
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 24 August 2022
Sec. Cancer Metabolism
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.938502
This article is part of the Research Topic Reviews in Cancer Metabolism View all 8 articles
Cholesterol metabolism is often dysregulated in cancer. Squalene monooxygenase (SQLE) is the second rate-limiting enzyme involved in cholesterol synthesis. Since the discovery of SQLE dysregulation in cancer, compelling evidence has indicated that SQLE plays a vital role in cancer initiation and progression and is a promising therapeutic target for cancer treatment. In this review, we provide an overview of the role and regulation of SQLE in cancer and summarize the updates of antitumor therapy targeting SQLE.
1. Squalene monooxygenase (SQLE) as the second rate-limiting enzyme involved in cholesterol synthesis, is dysregulated in tumors.
2. The dysregulation of SQLE is associated with poor prognosis and resistance to some therapies (radiation, hormone deprivation therapy).
3. SQLE promotes tumor growth, while inhibition SQLE can restrain tumor growth across various types of cancer. Targeting SQLE may be a potential direction for novel anti-cancer therapy.
Squalene monooxygenase (SQLE) catalyzes the oxidation of squalene to (S)-2,3-epoxysqualene (1). For quite a long time, it has been investigated as an anti-fungal target because the epoxysqualene derivative lanosterol is a component of the fungal membrane (2, 3). In human cells, the gene encoding SQLE is located at chromosome region 8q 24.1 (4). SQLE is a direct target of sterol regulatory element binding protein-2 (SREBP2), a transcription factor that regulates genes involved in cholesterol biosynthesis and homeostasis in a cholesterol-dependent manner; SQLE protein also contains a cholesterol sensing domain that can regulate proteasomal degradation of SQLE (5). Therefore, like 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), SQLE activity is also precisely regulated by intracellular cholesterol level in the form of feedback, which makes it a second rate-limiting step in cholesterol synthesis (2, 3).
Numerous studies have revealed that the deregulation of SQLE results in cholesterol metabolism disorder and is associated with many diseases, including Alzheimer’s disease, hypercholesterolemia, stroke, and cancer (6). Tumor cells exhibit a high requirement for energy and materials to meet the demands for rapid tumor growth. SQLE is crucial for cancer cells to meet the requirement for cholesterol. A high abundance of SQLE has been identified in multiple cancer types and has emerged as a hot topic in the field of cancer treatment (7).
In this review, we discuss the regulation of SQLE, its roles and its clinical relevance in cancers. Cholesterol metabolism centered on SQLE is described in brief. Finally, the latest developments in antitumor therapies targeting SQLE are summarized.
Cholesterol plays manifold roles in normal cells and tumor regression. As an essential lipid component of the mammalian cell membrane, cholesterol is vital for cell survival and proliferation. By maintaining the stability of lipid rafts, cholesterol can coordinate the signal transduction of multiple membrane receptors. In addition, cholesterol can act as a signaling molecule to directly regulate the activation of signaling pathways in cancer cells. The cellular cholesterol level is determined by a complex network, mainly including cholesterol biosynthesis, uptake, export, and esterification (7).
Almost all mammalian cells can de novo synthesize cholesterol from acetyl-CoA to cholesterol through more than 20 enzymatic reactions, including the mevalonate (MVA) pathway, squalene biosynthesis and subsequent reactions (Figure 1). In the cholesterol synthesis pathway, two rate-limiting enzymes, HMGCR and SQLE, are essential players (8). SQLE is responsible for the first oxygenation step in cholesterol synthesis, which converts squalene to 2,3-epoxysqualene (9). SQLE can also divert 2,3-epoxysqualene into 2,3(S),22(S),23-dioxidosqualene, especially when the activity of lanosterol synthase, the enzyme that converts 2,3-epoxysqualene to lanosterol, is low (10). The end product of this shunt pathway, 24(S),25-epoxycholesterol, is the ligand for liver X receptors, which can upregulate ATP-binding cassette transporter A1 (ABCA1) levels to promote cholesterol efflux. Loss of 24(S),25-epoxycholesterol can induce acute cholesterol synthesis by increasing HMGCR expression processed by SREBP2 (11). Collectively, reactions catalyzed by SQLE are crucial for cholesterol metabolism.
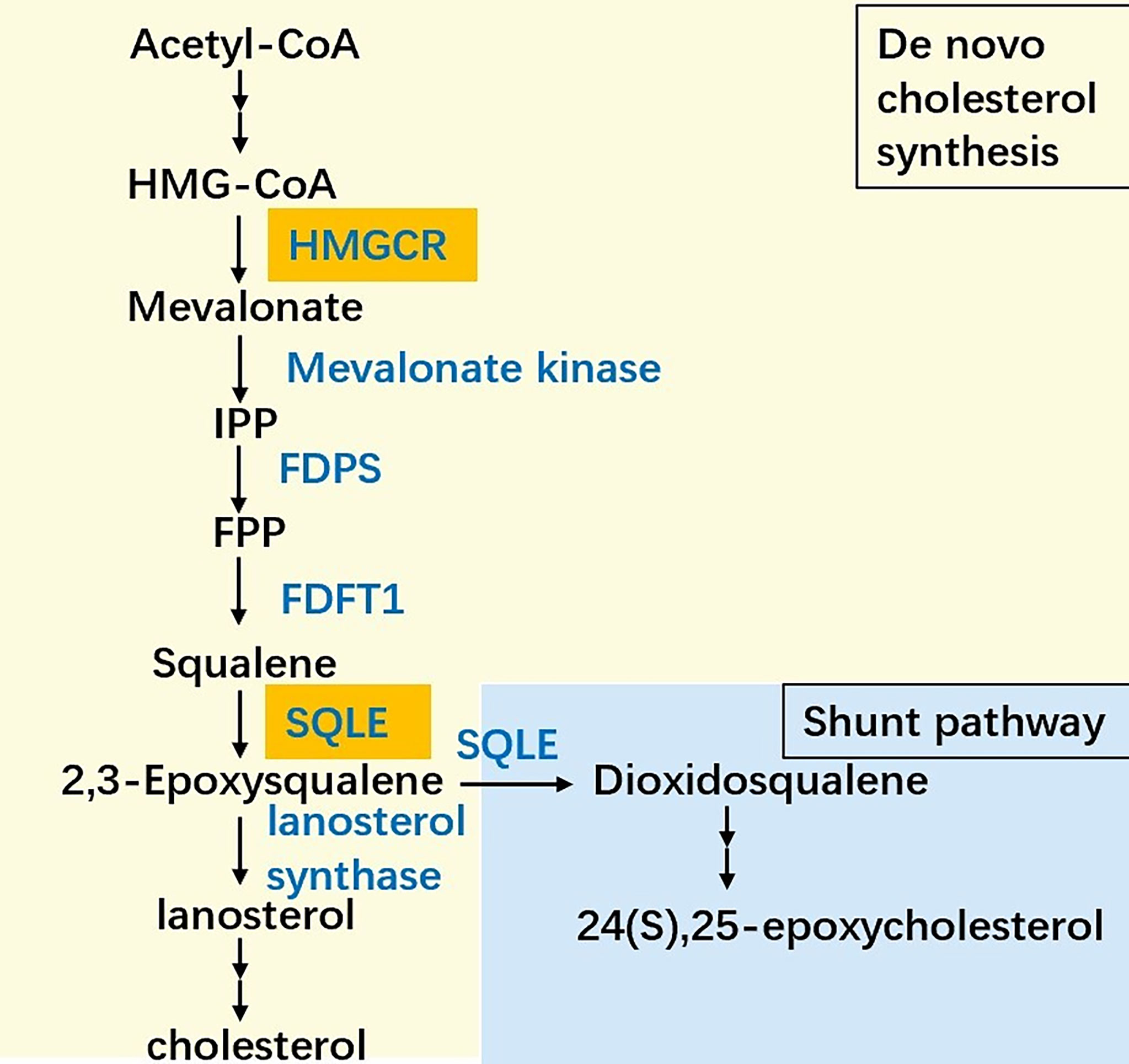
Figure 1 The simplified scheme of cholesterol biosynthesis. The biosynthesis pathway converts acetyl-CoA into cholesterol more than 20 enzymatic reactions, among which HMG-CoA reductase (HMGCR) and squalene epoxidase (SQLE) are the two key speed-limiting enzymes. Besides, SQLE can divert 2,3-epoxysqualene into dioxidosqualene. The end product of the shunt pathway, 24(S),25-epoxycholesterol can regulate the cholesterol metabolism in turn. IPP: Isopentenyl-PP, FPP: Farnesyl-PP, FDPS: Farnesyl-diphosphate farnesyltransferase 1, FDFT1: Farnesyl-diphosphate farnesyltransferase 1.
Cancer is a complex disease involving the dysregulation of cell proliferation, energy metabolism, angiogenesis, and immune surveillance (12). Given the characteristics of tumors and the function of cholesterol, cholesterol metabolic reprogramming is a milestone in cancer development. In recent years, SQLE has garnered increasing attention for its association with cancer. Various cancers have been found to exhibit high levels of SQLE protein/mRNA or have SQLE copy number alterations (13).
As an oncogene for breast cancer, gene amplification and overexpression of SQLE have been reported in cancer tissues/ductal carcinoma in situ tissues compared with normal tissues (14–17). Race expression differences have been reported: the expression of SQLE in African-Americans is higher than that in Caucasians in luminal A breast tumors and basal-like breast cancers (18, 19). In prostate cancer, the expression of SQLE is upregulated during the progression of cancer (20, 21). In pancreatic cancer, SQLE is upregulated and sqle gains in tumor tissues. In colorectal cancer, SQLE is upregulated in tumor tissues compared with normal tissues, but SQLE in stages I, II, and III is higher than that in stage IV (22, 23). Similarly, SQLE is upregulated in nasopharyngeal cancer (13), head and neck squamous cell carcinoma (24), leukemia (25), hepatocellular cancer (26–28), and squamous lung cancer (29, 30).
SQLE, as the second rate-limiting enzyme of cholesterol synthesis, is regulated exquisitely by a complex network, including the transcription program, posttranscriptional program, and posttranslational program. Recent studies have reported novel mechanisms that result in the high expression of SQLE in cancer (Figure 2).
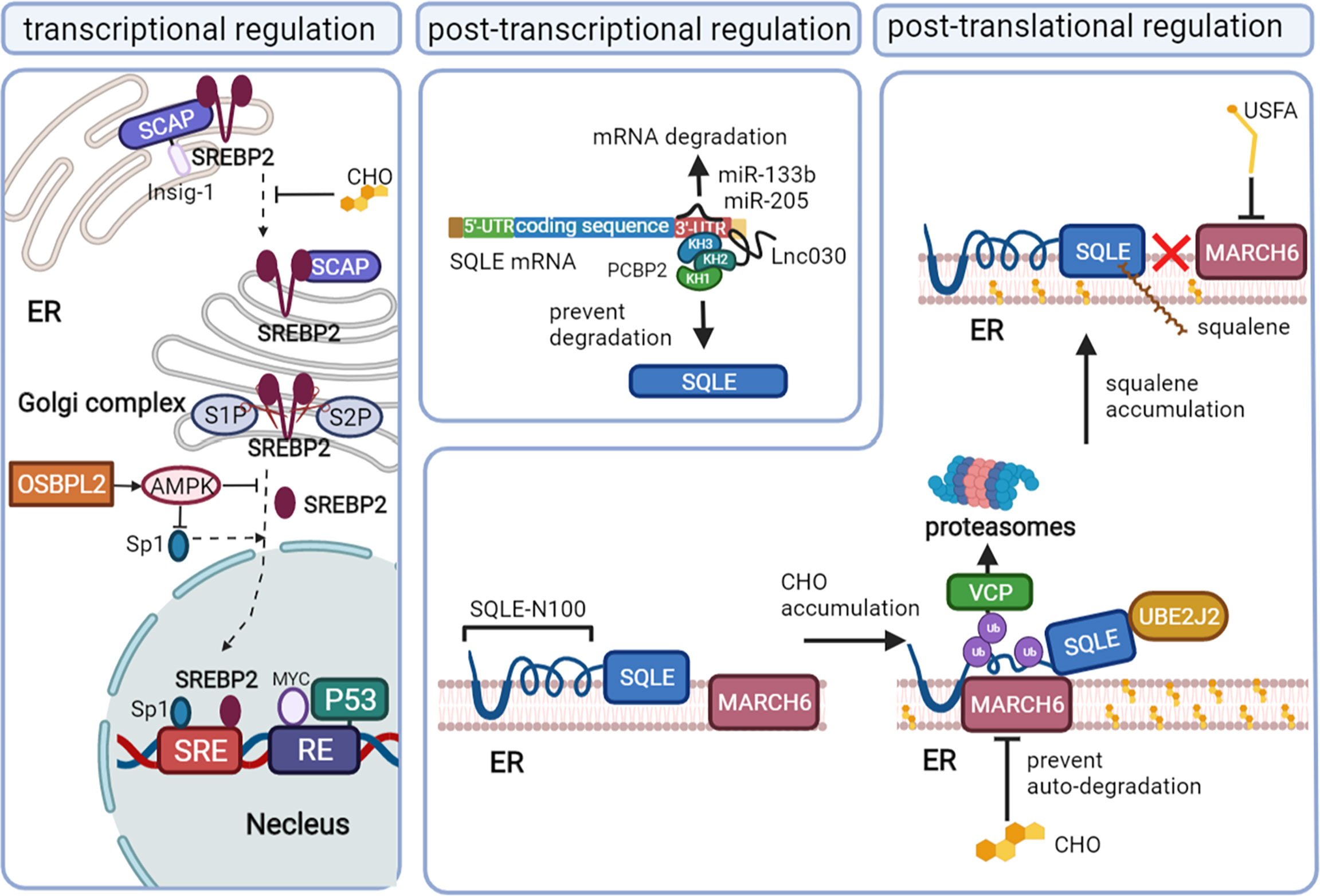
Figure 2 The mechanisms that regulate SQLE expression in cancer. Squalene monooxygenase (SQLE) can be regulated at the transcriptional level mainly via sterol regulatory element-binding protein 2 (SREBP2), the posttranscriptional level via microRNA (miRNA)/long noncoding RNA (lncRNA), and the posttranslational level via cholesterol feedback regulation. Mature SREBP2 promotes the transcription of SQLE by binding to the sterol-regulatory elements (SREs) of sqle (the gene encoding SQLE). When the cholesterol content in the endoplasmic reticulum membrane decreases, the SREBP2/SCAP complex dissociates from Insig-1 on the endoplasmic reticulum (ER) and is convoyed to the Golgi complex, where SREBP2 is cleaved by site-1 protease (S1P) and site-2 protease (S2P) and releases the mature form in the Golgi complex. Oxysterol binding protein like 2 (OSBPL2) deficiency can promote SREBP2 and specificity protein (Sp1) activation by inhibiting AMPK. P53 and MYC also regulate SQLE at the transcriptional level. MiR-133b and miR-205 promote SQLE mRNA degradation, while Lnc030 prevents SQLE mRNA degradation by forming a poly(rC) binding protein 2 (PCBP2)/Lnc030/SQLE 3’-UTR complex. Cholesterol (CHO) feedback regulation occurs via ubiquitin-proteasomal degradation of SQLE, which requires ubiquitin-conjugating enzyme E2 J2 (UBE2J2), membrane-associated RING finger 6 (MARCH6) and conformational change of SQLE N-100. Valosin-containing protein (VCP) extracts SQLE from the ER to proteasomes. Squalene and unsaturated fatty acids (USFAs) can disrupt the interaction between SQLE N-100 and MARCH6 by regulating SQLE and MARCH6, respectively, and suppress ubiquitin-proteasomal degradation.
The transcription factor SREBP2 directly regulates the mRNA levels of enzymes involved in sterol metabolism, including HMGCR, LDL, and SQLE, by binding the sterol-regulatory element (SRE) sequence in the promoters of target genes (31–33). The maturation of SREBP2 and its translocation depend on the intracellular cholesterol level. When the cholesterol level in the endoplasmic reticulum (ER) membrane increases, SREBP2 is retained at the ER due to its binding partner SREBP2 cleavage-activating protein (SCAP) interacting with an ER membrane anchor protein insulin-induced gene (Insig-1) protein. When the ER membrane cholesterol level decreases, SCAP undergoes a conformational change and dissociates from the Insig-1 protein and then convoys SREBP2 from the ER to the Golgi complex, where SREBP2 is proteolytically cleaved by site-1 protease (S1P) and site-2 protease (S2P) (34). The N-terminal domain of SREBP2 then enters the nucleus, binding with SRE to upregulate the mRNA level of SQLE (32).
In addition, specificity protein 1 (Sp1) and nuclear factor Y (NF-Y) may be required to coregulate SQLE levels with SREBP2, as evidenced by their binding sites on the SQLE gene (32, 33). The deficiency of oxysterol binding protein like 2 (OSBPL2) can also activate SREBP2 and Sp1, coregulating the expression of SQLE by inhibiting the adenosine 5’-monophosphate (AMP)-activated protein kinase (AMPK) pathway (35).
The gain of oncogenes and the loss of cancer suppressor genes are also involved in the transcriptional regulation of SQLE independent of SREBP2. MYC upregulates the transcription program of SQLE by binding the response element 1 (RE1) of the SQLE gene in cancer (36, 37). The cancer suppressor protein p53 suppresses the transcription of SQLE by directly binding to the RE of the SQLE gene in hepatocellular carcinoma cells. Loss of p53 can augment the expression of SQLE, even under normal or elevated ER membrane cholesterol levels (38).
Long noncoding RNA (lncRNA) and microRNA (miRNA) can interact with SQLE mRNA and then influence the stability of SQLE mRNA. In breast cancer, lnc030 is highly expressed to stabilize SQLE mRNA, especially in cancer stem cells. The stabilization function of lnc030 requires poly(rC) binding protein 2 (PCBP2), the 3’ untranslated region (3’UTR) of SQLE mRNA and lnc030 to form a complex. Lnc030 interacts with the K homology domains 2 of PCBP2, while the 3’UTR of SQLE mRNA binds to K homology domains 3 of PCBP2 (39).
MiR-133b is downregulated in esophageal squamous cell carcinoma and can bind directly to the 3’UTR of SQLE mRNA. In vitro, ectopic expression of miR-133b can decrease the mRNA and protein levels of SQLE (40). Another miRNA, miR-205, has also been reported to suppress the expression of SQLE by binding to the 3′-UTR of SQLE mRNA in progressive prostate cancer, where its expression is decreased (41).
Cholesterol, the end product of SQLE, plays an important role in regulating SQLE stability mainly via the cholesterol-membrane-associated RING finger 6 (MARCH6)-proteasomal degradation axis (42–44). The first 100 amino acids of SQLE (SQLE N-100), which can sense the cholesterol level in the cytoplasm (45, 46), can be attached to the ER membrane by a re-entrant loop. The Gln62–Leu73-sequence can form the amphipathic helix buried reversibly in the membrane, which is required for cholesterol-dependent degradation (47, 48). The accumulation of intracellular cholesterol thickens the anchoring of SQLE to the ER membrane, leading to the exposure of the hydrophobic core to the aqueous phase and triggering conformational changes in the re-entrant loop and amphipathic helix. Proteasomal degradation would be absent if these changes were disrupted (45).
The ubiquitin-proteasomal degradation system also requires the E2 ubiquitin-conjugating enzyme J2 (UBE2J2) and the E3 ubiquitin ligase MARCH6, unidentified deubiquitinases (48–51). MARCH6 ubiquitinates serine residues near the flanking amphipathic helix of SQLE deformed by excess cholesterol (51, 52). UBE2J2 is an important partner of MARCH6 in the cholesterol-stimulated degradation of SQLE. Valosin-containing protein (VCP), known to mediate the degradation of ubiquitinated endoplasmic reticulum-associated degradation (ERAD) substrates (53), is recruited downstream of ubiquitination, extracting SQLE from the ER and allowing proteasomal degradation (48). Excess cholesterol can stabilize MARCH6 by inhibiting its ubiquitination-proteasomal degradation, which in turn stimulates the degradation of SQLE (54).
Proteasomal degradation via MARCH6-VCP can also partially degrade SQLE from the N-terminus in a distinct ubiquitination pathway independent of cholesterol regulation, converting full-length SQLE to trunSQLE in various cell types. The enzymatic activity of trunSQLE is cholesterol-resistant, preventing the complete ablation of SQLE function under excess cholesterol levels (46). TrunSQLE might confer a supplemental route of cholesterol metabolism in pathophysiological contexts, especially in cancer.
Not only cholesterol but also squalene, the substrate of SQLE, can regulate SQLE expression by posttranslational modification. Squalene can directly bind to the SQLE N-100 domain, which is also squalene-sensitive, thereby inhibiting the interactions between MARCH6 and the SQLE N-100 domain. Thus, the accumulation of squalene can stabilize SQLE by preventing its proteasomal degradation (55–57). Unsaturated fatty acids (USFA) can also stabilize SQLE by blocking ubiquitination (58). The mechanism of USFA-mediated stabilization appears to occur through regulating MARCH6. In breast cancer, upregulation of the transmembrane microprotein cancer-associated small integral membrane open reading frame 1 (CASIMO1) increases the level of SQLE via interaction with SQLE proteins (59). The interaction of CASIMO1 and SQLE proteins may prevent the degradation of SQLE.
In summary, the cholesterol-dependent feedback regulation of SQLE via SREBP2 transcriptional regulation and ubiquitin-proteasomal degradation are the main mechanisms of SQLE regulation in various cells. In tumor tissues, the activation of SREBP2 leads to the high expression of SQLE. In addition, the activation of oncogenes, the loss of cancer suppressor genes, and the dysregulation of some lncRNAs, miRNAs and cancer-associated proteins also contribute to the upregulation of SQLE. The dysregulation of SQLE may be the result of multiple events in cancer, while a specific mechanism might prevail in a specific subset of tumor cells.
As mentioned above, cholesterol metabolic reprogramming is a hallmark in various cancer types and has been confirmed to promote tumor initiation and progression (7). As the key enzyme in the steps of cholesterol synthesis, the activity of SQLE determines the abundance of cholesterol and cholesterol derivatives in cancer (13, 39). SQLE can promote tumor growth via cholesterol/cholesteryl ester accumulation and the subsequent activation of multiple oncogenic pathways, such as PI3K/AKT signaling (13, 39). Some studies emphasized the role of cholesteryl ester rather than cholesterol in promoting tumor cell growth, as evidenced by the fact that the inhibition or knockdown of sterol O-acyltransferase (SOAT1/2) abolished the growth-promoting effect of SQLE (13, 28).
In addition to cholesterol-dependent effects, SQLE can also activate AKT by silencing PTEN in NAFLD-induced hepatocellular carcinoma (HCC). NADPH is required for the conversion of squalene to 2,3-epoxysqualene catalyzed by SQLE. The exhaustion of NADPH by SQLE induces oxidative stress, leading to the epigenetic modification of PTEN by the activation of DNA methyltransferase 3A (DNMT3A). Loss of PTEN activates AKT/mTOR pathways, subsequently activating the expression of SOAT1/2 and contributing to the accumulation of cholesteryl ester and NAFLD-induced HCC (28). Cholesteryl ester accumulation driven by the PTEN/PI3K/AKT/mTOR pathway in prostate cancer cells and subsequent activation of SOAT1 were reported previously (60). Collectively, these staggered cascade reactions amplify the cancer-promoting effect of SQLE.
In multiple cancer types, extracellular signal-regulated kinase (ERK), a crucial oncogenic signaling molecule, has been reported to be regulated by SQLE (26, 59, 61). Recently, HE L et al. revealed the underlying mechanism of ERK activation by SQLE. In colorectal cancer, knockdown of SQLE reduced calcitriol, the active metabolite of VitD3, leading to a reduction in cytochrome P450 family 24 subfamily A member 1 (CYP24A1) levels, which suppressed the phosphorylation and activation of ERK (22).
In addition to cell intrinsic effects, SQLE partially promotes tumor growth via host-microbiota interaction. The gut dysbiosis and altered the metabolism of the gut metabolism caused by elevated expression of SQLE, triggered the gut barrier defects and pro- inflammatory factors. Inquiringly, Transplantation of fecal bacteria from Sqle transgenic mice to germ-free mice can impair gut barrier function and stimulate cell proliferation, compared with fecal bacteria from control mice (62).
However, there is a report that cholesterol accumulation in colorectal cancer can downregulate SQLE and in turn promote tumor metastasis by activating epithelial-mesenchymal transition (EMT)-related pathways. Depletion of SQLE dissociates glycogen synthase 3b (GSK3β) and p53 and upregulates Mdm2, subsequently promoting the degradation of p53 and activation of β-catenin. Consequently, E-cadherin is downregulated, indicating the induction of EMT (23). However, many other studies revealed that EMT factors are upregulated with increased levels of SQLE in cancers, including colorectal cancer (63), pancreatic adenocarcinoma (64), and esophageal squamous cell carcinoma (40). These studies indicated that SQLE is associated with EMT but need to be further studied in certain tumor types (Figure 3).
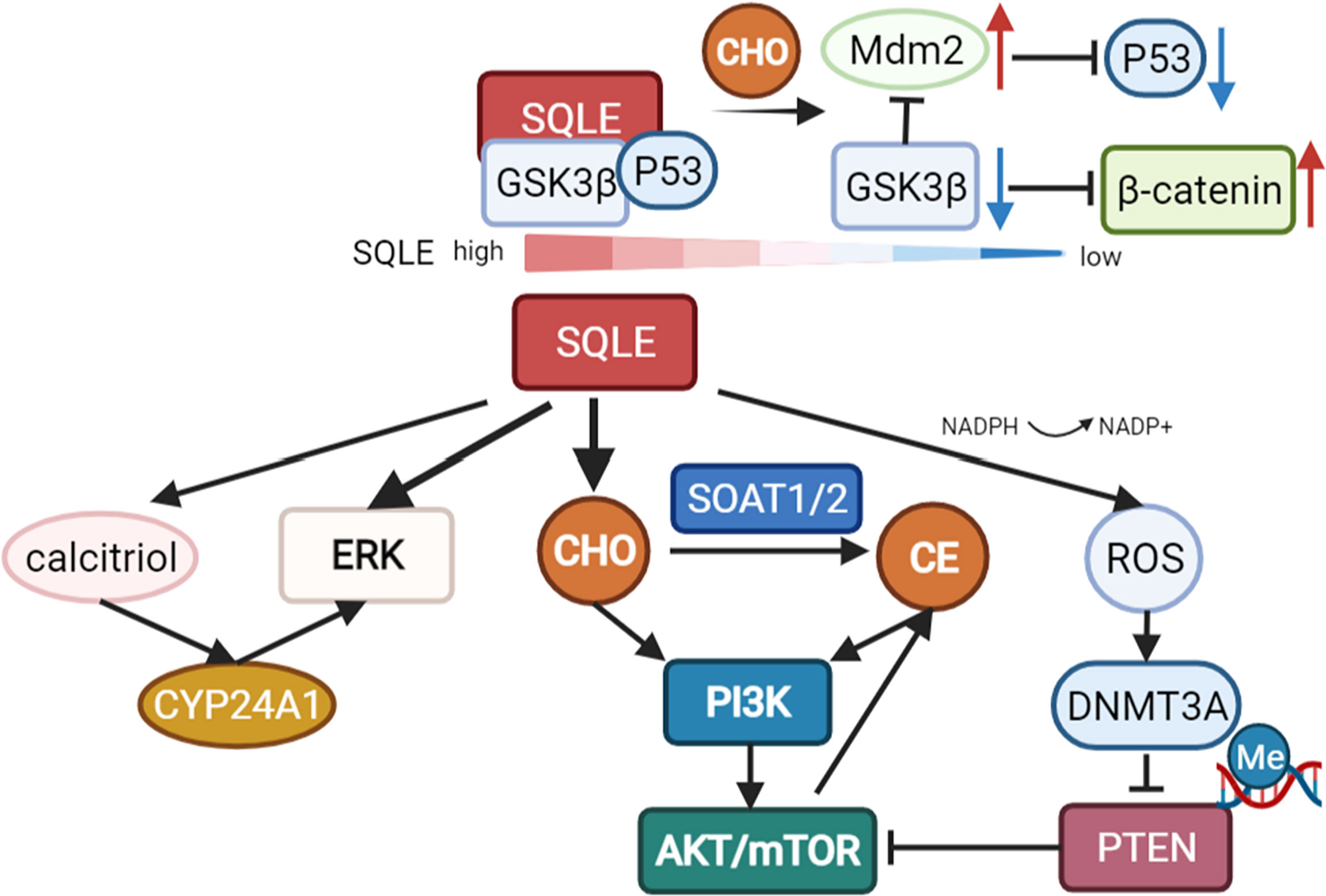
Figure 3 Pathways regulated by SQLE. SQLE can activate multiple oncogenic pathways, such as PI3K/AKT/mTOR signaling and the ERK pathway, via cholesterol/cholesteryl ester accumulation. Knockdown of SQLE can also reduce calcitriol, the active metabolite of VitD3, leading to a reduction in cytochrome P450 family 24 subfamily A member 1 (CYP24A1) levels, which suppresses the phosphorylation and activation of ERK. The exhaustion of NADPH during the conversion of squalene to 2,3-epoxysqualene by SQLE can induce oxidative stress and subsequently activate DNA methyltransferase 3A (DNMT3A), leading to the epigenetic silencing of PTEN. Loss of PTEN activates AKT/mTOR pathways, contributing to SOAT-mediated cholesteryl ester accumulation and NAFLD-induced HCC. However, there is also a report that depletion of SQLE can dissociate GSK3β and p53 and upregulate Mdm2, promoting the degradation of p53 and activation of β-catenin in colorectal cancer.
SQLE can promote cell proliferation (24) and cell migration (65), adjust the cell cycle, and repress cell apoptosis (13, 28), contributing to the different phenotypes of tumors. Deregulated SQLE is associated with tumor aggressiveness and therapy resistance, indicating the poor prognosis of cancers, as reported in previous studies (Table 1).
In prostate cancers, SQLE, which is involved in hormonal signaling, is associated with a high Gleason score, which implies poor biological behavior and prognosis of tumors (71, 72). After Gleason score matching, the expression of SQLE is higher in high-grade acinar cancer than in ductal carcinoma of the prostate, an unusual subtype of prostate cancer (73). This may imply that the role SQLE plays is different in different subtypes of cancer. High expression of SQLE was reported to be associated with poor outcomes in several case–control studies (72, 74). Higher SQLE is associated with metastasis (P = 1.1e-08, HR=3.7 [2.3−5.9]) in prostate tumors (65), and SQLE can predict metastasis combined with TPD52L2 (75). In locally advanced prostate cancer, the expression of SQLE is negatively associated with progression-free survival (PFS) (rs=-0.40) (76). Of note, both the primary tumor lesion and metastatic lesion overexpressed SQLE, which suggested that SQLE is involved in tumor growth and tumor metastasis (65). Androgen deprivation therapy (ADT) is the gold standard for hormone-sensitive metastatic prostate cancer (mHSPC) (77). However, mHSPC eventually develops into metastatic castration-resistant prostate cancer (mCRPC), which indicates poor prognosis after ADT. Overexpression of SQLE is involved in the resistance to castration mediated by metabolic reprogramming. Knockdown of SQLE can reverse this resistance (78).
In breast cancers, SQLE overexpression often indicates a more aggressive tumor (17) and is associated with tumor recurrence and short overall survival time, including estrogen receptor-positive (ER(+)) breast cancer and estrogen receptor-negative breast cancer or luminal A subtype and luminal B subtype (14, 16, 79, 80). Consistently, as early as 2007, it was reported that the amplification of 8q24.11-13 (regions including the SQLE gene) was associated with worse prognosis (81). BROWN D N et al. pinpointed that this amplification was associated with SQLE overexpression (17). The level of SQLE can also predict the response to estrogen deprivation therapy in ER (+) breast cancer. Tumor patients with SQLE overexpression commonly exhibited poor response to letrozole (P=0.38) and poor PFS under adjuvant tamoxifen (P<0.001, HR=2.02, 95% CI=1.5-2.7) (15). Larger tumor size, advanced TNM stage, HER2(+) status and lymph node metastasis are also associated with SQLE overexpression (16, 17).
In HCC, overexpression of SQLE is associated with poor prognosis (82). Similarly, copy number gains on 8q24.13-24.3 (area containing the SQLE gene) also indicated poor survival (83). Multivariate Cox analysis also suggested that SQLE was an independent biomarker of overall survival (28). High SQLE expression was associated with advanced TNM stage (p=0.045) and a-fetoprotein elevation (p=0.029) (82). For pancreatic cancer, the clinical relevance of SQLE is limited. SQLE overexpression is associated with poor overall survival (20), but no evidence has shown that SQLE is an independent prognostic factor for pancreatic cancer (20, 64). SQLE is associated with radio resistance in pancreatic cancer cells, but the correlation is much weaker than that of FDPS and IDI1 (84).
For colorectal cancer, a higher level of SQLE is associated with lymphovascular invasion, tumor budding, advanced pathological T stage, and regional lymph node metastasis (63). However, the prognostic prediction effect of SQLE seems to shift during tumor progression. A higher level of SQLE in tumors is associated with worse overall survival in a cohort mainly containing stage II and III patients (63). However, lower SQLE expression in T4 or stage IV tumors predicts worse prognosis. For lung squamous cell carcinoma, SQLE is associated with poor survival and clinicopathological variables, including poor differentiation and lymph node metastasis (30, 61). SQLE overexpression is also associated with worse survival in uveal melanoma (85), nasopharyngeal carcinoma (13), and head and neck squamous cell carcinoma (24). Daunorubicin-resistant leukemia cells express higher levels of SQLE than daunorubicin-sensitive leukemia cells (25). SQLE is associated with immune cell infiltration in tumors in head and neck squamous cell carcinoma based on bioinformatics analysis (24).
In most tumors, a high SQLE level predicts poor prognosis (except T4 or stage IV colorectal cancer), including tumor recurrence, tumor metastasis, higher-grade clinicopathological variables, and short overall survival time. In addition, as the key enzyme in cholesterol (the precursor of sex hormones) synthesis, overexpression of SQLE is associated with a poor response to hormone therapy. Collectively, considering the role of SQLE in tumorigenesis and tumor progression evidenced by basic studies and clinical analysis, SQLE may be a novel target for cancer therapy.
In the fungus, the inhibition of SQLE leads to a lack of ergosterol and the accumulation of squalene (86). Hence, SQLE inhibitors are widely used against fungal infections (87, 88). SQLE inhibitors can be classified into allylamines, squalene derivatives, natural compounds and derivatives according to their structure. Naftifine, the first antifungal agent and a representative allylamine inhibitor (89), paves the way for next-generation inhibitors: terbinafine, NB-598, Cmpd-4, FR194738, etc.
Given SQLE deregulation in cancers and its tumor promotion function, targeting SQLE is believed to be a novel and promising antitumor therapy. Allylamines, as pioneers of SQLE inhibitors, were investigated for antitumor therapy. In a retrospective cohort study, patients with prostate cancer receiving systemic use of terbinafine had a decreased risk of overall death (HR=0.64; 95% CI, 0.52–0.77) and a decreased risk of death from prostate cancer (HR, 0.64; 95% CI, 0.52–0.77), while the topical use of terbinafine seemed not to bring survival benefits (90). Recently, four patients with rapidly progressive end-stage metastatic prostate cancer after multiple prior treatment modalities, excluding ADT and radiation, were off-label administered terbinafine orally. A drop in prostate-specific antigen (PSA) levels in three of the four patients was observed, suggesting that SQLE blockade can reduce biochemical markers of disease progression in prostate cancer (41).
SQLE inhibitors (terbinafine and NB-598) have been confirmed to suppress cell proliferation, blunt cell viability, promote cell death in vitro, and retrain tumor growth in vivo across various types of cancers in a dosage-dependent manner (17, 22, 66). Retardation of the cell cycle at the G0/G1 phase and enhancement of apoptosis are involved in the antitumor function of terbinafine, while there is no impact on normal cells (91). For the arrest of the cell cycle at G0/G1 phase, cell cycle regulators exhibit corresponding changes: a decrease in the levels of CDK4, phosphorylation of Rb, CDK2, and the p53-activated signaling pathway is involved in terbinafine-induced cell cycle arrest (66, 91, 92). The evidence of apoptosis includes the increased expression of cleaved caspase-7 and cleaved caspase-9 and DNA strand breaks caused by endonuclease (13, 91).
The mechanism underlying the suppression of tumor growth by SQLE inhibitors was investigated in previous studies. In nonalcoholic fatty liver disease (NAFLD)-induced HCC, terbinafine enhanced the degradation of SQLE via autophagy and then reversed the expression of PTEN, which in turn inhibited AKT/mTOR signaling (28). Consistently, terbinafine can also downregulate AKT activity, probably by decreasing cholesteryl ester (13). In breast cancer cell lines, the inhibition of SQLE by terbinafine decreased the phosphorylation of ERK (59). The accumulation of squalene induced by SQLE inhibitors can also be toxic to tumor cells. The detrimental effect of squalene can be accentuated when the cholesterol biosynthetic flux is increased, while lipid droplets can constrict the toxic effects by storing squalene (68, 93). Squalene has been thought to be nontoxic to cells for quite a long time (94). This detrimental effect of squalene may largely depend on the metabolic status in tumors. This speculation was evidenced in a subset of lymphoma cells that lack SQLE, while squalene accumulation can prevent oxidative cell death (95).
It should be noted that the antitumor function of SQLE inhibitors might also be independent of SQLE. In HCC, terbinafine still exerts its anticancer function even after SQLE knockdown. Instead, terbinafine suppressed mTORC1 by activating AMPK in a SQLE-independent manner (66). In oral squamous cell carcinoma cells, terbinafine repressed cell growth by inhibiting the KSR1-Raf-MEK-ERK pathway, but the role of SQLE was not investigated in the study (67).
Preclinical studies have revealed the toxicity of SQLE inhibitors when used as antitumor agents. In small cell lung cancer, dogs and monkeys treated with allylamine inhibitors (NB-598 and cmpd-4’’) via oral gavage cannot tolerate predicted efficacious exposures, with dose-limiting toxicity due to dose-limiting gastrointestinal toxicity, accompanied by skin toxicity. When naftifine and terbinafine were used as antifungal agents, the reported adverse reaction profiles were similar to these preclinical toxicology profiles (96, 97). This toxicity might limit the potential therapeutic utility for cancer treatment (98). The IC50 values of allylamine inhibitors in mammalian cells are several orders of magnitude higher than those in fungi (2, 99, 100). With increasing dosage to achieve antitumor efficacy, the tolerability of adverse reactions needs to be assessed carefully. Insights into the structure of SQLE in complex with allylamines offer further understanding of the mechanism of inhibition and new drug development. For allylamines, the tertiary amine motif is a common feature that interacts with the hydroxyl moiety of Y195 in the catalytic domain to form a hydrogen bond and can explain its noncompetitive inhibition (5).
For other types of SQLE inhibitors, such as natural compounds and derivatives, their special properties may enable themselves to be potential antitumor agents or a starting point to develop clinically safe SQLE inhibitors. (-)-Epigallocatechin 3-O-gallate (EGCG) extracted from green tea has been proven to be a potent (IC50 = 0.69 μM) and safe SQLE inhibitor, and few side effects have been reported even when it is consumed at high doses (101). The antitumor effect of EGCG has been widely investigated (69, 70), but it is currently unknown whether the association between SQLE and EGCG contributes to the effect. Selenocystine (IC50 = 65 μM) and S-allylcysteine (IC50 = 110 μM), the components extracted from garlic (102), ellagitannins isolated from various plants (103), etc., might also be potent SQLE inhibitors.
Taken together, SQLE inhibitors are potential antitumor agents considering their antitumor effects in various cancers (Table 2). Diverse types of SQLE inhibitors offer different frameworks to develop new SQLE inhibitors to ablate side effects and improve affinity.
As an oncogenic gene in various cancers, the dysregulation of SQLE correlates with suppressing apoptosis and increasing cell proliferation and aggressiveness. The high abundance of SQLE in tumors indicates worse prognosis. Therefore, SQLE seems to be an attractive target for novel anticancer therapy. Delightfully, an increasing number of preclinical studies have revealed the antitumor effects and related mechanisms. However, the SQLE inhibitors applied in preclinical antitumor studies are mainly terbinafine, which is less selective to human SQLE, and the toxicity may limit antitumor therapy. The insights of the SQLE structure offer further understanding of new drug development to reduce adverse reactions and improve selectivity. Moreover, the metabolic status of tumor cells needs to be investigated before the application of SQLE inhibitors.
HX made contributions to the conception and design. YZ and HX wrote the manuscript. HZ and QT helped to revise the manuscript. FB helped to collect the data. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (82103364); the Sichuan Science and Technology Program (2022YFS0204); the Postdoctoral Innovative Talents Support Program of China (BX2021203); and the Sichuan University postdoctoral interdisciplinary Innovation Fund.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
3’UTR: 3’ untranslated region
ABCA1: ATP-binding cassette transporter A1
ADT: Androgen deprivation therapy;
AMPK: adenosine 5’-monophosphate (AMP)-activated protein kinase
CASIMO1: cancer-associated small integral membrane open reading frame 1
CYP24A1: cytochrome P450 family 24 subfamily A member 1
DNMT3A: DNA methyltransferase 3A;
EGCG: epigallocatechin-3-gallate
EMT: epithelialmesenchymal transition
ER(+): estrogen receptor-positive
ER: endoplasmic reticulum
ERAD: endoplasmic reticulumassociated degradation
ERK: extracellular signal-regulated kinase
GSK3b: glycogen synthase 3b
HCC: hepatocellular carcinoma
HMGCR: 3-hydroxy-3-methylglutaryl-CoA reductase
LncRNA: Long noncoding RNA
MARCH6: membrane-associated RING finger 6
mHSPC: hormonesensitive metastatic prostate cancer
miRNA: microRNA;
MVA: mevalonate
NAFLD: nonalcoholic fatty liver disease;
NF-Y: nuclear factor Y
OSBPL2: oxysterol binding protein like 2
PCBP2: poly(rC) binding protein 2
RE1: response element 1
S1P: site-1 protease
S2P: site-2 protease
SCAP: SREBP2 cleavage-activating protein
SOAT1/2: sterol Oacyltransferase;
Sp1: specificity protein 1
SQLE: squalene monooxygenase
SRE: sterol-regulatory elements
SREBP2: sterol regulatory element-binding protein 2
UBE2J2: E2 ubiquitin-conjugating enzyme J2
USFA: unsaturated fatty acids
VCP: Valosin-containing protein
1. Ryder NS, Dupont MC. Properties of a particulate squalene epoxidase from candida albicans. Biochim Biophys Acta (1984) 794(3):466–71. doi: 10.1016/0005-2760(84)90013-4
2. Ryder NS. Mechanism of action and biochemical selectivity of allylamine antimycotic agents. Ann New York Acad Sci (1988) 544:208–20. doi: 10.1111/j.1749-6632.1988.tb40405.x
3. Ryder NS. Squalene epoxidase as a target for the allylamines. Biochem Soc Trans (1991) 19(3):774–7. doi: 10.1042/bst0190774
4. Nagai M, Sakakibara J, Wakui K, Fukushima Y, Igarashi S, Tsuji S, et al. Localization of the squalene epoxidase gene (SQLE) to human chromosome region 8q24. 1 Genomics (1997) 44(1):141–3. doi: 10.1006/geno.1997.4825
5. Padyana AK, Gross S, Jin L, Cianchetta G, Narayanaswamy R, Wang F, et al. Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase. Nat Commun (2019) 10(1):97. doi: 10.1038/s41467-018-07928-x
6. Chua NK, Coates HW, Brown AJ. Squalene monooxygenase: A journey to the heart of cholesterol synthesis. Prog Lipid Res (2020) 79:101033. doi: 10.1016/j.plipres.2020.101033
7. Xu H, Zhou S, Tang Q, Xia H, Bi F. Cholesterol metabolism: New functions and therapeutic approaches in cancer. Biochim Biophys Acta Rev cancer (2020) 1874(1):188394. doi: 10.1016/j.bbcan.2020.188394
8. Göbel A, Rauner M, Hofbauer LC, Rachner TD. Cholesterol and beyond - the role of the mevalonate pathway in cancer biology. Biochim Biophys Acta (BBA) - Rev Cancer (2020) 1873(2):188351. doi: 10.1016/j.bbcan.2020.188351
9. Feltrin S, Ravera F, Traversone N, Ferrando L, Bedognetti D, Ballestrero A, et al. Sterol synthesis pathway inhibition as a target for cancer treatment. Cancer letters (2020) 493:19–30. doi: 10.1016/j.canlet.2020.07.010
10. Brown AJ, Chua NK, Yan N. The shape of human squalene epoxidase expands the arsenal against cancer. Nat Commun (2019) 10(1):888. doi: 10.1038/s41467-019-08866-y
11. Wong J, Quinn CM, Gelissen IC, Brown AJ. Endogenous 24(S),25-epoxycholesterol fine-tunes acute control of cellular cholesterol homeostasis. J Biol Chem (2008) 283(2):700–7. doi: 10.1074/jbc.M706416200
12. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
13. Li L, Zhang Q, Wang X, Li Y, Xie H, Chen X. Squalene epoxidase-induced cholesteryl ester accumulation promotes nasopharyngeal carcinoma development by activating PI3K/AKT signaling. Cancer science (2020) 111(7):2275–83. doi: 10.1111/cas.14426
14. Helms MW, Kemming D, Pospisil H, Vogt U, Buerger H, Korsching E, et al. Squalene epoxidase, located on chromosome 8q24.1, is upregulated in 8q+ breast cancer and indicates poor clinical outcome in stage I and II disease. Br J Cancer (2008) 99(5):774–80. doi: 10.1038/sj.bjc.6604556
15. Simigdala N, Gao Q, Pancholi S, Roberg-Larsen H, Zvelebil M, Ribas R, et al. Cholesterol biosynthesis pathway as a novel mechanism of resistance to estrogen deprivation in estrogen receptor-positive breast cancer. Breast Cancer Res BCR (2016) 18(1):58. doi: 10.1186/s13058-016-0713-5
16. Kim NI, Park MH, Kweon SS, Cho N, Lee JS. Squalene epoxidase expression is associated with breast tumor progression and with a poor prognosis in breast cancer. Oncol letters (2021) 21(4):259. doi: 10.3892/ol.2021.12520
17. Brown DN, Caffa I, Cirmena G, Piras D, Garuti A, Gallo M, et al. Squalene epoxidase is a bona fide oncogene by amplification with clinical relevance in breast cancer. Sci Rep (2016) 6:19435. doi: 10.1038/srep19435
18. D'Arcy M, Fleming J, Robinson WR, Kirk EL, Perou CM, Troester MA. Race-associated biological differences among luminal a breast tumors. Breast Cancer Res Treat (2015) 152(2):437–48. doi: 10.1007/s10549-015-3474-4
19. Parada H Jr., Sun X, Fleming JM, Williams-DeVane CR, Kirk EL, Olsson LT, et al. Race-associated biological differences among luminal a and basal-like breast cancers in the Carolina breast cancer study. Breast Cancer Res BCR (2017) 19(1):131. doi: 10.1186/s13058-017-0914-6
20. Bai R, Rebelo A, Kleeff J, Sunami Y. Identification of prognostic lipid droplet-associated genes in pancreatic cancer patients via bioinformatics analysis. Lipids Health Dis (2021) 20(1):58. doi: 10.1186/s12944-021-01476-y
21. Harada T, Chelala C, Crnogorac-Jurcevic T, Lemoine NR. Genome-wide analysis of pancreatic cancer using microarray-based techniques. Pancreatology (2009) 9(1-2):13–24. doi: 10.1159/000178871
22. He L, Li H, Pan C, Hua Y, Peng J, Zhou Z, et al. Squalene epoxidase promotes colorectal cancer cell proliferation through accumulating calcitriol and activating CYP24A1-mediated MAPK signaling. Cancer Commun (London England) (2021) 41(8):726–46. doi: 10.1002/cac2.12187
23. Jun SY, Brown AJ, Chua NK, Yoon JY, Lee JJ, Yang JO, et al. Reduction of squalene epoxidase by cholesterol accumulation accelerates colorectal cancer progression and metastasis. Gastroenterology (2021) 160(4):1194–207.e28. doi: 10.1053/j.gastro.2020.09.009
24. Liu Y, Fang L, Liu W. High SQLE expression and gene amplification correlates with poor prognosis in head and neck squamous cell carcinoma. Cancer Manage Res (2021) 13:4709–23. doi: 10.2147/CMAR.S305719
25. Stäubert C, Krakowsky R, Bhuiyan H, Witek B, Lindahl A, Broom O, et al. Increased lanosterol turnover: A metabolic burden for daunorubicin-resistant leukemia cells. Med Oncol (Northwood London England) (2016) 33(1):6. doi: 10.1007/s12032-015-0717-5
26. Sui Z, Zhou J, Cheng Z, Lu P. Squalene epoxidase (SQLE) promotes the growth and migration of the hepatocellular carcinoma cells. Tumour Biol J Int Soc Oncodevelopmental Biol Med (2015) 36(8):6173–9. doi: 10.1007/s13277-015-3301-x
27. Liang JQ, Teoh N, Xu L, Pok S, Li X, Chu ESH, et al. Dietary cholesterol promotes steatohepatitis related hepatocellular carcinoma through dysregulated metabolism and calcium signaling. Nat Commun (2018) 9(1):4490. doi: 10.1038/s41467-018-06931-6
28. Liu D, Wong CC, Fu L, Chen H, Zhao L, Li C, et al. Squalene epoxidase drives NAFLD-induced hepatocellular carcinoma and is a pharmaceutical target. Sci Trans Med (2018) 10(437):eaap9840. doi: 10.1126/scitranslmed.aap9840
29. Liu Y, Sun W, Zhang K, Zheng H, Ma Y, Lin D, et al. Identification of genes differentially expressed in human primary lung squamous cell carcinoma. Lung Cancer (Amsterdam Netherlands) (2007) 56(3):307–17. doi: 10.1016/j.lungcan.2007.01.016
30. Zhang HY, Li HM, Yu Z, Yu XY, Guo K. Expression and significance of squalene epoxidase in squamous lung cancerous tissues and pericarcinoma tissues. Thorac cancer (2014) 5(4):275–80. doi: 10.1111/1759-7714.12087
31. Kristiana I, Luu W, Stevenson J, Cartland S, Jessup W, Belani JD, et al. Cholesterol through the looking glass: Ability of its enantiomer also to elicit homeostatic responses. J Biol Chem (2012) 287(40):33897–904. doi: 10.1074/jbc.M112.360537
32. Nagai M, Sakakibara J, Nakamura Y, Gejyo F, Ono T. SREBP-2 and NF-y are involved in the transcriptional regulation of squalene epoxidase. Biochem Biophys Res Commun (2002) 295(1):74–80. doi: 10.1016/S0006-291X(02)00623-X
33. Howe V, Sharpe LJ, Prabhu AV, Brown AJ. New insights into cellular cholesterol acquisition: Promoter analysis of human HMGCR and SQLE, two key control enzymes in cholesterol synthesis. Biochim Biophys Acta Mol Cell Biol lipids (2017) 1862(7):647–57. doi: 10.1016/j.bbalip.2017.03.009
34. Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol (2020) 21(4):225–45. doi: 10.1038/s41580-019-0190-7
35. Zhang C, Zhang H, Zhang M, Lin C, Wang H, Yao J, et al. OSBPL2 deficiency upregulate SQLE expression increasing intracellular cholesterol and cholesteryl ester by AMPK/SP1 and SREBF2 signalling pathway. Exp Cell Res (2019) 383(2):111512. doi: 10.1016/j.yexcr.2019.111512
36. Yang F, Kou J, Liu Z, Li W, Du W. MYC enhances cholesterol biosynthesis and supports cell proliferation through SQLE. Front Cell Dev Biol (2021) 9:655889. doi: 10.3389/fcell.2021.655889
37. Yang F, Kou J, Liu Z, Li W, Du W. Corrigendum: MYC enhances cholesterol biosynthesis and supports cell proliferation through SQLE. Front Cell Dev Biol (2021) 9:705769. doi: 10.3389/fcell.2021.705769
38. Sun H, Li L, Li W, Yang F, Zhang Z, Liu Z, et al. p53 transcriptionally regulates SQLE to repress cholesterol synthesis and tumor growth. EMBO Rep (2021) 22(10):e52537. doi: 10.15252/embr.202152537
39. Qin Y, Hou Y, Liu S, Zhu P, Wan X, Zhao M, et al. A novel long non-coding RNA lnc030 maintains breast cancer stem cell stemness by stabilizing SQLE mRNA and increasing cholesterol synthesis. Advanced Sci (Weinheim Baden-Wurttemberg Germany) (2021) 8(2):2002232. doi: 10.1002/advs.202002232
40. Qin Y, Zhang Y, Tang Q, Jin L, Chen Y. SQLE induces epithelial-to-mesenchymal transition by regulating of miR-133b in esophageal squamous cell carcinoma. Acta Biochim Biophys Sinica (2017) 49(2):138–48. doi: 10.1093/abbs/gmw127
41. Kalogirou C, Linxweiler J, Schmucker P, Snaebjornsson MT, Schmitz W, Wach S, et al. MiR-205-driven downregulation of cholesterol biosynthesis through SQLE-inhibition identifies therapeutic vulnerability in aggressive prostate cancer. Nat Commun (2021) 12(1):5066. doi: 10.1038/s41467-021-25325-9
42. Gill S, Stevenson J, Kristiana I, Brown AJ. Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab (2011) 13(3):260–73. doi: 10.1016/j.cmet.2011.01.015
43. Sharpe LJ, Coates HW, Brown AJ. Post-translational control of the long and winding road to cholesterol. J Biol Chem (2020) 295(51):17549–59. doi: 10.1074/jbc.REV120.010723
44. Tan JME, van der Stoel MM, van den Berg M, van Loon NM, Moeton M, Scholl E, et al. The MARCH6-SQLE axis controls endothelial cholesterol homeostasis and angiogenic sprouting. Cell Rep (2020) 32(5):107944. doi: 10.1016/j.celrep.2020.107944
45. Howe V, Chua NK, Stevenson J, Brown AJ. The regulatory domain of squalene monooxygenase contains a re-entrant loop and senses cholesterol via a conformational change. J Biol Chem (2015) 290(46):27533–44. doi: 10.1074/jbc.M115.675181
46. Coates HW, Capell-Hattam IM, Brown AJ. The mammalian cholesterol synthesis enzyme squalene monooxygenase is proteasomally truncated to a constitutively active form. J Biol Chem (2021) 296:100731. doi: 10.1016/j.jbc.2021.100731
47. Chua NK, Howe V, Jatana N, Thukral L, Brown AJ. A conserved degron containing an amphipathic helix regulates the cholesterol-mediated turnover of human squalene monooxygenase, a rate-limiting enzyme in cholesterol synthesis. J Biol Chem (2017) 292(49):19959–73. doi: 10.1074/jbc.M117.794230
48. Chua NK, Scott NA, Brown AJ. Valosin-containing protein Mediates the ERAD of squalene monooxygenase and its cholesterol-responsive degron. Biochem J (2019) 476(18):2545–60. doi: 10.1042/BCJ20190418
49. Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS. E2 enzymes: more than just middle men. Cell Res (2016) 26(4):423–40. doi: 10.1038/cr.2016.35
50. Tan JME, Cook ECL, van den Berg M, Scheij S, Zelcer N, Loregger A. Differential use of E2 ubiquitin conjugating enzymes for regulated degradation of the rate-limiting enzymes HMGCR and SQLE in cholesterol biosynthesis. Atherosclerosis (2019) 281:137–42. doi: 10.1016/j.atherosclerosis.2018.12.008
51. Zelcer N, Sharpe LJ, Loregger A, Kristiana I, Cook EC, Phan L, et al. The E3 ubiquitin ligase MARCH6 degrades squalene monooxygenase and affects 3-hydroxy-3-methyl-glutaryl coenzyme a reductase and the cholesterol synthesis pathway. Mol Cell Biol (2014) 34(7):1262–70. doi: 10.1128/MCB.01140-13
52. Chua NK, Hart-Smith G, Brown AJ. Non-canonical ubiquitination of the cholesterol-regulated degron of squalene monooxygenase. J Biol Chem (2019) 294(20):8134–47. doi: 10.1074/jbc.RA119.007798
53. Meyer H, Weihl CC. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J Cell Sci (2014) 127(Pt 18):3877–83. doi: 10.1242/jcs.093831
54. Sharpe LJ, Howe V, Scott NA, Luu W, Phan L, Berk JM, et al. Cholesterol increases protein levels of the E3 ligase MARCH6 and thereby stimulates protein degradation. J Biol Chem (2019) 294(7):2436–48. doi: 10.1074/jbc.RA118.005069
55. Yoshioka H, Coates HW, Chua NK, Hashimoto Y, Brown AJ, Ohgane K. A key mammalian cholesterol synthesis enzyme, squalene monooxygenase, is allosterically stabilized by its substrate. Proc Natl Acad Sci USA (2020) 117(13):7150–8. doi: 10.1073/pnas.1915923117
56. Nathan JA. Squalene and cholesterol in the balance at the ER membrane. Proc Natl Acad Sci USA (2020) 117(15):8228–30. doi: 10.1073/pnas.2003388117
57. Chua NK, Brown AJ. The degron architecture of squalene monooxygenase and how specific lipids calibrate levels of this key cholesterol synthesis enzyme. Adv Exp Med Biol (2021) 21:1–12. doi: 10.1007/5584_2020_583
58. Stevenson J, Luu W, Kristiana I, Brown AJ. Squalene mono-oxygenase, a key enzyme in cholesterol synthesis, is stabilized by unsaturated fatty acids. Biochem J (2014) 461(3):435–42. doi: 10.1042/BJ20131404
59. Polycarpou-Schwarz M, Groß M, Mestdagh P, Schott J, Grund SE, Hildenbrand C, et al. The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene (2018) 37(34):4750–68. doi: 10.1038/s41388-018-0281-5
60. Yue S, Li J, Lee SY, Lee HJ, Shao T, Song B, et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab (2014) 19(3):393–406. doi: 10.1016/j.cmet.2014.01.019
61. Ge H, Zhao Y, Shi X, Tan Z, Chi X, He M, et al. Squalene epoxidase promotes the proliferation and metastasis of lung squamous cell carcinoma cells though extracellular signal-regulated kinase signaling. Thorac cancer (2019) 10(3):428–36. doi: 10.1111/1759-7714.12944
62. Li C, Wang Y, Liu D, Wong CC, Coker OO, Zhang X, et al. Squalene epoxidase drives cancer cell proliferation and promotes gut dysbiosis to accelerate colorectal carcinogenesis. Gut (2022). doi: 10.1136/gutjnl-2021-325851
63. Kim JH, Kim CN, Kang DW. Squalene epoxidase correlates e-cadherin expression and overall survival in colorectal cancer patients: The impact on prognosis and correlation to clinicopathologic features. J Clin Med (2019) 8(5):632. doi: 10.3390/jcm8050632
64. Xu F, Zhang Z, Zhao Y, Zhou Y, Pei H, Bai L. Bioinformatic mining and validation of the effects of ferroptosis regulators on the prognosis and progression of pancreatic adenocarcinoma. Gene (2021) 795:145804. doi: 10.1016/j.gene.2021.145804
65. Zhao SG, Evans JR, Kothari V, Sun G, Larm A, Mondine V, et al. The landscape of prognostic outlier genes in high-risk prostate cancer. Clin Cancer Res an Off J Am Assoc Cancer Res (2016) 22(7):1777–86. doi: 10.1158/1078-0432.CCR-15-1250
66. Zhang EB, Zhang X, Wang K, Zhang F, Chen TW, Ma N, et al. Antifungal agent terbinafine restrains tumor growth in preclinical models of hepatocellular carcinoma via AMPK-mTOR axis. Oncogene (2021) 40(34):5302–13. doi: 10.1038/s41388-021-01934-y
67. Li B, Lu L, Zhong M, Tan XX, Liu CY, Guo Y, et al. Terbinafine inhibits KSR1 and suppresses raf-MEK-ERK signaling in oral squamous cell carcinoma cells. Neoplasma (2013) 60(4):406–12. doi: 10.4149/neo_2013_052
68. Mahoney CE, Pirman D, Chubukov V, Sleger T, Hayes S, Fan ZP, et al. A chemical biology screen identifies a vulnerability of neuroendocrine cancer cells to SQLE inhibition. Nat Commun (2019) 10(1):96. doi: 10.1038/s41467-018-07959-4
69. Gan RY, Li HB, Sui ZQ, Corke H. Absorption, metabolism, anti-cancer effect and molecular targets of epigallocatechin gallate (EGCG): An updated review. Crit Rev Food Sci Nutr (2018) 58(6):924–41. doi: 10.1080/10408398.2016.1231168
70. Li F, Qasim S, Li D, Dou QP. Updated review on green tea polyphenol epigallocatechin-3-gallate as a cancer epigenetic regulator. Semin Cancer Biol (2022) 83:335–52. doi: 10.1016/j.semcancer.2020.11.018
71. Fromont G, Rozet F, Cathelineau X, Ouzzane A, Doucet L, Fournier G, et al. BCAR1 expression improves prediction of biochemical reccurence after radical prostatectomy. Prostate (2012) 72(12):1359–65. doi: 10.1002/pros.22485
72. Stopsack KH, Gerke TA, Andrén O, Andersson SO, Giovannucci EL, Mucci LA, et al. Cholesterol uptake and regulation in high-grade and lethal prostate cancers. Carcinogenesis (2017) 38(8):806–11. doi: 10.1093/carcin/bgx058
73. Jardel P, Debiais C, Godet J, Irani J, Fromont G. Ductal carcinoma of the prostate shows a different immunophenotype from high grade acinar cancer. Histopathology (2013) 63(1):57–63. doi: 10.1111/his.12129
74. Stopsack KH, Gerke TA, Sinnott JA, Penney KL, Tyekucheva S, Sesso HD, et al. Cholesterol metabolism and prostate cancer lethality. Cancer Res (2016) 76(16):4785–90. doi: 10.1158/0008-5472.CAN-16-0903
75. Zhao H, Logothetis CJ, Gorlov IP. Usefulness of the top-scoring pairs of genes for prediction of prostate cancer progression. Prostate Cancer Prostatic diseases (2010) 13(3):252–9. doi: 10.1038/pcan.2010.9
76. Pudova EA, Krasnov GS, Kobelyatskaya AA, Savvateeva MV, Fedorova MS, Pavlov VS, et al. Gene expression changes and associated pathways involved in the progression of prostate cancer advanced stages. Front Genet (2020) 11:613162. doi: 10.3389/fgene.2020.613162
77. Mohler JL, Antonarakis ES, Armstrong AJ, D'Amico AV, Davis BJ, Dorff T, et al. Prostate cancer, version 2.2019, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Network JNCCN (2019) 17(5):479–505. doi: 10.6004/jnccn.2019.0023
78. Xu Z, Huang L, Dai T, Pei X, Xia L, Zeng G, et al. SQLE mediates metabolic reprogramming to promote LN metastasis in castration-resistant prostate cancer. OncoTargets Ther (2021) 14:4285–95. doi: 10.2147/OTT.S315813
79. Kim S, Kon M, DeLisi C. Pathway-based classification of cancer subtypes. Biol direct (2012) 7:21. doi: 10.1186/1745-6150-7-21
80. Yu Z, He Q, Xu G. Screening of prognostic factors in early-onset breast cancer. Technol Cancer Res Treat (2020) 19:1533033819893670. doi: 10.1177/1533033819893670
81. Chin SF, Teschendorff AE, Marioni JC, Wang Y, Barbosa-Morais NL, Thorne NP, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol (2007) 8(10):R215. doi: 10.1186/gb-2007-8-10-r215
82. Shen T, Lu Y, Zhang Q. High squalene epoxidase in tumors predicts worse survival in patients with hepatocellular carcinoma: Integrated bioinformatic analysis on NAFLD and HCC. Cancer control J Moffitt Cancer Center (2020) 27(1):1073274820914663. doi: 10.1177/1073274820914663
83. Zhao K, Zhao Y, Zhu JY, Dong H, Cong WM, Yu Y, et al. A panel of genes identified as targets for 8q24.13-24.3 gain contributing to unfavorable overall survival in patients with hepatocellular carcinoma. Curr Med Sci (2018) 38(4):590–6. doi: 10.1007/s11596-018-1918-x
84. Souchek JJ, Baine MJ, Lin C, Rachagani S, Gupta S, Kaur S, et al. Unbiased analysis of pancreatic cancer radiation resistance reveals cholesterol biosynthesis as a novel target for radiosensitisation. Br J cancer (2014) 111(6):1139–49. doi: 10.1038/bjc.2014.385
85. Xu Y, Han W, Xu WH, Wang Y, Yang XL, Nie HL, et al. Identification of differentially expressed genes and functional annotations associated with metastases of the uveal melanoma. J Cell Biochem (2019) 120(11):19202–14. doi: 10.1002/jcb.29250
86. Barrett-Bee K, Dixon G. Ergosterol biosynthesis inhibition: A target for antifungal agents. Acta Biochim Polonica (1995) 42(4):465–79. doi: 10.18388/abp.1995_4900
87. Georgopapadakou NH, Bertasso A. Effects of squalene epoxidase inhibitors on candida albicans. Antimicrob Agents Chemother (1992) 36(8):1779–81. doi: 10.1128/AAC.36.8.1779
88. Favre B, Ryder NS. Characterization of squalene epoxidase activity from the dermatophyte trichophyton rubrum and its inhibition by terbinafine and other antimycotic agents. Antimicrob Agents Chemother (1996) 40(2):443–7. doi: 10.1128/AAC.40.2.443
89. Ryder NS, Dupont MC. Inhibition of squalene epoxidase by allylamine antimycotic compounds. a comparative study of the fungal and mammalian enzymes. Biochem J (1985) 230(3):765–70. doi: 10.1042/bj2300765
90. Ji J, Sundquist J, Sundquist K. Use of terbinafine and risk of death in patients with prostate cancer: A population-based cohort study. Int J cancer (2019) 144(8):1888–95. doi: 10.1002/ijc.31901
91. Lee WS, Chen RJ, Wang YJ, Tseng H, Jeng JH, Lin SY, et al. In vitro and in vivo studies of the anticancer action of terbinafine in human cancer cell lines: G0/G1 p53-associated cell cycle arrest. Int J cancer (2003) 106(1):125–37. doi: 10.1002/ijc.11194
92. Chien MH, Lee TS, Kao C, Yang SF, Lee WS. Terbinafine inhibits oral squamous cell carcinoma growth through anti-cancer cell proliferation and anti-angiogenesis. Mol carcinogenesis (2012) 51(5):389–99. doi: 10.1002/mc.20800
93. Xu X, Chen J, Li Y, Yang X, Wang Q, Wen Y, et al. Targeting epigenetic modulation of cholesterol synthesis as a therapeutic strategy for head and neck squamous cell carcinoma. Cell Death Dis (2021) 12(5):482. doi: 10.1038/s41419-021-03760-2
94. Chugh A, Ray A, Gupta JB. Squalene epoxidase as hypocholesterolemic drug target revisited. Prog Lipid Res (2003) 42(1):37–50. doi: 10.1016/S0163-7827(02)00029-2
95. Garcia-Bermudez J, Baudrier L, Bayraktar EC, Shen Y, La K, Guarecuco R, et al. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature (2019) 567(7746):118–22. doi: 10.1038/s41586-019-0945-5
96. Monk JP, Brogden RN. Naftifine. A review of its antimicrobial activity and therapeutic use in superficial dermatomycoses. Drugs (1991) 42(4):659–72. doi: 10.2165/00003495-199142040-00008
97. Darkes MJ, Scott LJ, Goa KL. Terbinafine: A review of its use in onychomycosis in adults. Am J Clin Dermatol (2003) 4(1):39–65. doi: 10.2165/00128071-200304010-00005
98. Nagaraja R, Olaharski A, Narayanaswamy R, Mahoney C, Pirman D, Gross S, et al. Preclinical toxicology profile of squalene epoxidase inhibitors. Toxicol Appl Pharmacol (2020) 401:115103. doi: 10.1016/j.taap.2020.115103
99. Belter A, Skupinska M, Giel-Pietraszuk M, Grabarkiewicz T, Rychlewski L, Barciszewski J. Squalene monooxygenase - a target for hypercholesterolemic therapy. Biol Chem (2011) 392(12):1053–75. doi: 10.1515/BC.2011.195
100. Abe I, Tomesch JC, Wattanasin S, Prestwich GD. Inhibitors of squalene biosynthesis and metabolism. Natural product Rep (1994) 11(3):279–302. doi: 10.1039/np9941100279
101. Abe I, Seki T, Umehara K, Miyase T, Noguchi H, Sakakibara J, et al. Green tea polyphenols: Novel and potent inhibitors of squalene epoxidase. Biochem Biophys Res Commun (2000) 268(3):767–71. doi: 10.1006/bbrc.2000.2217
102. Gupta N, Porter TD. Garlic and garlic-derived compounds inhibit human squalene monooxygenase. J Nutr (2001) 131(6):1662–7. doi: 10.1093/jn/131.6.1662
103. Abe I, Kashiwagi Y, Noguchi H, Tanaka T, Ikeshiro Y, Kashiwada Y. Ellagitannins and hexahydroxydiphenoyl esters as inhibitors of vertebrate squalene epoxidase. J Natural products (2001) 64(8):1010–4. doi: 10.1021/np010100y
Keywords: cancer treatment, cell proliferation and migration, cholesterol metabolism, drug target, squalene epoxidase
Citation: Zou Y, Zhang H, Bi F, Tang Q and Xu H (2022) Targeting the key cholesterol biosynthesis enzyme squalene monooxygenasefor cancer therapy. Front. Oncol. 12:938502. doi: 10.3389/fonc.2022.938502
Received: 18 May 2022; Accepted: 26 July 2022;
Published: 24 August 2022.
Edited by:
Richard David Feinman, Downstate Health Sciences University, United StatesReviewed by:
Paula Mariana Maloberti, University of Buenos Aires, ArgentinaCopyright © 2022 Zou, Zhang, Bi, Tang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiulin Tang, dGFuZ3FpdWxpbjE5ODFAMTYzLmNvbQ==; Huanji Xu, eHVodWFuamkxOTkwQHNjdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.