- 1Department of Hepatopancreatobiliary Surgery, The Second Affiliated Hospital of Harbin Medical University, Harbin, China
- 2Department of General Surgery, Tangdu Hospital, Air Force Medical University, Xi’an, China
Epigenetic changes are heritable changes in gene expression without changes in the nucleotide sequence of genes. Epigenetic changes play an important role in the development of cancer and in the process of malignancy metastasis. Previous studies have shown that abnormal epigenetic changes can be used as biomarkers for disease status and disease prediction. The reversibility and controllability of epigenetic modification changes also provide new strategies for early disease prevention and treatment. In addition, corresponding drug development has also reached the clinical stage. In this paper, we will discuss the recent progress and application status of tumor epigenetic biomarkers from three perspectives: DNA methylation, non-coding RNA, and histone modification, in order to provide new opportunities for additional tumor research and applications.
1 Introduction
Cancer is a disease characterized by the disruption of critical pathways that control cellular processes such as repairing DNA, ensuring cell survival, promoting cell proliferation, and inducing cell death (Halbrook et al., 2023). Mutations in certain genes, including tumor suppressor genes and proto-oncogenes, contribute to the development of cancer (Riaud et al., 2024; Tuval et al., 2024). However, recent research has indicated that epigenetic abnormalities can also lead to the inactivation of tumor suppressor genes and activation of proto-oncogenes, playing a crucial role in the initiation, progression, invasion, and spread of cancer (Feinberg and Levchenko, 2023) (Figure 1). Epigenetics refers to a phenomenon in which gene function produces inheritable variations without altering the DNA sequence, ultimately resulting in changes in an organism’s phenotype (Lee and Kim, 2022). Epigenetic processes mainly encompass three categories: DNA methylation, modification of histones, and regulation of non-coding RNA (ncRNA) (Fitz-James and Cavalli, 2022).
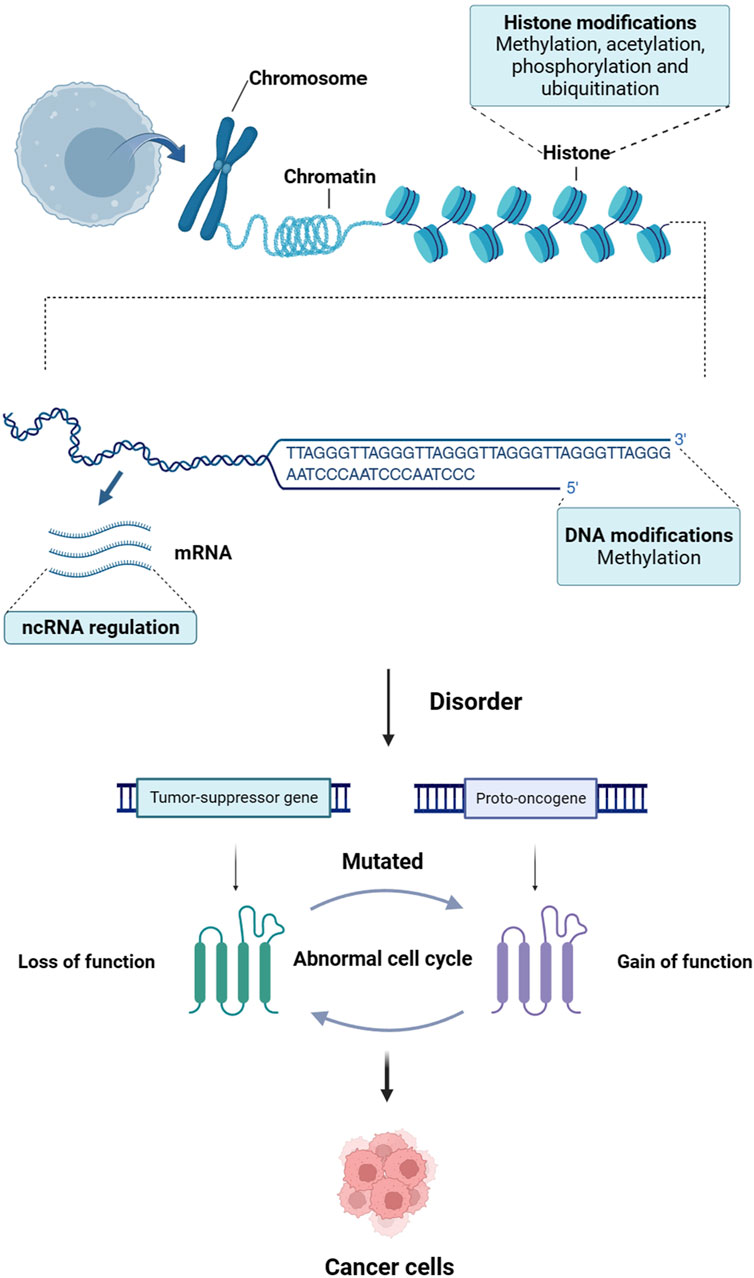
Figure 1. The link between epigenetics and cancer. Epigenetic disorders can lead to inactivation of tumor-suppressor genes and activation of proto-oncogenes, which play a crucial role in the initiation, progression, invasion and spread of cancer.
Advances in high-throughput sequencing techniques have enhanced our comprehension of epigenetic mechanisms in cancer development and revealed numerous cancer-specific epigenetic biomarkers that offer potential as effective indicators for assessing tumor high-risk, facilitating early diagnosis, informing treatment decisions, and predicting prognosis (Lauschke et al., 2019). Currently, several pivotal regulatory molecules involved in epigenetics have been integrated into clinical tumor diagnosis for metastasis, recurrence, and prognostic assessment, serving as targets for clinical oncology therapies (Kravitz et al., 2023). This paper aims to discuss recent advancements and applications of tumor epigenetic biomarkers from three perspectives: DNA methylation, ncRNA, and histone modification. Consequently, it presents novel opportunities for further refinement of tumor therapies.
2 DNA methylation
DNA methylation is a biochemical process that involves the addition of a methyl group to a DNA base (Galbraith and Snuderl, 2022). This methyl group is commonly added to the 5′position of CpG island cytosine. This process is catalyzed by enzymes called DNA methyltransferases (DNMTs), which utilize S-adenosylmethionine as a methyl donor (Deng et al., 2023). In mammalian cells, DNMTs can be classified into three main types: DNMT1, DNMT3a, and DNMT3b. DNMT1 is responsible for maintaining the existing DNA methylation state, as well as catalyzing hemimethylated DNA and participating in DNA methylation repair (Yan et al., 2023). On the other hand, DNMT3a and DNMT3b catalyze the addition of new methyl groups to unmethylated DNA, converting it into methylated DNA (Lyko, 2018). DNA methylation plays a crucial role in regulating various biological processes within the mammalian genome. These processes include transcription and post-transcriptional processing, post-translational modification, chromatin remodeling, genomic imprinting, X chromosome inactivation, and inhibition of repetitive DNA elements (Saghafinia et al., 2018).
During tumorigenesis, there is typically a decrease in the overall level of intracellular DNA methylation. However, there is abnormal elevation of DNA methylation in the CpG island regions of specific genes (Ghosh et al., 2022). This abnormal methylation pattern leads to genome-wide instability and altered gene expression profiles (Liang and Weisenberger, 2017). Interestingly, it has been observed that proto-oncogenes tend to have reduced DNA methylation levels during tumorigenesis. This reduction in methylation promotes the elevated expression of these proto-oncogenes, potentially contributing to tumor development (Casalino and Verde, 2020). Conversely, tumor suppressor genes often exhibit increased DNA methylation levels, which inhibits their expression and further promotes tumor growth. Recent studies have also shown that specific methylation patterns can be detected in circulating tumor cells, blood, urine, and other bodily fluids (Palanca-Ballester et al., 2021). These methylation patterns, known as tumor-specific methylation genes, have become valuable tools for early tumor diagnosis and prognosis (Oliver et al., 2022). For example, many studies have discussed DNA methylation as a potential diagnostic biomarker in early-stage ovarian cancer (Singh et al., 2019). Singh et al. (Singh et al., 2020) revealed that HOXA9 and HIC1 can be used as suitable diagnostic serum biomarkers for early screening of ovarian cancer. Moreover, researchers have utilized a phenomenon called CpG Island methylator phenotype (CIMP) to characterize different subtypes of gastric cancer (Tahara et al., 2019). In the field of breast cancer research, DNA methylation features have also proven useful in identifying different subtypes of breast cancer (Ma et al., 2023).
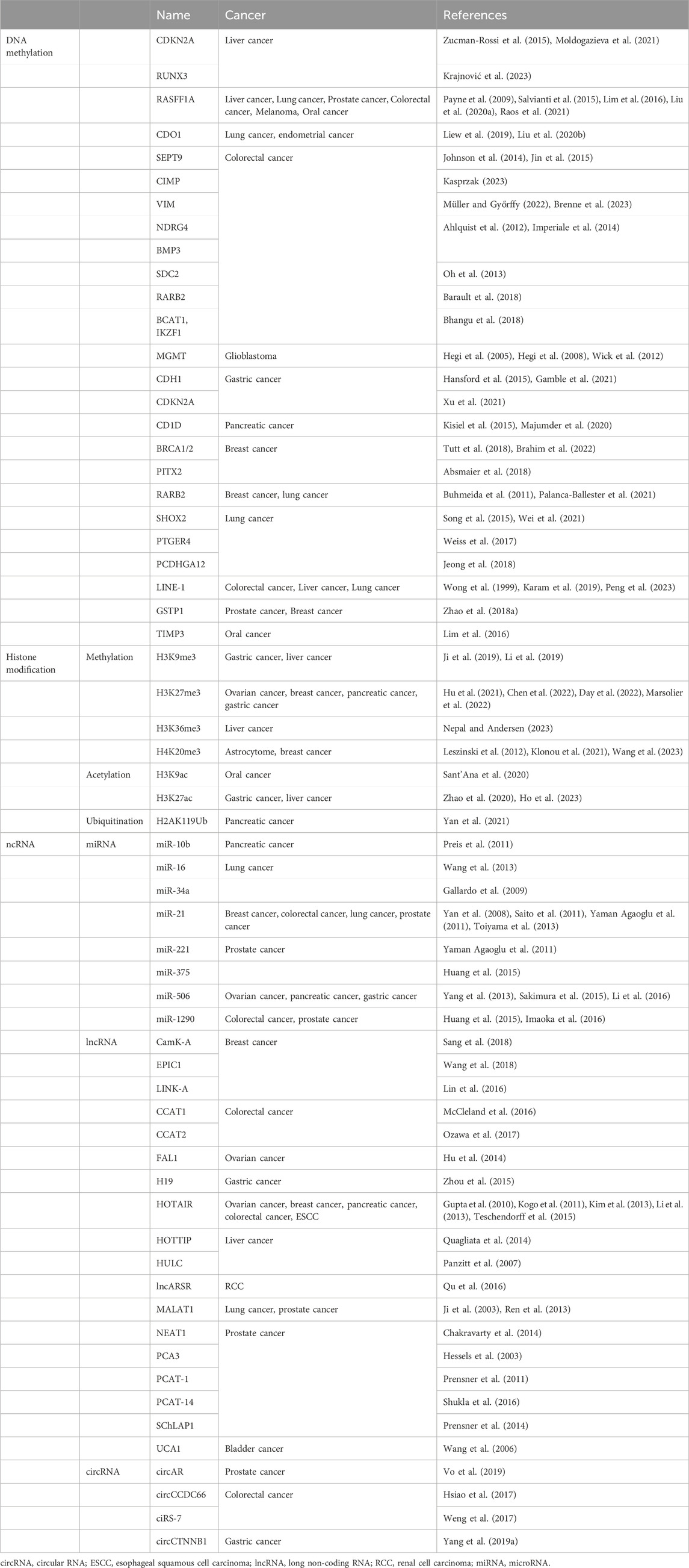
2.1 Common tumor biomarkers of DNA methylation
2.1.1 CIMP
CIMP is a CPG island that is abundant at CPG sites in the promoter region of DNA and exhibits a greatly methylated form (Suzuki et al., 2014). It can catalyze the transfer of DNMT from S-adenosine methionine to cause conformational changes in certain regions of DNA that affect protein-DNA interactions (Teodoridis et al., 2008). When methylation reaches a certain level, the regional DNA structure shrinks, the helix deepens, and the primordial regions on which numerous protein factors rely for binding shrink into large grooves, which is not conducive to transcription initiation (Sharma et al., 2010). It can cause the silencing of transcriptional genes or the inactivation of DNA repair genes and tumor suppressor genes, leading to tumor development. In 2006, Weisenberger’s research group in the United States recommended a simple and sensitive CIMP marker gene (CACNA1G, IGF2, NEUROG1, RUNX3 and SOCS1) to distinguish CIMP-positive colorectal cancer (Weisenberger et al., 2006). Moreover, CIMP is strongly associated with various molecular phenotypes such as elevated microsatellite instability, Tp53 wild-type, KRAS wild-type, BRAF variant, PIK3CA variant, and MLH1 methylation (Advani et al., 2018). The present study combines CIMP with additional tumor molecular biomarkers such as BRAF and KRAS for colorectal cancer diagnosis and prognosis prediction.
2.1.2 CDO1
The CDO1 gene, also known as cysteine dioxygenase 1, is a metalloenzyme that contains iron and lacks a heme structure (Chen M. et al., 2023). Its main function is to convert cysteine into cysteine sulfite, which is crucial for the synthesis of taurine (Chen M. et al., 2023). Cysteine metabolism plays a key role in cell drying by regulating reactive oxygen species, especially CDO1, which plays a crucial role in inducing apoptosis by significantly increasing the production of ROS (Yamashita et al., 2018). Moreover, the CDO1 protein can interact with peroxisome proliferation-activating receptor (PPAR) γ, leading to the activation of CCAAT-enhancer-binding proteins (CEBP) α, an essential onco-transcriptional factor (Deng et al., 2015; Zhao et al., 2016). Hence, it is possible that CDO1 has a significant impact on inhibiting tumor growth during tumorigenesis. Numerous recent studies have identified elevated frequencies of DNA methylation in the CDO1 gene across various cancer types (Yamashita et al., 2018). Methylation of the CDO1 gene can be readily detected in the plasma and urine of lung cancer patients. When combined with other highly correlated methylated genes, it exhibits a sensitivity of 73% and specificity of 92% for diagnosing lung cancer (Liu et al., 2020b). Furthermore, CDO1 hypermethylation in various body fluids, apart from plasma, may aid in preoperative disease diagnosis. For instance, cervical scratches are a crucial source of evidence for the molecular detection of endometrial cancer. CDO1 hypermethylation in cervical scratches demonstrates a sensitivity of 85% and specificity of 88% for detecting endometrial cancer (Liew et al., 2019). CDO1 hypermethylation detection can also be beneficial in intraoperative diagnosis during gastric cancer surgery (Harada et al., 2019) and in evaluating routine biopsy samples for determining tumor eradication after neoadjuvant therapy for esophageal cancer (Ushiku et al., 2017).
2.1.3 Septin 9
In the realm of cellular division and control of the cell cycle, the gene Septin 9 plays a crucial role (Sun et al., 2020). The methylation of Septin 9 gene promoter within plasma was initially confirmed in the year 2008 (Lofton-Day et al., 2008). To assess the viability of Septin 9 as a marker in colorectal cancer screening, a comprehensive investigation known as the PRESEPT study was carried out. The outcomes revealed an overall sensitivity of 50% and a specificity of 91% for the detection purposes (Church et al., 2010). In a study comparing the plasma Epi proColon test to a fecal immunochemical test (FIT), it was observed that Septin 9 exhibited identical sensitivity (73%) to FIT, but its specificity (81.5% vs 97%) was considerably lower (Johnson et al., 2014).
The second-generation Septin 9 assay, termed Epi proColon 2, displayed enhanced specificity of 87% when contrasted with 82% in the control group (Jin et al., 2015). Additionally, Epi proColon 2 showcased a remarkable detection sensitivity of 75%, surpassing that of the FIT trial (75% vs 58%) (Wu et al., 2016). In a 2016 study conducted by Song et al. (Song et al., 2016), three distinct detection methods for Septin 9 were compared, ultimately revealing that Epi proColon demonstrated superior sensitivity but relatively lower specificity, with both figures hovering around 82%. On the other hand, the effectiveness of Epi proColon 2.0 mirrored that of the commercially available Seni-Colon assay, which has been approved by the China Food and Drug Administration. These assays demonstrated sensitivity ranging from 75% to 77% and specificity ranging from 96% to 97% (Loktionov, 2020).
2.1.4 NDRG4 and BMP3
NDRG4, a member belonging to the N-myc downstream regulatory gene family, encodes a protein that is specifically expressed in brain and heart tissues (Zhou et al., 2001). The involvement of NDRG4 revolves around cell proliferation, apoptosis, and cell differentiation, and it has been linked to various types of cancer (Melotte et al., 2010). BMP3, which is a type of bone morphogenetic protein, was initially reported in 2005 as one of the tumor suppression factors in colorectal cancer (Koinuma et al., 2005). In a study conducted in 2012, where 252 patients with colorectal cancer and 293 subjects with negative results from colonoscopy were screened, Cologuard (KRAS mutation + fecal hemoglobin + NDRG4 and BMP3 methylation) exhibited a sensitivity of 85% and specificity of 90% (Ahlquist et al., 2012). Furthermore, a study involving 9,989 participants in 2014 demonstrated that the Cologuard test achieved a sensitivity of 92% for colorectal cancer, 42% for precancerous lesions, and a specificity of 90%. In comparison, the FIT test had lower sensitivity (74%) but higher specificity (96%) (Imperiale et al., 2014). At present, a multicenter prospective clinical trial based on Cologuard 2.0 is underway, and the results are currently pending (NCT04144751).
2.1.5 MGMT
MGMT is a greatly efficient DNA methyltransferase that transfers methyl groups from DNA molecules to their own amino acid residues (Fang, 2024). MGMT can repair damage to cellular DNA caused by alkylation of various agents and prevent DNA mutations caused by methylation, and thereby preventing cancer (Butler et al., 2020). Levels of MGMT expression in normal tissues are very low under physiological conditions, but increase under alkylating agents or radiation (Chen Y. et al., 2023). A 2005 randomized trial showed that patients with methylated MGMT promoters had a greater survival benefit than patients with unmethylated MGMT promoters in glioblastoma patients treated with combination radiotherapy and temozolomide chemotherapy (Hegi et al., 2005). Therefore, glioblastoma can be divided into two categories based on whether or not there is methylation of the MGMT promoter. Among them, the unmethylated patients were resistant to temozolomide and had poor efficacy (Hegi et al., 2008). The 2018 NCCN guidelines clearly suggest that MGMT is an important prognostic and outcome predictor of glioblastoma. In addition to predicting patient response to chemotherapy, MGMT methylation can also be used to predict radiotherapy response and prognosis for glioblastoma without adjuvant alkylating chemotherapy (Wick et al., 2012). At present, MGMT test kits have been developed and efforts are underway to find more sensitive and accurate means of detection.
2.1.6 LINE-1
LINE-1, also known as long-spread repeat 1, is a type of non-LTR retrotransposon (Mendez-Dorantes and Burns, 2023). Its transposition has the potential to cause changes and rearrangements in the DNA of host cells, which can lead to the development of various serious genetic diseases such as cancer (Kerachian and Kerachian, 2019). In normal tissues, LINE-1 is heavily methylated, but in tumor tissues, its methylation status is reduced (demethylated) (Lavia et al., 2022). Research has demonstrated that demethylated LINE-1 can serve as a reliable biomarker for detecting lung cancer (Stading et al., 2021). P16 is an important tumor suppressor gene located on human chromosome 9p21 (Serra and Chetty, 2018). It functions by inhibiting the growth of tumor-killing cells (Muss et al., 2020). Several studies have indicated that hypermethylation of the p16 gene is strongly associated with poor prognosis in patients diagnosed with colorectal cancer (Karam et al., 2019), liver cancer (Wong et al., 1999), and lung cancer (Peng et al., 2023). Another gene involved in cancer development is glutathione S-transferase 1 (GSTP1). Research has shown that the absence of GSTP1 promoter methylation expression in prostate cancer is associated with a sensitivity of 73%, specificity of 100%, positive predictive value of 100%, and negative predictive value of 78% (Miyake et al., 2012).
2.2 Inhibitors of DNA methylation
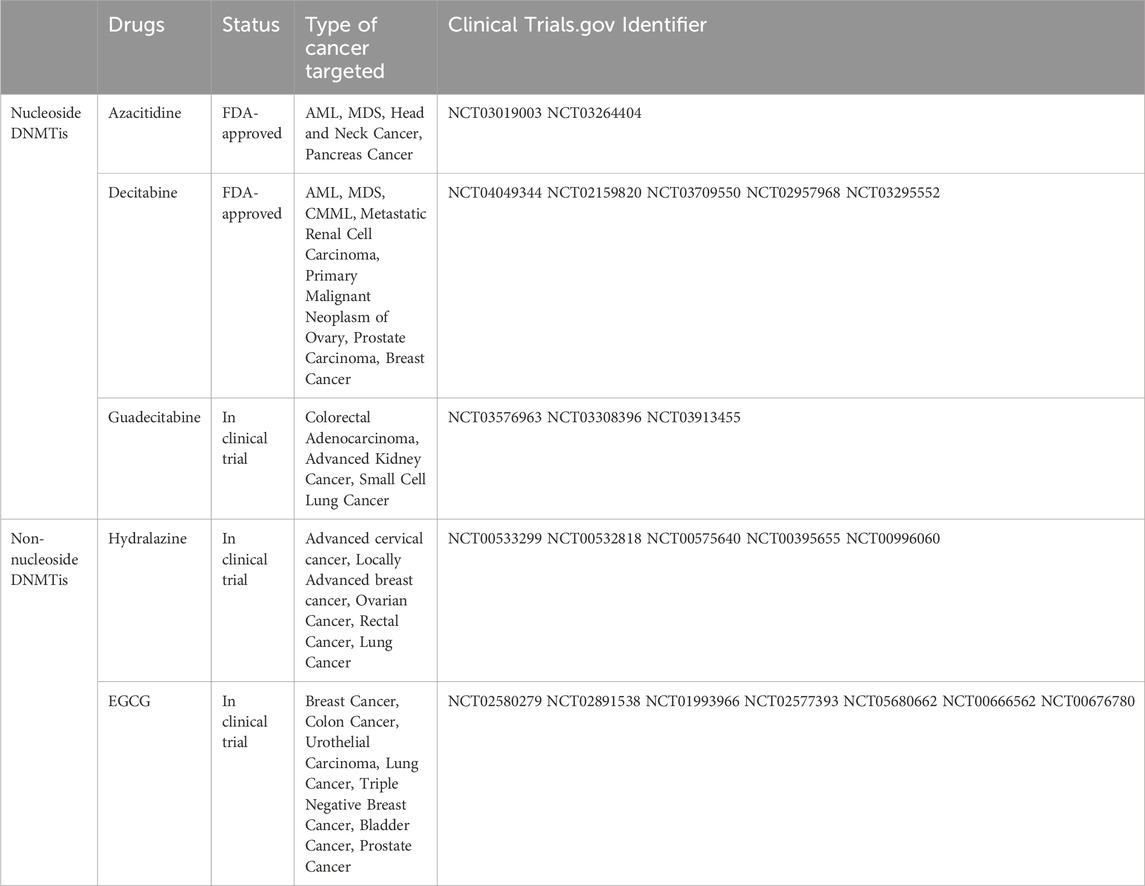
One of the earliest epigenetic drugs used to treat cancer were DNMT inhibitors (DNMTis) (Zhou et al., 2018). Currently, DNMTis developed for abnormal DNA methylation are divided into nucleoside inhibitors and non-nucleoside inhibitors.
2.2.1 Nucleoside inhibitors
Nucleoside inhibitors primarily include azacitidine, decitabine, zebularine and derivatives of cytosine nucleosides as substrates. Azacitidine is the first epigenetic drug approved by the FDA for the treatment of cancer (Myasoedova et al., 2019). It is a nucleoside analogue of cytidine that can specifically trap DNMT and thus achieve the effect of inhibiting DNA methylation (Raslan and Garcia-Horton, 2022). Azacitidine is mainly used to treat myelodysplastic syndromes (MDS) (Merz et al., 2024). The other drug approved by the FDA is decitabine (Myasoedova et al., 2019). It works by binding to DNA, causing DNA methylation and stopping DNA replication during the S phase (Dhillon, 2020). In 2020, the FDA approved an oral fixed-dose combination tablet containing deccitabine and cedazuridine (DEC-C) for the treatment of MDS (Kim et al., 2022). Zebularine is a cytidine DNA methylation inhibitor containing 2-(1H) pyrimidine cycloketone, which can effectively remove the DNA methylation modification of p16 gene in the hypermethylation modification state by covalently binding with DNMT (Andrade et al., 2017). Currently, Zebularine is in preclinical experiments for the treatment of breast cancer and certain blood tumors (Marquez et al., 2005; Chen et al., 2012). The next-generation of DNMTis includes guadecitabine, which is a dinucleotide prodrug of decitabine (Daher-Reyes et al., 2019). It combines decitabine and guanosine into one molecule (Wong et al., 2022). The longer half-life and better bioavailability make guadecitabine well tolerated in myelodysplastic syndrome and acute myeloid leukaemia patients with MDS (Issa et al., 2015; Kantarjian et al., 2017; Sébert et al., 2019). A phase Ⅰ dose-escalation trial demonstrated that guadelitabine combined with pembrolizumab is tolerated and has biological and anticancer activity in patients with advanced solid tumors (Papadatos-Pastos et al., 2022). In addition, azacitidine derivative CP-4200 contains hydrophobic fragments that improve its delivery to cancer cells, making it independent of the activity of the nucleoside transporter mechanism (Hummel-Eisenbeiss et al., 2013).
2.2.2 Non-nucleoside inhibitors
Non-nucleoside inhibitors do not contain the cytosine nucleoside skeleton structure and can bind directly to methylated regions in DNMT, rendering the enzyme ineffective (Del Castillo Falconi et al., 2022). Procaine is a benzoic acid compound with local anesthetic and anti-arrhythmic activity, but it has been found to inhibit the growth and demethylate liver cancer cells by treating them (Tada et al., 2007). Similar to Procaine, Procainamide is also a derivative of aminobenzoic acid, which is currently used to treat conditions such as anxiety and irregular heartbeat, and also has demethylation effects (Lee et al., 2005). Gao et al. found that procaine and procainamine may have potential use in preventing the development of lung cancer (Gao et al., 2009). Hydralazine is commonly used to treat high blood pressure and heart failure (Chaemsaithong et al., 2024). The study showed that hydrazine was able to restore the expression of tumor suppressor genes in cancer cell lines and primary tumors where promoter hypermethylation was silenced (Segura-Pacheco et al., 2003). In a phase II, single-arm study, hydralazine and valproate increased the chemotherapy efficacy of doxorubicin and cyclophosphamide (Arce et al., 2006). Then, a phase II study found that hydralazine and valproate could overcome chemotherapy resistance in patients with advanced refractory solid tumors (Candelaria et al., 2007). A randomized phase III demonstrates that epigenetic therapy (hydralazine and valproate) has a significant advantage over the current standard combination chemotherapy in terms of progression-free survival of cervical cancer (Coronel et al., 2011). In addition, a randomized, double-blind phase III trial comparing hydrazine valproate with placebo is ongoing (Saito et al., 2010). Epigallocatechin-3-gallate (EGCG) are bioactive polyphenol compounds that regulate epigenetic changes, including DNA methylation and histone modification (Włodarczyk et al., 2024). EGCG has become an important pathway for epigenetic therapy for leukemia and MDS, and it also has synergies when used in combination with conventional chemotherapy drugs (Della Via et al., 2023). In addition, in phase Ⅱ double-blind, placebo-controlled randomized clinical trial, preventive use of EGCG solution significantly reduced the incidence and severity of RID in patients with adjuvant radiotherapy for breast cancer (Zhao et al., 2022). And, in a randomized, double-blind trial, EGCG showed good tolerance, but across the study population as a whole, there was no statistically significant difference in adenoma rates between the EGCG and placebo groups (Seufferlein et al., 2022). Countless different anti-tumor applications of these drugs are still in the research stage, such as SGI-1027, RG108 analogues, Quinazoline, Propiophenone, and Pyrrolopyridine derivatives (Pechalrieu et al., 2017) (Table 1).
3 Histone modification
Histone proteins are the basic structural proteins of eukaryotic chromosomes and are a class of minor molecular alkaline proteins, with 5 types: H1, H2A, H2B, H3 and H4 (Fang et al., 2021). The histone amino terminal (n-terminal) domain extends out of the nucleosome and can interact with additional regulatory proteins and DNA (Robert and Jeronimo, 2023). Histone modifications include methylation, acetylation, ubiquitination, crotonylation and phosphorylation (Zaib et al., 2022). It can be passed from generation to generation in cells as a marker, thus constituting a “histone code” to effectively regulate specific genes. Imbalanced histone modification can lead to tumor development, and methylation and acetylation loss of histone H3 and H4 residues have been shown to be markers of tumor cells (Ray-Gallet and Almouzni, 2022). Histone modification is expected to be an effective epigenetic tumor marker.
3.1 Common tumor biomarkers of histone modification
3.1.1 Histone methylation modification
Histone methylation, which occurs on the amino terminal arginine or lysine residues of H3 and H4 histones, is a significant biochemical process (Huang et al., 2024). This methylation is mainly facilitated by histone methyltransferase (HMT), which can be further categorized into histone lysine methyltransferase (HKMT) and protein arginine methyltransferase (PRMT) (Patnaik et al., 2023). On the other hand, histone demethylases can be broadly classified into the lysine-specific demethylase (LSD) family and the JmjC domain-containing family (JMJD) (Tong et al., 2024). Methylation can occur at various sites, including H3 lysine (H3K) sites 4, 9, 27, 36, 79, and 20 of H4 lysine (H4K) (Hyun et al., 2017). According to several studies, specific methylation patterns, such as H3K9me3, H3K27me3, H3K36me3, and H4K20me3, serve as vital indicators for gastric, liver, breast, pancreatic, ovarian, and colon cancers (Sasidharan Nair et al., 2018; Sogutlu et al., 2022; An et al., 2023; Burlibasa et al., 2023; Tachaveeraphong and Phattarataratip, 2024). Furthermore, the expression of H3K9me2, H3K9me3, and H3K27me3 is significantly associated with clinical pathologies and may serve as independent risk factors for survival assessment in patients with gastric cancer (Li et al., 2019).
3.1.2 Histone acetylation modification
Histone acetylation is a reversible homeostatic process. The dynamical balance between histone acetylation and deacetylation is an influential factor in maintaining the stability of gene expression and chromatin structure, and disruption of this dynamical balance can lead to abnormal gene expression and lead to the occurrence and development of tumors (Zhao S. et al., 2018). This homeostasis is essentially co-regulated by histone acetyltransferase (HAT) and histone deacetylase (HDAC) (Audia and Campbell, 2016). HAT can add acetyl groups to lysine to make it positively charged, causing chromatin structures to open and promoting gene transcription, while HDAC can remove acetyl groups from lysine, reversing this process and inhibiting transcription (Dang and Wei, 2022). Studies have suggested that H3K9ac and H3K27ac may be markers of hepatocellular carcinoma and pancreatic cancer (Liu Y.-X. et al., 2020; Ono et al., 2021). Dou et al. (Dou et al., 2023) explained that H3K27 acetylation upregulates LINC00501 to promote gastric cancer metastasis through activation of epithelial-mesenchymal transformation and angiogenesis.
3.1.3 Histone phosphorylation modification
Histone phosphorylation is the phosphorylation of amino acid residues at the N terminal of a histone (Zhang et al., 2014). Its main types include histone H1 phosphorylation, histone H2A/H2B phosphorylation, histone H3 phosphorylation, and histone H4 phosphorylation, among others (Baker et al., 2010). The mechanism of histone phosphorylation may be due to the negative charge carried by the phosphate group neutralizing the positive charge on the histone, resulting in a decrease in the affinity between the histone and DNA (Rossetto et al., 2012). It is also possible to modify surfaces that can bind to protein recognition modules and interact with specific protein complexes. These two mechanisms affect the structure and function of chromosomes and are involved in physiological processes such as cell division (Ajiro et al., 2010). Currently, although histone phosphorylation has been extensively studied (Köhler et al., 2012; Abbaoui et al., 2017), its potential role as a tumor marker remains to be explored.
3.1.4 Histone ubiquitination and crotonylation modification
The modification sites for histone ubiquitination are located primarily on the C-terminal lysine residues of H2A, H2B, H3, and connexin H1 (Mattiroli and Penengo, 2021). Ubiquitination can initiate the transcription of target genes through interactions with other histone modifications and as a recruitment signal for transcription factors (Mattiroli and Penengo, 2021). Studies have found that an increase in H2AK119Ub1 and a decrease in H3K27Me3 may be markers of molecular staging of pancreatic cancer (Chen et al., 2014).
The concept of Crotonylation was first proposed in 2011, which can occur on lysine (K) residues of both histone and non-histone proteins and has similarities with acetylation in its structure, recognition proteins and regulated enzyme systems (Tan et al., 2011; Sabari et al., 2015). Although little research has been done, it plays an important role in regulating transcriptional activity, stress protection from kidney injury, and spermatogenesis and development (Ruiz-Andres et al., 2016) (Table 2).
3.2 Inhibitors of histone modification
Currently, histone methyltransferases inhibitors (HMTis) and histone deacetylases inhibitors (HDACis) are the main histone methyltransferases inhibitors in clinical trials (Epp et al., 2023). There are relatively few studies of additional types of histone-targeting inhibitors (Figure 2).
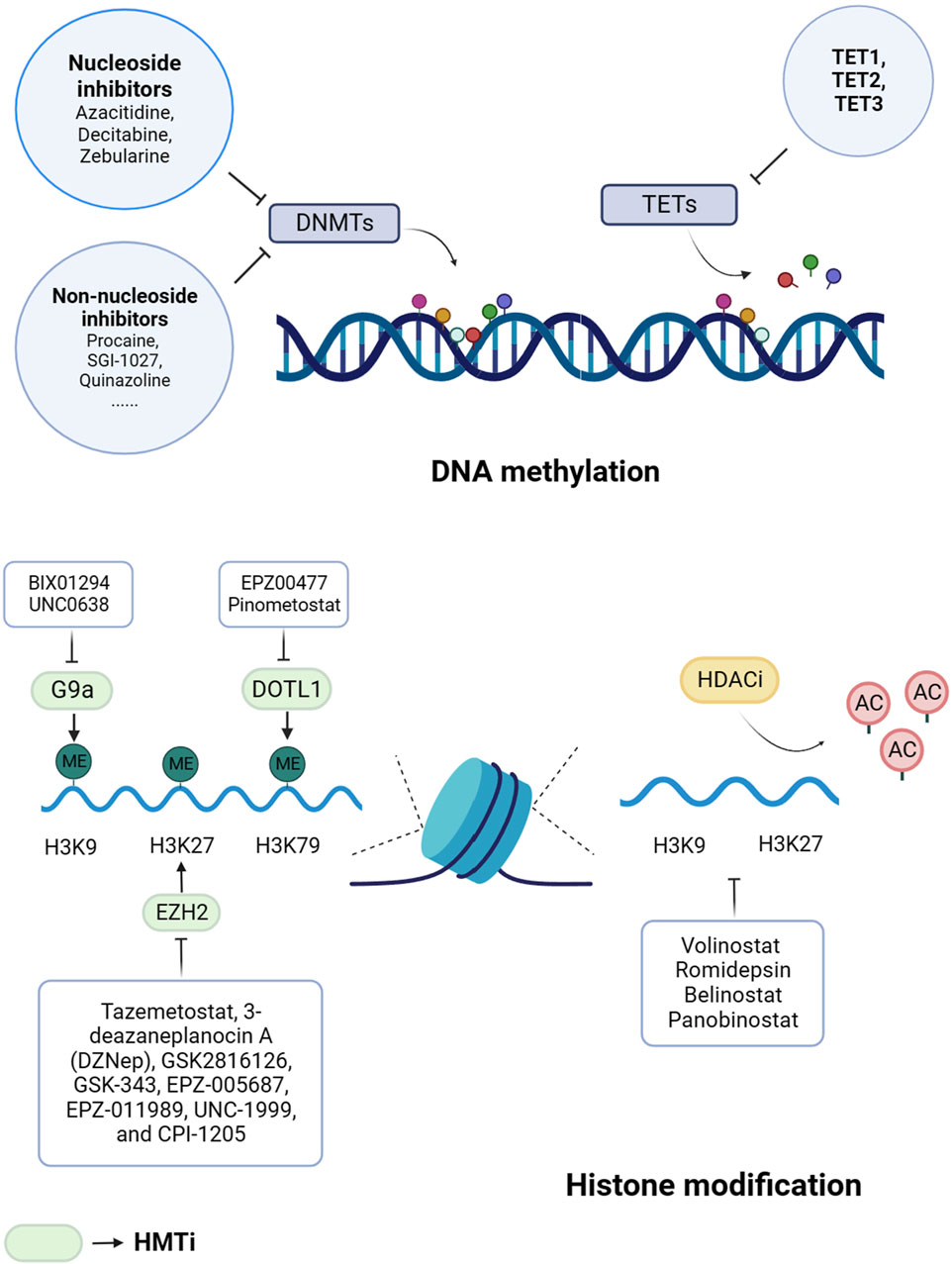
Figure 2. Epigenetic drugs for cancer treatment. Histone methyltransferases inhibitors (HMTis) and histone deacetylases inhibitors (HDACis) are the main histone methyltransferases inhibitors. DNMTs, DNA methyltransferases; TETs, ten-eleven translocation enzymes; H3, histone 3; H4, histone 4; K, lysine; ME, methylation; AC, acetylation; DOT1L, DOT1-like histone lysine methyltransferase; EZH2, enhancer of zeste homolog 2.
3.2.1 HMTi
3.2.1.1 EZH2
EZH2 is an important enzyme involved in the activity of the PRC2, which is responsible for the methylation of histone H3K27 (Guo et al., 2024). This methylation process helps in the formation of heterochromatin and the inhibition of transcription (Lee et al., 2022). EZH2 has the ability to promote the growth and spread of various types of tumors, including melanoma (White et al., 2021), oral squamous cell carcinoma (Zhou et al., 2023), and breast cancer (Velez et al., 2024). There are several EZH2 inhibitors, including Tazemetostat, 3-deazaneplanocin A (DZNep), GSK-343, EPZ-005687, EPZ-011989, UNC-1999, and CPI-1205, that have shown promising results in preclinical studies (Wan et al., 2023). However, their efficacy and safety need to be further evaluated in clinical trials. Currently, Tazemetostat is undergoing phase Ⅰ/II clinical trials for the treatment of various tumors (Italiano et al., 2018; Gounder et al., 2020; Morschhauser et al., 2020; Sarkozy et al., 2020; Zauderer et al., 2022). Additionally, CPI-1205 has shown strong antitumor effects in xenograft models and is undergoing phase I clinical trials (Vaswani et al., 2016). In a phase I study, GSK2816126 demonstrates moderate anticancer activity at tolerated doses in solid tumors or B-cell lymphomas (Yap et al., 2019).
3.2.1.2 DOT1L
DOT1L is responsible for the methylation modification of histone H3K79 and is also an important target for drug selection of methylase inhibitors (Alexandrova et al., 2022). Yang et al. (Yang L. et al., 2019) found that the use of DOT1L inhibitor EPZ004777 could induce cell cycle arrest of colorectal cancer cells. Another DOT1L inhibitor, EPZ-5676, showed promising results in a phase I clinical trial in MLL-rearranged leukemia (Stein et al., 2018) and was validated as ‘orphan drug’ toward MLL-rearranged leukemia by FDA (Cao et al., 2021).
3.2.1.3 G9a
G9a (also known as euchromatic histone-lysine N-methyltransferase 2, EHMT2) and G9A-like protein (GLP) are lysine methyltransferases that catalyze H3K9me1 and H3K9me2 and methylation modification of H3K27 (Haebe et al., 2021). G9a and GLP form a complex in the body that has been shown to promote tumor cell growth and affect cell cycle and metabolic pathways (Shinkai and Tachibana, 2011). Studies have found that G9a is overexpressed in esophageal squamous cell carcinoma, hepatocellular carcinoma, invasive lung cancer, brain cancer, multiple myeloma, and invasive ovarian cancer (Casciello et al., 2015). BIX01294 is the first substrate-competitive inhibitor that selectively inhibits G9a/GLP and downregulates H3K9me2 expression. However, BIX01294 has weak activity in cell assays and is cytotoxic at concentrations higher than 4.1 μmol/L (Kaniskan and Jin, 2015). UNC0638 is a further synthetic compound based on UNC0224 and UNC0321 that significantly downregulates H3K9me2 in a variety of cells. However, UNC0638 has poor pharmacokinetics in vivo and is not suitable for animal studies (Kaniskan et al., 2018).
3.2.2 HDACis
The HDACis have shown potential in inhibiting the growth of cancer cells, as well as inducing cancer cell differentiation and apoptosis by suppressing HDAC activity and promoting histone acetylation (Ramaiah et al., 2021). These inhibitors can be classified into four categories based on their chemical structure: short-chain fatty acids, hydroxamic acids, cyclic peptides, and benzamide derivatives (Shanmugam et al., 2022). In the United States, the FDA has currently approved four commonly used HDACis for the treatment of various hematologic tumors and solid tumors (Zhang et al., 2015).
3.2.2.1 Volinostat
Volinostat, also called nitrosamine hydroxamic acid (SAHA), is a drug that belongs to the hydroxamic acid category. Its approval by the US FDA in 2006 for treating cutaneous T-cell lymphoma (Siegel et al., 2009) marked a significant milestone. In the context of solid tumors, Volinostat is capable of inducing apoptosis in prostate tumor cells by blocking the Akt/FOXO3a signaling pathway (Shi et al., 2017). This inhibition can prevent the development of castration-resistant prostate cancer, which typically arises due to conventional therapies such as androgen deprivation therapy. Additionally, Volinostat reduces the adverse effects associated with drug resistance and dose toxicity induced by chemotherapy drugs such as Paclitaxel (Shi et al., 2017). Moreover, SAHA exhibits potential in improving the radio sensitivity of pancreatic cancer cells, making it a promising agent for enhancing the efficacy of radiotherapy against pancreatic cancer in the future (Wu et al., 2017). Notably, a clinical trial (NCT00731731) investigated the combination of Volinostat, Temozolomide, and radiotherapy for early treatment of glioblastoma multiforme has been reported. The positive anti-tumor effects of SAHA notwithstanding, it is crucial to highlight the elevated toxicity observed with high doses of the drug, resulting in adverse effects like fatigue, diarrhea, anorexia, dehydration, myelosuppression, thrombocytopenia, among others (Mrakovcic et al., 2017). As a consequence, enhancing the selectivity of HDACis or refining existing HDACis based on SAHA’s essential pharmacophore group holds great potential for future applications.
3.2.2.2 Romidepsin
FK228, also referred to as Romidepsin, is a member of the cyclic peptide class of inhibitors. In 2012, the approval for the treatment of cutaneous T cell lymphomas (CTCL) and peripheral T-Cell lymphomas (PTCL) was granted by the US FDA (Mottamal et al., 2015). HDAC1 and HDAC2 are directly inhibited by Romidepsin (Saijo et al., 2015). In cell culture experiments, it exhibits similar effects to SAHA, as it has the capability to impede cancer cell growth and induce both DNA double-strand break damage and cell apoptosis simultaneously (Pojani and Barlocco, 2021). Additionally, other research has demonstrated the effectiveness of combining Romidepsin with chemotherapy drugs for the treatment of non-small cell lung cancer (NSCLC) (Karthik et al., 2014).
3.2.2.3 Belinostat
Belinostat, an HDACis medication approved by the FDA in 2014 specifically for treating PTCL. It is a hydroxylate that effectively inhibits the activity of both class I and class II HDACs (McDermott and Jimeno, 2014). This drug carries significant anti-tumor properties, particularly against solid tumors. For instance, Belinostat has demonstrated its ability to induce apoptosis in PC3 cells, a human prostate cancer cell line, through a mitochondrial pathway during prostate cancer treatment (Molife and de Bono, 2011). This process effectively hinders the development and manifestation of prostate cancer (Gravina et al., 2012). Moreover, Belinostat disrupts the expression of ubiquitin-related proteins by triggering proteasomal degradation of the SOS protein and down-regulating downstream MAPK signal transduction. This interference effectively impacts the functioning of critical survival signals in lung squamous cell carcinoma, thereby promoting cellular apoptosis (Kong et al., 2017).
3.2.2.4 Panobinostat
In 2015, the FDA approved Panobinostat for clinical trials as a treatment for multiple myeloma (Laubach et al., 2015). As a hydroxylated derivative, Panobinostat exhibits inhibitory properties on HDACs from class Ⅰ, class Ⅱ, and class IV (Biersack et al., 2022). Currently, Panobinostat is being utilized for the treatment of other tumor types, including thyroid cancer (NCT01013597), colorectal cancer, and prostate cancer (NCT00663832) (Losson et al., 2016). In a study conducted by Ferrari et al. (Ferrari et al., 2019), the efficacy and safety of combining Panobinostat with Bicalutamide for hormone-resistant prostate cancer were evaluated. The results demonstrated that this combination therapy could effectively prolong the progression-free survival time of hormone-resistant prostate cancer, while remaining tolerable in terms of its toxic effects.
Moreover, anti-tumor research also emphasizes the significance of SIRTs (sirtuins), which belong to the family of NAD (+)-dependent class III histone deacetylases. Although the FDA has not yet approved any drugs targeting SIRTs, numerous studies have highlighted the anti-tumor effects of SIRT inhibitors. One such inhibitor is Suramin, which selectively targets SIRT1 and SIRT2. Currently, clinical trials are underway to explore its potential use in various cancers (Villalba and Alcaín, 2012).
4 ncRNA modification
ncRNA refers to RNA that does not code for proteins, including microRNA (miRNA), long non-coding RNA (lncRNA) and circular RNA (circRNA), etc., they have important functions in life activities (Yan and Bu, 2021). miRNAs are small RNAs with a total length of about 22 nucleotides (nt). The miRNAs bind to specific target miRNAs via complementary sequences, thereby inducing miRNA division, degradation, or translation blocking (Vos et al., 2019). Both lncRNAs and circRNAs are longer than 200nt, but lncRNAs are linear while circRNAs are circular (Wu et al., 2023). LncRNAs and circRNAs regulate gene expression through multiple mechanisms. They can act as decoys for miRNAs, preventing the degradation of the targeted mRNA (Batista and Chang, 2013). They can regulate the binding of transcription factors to promoters and thus regulate the expression of target genes (Fatica and Bozzoni, 2014). They can also serve as scaffolds to regulate protein-protein interactions and associated downstream signaling pathways (Flynn and Chang, 2014).
4.1 Common tumor biomarkers of ncRNA
Research has demonstrated the existence of diverse categories of ncRNAs, such as miRNA, lncRNA, and circRNA, which exhibit differential expression patterns in malignant tissues compared to adjacent tissues (Wang et al., 2019; Yan and Bu, 2021). Consequently, ncRNAs hold great potential as diagnostic and prognostic biomarkers for tumors. Up to now, urinary PCA3 represents the sole FDA-approved molecular tumor diagnostic assay derived from ncRNA (PRO-GENSA PCA3) (Borbiev et al., 2023). Investigations have revealed that PCA3 serves as a specific indicator that is upregulated in prostate cancer and can be identified in patient urine (Chen J.-Y. et al., 2023).
Numerous ongoing clinical trials are currently examining the feasibility of utilizing ncRNAs as indicators for tumors (Zhou et al., 2022). A diagnostic study, conducted in a prospective, longitudinal, blinded, observational manner, involved 400 patients who underwent low-dose computed tomography (LDCT) screening specifically for lung cancer. The main objective of this study was to assess if the miRNA profile (HMBDx, exclusively licensed to Hummingbird) outperforms LDCT in diagnosing lung cancer (NCT03452514). Previous investigations have indicated that miRNA-10b exhibits higher expression levels in glioblastomas compared to normal brain tissue (El Fatimy et al., 2017). A clinical trial, comprising of around 200 patients diagnosed with glioblastomas, aimed to evaluate if the expression levels of miR-10b in primary tumor, blood, and cerebrospinal fluid samples could serve as prognostic and diagnostic markers for glioblastomas (NCT01849952). Moreover, a case-control observational study utilized next-generation sequencing to analyze the differential expression of miRNAs and lncRNAs in blood samples obtained from 160 patients diagnosed with high-grade serous ovarian cancer and benign gynecologic disease (NCT03738319).
4.2 Therapeutic targeting of ncRNAs in cancer
In the field of tumorigenesis and development, ncRNAs play a crucial role as regulatory molecules. Hence, there has been a significant focus on the development of efficient therapeutic techniques for suppressing (proto-oncogenes) or increasing expression (tumor suppressor genes) of ncRNAs (Rupaimoole and Slack, 2017; Arun et al., 2018). Although most studies investigating ncRNA-targeted drugs for cancer are in the preclinical stage, they demonstrate the immense potential of this approach. For illustration, using antisense oligonucleotides, the deletion of lncRNA MALAT1 in animal models effectively impacts the growth and metastasis of lung and breast cancer cells (Gutschner et al., 2013; Arun et al., 2016). In the subsequent section, we present various clinical trials focusing on miRNAs, showcasing the therapeutic endeavors in this field.
Research findings suggest that miR-34, a miRNA that acts as a tumor suppressor, is directly controlled by p53 and has the ability to regulate the expression of multiple oncogenes (Liu et al., 2011; Adams et al., 2016a; Adams et al., 2016b). In a phase I study conducted in multiple centers (NCT01829971/NCT02862145), the safety of MRX34, liposomes containing MicroRNA miR-RX34, was assessed in patients with primary liver cancer, metastatic cancer, or hematologic malignancies involving the liver. Although MRX34 displayed some antitumor effects, immune-related toxicity became evident among several participants during the phase Ib examination. The final outcomes of the tests are yet to be published. Another study (NCT02369198) conducted a phase I trial to evaluate the maximum tolerance dose (MDT) of TargomiRs, minicells loaded with miR-16 mimic miRNA and aimed at EGFR, in patients with relapsed malignant pleural mesothelioma and NSCLC (Reid et al., 2013). 26 patients were treated with TargomiRs and it was determined that 5×109 TargomiRs once weekly was the MDT. The proportion of patients who achieved an objective response was one of 22 (5%). Therefore, further investigation into the use of TargomiRs in combination with chemotherapy or immune checkpoint inhibitors is warranted. H19, a frequently upregulated lncRNA in various cancer types, has been extensively utilized by researchers to achieve cancer-specific expression of downstream sequences (Bhan et al., 2017). BC-819, also known as DTA-H19, is a DNA plasmid that encodes diphtheria toxin A (DTA) under the control of the H19 promoter and can be administered as a complex with polyethylenimine (PEI) (Gofrit et al., 2014). Ongoing clinical trials involving BC-819 primarily focus on bladder cancer. A phase II, open-label, single-arm, monotherapy study conducted across multiple centers aims to examine 140 non-muscle-invasive bladder cancer patients who have shown resistance to Bacillus Calmette-Guerin (BCG) treatment (NCT03719300).
5 Discussion
Initially, cancer was thought to be an inherited disease, but over the past few decades, researchers have established a strong link between epigenetic factors and tumorigenesis. With the help of next-generation sequencing technology, people have successfully discovered changes in epigenome control genes that play a key role in tumor development. For example, single-cell chromatin overall omic-scale landscape sequencing (scCOOL-seq) revealed a new set of candidate biomarkers—ZNF667 and ZNF667-AS1, whose expression is associated with better prognosis in patients with pancreatic ductal adenocarcinoma by influencing the proliferation of cancer cells (Fan et al., 2022). In addition, in the early stages of the disease, people can detect epigenetic modification changes in blood or tissue marker genes for early diagnosis of the disease (Davalos and Esteller, 2023). For example, in 2014, the FDA approved Cologuard for screening for colorectal cancer. It is based on the detection of specific epigenetic changes in fecal DNA.
The epigenetic dysregulation that occurs in cancer makes epigenetic mechanisms a new target for drug development. There are only a few epigenetic drugs approved by the FDA, but more and more epigenetic drugs have been put into the clinical research of cancer treatment, and have achieved remarkable results. Epigenetic modifiers mainly regulate histone post-transcriptional modification and nucleosome assembly. In the future, epigenetic modifiers will also regulate key processes such as DNA repair, genome integrity and RNA splicing. Still, there are problems with epigenetic drugs that need to be addressed. For example, targeted delivery of drugs by siRNA has potential off-target problems. Drugs that interfere with DNA methylation modification, such as azacitidine and decitabine, can cause bone marrow suppression and gastrointestinal symptoms, while epigenetic drugs that can interfere with histone modification are generally cytotoxic. Answers to these questions may better promote the role of epigenetics in treating disease.
In the future, combination therapy based on epigenetic regulators may be the development direction of epigenetic therapy for solid tumors. First, epigenetic regulators can regulate NK cell function, restore depleted T cells and increase immune cell infiltration. Therefore, the combination of epigenetic drugs may enhance the efficacy of tumor immunity. Second, there is some resistance to traditional anticancer drugs. Drug-resistant cancer cells exhibit high chromatin inhibition and elevated levels of KDM5A. Epigenetic drugs can inhibit the development of drug resistance and enhance drug efficacy. In conclusion, the related research and development of epigenetic drugs will become an important way of cancer treatment.
Author contributions
JG: Writing–original draft, Writing–review and editing. WS: Writing–review and editing. JW: Data curation, Writing–review and editing. CG: Conceptualization, Writing–review and editing. QD: Methodology, Writing–review and editing. JS: Software, Writing–review and editing. XZo: Software, Writing–review and editing. ZX: Methodology, Writing–review and editing. YG: Investigation, Writing–review and editing. CY: Investigation, Writing–review and editing. JL: Conceptualization, Writing–review and editing. HB: Investigation, Writing–review and editing. XZh: Writing–review and editing. YC: Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (NO. 82270599) and Chen Xiao-ping Foundation for the Development of Science and Technology of Hubei Province, China (CXPJJH11900002-058).
Acknowledgments
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbaoui, B., Telu, K. H., Lucas, C. R., Thomas-Ahner, J. M., Schwartz, S. J., Clinton, S. K., et al. (2017). The impact of cruciferous vegetable isothiocyanates on histone acetylation and histone phosphorylation in bladder cancer. J. Proteomics 156, 94–103. doi:10.1016/j.jprot.2017.01.013
Absmaier, M., Napieralski, R., Schuster, T., Aubele, M., Walch, A., Magdolen, V., et al. (2018). PITX2 DNA-methylation predicts response to anthracycline-based adjuvant chemotherapy in triple-negative breast cancer patients. Int. J. Oncol. 52 (3), 755–767. doi:10.3892/ijo.2018.4241
Adams, B. D., Parsons, C., and Slack, F. J. (2016a). The tumor-suppressive and potential therapeutic functions of miR-34a in epithelial carcinomas. Expert Opin. Ther. Targets 20 (6), 737–753. doi:10.1517/14728222.2016.1114102
Adams, B. D., Wali, V. B., Cheng, C. J., Inukai, S., Booth, C. J., Agarwal, S., et al. (2016b). miR-34a silences c-SRC to attenuate tumor growth in triple-negative breast cancer. Cancer Res. 76 (4), 927–939. doi:10.1158/0008-5472.CAN-15-2321
Advani, S. M., Advani, P., DeSantis, S. M., Brown, D., VonVille, H. M., Lam, M., et al. (2018). Clinical, pathological, and molecular characteristics of CpG island methylator phenotype in colorectal cancer: a systematic review and meta-analysis. Transl. Oncol. 11 (5), 1188–1201. doi:10.1016/j.tranon.2018.07.008
Ahlquist, D. A., Zou, H., Domanico, M., Mahoney, D. W., Yab, T. C., Taylor, W. R., et al. (2012). Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology 142 (2), 248–256. doi:10.1053/j.gastro.2011.10.031
Ajiro, K., Scoltock, A. B., Smith, L. K., Ashasima, M., and Cidlowski, J. A. (2010). Reciprocal epigenetic modification of histone H2B occurs in chromatin during apoptosis in vitro and in vivo. Cell Death Differ. 17 (6), 984–993. doi:10.1038/cdd.2009.199
Alexandrova, E., Salvati, A., Pecoraro, G., Lamberti, J., Melone, V., Sellitto, A., et al. (2022). Histone methyltransferase DOT1L as a promising epigenetic target for treatment of solid tumors. Front. Genet. 13, 864612. doi:10.3389/fgene.2022.864612
An, X., Lan, X., Feng, Z., Li, X., and Su, Q. (2023). Histone modification: biomarkers and potential therapies in colorectal cancer. Ann. Hum. Genet. 87 (6), 274–284. doi:10.1111/ahg.12528
Andrade, A. F., Borges, K. S., Suazo, V. K., Geron, L., Corrêa, C. A. P., Castro-Gamero, A. M., et al. (2017). The DNA methyltransferase inhibitor zebularine exerts antitumor effects and reveals BATF2 as a poor prognostic marker for childhood medulloblastoma. Investig. New Drugs 35 (1), 26–36. doi:10.1007/s10637-016-0401-4
Arce, C., Pérez-Plasencia, C., González-Fierro, A., de la Cruz-Hernández, E., Revilla-Vázquez, A., Chávez-Blanco, A., et al. (2006). A proof-of-principle study of epigenetic therapy added to neoadjuvant doxorubicin cyclophosphamide for locally advanced breast cancer. PloS One 1 (1), e98. doi:10.1371/journal.pone.0000098
Arun, G., Diermeier, S., Akerman, M., Chang, K.-C., Wilkinson, J. E., Hearn, S., et al. (2016). Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes & Dev. 30 (1), 34–51. doi:10.1101/gad.270959.115
Arun, G., Diermeier, S. D., and Spector, D. L. (2018). Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol. Med. 24 (3), 257–277. doi:10.1016/j.molmed.2018.01.001
Audia, J. E., and Campbell, R. M. (2016). Histone modifications and cancer. Cold Spring Harb. Perspect. Biol. 8 (4), a019521. doi:10.1101/cshperspect.a019521
Baker, S. P., Phillips, J., Anderson, S., Qiu, Q., Shabanowitz, J., Smith, M. M., et al. (2010). Histone H3 Thr 45 phosphorylation is a replication-associated post-translational modification in S. cerevisiae. Nat. Cell Biol. 12 (3), 294–298. doi:10.1038/ncb2030
Barault, L., Amatu, A., Siravegna, G., Ponzetti, A., Moran, S., Cassingena, A., et al. (2018). Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut 67 (11), 1995–2005. doi:10.1136/gutjnl-2016-313372
Batista, P. J., and Chang, H. Y. (2013). Long noncoding RNAs: cellular address codes in development and disease. Cell 152 (6), 1298–1307. doi:10.1016/j.cell.2013.02.012
Bhan, A., Soleimani, M., and Mandal, S. S. (2017). Long noncoding RNA and cancer: a new paradigm. Cancer Res. 77 (15), 3965–3981. doi:10.1158/0008-5472.CAN-16-2634
Bhangu, J. S., Beer, A., Mittlböck, M., Tamandl, D., Pulverer, W., Schönthaler, S., et al. (2018). Circulating free methylated tumor DNA markers for sensitive assessment of tumor burden and early response monitoring in patients receiving systemic chemotherapy for colorectal cancer liver metastasis. Ann. Surg. 268 (5), 894–902. doi:10.1097/SLA.0000000000002901
Biersack, B., Polat, S., and Höpfner, M. (2022). Anticancer properties of chimeric HDAC and kinase inhibitors. Seminars Cancer Biol. 83, 472–486. doi:10.1016/j.semcancer.2020.11.005
Borbiev, T., Kohaar, I., and Petrovics, G. (2023). Clinical biofluid assays for prostate cancer. Cancers 16 (1), 165. doi:10.3390/cancers16010165
Brahim, S. M., Zein, E. E., Bonnet, C., Hamed, C. T., Salame, M., Zein, M. V., et al. (2022). Screening of BRCA1/2 variants in Mauritanian breast cancer patients. BMC Cancer 22 (1), 802. doi:10.1186/s12885-022-09903-8
Brenne, S. S., Madsen, P. H., Pedersen, I. S., Hveem, K., Skorpen, F., Krarup, H. B., et al. (2023). Colorectal cancer detected by liquid biopsy 2 years prior to clinical diagnosis in the HUNT study. Br. J. Cancer 129 (5), 861–868. doi:10.1038/s41416-023-02337-4
Buhmeida, A., Merdad, A., Al-Maghrabi, J., Al-Thobaiti, F., Ata, M., Bugis, A., et al. (2011). RASSF1A methylation is predictive of poor prognosis in female breast cancer in a background of overall low methylation frequency. Anticancer Res. 31 (9), 2975–2981. Available at: https://ar.iiarjournals.org/content/31/9/2975
Burlibasa, L., Nicu, A.-T., Chifiriuc, M. C., Medar, C., Petrescu, A., Jinga, V., et al. (2023). H3 histone methylation landscape in male urogenital cancers: from molecular mechanisms to epigenetic biomarkers and therapeutic targets. Front. Cell Dev. Biol. 11, 1181764. doi:10.3389/fcell.2023.1181764
Butler, M., Pongor, L., Su, Y. T., Xi, L., Raffeld, M., Quezado, M., et al. (2020). MGMT status as a clinical biomarker in glioblastoma. Trends Cancer 6 (5), 380–391. doi:10.1016/j.trecan.2020.02.010
Candelaria, M., Gallardo-Rincón, D., Arce, C., Cetina, L., Aguilar-Ponce, J. L., Arrieta, O., et al. (2007). A phase II study of epigenetic therapy with hydralazine and magnesium valproate to overcome chemotherapy resistance in refractory solid tumors. Ann. Oncol. Official J. Eur. Soc. For Med. Oncol. 18 (9), 1529–1538. doi:10.1093/annonc/mdm204
Cao, M., Li, T., Chen, Y., and Zhai, X. (2021). Nucleoside and non-nucleoside DOT1L inhibitors: dawn of MLLrearranged leukemia. Mini Rev. Med. Chem. 21 (11), 1337–1350. doi:10.2174/1389557521666210111144357
Casalino, L., and Verde, P. (2020). Multifaceted roles of DNA methylation in neoplastic transformation, from tumor suppressors to EMT and metastasis. Genes 11 (8), 922. doi:10.3390/genes11080922
Casciello, F., Windloch, K., Gannon, F., and Lee, J. S. (2015). Functional role of G9a histone methyltransferase in cancer. Front. Immunol. 6, 487. doi:10.3389/fimmu.2015.00487
Chaemsaithong, P., Biswas, M., Lertrut, W., Warintaksa, P., Wataganara, T., Poon, L. C., et al. (2024). Pharmacogenomics of Preeclampsia therapies: current evidence and future challenges for clinical implementation. Best Pract. Res. Clin. Obstetrics Gynaecol. 92, 102437. doi:10.1016/j.bpobgyn.2023.102437
Chakravarty, D., Sboner, A., Nair, S. S., Giannopoulou, E., Li, R., Hennig, S., et al. (2014). The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 5, 5383. doi:10.1038/ncomms6383
Chen, J.-Y., Wang, P.-Y., Liu, M.-Z., Lyu, F., Ma, M.-W., Ren, X.-Y., et al. (2023a). Biomarkers for prostate cancer: from diagnosis to treatment. Diagn. (Basel, Switz.) 13 (21), 3350. doi:10.3390/diagnostics13213350
Chen, M., Shabashvili, D., Nawab, A., Yang, S. X., Dyer, L. M., Brown, K. D., et al. (2012). DNA methyltransferase inhibitor, zebularine, delays tumor growth and induces apoptosis in a genetically engineered mouse model of breast cancer. Mol. Cancer Ther. 11 (2), 370–382. doi:10.1158/1535-7163.MCT-11-0458
Chen, M., Zhu, J.-Y., Mu, W.-J., and Guo, L. (2023b). Cysteine dioxygenase type 1 (CDO1): its functional role in physiological and pathophysiological processes. Genes & Dis. 10 (3), 877–890. doi:10.1016/j.gendis.2021.12.023
Chen, S., Chen, J., Zhan, Q., Zhu, Y., Chen, H., Deng, X., et al. (2014). H2AK119Ub1 and H3K27Me3 in molecular staging for survival prediction of patients with pancreatic ductal adenocarcinoma. Oncotarget 5 (21), 10421–10433. doi:10.18632/oncotarget.2126
Chen, Y., Qu, W., Tu, J., and Qi, H. (2023d). Implications of advances in studies of O6-methylguanine-DNA- methyltransferase for tumor prognosis and treatment. Front. Biosci. (Landmark Ed.) 28 (9), 197. doi:10.31083/j.fbl2809197
Chen, Z., Yuan, L., Li, X., Yu, J., and Xu, Z. (2022). BMP2 inhibits cell proliferation by downregulating EZH2 in gastric cancer. Cell Cycle (Georget. Tex) 21 (21), 2298–2308. doi:10.1080/15384101.2022.2092819
Church, T. R., Wandell, M., Lofton–Day, C., Mongin, S., Blumenstein, B. A., Allen, J. I., et al. (2010). Prospective clinical validation of an assay for methylated SEPT9 DNA in human plasma as a colorectal cancer screening tool in average risk men and women ≥50 years. Gastroenterology 139 (1), e18. doi:10.1053/j.gastro.2010.05.068
Coronel, J., Cetina, L., Pacheco, I., Trejo-Becerril, C., González-Fierro, A., de la Cruz-Hernandez, E., et al. (2011). A double-blind, placebo-controlled, randomized phase III trial of chemotherapy plus epigenetic therapy with hydralazine valproate for advanced cervical cancer. Preliminary results. Med. Oncol. N. Lond. Engl. 28 (Suppl. 1), S540–S546. doi:10.1007/s12032-010-9700-3
Daher-Reyes, G. S., Merchan, B. M., and Yee, K. W. L. (2019). Guadecitabine (SGI-110): an investigational drug for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Expert Opin. Investigational Drugs 28 (10), 835–849. doi:10.1080/13543784.2019.1667331
Dang, F., and Wei, W. (2022). Targeting the acetylation signaling pathway in cancer therapy. Seminars Cancer Biol. 85, 209–218. doi:10.1016/j.semcancer.2021.03.001
Davalos, V., and Esteller, M. (2023). Cancer epigenetics in clinical practice. CA a Cancer J. For Clin. 73 (4), 376–424. doi:10.3322/caac.21765
Day, C. A., Hinchcliffe, E. H., and Robinson, J. P. (2022). H3K27me3 in diffuse midline glioma and epithelial ovarian cancer: opposing epigenetic changes leading to the same poor outcomes. Cells 11 (21), 3376. doi:10.3390/cells11213376
Del Castillo Falconi, V. M., Torres-Arciga, K., Matus-Ortega, G., Díaz-Chávez, J., and Herrera, L. A. (2022). DNA methyltransferases: from evolution to clinical applications. Int. J. Mol. Sci. 23 (16), 8994. doi:10.3390/ijms23168994
Della Via, F. I., Alvarez, M. C., Basting, R. T., and Saad, S. T. O. (2023). The effects of green tea catechins in hematological malignancies. Pharm. (Basel, Switz.) 16 (7), 1021. doi:10.3390/ph16071021
Deng, J. Y., Wu, X. Q., He, W. J., Liao, X., Tang, M., and Nie, X. Q. (2023). Targeting DNA methylation and demethylation in diabetic foot ulcers. J. Adv. Res. 54, 119–131. doi:10.1016/j.jare.2023.01.009
Deng, P., Chen, Y., Ji, N., Lin, Y., Yuan, Q., Ye, L., et al. (2015). Cysteine dioxygenase type 1 promotes adipogenesis via interaction with peroxisome proliferator-activated receptor gamma. Biochem. Biophys. Res. Commun. 458 (1), 123–127. doi:10.1016/j.bbrc.2015.01.080
Dhillon, S. (2020). Decitabine/cedazuridine: first approval. Drugs 80 (13), 1373–1378. doi:10.1007/s40265-020-01389-7
Dou, R., Han, L., Yang, C., Fang, Y., Zheng, J., Liang, C., et al. (2023). Upregulation of LINC00501 by H3K27 acetylation facilitates gastric cancer metastasis through activating epithelial-mesenchymal transition and angiogenesis. Clin. Transl. Med. 13 (10), e1432. doi:10.1002/ctm2.1432
El Fatimy, R., Subramanian, S., Uhlmann, E. J., and Krichevsky, A. M. (2017). Genome editing reveals glioblastoma addiction to MicroRNA-10b. Mol. Ther. J. Am. Soc. Gene Ther. 25 (2), 368–378. doi:10.1016/j.ymthe.2016.11.004
Epp, S., Chuah, S. M., and Halasz, M. (2023). Epigenetic dysregulation in MYCN-amplified neuroblastoma. Int. J. Mol. Sci. 24 (23), 17085. doi:10.3390/ijms242317085
Fan, X., Lu, P., Wang, H., Bian, S., Wu, X., Zhang, Y., et al. (2022). Integrated single-cell multiomics analysis reveals novel candidate markers for prognosis in human pancreatic ductal adenocarcinoma. Cell Discov. 8 (1), 13. doi:10.1038/s41421-021-00366-y
Fang, Q. (2024). The versatile attributes of MGMT: its repair mechanism, crosstalk with other DNA repair pathways, and its role in cancer. Cancers 16 (2), 331. doi:10.3390/cancers16020331
Fang, Z., Wang, X., Sun, X., Hu, W., and Miao, Q. R. (2021). The role of histone protein acetylation in regulating endothelial function. Front. Cell Dev. Biol. 9, 672447. doi:10.3389/fcell.2021.672447
Fatica, A., and Bozzoni, I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15 (1), 7–21. doi:10.1038/nrg3606
Feinberg, A. P., and Levchenko, A. (2023). Epigenetics as a mediator of plasticity in cancer. Science 379 (6632), eaaw3835. doi:10.1126/science.aaw3835
Ferrari, A. C., Alumkal, J. J., Stein, M. N., Taplin, M.-E., Babb, J., Barnett, E. S., et al. (2019). Epigenetic therapy with Panobinostat combined with Bicalutamide rechallenge in castration-resistant prostate cancer. Clin. Cancer Res. Official J. Am. Assoc. For Cancer Res. 25 (1), 52–63. doi:10.1158/1078-0432.CCR-18-1589
Fitz-James, M. H., and Cavalli, G. (2022). Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 23 (6), 325–341. doi:10.1038/s41576-021-00438-5
Flynn, R. A., and Chang, H. Y. (2014). Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 14 (6), 752–761. doi:10.1016/j.stem.2014.05.014
Galbraith, K., and Snuderl, M. (2022). DNA methylation as a diagnostic tool. Acta Neuropathol. Commun. 10 (1), 71. doi:10.1186/s40478-022-01371-2
Gallardo, E., Navarro, A., Viñolas, N., Marrades, R. M., Diaz, T., Gel, B., et al. (2009). miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis 30 (11), 1903–1909. doi:10.1093/carcin/bgp219
Gamble, L. A., Heller, T., and Davis, J. L. (2021). Hereditary diffuse gastric cancer syndrome and the role of CDH1: a review. JAMA Surg. 156 (4), 387–392. doi:10.1001/jamasurg.2020.6155
Gao, Z., Xu, Z., Hung, M.-S., Lin, Y.-C., Wang, T., Gong, M., et al. (2009). Procaine and procainamide inhibit the Wnt canonical pathway by promoter demethylation of WIF-1 in lung cancer cells. Oncol. Rep. 22 (6), 1479–1484. doi:10.3892/or_00000590
Ghosh, S., Khetarpal, P., and Senapati, S. (2022). Functional implications of the CpG island methylation in the pathogenesis of celiac disease. Mol. Biol. Rep. 49 (10), 10051–10064. doi:10.1007/s11033-022-07585-w
Gofrit, O. N., Benjamin, S., Halachmi, S., Leibovitch, I., Dotan, Z., Lamm, D. L., et al. (2014). DNA based therapy with diphtheria toxin-A BC-819: a phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer. J. Urology 191 (6), 1697–1702. doi:10.1016/j.juro.2013.12.011
Gounder, M., Schöffski, P., Jones, R. L., Agulnik, M., Cote, G. M., Villalobos, V. M., et al. (2020). Tazemetostat in advanced epithelioid sarcoma with loss of INI1/SMARCB1: an international, open-label, phase 2 basket study. Lancet. Oncol. 21 (11), 1423–1432. doi:10.1016/S1470-2045(20)30451-4
Gravina, G. L., Marampon, F., Giusti, I., Carosa, E., Di Sante, S., Ricevuto, E., et al. (2012). Differential effects of PXD101 (belinostat) on androgen-dependent and androgen-independent prostate cancer models. Int. J. Oncol. 40 (3), 711–720. doi:10.3892/ijo.2011.1270
Guo, Y., Cheng, R., Wang, Y., Gonzalez, M. E., Zhang, H., Liu, Y., et al. (2024). Regulation of EZH2 protein stability: new mechanisms, roles in tumorigenesis, and roads to the clinic. EBioMedicine 100, 104972. doi:10.1016/j.ebiom.2024.104972
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464 (7291), 1071–1076. doi:10.1038/nature08975
Gutschner, T., Hämmerle, M., Eissmann, M., Hsu, J., Kim, Y., Hung, G., et al. (2013). The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73 (3), 1180–1189. doi:10.1158/0008-5472.CAN-12-2850
Haebe, J. R., Bergin, C. J., Sandouka, T., and Benoit, Y. D. (2021). Emerging role of G9a in cancer stemness and promises as a therapeutic target. Oncogenesis 10 (11), 76. doi:10.1038/s41389-021-00370-7
Halbrook, C. J., Lyssiotis, C. A., Pasca di Magliano, M., and Maitra, A. (2023). Pancreatic cancer: advances and challenges. Cell 186 (8), 1729–1754. doi:10.1016/j.cell.2023.02.014
Hansford, S., Kaurah, P., Li-Chang, H., Woo, M., Senz, J., Pinheiro, H., et al. (2015). Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 1 (1), 23–32. doi:10.1001/jamaoncol.2014.168
Harada, H., Hosoda, K., Moriya, H., Mieno, H., Ema, A., Ushiku, H., et al. (2019). Cancer-specific promoter DNA methylation of Cysteine dioxygenase type 1 (CDO1) gene as an important prognostic biomarker of gastric cancer. PLoS One 14 (4), e0214872. doi:10.1371/journal.pone.0214872
Hegi, M. E., Diserens, A.-C., Gorlia, T., Hamou, M.-F., de Tribolet, N., Weller, M., et al. (2005). MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 352 (10), 997–1003. doi:10.1056/NEJMoa043331
Hegi, M. E., Liu, L., Herman, J. G., Stupp, R., Wick, W., Weller, M., et al. (2008). Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J. Clin. Oncol. Official J. Am. Soc. Clin. Oncol. 26 (25), 4189–4199. doi:10.1200/JCO.2007.11.5964
Hessels, D., Klein Gunnewiek, J. M. T., van Oort, I., Karthaus, H. F. M., van Leenders, G. J. L., van Balken, B., et al. (2003). DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol. 44 (1), 8–16. doi:10.1016/s0302-2838(03)00201-x
Ho, S. W. T., Sheng, T., Xing, M., Ooi, W. F., Xu, C., Sundar, R., et al. (2023). Regulatory enhancer profiling of mesenchymal-type gastric cancer reveals subtype-specific epigenomic landscapes and targetable vulnerabilities. Gut 72 (2), 226–241. doi:10.1136/gutjnl-2021-326483
Hsiao, K.-Y., Lin, Y.-C., Gupta, S. K., Chang, N., Yen, L., Sun, H. S., et al. (2017). Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 77 (9), 2339–2350. doi:10.1158/0008-5472.CAN-16-1883
Hu, J., Zheng, Z., Lei, J., Cao, Y., Li, Q., Zheng, Z., et al. (2021). Targeting the EZH2-PPAR Axis is a potential therapeutic pathway for pancreatic cancer. PPAR Res. 2021, 5589342. doi:10.1155/2021/5589342
Hu, X., Feng, Y., Zhang, D., Zhao, S. D., Hu, Z., Greshock, J., et al. (2014). A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 26 (3), 344–357. doi:10.1016/j.ccr.2014.07.009
Huang, X., Chen, Y., Xiao, Q., Shang, X., and Liu, Y. (2024). Chemical inhibitors targeting histone methylation readers. Pharmacol. Ther. 108614, 108614. doi:10.1016/j.pharmthera.2024.108614
Huang, X., Yuan, T., Liang, M., Du, M., Xia, S., Dittmar, R., et al. (2015). Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 67 (1), 33–41. doi:10.1016/j.eururo.2014.07.035
Hummel-Eisenbeiss, J., Hascher, A., Hals, P.-A., Sandvold, M. L., Müller-Tidow, C., Lyko, F., et al. (2013). The role of human equilibrative nucleoside transporter 1 on the cellular transport of the DNA methyltransferase inhibitors 5-azacytidine and CP-4200 in human leukemia cells. Mol. Pharmacol. 84 (3), 438–450. doi:10.1124/mol.113.086801
Hyun, K., Jeon, J., Park, K., and Kim, J. (2017). Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 49 (4), e324. doi:10.1038/emm.2017.11
Imaoka, H., Toiyama, Y., Fujikawa, H., Hiro, J., Saigusa, S., Tanaka, K., et al. (2016). Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann. Oncol. Official J. Eur. Soc. For Med. Oncol. 27 (10), 1879–1886. doi:10.1093/annonc/mdw279
Imperiale, T. F., Ransohoff, D. F., Itzkowitz, S. H., Levin, T. R., Lavin, P., Lidgard, G. P., et al. (2014). Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 370 (14), 1287–1297. doi:10.1056/NEJMoa1311194
Issa, J.-P. J., Roboz, G., Rizzieri, D., Jabbour, E., Stock, W., O’Connell, C., et al. (2015). Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: a multicentre, randomised, dose-escalation phase 1 study. Lancet. Oncol. 16 (9), 1099–1110. doi:10.1016/S1470-2045(15)00038-8
Italiano, A., Soria, J.-C., Toulmonde, M., Michot, J.-M., Lucchesi, C., Varga, A., et al. (2018). Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet. Oncol. 19 (5), 649–659. doi:10.1016/S1470-2045(18)30145-1
Jeong, I. B., Yoon, Y. S., Park, S. Y., Cha, E. J., Na, M. J., Kwon, S. J., et al. (2018). PCDHGA12 methylation biomarker in bronchial washing specimens as an adjunctive diagnostic tool to bronchoscopy in lung cancer. Oncol. Lett. 16 (1), 1039–1045. doi:10.3892/ol.2018.8699
Ji, H., Zhou, Y., Zhuang, X., Zhu, Y., Wu, Z., Lu, Y., et al. (2019). HDAC3 deficiency promotes liver cancer through a defect in H3K9ac/H3K9me3 transition. Cancer Res. 79 (14), 3676–3688. doi:10.1158/0008-5472.Can-18-3767
Ji, P., Diederichs, S., Wang, W., Böing, S., Metzger, R., Schneider, P. M., et al. (2003). MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22 (39), 8031–8041. doi:10.1038/sj.onc.1206928
Jin, P., Kang, Q., Wang, X., Yang, L., Yu, Y., Li, N., et al. (2015). Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J. Gastroenterology Hepatology 30 (5), 830–833. doi:10.1111/jgh.12855
Johnson, D. A., Barclay, R. L., Mergener, K., Weiss, G., König, T., Beck, J., et al. (2014). Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: a prospective multicenter study. PloS One 9 (6), e98238. doi:10.1371/journal.pone.0098238
Kaniskan, H. Ü., and Jin, J. (2015). Chemical probes of histone lysine methyltransferases. ACS Chem. Biol. 10 (1), 40–50. doi:10.1021/cb500785t
Kaniskan, H. Ü., Martini, M. L., and Jin, J. (2018). Inhibitors of protein methyltransferases and demethylases. Chem. Rev. 118 (3), 989–1068. doi:10.1021/acs.chemrev.6b00801
Kantarjian, H. M., Roboz, G. J., Kropf, P. L., Yee, K. W. L., O’Connell, C. L., Tibes, R., et al. (2017). Guadecitabine (SGI-110) in treatment-naive patients with acute myeloid leukaemia: phase 2 results from a multicentre, randomised, phase 1/2 trial. Lancet. Oncol. 18 (10), 1317–1326. doi:10.1016/S1470-2045(17)30576-4
Karam, R. A., Zidan, H. E., Abd Elrahman, T. M., Badr, S. A., and Amer, S. A. (2019). Study of p16 promoter methylation in Egyptian colorectal cancer patients. J. Cell. Biochem. 120 (5), 8581–8587. doi:10.1002/jcb.28146
Karthik, S., Sankar, R., Varunkumar, K., and Ravikumar, V. (2014). Romidepsin induces cell cycle arrest, apoptosis, histone hyperacetylation and reduces matrix metalloproteinases 2 and 9 expression in bortezomib sensitized non-small cell lung cancer cells. Biomed. Pharmacother. = Biomedecine Pharmacother. 68 (3), 327–334. doi:10.1016/j.biopha.2014.01.002
Kasprzak, A. (2023). Prognostic biomarkers of cell proliferation in colorectal cancer (CRC): from immunohistochemistry to molecular biology techniques. Cancers 15 (18), 4570. doi:10.3390/cancers15184570
Kerachian, M. A., and Kerachian, M. (2019). Long interspersed nucleotide element-1 (LINE-1) methylation in colorectal cancer. Clin. Chimica Acta; Int. J. Clin. Chem. 488, 209–214. doi:10.1016/j.cca.2018.11.018
Kim, K., Jutooru, I., Chadalapaka, G., Johnson, G., Frank, J., Burghardt, R., et al. (2013). HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 32 (13), 1616–1625. doi:10.1038/onc.2012.193
Kim, N., Norsworthy, K. J., Subramaniam, S., Chen, H., Manning, M. L., Kitabi, E., et al. (2022). FDA approval summary: decitabine and cedazuridine tablets for myelodysplastic syndromes. Clin. Cancer Res. Official J. Am. Assoc. For Cancer Res. 28 (16), 3411–3416. doi:10.1158/1078-0432.CCR-21-4498
Kisiel, J. B., Raimondo, M., Taylor, W. R., Yab, T. C., Mahoney, D. W., Sun, Z., et al. (2015). New DNA methylation markers for pancreatic cancer: discovery, tissue validation, and pilot testing in pancreatic juice. Clin. Cancer Res. Official J. Am. Assoc. For Cancer Res. 21 (19), 4473–4481. doi:10.1158/1078-0432.CCR-14-2469
Klonou, A., Korkolopoulou, P., Gargalionis, A. N., Kanakoglou, D. S., Katifelis, H., Gazouli, M., et al. (2021). Histone mark profiling in pediatric astrocytomas reveals prognostic significance of H3K9 trimethylation and histone methyltransferase SUV39H1. Neurother. J. Am. Soc. For Exp. Neurother. 18 (3), 2073–2090. doi:10.1007/s13311-021-01090-x
Kogo, R., Shimamura, T., Mimori, K., Kawahara, K., Imoto, S., Sudo, T., et al. (2011). Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71 (20), 6320–6326. doi:10.1158/0008-5472.CAN-11-1021
Köhler, J., Erlenkamp, G., Eberlin, A., Rumpf, T., Slynko, I., Metzger, E., et al. (2012). Lestaurtinib inhibits histone phosphorylation and androgen-dependent gene expression in prostate cancer cells. PloS One 7 (4), e34973. doi:10.1371/journal.pone.0034973
Koinuma, K., Kaneda, R., Toyota, M., Yamashita, Y., Takada, S., Choi, Y. L., et al. (2005). Screening for genomic fragments that are methylated specifically in colorectal carcinoma with a methylated MLH1 promoter. Carcinogenesis 26 (12), 2078–2085. doi:10.1093/carcin/bgi184
Kong, L. R., Tan, T. Z., Ong, W. R., Bi, C., Huynh, H., Lee, S. C., et al. (2017). Belinostat exerts antitumor cytotoxicity through the ubiquitin-proteasome pathway in lung squamous cell carcinoma. Mol. Oncol. 11 (8), 965–980. doi:10.1002/1878-0261.12064
Krajnović, M., Kožik, B., Božović, A., and Jovanović-Ćupić, S. (2023). Multiple roles of the RUNX gene family in hepatocellular carcinoma and their potential clinical implications. Cells 12 (18), 2303. doi:10.3390/cells12182303
Kravitz, C. J., Yan, Q., and Nguyen, D. X. (2023). Epigenetic markers and therapeutic targets for metastasis. Cancer Metastasis Rev. 42 (2), 427–443. doi:10.1007/s10555-023-10109-y
Laubach, J. P., Moreau, P., San-Miguel, J. F., and Richardson, P. G. (2015). Panobinostat for the treatment of multiple myeloma. Clin. Cancer Res. Official J. Am. Assoc. For Cancer Res. 21 (21), 4767–4773. doi:10.1158/1078-0432.CCR-15-0530
Lauschke, V. M., Zhou, Y., and Ingelman-Sundberg, M. (2019). Novel genetic and epigenetic factors of importance for inter-individual differences in drug disposition, response and toxicity. Pharmacol. Ther. 197, 122–152. doi:10.1016/j.pharmthera.2019.01.002
Lavia, P., Sciamanna, I., and Spadafora, C. (2022). An epigenetic LINE-1-based mechanism in cancer. Int. J. Mol. Sci. 23 (23), 14610. doi:10.3390/ijms232314610
Lee, B. H., Yegnasubramanian, S., Lin, X., and Nelson, W. G. (2005). Procainamide is a specific inhibitor of DNA methyltransferase 1. J. Biol. Chem. 280 (49), 40749–40756. doi:10.1074/jbc.M505593200
Lee, J. E., and Kim, M.-Y. (2022). Cancer epigenetics: past, present and future. Seminars Cancer Biol. 83, 4–14. doi:10.1016/j.semcancer.2021.03.025
Lee, S. H., Li, Y., Kim, H., Eum, S., Park, K., and Lee, C.-H. (2022). The role of EZH1 and EZH2 in development and cancer. BMB Rep. 55 (12), 595–601. doi:10.5483/BMBRep.2022.55.12.174
Leszinski, G., Gezer, U., Siegele, B., Stoetzer, O., and Holdenrieder, S. (2012). Relevance of histone marks H3K9me3 and H4K20me3 in cancer. Anticancer Res. 32 (5), 2199–2205. Available at: https://ar.iiarjournals.org/content/32/5/2199.
Li, J., Wu, H., Li, W., Yin, L., Guo, S., Xu, X., et al. (2016). Downregulated miR-506 expression facilitates pancreatic cancer progression and chemoresistance via SPHK1/Akt/NF-κB signaling. Oncogene 35 (42), 5501–5514. doi:10.1038/onc.2016.90
Li, X., Wu, Z., Mei, Q., Li, X., Guo, M., Fu, X., et al. (2013). Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br. J. Cancer 109 (8), 2266–2278. doi:10.1038/bjc.2013.548
Li, Y., Guo, D., Sun, R., Chen, P., Qian, Q., and Fan, H. (2019). Methylation patterns of Lys9 and Lys27 on histone H3 correlate with patient outcome in gastric cancer. Dig. Dis. Sci. 64 (2), 439–446. doi:10.1007/s10620-018-5341-8
Liang, G., and Weisenberger, D. J. (2017). DNA methylation aberrancies as a guide for surveillance and treatment of human cancers. Epigenetics 12 (6), 416–432. doi:10.1080/15592294.2017.1311434
Liew, P. L., Huang, R. L., Wu, T. I., Liao, C. C., Chen, C. W., Su, P. H., et al. (2019). Combined genetic mutations and DNA-methylated genes as biomarkers for endometrial cancer detection from cervical scrapings. Clin. Epigenetics 11 (1), 170. doi:10.1186/s13148-019-0765-3
Lim, Y., Wan, Y., Vagenas, D., Ovchinnikov, D. A., Perry, C. F. L., Davis, M. J., et al. (2016). Salivary DNA methylation panel to diagnose HPV-positive and HPV-negative head and neck cancers. BMC Cancer 16 (1), 749. doi:10.1186/s12885-016-2785-0
Lin, A., Li, C., Xing, Z., Hu, Q., Liang, K., Han, L., et al. (2016). The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat. Cell Biol. 18 (2), 213–224. doi:10.1038/ncb3295
Liu, B., Ricarte Filho, J., Mallisetty, A., Villani, C., Kottorou, A., Rodgers, K., et al. (2020a). Detection of promoter DNA methylation in urine and plasma aids the detection of non-small cell lung cancer. Clin. Cancer Res. Official J. Am. Assoc. For Cancer Res. 26 (16), 4339–4348. doi:10.1158/1078-0432.CCR-19-2896
Liu, B., Ricarte Filho, J., Mallisetty, A., Villani, C., Kottorou, A., Rodgers, K., et al. (2020b). Detection of promoter DNA methylation in urine and plasma aids the detection of non-small cell lung cancer. Clin. Cancer Res. 26 (16), 4339–4348. doi:10.1158/1078-0432.Ccr-19-2896
Liu, C., Kelnar, K., Liu, B., Chen, X., Calhoun-Davis, T., Li, H., et al. (2011). The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 17 (2), 211–215. doi:10.1038/nm.2284
Liu, Y.-X., Li, Q.-Z., Cao, Y.-N., and Zhang, L.-Q. (2020c). Identification of key genes and important histone modifications in hepatocellular carcinoma. Comput. Struct. Biotechnol. J. 18, 2657–2669. doi:10.1016/j.csbj.2020.09.013
Lofton-Day, C., Model, F., Devos, T., Tetzner, R., Distler, J., Schuster, M., et al. (2008). DNA methylation biomarkers for blood-based colorectal cancer screening. Clin. Chem. 54 (2), 414–423. doi:10.1373/clinchem.2007.095992
Loktionov, A. (2020). Biomarkers for detecting colorectal cancer non-invasively: DNA, RNA or proteins? World J. Gastrointest. Oncol. 12 (2), 124–148. doi:10.4251/wjgo.v12.i2.124
Losson, H., Schnekenburger, M., Dicato, M., and Diederich, M. (2016). Natural compound histone deacetylase inhibitors (HDACi): synergy with inflammatory signaling pathway modulators and clinical applications in cancer. Mol. (Basel, Switz.) 21 (11), 1608. doi:10.3390/molecules21111608
Lyko, F. (2018). The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19 (2), 81–92. doi:10.1038/nrg.2017.80
Ma, L., Li, C., Yin, H., Huang, J., Yu, S., Zhao, J., et al. (2023). The mechanism of DNA methylation and miRNA in breast cancer. Int. J. Mol. Sci. 24 (11), 9360. doi:10.3390/ijms24119360
Majumder, S., Raimondo, M., Taylor, W. R., Yab, T. C., Berger, C. K., Dukek, B. A., et al. (2020). Methylated DNA in pancreatic juice distinguishes patients with pancreatic cancer from controls. Clin. Gastroenterology Hepatology Official Clin. Pract. J. Am. Gastroenterological Assoc. 18 (3), 676–683. doi:10.1016/j.cgh.2019.07.017
Marquez, V. E., Kelley, J. A., Agbaria, R., Ben-Kasus, T., Cheng, J. C., Yoo, C. B., et al. (2005). Zebularine: a unique molecule for an epigenetically based strategy in cancer chemotherapy. Ann. N. Y. Acad. Sci. 1058, 246–254. doi:10.1196/annals.1359.037
Marsolier, J., Prompsy, P., Durand, A., Lyne, A. M., Landragin, C., Trouchet, A., et al. (2022). H3K27me3 conditions chemotolerance in triple-negative breast cancer. Nat. Genet. 54 (4), 459–468. doi:10.1038/s41588-022-01047-6
Mattiroli, F., and Penengo, L. (2021). Histone ubiquitination: an integrative signaling platform in genome stability. Trends Genet. TIG 37 (6), 566–581. doi:10.1016/j.tig.2020.12.005
McCleland, M. L., Mesh, K., Lorenzana, E., Chopra, V. S., Segal, E., Watanabe, C., et al. (2016). CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J. Clin. Investigation 126 (2), 639–652. doi:10.1172/JCI83265
McDermott, J., and Jimeno, A. (2014). Belinostat for the treatment of peripheral T-cell lymphomas. Drugs Today (Barcelona, Spain 1998) 50 (5), 337–345. doi:10.1358/dot.2014.50.5.2138703
Melotte, V., Qu, X., Ongenaert, M., van Criekinge, W., de Bruïne, A. P., Baldwin, H. S., et al. (2010). The N-myc downstream regulated gene (NDRG) family: diverse functions, multiple applications. FASEB J. Official Publ. Fed. Am. Soc. For Exp. Biol. 24 (11), 4153–4166. doi:10.1096/fj.09-151464
Mendez-Dorantes, C., and Burns, K. H. (2023). LINE-1 retrotransposition and its deregulation in cancers: implications for therapeutic opportunities. Genes & Dev. 37 (21-24), 948–967. doi:10.1101/gad.351051.123
Merz, A. M. A., Sébert, M., Sonntag, J., Kubasch, A. S., Platzbecker, U., and Adès, L. (2024). Phase to phase: navigating drug combinations with hypomethylating agents in higher-risk MDS trials for optimal outcomes. Cancer Treat. Rev. 123, 102673. doi:10.1016/j.ctrv.2023.102673
Miyake, T., Nakayama, T., Naoi, Y., Yamamoto, N., Otani, Y., Kim, S. J., et al. (2012). GSTP1 expression predicts poor pathological complete response to neoadjuvant chemotherapy in ER-negative breast cancer. Cancer Sci. 103 (5), 913–920. doi:10.1111/j.1349-7006.2012.02231.x
Moldogazieva, N. T., Zavadskiy, S. P., Sologova, S. S., Mokhosoev, I. M., and Terentiev, A. A. (2021). Predictive biomarkers for systemic therapy of hepatocellular carcinoma. Expert Rev. Mol. Diagnostics 21 (11), 1147–1164. doi:10.1080/14737159.2021.1987217
Molife, L. R., and de Bono, J. S. (2011). Belinostat: clinical applications in solid tumors and lymphoma. Expert Opin. Investigational Drugs 20 (12), 1723–1732. doi:10.1517/13543784.2011.629604
Morschhauser, F., Tilly, H., Chaidos, A., McKay, P., Phillips, T., Assouline, S., et al. (2020). Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial. Lancet. Oncol. 21 (11), 1433–1442. doi:10.1016/S1470-2045(20)30441-1
Mottamal, M., Zheng, S., Huang, T. L., and Wang, G. (2015). Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Mol. (Basel, Switz.) 20 (3), 3898–3941. doi:10.3390/molecules20033898
Mrakovcic, M., Kleinheinz, J., and Fröhlich, L. F. (2017). Histone deacetylase inhibitor-induced autophagy in tumor cells: implications for p53. Int. J. Mol. Sci. 18 (9), 1883. doi:10.3390/ijms18091883
Müller, D., and Győrffy, B. (2022). DNA methylation-based diagnostic, prognostic, and predictive biomarkers in colorectal cancer. Biochimica Biophysica Acta. Rev. Cancer 1877 (3), 188722. doi:10.1016/j.bbcan.2022.188722
Muss, H. B., Smitherman, A., Wood, W. A., Nyrop, K., Tuchman, S., Randhawa, P. K., et al. (2020). p16 a biomarker of aging and tolerance for cancer therapy. Transl. Cancer Res. 9 (9), 5732–5742. doi:10.21037/tcr.2020.03.39
Myasoedova, V. A., Sukhorukov, V., Grechko, A. V., Zhang, D., Romanenko, E., Orekhov, V., et al. (2019). Inhibitors of DNA methylation and histone deacetylation as epigenetically active drugs for anticancer therapy. Curr. Pharm. Des. 25 (6), 635–641. doi:10.2174/1381612825666190405144026
Nepal, C., and Andersen, J. B. (2023). Alternative promoters in CpG depleted regions are prevalently associated with epigenetic misregulation of liver cancer transcriptomes. Nat. Commun. 14 (1), 2712. doi:10.1038/s41467-023-38272-4
Oh, T., Kim, N., Moon, Y., Kim, M. S., Hoehn, B. D., Park, C. H., et al. (2013). Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J. Mol. Diagnostics JMD 15 (4), 498–507. doi:10.1016/j.jmoldx.2013.03.004
Oliver, J., Garcia-Aranda, M., Chaves, P., Alba, E., Cobo-Dols, M., Onieva, J. L., et al. (2022). Emerging noninvasive methylation biomarkers of cancer prognosis and drug response prediction. Seminars Cancer Biol. 83, 584–595. doi:10.1016/j.semcancer.2021.03.012
Ono, H., Kato, T., Murase, Y., Nakamura, Y., Ishikawa, Y., Watanabe, S., et al. (2021). C646 inhibits G2/M cell cycle-related proteins and potentiates anti-tumor effects in pancreatic cancer. Sci. Rep. 11 (1), 10078. doi:10.1038/s41598-021-89530-8
Ozawa, T., Matsuyama, T., Toiyama, Y., Takahashi, N., Ishikawa, T., Uetake, H., et al. (2017). CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 ’gene desert’, serve as important prognostic biomarkers in colorectal cancer. Ann. Oncol. Official J. Eur. Soc. For Med. Oncol. 28 (8), 1882–1888. doi:10.1093/annonc/mdx248
Palanca-Ballester, C., Rodriguez-Casanova, A., Torres, S., Calabuig-Fariñas, S., Exposito, F., Serrano, D., et al. (2021). Cancer epigenetic biomarkers in liquid biopsy for high incidence malignancies. Cancers 13 (12), 3016. doi:10.3390/cancers13123016
Panzitt, K., Tschernatsch, M. M. O., Guelly, C., Moustafa, T., Stradner, M., Strohmaier, H. M., et al. (2007). Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 132 (1), 330–342. doi:10.1053/j.gastro.2006.08.026
Papadatos-Pastos, D., Yuan, W., Pal, A., Crespo, M., Ferreira, A., Gurel, B., et al. (2022). Phase 1, dose-escalation study of guadecitabine (SGI-110) in combination with pembrolizumab in patients with solid tumors. J. For Immunother. Cancer 10 (6), e004495. doi:10.1136/jitc-2022-004495
Patnaik, E., Madu, C., and Lu, Y. (2023). Epigenetic modulators as therapeutic agents in cancer. Int. J. Mol. Sci. 24 (19), 14964. doi:10.3390/ijms241914964
Payne, S. R., Serth, J., Schostak, M., Kamradt, J., Strauss, A., Thelen, P., et al. (2009). DNA methylation biomarkers of prostate cancer: confirmation of candidates and evidence urine is the most sensitive body fluid for non-invasive detection. Prostate 69 (12), 1257–1269. doi:10.1002/pros.20967
Pechalrieu, D., Etievant, C., and Arimondo, P. B. (2017). DNA methyltransferase inhibitors in cancer: from pharmacology to translational studies. Biochem. Pharmacol. 129, 1–13. doi:10.1016/j.bcp.2016.12.004
Peng, H., Fu, W., Chang, C., Gao, H., He, Q., Liu, Z., et al. (2023). The transcription activity of OTX2 on p16 expression is significantly blocked by methylation of CpG shore in non-promoter of lung cancer cell lines. Transl. Cancer Res. 12 (10), 2582–2595. doi:10.21037/tcr-23-909
Pojani, E., and Barlocco, D. (2021). Romidepsin (FK228), A histone deacetylase inhibitor and its analogues in cancer chemotherapy. Curr. Med. Chem. 28 (7), 1290–1303. doi:10.2174/0929867327666200203113926
Preis, M., Gardner, T. B., Gordon, S. R., Pipas, J. M., Mackenzie, T. A., Klein, E. E., et al. (2011). MicroRNA-10b expression correlates with response to neoadjuvant therapy and survival in pancreatic ductal adenocarcinoma. Clin. Cancer Res. Official J. Am. Assoc. For Cancer Res. 17 (17), 5812–5821. doi:10.1158/1078-0432.CCR-11-0695
Prensner, J. R., Iyer, M. K., Balbin, O. A., Dhanasekaran, S. M., Cao, Q., Brenner, J. C., et al. (2011). Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 29 (8), 742–749. doi:10.1038/nbt.1914
Prensner, J. R., Zhao, S., Erho, N., Schipper, M., Iyer, M. K., Dhanasekaran, S. M., et al. (2014). RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet. Oncol. 15 (13), 1469–1480. doi:10.1016/S1470-2045(14)71113-1
Qu, L., Ding, J., Chen, C., Wu, Z.-J., Liu, B., Gao, Y., et al. (2016). Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29 (5), 653–668. doi:10.1016/j.ccell.2016.03.004
Quagliata, L., Matter, M. S., Piscuoglio, S., Arabi, L., Ruiz, C., Procino, A., et al. (2014). Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatol. (Baltim. Md) 59 (3), 911–923. doi:10.1002/hep.26740
Ramaiah, M. J., Tangutur, A. D., and Manyam, R. R. (2021). Epigenetic modulation and understanding of HDAC inhibitors in cancer therapy. Life Sci. 277, 119504. doi:10.1016/j.lfs.2021.119504
Raos, D., Ulamec, M., Katusic Bojanac, A., Bulic-Jakus, F., Jezek, D., and Sincic, N. (2021). Epigenetically inactivated RASSF1A as a tumor biomarker. Bosnian J. Basic Med. Sci. 21 (4), 386–397. doi:10.17305/bjbms.2020.5219
Raslan, O., and Garcia-Horton, A. (2022). Azacitidine and its role in the upfront treatment of acute myeloid leukemia. Expert Opin. Pharmacother. 23 (8), 873–884. doi:10.1080/14656566.2022.2082284
Ray-Gallet, D., and Almouzni, G. (2022). H3-H4 histone chaperones and cancer. Curr. Opin. Genet. Dev. 73, 101900. doi:10.1016/j.gde.2022.101900
Reid, G., Pel, M. E., Kirschner, M. B., Cheng, Y. Y., Mugridge, N., Weiss, J., et al. (2013). Restoring expression of miR-16: a novel approach to therapy for malignant pleural mesothelioma. Ann. Oncol. Official J. Eur. Soc. For Med. Oncol. 24 (12), 3128–3135. doi:10.1093/annonc/mdt412
Ren, S., Wang, F., Shen, J., Sun, Y., Xu, W., Lu, J., et al. (2013). Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur. J. Cancer (Oxford, Engl. 1990) 49 (13), 2949–2959. doi:10.1016/j.ejca.2013.04.026
Riaud, M., Maxwell, J., Soria-Bretones, I., Dankner, M., Li, M., and Rose, A. A. N. (2024). The role of CRAF in cancer progression: from molecular mechanisms to precision therapies. Nat. Rev. Cancer 24 (2), 105–122. doi:10.1038/s41568-023-00650-x
Robert, F., and Jeronimo, C. (2023). Transcription-coupled nucleosome assembly. Trends Biochem. Sci. 48 (11), 978–992. doi:10.1016/j.tibs.2023.08.003
Rossetto, D., Avvakumov, N., and Côté, J. (2012). Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics 7 (10), 1098–1108. doi:10.4161/epi.21975
Ruiz-Andres, O., Sanchez-Niño, M. D., Cannata-Ortiz, P., Ruiz-Ortega, M., Egido, J., Ortiz, A., et al. (2016). Histone lysine crotonylation during acute kidney injury in mice. Dis. Models Mech. 9 (6), 633–645. doi:10.1242/dmm.024455
Rupaimoole, R., and Slack, F. J. (2017). MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16 (3), 203–222. doi:10.1038/nrd.2016.246
Sabari, B. R., Tang, Z., Huang, H., Yong-Gonzalez, V., Molina, H., Kong, H. E., et al. (2015). Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 58 (2), 203–215. doi:10.1016/j.molcel.2015.02.029
Saghafinia, S., Mina, M., Riggi, N., Hanahan, D., and Ciriello, G. (2018). Pan-cancer landscape of aberrant DNA methylation across human tumors. Cell Rep. 25 (4), 1066–1080. doi:10.1016/j.celrep.2018.09.082
Saijo, K., Imamura, J., Narita, K., Oda, A., Shimodaira, H., Katoh, T., et al. (2015). Biochemical, biological and structural properties of romidepsin (FK228) and its analogs as novel HDAC/PI3K dual inhibitors. Cancer Sci. 106 (2), 208–215. doi:10.1111/cas.12585
Saito, I., Kitagawa, R., Fukuda, H., Shibata, T., Katsumata, N., Konishi, I., et al. (2010). A phase III trial of paclitaxel plus carboplatin versus paclitaxel plus cisplatin in stage IVB, persistent or recurrent cervical cancer: gynecologic Cancer Study Group/Japan Clinical Oncology Group Study (JCOG0505). Jpn. J. Clin. Oncol. 40 (1), 90–93. doi:10.1093/jjco/hyp117
Saito, M., Schetter, A. J., Mollerup, S., Kohno, T., Skaug, V., Bowman, E. D., et al. (2011). The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin. Cancer Res. Official J. Am. Assoc. For Cancer Res. 17 (7), 1875–1882. doi:10.1158/1078-0432.CCR-10-2961
Sakimura, S., Sugimachi, K., Kurashige, J., Ueda, M., Hirata, H., Nambara, S., et al. (2015). The miR-506-induced epithelial-mesenchymal transition is involved in poor prognosis for patients with gastric cancer. Ann. Surg. Oncol. 22 (Suppl. 3), S1436–S1443. doi:10.1245/s10434-015-4418-2
Salvianti, F., Orlando, C., Massi, D., De Giorgi, V., Grazzini, M., Pazzagli, M., et al. (2015). Tumor-related methylated cell-free DNA and circulating tumor cells in melanoma. Front. Mol. Biosci. 2, 76. doi:10.3389/fmolb.2015.00076
Sang, L.-J., Ju, H.-Q., Liu, G.-P., Tian, T., Ma, G.-L., Lu, Y.-X., et al. (2018). LncRNA CamK-A regulates Ca2+-signaling-mediated tumor microenvironment remodeling. Mol. Cell 72 (3), 601. doi:10.1016/j.molcel.2018.10.024
Sant’Ana, J. M. D. A., Servato, J. P. S., Matsuo, F. S., Andrade, M. F., Pitorro, T. E. A., Moraes, A. d.S., et al. (2020). The role of the histones H3K9ac, H3K9me3, HP1γ, and H3K36me3 in oral squamous cell carcinoma loco-regional metastasis and relapse. Pathology, Res. Pract. 216 (11), 153201. doi:10.1016/j.prp.2020.153201
Sarkozy, C., Morschhauser, F., Dubois, S., Molina, T., Michot, J. M., Cullières-Dartigues, P., et al. (2020). A LYSA phase Ib study of tazemetostat (EPZ-6438) plus R-CHOP in patients with newly diagnosed diffuse large B-cell lymphoma (DLBCL) with poor prognosis features. Clin. Cancer Res. Official J. Am. Assoc. For Cancer Res. 26 (13), 3145–3153. doi:10.1158/1078-0432.CCR-19-3741
Sasidharan Nair, V., El Salhat, H., Taha, R. Z., John, A., Ali, B. R., and Elkord, E. (2018). DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clin. Epigenetics 10, 78. doi:10.1186/s13148-018-0512-1
Sébert, M., Renneville, A., Bally, C., Peterlin, P., Beyne-Rauzy, O., Legros, L., et al. (2019). A phase II study of guadecitabine in higher-risk myelodysplastic syndrome and low blast count acute myeloid leukemia after azacitidine failure. Haematologica 104 (8), 1565–1571. doi:10.3324/haematol.2018.207118
Segura-Pacheco, B., Trejo-Becerril, C., Perez-Cardenas, E., Taja-Chayeb, L., Mariscal, I., Chavez, A., et al. (2003). Reactivation of tumor suppressor genes by the cardiovascular drugs hydralazine and procainamide and their potential use in cancer therapy. Clin. Cancer Res. Official J. Am. Assoc. For Cancer Res. 9 (5), 1596–1603.
Serra, S., and Chetty, R. (2018). p16. J. Clin. Pathology 71 (10), 853–858. doi:10.1136/jclinpath-2018-205216
Seufferlein, T., Ettrich, T. J., Menzler, S., Messmann, H., Kleber, G., Zipprich, A., et al. (2022). Green tea extract to prevent colorectal adenomas, results of a randomized, placebo-controlled clinical trial. Am. J. Gastroenterology 117 (6), 884–894. doi:10.14309/ajg.0000000000001706
Shanmugam, G., Rakshit, S., and Sarkar, K. (2022). HDAC inhibitors: targets for tumor therapy, immune modulation and lung diseases. Transl. Oncol. 16, 101312. doi:10.1016/j.tranon.2021.101312
Sharma, S., Kelly, T. K., and Jones, P. A. (2010). Epigenetics in cancer. Carcinogenesis 31 (1), 27–36. doi:10.1093/carcin/bgp220
Shi, X.-Y., Ding, W., Li, T.-Q., Zhang, Y.-X., and Zhao, S.-C. (2017). Histone deacetylase (HDAC) inhibitor, suberoylanilide hydroxamic acid (SAHA), induces apoptosis in prostate cancer cell lines via the akt/FOXO3a signaling pathway. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 23, 5793–5802. doi:10.12659/msm.904597
Shinkai, Y., and Tachibana, M. (2011). H3K9 methyltransferase G9a and the related molecule GLP. Genes & Dev. 25 (8), 781–788. doi:10.1101/gad.2027411
Shukla, S., Zhang, X., Niknafs, Y. S., Xiao, L., Mehra, R., Cieślik, M., et al. (2016). Identification and validation of PCAT14 as prognostic biomarker in prostate cancer. Neoplasia (New York, N.Y.) 18 (8), 489–499. doi:10.1016/j.neo.2016.07.001
Siegel, D., Hussein, M., Belani, C., Robert, F., Galanis, E., Richon, V. M., et al. (2009). Vorinostat in solid and hematologic malignancies. J. Hematol. Oncol. 2, 31. doi:10.1186/1756-8722-2-31
Singh, A., Gupta, S., Badarukhiya, J. A., and Sachan, M. (2020). Detection of aberrant methylation of HOXA9 and HIC1 through multiplex MethyLight assay in serum DNA for the early detection of epithelial ovarian cancer. Int. J. Cancer 147 (6), 1740–1752. doi:10.1002/ijc.32984
Singh, A., Gupta, S., and Sachan, M. (2019). Epigenetic biomarkers in the management of ovarian cancer: current prospectives. Front. Cell Dev. Biol. 7, 182. doi:10.3389/fcell.2019.00182
Sogutlu, F., Pekerbas, M., and Biray Avci, C. (2022). Epigenetic signatures in gastric cancer: current knowledge and future perspectives. Expert Rev. Mol. Diagnostics 22 (12), 1063–1075. doi:10.1080/14737159.2022.2159381
Song, L., Li, Y., Jia, J., Zhou, G., Wang, J., Kang, Q., et al. (2016). Algorithm optimization in methylation detection with multiple RT-qPCR. PloS One 11 (11), e0163333. doi:10.1371/journal.pone.0163333
Song, L., Yu, H., and Li, Y. (2015). Diagnosis of lung cancer by SHOX2 gene methylation assay. Mol. Diagnosis Ther. 19 (3), 159–167. doi:10.1007/s40291-015-0144-5
Stading, R., Gastelum, G., Chu, C., Jiang, W., and Moorthy, B. (2021). Molecular mechanisms of pulmonary carcinogenesis by polycyclic aromatic hydrocarbons (PAHs): implications for human lung cancer. Seminars Cancer Biol. 76, 3–16. doi:10.1016/j.semcancer.2021.07.001
Stein, E. M., Garcia-Manero, G., Rizzieri, D. A., Tibes, R., Berdeja, J. G., Savona, M. R., et al. (2018). The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 131 (24), 2661–2669. doi:10.1182/blood-2017-12-818948
Sun, J., Zheng, M.-Y., Li, Y.-W., and Zhang, S.-W. (2020). Structure and function of Septin 9 and its role in human malignant tumors. World J. Gastrointest. Oncol. 12 (6), 619–631. doi:10.4251/wjgo.v12.i6.619
Suzuki, H., Yamamoto, E., Maruyama, R., Niinuma, T., and Kai, M. (2014). Biological significance of the CpG island methylator phenotype. Biochem. Biophysical Res. Commun. 455 (1-2), 35–42. doi:10.1016/j.bbrc.2014.07.007
Tachaveeraphong, W., and Phattarataratip, E. (2024). The significance of modified histone H3 in epithelial dysplasia and oral cancer. Int. Dent. J. doi:10.1016/j.identj.2024.01.011
Tada, M., Imazeki, F., Fukai, K., Sakamoto, A., Arai, M., Mikata, R., et al. (2007). Procaine inhibits the proliferation and DNA methylation in human hepatoma cells. Hepatol. Int. 1 (3), 355–364. doi:10.1007/s12072-007-9014-5
Tahara, T., Tahara, S., Horiguchi, N., Okubo, M., Terada, T., Yamada, H., et al. (2019). Molecular subtyping of gastric cancer combining genetic and epigenetic anomalies provides distinct clinicopathological features and prognostic impacts. Hum. Mutat. 40 (3), 347–354. doi:10.1002/humu.23700
Tan, M., Luo, H., Lee, S., Jin, F., Yang, J. S., Montellier, E., et al. (2011). Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146 (6), 1016–1028. doi:10.1016/j.cell.2011.08.008
Teodoridis, J. M., Hardie, C., and Brown, R. (2008). CpG island methylator phenotype (CIMP) in cancer: causes and implications. Cancer Lett. 268 (2), 177–186. doi:10.1016/j.canlet.2008.03.022
Teschendorff, A. E., Lee, S. H., Jones, A., Fiegl, H., Kalwa, M., Wagner, W., et al. (2015). HOTAIR and its surrogate DNA methylation signature indicate carboplatin resistance in ovarian cancer. Genome Med. 7, 108. doi:10.1186/s13073-015-0233-4
Toiyama, Y., Takahashi, M., Hur, K., Nagasaka, T., Tanaka, K., Inoue, Y., et al. (2013). Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J. Natl. Cancer Inst. 105 (12), 849–859. doi:10.1093/jnci/djt101
Tong, D., Tang, Y., and Zhong, P. (2024). The emerging roles of histone demethylases in cancers. Cancer Metastasis Rev. doi:10.1007/s10555-023-10160-9
Tutt, A., Tovey, H., Cheang, M. C. U., Kernaghan, S., Kilburn, L., Gazinska, P., et al. (2018). Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat. Med. 24 (5), 628–637. doi:10.1038/s41591-018-0009-7
Tuval, A., Strandgren, C., Heldin, A., Palomar-Siles, M., and Wiman, K. G. (2024). Pharmacological reactivation of p53 in the era of precision anticancer medicine. Nat. Rev. Clin. Oncol. 21 (2), 106–120. doi:10.1038/s41571-023-00842-2
Ushiku, H., Yamashita, K., Katoh, H., Ema, A., Minatani, N., Kikuchi, M., et al. (2017). Promoter DNA methylation of CDO1 gene and its clinical significance in esophageal squamous cell carcinoma. Dis. Esophagus Official J. Int. Soc. For Dis. Esophagus 30 (2), 1–9. doi:10.1111/dote.12496
Vaswani, R. G., Gehling, V. S., Dakin, L. A., Cook, A. S., Nasveschuk, C. G., Duplessis, M., et al. (2016). Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase I clinical trials for B-cell lymphomas. J. Med. Chem. 59 (21), 9928–9941. doi:10.1021/acs.jmedchem.6b01315
Velez, J., Dale, B., Park, K.-S., Kaniskan, H. Ü., Yu, X., and Jin, J. (2024). Discovery of a novel, highly potent EZH2 PROTAC degrader for targeting non-canonical oncogenic functions of EZH2. Eur. J. Med. Chem. 267, 116154. doi:10.1016/j.ejmech.2024.116154
Villalba, J. M., and Alcaín, F. J. (2012). Sirtuin activators and inhibitors. BioFactors Oxf. Engl. 38 (5), 349–359. doi:10.1002/biof.1032
Vo, J. N., Cieslik, M., Zhang, Y., Shukla, S., Xiao, L., Zhang, Y., et al. (2019). The landscape of circular RNA in cancer. Cell 176 (4), 869–881. doi:10.1016/j.cell.2018.12.021
Vos, P. D., Leedman, P. J., Filipovska, A., and Rackham, O. (2019). Modulation of miRNA function by natural and synthetic RNA-binding proteins in cancer. Cell. Mol. Life Sci. 76 (19), 3745–3752. doi:10.1007/s00018-019-03163-9
Wan, G., Feng, H., Su, C., Zhu, Y., Zhang, L., Zhang, Q., et al. (2023). A patent review of EZH2 inhibitors from 2017 and beyond. Expert Opin. Ther. Pat. 33 (4), 293–308. doi:10.1080/13543776.2023.2206018
Wang, B., Zhou, M., Gan, X.-L., Ren, Y.-X., Yang, Y.-Z., Weng, Z.-J., et al. (2023). Combined low levels of H4K16ac and H4K20me3 predicts poor prognosis in breast cancer. Int. J. Clin. Oncol. 28 (9), 1147–1157. doi:10.1007/s10147-023-02378-y
Wang, J., Zhu, S., Meng, N., He, Y., Lu, R., and Yan, G.-R. (2019). ncRNA-encoded peptides or proteins and cancer. Mol. Ther. J. Am. Soc. Gene Ther. 27 (10), 1718–1725. doi:10.1016/j.ymthe.2019.09.001
Wang, X.-S., Zhang, Z., Wang, H.-C., Cai, J.-L., Xu, Q.-W., Li, M.-Q., et al. (2006). Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin. Cancer Res. Official J. Am. Assoc. For Cancer Res. 12 (16), 4851–4858. doi:10.1158/1078-0432.CCR-06-0134
Wang, Y., Gu, J., Roth, J. A., Hildebrandt, M. A. T., Lippman, S. M., Ye, Y., et al. (2013). Pathway-based serum microRNA profiling and survival in patients with advanced stage non-small cell lung cancer. Cancer Res. 73 (15), 4801–4809. doi:10.1158/0008-5472.CAN-12-3273
Wang, Z., Yang, B., Zhang, M., Guo, W., Wu, Z., Wang, Y., et al. (2018). lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell 33 (4), 706–720.e9. doi:10.1016/j.ccell.2018.03.006
Wei, B., Wu, F., Xing, W., Sun, H., Yan, C., Zhao, C., et al. (2021). A panel of DNA methylation biomarkers for detection and improving diagnostic efficiency of lung cancer. Sci. Rep. 11 (1), 16782. doi:10.1038/s41598-021-96242-6
Weisenberger, D. J., Siegmund, K. D., Campan, M., Young, J., Long, T. I., Faasse, M. A., et al. (2006). CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 38 (7), 787–793. doi:10.1038/ng1834
Weiss, G., Schlegel, A., Kottwitz, D., König, T., and Tetzner, R. (2017). Validation of the SHOX2/PTGER4 DNA methylation marker panel for plasma-based discrimination between patients with malignant and nonmalignant lung disease. J. Thorac. Oncol. Official Publ. Int. Assoc. For Study Lung Cancer 12 (1), 77–84. doi:10.1016/j.jtho.2016.08.123
Weng, W., Wei, Q., Toden, S., Yoshida, K., Nagasaka, T., Fujiwara, T., et al. (2017). Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. Official J. Am. Assoc. For Cancer Res. 23 (14), 3918–3928. doi:10.1158/1078-0432.CCR-16-2541
White, J. R., Thompson, D. T., Koch, K. E., Kiriazov, B. S., Beck, A. C., van der Heide, D. M., et al. (2021). AP-2α-Mediated activation of E2F and EZH2 drives melanoma metastasis. Cancer Res. 81 (17), 4455–4470. doi:10.1158/0008-5472.CAN-21-0772
Wick, W., Platten, M., Meisner, C., Felsberg, J., Tabatabai, G., Simon, M., et al. (2012). Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: the NOA-08 randomised, phase 3 trial. Lancet. Oncol. 13 (7), 707–715. doi:10.1016/S1470-2045(12)70164-X
Włodarczyk, M., Ciebiera, M., Nowicka, G., Łoziński, T., Ali, M., and Al-Hendy, A. (2024). Epigallocatechin gallate for the treatment of benign and malignant gynecological diseases-focus on epigenetic mechanisms. Nutrients 16 (4), 559. doi:10.3390/nu16040559
Wong, I. H., Lo, Y. M., Zhang, J., Liew, C. T., Ng, M. H., Wong, N., et al. (1999). Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 59 (1), 71–73.
Wong, J., Gruber, E., Maher, B., Waltham, M., Sabouri-Thompson, Z., Jong, I., et al. (2022). Integrated clinical and genomic evaluation of guadecitabine (SGI-110) in peripheral T-cell lymphoma. Leukemia 36 (6), 1654–1665. doi:10.1038/s41375-022-01571-8
Wu, D., Zhou, G., Jin, P., Zhu, J., Li, S., Wu, Q., et al. (2016). Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J. Mol. Diagnostics 18 (4), 535–545. doi:10.1016/j.jmoldx.2016.02.005
Wu, S., Wu, Y., Deng, S., Lei, X., and Yang, X. (2023). Emerging roles of noncoding RNAs in human cancers. Discov. Oncol. 14 (1), 128. doi:10.1007/s12672-023-00728-w
Wu, Z., Jing, S., Li, Y., Gao, Y., Yu, S., Li, Z., et al. (2017). The effects of SAHA on radiosensitivity in pancreatic cancer cells by inducing apoptosis and targeting RAD51. Biomed. Pharmacother. = Biomedecine Pharmacother. 89, 705–710. doi:10.1016/j.biopha.2017.02.067
Xu, J., Li, N., Deng, W., and Luo, S. (2021). Discovering the mechanism and involvement of the methylation of cyclin-dependent kinase inhibitor 2A (CDKN2A) gene and its special locus region in gastric cancer. Bioengineered 12 (1), 1286–1298. doi:10.1080/21655979.2021.1887646
Yaman Agaoglu, F., Kovancilar, M., Dizdar, Y., Darendeliler, E., Holdenrieder, S., Dalay, N., et al. (2011). Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biol. J. Int. Soc. For Oncodevelopmental Biol. Med. 32 (3), 583–588. doi:10.1007/s13277-011-0154-9
Yamashita, K., Hosoda, K., Nishizawa, N., Katoh, H., and Watanabe, M. (2018). Epigenetic biomarkers of promoter DNA methylation in the new era of cancer treatment. Cancer Sci. 109 (12), 3695–3706. doi:10.1111/cas.13812
Yan, H., and Bu, P. (2021). Non-coding RNA in cancer. Essays Biochem. 65 (4), 625–639. doi:10.1042/EBC20200032
Yan, L., Geng, Q., Cao, Z., Liu, B., Li, L., Lu, P., et al. (2023). Insights into DNMT1 and programmed cell death in diseases. Biomed. Pharmacother. = Biomedecine Pharmacother. 168, 115753. doi:10.1016/j.biopha.2023.115753
Yan, L.-X., Huang, X.-F., Shao, Q., Huang, M.-Y., Deng, L., Wu, Q.-L., et al. (2008). MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA (New York, N.Y.) 14 (11), 2348–2360. doi:10.1261/rna.1034808
Yan, Q., Chen, B.-J., Hu, S., Qi, S.-L., Li, L.-Y., Yang, J.-F., et al. (2021). Emerging role of RNF2 in cancer: from bench to bedside. J. Cell. Physiology 236 (8), 5453–5465. doi:10.1002/jcp.30260
Yang, D., Sun, Y., Hu, L., Zheng, H., Ji, P., Pecot, C. V., et al. (2013). Integrated analyses identify a master microRNA regulatory network for the mesenchymal subtype in serous ovarian cancer. Cancer Cell 23 (2), 186–199. doi:10.1016/j.ccr.2012.12.020
Yang, F., Fang, E., Mei, H., Chen, Y., Li, H., Li, D., et al. (2019a). Cis-acting circ-CTNNB1 promotes β-catenin signaling and cancer progression via DDX3-mediated transactivation of YY1. Cancer Res. 79 (3), 557–571. doi:10.1158/0008-5472.CAN-18-1559
Yang, L., Lei, Q., Li, L., Yang, J., Dong, Z., and Cui, H. (2019b). Silencing or inhibition of H3K79 methyltransferase DOT1L induces cell cycle arrest by epigenetically modulating c-Myc expression in colorectal cancer. Clin. Epigenetics 11 (1), 199. doi:10.1186/s13148-019-0778-y
Yap, T. A., Winter, J. N., Giulino-Roth, L., Longley, J., Lopez, J., Michot, J.-M., et al. (2019). Phase I study of the novel enhancer of zeste homolog 2 (EZH2) inhibitor GSK2816126 in patients with advanced hematologic and solid tumors. Clin. Cancer Res. Official J. Am. Assoc. For Cancer Res. 25 (24), 7331–7339. doi:10.1158/1078-0432.CCR-18-4121
Zaib, S., Rana, N., and Khan, I. (2022). Histone modifications and their role in epigenetics of cancer. Curr. Med. Chem. 29 (14), 2399–2411. doi:10.2174/0929867328666211108105214
Zauderer, M. G., Szlosarek, P. W., Le Moulec, S., Popat, S., Taylor, P., Planchard, D., et al. (2022). EZH2 inhibitor tazemetostat in patients with relapsed or refractory, BAP1-inactivated malignant pleural mesothelioma: a multicentre, open-label, phase 2 study. Lancet. Oncol. 23 (6), 758–767. doi:10.1016/S1470-2045(22)00277-7
Zhang, B., Dong, Q., Su, H., Birchler, J. A., and Han, F. (2014). Histone phosphorylation: its role during cell cycle and centromere identity in plants. Cytogenet. Genome Res. 143 (1-3), 144–149. doi:10.1159/000360435
Zhang, L., Han, Y., Jiang, Q., Wang, C., Chen, X., Li, X., et al. (2015). Trend of histone deacetylase inhibitors in cancer therapy: isoform selectivity or multitargeted strategy. Med. Res. Rev. 35 (1), 63–84. doi:10.1002/med.21320
Zhao, F., Olkhov-Mitsel, E., Kamdar, S., Jeyapala, R., Garcia, J., Hurst, R., et al. (2018a). A urine-based DNA methylation assay, ProCUrE, to identify clinically significant prostate cancer. Clin. Epigenetics 10 (1), 147. doi:10.1186/s13148-018-0575-z
Zhao, H., Zhu, W., Zhao, X., Li, X., Zhou, Z., Zheng, M., et al. (2022). Efficacy of epigallocatechin-3-gallate in preventing dermatitis in patients with breast cancer receiving postoperative radiotherapy: a double-blind, placebo-controlled, phase 2 randomized clinical trial. JAMA Dermatol. 158 (7), 779–786. doi:10.1001/jamadermatol.2022.1736
Zhao, S., Zhang, X., and Li, H. (2018b). Beyond histone acetylation-writing and erasing histone acylations. Curr. Opin. Struct. Biol. 53, 169–177. doi:10.1016/j.sbi.2018.10.001
Zhao, X., Deng, P., Feng, J., Wang, Z., Xiang, Z., Han, X., et al. (2016). Cysteine dioxygenase type 1 inhibits osteogenesis by regulating wnt signaling in primary mouse bone marrow stromal cells. Sci. Rep. 6, 19296. doi:10.1038/srep19296
Zhao, Z., Song, J., Tang, B., Fang, S., Zhang, D., Zheng, L., et al. (2020). CircSOD2 induced epigenetic alteration drives hepatocellular carcinoma progression through activating JAK2/STAT3 signaling pathway. J. Exp. Clin. Cancer Res. CR 39 (1), 259. doi:10.1186/s13046-020-01769-7
Zhou, R.-H., Kokame, K., Tsukamoto, Y., Yutani, C., Kato, H., and Miyata, T. (2001). Characterization of the human NDRG gene family: a newly identified member, NDRG4, is specifically expressed in brain and heart. Genomics 73 (1), 86–97. doi:10.1006/geno.2000.6496
Zhou, W., Feng, Y., Lin, C., Chao, C. K., He, Z., Zhao, S., et al. (2023). Yin yang 1-induced long noncoding RNA DUXAP9 drives the progression of oral squamous cell carcinoma by blocking CDK1-mediated EZH2 degradation. Adv. Sci. (Weinheim, Baden-Wurttemberg, Ger.) 10 (25), e2207549. doi:10.1002/advs.202207549
Zhou, X., Ao, X., Jia, Z., Li, Y., Kuang, S., Du, C., et al. (2022). Non-coding RNA in cancer drug resistance: underlying mechanisms and clinical applications. Front. Oncol. 12, 951864. doi:10.3389/fonc.2022.951864
Zhou, X., Yin, C., Dang, Y., Ye, F., and Zhang, G. (2015). Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci. Rep. 5, 11516. doi:10.1038/srep11516
Zhou, Z., Li, H.-Q., and Liu, F. (2018). DNA methyltransferase inhibitors and their therapeutic potential. Curr. Top. Med. Chem. 18 (28), 2448–2457. doi:10.2174/1568026619666181120150122
Keywords: epigenetics, cancer, DNA methylation, histone modification, non-coding RNA
Citation: Gao J, Shi W, Wang J, Guan C, Dong Q, Sheng J, Zou X, Xu Z, Ge Y, Yang C, Li J, Bao H, Zhong X and Cui Y (2024) Research progress and applications of epigenetic biomarkers in cancer. Front. Pharmacol. 15:1308309. doi: 10.3389/fphar.2024.1308309
Received: 06 October 2023; Accepted: 25 March 2024;
Published: 12 April 2024.
Edited by:
Zhiqian Zhang, Southern University of Science and Technology, ChinaReviewed by:
Yuen Gao, Michigan State University, United StatesAlka Singh, The University of Chicago, United States
Copyright © 2024 Gao, Shi, Wang, Guan, Dong, Sheng, Zou, Xu, Ge, Yang, Li, Bao, Zhong and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangyu Zhong, emhvbmd4aWFuZ3l1QGhyYm11LmVkdS5jbg==; Yunfu Cui, eWZjdWk3QDE2My5jb20=
†These authors have contributed equally to this work
 Jianjun Gao
Jianjun Gao Wujiang Shi1†
Wujiang Shi1† Qingfu Dong
Qingfu Dong Zhaoqiang Xu
Zhaoqiang Xu Jiehan Li
Jiehan Li Yunfu Cui
Yunfu Cui