
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Pharmacol., 26 April 2021
Sec. Pharmacology of Anti-Cancer Drugs
Volume 12 - 2021 | https://doi.org/10.3389/fphar.2021.633453
This article is part of the Research TopicClinical Therapeutic Development Against Cancers Resistant to Targeted TherapiesView all 31 articles
Fibroblast growth factor receptor 4 (FGFR4) is a tyrosine kinase receptor that is a member of the fibroblast growth factor receptor family and is stimulated by highly regulated ligand binding. Excessive expression of the receptor and its ligand, especially FGF19, occurs in many types of cancer. Abnormal FGFR4 production explains these cancer formations, and therefore, this receptor has emerged as a potential target for inhibiting cancer development. This review discusses the diverse mechanisms of oncogenic activation of FGFR4 and highlights some currently available inhibitors targeting FGFR4.
FGFR1–4 and FGFR5 comprise the fibroblast growth factor receptor (FGFR) family (Wang and Ding, 2017). Among these members, FGFR1–4 are typical tyrosine kinase receptors, including a cell surface segment, a one-way cross-membrane section, and a protein–tyrosine kinase domain inside the membrane. FGFR5 is also called FGFRL1, and it differs from the others in that it is missing the intracellular kinase domain (Regeenes et al., 2018). In the process of FGF–FGFR binding, the receptor and the ligand combine to form a dimer stimulated and autophosphorylation complex leads to downstream pathways, including protein serine–threonine kinase (AKT), mitogen-activated protein kinase (MAPK), and signal transducer and activator of transcription 3 (STAT3)-activated pathways (Helsten et al., 2016). Current researchers have determined that the FGFR protein family participates in the generation of tumor cells, angiogenesis, immigration, differentiation, aggression, and drug resistance (Haugsten et al., 2010).
Among the FGFR family members, the role of FGFR4 in cancer has been expounded on by only a few studies. Herein, this review discusses the characteristics of FGFR4 signaling in tumor progression and features some small molecular inhibitors that target FGFR4, intending to increase our understanding of this pathway.
Genetic aberrations in FGFR4 are prevalent among various types of cancer like breast cancer, pancreatic cancer, and especially hepatocellular carcinoma (HCC) (Figure 1), and these aberrations are associated with poor prognoses (Shah et al., 2002; Sawey et al., 2011; Jain and Turner, 2012). The irregular expression of the FGFR4 pathway may be induced by gene amplification, posttranscriptional errors (Helsten et al., 2016), FGFR4 mutations (Futami et al., 2019), translocations, isoform switching, alternative splicing of FGFR4 (Kwiatkowski et al., 2008), and overexpression of specific ligands in cancer or stromal cells (Miura et al., 2012).
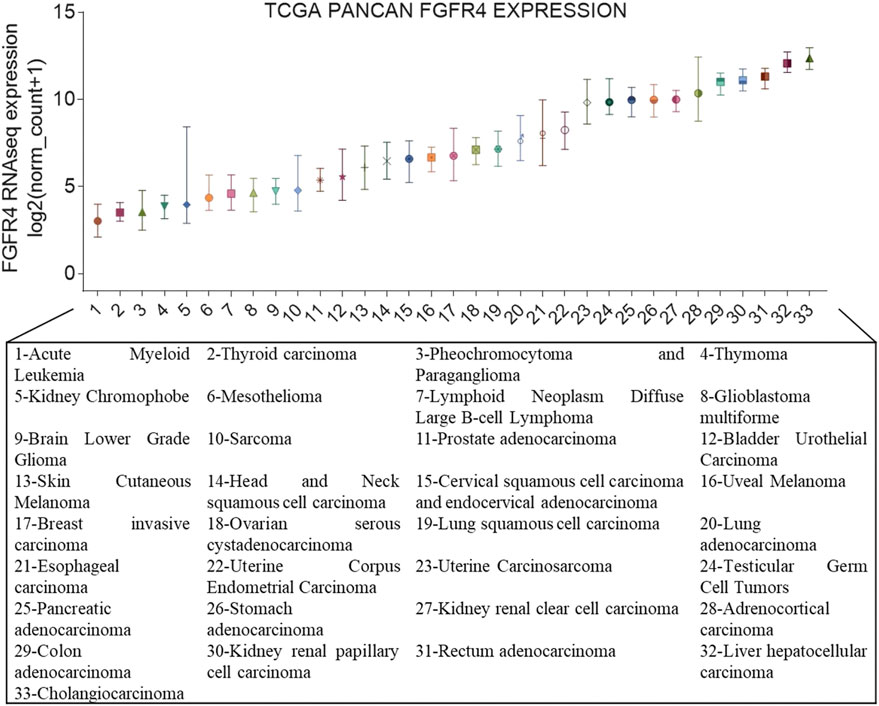
FIGURE 1. FGFR4 gene expression in cancer. Expression of FGFR4 in different cancer types from The Cancer Genome Atlas (TCGA). On the x-axis are the different cancer types in TCGA, and the y-axis depicts gene expression RNAseq of FGFR4 (IlluminaHiSeq), unit: log2 (norm_count+1).
Regarding the aberrations and abnormalities in the FGFR4 gene, except for point mutations, gene fusions, and splice variations, one crucial genetic mutation is the single nucleotide polymorphism (SNP). SNPs can exist in various regions of DNA and can affect the production of transcription factors, translation, and gene expression. Recently, the popularity and widespread use of SNP analysis platforms have made the identification of individual SNPs more convenient (Tsay et al., 2019). The SNPs from FGFR4 have been recognized as an essential participant in cancer occurrence and were associated with prognosis in patients. Examples are FGFR4 rs351855 in HCC (Sheu et al., 2015), FGFR4 SNP rs2011077 with rs1966265 in urothelial cell carcinoma (Tsay et al., 2019), and FGFR4 rs2011077 and rs1966265 in oral squamous cell carcinoma. FGFR4 can alter the production of relative transcript factors, which influences subsequent translation and gene expression (Su et al., 2018) and is directly related to patient survival (Chou et al., 2017).
FGFs function mainly in paracrine and autocrine metabolism (Li, 2019). However, FGF19 subfamilies, including FGF19, FGF21, and FGF23, act as endocrine factors or hormones that bind to specific receptors. FGF19 plays an essential role in metabolism under ordinary physiological conditions (Lin and Desnoyers, 2012). FGF19 subfamily proteins affect the enterohepatic circulation of bile involved in glucose and lipid metabolism and maintain homeostasis phosphorus and vitamin D3 (Dolegowska et al., 2019). Under normal circumstances, the intestinal tract secretes FGF19, and it binds to FGFR4 on liver cells through the hepatoenteral circulation to regulate metabolism (Liu et al., 2020) (Figure 2). In disease states, FGF19 might be crucial for the development and progression of multiple cancers. The specific binding of ligand FGF19 combined with co-receptor β-klotho activated FGFR4. FGF19-mediated activation of the phosphatidylinositol-3-kinase (PI3K)/AKT, MAPK, STAT3, and epithelial–mesenchymal transition (EMT) pathways might take part in the malignancy (Figure 3) (McKinnon et al., 2018; Xin et al., 2018).
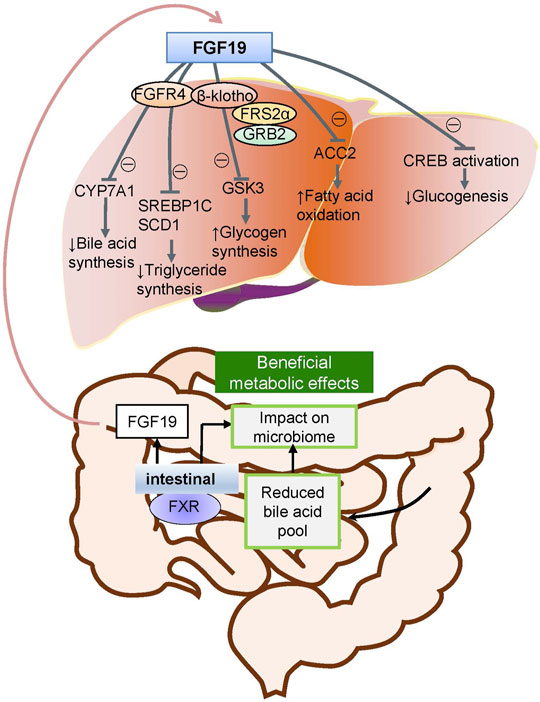
FIGURE 2. Metabolic pathways of FGF19 during normal conditions. Under normal conditions, FGF19 is secreted from the intestinal canal and enters into the enterohepatic circulation into the liver and subsequently binds to the specific receptor activating the FGF19/klotho/FGFR4 pathways. This results in the metabolic regulation of glucogenesis, fatty acid oxidation, glycogen synthesis, triglyceride synthesis, and bile acid synthesis.
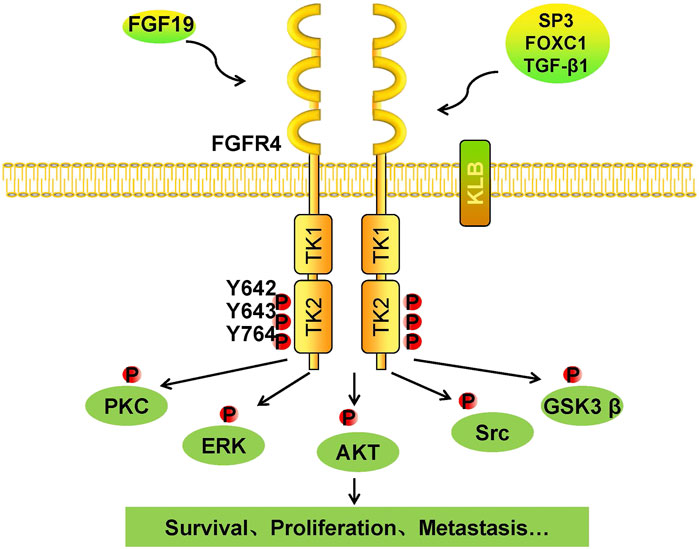
FIGURE 3. Tumorigenic mechanism of the FGF19/klotho/FGFR4 signaling pathways. Once FGF19 binds to FGFR4 with klotho, tyrosine residues in the intramembrane TK2 domain are phosphorylated, and FGFR4 is activated. Phosphorylated FGFR4 activates downstream kinases, including PKC, ERK, AKT, Src, and GSK3β. Cells respond to these activated kinases, and survival, proliferation, and metastasis result.
The FGF19 gene is located in 11q13.3, an amplified section of which is usually found in human HCC (Wu and Li, 2011). An orthotropic transplantation study confirmed that the transplanted hepatocytes overexpressing FGF19 developed tumors (Schwartz, 2011). A monoclonal FGF19-blocking antibody was created to prove the function of FGF19 in cancer development. When examining in vivo and chemically induced liver tumor models, it was observed that the FGF19 antibody suppressed tumor growth (Desnoyers et al., 2008). FGF19 gene amplification is common in several types of cancers, such as lung squamous cell carcinoma (Lang and Teng, 2019), breast cancer (Zhao et al., 2018), and esophageal cancer (Liu et al., 2020) (Figure 4). It is suggested that a potent approach to treating different types of cancer would involve targeting the FGF19 gene to silence it.

FIGURE 4. FGF19 gene copy number in cancer. The gene copy number of FGF19 in different cancer types from The Cancer Genome Atlas (TCGA). On the x-axis, there are 31different TCGA tumor types, and on the y-axis, there are the gene copy numbers of FGF19; unit: Gistic2 copy number.
Identifying the active mechanisms of FGFR4 can be an optimal strategy to develop new therapeutic inhibitors (Table 1). MicroRNAs (miRNAs) play an essential role in developing tumors because they can inhibit the transcription of corresponding target genes (Ueda et al., 2010). Recent research has claimed that miR-7-5p could bind to FGFR4 3’-UTR directly (Tian et al., 2020). It has been demonstrated that miR-491-5p suppresses tumor growth in certain cancers and can indirectly inhibit FGFR4, thus reducing the SNAIL level and weakening EMT-induced tumor migration (Yu et al., 2018). Another study demonstrated that overexpressed miR-29c-3p reduced the secretion of KIAA1199, a cell migration–inducing protein. Subsequently, suppressing the EGFR and FGFR4/AKT pathways’ excitation was ultimately harmful to EMT (Wang et al., 2019).
Interleukin-1β (IL-1β) acts as an essential pro-inflammatory cytokine mediating the innate immune response. It helps the host resist the invasion of microorganisms and is beneficial for body repair (Dinarello, 2011). Zhao et al. found that among the inflammatory cytokines released in response to lipopolysaccharide (LPS), an immune response resulted only when IL-1β specificity restricted the expression of β-klotho. After losing its co-receptor, FGF19 cannot successfully combine with FGFR4. Thus, IL-1β restrained FGF19/FGFR4-induced MAPK phosphorylation and tumor generation (Zhao et al., 2016b). The result suggests that inflammatory cytokines, especially IL-1β, can influence the activation of the FGF19/FGFR4 signaling pathway in the tumor microenvironment.
Specificity protein (Sp) transcription factors (TFs) play an essential role in promoting cancer. Accumulation of Sp1, Sp3, and Sp4 in cells leads to tumor development in organisms (Suske et al., 2005). One study showed that Sp TFs participate in the occurrence and development of a tumor, and they also support tumor resistance to drugs. Sp1, Sp3, and Sp4 are known as no oncogene addiction (NOA) genes and have become relevant drug targets (Safe et al., 2018). Another research demonstrated that the Sp TFs control the FGFR family’s activation (Cavanaugh and DiMario, 2017). Mutation analysis investigated the three Sp binding sites on the FGFR4 promoter, and chromatin immunoprecipitation and electromobility shift assays revealed that Sp3 binding occurred at the location of the FGFR4 promoter. After overexpression of Sp1 and Sp3, it was found that Sp1 inhibited FGFR4 expression, but Sp3 promoted FGFR4 expression (Cavanaugh and DiMario, 2017).
Forkhead box C1 (FOXC1), which belongs to the Forkhead box (FOX) transcription factor family, participates in neural crest, ocular, and mesodermal development. It performs a vital role in lymphatic vessel formation, angiogenesis, and metastasis (Elian et al., 2018). The data accumulated over the years demonstrate the unique behavior of FOXC1 in cancer, particularly in basal-like breast cancer (BLBC) (Wang et al., 2018a). Other studies determined that FOXC1 is significantly involved in breast cancer (BRCA) (Sabapathi et al., 2019) and also colon adenocarcinoma (COAD) (Zhang et al., 2020), pancreatic adenocarcinoma (PAAD) (Subramani et al., 2018), and non–small-cell lung cancer (NSCLC) (Chen et al., 2016). Using genetic epistasis analysis, Liu found that FOXC1 attaches to integrin α7 (ITGA7) and FGFR4 and then activates their expression in metastatic colorectal cancer (CRC). FOXC1 overexpression–mediated CRC metastasis can be reverted using an FGFR4 inhibitor (Liu et al., 2018). The research shows that targeting the FGFR4 signaling pathway might be a useful approach that can be used to treat FOXC1-driven CRC metastasis.
Transforming growth factor β1 (TGF-β1) is significantly associated with regulating cell multiplication, differentiation, invasion, and tumor promotion (Troncone et al., 2018). Among animal models, the TGF-β family significantly impacts metabolism and plays a critical role in tumor transformation, proliferation, invasion, extracellular matrix (ECM) production, and immune reaction (Xie et al., 2018). In the tumor microenvironment, TGF-β1 modulates and interferes with EMT progression, is associated with metastasis, and directly binds to membrane receptors TβR-1 and TβR-2 to exert its effect (Fransvea et al., 2009). Few studies have expounded on a correlation between TGF-β and FGFR4. The expression of FGFR4 is correlated with the diagnosis of HCC, which is related to TGF-β expression. The invasive and metastatic effects of TGF-β1 are realized by inducing FGFR4 and its downstream MAPK pathway (Huang et al., 2018).
The main reason cancer becomes resistant to chemotherapy is that cancer cells have formed antiapoptotic signaling pathways (Zhao et al., 2016a). Substantial studies have shown that the stimulation of the FGFR4 pathway endows cancer with the capacity to resist cancer therapies and chemotherapies (Prieto-Dominguez et al., 2018). A recent study found that breast cancer cell lines can express FGFR4 to gain the ability to resist apoptosis when treated with cyclophosphamide and doxorubicin, while this capacity disappears when the FGFR4 gene is silenced (Andre and Cortes, 2015). FGFR4 overexpression increased Bcl-x expression at the mRNA and protein level through the MAPK cascade, implying that FGFR4 inhibitors (e.g., opposing antibodies) combined with chemotherapeutic drugs should be used for treating FGFR4-overexpressing cancers (Roidl et al., 2009).
Another study showed that drug-resistant cells activate FGFR4 signaling to phosphorylate FGF receptor substrate 2 (FRS2) and then activate downstream MAPK/ERK signaling. Inhibitors that block the FGFR4-FRS2-ERK signaling pathway restrain the glycolytic phenotypes and chemoresistance of resistant cells (Xu et al., 2018). Ahmed et al. (2016) investigated CRC cells that can resist radiotherapy via expression of FGFR4 and discovered that inhibiting FGFR4 can weaken the RAD51-mediated double strain break (DSB) repair, hence attenuating the anti-radiation effect. FGFR4 may be an efficient target for combination therapies to improve radiation response.
FGF19 also plays a crucial role in resistance to therapies (Figure 5). In HCC, overexpression of FGF19 not only promotes EMT by activating the GSK3β/β-catenin and STAT3 pathways (Zhao et al., 2016b) but it also shields HCC cells against endoplasmic reticulum (ER) stress. In liver cancer, ER stress enhanced the transcriptional activation of FGF19 mediated by ATF4, and antiapoptotic ability was observed to increase during ER stress (Teng et al., 2017). Small nucleolar RNA host gene 16 (SNHG16) is a proto-oncogene common to various types of cancer (Gong et al., 2020). One recent study revealed that SNHG16 increased HCC growth and antiapoptosis through the SNHG16/miR-302a-3p/FGF19 pathway (Li et al., 2019). These studies indicate that FGF19 is associated with tumorigenesis, and targeting it may be useful as a form of cancer therapy.
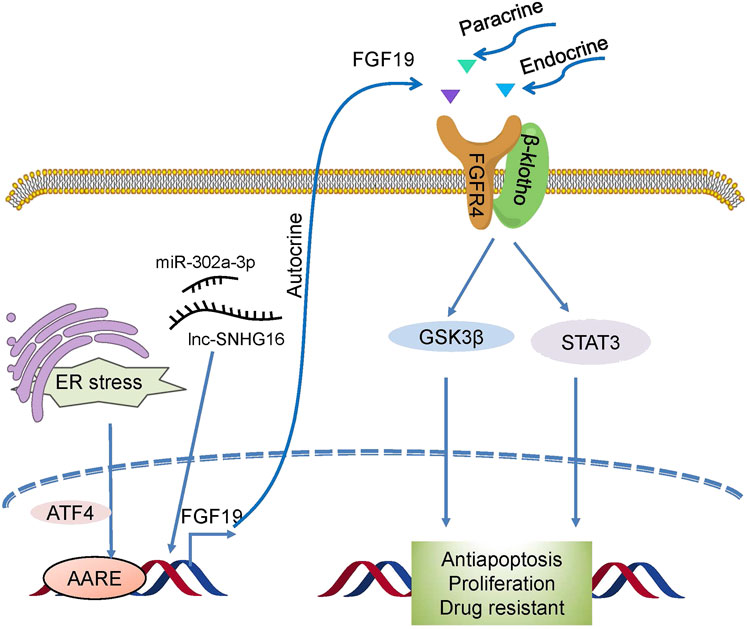
FIGURE 5. Drug resistance mechanism induced by FGF19/FGFR4. Novel activators act on FGF19/FGFR4, which then activates GSK3β/β-catenin with the STAT3 pathway and confers drug resistance to cells.
The FGF19/FGFR4 pathway participates in metabolism and maintaining cell processes such as growth and reproduction. Suppression of the expression of FGFR4 and its ligand or the impairment of its downstream activation has been known as the main reason for tumor growth (Liu et al., 2020). It has been reported that FGFR4 possesses three immunoglobulin-like domains (IgI, IgII, and IgIII) outside the membrane structure that is necessary for a particular ligand, which is also the case for the other three FGFRs (Dai et al., 2019). More importantly, unlike FGFR1–3, there are no splice variants of IgIII in FGFR4 (Touat et al., 2015), which may explain why pan-FGFR inhibitors have a low affinity for FGFR4 and suggests that developing selective FGFR4 inhibitors could be an effective therapeutic strategy. Numerous clinical trials have been performed to test several drugs that specifically target FGF19/FGFR4. Thus far, the most investigated approach in anticancer targeting of the FGF19/FGFR4 pathway has been the use of small molecular inhibitors of FGFR4 kinase.
In order to develop effective anticancer inhibitors of FGFR4, great efforts have been made. Several multi-targeting tyrosine kinase inhibitors (TKIs) have been developed for treatment, such as ponatinib (Massaro and Breccia, 2018), dovitinib (Andre et al., 2013), and lucitanib (Hui et al., 2020). Although these inhibitors have good kinase inhibitory activity against FGFR4, their therapeutic effects are limited by their inhibitory effects on other enzymes (Gavine et al., 2012). After analyzing the ATP domain of the FGFR family, a cysteine residue was found. It is possible to design some covalent inhibitors for this residue to inhibit the phosphorylation of FGFR. Pan inhibitors have been successfully reported as covalent inhibitors of FGFR. They all bind the cysteine residue position at position 477, such as TAS-120 (Goyal et al., 2019), FIIN-1 (Zhou et al., 2010), and FIIN-2 (Tan et al., 2014). However, when using these inhibitors, because of targeting both FGFR1 and FGFR3, severely toxic side effects occur in patients with hyperphosphatemia (Wang et al., 2018b). In view of the important fact that the FGFR family shares sequence homology about its kinase domain, developing a selective inhibitor of FGFR4 has been a daunting challenge. In 2015, Hagel et al. reported a selective FGFR4 inhibitor, BLU-9931, by binding the conserved Cys552 in the hinge region of the FGFR4 protein. This may be an effective treatment strategy. Herein, we reviewed the clinical trial results for FGFR4 inhibitors for different cancer types (Table 2).
According to Figure 1, we had already known that the abnormal expression of FGFR4 occurs obviously in cholangiocarcinoma and liver cancer. In fact, since the occurrence and development of liver cancer are more dependent on FGFR4, the current FGFR4 inhibitors are mainly aimed at the treatment of HCC (Lu et al., 2019). Cholangiocarcinoma is more commonly treated with FGFR2 inhibitors (Raggi et al., 2019). BLU-9931 is an irreversible kinase inhibitor that acts powerfully on FGFR4 but exhibits no sensitivity to other FGFRs, indicating promising kinase group selectivity (Lang et al., 2019). BLU-9931 has the potential to be used as an FGFR4-selective inhibitor to treat HCC patients with FGFR4 signaling abnormalities for the first time (Hagel et al., 2015). BLU-554 is a highly selective kinase inhibitor that inhibits FGFR4 with an IC50 of 5 nM; in contrast, the IC50 range for FGFR1–3 is 624–2,203 nM (Sarker et al., 2016). BLU-554 is currently being tested in ongoing clinical trials to treat HCC (NCT02508467, NCT04194801, etc.). FGF401 is a novel reversible covalent kinase inhibitor that is highly efficient and specific to FGFR4 while having little effect on the other FGFR members and other kinases in the kinome (Weiss et al., 2019). A clinical trial with FGF401 (NCT02325739) for HCC and other solid malignancies is now complete. Joshi et al. used a structure-guided drug design to create H3B-6527, a novel inhibitor selectively and covalently bound to FGFR4. A series of PDX models revealed that H3B-6527 has a beneficial therapeutic effect on patients with overexpression of FGF19, and clinical trials are currently being conducted (NCT03424577, NCT02834780, etc.). INCB062079 is a useful and discriminating irreversible inhibitor targeting FGFR4 (>250-fold vs. FGFR1/2/3) that suppresses the proliferation of HCC driven by increased expression of FGF19 (Ruggeri et al., 2017). Toxicological experiments are currently underway to investigate the safety and tolerability of INCB062079 in patients with a variety of malignancies (NCT03144661).
Although the current task of advancing our knowledge and the use of small-molecule FGFR4 inhibitors is highly interdisciplinary, inhibitors against FGFR4 must be carefully evaluated based on the clinical data. At present, only a few single-agent FGFR4 inhibitors have been confirmed to be efficient for therapy (Lu et al., 2019). Improving our understanding of the pathogenesis that occurs in tumors that overexpress FGFR4, as well as the elucidation of elements that alter the sensitivity to endurance of FGFR4 inhibitors, is vital for a more accurate selection of patients and for increasing the success rate of cancer treatment with FGFR4 inhibitors (Knights and Cook, 2010).
Many investigations and studies have demonstrated that the FGF19/FGFR4 pathway influences cells’ growth, development, and their differentiation in tumors (Katoh, 2016). Abnormal gene expression of FGFR4 with its ligand FGF19 has been determined as a vital factor in tumor growth (Babina and Turner, 2017).
The research on FGFR4 has focused on the exploitation of small molecular inhibitors (Mellor, 2014). Herein, we reviewed various inhibitors of FGFR4 in the cancer microenvironment, including immune evasion, paracrine signaling, and angiogenesis (Repana and Ross, 2015). However, the effectiveness of FGFR4 inhibitors is still being challenged (Wang et al., 2017). Compared to other RTKs, the selectivity of FGFR4 remains relatively new, and sometimes it needs to be used in combination with other adjuvant drugs in clinical treatment to be effective (Jiang et al., 2017). Moreover, their efficacy seems to apply to only a few cancers (Quintanal-Villalonga et al., 2019a). Additional preclinical studies are required to explore FGFR4 inhibitors further and increase their effectiveness during application.
FGFR4 inhibitors have currently attained remarkable potency, and the use of small-molecule inhibitors remains a powerful therapeutic approach (Heinzle et al., 2014). Multiple FGFR4 inhibitors can be applied to treat cancers where FGFR4 signaling is responsible for tumor development. Besides, the use of FGFR4 inhibitors remains a practical approach in cancer patients with high FGFR4 expression. Selective FGFR4 inhibitors are advantageous because of their low toxicity and their ability to be combined with other treatments for more optimal results (Quintanal-Villalonga et al., 2019b). Although FGFR4-based therapy is still relatively new, it should be completely utilized for treating human diseases. Various modern developments are required to explain FGFR4 biology and its treatment strategy further.
Cancer treatments' resistance to carcinogenic drivers remains a major clinical challenge (Zhao et al., 2016a; Zheng et al., 2019). Various mechanisms may induce this phenomenon, but the consequence is unavoidably relative to signaling reactions that promote cell antiapoptosis and survival (Gatenby and Brown, 2018; Yang et al., 2019). Most kinase inhibitors are hydrophobic with relatively small molecular weights, and they competitively bind to the ATP domain of the related kinase to cause inhibition. A typical resistance mechanism is that the kinase domain has a mutation that blocks the drug from combining with the active site (Jiao et al., 2018; Zhao et al., 2020). It is this mechanism that limits the application of FGFR4 inhibitors. One study found that the FGFR4 gene structure had been altered in patients who develop drug resistance to FGFR4 inhibitors in HCC (Hatlen et al., 2019). These changes were later confirmed by in vitro and in vivo assays. In the case of continuous dependence on oncogenes, the concept of differential resistance is of great significance for cancer treatment and contributes to the clinical progress of a new generation of FGFR4 inhibitors, which considers resistance mechanisms while maintaining selectivity for FGFR4.
YL and CW collected the articles and made the figures. JZ and CZ guided YL and CW in collecting articles and making figures. JL prerevised the manuscript. CZ and HX wrote the manuscript and made the tables.
This work was supported by the Natural Science Foundation of Zhejiang Province (LBY20H300001 and LY21H300005) and the Wenzhou Municipal Science and Technology Bureau (ZY2020025, Y20180854, and Y20190721).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ahmed, M. A., Selzer, E., Dörr, W., Jomrich, G., Harpain, F., Silberhumer, G. R., et al. (2016). Fibroblast growth factor receptor 4 induced resistance to radiation therapy in colorectal cancer. Oncotarget 7, 69976–69990. doi:10.18632/oncotarget.12099
André, F., Bachelot, T., Campone, M., Dalenc, F., Perez-Garcia, J. M., Hurvitz, S. A., et al. (2013). Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin. Cancer Res. 19, 3693–3702. doi:10.1158/1078-0432.CCR-13-0190
André, F., and Cortés, J. (2015). Rationale for targeting fibroblast growth factor receptor signaling in breast cancer. Breast Cancer Res. Treat. 150, 1–8. doi:10.1007/s10549-015-3301-y
Babina, I. S., and Turner, N. C. (2017). Advances and challenges in targeting FGFR signalling in cancer. Nat. Rev. Cancer 17, 318–332. doi:10.1038/nrc.2017.8
Cavanaugh, E., and DiMario, J. X. (2017). Sp3 controls fibroblast growth factor receptor 4 gene activity during myogenic differentiation. Gene 617, 24–31. doi:10.1016/j.gene.2017.03.039
Chen, S., Jiao, S., and Li, Y. Y. (2016). Effects of targeted silencing of FOXC1 gene on proliferation and in vitro migration of human non-small-cell lung carcinoma cells. Am. J. Transl Res. 8, 3309–3318.
Chou, C.-H., Hsieh, M.-J., Chuang, C.-Y., Lin, J.-T., Yeh, C.-M., Tseng, P.-Y., et al. (2017). Functional FGFR4 Gly388Arg polymorphism contributes to oral squamous cell carcinoma susceptibility. Oncotarget 8, 96225–96238. doi:10.18632/oncotarget.21958
Dai, S., Zhou, Z., Chen, Z., Xu, G., and Chen, Y. (2019). Fibroblast growth factor receptors (FGFRs): structures and small molecule inhibitors. Cells 8, 614. doi:10.3390/cells8060614
Desnoyers, L. R., Pai, R., Ferrando, R. E., Hötzel, K., Le, T., Ross, J., et al. (2008). Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene 27, 85–97. doi:10.1038/sj.onc.1210623
Dinarello, C. A. (2011). Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 117, 3720–3732. doi:10.1182/blood-2010-07-273417
Dolegowska, K., Marchelek-Mysliwiec, M., Nowosiad-Magda, M., and Dolegowska, M. B. (2019). FGF19 subfamily members: FGF19 and FGF21. J. Physiol. Biochem. 75, 229–240. doi:10.1007/s13105-019-00675-7
Elian, F. A., Yan, E., and Walter, M. A. (2018). FOXC1, the new player in the cancer sandbox. Oncotarget 9, 8165–8178. doi:10.18632/oncotarget.22742
Fransvea, E., Mazzocca, A., Antonaci, S., and Giannelli, G. (2009). Targeting transforming growth factor (TGF)-βRI inhibits activation of β1 integrin and blocks vascular invasion in hepatocellular carcinoma. Hepatology 49, 839–850. doi:10.1002/hep.22731
Futami, T., Kawase, T., Mori, K., Asaumi, M., Kihara, R., and Kuromitsu, N. S. (2019). Identification of a novel oncogenic mutation of FGFR4 in gastric cancer. Sci. Rep. 9, 14627. doi:10.1038/s41598-019-51217-6
Gatenby, R., and Brown, J. (2018). The evolution and ecology of resistance in cancer therapy. Cold Spring Harb Perspect. Med. 8, a033415. doi:10.1101/cshperspect.a033415
Gavine, P. R., Mooney, L., Kilgour, E., Thomas, A. P., Al-Kadhimi, K., Beck, S., et al. (2012). AZD4547: an orally bioavailable, potent, and selective inhibitor of the fibroblast growth factor receptor tyrosine kinase family. Cancer Res. 72, 2045–2056. doi:10.1158/0008-5472.CAN-11-3034
Gong, C.-Y., Tang, R., Nan, W., Zhou, K.-S., and Zhang, H.-H. (2020). Role of SNHG16 in human cancer. Clinica Chim. Acta 503, 175–180. doi:10.1016/j.cca.2019.12.023
Goyal, L., Shi, L., Liu, L. Y., Fece de la Cruz, F., Lennerz, J. K., Raghavan, S., et al. (2019). TAS-120 overcomes resistance to ATP-competitive FGFR inhibitors in patients with FGFR2 fusion-positive intrahepatic cholangiocarcinoma. Cancer Discov. 9, 1064–1079. doi:10.1158/2159-8290.CD-19-0182
Hagel, M., Miduturu, C., Sheets, M., Rubin, N., Weng, W., Stransky, N., et al. (2015). First selective small molecule inhibitor of FGFR4 for the treatment of hepatocellular carcinomas with an activated FGFR4 signaling pathway. Cancer Discov. 5, 424–437. doi:10.1158/2159-8290.CD-14-1029
Hatlen, M. A., Schmidt-Kittler, O., Sherwin, C. A., Rozsahegyi, E., Rubin, N., Sheets, M. P., et al. (2019). Acquired on-target clinical resistance validates FGFR4 as a driver of hepatocellular carcinoma. Cancer Discov. 9, CD–19. doi:10.1158/2159-8290.CD-19-0367
Haugsten, E. M., Wiedlocha, A., and Wesche, S. J. (2010). Roles of fibroblast growth factor receptors in carcinogenesis. Mol. Cancer Res. 8, 1439–1452. doi:10.1158/1541-7786.MCR-10-0168
Heinzle, C., Erdem, Z., Paur, J., Grasl-Kraupp, B., Holzmann, K., Grusch, M., et al. (2014). Is fibroblast growth factor receptor 4 a suitable target of cancer therapy? Cpd 20, 2881–2898. doi:10.2174/13816128113199990594
Helsten, T., Elkin, S., Arthur, E., Tomson, B. N., and Kurzrock, J. R. (2016). The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin. Cancer Res. 22, 259–267. doi:10.1158/1078-0432.CCR-14-3212
Huang, J., Qiu, M., Wan, L., Wang, G., Huang, T., Chen, Z., et al. (2018). TGF-β1 promotes hepatocellular carcinoma invasion and metastasis via ERK pathway-mediated FGFR4 expression. Cell Physiol Biochem 45, 1690–1699. doi:10.1159/000487737
Hui, R., Pearson, A., Cortes, J., Campbell, C., Poirot, C., Azim, H. A., et al. (2020). Lucitanib for the treatment of HR+/HER2− metastatic breast cancer: results from the multicohort phase II FINESSE study. Clin. Cancer Res. 26, 354–363. doi:10.1158/1078-0432.CCR-19-1164
Jain, V. K., and Turner, N. C. (2012). Challenges and opportunities in the targeting of fibroblast growth factor receptors in breast cancer. Breast Cancer Res. 14, 208. doi:10.1186/bcr3139
Jiang, D., Li, J., Li, J., Wang, M., Han, C., Wang, X., et al. (2017). Combination of FGFR4 inhibitor Blu9931 and 5-fluorouracil effects on the biological characteristics of colorectal cancer cells. Int. J. Oncol. 51, 1611–1620. doi:10.3892/ijo.2017.4143
Jiao, Q., Bi, L., Ren, Y., Song, S., Wang, Q., and Wang, Y.-s. (2018). Advances in studies of tyrosine kinase inhibitors and their acquired resistance. Mol. Cancer 17, 36. doi:10.1186/s12943-018-0801-5
Katoh, M. (2016). FGFR inhibitors: effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review). Int. J. Mol. Med. 38, 3–15. doi:10.3892/ijmm.2016.2620
Knights, V., and Cook, S. J. (2010). De-regulated FGF receptors as therapeutic targets in cancer. Pharmacol. Ther. 125, 105–117. doi:10.1016/j.pharmthera.2009.10.001
Kwiatkowski, B. A., Kirillova, I., Richard, R. E., Israeli, D., and Yablonka-Reuveni, Z. (2008). FGFR4 and its novel splice form in myogenic cells: interplay of glycosylation and tyrosine phosphorylation. J. Cel. Physiol. 215, 803–817. doi:10.1002/jcp.21365
Lang, L., Shull, A. Y., and Teng, Y. (2018). Interrupting the FGF19-FGFR4 Axis to therapeutically disrupt cancer progression. Ccdt 19, 17–25. doi:10.2174/1568009618666180319091731
Lang, L., and Teng, Y. (2019). Fibroblast growth factor receptor 4 targeting in cancer: new insights into mechanisms and therapeutic strategies. Cells 8, 31. doi:10.3390/cells8010031
Li, W., Xu, W., Song, J. S., Wu, T., and Wang, W. X. (2019). LncRNA SNHG16 promotes cell proliferation through miR-302a-3p/FGF19 axis in hepatocellular carcinoma. neo 66, 397–404. doi:10.4149/neo_2018_180720N504
Lin, B. C. R., and Desnoyers, L. R. (2012). FGF19 and cancer. Adv. Exp. Med. Biol. 728, 183–194. doi:10.1007/978-1-4614-0887-1_12
Liu, J., Zhang, Z., Li, X., Chen, J., Wang, G., Tian, Z., et al. (2018). Forkhead box C1 promotes colorectal cancer metastasis through transactivating ITGA7 and FGFR4 expression. Oncogene 37, 5477–5491. doi:10.1038/s41388-018-0355-4
Liu, Y., Cao, M., Cai, Y., Li, X., Zhao, C., and Cui, R. (2020). Dissecting the role of the FGF19-FGFR4 signaling pathway in cancer development and progression. Front. Cel Dev. Biol. 8, 95. doi:10.3389/fcell.2020.00095
Lu, X., Chen, H., Patterson, A. V., and Ding, J. B. K. (2019). Fibroblast growth factor receptor 4 (FGFR4) selective inhibitors as hepatocellular carcinoma therapy: advances and prospects. J. Med. Chem. 62, 2905–2915. doi:10.1021/acs.jmedchem.8b01531
Massaro, F., and Breccia, M. (2018). Ponatinib: a review of efficacy and safety. Ccdt 18, 847–856. doi:10.2174/1568009617666171002142659
McKinnon, T., Venier, R., Yohe, M., Sindiri, S., Gryder, B. E., Shern, J. F., et al. (2018). Functional screening of FGFR4-driven tumorigenesis identifies PI3K/mTOR inhibition as a therapeutic strategy in rhabdomyosarcoma. Oncogene 37, 2630–2644. doi:10.1038/s41388-017-0122-y
Mellor, H. R. (2014). Targeted inhibition of the FGF19-FGFR4 pathway in hepatocellular carcinoma; translational safety considerations. Liver Int. 34, e1–e9. doi:10.1111/liv.12462
Miura, S., Mitsuhashi, N., Shimizu, H., Kimura, F., Yoshidome, H., Otsuka, M., et al. (2012). Fibroblast growth factor 19 expression correlates with tumor progression and poorer prognosis of hepatocellular carcinoma. Bmc Cancer 12, 56. doi:10.1186/1471-2407-12-56
Prieto-Dominguez, N., Shull, A. Y., and Teng, Y. (2018). Making way for suppressing the FGF19/FGFR4 axis in cancer. Future Med. Chem. 10, 2457–2470. doi:10.4155/fmc-2018-0099
Quintanal-Villalonga, A., Ferrer, I., Molina-Pinelo, S., and Paz-Ares, L. (2019a). A patent review of FGFR4 selective inhibition in cancer (2007-2018). Expert Opin. Ther. Patents 29, 429–438. doi:10.1080/13543776.2019.1624720
Quintanal-Villalonga, A., Molina-Pinelo, S., Yagüe, P., Marrugal, Á., Ojeda-Márquez, L., Suarez, R., et al. (2019b). FGFR4 increases EGFR oncogenic signaling in lung adenocarcinoma, and their combined inhibition is highly effective. Lung Cancer 131, 112–121. doi:10.1016/j.lungcan.2019.02.007
Raggi, C., Fiaccadori, K., Pastore, M., Correnti, M., Piombanti, B., Forti, E., et al. (2019). Antitumor activity of a novel fibroblast growth factor receptor inhibitor for intrahepatic cholangiocarcinoma. Am. J. Pathol. 189, 2090–2101. doi:10.1016/j.ajpath.2019.06.007
Regeenes, R., Silva, P. N., Chang, H. H., Arany, E. J., Shukalyuk, A. I., Audet, J., et al. (2018). Fibroblast growth factor receptor 5 (FGFR5) is a co-receptor for FGFR1 that is up-regulated in beta-cells by cytokine-induced inflammation. J. Biol. Chem. 293, 17218–17228. doi:10.1074/jbc.RA118.003036
Repana, D., and Ross, P. (2015). Targeting FGF19/FGFR4 pathway: a novel therapeutic strategy for hepatocellular carcinoma. Diseases 3, 294–305. doi:10.3390/diseases3040294
Roidl, A., Berger, H.-J., Kumar, S., Bange, J., and Ullrich, P. A. (2009). Resistance to chemotherapy is associated with fibroblast growth factor receptor 4 up-regulation. Clin. Cancer Res. 15, 2058–2066. doi:10.1158/1078-0432.CCR-08-0890
Ruggeri, B., Stubbs, M., Yang, Y. O., JuvekarandP, A., and Liu, C. J. C. R. (2017). Abstract 1234: the novel FGFR4-selective inhibitor INCB062079 is efficacious in models of hepatocellular carcinoma harboring FGF19 amplification. Cancer Res. 77, 1234. doi:10.1158/1538-7445
Sabapathi, N., Sabarimurugan, S., Madurantakam Royam, M., Kumarasamy, C., Xu, X., Xu, G., et al. (2019). Prognostic significance of FOXC1 in various cancers: a systematic review and meta-analysis. Mol. Diagn. Ther. 23, 695–706. doi:10.1007/s40291-019-00416-y
Safe, S., Abbruzzese, J., and Hedrick, M. E. (2018). Specificity protein transcription factors and cancer: opportunities for drug development. Cancer Prev. Res. 11, 371–382. doi:10.1158/1940-6207.CAPR-17-0407
Sarker, D., Sung, M., Sharma, S., Wolf, B., and Choo, P. S (2016). First-in-human study of BLU-554, a potent, highly-selective FGFR4 inhibitor designed for hepatocellular carcinoma (HCC) with FGFR4 pathway activation. Cancer Discov. 9 (12), 1696–1707. doi:10.1158/2159-8290.CD-19-0555
Sawey, E. T., Chanrion, M., Cai, C., Wu, G., Zhang, J., Zender, L., et al. (2011). Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by Oncogenomic screening. Cancer Cell 19, 347–358. doi:10.1016/j.ccr.2011.01.040
Schwartz, S. (2011). Clinical utility of single nucleotide polymorphism arrays. Clin. Lab. Med. 31, 581–594. doi:10.1016/j.cll.2011.09.002
Shah, R. N., Ibbitt, J. C., Alitalo, K., and Hurst, H. C. (2002). FGFR4 overexpression in pancreatic cancer is mediated by an intronic enhancer activated by HNF1α. Oncogene 21, 8251–8261. doi:10.1038/sj.onc.1206020
Sheu, M. J., Hsieh, M. J., Chiang, W. L., Yang, S. F., Lee, H. L., LeeandC, L. M., et al. (2015). Fibroblast growth factor receptor 4 polymorphism is associated with liver cirrhosis in hepatocarcinoma. Plos One 10, e0122961. doi:10.1371/journal.pone.0122961
Su, S. C., Hsieh, M. J., Lin, C. W., Chuang, C. Y., Liu, Y. F., Yeh, C. M., et al. (2018). Impact of HOTAIR gene polymorphism and environmental risk on oral cancer. J. Dent Res. 97, 717–724. doi:10.1177/0022034517749451
Subramani, R., Camacho, F. A., Levin, C. I., Flores, K., Clift, A., Galvez, A., et al. (2018). FOXC1 plays a crucial role in the growth of pancreatic cancer. Oncogenesis 7, 52. doi:10.1038/s41389-018-0061-7
Suske, G., Bruford, E., and Philipsen, S. (2005). Mammalian SP/KLF transcription factors: bring in the family. Genomics 85, 551–556. doi:10.1016/j.ygeno.2005.01.005
Tan, L., Wang, J., Tanizaki, J., Huang, Z., Aref, A. R., Rusan, M., et al. (2014). Development of covalent inhibitors that can overcome resistance to first-generation FGFR kinase inhibitors. Proc. Natl. Acad. Sci. USA 111, E4869–E4877. doi:10.1073/pnas.1403438111
Teng, Y., Zhao, H., Gao, L., Zhang, W., Shull, A. Y., and Shay, C. (2017). FGF19 protects hepatocellular carcinoma cells against endoplasmic reticulum stress via activation of FGFR4-gsk3β-nrf2 signaling. Cancer Res. 77, 6215–6225. doi:10.1158/0008-5472.CAN-17-2039
Tian, S., Chen, M., Wang, B., Han, Y., and Chen, H. J. (2020). miR-7-5p promotes hepatic stellate cell activation by targeting fibroblast growth factor receptor 4. Gastroenterol. Res. Pract. 2020, 1. doi:10.1155/2020/5346573
Touat, M., Ileana, E., Postel-Vinay, S., André, F., and Soria, J.-C. (2015). Targeting FGFR signaling in cancer. Clin. Cancer Res. 21, 2684–2694. doi:10.1158/1078-0432.CCR-14-2329
Troncone, E., Marafini, I., Stolfi, C., and Monteleone, G. (2018). Transforming Growth Factor-beta1/Smad7 in Intestinal Immunity, Inflammation, and Cancer. Front Immunol. 9, 1407. doi:10.3389/fimmu.2018.01407
Tsay, M.-D., Hsieh, M.-J., Lee, C.-Y., Wang, S.-S., Chen, C.-S., Hung, S.-C., et al. (2019). Involvement of FGFR4 gene variants on the clinicopathological severity in urothelial cell carcinoma. Ijerph 17, 129. doi:10.3390/ijerph17010129
Ueda, T., Volinia, S., Okumura, H., Shimizu, M., Taccioli, C., Rossi, S., et al. (2010). Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 11, 136–146. doi:10.1016/S1470-2045(09)70343-2
Wang, J., Li, W., Zheng, X., and Du, X. G. (2018a). Research progress on the forkhead box C1. Oncotarget 9, 12471–12478. doi:10.18632/oncotarget.22527
Wang, L., Yu, T., Li, W., Li, M., Zuo, Q., Zou, Q., et al. (2019). The miR-29c-KIAA1199 axis regulates gastric cancer migration by binding with WBP11 and PTP4A3. Oncogene 38, 3134–3150. doi:10.1038/s41388-018-0642-0
Wang, S., and Ding, Z. (2017). Fibroblast growth factor receptors in breast cancer. Tumour Biol. 39, 101042831769837. doi:10.1177/1010428317698370
Wang, Y., Chen, Z., Dai, M., Sun, P., Wang, C., Gao, Y., et al. (2017). Discovery and optimization of selective FGFR4 inhibitors via scaffold hopping. Bioorg. Med. Chem. Lett. 27, 2420–2423. doi:10.1016/j.bmcl.2017.04.014
Wang, Y., Li, L., Fan, J., Dai, Y., Jiang, A., Geng, M., et al. (2018b). Discovery of potent irreversible pan-fibroblast growth factor receptor (FGFR) inhibitors. J. Med. Chem. 61, 9085–9104. doi:10.1021/acs.jmedchem.7b01843
Weiss, A., Adler, F., Buhles, A., Stamm, C., Fairhurst, R. A., Kiffe, M., et al. (2019). FGF401, A first-in-class highly selective and potent FGFR4 inhibitor for the treatment of FGF19-driven hepatocellular cancer. Mol. Cancer Ther. 18, 2194–2206. doi:10.1158/1535-7163.MCT-18-1291
Wu, X., and Li, Y. (2011). Therapeutic utilities of fibroblast growth factor 19. Expert Opin. Ther. Targets 15, 1307–1316. doi:10.1517/14728222.2011.624514
Xie, F., Ling, L., van Dam, H., ZhouandL, F., and Zhang, L. (2018). TGF-beta signaling in cancer metastasis. Acta Biochim. Biophys. Sin (Shanghai). 50, 121–132. doi:10.1093/abbs/gmx123
Xin, Z., Song, X., Jiang, B., Gongsun, X., Song, L., Qin, Q., et al. (2018). Blocking FGFR4 exerts distinct anti-tumorigenic effects in esophageal squamous cell carcinoma. Thorac. Cancer 9, 1687–1698. doi:10.1111/1759-7714.12883
Xu, M., Chen, S., Yang, W., Cheng, X., Ye, Y., Mao, J., et al. (2018). FGFR4 links glucose metabolism and chemotherapy resistance in breast cancer. Cel Physiol Biochem 47, 151–160. doi:10.1159/000489759
Yang, L. H., Lin, S. C., Kang, Y. T., Xiang, Y. Q., Xu, L. Y., Li, J. F., et al. (2019). Rhein sensitizes human pancreatic cancer cells to EGFR inhibitors by inhibiting STAT3 pathway. J. Exp. Clin. Cancer Res. 38. doi:10.1186/s13046-018-1015-9
Yu, T., Wang, L. N., Li, W., Zuo, Q. F., Li, M. M., ZouandB, Q. M., et al. (2018). Downregulation of miR-491-5p promotes gastric cancer metastasis by regulating SNAIL and FGFR4. Cancer Sci. 109, 1393–1403. doi:10.1111/cas.13583
Zhang, Y., Liao, Y., Chen, C., Sun, W., Sun, X., Liu, Y., et al. (2020). p38-regulated FOXC1 stability is required for colorectal cancer metastasis. J. Pathol. 250, 217–230. doi:10.1002/path.5362
Zhao, C., Li, H., Lin, H., Yang, S., Lin, J., and Liang, G. (2016a). Feedback activation of STAT3 as a cancer drug-resistance mechanism. Trends Pharmacol. Sci. 37, 47–61. doi:10.1016/j.tips.2015.10.001
Zhao, C., Yang, L., Zhou, F., Yu, Y., Du, X., Xiang, Y., et al. (2020). Feedback activation of EGFR is the main cause for STAT3 inhibition-irresponsiveness in pancreatic cancer cells. Oncogene 39, 3997–4013. doi:10.1038/s41388-020-1271-y
Zhao, H., Lv, F., Liang, G., Huang, X., Wu, G., Zhang, W., et al. (2016b). FGF19 promotes epithelial-mesenchymal transition in hepatocellular carcinoma cells by modulating the GSK3β/β- catenin signaling cascade via FGFR4 activation. Oncotarget 7, 13575–13586. doi:10.18632/oncotarget.6185
Zhao, X., Xu, F., Dominguez, N. P., Xiong, Y., Xiong, Z., Peng, H., et al. (2018). FGFR4 provides the conduit to facilitate FGF19 signaling in breast cancer progression. Mol. Carcinog 57, 1616–1625. doi:10.1002/mc.22884
Zheng, H., Yang, L., Kang, Y., Chen, M., Lin, S., Xiang, Y., et al. (2019). Alantolactone sensitizes human pancreatic cancer cells to EGFR inhibitors through the inhibition of STAT3 signaling. Mol. Carcinogenesis 58, 565–576. doi:10.1002/mc.22951
Keywords: FGFR4, FGF19, cancer, activator, inhibitor
Citation: Liu Y, Wang C, Li J, Zhu J, Zhao C and Xu H (2021) Novel Regulatory Factors and Small-Molecule Inhibitors of FGFR4 in Cancer. Front. Pharmacol. 12:633453. doi: 10.3389/fphar.2021.633453
Received: 25 November 2020; Accepted: 05 March 2021;
Published: 26 April 2021.
Edited by:
Hong Zhu, Zhejiang University, ChinaReviewed by:
Chen Ling, Fudan University, ChinaCopyright © 2021 Liu, Wang, Li, Zhu, Zhao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengguang Zhao, emhhb2NoZW5nZ3VhbmdAd211LmVkdS5jbg==; Huanhai Xu, eGhoMTk5OTIwMDBAMTYzLmNvbQ==; Jiandong Zhu, ODczMzMxNjY2QHFxLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.