
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 28 March 2025
Sec. Gastrointestinal Cancers: Hepato Pancreatic Biliary Cancers
Volume 15 - 2025 | https://doi.org/10.3389/fonc.2025.1523061
This article is part of the Research TopicAdvances in Epigenetic Inheritance Regulating Gastrointestinal CancerView all articles
 Jiayi Wang1,2†
Jiayi Wang1,2† Congcong Zhang1,2†
Congcong Zhang1,2† Yinghui Zhang1
Yinghui Zhang1 Jiaojiao Guo2
Jiaojiao Guo2 Chenyu Xie2
Chenyu Xie2 Yulu Liu1
Yulu Liu1 Lidian Chen2*
Lidian Chen2* Liangliang Ma1*
Liangliang Ma1*Liver cancer, characterized by its insidious nature, aggressive invasiveness, and propensity for metastasis, has witnessed a sustained increase in both incidence and mortality rates in recent years, underscoring the urgent need for innovative diagnostic and therapeutic approaches. Emerging research indicates that CircRNAs (circular RNAs) are abundantly and stably present within cells, with their expression levels closely associated with the progression of various malignancies, including hepatocellular carcinoma. In the context of liver cancer progression, circRNAs exhibit promising potential as highly sensitive diagnostic biomarkers, offering novel avenues for early detection, and also function as pivotal regulatory factors within the carcinogenic process. This study endeavors to elucidate the biogenesis, functional roles, and underlying mechanisms of circRNAs in hepatocellular carcinoma, thereby providing a fresh perspective on the pathogenesis of liver cancer and laying a robust foundation for the development of more precise and effective early diagnostic tools and therapeutic strategies.
Liver cancer ranks among the most common malignant tumors worldwide (1–4). Although various treatment methods exist, their therapeutic effects differ, and they each have specific side effects and limitations (5, 6). Despite therapeutic interventions, the rates of recurrence and metastasis remain elevated, resulting in a poor five-year survival rate (7–9). Additionally,liver cancer exhibits significant tumor heterogeneity and lacks early diagnostic methods, resulting in poor overall survival rates for patients with liver cancer (10–14). Consequently, it is imperative to identify novel therapeutic targets and biomarkers for effective early detection and personalized treatment strategies aimed at enhancing both survival outcomes and quality of life for individuals diagnosed with liver cancer.
Non-coding RNAs (ncRNAs) are integral to cell biology and physiological processes, playing pivotal roles in the pathogenesis of various diseases (15). Among these, circular RNAs (circRNAs), a distinct subclass of ncRNAs found in eukaryotes, have garnered significant attention due to their strong association with the progression of numerous diseases, particularly tumors (16–20). In recent years, a growing body of research has revealed that the abnormal expression of circRNA is intimately associated with the onset and progression of liver cancer. This alteration in expression patterns could have profound implications for the advancement of liver cancer and present novel, promising targets for its diagnosis and prognostic assessment.
This class of endogenous ncRNAs regulates gene expression in various biological organisms; circRNAs are covalently closed-loop RNA transcripts without polyadenylated tails or 5′–3′ polarity (21, 22). This configuration makes circRNA endogenous, abundant in cells (23), presenting higher stability than its linear counterpart and preventing nucleic acid exonuclease-mediated degradation (24). CircRNA exhibits specific expression patterns in tissues and organs, demonstrating significant potential as diagnostic, prognostic, and predictive biomarkers (25–29). With the rapid advancement of high-throughput sequencing technology, scientists have observed that the abundance of certain circRNAs exceeds that of their corresponding linear RNAs by more than tenfold, based on studies of thousands of circRNAs (30). This finding further affirms the crucial role of circRNAs in organisms and provides new insights for studying the mechanisms of circRNAs in liver cancer. Additionally, an extensive sequencing study of more than 2,000 clinical tissue samples and cell lines from over ten types of cancers has shown that circRNAs with stable structures can serve as cancer biomarkers in blood or urine (31).
This manuscript begins with a concise overview of the fundamental characteristics and biological roots of circRNAs. Next, it examines the roles of circRNAs in liver cancer. Lastly, it investigates the potential of circRNAs in diagnosing and treating liver cancer. By systematically studying circRNAs, we aim to offer innovative strategies and approaches for liver cancer diagnosis and treatment, ultimately enhancing patient prognosis and quality of life.
The investigation of CircRNAs commenced in 1976 when German researchers, led by Heinz L. Sanger, published findings in the journal PNAS. They confirmed the existence of viroids, which are organisms constituted of single-stranded, covalently closed circular RNA. This pivotal revelation represented humanity’s inaugural approach to understanding circRNA molecules (32). In 1979, Hsu and colleagues utilized electron microscopy to unveil circRNAs in eukaryotic cells (33). By 1993, the identification of circRNAs within human cells became a notable advancement in the field (34).
In the ensuing decades, most circRNAs were regarded as products of mis-splicing or by-products of pre-mRNA processing due to technological limitations and a research focus favoring protein-coding mRNAs (35). The study of circRNAs did not garner much attention. However, with the rapid advancement of high-throughput sequencing (Next-Generation Sequencing, NGS) and bioinformatics in the early 21st century, scientists gradually unveiled the prevalence of circRNAs in eukaryotes and their potential biological functions. After 2010, circRNA research entered a new phase. Researchers began to investigate the functions of circRNAs in depth and discovered their significant roles in gene expression regulation, disease development, and other areas, including serving as protein scaffolds or miR sponges and being translated into peptides (36–38). A circRNA can function as both a miR sponge and a protein template (39–41). In 2012, scientists identified numerous circRNAs in the human body, which prompted increased attention and rapid progress in circRNA research. As research advanced, circRNAs demonstrated close relationships with various biological processes, including development, physiological conditions, and many diseases, such as cancer (Table 1).
Thus, the utilization of circRNA in the realms of disease diagnosis, treatment, and prognosis evaluation holds significant value.
The biosynthesis of circRNAs is a complex and intricate mechanism centered on the reverse splicing of precursor mRNAs (pre-mRNAs). In this mechanism, the 5’ splice site located downstream of the intron connects with the 3’ splice site positioned upstream in reverse order. This results in the formation of a circular RNA structure, linked by a 3’,5′-phosphodiester bond between the reverse spliced exons (42). CircRNAs are typically derived from 1-5 exons and localize predominantly in the cytoplasm; however, nuclear-localized intron-containing circRNAs (which originate from distinct genomic loci) exhibit unique regulatory functions (43, 44). Based on their sequence types, circRNAs classify into three distinct categories: exonic circRNAs (EcRNAs), intronic circRNAs (CiRNAs), and exon-intronic circRNAs (EIcRNAs) (45–47).
The biosynthesis of circRNAs is a complex and intricate mechanism centered on the reverse splicing of precursor mRNAs (pre-mRNAs). Four primary mechanisms have been elucidated, each involving distinct molecular interactions and regulatory factors:
Mechanism I: “RNA-binding protein (RBPs)-driven cyclization.”
RBPs facilitate the interaction between upstream and downstream introns, promoting circRNA formation. For example, the RBP hnRNPA1 binds to flanking introns of SAMD4 and HIPK3 pre-mRNAs, facilitating Step 1: RBP binding → Step 2: Reverse splicing → Step 3: circRNA maturation (42, 48). This mechanism is particularly relevant in epithelial-mesenchymal transition (EMT) processes.
Mechanism II: “Intron pairing-driven cyclization.”
Reverse complementary sequences in flanking introns form secondary structures, enabling cyclization. The CDR1as locus exemplifies this mechanism, where 70 conserved Alu elements mediate circularization and miR-7 sequestration (49). Introns may subsequently be removed or retained, leading to the formation of ecircRNAs or EIciRNAs (50).
Mechanism III: “Lariat-driven cyclization.”
During canonical splicing, exon skipping generates a lariat intermediate containing exons and introns. This process can generate a lasso structure containing an exon and an intron. If the introns are subsequently removed, ecircRNA or EIciRNA forms (51).For instance, the SAMD4 gene produces circRNAs via this mechanism, with the lariat structure stabilized by U2AF65 and SF1 splicing factors (50).
Mechanism IV: “lasso intron-driven cyclization.”
CiRNAs (circular intronic RNAs) evade debranching enzyme degradation through specific motifs. The ci-ankrd52 locus retains a 7-nt GU-rich element near the 5′ splice site and an 11-nt C-rich element at the branchpoint, forming a stable lariat structure (44). This binding enables circRNAs to evade degradation by debranching enzymes, thereby forming stable ciRNAs (52). Together, these mechanisms highlight the complexity and diversity of circRNA biological origins.
Overall, circRNA biosynthesis constitutes a complex and delicate process involving multiple steps and regulatory factors. With ongoing research, we anticipate gaining a more comprehensive understanding of circRNA biosynthesis mechanisms and uncovering its significant functions in biology and medicine.
Circular RNAs exert multifaceted roles in modulating gene expression through diverse mechanisms:
CircRNAs act as ceRNAs or miR sponges and are one of their most notable functions (53, 54). miRNAs are a class of short non-coding RNA molecules that inhibit the transcription and translation process of target genes by binding to their mRNAs (55), whereas circRNAs have multiple miRNA-binding sites that compete for binding to miRNAs, thereby regulating miRNA activity and affecting miRNA-regulated target gene expression (56, 57). CDR 1as, for example, carries 63 conserved miR-7 binding sites, which significantly affect the expression of tumor-associated genes by enhancing the stability of miR-7 target mRNAs, which in turn are tightly linked to the process of tumor progression (49, 58, 59). In triple-negative breast cancer research, circCD 44 has been identified as a sponge for miR-502-5p, effectively sequestering and inhibiting its activity, thereby contributing to the initiation and progression of tumorigenesis (60). Similarly, the overexpression of circLRP 6 has also been empirically validated to accelerate the pathological progression of atherosclerosis by absorbing miR-145, further emphasizing the regulatory role of circRNAs in modulating disease processes (61).
RNA-binding proteins (RBPs) serve as critical regulators of RNA metabolism and play significant roles in various RNA processes (62). Recent investigations have revealed that circRNAs can bind to RBPs, thereby co-regulating gene expression (42, 63–65). For instance, both two circRNAs—circBACH1 and circZKSCAN1—have been implicated in liver cancer progression; they modulate the expression of oncogenic genes and associated signaling pathways through interactions with distinct RBPs (66–70). Additionally, specific circRNAs can modify the cellular localization of RBPs, enabling these proteins to execute specialized functions in non-canonical contexts, which provides novel mechanistic insights into their regulatory mechanisms (66).
CircRNAs interact with RBPs to modulate RNA transcription and translation processes. These circular RNAs possess unique structures that allow them to specifically bind RBPs and forming stable protein-RNA complexes. Such interactions can modulate key aspects of RBP function, stability, and activity of these proteins, further impacting RNA transcription and translation. Consequently, circRNAs serve as critical regulators of RNA metabolism and global gene expression.
Circular RNAs, specifically circSEP3 and circSMARCA5, act as transcriptional regulators. They induce pauses and termination in transcription at specific exons by binding directly to DNA, which alters gene expression patterns (71, 72). In contrast, EIciRNA enhances gene expression through its interaction with U1 small nuclear ribonucleoprotein (46). Additionally, cyclic intronic RNAs (ciRNAs) accumulate at their production sites, boosting gene expression by regulating the activity of RNA polymerase II (73). CircRNAs function as crucial transcriptional regulators, engaging with transcription factors and regulatory elements. This interaction influences the formation and activity of transcription complexes, thereby impacting the transcription levels of targeted genes. Moreover, circRNAs can directly modulate gene expression via chromatin interactions. These multifaceted roles position circRNAs as vital players in cell fate determination, tissue development, and the initiation of diseases.
Circular RNAs directly influence protein activity, localization, and translational dynamics through structural and scaffolding mechanisms:
Researchers have identified translatable circRNAs (74, 75), and recent investigations have confirmed their widespread existence (76, 77). Although circRNAs do not directly encode proteins, they play a role in protein translation by interacting with proteins. They function as “molecular adsorbents,” binding proteins to form complexes. This interaction can modify the stability, subcellular localization, and activity of proteins. Such associations may influence protein synthesis, folding, transport, and degradation, thereby regulating the intracellular protein balance and functionality.
CircRNAs function as scaffolds for protein complexes by interacting with a range of proteins. This scaffolding role enhances protein interactions and signal transduction, thereby modulating intracellular signaling pathways and biological processes. By forming these protein assemblies, circRNAs coordinate the actions of multiple proteins, facilitating intricate cellular regulation. Moreover, circRNAs are instrumental in fostering interactions between two or more proteins. Notable examples include circ-Amotl1 and circ-Foxo3, which act as protein scaffolds to help align enzymes with their substrates. For instance, circ-Amotl1 has been recognized as a scaffold that promotes interactions with PDK1 and AKT1 (78), facilitating their movement into the nucleus, a vital process for cell growth and survival. Furthermore, a study by Du et al. underscored the role of circ-Foxo3 as a scaffold for a variety of proteins, demonstrating its ability to bind with p53 and the E3 ubiquitin-protein ligase Mdm2 (79). This interaction enhances Mdm2-driven ubiquitination and the subsequent degradation of p53.
In conclusion, circRNAs are vital in functions such as miRNA sponging, RNA-binding protein interactions, gene transcription regulation, protein translation, and acting as binding protein scaffolds (Figure 1). These roles position circRNAs as key players in managing intracellular gene expression and protein functionality, crucial for insights into cell biology and disease mechanisms. As research progresses, we anticipate uncovering additional functions and regulatory pathways of circRNAs, offering novel perspectives and approaches for disease diagnosis and treatment
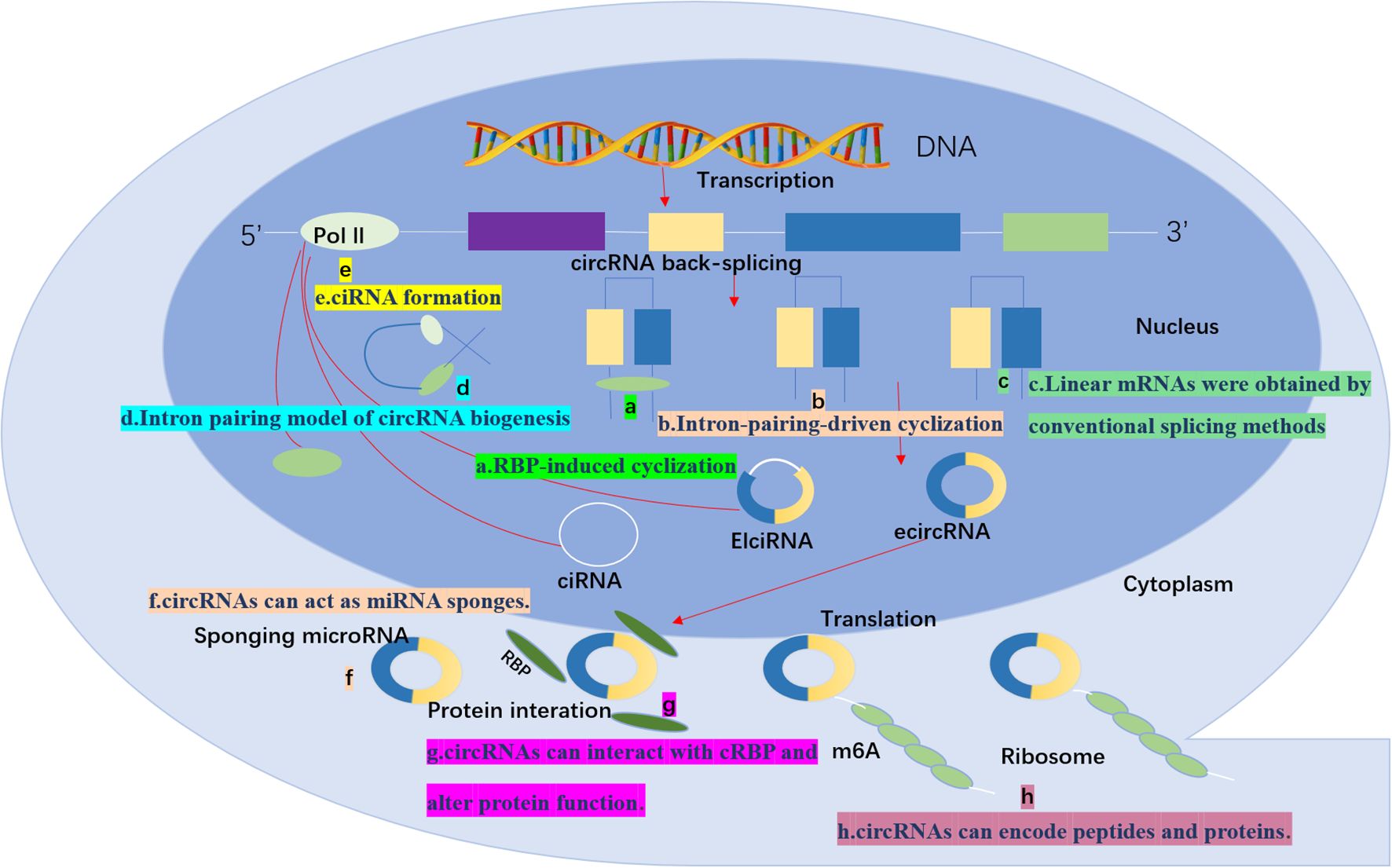
Figure 1. CircRNA biogenesis and function: Typical reverse splicing produces three types of circRNAs: ciRNAs, EIciRNAs, and ecircRNAs. function (a) RBP-induced cyclization: the RBP facilitates a reverse splicing event in which introns are removed to form circRNAs. (b) Intron-pairing-driven cyclization; reverse complementary sequences (Alu sequences) direct the insertion and by removing or retaining the introns generates EIciRNA or ecircRNA. (c) Linear mRNAs were obtained by conventional splicing methods (d) Intron pairing model of circRNA biogenesis: a 3’ splice donor site in an exon binds to a 5’ splice acceptor site in an upstream intron to form circRNAs or ecircRNAs by removing introns between exons. ecircRNA is formed by removing the intron between the exons. (e) ciRNA formation; ciRNA is derived from the lactinuclear intron. (f) circRNAs can act as miRNA sponges. (g) circRNAs can interact with cRBP and alter protein function. (h) circRNAs can encode peptides and proteins. This image was made using Microsoft PowerPoint.
Non-alcoholic fatty liver disease (NAFLD) is characterized by the abnormal buildup of fat in the liver. If left untreated, NAFLD can advance to nonalcoholic steatohepatitis (NASH), which represents a more severe state (80). The frequency of both NAFLD and NASH has notably increased due to shifts in lifestyle and contemporary dietary practices. These liver conditions are closely associated with cirrhosis and liver cancer (81). Day and James hypothesized that steatosis serves as a possible second insult leading to NASH (82). Recent research has unveiled abnormal miRNA expression, excessive triglyceride build-up, and lipid peroxidation dysregulation as key contributors to hepatic steatosis. Importantly, miR-34a is crucial in this context; circRNA_0046367 finely tunes its function (83). The expression of circRNA_0046367 declines significantly in the context of hepatic steatosis. Restoring its levels can effectively diminish lipid peroxidation, reduce apoptosis, and ameliorate mitochondrial dysfunction. This recovery lifts the inhibitory action of miR-34a on peroxisome proliferator-activated receptor α (PPARα), subsequently enhancing the expression of genes involved in lipid metabolism (84). Additionally, studies indicate that circ_0046366 is another circRNA linked to NAFLD pathogenesis, functioning as an antagonist of miR-34a. The absence of this circRNA prominently contributes to hepatocellular steatosis induced by a high-fat diet. Fortunately, reinstating circ_0046366 expression effectively hampers the steatosis progression, deactivates miR-34a, and spurs the transcriptional activation of lipid metabolism-related genes. This process restores compromised PPARα activity, representing a novel strategic approach for NAFLD treatment. While a direct link between circRNAs and liver disease metabolism has yet to be established, the miRNAs discovered in this study, which interact with differentially expressed circRNAs, undeniably present valuable insights for future investigations into these non-coding RNAs within the NAFLD/NASH pathology.
Liver fibrosis is an end-stage liver disease that develops from chronic liver injury and can lead to cirrhosis with a poor prognosis. Studies have shown that one of the key mechanisms of liver fibrosis is the activation of hepatic stellate cells (HSCs), which transform from a resting state to myofibroblasts and promote fiber formation (85). It was shown that 179 circRNAs were found to be up-regulated and 630 down-regulated in radiation-induced activation of HSCs, indicating significant changes in circRNA expression, and in particular, silencing of hsa_circ_0071410 increased miR-9-5p expression and inhibited HSCs activation (86). These data suggest that circRNAs may play an important role in NAFLD/NASH-associated hepatic fibrosis, but the specific mechanisms need to be thoroughly investigated.
CircRNA is a closed ring RNA molecule that exhibits greater stability and resistance to RNase degradation compared to linear RNA. In liver cancer tissues and cells, circRNA expression patterns often show tissue specificity and tumor relevance. Studies reveal that the expression levels of numerous circRNAs in liver cancer differ significantly from those in normal liver tissues, closely linking them to the clinicopathologic features of liver cancer. While the specific functions and mechanisms of circRNAs in liver cancer require further exploration, many reports confirm their pivotal roles in this disease (87–97).
In terms of cell proliferation and carcinogenesis, CircRNAs affect cell proliferation and tumorigenesis in liver cancer progression. Some CircRNAs express highly at the early stage of liver cancer and accelerate the malignant proliferation of tumor cells by regulating specific genes and signaling pathways. One study summarized the following signaling pathways through which CircRNAs regulate tumor proliferation and metastasis (98).
The specifics are as follows:
1. The Wnt/β-catenin pathway is a signaling pathway closely related to tumor progression. In liver cancer, circMTO1 activates the Wnt/β-catenin signaling pathway by regulating the miR-541-5p/ZIC1 axis, which in turn inhibits tumor proliferation. In contrast, hsa_circRNA_104348 activated the pathway through the miR-187-3p/RTKN2 axis and promoted cell proliferation. CircRNA-SORE acted as a sponge for miR-103a-2-5p and miR-660-3p, and similarly competed to activate the Wnt/β-catenin signaling pathway. In contrast, circZFR further enhances the activation of the Wnt/β-catenin signaling pathway and promotes proliferation by regulating miR-3619-5p/CTNNB1 (99–102).
2. The nuclear factor-κB (NF-κB) signaling pathway is also closely related to proliferation. circZFR promotes liver cancer development by inhibiting the STAT3/NF-κB pathway (103). CircLIFR, on the other hand, promotes cell proliferation by interacting with TBK1, a serine/threonine kinase that regulates the NF-κB pathway (104); In contrast, circCORO1C further promotes liver cancer proliferation and metastasis by upregulating NF-κB pathway-induced PD-L1 expression (105).
3. The PI3K/AKT signaling pathway plays a critical role in regulating multiple cellular functions. In liver cancer, circCDYL, circCDK13, and circEPHB4 regulate the proliferative capacity of liver cancer cells through the PI3K/AKT signaling pathway (89, 106, 107).
4. In the context of liver cancer, the mitogen-activated protein kinase (MAPK) signaling pathway, as a series of highly conserved enzymatic cascades, plays a crucial role in physiological activities such as cell proliferation, differentiation, and apoptosis. CircDHPR targets the RASGEF1B/RAS/MAPK signaling pathway by interacting with miR-3194-5p, which in turn promotes tumor growth and metastasis (108). In addition, circASAP1 acts as a sponge for miR-326 and miR-532-5p and regulates MAPK1 expression, thereby enhancing cell proliferation and invasion (88). In contrast, circSETD3 exerts an inhibitory effect on tumor growth by targeting the miR-421/MAPK14 signaling pathway (109).
Many circRNAs also regulate pathways that impact liver cancer progression. By influencing tumor-related signaling pathways, circRNAs ultimately affect tumor progression, making this a promising research direction. circIPO11, for instance, is a conserved circRNA that shows high expression in liver cancer tumors and hepatic CSCs, and plays a role in maintaining hepatic CSC self-renewal. It promotes liver CSC self-renewal and advances liver cancer proliferation through the activation of the Hedgehog signaling pathway. Antisense oligonucleotides (ASOs) that target circIPO11 exhibit synergistic antitumor effects when combined with the TOP1 inhibitor camptothecin (CPT) (110). The nuclear circRNA circASH2 is absent in liver cancer tissues and suppresses liver cancer metastasis by modifying the structure of the tumor cytoskeleton. Proto-myosin 4 (TPM4), the primary target of circASH2, is repressed post-transcriptionally. CircASH2 enhances the liquid-liquid phase separation of nuclear Y box binding protein 1 (YBX1), thus promoting the decay of TPM4 transcripts, highlighting the role of tumor suppressor circRNAs and their intricate regulatory mechanisms in liver cancer progression (111).
Second, circRNAs play a key role in cell movement and infiltration during liver cancer progression. Cellular transformation is a complex process that enables cancer cells to gain enhanced movement and infiltration capabilities. Certain circRNAs promote cancer cell migration and invasion by regulating associated genes and signaling pathways. For example, circARFGEF2 significantly accelerates the metastasis of liver cancer in the liver and lung by regulating specific signaling pathways (96).
In addition, circRNAs contribute to the regulation of immune system function and influence liver cancer development. Immune dysfunction closely relates to liver cancer progression, and circRNAs can regulate the activity and function of immune cells, affecting the immune evasion of cancer cells. For example, circUHRF1 inhibits the immune function of NK cells by upregulating TIM-3 expression in NK cells, thus promoting immune evasion by cancer cells (112).
Finally, circRNAs may play an important role in drug response. Although researchers have not fully understood the specific mechanisms of circRNAs in drug response, evidence shows that certain circRNAs may regulate drug effects and contribute to drug resistance. Studies in this area will deepen our understanding of the mechanisms of drug response in liver cancer and provide new insights for future therapeutic strategies.In summary, the pathogenesis of liver cancer is closely related to the function of circRNAs (Figure 2).
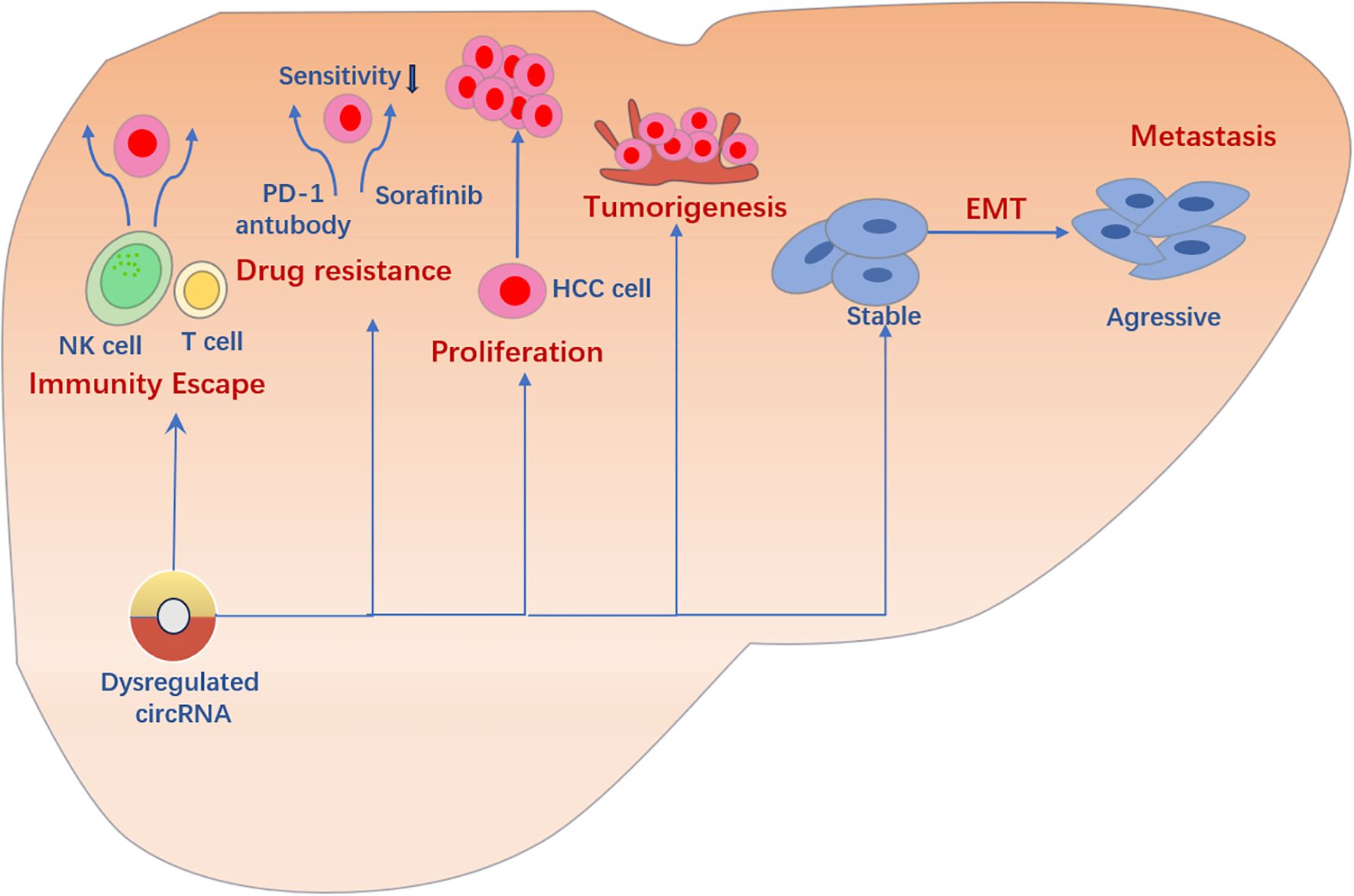
Figure 2. Multiple roles of CircRNA in liver cancer:circRNA plays multiple roles in liver cancer. It regulates cell proliferation and tumor initiation, participates in the EMT process, and promotes invasion and metastasis of liver cancer cells. In addition, circRNA affects the progression and prognosis of liver cancer by regulating the immune system. Notably, some circRNAs may induce and maintain drug resistance in liver cancer treatment, challenging therapeutic strategies.This image was made using Microsoft PowerPoint.
Due to the tissue specificity and stability of circRNA expression, many studies now explore its potential in the early diagnosis of liver cancer. Compared with traditional serum markers like AFP, circRNAs demonstrate higher specificity and sensitivity. Researchers found that changes in the expression levels of specific circRNAs in liver cancer tissues and blood samples effectively differentiate liver cancer patients from healthy controls, providing better diagnostic value. For instance, Conn et al. (48) discovered that circRNA_circFOXP1 promoted the infiltration and migration of liver cancer by regulating cell-cell interactions within the liver cancer microenvironment. Its high expression in liver cancer tissues closely linked to deep tumor infiltration and poor prognosis. Hence, circRNA can serve as a novel diagnostic marker for liver cancer, potentially improving the accuracy and efficiency of early diagnosis.
Moreover, numerous studies on liver cancer patients reveal that hsa_circ_0000798 (113), hsa_circ_0027089 (114), and hsa_circ_0058124 (115) are up-regulated in liver cancer tissues. In contrast, some circRNAs down-regulate, including hsa_circSMARCA5 (116), hsa_circ_0068669 (117), hsa_circ_0028502 (118), and hsa_circ_0076251 (118). These circRNAs are regarded as potential biomarkers for liver cancer. Additionally, some circRNAs play roles in several hallmarks of cancer, including cell death and survival, invasion, metastasis, and angiogenesis. Various validation tests, such as northern blotting, spot blotting, RNA-seq, and circRNA-specific microarrays, show significant dysregulation of many key circRNAs in liver cancer cells, tissues, blood, and exosomes (Figure 3).
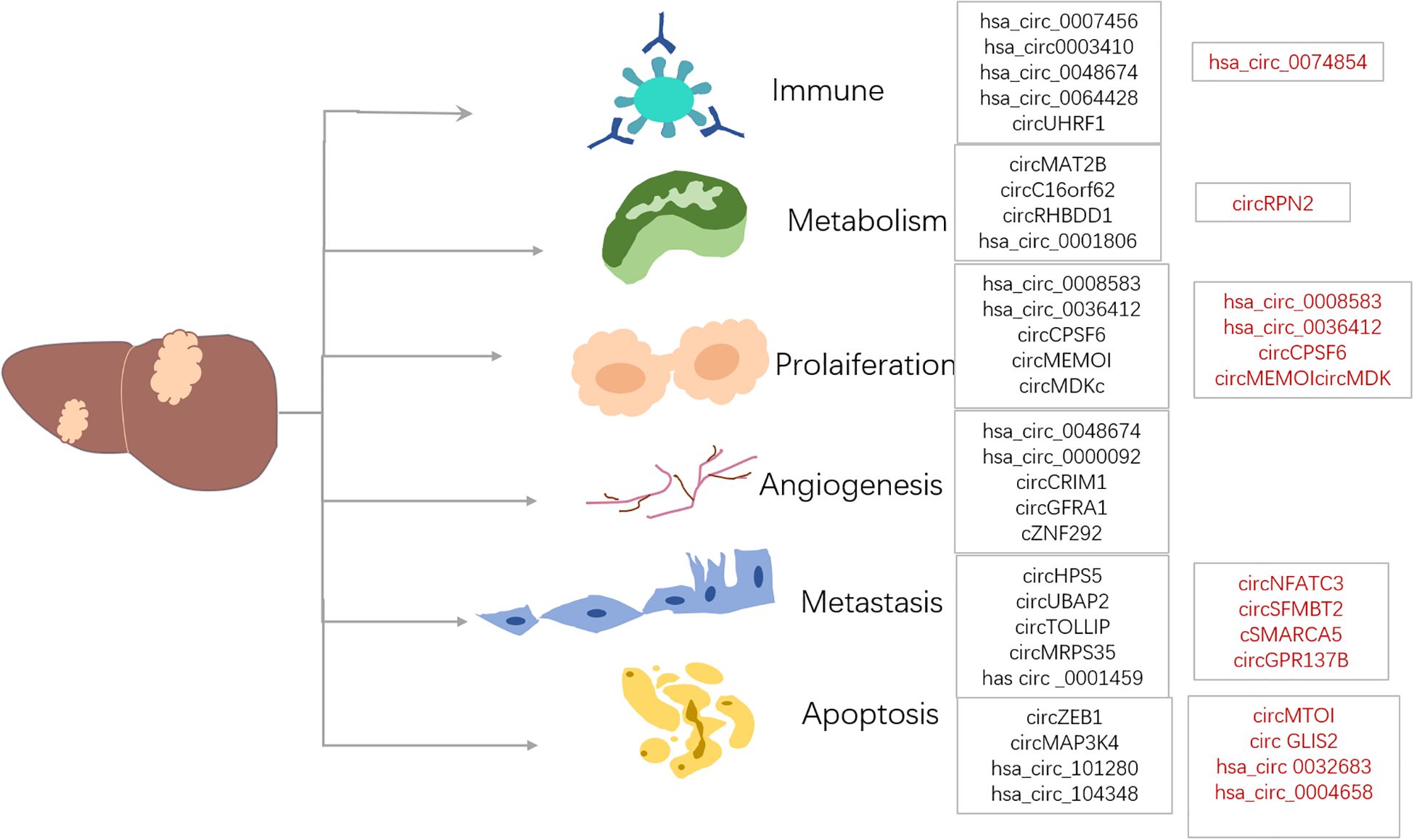
Figure 3. CircRNAs are closely related to liver cancer, and they are associated with processes such as proliferation, metabolism, angiogenesis, metastasis, immune regulation and apoptosis. In these processes, CircRNAs may either act as oncogenes (marked in black font), promoting tumor development, or play the role of suppressor genes (marked in red font), inhibiting the tumor process, demonstrating their complex and diverse functions. This image was made using Microsoft PowerPoint.
In liver cancer, a variety of specific circRNAs show aberrant expression, and these molecules occupy a central role in the progression of liver cancer, and thus are considered as potential diagnostic indicators and therapeutic targets for liver cancer (119–121). circRNAs can be found in tissues (122), exosomes (123–125), plasma (31), serum (126), cerebrospinal fluid (127), urine (31), saliva (128), and other biological samples were detected. Due to the structural property of reverse splicing of circRNAs, they are able to avoid recognition by nucleic acid exonucleases and therefore have a longer half-life compared to linear RNAs. This stability makes circRNAs ideal candidates for cancer diagnosis, monitoring and treatment (129–131).
Numerous studies have revealed the potential application of circRNAs as cancer biomarkers (132–137). However, in-depth evaluation of their diagnostic accuracy is lacking. For instance, high expression of circASAP 1 strongly associates with poor prognosis, lower survival, and higher recurrence rates in liver cancer (88). Additionally, the expression levels of hsa_circ_0001955 (138) and circ_104075 (139) increase in the tissues, serum, and plasma of liver cancer patients. Notably, the increased expression of circ_104075 is specific to liver cancer, as similar increases do not occur in other liver diseases or tumors (139). Serum levels of both circRNAs significantly reduce after surgical resection of the tumor tissue, suggesting that hsa_circ_0001955 and circ_104075 may serve as biomarkers for assessing the efficacy of surgical treatment and the risk of tumor recurrence. Moreover, DHX 9 expression significantly upregulates in liver cancer, inhibiting the production of cSMARCA 5 (hsa_circ_0001445) by binding and inhibiting the pairing of flanking reverse-complementary sequences. This, in turn, hampers the production of cSMARCA 5 (hsa_circ_0001445) (140). As a tumor suppressor, cSMARCA 5 inhibits liver cancer growth through the cSMARCA 5/miR-17-3p/miR-181b-5p/TIMP3 pathway. A decrease in cSMARCA 5 in liver cancer tissues correlates with increased tumor growth and metastasis, making it an independent prognostic indicator for patients post-tumor resection (87).
In studies exploring the relationship between circRNAs in exosomes and liver cancer, we observed a significant overexpression phenomenon of circRNA Cdr1as in exosomes released by liver cancer cells, a phenomenon that promotes the proliferation and migration ability of neighboring normal cells. circ-ZEB 1 and circ-AFAP 1 are associated with stemness and prognosis of liver cancer poorly and can regulate the epithelial-mesenchymal transition (EMT) process. Our novel exosome-derived circRNAs play crucial roles as key components of various intercellular crosstalk and communication systems in malignant transmission. This finding provides valuable support for the use of plasma exosomal circRNA as a clinical prognostic indicator for liver cancer patients (141). zhang et al. found that adipocyte-derived circ-DB promoted tumor growth and affected DNA damage repair through miR-34a and USP7 regulation (142), and Lai et al. indicated that circFBLIM1 was found in serum exosomes and liver cancer cells are highly expressed, and its inhibition limits glycolysis and liver cancer progression (143), and Huang et al. showed that circRNA-100338 in MHCC97H exosomes promotes liver cancer cell invasion and metastasis (144). The selective sorting of circRNAs into exosomes involves interactions with RNA-binding proteins (RBPs) such as GW182 and ELAVL1, which recognize specific sequence motifs (e.g., GGAG/CCCU) in circRNA loops (141). The nSMase2/sphingomyelinase pathway regulates ceramide production, promoting exosome biogenesis and circRNA loading (142). The exosome circANTXR 1 correlates with clinical characteristics (TNM stage and tumor size) and poor prognosis of liver cancer patients, and the circANTXR 1/miR-532- 5 p/XRCC 5 axis-mediated inhibition of liver cancer progression may be an effective strategy for treatment (145). For the association between high expression of exosomal circAKT 3 and poor prognosis, circAKT 3 could be used as a surveillance biomarker for early detection of recurrence (146). sun et al. identified three circRNAs (hsa_circ_0004001, hsa_circ_0004123, hsa_circ_0075792) as highly sensitive and specific biomarkers for liver cancer diagnosis (147), and Chen et al. found that circ-0051443 was delivered to liver cancer cells via exosomes, regulated miR-331-3p and BAK1, and inhibited the malignant behavior of liver cancer (148). The above studies not only deepened our understanding of the mechanism of circRNA action in liver cancer, but also suggested the important role of exosomes as circRNA delivery mediators in the regulation of tumor microenvironment. These findings provide new perspectives and potential targets for the development of circRNA-based therapeutic strategies for liver cancer, and are expected to provide new ideas and approaches for clinical intervention. A growing number of studies have indicated that some serum exosomal circRNAs also have potential as biomarkers for prognosis and tumor monitoring. As shown in the Table 2.
Emerging evidence from liver cancer research underscores the pivotal role of circRNAs in modulating gene expression networks and cellular signaling pathways. These regulatory mechanisms not only elucidate the pathophysiological basis of hepatocellular carcinoma but also provide actionable insights for developing circRNA-based therapeutics, including innovative biomarkers and precision-targeted anticancer interventions (149).It has gradually attracted extensive attention from researchers. Despite the remarkable progress in circRNA research related to liver cancer, several challenges still remain.
While single-cell RNA-seq and spatial transcriptomics have revolutionized our understanding of circRNA heterogeneity, their application remains constrained by:
Current research on circRNA function primarily relies on bioinformatics predictions and in vitro experiments but lacks in vivo validation. Meanwhile, researchers have not yet completely clarified the target genes and mechanisms of action of circRNAs, and the specific interaction network between circRNAs and liver cancer requires further elucidation. The multiple cell types within the liver, such as hepatocytes, hepatic stellate cells, blast cells, and immune cells, also complicate circRNA studies in liver disease. Future studies should aim to expand the functional spectrum of circRNAs, thoroughly resolve their regulatory networks, and utilize advanced technological tools like single-cell sequencing and functional genomics to uncover the mechanisms of their roles in liver cancer development and progression. More in-depth studies are necessary.
Future directions:
1.Develop integrative pipelines combining single-cell circRNA profiling with patient-derived organoids to bridge preclinical and clinical data gaps.
2. Leverage AI-driven models (e.g., Transformer-based architectures) to predict circRNA-protein interaction networks.
Existing studies mainly rely on small sample sizes and heterogeneous datasets, which restrict the broad application and dissemination of the results. Large-scale, standardized clinical samples and datasets will enhance the reliability and accuracy of the studies.
Future directions:
1. Large-Scale Cohort Studies: Initiatives like the Liver Cancer Precision Medicine Consortium provide unparalleled opportunities to analyze circRNA signatures across diverse ethnicities and disease stages.
2. Standardized Data Repositories: Expanding public databases with uniformly processed RNA-seq and clinical metadata will accelerate meta-analytic discoveries.
Although circRNA has shown great potential in laboratory research, its translation into clinical applications still faces many challenges, such as drug delivery, stability, and specificity.
Circular RNA, as a novel non-coding RNA, holds significant theoretical and applied value in the study of liver cancer. Future studies can further explore the potential of circRNA for early diagnosis and prognostic assessment of liver cancer, develop new strategies for circRNA-targeted therapy, and utilize circRNA in individualized therapy. Additionally, by combining bioinformatics analysis with clinical data, researchers can establish a multidimensional database of circRNA to facilitate its application as a biomarker. In summary, further in-depth exploration of circRNA in liver cancer research will lead to new breakthroughs and advancements in liver cancer treatment and management.
JW: Conceptualization, Investigation, Writing – original draft, Methodology. CZ: Conceptualization, Funding acquisition, Investigation, Writing – original draft. YZ: Investigation, Writing – original draft. JG: Methodology, Writing – original draft. CX: Methodology, Writing – original draft. YL: Writing – review & editing, Investigation. LC: Conceptualization, Investigation, Writing – review & editing. LM: Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Natural Science Foundation of China (grant no. 82204900), Henan Province Science and Technology Research and Development Plan Joint Fund Project (no. 242301420101) and the HSRP-DFCTCM-2023 (grant no. 15102046-2023).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Singh SP, Madke T, Chand P. Global epidemiology of hepatocellular carcinoma. J Clin Exp Hepatol. (2025) 15:102446. doi: 10.1016/j.jceh.2024.102446
2. Siegel RL, Kratzer TB, Giaquinto AN, Sung H, Jemal A. Cancer statistics, 2025. CA Cancer J Clin. (2025) 75:10–45. doi: 10.3322/caac.21871
3. McGlynn KA, Petrick JL, Groopman JD. Liver cancer: progress and priorities. Cancer Epidemiol Biomarkers Prev. (2024) 33:1261–72. doi: 10.1158/1055-9965.EPI-24-0686
4. Singal AG, Kanwal F, Llovet JM. Global trends in hepatocellular carcinoma epidemiology: implications for screening, prevention and therapy. Nat Rev Clin Oncol. (2023) 20:864–84. doi: 10.1038/s41571-023-00825-3
5. Childs A, Aidoo-Micah G, Maini MK, Meyer T. Immunotherapy for hepatocellular carcinoma. JHEP Rep. (2024) 6:101130. doi: 10.1016/j.jhepr.2024.101130
6. Llovet JM, Pinyol R, Yarchoan M, Singal AG, Marron TU, Schwartz M, et al. Adjuvant and neoadjuvant immunotherapies in hepatocellular carcinoma. Nat Rev Clin Oncol. (2024) 21:294–311. doi: 10.1038/s41571-024-00868-0
7. Koshy A. Evolving global etiology of hepatocellular carcinoma (HCC): insights and trends for 2024. J Clin Exp Hepatol. (2025) 15:102406. doi: 10.1016/j.jceh.2024.102406
8. Liu Y, Yang H, Li T, Zhang N. Immunotherapy in liver cancer: overcoming the tolerogenic liver microenvironment. Front Immunol. (2024) 15:1460282. doi: 10.3389/fimmu.2024.1460282
9. Xue Y, Ruan Y, Wang Y, Xiao P, Xu J. Signaling pathways in liver cancer: pathogenesis and targeted therapy. Mol Biomed. (2024) 5:20. doi: 10.1186/s43556-024-00184-0
10. Ito T, Nguyen MH. Perspectives on the underlying etiology of HCC and its effects on treatment outcomes. J Hepatocell Carcinoma. (2023) 10:413–28. doi: 10.2147/jhc.S347959
11. Motta BM, Masarone M, Torre P, Persico M. From non-alcoholic steatohepatitis (NASH) to hepatocellular carcinoma (HCC): epidemiology, incidence, predictions, risk factors, and prevention. Cancers (Basel). (2023) 15. doi: 10.3390/cancers15225458
12. Jiang D, Ma X, Zhang X, Cheng B, Wang R, Liu Y, et al. New techniques: a roadmap for the development of HCC immunotherapy. Front Immunol. (2023) 14:1121162. doi: 10.3389/fimmu.2023.1121162
13. Wang Y, Deng B. Hepatocellular carcinoma: molecular mechanism, targeted therapy, and biomarkers. Cancer Metastasis Rev. (2023) 42:629–52. doi: 10.1007/s10555-023-10084-4
14. Cranford HM, Jones PD, Wong RJ, Liu Q, Kobetz EN, Reis IM, et al. Hepatocellular carcinoma etiology drives survival outcomes: A population-based analysis. Cancer Epidemiol Biomarkers Prev. (2024) 33:1717–26. doi: 10.1158/1055-9965.EPI-24-0626
15. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. (2021) 21:22–36. doi: 10.1038/s41568-020-00306-0
16. Misir S, Wu N, Yang BB. Specific expression and functions of circular RNAs. Cell Death Differ. (2022) 29:481–91. doi: 10.1038/s41418-022-00948-7
17. Chen L, Huang C, Shan G. Circular RNAs in physiology and non-immunological diseases. Trends Biochem Sci. (2022) 47:250–64. doi: 10.1016/j.tibs.2021.11.004
18. Sayad A, Najafi S, Kashi AH, Hosseini SJ, Akrami SM, Taheri M, et al. Circular RNAs in renal cell carcinoma: Functions in tumorigenesis and diagnostic and prognostic potentials. Pathol Res Pract. (2022) 229:153720. doi: 10.1016/j.prp.2021.153720
19. Du Y, Zuber PK, Xiao H, Li X, Gordiyenko Y, Ramakrishnan V. Efficient circular RNA synthesis for potent rolling circle translation. Nat BioMed Eng. (2024) 10:1–28. doi: 10.1038/s41551-024-01306-3
20. Drula R, Braicu C, Neagoe IB. Current advances in circular RNA detection and investigation methods: Are we running in circles? Wiley Interdiscip Rev RNA. (2024) 15:e1850. doi: 10.1002/wrna.1850
21. Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. (2013) 340:440–1. doi: 10.1126/science.1238522
22. Liu CX, Chen LL. Circular RNAs: Characterization, cellular roles, and applications. Cell. (2022) 185:2016–34. doi: 10.1016/j.cell.2022.04.021
23. He AT, Liu J, Li F, Yang BB. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther. (2021) 6:185. doi: 10.1038/s41392-021-00569-5
24. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. (2013) 19:141–57. doi: 10.1261/rna.035667.112
25. Nemeth K, Bayraktar R, Ferracin M, Calin GA. Non-coding RNAs in disease: from mechanisms to therapeutics. Nat Rev Genet. (2024) 25:211–32. doi: 10.1038/s41576-023-00662-1
26. Chen D, Gu X, Nurzat Y, Xu L, Li X, Wu L, et al. Writers, readers, and erasers RNA modifications and drug resistance in cancer. Mol Cancer. (2024) 23:178. doi: 10.1186/s12943-024-02089-6
27. De Palma FDE, Salvatore F, Pol JG, Kroemer G, Maiuri MC. Circular RNAs as potential biomarkers in breast cancer. Biomedicines. (2022) 10(3):725. doi: 10.3390/biomedicines10030725
28. Pisignano G, Michael DC, Visal TH, Pirlog R, Ladomery M, Calin GA. Going circular: history, present, and future of circRNAs in cancer. Oncogene. (2023) 42:2783–800. doi: 10.1038/s41388-023-02780-w
29. Muskan M, Abeysinghe P, Cecchin R, Branscome H, Morris KV, Kashanchi F. Therapeutic potential of RNA-enriched extracellular vesicles: The next generation in RNA delivery via biogenic nanoparticles. Mol Ther. (2024) 32:2939–49. doi: 10.1016/j.ymthe.2024.02.025
30. Gao Y, Shang S, Guo S, Li X, Zhou H, Liu H, et al. Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. (2021) 49:D1251–d8. doi: 10.1093/nar/gkaa1006
31. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. (2019) 176:869–81.e13. doi: 10.1016/j.cell.2018.12.021
32. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. (1976) 73:3852–6. doi: 10.1073/pnas.73.11.3852
33. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. (1979) 280:339–40. doi: 10.1038/280339a0
34. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. (1993) 7:155–60. doi: 10.1096/fasebj.7.1.7678559
35. Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, et al. Scrambled exons. Cell. (1991) 64:607–13. doi: 10.1016/0092-8674(91)90244-s
36. Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. (2020) 21:475–90. doi: 10.1038/s41580-020-0243-y
37. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. (2019) 20:675–91. doi: 10.1038/s41576-019-0158-7
38. Xiao MS, Ai Y, Wilusz JE. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. (2020) 30:226–40. doi: 10.1016/j.tcb.2019.12.004
39. Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou X, et al. circFBXW7 inhibits Malignant progression by sponging miR-197-3p and encoding a 185-aa protein in triple-negative breast cancer. Mol Ther Nucleic Acids. (2019) 18:88–98. doi: 10.1016/j.omtn.2019.07.023
40. Xu Y, Leng K, Yao Y, Kang P, Liao G, Han Y, et al. Cholangiocarcinoma-associated circular RNA 1, contributes to cholangiocarcinoma progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. (2021) 73:1419–35. doi: 10.1002/hep.31493
41. Huang S, Li X, Zheng H, Si X, Li B, Wei G, et al. Loss of super-enhancer-regulated circRNA nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. (2019) 139:2857–76. doi: 10.1161/circulationaha.118.038361
42. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. (2014) 56:55–66. doi: 10.1016/j.molcel.2014.08.019
43. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. (2014) 15:409. doi: 10.1186/s13059-014-0409-z
44. Li X, Liu S, Zhang L, Issaian A, Hill RC, Espinosa S, et al. A unified mechanism for intron and exon definition and back-splicing. Nature. (2019) 573:375–80. doi: 10.1038/s41586-019-1523-6
45. Meng X, Li X, Zhang P, Wang J, Zhou Y, Chen M. Circular RNA: an emerging key player in RNA world. Brief Bioinform. (2017) 18:547–57. doi: 10.1093/bib/bbw045
46. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. (2015) 22:256–64. doi: 10.1038/nsmb.2959
47. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. (2018) 18:5–18. doi: 10.1038/nrc.2017.99
48. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. (2015) 160:1125–34. doi: 10.1016/j.cell.2015.02.014
49. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. doi: 10.1038/nature11928
50. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. (2014) 159:134–47. doi: 10.1016/j.cell.2014.09.001
51. Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. (2014) 15:9331–42. doi: 10.3390/ijms15069331
52. Tang L, Li P, Jang M, Zhu W. Circular RNAs and cardiovascular regeneration. Front Cardiovasc Med. (2021) 8:672600. doi: 10.3389/fcvm.2021.672600
53. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. (2018) 1087:67–79. doi: 10.1007/978-981-13-1426-1_6
54. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. (2016) 17:272–83. doi: 10.1038/nrg.2016.20
55. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. (2016) 96:1297–325. doi: 10.1152/physrev.00041.2015
56. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
57. Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. (2011) 147:370–81. doi: 10.1016/j.cell.2011.09.041
58. Zhong Q, Huang J, Wei J, Wu R. Circular RNA CDR1as sponges miR-7-5p to enhance E2F3 stability and promote the growth of nasopharyngeal carcinoma. Cancer Cell Int. (2019) 19:252. doi: 10.1186/s12935-019-0959-y
59. Zou Y, Zheng S, Deng X, Yang A, Xie X, Tang H, et al. The role of circular RNA CDR1as/ciRS-7 in regulating tumor microenvironment: A pan-cancer analysis. Biomolecules. (2019) 9:429. doi: 10.3390/biom9090429
60. Li J, Gao X, Zhang Z, Lai Y, Lin X, Lin B, et al. CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502-5p/KRAS and IGF2BP2/Myc axes. Mol Cancer. (2021) 20:138. doi: 10.1186/s12943-021-01444-1
61. Hall IF, Climent M, Quintavalle M, Farina FM, Schorn T, Zani S, et al. Circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ Res. (2019) 124:498–510. doi: 10.1161/CIRCRESAHA.118.314240
62. Gebauer F, Schwarzl T, Valcárcel J, Hentze MW. RNA-binding proteins in human genetic disease. Nat Rev Genet. (2021) 22:185–98. doi: 10.1038/s41576-020-00302-y
63. Okholm TLH, Sathe S, Park SS, Kamstrup AB, Rasmussen AM, Shankar A, et al. Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. (2020) 12:112. doi: 10.1186/s13073-020-00812-8
64. Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. (2017) 14:361–9. doi: 10.1080/15476286.2017.1279788
65. Schneider T, Hung LH, Schreiner S, Starke S, Eckhof H, Rossbach O, et al. CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep. (2016) 6:31313. doi: 10.1038/srep31313
66. Liu B, Yang G, Wang X, Liu J, Lu Z, Wang Q, et al. CircBACH1 (hsa_circ_0061395) promotes hepatocellular carcinoma growth by regulating p27 repression via HuR. J Cell Physiol. (2020) 235:6929–41. doi: 10.1002/jcp.v235.10
67. Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. (2019) 9:3526–40. doi: 10.7150/thno.32796
68. Sun J, Gu X, Wu N, Zhang P, Liu Y, Jiang S. Human antigen R enhances the epithelial-mesenchymal transition via regulation of ZEB-1 in the human airway epithelium. Respir Res. (2018) 19:109. doi: 10.1186/s12931-018-0805-0
69. Kullmann M, Göpfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5’UTR. Genes Dev. (2002) 16:3087–99. doi: 10.1101/gad.248902
70. Alpatov R, Lesch BJ, Nakamoto-Kinoshita M, Blanco A, Chen S, Stützer A, et al. A chromatin-dependent role of the fragile X mental retardation protein FMRP in the DNA damage response. Cell. (2014) 157:869–81. doi: 10.1016/j.cell.2014.03.040
71. Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. (2017) 3:17053. doi: 10.1038/nplants.2017.53
72. Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen W, et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol Cancer. (2020) 19:128. doi: 10.1186/s12943-020-01246-x
73. Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. (2013) 51:792–806. doi: 10.1016/j.molcel.2013.08.017
74. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. (1995) 268:415–7. doi: 10.1126/science.7536344
75. Perriman R, Ares M Jr. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. Rna. (1998) 4:1047–54. doi: 10.1017/S135583829898061X
76. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of circRNAs. Mol Cell. (2017) 66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021
77. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. (2020) 19:30. doi: 10.1186/s12943-020-1135-7
78. Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. (2017) 7:3842–55. doi: 10.7150/thno.19764
79. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. (2017) 24:357–70. doi: 10.1038/cdd.2016.133
80. Pouwels S, Sakran N, Graham Y, Leal A, Pintar T, Yang W, et al. Non-alcoholic fatty liver disease (NAFLD): a review of pathophysiology, clinical management and effects of weight loss. BMC Endocr Disord. (2022) 22:63. doi: 10.1186/s12902-022-00980-1
81. de Alwis NM, Day CP. Non-alcoholic fatty liver disease: the mist gradually clears. J Hepatol. (2008) 48 Suppl 1:S104–12. doi: 10.1016/j.jhep.2008.01.009
82. Day CP, James OF. Steatohepatitis: a tale of two “hits”? Gastroenterology. (1998) 114:842–5. doi: 10.1016/s0016-5085(98)70599-2
83. Guo XY, Chen JN, Sun F, Wang YQ, Pan Q, Fan JG. circRNA_0046367 prevents hepatoxicity of lipid peroxidation: an inhibitory role against hepatic steatosis. Oxid Med Cell Longev. (2017) 2017:3960197. doi: 10.1155/2017/3960197
84. Guo XY, Sun F, Chen JN, Wang YQ, Pan Q, Fan JG. circRNA_0046366 inhibits hepatocellular steatosis by normalization of PPAR signaling. World J Gastroenterol. (2018) 24:323–37. doi: 10.3748/wjg.v24.i3.323
85. Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. (2017) 14:397–411. doi: 10.1038/nrgastro.2017.38
86. Chen Y, Yuan B, Wu Z, Dong Y, Zhang L, Zeng Z. Microarray profiling of circular RNAs and the potential regulatory role of hsa_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene. (2017) 629:35–42. doi: 10.1016/j.gene.2017.07.078
87. Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. (2018) 68:1214–27. doi: 10.1016/j.jhep.2018.01.012
88. Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, et al. Circular RNA sequencing identifies circASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. (2020) 72:906–22. doi: 10.1002/hep.31068
89. Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, et al. A noncoding regulatory RNAs network driven by circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. (2020) 71:130–47. doi: 10.1002/hep.30795
90. Wang H, Chen W, Jin M, Hou L, Chen X, Zhang R, et al. CircSLC3A2 functions as an oncogenic factor in hepatocellular carcinoma by sponging miR-490-3p and regulating PPM1F expression. Mol Cancer. (2018) 17:165. doi: 10.1186/s12943-018-0909-7
91. Bai N, Peng E, Qiu X, Lyu N, Zhang Z, Tao Y, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res. (2018) 37:172. doi: 10.1186/s13046-018-0838-8
92. Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. (2018) 9:175. doi: 10.1038/s41419-017-0204-3
93. Li S, Weng J, Song F, Li L, Xiao C, Yang W, et al. Circular RNA circZNF566 promotes hepatocellular carcinoma progression by sponging miR-4738-3p and regulating TDO2 expression. Cell Death Dis. (2020) 11:452. doi: 10.1038/s41419-020-2616-8
94. Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B promotes glycolysis and Malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. (2019) 70:1298–316. doi: 10.1002/hep.30671
95. Wang ZY, Zhu Z, Wang HF, Qin B, Liu J, Yao XH, et al. Downregulation of circDYNC1H1 exhibits inhibitor effect on cell proliferation and migration in hepatocellular carcinoma through miR-140-5p. J Cell Physiol. (2019) 234:17775–85. doi: 10.1002/jcp.v234.10
96. Song LN, Qiao GL, Yu J, Yang CM, Chen Y, Deng ZF, et al. Hsa_circ_0003998 promotes epithelial to mesenchymal transition of hepatocellular carcinoma by sponging miR-143-3p and PCBP1. J Exp Clin Cancer Res. (2020) 39:114. doi: 10.1186/s13046-020-01576-0
97. Vromman M, Vandesompele J, Volders PJ. Closing the circle: current state and perspectives of circular RNA databases. Brief Bioinform. (2021) 22:288–97. doi: 10.1093/bib/bbz175
98. Meng L, Wu H, Wu J, Ding P, He J, Sang M, et al. Mechanisms of immune checkpoint inhibitors: insights into the regulation of circular RNAS involved in cancer hallmarks. Cell Death Dis. (2024) 15:3. doi: 10.1038/s41419-023-06389-5
99. Li D, Zhang J, Yang J, Wang J, Zhang R, Li J, et al. CircMTO1 suppresses hepatocellular carcinoma progression via the miR-541-5p/ZIC1 axis by regulating Wnt/β-catenin signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis. (2021) 13:12. doi: 10.1038/s41419-021-04464-3
100. Huang G, Liang M, Liu H, Huang J, Li P, Wang C, et al. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell Death Dis. (2020) 11:1065. doi: 10.1038/s41419-020-03276-1
101. Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, et al. N(6)-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer. (2020) 19:163. doi: 10.1186/s12943-020-01281-8
102. Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/β-catenin pathway. Arch Biochem Biophys. (2019) 661:196–202. doi: 10.1016/j.abb.2018.11.020
103. Zhou Y, Tang W, Zhuo H, Zhu D, Rong D, Sun J, et al. Cancer-associated fibroblast exosomes promote chemoresistance to cisplatin in hepatocellular carcinoma through circZFR targeting signal transducers and activators of transcription (STAT3)/nuclear factor -kappa B (NF-κB) pathway. Bioengineered. (2022) 13:4786–97. doi: 10.1080/21655979.2022.2032972
104. Ji J, Tang J, Ren P, Cai W, Shen M, Wang Q, et al. Downregulation of circLIFR exerts cancer-promoting effects on hepatocellular carcinoma in vitro. Front Genet. (2022) 13:986322. doi: 10.3389/fgene.2022.986322
105. Wu F, Sun G, Zheng W, Tang W, Cheng Y, Wu L, et al. circCORO1C promotes the proliferation and metastasis of hepatocellular carcinoma by enhancing the expression of PD-L1 through NF-κB pathway. J Clin Lab Anal. (2021) 35:e24003. doi: 10.1002/jcla.v35.12
106. Lin Q, Ling YB, Chen JW, Zhou CR, Chen J, Li X, et al. Circular RNA circCDK13 suppresses cell proliferation, migration and invasion by modulating the JAK/STAT and PI3K/AKT pathways in liver cancer. Int J Oncol. (2018) 53:246–56. doi: 10.3892/ijo.2018.4371
107. Tan Y, Du B, Zhan Y, Wang K, Wang X, Chen B, et al. Antitumor effects of circ-EPHB4 in hepatocellular carcinoma via inhibition of HIF-1α. Mol Carcinog. (2019) 58:875–86. doi: 10.1002/mc.22976
108. Guo Z, Xie Q, Wu Y, Mo H, Zhang J, He G, et al. Aberrant expression of circular RNA DHPR facilitates tumor growth and metastasis by regulating the RASGEF1B/RAS/MAPK axis in hepatocellular carcinoma. Cell Oncol (Dordr). (2023) 46:1333–50. doi: 10.1007/s13402-023-00814-9
109. Xu L, Feng X, Hao X, Wang P, Zhang Y, Zheng X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. (2019) 38:98. doi: 10.1186/s13046-019-1041-2
110. Gu Y, Wang Y, He L, Zhang J, Zhu X, Liu N, et al. Circular RNA circIPO11 drives self-renewal of liver cancer initiating cells via Hedgehog signaling. Mol Cancer. (2021) 20:132. doi: 10.1186/s12943-021-01435-2
111. Liu B, Shen H, He J, Jin B, Tian Y, Li W, et al. Cytoskeleton remodeling mediated by circRNA-YBX1 phase separation suppresses the metastasis of liver cancer. Proc Natl Acad Sci U S A. (2023) 120:e2220296120. doi: 10.1073/pnas.2220296120
112. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. (2020) 19:110. doi: 10.1186/s12943-020-01222-5
113. Lei B, Zhou J, Xuan X, Tian Z, Zhang M, Gao W, et al. Circular RNA expression profiles of peripheral blood mononuclear cells in hepatocellular carcinoma patients by sequence analysis. Cancer Med. (2019) 8:1423–33. doi: 10.1002/cam4.2019.8.issue-4
114. Zhu K, Zhan H, Peng Y, Yang L, Gao Q, Jia H, et al. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis. (2020) 41:296–302. doi: 10.1093/carcin/bgz154
115. Yang C, Dong Z, Hong H, Dai B, Song F, Geng L, et al. circFN1 mediates sorafenib resistance of hepatocellular carcinoma cells by sponging miR-1205 and regulating E2F1 expression. Mol Ther Nucleic Acids. (2020) 22:421–33. doi: 10.1016/j.omtn.2020.08.039
116. Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang L, et al. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin Chim Acta. (2019) 492:37–44. doi: 10.1016/j.cca.2019.02.001
117. Yao T, Chen Q, Shao Z, Song Z, Fu L, Xiao B. Circular RNA 0068669 as a new biomarker for hepatocellular carcinoma metastasis. J Clin Lab Anal. (2018) 32:e22572. doi: 10.1002/jcla.2018.32.issue-8
118. Jiang Z, Shen L, Wang S, Wu S, Hu Y, Guo J, et al. Hsa_circ_0028502 and hsa_circ_0076251 are potential novel biomarkers for hepatocellular carcinoma. Cancer Med. (2019) 8:7278–87. doi: 10.1002/cam4.v8.17
119. Wang M, Yu F, Li P. Circular RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers (Basel). (2018) 10(8):258. doi: 10.3390/cancers10080258
120. Yao R, Zou H, Liao W. Prospect of circular RNA in hepatocellular carcinoma: A novel potential biomarker and therapeutic target. Front Oncol. (2018) 8:332. doi: 10.3389/fonc.2018.00332
121. Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. (2020) 6:319–36. doi: 10.1016/j.trecan.2020.01.012
122. Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl). (2017) 95:1179–89. doi: 10.1007/s00109-017-1582-9
123. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stabl e in exosomes: a promising biomarker for cancer diagnosis. Cell Res. (2015) 25:981–4. doi: 10.1038/cr.2015.82
124. Shi X, Wang B, Feng X, Xu Y, Lu K, Sun M. circRNAs and exosomes: A mysterious frontier for human cancer. Mol Ther Nucleic Acids. (2020) 19:384–92. doi: 10.1016/j.omtn.2019.11.023
125. Cheng J, Meng J, Zhu L, Peng Y. Exosomal noncoding RNAs in Glioma: biological functions and potential clinical applications. Mol Cancer. (2020) 19:66. doi: 10.1186/s12943-020-01189-3
126. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. (2019) 10:4695. doi: 10.1038/s41467-019-12651-2
127. Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. (2018) 34:267–74. doi: 10.1016/j.ebiom.2018.07.036
128. Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, et al. and circular RNA in human saliva. Clin Chem. (2015) 61:221–30. doi: 10.1373/clinchem.2014.230433
129. Cui X, Wang J, Guo Z, Li M, Li M, Liu S, et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer. (2018) 17:123. doi: 10.1186/s12943-018-0877-y
130. Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. (2019) 16:503–14. doi: 10.1038/s41569-019-0185-2
131. Li S, Han L. Circular RNAs as promising biomarkers in cancer: detection, function, and beyond. Genome Med. (2019) 11:15. doi: 10.1186/s13073-019-0629-7
132. Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y, et al. Circular RNAs in Cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. (2019) 18:90. doi: 10.1186/s12943-019-1002-6
133. Zhang Y, Liang W, Zhang P, Chen J, Qian H, Zhang X, et al. Circular RNAs: emerging cancer biomarkers and targets. J Exp Clin Cancer Res. (2017) 36:152. doi: 10.1186/s13046-017-0624-z
134. Naeli P, Pourhanifeh MH, Karimzadeh MR, Shabaninejad Z, Movahedpour A, Tarrahimofrad H, et al. Circular RNAs and gastrointestinal cancers: Epigenetic regulators with a prognostic and therapeutic role. Crit Rev Oncol Hematol. (2020) 145:102854. doi: 10.1016/j.critrevonc.2019.102854
135. Shan C, Zhang Y, Hao X, Gao J, Chen X, Wang K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol Cancer. (2019) 18:136. doi: 10.1186/s12943-019-1069-0
136. Pardini B, Sabo AA, Birolo G, Calin GA. Noncoding RNAs in extracellular fluids as cancer biomarkers: the new frontier of liquid biopsies. Cancers (Basel). (2019) 11(8):1170. doi: 10.3390/cancers11081170
137. Ma J, Yang BB. A novel prognostic prediction tool for postoperative recurrence in patients with stage II/III colon cancer. Cancer Commun (Lond). (2019) 39:52. doi: 10.1186/s40880-019-0397-1
138. Yao Z, Xu R, Yuan L, Xu M, Zhuang H, Li Y, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. (2019) 10:945. doi: 10.1038/s41419-019-2176-y
139. Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. (2018) 9:1091. doi: 10.1038/s41419-018-1132-6
140. Lee T, Paquet M, Larsson O, Pelletier J. Tumor cell survival dependence on the DHX9 DExH-box helicase. Oncogene. (2016) 35:5093–105. doi: 10.1038/onc.2016.52
141. Han T, Chen L, Li K, Hu Q, Zhang Y, You X, et al. Significant CircRNAs in liver cancer stem cell exosomes: mediator of Malignant propagation in liver cancer? Mol Cancer. (2023) 22:197. doi: 10.1186/s12943-023-01891-y
142. Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. (2019) 38:2844–59. doi: 10.1038/s41388-018-0619-z
143. Lai Z, Wei T, Li Q, Wang X, Zhang Y, Zhang S. Exosomal circFBLIM1 Promotes Hepatocellular Carcinoma Progression and Glycolysis by Regulating the miR-338/LRP6 Axis. Cancer Biother Radiopharm. (2023) 38:674–83. doi: 10.1089/cbr.2020.3564
144. Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. (2020) 39:20. doi: 10.1186/s13046-020-1529-9
145. Huang C, Yu W, Wang Q, Huang T, Ding Y. CircANTXR1 contributes to the Malignant progression of hepatocellular carcinoma by promoting proliferation and metastasis. J Hepatocell Carcinoma. (2021) 8:1339–53. doi: 10.2147/jhc.S317256
146. Luo Y, Liu F, Gui R. High expression of circulating exosomal circAKT3 is associated with higher recurrence in HCC patients undergoing surgical treatment. Surg Oncol. (2020) 33:276–81. doi: 10.1016/j.suronc.2020.04.021
147. Sun XH, Wang YT, Li GF, Zhang N, Fan L. Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int. (2020) 20:226. doi: 10.1186/s12935-020-01302-y
148. Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. (2020) 475:119–28. doi: 10.1016/j.canlet.2020.01.022
149. Conn VM, Chinnaiyan AM, Conn SJ. Circular RNA in cancer. Nat Rev Cancer. (2024) 24:597–613. doi: 10.1038/s41568-024-00721-7
150. Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. (2019) 40:432–45. doi: 10.1016/j.ebiom.2018.12.062
151. Su Y, Lv X, Yin W, Zhou L, Hu Y, Zhou A, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY). (2019) 11:8183–203. doi: 10.18632/aging.102312
152. Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X, et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. (2020) 5:298. doi: 10.1038/s41392-020-00375-5
153. Zhang T, Jing B, Bai Y, Zhang Y, Yu H. Circular RNA circTMEM45A acts as the sponge of microRNA-665 to promote hepatocellular carcinoma progression. Mol Ther Nucleic Acids. (2020) 22:285–97. doi: 10.1016/j.omtn.2020.08.011
154. Liu L, Gu M, Ma J, Wang Y, Li M, Wang H, et al. CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol Cancer. (2022) 21:149. doi: 10.1186/s12943-022-01619-4
155. Li J, Hu ZQ, Yu SY, Mao L, Zhou ZJ, Wang PC, et al. CircRPN2 inhibits aerobic glycolysis and metastasis in hepatocellular carcinoma. Cancer Res. (2022) 82:1055–69. doi: 10.1158/0008-5472.CAN-21-1259
156. Zhou L, Wang Q, Hou J, Wu X, Wang L, Chen X. Upregulation of hsa_circ_0002003 promotes hepatocellular carcinoma progression. BMC Cancer. (2023) 23:611. doi: 10.1186/s12885-023-11086-9
Keywords: liver cancer, circRNA, diagnostic biomarkers, therapeutic targets, cancer
Citation: Wang J, Zhang C, Zhang Y, Guo J, Xie C, Liu Y, Chen L and Ma L (2025) Circular RNA in liver cancer research: biogenesis, functions, and roles. Front. Oncol. 15:1523061. doi: 10.3389/fonc.2025.1523061
Received: 07 November 2024; Accepted: 11 March 2025;
Published: 28 March 2025.
Edited by:
Liyong Zeng, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Naren Li, Baylor College of Medicine, United StatesCopyright © 2025 Wang, Zhang, Zhang, Guo, Xie, Liu, Chen and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lidian Chen, Y2xkQGZqdGNtLmVkdS5jbg==; Liangliang Ma, bWxsMDcwOEBmb3htYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.