
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Pharmacol., 22 January 2019
Sec. Experimental Pharmacology and Drug Discovery
Volume 9 - 2018 | https://doi.org/10.3389/fphar.2018.01477
This article is part of the Research TopicPeptidyl-prolyl Isomerases in Human PathologiesView all 8 articles
 Maguie El Boustani1,2†
Maguie El Boustani1,2† Lucia De Stefano1,3†
Lucia De Stefano1,3† Isabella Caligiuri1
Isabella Caligiuri1 Nayla Mouawad1,4
Nayla Mouawad1,4 Carlotta Granchi4
Carlotta Granchi4 Vincenzo Canzonieri1
Vincenzo Canzonieri1 Tiziano Tuccinardi4
Tiziano Tuccinardi4 Antonio Giordano5
Antonio Giordano5 Flavio Rizzolio1,6*
Flavio Rizzolio1,6*PIN1 is a member of a family of peptidylprolyl isomerases that bind phosphoproteins and catalyze the rapid cis–trans isomerization of proline peptidyl bonds, resulting in an alteration of protein structure, function, and stability. PIN1 is overexpressed in human cancers, suggesting it promotes tumorigenesis, but depending on the cellular context, it also acts as a tumor suppressor. Here, we review the role of PIN1 in cancer and the regulation of PIN1 expression, and catalog the single nucleotide polymorphisms, and mutations in PIN1 gene associated with cancer. In addition, we provide a 3D model of the protein to localize the mutated residues.
Proline is the only amino acid with the ability to adopt either a cis or trans conformation, and this isomerization is catalyzed by peptidylprolyl isomerases (PPIases). The cis-trans isomerization of proline in phosphorylated Ser/Thr-Pro motifs is catalyzed exclusively by PIN1 (peptidylprolyl cis/trans isomerase, NIMA-interacting 1) (Liou et al., 2011).
PIN1-mediated isomerization is an important regulatory mechanism in human physiology and pathology: the conformational change regulates various protein functions, including the catalytic activity, the phosphorylation status, protein interaction, subcellular location, and/or protein stability (Lu et al., 1996).
Structurally, PIN1 has two domains connected by a flexible linker: the N-terminal domain is called “WW” (referring to two invariant Trp residues) and targets the enzyme to pSer/Thr-Pro motifs in substrates; the C-terminal PPIase domain has the catalytic activity (Lu et al., 1996).
PIN1 is involved in cellular processes such as the cell cycle, the folding of newly synthesized proteins, responses to DNA damage and stress, and immune responses (Lu et al., 1996). It is overexpressed in several human cancers (Lee et al., 2011), including prostate cancer (Ayala et al., 2003; La Montagna et al., 2012), breast cancer (Wulf et al., 2001; Ryo et al., 2002; Lucchetti et al., 2013), and oral squamous carcinomas (Miyashita et al., 2003). However, it is still not fully understood how this enzyme participates in cancer development and progression. Several studies showed that some single nucleotide polymorphisms (SNPs) in PIN1 gene increase the risk of cancer whereas other variants operate as protective factors (Segat et al., 2007; Lu et al., 2009; Han et al., 2010; Li et al., 2013; Huang et al., 2016). Little has been reported so far about PIN1 somatic mutations and cancer. This review summarizes the role of PIN1 in cancer and the regulation of PIN1 expression, and is an exhaustive guide to PIN1 SNPs and mutations across cancers.
PIN1 has been shown to be a proto-oncogene whose protein product regulates several proteins involved in cancer initiation and progression (Zhou and Lu, 2016; Russo Spena et al., 2018). For example, PIN1 upregulates the expression of cyclin D1 at both the transcriptional and post-translational levels. At the transcriptional level, PIN1 activates transcription of the gene encoding cyclin D1 (CCND1) via two signal transduction pathways. In the Ras signaling pathway, activation of a kinase cascade leads to phosphorylation, and activation of JNK (c-Jun N-terminal kinase), which phosphorylates and activates the transcription factor c-Jun. PIN1 can bind and isomerize both phosphorylated JNK and phosphorylated c-Jun to potentiate c-Jun transcriptional activity at the CCND1 promoter (Wulf et al., 2001).
PIN1 also stimulates cyclin D1 expression via the Wnt / β-catenin pathway. Briefly, in unstimulated cells, a complex composed of adenomatous polyposis coli (APC), glycogen synthase kinase 3β (GSK-3β), and other proteins keeps cytosolic levels of β-catenin low by triggering this protein’s phosphorylation, ubiquitination and degradation. When extracellular Wnt proteins activate their receptor (composed of a Frizzled receptor and other proteins), GSK-3β is displaced from the complex so β-catenin can accumulate and translocate to the nucleus. There, β-catenin binds transcription factors and other co-activators in a transcription complex that activates CCND1 and other Wnt target genes (MacDonald et al., 2009). PIN1 and β-catenin levels are strictly correlated. PIN1 inhibits the APC-dependent exporting of β-catenin from the nucleus to the cytoplasm and cytoplasmic degradation of β-catenin, thereby β-catenin accumulates in the nucleus where it activates the transcription of genes such as CCND1 (Ryo et al., 2001).
At the protein level, PIN1 isomerizes cyclin D1; this protein modification has a stabilizing effect (Liou et al., 2002). Cyclin D1 then accumulates in the nucleus, where in concert with other proteins it drives cell cycle progression (Liou et al., 2002; Ryo et al., 2002; Gladden and Diehl, 2005). The cyclin D1 activation as downstream target suggests that PIN1 coordinates different events of cell cycle, by acting as molecular timer, and that the overexpression of PIN1 in cancer leads to uncontrolled cell cycle.
Other oncogenic proteins stabilized by being isomerized by PIN1 are Akt (also called protein kinase B), retinoblastoma-associated protein (pRb), and myeloid cell leukemia 1 protein (MCL-1). PIN1 isomerization of Akt is critical for activation of the Akt signaling cascade that in turn activates the transcription of genes encoding cyclin D1, p53 and IKK-NFκB. In cancer cells, high levels of PIN1 amplify the activation of the Akt cascade and thus enhance tumor progression (Liao et al., 2009). PIN1 isomerization of pRb facilitates its binding to CDK–cyclin complexes in mid- to late G1. As a result, pRb is hyperphosphorylated and orchestrates cell proliferation by allowing the expression of genes that mediate entry into the S phase via the E2F transcription factor. In cancer, PIN1 overexpression leads to pRb pathway iperactivation (Rizzolio et al., 2012, 2013). Finally, isomerization of MCL-1 causes a conformational change that may stabilize the protein and enhance its anti-apoptotic function. Briefly, MCL-1 is phosphorylated by GSK-3β, facilitating MCL-1 association with the E3 ligase β-TrCP. The interaction between MCL-1 and the GSK-3β–E3 ligase β-TrCP complex leads to MCL-1 ubiquitination and degradation (Ding et al., 2007). PIN1-mediated isomerization may prevent MCL-1 association with the GSK-3β–E3 ligase β-TrCP complex, blocking MCL-1 degradation, but further studies are required (Ding et al., 2008).
Finally, PIN1 isomerizes two transcription factors, namely NF-κB, increasing its nuclear retention (Ryo et al., 2003), and STAT3, promoting its transactivation (Ryo et al., 2003; Lufei et al., 2007). These two proteins are involved in inflammation-induced carcinogenesis and are constitutively activated in several cancers (Grivennikov and Karin, 2010). PIN1’s action on these transcription factors enhances the transcription of genes encoding cyclin D1, c-Myc and Bcl-2 (Ryo et al., 2003; Lufei et al., 2007).
Altogether, these results feature PIN1 as a tumor promoter, but Yeh and Means described PIN1 as a “conditional” tumor suppressor (Yeh and Means, 2007) and successive studies support this theory. Indeed, PIN1 can induce apoptosis, prevent genomic instability, and promote the ubiquitin-dependent proteolysis of many oncogenic proteins. All these processes limit tumor progression.
In stress conditions, PIN1 induces apoptosis via p53 and p73 (Mantovani et al., 2004). Moreover, a study on murine embryonic fibroblasts showed that PIN1 prevents p53-dependent genomic instability (Wulf et al., 2004).
PIN1 is involved in the ubiquitin-dependent proteolysis of Myc, Bcl-6 and cyclin E (Yeh et al., 2004; Yi et al., 2005; Phan et al., 2007; Farrell et al., 2013). For example, PIN1 binds to doubly phosphorylated Myc on Thr58 and Ser62. The conformational change facilitates Myc dephosphorylation on Ser62 by protein phosphatase 2 (PP2A), which allows Myc ubiquitination and degradation by the proteasome (Farrell et al., 2013). Additionally, upon DNA damage, the kinase ATM phosphorylates Bcl-6 that in turn becomes a substrate for PIN1. Bcl-6 isomerization signals its degradation by the ubiquitin-proteasome system (Phan et al., 2007).
Finally, PIN1 regulates the degradation of cyclin E during the G0/G1-S phase transition of the cell cycle permitting to cell cycle to proceed correctly (Yeh et al., 2006). Experiments in which PIN1 expression was down-regulated showed increased steady-state levels of cyclin E, the arrest of cells in G1/S phase, genomic instability, and tumoral transformation (Yeh et al., 2006).
PIN1 transcription can be activated by the E2F transcription factor or by Notch1 binding to the PIN1 promoter (Ryo et al., 2002; Rustighi et al., 2009). In this way PIN1 sustains the transformed phenotype induced by E2F or Notch1 activation.
PIN1 transcription can also be suppressed by the tumor suppressor gene BRCA1 (MacLachlan et al., 2000). BRCA1 associates with several proteins to regulate DNA repair response. In cancer, BRCA1 is often mutated and lost such function, thereby cells accumulates DNA damage (Mersch et al., 2015).
Recently, micro-RNAs (miRNAs) have been identified as regulators of PIN1 expression. For instance, miR200c binds to a conserved region in the 3′-untranslated region (UTR) of PIN1 mRNA and prevents its translation (Luo et al., 2014). Mutations in this region of PIN1 can prevent the repressive effects of miR200c (Luo et al., 2014). miRNA-200b, and miR-296-5p also bind the 3′ UTR of PIN1 mRNA and down-regulate its expression. In cancer cells, both these miRNAs were found to be underexpressed, allowing PIN1 to sustain tumor progression (Zhang et al., 2013; Lee et al., 2014).
Depending on the physiological or pathological conditions, the activity of proteins is regulated by post-translational modifications. For PIN1, there is evidence of post-translational modification by phosphorylation, ubiquitination, SUMOylation and oxidization at specific sites.
Phosphorylation of PIN1 on Ser16 in the WW domain suppresses its ability to interact with its substrates (Lu et al., 2002). At least three kinases can phosphorylate this residue: protein kinase A (Lu et al., 2002), ribosomal S6 kinase 2 (Cho et al., 2012), and aurora kinase A (Lee et al., 2013). Phosphorylation on Ser71 in the PPIase domain inhibits the protein’s enzymatic activity (Lee et al., 2013). Phosphorylation on Ser65 by polo-like kinase (Plk1) (Eckerdt et al., 2005) and on Ser138 by mixed-lineage kinase 3 increases PIN1’s catalytic activity and nuclear translocation (Rangasamy et al., 2012). Plk1-mediated Ser65 phosphorylation is suggested to regulate PIN1 turnover. It induced PIN1 deubiquitination and stabilization, while the absence of Plk1 enhanced the ubiquitination and degradation of PIN1 (Eckerdt et al., 2005).
SUMOylation of Lys6 in the WW domain and Lys63 in the PPIase domain abolishes PIN1’s enzymatic activity and oncogenic functions (Chen et al., 2013). However, deSUMOylation of these two domains by SUMO1/sentrin specific peptidase 1 (SENP1) restores PIN1’s activity. SENP1 overexpression increases the levels of deSUMOylated PIN1 and in turn the ability of PIN1 to induce centrosome amplification and cell transformation (Chen et al., 2013).
Finally, under conditions of oxidative stress, PIN1 is oxidized on Cys113 in the catalytic site, inhibiting its enzymatic activity (Chen et al., 2015).
Several SNPs, located in the promoter or coding region of PIN1, are associated with cancer risk. The PIN1 variants rs2233678 (c.–842G>C) and rs2233679 (c.–667T>C), both located in the promoter, and the synonymous change rs2233682 (G>A; p.Gln33Gln) in exon 2 of the coding region, have been widely investigated.
The −842C allele of rs2233678 was found to confer a significantly lower risk of cancer (odds ratio, 0.75) in a meta-analysis of 11 studies (9280 participants) of patients with esophageal carcinoma, nasopharyngeal carcinoma, laryngeal squamous cell carcinoma, lung cancer, breast cancer, squamous cell carcinoma of the head, and neck or hepatocellular carcinoma (HCC), and matched healthy controls (Li et al., 2013). Seven of the included studies had found that the allele reduced risk, while four found no association with cancer risk.
A study of 209 patients with oral squamous cell carcinoma and 444 controls did not find an association between the −842G>C polymorphism and cancer risk (Yao et al., 2014).
The −667T>C polymorphism has been found to not associate with esophageal carcinoma (You et al., 2013), breast cancer (Han et al., 2010), or squamous cell carcinoma of the head and neck (Lu et al., 2009), whereas it did associate with a lower risk of nasopharyngeal carcinoma (Lu et al., 2013), and a higher risk of oral squamous cell carcinoma (Yao et al., 2014).
Finally, the synonymous change Gln33Gln was not found to associate with the risk of breast cancer (Han et al., 2010), or squamous cell carcinoma of the head and neck (Lu et al., 2009). However, a higher risk of HCC was found among carriers of the Gln33Gln variant in a Chinese population (Huang et al., 2016).
Because of the lack of published papers reporting on PIN1 somatic mutations, we obtained deposited genetic data on these mutations in different tumor types from the cBioPortal for Cancer Genomics and COSMIC (Catalog of Somatic Mutations in Cancer). cBioPortal is the main resource for the analysis of large-scale cancer genomics datasets (Cerami et al., 2012; Gao et al., 2013). COSMIC, a database of mutations reported in the scientific literature or from the Cancer Genome Project, permits researchers to explore the effects of somatic mutations in cancer (Forbes et al., 2017).
The data obtained from cBioPortal regarded 11,000 cancer cases collected for genomic characterization in December 2013 and 32,555 cases in 61 primary sites of cancer (retrieved on June 13, 2018) (Cerami et al., 2012; Gao et al., 2013). The data from COSMIC refer to 41,924 unique samples from patients with different types of cancer including skin, breast, intestinal, lung, liver, prostate, and stomach cancer (retrieved on June 14, 2018) (Forbes et al., 2017). Altogether, the data revealed the existence of 32 somatic mutations affecting 29 unique residues in the coding region of PIN1 gene (Table 1). Five mutations affect the WW domain (residues 1–39), two are in the flexible linker (residues 35–53), and 25 affect the PPIase domain (residues 50–163). Twenty-three are missense mutations, four are synonymous mutations (they have no effect on PIN1 function), and four are nonsense mutations (R21∗, S71∗, S108∗, and E163∗). Overall, 17 mutations were predicted to be pathogenic by the Functional Analysis through Hidden Markov Models (FATHMM) filter in COSMIC and three to be deleterious by the Sorting Intolerant from Tolerant (SIFT) algorithm in cBioPortal. The others are predicted to be tolerated.
Figure 1 illustrates the positions of PIN1 somatic mutations that alter the protein’s primary sequence and are predicted to be pathogenic or deleterious. This model is based on the X-ray crystal structure of PIN1 bound to a non-natural peptide inhibitor (Protein Data Bank accession code, 2ITK) (Zhang et al., 2007). The Q33K and R36P mutations (orange) are in the WW domain, as is G39C that is not shown because it belongs to a peptide loop missing from the X-ray structure. All the other mutations are found in the PPIase domain: F134S, S154F, and H157Y (magenta) interact with the enzyme’s substrates (Ranganathan et al., 1997; Wilson et al., 2013) while S71L, D112N, P133L, and T152M (yellow) are indirectly involved in the interaction with substrates (Ranganathan et al., 1997; Behrsin et al., 2007; Namanja et al., 2011). The other mutations (green) do not interact with substrates and thus could have a role in interactions with other proteins or in PIN1 protein folding. Among them, F139S is within the PPIase domain interface (S138 to R142) that is involved in interdomain communication and regulates the function of PIN1 upon substrate binding (Behrsin et al., 2007; Namanja et al., 2011).
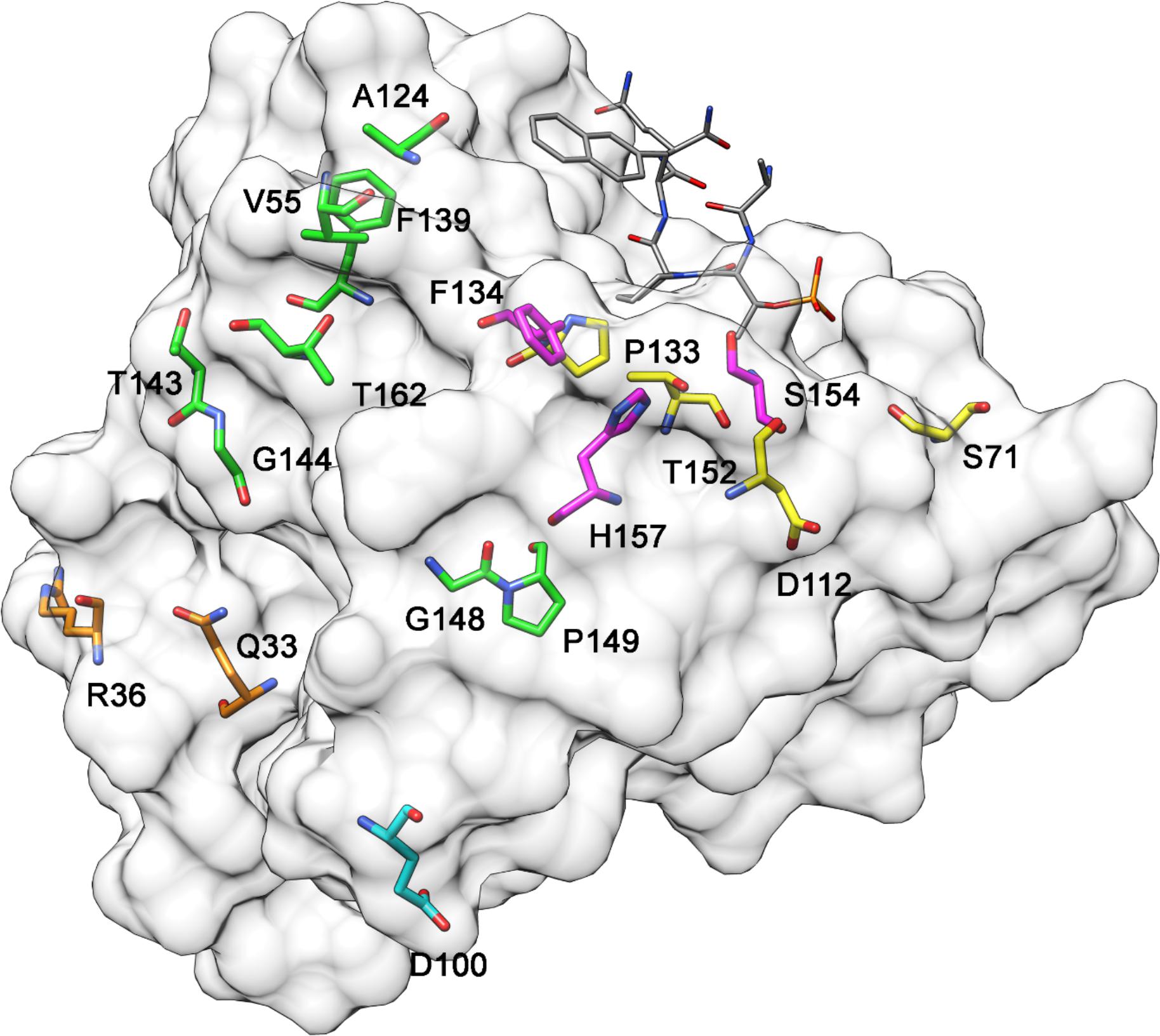
Figure 1. 3D model of PIN1 protein highlighting amino acids that have undergone pathogenic mutation in cancer cases. Orange, mutated residues in the WW domain; magenta, residues in the PPIase domain that interact directly with substrates; yellow, residues in the PPIase domain that interact indirectly with substrates; green, residues in the PPIase domain that do not interact with substrates The gray molecule is a non-natural peptide inhibitor bound to PIN1 in the X-ray crystal structure (Protein Data Bank accession code, 2ITK) (Zhang et al., 2007).
In the last decade, some PIN1 inhibitors were discovered by industries and academic research groups. Compounds 1 (Guo et al., 2009) and 2 (Dong et al., 2010; Figure 2), developed by Pfizer Worldwide Research & Development, are nanomolar PIN1 inhibitors (Ki = 6 and 890 nM for compounds 1 and 2, respectively), as confirmed by X-ray structures that showed their binding to the enzyme. Unfortunately, both of them failed to be active in cell-based assays, likely due to the phosphate or carboxylate groups which were necessary to properly interact in a charged pocket of the enzyme, but strongly limited cell membrane permeability.
The α-Amino acid-derived compounds 3 and 4 (Figure 2) were discovered by researchers at Vernalis (R&D) Ltd. (Potter A.J. et al., 2010). These compounds maintained the carboxylate group, which is the moiety necessary for an optimal interaction in the enzyme binding site, and the aromatic portions were varied, inserting benzimidazole (3), or naphthalene (4), since they lied in a hydrophobic region of the protein. Compound 3 showed an IC50 value in the submicromolar range (0.13 μM), but it was unable to be active in cellular assays. Nevertheless, with the aim of reducing the polar surface area, its analog 4 was about twenty-fold less potent on the isolated enzyme (IC50 = 2.6 μM), but it gained activity in PC3 prostate cancer cells in which it reduced the proliferation.
Due to the problems encountered with the benzimidazole/naphthyl-based compounds, a further development in the search for PIN1 inhibitors at Vernalis (R&D) Ltd, consisted in the phenyl-imidazole derivatives. As a result, compound 5 (Figure 2) reached an optimal balance between inhibition activity on PIN1 (IC50 = 0.83 μM) and antiproliferative activity in PC3 cells (GI50 = 13 μM) (Potter A. et al., 2010).
The dihydrothiazole series, exemplified by compound 6 (Figure 2), was discovered in 2014 at Pfizer Worldwide Research & Development: in this chemical class, the amide group present in all the PIN1 inhibitors previously reported in literature was substituted by a dihydrothiazole ring, bearing a carboxylic acid moiety, with aim of decreasing polar surface area, which is a factor strongly influencing cellular permeability (Asso et al., 2008; Mikami et al., 2017). They showed micromolar inhibition potencies (IC50 values ranging from 1.9 to 27 μM) on PIN1 and they were able to reduce the proliferation of colon cancer cells (HT29) (Guo et al., 2014).
A new class of PIN1 inhibitors developed by Zhao et al. consisted in thiazole derivatives bearing oxalic or acetic acid group at 4-position and, according to modeling studies, this last portion was found to be located in the charged pocket of the enzyme. Compound 7 (Figure 2) showed an IC50 value of 2.93 μM, but unfortunately no data about its activity in cancer cells was reported (Zhao et al., 2016). Among the recently developed PIN1 inhibitors, a selenium containing compound (compound 8, Figure 2) was identified by a novel high-throughput screening study. Compound 8 efficiently inhibited PIN1 (IC50 = 0.43 μM) and it was able to affect the proliferation of breast MDA-MB-231 cancer cells in which PIN1 is overexpressed, also reducing the viability of induced cancer stem cell-like cells (Subedi et al., 2016).
Wei et al. (2015) identified PIN1 as a target of all-trans retinoic acid (ATRA), compound 9 (Figure 2). ATRA inhibited and degraded PIN1 (Ki value of 0.82 μM), as confirmed by the co-crystal structure of ATRA with PIN1. Furthermore, ATRA was able to suppress the growth of triple-negative breast cancer and acute promyelocytic leukemia cells, both in humans and in animal models.
It is also noteworthy to mention two covalent PIN1 inhibitors. The α,β-unsaturated isothiazolone derivative 10 (Figure 3) showed a micromolar inhibition activity on the enzyme (IC50 = 6.1 μM); however, it was not selective for PIN1 also exerting a similar activity on cyclophilin (IC50 = 13.7 μM) (Mori et al., 2011). The quinone-sulfonamide derivative 11 (Figure 3) covalently bound Cys 113 (IC50 value of 0.64 μM), ultimately leading to PIN1 degradation. Moreover, 11 impaired PIN1-dependent invasive behavior of breast (MDA-MB-231) and prostate (PC3) cancer cells (Campaner et al., 2017).
PIN1 is overexpressed in various cancer types and is associated with a malignant phenotype and tumor progression (Bao et al., 2004; Yeh and Means, 2007; Zhou and Lu, 2016). PIN1 may also have an anti-cancer role depending on the cellular context; therefore PIN1 has been called a conditional tumor suppressor gene (Yeh et al., 2006; Yeh and Means, 2007). Some SNPs in PIN1 gene were found to associate with cancer risk. Carriers of the −842C variant in the PIN1 promoter have low PIN1 protein levels and low risk for developing cancer (Li et al., 2013). Contrasting evidence has been reported for the −667C variant in the PIN1 promoter, which was found to associate with a low risk of developing nasopharyngeal carcinoma (Li et al., 2013), but a high risk for oral squamous cell carcinoma and HCC (Yao et al., 2014; Huang et al., 2016).
So far, 32 somatic mutations in PIN1 gene have been found in different types of cancer. Of these, 20 are predicted to be pathogenic or deleterious. Although further studies are required, we believe that investigating the complex pattern of PIN1 gene alterations and their effects on PIN1 protein structure and function is a valid strategy for identifying new biomarkers for susceptibility to cancer and response to anti-PIN1 inhibitors.
All the authors wrote and approved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to and would like to recognize the Associazione Italiana per la Ricerca sul Cancro – My First AIRC (n. 15639). Valerie Matarese provided editorial advice and scientific editing.
Asso, V., Ghilardi, E., Bertini, S., Digiacomo, M., Granchi, C., Minutolo, F., et al. (2008). α-Naphthylaminopropan-2-ol derivatives as BACE1 inhibitors. ChemMedChem 3, 1530–1534. doi: 10.1002/cmdc.200800162
Ayala, G., Wang, D., Wulf, G., Frolov, A., Li, R., Sowadski, J., et al. (2003). The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer Res. 63, 6244–6251.
Bao, L., Kimzey, A., Sauter, G., Sowadski, J. M., Lu, K. P., and Wang, D. G. (2004). Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am. J. Pathol. 164, 1727–1737. doi: 10.1016/S0002-9440(10)63731-5
Behrsin, C. D., Bailey, M. L., Bateman, K. S., Hamilton, K. S., Wahl, L. M., Brandl, C. J., et al. (2007). Functionally important residues in the peptidyl-prolyl isomerase Pin1 revealed by unigenic evolution. J. Mol. Biol. 365, 1143–1162. doi: 10.1016/j.jmb.2006.10.078
Beltran, H., Prandi, D., Mosquera, J. M., Benelli, M., Puca, L., Cyrta, J., et al. (2016). Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305. doi: 10.1038/nm.4045
Campaner, E., Rustighi, A., Zannini, A., Cristiani, A., Piazza, S., Ciani, Y., et al. (2017). A covalent PIN1 inhibitor selectively targets cancer cells by a dual mechanism of action. Nat. Commun. 8:15772. doi: 10.1038/ncomms15772
Campbell, J. D., Alexandrov, A., Kim, J., Wala, J., Berger, A. H., Pedamallu, C. S., et al. (2016). Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 48, 607–616. doi: 10.1038/ng.3564
Cancer Genome Atlas Network (2015). Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582. doi: 10.1038/nature14129
Cancer Genome Atlas Research Network, A. J., Thorsson, V., Shmulevich, I., and Reynolds, S. M., (2014). Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209. doi: 10.1038/nature13480
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data: figure 1. Cancer Discov. 2, 401–404. doi: 10.1158/2159-8290.CD-12-0095
Chen, C.-H., Chang, C.-C., Lee, T. H., Luo, M., Huang, P., Liao, P.-H., et al. (2013). SENP1 deSUMOylates and regulates Pin1 protein activity and cellular function. Cancer Res. 73, 3951–3962. doi: 10.1158/0008-5472.CAN-12-4360
Chen, C.-H., Li, W., Sultana, R., You, M.-H., Kondo, A., Shahpasand, K., et al. (2015). Pin1 cysteine-113 oxidation inhibits its catalytic activity and cellular function in Alzheimer’s disease. Neurobiol. Dis. 76, 13–23. doi: 10.1016/j.nbd.2014.12.027
Cho, Y. S., Park, S. Y., Kim, D. J., Lee, S.-H., Woo, K.-M., Lee, K.-A., et al. (2012). TPA-induced cell transformation provokes a complex formation between Pin1 and 90 kDa ribosomal protein S6 kinase 2. Mol. Cell. Biochem. 367, 85–92. doi: 10.1007/s11010-012-1322-y
Ding, Q., He, X., Hsu, J.-M., Xia, W., Chen, C.-T., Li, L.-Y., et al. (2007). Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol. Cell. Biol. 27, 4006–4017. doi: 10.1128/MCB.00620-06
Ding, Q., Huo, L., Yang, J.-Y., Xia, W., Wei, Y., Liao, Y., et al. (2008). Down-regulation of myeloid cell leukemia-1 through inhibiting Erk/Pin 1 pathway by sorafenib facilitates chemosensitization in breast cancer. Cancer Res. 68, 6109–6117. doi: 10.1158/0008-5472.CAN-08-0579
Dong, L., Marakovits, J., Hou, X., Guo, C., Greasley, S., Dagostino, E., et al. (2010). Structure-based design of novel human Pin1 inhibitors (II). Bioorg. Med. Chem. Lett. 20, 2210–2214. doi: 10.1016/j.bmcl.2010.02.033
Durinck, S., Ho, C., Wang, N. J., Liao, W., Jakkula, L. R., Collisson, E. A., et al. (2011). Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 1, 137–143. doi: 10.1158/2159-8290.CD-11-0028
Eckerdt, F., Yuan, J., Saxena, K., Martin, B., Kappel, S., Lindenau, C., et al. (2005). Polo-like kinase 1-mediated phosphorylation stabilizes Pin1 by inhibiting its ubiquitination in human cells. J. Biol. Chem. 280, 36575–36583. doi: 10.1074/jbc.M504548200
Farrell, A. S., Pelz, C., Wang, X., Daniel, C. J., Wang, Z., Su, Y., et al. (2013). Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol. Cell. Biol. 33, 2930–2949. doi: 10.1128/MCB.01455-12
Forbes, S. A., Beare, D., Boutselakis, H., Bamford, S., Bindal, N., Tate, J., et al. (2017). COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D777–D783. doi: 10.1093/nar/gkw1121
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:l1. doi: 10.1126/scisignal.2004088
Giannakis, M., Hodis, E., Jasmine Mu, X., Yamauchi, M., Rosenbluh, J., Cibulskis, K., et al. (2014). RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet. 46, 1264–1266. doi: 10.1038/ng.3127
Giannakis, M., Mu, X. J., Shukla, S. A., Qian, Z. R., Cohen, O., Nishihara, R., et al. (2016). Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 15, 857–865. doi: 10.1016/j.celrep.2016.03.075
Gladden, A. B., and Diehl, J. A. (2005). Location, location, location: the role of cyclin D1 nuclear localization in cancer. J. Cell. Biochem. 96, 906–913. doi: 10.1002/jcb.20613
Grivennikov, S. I., and Karin, M. (2010). Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth. Factor. Rev. 21, 11–19. doi: 10.1016/j.cytogfr.2009.11.005
Guo, C., Hou, X., Dong, L., Dagostino, E., Greasley, S., Ferre, R., et al. (2009). Structure-based design of novel human Pin1 inhibitors (I). Bioorg. Med. Chem. Lett. 19, 5613–5616. doi: 10.1016/j.bmcl.2009.08.034
Guo, C., Hou, X., Dong, L., Marakovits, J., Greasley, S., Dagostino, E., et al. (2014). Structure-based design of novel human Pin1 inhibitors (III): optimizing affinity beyond the phosphate recognition pocket. Bioorg. Med. Chem. Lett. 24, 4187–4191. doi: 10.1016/j.bmcl.2014.07.044
Han, C. H., Lu, J., Wei, Q., Bondy, M. L., Brewster, A. M., Yu, T.-K., et al. (2010). The functional promoter polymorphism (-842G & gt;C) in the PIN1 gene is associated with decreased risk of breast cancer in non-hispanic white women 55 years and younger. Breast Cancer Res. Treat. 122, 243–249. doi: 10.1007/s10549-009-0682-9
Hodis, E., Watson, I. R., Kryukov, G. V., Arold, S. T., Imielinski, M., Theurillat, J.-P., et al. (2012). A landscape of driver mutations in melanoma. Cell 150, 251–263. doi: 10.1016/j.cell.2012.06.024
Huang, L., Mo, Z., Lao, X., Zhang, X., Liu, Y., Sui, J., et al. (2016). PIN1 genetic polymorphisms and the susceptibility of HBV-related hepatocellular carcinoma in a Guangxi population. Tumour Biol. 37, 6599–6606. doi: 10.1007/s13277-015-4539-z
Krauthammer, M., Kong, Y., Bacchiocchi, A., Evans, P., Pornputtapong, N., Wu, C., et al. (2015). Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat. Genet. 47, 996–1002. doi: 10.1038/ng.3361
La Montagna, R., Caligiuri, I., Maranta, P., Lucchetti, C., Esposito, L., Paggi, M. G., et al. (2012). Androgen receptor serine 81 mediates Pin1 interaction and activity. Cell Cycle 11, 3415–3420. doi: 10.4161/cc.21730
Lee, K.-H., Lin, F.-C., Hsu, T.-I., Lin, J.-T., Guo, J.-H., Tsai, C.-H., et al. (2014). MicroRNA-296-5p (miR-296-5p) functions as a tumor suppressor in prostate cancer by directly targeting Pin1. Biochim. Biophys. Acta 1843, 2055–2066. doi: 10.1016/j.bbamcr.2014.06.001
Lee, T. H., Pastorino, L., and Lu, K. P. (2011). Peptidyl-prolyl cis-trans isomerase Pin1 in ageing, cancer and Alzheimer disease. Expert Rev. Mol. Med. 13:e21. doi: 10.1017/S1462399411001906
Lee, Y.-C., Que, J., Chen, Y.-C., Lin, J.-T., Liou, Y.-C., Liao, P.-C., et al. (2013). Pin1 acts as a negative regulator of the G2/M transition by interacting with the aurora-a-bora complex. J. Cell Sci. 126, 4862–4872. doi: 10.1242/jcs.121368
Li, Q., Dong, Z., Lin, Y., Jia, X., Li, Q., Jiang, H., et al. (2013). The rs2233678 polymorphism in PIN1 promoter region reduced cancer risk: a meta-analysis. PLoS One 8:e68148. doi: 10.1371/journal.pone.0068148
Liao, Y., Wei, Y., Zhou, X., Yang, J. Y., Dai, C., Chen, Y. J., et al. (2009). Peptidyl-prolyl cis/trans isomerase Pin1 is critical for the regulation of PKB/Akt stability and activation phosphorylation. Oncogene 28, 2436–2445. doi: 10.1038/onc.2009.98
Liou, Y.-C., Ryo, A., Huang, H.-K., Lu, P.-J., Bronson, R., Fujimori, F., et al. (2002). Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc. Natl. Acad. Sci. U.S.A. 99, 1335–1340. doi: 10.1073/pnas.032404099
Liou, Y.-C., Zhou, X. Z., and Lu, K. P. (2011). Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem. Sci. 36, 501–514. doi: 10.1016/j.tibs.2011.07.001
Lu, J., Hu, Z., Wei, S., Wang, L.-E., Liu, Z., El-Naggar, A. K., et al. (2009). A novel functional variant (-842G & gt;C) in the PIN1 promoter contributes to decreased risk of squamous cell carcinoma of the head and neck by diminishing the promoter activity. Carcinogenesis 30, 1717–1721. doi: 10.1093/carcin/bgp171
Lu, K. P., Hanes, S. D., and Hunter, T. (1996). A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380, 544–547. doi: 10.1038/380544a0
Lu, P.-J., Zhou, X. Z., Liou, Y.-C., Noel, J. P., and Lu, K. P. (2002). Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J. Biol. Chem. 277, 2381–2384. doi: 10.1074/jbc.C100228200
Lu, Y., Huang, G.-L., Pu, X.-X., He, Y.-X., Li, B.-B., Liu, X.-Y., et al. (2013). Association between PIN1 promoter polymorphisms and risk of nasopharyngeal carcinoma. Mol. Biol. Rep. 40, 3777–3782. doi: 10.1007/s11033-012-2454-6
Lucchetti, C., Caligiuri, I., Toffoli, G., Giordano, A., and Rizzolio, F. (2013). The prolyl isomerase Pin1 acts synergistically with CDK2 to regulate the basal activity of estrogen receptor α in breast cancer. PLoS One 8:e55355. doi: 10.1371/journal.pone.0055355
Lufei, C., Koh, T. H., Uchida, T., and Cao, X. (2007). Pin1 is required for the Ser727 phosphorylation-dependent Stat3 activity. Oncogene 26, 7656–7664. doi: 10.1038/sj.onc.1210567
Luo, M.-L., Gong, C., Chen, C.-H., Lee, D. Y., Hu, H., Huang, P., et al. (2014). Prolyl isomerase Pin1 acts downstream of miR200c to promote cancer stem-like cell traits in breast cancer. Cancer Res. 74, 3603–3616. doi: 10.1158/0008-5472.CAN-13-2785
MacDonald, B. T., Tamai, K., and He, X. (2009). Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26. doi: 10.1016/j.devcel.2009.06.016
MacLachlan, T. K., Somasundaram, K., Sgagias, M., Shifman, Y., Muschel, R. J., Cowan, K. H., et al. (2000). BRCA1 effects on the cell cycle and the DNA damage response are linked to altered gene expression. J. Biol. Chem. 275, 2777–2785. doi: 10.1074/jbc.275.4.2777
Mantovani, F., Piazza, S., Gostissa, M., Strano, S., Zacchi, P., Mantovani, R., et al. (2004). Pin1 links the activities of c-Abl and p300 in regulating p73 function. Mol. Cell 14, 625–636. doi: 10.1016/j.molcel.2004.05.007
Mersch, J., Jackson, M. A., Park, M., Nebgen, D., Peterson, S. K., Singletary, C., et al. (2015). Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer 121, 269–275. doi: 10.1002/cncr.29041
Mikami, S., Sasaki, S., Asano, Y., Ujikawa, O., Fukumoto, S., Nakashima, K., et al. (2017). Discovery of an orally bioavailable, brain-penetrating, in vivo active phosphodiesterase 2A inhibitor lead series for the treatment of cognitive disorders. J. Med. Chem. 60, 7658–7676. doi: 10.1021/acs.jmedchem.7b00709
Miyashita, H., Mori, S., Motegi, K., Fukumoto, M., and Uchida, T. (2003). Pin1 is overexpressed in oral squamous cell carcinoma and its levels correlate with cyclin D1 overexpression. Oncol. Rep. 10, 455–461. doi: 10.3892/or.10.2.455
Mori, T., Hidaka, M., Lin, Y.-C., Yoshizawa, I., Okabe, T., Egashira, S., et al. (2011). A dual inhibitor against prolyl isomerase Pin1 and cyclophilin discovered by a novel real-time fluorescence detection method. Biochem. Biophys. Res. Commun. 406, 439–443. doi: 10.1016/j.bbrc.2011.02.066
Mouradov, D., Sloggett, C., Jorissen, R. N., Love, C. G., Li, S., Burgess, A. W., et al. (2014). Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 74, 3238–3247. doi: 10.1158/0008-5472.CAN-14-0013
Namanja, A. T., Wang, X. J., Xu, B., Mercedes-Camacho, A. Y., Wilson, K. A., Etzkorn, F. A., et al. (2011). Stereospecific gating of functional motions in Pin1. Proc. Natl. Acad. Sci. U.S.A. 108, 12289–12294. doi: 10.1073/pnas.1019382108
Nikolaev, S. I., Rimoldi, D., Iseli, C., Valsesia, A., Robyr, D., Gehrig, C., et al. (2011). Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat. Genet. 44, 133–139. doi: 10.1038/ng.1026
Phan, R. T., Saito, M., Kitagawa, Y., Means, A. R., and Dalla-Favera, R. (2007). Genotoxic stress regulates expression of the proto-oncogene Bcl6 in germinal center B cells. Nat. Immunol. 8, 1132–1139. doi: 10.1038/ni1508
Pickering, C. R., Zhou, J. H., Lee, J. J., Drummond, J. A., Peng, S. A., Saade, R. E., et al. (2014). Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin. Cancer Res. 20, 6582–6592. doi: 10.1158/1078-0432.CCR-14-1768
Potter, A. J., Ray, S., Gueritz, L., Nunns, C. L., Bryant, C. J., Scrace, S. F., et al. (2010). Structure-guided design of α-amino acid-derived Pin1 inhibitors. Bioorg. Med. Chem. Lett. 20, 586–590. doi: 10.1016/j.bmcl.2009.11.090
Potter, A., Oldfield, V., Nunns, C., Fromont, C., Ray, S., Northfield, C. J., et al. (2010). Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution. Bioorg. Med. Chem. Lett. 20, 6483–6488. doi: 10.1016/j.bmcl.2010.09.063
Ranganathan, R., Lu, K. P., Hunter, T., and Noel, J. P. (1997). Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell 89, 875–886. doi: 10.1016/S0092-8674(00)80273-1
Rangasamy, V., Mishra, R., Sondarva, G., Das, S., Lee, T. H., Bakowska, J. C., et al. (2012). Mixed-lineage kinase 3 phosphorylates prolyl-isomerase Pin1 to regulate its nuclear translocation and cellular function. Proc. Natl. Acad. Sci. U.S.A. 109, 8149–8154. doi: 10.1073/pnas.1200804109
Rizzolio, F., Caligiuri, I., Lucchetti, C., Fratamico, R., Tomei, V., Gallo, G., et al. (2013). Dissecting Pin1 and phospho-pRb regulation. J. Cell. Physiol. 228, 73–77. doi: 10.1002/jcp.24107
Rizzolio, F., Lucchetti, C., Caligiuri, I., Marchesi, I., Caputo, M., Klein-Szanto, A. J., et al. (2012). Retinoblastoma tumor-suppressor protein phosphorylation and inactivation depend on direct interaction with Pin1. Cell Death. Differ. 19, 1152–1161. doi: 10.1038/cdd.2011.202
Robinson, D. R., Wu, Y.-M., Vats, P., Su, F., Lonigro, R. J., Cao, X., et al. (2013). Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 45, 1446–1451. doi: 10.1038/ng.2823
Rudin, C. M., Durinck, S., Stawiski, E. W., Poirier, J. T., Modrusan, Z., Shames, D. S., et al. (2012). Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 44, 1111–1116. doi: 10.1038/ng.2405
Russo Spena, C., De Stefano, L., Palazzolo, S., Salis, B., Granchi, C., Minutolo, F., et al. (2018). Liposomal delivery of a Pin1 inhibitor complexed with cyclodextrins as new therapy for high-grade serous ovarian cancer. J. Control. Release 281, 1–10. doi: 10.1016/j.jconrel.2018.04.055
Rustighi, A., Tiberi, L., Soldano, A., Napoli, M., Nuciforo, P., Rosato, A., et al. (2009). The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat. Cell Biol. 11, 133–142. doi: 10.1038/ncb1822
Ryo, A., Liou, Y. C., Wulf, G., Nakamura, M., Lee, S. W., and Lu, K. P. (2002). PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol. Cell. Biol. 22, 5281–5295. doi: 10.1128/MCB.22.15.5281-5295.2002
Ryo, A., Nakamura, M., Wulf, G., Liou, Y. C., and Lu, K. P. (2001). Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat. Cell Biol. 3, 793–801. doi: 10.1038/ncb0901-793ncb0901-793
Ryo, A., Suizu, F., Yoshida, Y., Perrem, K., Liou, Y.-C., Wulf, G., et al. (2003). Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol. Cell 12, 1413–1426. doi: 10.1016/S1097-2765(03)00490-8
Schulze, K., Imbeaud, S., Letouzé, E., Alexandrov, L. B., Calderaro, J., Rebouissou, S., et al. (2015). Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 47, 505–511. doi: 10.1038/ng.3252
Segat, L., Milanese, M., and Crovella, S. (2007). Pin1 promoter polymorphisms in hepatocellular carcinoma patients. Gastroenterology 132, 2618–9; authorrely 2619–20. doi: 10.1053/j.gastro.2007.04.037
Shain, A. H., Garrido, M., Botton, T., Talevich, E., Yeh, I., Sanborn, J. Z., et al. (2015). Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat. Genet. 47, 1194–1199. doi: 10.1038/ng.3382
Subedi, A., Shimizu, T., Ryo, A., Sanada, E., Watanabe, N., and Osada, H. (2016). Discovery of novel selenium derivatives as Pin1 inhibitors by high-throughput screening. Biochem. Biophys. Res. Commun. 474, 528–533. doi: 10.1016/j.bbrc.2016.04.124
Tahara, T., Yamamoto, E., Madireddi, P., Suzuki, H., Maruyama, R., Chung, W., et al. (2014). Colorectal carcinomas with CpG island methylator phenotype 1 frequently contain mutations in chromatin regulators. Gastroenterology 146, 530.e–538.e. doi: 10.1053/j.gastro.2013.10.060
The Cancer Genome Atlas [TCGA] (2012). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. doi: 10.1038/nature11252
Wei, S., Kozono, S., Kats, L., Nechama, M., Li, W., Guarnerio, J., et al. (2015). Active Pin1 is a key target of all-trans retinoic acid in acute promyelocytic leukemia and breast cancer. Nat. Med. 21, 457–466. doi: 10.1038/nm.3839
Wei, X., Walia, V., Lin, J. C., Teer, J. K., Prickett, T. D., Gartner, J., et al. (2011). Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat. Genet. 43, 442–446. doi: 10.1038/ng.810
Wilson, K. A., Bouchard, J. J., and Peng, J. W. (2013). Interdomain interactions support interdomain communication in human Pin1. Biochemistry 52, 6968–6981. doi: 10.1021/bi401057x
Wulf, G., Garg, P., Liou, Y. C., Iglehart, D., and Lu, K. P. (2004). Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. 23, 3397–3407. doi: 10.1038/sj.emboj.76003237600323
Wulf, G. M., Ryo, A., Wulf, G. G., Lee, S. W., Niu, T., Petkova, V., et al. (2001). Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 20, 3459–3472. doi: 10.1093/emboj/20.13.3459
Yao, J., Wang, J.-M., Wang, Z.-L., and Wu, Y.-N. (2014). The Pin1 gene polymorphism and the susceptibility of oral squamous cell carcinoma in East China. Cancer Biomark. 14, 441–447. doi: 10.3233/CBM-140421
Yeh, E., Cunningham, M., Arnold, H., Chasse, D., Monteith, T., Ivaldi, G., et al. (2004). A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6, 308–318. doi: 10.1038/ncb1110
Yeh, E. S., Lew, B. O., and Means, A. R. (2006). The loss of PIN1 deregulates cyclin E and sensitizes mouse embryo fibroblasts to genomic instability. J. Biol. Chem. 281, 241–251. doi: 10.1074/jbc.M505770200
Yeh, E. S., and Means, A. R. (2007). PIN1, the cell cycle and cancer. Nat. Rev. Cancer 7, 381–388. doi: 10.1038/nrc2107
Yi, P., Wu, R. C., Sandquist, J., Wong, J., Tsai, S. Y., Tsai, M. J., et al. (2005). Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1). Mol. Cell. Biol. 25, 9687–9699. doi: 10.1128/MCB.25.21.9687-9699.2005
You, Y., Deng, J., Zheng, J., Jiang, L., Li, N., Wu, H., et al. (2013). Functional polymorphisms in PIN1 promoter and esophageal carcinoma susceptibility in Chinese population. Mol. Biol. Rep. 40, 829–838. doi: 10.1007/s11033-012-2122-x
Zhang, X., Zhang, B., Gao, J., Wang, X., and Liu, Z. (2013). Regulation of the microRNA 200b (miRNA-200b) by transcriptional regulators PEA3 and ELK-1 protein affects expression of Pin1 protein to control anoikis. J. Biol. Chem. 288, 32742–32752. doi: 10.1074/jbc.M113.478016
Zhang, Y., Daum, S., Wildemann, D., Zhou, X. Z., Verdecia, M. A., Bowman, M. E., et al. (2007). Structural basis for high-affinity peptide inhibition of human Pin1. ACS Chem. Biol. 2, 320–328. doi: 10.1021/cb7000044
Zhao, H., Cui, G., Jin, J., Chen, X., and Xu, B. (2016). Synthesis and Pin1 inhibitory activity of thiazole derivatives. Bioorg. Med. Chem. 24, 5911–5920. doi: 10.1016/j.bmc.2016.09.049
Keywords: PIN1, cancer, mutations, SNP, 3D modeling
Citation: El Boustani M, De Stefano L, Caligiuri I, Mouawad N, Granchi C, Canzonieri V, Tuccinardi T, Giordano A and Rizzolio F (2019) A Guide to PIN1 Function and Mutations Across Cancers. Front. Pharmacol. 9:1477. doi: 10.3389/fphar.2018.01477
Received: 30 October 2018; Accepted: 03 December 2018;
Published: 22 January 2019.
Edited by:
Filippo Caraci, Università degli Studi di Catania, ItalyReviewed by:
Fabrizio Carta, Università degli Studi di Firenze, ItalyCopyright © 2019 El Boustani, De Stefano, Caligiuri, Mouawad, Granchi, Canzonieri, Tuccinardi, Giordano and Rizzolio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Flavio Rizzolio, ZmxhdmlvLnJpenpvbGlvQHVuaXZlLml0
†Co-first authors
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.