- 1Department of Orthopedics, Liyuan Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Department of Nephropathy, Huanggang Hospital of Traditional Chinese Medicine, Hubei University of Traditional Chinese Medicine, Huanggang, China
Osteosarcoma (OS) is a malignancy characterized by the proliferation of osteoblasts that predominantly affects pediatric and adolescent populations. At present, early detection of OS is significantly lacking, coupled with treatment challenges such as high recurrence rates, increased side effects, and the development of drug resistance. Therefore, developing new diagnostic and therapeutic modalities is clinically significant. Exosomes are naturally occurring nanoparticles found in the body that contain various materials, including DNA, RNA, and proteins. Owing to their numerous beneficial properties, including histocompatibility and in vivo stability, they can be useful as drug carriers. With the development of competitive endogenous non-coding RNA (ncRNA) networks, the role of ncRNA in OS cell control has been increasingly studied. This review provides a thorough summary of multiple potential biogenetic pathways of different ncRNAs in exosomes, including microRNAs, long ncRNAs, and circular RNAs. Moreover, the review highlights their effects on OS cells and their potential applications in the diagnosis, treatment, and control of OS drug resistance. The interplay between different types of ncRNAs, which collectively affect OS through the networks of competing endogenous ncRNAs, is the primary focus of this research.
1 Introduction
In osteosarcoma (OS), a type of osteoblastosis, tumor cells generate neoplastic osteogenesis or neoplastic osteoid matrix. The disease primarily affects children and adolescents, typically occurring proximal humerus and near the knee joint of the distal femur, proximal tibia (1–3). Currently, routine radiography and other imaging procedures, as well as the clinical manifestations of patients with frequent bone pain without induction at night, are the primary means of diagnosing OS. Additional diagnostic methods include biopsy, pathology, immunohistochemistry, and auxiliary examinations (4). Before 1970, treatment of OS primarily involved surgical resection. The current MDT strategy involves adjuvant chemoradiotherapy, surgical resection, and preoperative neoadjuvant chemotherapy using a combination of high-dose methotrexate, adriamycin, and cisplatin (5, 6). Unfortunately, these treatments have numerous disadvantages, including incomplete surgical resection, chemoradiotherapy side effects, and tumor resistance development (7).
Exosomes are extracellular vesicles with diameters ranging from 30 to 100 nm. Moreover, they are packed with various RNAs, proteins, lipids, and DNA (8). Exosome research has grown rapidly since 2012, mostly concentrating on the fundamentals of tumor-targeted therapy. With the advancement in exosome research, the non-coding RNA (ncRNA) in exosomes has attracted significant attention. This type of RNA does not translate proteins. There are numerous types of ncRNA, including miRNA, lncRNA, circRNA, tRNA, snRNA, siRNA, and piRNA (9). Exosomal ncRNAs have been explored as potential tools for the early detection and treatment of bone tumors. These ncRNAs are involved in OS cell proliferation, metastasis, apoptosis, and drug resistance (10).
However, few studies on the function of ncRNAs in exosomes in OS have discussed the interactions between different forms of ncRNAs, which collectively affect OS proliferation, metastasis, and death. This can be attributed to our inadequate knowledge of their critical role in these processes. However, they have various effects on the onset and progression of OS through synergistic or competitive antagonistic interactions. Moreover, they play a major role in determining the course of tumor treatment, including therapeutic targets and drug resistance (11). In this study, three extensively researched RNAs, circRNAs, lncRNA, and miRNAs, were chosen for evaluation. The study emphasized their roles in the diagnosis and treatment of OS, particularly their interactions that jointly affect growth, proliferation, apoptosis and drug resistance of OS cells.
2 ncRNA in exosomes
Exosomes are a component of intracellular vesicles, which are mainly produced through the endocytosis pathway. In the formation of multivesicular bodies (MVB), exosomes, which serve as intraluminal vesicles(ILV), are gradually formed and accumulated in endosomal sorting complexes for transport (ESCRT)-dependent pathways and ESCRT-independent pathways through multiple deslamination. Following lysosome-mediated selective transport screening and other modifications, MVB fusion with the cell membrane through exocytosis ultimately releases ILV into the body fluids (12–14). With the progress in research on exosomes, they have demonstrated significant potential in the diagnosis and treatment of OS. Exosomes have the potential to be used as OS biomarkers that could help in early and individualized tumor diagnosis. Moreover, owing to its numerous advantages, including membrane stability and biocompatibility, it can be utilized as a natural drug delivery system to pave a new path in the field of tumor-targeted therapy (15, 16).
Nucleic acids, proteins, and lipids are among the numerous components of exosomes. These vesicles are also intimately engaged in the growth, development, and spread of tumors (17). Among these are the ncRNAs that this study mentions. RNAs that aid in the expression of genes in vivo are known as non-coding RNAs (ncRNAs), and they can be categorized into different forms based on factors such as nucleotide length, shape, and function. These categories include miRNA, lncRNA, circRNA, tRNA, rRNA, siRNA, snRNA, piRNA, and antisense RNA (18). circRNAs, miRNAs, and lncRNAs have been thoroughly investigated. They are also closely associated with OS (19). As a result, the purpose of the three and their interactions are the main topics of this essay. First, miRNAs influence gene expression, cell division, and apoptosis by terminating translation or directly degrading the target mRNA at the microscopic level by complementary pairing with the target mRNA (20). Macro-level manifestations include the regulation of body development and growth (21), fat metabolism (22), hematopoiesis (23), and anti-inflammatory processes (24). Second, lncRNAs influence epigenetic, transcriptional translation, and post-translational modification of genes in cells, thereby affecting the cell cycle, differentiation, growth, and development of cells and the body (25, 26). Ultimately, circRNAs function as competitive endogenous RNA or miRNA sponges that regulate gene transcription and affect the cell cycle, contributing to the development and occurrence of malignancies (27) and illnesses of the cardiovascular system (28–30). More significantly, a substantial body of research demonstrates that all three of these elements are interdependent and inseparable from the body and have an impact on the OS life cycle.
3 Exosomal miRNAs
3.1 Overview of exosomal miRNAs
With 19–25 nucleotides, miRNAs are a type of ncRNAs that primarily assist in mRNA processing and translation in vivo. By destroying mRNA or regulating the initial phase of transcription, miRNAs primarily prevent their translation into proteins (31). In the natural state of the body, exosomal miRNAs are primarily formed in the following ways. Prior to maturation, the RNApol II-catalyzed translation of the miRNA-encoding gene results in pri-miRNA (19), which is further processed into pre-miRNA by the Drosha complex. Once the pre-miRNA enters the cytoplasm via the exportin5 complex (32), it gets processed by the dicer complex (33). Tang et al. (34) and Zhang et al. (35) summarized several potential sorting mechanisms in the cytoplasm that may be used to move mature miRNAs from the cytoplasm to exosomes. These include pathways that rely on particular miRNA sequences, such as the hnRNP pathways (36) and miRNA3 ‘-terminal sequence dependent pathways (37). Additionally, there are pathways that are independent of particular miRNA sequences, such as the nSMase2 pathway (38) and potential miRNA-induced silencing complex (miRISC)/AGO 2 protein-related pathways (39). When the micronucleus (MN) carrying DNA ruptures and MVB wraps the genomic DNA that is dispersed into the cytoplasm, it is transferred to exosomal vesicles during exosome formation (40, 41). This exosome-transferred DNA has the ability to encode miRNAs and other substances which could serve as targets for treatments as well as biomarkers for the identification of specific diseases (42, 43). Engineering methods such as artificial cell modification and transfection, ultrasound, electroporation, and calcium chloride heat shock (44) can also be used to artificially load miRNAs into exosomes under unnatural conditions. (Figure 1) Because of space constraints, these methods are not covered in detail in this study.
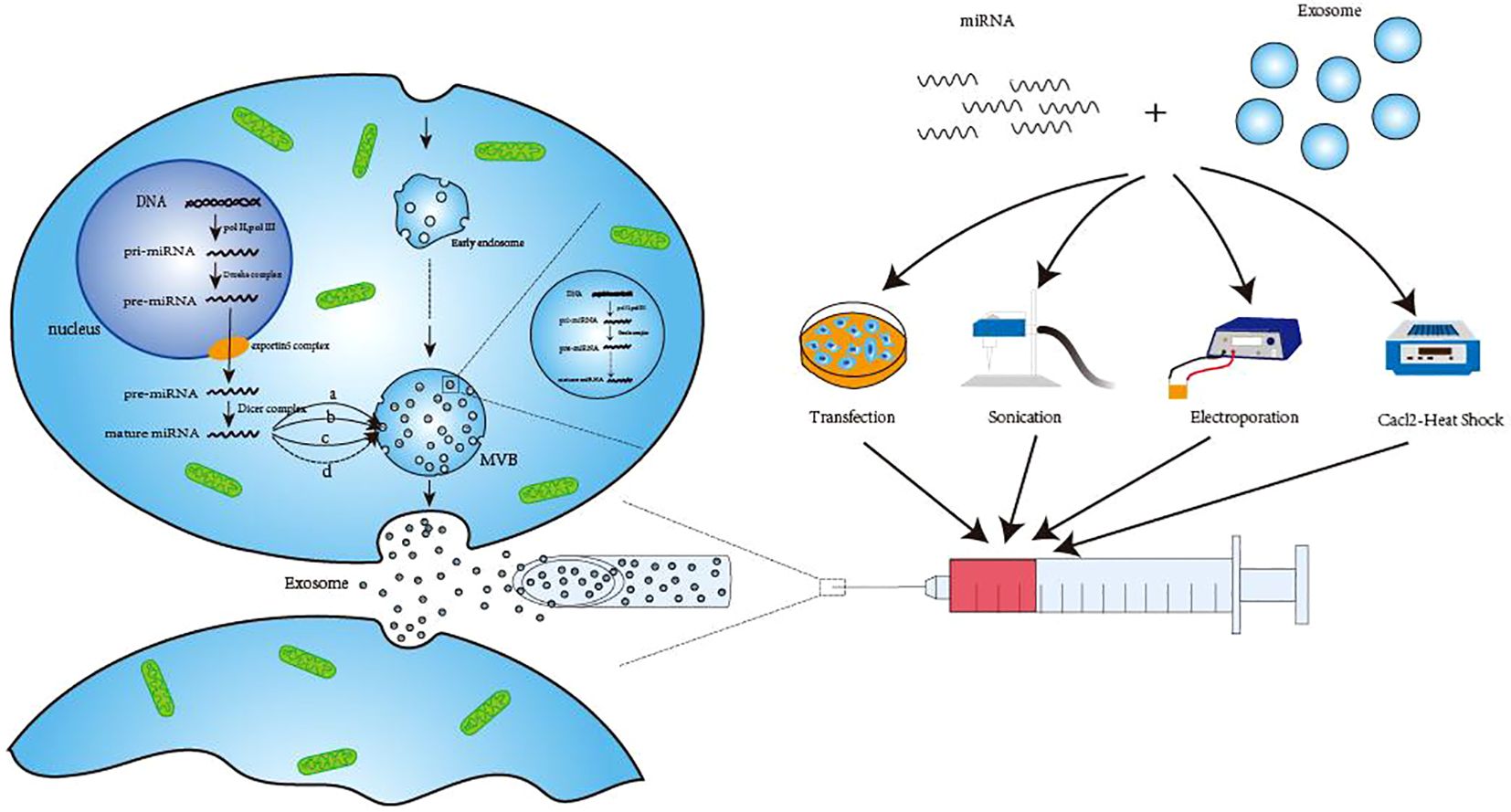
Figure 1. Three ways of exosomal miRNA production. The first way to generate pre-miRNA in the nucleus and then transport it to the cytoplasm to form mature miRNA after processing. Finally, hnRNP pathway (A), miRNA 3’-terminal sequence-dependent pathway (B), nSMase2 pathway (C) and possible miRNA-induced silencing complex (miRISC)/AG02 protein-related pathway (D) are transported to exosomes to perform corresponding functions. The second method is to use the raw material in the exosomes to directly undergo enzymatic shear processing to form mature miRNA. The third way is generated by engineering techniques, such as transfection, sonication, electroporation and Cacl-·Heat Shock. DNA, Deoxyribonucleic Acid; pri-miRNA, primary microRNA; pre-miRNA, precursor microRNA; MVB, Multivesicular body; miRNA, microRNA.
3.2 The role of exosomal miRNAs in the life cycle of OS cells
Exosomal miRNAs affect OS cells in various phases of their life cycle. According to Zhang et al., exosomal miRNAs have two primary functions. First, tumor-derived miR-21 and miR-29a can act as ligands and bind to murine TLR7. Second, they regulate intracellular target genes and bind to target cell mRNA or other receptors as ligands to facilitate intercellular communication (35). For instance, Fabbri et al. (45) discovered human TLR8 receptors, which belong to the Toll-like receptor (TLR) family, in immune cells. This in turn causes inflammation, which stimulates tumor growth and migration. As a result, miRNAs can affect OS cells at various points during their existence. First, research indicates that certain exosomal miRNAs may facilitate OS growth and multiplication. For example, Raimondi et al. (46) showed that the OS cell-derived exosomes miR148a-3p and miR21-5p could be overexpressed to significantly enhance OS activity and differentiation. Overexpression of both can stimulate the production of angiogenic factors in HUVEC, which facilitate the creation of blood vessels. Interestingly, this was not the outcome of the co-transfection. Second, exosomes containing miR-675 (47) and miR-1228 (48) can promote OS migration and invasion. Previous studies have demonstrated that a few exosomal miRNAs, including exosomes miR-208a (49), miR-221-3p (50), and miR-1307 (51), promote OS growth, metastasis, and invasion. Finally, exosomal miR-206 is a classic example of an exosomal miRNA that can cause OS cells to undergo apoptosis or suppress their ability to proliferate and spread. Zhang et al. (52) demonstrated that the targeted suppression of TRA 2B and overexpression of miR-206 can dramatically reduce the growth and metastasis of OS and induce apoptosis. The other examples of exosomal miRNAs are miR-144-3p (53), miR-15a (54), and miR-371b-5p (55).
In summary, exosomal miRNAs significantly affect the onset, growth, multiplication, metastasis, invasion, and apoptosis of OS cells. They offer a wide range of potential applications as therapeutic targets and clinical diagnostic indicators.
3.3 The significance of exosomal miRNAs in the diagnosis, treatment, and prognosis of osteosarcoma, as well as their role in drug resistance
Numerous studies have demonstrated a connection between aberrant miRNAs and a wide range of illnesses, including cancer, cardiovascular system disorders, and endocrine and metabolic disorders (56). In vivo, OS formation, proliferation, migration, and apoptosis are significantly influenced by exosomal miRNAs (57). As a result, it has a significant application value in OS diagnosis and therapy. First, Ye et al. (58) determined that patients with OS had upregulated levels of mir-195-3p, let-7i-3p, miR-92a-3p, and miR-130a-3p using high-throughput sequencing of numerous exosomal miRNAs. In particular, the expression of miR-195-3p. These exosomal miRNAs may serve as biomarkers for early diagnosis of OS. However, further investigation is required. By inhibiting BCL6 expression, exosome miR-101 produced from adipose mesenchymal stromal cells may reduce the incidence and metastasis of OS cells, as shown by Zhang et al. (59). Therefore, it is anticipated to be a useful biomarker of metastatic OS. Second, regarding therapy, McNamara et al. (60) discovered that chemotherapy medications were administered to patients with Kaposi’s sarcoma via exosomes and that exosomal miRNAs may encourage the aggregation of chemotherapy medications within the exosomes originating from tumors. This discovery raises the possibility of poisoning certain tumor cells and the surrounding tissue using chemotherapeutic medicines. In addition, the utilization of exosomes laden with chemotherapy medications might also alter tumor migration, improving the prognosis of the tumor, as demonstrated in the McNamara et al. publication. Prognosis and treatment are related, and a patient’s treatment course has a significant impact on the prognosis.
Lastly, despite minor advancements in immunotherapy and other areas, the primary treatment for OS remains the administration of methotrexate, doxorubicin, and cisplatin (MAP) as neoadjuvant chemotherapy drugs in conjunction with surgery (61, 62). As a result, tumor resistance to chemotherapeutic medications has inevitably drawn the attention of researchers and is challenging. Exosomes provide a possible solution for tumor resistance, prompting researchers to consider them because of the ncRNAs found within them. Exosomal miRNAs affect drug efflux or inactivation, metabolic regulation, tumor microenvironment, classical DNA damage repair mechanisms, and cancer cell apoptosis; therefore, they can affect tumor resistance to drugs (63, 64). Meng et al. (65), for instance, discovered that osteosarcoma cells can enhance their cisplatin resistance through autophagy and exosomal miR-331-3p secretion. Additionally, they observed that drug-resistant osteosarcoma cells can employ exosomal transmission to impart drug resistance to neighboring osteosarcoma cells.Contrary to the findings of earlier investigations on this miRNA (66), the study by Cai et al. (67) also discovered that exosome miR-143-3p was up-regulated in doxorubicin-resistant osteosarcoma cells.Yoshida et al. (68) showed that intracellular miR-25-3p increases OS medication resistance.
In conclusion, Exosomal miRNAs have a major influence on overall OS diagnosis, therapy and drug resistance, although their implementation is hampered by a lack of clarity surrounding some of the relevant mechanisms. As a result, we hope to used exosomal miRNAs as a starting point to eventually release their huge application potential.
4 Exosomal lncRNA
4.1 Overview of exosomal lncRNAs
Long non-coding RNA (LncRNAs) are RNA molecules longer than 200nt that often do not encode proteins and have conserved secondary structures (69). They share a similar biogenesis with miRNAs. First, the transcriptional splicing of genomic DNA is catalyzed by RNA polymerase II in the nucleus. Next, 5’end-capping and 3’ end-polyadenylation occur. The effectively processed lncRNA is exported to the cytoplasm by NXF1 (70, 71). The majority of lncRNAs that enter the cytoplasm are polyribosomal components, but only a small percentage are sorted into exosomes to produce exosomal lncRNAs (72). Unfortunately, the exact mechanism of this sorting process is still unknown, however, Statello et al. (73) showed that it may be connected to several RNA-binding proteins, most notably the Major Vault Protein (MVP), which is able to bind to RNA during the transfer of RNA from the cytoplasm to exosomes and from exosomes to recipient cells, thus maintaining the stability of RNA. On the other hand, exosome lncRNAs may potentially be directly transcribed by DNA in exosomes because exosomes also include DNA derived from nuclei (40),the majority of which are non-coding sections that cannot be expressed. Nevertheless, there aren’t many studies in this field currently, more research is necessary. Finally, similar to miRNAs, exosomal lncRNAs can also be produced artificially through genetic engineering methods such as transfection (74). The development of this field of study opens the door to the possibility of loading exosomal lncRNAs with medications for targeted therapy.
4.2 The role of exosomal lncRNA in the life cycle of OS cells
Exosomal long noncoding RNAs are present in many different cell types throughout the body, and as research progresses, more information is being revealed about their roles. It is involved in various biological processes in the human body. First, considering the inherent characteristics of lncRNAs, they can control gene expression by influencing transcriptional and post-transcriptional modification, epigenetic inheritance, and serve as a sponge for miRNAs (72, 75). Alternatively, attachment to proteins influences the functions of related proteins (76). Second, exosomal long noncoding RNAs (lncRNAs) have the ability to act as mediators in intercellular communication; Zhang et al. (77) discovered that when exosomes containing HOTAIR were added to A549 and H1299 cells, there was an increase in HOTAIR expression as well as an improvement in cell proliferation and invasion capacity. These results suggest that lung cancer exosomes mediate intercellular communication via HOTAIR. And then promote the proliferation and migration of tumor cells. Lastly, exosomal long noncoding RNA are involved in the genesis, proliferation, metastasis, and death of OS cells. Wang et al. (78), for instance, discovered that the lncRNA ELFN 1-AS1, an exosome generated from OS cells, can function as an miR-138-5p and miR-1291 sponge to cause macrophages to polarize towards the M2 type, thereby encouraging the formation of OS. Zhao et al. (79) found that by boosting the expression of the oncogenic protein ERG and preventing the ubiquitination of cancer cells, BMSC-derived exosomes loaded with lncRNA PVT1 could encourage the proliferation and migration of OS cells. By studying the miR-29/NFIA axis, Zhang et al. (80) showed that the exosomal long noncoding RNA LIR-AS1 increased the proliferation and invasion capacity of OS cells and prevented cancer cell apoptosis. Therefore, it may be a potential therapeutic target for OS. Li et al. (81) showed that exosome-related lncRNAs influence miR-153 and autophagy-related protein 5 (ATG5), which, in turn, regulate angiogenesis, migration, and autophagy in OS cells.
4.3 The significance of exosomal lncRNA in the diagnosis, treatment and drug resistance of OS
The primary functions of lncRNAs in vivo include gene expression, mRNA shearing, epigenetic regulation, cell cycle, differentiation regulation, and many other processes (82). These proteins have diverse effects on OS cell activity, proliferation, metastasis, and death. Consequently, it offers a wide range of potential applications in tumor diagnosis and treatment. Exosomal lncRNAs are mostly used as drug delivery vehicles to treat OS and as diagnostic biomarkers. For instance, Yuan et al. (83) evaluated the lncRNA DANCR content of exosomes in benign bone tumors and healthy controls and found that OS patients had considerably higher DANCR expression in exosomes, with a statistically significant difference. Consequently, in OS patients, the exosome lncRNA DANCR could be employed as a potential tumor biomarker. Huang et al. (84) created the engineered exosome cRGD-Exo-MEG 3 by transfecting lncRNA MEG3 into exosomes and altering the targeting ligand. This exosome demonstrated a noteworthy inhibitory effect on OS. This approach is expected to be useful in the clinical treatment of OS. Lastly, exosomal lncRNAs exhibit the potential for use in the treatment of tumor drug resistance. For instance, Hu et al. (85) demonstrated that exosome lncRNA ANCR expression may increase patient overall survival rates in chemotherapy-resistant OS. Furthermore, and because its blood source is readily available and non-invasive, ANCR has the potential to be a prognostic biomarker for OS patients. Additionally, Tao et al. (86) discovered that exosome lncRNA EWSAT1 might influence tumor growth by causing an increase in angiogenic factor secretion and enhancing the sensitivity of vascular endothelial cells, a process known as the “double stacking effect.”
Exosomal lncRNAs are currently under active investigation for their potential in diagnosing and treating OS, with further studies planned for the future.
5 Exosomal circRNA
5.1 Overview of circRNAs in exosomes
CircRNAs, a unique type of ncRNA with nucleotide lengths of more than 200nt in vivo. Its closed-ring structure allows persistent expression and resistance to degradation (87). And it is difficult to be degraded by RNA exonuclease. With advances in research, the function of exosomal circRNAs in vivo has drawn increasing attention. Exosomal circRNAs can be produced in three different ways, similar to the previously mentioned lncRNAs and miRNAs. First, a procedure known as “reverse splicing” occurs in the nucleus, synthesizing circRNAs. This procedure is classified into two models: lariat splicing and direct reverse splicing, depending on the sequence of circRNA cyclization and classical splicing (88). In lariat splicing, a segment comprising certain exons is spliced out first, followed by transcription of the pre-mRNA from the DNA in the nucleus. This segment then targets the 3’terminal position of the exon downstream of the disconnected mRNA and links with it. The remaining ends of the mRNA are joined end-to-end to create a cyclization, which joins the ends to produce a linear and circular double-splicing structure. The latter is achieved by coupling intermediate bases in a complementary manner to produce a Y-shaped structure. The two ends are then split, and the separated ends are linked to produce a two-part construction that is both circular and linear. RNA helicases DDX 39A and 39 B then transport the circRNA synthesized in the nucleus to the cytoplasm, and this process is dependent on the length of the RNA (27). However, the precise procedure remains unclear. Research by Li et al. (89) on the sorting of circRNAs into exosomes revealed a relationship with miR-7, suggesting that the process is mostly controlled by the miRNA content present in cells. Additionally, Dou et al. (90) discovered that this sorting process is highly intricate and may be performed by RNA-binding proteins in exosomes. However, the precise mechanism governing this process remains unknown.
Second, although exosomes are known to contain DNA and certain associated transcriptases, there is no concrete evidence that DNA transcription occurring in exosomes can directly produce circRNAs. Nonetheless, given the intracellular origin of exosomes, it is conceivable that circRNAs may be produced directly from DNA in exosomes via several intricate processes (14). Finally, exosomal circRNAs can be produced in vitro using artificial modification techniques such as transfection and artificial cyclization. These techniques can be used in relevant experiments and serve as prospective clinical diagnostic markers (91, 92).
5.2 The role of exosome circRNA in the life cycle of OS cells
Exosomal circRNAs perform various functions in vivo. circRNAs have the ability to control the molecular expression level of downstream target genes by acting as a miRNA sponge. For example, circRNA Rtn4-modified BMSC exosomes function as miR-146a sponges to prevent TNF-α-induced MC3T3-E1 cytotoxicity and death in mice (93).
Additionally, circRNAs can influence gene expression by regulating transcription, interacting with RNA-binding proteins, and acting as protein sponges to influence their function (94). A small fraction of circRNA is translated into proteins that control biological processes within cells (95). Furthermore, circRNAs can act as intercellular communication media by assuming the role of exosomal content in information transfer between cells. For instance, Lin et al. (96) summarized the role of exosomal circRNA in cell-to-cell communication in cancer biology. Exosomal circRNAs may play a role in the intricate intercellular communication that occurs in the tumor microenvironment between tumor, stromal, and normal cells. Exosomal circRNAs also play significant roles in OS, including drug resistance, angiogenesis, metastasis, invasion, apoptosis, and cell proliferation. It significantly affect OS cell division, migration, invasion, and apoptosis. Li et al. (97) demonstrated that circRNA circ-0000190 nanovesicles prevent cell proliferation, migration, and invasion. Additional analyses revealed that low circRNA expression is associated with tumor growth and migration. Their study showed that the expression of circ-0000190 was considerably lower in OS cell lines than in normal cells. According to Yang et al. (98), circKEAP1 can stimulate the growth and migration of tumor cells by specifically targeting exosomal miR-486-3p in OS cells, resulting in the overexpression of MARCH1. These occurrences imply that exosomal circRNAs affect the OS cell life cycle, and could serve as diagnostic markers and potential therapeutic targets for OS.
5.3 The role of exosomal circRNA in OS diagnosis, treatment, and drug resistance
Owing to their relatively stable ring structure and long half-life in human serum, circRNAs can serve as serum biomarkers to aid in the identification of cancers. Numerous studies have been conducted, and while its clinical applications are still in early stages, efforts to refine and promote their use are actively underway. Because exosomal circRNAs and OS cells are intimately connected, they are expected to aid in clinical diagnosis and treatment. Li et al. discovered that the content of circ-0000190 was considerably lower in the extracellular vesicles and tissues of OS patients, with the majority being encased in extracellular vesicles, as previously described. As a result, it is anticipated that this RNA will serve as a novel biomarker for OS diagnosis. Regarding therapy, exosomal circRNAs have been studied in relation to treating OS; however, owing to numerous obstacles, their useful clinical application has not yet been realized. It is conceivable that exosomal circRNAs that can impact the life cycle of OS cells will serve as therapeutic entry points in the future, and that medications targeting these characteristics may be developed for the treatment of OS. It is theoretically feasible to prevent tumor development and migration by creating tailored exosomes that block circKEAP1, a miRNA sponge known to promote the proliferation and metastasis of OS cells, as previously discussed. This would result in anti-tumor effects. Ultimately, a study by Pan et al. (99) revealed that exosomes can upregulate their receptor cells’ resistance to cisplatin by mediating exosomal circRNA circ_103801. Circ_103801 was overexpressed in cisplatin-resistant cells compared to normal MG63 cells and was abundantly present in a significant number of exosomes. This overexpression reduced the sensitivity of MG63 and U2 cells to cisplatin, increased the expression of P-glycoprotein and multidrug resistance related protein 1, and decreased apoptosis. Considering these outcomes, circ_103801 may be employed as a predictive biomarker to assess the effectiveness of chemotherapy for OS. Moreover, further studies are required to determine its usefulness in clinical settings as a target to overcome drug resistance.
6 Effects of miRNAs, lncRNAs, and circRNAs interactions within exosomes on OS
Since Salmena et al. (100) proposed competitive endogenous RNA (ceRNA) regulation networks, ncRNAs have received increasing attention. Gene expression and cell cycle are regulated by interactions between various types of ncRNAs. Given that exosomes originate from the cytoplasm and include a diverse array of ncRNAs, it is extremely likely that this regulatory network also exists within exosomes. Therefore, the interactions between exosomal ncRNAs, their impact on OS cell invasion and proliferation, and their use in cancer cell diagnosis and treatment are discussed in this study.
6.1 Interaction between miRNA and LncRNA in exosomes
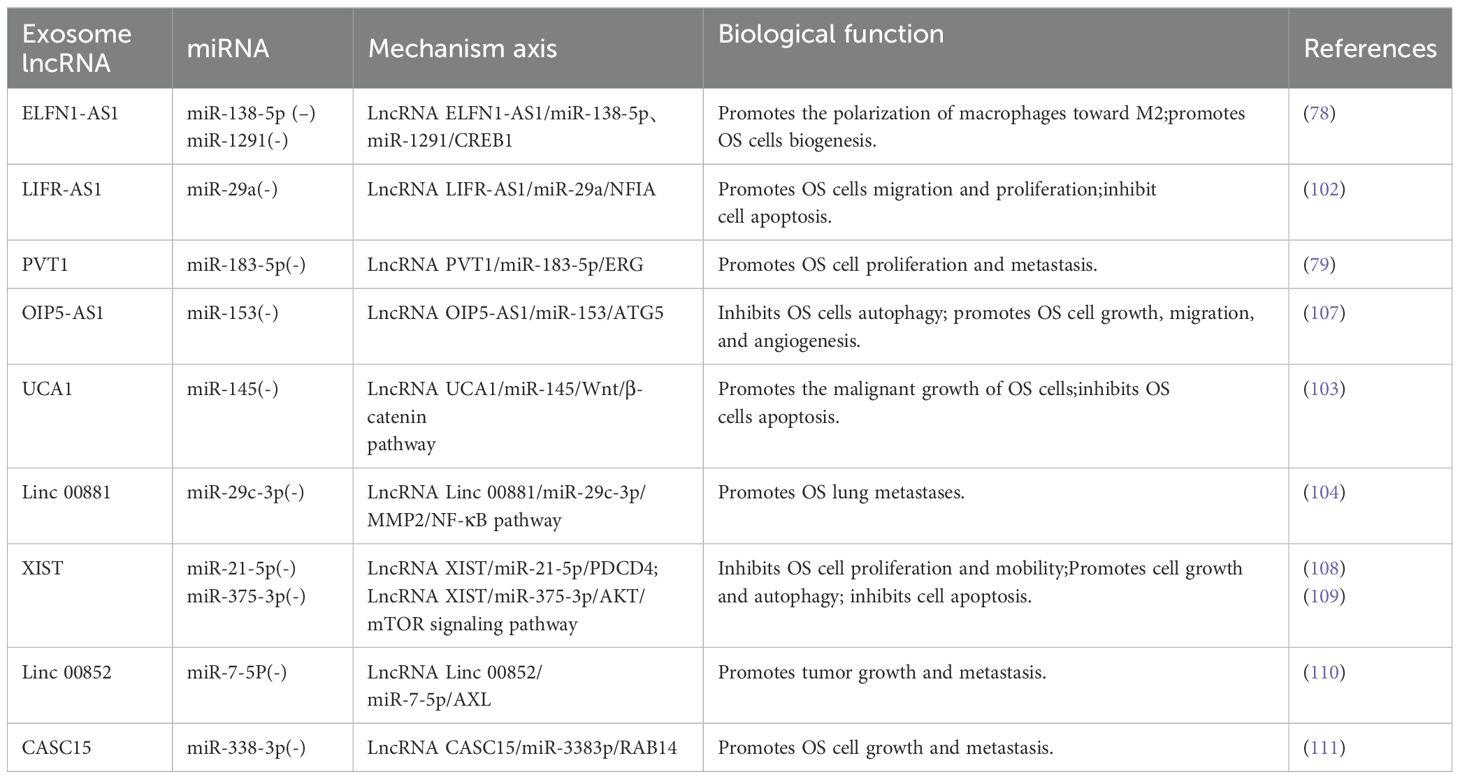
As our understanding of this subject has grown, we have discovered that miRNAs and lncRNAs in exosomes are closely linked and engage in various interactions. Yin et al. (101) summarized the relationship between lncRNAs and miRNAs, and subsequent research has focused on lncRNAs acting as miRNA sponges to competitively inhibiting miRNAs (79). Various studies have discussed the effects of lncRNAs and miRNAs on OS cells and their use in diagnosis and treatment. For instance, Wang et al. (78) found that the OS cell-derived exosome lncRNA ELFN1-AS1 can upregulates CREB1 by suppressing miR-138-5p and miR-1291, promoting macrophage polarization toward M2 and ultimately enhancing OS cell biogenesis. In a different study, Zhang et al. (102) also showed that the macrophage-derived exosome lncRNA LIFR-AS1 suppresses NFIA by downregulating miR-29a, promoting cell migration and proliferation while suppressing apoptosis. According to Zhao et al. (79), exosome-coated lncRNA PVT1 suppresses miR-183-5p and raises ERG expression, thereby boosting OS cell proliferation and metastasis. According to Li et al. (81), by downregulating miR-153 and upregulating autophagy-related protein 5, the enrichment of exosome lncRNA OIP5-AS1 may block autophagy and promote the growth, migration, and angiogenesis of OS cells. As a result, it may be used as an OS therapeutic target. Guan et al. (103) discovered that cutting down lncRNA UCA1 boosted the production of miR-145, which inhibited the malignant growth of OS cells and triggering apoptosis, thereby achieving anti-tumor effects. Chang et al. (104) discovered that lncRNA linc00881, an exosome produced by OS cells, acts as an miR-29c-3p sponge, facilitating intercellular crosstalk between lung fibroblasts and OS cells, regulating associated proteases and signaling cascades, and eventually encouraging OS lung metastases (Table 1). These pathways merit further investigation because they may be employed as viable targets in the management of OS. A negative feedback regulatory loop exists between miRNAs and lncRNA (101). Moreover, lncRNAs control the production of miRNAs by functioning as precursors to miRNAs (105), modifying chromatin (106), and utilizing other mechanisms. This study does not delve into specifics due to constraints related to the length of the paper.
6.2 Interaction between miRNA and circRNA in exosomes
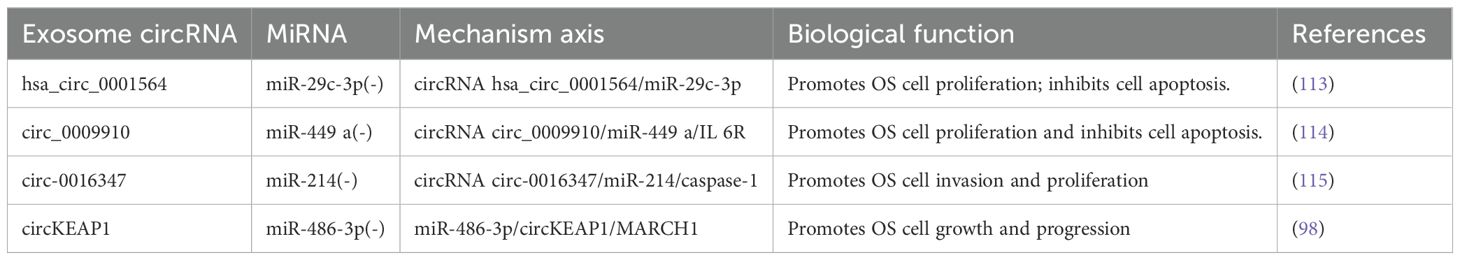
The discovery revealing that circRNAs function as miRNA sponges with competitive inhibitory properties (112) has sparked a rapid increase in circRNA research. The main linkage between miRNAs and circRNAs involves circRNAs acting as miRNA sponges to inhibit miRNA activity and regulate the expression of downstream target genes. This mechanism can be utilized in diverse ways for tumor diagnosis and treatment, indirectly influencing osteosarcoma proliferation, metastasis, and apoptosis. Li et al. (113) discovered that through complimentary binding with miR-29c-3p, the circRNA hsa_circ_0001564 may suppress OS cell proliferation, end the cell cycle, and cause cell death. In the circ_0009910/miR-449 a/IL 6R axis, Deng et al. (114) demonstrated that circ_0009910 upregulates the mRNA expression of the IL-6R gene by suppressing miR-449. This, in turn, alters the OS cell cycle, thereby encouraging the proliferation of cancer cells and preventing their apoptosis. Therefore, circ_0009910 may be a viable target for practical assistance in the diagnosis and treatment of OS. According to Jin et al. (115), circ-0016347 functions as an miR-214 sponge to control the expression of caspase-1, thereby enhancing OS cell invasion and proliferation. Yang et al. (98) demonstrated that circKEAP1 functions as an exosomal miR-486-3p sponge and indirectly upregulates miR-214 expression. This promotes the growth of OS cells; thus, upregulating the expression of miR-486-3p or downregulating the expression of circKEAP1 can prevent the growth of OS and stop its cell cycle (Table 2). However, additional attention is required because what is known in this field is insufficient, and there are currently few relevant clinical application transformations.
6.3 Interaction between LncRNA and circRNA in exosomes
As previously mentioned, circRNAs are a unique class of lncRNAs that function as miRNA sponges and interact with miRNAs to regulate metabolism and gene expression. Through RNA–RNA interactions, lncRNAs and circRNAs collectively establish a competitive endogenous RNA network with miRNAs and mRNAs to control apoptosis, cell metabolism, and body growth and development (116). Research on the regulatory pathways between circRNAs and lncRNAs in the OS cell cycle is limited, and the underlying mechanisms are still unknown. But we can hypothesize that, in ways similar to that of miRNAs, lncRNAs can also impact their function in vivo by functioning as circRNAs sponges and so influencing the OSc cells’ life cycle. Conversely, circRNAs has the same effect on lncRNAs as described above.
6.4 lncRNA/circRNA-miRNA-mRNA axis in exosomes
As previously discussed, research in this field has steadily grown since the introduction of competitive endogenous RNA networks. The two primary axes are circRNA-miRNA-mRNA and lncRNA-miRNA-mRNA. Through various mechanisms, they affect treatment resistance, tumor growth, metastasis, the tumor microenvironment, and cell cycle events such as OS cell proliferation and apoptosis. Wang et al. (117) demonstrated that the Notch signaling pathway, PI3K/AKT pathway, JNK and Wnt pathways, SNHG20/miR-139/RUNX2 axis, and other signaling pathways are various signaling pathways through which the lncRNA-miRNA-mRNA axis can influence the growth and metastasis of OS. For instance, by blocking miR-195-5p, lncRNA SNHG12 enhances Notch2 expression and promotes the development and spread of OS (118). As an miR-33a-5p sponge, lncRNA DANCR promotes the AKT signaling pathway, upregulates the expression of AXL, and accelerates the development of OS (119). The miR-39/RUNX2 axis can be targeted by the downregulation of lncRNA SNHG20, which can also trigger the mitochondrial apoptosis pathway, limit OS development, and cause OS apoptosis (120).
Additionally, the circRNA-miRNA-mRNA axis affects the growth, metastasis, and treatment resistance of OS cells. Liu et al. (121) discovered that by influencing the three axes of miRNA-TRIM21, MAP3K5, and PRKX, the downregulation of circ_0001060 may ultimately decrease the proliferation and metastasis of OS cells. In their research, Qin et al. (122) concluded that competitive endogenous RNA pathways can impact tumor resistance to chemotherapeutic treatments such as doxorubicin and cisplatin through various signaling pathways, including the Wnt/β-catenin pathway and the miRNA-RASSF6 axis. In summary, competitive endogenous RNA networks have a significant impact on OS, and further research is essential to explore their potential clinical applications (Figure 2).
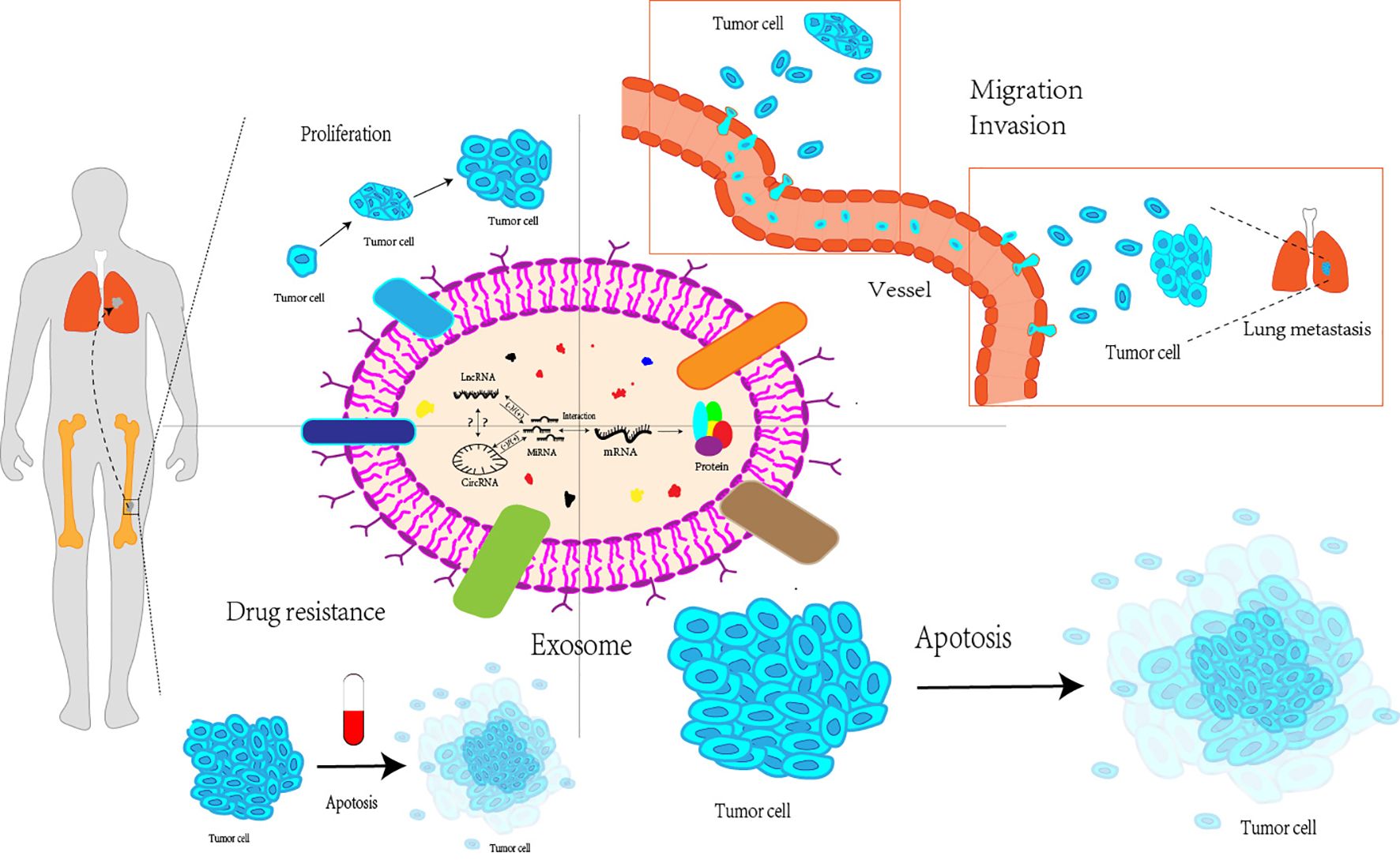
Figure 2. Exosomal miRNAs, lncRNAs and circRNAs interact with each other to affect proliferation, migration and invasion, apoptosis and drug resistance of osteosarcoma cells. Exosomal miRNAs, lncRNAs, and circRNAs can interact with each to affect various processes of the life cycle of osteosarcoma cells, including their proliferation stage, invasion and metastasis to distant regions, apoptosis, as well as the resistance of osteosarcoma cells to multiple drugs LncRNA, Long non-coding RNA; CircRNA, Circular RNA; MiRNA, MicroRNA; mRNA, messenger RNA.
7 Interactions of other ncRNAs and their significance in the diagnosis and treatment of OS
ncRNAs encompass not only the three RNA types mentioned above but also siRNAs, piRNAs, snoRNA, tRNA, and long mRNA-like ncRNAs. These RNAs can also influence other RNAs, such as miRNAs and lncRNAs, to change the course of the OS cell life cycle. The characteristics and applications of siRNAs, piRNAs, and snoRNAs are briefly described in the following section.
First, cells create a class of double-stranded RNA called small interfering RNA (siRNAs), which are 21–25nt in length. Dicer nuclease cuts RNA and depends on the Argonaute family of proteins to accomplish its function (123). It controls the post-transcriptional degradation of mRNA generated by target genes. Zhao et al. (124) synthesized biomimetic nanoparticles by co-polymerizing survivin siRNA and doxorubicin hydrochloride into tumor cells. They observed that these nanoparticles had good tumor-targeting properties, were safe in vivo, and possibly have antitumor therapeutic effects in vivo. After producing chemically altered siRad 18 loaded with engineered RGD-exosomes (RGD-EXOs), Du et al. (125) discovered that lowering the expression of Rad 18 significantly boosted the susceptibility of OS cells to adriamycin. By altering the host liver, Yu et al. (126) were able to transfer small cell vesicles containing VEGFR2 siRNA to the lung. By silencing the VEGFR2 gene, siRNA decreased OS cell lung metastasis. Moreover, siRNAs contributed to drug resistance in OS cells. For instance, siRNA knockdown of lncRNA ANCR affects exosomal ANCR in patients with OS and doxorubicin resistance (127). These findings are expected to be applicable in clinical settings, providing an alternative treatment approach for OS.
Second, a family of short ncRNAs, called PIWI-interacting RNAs (piRNAs), with a length of approximately 30nt interacts with Piwi proteins in a functional manner. It performs biological tasks, including maintaining germline and stem cell functions, controlling translation and mRNA stability, and silencing transcribed genes (128). This type of RNA can also affect OS cell growth and metastasis. According to Das et al. (129), piRNA-39980 can activate MMP-2 and impede SERPINB1 to increase the invasion and metastasis of OS cells while suppressing their death. Wang et al. (130) also discovered notable alterations in the expression of numerous short ncRNAs, such as piRNAs, snoRNAs, and miRNAs, in MG63 OS cells using sequencing and other methods. Tumor cell growth and metastasis have been reported to be inhibited by piRNA DQ596225, snoRNA ENST00000364830.2, snRNA ENST00000410533.1, and miRNA hsa-miR-369-5p. Some medications act on piRNAs and can alter the life cycle of OS cells. Cui et al. (131) discovered that butorphanol suppresses the expression of mRNA FN1 and increases the expression of piRNA hsa_piR_006613, thereby preventing OS cell growth and metastasis. This suggests that butorphanol could serve as a novel therapeutic target for OS. Finally, a class of tiny ncRNAs called small nucleolar RNA (snoRNA) is found in the nucleolus of eukaryotic cells and is involved in the processing of mRNA3 ‘terminal and ribosomal RNA in vivo (132). Regarding its impact on OS cells, Xu et al. (133) discovered that lncRNA H19 suppresses the development of OS by controlling snoRNA expression. This suppression occurs by regulating snoRNA expression, including the suppression of SNORA 7A, and influencing the DNA damage response and repair protein complex. As mentioned above, snoRNA ENST00000364830.2, discovered by Wang et al. (130), can also be utilized as a biomarker for OS diagnosis. Other ncRNAs may also have an impact on OS cell growth, metastasis, and other functions; however space constraints prevent a more detailed exploration of this study. In conclusion, these ncRNAs have a significant effect on the pathophysiology of OS and hold considerable promise for use in both diagnosis and treatment.
8 Conclusions and future prospects
At present, there are several issues associated with primary chemotherapy and surgical treatment of OS, such as drug-resistant tumor cells and postoperative recurrence. Targeted tumor therapy has been widely pursued in the treatment of OS. Recent discoveries in the field of exosomes have led to advancements in this area. Various studies have demonstrated that ncRNA depth influences cell developmental cycles, including in OS cells. Thus, artificial manipulation of these ncRNAs for diagnosing and treating OS is a highly appropriate large entry point. However, ncRNAs are fragile in both serum and intracellular environments, and their widespread distribution poses a challenge in therapeutic applications, limiting their efficiency. Addressing these challenges could provide a valuable solution. Exosomes can be used as natural carriers of ncRNA and medications to treat OS, showing promise due to their relatively stable presence in the blood and cells and excellent tissue targeting. Previous research has focused on the function of a single ncRNA, neglecting the endogenous competitive network that ncRNAs form in vivo. This makes it difficult to avoid the effects of other substances in vivo when the research results are applied. However, progress has been made since the discovery of competitive endogenous RNA networks. In addition, the study of single RNAs in RNA networks is gradually transitioning toward exploring pathways involving multiple RNAs.
Despite these advancements, several challenges must be addressed before exosomes can effectively serve as carriers for ncRNAs in OS therapy. These challenges include difficulties in exosome enrichment, which prevents their widespread application in therapy, inadequate understanding of endogenous RNA networks to enable the development of more reliable molecular pathways that impede OS cells, and it is difficult that the carrier of loading effective ncRNAs and drugs into exosomes in a stable manner. Furthermore, there is a lack of sufficient clinical trials to confirm the efficacy of using exosomes for delivering medicines. Addressing these issues through comprehensive studies is crucial to making exosome-based therapies applicable in clinical practice, offering potential solutions to combat cancer effectively.
Author contributions
WZ: Funding acquisition, Supervision, Writing – review & editing. XL: Visualization, Writing – original draft, Writing – review & editing. YW: Supervision, Visualization, Writing – review & editing. CW: Supervision, Visualization, Writing – review & editing. XW: Writing – review & editing. GT: Writing – review & editing. ZX: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Hubei Provincial Natural Science Foundation. (Grant No.2021CFB333).
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Chen H, Gong Z, Zhou H, Han Y. Deciphering chemoresistance in osteosarcoma: Unveiling regulatory mechanisms and function through the lens of noncoding RNA. Drug Dev Res. (2024) 85:e22167. doi: 10.1002/ddr.22167
2. Wang L, Fan Y, Zhou Yu, Zhong G. Prognosis of limb−salvage treatment of osteosarcoma in adolescent patients: a meta−analysis. Oncol Lett. (2023) 26. https://kns.cnki.net/kcms2/article/abstract?v=M7N75Hb03FVEC2bJShYeNoc0zTRqm4W_YIyuTyDGvgEQW3-Jz4MekN7EmEG2ypvuTtj2tUFM-EgTyNwsRXraVq15Yj8k321HyT3PeIXBToLkzujIpHYAu1V-zvVIU0ogikvUMGtA_iOGCL_xPOUBtQ==&uniplatform=NZKPT&language=gb.
3. Gill J, Gorlick R. Advancing therapy for osteosarcoma. Nat Rev Clin Oncol. (2021) 18:609–24. https://www.nature.com/articles/s41571-021-00519-8.
4. Strauss SJ, Frezza AM, Abecassis N, Bajpai J, Bauer S, Biagini R, et al. Bone sarcomas: ESMO–EURACAN–GENTURIS–ERN PaedCan Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. (2021) 32:1520–36. https://linkinghub.elsevier.com/retrieve/pii/S0923753421042800.
5. Whelan JS, Bielack SS, Marina N, Smeland S, Jovic G, Hook JM, et al. EURAMOS-1, an international randomised study for osteosarcoma: results from pre-randomisation treatment. Ann Oncol. (2015) 26:407–14. https://linkinghub.elsevier.com/retrieve/pii/S0923753419313778.
6. Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma — connecting aetiology, biology and therapy. Nat Rev Endocrinol. (2017) 13:480–91. https://www.nature.com/articles/nrendo.2017.16.
7. Janeway KA, Grier HE. Sequelae of osteosarcoma medical therapy: a review of rare acute toxicities and late effects. Lancet Oncol. (2010) 11:670–8. https://linkinghub.elsevier.com/retrieve/pii/S1470204510700620.
8. Isola AL, Chen S. Exosomes: the messengers of health and disease. CN. (2017) 15:157–65. https://www.eurekaselect.com/145079/article.
9. Tu C, He J, Chen R, Li Z. The emerging role of exosomal non-coding RNAs in musculoskeletal diseases. CPD. (2020) 25:4523–35. http://www.eurekaselect.com/176629/article.
10. Qin S, Wang Y, Ma C, Lv Q. Competitive endogenous network of circRNA, lncRNA, and miRNA in osteosarcoma chemoresistance. Eur J Med Res. (2023) 28:354. doi: 10.1186/s40001-023-01309-x
11. Wang JY, Yang Y, Ma Y, Wang F, Xue A, Zhu J, et al. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomedicine Pharmacotherapy. (2020) 121:109627. https://linkinghub.elsevier.com/retrieve/pii/S0753332219352497.
12. Yáñez-Mó M, Siljander PR -M, Andreu Z, Bedina Zavec A, Borràs FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracellular Vesicle. (2015) 4:27066. doi: 10.3402/jev.v4.27066
13. Tian W, Niu X, Feng F, Wang X, Wang J, Yao W, et al. The promising roles of exosomal microRNAs in osteosarcoma: A new insight into the clinical therapy. Biomedicine Pharmacotherapy. (2023) 163:114771. https://linkinghub.elsevier.com/retrieve/pii/S0753332223005607.
14. Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. (2014) 30:255–89. doi: 10.1146/annurev-cellbio-101512-122326
15. Gilligan K, Dwyer R. Engineering exosomes for cancer therapy. IJMS. (2017) 18:1122. https://www.mdpi.com/1422-0067/18/6/1122.
16. Nazimek K, Bryniarski K. Perspectives in manipulating EVs for therapeutic applications: focus on cancer treatment. IJMS. (2020) 21:4623. https://www.mdpi.com/1422-0067/21/13/4623.
17. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. (2019) 18:75. doi: 10.1186/s12943-019-0991-5
18. Yan H, Bu P. Non-coding RNA in cancer. In: Hon CC, editor. Essays in biochemistry, vol. 65 (2021). p. 625–39. Available at: https://portlandpress.com/essaysbiochem/article/65/4/625/228358/Non-coding-RNA-in-cancer.
19. Yang G, Wu Y, Wan R, Sang H, Liu H, Huang W. The role of non−coding RNAs in the regulation, diagnosis, prognosis and treatment of osteosarcoma (Review). Int J Oncol. (2021) 59:69. doi: 10.3892/ijo.2021.5249
20. Hausser J, Syed AP, Bilen B, Zavolan M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. (2013) 23:604–15. doi: 10.1101/gr.139758.112
21. Maccani MA, Marsit CJ. Exposure and fetal growth-associated miRNA alterations in the human placenta. Clin Epigenet. (2011) 2:401–4. doi: 10.1007/s13148-011-0046-2
22. Luo G, Chen J, Ren Z. Regulation of methylase METTL3 on fat deposition. DMSO. (2021) 14:4843–52. https://www.dovepress.com/regulation-of-methylase-mettl3-on-fat-deposition-peer-reviewed-fulltext-article-DMSO.
23. Petriv OI, Hansen CL, Humphries RK, Kuchenbauer F. Probing the complexity of miRNA expression across hematopoiesis. Cell Cycle. (2011) 10:2–3. doi: 10.4161/cc.10.1.14289
24. Jiang Y, Xu X, Xiao L, Wang L, Qiang S. The role of microRNA in the inflammatory response of wound healing. Front Immunol. (2022) 13:852419/full. doi: 10.3389/fimmu.2022.852419/full
25. Kazimierczyk M, Wrzesinski J. Long non-coding RNA epigenetics. IJMS. (2021) 22:6166. https://www.mdpi.com/1422-0067/22/11/6166.
26. Pan J-F, Zhang Y-J, Li J-Q. LncRNA in different subcellular structures. Zhongguo Shengwu Huaxue yu Fenzi Shengwu Xuebao. (2022) 38:165–74.
27. Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. (2022) 19:188–206. https://www.nature.com/articles/s41571-021-00585-y.
28. Liu CX, Chen LL. Circular RNAs: Characterization, cellular roles, and applications. Cell. (2022) 185:2016–34. https://linkinghub.elsevier.com/retrieve/pii/S0092867422004688.
29. Huang X, Zhao Y, Zhou H, Li Y. Circular RNAs in atherosclerosis. Clinica Chimica Acta. (2022) 531:71–80. https://linkinghub.elsevier.com/retrieve/pii/S0009898122001073.
30. Xiao J. Circular RNAs. In: Biogenesis and functions, vol. 1087. Springer Singapore, Singapore (2018). Advances in Experimental Medicine and Biology. doi: 10.1007/978-981-13-1426-1
31. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. (2011) 12:861–74. https://www.nature.com/articles/nrg3074.
32. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. (2003) 425:415–9. https://www.nature.com/articles/nature01957.
33. Foulkes WD, Priest JR, Duchaine TF. DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer. (2014) 14:662–72. https://www.nature.com/articles/nrc3802.
34. Tang J, He J, Feng C, Tu C. Exosomal miRNAs in osteosarcoma: biogenesis and biological functions. Front Pharmacol. (2022) 13:902049/full. doi: 10.3389/fphar.2022.902049/full
35. Zhang J, Li S, Li L, Li M, Guo C, Yao J, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinf. (2015) 13:17–24. https://linkinghub.elsevier.com/retrieve/pii/S167202291500011X.
36. Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. (2013) 4:2980. https://www.nature.com/articles/ncomms3980.
37. Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MAJ, Sadek P, Sie D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. (2014) 8:1649–58. https://linkinghub.elsevier.com/retrieve/pii/S2211124714007037.
38. Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. (2013) 288:10849–59. https://linkinghub.elsevier.com/retrieve/pii/S0021925820673070.
39. Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. (2014) 8:1432–46. https://linkinghub.elsevier.com/retrieve/pii/S2211124714006196.
40. Yokoi A, Villar-Prados A, Oliphint PA, Zhang J, Song X, De Hoff P, et al. Mechanisms of nuclear content loading to exosomes. Sci Adv. (2019) 5:eaax8849. doi: 10.1126/sciadv.aax8849
41. Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. (2013) 154. https://kns.cnki.net/kcms2/article/abstract?v=9hl5eXOdJcZurYE5F_0mcZqJD8ZfCmqRF_wb0vtyhnGkqcPkA8uI1eMVCNH3neBCJ-pxmEDntu8PFeNJBbe0cAoGtWc_6pXaTY8kp6s-PZzAsjmvbaL0yw_Ehd73zn7l&uniplatform=NZKPT&language=gb.
42. Xiang Z, Xie Q, Yu Z. Exosomal DNA: role in reflecting tumor genetic heterogeneity, diagnosis, and disease monitoring. Cancers. (2023) 16:57. https://www.mdpi.com/2072-6694/16/1/57.
43. Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. (2014) 24. https://kns.cnki.net/kcms2/article/abstract?v=9hl5eXOdJcaJYA8cUXOdT4kB6XkX0yVzj2e1ROL6tuXHISiJEU0CrBlpWWDr9HPUXg3cDbIYO4IkEFRyS-nM6Hf89grnSBLpWHdCSgwLwkIi9j0HUrqgYWid12iZfV8H_cYsRVVNfKhBRV0MZ_knUw==&uniplatform=NZKPT&language=gb.
44. Foo JB, Looi QH, How CW, Lee SH, Al-Masawa ME, Chong PP, et al. Mesenchymal Stem Cell-Derived Exosomes and MicroRNAs in Cartilage Regeneration: Biogenesis, Efficacy, miRNA Enrichment and Delivery. Pharmaceuticals. (2021) 14:1093. https://www.mdpi.com/1424-8247/14/11/1093.
45. Muller F, Alessio P, Federica C, Roberta G, Eugenio G, Ramasamy S, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. (2012) 109. doi: 10.1073/pnas.1209414109
46. Lavinia R, Angela DL, Alessia G, Viviana C, Giovanna R, Nicola C, et al. Osteosarcoma cell-derived exosomes affect tumor microenvironment by specific packaging of microRNAs. Carcinogenesis. (2020) 41:666–77. https://academic.oup.com/carcin/article/41/5/666/5531022.
47. Gong L, Bao Q, Hu C, Wang J, Zhou Q, Wei L, et al. Exosomal miR-675 from metastatic osteosarcoma promotes cell migration and invasion by targeting CALN1. Biochem Biophys Res Commun. (2018) 500:170–6. https://linkinghub.elsevier.com/retrieve/pii/S0006291X18307794.
48. Wang JW, Wu XF, Gu XJ, Jiang XH. Exosomal miR-1228 from cancer-associated fibroblasts promotes cell migration and invasion of osteosarcoma by directly targeting SCAI. Oncol Res. (2019) 27:979–86. doi: 10.3727/096504018X15336368805108
49. Qin F, Tang H, Zhang Y, Zhang Z, Huang P, Zhu J. Bone marrow-derived mesenchymal stem cell-derived exosomal microRNA-208a promotes osteosarcoma cell proliferation, migration, and invasion. J Cell Physiol. (2020) 235:4734–45. doi: 10.1002/jcp.29351
50. Liu W, Long Q, Zhang W, Zeng D, Hu B, Liu S, et al. miRNA-221-3p derived from M2-polarized tumor-associated macrophage exosomes aggravates the growth and metastasis of osteosarcoma through SOCS3/JAK2/STAT3 axis. Aging. (2021) 13:19760–75. doi: 10.18632/aging.203388
51. Han F, Pu P, Wang C, Ding X, Zhu Z, Xiang W, et al. Osteosarcoma Cell-Derived Exosomal miR-1307 Promotes Tumorgenesis via Targeting AGAP1. In: Chutipongtanate S, editor. BioMed research international, vol. 2021 (2021). p. 1–17. Available at: https://www.hindawi.com/journals/bmri/2021/7358153/.
52. Zhang H, Wang J, Ren T, Huang Y, Liang X, Yu Y, et al. Bone marrow mesenchymal stem cell-derived exosomal miR-206 inhibits osteosarcoma progression by targeting TRA2B. Cancer Lett. (2020) 490:54–65. https://linkinghub.elsevier.com/retrieve/pii/S0304383520303633.
53. Jiang M, Jike Y, Liu K, Gan F, Zhang K, Xie M, et al. Exosome-mediated miR-144-3p promotes ferroptosis to inhibit osteosarcoma proliferation, migration, and invasion through regulating ZEB1. Mol Cancer. (2023) 22:113. doi: 10.1186/s12943-023-01804-z
54. Wu C, Li Z, Feng G, Wang L, Xie J, Jin Y, et al. Tumor suppressing role of serum-derived exosomal microRNA-15a in osteosarcoma cells through the GATA binding protein 2/murine double minute 2 axis and the p53 signaling pathway. Bioengineered. (2021) 12:8378–95. doi: 10.1080/21655979.2021.1987092
55. Xue Q, Yang Y, Yang L, Yan X, Shen Z, Liu J, et al. miR-371b-5p-engineered exosomes enhances tumor inhibitory effect. Front Cell Dev Biol. (2021) 9:750171/full. doi: 10.3389/fcell.2021.750171/full
56. Beyer C, Zampetaki A, Lin NY, Kleyer A, Perricone C, Iagnocco A, et al. Signature of circulating microRNAs in osteoarthritis. Ann Rheum Dis. (2015) 74:e18–8. doi: 10.1136/annrheumdis-2013-204698
57. Cai X, Yin W, Tang C, Lu Y, He Y. Molecular mechanism of microRNAs regulating apoptosis in osteosarcoma. Mol Biol Rep. (2022) 49:6945–56. doi: 10.1007/s11033-022-07344-x
58. Ye Z, Zheng Z, Peng L. Microrna profiling of serum exosomes in patients with osteosarcoma by high-throughput sequencing. J Invest Med. (2020) 68:893–901. doi: 10.1136/jim-2019-001196
59. Zhang K, Dong C, Chen M, Yang T, Wang X, Gao Y, et al. Extracellular vesicle-mediated delivery of miR-101 inhibits lung metastasis in osteosarcoma. Theranostics. (2020) 10:411–25. http://www.thno.org/v10p0411.htm.
60. McNamara RP, Eason AB, Zhou Y, Bigi R, Griffith JD, Costantini LM, et al. Exosome-encased nucleic acid scaffold chemotherapeutic agents for superior anti-tumor and anti-angiogenesis activity. ACS Bio Med Chem Au. (2022) 2:140–9. doi: 10.1021/acsbiomedchemau.1c00030
61. Chen C, Xie L, Ren T, Huang Y, Xu J, Guo W. Immunotherapy for osteosarcoma: Fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett. (2021) 500:1–10. https://linkinghub.elsevier.com/retrieve/pii/S0304383520306844.
62. Smrke A, Anderson PM, Gulia A, Gennatas S, Huang PH, Jones RL. Future directions in the treatment of osteosarcoma. Cells. (2021) 10:172. https://www.mdpi.com/2073-4409/10/1/172.
63. Guo Q, Wang H, Yan Y, Liu Y, Su Cy, Chen Hb, et al. The role of exosomal microRNA in cancer drug resistance. Front Oncol. (2020) 10:472/full. doi: 10.3389/fonc.2020.00472/full
64. Yang Z, Ma R, Li J, Zhao L. Noncoding RNAs in esophageal cancer: A glimpse into implications for therapy resistance. Pharmacol Res. (2023) 188:106678. https://linkinghub.elsevier.com/retrieve/pii/S1043661823000348.
65. Meng C. Exosomal miR-331-3p derived from chemoresistant osteosarcoma cells induces chemoresistance through autophagy. (2023). doi: 10.1186/s13018-023-04338-8
66. Zhou J, Wu S, Chen Y, Zhao J, Zhang K, Wang J, et al. microRNA-143 is associated with the survival of ALDH1 + CD133 + osteosarcoma cells and the chemoresistance of osteosarcoma. Exp Biol Med (Maywood). (2015) 240:867–75. doi: 10.1177/1535370214563893
67. Cai T, Zhang C, Zhan T. Transfer of exosomal microRNAs confers doxorubicin resistance in osteosarcoma cells. Mol Med Rep. (2023) 27:86. doi: 10.3892/mmr.2023.12973
68. Yoshida A, Fujiwara T, Uotani K, Morita T, Kiyono M, Yokoo S, et al. Clinical and functional significance of intracellular and extracellular microRNA-25-3p in osteosarcoma. Acta Med Okayama. (2018) 72.
69. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. (2023) 24:430–47. https://www.nature.com/articles/s41580-022-00566-8.
70. Choudhuri S. Long noncoding RNAs: biogenesis, regulation, function, and their emerging significance in toxicology. Toxicol Mech Methods. (2023) 33:541–51. doi: 10.1080/15376516.2023.2197489
71. Nojima T, Proudfoot NJ. Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat Rev Mol Cell Biol. (2022) 23:389–406. https://www.nature.com/articles/s41580-021-00447-6.
72. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. (2021) 22:96–118. https://www.nature.com/articles/s41580-020-00315-9.
73. Statello L, Maugeri M, Garre E, Nawaz M, Wahlgren J, Papadimitriou A, et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PloS One. (2018) 13:e0195969. doi: 10.1371/journal.pone.0195969
74. Huang X, Wu W, Jing D, Yang L, Guo H, Wang L, et al. Engineered exosome as targeted lncRNA MEG3 delivery vehicles for osteosarcoma therapy. J Controlled Release. (2022) 343:107–17. https://linkinghub.elsevier.com/retrieve/pii/S0168365922000402.
75. Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. (2015) 6:22513–25. doi: 10.18632/oncotarget.4154
76. Li Q, Dong C, Cui J, Wang Y, Hong X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res. (2018) 37:265. doi: 10.1186/s13046-018-0941-x
77. Zhang C, Xu L, Deng G, Ding Y, Bi K, Jin H, et al. Exosomal HOTAIR promotes proliferation, migration and invasion of lung cancer by sponging miR-203. Sci China Life Sci. (2020) 63:1265–8. doi: 10.1007/s11427-019-1579-x
78. Wang B, Wang X, Li P, Niu X, Liang X, Liu G, et al. Osteosarcoma Cell-Derived Exosomal ELFN1-AS1 Mediates Macrophage M2 Polarization via Sponging miR-138-5p and miR-1291 to Promote the Tumorgenesis of Osteosarcoma. Front Oncol. (2022) 12:881022/full. doi: 10.3389/fonc.2022.881022/full
79. Zhao W, Qin P, Zhang D, Cui X, Gao J, Yu Z, et al. Long non-coding RNA PVT1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing ERG and sponging miR-183-5p. Aging. (2019) 11:9581–96. doi: 10.18632/aging.102406
80. Zhang H, Yu Y, Wang J, Han Y, Ren T, Huang Y, et al. Macrophages-derived exosomal lncRNA LIFR-AS1 promotes osteosarcoma cell progression via miR-29a/NFIA axis. Cancer Cell Int. (2021) 21. https://kns.cnki.net/kcms2/article/abstract?v=2dRWw02s3kxjUlbrG3RLSQeFxq0AXg8sj2SNK-x3i-RzhbhEj-Kt3hHs9-qW4DWbT3tDbqEkJXGYh30reLuORm0-F53h_k5onxY13QRfT98Ho_-JGHEkS9bUz63HDxL9tSJJY7qCHr0-vMo4idvK8A==&uniplatform=NZKPT&language=gb.
81. Li Y, Lin S, Xie X, Zhu H, Fan T, Wang S. Highly enriched exosomal lncRNA OIP5-AS1 regulates osteosarcoma tumor angiogenesis and autophagy through miR-153 and ATG5. Am J Trans Res. (2021) 13:4211–23.
82. Arab I, Park J, Shin JJ, Shin HS, Suk K, Lee WH. Macrophage lncRNAs in cancer development: Long-awaited therapeutic targets. Biochem Pharmacol. (2023) 218:115890. https://linkinghub.elsevier.com/retrieve/pii/S0006295223004811.
83. Yuan T, Liu J, Chen S, Jiang L, Chen Z, Chen J, et al. Clinical significance of exosomal long noncoding RNA DANCR as a novel serum-based diagnostic and prognostic biomarker in osteosarcoma. Int J Clin Exp Med. (2019) 12:423–32.
84. Huang X, Wu W, Jing D, Yang L, Guo H, Wang L, et al. Engineered exosome as targeted lncRNA MEG3 delivery vehicles for osteosarcoma therapy. J Controlled Release. (2022) 343:107–17. https://linkinghub.elsevier.com/retrieve/pii/S0168365922000402.
85. Hu X, Wen Y, Tan LY, Wang J, Tang F, Wang YT, et al. Exosomal long non-coding RNA ANCR mediates drug resistance in osteosarcoma. Front Oncol. (2022) 11:735254/full. doi: 10.3389/fonc.2021.735254/full
86. Tao S, Huang J, Wei Z, Li Z, Guo S. EWSAT1 acts in concert with exosomes in osteosarcoma progression and tumor-induced angiogenesis: the “Double stacking effect. Adv Biosys. (2020) 4:2000152. doi: 10.1002/adbi.202000152
87. Fanale D, Taverna S, Russo A, Bazan V. Circular RNA in exosomes. In: Xiao J, editor. Advances in experimental medicine and biology, vol. 1087 . Springer Singapore, Singapore (2018). p. 109–17. Circular RNAs. doi: 10.1007/978-981-13-1426-1_9
88. Eger N, Schoppe L, Schuster S, Laufs U, Boeckel JN. Circular RNA splicing. In: Xiao J, editor. Advances in experimental medicine and biology, vol. 1087 . Springer Singapore, Singapore (2018). p. 41–52. Circular RNAs. doi: 10.1007/978-981-13-1426-1_4
89. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. (2015) 25:981–4. https://www.nature.com/articles/cr201582.
90. Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. (2016) 6:37982. https://www.nature.com/articles/srep37982.
91. Hu N, Cai Z, Jiang X, Wang C, Tang T, Xu T, et al. Hypoxia-pretreated ADSC-derived exosome-embedded hydrogels promote angiogenesis and accelerate diabetic wound healing. Acta Biomaterialia. (2023) 157:175–86. https://linkinghub.elsevier.com/retrieve/pii/S1742706122007851.
92. Müller S, Appel B. In vitro circularization of RNA. RNA Biol. (2017) 14:1018–27. doi: 10.1080/15476286.2016.1239009
93. Cao G, Meng X, Han X, Li J. Exosomes derived from circRNA Rtn4-modified BMSCs attenuate TNF-α-induced cytotoxicity and apoptosis in murine MC3T3-E1 cells by sponging miR-146a. Bioscience Rep. (2020) 40. doi: 10.1042/bsr20193436
94. Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and Characterizing circRNA-Protein Interaction. Theranostics. (2017) 7:4183–91. http://www.thno.org/v07p4183.htm.
95. Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. (1995) 268:415–7. doi: 10.1126/science.7536344
96. Lin H, Yu J, Gu X, Ge S, Fan X. Novel insights into exosomal circular RNAs: Redefining intercellular communication in cancer biology. Clin Trans Med. (2021) 11:e636. doi: 10.1002/ctm2.636
97. Li S, Pei Y, Wang W, Liu F, Zheng K, Zhang X. Extracellular nanovesicles-transmitted circular RNA has_circ_0000190 suppresses osteosarcoma progression. J Cell Mol Medi. (2020) 24:2202–14. doi: 10.1111/jcmm.14877
98. Yang H, He C, Feng Y, Jin J. Exosome−delivered miR−486−3p inhibits the progression of osteosarcoma via sponging CircKEAP1/MARCH1 axis components. Oncol Lett. (2023) 27:24. doi: 10.3892/ol.2023.14157
99. Pan Y, Lin Y, Mi C. Cisplatin-resistant osteosarcoma cell-derived exosomes confer cisplatin resistance to recipient cells in an exosomal circ_103801-dependent manner. Cell Biol Int. (2021) 45:858–68. doi: 10.1002/cbin.11532
100. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell. (2011) 146:353–8. https://linkinghub.elsevier.com/retrieve/pii/S0092867411008129.
101. Yin LD, Sun Q, Sun M, De W. Advances in the Interaction between long non-coding RNAs and miRNAs in human cancer. J Clin Oncol. (2014) 19:662–6. https://kns.cnki.net/kcms2/article/abstract?v=C06iYwc_NfQJfZ7S5u2EMxlbSfeTe-70T_8eDuVsWIZsPwoeD6GspejBl6chmOWREahiEx1TK17V-t4eekH0hjA-0YQQKmXJKzhUlwyfFgUIlFMNCE0-Qw==&uniplatform=NZKPT&language=gb.
102. Zhang H, Yu Y, Wang J, Han Y, Ren T, Huang Y, et al. Macrophages-derived exosomal lncRNA LIFR-AS1 promotes osteosarcoma cell progression via miR-29a/NFIA axis. Cancer Cell Int. (2021) 21. https://kns.cnki.net/kcms2/article/abstract?v=b4E8SuETvlKck8wFFLFZuMResIGnYoAuUrRBgVoxwQZOcqmbX3A2xLXC753f3j121Pki9a8UzW90nUPCvwKh1WnAIsYek2bKtdpS_t5gWurVTOIa9-5ug4Kx41y2X-vgcEtQpZGkWluQc68ASnBUZg==&uniplatform=NZKPT&language=gb.
103. Guan J, He J, Liao S, Wu Z, Lin X, Liu B, et al. LncRNA UCA1 accelerates osteosarcoma progression via miR-145 and Wnt/β-catenin pathway. Am J Trans Res. (2022) 14:6029–42.
104. Chang X, Tan Q, Xu J, Wu X, Wang Y, Zhang Y, et al. Tumor-derived exosomal linc00881 induces lung fibroblast activation and promotes osteosarcoma lung migration. Cancer Cell Int. (2023) 23:287. doi: 10.1186/s12935-023-03121-3
105. Robson JE, Eaton SA, Underhill P, Williams D, Peters J. MicroRNAs 296 and 298 are imprinted and part of the GNAS/Gnas cluster and miR-296 targets IKBKE and Tmed9. RNA. (2012) 18:135–44. doi: 10.1261/rna.029561.111
106. Neveu P, Kye MJ, Qi S, Buchholz DE, Clegg DO, Sahin M, et al. MicroRNA Profiling Reveals Two Distinct p53-Related Human Pluripotent Stem Cell States. Cell Stem Cell. (2010) 7:671–81. https://linkinghub.elsevier.com/retrieve/pii/S193459091000634X.
107. Li Y, Lin S, Xie X, Zhu H, Fan T, Wang S. Highly enriched exosomal lncRNA OIP5-AS1 regulates osteosarcoma tumor angiogenesis and autophagy through miR-153 and ATG5. Am J Trans Res. (2021) 13:4211–23.
108. Zhang R, Xia T. Long non-coding RNA XIST regulates PDCD4 expression by interacting with miR-21-5p and inhibits osteosarcoma cell growth and metastasis. Int J Oncol. (2017) 51:1460–70. doi: 10.3892/ijo.2017.4127
109. Sun X, Wei B, Peng ZH, Fu QL, Wang CJ, Zheng JC, et al. Knockdown of lncRNA XIST suppresses osteosarcoma progression by inactivating AKT/mTOR signaling pathway by sponging miR-375-3p. Int J Clin Exp Pathology. (2019) 12:1507–17.
110. Li Q, Wang X, Jiang N, Xie X, Liu N, Liu J, et al. Exosome-transmitted linc00852 associated with receptor tyrosine kinase AXL dysregulates the proliferation and invasion of osteosarcoma. Cancer Med. (2020) 9:6354–66. doi: 10.1002/cam4.3303
111. Zhang H, Wang J, Ren T, Huang Y, Yu Y, Chen C, et al. LncRNA CASC15 is Upregulated in Osteosarcoma Plasma Exosomes and CASC15 Knockdown Inhibits Osteosarcoma Progression by Regulating miR-338-3p/RAB14 Axis. OTT. (2020) 13:12055–66. https://www.dovepress.com/lncrna-casc15-is-upregulated-in-osteosarcoma-plasma-exosomes-and-casc1-peer-reviewed-article-OTT.
112. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. https://www.nature.com/articles/nature11993.
113. Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun. (2018) 495:2369–75. https://linkinghub.elsevier.com/retrieve/pii/S0006291X17324415.
114. Deng N, Li L, Gao J, Zhou J, Wang Y, Wang C, et al. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun. (2018) 495:189–96. https://linkinghub.elsevier.com/retrieve/pii/S0006291X17322052.
115. Jin H, Jin X, Zhang H, Wang W. Circular RNA hsa-circ-0016347 promotes proliferation, invasion and metastasis of osteosarcoma cells. Oncotarget. (2017) 8:25571–81. doi: 10.18632/oncotarget.16104
116. Zhu K, Wang Q, Wang L. Analysis of competitive endogenous RNA regulatory network of exosomal breast cancer based on exoRBase. Evol Bioinform Online. (2022) 18:117693432211132. doi: 10.1177/11769343221113286
117. Wang J, Yang Y, Ma Y, Wang F, Xue A, Zhu J, et al. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomedicine Pharmacotherapy. (2020) 121. doi: 10.1016/j.biopha.2019.109627
118. Zhou S, Yu L, Xiong M, Dai G. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res Commun. (2018) 495:1822–32. https://linkinghub.elsevier.com/retrieve/pii/S0006291X17324397.
119. Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu J, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. (2017) 405:46–55. https://linkinghub.elsevier.com/retrieve/pii/S0304383517303907.
120. Wang W, Luo P, Guo W, Shi Y, Xu D, Zheng H, et al. LncRNA SNHG20 knockdown suppresses the osteosarcoma tumorigenesis through the mitochondrial apoptosis pathway by miR-139/RUNX2 axis. Biochem Biophys Res Commun. (2018) 503:1927–33. https://linkinghub.elsevier.com/retrieve/pii/S0006291X18316413.
121. Liu H, Huang Q, Tang H, Luo K, Qin Y, Li F, et al. Circ_0001060 upregulates and encourages progression in osteosarcoma. DNA Cell Biol. (2023) 42:53–64. doi: 10.1089/dna.2022.0500
122. Qin S, Wang Y, Ma C, Lv Q. Competitive endogenous network of circRNA, lncRNA, and miRNA in osteosarcoma chemoresistance. Eur J Med Res. (2023) 28. doi: 10.1186/s40001-023-01309-x
123. Lee IN, Lu YP, Chen JC, Yang CH, Huang JW, Yang JT, et al. Emerging Physical Approaches for Promoting siRNA Intracellular Delivery for Cancer Therapy. COC. (2016) 20:2960–70. http://www.eurekaselect.com/openurl/content.php?genre=article&issn=1385-2728&volume=20&issue=28&spage=2960.
124. Zhao J, Mu X, Hou X, Zhang X, Li P, Jiang J. Synergistic treatment of osteosarcoma with biomimetic nanoparticles transporting doxorubicin and siRNA. Front Oncol. (2023) 13:1111855/full. doi: 10.3389/fonc.2023.1111855/full
125. Du M, Gu J, Liu C, Liu N, Yu Z, Zhou C, et al. Genome-wide CRISPR screen identified Rad18 as a determinant of doxorubicin sensitivity in osteosarcoma. J Exp Clin Cancer Res. (2022) 41:154. doi: 10.1186/s13046-022-02344-y
126. Yu L, Fan G, Wang Q, Zhu Y, Zhu H, Chang J, et al. In vivo self-assembly and delivery of VEGFR2 siRNA-encapsulated small extracellular vesicles for lung metastatic osteosarcoma therapy. Cell Death Dis. (2023) 14. doi: 10.1038/s41419-023-06159-3
127. Hu X, Wen Y, Tan L, Wang J, Tang F, Wang Y, et al. Exosomal long non-coding RNA ANCR mediates drug resistance in osteosarcoma. Front Oncol. (2022) 11. doi: 10.3389/fonc.2021.735254
128. Zhang J, Chen S, Liu K. Structural insights into piRNA biogenesis. Biochim Biophys Acta (BBA) - Gene Regul Mech. (2022) 1865:194799. https://linkinghub.elsevier.com/retrieve/pii/S1874939922000141.
129. Das B, Jain N, Mallick B. piR-39980 promotes cell proliferation, migration and invasion, and inhibits apoptosis via repression of SERPINB1 in human osteosarcoma. Biol Cell. (2020) 112:73–91. doi: 10.1111/boc.201900063
130. Wang H, Cai G, Yu F, Li D, Wang C, Ma D, et al. Changes in the small noncoding RNA transcriptome in osteosarcoma cells. J Orthopaedic Surg Res. (2023) 18. doi: 10.1186/s13018-023-04362-8
131. Cui P, Xin D, Li F, Deng L, Gao Y. Butorphanol Suppresses the Proliferation and Migration of Osteosarcoma by Promoting the Expression of piRNA hsa_piR_006613. Front Oncol. (2022) 12. doi: 10.3389/fonc.2022.775132
132. Liang J, Wen J, Huang Z, Chen XP, Zhang BX, Chu L. Small nucleolar RNAs: insight into their function in cancer. Front Oncol. (2019) 9:587/full. doi: 10.3389/fonc.2019.00587/full
Keywords: exosome, osteosarcoma, non-coding RNA, micro-RNA, long non-coding RNA, circular RNA
Citation: Liu X, Wang Y, Wang C, Wang X, Tang G, Xiong Z and Zhou W (2024) Role of non-coding RNA in exosomes for the diagnosis and treatment of osteosarcoma. Front. Oncol. 14:1469833. doi: 10.3389/fonc.2024.1469833
Received: 24 July 2024; Accepted: 08 October 2024;
Published: 24 October 2024.
Edited by:
Yujing Li, Emory University, United StatesReviewed by:
Francesca Lovat, The Ohio State University, United StatesZhen Mei, University of California, Davis, United States
Copyright © 2024 Liu, Wang, Wang, Wang, Tang, Xiong and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Zhou, ODcxODQ2OTk4QHFxLmNvbQ==
 Xin Liu
Xin Liu Yaling Wang2
Yaling Wang2