- 1College of Agronomy, Liaocheng University, Liaocheng, China
- 2Burns and Plastic Surgery Department, The 960th Hospital of the PLA Joint Logistics Support Force, Jinan, China
- 3Laizhou City Laiyu Chemical Co., Ltd., Laizhou, China
- 4School of Biological Science and Technology, University of Jinan, Jinan, China
Three type III secretion system (T3SS) inhibitors (compounds 5, 19, and 32) were identified by virtual screening and biological evaluation. These three compounds were evaluated against a panel of Salmonella species strains including S. enteritidis, S. typhi, S. typhimurium, S. paratyphi, and S. abortus equi, and their minimum inhibitory concentrations ranged from 1 to 53 μg/ml. Especially, these compounds showed comparable activity as the of the positive control gatifloxacin towards S. abortus equi. The present results suggest that these new T3SS inhibitors could be used as a potential lead molecule for drug development of anti-Salmonella.
Introduction
Salmonellosis is a general term for infectious diseases of livestock and poultry caused by Salmonella bacteria. Salmonella, belonging to the Enterobacteriaceae family, are foodborne zoonotic enteric and facultative intracellular pathogenic bacteria, which are one of the most common causes of intestinal infections worldwide (Akiba et al., 2011). Salmonella are mainly categorized into two species including Salmonella bongori and Salmonella enterica, especially the last of which has a large number of subspecies and serovars, and some of them, such as serovar Enteritidis, are highly pathogenic (Vullo et al., 2011). It mainly causes abortion or death of pregnant female animals, such as pigs, chickens, cattle, donkey and other animals, with the symptoms of fever, diarrhea, sepsis, gastroenteritis and meningitis, as well as intestinal damage in both humans and animals (Zha et al., 2019). The extensive resistance of Salmonella to a variety of antibiotics (ampicillin, chloramphenicol, streptomycin, antimicrobial sulfonamides and tetracycline) has been reported worldwide (Arcangioli et al., 2000; Mąka et al., 2015; Tran-Dien et al., 2018; Vilela et al., 2019; Mengistu et al., 2020). Thus there is an urgent need for the discovery of new anti-Salmonella drugs.
The type III secretion system (T3SS) is a nanomachinery protein utilized by many Gram-negative bacteria to inject bacterial effector proteins from the bacterial cytoplasm directly into the eukaryotic host cells. Although the detailed molecular mechanisms of the bacterial effector functions and the biogenesis of T3SSs are still need extensive research, biochemical and genetic studies of lots of Gram-negative bacterial pathogens have indicated that effector translocation by T3SSs is vital for successful infection (Galan, 2001). T3SS is required to initiate infections and central to the virulence of many pathogenic bacteria, such as Salmonella and Shigella (Tsou, et al., 2013). The mainly structure of T3SS is a needle apparatus composed of more than 20 different proteins (Cornelis, 2006), of which the tip complex are essential component consisted of tip proteins and translocon proteins, and tip proteins bound on the top of the needle, while the translocon proteins are membrane-spanning proteins. The T3SS tip proteins are SipD (Lara-Tejero and Galan, 2009) in the Salmonella typhimurium pathogenicity island 1 (SPI-1). T3SS has been regarded as promising targets for the development of anti-virulence agents, and some T3SS inhibitors have been identified (Hussain et al., 2021). However, there is no T3SS inhibitor reported targeting SipD protein.
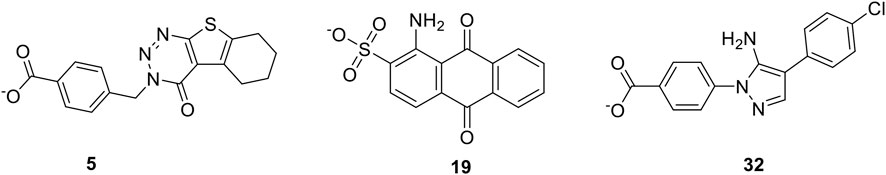
In this study, the discovery of T3SS inhibitors targeting SipD protein was performed using molecular docking-based virtual screening on Specs database. After the biological evaluation of forty-six purchased compounds that were selected according to the docking screening and structural clustering, three of them, namely, compounds 5, 19, and 32 (Figure 1) with different scaffolds, were found to have effect on the Salmonella invasion, which indicated that they were new T3SS inhibitors. The binding model of these compounds with T3SS were investigated and the results indicated that all of them could form tight binding affinity with T3SS tip protein Sip D. Then all these T3SS inhibitors were evaluated for their inhibitory activities towards against a panel of Salmonella species strains including S. enteritidis, S. typhi, S. typhimurium, S. paratyphi, and S. abortus equi. The present study provided new chemotypes for the development of novel T3SS inhibitors targeting SipD protein, which could serve as lead compounds for developing novel medications against Salmonella bacteria.
Results and Discussion
Molecular Docking Based Virtual Screening Yielded 46 T3SS Inhibitor Candidates
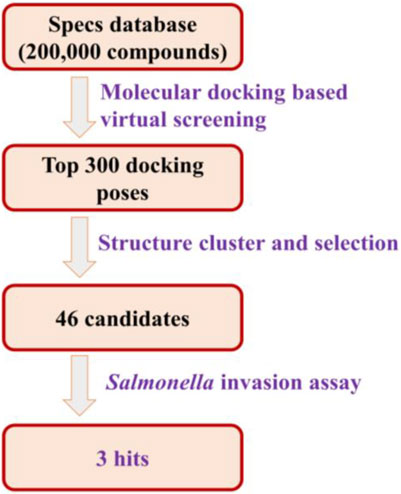
In order to obtain T3SS inhibitors candidates targeting the tip protein SipD of T3SS, we chose SPECS database to perform molecular docking based virtual screening. The function of SipD protein was documented to promote secretion of effectors and functions at the post-transcriptional and post-translational levels (Glasgow et al., 2017). The needle tip complex in Salmonella enterica consists of three translocon proteins: SipB, SipC, and SipD. It is known that knocking out SipD disrupts T3SS regulation to cause constitutive secretion of native proteins (Glasgow et al., 2017). Thus, we used SipD protein to perform virtual screening. According to the docking results, the top 300 docking poses ranked on the docking score were selected for the following cluster analysis and visual selection. Finally, we selected 46 compounds as T3SS inhibitor candidates for the following Salmonella invasion assay test. The workflow of the virtual screening was shown in Figure 2. And the detailed method was provided in Supplementary Material.
The Salmonella Invasion Assay Identified three Novel T3SS Inhibitors
To assess the effects of T3SS inhibitor candidates on bacterial invasiveness, we used the Salmonella invasion assay to determine the effect of T3SS inhibitor candidates on the ability of Salmonella to invade cultured human epithelial cells. Results of Salmonella invasion assay with respect to T3SS inhibitors are shown in Figure 3. Overall, most of the 46 compounds could decrease invasiveness, and three of them (5, 19, and 32) showed most drastic effect on the invasiveness.
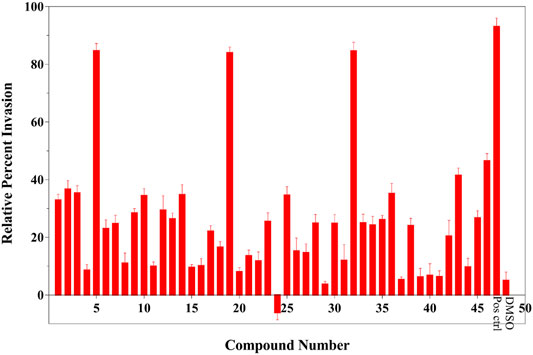
FIGURE 3. Results of Salmonella invasion assay (legends: Pos ctrl: positive control, DMSO: negative control). This result was shown the average of triplicate assay.
Binding Mode Analysis Exhibited Tight Binding Affinity Between 5, 19, and 32 With SipD Protein
The probable binding modes of the three compounds were provided to show the detailed interaction mechanism. As shown in Figure 4, all of the three compounds could occupy the deoxycholate binding site in SipD, with the binding energy showing in Table 1. Compound 5 displayed the most binding interactions than 19, and 32. Compound 5 established hydrophobic interactions with residues Arg41, Ile45, Asn104, Ala108, Leu318, Val325, and Lys338, and formed H-bond and π-π interactions with Asn321 and Arg41, respectively. Compound 19 displayed two H-bond interactions with Asn321 and Lys338, and also established hydrophobic interactions with residues Arg41, Ile45, Ala108, and Leu318. Compound 32 formed H-bond interaction with Asn321, and established hydrophobic interactions with residues Arg41, Ile45, Ala108, Leu318, and Val325. In brief, residues Arg41, Ile45, Ala108, Leu318, and Asn321 were the key residues that showed interactions with all of the three compounds. From the binding mode of the active compounds 5, 19, and 32 with SipD protein, we could see that there was a lot of space in the binding pocket (Supplementary Figure S1) which could be occupied by the inhibitor. Therefore, further structure modification can be performed from the following direction: while maintaining the hydrogen bond interactions, large substituents may be added to the molecular structure of 5, 19, and 32 to increase binding affinity.
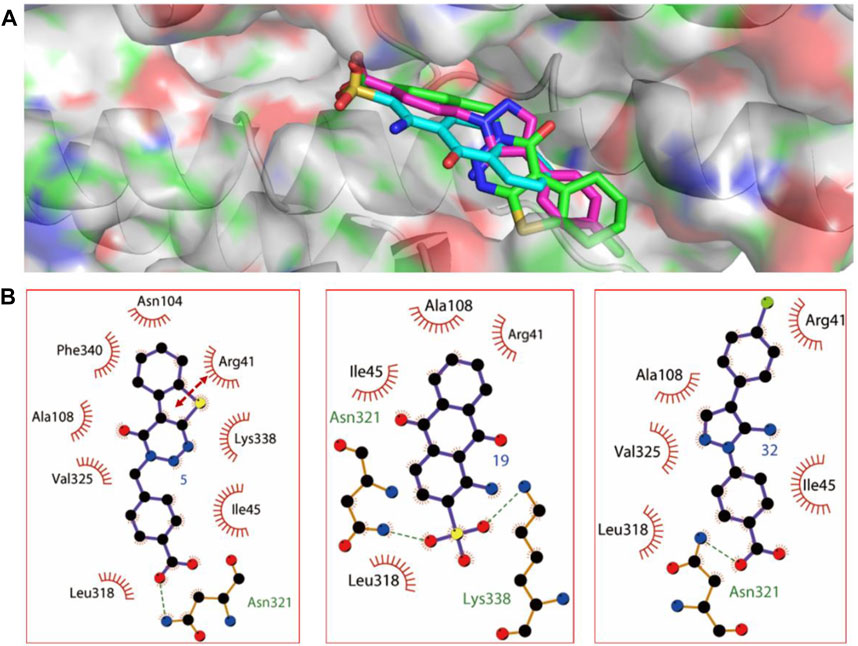
FIGURE 4. The predicted binding modes of compounds 5, 19, and 32. (A) The three dimensional binding mode of compounds 5, 19, and 32. (B) The relative schematic diagram showed the hydrophobic interactions (shown as starbursts), H-bond interactions (denoted by dotted green lines), and π-π interactions (displayed as double-head arrow) between compounds 5, 19, and 32 with T3SS tip protein SipD.
Anti-Salmonella Activity Assay of Compounds 5, 19, and 32
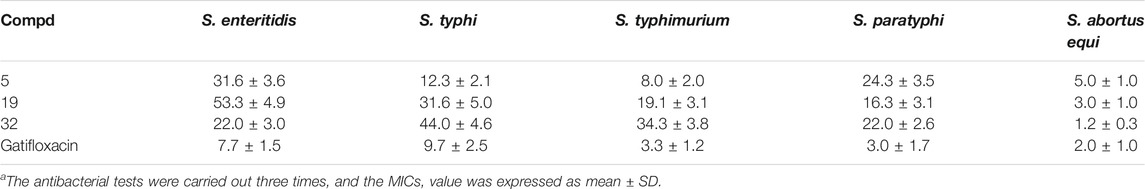
The in vitro anti-Salmonella activity of these found compounds are shown in Table 2. All of them showed potent inhibitory activity against bacteria in Salmonella sp., including S. enteritidis, S. typhi, S. typhimurium, S. paratyphi, S. abortus equi, with MICs ranging from 1 to 53 μg/mL. S. abortus equi is well known as the aetiological agent of equus abortion, and interestingly, almost all of these compounds exhibited the most sensitive activity towards S. abortus equi, with MICs ranging from 1 to 8 μg/ml, almost as similar as that of positive drug (gatifloxacin). In addition, barring S. abortus equi, for all other strains, the three compounds exhibited lesser potential as compared gatifloxacin. The reason for such results may due to the reduced binding to SipD orthologues.
In vitro Cytotoxicity of Compounds 5, 19, and 32
The cytotoxicity of compounds 5, 19, 32 against RAW 264.7 cells at the concentrations based their respective MICs towards S. typhimurium was initially evaluated using MTT assay (Zhang et al., 2020). The results in Figure 5 showed that none of these compounds was toxic towards RAW 264.7 cells, with the cell viability of 84.24 ± 5.45% (5, 8 μg/ml), 91.50 ± 7.09% (9, 19 μg/ml), and 86.36 ± 6.04% (32, 34 μg/ml)), respectively, compared with untreated cell group.
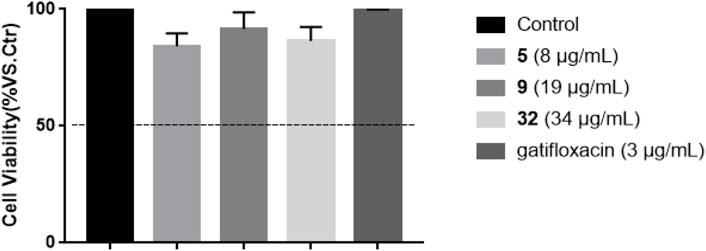
FIGURE 5. Effects of compounds 5, 19, 32, and gatifloxacin on the cell viability of RAW 264.7 cells for 24 h. Cell viability was expressed as percent cell viability compared to that of DMSO vehicle control cells (100%), and cell viability more than 50% at their respective concentration was considered to be non-toxic.
Intracellular Killing Assay of Compounds 5, 19, and 32
The intracellular anti-Salmonella activity of compounds 5, 19, and 32 was then evaluated in the model of RAW 264.7 cells infected with S. typhimurium according to the protected protocol (Birhanu et al., 2018). The concentrations of these tested compounds were selected on the basis of their MIC values. Based on the results of intracellular killing assay shown in Figure 6, All these compounds 5, 19, and 32 could decrease the intracellular-survival of S. typhimurium by 44.4, 32.5, and 52.2%, respectively, compared with the non-treated S. typhimurium group. The intracellular survival suppression by the positive control gatifloxacin was 60.2%.
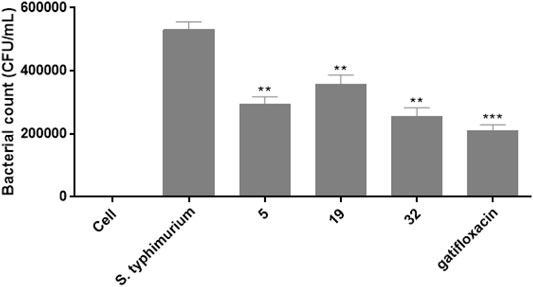
FIGURE 6. The effects of compounds 5, 19, 32, and gatifloxacin on the survival of S. typhimurium in RAW 264.7 cells.
Prediction of ADMET Properties
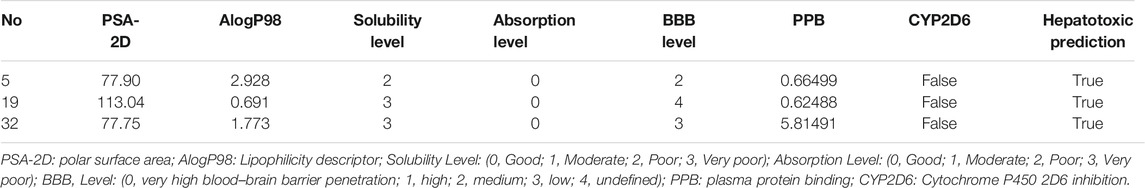
The ADMET profiling for the identified three compounds were predicted. From the results (Table 3), we can see that all of the three compounds were predicted to have acceptable solubility and good absorption level, and displayed moderate BBB penetration level. Importantly, both of them may not bind to PPB and CYP2D6, which possibly indicated fewer side effects of these three compounds.
Conclusion
In this study, three novel T3SS inhibitors 5, 19, and 32 with different structural scaffold were first discovered based on virtual screening, and the in vitro anti-bacterial activities of these inhibitors against five stains of Salmonella sp. were evaluated. Finally, the activities of these compounds against a panel of Salmonella bacteria were tested in anti-bacterial bioassay, and the results showed all of them exhibited promising anti-Salmonella activity with MICs values ranging from 1 to 53 μg/ml. Further experiment revealed intracellular-inhibition of these compounds against S. typhimurium. It was worth to note that all these three inhibitors showed potent activity towards S. abortus equi as that of gatifloxacin. Based on these results, we first confirmed that the T3SS tip protein SipD is a potential target for T3SS inhibitor discovery, and these new T3SS inhibitors 5, 19, and 32 could be lead compounds used in the discovery of drugs against the infection of Salmonella bacteria, especially S. abortus equi, and their further structural modification and bioactive optimization are deserved. And when we performed structural modification, we would first predict the drug-likeness properties of the designed compounds.
Materials and Methods
Molecular Docking Based Virtual Screening
Molecular docking was performed using the Autodock 4.2 program (; Morris et al., 1998; Huey et al., 2007). The crystal structure of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate (PDB ID: 3O01) (Chatterjee et al., 2011) was used to construct the docking model. The missing hydrogen atoms were added, Gasteiger charges was assigned, and the protein were parameterized with AD4 type by Autodock Tools 1.5.6. Finally, the protein structure was used as an input for the Autogrid program. Grid map with 60 × 60 × 60 points was made according to the conformation of ligand, and the grid spacing was set to 0.375 Å. Rigid ligand docking was performed for prepared SPECS database compounds. Docking calculations were carried out using the Lamarckian genetic algorithm (LGA). “Clustering Molecules” protocols embedded in Pipeline Pilot 7.5 (Pipeline Pilot; Accelrys Software Inc., San Diego, CA) was used to do the cluster analysis. Finally, according to the cluster analysis results, the candidate compounds were selected and purchased from SPECS database supplier.
Screening of Small Molecules for Inhibition of Salmonella Invasion
The effect of T3SS inhibitor candidates on the ability of S. typhimurium to invade a cultured human epithelial cell line (Henle 407) was performed in a method similar to one previously described (Chatterjee et al., 2011). Henle 407 cells were grown in DMEM with 10% fetal calf serum at 37°C in 5% CO2 in 24-well plates. The plasmid pRK2-SipD with WT SipD was electroporated into the S. typhimurium SipD− strain and single colonies were inoculated into LB media supplemented with 25 mg/L trimethoprim, 50 mg/L ampicillin, 50 mg/L kanamycin and grown in standing cultures overnight. A 10 ml LB culture with 1 mM IPTG was inoculated with 1 ml of overnight bacterial growth, and incubated at 37°C on standing for 2.5–3 h. Approximately 15–30 uL of bacterial suspension was added with 300 uL DMEM into the Henle 407 cells and incubated at 37°C for 60 min to allow invasion before the suspension was removed by aspiration. The Henle 407 cells were incubated with fresh DMEM with 100 mg/L of gentamycin for 1.5 h, aspirated, rinsed with DMEM, and lysed with 1% Triton X-100 to free the entrapped bacteria. The number of bacterial colonies, which correlated with invasiveness, was estimated by serial dilution and plating. The Salmonella invasion assay was done in triplicate.
In vitro anti-Salmonella Bioassay
The anti-Salmonella activities of these compounds were performed according to the previous reported protocol (Wei et al., 2016), using the minimum inhibitory concentration (MIC) with different strains, including S. enteritidis, S. typhi, S. typhimurium, S. paratyphi, S. and abortus equi. Gatifloxacin was used as positive controls. The test compounds 5, 19, and 32 in DMSO were prepared and then poured into 96-well plates. The final concentration of o.39–100 μg/ml underwent a twofold serial dilution. The bacteria were incubated with a series of different concentrations of compounds at 37°C for 24 h. The microbacterial growth was measured at the absorption of 630 nm. All experiments were carried out in triplicate.
In vitro Cytotoxicity Assay
To study the cytotoxic effects of compounds on cell viability, the RAW 264.7 cells were seeded into 96-well plates at 1 ×104 cells/well and allowed to attach for 24 h. The medium was replaced with 100 μL medium containing the indicated concentrations of compounds and further incubated for 24 h. Each well was added 10 μL MTT (5 mg/ml in PBS) and the plates were incubated for 4 h at 37°C. Supernatants were aspirated and formed formazan was dissolved in 100 μL of dimethyl sulfoxide (DMSO). The optical density (OD) was measured at an absorbance wavelength of 490 nm using a Microplate Reader (Tecan, Switzerland).
Intracellular Killing Assay
The intracellular killing experiment was performed according to the previous reported protocol (Birhanu et al., 2018). RAW 264.7 cells (105 cells/ml) were cultured in 24-well plates, and then treated with S. Typhimurium (107 CFU/ml) and further incubated for 45 min. After the cells were washed, the compound 5 (8 μg/ml), 9 (19 μg/ml) and 32 (34 μg/ml) or gatifloxacin (3 μg/ml) were respectively added and incubated for 1 h at 37 °C. Finally, cells were treated with gentamicin (100 μg/ml) for 1 h and lysed with 0.1% of trition × 100 before being serially diluted and plated on LB agar. The cells infected with S. Typhimurium without treatment was used as the control.
Statistical Analysis
All data are presented as the mean ± standard deviation. Data were processed using 17.0 SPSS software (SPSS Inc., Chicago, IL, United States ). Statistical comparisons were analyzed using one-way analysis of variance (ANOVA). p values of less than 0.05 were considered to be statistically significant. *p < 0.05, **p < 0.01, and ***p < 0.001.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This research was funded by the National Natural Science Foundation of China (No. 31671287), Taishan Leading Industry Talents–Agricultural Science of Shandong Province (No. LJNY201713), Shandong Province Modern Agricultural Technology System Donkey Industrial Innovation Team (No. SDAIT-27), and the Open Project of Shandong Collaborative Innovation Center for Donkey Industry Technology (No. 3193308).
Conflict of Interest
Authors ZK, GZ, and YL were employed by the company Laiyu Chemical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2021.764191/full#supplementary-material
References
Akiba, M., Kusumoto, M., and Iwata, T. (2011). Rapid Identification of Salmonella enterica Serovars, Typhimurium, Choleraesuis, Infantis, Hadar, Enteritidis, Dublin and Gallinarum, by Multiplex PCR. J. Microbiol. Methods 85, 9–15. doi:10.1016/j.mimet.2011.02.002
Arcangioli, M. A., Leroy-Setrin, S., Martel, J. L., and Chaslus-Dancla, E. (2000). Evolution of Chloramphenicol Resistance, with Emergence of Cross-Resistance to Florfenicol, in Bovine Salmonella Typhimurium Strains Implicates Definitive Phage Type (DT) 104. J. Med. Microbiol. 49, 103–110. doi:10.1099/0022-1317-49-1-103
Birhanu, B. T., Park, N. H., Lee, S. J., Hossain, M. A., and Park, S. C. (2018). Inhibition of Salmonella Typhimurium Adhesion, Invasion, and Intracellular Survival via Treatment with Methyl Gallate Alone and in Combination with Marbofloxacin. Vet. Res. 49, 101. doi:10.1186/s13567-018-0597-8
Chatterjee, S., Zhong, D., Nordhues, B. A., Battaile, K. P., Lovell, S., and De Guzman, R. N. (2011). The crystal Structures of the Salmonella Type III Secretion System Tip Protein SipD in Complex with Deoxycholate and Chenodeoxycholate. Protein Sci. 20, 75–86. doi:10.1002/pro.537
Cornelis, G. R. (2006). The Type III Secretion Injectisome. Nat. Rev. Microbiol. 4, 811–825. doi:10.1038/nrmicro1526
Galán, J. E. (2001). Salmonella Interactions with Host Cells: Type III Secretion at Work. Annu. Rev. Cel Dev. Biol. 17, 53–86. doi:10.1146/annurev.cellbio.17.1.53
Glasgow, A. A., Wong, H. T., and Tullman-Ercek, D. (2017). A Secretion-Amplification Role for Salmonella enterica Translocon Protein SipD. ACS Synth. Biol. 6, 1006–1015. doi:10.1021/acssynbio.6b00335
Huey, R., Morris, G. M., Olson, A. J., and Goodsell, D. S. (2007). A Semiempirical Free Energy Force Field with Charge-Based Desolvation. J. Comput. Chem. 28, 1145–1152. doi:10.1002/jcc.20634
Hussain, S., Ouyang, P., Zhu, Y., Khalique, A., He, C., Liang, X., et al. (2021). Type 3 Secretion System 1 of Salmonella typhimurium and its Inhibitors: a Novel Strategy to Combat Salmonellosis. Environ. Sci. Pollut. Res. Int. 28, 34154–34166. doi:10.1007/s11356-021-13986-4
Lara-Tejero, M., and Galán, J. E. (2009). Salmonella enterica Serovar Typhimurium Pathogenicity Island 1-encoded Type III Secretion System Translocases Mediate Intimate Attachment to Nonphagocytic Cells. Infect. Immun. 77, 2635–2642. doi:10.1128/IAI.00077-09
Mąka, Ł., Maćkiw, E., Ścieżyńska, H., Modzelewska, M., and Popowska, M. (2015). Resistance to Sulfonamides and Dissemination of Sul Genes Among Salmonella Spp. Isolated from Food in Poland. Foodborne Pathog. Dis. 12, 383–389. doi:10.1089/fpd.2014.1825
Mengistu, G., Dejenu, G., Tesema, C., Arega, B., Awoke, T., Alemu, K., et al. (2020). Epidemiology of Streptomycin Resistant Salmonella from Humans and Animals in Ethiopia: A Systematic Review and Meta-Analysis. PLoS One 15, e0244057. doi:10.1371/journal.pone.0244057
Morris, G. M., Goodsell, D. S., Halliday, R. S., Huey, R., Hart, W. E., Belew, R. K., et al. (1998). Automated Docking Using a Lamarckian Genetic Algorithm and an Empirical Binding Free Energy Function. J. Comput. Chem. 19, 1639–1662. doi:10.1002/(SICI)1096-987X(19981115)19:14<1639:AID-JCC10>3.0.CO;2-B
Tran-Dien, A., Le Hello, S., Bouchier, C., and Weill, F. X. (2018). Early Transmissible Ampicillin Resistance in Zoonotic Salmonella enterica Serotype Typhimurium in the Late 1950s: a Retrospective, Whole-Genome Sequencing Study. Lancet Infect. Dis. 18, 207–214. doi:10.1016/S1473-3099(17)30705-3
Tsou, L. K., Dossa, P. D., and Hang, H. C. (2013). Small Molecules Aimed at Type III Secretion Systems to Inhibit Bacterial Virulence. Medchemcomm 4, 68–79. doi:10.1039/C2MD20213A
Vilela, F. P., Gomes, C. N., Passaglia, J., Rodrigues, D. P., Costa, R. G., Tiba Casas, M. R., et al. (2019). Genotypic Resistance to Quinolone and Tetracycline in Salmonella Dublin Strains Isolated from Humans and Animals in Brazil. Microb. Drug Resist. 25, 143–151. doi:10.1089/mdr.2017.0329
Vullo, D., Nishimori, I., Minakuchi, T., Scozzafava, A., and Supuran, C. T. (2011). Inhibition Studies with Anions and Small Molecules of Two Novel β-carbonic Anhydrases from the Bacterial Pathogen Salmonella enterica Serovar Typhimurium. Bioorg. Med. Chem. Lett. 21, 3591–3595. doi:10.1016/j.bmcl.2011.04.105
Wei, Z. Y., Chi, K. Q., Yu, Z. K., Liu, H. Y., Sun, L. P., Zheng, C. J., et al. (2016). Synthesis and Biological Evaluation of Chalcone Derivatives Containing Aminoguanidine or Acylhydrazone Moieties. Bioorg. Med. Chem. Lett. 26, 5920–5925. doi:10.1016/j.bmcl.2016.11.001
Zha, L., Garrett, S., and Sun, J. (2019). Salmonella Infection in Chronic Inflammation and Gastrointestinal Cancer. Diseases 7, 28. doi:10.3390/diseases7010028
Zhang, Y. Y., Zhang, Q. Q., Zhang, J., Song, J. L., Li, J. C., Han, K., et al. (2020). Synthesis and Evaluation of 1,2,4-oxadiazole Derivatives as Potential Anti-inflammatory Agents by Inhibiting NF-κB Signaling Pathway in LPS-Stimulated RAW 264.7 Cells. Bioorg. Med. Chem. Lett. 30, 127373. doi:10.1016/j.bmcl.2020.127373
Keywords: type III secretion system inhibitor, virtual screening, molecular docking, Salmonella species, anti-bacterial activity
Citation: Wang Y, Hou M, Kan Z, Zhang G, Li Y, Zhou L and Wang C (2021) Identification of Novel Type Three Secretion System (T3SS) Inhibitors by Computational Methods and Anti-Salmonella Evaluations. Front. Pharmacol. 12:764191. doi: 10.3389/fphar.2021.764191
Received: 25 August 2021; Accepted: 01 November 2021;
Published: 16 November 2021.
Edited by:
Junyan Lu, European Molecular Biology Laboratory Heidelberg, GermanyReviewed by:
Arijit Bhattacharya, Adamas University, IndiaMohamed Abdo Rizk, Mansoura University, Egypt
Copyright © 2021 Wang, Hou, Kan, Zhang, Li, Zhou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changfa Wang, d2FuZ2NmMTk2N0AxNjMuY29t; Lei Zhou, YmlvX3pob3VsQHVqbi5lZHUuY24=
 Yonghui Wang
Yonghui Wang Meihui Hou2
Meihui Hou2 Lei Zhou
Lei Zhou Changfa Wang
Changfa Wang