- 1Analysis of Circulating Tumor Cells Lab, Lab of Analytical Chemistry, Department of Chemistry, National and Kapodistrian University of Athens, Athens, Greece
- 2Laboratory of Biochemistry/Metastatic Signaling, Section of Genetics, Cell Biology and Development, Department of Biology, University of Patras, Patras, Greece
- 3Department of Medical Oncology, General University Hospital of Larissa, Larissa, Greece
- 4First Department of Medical Oncology, Metropolitan General Hospital of Athens, Cholargos, Greece
Background: The heterogeneous and complex genetic landscape of NSCLC impacts the clinical outcomes of patients who will eventually develop resistance to osimertinib. Liquid biopsy (LB) analysis as a minimally invasive approach is a key step to efficiently identify resistance mechanisms and adjust to proper subsequent treatments.
Materials and methods: In the present study, we combined plasma-cfDNA and CTC analysis from 30 NSCLC patients in samples collected before treatment and at the progression of disease (PD). We detected molecular alterations at the DNA mutation (EGFR, PIK3CA, KRAS G12C, BRAF V600E), DNA methylation (RASSF1A, BRMS1, FOXA1, SLFN1, SHISA3, RARβ,, WIF-1, RASSF10 and APC), gene expression (CK-19, CK-18, CK-8, AXL, TWIST-1, PD-L1, PIM-1, Vimentin, ALDH-1, and B2M) and chromosomal level (HER2 and MET amplification) as possible resistance mechanisms and druggable targets. We also studied the expression of PD-L1 in single CTCs using immunofluorescence.
Results: In some cases, T790M resistance EGFR mutation was detected at baseline in CTCs but not in the corresponding plasma cfDNA. PIK3CA mutations were detected only in plasma-cfDNA but not in corresponding CTCs. KRAS G12C and BRAF V600E mutations were not detected in the samples analyzed. MET amplification was detected in the CTCs of two patients before treatment whereas HER2 amplification was detected in the CTCs of three patients at baseline and in one patient at PD. DNA methylation analysis revealed low concordance between CTCs and cfDNA, indicating the complementary information obtained through parallel LB analysis. Results from gene expression analysis indicated high rates of vimentin-positive CTCs detected at all time points during osimertinib. Moreover, there was an increased number of NSCLC patients at PD harboring CTCs positive in PD-L1. AXL and PIM-1 expression detected in CTCs during treatment suggesting new possible therapeutic strategies.
Discussion: Our results reveal that comprehensive liquid biopsy analysis can efficiently represent the heterogeneous molecular landscape and provide prominent information on subsequent treatments for NSCLC patients at PD since druggable molecular alterations were detected in CTCs.
1 Introduction
During the last twenty years, the emergence of molecular targeted therapies has significantly changed non-small cell lung cancer (NSCLC) treatments since it was shown that they are more effective over chemotherapeutic regimens (1, 2). Several clinical trials have clearly demonstrated that tyrosine kinase inhibitors (TKI) of the epidermal growth factor receptor (EGFR) have achieved improved clinical outcomes for EGFR mutant (EGFRm) NSCLC patients (3–6). Osimertinib, a third-generation EGFR TKI, has changed the therapeutic management of NSCLC patients (7, 8). Initially, only EGFRm NSCLC patients that were previously treated with first- or second-generation EGFR TKIs (9–12) were subjected to osimertinib treatment. Still, this drug is now the standard of care for first-line therapy (13, 14). Recently osimertinib has been administered as an adjuvant treatment since clinical outcomes are significantly improved (15–17). The clinical benefit of this drug is now being investigated in the neoadjuvant setting (18, 19).
Acquisition of new mutations or pre-existing genetic alterations is linked to disease progression in NSCLC patients with EGFR mutations (20–23), and the highly heterogeneous and complex genetic landscape of this type of cancer impacts clinical outcomes. This is a major reason for the emergence of resistance mechanisms to this type of therapy in these patients (24–26). The most common resistance mechanism to first- and second-generation EGFR TKIs is the T790M EGFR mutation that is now therapeutically targeted by osimertinib (27). The heterogeneity of resistance mechanisms is the main reason that it is such a difficult challenge to overcome resistance to osimertinib (28). These acquired resistance mechanisms can be either EGFR-dependent or EGFR-independent (25, 29). We now know that activation of MET or HER2 amplification, acquisition of mutations in BRAF, PIK3CA, and KRAS, histological transformation to small cell lung cancer (SCLC), and epithelial-to-mesenchymal transition (EMT) are major reasons (30–35). Results from the AURA clinical trials underlined the use of ctDNA analysis in reflecting tumor heterogeneity, monitoring the efficacy of EGFR TKIs as well as the early detection of resistance mechanisms (7, 8, 10, 36).
The key step to efficiently overcome resistance mechanisms and adjust to proper subsequent treatments is to identify the resistance much earlier than conventional strategies (37). Liquid biopsy (LB) analysis as a minimally invasive approach to longitudinally and regularly monitoring NSCLC patients plays a pivotal role in early tracking tumor evolution and relapse (1, 28, 38). The increasing number of LB tests that have been cleared by the US Food and Drug Administration (FDA) has led to the application of circulating tumor DNA (ctDNA) analysis in clinical routine testing in NSCLC) (39), and recent recommendations suggest these tests for patients with advanced or metastatic NSCLC especially when tissue sampling cannot be performed (40–46). A recent comprehensive genomic profiling (CGP) of osimertinib resistance mechanisms performed in primary tumors or peripheral blood of NSCLC patients concluded that this type of analysis could help select therapies (47). Conversely, many studies have shown that circulating tumor cell (CTC) analysis in EGFRm NSCLC could contribute to the management of patients in a complementary way (48–52). It has been clearly shown that CTC analysis gives significant information on tumor heterogeneity and clonal evolution occurring under treatment (48–51, 53). Thus, CTC analysis would aid in adjusting targeted therapy for EGFRm NSCLC patients based on the resistance mechanisms identified.
It is now evident that CTCs when compared to ctDNA provide complementary information, thus comprehensive LB analysis is highly essential for the management of cancer patients. Moreover, CTCs better depict tumor heterogeneity and provide unique information derived from many different cellular components that cannot be revealed by ctDNA analysis (54). Lately, few but still important studies have shown the clinical significance of combining the information derived from different LB analytes in various types of cancer, such as breast (55–61), NSCLC (49, 62, 63), prostate (64), and very recently in melanoma (65). Regarding the treatment monitoring of NSCLC patients, a comprehensive analysis of EGFR mutations in cfDNA and CTCs could be more informative as this was recently demonstrated in studies that included both LB analytes (66, 67). Besides, it has already been demonstrated that several distinct molecular features contribute to the heterogeneity of NSCLC apart from EGFR tumor clonality that arises during cancer development and treatment (21). Tumor clonality in conjunction with the highly heterogeneous landscape of resistance in NSCLC patients under osimertinib claims for the identification of new molecular biomarkers that could be potential therapeutic targets (23, 28, 68). Therefore, analyzing more than one LB analytes in patients’ samples as an integrated approach could be more beneficial for the early detection of resistance and efficient treatment management. Our group has extensively studied the combination of ctDNA and CTC analysis in NSCLC patients during osimertinib therapy (69–72).
In the current study, we performed a comprehensive LB analysis for monitoring NSCLC patients under second-line osimertinib treatment, by combining plasma-cfDNA and matched CTC to identify molecular alterations (DNA mutations, DNA methylation, and gene expression) as well as chromosomal alterations that could be early indicators of resistance and provide potential targets for subsequent treatments.
2 Materials and methods
2.1 Patients
Thirty patients, recruited through a multicenter Phase II clinical study [ClinicalTrials.gov number: NCT02771314], all diagnosed with EGFR mutated lung adenocarcinomas, and treated with osimertinib (AZD9291; Astra Zeneca, UK) were included. Peripheral blood from ten healthy donors (HD) was used as a control group. The study was conducted following the Declaration of Helsinki, and all patients and HD gave their written informed consent. The study was approved by the National Drug Administration of Greece (EOF), the National Ethics Committee (35/00-03/16, 35/03-11/16) and the Institutional Ethical Committees of the HORG’s participating centers.
2.2 Collection of peripheral blood samples
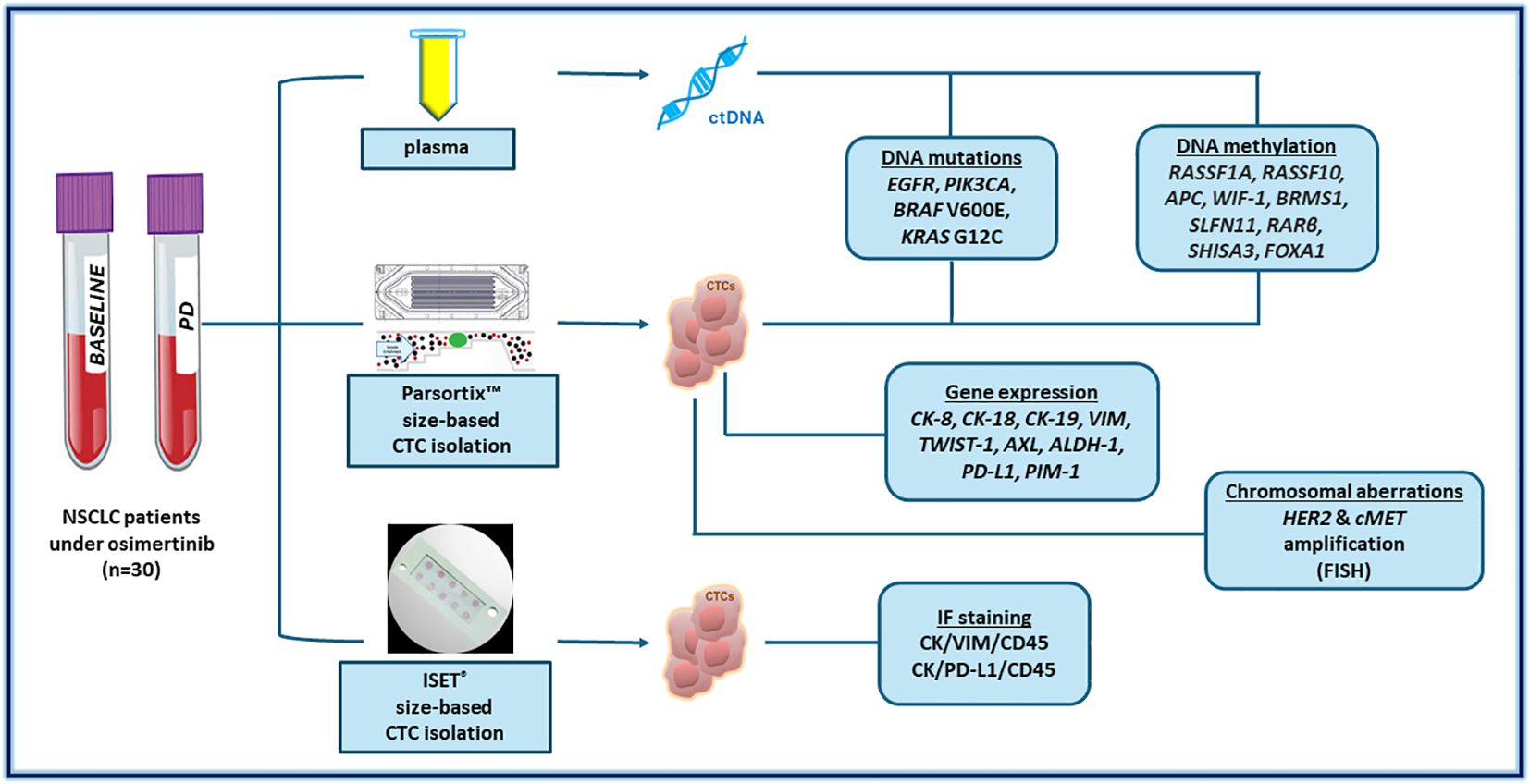
Peripheral blood (PB) was collected at baseline before treatment initiation and at disease progression (PD). PB (30mL) was collected in EDTA tubes and the first 5mL of blood was discarded to avoid contamination of skin epithelial cells. Plasma separation from buffy coat and erythrocytes was performed as previously described (69, 70, 73) (Figure 1).
2.3 Plasma-cfDNA extraction and CTC enrichment
For each blood draw, plasma samples were aliquoted; 2.00 mL plasma was used for cfDNA isolation, using silica-based membrane extraction kits: cobas® cfDNA Sample Preparation Kit (Roche Molecular Systems, Inc.) for downstream EGFR mutation analysis. Another aliquot of 2.00 mL plasma was used for cfDNA isolation for downstream Crystal Digital PCR analysis with the naica® system (Stilla Technologies, Villejuif, France) (70). Another identical plasma aliquot was used for cfDNA extraction for downstream mutation analysis of PIK3CA, KRAS, and BRAF, as reported below, using the QIAamp Circulating Nucleic Acid Kit (Qiagen®, Hilden, Germany) was used.
Following plasma separation, CTC enrichment was performed, using the FDA-cleared Parsortix™ (ANGLE plc, UK) device (73), and the CTC-enriched fraction was collected in 200 μL of PBS. Total RNA was extracted from the CTC-enriched fraction using TRIZOL followed by cDNA synthesis (73). Genomic DNA (gDNA) was extracted from the CTC-enriched fraction using TRIZOL as previously described (70). Ten mL of peripheral blood in EDTA using identical blood draws was used for CTC enrichment with ISET (Rarecells Diagnostics, France, and downstream molecular characterization by triple immunofluorescence.
2.4 Whole genome amplification
Whole genome amplification of gDNA extracted from the enriched CTCs was performed using a commercially available kit (Ampli1™ Whole Genome Amplification, Menarini Silicon Biosystems, Italy) (70).
2.5 Plasma-cfDNA and paired CTC analysis: DNA mutations
2.5.1 EGFR mutations
All plasma-cfDNA samples were analyzed by the FDA-cleared cobas® EGFR Mutation Test v2 in the cobas® z 480 analyzer (Roche) (70), a test for which our lab has an ISO-15189 accreditation (74). Plasma-cfDNA isolated from identical plasma aliquots and paired CTC-derived gDNA were further analyzed by Crystal digital PCR™ as previously described (70).
2.5.2 PIK3CA mutations
All plasma-cfDNA samples and paired CTC-derived gDNA samples were analyzed for the presence of three PIK3CA hotspot mutations (p.E545K exon 9, p.E542K exon 9 and p.H1047R exon 20) using our previously described ultrasensitive real-time PCR methodology (75).
2.5.3 KRAS G12C and BRAF V600E
Droplet digital PCR (ddPCR) was used for the analysis of cfDNA and paired CTC-derived gDNA samples for KRAS G12C and BRAF V600E mutations in a BioRad QX200 ddPCR System using a commercially available ddPCR reaction mix and specific primers and probes (Bio-Rad Laboratories).
2.6 DNA methylation analysis in plasma-cfDNA and paired CTC-derived gDNA
All cfDNA samples and paired CTC-derived gDNA samples were subjected to Sodium Bisulfite (SB) treatment as previously described (69). SB-treated samples were subsequently analyzed for the DNA methylation of RASSF10, WIF-1, APC, RARβ, RASSF1A, BRMS1, FOXA1, SLFN1, SHISA3 genes with our previously developed and analytically validated real-time methylation-specific PCR (MSP) assays (69).
2.7 CTC analysis: gene expression
Gene expression was studied in CTC-derived total RNA by RT-qPCR for the following genes: CK-8, CK-18, CK-19, Vimentin, TWIST-1, AXL, ALDH-1, PD-L1, PIM-1 and B2M as previously reported (73).
2.8 CTC analysis: triple immunofluorescence
CTCs captured in the ISET filters were subsequently analyzed by triple immunofluorescence for CK/VIM/CD45 using the Confocal laser Scanning microscopy (LEICA), as previously described (73). For CK/PD-L1/CD45, the process was according to our previous report (71).
2.9 CTC analysis: fluorescent in situ hybridization
2.9.1 Detection of HER2
Amplification by FISH was performed on enriched CTCs, using PathVysion HER2 DNA Probe Kit (Abbott Molecular, Inc). FISH signal patterns were determined for the HER2 gene and the centromere of chromosome 17 in a fluorescent microscope (Axioplan 2, Zeiss, and Leica GSL120) equipped with Cytovision Image Analysis Software. The ratio of the total number of HER-2/neu (red signals) as compared to the total number of CEP 17 (green signals) was calculated for every nucleus. When the HER2/CEP17 ratio was ≥2 a cell was considered as HER2-amplified according to the manufacturer and previous reports (76, 77).
2.9.2 C-MET amplification
FISH analysis for the detection of c-MET amplification was performed on enriched CTCs, using c-MET (MET) Amplification Probe (Cytocell). The evaluation of MET amplification is based on the determination of FISH signal patterns for MET and the centromere of chromosome 7, in a fluorescent microscope (Axioplan 2, Zeiss and Leica GSL120) equipped with Cytovision Image Analysis Software. When the C-MET/CEP7 ratio was ≥2 the cell was considered as c-MET-amplified according to the manufacturer and previous reports (78).
3 Results
3.1 Plasma-cfDNA and paired CTC analysis: DNA gene mutations
We have previously analyzed plasma-cfDNA samples for the detection of EGFR mutations both with crystal dPCR and with the FDA-cleared assay cobas EGFR mutation test v2 (70) and reported high concordance rates between the two methodologies. By combining the results for the subgroup of patients included in the current study, T790M mutation was detected in 10/30 (33.3%), exon19 deletions in 13/30 (44.3%), L858R in 6/30 (20%), S768I and G719X in 2/30 (6.7%) patients, at baseline. At PD, T790M was detected in 2/27 (7.4%), exon19 deletions in 8/27 (29.6%), L858R in 6/27 (22.2%), S768I in 1/27 (3.7%) and G719X in 2/27 (7.4%) (Figure 2). In addition, C797S was found in trans configuration with T790M in patient #1 (P#1) at PD. In parallel, we have also analyzed CTC-gDNA samples for EGFR mutations using crystal dPCR. In baseline, one patient was found positive for T790M and 4/21 (19%) patients for L858R. P#10 was found positive for S768I and G719X mutations both in baseline and PD samples. T790M was detected only in 3/19 (15.8%) CTC-gDNA samples at PD (Figure 2).
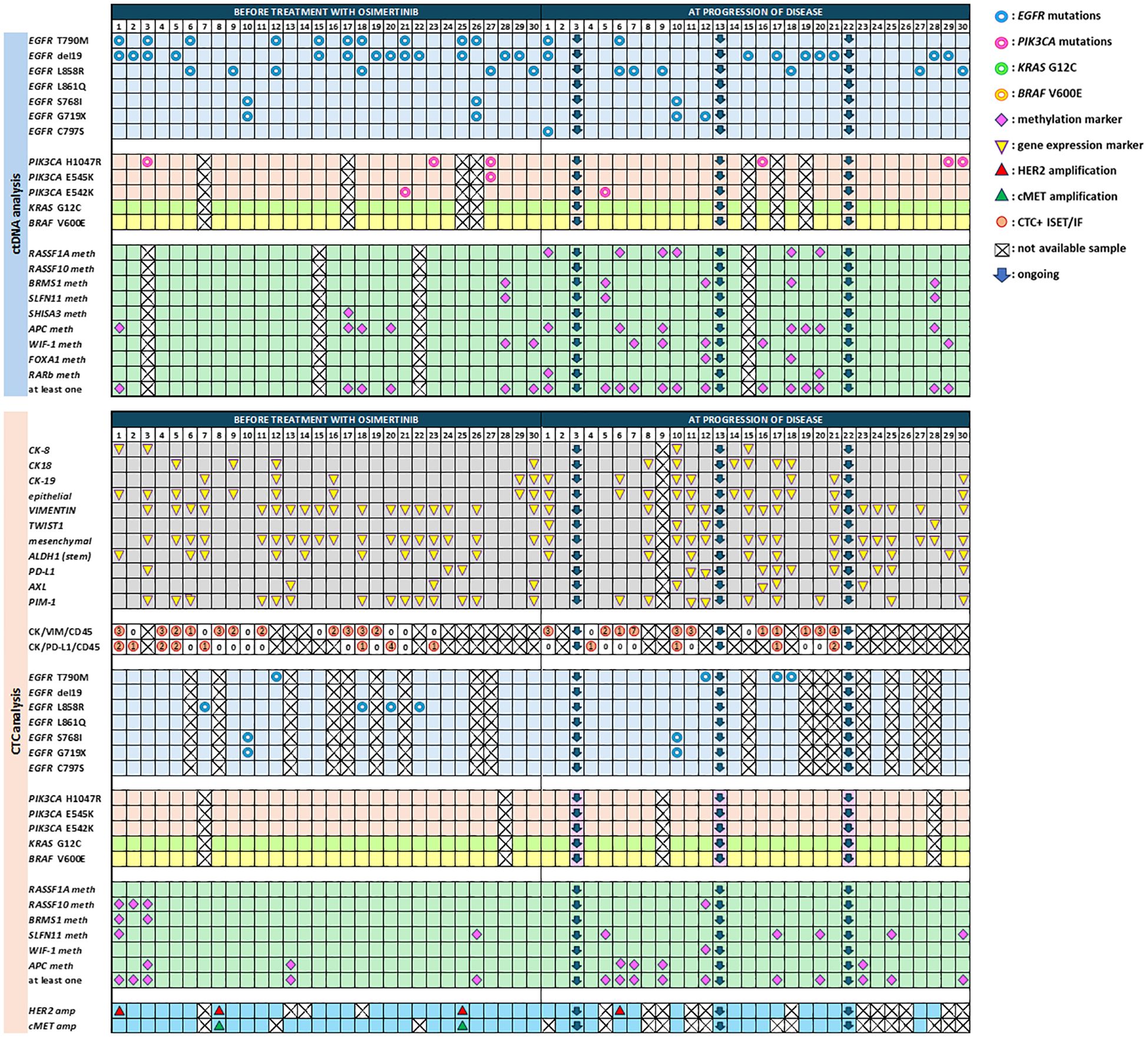
Figure 2. Comprehensive analysis in cfDNA and CTCs of NSCLC patients before treatment with osimertinib and at progression of disease.
In this study, we have analyzed plasma-cfDNA samples (n=50) and paired CTC-gDNA samples (n=53) for the detection of PIK3CA mutations (H1047R, E542K, E545K). at two time points. PIK3CA mutations were detected in 4/26 (15.4%) cfDNA samples at baseline, and 4/24 (16.7%) at PD. More precisely, H1047R was found only in the baseline samples of three patients (P#3, P#23, P#27), E545K was found concomitantly with H1047R in patient P#27, and E542K was found in P#21 (Figure 2). At PD, H1047R has emerged in three patients (P#16, P#29, P#30), E542K was found only in P#5, and E545K was not detected in any sample. In CTC-gDNA samples, no PIK3CA mutations were detected at any time point (Figure 2). When these 50 plasma-cfDNA and 53 paired CTC-derived gDNA samples were analyzed for the detection of KRAS G12C and BRAF V600E mutations with ddPCR, they were all found negative for both mutations.
3.2 Plasma-cfDNA and paired CTC analysis: DNA methylation
At baseline one methylation marker was detected at least in 6/27 (22.2%) patients at PD in 13/26 (50%). Even if methylation for these markers was detected in more samples at PD in respect to baseline there was no statistically significant difference (Figure 2). CTC analysis also revealed an overall increase in the detection of DNA methylation markers at PD. More specifically, at baseline, 5/30 (16.7%) samples and at PD 10/27 (37%) were found positive for at least one methylation marker (Figure 2).
3.3 CTC analysis: gene expression
In CTC-enriched fractions before treatment, 9/30 (30%) patients were positive for at least one epithelial marker in CTCs (CK-8, and/or CK-18, and/or CK-19) whereas at PD more samples were positive for the expression of epithelial markers (Figure 2), but there was no statistically significant difference between these two time points. The same was seen for the mesenchymal/EMT markers tested (VIM, AXL, and TWIST-1) [18/30 (60%) versus 15/26 (57.7%)], for ALDH-1, that was detected in 9/30 (30%) samples at baseline and 10/26 (38.5%) at PD and for PIM-1 overexpression that was detected in 14/30 (46.7%) baseline samples and 9/26 (34.6%) at PD. PD-L1 expression levels were significantly different between baseline and disease progression [3/30(10%) patients versus 9/26 (34.6%), McNemar test, p = 0.016], as this was previously described (73), (Figure 2).
3.4 CTC analysis: characterization of ISET-enriched CTCs by triple IF
CTC molecular characterization at the single-cell level was performed by using a combination of the size-based isolation platform, ISET, and confocal microscopy in 31 CTC samples, at baseline and PD. A direct comparison between RT-qPCR and IF staining for the presence of CTC positive for CK (CK-8, CK-18, CK-19) and/or VIM revealed an agreement of 54.8% (17/31 samples) (Figure 2).
The same ISET filters were analyzed for the phenotype CK+PD-L1+CD45− in 16 baseline samples and 11 PD samples matched with those analyzed for the mRNA expression of PD-L1 with RT-qPCR. A representative image from PD-L1 positive CTCs enriched with the ISET is shown in Figure 3. Combining the results from RT-qPCR and IF for the presence of PD-L1 in CTCs, we found 5/11 (45.4%) patients at baseline and 8/17 (47%) patients at PD with CTCs and/or CTC-gDNA positive for PD-L1 (Figure 2).
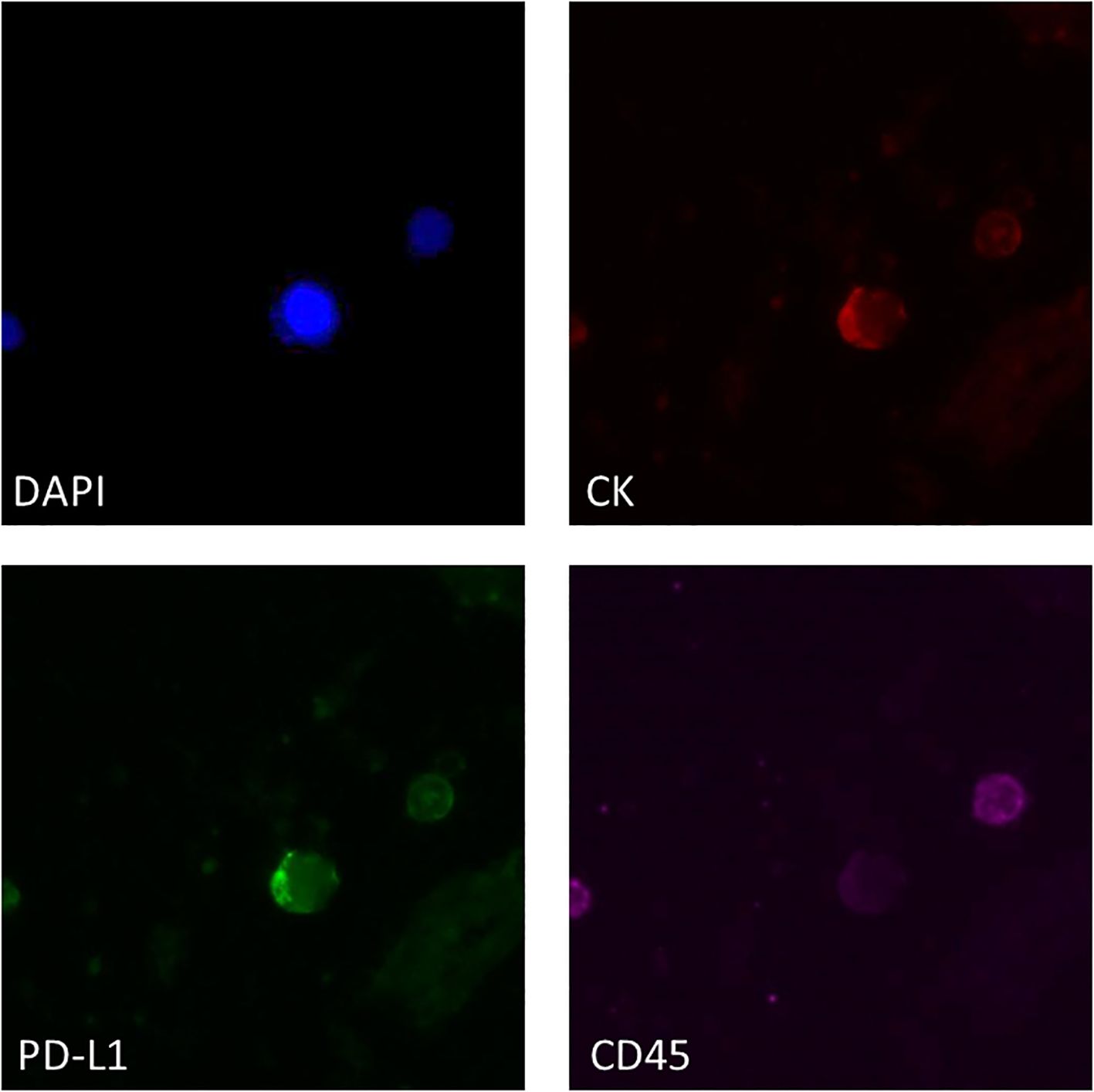
Figure 3. Representative IF image of PD-L1 expression in ISET-enriched CTCs. CTCs stained with CK (red), PD-L1 (green) and CD45 (purple). Nuclei (blue) were stained with DAPI.
3.5 CTC analysis: fluorescence in situ hybridization
FISH analysis was performed on enriched CTCs from these 30 NSCLC patients for the detection of HER2 amplification at two time points: a) baseline (n=26) and b) PD (n=15) (Figure 4). HER2 amplification was found in 3/26 (11.5%) patients at baseline and 1/15 (6.7%) at PD. For the detection of MET amplification, FISH analysis was performed on enriched CTCs also at two time points: a) baseline (n=27) and b) PD (n=12). MET amplification was found in 2/27 (7.4%) patients at baseline whereas there was no positive sample at PD. Patient#8 (P#8) and P#25 had concomitant HER2 and MET amplification in their enriched CTCs at baseline.
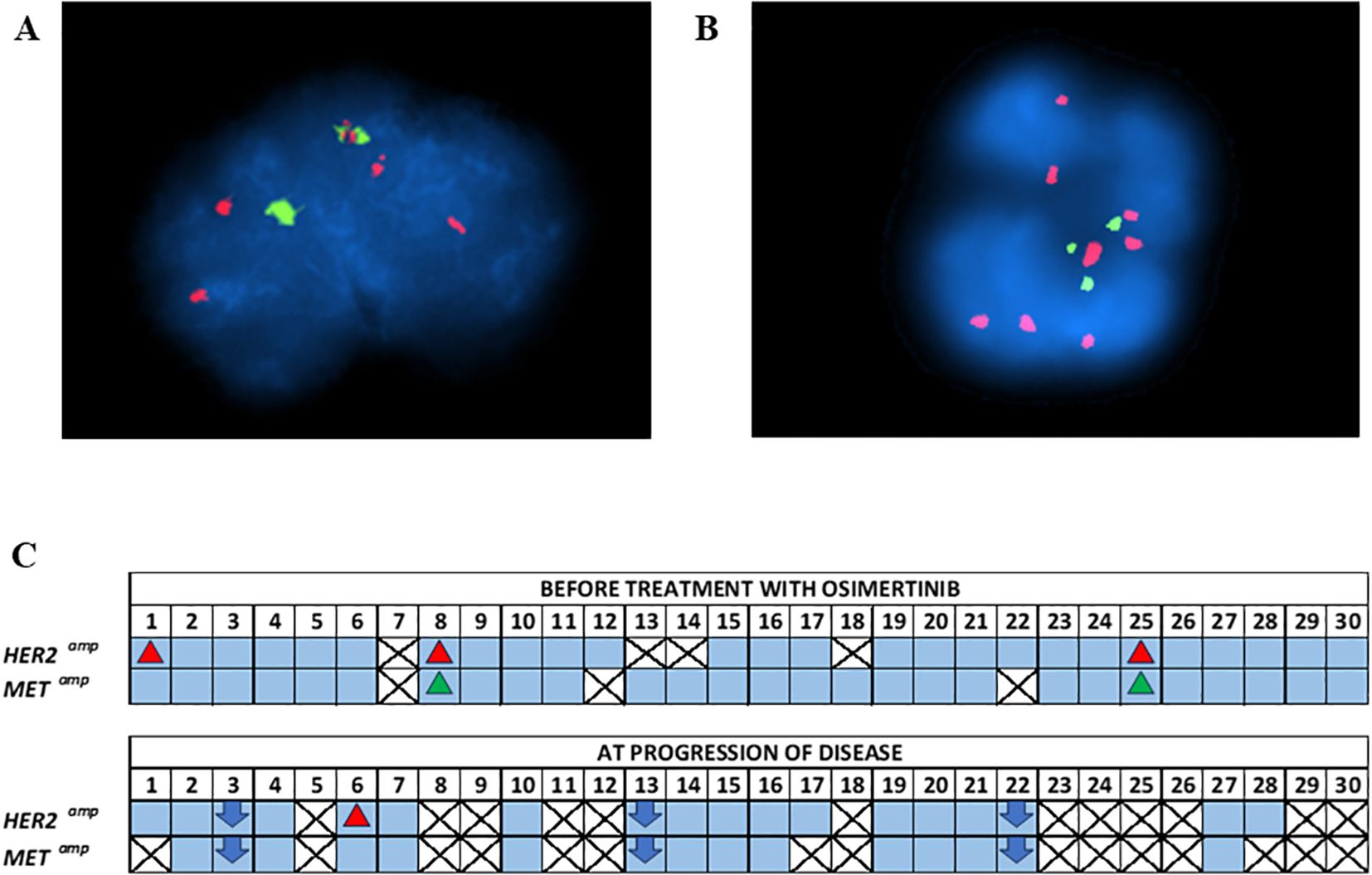
Figure 4. (A) Representative FISH image of CTC with HER2 amplification. Red and green signals within nuclei indicate the HER2 gene and the centromere 17 (CEP17), respectively, (B) Representative FISH image of CTC with cMET amplification. Red and green signals within nuclei indicate the cMET gene and the centromere 7 (CEP7), respectively, (C) FISH analysis of enriched CTCs of NSCLC patients under osimertinib treatment at baseline and at PD.
3.6 Molecular alterations at PD
Molecular alterations detected at PD either in plasma-cfDNA samples or in paired CTCs of NSCLC patients (n=27) treated with osimertinib and who finally progressed during this analysis in DNA mutation, DNA methylation, gene expression, and chromosomal level are summarized in detail in Table 1. Figure 5 depicts the presence of the molecular alterations in correlation with their time to progression.
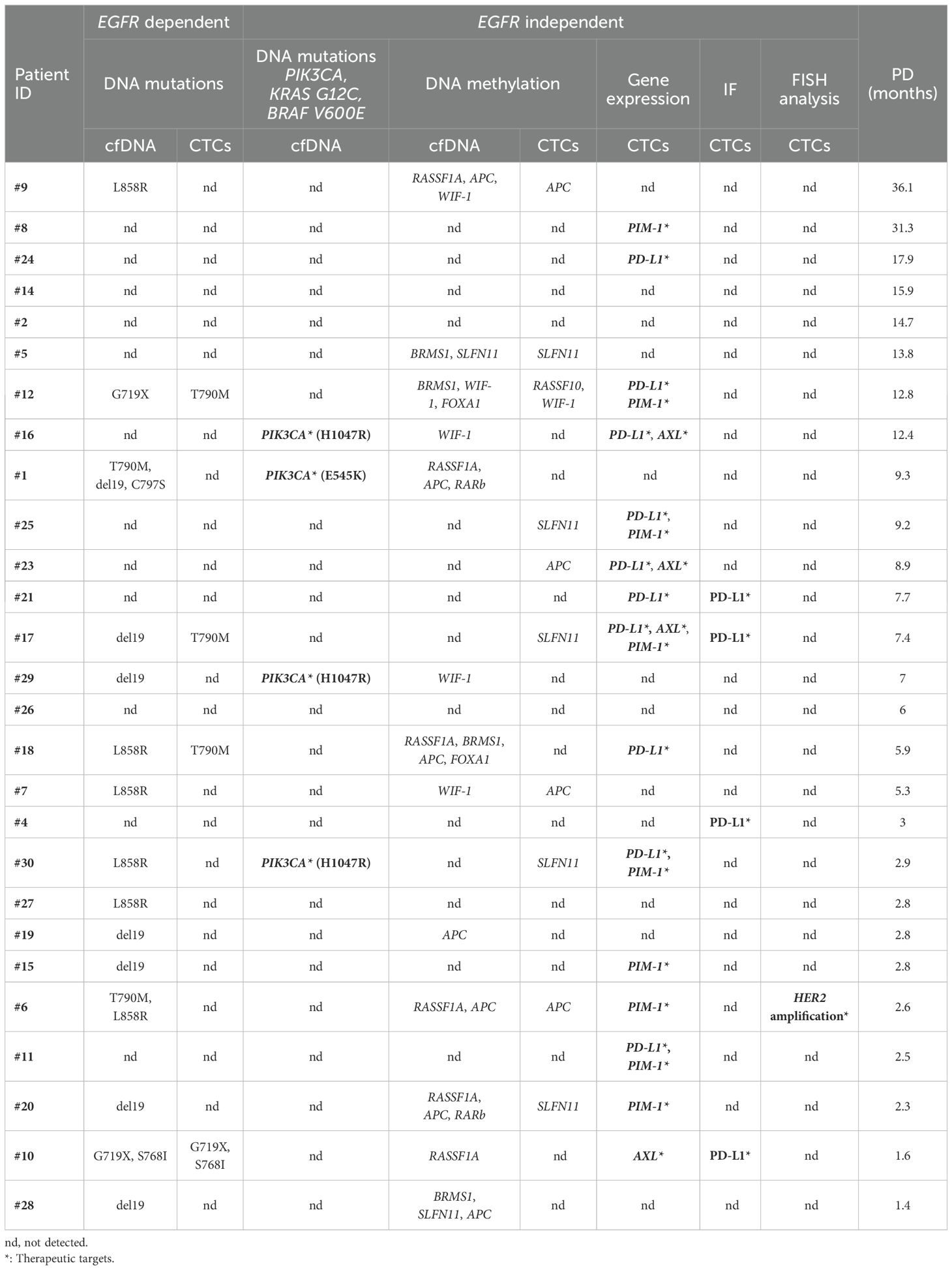
Table 1. EGFR-dependent and independent molecular alterations detected in cfDNA and/or CTCs of NSCLC patients treated with osimertinib at PD, ranked according to reducing the time of PD.
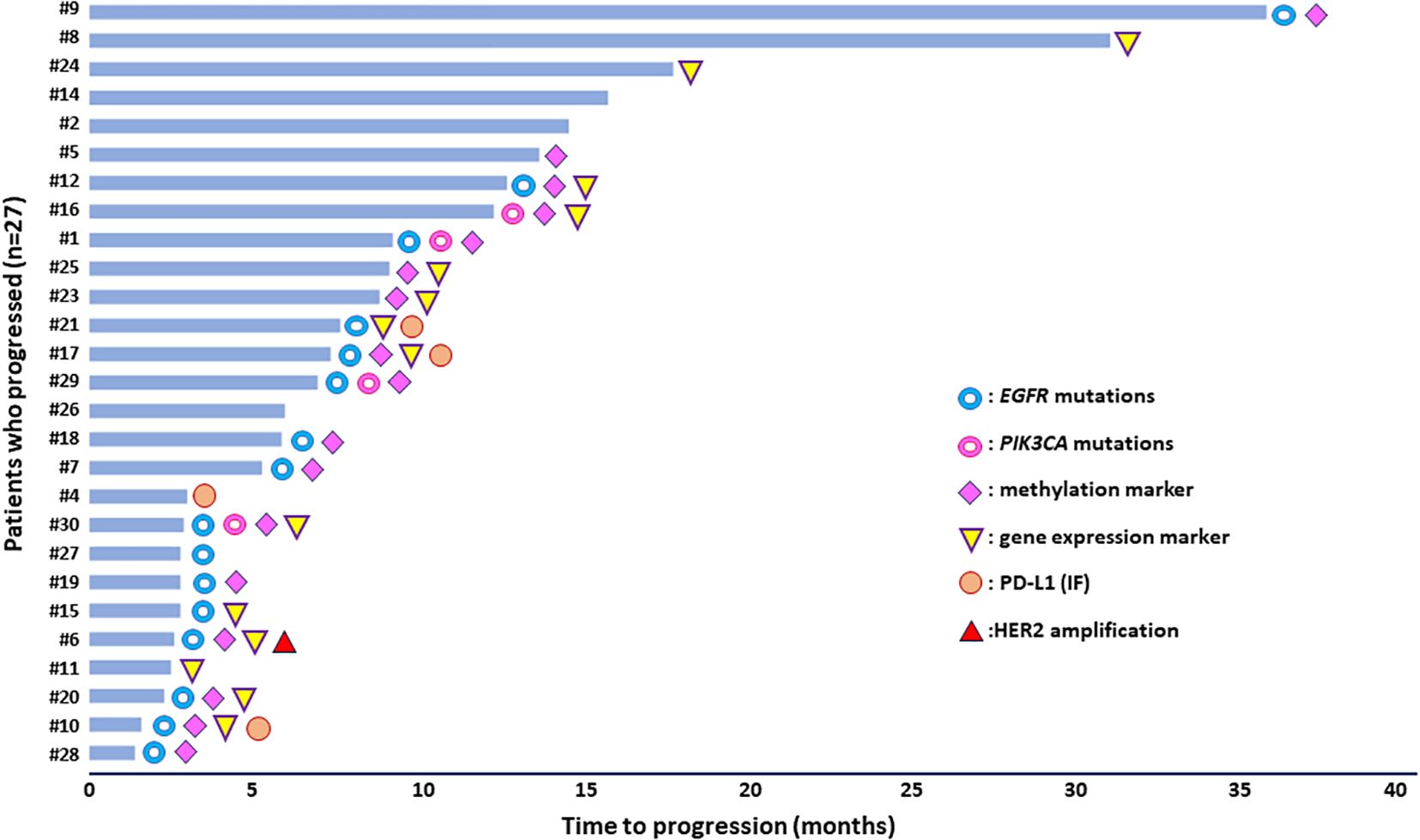
Figure 5. Molecular alterations detected in plasma-cfDNA and/or paired CTCs of NSCLC patients under osimertinib treatment in correlation with time to progression.
4 Discussion
Comprehensive analysis including alternative molecular alterations that may occur at resistance to osimertinib is important for guiding subsequent treatments, especially for those patients who relapse early. The present study highlights the potential of combining liquid biopsy analytes to elucidate NSCLC molecular heterogeneity in patients under osimertinib treatment. Comprehensive liquid biopsy analysis for DNA mutations, DNA methylation markers, gene expression and chromosomal level in both plasma-cfDNA and CTCs revealed druggable molecular alterations and resistance mechanisms that may occur under selective therapy pressure. Discrepancies observed between plasma-cfDNA and paired CTCs are representing tumor heterogeneity, a well-known characteristic of NSCLC. Our results indicate that complementary information derived from plasma-cfDNA and CTC analysis proved to be very informative for treatment monitoring of NSCLC patients.
There are only a few direct comparison studies between CTC and plasma-cfDNA up to now, that use identical same blood draws and identical methodologies to detect mutations, DNA methylation markers, or gene expression not only in NSCLC (49, 69, 70, 79) but also in other types of cancer (59–61, 64). These few studies have shown that there are significant discrepancies in information derived from CTC and plasma-cfDNA material, especially when very low target concentrations are present in the sample. Currently, ongoing clinical trials include parallel analysis of CTCs and ctDNA in many types of cancer to test if this dual analysis is more sensitive for disease monitoring (80).
EGFR mutation analysis in the context of the current comprehensive study confirmed the significance of combined liquid biopsy analysis. Our previous results (70), have shown that T790M resistance mutation was detected at baseline in gDNA isolated from CTC but not in the corresponding plasma cfDNA (P#11) indicating that this may be a case of subclonal T790M, that is potent to lead to early PD. This finding is consistent with data from the AURA3 phase III trial (22). EGFR T790M mutation was detected at PD only in gDNA samples isolated from the CTCs of three patients but not in the corresponding plasma-cfDNA. There was only one case (P#1) of acquired resistance that was due to EGFR p.C797S mutation, which is one of the most frequent resistance mechanisms to second-line osimertinib treatment (81). Importantly, C797S mutation was detected in cis configuration with T790M, suggesting that the administration of brigatinib with an anti-EGFR antibody would be beneficial to the patient (82, 83) or fourth-generation EGFR TKIs (26). BLU-945 is another EGFR inhibitor, and the benefits when combined with osimertinib are being investigated in the phase I clinical trial SYMPHONY (NCT04862780). The rechallenge with first or second EGFR TKIs to NSCLC patients who acquired the C797S resistance mutation during osimertinib treatment has also been proposed, delaying the use of platinum-based chemotherapy since there is no approved targeted therapeutic strategies in this setting, so far (84).
The presence of the BRAF V600E mutation may limit the activity of EGFR TKIs in EGFRm NSCLC, as this was previously shown (85). BRAF V600E mutation as a resistance mechanism to osimertinib has been identified in 3% of NSCLC patients positive for EGFR mutations, irrelevant of the presence of a T790M mutation (25, 29, 86). Concomitant detection of BRAF and EGFR mutations has been reported in a few cases (87, 88), and in these cases, patients progressed more rapidly upon EGFR TKIs (89, 90). For this reason, different combination therapies have been proposed for patients carrying mutations in BRAF in EGFRm lung cancer (91–94). In this study, we did not detect any sample positive for BRAF V600E.
KRAS G12C was not detected in the samples analyzed. Although alterations in the RAS-MAPK pathway have been shown to lead to osimertinib resistance, cases reporting the presence of KRAS G12C mutation are limited (26). However, there is only a recent case report study of acquired KRAS G12C mutations during first-line osimertinib resistance which showed the effective and well-tolerated combination of osimertinib and sotorasib (95). Sotorasib which has been recently approved by the FDA for patients with locally advanced or metastatic NSCLC, carrying KRAS G12C mutations could be an option for those who progress to osimertinib.
PIK3CA mutations were detected only in plasma-cfDNA but not in corresponding CTCs. PIK3CA mutations have been detected in a frequency of 4%-14%, as a resistance mechanism against second-line osimertinib treatment. The most common are the PIK3CA hotspot mutations H1047R, E545K, and E542K (81, 96, 97). Results from the AURA3 trial have shown that PIK3CA alterations were detected more frequently in the T790M positive NSCLC patients (98). PIK3CA concurrent mutations confer resistance to osimertinib as confirmed with in vitro experiments (22). Based on previously reported evidence, these patients could have been treated with appropriate combination therapies against the PIK3/AKT/mTOR pathway (22, 99). Han et al. reported that both PIK3CG (L468M) and PIK3CA (H1047R) mutations could induce osimertinib resistance through PI3K/Akt/mTOR pathway-dependent mechanisms. They proposed the administration of aspirin which could effectively reverse in vitro and in vivo osimertinib resistance as a treatment strategy for NSCLC patients who develop these mutations (100).
In our comprehensive analysis, we used FISH analysis to assess the presence of cMET and HER2 amplification in CTCs enriched with the microfluidic platform Parsortix. To the best of our knowledge, this is the first study that detects these two gene alterations in NSCLC patients before and after osimertinib treatment using FISH. In our study, MET amplification was detected in the CTCs of two patients (P#8, P#25) before treatment with osimertinib. MET amplification in NSCLC may be a primary oncogenic alteration or may arise as a secondary driver resistance mechanism to EGFR TKI treatment through the activation of downstream signaling pathways, like MAPK or PI3K-Akt (98, 101). Up to date, the standard of care for EGFRm patients with MET alterations is platinum-based chemotherapy with limited efficacy. However, recent data from the TATTON trial have shown that combination therapy of osimertinib with the MET inhibitor, savolitinib, presented acceptable tolerability and clinical activity (102). Consequently, patients in our study positive for MET amplification at baseline could have benefited from such a combinatorial therapeutic strategy. Currently, the ongoing SAVANNAH study aims to examine the efficacy of this treatment combination in patients with MET-mediated who progressed following treatment with osimertinib whereas the SAFFRON study (NCT05261399) is designed to assess savolitinib in combination with osimertinib versus platinum-based chemotherapy in the same group of patients (26).
Based on our results, HER2 amplification was detected in the CTCs of three patients before treatment with osimertinib (P#1, #8, #25) and in one patient at PD (P#6). HER2 gene amplification is another common osimertinib resistance mechanism that leads to the activation of HER2 signaling followed by the downstream activation of the PI3K-Akt pathway. Results from the AURA3 trial demonstrated that 5% of patients who progressed on second-line osimertinib treatment had HER2 amplification (98). TRAEMOS is the first trial testing the combination of trastuzumab-emtansine and osimertinib to target HER2-mediated resistance in patients with EGFRm NSCLC. Despite the favorable safety profile, this combination revealed limited efficacy to patients (103).
It is now well known that dominant tumor cancer cells are subjected to epigenetic modifications and switch to drug-resistant cancer cells in various types of cancer (104–106). DNA methylation provides useful insights into lung cancer development and is correlated with early detection, prognosis, and prediction of response to specific treatments (107). Liquid biopsy is a very powerful tool for identifying circulating DNA methylation markers that could be of clinical importance (107, 108). Concerning the role of methylation in resistance to EGFR ΤΚΙ therapy, there are now studies performed in lung cancer cell lines or primary tissues showing that epigenetic modifications negatively affect EGFR TKI treatment outcome and that their combination with epigenetic drugs could be very promising (109–115). Intriguingly, a methylation-associated mechanism behind the acquisition of T790M mutation was previously described (113). Recent advances in DNA methylation modifications linked to TKI resistance mechanisms in EGFRm patients have been previously reported (108). However, only a few studies focused on DNA methylation in ctDNA or CTCs of NSCLC patients receiving osimertinib (69, 116, 117).
We have previously reported results on DNA methylation of RASSF1A, RASSF10, APC, WIF-1, BRMS1, SLFN11, RARβ, SHISA3, and FOXA1 in plasma-cfDNA and paired CTCs of NSCLC patients during osimertinib therapy. There was a low concordance of DNA methylation markers in CTCs and cfDNA, indicating the importance of complementary information obtained through parallel CTCs and cfDNA analysis. A predictive role of DNA methylation as a potential resistance mechanism was shown in this study where patients with at least one methylated marker in liquid biopsy samples at PD eventually progressed earlier than those negative for methylation (69).
CTC analysis at the gene expression level provides important information on tumor heterogeneity and can reveal differential gene expressions related to metastasis or treatment sensitivity and resistance (72). The first gene expression study in CTCs of NSCLC patients during osimertinib treatment revealed heterogeneous patterns of gene expression of epithelial, mesenchymal/EMT, and stem cell markers among patients. A potential role of EMT was shown based on the high rates of vimentin-positive CTCs detected at all time points during osimertinib treatment (118).
The increased number of NSCLC patients at PD harboring CTCs positive in PD-L1 suggests a theoretical background for immune checkpoint inhibition (ICI) therapy in EGFRm NSCLC patients resistant to osimertinib (73). However, the combination of data from IF and RT-qPCR for the presence of PD-L1 positive CTCs in matched samples revealed high detection rates both at baseline and at PD. Immunotherapy treatments for EGFRm NSCLC are still a big challenge since to date numerous studies have shown the confined efficacy of immunotherapy either as monotherapy or in combination with chemotherapy (118).
Our study was the first to evaluate AXL gene expression levels in CTCs of patients during osimertinib therapy (73). Several preclinical studies on AXL inhibition suggest this approach as a new additional tool for personalized therapy of NSCLC patients with EGFR mutations, since these patients may benefit from AXL inhibitors (119–122). EGFRm NSCLC patients harboring high levels of AXL expression had significantly shorter PFS and OS after ICI-based therapy (123).
Herein, we detected PIM-1 expression in CTCs of EGFRm NSCLC patients before osimertinib and at PD, suggesting that concurrent use of PIM-1 inhibitors with osimertinib could be a possible therapeutic strategy. The synergistic effects of PIM inhibitor in combination with osimertinib acting through the inhibition of oncogenic signaling pathways have previously been reported (124, 125). EGFR signaling is indirectly affected by PIM-1 suggesting that PIM-1 inhibition can improve patient’s outcomes (124, 126).
The current study highlights the potential of analyzing both CTC and ctDNA derived from a single blood draw to identify molecular biomarkers clinically significant for the patient’s outcome or alternative treatment approaches upon osimertinib treatment. We would like to point out that in our study we have included different methodologies for the identification of molecular alterations at the DNA, RNA, and epigenetic level. According to recent guidelines for NSCLC, NGS approaches are the most suitable for the identification of multiple molecular alterations in LB samples but only at the DNA level (41, 127, 128). Commercially available technologies such as nanopore DNA sequencing offer now analysis of whole genome sequencing and the identification of DNA methylation aberrations, simultaneously (129–131). However, such a commercially available integrated approach before and after treatment with osimertinib has not been performed so far.
Treatment of NSCLC patients with osimertinib is very challenging if we consider the high molecular heterogeneity of this disease and also the clonal evolution that arises through selective therapy pressure. These two significant factors compose the wide spectrum of resistance mechanisms that claim early identification and proper therapeutic interventions. In the present study, it was clearly highlighted the potential of comprehensive liquid biopsy analysis to efficiently represent the heterogeneous molecular landscape and provide prominent information on subsequent treatments for NSCLC patients based on the druggable molecular alterations found at PD. Epigenetic alterations give additional information to DNA mutation analysis to identify patients who are unlikely to benefit from EGFR TKI therapy. Studies have shown that targeting epigenetic alterations might be a therapeutic intervention to reverse EGFR TKI resistance. Complementary information obtained from cfDNA and CTC analysis is of utmost importance during the management of NSCLC patients.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by National Ethics Committee (35/00-03/16, 35/03-11/16). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
AN: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. TM: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. GK: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. AK: Writing – original draft, Writing – review & editing. VG: Writing – original draft, Writing – review & editing. EL: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Stavros Niarchos Foundation within the framework of a grant to the National and Kapodistrian University of Athens (No.16785).
Acknowledgments
We would like to thank ANGLE for providing the Parsortix system for this analysis and GENOTYPOS Science Labs, Greece for their contribution to FISH analysis. We would like to thank all patients who participated in this clinical study. Osimertinib (AZD9291; Astra Zeneca, UK) was administered to the patients in the context of a multicenter Phase II clinical study [ClinicalTrials.gov No., NCT02771314] sponsored by Astra Zeneca and conducted by the Hellenic Oncology Research Group (HORG).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Buszka K, Ntzifa A, Owecka B, Kamińska P, Kolecka-Bednarczyk A, Zabel M, et al. Liquid biopsy analysis as a tool for TKI-based treatment in non-small cell lung cancer. Cells. (2022) 11:2871. doi: 10.3390/cells11182871
2. de Jong D, Das JP, Ma H, Pailey Valiplackal J, Prendergast C, Roa T, et al. Novel targets, novel treatments: The changing landscape of non-small cell lung cancer. Cancers (Basel). (2023) 15:2855. doi: 10.3390/cancers15102855
3. Mok TS, Wu Y, Thongprasert S, Yang C, Saijo N, Sunpaweravong P, et al. Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med. (2009) 361:947–57. doi: 10.1056/NEJMoa0810699
4. Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. (2010) 362:2380–8. doi: 10.1056/NEJMoa0909530
5. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for european patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. (2012) 13:239–46. doi: 10.1016/S1470-2045(11)70393-X
6. Park K, Tan EH, O’Byrne K, Zhang L, Boyer M, Mok T, et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-lung 7): A phase 2B, open-label, randomised controlled trial. Lancet Oncol. (2016) 17:577–89. doi: 10.1016/S1470-2045(16)30033-X
7. Jänne PA, Chih-Hsin Yang J, Kim DW, Planchard D, Ohe Y, Ramalingam SS, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. (2015) 372:1689–99. doi: 10.1056/NEJMoa1411817
8. Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discovery. (2014) 4:1046–61. doi: 10.1158/2159-8290.CD-14-0337.AZD9291
9. Goss G, Tsai CM, Shepherd FA, Bazhenova L, Lee JS, Chang GC, et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol. (2016) 17:1643–52. doi: 10.1016/S1470-2045(16)30508-3
10. Mok TS, Wu Y-L, Ahn M-J, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or platinum–pemetrexed in EGFR T790M–positive lung cancer. N Engl J Med. (2017) 376:629–40. doi: 10.1056/nejmoa1612674
11. Yang JCH, Ahn MJ, Kim DW, Ramalingam SS, Sequist LV, Su WC, et al. Osimertinib in pretreated T790M-positive advanced non-small-cell lung cancer: AURA study phase II extension component. J Clin Oncol. (2017) 35:1288–96. doi: 10.1200/JCO.2016.70.3223
12. Papadimitrakopoulou VA, Mok TS, Han JY, Ahn MJ, Delmonte A, Ramalingam SS, et al. Osimertinib versus platinum–pemetrexed for patients with EGFR T790M advanced NSCLC and progression on a prior EGFR-tyrosine kinase inhibitor: AURA3 overall survival analysis. Ann Oncol. (2020) 31:1536–44. doi: 10.1016/j.annonc.2020.08.2100
13. Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Y, et al. Overall survival with osimertinib in untreated, EGFR -mutated advanced NSCLC. N Engl J Med. (2020) 382:41–50. doi: 10.1056/nejmoa1913662
14. Soria J-C, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR -mutated advanced non–Small-Cell lung cancer. N Engl J Med. (2018) 378:113–25. doi: 10.1056/nejmoa1713137
15. Wu Y-L, Tsuboi M, He J, John T, Grohe C, Majem M, et al. Osimertinib in resected EGFR -mutated non–Small-Cell lung cancer. N Engl J Med. (2020) 383:1711–23. doi: 10.1056/nejmoa2027071
16. Wu Y-L, John T, Grohe C, Majem M, Goldman JW, Kim S-W, et al. Postoperative chemotherapy use and outcomes from ADAURA: Osimertinib as adjuvant therapy for resected EGFR-mutated NSCLC. J Thorac Oncol. (2021) S1556-0864:03285–8. doi: 10.1016/j.jtho.2021.10.014
17. Herbst RS, Wu Y-L, John T, Grohe C, Majem M, Wang J, et al. Adjuvant osimertinib for resected EGFR-mutated stage IB-IIIA non–Small-Cell lung cancer: Updated results from the phase III randomized ADAURA trial. J Clin Oncol. (2023) 41:1830–41. doi: 10.1200/jco.22.02186
18. Rotow J, Urisman A, Mccoach C, Jahan T, Bivona T, Jones K, et al. P1.14-58 a phase II study to evaluate neoadjuvant osimertinib for surgically resectable, EGFR-mutant non-small cell lung cancer. J Thorac Oncol. (2019) 14:S578. doi: 10.1016/j.jtho.2019.08.1209
19. Tsuboi M, Weder W, Escriu C, Blakely C, He J, Dacic S, et al. Neoadjuvant osimertinib with/without chemotherapy versus chemotherapy alone for EGFR-mutated resectable non-small-cell lung cancer: NeoADAURA. Futur Oncol. (2021) 17:4045–55. doi: 10.2217/fon-2021-0549
20. Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. (2016) 22:262–9. doi: 10.1038/nm.4040.Tumor
21. Passaro A, Malapelle U, Del R, Attili I, Fumagalli C, Russo A, et al. Understanding EGFR heterogeneity in lung cancer. ESMO Open. (2020) 5:e000919. doi: 10.1136/esmoopen-2020-000919
22. Vaclova T, Grazini U, Ward L, O’Neill D, Markovets A, Huang X, et al. Clinical impact of subclonal EGFR T790M mutations in advanced-stage EGFR-mutant non-small-cell lung cancers. Nat Commun. (2021) 12:1780. doi: 10.1038/s41467-021-22057-8
23. Kohsaka S, Petronczki M, Solca F, Maemondo M. Tumor clonality and resistance mechanisms in EGFR mutation-positive non-small-cell lung cancer: Implications for therapeutic sequencing. Futur Oncol. (2019) 15:637–52. doi: 10.2217/fon-2018-0736
24. Oxnard GR, Thress KS, Alden RS, Lawrance R, Paweletz CP, Cantarini M, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. (2016) 34:3375–82. doi: 10.1200/JCO.2016.66.7162
25. Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer. (2019) 121:725–37. doi: 10.1038/s41416-019-0573-8
26. Ntzifa A, Marras T, Georgoulias V, Lianidou E. Liquid biopsy for the management of NSCLC patients under osimertinib treatment treatment. Crit Rev Clin Lab Sci. (2024) 0:1–23. doi: 10.1080/10408363.2024.2302116
27. Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. (2013) 19:2240–7. doi: 10.1158/1078-0432.CCR-12-2246.Analysis
28. Blaquier JB, Ortiz-Cuaran S, Ricciuti B, Mezquita L, Cardona AF, Recondo G. Tackling osimertinib resistance in EGFR mutant non-small cell lung cancer. Clin Cancer Res. (2023) 29:3579–91. doi: 10.1158/1078-0432.CCR-22-1912
29. Passaro A, Jänne PA, Mok T, Peters S. Overcoming therapy resistance in EGFR-mutant lung cancer. Nat Cancer. (2021) 2:377–91. doi: 10.1038/s43018-021-00195-8
30. Le X, Puri S, Negrao MV, Nilsson MB, Boyle T, Hicks JK, et al. Landscape of EGFR -dependent and -independent resistance mechanisms to osimertinib and continuation therapy post- progression in EGFR-mutant NSCLC. Clin Cancer Res. (2019) 24:6195–203. doi: 10.1158/1078-0432.CCR-18-1542
31. Santoni-Rugiu E, Melchior LC, Urbanska EM, Jakobsen JN, De Stricker K, Grauslund M, et al. Intrinsic resistance to EGFR-tyrosine kinase inhibitors in EGFR-mutant non-small cell lung cancer: Differences and similarities with acquired resistance. Cancers (Basel). (2019) 11:923. doi: 10.3390/cancers11070923
32. Lazzari C, Gregorc V, Karachaliou N, Rosell R, Santarpia M. Mechanisms of resistance to osimertinib. J Thorac Dis. (2020) 12:2851–8. doi: 10.21037/jtd.2019.08.30
33. Fu K, Xie F, Wang F, Fu L. Therapeutic strategies for EGFR-mutated non-small cell lung cancer patients with osimertinib resistance. J Hematol Oncol. (2022) 15:173. doi: 10.1186/s13045-022-01391-4
34. Gomatou G, Syrigos N, Kotteas E. Osimertinib resistance: Molecular mechanisms and emerging treatment options. Cancers (Basel). (2023) 15:841. doi: 10.3390/cancers15030841
35. Schmid S, Li JJN, Leighl NB. Mechanisms of osimertinib resistance and emerging treatment options. Lung Cancer. (2020) 147:123–9. doi: 10.1016/j.lungcan.2020.07.014
36. Remon J, Caramella C, Jovelet C, Lacroix L, Lawson A, Smalley S, et al. Osimertinib benefit in EGFR -mutant NSCLC patients with T790M -mutation detected by circulating tumour DNA. Ann Oncol. (2017) 28:784–90. doi: 10.1093/annonc/mdx017
37. Russo A, Lee JK, Pasquina LW, Del R, Dilks HH. Liquid biopsy of lung cancer before pathological diagnosis is associated with shorter time to treatment. JCO Precis Oncol. (2024) 8:e2300535. doi: 10.1200/PO.23.00535
38. Del Re M, Luculli GI, Petrini I, Sbrana A, Scotti V, Perez D de M, et al. Clinical utility of next generation sequencing of plasma cell-free DNA for the molecular profiling of patients with NSCLC at diagnosis and disease progression. Transl Oncol. (2024) 41:101869. doi: 10.1016/j.tranon.2023.101869
39. Ntzifa A, Lianidou E. Pre-analytical conditions and implementation of quality control steps in liquid biopsy analysis. Crit Rev Clin Lab Sci. (2023) 60:573–94. doi: 10.1080/10408363.2023.2230290
40. Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from. J Mol Diagnostics. (2018) 20:129–59. doi: 10.1016/j.jmoldx.2017.11.004
41. Rolfo C, Mack P, Scagliotti GV, Aggarwal C, Arcila ME, Barlesi F, et al. Liquid biopsy for advanced NSCLC: A consensus statement from the international association for the study of lung cancer. J Thorac Oncol. (2021) 16:1647–62. doi: 10.1016/j.jtho.2021.06.017
42. Planchard D, Popat S, Kerr K, Novello S, Smit EF, Faivre-Finn C, et al. Metastatic non-small cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. (2018) 29:iv192–237. doi: 10.1093/annonc/mdy275
43. Aggarwal C, Rolfo CD, Oxnard GR, Gray JE, Sholl LM, Gandara DR. Strategies for the successful implementation of plasma-based NSCLC genotyping in clinical practice. Nat Rev Clin Oncol. (2021) 18:56–62. doi: 10.1038/s41571-020-0423-x
44. Malapelle U, Tiseo M, Vivancos A, Kapp J, Serrano MJ, Tiemann M. Liquid biopsy for biomarker testing in non-small cell lung cancer: A european perspective. J Mol Pathol. (2021) 2:255–73. doi: 10.3390/jmp2030022
45. National comprehensive cancer network (NCCN) NCCN clinical practice guidelines in oncology (NCCN guidelines®): Non-small cell lung cancer. version: 7.2021. (2021). Available at: https://www.nccn.org/professionals/physician_gls/PDF/nscl.pdf (Accessed May 20, 2024).
46. Rolfo C, Mack PC, Scagliotti GV, Baas P, Barlesi F, Bivona TG, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): A statement paper from the IASLC. J Thorac Oncol. (2018) 13:1248–68. doi: 10.1016/j.jtho.2018.05.030
47. Rotow JK, Lee JK, Madison RW, Oxnard GR, Pasi A. Jänne ABS. Real-world genomic profile of EGFR second-site mutations and other osimertinib resistance mechanisms, and clinical landscape of non-small cell lung cancer (NSCLC) post-osimertinib. J Thorac Oncol. (2023) S1556-0864:02263–3. doi: 10.1016/j.jtho.2023.09.1453
48. Punnoose EA, Atwal S, Liu W, Raja R, Fine BM, Hughes BGM, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: Association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res. (2012) 18:2391–401. doi: 10.1158/1078-0432.CCR-11-3148
49. Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. (2008) 24:366–77. doi: 10.1056/NEJMoa0800668
50. Marchetti A, Del Grammastro M, Felicioni L, Malatesta S, Filice G, Centi I, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: Toward a real-time liquid biopsy for treatment. PloS One. (2014) 9:e103883. doi: 10.1371/journal.pone.0103883
51. Sundaresan TK, Sequist LV, Heymach JV, Riely GJ, Jänne PA, Koch WH, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood-based analyses. Clin Cancer Res. (2016) 22:1103–10. doi: 10.1158/1078-0432.CCR-15-1031
52. Kapeleris J, Kulasinghe A, Warkiani ME, Oleary C, Vela I, Leo P, et al. Ex vivo culture of circulating tumour cells derived from non-small cell lung cancer. Transl Lung Cancer Res. (2020) 9:1795–809. doi: 10.21037/tlcr-20-521
53. Owen S, Lo TW, Fouladdel S, Zeinali M, Keller E, Azizi E, et al. Simultaneous single cell gene expression and EGFR mutation analysis of circulating tumor cells reveals distinct phenotypes in NSCLC. Adv Biosyst. (2020) 4:e2000. doi: 10.1002/adbi.202000110
54. Strati A, Markou A, Kyriakopoulou E, Lianidou E. Detection and molecular characterization of circulating tumour cells: Challenges for the clinical setting. Cancers (Basel). (2023) 15:1–20. doi: 10.3390/cancers15072185
55. Mastoraki S, Strati A, Tzanikou E, Chimonidou M, Politaki E, Voutsina A, et al. ESR1 methylation: A liquid biopsy-based epigenetic assay for the follow up of patients with metastatic breast cancer receiving endocrine treatment: Short running title: Liquid biopsy: ESR1 methylation in CTCs and paired ctDNA. Clin Cancer Res. (2018) 24:1500–10. doi: 10.1158/1078-0432.CCR-17-1181
56. Chimonidou M, Strati A, Malamos N, Georgoulias V, Lianidou ES. SOX17 promoter methylation in circulating tumor cells and matched cell-free DNA isolated from plasma of patients with breast cancer. Clin Chem. (2013) 59:270–9. doi: 10.1373/clinchem.2012.191551
57. Beije N, Sieuwerts AM, Kraan J, Van NM, Onstenk W, Vitale SR, et al. Estrogen receptor mutations and splice variants determined in liquid biopsies from metastatic breast cancer patients. Mol Oncol. (2018) 12:48–57. doi: 10.1002/1878-0261.12147
58. Paolillo C, Mu Z, Rossi G, Schiewer MJ, Nguyen T, Austin L, et al. Detection of activating estrogen receptor gene (ESR1) mutations in single circulating tumor cells HHS public access. Clin Cancer Res. (2017) 23:6086–93. doi: 10.1158/1078-0432.CCR-17-1173.Detection
59. Stergiopoulou D, Markou A, Strati A, Zavridou M, Tzanikou E, Mastoraki S, et al. Comprehensive liquid biopsy analysis as a tool for the early detection of minimal residual disease in breast cancer. Sci Rep. (2023) 13:1258. doi: 10.1038/s41598-022-25400-1
60. Tzanikou E, Markou A, Politaki E, Koutsopoulos A, Psyrri A, Mavroudis D, et al. PIK3CA hotspot mutations in circulating tumor cells and paired circulating tumor DNA in breast cancer: a direct comparison study. Mol Oncol. (2019) 13:2515–30. doi: 10.1002/1878-0261.12540
61. Keup C, Suryaprakash V, Hauch S, Storbeck M, Hahn P, Sprenger-Haussels M, et al. Integrative statistical analyses of multiple liquid biopsy analytes in metastatic breast cancer. Genome Med. (2021) 13:1–14. doi: 10.1186/s13073-021-00902-1
62. Eslami-S Z, Cortés-Hernández LE, Sinoquet L, Gauthier L, Vautrot V, Cayrefourcq L, et al. Circulating tumour cells and PD-L1-positive small extracellular vesicles: the liquid biopsy combination for prognostic information in patients with metastatic non-small cell lung cancer. Br J Cancer. (2024) 130:63–72. doi: 10.1038/s41416-023-02491-9
63. Kapeleris J, Müller Bark J, Ranjit S, Irwin D, Hartel G, Warkiani ME, et al. Prognostic value of integrating circulating tumour cells and cell-free DNA in non-small cell lung cancer. Heliyon. (2022) 8:e09971. doi: 10.1016/j.heliyon.2022.e09971
64. Zavridou M, Strati A, Bournakis E, Smilkou S, Tserpeli V LE. Prognostic significance of gene expression and DNA methylation markers in circulating tumor cells and paired plasma derived exosomes in metastatic castration resistant prostate cancer. Cancers (Basel). (2021) 13:780. doi: 10.3390/cancers13040780
65. Sementsov M, Ott L, Kött J, Sartori A, Lusque A, Degenhardt S, et al. Mutation analysis in individual circulating tumor cells depicts intratumor heterogeneity in melanoma. EMBO Mol Med. (2024), 1–19. doi: 10.1038/s44321-024-00082-6
66. Yanagita M, Redig AJ, Paweletz CP, Dahlberg SE, O’Connell A, Feeney N, et al. A prospective evaluation of circulating tumor cells and cell-free DNA in EGFR-mutant non-small cell lung cancer patients treated with erlotinib on a phase II trial. Clin Cancer Res. (2016) 22:6010–20. doi: 10.1158/1078-0432.CCR-16-0909
67. Liu HE, Vuppalapaty M, Wilkerson C, Renier C, Chiu M, Lemaire C, et al. Detection of EGFR mutations in cfDNA and CTCs, and comparison to tumor tissue in non-Small-Cell-Lung-Cancer (NSCLC) patients. Front Oncol. (2020) 10:572895. doi: 10.3389/fonc.2020.572895
68. Cheng X, Chen H. Tumor heterogeneity and resistance to EGFR-targeted therapy in advanced nonsmall cell lung cancer: Challenges and perspectives. Onco Targets Ther. (2014) 7:1689–704. doi: 10.2147/OTT.S66502
69. Ntzifa A, Londra D, Rampias T, Kotsakis A, Georgoulias V, Lianidou E. DNA methylation analysis in plasma cell-free DNA and paired CTCs of NSCLC patients before and after osimertinib treatment. Cancers (Basel). (2021) 13:5974. doi: 10.3390/cancers13235974
70. Ntzifa A, Kotsakis A, Georgoulias V, Lianidou E. Detection of EGFR mutations in plasma cfDNA and paired CTCs of NSCLC patients before and after osimertinib therapy using crystal digital PCR. Cancers (Basel). (2021) 13:2736. doi: 10.3390/cancers13112736
71. Pantazaka E, Ntzifa A, Roumeliotou A, Lianidou E, Georgoulias V, Kotsakis A KG. PD-L1/pS6 in circulating tumor cells (CTCs) during osimertinib treatment in patients with non-small cell lung cancer (NSCLC). Biomedicines. (2022) 10:1893. doi: 10.3390/biomedicines10081893
72. Kallergi G, Kontopodis E, Ntzifa A, Jordana-Ariza N, Karachaliou N, Pantazaka E, et al. Effect of osimertinib on CTCs and ctDNA in EGFR mutant non-small cell lung cancer patients: The prognostic relevance of liquid biopsy. Cancers (Basel). (2022) 14:1574. doi: 10.3390/cancers14061574
73. Ntzifa A, Strati A, Kallergi G, Kotsakis A, Georgoulias V, Lianidou E. Gene expression in circulating tumor cells reveals a dynamic role of EMT and PD-L1 during osimertinib treatment in NSCLC patients. Sci Rep. (2021) 11:2313. doi: 10.1038/s41598-021-82068-9
74. Ntzifa A, Kroupis C, Haliassos A, Lianidou E. A pilot plasma-ctDNA ring trial for the cobas® EGFR mutation test in clinical diagnostic laboratories. Clin Chem Lab Med. (2019) 57:E97–E101. doi: 10.1515/cclm-2018-0676
75. Markou A, Farkona S, Schiza C, Efstathiou T, Kounelis S, Malamos N, et al. PIK3CA mutational status in circulating tumor cells can change during disease recurrence or progression in patients with breast cancer. Clin Cancer Res. (2014) 20:5823–34. doi: 10.1158/1078-0432.CCR-14-0149
76. Flores LM, Kindelberger DW, Ligon AH, Capelletti M, Fiorentino M, Loda M, et al. Improving the yield of circulating tumour cells facilitates molecular characterisation and recognition of discordant HER2 amplification in breast cancer. Br J Cancer. (2010) 102:1495–502. doi: 10.1038/sj.bjc.6605676
77. Cohen EN, Jayachandran G, Moore RG, Cristofanilli M, Lang JE, Khoury JD, et al. A multi-center clinical study to harvest and characterize circulating tumor cells from patients with metastatic breast cancer using the parsortix ® PC1 system. Cancers (Basel). (2022) 14:5238. doi: 10.3390/cancers14215238
78. Schildhaus HU, Schultheis AM, Rüuschoff J, Binot E, Merkelbach-Bruse S, Fassunke J, et al. Met amplification status in therapy-naïve adeno- and squamous cell carcinomas of the lung. Clin Cancer Res. (2015) 21:907–15. doi: 10.1158/1078-0432.CCR-14-0450
79. Markou AN, Londra D, Stergiopoulou D, Vamvakaris I, Potaris K, Pateras IS, et al. Preoperative mutational analysis of circulating tumor cells (CTCs) and plasma-cfDNA provides complementary information for early prediction of relapse: A pilot study in early-stage non-small cell lung cancer. Cancers (Basel). (2023) 15:1877. doi: 10.3390/cancers15061877
80. Wishart G, Templeman A, Hendry F, Miller K. Molecular profiling of circulating tumour cells and circulating tumour DNA : Complementary insights from a single blood sample utilising the parsortix ® system. Curr Issues Mol Biol. (2024) 46:773–87. doi: 10.3390/cimb46010050
81. Oxnard GR, Hu Y, Mileham KF, Husain H, Costa DB, Tracy P, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. (2018) 4:1527–34. doi: 10.1001/jamaoncol.2018.2969
82. Uchibori K, Inase N, Araki M, Kamada M, Sato S, Okuno Y, et al. Brigatinib combined with anti-EGFR antibody overcomes osimertinib resistance in EGFR-mutated non-small-cell lung cancer. Nat Commun. (2017) 8:1–16. doi: 10.1038/ncomms14768
83. Wang X, Zhou L, Yin JC, Wu X, Shao YW, Gao B. Lung adenocarcinoma harboring EGFR 19del/C797S/T790M triple mutations responds to brigatinib and anti-EGFR antibody combination therapy. J Thorac Oncol. (2019) 14:e85–8. doi: 10.1016/j.jtho.2019.01.015
84. Russo A, Scilla KA, Mehra R, Gittens A, McCusker MG, de Miguel-Perez D, et al. Tracking clonal evolution of EGFR-mutated non-small cell lung cancer through liquid biopsy: Management of C797S acquired mutation. Clin Lung Cancer. (2023) 24:660–5. doi: 10.1016/j.cllc.2023.07.003
85. Schaufler D, Ast DF, Tumbrink HL, Abedpour N, Maas L, Schwäbe AE, et al. Clonal dynamics of BRAF-driven drug resistance in EGFR-mutant lung cancer. NPJ Precis Oncol. (2021) 5:102. doi: 10.1038/s41698-021-00241-9
86. Ho CC, Liao WY, Lin CA, Shih JY, Yu CJ, Chih-Hsin Yang J. Acquired BRAF V600E mutation as resistant mechanism after treatment with osimertinib. J Thorac Oncol. (2017) 12:567–72. doi: 10.1016/j.jtho.2016.11.2231
87. Martín Martorell P, Huerta M, Compañ Quilis A, Abellán R, Seda E, Blesa S, et al. Coexistence of EGFR, KRAS, BRAF, and PIK3CA mutations and ALK rearrangement in a comprehensive cohort of 326 consecutive spanish nonsquamous NSCLC patients. Clin Lung Cancer. (2017) 18:e395–402. doi: 10.1016/j.cllc.2017.04.006
88. Zhuang X, Zhao C, Li J, Su C, Chen X, Ren S, et al. Clinical features and therapeutic options in non-small cell lung cancer patients with concomitant mutations of EGFR, ALK, ROS1, KRAS or BRAF. Cancer Med. (2019) 8:2858–66. doi: 10.1002/cam4.2183
89. Ohashi K, Sequist LV, Arcila ME, Moran T, Chmielecki J, Lin YL, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci USA. (2012) 109:1–7. doi: 10.1073/pnas.1203530109
90. Chen M, Xu Y, Zhao J, Zhong W, Zhang L, Bi Y, et al. Concurrent driver gene mutations as negative predictive factors in epidermal growth factor receptor-positive non-small cell lung cancer. EBioMedicine. (2019) 42:304–10. doi: 10.1016/j.ebiom.2019.03.023
91. Huang Y, Gan J, Guo K, Deng Y, Fang W. Acquired BRAF V600E mutation mediated resistance to osimertinib and responded to osimertinib, dabrafenib, and trametinib combination therapy. J Thorac Oncol. (2019) 14:e236–7. doi: 10.1016/j.jtho.2019.05.040
92. Meng P, Koopman B, Kok K, ter Elst A, Schuuring E, van Kempen LC, et al. Combined osimertinib, dabrafenib and trametinib treatment for advanced non-small-cell lung cancer patients with an osimertinib-induced BRAF V600E mutation. Lung Cancer. (2020) 146:358–61. doi: 10.1016/j.lungcan.2020.05.036
93. Ding H, Zhuang Z, Xie J, Huang H, Tao Z, Liu Z. Durable clinical response of advanced lung adenocarcinoma harboring egfr-19del/T790M/ brafv600e mutations after treating with osimertinib and dabrafenib plus trametinib: A case report. Onco Targets Ther. (2020) 13:7933–9. doi: 10.2147/OTT.S240775
94. Fernando M, Almeida S, Knebel FH, Bettoni F, Saddi R, Sacardo KP, et al. Impressive response to dabrafenib, trametinib, and osimertinib in a metastatic EGFR-mutant/ BRAF V600E lung adenocarcinoma patient. NPJ Precis Oncol. (2021) 5:5. doi: 10.1038/s41698-021-00149-4
95. Ernst SM, Uzun S, Paats MS, van Marion R, Atmodimedjo PN, de Jonge E, et al. Efficacy and tolerability of osimertinib and sotorasib combination treatment for osimertinib resistance caused by KRAS G12C mutation: A report of two cases. JCO Precis Oncol. (2023) 7:e2300451. doi: 10.1200/PO.23.00451
96. Suryavanshi M, Jaipuria J, Mattoo S, Dhandha S, Khatri M. Audit of molecular mechanisms of primary and secondary resistance to various generations of tyrosine kinase inhibitors in known epidermal growth factor receptor-mutant non-small cell lung cancer patients in a tertiary centre. Clin Oncol. (2022) 34:e451–62. doi: 10.1016/j.clon.2022.06.003
97. Shi Y, Xing P, Han X, Wang S, Liu Y, Liu P, et al. P1.13-18 exploring the resistance mechanism of osimertinib and monitoring the treatment response using plasma ctDNA in chinese NSCLC patients. J Thorac Oncol. (2018) 13:S589. doi: 10.1016/j.jtho.2018.08.875
98. Chmielecki J, Mok T, Wu YL, Han JY, Ahn MJ, Ramalingam SS, et al. Analysis of acquired resistance mechanisms to osimertinib in patients with EGFR-mutated advanced non-small cell lung cancer from the AURA3 trial. Nat Commun. (2023) 14:1071. doi: 10.1038/s41467-023-35962-x
99. Fang W, Huang Y, Gu W, Gan J, Wang W, Zhang S, et al. PI3K-AKT-mTOR pathway alterations in advanced NSCLC patients after progression on EGFR-TKI and clinical response to EGFR-TKI plus everolimus combination therapy. Transl Lung Cancer Res. (2020) 9:1258–67. doi: 10.21037/tlcr-20-141
100. Han R, Lin C, Zhang C, Kang J, Lu C, Zhang Y, et al. The potential therapeutic regimen for overcoming resistance to osimertinib due to rare mutations in NSCLC. iScience. (2023) 26:107105. doi: 10.1016/j.isci.2023.107105
101. Guo R, Luo J, Chang J, Rekhtman N, Arcila M DA. MET-dependent solid tumours - molecular diagnosis and targeted therapy. Nat Rev Clin Oncol. (2020) 17:569–87. doi: 10.1038/s41571-020-0377-z.MET-Dependent
102. Hartmaier RJ, Markovets AA, Ahn MJ, Sequist LV, Han JY, Cho BC, et al. Osimertinib + savolitinib to overcome acquired MET-mediated resistance in epidermal growth factor receptor–mutated, MET-amplified non–small cell lung cancer: TATTON. Cancer Discovery. (2023) 13:98–113. doi: 10.1158/2159-8290.CD-22-0586
103. Jebbink M, De Langen AJ, Monkhorst K, Boelens MC, Van Den BD, Van Der NV, et al. Trastuzumab-emtansine and osimertinib combination therapy to target HER2 bypass track resistance in EGFR mutation-positive NSCLC. JTO Clin Res Rep. (2023) 4:100481. doi: 10.1016/j.jtocrr.2023.100481
104. Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. (2017) 17:637–58. doi: 10.1038/nrc.2017.84
105. Romero-Garcia S, Prado-Garcia H, Carlos-Reyes A. Role of DNA methylation in the resistance to therapy in solid tumors. Front Oncol. (2020) 10:1152. doi: 10.3389/fonc.2020.01152
106. Sharma SV, Lee DY, Li B, Quinlan MP, Maheswaran S, Mcdermott U, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. (2010) 141:69–80. doi: 10.1016/j.cell.2010.02.027.A
107. Lianidou E. Detection and relevance of epigenetic markers on ctDNA: recent advances and future outlook. Mol Oncol. (2021) 15:1683–700. doi: 10.1002/1878-0261.12978
108. Fabrizio FP, Sparaneo A, Muscarella LA. Monitoring EGFR -lung cancer evolution : a possible beginning of a “ methylation era ” in TKI resistance prediction. Front Oncol. (2023) 13:1137384. doi: 10.3389/fonc.2023.1137384
109. Maeda M, Murakami Y, Watari K, Kuwano M, Izumi H, Ono M. CpG hypermethylation contributes to decreased expression of PTEN during acquired resistance to gefitinib in human lung cancer cell lines. Lung Cancer. (2015) 87:265–71. doi: 10.1016/j.lungcan.2015.01.009
110. Zhu J, Wang Y, Duan J, Bai H, Wang Z, Wei L, et al. DNA methylation status of wnt antagonist SFRP5 can predict the response to the EGFR-tyrosine kinase inhibitor therapy in non-small cell lung cancer. J Exp Clin Cancer Res. (2012) 31:80. doi: 10.1186/1756-9966-31-80
111. Greve G, Schiffmann I, Pfeifer D, Pantic M, Schüler J, Lübbert M. The pan-HDAC inhibitor panobinostat acts as a sensitizer for erlotinib activity in EGFR-mutated and -wildtype non-small cell lung cancer cells. BMC Cancer. (2015) 15:947. doi: 10.1186/s12885-015-1967-5
112. Hou T, Ma J, Hu C, Zou F, Jiang S, Wang Y, et al. Decitabine reverses gefitinib resistance in PC9 lung adenocarcinoma cells by demethylation of RASSF1A and GADD45β promoter. Int J Clin Exp Pathol. (2019) 12:4002–10. http://www.ncbi.nlm.nih.gov/pubmed/31933796%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC6949799.
113. El Kadi N, Wang L, Davis A, Korkaya H, Cooke A, Vadnala V, et al. The EGFR T790M mutation is acquired through AICDA-mediated deamination of 5-methylcytosine following TKI treatment in lung cancer. Cancer Res. (2018) 78:6728–35. doi: 10.1158/0008-5472.CAN-17-3370
114. Su S-F, Liu C-H, Cheng C-L, Ho C-C, Yang T-Y, Chen K-C, et al. Genome-wide epigenetic landscape of lung adenocarcinoma links HOXB9 DNA methylation to intrinsic EGFR-TKI resistance and heterogeneous responses. JCO Precis Oncol. (2021) 19:418–31. doi: 10.1200/po.20.00151
115. Lin S, Ruan H, Qin L, Zhao C, Gu M, Wang Z, et al. Acquired resistance to EGFR-TKIs in NSCLC mediates epigenetic downregulation of MUC17 by facilitating NF-κB activity via UHRF1/DNMT1 complex. Int J Biol Sci. (2023) 19:832–51. doi: 10.7150/ijbs.75963
116. Xia S, Ye J, Chen Y, Lizaso A, Huang L, Shi L, et al. Parallel serial assessment of somatic mutation and methylation profile from circulating tumor DNA predicts treatment response and impending disease progression in osimertinib-treated lung adenocarcinoma patients. Transl Lung Cancer Res. (2019) 8:1016–28. doi: 10.21037/tlcr.2019.12.09
117. Nguyen HN, Cao NPT, Van Nguyen TC, Le KND, Nguyen DT, Nguyen QTT, et al. Liquid biopsy uncovers distinct patterns of DNA methylation and copy number changes in NSCLC patients with different EGFR-TKI resistant mutations. Sci Rep. (2021) 11:16436. doi: 10.1038/s41598-021-95985-6
118. Lee ATM, Nagasaka M. CheckMate-722: The rise and fall of nivolumab with chemotherapy in TKI-refractory EGFR-mutant NSCLC. Lung Cancer Targets Ther. (2023) 14:41–6. doi: 10.2147/LCTT.S408886
119. Kim D, Bach DH, Fan YH, Luu TTT, Hong JY, Park HJ, et al. AXL degradation in combination with EGFR-TKI can delay and overcome acquired resistance in human non-small cell lung cancer cells. Cell Death Dis. (2019) 10:361. doi: 10.1038/s41419-019-1601-6
120. Namba K, Shien K, Takahashi Y, Torigoe H, Sato H, Yoshioka T, et al. Activation of AXL as a preclinical acquired resistance mechanism against osimertinib treatment in EGFR-mutant non-small cell lung cancer cells. Mol Cancer Res. (2019) 17:499–507. doi: 10.1158/1541-7786.MCR-18-0628
121. Taniguchi H, Yamada T, Wang R, Tanimura K, Adachi Y, Nishiyama A, et al. AXL confers intrinsic resistance to osimertinib and advances the emergence of tolerant cells. Nat Commun. (2019) 10:259. doi: 10.1038/s41467-018-08074-0
122. Karachaliou N, Chaib I, Cardona AF, Berenguer J, Bracht JWP, Yang J, et al. Common co-activation of AXL and CDCP1 in EGFR-mutation-positive non-small cell lung cancer associated with poor prognosis. EBioMedicine. (2018) 29:112–27. doi: 10.1016/j.ebiom.2018.02.001
123. Morimoto K, Yamada T, Sawada R, Azuma K, Goto Y, Harada T, et al. Predictive value of p53 and AXL immunostaining for the efficacy of immune checkpoint inhibitor-based therapy after osimertinib treatment in patients with epidermal growth factor-mutant non-small cell lung cancer. Cancer Immunol Immunother. (2023) 72:1699–707. doi: 10.1007/s00262-023-03370-1
124. Bracht JWP, Karachaliou N, Berenguer J, Fernandez-Bruno M, Filipska M, Pedraz-Valdunciel C, et al. PIM-1 inhibition with AZD1208 to prevent osimertinib-induced resistance in EGFR-mutation positive non-small cell lung cancer. J Cancer Metastasis Treat. (2019) 5:22. doi: 10.20517/2394-4722.2018.111
125. Sun Z, Zeng L, Zhang M, Zhang Y, Yang N. PIM1 inhibitor synergizes the anti-tumor effect of osimertinib via STAT3 dephosphorylation in EGFR-mutant non-small cell lung cancer. Ann Transl Med. (2020) 8:366–6. doi: 10.21037/atm.2020.02.43
126. Cao L, Wang F, Li S, Wang X, Huang D, Jiang R. PIM1 kinase promotes cell proliferation, metastasis and tumor growth of lung adenocarcinoma by potentiating the c-MET signaling pathway. Cancer Lett. (2019) 444:116–26. doi: 10.1016/j.canlet.2018.12.015
127. Malapelle U, Leprieur EG, Kamga PT, Chiasseu MT, Rolfo C. Editorial: Emerging biomarkers for NSCLC: Recent advances in diagnosis and therapy. Front Oncol. (2021) 11:694578. doi: 10.3389/fonc.2021.694578
128. Pascual J, Attard G, Bidard FC, Curigliano G, De Mattos-Arruda L, Diehn M, et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: a report from the ESMO precision medicine working group. Ann Oncol. (2022) 33:750–68. doi: 10.1016/j.annonc.2022.05.520
129. Martignano F, Munagala U, Crucitta S, Mingrino A, Semeraro R, Del Re M, et al. Nanopore sequencing from liquid biopsy: analysis of copy number variations from cell-free DNA of lung cancer patients. Mol Cancer. (2021) 20:4–9. doi: 10.1186/s12943-021-01327-5
130. Lau BT, Almeda A, Schauer M, McNamara M, Bai X, Meng Q, et al. Single-molecule methylation profiles of cell-free DNA in cancer with nanopore sequencing. Genome Med. (2023) 15:1–13. doi: 10.1186/s13073-023-01178-3
Keywords: NSCLC, osimertinib, liquid biopsy, ctDNA, CTC, resistance
Citation: Ntzifa A, Marras T, Kallergi G, Kotsakis A, Georgoulias V and Lianidou E (2024) Comprehensive liquid biopsy analysis for monitoring NSCLC patients under second-line osimertinib treatment. Front. Oncol. 14:1435537. doi: 10.3389/fonc.2024.1435537
Received: 20 May 2024; Accepted: 09 September 2024;
Published: 21 October 2024.
Edited by:
Chiara Nicolazzo, Sapienza University of Rome, ItalyReviewed by:
Francesco Pepe, University of Naples Federico II, ItalyKulbhushan Thakur, University of Delhi, India
Copyright © 2024 Ntzifa, Marras, Kallergi, Kotsakis, Georgoulias and Lianidou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Evi Lianidou, bGlhbmlkb3VAY2hlbS51b2EuZ3I=
 Aliki Ntzifa
Aliki Ntzifa Theodoros Marras1
Theodoros Marras1 Galatea Kallergi
Galatea Kallergi Vasilis Georgoulias
Vasilis Georgoulias Evi Lianidou
Evi Lianidou