- 1School of Laboratory Medicine, Chengdu Medical College, Chengdu, Sichuan, China
- 2School of Dental Medicine, Dalian University, Dalian, Liaoning, China
- 3Department of Laboratory Medicine and Sichuan Provincial Key Laboratory for Human Disease Gene Study, Sichuan Provincial People’s Hospital, University of Electronic Science and Technology of China, Chengdu, China
- 4Department of Hematopathology, The Second Affiliated Hospital of Chengdu Medical College, China National Nuclear Corporation 416 Hospital, Chengdu, Sichuan, China
Multiple Myeloma (MM) is the second most common malignancy of the hematopoietic system, accounting for approximately 10% of all hematological malignancies, and currently, there is no complete cure. Existing research indicates that exosomal long non-coding RNAs (lncRNAs) play a crucial regulatory role in the initiation and progression of tumors, involving various interactions such as lncRNA-miRNA, lncRNA-mRNA, and lncRNA-RNA binding proteins (RBP). Despite the significant clinical application potential of exosomal lncRNAs, research in this area still faces challenges due to their low abundance and technical limitations. To our knowledge, this review is the first to comprehensively integrate and elucidate the three mechanisms of action of exosomal lncRNAs in MM, and to propose potential therapeutic targets and clinical cases based on these mechanisms. We highlight the latest advancements in the potential of exosomal lncRNAs as biomarkers and therapeutic targets, offering not only a comprehensive analysis of the role of exosomal lncRNAs in MM but also new perspectives and methods for future clinical diagnosis and treatment of multiple myeloma.
1 Introduction
Multiple myeloma (MM) predominantly affects middle-aged and elderly populations and is characterized by malignant proliferation of bone marrow plasma cells and abnormal secretion of immunoglobulins and their peptide chains (1). It ranks second among hematologic malignancies, comprising approximately 10% of all hematologic tumors in high-income countries (2). MM-associated syndromes include recurrent infections, anemia, bone pain, bone lesions, limb weakness, and kidney disease (3). Despite significant improvements in survival rates due to advances in autologous hematopoietic stem cell transplantation and the emergence of new drugs such as bortezomib, carfilzomib, lenalidomide, and pomalidomide (4), patients with MM often experience relapse, develop drug resistance post-treatment, and remain incurable. With the rapid development of novel detection technologies, such as fluorescence in situ hybridization, high-throughput sequencing, gene chips, and quantitative polymerase chain reaction, numerous studies have indicated the crucial role of long non-coding RNAs (lncRNAs) in the pathogenesis and progression of MM (5). Investigating the formation and evolution of myeloma from the molecular and genetic perspectives can provide a key theoretical foundation and practical value for its prevention and treatment.
Extracellular vesicles (EVs) serve as bioactive transport substances released by various cell types and play an important role in the occurrence and development of various diseases (6–8). Recent domestic and international studies have found correlations between the types and quantities of EVs and the disease status and prognosis of MM patients, providing new insights into the diagnosis, prognostic assessment, and targeted therapy of MM (9, 10). In recent years, EVs have shown promising potential in the pathogenesis, diagnosis, prognosis, and treatment of MM (10).
Long noncoding RNAs (lncRNAs) are RNAs that are longer than 200 nucleotides, possess diverse regulatory functions and are transcribed by RNA polymerase II. They undergo splicing and processing to form mature lncRNAs, which, however, are not translated into proteins. lncRNAs regulate genes at the epigenetic, transcriptional, and post-transcriptional levels (11, 12). Increasing evidence suggests that dysregulation of lncRNA function is implicated in various diseases, particularly cancer (5, 13). LncRNAs in EVs associated with MM primarily regulate the expression of genes and proteins within tumor cells through mechanisms such as “competitive endogenous RNA (ceRNA)”, “RNA duplex,” and “lncRNA-RNA-binding proteins (RBP),” thereby affecting the occurrence, development, and drug resistance of MM.
In recent years, there has been a plethora of research on the role of extracellular vesicles (EVs) in multiple myeloma (MM); however, the majority of studies have focused on microRNAs (miRNAs) within EVs, with long non-coding RNAs (lncRNAs) constituting only a small fraction. Yet, due to their sheer number, diverse regulatory mechanisms, and significant functionality, lncRNAs hold the potential to become the largest repository of targets for the development of gene therapy drugs. Moreover, although some exosomal lncRNAs associated with tumor development have been identified, there are currently no anticancer drugs targeting these exosomal lncRNAs on the market. Additionally, membrane vesicles derived from cells, including exosomes and microvesicles, are considered ideal delivery systems due to their low antigenicity, low cytotoxicity, and ability to bypass endocytic pathways and phagocytosis. The biodistribution of these membrane vesicles can be tailored through their specific composition and cells of origin to meet particular needs. Therefore, there is an urgent need to identify exosomal lncRNAs associated with disease biology, to delve into their cellular functions and biological mechanisms, in order to establish their basis as therapeutic targets for multiple myeloma. This review aims to summarize the mechanisms and clinical significance of exosomal lncRNAs in MM, emphasizing their mechanisms and the latest advancements, and to reveal their potential as effective therapeutic targets and diagnostic biomarkers, in the hope of providing new therapeutic strategies and clinical applications for MM.
2 Role of exosomes in the pathogenesis of MM
EVs play crucial roles in MM pathogenesis and have received significant attention in cancer research. Recent studies revealed various aspects of EV biogenesis, regulation, and function in cancer cells. Han et al. (14) have provided a comprehensive overview of EV biogenesis mechanisms, emphasizing the therapeutic significance of targeting EV biogenesis in cancer treatment. Guo et al. (15) focused on the impact of EVs on the formation of premetastatic niches in tumors, highlighting their effects on inflammation, immune responses, and angiogenesis. Yang et al. (16) explored the link between EVs and metabolic reprogramming in tumors and offered new insights into cancer prevention and treatment. Additionally, Paskeh et al. (17) discussed the novel roles of EVs in cancer progression and reshaping the tumor microenvironment, emphasizing their therapeutic value in cancer treatment. Zhang et al. (18) have designed neutrophil-derived EV-like vesicles for targeted cancer therapy, providing a unique approach for precise cancer treatment using EVs. Collectively, research advancements in tumor-derived EVs indicate that these EVs play multifaceted roles in cancer progression, microenvironment remodeling, and targeted therapy. These findings collectively enhance our understanding of the complex interactions between EVs and tumors, and pave the way for innovative strategies for cancer diagnosis and treatment.
Numerous studies have demonstrated that EVs released by the host or cancer cells are involved in the initiation, growth, infiltration, and metastasis of tumors. Furthermore, EVs play a dual role in the communication between immune and cancer cells by regulating tumor immune responses. EVs contain proteins, cytokines, lipids, miRNAs, lncRNAs, and circular RNAs (circRNAs) that play crucial roles in intercellular communication and are involved in various physiological and pathological processes (10, 19). Therefore, EVs play significant roles in the pathogenesis of MM and serve as novel biomarkers and therapeutic tools in MM (20). Different types of cells can release different types of EVs that play important roles in the occurrence and development of various diseases, with varying levels in patients with MM and healthy individuals. Roccaro et al. demonstrated that BMSC-EXs derived from patients with MM promote tumor growth, whereas BMSC-EXs obtained from healthy donors inhibit MM cell proliferation (21, 22). Wang et al. (23) demonstrated that EVs derived from bone marrow adipocytes lead to MM drug resistance by inhibiting chemotherapy-induced tumor cell apoptosis. Liu et al. (24) showed that EVs from MM cells inhibit osteoblast differentiation and enhance IL-6 secretion, leading to poor bone health. Additionally, Frassanito et al. (25) emphasized the role of MM cell-mediated, EV-mediated miRNA regulation in altering the bone marrow microenvironment and affecting disease progression. Long et al. (26) demonstrated in vitro experiments using cell lines that BMSC-derived EV miR-182 can be transferred to MM cells to regulate their proliferation, metastasis, and resistance to carfilzomib. Sun et al. (27) reported that cancer-associated fibroblast (CAFs)-derived EV miR-21 entering MM endothelial cells (MMECs) promotes MM proliferation, invasion, and vascular formation. Liu et al. (28) revealed that C6-ceramide treatment inhibited the proangiogenic activity of MM EVs, providing insights into potential therapeutic strategies targeting the EV pathway. EVs have a significant impact on the pathogenesis of MM, owing to their influence on the bone marrow microenvironment and their involvement in regulating immune responses. Understanding the complex interactions of EVs in MM may facilitate the development of new diagnostic and therapeutic strategies.
3 Role of lncRNA in the pathogenesis of MM
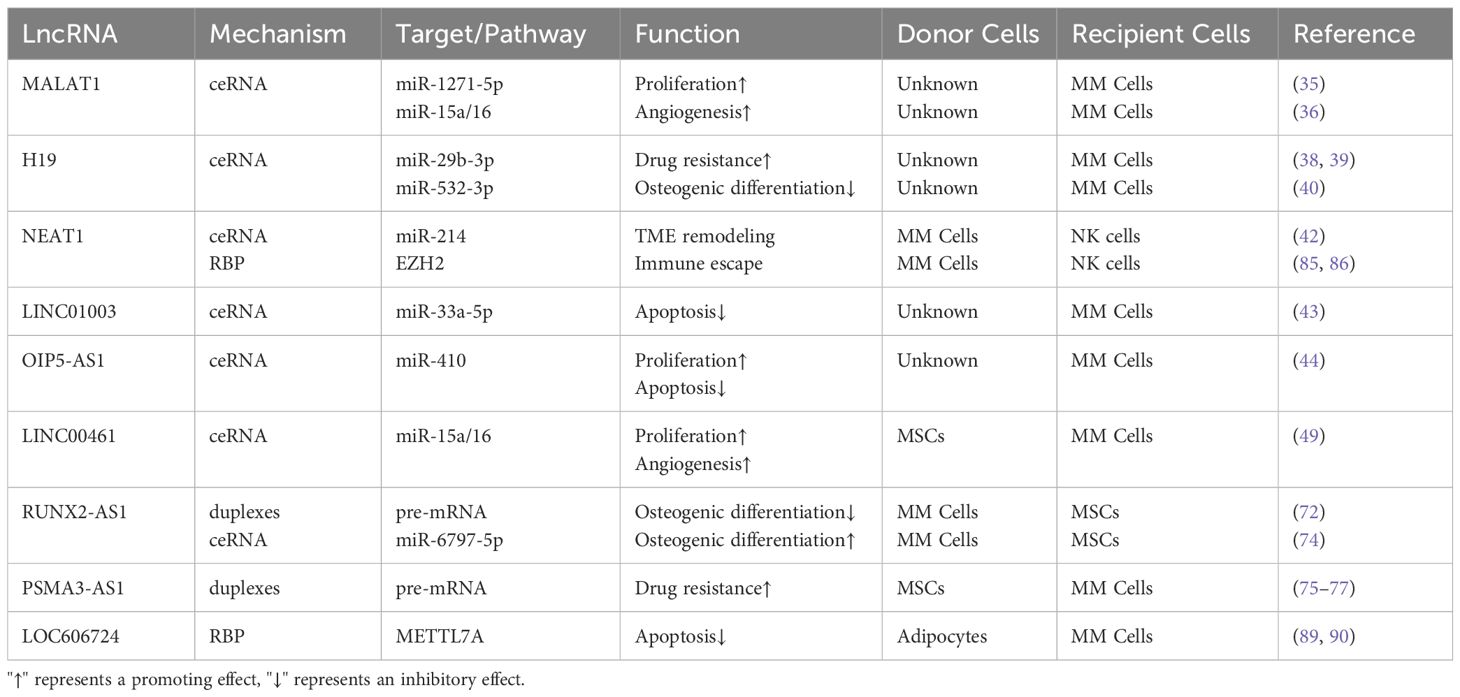
Recent studies have shown that lncRNAs participate in the occurrence, progression, and metastasis of tumors (29). First, lncRNAs can regulate the gene expression by interacting with other RNAs such as mRNA and miRNAs, among which lncRNAs act as large “sponges” that bind to miRNAs and form ceRNA networks (30, 31). In this network, lncRNAs attract miRNAs, reducing the available concentration of miRNAs and thereby decreasing miRNA binding to the target mRNA, ultimately leading to increased stability of the target mRNA and higher transcriptional levels. Second, lncRNAs can interact with proteins or DNA, affecting the chromatin structure and transcriptional regulation (32, 33). lncRNAs regulate the chromatin status by interacting with chromatin-modifying enzymes or transcription factors, thereby influencing the target gene expression. Additionally, lncRNAs can serve as molecular bridges, regulate protein-protein interactions, and affect cell signaling and gene expression (33). These modes of action indicate that lncRNAs participate in key processes of tumor cell transformation, proliferation, apoptosis, and migration in multiple ways. We have organized the lncRNAs mentioned in this article that affect the occurrence and development of multiple myeloma into a table, as shown in Table 1.
lncRNAs promote the occurrence and development of MM by promoting MM cell proliferation and invasion, maintaining the cell cycle, drug resistance, inhibiting osteogenesis, and reshaping the TME. lncRNAs can directly mediate protein expression, activate signaling pathways, or act as ceRNAs to regulate the miRNA expression by acting on downstream genes, thereby promoting the occurrence and development of MM (34). Conversely, lncRNAs inhibit the occurrence and development of MM by inhibiting the MM cell proliferation and cell cycle arrest and promoting cell apoptosis.
Recent studies have shown that lncRNAs can directly promote the proliferation, invasion, and regulation of the cell cycle, inhibition of bone formation, drug resistance, and TME remodeling of MM tumor cells by targeting miRNAs, thereby promoting the occurrence and development of MM and leading to a poor prognosis. Liu et al. (35) demonstrated that MALAT1 regulates the expression of SOX13 by targeting miR-1271-5p through a ceRNA mechanism, thereby promoting MM cell proliferation, invasion, and glycolytic ability. In addition, lncRNA MALAT1 is highly expressed in MM and acts as a competitive endogenous RNA for microRNA-15a/16 to promote the expression of vascular endothelial growth factor A (VEGFA), facilitating angiogenesis in MM (36). Shen et al. (37) demonstrated that knockdown of lncRNA AL928768.3 significantly inhibited the MM cell proliferation and colony formation, induced MM cell cycle arrest in the G0/G1 phase, inhibited the expression of CDK2 and CCND1, and promoted P21 expression. Additionally, lncRNA H19 is reported to significantly upregulate in the serum of MM patients. H19 indirectly regulates the MCL-1/Akt pathway by targeting miR-29b-3p to induce MM resistance to bortezomib or doxorubicin (38, 39). In a recent study, H19 was shown to act as a sponge for miR-532-3p to upregulate E2F7 and inhibit tumor suppressor gene (PTEN) epigenetics. Guo et al. (40) concluded after in vivo experiments that H19 disrupts the balance between osteogenesis and osteolysis through Akt/mTOR signal transduction, leading to reduced osteogenic activity and increased osteoclast activity, promoting the occurrence and development of MM. Additionally, EV-derived lncRNA RUNX2-AS1 can be delivered to mesenchymal stem cells (MSCs) through vesicle transfer, inhibiting their osteogenic activity (41). lncRNAs can also participate in the remodeling of the TME in MM cells. For example, lncRNA NEAT1, highly expressed, inhibits miR-214, upregulates the expression and release of B7-H3, promotes M2 macrophage polarization, and accelerates MM progression (42).
Few studies have shown that lncRNAs can also inhibit the occurrence and development of MM through various pathways, such as cell cycle arrest and the promotion of cell apoptosis. Using the dual-luciferase reporter gene method, Wu et al. (43) demonstrated that LINC01003 inhibits the MM cell vitality and adhesion by upregulating the expression of the miR-33a-5p target gene, PIM1, as a sponge for miR-33a-5p, promoting MM cell apoptosis. In addition, OIP5-AS1 negatively regulates miR-410, and miR-410 further directly targets KLF10, thereby negatively regulating KLF10 expression. KLF10 mediates the PTEN/AKT signaling pathway, forming the OIP5-AS1-miR-410-KLF10/PTEN/AKT signaling axis. Compared to normal tissues, the lncRNA OIP5-AS1 is downregulated in MM tissues and miR-410 expression is upregulated, promoting MM cell proliferation, cell cycle progression, and inhibition of cell apoptosis (44). In addition, Jiang et al. (45) downregulated the expression of the lncRNA IRAIN in vitro, promoting MM cell proliferation, and revealing its potential as a new therapeutic target for MM. These lncRNAs are expected to become new targets and biomarkers for MM treatment with broad prospects for cancer treatment, as shown in Figure 1.
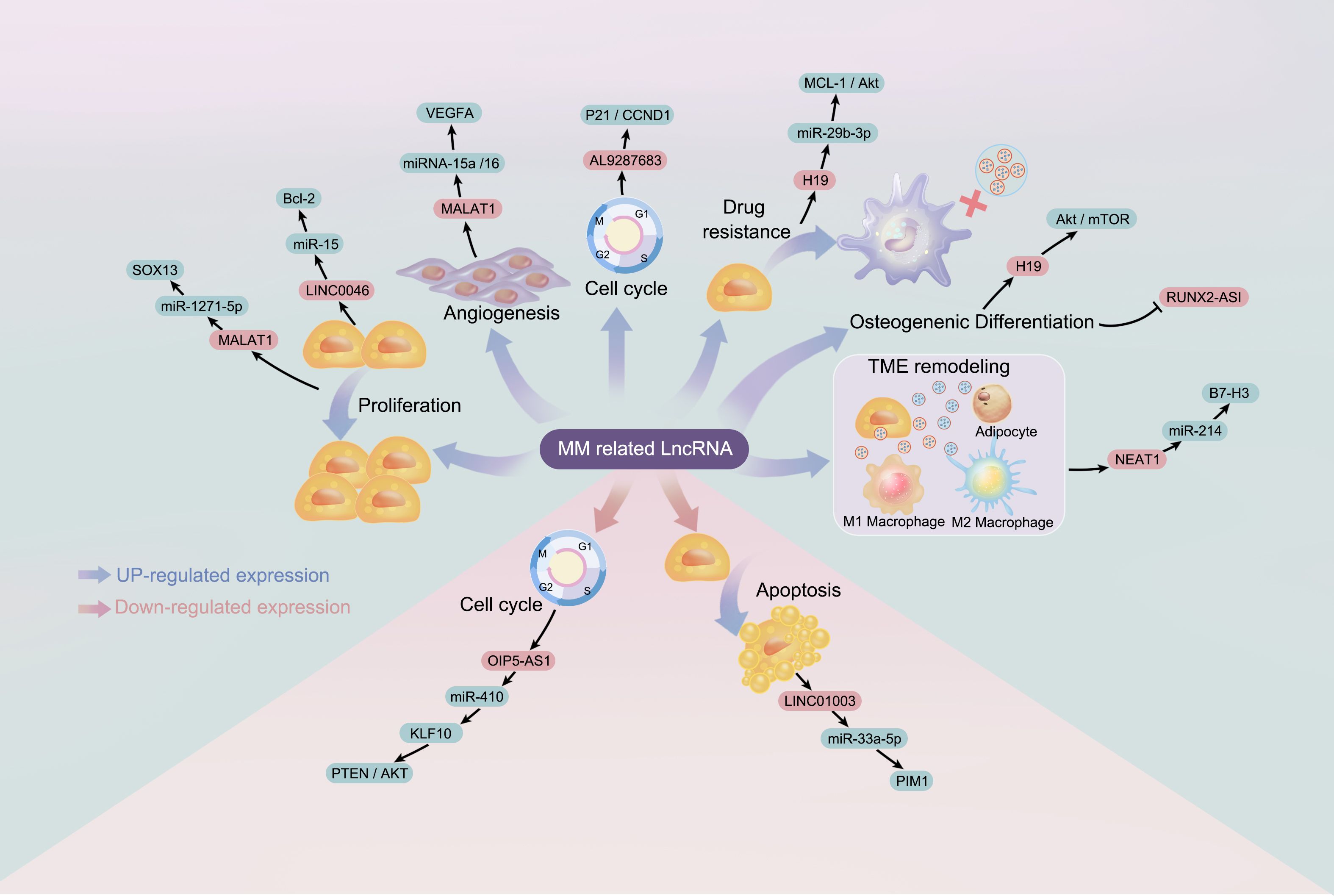
Figure 1. The regulatory role of long non-coding RNA (lncRNA) in the pathogenesis of multiple myeloma: lncRNA refers to long non-coding RNA; miRNA refers to microRNA.
4 Role of exosomal lncRNA in MM
Exosomes associated with MM mostly carry proteins and RNA, including non-coding genetic material, which are closely related to various biological processes involved in the disease. Currently, miRNAs are the most extensively studied substances in exosomes. However, in recent years, there has been growing interest in the lncRNA, both domestically and internationally, especially in the field of MM. lncRNAs are excellent biomarkers, and their detection in exosomes may have significant implications for patients with MM. In addition, exosomes can be viewed as potential drug carriers and are promising candidates for clinical therapies.
The role of lncRNAs in the pathogenesis of MM has attracted increasing attention because of their potential impact on the disease process. Exosomal lncRNAs have been found to be involved in various key mechanisms of MM, such as drug resistance, immune escape, and tumor progression (23, 41, 46). For instance, Wang et al. elucidated the inducing effect of m6A methylation in adipocyte-derived exosomal lncRNAs on drug resistance in MM, providing insights into previously unexplored mechanisms exacerbating MM drug resistance and suggesting potential strategies to enhance therapeutic efficacy (23). Their work also revealed the role of the exosomal lncRNA NEAT1 in inhibiting NK cell activity and promoting the immune escape of MM cells, offering a new therapeutic potential for MM by targeting exosomal lncRNAs (47). Long noncoding RNA 00461 (LINC00461) is an important member of the lncRNA family that plays a role in the occurrence and development of various cancers. Knockdown of LINC00461 enhances the antitumor effects of dexamethasone and promotes apoptosis in MM cells. Similar effects were observed in studies involving MSCs, and exosomes derived from MSCs promoted MM progression of MM, indicating a possible association with LINC00461 (48). Furthermore, other studies have demonstrated the broader relevance of exosomal lncRNAs in cancer biology and clinical management, as exosomal lncRNAs promote proliferation, metastasis, and acquisition of drug resistance in various cancers, including MM (36, 49). These findings collectively underscore the importance of exosomal lncRNAs in the pathogenesis of MM and highlight their potential as diagnostic markers and therapeutic targets for disease management. Building on this foundation, further analyses and discussions on the mechanisms of action of exosomal lncRNAs in MM will be conducted.
4.1 “ceRNA”
ceRNA networks have attracted extensive research interest in various cancers, including gastric cancer, intervertebral disc degeneration, epithelial ovarian cancer, cervical cancer, and hepatocellular carcinoma (50–54). Different RNA transcripts compete for miRNA response elements to regulate free miRNA expression, thereby facilitating mutual regulation. lncRNAs and miRNAs are interrelated in cancer, and lncRNA-mRNA-miRNA networks have been constructed in MM (30, 55–60). These findings provide new hope for the diagnosis, prognosis, and treatment of MM, and pave the way for the development of precision medicine. lncRNAs regulate MM progression through a ceRNA mechanism, demonstrating their potential clinical applications and biomarker values (36). Exosomal lncRNAs are considered therapeutic targets and biomarkers for malignant tumors, aiding in diagnosis, prognostic assessment, and drug treatment research. The ceRNA mechanism is illustrated in Figure 2.
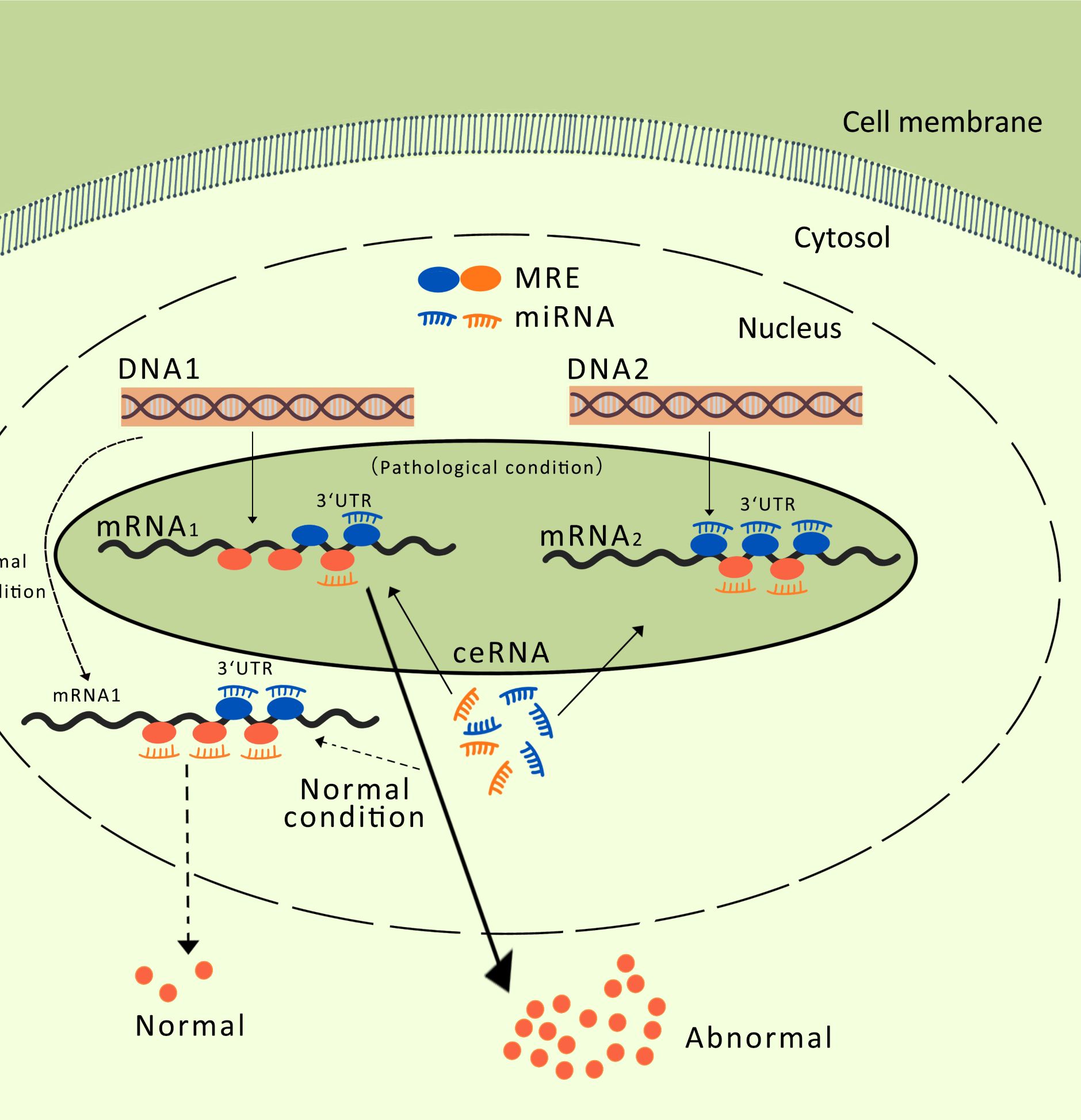
Figure 2. MREs, miRNA Response Elements. The competitive endogenous RNA mechanism: When an RNA2 molecule binds miRNA through a miRNA binding site, it can prevent miRNA from binding to other RNAs that share the same binding site, thereby reducing the inhibitory effect of miRNA on those other RNAs (such as RNA1), resulting in an increased expression level of these other RNAs (RNA1). This interaction, regulated by miRNA, creates a complex regulatory network that impacts the balance and regulation of gene expression.
Exosomal LINC00461 promotes the proliferation of MM cells. MSCs, a major member of the stem cell family, play crucial roles in tissue repair, cancer treatment, and immune regulation and are widely used in the study and treatment of various human diseases (61). Exosomal transfer of regulatory RNAs mediated by MSCs and MM cell sources is a key feature of cancer cell formation, promoting the tumor microenvironment, and regulating MM cell proliferation, spread, invasion, and drug resistance (62, 63). The role of lncRNAs in MM proliferation and progression is increasingly being recognized, with one lncRNA, LINC00461, promoting MM cell proliferation (64). LINC00461 is located in the intergenic region between the human chromosome 5 protein-coding genes MEF2C and TMEM161B. It is a ceRNA associated with 18 miRNAs and is overexpressed in respiratory, digestive, urinary, nervous system, and hematological malignancies (65). Deng et al. (49) demonstrated that the expression of LINC00461 in plasma cells from patients with MM was significantly higher in MSCs from adjacent tissues than in those from the control group. Moreover, LINC00461 is enriched in exosomes derived from MSCs of adjacent tissues and transferred to MM cells, affecting their proliferative ability. In gene knockout experiments, LINC00461 knockdown resulted in MM cell apoptosis and G1 cell cycle arrest, significantly inhibiting cell proliferation and migration. Overall, MSC-derived LINC00461 plays an important role in regulating MM cell proliferation, and its high expression significantly correlates with patient prognosis and poor survival.
LINC00461 acts as a ceRNA to regulate the miRNA expression. RNA structure analysis showed that miR-15a and miR-16 had two binding sites for each phenotype of LINC00461. In MS2-RIP radiolabeling immunoprecipitation assays, compared to the empty vector MS2, miR-15a/16 directly bound to LINC00461 and was enriched in MS2-LINC00461. Moreover, in cells overexpressing LINC00461, the expression levels of miR-15a and miR-16 were significantly lower than those in control group (49). In conclusion, exosomal LINC00461 has a sponge effect on miR-15a and miR-16, which is capable of binding and downregulating their expression. Zhang et al. (66) showed that low expression of miR-15a/16 in patients leads to increased expression of CABIN1 mRNA, promoting tumor proliferation. Essentially, miR-15a/16 directly targets CABIN1 mRNA and negatively regulates CABIN1 expression at both the mRNA and protein levels in MM cells. In another study, VEGFA, a target gene of miR-15a/16, affected angiogenesis in MM by regulating the expression of VEGFA (67). Calin et al. (68) showed that miR-15a/16, a natural antisense transcripts of BCL2, directly interact with BCL2 and negatively regulates its expression at the transcriptional level. BCL2, as an important gene that regulates tumor cell apoptosis, plays a crucial role in mediating MM cell apoptosis and drug resistance.
In summary, exosomal LINC00461 has a sponge effect on miR-15a/16, affecting the expression of downstream genes such as CABIN1 mRNA, VEGFA, and BCL2, and forming a ceRNA regulatory network centered on LINC00461-miR-15a/16, thereby influencing MM proliferation, invasion, and apoptosis. The ceRNA network centered on exosomal LINC00461 is still in the early stages of research, and its specific relationship with MM has not yet been elucidated. The interaction between LINC00461-miR-15a/16 may be influenced by other molecular stress conditions, making it difficult to elucidate the intrinsic regulatory mechanisms of LINC00461. In conclusion, the complex interaction between lncRNAs and MM cell proliferation emphasizes the importance of further studies on the regulatory roles of these ncRNAs in the pathogenesis of MM. Understanding the mechanism by which lncRNAs such as LINC00461 promote MM cell proliferation may pave the way for the development of new therapeutic interventions against this hematological malignancy.
4.2 RNA duplexes
Extensive studies have demonstrated the complex role of exosomal lncRNAs in MM. In addition to the ceRNA mechanism mentioned above, lncRNAs can bind to mRNA to form RNA duplexes. The molecular interactions of lncRNAs play crucial roles in tumors (69, 70). Understanding the role of RNA duplexes in MM is key to understanding the molecular mechanisms underlying this complex disease. In MM, RNA duplexes play important roles in regulating gene expression, cell signaling, and intercellular communication. Understanding the various roles of exosomal lncRNAs in MM, from intercellular communication to targeted therapy through the formation of RNA duplexes with transcripts, will contribute to a better understanding of the molecular mechanisms underlying the occurrence and progression of MM. The lncRNA lncRUNX2-AS1 formed an RNA duplex with the precursor mRNA of RUNX2, reducing the osteogenic potential of MSCs (Figure 3). The exosome-mediated transfer of lncRUNX2-AS1 from MM cells to MSCs has been identified as a potential mechanism for inhibiting osteogenesis. Li et al. (41) demonstrated that exosomes derived from U266 or MM1S MM cells significantly reduced bone nodule formation in cocultured MSCs. Exosomes derived from MM cells with knocked-down RUNX2-AS1 reduced the intracellular levels of RUNX2-AS1 in MSCs and diminished their osteogenic capacity. Therefore, high levels of RUNX2-AS1 in MSCs were significantly correlated with the osteogenic activity. Antisense lncRNA can bind to mRNA through complementary base pairing, affecting the mRNA alternative splicing (71). Fluorescence in situ hybridization and reverse transcription PCR showed gene overlap between RUNX2 and RUNX2-AS1, but in opposite transcriptional directions. Overexpression of RUNX2-AS1 in MM cells significantly reduced the RUNX2/pre-RUNX2 ratio. After RNase protection experiments on RNA in bone marrow stromal cells, followed by reverse transcription PCR probing for the products, it was found that the overlapping portion of the transcripts was protected and not degraded (41). Therefore, RUNX-2AS1 acts as a highly specific sensor for mRNA and forms an RNA duplex with RUNX2 through overlapping base pairs. This interaction blocks the splicing of the RUNX2 pre-mRNA, thereby negatively regulating RUNX2 mRNA. Unlike the lncRNA-miRNA axis, RUNX2 is a direct target gene of RUNX2-AS1, which directly regulates osteoblast differentiation and indirectly regulates osteoclast differentiation (72). In mouse models, Xu et al. (73) found that osteoblasts lacking RUNX2 produce a highly chemoattractive and immunosuppressive bone marrow microenvironment, affecting the localization and progression of MM to new bone sites. In summary, the exosomal lncRNA RUNX2-AS1 derived from MM cells acts as a natural antisense transcript of RUNX2 and directly binds to and downregulates the expression of RUNX2 in bone marrow stromal cells, thereby inhibiting bone formation.
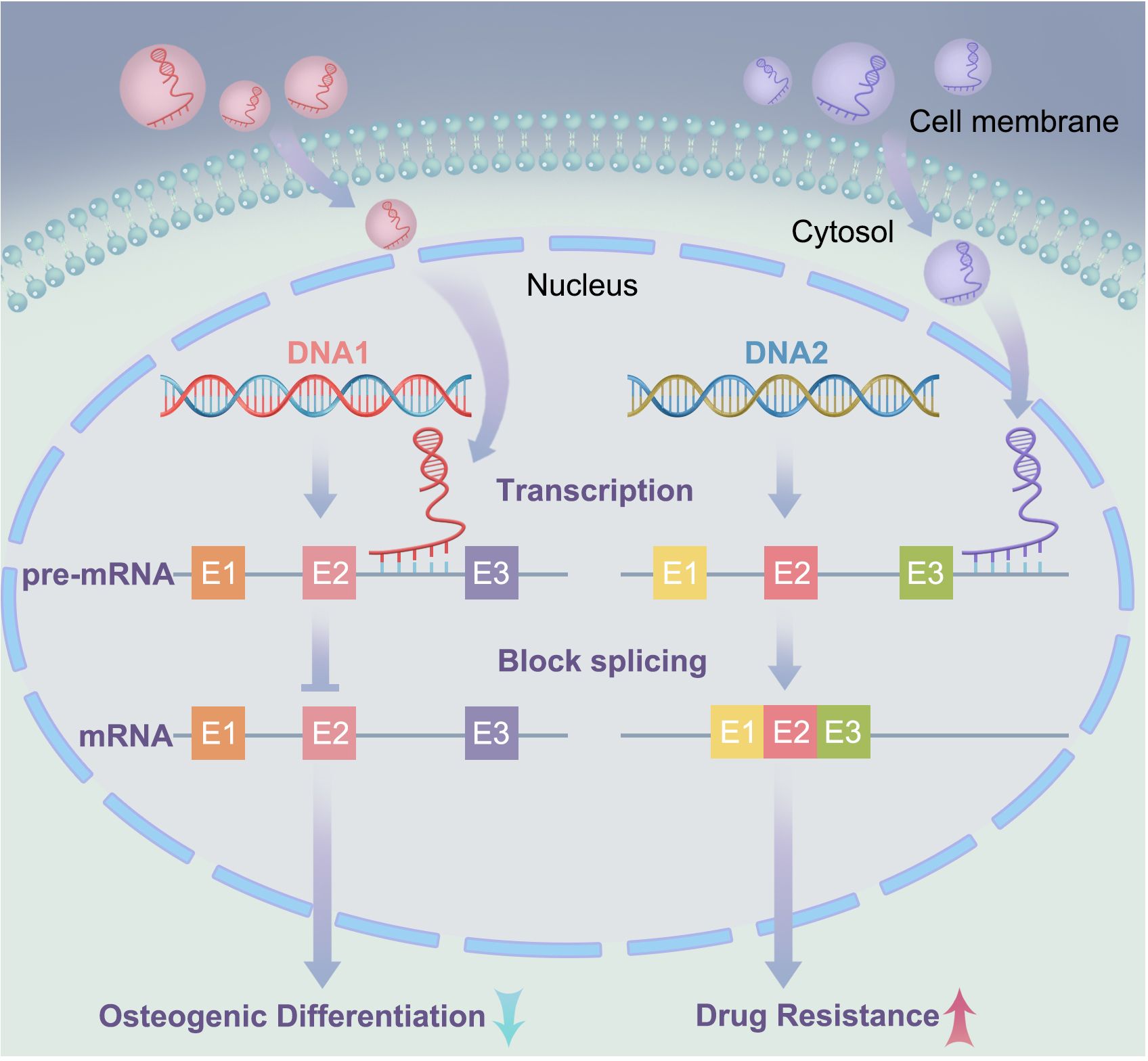
Figure 3. IncRNA forms RNA duplexes with mRNA: In the left signaling pathway, exosomal lncRNA binds to pre-mRNA through base complementary pairing, forming RNA duplexes that block alternative splicing of mRNA, thereby reducing the osteogenic potential of mesenchymal stem cells (MSCs); In the right signaling pathway, exosomal lncRNA forms RNA duplexes with pre-mRNA through base complementary pairing, enhancing the stability of mRNA. E1, E2, and E3 represent exons.
Furthermore, in studies by Arumugam et al. (74), the lncRNA RUNX2-AS1 also had a sponge effect on miR-6797-5p, forming the RUNX2-AS1/miR-6797-5p/RUNX2 axis, negatively regulating the expression of RUNX2, and promoting osteoblast differentiation. Most studies reporting ceRNA communication between RNA molecules have focused on binary interactions between two transcripts, but these interactions are mediated by one or more miRNAs. The two regulatory modes of the exosomal lncRNA RUNX2-AS1 illustrate the complexity of the relationships between the components in ceRNA regulatory networks.
The lncRNA, lncPSMA3-AS1, forms an RNA duplex with the precursor mRNA of PSMA3, promoting resistance to proteasome inhibitors in MM. In previous studies, lncRNA PSMA3-AS1 was shown to regulate the expression of corresponding miRNAs in various tumors, such as cholangiocarcinoma, gliomas, and pancreatic ductal adenocarcinoma (75–77). Xu et al. (78) showed that the mRNA levels of PSMA3 and PSMA3-AS1 in CD138+ MM cells were significantly elevated in patients with bortezomib-resistant MM compared with those in patients with bortezomib-sensitive MM. PSMA3-AS1 is mainly encapsulated in MSC-derived exosomes, transferred to MM cells, and enriched therein, further affecting their resistance to proteasome inhibitors. Kaplan-Meier analysis showed that high levels of PSMA3 in CD138+ MM cells correlated with decreased progression-free survival and overall survival in patients with MM, and high levels of PSMA3 could also serve as an independent prognostic factor for patients with MM receiving bortezomib treatment. All the above findings indicate a correlation between PSMA3-AS1 and disease progression and proteasome inhibitor resistance in patients with MM.
The target gene of PSMA3-AS1 is the sense transcript PSMA3. Fluorescence in situ hybridization and nuclear-cytoplasmic fractionation analyses showed that PSMA3-AS1 coexists in both the nucleus and cytoplasm. PSMA3 and PSMA3-AS1 are located on chromosome 14p23.1, and PSMA3-AS1 overlaps with intron 7 of PSMA3 by 2029 nucleotides. Previous studies have shown that antisense transcripts can regulate the sense transcript in two ways: first, by positively regulating the expression of the sense transcript, and second, by negatively regulating its expression (79). After blocking new RNA synthesis with α-amanitin, the loss of PSMA3, PSMA3-AS1, GAPDH, and 18s RNA was measured. The results showed that knocking out PSMA3-AS1 reduced the stability of PSMA3, whereas PSMA3-AS1 overexpression increased the stability of PSMA3. This finding suggests that PSMA3-AS1 can form a duplex with PSMA3, increasing its stability of PSMA3 by reducing its degradation and thereby positively regulating the expression of PSMA3 (78).
Previous studies have shown that PSMA3 encodes the alpha 7 subunit of the proteasome, which plays an important role in its formation and function (80). PSMA3 and PSMA3-AS1 knockout resulted in decreased proteasome activity in MM cells, whereas overexpression increased proteasome activity. In summary, PSMA3-AS1 can bind to PSMA3-AS1 pre-mRNA and increase its stability, promoting the proliferation of MM cells and proteasome activity, and conferring resistance to proteasome inhibitors in MM cells, as shown in Figure 3. In vivo experiments by Xu et al. (78) showed that exogenously injected therapeutic PSMA3-AS1 siRNA effectively increases the sensitivity of U266 xenografts to carfilzomib, significantly prolonging overall survival when combined with carfilzomib treatment. These experiments further demonstrate how the biological activity of the exosomal lncRNA PSMA3-AS1 can be transmitted between different cell types, affecting cell function, and may serve as a therapeutic target for MM bone lesions. RNA-based therapies, particularly siRNA, have shown great potential in cancer treatment because they can silence key genes in tumor progression (81). In vivo siRNA delivery can be achieved through various methods such as liposomes, lipid nanoparticles, polymeric nanoparticles, viral vectors, proteins and peptides, and exosomes, to enhance the stability of siRNA and cellular uptake efficiency, and to overcome delivery barriers (82). Preclinical and clinical studies have demonstrated the potential for treating solid tumors and hematological malignancies, as well as cancer immunotherapy. This indicates a novel signaling pathway involved in drug resistance and highlights the role of exosomes in intercellular communication in the context of this disease.
4.3 IncRNA RBP
Complex interactions between exosomal lncRNAs and RBPs have garnered significant attention. Exosomes are EVs involved in intercellular communication and are associated with the progression and drug resistance of MM (83). RBPs have been identified as promising biomarkers for MM and the construction of RBP signatures can effectively predict the prognosis of patients with MM. The interaction between lncRNAs and RBPs plays a crucial role in the occurrence and development of cancer (84–86), such as regulating target gene expression, RBP activity and stability, and lncRNA expression levels (11). These findings emphasize the importance of further exploration of these molecular mechanisms to develop new therapeutic strategies and improve patient prognosis.
Exosomal lncRNA NEAT1 binds to EZH2 and promotes immune escape of MM cells. NEAT1 is a long non-coding RNA that binds to EZH2 and regulates the expression of its downstream effectors (87, 88). EZH2 is a histone methyltransferase involved in the occurrence and development of various tumors (89, 90). The interaction between NEAT1 and EZH2 enables EZH2 to bind to the promoter region of PBX1, thereby inhibiting PBX1 expression through H3K27 trimethylation, as shown in Figure 4. Studies have shown that NEAT1 can affect EZH2 modification of target gene promoter regions by interacting with EZH2, thereby regulating gene expression (47). In this study, NEAT1 inhibited PBX1 expression by recruiting EZH2, thereby suppressing NK cell activity and promoting the immune escape of MM cells. Natural killer (NK) cells are important immune cells with cytotoxic and inhibitory activities against cancer cells. However, MM cells can evade immune surveillance by inhibiting NK cell activity. These findings provide new strategies for the treatment of MM, and exosomal lncRNA NEAT1 has the potential to become a new therapeutic target for MM.
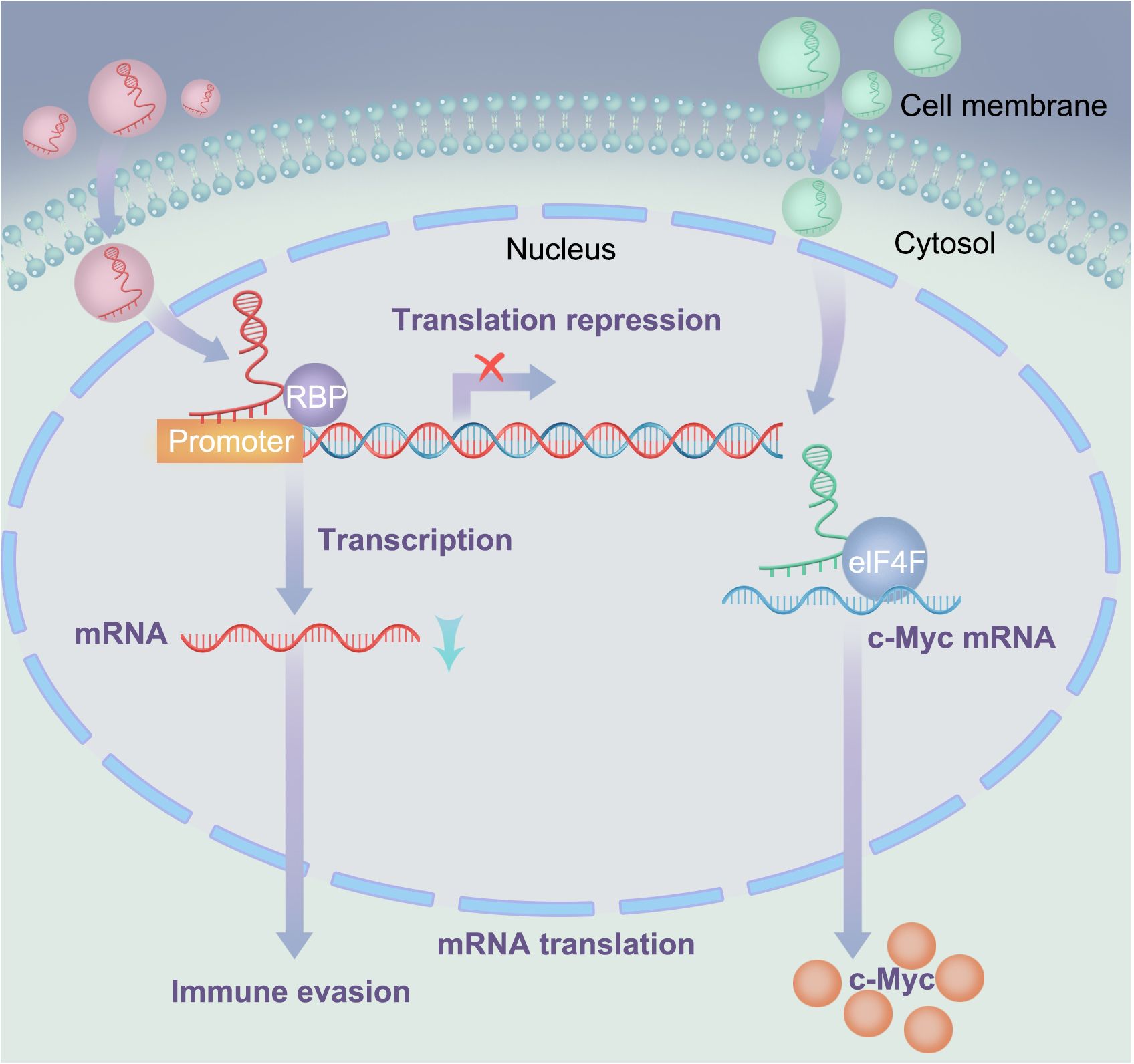
Figure 4. Interactions between lncRNA and RNA-binding proteins: In the left signaling pathway, lncRNA suppresses the expression of downstream effectors (mRNA) by recruiting RNAbinding proteins; In the right signaling pathway, LOC606724 acts as a bridge connecting eIF4E and c-Myc, upregulating c-Myc protein at the post-transcriptional level in MM cells.
Exosomal LOC606724 binds to METTL7A and inhibits apoptosis in MM cells. In current research, lncRNAs play important regulatory roles in the interaction between MM cells and adipocytes, with adipocytes promoting obesity-induced myeloma (91, 92). Studies have also identified interactions between lncRNAs and RNA-binding proteins and identified METTL7A as an RNA methyltransferase. Further experiments showed that MM cells promoted the packaging of lncRNAs into adipocyte-derived exosomes via METTL7A-mediated lncRNA m6A methylation. Previous experiments have demonstrated that MM cells can reprogram adipocytes through direct intercellular interactions, upregulating EZH2 expression and activating histone methylation in adipogenic factor promoters (93). This suggests that the methylation modifications of lncRNAs play an important regulatory role in MM. Conversely, adipocyte-derived exosomes protect MM cells from chemotherapy-induced apoptosis. Wang et al. (23) showed that two lncRNAs, LOC606724 and SNHG1, were significantly upregulated in MM cells exposed to adipocyte-derived exosomes. LOC606724, as a bridge connecting eIf4E and c-Myc, upregulated the c-Myc protein in MM cells at the post-transcriptional level (Figure 4). This indicates the existence of a malignant cycle between MM cells and adipocytes. Blocking this exosome-mediated malignant cycle may be a potential strategy to improve treatment efficacy.
In summary, exosomal lncRNAs have multiple mechanisms in MM, including the regulation of cell apoptosis, methylation modifications, and protein transformation. These findings provide important clues for a deeper understanding of MM pathogenesis, and offer potential targets for developing new therapeutic strategies. However, further research is needed to validate these findings and explore the association between lncRNAs and MM, which will greatly assist in devising more effective treatment strategies for patients with MM.
5 Exosomal lncRNAs as therapeutic targets in MM: clinical implications
Currently, research on exosomal lncRNA in multiple myeloma (MM) is still in its infancy, and as a novel diagnostic and therapeutic approach, it faces several challenges. Firstly, the techniques for exosome isolation and purification vary at present, and the yield of native exosomes is too low to meet the demands of clinical applications. It is crucial to optimize and upgrade the isolation and purification technologies for exosomes to enhance their purity and yield (94). Secondly, the mechanism of action of exosomal lncRNA in MM is not yet fully understood, necessitating further basic and clinical research, as well as multicenter collaborative studies, to fully explore the role of exosomes in aiding the diagnosis, prognosis assessment, and treatment guidance of MM. Thirdly, the high heterogeneity of multiple myeloma complicates the study of the role of exosomal lncRNA in MM (95). Fourthly, the identification of biomarkers: although the abnormal cargo in exosomes can be used as cancer biomarkers for the detection or screening of early prognosis in MM patients, identifying these biomarkers remains a challenge. Combining DNA signal amplification techniques with the signal enhancement characteristics of nanomaterials offers a promising solution to address these limitations and improve the efficiency and accuracy of exosome detection (96). Fifthly, the determination of therapeutic targets: identifying exosomal lncRNA as therapeutic targets requires in-depth research into their specific roles and mechanisms in the progression of MM. In summary, we believe that exosomal lncRNA may play a significant role in the diagnosis and treatment of multiple myeloma in the future.
Compared to traditional drug delivery systems, exosomes offer outstanding biocompatibility, high specificity, and effective drug release capabilities (97–99). Loading drugs into exosomes enables precise delivery to affected sites, thereby enhancing treatment efficacy and reducing adverse reactions. Additionally, exosomes have potential as diagnostic tools because they contain rich information on biomarkers that can be utilized for early disease diagnosis and treatment prediction (100, 101). For example, circulating exosomes can serve as cancer biomarkers and aid in early detection and monitoring. Furthermore, recent studies have suggested that ceRNAs can also serve as potential therapeutic targets and biomarkers for analyzing the pathogenesis of malignant tumors (57, 102, 103), demonstrating significant clinical application and research significance. There are also abundant data suggesting that exosomal lncRNAs can serve as potential therapeutic targets for malignant tumors (104–106), aiding in diagnosis, prognosis prediction, early assessment of specific drug treatment effects, and drug resistance issues, and even indicating directions for exploring the mechanisms of malignant tumor formation.
An increasing number of studies have revealed the clinical significance of exosomal-derived lncRNAs in MM treatment, which play a key role in the diagnosis, treatment, and monitoring of various cancers (107–109). In acute myeloid leukemia (AML), extracellular lncRNAs LINC00265, LINC00467, UCA1, and SNHG1 are promising biomarkers for disease diagnosis and treatment monitoring (107). Similarly, Zhao et al. emphasized the predictive value of the lncRNA PCAT1 in patients undergoing MM-induced therapy, linking it to prognosis and treatment response (110). In MM, the interaction between exosomal lncRNAs and drug resistance mechanisms has attracted attention. Exosome-mediated SNHG16 promotes dexamethasone resistance by transferring side population cells to main population cells, revealing a new mechanism for MM exosome transfer (109). Similarly, the exosomal lncRNA NEAT1 inhibits the activity of natural killer (NK) cells, promoting MM immune escape via the EZH2/PBX1 axis (47). These findings highlight the diagnostic, prognostic, and therapeutic potential of exosomal lncRNAs in the treatment of MM. Additionally, the exploration of extracellular vesicle proteins as MM biomarkers emphasizes the development prospects of liquid biopsy to improve patient stratification and prognosis (111, 112). In summary, exosome-derived lncRNAs represent valuable targets for MM treatment and provide insights into disease monitoring, drug resistance mechanisms, and prognostic stratification. Elucidation of these molecular mechanisms holds great promise for advancing personalized treatment strategies for patients with MM and improving clinical outcomes.
6 Prospects and limitations
The mechanisms of action of exosomal lncRNAs are highly complex and diverse. According to current research findings, the mechanisms of exosomal lncRNAs primarily include the following aspects: epigenetic regulation, transcriptional regulation, post-transcriptional regulation (113), miRNA sponge function, molecular decoy and guide functions, scaffold functions, interactions with proteins, regulation of cellular localization, and serving as precursors to small non-coding RNAs (114). These mechanisms demonstrate the multifunctionality and importance of lncRNAs in cell biology and disease pathogenesis. They are not only involved in a variety of physiological and pathological processes but also provide new perspectives for disease diagnosis and treatment. In multiple myeloma, the interactions of lncRNA with RNA (ceRNA and RNA duplexes), lncRNA with DNA (IncRNA RBP), and lncRNA with proteins (IncRNA RBP) are more representative and play a very important role in the occurrence and development of multiple myeloma. In comparison, the mechanisms of other exosomal lncRNAs in multiple myeloma warrant further exploration.
The role of exosomal lncRNAs in multiple myeloma (MM) remains a relatively new field of study, particularly in terms of their application in the diagnosis and treatment of MM. Exosomal lncRNAs hold the potential to serve as biomarkers, which could be employed as monitoring items for the diagnosis and prognostic assessment of MM, a key aspect of personalized medicine and precision therapy. We have explored the possibility of exosomal lncRNAs as therapeutic targets, including their roles in drug resistance, immune evasion, and tumor progression, providing a theoretical foundation for the development of new treatment strategies. The review integrates the latest research findings, including new discoveries regarding exosomal lncRNAs in MM, as well as their potential applications in disease monitoring, mechanisms of drug resistance, and prognostic stratification. By analyzing the role of exosomal lncRNAs in MM from multiple perspectives, including molecular biology, genetics, and epigenetics, the review offers new insights into the field of MM research and potential new tools for clinical diagnosis and treatment.
7 Conclusion
Although an increasing number of genetic and epigenetic events that lead to the occurrence and development of MM have been identified, their roles in the diagnosis, treatment, and clinical outcomes of MM remain unclear. Recent studies have confirmed the involvement of lncRNA dysregulation in the transcriptional, post-transcriptional, and epigenetic regulation of MM. Multiple lncRNA-based diagnostic and prognostic models have been established (58). Compared to traditional sources of lncRNAs, exosomal lncRNAs are widely present in various body fluids and exhibit high cell and tissue specificity, making it possible to construct novel, minimally invasive biomarkers for cancer diagnosis and prognosis using exosomal lncRNAs (115, 116). Furthermore, combined with the continuously evolving ceRNA network, RNA duplexes, RNA-binding proteins, and other molecular mechanisms centered on MM, establishing reliable prediction models based on exosomal lncRNA expression characteristics, assisting in identifying new drug targets through ceRNA regulatory networks, and utilizing nucleic acid-targeted therapy to avoid MM drug resistance and disease progression will have significant implications for the diagnosis and treatment of MM patients.
Author contributions
HY: Writing – original draft, Conceptualization. NJ: Conceptualization, Writing – original draft. XL: Writing – original draft. CL: Writing – original draft. FW: Writing – original draft. LC: Writing – review & editing. JZ: Writing – review & editing. DL: Conceptualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Open Fund of Development and Regeneration Key Laboratory of Sichuan Province (SYS23-08), the Joint Fund of Chengdu Medical College and The Second Affiliated Hospital of Chengdu Medical College (23LHHGYMP15), and the Special Project of Liyan Workshop Aesthetic Medicine Research Center of Chengdu Medical College (23YM12 and 23YM08).
Acknowledgments
We extend our gratitude to the authors for their contributions to this review and acknowledge the financial support from the Open Fund of Development and Regeneration Key Laboratory of Sichuan Province and the Joint Fund of Chengdu Medical College and The Second Affiliated Hospital of Chengdu Medical College.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Das S, Juliana N, Yazit NAA, Azmani S, Abu IF. Multiple myeloma: challenges encountered and future options for better treatment. Int J Mol Sci. (2022) 23:1649. doi: 10.3390/ijms23031649
2. van de Donk N, Pawlyn C, Yong KL. Multiple myeloma. Lancet. (2021) 397:410–27. doi: 10.1016/s0140-6736(21)00135-5
3. Silberstein J, Tuchman S, Grant SJ. What is multiple myeloma? Jama. (2022) 327:497. doi: 10.1001/jama.2021.25306
4. Cowan AJ, Green DJ, Kwok M, Lee S, Coffey DG, Holmberg LA, et al. Diagnosis and management of multiple myeloma: A review. Jama. (2022) 327:464–77. doi: 10.1001/jama.2022.0003
5. Yao ZT, Yang YM, Sun MM, He Y, Liao L, Chen KS, et al. New insights into the interplay between long non-coding rnas and rna-binding proteins in cancer. Cancer Commun (Lond). (2022) 42:117–40. doi: 10.1002/cac2.12254
6. Liu J, Ren L, Li S, Li W, Zheng X, Yang Y, et al. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B. (2021) 11:2783–97. doi: 10.1016/j.apsb.2021.01.001
7. Li W, Li C, Zhou T, Liu X, Liu X, Li X, et al. Role of exosomal proteins in cancer diagnosis. Mol Cancer. (2017) 16:145. doi: 10.1186/s12943-017-0706-8
8. Wang X, Huang J, Chen W, Li G, Li Z, Lei J. The updated role of exosomal proteins in the diagnosis, prognosis, and treatment of cancer. Exp Mol Med. (2022) 54:1390–400. doi: 10.1038/s12276-022-00855-4
9. Menu E, Vanderkerken K. Exosomes in multiple myeloma: from bench to bedside. Blood. (2022) 140:2429–42. doi: 10.1182/blood.2021014749
10. Moloudizargari M, Abdollahi M, Asghari MH, Zimta AA, Neagoe IB, Nabavi SM. The emerging role of exosomes in multiple myeloma. Blood Rev. (2019) 38:100595. doi: 10.1016/j.blre.2019.100595
11. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen L-L, et al. Long non-coding rnas: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. (2023) 24:430–47. doi: 10.1038/s41580-022-00566-8
12. Ferrer J, Dimitrova N. Transcription regulation by long non-coding rnas: mechanisms and disease relevance. Nat Rev Mol Cell Biol. (2024) 25(5):396–415. doi: 10.1038/s41580-023-00694-9
13. Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. Lncrna-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond). (2021) 41:109–20. doi: 10.1002/cac2.12108
14. Han QF, Li WJ, Hu KS, Gao J, Zhai WL, Yang JH, et al. Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer. Mol Cancer. (2022) 21:207. doi: 10.1186/s12943-022-01671-0
15. Guo Y, Ji X, Liu J, Fan D, Zhou Q, Chen C, et al. Effects of exosomes on pre-metastatic niche formation in tumors. Mol Cancer. (2019) 18:39. doi: 10.1186/s12943-019-0995-1
16. Yang E, Wang X, Gong Z, Yu M, Wu H, Zhang D. Exosome-mediated metabolic reprogramming: the emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct Target Ther. (2020) 5:242. doi: 10.1038/s41392-020-00359-5
17. Paskeh MDA, Entezari M, Mirzaei S, Zabolian A, Saleki H, Naghdi MJ, et al. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J Hematol Oncol. (2022) 15:83. doi: 10.1186/s13045-022-01305-4
18. Zhang J, Ji C, Zhang H, Shi H, Mao F, Qian H, et al. Engineered neutrophil-derived exosome-like vesicles for targeted cancer therapy. Sci Adv. (2022) 8:eabj8207. doi: 10.1126/sciadv.abj8207
19. Allegra A, Di Gioacchino M, Tonacci A, Petrarca C, Musolino C, Gangemi S. Multiple myeloma cell-derived exosomes: implications on tumorigenesis, diagnosis, prognosis and therapeutic strategies. Cells. (2021) 10:2865. doi: 10.3390/cells10112865
20. Stefańska K, Józkowiak M, Angelova Volponi A, Shibli JA, Golkar-Narenji A, Antosik P, et al. The role of exosomes in human carcinogenesis and cancer therapy-recent findings from molecular and clinical research. Cells. (2023) 12:356. doi: 10.3390/cells12030356
21. Li M, Xia B, Wang Y, You MJ, Zhang Y. Potential therapeutic roles of exosomes in multiple myeloma: A systematic review. J Cancer. (2019) 10:6154–60. doi: 10.7150/jca.31752
22. Roccaro AM, Sacco A, Maiso P, Azab AK, Tai YT, Reagan M, et al. Bm mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J Clin Invest. (2013) 123:1542–55. doi: 10.1172/jci66517
23. Wang Z, He J, Bach DH, Huang YH, Li Z, Liu H, et al. Induction of M(6)a methylation in adipocyte exosomal lncrnas mediates myeloma drug resistance. J Exp Clin Cancer Res. (2022) 41:4. doi: 10.1186/s13046-021-02209-w
24. Liu Z, Liu H, Li Y, Shao Q, Chen J, Song J, et al. Multiple myeloma-derived exosomes inhibit osteoblastic differentiation and improve il-6 secretion of bmscs from multiple myeloma. J Investig Med. (2020) 68:45–51. doi: 10.1136/jim-2019-001010
25. Frassanito MA, Desantis V, Di Marzo L, Craparotta I, Beltrame L, Marchini S, et al. Bone marrow fibroblasts overexpress mir-27b and mir-214 in step with multiple myeloma progression, dependent on tumour cell-derived exosomes. J Pathol. (2019) 247:241–53. doi: 10.1002/path.5187
26. Long S, Long S, He H, Luo L, Liu M, Ding T. Exosomal mir-182 derived from bone marrow mesenchymal stem cells drives carfilzomib resistance of multiple myeloma cells by targeting sox6. J Orthop Surg Res. (2023) 18:937. doi: 10.1186/s13018-023-04399-9
27. Miaomiao S, Xiaoqian W, Yuwei S, Chao C, Chenbo Y, Yinghao L, et al. Cancer-associated fibroblast-derived exosome microrna-21 promotes angiogenesis in multiple myeloma. Sci Rep. (2023) 13:9671. doi: 10.1038/s41598-023-36092-6
28. Liu L, Ye Q, Liu L, Bihl JC, Chen Y, Liu J, et al. C6-ceramide treatment inhibits the proangiogenic activity of multiple myeloma exosomes via the mir-29b/akt pathway. J Transl Med. (2020) 18:298. doi: 10.1186/s12967-020-02468-9
29. Ahmad M, Weiswald LB, Poulain L, Denoyelle C, Meryet-Figuiere M. Involvement of lncrnas in cancer cells migration, invasion and metastasis: cytoskeleton and ecm crosstalk. J Exp Clin Cancer Res. (2023) 42:173. doi: 10.1186/s13046-023-02741-x
30. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. Cerna in cancer: possible functions and clinical implications. J Med Genet. (2015) 52:710–8. doi: 10.1136/jmedgenet-2015-103334
31. Zhu Y, Huang C, Zhang C, Zhou Y, Zhao E, Zhang Y, et al. Lncrna mir200chg inhibits emt in gastric cancer by stabilizing mir-200c from target-directed mirna degradation. Nat Commun. (2023) 14:8141. doi: 10.1038/s41467-023-43974-w
32. Agirre X. Rrol lncrna role in multiple myeloma. Blood. (2023) 141:328–30. doi: 10.1182/blood.2022018471
33. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding rnas and its biological functions. Nat Rev Mol Cell Biol. (2021) 22:96–118. doi: 10.1038/s41580-020-00315-9
34. Yang C, Liang Y, Shu J, Wang S, Hong Y, Chen K, et al. Long non−Coding rnas in multiple myeloma (Review). Int J Oncol. (2023) 62:69. doi: 10.3892/ijo.2023.5517
35. Liu N, Feng S, Li H, Chen X, Bai S, Liu Y. Long non-coding rna malat1 facilitates the tumorigenesis, invasion and glycolysis of multiple myeloma via mir-1271-5p/sox13 axis. J Cancer Res Clin Oncol. (2020) 146:367–79. doi: 10.1007/s00432-020-03127-8
36. Yan H, Gao S, Xu A, Zuo L, Zhang J, Zhao Y, et al. Malat1 regulates network of microrna-15a/16-vegfa to promote tumorigenesis and angiogenesis in multiple myeloma. Carcinogenesis. (2023) 44:760–72. doi: 10.1093/carcin/bgad053
37. Shen Q, Jiang Q, Cong Z, Zhou Y, Huang X, Zhu L, et al. Knockdown of lncrna al928768.3 inhibits multiple myeloma cell proliferation by inducing cell cycle arrest in G0/G1 phase. Ann Transl Med. (2022) 10:172. doi: 10.21037/atm-21-6710
38. Pan Y, Zhang Y, Liu W, Huang Y, Shen X, Jing R, et al. Lncrna H19 overexpression induces bortezomib resistance in multiple myeloma by targeting mcl-1 via mir-29b-3p. Cell Death Dis. (2019) 10:106. doi: 10.1038/s41419-018-1219-0
39. Wang Y, Xu S, Wang L, Liu J, Wang J, Zhou J. Long noncoding rna H19 promotes vincristine resistance in multiple myeloma by targeting akt. Cell Mol Biol (Noisy-le-grand). (2020) 66:76–80. doi: 10.14715/cmb/2020.66.6.14
40. Guo N, Song Y, Zi F, Zheng J, Cheng J. Abnormal expression pattern of lncrna H19 participates in multiple myeloma bone disease by unbalancing osteogenesis and osteolysis. Int Immunopharmacol. (2023) 119:110058. doi: 10.1016/j.intimp.2023.110058
41. Li B, Xu H, Han H, Song S, Zhang X, Ouyang L, et al. Exosome-mediated transfer of lncrunx2-as1 from multiple myeloma cells to mscs contributes to osteogenesis. Oncogene. (2018) 37:5508–19. doi: 10.1038/s41388-018-0359-0
42. Gao Y, Fang P, Li WJ, Zhang J, Wang GP, Jiang DF, et al. Lncrna neat1 sponges mir-214 to regulate M2 macrophage polarization by regulation of B7-H3 in multiple myeloma. Mol Immunol. (2020) 117:20–8. doi: 10.1016/j.molimm.2019.10.026
43. Wu L, Xia L, Chen X, Ruan M, Li L, Xia R. Long non-coding rna linc01003 suppresses the development of multiple myeloma by targeting mir-33a-5p/pim1 axis. Leuk Res. (2021) 106:106565. doi: 10.1016/j.leukres.2021.106565
44. Yang N, Chen J, Zhang H, Wang X, Yao H, Peng Y, et al. Lncrna oip5-as1 loss-induced microrna-410 accumulation regulates cell proliferation and apoptosis by targeting klf10 via activating pten/pi3k/akt pathway in multiple myeloma. Cell Death Dis. (2017) 8:e2975. doi: 10.1038/cddis.2017.358
45. Jiang Y, Chen J, Chen G. Long noncoding rna irain acts as tumor suppressor via mir-125b in multiple myeloma. Oncol Lett. (2019) 18:6787–94. doi: 10.3892/ol.2019.11012
46. Li J, Zou J, Wan X, Sun C, Chu Z, Hu Y. Roles of noncoding rnas in drug resistance in multiple myeloma. J Cell Physiol. (2020) 235:7681–95. doi: 10.1002/jcp.29726
47. Wang QM, Lian GY, Sheng SM, Xu J, Ye LL, Min C, et al. Exosomal lncrna neat1 inhibits nk-cell activity to promote multiple myeloma cell immune escape via an ezh2/pbx1 axis. Mol Cancer Res. (2024) 22:125–36. doi: 10.1158/1541-7786.Mcr-23-0282
48. Deng M, Yuan H, Peng H, Liu S, Xiao X, Wang Z, et al. Mesenchymal stem cells inhibit the effects of dexamethasone in multiple myeloma cells. Stem Cells Int. (2022) 2022:4855517. doi: 10.1155/2022/4855517
49. Deng M, Yuan H, Liu S, Hu Z, Xiao H. Exosome-transmitted linc00461 promotes multiple myeloma cell proliferation and suppresses apoptosis by modulating microrna/bcl-2 expression. Cytotherapy. (2019) 21:96–106. doi: 10.1016/j.jcyt.2018.10.006
50. Ding W, Wu L, Li X, Chang L, Liu G, Du H. Comprehensive analysis of competitive endogenous rnas network: identification and validation of prediction model composed of mrna signature and mirna signature in gastric cancer. Oncol Lett. (2022) 23:150. doi: 10.3892/ol.2022.13270
51. Cao S, Liu H, Fan J, Yang K, Yang B, Wang J, et al. An oxidative stress-related gene pair (Ccnb1/pkd1), competitive endogenous rnas, and immune-infiltration patterns potentially regulate intervertebral disc degeneration development. Front Immunol. (2021) 12:765382. doi: 10.3389/fimmu.2021.765382
52. Wu W, Gao C, Chen L, Zhang D, Guo S. Comprehensive analysis of competitive endogenous rnas networks reveals potential prognostic biomarkers associated with epithelial ovarian cancer. Oncol Lett. (2021) 22:843. doi: 10.3892/ol.2021.13104
53. Chen Y, Chen D, Wang J, Zhang Y, Zhang J, Chen B, et al. Dysregulated lncrnas act as competitive endogenous rnas and are associated with cervical cancer development in uyghur women. Technol Cancer Res Treat. (2021) 20:1533033821989711. doi: 10.1177/1533033821989711
54. Song M, Zhong A, Yang J, He J, Cheng S, Zeng J, et al. Large-scale analyses identify a cluster of novel long noncoding rnas as potential competitive endogenous rnas in progression of hepatocellular carcinoma. Aging (Albany NY). (2019) 11:10422–53. doi: 10.18632/aging.102468
55. Karreth FA, Pandolfi PP. Cerna cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discovery. (2013) 3:1113–21. doi: 10.1158/2159-8290.Cd-13-0202
56. Long J, Bai Y, Yang X, Lin J, Yang X, Wang D, et al. Construction and comprehensive analysis of a cerna network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int. (2019) 19:90. doi: 10.1186/s12935-019-0817-y
57. Mao M, Zhang J, Xiang Y, Gong M, Deng Y, Ye D. Role of exosomal competitive endogenous rna (Cerna) in diagnosis and treatment of Malignant tumors. Bioengineered. (2022) 13:12156–68. doi: 10.1080/21655979.2022.2073130
58. Zhou S, Fang J, Sun Y, Li H. Integrated analysis of a risk score system predicting prognosis and a cerna network for differentially expressed lncrnas in multiple myeloma. Front Genet. (2020) 11:934. doi: 10.3389/fgene.2020.00934
59. Lu M, Wu Y, Gao W, Tian Y, Wang G, Liu A, et al. Novel non-coding rna analysis in multiple myeloma identified through high-throughput sequencing. Front Genet. (2021) 12:625019. doi: 10.3389/fgene.2021.625019
60. Watanabe T. Realization of osteolysis, angiogenesis, immunosuppression, and drug resistance by extracellular vesicles: roles of rnas and proteins in their cargoes and of ectonucleotidases of the immunosuppressive adenosinergic noncanonical pathway in the bone marrow niche of multiple myeloma. Cancers (Basel). (2021) 13:2969. doi: 10.3390/cancers13122969
61. Timaner M, Tsai KK, Shaked Y. The multifaceted role of mesenchymal stem cells in cancer. Semin Cancer Biol. (2020) 60:225–37. doi: 10.1016/j.semcancer.2019.06.003
62. Wang X, He L, Huang X, Zhang S, Cao W, Che F, et al. Recent progress of exosomes in multiple myeloma: pathogenesis, diagnosis, prognosis and therapeutic strategies. Cancers (Basel). (2021) 13:1635. doi: 10.3390/cancers13071635
63. Tang JX, Chen Q, Li Q, He YH, Xiao D. Exosomal mrnas and lncrnas involved in multiple myeloma resistance to bortezomib. Cell Biol Int. (2021) 45:965–75. doi: 10.1002/cbin.11540
64. Deng M, Yuan H, Peng H, Liu S, Xiao X, Wang Z, et al. Linc00461 knockdown enhances the effect of ixazomib in multiple myeloma cells. Curr Cancer Drug Targets. (2023) 23:643–52. doi: 10.2174/1568009623666230316152713
65. Zhang Q, Zhong C, Shen J, Chen S, Jia Y, Duan S. Emerging role of linc00461 in cancer. BioMed Pharmacother. (2022) 152:113239. doi: 10.1016/j.biopha.2022.113239
66. Zhang L, Zhou L, Shi M, Kuang Y, Fang L. Downregulation of mirna-15a and mirna-16 promote tumor proliferation in multiple myeloma by increasing cabin1 expression. Oncol Lett. (2018) 15:1287–96. doi: 10.3892/ol.2017.7424
67. Sun CY, She XM, Qin Y, Chu ZB, Chen L, Ai LS, et al. Mir-15a and mir-16 affect the angiogenesis of multiple myeloma by targeting vegf. Carcinogenesis. (2013) 34:426–35. doi: 10.1093/carcin/bgs333
68. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. Mir-15 and mir-16 induce apoptosis by targeting bcl2. Proc Natl Acad Sci U.S.A. (2005) 102:13944–9. doi: 10.1073/pnas.0506654102
69. Teng L, Feng YC, Guo ST, Wang PL, Qi TF, Yue YM, et al. The pan-cancer lncrna plane regulates an alternative splicing program to promote cancer pathogenesis. Nat Commun. (2021) 12:3734. doi: 10.1038/s41467-021-24099-4
70. Wang J, Hou F, Tang L, Xiao K, Yang T, Wang Z, et al. The interaction between long non-coding rna linc01564 and pou2f1 promotes the proliferation and metastasis of gastric cancer. J Transl Med. (2022) 20:220. doi: 10.1186/s12967-022-03391-x
71. Khorkova O, Stahl J, Joji A, Volmar CH, Zeier Z, Wahlestedt C. Natural antisense transcripts as drug targets. Front Mol Biosci. (2022) 9:978375. doi: 10.3389/fmolb.2022.978375
72. Trotter TN, Li M, Pan Q, Peker D, Rowan PD, Li J, et al. Myeloma cell-derived runx2 promotes myeloma progression in bone. Blood. (2015) 125:3598–608. doi: 10.1182/blood-2014-12-613968
73. Xu X, Zhang C, Trotter TN, Gowda PS, Lu Y, Ponnazhagan S, et al. Runx2 deficiency in osteoblasts promotes myeloma progression by altering the bone microenvironment at new bone sites. Cancer Res. (2020) 80:1036–48. doi: 10.1158/0008-5472.Can-19-0284
74. Arumugam B, Vishal M, Shreya S, Malavika D, Rajpriya V, He Z, et al. Parathyroid hormone-stimulation of runx2 during osteoblast differentiation via the regulation of lnc-supt3h-1:16 (Runx2-as1:32) and mir-6797-5p. Biochimie. (2019) 158:43–52. doi: 10.1016/j.biochi.2018.12.006
75. Sun D, Li F, Liu L, Yu S, Wang H, Gao X, et al. Psma3-as1 induced by transcription factor pax5 promotes cholangiocarcinoma proliferation, migration and invasion by sponging mir-376a-3p to up-regulate lamc1. Aging (Albany NY). (2022) 14:509–25. doi: 10.18632/aging.203828
76. Huang T, Chen Y, Zeng Y, Xu C, Huang J, Hu W, et al. Long non-coding rna psma3-as1 promotes glioma progression through modulating the mir-411-3p/hoxa10 pathway. BMC Cancer. (2021) 21:844. doi: 10.1186/s12885-021-08465-5
77. Bi J, Liang W, Wang Y, Tian W, Cao S, Liu P. Long noncoding rna psma3 antisense rna 1 promotes cell proliferation, migration, and invasion in pancreatic ductal adenocarcinoma via targeting microrna-154-5p to positively modulate karyopherin subunit alpha 4. Pancreas. (2022) 51:1037–46. doi: 10.1097/mpa.0000000000002136
78. Xu H, Han H, Song S, Yi N, Qian C, Qiu Y, et al. Exosome-transmitted psma3 and psma3-as1 promote proteasome inhibitor resistance in multiple myeloma. Clin Cancer Res. (2019) 25:1923–35. doi: 10.1158/1078-0432.Ccr-18-2363
79. Rehman SU, Ullah N, Zhang Z, Zhen Y, Din AU, Cui H, et al. Recent insights into the functions and mechanisms of antisense rna: emerging applications in cancer therapy and precision medicine. Front Chem. (2023) 11:1335330. doi: 10.3389/fchem.2023.1335330
80. Mittenberg AG, Kuzyk VO, Shabelnikov SV, Gorbach DP, Shatrova AN, Fedorova OA, et al. Combined treatment of human multiple myeloma cells with bortezomib and doxorubicin alters the interactome of 20s proteasomes. Cell Cycle. (2018) 17:1745–56. doi: 10.1080/15384101.2018.1496742
81. El Moukhtari SH, Garbayo E, Amundarain A, Pascual-Gil S, Carrasco-León A, Prosper F, et al. Lipid nanoparticles for sirna delivery in cancer treatment. J Control Release. (2023) 361:130–46. doi: 10.1016/j.jconrel.2023.07.054
82. Zhang MM, Bahal R, Rasmussen TP, Manautou JE, Zhong XB. The growth of sirna-based therapeutics: updated clinical studies. Biochem Pharmacol. (2021) 189:114432. doi: 10.1016/j.bcp.2021.114432
83. Yamamoto T, Yamamoto Y. Extracellular vesicle-mediated drug resistance in multiple myeloma. Rinsho Ketsueki. (2023) 64:1092–8. doi: 10.11406/rinketsu.64.1092
84. Wang S, Sun Z, Lei Z, Zhang HT. Rna-binding proteins and cancer metastasis. Semin Cancer Biol. (2022) 86:748–68. doi: 10.1016/j.semcancer.2022.03.018
85. Qin H, Ni H, Liu Y, Yuan Y, Xi T, Li X, et al. Rna-binding proteins in tumor progression. J Hematol Oncol. (2020) 13:90. doi: 10.1186/s13045-020-00927-w
86. Zhang J, Li S, Zhang L, Xu J, Song M, Shao T, et al. Rbp eif2s2 promotes tumorigenesis and progression by regulating myc-mediated inhibition via fhit-related enhancers. Mol Ther. (2020) 28:1105–18. doi: 10.1016/j.ymthe.2020.02.004
87. Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, et al. Long noncoding rna neat1, regulated by the egfr pathway, contributes to glioblastoma progression through the wnt/β-catenin pathway by scaffolding ezh2. Clin Cancer Res. (2018) 24:684–95. doi: 10.1158/1078-0432.Ccr-17-0605
88. Ge Z, Yin C, Li Y, Tian D, Xiang Y, Li Q, et al. Long noncoding rna neat1 promotes cardiac fibrosis in heart failure through increased recruitment of ezh2 to the smad7 promoter region. J Transl Med. (2022) 20:7. doi: 10.1186/s12967-021-03211-8
89. Yu M, Su Z, Huang X, Zhou Y, Zhang X, Wang B, et al. Histone methyltransferase ezh2 negatively regulates nk cell terminal maturation and function. J Leukoc Biol. (2021) 110:1033–45. doi: 10.1002/jlb.1ma0321-155rr
90. Dhar S, Gadd S, Patel P, Vaynshteyn J, Raju GP, Hashizume R, et al. A tumor suppressor role for ezh2 in diffuse midline glioma pathogenesis. Acta Neuropathol Commun. (2022) 10:47. doi: 10.1186/s40478-022-01336-5
91. Li Z, Liu H, He J, Wang Z, Yin Z, You G, et al. Acetyl-coa synthetase 2: A critical linkage in obesity-induced tumorigenesis in myeloma. Cell Metab. (2021) 33:78–93.e7. doi: 10.1016/j.cmet.2020.12.011
92. Wu YL, Lin ZJ, Li CC, Lin X, Shan SK, Guo B, et al. Adipose exosomal noncoding rnas: roles and mechanisms in metabolic diseases. Obes Rev. (2024) 25(6):e13740. doi: 10.1111/obr.13740
93. Liu H, He J, Koh SP, Zhong Y, Liu Z, Wang Z, et al. Reprogrammed marrow adipocytes contribute to myeloma-induced bone disease. Sci Transl Med. (2019) 11:eaau9807. doi: 10.1126/scitranslmed.aau9087
94. Gao J, Li A, Hu J, Feng L, Liu L, Shen Z. Recent developments in isolating methods for exosomes. Front Bioeng Biotechnol. (2022) 10:1100892. doi: 10.3389/fbioe.2022.1100892
95. Liang Y, He H, Wang W, Wang H, Mo S, Fu R, et al. Malignant clonal evolution drives multiple myeloma cellular ecological diversity and microenvironment reprogramming. Mol Cancer. (2022) 21:182. doi: 10.1186/s12943-022-01648-z
96. Zhang Q, Wang H, Liu Q, Zeng N, Fu G, Qiu Y, et al. Exosomes as powerful biomarkers in cancer: recent advances in isolation and detection techniques. Int J Nanomedicine. (2024) 19:1923–49. doi: 10.2147/ijn.S453545
97. Lyu C, Sun H, Sun Z, Liu Y, Wang Q. Roles of exosomes in immunotherapy for solid cancers. Cell Death Dis. (2024) 15:106. doi: 10.1038/s41419-024-06494-z
98. Li J, Zhang G, Liu CG, Xiang X, Le MTN, Sethi G, et al. The potential role of exosomal circrnas in the tumor microenvironment: insights into cancer diagnosis and therapy. Theranostics. (2022) 12:87–104. doi: 10.7150/thno.64096
99. Xu Z, Chen Y, Ma L, Chen Y, Liu J, Guo Y, et al. Role of exosomal non-coding rnas from tumor cells and tumor-associated macrophages in the tumor microenvironment. Mol Ther. (2022) 30:3133–54. doi: 10.1016/j.ymthe.2022.01.046
100. Wang Z, Wang Q, Qin F, Chen J. Exosomes: A promising avenue for cancer diagnosis beyond treatment. Front Cell Dev Biol. (2024) 12:1344705. doi: 10.3389/fcell.2024.1344705
101. Jo H, Shim K, Jeoung D. Exosomes: diagnostic and therapeutic implications in cancer. Pharmaceutics. (2023) 15:1465. doi: 10.3390/pharmaceutics15051465
102. Khashkhashi Moghadam S, Bakhshinejad B, Khalafizadeh A, Mahmud Hussen B, Babashah S. Non-coding rna-associated competitive endogenous rna regulatory networks: novel diagnostic and therapeutic opportunities for hepatocellular carcinoma. J Cell Mol Med. (2022) 26:287–305. doi: 10.1111/jcmm.17126
103. Shen X, Bai H, Zhu H, Yan Q, Yang Y, Yu W, et al. Long non-coding rna meg3 functions as a competing endogenous rna to regulate hoxa11 expression by sponging mir-181a in multiple myeloma. Cell Physiol Biochem. (2018) 49:87–100. doi: 10.1159/000492846
104. Zang X, Gu J, Zhang J, Shi H, Hou S, Xu X, et al. Exosome-transmitted lncrna ufc1 promotes non-small-cell lung cancer progression by ezh2-mediated epigenetic silencing of pten expression. Cell Death Dis. (2020) 11:215. doi: 10.1038/s41419-020-2409-0
105. Chai Y, Wu HT, Liang CD, You CY, Xie MX, Xiao SW. Exosomal lncrna ror1-as1 derived from tumor cells promotes glioma progression via regulating mir-4686. Int J Nanomedicine. (2020) 15:8863–72. doi: 10.2147/ijn.S271795
106. Li Z, Cao Z, Li N, Wang L, Fu C, Huo R, et al. M2 macrophage-derived exosomal lncrna mir4435-2hg promotes progression of infantile hemangiomas by targeting hnrnpa1. Int J Nanomedicine. (2023) 18:5943–60. doi: 10.2147/ijn.S435132
107. Xiao Q, Lin C, Peng M, Ren J, Jing Y, Lei L, et al. Circulating plasma exosomal long non-coding rnas linc00265, linc00467, uca1, and snhg1 as biomarkers for diagnosis and treatment monitoring of acute myeloid leukemia. Front Oncol. (2022) 12:1033143. doi: 10.3389/fonc.2022.1033143
108. Saltarella I, Lamanuzzi A, Apollonio B, Desantis V, Bartoli G, Vacca A, et al. Role of extracellular vesicle-based cell-to-cell communication in multiple myeloma progression. Cells. (2021) 10:3185. doi: 10.3390/cells10113185
109. Yang X, Lin Z, Liu H, Wang X, Liu H. Multiple myeloma side population cells promote dexamethasone resistance of main population cells through exosome metastasis of lncrna snhg16. J Oncol. (2023) 2023:5135445. doi: 10.1155/2023/5135445
110. Zhao P, Zhao X. Baseline lncrna pcat1 high expression and its longitude increment during induction therapy predict worse prognosis in multiple myeloma patients. J Clin Lab Anal. (2021) 35:e23924. doi: 10.1002/jcla.23924
111. Ferreira BV, Carneiro EA, Pestana C, Barahona F, Caetano J, Lopes R, et al. Patient-derived extracellular vesicles proteins as new biomarkers in multiple myeloma - a real-world study. Front Oncol. (2022) 12:860849. doi: 10.3389/fonc.2022.860849
112. Nielsen T, Kristensen SR, Gregersen H, Teodorescu EM, Christiansen G, Pedersen S. Extracellular vesicle-associated procoagulant phospholipid and tissue factor activity in multiple myeloma. PloS One. (2019) 14:e0210835. doi: 10.1371/journal.pone.0210835
113. Hao L, Wu W, Xu Y, Chen Y, Meng C, Yun J, et al. Lncrna-malat1: A key participant in the occurrence and development of cancer. Molecules. (2023) 28:2126. doi: 10.3390/molecules28052126
114. Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular vesicle-packaged hif-1α-stabilizing lncrna from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. (2019) 21:498–510. doi: 10.1038/s41556-019-0299-0
115. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. (2019) 18:75. doi: 10.1186/s12943-019-0991-5
Keywords: lncRNA, ceRNA, exosomes, multiple myeloma, biological markers, dsRNA, RNA binding proteins
Citation: Yan H, Jiang N, Li X, Lin C, Wang F, Zhang J, Chen L and Li D (2025) Exosomal lncRNAs as diagnostic and therapeutic targets in multiple myeloma. Front. Oncol. 14:1522491. doi: 10.3389/fonc.2024.1522491
Received: 04 November 2024; Accepted: 16 December 2024;
Published: 16 January 2025.
Edited by:
Cyrus Khandanpour, Klinik für Hämatologie und Onkologie, GermanyReviewed by:
Zhe Li, Fudan University, ChinaJinjun Qian, Nanjing University of Chinese Medicine, China
Copyright © 2025 Yan, Jiang, Li, Lin, Wang, Zhang, Chen and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijuan Chen, NDQ0NjgyODFAcXEuY29t; Dan Li, MTA2NDA4MDgyQHFxLmNvbQ==
 Hong Yan
Hong Yan Nan Jiang
Nan Jiang Xiaoying Li1
Xiaoying Li1 Juan Zhang
Juan Zhang Dan Li
Dan Li