
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 30 May 2022
Sec. Cancer Molecular Targets and Therapeutics
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.897895
This article is part of the Research Topic Targeted Cancer Therapies, from Small Molecules to Antibodies, Volume II View all 33 articles
N6-methyladenosine (m6A) is a major internal epigenetic modification in eukaryotic mRNA, which is dynamic and reversible. m6A is regulated by methylases (“writers”) and demethylases (“erasers”) and is recognized and processed by m6A-binding proteins (“readers”), which further regulate RNA transport, localization, translation, and degradation. It plays a role in promoting or suppressing tumors and has the potential to become a therapeutic target for malignant tumors. In this review, we focus on the mutual regulation of m6A and coding and non-coding RNAs and introduce the molecular mechanism of m6A methylation involved in regulation and its role in cancer treatment by taking common female malignant tumors as an example.
N6-methyladenosine (m6A) alteration is a methylation modification located on the 6th nitrogen atom of adenine. m6A is the most abundant form of epigenetic modification in eukaryotic RNA, which exists in several different types of RNA, including mRNA and non-coding RNA. At present, the research on m6A methylation and malignant tumors mainly focuses on the influence of m6A methylation on tumor cell proliferation, invasion, resistance to radiotherapy and chemotherapy, and prognosis of patients (1). The detection rate of malignant tumors is growing year by year, and the age of onset tends to be younger, thanks to the popularization of associated cancer screening tools and an increase in people’s health awareness (2). The primary clinical treatment for malignant tumors is to select individualized surgery combined with postoperative radiotherapy and chemotherapy according to the patient’s condition (3). However, for some patients with relapsed and refractory tumors, the treatment effect is often difficult to achieve the expected (4). Therefore, one of the most pressing issues to be addressed is elucidating the etiology of malignant tumors and finding novel therapeutic medications to overcome tumor resistance. In this review, we focus on the effect of m6A methylation on the occurrence and development of malignant tumors and introduce the molecular mechanisms involved in the regulation of m6A methylation and its role in cancer treatment by taking common female malignancies as an example.
The m6A methylation modification of RNA is the most common internal modification in RNA modification. m6A is the methylation modification of RNA on the 6th nitrogen atom of adenosine, and the process is dynamic and reversible. The proteins involved in the methylation and demethylation of m6A are divided into three categories, namely methyltransferases (“writers”), demethylases (“erasers”), and m6A recognition proteins (“readers”) (Figure 1). Through RNA transcription, splicing, processing, translation, and degradation, it plays a role in the formation and spread of numerous malignant cancers. (5). Furthermore, studies have shown that m6A methylation modification is closely related to the activation and inhibition of cancer-related signaling pathways, which mainly affect tumor progression by regulating related tumor biological functions (6). At present, m6A RNA modification is increasingly used in cancer detection and related targeted molecules (7, 8).
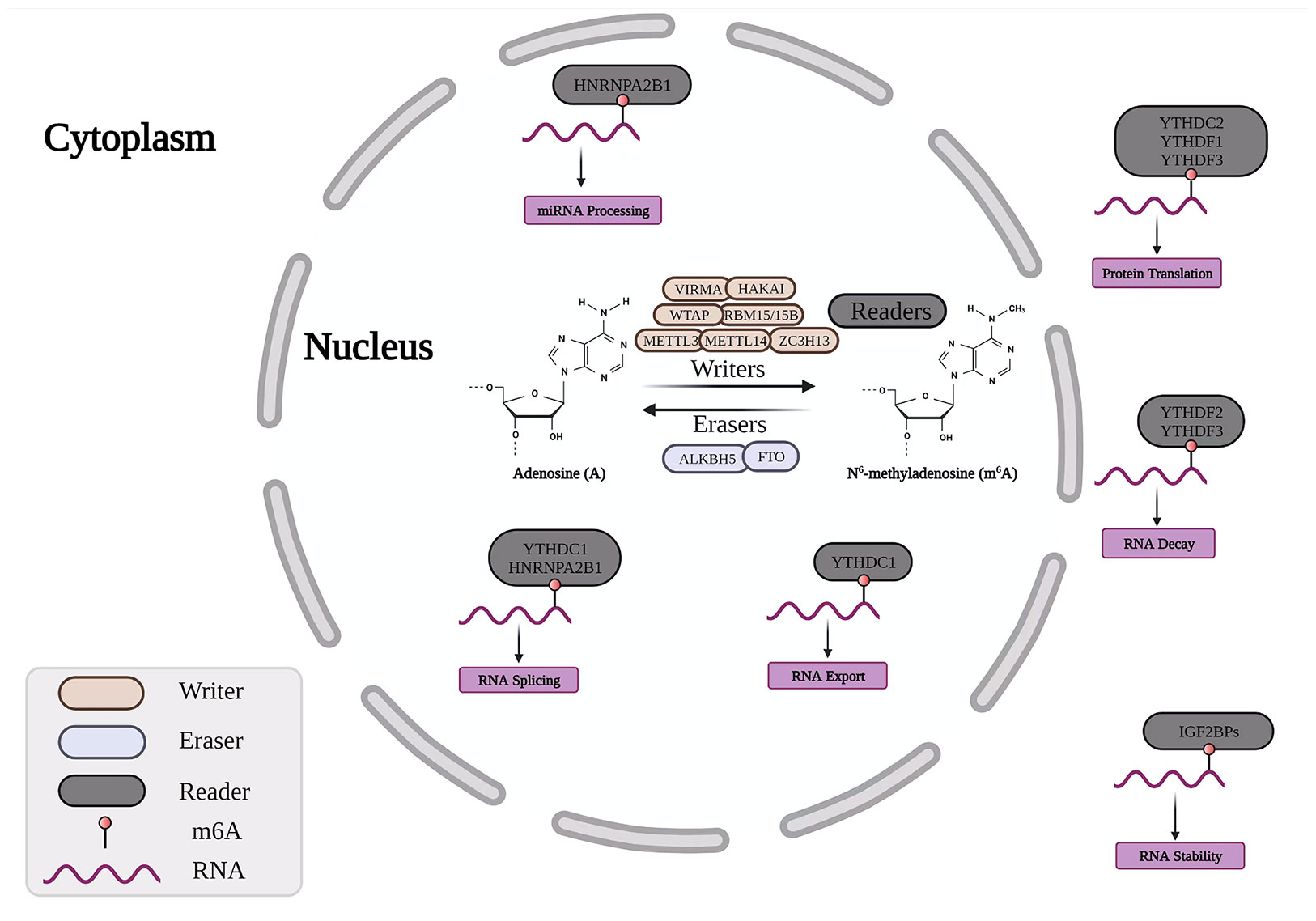
Figure 1 Dynamic m6A RNA modifying process. The RNA modification of m6A is regulated by methyltransferases (“writers”), demethylases (“erasers”) and m6A-binding proteins (“readers”). VIRMA, Vir Like m6A Methyltransferase Associated; HAKAI, Cbl Proto-Oncogene Like 1; WTAP, Wilms Tumor 1 Associated Protein; RBM15, RNA Binding Motif Protein 15; METTL3, Methyltransferase Like 3; METTL14, Methyltransferase Like 14; ZC3H13, Zinc Finger CCCH-Type Containing 13; ALKBH5, AlkB Homolog 5; FTO, Fat Mass And Obesity-Associated Protein; HNRNP, Heterogeneous Nuclear Ribonucleoprotein; YTHDF, YTH domain protein family; IGF2BP, Insulin Like Growth Factor 2 MRNA Binding Protein.
For m6A-modified mRNA to perform specific biological functions, a specific RNA-binding protein, methylation reader protein, is required. It mainly includes YTH m6A RNA-binding protein (YTH) domain proteins (including YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2) (9), heterogeneous nuclear ribonucleoproteins (HNRNPC, HNRNPG, and HNRNPA2B1) and eukaryotic initiation factor (eIF). The functions of these reader proteins are mainly to alter protein-RNA interactions by impairing the homologous binding of m6A to RNA-binding proteins and altering RNA secondary structure (10). Studies have found that YTHDF1 has a clear oncogenic role, and its high expression in cancer genes can accelerate the transformation of important oncogenic drivers in cancer via numerous methods, impacting cancer progression and prognosis. For example, in gastric cancer progression, mutated YTHDF1 enhances the expression of the key oncogenic factor Wnt receptor Frizzled7 (FZD7), leading to gastric cancer progression and poor prognosis (11).
The m6A methyltransferase consists of methyltransferase-like 3 (METTL3), METTL14, WTAP, RBM15, ZC3H13, VIRMA, and the newly discovered METTL16, which are also called writers. Its primary function is to catalyze the m6A modification of adenylate on mRNA. METTL3 and METTL14 have a critical catalytic domain (12), and the METTL3-METTL14 methyltransferase complex assembly during m6A modification is a crucial factor initiating the modification. In addition, methyltransferases also include associated protein (WTAP), RBM15, ZC3H13, vir-like m6A methyltransferase associated (VIRMA), and METTL16. These methyltransferases all play an important role in forming METTL3-METTL14 complexes in different links, affecting cancer cell proliferation and migration (13–16).
m6A modification is the earliest reversible modification found among many RNA modifications, and the reversibility of its modification is due to the existence of demethylases. In addition, their encoding genes are called “erasers”. Fat mass and obesity-associated protein (FTO) is the first demethylase discovered and is a member of the Alkb protein family, which can affect the RNA-binding ability of the splicing factor serine and arginine-rich splicing factor 2 (SRSF2), thereby regulating the splicing process of pre-mRNA (17). ALKBH5, another member of the Alkb protein family, was found to have a demethylation function, which can directly catalyze m6A-methylated adenosine to remove methyl groups, different from the oxidative demethylation of FTO (18).
In recent years, m6A has been found to exist in various ncRNAs such as miRNAs, long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), ribosomal RNAs (rRNAs), and small nuclear RNAs (snRNAs), essential for its metabolism and function (19). Moreover, some m6A regulatory proteins associated with aberrant m6A modification of ncRNAs are also involved in cancer cells proliferation, invasion, and drug resistance, suggesting a potential link between cancer and m6A-ncRNA modification (20).
LncRNAs, a type of ncRNAs of 200 or more nucleotides, can regulate gene expression at multiple levels (21). Many m6A-modified lncRNAs have been found recently, and they can control gene expression and function in a variety of ways. They can act as transcriptional regulators, acting in cis or trans, regulating the transcription of adjacent genes (22). XIST, one of the first functionally annotated lncRNAs, plays a key role in X chromosome inactivation by recruiting multiple factors (23). A study shows that in head and neck squamous cell carcinoma, METTL3- and METTL14-mediated m6A methylation contributes to the stability of LNCAROD, and LNCAROD overexpression promotes the malignant development of HNSCC by promoting YBX1-hspa1a interaction, thereby enhancing the stability of the YBX1 protein (24). Another study found that modifying lncRNAs with m6A had the opposite effect on cancers. METTL14 suppressed colorectal cancer growth and metastasis by downregulating oncogenic long non-coding RNA XIST in the METTL14-YTHDF2-lncRNA regulatory axis (25). LncRNA GAS5 binds directly to Yes-associated protein (YAP), promoting its phosphorylation and ubiquitin-mediated degradation, thereby attenuating YAP-mediated transcription of YTHDF3 and inhibiting the progression of rectal cancer (26). LincRNA-p21 acts as a tumor inhibitor in the development of esophageal squamous cell carcinoma (27). In hepatocellular carcinoma, the m6A methylation modification of LINC00152 is involved in the prognosis of LIHC patients through the cytoskeleton regulation pathway (28). Furthermore, in rectal cancer, m6A modification of lncRNA RP11 can upregulate the translation of Zeb1 to trigger cancer cell dissemination (29); lncRNA-THOR enhances IGF2BP1-targeted mRNA expression and promotes human osteosarcoma cell survival and proliferation (30).
MiRNAs are non-coding single-stranded RNAs of 21-25 nucleotides in length that regulate gene expression at the post-transcriptional level by building RNA-induced silencing complexes (RISCs) that bind to the 3’untranslated region of target mRNAs (3’UTR) to regulate gene expression (31). In the nucleus, miRNAs are first transcribed into longer primary miRNAs (pri-miRNAs) and subsequently processed into precursor miRNAs (pre-miRNAs). It is then cleaved into mature single-stranded miRNAs by Dicer in the cytoplasm, and the participation and processing of such pri-miRNAs are m6A-dependent. METTL3 tags pre-miRNAs through m6A modification, enabling DGCR8 to recognize and bind its specific substrates, thereby promoting miRNA maturation and increased miRNA levels in cells (32).
Circular RNAs (circRNAs) are a class of single-stranded covalently closed RNA molecules that participate in many physiological processes, including competing with endogenous RNAs as sponge miRNAs, forming RNA-protein complexes, regulating gene transcription, and even encoding proteins (33). In most cases, abnormal m6A modification contributes to tumorigenesis and tumor progression. However, m6A modification on circRNAs can suppress innate immunity; YTHDF2 sequesters m6A-circRNA and is essential for suppressing innate immunity (34). Chen et al. found that m6A modification of circNSUN2 promoted liver metastasis of colorectal cancer by promoting cytoplasmic export and forming a circNSUN2/IGF2BP2/HMGA2 RNA-protein triple complex to stabilize HMGA2 mRNA (35).
m6A affects all physiological processes such as mRNA processing, nuclear export, translation, and degradation. It mainly affects mRNA stability, which is also closely related to the occurrence and development of malignant tumors. At present, the research on m6A and its participants in the reversible regulation process (m6A-modifying enzymes and m6A-binding proteins) and the mechanism of tumorigenesis and development has gradually become a hot spot.
Ries et al. found that m6A-mRNA is regulated by compartments, including mRNA stability and reduced translation. This study demonstrates that the number and distribution of m6A sites in cellular mRNA can modulate and influence the composition of the phase-separated transcriptome (36). Li et al. first elucidated the in vivo biological role of m6A modification in T cell-mediated pathogenesis. They revealed a novel mechanism for T cell homeostasis and signal-dependent induction of mRNA degradation (37). RNA methyltransferase (METTL3) acts as a translation initiation complex, thereby enhancing the translation of target mRNAs (38). In addition, at different intracellular locations, m6A exerts methyltransferase activity-dependent and -independent functions in gene regulation. Besides, the RNA methyltransferase METTL16 is in the nucleus, acting as an m6A writer, depositing m6A into its hundreds of a specific messenger RNA target. In the cytosol, METTL16 promotes translation in an m6A-independent manner (39).
Common malignant tumors in women mainly include breast cancer (BC), ovarian cancer (OC), cervical cancer (CC), and endometrial cancer (EC). Despite advancements in examination methods in the prevention and treatment of common female malignant tumors, most patients are in the middle and late stages of their disease due to difficulties in early diagnosis and localization of tumors and a lack of effective efficacy evaluation and prognosis monitoring methods. Therefore, the mortality rate of common malignant tumors in women continues to increase. Genetic, epigenetic, and environmental factors drive its occurrence, development, and transfer, and epigenetic factors play an important role as a bridge between genetic and environmental factors. Epigenetics has multiple forms of expression, of which m6A is the most abundant form of internal modification. In this review, we take common female malignant tumors as examples to introduce the molecular mechanism of m6A modification in the occurrence and development of cancer and its application in cancer treatment (Table 1).
m6A methyltransferases (“Writers”) are an essential class of catalytic enzymes. METTL3 is a key regulator that promotes m6A modification, and the abnormal regulation of METTL3 is also inextricably linked to tumor development. Some studies have confirmed that the overexpression of METTL3 may be an important factor in promoting the development of common malignant tumors in women. Pan et al. found that the expression of RBM15 and METTL3 in CESC (cervical squamous cell carcinoma) tissues was higher than that in normal tissues (49). In addition, Hua et al. found that METTL3 promoted the epithelial-to-mesenchymal transition of ovarian cancer cells and the proliferation, invasion, and tumor formation of ovarian cancer cells, affecting their prognosis and overall survival (50). Moreover, Ma et al. compared the expressions of METTL14, WTAP, and METTL3 in ovarian cancer and found that METTL3 independently regulates m6A modification and thus affects the proliferation and metastasis (51). Besides, Li et al. found that METTL3 inhibited the viability of cervical cancer cells and enhanced their sensitivity to the chemotherapeutic drug cisplatin by downregulating the expression of the receptor for advanced glycation and its product in cervical cancer tissues (40); METTL3 modulated the m6A modification of MALAT1. The expression of MALAT1 is upregulated, and MALAT1 can promote the expression of high mobility group A2(HMGA2) by sponge miR-26b, thereby promoting the development of breast cancer (41). Regarding this, Wang et al. proposed that WTAP may promote the proliferation, invasion, and migration of ovarian cancer through two gene sequences of FAM76A and HBS1 (42).
As m6A methylation reading proteins, “Readers” can recognize the information of RNA methylation modification and participate in downstream mRNA translation, degradation, and miRNA processing. It mainly changes the interaction between protein and RNA by weakening the homologous binding of m6A to RNA-binding protein and changing the secondary structure of RNA.
YTHDF1 acts as an important reading element in m6A modification by recognizing m6A-containing mRNAs and promoting their translation initiation and elongation (52). Wang et al. applied online data analysis to identify RANBP2 as a critical target of YTHDF1 in cervical cancer cells, and subsequent reduction of RANBP2 decreased cervical cancer cell proliferation, migration, and invasion (43). Overexpression of YTHDF1 promoted the growth, migration, and invasion of Hela and Siha cells. At the same time, knockdown of RANBP2 reversed the effect of overexpression of YTHDF1 on cervical cancer progression, indicating that YTHDF1 promotes cervical cancer progression by regulating RANBP2 expression in an m6A-dependent manner. Some scholars have proposed that YTHDF1 directly targets eIF3C (a subunit of EIF3) and promotes ovarian cancer’s occurrence, metastasis, and prognosis (51).
Heterogeneous ribonucleoproteins (hnRNPs) are a diverse family of RNA-binding proteins that function in most stages of RNA metabolism (53). Studies have shown that HNRNPC regulates target transcripts’ abundance and alternative splicing through mA binding to RNA. Other researchers have proposed that hnRNPA2B1 can inhibit the growth of ovarian cancer cells, reduce the mobility of ovarian cancer cells in vitro, and hinder the formation of xenograft tumors in vivo. In addition, hnRNPA2B1 promotes the occurrence and development of malignant phenotypes of ovarian cancer by activating the expression of Lin28B (54). Moreover, Shi et al. found that lobaplatin induced apoptosis and cell cycle arrest by downregulating hnRNP A2/B1 in cervical cancer cells, and knockdown of hnRNP A2/B1 significantly reduced tumor growth in nude mice xenografts and increased cervical cancer Cellular sensitivity to lobaplatin and irinotecan (55). Eukaryotic initiation factor 3 (eIF3) can bind to m6A-modified bases in the 5’ UTR of RNA, promoting mRNA translation (56). The high expression of eLF3 in ovarian cancer is closely related to its poor prognosis (44); Zhu et al. concluded that eIF3B is highly expressed in cervical cancer tissues and is closely related to advanced FIGO in cervical cancer patients staging, shorter overall survival and lymph node metastasis (57).
Demethylase is an integral part of the reversible modification of m6A, and FTO, as the first discovered demethylase, is widely present in adult and embryonic tissues, and its expression is exceptionally high in the brain. Moreover, recent studies have shown that FTO has an important effect on glioblastoma growth and self-renewal (58).
The expression level of FTO is also elevated in cervical squamous cell carcinoma, which can enhance chemoradiotherapy resistance in vitro and in vivo by reducing m6A-regulated β-catenin expression (59). Zhao et al. believed that FTO accelerated the growth of cancer cells by promoting proliferation, inhibiting apoptosis, and activating autophagy in ovarian cancer (60). In the latest study, Huang et al. found that FTO expression in high-grade serous ovarian cancer (HGSOC) tumor cells were significantly lower than that in other tissues, and it had a significant inhibitory effect on ovarian cancer cells (47). It can be seen that FTO may have a bidirectional regulatory impact on ovarian cancer tissue, and the specific mechanism needs to be further studied. Furthermore some researchers believe that FTO primarily stimulates the oncogenic activity of breast cancer cell invasion and migration through the FTO/miR-181b-3p/ARL5B signaling pathway, promoting tumor proliferation (61). The tumor suppressor BNIP3 is a downstream target of FTO-mediated m6A modification. FTO mediates m6A demethylation in the 3’UTR of BNIP3 mRNA and induces its degradation through a YTHDF2-independent mechanism, promoting breast cancer cell proliferation, colony formation, and in vitro and in vivo transfer (46).
Among them, ALKBH5, another member of the Alkb protein family, was found to have demethylation, which can directly catalyze m6A-methylated adenosine to remove methyl groups is different from the oxidative demethylation of FTO (62). ALKBH5 affects tumor growth by regulating cell proliferation, migration, invasion, and metastasis. Recently, some scholars have proposed that ALKBH5 has dual roles in various cancers. ALKBH5 can reverse METTL3 autophagy in cells through down-regulation of mRNA stability. In epithelial ovarian cancer cells (63), overexpressed ALKBH5 in human ovarian cancer (SKOV3) cells enhanced the stability of BCL-2 mRNA and inhibited tumor cell growth and metastasis. In contrast, overexpressed ALKBH5 in A2780 cells had the exact opposite effect. In cervical cancer tissues, GAS5-AS1 was also low-expressed and inhibited the proliferation and metastasis of cervical cancer cells. And regulate GAS5 expression by interacting with RNA demethylase ALKBH5, thereby inhibiting CC cell proliferation, migration, and invasion (64).
Targeted therapy is at the cellular and molecular level to design corresponding therapeutic drugs for the already defined carcinogenic sites. The drug enters the body and will specifically select the carcinogenic sites to combine and act so that the tumor cells specifically die instead of normal tissue cells surrounding the tumor are affected. Early studies of targeting strategies based on m6A modulators have focused on demethylases. Besides, previous studies have shown that m6A plays an essential role in the occurrence and development of tumors. Therefore, it is of great scientific significance and clinical value to develop specific inhibitors of m6A-related proteins. As the first discovered RNA-modifying demethylase, FTO is widely involved in various physiological processes, and its dysregulation is associated with multiple human diseases.
Due to its involvement in obesity and obesity-induced metabolic diseases and the occurrence, development and prognosis of various cancers, such as melanoma, acute myeloid leukemia, glioblastoma, lung cancer, hepatocellular carcinoma and breast cancer, studies have shown that rhein can induce apoptosis (65). Huang et al.. systematically investigated the effect of rhein on adipogenesis by transcriptional and post-transcriptional approaches and found that rhein regulates m6A methylation rearrangement and adipogenesis in an independent manner, inhibiting fat mass and obesity-related (FTO) demethylase activity (66). It is indicated that rhein can inhibit the demethylation activity of FTO on m6A on mRNA in vitro and in vivo, thereby increasing the level of m6A in cells. ALKBH5 and FTO are both m6A demethylases. Studies have found that in ovarian cancer, the core cytokine NANOG is a key target to promote the development of ovarian cancer (48). On the contrary, there is much evidence to prove the overexpression of METTL3 in tumor tissues (67), while studies targeting METTL3 have shown that it can effectively inhibit tumor growth, proliferation, and metastasis (68).
In the progression of common malignant tumors in women, drug resistance that often occurs in the later stage is also a significant difficulty in its treatment. Chemotherapy resistance, especially platinum resistance, is a major cause of poor prognosis in ovarian cancer. Bowen Li et al. found that m6A can modulate the modification of anticancer drug resistance by modulating drug-target interactions and drug-mediated cell death signaling (69). The ethyl ester form of the FTO inhibitor Meclofenaic (MA2) inhibits FTO and enhances the effect of the chemotherapeutic drug temozolom by targeting the MYC-miR-155/23a cluster-MXI1 feedback circuit in gliomas anti-tumor effects (45) (Table 2).
The tumor microenvironment (TME) is primarily responsible for mediating immunotherapy responses in tumor progression, and bioinformatics research has shown that m6A alteration and its regulators may regulate the TME and are linked to immune checkpoint inhibition (ICB) (7, 72).
Yi et al. systematically studied head and neck squamous cell carcinoma (HNSCC) compared with adjacent normal pairs, concluded that m6A regulators were upregulated in HNSCC, and found that m6A regulators were associated with PDL in the tumor immune microenvironment (TIME) (73). The expression of -1 was positively correlated, which may provide a promising target for improving the responsiveness of HNSCC to immunotherapy. In addition, He et al. systematically analyzed RNA-sequencing data of 24 major m6A methylation regulators in 775 breast cancer patients from the TCGA database and classified them for overall survival in the lower RNA methylation status group (RM1). The higher methylation status (RM2) group was significantly reduced (70). Moreover, the RM2 group displayed higher expression and higher numbers of tumor-infiltrating CD8+ T cells, helper T cells, and activated NK cells. The expressions of PD-L2, TIM3, and CCR4 were lower than those of the RM1 group, so it can be considered that the regulator of m6A is closely related to the malignant degree, prognosis, and anti-tumor immune response of breast cancer and can be used as a potential target and biological target for breast cancer immunotherapy.
In addition, anti-PD-1 immunotherapy is effective initially, but its efficacy is significantly reduced later due to FTO-mediated resistance (71). However, recent studies have shown that FTO knockdown can increase tumor sensitivity to anti-PD-1 immunotherapy, thereby improving efficacy (69). Therefore, the combined use of ICB and FTO inhibitors may block the development of drug resistance in individuals who develop adaptive immunity.
With the rapid development of high-throughput sequencing technology and bioinformatics, m6A has been gradually revealed as an important epigenetic modification with reversible properties, modification-related enzyme system, and role in different disease processes. It provides infinite possibilities for subsequent tumor diagnosis and treatment. These m6A-modified molecules are expected to become effective early diagnosis and prognostic markers for tumors and potential therapeutic targets, providing new ideas for tumor diagnosis and treatment.
Since m6A research provides a new understanding of the molecular mechanisms of tumorigenesis, metastasis, immune response, and drug resistance and promotes the development of new therapeutics, the process from theory to clinical translation still needs to be explored. Currently, the understanding of how m6A modification affects immune phenotype is still in its infancy. Although some methylase inhibitors have been discovered so far and provide new targets for tumor drugs, their mechanisms of action in vitro and in vivo are not fully understood and lack specificity. Therefore, the development of more inhibitors against m6A-related proteins brings a new dawn for guiding tumor-targeted therapy based on RNA epigenetics. Targeted intervention in m6A modification can promote basic research in related fields, show excellent application prospects in tumor treatment and other disease-related fields, and show important scientific significance in life sciences and new drug discovery.
HS and X-xH conceived and designed the study. Y-fZ, S-jW, JZ, Y-hS, JM, and Y-mC collected data and aided in writing the manuscript. HS and Y-fZ edited the manuscript. All authors read and approved the final manuscript.
This study was supported by the National Natural Science Foundation of China (No. 81802103, 81803938), Project of High-Level Talents in AHUTCM (Project code: 2019rcZD001), Excellent Young Scholars Project of Natural Science Foundation of Anhui Province in China (grant No. 2108085Y29), Natural Science Research Project of Colleges and Universities in Anhui Province (No. KJ2021A0557), Opening Project of Zhejiang Provincial Preponderant and Characteristic Subject of Key University (Chinese Traditional Medicine), and Zhejiang Chinese Medical University (No.ZYXZD2019004).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
3’ -UTR, 3’-untranslation regions; 5’-UTR, 5’-untranslated region; YTH, YTH m6A RNA-binding protein; ALKBH5, AlkB homolog 5; ALKBH3, AlkB homolog 3; FZD7, Wnt receptor frizzled7; WTAP, associated protein; VIRMA, vir-like m6A methyltransferase associated; CC, Cervical cancer; circRNAs, Circular RNAs; EC, Endometrial cancer; eIF3, Eukaryotic initiation factor 3; FTO, Fat mass and obesity-associated protein; SRSF2, Serine and arginine-rich splicing factor 2; HNSCCs, Head and neck squamous cell carcinoma; HNRNPC, HNRNPG and HNRNPA2B1, Heterogeneous nuclear ribonucleoproteins; HGSOC, High-grade serous ovarian cancer; ICB, Immune checkpoint blockade; YAP, Yes-associated protein; IGFBP7, Insulin-like growth factor-binding protein-7; lncRNAs, Long non-coding RNAs; MA, Meclofenaic; mRNA, Messenger RNA; METTL3, Methyltransferase-like 3; METTL14, Methyltransferase-like 14; SKOV3, human ovarian cancer; m6A, N6-Methyladenosine; NPC, Nasopharyngeal carcinoma; ncRNAs, Non-coding RNAs; OC, Ovarian cancer; HMGA2, high mobility group A2; pre-miRNAs, Precursor miRNAs; pri-miRNAs, Primary miRNAs; PD-1, Programmed cell death protein-1; RBM15, RNA-binding motif protein 15; RBM15B, RNA-binding motif protein 15B; RISCs, RNA-induced silencing complexes; rRNAs, Ribosomal RNAs; snRNAs, Small nuclear RNAs; snoRNAs, Small nucleolar RNAs; TME, Tumor microenvironment; TIME, Tumor immune microenvironment; YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3, YTH domain protein family.
1. Han X, Liu J, Cheng G, Cui S. Gene Signatures and Prognostic Values of M6a RNA Methylation Regulators in Ovarian Cancer. Cancer Control (2020) 27:1073274820960460. doi: 10.1177/1073274820960460
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
3. Byrd DR, Brierley JD, Baker TP, Sullivan DC, Gress DM. Current and Future Cancer Staging After Neoadjuvant Treatment for Solid Tumors. CA Cancer J Clin (2021) 71:140–8. doi: 10.3322/caac.21640
4. Graillon T, Tabouret E, Chinot O. Chemotherapy and Targeted Therapies for Meningiomas: What Is the Evidence? Curr Opin Neurol (2021) 34:857–67. doi: 10.1097/WCO.0000000000001002
5. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-Methyladenosine and its Role in Cancer. Mol Cancer (2019) 18:176. doi: 10.1186/s12943-019-1109-9
6. Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D, et al. Mechanism of RNA Modification N6-Methyladenosine in Human Cancer. Mol Cancer (2020) 19:104. doi: 10.1186/s12943-020-01216-3
7. Kuai D, Zhu S, Shi H, Yang R, Liu T, Liu H, et al. Aberrant Expression of M(6)A mRNA Methylation Regulators in Colorectal Adenoma and Adenocarcinoma. Life Sci (2021) 273:119258. doi: 10.1016/j.lfs.2021.119258
8. Zhang B, Chen Z, Tao B, Yi C, Lin Z, Li Y, et al. M(6)A Target microRNAs in Serum for Cancer Detection. Mol Cancer (2021) 20:170. doi: 10.1186/s12943-021-01477-6
9. Liu S, Li G, Li Q, Zhang Q, Zhuo L, Chen X, et al. The Roles and Mechanisms of YTH Domain-Containing Proteins in Cancer Development and Progression. Am J Cancer Res (2020) 10(4):1068–84.
10. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-Methyladenosine Alters RNA Structure to Regulate Binding of a Low-Complexity Protein. Nucleic Acids Res (2017) 45:6051–63. doi: 10.1093/nar/gkx141
11. Pi J, Wang W, Ji M, Wang X, Wei X, Jin J, et al. YTHDF1 Promotes Gastric Carcinogenesis by Controlling Translation of FZD7. Cancer Res (2021) 81:2651–65. doi: 10.1158/0008-5472.CAN-20-0066
12. Zaccara S, Ries RJ, Jaffrey SR. Reading, Writing and Erasing mRNA Methylation. Nat Rev Mol Cell Biol (2019) 20:608–24. doi: 10.1038/s41580-019-0168-5
13. Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 Regulates Nuclear RNA M(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell (2018) 69:1028–1038 e1026. doi: 10.1016/j.molcel.2018.02.015
14. Lan Y, Liu B, Guo H. The Role of M(6)A Modification in the Regulation of Tumor-Related lncRNAs. Mol Ther Nucleic Acids (2021) 24:768–79. doi: 10.1016/j.omtn.2021.04.002
15. Mendel M, Delaney K, Pandey RR, Chen KM, Wenda JM, Vagbo CB, et al. Splice Site M(6)A Methylation Prevents Binding of U2AF35 to Inhibit RNA Splicing. Cell (2021) 184:3125–3142 e3125. doi: 10.1016/j.cell.2021.03.062
16. Zhu W, Wang JZ, Wei JF, Lu C. Role of M6a Methyltransferase Component VIRMA in Multiple Human Cancers (Review). Cancer Cell Int (2021) 21:172. doi: 10.1186/s12935-021-01868-1
17. Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell (2020) 38:79–96 e11. doi: 10.1016/j.ccell.2020.04.017
18. Wu X, Dai M, Li J, Cai J, Zuo Z, Ni S, et al. M(6)A Demethylase ALKBH5 Inhibits Cell Proliferation and the Metastasis of Colorectal Cancer by Regulating the FOXO3/miR-21/SPRY2 Axis. Am J Transl Res (2021) 13(10):11209–22.
19. Li Y, Xiao J, Bai J, Tian Y, Qu Y, Chen X, et al. Molecular Characterization and Clinical Relevance of M(6)A Regulators Across 33 Cancer Types. Mol Cancer (2019) 18:137. doi: 10.1186/s12943-019-1066-3
20. Huang H, Weng H, Chen J. M(6)A Modification in Coding and Non-Coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell (2020) 37:270–88. doi: 10.1016/j.ccell.2020.02.004
21. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell (2018) 172:393–407. doi: 10.1016/j.cell.2018.01.011
22. Furlan G, Gutierrez Hernandez N, Huret C, Galupa R, Van Bemmel JG, Romito A, et al. The Ftx Noncoding Locus Controls X Chromosome Inactivation Independently of Its RNA Products. Mol Cell (2018) 70:462–472 e468. doi: 10.1016/j.molcel.2018.03.024
23. Lee JT, Bartolomei MS. X-Inactivation, Imprinting, and Long Noncoding RNAs in Health and Disease. Cell (2013) 152:1308–23. doi: 10.1016/j.cell.2013.02.016
24. Ban Y, Tan P, Cai J, Li J, Hu M, Zhou Y, et al. LNCAROD is Stabilized by M6a Methylation and Promotes Cancer Progression via Forming a Ternary Complex With HSPA1A and YBX1 in Head and Neck Squamous Cell Carcinoma. Mol Oncol (2020) 14:1282–96. doi: 10.1002/1878-0261.12676
25. Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 Suppresses Proliferation and Metastasis of Colorectal Cancer by Down-Regulating Oncogenic Long Non-Coding RNA XIST. Mol Cancer (2020) 19:46. doi: 10.1186/s12943-020-1146-4
26. Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long Noncoding RNA GAS5 Inhibits Progression of Colorectal Cancer by Interacting With and Triggering YAP Phosphorylation and Degradation and is Negatively Regulated by the M(6)A Reader YTHDF3. Mol Cancer (2019) 18:143. doi: 10.1186/s12943-019-1079-y
27. Zhang Y, Miao Y, Shang M, Liu M, Liu R, Pan E, et al. LincRNA-P21 Leads to G1 Arrest by P53 Pathway in Esophageal Squamous Cell Carcinoma. Cancer Manag Res (2019) 11:6201–14. doi: 10.2147/CMAR.S197557
28. Zhu HX, Lu WJ, Zhu WP, Yu S. Comprehensive Analysis of N(6) -Methyladenosine-Related Long Non-Coding RNAs for Prognosis Prediction in Liver Hepatocellular Carcinoma. J Clin Lab Anal (2021) 35:e24071. doi: 10.1002/jcla.24071
29. Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, et al. M(6)A-Induced lncRNA RP11 Triggers the Dissemination of Colorectal Cancer Cells via Upregulation of Zeb1. Mol Cancer (2019) 18:87. doi: 10.1186/s12943-019-1014-2
30. Chen W, Chen M, Xu Y, Chen X, Zhou P, Zhao X, et al. Long Non-Coding RNA THOR Promotes Human Osteosarcoma Cell Growth In Vitro and In Vivo. Biochem Biophys Res Commun (2018) 499:913–9. doi: 10.1016/j.bbrc.2018.04.019
31. Correia De Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs' Action Through miRNA Editing. Int J Mol Sci (2019) 20(24):6249. doi: 10.3390/ijms20246249
32. Chen Y, Lin Y, Shu Y, He J, Gao W. Interaction Between N(6)-Methyladenosine (M(6)A) Modification and Noncoding RNAs in Cancer. Mol Cancer (2020) 19:94. doi: 10.1186/s12943-020-01207-4
33. Li X, Ding J, Wang X, Cheng Z, Zhu Q. NUDT21 Regulates circRNA Cyclization and ceRNA Crosstalk in Hepatocellular Carcinoma. Oncogene (2020) 39:891–904. doi: 10.1038/s41388-019-1030-0
34. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell (2019) 76:96–109 e109. doi: 10.1016/j.molcel.2019.07.016
35. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-Methyladenosine Modification of Circnsun2 Facilitates Cytoplasmic Export and Stabilizes HMGA2 to Promote Colorectal Liver Metastasis. Nat Commun (2019) 10:4695. doi: 10.1038/s41467-019-12651-2
36. Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, et al. M(6)A Enhances the Phase Separation Potential of mRNA. Nature (2019) 571:424–8. doi: 10.1038/s41586-019-1374-1
37. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. M(6)A mRNA Methylation Controls T Cell Homeostasis by Targeting the IL-7/STAT5/SOCS Pathways. Nature (2017) 548:338–42. doi: 10.1038/nature23450
38. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The M(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell (2016) 62:335–45. doi: 10.1016/j.molcel.2016.03.021
39. Su R, Dong L, Li Y, Gao M, He PC, Liu W, et al. METTL16 Exerts an M(6)A-Independent Function to Facilitate Translation and Tumorigenesis. Nat Cell Biol (2022) 24:205–16. doi: 10.1038/s41556-021-00835-2
40. Li R, Song Y, Chen X, Chu M, Wang ZW, Zhu X. METTL3 Increases Cisplatin Chemosensitivity of Cervical Cancer Cells via Downregulation of the Activity of RAGE. Mol Ther Oncolytics (2021) 22:245–55. doi: 10.1016/j.omto.2021.05.013
41. Zhao C, Ling X, Xia Y, Yan B, Guan Q. The M6a Methyltransferase METTL3 Controls Epithelial-Mesenchymal Transition, Migration and Invasion of Breast Cancer Through the MALAT1/miR-26b/HMGA2 Axis. Cancer Cell Int (2021) 21:441. doi: 10.1186/s12935-021-02113-5
42. Wang J, Xu J, Li K, Huang Y, Dai Y, Xu C, et al. Identification of WTAP-Related Genes by Weighted Gene Co-Expression Network Analysis in Ovarian Cancer. J Ovarian Res (2020) 13:119. doi: 10.1186/s13048-020-00710-y
43. Wang H, Luo Q, Kang J, Wei Q, Yang Y, Yang D, et al. YTHDF1 Aggravates the Progression of Cervical Cancer Through M(6)A-Mediated Up-Regulation of RANBP2. Front Oncol (2021) 11:650383. doi: 10.3389/fonc.2021.650383
44. Xu H, Wang H, Li G, Jin X, Chen B. The Immune-Related Gene ELF3 Is a Novel Biomarker for the Prognosis of Ovarian Cancer. Int J Gen Med (2021) 14:5537–48. doi: 10.2147/IJGM.S332320
45. Xiao L, Li X, Mu Z, Zhou J, Zhou P, Xie C, et al. FTO Inhibition Enhances the Antitumor Effect of Temozolomide by Targeting MYC-miR-155/23a Cluster-MXI1 Feedback Circuit in Glioma. Cancer Res (2020) 80:3945–58. doi: 10.1158/0008-5472.CAN-20-0132
46. Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, et al. RNA N6-Methyladenosine Demethylase FTO Promotes Breast Tumor Progression Through Inhibiting BNIP3. Mol Cancer (2019) 18:46. doi: 10.1186/s12943-019-1004-4
47. Huang H, Wang Y, Kandpal M, Zhao G, Cardenas H, Ji Y, et al. FTO-Dependent N (6)-Methyladenosine Modifications Inhibit Ovarian Cancer Stem Cell Self-Renewal by Blocking cAMP Signaling. Cancer Res (2020) 80:3200–14. doi: 10.1158/0008-5472.CAN-19-4044
48. Jiang Y, Wan Y, Gong M, Zhou S, Qiu J, Cheng W. RNA Demethylase ALKBH5 Promotes Ovarian Carcinogenesis in a Simulated Tumour Microenvironment Through Stimulating NF-kappaB Pathway. J Cell Mol Med (2020) 24:6137–48. doi: 10.1111/jcmm.15228
49. Pan J, Xu L, Pan H. Development and Validation of an M6a RNA Methylation Regulator-Based Signature for Prognostic Prediction in Cervical Squamous Cell Carcinoma. Front Oncol (2020) 10:1444. doi: 10.3389/fonc.2020.01444
50. Hua W, Zhao Y, Jin X, Yu D, He J, Xie D, et al. METTL3 Promotes Ovarian Carcinoma Growth and Invasion Through the Regulation of AXL Translation and Epithelial to Mesenchymal Transition. Gynecol Oncol (2018) 151:356–65. doi: 10.1016/j.ygyno.2018.09.015
51. Ma Z, Li Q, Liu P, Dong W, Zuo Y. METTL3 Regulates M6a in Endometrioid Epithelial Ovarian Cancer Independently of METTl14 and WTAP. Cell Biol Int (2020) 44:2524–31. doi: 10.1002/cbin.11459
52. Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, et al. RNA M(6)A Methylation Regulates the Epithelial Mesenchymal Transition of Cancer Cells and Translation of Snail. Nat Commun (2019) 10:2065. doi: 10.1038/s41467-019-09865-9
53. Levengood JD, Tolbert BS. Idiosyncrasies of hnRNP A1-RNA Recognition: Can Binding Mode Influence Function. Semin Cell Dev Biol (2019) 86:150–61. doi: 10.1016/j.semcdb.2018.04.001
54. Yang Y, Wei Q, Tang Y, Yuanyuan W, Luo Q, Zhao H, et al. Loss of Hnrnpa2b1 Inhibits Malignant Capability and Promotes Apoptosis via Down-Regulating Lin28B Expression in Ovarian Cancer. Cancer Lett (2020) 475:43–52. doi: 10.1016/j.canlet.2020.01.029
55. Shi X, Ran L, Liu Y, Zhong SH, Zhou PP, Liao MX, et al. Knockdown of hnRNP A2/B1 Inhibits Cell Proliferation, Invasion and Cell Cycle Triggering Apoptosis in Cervical Cancer via PI3K/AKT Signaling Pathway. Oncol Rep (2018) 39:939–50. doi: 10.3892/or.2018.6195
56. Bhardwaj U, Powell P, Goss DJ. Eukaryotic Initiation Factor (eIF) 3 Mediates Barley Yellow Dwarf Viral mRNA 3'-5' UTR Interactions and 40S Ribosomal Subunit Binding to Facilitate Cap-Independent Translation. Nucleic Acids Res (2019) 47:6225–35. doi: 10.1093/nar/gkz448
57. Zhu P, Tan Q, Jiang W, Ou Y, Xu P, Yuan L. Eukaryotic Translation Initiation Factor 3B is Overexpressed and Correlates With Deteriorated Tumor Features and Unfavorable Survival Profiles in Cervical Cancer Patients. Cancer biomark (2019) 26:123–30. doi: 10.3233/CBM-182114
58. Huff S, Tiwari SK, Gonzalez GM, Wang Y, Rana TM. M(6)A-RNA Demethylase FTO Inhibitors Impair Self-Renewal in Glioblastoma Stem Cells. ACS Chem Biol (2021) 16:324–33. doi: 10.1021/acschembio.0c00841
59. Zhou S, Bai ZL, Xia D, Zhao ZJ, Zhao R, Wang YY, et al. FTO Regulates the Chemo-Radiotherapy Resistance of Cervical Squamous Cell Carcinoma (CSCC) by Targeting Beta-Catenin Through mRNA Demethylation. Mol Carcinog (2018) 57:590–7. doi: 10.1002/mc.22782
60. Zhao L, Kong X, Zhong W, Wang Y, Li P. FTO Accelerates Ovarian Cancer Cell Growth by Promoting Proliferation, Inhibiting Apoptosis, and Activating Autophagy. Pathol Res Pract (2020) 216:153042. doi: 10.1016/j.prp.2020.153042
61. Xu Y, Ye S, Zhang N, Zheng S, Liu H, Zhou K, et al. The FTO/miR-181b-3p/ARL5B Signaling Pathway Regulates Cell Migration and Invasion in Breast Cancer. Cancer Commun (Lond) (2020) 40:484–500. doi: 10.1002/cac2.12075
62. Gao R, Ye M, Liu B, Wei M, Ma D, Dong K. M6a Modification: A Double-Edged Sword in Tumor Development. Front Oncol (2021) 11:679367. doi: 10.3389/fonc.2021.679367
63. Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 Inhibited Autophagy of Epithelial Ovarian Cancer Through miR-7 and BCL-2. J Exp Clin Cancer Res (2019) 38:163. doi: 10.1186/s13046-019-1159-2
64. Wang X, Zhang J, Wang Y. Long Noncoding RNA GAS5-AS1 Suppresses Growth and Metastasis of Cervical Cancer by Increasing GAS5 Stability. Am J Transl Res (2019) 11(8):4909–21.
65. Heo SK, Noh EK, Kim JY, Jegal S, Jeong Y, Cheon J, et al. Rhein Augments ATRA-Induced Differentiation of Acute Promyelocytic Leukemia Cells. Phytomedicine (2018) 49:66–74. doi: 10.1016/j.phymed.2018.06.027
66. Huang L, Zhang J, Zhu X, Mi X, Li Q, Gao J, et al. The Phytochemical Rhein Mediates M(6)A-Independent Suppression of Adipocyte Differentiation. Front Nutr (2021) 8:756803. doi: 10.3389/fnut.2021.756803
67. Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-Molecule Inhibition of METTL3 as a Strategy Against Myeloid Leukaemia. Nature (2021) 593:597–601. doi: 10.1038/s41586-021-03536-w
68. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-Mediated M(6)A Modification of HDGF mRNA Promotes Gastric Cancer Progression and has Prognostic Significance. Gut (2020) 69:1193–205. doi: 10.1136/gutjnl-2019-319639
69. Li B, Jiang J, Assaraf YG, Xiao H, Chen ZS, Huang C. Surmounting Cancer Drug Resistance: New Insights From the Perspective of N(6)-Methyladenosine RNA Modification. Drug Resist Update (2020) 53:100720. doi: 10.1016/j.drup.2020.100720
70. He X, Tan L, Ni J, Shen G. Expression Pattern of M(6)A Regulators Is Significantly Correlated With Malignancy and Antitumor Immune Response of Breast Cancer. Cancer Gene Ther (2021) 28:188–96. doi: 10.1038/s41417-020-00208-1
71. Fukumoto T, Zhu H, Nacarelli T, Karakashev S, Fatkhutdinov N, Wu S, et al. N(6)-Methylation of Adenosine of FZD10 mRNA Contributes to PARP Inhibitor Resistance. Cancer Res (2019) 79:2812–20. doi: 10.1158/0008-5472.CAN-18-3592
72. Gu J, Bi F. Significance of N6-Methyladenosine RNA Methylation Regulators in Immune Infiltrates of Ovarian Cancer. Front Genet (2021) 12:671179. doi: 10.3389/fgene.2021.671179
Keywords: RNA modifications, N6-methyladenosine, female malignancies, molecular mechanisms, immunotherapy
Citation: Zhu Y-f, Wang S-J, Zhou J, Sun Y-h, Chen Y-m, Ma J, Huo X-x and Song H (2022) Effects of N6-Methyladenosine Modification on Cancer Progression: Molecular Mechanisms and Cancer Therapy. Front. Oncol. 12:897895. doi: 10.3389/fonc.2022.897895
Received: 16 March 2022; Accepted: 21 April 2022;
Published: 30 May 2022.
Edited by:
Yan-yan Yan, Shanxi Datong University, ChinaReviewed by:
Zexian Liu, Sun Yat-sen University Cancer Center (SYSUCC), ChinaCopyright © 2022 Zhu, Wang, Zhou, Sun, Chen, Ma, Huo and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hang Song, aGFuZ3NvbmdAYWh0Y20uZWR1LmNu; Xing-xing Huo, aHh4MDUyMEBhaHRjbS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.