- 1Department of Central Laboratory, Liaocheng People’s Hospital, Liaocheng, Shandong, China
- 2Department of Clinical Medicine, Shandong First Medical University, Jinan, Shandong, China
Background: The status of the sentinel lymph nodes (SLNs) was an important prognostic factor in varies cancers. A one-step nucleic acid amplification (OSNA) assay, a molecular-based whole-node analysis method based on CK19 mRNA copy number, was developed to diagnose lymph node metastases. We aimed to evaluate the value of OSNA for the diagnosis of sentinel lymph node metastasis in CK19 positive cancers. CK19 mRNA and protein expression for pan-caner analysis were obtained from TCGA and the Human protein atlas database.
Methods: Two researchers independently searched the PubMed, Cochrane Library and Web of Science databases for qualified articles published before December 1, 2023. A meta-analysis was performed using MetaDisc and STATA. Risk bias and quality assessments of the included studies were evaluated, and a subgroup analysis was performed. Ten cancer types were found to be CK19 positively expressed and 7 of 10 had been reported to use OSNA for SLN detection.
Results: After literature review, there were 61 articles included in the meta-analysis, which consisted of 7115 patients with 18007 sentinel lymph nodes. The pooled sensitivity and specificity of OSNA were 0.87 and 0.95 in overall patients. Moreover, we found the background CK19 expression in normal tissue affected the diagnostic accuracy of OSNA. In breast cancer, we performed subgroup analysis. OSNA exhibited to be a stable method across different population groups and various medical centers. In addition, when 250 copies/μl was chosen as the cutoff point of CK19 mRNA, there were a relatively higher sensitivity and AUC in detecting SLN micro-metastasis than 5000 copies/μl.
Discussion: OSNA can predict the occurrence of SLN metastasis accurately in CK19 positive cancers, especially in breast cancer, colorectal cancer, lung cancer, gastric cancer and endometrial cancer. Our study warrants future studies investigating the clinical application of OSNA in pancreatic, ovarian and bladder cancers.
1 Introduction
Sentinel lymph nodes (SLNs) are the first regional LNs to which tumor cells metastasize through lymphatic vessels (1). The importance of tumor-associated lymphatic vessels and lymphangiogenesis in the formation of LNs metastasis has been emphasized and described during the last two decades (2). Lymphatic vessels from the tumor into LNs is thought to provide a route for metastatic cancer spread, which is prognostic of distant organ metastasis and poor survival (3). Once established in the SLNs, in most cases such as melanoma and breast cancer, tumor cells can migrate to non-SLNs in an orderly sequence. The analysis of SLNs consists of intraoperative evaluation and postoperative pathological examination (4). In terms of the postoperative evaluation, multistep formalin-fixed tissue sections stained by hematoxylin and eosin (H&E) with or without immunohistochemistry (IHC) are commonly used (5). In the intraoperative evaluation, frozen section (FS) and touch imprint cytology (TIC) are recommended (6). However, these methods all have some weaknesses, such as inaccuracy and their required time. Therefore, a novel and more efficient approach for intraoperative detection of SLN metastasis is urgently needed.
Cytokeratin 19 (CK19) belongs to a family of keratins, which are widely used as an epithelial marker in clinical practice and served as a useful research tool in diagnosis, management, and prognosis of the tumors (7). CK19 is reported to be overexpressed in a variety of epithelial malignancies because of its metastatic potential, such as gastrointestinal cancers, lung cancer and breast cancer (8). Recently, a one-step nucleic acid amplification (OSNA) assay, a molecular-based whole-node analysis method, was a rapid intraoperative molecular detection technique and developed to more sensitively diagnose lymph node metastasis via the quantitative measurement of target CK19 mRNA (9). The OSNA assay can quantify the total metastatic volume in a whole lymph node based on CK19 mRNA copy number (10). To date, numerous studies have focused on the diagnostic value of OSNA for detecting SLNs in different cancer types, but the results have been inconclusive. The anticipated drawbacks of OSNA method are false-negative results due to unstable CK19 expression.
In this study, we firstly explored the CK19 expression in different cancers and identified CK19 positive cancers. Next, we performed meta-analysis to investigate the diagnostic value of OSNA for detecting SLN metastasis in different cancers with CK19 positive expression. Our study provided evidence for patient selection when using OSNA in clinical practice.
2 Materials and methods
2.1 CK19 mRNA and protein expression data in pan-cancer
The expression data (mRNA-seq) were obtained from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) was analyzed using TCGA-biolinks in R software (R×64 3.5.1). The raw data were integrated, CK19 expression in 31 cancer types including 9518 tumor samples and 5540 non-tumor samples was analyzed and compared with the normal tissue using the transcripts per million (TPM) values. Meanwhile, CK19 protein expression data in 20 cancer types containing 422 patients were obtained from the Human Protein Atlas database (http://www.proteinatlas.org/). The CK19 protein expression level was defined according to the results of IHC staining using anti-CK19 monoclonal antibody (Agilent Cat# M0888, RRID: AB_2234418).
2.2 Data sources and search strategy
Two investigators systematically and independently searched the PubMed, Cochrane Library and Web of Science databases for articles published before December 1, 2023. Quality studies were needed to provide information on the diagnostic accuracy of OSNA for SLN metastasis in patients with cancer. Subject terms used for the literature search included “(molecular intraoperative) AND cancer”, “(intraoperative molecular analysis) AND cancer”, “(intraoperative nucleic acid amplification) AND cancer”, “(intraoperative nucleic acid amplification) AND cancer”, “(intraoperative nucleic acid amplification) AND cancer”, “(intraoperative nucleic acid amplification) AND cancer”, “(one step nucleic acid amplification) AND cancer”, “(one-step nucleic acid amplification) AND cancer” and “(OSNA) AND cancer”. The range of the search was also extended to the reference lists of retrieved original and review articles. No further ethical approval is required since the program does not require the recruitment of patients and the collection of personal information.
2.3 Study selection
All articles were screened according to the inclusion and exclusion criteria by two independent reviewers. The inclusion criteria were as follows: (1) patients who were diagnosed with cancer; (2) the specimens collected were fresh SLNs; (3) the study’s purpose was to investigate the performance of the OSNA assay for detecting SLN metastasis in cancer patients; (4) the reference method for detecting SLN metastasis was postoperative pathology; (5) the study adopted identical machines and thresholds recommended by the OSNA manufacturer, Sysmex company; (6) the method of pathological examination was described in detail; (7) the study analysis was based on per node; and (8) extracted data were available for obtaining true-positive, false-positive, false negative, and true-negative values. The exclusion criteria were as follows: (1) non-English articles; (2) nonclinical research literature, including basic experiments, reviews, conference abstracts and letters to journal editors; and (3) intraoperative pathology, such as frozen section or touch imprint cytology (11).
2.4 Data extraction and synthesis
Two investigators independently evaluated the eligibility and quality of the studies. The data extracted were the first author, year of publication, country, type of study design, number of patients, number of lymph nodes, number of study centers, section interval, reference standard method, and type of samples. Diagnostic accuracy estimates included true positives (TPs), true negatives (TNs), false positives (FPs) and false negatives (FNs).
2.5 Assessment of diagnostic test accuracy
The diagnostic accuracy of OSNA for SLN metastasis was quantified by the area under the summary receiver operating characteristic (AUC), summary diagnostic odds ratio (DOR), summary sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR) and their 95% confidence interval (CI). A subgroup analysis was further performed to identify possible sources of heterogeneity.
2.6 Assessment of heterogeneity and publication bias
Heterogeneity was explored further by subgroup analyses. In addition, publication bias was assessed by the asymmetry of the Deek’s funnel plot (12). All statistical tests were two sided, and P < 0.05 was considered significant unless otherwise indicated. The above statistical analyses were completed with Stata 16.0.
3 Results
3.1 Selection of CK19 positively expressed cancers
CK19 is well acknowledged as a biomarker of epithelial tumors. Tumor cells carrying high CK19 expression is found to be associated with high invasive phenotype. Therefore, we screened the CK19 protein expression in 20 cancer types (Figure 1A). The IHC staining results showed that the protein levels of CK19 were elevated in colorectal cancer, pancreatic cancer, gastric cancer, urothelial cancer, thyroid cancer, lung cancer, cervical cancer, ovarian cancer, endometrial cancer and breast cancer (mean IHC score >2) (Figure 1B). Considering the OSNA method is based on CK19 mRNA copy number, we next explored the mRNA expression of CK19 in 31 cancer types based on TCGA-sequencing data. The CK19 mRNA expression in tumor and normal tissues were presented in Figure 1C. After matching the mRNA expression and its corresponding protein expression in each cancer type, we found the cancer types with elevated CK19 IHC scores were always carrying high CK19 mRNA expression levels (r=0.778, P=0.0002). Finally, we identified ten cancer types, including breast cancer, bladder cancer, cervical cancer, colorectal cancer, endometrial cancer, gastric cancer, lung cancer, ovarian cancer, pancreatic cancer and thyroid cancer, as the CK19 positively expressed cancers (Figure 1D).
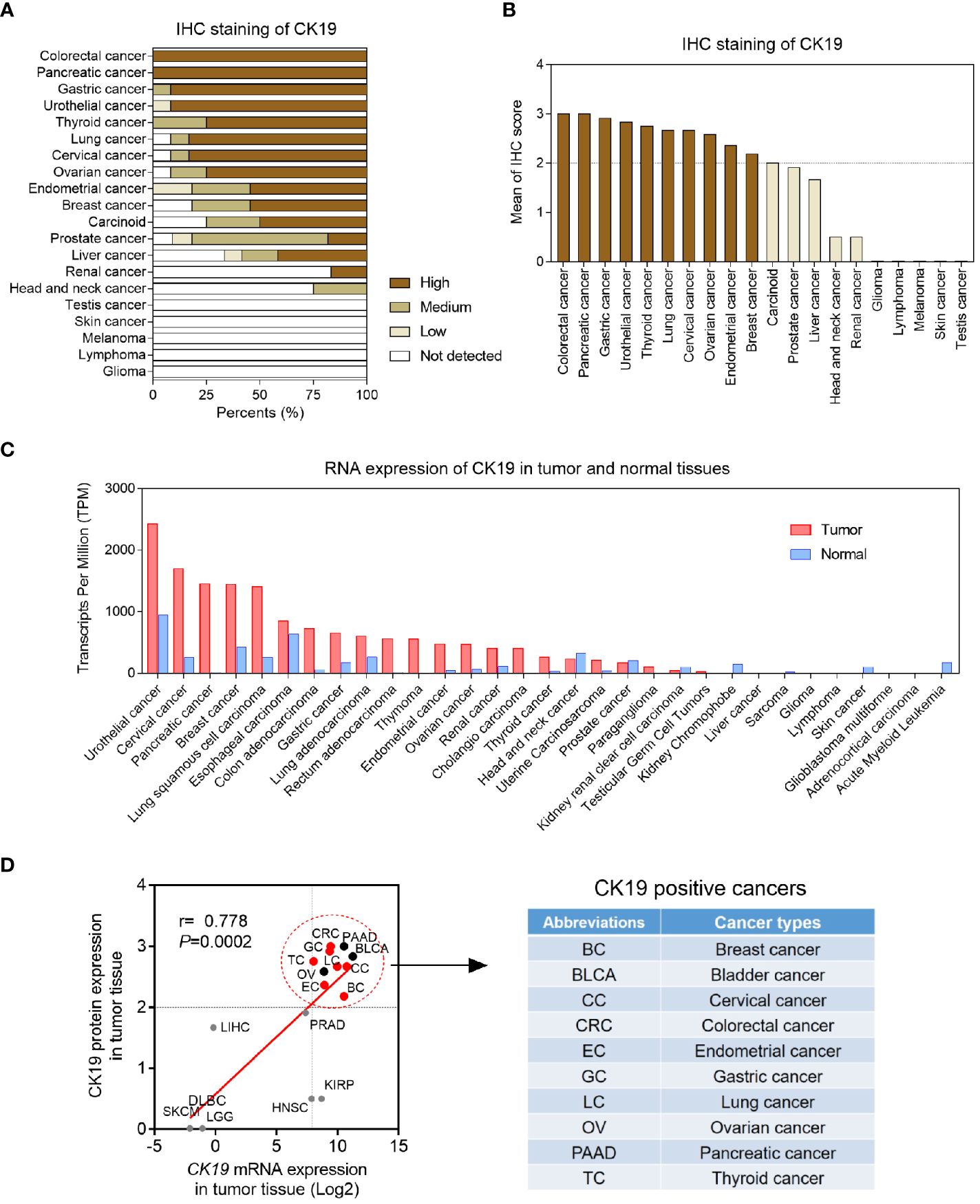
Figure 1 Pan-cancer CK19 protein and mRNA levels. (A) Pan-cancer CK19 protein levels based on IHC scores according to Human Protein Atlas (http://www.proteinatlas.org/). (B) Organized IHC scores of pan-cancer CK19 protein levels. High, medium, low and not detected protein levels were weighted as 3, 2, 1 and 0, respectively. Mean IHC score >2 was identified as high CK19 expressed tumors. (C) Pan-cancer and para-tumoral CK19 RNA levels according to TCGA (https://www.cancer.gov/). (D) Correlation CK19 mRNA expression and protein in different tumor tissues.
3.2 Characteristics of included studies for meta-analysis
Next, we investigated the diagnostic accuracies of OSNA in CK19 positive cancers using meta-analysis method. Figure 2 is a flow chart that schematizes the exclusion of relevant articles for specific reasons. An initial search using predetermined key terms found 1290 potentially relevant articles, but 636 articles were duplications. Studies were also excluded because 417 articles were not related to the topic, 64 articles only focused on the non-SLNs and 53 articles were not clinical studies. 54 articles did not have sufficient data for diagnostic testing. In summary, 61 articles involving 7 cancer types were included in the meta-analysis (9, 10, 13–71). Four cancer types, including bladder cancer, head and neck squamous cell carcinoma, ovarian cancer and pancreatic cancer, were not included for meta-analysis, due to limited literature.
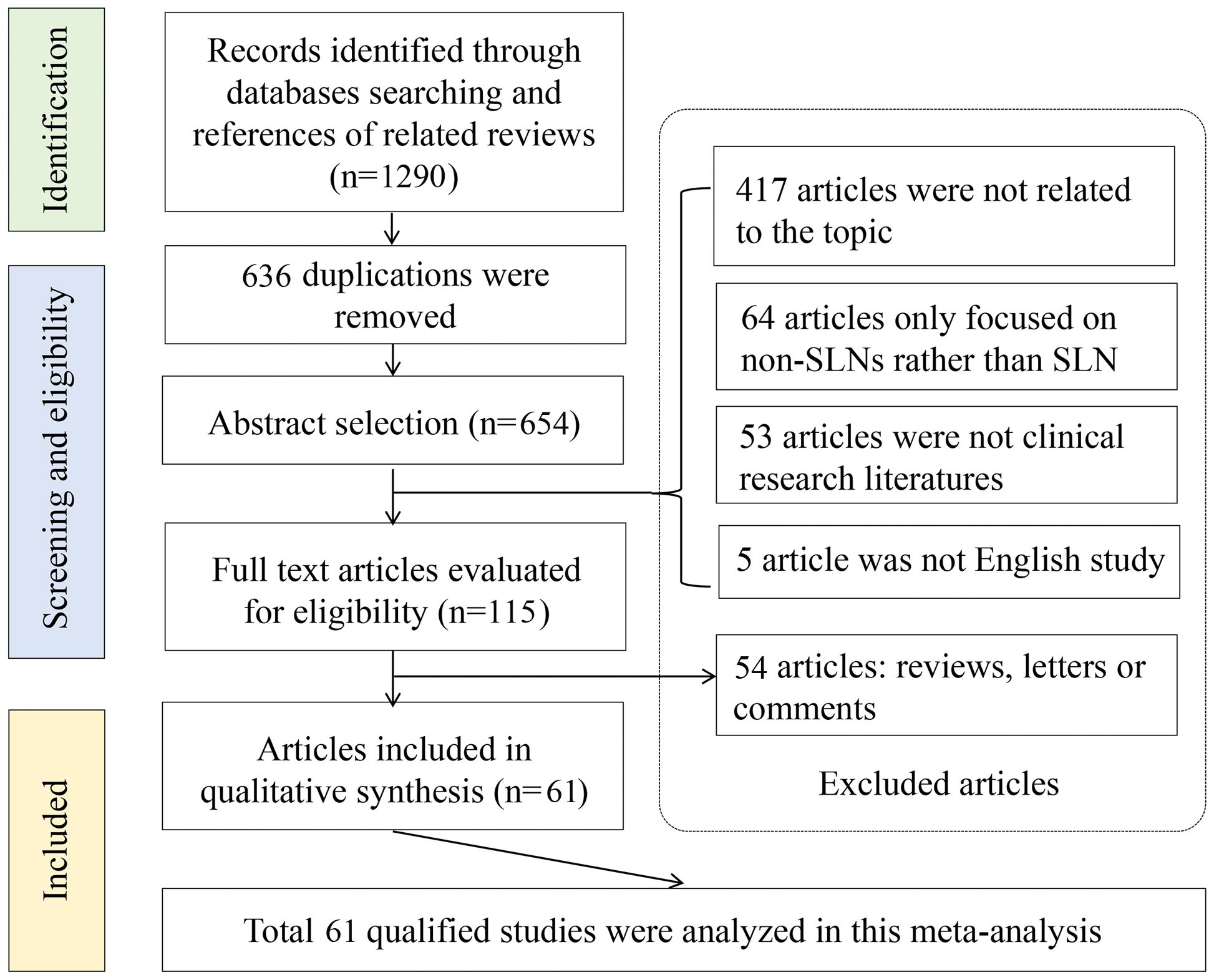
Figure 2 Flowchart of meta-analysis. A total of 1290 records were identified through databases, after screening, 636 articles were duplications. Six hundred and thirty-six articles were excluded, including 417 articles were not related to the topic, 64 articles only focused on the non-SLNs and 53 articles were not clinical studies. 54 articles did not have sufficient data for diagnostic testing. Finally, 61 articles were included in the meta-analysis.
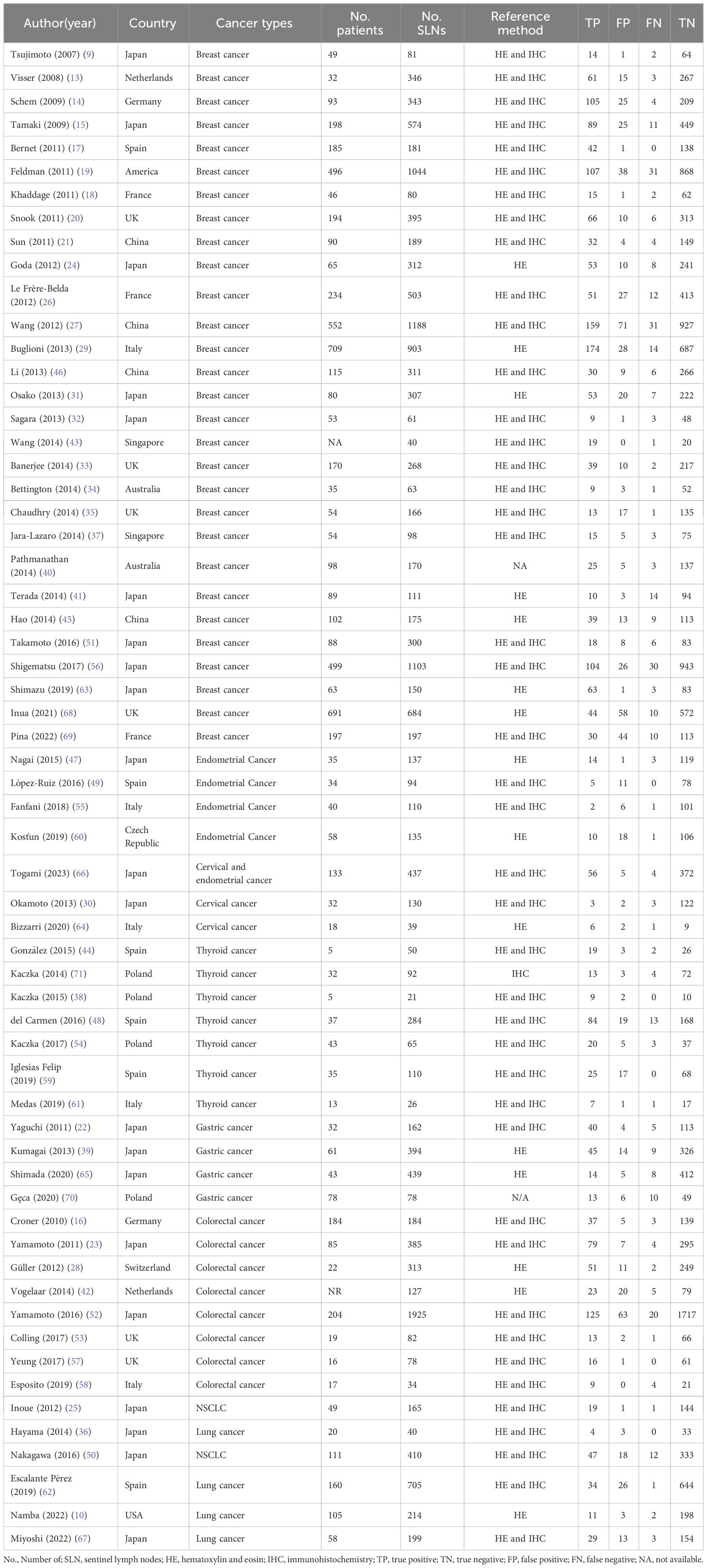
The basic characteristics of the 61 included studies are shown in Table 1. Our study consisted of 7115 patients with 18007 sentinel lymph nodes. Among them, 29 studies enrolled patients of breast cancer, 8 studies involving colorectal cancer, 7 studies involving thyroid cancer, 6 studies involving lung cancer, 4 studies involving gastric cancer, 4 studies involving endometrial cancer and 2 study enrolled patients of cervical cancer. The reference standards of all studies were assessed by postoperative pathology, but the detailed approaches were different, because 43 studies were taken with serial sections with HE staining and IHC and 15 studies undertook serial sections with HE staining only.
3.3 Diagnostic accuracy of OSNA for SLNs in CK19 positive cancers
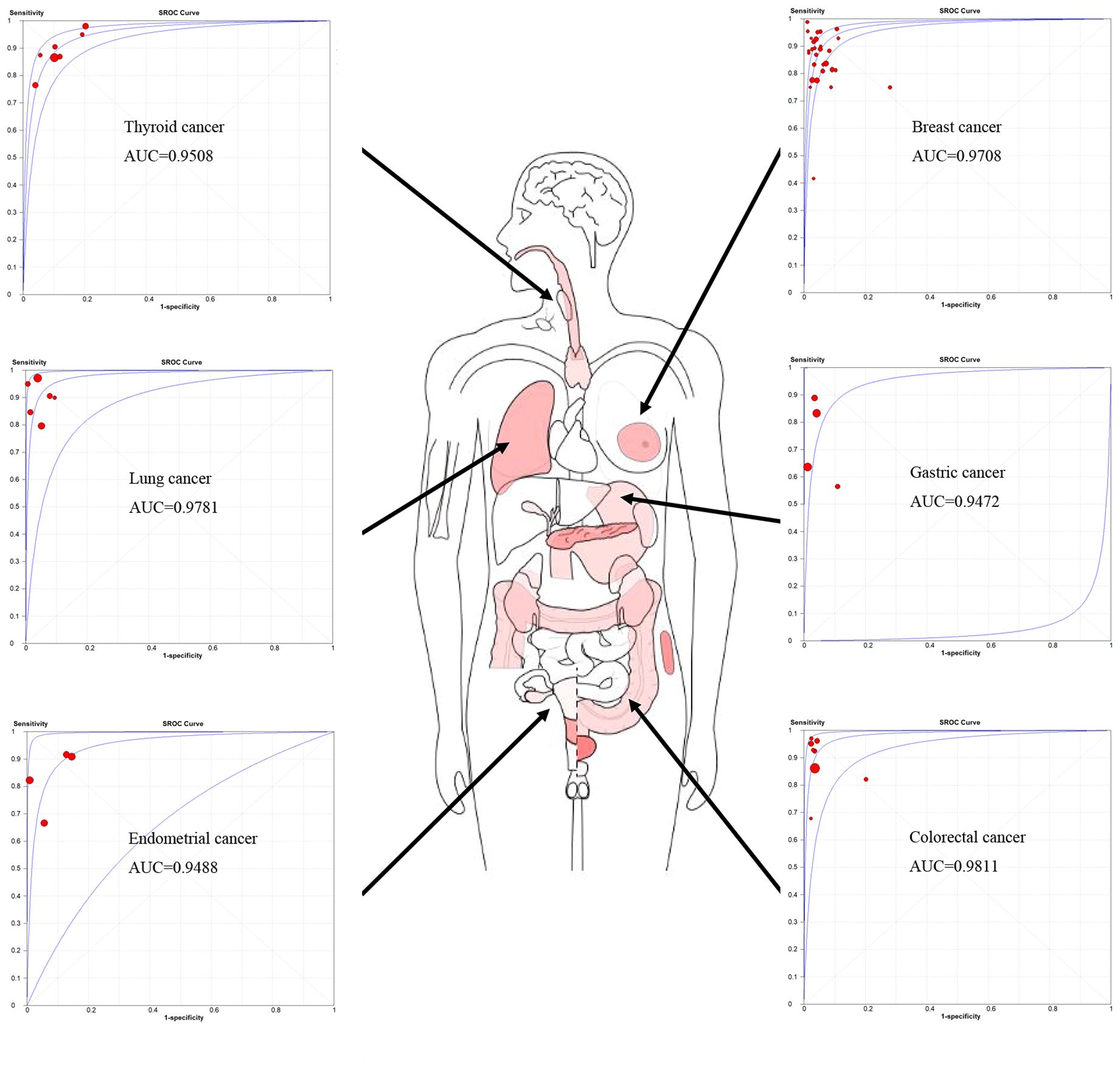
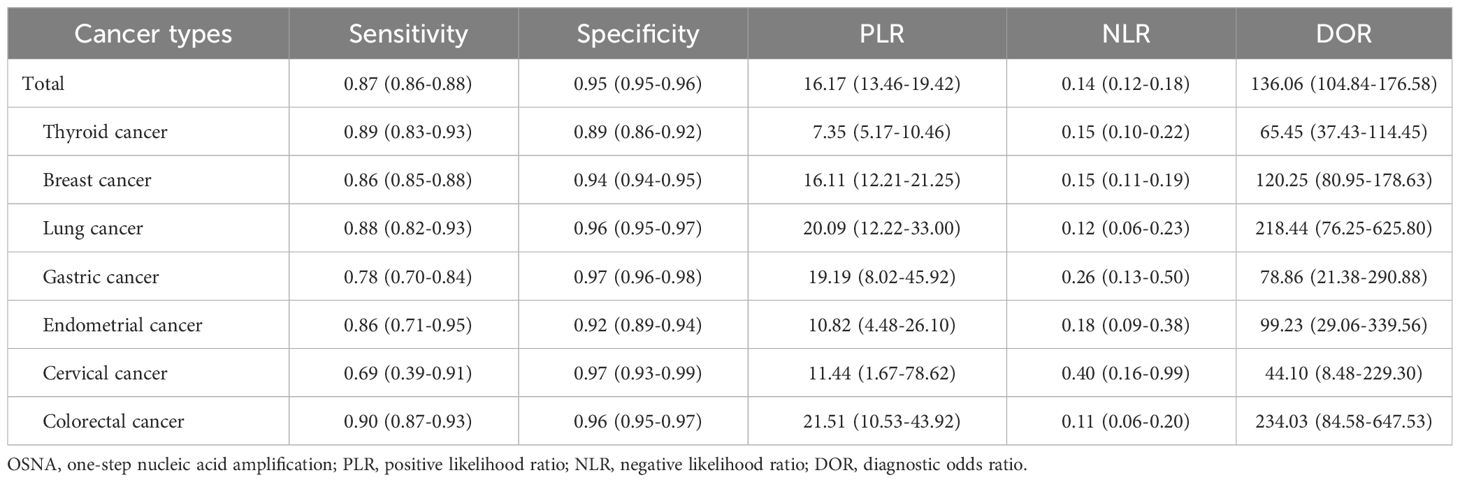
A total of 61 studies assessed the diagnostic accuracy of OSNA for SLNs. Overall, the pooled AUC of OSNA to diagnose SLN metastasis in CK19 positive cancer was 0.9696 (Figure 3A), with a sensitivity of 0.87 (95% CI from 0.85 to 0.88) and a specificity of 0.95 (95% CI from 0.94 to 0.95) (Figures 3B, C). Moreover, the diagnostic accuracy of OSNA for SLN metastasis was assessed across different cancer subtypes. The AUCs of thyroid cancer, breast cancer, lung cancer, gastric cancer, endometrial cancer and colorectal cancer were 0.9508, 0.9708, 0.9781, 0.9472, 0.9488 and 0.9811, respectively (Figure 4). In detail, the sensitivity for detecting SLN metastasis was highest in colorectal cancer (0.90, 95% CI=0.87-0.93) and lowest in cervical cancer (0.69, 95% CI=0.39-0.91). Meanwhile, the specificity was found to be highest in both cervical cancer (0.97, 95% CI=0.93-0.99) and gastric cancer (0.97, 95% CI=0.96-0.98). In contrast, the lowest specificity was shown in thyroid cancer (0.89, 95% CI=0.86-0.92) (Table 2).
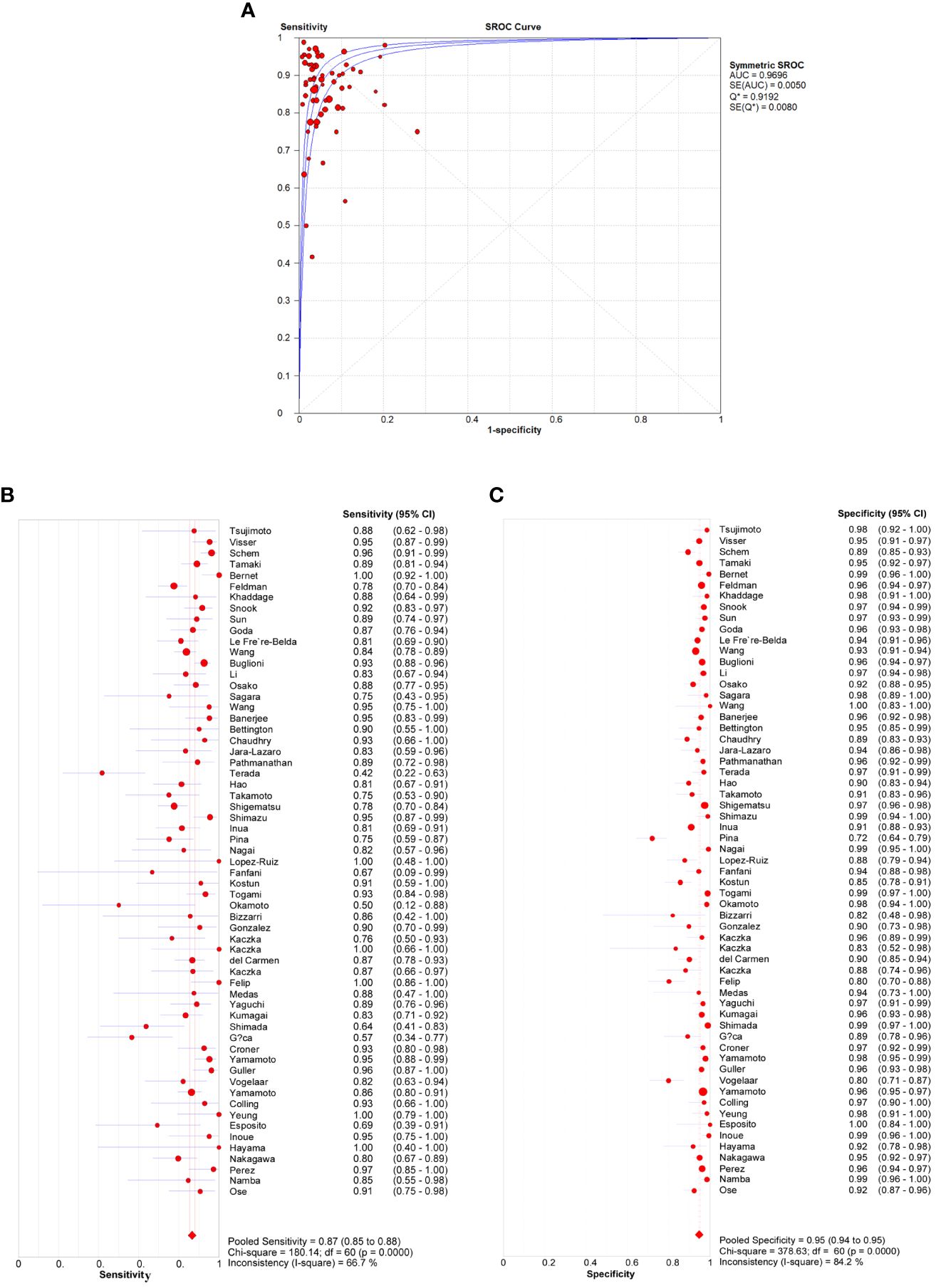
Figure 3 Diagnosis accuracy of OSNA through all studies. (A) ROC curve of OSNA by meta-analysis. The AUC was 0.9696. Sensitivity (B) and specificity (C) of each studies, pooled sensitivity and specificity were 0.87 and 0.95.
3.4 Comparison of diagnostic accuracies and CK19 expression levels in seven cancer types
Considering the principle of OSNA method was based on CK19 mRNA copy number detection, we hypothesized that the diagnostic accuracy of OSNA might affected by CK19 mRNA expression in different cancer types. Therefore, we compared the sensitivity and specificity with CK19 mRNA expression, respectively (Figure 5A). Interestingly, we found that the sensitivity was negatively associated with tumor CK19 expression, whereas the specificity was positively associated with tumor CK19 expression (Figure 5B). Similarly, we observed a negative association between sensitivity and CK19 expression in normal tissue and a positive association between specificity and CK19 expression in normal tissue, indicating the background expression of CK19 might affect the diagnostic accuracy of OSNA (Figure 5C). Furthermore, we compared the background CK19 expression (normal tissue) and tumor CK19 expression. Our results confirmed that the background CK19 expression was elevated along with high expression of CK19 in tumor tissue, especially in CK19 positive cancers (Figure 5D).
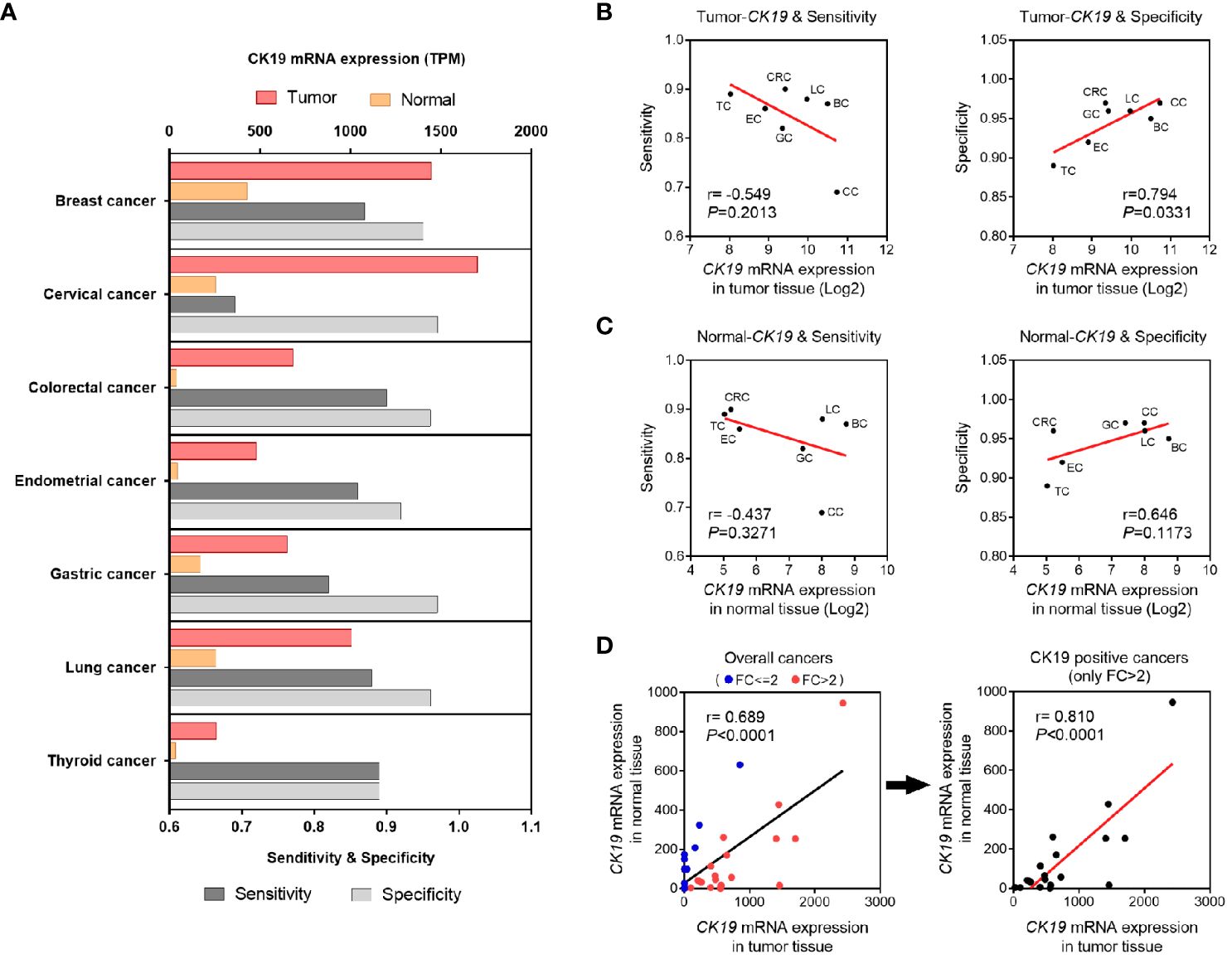
Figure 5 Association of CK19 expression and diagnosis accuracy of OSNA. (A) CK19 expression and diagnosis accuracy of OSNA in different cancer types. (B, C) OSNA diagnosis accuracy variants according to different CK19 expression levels in different types of tumor (B) or normal tissues (C). (D) Correlation of CK19 mRNA expression in tumor tissues and normal tissues.
3.5 Subgroup analysis of diagnostic accuracies of OSNA in breast cancer
Furthermore, we performed subgroup analysis in breast cancer, which included the largest number of studies (n=29). Our results showed a slightly difference of AUC among different subgroups, such as ethnic origins, patient number, LN number, center number, reference method and section interval (Table 3). Interestingly, we found the different OSNA cut-off values (250 or 5000 copies/μl) affected detecting micro-metastasis in breast cancer. In per-node micro-metastasis analysis, the sensitivity and specificity of 250 copies/μl were 0.69 and 0.95, respectively, whereas the sensitivity and specificity of 5000 copies/μl were 0.26 and 0.97, respectively. The diagnostic value of OSNA using 250 copies/μl to predict SLN micro metastasis (DOR, 44; AUC, 0.93) was substantially higher than using 5000 copies/μl (DOR, 16; AUC, 0.41). At the same time, there was no significantly difference between two cutoff values in diagnosing macro-metastasis. Thus, measurements of SLN micro-metastasis using 250 copies/μl may have a higher diagnostic accuracy.
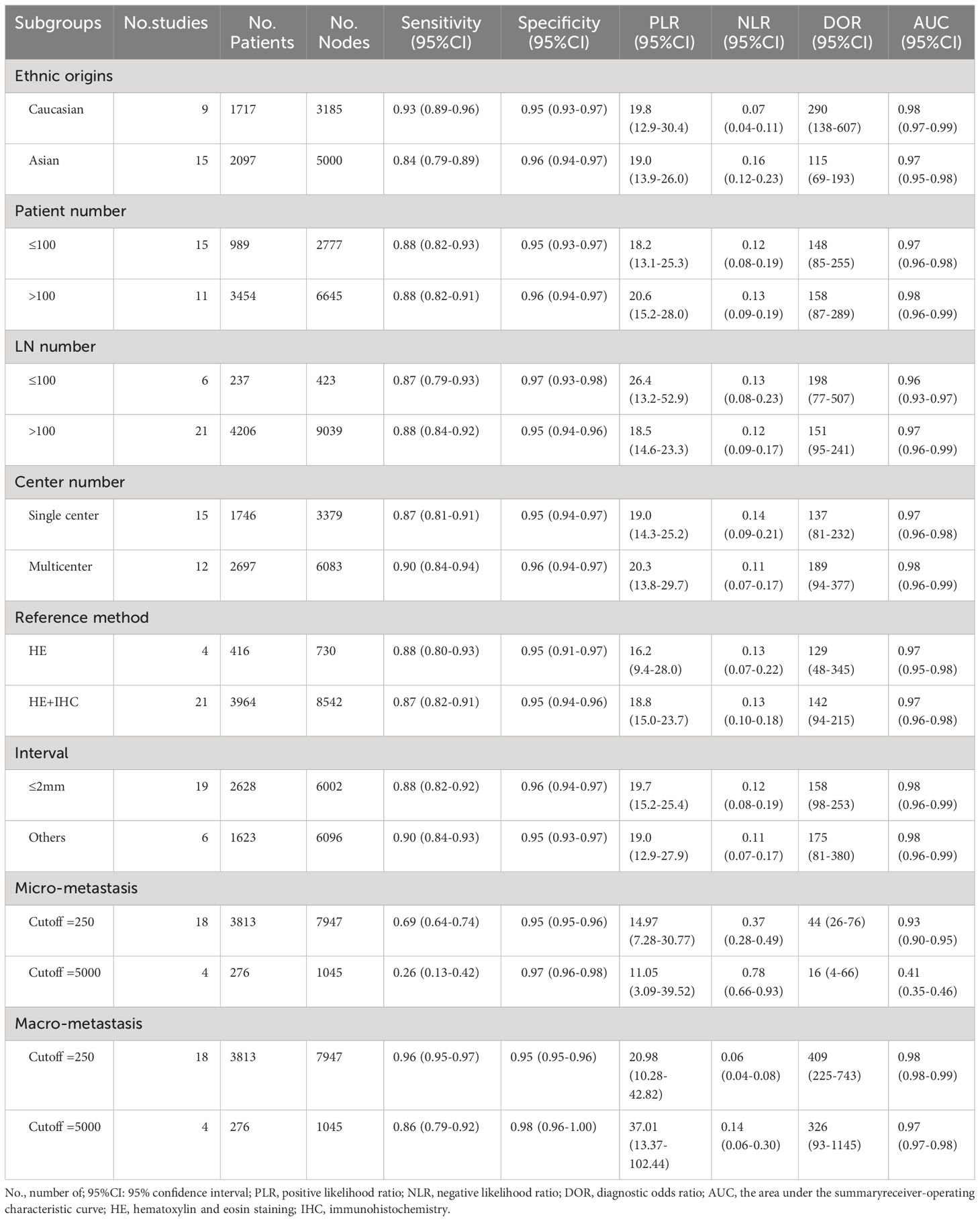
Table 3 Subgroup analysis of diagnostic accuracies of OSNA in breast cancer based on a per-node analysis.
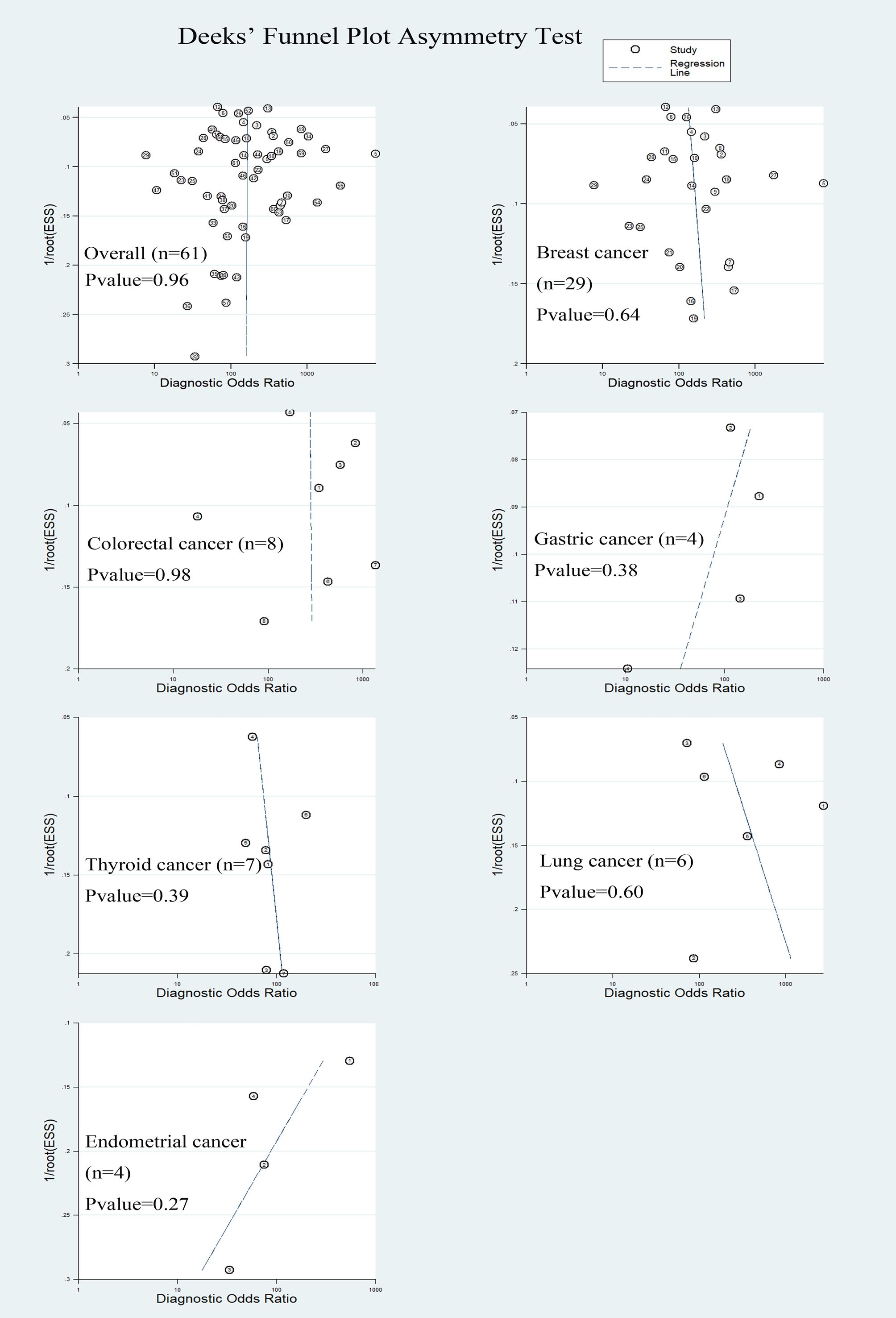
3.6 Heterogeneity and risk of publication bias
The Deek’s funnel chart was used to analyze any potential publication bias. The results are shown in Figure 6. All funnel charts were symmetric, and P > 0.05, suggesting that no significant publication bias existed in this study.
4 Discussion
The formation of distant lymph node metastasis is the deadliest step of cancer progression and influence surgical decision making, which is an essential prognostic indicator in many different types of cancer (72). Therefore, LNM affects the prognosis and therapy of cancer patients in order to provide accurate assessment and effective treatment strategies (73). Classic intraoperative SLN detection methods, including FS and TIC, are limited due to a rather low sensitivity and no unified standardization. A meta-analysis based on intraoperative FS for SLNs suggested a pooled sensitivity of 0.73 (74). The poor sensitivity is related to the limited amount of tissue detected by FS and to the destructive, freezing, and compression artifacts of samples (75). To date, the diagnostic performance of OSNA for SLN metastases has been shown in a few studies. OSNA is shown to have a higher diagnostic performance than classic method. In addition, the turnaround time of the OSNA assay was less than 40 min for detecting one node, supporting OSNA to become a routine intraoperative SLN detection method. Nowadays, OSNA has been an established technique in breast cancer. In light of the high diagnostic performance in breast cancer, its accuracy for detecting SLN metastases in other CK19 positive cancers is not clear. To fill this void in knowledge, we attempted to conduct a meta-analysis to quantify the diagnostic accuracy of OSNA for the detection of SLN metastases in cancer patients carrying positive CK19 expression.
In this study, we firstly identified CK19 positive cancer by combining RNA sequencing and protein expression database. A total of 18 cancer types were shown to exhibit higher CK19 mRNA expression (fold-change >2), and CK19 IHC staining method confirmed 10 cancer types (mean IHC score >2). When performing literature review, we found OSNA method was reported to be used in seven cancer types, including breast cancer, cervical cancer, colorectal cancer, endometrial cancer, gastric cancer, lung cancer and thyroid cancer. However, three kinds of cancer, bladder cancer, ovarian cancer and pancreatic cancer, which also showed high CK19 expression and high capacities of LN metastasis, have no report of using OSNA in detecting LN metastasis. Therefore, our study suggested a pilot clinical study for evaluating the diagnostic accuracy of OSNA in above three cancers.
According to this meta-analysis, which included 61 related studies, we concluded that the pooled sensitivity, specificity and AUC of OSNA were 0.87, 0.95 and 0.97, respectively. By analyzing each subtype, we found the AUC of OSNA was higher than 0.95 in nearly all CK19 positive cancers. The high estimates suggested that OSNA appears to be a useful tool in assessing SLN metastasis for CK19 positive cancer patients. Next, we investigated that whether high CK19 expression would increase OSNA diagnostic accuracy. Interestingly, our data demonstrated that higher tumor CK19 mRNA expression increased specificity but decreased sensitivity of OSNA. We found that the CK19 expression in tumor tissues was strongly associated with its expression in normal tissues, indicating that a high background CK19 expression existed in CK19 positive cancers. Above findings might affect the diagnostic estimates of OSNA.
We selected breast cancer to perform subgroup analysis, mainly due to its largest study number. There is no significant difference between different subgroups, indicating that the OSNA was quite a stable method. We found that the cut-off values of OSNA affected its diagnostic accuracy to predict SLN micro-metastasis. Both sensitivity and AUC of OSNA using 5000 copies/μl were substantially lower than 250copies/μl. In clinical practice, the cut-off value of 250 copies/μl has been widely used for diagnosing micro-metastasis in breast cancer patients. However, we noticed that a few cancers, such as cervical cancer, had significantly higher CK19 expression than breast cancer. How to select appropriate cut-off values warrant further studies.
Given the potential benefits derived from the OSNA assay, OSNA is still proposed for use as a diagnostic tool in the intraoperative clinical setting. A reliable intraoperative SLN assessment is valuable for the optimal surgical treatment of breast cancer. The intraoperative analysis of the SLN is economically advantageous, and the implicit savings, resulting from the reduced number of hospitalizations and the avoidance of additional surgeries, compensates for the cost of the intraoperative OSNA assay (76). However, there are some limitations in our work. Firstly, we did not include studies with cancer patients receiving neoadjuvant chemotherapy and radiotherapy because of the insufficient data on this topic. Moreover, due to the lack of genotype information, we did not analyze the diagnostic values of OSNA in different cancer genotypes. Secondly, this meta-analysis mainly focused on the diagnostic value of OSNA, and its prognostic value could be evaluated in future studies (77).
In summary, this meta-analysis provides evidence that OSNA can predict the occurrence of SLN metastasis in patients with CK19 positive tumors. The diagnostic value of OSNA to predict SLN macro-metastasis was substantially higher than that of micro-metastasis. However, except for breast cancer, the number of studies involved in this analysis is limited, which may lead to insufficient evaluation. Therefore, these results need to be treated with caution, and should be further validated by more high-quality and multi-center clinical trials.
Data availability statement
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding authors.
Author contributions
KL: Writing – original draft, Writing – review & editing, Investigation. MM: Writing – original draft, Writing – review & editing, Conceptualization. WZ: Investigation, Resources, Writing – original draft. JL: Data curation, Project administration, Writing – original draft. YW: Conceptualization, Investigation, Supervision, Writing – review & editing. CZ: Conceptualization, Formal analysis, Investigation, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the Natural Science Foundation of Shandong (No. 2015ZRA15027).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Leong SP, Pissas A, Scarato M, Gallon F, Pissas MH, Amore M, et al. The lymphatic system and sentinel lymph nodes: conduit for cancer metastasis. Clin Exp Metastasis. (2021) 39:139–57. doi: 10.1007/s10585-021-10123-w
2. Dieterich LC, Tacconi C, Ducoli L, Detmar M. Lymphatic vessels in cancer. Physiol Rev. (2022) 102:1837–79. doi: 10.1152/physrev.00039.2021
3. Farnsworth RH, Achen MG, Stacker SA. The evolving role of lymphatics in cancer metastasis. Curr Opin Immunol. (2018) 53:64–73. doi: 10.1016/j.coi.2018.04.008
4. Uno Y, Akiyama N, Yuzawa S, Kitada M, Takei H. The value and practical utility of intraoperative touch imprint cytology of sentinel lymph node(s) in patients with breast cancer: A retrospective cytology-histology correlation study. CytoJournal. (2020) 17:11. doi: 10.25259/Cytojournal_80_2019
5. Cserni G. Discrepancies in current practice of pathological evaluation of sentinel lymph nodes in breast cancer. Results of a questionnaire based survey by the European Working Group for Breast Screening Pathology. J Clin Pathol. (2004) 57:695–701. doi: 10.1136/jcp.2003.013599
6. Ahmed S, Ahmad M. Comparison of diagnostic accuracy of touch imprint cytology and frozen section techniques in detecting breast Malignancies. JPMA J Pakistan Med Assoc. (2016) 66:292–5.
7. Mehrpouya M, Pourhashem Z, Yardehnavi N, Oladnabi M. Evaluation of cytokeratin 19 as a prognostic tumoral and metastatic marker with focus on improved detection methods. J Cell Physiol. (2017) 234:21425–35. doi: 10.1002/jcp.28768
8. Alix-Panabières C, Vendrell J-P, Slijper M, Pellé O, Barbotte E, Mercier G, et al. Full-length cytokeratin-19 is released by human tumor cells: a potential role in metastatic progression of breast cancer. Breast Cancer Res. (2009) 11:R39. doi: 10.1186/bcr2326
9. Tsujimoto M, Nakabayashi K, Yoshidome K, Kaneko T, Iwase T, Akiyama F, et al. One-step nucleic acid amplification for intraoperative detection of lymph node metastasis in breast cancer patients. Clin Cancer Res. (2007) 13:4807–16. doi: 10.1158/1078-0432.ccr-06-2512
10. Namba K, Suzawa K, Shien K, Miura A, Takahashi Y, Miyauchi S, et al. One-step nucleic acid amplification for intraoperative diagnosis of lymph node metastasis in lung cancer patients: a single-center prospective study. Sci Rep. (2022) 12:7297. doi: 10.1038/s41598-022-11064-4
11. Shi F, Liang Z, Zhang Q, Wang C, Liu X. The performance of one-step nucleic acid amplification assay for intraoperative detection of sentinel lymph node macro metastasis in breast cancer: An updated meta-analysis. Breast. (2018) 39:39–45. doi: 10.1016/j.breast.2018.03.005
12. Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. (2005) 58:882–93. doi: 10.1016/j.jclinepi.2005.01.016
13. Visser M, Jiwa M, Horstman A, Brink AATP, Pol RP, van Diest P, et al. Intra-operative rapid diagnostic method based on CK19 mRNA expression for the detection of lymph node metastases in breast cancer. Int J Cancer. (2008) 122:2562–7. doi: 10.1002/ijc.23451
14. Schem C, Maass N, Bauerschlag DO, Carstensen MH, Löning T, Roder C, et al. One-step nucleic acid amplification—a molecular method for the detection of lymph node metastases in breast cancer patients; results of the German study group. Virchows Archiv. (2009) 454:203–10. doi: 10.1007/s00428-008-0703-9
15. Tamaki Y, Akiyama F, Iwase T, Kaneko T, Tsuda H, Sato K, et al. Molecular detection of lymph node metastases in breast cancer patients: results of a multicenter trial using the one-step nucleic acid amplification assay. Clin Cancer Res. (2009) 15:2879–84. doi: 10.1158/1078-0432.ccr-08-1881
16. Croner RS, Schellerer V, Demund H, Schildberg C, Papadopulos T, Naschberger E, et al. One Step Nucleic Acid Amplification (OSNA) - a new method for lymph node staging in colorectal carcinomas. J Trans Med. (2010) 8:83. doi: 10.1186/1479-5876-8-83
17. Bernet L, Cano R, Martinez M, Dueñas B, Matias-Guiu X, Morell L, et al. Diagnosis of the sentinel lymph node in breast cancer: a reproducible molecular method: a multicentric Spanish study. Histopathology. (2011) 58:863–9. doi: 10.1111/j.1365-2559.2011.03836.x
18. Khaddage A, Berremila SA, Forest F, Clemenson A, Bouteille C, Seffert P, et al. Implementation of molecular intra-operative assessment of sentinel lymph node in breast cancer. Anticancer Res. (2011) 31:585–90.
19. Feldman S, Krishnamurthy S, Gillanders W, Gittleman M, Beitsch PD, Young PR, et al. A novel automated assay for the rapid identification of metastatic breast carcinoma in sentinel lymph nodes. Cancer. (2011) 117:2599–607. doi: 10.1002/cncr.25822
20. Snook KL, Layer GT, Jackson PA, de Vries CS, Shousha S, Sinnett HD, et al. Multicenter evaluation of intraoperative molecular analysis of sentinel lymph nodes in breast carcinoma. Br J Surg. (2011) 98:527–35. doi: 10.1002/bjs.7347
21. Sun X, Fu X, Han W, Zhao Y, Liu H, Sheng Z. Dedifferentiation of human terminally differentiating keratinocytes into their precursor cells induced by basic fibroblast growth factor. Biol Pharm Bull. (2011) 34:1037–45. doi: 10.1248/bpb.34.1037
22. Yaguchi Y, Sugasawa H, Tsujimoto H, Takata H, Nakabayashi K, Ichikura T, et al. One-step nucleic acid amplification (OSNA) for the application of sentinel node concept in gastric cancer. Ann Surg Oncol. (2011) 18:2289–96. doi: 10.1245/s10434-011-1591-9
23. Yamamoto H, Sekimoto M, Oya M, Yamamoto N, Konishi F, Sasaki J, et al. OSNA-based novel molecular testing for lymph node metastases in colorectal cancer patients: results from a multicenter clinical performance study in Japan. Ann Surg Oncol. (2011) 18:1891–8. doi: 10.1245/s10434-010-1539-5
24. Goda H, Nakashiro K-i, Oka R, Tanaka H, Wakisaka H, Hato N, et al. One-step nucleic acid amplification for detecting lymph node metastasis of head and neck squamous cell carcinoma. Oral Oncol. (2012) 48:958–63. doi: 10.1016/j.oraloncology.2012.03.026
25. Inoue M, Hiyama K, Nakabayashi K, Morii E, Minami M, Sawabata N, et al. An accurate and rapid detection of lymph node metastasis in non-small cell lung cancer patients based on one-step nucleic acid amplification assay. Lung Cancer (Amsterdam Netherlands). (2012) 78:212–8. doi: 10.1016/j.lungcan.2012.08.018
26. Le Frère-Belda MA, Bats AS, Gillaizeau F, Poulet B, Clough KB, Nos C, et al. Diagnostic performance of one-step nucleic acid amplification for intraoperative sentinel node metastasis detection in breast cancer patients. Int J Cancer. (2012) 130:2377–86. doi: 10.1002/ijc.26291
27. Wang Ys, Ou-yang T, Wu J, Yh L, Xc C, Sun X, et al. Comparative study of one-step nucleic acid amplification assay, frozen section, and touch imprint cytology for intraoperative assessment of breast sentinel lymph node in Chinese patients. Cancer Sci. (2012) 103:1989–93. doi: 10.1111/cas.12001
28. Güller U, Zettl A, Worni M, Langer I, Cabalzar-Wondberg D, Viehl CT, et al. Molecular investigation of lymph nodes in colon cancer patients using one-step nucleic acid amplification (OSNA). Cancer. (2012) 118:6039–45. doi: 10.1002/cncr.27667
29. Buglioni S, Di Filippo F, Terrenato I, Casini B, Gallo E, Marandino F, et al. Quantitative molecular analysis of sentinel lymph node may be predictive of axillary node status in breast cancer classified by molecular subtypes. PloS One. (2013) 8:e58823. doi: 10.1371/journal.pone.0058823
30. Okamoto S, Niikura H, Nakabayashi K, Hiyama K, Matoda M, Takeshima N, et al. Detection of sentinel lymph node metastases in cervical cancer: Assessment of KRT19 mRNA in the one-step nucleic acid amplification (OSNA) method. Gynecol Oncol. (2013) 130:530–6. doi: 10.1016/j.ygyno.2013.06.027
31. Osako T, Iwase T, Kimura K, Horii R, Akiyama F. Sentinel node tumor burden quantified based on cytokeratin 19 mRNA copy number predicts non-sentinel node metastases in breast cancer: Molecular whole-node analysis of all removed nodes. Eur J Cancer. (2013) 49:1187–95. doi: 10.1016/j.ejca.2012.11.022
32. Sagara Y, Ohi Y, Matsukata A, Yotsumoto D, Baba S, Tamada S, et al. Clinical application of the one-step nucleic acid amplification method to detect sentinel lymph node metastasis in breast cancer. Breast Cancer (Tokyo Japan). (2013) 20:181–6. doi: 10.1007/s12282-011-0324-z
33. Banerjee SM, Michalopoulos NV, Williams NR, Davidson T, El Sheikh S, McDermott N, et al. Detailed evaluation of one step nucleic acid (OSNA) molecular assay for intra-operative diagnosis of sentinel lymph node metastasis and prediction of non-sentinel nodal involvement: Experience from a London Teaching Hospital. Breast. (2014) 23:378–84. doi: 10.1016/j.breast.2014.02.001
34. Bettington M, Lakhani SR, Ung OA. Is the one-step nucleic acid amplification assay better for intra-operative assessment of breast sentinel nodes? ANZ J Surg. (2014) 84:725–9. doi: 10.1111/ans.12497
35. Chaudhry A, Williams S, Cook J, Jenkins M, Sohail M, Calder C, et al. The real-time intra-operative evaluation of sentinel lymph nodes in breast cancer patients using One Step Nucleic Acid Amplification (OSNA) and implications for clinical decision-making. Eur J Surg Oncol (EJSO). (2014) 40:150–7. doi: 10.1016/j.ejso.2013.12.007
36. Hayama M, Chida M, Karube Y, Tamura M, Kobayashi S, Oyaizu T, et al. One-step nucleic acid amplification for detection of lymph node metastasis in lung cancer. Ann Thorac Cardiovasc Surg. (2014) 20:181–4. doi: 10.5761/atcs.oa.12.02224
37. Jara-Lazaro AR, Hussain IHM, Thike AA, Wong CY, Ho GH, Yong WS, et al. Assessment of suitability of the one step nucleic acid amplification (OSNA) assay as an intraoperative procedure for detection of metastasis in sentinel lymph nodes of breast cancer. J Clin Pathol. (2014) 67:1032–7. doi: 10.1136/jclinpath-2014-202361
38. Kaczka K, Fendler W, Borowiec M, Młynarski W, Pomorski L. Badanie metodą jednostopniowej amplifikacji kwasu nukleinowego węzłów chłonnych w raku brodawkowatym tarczycy — porównanie z badaniem histopatologicznym i badaniem PCR w czasie rzeczywistym. Endokrynol Polska. (2015) 65:422–30. doi: 10.5603/ep.2014.0059
39. Kumagai K, Yamamoto N, Miyashiro I, Tomita Y, Katai H, Kushima R, et al. Multicenter study evaluating the clinical performance of the OSNA assay for the molecular detection of lymph node metastases in gastric cancer patients. Gastric Cancer. (2013) 17:273–80. doi: 10.1007/s10120-013-0271-9
40. Pathmanathan N, Renthawa J, French JR, Edstrom-Elder E, Hall G, Mahajan H, et al. Intraoperative sentinel lymph node assessment in breast cancer: a comparison of rapid diagnostic method based on CK19 mRNA expression and imprint cytology. ANZ J Surg. (2014) 84:730–4. doi: 10.1111/ans.12668
41. Terada M, Niikura N, Tsuda B, Masuda S, Kumaki N, Tang X, et al. Comparative study of the one-step nucleic acid amplification assay and conventional histological examination for the detection of breast cancer sentinel lymph node metastases. Tokai J Exp Clin Med. (2014) 39:122–7.
42. Vogelaar FJ, Reimers MS, van der Linden RLA, van der Linden JC, Smit VTHBM, Lips DJ, et al. The diagnostic value of one-step nucleic acid amplification (OSNA) for sentinel lymph nodes in colon cancer patients. Ann Surg Oncol. (2014) 21:3924–30. doi: 10.1245/s10434-014-3820-5
43. Wang Y-s, Liu Y-h, Ou-yang T, Yang X-h, Wu J, Su F-x, et al. GeneSearch™ BLN Assay could replace frozen section and touch imprint cytology for intra-operative assessment of breast sentinel lymph nodes. Breast Cancer (Tokyo Japan). (2014) 21:583–9. doi: 10.1007/s12282-012-0437-z
44. González O, Iglesias C, Zafon C, Castellví J, García-Burillo A, Temprana J, et al. Detection of thyroid papillary carcinoma lymph node metastases usingOne step nucleic acid amplification(OSNA): preliminary results. J Invest Surg. (2015) 28:153–9. doi: 10.3109/08941939.2014.990123
45. Hao X, Liu Y, Li X, Kang H, Qu X, He J, et al. An intra-operative RT-LAMP method allows rapid and reliable detection of sentinel lymph node metastasis in breast cancer patients. Virchows Archiv. (2014) 466:169–76. doi: 10.1007/s00428-014-1693-4
46. Li D, Xu X, Chen J, Chen J, Yang B, Yang W, et al. Utility of one-step nucleic acid amplification (OSNA) assay in detecting breast cancer metastases of sentinel lymph nodes in a Chinese population. Breast Cancer (Tokyo Japan). (2013) 22:135–40. doi: 10.1007/s12282-013-0461-7
47. Nagai T, Niikura H, Okamoto S, Nakabayashi K, Matoda M, Utsunomiya H, et al. A new diagnostic method for rapid detection of lymph node metastases using a one-step nucleic acid amplification (OSNA) assay in endometrial cancer. Ann Surg Oncol. (2015) 22:980–6. doi: 10.1245/s10434-014-4038-2
48. del Carmen S, Gatius S, Franch-Arcas G, Baena JA, Gonzalez O, Zafon C, et al. Concordance study between one-step nucleic acid amplification and morphologic techniques to detect lymph node metastasis in papillary carcinoma of the thyroid. Hum Pathol. (2016) 48:132–41. doi: 10.1016/j.humpath.2015.09.020
49. López-Ruiz ME, Diestro MD, Yébenes L, Berjón A, Díaz de la Noval B, Mendiola M, et al. One-step nucleic acid amplification (OSNA) for the detection of sentinel lymph node metastasis in endometrial cancer. Gynecol Oncol. (2016) 143:54–9. doi: 10.1016/j.ygyno.2016.07.106
50. Nakagawa K, Asamura H, Tsuta K, Nagai K, Yamada E, Ishii G, et al. The novel one-step nucleic acid amplification (OSNA) assay for the diagnosis of lymph node metastasis in patients with non-small cell lung cancer (NSCLC): Results of a multicenter prospective study. Lung Cancer (Amsterdam Netherlands). (2016) 97:1–7. doi: 10.1016/j.lungcan.2016.03.015
51. Takamoto K, Shimazu K, Naoi Y, Shimomura A, Shimoda M, Kagara N, et al. One-step nucleic acid amplification assay for detection of axillary lymph node metastases in breast cancer patients treated with neoadjuvant chemotherapy. Ann Surg Oncol. (2016) 23:78–86. doi: 10.1245/s10434-015-4693-y
52. Yamamoto H, Tomita N, Inomata M, Furuhata T, Miyake Y, Noura S, et al. OSNA-assisted molecular staging in colorectal cancer: A prospective multicenter trial in Japan. Ann Surg Oncol. (2016) 23:391–6. doi: 10.1245/s10434-015-4880-x
53. Colling R, Yeung T, Hompes R, Kraus R, Cahill R, Mortensen N, et al. OSNA testing for lymph node staging in colorectal cancer. J Clin Pathol. (2017) 70:638–9. doi: 10.1136/jclinpath-2016-204299
54. Kaczka KA, Pomorski L. One-step nucleic acid amplification analysis of sentinel lymph nodes in papillary thyroid cancer patients. Arch Med Sci. (2017) 6:1416–26. doi: 10.5114/aoms.2017.65466
55. Fanfani F, Monterossi G, Ghizzoni V, Rossi ED, Dinoi G, et al. One-Step Nucleic Acid Amplification (OSNA): A fast molecular test based on CK19 mRNA concentration for assessment of lymph-nodes metastases in early stage endometrial cancer. PloS One. (2018) 13:e0195877. doi: 10.1371/journal.pone.0195877
56. Shigematsu H, Ozaki S, Yasui D, Zaitsu J, Taniyama D, Saitou A, et al. Comparison of CK-IHC assay on serial frozen sections, the OSNA assay, and in combination for intraoperative evaluation of SLN metastases in breast cancer. Breast Cancer (Tokyo Japan). (2017) 25:191–7. doi: 10.1007/s12282-017-0811-y
57. Yeung TM, Wang LM, Colling R, Kraus R, Cahill R, Hompes R, et al. Intraoperative identification and analysis of lymph nodes at laparoscopic colorectal cancer surgery using fluorescence imaging combined with rapid OSNA pathological assessment. Surg Endoscopy. (2017) 32:1073–6. doi: 10.1007/s00464-017-5644-4
58. Esposito F, Noviello A, Moles N, Coppola Bottazzi E, Baiamonte M, Macaione I, et al. Sentinel lymph node analysis in colorectal cancer patients using one-step nucleic acid amplification in combination with fluorescence and indocyanine green. Ann Coloproctol. (2019) 35:174–80. doi: 10.3393/ac.2018.07.21.1
59. Iglesias Felip C, Zafon Llopis C, Temprana-Salvador J, García-Burillo A, Serres Créixams X, Caubet Busquet E, et al. One-step nucleic acid amplification for intraoperative analysis of sentinel lymph node in papillary thyroid carcinoma. Eur J Endocrinol. (2019) 180:21–9. doi: 10.1530/eje-18-0624
60. Kosťun J, Pešta M, Sláma J, Slunéčko R, Vlasák P, Bouda J, et al. One-step nucleic acid amplification vs ultrastaging in the detection of sentinel lymph node metastasis in endometrial cancer patients. J Surg Oncol. (2019) 119:361–9. doi: 10.1002/jso.25322
61. Medas F, Coni P, Podda F, Salaris C, Cappellacci F, Faa G, et al. Evaluation of accuracy of one-step nucleic acid amplification (OSNA) in diagnosis of lymph node metastases of papillary thyroid carcinoma. Diagnostic study. Ann Med Surg. (2019) 46:17–22. doi: 10.1016/j.amsu.2019.08.006
62. Escalante Pérez M, Hermida Romero MT, Otero Alén B, Álvarez Martínez M, Fernández Prado R, de la Torre Bravos M, et al. Detection of lymph node metastasis in lung cancer patients using a one-step nucleic acid amplification assay: a single-center prospective study. J Trans Med. (2019) 17:233. doi: 10.1186/s12967-019-1974-4
63. Shimazu K, Tanei T, Tamaki Y, Saeki T, Osaki A, Hasebe T, et al. Performance of a new system using a one-step nucleic acid amplification assay for detecting lymph node metastases in breast cancer. Med Oncol. (2019) 36:54. doi: 10.1007/s12032-019-1277-x
64. Bizzarri N, Pedone Anchora L, Zannoni GF, Santoro A, Valente M, Inzani F, et al. Role of one-step nucleic acid amplification (OSNA) to detect sentinel lymph node low-volume metastasis in early-stage cervical cancer. Int J Gynecol Cancer. (2020) 30:364–71. doi: 10.1136/ijgc-2019-000939
65. Shimada A, Takeuchi H, Nishi T, Mayanagi S, Fukuda K, Suda K, et al. Utility of the one-step nucleic acid amplification assay in sentinel node mapping for early gastric cancer patients. Gastric Cancer. (2020) 23:418–25. doi: 10.1007/s10120-019-01016-9
66. Togami S, Tanimoto A, Yanazume S, Tokunaga H, Nagai T, Watanabe M, et al. Evaluation of the one-step nucleic acid amplification assay for detecting lymph node metastasis in patients with cervical and endometrial cancer: A multicenter prospective study. Gynecol Oncol. (2023) 170:70–6. doi: 10.1016/j.ygyno.2022.12.016
67. Miyoshi N, Ose N, Takeuchi Y, Sakamaki Y, Kadota Y, Urasaki K, et al. Detection of lymph node metastasis in non-small cell lung cancer using the new system of one-step nucleic acid amplification assay. PloS One. (2022) 17:e0265603. doi: 10.1371/journal.pone.0265603
68. Inua B, Fung V, Al−Shurbasi N, Howells S, Hatsiopoulou O, Somarajan P, et al. Sentinel lymph node biopsy with one−step nucleic acid assay relegates the need for preoperative ultrasound−guided biopsy staging of the axilla in patients with early stage breast cancer. Mol Clin Oncol. (2021) 14:51. doi: 10.3892/mco.2021.2213
69. Pina H, Salleron J, Gilson P, Husson M, Rouyer M, Leroux A, et al. Intraoperative prediction of non−sentinel lymph node metastases in breast cancer using cytokeratin 19 mRNA copy number: A retrospective analysis. Mol Clin Oncol. (2022) 16:58. doi: 10.3892/mco.2022.2491
70. Gęca K, Rawicz-Pruszyński K, Mielko J, Mlak R, Sędłak K, Polkowski WP. Rapid detection of free cancer cells in intraoperative peritoneal lavage using one-step nucleic acid amplification (OSNA) in gastric cancer patients. Cells. (2020) 9:2168. doi: 10.3390/cells9102168
71. Kaczka K, Fendler W, Borowiec M, Młynarski W, Pomorski L. First one-step nucleic acid amplification testing in papillary thyroid cancer lymph nodes - a comparison with histopathology and real-time PCR. Endokrynol Polska. (2014) 65:422–30. doi: 10.5603/ep.2014.0059
72. Urrutia G, Laurito S, Marzese DM, Gago F, Orozco J, Tello O, et al. Epigenetic variations in breast cancer progression to lymph node metastasis. Clin Exp Metastasis. (2015) 32:99–110. doi: 10.1007/s10585-015-9695-4
73. Ji H, Hu C, Yang X, Liu Y, Ji G, Ge S, et al. Lymph node metastasis in cancer progression: molecular mechanisms, clinical significance and therapeutic interventions. Signal Transduction Targeted Ther. (2023) 8:367. doi: 10.1038/s41392-023-01576-4
74. Liu LC, Lang JE, Lu Y, Roe D, Hwang SE, Ewing CA, et al. Intraoperative frozen section analysis of sentinel lymph nodes in breast cancer patients. Cancer. (2010) 117:250–8. doi: 10.1002/cncr.25606
75. Olson TP, Harter J, Muñoz A, Mahvi DM, Breslin TM. Frozen section analysis for intraoperative margin assessment during breast-conserving surgery results in low rates of re-excision and local recurrence. Ann Surg Oncol. (2007) 14:2953–60. doi: 10.1245/s10434-007-9437-1
76. Cutress RI, McDowell A, Gabriel FG, Gill J, Jeffrey MJ, Agrawal A, et al. Observational and cost analysis of the implementation of breast cancer sentinel node intraoperative molecular diagnosis. J Clin Pathol. (2010) 63:522–9. doi: 10.1136/jcp.2009.072942
Keywords: OSNA, CK19, sentinel lymph nodes, metastasis, meta-analysis
Citation: Li K, Meng M, Zhang W, Li J, Wang Y and Zhou C (2024) Diagnostic value of one-step nucleic acid amplification for sentinel lymph node metastasis in cytokeratin 19-positive tumors: evidence from bioinformatics and meta-analysis. Front. Oncol. 14:1370709. doi: 10.3389/fonc.2024.1370709
Received: 16 January 2024; Accepted: 19 March 2024;
Published: 08 April 2024.
Edited by:
Daniel Matias Regiart, University of São Paulo, BrazilReviewed by:
Akira Sugawara, Tohoku University, JapanLuigi Pedone Anchora, Agostino Gemelli University Polyclinic (IRCCS), Italy
Copyright © 2024 Li, Meng, Zhang, Li, Wang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changhui Zhou, emhvdWNoYW5naHVpMDA4QDE2My5jb20=; Yiting Wang, d3l0MTM2ODYzNTAwMDVAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Ke Li1†
Ke Li1† Min Meng
Min Meng