- 1Oral Microbiology Research Laboratory, Department of Bioclinical Sciences, Faculty of Dentistry, Health Sciences Center, Kuwait University, Kuwait, Kuwait
- 2Department of Surgical Sciences, Faculty of Dentistry, Health Sciences Center, Kuwait University, Kuwait, Kuwait
Recent studies have shown that antimicrobial treatment results in up- or down regulation of several virulence-associated genes in bacterial biofilms. The genes encoding NADH oxidase (nox) and fibronectin-binding protein (fbp) are known to play important roles in biofilm growth of some oral bacterial species. The objective was to study the effect of benzyl isothiocyanate (BITC), an antimicrobial agent from Miswak plant, on the expression of nox and fbp genes in some oral streptococci. The biofilms were treated with BITC and mRNA expression of nox and fbp genes was measured by comparative ΔΔCt method. The highest amount of biofilm mass was produced by A. defectiva, followed by S. gordonii, S. mutans, G. elegans and G. adiacens. Upon treatment with BITC, S. gordonii biofilms showed highest folds change in mRNA expression for both fbp and nox genes followed by S. mutans, A. defectiva, and G. adiacens. G. elegans mRNA levels for nox were extremely low. In conclusion, BITC treatment of the biofilms caused an upregulation of biofilm-associated genes fbp and nox genes in most of the tested species suggesting the significance of these genes in biofilm lifestyle of these oral bacteria and needs further investigation to understand if it contributes to antimicrobial resistance.
Introduction
Biofilms are a complex and functional community of microorganisms encased in a polymeric matrix and attached to one another or to a solid surface. A characteristic feature of microbial biofilms is the secretion of an adhesive matrix of highly hydrated extracellular polymeric substances (EPSs), comprised of polysaccharides, proteins, lipids, and nucleic acids [1]. Biofilms are structurally and physiologically complex entities. Within the biofilms, microorganisms coordinate and cooperate like multicellular organisms [2]. Ability of bacterial species to form biofilm is regarded a key virulence factor. Biofilm-associated bacterial infections in medicine have been extensively studied [3, 4].
Dental plaque develops naturally, but it is also associated with two of the most prevalent diseases affecting the people worldwide, which are caries and periodontal diseases [5]. Dental plaque is the diverse microbial community that develops on the tooth surface embedded in a matrix of polymers of bacterial and salivary origin [6]. Plaque formation starts once a tooth surface is cleaned and the early colonizers adhere to the acquired enamel pellicle, which is a thin layer of salivary glycoproteins deposited on tooth surface. Subsequently, secondary colonizers attach to the early colonizers through specific molecular interactions known as co-aggregation [6, 7]. These can involve protein-protein or carbohydrate-protein interactions, and this process determines the pattern of bacterial succession.
The oral cavity contains complex, multispecies microbial communities [8, 9]. This indicates that the residents in this community should exhibit extensive interactions while forming biofilm structures, carrying out physiological functions, and inducing microbial pathogenesis. Interspecies interactions in biofilms are competitive, cooperative and antagonistic [10]. In addition, such interactions may have specific effects in terms of the virulence properties of biofilm residents, which could influence the overall pathogenicity of biofilms.
Even though it is known that biofilms generally are more resistant to antibiotics, they can affect bacterial biofilms at different levels, including changes in the overall structure of the population, selection of resistant organisms, or alterations in bacterial physiology. Recent studies have shown that antibiotic treatment results in up- or down regulation of several genes [11]. However, little is known how antimicrobials, which have a general and broader mechanism of antibacterial action, influence the expression of biofilm-associated genes [12, 13]. The genes encoding NADH oxidase (nox) [14] and fibronectin-binding protein (fbp) [15] are known to play important roles in biofilm formation of some oral bacterial species. In this study, we aim to study the effect of benzyl isothiocyanate [16], an antimicrobial compound from Miswak plant, on the expression of nox and fbp genes in oral streptococci.
Methods
Bacterial Strains and Culture Conditions
Granulicatella elegans CCUG 38949, Granulicatella adiacens CCUG 27809, and Abiotrophia defectiva CCUG 27639 were cultured on chocolate blood agar with 0.001% pyridoxal HCl [17] for 2 days at 37°C and 5% CO2. in air. While Streptococcus mutans CCUG 11877 and Streptococcus gordonii CCUG 33482 were cultured on brucella blood agar containing 5% sheep blood and incubated as above. Prior to harvesting the growth for each experiment, purity of cultures was checked by observing the plates under stereo microscope.
In silico- and PCR-Confirmation of the Target Genes
Presence of the target genes encoding NADH oxidase and fibronectin binding protein was checked in the whole genome sequences of each species by using NCBI BLAST. Once confirmed, the two genes were PCR-amplified from each species. For PCR, primers were designed based on the obtained nucleotide sequences using the Bioinformatics Software Laser Gene Core Suite vs. 12 (Table 1). PCR reactions were performed on VeritiTM PCR machine (Applied Biosystems) using Ready-To-Go PCR beads (GE Life Sciences) and reaction conditions mentioned as earlier [17]. Agarose gel electrophoresis of the PCR amplicons was performed as described [17] and the amplicons were finally visualized on a G:Box Imaging system (SynGene).
Biofilm Culture and Quantification
Biofilm cultures were set up as described previously [17]. Briefly, from the agar-grown cultures, colonies were harvested, suspended in brucella broth and the cell suspensions adjusted to OD600 = 1. A 100-μl aliquot from each bacterial cell suspension was added separately into wells of a 24-well cell culture plate containing 900-μl brucella broth. For Granulicatella and Abiotrophia species, 0.001% pyridoxal was added. Wells containing only broth was served as a negative control. Biofilm cultures were incubated for 3 days in 5% CO2 atmosphere at 37°C.
Crystal Violet Staining of Biofilms
Following the incubation, broth supernatant was removed, and biofilms were washed twice gently with 1 ml sterile Phosphate Buffer Saline (PBS). One ml of crystal violet stain (2%) was applied to each well and incubated for 10 min. The stain was removed, and the wells were washed 6 times using PBS and airdried before taking pictures.
Confocal Microscopy
After growing biofilms as described above, 1 ml of 4% paraformaldehyde solution (pH = 7.4) was added to each well and incubated for 30 min at room temperature for fixation. Supernatant was removed and the wells were washed twice with 1 ml of sterile PBS. To each well, 500-μl of a mixture containing 1.5 ml PBS and 4.5 μl Syto-9 dye was added. The plate was incubated in dark for 15 min and then the supernatant was removed and washed with PBS. The plate was dried, and a drop of mounting oil was added to each well and covered with 24 × 50 mm coverslip. Biofilms were visualized by confocal laser scanning microscopy (LSM 700, Carl-Zeiss).
Effect of BITC on Biofilm Formation
The effect of BITC on the biofilm forming abilities of the bacterial species was studied by growing the biofilms in the presence of 10 μM BITC for 2 days under the same culture conditions as above. After incubation, 1 ml methanol was added to each well and the plate was kept undisturbed for 15 min at room temperature for fixing of the biofilms. Methanol was removed and the plate was air-dried for 45 min at room temperature. One ml of 0.1% aqueous crystal violet stain was added to each well and allowed to stay at room temperature for 20 min. Excess water was removed from the wells by tapping the plate upside down on a dry tissue paper. The plate was completely dried by keeping it open for 5–10 min in the fume hood. To each well, 500 μl 33% acetic acid was added and incubated at room temperature on a shaker for 5 min. One hundred microliter from each well was added into wells of a 96-well plate in triplicates and the absorbance was read at 590 nm using a microplate reader.
Treatment of Biofilms With BITC
Biofilms of each bacterial species were grown as described above and benzyl isothiocyanate (Sigma) was added at a concentration of 10 μM [16] prepared in DMSO and allowed for 2 h at the same incubation conditions as above. For negative control, biofilms were treated with DMSO alone. Following the incubation, broth supernatants were removed, and the biofilms were scrapped off and collected into sterile micro centrifuge tubes. Immediately, RNA later® (~10 volumes of the approximate weight of biofilms) was added and all samples were stored at −80°C until used.
RNA Isolation
For RNA isolation, the samples were taken out from the freezer and thawed on ice. After centrifuging at 5,000 × g for 5 min, the pellets were subjected to RNA isolation. Equal volume of RLT buffer (350 μl) and 70% ethanol (350 μl) were added and mixed gently with a pipet tip without vortexing. All the contents were transferred to mini spin columns and centrifuged at 8,000 × g for 15 s. The flow-through was discarded and the RNA bound to the membrane was washed with 700 μl wash buffers RW1. Five hundred microliter elution buffer RPE was added and centrifuged at 8,000 × g for 15 s and the flow-through was discarded. Another 500 μl elution buffer RPE was also added and centrifuged at 8,000 × g for 2 min. Finally, to elute the bounded RNA, 50 μl RNase-free water was added directly to the spin column membrane and centrifuged for 1 min at 8,000 × g. The concentration of RNA was determined by Nanodrop and the purity was assessed by A260/A280 ratio.
Reverse Transcription Real-Time PCR
From the purified RNA stored at −20°C, cDNA was synthesized by using a High-Capacity cDNA Reverse Transcription Kit (ABI Systems) according to manufacturer's instructions. Expression levels of all target genes were normalized using the 16S rRNA gene. The reactions were performed on ABI 7500 Fast Real-Time PCR machine. The reaction mixture was as follows: 10 μl SYBR Green master mix (Power SYBR Green® Kit, Applied Biosystems), 1 μl each of forward and reverse primer (0.2 μM), 7 μl H2O and 1 μl DNA template. Temperature profile was as follows: a 10-min initial denaturation at 95°C followed by 40 cycles of 95°C for 15 s, 50–60°C (depending on the primer pair) for 30 s and 72°C for 30 s. Primers and conditions used in real-time PCR for mRNA gene expression analysis are mentioned in Table 1. Data from fluorescent signal was acquired at the elongation step and analyzed using the software SDS 2.3.0v. Analysis of the expression of nox and fbp genes was performed on cells from untreated control and biofilm treated groups by comparative ΔΔCt method in ABI 7500 SDS system (Applied Biosystems). The amount of target, normalized to endogenous (16S rRNA gene) and relative to calibrator (untreated), by 2−ΔΔCt and the expression fold change was presented graphically.
Statistics
All experiments were repeated 3 times. Biological and technical replicates were included in each experiment. Differences between groups were compared by Mann Whitney U test. A P-value <0.05 was regarded significant. SPSS vs25 for windows was used for statistical analysis.
Results
PCR Optimization for the Target Genes nox and fbp
Prior to quantification of mRNA expression from nox and fbp genes, PCR conditions and the amplicon sizes were standardized. PCR products when analyzed on agarose gel electrophoresis showed single bands for all the five species. Expected sizes of the amplicons for the fbp gene visualized on the agarose gels were for G. elegans 127 bp, G. adiacens 118 bp, A. defectiva 137 bp, S. mutans 106 bp, and S. gordonii 102 bp. Similarly, expected sizes of the amplicons for the nox gene were for G. elegans 127 bp, G. adiacens 109 bp, A. defectiva 108 bp, S. mutans 127 bp, and S. gordonii 120 bp (Table 1). The absence of non-specific amplifications in all the tested species was evident from the single bands (Figure 1).
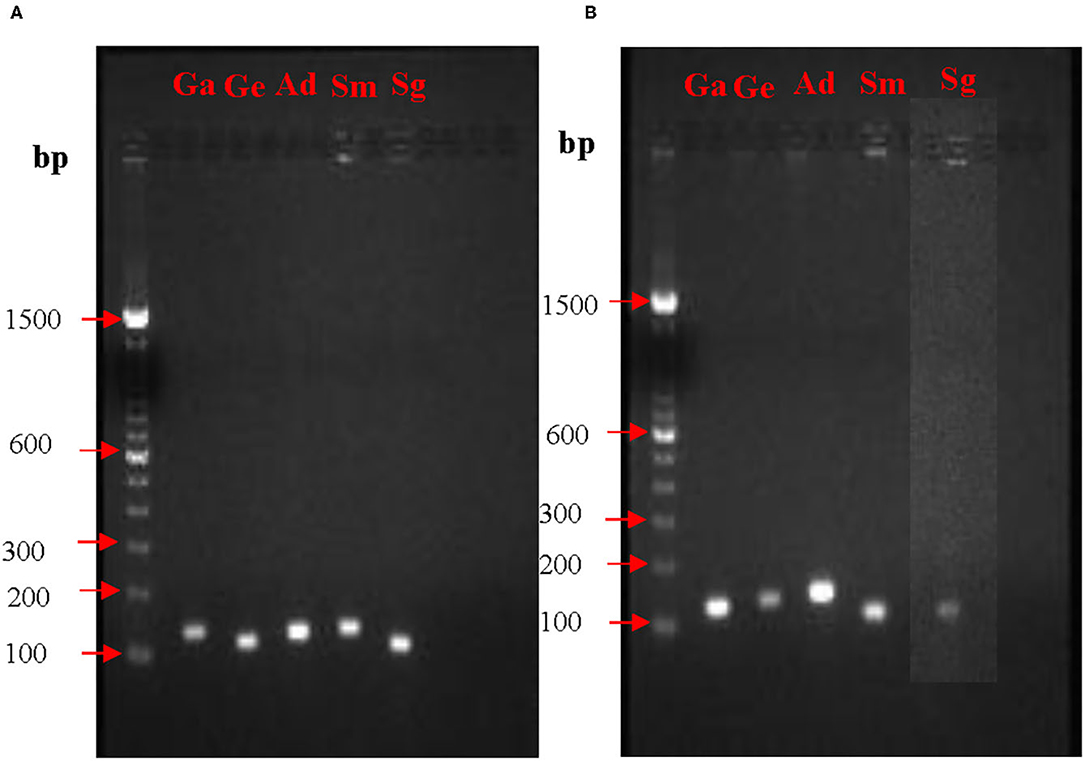
Figure 1. PCR optimization of nox (A) and fbp (B) genes. To confirm the amplification of nox and fbp genes from each strain, PCR and agarose gel electrophoresis were performed prior to RT-PCR experiments. bp, base pair; Ga, G. adiacens; Ge, G. elegans; Ad, A. defectiva; Sm, S. mutans; Sg, S. gordonii.
Biofilm Visualization and the Effect of BITC on Biofilm Formation Ability of the Bacterial Species
As evident from confocal microscopy and visual inspection of crystal violet-stained biofilm preparations (Figure 2), all five bacterial species formed biofilm after 3 days of incubation. G. adiacens and A. defectiva formed lowest and highest amounts of biofilm, respectively (Figure 2). Further, A. defectiva biofilm mass was significantly higher than that of all other species except S. gordonii. Interestingly, crystal violet staining results were different from Syto-9 staining. This discrepancy is probably because crystal violet mainly stains complete EPS in the biofilm matrix while Syto-9 is a nucleic acid stain. Further, when we tested the effect of BITC on biofilm formation of the species, the inhibitory effect was not evident on the biofilms (Figure 3). No significant difference was seen between the quantities of BITC-treated and -untreated biofilms.
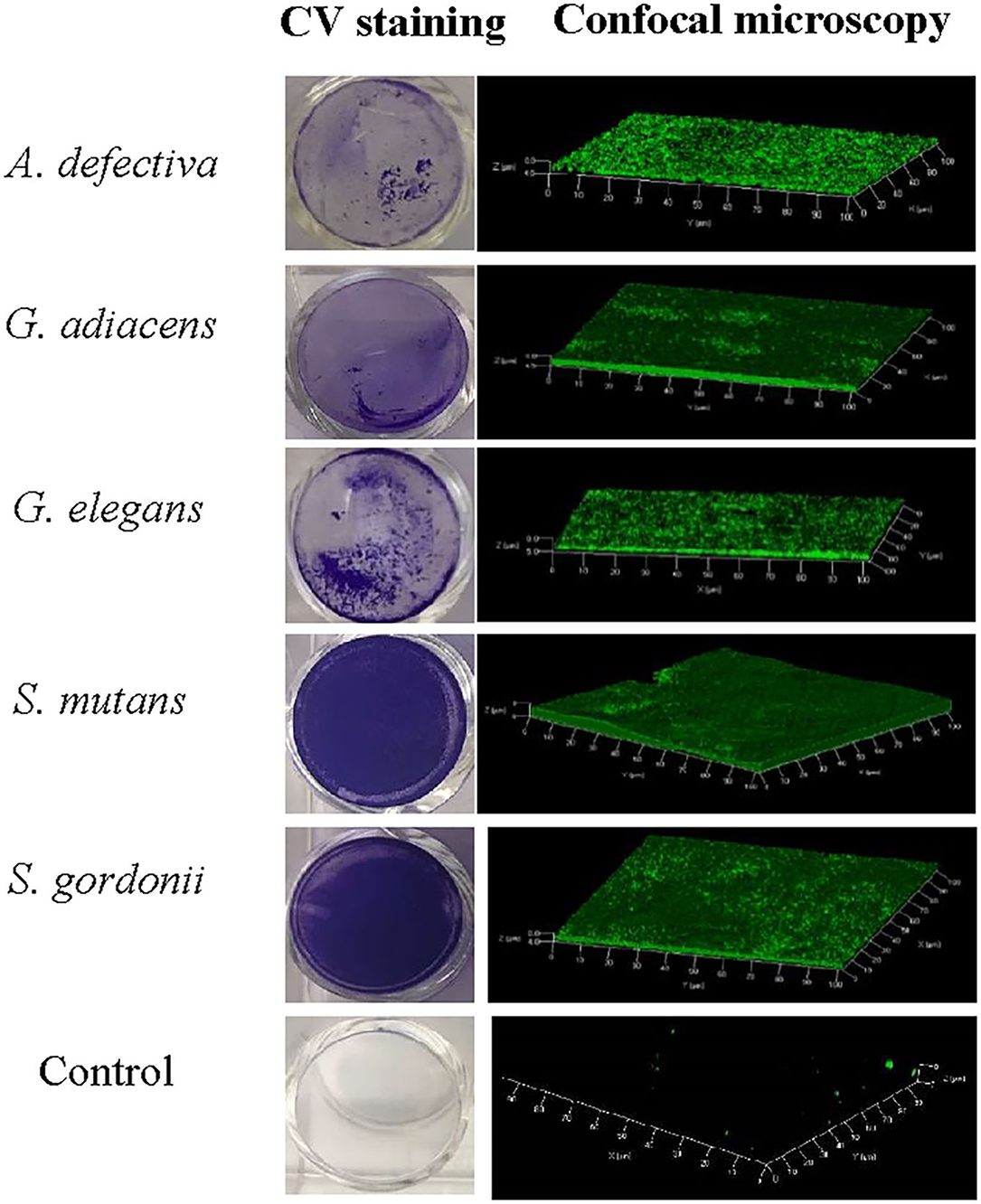
Figure 2. Confocal microscopy and crystal violet staining of monospecies biofilms of five oral streptococcal species. For confocal microscopy, bacteria were cultured in brucella broth in the wells of Millicell® EZ slides (Millipore) and stained with Syto9®. Images were acquired on Carl-Zeiss LSM 700 at 630× magnification and 3D view reconstructed using the software ZEN 2012. The bacteria were grown similarly for crystal violet staining except that 24-well cell culture plates (BD Bioscience) were used.
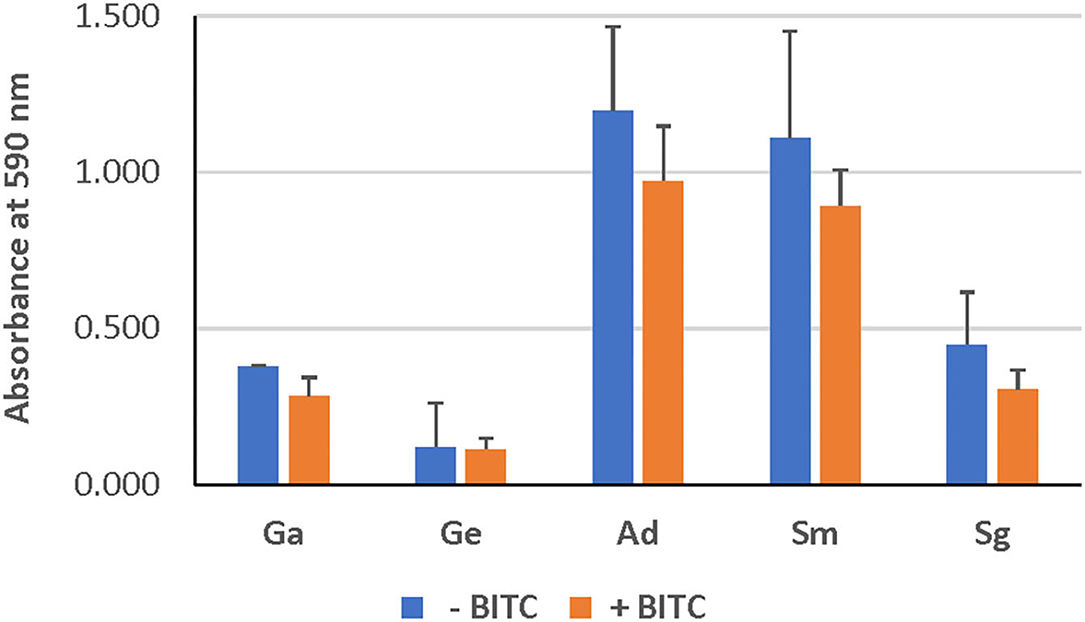
Figure 3. Effect of BITC treatment on the biofilms. The biofilm biomass was quantified by crystal violet staining after treating with BITC for 2 h in 5% CO2 at 37°C. The absorbance was measured at 590 nm wavelength. Ga, G. adiacens; Ge, G. elegans; Ad, A. defectiva; Sm, S. mutans; Sg, S. gordonii.
Effect of BITC Treatment on the Expression of Biofilm-Associated Genes
Following treatment of the biofilms with BITC, nox mRNA expression in A. defectiva, S. mutans, and S. gordonii increased by 1.03, 1.25, 1.98 folds, respectively, compared to the untreated control after normalization with the housekeeping gene 16S rRNA. However, in G. adiacens and G. elegans, the expression decreased by 0.73 and 0.06-fold, respectively (Figure 4A). The nox mRNA expression in S. gordonii and S. mutans was significantly higher than in G. adiacens and G. elegans (P < 0.05). The expression of fbp mRNA was 1.61, 1.57, 2.09, and 2.65-folds higher in G. elegans, S. mutans, A. defectiva, and S. gordonii, respectively, when compared to the untreated control.
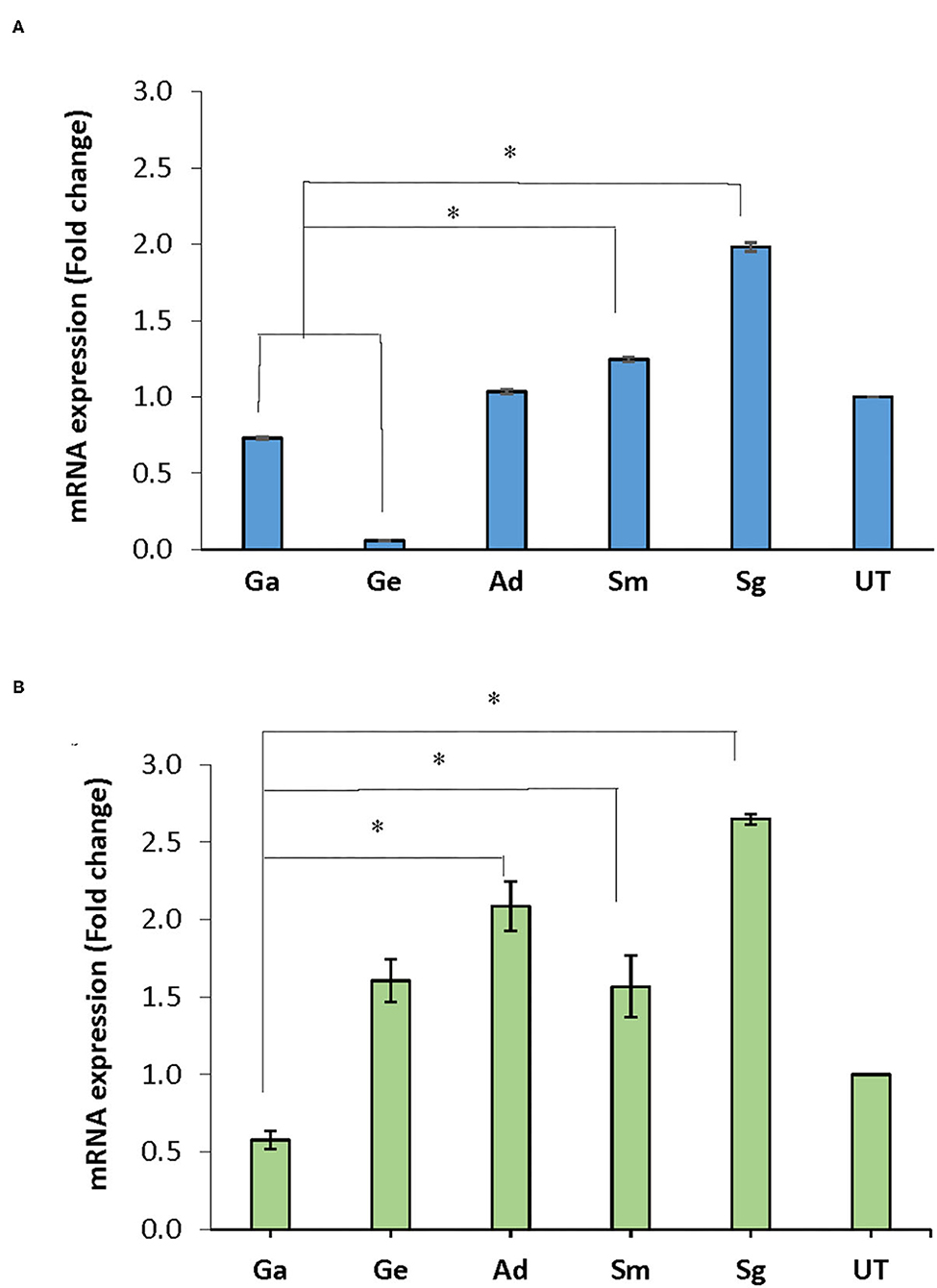
Figure 4. mRNA expression of nox and fbp genes. Mature biofilms of all strains were treated with BITC (10 μM) for 2 h in 5% CO2 at 37°C. The biofilms were scraped off from the wells, RNA later® was added to the biofilms and preserved at −80°C until used. RNA purified from the samples was converted to cDNA and expression of nox (A) and fbp (B) genes was assessed by Real-time PCR quantification. Ga, G. adiacens; Ge, G. elegans; Ad, A. defectiva; Sm, S. mutans; Sg, S. gordonii; UT, untreated. *P < 0.05.
Similar to nox, fbp expression decreased 0.58-folds in G. adiacens (Figure 4B). The fbp mRNA levels in A. defectiva, S. mutans, and S. gordonii were significantly (P < 0.05) higher than in G. adiacens.
Discussion
Dental plaque biofilms are the important etiologic factor in oral infectious diseases. Bacteria embedded in multilayered biofilms are difficult to kill because of the inherent resistance of the biofilm matrix toward antimicrobials. The present study demonstrated that the biofilms of three of the five bacterial species tested showed an increase in expression of the nox gene, which encodes for NADH oxidase and four bacterial species showed an increase of the fbp gene, encoding fibronectin binding protein following treatment with BITC. Altered gene expression due to antbiotic treatment of biofilms has been reported previously [11, 21, 22]. Further, like antibiotics, antimicrobials are also known to modify the expression of biofilm and virulence-associated genes in bacteria [12, 13, 23]. The natural antimicrobial BITC has been shown to exert potential health benefits on humans [22, 24] in addition to its antimicrobial activity [25, 26]. Recently, the expression differences of pathogenicity-related genes in Salmonella typhimurium upon BITC treatment was explored using transcriptome analysis and phenotypic validation like motility and biofilm formation characteristics. Differentially expressed genes including down- and upregulated were noticed in BITC treated group in comparison to the control group [27]. Similarly, downregulated expression of virulence factors was also observed in BITC treated Staphylococcus aureus experimental group [28].
We subjected all strains to the same concentration (10 μM) of BITC, which was chosen based on previous studies [16]. Even though our screening experiments showed that the inhibitory zones ranged 10–15 mm, no significant reduction in biofilm mass was seen at this concentration of BITC (Figure 3). Thus, we were prompted to study the expression of the biofilm-associated genes nox and fbp since the biofilm growth was not inhibited. Importantly, this eliminates the possibility that the reduced gene expression observed in some species in this study may be due to cell death.
We chose NADH oxidase (nox) and fibronectin (fbp) since their role in biofilm formation and virulence has been known in other bacterial species [14, 15]. In mutants lacking nox and fbp, biofilm formation was significantly decreased, which appeared to be due to reduced expression of a certain biofilm-associated genes [14]. In our study, when biofilms were treated with BITC, the overall structure of the biofilms was intact and no signs of biofilm detachment were seen upon visual inspection by stereo microscopy. While the expression of nox genes increased in S. gordonii, S. mutans and A. defectiva, a decreased expression was found in G. adiacens and G. elegans compared to the untreated control. In case of fbp genes, the increased expression was noticed in S. gordonii, S. mutans, A. defective and G. elegans while the expression was found to be decreased in G. adiacens alone compared to the untreated control. In the human oral cavity, bacteria need to cope with oxidative stress caused by diatomic oxygen as well as large amounts of H2O2 produced by competitor species. NADH oxidase is an important oxygen-metabolizing enzyme that plays a critical role in reducing dissolved oxygen in dental plaque [29]. Further, nox is also involved in the regulation of competence in some streptococci [30]. The increased expression of nox genes was reported in biofilm cells and under aerated and static aerobic conditions when compared to planktonic cells and anaerobic conditions [19]. Thus, our results suggest that S. gordonii, S. mutans and A. defectiva and G. elegans in which the gene expressions were increased, are possibly better equipped against antimicrobial assault. Increased expression of nox may indicate that these species produce higher amounts of NADH oxidase to combat an increased oxygen stress upon treatment with antimicrobials [31, 32]. Similarly, antibiotic treatment is also known to cause an increased expression of fbp genes in some bacterial species, enhancing adhesion efficacy of the bacterial cells [33, 34]. On the contrary, G. elegans and G. adiacens, which showed reduced expression of nox and G. adiacens of fbp upon BITC treatment, possibly are less capable of tailoring their gene expression upon treatment with antimicrobials.
A major limitation of this study is the lack of overall effect of BITC treatment on gene expression by microarray or transcriptome analysis, which may throw light on more interesting genes. Nevertheless, the two genes we studied, nox and fbp are known to be important for biofilm life in other species. In our ongoing study, we are aiming to show causality between upregulation of the expression of these two and several other genes upon antimicrobial treatment by constructing specific deletion mutants.
Conclusion
Biofilms of the studied oral bacteria were not significantly killed in the presence of BITC.
Increased expression of the biofilm-associated genes nox and fbp following BITC treatment suggests a possible biological significance of the genes in the biofilm life style of these species. Further investigation is needed to understand the role of nox and fbp genes when oral bacterial biofilms are exposed to BITC. Further, molecular mechanisms of species-specific regulatory effects of BITC treatment on the expression of the biofilm-associated genes may offer a greater insight.
Data Availability Statement
The raw data used in preparing this article will be made available by the authors, without undue reservation.
Author Contributions
HA: investigation, formal analysis, and original draft. EA: investigation, formal analysis, and original draft-review and editing. RB: methodology, data curation, and formal analysis. AA: project administration, resources, and original draft-review and editing. MK: conceptualization, funding acquisition, supervision, and original draft-review and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Kuwait University Grant SRUL 01/14.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Oral Microbiology Teaching and Research Laboratory (SRUL 01/14) for utilizing their resources.
References
1. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. (2010) 8:623–33. doi: 10.1038/nrmicro2415
2. Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. (2009) 33:206–24. doi: 10.1111/j.1574-6976.2008.00150.x
3. Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest. (2003) 112:1466–77. doi: 10.1172/JCI200320365
4. Sun F, Qu F, Ling Y, Mao P, Xia P, Chen H, et al. Biofilm-associated infections: antibiotic resistance and novel therapeutic strategies. Future Microbiol. (2013) 8:877–86. doi: 10.2217/fmb.13.58
5. Listl S, Galloway J, Mossey PA, Marcenes W. Global economic impact of dental diseases. J Dent Res. (2015) 94:1355–61. doi: 10.1177/0022034515602879
6. Kolenbrander PE, Palmer RJ, Jr., Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. (2010) 8:471–80. doi: 10.1038/nrmicro2381
7. Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. (2003) 11:94–100. doi: 10.1016/S0966-842X(02)00034-3
8. Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. (2005) 13:589–95. doi: 10.1016/j.tim.2005.09.006
9. Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. (2006) 42:80–7. doi: 10.1111/j.1600-0757.2006.00174.x
10. Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. (2016) 14:589–600. doi: 10.1038/nrmicro.2016.84
11. Redelman CV, Chakravarty S, Anderson GG. Antibiotic treatment of Pseudomonas aeruginosa biofilms stimulates expression of the magnesium transporter gene mgtE. Microbiology. (2014) 160(Pt 1):165–78. doi: 10.1099/mic.0.070144-0
12. Kot B, Sytykiewicz H, Sprawka I, Witeska M. Effect of manuka honey on biofilm-associated genes expression during methicillin-resistant Staphylococcus aureus biofilm formation. Sci Rep. (2020) 10:13552. doi: 10.1038/s41598-020-70666-y
13. Vaishampayan A, de Jong A, Wight DJ, Kok J, Grohmann E. A novel antimicrobial coating represses biofilm and virulence-related genes in methicillin-resistant Staphylococcus aureus. Front Microbiol. (2018) 9:221. doi: 10.3389/fmicb.2018.00221
14. Ge X, Shi X, Shi L, Liu J, Stone V, Kong F, et al. Involvement of NADH oxidase in biofilm formation in Streptococcus sanguinis. PLoS ONE. (2016) 11:e0151142. doi: 10.1371/journal.pone.0151142
15. McCourt J, O'Halloran DP, McCarthy H, O'Gara JP, Geoghegan JA. Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol Lett. (2014) 353:157–64. doi: 10.1111/1574-6968.12424
16. Sofrata A, Santangelo EM, Azeem M, Borg-Karlson AK, Gustafsson A, Putsep K. Benzyl isothiocyanate, a major component from the roots of Salvadora persica is highly active against Gram-negative bacteria. PLoS ONE. (2011) 6:e23045. doi: 10.1371/journal.pone.0023045
17. Karched M, Bhardwaj RG, Asikainen SE. Coaggregation and biofilm growth of Granulicatella spp. with Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans. BMC Microbiol. (2015) 15:114. doi: 10.1186/s12866-015-0439-z
18. Kajfasz JK, Rivera-Ramos I, Scott-Anne K, Gregoire S, Abranches J, Lemos JA. Transcription of oxidative stress genes is directly activated by SpxA1 and, to a lesser extent, by SpxA2 in Streptococcus mutans. J Bacteriol. (2015) 197:2160–70. doi: 10.1128/JB.00118-15
19. Loo CY, Mitrakul K, Jaafar S, Gyurko C, Hughes CV, Ganeshkumar N. Role of a nosX homolog in Streptococcus gordonii in aerobic growth and biofilm formation. J Bacteriol. (2004) 186:8193–206. doi: 10.1128/JB.186.24.8193-8206.2004
20. Maeda H, Fujimoto C, Haruki Y, Maeda T, Kokeguchi S, Petelin M, et al. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol Med Microbiol. (2003) 39:81–6. doi: 10.1016/S0928-8244(03)00224-4
21. Anderson GG, Moreau-Marquis S, Stanton BA, O'Toole GA. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect Immun. (2008) 76:1423–33. doi: 10.1128/IAI.01373-07
22. Ranjan A, Ramachandran S, Gupta N, Kaushik I, Wright S, Srivastava S, et al. Role of phytochemicals in cancer prevention. Int J Mol Sci. (2019) 20:4981. doi: 10.3390/ijms20204981
23. Loi VV, Busche T, Preuss T, Kalinowski J, Bernhardt J, Antelmann H. The AGXX(R) antimicrobial coating causes a thiol-specific oxidative stress response and protein S-bacillithiolation in Staphylococcus aureus. Front Microbiol. (2018) 9:3037. doi: 10.3389/fmicb.2018.03037
24. Han KWW, Po WW, Sohn UD, Kim HJ. Benzyl isothiocyanate induces apoptosis via reactive oxygen species-initiated mitochondrial dysfunction and DR4 and DR5 death receptor activation in gastric adenocarcinoma cells. Biomolecules. (2019) 9:1–15. doi: 10.3390/biom9120839
25. Borges A, Abreu AC, Ferreira C, Saavedra MJ, Simoes LC, Simoes M. Antibacterial activity and mode of action of selected glucosinolate hydrolysis products against bacterial pathogens. J Food Sci Technol. (2015) 52:4737–48. doi: 10.1007/s13197-014-1533-1
26. Kaiser SJ, Mutters NT, Blessing B, Gunther F. Natural isothiocyanates express antimicrobial activity against developing and mature biofilms of Pseudomonas aeruginosa. Fitoterapia. (2017) 119:57–63. doi: 10.1016/j.fitote.2017.04.006
27. Niu TX, Wang XN, Wu HY, Bi JR, Hao HS, Hou HM, et al. Transcriptomic analysis, motility and biofilm formation characteristics of Salmonella typhimurium exposed to benzyl isothiocyanate treatment. Int J Mol Sci. (2020) 21:1–13. doi: 10.3390/ijms21031025
28. Wang X, Wu H, Niu T, Bi J, Hou H, Hao H, et al. Downregulated expression of virulence factors induced by benzyl isothiocyanate in Staphylococcus aureus: a transcriptomic analysis. Int J Mol Sci. (2019) 20:1–13. doi: 10.3390/ijms20215441
29. Derr AM, Faustoferri RC, Betzenhauser MJ, Gonzalez K, Marquis RE, Quivey RG, Jr. Mutation of the NADH oxidase gene (nox) reveals an overlap of the oxygen- and acid-mediated stress responses in Streptococcus mutans. Appl Environ Microbiol. (2012) 78:1215–27. doi: 10.1128/AEM.06890-11
30. Auzat I, Chapuy-Regaud S, Le Bras G, Dos Santos D, Ogunniyi AD, Le Thomas I, et al. The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol Microbiol. (1999) 34:1018–28. doi: 10.1046/j.1365-2958.1999.01663.x
31. Wang X, Zhao X. Contribution of oxidative damage to antimicrobial lethality. Antimicrob Agents Chemother. (2009) 53:1395–402. doi: 10.1128/AAC.01087-08
32. Goswami M, Mangoli SH, Jawali N. Involvement of reactive oxygen species in the action of ciprofloxacin against Escherichia coli. Antimicrob Agents Chemother. (2006) 50:949–54. doi: 10.1128/AAC.50.3.949-954.2006
33. Rasigade JP, Moulay A, Lhoste Y, Tristan A, Bes M, Vandenesch F, et al. Impact of sub-inhibitory antibiotics on fibronectin-mediated host cell adhesion and invasion by Staphylococcus aureus. BMC Microbiol. (2011) 11:263. doi: 10.1186/1471-2180-11-263
Keywords: biofilm, Miswak, antibacterial agents, oral health, gram-positive bacteria, gene expression, streptococci
Citation: Alhandal H, Almesaileikh E, Bhardwaj RG, Al Khabbaz A and Karched M (2022) The Effect of Benzyl Isothiocyanate on the Expression of Genes Encoding NADH Oxidase and Fibronectin-Binding Protein in Oral Streptococcal Biofilms. Front. Oral. Health 3:863723. doi: 10.3389/froh.2022.863723
Received: 27 January 2022; Accepted: 15 March 2022;
Published: 11 April 2022.
Edited by:
Prasanna Neelakantan, The University of Hong Kong, Hong Kong SAR, ChinaReviewed by:
Vivek Babu C. S., Central Food Technological Research Institute (CSIR), IndiaAnjali Y. Bhagirath, University of Alberta, Canada
Copyright © 2022 Alhandal, Almesaileikh, Bhardwaj, Al Khabbaz and Karched. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maribasappa Karched, bWFyaWJhc2FwcGEua2FyY2hlZEBrdS5lZHUua3c=
 Hawraa Alhandal1
Hawraa Alhandal1 Radhika G. Bhardwaj
Radhika G. Bhardwaj Maribasappa Karched
Maribasappa Karched