
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Oncol., 23 February 2022
Sec. Cancer Molecular Targets and Therapeutics
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.821958
Background: Aldehyde dehydrogenase (ALDH) 1 is an important enzyme involved in the regulation of several cellular mechanisms via aldehyde detoxification. High ALDH1 levels were correlated with tumorigenesis and stemness maintenance in cancer.
Methods: We used UALCAN, Human Protein Atlas, Kaplan–Meier plotter, TISIDB, TIMER, and KOBAS databases to investigate the expression and role of ALDH1 in thyroid cancer progression. In addition, quantitative real-time polymerase chain reaction was performed to detect the expression of the target genes in thyroid cancer cell lines and cancer tissues.
Results: Expression of ALDH1A1/B1 was significantly decreased based on individual cancer stages and tumor histology, and high levels of ALDH1A1/B1 were associated with poor overall survival in thyroid cancer patients. Moreover, ALDH1A1/B1 expression was negatively correlated with immune-stimulating genes, major histocompatibility complex, chemokines, and receptors.
Conclusions: These results suggest that ALDH1A1/B1 might serve as potential prognostic biomarkers for thyroid cancer diagnosis.
Thyroid cancer is the most prevalent endocrine malignancy worldwide, with a global incidence of over 500,000 per year (1). The number of endocrine cases has been growing rapidly for decades (2), resulting in approximately 90% of the endocrine malignancies and 70% of deaths (3). Thyroid cancer was reported to be prevalent in women (4), but more advanced-stage cases were found in men (5). Recently, different research groups have explored prognostic markers and therapeutic targets for thyroid cancer, and some progress has been made (1, 4, 6). However, current diagnosis of thyroid cancer is still far from sufficient owing to the lack of appropriate biomarkers.
The aldehyde dehydrogenase 1 (ALDH1) family consists of conserved enzymes that oxidize aldehydes to carboxylic acids (7). The ALDH1 family includes six members: ALDH1A1 to ALDH1A3, ALDH1B1, ALDH1L1, and ALDH1l2 (8), which are localized in cytosol and mitochondria and can be found in almost all important organs (8). Recent studies demonstrated that Nuclear factor E2-related factor 2 (Nrf2) activation is involved in cancer stem cell-like properties’ exposure to high ALDH1A1 concentrations (9) that result in shorter overall survival (OS) and progression-free survival in ovarian cancer patients (10). In addition, targeting ALDH1 may facilitate the elimination of cancer stem cells and inhibit breast cancer progression (11, 12). Cumulative evidence has revealed that aberrant expression of ALDH1 is being evaluated as a potential novel prognostic marker in breast cancer (13), head and neck squamous cell carcinoma (7), ovarian cancer (9), and lung cancer (14). However, the role of the ALDH1 family in thyroid cancer remains poorly understood.
In this study, we systematically investigated the role of the ALDH1 family in thyroid cancer patients by searching public databases. We demonstrated that the levels of ALDH1A1/B1 had been significantly decreased in thyroid cancer patients. Aberrantly elevated ALDH1A1/B1 levels in cancer tissues were negatively correlated with OS in thyroid cancer patients. Besides, the perturbation of ALDH1A1/A3/B1 expression was significantly correlated with pathological stage. Finally, we elucidated the important but overlooked role of ALDH1A1/B1 in tumor-infiltrating lymphocytes (TILs), and we revealed a non-negligible immune regulatory role of ALDH1A1/B1 by drawing gene expression heat maps. Overall, our data provide motivation to better understand the tumor promotion and immune inhibition role of ALDH1A1/B1 in thyroid cancer patients, suggesting that ALDH1A1/B1 might serve as potential prognostic biomarkers for thyroid cancer treatment.
The Gene Expression Profiling Interactive Analysis (GEPIA) dataset is a web-based tool exploited by Tang et al. (15) to analyze the RNA sequencing (RNA-seq) expression data in cancer tissues and normal samples. Here, we used GEPIA to investigate the target genes’ expression level in thyroid cancer tissues and normal control and analyzed the relationship between target genes and pathological stage in thyroid cancer patients. And search term was limited to thyroid cancer. In addition, target proteins expressed in thyroid cancer tissues were confirmed by immunohistochemistry data acquired from The Human Protein Atlas (https://www.proteinatlas.org/).
UALCAN (http://ualcan.path.uab.edu) is a free web database based on The Cancer Genome Atlas (TCGA) level 3 RNA-seq and 31 different types of cancer (16). In the current study, we investigated the expression level of target genes in thyroid cancer tissues and corresponding adjacent tissues based on UALCAN platform, and the search term was limited to thyroid cancer. Finally, we analyzed data based on individual cancer stages, patient’s gender, patient’s age, tumor histology, and nodal metastasis. P-value was determined based on Student’s t-test, and P < 0.05 was considered as statistically significant.
Initiation and progression of many cancers could be characterized by aberrant change of gene expression (17). Here, we used Kaplan–Meier plotter (http://kmplot.com/analysis/) to access the prognostic value of ALDH1 level in thyroid cancer patients. Kaplan–Meier plotter can investigate the gene expression level in pan-caner under TCGA consortium support (18). We investigated expression levels of target genes in thyroid cancer tissues and corresponding adjacent tissues, and the search term was limited to thyroid cancer. Finally, we analyzed data based on patient’s gender to plot the OS, first progression (FP), and post progression survival (PPS) of patients with thyroid cancer, with the hazard ratio (HR) (95% confidence intervals) and log rank P-value.
Tumor IMmune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) contains 10,897 tumors from 32 cancer types (19), which could be used to systematically analyze the association between the target genes and tumor-infiltrating immune cells. Here, we investigated the somatic copy number alterations of target genes with the Somatic copy number alterations (SCNA) module. And correlation between target genes and infiltrating immune cells also was analyzed by limiting the search term to thyroid cancer.
In addition, the correlation between target genes with lymphocytes and immunomodulators was investigated by using the TISIDB database (http://cis.hku.hk/TISIDB/index.php).
inBio Discover (https://inbio-discover.com/) could visualize protein–protein interaction networks under genome scale in humans (20). inBio Discover contains over 500,000 interactions from 8 source databases. Here, we used inBio Discover to construct target gene networks by limiting the search terms to ALH1A1, ALDH1A3, and ALDH1B1.
Thyroid epithelial cell line nthy-ori 3-1 and thyroid cancer cell lines (CHO-K1 and TPC-1) were purchased from the Shanghai Zhong Qiao Xin Zhou Biotechnology Co., Ltd. Cell lines were mycoplasma negative, and nthy-ori 3-1 was cultured in RPMI 1640 medium, and thyroid cancer cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM). All cell lines were supplemented with 10% fetal bovine serum (Gibco, USA).
Five thyroid cancer tissues and their corresponding adjacent tissues were collected from the First Hospital of Jilin University and stored at -80°C conditions. All participants were given written informed consent, and the current study has been approved by the ethics committee of the First Hospital of Jilin University.
We used TransZol Up (Transgen, CN) to extract total RNA from the cell lines and cancer tissues according to manufacturer’s instructions. cDNA was synthesized by using the Hifair® II 1st Strand cDNA Synthesis Kit (Yeasen, CN) according to manufacturer’s instructions. Then, quantitative real-time PCR (qRT-PCR) was performed using a Hieff® qPCR SYBR Green Master Mix (Yeasen, CN) according to the manufacturer’s instructions. Here, β-actin was used as endogenous control. The primer sequence sets were listed as follows: β-actin: F: 5′TTCAACACCCCAGCCATG3′, R: 5′CCTCGTAGATGGGCACAGT3′; ALDH1A1: F: 5′ CCGTGGCGTACTATGGATGC3′, R: 5′GCAGCAGACGATCTCTTTCGAT3′; ALDH1B1: F: 5′AGAGTCTTACGCCTTGGACTT3′, R: 5′ GTCTTGCCATGCCACTTGTC3′.
Significance was determined with the either HR and P-values for plotting survival curves. The independent-samples Student’s t-test analysis was performed on GraphPad Prism 7.0, and P < 0.05 was considered statistically significant.
The members of the ALDH1 family have been proposed as potential therapeutic targets of ovarian cancer (21) and have shown the prognostic value in lung cancer (14) and breast carcinomas (22). However, the role of the ALDH1 family in thyroid cancer is still ill-defined. Here, we investigated the expression of the ALDH1 family in thyroid cancer, and we found that all ALDH1 family members could be detected in thyroid cancer tissues at the transcriptional level (Figure 1A). Interestingly, the expressions of ALDH1A1 and ALDH1B1 were higher in the normal thyroid tissues and decreased in the thyroid cancer tissues (Figure 1A). In contrast, the level of ALDH1A3 was slightly increased in cancer tissues (Figure 1A). The low expression levels of other genes in the thyroid tissues were negligible. Furthermore, immunohistochemical results verified that ALDH1A1, ALDH1A3, and ALDH1B1 were highly expressed in thyroid cancer at the protein level (Figures 1B–D), suggesting the role of ALDH1A1/A3/B1 in thyroid cancer development. Therefore, we determined the expressions of ALDH1A1/A3/B1 in thyroid cancer tissues.
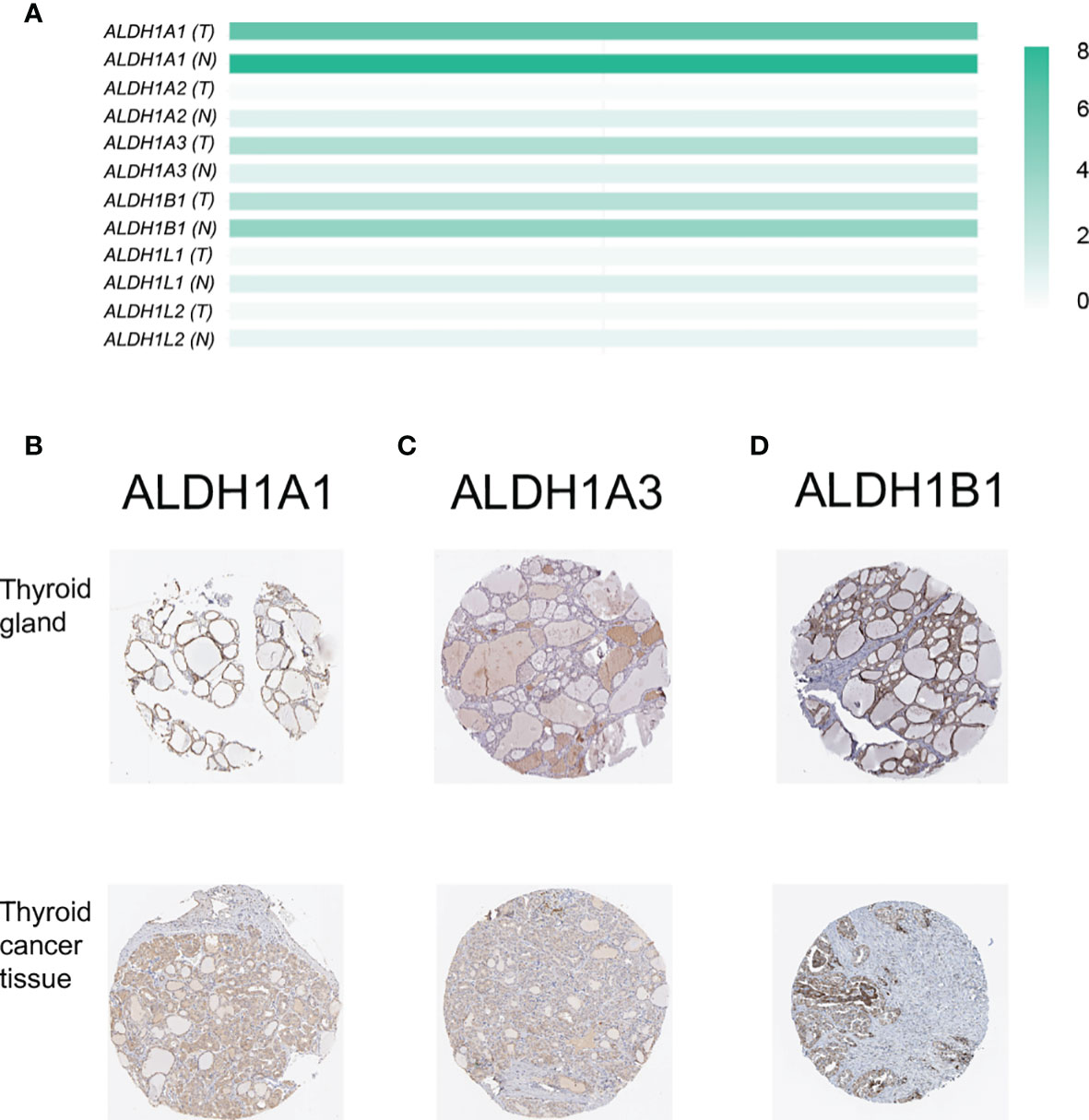
Figure 1 Confirmation of the expression of ALDH1 family members in thyroid cancer tissues. (A) ALDH1A1/A3/B1 were the top 3 genes expressed in thyroid cancer tissues (T, tumor tissues, n = 503; N, normal tissues, n = 59). (B–D) ALDH1A1/A3/B1 expression was confirmed by immunohistochemistry data from The Human Protein Atlas. Data in panel (A) were collected from the Gene Expression Profiling Interactive Analysis (GEPIA) database. Data in panels (B–D) were collected from The Human Protein Atlas database.
As the expressions of ALDH1A1/A3/B1 were relatively high in thyroid cancer tissues, we assessed the level of ALDH1A1/A3/B1 in thyroid tumors and normal tissues. Our results showed that the transcriptional levels of ALDH1A1 and ALDH1B1 significantly decreased in thyroid cancer tissues, whereas that of ALDH1A3 increased (Figures 2A–C). Next, we further assessed the changes in ALDH1A1/A3/B1 based on different classifications. As shown in Figure 2, mRNA expressions of ALDH1A1 and ALDH1B1 significantly decreased in thyroid cancer tissues compared to that in normal groups based on individual cancer stages (Figures 2D–F), gender (Figures 2G–I), tumor histology (Figures 2J–L), and nodal metastasis (Figures 2M–O). Meanwhile, the levels of ALDH1A3 significantly increased (Figures 2B, E, H, K, N). These results indicate that ALDH1A1/A3/B1 may be correlated with and may play a significant role in thyroid cancer progression.
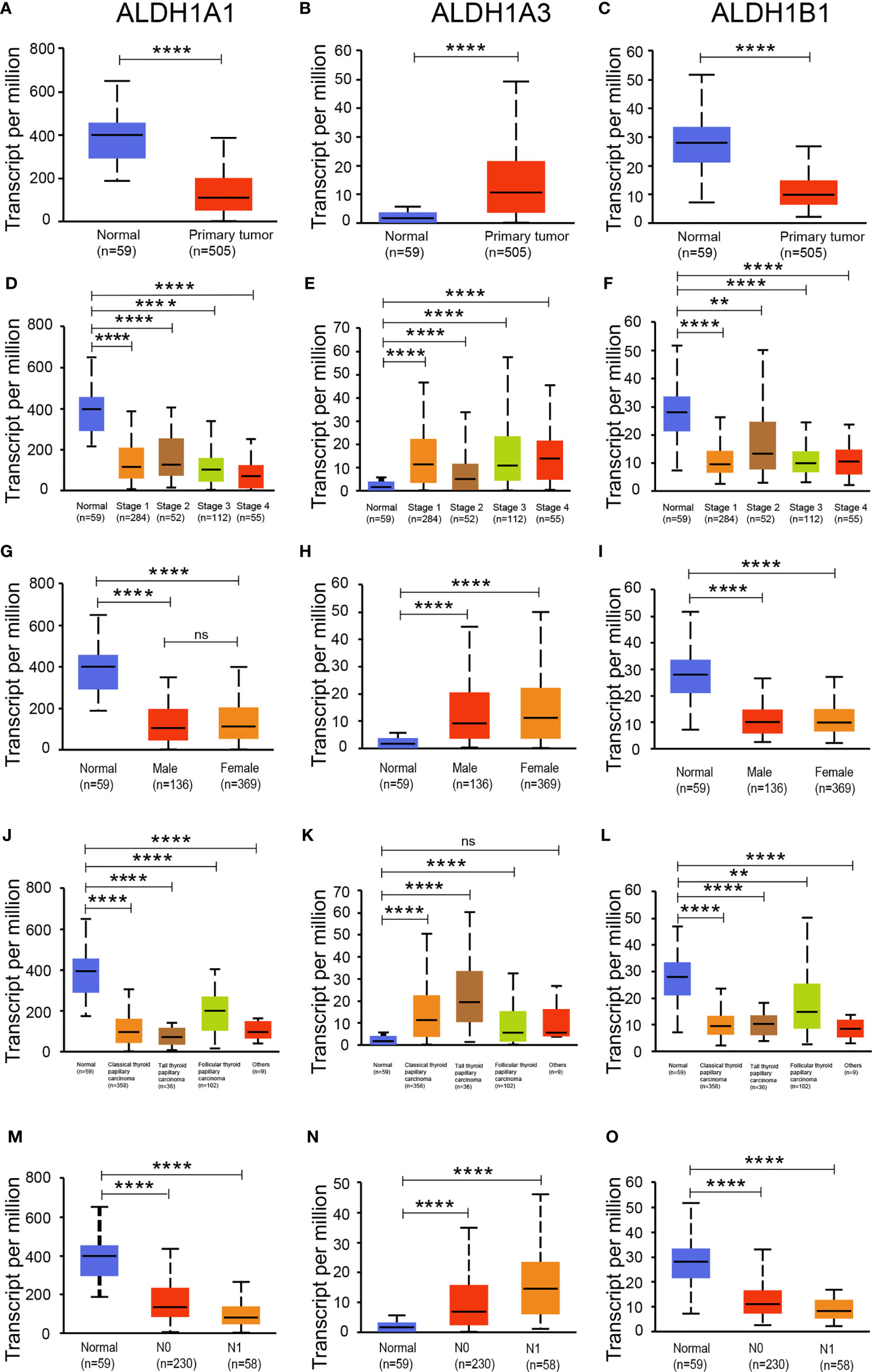
Figure 2 Expression of ALDH1A1/B1 was significantly decreased in thyroid cancer tissues. Differential expression of ALDH1A1 (A), ALDH1A3 (B), and ALDH1B1 (C) in thyroid cancer tissues and normal tissues based on individual cancer stage (D–F), patient’s gender (G–I), histological subtype (J–L), and nodal metastasis status (M–O) (N0, no regional lymph node metastasis; N1, metastases in 1–3 axillary lymph nodes; N2, metastases in 4–9 axillary lymph nodes; N3, metastases in 10 or more axillary lymph nodes). **P < 0.01; ****P < 0.0001. Data were collected from the UALCAN database. ns, No significance.
To determine if ALDH1A1/A3/B1 is associated with tumor progression and clinical outcome of thyroid cancer, we assessed the correlation between the expression of ALDH1A1/A3/B1 and the pathological stage of the patients. Results showed that there was a significant correlation between the expression of ALDH1A1, ALDH1A3, and ALDH1B1 with the pathological stage (Figures 3A–C). These results indicate that the aberrant expression of ALDH1A1/A3/B1 may correlate with tumorigenesis and development.
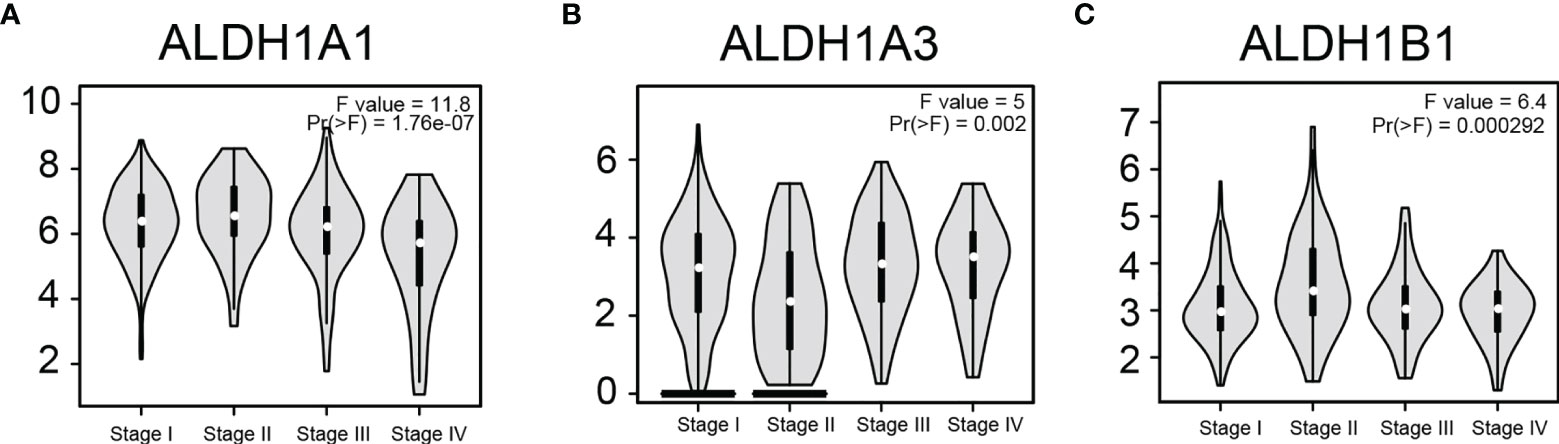
Figure 3 Correlation between differentially expressed ALDH1A1/A3/B1 and the pathological stage of thyroid cancer patients. Violin plots showed the distribution of ALDH1A1/A3/B1 [(A–C), respectively] mRNA expression correlated with tumor stage in thyroid cancer patients (Stage I, n = 284; stage II, n = 52; stage III, n = 112; stage IV, n = 55). Data were collected from the Gene Expression Profiling Interactive Analysis (GEPIA) database.
Furthermore, we assessed the prognostic significance of ALDH1A1/A3/B1 in patients using Kaplan–Meier plotter analysis. As shown in Figure 4, patients were divided into low- and high-risk groups based on the expression levels of ALDH1A1/A3/B1. Our results showed that changes in the expression level of ALDH1A1/A3 had no influence over the OS of female patients with thyroid cancer (Figures 4A, B, D, E); however, increased mRNA level of ALDH1A1 impaired OS in male patients with thyroid cancer (Figure 4C). Moreover, high mRNA expression of ALDH1A3 only enhanced OS in male patients with thyroid cancer (Figure 4F). And high levels of ALDH1B1 also impaired OS in patients with thyroid cancer (Figures 4G–I). Taken together, we identified that ALDH1A1/A3/B1 in thyroid cancer patients were significantly correlated with OS in male patients, supporting the possible prognostic value of these enzymes particularly for male patients with thyroid cancer.
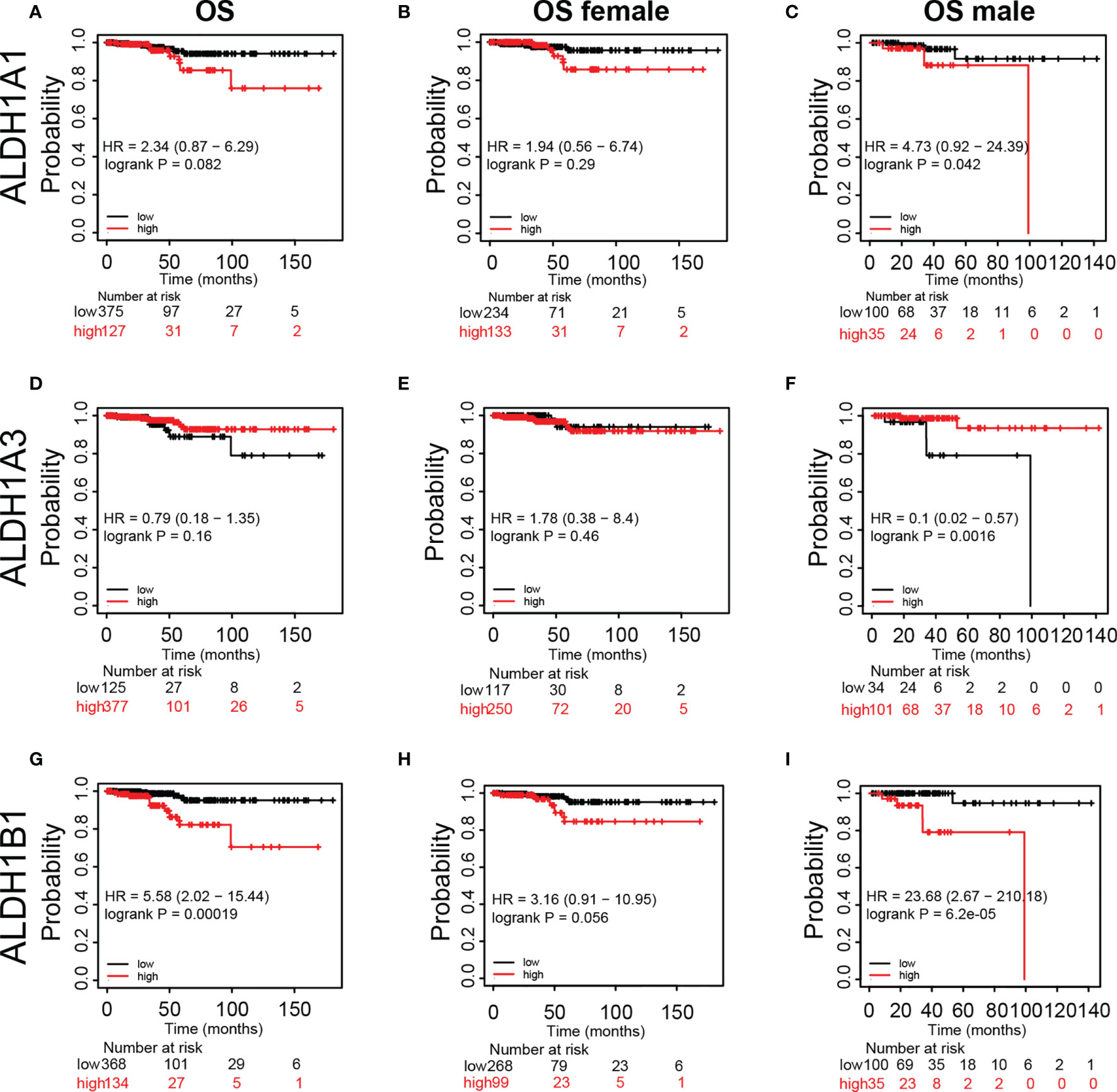
Figure 4 Kaplan–Meier plot revealing the correlation between ALDH1A1/A3/B1 level and overall survival (OS) in thyroid cancer patients. Patients were divided into two groups based on ALDH1A1/A3/B1 level [(A, D, G), respectively], and Kaplan–Meier plot of ALDH1A1/A3/B1 in female patients (B, E, H) and male patients (C, F, I) with thyroid cancer (total number of patients with thyroid cancer, n = 502; total number of female patients with thyroid cancer, n = 367; total number of male patients with thyroid cancer, n = 135). Data were collected from the Kaplan–Meier plotter database.
Tumor-infiltrating lymphocytes (TILs) are shaped by interactions between different types of killer cell, regulatory cell, stroma cell, and tumor cell in the tumor microenvironment (23). Recent studies have shown that tumor progression or suppression is closely associated with TIL response in the tumor microenvironment (24). Thus, we investigated the relationship between ALDH1A1/A3/B1 expression and TILs using TIMER. ALDH1A1/B1 showed weak correlation with the six key types of immune cell in the tumor microenvironment (Figures 5A, C). Meanwhile, a significant correlation between ALDH1A3 expression and neutrophils, CD4+ T cells, dendritic cells, macrophages, and B cells (Figure 5B) was observed. These results suggest that ALDH1A1/A3/B1 may affect multiple cells in the tumor microenvironment that may participate in the tumor immune response.
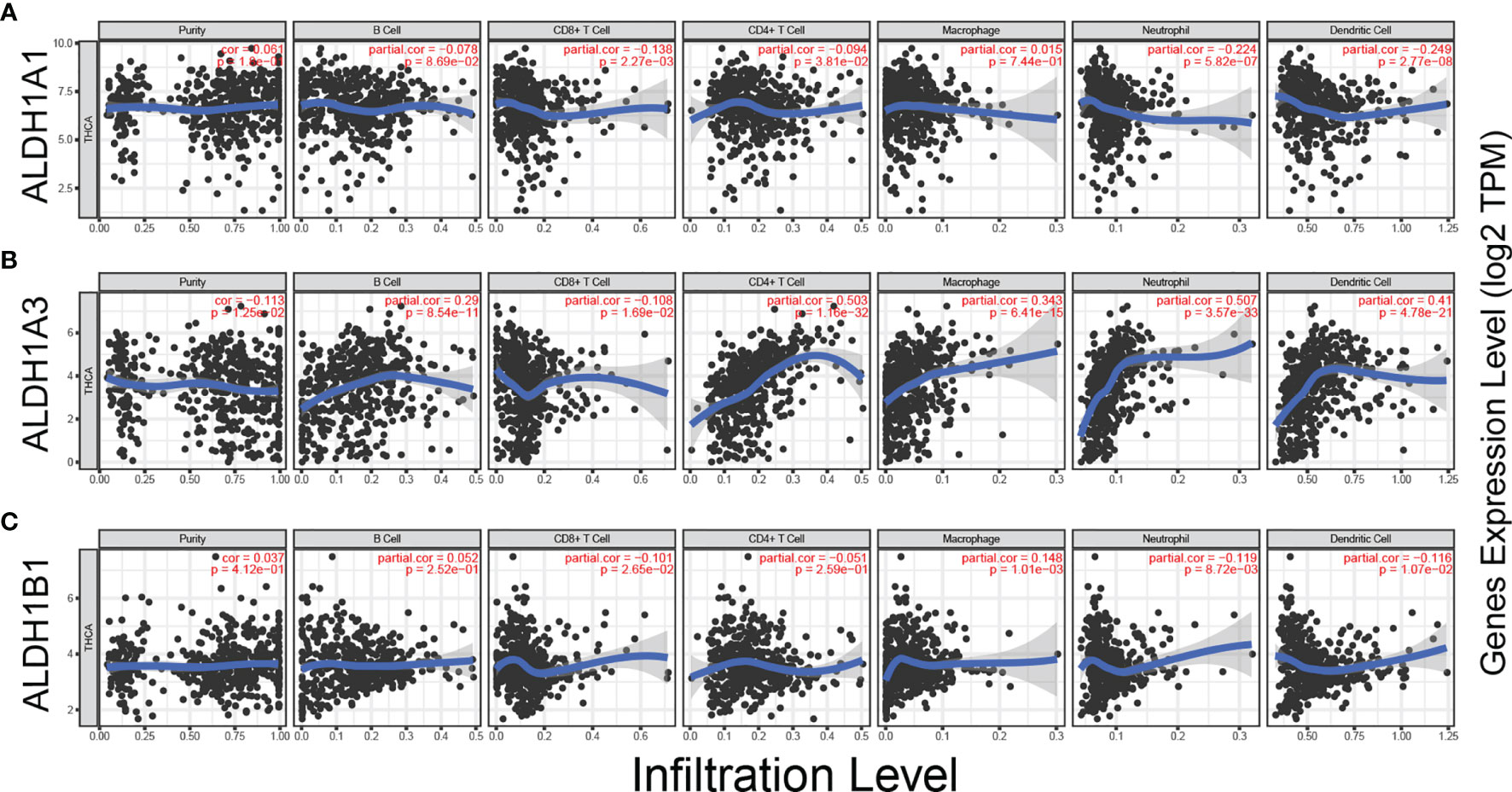
Figure 5 Correlation between ALDH1A1/A3/B1 expression and immune infiltration in thyroid cancer. (A–C) Correlation of ALDH1A1/A3/B1 expression with infiltration levels of CD8+ T cell, CD4+ T cell, Treg cell, B cell, neutrophil, macrophage, dendritic cell, natural killer cell, and monocyte in thyroid cancer (total number of patients with thyroid cancer, n = 223). Data were collected from the Tumor IMmune Estimation Resource.
Furthermore, to study the role of ALDH1A1/A3/B1 in thyroid cancer, we constructed protein–protein interaction networks by using the tool of inBio Discover. The results showed that ALDH1A1/A3/B1 could interact with various proteins (Figure 6), most of which were identified as enzymes. Besides, we showed that ALDH1A1 and ALDH1B1 could interact with transcription factors [CCAAT/enhancer-binding protein β (CEBPB) and CCHC-type zinc finger nucleic acid-binding protein (CNBP), respectively] and membrane-bound proteins [ATP-binding cassette subfamily C member 6 (ABCC6) and mucin 1 (MUC1)], suggesting that the aberrant changes of ALDH1A1/B1 may alter multiple protein expressions and intracellular signal transduction. Indeed, our Gene Ontology (GO) enrichment analysis revealed a close relationship between ALDH1A1/A3/B1 and ethanol oxidation (GO:0006069), as well as retinol metabolic process (GO:0042572), which are known to be important in detoxification (8), tumorigenesis (25), and thyroid tumor progression (Supplementary Material) (26). Specifically, we found that ALDH1A1/A3/B1 were enriched in thyroid hormone binding (GO:0070324). Similarly, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis also showed that ALDH1A1/A3/B1 were associated with metabolic pathways (Supplementary Material). Collectively, these results indicate that ALDH1A1/A3/B1 may influence thyroid cancer via metabolism and interaction with various enzymes and transcription factors.
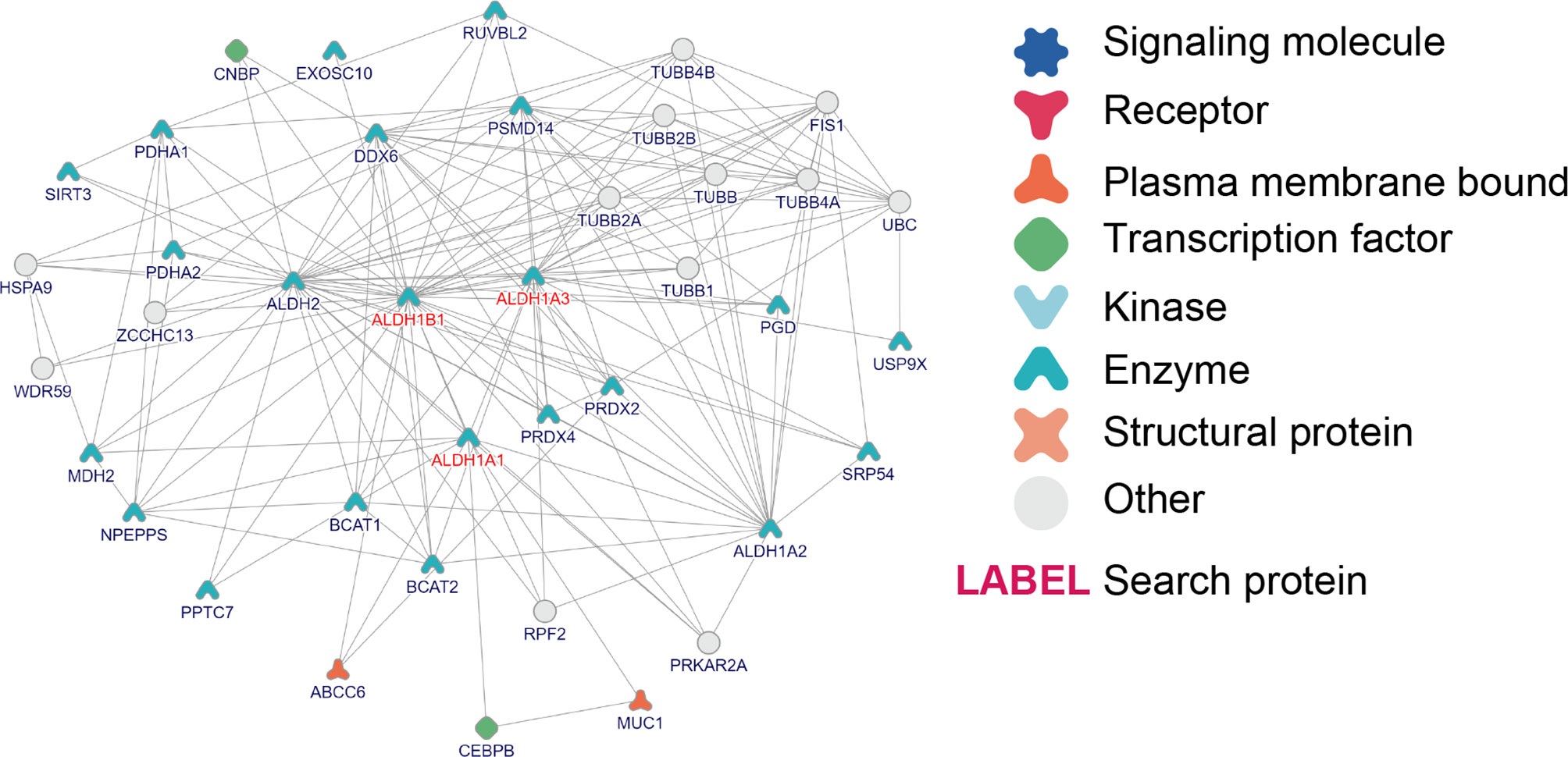
Figure 6 Interaction and Gene Ontology (GO)/pathway analysis of ALDH1A1/A3/B1. Protein–protein interaction network of ALDH1A1/A3/B1, constructed by the online tool inBio Discover.
Immune modulation is a critical factor that should be considered in cancer therapy, as numerous genes play a role in response to cancer immunotherapy (27). Using the TISIDB database, we demonstrated that ALDH1A1/A3/B1 had significant negative correlations with immune-stimulating genes, such as CD276, TMEM173, TNFSF9, CD70, TNFRSF25, and CD40 in thyroid cancer (Figure 7A). Moreover, high levels of ALDH1A1/B1 were also associated with restrained expression of major histocompatibility complex (MHC) genes (HLA-G, HLA-B, HLA-DOB, and TAP1) (Figure 7B). Notably, previous studies have demonstrated that higher levels of immune stimulators and MHC promote better therapeutic effect in cancer patients (28, 29). In addition, the expressions of ALDH1A1/B1 were found to be negatively correlated with chemokines (CXCL5, CXCL16, and CCL13; and CXCL14, respectively) (Figure 7C) and receptors (CXCR2, CCR4, and CCR8, and CCR9, CCR10, respectively) (Figure 7D). Therefore, ALDH1A1/B1 may be involved in regulating the above immune molecules.
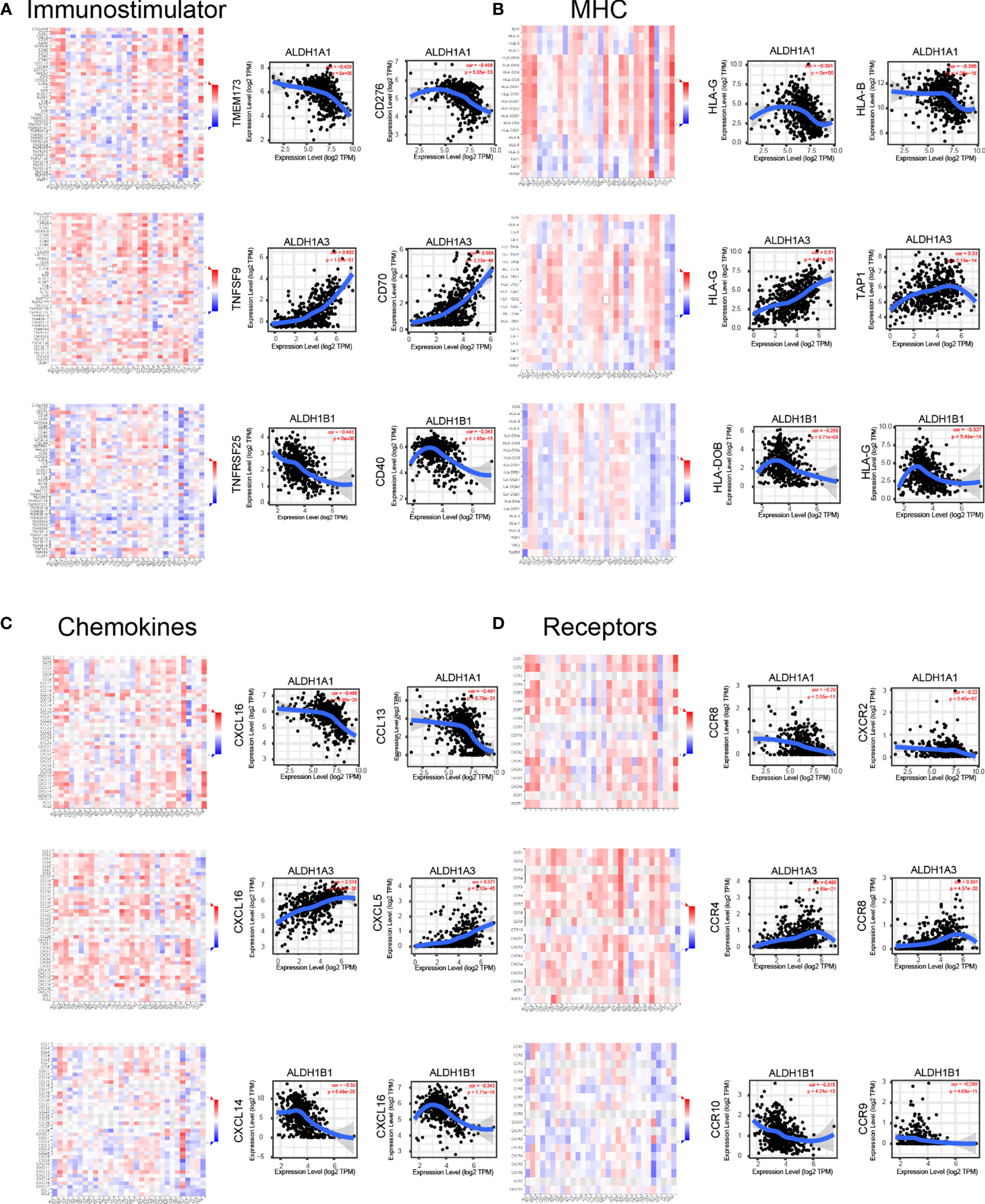
Figure 7 Correlation of ALDH1A1/A3/B1 with lymphocytes and immunomodulators. Relations between the abundance of immunostimulators (A), MHC (B), chemokines (C), and receptors (D) and ALDH1A1/A3/B1 expression and the top 2 genes displaying the greatest Spearman’s correlation with ALDH1A1/A3/B1 expression. Red and blue cells indicate positive and negative correlations, respectively. The color intensity is directly proportional to the strength of the correlations. Data were collected from the TISIDB and TIMER database (total number of patients with thyroid cancer, n = 223). TILs, tumor-infiltrating lymphocytes, MHC major histocompatibility complex; TIMER, Tumor IMmune Estimation Resource.
We have observed a decreased expression of ALDH1A1/B1 in thyroid cancer tissues (Figures 1A–D), which was correlated with poor survival of patients (Figures 4C, G, I). Thus, we aimed to verify the levels of ALDH1A1/B1 in thyroid cancer cell lines and thyroid cancer tissues in vitro. We found that ALDH1A1/B1 expression was significantly reduced in CHO-K1 and TPC-1 cells (Figures 8A, B). In addition, we also detected a decreased expression of ALDH1A1/B1 in thyroid cancer tissues (Figures 8C, D). These results confirmed the effectiveness and reliability of the results in bioinformatic analysis.
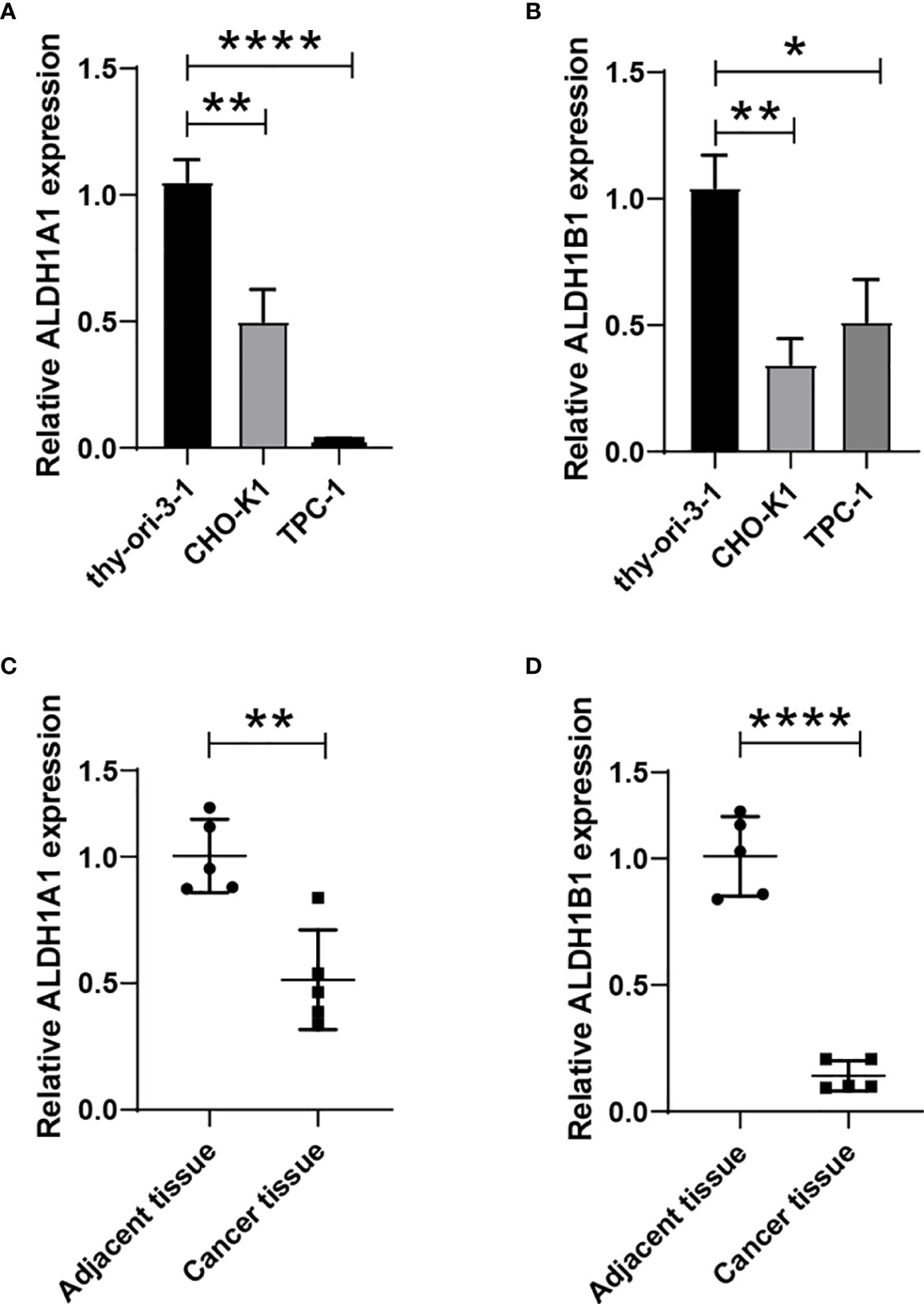
Figure 8 Reduced expression of ALDH1A1/B1 in thyroid cancer cell lines and thyroid cancer tissues. Thyroid epithelial cell line nthy-ori 3-1 and thyroid cancer cell lines (CHO-K1 and TPC-1) were collected, and the levels of ALDH1A1 (A) and ALDH1B1 (B) were detected by using qRT-PCR. Five thyroid cancer tissues and corresponding adjacent tissues were collected from the First Hospital of Jilin University to verify the expression of ALDH1A1 (C) and ALDH1B1 (D) in patients. *P < 0.05; **P < 0.01; ****P < 0.0001.
ALDH1 is a critical gene mediating the enzymatic detoxification of aldehydes (8), and it is also required in cell proliferation (30) and stem cell differentiation (31) in cancers. Although all six ALDH1 members have been reported to be intracellularly involved in various biological processes, ALDH1A1/A3/B1 were the highly expressed genes in thyroid cancer tissues in this study. We observed that the regulatory functions of ALDH1A1 and ALDH1B1 in thyroid cancer were involved in poor outcome in patients and inhibition of TILs.
ALDH1 has been reported to play a role in promoting triple-negative breast cancer progression and enhanced drug resistance in a Notch receptor 4 (NOTCH4)-dependent manner (32, 33). Ciccone et al. (31) observed that overexpression of ALDH1A1 was significantly associated with Vascular endothelial growth factor (VEGF)-mediated angiogenesis by retinoic acid/ Hypoxia-inducible factor (HIF)-1α signal. Similarly, a study on esophageal cancer demonstrated that ALDH1 was positively correlated with higher clinical stage and poor prognosis (34). Here, we showed that ALDH1A1/B1 expressions were significantly decreased in thyroid cancer patients, but ALDH1A3 was elevated. However, the level of ALDH1A3 had no significant changes. These results indicated the different roles of ALDH1 members in thyroid cancer progression. ALDH1A1 could regulate retinoid signaling via the CEBPB to promote stemness maintenance in cancer stem cells (35). In addition, ALDH1A1 also profoundly affected acetaldehyde metabolism and drug resistance in chemotherapy (8), suggesting that ALDH1A1 could broadly regulate biological processes in cancer. Indeed, our results showed that elevated ALDH1A1 was negatively correlated with OS of thyroid cancer patients in male patients but was beneficial to Relapse-free survival (RFS) in female patients; it indicated that differently expressed gonadal hormones may also be involved in ALDH1A1 regulation in thyroid cancer development.
Although the tumorigenic property of ALDH1A3 in cancer has been observed (8, 36), it is suggested that upregulated epithelial-to-mesenchymal transition and cancer stem cell-like properties related genes, including ALDH13, may be involved in the lymphatic spread of papillary thyroid microcarcinoma (37). However, another study found that administration of ALDH inhibitor had no impact on proliferation or spherogenicity in several thyroid cancer cell lines (38). Here, we showed that aberrant expression of ALDH1A3 was not associated with OS in women. Meanwhile, we noted that there was a significant correlation between the expression of ALDH1A3 with the pathological stage in patients. These data suggest that increased ALDH1A3 may only affect alternative subtypes of thyroid cancer. However, the role of ALDH1A3 in thyroid cancer development still needs further study.
Upregulation of ALDH1B1 by Kras is associated with pancreatic lineage homeostasis and initiation/development of pancreatic ductal adenocarcinoma, which are characteristics of enhanced production of reactive oxygen species by altering mitochondrial metabolism (39). ALDH1B1 is an enzyme that has been proposed to play an important role in acetaldehyde detoxification. Increased ALDH1B1 level was involved in the regulation of the Wnt-, Notch-, and Phosphatidylinositol-3-kinase (PI3K)/Akt signaling pathways in colon tumorigenesis (40). In this study, high ALDH1B1 expression in thyroid cancer was associated with poor OS in male patients, suggesting a correlation between ALDH1B1 and androgen. Notably, a study in prostate cell lines showed that cells with elevated ALDH1B1 expression frequently expressed androgen receptor (41). In addition, a study in prostate cancer cell line LNCaP reported that androgen dihydrotestosterone was involved in aldehyde dehydrogenase regulation (42). Also, a high level of aldehyde dehydrogenase promoted metastatic growths in stem-like prostate cancer cells (43). Therefore, androgen receptor expression may be involved in ALDH1B1-induced thyroid cancer progression.
We have shown the negative correlation between ALDH1A1/B1 and several immune-stimulating genes. In addition, we observed an opposite expression of the ALDH1A1/B1 and MHC genes. During thyroid cancer progression, the immune response relies on MHCs to recognize and present antigens to cytotoxic T lymphocytes. Low levels of immune-stimulating genes and MHCs would inhibit immune cell activation and induce a weaker response or promote immunological unresponsiveness (44). Recently, several chemokines were reported to be highly expressed in thyroid cancer tissues, which might be involved in cancer progression and invasion by activating Akt and Erk signaling pathway (45). Our results revealed the importance of chemokines and receptors in thyroid cancer, which were in agreement with the study that emphasized the crucial role of chemokine receptors during papillary thyroid carcinoma development (45). As soluble mediators secreted by cells in tumor microenvironments, different chemokines recruit various types of immune cells by binding to its corresponding receptors (46). Moreover, targeting chemokine and chemokine receptor has been implicated in non-small cell lung cancer (47), gastric cancer (48), and breast cancer therapy (49). Of note, our results showed a negative correlation between ALDH1A1/B1 and several chemokines/receptors in thyroid cancer tissues. Thus, some chemokines/receptors that showed decreased expression in thyroid cancer tissues may be avoided as potential therapeutic targets.
In conclusion, we have revealed the aberrant expressions of ALDH1A1/B1 in thyroid cancer tissues that were significantly correlated with the OS of patients and with the pathological stage. In addition, we found that elevated ALDH1A1/B1 expression was negatively associated with chemokine/receptor expression and involved in several TILs. Thus, ALDH1A1/B1 are potential biomarkers for thyroid cancer and might be valuable therapeutic targets for diagnosis and therapy. However, future studies are required to validate our findings and promote the clinical utility of ALDH1A1/B1 in thyroid cancer treatment.
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
All participants were given written informed consent, and the current study has been approved by the ethics committee of the First Hospital of Jilin University. The patients/participants provided their written informed consent to participate in this study.
YC performed the experiments. YC and YLiu analyzed the data and wrote the article. LM collected samples. YC, YLiu, YLi, and GW revised the article. GW conceived the idea and supervised the project. All authors contributed to the article and approved the submitted version.
This work was supported by Scientific Research Funds of the Education Department of Liaoning Province (JYTQN2020018).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.821958/full#supplementary-material.
1. Ancker OV, Krüger M, Wehland M, Infanger M, Grimm D. Multikinase Inhibitor Treatment in Thyroid Cancer. Int J Mol Sci (2019) 21(1):10–29. doi: 10.3390/ijms21010010
2. Raue F, Frank-Raue K. Thyroid Cancer: Risk-Stratified Management and Individualized Therapy. Clin Cancer Res an Off J Am Assoc Cancer Res (2016) 22(20):5012–21. doi: 10.1158/1078-0432.ccr-16-0484
3. Varricchi G, Loffredo S, Marone G, Modestino L, Fallahi P, Ferrari SM, et al. The Immune Landscape of Thyroid Cancer in the Context of Immune Checkpoint Inhibition. Int J Mol Sci (2019) 20(16):3934–65. doi: 10.3390/ijms20163934
4. Cabanillas ME, McFadden DG, Durante C. Thyroid Cancer. Lancet (London England) (2016) 388(10061):2783–95. doi: 10.1016/s0140-6736(16)30172-6
5. Nilubol N, Zhang L, Kebebew E. Multivariate Analysis of the Relationship Between Male Sex, Disease-Specific Survival, and Features of Tumor Aggressiveness in Thyroid Cancer of Follicular Cell Origin. Thyroid Off J Am Thyroid Assoc (2013) 23(6):695–702. doi: 10.1089/thy.2012.0269
6. Abdullah MI, Junit SM, Ng KL, Jayapalan JJ, Karikalan B, Hashim OH. Papillary Thyroid Cancer: Genetic Alterations and Molecular Biomarker Investigations. Int J Med Sci (2019) 16(3):450–60. doi: 10.7150/ijms.29935
7. Zhou C, Sun B. The Prognostic Role of the Cancer Stem Cell Marker Aldehyde Dehydrogenase 1 in Head and Neck Squamous Cell Carcinomas: A Meta-Analysis. Oral Oncol (2014) 50(12):1144–8. doi: 10.1016/j.oraloncology.2014.08.018
8. Tomita H, Tanaka K, Tanaka T, Hara A. Aldehyde Dehydrogenase 1A1 in Stem Cells and Cancer. Oncotarget (2016) 7(10):11018–32. doi: 10.18632/oncotarget.6920
9. Kim D, Choi BH, Ryoo IG, Kwak MK. High NRF2 Level Mediates Cancer Stem Cell-Like Properties of Aldehyde Dehydrogenase (ALDH)-High Ovarian Cancer Cells: Inhibitory Role of All-Trans Retinoic Acid in ALDH/NRF2 Signaling. Cell Death Disease (2018) 9(9):896. doi: 10.1038/s41419-018-0903-4
10. Ruscito I, Darb-Esfahani S, Kulbe H, Bellati F, Zizzari IG, Rahimi Koshkaki H, et al. The Prognostic Impact of Cancer Stem-Like Cell Biomarker Aldehyde Dehydrogenase-1 (ALDH1) in Ovarian Cancer: A Meta-Analysis. Gynecologic Oncol (2018) 150(1):151–7. doi: 10.1016/j.ygyno.2018.05.006
11. Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell (2007) 1(5):555–67. doi: 10.1016/j.stem.2007.08.014
12. Lin CC, Lo MC, Moody RR, Stevers NO, Tinsley SL, Sun D. Doxycycline Targets Aldehyde Dehydrogenase−Positive Breast Cancer Stem Cells. Oncol Rep (2018) 39(6):3041–7. doi: 10.3892/or.2018.6337
13. Li J, Zhang B, Yang YF, Jin J, Liu YH. Aldehyde Dehydrogenase 1 as a Predictor of the Neoadjuvant Chemotherapy Response in Breast Cancer: A Meta-Analysis. Medicine (2018) 97(34):e12056. doi: 10.1097/md.0000000000012056
14. Wei D, Peng JJ, Gao H, Zhang T, Tan Y, Hu YH. ALDH1 Expression and the Prognosis of Lung Cancer: A Systematic Review and Meta-Analysis. Heart Lung Circulation (2015) 24(8):780–8. doi: 10.1016/j.hlc.2015.03.021
15. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res (2017) 45(W1):W98–102. doi: 10.1093/nar/gkx247
16. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (New York NY) (2017) 19(8):649–58. doi: 10.1016/j.neo.2017.05.002
17. Prevarskaya N, Skryma R, Shuba Y. Ion Channels in Cancer: Are Cancer Hallmarks Oncochannelopathies? Physiol Rev (2018) 98(2):559–621. doi: 10.1152/physrev.00044.2016
18. Nagy Á, Munkácsy G, Győrffy B. Pancancer Survival Analysis of Cancer Hallmark Genes. Sci Rep (2021) 11(1):6047. doi: 10.1038/s41598-021-84787-5
19. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res (2017) 77(21):e108–10. doi: 10.1158/0008-5472.can-17-0307
20. Li T, Wernersson R, Hansen RB, Horn H, Mercer J, Slodkowicz G, et al. A Scored Human Protein-Protein Interaction Network to Catalyze Genomic Interpretation. Nat Methods (2017) 14(1):61–4. doi: 10.1038/nmeth.4083
21. Chefetz I, Grimley E, Yang K, Hong L, Vinogradova EV, Suciu R, et al. A Pan-ALDH1A Inhibitor Induces Necroptosis in Ovarian Cancer Stem-Like Cells. Cell Rep (2019) 26(11):3061–3075.e6. doi: 10.1016/j.celrep.2019.02.032
22. Demir H, Dulgar O, Gulle BT, Turna H, Ilvan S. Prognostic Value of Aldehyde Dehydrogenase 1 (ALDH1) in Invasive Breast Carcinomas. Bosnian J basic Med Sci (2018) 18(4):313–9. doi: 10.17305/bjbms.2018.3094
23. Petitprez F, Meylan M, de Reyniès A, Sautès-Fridman C, Fridman WH. The Tumor Microenvironment in the Response to Immune Checkpoint Blockade Therapies. Front Immunol (2020) 11:784. doi: 10.3389/fimmu.2020.00784
24. Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive Analyses of Tumor Immunity: Implications for Cancer Immunotherapy. Genome Biol (2016) 17(1):174. doi: 10.1186/s13059-016-1028-7
25. Wang W, Wang C, Xu H, Gao Y. Aldehyde Dehydrogenase, Liver Disease and Cancer. Int J Biol Sci (2020) 16(6):921–34. doi: 10.7150/ijbs.42300
26. Kurashige T, Shimamura M, Yasui K, Mitsutake N, Matsuse M, Nakashima M, et al. Studies on Expression of Aldehyde Dehydrogenase in Normal and Cancerous Tissues of Thyroids. Hormone Metab Res = Hormon- und Stoffwechselforschung = Hormones metabolisme (2015) 47(3):194–9. doi: 10.1055/s-0034-1387770
27. Nixon AB, Schalper KA, Jacobs I, Potluri S, Wang IM, Fleener C. Peripheral Immune-Based Biomarkers in Cancer Immunotherapy: Can We Realize Their Predictive Potential? J Immunotherapy Cancer (2019) 7(1):325. doi: 10.1186/s40425-019-0799-2
28. Huang Y, Kim BYS, Chan CK, Hahn SM, Weissman IL, Jiang W. Improving Immune-Vascular Crosstalk for Cancer Immunotherapy. Nat Rev Immunol (2018) 18(3):195–203. doi: 10.1038/nri.2017.145
29. Axelrod ML, Cook RS, Johnson DB, Balko JM. Biological Consequences of MHC-II Expression by Tumor Cells in Cancer. Clin Cancer Res an Off J Am Assoc Cancer Res (2019) 25(8):2392–402. doi: 10.1158/1078-0432.ccr-18-3200
30. Fischer AK, Pham DL, Bösmüller H, Lengerke C, Wagner P, Bachmann C, et al. Comprehensive in Situ Analysis of ALDH1 and SOX2 Reveals Increased Expression of Stem Cell Markers in High-Grade Serous Carcinomas Compared to Low-Grade Serous Carcinomas and Atypical Proliferative Serous Tumors. Virchows Archiv an Int J Pathology (2019) 475(4):479–88. doi: 10.1007/s00428-019-02647-0
31. Ciccone V, Terzuoli E, Donnini S, Giachetti A, Morbidelli L, Ziche M. Stemness Marker ALDH1A1 Promotes Tumor Angiogenesis via Retinoic Acid/HIF-1α/VEGF Signalling in MCF-7 Breast Cancer Cells. J Exp Clin Cancer Res CR (2018) 37(1):311. doi: 10.1186/s13046-018-0975-0
32. Simões BM, O'Brien CS, Eyre R, Silva A, Yu L, Sarmiento-Castro A, et al. Anti-Estrogen Resistance in Human Breast Tumors Is Driven by JAG1-NOTCH4-Dependent Cancer Stem Cell Activity. Cell Rep (2015) 12(12):1968–77. doi: 10.1016/j.celrep.2015.08.050
33. Panigoro SS, Kurnia D, Kurnia A, Haryono SJ, Albar ZA. ALDH1 Cancer Stem Cell Marker as a Prognostic Factor in Triple-Negative Breast Cancer. Int J Surg Oncol (2020) 2020:7863243. doi: 10.1155/2020/7863243
34. Chen MF, Chen PT, Lu MS, Chen WC. Role of ALDH1 in the Prognosis of Esophageal Cancer and its Relationship With Tumor Microenvironment. Mol Carcinogenesis. (2018) 57(1):78–88. doi: 10.1002/mc.22733
35. Wang J, Wang L, Ho CT, Zhang K, Liu Q, Zhao H. Garcinol From Garcinia Indica Downregulates Cancer Stem-Like Cell Biomarker ALDH1A1 in Nonsmall Cell Lung Cancer A549 Cells Through DDIT3 Activation. J Agric Food Chem (2017) 65(18):3675–83. doi: 10.1021/acs.jafc.7b00346
36. Nie S, Qian X, Shi M, Li H, Peng C, Ding X, et al. ALDH1A3 Accelerates Pancreatic Cancer Metastasis by Promoting Glucose Metabolism. Front Oncol (2020) 10:915. doi: 10.3389/fonc.2020.00915
37. Lee S, Bae JS, Jung CK, Chung WY. Extensive Lymphatic Spread of Papillary Thyroid Microcarcinoma is Associated With an Increase in Expression of Genes Involved in Epithelial-Mesenchymal Transition and Cancer Stem Cell-Like Properties. Cancer Med (2019) 8(15):6528–37. doi: 10.1002/cam4.2544
38. Shimamura M, Kurashige T, Mitsutake N, Nagayama Y. Aldehyde Dehydrogenase Activity Plays No Functional Role in Stem Cell-Like Properties in Anaplastic Thyroid Cancer Cell Lines. Endocrine (2017) 55(3):934–43. doi: 10.1007/s12020-016-1224-y
39. Mameishvili E, Serafimidis I, Iwaszkiewicz S, Lesche M, Reinhardt S, Bölicke N, et al. Aldh1b1 Expression Defines Progenitor Cells in the Adult Pancreas and is Required for Kras-Induced Pancreatic Cancer. Proc Natl Acad Sci USA (2019) 116(41):20679–88. doi: 10.1073/pnas.1901075116
40. Singh S, Arcaroli J, Chen Y, Thompson DC, Messersmith W, Jimeno A, et al. ALDH1B1 Is Crucial for Colon Tumorigenesis by Modulating Wnt/β-Catenin, Notch and PI3K/Akt Signaling Pathways. PloS One (2015) 10(5):e0121648. doi: 10.1371/journal.pone.0121648
41. Gangavarapu KJ, Azabdaftari G, Morrison CD, Miller A, Foster BA, Huss WJ. Aldehyde Dehydrogenase and ATP Binding Cassette Transporter G2 (ABCG2) Functional Assays Isolate Different Populations of Prostate Stem Cells Where ABCG2 Function Selects for Cells With Increased Stem Cell Activity. Stem Cell Res Ther (2013) 4(5):132. doi: 10.1186/scrt343
42. Trasino SE, Harrison EH, Wang TT. Androgen Regulation of Aldehyde Dehydrogenase 1A3 (ALDH1A3) in the Androgen-Responsive Human Prostate Cancer Cell Line LNCaP. Exp Biol Med (Maywood NJ) (2007) 232(6):762–71. doi: 10.3181/00379727-232-2320762
43. Miftakhova R, Hedblom A, Semenas J, Robinson B, Simoulis A, Malm J, et al. Cyclin A1 and P450 Aromatase Promote Metastatic Homing and Growth of Stem-Like Prostate Cancer Cells in the Bone Marrow. Cancer Res (2016) 76(8):2453–64. doi: 10.1158/0008-5472.can-15-2340
44. Rock KL, Reits E, Neefjes J. Present Yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol (2016) 37(11):724–37. doi: 10.1016/j.it.2016.08.010
45. Zhu X, Bai Q, Lu Y, Lu Y, Zhu L, Zhou X, et al. Expression and Function of CXCL12/CXCR4/CXCR7 in Thyroid Cancer. Int J Oncol (2016) 48(6):2321–9. doi: 10.3892/ijo.2016.3485
46. Mollica Poeta V, Massara M, Capucetti A, Bonecchi R. Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy. Front Immunol (2019) 10:379. doi: 10.3389/fimmu.2019.00379
47. Wang CL, Sun BS, Tang Y, Zhuang HQ, Cao WZ. CCR1 Knockdown Suppresses Human Non-Small Cell Lung Cancer Cell Invasion. J Cancer Res Clin Oncol (2009) 135(5):695–701. doi: 10.1007/s00432-008-0505-0
48. Aldinucci D, Casagrande N. Inhibition of the CCL5/CCR5 Axis Against the Progression of Gastric Cancer. Int J Mol Sci (2018) 19(5):1477–93. doi: 10.3390/ijms19051477
Keywords: ALDH1, thyroid cancer, prognostic biomarker, tumor-infiltrating lymphocytes, cancer progression
Citation: Cui Y, Liu Y, Mu L, Li Y and Wu G (2022) Transcriptional Expressions of ALDH1A1/B1 as Independent Indicators for the Survival of Thyroid Cancer Patients. Front. Oncol. 12:821958. doi: 10.3389/fonc.2022.821958
Received: 25 November 2021; Accepted: 24 January 2022;
Published: 23 February 2022.
Edited by:
Yun Dai, First Affiliated Hospital of Jilin University, ChinaReviewed by:
Tao Yi, Hong Kong Baptist University, Hong Kong, SAR ChinaCopyright © 2022 Cui, Liu, Mu, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Wu, eWluZ3d1MjAwMkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.