- 1Genetics and Bioinformatics Department, Dasman Diabetes Institute, Kuwait City, Kuwait
- 2Narcotic and Psychotropic Department, Ministry of Interior, Farwaniya, Kuwait
- 3College of Medical Veterinary and Life Sciences, University of Glasgow, Glasgow, United Kingdom
Objective: This study explores the frequency of human leukocyte antigen (HLA) genes, particularly HLA-B alleles, within the Kuwaiti population. We aim to identify alleles with known associations to adverse drug reactions (ADRs) based on existing literature. We focus on the HLA-B gene due to its well-documented associations with severe cutaneous adverse reactions and the extensive pharmacogenetic research supporting its clinical relevance.
Methods: We utilized the HLA-HD tool to extract, annotate, and analyse HLA-B alleles from the exome data of 561 Kuwaiti individuals, sequenced on the Illumina HiSeq platform. HLA typing was conducted using the HLA-HD tool with a reference panel from the IPD-IMGT/HLA database. The major HLA-B pharmacogenetic markers were obtained from the HLA Adverse Drug Reaction Database, focusing on alleles with significant ADR associations in published literature.
Results: The distribution of HLA-B alleles in the Kuwaiti population revealed that the most frequent alleles were HLA-B*50:01 (10.52%), HLA-B*51:01 (9.89%), HLA-B*08:01 (6.06%), HLA-B*52:01 (4.55%), HLA-B*18:01 (3.92%), and HLA-B*41:01 (3.65%). Notably, alleles HLA-B*13:01, HLA-B*13:02, HLA-B*15:02, HLA-B*15:13, HLA-B*35:02, HLA-B*35:05, HLA-B*38:01, HLA-B*40:02, HLA-B*44:03, HLA-B*51:01, HLA-B*57:01 and HLA-B*58:01 were identified with known associations to various ADRs. For example, HLA-B*51:01 was associated with clindamycin, phenobarbital, and phenytoin, and was found in 18% of individuals.
Conclusion: Our study enriches the regional genetic landscape by delineating HLA-B allele variations within Kuwait and across the Arabian Peninsula. This genetic insight, along with the identification of markers previously linked to drug hypersensitivity, provides a foundation for future pharmacogenetic research and potential personalized medicine strategies in the region.
Introduction
Adverse drug reactions (ADRs) manifesting as hypersensitivity drug reactions are significant health concerns, often leading to hospitalizations and fatalities (Lazarou et al., 1998; Pirmohamed et al., 2004; Davies et al., 2009). These reactions, triggered by various chemicals, involve the immune system, particularly delayed hypersensitivity responses mediated by T cells (Shapiro and Shear, 1996; Pichler, 2003).
The major histocompatibility complex (MHC), located on chromosome 6, plays a crucial role in both innate and adaptive immunity due to its high degree of polymorphism and linkage disequilibrium (The MHC sequencing consortium, 1999; Mungall et al., 2003). The human leukocyte antigen (HLA) system, a part of the MHC, consists of genes inherited from both parents, which are expressed on the surface of antigen-presenting cells. HLA molecules are classified into three classes (I, II, and III) based on their gene location, function, expression patterns, and biochemical properties (Howell et al., 2010). Class I molecules (HLA-A, HLA-B, HLA-C) present intracellular peptides to cytotoxic T cells (CD8+), while class II molecules (HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQB1, HLA-DRA, HLA-DRB1) present exogenous peptides to helper T cells (CD4+) (Dendrou et al., 2018).
The HLA-B gene, characterized by a high frequency of polymorphisms and complex linkage disequilibrium, is particularly challenging for traditional genotyping techniques. Next-generation sequencing (NGS) offers high-throughput and accurate HLA typing, essential for studying genetic diversity and phenotypic correlations worldwide (Claeys et al., 2023). Studies have identified HLA-B as a key genetic factor in ADRs, particularly severe cutaneous adverse reactions (SCARs) such as Stevens-Johnson syndrome (SJS), toxic epidermal necrolysis (TEN), and drug rash with eosinophilia and systemic symptoms (DRESS) (Jantararoungtong et al., 2021; Kloypan et al., 2021). For instance, HLA-B*15:02 is linked to carbamazepine-induced SJS/TEN (Ferrell and McLeod, 2008; Chang et al., 2011; Wei et al., 2012), and HLA-B*58:01 is associated with allopurinol-induced SCARs (Gonçalo et al., 2013). Screening for these alleles before prescribing medications can significantly reduce severe reactions, underscoring the clinical utility of pharmacogenetic testing (Chen et al., 2018).
Our focus on the HLA-B gene is based on the extensive body of pharmacogenetic research available. According to the HLA Adverse Drug Reaction Database (HLA-ADR) on the Allele Frequency Net Database (allelefrequencies.net), HLA-B alleles are more extensively studied compared to HLA-A and HLA-C genes. Other studies have similarly focused on the HLA-B gene in various populations due to its strong associations with pharmacogenomics (Koomdee et al., 2022; Yuliwulandari et al., 2024) and immunogenetics (Sajulga et al., 2022; Darbas et al., 2023). The Arabian Peninsula populations are underrepresented in global studies, and data on HLA-B allele frequencies can aid in understanding drug hypersensitivity in these populations (Arnaiz-Villena et al., 2019; Jawdat et al., 2020; Alfraih et al., 2021; Hajjej et al., 2020; Albalushi et al., 2014; Dashti et al., 2022; Ameen et al., 2020).
The latest attempt to explore HLA-B alleles in the Kuwaiti population was conducted by Ameen et al. (2020), focusing on reporting the most frequent alleles of classical HLA class I and class II genes using low-resolution typing. The most common group of HLA-B alleles reported was B*50:01, with a frequency of 12% (Ameen et al., 2020). Additionally, neither the study by Ameen et al. (2020) nor other studies have explored HLA-B alleles as pharmacogenetic markers in Kuwait (Moussa et al., 1985; Al-Bader et al., 2019)
Therefore, this study aims to explore the frequency of HLA-B alleles in the Kuwaiti population using high-resolution typing and to identify alleles with known associations to ADRs based on existing literature. Our goal is to determine the prevalence of these pharmacogenetically relevant HLA-B alleles in Kuwait and compare our data with those from other Gulf countries, contributing to a foundational understanding that may inform future personalized medicine initiatives in the region.
In a previous study, we ranked NGS-based HLA typing tools, focusing on those that are alignment-based and utilize the genetic diversity catalogued in the IPD-IMGT/HLA database (Robinson et al., 2020) for accurate allele calling. Our ranking was based on multiple independent benchmarking studies (Chen et al., 2021; Thuesen et al., 2022; Claeys et al., 2023), where we prioritized the top tools based on their performance (Dashti et al., 2024). We then evaluated the computational efficiency and capabilities of these top HLA typing tools on whole exome sequencing (WES) data, identifying HLA-HD (Kawaguchi et al., 2017) as one of the top performers. Additionally, we compared the performance of the HLA-HD tool against clinical grade HLA typing tool using various NGS datasets, confirming its reliability and consistency across multiple HLA loci (Dashti et al., 2024).
This work provides a solid foundation for using the HLA-HD tool in our current research, ensuring that our findings are both accurate and relevant to population-scale studies of HLA-B allele frequencies and their potential implications for drug hypersensitivity.
Methods and materials
Ethics Statement
The study was approved by the Ethical Review Committee at Dasman Diabetes Institute in Kuwait, in accordance with the guidelines outlined in the Declaration of Helsinki. The project reference number is RAHM 2019-025.
Study samples
Whole exome sequence data from 561 Kuwaiti individuals used in this study were sequenced on the Illumina HiSeq platform using the TruSeq Exome Enrichment kit and the Nextera Rapid Capture Exome kit (Illumina Inc., United States). A total of 561 Kuwaitis, including 271 males and 290 females, with an average age of 52 years, participated in the study. All participants provided informed consent prior to recruitment. These samples are part of an ongoing project on the Kuwaiti population aimed at capturing the extent of exome variation within the population, involving a larger cohort than previously reported (John et al., 2018). All participants were healthy and free of Mendelian or rare genetic disorders. For more details about the sequencing protocol used in the initial phase of this project, please refer to John et al., 2018.
HLA-B typing
Raw sequencing data in BCL format obtained from the Illumina sequencing platform were converted to Fastq format using the bcl2fastq v2.20 Conversion Software (Illumina, United States). The converted raw paired-end reads of 561 Kuwaiti individuals were then processed with the HLA-HD tool version 1.4.0 (Kawaguchi et al., 2017) to determine the HLA-B alleles. This was achieved by mapping the reads to the relevant region of the human genome reference using Bowtie 2 tool version 2.5.2 (Langmead and Salzberg, 2012). A comprehensive reference panel from the IPD-IMGT/HLA database version 3.46 (accessible at http://hla.alleles.org and https://www.ebi.ac.uk/ipd/imgt/hla/licence/) (Robinson et al., 2020) was used for genomic imputation, and a score based on weighted read counts was calculated to select the most suitable pair of alleles.
HLA-B pharmacogenomic markers
The major HLA-B pharmacogenetic markers were obtained from the HLA Adverse Drug Reaction Database website (http://www.allelefrequencies.net/) using a p-value filter of <0.01 across all ethnicities (accessed on 15 July 2024). Given that the database is continually updated by researchers, a comprehensive manual review was performed to identify relevant markers. This review aimed to confirm the association of each marker with drug hypersensitivity, ensuring they met the criteria of being risk alleles, having passed multivariate analysis with significant adjusted p-values, and being correctly typed. This process resulted in the identification of 17 unique HLA-B alleles associated with pharmacogenetic risk: HLA-B*13:01, HLA-B*13:02, HLA-B*15:02, HLA-B*15:11, HLA-B*15:13, HLA-B*15:27, HLA-B*35:02, HLA-B*35:05, HLA-B*38:01, HLA-B*39:05, HLA-B*40:02, HLA-B*44:03, HLA-B*51:01, HLA-B*57:01, HLA-B*58:01, HLA-B*58:05, and HLA-B*59:01.
Comparison of HLA-B top alleles with Arab Gulf countries and other ethnic groups
In addition to analysing the HLA-B allele frequencies within the Kuwaiti population, we compared these frequencies with those reported in other Arab Gulf countries and in various continental ethnic groups.
For the regional comparison, we utilized published literature on HLA-B alleles in Gulf countries, including Saudi Arabia (Jawdat et al., 2020), Qatar (Dashti et al., 2022), Bahrain (Hajjej et al., 2020), the United Arab Emirates (Arnaiz-Villena et al., 2019), and Oman (Albalushi et al., 2014). We extracted and compared the 10 most frequent HLA-B alleles in each population with the top 10 most frequent HLA-B alleles identified in the Kuwaiti population.
For the broader comparison with other ethnic groups, we utilized the Allele Frequency Net Database (accessed on 13 August 2024). This database provides comprehensive allele frequency data from a variety of ethnic groups. We queried the top 10 frequent HLA-B alleles in the Kuwaiti population and compared them with those in regions such as Europe, North Africa, North America, South Asia, Western Asia, and Sub-Saharan Africa. The data sources were filtered based on literature, and the study type was set to anthropology. We sorted the studies based on cohort size, selecting the most representative studies for each region. In cases where a specific allele was not investigated in the primary study, we used the next best study by cohort size for our comparative analysis.
Statistical analysis
The HLA-B allele frequencies were calculated by manually counting the occurrences of each allele and dividing them by the total number of HLA-B alleles in the cohort. For a diploid cohort, this total is twice the number of individuals, as each individual has two HLA-B alleles.
To assess the deviation from Hardy-Weinberg equilibrium (HWE), we utilized the R software, version 3.6.2 (R Core Team, 2023). The observed genotype frequencies were compared to the expected frequencies under HWE assumptions. Expected genotype counts were estimated based on the observed allele frequencies, while the actual genotype counts represented the genotypes observed in the cohort. Genotype frequencies were calculated as the proportion of each genotype among the total number of observed genotypes. The significance of the deviation from HWE was evaluated using p-values, with a threshold of p < 0.05 indicating significant deviation.
For the comparison of the most frequent HLA-B alleles in the Kuwaiti population with those in other ethnic groups, we calculated 95% confidence intervals using the R software. The confidence intervals were derived based on the extracted allele frequencies and the corresponding sample sizes of each ethnic group. This statistical approach allowed us to determine the range within which the true allele frequency is likely to fall, with a 95% level of confidence. Comparison of allele frequencies, along with their confidence intervals, was then visualized using stacked bar chart generated in R, facilitating the assessment of genetic similarities and differences across the regions.
Results
HLA-B allele frequencies
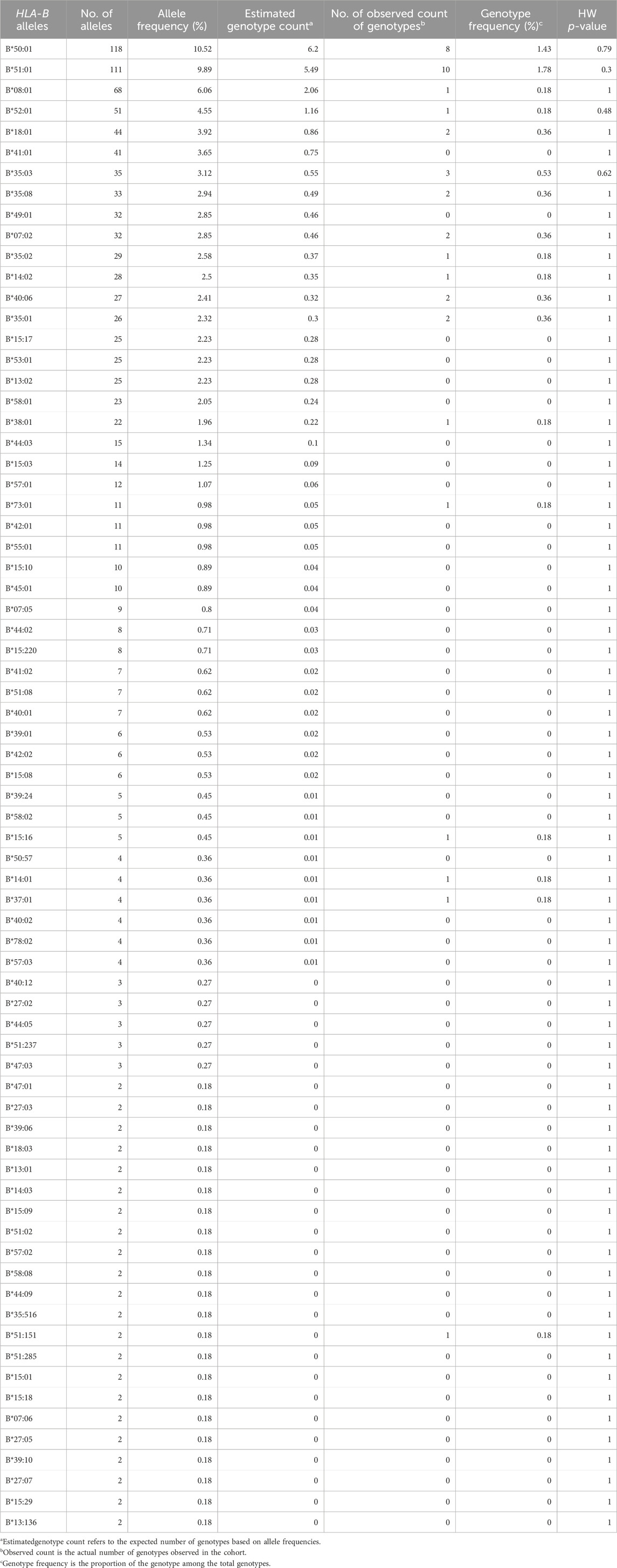
In total, we identified 160 unique HLA-B alleles in our study of 561 Kuwaiti individuals (Supplementary Table S1). All the identified HLA-B alleles observed in more than one individual (n > 1) are presented in Table 1. The frequency of the observed 143 distinct HLA-B alleles among the 561 Kuwaiti individuals is listed in Table 1. The most frequent HLA-B alleles identified were HLA-B*50:01 (10.52%), HLA-B*51:01 (9.89%), HLA-B*08:01 (6.06%), HLA-B*52:01 (4.55%), HLA-B*18:01 (3.92%), and HLA-B*41:01 (3.65%). The HLA-B alleles passed the quality control for HWE >10−3.
HLA-B genotype frequencies
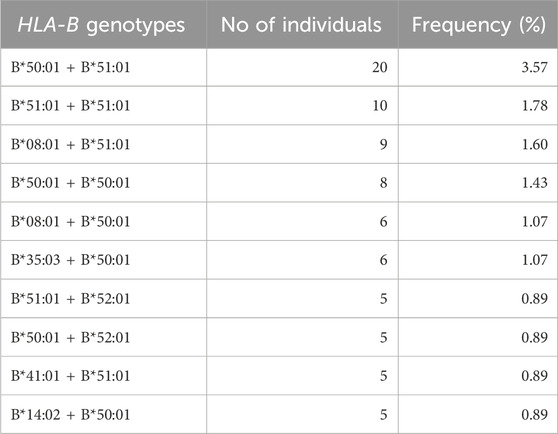
Examining the HLA-B genotypes of 561 Kuwaiti individuals revealed 370 distinct genotypes in total. The most frequently observed genotype among the population, as listed in Table 2, was B*50:01 + B*51:01, which was the most common at a rate of 3.57%. The frequencies of the rest of the frequent genotypes were under 3% in the Kuwaiti population.
Prevalence of HLA-B pharmacogenomic markers in the Kuwaiti population
We identified twelve HLA-B pharmacogenetic markers that associated with ADRs in 235 of the 561 Kuwaiti individuals (41.1%) (Table 3). The most prevalent pharmacogenetic markers were HLA-B*51:01, found in 18% of individuals and associated with phenytoin, phenobarbital, carbamazepine, and clindamycin (Niihara et al., 2012; Kaniwa et al., 2013; Manuyakorn et al., 2020; John et al., 2021), HLA-B*35:02, present in 5% and associated with minocycline (Urban et al., 2017), HLA-B*13:02, present in 4.5% and associated with allopurinol, lamotrigine, and oxcarbazepine (He et al., 2012; Kim et al., 2017; Wu et al., 2018), and HLA-B*58:01, present in 4.1% and associated with allopurinol (Lonjou et al., 2008; Gonçalo et al., 2013; Sukasem et al., 2016; Fontana et al., 2021). Other identified markers included HLA-B*38:01 (3.9%), associated with lamotrigine and other aromatic antiepileptic drugs (Ramírez et al., 2017), HLA-B*44:03 (2.7%), associated with phenytoin (Ueta et al., 2014; Park et al., 2016; Wakamatsu et al., 2021), HLA-B*57:01 (2.1%), associated with abacavir (Mallal et al., 2002), HLA-B*40:02 (0.7%), associated with oxcarbazepine (Moon et al., 2016), HLA-B*13:01 (0.4%), associated with dapsone, salazosulfapyridine, and phenytoin (Yang et al., 2014; Wu et al., 2018; Su et al., 2019; Ahmed et al., 2021), HLA-B*15:02 (0.2%), associated with carbamazepine and phenytoin (Ferrell and McLeod, 2008; Chang et al., 2011; Wei et al., 2012; Ahmed et al., 2021), HLA-B*15:13 (0.2%), associated with phenytoin (Chang et al., 2017), and HLA-B*35:05 (0.2%), associated with nevirapine (Ahmed et al., 2021).
Comparison of HLA-B top alleles across Arab Gulf countries and other regions
Table 4 presents a comparative analysis of the top 10 most frequent HLA-B alleles in the Kuwaiti population with those observed in other Arab Gulf countries, including Saudi Arabia, Qatar, Bahrain, the United Arab Emirates, and Oman. This comparison highlights the similarities and differences in HLA-B allele distribution across these closely related regions. The data indicate that many of the most prevalent HLA-B alleles in the Kuwaiti population are also commonly found in neighbouring Gulf countries, suggesting shared genetic backgrounds and potential regional influences on allele frequencies.
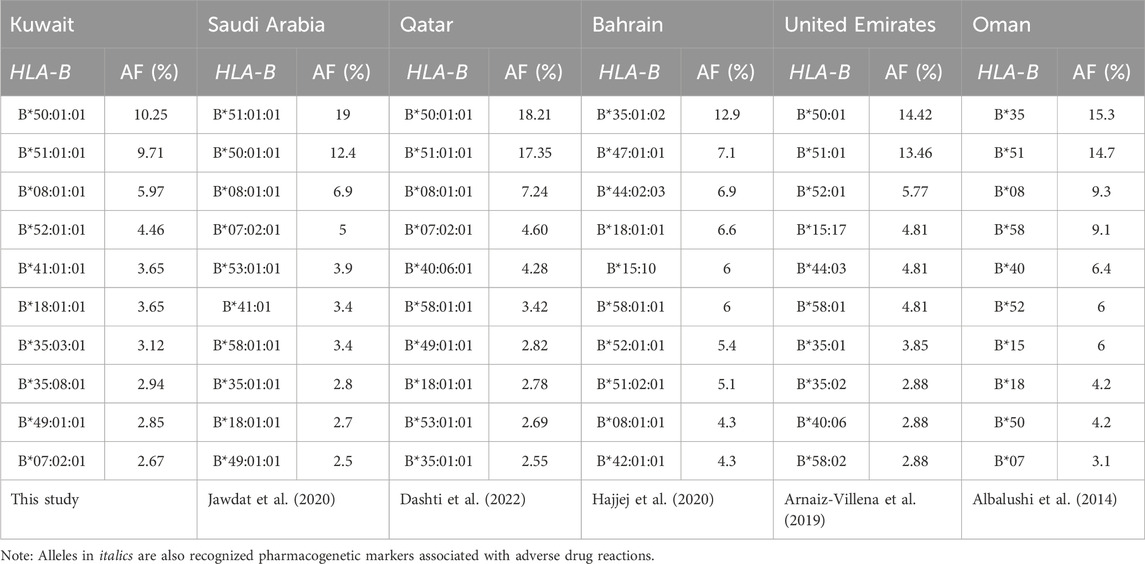
Table 4. Top 10 frequent HLA-B alleles, as ordered considering allele frequency (AF), in the Arab populations from the Gulf region.
Expanding beyond the Gulf region, Figure 1 illustrates the differences in the frequencies of the top HLA-B alleles between the Kuwaiti population and various other ethnic groups, including populations from Europe, North Africa, North America, South Asia, Western Asia, and Sub-Saharan Africa.
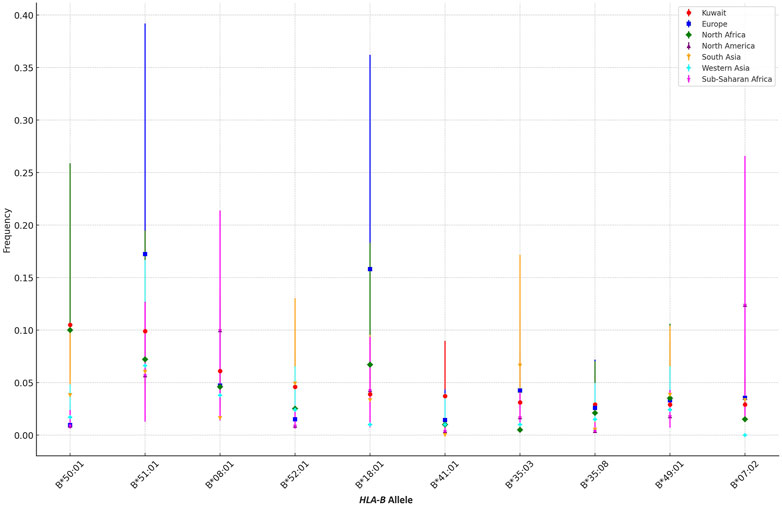
Figure 1. Comparison of the Top 10 HLA-B Allele Frequencies in the Kuwaiti Population Across Different Ethnic Groups. A comparative analysis of the top 10 most frequent HLA-B alleles in the Kuwaiti region and various other regions, including Europe, North Africa, North America, South Asia, Western Asia, and Sub-Saharan Africa. The frequencies of these alleles are presented with 95% confidence intervals to highlight genetic similarities and differences.
The analysis reveals that certain HLA-B alleles in the Kuwaiti population, such as B*50:01 and B*18:01, are more closely aligned with frequencies observed in other Middle Eastern regions, including Western Asia. However, these alleles show significant differences when compared to populations from Europe and North America, where these alleles are much less common. For instance, B*50:01, with a frequency of 0.105 in Kuwait, is almost absent in European (0.0094) and North American (0.009) populations, as indicated by non-overlapping confidence intervals, suggesting a distinct genetic profile in these regions.
Conversely, alleles like B*07:02, which is present in the Kuwaiti population, show more similarity in frequency with North African and Sub-Saharan African populations, indicating a shared genetic background or historical gene flow between these regions. In contrast, alleles such as B*51:01 demonstrate variability across all regions, with Kuwait showing closer frequencies to South Asia and Western Asia compared to other regions.
Discussion
In total, we identified 160 unique HLA-B alleles at high resolution in our study of 561 Kuwaiti individuals. High-resolution typing can be beneficial for pharmacogenetic studies, as it has the potential to increase the statistical power and accuracy in associating specific alleles with diseases and ADRs. This higher level of detail may help to better understand the variability in drug responses among individuals. Recent studies suggest that some synonymous variants, while not altering the protein sequence, may still impact splicing, RNA stability, RNA folding, translation, or co-translational protein folding, and could be implicated in various human diseases (Lin et al., 2023; Sharma et al., 2019). However, the risk of manifesting an ADR is also likely influenced by a combination of genetic factors, such as specific HLA alleles, and environmental variables, reflecting a multifactorial nature to these outcomes.
The most frequent alleles identified were HLA-B*50:01 (10.52%), HLA-B*51:01 (9.89%), HLA-B*08:01 (6.06%), HLA-B*52:01 (4.55%), HLA-B*18:01 (3.92%), and HLA-B*41:01 (3.65%). These findings align with the previously reported distribution of the most frequent HLA-B alleles in Kuwait (Ameen et al., 2020), which focused on the top alleles within the classical HLA class I and class II genes. Our study examines all HLA-B alleles present in our cohort, with particular emphasis on their relevance as pharmacogenetic markers. This demonstrates that HLA typing using WES data can effectively capture the same allele frequencies identified by the combination of sequence-specific oligonucleotide (SSO) probe-based hybridization and high-resolution HLA genotyping, as employed by Ameen et al. (2020).
Furthermore, twelve HLA-B pharmacogenomic markers were identified in 235 of the 561 (41.1%) Kuwaiti individuals. The most frequent marker, accounting for 18% of the Kuwaiti individuals, is HLA-B*51:01. This allele has been previously reported to be involved in the pathogenesis of SJS/TEN associated with phenobarbital (an antiepileptic drug used to control seizures) in the Japanese population (Kaniwa et al., 2013), phenytoin (another antiepileptic drug) in the South Indian Tamil (John et al., 2021) and Thai (Tassaneeyakul et al., 2016; Manuyakorn et al., 2020) populations, carbamazepine (an anticonvulsant and mood stabilizer) in the Japanese population (Niihara et al., 2012), and clindamycin (an antibiotic) in the Han Chinese population (Yang et al., 2017). Therefore, HLA-B*51:01 may serve as a susceptibility factor for SJS/TEN in Asian populations. Our previous study also revealed that HLA-B*51:01 is the most frequent pharmacogenetic HLA-B marker, carried by 26.67% of Qatari individuals (Dashti et al., 2022). Additionally, the allele frequency of HLA-B*51:01 has been shown to be high in other Arab Gulf countries (Table 4). These similarities in allele frequencies may be due to a shared gene pool, potentially influenced by historical migrations, geographic proximity, and common ancestry among the Gulf Cooperation Council (GCC) countries.
The second most frequent pharmacogenetic marker identified is HLA-B*35:02, which is carried by 5% of the studied Kuwaiti individuals. This allele has been associated with minocycline (an antibiotic commonly used to treat bacterial infections)-induced drug-induced liver injury in a Caucasian cohort in the United States (Urban et al., 2017). In the Qatari population, the allele frequency of HLA-B*35:02 is 1.59% (Dashti et al., 2022), and in the Emirati population, it is 2.88% (Arnaiz-Villena et al., 2019), which is very similar to the Kuwaiti population’s allele frequency of 2.58%.
HLA-B*13:02 allele is the third most prevalent pharmacogenetic marker, at 4.5%, in the Kuwaiti cohort. The allele frequency of this marker is higher in the Kuwaiti population compared to other Arab populations in the Gulf region, where it is less than 1% in Qatar and not among the top ten HLA-B alleles in the Arabs of the Gulf countries (Table 4). HLA-B*13:02 allele has been nominally associated with lamotrigine (an antiepileptic drug used to treat epilepsy and bipolar disorder)-induced SCAR in the Korean population (Kim et al., 2017). It is also a marker for oxcarbazepine (an antiepileptic drug used to treat partial seizures)-induced maculopapular eruption in the Southern Han Chinese population (He et al., 2012). Additionally, HLA-B*13:02 has been associated with allopurinol (a medication used to treat gout and hyperuricemia)-induced DRESS in the Shanghai population (Wu et al., 2018).
The fourth prevalent pharmacogenetic marker is HLA-B*58:01, which is carried by 4.1% of the Kuwaiti cohort. Upon examining the frequencies of this allele in neighbouring countries (Table 4), we found that HLA-B*58:01 is among the top ten most frequent alleles in the Qatari, Saudi, Bahraini, and Emirati populations. The same can be said for Oman; however, the available HLA typing in Oman was conducted at low resolution, where HLA-B*58 is frequent. Nevertheless, higher resolution typing is needed to confirm the exact allele. Pharmacogenetic studies have demonstrated an association between HLA-B*58:01 and allopurinol-induced SCARs across diverse ethnicities, including African American (Fontana et al., 2021), European (Lonjou et al., 2008; Gonçalo et al., 2013), and Asian (Sukasem et al., 2016) populations. The risk of allopurinol-induced SCARs is associated with a gene dosage effect of HLA-B*58:01 on renal function (Chung et al., 2015; Ng et al., 2016), as well as with increased plasma levels of the allopurinol metabolite (Ng et al., 2016). Allopurinol is used to lower blood uric acid levels induced by chemotherapy and to prevent the formation of certain types of kidney stones (Jung et al., 2015). Nevertheless, it has been suggested that additional genetic variations beyond the HLA region might also contribute to the risk (Tohkin et al., 2013).
The fifth most prevalent pharmacogenetic marker in our study is HLA-B*38:01, which is associated with SCARs induced by lamotrigine and phenytoin in the Spanish Caucasian population (Ramírez et al., 2017). This allele is carried by 3.9% of the Kuwaiti cohort. However, it has a low allele frequency in the Qatari population (Dashti et al., 2022) and is not among the top frequent alleles in Arab populations from the Gulf region (Table 4).
Another pharmacogenetic marker identified is HLA-B*44:03, which is carried by 2.7% of the Kuwaiti cohort, a percentage similar to that observed in the Qatari population (2.8%) (Dashti et al., 2022), and is very frequent in the Emirati population (Arnaiz-Villena et al., 2019). This allele has been associated with cold-medicine (multi-ingredient cold and anti-inflammatory drug remedies)-induced SJS/TEN in Japanese (Ueta et al., 2014) and Brazilian (Wakamatsu et al., 2021) populations. Additionally, another study suggests a potential correlation between HLA-B*44:03 and lamotrigine-induced SJS/TEN in Koreans (Park et al., 2016).
In addition, the HLA-B*57:01 pharmacogenetic marker is carried by 2.1% of the Kuwaiti individuals in this study. This allele is also present at a similar percentage in the Qatari population (Dashti et al., 2022); however, it is not among the top 10 most common HLA-B alleles in any of the Gulf countries (Table 4). As a pharmacogenetic marker, HLA-B*57:01 is known to be associated with abacavir (an antiretroviral medication)-induced hypersensitivity (Mallal et al., 2002) and is more prevalent in Caucasian populations compared to Asian populations (Jung et al., 2018).
Moreover, we have identified several pharmacogenetic markers, each carried by less than 1% of the Kuwaiti cohort, and none are among the top frequent alleles of Arabs from the Gulf region (Table 4). Among these are the HLA-B*40:02 allele, associated with oxcarbazepine-induced maculopapular eruption in the Korean population (Moon et al., 2016), and the HLA-B*13:01 allele, linked to dapsone-induced SCARs in Thai and Han Chinese populations (Ahmed et al., 2021), salazosulfapyridine-induced drug rash with DRESS in the Shanghai and Han Chinese populations (Yang et al., 2014; Wu et al., 2018), and phenytoin-related SCARs in East Asians (Su et al., 2019). Additionally, the HLA-B*15:02 allele is known for its association with carbamazepine and phenytoin-induced SJS/TEN, particularly in Southeast Asian populations (Ferrell and McLeod, 2008; Chang et al., 2011; Wei et al., 2012; Ahmed et al., 2021), while the HLA-B*15:13 allele is associated with phenytoin-induced SCARs in the Malay population (Chang et al., 2017). Furthermore, the HLA-B*35:05 allele has been linked to nevirapine-induced hypersensitivity reactions in various ethnic groups (Ahmed et al., 2021).
In general, our data shows that the majority of the most prevalent HLA-B alleles in the Kuwaiti population are common in other Gulf countries (Table 4). This demonstrates that repurposing WES datasets for HLA typing to explore the frequency of HLA genes relevant to disease or pharmacology on a population scale is feasible. Additionally, some of the frequent HLA-B alleles serve as pharmacogenetic markers, indicating potential opportunities for collaborative regional health strategies to address shared pharmacogenetic risks. However, there are slight variations in the top frequent HLA-B alleles among these countries, reflecting genetic diversity influenced by factors such as genetic drift, selection pressure, or historical migration.
Our study highlights significant differences in the frequencies of various HLA-B alleles between the Kuwaiti population and other regions, underscoring the unique genetic heritage of Kuwait, particularly when compared to Europe, North America, and even neighbouring regions like Western Asia. The distinct allele frequencies observed, such as those of B*50:01 and B*18:01, reflect true population-specific patterns, as indicated by non-overlapping confidence intervals.
Incorporating other ethnic groups into the analysis further enriches our understanding of the genetic diversity across regions. For example, while alleles like B*07:02 show similar frequencies across diverse populations, others, such as B*51:01, exhibit considerable variability. These findings emphasize the influence of regional and ethnic factors in shaping HLA-B allele distribution, which may have important implications for disease susceptibility, transplantation compatibility, and other health-related outcomes in the Kuwaiti population.
The current study has a few limitations. First, the relatively small sample size in our investigation may have influenced the accuracy of frequency estimates for the loci examined, and there is a possibility of overlooking low-frequency alleles due to this limitation. Additionally, it is important to note that the HLA-B pharmacogenetic markers analysed in this study are reference markers derived from databases and studies conducted in other populations, such as those from the HLA Adverse Drug Reaction Database. These markers may be ethnicity-specific and might not be causative of ADRs in our Kuwaiti population. As a result, we may have missed additional potential markers specific to the Kuwaiti population that reflect differences in genetic backgrounds. Furthermore, this study used the HLA-HD tool for HLA typing. While benchmark studies (Chen et al., 2021; Thuesen et al., 2022; Claeys et al., 2023) have shown that other top-performing tools, such as HLA*LA (Dilthey et al., 2019) and HISAT-genotype (Kim et al., 2019), might be more consistent and accurate in typing HLA Class I genes, the difference in consistency is marginal. Additionally, some of these tools are computationally intensive, making them less suitable for population-scale projects. Importantly, the HLA-HD tool does not detect novel alleles; this limitation was known prior to the study’s design, as our aim was to analyse the distribution of known HLA-B alleles rather than to discover novel ones. Therefore, further studies are needed to confirm the association of the current pharmacogenetic markers with ADRs in our population and to identify additional pharmacogenetic markers that may be relevant.
Conclusion
Our study enriches the regional genetic landscape by delineating HLA-B allele variations within Kuwait and across the Arabian Peninsula. This detailed characterization is invaluable for future studies on genetic diversity, disease risk, and pharmacogenetics, ultimately contributing to personalized medicine strategies in the region. By determining the frequency of pharmacogenetic markers, previously reported in different populations, within the Kuwaiti population, we provide a solid foundation for future pharmacogenetic research. While these markers are not necessarily causative of ADRs in our population, they offer valuable insights. Future research should focus on hypersensitivity studies involving different drugs and HLA-B alleles, as well as exploring additional HLA genes variations, to further advance personalized healthcare strategies in the Gulf region.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: SRA repository, PRJNA1166832 (https://www.ncbi.nlm.nih.gov/sra/PRJNA1166832).
Ethics statement
The studies involving humans were approved by Ethical Review Committee at Dasman Diabetes Institute. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
MD: Writing–review and editing, Writing–original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal Analysis, Data curation, Conceptualization. MM: Writing–review and editing, Writing–original draft. AA-M: Writing–original draft, Writing–review and editing. SB: Writing–review and editing, Writing–original draft. RN: Data curation, Writing–review and editing. SJ: Data curation, Writing–review and editing. FA-M: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing–review and editing, Writing–original draft, Visualization. TT: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by institutional funding from the Kuwait Foundation for the Advancement of Sciences (KFAS).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2024.1423636/full#supplementary-material
SUPPLEMENTARY TABLE S1 | Raw HLA-B alleles identified in 561 Kuwaiti individuals.
References
Ahmed, A. F., Sukasem, C., Sabbah, M. A., Musa, N. F., Mohamed Noor, D. A., and Daud, N. A. A. (2021). Genetic determinants in HLA and cytochrome P450 genes in the risk of aromatic antiepileptic-induced severe cutaneous adverse reactions. J. Pers. Med. 11 (5), 383. doi:10.3390/jpm11050383
Al-Bader, S., Al-Bader, K., and El-Reshaid, K. (2019). The spectrum of adverse drug reactions in a multidisciplinary kidney clinic. J. Drug Deliv. and Ther. 9 (1), 61–67. doi:10.22270/jddt.v9i1.2160
Albalushi, K. R., Sellami, M. H., AlRiyami, H., Varghese, M., Boukef, M. K., and Hmida, S. (2014). The investigation of the evolutionary history of the Omani population by analysis of HLA class I polymorphism. Anthropol. 18 (1), 205–210. doi:10.1080/09720073.2014.11891537
Alfraih, F., Alawwami, M., Aljurf, M., Alhumaidan, H., Alsaedi, H., El Fakih, R., et al. (2021). High-resolution HLA allele and haplotype frequencies of the Saudi Arabian population based on 45,457 individuals and corresponding stem cell donor matching probabilities. Hum. Immunol. 82 (2), 97–102. doi:10.1016/j.humimm.2020.12.006
Ameen, R., Al Shemmari, S. H., and Marsh, S. G. E. (2020). HLA haplotype frequencies and genetic profiles of the Kuwaiti population. Med. Princ. Pract. 29 (1), 39–45. doi:10.1159/000499593
Arnaiz-Villena, A., Yafei, Z. A., Juarez, I., Palacio-Gruber, J., Mahri, A. A., Alvares, M., et al. (2019). HLA genetic study from United Arab Emirates (UAE), Abu Dhabi. Hum. Immunol. 80 (7), 421–422. doi:10.1016/j.humimm.2019.04.013
Chang, C. C., Ng, C. C., Too, C. L., Choon, S. E., Lee, C. K., Chung, W. H., et al. (2017). Association of HLA-B*15:13 and HLA-B*15:02 with phenytoin-induced severe cutaneous adverse reactions in a Malay population. Pharmacogenomics J. 17 (2), 170–173. doi:10.1038/tpj.2016.10
Chang, C. C., Too, C. L., Murad, S., and Hussein, S. H. (2011). Association of HLA-B*1502 allele with carbamazepine-induced toxic epidermal necrolysis and Stevens-Johnson syndrome in the multi-ethnic Malaysian population. Int. J. Dermatol 50 (2), 221–224. doi:10.1111/j.1365-4632.2010.04745.x
Chen, C. B., Abe, R., Pan, R. Y., Wang, C. W., Hung, S. I., Tsai, Y. G., et al. (2018). An updated review of the molecular mechanisms in drug hypersensitivity. J. Immunol. Res. 2018, 6431694. doi:10.1155/2018/6431694
Chen, J., Madireddi, S., Nagarkar, D., Migdal, M., Vander Heiden, J., Chang, D., et al. (2021). In silico tools for accurate HLA and KIR inference from clinical sequencing data empower immunogenetics on individual-patient and population scales. Brief. Bioinform 22 (3), bbaa223. doi:10.1093/bib/bbaa223
Chung, W. H., Chang, W. C., Stocker, S. L., Juo, C. G., Graham, G. G., Lee, M. H., et al. (2015). Insights into the poor prognosis of allopurinol-induced severe cutaneous adverse reactions: the impact of renal insufficiency, high plasma levels of oxypurinol and granulysin. Ann. Rheum. Dis. 74 (12), 2157–2164. doi:10.1136/annrheumdis-2014-205577
Claeys, A., Merseburger, P., Staut, J., Marchal, K., and Van den Eynden, J. (2023). Benchmark of tools for in silico prediction of MHC class I and class II genotypes from NGS data. BMC Genomics 24 (1), 247. doi:10.1186/s12864-023-09351-z
Darbas, S., Inan, D., Kilinc, Y., Arslan, H. S., Ucar, F., Boylubay, O., et al. (2023). Relationship of HLA-B alleles on susceptibility to and protection from HIV infection in Turkish population. North Clin. Istanb 10 (1), 67–73. doi:10.14744/nci.2021.00018
Dashti, M., Al-Matrouk, A., Channanath, A., Hebbar, P., Al-Mulla, F., and Thanaraj, T. A. (2022). Distribution of HLA-B alleles and haplotypes in Qatari: recommendation for establishing pharmacogenomic markers screening for drug hypersensitivity. Front. Pharmacol. 13, 891838. doi:10.3389/fphar.2022.891838
Dashti, M., Malik, M. Z., Nizam, R., Jacob, S., Al-Mulla, F., and Thanaraj, T. A. (2024). Evaluation of HLA typing content of next-generation sequencing datasets from family trios and individuals of arab ethnicity. Front. Genet. 15, 1407285. doi:10.3389/fgene.2024.1407285
Davies, E. C., Green, C. F., Taylor, S., Williamson, P. R., Mottram, D. R., and Pirmohamed, M. (2009). Adverse drug reactions in hospital in-patients: a prospective analysis of 3695 patient-episodes. PLoS One 4 (2), e4439. doi:10.1371/journal.pone.0004439
Dendrou, C. A., Petersen, J., Rossjohn, J., and Fugger, L. (2018). HLA variation and disease. Nat. Rev. Immunol. 18 (5), 325–339. doi:10.1038/nri.2017.143
Dilthey, A. T., Mentzer, A. J., Carapito, R., Cutland, C., Cereb, N., Madhi, S. A., et al. (2019). HLA*LA-HLA typing from linearly projected graph alignments. Bioinformatics 35 (21), 4394–4396. doi:10.1093/bioinformatics/btz235
Ferrell, P. B., and McLeod, H. L. (2008). Carbamazepine, HLA-B*1502 and risk of Stevens-Johnson syndrome and toxic epidermal necrolysis: US FDA recommendations. Pharmacogenomics 9 (10), 1543–1546. doi:10.2217/14622416.9.10.1543
Fontana, R. J., Li, Y. J., Phillips, E., Saeed, N., Barnhart, H., Kleiner, D., et al. (2021). Allopurinol hepatotoxicity is associated with human leukocyte antigen Class I alleles. Liver Int. 41 (8), 1884–1893. doi:10.1111/liv.14903
Gonçalo, M., Coutinho, I., Teixeira, V., Gameiro, A. R., Brites, M. M., Nunes, R., et al. (2013). HLA-B*58:01 is a risk factor for allopurinol-induced DRESS and Stevens-Johnson syndrome/toxic epidermal necrolysis in a Portuguese population. Br. J. Dermatol 169 (3), 660–665. doi:10.1111/bjd.12389
Hajjej, A., Saldhana, F. L., Dajani, R., and Almawi, W. Y. (2020). HLA-A, -B, -C, -DRB1 and -DQB1 allele and haplotype frequencies and phylogenetic analysis of Bahraini population. Gene 735, 144399. doi:10.1016/j.gene.2020.144399
He, N., Min, F. L., Shi, Y. W., Guo, J., Liu, X. R., Li, B. M., et al. (2012). Cutaneous reactions induced by oxcarbazepine in Southern Han Chinese: incidence, features, risk factors and relation to HLA-B alleles. Seizure 21 (8), 614–618. doi:10.1016/j.seizure.2012.06.014
Howell, W. M., Carter, V., and Clark, B. (2010). The HLA system: immunobiology, HLA typing, antibody screening and crossmatching techniques. J. Clin. Pathol. 63 (5), 387–390. doi:10.1136/jcp.2009.072371
Jantararoungtong, T., Tempark, T., Koomdee, N., Medhasi, S., and Sukasem, C. (2021). Genotyping HLA alleles to predict the development of Severe cutaneous adverse drug reactions (SCARs): state-of-the-art. Expert Opin. Drug Metab. Toxicol. 17 (9), 1049–1064. doi:10.1080/17425255.2021.1946514
Jawdat, D., Uyar, F. A., Alaskar, A., Müller, C. R., and Hajeer, A. (2020). HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 allele and haplotype frequencies of 28,927 Saudi stem cell donors typed by next-generation sequencing. Front. Immunol. 11, 544768. doi:10.3389/fimmu.2020.544768
John, S., Balakrishnan, K., Sukasem, C., Anand, T. C. V., Canyuk, B., and Pattharachayakul, S. (2021). Association of HLA-B*51:01, HLA-B*55:01, CYP2C9*3, and phenytoin-induced cutaneous adverse drug reactions in the South Indian Tamil population. J. Pers. Med. 11 (8), 737. doi:10.3390/jpm11080737
John, S. E., Antony, D., Eaaswarkhanth, M., Hebbar, P., Channanath, A. M., Thomas, D., et al. (2018). Assessment of coding region variants in Kuwaiti population: implications for medical genetics and population genomics. Sci. Rep. 8 (1), 16583. doi:10.1038/s41598-018-34815-8
Jung, J. W., Kim, D. K., Park, H. W., Oh, K. H., Joo, K. W., Kim, Y. S., et al. (2015). An effective strategy to prevent allopurinol-induced hypersensitivity by HLA typing. Genet. Med. 17 (10), 807–814. doi:10.1038/gim.2014.195
Jung, J. W., Kim, J. Y., Park, I. W., Choi, B. W., and Kang, H. R. (2018). Genetic markers of severe cutaneous adverse reactions. Korean J. Intern Med. 33 (5), 867–875. doi:10.3904/kjim.2018.126
Kaniwa, N., Sugiyama, E., Saito, Y., Kurose, K., Maekawa, K., Hasegawa, R., et al. (2013). Specific HLA types are associated with antiepileptic drug-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in Japanese subjects. Pharmacogenomics 14 (15), 1821–1831. doi:10.2217/pgs.13.180
Kawaguchi, S., Higasa, K., Shimizu, M., Yamada, R., and Matsuda, F. (2017). HLA-HD: an accurate HLA typing algorithm for next-generation sequencing data. Hum. Mutat. 38 (7), 788–797. doi:10.1002/humu.23230
Kim, B. K., Jung, J. W., Kim, T. B., Chang, Y. S., Park, H. S., Moon, J., et al. (2017). HLA-A*31:01 and lamotrigine-induced severe cutaneous adverse drug reactions in a Korean population. Ann. Allergy Asthma Immunol. 118 (5), 629–630. doi:10.1016/j.anai.2017.02.011
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37 (8), 907–915. doi:10.1038/s41587-019-0201-4
Kloypan, C., Koomdee, N., Satapornpong, P., Tempark, T., Biswas, M., and Sukasem, C. (2021). A A comprehensive review of HLA and severe cutaneous adverse drug reactions: implication for clinical pharmacogenomics and precision medicine. Pharm. (Basel) 14 (11), 1077. doi:10.3390/ph14111077
Koomdee, N., Kloypan, C., Jinda, P., Rachanakul, J., Jantararoungtong, T., Sukprasong, R., et al. (2022). Evolution of HLA-B pharmacogenomics and the importance of PGx data integration in health care system: a 10 Years retrospective study in Thailand. Front. Pharmacol. 13, 866903. doi:10.3389/fphar.2022.866903
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. doi:10.1038/nmeth.1923
Lazarou, J., Pomeranz, B. H., and Corey, P. N. (1998). Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA 279 (15), 1200–1205. doi:10.1001/jama.279.15.1200
Lin, B. C., Katneni, U., Jankowska, K. I., Meyer, D., and Kimchi-Sarfaty, C. (2023). In silico methods for predicting functional synonymous variants. Genome Biol. 24 (1), 126. doi:10.1186/s13059-023-02966-1
Lonjou, C., Borot, N., Sekula, P., Ledger, N., Thomas, L., Halevy, S., et al. (2008). A European study of HLA-B in Stevens-Johnson syndrome and toxic epidermal necrolysis related to five high-risk drugs. Pharmacogenet Genomics 18 (2), 99–107. doi:10.1097/FPC.0b013e3282f3ef9c
Mallal, S., Nolan, D., Witt, C., Masel, G., Martin, A. M., Moore, C., et al. (2002). Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet 359 (9308), 727–732. doi:10.1016/s0140-6736(02)07873-x
Manuyakorn, W., Likkasittipan, P., Wattanapokayakit, S., Suvichapanich, S., Inunchot, W., Wichukchinda, N., et al. (2020). Association of HLA genotypes with phenytoin induced severe cutaneous adverse drug reactions in Thai children. Epilepsy Res. 162, 106321. doi:10.1016/j.eplepsyres.2020.106321
Moon, J., Kim, T. J., Lim, J. A., Sunwoo, J. S., Byun, J. I., Lee, S. T., et al. (2016). HLA-B*40:02 and DRB1*04:03 are risk factors for oxcarbazepine-induced maculopapular eruption. Epilepsia 57 (11), 1879–1886. doi:10.1111/epi.13566
Moussa, M. A., Bayoumi, A., Al-Khars, A., and Thulesius, O. (1985). Adverse drug reaction monitoring in Kuwait (1981-1984). J. Clin. Pharmacol. 25 (3), 176–181. doi:10.1002/j.1552-4604.1985.tb02821.x
Mungall, A. J., Palmer, S. A., Sims, S. K., Edwards, C. A., Ashurst, J. L., Wilming, L., et al. (2003). The DNA sequence and analysis of human chromosome 6. Nature 425 (6960), 805–811. doi:10.1038/nature02055
Ng, C. Y., Yeh, Y. T., Wang, C. W., Hung, S. I., Yang, C. H., Chang, Y. C., et al. (2016). Impact of the HLA-B(*)58:01 allele and renal impairment on allopurinol-induced cutaneous adverse reactions. J. Invest Dermatol 136 (7), 1373–1381. doi:10.1016/j.jid.2016.02.808
Niihara, H., Kakamu, T., Fujita, Y., Kaneko, S., and Morita, E. (2012). HLA-A31 strongly associates with carbamazepine-induced adverse drug reactions but not with carbamazepine-induced lymphocyte proliferation in a Japanese population. J. Dermatol 39 (7), 594–601. doi:10.1111/j.1346-8138.2011.01457.x
Park, H. J., Kim, Y. J., Kim, D. H., Kim, J., Park, K. H., Park, J. W., et al. (2016). HLA allele frequencies in 5802 Koreans: varied allele types associated with SJS/TEN according to culprit drugs. Yonsei Med. J. 57 (1), 118–126. doi:10.3349/ymj.2016.57.1.118
Pichler, W. J. (2003). Delayed drug hypersensitivity reactions. Ann. Intern Med. 139 (8), 683–693. doi:10.7326/0003-4819-139-8-200310210-00012
Pirmohamed, M., James, S., Meakin, S., Green, C., Scott, A. K., Walley, T. J., et al. (2004). Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ 329 (7456), 15–19. doi:10.1136/bmj.329.7456.15
Ramírez, E., Bellón, T., Tong, H. Y., Borobia, A. M., de Abajo, F. J., Lerma, V., et al. (2017). Significant HLA class I type associations with aromatic antiepileptic drug (AED)-induced SJS/TEN are different from those found for the same AED-induced DRESS in the Spanish population. Pharmacol. Res. 115, 168–178. doi:10.1016/j.phrs.2016.11.027
R Core Team (2023). R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available at: https://www.R-project.org/.
Robinson, J., Barker, D. J., Georgiou, X., Cooper, M. A., Flicek, P., and Marsh, S. G. E. (2020). IPD-IMGT/HLA database. Nucleic Acids Res. 48 (D1), D948–D955. doi:10.1093/nar/gkz950
Sajulga, R., Bolon, Y. T., Maiers, M. J., and Petersdorf, E. W. (2022). Assessment of HLA-B genetic variation with an HLA-B leader tool and implications in clinical transplantation. Blood Adv. 6 (1), 270–280. doi:10.1182/bloodadvances.2021004561
Shapiro, L. E., and Shear, N. H. (1996). Mechanisms of drug reactions: the metabolic track. Semin. Cutan. Med. Surg. 15 (4), 217–227. doi:10.1016/s1085-5629(96)80034-4
Sharma, Y., Miladi, M., Dukare, S., Boulay, K., Caudron-Herger, M., Groß, M., et al. (2019). A pan-cancer analysis of synonymous mutations. Nat. Commun. 10 (1), 2569. doi:10.1038/s41467-019-10489-2
Su, S. C., Chen, C. B., Chang, W. C., Wang, C. W., Fan, W. L., Lu, L. Y., et al. (2019). HLA alleles and CYP2C9*3 as predictors of phenytoin hypersensitivity in East Asians. Clin. Pharmacol. Ther. 105 (2), 476–485. doi:10.1002/cpt.1190
Sukasem, C., Jantararoungtong, T., Kuntawong, P., Puangpetch, A., Koomdee, N., Satapornpong, P., et al. (2016). HLA-B (*) 58:01 for allopurinol-induced cutaneous adverse drug reactions: implication for clinical interpretation in Thailand. Front. Pharmacol. 7, 186. doi:10.3389/fphar.2016.00186
Tassaneeyakul, W., Prabmeechai, N., Sukasem, C., Kongpan, T., Konyoung, P., Chumworathayi, P., et al. (2016). Associations between HLA class I and cytochrome P450 2C9 genetic polymorphisms and phenytoin-related severe cutaneous adverse reactions in a Thai population. Pharmacogenet Genomics 26 (5), 225–234. doi:10.1097/FPC.0000000000000211
The MHC sequencing consortium (1999). Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature 401 (6756), 921–923. doi:10.1038/44853
Thuesen, N. H., Klausen, M. S., Gopalakrishnan, S., Trolle, T., and Renaud, G. (2022). Benchmarking freely available HLA typing algorithms across varying genes, coverages and typing resolutions. Front. Immunol. 13, 987655. doi:10.3389/fimmu.2022.987655
Tohkin, M., Kaniwa, N., Saito, Y., Sugiyama, E., Kurose, K., Nishikawa, J., et al. (2013). A whole-genome association study of major determinants for allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis in Japanese patients. Pharmacogenomics J. 13 (1), 60–69. doi:10.1038/tpj.2011.41
Ueta, M., Kaniwa, N., Sotozono, C., Tokunaga, K., Saito, Y., Sawai, H., et al. (2014). Independent strong association of HLA-A*02:06 and HLA-B*44:03 with cold medicine-related Stevens-Johnson syndrome with severe mucosal involvement. Sci. Rep. 4, 4862. doi:10.1038/srep04862
Urban, T. J., Nicoletti, P., Chalasani, N., Serrano, J., Stolz, A., Daly, A. K., et al. (2017). Minocycline hepatotoxicity: clinical characterization and identification of HLA-B∗35:02 as a risk factor. J. Hepatol. 67 (1), 137–144. doi:10.1016/j.jhep.2017.03.010
Wakamatsu, T. H., Dos Santos, M. S., Barreiro, T. P., Sant'Anna, A. E. B. P., Murta, F., da Costa, A. X., et al. (2021). Clinical aspects of stevens-johnson syndrome and toxic epidermal necrolysis with severe ocular complications in Brazil. Front. Med. (Lausanne) 8, 649369. doi:10.3389/fmed.2021.649369
Wei, C. Y., Chung, W. H., Huang, H. W., Chen, Y. T., and Hung, S. I. (2012). Direct interaction between HLA-B and carbamazepine activates T cells in patients with Stevens-Johnson syndrome. J. Allergy Clin. Immunol. 129 (6), 1562–1569. doi:10.1016/j.jaci.2011.12.990
Wu, X., Yang, F., Chen, S., Xiong, H., Zhu, Q., Gao, X., et al. (2018). Clinical, viral and genetic characteristics of drug reaction with eosinophilia and systemic symptoms (DRESS) in Shanghai, China. Acta Derm. Venereol. 98 (4), 401–405. doi:10.2340/00015555-2867
Yang, F., Gu, B., Zhang, L., Xuan, J., Luo, H., Zhou, P., et al. (2014). HLA-B*13:01 is associated with salazosulfapyridine-induced drug rash with eosinophilia and systemic symptoms in Chinese Han population. Pharmacogenomics 15 (11), 1461–1469. doi:10.2217/pgs.14.69
Yang, Y., Chen, S., Yang, F., Zhang, L., Alterovitz, G., Zhu, H., et al. (2017). HLA-B*51:01 is strongly associated with clindamycin-related cutaneous adverse drug reactions. Pharmacogenomics J. 17 (6), 501–505. doi:10.1038/tpj.2016.61
Keywords: HLA-B alleles, pharmacogenetics, NGS-HLA typing, Kuwaiti population, precision medicine
Citation: Dashti M, Malik MZ, Al-Matrouk A, Bhatti S, Nizam R, Jacob S, Al-Mulla F and Thanaraj TA (2024) HLA-B allele frequencies and implications for pharmacogenetics in the Kuwaiti population. Front. Pharmacol. 15:1423636. doi: 10.3389/fphar.2024.1423636
Received: 26 April 2024; Accepted: 16 September 2024;
Published: 11 October 2024.
Edited by:
Mounir Tilaoui, Waterford Institute of Technology, IrelandReviewed by:
Mathijs Groeneweg, Maastricht University Medical Centre, NetherlandsKaren Sherwood, BC Provincial Immunology Laboratory, Vancouver Coastal Health, Canada
M. Carmen Martin, Centro de Hemoterapia y Hemodonación de Castilla y León, Spain
Copyright © 2024 Dashti, Malik, Al-Matrouk, Bhatti, Nizam, Jacob, Al-Mulla and Thanaraj. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fahd Al-Mulla, ZmFoZC5hbG11bGxhQGRhc21hbmluc3RpdHV0ZS5vcmc=; Thangavel Alphonse Thanaraj, YWxwaG9uc2UudGhhbmdhdmVsQGRhc21hbmluc3RpdHV0ZS5vcmc=
†These authors have contributed equally to this work
 Mohammed Dashti
Mohammed Dashti Md Zubbair Malik
Md Zubbair Malik Abdullah Al-Matrouk
Abdullah Al-Matrouk Saeeda Bhatti3
Saeeda Bhatti3 Fahd Al-Mulla
Fahd Al-Mulla Thangavel Alphonse Thanaraj
Thangavel Alphonse Thanaraj