- 1Center for Perinatal Research, The Research Institute at Nationwide Children's Hospital, Columbus, OH, United States
- 2College of Pharmacy, The Ohio State University, Columbus, OH, United States
Neural circuits are the underlying functional units of the human brain that govern complex behavior and higher-order cognitive processes. Disruptions in neural circuit development have been implicated in the pathogenesis of multiple neurodevelopmental disorders such as autism spectrum disorder (ASD), attention deficit hyperactivity disorder (ADHD), and schizophrenia. Until recently, major efforts utilizing neurological disease modeling platforms based on human induced pluripotent stem cells (hiPSCs), investigated disease phenotypes primarily at the single cell level. However, recent advances in brain organoid systems, microfluidic devices, and advanced optical and electrical interfaces, now allow more complex hiPSC-based systems to model neuronal connectivity and investigate the specific brain circuitry implicated in neurodevelopmental disorders. Here we review emerging research advances in studying brain circuitry using in vitro and in vivo disease modeling platforms including microfluidic devices, enhanced functional recording interfaces, and brain organoid systems. Research efforts in these areas have already yielded critical insights into pathophysiological mechanisms and will continue to stimulate innovation in this promising area of translational research.
Introduction
Neural circuits can be detected as early as 18–22 weeks gestational age (GA) during human brain development. Transient circuitries in the brain are initially established in the subplate, formed by afferent inputs from multiple brain regions, including the basal forebrain, brainstem, and somatosensory thalamus (1–4). Then through a process of synaptic refinement and dissipation of the subplate occurring between weeks 24 and 28 GA, more permanent connections are established within the cortical plate that include longer range thalamo-cortical and cortico-cortico circuits (1–4). During this early and vulnerable period, exposure to medications, drugs of abuse, or extrinsic factors such as maternal stress and infections has the potential to alter the establishment of neural circuitry. Post-natally, local circuits are fine-tuned and longer-range circuits such as cortico-striatal, meso-cortical, and cortico-hippocampal connections become increasingly organized and unified (4–7). The long-range neural circuits regulating motivation and reward, which have been implicated in drug addiction and mood disorders, are further refined at postnatal stages when neurons of the nucleus accumbens receive excitatory inputs from the prefrontal cortex (PFC), hippocampus, and amygdala as well as dopaminergic neuron inputs from the midbrain (8, 9). Neural circuitry dysfunction is also linked to neurodevelopmental disorders such as autism spectrum disorder (ASD) with deficits in cortico-cortico, cortico-striatal, and cortico-thalamic circuits (10, 11). In a mouse model of schizophrenia, genetic engineering of the human microdeletion 22q11.2 equivalent in mouse, resulted in alterations in functional connectivity between the hippocampus and the PFC (12–14). Modeling the specific brain circuits implicated in these disorders would allow for a basic understanding of circuit function and could provide mechanistic insight into how these circuits malfunction in the disease state.
Apart from understanding how neural circuits malfunction in disease, studying neural circuits also provides multiple benefits from a basic biological perspective. For example, with the development of sophisticated optogenetic approaches, it is possible to functionally map both short and long-range cortical connections at single neuron resolution within the mouse brain, or map microcircuit connections in vitro (15). In addition, understanding how neuronal circuits process information can be applied to the development of more advanced silicon-based electronic circuits. For example, by studying how neurons process information within the neocortex, an electronic circuit was conceptualized and designed that models a phenomenon called gain modulation, which is a non-linear method by which neurons process information from multiple sources (15).
Due to the experimental challenges associated with research on post-mortem human brain tissue and the inability of neuroimaging to provide cellular-level mechanistic insights, our current understanding of neural circuit function has largely been deduced from animal models. Recently, more physiologically relevant and tractable model systems based on human induced pluripotent stem cells (hiPSCs) are emerging to complement these animal models. Since hiPSCs were first generated (16–21), patient-specific stem cell lines have now been produced for a wide array of neurodevelopmental disorders such as ASD, schizophrenia, epilepsy, and ADHD (16, 17, 22–24). Lately, more complex brain organoid systems derived from hiPSCs have been developed, providing a more realistic three-dimensional model of human brain development with significant potential for modeling neural circuitry dysfunctions in neurodevelopmental disorders (25–33). In parallel, the development of microfluidic brain-on-a-chip devices that model defective neural circuits in disease may facilitate the investigation of pathogenic mechanisms (34–36). In this review, we discuss recent advances in the development of platforms using hiPSCs to model human brain circuitry, their advantages and challenges, as well as the use of microfluidic devices and other technological approaches to enhance their application in basic and translational research.
Models for Reconstructing Human Brain Circuitry
The field of in vitro disease modeling has significantly accelerated following the generation of hiPSCs in 2007. The ability to harness patient-specific somatic cells and reprogram them into pluripotent stem cells indistinguishable from human embryonic stem cells (hESCs) has opened a novel area of research for modeling neurodevelopmental disorders. Development of robust hiPSC reprogramming methodologies has been an area of intense research with more than a dozen methods currently available (22). Although, the choice of reprogramming methodology ultimately depends on the research objectives, protocols that are rapid, low cost, and limit genomic integrations are the preferred methods of choice when deriving hiPSCs for disease modeling applications. A highly-efficient and RNA-based hiPSC reprogramming method utilizing primary neonatal fibroblasts recently achieved an 800% reprogramming efficiency, which is equivalent to 8 hiPSC colonies generated per fibroblast cell (37). Other methods allow simultaneous reprogramming and gene editing (to generate control isogenic hiPSC lines) from either fibroblasts or erythroblasts (38, 39). With improved methods of reprogramming and gene editing, hiPSC-based disease model platforms are increasingly being adopted.
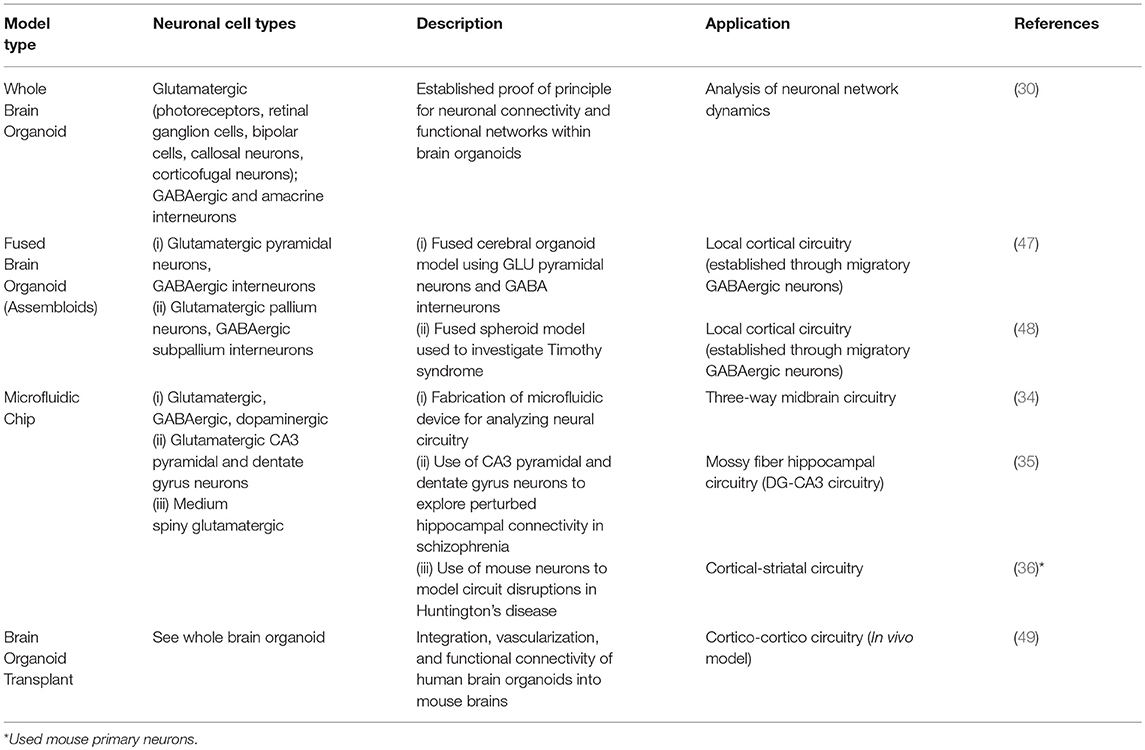
Multiple hiPSC lines from patients with neurodevelopmental disorders have been generated that manifest similar functional deficits in neuronal cultures to those seen in the patients (31–33, 40, 41). For example, hiPSC-derived cortical neurons generated from patients with Rett syndrome display altered neuronal networks and synaptic deficiencies (42). Similar phenotypes have been described in cortical neurons generated from Fragile X syndrome patients containing mutations within the FMR1 gene (43). A more comprehensive summary of neuronal functional deficits generated from hiPSC-based neurological disease models can be found in several recent reviews (24, 44, 45). Alternative non-hiPSC sources such as the use of organotypic explant cultures or dissociated neuronal cultures to model three-dimensional neural circuits on a chip are not the focus of this review and have also been recently described elsewhere (46).Here we focus instead on the recent technological innovations in developing platforms to model specific brain circuits associated with neurological disease utilizing hiPSCs. We will also highlight opportunities to couple emerging technologies thereby increasing their utility in disease and disorder modeling (Table 1).
Brain Organoid Systems
Organoids are three-dimensional in vitro structures derived either from hiPSCs or adult stem cells (32, 40, 41). They are generated as a result of the ability of stem cells to self-organize and develop to form complex structures with organ-like features. Brain organoids, or “cerebral organoids” are hiPSC-derived tissue structures that recapitulate the morphological features and developmental processes of the developing human brain. Two main methods are utilized to generate brain organoids: (i) the un-directed or whole-brain organoid protocol that is based on the self-patterning ability of stem cells to develop with minimal extrinsic differentiation cues, or (ii) the directed differentiation protocol, which guides hiPSCs toward defined brain lineages using small molecules and extrinsic factors. The latter protocol can generate organoids with structures resembling the hippocampus, midbrain, cerebellum, and other brain regions. In contrast, the un-directed protocol generates brain organoids with greater cellular diversity and regional brain identities, but at the expense of increased inter-organoid variability. Drop-seq single-cell mRNA sequencing studies have demonstrated whole brain organoids contain considerable cellular diversity (Table 1). Six-month-old developed whole brain organoids were shown to contain cells predominately belonging to the neuroectodermal lineage such as astrocytes, oligodendrocyte precursor cells, neural progenitor cells, dopaminergic neurons, cortical neurons such as GABAergic and glutamatergic neurons, interneurons, retinal cells, and mesodermal progenitor cells (30).
Despite significant advances in our understanding of the cellular diversity and morphological complexity of brain organoids, only a few studies have investigated their electrical properties and neuronal connectivity. By performing extracellular recordings with high-density silicon electrodes implanted into 8-month-old brain organoids, researchers were able to identify spontaneous action potentials and coordinated bursting activity, suggesting that the brain organoids had established functional neuronal networks (30). Others have analyzed the electrical properties of neurons within brain organoid slices using whole-cell patch clamp techniques (29, 48, 50). Recent studies have demonstrated nested oscillatory network activity emerges within cortical organoids initiating at 6 months in culture, highlighting the potential to model the development of neural networks (51). Nonetheless, no study to date has investigated whether brain organoids can be coaxed to form local or long-range neural circuits.
Developing in vitro platforms for high resolution probing and manipulation of brain circuits remains challenging. New methods have generated regionally-specified brain organoids through the ability of two or more organoids to spontaneously fuse together. For example, several methods have been developed for generating dorsal and ventral forebrain organoids or spheroids (48) using parallel directed differentiation protocols followed by spontaneous fusion of the organoids or spheroids to study interneuron migration (47, 48, 52) and functional activity (48, 52). Fused regionally specified brain organoids, named “assembloids,” may reconstruct multiple types of brain circuits that have been implicated in neurological diseases, such as cortico-striatal, cortical-spinal, meso-cortical, and cortico-hippocampal circuits (33). This technology could model brain circuitry dysfunctions across a large number of neurodevelopmental disorders by combining the differentiation capacity of hiPSCs to generate diverse neuronal subtypes with their ability to produce arrays of assembloids.
Beyond in vitro studies, a new in vivo model of human brain neuronal connectivity was recently developed by transplanting human brain organoids into mouse brains (49). In this study, human brain organoids became vascularized, were integrated with microglia, extended long-range axons, demonstrated synchronized neural activity, and showed functional connectivity within the mouse brain. The ability of human brain organoids to integrate and generate functional neural circuits within the mouse brain opens up multiple opportunities for future exploration. One of the more compelling lines of research would be to investigate whether this transplantation method could restore neural circuit dysfunction in mouse models of neurodevelopmental disorders.
Disease-on-a-Chip Microfluidic Platforms
The development of compartmentalized microfluidic devices has further enabled innovative methods to model human brain circuitry with an unmatched level of control. Microfluidic chip devices provide several novel features as platforms to model circuit-level connectivity in vitro such as: (i) the ability to physically segregate cell bodies of different neural subtypes while still allowing axonal growth between compartments (ii) compatibility with high-resolution video microscopy to investigate morphological and functional connectivity (iii) the ability to control axonal growth and monitor stages from early synaptic formation through late synaptic maturation. Thus, microfluidic chip platforms provide compelling systems to investigate neural circuit dysfunction in neurodevelopmental disorders.
As a proof of concept, a five-compartment microfluidic device was developed to model a mid-brain neural circuit composed of GABAergic, glutamatergic, and dopaminergic (DA) neurons (34). Using distinct fluorescent tags to label neuronal subtypes, allowed the visualization of glutamatergic axons projecting to a centralized compartment and forming synaptic connections with DA neurons. In addition, calcium imaging techniques demonstrated functional connectivity between neuronal compartments while whole-cell recordings and optogenetic stimulation techniques revealed circuit function.
In another study, a microfluidic chip platform modeled the hippocampal dentate gyrus (hDG)-Cornu Ammonis region 3 (CA3) circuit of schizophrenic patients (35). An efficient differentiation protocol was used to generate hDG and pyramidal human CA3 neuronal subtypes using hiPSCs from schizophrenic (SZ) and healthy patients (35, 53). Utilizing a two-compartment, microfluidic device connected by narrow channels, pre-synaptic hDGs were seeded into one compartment and post-synaptic hCA3s were seeded into the opposing compartment. A rabies virus tracing assay confirmed direct synaptic connections were established within this microfluidic system. Multi-electrode array (MEA) recordings and whole-cell patch clamp techniques established that the SZ hCA3 neurons had defects in both spontaneous and evoked electrophysiological activity.
A similar two-compartment microfluidic device reconstructed cortico-striatal circuits using primary neurons from a mouse model of Huntington's disease (HD). Though the main focus of this review is based on hiPSCs, this innovative system was included to emphasize the value of microfluidic systems in modeling brain circuitry. Data from this study suggested that presynaptic cortical dysfunction was both necessary and sufficient to induce pathogenic properties within post-synaptic striatal neurons, thus highlighting the critical value of these systems in providing mechanistic insights into disease pathogenesis (36). Through the combination of sophisticated optical tools such as glutamate and calcium reporter systems, HD cortical neurons demonstrated decreased glutamate release and overall reduced, but hypersynchronized, bursting episodes. An extension of this study should determine whether HD hiPSC derived cortical and striatal neurons show similar pathological neural connectivity deficits. Other neurodevelopmental disorders manifesting hyper-connected circuits such as ASD and ADHD could also be modeled using this microfluidic system.
As a potential extension of these studies and as a means to further understand the impact of multiple organ system interactions on human brain circuits, the linking of multiple organoid tissue types within a single microfluidic chip would provide for greater mechanistic understanding of neural circuit dynamics within a context of increased resemblance to in vivo conditions (54, 55). For example, recent studies have highlighted the complex signaling interactions between the gastrointestinal tract and the brain mediated through the gut microbiota (56, 57). To model these interactions, the development of an organ-on-a-chip platform that interlinks gastrointestinal organoids with an assembloid-derived brain circuit separated by an endothelial cell layer to mimic the blood-brain barrier would represent a further improvement in modeling accurate physiological responses on brain circuits.
Current Challenges and Limitations
Despite the many advances, several challenges still exist to increase the physiological relevance of brain organoid systems and microfluidic platforms in modeling human brain circuitry. First, brain organoid cultures have variable and inconsistent internal morphological features, which limit their full potential. A recent study has shown that cellular diversity within brain organoids could vary within one-third of organoids of a given batch for a particular cell type such as forebrain neurons (30). Further, within an individual organoid, morphological structures such as ventricular zone-like and cortical plate-like structures can vary in size and organization. Recent improvements in bioreactor designs and organoid protocols have reduced this intra-organoid variability (29). Strategies that couple the use of microfluidics by precisely controlling media flow on brain organoids as they develop may minimize organoid variability. A recent study investigated whether a perfusable organ-on-a-chip system could be used to generate brain organoids (58). Although this novel system yielded organoids with enhanced neuronal organization, it is unknown whether this system could reduce organoid variability. Another source of variability stems from the use of different hiPSC lines, as they may contain significant genetic heterogeneity. In designing experimental studies, this issue can be circumvented by using isogenic or closely genetically matched hiPSC lines (59). Second, current protocols generate brain organoids that lack some of the brain cell types represented in the human brain such as microglia, endothelial cells, and oligodendrocytes, further decreasing their ability to model complex signaling events and neurodevelopmental processes. However, recent improvements to conventional protocols have yielded the generation of microglia within developing brain organoids (60), which are critical for the formation and maintenance of neural circuits (61). Thus, utilizing protocols that generate innate microglia within developing brain organoids would provide a model system with greater in vivo relevance to study neural circuits. Similarly, although current protocols generate brain organoids that lack functional blood vessels, it may be possible to bioengineer blood vessels within brain organoids or fuse them with blood vessel organoids based on recent advances in three-dimensional vascular organoid methods (62). Implementing this strategy may not only enhance nutrient diffusion throughout brain organoids, but would also provide relevant vascular cell types critical for maintaining neural circuit homeostasis through neurovascular signaling interactions (63–65).
Microfluidics approaches also have several intrinsic limitations in modeling human brain circuitry. Technical limitations include shear stress generated by microfluidic channels that can potentially harm cells or the microchannels can become easily trapped with cellular debris and air bubbles (66). In addition, the volumetric flow rate of microfluidic devices is considerably low throughput, thereby precluding the analysis of large sample volumes or assays that require rapid reaction kinetics such as exposing neurons to compounds that modulate their electrophysiological responses (66). In order to realize the full potential of organoid and microfluidics systems to model brain circuitry in neurological disease, future studies will need to overcome these obstacles.
Future Directions and Applications
In the last decade, hiPSC research has continuously advanced the elucidation of neurological disease mechanisms and the search for potential treatments. More recently, advances in disease modeling techniques have led to the introduction of organoid platforms, which provide a more realistic model of organ development and have the potential to be more predictive drug screening platforms. In particular, brain organoids recapitulate many of the morphological features of the developing human brain. Despite significant advances in our understanding of the cellular diversity and morphological complexity of brain organoids, only a few studies have examined their electrical properties, neural oscillations or neural networks. With the development of new technologies to record extracellular field potentials from multiple neurons with high spatial and temporal resolution using multi-electrode arrays (MEAs), assays could be developed to record the electrophysiological properties of brain organoids in the context of normal development or exposure to environmental factors. Further, MEAs could be utilized as functional readout tools to phenotypically profile abnormal electrical activity arising from diseased brain organoids, thereby allowing for drug screening approaches to identify potential therapies that rescue electrophysiological deficits. Additional applications for MEAs include analysis of network topological properties and functional connectivity graphs within brain organoids based on spike train data, which could be derived using information-theory based network analysis (67–69). Additionally, the coupling of MEAs with optogenetics techniques would allow a greater level of analysis of neural circuit function by specifically controlling the activity of inhibitory or excitatory neuron subtypes. To further probe the function of neural circuits and also provide a phenotypic profiling tool in monitoring brain organoid neural activity, genetically encoded calcium or voltage indicators could also be integrated allowing for multi-level readouts of functional activity.
In conjunction with neural circuit analysis within single brain organoids, regionally diverse brain organoids could be combined into assembloids to model long-range circuits such as those implicated in neurodevelopmental disorders and drug addiction. The same optical and electrical interface tools could then also be applied to these long-range circuit models. To further enhance the fidelity and reproducibility of assembloid-based platforms in modeling long-range circuits, microfluidic devices could segregate organoids into different compartments, while still allowing inter-compartmental projections and synaptic contacts between the separated organoids (Figure 1). Thus, combining the advantages of spatiotemporal control provided by a microfluidic device together with brain organoid cultures would allow a greater level of control and predictability in modeling brain circuitry dysfunction in neurodevelopmental disorders.
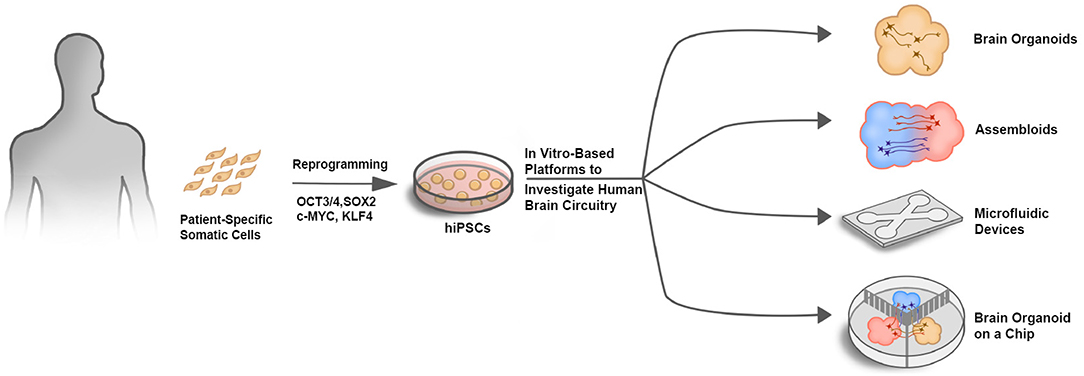
Figure 1. In vitro-based platforms to investigate human brain circuitry. Schematic illustrating different approaches to investigate human brain circuitry in vitro. Patient-specific somatic cells such as skin fibroblasts or peripheral blood mononuclear cells (PBMCs) that are easily isolated can be reprogrammed into hiPSCs using the Yamanka factors (OCT3/4, SOX2, c-MYC, and KLF4). hiPSCs can then be used as a source for generating multiple cell types in various platforms to investigate human brain circuitry.
Finally, when considering the clinical translational potential of these in vitro platforms for modeling human brain circuitry, it is critical to remember the developmental context of many neurological disorders. Some neurodevelopmental disorders may be triggered early, potentially resulting from environmental exposures in combination with a susceptible genetic makeup; common examples are cases of parenteral heavy metal (e.g., mercury, lead, manganese) (70) or intrauterine drug (e.g., opioids, antiepileptics) exposures at key phases of neurodevelopment (71–78). Other disorders, such as cerebral palsy, start with a discrete lesion early in development and express varied developmental phenotypes, with neuroplasticity occurring in response to use-driven rewiring of circuits (79–81) or the introduction of neuroprotective agents (e.g., erythropoietin, caffeine) (82–84). New model systems accounting for the complexity of chemical, physiological, neuroelectrical, and genetic interactions with developmental timelines relevant to human processes will be invaluable in achieving true progress in clinically relevant neuroscience. Regardless of the many challenges that lie ahead for developing robust hiPSC-based models of brain circuitry, current systems have already provided key mechanistic insights into many neurological diseases, which is the essential first step toward developing efficacious therapies.
Author Contributions
AH and MH conceived the main ideas and developed the structure of the paper. AH, MH, NM, and CM wrote the manuscript. AH prepared the figure and the table. All authors critically reviewed and approved the final manuscript for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling Editor declared a past co-authorship with one of the authors MH.
Acknowledgments
We are grateful to Arelis Berrios Hester for critically reading and providing insights and edits to this manuscript. We also thank Dr. Rodney Britt and members of the Buhimschi laboratory for providing helpful feedback and support. This work was supported by Internal Research Funds from The Research Institute at Nationwide Children's Hospital.
References
1. McConnell SK, Ghosh A, Shatz CJ. Subplate neurons pioneer the first axon pathway from the cerebral cortex. Science. (1989) 245:978–82. doi: 10.1126/science.2475909
2. McConnell SK, Ghosh A, Shatz CJ. Subplate pioneers and the formation of descending connections from cerebral cortex. J Neurosci. (1994) 14:1892–907. doi: 10.1523/JNEUROSCI.14-04-01892.1994
3. Supèr H, Soriano E, Uylings HB. The functions of the preplate in development and evolution of the neocortex and hippocampus. Brain Res Brain Res Rev. (1998) 27:40–64. doi: 10.1016/S0165-0173(98)00005-8
4. Tau GZ, Peterson BS. Normal development of brain circuits. Neuropsychopharmacology. (2010) 35:147–68. doi: 10.1038/npp.2009.115
5. Huisman TA, Martin E, Kubik-Huch R, Marincek B. Fetal magnetic resonance imaging of the brain: technical considerations and normal brain development. Eur Radiol. (2002) 12:1941–51. doi: 10.1007/s00330-001-1209-x
6. Huisman TA, Wisser J, Martin E, Kubik-Huch R, Marincek B. Fetal magnetic resonance imaging of the central nervous system: a pictorial essay. Eur Radiol. (2002) 12:1952–61. doi: 10.1007/s00330-001-1281-2
7. Kostović I, Judas M, Rados M, Hrabac P. Laminar organization of the human fetal cerebrum revealed by histochemical markers and magnetic resonance imaging. Cereb Cortex. (2002) 12:536–44. doi: 10.1093/cercor/12.5.536
8. Kauer JA, Malenka RC. Synaptic plasticity and addiction. Nat Rev Neurosci. (2007) 8:844–58. doi: 10.1038/nrn2234
9. Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nat Rev Neurosci. (2013) 14:609–25. doi: 10.1038/nrn3381
10. Courchesne E. Brainstem, cerebellar and limbic neuroanatomical abnormalities in autism. Curr Opin Neurobiol. (1997) 7:269–78. doi: 10.1016/S0959-4388(97)80016-5
11. Nair A, Treiber JM, Shukla DK, Shih P, Müller RA. Impaired thalamocortical connectivity in autism spectrum disorder: a study of functional and anatomical connectivity. Brain. (2013) 136(Pt 6):1942–55. doi: 10.1093/brain/awt079
12. Lawrie SM, Buechel C, Whalley HC, Frith CD, Friston KJ, Johnstone EC. Reduced frontotemporal functional connectivity in schizophrenia associated with auditory hallucinations. Biol Psychiatry. (2002) 51:1008–11. doi: 10.1016/S0006-3223(02)01316-1
13. Meyer-Lindenberg AS, Olsen RK, Kohn PD, Brown T, Egan MF, Weinberger DR, et al. Regionally specific disturbance of dorsolateral prefrontal-hippocampal functional connectivity in schizophrenia. Arch Gen Psychiatry. (2005) 62:379–86. doi: 10.1001/archpsyc.62.4.379
14. Steel RM, Whalley HC, Miller P, Best JJ, Johnstone EC, Lawrie SM. Structural MRI of the brain in presumed carriers of genes for schizophrenia, their affected and unaffected siblings. J Neurol Neurosurg Psychiatry. (2002) 72:455–8. doi: 10.1136/jnnp.72.4.455
15. Hahnloser RH, Sarpeshkar R, Mahowald MA, Douglas RJ, Seung HS. Digital selection and analogue amplification coexist in a cortex-inspired silicon circuit. Nature. (2000) 405:947–51. doi: 10.1038/35016072
16. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. (2007) 131:861–72. doi: 10.1016/j.cell.2007.11.019
17. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. (2007) 318:1917–20. doi: 10.1126/science.1151526
18. Hester ME, Song S, Miranda CJ, Eagle A, Schwartz PH, Kaspar BK. Two factor reprogramming of human neural stem cells into pluripotency. PLoS ONE. (2009) 4:e7044. doi: 10.1371/journal.pone.0007044
19. Kim JB, Zaehres H, Araúzo-Bravo MJ, Schöler HR. Generation of induced pluripotent stem cells from neural stem cells. Nat Protoc. (2009) 4:1464–70. doi: 10.1038/nprot.2009.173
20. Deleidi M, Cooper O, Hargus G, Levy A, Isacson O. Oct4-induced reprogramming is required for adult brain neural stem cell differentiation into midbrain dopaminergic neurons. PLoS ONE. (2011) 6:e19926. doi: 10.1371/journal.pone.0019926
21. Tsai SY, Bouwman BA, Ang YS, Kim SJ, Lee DF, Lemischka IR, et al. Single transcription factor reprogramming of hair follicle dermal papilla cells to induced pluripotent stem cells. Stem Cells. (2011) 29:964–71. doi: 10.1002/stem.649
22. Omole AE, Fakoya OJ. Ten years of progress and promise of induced pluripotent stem cells: historical origins, characteristics, mechanisms, limitations, and potential applications. PeerJ. (2018) 6:e4370 doi: 10.7717/peerj.4370
23. Shen X, Yeung HT, Lai KO. Application of human-induced pluripotent stem cells (hiPSCs) to study synaptopathy of neurodevelopmental disorders. Dev Neurobiol. (2018) 79:20–35. doi: 10.1002/dneu.22644
24. Vitrac A, Cloez-Tayarani I. Induced pluripotent stem cells as a tool to study brain circuits in autism-related disorders. Stem Cell Res Ther. (2018) 9:226. doi: 10.1186/s13287-018-0966-2
25. Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci USA. (2015) 112:15672–7. doi: 10.1073/pnas.1520760112
26. Hester ME, Snyder EY, Teng YD, Srivastava AK. (Ed). Generation of cerebral organoids derived from human pluripotent stem cells. In: Stem Cell Technologies in Neuroscience. New York, NY: Neuromethods: Springer Protocols; Human Press (2017). p. 123–34.
27. Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. (2014) 9:2329–40. doi: 10.1038/nprot.2014.158
28. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. (2013) 501:373–9. doi: 10.1038/nature12517
29. Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. (2016) 165:1238–54. doi: 10.1016/j.cell.2016.04.032
30. Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. (2017) 545:48–53. doi: 10.1038/nature22047
31. Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O. Engineering stem cell organoids. Cell Stem Cell. (2016) 18:25–38. doi: 10.1016/j.stem.2015.12.005
32. Di Lullo E, Kriegstein AR. The use of brain organoids to investigate neural development and disease. Nat Rev Neurosci. (2017) 18:573–84. doi: 10.1038/nrn.2017.107
33. Pasca SP. The rise of three-dimensional human brain cultures. Nature. (2018) 553:437–45. doi: 10.1038/nature25032
34. Fantuzzo JA, De Filippis L, McGowan H, Yang N, Ng YH, Halikere A, et al. muNeurocircuitry: establishing in vitro models of neurocircuits with human neurons. Technology. (2017) 5:87–97. doi: 10.1142/S2339547817500054
35. Sarkar A, Mei A, Paquola ACM, Stern S, Bardy C, Klug JR, et al. Efficient generation of CA3 neurons from human pluripotent stem cells enables modeling of hippocampal connectivity in vitro. Cell Stem Cell. (2018) 22:684–697 e9. doi: 10.1016/j.stem.2018.04.009
36. Virlogeux A, Moutaux E, Christaller W, Genoux A, Bruyère J, Fino E, et al. Reconstituting corticostriatal network on-a-chip reveals the contribution of the presynaptic compartment to Huntington's disease. Cell Rep. (2018) 22:110–22. doi: 10.1016/j.celrep.2017.12.013
37. Kogut I, McCarthy SM, Pavlova M, Astling DP, Chen X, Jakimenko A, et al. High-efficiency RNA-based reprogramming of human primary fibroblasts. Nat Commun. (2018) 9:745. doi: 10.1038/s41467-018-03190-3
38. Howden SE, Thomson JA, Little MH. Simultaneous reprogramming and gene editing of human fibroblasts. Nat Protoc. (2018) 13:875–98. doi: 10.1038/nprot.2018.007
39. Melo US, de Souza Leite F, Costa S, Rosenberg C, Zatz M. A fast method to reprogram and CRISPR/Cas9 gene editing from erythroblasts. Stem Cell Res. (2018) 31:52–4. doi: 10.1016/j.scr.2018.07.002
40. Dutta D, Heo I, Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med. (2017) 23:393–410. doi: 10.1016/j.molmed.2017.02.007
41. Shi Y, Inoue H, Wu JC, Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov. (2017) 16:115–30. doi: 10.1038/nrd.2016.245
42. Nageshappa S, Carromeu C, Trujillo CA, Mesci P, Espuny-Camacho I, Pasciuto E, et al. Altered neuronal network and rescue in a human MECP2 duplication model. Mol Psychiatry. (2016) 21:178–88. doi: 10.1038/mp.2015.128
43. Doers ME, Musser MT, Nichol R, Berndt ER, Baker M, Gomez TM, et al. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev. (2014) 23:1777–87. doi: 10.1089/scd.2014.0030
44. Chinchalongporn V, Koppensteiner P, Prè D, Thangnipon W, Bilo L, Arancio O. Connectivity and circuitry in a dish versus in a brain. Alzheimers Res Ther. (2015) 7:44. doi: 10.1186/s13195-015-0129-y
45. Weick JP. Functional properties of human stem cell-derived neurons in health and disease. Stem Cells Int. (2016) 2016:419043. doi: 10.1155/2016/4190438
46. Hasan MF, Berdichevsky Y. Neural circuits on a chip. Micromachines. (2016) 7:9. doi: 10.3390/mi7090157
47. Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Methods. (2017) 14:743–51. doi: 10.1038/nmeth.4304
48. Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, et al. Assembly of functionally integrated human forebrain spheroids. Nature. (2017) 545:54–9. doi: 10.1038/nature22330
49. Mansour AA, Gonçalves JT, Bloyd CW, Li H, Fernandes S, Quang D, et al. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol. (2018) 36:432–41. doi: 10.1038/nbt.4127
50. Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. (2015) 12:671–8. doi: 10.1038/nmeth.3415
51. Trujillo CA, Gao R, Negraes D, Chaim IA, Domissy A, Vandenberghe M, et al. Nested oscillatory dynamics in cortical organoids model early human brain network development. bioRxiv. (2018). doi: 10.1101/358622
52. Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, et al. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. (2017) 21:383–398 e7. doi: 10.1016/j.stem.2017.07.007
53. Yu DX, Di Giorgio FP, Yao J, Marchetto MC, Brennand K, Wright R, et al. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Reports. (2014) 2:295–310. doi: 10.1016/j.stemcr.2014.01.009
54. Edington CD, Chen WLK, Geishecker E, Kassis T, Soenksen LR, Bhushan BM, et al. Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci Rep. (2018) 8:4530. doi: 10.1038/s41598-018-22749-0
55. Truskey GA. Human microphysiological systems and organoids as in vitro models for toxicological studies. Front Public Health. (2018) 6:185. doi: 10.3389/fpubh.2018.00185
56. Chesnokova V, Pechnick RN. New signaling pathway for gut-brain interactions. Neuropsychopharmacology. (2016) 41:372–3. doi: 10.1038/npp.2015.237
57. Dinan TG, Cryan JF. Gut-brain axis in 2016: brain-gut-microbiota axis - mood, metabolism and behaviour. Nat Rev Gastroenterol Hepatol. (2017) 14:69–70. doi: 10.1038/nrgastro.2016.200
58. Wang Y, Wang L, Guo Y, Zhuac Y, Qin J. Engineering stem cell-derived 3D brain organoids in a perfusable organ-on-a-chip system. R Soc Chem. (2018) 8:1677. doi: 10.1039/C7RA11714K
59. Fossati V, Jain T, Sevilla A. The silver lining of induced pluripotent stem cell variation. Stem Cell Investig. (2016) 3:86. doi: 10.21037/sci.2016.11.16
60. Ormel PR, Vieira de Sá R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, et al. Microglia innately develop within cerebral organoids. Nat Commun. (2018) 9:4167. doi: 10.1038/s41467-018-06684-2
61. Wake H, Moorhouse AJ, Miyamoto A, Nabekura J. Microglia: actively surveying and shaping neuronal circuit structure and function. Trends Neurosci. (2013) 36:209–17. doi: 10.1016/j.tins.2012.11.007
62. Wimmer RA, Leopoldi A, Aichinger M, Wick N, Hantusch B, Novatchkova M, et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. (2019) 565:505–10. doi: 10.1038/s41586-018-0858-8
63. Li S, Kumar TP, Joshee S, Kirschstein T, Subburaju S, Khalili JS, et al. Endothelial cell-derived GABA signaling modulates neuronal migration and postnatal behavior. Cell Res. (2018) 28:221–48. doi: 10.1038/cr.2017.135
64. Tan C, Lu NN, Wang CK, Chen DY, Sun NH, Lyu H, et al. Endothelium-derived semaphorin 3G regulates hippocampal synaptic structure and plasticity via neuropilin-2/PlexinA4. Neuron. (2019) 101:1–18. doi: 10.1016/j.neuron.2018.12.036
65. Tillo M, Ruhrberg C, Mackenzie F. Emerging roles for semaphorins and VEGFs in synaptogenesis and synaptic plasticity. Cell Adh Migr. (2012) 6:541–6. doi: 10.4161/cam.22408
66. Huang Y, Williams JC, Johnson SM. Brain slice on a chip: opportunities and challenges of applying microfluidic technology to intact tissues. Lab Chip. (2012) 12:2103–17. doi: 10.1039/c2lc21142d
67. Pastore VP, Poli D, Martinoia S, Massobrio P. A new connectivity toolbox to infer topological features of in-vitro neural networks. Conf Proc IEEE Eng Med Biol Soc. (2015) 2015:2832–5. doi: 10.1109/EMBC.2015.7318981
68. Pastore VP, Poli D, Godjoski A, Martinoia S, Massobrio P. Toolconnect: a functional connectivity toolbox for in vitro networks. Front Neuroinform. (2016) 10:13. doi: 10.3389/fninf.2016.00013
69. Pastore VP, Godjoski A, Martinoia S, Massobrio P. SPICODYN: a toolbox for the analysis of neuronal network dynamics and connectivity from multi-site spike signal recordings. Neuroinformatics. (2018) 16:15–30. doi: 10.1007/s12021-017-9343-z
70. Maitre NL, Aschner JL, Aschner M. Manganese neurotoxicity may underlie the association between early life iron deficiency and impaired spatial cognition in neonatal piglets. J Nutr. (2013) 143:548. doi: 10.3945/jn.112.173377
71. Kern JK, Geier DA, Homme KG, King PG, Bjørklund G, Chirumbolo S, et al. Developmental neurotoxicants and the vulnerable male brain: a systematic review of suspected neurotoxicants that disproportionally affect males. Acta Neurobiol Exp. (2017) 77:269–6. doi: 10.21307/ane-2017-061
72. Sanders AP, Claus Henn B, Wright RO. Perinatal and childhood exposure to cadmium, manganese, and metal mixtures and effects on cognition and behavior: a review of recent literature. Curr Environ Health Rep. (2015) 2:284–94. doi: 10.1007/s40572-015-0058-8
73. Cattane N, Richetto J, Cattaneo A. Prenatal exposure to environmental insults and enhanced risk of developing schizophrenia and autism spectrum disorder: focus on biological pathways and epigenetic mechanisms. Neurosci Biobehav Rev. (2018). doi: 10.1016/j.neubiorev.2018.07.001
74. Bellinger DC. Prenatal exposures to environmental chemicals and children's neurodevelopment: an update. Saf Health Work. (2013) 4:1–11. doi: 10.5491/SHAW.2013.4.1.1
75. Keil KP, Lein PJ. DNA methylation: a mechanism linking environmental chemical exposures to risk of autism spectrum disorders? Environ Epigenet. (2016) 2:dvv012. doi: 10.1093/eep/dvv012
76. Rauh VA, Margolis AE. Research Review: Environmental exposures, neurodevelopment, and child mental health - new paradigms for the study of brain and behavioral effects. J Child Psychol Psychiatry. (2016) 57:775–93. doi: 10.1111/jcpp.12537
77. Kalkbrenner AE, Schmidt RJ, Penlesky AC. Environmental chemical exposures and autism spectrum disorders: a review of the epidemiological evidence. Curr Probl Pediatr Adolesc Health Care. (2014) 44:277–318. doi: 10.1016/j.cppeds.2014.06.001
78. Norman RE, Carpenter DO, Scott J, Brune MN, Sly PD. Environmental exposures: an underrecognized contribution to noncommunicable diseases. Rev Environ Health. (2013) 28:59–65. doi: 10.1515/reveh-2012-0033
79. Maitre NL, Key AP, Chorna OD, Slaughter JC, Matusz PJ, Wallace MT, et al. The dual nature of early-life experience on somatosensory processing in the human infant brain. Curr Biol. (2017) 27:1048–54. doi: 10.1016/j.cub.2017.02.036
80. Maitre NL, Henderson G, Gogliotti S, Pearson J, Simmons A, Wang L, et al. Feasibility of event-related potential methodology to evaluate changes in cortical processing after rehabilitation in children with cerebral palsy: a pilot study. J Clin Exp Neuropsychol. (2014) 36:669–79. doi: 10.1080/13803395.2014.925094
81. Maitre NL, Slaughter JC, Aschner JL, Key AP. Hemisphere differences in speech-sound event-related potentials in intensive care neonates: associations and predictive value for development in infancy. J Child Neurol. (2014) 29:903–11. doi: 10.1177/0883073813493502
82. Maitre NL, Chan J, Stark AR, Lambert WE, Aschner JL, Key AP. Effects of caffeine treatment for apnea of prematurity on cortical speech-sound differentiation in preterm infants. J Child Neurol. (2015) 30:307–13. doi: 10.1177/0883073814538500
83. Maitre NL, Stark AR. Neuroprotection for premature infants?: another perspective on caffeine. JAMA. (2012) 307:304–5. doi: 10.1001/jama.2011.2026
Keywords: human induced pluripotent stem cell (hiPSC), cerebral organoid, brain organoid, neurodevelopment, microfluidic, neural circuit
Citation: Hartlaub AM, McElroy CA, Maitre NL and Hester ME (2019) Modeling Human Brain Circuitry Using Pluripotent Stem Cell Platforms. Front. Pediatr. 7:57. doi: 10.3389/fped.2019.00057
Received: 06 December 2018; Accepted: 13 February 2019;
Published: 05 March 2019.
Edited by:
Sofia Beatriz Lizarraga, University of South Carolina, United StatesReviewed by:
Rohit Kumar Srivastava, Johns Hopkins University, United StatesThomas M. Durcan, Montreal Neurological Institute, Mcgill University, Canada
Copyright © 2019 Hartlaub, McElroy, Maitre and Hester. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark E. Hester, bWFyay5oZXN0ZXJAbmF0aW9ud2lkZWNoaWxkcmVucy5vcmc=
 Annalisa M. Hartlaub1
Annalisa M. Hartlaub1 Craig A. McElroy
Craig A. McElroy Mark E. Hester
Mark E. Hester