
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol., 13 October 2022
Sec. Molecular and Cellular Oncology
Volume 12 - 2022 | https://doi.org/10.3389/fonc.2022.1018279
This article is part of the Research TopicReviews in Molecular and Cellular OncologyView all 47 articles
Ribonuclease P protein subunit p30 (RPP30) is a highly conserved housekeeping gene that exists in many species and tissues throughout the three life kingdoms (archaea, bacteria, and eukaryotes). RPP30 is closely related to a few types of tumors in human diseases but has a very stable transcription level in most cases. Based on this feature, increasing number of studies have used RPP30 as an internal reference gene. Here, the structure and basic functions of RPP30 are summarized and the likely relationship between RPP30 and various diseases in plants and human is outlined. Finally, the current application of RPP30 as an internal reference gene and its advantages over traditional internal reference genes are reviewed. RPP30 characteristics suggest that it has a good prospect of being selected as an internal reference; more work is needed to develop this research avenue.
The ribonuclease P protein subunit P30 (RPP30) gene is included in the National Center for Biotechnology Information (NCBI ID#10556) database. RPP30 has been shown to be highly conserved in gene pool data, and many studies have shown that there are 16 homologous genes of RPP30 contained in many species from the three life kingdoms (archaea, bacteria, and eukaryotes) (1, 2). As a housekeeping gene, the protein encoded by RPP30 is one of shared protein subunits of ribonuclease P (RNase P) and ribonuclease MRP (RMRP), which are widely expressed in various tissues and participate in many life processes of microscopic and macroscopic organisms. It should be noted that as a protein subunit, detection of RPP30 in different tissues is not uniform and stable, possibly because of the complex modification process after translation (3–6).
In this review, the diseases associated with RPP30 and the factors that may influence its expression are introduced for reference in further studies and in quality control. Abnormal gene expression or mutation studies have made some progress with regard to botanical diseases (5–7). At present, studies on human diseases mainly involve tumors, but only a few of them have demonstrated RPP30 overexpression (8). In addition, RPP30 is associated with glioblastoma (GBM) pathogenesis and low bone mineral density (LBMD) (9)
Reports that RPP30 expression level is affected by other factors are very limited, such as aging (10). Given the relatively high and stable ribose nucleic acid (RNA) expression of RPP30 in human tissues, increasing studies have recently used RPP30 as an internal reference gene in reverse transcription-polymerase chain reaction (RT-PCR) protocols. Thus, the use of RPP30 as an internal reference gene for many applications, including detection of pathogens, calculation of the number of tumor cells, diagnosis of tumors, and some childhood diseases are discussed. In particular, the application of this gene in nucleic acid detection of SARS-CoV-2 demonstrates its great value as an internal reference (11).
Finally, the advantages of RPP30 over conventional reference genes, such as β-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are discussed. Ideal reference genes should be stably expressed in different tissues and different life cycles. With increased research and more applications, the expression of β-actin and GAPDH has been observed to be related to physiological/pathological states, experimental conditions, and tissue type (12–14). In contrast, changes in RPP30 expression seem less likely to be reported in the many conditions described above. These data suggest RPP30 may be used as an internal reference gene in further studies. Of course, the reliability of RPP30 as an internal reference is required to be verified by more comprehensive experiments.
The highly conserved RPP30 genome sequence is located on human chromosome 10 (10Q23.31) at 90,871,974–90,908,556 and is 36,582 nucleotides in length, with 14 exons (https://www.ncbi.nlm.nih.gov/gene/). There are 16 homologous genes in primates, canine, bovine, Rodentia, Amphibia, Drosophila, Arthropoda, and Saccharomycetes. These data are obtained from NCBI (ID#10556). The highly conserved sequence and other characteristics of RPP30 are illustrated in a gene evolutionary tree in Figure 1A.
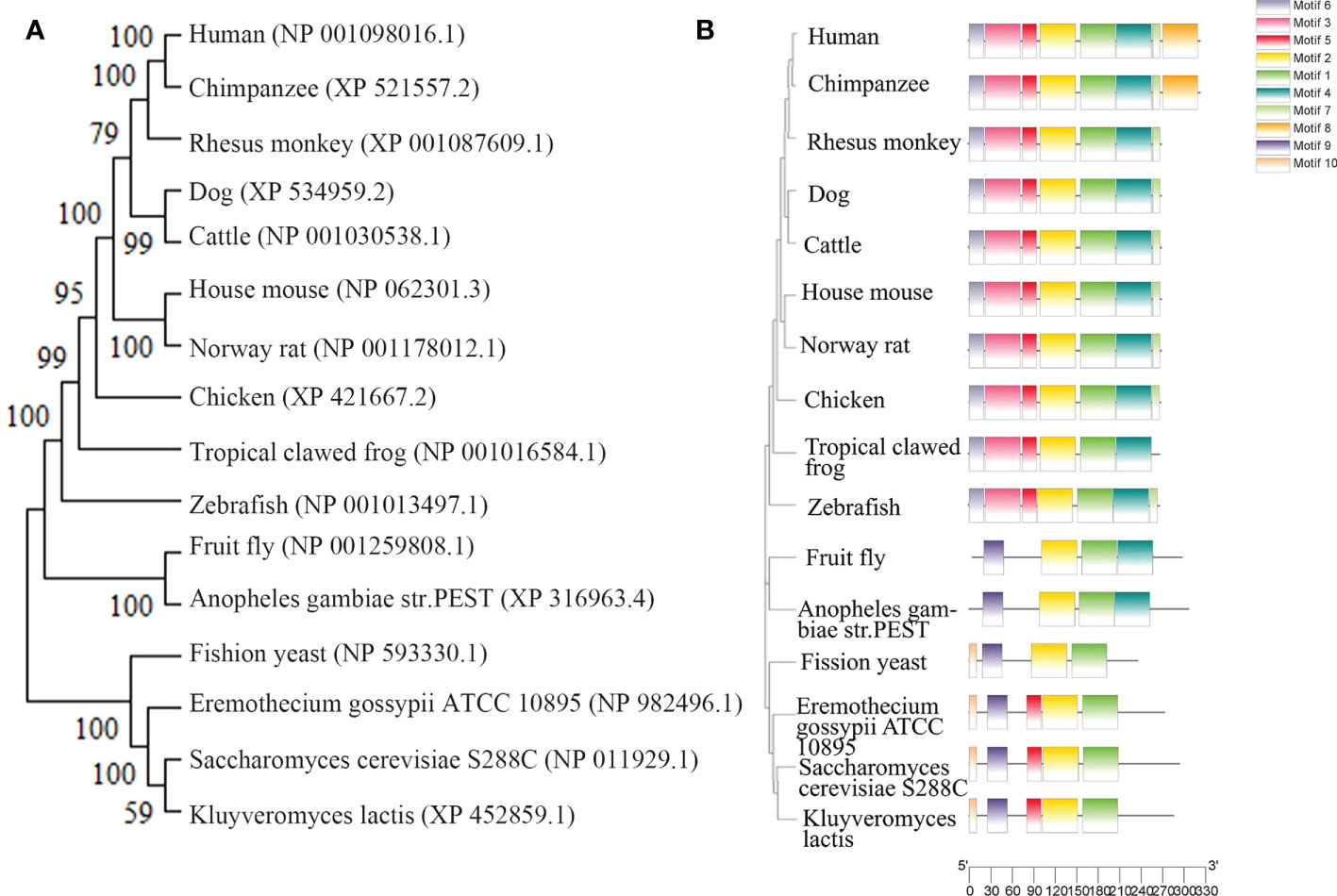
Figure 1 Homology of ribonuclease P protein (RPP) in various species. (A) Homologous evolutionary tree, (B) Human RPP30 protein subunit, and homologous protein subunits of other organisms.
Human nuclear RNase P consists of 1 RNA subunit H1 and 10 conserved proteins, and the complex has a slender conformation similar to the overall shape of yeast RNase P shown by cryo-negative staining electron microscopy (2). The human RNase P protein consists of a single protein, Pop1, and three subcomplexes, which include the RPP20-RPP25 heterodimer, Pop5-RPP14-(RPP30)2-RPP40 heteropentamer, and RPP21-RPP29-RPP38 heterotrimer (2). The proteins are tightly attached to each other, forming a structure similar to a right-handed clip with three modules: finger, palm, and wrist. The POP5-RPP14-(RPP30)2-RPP40 heteropentamer becomes the palm module of the protein clamp. Two copies of the RPP30 molecule bind to the central POP5-RPP14 from opposite sides, forming a typical trisose phosphate isomerase (TIM) barrel fold. The molecule of RPP30 that interacts with RPP40 is called RPP30B, and the other molecule is called RPP30A. The secondary and tertiary structures of RPP30 have not been analyzed at this stage (2).
The RPP30 protein subunit is homologous to the RNase P protein subunit of archaea and other eukaryotes, as shown in Figure 1B. By comparing the amino acid sequence of homologous genes of RPP30, the domain and conserved site of the RPP30 protein may be identified, which will illustrate how conserved this protein is.
RPP30 mainly functions in catalysis, nuclear localization, assembly, and/or regulation of holoenzyme activity (3). The GeneCards (https://www.genecards.org/) and Gene Ontology (GO; http://geneontology.org/) databases were searched for the RPP30 gene to identify its basic functions. The gene’s molecular functions include binding proteins, catalyzing reactions, and so on (15). In archaea, two RPP30 copies bind with ribonuclease P/MRP protein subunit Pop5 dimers to form the Pop5•RPP30 heterodimer. The Pop5•RPP30 heterodimer is anchored on the catalytic domain of RNase P RNA(RPR), which is necessary for pre-tRNA cleavage (16). Cellular components encoded by the gene include RNase P complex, RNase MRP complex, and the multimeric ribonuclease P complex. Biological processes mediated by the gene include rRNA and tRNA processing (15). RPP30 gene encodes a type of ribonuclease that achieves RNaseP RNA binding activity, contributes to ribonuclease P activity, and participates in the removal of tRNA5′- precursor, as well as the formation of polynuclease P complex, and ribonuclease MRP complex, which is necessary for the gene transcription of RNA polymerase III (17). RPP30 also facilitates immunity in rice and reproduction in Arabidopsis and Drosophila (5–7). Therefore, as the most conserved gene in various types of organisms, RPP30 is also involved in the most basic life processes, which drive almost all life functions and activities.
RNase P and RMRP are both small nucleolar ribonucleoprotein complexes (snoRNPs) that are classified into three major classes (box H/ACA snoRNPs, box C/D snoRNPs, RNase P and RnaseMRP) (18). RMRP has only been found in eukaryotes, located mostly in the nucleolus, and has many functions, including cleaving the pre-rRNA at site A3 in vivo and in vitro to mature the 5′ end of the 5.8S rRNA, cleaving an RNA transcript to generate RNA primers for mitochondrial DNA duplication, cleaving the B-type cyclin, Clb2, mRNA, recognizing and cutting pre-tRNA, and is required to turn over cell cycle mRNA (19–21). RNase P, located both in the nucleoplasm and nucleolus, is necessary for Mg2+ dependent 5′ maturation of tRNAs in archaeal, bacterial, and eukaryotic kingdoms (22). RNase P may also act in the stress response and be a transcription factor that regulates polymerase I and III (23). RNAs containing N6 methyladenosine (m6A), 4.5S pre-rRNA, operon mRNAs, box C/D small nucleolar RNAs that reassemble tRNAs are also substrates of RNase P (15, 24–30). RNase P and RMRP have similar functional and structural characteristics (18, 31). These two enzymes share at least ten protein subunits, including RPP14, RPP20, RPP21, RPP25, RPP29, RPP30, RPP38, RPP40, Pop1, and Pop5 (4, 15). RPP30, with a highly conserved amino acid sequence, has an important role in joining the RNase P and RMRP complexes (32). RPP30, as one of the common subunits between the RNase P and RMRP complexes, contributes to an increased number of RNA substrates and atypical functions of eukaryotes (4, 33). RNase P H1 and RMRP RNAs may crosstalk with miRNAs that are related to stability and translation of mRNAs (34). Stolc and Altman reveal that reduction RPP1 (homologous to human RPP30) in S. cerevisiae causes disruptions in both RNase P and RMRP by inhibiting correct cleavage of the internal transcribed spacer I of rRNA surrounding the A3 site (35).
Although RNase P RNA (RPR) is suggested to have activity in vitro, its activity in vivo requires protein cofactors (36). In 2006, Welting et al. used glycerol gradient sedimentation and coimmunoprecipitation to determine that RPP30 is related to the RNA subunit of RNase P and RMRP (18). UV–crosslinking studies also show that RPP30 interacts directly with H1 RNA, an RNA subunit of RNase P (37, 38). Isothermal titration calorimetry has been used to explore interactions among the protein subunits of RNase P and RMRP (22, 39, 40). In archaea, bacteria, and yeast, RPP30/RPP30 paired with Pop5/Pop5, may be functionally reconstituted with the phylogenetically-conserved core catalytic domain (C domain) of the RNA subunit to promote the assembly of RNase P providing substrate RNA binding sites and activating the RNA subunit (probably by RNA annealing and strand displacement (41) and stabilize ionic interactions with the RNA subunit or the substrate pre-tRNA at a relatively lower salt concentration (1, 22, 42–44). In the hyperthermophilic archaeon Pyrococcushorikoshii, PhoRPP30 is homologous to human RPP30 and acts as a molecular chaperone of PhoPop5, which recognizes the stem-loop containing the P3 helix in PhopRNA (45). RPP30-Pop5 is a tight heterotetrameric complex that increases the affinity of the holoenzyme for Mg2+ and protects the RNase P M1 RNA’s C domain from RNase T1 cleavage, especially near conserved nucleotides of RNase P in archaea whose RNase P protein is homologous to eukaryotic counterparts (36, 46–48). The RPP30-Pop5 complex also increases the RPR cleavage rate of pre-tRNA and may be activated by the RPP21-RPP29 complex reflecting indirect effects (36). In Dictyosteliumdiscoideum, RPP30 adopts a TIM-barrel fold that stabilizes the structure and enhances the affinity of pre-tRNA of RNase P to promote the formation of a native fold (46, 49, 50). In humans, RPP30 interacts with RPP14, RPP40, RPP20, RPP21, Pop1, RPP29, 4pp38, and RPP30 itself (15, 37, 51). Moreover, Stolc and Altman have shown that the RPP30 and RPP38 cDNA code for proteins related to catalytic complexes of RNase P from HeLa cells (35). Additionally, RPP30 may interact with other RNAs; as an important subunit of RNase P, RPP30 may be involved in the cleavage of hepatitis C virus RNA (52).
In Arabidopsis, the RPP30 domain is present from 98–248 amino acids in gametophyte defective 1 (GAF1), which is important in female gametophyte development and male competence and has a universal contribution to plant development (5). In Drosophila, RPP30 is necessary for female oogenesis because of its relationship with tRNA processing, DNA replication, and piRNA transcription (7). RPP30 also positively regulates rice immunity by interacting with histone deacetylase 701 (HDT701, RPP30 may be a substrate of HDT701), which functions in suppressing innate immunity in rice and may upregulate expression of defense genes (6).
Although many functions of RPP30 have recently been identified, the specific role of RPP30 in basic life processes requires further research.
RNase P and RMRP play an important role in RNA or non-RNA processing that are universal programs closely related to many life activities. As an important subunit, the mutation and abnormal expression of RPP30 leads to many diseases.
In Arabidopsis, GAF1 mutations result in decreased RPP30 levels that induce defects in mitosis during female gametophyte development, arrest embryo sacs at stages FG1–FG7 and also cause defects in male competence (5). In Drosophila, an isolated mutation that inserts the P-element P(lacW)k01901 into RPP30 leads to complete sterility in females (7, 49). The pathogenic mechanisms that have been uncovered include a mutation in RPP30 that arrests oogenesis by decreasing tRNA processing, which leads to transcription-replication conflicts (7). This includes decreases in transposon expression, accumulation of the polymerase III subunit Brf, and the collapse of Proliferating Cell Nuclear Antigen (PCNA), which increases DNA replication stress and gene defense by small RNAs and activates several DNA duplication checkpoint proteins, including p53, claspin, and checkpoint kinase 2 that decrease piRNA transcription and piRNAclusterpopulations (7). piRNAs are native defenders of germline cell genomes whose mature structure called a “nuage” surrounds the nurse cells that provide nutrients to oocytes (53). Additionally, downregulation of piRNA levels leads to derepression of transposable elements and activates DNA checkpoints to promote positive feedback of defective oogenesis (7, 53–56).
Li et al. have identified OsRPP30, a cellular protein that may regulate the biological function of rice HDT701 (6). HDT701 negatively regulates defense mechanisms in rice by increasing histone H4 deacetylation and increasing the sensitivity to Magnaporthe grisea and Xanthomonas oryzaepv.oryzae (57). When rice is infected with Pyriculariaoryzae (syn. Magnaportheoryzae), RPP30 expression increases, which activates the transcription of defense genes (6). The overexpression of OsRPP30 in genetically modified rice increases expression of defense genome and the production of reactive oxygen species, resulting in resistance to Magnaporthe grisea and Xanthomonas oryzae. OsRPP30 is located at the top of the immune pathway triggered by HDT701-mediated pathogen-associated molecular patterns, which may overcome the negative effects of HTD701 and provide a new direction for the cultivation of pathogen-resistant food in the future (6).
Anti-Th/To is one of the rarer antinuclear antibodies identified in patients with systemic sclerosis (SSc) and is composed of hPOP1, RPP25, RPP30, and RPP40 (58, 59). Researchers refer to “anti-Th” and “anti-To” in the cases of RNase MRP and RNase P, respectively (60, 61). Recombinant RPP30 and RPP38 cross-react with anti-Th/To antibodies of patients afflicted with SSc (3, 32). In addition, people with positive anti-RPP30 antibodies have a lower risk of tendon friction rubs and cancer, but more likely to have severe lung diseases and pulmonary hypertension (59, 62). However, the positivity of anti-RPP30 antibodies only represents the antigenicity of RPP30 protein, and does not suggest the existence of abnormal expression or a RPP30 gene defect, which requires further research.
The nucleophosmin (NPM1) gene, located at human chromosome 5Q35, contains 12 exons and encodes a multifunctional shuttling protein that shuttles between the nucleolus and cytoplasm. NPM1 mutations happen in approximately one-third of acute myeloid leukemias (AMLs) (63). Martelli et al. have shown that the NPM1/RPP30 complex serves as one of three NPM1 rearrangements that have been found and analyzed in 13,979 AML samples (64). In patients with AML that have a NPM1 rearrangement, RPP30 is rearranged with NPM1 at exon 11, whereas the rearrangement of NPM1 with RPP30 is at the end of exon 9 (64). These data indicate RPP30 may help detect AML and monitor NPM1-mutated AML. A new study found that RPP30 may be a transcriptional regulator in glioblastoma (GBM) and the decreased RPP30 expression in elderly people could be a risk factor for GBM (10). This study showed that RPP30 was related to RNA and post-transcriptional modification in non-tumor tissues, and RNA modification in GBM. RPP30 regulates protein expression in GBM by affecting post-transcriptional modification of proteins and functional accumulation of these proteins indicates that these proteins are mainly involved in the activation of cancer signaling pathways (10). In addition, downregulation of RPP30 expression in human astrocyte (HA) cells promotes the proliferation of HA cells, while overexpression inhibits the activation of tumor-related pathways and the proliferation of HA cells, further confirming the close relationship between RPP30 and the occurrence and development of GBM (10). Correlation analysis of RPP30 expression levels with gene expression in cancer-related pathways, such as cancer, Wnt, and mitogen-activated protein kinase pathways in the Chinese Glioma Genome Atlas and The Cancer Genome Atlas databases show significant correlation (10). The Gene Expression Profiling Interactive Analysis (GEPIA2) database has been used to obtain broad knowledge of the relationship between RPP30 (Ensembl ID: ENSG00000148688.13) and tumors (Figure 2) (8). RPP30 expression was significantly different in tumor tissues (higher) and non-tumor tissues in diffuse large B-cell lymphoma, pancreatic adenocarcinoma (PAAD) and thymoma (THYM) (Figure 3). These data also show that there is no significant difference in RPP30 expression levels in different stages of those tumors while high expression of RPP30 is correlated with lower overall survival in PAAD using data from the GEPIA2 public database (http://gepia2.cancer-pku.cn/#index). RPP30 gene expression is high under epidermal development, cell differentiation, and keratinocyte differentiation processes, which play important roles in the differentiation of gastric epithelial cells. Recently, Kan et al. used TGCA RNA-seq to explore the role of RPP30 expression in gastric cancer. They found that RPP30 protein expression was positively correlated with the number of T helper 2 cells, active dendritic cells, and T helper 1 cells, and negatively correlated with the number of T helper 17 cells. They also found that RPP30 RNA expression in gastric cancer (GC) tissue is higher than that in normal tissue and higher RPP30 RNA expression is related to worse overall survival (OS) at the T1, T2, and N0 stages of the tumor. The mechanism may be that RPP30 RNA expression is upregulation via the G alpha S signaling pathway, neuronal system, and olfactory transduction, in addition to increasing cAMP levels, which are tightly correlated with GC histopathology. RPP30 could regulate tRNA modification, transcriptional replication, DNA repair, replication fork stagnation, and protein expression, which are correlated with cancer cell proliferation (65, 66).
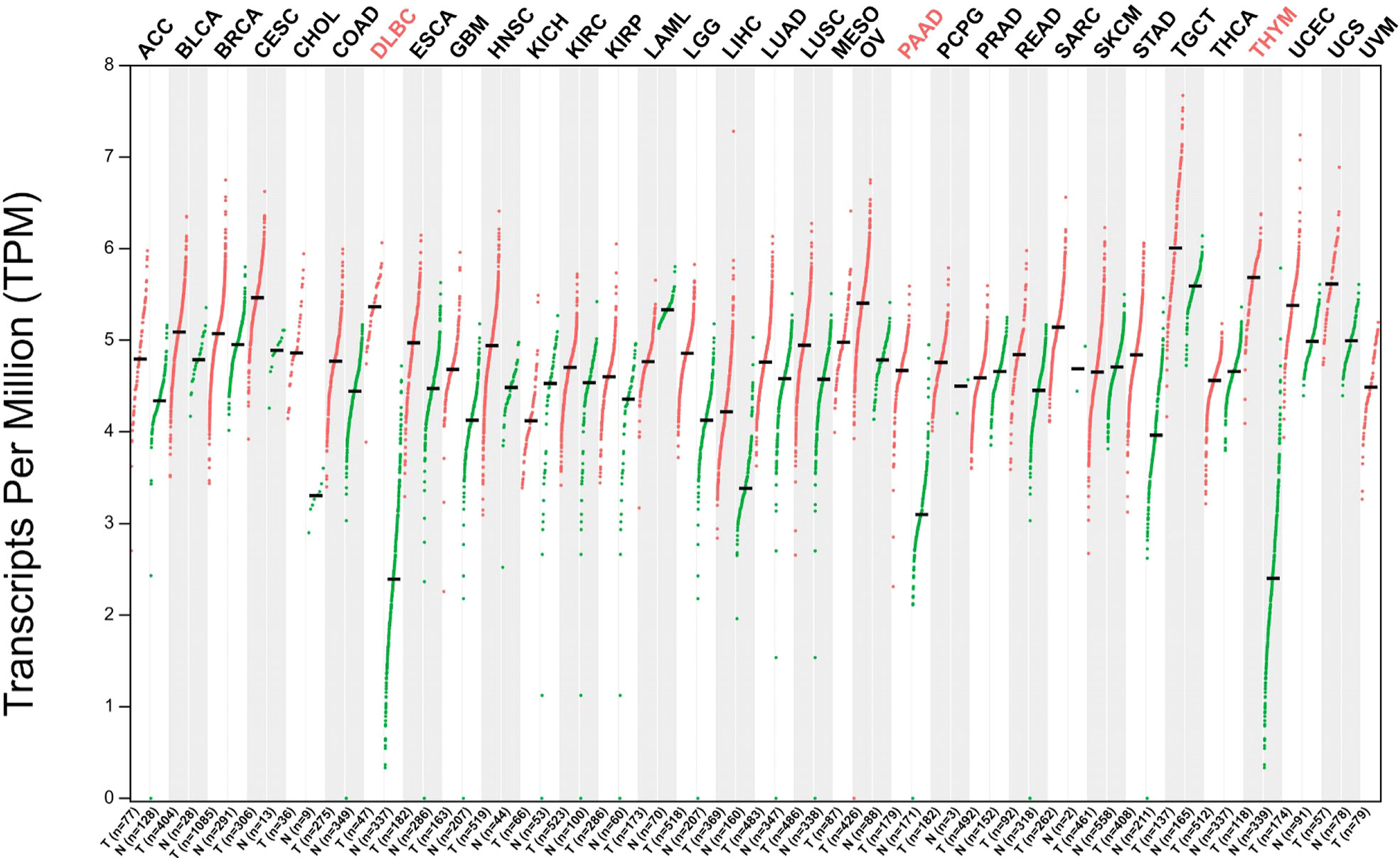
Figure 2 Gene expression profile of ribonuclease P protein subunit p30 (RPP30) in all tumor samples and paired normal tissues. Red words: significant difference; Black words: No significant difference. Abbreviations: ACC: Adrenocortical carcinoma, Bladder Urothelial Carcinoma, Breast invasive carcinoma, Cervical squamous cell carcinoma and endocervical adenocarcinoma, Cholangio carcinoma, Colon adenocarcinoma, Diffuse Large B-cell Lymphoma, Esophageal carcinoma, Glioblastoma multiforme, Head and Neck squamous cell carcinoma, Kidney Chromophobe, Kidney renal clear cell carcinoma, Kidney renal papillary cell carcinoma, Acute Myeloid Leukemia, Brain Lower Grade Glioma, Liver hepatocellular carcinoma, Lung adenocarcinoma, Lung squamous cell carcinoma, Mesothelioma, Ovarian serous cystadenocarcinoma, Pancreatic adenocarcinoma, Pheochromocytoma and Paraganglioma, Prostate adenocarcinoma, Rectum adenocarcinoma, Sarcoma, Skin Cutaneous Melanoma, Stomach adenocarcinoma, Stomach and Esophageal carcinoma, Testicular Germ Cell Tumors, Thyroid carcinoma, Thymoma, Uterine Corpus Endometrial Carcinoma, Uterine Carcinosarcoma, Uveal Melanoma, in turn.
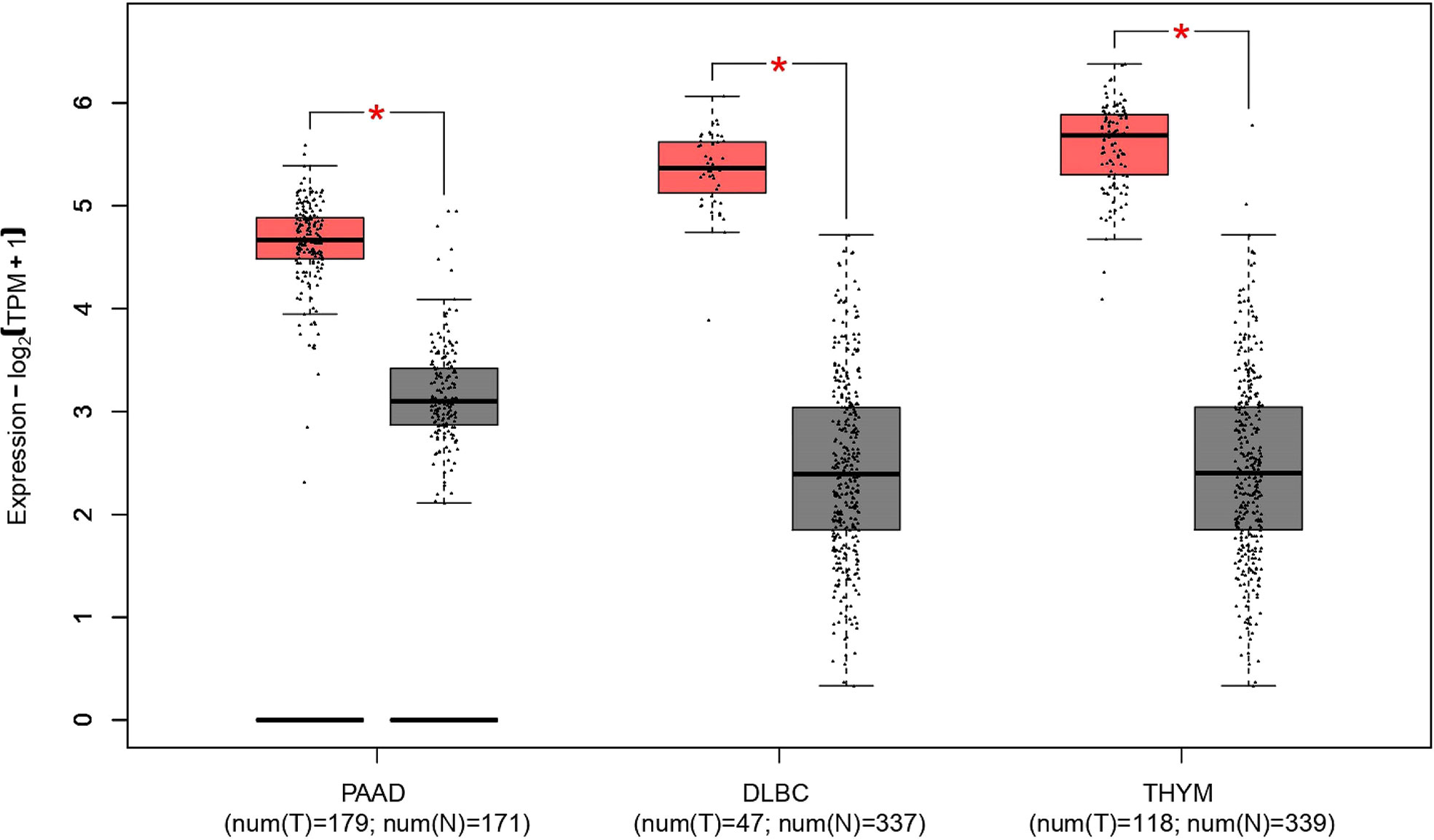
Figure 3 Differences in ribonuclease P protein subunit p30 (RPP30) expression between DLBC, PAAD, THYM, and normal tissues. *Significant difference between tumor and normal tissues; DLBC, Diffuse large B-cell lymphoma; PAAD, Pancreatic adenocarcinoma; THYM, Thymoma. Red represents tumor group, gray represents normal group.
Lee et al. Have found that rpp30 may be related to genetic factors of LBMD through genome-wide association studies involving two signaling pathways of eight related diseases (9). No further association between rpp30 and LBMD has been reported. RPP30 is indirectly related to some diseases, including lung diseases and pulmonary hypertension, secondary to autoimmune diseases (62).
Currently, the research on genes and diseases is extremely in-depth and making rapid progress. Although rpp30 is involved in basic life activities, only a few human diseases have been confirmed to be related to rpp30, and even fewer have been confirmed to have abnormal expression. These results further reflect the stable expression of rpp30 and how well conserved it is.
Although RPP30 is associated with appellate disease, the expression of RPP30 in normal tissues and most tumor cells is stable. Approximately 3647 species have RPP30 subunits and 424 organisms have orthologs of human RPP30. RPP30 RNA is widely expressed in 27 human tissues, including testis, heart, kidney, lung, thymus, and lymph nodes, and more, among which testes and lymph nodes show the most expression and pancreas shows the least expression using data from the NCBI, InterPro (https://www.ebi.ac.uk/interpro/), and GeneCards public databases. In Figure 4A, although there are differences in RPP30 RNA expression levels calculated by different databases, RPP30 RNA expression levels of different organizations calculated by the same database are basically the same, which is consistent with the results obtained by Bgee involving gene expression data in animals. However, the expression of human RPP30 protein is not as stable as RPP30 RNA. There are differences in the expression of RPP30 protein among different tissues or cells and the RPP30 protein has weak expression in some tissues or cells, such as lymph node, brain, spinal cord, ovary, bone, colon, and liver secretion (Figure 4B). There have also been no reports using RPP30 protein as an internal reference for western blotting. To summarize, the expression level of RPP30 RNA in human tissue is relatively high and stable and is suitable to be used as an internal reference gene (11, 67, 68). Mouse RPP30 RNA is widely expressed in the central nervous system, bladder, brain, liver, and testis, etc., with higher expression in the central nervous system and lower expression in the adrenal gland and stomach.
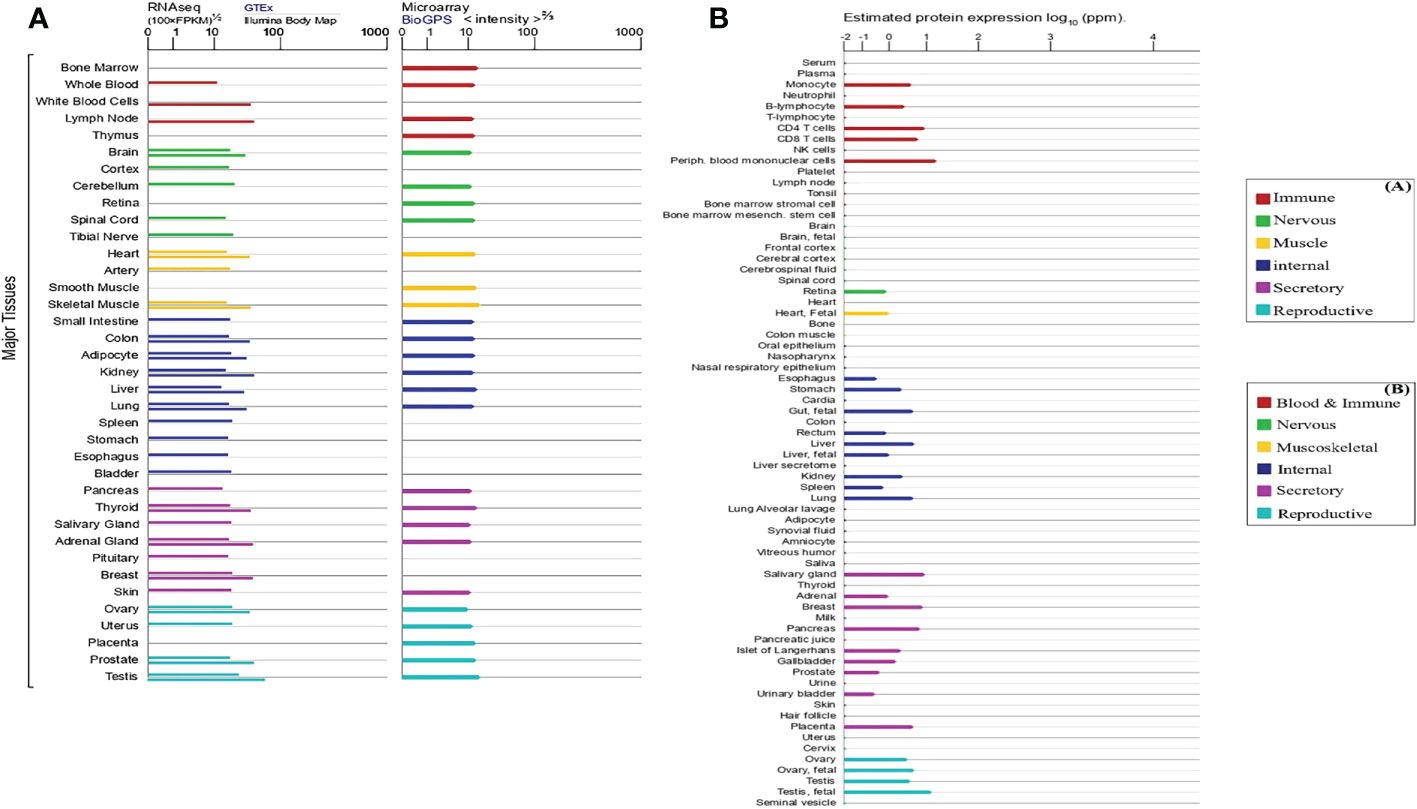
Figure 4 Expression level of human ribonuclease P protein subunit p30 (RPP30) RNA and protein in different tissues and cells (line colors indicate tissue types and length indicates levels of expression). (A) Expression of RPP30 RNA using RNA sequencing (left: expression results above gray horizontal lines are from the Genotype-Tissue Expression (GTEx) database and those below are from the Illumina Body Map) and microarray (right: from the BioGPS database); (B) Protein expression in normal tissues and cell lines from Proteomics DB, the MaxQuantDataBase (MaxQB), and Multi-Omics Profiling Expression Database (MOPED).
At present, there are relatively few reports on factors affecting RPP30 expression levels. Li, Zhai (10) have found that RPP30 expression is affected by age-related factors. Using analysis of age-related genes, RPP30 expression was negatively correlated with increased age, indicating that the change in RPP30 expression may be related to cell senescence. Li, Xiong (6) have found that RPP30 expression is upregulated after rice has been infected with fungal and bacterial pathogens. Mattijssen, Welting (69) have speculated that the expression of housekeeping genes may be altered in the growth plates of patients with cartilage-hair hypoplasia.
RPP30 has been used as an internal reference gene in the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Coronavirus disease 2019 (COVID-19) broke out in Wuhan, China in December 2019 and then spread widely around the world, with strong infectivity (70). Despite its reputation as the gold standard for the detection of SARS-CoV-2, RT-PCR often produces false negative results in detection and diagnosis (71). This may be related to sample quality or changes of primer/probe binding site sequences, but the latter is less likely (72, 73). RPP30 is a single copy sequence gene stably expressed in the human genome, which has a good amplification efficiency, shows 100% sensitivity and specificity, and is not affected by swabs and methodology (74). Compared to other internal parameters, only RPP30 exists in all types of SARS-COV-2 infection samples (67). Figure 5A is the flow of RT-PCR. Both RPP30 RNA and viral RNA were present in epithelial cells (Figure 5B), and RPP30 RNA levels were closely related to SARS-CoV-2 RNA levels in respiratory tract samples (Figure 5C). Thus, RPP30 RNA may be used to control sample quality and the RPP30 Ct cutoff value may effectively identify false negative results (11, 72), which may increase sensitivity and reduce the spread of SARS-CoV-2. In addition to evaluating the quality of the sample, RPP30 may also determine whether mRNA has been extracted successfully and whether there is inhibition in the PCR (75).
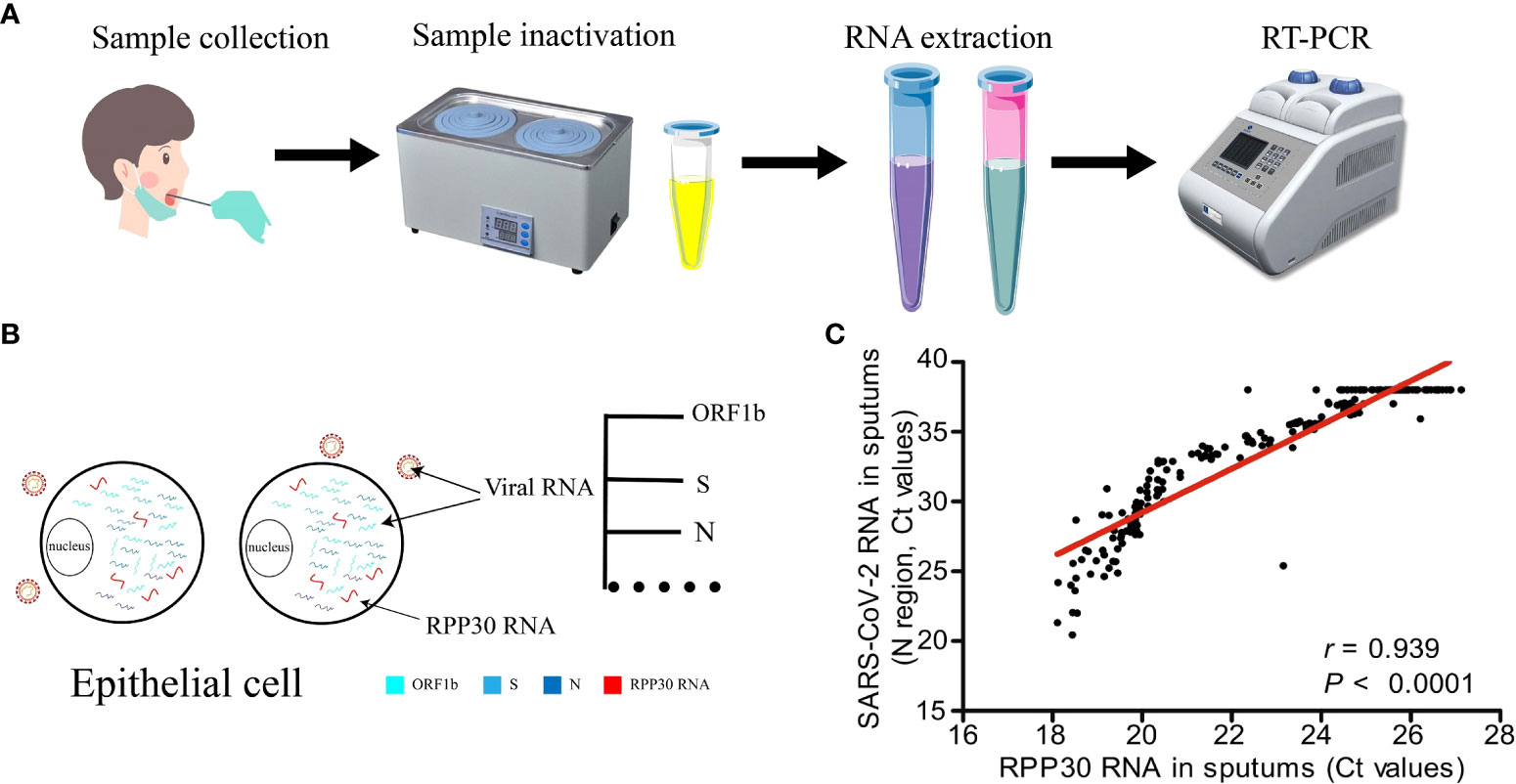
Figure 5 Ribonuclease P protein subunit p30 (RPP30) is used as an internal reference gene in the detection of SARS-CoV-2. (A) Sample collection and reverse transcription-polymerase chain reaction (RT-PCR), (B) Viral RNA coexists with RPP30 in epithelial cells, (C) Detection results of RPP30 and SARS-COV-2 are significantly positively correlated (11).
RPP30 is used as an internal reference gene to determine the best effective drug concentration for tumor treatment (76). The efficacy of traditional antineoplastic drugs is evaluated by calculating the lethality of drugs to all cells in vitro, so it is impossible to measure the lethality of antineoplastic drugs to normal cells, which is often accompanied by unpredictable side effects. Because RPP30 is stably expressed in the vast majority of tumor cells and non-tumor cells, while the neurofibromatosis type 1 (NF1) gene loses heterozygosity in tumor cells, the number of tumor cells may be evaluated by the quantitative RT-PCR ratio of NF1 to RPP30, which may be used to evaluate the efficacy and side effects of tumor drugs, and may also be used in personalized adjuvant chemotherapy. Due to the different behavior of cells in vivo and in vitro, this method has some limitations (76–78). As an internal reference gene, RPP30 may also accurately and effectively evaluate the concentration of antiretroviral drugs in cells (79).
RPP30 is used as an internal reference gene to analyze the feasibility of HIV DNA detection in cerebrospinal fluid (CSF) (80). RPP30, as a housekeeper gene, is highly conserved and widely expressed in human tissues and may be used as an internal reference gene to detect the number of leukocytes in CSF. Using droplet digital PCR (dd-PCR) detection, the level of HIV DNA in CSF cells is not correlated with RPP30 levels, indicating that the detectability of HIV DNA does not depend entirely on the number of cells available in each sample. Then, the correlation between the level of HIV DNA in the CSF and the level of HIV RNA in peripheral blood cells, as well as the relationship between the virus inhibition and non-inhibition subgroups, may be analyzed to explore the detectability of HIV DNA level in CSF.
RPP30 is used as an internal reference gene in diagnostic experiments (68). At least 5.5% of all pathogenic genetic changes in humans are large genome deletions or duplicates (81). With the discovery of disease-related genes, dd-PCR has been used to quantify the copy number of genes to diagnose diseases (82). Because of its conserved sequence and stable expression in almost all cells, RPP30 is widely used as an internal reference gene (83). For example, RPP30 has been used as an internal reference in real-time fluorescent PCR or dd-PCR to quantify the survival of motor neuron 1 gene, T-cell receptor excision circles, and Kappa-deleting recombination excision circles; to screen neonatal spinal muscular atrophy, severe combined immunodeficiency disease, and detect immune remodeling of the thymus and bone marrow (84–86); and to quantify the sex-determining region Y gene to detect male/female chimerism, which may track chimerism after hematopoietic stem cell transplantation (87). RPP30 has also been used as an internal reference in single-cell dd-PCR to evaluate the genomic DNA of rare circulating fetal cells in peripheral blood samples of pregnant women with male fetuses and validate the concept of non-invasive prenatal diagnosis (88). There are also many reports on the use of RPP30 as an internal reference in different molecular biology techniques for the diagnosis of human diseases, as shown in Table 1.
Dyavar et al. (79) used human and rhesus macaque (RM) gDNA templates to quantitate RPP30 copies, and found a low coefficient of variation and strong correlation between human and RM gDNA templates and the number of RPP30 copies in intra-laboratory (R2 = 0.996, p < 0.001; R2 = 0.975, p < 0.001), inter-laboratory (R2 = 0.997, p < 0.001; R2 = 0.989, p < 0.001), and inter-operational (R2 = 0.994, p < 0.001; R2 = 0.986, p < 0.001) studies, which confirms the high accuracy and precision of the RPP30 dd-PCR assay. In addition, Profaizer and Slev (86) observed that RPP30 dd-PCR could detect 2 copies/µL of genes, which is more accurate than the previous 24 copies/µL for qPCR.
Housekeeping genes are mainly involved in the maintenance of basic cell functions and are thought to be expressed in all cells (98), They are widely used as internal controls to standardize the expression of genes in western blotting, northern blotting, and RT-PCR. The ideal housekeeping gene should be expressed at the same level in all tissues (99). At present, frequently used housekeeping genes are β-actin and GAPDH, in which β-actin has a molecular weight of approximately 42–43 kDa and is composed of 375 amino acids. It is widely distributed in the cytoplasm and is involved in cell movement, structure, and integrity (100), whereas GAPDH is an enzyme with a molecular weight of approximately 37 kDa and is involved in glycolysis, DNA repair, tRNA output, membrane fusion, and transport (101). However, there are increasing reports that the RNA expression level of these genes is affected by the physiological/pathological state, experimental conditions, and tissue types (12–14). Thus, the factors affecting mRNA expression levels of RPP30, GAPDH, and β-actin were compared.
Table 2 shows that the length of the RPP30 amplification product is smaller than that of β-actin and GAPDH, which reduces errors and improves efficiency during the process of RPP30 amplification. Table 3 lists the pseudogenes found in β-actin and GAPDH, but, to date, no pseudogenes have been found in RPP30. The existence of pseudogenes reduces the amplification efficiency of genes and reduces the accuracy of their use as internal reference genes for standardization (122, 123). Therefore, using RPP30 as the internal reference gene may be more accurate.
Numerous reports suggest that gene expression levels of β-actin and GAPDH are affected by many factors under different pathological conditions, such as tumor cells and non-tumor cells (103–105, 124), steatosis and alcoholic hepatitis (106), and Alzheimer’s disease (107). Under different experimental conditions, expression levels of the traditional internal reference genes, β-actin and GAPDH, vary greatly, such as in serum-stimulated fibroblasts (108), miR-644a (109), dietary conditions (125), and other conditions (110–114, 119–121). In addition, β-actin has extensive variation in mouse lymphocytes and is not appropriate for use as an internal reference gene for the quantitative PCR analysis of mouse lymphocytes (126), since such changes may lead to data divergence and inaccuracy. At present, there are few reports on factors affecting RPP30 expression levels, which may be related to the existence of RPP30 in all three fields of life (archaea, bacteria, and eukaryotes), and because it is widely expressed in different tissues whose gene sequences are conserved and homologous, such as in humans, chimpanzees, rhesus monkeys, mice, fruit flies, Saccharomyces cerevisiae, and archaea (127). In addition, currently, research on RPP30 is scant.
The mRNA of β-actin and GAPDH are not highly expressed in all cell types or tissues of chicken embryos, and the expression levels are different in different tissues (115), which is similar to the 15-fold difference between the highest and lowest expression levels of GAPDH in different human tissues observed by Barber et al. (128). GAPDH expression levels also vary in different varieties of the same plant (129). Furthermore, β-actin and GAPDH expression levels fluctuate significantly at different stages of lymphocyte activation (116), which may be related to their participation in other cellular biology functions. RPP30 mRNA expression in different tissues is more stable than those of β-actin and GAPDH. In addition, RPP30 is widely expressed in 27 human tissues, is relatively conserved in structure and function, is not correlated with DNA content in the sample, and is not affected by the content of genes to be tested, resulting in high application value in a series of samples with scarce and uneven DNA content (91). Currently, to reduce the inaccurate data caused by differences in the expression of internal reference genes among different tissue types, RPP30 has become the main internal reference gene for quantitative detection of genes (130).
Moreover, the expression level of the three genes is affected by age (10, 117); their expression level decrease with age, but it is not known whether the specific mechanism is the same. There are also differences in the expression level of β-actin at different developmental stages (131). The factors affecting the expression level of RPP30 RNA in different pathological states, experimental conditions, and tissue types is lower than that of the commonly used internal reference genes, β-actin and GAPDH. RPP30 has good amplification efficiency and may be better used in RT-PCR experiments. Currently, there is no housekeeping gene that has stable expression, is abundant, and consistent under any condition (132). Therefore, specific reference genes should be verified and selected according to experimental conditions and sample type (102, 118).
RPP30 is a highly conserved gene that has homologous genes in 16 species. Although RPP30 is a housekeeping gene and its encoding protein is a key subunit that maintains basic life activities, it is rarely reported to be associated with human diseases, and is overexpressed only in a few patients with cancer. In addition, compared to traditional reference genes, RPP30 has advantages of short sequence length, is widely and uniformly expressed in various tissues, and its expression level is rarely disturbed by external factors. Overall, RPP30 has great prospects and value as an internal reference gene. To date, RPP30 has been used as an internal reference for nucleic acid tests of Sars-CoV-2, evaluation of therapeutic drugs and drug side effects, analysis of the feasibility of HIV detection, and many other diagnostic experiments. However, due to the unstable detection results of the RPP30 protein, there are no studies that have used RPP30 as a reference in western blotting.
In such conditions, RPP30 may not be the first choice as a reference gene for these tests. Regardless of which kind of reference that is chosen, it may be affected by a few inevitable conditions. Thus, the correct reference to be used for these tests should be further explored.
JW, SY and YW collected literature and wrote the original draft and prepared the figures and tables. JZ revised the original draft. ZZ conceived the idea and revised the manuscript. All authors contributed to the article and approved the submitted version.
The study was supported by Anhui Provincial Natural Science Foundation (grant number 2108085MH298) and the Scientific research project of Anhui Medical University (grant number 2019GMFY02, 2021lcxk027). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We would like to thank Editage (www.editage.cn) for English language editing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Pulukkunat DK, Gopalan V. Studies on methanocaldococcus jannaschii rnase p reveal insights into the roles of rna and protein cofactors in rnase p catalysis. Nucleic Acids Res (2008) 36(12):4172–80. doi: 10.1093/nar/gkn360
2. Wu J, Niu S, Tan M, Huang C, Li M, Song Y, et al. Cryo-em structure of the human ribonuclease p holoenzyme. Cell (2018) 175(5):1393–404.e11. doi: 10.1016/j.cell.2018.10.003
3. Jarrous N, Eder PS, Guerrier-Takada C, Hoog C, Altman S. Autoantigenic properties of some protein subunits of catalytically active complexes of human ribonuclease p. Rna (1998) 4(4):407–17.
4. Jarrous N, Gopalan V. Archaeal/Eukaryal rnase p: Subunits, functions and rna diversification. Nucleic Acids Res (2010) 38(22):7885–94. doi: 10.1093/nar/gkq701
5. Wang SQ, Shi DQ, Long YP, Liu J, Yang WC. Gametophyte defective 1, a putative subunit of rnases P/Mrp, is essential for female gametogenesis and Male competence in arabidopsis. PloS One (2012) 7(4):e33595. doi: 10.1371/journal.pone.0033595
6. Li W, Xiong Y, Lai LB, Zhang K, Li Z, Kang H, et al. The rice rnase p protein subunit RPP30 confers broad-spectrum resistance to fungal and bacterial pathogens. Plant Biotechnol J (2021) 19(10):1988–99. doi: 10.1111/pbi.13612
7. Molla-Herman A, Valles AM, Ganem-Elbaz C, Antoniewski C, Huynh JR. Trna processing defects induce replication stress and Chk2-dependent disruption of pirna transcription. EMBO J (2015) 34(24):3009–27. doi: 10.15252/embj.201591006
8. Tang Z, Kang B, Li C, Chen T, Zhang Z. Gepia2: An enhanced web server for Large-scale expression profiling and interactive analysis. Nucleic Acids Res (2019) 47(W1):W556–w60. doi: 10.1093/nar/gkz430
9. Lee YH, Choi SJ, Ji JD, Song GG. Pathway analysis of genome-wide association study for bone mineral density. Mol Biol Rep (2012) 39(8):8099–106. doi: 10.1007/s11033-012-1657-1
10. Li G, Zhai Y, Liu H, Wang Z, Huang R, Jiang H, et al. RPP30, a transcriptional regulator, is a potential pathogenic factor in glioblastoma. Aging (Albany NY) (2020) 12(16):16155–71. doi: 10.18632/aging.103596
11. Zhang Y, Wang C, Han M, Ye J, Gao Y, Liu Z, et al. Discrimination of false negative results in rt-pcr detection of sars-Cov-2 rnas in clinical specimens by using an internal reference. Virol Sin (2020) 35(6):758–67. doi: 10.1007/s12250-020-00273-8
12. Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, et al. Housekeeping genes as internal standards: Use and limits. J Biotechnol (1999) 75(2-3):291–5. doi: 10.1016/s0168-1656(99)00163-7
13. Yin R, Liu X, Liu C, Ding Z, Zhang X, Tian F, et al. Systematic selection of housekeeping genes for gene expression normalization in chicken embryo fibroblasts infected with Newcastle disease virus. Biochem Biophys Res Commun (2011) 413(4):537–40. doi: 10.1016/j.bbrc.2011.08.131
14. Li R, Shen Y. An old method facing a new challenge: Re-visiting housekeeping proteins as internal reference control for neuroscience research. Life Sci (2013) 92(13):747–51. doi: 10.1016/j.lfs.2013.02.014
15. Jarrous N. Roles of rnase p and its subunits. Trends Genet (2017) 33(9):594–603. doi: 10.1016/j.tig.2017.06.006
16. Phan HD, Norris AS, Du C, Stachowski K, Khairunisa BH, Sidharthan V, et al. Elucidation of structure-function relationships in methanocaldococcus jannaschii rnase p, a multi-subunit catalytic ribonucleoprotein. Nucleic Acids Res (2022) 50(14):8154–67. doi: 10.1093/nar/gkac595
17. Reiner R, Ben-Asouli Y, Krilovetzky I, Jarrous N. A role for the catalytic ribonucleoprotein rnase p in rna polymerase iii transcription. Genes Dev (2006) 20(12):1621–35. doi: 10.1101/gad.386706
18. Welting TJ, Kikkert BJ, van Venrooij WJ, Pruijn GJ. Differential association of protein subunits with the human rnase mrp and rnase p complexes. RNA (2006) 12(7):1373–82. doi: 10.1261/rna.2293906
19. Chang DD, Clayton DA. A mammalian mitochondrial rna processing activity contains nucleus-encoded rna. Science (1987) 235(4793):1178–84. doi: 10.1126/science.2434997
20. Lygerou Z, Allmang C, Tollervey D, Seraphin B. Accurate processing of a eukaryotic precursor ribosomal rna by ribonuclease mrp in vitro. Science (1996) 272(5259):268–70. doi: 10.1126/science.272.5259.268
21. Cai T, Aulds J, Gill T, Cerio M, Schmitt ME. The saccharomyces cerevisiae rnase mitochondrial rna processing is critical for cell cycle progression at the end of mitosis. Genetics (2002) 161(3):1029–42. doi: 10.1093/genetics/161.3.1029
22. Guerriertakada C, Gardiner K, Marsh T, Pace N, Altman S. The rna moiety of ribonuclease-p is the catalytic subunit of the enzyme. Cell (1983) 35(3):849–57. doi: 10.1016/0092-8674(83)90117-4
23. Li N, Yuan L, Liu N, Shi D, Li X, Tang Z, et al. Slow Walker2, a Noc1/Mak21 homologue, is essential for coordinated cell cycle progression during female gametophyte development in arabidopsis. Plant Physiol (2009) 151(3):1486–97. doi: 10.1104/pp.109.142414
24. Guerrier-Takada C, Altman S. Catalytic activity of an rna molecule prepared by transcription in vitro. Science (1984) 223(4633):285–6. doi: 10.1126/science.6199841
25. Peck-Miller KA, Altman S. Kinetics of the processing of the precursor to 4.5 s rna, a naturally occurring substrate for rnase p from escherichia coli. J Mol Biol (1991) 221(1):1–5. doi: 10.1016/0022-2836(91)80194-y
26. Alifano P, Rivellini F, Piscitelli C, Arraiano CM, Bruni CB, Carlomagno MS. Ribonuclease e provides substrates for ribonuclease p-dependent processing of a polycistronic mrna. Genes Dev (1994) 8(24):3021–31. doi: 10.1101/gad.8.24.3021
27. Li Y, Altman S. A specific endoribonuclease, rnase p, affects gene expression of polycistronic operon mrnas. Proc Natl Acad Sci U.S.A. (2003) 100(23):13213–8. doi: 10.1073/pnas.2235589100
28. Li Y, Altman S. Polarity effects in the lactose operon of escherichia coli. J Mol Biol (2004) 339(1):31–9. doi: 10.1016/j.jmb.2004.03.041
29. Coughlin DJ, Pleiss JA, Walker SC, Whitworth GB, Engelke DR. Genome-wide search for yeast rnase p substrates reveals role in maturation of intron-encoded box C/D small nucleolar rnas. Proc Natl Acad Sci U.S.A. (2008) 105(34):12218–23. doi: 10.1073/pnas.0801906105
30. Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of M(6)a-containing rnas by rnase P/Mrp complex. Mol Cell (2019) 74(3):494–507.e8. doi: 10.1016/j.molcel.2019.02.034
31. Van Eenennaam H, Jarrous N, . van Venrooij WJ, . Pruijn GJM. Architecture and function ofthe human endonucleasesrnase p and rnase mrp. IUBMB Life (2000) 49(4):265–72. doi: 10.1080/15216540050033113
32. Eder PS, Kekuda R, Stolc V, Altman S. Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease p. Proc Natl Acad Sci U.S.A. (1997) 94(4):1101–6. doi: 10.1073/pnas.94.4.1101
33. Engelke DR, Fierke CA. The evolution of rnase p. RNA (2015) 21(4):517–8. doi: 10.1261/rna.050732.115
34. Shao Y, Ye M, Li Q, Sun W, Ye G, Zhang X, et al. Lncrna-rmrp promotes carcinogenesis by acting as a mir-206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget (2016) 7(25):37812–24. doi: 10.18632/oncotarget.9336
35. Stolc V, Altman S. Rpp1, an essential protein subunit of nuclear rnase p required for processing of precursor trna and 35s precursor rrna in saccharomyces cerevisiae. Genes Dev (1997) 11(21):2926–37. doi: 10.1101/gad.11.21.2926
36. Chen WY, Pulukkunat DK, Cho IM, Tsai HY, Gopalan V. Dissecting functional cooperation among protein subunits in archaeal rnase p, a catalytic ribonucleoprotein complex. Nucleic Acids Res (2010) 38(22):8316–27. doi: 10.1093/nar/gkq668
37. Jiang T, Altman S. Protein-protein interactions with subunits of human nuclear rnase p. Proc Natl Acad Sci U.S.A. (2001) 98(3):920–5. doi: 10.1073/pnas.98.3.920
38. Jiang T, Guerrier-Takada C, Altman S. Protein-rna interactions in the subunits of human nuclear rnase p. Rna (2001) 7(7):937–41. doi: 10.1017/s1355838201010299
40. Xu Y, Oruganti SV, Gopalan V, Foster MP. Thermodynamics of coupled folding in the interaction of archaeal rnase p proteins Rpp21 and Rpp29. Biochemistry (2012) 51(4):926–35. doi: 10.1021/bi201674d
41. Ishihara M, Nishimoto E, Yamashita S, Kakuta Y, Kimura M. A distinct binding mode of archaeal ribonuclease p proteins to rna. Biosci Biotechnol Biochem (2012) 76(12):2335–7. doi: 10.1271/bbb.120546
42. Ziehler WA, Day JJ, Fierke CA, Engelke DR. Effects of 5 ‘ leader and 3 ‘ trailer structures on pre-trna processing by nuclear rnase p. Biochemistry (2000) 39(32):9909–16. doi: 10.1021/bi000603n
43. Wilson RC, Bohlen CJ, Foster MP, Bell CE. Structure of pfu Pop5, an archaeal rnase p protein. Proc Natl Acad Sci U.S.A. (2006) 103(4):873–8. doi: 10.1073/pnas.0508004103
44. Amero CD, Boomershine WP, Xu Y, Foster M. Solution structure of pyrococcus furiosus Rpp21, a component of the archaeal rnase p holoenzyme, and interactions with its Rpp29 protein partner. Biochemistry (2008) 47(45):11704–10. doi: 10.1021/bi8015982
45. Hamasaki M, Hazeyama K, Iwasaki F, Ueda T, Nakashima T, Kakuta Y, et al. Functional implication of archaeal homologues of human rnase p protein pair Pop5 and RPP30. J Biochem (2016) 159(1):31–40. doi: 10.1093/jb/mvv067
46. Smith JK, Hsieh J, Fierke CA. Importance of rna-protein interactions in bacterial ribonuclease p structure and catalysis. Biopolymers (2007) 87(5-6):329–38. doi: 10.1002/bip.20846
47. Xu Y, Amero CD, Pulukkunat DK, Gopalan V, Foster MP. Solution structure of an archaeal rnase p binary protein complex: Formation of the 30-kda complex between pyrococcus furiosus Rpp21 and Rpp29 is accompanied by coupled protein folding and highlights critical features for protein-protein and protein-rna interactions. J Mol Biol (2009) 393(5):1043–55. doi: 10.1016/j.jmb.2009.08.068
48. Crowe BL, Bohlen CJ, Wilson RC, Gopalan V, Foster MP. Assembly of the complex between archaeal rnase p proteins RPP30 and Pop5. Archaea (2011) 2011:891531. doi: 10.1155/2011/891531
49. Vourekas A, Kalavrizioti D, Zarkadis IK, Spyroulias GA, Stathopoulos C, Drainas D. A 40.7 kda RPP30/Rpp1 homologue is a protein subunit of dictyostelium discoideum rnase p holoenzyme. Biochimie (2007) 89(3):301–10. doi: 10.1016/j.biochi.2006.11.006
50. Honda T, Hara T, Nan J, Zhang X, Kimura M. Archaeal homologs of human rnase p protein pairs Pop5 with RPP30 and Rpp21 with Rpp29 work on distinct functional domains of the rna subunit. Biosci Biotechnol Biochem (2010) 74(2):266–73. doi: 10.1271/bbb.90550
51. Welting TJ, van Venrooij WJ, Pruijn GJ. Mutual interactions between subunits of the human rnase mrp ribonucleoprotein complex. Nucleic Acids Res (2004) 32(7):2138–46. doi: 10.1093/nar/gkh539
52. Ariza-Mateos A, Briones C, Perales C, Domingo E, Gómez J. The archaeology of coding rna. Ann New York Acad Sci (2019) 1447(1):119–34. doi: 10.1111/nyas.14173
53. Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, et al. Pirna-mediated nuclear accumulation of retrotransposon transcripts in the drosophila female germline. Proc Natl Acad Sci U.S.A. (2008) 105(39):14964–9. doi: 10.1073/pnas.0805943105
54. Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small rna-generating loci as master regulators of transposon activity in drosophila. Cell (2007) 128(6):1089–103. doi: 10.1016/j.cell.2007.01.043
55. Pane A, Wehr K, Schupbach T. Zucchini and squash encode two putative nucleases required for rasirna production in the drosophila germline. Dev Cell (2007) 12(6):851–62. doi: 10.1016/j.devcel.2007.03.022
56. Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, et al. Uap56 couples pirna clusters to the perinuclear transposon silencing machinery. Cell (2012) 151(4):871–84. doi: 10.1016/j.cell.2012.09.040
57. Ding B, Bellizzi Mdel R, Ning Y, Meyers BC, Wang GL. Hdt701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell (2012) 24(9):3783–94. doi: 10.1105/tpc.112.101972
58. Okano Y, Medsger TA Jr. Autoantibody to Th ribonucleoprotein (Nucleolar 7-2 rna protein particle) in patients with systemic sclerosis. Arthritis Rheum (1990) 33(12):1822–8 doi: 10.1002/art.1780331210
59. Mecoli CA, Adler BL, Yang Q, Hummers LK, Rosen A, Casciola-Rosen L, et al. Cancer in systemic sclerosis: Analysis of antibodies against components of the th/to complex. Arthritis Rheumatol (2021) 73(2):315–23. doi: 10.1002/art.41493
60. Hardin JA, Rahn DR, Shen C, Lerner MR, Wolin SL, Rosa MD, et al. Antibodies from patients with connective tissue diseases bind specific subsets of cellular rna-protein particles. J Clin Invest (1982) 70(1):141–7. doi: 10.1172/jci110587
61. Reddy R, Tan EM, Henning D, Nohga K, Busch H. Detection of a nucleolar 7-2 ribonucleoprotein and a cytoplasmic 8-2 ribonucleoprotein with autoantibodies from patients with scleroderma. J Biol Chem (1983) 258(3):1383–6. doi: 10.1016/S0021-9258(18)32991-0
62. Nunes JPL, Cunha AC, Meirinhos T, Nunes A, Araujo PM, Godinho AR, et al. Prevalence of auto-antibodies associated to pulmonary arterial hypertension in scleroderma - a review. Autoimmun Rev (2018) 17(12):1186–201. doi: 10.1016/j.autrev.2018.06.009
63. Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med (2005) 352(3):254–66. doi: 10.1056/NEJMoa041974
64. Martelli MP, Rossi R, Venanzi A, Meggendorfer M, Perriello VM, Martino G, et al. Novel Npm1 exon 5 mutations and gene fusions leading to aberrant cytoplasmic nucleophosmin in aml. Blood (2021) 138(25):2696–701. doi: 10.1182/blood.2021012732
65. Kovacs D, Cota C, Cardinali G, Aspite N, Bolasco G, Amantea A, et al. Expression of keratinocyte growth factor and its receptor in clear cell acanthoma. Exp Dermatol (2006) 15(10):762–8. doi: 10.1111/j.1600-0625.2006.00459.x
66. Kan Y, Lu X, Feng L, Yang X, Ma H, Gong J, et al. RPP30 is a novel diagnostic and prognostic biomarker for gastric cancer. Front Genet (2022) 13:888051. doi: 10.3389/fgene.2022.888051
67. Yan Y, Chang L, Wang L. Laboratory testing of sars-cov, mers-cov, and sars-Cov-2 (2019-ncov): Current status, challenges, and countermeasures. Rev Med Virol (2020) 30(3):e2106. doi: 10.1002/rmv.2106
68. Nyaruaba R, Li X, Mwaliko C, Li C, Mwau M, Odiwour N, et al. Two-step reverse transcription droplet digital pcr protocols for sars-Cov-2 detection and quantification. J Vis Exp (2021) 169:e62295. doi: 10.3791/62295
69. Mattijssen S, Welting TJ, Pruijn GJ. Rnase mrp and disease. Wiley Interdiscip Rev RNA (2010) 1(1):102–16. doi: 10.1002/wrna.9
70. Dong E, Du H, Gardner L. An interactive web-based dashboard to track covid-19 in real time. Lancet Infect Dis (2020) 20(5):533–4. doi: 10.1016/s1473-3099(20)30120-1
71. Li D, Wang D, Dong J, Wang N, Huang H, Xu H, et al. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: Role of deep-Learning-Based ct diagnosis and insights from two cases. Korean J Radiol (2020) 21(4):505–8. doi: 10.3348/kjr.2020.0146
72. Reijns MAM, Thompson L, Acosta JC, Black HA, Sanchez-Luque FJ, Diamond A, et al. A sensitive and affordable multiplex rt-qpcr assay for sars-Cov-2 detection. PloS Biol (2020) 18(12):e3001030. doi: 10.1371/journal.pbio.3001030
73. de Kock R, Baselmans M, Scharnhorst V, Deiman B. Sensitive detection and quantification of sars-Cov-2 by multiplex droplet digital rt-pcr. Eur J Clin Microbiol Infect Dis (2021) 40(4):807–13. doi: 10.1007/s10096-020-04076-3
74. Deiana M, Mori A, Piubelli C, Scarso S, Favarato M, Pomari E. Assessment of the direct quantitation of sars-Cov-2 by droplet digital pcr. Sci Rep (2020) 10(1):18764. doi: 10.1038/s41598-020-75958-x
75. Rowan AG, May P, Badhan A, Herrera C, Watber P, Penn R, et al. Optimized protocol for a quantitative sars-Cov-2 duplex rt-qpcr assay with internal human sample sufficiency control. J Virol Methods (2021) 294:114174. doi: 10.1016/j.jviromet.2021.114174
76. Kluwe L. Digital pcr for discriminating mosaic deletions and for determining proportion of tumor cells in specimen. Eur J Hum Genet (2016) 24(11):1644–8. doi: 10.1038/ejhg.2016.56
77. Kluwe L. Assessing specificity of anticancer drugs in vitro. J Vis Exp (2016) 109):e53752. doi: 10.3791/53752
78. Kluwe L, Jiang W, Alster I, Hanken H. A novel genetic- and cell-based tool for assessing the efficacy and toxicity of anticancer drugs in vitro. BioMed Pap Med Fac Univ Palacky Olomouc Czech Repub (2016) 160(1):64–9. doi: 10.5507/bp.2015.057
79. Dyavar SR, Ye Z, Byrareddy SN, Scarsi KK, Winchester LC, Weinhold JA, et al. Normalization of cell associated antiretroviral drug concentrations with a novel RPP30 droplet digital pcr assay. Sci Rep (2018) 8(1):3626. doi: 10.1038/s41598-018-21882-0
80. de Oliveira MF, Gianella S, Letendre S, Scheffler K, Kosakovsky Pond SL, Smith DM, et al. Comparative analysis of cell-associated hiv DNA levels in cerebrospinal fluid and peripheral blood by droplet digital pcr. PloS One (2015) 10(10):e0139510. doi: 10.1371/journal.pone.0139510
81. Krawczak M, Cooper DN. The human gene mutation database. Trends Genet (1997) 13(3):121–2. doi: 10.1016/s0168-9525(97)01068-8
82. Härmälä SK, Butcher R, Roberts CH. Copy number variation analysis by droplet digital pcr. Methods Mol Biol (2017) 1654:135–49. doi: 10.1007/978-1-4939-7231-9_9
83. Imaizumi T, Yamamoto-Shimojima K, Yamamoto H, Yamamoto T. Establishment of a simple and rapid method to detect Mecp2 duplications using digital polymerase chain reaction. Congenit Anom (Kyoto) (2020) 60(1):10–4. doi: 10.1111/cga.12325
84. Taylor JL, Lee FK, Yazdanpanah GK, Staropoli JF, Liu M, Carulli JP, et al. Newborn blood spot screening test using multiplexed real-time pcr to simultaneously screen for spinal muscular atrophy and severe combined immunodeficiency. Clin Chem (2015) 61(2):412–9. doi: 10.1373/clinchem.2014.231019
85. Gutierrez-Mateo C, Timonen A, Vaahtera K, Jaakkola M, Hougaard DM, Bybjerg-Grauholm J, et al. Development of a multiplex real-time pcr assay for the newborn screening of scid, sma, and xla. Int J Neonatal Screen (2019) 5(4):39. doi: 10.3390/ijns5040039
86. Profaizer T, Slev P. A multiplex, droplet digital pcr assay for the detection of T-cell receptor excision circles and kappa-deleting recombination excision circles. Clin Chem (2020) 66(1):229–38. doi: 10.1373/clinchem.2019.308171
87. Okano T, Tsujita Y, Kanegane H, Mitsui-Sekinaka K, Tanita K, Miyamoto S, et al. Droplet digital pcr-based chimerism analysis for primary immunodeficiency diseases. J Clin Immunol (2018) 38(3):300–6. doi: 10.1007/s10875-018-0497-8
88. Sato T, Ito Y, Samura O, Aoki H, Uchiyama T, Okamoto A, et al. Direct assessment of single-cell DNA using crudely purified live cells: A proof of concept for noninvasive prenatal definitive diagnosis. J Mol Diagn (2020) 22(2):132–40. doi: 10.1016/j.jmoldx.2019.10.006
89. Oscorbin I, Kechin A, Boyarskikh U, Filipenko M. Multiplex ddpcr assay for screening copy number variations in Brca1 gene. Breast Cancer Res Treat (2019) 178(3):545–55. doi: 10.1007/s10549-019-05425-3
90. Wang J, Ramakrishnan R, Tang Z, Fan W, Kluge A, Dowlati A, et al. Quantifying egfr alterations in the lung cancer genome with nanofluidic digital pcr arrays. Clin Chem (2010) 56(4):623–32. doi: 10.1373/clinchem.2009.134973
91. Chamizo C, Rojo F, Madoz-Gurpide J. Determination of true Erbb2 gene amplification in breast cancer by quantitative pcr using a reference and a novel control gene. Appl Immunohistochem Mol Morphol (2016) 24(3):179–87. doi: 10.1097/PAI.0000000000000160
92. Vidal-Folch N, Milosevic D, Majumdar R, Gavrilov D, Matern D, Raymond K, et al. A droplet digital pcr method for severe combined immunodeficiency newborn screening. J Mol Diagn (2017) 19(5):755–65. doi: 10.1016/j.jmoldx.2017.05.011
93. Adams SP, Gravett E, Kent N, Kricke S, Ifederu A, Scoto M, et al. Screening of neonatal uk dried blood spots using a duplex Smn1 screening assay. Int J Neonatal Screen (2021) 7(4):69. doi: 10.3390/ijns7040069
94. Bao X, Qin D, Ma J, Zhou X, Wang J, Yao C, et al. Accurate detection of α-globin gene copy number variants with two reactions using droplet digital pcr. Hematology (2022) 27(1):198–203. doi: 10.1080/16078454.2022.2030885
95. Wen T, Zhang X, Lippuner C, Schiff M, Stuber F. Development and evaluation of a droplet digital pcr assay for 8p23 beta-defensin cluster copy number determination. Mol Diagn Ther (2021) 25(5):607–15. doi: 10.1007/s40291-021-00546-2
96. Imaizumi T, Yamamoto-Shimojima K, Yamamoto T. Advantages of ddpcr in detection of Plp1 duplications. Intractable Rare Dis Res (2019) 8(3):198–202. doi: 10.5582/irdr.2019.01067
97. Ito T, Kawashima Y, Fujikawa T, Honda K, Makabe A, Kitamura K, et al. Rapid screening of copy number variations in strc by droplet digital pcr in patients with mild-to-Moderate hearing loss. Hum Genome Var (2019) 6:41. doi: 10.1038/s41439-019-0075-5
98. Chang CW, Cheng WC, Chen CR, Shu WY, Tsai ML, Huang CL, et al. Identification of human housekeeping genes and tissue-selective genes by microarray meta-analysis. PloS One (2011) 6(7):e22859. doi: 10.1371/journal.pone.0022859
99. Mehta R, Birerdinc A, Hossain N, Afendy A, Chandhoke V, Younossi Z, et al. Validation of endogenous reference genes for qrt-pcr analysis of human visceral adipose samples. BMC Mol Biol (2010) 11:39. doi: 10.1186/1471-2199-11-39
100. Ampe C, Van Troys M. Mammalian actins: Isoform-specific functions and diseases. Handb Exp Pharmacol (2017) 235:1–37. doi: 10.1007/164_2016_43
101. Tristan C, Shahani N, Sedlak TW, Sawa A. The diverse functions of gapdh: Views from different subcellular compartments. Cell Signal (2011) 23(2):317–23. doi: 10.1016/j.cellsig.2010.08.003
102. Sun Y, Li Y, Luo D, Liao DJ. Pseudogenes as weaknesses of actb (Actb) and gapdh (Gapdh) used as reference genes in reverse transcription and polymerase chain reactions. PloS One (2012) 7(8):e41659. doi: 10.1371/journal.pone.0041659
103. Chang TJ, Juan CC, Yin PH, Chi CW, Tsay HJ. Up-regulation of beta-actin, cyclophilin and gapdh in N1s1 rat hepatoma. Oncol Rep (1998) 5(2):469–71. doi: 10.3892/or.5.2.469
104. Ferguson RE, Carroll HP, Harris A, Maher ER, Selby PJ, Banks RE. Housekeeping proteins: A preliminary study illustrating some limitations as useful references in protein expression studies. Proteomics (2005) 5(2):566–71. doi: 10.1002/pmic.200400941
105. Ruan W, Lai M. Actin, a reliable marker of internal control? Clin Chim Acta (2007) 385(1-2):1–5. doi: 10.1016/j.cca.2007.07.003
106. Boujedidi H, Bouchet-Delbos L, Cassard-Doulcier AM, Njiké-Nakseu M, Maitre S, Prévot S, et al. Housekeeping gene variability in the liver of alcoholic patients. Alcohol Clin Exp Res (2012) 36(2):258–66. doi: 10.1111/j.1530-0277.2011.01627.x
107. Gebhardt FM, Scott HA, Dodd PR. Housekeepers for accurate transcript expression analysis in alzheimer’s disease autopsy brain tissue. Alzheimers Dement (2010) 6(6):465–74. doi: 10.1016/j.jalz.2009.11.002
108. Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative rt-pcr. J Biochem Biophys Methods (2000) 46(1-2):69–81. doi: 10.1016/s0165-022x(00)00129-9
109. Sikand K, Singh J, Ebron JS, Shukla GC. Housekeeping gene selection advisory: Glyceraldehyde-3-Phosphate dehydrogenase (Gapdh) and β-actin are targets of mir-644a. PloS One (2012) 7(10):e47510. doi: 10.1371/journal.pone.0047510
110. Wang XT, Cheng K, Zhu L. [Hypoxia accelerate β-actin expression through transcriptional activation of actb by nuclear respiratory factor-1]. Mol Biol (Mosk) (2021) 55(3):460–7. doi: 10.31857/s0026898421030186
111. Nyström K, Biller M, Grahn A, Lindh M, Larson G, Olofsson S. Real time pcr for monitoring regulation of host gene expression in herpes simplex virus type 1-infected human diploid cells. J Virol Methods (2004) 118(2):83–94. doi: 10.1016/j.jviromet.2004.01.019
112. Jemiolo B, Trappe S. Single muscle fiber gene expression in human skeletal muscle: Validation of internal control with exercise. Biochem Biophys Res Commun (2004) 320(3):1043–50. doi: 10.1016/j.bbrc.2004.05.223
113. Yamada H, Chen D, Monstein HJ, Håkanson R. Effects of fasting on the expression of gastrin, cholecystokinin, and somatostatin genes and of various housekeeping genes in the pancreas and upper digestive tract of rats. Biochem Biophys Res Commun (1997) 231(3):835–8. doi: 10.1006/bbrc.1997.6198
114. Bémeur C, Ste-Marie L, Desjardins P, Hazell AS, Vachon L, Butterworth R, et al. Decreased beta-actin mrna expression in hyperglycemic focal cerebral ischemia in the rat. Neurosci Lett (2004) 357(3):211–4. doi: 10.1016/j.neulet.2003.12.081
115. Lin J, Redies C. Histological evidence: Housekeeping genes beta-actin and gapdh are of limited value for normalization of gene expression. Dev Genes Evol (2012) 222(6):369–76. doi: 10.1007/s00427-012-0420-x
116. Bas A, Forsberg G, Hammarström S, Hammarström ML. Utility of the housekeeping genes 18s rrna, beta-actin and glyceraldehyde-3-Phosphate-Dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand J Immunol (2004) 59(6):566–73. doi: 10.1111/j.0300-9475.2004.01440.x
117. Hernandez-Segura A, Rubingh R, Demaria M. Identification of stable senescence-associated reference genes. Aging Cell (2019) 18(2):e12911. doi: 10.1111/acel.12911
118. Liu YJ, Zheng D, Balasubramanian S, Carriero N, Khurana E, Robilotto R, et al. Comprehensive analysis of the pseudogenes of glycolytic enzymes in vertebrates: The anomalously high number of gapdh pseudogenes highlights a recent burst of retrotrans-positional activity. BMC Genomics (2009) 10:480. doi: 10.1186/1471-2164-10-480
119. Alexander MC, Lomanto M, Nasrin N, Ramaika C. Insulin stimulates glyceraldehyde-3-Phosphate dehydrogenase gene expression through cis-acting DNA sequences. Proc Natl Acad Sci U.S.A. (1988) 85(14):5092–6. doi: 10.1073/pnas.85.14.5092
120. Graven KK, Troxler RF, Kornfeld H, Panchenko MV, Farber HW. Regulation of endothelial cell glyceraldehyde-3-Phosphate dehydrogenase expression by hypoxia. J Biol Chem (1994) 269(39):24446–53.
121. Bereta J, Bereta M. Stimulation of glyceraldehyde-3-Phosphate dehydrogenase mrna levels by endogenous nitric oxide in cytokine-activated endothelium. Biochem Biophys Res Commun (1995) 217(1):363–9. doi: 10.1006/bbrc.1995.2785
122. Wimmer K, Wernstedt A. Pms2 gene mutational analysis: Direct cdna sequencing to circumvent pseudogene interference. Methods Mol Biol (2014) 1167:289–302. doi: 10.1007/978-1-4939-0835-6_20
123. Balaji S, Vanniarajan A. Implication of pseudo reference genes in normalization of data from reverse transcription-quantitative pcr. Gene (2020) 757:144948. doi: 10.1016/j.gene.2020.144948
124. Zhang JY, Zhang F, Hong CQ, Giuliano AE, Cui XJ, Zhou GJ, et al. Critical protein gapdh and its regulatory mechanisms in cancer cells. Cancer Biol Med (2015) 12(1):10–22. doi: 10.7497/j.issn.2095-3941.2014.0019
125. Janovick-Guretzky NA, Dann HM, Carlson DB, Murphy MR, Loor JJ, Drackley JK. Housekeeping gene expression in bovine liver is affected by physiological state, feed intake, and dietary treatment. J Dairy Sci (2007) 90(5):2246–52. doi: 10.3168/jds.2006-640
126. Albershardt TC, Iritani BM, Ruddell A. Evaluation of reference genes for quantitative pcr analysis of mouse lymphocytes. J Immunol Methods (2012) 384(1-2):196–9. doi: 10.1016/j.jim.2012.07.020
127. Jarrous N. Human ribonuclease p: Subunits, function, and intranuclear localization. Rna (2002) 8(1):1–7. doi: 10.1017/s1355838202011184
128. Barber RD, Harmer DW, Coleman RA, Clark BJ. Gapdh as a housekeeping gene: Analysis of gapdh mrna expression in a panel of 72 human tissues. Physiol Genomics (2005) 21(3):389–95. doi: 10.1152/physiolgenomics.00025.2005
129. Kim BR, Nam HY, Kim SU, Kim SI, Chang YJ. Normalization of reverse transcription quantitative-pcr with housekeeping genes in rice. Biotechnol Lett (2003) 25(21):1869–72. doi: 10.1023/a:1026298032009
130. Mancini C, Messana E, Turco E, Brussino A, Brusco A. Gene-targeted embryonic stem cells: Real-time pcr assay for estimation of the number of neomycin selection cassettes. Biol Proced Online (2011) 13:10. doi: 10.1186/1480-9222-13-10
131. Sellars MJ, Vuocolo T, Leeton LA, Coman GJ, Degnan BM, Preston NP. Real-time rt-pcr quantification of kuruma shrimp transcripts: A comparison of relative and absolute quantification procedures. J Biotechnol (2007) 129(3):391–9. doi: 10.1016/j.jbiotec.2007.01.029
Keywords: RPP30, protein structure, tumor, internal reference gene, PCR diagnosis
Citation: Wu J, Yu S, Wang Y, Zhu J and Zhang Z (2022) New insights into the role of ribonuclease P protein subunit p30 from tumor to internal reference. Front. Oncol. 12:1018279. doi: 10.3389/fonc.2022.1018279
Received: 17 August 2022; Accepted: 28 September 2022;
Published: 13 October 2022.
Edited by:
Daniel P. Bezerra, Oswaldo Cruz Foudantion (FIOCRUZ), BrazilReviewed by:
Shetty Ravi Dyavar, Adicet Bio, Inc, United StatesCopyright © 2022 Wu, Yu, Wang, Zhu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenhua Zhang, enpoMTk3NGNuQDE2My5jb20=
†These authors have contributed equally to this article
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.