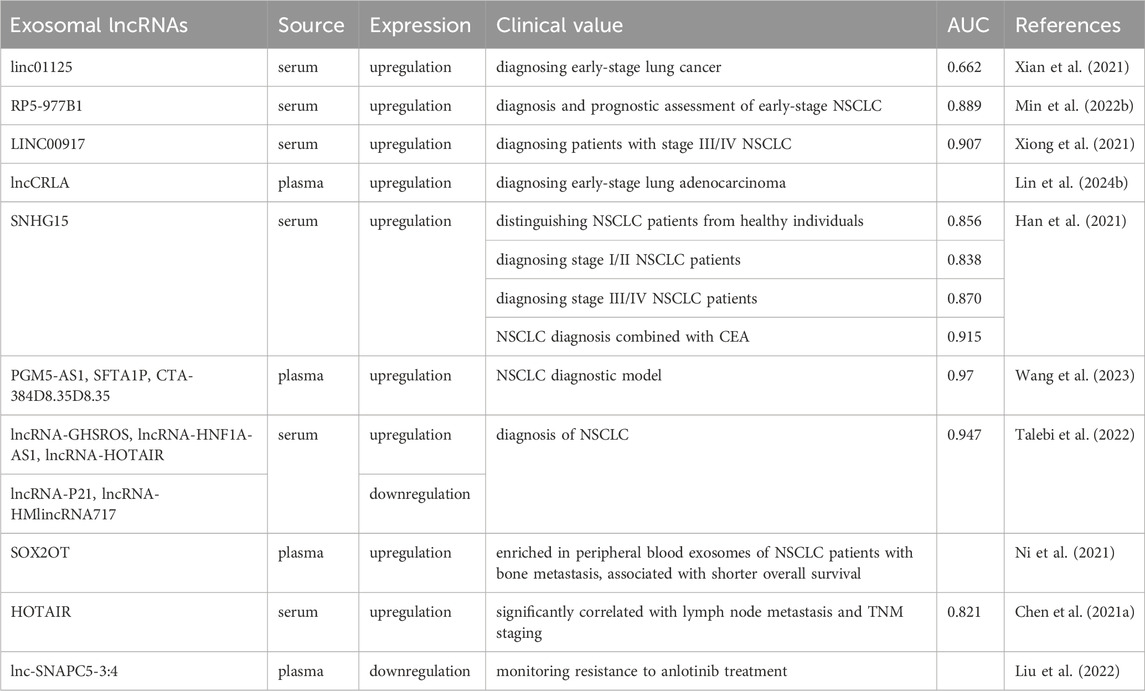
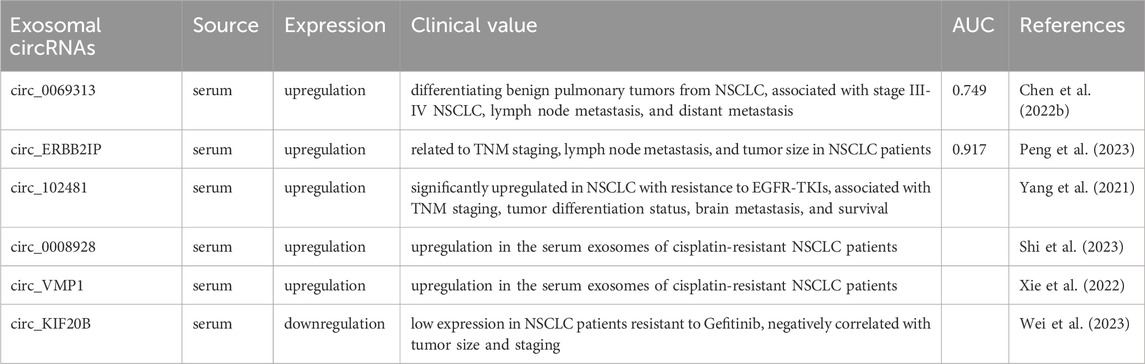
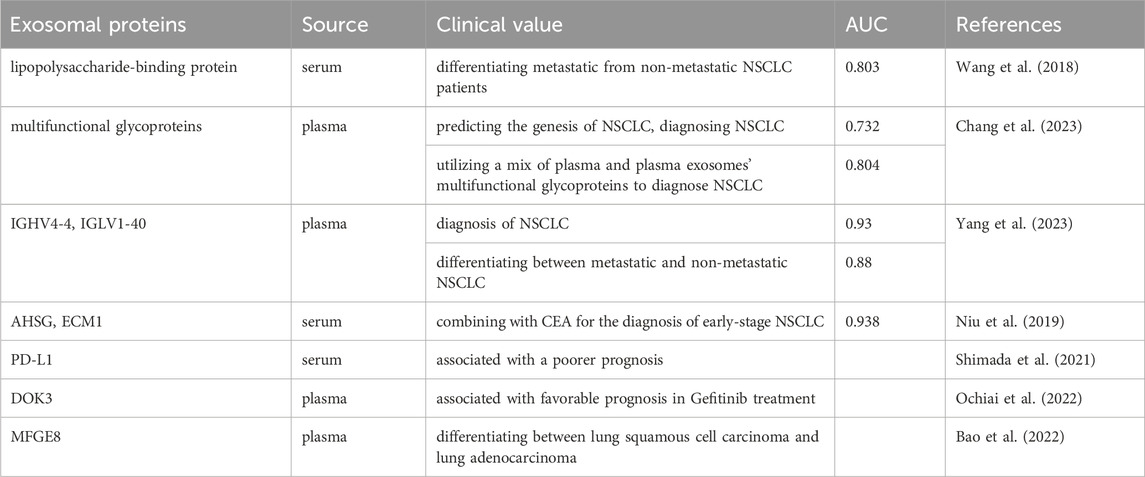
In the era of liquid biopsy, using exosomes as biomarkers for NSCLC is a promising approach. Exosomes, which are directly secreted into bodily fluids by tumor cells, encompass components such as ncRNAs and protein alterations (including EGFR mutations), thereby rendering highly distinctive and representative information (Casagrande et al., 2023; Vasu et al., 2025). Exosomes exhibit a ubiquitous distribution, possess remarkable permeability, are easily accessible, and are encapsulated by different lipid bilayers and are not easily degraded (Cheng et al., 2022; Boukouris and Mathivanan, 2015). The identification of exosomes with differential expression patterns in liquid biopsy exhibits promising applications in various medical domains, including the diagnosis of cancer, prognostic evaluation, monitoring of disease progression, and assessment of chemotherapy resistance (Figure 2) (Rezaie et al., 2022). We have conducted a comprehensive summary of extracellular vesicle RNA and protein as potential biomarkers for the diagnosis, prognosis, and prediction of treatment response in NSCLC, as presented in Tables 1–4.
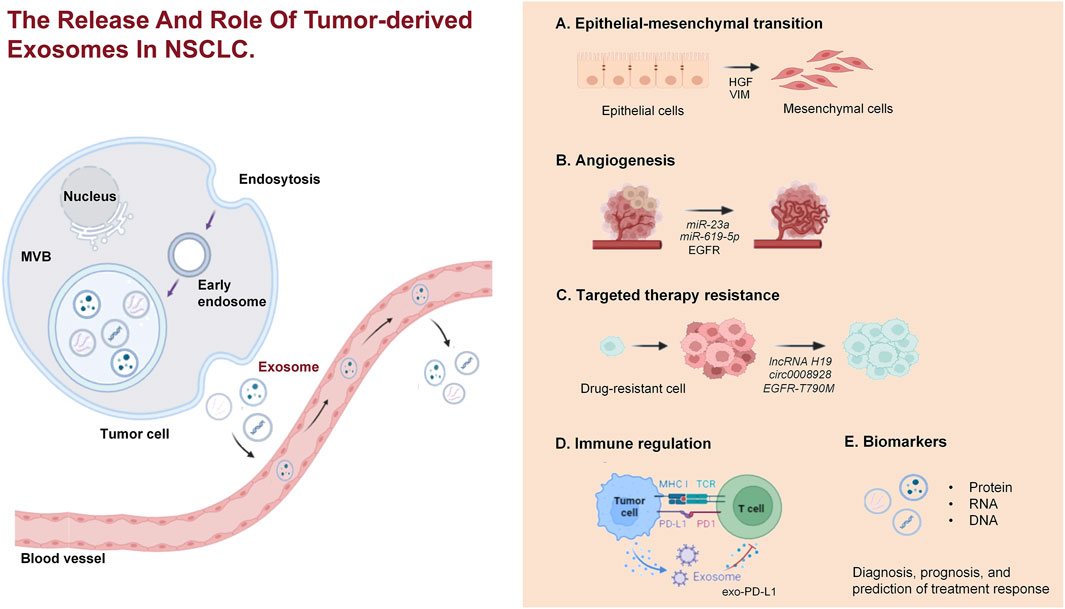
Figure 2. The release and role of TEXs in NSCLC. TEXs in NSCLC are released from tumor cells via the endocytic pathway, involving the formation of early endosomes, which mature into multivesicular bodies. These MVBs then fuse with the plasma membrane, releasing exosomes into the extracellular environment. The exosomes contribute to various processes in NSCLC, including EMT, angiogenesis, drug resistance, immune regulation, and they serve as biomarkers for diagnosis, prognosis, and treatment response prediction.
5.1 TEXs as biomarkers for NSCLC diagnosis
Owing to the enhanced production of exosomes by tumor cells, exosomes present a promising role as novel diagnostic biomarkers. The diagnostic potential of exosomes in plasma or serum of NSCLC patients can be determined by analyzing the area under the gene expression curve (AUC). Exosomal miR-3182 (Visan et al., 2022), miR-1290, and miR-29c-3p (Zhang et al., 2023) have been shown to be useful in the early detection of lung cancer. In comparison to conventional tumor biomarkers, exosomal miR-1290 and miR-29c-3p exhibit superior diagnostic efficacy in discerning benign lung diseases from lung cancer, achieving AUC values of 0.934 and 0.868 respectively. These miRNAs demonstrate higher diagnostic accuracy for early-stage lung cancer, with AUC values of 0.947 and 0.895, compared to traditional markers (Zhang et al., 2023). Circulating exosomal miR-342-5p and miR-574-5p were significantly elevated in early-stage lung adenocarcinoma (LA) patients compared to healthy controls and decreased after tumor resection. The combination of these miRNAs achieved an AUC of 0.813, with 80% sensitivity and 73.2% specificity, underscoring their potential as novel biomarkers for early stage LA diagnosis (Han et al., 2020).
miR-1169 and miR-260 can effectively distinguish between EGFR wild-type and mutant NSCLC patients (Xia et al., 2021). Additionally, miR-181-5p, miR-30a-3p, miR-30e-3p, and miR-320b have emerged as key biomarkers for differentiating LA from squamous cell carcinoma (SCC) in NSCLC, with a diagnostic accuracy demonstrated by an AUC value of 0.936 for detecting SCC (Jin et al., 2017). Furthermore, the combined application of multiple exosomal miRNAs improves the accuracy of NSCLC diagnosis. Specifically, the combination of exosomal miR-382 and CEA in serum (AUC: 0.953) (Luo et al., 2021) and plasma exosomal miR-let-7f-5p combined with CEA and CYFRA21-1 (AUC: 0.981), have notable advantages in the diagnosis of NSCLC (Wang et al., 2020).
LINC00917 in exosomes showed stronger predictive ability for stage III/IV NSCLC (AUC: 0.907) compared to stage I/II (IUC: 0.773) (Xiong et al., 2021). LASSO regression analysis was used to screen biomarkers from exosomal lncRNAs in a large clinical population through exosomes in plasma. Then, a multi-marker diagnostic model was constructed using logistic regression, which integrates specific exosomal lncRNAs (PGM5-AS1, SFTA1P, and CTA-384D8.35), achieving a high prediction accuracy with an AUC of 0.97 (Wang et al., 2023). Similarly, a ncRNA profile consisting of five specific lncRNAs was found to improve the diagnosis of NSCLC with an AUC of 0.947 (Talebi et al., 2022), indicating that exosomal lncRNA patterns constructed through histological research and data analysis techniques have higher diagnostic value compared to previous single biomarkers.
One proteomic analysis revealed that plasma exosomal MFGE8 has a high diagnostic effect in distinguishing between squamous cell carcinoma and lung adenocarcinoma, with an AUC of 0.812 (Bao et al., 2022). Another identical methodology was employed, and discovered that the concurrent utilization of AHSG, ECM1, and CEA substantially increased the diagnostic accuracy for NSCLC. Specifically, the AUC values for distinguishing NSCLC from healthy individuals were 0.938 for overall NSCLC and 0.911 for early-stage NSCLC (Niu et al., 2019).
5.2 TEXs as prognostic markers for NSCLC
Increasing research has shown that exosomal proteins and miRNAs are closely related with tumor progression, highlighting that exosome can be utilized as prognostic markers to enhance the treatment options available for NSCLC patients (Niloufar et al., 2024). For instance, phenotypic analysis of exosomes from the plasma of 276 NSCLC patients revealed that exosomal membrane-bound proteins, such as EGFR, NY-ESO-1, ALIX, PLAP and EPCAM, are significantly associated with overall survival (OS) in patients, suggesting that exosomal membrane-bound proteins can be used as prognostic biomarkers for NSCLC (Sandfeld-Paulsen et al., 2016).
In addition, the downregulation of miR-503 in NSCLC tissues, compared to non-malignant lung tissue, has been linked to advanced tumor stages and poor prognosis. Kaplan-Meier analysis further indicated worse survival outcomes in patients with lower miR-503 expression, suggesting that miR-503 could be a valuable prognostic biomarker for survival in NSCLC patients (Liu et al., 2015). Another study found that deregulated expression of miR-21, miR-143, and miR-181a in NSCLC is associated with clinicopathological characteristics and poor prognosis, with elevated miR-21 expression being linked to worse survival outcomes (Gao et al., 2010).
Additionally, studies have demonstrated the potential of exosomal biomarkers such as PLA2G10 mRNA and RP5-977B1 lncRNA for both diagnostic and prognostic purposes in NSCLC, enhancing tumor detection, prognosis assessment, and early-stage diagnosis (Chen Yinfeng et al., 2022; Min Ling et al., 2022). Moreover, a study found that 84 plasma exosomal miRNAs from patients with LA and healthy controls and found that elevated levels of exosomal miR-10b-5p, miR-21-5p and miR-23b-3p are associated with worse overall survival, indicating that exosomal miRNAs can also be used as prognostic biomarkers for NSCLC (Liu et al., 2017). In recurrent cases of NSCLC patients, elevated levels of exosomal miR-203-3a-3p (Han B. et al., 2022) and miR-124 (Sanchez-Cabrero et al., 2023) reveal the potential for exosomal miRNAs to predict disease progression.
Furthermore, studies have reported significant associations between exosomal lncRNA and the prognosis, lymph node metastasis, TNM stage, and tumor invasion (Lin Shuai et al., 2024; Yin Cunli et al., 2024; Zhang et al., 2025) in NSCLC patients. In summary, exosomes offer a promising, non-invasive approach for prognostic biomarker development in NSCLC.
5.3 TEXs as markers of targeted therapy resistance in NSCLC
In recent years, targeted therapy has garnered significant attention and yielded remarkable outcomes in the treatment of NSCLC patients. Nonetheless, despite an initial positive response to targeted therapy, the eventual development of acquired resistance is inevitable, resulting in deteriorated treatment outcomes and prognosis. Consequently, it is imperative to unravel the fundamental mechanisms underlying targeted resistance and identify potential biomarkers and targets that contribute to the resistance to tumor-specific targeted therapy. Increasing research suggests that exosomes can promote resistance through various mechanisms. Exosomes exhibit the ability to convey miRNA, lncRNAs, and proteins to targeted cells, facilitating the transmission of signals between resistant and sensitive cells, as well as between stromal and tumor cells, ultimately leading to the induction of drug resistance in tumor cells (Shedden et al., 2003; Bach et al., 2017; Yu et al., 2015).
Exosomal miRNAs have been shown to play a significant role in drug resistance, particularly in EGFR-TKIs. TEXs contribute to EGFR-TKI resistance by transferring active cargoes, including miRNAs. Research has demonstrated that exosomal RNA can detect EGFR-T790M and activated EGFR mutations with sensitivities of 90% and 98%, respectively (Krug et al., 2018). In addition, Nano-LC-MS/MS analysis of gefitinib-resistant PC9R cells, due to the EGFR-T790M mutation, revealed the enrichment of specific exosomal proteins (Choi et al., 2014). Extensive research has demonstrated that the level of expression of lncRNA H19 is elevated in gefitinib-resistant NSCLC. Specifically, lncRNA H19 is encapsulated within exosomes, facilitated by the mediation of hnRNPA2B1, and transmitted to non-resistant NSCLC cells to induce gefitinib resistance (Lei et al., 2018). Moreover, nine exosomal miRNAs were found to be upregulated in patients resistant to Osimertinib, providing a predicting basis for treatment response (Janpipatkul et al., 2021; Han et al., 2023; Kaźmierczak et al., 2022).
Additionally, exosomal circRNAs, such ascirc0008928 (Shi et al., 2023) and circVMP1 (Xie et al., 2022),are upregulated in the serum of cisplatin-resistant NSCLC patients, suggesting a potential role in resistance to chemotherapy (Pérez-Ruiz et al., 2020). Furthermore, exosomes from an ALK-TKI-resistant NSCLC subclone have been shown to induce drug resistance in a previously sensitive subclone. Differential expressions of miRNAs, including miR-21-5p and miR-486-3p, and lncRNAs like MEG3 and XIST were identified in exosomes secreted by resistant subclones (Kwok et al., 2019).
These findings underscore the potential of TEXs as biomarkers for assessing the efficacy of targeted therapies through liquid biopsy. TEXs could also serve as indicators of resistance to targeted therapy in NSCLC, providing valuable insights for monitoring treatment response and predicting resistance.
5.4 TEXs as markers for immunotherapy in NSCLC
Immunotherapy has significantly transformed the treatment of NSCLC with immune checkpoint inhibitors (ICIs) playing a central role (Addeo et al., 2021; Li et al., 2011; Sharma and Allison, 2015). These therapies, including antibodies targeting the PD-1/PD-L1 pathway and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), have significantly improved patient outcomes, especially when combined with chemotherapy (Yu et al., 2016). These therapies, including antibodies targeting the PD-1/PD-L1 pathway and CTLA-4, have significantly improved patient outcomes, especially when combined with chemotherapy.
However, beyond PD-1/PD-L1 and CTLA-4, other immune checkpoints are emerging as important therapeutic targets. These include lymphocyte activation gene-3 (LAG-3), T cell immunoglobulin and mucin domain-containing protein 3 (TIM-3), T cell immunoreceptor with Ig and ITIM domains (TIGIT), V-type immunoglobulin domain-containing suppressor of T cell activation (VISTA), and CD276, each playing distinct roles in immune regulation and contributing to tumor immune evasion (Yin Nanhao et al., 2024). Currently, these checkpoints are under active investigation for their potential in enhancing immunotherapy responses, often in combination with existing PD-1/PD-L1 therapies. Exosome-based biomarker research in NSCLC has primarily focused on the PD-1/PD-L1 pathway, with exosomal PD-L1 demonstrating significant potential as a non-invasive marker for monitoring immunotherapy responses. While studies on exosomal PD-L1 have shown a correlation with treatment outcomes, research into other immune checkpoint markers, such as CTLA-4, is still limited.
PD-L1 is a key protein in tumor cells that binds to the PD-1 receptor on T cells, inhibiting their activation and promoting immune evasion by suppressing T cell activity. This interaction allows tumor cells to escape immune surveillance, making it harder for the immune system to attack them (Xia et al., 2019). PD-L1 is present not only on the surface of numerous tumor cell types, but also on the surface of exosomes, known as exosomal programmed death-ligand 1 (exo-PD-L1) (Ayala-Mar et al., 2021). Tumor-derived Exo-PD-L1 has the capability to competitively interact with PD-1 receptors present on the surface of T cells, inhibiting T cell activity and cytokine release, thereby mediating immune escape of tumor cells and the efficacy of immunotherapy (Figure 3). A study involving 85 patients with NSCLC demonstrated a significant correlation between the expression of exo-PD-L1 in serum and key clinical parameters, including tumor size, lymph node status, metastasis, and disease progression, highlighting its potential as a clinically relevant biomarker for NSCLC management (Li et al., 2019). Additionally, Peng et al. suggested that high levels of exosomal miR-320d, miR-320c, and miR-320b were associated with poor response to anti-PD-1 treatment in NSCLC patients, while exosomal miR-125b-5p was identified as a potential target for improving the effectiveness of anti-PD-1 therapy (Peng et al., 2020a).
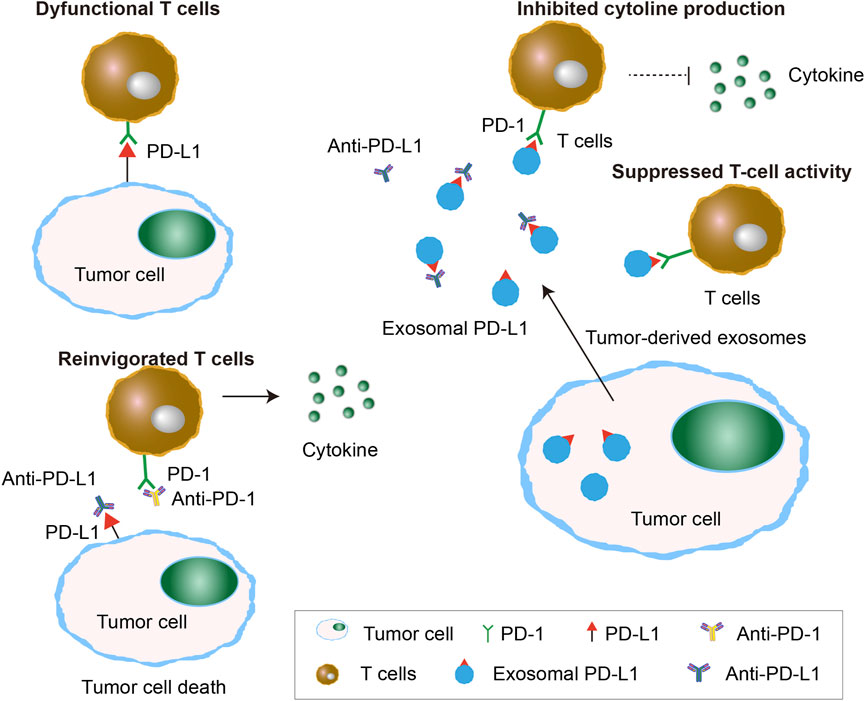
Figure 3. Exosomal PD-L1 caused failure of immune check-point therapy. Exosomal PD-L1 released by tumor cells causes the failure of immune checkpoint therapy by binding to PD-1 receptors on T cells, leading to suppressed T-cell activity and inhibited cytokine production. This prevents T-cell reinvigoration despite the use of anti-PD-1/PD-L1 therapies, allowing the tumor to evade immune detection.
CTLA-4, a negative regulatory receptor on effector and regulatory T cells, suppresses T cell activity and allows tumor cells to evade immune detection. The CheckMate-227 trial showed that simultaneous blockade of PD-1 and CTLA-4 significantly improved overall survival in NSCLC patients (Théry et al., 2018). In addition, a study aimed at elucidating the prognostic relevance of exo-PD-L1 and CD28 in NSCLC patients subjected to ICI treatment uncovered that patients with elevated exo-PD-L1 expression coupled with reduced CD28 levels displayed a shorter progression-free survival, underscoring the importance of considering baseline exo-PD-L1 and CD28 levels as potential prognostic indicators for the outcomes of PD-1-based therapeutic interventions (Zhang C. et al., 2020).
6 Conclusion
Exosomes function as pivotal “messengers” among cells, efficiently facilitating the transfer of critical signals and substances, thus enhancing intercellular communication. TEXs play a crucial role in almost every step of the invasion and metastasis process in NSCLC, such as immune regulation, angiogenesis, drug resistance, EMT, and pre-metastatic niche formation. By coordinating these complex interactions, exosomes significantly influence the progression and distant metastasis of NSCLC, highlighting their importance in the biology and dynamics of the tumor microenvironment. Moreover, the structural integrity of their lipid bilayer ensures stability both in vivo and in vitro, while protecting enclosed bioactive molecules, further supporting their potential as clinical diagnostic and prognostic tools (Kimiz-Gebologlu and Oncel, 2022).
Despite these promising attributes, the clinical translation of exosome-based liquid biopsies and therapeutics faces several key challenges. The lack of standardized and scalable isolation methods results in inconsistent purity and recovery, necessitating the development of cost-effective, high-throughput technologies with robust quality control measures. Additionally, the heterogeneity of exosomes and the complexity of their cargo complicate the identification of tumor-specific biomarkers, emphasizing the need for advanced single-vesicle analysis and omics-driven approaches. Furthermore, the limited sensitivity and specificity of exosome-based assays for early cancer detection require large-scale validation studies to establish reliable biomarker panels. The current infrastructure of conventional clinical laboratories is insufficient to handle the analytical demands of exosomal data, highlighting the need for automated and user-friendly platforms. Lastly, regulatory and logistical barriers, such as the lack of clear guidelines and extensive approval processes, delay the widespread adoption of exosome-based applications. Currently, the utilization of exosomes in the diagnostic and therapeutic of NSCLC remains in its nascent stage. Anticipated advancements in exosome research encompassing their biosynthesis, secretion processes, interactions with targeted cells, and the functional significance of exosomal constituents, have the potential to enhance their application in medical practice and elevate the survival prospects for patients afflicted with NSCLC.
Author contributions
YG: Investigation, Validation, Writing–original draft. JX: Data curation, Software, Visualization, Writing–review and editing. ZY: Data curation, Investigation, Writing–review and editing. ML: Data curation, Formal Analysis, Writing–review and editing. HY: Conceptualization, Project administration, Supervision, Validation, Visualization, Writing–review and editing. RL: Conceptualization, Funding acquisition, Project administration, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Fundamental Research Funds for the Central Universities (University of Electronic Science and Technology of China) (No. ZYGX2021YGCX020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Addeo, A., Passaro, A., Malapelle, U., Banna, G. L., Subbiah, V., and Friedlaender, A. (2021). Immunotherapy in non-small cell lung cancer harbouring driver mutations. Cancer Treat. Rev. 96, 102179. doi:10.1016/j.ctrv.2021.102179
Agrawal, P., Olgun, G., Singh, A., Gopalan, V., and Hannenhalli, S. (2024). Characterizing the pan-cancer role of exosomal miRNAs in metastasis across cancers. Comput. Struct. Biotechnol. J. 27, 252–264. doi:10.1016/j.csbj.2024.12.025
Al-Nedawi, K., Meehan, B., Kerbel, R. S., Allison, A. C., and Rak, J. (2009). Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc. Natl. Acad. Sci. U. S. A. 106, 3794–3799. doi:10.1073/pnas.0804543106
Amicone, L., Marchetti, A., and Cicchini, C. (2022). Exosome-associated circRNAs as key regulators of EMT in cancer. Cells 11, 1716. doi:10.3390/cells11101716
Andrey, T., Ludmila, W., Anne, L., and Barbara, B. (2011). Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39 (16), 7223–7233. doi:10.1093/nar/gkr254
Arya, S. B., Collie, S. P., and Parent, C. A. (2024). The ins-and-outs of exosome biogenesis, secretion, and internalization. Trends Cell Biol. 34 (2), 90–108. doi:10.1016/j.tcb.2023.06.006
Ayala-Mar, S., Donoso-Quezada, J., and González-Valdez, J. (2021). Clinical implications of exosomal pd-l1 in cancer immunotherapy. J. Immunol. Res. 2021, 8839978. doi:10.1155/2021/8839978
Bach, D. H., Hong, J. Y., Park, H. J., and Lee, S. K. (2017). The role of exosomes and miRNAs in drug-resistance of cancer cells. Int. J. Cancer 141, 220–230. doi:10.1002/ijc.30669
Bao, M., Huang, Y., Lang, Z., Zhao, H., Saito, Y., Nagano, T., et al. (2022). Proteomic analysis of plasma exosomes in patients with non-small cell lung cancer. Transl. Lung Cancer Res. 11, 1434–1452. doi:10.21037/tlcr-22-467
Bertoli, E., De Carlo, E., Basile, D., Zara, D., Stanzione, B., Schiappacassi, M., et al. (2023). Liquid biopsy in NSCLC: an investigation with multiple clinical implications. Int. J. Mol. Sci. 24, 10803. doi:10.3390/ijms241310803
Boukouris, S., and Mathivanan, S. (2015). Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin. Appl. 9 (3–4), 358–367. doi:10.1002/prca.201400114
Bridges, M. C., Daulagala, A. C., and Kourtidis, A. (2021). LNCcation: lncRNA localization and function. J. Cell Biol. 220, e202009045. doi:10.1083/jcb.202009045
Buszka, K., Ntzifa, A., Owecka, B., Kamińska, P., Kolecka-Bednarczyk, A., Zabel, M., et al. (2022). Liquid biopsy analysis as a tool for TKI-based treatment in non-small cell lung cancer. Cells 11, 2871. doi:10.3390/cells11182871
Can, C., Yang, X., Jia, H., Wu, H., Guo, X., Wei, Y., et al. (2025). Exosomal circ_0006896 promotes AML progression via interaction with HDAC1 and restriction of antitumor immunity. Mol. Cancer 24 (1), 4. doi:10.1186/s12943-024-02203-8
Casagrande, G. M. S., Silva, M. O., Reis, R. M., and Leal, L. F. (2023). Liquid biopsy for lung cancer: up-to-date and perspectives for screening programs. Int. J. Mol. Sci. 24, 2505. doi:10.3390/ijms24032505
Chalmin, F., Ladoire, S., Mignot, G., Vincent, J., Bruchard, M., Remy-Martin, J. P., et al. (2010). Membrane-associated Hsp 72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J. Clin. Invest 120, 457–471. doi:10.1172/JCI40483
Chang, W., Zhu, J., Yang, D., Shang, A., Sun, Z., Quan, W., et al. (2023). Plasma versican and plasma exosomal versican as potential diagnostic markers for non-small cell lung cancer. Respir. Res. 24, 140. doi:10.1186/s12931-023-02423-4
Chen, L., Huang, S., Huang, J., Chen, Q., and Zhuang, Q. (2021a). Role and mechanism of exosome-derived long noncoding RNA HOTAIR in lung cancer. ACS Omega 6, 17217–17227. doi:10.1021/acsomega.1c00905
Chen, P. M., Wu, T. C., Shieh, S. H., Wu, Y. H., Li, M. C., Sheu, G. T., et al. (2013). MnSOD promotes tumor invasion via upregulation of FoxM1-MMP2 axis and related with poor survival and relapse in lung adenocarcinomas. Mol. cancer Res. MCR 11, 261–271. doi:10.1158/1541-7786.MCR-12-0527
Chen, S. W., Zhu, S. Q., Pei, X., Qiu, B. Q., Xiong, D., Long, X., et al. (2021b). Cancer cell-derived exosomal circUSP7 induces CD8(+) T cell dysfunction and anti-PD1 resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol. Cancer 20, 144. doi:10.1186/s12943-021-01448-x
Chen, S. W., Zhu, S. Q., Pei, X., Qiu, B. Q., Xiong, D., Long, X., et al. (2021c). Cancer cell-derived exosomal circUSP7 induces CD8+ T cell dysfunction and anti-PD1 resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol. Cancer 20 (1), 144. doi:10.1186/s12943-021-01448-x
Chen, Y., Lou, C., Ma, X., Zhou, C., Zhao, X., Li, N., et al. (2022b). Serum exosomal hsa_circ_0069313 has a potential to diagnose more aggressive non-small cell lung cancer. Clin. Biochem. 102, 56–64. doi:10.1016/j.clinbiochem.2022.01.005
Chen, Y., Ma, X., Lou, C., Zhou, C., Zhao, X., Li, N., et al. (2022a). PLA2G10 incorporated in exosomes could be diagnostic and prognostic biomarker for non-small cell lung cancer. Clin. Chim. Acta 530, 55–65. doi:10.1016/j.cca.2022.02.016
Cheng, J., Wang, X., Yuan, X., Liu, G., and Chu, Q. (2022). Emerging roles of exosome-derived biomarkers in cancer theranostics: messages from novel protein targets. Am. J. Cancer Res. 12 (5), 2226–2248.
Choi, D. Y., You, S., Jung, J. H., Lee, J. C., Rho, J. K., Lee, K. Y., et al. (2014). Extracellular vesicles shed from gefitinib-resistant nonsmall cell lung cancer regulate the tumor microenvironment. Proteomics 14 (16), 1845–1856. doi:10.1002/pmic.201400008
Conde-Vancells, J., Rodriguez-Suarez, E., Embade, N., Gil, D., Matthiesen, R., Valle, M., et al. (2008). Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. Proteome Res. 7 (2), 5157–5166. doi:10.1021/pr8004887
Daßler-Plenker, J., Küttner, V., and Egeblad, M. (2020). Communication in tiny packages: exosomes as means of tumor-stroma communication. Biochim. Biophys. Acta Rev. Cancer 1873 (2), 188340. doi:10.1016/j.bbcan.2020.188340
Dong, Z., Fu, Y., Cai, Z., Dai, H., and He, Y. (2025). Recent advances in adipose-derived mesenchymal stem cell-derived exosomes for regulating macrophage polarization. Front. Immunol. 16, 1525466. doi:10.3389/fimmu.2025.1525466
Du, X., Zhang, J., Wang, J., Lin, X., and Ding, F. (2018). Role of miRNA in lung cancer-potential biomarkers and therapies. Curr. Pharm. Des. 23 (39), 5997–6010. doi:10.2174/1381612823666170714150118
Gao, L., Wang, L., Dai, T., Jin, K., Zhang, Z., Wang, S., et al. (2018). Tumor-derived exosomes antagonize innate antiviral immunity. Nat. Immunol. 19, 233–245. doi:10.1038/s41590-017-0043-5
Gao, W., Yu, Y., Cao, H., Shen, H., Li, X., Pan, S., et al. (2010). Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed. Pharmacother. 64 (6), 399–408. doi:10.1016/j.biopha.2010.01.018
Gasparics, A., and Sebe, A. (2022). Forward genetic screens as tools to investigate role and mechanisms of EMT in cancer. Cancers (Basel) 14 (23), 5928. doi:10.3390/cancers14235928
Goudar, R. K., and Vlahovic, G. (2008). Hypoxia, angiogenesis, and lung cancer. Curr. Oncol. Rep. 10 (4), 277–282. doi:10.1007/s11912-008-0043-6
Greco, L., Rubbino, F., and Laghi, L. (2022). Epithelial to mesenchymal transition as mechanism of progression of pancreatic cancer: from mice to men. Cancers (Basel) 14 (23), 5797. doi:10.3390/cancers14235797
Greening, D. W., Gopal, S. K., Xu, R., Simpson, R. J., and Chen, W. (2015). Exosomes and their roles in immune regulation and cancer. Semin. Cell Dev. Biol. 40, 72–81. doi:10.1016/j.semcdb.2015.02.009
Gu, G., Hu, C., Hui, K., Zhang, H., Chen, T., Zhang, X., et al. (2021). Exosomal miR-136-5p derived from anlotinib-resistant NSCLC cells confers anlotinib resistance in non-small cell lung cancer through targeting PPP2R2A. Int. J. Nanomedicine 16, 6329–6343. doi:10.2147/IJN.S321720
Gurung, S., Perocheau, D., Touramanidou, L., and Baruteau, J. (2021). The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun. Signal 19, 47. doi:10.1186/s12964-021-00730-1
Hamid, Y., Rabbani, R. D., Afsara, R., Nowrin, S., Ghose, A., Papadopoulos, V., et al. (2025). Exosomal liquid biopsy in prostate cancer: a systematic review of biomarkers for diagnosis, prognosis, and treatment response. Int. J. Mol. Sci. 26 (2), 802. doi:10.3390/ijms26020802
Han, B., Molins, L., He, Y., Viñolas, N., Sánchez-Lorente, D., Boada, M., et al. (2022a). Characterization of the MicroRNA cargo of extracellular vesicles isolated from a pulmonary tumor-draining vein identifies miR-203a-3p as a relapse biomarker for resected non-small cell lung cancer. Int. J. Mol. Sci. 23, 7138. doi:10.3390/ijms23137138
Han, J., Meng, J., Chen, S., Wang, X., Yin, S., Zhang, Q., et al. (2022b). Correction: YY1 complex promotes quaking expression via super-enhancer binding during EMT of hepatocellular carcinoma. Cancer Res. 82 (24), 4694. doi:10.1158/0008-5472.CAN-22-3444
Han, P., Zhao, J., and Gao, L. (2021). Increased serum exosomal long non-coding RNA SNHG15 expression predicts poor prognosis in non-small cell lung cancer. J. Clin. Lab. Anal. 35, e23979. doi:10.1002/jcla.23979
Han, R., Guo, H., Shi, J., Wang, H., Zhao, S., Jia, Y., et al. (2023). Tumour microenvironment changes after osimertinib treatment resistance in non-small cell lung cancer. Eur. J. Cancer 189, 112919. doi:10.1016/j.ejca.2023.05.007
Han, Z., Li, Y., Zhang, J., Guo, C., Li, Q., Zhang, X., et al. (2020). Tumor-derived circulating exosomal miR-342-5p and miR-574-5p as promising diagnostic biomarkers for early-stage Lung Adenocarcinoma. Int. J. Med. Sci. 17 (10), 1428–1438. doi:10.7150/ijms.43500
Hisakane, K., Seike, M., Sugano, T., Matsuda, K., Kashiwada, T., Nakamichi, S., et al. (2023). Serum-derived exosomal miR-125a-3p predicts the response to anti-programmed cell death-1/programmed cell death-ligand 1 monotherapy in patients with non-small cell lung cancer. Gene 857, 147177. doi:10.1016/j.gene.2023.147177
Hoshino, A., Costa-Silva, B., Shen, T. L., Rodrigues, G., Hashimoto, A., Tesic Mark, M., et al. (2015b). Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335. doi:10.1038/nature15756
Hoshino, A., Costa-Silva, B., Shen, T. L., Rodrigues, G., Hashimoto, A., Tesic Mark, M., et al. (2015a). Tumour exosome integrins determine organotropic metastasis. Nature 527 (7578), 329–335. doi:10.1038/nature15756
Hsu, Y. L., Hung, J. Y., Chang, W. A., Lin, Y. S., Pan, Y. C., Tsai, P. H., et al. (2017). Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene 36 (34), 4929–4942. doi:10.1038/onc.2017.105
Huang, M., Li, T., Wang, Q., Li, C., Zhou, H., Deng, S., et al. (2021). Silencing circPVT1 enhances radiosensitivity in non-small cell lung cancer by sponging microRNA-1208. Cancer Biomark. 31 (3), 263–279. doi:10.3233/CBM-203252
Huang, W., Yan, Y., Liu, Y., Lin, M., Ma, J., Zhang, W., et al. (2020). Exosomes with low miR-34c-3p expression promote invasion and migration of non-small cell lung cancer by upregulating integrin α2β1. Signal Transduct. Target Ther. 5, 39. doi:10.1038/s41392-020-0133-y
Hussain, M. S., Afzal, O., Gupta, G., Goyal, A., Almalki, W. H., Kazmi, I., et al. (2024). Unraveling NEAT1's complex role in lung cancer biology: a comprehensive review. EXCLI J. 23, 34–52. doi:10.17179/excli2023-6553
Janpipatkul, K., Trachu, N., Watcharenwong, P., Panvongsa, W., Worakitchanon, W., Metheetrairut, C., et al. (2021). Exosomal microRNAs as potential biomarkers for osimertinib resistance of non-small cell lung cancer patients. Cancer Biomark. 31, 281–294. doi:10.3233/CBM-203075
Jin, X., Chen, Y., Chen, H., Fei, S., Chen, D., Cai, X., et al. (2017). Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin. Cancer Res. 23 (17), 5311–5319. doi:10.1158/1078-0432.CCR-17-0577
Kalluri, R. (2016). The biology and function of exosomes in cancer. J. Clin. Invest 126 (4), 1208–1215. doi:10.1172/JCI81135
Kalluri, R., and LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science 367 (6478), eaau6977. doi:10.1126/science.aau6977
Kaźmierczak, D., Eide, I. J. Z., Gencheva, R., Lai, Y., Lewensohn, R., Tsakonas, G., et al. (2022). Elevated expression of miR-494-3p is associated with resistance to osimertinib in EGFR T790M-positive non-small cell lung cancer. Transl. Lung Cancer Res. 11, 722–734. doi:10.21037/tlcr-21-955
Kim, D. H., Park, H., Choi, Y. J., Kang, M. H., Kim, T. K., Pack, C. G., et al. (2021). Exosomal miR-1260b derived from non-small cell lung cancer promotes tumor metastasis through the inhibition of HIPK2. Cell Death Dis. 12, 747. doi:10.1038/s41419-021-04024-9
Kim, D. H., Park, S., Kim, H., Choi, Y. J., Sung, K. J., Kim, Y., Sung, K. j., et al. (2020). Tumor-derived exosomal miR-619-5p promotes tumor angiogenesis and metastasis through the inhibition of RCAN1.4. Cancer Lett. 475, 2–13. doi:10.1016/j.canlet.2020.01.023
Kim, J. (2022). Cell dissemination in pancreatic cancer. Cells 11 (22), 3683. doi:10.3390/cells11223683
Kim, J. W., Wieckowski, E., Taylor, D. D., Reichert, T. E., Watkins, S., and Whiteside, T. L. (2005). Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin. Cancer Res. 11, 1010–1020. doi:10.1158/1078-0432.1010.11.3
Kimiz-Gebologlu, I., and Oncel, S. S. (2022). Exosomes: large-scale production, isolation, drug loading efficiency, and biodistribution and uptake. J. Control Release 347, 533–543. doi:10.1016/j.jconrel.2022.05.027
Kogure, T., Lin, W. L., Yan, I. K., Braconi, C., and Patel, T. (2011). Inter-cellular nanovesicle mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology 54 (4), 1237–1248. doi:10.1002/hep.24504
Krug, A. K., Enderle, D., Karlovich, C., Priewasser, T., Bentink, S., Spiel, A., et al. (2018). Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann. Oncol. 29 (3), 700–706. doi:10.1093/annonc/mdx765
Krylova, S. V., and Feng, D. (2023). The machinery of exosomes: biogenesis, release, and uptake. Int. J. Mol. Sci. 24, 1337. doi:10.3390/ijms24021337
Kwok, H. H., Ning, Z., Chong, P. W., Wan, T. S., Ng, M. H., Ho, G. Y. F., et al. (2019). Transfer of extracellular vesicle-associated-RNAs induces drug resistance in ALK-translocated lung adenocarcinoma. Cancers (Basel) 11 (1), 104. doi:10.3390/cancers11010104
Lee, E., Kim, G., Jang, S. Y., Choi, Y. H., Yu, R. L., Tak, W. Y., et al. (2017). Abstract 3504: high expression of lncRNA-ATB and miR-21 as a biomarker in human hepatocellular carcinoma. Canc. Res. 77 (13 Suppl. ment), 3504. doi:10.1158/1538-7445.am2017-3504
Lei, Y., Guo, W., Chen, B., Chen, L., Gong, J., and Li, W. (2018). Tumorreleased lncRNA H19 promotes gefitinib resistance via packaging into exosomes in nonsmall cell lung cancer. Oncol. Rep. 40, 3438–3446. doi:10.3892/or.2018.6762
Levy, B., Hu, Z. I., Cordova, K. N., Close, S., Lee, K., and Becker, D. (2016). Clinical utility of liquid diagnostic platforms in non-small cell lung cancer. Oncologist 21 (9), 1121–1130. doi:10.1634/theoncologist.2016-0082
Li, C., Li, C., Zhi, C., Liang, W., Wang, X., Chen, X., et al. (2019). Clinical significance of PD-L1 expression in serum-derived exosomes in NSCLC patients. J. Transl. Med. 17 (1), 355. doi:10.1186/s12967-019-2101-2
Li, D., Chu, X., Ma, Y., Zhang, F., Tian, X., Yang, Y., et al. (2024a). Tumor-derived exosomes: unravelling the pathogenesis of pancreatic cancer with liver metastases and exploring the potential for clinical translation. Cancer Lett. 611, 217403. doi:10.1016/j.canlet.2024.217403
Li, J., Zhou, W., Wang, H., Huang, M., and Deng, H. (2024b). Exosomal circular RNAs in tumor microenvironment: an emphasis on signaling pathways and clinical opportunities. MedComm 5 (12), e70019. doi:10.1002/mco2.70019
Li, M., Liu, T., Cheng, W., Jin, H., and Wang, X. (2023). A test of miR-128-3p and miR-33a-5p in serum exosome as biomarkers for auxiliary diagnosis of non-small cell lung cancer. J. Thorac. Dis. 15, 2616–2626. doi:10.21037/jtd-23-398
Li, Y., Zhang, Y., Qiu, F., and Qiu, Z. (2011). Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis 32 (15), 1976–1983. doi:10.1002/elps.201000598
Liao, H., Wang, Z., Deng, Z., Ren, H., and Li, X. (2015). Curcumin inhibits lung cancer invasion and metastasis by attenuating GLUT1/MT1-MMP/MMP2 pathway. Int. J. Clin. Exp. Med. 8, 8948–8957.
Lin, H., Li, J., Wang, M., Zhang, X., and Zhu, T. (2023). Exosomal long noncoding RNAs in NSCLC: dysfunctions and clinical potential. J. Cancer 14, 1736–1750. doi:10.7150/jca.84506
Lin, S., He, C., Song, L., Sun, L., Zhao, R., Min, W., et al. (2024a). Exosomal lncCRLA is predictive for the evolvement and development of lung adenocarcinoma. Cancer Lett. 582, 216588. doi:10.1016/j.canlet.2023.216588
Lin, S., He, C., Song, L., Sun, L., Zhao, R., Min, W., et al. (2024b). Exosomal lncCRLA is predictive for the evolvement and development of lung adenocarcinoma. Cancer Lett. 582, 216588. doi:10.1016/j.canlet.2023.216588
Lin, Z., Wu, Y., Xu, Y., Li, G., Li, Z., and Liu, T. (2022). Mesenchymal stem cell-derived exosomes in cancer therapy resistance: recent advances and therapeutic potential. Mol. Cancer 21 (1), 179. doi:10.1186/s12943-022-01650-5
Liu, B., Zhang, R., Zhu, Y., and Hao, R. (2021). Exosome-derived microRNA-433 inhibits tumorigenesis through incremental infiltration of CD4 and CD8 cells in non-small cell lung cancer. Oncol. Lett. 22, 607. doi:10.3892/ol.2021.12868
Liu, C., Hu, C., Chen, T., Jiang, Y., Zhang, X., Liu, H., et al. (2022). The role of plasma exosomal lnc-SNAPC5-3:4 in monitoring the efficacy of anlotinib in the treatment of advanced non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 148, 2867–2879. doi:10.1007/s00432-022-04071-5
Liu, L., Qu, W., and Zhong, Z. (2015). Down-regulation of miR-503 expression predicate advanced mythological features and poor prognosis in patients with NSCLC. Int. J. Clin. Exp. Pathol. 8 (5), 5609–5613. PMID: 26191272; PMCID: PMC4503143.
Liu, Q., Yu, Z., Yuan, S., Xie, W., Li, C., Hu, Z., et al. (2017). Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 8 (8), 13048–13058. doi:10.18632/oncotarget.14369
Liu, Y., Wang, Y., Lv, Q., and Li, X. (2020). Exosomes: from garbage bins to translational medicine. Int. J. Pharm. 583, 119333. doi:10.1016/j.ijpharm.2020.119333
Liu, Z. L., Chen, H. H., Zheng, L. L., Sun, L. P., and Shi, L. (2023). Angiogenic signaling pathways and anti-angiogenic therapy for cancer. Signal Transduct. Target Ther. 8 (1), 198. doi:10.1038/s41392-023-01460-1
Luo, R., Liu, H., and Chen, J. (2021). Reduced circulating exosomal miR-382 predicts unfavorable outcome in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 14, 469–474.
Luo, X., Li, Y., Hua, Z., Xue, X., Wang, X., Pang, M., et al. (2023). Exosomes-mediated tumor metastasis through reshaping tumor microenvironment and distant niche. J. Control Release 353, 327–336. doi:10.1016/j.jconrel.2022.11.050
Ma, L., Guo, H., Zhao, Y., Liu, Z., Wang, C., Bu, J., et al. (2024). Liquid biopsy in cancer: current status, challenges and future prospects. Signal Transduct. Target Ther. 9, 336. doi:10.1038/s41392-024-02021-w
Ma, Z., Wei, K., Yang, F., Guo, Z., Pan, C., He, Y., et al. (2021). Tumor-derived exosomal miR-3157-3p promotes angiogenesis, vascular permeability and metastasis by targeting TIMP/KLF2 in non-small cell lung cancer. Cell Death Dis. 12, 840. doi:10.1038/s41419-021-04037-4
Mederer, T., Elsner, F., Robold, T., Großer, C., Neu, R., Ried, M., et al. (2022). EpCAM-positive disseminated cancer cells in bone marrow impact on survival of early-stage NSCLC patients. Lung Cancer 167, 73–77. doi:10.1016/j.lungcan.2022.02.008
Meng, W., Hao, Y., He, C., Li, L., and Zhu, G. (2019). Exosome-orchestrated hypoxic tumor microenvironment. Mol. Cancer 18 (1), 57. doi:10.1186/s12943-019-0982-6
Milane, L., Singh, A., Mattheolabakis, G., Suresh, M., and Amiji, M. M. (2015). Exosome mediated communication within the tumor microenvironment. J. Control Release 219, 278–294. doi:10.1016/j.jconrel.2015.06.029
Min, K., Ren, M., Yan, L., Fu, Y., Deng, M., and Li, C. (2018). Exosome-mediated transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell carcinoma via functioning as a competing endogenous RNA. J. Exp. Clin. Canc. Res. 37 (1), 171. doi:10.1186/s13046-018-0845-9
Min, L., Zhu, T., Lv, Bo, An, T., Zhang, Q., Shang, Y., et al. (2022a). Exosomal LncRNA RP5-977B1 as a novel minimally invasive biomarker for diagnosis and prognosis in non-small cell lung cancer. Int. J. Clin. Oncol. 27 (6), 1013–1024. doi:10.1007/s10147-022-02129-5
Min, L., Zhu, T., Lv, B., An, T., Zhang, Q., Shang, Y., et al. (2022b). Exosomal LncRNA RP5-977B1 as a novel minimally invasive biomarker for diagnosis and prognosis in non-small cell lung cancer. Int. J. Clin. Oncol. 27, 1013–1024. doi:10.1007/s10147-022-02129-5
Molina-Vila, M. A., Mayo-de-Las-Casas, C., Giménez-Capitán, A., Jordana-Ariza, N., Garzón, M., Balada, A., et al. (2016). Liquid biopsy in non-small cell lung cancer. Front. Med. (Lausanne) 3, 69. doi:10.3389/fmed.2016.00069
Mortezaee, K., Majidpoor, J., and Fathi, F. (2022). Extracellular vesicle isolation, purification and evaluation in cancer diagnosis. Expert Rev. Mol. Med. 24, e41–e44. doi:10.1017/erm.2022.34
Najafi, S. (2022). The emerging roles and potential applications of circular RNAs in ovarian cancer: a comprehensive review. J. Cancer Res. Clin. Oncol. 149, 2211–2234. doi:10.1007/s00432-022-04328-z
Nazarenko, I. (2020). Extracellular vesicles: recent developments in technology and perspectives for cancer liquid biopsy. Recent Results Cancer Res. 215, 319–344. doi:10.1007/978-3-030-26439-0_17
Ni, J., Zhang, X., Li, J., Zheng, Z., Zhang, J., Zhao, W., et al. (2021). Tumour-derived exosomal lncRNA-SOX2OT promotes bone metastasis of non-small cell lung cancer by targeting the miRNA-194-5p/RAC1 signalling axis in osteoclasts. Cell death and Dis. 12, 662. doi:10.1038/s41419-021-03928-w
Niloufar, O., Fadaee, M., Kazemi, T., and Yousefi, B. (2024). Exosome therapeutics for non-small cell lung cancer tumorigenesis. Cancer Cell Int. 24 (1), 360. doi:10.1186/s12935-024-03544-6
Niu, L., Song, X., Wang, N., Xue, L., Song, X., and Xie, L. (2019). Tumor-derived exosomal proteins as diagnostic biomarkers in non-small cell lung cancer. Cancer Sci. 110, 433–442. doi:10.1111/cas.13862
Obenauf, A. C., and Massagué, J. (2015). Surviving at a distance: organ specific metastasis. Trends Cancer 1 (1), 76–91. doi:10.1016/j.trecan.2015.07.009
Ochiai, R., Hayashi, K., Yamamoto, H., Fujii, R., Saichi, N., Shinchi, H., et al. (2022). Plasma exosomal DOK3 reflects immunological states in lung tumor and predicts prognosis of gefitinib treatment. Cancer Sci. 113, 3960–3971. doi:10.1111/cas.15512
Oh, H.-Ji, Imam-Aliagan, A. B., Kim, Y.-B., Kim, H. J., Izaguirre, I. A., Sung, C. K., et al. (2024). Clinical applications of circulating biomarkers in non-small cell lung cancer. Front. Cell Dev. Biol. 21 (12), 1449232. doi:10.3389/fcell.2024.1449232
Olejarz, W., Kubiak-Tomaszewska, G., Chrzanowska, A., and Lorenc, T. (2020). Exosomes in angiogenesis and anti-angiogenic therapy in cancers. Int. J. Mol. Sci. 21 (16), 5840. doi:10.3390/ijms21165840
Padinharayil, H., and George, A. (2024). Small extracellular vesicles: multi-functional aspects in non-small cell lung carcinoma. Crit. Rev. Oncol. Hematol. 198, 104341. doi:10.1016/j.critrevonc.2024.104341
Paget, S. (1889). The distribution of secondary growths in cancer of the breast. Lancet 133, 571–573. doi:10.1016/s0140-6736(00)49915-0
Pan, B. T., Teng, K., Wu, C., Adam, M., and Johnstone, R. M. (1985). Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 101, 942–948. doi:10.1083/jcb.101.3.942
Peng, X., Zhao, L., Yao, L., Dong, J., Wu, W., and Luo, T. (2023). Exosomal ERBB2IP contributes to tumor growth via elevating PSAT1 expression in non-small cell lung carcinoma. Thorac. Cancer 14, 1812–1823. doi:10.1111/1759-7714.14926
Peng, X. X., Yu, R., Wu, X., Wu, S. Y., Pi, C., Chen, Z. H., et al. (2020a). Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wildtype advanced non-small cell lung cancer. J. Immunother. Cancer 8, e000376. doi:10.1136/jitc-2019-000376
Peng, X. X., Yu, R., Wu, X., Wu, S. Y., Pi, C., Chen, Z. H., et al. (2020b). Correlation of plasma exosomal microRNAs with the efficacy of immunotherapy in EGFR/ALK wild-type advanced non-small cell lung cancer. J. Immunother. Cancer 8, e000376. doi:10.1136/jitc-2019-000376
Pérez-Ruiz, E., Melero, I., Kopecka, J., Sarmento-Ribeiro, A. B., García-Aranda, M., and De Las Rivas, J. (2020). Cancer immunotherapy resistance based on immune checkpoints inhibitors: targets, biomarkers, and remedies. Drug Resist Updat 53, 100718. doi:10.1016/j.drup.2020.100718
Qiao, Z., Zhang, Y., Ge, M., Liu, S., Jiang, X., Shang, Z., et al. (2019). Cancer cell derived small extracellular vesicles contribute to recipient cell metastasis through promoting HGF/c-met pathway. Mol. Cell Proteomics 18, 1619–1629. doi:10.1074/mcp.RA119.001502
Rahman, M. A., Barger, J. F., Lovat, F., Gao, M., Otterson, G. A., and Nana-Sinkam, P. (2016). Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget 7 (34), 54852–54866. doi:10.18632/oncotarget.10243
Rana, S., and Zöller, M. (2011). Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochem. Soc. Trans. 39 (2), 559–562. doi:10.1042/BST0390559
Reck, M., Remon, J., and Hellmann, M. D. (2022). First-line immunotherapy for non-small-cell lung cancer. J. Clin. Oncol. 40, 586–597. doi:10.1200/JCO.21.01497
Rehulkova, A., Chudacek, J., Prokopova, A., Vidlarova, M., Stranska, J., Drabek, J., et al. (2023). Clinical and prognostic significance of detecting CEA, EGFR, LunX, c-met and EpCAM mRNA-positive cells in the peripheral blood, tumor-draining blood and bone marrow of non-small cell lung cancer patients. Transl. Lung Cancer Res. 12, 1034–1050. doi:10.21037/tlcr-22-801
Ren, Z., Cao, J., Ding, L., Xin, L., and Yan, Z. (2024). Exosomes in esophageal cancer: a promising frontier for liquid biopsy in diagnosis and therapeutic monitoring. Front. Pharmacol. 15, 1459938. doi:10.3389/fphar.2024.1459938
Rezaie, J., Feghhi, M., and Etemadi, T. (2022). A review on exosomes application in clinical trials: perspective, questions, and challenges. Cell Commun. Signal 20, 145. doi:10.1186/s12964-022-00959-4
Ridder, K., Sevko, A., Heide, J., Dams, M., Rupp, A. K., Macas, J., et al. (2015). Extracellular vesicle-mediated transfer of functional RNA in the tumor microenvironment. Oncoimmunology 4 (6), e1008371. doi:10.1080/2162402X.2015.1008371
Rizwan, M. N., Ma, Y., Nenkov, M., Jin, L., Schröder, D. C., Westermann, M., et al. (2022). Tumor-derived exosomes: key players in non-small cell lung cancer metastasis and their implication for targeted therapy. Mol. Carcinog. 61, 269–280. doi:10.1002/mc.23378
Rodrigues, G., Hoshino, A., Kenific, C. M., Matei, I. R., Steiner, L., Freitas, D., et al. (2019). Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat. Cell Biol. 21, 1403–1412. doi:10.1038/s41556-019-0404-4
Rolfo, C., Mack, P., Scagliotti, G. V., Aggarwal, C., Arcila, M. E., Barlesi, F., et al. (2021). Liquid biopsy for advanced NSCLC: a consensus statement from the international association for the study of lung cancer. J. Thorac. Oncol. 16, 1647–1662. doi:10.1016/j.jtho.2021.06.017
Saliminejad, K., Khorram Khorshid, H. R., Soleymani Fard, S., and Ghaffari, S. H. (2019). An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 234, 5451–5465. doi:10.1002/jcp.27486
Sanchez-Cabrero, D., Garcia-Guede, Á., Burdiel, M., Pernía, O., Colmenarejo-Fernandez, J., Gutierrez, L., et al. (2023). miR-124 as a liquid biopsy prognostic biomarker in small extracellular vesicles from NSCLC patients. Int. J. Mol. Sci. 24, 11464. doi:10.3390/ijms241411464
Sandfeld-Paulsen, B., Aggerholm-Pedersen, N., Bæk, R., Jakobsen, K. R., Meldgaard, P., Folkersen, B. H., et al. (2016). Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol. Oncol. 10 (10), 1595–1602. doi:10.1016/j.molonc.2016.10.003
Sandúa, A., Alegre, E., and González, Á. (2021). Exosomes in lung cancer: actors and heralds of tumor development. Cancers (Basel) 13 (17), 4330. doi:10.3390/cancers13174330
Shanehbandi, D., Asadi, M., Seyedrezazadeh, E., Zafari, V., Shekari, N., Akbari, M., et al. (2023). MicroRNA-based biomarkers in lung cancer: recent advances and potential applications. Curr. Mol. Med. 23, 648–667. doi:10.2174/2772432817666220520085719
Sharma, P., and Allison, J. P. (2015). Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 161 (2), 205–214. doi:10.1016/j.cell.2015.03.030
Shedden, K., Xie, X. T., Chandaroy, P., Chang, Y. T., and Rosania, G. R. (2003). Expulsion of small molecules in vesicles shed by cancer cells: association with gene expression and chemosensitivity profiles. Cancer Res. 63, 4331–4337.
Shi, Q., Ji, T., Ma, Z., Tan, Q., and Liang, J. (2023). Serum exosomes-based biomarker circ_0008928 regulates cisplatin sensitivity, tumor progression, and glycolysis metabolism by miR-488/HK2 Axis in cisplatin-resistant nonsmall cell lung carcinoma. Cancer Biother Radiopharm. 38, 558–571. doi:10.1089/cbr.2020.4490
Shields, M. D., Chen, K., Dutcher, G., Patel, I., and Pellini, B. (2022). Making the rounds: exploring the role of circulating tumor DNA (ctDNA) in non-small cell lung cancer. Int. J. Mol. Sci. 23, 9006. doi:10.3390/ijms23169006
Shimada, Y., Matsubayashi, J., Kudo, Y., Maehara, S., Takeuchi, S., Hagiwara, M., et al. (2021). Serum-derived exosomal PD-L1 expression to predict anti-PD-1 response and in patients with non-small cell lung cancer. Sci. Rep. 11, 7830. doi:10.1038/s41598-021-87575-3
Siedlecki, E., Remiszewski, P., and Stec, R. (2024). The role of circHIPK3 in tumorigenesis and its potential as a biomarker in lung cancer. Cells 13 (17), 1483. doi:10.3390/cells13171483
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2022. CA Cancer J. Clin. 72, 7–33. doi:10.3322/caac.21708
Song, Q., Yu, H., Cheng, Y., Han, J., Li, K., Zhuang, J., et al. (2022). Bladder cancer-derived exosomal KRT6B promotes invasion and metastasis by inducing EMT and regulating the immune microenvironment. J. Transl. Med. 20 (1), 308. doi:10.1186/s12967-022-03508-2
Su, C., Zhang, J., Yarden, Y., and Fu, L. (2021). The key roles of cancer stem cell-derived extracellular vesicles. Signal Transduct. Target Ther. 6, 109. doi:10.1038/s41392-021-00499-2
Talebi, S., Abadi, A. J., Kazemioula, G., Hosseini, N., Taheri, F., Pourali, S., et al. (2022). Expression analysis of five different long non-coding ribonucleic acids in nonsmall-cell lung carcinoma tumor and tumor-derived exosomes. Diagn. (Basel) 12, 3209. doi:10.3390/diagnostics12123209
Tang, L., Pei, H., Yang, Y., Wang, X., Wang, T., Gao, E., et al. (2016). The inhibition of calpains ameliorates vascular restenosis through MMP2/TGF-β1 pathway. Sci. Rep. 6, 29975. doi:10.1038/srep29975
Tang, T., Yang, T., Xue, H., Liu, X., Yu, J., Liang, C., et al. (2024). Breast cancer stem cell-derived exosomal lnc-PDGFD induces fibroblast-niche formation and promotes lung metastasis. Oncogene 44, 601–617. doi:10.1038/s41388-024-03237-4
Théry, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (Misev2018): a position statement of the international society for extracellular vesicles and update of the Misev2014 guidelines. J. Extracell. Vesicles 7 (1), 1535750. doi:10.1080/20013078.2018.1535750
Théry, C., Zitvogel, L., and Amigorena, S. (2002). Exosomes: composition, biogenesis andfunction. Nat. Rev. Immunol. 2 (8), 569–579. doi:10.1038/nri855
Valadi, H., Ekström, K., Bossios, A., Sjöstrand, M., Lee, J. J., and Lötvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. cell Biol. 9, 654–659. doi:10.1038/ncb1596
van der Pol, E., Böing, A. N., Harrison, P., Sturk, A., and Nieuwland, R. (2012). Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 64 (3), 676–705. doi:10.1124/pr.112.005983
Vasu, S., Johnson, V., Archana, M., Reddy, K. A., and Sukumar, U. K. (2025). Circulating extracellular vesicles as promising biomarkers for precession diagnostics: a perspective on lung cancer. ACS Biomater. Sci. Eng. 11 (1), 95–134. doi:10.1021/acsbiomaterials.4c01323
Visan, K. S., Lobb, R. J., Wen, S. W., Bedo, J., Lima, L. G., Krumeich, S., et al. (2022). Blood-derived extracellular vesicle-associated miR-3182 detects non-small cell lung cancer patients. Cancers (Basel) 14, 257. doi:10.3390/cancers14010257
Wang, N., Guo, W., Song, X., Liu, L., Niu, L., Song, X., et al. (2020). Tumor-associated exosomal miRNA biomarkers to differentiate metastatic vs. nonmetastatic non-small cell lung cancer. Clin. Chem. Lab. Med. 58, 1535–1545. doi:10.1515/cclm-2019-1329
Wang, N., Song, X., Liu, L., Niu, L., Wang, X., Song, X., et al. (2018). Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer. Cancer Sci. 109, 1701–1709. doi:10.1111/cas.13581
Wang, N., Yao, C., Luo, C., Liu, S., Wu, L., Hu, W., et al. (2023). Integrated plasma and exosome long noncoding RNA profiling is promising for diagnosing non-small cell lung cancer. Clin. Chem. Lab. Med. 61, 2216–2228. doi:10.1515/cclm-2023-0291
Wang, Z., Lin, M., He, L., Qi, H., Shen, J., and Ying, K. (2021). Exosomal lncRNA SCIRT/miR-665 transferring promotes lung cancer cell metastasis through the inhibition of HEYL. J. Oncol. 2021, 9813773. doi:10.1155/2021/9813773
Wei, S. L., Ye, J. J., Sun, L., Hu, L., Wei, Y. Y., Zhang, D. W., et al. (2023). Exosome-derived circKIF20B suppresses gefitinib resistance and cell proliferation in non-small cell lung cancer. Cancer Cell Int. 23, 129. doi:10.1186/s12935-023-02974-y
Whiteside, T. L. (2016a). Exosomes and tumor-mediated immune suppression. J. Clin. Invest 126 (4), 1216–1223. doi:10.1172/JCI81136
Whiteside, T. L. (2016b). Exosomes and tumor-mediated immune suppression. J. Clin. Invest 126, 1216–1223. doi:10.1172/JCI81136
Wood, S. L., Pernemalm, M., Crosbie, P. A., and Whetton, A. D. (2014). The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat. Rev. 40 (4), 558–566. doi:10.1016/j.ctrv.2013.10.001
Wu, D., Deng, S., Li, L., Liu, T., Zhang, T., Li, J., et al. (2021). TGF-β1-mediated exosomal lnc-MMP2-2 increases blood-brain barrier permeability via the miRNA-1207-5p/EPB41L5 axis to promote non-small cell lung cancer brain metastasis. Cell death and Dis. 12, 721. doi:10.1038/s41419-021-04004-z
Wu, D. M., Deng, S. H., Liu, T., Han, R., Zhang, T., and Xu, Y. (2018). TGF-β-mediated exosomal lnc-MMP2-2 regulates migration and invasion of lung cancer cells to the vasculature by promoting MMP2 expression. Cancer Med. 7, 5118–5129. doi:10.1002/cam4.1758
Xia, J., Luo, M., Dai, L., Wang, L., Wang, L., and Zhu, J. (2021). Serum exosomal microRNAs as predictive markers for EGFR mutations in non-small-cell lung cancer. J. Clin. Lab. Anal. 35, e23743. doi:10.1002/jcla.23743
Xia, L., Liu, Y., and Wang, Y. (2019). PD-1/PD-L1 Blockade therapy in advanced non-small-cell lung cancer: current status and future directions. Oncologist 24 (Suppl. 1), S31–S41. doi:10.1634/theoncologist.2019-IO-S1-s05
Xian, J., Zeng, Y., Chen, S., Lu, L., Liu, L., Chen, J., et al. (2021). Discovery of a novel linc01125 isoform in serum exosomes as a promising biomarker for NSCLC diagnosis and survival assessment. Carcinogenesis 42, 831–841. doi:10.1093/carcin/bgab034
Xie, F., Zhou, X., Fang, M., Li, H., Su, P., Tu, Y., et al. (2019). Extracellular vesicles in cancer immune microenvironment and cancer immunotherapy. Adv. Sci. 6 (24), 1901779. doi:10.1002/advs.201901779
Xie, H., Yao, J., Wang, Y., and Ni, B. (2022). Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Deliv. 29, 1257–1271. doi:10.1080/10717544.2022.2057617
Xiong, D., Wang, C., Yang, Z., Han, F., and Zhan, H. (2021). Clinical significance of serum-derived exosomal LINC00917 in patients with non-small cell lung cancer. Front. Genet. 12, 728763. doi:10.3389/fgene.2021.728763
Xu, Z., Zeng, S., Gong, Z., and Yan, Y. (2020). Exosome-based immunotherapy: a promising approach for cancer treatment. Mol. Cancer 19, 160. doi:10.1186/s12943-020-01278-3
Yang, B., Teng, F., Chang, L., Wang, J., Liu, D. L., Cui, Y. S., et al. (2021). Tumor-derived exosomal circRNA_102481 contributes to EGFR-TKIs resistance via the miR-30a-5p/ROR1 axis in non-small cell lung cancer. Aging (Albany NY) 13, 13264–13286. doi:10.18632/aging.203011
Yang, P., Zhang, Y., Zhang, R., Wang, Y., Zhu, S., Peng, X., et al. (2023). Plasma-derived exosomal immunoglobulins IGHV4-4 and IGLV1-40 as new non-small cell lung cancer biomarkers. Am. J. Cancer Res. 13, 1923–1937.
Yi, J., Ye, Z., Xu, H., Zhang, H., Cao, H., Li, X., et al. (2024). EGCG targeting STAT3 transcriptionally represses PLXNC1 to inhibit M2 polarization mediated by gastric cancer cell-derived exosomal miR-92b-5p. Phytomedicine 135, 156137. doi:10.1016/j.phymed.2024.156137
Yin, C., Li, J., Li, S., Yang, X., Lu, Y., Wang, C., et al. (2024a). LncRNA-HOXC-AS2 regulates tumor-associated macrophage polarization through the STAT1/SOCS1 and STAT1/CIITA pathways to promote the progression of non-small cell lung cancer. Cell Signal 115, 111031. doi:10.1016/j.cellsig.2023.111031
Yin, N., Li, X., Zhang, X., Xue, S., Cao, Y., Niedermann, G., et al. (2024b). Development of pharmacological immunoregulatory anti-cancer therapeutics: current mechanistic studies and clinical opportunities. Signal Transduct. Target Ther. 9 (1), 126. doi:10.1038/s41392-024-01826-z
Yu, D. D., Wu, Y., Shen, H. Y., Lv, M. M., Chen, W. X., Zhang, X. H., et al. (2015). Exosomes in development, metastasis and drug resistance of breast cancer. Cancer Sci. 106, 959–964. doi:10.1111/cas.12715
Yu, H., Boyle, T. A., Zhou, C., Rimm, D. L., and Hirsch, F. R. (2016). PD-L1 expression in lung cancer. J. Thorac. Oncol. 11 (7), 964–975. doi:10.1016/j.jtho.2016.04.014
Yuana, Y., Sturk, A., and Nieuwland, R. (2013). Extracellular vesicles in physiological and pathological conditions. Blood Rev. 27 (1), 31–39. doi:10.1016/j.blre.2012.12.002
Yue, S., Fang, Z., Wang, H., Liang, X., Wang, F., Wang, F., et al. (2024). Exosomal long non-coding RNA-LINC00839 promotes lung adenocarcinoma progression by activating NF-κB signaling pathway. Ann. Med. 56 (1), 2430029. doi:10.1080/07853890.2024.2430029
Zang, X., Gu, J., Zhang, J., Shi, H., Hou, S., Xu, X., et al. (2020). Exosome-transmitted lncRNA UFC1 promotes non-small-cell lung cancer progression by EZH2-mediated epigenetic silencing of PTEN expression. Cell Death Dis. 11, 215. doi:10.1038/s41419-020-2409-0
Zhang, C., Fan, Y., Che, X., Zhang, M., Li, Z., Li, C., et al. (2020b). Anti-PD-1 Therapy response predicted by the combination of exosomal PD-L1 and CD28. Front. Oncol. 10, 760. doi:10.3389/fonc.2020.00760
Zhang, J., Hu, Z., Horta, C. A., and Yang, J. (2022). Regulation of epithelial-mesenchymal transition by tumor microenvironmental signals and its implication in cancer therapeutics. Semin. Cancer Biol. 88, 46–66. doi:10.1016/j.semcancer.2022.12.002
Zhang, M., Wen, F., and Zhao, K. (2021). Circular RNA_0001946 is insufficiently expressed in tumor tissues, while its higher expression correlates with less lymph node metastasis, lower TNM stage, and improved prognosis in NSCLC patients. J. Clin. Lab. Anal. 35 (8), e23625. doi:10.1002/jcla.23625
Zhang, N., Nan, A., Chen, L., Li, X., Jia, Y., Qiu, M., et al. (2020a). Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol. Cancer 19, 101. doi:10.1186/s12943-020-01221-6
Zhang, Q., Zheng, K., Gao, Y., Zhao, S., Zhao, Y., Li, W., et al. (2023). Plasma exosomal miR-1290 and miR-29c-3p as diagnostic biomarkers for lung cancer. Heliyon 9, e21059. doi:10.1016/j.heliyon.2023.e21059
Zhang, R., Jiang, Y., Gu, J., Zhang, X., and Xie, Y. (2025). Diagnostic role of circulating long non-coding RNA LINC00312 in patients with non-small cell lung cancer: a retrospective study. BMC Cancer 25 (1), 47. doi:10.1186/s12885-024-13393-1
Zhang, R., Xia, Y., Wang, Z., Zheng, J., Chen, Y., Li, X., et al. (2017). Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 490 (2), 406–414. doi:10.1016/j.bbrc.2017.06.055
Zhang, W., He, P., Wang, S., Adili, A., Chen, Z., Zhang, C. Y., et al. (2019b). Characterization of protein profiling and mRNA expression of LLC exosomes. Protein J. 38, 586–597. doi:10.1007/s10930-019-09849-0
Zhang, X., Yuan, X., Shi, H., Wu, L., Qian, H., and Xu, W. (2015). Exosomes in cancer: small particle, big player. J. Hematol. Oncol. 8, 83. doi:10.1186/s13045-015-0181-x
Zhang, Y., Liu, Y., Liu, H., and Tang, W. H. (2019a). Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 9, 19. doi:10.1186/s13578-019-0282-2
Zhao, L., Ma, X., and Yu, J. (2021). Exosomes and organ-specific metastasis. Mol. Ther. Methods Clin. Dev. 22, 133–147. doi:10.1016/j.omtm.2021.05.016
Zheng, B., Peng, M., Gong, J., Li, C., Cheng, H., Li, Y., et al. (2023). Circulating exosomal microRNA-4497 as a potential biomarker for metastasis and prognosis in non-small-cell lung cancer. Exp. Biol. Med. (Maywood) 248, 1403–1413. doi:10.1177/15353702231184223
Keywords: exosomes, NSCLC, tumor progression and metastasis, tumor drug resistance, immune regulation
Citation: Gao Y, Xie J, Yang Z, Li M, Yuan H and Li R (2025) Functional tumor-derived exosomes in NSCLC progression and clinical implications. Front. Pharmacol. 16:1485661. doi: 10.3389/fphar.2025.1485661
Received: 20 September 2024; Accepted: 27 February 2025;
Published: 19 March 2025.
Edited by:
Slawomir Jakiela, Warsaw University of Life Sciences, PolandReviewed by:
Ketki Bhise, Xsphera Biosciences Inc., United StatesTarik Aanniz, Mohammed V University in Rabat, Morocco
Copyright © 2025 Gao, Xie, Yang, Li, Yuan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Li, bGlydWkxQHNjc3pseXkub3JnLmNu; Hongfan Yuan, eXVhbmhvbmdmYW5Ac2Nzemx5eS5vcmcuY24=
†ORCID: Rui Li, orcid.org/0009-0007-2684-1958
‡These authors have contributed equally to this work
 Yuxin Gao1‡
Yuxin Gao1‡ Rui Li
Rui Li